- 1Beijing Advance Innovation Center for Food Nutrition and Human Health, Beijing Laboratory of Food Quality and Safety, Beijing Engineering and Technology Research Center of Food Additives, Beijing Technology and Business University, Beijing, China
- 2School of Control and Computer Engineering, North China Electric Power University, Beijing, China
Plantaricin is a kind of bacteriocin with broad-spectrum antibacterial activity on several food pathogens and spoilage microorganisms, showing potential in biopreservation applications. However, the low yield of plantaricin limits its industrialization. In this study, it was found that the co-culture of Wickerhamomyces anomalus Y-5 and Lactiplantibacillus paraplantarum RX-8 could enhance plantaricin production. To investigate the response of L. paraplantarum RX-8 facing W. anomalus Y-5 and understand the mechanisms activated when increasing plantaricin yield, comparative transcriptomic and proteomic analyses of L. paraplantarum RX-8 were performed in mono-culture and co-culture. The results showed that different genes and proteins in the phosphotransferase system (PTS) were improved and enhanced the uptake of certain sugars; the key enzyme activity in glycolysis was increased with the promotion of energy production; arginine biosynthesis was downregulated to increase glutamate mechanism and then promoted plantaricin yield; and the expression of several genes/proteins related to purine metabolism was downregulated and those related to pyrimidine metabolism was upregulated. Meanwhile, the increase of plantaricin synthesis by upregulation of plnABCDEF cluster expression under co-culture indicated that the PlnA-mediated quorum sensing (QS) system took part in the response mechanism of L. paraplantarum RX-8. However, the absence of AI-2 did not influence the inducing effect on plantaricin production. Mannose, galactose, and glutamate were critical metabolites and significantly simulate plantaricin production (p < 0.05). In summary, the findings provided new insights into the interaction between bacteriocin-inducing and bacteriocin-producing microorganisms, which may serve as a basis for further research into the detailed mechanism.
Introduction
Bacteriocins are proteins or peptides that display antibacterial activity against various spoilage and pathogenic bacteria (Bu et al., 2021b). Bacteriocins from lactic acid bacteria (LAB) are regarded as safe, natural antimicrobials for food biological preservatives (Bu et al., 2021a). However, the manufacturing and secretion of bacteriocin have high metabolic costs; therefore, bacteria have developed regulatory mechanisms that relay quorum sensing (QS) signals to generate bacteriocins only upon necessity (Kern et al., 2021). As they lack competitors, bacteriocin production is low and may be gradually lost in ideal laboratory conditions. To increase bacteriocin production, it is useful to add certain microorganisms to induce bacteriocin-producing bacteria to produce more bacteriocins. This approach uses the interaction between different strains in mixed culture and was widespread in the area of natural bacteriocin production (Chanos and Mygind, 2016).
Plantaricin, a class IIb bacteriocin produced by Lactiplantibacillus plantarum, is often induced by the bacteria–bacteria interactions. The mechanism of induction regulation behind plantaricin production under co-culturing with other bacteria is associated with QS, the regulation of specific genes in a bacterial population. Specifically, it was reported that the plantaricin production of L. plantarum NC8 by co-culture is mediated by the three-component regulatory system, compounded by an autoinduction factor (PlNC8IF), the histidine kinase (PlNC8HK), and the response regulator (RR; PlnD) (Maldonado-Barragán et al., 2013). Furthermore, the interspecies QS molecule AI-2 and its related gene LuxS both increased in L. plantarum DC400 under co-culture (Calasso et al., 2013). Meanwhile, the two QS signals (PlnA and AI-2) independently participate in the plantaricin induction mechanism of the co-culture system (L. paraplantarum RX-8 and Bacillus subtilis BS-15) (Liu et al., 2022).
However, the inducing mechanism of bacteriocin production when bacteriocin-producing strains co-culture with fungi remains unknown, especially for plantaricin. It has been reported that undissociated lactate and a low pH strongly inhibit the growth of Lactococcus lactis and lead to a low yield of nisin, while Kluyveromyces marxianus MS1 (Wardani et al., 2006), Yarrowia lipolytica ATCC 18942 (Ariana and Hamedi, 2017), and Saccharomyces cerevisiae W303-1A (Liu et al., 2021) could increase nisin production by consuming lactate. However, research involving a detailed mechanism at the gene level is still inadequate. Unlike that in bacteria, the QS system in fungi has different models and the bacteria–fungi interaction has yet to be studied in detail. A deeper understanding of interaction networks between microbes, including bacteria–bacteria, bacteria–fungi, and fungi–fungi, will help in the exploration of the induction mechanism of yeast, which has an inducing effect on bacteriocin production, and provide a new method for control.
Plantaricin RX-8 produced by L. paraplantarum RX-8 was proven to exhibit broad-spectrum antibacterial activity, and the yield of plantaricin RX-8 was improved by Wickerhamomyces anomalus Y-5. In this study, interactions between L. paraplantarum RX-8 and W. anomalus Y-5, the pH, the plantaricin production, and the transcription levels of related bacteriocin biosynthesis genes from 4 to 32 h in co-culture/mono-culture were investigated. Furthermore, transcriptomic and proteomic technologies were used to compare the difference in gene and protein expression between the L. paraplantarum RX-8 cells under a co-culture and mono-culture.
Methods and materials
2.1. Co-culture experiment
L. paraplantarum RX-8 (CGMCC 20852) was isolated from the traditional pickle in Chengdu, Sichuan Province, China. W. anomalus Y-5 was isolated from fermented grains of Chinese liquor and stored in the food quality and safety laboratory at Beijing Technology and Business University (Beijing, China). In brief, L. paraplantarum RX-8 and W. anomalus Y-5 were cultured overnight in De Man–Rogosa–Sharp (MRS) medium at 37°C and Yeast Peptone Dextrose (YPD) medium at 30°C under aerobic conditions, respectively. Then, 1 ml of L. paraplantarum RX-8 and W. anomalus Y-5 were singly inoculated into 100 ml of MRS liquid media for 24 h at 37°C as mono-cultured samples. Then, 1 ml of L. paraplantarum RX-8 and W. anomalus Y-5 were simultaneously inoculated into 100 ml of MRS liquid media for 24 h at 37°C as co-cultured samples.
2.2. Determination of viable cells and pH
The viable cells were counted every 4 h from 0 to 32 h, and the assay of viable cell counts was carried out according to the method of Zhang et al. (2021). At the same fermentation time points, the cell-free supernatants (CFSs) were obtained by removing cells through centrifugation (10,000 rpm, 10 min, 4°C). Then, the pH value of the CFSs was measured with a pH meter (PH400, Alalis, USA).
2.3. Plantaricin production assay
The yield of plantaricin in mono-culture/co-culture was assayed through agar well diffusion. After every 4 h (4–32 h) fermentation, the CFSs were obtained by centrifugation at 10,000 rpm for 10 min at 4°C; then, the pH of CFSs was adjusted to 6.0 with 1 mol/L NaOH, and these CFS samples were concentrated by vacuum centrifugal concentration (1,500 rpm, 45°C, 5 h). Wells of 6 mm were loaded with 100 μl of 5-fold concentrated plantaricin in the agar, which was inoculated at about 107 CFU Listeria monocytogenes 35152. The plates were stored at 4°C overnight to allow for the plantaricin to completely diffuse; then, these plates were cultured for 8 h at 37°C. The bacteriocin activity was expressed in arbitrary units (AU/ml), which are represented as the reciprocal of the highest dilution, showing distinct zones of inhibition, and calculated according to the equation:
where a is 2 (dilution factor), n is the reciprocal of the highest dilution that resulted in inhibition of the indicator strain, b is 100 μl (sample volume in each well), and c is 5 (sample concentration fold). Furthermore, the relative bacteriocin activity was defined as the ratio of bacteriocin activity in co-culture to that in mono-culture. This was used to evaluate the influence of W. anomalus Y-5 on plantaricin production in co-culture.
To verify the role of bacteriocin in the competition of bacteria and yeast, the inhibiting effect of plantaricin on W. anomalus Y-5 was determined according to Section 2.3.
2.4. RT-qPCR analysis of plantaricin locus and AI-2 synthesis-related gene
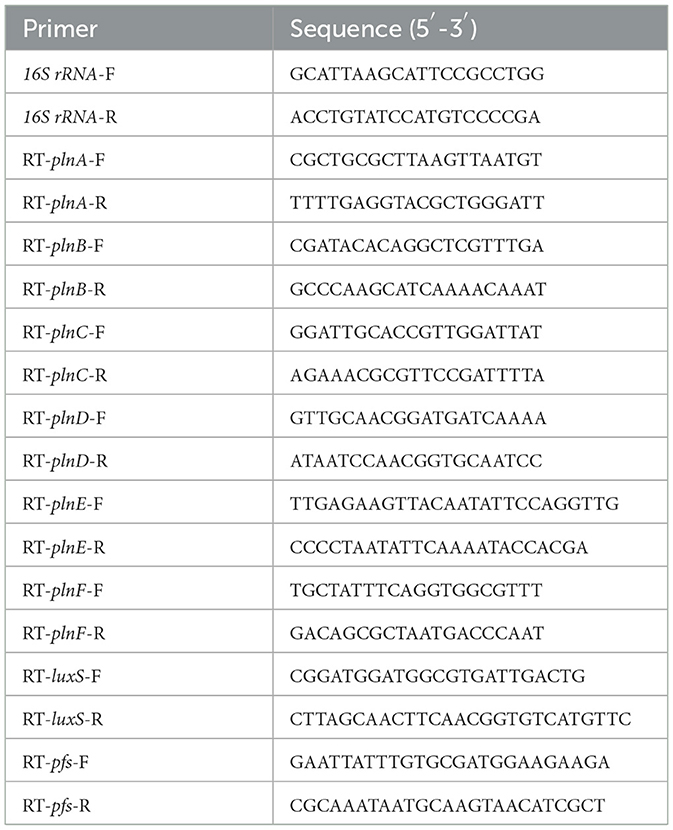
To determine the expression level of plantaricin biosynthesis gene clusters (plnABCDEF) and AI-2 synthesis-related genes (pfs and luxS), cells of L. paraplantarum RX-8 were harvested through centrifugation (10,000 rpm, 2 min, 4°C) at every 4 h in mono-culture/co-culture (4–32 h). RNA was extracted from cells using the RNAprep Pure Cell/Bacteria Kit according to the manufacturer's instructions (DP430, Tiangen Biotech, China). The extracted RNA was reverse-transcribed into cDNA using the FastQuant RT Kit (with gDNase) (KR116, Tiangen Biotech, China). The transcriptional levels of plnABCDEF, pfs, and luxS were realized by CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) using SYBR Green PCR master mix (FP205, Tiangen Biotech, China). The reaction mixture consists of 10 μl 2×SuperReal PreMix Plus, 0.6 μl each of the forward and reverse primer, 1.0 μl cDNA (100 ng), and 7.8 μl nuclease-free H2O. The amplification program was performed at 95°C for 15 min, followed by 39 cycles of 95°C for 10 s, 55°C for 20 s, and 72°C for 30 s. Primers were designed based on the genome sequence of L. paraplantarum RX-8 and synthesized by Beijing Genomics Institute (Table 1). 16S rRNA genes were used as the reference gene. The relative expression levels of target genes at each sampling point for L. paraplantarum RX-8 in mono-culture/co-culture were calculated by the ΔΔCt method.
