- 1College of Plant Health and Medicine, Engineering Research Center for Precision Pest Management for Fruits and Vegetables of Qingdao, Shandong Engineering Research Center for Environment-Friendly Agricultural Pest Management, Shandong Province Key Laboratory of Applied Mycology, Qingdao Agricultural University, Qingdao, China
- 2College of Science and Information, Qingdao Agricultural University, Qingdao, China
Pathogens utilize secretory effectors to manipulate plant defense. Fusarium oxysporum f. sp. lycopersici (Fol) is the causal agent of Fusarium wilt disease in tomatoes. We previously identified 32 secreted effector candidates by LC-MS analysis. In this study, we functionally identified one of the secreted proteins, FolAsp, which belongs to the aspartic proteases (Asp) family. The FolAsp was upregulated with host root specifically induction. Its N-terminal 1–19 amino acids performed the secretion activity in the yeast system, which supported its secretion in Fol. Phenotypically, the growth and conidia production of the FolAsp deletion mutants were not changed; however, the mutants displayed significantly reduced virulence to the host tomato. Further study revealed the FolAsp was localized at the apoplast and inhibited INF1-induced cell death in planta. Meanwhile, FolAsp could inhibit flg22-mediated ROS burst. Furthermore, FolAsp displayed protease activity on host protein, and overexpression of FolAsp in Fol enhanced pathogen virulence. These results considerably extend our understanding of pathogens utilizing secreted protease to inhibit plant defense and promote its virulence, which provides potential applications for tomato improvement against disease as the new drug target.
Introduction
Plants are challenged by a wide variety of microbes and parasites. Thanks to their multilayered physical barriers, preformed defenses, and innate immune system, plants can defeat most potential microbial invaders (Zhang et al., 2017; Cao et al., 2018). However, pathogens can secrete abundant proteins to suppress plants’ innate immune in order to effectively infect the hosts (Gawehns et al., 2015). These proteins are mostly secreted extracellularly via the endoplasmic reticulum (ER)/Golgi pathway, which depends on their N-terminal signal peptides. These protein functions range from altering plant cellular metabolic pathways and signaling cascades, RNA silencing, antimicrobial inhibition, and interfering with recognition machinery (Sharpee and Dean, 2016). The importance of understanding the function of secreted proteins has given rise to the field of pathogen–host interaction.
Proteins secreted from pathogens can be divided into two classes according to their functions: enzymatic functional proteins and non-enzymatic functional proteins (Qian et al., 2022b). Cell wall-degrading enzymes (CWDEs), cutinases, proteases, peptidases, and ribonucleases can be secreted from pathogens to proceed with enzymatic function. For instance, the glycoside hydrolase family 12 (GH12) protein, XEG1, produced by the soybean pathogen Phytophthora sojae exhibits xyloglucanase and beta-glucanase activity (Ma et al., 2015). In addition, another two GH12 proteins, FoEG1 and VdEG1, secreted by Fusarium oxysporum and Verticillium dahliae, respectively, are essential for fungal virulence via its cellulase activity (Gui et al., 2017; Zhang L. et al., 2021). In Botrytis cinerea, Xyn11A, xylanase, is required for virulence in host plants (Noda et al., 2010). Another class is composed of many low-molecular-weight proteins, which are rich in cysteine and lack known enzyme activity (Hacquard et al., 2012; Lyu et al., 2016). Some of these secreted proteins are among a repertoire of pathogenicity effectors.
The aspartic protease was first isolated from Aspergillus niger, named aspergillopepsin I (EC 3.4.23.18), which is a typical pepsin aspartic protease active between pH 2 and 4 and is inhibited by pepstatin (Song et al., 2020). Some of the aspergillopepsin-like family proteins from various fungal species may serve as virulence factors beyond its wide range of commercial applications as industrial aids (Song et al., 2020). For example, the aspergillopepsin-like proteins (cd06097) contribute to virulence in the human pathogen Candida (Monika and Zbigniew, 2017). A type III-independent extracellular aspartic protease Rsa1e is identified as a virulence factor of Ralstonia solanacearum in potatoes (Jeong et al., 2011). Aspergillopepsin from Aspergillus fumigatus is involved in invasive aspergillosis owing to its elastolytic activity.
Fusarium oxysporum f. sp. lycopersici (Fol), the causal agent of Fusarium wilt limited to tomato, invades the roots and subsequently colonizes the xylem vessels, thereby compromising water transport resulting in wilting (Michielse and Rep, 2009). During the colonization of the host xylem vessels, Fol secretes small effectors, including 14 different “Secreted-in-Xylem” (Six) proteins, and some of them play critical roles in determining plant immunity (Houterman et al., 2007; Lievens et al., 2009; Ma et al., 2013; Gawehns et al., 2015). In our previous studies, we have identified 32 secreted effector candidates in Fol by mass spectrometry, which gives the first clues into the establishment of Fol early infection (Li J. et al., 2020). To date, the FolSvp1 is characterized to contribute virulence by translocating a key defense protein PR1 to dampen the host defense (Li et al., 2022). FoRnt2, a secreted ribonuclease T2 protein, contributes to the full virulence of F. oxysporum in tomato plants depending on its RNase activity (Qian et al., 2022a). The FoAYP1 contributes to the virulence of Fol through peptidase activity on host protein (Qian et al., 2022b). However, the function of some other secreted proteins in fungal virulence remains largely unknown.
In this study, we identified one Fol-secreted aspartic protease FolAsp, which belongs to the aspergillopepsin-like protein family. FolAsp was found to contribute to the full virulence of Fol in tomato seedlings and exhibited protease activity to host roots. FolAsp was located at the apoplast in planta, which displayed the inhibition activity of plant ROS burst. These findings provide another example in which the apoplast effector contributed virulence via its protease activity, directly facilitating the successful outcome of a fungal plant pathogen invasion.
Materials and methods
Plants, fungal strains, and cultural conditions
The WT tomato (Solanum lycopersicum cv. Alisa Craig [AC]) was used and potted in a soil mix (vermiculite:humus = 1:2) and placed in a climatized greenhouse at 28°C with 65% relative humidity and 16-h light/8-h dark cycle at 25°C (Cao et al., 2018). The Fol strain Fol4287 (Li J. et al., 2020) was used and cultured on potato dextrose agar (PDA) plates at 25°C. To harvest conidia, mycelial plugs were inoculated in 150 ml of potato dextrose broth (PDB) liquid medium with shaking at 180 rpm at 25°C overnight.
RNA extraction and qRT-PCR assay
For qRT-PCR of FolAsp in response to root induction, the harvested Fol conidia (108 conidia) were suspended in 100 ml of 10% liquid YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) amended with 50 mg aseptic tomato root or not, and cultured at 25°C with shaking at 150 rpm. The aseptic tomato seedlings were grown in half-strength MS media (1/2 MS). Then, Fol cultures at different time points (0, 3, 6, 9, and 12) were harvested by pelleting the developed conidia in the centrifuge at 10,000 rpm for 5 min and finally ground in liquid nitrogen. Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA (2 μg) was used for reverse transcription with the PrimeScript™ RT reagent Kit (TaKaRa). Quantitative expression assays were performed by using the 2x M5 HiPer SYBR Premix EsTag kit (Mei5bio) and a LightCycle®96 (Roche) real-time PCR detection system. The relative quantification of gene expression was performed using the 2–ΔΔct method (Li J. et al., 2018). RNA samples were extracted from three biological replicates. Data were normalized against the Fol histone H4-specific gene FolH4. The primer pairs used for real-time PCR are listed in Supplementary Table 1.
Constructs for gene deletion, complementation, and overexpression in Fol
To generate a FolAsp gene deletion mutant, two 0.7-kb fragments flanking the target gene were PCR amplified with the primer pairs FolAsp-up-F/R and FolAsp-down-F/R. The 5’ and 3’ flanking sequences of the target gene and the hygromycin resistance gene cassette (Hph) were amplified with primer pair Hph-F/R by overlap PCR. The resulting PCR fragment was purified for protoplast transformation of Fol. To construct the complementation vectors, a genomic region encompassing the entire FolAsp coding sequence and its native promoter region was amplified using the primers FolAsp-C-F/R and inserted into the pYF11 (G418 resistance) vector using the ClonExpress II One Step Cloning Kit (Vazyme). The resulting vectors were transformed into the corresponding knockout mutants ΔFolAsp-KO11 by protoplast transformation. To overexpress FolAsp1-GFP, we amplified FolAsp using the primers FolAsp(OE)-F and FolAsp-C-R and inserted them into the XhoI-digested pYF11 vector carrying the RP27 promoter. Then the construct was used for the protoplast transformation of WT Fol. All the primers used in this study are listed in Supplementary Table 1.
Functional verification of FolAsp signal peptide
The predicted N-terminal 19-amino-acid signal peptide sequences of FolAsp were fused in frame to the invertase gene in the pSUC2 vector by two complementary primer sequences (Supplementary Table 1). EcoRI and XhoI restriction enzymes were used to insert the signal peptide sequences into the pSUC2 vector (Yin et al., 2018). The recombinant plasmids were then transformed into invertase secretion-deficient yeast strain YTK12 by lithium acetate-mediated transformation. The function of the signal peptides can be evaluated by using different selective media and color reactions to verify the secretion of invertase as described previously (Yin et al., 2018).
