
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 March 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1112941
This article is part of the Research Topic Global Dissemination and Evolution of Epidemic Multidrug-Resistant Gram-Negative Bacterial Pathogens: Surveillance, Diagnosis and Treatment Volume II View all 11 articles
Aeromonas veronii is a Gram-negative bacterium ubiquitously found in aquatic environments. It is a foodborne pathogen that causes diarrhea in humans and hemorrhagic septicemia in fish. In the present study, we used whole-genome sequencing (WGS) to evaluate the presence of antimicrobial resistance (AMR) and virulence genes found in A. veronii Ah5S-24 isolated from catfish pond sediments in South-East, United States. We found cphA4, dfrA3, mcr-7.1, valF, blaFOX-7, and blaOXA-12 resistance genes encoded in the chromosome of A. veronii Ah5S-24. We also found the tetracycline tet(E) and tetR genes placed next to the IS5/IS1182 transposase, integrase, and hypothetical proteins that formed as a genetic structure or transposon designated as IS5/IS1182/hp/tet(E)/tetR/hp. BLAST analysis showed that a similar mobile gene cassette (MGC) existed in chromosomes of other bacteria species such as Vibrio parahaemolyticus isolated from retail fish at markets, Aeromonas caviae from human stool and Aeromonas media from a sewage bioreactor. In addition, the IS5/IS1182/hp/tet(E)/tetR/hp cassette was also found in the plasmid of Vibrio alginolyticus isolated from shrimp. As for virulence genes, we found the tap type IV pili (tapA and tapY), polar flagellae (flgA and flgN), lateral flagellae (ifgA and IfgL), and fimbriae (pefC and pefD) genes responsible for motility and adherence. We also found the hemolysin genes (hylII, hylA, and TSH), aerA toxin, biofilm formation, and quorum sensing (LuxS, mshA, and mshQ) genes. However, there were no MGCs encoding virulence genes found in A. veronii AhS5-24. Thus, our findings show that MGCs could play a vital role in the spread of AMR genes between chromosomes and plasmids among bacteria in aquatic environments. Overall, our findings are suggesting that MGCs encoding AMR genes could play a vital role in the spread of resistance acquired from high usage of antimicrobials in aquaculture to animals and humans.
Aeromonas veronii is a Gram-negative bacterium ubiquitously found in different aquatic environments. It was first reported by Hickman-Brenner et al. (1987) as a new species in 1983. It is pathogenic to several fish species that include the top farmed species such as common carp (Cyprinus carpio), channel catfish (Ictalurus punctatus), tilapia (Oreochromis niloticus), and pangasius (Pangasius hypophthalmus) (González-Serrano et al., 2002; Smyrli et al., 2017, 2019; Wang et al., 2022). It causes hemorrhagic septicemia and skin ulcers in fish (Hoai et al., 2019; Tekedar et al., 2020) and diarrhea in humans (Roberts et al., 2006). Strain variations have been linked to virulence leading to studies aimed at identifying the virulence factors associated with mortalities (González-Serrano et al., 2002; Smyrli et al., 2017, 2019; Wang et al., 2022). The high mortalities experienced in aquaculture have led to use of antibiotics, thereby contributing to increase of antimicrobial resistance (AMR) (Roberts et al., 2006). As mentioned in our previous studies (Dubey et al., 2022a,b), the major limitation with most studies aimed at identifying AMR genes in bacteria is that they are mostly done by PCR that only detects AMR genes based on the primers used in the assay. This poses the risk of omitting important AMR genes whose primers are not included in PCR assays. Besides, PCR-based assays do not determine whether the AMR genes are intrinsically encoded in the chromosomes or extrinsically in plasmids. So, the use of whole-genome sequencing (WGS) able to detect all genes and their location in bacteria genomes is a better approach for elucidating the role of different bacteria species in the spread of AMR and virulence genes than PCR-based assays.
The spreading of AMR genes by horizontal transfer is contributing to involvement of bacteria species outside the 12 bacteria families enlisted to pose the greatest AMR threat to human health by the World Health Organization (WHO) (Willyard, 2017). As pointed out by White et al. (2001), the spread of AMR genes is enhanced when they form part of mobile gene cassettes (MGCs) or transposons. The MGCs were first identified as integrated AMR genes found in integrons in the early 1980s (Ward and Grinsted, 1982; Meyer et al., 1983; White et al., 2001). Although studies done this far have focused on cassettes carrying AMR genes, it is likely that the packaging in cassettes includes other genes such as virulence factors. As stated by White et al. (2001), MGCs facilitate horizontal gene transfer using various mechanisms that include mobilization of individual cassettes by integrons (Collis and Hall, 1992), movement of integrons having cassettes by transposases (Brown et al., 1996; Craig, 1996; Minakhina et al., 1999), dissemination of larger transposons carrying integrases (Liebert et al., 1999), and translocation of conjugative plasmids having integrases among bacteria (White et al., 2001). It is likely that most of the AMR genes associated with infections in aquaculture, livestock and humans are part of MGCs (Recchia and Hall, 1995). Yet, gene cassettes conferring resistance to antibiotics used in aquaculture have not been widely investigated as done in mammalian studies. Hence, it is unknown whether the AMR genes selected against drugs like tetracycline, sulphonamide, and trimethoprim widely used in aquaculture are packaged in MGCs. Thus, although previous studies have focused on identifying individual genes associated with resistance, the cassettes responsible for the spread of AMR genes has not been widely investigated for bacteria found in aquaculture.
In the present study we used WGS to profile all AMR and virulence genes found in A. veronii Ah5S-24 isolated from pond sediment obtained from the South East, USA by DePaola et al. (1988). Although in the previous study, they detected presence of Oxytetracycline-resistance (OTcr) and tetracycline-resistance (Tcr) by selecting for isolates that replicated on MacConkey agar containing oxytetracycline or tetracycline antibiotics, they did not determine whether the resistance gene was located in the chromosome or plasmids. Even though they showed the transfer of OTcr and Tcr resistance from the Aeromonas isolate to Escherichia coli, they did not determine whether the transfer was plasmid mediated or MGC. Thus, we wanted to determine whether the OTcr and Tcr resistance in the isolate was encoded in the chromosome or plasmid. We also wanted to determine whether the resistance detected was associated with a tetracycline genetic structure similar to that found in other bacteria species. We anticipate that data presented herein will underscore the importance of screening for MGCs carrying AMR genes from aquatic organisms with potential transmission to animals and humans.
A suspected Aeromonas hydrophila isolated from pond sediments in the South-Eastern USA by DePaola et al. (1988) in 1988 was retrieved from the –80°C freezer at the Norwegian University of Life Sciences (NMBU), Ås, Norway. The isolate was kindly provided by Dr. Angelo DePaola, Gulf Coast Seafood Laboratory, United States. After thawing, the bacteria isolate was streaked on blood agar and incubated at 10°C for 5–7 days. Single colonies were streaked on tryptone soy agar (TSA) for purification followed by characterization using the Matrix-assisted laser Desorption/Ionization-Time of Flight (MALDI-TOF) mass spectrometry while DNA was extracted based on manufacturer’s protocol (Qiagen, Germany). Identification of the bacteria species was done by PCR using universal 16S rRNA primers 27F and 1492R. Phenotypic characterization of antibiotic resistance was done using the Kirby-Bauer disk diffusion test (Joseph et al., 2011). The commercial antibiotic discs (Neo-Sensitabs™, Rosco) used consisted of Penicillin (PEN-10 μg), Amoxicillin (AMOXY-30 μg), Ampicillin (AMP-10 μg), Ciprofloxacin (CIPR-5 μg), Cefoxitin (CFO-30 μg), Cephalothin (CEP-30 μg), Tetracycline (TET-30 μg), Gentamycin (GEN-10 μg), Rifampicin (RIF-5 μg), Sulfonamide (SULFA-240 μg), Trimethoprim (TRIM-5 μg), Erythromycin (Ery-15 μg), Nitrofurantoin (NI-300 μg), and (Colistin-CO-150 μg) (Table 1). A volume of 100 μl containing freshly cultured bacteria diluted at McFarland concentration of 108 CFU/ml was spread on Müller Hinton agar followed by putting the antibiotic discs on the bacteria lawn. Next, the plates were incubated at 30°C overnight followed by measuring the susceptibility or resistance based on the Clinical and Laboratory Standards Institute (CLSI) guidelines (Kahlmeter et al., 2006; Cockerill et al., 2012).