2.5. Transcriptomic analysis
2.5.1. RNA extraction, library construction, and transcriptome sequencing
Cell samples of L. paraplantarum RX-8 were obtained in co-culture and mono-culture at 24 h by centrifugation (10,000 rpm, 10 min, 4°C). After removing the supernatant, cells were immediately cleaned by phosphate buffered saline (PBS) two times, frozen in liquid nitrogen for 15 min, and then stored at −80°C.
Total RNA extractions were performed using Magen HiPure Universal RNA Kit (Magen, China). RNA concentration and purity were measured using Qubit 3.0 (Thermo Fisher Scientific, MA, USA) and Nanodrop One (Thermo Fisher Scientific, MA, USA), and integrity was confirmed using the Agilent 4200 system (Agilent Technologies, Waldbron, Germany). The RNA samples with an RNA integrity number (RIN) >6.5, and 280/260 ratios >1.5 were further used for RNA-sequencing purposes. Libraries were generated from three replicates using NEB Next®Ultra™ Directional RNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA). From these libraries, the PE150 reads were produced with the Illumina Novaseq6000 platform by Guangdong Magigene Biotechnology Co., Ltd. (Guangzhou, China). All raw RNA-Seq data were submitted to NCBI under BioProject PRJNA901346.
2.5.2. Sequencing data processing
The raw reads were trimmed using Trimmomatic (version 0.36) to obtain the qualified reads (Bolger et al., 2014). Then, the rRNA sequences were removed in the qualified reads mapping with NCBI Rfam datasets using Bowtie2 (version 2.33) (Langdon, 2015). Initially, we mapped the mRNA reads of these samples to the reference genome L. plantarum WCFS1 downloaded from NCBI, which was the strain that was most closely related to L. paraplantarum RX-8, but the acquired alignment rate was mostly low at around 50% and limited to transcriptomic analysis. Therefore, to obtain a higher mapping rate, we assembled the draft genome of L. paraplantarum RX-8 using Unicycler (version 0.4.7) to assemble the combined Illumina NovaSeq PE150 data and Nanopore PromethION data (Wick et al., 2017). The new reference genome was uploaded to the NCBI shared database for research retrieval in NCBI GenBank CP111117-CP111119. Eventually, the transcriptomic data were mapped to the new reference genome using Hisat2 (version 2.1.0) (Kim et al., 2019), and the mapping rate was improved to above 90%. Read counts per genome assembly annotated transcript were calculated using HTseq (version 0.9.1) (Anders et al., 2015).
2.5.3. Differential expression genes detection
The differentially expressed genes (DEGs) between groups (RY and R, three biological replicates) were identified from all genes according to the expression levels of the transcripts, based on the statistics of the fragments per kilobase of each reading, per million mapped reads (FPKM). The DEGs were detected using edgeR (version 3.16.5) with default parameters (Robinson et al., 2010). We then used Benjamini and Hochberg's correction approach to adjust the p-values to control the false discovery rate (FDR). The genes were regarded as DEGs if they satisfied the threshold with FDR ≤ 0.05 and |Log2FC| ≥ 1. These genes were used for further enrichment analysis.
2.5.4. Enrichment analysis
The enrichment analyses of DEGs with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were implemented using clusterProfiler (version 3.4.4), in which gene length bias was corrected. The GO terms and KEGG pathways with FDR ≤ 0.05 were regarded as candidates for enrichment functional annotation via the DEGs.
2.6. Proteomic analysis
2.6.1. Cytosolic protein extraction and in-solution protein digestion
Cell samples of L. paraplantarum RX-8 in co-culture and mono-culture were collected at 24 h according to Section 2.5.1. The protein was extracted by using a lysis buffer (8 M urea, 50 mM Tris8.0, 1% NP40, 1% sodium deoxycholate, 5 mM dithiothreitol (DTT), 2 mM EDTA, 30 mM nicotinamide, and 3 μm trichostatin A), and, after sonication on ice, the total protein concentration of the supernatant, which was obtained by centrifugation (20,000 rpm, 10 min, 4°C), was determined by using a BCA Protein Assay kit. The protein sample was reduced by DTT (5 mM, 45 min, 30°C), later alkylated with 30 mM iodoacetamide (30 mM, 1 h, RT) in darkness, and then precipitated with ice-cold acetone. After being washed thrice with acetone, the precipitate was suspended in 0.1 M triethylammonium bicarbonate (TEAB) and digested with trypsin (1/25 protein mass, Promega) for 12 h at 37°C. Finally, the reaction was ended with 1% trifluoroacetic acid (TFA), and the resulting peptide was desalted with Strata X C18 SPE column (Phenomenex, Torrance, CA, USA) and vacuum-dried in Scanvac maxi-beta (Labogene, Alleroed, Denmark).
2.6.2. TMT labeling and HPLC fractionation
After reduction and alkylation, six digested peptide samples (RY and R, three biological replicates, 100 μg/sample) were transferred to the TMT Reagent vial (Thermo Fisher Scientific, 90066) and incubated for 2 h at room temperature. Then, 8 μl of 5% hydroxylamine was added to the mixture and incubated for 15 min to quench the reaction. Finally, the labeled samples were fractionated into 15 fractions by XBridge Shield C18 RP column (Waters, Milford, MA, USA) in an LC20AD HPLC system (Shimadzu, Kyoto, Japan) for proteomic analysis.
2.6.3. LC-MS analysis
The sample was loaded to the column equilibrated with buffers A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). The Acclaim PepMap 100 C18 trap column (75 μm × 2 cm, Dionex) was equilibrated with liquid A by Ultimate 3000 nanoUPLC (Dionex), and the sample was eluted onto an Acclaim PepMap RSLC C18 analytical column (75 μm × 25 cm, Dionex) at a flow rate of 300 nl/min. The liquid-phase gradient was given as follows: 0–6 min, where the linear gradient of buffer B was from 2 to 10%; 7–51 min, where the linear gradient of buffer B was from 10 to 20%; 51–53 min, where the linear gradient of buffer B was from 20 to 80%; 53–57 min, where the linear gradient of buffer B solution was held at 80%.
Mass analysis was performed by nano-spray ionization-mass spectrometry (NSI-MS) and Q-Exactive HF mass spectrometry (Thermo Scientific). The intact peptide was detected by the Orbitrap, the scanning range was set to 250–1,500 m/z, the automatic gain control (AGC) target was 3E6, the resolution was 70,000, the max injection time (IT) was 250 ms, and the dynamic exclusion time was 15 s. The peptide was selected and fragmented for MS/MS using 28% NCE; ion fragments were detected in the Orbitrap, the resolution was 17,500, AGC was 1E5 or 5E4, and the maximum IT was 100 or 200 ms. LC-MS was performed by Micrometer Biotech (Hangzhou, China).
2.6.4. Protein database search and protein quantification
To improve the accuracy of protein identification, RNA-Seq data as mentioned earlier were used to construct specific reference protein databases using DIAMOND (version 2.0.15) (Buchfink et al., 2015). The MS files were searched against the sample-specific L. paraplantarum RX-8 database using MaxQuant (version 1.5.2.8) (Tyanova et al., 2016). The mass error of precursor and fragment ions was set as 10 ppm and 0.02 Da, respectively. Trypsin was selected for enzyme specificity and two missed cleavages were allowed. Carbamidomethylation on Cys, TMT 6-plex tag of Lys, and peptide N-terminal was specified as a fixed modification. The variable modifications were oxidation on Met and TMT 6-plex tag on Tyr. FDR was estimated by reverse-decoy strategy, percolator algorithm, and peptide–spectrum match (PSM). P < 0.05 and e < 0.05 were used to calculate the results. For quantitation, a protein must have at least two unique peptides with the above identity. The protein ratio type was median, and the normalization method was median. All the raw files were uploaded to PRIDE with the accession number PXD038315.
2.6.5. Bioinformatics analysis
The proteins identified as differentially expressed proteins (DEPs) should satisfy the threshold of p ≤ 0.05 and |Fold Change| ≥ 1.3. The enrichment analysis of DEPs used the clusterProfiler (version 3.4.4) for GO function annotation and KEGG pathways (Yu et al., 2012). The GO terms and KEGG pathways were regarded as significant protein enrichment annotations using the DEPs.
2.7. Key differential genes verification
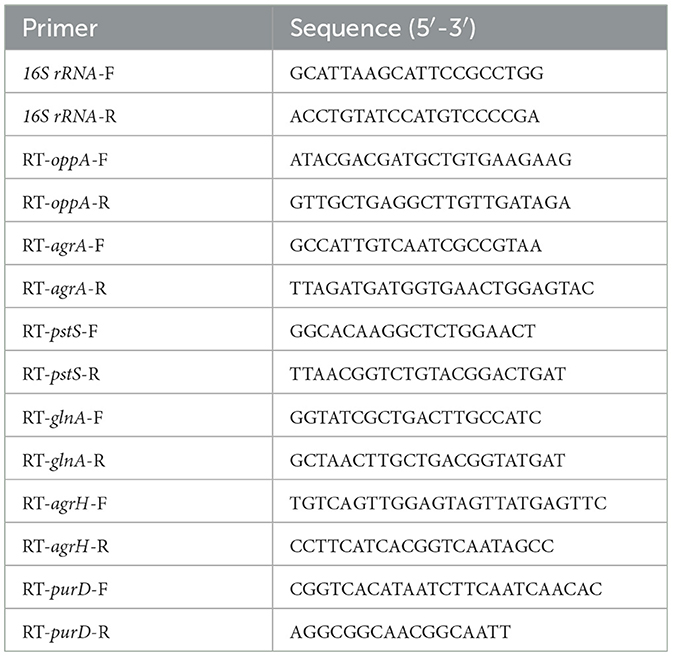
To validate the transcriptomic and proteomic data, RT-qPCR was employed to measure the expression of selected genes in mRNA, according to Section 2.4. The primers for selected genes were synthesized by the Beijing Genomics Institute, and the sequences are listed in Table 2.
2.8. Key inducing metabolite verification
According to the results of transcriptomic and proteomic analyses, the carbohydrate substance (mannose, galactose, cellobiose, and D-ribose) was added to the mono-culture of L. paraplantarum RX-8 at the concentrations of 2, 20, and 200 mM. Meanwhile, the amino acid substance (arginine, cysteine, glutamate, and glutamine) was added to the mono-culture of L. paraplantarum RX-8 at the concentrations of 0.5, 1.25, and 2.5 g/L. After being cultured for 24 h at 37°C, the supernatants of each sample were collected for the plantaricin production assay. The mono-culture of L. paraplantarum RX-8 was used as a control.
2.9. Determination of AI-2 activity on inducing effect
To determine AI-2's role in inducing effect, AI-2 inhibitor D-ribose (0, 200, 300, and 400 mM) was added to the co-culture system; then, the plantaricin production of these samples at 24 h was detected according to Section 2.2, as described earlier. The plantaricin production in co-culture was used as the positive control, and that in mono-culture was used as the negative control.
