
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 02 March 2023
Sec. Extreme Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1110084
This article is part of the Research TopicMicrobiology of Radioactive EnvironmentsView all 6 articles
Deinococcus radiopugnans DY59 (formerly Deinococcus swuensis DY59) is a radiation-resistant bacterium isolated from soil. From the 3.5 Mb genomic DNA sequence of strain DY59 (December 2014), 31 insertion sequence (IS) elements of six IS families including IS1, IS4, IS5, IS66, IS630, and IS701 and five unclassified IS elements were detected. Upon induction of oxidative stress with 80 and 100 mM H2O2, the unique ISs of the IS4 family member were actively translocated into a carotenoid biosynthesis gene phytoene desaturase (QR90_10400), resulting in non-pigment phenotypic selection. Therefore, these active transpositions of a specific IS family member were induced by oxidative stress at 80 and 100 mM H2O2. Furthermore, D. radiopugnans DY59 exhibited extremely higher MIC values against H2O2 treatment. To explain this phenomenon, qRT-PCR was conducted to assess the expression levels of catalase and three LysR family regulators. Our findings indicated that the ISDrpg2 and ISDrpg3 elements of the IS4 family were actively transposed into the phytoene desaturase gene by H2O2 treatment via replicative transposition. However, high H2O2 resistance did not originate from H2O2-induced expression of catalase and LysR family regulators.
Most species belonging to the genus Deinococcus exhibit extreme resistance to gamma irradiation, ultraviolet ray exposure, and desiccation, which makes them ideal model microorganisms for studying anti-oxidation mechanisms. Particularly, these models allow for the assessment of the specific DNA repair and redox-sensing regulation mechanisms of cells based on the functional roles of genus-specific proteins for DNA damage responses (Daly, 2009; Slade and Radman, 2011; Luan et al., 2014; Lim et al., 2019).
Deinococcus swuensis DY59T was isolated by separating gamma-ray-resistant microorganisms from a soil sample obtained from the Deogyusan mountain (Jeonbuk Province, South Korea) at 1,500 m altitude (Lee et al., 2013). The soil was irradiated with 5 kGy gamma rays using a cobalt-60 gamma irradiator. Strain DY59 exhibited survival rates of 21 and 1% when exposed to 5 and 10 kGy of gamma radiation, respectively (Lee et al., 2013). The DY59 cells were found to be aerobic, coccus-shaped, had little mobility, and formed pink colonies in Luria-Bertani agar plates. The published genome of DY59 consisted of a single chromosome of 3,531,443 bp with a G + C content of 67.4%, which included 3,305 protein-encoding genes and 58 RNA genes (GenBank accession number GCA_000800395.1, December 2014; Kim et al., 2015). Upon sequencing of the 16S rRNA gene, strain DY59T showed high sequence similarity (99%) with the Deinococcus radiopugnans type strain (ATCC19172) as well as to Deinococcus marmoris KACC12218T (97.9%), Deinococcus saxicola KACC12240T (97.0%), Deinococcus aerolatus KACC12745T (96.2%), and Deinococcus frigens KACC12220T (96.1%; Lee et al., 2013; Kim et al., 2015). Recently, D. swuensis DY59 was reclassified as Deinococcus radiopugnans following genome data analysis including average nucleotide and amino acid identity and digital DNA–DNA hybridization on July 2021 (Lakra et al., 2021).
Prokaryotic genomes exhibit the smallest transposable element insertion sequences (ISs). They have important roles in genomic evolution including the enhancement of gene inactivation and genome plasticity (Touchon and Rocha, 2007; Siguier et al., 2014, 2015). IS abundance is positively correlated with the frequency of horizontal gene transfer (HGT), genome size, pathogenicity, non-obligatory ecological associations, and human association (Touchon and Rocha, 2007; Vandecraen et al., 2017). Moreover, recent mobilome studies have characterized IS distribution at the genus level (Blesa et al., 2019; Fayad et al., 2019). IS elements are transferred between genomes through all classical mechanisms of HGT (Frost et al., 2005). IS elements generally consist of one or two transposase ORFs and a terminal inverted repeat (TIR) sequence. Transposases are the most prevalent genes in nature. When IS element integrated into genomic DNA, the direct repeat (DR) sequences were produced generally. Unique and random DR sequences in border of IS elements indicated a specific and random sequence recognition and integration. Their mobile nature not only promotes the dissemination of transposable elements within and between genomes but also leads to mutations and rearrangements that can accelerate biological diversification and consequent evolution (Aziz et al., 2010).
In the case of Deinococcus geothermalis (Dgeo), 73 full-length IS elements belonging to nine IS families were distributed across the different molecules of the genome including two mega plasmids. Several IS elements were actively transposed into other genome loci through the copy-and-paste mechanism of ISDge5, ISDge6, and ISDge7 members under hydrogen peroxide (H2O2)-induced oxidative stress. Furthermore, we reported the occurrence of a colorless phenotype through the loss of carotenoid biosynthesis via IS transposition, which was supported by several lines of evidence (Tian and Hua, 2010; Lee et al., 2019, 2020; Ye et al., 2021; Shin et al., 2022). Research on the insertion sequence of bacteria can be conducted from a molecular evolution perspective, including cutting-edge mutagenesis using transposable elements to examine the plasticity of bacterial genomes induced by long-term culture or oxidative stress. It can also serve as a basic study of genome stability for the potential application of bioremediation in the genus Deinococcus species (Gerber et al., 2015; Wright et al., 2017; Consuegra et al., 2021).
In this work, we describe that the genome of D. radiopugnans DY59 has a total of 36 IS elements with six IS families including five unclassified IS elements. Interestingly, strain DY59_IS elements have multiple copies of the IS66, IS630, IS701, IS4, and IS5 families. Nevertheless, when strain DY59 was exposed to 5 kGy of gamma irradiation, non-pigmented mutants were not observed (Ye et al., 2022). Here, we evaluated the occurrence of active transposition induced by short-term oxidative stress conditions and its effects on the genomic plasticity in the radiation-resistant bacterium D. radiopugnans DY59. When oxidative stress was induced via hydrogen peroxide treatment with 80 mM, the ISDrpg2 and ISDrpg3 IS elements of the IS4 family were transposed into a carotenoid synthesis gene encoding a phytoene dehydrogenase (QR90_10400), resulting in a non-pigment phenotype. Furthermore, strain DY59 exhibited a high minimum inhibitory concentration (MIC) of 300 mM H2O2. Thus, the expression levels of catalase and LysR family members (i.e., its putative controlling regulators) were determined by qRT-PCR.
Deinococcus radiopugnans DY59 was kindly provided by Dr. M. Kim at Seoul Women’s University. This strain was deposited into the Korean Collection for Type Cultures (KCTC33033T). Deinococcus geothermalis DSM11300T and D. radiodurans R1 were used as a positive control for the active transposition of ISs. Deinococcus geothermalis and D. radiodurans R1 were cultured on TGY medium containing 1% tryptone, 0.5% yeast extract, and 0.1% glucose with 150 rpm for broth culture at 48 and 30°C, respectively. Strain DY59 is known to grow well in R2A or LB medium. Therefore, the cells were cultured for 3–4 days at 30°C in R2A medium, whereas the cells in the TGY medium required only 2–3 days to grow. All downstream Deinococcus cultures were thus conducted using TGY culture medium.
Deinococcus radiopugnans DY59 cells were typically grown to a maximum optical density (OD600) for 2–3 days. When the culture reached an absorbance of 2.0 or 4.0 at OD600 as measured by a UV–VIS spectrophotometer, the cells were harvested by centrifugation at 3,000 rpm and resuspended in a 0.9% NaCl solution to an OD600 of 2.0 for exposure to H2O2.
Next, the cells were treated with final H2O2 concentration ranges of 80 and 100 mM/200 and 300 or 0 mM as a control and continuously cultured for 1 h at 30°C with 150 rpm shaking. The samples were then directly diluted to 10−4–10−5 with 0.9% NaCl solution and 100 μL of the cell suspensions were spread on TGY agar medium using sterilized glass beads. After discarding the glass beads, the TGY plates were incubated at 30°C for 2–3 days. Non-pigmented colonies were detected and cell culture dilutions were streaked on TGY agar plates for pure cultivation. Non-pigment forming frequency was calculated the ratio of number of non-pigment colony and CFU. We also evaluated whether hypochlorite and gamma irradiation induced the occurrence of IS transposition. Active transposition was detected via PCR amplification of four selected target genes related to carotenoid biosynthesis (Supplementary Table S1).
