- 1Jiangxi Provincial Engineering Research Center for Seed-Breeding and Utilization of Camphor Trees, School of Hydraulic and Ecological Engineering, Nanchang Institute of Technology, Nanchang, China
- 2College of Agriculture, Yangtze University, Jingzhou, China
- 3Jiangxi Academic of Forestry, Nanchang, China
Cinnamomum camphora (C. camphora) is a broad-leaved evergreen tree cultivated in subtropical China. Currently, the use of C. camphora clonal cuttings for coppice management has become popular. However, the effects of C. camphora coppice planting on soil abiotic and biotic variances remained unclear. In this study, we collected soil from three points in the seven-year C. camphora coppice planting land: under the tree canopy (P15), between trees (P50), and abandoned land (Control) to investigate the effects of C. camphora coppice planting on soil fertility, microbial community structure and enzyme activity. The results revealed that C. camphora coppice planting significantly increased soil fertility in the point under the tree canopy (P15) and point between trees (P50), and P15 had more significant effects than P50. Meanwhile, in P15 and P50, soil bacterial, fungal alpha-diversity were improved and microbial community structures were also changed. And the changes of soil organic carbon and total nitrogen promote the transformation of soil bacterial, fungal community structures, respectively. In addition, C. camphora coppice planting significantly (p < 0.05) increased soil urease (UE), polyphenol oxidase, and peroxidase activities, while significantly decreased soil ACP activity. This study demonstrated that the C. camphora coppice planting could improve soil fertility in subtropical China, which promoted the transformation of soil microbial community from oligotrophs (K-strategist) to copiotrophs (r-strategist). Thus, this work can provide a theoretical basis for soil nutrient variation and productive management of C. camphora coppice plantation in subtropical China.
Introduction
Tree planting has been proposed as a practical method to prevent soil degradation. It can increase vegetation coverage, reduce rain erosion and enhance soil water and fertilizer retention ability (Speranza et al., 2019; Kumar et al., 2021). Meanwhile, the litterfall will return generous amounts of organic matter to the soil and the well-developed roots can activate the soil nutrients elements (Asigbaase et al., 2021). All of these are vital in the formation and evolution of planting land soil fertility (Feng et al., 2019). Various studies proved that tree planting can improve the soil nutrient stocks and availabilities in the wasteland and farmland (Zarafshar et al., 2020). For instance, relative to the abandoned land, Robinia pseudoacacia planting significantly increased the soil total C, N, and P nutrients in Calcaric Cambisol of the Loess Plateau (Zhang et al., 2019). And Shorea robusta planting significantly enhanced the available N and P contents in the topsoil compared with the agricultural land used for Oryza sativa L. cultivation in tropical areas (Ahirwal and Maiti, 2016). Nevertheless, some researches declared that tree planting has the negative impact on soil fertility. Teng et al. (2020) summarized that the 19-year-old Pinus sylvestris var. mongolica planting decreased the soil organic carbon (SOC) and TN contents in aeolian sandy soil of northern China. Xu et al. (2021a) also recorded that in subtropical China, the Manglietia glauca Blume planting reduced the soil TN and AP contents relative to the abandoned land with 10.42 and 58.77%, respectively. Thus, soil fertility under tree planting may vary with tree species, soil types, and so on.
Soil microorganisms are highly sensitive to changes in soil nutrients and are regarded as one critical biological indicator of soil fertility (Carrara et al., 2018; Liu et al., 2018a; Sun et al., 2020). Tree planting will exert extensive and profound impacts on soil microorganisms (Thoms et al., 2010). A previous study recorded that 20-year-old Ormosia hosiei planting obviously improved the soil microbial (bacterial and fungal) biomass and diversity in red-yellow soil (Wan et al., 2021). While the rubber plantation caused the degradation of the soil microbial community structure relative to the natural forests in Latosols and Ferralsols in tropical regions (Monkai et al., 2018). The soil fertility variations caused by tree planting are the dominant factors that drive variations in soil microbial community (Pereira et al., 2019). Xu et al. (2021b) believed that the increase of SOC and TN in R. pseudoacacia planting improved the microbial biomass and nutrients metabolism capability. On the contrary, Wu et al. (2015) discovered that with the increase of stand age in Pinus elliottii planting, decreased soil fertility leads to the decline of total PLFA amounts and carbon metabolic capability. Hence, to verify the changes of soil microbial community and determine their driving factors after tree planting are conducive to timely and efficiently adjusting the management strategies, thereby maintaining the stability of the forest ecosystems and promoting the sustainable development of forest resources.
Soil enzymes are secreted by soil microorganisms, and they have a pivotal influence on biochemical processes such as litter decomposition and element cycling (Rosinger et al., 2019). Tree planting induces strong effects on soil enzyme activities, and their activities may vary among different tree species (Bueno de Mesquita et al., 2017). Chen et al. (2022) reported that the Hippophae rhamnoides (broadleaf) planting increased enzyme activities of soil catalase, urease, and polyphenol oxidase (PPO) compared with the Larix gmelina (conifer) planting. Wang et al. (2020) proved that broadleaved tree planting increased soil PPO and peroxidase (POD) activities, while coniferous tree planting increased soil acid phosphatase activity (Moghimian et al., 2017). Cause broadleaved trees have higher litterfall quantity and quality (with low carbon to nitrogen ratio), which can produce large amounts of easily decomposed organic matter to stimulate microbial reproduction and thus secrete more extracellular enzymes (Nakayama et al., 2019). On the contrary, coniferous trees have lower litterfall quantity and quality (with high carbon to nitrogen ratio) with a great deal of hardly decomposed tannin, resin, and wax, which is unfavorable for metabolism of soil microorganisms (Ushio et al., 2008). Therefore, effects on soil enzyme activities will influence by different tree species.
Cinnamomum camphora (L.) Presl is a broad-leaved evergreen tree belonging to the Lauraceae family, which is widely distributed in subtropical China (Liu et al., 2022). C. camphora tree can produce large amounts of secondary metabolites such as essential oil (Zhong et al., 2022). Traditionally, the whole C. camphora tree was cut down to extract essential oil from its roots, stems, and leaves, and this disposable production method will cause great damage to the ecological environment (Liu et al., 2018b). Currently, using the C. camphora clonal cuttings for coppice management has become popular in southern China (Zhao, 2021). From the second year after transplanting, its aerial parts are felled down at 20 cm away from the ground to extract essential oil during July to September every year. Then the remaining C. camphora tree stump will sprout year after year, so as to realize the circular production. This management method is similar to regular harvest of crops, which is fundamentally different from traditional tree planting. However, the effects of C. camphora coppice planting patterns on soil quality and its ecological effects are still unclear.
In this study, one 5-year-old camphor coppice plantation land was selected to compare with the adjacent abandoned land in subtropical China. We further chose the point under the tree canopy and the point between trees. The former is greatly affected by nutrients transfer and tree growth, while the latter is the opposite. The major objectives of the present study were: (1) to determine how the C. camphora coppice planting affected soil fertility. (2) To reveal how the soil microbial community structure varied with the C. camphora coppice planting, and confirm the key driving factors. (3) To explore how soil enzyme activity responded to the C. camphora coppice planting and clarify the main reasons. This study can offer a theoretical basis for scientifically guiding the production and management of C. camphora coppice plantations in subtropical China.
Materials and methods
Study area
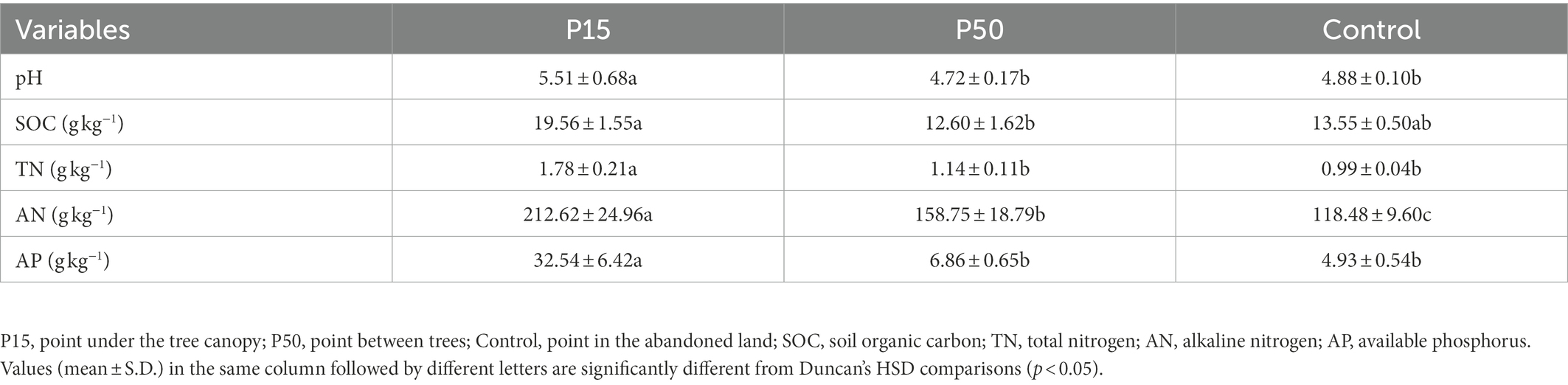
The field experiment was conducted on C. camphora coppice plantation land in Guixi City, Jiangxi Province, southern China (28°17′46″ N, 117°13′28″E). This research region has the subtropical monsoon climate in which the mean annual temperature precipitation and sunlight hours are 18.8°C, 1980.8 mm, and 1611.5 h at this site, respectively. The soils derived from the quaternary red clay, was abandoned land before the experiment. The initial properties of the topsoil (0–15 cm) with soil pH of 4.83, and with SOC, total nitrogen (TN), available nitrogen (AN), available phosphorus (AP) of 12.95, 0.96, 102.93, 5.21 g·kg−1, respectively.
