
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Microbiol., 30 January 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1094877
This article is part of the Research TopicLegionella pneumophila-Transmission, Pathogenesis, Host-pathogen interaction, Prevention and TreatmentView all 7 articles
Legionella pneumophila is a waterborne pathogen and, as the causative agent of Legionnaires’ disease, a significant public health concern. Exposure to environmental stresses, and disinfection treatments, promotes the formation of resistant and potentially infectious viable but non-culturable (VBNC) Legionella. The management of engineered water systems to prevent Legionnaires’ disease is hindered by the presence of VBNC Legionella that cannot be detected using the standard culture (ISO11731:2017-05) and quantitative polymerase reaction (ISO/TS12869:2019) methods. This study describes a novel method to quantify VBNC Legionella from environmental water samples using a “viability based flow cytometry-cell sorting and qPCR” (VFC + qPCR) assay. This protocol was then validated by quantifying the VBNC Legionella genomic load from hospital water samples. The VBNC cells were unable to be cultured on Buffered Charcoal Yeast Extract (BCYE) agar; however, their viability was confirmed through their ATP activity and ability to infect amoeba hosts. Subsequently, an assessment of the ISO11731:2017-05 pre-treatment procedure demonstrated that acid or heat treatment cause underestimation of alive Legionella population. Our results showed that these pre-treatment procedures induce culturable cells to enter a VBNC state. This may explain the observed insensitivity and lack of reproducibility often observed with the Legionella culture method. This study represents the first time that flow cytometry-cell sorting in conjunction with a qPCR assay has been used as a rapid and direct method to quantify VBNC Legionella from environmental sources. This will significantly improve future research evaluating Legionella risk management approaches for the control of Legionnaires’ disease.
Legionella is an opportunistic premise plumbing pathogen associated with Legionnaires’ disease (LD) and Pontiac fever. It is ubiquitously present in natural and engineered water systems (Fields et al., 2002). Aerosols generated from cooling towers, showers, humidifiers and other fabricated water distribution systems are major sources of LD (Bartram et al., 2007). Within these engineered water sources, microbial biofilms and host protozoa protect Legionella from environmental stresses and disinfection procedures. Furthermore, disinfection protocols (Allegra et al., 2008; Casini et al., 2018) and environmental stresses (Li et al., 2014) cause culturable Legionella to enter a viable but non-culturable (VBNC) state. VBNC Legionella tolerate environmental stresses and are still able to infect human macrophages (Dietersdorfer et al., 2018) and alveolar epithelial cells (Epalle et al., 2015). Under favorable conditions, VBNC Legionella can be resuscitated using protozoan hosts to generate culturable Legionella (Steinert et al., 1997; Garcia et al., 2007).
Over the last decade, in the United States and across Europe the number of reported LD cases has been significantly increasing (The European Centre for Disease Prevention and Control, n.d.; Smith et al., 2007; National Notifiable Diseases Surveillance System and Centers for Disease Control and Prevention, 2018). In 2022, the European Centre for Disease Prevention and Control reported 8,372 confirmed cases of LD, of which 66.9% were community associated and 5.1% were nosocomial infections (The European Centre for Disease Prevention and Control, n.d.). During COVID-19 pandemic, lockdown resulted in water stagnation in some buildings, which ultimately increased Legionella pneumophila contamination (Liang et al., 2021; Chao and Lai, 2022). Furthermore, climate change, urbanization and new energy conservation approaches are also increasing the risk of legionellosis (both LD and Pontiac fever; Connolly et al., 2021; Gattuso et al., 2022). As such there is a need for improved risk management protocols to reduce the risk of legionellosis.
One of the challenges preventing the improved control of Legionella in engineered water systems is the uncertainty associated with standard detection methods (Whiley, 2016). The International Organization for Standardization (ISO) provides two protocols for the detection and quantification of Legionella contamination in potable water. ISO11731:2017-05 is a culture based method that detects only culturable Legionella (International Organization for Standardization, 2017); whereas, ISO/TS12869:2019 is a quantitative PCR (qPCR) based assay which estimates the bacterial genomic load (International Organization for Standardization, 2019). The culture method is considered the gold standard for Legionella detection and characterization. However, it is time consuming, taking 10–14 days, and does not detect VBNC Legionella resulting in underestimation of Legionella numbers and false negatives (Whiley and Taylor, 2016; International Organization for Standardization, 2017). In contrast, the qPCR method is faster (International Organization for Standardization, 2019); however, it quantifies both the live, VBNC and dead Legionella, resulting in overestimations and false positive results (Whiley and Taylor, 2016).
The concentration of culturable Legionella is typically low in potable water, however, previous studies have found high concentration of Legionella DNA using qPCR quantification (e.g., 103–107 GU/L) in culture negative potable water samples (Casini et al., 2018; Dai et al., 2019; Hayes-Phillips et al., 2019). Several methods have been used to try to overcome this discrepancy observed between the two standard methods (culture and qPCR). Ethidium monoazide (EMA) or propidium monoazide (PMA) based viability qPCR (Delgado-Viscogliosi et al., 2009; Taylor et al., 2014; Zacharias et al., 2015) and fluorescence in situ hybridization (FISH; Whiley et al., 2011) assays are alternative approaches designed to differentiate between viable and non-viable Legionella from environmental samples. However, background microbial populations compromise the validity of both techniques (Zacharias et al., 2015) and PMA and EMA have a concentration depended cytotoxic effect that can make it challenging to use in environmental samples of with unknown cell concentrations (Fittipaldi et al., 2012). Also, VBNC bacterial cells show low metabolic activity and rRNA contents, as such, FISH is not an effective technique to detect and characterize VBNC Legionella (Satoh et al., 2002). Catalyzed reporter deposition FISH (CARD-FISH) technique was designed to overcome lower contents of rRNA. It was used for direct enumeration of L. pneumophila from hospital water system (Kirschner et al., 2012). However, CARD-FISH detect and enumerate non-viable cells as well. Alternatively, combined microcolonies cultivation-FISH-solid phase cytometry assay was designed to detect and quantify viable Legionella (Baudart et al., 2015). However, this method cannot detect VBNC Legionella. Legionella-amoebae coculture assay is another technique used to detect VBNC Legionella from environmental samples (Conza et al., 2013). However, it is very time-consuming and is unable to quantify the population of VBNC Legionella.
Flow cytometric analysis of environmental samples has previously been used to characterize total bacterial VBNC populations (Sachidanandham et al., 2005; Khan et al., 2010). However, this method does not enable the discrimination and quantification of specific species from mixed bacterial population (Zacharias et al., 2015). Previous studies have also used flow cytometry to characterize VBNC Legionella generated by physical and chemical disinfection procedures, but these studies used pure cultures of L. pneumophila (Allegra et al., 2008; Mustapha et al., 2015). Another method used flow cytometry with specific fluorogenic antibodies to detect L. pneumophila from environmental samples (2010). In this method, FITC (ab20818, abcam) and Alexa (GTX40943, GeneTex) conjugated L. pneumophila sg1 (ATCC® 33152™) specific polyclonal antibodies were applied to detected L. pneumophila sg1 (ATCC® 33152™) which had been used to spike a water sample. Antibodies required for a universal assay must target epitope(s) that are well expressed in all serogroups and VBNC Legionella. In our study, to overcome this problem, instead of bacteria specific antibodies, dual staining flow cytometry-cell sorting was used, and genomic load estimated by a standard qPCR assay.
The aim of this study was to develop a rapid and robust “viability based flow cytometry-cell sorting and qPCR” (VFC + qPCR) assay to detect and quantify VBNC Legionella from engineered water systems. Following the optimization of the assay using pure Legionella cultures, its performance was validated using environmental water samples. It was also used to assess the impact of the pre-treatments described in the ISO11731:2017-05 culture method on Legionella recovery.
Legionella pneumophila subsp. pneumophila Philadelphia serogroup 1 (ATCC® 33152™) was cultured on BCYE (buffered charcoal yeast extract: CM0655, Oxoid Ltd.) agar supplemented with GVPC (glycine, vancomycin, polymyxin B and cycloheximide: SR0152, Oxoid Ltd.) and Legionella growth supplement (buffer/potassium hydroxide, ferric pyrophosphate, L-cysteine and α-ketoglutarate: SR0110C, Oxoid Ltd.); and incubated at 37 ± 1°C for 3–4 days. Escherichia coli HS (pFamp) R (ATCC® 700891™) and Acinetobacter calcoaceticus (environmental isolate) were cultured on nutrient and MacConkey agar, respectively; and incubated at 37 ± 1°C for 24 h. Acanthamoeba polyphaga (ATCC® 30461™) cells were cultured in a T25 flask (156367, Nunc™ EasYFlask™, Thermo Fisher Scientific) containing 4.5 mL PYG broth (2% peptone, 0.2% yeast extract and 1.8% D-glucose) supplemented with 10% heat inactivated fetal bovine serum (FBS: 10100139, Gibco®, Thermo Fisher Scientific) and incubated at 25 ± 1°C for 4–5 days. Whenever washing was required 1X phosphate-buffered saline (PBS: 003002, Invitrogen™, Thermo Fisher Scientific) and 1X Page’s saline (0.12 g NaCl, 0.004 g MgSO4.5H2O, 0.004 g CaCl2.2H2O, 0.142 g Na2HPO4, and 0.136 g KH2PO4 per liter distilled water, pH 6.8 ± 0.2) were used for bacterial strains and amoebae, respectively.