The D. radiopugnans DY59 genome sequence was obtained from the NCBI database (GenBank accession number NZ_CP010028.1). IS elements were detected from the annotated transposase genes using protein profiling data obtained from the GenBank database. We first extracted sequences from upstream and downstream of a transposase (1 kb region; 3 kb total length). Then, terminal inverted repeat (TIR) sequences and direct repeat (DR) sequences were determined using DNA sequence alignment software such as BLAST, ClustalW, and BioEdit. All detected IS elements were sorted into IS family members. Unfortunately, the IS detection platform ISfinder (https://isfinder.biotoul.fr; Siguier et al., 2006) still does not support the detection and classification of D. radiopugnans DY59 IS elements with calling ID as of September 2022. Therefore, we classified the IS ourselves based on IS structure analysis and nomenclature rules (Supplementary Table S2).
Afterward, 10 ml of cultured cells till to an OD600 of 2.0 and 4.0 were harvested, resuspended with 0.9% NaCl, and separately exposed for 1 h to various H2O2 concentrations or 100 μg/mL sodium hypochlorite treatment or for in total 5 kGy gamma-irradiation (160 Gy/min). Then, to measure the cell viability using colony forming units, samples were diluted to a 10−5 ratio and spread on TGY agar plates. After 5 kGy gamma-irradiation, DY59 strain was exhibited 0.5% viability comparing to non-radiation. The viability of hypochlorite treatment was not analytical determined. To measure the minimum inhibitory concentration (MIC) of antibiotics, the strain DY59 cells of 100 μL on OD600 = 4.0 were spread on TGY agar plates, and the disk diffusion assay was conducted with different concentrations of 10 μL antibiotic solutions, such as streptomycin, kanamycin, and ampicillin. The MIC values were determined based on the formation of a clear zone.
Strain DY59 cells were harvested at OD600 values of 2.0 and 4.0, resuspended to OD600 of 2.0 in 5 mL 0.9% NaCl, and then treated with 50–150 mM H2O2 for 1 h at 150 rpm. After the H2O2 challenge, the samples were centrifuged at 10,000 rpm for 5 min and washed once more with 0.9% NaCl. The supernatants were discarded and the pellets were stored at −20°C. The cell wall was broken using phenol and DNA digestion was performed using DNase I. RNA was extracted using an RNA prep kit for RNA isolation (RNeasy mini purification kit; Qiagen, Germany). After measuring the extracted RNA concentration, the concentration was normalized to 1,000 ng in an 8 μL volume for all samples. cDNA synthesis was performed with a dNTP mixture and 6-mer random primers using the following protocol: 60°C for 5 min, 4°C for 3 min, 30°C for 10 min, 42°C for 60 min, and finally 95°C for 5 min (PrimeScript™ first strand cDNA Synthesis Kit; TaKaRa, Japan). In the step at 4°C for 3 min, 4 μL 5× buffer, 4.5 μL RNase free water, 1 μL RTase, and 0.5 μL RNase inhibitor were added. qRT-PCR analysis was performed using TB Green® Premix Ex Taq™ (TaKaRa, Japan) on a Bio-RAD RT-PCR model CFX96™ Optics Module (Bio-RAD, United States). The expression of catalase (QR90_06310) and three LysR family regulators (QR90_13110, QR90_14595, and QR90_15105) was normalized to that of GAPDH, a gene that is constitutively expressed throughout all growth phases. The related expression levels of both catalase and LysR genes were calculated as described in a previous study (Choo et al., 2020). Differences between samples were determined via the Student’s t-test using the Prism™ software (ver. 8.0). Differences were deemed statistically significant at p < 0.05 (*) and p < 0.01 (**). The primer sequences for qRT-PCR were shown in Supplementary Table S3.
Generally, D. radiopugnans DY59 formed reddish-colored colonies after being cultured for 2–3 days on TGY medium at 30°C. The strain also exhibited a stronger viscosity compared to D. geothermalis. Here, we observed different MIC values, which were indicative of different levels of antibiotic resistance. The MIC of streptomycin was less than 25 μg/mL, whereas those of ampicillin, kanamycin, and chloramphenicol were 30, 300, and 60 μg/mlL, respectively (Figure 1A; Supplementary Figure S1). Therefore, we conclude that strain DY59 displays a resistant phenotype to kanamycin and chloramphenicol, and a tolerant phenotype to streptomycin and ampicillin when compared to the typical working concentrations of antibiotics.
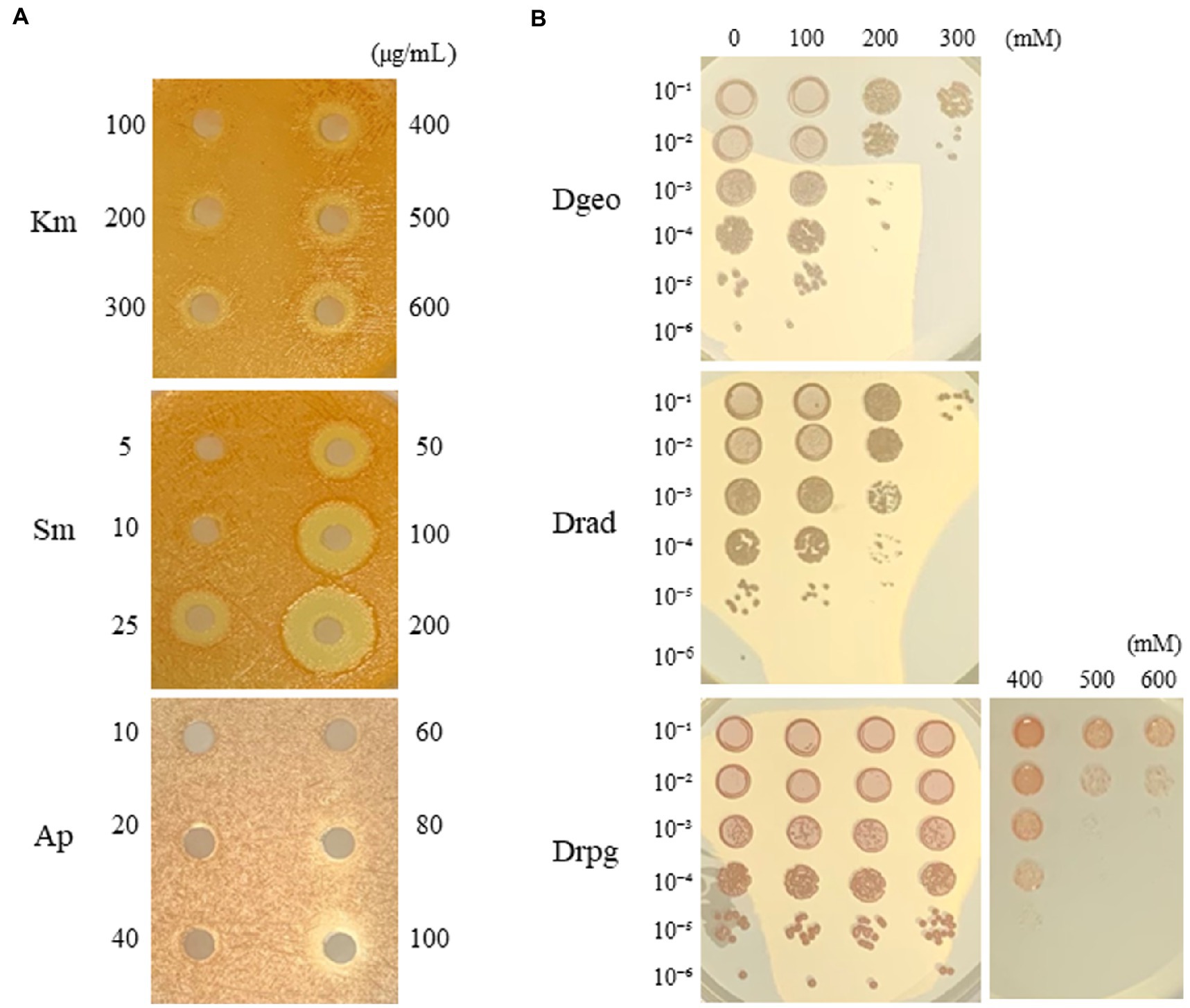
Figure 1. Minimum inhibitory concentration (MIC) measurement of kanamycin, streptomycin, and ampicillin via the disk diffusion method on D. radiopugnans DY59 (A) and viability assay under different concentrations of H2O2 among Drad, D. radiodurans; Dgeo, D. geothermalis; Drpg, D. radiopugnans (B). Three Deinococcus species were grown to OD600 of 4.0, harvested, and resuspended by 0.9% NaCl to OD600 of 2.0. H2O2 treatment was performed on different final concentrations for 1 h then, serially diluted and spotted on TGY medium.