Experiment design and soil sampling
The 5,000 m2 C. camphora coppice plantation land selected for this study was established in 2015. In early September of each year since 2016, the aboveground part of the C. camphora tree was felled at 20 cm away from the ground for essential oil extraction. The row and inter-plant spacings of the C. camphora tree are 1 m × 1 m, and the planting density is 10,000 tree·hm−2. Urea (contains 46% N), calcium magnesium phosphate (contains 12% P2O5) and potassium chloride (contains 60% K2O) were used for the nitrogen, phosphorus, potassium fertilizers respectively, and their application rate were all 150 g tree−1 year−1. 50% of the fertilizers were applied in early March and the remaining 50% were applied in late September after the C. camphora tree felling. The fertilization point (FP) was a 15 cm deep circle 25 cm from the center of the tree (Supplementary Figure S1).
In this study, we selected three different points as three treatments, each with four replicates: (1) Point under the tree canopy, 15 cm from the center of the tree (P15, inside the fertilization point); (2) point between trees, 50 cm from the center of the tree (P50, outside the fertilization point); (3) point in the abandoned land (control, next to the selected C. camphora coppice). The specific sampling scheme is shown (Supplementary Figure S1). Soil samples were randomly collected using the S-shaped sampling method after the C. camphora tree felling on September 12th, 2021. Each sample was carried out using a soil core sampler (diameter of 2.0 cm) from the depth of 0–15 cm. Each core was taken in 50 m2 areas, and twenty cores were mixed into one soil sample. Then pass the soils through the 2 mm sieve to remove the impurities and divided into three parts: one part was air-dried for soil fertility measurement, the second part was preserved at 4°C for enzyme activity measurement, and the remaining part was kept at −80°C for DNA extraction.
Soil properties and enzyme activity
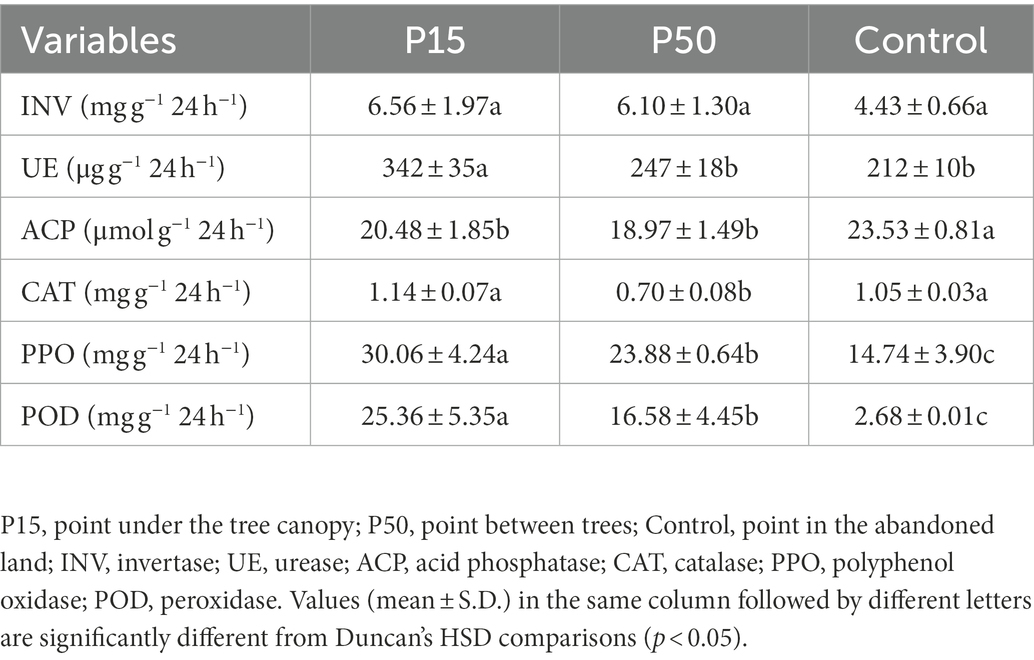
Soil pH value (soil to water ratio of 1:2.5) was measured with a pH meter (FE28-Standard, METTLER-TOLEDO, Switzerland; Sun et al., 2022). SOC was determined by dichromate oxidation and titration with ferrous sulfate. TN was determined using the Kjeldahl digestion distillation method. Available nitrogen (AN) and phosphorus (AP) were measured using alkali hydrolyzation and Bray method, respectively (Akhtar et al., 2018). Soil enzyme activities of invertase (INV), urease (UE), acid phosphatase (ACP), catalase (CAT), PPO, and POD were determined using kits from Beijing Solarbio Technology Co. Ltd. according to the test instructions (Tabatabai, 1994; Saiya-Cork et al., 2002).
Soil microbial DNA extraction and Illumina MiSeq sequencing
DNA was extracted from half a gram of fresh soil using the FastDNA® SPIN Kit for soil. The purification of DNA was tested by Power Clean DNA Clean-Up Kit to remove PCR inhibitors. The eluted DNA was examined by 1% (m/v) agarose gel electrophoresis and quantified with NanoDrop® 2000 spectrophotometer. Aliquots of the DNA were stored in a −20°C freezer for subsequent analyses. Primer sets Eub338 and Eub518 were used to determine the bacterial biomass by quantitative real-time PCR (qPCR) on the ABI Prism® 7300 Real-Time detection system. Fungal abundance was determined by Primer set SSU_0817 and SSU_1196 of the 18S rRNA gene in the same method with bacteria. The V4-V5 highly variable regions of the bacterial 16S rRNA genes were amplified with barcoded universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCCTTTGAGTTT-3′; Biddle et al., 2008). And the ITS regions of the fungal rRNA genes were amplified by primer sets ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5-GCTGCGTTCTTCATCGATGC-3′; Adams et al., 2013). The qPCR parameters were referred to Xiang et al. (2021) and Jin et al. (2021) methods.
Bioinformatics analysis for raw sequences
The original raw sequences were analyzed using the QIIME v.1.91 pipeline (Caporaso et al., 2010). And short (<200 bp) and low quality (average scores <25) or sequences were removed before downstream analysis (Magoč and Salzberg, 2011). High-quality sequences at a 3% dissimilarity level were selected as OTUs using UNOISE algorithm (Edgar, 2016). Representative set sequences were analyzed by RDP classifier using the SILVA 132 database with a confidence threshold of 80% (Wang et al., 2007). The representative sequences of ITS were annotated for species using the UNITE database with the blast method (Kõljalg et al., 2013). For downstream analyses, the number of bacterial and fungal sequences was rarefied to 30, 772 and 36, 205 for each sample to uniform the sequencing depths, respectively. A total of 4,427 bacteria OTUs and 1,690 fungal OTUs were identified for subsequent analysis.
Statistical analysis
Firstly, we distinguished whether the initial data were accord with the normal distribution. The non-normally distributed data were logarithm or square root-transformed to make them close to normal distribution. And the mean and standard deviations (e.g., soil fertility indexes and enzyme activities, microbial abundance, and alpha diversity indexes) of different treatments were calculated. One-way analysis of variance (ANOVA) by Duncan’s HSD test (p < 0.05) was used for multiple comparisons. Pearson correlation analysis was used to test the relationship between soil microbial abundance and alpha diversity indexes or enzyme activities and the soil fertility indexes. Constrained Principal of Coordinate Analysis (CAP) with Bray-Curtis distance was used to distinguish the differences of soil microbial communities or the enzyme profiling in different treatments, and analysis of similarity (ANOSIM) test was performed to verify whether the difference between treatments is significant. Using an Bray-Curtis distance matrix, the correlation between the soil microbial community structure or the enzyme profiling and soil fertility indexes was calculated using Mantel test with 999 permutations. The relationships between the soil microbial community composition or the enzyme profiling and soil fertility indexes were determined by redundancy analysis (RDA). Multivariate regression tree (MRT) was used to identify the key factors that shift the soil microbial community composition. Procrustes analysis was used to confirm the correlation between the soil enzyme profiling and microbial community composition in different treatments.
ANOVA and Pearson correlation analysis were conducted using the statistical software SPSS 20.0. ANOSIM, CAP, Mantel test, RDA, and Procrustes analysis were all done using the “vegan” package (Dixon, 2003), and MRT analysis was performed using the “mvpart” package of the R software (ver. 4.0.1; De’ath, 2003). The differences in soil microbial taxon and enzyme activities were assessed using STAMP software (Parks and Beiko, 2010).
Results
Soil fertility
Significant differences (p < 0.05) in soil fertility among different treatments were tested (Table 1). Compared with the Control, soil pH, SOC, TN, AN, and AP of the P15 were increased by 12.96% (p < 0.05), 44.29, 80.06% (p < 0.05), 79.46% (p < 0.05), and 560.16% (p < 0.05), respectively. Moreover, soil AN in P50 was significantly greater than Control with 33.99% (p < 0.05) increase, but there were no significant changes with other soil fertility indexes. Therefore, C. camphora coppice planting can increase soil fertility to a certain extent.
Soil bacterial and fungal community structure
Cinnamomum camphora coppice planting can increase the soil microbial biomass and alpha-diversity (Figure 1). Soil bacterial biomass in P15 and P50 increased by 45.68% (p < 0.05) and 26.05% (p > 0.05) in comparison with Control, respectively (Figure 1A). The soil bacterial alpha-diversity in P15 and P50 were significantly higher relative to Control, except the pielou index in P50 (Figures 1B–D). Similarly, the P15 and P50 increased soil fungal biomass by 25.00 and 9.79% compared with the Control (Figure 1F). Furthermore, the four fungal alpha-diversity indexes increased significantly (p < 0.05) in P15 and P50 relative to the control (Figures 1G–J). The increase of bacterial or fungal biomass and diversity indexes in P15 was greater than that in P50.
