- 1Department of Epidemiology and Health Statistics, School of Public Health, Guangdong Pharmaceutical University, Guangzhou, China
- 2The Sixth People's Hospital of Dongguan, Dongguan, China
- 3Department of Acquired Immune Deficiency Syndrome/Sexually Transmitted Disease Control and Prevention, Guangzhou Center for Disease Control and Prevention, Guangzhou, China
Klebsiella aerogenes is a common infectious bacterium that poses a threat to human health. Nevertheless, there are limited data on the population structure, genetic diversity, and pathogenicity of K. aerogenes, especially among men who have sex with men (MSM). The present study aimed to clarify the sequence types (STs), clonal complexes (CCs), resistance genes, and virulence factors of popular strains. Multilocus sequence typing was used to describe the population structure of K. aerogenes. The Virulence Factor Database and Comprehensive Antibiotic Resistance Database were used to assess the virulence and resistance profiles. In this study, next-generation sequencing was performed on nasal swabs specimens collected in an HIV Voluntary Counseling Testing outpatient department in Guangzhou, China, from April to August 2019. The identification results showed that a total of 258 K. aerogenes isolates were collected from 911 participants. We found that the isolates were most resistant to furantoin (89.53%, 231/258) and ampicillin (89.15%, 230/258), followed by imipenem (24.81%, 64/258) and cefotaxime (18.22%, 47/258). The most common STs in carbapenem-resistant K. aerogenes were ST4, ST93, and ST14. The population has at least 14 CCs, including several novel ones identified in this study (CC11-CC16). The main mechanism of drug resistance genes was antibiotic efflux. Based on the presence of the iron carrier production genes irp and ybt, we identified two clusters according to virulence profiles. In cluster A, CC3 and CC4 carry the clb operator encoding the toxin. Increased monitoring is needed for the three main ST type strains carried by MSM. The main clone group CC4 has a large number of toxin genes, and it spreads among MSM. Caution is needed to prevent further spread of this clone group in this population. In sum, our results may provide a foundation for the development of new therapeutic and surveillance strategies for treating MSM.
1. Introduction
Klebsiella aerogenes, previously known as Enterobacter aerogenes, is a Gram-negative bacterium (Tindall et al., 2017). With the development of whole-genome sequencing (WGS), comparative bacterial phylogeny shows that K. aerogenes is more closely related to Klebsiella pneumonia (Chavda et al., 2016). K. aerogenes, an opportunistic pathogen that is widely found in environments such as soil, air, and hospitals, can cause hospital-acquired infections, such as bacteremia and infections of the skin and soft tissue, the respiratory tract, and the urinary tract (Davin-Regli and Pagès, 2015). One of the most common enterobacteria, K. aerogenes, has been associated with hospital infections since 1992 and was once the main enterobacterium responsible for these infections (Chow et al., 1991; De Gheldre et al., 1997). Multidrug-resistant (MDR) K. aerogenes infections are associated with a high mortality rate among patients in intensive care (Davin-Regli and Pagès, 2015). In particular, between 1993 and 2003, K. aerogenes was widespread in several Western European countries, including France, and was considered one of the most dangerous MDR bacteria in Europe (Galdbart et al., 2000; Salso et al., 2003; Chevalier et al., 2008; Brennan et al., 2014). In 2010, K. aerogenes was overtaken by Escherichia coli and Klebsiella pneumoniae as the predominant hospital pathogen because of the prevalence of antibiotic-resistant strains of these bacteria (Davin-Regli and Pagès, 2015). However, K. aerogenes causes more severe septic shock in patients, resulting in higher mortality (Song et al., 2010; Lavigne et al., 2013).
Owing to the widespread and inappropriate use of antibiotics, the emergence of antibiotic-resistant bacteria has accelerated (Martinez et al., 2009). As early as the 1990s, a K. pneumoniae strain resistant to imipenem was found in the United Kingdom (Lavigne et al., 2013). Subsequently, similar resistant strains were reported in France, Spain, the United States, and other European and North American countries (de Champs et al., 1993; Langley et al., 2001; Biendo et al., 2008). Although its prevalence in hospitals has decreased, K. pneumoniae is the most widely expressed carbapenemase among Enterobacteriaceae. It can cause septic shock in patients, thus leading to a high mortality rate of 39% due to its strong toxicity (Song et al., 2010). Carbapenem was once considered the most-effective treatment for Enterobacteriaceae isolates (Tzouvelekis et al., 2012). However, as it is increasingly used in clinical treatment, carbapenem-resistant Enterobacteriaceae is on the rise, and there are severe challenges to clinical treatment. Carbapenem-resistant K. aerogenes (CRKA) is related to nosocomial infections (Chen et al., 2008). The genes encoding carbapenemases, such as IMP, NDM, OXA, and others, typically cause phenotypic resistance to carbapenems (Logan and Weinstein, 2017). Hypervirulence is generally linked to iuc and iro gene loci carried by plasmids, which encode aerobactin and salmochelin. As antibiotic resistance is often associated with virulence (Beceiro et al., 2013), the genomic analysis may be a powerful tool to help control the spread of MDR and virulent strains.
Men who have sex with men (MSM) often engage in high-risk behaviors and have a large number of sexual partners (Zhang et al., 2013; Ahmed et al., 2017). Previous research indicated that they have a higher risk of bacterial infection. Data from England showed an intensified shigellosis epidemic in MSM (Simms et al., 2015). In Italy, the prevalence of nasal colonization with Streptococcus pneumoniae is as high as 15.6% in MSM (Rossotti et al., 2019). In the United States, the prevalence of methicillin-resistant Staphylococcus aureus colonization in community-based MSM is 3.0%, which is two times higher than that in the general population (Leung et al., 2017).
To the best of our knowledge, there is no published genomic analysis of K. aerogenes among MSM. We thus undertook a genomic analysis of K. aerogenes to elucidate the genomic features associated with resistance and virulence and to provide evidence to help prevent outbreaks and limit the spread of this bacterium, mainly among MSM.