The AI-2 activity was detected by the bioluminescence of Vibrio harveyi BB170. After overnight culture at 30°C, V. harveyi BB170 was diluted in a ratio of 1:5,000 with fresh AB medium. The CFSs of the above samples were adjusted to pH 7.0, then filtrated with a 0.22-μm sterile filter, and added to the diluted BB170 culture at the percentage of 10%. The mixture was incubated at 30°C for 4 h under aerobic conditions (180 rpm), and 200 μl of aliquots were added to white 96-well plates (Thermo, USA) to measure relative luminescence units (RLUs) using the Multi-Detection Plate Reader (SpectraMax i3, Molecular Devices, USA). The suspension of strain BB170 in AB medium (1:5,000) was used as a blank control.
Results and discussion
3.1. Growth dynamics and bacteriocin production of L. paraplantarum RX-8 and W. anomalus Y-5 under co-culture
The growths of L. paraplantarum RX-8/W. anomalus Y-5, the pH, and bacteriocin production in co-culture and every mono-culture from 4 to 32 h are shown in Figure 1. The viable cell numbers for the bacteria and yeast in co-culture were tested using different antibiotic susceptibilities of L. paraplantarum RX-8/W. anomalus Y-5. Up to 16 h, the growth rate of L. paraplantarum RX-8 was very consistent between the mono-culture and co-culture. However, a decline in L. paraplantarum RX-8 cell number was evident during 20–32 h. W. anomalus Y-5 grew poorly in liquid MRS media, as it lacked adequate oxygen and an appropriate temperature. The growth of W. anomalus Y-5 was also significantly reduced when L. paraplantarum RX-8 was present for 4–20 h, while W. anomalus Y-5 began to regrow over 24–32 h. The results of the antimicrobial assay show that plantaricin from L. paraplantarum RX-8 could not inhibit the growth of W. anomalus Y-5 (Supplementary Figure 1). A suitable pH for W. anomalus Y-5 growth was about 5.5. It was also reported that W. anomalus could not grow well under low pH conditions (Tian et al., 2020). The competitive effect of the two strains may be caused by nutrient competition and acid stress.
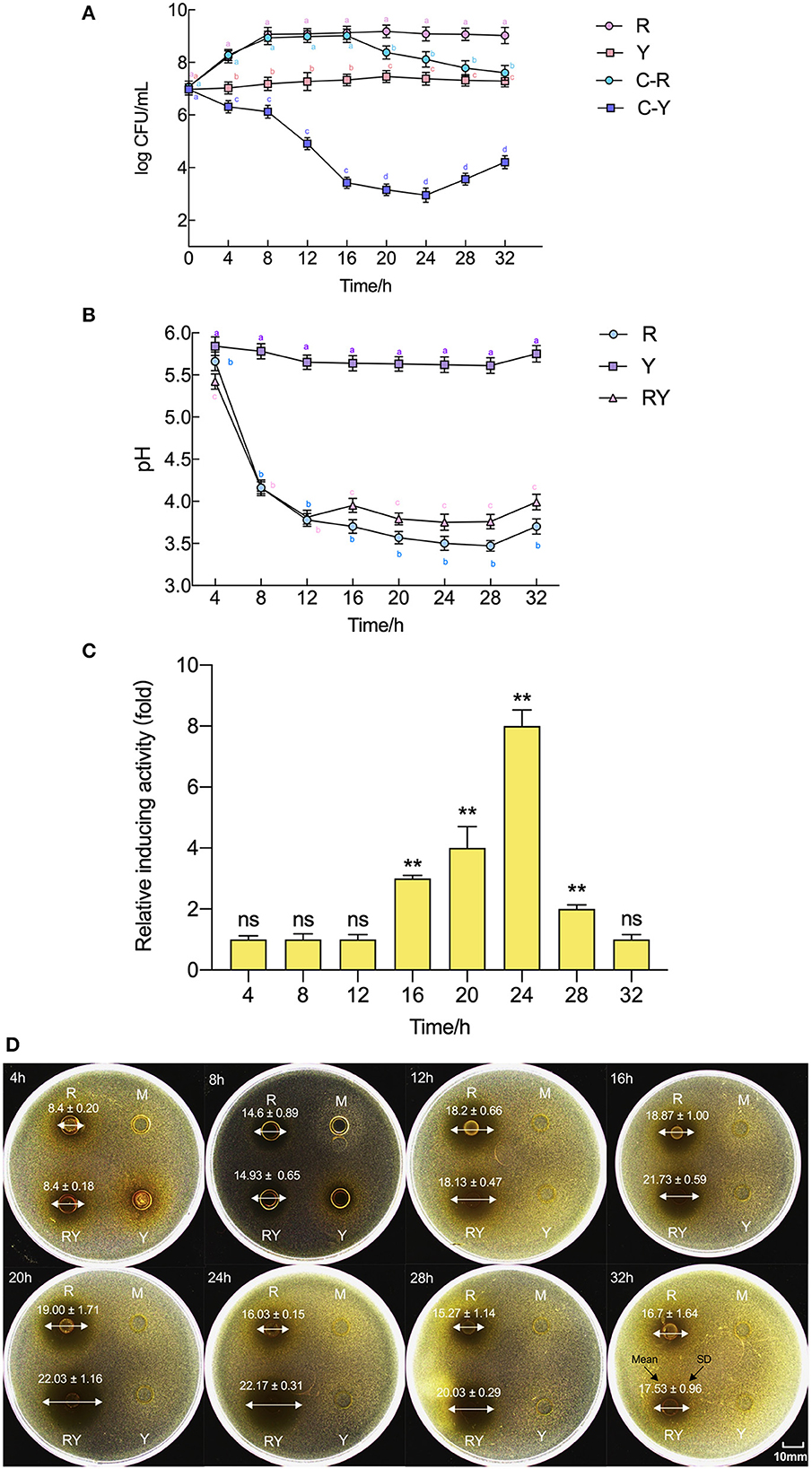
Figure 1. Evolution of pH, plantaricin production, and viable cell counts of L. paraplantarum RX-8/W. anomalus Y-5 in co-culture. (A) Indicates viable counts of L. paraplantarum RX-8/W. anomalus Y-5 at every 4 h from 4 to 32 h in mono-culture and co-culture, (B) indicates the pH value of CFS from mono-culture and co-culture at every 4 h from 4 to 32 h, (C) indicates the relative inducing activity in co-culture at every 4 h from 4 to 32 h compared with the mono-culture, (D) were images of inhibition zone of CFSs from mono-culture and co-culture against L. monocytogenes 35152. Mean diameter of inhibition zone (mm) ± standard deviation. R means mono-culture of L. paraplantarum RX-8 at 37°C in 4–32 h, Y means mono-culture of W. anomalus Y-5 at 37°C in 4–32 h, RY means bacteriocin-inducing co-culture of L. paraplantarum RX-8 and W. anomalus Y-5 at 37°C in 4–32 h, and M means blank control (MRS, pH 6.0). C-R means the viable cell count of L. paraplantarum RX-8 in co-culture, and C-Y means the viable cell count of W. anomalus Y-5 in co-culture. ns, not significant, *p < 0.05, **p < 0.01. Bars with no letter in common for each treatment are significantly different (p < 0.01).
In the fermentation, L. paraplantarum RX-8 produced lactic acid and other organic acids, which rapidly decreased the pH value. The pH value of L. paraplantarum RX-8 under co-culture and mono-culture was similar from 4 to 12 h and rapidly declined from pH 5.7 to pH 3.8. The pH value in co-culture (pH 3.79–3.99) was slightly higher than that in mono-culture (pH 3.57–3.70) of L. paraplantarum RX-8 from 16 to 32 h. The pH value of W. anomalus Y-5 under mono-culture was maintained at 5.5–6.0. A reason for the increased pH in co-culture from 16 to 32 h may be that less organic acid is produced by L. paraplantarum RX-8, or it may be due to ammonia and proton production via W. anomalus Y-5.
Similarly, the bacteriocin production in co-culture did not increase until 16 h compared with that in mono-culture, then reached the highest point at 20 h, and stabilized in the 20–24 h period before declining at 28 h. Compared with the decrease in the population of L. paraplantarum RX-8, the bacteriocin activity of L. paraplantarum RX-8 in co-culture was promoted effectively. This suggests that the inducing effect of bacteriocin production is not due to an increase in cell number. Specifically, bacteriocin production at 24 h in co-culture finally reached 512 AU/ml and showed the most significant increase (8-fold) compared to that in mono-culture. Therefore, the sampling point (24 h) was selected for transcriptomic and proteomic analyses to determine the interaction between the two strains.
3.2. Effects of W. anomalus Y-5 on the transcription of plantaricin biosynthesis and AI-2 synthesis-related gene
To explore L. paraplantarum RX-8's response to the inducing effect from W. anomalus Y-5, the plantaricin gene cluster plnABCDEF and AI-2 biosynthesis genes pfs and luxS in L. paraplantarum RX-8 were detected by RT-qPCR from 4 to 32 h. As shown in Figure 2, the transcriptional levels of the L. paraplantarum RX-8 genes in mono-culture (R) and co-culture (RY) increased over time and then decreased toward the end of their growth. The QS signal PlnA was encoded by plnA, which markedly increased in RY compared to that in sample R at 12 h, while the expression of plnA showed a decrease at 28 and 32 h in RY (Figure 2A). The reason for these phenomena is that the autoinducing peptide PlnA was secreted at an early stage before the production of bacteriocin. When the concentration of PlnA reached a critical threshold, it would activate the histidine protein kinase (HPK, encoded by plnB) on the cell membrane (Liu et al., 2022). However, the plnB trend was not consistent with the change of plnA in RY (Figure 2B). This indicates that some factors may induce an increase in plnA, and signal-transduction proteins may work with plnB on plantaricin production under co-culture. For instance, the overexpression of gene Lp_2642, belonging to the TetR family, and its regulatory factors could enhance plantaricin EF synthesis in L. plantarum 163, and the Lp_2642 protein can bind to the promoter sequence of the plnA gene in vitro experiments (Zhao et al., 2022). It was predicted that gene lamK (HPK) and lamR (RRs) would enhance the transcription of genes plnB and plnD, resulting in an increase in plantaricin EF production (Zhao et al., 2021). Then, the two RRs PlnC and PlnD were phosphorylated by HPK and bound to the promoters of plnEF to activate (by plnC) or repress (by plnD) the production of plantaricin EF (Straume et al., 2009). The transcriptional level of plnC in RY was higher than that in R over 4–32 h, and the transcriptional level of plnD in RY was lower than that in R over 12–20 h (Figures 2C, D). As for plnEF, the transcription expressions of plnE and plnF in RY were both obviously higher than those in R over 20–32 h, which was consistent with the changes in the plantaricin production (Figures 2E, F). The maximum transcriptional level of plnE in R was reached at 12 h; then, it was downregulated from 24 to 32 h. However, the expression of plnE in RY was the highest at 16 h, and it remained upregulated at 24–32 h. The maximum transcriptional level of plnF in R was reached at 12 h, while it reached the maximum point at 28 h in RY. For the gene of AI-2 biosynthesis' key enzymes, the transcriptional level of pfs and luxS decreased more in RY compared to R from 8 to 20 h, while they increased in RY at 24 and 28 h (Figures 2G, H). The AI-2 activity at 4–32 h showed the same trend in R and RY (Supplementary Figure 2). However, the effect of AI-2 on the induction process under co-culture is still unclear.