Construction of binary vectors and transient expression
For the subcellular localization of FolAsp in planta, the full coding sequence of FolAsp was PCR amplified using primer pairs FolAsp-pQB-F/R. The forward primer FolAsp1SlPR1SP-PQB-F was used to generate an artificial form of FolAsp containing the tomato SlPR1 signal peptide (1–24 amino acids) instead of its endogenous signal peptide (1–19 amino acids) (Li et al., 2022). These constructs were cloned into a pQB-V3 vector (Chen et al., 2020) and then transferred to the C-terminal GFP-tagged pK7FWG2 (Karimi et al., 2002) using Gateway LR Clonase Enzyme Mix (Invitrogen). Later, these binary constructs were transformed into Agrobacterium LBA4404 for transient expression in N. benthamiana as described previously (Li H. et al., 2018). The oomycete PAMP INF1-expressed INF1 Agrobacterium was used as a positive control to trigger cell death (Qiu et al., 2022). A strain containing the silencing suppressor P19 was co-infiltrated at an OD600 of 0.3 for enhancing the expression of the target genes. For plasmolysis, leaf tissues were cut and incubated for 6 h with 800 mM mannitol and then imaged under the Olympus BX53 fluorescence microscope (Melville, NY, USA).
Protein extraction and western blotting
The plant tissues were frozen and ground in liquid nitrogen and then homogenized in a protein extraction buffer and a protease inhibitor mixture (Solarbio) as described previously (Xian et al., 2020). The obtained proteins were separated by 12% SDS-PAGE gel and immunoblotted using anti-GFP (1:5000 dilution, Abcam, ab183734). The signals in the blots were visualized with ECL kits (Millipore) and photographed using an Amersham Imager 680 (GE Healthcare). SDS-PAGE was performed with Coomassie brilliant blue (CBB) straining to verify equal loading.
Prokaryotic expression and purification of FolAsp
FolAsp sequences lacking the N-terminal signal peptide were cloned into the pET-28a expression vector via the overhangs added by the primers during amplification (Supplementary Table 1). The His-FolAsp fusion proteins were expressed in E. coli BL21 cells as described previously (Gamir et al., 2016). Briefly, isopropyl β-D-thiogalactoside (IPTG) was added into the medium to induce target protein expression and then cultured at 25°C and 180 rpm for 16 h. The harvested cells were resuspended in lysis buffer containing 20 mM Tris-HCl, 500 mM NaCl, 20 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and lysed with a microfluidizer. The proteins were purified using Ni-nitrilotriacetic acid (Ni-NTA) affinity resin (Qiagen) following the manufacturer’s instructions. The eluted fraction was desalted on a Sephadex G-25 column (PD-10, GE Healthcare) equilibrated with 50 mM Tris-HCl (pH 8). The protein concentrations were determined with an Omni-Easy Instant BCA Protein Assay Kit (Epizyme). The purified protein was concentrated with a 10-kDa MWCO Amicon Ultra Centrifugal Filter (Merck Millipore) and visualized by Coomassie-stained SDS-PAGE.
Protease activity assay
To determine whether FolAsp could degrade host protein, root protein was extracted and inoculated with purified FolAsp or a spore suspension (5 × 106 spores/ml) of the corresponding Fol strains for 2 or 4 h, respectively. The treated root protein was subsequently used for SDS-PAGE and CBB staining to evaluate the protein change induced by FolAsp.
Disease assays
Pathogenicity assays of Fol were performed using the modified root dip method (Di et al., 2016; Zhang N. et al., 2021). The conidia at the optimum concentration of 5 × 106 conidia/ml (Vitale et al., 2019) were used for the infection assay and antifungal assay. Briefly, 14-day-old tomato seedlings were uprooted from the soil, inoculated with a spore suspension (5 × 106 spores/ml) of Fol for 10 min, and subsequently potted and kept at 25°C. Two weeks later, disease symptoms of plants were scored using a disease index ranging from 0 to 5 (0, no symptoms; 1, 0–20% defoliated leaves; 2, 20–40% defoliated leaves; 3, 40–60% defoliated leaves; 4, 60–80% defoliated leaves; and 5, 80–100% defoliated leaves).
A burst of reactive oxygen species (ROS)
Reactive oxygen species production was monitored with a luminol/peroxidase-based assay as described previously (Sang and Macho, 2017). In brief, the sterile H2O-balanced tomato leaf discs in a 96-well white plate were replaced with the luminol (100 μM/L)/peroxidase (20 μg/ml) reaction solution supplied with sterile water or 50 nM flg22 (Genscript Biotech Corporation, China). Luminescence was measured using the Multimode Microplate Reader (Varioskan LUX, Thermo Scientific, USA).
DAB staining and H2O2 quantification
Detection of hydrogen peroxide was conducted using 3,3-diaminobenzidine (DAB) from Sigma-Aldrich. Briefly, the detached tobacco leaves were immersed in a 1 mg/ml solution of DAB in buffer (pH 3.8) and incubated at room temperature for 8 h in the dark. Then, the leaves were bleached with 95% ethanol until the samples became colorless (Yang et al., 2022).
The content of H2O2 was analyzed with a hydrogen peroxide assay kit (Beyotime, S0038) according to the manufacturer’s protocols. In brief, 10 mg of plant tissue was ground into a homogenate with 200 μl of lysate and centrifuged at 12,000 × g for 5 min at 4°C. The reaction mixture containing 50 μl of supernatant and 100 μl of H2O2 detection reagent was left at room temperature (20–25°C) for 30 min and then immediately measured with a spectrometer at a wavelength of 560 nm. Absorbance values were calibrated to a standard curve generated with known concentrations of H2O2.
Statistical analysis
All graphs were exported using the GraphPad Prism 6 software (La Jolla, CA, USA). Statistical analysis was performed by analysis of variance (ANOVA) or unpaired Student’s t-test methods.
Results
Expression profiling of FolAsp in response to the host root
From the previously identified Fol secretome, we found that a hypothetical protein (FOXG_03994, XP_018238471.1) consists of 408 amino acids with a signal peptide at the N-terminus, which is homologous to aspartic protease protein (ASP) from other fungal species (Li J. et al., 2020). This protein was then termed as FolAsp in Fol, which was predicted to contain an ASP domain at the C-terminus by SmartBLAST (Figure 1A). The SmartBLAST analysis results displayed a wide taxonomic diversity in mycetozoa (Dictyostelium discoideum), fruit fly (Drosophila melanogaster), and animal species (Mus musculus, Homo sapiens, and Caenorhabditis elegans). Meanwhile, phylogenetic analysis of the putative amino acid sequence of Asp homologs showed that FolAsp was much more closely related to Fusarium taxa and other pathogenic fungi, compared with metazoan species (Figure 1B). However, the FolAsp was far away with its homologs from plants, yeast, or oomycete, which is consistent with no conserved homologs in these species found by SmartBLAST searching. These results suggested that FolAsp was conserved in the fungi and metazoan.
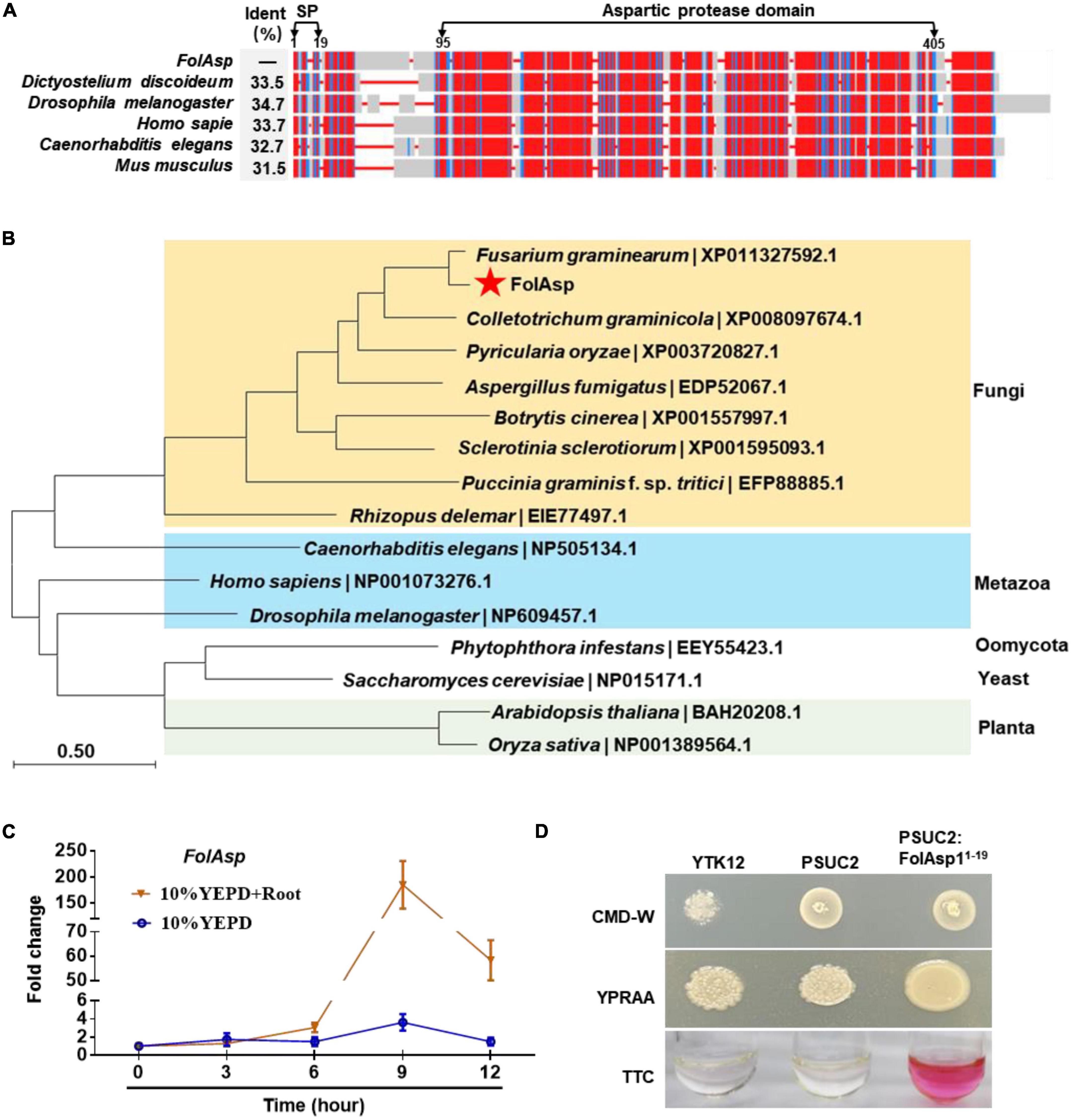
Figure 1. Conservation, expression, and secretion analysis of FolAsp protein. (A) SmartBLAST was used to locate the regions of high sequence conservation in a graphical view among different model organisms. The identity (Ident) of the FolAsp and its homologs were as indicated. The predicted signal peptide (SP) and aspartic protease domain were marked on the top of the graph. (B) Phylogenetic analysis of Asp proteins in Fol and other species. The phylogenetic tree was generated by NJ method in MEGA. (C) Expression profile of FolAsp in Fol with tomato root induction at the indicated times measured by qRT-PCR. The expression levels were normalized to that of the histone H4 gene FolH4. Data represent means ± SE of three independent replicates. (D) Functional validation of the signal peptide (1–19 amino acids) of FolAsp. The indicated strains were grown on CMD-W or YPRAA medium for 3 days. TTC was used to test the enzymatic activity of reducing TTC to red TPF.