2. Material and methods
2.1. Study population
Participants were recruited from the Guangzhou Municipal Center for Disease Prevention and Control and the Lingnan Partner Community Support Center's HIV Voluntary Counseling Testing outpatient department between April 2019 and August 2019. Convenience sampling was used. The study included men above 18 years of age who have had penetrative oral or anal sex with another man in the past 12 months and consented to participate. All participants were asymptomatic colonizers. The study was approved by the Ethics Committee of Guangdong Pharmaceutical University. Patient records and information were anonymized.
2.2. Sample collection
After obtaining informed consent from the study subjects, trained investigators conducted nasal swab sampling. They disinfected their hands, donned sterile gloves, and inserted two sterile cotton swabs moistened with sterile physiological saline into both nasal cavities of the study subject, rotating the swabs for 3–5 times. The cotton swabs were then removed, and the handheld portion was snapped off. The cotton swabs were placed in a 5-ml sampling tube of common meat enrichment broth and were shaken to ensure that the head was fully immersed. The investigator labeled the samples and placed them in a low-temperature storage box. When sampling was complete, the samples were returned to the Molecular Epidemiology Laboratory of Guangdong Pharmaceutical University for culture on the same day.
2.3. Identification of bacterial isolates
The swabs were soaked in a 5-ml enrichment broth containing 1% tryptone, 7.5% sodium chloride, 1% mannitol, and 0.25% yeast extract at 36 ± 1°C for 24 h. The enrichment broth was then inoculated onto mannitol salt agar plates using a loop and incubated at 37°C for 24 h. Samples with pink and viscous colonies with red diffusion in the surrounding medium were considered positive; negative samples were observed again after an additional 48 h of incubation. Gram staining of positive single colonies on mannitol salt agar plates was performed, and the colonies were observed under a microscope; those that were recorded as red were considered Gram negative. The corresponding single colonies of negative samples were then inoculated onto standard nutrient agar plates for bacterial isolation and cultivation.
After the bacterial isolation culture, the sample was sent to the Guangdong Provincial Center for Disease Control and Prevention for species identification with the VITEK MS Clinical Microbial Identification Mass Spectrometry System and the VITEK 2 COMPACT automatic bacteria identification instrument (bioMérieux, Marcy-l'Étoile, Rhône, France) following the manufacturer's instructions. Based on the identification results, the strain initially identified as producing Clostridium perfringens was confirmed as the final result. A total of 258 K. aerogenes isolates were collected from 911 participants.
2.4. Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed using the Kirby–Bauer disk diffusion method according to the guidelines of the Clinical Laboratory Standards Institute (Humphries et al., 2021). A total of 16 antibiotics were tested: imipenem (10 μg), meropenem (10 μg), ertapenem (10 μg), ceftazidime (30 μg), cefepime (30 μg), cefotaxime sodium (30 μg), ampicillin (30 μg), aztreonam (30 μg), gentamicin (10 μg), amikacin (30 μg), tobramycin (30 μg), levofloxacin (5 μg), furantoin (5 μg), tetracycline (30 μg), cotrimoxazole (1.25/23.75 μg), and chloramphenicol (30 μg). K. aerogenes that were not sensitive (drug-resistant or intermediate) to any carbapenem antibiotics (imipenem, meropenem, and ertapenem in this study) were defined as CRKA. All others were defined as carbapenem-susceptible Klebsiella aerogenes (CSKA). Isolates were identified as MDR if they were resistant to ≥3 antibiotic classes.
2.5. Bacterial DNA extraction and WGS
All isolates were cultured on general nutrient agar plates at 36 ± 1°C for 24 h. DNA was extracted with a HiPurA Bacterial DNA kit (Magen, Guangzhou, China) based on the manufacturer's instructions. The extracted DNA was measured for DNA concentration using a NanoDrop ultraviolet (UV)-visible spectrophotometer. The required concentration was >20 ng/μl, with a total amount of 1–2 μg. The obtained DNA was used for WGS. DNA-constructed paired-end libraries were generated for WGS (Illumina HiSeq, San Diego, CA, United States) with a target coverage of 100× and 300 bp length. The quality of the raw reads was assessed with FastQC 0.11.9 (Andrews, 2010). Kraken 2 was used to classify and identify the QC sequence (Wood et al., 2019). Raw reads were assembled with SPAdes (version 3.14) with read error correction enabled (Bankevich et al., 2012).
2.6. Genomic analysis
The Comprehensive Antibiotic Resistance Database was used to assess determinants of resistance in all isolates. Virulence factors were analyzed using the Virulence Factor Database (Liu et al., 2019). To determine the sequence type (ST) of each isolate, we used the pubMLST database of K. aerogenes to identify single-nucleotide polymorphisms (SNPs) in seven housekeeping loci: dnaA, fusA, gyrB, leuS, pryG, rplB, and rpoB (Jolley et al., 2018). Clonal complexes (CCs) were determined by using the eBURST algorithm (Feil et al., 2004). Minimum spanning tree-like structures were illustrated with PHYLOVIZ (version 2.0) via goeBURST Full MST (Francisco et al., 2012).
2.7. Phylogenetic analysis
The maximum-likelihood phylogenetic tree was produced in kSNP3.0 to determine the phylogenetic relationships (Gardner et al., 2015). SNPs within each genome were combined into a single contiguous sequence and subsequently aligned. The final tree was generated and visualized in iTOL (Letunic and Bork, 2021).
To place our circulating strains in the broader context of K. aerogenes strains globally, we included 78 isolates of K. aerogenes with WGS reads available in GenBank. The inclusion criteria were as follows: (1) the assembly level was contig; (2) the assembly submission date was before 2021; and (3) the isolate belonged to CC1, CC3, and CC4. Isolates collected after 2019 were excluded.
2.8. Statistical analysis and visualization of the data
All data were entered into EpiData 3.1 (EpiData Association, Odense, Denmark). We compared the resistance genes of CRKA and CSKA using the Chi-squared test and Fisher's exact test by determining odds ratios and 95% confidence intervals. The heatmap was visualized with the pheatmap package in R (version 4.1.3). Statistical analyses were performed with STATA 16.0 (StataCorp, College Station, TX, USA).