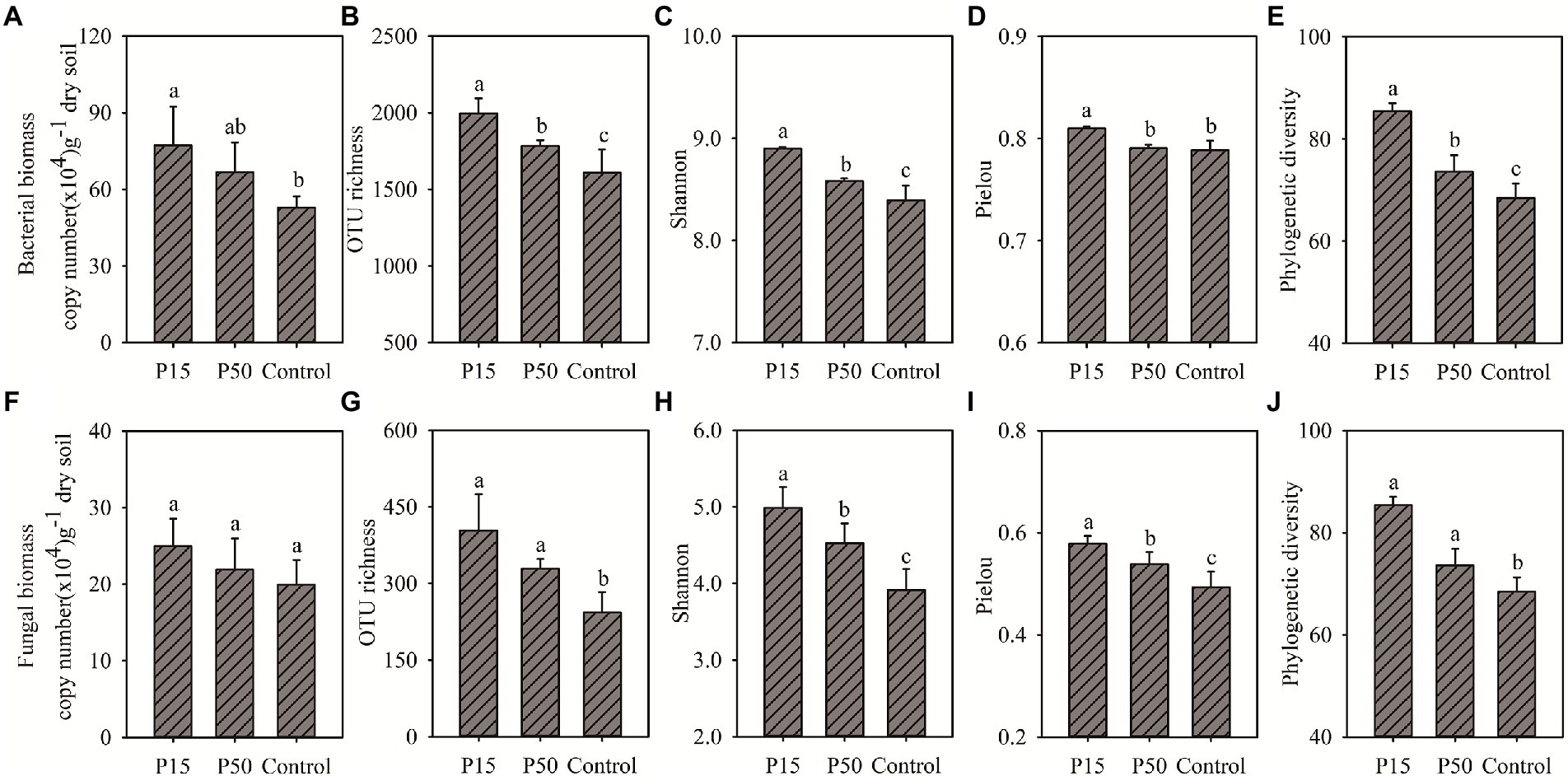
Figure 1. (A) Bacterial biomass, (B) Bacterial OTU richness, (C) Bacterial Shannon index, (D) Bacterial Pielou index, (E) Bacterial Phylogenetic diversity, (F) Fungal biomass, (G) Fungal OTU richness, (H) Fungal Shannon index, (I) Fungal Pielou index, (J) Fungal Phylogenetic diversity. Soil bacterial, fungal abundance and alpha-diversity under different treatments in the Cinnamomum camphora coppice planting. P15, point under the tree canopy; P50, point between trees; Control, point in the abandoned land. Bars represent mean; error bars denote standard deviation. Letters above bars represent differences from Duncan’s HSD comparisons (p < 0.05).
For bacterial, in comparison with the Control, the P15 and P50 significantly (p < 0.05) improved relative abundance of Proteobacteria (from 29.35 to 40.95 and 36.82%, respectively), Bacteroidetes (from 1.30 to 5.33 and 5.68%, respectively) and Gemmatimonadetes (from 1.26 to 2.51 and 1.47%, respectively). Adversely, relative abundance of Chloroflexi in P15 and P50 was significantly (p < 0.05) decreased by 71.49 and 59.46% relative to the Control (Supplementary Figure S2). STAMP analysis was used to analyze significantly differentiated bacterial genera between treatments (Supplementary Figure S3). There were thirteen bacterial genera in P15 and six bacterial genera in P50 significant differential with Control (p < 0.05, effect size >1). Specifically, relative to Control, P15 reduced relative abundance of Conexibacter, Ktedonobacter, Edaphobacter, Fimbriiglobus, Rhodanobacter and so forth, but enhanced relative abundance of Gaiella and GP6. Similarly, in comparison with Control, the P50 decreased relative abundance of Conexibacter and Ktedonobacter, whereas increased relative abundance of Flavitalea and Chryseolinea.
For fungal, in comparison with Control, the P15 (p < 0.05) and P50 increased relative abundance of Zygomycota (by 61.31 and 34.70%) and Basidiomycota (by 149.59 and 36.60%), but reduced the relative abundance of Ascomycota by 35.37 and 18.54% (Supplementary Figure S2). Significantly differentiated fungal genera between treatments were also analyzed (Supplementary Figure S3). Correspondingly, four fungal genera in P15 and five fungal genera in P50 were significantly different with the Control (p < 0.05, effect size >1). The relative abundance of Talaromyces and Hyaloscyphaceae were reduced in both the P15 and P50 relative to Control. In addition, compared with Control, P15 increased relative abundance of Westerdykella and Trichocomaceae, whereas P50 enhanced relative abundance of Fusarium, Archaeorhizomyces and Flagellospora.
Soil enzyme activities
Cinnamomum camphora coppice planting significantly affected the soil enzyme activities (Table 2). Soil UE, PPO, and POD activities were significant (p < 0.05) increased in P15 and P50 relative to Control, except the soil UE activity in the P50 (p > 0.05). Compared with Control, soil INV activity in P15 and P50 was increased (p > 0.05) by 47.98 and 37.58%. In addition, P15 and P50 significantly (p < 0.05) decreased the soil ACP activity, and P50 significantly (p < 0.05) decreased soil CAT activity in comparison with Control. STAMP analysis (p < 0.05, effect size >1) analyzed significantly different soil enzymes between treatments (Supplementary Figure S3). Specifically, in comparison with Control, P15 and P50 reduced the soil ACP activity, but enhanced the soil UE activity. Moreover, P15 also enhanced the soil POD activity.
Correlations between soil microbial community composition or enzyme activities and soil fertility indexes
Pearson correlation analysis was performed to further characterize the relationships between the soil microbial biomass, alpha-diversity, and dominant genera with soil fertility (Figure 2). The soil bacterial biomass was significantly positive correlated to SOC (r = 0.705, p < 0.05) and AP (r = 0.687, p < 0.05). As a whole, the four bacterial alpha-diversity indexes had significant positive correlation with all the soil fertility indexes (p < 0.01). For the dominant bacteria phyla, the Proteobacteria was significantly positive correlated with all the soil fertility indexes except pH, and the Gemmatimonadetes had significantly positive correlation with soil pH, SOC, TN and AP. However, the Chloroflexi was significantly negative correlated with soil TN (r = −0.701, p < 0.05), AN (r = −0.795, p < 0.01) and AP (r = −0.645, p < 0.05).
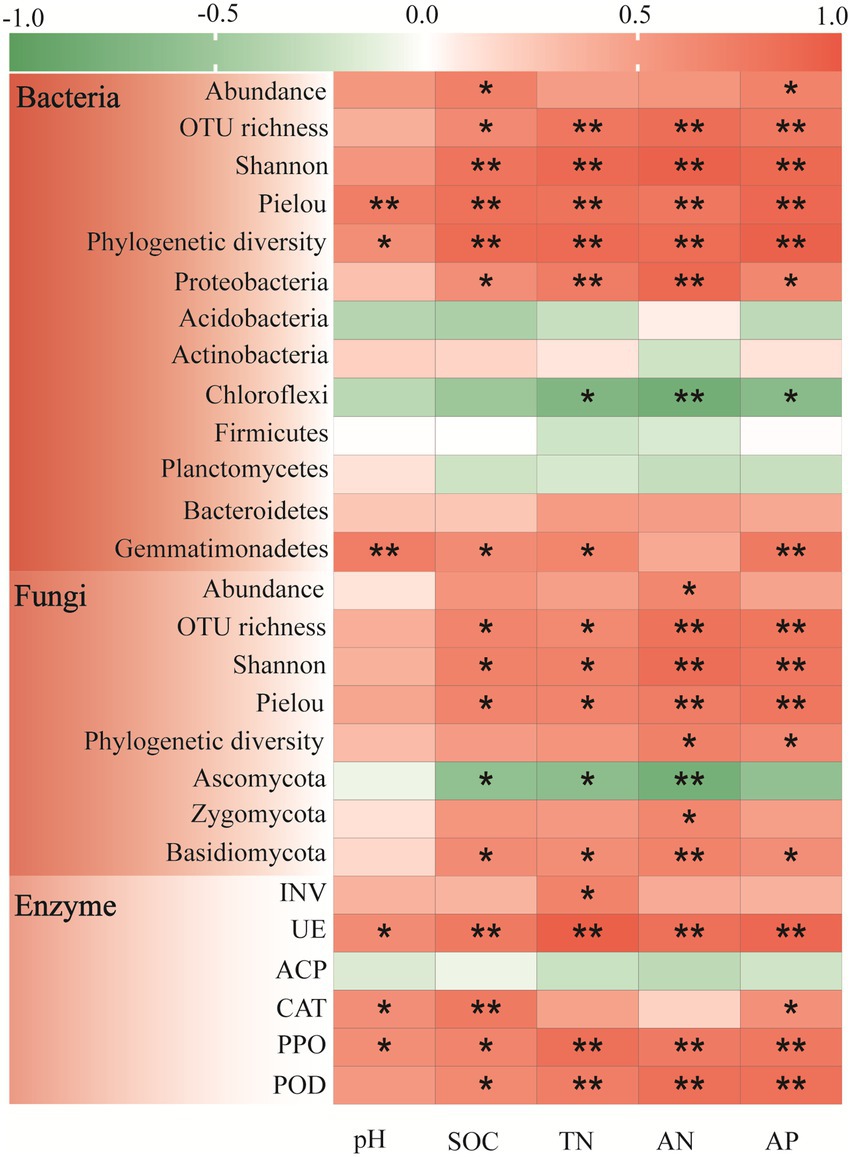
Figure 2. Pearson correlation analysis of soil fertility indexes with microbial community composition (abundance, alpha-diversity and dominant phyla) and enzyme activities in the Cinnamomum camphora coppice planting. Red represents a positive correlation, and green represents a negative correlation. (*p < 0.05; **p < 0.01).