Inducing with tomato root, FolAsp is upregulated expressing during conidia generation (Li J. et al., 2020). To more precisely define the regulation of FolAsp expression, we examined its transcript accumulation under different cultured conditions by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The expression profile revealed transcript accumulation of FolAsp was significantly higher across developmental stages in 10% YEPD medium with tomato root, compared with in 10% YEPD medium alone (Figure 1C). This indicates that the host root could specifically enhance the upregulation expression of FolAsp gene.
Secretory activity analysis of FolAsp N-terminal signal peptide
FolAsp is believed to contain a predicted 19-amino-acid signal peptide at the N-terminus according to SignalP-5.0,1 which indicates that FolAsp is a secreted protein (Figure 1A), in accordance with its existence in the Fol secretome (Li et al., 2022). Here, we further performed a yeast signal trap assay to validate the secretory function of the signal peptide (Yin et al., 2018). As shown in Figure 1D, the YTK12 strain carrying pSUC2-FolAsp1–19 was able to grow on YPRAA medium with raffinose as the sole carbon source and reduce the triphenyltetrazolium chloride (TTC) to the red-colored insoluble triphenylformazan (TPF), indicating that the N-terminal signal peptide of FolAsp is functional for its secretion.
FolAsp is not involved in mycelial growth or conidiation
To investigate the biological function of the FolAsp, we used the split-marker method to generate a gene replacement construct containing an Hph gene (Supplementary Figure 1A) and transformed the construct into protoplasts of the wild-type (WT) Fol. Five deletion mutants (ΔFolAsp-10, ΔFolAsp-11, ΔFolAsp-15, ΔFolAsp-28, and ΔFolAsp-29) were obtained on 100 mg/ml hygromycin plates. Homologous recombination events were verified by PCR (Supplementary Figure 1B), with the primers listed in Supplementary Table 1. To further confirm that the change in ΔFolAsp strains occurred because of FolAsp gene deletion, FolAsp gene complementation transformants were produced from ΔFolAsp-10 protoplasts following transformation with the full-length FolAsp gene including its upstream 1,000 bp sequences as the native promoter. Transformants were selected on potato dextrose agar (PDA) media with 100 mg/ml G418, and two PCR-positive transformants were purified and verified by PCR. One recovered strain ΔFolAsp-C (Supplementary Figure 1C) was chosen for detailed phenotypic characterization.
We first compared the growth phenotypes of the knockout strain with those of the WT and complementary (ΔFolAsp-C) strains to determine the potential function of FolAsp. No differences in colony morphology were found among the WT, ΔFolAsp, and ΔFolAsp-C strains on PDA media, complete media (CM), and minimal media (MM) (Figure 2A). In addition, the growth rate of the ΔFolAsp strains was similar to that of the WT and ΔFolAsp-C strains on media under the same conditions (Figure 2B). To evaluate the role of FolAsp in conidiation, we also inoculated mycelial plugs of all strains in carboxymethyl cellulose (CMC) liquid media for different time series. Our quantitative data showed that loss of the FolAsp gene did not affect the conidiation of the ΔFolAsp strain compared with the WT and ΔFolAsp-C strains (Figure 2C), suggesting that FolAsp is not required for the conidiation of F. oxysporum. Taken together, our results revealed that FolAsp did not affect vegetative hypha growth or conidia production.
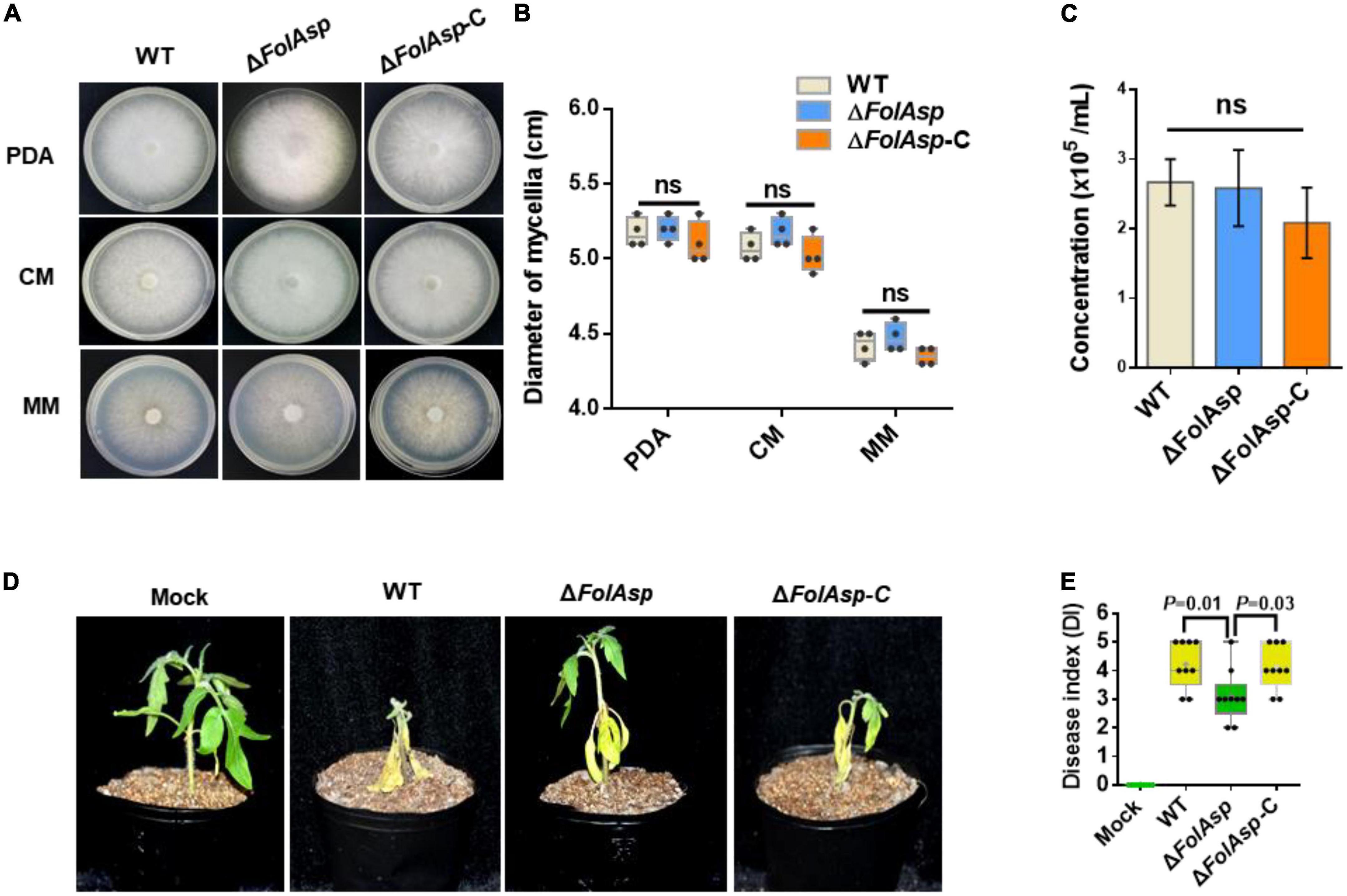
Figure 2. Phenotypic analysis of FolAsp mutant strains. (A) Mycelial growth of the WT and ΔFolAsp (ΔFolAsp-KO10) mutant strains on the indicated plates after 5 days of cultivation. (B) Mycelium diameters of colonies of the strains in panel (A) were measured at 5 days of cultivation. (C) Conidia production of the WT and ΔFolAsp mutant strains in PDB liquid medium at the indicated times. No significant difference (ns) was indicated according to multiple t-test. (D) Virulence of the wild-type (WT) and FolAsp mutant strains on tomato. The disease symptoms were observed, and photographs were taken at 14 days after inoculation (DAI). Mock, inoculation with water. Experiments were repeated three times with similar results. (E) Disease index scored at 14 DAI. Significant differences were determined according to one-way ANOVA, and p value was shown. For panels (B,E) whiskers of the boxplots display the upper and lower quartiles, the boxes display the interquartile range, and the plus displays the mean.
FolAsp contributes full virulence of Fol
To investigate the possible function of FolAsp in Fol virulence, we observed changes in symptoms of tomato seedlings during inoculation. Three-week-old seedlings were inoculated with conidia of the WT and ΔFolAsp strains (KO-10; 11, 15, 28, and 29) at 25°C for 14 days. Obvious disease symptoms, such as stunted plant growth and leaf yellowing, occurred for tomato seedlings inoculated with the WT strain, which showed higher infection ability, whereas significantly reduced disease symptoms were detected in tomato seedlings inoculated with all the five knockout strains (Supplementary Figure 2). Furthermore, we confirmed its virulence function on tomato seedlings by comparing WT, ΔFolAsp, and complementary ΔFolAsp-C strains. Obviously, the virulence of the knockout mutant was strongly reduced in tomato seedlings (Supplementary Figure 2) at 14 days after inoculation (DAI). However, the ΔFolAsp-C strain displayed a higher disease index, which was similar to WT under the same inoculation conditions (Figures 2D, E). These results indicate that FolAsp is required for the full virulence of Fol.