3. Results
3.1. Prevalence of nasal colonization with K. aerogenes
Nasal sampling was performed on 911 MSM in Guangzhou, followed by bacterial cultivation and isolation. A total of 258 strains of K. aerogenes were detected, with a nasal colonization rate of 28.32% (258/911) in this population. Of these strains, 71 (27.52%) of them were CRKA, representing a colonization rate of 7.79% (71/911; Supplementary Figure S1).
3.2. Antimicrobial resistance
Most of the isolates were resistant to furantoin (89.53%, 231/258) and ampicillin (89.53%, 230/258). The resistance rates of six drugs (gentamicin, chloramphenicol, ceftazidime, tobramycin, levofloxacin, and meropenem) were <5%. Approximately 20.16% (52/258) of the isolates were resistant to ≥3 antibiotic classes. In total, 71 strains were determined to be CRKA. Among all CRKA isolates, the most significant resistance was to ampicillin (98.59%, 70/71), and the least resistance was to tobramycin (1.41%, 1/71). Except furantoin (87.70%, 164/187) and ampicillin (85.56%, 160/187), all other antimicrobials of CSKA strains had resistance rates of <10%. The strain was resistant to both carbapenem antibiotics and levofloxacin. Compared to the CSKA strains, CRKA strains were significantly more resistant to imipenem, ertapenem, cefepime, cefotaxime sodium, ampicillin, and aztreonam (Fisher's exact test, p < 0.05). It should be noted that the proportion of multidrug resistance was significantly higher in CRKA isolates than in CSKA isolates (Fisher's exact test, p < 0.05; Table 1; Supplementary Table S1).
3.3. Identification of new K. aerogenes CCs
A total of 135 different sequences were detected among the K. aerogenes isolates. A total of 27 isolates were ST4 (10.47%), 18 were ST93 (6.98%), and 15 were ST14 (5.81%). Of the 135 reported STs, 85.63% (n = 149) of them belonged to a known CC. According to our updated multilocus sequence typing (MLST) analysis of sequenced isolates, at least 14 CCs compose the K. aerogenes population structure. We updated this database locally with 91 new STs and six new CCs, which we named CC11 to CC16 (Figure 1). It was impossible to assign a CC to 84 isolates, including 12 isolates that were not assigned to any STs, which were called NOCC (Supplementary Table S2).
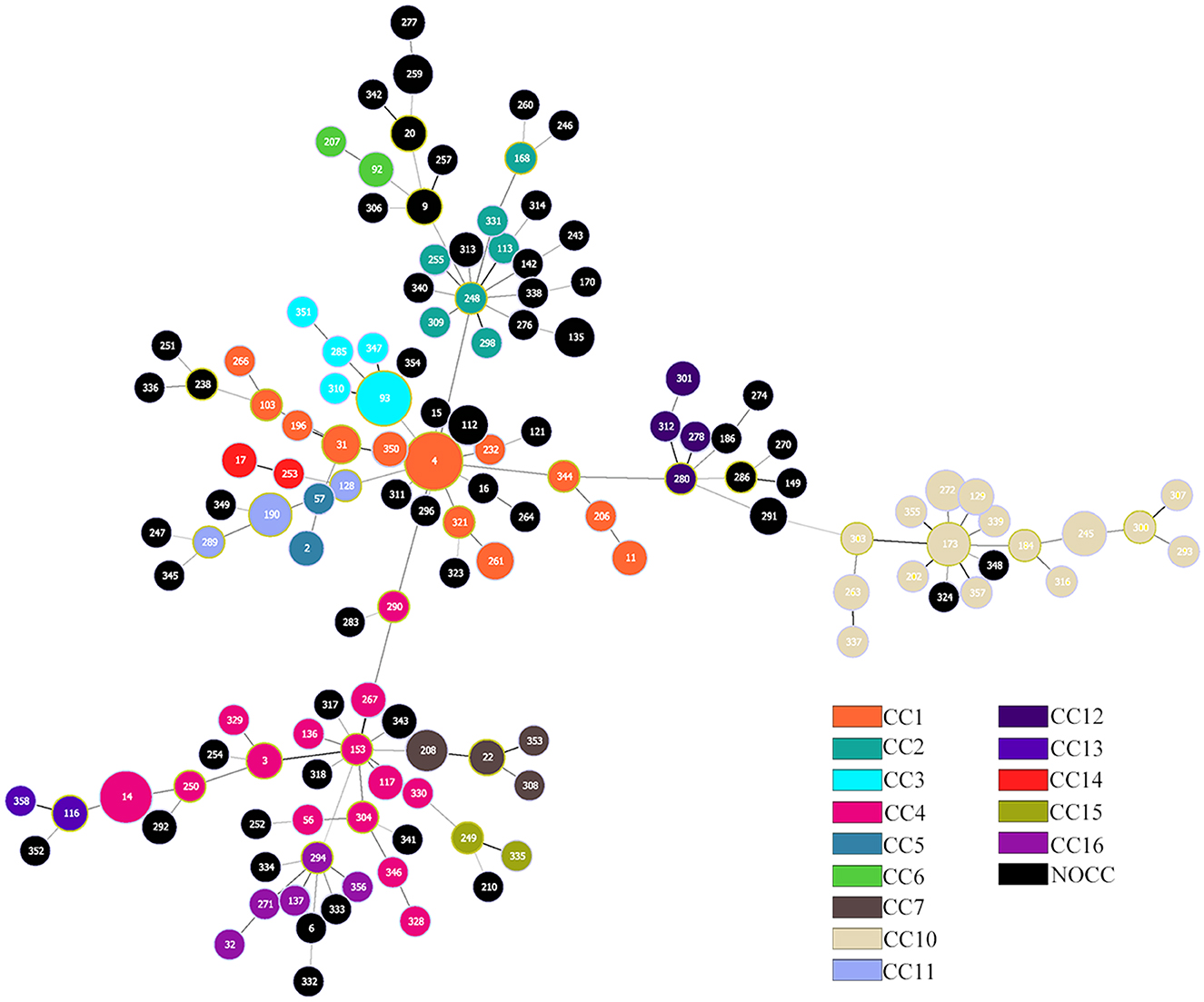
Figure 1. Population structure of Klebsiella aerogenes based on multilocus sequence typing (MLST). Each circle represents a sequence type, and each color represents a different clonal complex (CC). The size of the circle is proportional to the frequency of the sequence type. A total of 258 K. aerogenes isolates are included.