Further analyses illustrated that the soil fungal biomass only had a significant positive correlation with AN (Figure 2; r = 0.659, p < 0.05). Notably, the fungal OTU richness, Shannon index, and pielou indexes were all significantly positively correlated with all the soil fertility indexes except pH, whereas phylogenetic diversity was only significantly positively correlated with AN (r = 0.696, p < 0.05) and AP (r = 0.632, p < 0.05). And the Zygomycota was only significantly positive correlated with AN (r = 0.651, p < 0.05). Overall, the Basidiomycota was positively correlated with all the soil fertility indexes, while the Ascomycota showed the opposite trend.
Correlation analysis was also detected between soil enzyme activities and soil fertility (Figure 2). In general, the activities of soil UE, PPO, and POD were significantly positive correlated to all the soil fertility indexes. The soil CAT activity had significant positive correlation with pH (r = 0.599, p < 0.05), SOC (r = 0.751, p < 0.01), and AP (r = 0.584, p < 0.05), while the soil INV activity only positive correlated with TN (r = 0.677, p < 0.05). Nevertheless, the soil ACP activity was slightly negative correlated with all the soil fertility indexes.
Primary factors for changes in soil microbial community and the enzyme profiling
The CAP analysis showed significant differences in soil microbial community between treatments (Supplementary Figure S4). Remarkably, we found that the soil bacterial community clearly separated (p = 0.001) among the three treatments (Supplementary Figure 4A; 43.1% of total variance was explained, p = 0.001). ANOSIM tests (Supplementary Table S1) revealed significantly (p < 0.05) different soil bacterial community among each other. Mantel test exhibited that soil bacterial community had significant (p < 0.01) correlation with all the soil fertility indexes (Supplementary Figure S5). In addition, RDA analysis pointed that the SOC (F = 2.450, p = 0.036) significantly affected soil bacterial community composition (Figure 3A). Furthermore, the MRT analysis performed that the SOC produced the largest deterministic effects on soil bacterial community variation (31.91%), with a threshold of 15.91 g kg−1 (Figure 3C).
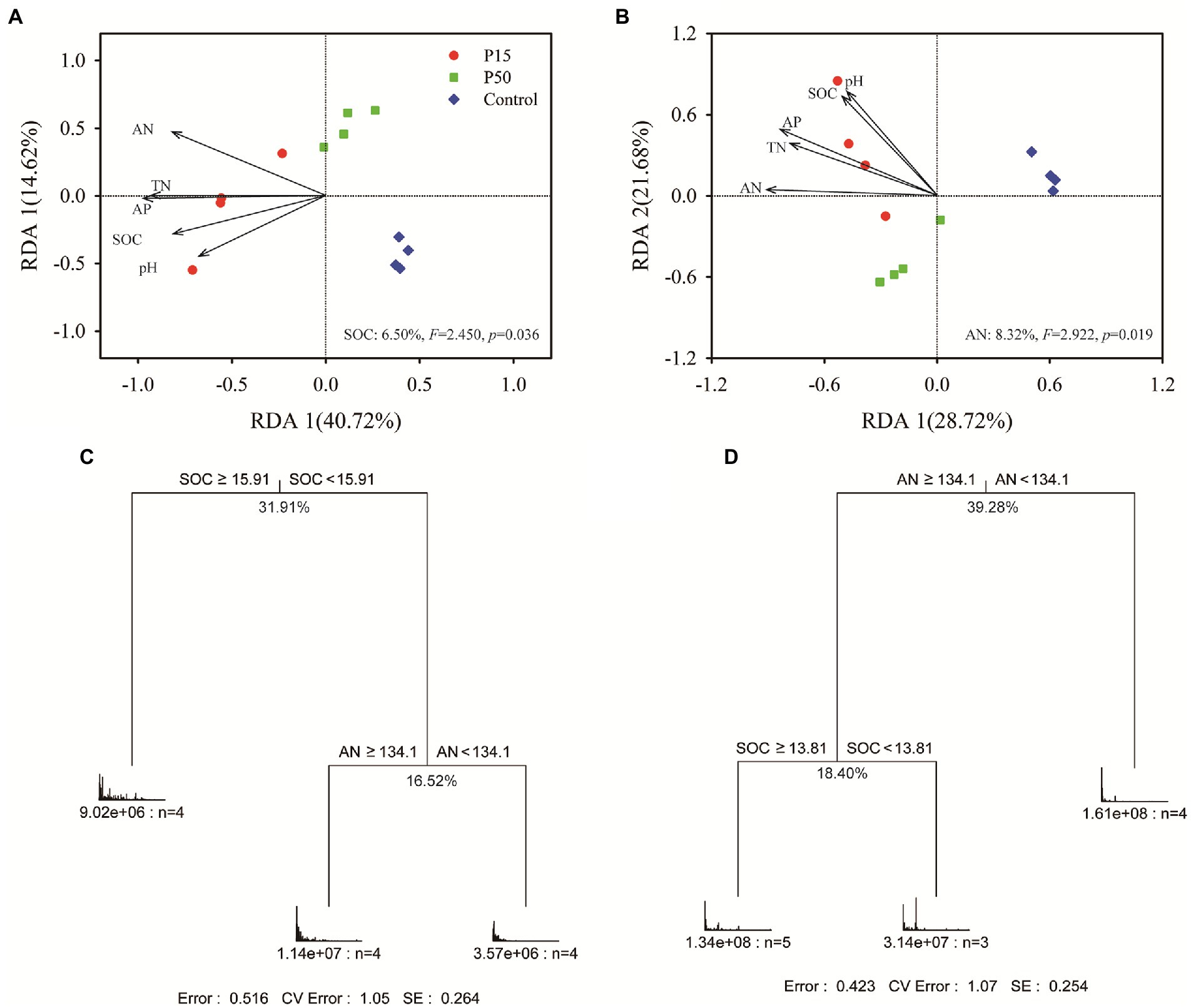
Figure 3. Redundancy analysis (RDA) between the soil bacterial (A), fungal (B) community and fertility indexes at the OTU level in the Cinnamomum camphora coppice planting. P15, point under the tree canopy; P50, point between trees; Control, point in the abandoned land. Multivariate regression trees (MRT) of soil bacteria (C), fungi (D) community composition and fertility indexes. Numbers under the crosses of each split indicate percentages of variance explained by the split.
Correspondingly, the CAP analysis corroborated obvious differences in soil fungal community (Supplementary Figure S4B, 39.6% of total variance was explained, p = 0.001), and the ANOSIM tests indicated there were significantly different (p < 0.05) between treatments (Supplementary Table S1). Mantel test (Supplementary Figure S5) further verified that soil fungal community had significant correlation with soil fertility indexes (p < 0.05), especially the soil AN and AP (p < 0.01). Similarly, RDA analysis suggested that the soil AN (F = 2.922, p = 0.019) was the primary factor shifting the fungal community composition. And MRT analysis showed that the soil AN produced the largest deterministic effects on soil fungal community variation (39.28%), with a threshold of 134.1 g kg−1 (Figure 3D).
In addition, CAP plot demonstrated that the soil enzyme profiling was also clearly segregated (p = 0.007) in three different treatments, which explains 50.4% of the variance in the data (Supplementary Figure 4C). ANOSIM tests pointed that the soil enzyme profiling in P15 and P50 were altered significantly (p < 0.05) compared with Control, but there was insignificant difference between P15 and P50 (r = 0.0625, p = 0.318). Unlike the soil microbial community, Mantel test (Supplementary Figure S5) revealed that there were no significant relationships between the soil enzyme profiling and fertility indexes.
Correlations between soil enzyme activities and microbial community
Due to the fact that there had no significant correlation between the soil enzyme profiling and fertility indexes, we further performed our analysis with Procrustes tests to find the linkages between the soil enzyme profiling and microbial community. Procrustes tests corroborated that the soil enzyme profiling had significant correlation with the soil bacterial (Figure 4A, Correlation r = 0.9475, M2 = 0.1022, p = 0.001) and fungal community (Figure 4B, Correlation r = 0.9584, M2 = 0.0814, p = 0.001).