As FolAsp deletion reduced Fol virulence in tomatoes, it is believed that overexpressing this protein should promote more severe Fusarium wilt disease. To verify this hypothesis, we overexpressed FolAsp-GFP fusion protein [FolAsp (OE)] in Fol (Figure 3A and Supplementary Figure 1C). Phenotypically, FolAsp overexpression did not affect vegetative hypha growth on PDA, CM, and MM (Figures 3B, C). As expected, the conidia production of FolAsp (OE) was not affected compared with the WT (Figure 3D). These results further indicated that FolAsp was not involved in the growth development of Fol. Then, we performed the disease assay inoculated with WT- or FolAsp-overexpressing strain FolAsp (OE). As shown in Figures 3E, F, overexpression of FolAsp significantly caused more severe disease symptoms compared with the WT, which was consistent with the reduced pathogenicity by the FolAsp deletion mutant.
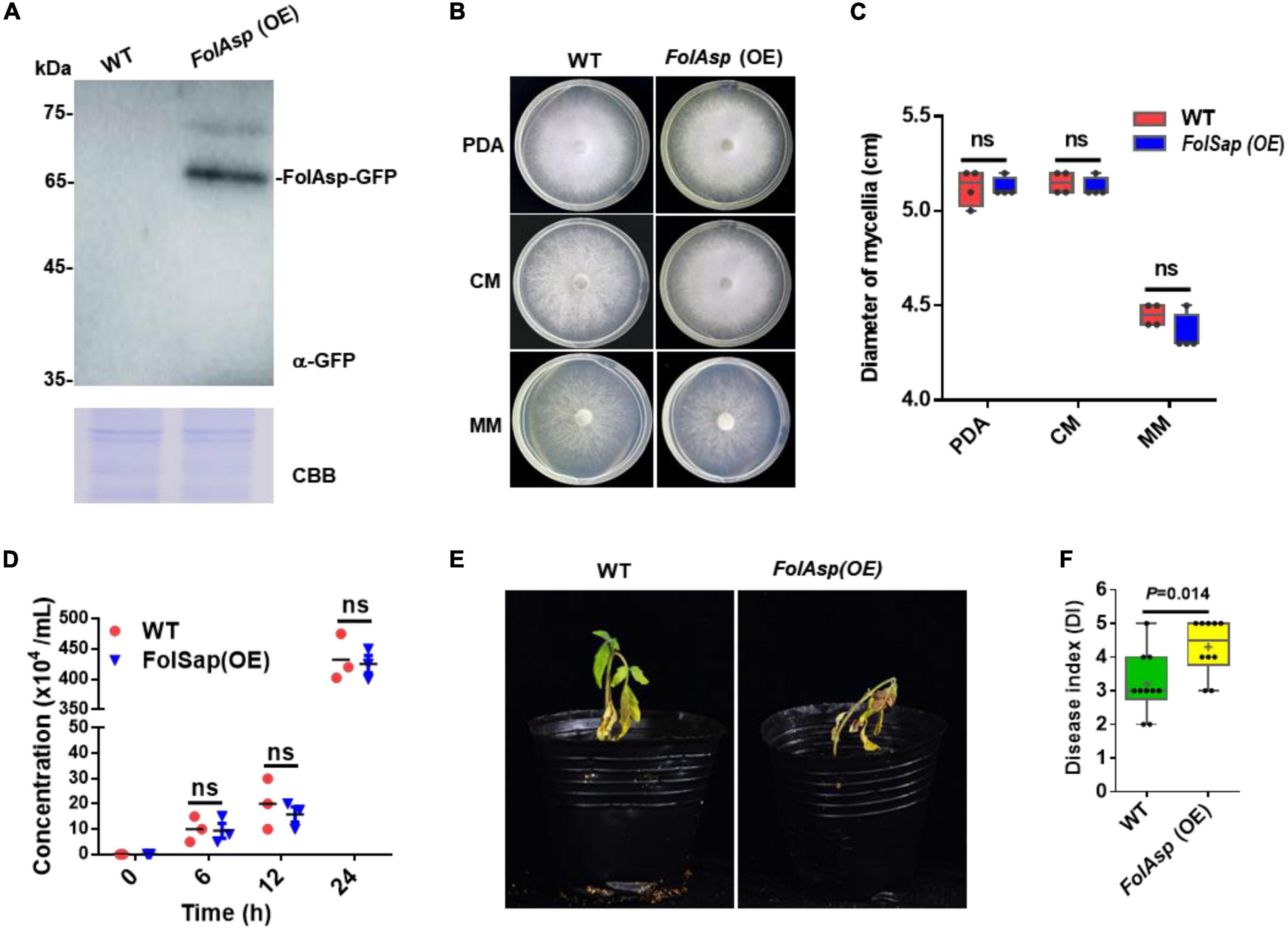
Figure 3. FolAsp-GFP overexpression increases virulence. (A) FolAsp-GFP overexpression [FolAsp (OE)] in Fol was determined by Western blotting with anti-GFP antibody. The WT strain was used as the control. Protein loading is indicated by CBB staining. (B) Mycelial growth of the WT and FolAsp (OE) strains on PDA plates after 5 days of cultivation. (C) Mycelium diameters of colonies of the strains in panel (B) were measured at 5 days of cultivation. (D) Conidia production of the WT and FolAsp (OE) strains in PDB liquid medium at the indicated times. No significant difference (ns) was indicated according to multiple t-test. (E) Virulence of the WT- and FolAsp-overexpressed mutant strains on tomato. (F) Disease index scored at 14 DAI. For panels (E,F) the virulence and disease index were determined as in figure.
FolAsp locates at the apoplast in plant cells
To explore the localization and biological function of FolAsp, we transiently expressed FolAsp-GFP fusion proteins, in which green fluorescent protein (GFP) was added to the C-terminus of FolAsp protein. Nicotiana benthamiana leaves were used to determine subcellular localization using Agrobacterium tumefaciens infiltration by observing the GFP signal at 48 h. To increase protein levels for visualization, these constructs were expressed by the 35S promoter. Using a fluorescence microscope, FolAsp-GFP was detected in the plant cytoplasm with a stronger fluorescent signal (Figure 4). As FolAsp was identified to be secreted, it might function in the apoplast. Though FolAsp was transiently expressed with its native N-terminal signal peptide, FolAsp might not be faithfully secreted in planta, as described previously (Li et al., 2022).
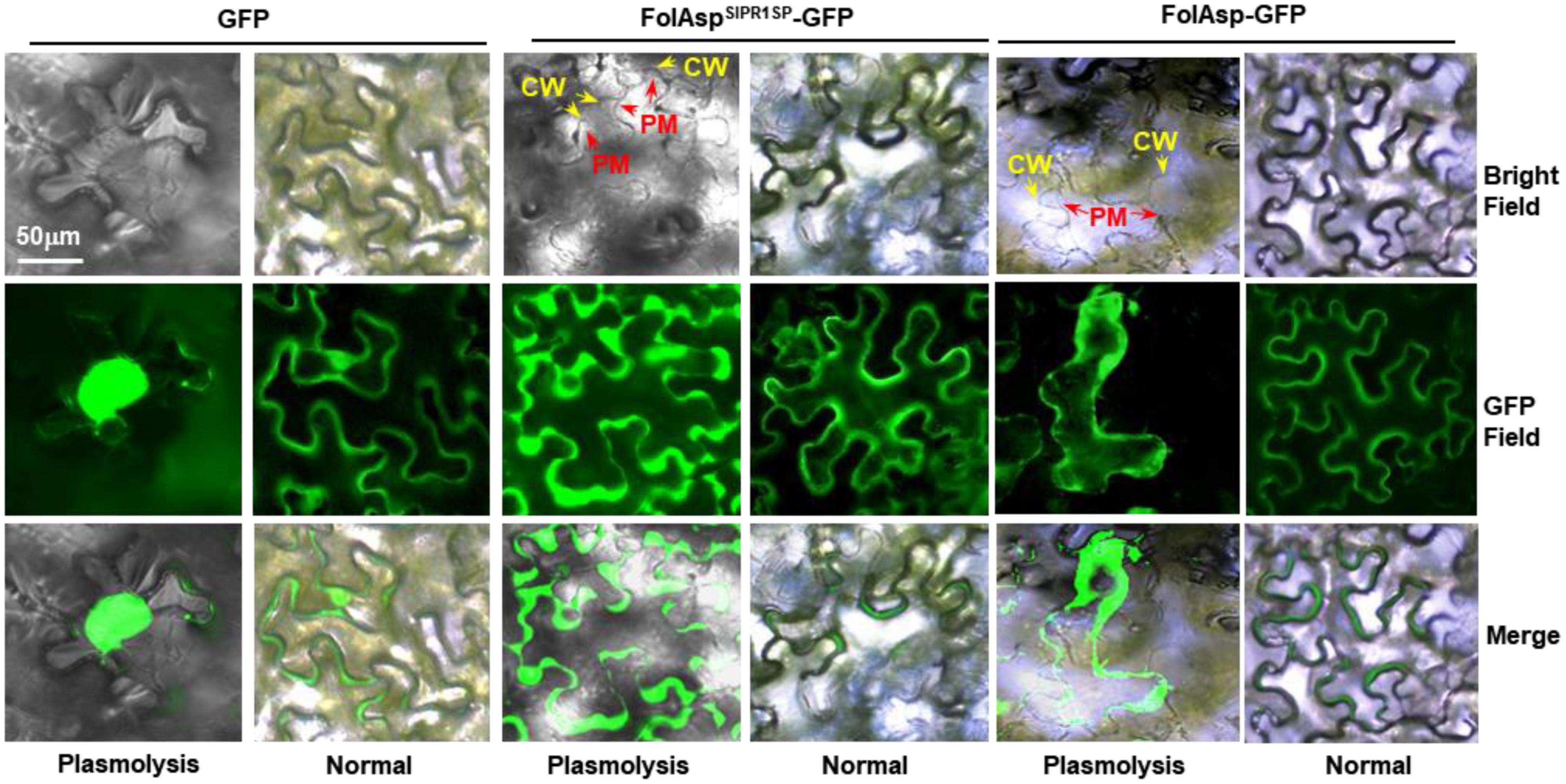
Figure 4. Subcellular localization of FolAsp in tobacco leaves. FolAsp-GFP indicates the fusion protein with its native N-terminal signal peptide. FolAspSlPR1SP-GFP indicates replacement of its native signal peptide with the tomato PR1 signal peptide (N-terminal 1–24 amino acids). Images were taken at 3 DAI. The plasmolysis was visualized after treatment with 800 mM mannitol for 6 h. CM, cell wall; PM, plasma membrane.