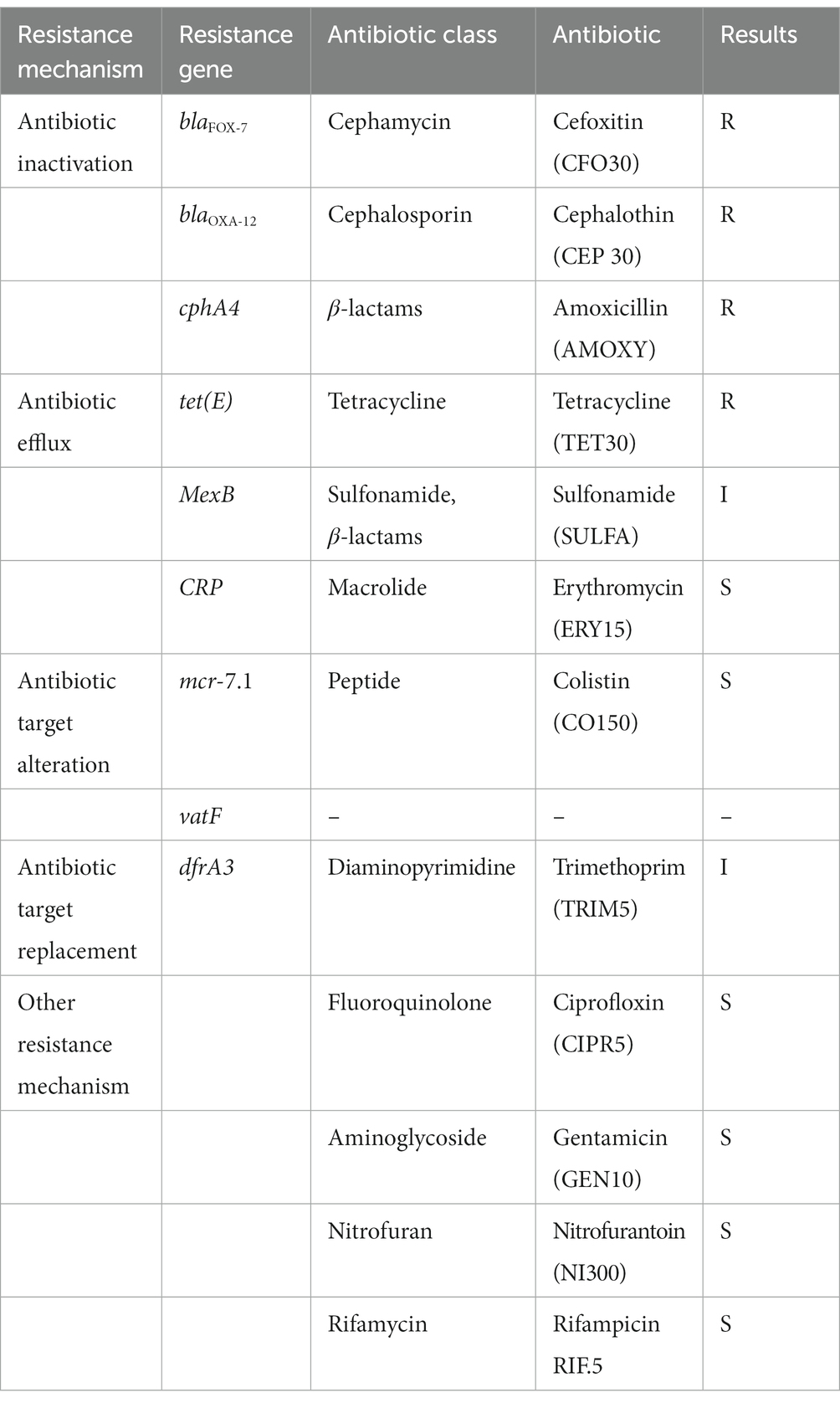
Table 1. Overview of antibiotic resistence genes detected in the draft genome of Aeromonas veronii AhS5-24 together with phenotypic antibiotic susceptibility testing results using disk diffusion assay.
Genomic DNA (gDNA) was extracted as previously described (Becker et al., 2016) using the MagAttract® HMW DNA kit based on the manufacturer protocols (Qiagen GmbH, Hilden, Germany). Briefly, a 1 ml volume of approximately 2 × 109 CFU/ml of freshly overnight cultured bacteria was spanned in 2 ml Eppendorf tubes followed by suspending the pellets in 180 μl buffer ATL (tissue lysis buffer, Qiagen GmbH, Hilden, Germany). Next, Proteinase K was added to each vial at a concentration of 20 mg/ml followed by incubation at 56°C in an Eppendorf thermomixer for 30 min. Afterward, 4 μl RNase was added and the vials were pulse vortexed followed by adding 15 μl of MagAttract Suspension G and 280 μl Buffer MB to each vial (Tarumoto et al., 2017). The suspension from each tube was transferred onto the MagAttract holder followed by mixing for 1 min on an Eppendorf thermomixer. The magnetic beads having the gDNA were separated on the MagAttract magnetic rack for approximately 1 min. Supernatants were removed without disturbing the beads followed by washing the magnetic beads twice using MW1 and PE buffer (Becker et al., 2016; Tarumoto et al., 2017). The remaining suspension was removed by washing the beads twice using 1 ml RNAase free water (Qiagen GmbH, Hilden, Germany) (Becker et al., 2016). The gDNA was harvested by eluting in 100 μl buffer EB while purity was evaluated using the NanoDrop (Thermo Fisher, United States) and gel electrophoresis using 1% agarose. Quantification of gDNA was carried out using the Qubit double-stranded DNA high-CHS kit following the manufacturer’s guidelines (Life Technologies Inc., Carlsbad, CA, United States) (Guan et al., 2020).
Library preparation was carried out using Nextera DNA Flex (Tagmentation Illumina Inc. San Diego, CA, United States) while Illumina MiSeq were used with a paired-end read length of 2 × 300 bp. The bacterial raw DNA reads were analyzed using the online Galaxy platform1 version 21.05. Quality of both forward and reverse raw reads were analyzed using the FastQC Version 0.11.9 software (Bioinformatics B, 2011), while the Trimmometric version 0.38.1 was used to remove the adapters and low-quality reads from paired-end sequences (Bolger et al., 2014). The resulting paired-end sequence reads were de novo assembled using SPAdes v. 3.12.0 (Coil et al., 2015) with 33 to 91 k-mers (Bankevich et al., 2012) while genome annotation was done using the prokaryotic genome annotation pipeline (PGAP) (Tatusova et al., 2016) from the National Center for Biotechnology and Information (NCBI) and Prokka (Seemann, 2014). Online Galaxy platform (see Footnote 1) version 21.05 was used for bioinformatic analysis.
Pangenome analysis of A. veronii AhS5-24 together with 30 complete genomes of other A. veronii isolates retrieved from the NCBI was carried out using Roary v. 3.13.0 using general feature files 3 (.gff) file generated from Prokka v. 1.14.5. The phylogenetic tree was made using the Phandango software using Gene_presence_absence and Newick files obtained from Roary v. 3.13.0. The average nucleotide identity (ANI) of all 31 A. veronii genomes was computed using FastANI v1.3 using A. veronii FC951 (CP032839) as a reference strain. Antimicrobial resistance (AMR) genes were identified using staramr version 0.7.2 (Tran et al., 2021) and ABRicate v1.0.1 (Seemann, 2016) in the Comprehensive antimicrobial resistance database (CARD) (Alcock et al., 2020) and staramr v. 0.7.2 with the identification threshold set at 80%. Plasmidfinder v 2.0 (Ullah et al., 2020) was used to identify plasmids in the bacterial genomes while virulence genes were identified using virulence factors database (VFDB). Genome circular maps were created using Proksee.3
Phylogenetic comparison of the tet(E) and tetR genes from strain AhS5-24 with other A. veronii isolates was done using the Molecular Evolutionary Genetic Analysis version 7 (MEGA-7) software (Kumar et al., 2016). The tet(E) and tetR sequences from strain AhS5-24 were retrieved after screening using ABRicate version 1.0.1 followed by comparison with tet(E) and tetR sequences from other A. veronii isolates retrieved from NCBI. Phylogenetic trees were produced using the Neighbor-joining and BioNJ algorithm to a pairwise matrix estimated using JTT model and expressed as number of base substitution per site (Jones et al., 1992).
The draft genome of A. veronii AhS5-24 showed a high similarity with other A. veronii genomes, as shown in Figure 1. The draft genome of strain AhS5-24 had a size of 4,748,224 bp with G + C content of 58.48%. It contained 157 contigs with an N50 value of 115,408. A total of 4,493 genes were predicted with 4,334 genes coding for proteins. The genome contained a total of 108 genes of RNA consisting of 99 tRNA and 5 rRNAs. The total number of genes detected from the 31 A. veronii genomes based on pangenome analysis was 20,352 of which 1,429 genes were core-, 875 softcore-, 2,241 shell-, and 15,807 cloud genes. The phylogenetic tree divided the genomes into three groups of which strain AhS5-24 was closely related to the human CP032839 (FC951) and hospital sewage CP079823 (HD6454) isolates (Figure 1). The average nucleotide identity (ANI) analysis using FastANI showed high similarity (>93%) of all 31 A. veronii isolates despite coming from different host species and geographic locations. The ANI of strain AhS5-24 was 96.31% similar with the A. veronii CF951 (CP032839) human clinical isolate and 96.20% similar with A. veronii HD6454 (CP079823) from hospital sewage.