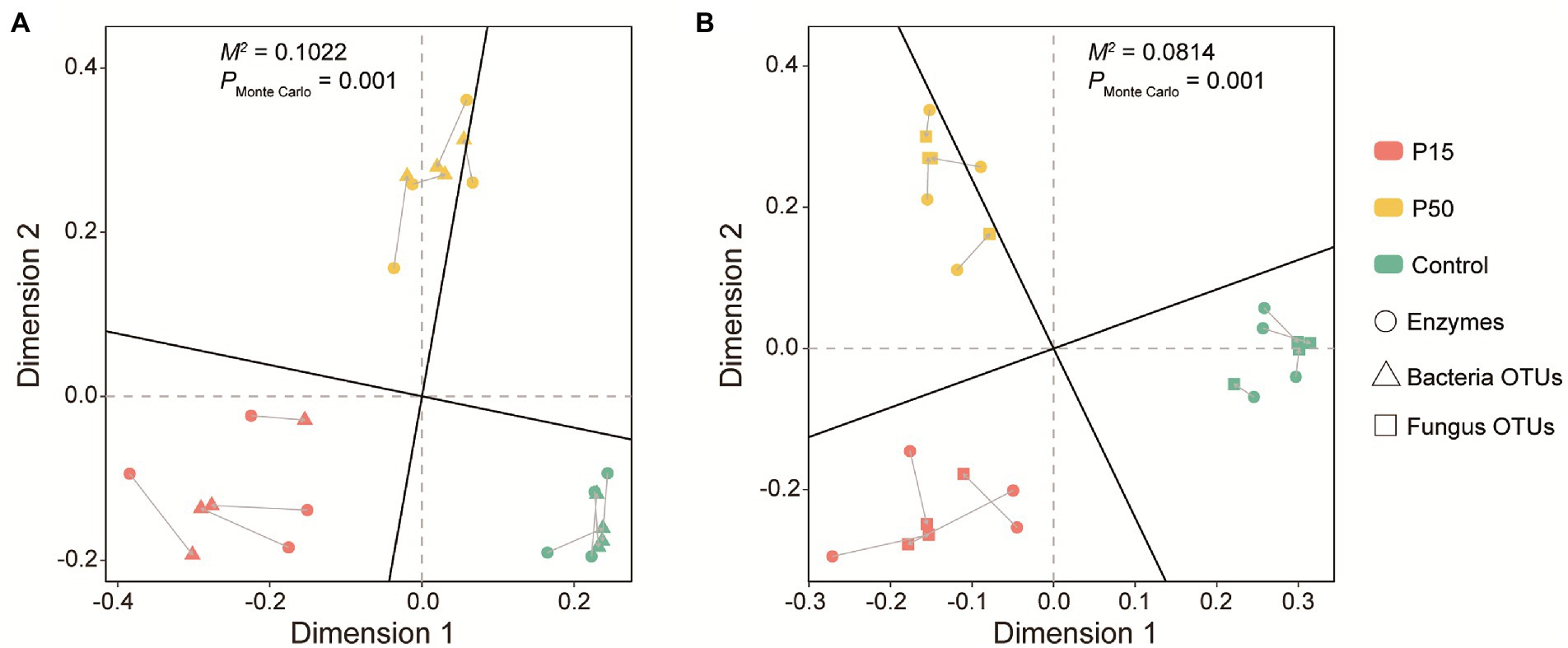
Figure 4. Procrustes analysis between soil bacterial (A), fungal (B) community composition and enzyme profiling in the Cinnamomum camphora coppice planting. P15, point under the tree canopy; P50, point between trees; Control, point in the abandoned land. The lengths of connecting lines represent the dissimilarities of soil bacterial, fungal community composition and enzyme profiling.
Furthermore, relationships between soil enzyme activity and microorganism properties were testified by Pearson correlation analysis (Figure 5). In general, the soil UE, PPO, and POD activities had significantly positive correlation with soil bacterial biomass and alpha diversity, whereas the soil INV and CAT activities had no significant correlation with the above indexes. Notably, soil ACP activity only had significant negative relevance with the bacterial biomass (r = −0.635, p < 0.05). The soil UE, PPO, and POD activities were significantly positive connected with Proteobacteria, Bacteroidetes and Gemmatimonadetes, but negative connected with Chloroflexi. While the soil ACP activity showed the completely opposite tendency. Meanwhile, the soil INV activity had significantly positive relationship with Bacteroidetes (r = 0.665, p < 0.05) and significantly negative relationship with Chloroflexi (r = −0.665, p < 0.05). And the soil CAT activity was significant and positive connected with Actinobacteria (r = 0.579, p < 0.05) whereas significant and negative connected with Acidobacteria (r = −0.773, p < 0.01).
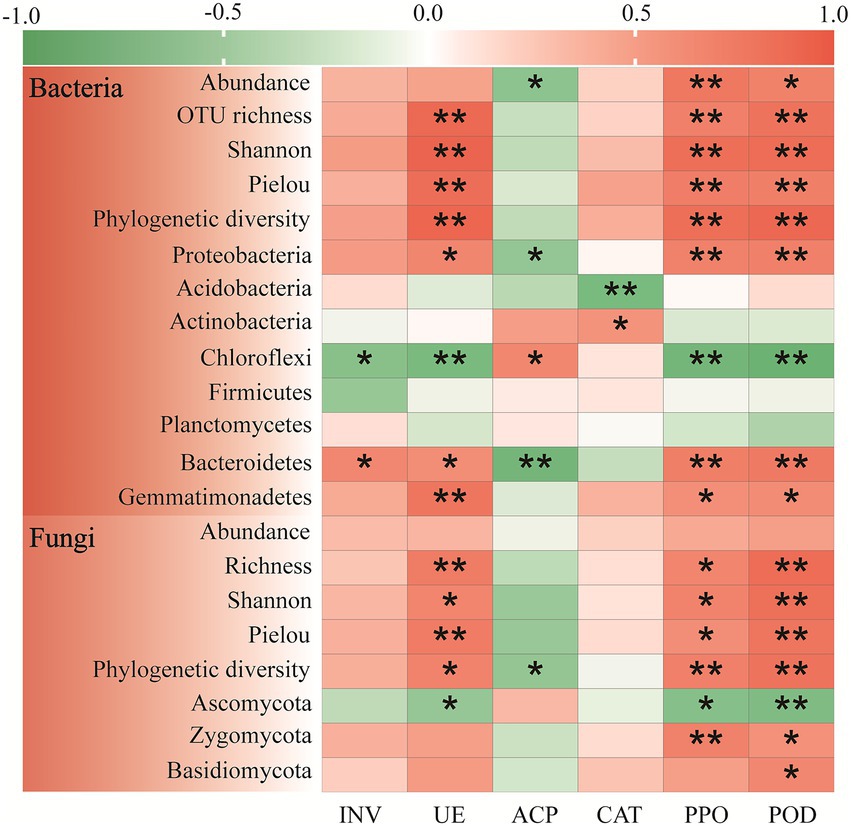
Figure 5. Pearson correlation heat maps between soil bacterial, fungal community composition (abundance, alpha-diversity and dominant phyla) and enzyme profiling in the Cinnamomum camphora coppice planting. Red represents a positive correlation, and green represents a negative correlation. (*p < 0.05; **p < 0.01).
Correspondingly, the soil UE, PPO, and POD activities were significant and positive related to the four fungal alpha-diversity indexes, while the soil ACP activity was only significantly negative related to fungal phylogenetic diversity (r = −0.596, p < 0.05). The soil UE, PPO, and POD activities had significantly negative correlation with Ascomycota. In the meantime, the soil POD activity had significantly positive correlation with Zygomycota (r = 0.593, p < 0.05) and Basidiomycota (r = 0.661, p < 0.05), whereas the soil PPO activity only significantly positively correlated with Zygomycota (r = 0.713, p < 0.01). Remarkably, the soil INV and CAT activities had no significant correlation with all the soil fungal properties.
Discussion
Effects of Cinnamomum camphora coppice planting on soil fertility
In this study, the C. camphora tree was cultivated as coppice and its aerial part were annually cut down to extract the essential oil. This will inevitably take away nutrients from the soil. However, we found that relative to the abandoned land, the point under the tree canopy (P15) substantially increased the soil pH and N, P contents. Firstly, fertilization can compensate for the high quantities of soil nutrients consumed by the C. camphora trees (Selvaraj et al., 2017). Secondly, though C. camphora coppices were felled during July to September every year, they sprouted and form the canopy rapidly, which can effectively reduce the erosion caused by the concentrated rainfall season (March to July) in south China (Hinojosa et al., 2021; Tang et al., 2021). Moreover, the continuously growing roots of the C. camphora tree can promote the activation and transportation of soil mineral elements (Männistö et al., 2016). Earlier study had suggested that the soil nutrients content under the tree canopy was higher than the point outside the tree canopy in the Artemisia ordosica Krasch planting (Qi et al., 2010). On account of the “fertility islands” effects that the soil fertility gradually decreased from the inside to the outside under the tree canopy (San Emeterio et al., 2021). Nevertheless, our research revealed that even the point between trees (P50), which was weakly affected by fertilization and tree growth, can somehow enhance the soil nutrients availability (Han et al., 2021). This further corroborated that the C. camphora tree planting can improve the soil fertility.
Effects of Cinnamomum camphora coppice planting on soil microbial structure
Soil microorganisms are important regulatory factors in soil organic matter transformation, and they may influence by changes in soil fertility after tree planting (Thébault et al., 2014). Huang et al. (2022) recorded that tree planting enhanced the SOC and TN contents, and they had notable correlation with soil microbial biomass and diversity. A previous study on Cryptomeria japonica also verified that the increase of the soil C and N nutrients stimulated the improvement of soil microbial biomass, OTU richness, and Shannon index (Sawada et al., 2021). Likewise, our results proved that the C. camphora tree coppice planting enhanced the bacterial, fungal biomass and diversity (Figure 1), and their change was induced by increased soil fertility (Figure 2). One possible reason was that the C. camphora tree coppice planting can produce a large amount of soil N, P nutrients and C substratum, which in turn provide more diverse niches available for microorganisms (Pereira et al., 2019; Asigbaase et al., 2021).