The signal peptide of PR1 is highly effective in targeting proteins/peptides to the apoplast via the ER/Golgi secretory pathway (Kloppholz et al., 2011). To manipulate FolAsp secretion in tobacco, we replaced its native signal peptide with that of tomato SlPR1 (Li et al., 2022), named FolAspSlPR1SP-GFP, and found that FolAspSlPR1SP-GFP location was dramatically changed, but it was difficult to distinguish between apoplast and cytoplasm. Therefore, we adopted the plasmolysis assay. Following treatment with 800 mM mannitol for 6 h, plasmolysis was greatly observed, with a clear observation of the FolAspSlPR1SP protein in the apoplast. However, both the FolAsp-GFP and GFP were located in the cytoplasm (Figure 4). These results indicated that FolAsp might locate at apoplast in planta after secreting from Fol.
FolAsp inhibits plant cell death (PCD)
To elucidate whether the transient expression of FolAsp alters plant immune responses, the local hypersensitive response (HR) was analyzed in N. benthamiana-using A. tumefaciens-mediated transient expression assay. The FolAsp-GFP protein was infiltrated into the leaves of tobacco through the stoma. A secreted FolAsp (FolAspSlPR1SP-GFP) was also expressed to determine whether apoplastic localization works for the induction of plant immune responses. After 3 days of infiltration, there was no visible cell death detected in GFP-expressing control, FolAsp-GFP, and FolAspSlPR1SP-GFP-expressing plants on the tobacco leaves (Figure 5A). By contrast, the transient expression of a pathogen-associated molecular pattern (PAMP) protein INF1 triggered significant PCD at the infiltration site, suggesting that the transient expression of FolAsp does not trigger the plant HR activity in tobacco. The mature protein was also determined by Western blotting analysis using an anti-GFP antibody (Figure 5B).
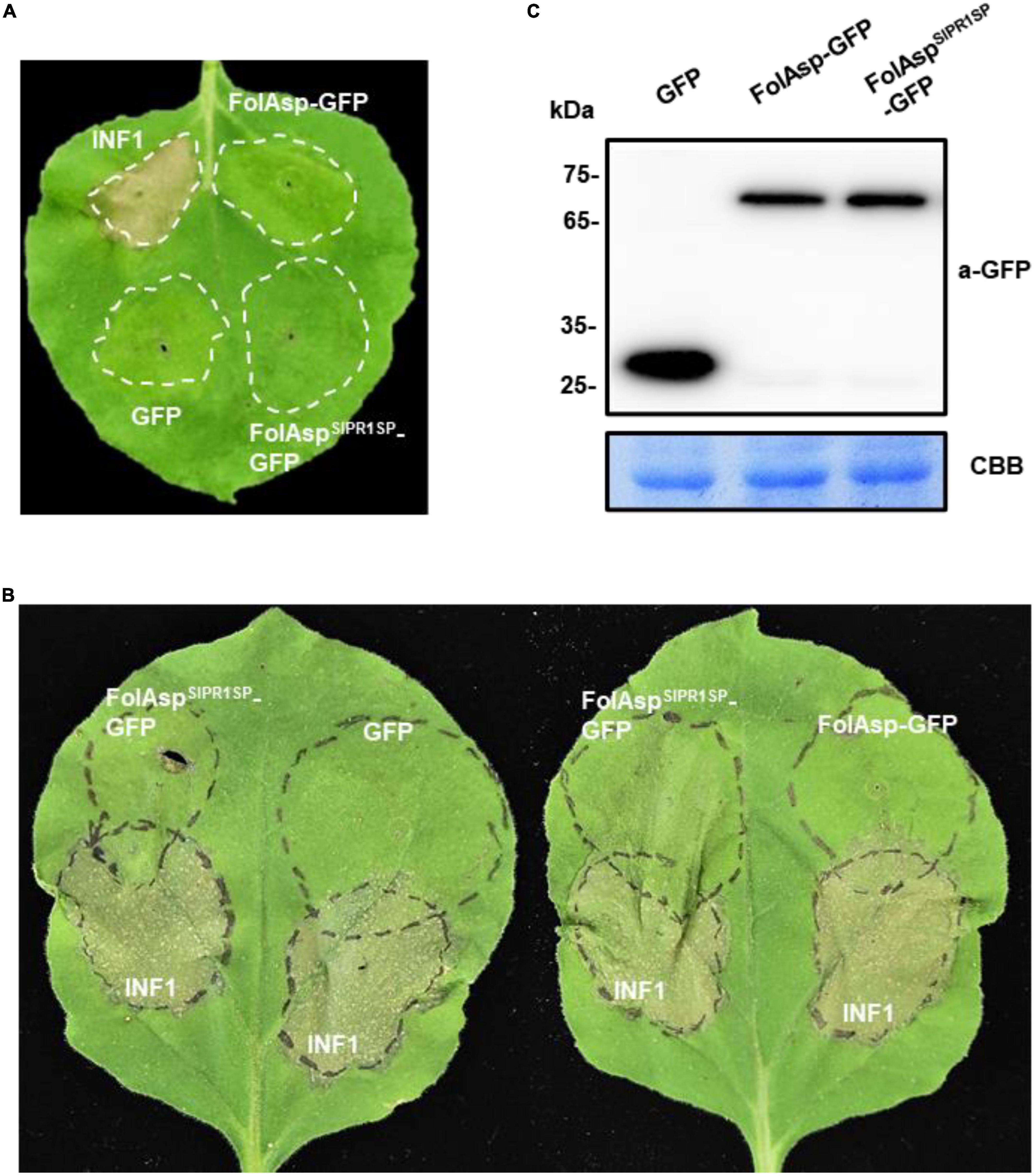
Figure 5. FolAsp inhibits INF1-mediated cell death. (A) Validation of FolAsp in induced cell death in N. benthamiana. Leaves were with agroinfiltration expressing GFP, FolAsp-GFP, and FolAspSlPR1SP-GFP, or the cell death inducer INF1. Photographs were taken 3 DAI. (B) FolAsp is involved in suppression of cell death. Representative leaves showed cell death induced by transient expression of INF1 in N. benthamiana leaves pretreated with agroinfiltration expressing GFP, FolAsp-GFP, or FolAspSlPR1SP-GFP for 2 days. The INF1-infiltrated leaves were photographed at 60 HAI. (C) Protein expression at 2 DAI prior to INF1 infiltration was monitored by Western blotting with anti-GFP antibody. Protein loading is indicated by CBB staining. Experiments were repeated at least three times with similar results.
To examine whether FolAsp acts as an effector to inhibit defense responses during host–pathogen interactions, we determined the effector virulence function in suppressing PCD induced by INF1 in N. benthamiana. Strikingly, the sites infiltrated with FolAspSlPR1SP-GFP 2 days and then challenged with the bacteria suspension carrying INF1 exhibited inhibited PCD 60 h later at the overlapped infiltration area, compared with agroinfiltration of GFP or FolAsp-GFP prior to INF1 incubation (Figure 5B). The mature protein was determined by Western blotting analysis using an anti-GFP antibody (Figure 5C). These results suggested that apoplast FolAsp (FolAspSlPR1SP-GFP) suppresses PCD induced by INF1 and alludes to the possible virulence function of FolAsp, resulting in enhanced susceptible to the fungal pathogen Fol.
FolAsp inhibits plant ROS production
To further expand the bioactivity of FolAsp in response to plant immunity, we determined reactive oxygen species (ROS) levels in tobacco leaves transiently expressing GFP, FolAsp-GFP, and FolAspSlPR1SP-GFP or of the control leaves (Mock). First, we determined whether FolAspSlPR1SP-GFP inhibits PAMP-triggered immunity (PTI) in tobacco. In this instance, the bacterial flg22 was used, since it has been widely shown to induce ROS production as the MAMP (Ngou et al., 2022). In tobacco leaves treated with the flg22, ROS burst was significantly observed in the GFP-, FolAsp- GFP-, or FolAspSlPR1SP-GFP-expressing leaves compared with the control leaves (Figure 6A). However, the ROS burst was markedly suppressed in the FolAspSlPR1SP-GFP-expressed leaves compared with GFP- or FolAsp-GFP-expressed leaves. Furthermore, the effector-mediated ROS inhibition was determined by DAB staining and H2O2 quantification. As can be seen in Figures 6B, C, flg22-mediated ROS accumulation in tobacco could be significantly suppressed with expressing FolAspSlPR1SP-GFP; however, it is not altered on the GFP- or FolAsp-GFP-expressing leaves, compared with the control leaves (Mock). This result indicated that apoplast FolAspSlPR1SP-GFP effectively decreased H2O2 production. This suggested that FolAsp was likely an important apoplast protein to inhibit MAMP-induced basal immunity.