The main CCs of CRKA were CC4 (n = 16, 22.54%), CC1 (n = 11, 15.49%), and CC3 (n = 9, 12.68%). The main STs were ST14 (n = 14, 19.7%), ST4 (n = 7, 9.95%), and ST93 (n = 7, 9.95%), corresponding to CC4, CC1, and CC3. For CSKA, the main STs were ST4 (n = 20, 10.70%) and ST93 (n = 11, 5.88%), corresponding to CC1 and CC3.
3.4. Distribution of resistance gene distribution across K. aerogenes CCs
Using the Comprehensive Antibiotic Resistance Database, we uncovered the presence of 41 different unique resistance genes. The detected genes represented seven resistance mechanisms, including different classes (Figure 2). KpnE, KpnF, rsmA, and adeF were detected in all K. aerogenes isolates. Ten genes were detected in all CRKA isolates: KpnE, KpnF, rsmA, H-NS, CRP, blaampH, eptB, baeR, ArnT, and adeF. Six genes were detected in all CSKA isolates: KpnE, KpnF, rsmA, adeF, marA, and marR. Furthermore, it was found that blaampH, a beta-lactam antibiotic, was present in all CRKA samples. This finding is consistent with the results of the drug sensitivity test (Figure 2). The primary mechanism was antibiotic efflux, accounting for 36.59% (15/41) of all identified genes, which included adeF, baeR, CRP, H-NS, KpnE, KpnF, KpnG, KpnH, and so on. These genes potentially confer resistance to different antibiotic classes, such as aminoglycoside antibiotics and tetracycline antibiotics. The antibiotic inactivation mechanism accounted for 24.39% (10/41) of all identified genes.

Figure 2. Detected antibiotic resistance genes. The prediction of antimicrobial resistance genes was performed using a Resistance Gene Identifier (RGI) search of the Comprehensive Antibiotic Resistance Database. Columns and rows represent antibiotic resistance genes and strains, respectively. Blue means that the gene was detected, and tan means that it was not detected. CRKA, carbapenem-resistant K. aerogenes; CSKA, carbapenem-susceptible K. aerogenes.
By analyzing the frequency of each gene based on the class of antibiotics to which they conferred resistance, we found that only CC1, CC3, CC4, CC10, and NOCC detected genes resistant to sulfonamide and tetracycline (Figure 3).

Figure 3. Relative frequencies of antibiotic genes per clonal complex (CC). Each box shows the percentage of genes associated with resistance to each class of antibiotics. For better visualization, the “multidrug-resistant” category is not shown here.
3.5. Virulence profile analysis reveals two clusters of K. aerogenes strains
We utilized the Virulence Factor Database to identify virulence loci in K. aerogenes, incorporating genes related to adherence, antiphagocytosis, autotransport, and biofilm formation, among others. Upon analysis, we observed that non-pillar adhesion factors pilU, pilW, and mrkC exhibited low detection rates. In contrast, hofC and mrkH had a detection rate of 50%, while the remaining virulence factors, such as the fim gene cluster, pil gene cluster, cafB, and mrk gene cluster, were detected at almost 100%. Virulence factors were not common for all isolates and were highly variable in terms of frequency across genomes. Apparently, cluster A has more iron uptake genes than cluster B. We also found that CC3 and CC4 had more toxin genes than the other clusters in cluster A (Figure 4). Overall, ~40.64% (76/187) of CSKA and 46.48% (33/71) of CRKA encoded 106 or more of the 212 virulence-associated genes tested.

Figure 4. Prevalence of acquired virulence factors. The prediction of virulence genes was performed using a BLAST search of the Virulence Factor Database. Columns and rows represent virulence factors and strains, respectively. Blue means that the gene was detected, and tan means that it was not detected.
3.6. Phylogenetic analysis shows the importance of CCs
Maximum likelihood phylogenetic reconstruction using 33,153 SNPs found in the core genome (Figure 5) indicated that most isolates clustered together according to CC.

Figure 5. Maximum likelihood phylogenomic analysis of Klebsiella aerogenes in our circulating strains. Discriminatory single-nucleotide polymorphisms (SNPs) based on core-genome comparisons were used to plot the tree. Carbapenem-resistant K. aerogenes (CRKA) strains are shown as solid blue squares; multidrug-resistant (MDR) strains are shown as solid red squares. Different colored squares represent different clonal complexes (CCs). Resistant species are shown in brown, and resistance genes are shown in orange. The scale bar indicates their number.
We divided the maximum likelihood tree into six main branches. Some clusters (e.g., CC10) were distant from the main population, which supported the intraspecific diversity of K. aerogenes. In addition, CC4, the primary isolate of CRKA, showed more resistant species and resistance genes than other CCs.
We performed phylogenomic analyses to better understand the emergence and epidemiology of K. aerogenes outbreak clones, and to place our circulating strains in the broader context of K. aerogenes strains globally (Figure 6).