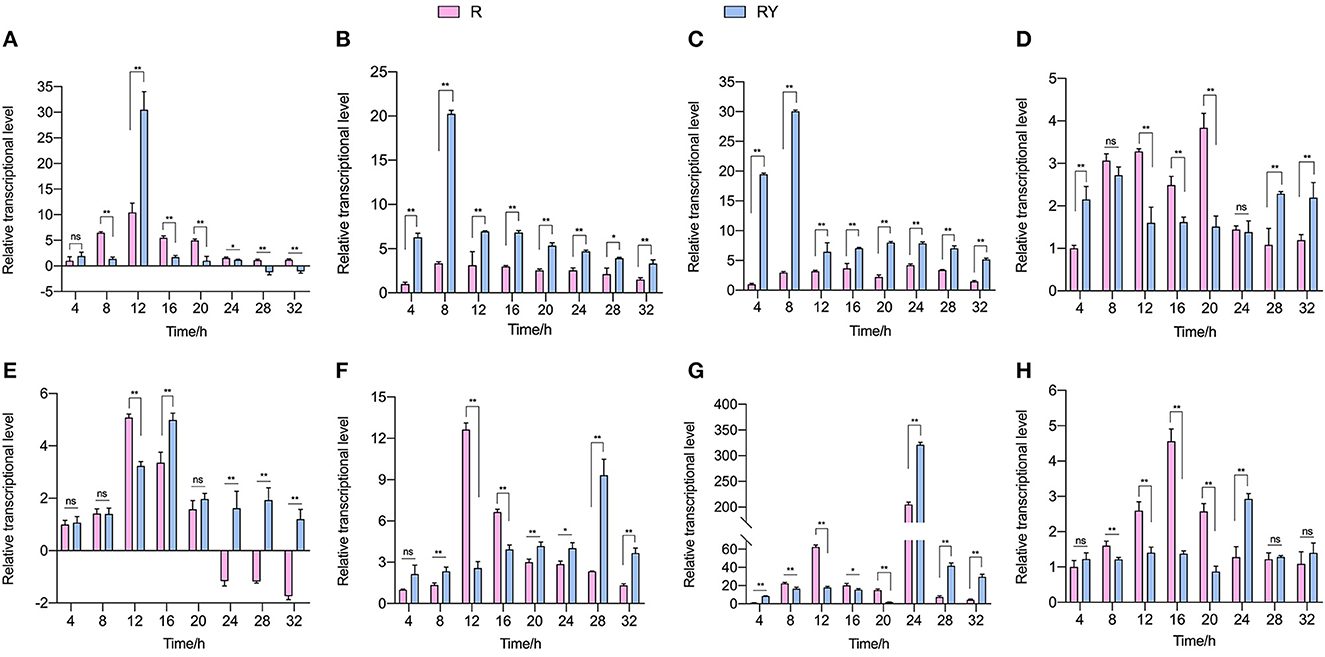
Figure 2. Transcriptional level of plantaricin biosynthesis (plnABCDEF; A–F) and AI-2 synthesis-related gene (pfs and luxS; G, H). R means mono-culture of L. paraplantarum RX-8 at 37°C in 4–32 h, and RY means bacteriocin-inducing co-culture of L. paraplantarum RX-8 and W. anomalus Y-5 at 37°C in 4–32 h. ns, not significant, *p < 0.05, **p < 0.01.
3.3. Transcriptomic and proteomic analyses of L. paraplantarum RX-8 in co-culture
To identify the DEGs and DEPs of L. paraplantarum RX-8 in response to W. anomalus Y-5 under co-culture, the cells of L. paraplantarum RX-8 in co-culture and mono-culture were harvested at 24 h for transcriptomic and proteomic analyses. Due to a large amount of transcriptomic data and to reduce the false-positive rate, candidate genes of L. paraplantarum RX-8 involved in co-culture were chosen according to the following criteria: (1) more than 2-fold change after normalization; (2) Benjamini and Hochberg's correction approach was used to adjust the p-values; and (3) statistically significant level of p < 0.05. Finally, the transcription of 580 genes was detected, including 199 upregulated genes and 381 downregulated genes (Figure 3). In addition, iTRAQ was further performed to identify the DEPs of L. paraplantarum RX-8 in co-culture and mono-culture. Compared with the transcriptomic data, the amount of proteomic data was relatively small; therefore, FDR verification of the p-value was not needed. Compared to the mono-culture group, 339 DEPs (fold change ≥ 1.3, p < 0.05) were identified, including 244 upregulated proteins and 95 downregulated proteins (Figure 3). In the Venn diagrams, consistency between the significantly expressed genes and proteins was low. The correlation coefficient between mRNAs and proteins was 0.44 (Figure 3). This can be explained by some biological reasons, including weak ribosome-binding sites, regulatory proteins, codon usage bias, and the half-life difference between protein and mRNA, resulting in gene and protein expression levels that are not always perfectly correlated (Kumar et al., 2016). The 580 DEGs and 339 DEPs were subsequently applied to functional analysis in the functional enrichment of the KEGG pathway. After KEGG analysis, these pathways were remarkably upregulated under co-culture, which included the phosphotransferase system (PTS), glycolysis, galactose metabolism, and pyruvate metabolism (Figure 4). Other pathways were significantly downregulated under co-culture, which included purine metabolism and arginine biosynthesis (Figure 4).
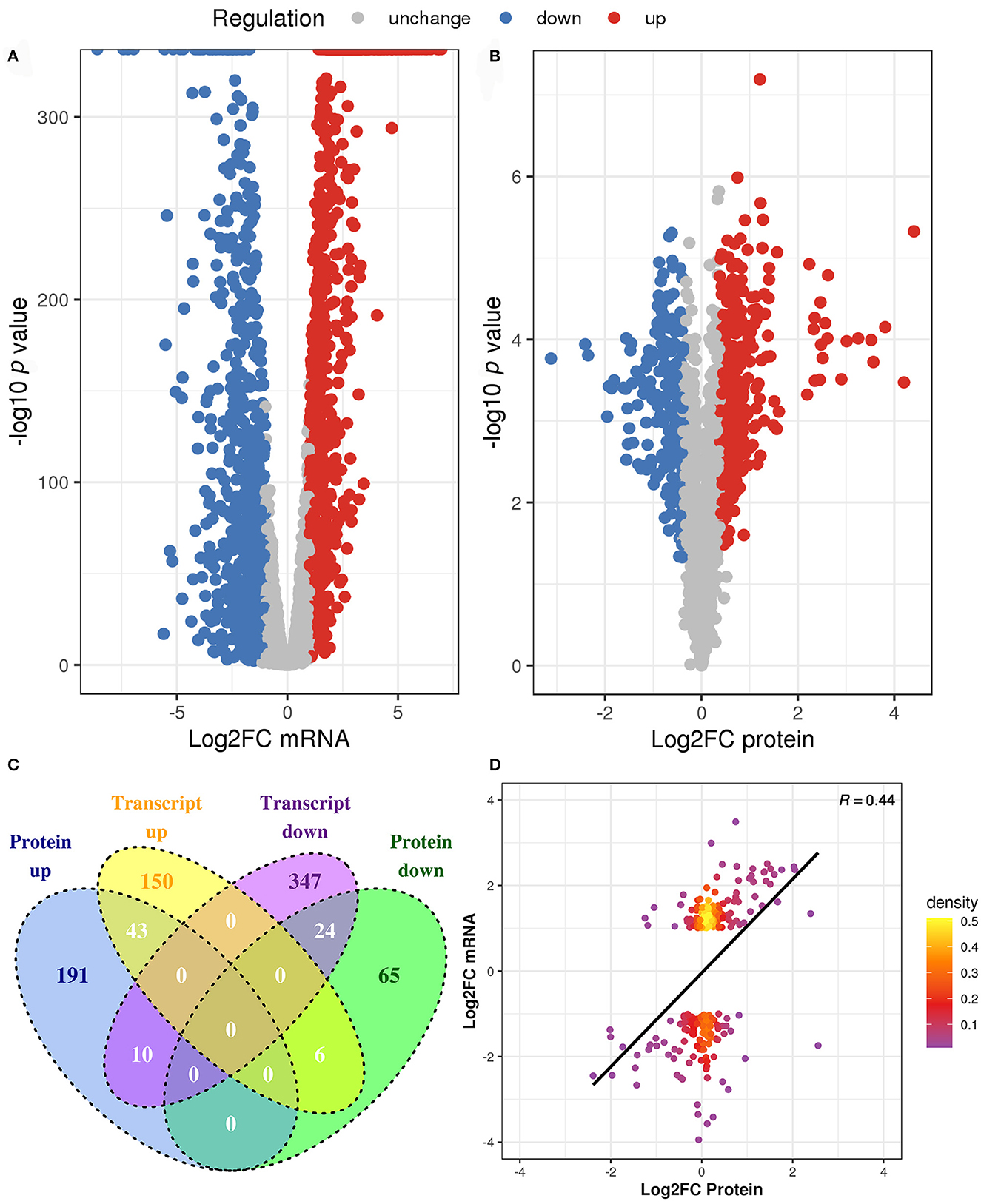
Figure 3. The changes of L. paraplantarum RX-8 under co-culture in transcriptional and protein levels. (A) Indicates volcano plots in the DEGs of L. paraplantarum RX-8 under co-culture. (B) Indicates volcano plots in the DEPs of L. paraplantarum RX-8 under co-culture. (C) Venn diagram illustrates the count of DEGs and DEPs coherence regulation, when comparing L. paraplantarum RX-8 in co-culture and mono-culture. (D) Correlation between DEGs and DEPs L. paraplantarum RX-8 under co-culture.