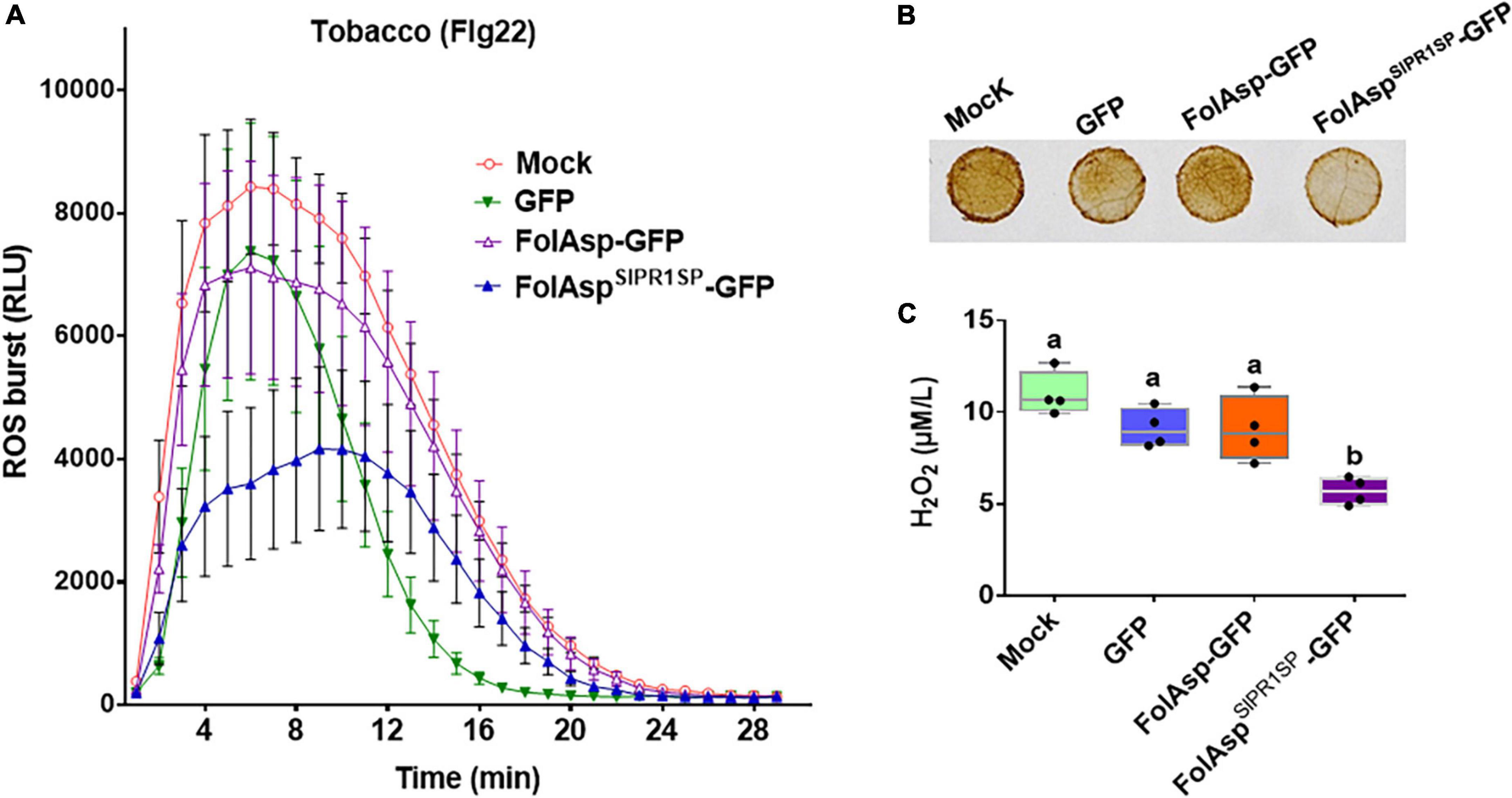
Figure 6. FolAsp inhibits ROS production. (A) Burst of ROS in GFP, FolAsp-GFP, or FolAspSlPR1SP-GFP transiently expressed tobacco leaves was measured with 50 nM flg22. Mean values (±SEM) of eight replicates are shown. (B) FolAsp inhibits leaf H2O2 production induced by flg22. The GFP, FolAsp-GFP, or FolAspSlPR1SP-GFP transiently expressed tobacco leaves and the control leaves (Mock) were treated with the DAB following 50 nM flg22 treatment for 30 min to visualize H2O2 accumulation. (C) Quantification of H2O2 was measured with samples from (B). Statistical significance was revealed by one-way ANOVA (mean ± SE of 3 independent biological replicates, p < 0.05). Experiments were repeated three times with similar results.
FolAsp possesses protease activity
The predicted aspartic protease domain indicated FolAsp might possess protease activity. To test this hypothesis, we expressed the protein in Escherichia coli BL21 (DE3) to purify it for enzyme activity analysis. The FolAsp protein was expressed in a supernatant culture and used for protein purification. The results of Coomassie brilliant blue staining revealed the obtained FolAsp protein (Figure 7A). Subsequently, root protein was used as the substrate to detect the protease activity of FolAsp in vitro. As can be seen in Figures 7B, C, adding FolAsp could remarkably promote host protein degradation. The results revealed that the FolAsp possessed protease activity on host protein.
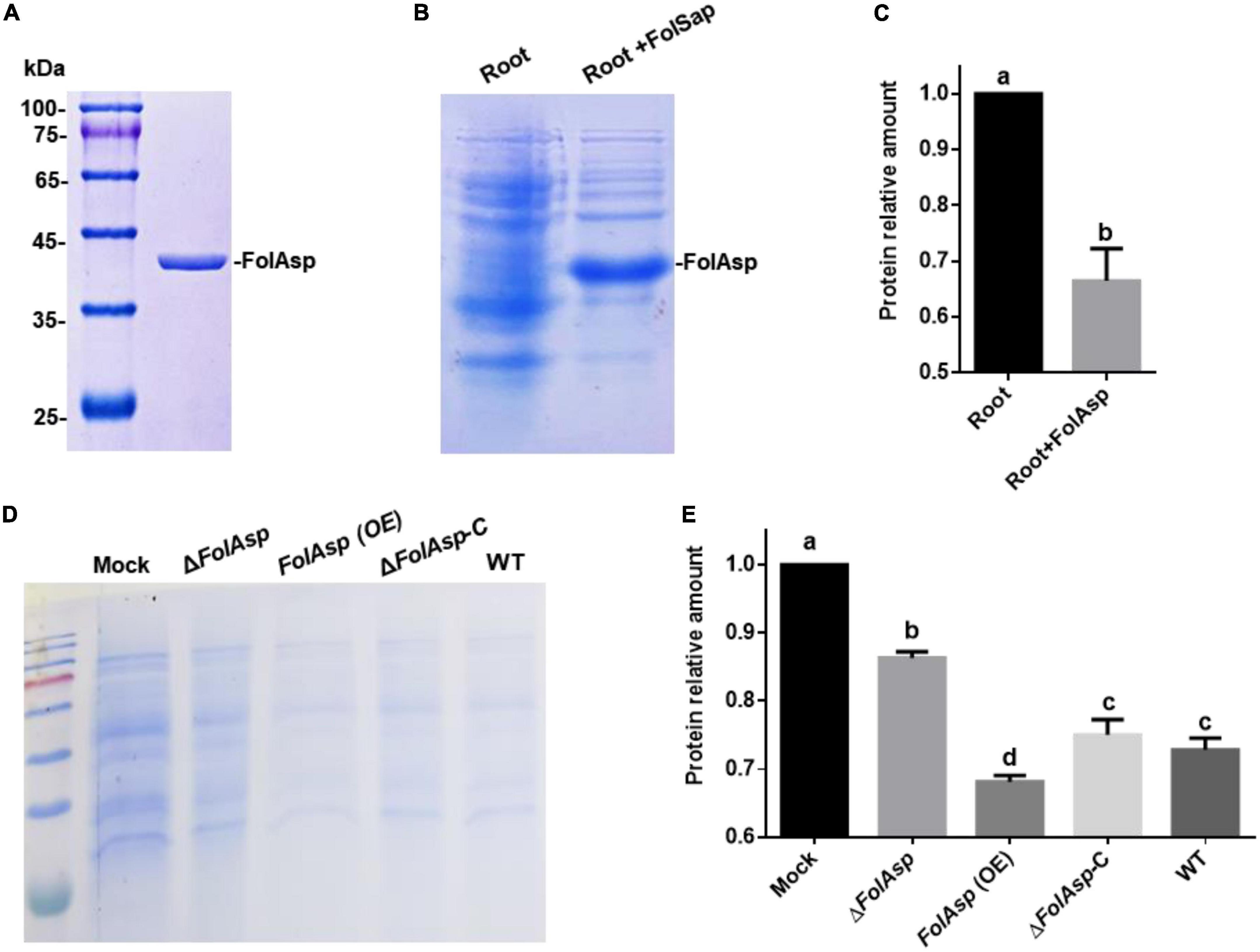
Figure 7. FolAsp possesses protease activity on host protein. (A) CBB-stained SDS-PAGE gel of purified FolAsp-his protein is shown. Heterologous expression of recombinant FolAsp-his protein using Escherichia coli BL21. Extracted protein was incubated with Ni-NTA agarose and then eluted with imidazole. (B) FolSap promoted degradation of host protein. The extracted root protein was inoculated with 1 μg purified FolSap for 2 h. CBB-stained SDS-PAGE gel of supernatant fractions is shown. (C) Quantification of the relative root protein abundance after FolAsp treatment, as in (B). (D) Amount of tomato protein following treatment with the indicated strains or without any treatment as the control (Mock). The root protein was extracted and incubated with the spores of the indicated strains for 4 h. CBB-stained SDS-PAGE gel of supernatant fractions is shown. (E) Quantification of root proteins treated with Fol strains relative to Mock, as in (D). For (C,E), statistical significance was revealed by one-way ANOVA (mean ± SE of 3 independent biological replicates, p < 0.05).