A water sample spiked with 106 CFU/L L. pneumophila was prepared. The sample was concentrated through filtration. After filtration, the contents present on the membrane were carefully resuspended in 1X PBS. This concentrated sample was processed according to the ISO11731:2017-05 standard culture method, which recommends heat or acid pretreatment to remove background microbes from water samples. For the heat treated sample, 2 mL of sample was heated at 50 ± 1°C for 30 ± 2 min, whereas for the acid pretreatment procedure, 1 mL sample was mixed with 9 mL of acid buffer (3.5 mL 0.2 M HCl and 25 mL 0.2 M KOH, final pH 2.2) and incubated at room temperature for 5 ± 0.5 min (International Organization for Standardization, 2017). Then untreated, heat and acid treated samples were cultured on BCYE-GVPC agar and tested using the VFC + qPCR assay described below. The data was analyzed using R (version 4.2.2) package agricolae (version 1.3-5; De Mendiburu, 2021). Firstly, normality was assessed using the Shapiro–Wilk test, then one way ANOVA (analysis of variance) was performed followed by Tukey’s HSD (honestly significant difference) test. Finally, graph was designed using ggplot2 (version 3.3.6) in the R environment (Wickham, 2016).
To prepare Legionella suspension, 5 mL of 1X PBS (temperature preadjusted to 37 ± 1°C) was pipetted onto an agar plate containing Legionella colonies. These colonies were harvested by gently scrapping with a spreader to resuspended them in the PBS. The suspension containing harvested cells was then pipetted into a sterile 10 ml tube and repeatedly pipette mixed. Finally, 2.5 mL of the cell suspension was transferred to a 10 mL sterile tube containing 2.5 mL of PBS and again pipette mixed thoroughly. This was used as bacterial stock solution and diluted accordingly.
The BD™ cell viability kit (349480, BD™) was used for staining and estimation of alive, dead and injured cell populations (Alsharif and Godfrey, 2002). Briefly, 100 μL of Legionella cells were mixed in 400 μL filter sterilized staining buffer (1 mM EDTA and 0.01% tween-20 in 1X PBS, pH 7.4 ± 0.1). In this mixture 420 nM of thiazole orange (TO; λ(excitation)/λ(emission): 512/533 nm) and 48 μM propidium iodide (PI; λ(excitation)/λ(emission): 537/618 nm) were added and vortexed. Sample tubes were incubated at 5°C for 15 min, then 50 μL of counting beads were added in each tube. Using a BD™ FACSAria™ Fusion instrument (BD Biosciences), cells were analyzed and the TO (FL1) vs. PI (FL3) fluorescence plots were used to distinguish alive, dead and injured cells. Untreated cells and heat killed cell (75°C for 10 min) were used as controls to define gates distinguishing alive and dead cell populations, respectively. The bacterial populations were first gated based on their forward (FSC) and side scatter (SSC) properties (Supplementary Figure S2). During optimization of the protocol, control samples containing predominantly dead or alive Legionella cells were used to position the quadrant gate. A blank control of buffer containing dyes was also included to confirm that debris was not being stained with the dyes and was not included in the gated populations (Supplementary Figure S1). All gated cell populations were examined to monitor viability and culturability of Legionella. Depending upon samples type and cell number, approximately 103–106 alive, dead, and injured cell fractions were then sorted into different tubes for further characterization.
To confirm that TO and PI dyes did not interfere with the qPCR assay, sorted cell fractions (103–105 cells from each fraction) were subjected to DNA extraction and qPCR. The results of TO and PI dyed cell fractions were compared with the unstained Legionella and gBlocks gene fragments (IDT™). DNA was extracted using the Aquadien™ DNA extraction and purification kit (3578121, BIO-RAD Laboratories Ltd.). According to the guidelines of ISO/TS12869:2019 (International Organization for Standardization, 2019), the 16S rDNA gene was amplified and quantified. Briefly, gBlocks gene fragments (IDT™) were used as standard DNA with 10-fold serial dilution with concentrations ranging from 100 pg/μL to 0.00001 pg/μL. Using 2X SsoAdvanced™ universal probes supermix (172–5281, BIO-RAD Laboratories Ltd.) and 16S rDNA specific oligos (BIO-RAD Laboratories Ltd., Table 1), the reaction mix was prepared and subjected to a Rotor-Gene Q thermal cycler (QIAGEN Ltd.) for qPCR assay. In the assays, a channel with λ(source) 470 nm and λ(detector) 510 nm was used for detection of 6-carboxyfluorescein (λ(excitation)/λ(emission) 495/520 nm) and Iowa Black® FQ quencher labeled fluorogenic probe (BIO-RAD Laboratories Ltd.). Based on 10-fold serial dilution and qPCR assay, the limit of detection and limit of quantification were estimated. A standard curve was used to estimate the Legionella GU/L from alive, dead and injured cell fractions. All qPCR assays were performed in triplicate and mean CT values were used for all calculations.
To study thermal tolerance, L. pneumophila was resuspended in 1X PBS at ≈ 108 CFU/mL (λ600nm 0.2) and heat treated at 37 to 75°C for 10 min, with gentle shaking after regular intervals of 3 min. To investigate effect of exposure time, cells were heated at 55°C in 1 min increments from 0 to 10 min. The time required to attain desired temperature within the bacterial vials was also considered. Following the thermal treatment vials were placed at 5°C for 60 min. All treated cells were analyzed by flow cytometry and sorted into alive, dead and injured (suspected VBNC) cell fractions (as described in section Flow cytometry-cell sorting assay).
Culturability of sorted cell fractions was determined using BCYE agar (CM0655, Oxoid Ltd.) containing Legionella growth supplement (SR0110C, Oxoid Ltd.) and Legionella enrichment broth (M1399-100G, Himedia®) containing Legionella growth supplement (FD016A-5VL, Himedia®). For culturing, from 100 to 300 sorted cells from each fraction was inoculated onto each growth medium and incubated at 37 ± 1°C for 5–7 days.
The viability of non-culturable cells was determined using two methods: an ATP estimation assay and an amoebae coculture assay.
ATP contents were estimated using a luciferase-based ATP determination kit (A22066, Thermo Fisher scientific) according to the manufacturer’s instructions. The quantity of ATP in sorted cell fractions was estimated as a function of luminescence measured at 540 nm using a CLARIOstar® microplate reader (BMG Labtech GmbH).
In this assay, 100 μL (107 Legionella) of each of the sorted fractions, or a filter concentrated environmental water sample, were inoculated on 106 A. polyphaga cells growing in infection medium (FBS-PYG broth and Page’s saline in 1: 10 ratio) and incubated at 25 ± 1°C for 7 days (multiplicity of infection of 10; Moffat and Tompkins, 1992). Internalization and resuscitation of Legionella within amoeba hosts was confirmed through FISH using Alexa Fluor 488 labeled Legionella LEG705 and Alexa Fluor 647 labeled eukaryotic universal EUK1209 fluorogenic probes (Invitrogen™, Table 1; Amann et al., 1990; Manz et al., 1995; Whiley et al., 2011). Briefly, infected cells resuspended in 1X PBS were fixed with 4% paraformaldehyde. Fixed cells were placed on a slide, air dried and dehydrated in an ethanol series: 50% ethanol/2 min, 80% ethanol/2 min and 96% ethanol/2 min. Fixed, and dehydrated cells were covered with 200 μL hybridization buffer (0.9 M NaCl, 0.01% SDS, 20 mM Tris–HCl pH 7.6) containing 100 ng of each probe and placed in a dark humidified chamber at 55 ± 1°C for 100 min. To remove unbound probes, cells were washed with warm wash buffer (0.9 M NaCl, 0.01% SDS, 20 mM Tris–HCl pH 7.6). Finally, the slide was washed with Milli-Q® water, air dried, mounted with 20 μL of CitiFluor™ AF1 (17970–25, Electron Microscopy Sciences) and examined under fluorescence microscope (AX70, Olympus). Nonspecific binding of these probes was tested on E. coli.
Four water samples spiked with different concentrations of L. pneumophila, E. coli, A. calcoaceticus and A. polyphaga were prepared (Table 2). All water samples were heated at 50°C for 15 min, then vacuum filtered onto a 47 mm diameter 0.2 μm polycarbonate membrane (GTTP04700, Isopore™). The filter membrane was transferred into a sterile tube containing 1,000 μL 1X PBS and vortexed. Presence of culturable Legionella in all four samples were confirmed using standard culturing method (International Organization for Standardization, 2017). From each resuspended sample, 200 μL was mixed in 300 μL filter sterilized staining buffer and used for flow cytometry-cell sorting. The sorted VBNC fractions were subjected to qPCR to estimate Legionella GU/L (as described in section Interference of viability dyes in quantitative PCR).
From an Australian hospital, shower water (1.15 L) samples were collected in sterile screw capped wide-mouth plastic bottle. Culturable Legionella was detected using Legiolert kit (IDEXX) and standard culture method (International Organization for Standardization, 2017). For the standard culture method, 1 L water sample was concentrated through filtration. The concentrated residues were resuspended in 5 mL distilled water and subjected to both acid and heat pre-treatment steps. The treated sample was plated on BCYE-GVPC agar and incubated under standard culturing conditions (as described in section Microbial strains, culture media and growth conditions). For the Legiolert kit method, 100 mL sample water was processed according to the manufacturer’s protocols. Using the Aquadien™ DNA extraction and purification kit DNA was extracted from the water sample and Legionella GU/L was estimated. To detect presence of any VBNC Legionella, the PCR positive and culture negative samples were subjected to amoeba coculture assay. Finally, these Legionella PCR positive and culture negative shower water samples were processed to detect and quantify VBNC Legionella as described in section Flow cytometry-cell sorting and qPCR assay development.