Alterations in soil microorganisms after tree planting were closely correlated with their ecological strategies (Fierer et al., 2007). The C. camphora coppice planting significantly influenced the soil microbial dominant phyla (Supplementary Figure S2). Proteobacteria was the typical copiotrophs (r-strategist), which generally exists in high C and N soils (Trivedi et al., 2013). An earlier study indicated that mixed Eucalyptus grandis plantings significantly improved soil fertility, and relative abundance of Proteobacteria increased from 14.29 to 27.90% (Zhang et al., 2022). In the present study, relative abundance of Proteobacteria increased significantly by 7.47 and 11.60% in P15 and P50, respectively. This also reflected that the C. camphora coppice planting can increase soil fertility. Similarly, Zygomycota and Basidiomycota were also copiotrophs, which can form the mycorrhiza with tree roots and use readily available nutrients in a short period (Wang et al., 2017; Zhu et al., 2019). Their relative abundance was also significantly improved in the C. camphora coppice planting (Supplementary Figure S2). And the correlation analysis further identified that Proteobacteria, Zygomycota, and Basidiomycota were significantly positively correlated with soil fertility indexes (Figure 2).
On the contrary, Chloroflexi and Ascomycota were the typical oligotrophs (K-strategist), early studies reported that with increase of soil fertility, relative abundance of Chloroflexi and Ascomycota were decreased (Zhu et al., 2019; Idbella et al., 2022). This further confirmed by our research that relative abundance of Chloroflexi and Ascomycota were significantly negative correlated with soil fertility indexes in general (Figure 2). Additionally, Ktedonobacter was the member of Chloroflexi, and Talaromyces and Hyaloscyphaceae were the members of Ascomycota. STAMP analysis indicated that their relative abundance was declining when soil fertility was improved. And in the low fertile soil, such as P15 vs Control, Ktedonobacter, Talaromyces and Hyaloscyphaceae were the significantly different species between treatments (Supplementary Figure S3). In conclusion, the C. camphora coppice planting shifted the soil microbial community from oligotrophs to copiotrophs, and relative abundance of copiotrophs microorganisms in P15 were increased from 37.78% (bacteria), 18.09% (fungi) to 53.80% (bacteria), 29.24% (fungi), respectively (Supplementary Table S2).
Various studies have summarized that soil microbial community compositions changed under tree planting. The conversion from natural forest to tree planting had significantly changed the community of soil bacteria (Jiang et al., 2021; Li et al., 2022), and SOC was the primary factor for its change (Zhang et al., 2016). This was consistent with our results, indicating that SOC can provide resources for soil bacterial community and modify its ecological strategy, thus affecting soil bacterial community composition (Qu et al., 2020; Yang et al., 2021a). Additionally, soil fungal community significantly shifted in the C. camphora coppice planting (Supplementary Figure S4), and RDA, MRT analysis exhibited that AN produced the largest deterministic effects to them (Figure 3). This had also confirmed by Wang et al. (2021) who recorded that soil fungal community composition had significant positive relation to soil AN content in Cunninghamia lanceolate (Lamb.) Hook plantings.
Effects of Cinnamomum camphora coppice planting on soil enzyme activities
Soil enzymes, as the participants in soil organic matter transformation, can also make vital functions in carbon (C), nitrogen (N), and phosphorus (P) cycling of soil ecosystems (Feyissa et al., 2022). our results declared the C. camphora coppice planting significantly enhanced the soil INV, UE, PPO and POD activities (Table 2). And correlation analysis suggested that soil enzyme activities except soil ACP showed positive relationship with soil pH and fertility indexes in general (Figure 2). Many previous studies pointed out that soil INV and UE activities were involved in soil C and N transformation, respectively (Boughattas et al., 2019; Zhu et al., 2019). Wang et al. (2018) acknowledge that Camellia sinensis L. planting increased soil C, N concentration, and soil INV, UE activities were also improved with the stand age. Moreover, soil PPO and POD were involved in the oxidation and degradation of soil lignin and SOM (Sinsabaugh, 2010), and their increase illustrated that C. camphora coppice planting can improve soil nutrients availability. Nevertheless, in the present study, soil ACP activity under the tree canopy (P15) and between the trees (P50) was significantly decreased. Though the soil ACP is involved in the decomposition of soil organic phosphorus to promote its metabolic turnover and reuse efficiency, the increasing soil available phosphorus content will reduce the ACP activity (Tran et al., 2010). In other words, high nutrient availability may inhibit soil ACP activity (Janes-Bassett et al., 2022), and we also found that there were negative correlations between soil ACP activity and soil fertility (Figure 2). And STAMP analysis further revealed that soil ACP activity was higher in the low fertility soil (Supplementary Figure S3).
Our work corroborated that soil enzyme profiling was weakly (p > 0.05) affected by soil fertility (Supplementary Figure S5), while they were highly connected with soil bacterial and fungal community (Figure 4). Further correlation analysis identified that soil UE, PPO, and POD activity was significantly positively correlated with soil microbial biomass and diversity (Figure 5). Earlier studies declared that soil microbial biomass, diversity (e.g., OTUs, Shannon) and enzyme activities (e.g., UE and POD) were all significantly increased after afforestation. (Thoms et al., 2010; Huang et al., 2022). For one thing, soil microorganisms have their unique ecological functions, and the increasing soil microbial diversity may enhance the quantities of specific-species involved in soil nutrient cycling (Weand et al., 2010). For another, the enhancement of soil microbial biomass will promote the secretion of soil extracellular enzymes (van der Heijden et al., 2008). As for the dominant microbial phylum, we found that soil UE, PPO, and POD activities had significantly positive correlation with copiotrophs (e.g., Proteobacteria, Bacteroidetes and Gemmatimonadetes), while they had significantly negative correlation with oligotrophs (e.g., Chloroflexi and Ascomycota). As soil UE, PPO and POD were conducive to recalcitrant carbon decomposition, their increase may signify the enhancement of soil nutrient supplying capacity (Yang et al., 2021b). This also confirmed that the C. camphora coppice planting can improve soil fertility. Conversely, soil ACP activity was significantly positive connected with Chloroflexi (oligotrophs), whereas significantly negatively related to Proteobacteria (copiotrophs). These findings suggested that Chloroflexi have more survival advantages in low soil fertility (abandoned land), which may promote the secretion of soil ACP (Kaiser et al., 2010; Gong et al., 2019).
Conclusion
The present study indicated that the C. camphora coppice planting significantly increased the soil fertility, and therefore cause a corresponding alteration in soil microbial community composition. Compared with the abandoned land, the microbial biomass and diversity were significantly higher under the tree canopy (P15) and between the trees (P50). The C. camphora coppice planting shifted the soil microbial community from oligotrophs (K-strategist) to copiotrophs (r-strategist). SOC and AN were the primary factors that drive changes in soil bacterial, fungal community composition, respectively. In addition, soil UE, PPO, and POD activities were significantly increased, while soil ACP activity was significantly decreased. And soil bacterial and fungal community significantly influenced the soil enzyme profiling. Soil UE, PPO, and POD activities were significantly positive correlated with copiotrophic microorganisms, while significantly negative correlated with oligotrophic microorganisms, and soil ACP activity show the opposite trend. Overall, the C. camphora coppice planting led to significant changes in soil fertility, microbial community structure, and enzyme activities, at which the point under the tree canopy (P15) had more significant effects than the point between trees (P50). This work can provide a theoretical basis for soil sustainable utilization and productive management of C. camphora coppice plantation in subtropical China.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
LS, JieZ, and YL contributed to conception and design of the study. LS, JieZ, JiaZ, XL, YG, HS, HL, and YL completed the field sampling. LS, JieZ, CX, ZX, TZ, and YL performed the statistical analysis. LS, JieZ, and YL wrote the manuscript. LS, JieZ, JiaZ, XL, CX, ZX, TZ, and YL contributed to the revision of manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by Jiangxi Provincial Science and Technology Program (20204BCJL23046, 20181ACF60002), the National Natural Science Foundation of China (32060333, 52269013), and the National Key Research and Development Program of China (no. 2021YFD1901201-05).