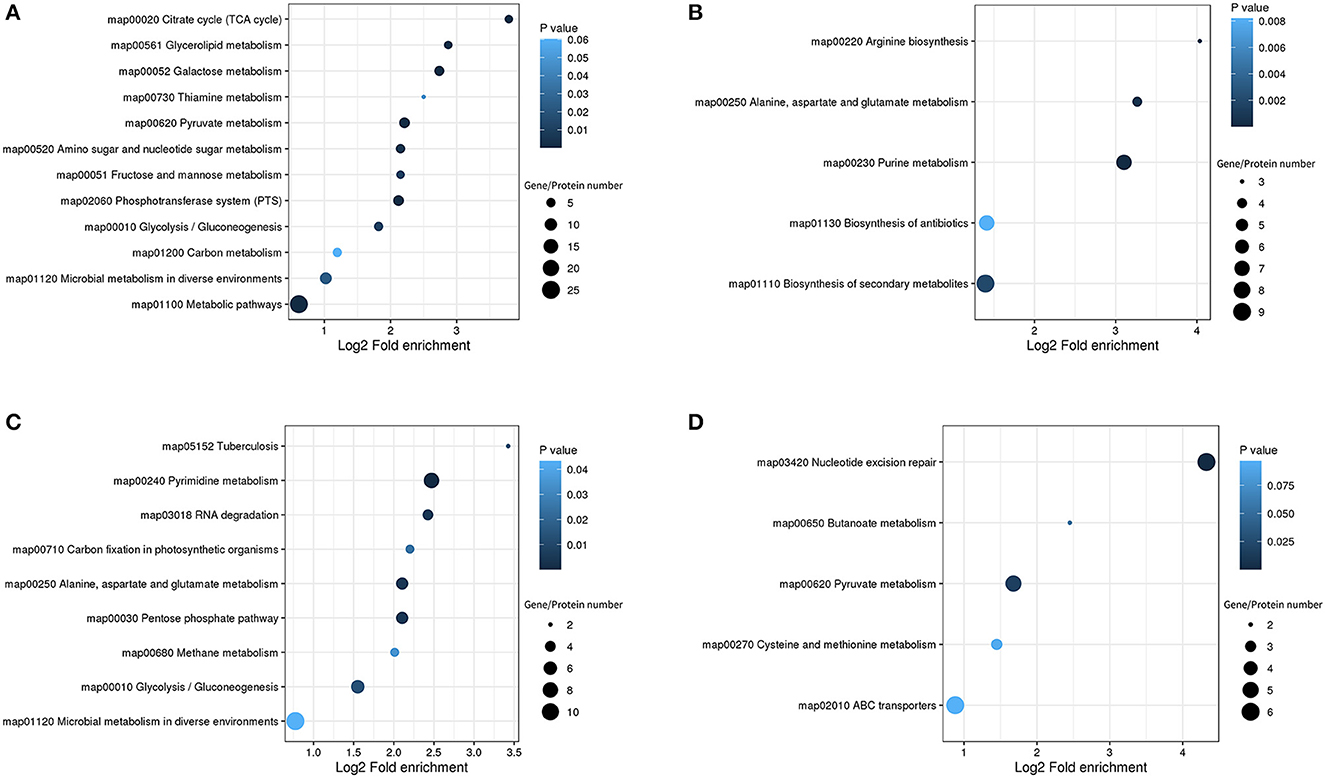
Figure 4. KEGG pathway enrichment bubble diagrams in the DEGs/DEPs of L. paraplantarum RX-8 under co-culture. (A) Indicates KEGG pathway enrichment of upregulated genes and upregulated proteins, (B) indicates KEGG pathway enrichment of downregulated genes and downregulated proteins, (C) indicates KEGG pathway enrichment of upregulated genes and unchanged proteins, and (D) indicates KEGG pathway enrichment of downregulated genes and unchanged proteins.
3.3.1 Carbohydrate transport and metabolism of L. paraplantarum RX-8 in co-culture
Compared with L. paraplantarum RX-8 in mono-culture, L. paraplantarum RX-8 in co-culture presented a much stronger carbohydrate utilization ability, which contributes to its great adaptability in competition with W. anomalus Y-5. In the process of carbohydrate transport and metabolism, the following metabolic pathways are involved in L. paraplantarum RX-8, with or without co-culture: PTS, fructose and mannose metabolism, galactose metabolism, glycolysis, TCA cycle, and amino sugar and nucleotide sugar metabolism.
The phosphotransferase system is responsible for the absorption of sugars, which depends on ATP and the 50 DEGs and 13 DEPs enriched in the PTS. In the PTS, the genes and proteins of mannose transport (manXaYZ) and galactitol transport (gatABC) are upregulated, while genes and proteins of fructose transport (FruA/B) are downregulated. In Lactobacillus pentosus, the mannose PTS EIIB participates in the inhibition of the fructose PTS (Chaillou et al., 2001). The mannose PTS in L. paraplantarum RX-8 may have a similar effect on inhibiting fructose transport (FruA/B). Aside from mannose, other carbohydrates, such as glucose, cellobiose, and maltose, could be transported by the mannose PTS. The sensing of exogenous AI-2 in Streptococcus pneumoniae is dependent on FruA; then, AI-2 promotes the transition of the pneumococcus from colonization to invasion by facilitating the utilization of galactose (Trappetti et al., 2017). In Escherichia coli, the internalization of AI-2 also depends on the PTS (Ha et al., 2018). AI-2 plays an important role in the inducing effect under a bacteria–bacteria co-culture (Chanos and Mygind, 2016; Liu et al., 2022), while AI-2's role in inducing this effect under fungi–bacteria co-culture needs further investigation. The proteins celA, celB, and celC, comprising the cellobiose transport system permease, take charge of the cellobiose utilization in L. paraplantarum RX-8, and are significantly upregulated, by 2.639-, 1.375-, and 2.71-fold, compared to that in mono-culture. However, the gene celA showed the opposite expression trend and the gene celC did not change. This indicates that post-transcriptional regulation may occur in L. paraplantarum RX-8. The gene crr encodes the sugar PTS EIIA component; when it is non-phosphorylated, Crr can inhibit the uptake of certain sugars, such as maltose, melibiose, lactose, and glycerol. However, when Crr is phosphorylated, it may activate adenylate cyclase (Saier and Roseman, 1976). The downregulation of the gene crr suggests that sugar uptake restrictions are lifted. Moreover, the mannose PTS EIIC acts as a target protein to confer class II bacteriocin sensitivity in L. monocytogenes, Enterococcus faecalis, and Latilactobacillus sakei (Kjos et al., 2009; Jeckelmann and Erni, 2020). The class IIa bacteriocin, immunity protein, and receptor proteins (mannose PTS EIIC and EIID) build up a tight complex to prevent pore formation and block cell death (Diep et al., 2007). However, the relationship between the mannose PTS and the immunity of class IIb bacteriocin produced by L. plantarum has not been reported to date (Kjos et al., 2010). Apart from its dual role in carbohydrate transport, the PTS is involved in the coupling of the sensory and regulatory mechanisms (Lengeler and Jahreis, 2009). The PTS transporter subunit IIB may be related to the effect of β-glucooligosaccharides on enhancing nisin production in L. lactis I2 (Lee et al., 2020). According to the results of transcriptomic and proteomic analyses, the PTS seems to be involved in the regulation of the response of L. paraplantarum to W. anomalus Y-5, leading to an increase in bacteriocin production.
Under co-culture conditions, the genes and proteins responsible for the utilization of cellobiose, lactose, galactose, fructose, sorbitol, galactitol, and ribose were also upregulated in L. paraplantarum RX-8. These sugars undergo glycolysis to pyruvate and ATP. Glucose-6-phosphate isomerase (GPI) was upregulated by 2.63-fold, which catalyzed the mutual conversion of glucose-6-phosphate and fructose-6-phosphate and helped sugars to enter the glycolysis pathway. The upregulated phosphoglycerate kinase (PGK), which directly generates ATP by phosphorylating the ADP, suggested that L. paraplantarum RX-8 produced more energy under co-culture. The pyruvate produced by glycolysis is a major metabolic intersection connecting the utilization and biosynthetic pathways of carbohydrates or amino acids; it linked the L-cysteine and carbon metabolism (Figure 5). After pyruvate dehydrogenase complex catalysis, pyruvate is oxidatively decarboxylated with the loss of one carbon and converted into acetyl-CoA, before entering the TCA cycle (Tian et al., 2022). The upregulation of DEPs in the TCA cycle enhances glucose oxidation and energy production. However, the increase in TCA decreased the transformation from phosphoenolpyruvate to aspartate, thereby weakening the serine synthesis. This leads to the downregulation of aspartate, serine, and L-cysteine metabolism.
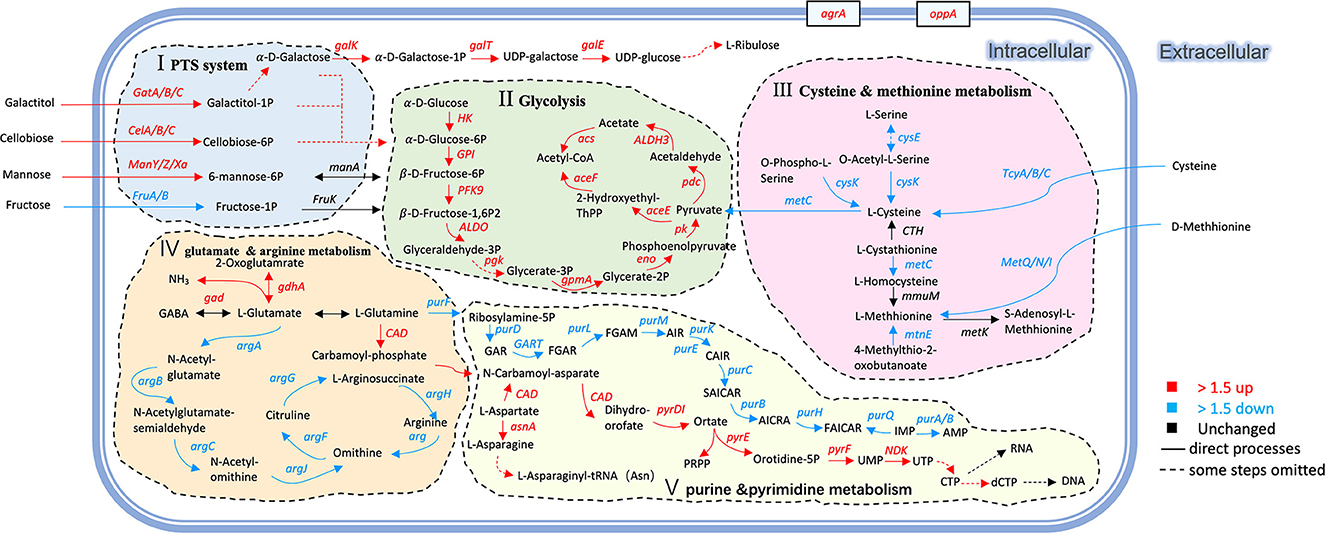
Figure 5. Changes of genes and proteins in L. paraplantarum RX-8 response to inducing effect from W. anomalus Y-5. The serial number (I, II, III, IV, V) indicates the response pathways of L. paraplantarum RX-8 under co-culture. (I) improving PTS and enhancing the uptake of certain sugars; (II) increasing the key enzyme activity in glycolysis, then promoting the energy production; (III) repressing the precursor of QS signal AI-2; (IV) decreasing arginine biosynthesis to increase glutamate mechanism, then promote plantaricin yield; (V) changing the expression of several genes/proteins related to purine metabolism (downregulation) and pyrimidine metabolism (upregulation). Pathways were constructed based on information provided in the KEGG database and previous studies.