L. pneumophila spiked water samples were assayed according to ISO11731:2017-05 (International Organization for Standardization, 2017). Figure 1 shows that the highest culturable Legionella numbers were obtained by directly inoculating the sample after membrane filtration (1.136 ± 0.087 CFU/L). There were statistically significant differences between the control sample (no pre-treatment) and the two pre-treatments (p < 0.001). Both acid and heat decontamination steps transformed culturable Legionella to non-culturable cells and decreased the cell numbers to 0.774 ± 0.043 CFU/L and 0.766 ± 0.097 CFU/L, respectively. These results demonstrate that both sample pre-treatment procedures also effect the reliability of the ISO culturing assay (ISO11731:2017-05).
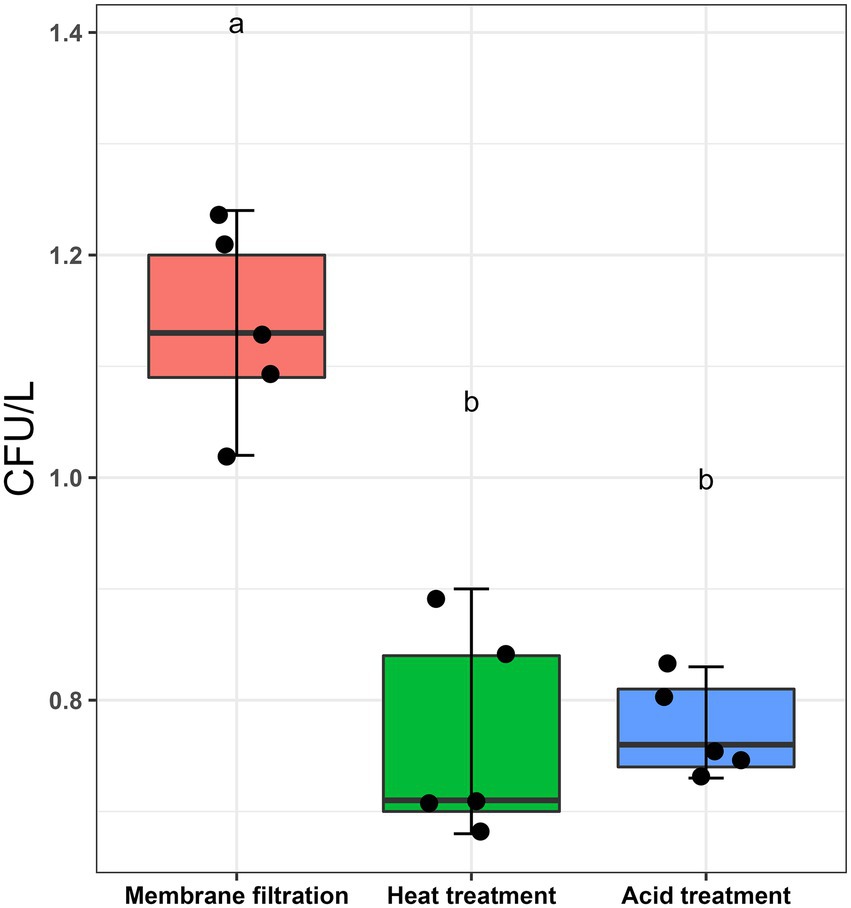
Figure 1. Effect of sampling processing methods (ISO11731:2017-05) on L. pneumophila culturability. L. pneumophila spiked water samples were membrane filtered (no pre-treatment used as control), followed by heat (50°C/30 min) and acid (KCl-HCl with pH 2.2/5 min) pre-treatment method. These pretreatment methods transformed ≈30% culturable Legionella into a non-culturable state. Data are the mean with standard deviation of five replicates, while the same letter within the same treatment is statistically similar according to Tukey’s HSD test (p < 0.001).
Three distinct regions can be distinguished on the cytograms: PI-stained dead cells, TO-stained alive cells and TO-stained injured (transition state) cells (Figure 2A). Injured or intermediate cells were observed in between the alive and dead cell regions. The principle of the PI/TO assay is to selectively stain dead bacteria (damaged membrane) with the dye PI, whereas TO can enter into all cells (Alsharif and Godfrey, 2002). The PI staining divides cells into two major populations, i.e., PI-staining (dead) and PI-unstained (alive and injured) cells. In PI unstained cell population, fluorescence of TO dye become very prominent. Injured cells are in a transitional physiological state and stain positive with TO dye. In these experiments Legionella was grown at 37°C on agar for 3–4 days and all samples consisted of some dead, injured, and alive cells although the proportion of alive cells was the highest. It is possible that the washing, and centrifugation steps could have generated these small number of injured and dead cells. Different handling and processing steps have been shown to cause Legionella to lose culturability (Whiley, 2016). Figure 2A shows that thermal treatment at 75°C for 10 min completely inactivated the cells and these dead cells are stained with PI. These results clear demonstrated that using flow cytometry-cell sorting, it was possible to isolate and characterize all three cell fractions.
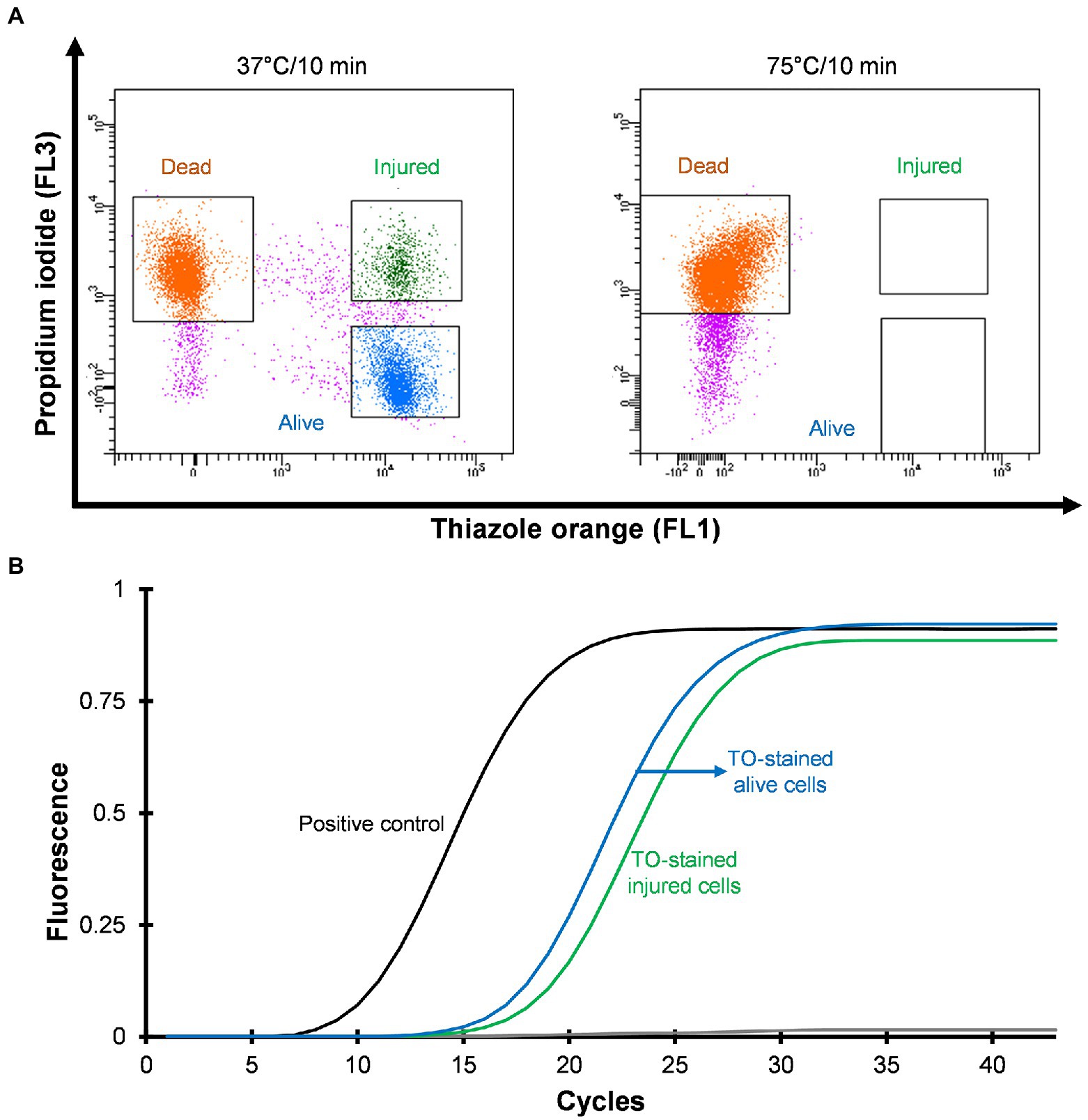
Figure 2. Viability based flow cytometry-cell sorting and qPCR assay. L. pneumophila grown on BCYE-GVPC agar at 37°C were used as a positive control and cells treated at 75°C for 10 min were used as a negative control. (A) Cytograms represent three populations: thiazole orange-stained alive (blue), thiazole orange-stained injured (green) and propidium iodide-stained dead (orange) cells. Propidium iodide selectively stained dead cells. Pink colored events are debris. (B) qPCR assay of thiazole orange stained alive and injured cells. The results showed that thiazole orange (TO) does not interfere with DNA extraction and qPCR.