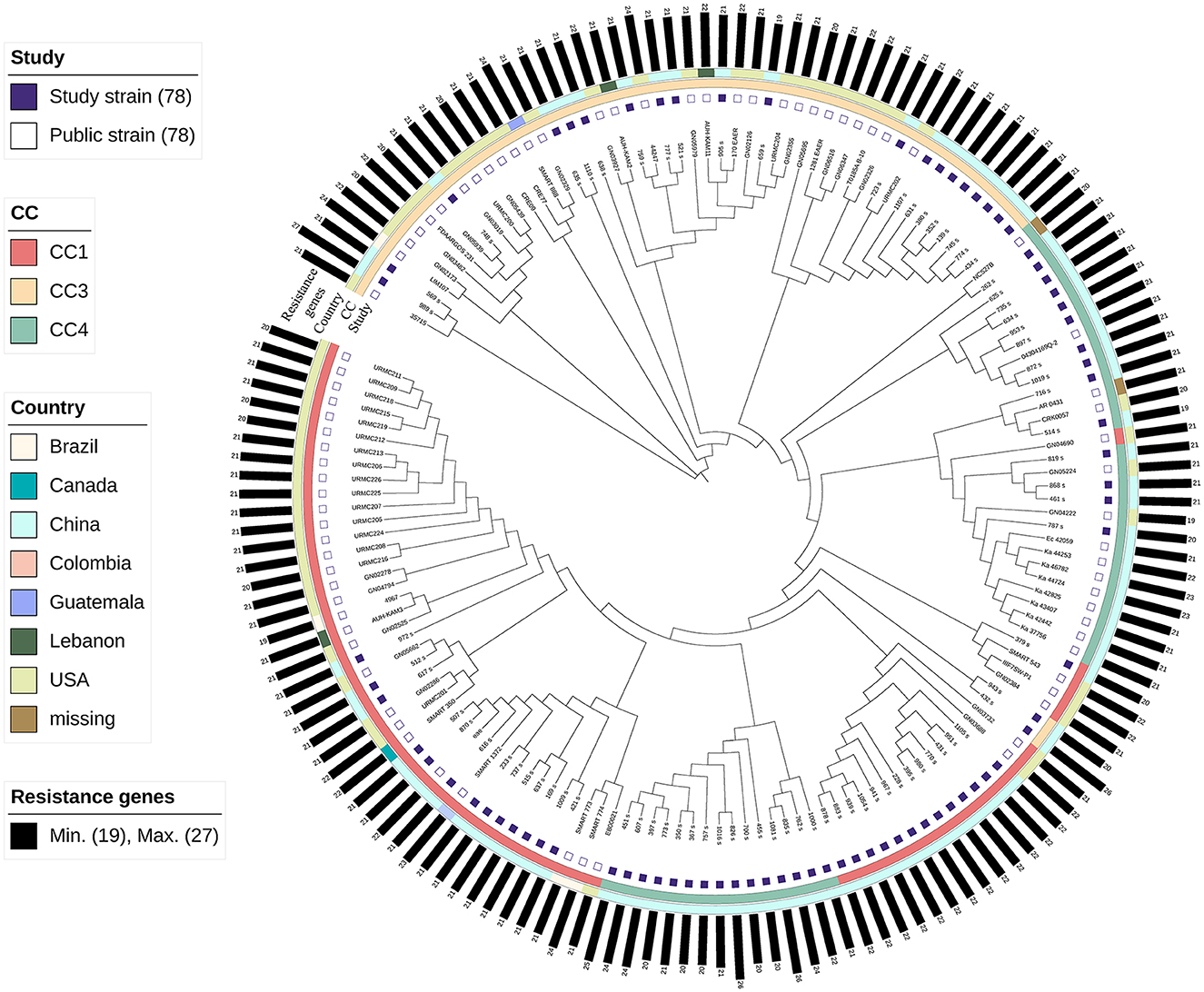
Figure 6. Maximum likelihood phylogenomic comparison of Klebsiella aerogenes genomes to global K. aerogenes from GenBank. Discriminatory single-nucleotide polymorphisms (SNPs) based on core-genome comparisons were used to plot the tree. Different color squares represent different clonal complexes (CCs) and regions. Carbapenem-resistant K. aerogenes strains are shown as green squares.
Because CC3 and CC4 in MSM have particular virulence factors compared to other CCs, and because ST4 is the primary epidemic strain in this population and globally, we included all 79 isolates in our analysis. In addition, we included 79 publicly available genome assemblies of K. aerogenes (Supplementary Table S3).
The results showed that the strains clustered together according to CC. The strains of CC3 were closer than those of CC1 and CC4. Almost all the CC3 strains, except 943_s and 432_s, were clustered. Most of the strains of CC4 originated from China. In CC4, one cluster (n = 15) was closer to CC1, and all belonged to ST14. In CC1, the strains were mainly clustered, except GN04690, representing strains derived from clinical specimens from patients of the United States in 2012.
4. Discussion
In this WGS study, we performed the molecular characterization of 258 K. aerogenes isolates from nasal swabs from MSM. To the best of our knowledge, this is the first study to explore K. aerogenes in MSM by WGS.
The site of highest risk for transmission of bacteria in this population is the rectal or perianal area. Considering that sampling from such areas is not easily acceptable to participants, we chose nasal swabs or throat swabs instead. Studies conducted in Germany have shown that K. aerogenes shows long-term colonization of the nasal vestibule (Köck et al., 2016). To unify the sample source and improve sampling efficiency, we ultimately chose to use nasal swabs. We investigated the nasal colonization in the MSM population and found a prevalence of K. aerogenes of 28.32%, of which 27.52% were CRKA. In a study in Germany of long-term colonization of nasal pathogens in 1,878 healthy people from a community reported the K. aerogenes prevalence of 2.6% with no CRKA detected (Köck et al., 2016). A survey of Gram-negative bacilli in the nasal cavity and hands of medical workers in Tianjin City, China reported a detection rate of 6.41% for K. aerogenes and 1.35% for MDR K. aerogenes (Liu et al., 2017). After 5 months of monitoring at a hospital in Kathmandu, Nepal, we observed that the carrying rate of K. aerogenes in hospitalized patients was 0.9% (Pandey et al., 2021). This is significantly lower than that in the nasal cavity of our population. Previous CRKA studies have mostly focused on hospital strains. The detection rate of CRKA in a hospital in Shanghai was 21% (Qin et al., 2014), and the detection rate of CRKA in a hospital in the United States was 12.5% (Guh et al., 2015). The reason for the differences among these studies may be because of the heavy use of antibiotics, which leads to the emergence of more CRKA over time.