Furthermore, we detected the Fol transformants promoted protein degradation on host protein to evaluate the contribution of FolSap on the protease activity. Following incubation with extracted root protein as the substrate, the results of Coomassie brilliant blue staining revealed that all Fol strains could promote root protein degradation compared with the control (Mock) (Figures 7D, E). However, FolAsp overexpression (Supplementary Figure 1A) significantly increased protein degradation compared with the WT and ΔFolAsp-C strains. Conversely, FolAsp deletion remarkably decreased protein degradation. These results indicated that FolAsp can degrade host protein through its protease activity.
Taken together, these results suggested that secreted protease FolAsp might inhibit plant basal defense or possess host protein degradation in apoplast space to facilitate its virulence on the host.
Discussion
Most effectors identified from Fol were originally identified in the xylem sap of Fol-infected tomato plants, such as secreted in xylem (Six) proteins, which are important for pathogenicity (Houterman et al., 2007; Gawehns et al., 2015). However, the identification of secreted proteins beyond the six proteins is poorly understood in Fol, especially during the initial infection stages. We previously performed a secretome-wide analysis in Fol to provide new sight of infection-related proteins and found that most of them with an N-terminal SP were annotated as enzymes (Li J. et al., 2020). To date, three secreted proteins are shown to contribute virulence, and two of them encode the ribonuclease and peptidase, respectively (Li et al., 2022; Qian et al., 2022a,b). In this study, we characterized one aspartic protease FolAsp from the Fol secretome. Deletion of the FolAsp significantly decreases its pathogenicity to host tomato, indicating that secreted FolAsp is involved in the virulence of pathogen Fol.
Secreted proteins/effectors play important roles in the invading pathogen by blocking plant immunity or altering specific plant processes (Zhang et al., 2017; Cao et al., 2018). The N-terminal signal peptide of FolAsp displayed secretory activity in the yeast system, indicating that FolAsp is possibly secreted via the ER/Golgi secretory pathway (Kloppholz et al., 2011). In the biotrophic fungus Fol, whose growth is confined to the apoplast space, the signal peptide sequences might dominate the secretion and location of FolAsp in the apoplastic space of the host. To support it, the transiently expressed FolAsp with a plant signal peptide revealed its location at apoplastic space in planta. Therefore, FolAsp probably functions at the plant apoplast during Fol infection.
Proteases have been described as playing an important role in the host invasion process in many pathogenic microorganisms. Extracellular proteases from pathogenic fungi fulfill a number of specialized functions during the infective process in addition to the simple role of digesting molecules for nutrient acquisition (de Assis Tacco et al., 2009). Some studies investigating the role of extracellular aspartyl proteases in pathogenesis have focused on fungi (Li C. et al., 2020). In A. fumigatus, the aspergillopepsin aspartic protease is secreted in large amounts during infection of the mouse lung, facilitating fungus penetration (Lee and Kolattukudy, 1995). The FolAsp, identified from root-induced secretome (Li J. et al., 2020), is specifically upregulated induced with host root. Similarly, two ASP domain-containing proteases were also upregulated during infection of Sclerotinia sclerotiorum. An aspartyl (acid) protease, Sscle04g035550, was upregulated at all time points commonly associated with plant pathogenic protease activity (de Assis Tacco et al., 2009). Another aspartate protease, Sscle07g058540, was the second most upregulated protease at 24 hpi and shows homology to several aspergillopepsin-like proteins whose activity is important in invasive aspergillosis of humans (Rementeria et al., 2005). In Monilinia laxa, which is responsible for brown rots disease of stone and pome fruits, a secreted pepsin-like protein with a conserved domain for aspergillopepsin-like aspartic proteases was significantly upregulated by transcriptome analyses (De Miccolis Angelini et al., 2018). There results implied its important function in initial pathogenesis. As expected, deletion of FolAsp decreased its virulence in tomatoes; however, its growth phenotype was not affected. Furthermore, overexpression of FolAsp significantly promoted invasion and enhanced tomato susceptibility, suggesting that FolAsp may suppress the defense response of host plants upon Fol infection.
The identified FolAsp (FOXG_03994) had widely conserved homologs in fungi, indicating that this protein might be involved in general pathogenicity by conserved mechanisms. The degradative properties of secreted proteases have attracted much attention as potential mediators of fungal invasion in infected tissue (Naglik et al., 2003). Numerous functions have been attributed to aspartyl proteases in microorganisms, which range from nutrient degradation to the activation of signaling molecules (de Assis Tacco et al., 2009). In addition, aspartyl proteases from fungi serve to activate other zymogens such as alkaline phosphatase, chitin synthase, and other proteases. Aspartyl proteases from Schistosoma species are known to be responsible for host-specific proteolytic degradation of mammalian hemoglobin (Koehler et al., 2007). In this regard, the fact that the host protein is less abundant treated with FolAsp protein or FolAsp overexpression strain would point to its importance of enzymatic activity in the pathogenesis of the organism. The biochemical functions of many Avr proteins identified from various plant pathogens mainly constitute enzymatic activities (Song et al., 2020). However, the biochemical role of the aspartyl protease FolAsp in Fol still remains unclear. Future work will focus on its biochemical characterization.
When pathogens infect and secrete effectors to host cells, the basal defense responses of plants, such as ROS bursts, resistance gene expression, and cell death, are activated to inhibit the pathogen growth and development (Lyu et al., 2016; Kettles et al., 2018; Yang et al., 2018). In our study, transient expression of FolAsp protein in N. benthamiana did not induce cell death, similar to the recent findings of a peptidase effector FoAPY1 from Fol, which does not have the ability to induce cell death (Qian et al., 2022b). This family of aspartate proteases is also annotated by the MEROPS database as the peptidase family A1 (pepsin A, clan AA) (Santos et al., 2013). In addition to degrading host proteins into amino acids for nutrient acquisition or catabolic activities (Lowe et al., 2015), peptidases are reported to possess other functions in host plant–pathogen interactions. HopN1, a secreted protein from the bacterium Pseudomonas syringae, is a cysteine peptidase that cleaves the host PsbQ protein, which is a key photosynthesis enzyme, to block programmed cell death (Rodríguez-Herva et al., 2012). Similarly, the FolAsp could inhibit INF1, a MAMP identified from Phytophthora species, and induce cell death (Qiu et al., 2022). Further study revealed that FolAsp could suppress flg22-activated ROS production and H2O2 accumulation. Plants produce ROS to restrict pathogen invasion. A recent report revealed that ROS detoxification employed by Botrytis cinerea facilitates fungal invasion (Yang et al., 2022). The results indicated that the FolAsp protein might not be similar to other Avr effectors in terms of its cytotoxic effects in the plant or may not be recognized in the host cell. Instead, FolAsp likely suppresses the defense response of host plants by arresting ROS production to promote pathogen invasion.
Conclusion
The results showed that FolAsp significantly promoted invasion and enhanced host susceptibility to fungal pathogens, suggesting that FolAsp may suppress the defense response of host plants during F. oxysporum infection. These findings provide insight into the interactions between F. oxysporum and host plants, contributing to the understanding of the pathogenic mechanism of this fungus. With the extensive study on the structure and function of secreted aspartic protease, this kind of virulence factor was supposed to be a potential target for the development of novel antifungal agents.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
CW, YZ, ZL, and YQ carried out the experiments. YL, LY, and SL performed technical supports and statistical analysis. WL and JL initiated the project, designed the experiments, analyzed the data, and provided funding. JL, CW, and WL wrote the manuscript with inputs from all the authors. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the National Natural Science Foundation of China (32272489, 31901830, and 31972213), Shandong Province “Double-Hundred Talent Plan” (WST2018008), and Taishan Scholar Construction Foundation of Shandong Province (tshw20130963).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1103418/full#supplementary-material
Footnotes
References
Cao, L., Blekemolen, M. C., Tintor, N., Cornelissen, B. J. C., and Takken, F. L. W. (2018). The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol. Plant 11, 691–705. doi: 10.1016/j.molp.2018.02.011
Chen, C., He, B., Liu, X., Ma, X., Liu, Y., Yao, H.-Y., et al. (2020). Pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PFP1) regulates starch biosynthesis and seed development via heterotetramer formation in rice (Oryza sativa L.). Plant Biotechnol. J. 18, 83–95. doi: 10.1111/pbi.13173
de Assis Tacco, B. A. C., Parente, J. A., Santiago Barbosa, M., Bao, S. N., de Souza Góes, T., Pereira, M., et al. (2009). Characterization of a secreted aspartyl protease of the fungal pathogen Paracoccidioides brasiliensis. Med. Mycol. 47, 845–854. doi: 10.3109/13693780802695512
De Miccolis Angelini, R. M., Abate, D., Rotolo, C., Gerin, D., Pollastro, S., and Faretra, F. (2018). De novo assembly and comparative transcriptome analysis of Monilinia fructicola, Monilinia laxa and Monilinia fructigena, the causal agents of brown rot on stone fruits. BMC Genomics 19:436.
Di, X., Gomila, J., Ma, L., van den Burg, H. A., and Takken, F. L. W. (2016). Uptake of the Fusarium effector Avr2 by tomato is not a cell autonomous event. Front. Plant Sci. 7:1915.
Gamir, J., Darwiche, R., van’t Hof, P., Choudhary, V., Stumpe, M., Schneiter, R., et al. (2016). The sterol-binding activity of pathogenesis-related protein 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. doi: 10.1111/tpj.13398
Gawehns, F., Ma, L., Bruning, O., Houterman, P. M., Boeren, S., Cornelissen, B. J. C., et al. (2015). The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 6:967.