Mean CT values obtained from qPCR assays and logarithmic concentration of 10-fold dilutions of Legionella 16S rDNA gBlocks were used to plot a standard curve with a 0.9995 regression coefficient and 88.9912% amplification efficiency. In this study, the limit of detection and limit of quantification were 5 GU and 25 GU, respectively. To investigate the effect of the TO dye, 103–105 sorted Legionella cells were used for DNA extraction. The extracted DNA was subjected to qPCR assay. Figure 2B illustrates that the TO dye did not interfere with DNA extraction and amplification.
In order to study thermal tolerance of Legionella, bacteria were heat shocked sequentially in 5°C increments from 40 to 70°C for 10 min and examined by flow cytometry (Figure 3A). From the cytograms, PI-stained cells were non-culturable and non-viable Legionella and defined as dead cells. TO-stained alive cells grew on culture medium, whereas TO-stained injured cells obtained from ≤45°C treatment was unable to grow on both BCYE agar and Legionella enrichment broth (Figure 3B). The 50°C treatment triggered the formation of non-culturable injured cells, which were considered VBNC Legionella. Heat treatment at 55°C for 0–3 min increased the rate of transformation of alive cells into VBNC cells (Figure 3C). It is worth noting that a considerable number of Legionella converted into VBNC cells after 3 min of treatment. During the 55°C thermal treatment for 0–10 min, the number of PI-stained dead cells increased proportionally with the duration of thermal treatment. After 10 min of heat shock treatment at 55°C, all bacteria clustered in the dead cell region on the cytogram and were no longer culturable on BCYE agar or in Legionella enrichment broth. Thus, according to the cytograms (Figures 3A,C), the PI/TO membrane integrity assay is an effective way to study the effect of disinfection treatment on Legionella.
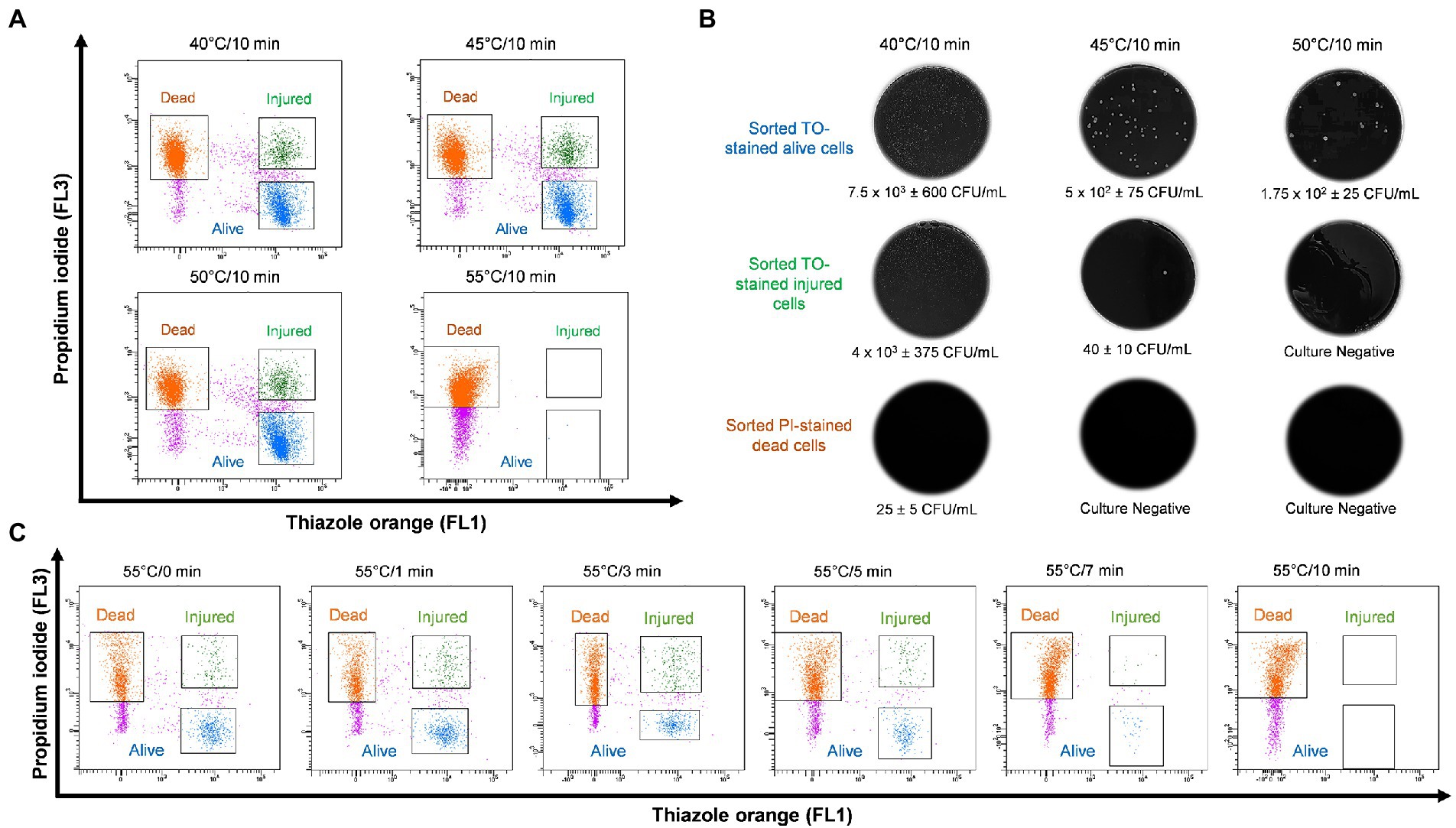
Figure 3. Legionella pneumophila thermal profile and culturability assay. (A) Bacterial cells were heat shocked at 40–55°C for 10 min and analyzed using flow cytometry. (B) Sorted cell fractions cultured on BCYE agar. Thermal shock at 50°C for 10 min transformed TO-stained injured cells into VBNC Legionella, as indicated by the lack of growth on the BCYE agar. (C) Pure culture of L. pneumophila subjected to 55°C thermal treatment in increments from 0 to 10 min. The population of VBNC L. pneumophila increased gradually with time 9% at 1 min, 20% at 3 min and then decreased to 5% at 5 min.
The injured cells isolated following 50°C heat shock was unable to grow on either agar or in broth medium (Figures 4A,B). The viability of these cells was confirmed by quantification of ATP contents and resuscitation in A. polyphaga. According to the luciferase-based ATP detection assay, TO-stained alive culturable cells produced the highest amount of ATP (11,030 ± 860 units), whereas the least amount of ATP was detected from PI-stained dead cells (85 ± 10 units). TO-stained injured non-culturable cells were found to be metabolically active and able to produce 4,645 ± 345 units ATP.
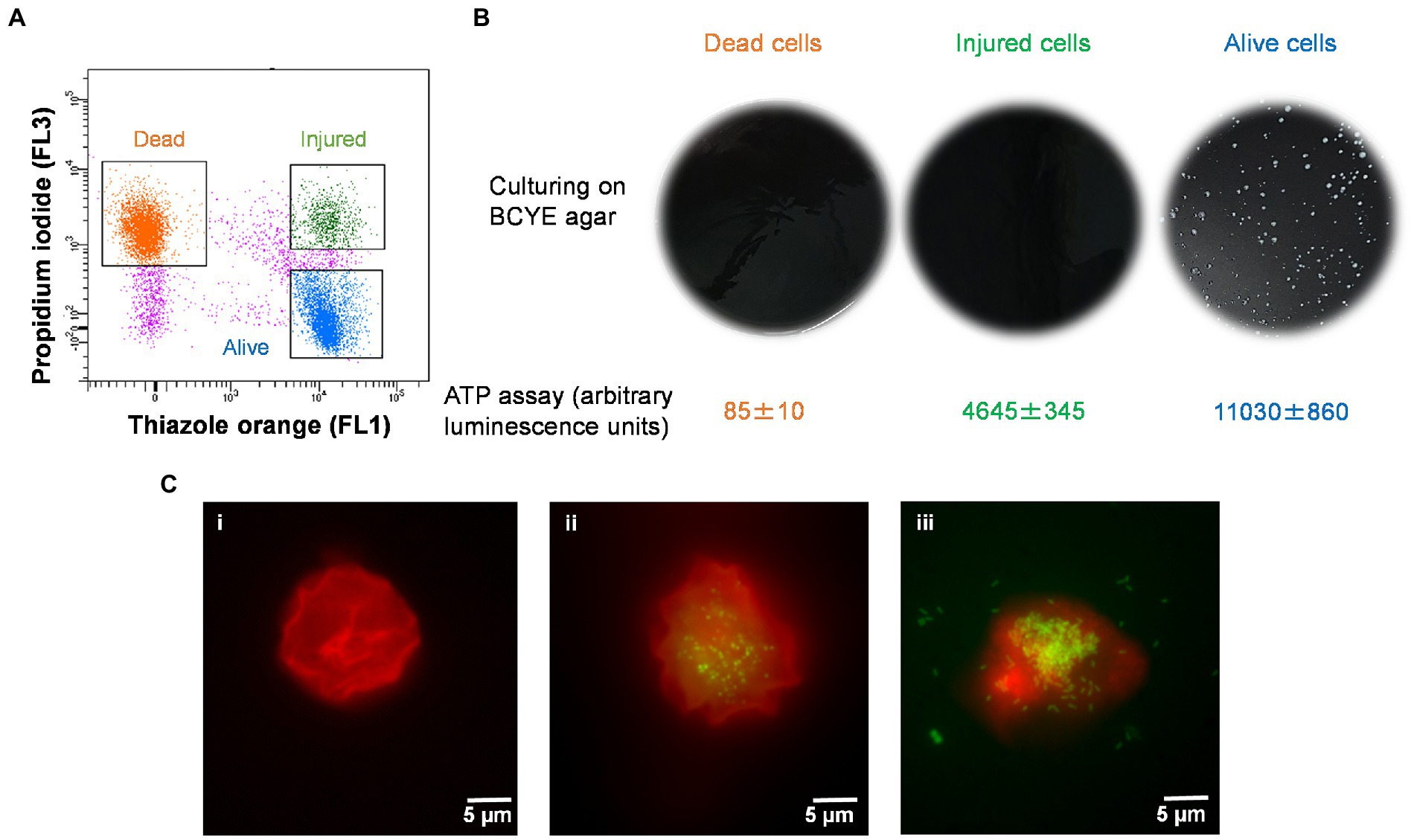
Figure 4. Characterization of thermally generated (50°C/10 min) VBNC L. pneumophila. (A) Cytogram representing thiazole orange-stained alive (blue), thiazole orange-stained injured (green) and propidium iodide-stained dead (orange) cells generated by 50°C heat shock for 10 min. (B) These three fractions were cultured on BCYE agar, and their ATP contents measured using a luciferase-based ATP estimation assay. (C): FISH of A. polyphaga with Alexa Fluor 647 labeled EUK1209 (red) and Legionella with Alexa Fluor 488 labeled LEG705 (green) probes. (Ci) uninfected A. polyphaga. (Cii,iii) These micrographs show intracellular proliferation of VBNC Legionella within A. polyphaga after 7 and 10 days of infection, respectively.