MLST enriched epidemiological and evolutionary studies of K. aerogenes. In this study, we identified 91 new STs, showing epidemic strains ST4, ST93, and ST14. Malek et al. showed that ST4 and ST93 were dominant global clones associated with K. aerogenes infections (Malek et al., 2019). Our study showed that ST4 and ST93 were common in a community environment. In our study, ST14 was mainly found in CRKA, and few studies mentioned it. More large-scale studies are needed to examine strains from diverse global locations. This will help elucidate the emergence of K. aerogenes antibiotic resistance and the evolution of pathogenicity. Several recent studies have identified new ST types (Passarelli-Araujo et al., 2019; Shen et al., 2019; Pan et al., 2021) and have reported a diversity of K. aerogenes isolates. It is expected that several new ST types will be identified as samples from different countries and sequenced. Besides MLST, CCs are important in evolutionary studies. In our study, CC1 was the most common strain, and the founder genotype was ST4. Phylogenomic analyses indicated that identifying STs and CCs is useful for epidemiological and genetic studies of K. aerogenes. The same CCs were generally clustered together.
K. aerogenes poses complex challenges because of its drug resistance. There are many types of antibiotic-resistance genes. The International Antibiotic Resistance Gene Database has reported 13,000 resistance genotypes (Liu and Pop, 2008). Among them, bacterial efflux systems are the determinant of multidrug resistance (Sharma et al., 2019), which are consistent with our findings. The primary efflux pump KpnEFGH gene cluster, which belongs to the small multidrug resistance group, uses proton motive force or ATP energy to flush antibiotics from cells (Srinivasan and Rajamohan, 2013). An analysis revealed kpnEF in >70% of isolates (Srinivasan and Rajamohan, 2013). In our study, KpnE and KpnF were detected in all isolates. This may mean that KpnEF is increasingly important in drug resistance over time.
We found that most isolates of K. aerogenes (n = 224, 86.82%) carried a salmochelin locus. This result is similar to the findings of a small-scale study and is consistent with global research data (Passarelli-Araujo et al., 2019; Spadar et al., 2023). This indicates that salmochelin toxin-producing E. coli is widely present in K. aerogenes, and the results from the general population are consistent with those from the MSM population. We found differences in the resistance genes carried by different clone groups. CC3 and CC4 carried more drug resistance species than other clone groups, especially when the detection rate of tetracycline and sulfonamide drug-related resistance genes was higher than that of the other clone groups. The CC3 result is similar to the results of a 2019 study, but that study did not include CC4, and CC10 carried more aminoglycoside resistance genes (Passarelli-Araujo et al., 2019). One possible reason for the difference is that the strains in that study were mainly from hospitals in Europe and the United States, unlike the strains in our study.
We created a presence/absence matrix to assess the virulence profile to investigate whether different virulence factors are present in different CCs. The results showed a clear separation between less virulent and more virulent strains. Cluster A carried ybt operon, which is related to the production of yersiniabactin siderophores. Ybt is the main virulence factor produced during K. pneumoniae lung infection (Holden et al., 2016). Within cluster A, we found that CC3 and CC4 carried the clb operon, which is required for the biosynthesis of colibactin (Lu et al., 2017). Our study is consistent with previous research that indicates that ST93, which belongs to CC3, has a higher propensity for the production of enterobacterial toxins (Malek et al., 2019). Our study recommends that a matrix can effectively differentiate the virulence profiles of different CCs. This finding is significant, as the identification of CCs can be crucial for controlling the spread of K. aerogenes strains by enabling targeted measures to be taken against highly virulent strains compared to the less-virulent ones.
The comparison of global strains revealed that the CC3 distance is longer than the other strains, similar to previous studies (Passarelli-Araujo et al., 2019). It is crucial to give special consideration to the CC4 strains because the majority of these strains originated from China. Additionally, almost all of the CC4 strains identified in this study are MDRKA and CRKA, with a higher number of clb virulence factors compared to other clonal strains. This may indicate that CC4 is a unique K. aerogenes strain type prevalent in China. Alternatively, it may imply that there is low diversity within this group and that the variation among strains from different origins is minimal. Therefore, vigilance is essential to monitor for any potential transmission among this population or others.
This study has several limitations. First, it was a cross-sectional study. Long-term monitoring and follow-up of MSM were not performed. Second, only a single Voluntary Counseling Testing outpatient clinic was involved. Third, the effect of oral sex on bacterial carriage in the MSM population was not considered, and throat swab sampling was not used.
In summary, to the best of our knowledge, this study is the first to report a genomic analysis of K. aerogenes in MSMs. This comprehensive strain profiling analysis reveals the bacterial STs, phylogenetic relationships, and structure of K. aerogenes strains recovered from MSMs. In addition, our study demonstrated that identifying CCs has powerful implications for the biological aspects of K. aerogenes (e.g., virulence and resistance). Our results provide new insights for the future applications of WGS in molecular epidemiology and culture-independent genome monitoring of related pathogens as more samples from different countries are sequenced.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA937033.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of Guangdong Pharmaceutical University. The participants provided their written informed consent to participate in this study.
Author contributions
QC and ZM conceptualized, designed the methodology, did a formal analysis, and wrote the original draft. ZG scrubbed data and maintained research data. YL and JG collected literature and visualized the data. XY, ZH, and ZY reviewed and revised the article, obtained funding, and supervised this study. All authors read and approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant no. 81973069).