Surprisingly, the results of our H2O2 viability assays indicated that D. radiopugnans DY59 could form colonies after being exposed to H2O2 concentrations of up to 600 mM for 1 h, which is two times higher than the resistance of two well-studied control strains of D. radiodurans and D. geothermalis (300 mM H2O2; Figure 1B).
Genes related to carotenoid biosynthesis in genus Deinococcus, which were linked to the phenotypic reddish color of the colonies, were detected via KEGG pathway analysis and genome-wide genomics studies (Tian and Hua, 2010; Lim et al., 2019). We selected four genes that are involved in the carotenoid biosynthesis pathway, which were marked in a simplified pathway schematic (Figure 2). The selected four carotenoid biosynthesis genes of strain DY59 exhibited amino acid sequence similarity with more than 60% to D. geothermalis genes: QR90_03795, a phytoene synthase, is 69.4% similar to Dgeo_0523; QR90_10400, a phytoene dehydrogenase (desaturase), is 78.9% similar to Dgeo_0524; QR90_14380, a carotenoid hydratase, is 58.6% similar to Dgeo_2309; and QR90_14400, a FAD-dependent oxidoreductase, is 74.5% similar to Dgeo_2306 (Supplementary Table S1). Interestingly, some carotenoid biosynthesis-related genes of D. geothermalis were adjacently clustered in the genome, for examples crtB-crtI and cruC-cruD-cruF-crtO. In contrast, each carotenoid biosynthesis gene in D. radiopugnans was separated.
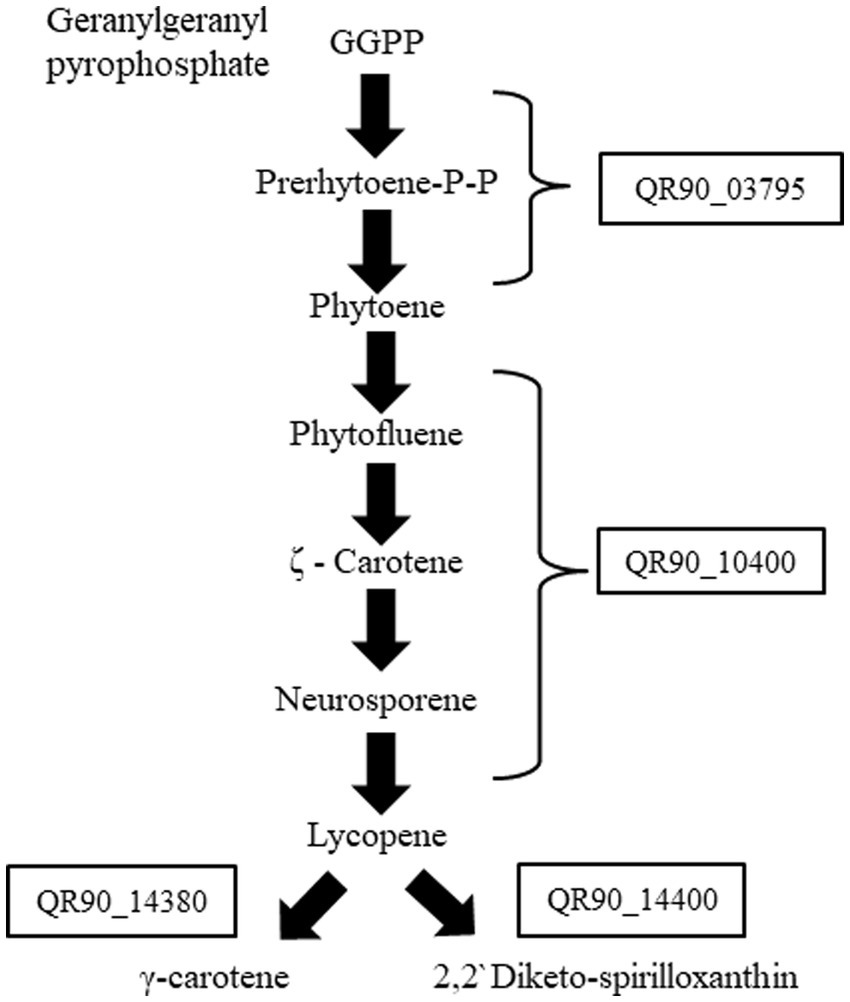
Figure 2. Simplified schematic of the carotenoid biosynthesis pathway in DY59 strain from KEGG carotenoid pathway analysis.
From the NCBI genome sequence data (submitted at December 2014) and protein information of DY59 (Kim et al., 2015), we first selected transposases and their genomic loci and defined full-length IS elements. The genome of strain DY59 contains six defined IS family members with DDE type transposase (Tpase) including IS1, IS4, IS5, IS66, IS630, and IS701, as well as five unclassified Tpases within a total of 36 ISs (Supplementary Table S2; Supplementary Figure S2). In general, Tpase is a major component of IS elements and simplified schematics of the IS structures except for the IS1 family member, which has only a partial fragment, are illustrated in Supplementary Figure S3.
The IS1 family member is an IS element (QR90_RS04110) with a 119 aa-length Tpase. In general, the IS1 family IS element is composed of two transposase-related open reading frames (ORFs) with the same transcriptional direction. Therefore, TIR and DR sequences for this IS region of the IS1 family could not be easily determined because it was only a partial fragment.
The IS4 family members had five copies in the genome. The IS elements including transposases were separated into two subtypes. QR90_RS09690, QR90_04660, and QR90_01215 have identical IS elements with a 320 aa-long transposase, a conserved TIR sequence (CTCTGTACCGGACAACT), and DR sequences of variable sequences with 9 nt (“GCCGTGATC,” “GTCACGCCC,” and “GAAAGCAAT,” respectively). QR90_RS10860 and QR90_04150 possess another identical IS element with an identical transposase with 327 aa-length. Both IS elements have a conserved TIR sequence “CTCGGTAGCTGACAACTTCA” and DR sequences “AGATTGAAC” and “CAGGGTCAG.” Thus, these IS4 family members were divided into the two subtypes ISDrpg2 and ISDrpg3, with their Tpase sequences having a 67.19% amino acid identity.
The IS5 family members have four copies in the genome with two subtypes elements: QR90_RS06350 with a 277 aa-length Tpase and QR90_RS04355, QR90_RS07350, and QR90_06585 with 265 aa-length Tpase sequences. The three 265 amino acid-long transposase genes are highly similar, with similarity rates of 97.7–100%. However, the Tpase of QR90_RS06350 with a 277 amino acid length exhibited a quite low similarity of 12–12.8% to the 265 aa-length Tpase in the IS5 family. Therefore, the IS5 family members were separated into two subtypes: ISDrpg4 and ISDrpg5. The TIR and DR sequences of three IS elements with a 265 aa length Tpase were “AGGCTG” and “TAG,” respectively. The TIR and DR sequences of QR90_RS06350 were “ACCTCCTGCGAAAGTC” and “TAG,” respectively. The structural schemes for the forward and reverse area of the Tpase are identical, with a 54 and 9 nt distance, respectively. One interesting finding is that the loci of the IS5 family IS elements are positioned close to the IS701 family member, except for QR_ RS06350.
The IS66 family IS elements have five copies in the DY59 genome: QR90_07275, QR90_RS09525, QR90_07340, QR90_11595, and QR90_09840, with a 472 aa-long Tpase named ISDrpg6. The IS elements have a conserved terminal inverted repeat (TIR) sequence “GTCTGTGATTAGCGGTCG” and 8 nt variable direct repeat (DR) sequences (“GATGGGGG,” “GGTGCAGG,” “ATGTCGTC,” “GGC GAGAG,” and “TATTTTTG”).
The IS630 family members were divided into the two subtypes ISDrpg7 and ISDrpg8: QR90_RS17010 with a 181 aa-long Tpase, QR90_RS08750, QR90_RS17180, QR90_RS17220, QR90_RS17305, and QR90_RS17410 with a 187 aa-long Tpase. Although QR90_17010 and QR90_RS17410 exhibited different Tpase lengths, the amino acid sequence similarities were 100% identical, whereas the 187 aa-length Tpases exhibited sequence identities ranging from 92.8 to 98.9%. In contrast, the Tpase of QR90_RS08750 exhibited a 64.7–67.9% identity when compared to the 187 aa-long Tpase. Thus, ISDrpg7 included five IS elements except QR90_RS08750 for ISDrpg8. All of the examined IS630 family members exhibited three-nucleotide DR sequences “TGA/TAA/TCA” and their TIR sequence was “TACGGACTCCGATTAA.”