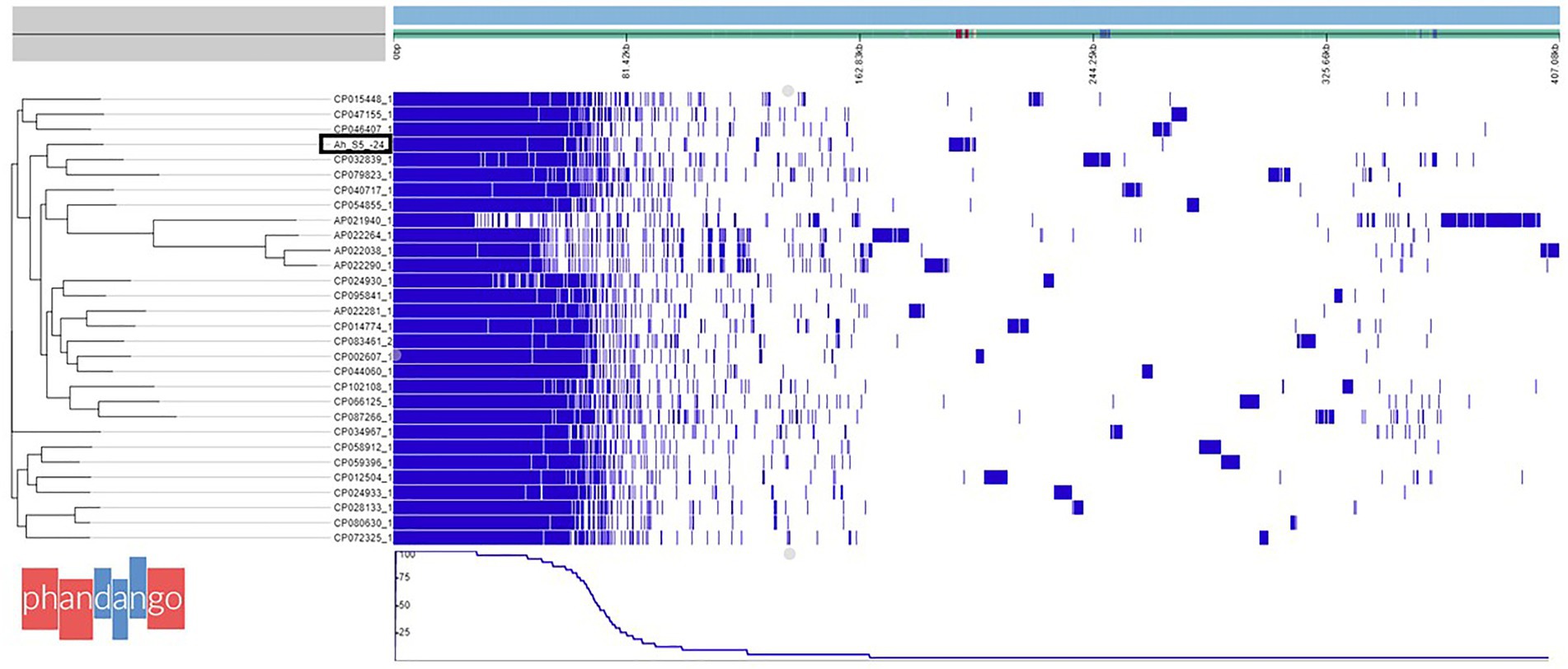
Figure 1. Phylogenetic tree based on pangenome analysis of Aeromonas veronii AhS5-24 together with 30 complete genomes of other A. veronii strains obtained from the National Center for Biotechnology and Information (NCBI). Note that A. veronii AhS5-24 is clustered together with A. veronii strains FC951 (CP032839) isolated from hospital sewage. A total of 20352 genes were detected in all 31 A. veronii genomes by pangenome analysis using the Roary software of which 1,429 genes were core genes, 875 soft core genes while the number of shell and cloud genes was 2,241 and 15,807 genes, respectively.
The virulence genes found in A. veronii AhS5-24 comprised the motility and adherence genes that included the (i) lateral flagella proteins consisting of lfgA and lfgL, (ii) polar flagella that were represented by flgA and flgN, (iii) members of the tap type IV pili that included tapA, tapW and tapY, and (iv) fimbrial adherence determinants that included pefC and pefD genes (Figure 2). The mannose-sensitive hemagglutinin (MHSA) is encoded by the genes mshA and mshQ (Figure 2). Genes associated with capsule formation and immune evasion included ddhA, ddhC, and wcaG1. The hemolysin genes detected were hlyA, hylIII, and thermostable hemolysin (TSH) while toxin genes consisted of aerolysin aerA. Genes associated with iron acquisition consisted of the Iron ABC transporter while biofilm formation and quorum sensing genes were represented by luxS and MshA-Q pilus. We detected genes belonging to the type II secretion systems (T2SS) represented by exeA to exeN (Supplementary Table S1) and vgrG, which is part of T6SS (Figure 2). Overall, the virulence genes detected belonged to motility, adherence, secretion systems, iron acquisition, biofilm formation, quorum sensing, and immune evasion groups.
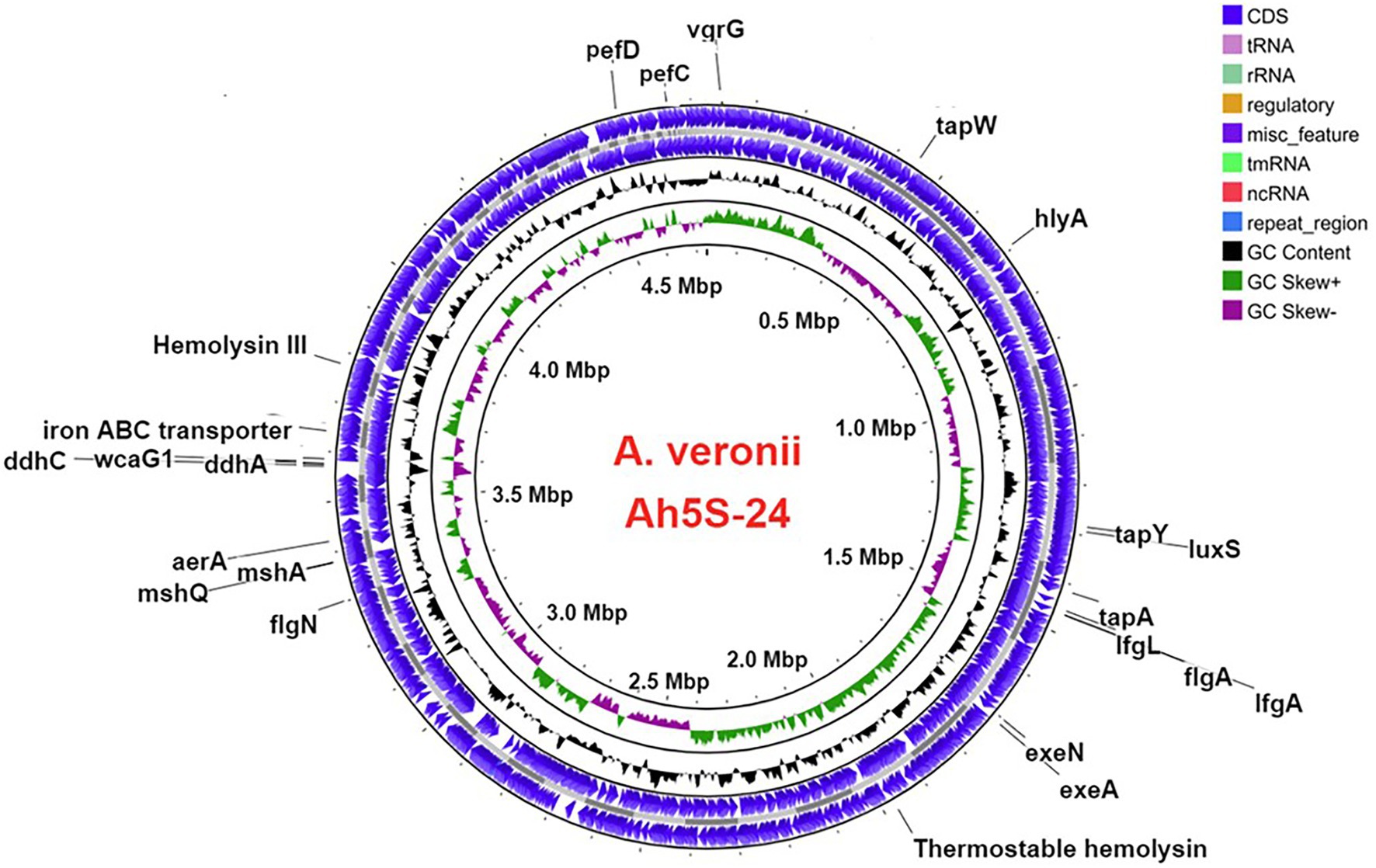
Figure 2. Circular map showing the loci for different virulence genes in the genome of Aeromonas veronii strain AhS5-24. Motility and adherence genes detected included the lateral flagella ifgA and IfgL genes; polar flagella flaA and flgN genes; tap type IV pili tapA and tapY; fimbriae pefC and pefD genes. The mannose-sensitive hemagglutinin (MHSA) genes were represented by mshA and mshQ while the hemolysin genes included hylA, hylIII, and TSH. Iron acquisition was represented by the iron ABC transporter protein and genes associated with capsule formation included the ddhA, ddhC, and wcaG1 genes. Quorum sensing and biofilm formation genes were represented by luxS. The T2SS was represented by exeA and exeN genes while T6SS was represented by the vgrG gene.
Results of the disk diffusion test showed that strain Ah5S-24 was resistant to CFO30, CEP30, AMOXY30, and TET30, whereas it showed intermediate resistance against SULFA240 and TRIM5 (Table 1). However, it was susceptible to ERY15, CO150, CIPR5, GEN10, NI300, and RIF5. We found an overall correlation kappa score of 82% (Cohen’s k = 0.8235) with a specificity of 91.66% and sensitivity of 93% between the phenotypic profile based on the disk diffusion test and genotypic profile based on the genes identified using the CARD (Alcock et al., 2020).