Gui, Y. J., Chen, J. Y., Zhang, D. D., Li, N. Y., Li, T. G., Zhang, W. Q., et al. (2017). Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 19, 1914–1932. doi: 10.1111/1462-2920.13695
Hacquard, S., Joly, D. L., Lin, Y.-C., Tisserant, E., Feau, N., Delaruelle, C., et al. (2012). A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol. Plant Microbe Interact. 25, 279–293. doi: 10.1094/MPMI-09-11-0238
Houterman, P. M., Speijer, D., Dekker, H. L., De Koster, C. G., Cornelissen, B. J. C., and Rep, M. (2007). The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 8, 215–221. doi: 10.1111/j.1364-3703.2007.00384.x
Jeong, H.-J., Shin, J. S., and Ok, S. H. (2011). Barley DNA-binding methionine aminopeptidase, which changes the localization from the nucleus to the cytoplasm by low temperature, is involved in freezing tolerance. Plant Sci. 180, 53–60. doi: 10.1016/j.plantsci.2010.09.004
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195.
Kettles, G. J., Bayon, C., Sparks, C. A., Canning, G., Kanyuka, K., and Rudd, J. J. (2018). Characterization of an antimicrobial and phytotoxic ribonuclease secreted by the fungal wheat pathogen Zymoseptoria tritici. New Phytol. 217, 320–331. doi: 10.1111/nph.14786
Kloppholz, S., Kuhn, H., and Requena, N. (2011). A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 21, 1204–1209. doi: 10.1016/j.cub.2011.06.044
Koehler, J. W., Morales, M. E., Shelby, B. D., and Brindley, P. J. (2007). Aspartic protease activities of schistosomes cleave mammalian hemoglobins in a host-specific manner. Mem. Inst. Oswaldo Cruz 102, 83–85. doi: 10.1590/s0074-02762007000100014
Lee, J., and Kolattukudy, P. (1995). Molecular cloning of the cDNA and gene for an elastinolytic aspartic proteinase from Aspergillus fumigatus and evidence of its secretion by the fungus during invasion of the host lung. Infect. Immun. 63, 3796–3803. doi: 10.1128/iai.63.10.3796-3803.1995
Li, J., Gao, M., Gabriel, D. W., Liang, W., and Song, L. (2020). Secretome-wide analysis of lysine acetylation in Fusarium oxysporum f. sp. lycopersici provides novel insights into infection-related proteins. Front. Microbiol. 11:559440. doi: 10.3389/fmicb.2020.559440
Li, C., Liu, Y., Wu, S., Han, G., Tu, J., Dong, G., et al. (2020). Targeting fungal virulence factor by small molecules: Structure-based discovery of novel secreted aspartic protease 2 (SAP2) inhibitors. Eur. J. Med. Chem. 201:112515.
Li, J., Mu, W., Veluchamy, S., Liu, Y., Zhang, Y., Pan, H., et al. (2018). The GATA-type IVb zinc-finger transcription factor SsNsd1 regulates asexual–sexual development and appressoria formation in Sclerotinia sclerotiorum. Mol. Plant Pathol. 19, 1679–1689. doi: 10.1111/mpp.12651
Li, H., Wang, H., Jing, M., Zhu, J., Guo, B., Wang, Y., et al. (2018). A Phytophthora effector recruits a host cytoplasmic transacetylase into nuclear speckles to enhance plant susceptibility. eLife 7:e40039. doi: 10.7554/eLife.40039
Li, J., Ma, X., Wang, C., Liu, S., Yu, G., Gao, M., et al. (2022). Acetylation of a fungal effector that translocates host PR1 facilitates virulence. eLife 11:e82628. doi: 10.7554/eLife.82628
Lievens, B., Houterman, P. M., and Rep, M. (2009). Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300, 201–215. doi: 10.1111/j.1574-6968.2009.01783.x
Lowe, R. G., McCorkelle, O., Bleackley, M., Collins, C., Faou, P., Mathivanan, S., et al. (2015). Extracellular peptidases of the cereal pathogen Fusarium graminearum. Front. Plant Sci. 6:962. doi: 10.3389/fpls.2015.00962
Lyu, X., Shen, C., Fu, Y., Xie, J., Jiang, D., Li, G., et al. (2016). A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathol. 12:e1005435. doi: 10.1371/journal.ppat.1005435
Ma, L.-J., Geiser, D. M., Proctor, R. H., Rooney, A. P., O’Donnell, K., Trail, F., et al. (2013). Fusarium pathogenomics. Annu. Rev. Microbiol. 67, 399–416. doi: 10.1146/annurev-micro-092412-155650
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Michielse, C. B., and Rep, M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10, 311–324.
Monika, S., and Zbigniew, O. (2017). Contribution of aspartic proteases in Candida virulence. Protease inhibitors against Candida infections. Curr. Protein Pept. Sci. 18, 1050–1062.
Naglik, J. R., Challacombe, S. J., and Hube, B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67, 400–428.
Ngou, B. P. M., Ding, P., and Jones, J. D. (2022). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Noda, J., Brito, N., and González, C. (2010). The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol. 10:38. doi: 10.1186/1471-2229-10-38.15
Qian, H., Song, L., Wang, L., Wang, B., and Liang, W. (2022b). The secreted FoAPY1 peptidase promotes Fusarium oxysporum invasion. Front. Microbiol. 13:1040302.
Qian, H., Wang, L., Wang, B., and Liang, W. (2022a). The secreted ribonuclease T2 protein FoRnt2 contributes to Fusarium oxysporum virulence. Mol. Plant Pathol. 23, 1346–1360. doi: 10.1111/mpp.13237
Qiu, X., Kong, L., Chen, H., Lin, Y., Tu, S., Wang, L., et al. (2022). The Phytophthora sojae nuclear effector PsAvh110 targets a host transcriptional complex to modulate plant immunity. Plant Cell 35, 574–597. doi: 10.1093/plcell/koac300
Rementeria, A., López-Molina, N., Ludwig, A., Vivanco, A. B., Bikandi, J., Pontón, J., et al. (2005). Genes and molecules involved in Aspergillus fumigatus virulence. Rev. Iberoam. Micol. 22, 1–23.
Rodríguez-Herva, J. J., González-Melendi, P., Cuartas-Lanza, R., Antúnez-Lamas, M., Río-Alvarez, I., Li, Z., et al. (2012). A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell. Microbiol. 14, 669–681. doi: 10.1111/j.1462-5822.2012.01749.x
Sang, Y., and Macho, A. P. (2017). “Analysis of PAMP-triggered ROS burst in plant immunity,” in Plant pattern recognition receptors, eds L. Shan and P. He (New York, NY: Humana Press), 143–153.
Santos, L. O., Garcia-Gomes, A. S., Catanho, M., Sodre, C. L., Santos, A. L., Branquinha, M. H., et al. (2013). Aspartic peptidases of human pathogenic trypanosomatids: Perspectives and trends for chemotherapy. Curr. Med. Chem. 20, 3116–3133.
Sharpee, W. C., and Dean, R. A. (2016). Form and function of fungal and oomycete effectors. Fungal Biol. Rev. 30, 62–73.
Song, P., Cheng, L., Tian, K., Zhang, M., McHunu, N. P., Niu, D., et al. (2020). Biochemical characterization of two new Aspergillus niger aspartic proteases. 3 Biotech 10:303. doi: 10.1007/s13205-020-02292-4
Vitale, S., Di Pietro, A., and Turrà, D. (2019). Autocrine pheromone signalling regulates community behaviour in the fungal pathogen Fusarium oxysporum. Nat. Microbiol. 4, 1443–1449. doi: 10.1038/s41564-019-0456-z
Xian, L., Yu, G., Wei, Y., Rufian, J. S., Li, Y., Zhuang, H., et al. (2020). A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe 28, 548–557.e7. doi: 10.1016/j.chom.2020.07.003
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Yang, Q., Yang, J., Wang, Y., Du, J., Zhang, J., Luisi, B. F., et al. (2022). Broad-spectrum chemicals block ROS detoxification to prevent plant fungal invasion. Curr. Biol. 32, 3886–3897.e6. doi: 10.1016/j.cub.2022.07.022
Yin, W., Wang, Y., Chen, T., Lin, Y., and Luo, C. (2018). Functional evaluation of the signal peptides of secreted proteins. Bio Protoc. 8:e2839.
Zhang, L., Ni, H., Du, X., Wang, S., Ma, X.-W., Nürnberger, T., et al. (2017). The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 215, 368–381. doi: 10.1111/nph.14537
Zhang, L., Yan, J., Fu, Z., Shi, W., Ninkuu, V., Li, G., et al. (2021). FoEG1, a secreted glycoside hydrolase family 12 protein from Fusarium oxysporum, triggers cell death and modulates plant immunity. Mol. Plant Pathol. 22, 522–538. doi: 10.1111/mpp.13041
Keywords: Fusarium oxysporum, effector, virulence, plant defense, protease activity
Citation: Wang C, Zheng Y, Liu Z, Qian Y, Li Y, Yang L, Liu S, Liang W and Li J (2023) The secreted FolAsp aspartic protease facilitates the virulence of Fusarium oxysporum f. sp. lycopersici. Front. Microbiol. 14:1103418. doi: 10.3389/fmicb.2023.1103418
Received: 20 November 2022; Accepted: 03 January 2023;
Published: 25 January 2023.
Edited by:
Haitao Cui, Fujian Agriculture and Forestry University, ChinaCopyright © 2023 Wang, Zheng, Liu, Qian, Li, Yang, Liu, Liang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxing Liang,  d2xpYW5nMUBxYXUuZWR1LmNu; Jingtao Li,
d2xpYW5nMUBxYXUuZWR1LmNu; Jingtao Li,  TGlqaW5ndGFvQHFhdS5lZHUuY24=
TGlqaW5ndGFvQHFhdS5lZHUuY24=
†These authors have contributed equally to this work
 Chenyang Wang1†
Chenyang Wang1† Wenxing Liang
Wenxing Liang Jingtao Li
Jingtao Li