The IS701 family members included the 10 IS elements QR90_RS00720, QR90_04350, QR90_RS06590, QR90_05955, QR90_10425, QR90_RS17070, QR90_RS17080, QR90_RS17170, QR90_RS17235, and QR90_RS17395, with a 432 aa-length. Two distinct subtypes were identified according to Tpase identity: QR90_RS17070, QR90_RS17080, QR90_RS17170, QR90_RS17235, and QR90_RS17395 had a DNA sequence of 100% identity, whereas QR90_RS00720, QR90_04350, QR90_RS06590, QR90_05955, and QR90_10425 had a 99.5% identity. In contrast, the two subtypes exhibited only a 89.12% Tpase identity between each other. All IS701 family members had a unique DR sequence (“nTAG”) and TIR sequence (“CTGTACTTTG GGGATATTCA”). Interestingly, the 3′ end of the TIR sequence of all IS701 family members overlapped into the Tpase ORF.
In this study, we identified five unclassified IS members: QR90_08760 with a 270 aa-long Tpase; QR90_RS08625 with a 477 aa-long Tpase; QR90_08735 with a 434 aa-long Tpase; QR90_RS05950 with a 120 aa-long Tpase; and QR90_04880 with an 89 aa-long Tpase.
Next, the active transposition of IS elements was detected on non-pigment phenotypic selection under oxidative stress conditions induced by H2O2 treatment in the present study.
A total of 24 and one non-pigmented colonies were detected after low concentration (80 and 100 mM) and high concentration (200 and 300 mM) H2O2 treatment for 1 h, respectively. It seems that generation of non-pigmented mutants is less efficient with higher H2O2 concentration.
In the high-concentration treatment, one is a complete non-pigmented strain (w3) and two isolates exhibited a pale reddish color on TGY plates (w1 and w2; Figure 3A). When carotenoid biosynthesis genes were amplified by PCR with the appropriate primers, the complete non-pigmented strain only exhibited an enlarged PCR product from QR90_10400, a phytoene desaturase (Figure 3B). However, other carotenoid biosynthesis genes were not affected. The ISDrpg3 of the IS4 family was integrated at the 275th nucleotide of QR90_10400 (Figure 4). The TIR sequence of this IS element was “CTCGGTAGCTGACAACTTCA” and the DR sequence was “ACCCGCCCC.”
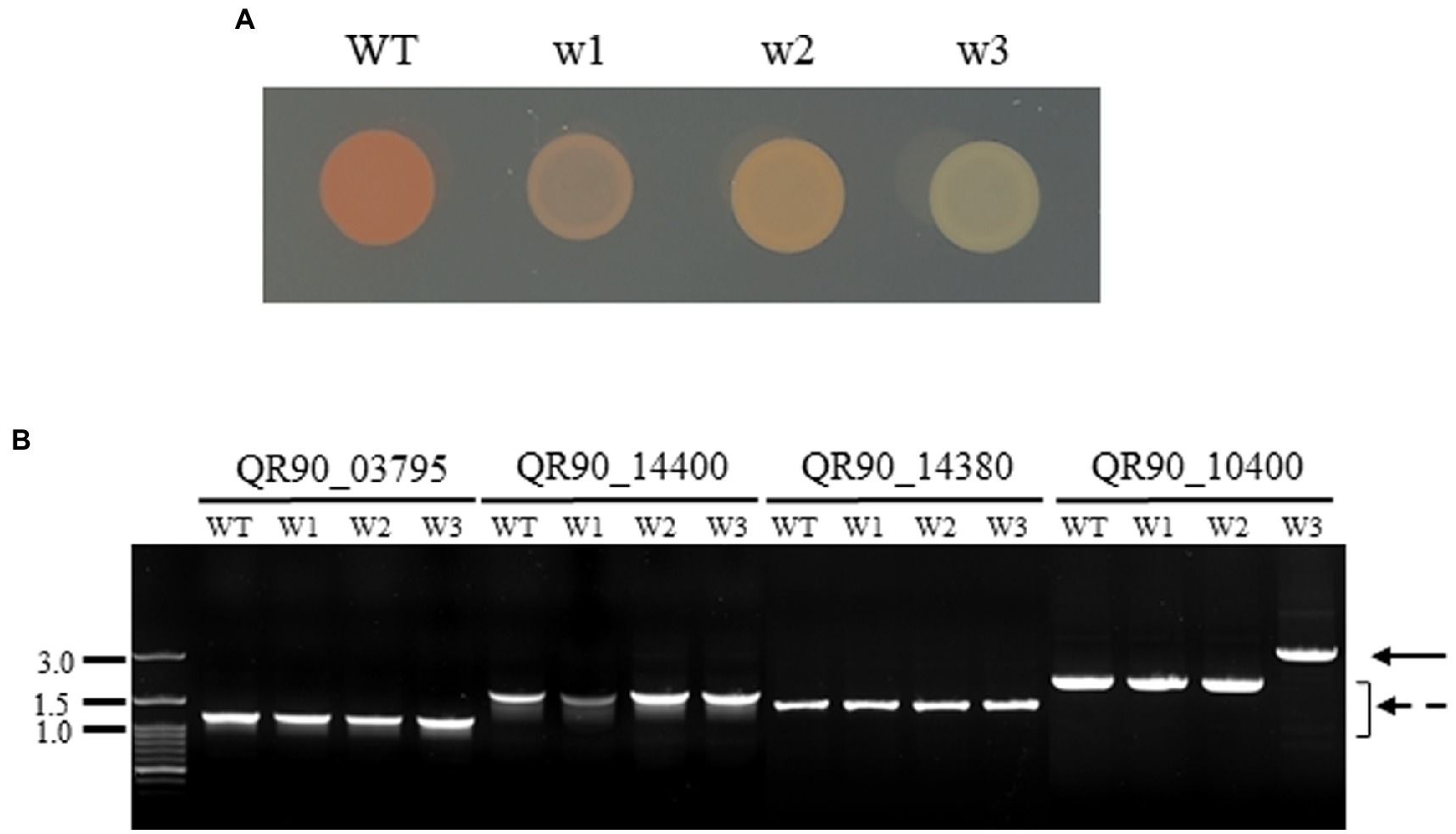
Figure 3. Phenotypic diversity in the high concentration of H2O2 treatment (200 mM for w1 and w2 or 300 mM for w3) for 1 h (A) and detection of gene disruption by PCR of genes involved in the carotenoid pathway (B). QR90_03795, Phytoene synthesis; QR90_14400, FAD-dependent oxidoreductase; QR90_14380, carotenoid 1,2-hydratase; QR90_10400, phytoene dehydrogenase. Lanes: M, size marker; 1, 5, 9: WT; 2, 6, 10: w1; 3, 7, 11: w2; 4, 8, 12: w3. The dotted arrow indicates the wild-type gene PCR products. The solid arrow indicates the IS-integrated PCR product.