3.3.2. Amino acid metabolism of L. paraplantarum RX-8 in co-culture
Amino acids could serve as the materials in plantaricin synthesis. Aminoacyl-tRNA was responsible for the transportation of amino acids to ribosomes for incorporation into polypeptide chains of plantaricin. Under co-culture, the aspartate–ammonia ligase (asnA) and asparagine–tRNA ligase (asnS) increased by 2.02- and 2.35-fold, respectively. The aspartate–ammonia ligase catalyzed the production of L-asparagine; then, it was converted by asparaginyl–tRNA synthetase into L-asparaginyl–tRNA, which played a vital role in the translation of RNA into protein (Yeom et al., 2020). In addition, the enzymes that catalyzed the conversion of L-aspartate into fumarate were downregulated. The L-aspartate concentration in Lactobacillus casei increased when it faced acid resistance, and the transcriptional levels of argG and argH in L. casei increased with the decrease in pH (Wu et al., 2013). In our study, the pH of L. paraplantarum RX-8 in mono-culture was lower than that in co-culture, and a higher expression of argG and argH was detected in mono-culture.
At the same time, glutamate decarboxylase (gad), which was responsible for the mutual conversion of L-glutamate and γ-aminobutyrate (GABA), increased by 2.02-fold. The gad process converted L-glutamate into GABA, accompanied by the consumption of H+. This may explain why the pH of the co-culture was higher than that of the mono-culture of L. paraplantarum RX-8. Considering the upregulation of glutamate decarboxylase (gdhA), it is possible that L. paraplantarum RX-8 under co-culture produces more ammonia or proton consumption via glutamate decarboxylase (Teixeira et al., 2014). Moreover, the repression of GlnR could increase the expression of glutamine- and glutamate-metabolism-related genes (glnP, glnQ, amtB, glnK, gltB, and gad), promoting cell growth, acid resistance, and nisin yield (Miao et al., 2018). However, direct interactions between the anti41, an sRNA with a strong inhibitory effect on glnR, and the nisin gene cluster were not found (Miao et al., 2018). In addition, purine metabolism was weakened when the gene purF was downregulated, while pyrimidine metabolism was enhanced when the gene CAD was upregulated (Figure 5). The expression of carbamoyl–phosphate synthase (CAD) converted L-glutamine into carbamoyl phosphate, which is an intermediate in the biosynthesis of nucleotides through the orotate pathway (Namrak et al., 2022).
Meanwhile, the related arginine biosynthesis (argACDJGH) genes were downregulated, especially the genes controlling the transportation from glutamate to arginine (Figure 5). It was indicated that L. paraplantarum RX-8, under co-culture, led to the accumulation of glutamate by reducing arginine biosynthesis. The gene metC, which converted L-cysteine into L-methionine and was a precursor of S-ribosomal homocysteine (SRH), was downregulated. The gene luxS, a key enzyme, converted SRH into 4,5-dihydroxy-2,3-pentanedione (DPD). Then, DPD underwent spontaneous rearrangements to form AI-2. The downregulation of luxS influences the biosynthesis of AI-2, and a similar tendency was observed in co-culture AI-2 activity (Supplementary Figure 2). It was reported that the decrease in the L-methionine and L-cysteine cycling pathways was caused by the knockout of the luxS gene (Qian et al., 2022b). Similar results of changes in L-methionine- and L-cysteine-related genes were found in co-culture.
3.3.3. Quorum sensing system of L. paraplantarum RX-8 in co-culture
Some genes related to the quorum-sensing system and ABC-transport system were upregulated in L. paraplantarum RX-8 under co-culture. OppA is an oligopeptide-binding protein that is responsible for peptides' capture (Kieliszek et al., 2021). Therefore, the strong upregulation of oppA suggested an increase in the transportation of oligopeptides. OppA could increase Ox-bile resistance in L. salivarius Ren and salt resistance in Lactobacillus paracasei ATCC 334 (Ma et al., 2018). It is possible that OppA helps L. paraplantarum RX-8 to survive in the presence of yeast. When PlnA or Lactobacillus sanfranciscensis DPPMA174 is co-cultured with L. plantarum DC400, OppA is upregulated compared with L. plantarum DC400 mono-culture (Calasso et al., 2013). Although oppA did not show a difference in the plantaricin Q7 biosynthesis of L. plantarum Q7, oppD-, and oppF-encoding intracellular ATPases were upregulated when plantaricin Q7 production increased and downregulated when plantaricin Q7 was hydrolyzed to support the nutrients needed to promote the growth of L. plantarum Q7 (Bu et al., 2021a). Moreover, the Opp system in E. faecalis and Streptococcus mutans has specific pheromone binding activity (Leonard et al., 1996; Li and Tian, 2012). It can be hypothesized that L. plantarum DC400 transports PlnA via the Opp system (Calasso et al., 2013). The upregulation of phosphate-inducible transport system (pstSCAB) suggested that the phosphate uptake was increased to meet the increase in the phosphorylated nucleotides and proteins, which control the biosynthesis of many secondary metabolites. In addition, fructose, galactose, or mannose in media could promote the expression of the pstSCAB operon and significantly increase the formation of PstS (Martín and Liras, 2021). In this result, the mannose transport system and galactose metabolism were also upregulated.
The production of plantaricin was regulated by the quorum-sensing system (encoded by plnABCDEF gene clusters) (Zhao et al., 2021; Liu et al., 2022). Although plnABCDEF gene clusters synthesizing bacteriocin did not show significant differences (|Log2FC| ≥ 1, p < 0.05) in transcriptomic analysis, the plnABCDEF gene clusters were found to show 2.51-, 1.61-, 1.45-, 1.27-, 1.03-, and 1.41-fold changes at 24 h in transcriptomic analysis, and the results of RT-qPCR indicated that W. anomalus Y-5 stimulated plantaricin production via plnABCDEF. In the QS system, the gene agrA, an RR, was upregulated under co-culture. The Agr-mediated QS system (agrBDCA) might participate in commensal host–microbe interactions, especially agrA-encoded RR, which performs important functions in cell adhesion and biofilm formation in L. plantarum WCFS1 (Sturme et al., 2005). In addition, the expression of agrA was related to a global cellular response, such as the upregulated pyrimidine biosynthesis genes (pyrB, pyrD, pyrF, and pyrR) in L. plantarum WCFS1 (Sturme et al., 2005). Interestingly, similar results were also found for L. paraplantarum RX-8 in co-culture, suggesting that agrA controls pyrimidine biosynthesis under the interactions of L. paraplantarum RX-8 and W. anomalus Y-5. agrBDCA may influence plantaricin Q7 biosynthesis; specifically, agrC and agrA were upregulated in the synthesis of plantaricin Q7 (Bu et al., 2021a). The gene agrB of L. paraplantarum RX-8 was upregulated by 1.5-fold under co-culture. Protein AgrB also increased when L. plantarum DC400 was co-cultured with L. plantarum DPPMA20 or L. sanfranciscensis DPPMA174 (Calasso et al., 2013). This indicates that the agrBCDA quorum-sensing system might enhance plantaricin production when L. paraplantarum RX-8 is induced by W. anomalus Y-5.
3.3.4. RNA degradation and nucleotide repair of L. paraplantarum RX-8 in co-culture
To repair DNA damage, there are different classes of DNA repair systems. The nucleotide excision repair (uvrABCD) system recognizes the damaged strand, removes it, and then fills the gap (Goosen and Moolenaar, 2001). The gene uvrABCD was downregulated, which indicates that there is less DNA synthesis or less DNA damage when L. paraplantarum RX-8 is under co-culture. It is important for a tightly regulated RNA metabolism to adapt to environmental change and utilize nutrients. RNA helicases could locally unwind double-stranded RNA, or they can clamp protein complexes to a substrate RNA in an ATP-dependent manner. Gene cshA-encoded RNA helicase was downregulated in L. paraplantarum RX-8 under co-culture. It was reported that the cshA mutant of Staphylococcus aureus SA564 led to increased stability in agr mRNA, and it was hypothesized that the agrBDCA mRNAs were unable to correctly degrade in the absence of cshA (Oun et al., 2013). This would account for the lower cshA and higher agrA expression in L. paraplantarum RX-8 under co-culture. GroEL could partially unfold the misfolded RNA or proteins in an ATP-dependent manner. DnaK is a major chaperone, and the binding and release of protein clients by DnaK are regulated by ATP's hydrolytic activity. The downregulation of groEL and dnaK may indicate that there is less RNA degradation and a more stable RNA in co-culture.
3.4. Transcription- and protein-level validation
Based on the results of transcriptomic and proteomic analyses, oppA, agrA, pstS, glnA, argH, and purD (four upregulated and two downregulated) were selected for validation by RT-qPCR. Although the value of the obtained fold-change was different between RT-qPCR and transcriptomic and proteomic data, the expressions of the tested genes were entirely consistent in the direction of regulation (Figure 6). The concordance between RT-qPCR and transcriptomic and proteomic data suggests high reliability in transcriptomic and proteomic data.
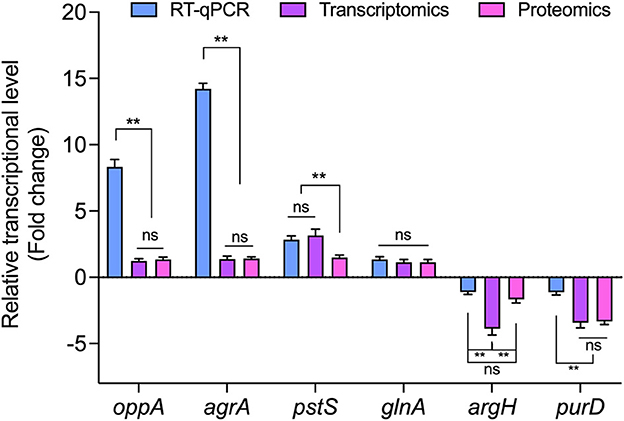
Figure 6. The relative expression of genes encoding DEPs in L. paraplantarum RX-8 under co-culture. ns, not significant, **p < 0.01.
3.5. Inducing effects of key metabolites on plantaricin production
The effects of key-inducing metabolites on the plantaricin production of L. paraplantarum RX-8 were studied. It was found that the addition of galactose, mannose, arginine, cysteine, glutamate, and glutamine could induce plantaricin production, while cellobiose and D-ribose had little less effect on plantaricin production (Figure 7). It was indicated that galactose, mannose, arginine, cysteine, glutamate, and glutamine could be essential to the plantaricin production of L. paraplantarum RX-8. The production of plantaricin was significantly enhanced in mono-culture with the addition of 200 mM, but lower concentrations of galactose (2 and 20 mM) did not induce any effects on plantaricin yield (Figure 7A). The exogenous addition of mannose could increase the plantaricin synthesis at concentrations of 2, 20, and 200 mM (Figure 7B). The yield of plantaricin was strengthened at 24 h by adding arginine to mono-culture (Figure 7E). The addition of arginine, 2.5 g/L of cysteine, 0.5 g/L of glutamate, and 0.5 g/L of glutamine had significant effects on plantaricin production (Figures 7E, F, H), while cysteine, glutamate, and glutamine at other concentrations did not show a significant impact on plantaricin production.