Identification of AMR genes using the CARD (Alcock et al., 2020) showed that strain Ah5S-24 encoded multiple AMR genes that included the β-lactamase like blaFOX-7, blaOXA-12, and cphA4. Other genes detected included the colistin crp and mcr-7.1 genes as well as the streptogramin A acetyl transferase vatF gene (Figure 3). There were no integrase and transposases located near the blaFOX-7, blaOXA-12, cphA4, crp, mcr-7.1, and vatF genes. The trimethoprim dfrA3 gene was placed together with the sulfurtransferase, DUF2541 family protein, mog, DUF3135 domain-containing protein, threonine exporter protein, and phosphoadenyl-sulfate reductase (Figure 3). The efflux pumps detected included the resistance-nodulation-cell division (RND) mexB and smeD that were placed next to each other together with the IS5 transposase (Figure 3).
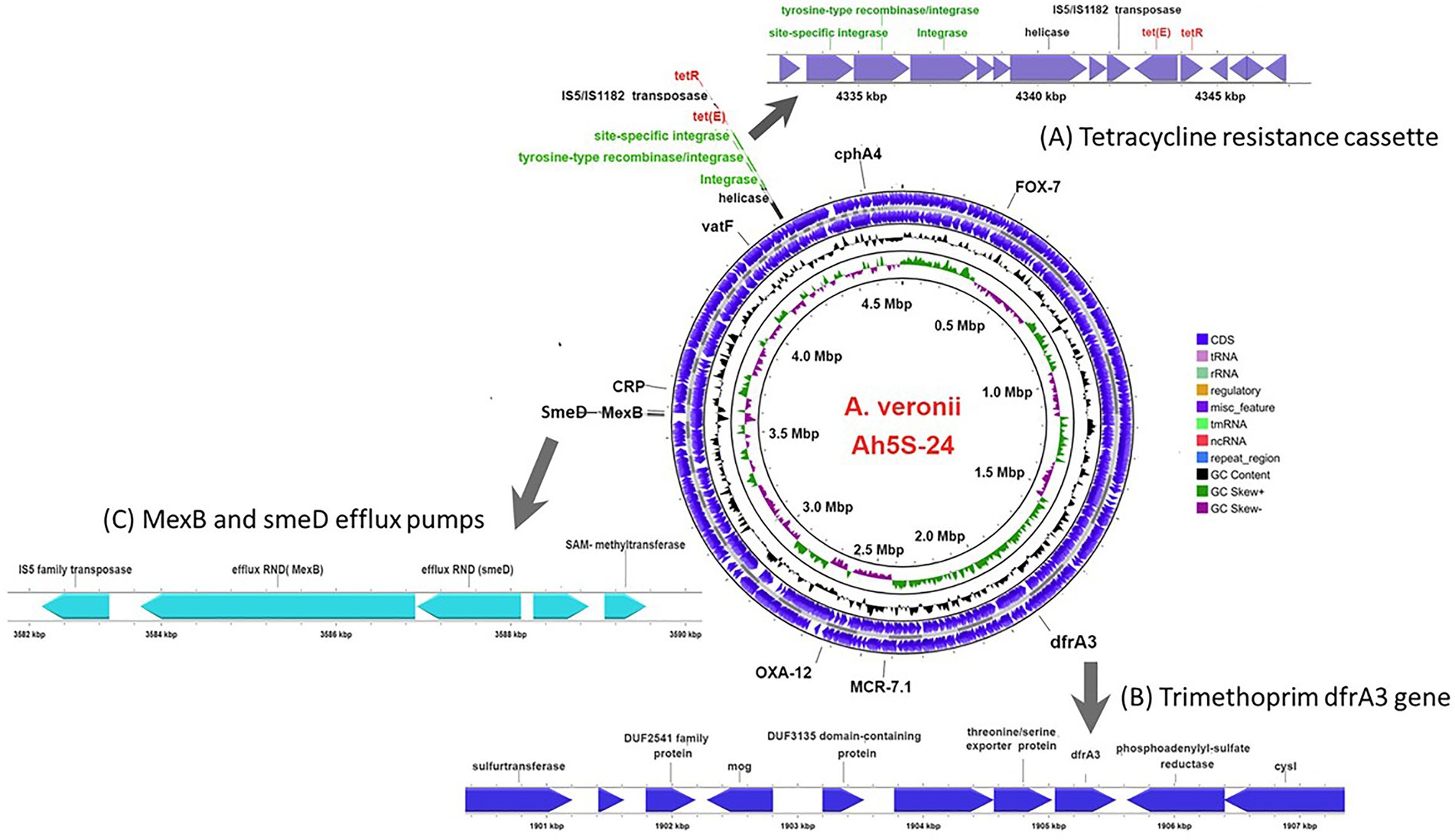
Figure 3. Circular genomic map of Aeromonas veronii AhS5-24 showing the loci for antimicrobial resistance (AMR) genes. The AMR genes detected included the β-lactam blaOXA-12 and blaFOX-7 genes together with cphA4, dfrA3, mcr-7.1, and vatF genes while the efflux pump proteins detected were CRP, smedD, and mexB. The extended linear map (A) shows the cassette encoding the site-specific integrase, tyrosine-type recombinase/integrase, integrase, helicase, IS5/IS1182 transposase, tet(E) efflux pump protein gene and tetR gene designated as IS5/IS1182/hp/tet(E)/tetR/hp. The extended line map (B) shows the linear relationship between the trimethoprim dfrA3 gene and other genes that includes sulfurtransferase, DUF2541 family protein, mog, DUF3135 domain-containing protein, threonine/serine exporter protein, dfrA3 and phosphoadenyl-sulfate reductase in the genome of A. veronii AhS5-24. The extended map (C) shows the linear relationship between the smeD and mexB efflux pumps in the genome of A. veronii AhS5-24.
Our findings show that the repressor of the tetracycline resistance element gene tetR was placed next to tet(E) together with the IS5/IS1182 transposase, helicase, integrase, tyrosine type recombinase/integrase, and the site-specific integrase all in one cassette (Figure 3). The cassette found in A. veronii AhS5-24 showed a high similarity with cassettes found in Vibrio parahaemolyticus (MN199028.1) isolated from a fish market, Vibrio alginolyticus plasmid (MN865127.1) from shrimp, Aeromonas caviae (CP110176) from human stool, and Aeromonas media (CP03844.1) from a sewage bioreactor (Figure 4). They all had a similar genetic structure or transposon consisting of the IS5/IS1182 transposase followed by a gene encoding a hypothetical protein (hp), Tet(E), tetR, and another hypothetical protein (hp), thereby forming a MGC designated as 1S5/IS1182/hp/tet(E)/tetR/hp (Figure 4). Suffice to point out that the cassette from Vibrio alginolyticus (MN865127.1) was from a plasmid, while the cassettes from A. veronii AhS5-24, Vibrio parahaemolyticus (MN199028.1), A. media (CP038444.1), and Aeromonas caviae (CP110176) were from chromosomes. This findings demonstrate that the IS5/IS1182/hp/tet(E)/tetR/hp cassette can be found both in chromosomes and plasmids of different bacteria species. It is noteworthy that the cassette for Vibrio alginolyticus plasmid (MN865127.1) had the IShfr9 transposase, and not the IS5/IS1182 transposase, despite having a similar hp/tet(E)/tetR/hp component with other bacteria species used in the comparison (Figure 4).
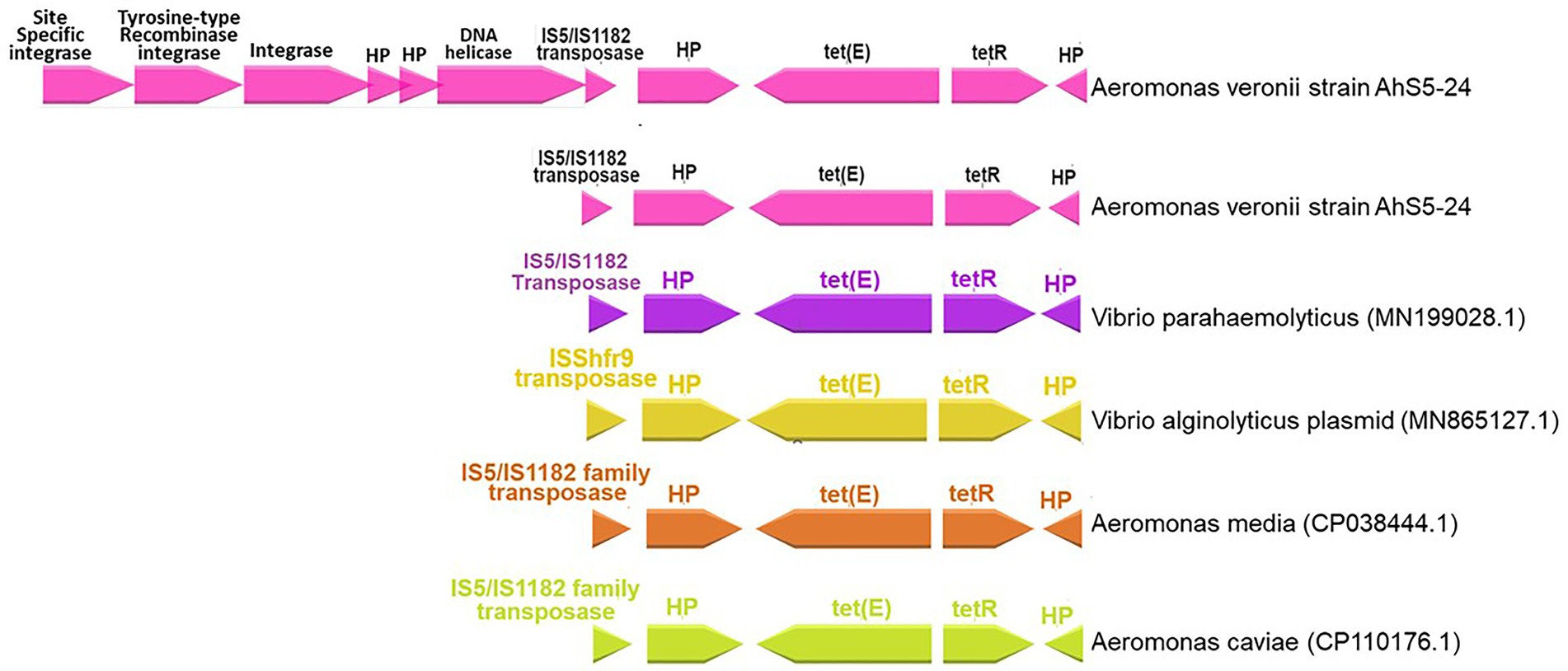
Figure 4. Comparison of the IS5/IS1182/hp/tet(E)/tetR/hp gene cassettes for Aeromonas veronii strain AhS5-24 from pond sediments, Vibrio parahaemolyticus (MN199028) isolated from retail fish from a market, Vibrio alginolyticus plasmid (MN865127.1) from shrimp, Aeromonas media (CP038444.1) from sewage bioreactor and Aeromonas caviae (CP110176.1) from human stool. Note that all isolates had the hypothetical proteins (hp), tetR, tet(E), and IS5/IS1182 transposase forming a gene cassette designated as IS5/IS1182/hp/tet(E)/tetR/hp. The uppermost linear map shows Aeromonas veronii strain AhS5-24 having the IS5/IS1182/hp/tet(E)/tetR/hp cassette linked to the DNA helicase, two hypothetical proteins, integrase, tyrosine-type recombinase/integrase and site-specific integrase.