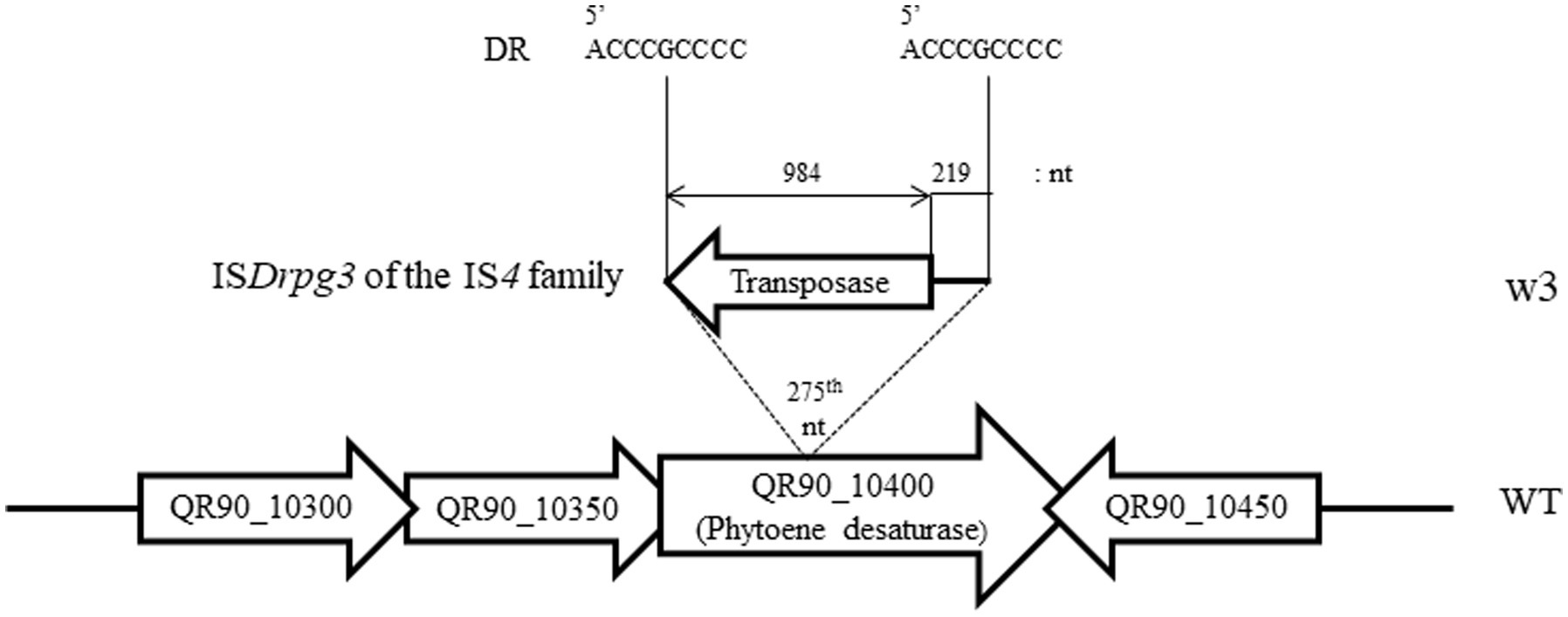
Figure 4. Detection of IS integration loci and composition of IS element in QR90_10400 phytoene desaturase in w3 mutant obtained after 300 mM H2O2 treatment. The ISDrpg3 of the IS4 family member was integrated at the 275th nt locus of the QR90_10400 gene with reverse transcriptional direction.
In the low-concentration H2O2 treatment, five out of 24 non-pigmented strains exhibited PCR amplicons with QR90_10400 gene enlargement: two strains with an OD600 of 2.0 and three strains with an OD600 of 4.0 at 80 mM H2O2 (Figure 5A). The frequency of IS transposition in gene QR90_10400 exhibited 5.1 × 10−4 and 1.8 × 10−4 from OD600 of 2.0 and 4.0, respectively (Supplementary Table S4). All five of these IS-integrated mutants exhibited IS4 family transposition (Figure 5B; Supplementary Figure S4). Other carotenoid-related genes QR90_03795, QR90_14380, and QR90_14400 were not affected (Figure 5C). The 19 remaining clones did not exhibit any changes in the size of the PCR products, indicating that there was no IS transposition in the selected four carotenoid biosynthesis genes. However, there might be point mutations in carotenoid-related genes or perhaps IS transposition in non-analyzed carotenoid biosynthesis-related genes.
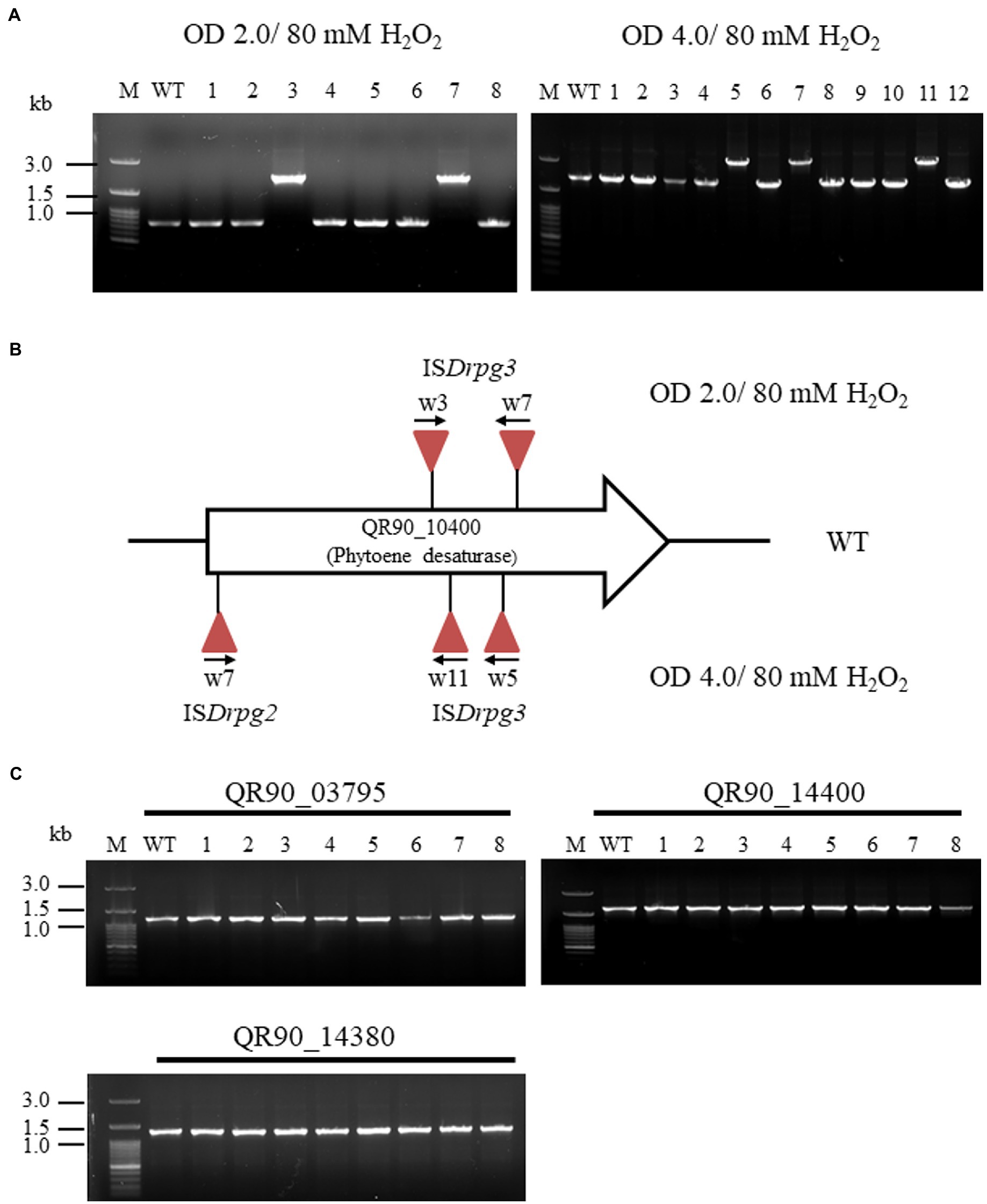
Figure 5. Insertion sequence transposition triggering at low concentration of H2O2 treatment with 80 mM. (A), Detection of IS transposition at two growth phases (OD600 2.0 and 4.0) in gene QR90_10400. (B), Analysis of IS integrational loci at both detection conditions. All transposed IS elements are both ISDrpg2 and ISDrpg3 of the IS4 family members. (C), Analysis of IS transposition in three carotenoid biosynthesis-related genes. These genes were not affected by IS integration under H2O2 oxidative stress conditions.
At an OD600 of 2.0, one IS element (w3) was integrated in the forward direction at the 401st nt of phytoene desaturase. The DR sequence of this element was “CTTCTTCGA” and the TIR sequence was “CTCGGTAGCTGACAACTTCA.” Another (w7) integrated in the reverse direction at the 617th nt. The DR sequence of this element was “GTAAACGAG” and the TIR sequence was “CTCGGTA GCTGACAACTTCA” (Figure 5B; Supplementary Figure S4A). Both IS elements were belonged to ISDrpg3 of QR90_04150 or QR90_RS10860 (Supplementary Table S2).
At an OD600 of 4.0, one IS element (w5) integrated in the reverse direction at the 587th nt of phytoene desaturase. The DR sequence of this element was “AGGCGCTC” and the TIR sequence was “CTCGGTAGCTGACAACTTCA.” Another (w11) integrated in the reverse direction at the 474th nt; its DR sequence was “GCTCGTAGC” and its TIR sequence was identical to that of w5. Both IS elements were identical to ISDrpg3 (Supplementary Figure S4B). The last IS element (w7) integrated in the forward direction at the 22nd nt with DR sequence “CCAGCAGGC” and TIR sequence “CTCT GTACCGGACAACT” (Figure 5B; Supplementary Figure S4C). This IS element was identical to ISDrpg2 of QR90_01215, QR90_04660, or QR90_09690. Therefore, D. radiopugnans DY59 exhibits active transposition of IS4 family members in H2O2 treatment conditions. The PCR detection of five copies of IS4 family members at the location as found in the genome sequence indicates that the active transposition occurred through replicative mode in present (Supplementary Figure S5).