Acknowledgments
The authors are grateful to all of the study participants.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1102907/full#supplementary-material
References
Ahmed, N., Chung, E., Morris-Jones, S., and Miller, R. F. (2017). Correspondence to invasive shigellosis in MSM. Int. J. STD AIDS 28, 421–422. doi: 10.1177/0956462417691441
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Bioinformatics, Babraham Institute.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beceiro, A., Tomás, M., and Bou, G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26, 185–230. doi: 10.1128/CMR.00059-12
Biendo, M., Canarelli, B., Thomas, D., Rousseau, F., Hamdad, F., Adjide, C., et al. (2008). Successive emergence of extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacter aerogenes isolates in a university hospital. J. Clin. Microbiol. 46, 1037–1044. doi: 10.1128/JCM.00197-07
Brennan, B. M., Coyle, J. R., Marchaim, D., Pogue, J. M., Boehme, M., Finks, J., et al. (2014). Statewide surveillance of carbapenem-resistant Enterobacteriaceae in Michigan. Infect. Control Hosp. Epidemiol. 35, 342–349. doi: 10.1086/675611
Chavda, K. D., Chen, L., Fouts, D. E., Sutton, G., Brinkac, L., Jenkins, S. G., et al. (2016). Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. MBio 7, e02093–e02016. doi: 10.1128/mBio.02093-16
Chen, Y. G., Zhang, Y., Yu, Y. S., Qu, T. T., Wei, Z. Q., Shen, P., et al. (2008). In vivo development of carbapenem resistance in clinical isolates of Enterobacter aerogenes producing multiple beta-lactamases. Int. J. Antimicrob. Agents 32, 302–307. doi: 10.1016/j.ijantimicag.2008.02.014
Chevalier, J., Mulfinger, C., Garnotel, E., Nicolas, P., Davin-Regli, A., and Pages, J. M. (2008). Identification and evolution of drug efflux pump in clinical Enterobacter aerogenes strains isolated in 1995 and 2003. PLoS ONE 3, e3203. doi: 10.1371/journal.pone.0003203
Chow, J. W., Fine, M. J., Shlaes, D. M., Quinn, J. P., Hooper, D. C., Johnson, M. P., et al. (1991). Enterobacter bacteremia: clinical features and emergence of antibiotic resistance during therapy. Ann. Intern. Med. 115, 585–590. doi: 10.7326/0003-4819-115-8-585
Davin-Regli, A., and Pagès, J. M. (2015). Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6, 392. doi: 10.3389/fmicb.2015.00392
de Champs, C., Henquell, C., Guelon, D., Sirot, D., Gazuy, N., and Sirot, J. (1993). Clinical and bacteriological study of nosocomial infections due to Enterobacter aerogenes resistant to imipenem. J. Clin. Microbiol. 31, 123–127. doi: 10.1128/jcm.31.1.123-127.1993
De Gheldre, Y., Maes, N., Rost, F., De Ryck, R., Clevenbergh, P., Vincent, J. L., et al. (1997). Molecular epidemiology of an outbreak of multidrug-resistant Enterobacter aerogenes infections and in vivo emergence of imipenem resistance. J. Clin. Microbiol. 35, 152–160. doi: 10.1128/jcm.35.1.152-160.1997
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Francisco, A. P., Vaz, C., Monteiro, P. T., Melo-Cristino, J., Ramirez, M., and Carriço, J. A. (2012). PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformat. 13, 87. doi: 10.1186/1471-2105-13-87
Galdbart, J. O., Lemann, F., Ainouz, D., Feron, P., Lambert-Zechovsky, N., and Branger, C. (2000). TEM-24 extended-spectrum beta-lactamase-producing Enterobacter aerogenes: long-term clonal dissemination in French hospitals. Clin. Microbiol. Infect. 6, 316–323. doi: 10.1046/j.1469-0691.2000.00092.x
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3. 0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Guh, A. Y., Bulens, S. N., Mu, Y., Jacob, J. T., Reno, J., Scott, J., et al. (2015). Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA 314, 1479–1487. doi: 10.1001/jama.2015.12480
Holden, V. I., Breen, P., Houle, S., Dozois, C. M., and Bachman, M. A. (2016). Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. MBio 7, e01397–e01316. doi: 10.1128/mBio.01397-16
Humphries, R., Bobenchik, A. M., Hindler, J. A., and Schuetz, A. N. (2021). Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100. J. Clin. Microbiol. 59, e00213–00221. doi: 10.1128/JCM.00213-21
Jolley, K. A., Bray, J. E., and Maiden, M. C. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST. org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Köck, R., Werner, P., Friedrich, A., Fegeler, C., Becker, K., Bindewald, O., et al. (2016). Persistence of nasal colonization with human pathogenic bacteria and associated antimicrobial resistance in the German general population. New Microb. New Infect. 9, 24–34. doi: 10.1016/j.nmni.2015.11.004
Langley, J. M., Hanakowski, M., and Leblanc, J. C. (2001). Unique epidemiology of nosocomial urinary tract infection in children. Am. J. Infect. Control 29, 94–98. doi: 10.1067/mic.2001.111537
Lavigne, J. P., Sotto, A., Nicolas-Chanoine, M. H., Bouziges, N., Pages, J. M., and Davin-Regli, A. (2013). An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int. J. Antimicrob. Agents 41, 130–136. doi: 10.1016/j.ijantimicag.2012.10.010
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Leung, N. S., Vidoni, M. L., Robinson, D. A., Padgett, P., and Brown, E. L. (2017). A community-based study of Staphylococcus aureus nasal colonization and molecular characterization among men who have sex with men. LGBT Health 4, 345–351. doi: 10.1089/lgbt.2017.0016
Liu, B., and Pop, M. (2008). ARDB—Antibiotic resistance genes database. Nucl. Acids Res. 37(suppl_1), D443–D447. doi: 10.1093/nar/gkn656
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucl. Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Liu, H., Fei, C. N., Zhang, Y., Liu, G. W., Liu, J., and Dong, J. (2017). Presence, distribution and molecular epidemiology of multi-drug-resistant Gram-negative bacilli from medical personnel of intensive care units in Tianjin, China, 2007-2015. J. Hosp. Infect. 96, 101–110. doi: 10.1016/j.jhin.2017.01.012