Acknowledgments
We thank the reviewers and editors for their helpful comments regarding the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1104077/full#supplementary-material
References
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D. (2013). Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7, 1262–1273. doi: 10.1038/ismej.2013.28
Ahirwal, J., and Maiti, S. K. (2016). Assessment of soil properties of different land uses generated due to surface coal mining activities in tropical Sal (Shorea robusta) forest, India. Catena 140, 155–163. doi: 10.1016/j.catena.2016.01.028
Akhtar, K., Wang, W. Y., Ren, G. X., Khan, A., Feng, Y. Z., and Yang, G. H. (2018). Changes in soil enzymes, soil properties, and maize crop productivity under wheat straw mulching in Guanzhong, China. Soil Tillage Res. 182, 94–102. doi: 10.1016/j.still.2018.05.007
Asigbaase, M., Dawoe, E., Sjogersten, S., and Lomax, B. H. (2021). Decomposition and nutrient mineralisation of leaf litter in smallholder cocoa agroforests: a comparison of organic and conventional farms in Ghana. J. Soils Sediments 21, 1010–1023. doi: 10.1007/s11368-020-02844-4
Biddle, J. F., Fitz-Gibbon, S., Schuster, S. C., Brenchley, J. E., and House, C. H. (2008). Metagenomic signatures of the Peru margin subseafloor biosphere show a genetically distinct environment. PNAS Nexus 105, 10583–10588. doi: 10.1073/pnas.0709942105
Boughattas, I., Hattab, S., Alphonse, V., Livet, A., Giusti-Miller, S., Boussetta, H., et al. (2019). Use of earthworms Eisenia andrei on the bioremediation of contaminated area in north of Tunisia and microbial soil enzymes as bioindicator of change on heavy metals speciation. J. Soils Sediments 19, 296–309. doi: 10.1007/s11368-018-2038-8
Bueno de Mesquita, C. P., Knelman, J. E., King, A. J., Farrer, E. C., Porazinska, D. L., Schmidt, S. K., et al. (2017). Plant colonization of moss-dominated soils in the alpine: microbial and biogeochemical implications. Soil Biol. Biochem. 111, 135–142. doi: 10.1016/j.soilbio.2017.04.008
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carrara, J. E., Walter, C. A., Hawkins, J. S., Peterjohn, W. T., Averill, C., and Brzostek, E. R. (2018). Interactions among plants, bacteria, and fungi reduce extracellular enzyme activities under long-term N fertilization. Glob. Chang. Biol. 24, 2721–2734. doi: 10.1111/gcb.14081
Chen, Y. X., Wei, T. X., Sha, G. L., Zhu, Q. K., Liu, Z., Ren, K., et al. (2022). Soil enzyme activities of typical plant communities after vegetation restoration on the loess plateau, China. Appl. Soil Ecol. 170:104292. doi: 10.1016/j.apsoil.2021.104292
De’ath, G. (2003). Multivariate regression trees: a new technique for modeling species–environment relationships. Ecology 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edgar, R. C. (2016). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv. :081257. doi: 10.1101/081257
Feng, J. X., Cui, X. W., Zhou, J., Wang, L. M., Zhu, X. S., and Lin, G. H. (2019). Effects of exotic and native mangrove forests plantation on soil organic carbon, nitrogen, and phosphorus contents and pools in Leizhou, China. Catena 180, 1–7. doi: 10.1016/j.catena.2019.04.018
Feyissa, A., Gurmesa, G. A., Yang, F., Long, C. Y., Zhang, Q., and Cheng, X. L. (2022). Soil enzyme activity and stoichiometry in secondary grasslands along a climatic gradient of subtropical China. Sci. Total Environ. 825:154019. doi: 10.1016/j.scitotenv.2022.154019
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Gong, X. W., Liu, C. J., Li, J., Luo, Y., Yang, Q. H., Zhang, W. L., et al. (2019). Responses of rhizosphere soil properties, enzyme activities and microbial diversity to intercropping patterns on the loess plateau of China. Soil Tillage Res. 195:104355. doi: 10.1016/j.still.2019.104355
Han, Z. Q., Wang, J. Y., Xu, P. S., Sun, Z. R., Ji, C., Li, S. Q., et al. (2021). Greater nitrous and nitric oxide emissions from the soil between rows than under the canopy in subtropical tea plantations. Geoderma 398:115105. doi: 10.1016/j.geoderma.2021.115105
Hinojosa, M. B., Albert-Belda, E., Gómez-Muñoz, B., and Moreno, J. M. (2021). High fire frequency reduces soil fertility underneath woody plant canopies of Mediterranean ecosystems. Sci. Total Environ. 752:141877. doi: 10.1016/j.scitotenv.2020.141877
Huang, H. Y., Tian, D., Zhou, L. H., Su, H. J., Ma, S. H., Feng, Y. H., et al. (2022). Effects of afforestation on soil microbial-diversity and enzyme activity: a meta-analysis. Geoderma 423:115961. doi: 10.1016/j.geoderma.2022.115961
Idbella, M., De Filippis, F., Zotti, M., Sequino, G., Abd-ElGawad, A. M., Fechtali, T., et al. (2022). Specific microbiome signatures under the canopy of Mediterranean shrubs. Appl. Soil Ecol. 173:104407. doi: 10.1016/j.apsoil.2022.104407
Janes-Bassett, V., Blackwell, M. S. A., Blair, G., Davies, J., Haygarth, P. M., Mezeli, M. M., et al. (2022). A meta-analysis of phosphatase activity in agricultural settings in response to phosphorus deficiency. Soil Biol. Biochem. 165:108537. doi: 10.1016/j.soilbio.2021.108537
Jiang, S., Xing, Y. J., Liu, G. C., Hu, C. Y., Wang, X. C., Yan, G. Y., et al. (2021). Changes in soil bacterial and fungal community composition and functional groups during the succession of boreal forests. Soil Biol. Biochem. 161:108393. doi: 10.1016/j.soilbio.2021.108393
Jin, L. L., Xiang, X. J., Zhang, J. Y., Zhang, J., Liu, M., Qin, W. J., et al. (2021). Dramatic shifts in fungal communities following application of hairy vetch (Vicia villosa Roth L.) in upland of Ultisol. Eur. J. Soil Biol. 106:103349. doi: 10.1016/j.ejsobi.2021.103349
Kaiser, C., Koranda, M., Kitzler, B., Fuchslueger, L., Schnecker, J., Schweiger, P., et al. (2010). Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 187, 843–858. doi: 10.1111/j.1469-8137.2010.03321.x
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Kumar, R., Bhardwaj, A. K., Rao, B. K., Vishwakarma, A. K., Kakade, V., Dinesh, D., et al. (2021). Soil loss hinders the restoration potential of tree plantations on highly eroded ravine slopes. J. Soils Sediments 21, 1232–1242. doi: 10.1007/s11368-020-02833-7
Li, J., Li, S. F., Huang, X. B., Tang, R., Zhang, R., Li, C., et al. (2022). Plant diversity and soil properties regulate the microbial community of monsoon evergreen broad-leaved forest under different intensities of woodland use. Sci. Total Environ. 821:153565. doi: 10.1016/j.scitotenv.2022.153565
Liu, Z. Z., Deng, B. Q., Li, S. L., and Zou, Z. R. (2018b). Optimization of solvent-free microwave assisted extraction of essential oil from Cinnamomum camphora leaves. Ind. Crop. Prod. 124, 353–362. doi: 10.1016/j.indcrop.2018.08.016
Liu, Z. Z., Li, H. L., Zhu, Z., Huang, D., Qi, Y. L., Ma, C. H., et al. (2022). Cinnamomum camphora fruit peel as a source of essential oil extracted using the solvent-free microwave-assisted method compared with conventional hydrodistillation. Lebensm. Wiss. Technol. 153:112549. doi: 10.1016/j.lwt.2021.112549
Liu, J., Liu, M., Wu, M., Jiang, C. Y., Chen, X. F., Cai, Z. J., et al. (2018a). Soil pH rather than nutrients drive changes in microbial community following long-term fertilization in acidic Ultisols of southern China. J. Soils Sediments 18, 1853–1864. doi: 10.1007/s11368-018-1934-2
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Männistö, M., Ganzert, L., Tiirola, M., Häggblom, M. M., and Stark, S. (2016). Do shifts in life strategies explain microbial community responses to increasing nitrogen in tundra soil? Soil Biol. Biochem. 96, 216–228. doi: 10.1016/j.soilbio.2016.02.012
Moghimian, N., Hosseini, S. M., Kooch, Y., and Darki, B. Z. (2017). Impacts of changes in land use/cover on soil microbial and enzyme activities. Catena 157, 407–414. doi: 10.1016/j.catena.2017.06.003
Monkai, J., Goldberg, S. D., Hyde, K. D., Harrison, R. D., Mortimer, P. E., and Xu, J. C. (2018). Natural forests maintain a greater soil microbial diversity than that in rubber plantations in Southwest China. Agric. Ecosyst. Environ. 265, 190–197. doi: 10.1016/j.agee.2018.06.009
Nakayama, M., Imamura, S., Taniguchi, T., and Tateno, R. (2019). Does conversion from natural forest to plantation affect fungal and bacterial biodiversity, community structure, and co-occurrence networks in the organic horizon and mineral soil? For. Ecol. Manag. 446, 238–250. doi: 10.1016/j.foreco.2019.05.042
Parks, D. H., and Beiko, R. G. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26, 715–721. doi: 10.1093/bioinformatics/btq041
Pereira, A. P. A., Durrer, A., Gumiere, T., Gonçalves, J. L. M., Robin, A., Bouillet, J. P., et al. (2019). Mixed eucalyptus plantations induce changes in microbial communities and increase biological functions in the soil and litter layers. For. Ecol. Manag. 433, 332–342. doi: 10.1016/j.foreco.2018.11.018
Qi, Y. C., Dong, Y. S., Jin, Z., Peng, Q., Xiao, S. S., and He, Y. T. (2010). Spatial heterogeneity of soil nutrients and respiration in the Desertified grasslands of Inner Mongolia, China. Pedosphere 20, 655–665. doi: 10.1016/S1002-0160(10)60055-0
Qu, Z. L., Liu, B., Ma, Y., Xu, J., and Sun, H. (2020). The response of the soil bacterial community and function to forest succession caused by forest disease. Funct. Ecol. 34, 2548–2559. doi: 10.1111/1365-2435.13665
Rosinger, C., Rousk, J., and Sandén, H. (2019). Can enzymatic stoichiometry be used to determine growth-limiting nutrients for microorganisms? - a critical assessment in two subtropical soils. Soil Biol. Biochem. 128, 115–126. doi: 10.1016/j.soilbio.2018.10.011
Saiya-Cork, K. R., Sinsabaugh, R. L., and Zak, D. R. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34, 1309–1315. doi: 10.1016/S0038-0717(02)00074-3
San Emeterio, L., Durán, M., Múgica, L., Jiménez, J. J., and Canals, R. M. (2021). Relating the spatial distribution of a tall-grass to fertility islands in a temperate mountain grassland. Soil Biol. Biochem. 163:108455. doi: 10.1016/j.soilbio.2021.108455
Sawada, K., Inagaki, Y., Sugihara, S., Funakawa, S., Ritz, K., and Toyota, K. (2021). Impacts of conversion from natural forest to cedar plantation on the structure and diversity of root-associated and soil microbial communities. Appl. Soil Ecol. 167:104027. doi: 10.1016/j.apsoil.2021.104027
Selvaraj, S., Duraisamy, V., Huang, Z. J., Guo, F. T., and Ma, X. Q. (2017). Influence of long-term successive rotations and stand age of Chinese fir (Cunninghamia lanceolata) plantations on soil properties. Geoderma 306, 127–134. doi: 10.1016/j.geoderma.2017.07.014
Sinsabaugh, R. L. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42, 391–404. doi: 10.1016/j.soilbio.2009.10.014
Speranza, C. I., Adenle, A., and Boillat, S. (2019). Land degradation neutrality—potentials for its operationalisation at multi-levels in Nigeria. Environ. Sci. Pol. 94, 63–71. doi: 10.1016/j.envsci.2018.12.018
Sun, R. B., Chen, Y., Han, W. X., Dong, W. X., Zhang, Y. M., Hu, C., et al. (2020). Different contribution of species sorting and exogenous species immigration from manure to soil fungal diversity and community assemblage under long-term fertilization. Soil Biol. Biochem. 151:108049. doi: 10.1016/j.soilbio.2020.108049
Sun, R. B., Zhang, W. J., Liu, Y. B., Yun, W. J., Luo, B. B., Chai, R. S., et al. (2022). Changes in phosphorus mobilization and community assembly of bacterial and fungal communities in rice rhizosphere under phosphate deficiency. Front. Microbiol. 13:953340. doi: 10.3389/fmicb.2022.953340
Tabatabai, M. A. (1994). “Soil enzymes” in Methods of Soil Analysis: Part 2 Microbiological and Biochemical Properties. ed. A. L. Page, vol. 5 (Madison, WI: The American Society of Agronomy), 775–833.