The 100 μM sodium hypochlorite does not affected viability of the strain DY59. 5 kGy irradiated strain DY59 exhibits 99.5% reduction of CFU. The selection of non-pigmented colonies was performed via gamma irradiation exposure and sodium hypochlorite treatment. Unlike the lack of pigment production of D. geothermalis, gamma irradiation of total 5 kGy did not induce non-pigment phenotypes in the D. radiopugnans DY59 wild-type strain (Ye et al., 2022). Moreover, although hypochlorite treatment induced the non-pigment phenotype, the non-pigmented colony did not exhibit IS element integration in the four analyzed carotenoid biosynthesis genes (data not shown). This may be the same explanation for the absence of transposition of IS in phenotypic changes due to point mutations in four carotenoid biosynthetic genes and the decay of other genes related to pigment formation.
To determine the mechanisms underlying the high H2O2 resistance of DY59, the expression levels of catalase and three LysR family regulators including possible oxyR were first measured by qRT-PCR analysis. The chromosome of DY59 strain has a single catalase QR90_06310 with 72.17 and 74.17% amino acid sequence similarity to D. radiodurans catalase KatE1 and D. geothermalis KatE, respectively. After exposure to various concentrations of H2O2 (50, 100, 150 mM, and an unexposed control) for 1 h and growing the cells to OD600 values of 2.0 and 4.0, the relative expression levels of catalase and LysR family members were measured via qRT-PCR by the basal expression level of OD600 of 2.0 with unexposed control as 1.0. Unexpectedly, catalase was not dramatically induced at OD600 2.0 or 4.0 in any of the tested H2O2 concentrations. Nevertheless, there was a 2-fold increase in catalase expression at OD600 4.0 when the cells were challenged with 50 and 100 mM H2O2 (Figure 6A).
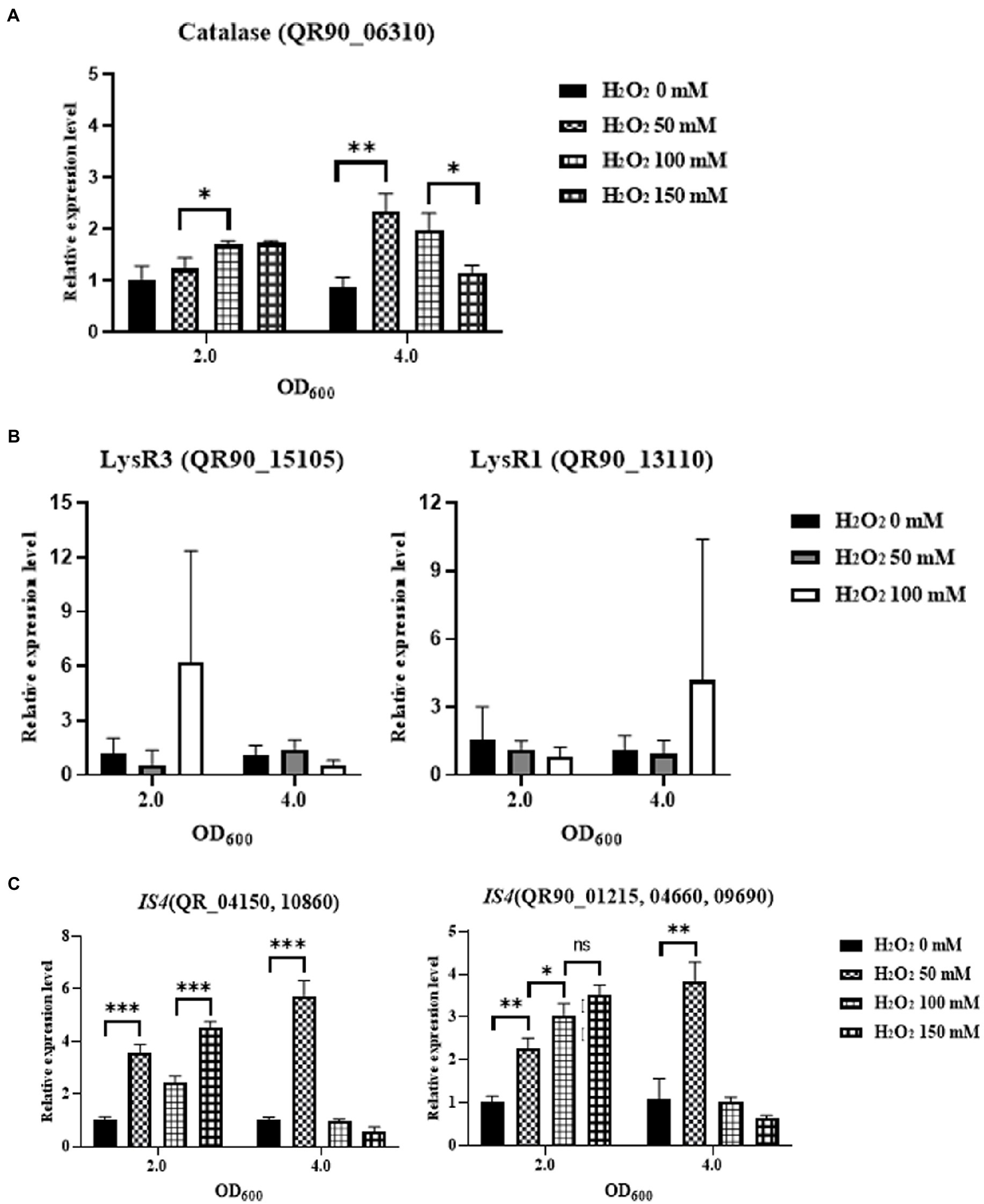
Figure 6. Comparison of the gene expression levels of catalase (A), two LysR family regulators (B) and two transposases in ISDrpg2 and ISDrpg3 (C) by qRT-PCR under different final concentrations of H2O2 (0–150 mM). Pair-wise comparisons between experimental groups were conducted via Student’s t-test with the Prism™ ver. 8.0 software (*p < 0.05; **p < 0.01; ***p < 0.001).
LysR1 QR90_13110 exhibited a more than 6-fold induction at an OD600 of 4.0 upon exposure to 100 mM H2O2 and LysR3 QR90_15105 exhibited a more than 6-fold induction at an OD600 of 2.0 in the 100 mM H2O2 condition. Our findings thus indicated that 100 mM H2O2 controlled the expression of LysR1 and LysR3 at two different growth phases (Figure 6B). However, LysR2 (QR90_14595) was not affected regardless of growth phase and oxidation condition (data not shown). LysR2 was exhibited 70.3 and 75.7% identities of amino acid sequence to the proposed OxyR of D. radiodurans and D. geothermalis, respectively. Therefore, the high H2O2 resistance phenotype of the DY59 strain cannot be attributed to changes in the expression of catalase and its three potential LysR family regulator members, as determined by RNA production levels measured by qRT-PCR analysis.
There are two subtypes of IS4 family which were actively transposed to other loci. Tpase of ISDrpg3 such as QR90_04150 and QR90_10860/ISDrpg2 such as QR90_01215, QR90_04660, and QR90_09690 exhibited a more than 5-6-fold induction at an OD600 of 4.0 in the 50 mM H2O2 condition. The related expression levels were reduced at an OD600 of 4.0 in the 100 and 150 mM H2O2 condition (Figure 6C). However, both ISDrpg2 and ISDrpg3 Tpases were gradually induced over 2-fold at an OD600 of 2.0 in the different H2O2 conditions. Thus, IS elements of the IS4 family were induced by the H2O2 and actively transposed into other genomic loci with replicated mode.
Bacillus subtilis, a widely known Gram-positive bacterial model organism, lacks IS or any other transposable element excluding several bacteriophages and other remnants of horizontal gene transfer (HGT) events from genome study of wild-type strain. The IS identification platform “ISfinder” revealed four IS families in the B. subtilis genome including IS4Bsu1 (IS4 family), ISBsu1 (IS3 family), ISBsu2 (IS256 family), and ISBsu3 (IS1595 family; Siguier et al., 2006). Interestingly, pathogenic Bacillus species such as B. cereus, B. anthracis, B. thuringiensis, and others have many IS families and Tn3 elements (Fayad et al., 2019). Therefore, the distribution of transposable elements may be associated with pathogenicity. Additionally, the genomes of the pathogenic bacteria may have undergone multiple acquisition of transposable elements through HGT in response to environmental stimuli (Fayad et al., 2019).