Phylogenetic analysis showed that the tet(E) gene from A. veronii AhS5-24 had a 100% similarity with tet(E) genes from different bacteria species that included Escherichia coli (AIL23572.1, CAC20135.1, and WP_20194468.1), Aeromonas caviae (BBR12376.1, WP_244056220.1, and WP_201964468.1), Yersinia ruckeri (APO36645.1, APO36648.1, and APO36646.1), Klebsiella pneumoniae (EIW8806435.1), Aeromonas spp. (QEV84027.1 and WP_017780889.1), and Enterobacter cloacae (ASF90526.1) (Figure 5). Phylogenetic analysis also showed that the tetR gene from A. veronii AhS5-24 had a 100% similarity with tetR genes from different bacteria species that included E. coli (AAA98409.1), Gammaproteobacteria (W_P011899269.1 and WP_017411289.1), Aeromonas salmonicida (QJR83010.1), Aliivibrio salmonicida (CAC81917.1), and A. caviae (WP_223946105.1 and WP_223945860.1) (Figure 6). Altogether, our findings show that tet(E) and tetR genes were highly similar with those found in different bacteria species.
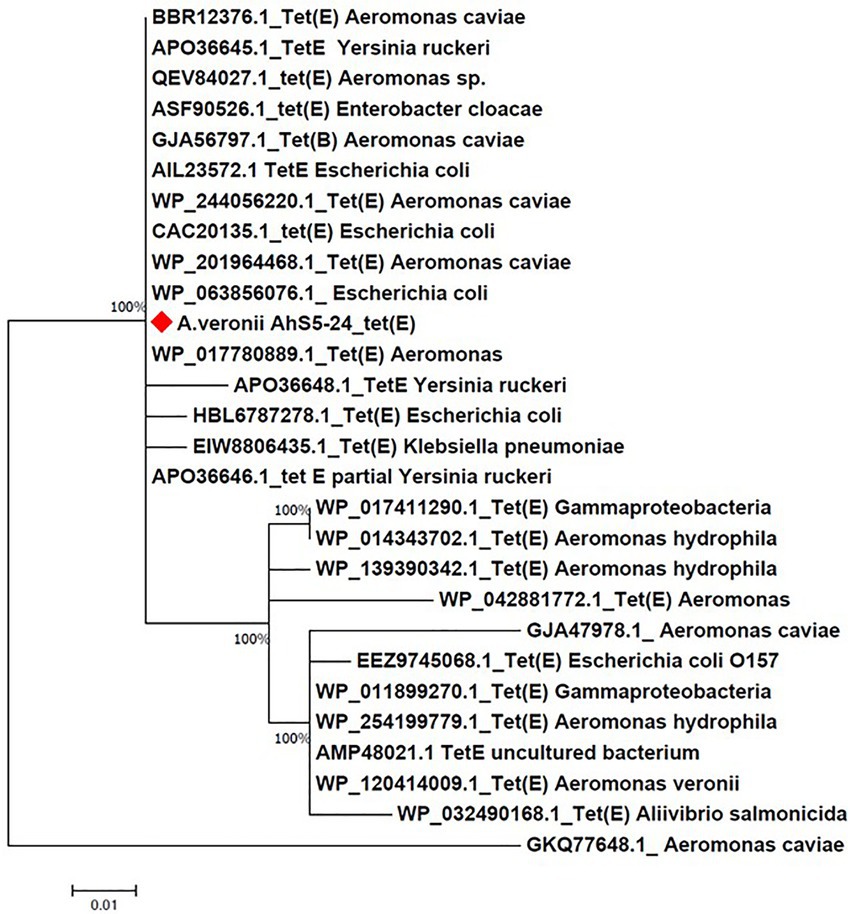
Figure 5. Phylogenetic tree comparing the tet(E) gene from different bacteria species. Note that the tet(E) Aeromonas veronii strain AhS5-24 had 100% similarity with tet(E) genes from other bacteria species that include Aeromonas caviae (BBR12376.1, GJA56797.1, and WP/244056220.1 and WP_20964468.1), Yersinia ruckeri (Apo036645.1, APO36646.1, and APO36648.1), Enterobacter cloacae (ASF90526.1), Escherichia coli (AIL23572.1, CAC20135.1, WP_063856076.1 and HBL6787278.1), and Klebsiella pneumoniae (EIW8806435.1).
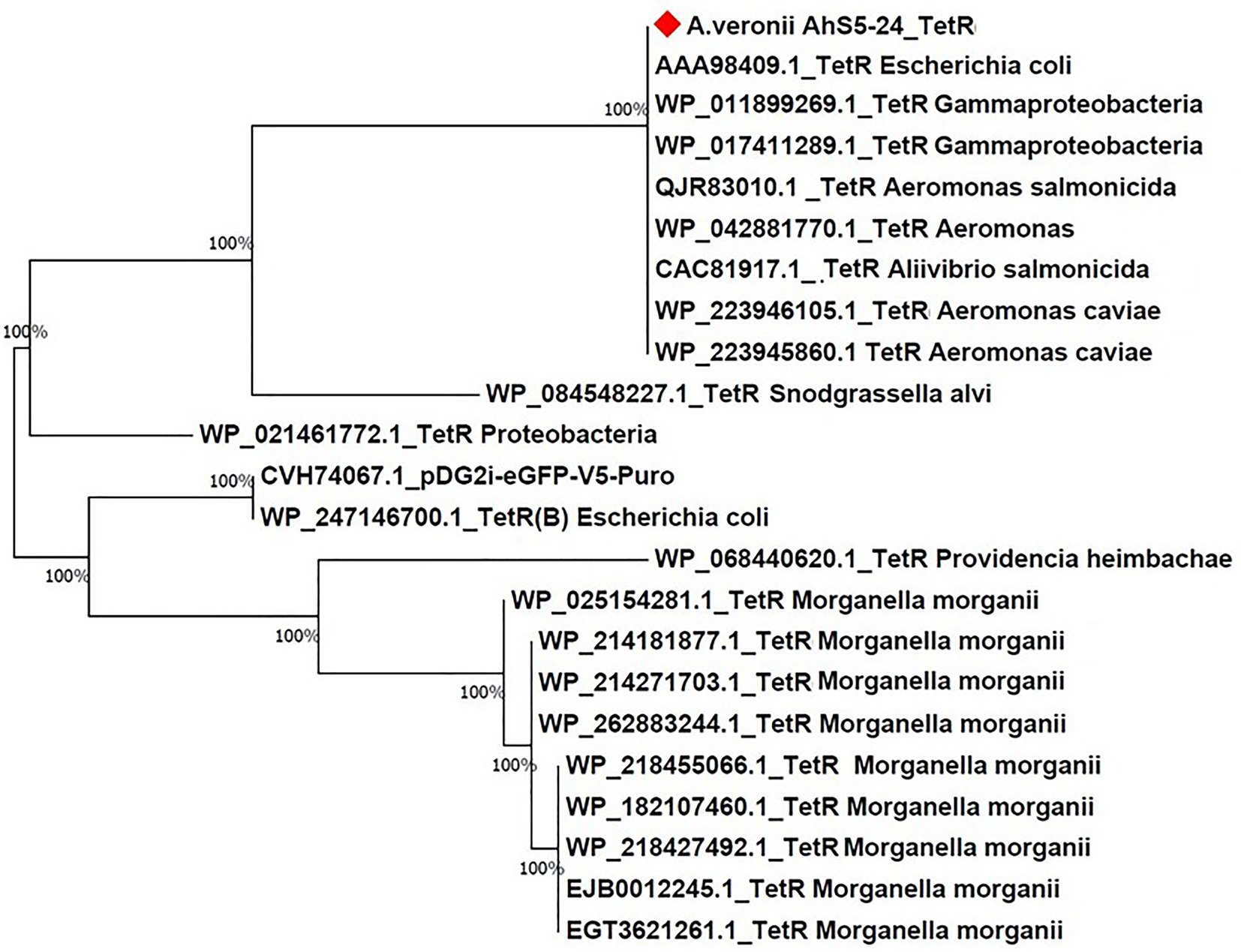
Figure 6. Phylogenetic tree comparing the tetR gene from different bacteria species. Note the high (100%) similarity between the tetR gene of Aeromonas veronii strain AhS5-24 with tetR genes of Escherichia coli (AAA98409.1), Gammaproteobacteria (WP_011899269.1 and WP_017411289), Aeromonas salmonicida (QJR83010.1), Aeromonas spp. (WP_04288770.1), and Aeromonas caviae (WP_223946105.1 and WP_223945860.1).