Tang, C. J., Liu, Y., Li, Z. W., Guo, L. P., Xu, A. Z., and Zhao, J. D. (2021). Effectiveness of vegetation cover pattern on regulating soil erosion and runoff generation in red soil environment, southern China. Ecol. Indic. 129:107956. doi: 10.1016/j.ecolind.2021.107956
Teng, Y. M., Zhan, J. Y., Liu, W., Zhang, F., Wang, C., and Dong, S. K. (2020). Larch or Mongolian pine? Effects of tree species on soil properties and microbial biomass with the consideration of afforestation time. Ecol. Eng. 158:106074. doi: 10.1016/j.ecoleng.2020.106074
Thébault, A., Clément, J. C., Ibanez, S., Roy, J., Geremia, R. A., Pérez, C. A., et al. (2014). Nitrogen limitation and microbial diversity at the treeline. Oikos 123, 729–740. doi: 10.1111/j.1600-0706.2013.00860.x
Thoms, C., Gattinger, A., Jacob, M., Thomas, F. M., and Gleixner, G. (2010). Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biol. Biochem. 42, 1558–1565. doi: 10.1016/j.soilbio.2010.05.030
Tran, H. T., Hurley, B. A., and Plaxton, W. C. (2010). Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci. 179, 14–27. doi: 10.1016/j.plantsci.2010.04.005
Trivedi, P., Anderson, I. C., and Singh, B. K. (2013). Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol. 21, 641–651. doi: 10.1016/j.tim.2013.09.005
Ushio, M., Wagai, R., Balser, T. C., and Kitayama, K. (2008). Variations in the soil microbial community composition of a tropical montane forest ecosystem: does tree species matter? Soil Biol. Biochem. 40, 2699–2702. doi: 10.1016/j.soilbio.2008.06.023
Van Der Heijden, M. G., Bardgett, R. D., and Van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Wan, P., Peng, H., Ji, X. L., Chen, X. L., and Zhou, H. M. (2021). Effect of stand age on soil microbial communities of a plantation Ormosia hosiei forest in southern China. Ecol. Inform. 62:101282. doi: 10.1016/j.ecoinf.2021.101282
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, S. Q., Li, T. X., and Zheng, Z. C. (2018). Effects of tea plantation age on soil aggregate-associated C-and N-cycling enzyme activities in the hilly areas of Western Sichuan, China. Catena 171, 145–153. doi: 10.1016/j.catena.2018.07.010
Wang, J. C., Song, Y., Ma, T. F., Raza, W., Li, J., Howland, J. G., et al. (2017). Impacts of inorganic and organic fertilization treatments on bacterial and fungal communities in a paddy soil. Appl. Soil Ecol. 112, 42–50. doi: 10.1016/j.apsoil.2017.01.005
Wang, C. Q., Xue, L., Dong, Y. H., and Jiao, R. Z. (2021). Effects of stand density on soil microbial community composition and enzyme activities in subtropical Cunninghamia lanceolate (lamb.) hook plantations. For. Ecol. Manag. 479:118559. doi: 10.1016/j.foreco.2020.118559
Wang, J. C., Zou, Y. K., Gioia, D. D., Singh, B. K., and Li, Q. F. (2020). Conversion to agroforestry and monoculture plantation is detrimental to the soil carbon and nitrogen cycles and microbial communities of a rainforest. Soil Biol. Biochem. 147:107849. doi: 10.1016/j.soilbio.2020.107849
Weand, M. P., Arthur, M. A., Lovett, G. M., McCulley, R. L., and Weathers, K. C. (2010). Effects of tree species and N additions on forest floor microbial communities and extracellular enzyme activities. Soil Biol. Biochem. 42, 2161–2173. doi: 10.1016/j.soilbio.2010.08.012
Wu, Z. Y., Haack, S. E., Lin, W. X., Li, B. L., Wu, L. K., Fang, C. X., et al. (2015). Soil microbial community structure and metabolic activity of Pinus elliottii plantations across different stand ages in a subtropical area. PLoS One 10:e0135354. doi: 10.1371/journal.pone.0135354
Xiang, X. J., Adams, J. M., Qiu, C. F., Qin, W. J., Chen, J. R., Jin, L. L., et al. (2021). Nutrient improvement and soil acidification inducing contrary effects on bacterial community structure following application of hairy vetch (Vicia villosa Roth L.) in Ultisol. Agric. Ecosyst. Environ. 312:107348. doi: 10.1016/j.agee.2021.107348
Xu, M. P., Jian, J. N., Wang, J. Y., Zhang, Z. J., Yang, G. H., Han, X. H., et al. (2021b). Response of root nutrient resorption strategies to rhizosphere soil microbial nutrient utilization along Robinia pseudoacacia plantation chronosequence. For. Ecol. Manag. 489:119053. doi: 10.1016/j.foreco.2021.119053
Xu, Y. X., Ren, S. Q., Liang, Y. F., Du, A. P., Li, C., Wang, Z. C., et al. (2021a). Soil nutrient supply and tree species drive changes in soil microbial communities during the transformation of a multi-generation eucalyptus plantation. Appl. Soil Ecol. 166:103991. doi: 10.1016/j.apsoil.2021.103991
Yang, Y., Li, T., Wang, Y. Q., Cheng, H., Chang, S. X., Liang, C., et al. (2021b). Negative effects of multiple global change factors on soil microbial diversity. Soil Biol. Biochem. 156:108229. doi: 10.1016/j.soilbio.2021.108229
Yang, C. B., Zhang, X. P., Ni, H. J., Gai, X., Huang, Z. C., Du, X. H., et al. (2021a). Soil carbon and associated bacterial community shifts driven by fine root traits along a chronosequence of Moso bamboo (Phyllostachys edulis) plantations in subtropical China. Sci. Total Environ. 752:142333. doi: 10.1016/j.scitotenv.2020.142333
Zarafshar, M., Bazot, S., Matinizadeh, M., Bordbar, S. K., Rousta, M. J., Kooch, Y., et al. (2020). Do tree plantations or cultivated fields have the same ability to maintain soil quality as natural forests? Appl. Soil Ecol. 151:103536. doi: 10.1016/j.apsoil.2020.103536
Zhang, D. J., Li, J. J., Huang, Y. M., Gao, S., and Zhang, J. (2022). Root-soil facilitation in mixed Eucalyptus grandis plantations including nitrogen-fixing species. For. Ecol. Manag. 516:120215. doi: 10.1016/j.foreco.2022.120215
Zhang, C., Liu, G. B., Xue, S., and Wang, G. L. (2016). Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the loess plateau. Sci. Total Environ. 97, 40–49. doi: 10.1016/j.soilbio.2016.02.013
Zhang, W., Xu, Y., Gao, D., Wang, X., Liu, W., Deng, J., et al. (2019). Ecoenzymatic stoichiometry and nutrient dynamics along a revegetation chronosequence in the soils of abandoned land and Robinia pseudoacacia plantation on the loess plateau, China. Soil Biol. Biochem. 134, 1–14. doi: 10.1016/j.soilbio.2019.03.017
Zhao, J. (2021). Dynamic changes of the contents of essential oil and nutrients of Cinnamomum camphora var. linaloolifera and their correlation. Scientia Silvae Sinicae 57, 57–67. doi: 10.11707/j.1001-7488.20211206
Zhong, Y., Chen, C., Gong, X., Luan, X., Wu, Z., Li, H., et al. (2022). Transcriptome and metabolome analyses reveal a key role of the anthocyanin biosynthetic pathway cascade in the pigmentation of a Cinnamomum camphora red bark mutant (‘Gantong 1’). Ind. Crop. Prod. 175:114236. doi: 10.1016/j.indcrop.2021.114236
Keywords: Cinnamomum camphora coppice planting, soil fertility, bacterial community structure, fungal community structure, enzyme activity
Citation: Sun L, Zhang J, Zhao J, Lu X, Xiao C, Xiao Z, Zhang T, Gu Y, Sun H, Liu H and Li Y (2023) Effects of Cinnamomum camphora coppice planting on soil fertility, microbial community structure and enzyme activity in subtropical China. Front. Microbiol. 14:1104077. doi: 10.3389/fmicb.2023.1104077
Edited by:
Muhammad Saleem, Alabama State University, United StatesReviewed by:
Kailou Liu, Jiangxi Institute of Red Soil, ChinaJiang Nan, China National Rice Research Institute (CAAS), China
Ruibo Sun, Chinese Academy of Sciences, China
Copyright © 2023 Sun, Zhang, Zhao, Lu, Xiao, Xiao, Zhang, Gu, Sun, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Zhang, ✉ bW5femhhbmdqaWVAMTYzLmNvbQ==; Yanli Li, ✉ eWFubGkyMDIyMTFAMTI2LmNvbQ==
 Luyuan Sun
Luyuan Sun Jie Zhang
Jie Zhang Jiao Zhao1
Jiao Zhao1