Here, we detected active transposition of IS elements under oxidative stress conditions of H2O2 treatment in the radiation-resistant bacterium D. radiopugnans DY59, which was isolated from mountain soil collected in South Korea. The DY59 isolate was phylogenetically clustered near three Deinococcus species from Antarctic marine environments (Hirsch et al., 2004; Kim et al., 2015). The genome of D. radiopugnans DY59 has a total of 36 IS elements encompassing six IS families. Interestingly, these six family members belonged to D. geothermalis IS families that also contained the IS6, IS982, and IS200/605 families (Lee et al., 2020). Unfortunately, the IS names cannot be classified based on the IS distribution in the D. radiopugnans genome from “ISfinder” platform (at Sep. 2022; Siguier et al., 2006). The transposase annotation as a key component of IS element was varied among the genome data and between prediction and analysis.
When wild-type DY59 cells were treated with 80 or 300 mM H2O2, the active transposition of the IS4 family members was only detected on phytoene desaturase (QR90_10400; Figures 4, 5). The active transposition of the IS4 family was also particularly detected in wild-type D. geothermalis upon H2O2 treatment and dielectric bilayer discharge (DBD) plasma radiation (Lee et al., 2020; Ye et al., 2022).
When the D. radiodurans strains were exposed to gamma irradiation, there were two particular active transposition events on a trimethoprim-resistant selection. One was an IS integrated into a thyA gene; the other was an uvrA gene disruption that resulted in mitomycin-resistant phenotypic selection (Narumi et al., 1997; Mennecier et al., 2006; Pasternak et al., 2010). When the cells were treated with 5 kGy of gamma irradiation, the wild-type D. geothermalis strain exhibited active transposition of IS families, (e.g., IS1 and IS5 family), whereas the wild-type DY59 strain and D. radiodurans strain did not exhibit non-pigment phenotypic mutations (Ye et al., 2022).
Insertion sequence transposition has been detected under various environmental stressors, such as nutrient deprivation, temperature changes, metal ion exposure, and oxidative stress caused by UV irradiation, gamma irradiation, and H2O2 treatment (Ohtsubo et al., 2005; Twiss et al., 2005; Kharat et al., 2006; Mijnendonckx et al., 2011; Suzuki et al., 2021). Interestingly, this active transposition of IS elements was found to vary in a species-specific manner. D. geothermalis wild-type and several particular gene-disrupted mutants have been reported to exhibit different types of IS transposition. For example, ISDge3 of the IS1 family was actively transposed under gamma irradiation, whereas ISDge11 of the IS4 family was transposed under H2O2 treatment and DBD plasma radiation. Moreover, ISDge5 and ISDge6 of the IS5 family were found to be transposed in dps-, oxyR-, cystine importer-, and lysR-deficient mutants (Lee et al., 2020, 2022; Ye et al., 2021).
Here, we explored the IS distribution in the genome size of 3.54 Mb of the DY59 strain. Our findings suggested that DY59 exhibits a less complex IS distribution than that of D. geothermalis. In this study, a strict IS element of the IS4 family was actively transposed into a carotenoid biosynthesis gene QR90_10400 phytoene desaturase under H2O2 treatment (Figures 4, 5B; Supplementary Figure S4). Therefore, the wild-type D. radiopugnans DY59 may serve as a suitable model organism for studying the active transposition of unique IS element using a single oxidation inducer such as H2O2. These experiments provide the opportunity to determine unique IS transposition events in organisms that exhibit genomic plasticity. However, there are still many challenges associated with IS naming and classification from gene annotation, emphasizing the need for more accurate algorithms and additional criteria including machine learning tools for IS identification and assignment, as well as the creation of a global network of research groups working together.
The genome sequence information of D. radiopugnans ATCC19172 was updated twice on June 2019 and August 2020, with contig assembly lengths of 4.33 and 4.3 Mb, respectively (NCBI genomes). Deinococcus radiopugnans ATCC19172 has three catalases, FHR04_11220, FHR04_17100, and FHR04_17320, sharing, respectively, 99, 26.6, and 47.6% identity with the single identified catalase (QR90_06310) from strain DY59. In case the published genome sequence of DY59 is incomplete, this strain might also possess homologs of FHR04_17100 and FHR04_17320. Thus, we performed qRT-PCR using primer sets corresponding to FHR04_17100 and FHR04_17320 to investigate expression of possible homologs in strain DY59. The possible FHR04_17100 homolog was not induced by the H2O2 treatment and the possible FHR04_17320 homolog exhibited more than 2-fold induction at OD600 of 2.0 with 50 and 100 mM H2O2 treatment (Supplementary Figure S6). When 150 mM H2O2 treatment at OD600 of 4.0 was applied, the possible FHR04_17320 homolog exhibited nearby 3-fold induction. In the present additional catalase expression data, the high hydrogen peroxide resistance of DY59 is not explained by induced expression of catalase genes. DY59 strain may have a high level of constitutive catalase enzyme activity, or it may employ other mechanisms for hydrogen peroxide resistance, such as unidentified protective pathways and physiological defense systems using extracellular matrixe components such as extracellular polymeric substances (EPS), proteins, and eDNA, which aid in biofilm formation and enhance tolerance to oxidative stress, and certain transporters (Li et al., 2013; Molina-Santiago et al., 2021).
The genome sequence of strain ATCC19172 is 0.8 Mb larger than that of DY59. Although the reported genome sequence of strain DY59 consist of only a chromosome, we consider the possibility that this strain might have one or more additional genome molecules such as plasmids. If the strain DY59 has plasmids, the IS family and number will expand. IS elements are commonly known to exhibit random movement. However, experiments conducted on Deinococcus geothermalis have shown that the transposition of unique IS elements follows a specific pattern. The exact pattern is still unknown, but this experiment provides a starting point for further research. Specifically, it was observed that the IS4 family of IS element only transposed under hydrogen peroxide conditions in D. radiopugnans DY59. Additionally, the ability of Deinococcus species to endure various forms of stress such as radiation, drying, and toxic chemicals is attempted to be explained from a molecular evolutionary perspective through the transposition of IS elements. Further exploration into this research area presents a significant challenge for the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary material.
ES, HN, QY, and S-JL performed conception, designed experiments, and performed and analyzed data. ES, HN, and S-JL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MS&ICT; 2022R1A2C1010233).