Axenic A. polyphaga cultures were used to study infectivity of suspected VBNC (TO-stained injured) and culturable Legionella. This was achieved by labeling Legionella infected A. polyphaga cells with fluorescent probes and examining them under a fluorescent microscope. In the FISH images (Figure 4C), the LEG705 Legionella probe appears green and the EUK1209 eukaryotic probe is colored red. A. polyphaga appeared as red colored spherical bodies (Figure 4C), whereas Legionella appears as distinct cells localized within the amoebae cytoplasm (Figures 4Cii,iii). Unlike the VBNC Legionella, culturable Legionella were able to proliferate within A. polyphaga after 60 h of infection. The results from 7 days post-infection showed internalization and multiplication of suspected VBNC Legionella in A. polyphaga (Figure 4Cii). After 10 days of infection, host amoebae lost its cellular integrity and Legionella egressed (Figure 4Ciii). This demonstrates that thermally generated non-culturable injured cells exhibit intracellular proliferation within A. polyphaga. After 120 h of infection, the alive Legionella infected a higher number of A. polyphaga ≈ 23%, whereas the VBNC Legionella infected only ≈ 7% of A. polyphaga. Unlike alive and VBNC Legionella cells, the thermally inactivated Legionella (75°C for 10 min) were unable to proliferate within A. polyphaga cells. Furthermore, the differences with proliferation and infection rates suggest that cellular behavior and pathogenicity of culturable and VBNC Legionella is not identical. Both assays clearly establish that TO-stained injured non-culturable cells are metabolically active and infect A. polyphaga and can be regarded as VBNC Legionella.
Four different water samples spiked with L. pneumophila were prepared and heat treated at 50°C for 15 min. Standard culturing method confirmed that all samples were negative for culturable Legionella. Using the flow cytometry-cell sorting assay these water samples were examined. It was observed that water samples (sample 3 and 4) consisted of mixed bacterial population generated a large population of TO-stained injured cells and a smaller population of PI-stained dead cells (Figure 5A). The qPCR results of injured cell fractions showed that water samples containing mixed bacterial populations harbored the highest amount of TO-stained injured Legionella cells (potentially VBNC cells ≈2 × 106 GU/L, Figure 5B). It demonstrated that heat treatment of Legionella spiked water samples resulted in generation of VBNC Legionella and mixed bacterial cultures significantly increased the numbers of VBNC Legionella. This could potentially be due to the additional mixed bacteria cells providing a slight protection to the Legionella during the heat treatment or the mixed bacterial population harbored more biomass which increased the efficiency of DNA extraction and artificially increased the counts of Legionella. More importantly, in combination the PI/TO staining flow cytometry-cell sorting and qPCR assays can detect and quantify VBNC Legionella from potable water.
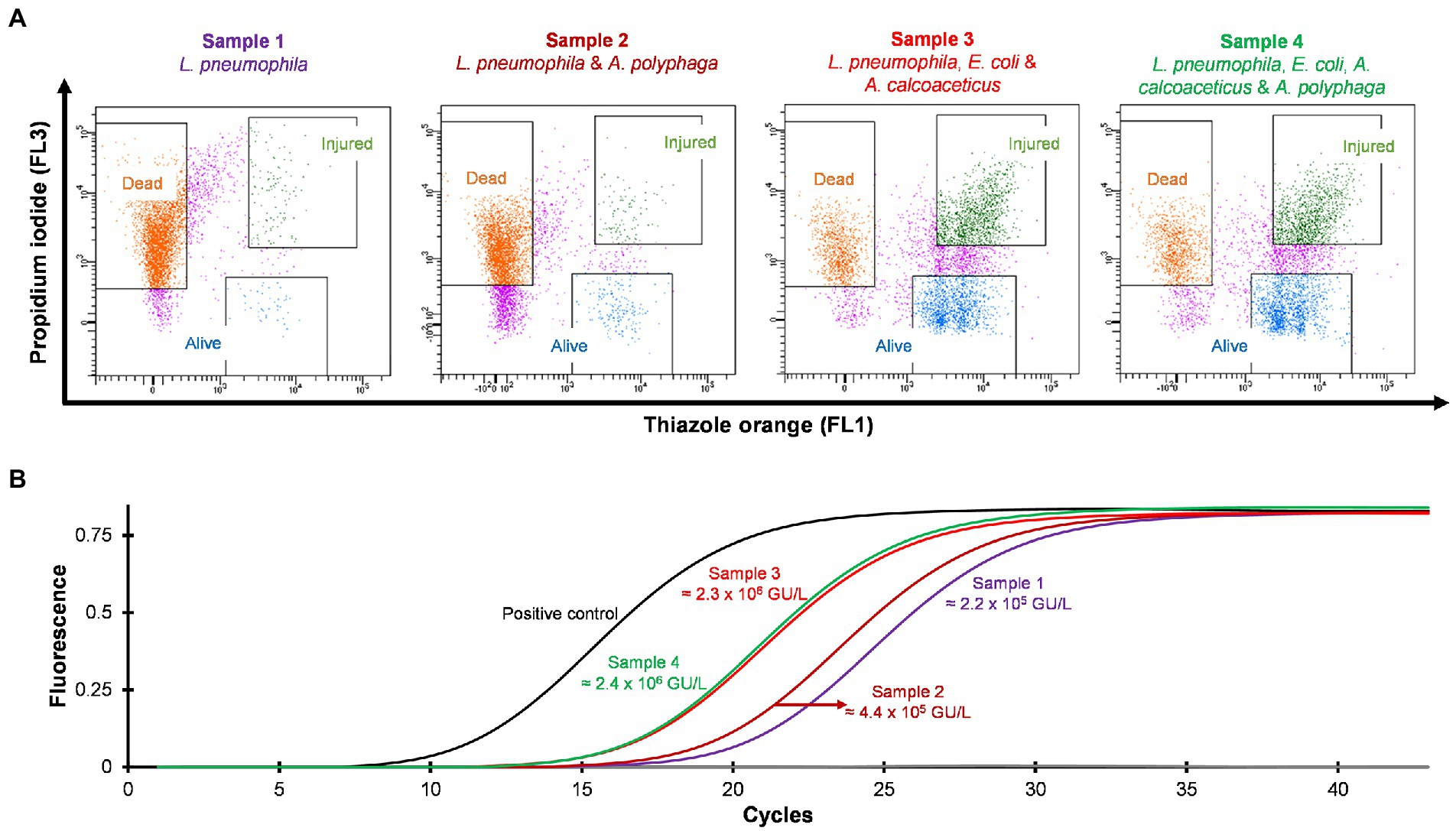
Figure 5. Detection and quantification of VBNC L. pneumophila from spiked water samples. All four spiked water samples were heated at 50°C for 15 min to generate VBNC Legionella. (A) Cytograms of four different water samples spiked with 4.5 × 106 CFU/L L. pneumophila. Samples 3 and 4 generated the highest amount of VBNC cells. (B) qPCR of VBNC (collected injured fraction) Legionella taken from the four samples.