In this study, we have shown that the bacteria isolated from pond sediments in the South East USA previously classified as A. hydrophila using the API 20E system in DePaola et al. (1988) was characterized as A. veronii Ah5S-24 using WGS and pangenome analysis. We have also shown that the Tr and TOr detected by DePaola et al. (1988) could be linked to the tetR and tet(E) genes found in the same isolate designated as strain Ah5S-24 in this study. In addition, strain Ah5S-24 encoded several virulence and AMR genes of which tetracycline resistance genes were placed in the same genetic structure with an integrase, transposase and recombinase and can be defined as a transposon. These findings demonstrate that Aeromonas spp. isolated from aquatic environments have the potential to transmit AMR genes to other bacteria using transposons carrying different AMR genes.
Pangenome analysis showed a high similarity of strain Ah5S-24 with other A. veronii strains linked to different diseases in aquatic organism and humans. For example, strains CP032839.1 and CP046407.1 shown to be closely related with A. veronii Ah5S-24 were from human clinical cases (Ragupathi et al., 2020) and diseased rohu (Labeo rohita) (Tyagi et al., 2022), respectively. Besides, A. veronii Ah5S-24 had several virulence genes linked to adherence, biofilm formation, quorum sensing, immune evasion, toxins and intracellular secretion systems (TSS) found in other pathogenic A. veronii strains (Arechaga and Cascales, 2022). Detection of the Msh pili, tap type IV pili, lateral-and polar flagellar genes associated with intestinal adherence, colonization, and biofilm formation (Ro, 2006; Hadi et al., 2012) is suggestive that these genes play a vital role in the pathogenicity of strain Ah5S-24. The presence of the LuxS and mshQ genes is suggestive that strain Ah5S-24 has the capacity for biofilm formation and quorum sensing as seen in other bacteria species (Enos-Berlage et al., 2005; Trappetti et al., 2011) while presence of the iron ABC transporter is suggestive that strain Ah5S-24 uses this protein in acquiring iron from infected hosts (Delepelaire, 2019). Detection of ddhA, ddhC, and wcaG1 associated with capsule formation (Mobine, 2008) is suggestive strain Ah5S-24 has the ability to form a capsule as a defense mechanism against host immune responses while presence of hylA, hylII,I, and TSH together with aerolysin aerA is suggestive that these genes might be linked to pore formation and intracellular release of enterotoxins by strain Ah5S-24 as seen in other bacteria species (Honda et al., 1992; Baida and Kuzmin, 1996; Abrami et al., 2000; Maté et al., 2014). Besides, several scientists (Wong et al., 1998; Heuzenroeder et al., 1999; Wu et al., 2007; Castilho et al., 2009) have shown that a combination of hylA(+) and aerA(+) is a major virulence determinant in Aeromonas spp. Castilho et al. (2009) found a high prevalence of hemolytic and cytotoxic Aeromonas spp. that had both hylA(+) and aerA+ from human clinical, food, and environmental samples in Brazil while Wu et al. (2007) showed that the absence of hlyA(−) and aerA(−) in Aeromonas spp. from fish and human samples in Taiwan was associated with low virulence. Heuzenroeder et al. (1999) and Wong et al. (1998) showed that deletion or attenuation of the hlyA(+) and aerA(+) double mutant significantly reduced the pathogenicity of A. hydrophila in mice. They also showed that cytotoxicity to buffalo green monkey kidney cells and hemolysis on horse blood agar was only eliminated in the double and not in the single mutants of A. veronii, A. hydrophila, and A. caviae. Our findings show that A. veronii Ah5S-24 had both hlyA(+) and aerA(+), indicating that it shares the two key virulence determinants with other pathogenic Aeromonas spp.
Several studies have shown that Aeromonas spp. intrinsically carry various blaOXA genes in their genomes that include the blaOXA-12 gene (Dubey et al., 2022a,b) previously detected in A. media, A. jandaei, A. sobria, A. dhakensis, and A. hydrophila (Dubey et al., 2022a; Rasmussen et al., 1994; Alksne and Rasmussen, 1997; Hilt et al., 2020; Huang et al., 2020) being in line with its presence in strain Ah5S-24 while blaFOX-7 previously reported in A. media and A. allosaccharophila was also found in strain Ah5S-24 (Ebmeyer et al., 2019). Other AMR genes detected included the cphA4 gene known to be intrinsically encoded in various Aeromonas spp. (Dubey et al., 2022a,b) as well as the colistin-resistance mcr-7.1 gene also reported from different Aeromonas spp. (Dubey et al., 2022a,b). Despite so, the blaOXA-12, blaFOX-7, and mcr-7.1 genes detected in strain Ah5S-24 were not associated with integrases, recombinases or transposases suggesting that these genes could not be easily transferred or acquired from other bacteria species. Similarly, although trimethoprim and sulfonamide are among the most widely used antibiotics linked to AMR in aquaculture (Gao et al., 2012; Muziasari et al., 2014; Phu et al., 2015), the trimethoprim resistance gene dfrA3 detected in the present study was not linked to integrases and transposases. Thus, the sulfonamide and trimethoprim resistance observed in the disc diffusion test could have been mediated by the MexB and smeD pumps that have been associated with resistance of several drugs that include sulfonamide, fluoroquinolone, cephalosporins, carbapenem, and trimethoprim. The trimethoprim and sulfonamide resistance observed on the disc diffusion test was intermediate (I) unlike the tetracycline resistance (R), which was highly expressed suggesting that the impact of trimethoprim and sulfonamide in conferring resistance was not as high as tetracycline in strain AhS5-24. Despite so, we found a high correlation of kappa score of 82% (Cohen’s k = 0.8235) with a specificity of 91.66% and sensitivity of 93% between the phenotype characterization based on the disc diffusion test and genotypic characterization based on the CARD (Alcock et al., 2020), indicating that the two diagnostic tests were highly in agreement.
Tetracycline is one of the most widely used antibiotics in aquaculture, which has been linked to resistance in farmed aquatic organisms (Seyfried et al., 2010; Tamminen et al., 2011). Thus, it is likely that selection of the Tet E operon in strain AhS5-24 occurred in pond sediments used for aquaculture where tetracycline was used for the treatment of fish diseases. Although the absence of plasmids is suggestive that strain Ah5S-24 had lesser chances of transferring AMR genes to other bacteria, detection of the Tet E operon together with the integrase and IS5/IS1182 transposase suggests that tetR and tet(E) genes could be transferred or acquired from other bacteria using the IS5/IS1182/hp/tet(E)/tetR/hp cassette encoded in strain Ah5S-24. Besides, DePaola et al. (1988) used the same isolate to transfer the OTr and Tr resistance to E. coli suggesting that the IS5/IS1182/hp/tet(E)/tetR/hp cassette found in strain AhS5-24 could have been responsible for transferring the tetracycline resistance to E. coli. Also, detection of the same cassette in V. parahaemolyticus, V. alginolyticus (MN199028.1), A. media (CP038444.1), A. caviae (CP110176.1), and A. caviae (CP038445.1) emanating from fish market, shrimp, sewage bioreactor and human stool is suggesting that the IS5/IS1182/hp/tet(E)/tetR/hp transposon could be involved in interspecies transmission of the tet(E) and tetR genes in different bacteria species. These findings also suggest that the IS5/IS1182/hp/tet(E)/tetR/hp transposon might be in existence in different bacteria species found in different aquatic environments hosted by species that include shrimps, fish, animals and humans. Its presence in V. parahaemolyticus (MN199028.1) isolated from retail fish at markets and V. alginolyticus (MN865127.1) from shrimps is indicative that it could play a vital role in transmission of tet(E) and tetR genes to humans through food.