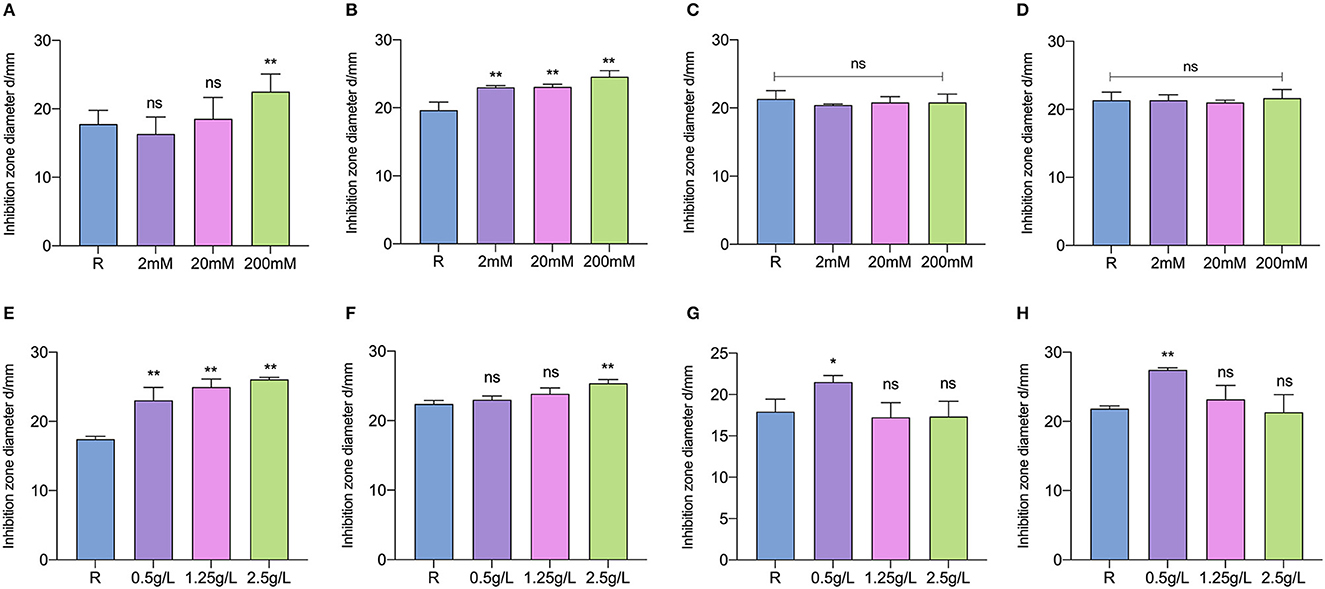
Figure 7. Inducing effects of different concentrations of galactose (A), mannose (B), cellobiose (C), D-ribose (D), arginine (E), cysteine (F), glutamate (G), and glutamine (H) on plantaricin production. ns: not significant, *p < 0.05, **p < 0.01.
Interestingly, the stimulus effects of glutamate and glutamine were not exhibited at higher concentrations. Considering the upregulation of related genes and proteins in galactose metabolism, this could because the significant induction that occurs at low concentrations of glutamate and glutamine makes them the most economic strategy for enhancing plantaricin production in L. paraplantarum RX-8. Bu et al. (2021b) reported that the plantaricin synthesis of L. plantarum Q7 was promoted by the exogenous addition of glutamate at 16 h, but plantaricin Q7 production was not stimulated by glutamate at 2, 6, and 12 h. Although S. cerevisiae could secrete different kinds of amino acids that are crucial to the survival of L. plantarum, the supplementing CDM35 medium with glutamine, threonine, phenylalanine, tryptophan, and serine, which were under the observed concentrations, had only a minimal impact on L. plantarum growth (Ponomarova et al., 2017). This is because the synergistic nature of multi-component cross-feeding was challenging to characterize. In addition, the nutritional competition was one of the reasons that L. plantarum SS-128 inhibits the growth of Shewanella baltica (Qian et al., 2022a). To further demonstrate the advantage of using W. anomalus Y-5 to induce bacteriocin production, metabolomics would be performed on quantified amino acids, sugar, and other metabolites secreted by L. paraplantarum RX-8 and W. anomalus Y-5. Then, the induction effect of the target metabolite at the observed concentrations in mono-culture would be analyzed.
3.6. The role of AI-2 activity in inducing effect
D-ribose, as a competitive inhibitor of AI-2, could cut the intracellular communication mediated by AI-2 among bacteria. AI-2 activity in co-culture was inhibited by D-ribose at all selected concentrations (Figure 8A). All samples whose AI-2 activity was cut still showed antibacterial activity and did not show a significant difference compared with co-culture samples without D-ribose (Figure 8B). It was suggested that AI-2 does not play an important role in inducing bacteriocin production when the bacteriocin-producing strain is co-cultured with W. anomalus Y-5.
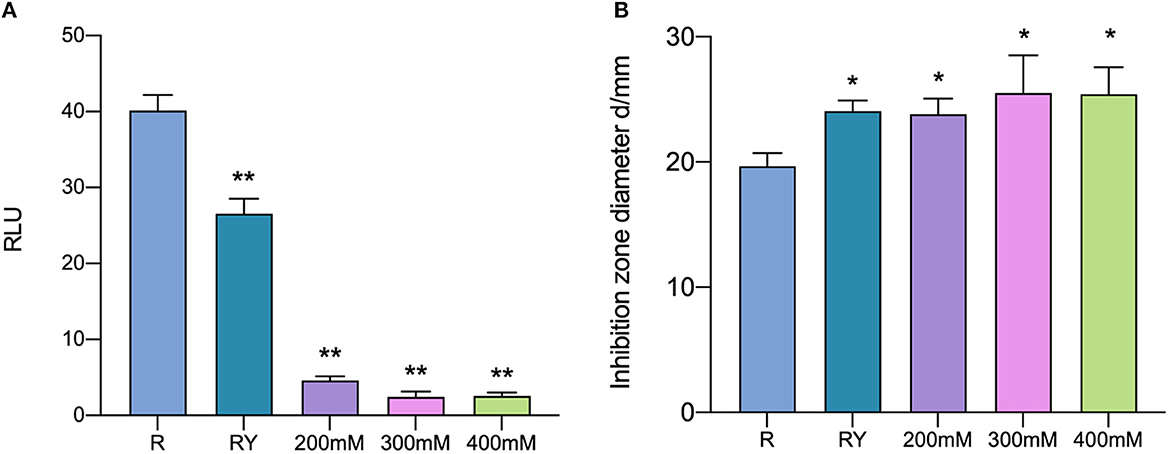
Figure 8. AI-2 activity (A) and the antibacterial activity (B) in co-culture with the addition of D-ribose. R means mono-culture of L. paraplantarum RX-8 at 37°C; RY means bacteriocin-inducing co-culture of L. paraplantarum RX-8 and W. anomalus Y-5 at 37°C as a positive control; 200/300/400 mM means co-culture of L. paraplantarum RX-8 and W. anomalus Y-5 at 37°C with 200, 300, and 400 mM D-ribose. *p < 0.05, **p < 0.01.
Conclusion
In this study, the plantaricin production and response mechanisms that occur when L. paraplantarum RX-8 is co-cultured with W. anomalus Y-5 were investigated. The results indicated that W. anomalus Y-5 could enhance plantaricin production by activating the expression of plnABCDEF cluster in L. paraplantarum RX-8. Then, the differences between L. paraplantarum RX-8 at transcriptomic and protein levels were analyzed under co-culture and mono-culture. The data showed that multiple metabolic pathways of L. paraplantarum RX-8 were significantly affected in the co-culture including (i) upregulation of the PTS in mannose, galactitol, and cellobiose transport, and downregulation of the PTS in fructose transport; (ii) modulation of the expression of several enzymes involved in glycolysis and other carbohydrate pathways; (iii) upregulation of key enzymes involved in glutamate metabolism and downregulation of multiple enzymes involved in aspartate, arginine, and cysteine metabolisms; (iv) modulation of the expression of several enzymes involved in the quorum-sensing system and ABC-transport system; and (v) upregulation of several enzymes involved in pyrimidine metabolism and downregulation of several enzymes involved in purine metabolism. In combination with the induction of metabolites' validation during plantaricin production and previous studies, we predicted that L. paraplantarum RX-8 mechanisms would occur in response to W. anomalus Y-5; that the changes in the PTS led to an increase in mannose and galactose uptake; that the glycolysis and fermentative pathways would be promoted and generated energy to support plantaricin production. The increase in glutamate metabolism and decrease in arginine biosynthesis may replenish the glutamate and then enhance the plantaricin yield, and the upregulation of the agrA and oppA genes may help transport PlnA and regulate plantaricin production. Our study provides a novel insight into the mechanisms of bacteriocin-producing bacteria's response to bacteriocin-inducing yeast and offers a reference for the interaction between L. paraplantarum and W. anomalus. In the future, a detailed study of the bacteriocin-inducing regulatory mechanism in co-culture could be undertaken using metabolomics methods.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
RN and GL conceptualized the idea and designed the experiments. RN wrote the manuscript. GL, RN, and YQ revised, make corrections, and approved the research article for publication. RN, ZZ, ZW, and HS performed the experiments. RN and ZZ analyzed the results. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by the Natural Science Foundation of China (Grant Nos. 31871772 and 31671832), the Beijing Natural Science Foundation (Grant No. 6192003), the Natural Science Foundation of Guangdong Province, China (Grant No. 2022A1515140021), and the Cultivation Project of Double First-Class Disciplines of Food Science and Engineering, Beijing Technology & Business University (Grant No. BTBUYXTD202208).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1111516/full#supplementary-material
References
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Ariana, M., and Hamedi, J. (2017). Enhanced production of nisin by co-culture of Lactococcus lactis sub sp. lactis and Yarrowia lipolytica in molasses based medium. J. Biotechnol. 256, 21–26. doi: 10.1016/j.jbiotec.2017.07.009
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bu, Y., Liu, Y., Li, J., Liu, T., Gong, P., Zhang, L., et al. (2021a). Analyses of plantaricin Q7 synthesis by Lactobacillus plantarum Q7 based on comparative transcriptomics. Food Control 124, 107909. doi: 10.1016/j.foodcont.2021.107909
Bu, Y., Yang, H., Li, J., Liu, Y., Liu, T., Gong, P., et al. (2021b). Comparative metabolomics analyses of plantaricin Q7 production by Lactobacillus plantarum Q7. J. Agric. Food Chem. 69, 10741–10748. doi: 10.1021/acs.jafc.1c03533
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Calasso, M., Di Cagno, R., De Angelis, M., Campanella, D., Minervini, F., and Gobbetti, M. (2013). Effects of the peptide pheromone plantaricin A and cocultivation with Lactobacillus sanfranciscensis DPPMA174 on the exoproteome and the adhesion capacity of Lactobacillus plantarum DC400. Appl. Environ. Microbiol. 79, 2657–2669. doi: 10.1128/AEM.03625-12
Chaillou, S., Postma, P. W., and Pouwels, P. H. (2001). Contribution of the phosphoenolpyruvate:mannose phosphotransferase system to carbon catabolite repression in Lactobacillus pentosus. Microbiology 147, 671–679. doi: 10.1099/00221287-147-3-671
Chanos, P., and Mygind, T. (2016). Co-culture-inducible bacteriocin production in lactic acid bacteria. Appl. Microbiol. Biotechnol. 100, 4297–4308. doi: 10.1007/s00253-016-7486-8
Diep, D. B., Skaugen, M., Salehian, Z., Holo, H., and Nes, I. F. (2007). Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104, 2384–2389. doi: 10.1073/pnas.0608775104
Goosen, N., and Moolenaar, G. F. (2001). Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair. Res. Microbiol. 152, 401–409. doi: 10.1016/S0923-2508(01)01211-6
Ha, J.-H., Hauk, P., Cho, K., Eo, Y., Ma, X., Stephens, K., et al. (2018). Evidence of link between quorum sensing and sugar metabolism in Escherichia coli revealed via cocrystal structures of LsrK and HPr. Sci. Adv. 4, eaar7063. doi: 10.1126/sciadv.aar7063
Jeckelmann, J.-M., and Erni, B. (2020). The mannose phosphotransferase system (Man-PTS) - Mannose transporter and receptor for bacteriocins and bacteriophages. Biochim. Biophys. Acta Biomembr. 1862, 183412. doi: 10.1016/j.bbamem.2020.183412
Kern, L., Abdeen, S. K., Kolodziejczyk, A. A., and Elinav, E. (2021). Commensal inter-bacterial interactions shaping the microbiota. Curr. Opin. Microbiol. 63, 158–171. doi: 10.1016/j.mib.2021.07.011
Kieliszek, M., Pobiega, K., Piwowarek, K., and Kot, A. M. (2021). Characteristics of the proteolytic enzymes produced by lactic acid bacteria. Molecules 26, 1858. doi: 10.3390/molecules26071858
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kjos, M., Nes, I. F., and Diep, D. B. (2009). Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology 155, 2949–2961. doi: 10.1099/mic.0.030015-0
Kjos, M., Snipen, L., Salehian, Z., Nes, I. F., and Diep, D. B. (2010). The abi proteins and their involvement in bacteriocin self-immunity. J. Bacteriol. 192, 2068–2076. doi: 10.1128/JB.01553-09
Kumar, D., Bansal, G., Narang, A., Basak, T., Abbas, T., and Dash, D. (2016). Integrating transcriptome and proteome profiling: strategies and applications. Proteomics 16, 2533–2544. doi: 10.1002/pmic.201600140
Langdon, W. B. (2015). Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Mining 8, 1. doi: 10.1186/s13040-014-0034-0
Lee, J. M., Jang, W. J., Lee, E.-W., and Kong, I.-S. (2020). β-glucooligosaccharides derived from barley β-glucan promote growth of lactic acid bacteria and enhance nisin Z secretion by Lactococcus lactis. LWT 122, 109014. doi: 10.1016/j.lwt.2020.109014
Lengeler, J. W., and Jahreis, K. (2009). “Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms,” in Contributions to Microbiology, eds. M. Collin and R. Schuch (Basel: KARGER), 65–87.