Hospital shower water samples were screened for Legionella using the Legiolert kit, standard Legionella culturing and qPCR assays. The two samples were selected that were negative for culturable Legionella, but the qPCR assay demonstrated the presence of Legionella DNA (Supplementary Table S1 shows the concentrations of the different Legionella populations enumerated using each method). Furthermore, the amoebae coculture assay and FISH analysis confirmed that non-culturable Legionella present in both samples effectively propagate in A. polyphaga after 7 days of infection (≈2% infected cells; Figure 6). To test efficacy of the VFC + qPCR assay both samples were stained with PI/TO dyes and analyzed by flow cytometry and injured cell fractions were sorted out. Figure 7A shows that both samples contained a significant amount of TO-stained injured cells, with environmental sample 1 having 1.47 × 105 TO-stained injured cells and sample 2 contained 7.4 × 104 TO-stained injured cells. DNA was extracted from the whole water sample and TO-stained injured cells and subjected to qPCR assay for the estimation of Legionella GU/L. Figure 7B clearly shows the difference between Legionella GU/L of the whole water sample and TO-stained injured cells. Environmental sample 1 contained 3.9 × 104 GU/L Legionella whereas in TO-stained injured cells it was 3.2 × 103 GU/L. Similarly, in sample 2, the Legionella concentration was 3.5 × 104 GU/L in the whole sample whereas TO-stained injured cells contained 1 × 103 GU/L Legionella. The difference (1 to 1.5 Log10) between Legionella genomic load of whole potable water sample and TO-stained injured cells indicated presence of dead Legionella or its residual DNA. These results challenge the efficacy and reliability of the culturing (ISO11731:2017-05) and standard qPCR assays (ISO/TS12869:2019).
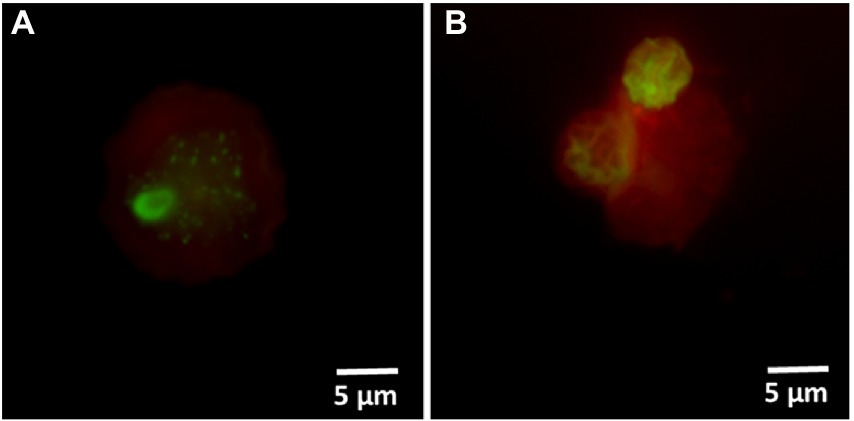
Figure 6. FISH of A. polyphaga stained with Alexa Fluor 647 labeled EUK1209 (red) and Legionella stained with Alexa Fluor 488 labeled LEG705 (green) probes. These micrographs show intracellular proliferation of environmental (shower water sample 1: A and sample 2: B) non-culturable Legionella within A. polyphaga.
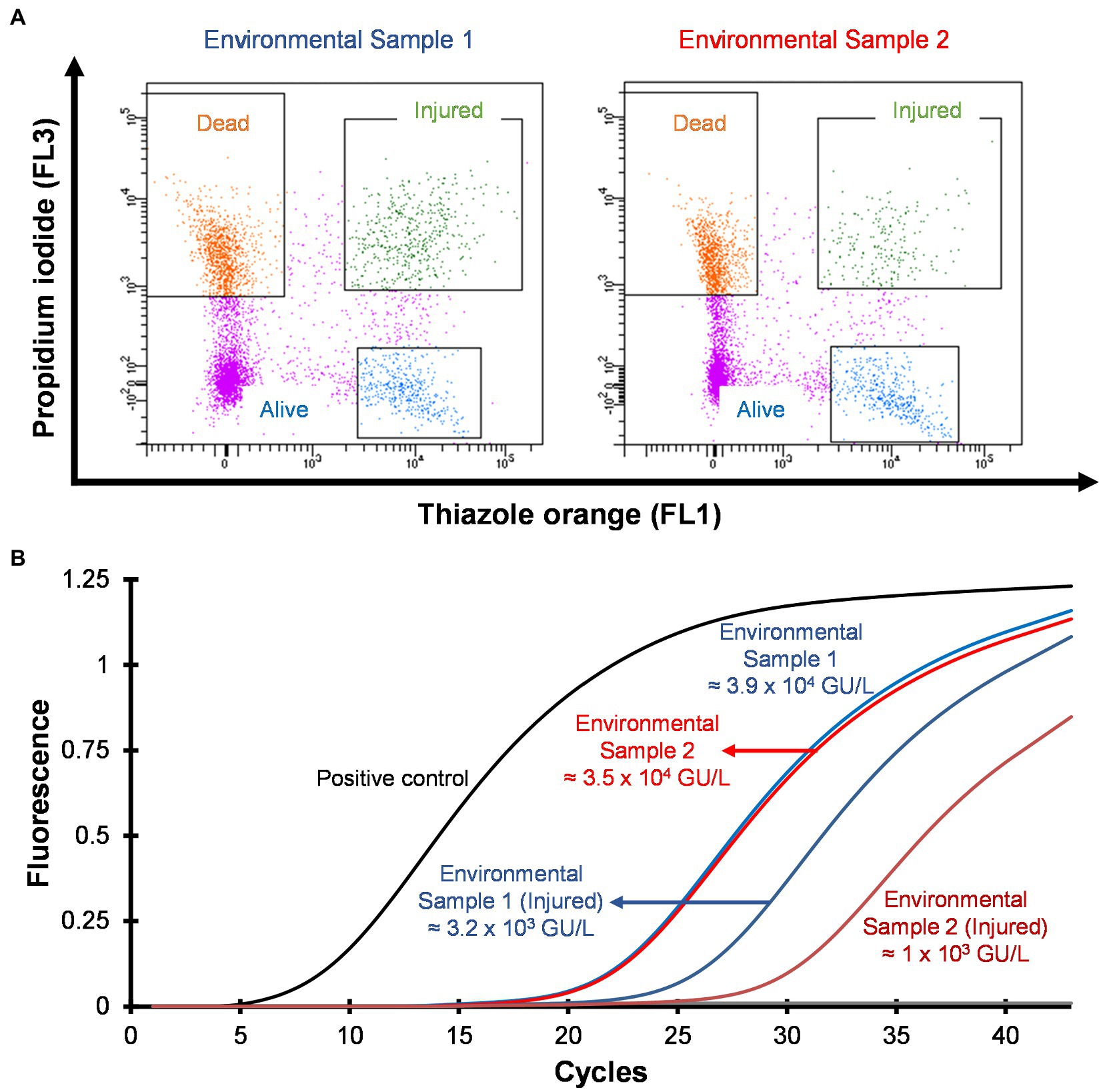
Figure 7. Detection and quantification of VBNC L. pneumophila from potable water samples. (A) Cytograms of culture negative potable water samples. (B) qPCR of whole water sample and VBNC Legionella and estimation of GU/L after viability cell sorting using flow cytometry shown in (A). This image clearly demonstrates the difference in the concentration of Legionella, indicating presence of dead Legionella of its DNA in the water sample.
VBNC state is a physiological condition in which Legionella lose its ability to grow on standard microbiological culture media but retain cellular viability and pathogenicity (Epalle et al., 2015). It is unclear whether the VBNC state is an adaptive response supporting survival and persistence of Legionella under unfavorable conditions or a form of cellular damage, which results in loss of culturability. This presents an important challenge in Legionella research, as it is vital to understand the role of VBNC Legionella within manufactured water systems to improve control and risk management strategies to prevent LD. One cause of long-term persistence of Legionella contamination in engineered water systems is regular reseeding of Legionella and VBNC Legionella from the supply/source water (Whiley et al., 2017; Nisar et al., 2020). Common building water disinfection procedures including pasteurization, chlorination, and chlorine dioxide treatment, induces and selects VBNC Legionella (Allegra et al., 2008; Mustapha et al., 2015; Whiley et al., 2017). Importantly, in hospital buildings thermal disinfection can act as a selection pressure that results in the persistence of heat resistant Legionella (Allegra et al., 2011). The method described in this study successfully detected VBNC Legionella from environmental water samples (hospital showers). The viability-based flow cytometry assay clearly discriminated Legionella into dead and viable fractions (Figure 2A). The injured cells sorted in this assay, met the definition of VBNC Legionella as they were unculturable on standard BCYE agar (Figure 4B); however, were still viable as determined by ATP quantification and the ability to infect amoebae hosts (Figure 4C). These are accepted methods to measure viability, with ATP estimation kits widely used to discriminate metabolically active (alive or injured) cells from metabolically inactive (dead) cells (Allegra et al., 2008). Amoebae coculture assay is described as most suitable system to determine pathogenicity of VBNC Legionella (La Scola et al., 2001; Cervero-Arago et al., 2019). According to these results, the viability-based flow cytometry assay is an effective parameter to study behavior and physiology of Legionella under different disinfection treatments.
Complete eradication of Legionella from manufactured water systems is not practically possible; therefore, environmental monitoring and risk assessment of Legionella on a regular basis are crucial to reduce the risk of legionellosis outbreaks. Despite recent advances in molecular biology and biotechnology, the culture based approach is regarded as the standard method to detect Legionella contamination in engineered water systems (International Organization for Standardization, 2017; Standards Australia, 2017). Importantly in engineered water systems, the majority of the Legionella population exists in the VBNC state (Diederen et al., 2007). Routine detection and quantification methods, culture based or qPCR assay, either over or underestimate Legionella contamination and do not provide information about VBNC Legionella (Whiley, 2016).