The similarity of the IS5/IS1182/hp/tet(E)/tetR/hp cassette found in the chromosomes of strain A. veronii AhS5-24, V. parahaemolyticus (MN199028.1) and A. media (CP038445.1), with the transposon found in the plasmid of V. alginolyticus (MN865127.1) is suggesting that the IS5/IS1182/hp/tet(E)/tetR/hp transposon can be transferable between chromosomes and plasmids of different bacteria species. Also, the high similarity of the tet(E) and tetR genes detected in strain AhS5-24 with those found in E. coli, K. pneumoniae, and Aeromonas spp. shown in the phylogenetic analysis consolidates our view that tet(E) and tetR genes could be transmissible between different bacteria species using MGCs. Thus, it is likely that the transfer of the OTr and Tr resistance to E. coli observed by DePaola et al. (1988) was not plasmid mediated but it was done by the IS5/IS1182/hp/tet(E)/tetR/hp transposon found in strain Ah5S-24. Therefore, our findings indicate that the resistance acquired by different Aeromonas spp. in aquatic environments could play a vital role in the transfer of AMR genes to foodborne, environmental, nosocomial and other bacteria species using MGCs. However, future studies should seek to demonstrate the transfer of tet(E) and tetR genes using the IS5/IS1182/hp/tet(E)/tetR/hp cassette to other bacteria spp. including nosocomial, foodborne and environmental bacteria.
In this study, we have shown that A. veronii AhS5-24 is a multidrug-resistant bacterium encoding several AMR and virulence genes. It encoded a tetracycline resistance operon Tet E placed in a transposon designated as IS5/IS1182/hp/tet(E)/tetR/hp found in different bacteria species inhabiting different aquatic environments and infecting different host species suggesting that the Tet E operon could be transferred to other bacteria. Overall, this study shows that MGCs encoding AMR genes found in bacteria inhabiting aquatic environments could play a vital role in the spread of AMR genes to other bacteria infecting animals and humans.
The Aeromonas veronii whole genome shotgun (WGS) project has the project accession JAJVCX000000000. This version of the project (01) has the accession number JAJVCX010000000 and consists of sequences JAJVCX010000001-JAJVCX010000157.
SD, HS, and HM: conceptualization, methodology, mobilizing resources, supervision, data curation, and bioinformatics analysis. SD, EA-W, BP, AD, HS, and HM: manuscript preparation, editing, and submission. All authors contributed to the article and approved the submitted version.
This study was financed by the Research Council of Norway (FIFOSA-21 Project) Grant Number 320692 and the National Natural Science Foundation of China (NSFC) Grant Number 32061133007.
The authors are grateful to Erik Hjerde from the Arctic University of Norway and ELIXIR Norway for guidance on Bioinformatics. Sofie Persdatter Sangnæs, at the Norwegian University of Life Sciences (NMBU) for technical support.
AD was employed by Angelo DePaola Consulting LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1112941/full#supplementary-material
Abrami, L., Fivaz, M., and Van Der Goot, F. G. (2000). Adventures of a pore-forming toxin at the target cell surface. Trends Microbiol. 8, 168–172. doi: 10.1016/S0966-842X(00)01722-4
Alcock, B. P., Raphenya, A. R., Lau, T. T., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alksne, L. E., and Rasmussen, B. A. (1997). Expression of the AsbA1, OXA-12, and AsbM1 beta-lactamases in Aeromonas jandaei AER 14 is coordinated by a two-component regulon. J. Bacteriol. 179, 2006–2013. doi: 10.1128/jb.179.6.2006-2013.1997
Arechaga, I., and Cascales, E. (2022). Editorial: bacterial secretion systems, volume II. Front. Microbiol. 13:13. doi: 10.3389/fmicb.2022.917591
Baida, G. E., and Kuzmin, N. P. (1996). Mechanism of action of hemolysin III from Bacillus cereus. Biochim Biophysica Acta 1284, 122–124. doi: 10.1016/S0005-2736(96)00168-X
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Becker, L., Steglich, M., Fuchs, S., Werner, G., and Nübel, U. (2016). Comparison of six commercial kits to extract bacterial chromosome and plasmid DNA for MiSeq sequencing. Sci. Rep. 6:28063, 1–5. doi: 10.1038/srep28063
Bioinformatics B. (2011). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Institute.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brown, H. J., Stokes, H., and Hall, R. M. (1996). The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178, 4429–4437. doi: 10.1128/jb.178.15.4429-4437.1996
Castilho, M. C., Castro, T. L., Araújo, V. S., Trajano, R. S., Santos, P. A., Pimenta, P., et al. (2009). High frequency of hemolytic and cytotoxic activity in Aeromonas spp. isolated from clinical, food and environmental in Rio de Janeiro, Brazil. Antonie Van Leeuwenhoek 96, 53–61. doi: 10.1007/s10482-009-9335-6
Cockerill, F. R., Wikler, M., Bush, K., Dudley, M., Eliopoulos, G., and Hardy, D.. (2012) Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement. CLSI, Pennsylvania USA.
Coil, D., Jospin, G., and Darling, A. E. (2015). A5-miseq: an updated pipeline to assemble microbial genomes from illumina MiSeq data. Bioinformatics 31, 587–589. doi: 10.1093/bioinformatics/btu661
Collis, C. M., and Hall, R. M. (1992). Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6, 2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x
Craig, N. (1996). Transposon Tn7. Curr Top Microbiol Immunol 204, 27–48. doi: 10.1007/978-3-642-79795-8_2
Delepelaire, P. (2019). Bacterial ABC transporters of iron containing compounds. Res. Microbiol. 170, 345–357. doi: 10.1016/j.resmic.2019.10.008
DePaola, A., Flynn, P., McPhearson, R. M., and Levy, S. (1988). Phenotypic and genotypic characterization of tetracycline-and oxytetracycline-resistant Aeromonas hydrophila from cultured channel catfish (Ictalurus punctatus) and their environments. Appl. Environ. Microbiol. 54, 1861–1863. doi: 10.1128/aem.54.7.1861-1863.1988
Dubey, S., Ager-Wiick, E., Peng, B., Evensen, O., Sørum, R. H., and Munang'andu, H. M. (2022a). Characterization and comparative analysis of virulence and antimicrobial resistance genes of Aeromonas media strain SD/21-15 from marine sediments with other Aeromonas spp. Front. Microbiol. 4217. doi: 10.3389/fmicb.2022.1022639
Dubey, S., Anger-Wick, E., Kumar, J., Karunasagar, I., Karunasagar, I., Peng, B., et al. (2022b). Aeromonas species isolated from aquatic organisms, insects, chicken, and humans in India show similar antimicrobial resistance profiles. Front. Microbiol. 4364. doi: 10.3389/fmicb.2022.1008870
Ebmeyer, S., Kristiansson, E., and Larsson, D. J. (2019). The mobile FOX AmpC beta-lactamases originated in Aeromonas allosaccharophila. Int. J. Antimicrob. Agents 54, 798–802. doi: 10.1016/j.ijantimicag.2019.09.017
Enos-Berlage, J. L., Guvener, Z. T., Keenan, C. E., and McCarter, L. L. (2005). Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55, 1160–1182. doi: 10.1111/j.1365-2958.2004.04453.x
Gao, P., Mao, D., Luo, Y., Wang, L., Xu, B., and Xu, L. (2012). Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 46, 2355–2364. doi: 10.1016/j.watres.2012.02.004
González-Serrano, C., Santos, J., García-López, M., and Otero, A. (2002). Virulence markers in Aeromonas hydrophila and Aeromonas veronii biovar sobria isolates from freshwater fish and from a diarrhoea case. J. Appl. Microbiol. 93, 414–419. doi: 10.1046/j.1365-2672.2002.01705.x
Guan, G., He, X., Chen, J., Bin, L., and Tang, X. (2020). Identifying the mechanisms underlying the protective effect of tetramethylpyrazine against cisplatin-induced in vitro ototoxicity in HEI-OC1 auditory cells using gene expression profiling. Mol. Med. Rep. 22, 5053–5068. doi: 10.3892/mmr.2020.11631
Hadi, N., Yang, Q., Barnett, T. C., Tabei, S. M. B., Kirov, S. M., and Shaw, J. G. (2012). Bundle-forming pilus locus of Aeromonas veronii bv sobria. Infect. Immun. 80, 1351–1360. doi: 10.1128/IAI.06304-11
Heuzenroeder, M. W., Wong, C. Y., and Flower, R. L. (1999). Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: correlation with virulence in a suckling mouse model. FEMS Microbiol. Lett. 174, 131–136. doi: 10.1111/j.1574-6968.1999.tb13559.x
Hickman-Brenner, F., Mac Donald, K., Steigerwalt, A., Fanning, G., Brenner, D. J., and Farmer, J. 3rd. (1987). Aeromonas veronii, a new ornithine decarboxylase-positive species that may cause diarrhea. J. Clin. Microbiol. 25, 900–906. doi: 10.1128/jcm.25.5.900-906.1987
Hilt, E. E., Fitzwater, S. P., Ward, K., de St Maurice, A., Chandrasekaran, S., Garner, O. B., et al. (2020). Carbapenem resistant Aeromonas hydrophila carrying Bla(cphA7) isolated from two solid organ transplant patients. Front. Cell. Infect. Microbiol. 10:563482. doi: 10.3389/fcimb.2020.563482
Hoai, T. D., Trang, T. T., Van Tuyen, N., Giang, N. T. H., and Van Van, K. (2019). Aeromonas veronii caused disease and mortality in channel catfish in Vietnam. Aquaculture 513:734425. doi: 10.1016/j.aquaculture.2019.734425
Honda, T., Ni, Y., Miwatani, T., Adachi, T., and Kim, J. (1992). The thermostable direct hemolysin of Vibrio parahaemolyticus is a pore-forming toxin. Can. J. Microbiol. 38, 1175–1180. doi: 10.1139/m92-192
Huang, M., Chen, H., Li, C., Liu, Y., Gan, C., MAE-G, E.-S. A., et al. (2020). Rapid fulminant progression and mortality secondary to Aeromonas dhakensis septicemia with hepatitis B virus infection following the ingestion of snakehead fish in mainland China: a case report. Foodborne Pathog. Dis. 17, 743–749. doi: 10.1089/fpd.2019.2780
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Joseph, N. M., Sistla, S., Dutta, T. K., Badhe, A. S., Rasitha, D., and Parija, S. C. (2011). Reliability of Kirby-Bauer disk diffusion method for detecting meropenem resistance among non-fermenting gram-negative bacilli. Indian J. Pathol. Microbiol. 54, 556–560. doi: 10.4103/0377-4929.85092
Kahlmeter, G., Brown, D. F., Goldstein, F. W., MacGowan, A. P., Mouton, J. W., Odenholt, I., et al. (2006). European Committee on Antimicrobial Susceptibility Testing (EUCAST) Technical Notes on antimicrobial susceptibility testing. Clin Microbiol Infect. 12, 501–503. doi: 10.1111/j.1469-0691.2006.01454.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liebert, C. A., Hall, R. M., and Summers, A. O. (1999). Transposon Tn 21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522. doi: 10.1128/MMBR.63.3.507-522.1999
Maté, S. M., Vázquez, R. F., Herlax, V. S., Millone, M. A. D., Fanani, M. L., Maggio, B., et al. (2014). Boundary region between coexisting lipid phases as initial binding sites for Escherichia coli alpha-hemolysin: a real-time study. Biochim Biophys Acta 1838, 1832–1841. doi: 10.1016/j.bbamem.2014.02.022
Meyer, J. F., Nies, B. A., and Wiedemann, B. (1983). Amikacin resistance mediated by multiresistance transposon Tn2424. J. Bacteriol. 155, 755–760. doi: 10.1128/jb.155.2.755-760.1983
Minakhina, S., Kholodii, G., Mindlin, S., Yurieva, O., and Nikiforov, V. (1999). Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33, 1059–1068. doi: 10.1046/j.1365-2958.1999.01548.x
Mobine, H. R. (2008). Pheochromocytoma-Induced Cardiomyopathy: A Model of Synergistic Effects of Multifactorial Tumor Secretions Massachusetts Institute of Technology. Doctoral dissertation, MIT, USA.