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215(suppl_1), S28–S36. doi: 10.1093/infdis/jiw282
Lu, M. C., Chen, Y. T., Chiang, M. K., Wang, Y. C., Hsiao, P. Y., Huang, Y. J., et al. (2017). Colibactin contributes to the hypervirulence of pks(+) K1 CC23 Klebsiella pneumoniae in mouse meningitis infections. Front. Cell. Infect. Microbiol. 7, 103. doi: 10.3389/fcimb.2017.00103
Malek, A., McGlynn, K., Taffner, S., Fine, L., Tesini, B., Wang, J., et al. (2019). Next-generation-sequencing-based hospital outbreak investigation yields insight into Klebsiella aerogenes population structure and determinants of carbapenem resistance and pathogenicity. Antimicrob. Agents Chemother. 63, e02577–e02518. doi: 10.1128/AAC.02577-18
Martinez, J. L., Fajardo, A., Garmendia, L., Hernandez, A., Linares, J. F., Martínez-Solano, L., et al. (2009). A global view of antibiotic resistance. FEMS Microbiol. Rev. 33, 44–65. doi: 10.1111/j.1574-6976.2008.00142.x
Pan, F., Xu, Q., and Zhang, H. (2021). Emergence of NDM-5 producing carbapenem-resistant Klebsiella aerogenes in a pediatric hospital in Shanghai, China. Front. Public Health 9, 621527. doi: 10.3389/fpubh.2021.621527
Pandey, R., Mishra, S. K., and Shrestha, A. (2021). Characterisation of ESKAPE pathogens with special reference to multidrug resistance and biofilm production in a Nepalese hospital. Infect. Drug Resist. 14, 2201–2212. doi: 10.2147/IDR.S306688
Passarelli-Araujo, H., Palmeiro, J. K., Moharana, K. C., Pedrosa-Silva, F., Dalla-Costa, L. M., and Venancio, T. M. (2019). Genomic analysis unveils important aspects of population structure, virulence, and antimicrobial resistance in Klebsiella aerogenes. FEBS J. 286, 3797–3810. doi: 10.1111/febs.15005
Qin, X., Yang, Y., Hu, F., and Zhu, D. (2014). Hospital clonal dissemination of Enterobacter aerogenes producing carbapenemase KPC-2 in a Chinese teaching hospital. J. Med. Microbiol. 63, 222–228. doi: 10.1099/jmm.0.064865-0
Rossotti, R., Merli, M., Cantone, M., Moioli, M. C., Motta, D., Orcese, C., et al. (2019). Risk factors for community-acquired bacterial meningitis: men who have sex with men (MSM) as a population at risk for meningococcal nasopharyngeal carriage. Infect Dis. 51, 714–717. doi: 10.1080/23744235.2019.1642512
Salso, S., Culebras, E., Andrade, R., and Picazo, J. J. (2003). Outbreak of TEM-24-producing Enterobacter aerogenes in a Spanish hospital. Microb. Drug Resist. 9, 299–305. doi: 10.1089/107662903322286517
Sharma, A., Gupta, V. K., and Pathania, R. (2019). Efflux pump inhibitors for bacterial pathogens: from bench to bedside. Indian J. Med. Res. 149, 129–145. doi: 10.4103/ijmr.IJMR_2079_17
Shen, X., Liu, L., Yu, J., Cao, X., Zhan, Q., Guo, Y., et al. (2019). Coexistence of bla NDM-1 and rmtC on a transferrable plasmid of a novel ST192 Klebsiella aerogenes clinical isolate. Infect. Drug Resist. 3883–3891. doi: 10.2147/IDR.S228130
Simms, I., Field, N., Jenkins, C., Childs, T., Gilbart, V. L., Dallman, T. J., et al. (2015). Intensified shigellosis epidemic associated with sexual transmission in men who have sex with men - Shigella flexneri and S. sonnei in England, 2004 to end of February 2015. Eurosurveillance 20, 21097. doi: 10.2807/1560-7917.ES2015.20.15.21097
Song, E. H., Park, K. H., Jang, E. Y., Lee, E. J., Chong, Y. P., Cho, O. H., et al. (2010). Comparison of the clinical and microbiologic characteristics of patients with Enterobacter cloacae and Enterobacter aerogenes bacteremia: a prospective observation study. Diagn. Microbiol. Infect. Dis. 66, 436–440. doi: 10.1016/j.diagmicrobio.2009.11.007
Spadar, A., Perdigão, J., Campino, S., and Clark, T. G. (2023). Large-scale genomic analysis of global Klebsiella pneumoniae plasmids reveals multiple simultaneous clusters of carbapenem-resistant hypervirulent strains. Genome Med. 15, 1–12. doi: 10.1186/s13073-023-01153-y
Srinivasan, V. B., and Rajamohan, G. (2013). KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 57, 4449–4462. doi: 10.1128/AAC.02284-12
Tindall, B. J., Sutton, G., and Garrity, G. M. (2017). Enterobacter aerogenes Hormaeche and Edwards 1960 (Approved Lists 1980) and Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980) share the same nomenclatural type (ATCC 13048) on the Approved Lists and are homotypic synonyms, with consequences for the name Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980). Int. J. Syst. Evol. Microbiol. 67, 502–504. doi: 10.1099/ijsem.0.001572
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 1–13. doi: 10.1186/s13059-019-1891-0
Keywords: antimicrobial resistance, Enterobacteriaceae, pathogens, whole genome sequencing (WGS), multidrug-resistant (MDR)
Citation: Cheng Q, Ma Z, Gong Z, Liang Y, Guo J, Ye X, Han Z and Yao Z (2023) Whole-genome sequencing analysis of Klebsiella aerogenes among men who have sex with men in Guangzhou, China. Front. Microbiol. 14:1102907. doi: 10.3389/fmicb.2023.1102907
Received: 19 November 2022; Accepted: 09 May 2023;
Published: 02 June 2023.
Edited by:
Ben Pascoe, University of Oxford, United KingdomReviewed by:
Gabriel Augusto Marques Rossi, Vila Velha University, BrazilPolly Soo Xi Yap, Monash University Malaysia, Malaysia
Copyright © 2023 Cheng, Ma, Gong, Liang, Guo, Ye, Han and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Han, ODYzODMzNzI4QHFxLmNvbQ==; Zhenjiang Yao, NDk3MzQ1MjQwQHFxLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Qi Cheng
Qi Cheng Zheng Ma2†
Zheng Ma2† Yuelang Liang
Yuelang Liang