There are previously described alive/dead dye-based flow cytometry protocols used to differentiate between alive and VBNC Legionella (Allegra et al., 2008; Zacharias et al., 2015; Braun et al., 2019). However, these previous protocols are designed to study pure cultures of Legionella and are not designed for analyzing environmental samples in which Legionella are present in complex microbial communities. When these previously described methods are applied to environmental samples they can differentiate the dead, VBNC, and alive total bacterial populations, but they cannot determine if Legionella is part of these populations (Li et al., 2014; Ramamurthy et al., 2014). One approach that has been previously used to overcome this limitation is the use of specific fluorogenic antibodies; however, this approach is currently strain specific and cannot be used to detect different environmental strains of Legionella (Füchslin et al., 2010). In contrast, the method described in this paper (VFC + qPCR) can detect and quantify VBNC Legionella from environmental samples. It combines alive/dead dye-based flow cytometry with cell sorting of the separate dead, VBNC and alive bacterial populations followed by qPCR enumeration of Legionella spp. and L. pneumophila present in the VBNC and alive populations. This study used PI and TO dyes to differentiate the alive, dead and VBNC bacterial populations. The PI selectively stains dead cells with damaged cell membranes (Crowley et al., 2016). The TO dye is then used to characterize the cells which do not uptake the PI (Alsharif and Godfrey, 2002; Allegra et al., 2008). A limitation of this approach is that PI has been shown to have reduce penetration of biofilm associated bacterial cells, which can lead to and overestimation of bacterial viability (Rosenberg et al., 2019).
This study explored the potential of thermal disinfection to induce the VBNC state. The findings demonstrated that culturability on the standard medium (BCYE and BCYE-GVPC agar) was the least reliable cellular viability indicator (Figures 3B, 4B). In addition, the effect of the acid and heat pre-treated methods (prescribed in the ISO11731:2017-05 culture method) on Legionella recovery was examined. These pre-treatments are included in the standard culture method to selectively kill other environmental bacteria and prevent plates from becoming overgrown (International Organization for Standardization, 2017). However, this study (Figure 1), demonstrated that both selective decontamination methods statistically significantly reduced the recovery of culturable Legionella and analysis using this described method showed that this reduction was caused by the induction of the VBNC state. This could potentially explain some of the unreliability and lack of reproducibility often observed with the ISO11731:2017-05 method (De Luca et al., 1999; Lucas et al., 2011). Other factors which compromise the validity of culture-based techniques are sample holding time (McCoy et al., 2012) and residual disinfectant (Wiedenmann et al., 2001) present in collected water sample. The limit of detection for this described method (VFC + qPCR) when applied to the environmental water samples was 102 GU/L. This is comparable to the limit of detections for the standard ISO methods (102 CFU/L for ISO11731:2017-05 and 102 GU/L for ISO/TS12869:2019). Supplementary Figure S3 presents a comparison of Legionella CFU/L and GU/L and shows they were comparable at concentrations from 103 to 108 CFU/L or GU/L.
In conclusion, this study describes a direct and rapid (results within 5–6 h) assay to detect and quantify VBNC Legionella from potable water samples. This assay offers following advantages: (a) viable and dead Legionella quantification without culturing (b) quantification of viable and dead Legionella; and (c) rapid and direct screening of VBNC Legionella.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
MN, HW, and RB conceived and designed the research. MN performed the experiments. GB and MN conducted flow cytometry-cell sorting assay. KR, MB, HW, and RB provided technical assistance. MN and HW drafted and edited the manuscript. HW, KR, MB, GB, and RB corrected and contributed to the manuscript. All authors contributed to the article and approved the submitted version.
The authors thank Pat Vilimas and Nicholas Eyre of Flinders Microscopy and Microanalysis for the expert technical advice and assistance with fluorescence microscopy imaging. The authors are also thankful to Catherine Abbott of College of Science and Engineering, Flinders University for the assistance in the ATP quantification assay. MN is the recipient of a doctoral scholarship by the Australian Government Research Training Program Scholarship (AGRTP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1094877/full#supplementary-material
Allegra, S., Berger, F., Berthelot, P., Grattard, F., Pozzetto, B., and Riffard, S. (2008). Use of flow cytometry to monitor Legionella viability. Appl. Environ. Microbiol. 74, 7813–7816. doi: 10.1128/AEM.01364-08
Allegra, S., Grattard, F., Girardot, F., Riffard, S., Pozzetto, B., and Berthelot, P. (2011). Longitudinal evaluation of the efficacy of heat treatment procedures against Legionella spp. in hospital water systems by using a flow cytometric assay. Appl. Environ. Microbiol. 77, 1268–1275. doi: 10.1128/AEM.02225-10
Alsharif, R., and Godfrey, W. (2002). Bacterial detection and live/dead discrimination by flow cytometry. BD Biosciences, Immunocytometry Systems, San Jose, CA.
Amann, R. I., Binder, B. J., Olson, R. J., Chisholm, S. W., Devereux, R., and Stahl, D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990
Bartram, J., Chartier, Y., Lee, J.V., Pond, K., and Surman-Lee, S. (2007). Legionella and the prevention of legionellosis. Geneva: World Health Organization.
Baudart, J., Guillaume, C., Mercier, A., Lebaron, P., and Binet, M. (2015). Rapid quantification of viable Legionella in nuclear cooling tower waters using filter cultivation, fluorescent in situ hybridization and solid-phase cytometry. J. Appl. Microbiol. 118, 1238–1249. doi: 10.1111/jam.12783
Braun, R. S., Mendis, N., Li, L., and Faucher, S. P. (2019). Quantification of viable but non-culturable cells of Legionella pneumophila. Methods Mol. Biol. 1921, 45–53. doi: 10.1007/978-1-4939-9048-1_3
Casini, B., Baggiani, A., Totaro, M., Mansi, A., Costa, A. L., Aquino, F., et al. (2018). Detection of viable but non-culturable Legionella in hospital water network following monochloramine disinfection. J. Hosp. Infect. 98, 46–52. doi: 10.1016/j.jhin.2017.09.006
Cervero-Arago, S., Schrammel, B., Dietersdorfer, E., Sommer, R., Luck, C., Walochnik, J., et al. (2019). Viability and infectivity of viable but nonculturable Legionella pneumophila strains induced at high temperatures. Water Res. 158, 268–279. doi: 10.1016/j.watres.2019.04.009
Chao, C. M., and Lai, C. C. (2022). Increasing Legionella in Taiwan during COVID-19 pandemic. Am. J. Infect. Control 50, 237–238. doi: 10.1016/j.ajic.2021.10.024
Connolly, C., Keil, R., and Ali, S. H. (2021). Extended urbanisation and the spatialities of infectious disease: demographic change, infrastructure and governance. Urban Stud. 58, 245–263. doi: 10.1177/0042098020910873
Conza, L., Casati, S., and Gaia, V. (2013). Detection limits of Legionella pneumophila in environmental samples after co-culture with Acanthamoeba polyphaga. BMC Microbiol. 13:49. doi: 10.1186/1471-2180-13-49
Crowley, L. C., Scott, A. P., Marfell, B. J., Boughaba, J. A., Chojnowski, G., and Waterhouse, N. J. (2016). Measuring cell death by propidium iodide uptake and flow cytometry. Cold Spring Harb. Protoc. 2016. doi: 10.1101/pdb.prot087163
Dai, D. J., Rhoads, W. J., Katner, A., Strom, L., Edwards, M. A., Pruden, A., et al. (2019). Molecular survey of legionella and Naegleria fowleri in private well water and premise plumbing following the 2016 Louisiana flood. Environ. Sci. Wat. Res. Technol. 5, 1464–1477. doi: 10.1039/C9EW00109C
De Luca, G., Stampi, S., Lezzi, L., and Zanetti, F. (1999). Effect of heat and acid decontamination treatments on the recovery of Legionella pneumophila from drinking water using two selective media. New Microbiol. 22, 203–208.