Leonard, B. A., Podbielski, A., Hedberg, P. J., and Dunny, G. M. (1996). Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. U. S. A. 93, 260–264. doi: 10.1073/pnas.93.1.260
Li, Y.-H., and Tian, X. (2012). Quorum sensing and bacterial social interactions in biofilms. Sensors 12, 2519–2538. doi: 10.3390/s120302519
Liu, G., Nie, R., Liu, Y., Li, X., Duan, J., Hao, X., et al. (2022). Bacillus subtilis BS-15 effectively improves plantaricin production and the regulatory biosynthesis in Lactiplantibacillus plantarum RX-8. Front. Microbiol. 12, 772546. doi: 10.3389/fmicb.2021.772546
Liu, W., Zhou, J., Tan, F., Yin, H., Yang, C., and Lu, K. (2021). Improvement of nisin production by using the integration strategy of co-cultivation fermentation, foam fractionation and pervaporation. LWT 142, 111093. doi: 10.1016/j.lwt.2021.111093
Ma, X., Wang, G., Zhai, Z., Zhou, P., and Hao, Y. (2018). Global transcriptomic analysis and function identification of malolactic enzyme pathway of Lactobacillus paracasei L9 in response to bile stress. Front. Microbiol. 9, 1978. doi: 10.3389/fmicb.2018.01978
Maldonado-Barragán, A., Caballero-Guerrero, B., Lucena-Padrós, H., and Ruiz-Barba, J. L. (2013). Induction of bacteriocin production by coculture is widespread among plantaricin-producing Lactobacillus plantarum strains with different regulatory operons. Food Microbiol. 33, 40–47. doi: 10.1016/j.fm.2012.08.009
Martín, J. F., and Liras, P. (2021). Molecular mechanisms of phosphate sensing, transport and signalling in streptomyces and related actinobacteria. IJMS 22, 1129. doi: 10.3390/ijms22031129
Miao, S., Wu, H., Zhao, Y., Caiyin, Q., Li, Y., and Qiao, J. (2018). Enhancing nisin yield by engineering a small noncodding RNA anti41 and inhibiting the expression of glnR in Lactococcus lactis F44. Biotechnol. Lett. 40, 941–948. doi: 10.1007/s10529-018-2550-3
Namrak, T., Raethong, N., Jatuponwiphat, T., Nitisinprasert, S., Vongsangnak, W., and Nakphaichit, M. (2022). Probing genome-scale model reveals metabolic capability and essential nutrients for growth of probiotic Limosilactobacillus reuteri KUB-AC5. Biology 11, 294. doi: 10.3390/biology11020294
Oun, S., Redder, P., Didier, J.-P., François, P., Corvaglia, A.-R., Buttazzoni, E., et al. (2013). The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol. 10, 157–165. doi: 10.4161/rna.22899
Ponomarova, O., Gabrielli, N., Sévin, D. C., Mülleder, M., Zirngibl, K., Bulyha, K., et al. (2017). Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 5, 345–357.e6. doi: 10.1016/j.cels.2017.09.002
Qian, Y., Li, Y., Tang, Z., Wang, R., Zeng, M., and Liu, Z. (2022a). The role of AI-2/LuxS system in biopreservation of fresh refrigerated shrimp: enhancement in competitiveness of Lactiplantibacillus plantarum for nutrients. Food Res. Int. 161, 111838. doi: 10.1016/j.foodres.2022.111838
Qian, Y., Li, Y., Xu, T., Zhao, H., Zeng, M., and Liu, Z. (2022b). Dissecting of the AI-2/LuxS mediated growth characteristics and bacteriostatic ability of Lactiplantibacillus plantarum SS-128 by integration of transcriptomics and metabolomics. Foods 11, 638. doi: 10.3390/foods11050638
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Saier, M. H., and Roseman, S. (1976). Sugar transport. The crr mutation: its effect on repression of enzyme synthesis. J. Biol. Chem. 251, 6598–6605. doi: 10.1016/S0021-9258(17)32988-5
Straume, D., Johansen, R. F., Bjørås, M., Nes, I. F., and Diep, D. B. (2009). DNA binding kinetics of two response regulators, PlnC and PlnD, from the bacteriocin regulon of Lactobacillus plantarum C11. BMC Biochem. 10, 17. doi: 10.1186/1471-2091-10-17
Sturme, M. H. J., Nakayama, J., Molenaar, D., Murakami, Y., Kunugi, R., Fujii, T., et al. (2005). An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187, 5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005
Teixeira, J. S., Seeras, A., Sanchez-Maldonado, A. F., Zhang, C., Su, M. S.-W., and Gänzle, M. G. (2014). Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol. 42, 172–180. doi: 10.1016/j.fm.2014.03.015
Tian, S., Liang, X., Chen, J., Zeng, W., Zhou, J., and Du, G. (2020). Enhancement of 2-phenylethanol production by a wild-type Wickerhamomyces anomalus strain isolated from rice wine. Bioresour. Technol. 318, 124257. doi: 10.1016/j.biortech.2020.124257
Tian, Y., Wang, Y., Zhang, N., Xiao, M., Zhang, J., Xing, X., et al. (2022). Antioxidant mechanism of Lactiplantibacillus plantarum KM1 under H2O2 stress by proteomics analysis. Front. Microbiol. 13, 897387. doi: 10.3389/fmicb.2022.897387
Trappetti, C., McAllister, L. J., Chen, A., Wang, H., Paton, A. W., Oggioni, M. R., et al. (2017). Autoinducer 2 signaling via the phosphotransferase FruA drives galactose utilization by Streptococcus pneumoniae, resulting in hypervirulence. mBio 8, e02269–e02216. doi: 10.1128/mBio.02269-16
Tyanova, S., Temu, T., and Cox, J. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319. doi: 10.1038/nprot.2016.136
Wardani, A. K., Egawa, S., Nagahisa, K., Shimizu, H., and Shioya, S. (2006). Robustness of cascade pH and dissolved oxygen control in symbiotic nisin production process system of Lactococcus lactis and Kluyveromyces marxianus. J. Biosci. Bioeng. 101, 274–276. doi: 10.1263/jbb.101.274
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, C., Zhang, J., Du, G., and Chen, J. (2013). Aspartate protects Lactobacillus casei against acid stress. Appl. Microbiol. Biotechnol. 97, 4083–4093. doi: 10.1007/s00253-012-4647-2
Yeom, E., Kwon, D.-W., Lee, J., Kim, S.-H., Lee, J.-H., Min, K.-J., et al. (2020). Asparaginyl-tRNA synthetase, a novel component of hippo signaling, binds to salvador and enhances yorkie-mediated tumorigenesis. Front. Cell Dev. Biol. 8, 32. doi: 10.3389/fcell.2020.00032
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, G., Qi, Q., Sadiq, F. A., Wang, W., He, X., and Wang, W. (2021). Proteomic analysis explores interactions between Lactiplantibacillus plantarum and Saccharomyces cerevisiae during sourdough fermentation. Microorganisms 9, 2353. doi: 10.3390/microorganisms9112353
Zhao, D., Meng, F., Zhou, L., Lu, F., Bie, X., Sun, J., et al. (2021). Maltose effective improving production and regulatory biosynthesis of plantaricin EF in Lactobacillus plantarum 163. Appl. Microbiol. Biotechnol. 105, 2713–2723. doi: 10.1007/s00253-021-11218-w
Keywords: bacteriocin, co-culture, microorganism interaction, quorum sensing, response mechanism
Citation: Nie R, Zhu Z, Qi Y, Wang Z, Sun H and Liu G (2023) Bacteriocin production enhancing mechanism of Lactiplantibacillus paraplantarum RX-8 response to Wickerhamomyces anomalus Y-5 by transcriptomic and proteomic analyses. Front. Microbiol. 14:1111516. doi: 10.3389/fmicb.2023.1111516
Received: 29 November 2022; Accepted: 30 January 2023;
Published: 24 February 2023.
Edited by:
Paloma López, Margarita Salas Center for Biological Research (CSIC), SpainReviewed by:
John Renye, Agricultural Research Service (USDA), United StatesRodney Honrada Perez, University of the Philippines Los Baños, Philippines
Copyright © 2023 Nie, Zhu, Qi, Wang, Sun and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guorong Liu,  bGl1Z3Vvcm9uZzE5ODNAMTI2LmNvbQ==
bGl1Z3Vvcm9uZzE5ODNAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Rong Nie1†
Rong Nie1† Yanwei Qi
Yanwei Qi Guorong Liu
Guorong Liu