The authors would like to thank M. Kim for providing the DY59 strain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1110084/full#supplementary-material
Aziz, R. K., Breitbart, M., and Edwards, R. A. (2010). Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 38, 4207–4217. doi: 10.1093/nar/gkq140
Blesa, A., Sanchez, M., Sacristan-Horcajada, E., Fuente, S. G., Peiro, R., and Berenguer, J. (2019). Into the Thermus mobilome: presence, diversity and recent activities of insertion sequences across Thermus spp. Microorganisms 7:25. doi: 10.3390/microorganisms7010025
Choo, K., Kim, M., Abdi Nansa, S., Bae, M. K., Lee, C., and Lee, S.-J. (2020). Redox potential change by the cystine importer affected on enzymatic antioxidant protection in Deinococcus geothermalis. Antonie Van Leeuwenhoek 113, 779–790. doi: 10.1007/s10482-020-01388-4
Consuegra, J., Gaffé, J., Lenski, R. E., Hindré, T., Barrick, J. E., Tenaillon, O., et al. (2021). Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria. Nat. Commun. 12:980. doi: 10.1038/s41467-021-21210-7
Daly, M. J. (2009). A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7, 237–245. doi: 10.1038/nrmicro2073
Fayad, N., Awad, M. K., and Mahillon, J. (2019). Diversity of Bacillus cereus sensu lato mobilome. BMC Genom. 20:436. doi: 10.1186/s12864-019-5764-4
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Gerber, E., Bernard, R., Castang, S., Chabot, N., Coze, F., Dreux-Zigha, A., et al. (2015). Deinococcus as new chassis for industrial biotechnology: biology, physiology and tools. J. Appl. Microbiol. 119, 1–10. doi: 10.1111/jam.12808
Hirsch, P., Gallikowski, C. A., Siebert, J., Peissl, K., Kroppenstedt, R., Schumann, P., et al. (2004). Deinococcus frigens sp. nov., Deinococcus saxicola sp. nov., and Deinococcus marmoris sp. nov., low temperature and draught-tolerating, UV-resistant bacteria from continental Antarctica. Syst. Appl. Microbiol. 27, 636–645. doi: 10.1078/0723202042370008
Kharat, A., Coursange, E., Noirclerc-Savoye, M., Lacoste, J., and Blot, M. (2006). IS1 transposition IS enhanced by translation errors and by bacterial growth at extreme glucose levels. Acta Biochim. Pol. 53, 729–738. doi: 10.18388/abp.2006_3300
Kim, M. K., Srinivasan, S., Back, C.-G., Joo, E. S., Lee, S.-Y., and Jung, H.-Y. (2015). Complete genome sequence of Deinococcus swuensis, a bacterium resistant to radiation toxicity. Mol. Cell Toxicol. 11, 315–321. doi: 10.1007/s13273-015-0031-5
Lakra, P., Verma, H., Talwar, C., Singh, D. N., Singhvi, N., Lal, R., et al. (2021). Genome based reclassification of Deinococcus swuensis as a heterotypic synonym of Deinococcus radiopugnans. Int. J. Syst. Evol. Microbiol. 71:004879. doi: 10.1099/ijsem.0.004879
Lee, C., Choi, N., Bae, M. K., Choo, K., and Lee, S.-J. (2019). Transposition of insertion sequences was triggered by oxidative stress in radiation-resistant bacterium Deinococcus geothermalis. Microorganisms 7:446. doi: 10.3390/microorganisms7100446
Lee, C., Choo, K., and Lee, S.-J. (2020). Active transposition of insertion sequences by oxidative stress in Deinococcus geothermalis. Front. Microbiol. 11:558747. doi: 10.3389/fmicb.2020.558747
Lee, J.-J., Lee, H. J., Jang, G. S., Yu, J. M., Cha, J. Y., Kim, S. J., et al. (2013). Deinococcus swuensis sp. nov., a gamma-radiation-resistant bacterium isolated from soil. J. Microbiol. 51, 305–311. doi: 10.1007/s12275-013-3023-y
Lee, C., Ye, Q., Shin, E., Lee, C., Choi, N., Kim, Y., et al. (2022). Transposition of insertion sequences by dielectric barrier discharge plasma and gamma irradiation in the radiation-resistant bacterium Deinococcus geothermalis. J. Microbiol. Methods 196:106473. doi: 10.1016/j.mimet.2022.106473
Li, M., Sun, H., Feng, Q., Lu, H., Zhao, Y., Zhang, H., et al. (2013). Extracellular dGMP enhances Deinococcus radiodurans tolerance to oxidative stress. PLoS One 8:e54420. doi: 10.1371/journal.pone.0054420
Lim, S., Jung, J.-H., Blanchard, L., and de Groot, A. (2019). Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 43, 19–52. doi: 10.1093/femsre/fuy037
Luan, H., Meng, N., Fu, J., Chen, X., Xu, X., Feng, Q., et al. (2014). Genome-wide transcriptome and antioxidant analyses on gamma-irradiated phases of Deinococcus radiodurans R1. PLoS One 9:e85649. doi: 10.1371/journal.pone.0085649
Mennecier, S., Servant, P., Coste, G., Bailone, A., and Sommer, S. (2006). Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 59, 317–325. doi: 10.1111/j.1365-2958.2005.04936.x
Mijnendonckx, K., Provoost, A., Monsieurs, P., Leys, N., Mergeay, M., Mahillon, J., et al. (2011). Insertion sequence elements in Cupriavidus metallidurans CH34: distribution and role in adaptation. Plasmid 65, 193–203. doi: 10.1016/j.plasmid.2010.12.006
Molina-Santiago, C., de Vicente, A., and Romero, D. (2021). Bacterial extracellular matrix as a natural source of biotechnologically multivalent materials. Comput. Struct. Biotechnol. J. 19, 2796–2805. doi: 10.1016/j.csbj.2021.05.008
Narumi, I., Cherdchu, K., Kitayama, S., and Watanabe, H. (1997). The Deinococcus radiodurans uvrA gene: identification of mutation sites in two mitomycin-sensitive strains and the first discovery of insertion sequence element from deinobacteria. Gene 198, 115–126. doi: 10.1016/s0378-1119(97)00301-6
Ohtsubo, Y., Genka, H., Komatsu, H., Nagata, Y., and Tsuda, M. (2005). High-temperature-induced transposition of insertion elements in Burkholderia multivorans. Microbiology 71, 1822–1828. doi: 10.1128/AEM.71.4.1822-1828.2005
Pasternak, C., Ton-Hoang, B., Coste, G., Bailone, A., Chandler, M., and Sommer, S. (2010). Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 6:e1000799. doi: 10.1371/journal.pgen.1000799
Shin, E., Ye, Q., and Lee, S.-J. (2022). Active transposition of insertion sequences in prokaryotes: insights from the response of Deinococcus geothermalis to oxidative stress. Antioxidants 11:481. doi: 10.3390/antiox11030481
Siguier, P., Gourbeyre, E., and Chandler, M. (2014). Bacterial insertion sequences: their genomic impact and diversity. FEMS Rev. 38, 865–891. doi: 10.1111/1574-6976.12067
Siguier, P., Gourbeyre, E., Varani, A., Ton-Hoang, B., and Chandler, M. (2015). Everyman’s guide to bacterial insertion sequences. Microbiol. Spectr. 3:MDNA3-0030-2014. doi: 10.1128/microbiolspec.MDNA3-0030-2014
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Slade, D., and Radman, M. (2011). Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 75, 133–191. doi: 10.1128/MMBR.00015-10
Suzuki, H., Taketani, T., Tanabiki, M., Ohara, M., Kobayashi, J., and Ohshiro, T. (2021). Frequent transposition of multiple insertion sequences in Geobacillus kaustophilus HTA426. Front. Microbiol. 12:650461. doi: 10.3389/fmicb.2021.650461
Tian, B., and Hua, Y. (2010). Carotenoid biosynthesis in extremeophilic Deinococcus-Thermus bacteria. Trends Microbiol. 18, 512–520. doi: 10.1016/j.tim.2010.07.007
Touchon, M., and Rocha, E. P. C. (2007). Causes of insertion sequences abundance in prokaryotic genomes. Mol. Biol. Evol. 24, 969–981. doi: 10.1093/molbev/msm014
Twiss, E., Coros, A. M., Tavakoli, N. P., and Derbyshire, K. M. (2005). Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 57, 1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x
Vandecraen, J., Chandler, M., Aertsen, A., and Van Houdt, R. (2017). The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 43, 709–730. doi: 10.1080/1040841X.2017.1303661
Wright, M. S., Mountain, S., Beeri, K., and Adams, M. D. (2017). Assessment of insertion sequence mobilization as an adaptive response to oxidative stress in Acinetobacter baumannii using IS-seq. J. Bacteriol. 199, e00833–e00816. doi: 10.1128/JB.00833-16
Ye, Q., Lee, C., Shin, E., and Lee, S.-J. (2021). Influence of redox imbalances on the transposition of insertion sequences in Deinococcus geothermalis. Antioxidants 10:1623. doi: 10.3390/antiox10101623
Ye, Q., Shin, E., Lee, C., Choi, N., Kim, Y., Yoon, K. S., et al. (2022). Transposition of insertion sequences by dielectric barrier discharge plasma and gamma irradiation in the radiation-resistant bacterium Deinococcus geothermalis. J. Microbiol. Methods 196:106473. doi: 10.1016/j.mimet.2022.106473
Keywords: Deinococcus radiopugnans DY59, genome plasticity, insertion sequences, oxidative stress, phenotypic selection, transposition
Citation: Shin E, Noh HS, Ye Q and Lee S-J (2023) Hydrogen peroxide treatment induces the transposition of an insertion sequence in Deinococcus radiopugnans DY59. Front. Microbiol. 14:1110084. doi: 10.3389/fmicb.2023.1110084
Received: 28 November 2022; Accepted: 13 February 2023;
Published: 02 March 2023.
Edited by:
Sudhir K. Shukla, Bhabha Atomic Research Center (BARC), IndiaReviewed by:
Arjan De Groot, Commissariat à l'Energie Atomique et aux Energies Alternatives (CEA), FranceCopyright © 2023 Shin, Noh, Ye and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung-Jae Lee, c3VuZ2phZWxlZUBraHUuYWMua3I=
‡Present address: Qianying Ye, Department of Biomedical science and technology, Kyung Hee University, Seoul, Republic of Korea
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.