De Mendiburu, F. (2021). agricolae: statistical procedures for agricultural research. R package version 1.3-5, 155
Delgado-Viscogliosi, P., Solignac, L., and Delattre, J. M. (2009). Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples. Appl. Environ. Microbiol. 75, 3502–3512. doi: 10.1128/AEM.02878-08
Diederen, B. M., De Jong, C. M., Aarts, I., Peeters, M. F., and Van Der Zee, A. (2007). Molecular evidence for the ubiquitous presence of Legionella species in Dutch tap water installations. J. Water Health 5, 375–383. doi: 10.2166/wh.2007.033
Dietersdorfer, E., Kirschner, A., Schrammel, B., Ohradanova-Repic, A., Stockinger, H., Sommer, R., et al. (2018). Starved viable but non-culturable (VBNC) Legionella strains can infect and replicate in amoebae and human macrophages. Water Res. 141, 428–438. doi: 10.1016/j.watres.2018.01.058
Epalle, T., Girardot, F., Allegra, S., Maurice-Blanc, C., Garraud, O., and Riffard, S. (2015). Viable but not culturable forms of Legionella pneumophila generated after heat shock treatment are infectious for macrophage-like and alveolar epithelial cells after resuscitation on Acanthamoeba polyphaga. Microb. Ecol. 69, 215–224. doi: 10.1007/s00248-014-0470-x
Fields, B. S., Benson, R. F., and Besser, R. E. (2002). Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526. doi: 10.1128/CMR.15.3.506-526.2002
Fittipaldi, M., Nocker, A., and Codony, F. (2012). Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 91, 276–289. doi: 10.1016/j.mimet.2012.08.007
Füchslin, H. P., Kötzsch, S., Keserue, H. A., and Egli, T. (2010). Rapid and quantitative detection of Legionella pneumophila applying immunomagnetic separation and flow cytometry. Cytometry A 77a, 264–274. doi: 10.1002/cyto.a.20858
Garcia, M. T., Jones, S., Pelaz, C., Millar, R. D., and Abu Kwaik, Y. (2007). Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. 9, 1267–1277. doi: 10.1111/j.1462-2920.2007.01245.x
Gattuso, G., Rizzo, R., Lavoro, A., Spoto, V., Porciello, G., Montagnese, C., et al. (2022). Overview of the clinical and molecular features of Legionella pneumophila: focus on novel surveillance and diagnostic strategies. Antibiotics 11:370. doi: 10.3390/antibiotics11030370
Hayes-Phillips, D., Bentham, R., Ross, K., and Whiley, H. (2019). Factors influencing Legionella contamination of domestic household showers. Pathogens 8:27. doi: 10.3390/pathogens8010027
International Organization for Standardization (2017). ISO11731:2017-05 Water Quality – Enumeration of Legionella. Geneva: ISO.
International Organization for Standardization (2019). ISO/TS12869:2019 Water Quality – Detection and Quantification of Legionella spp. and/or Legionella pneumophila by Concentration and Genic Amplification by Quantitative Polymerase Chain Reaction (qPCR). Geneva: ISO.
Khan, M. M. T., Pyle, B. H., and Camper, A. K. (2010). Specific and rapid enumeration of viable but nonculturable and viable-culturable gram-negative bacteria by using flow cytometry. Appl. Environ. Microbiol. 76, 5088–5096. doi: 10.1128/AEM.02932-09
Kirschner, A. K., Rameder, A., Schrammel, B., Indra, A., Farnleitner, A. H., and Sommer, R. (2012). Development of a new CARD-FISH protocol for quantification of Legionella pneumophila and its application in two hospital cooling towers. J. Appl. Microbiol. 112, 1244–1256. doi: 10.1111/j.1365-2672.2012.05289.x
La Scola, B., Mezi, L., Weiller, P. J., and Raoult, D. (2001). Isolation of Legionella anisa using an amoebic coculture procedure. J. Clin. Microbiol. 39, 365–366. doi: 10.1128/JCM.39.1.365-366.2001
Li, L., Mendis, N., Trigui, H., Oliver, J. D., and Faucher, S. P. (2014). The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5:258. doi: 10.3389/fmicb.2014.00258
Liang, J., Swanson, C. S., Wang, L., and He, Q. (2021). Impact of building closures during the COVID-19 pandemic on Legionella infection risks. Am. J. Infect. Control 49, 1564–1566. doi: 10.1016/j.ajic.2021.09.008
Lim, E. L., Amaral, L. A., Caron, D. A., and Delong, E. F. (1993). Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl. Environ. Microbiol. 59, 1647–1655. doi: 10.1128/aem.59.5.1647-1655.1993
Lucas, C. E., Taylor, T. H., and Fields, B. S. (2011). Accuracy and precision of Legionella isolation by US laboratories in the ELITE program pilot study. Water Res. 45, 4428–4436. doi: 10.1016/j.watres.2011.05.030
Manz, W., Amann, R., Szewzyk, R., Szewzyk, U., Stenström, T.-A., Hutzler, P., et al. (1995). In situ identification of Legionellaceae using 16S rRNA-targeted oligonucleotide probes and confocal laser scanning microscopy. Microbiology 141, 29–39. doi: 10.1099/00221287-141-1-29
Mccoy, W. F., Downes, E. L., Leonidas, L. F., Cain, M. F., Sherman, D. L., Chen, K., et al. (2012). Inaccuracy in Legionella tests of building water systems due to sample holding time. Water Res. 46, 3497–3506. doi: 10.1016/j.watres.2012.03.062
Moffat, J. F., and Tompkins, L. S. (1992). A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60, 296–301. doi: 10.1128/iai.60.1.296-301.1992
Mustapha, P., Epalle, T., Allegra, S., Girardot, F., Garraud, O., and Riffard, S. (2015). Monitoring of Legionella pneumophila viability after chlorine dioxide treatment using flow cytometry. Res. Microbiol. 166, 215–219. doi: 10.1016/j.resmic.2015.01.004
National Notifiable Diseases Surveillance System and Centers for Disease Control and Prevention (2018). National Notifiable Infectious Diseases: Weekly Tables. U.S. Department of Health and Human Services.
Nisar, M. A., Ross, K. E., Brown, M. H., Bentham, R., and Whiley, H. (2020). Water stagnation and flow obstruction reduces the quality of potable water and increases the risk of Legionelloses. Front. Environ. Sci. 8:611611. doi: 10.3389/fenvs.2020.611611
Ramamurthy, T., Ghosh, A., Pazhani, G. P., and Shinoda, S. (2014). Current perspectives on viable but non-culturable (VBNC) pathogenic bacteria. Front. Public Health 2:103. doi: 10.3389/fpubh.2014.00103
Rosenberg, M., Azevedo, N. F., and Ivask, A. (2019). Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 9:6483. doi: 10.1038/s41598-019-42906-3
Sachidanandham, R., Yew-Hoong Gin, K., and Laa Poh, C. (2005). Monitoring of active but non-culturable bacterial cells by flow cytometry. Biotechnol. Bioeng. 89, 24–31. doi: 10.1002/bit.20304
Satoh, W., Nakata, M., Yamamoto, H., Ezaki, T., and Hiramatsu, K. (2002). Enumeration of Legionella CFU by colony hybridization using specific DNA probes. Appl. Environ. Microbiol. 68, 6466–6470. doi: 10.1128/AEM.68.12.6466-6470.2002
Smith, P., Moore, M., Alexander, N., Hicks, L., and O'loughlin, R. (2007). Surveillance for travel-associated Legionnaires’ disease - United States, 2005-2006. Morb. Mortal Wkly Rep. 56, 1261–1263.
Standards Australia (2017). AS5132:2017 Waters – Examination for Legionella spp. Including Legionella pneumophila – Using Concentration. Sydney, NSW: Standards Australia
Steinert, M., Emody, L., Amann, R., and Hacker, J. (1997). Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl. Environ. Microbiol. 63, 2047–2053. doi: 10.1128/aem.63.5.2047-2053.1997
Taylor, M. J., Bentham, R. H., and Ross, K. E. (2014). Limitations of using propidium monoazide with qPCR to discriminate between live and dead Legionella in biofilm samples. Microb. Insights 7, 15–24. doi: 10.4137/MBI.S17723
The European Centre for Disease Prevention and Control (n.d.). Surveillance atlas of infectious diseases. Available at: https://atlas.ecdc.europa.eu/public/index.aspx (Accessed August 2022).
Whiley, H. (2016). Legionella risk management and control in potable water systems: argument for the abolishment of routine testing. Int. J. Environ. Res. Public Health 14:12. doi: 10.3390/ijerph14010012
Whiley, H., Bentham, R., and Brown, M. H. (2017). Legionella persistence in manufactured water systems: pasteurization potentially selecting for thermal tolerance. Front. Microbiol. 8:1330. doi: 10.3389/fmicb.2017.01330
Whiley, H., and Taylor, M. (2016). Legionella detection by culture and qPCR: comparing apples and oranges. Crit. Rev. Microbiol. 42, 65–74. doi: 10.3109/1040841X.2014.885930
Whiley, H., Taylor, M., and Bentham, R. (2011). Detection of Legionella species in potting mixes using fluorescent in situ hybridisation (FISH). J. Microbiol. Methods 86, 304–309. doi: 10.1016/j.mimet.2011.05.023
Wickham, H. (2016). ggplot2: elegant Graphics for Data Analysis. New York: Springer-Verlag New York.
Wiedenmann, A., Langhammer, W., and Botzenhart, K. (2001). A case report of false negative Legionella test results in a chlorinated public hot water distribution system due to the lack of sodium thiosulfate in sampling bottles. Int. J. Hyg. Environ. Health 204, 245–249. doi: 10.1078/1438-4639-00101
Keywords: Legionnaires’ disease, premise plumbing, engineered water system, potable water, ISO11731:2017-05, ISO/TS12869:2019
Citation: Nisar MA, Ross KE, Brown MH, Bentham R, Best G and Whiley H (2023) Detection and quantification of viable but non-culturable Legionella pneumophila from water samples using flow cytometry-cell sorting and quantitative PCR. Front. Microbiol. 14:1094877. doi: 10.3389/fmicb.2023.1094877
Received: 10 November 2022; Accepted: 03 January 2023;
Published: 30 January 2023.
Edited by:
Hector Mora Montes, University of Guanajuato, MexicoReviewed by:
Maria Luisa Ricci, National Institute of Health (ISS), ItalyCopyright © 2023 Nisar, Ross, Brown, Bentham, Best and Whiley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harriet Whiley, ✉ aGFycmlldC53aGlsZXlAZmxpbmRlcnMuZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.