Muziasari, W. I., Managaki, S., Pärnänen, K., Karkman, A., Lyra, C., Tamminen, M., et al. (2014). Sulphonamide and trimethoprim resistance genes persist in sediments at Baltic Sea aquaculture farms but are not detected in the surrounding environment. PLoS One 9:e92702. doi: 10.1371/journal.pone.0092702
Phu, T. M., Scippo, M.-L., Phuong, N. T., Tien, C. T. K., Son, C. H., and Dalsgaard, A. (2015). Withdrawal time for sulfamethoxazole and trimethoprim following treatment of striped catfish (Pangasianodon hypophthalmus) and hybrid red tilapia (Oreochromis mossambicus × Oreochromis niloticus). Aquaculture 437, 256–262. doi: 10.1016/j.aquaculture.2014.12.009
Ragupathi, N. K. D., Sethuvel, D. P. M., Anandan, S., Murugan, D., Asokan, K., Mohan, R. G. N., et al. (2020). First hybrid complete genome of Aeromonas veronii reveals chromosome-mediated novel structural variant mcr-3.30 from a human clinical sample. Access Microbiol. 2:acmi000103. doi: 10.1099/acmi.0.000103
Rasmussen, B. A., Keeney, D., Yang, Y., and Bush, K. (1994). Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob. Agents Chemother. 38, 2078–2085. doi: 10.1128/AAC.38.9.2078
Recchia, G. D., and Hall, R. M. (1995). Gene cassettes: a new class of mobile element. Microbiology 141, 3015–3027. doi: 10.1099/13500872-141-12-3015
Ro, C. (2006). Altarriba M, Vilches S, Horsburgh G, Shaw JG, Tomás JM, merino S: analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 188, 852–862. doi: 10.1128/JB.188.3.852-862.2006
Roberts, M., Enoch, D., Harris, K., and Karas, J. (2006). Aeromonas veronii biovar sobria bacteraemia with septic arthritis confirmed by 16S rDNA PCR in an immunocompetent adult. J. Med. Microbiol. 55, 241–243. doi: 10.1099/jmm.0.46295-0
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seemann, T. (2016). ABRicate: Mass screening of contigs for antibiotic resistance genes. GitHub. Available at: https://github.com/tseemann/abricate
Seyfried, E. E., Newton, R. J., Rubert, K. F., Pedersen, J. A., and McMahon, K. D. (2010). Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb. Ecol. 59, 799–807. doi: 10.1007/s00248-009-9624-7
Smyrli, M., Prapas, A., Rigos, G., Kokkari, C., Pavlidis, M., and Katharios, P. (2017). Aeromonas veronii infection associated with high morbidity and mortality in farmed European seabass Dicentrarchus labrax in the Aegean Sea. Greece. Fish Pathol. 52, 68–81. doi: 10.3147/jsfp.52.68
Smyrli, M., Triga, A., Dourala, N., Varvarigos, P., Pavlidis, M., Quoc, V. H., et al. (2019). Comparative study on a novel pathogen of European seabass. Diversity of Aeromonas veronii in the Aegean Sea. Microorganisms 7:504. doi: 10.3390/microorganisms7110504
Tamminen, M., Karkman, A., Lohmus, A., Muziasari, W. I., Takasu, H., Wada, S., et al. (2011). Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45, 386–391. doi: 10.1021/es102725n
Tarumoto, N., Sakai, J., Sujino, K., Yamaguchi, T., Ohta, M., Yamagishi, J., et al. (2017). Use of the Oxford Nanopore MinION sequencer for MLST genotyping of vancomycin-resistant enterococci. J. Hosp. Infect. 96, 296–298. doi: 10.1016/j.jhin.2017.02.020
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tekedar, H. C., Arick, M. A., Hsu, C.-Y., Thrash, A., Blom, J., Lawrence, M. L., et al. (2020). Identification of antimicrobial resistance determinants in Aeromonas veronii strain MS-17-88 recovered from channel catfish (Ictalurus punctatus). Front. Cell. Infect. Microbiol. 10:348. doi: 10.3389/fcimb.2020.00348
Tran, T. T., Scott, A., Tien, Y.-C., Murray, R., Boerlin, P., Pearl, D. L., et al. (2021). On-farm anaerobic digestion of dairy manure reduces the abundance of antibiotic resistance-associated gene targets, and the potential for plasmid transfer. Appl. Environ. Microbiol. 87, 02980–02920. doi: 10.1128/AEM.02980-20
Trappetti, C., Potter, A. J., Paton, A. W., Oggioni, M. R., and Paton, J. C. (2011). LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect. Immun. 79, 4550–4558. doi: 10.1128/IAI.05644-11
Tyagi, A., Sharma, C., Srivastava, A., Kumar, B. N., Pathak, D., and Rai, S. (2022). Isolation, characterization and complete genome sequencing of fish pathogenic Aeromonas veronii from diseased Labeo rohita. Aquaculture 553:738085:738085. doi: 10.1016/j.aquaculture.2022.738085
Ullah, S. R., Majid, M., and Andleeb, S. (2020). Draft genome sequence of an extensively drug-resistant neonatal Klebsiella pneumoniae isolate harbouring multiple plasmids contributing to antibiotic resistance. J. Glob. Antimicrob. Resist. 23, 100–101. doi: 10.1016/j.jgar.2020.08.008
Wang, H., Lin-Zhao, Z., Jie-An, D., Lin-Wang, J., Tong-Yang, B., Huan-Kang, Y., et al. (2022). The lip gene contributes to the virulence of Aeromonas veronii strain TH0426. Microb. Pathog. 167:105566. doi: 10.1016/j.micpath.2022.105566
Ward, J., and Grinsted, J. (1982). Physical and genetic analysis of the Inc-W group plasmids R388, Sa, and R7K. Plasmid 7, 239–250. doi: 10.1016/0147-619X(82)90005-1
White, P. A., McIver, C. J., and Rawlinson, W. D. (2001). Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45, 2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001
Wong, C. Y., Heuzenroeder, M. W., and Flower, R. L. (1998). Inactivation of two haemolytic toxin genes in Aeromonas hydrophila attenuates virulence in a suckling mouse model. Microbiology 144, 291–298. doi: 10.1099/00221287-144-2-291
Keywords: Aeromonas veronii, antimicrobial resistance, mobile gene cassette, virulence, tetracycline, environment, foodborne
Citation: Dubey S, Ager-Wiick E, Peng B, DePaola A, Sørum H and Munang’andu HM (2023) The mobile gene cassette carrying tetracycline resistance genes in Aeromonas veronii strain Ah5S-24 isolated from catfish pond sediments shows similarity with a cassette found in other environmental and foodborne bacteria. Front. Microbiol. 14:1112941. doi: 10.3389/fmicb.2023.1112941
Received: 30 November 2022; Accepted: 13 February 2023;
Published: 15 March 2023.
Edited by:
Zhi Ruan, Zhejiang University, ChinaReviewed by:
Tingting Xu, Jinan University, ChinaCopyright © 2023 Dubey, Ager-Wiick, Peng, DePaola, Sørum and Munang’andu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hetron Mweemba Munang’andu, aGV0cm9uLm0ubXVuYW5nYW5kdUBub3JkLm5v
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.