
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 February 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1092273
 Yuan Chen1†
Yuan Chen1† Shile Luo1†
Shile Luo1† Jianmei Tan1
Jianmei Tan1 Luhua Zhang1
Luhua Zhang1 Shengwu Qiu1
Shengwu Qiu1 Zhiyou Hao2
Zhiyou Hao2 Naidong Wang1
Naidong Wang1 Zhibang Deng1
Zhibang Deng1 Aibing Wang1
Aibing Wang1 Qing Yang1
Qing Yang1 Yi Yang1
Yi Yang1 Changjian Wang3*
Changjian Wang3* Yang Zhan1*
Yang Zhan1*Many pathogens cause reproductive failure in sows suffering a broad spectrum of sequelae, including abortions, stillbirth, mummification, embryonic death, and infertility. Although various detection methods, such as polymerase chain reaction (PCR) and real-time PCR, have been widely used for molecular diagnosis, mainly for a single pathogen. In this study, we developed a multiplex real-time PCR method for the simultaneous detection of porcine circovirus type 2 (PCV2), porcine circovirus type 3 (PCV3), porcine parvovirus (PPV) and pseudorabies virus (PRV) associated with porcine reproductive failure. The R2 values for the standard curve of multiplex real-time PCR of PCV2, PCV3, PPV, and PRV reached to 0.996, 0.997, 0.996, and 0.998, respectively. Importantly, the limit of detection (LoD) of PCV2, PCV3, PPV, and PRV, were 1, 10, 10, 10 copies/reaction, respectively. Meanwhile, specificity test results indicated that multiplex real-time PCR for simultaneous detection is specific for these four target pathogens and does not react with other pathogens, such as classical swine fever virus, porcine reproductive and respiratory syndrome virus, and porcine epidemic diarrhea virus. Besides, this method had good repeatability with coefficients of variation of intra- and inter-assay less than 2%. Finally, this approach was further evaluated by 315 clinical samples for its practicality in the field. The positive rates of PCV2, PCV3, PPV, and PRV were 66.67% (210/315), 8.57% (27/315), 8.89% (28/315), and 4.13% (13/315), respectively. The overall co-infection rates of two or more pathogens were 13.65% (43/315). Therefore, this multiplex real-time PCR provides an accurate and sensitive method for the identification of those four underlying DNA viruses among potential pathogenic agents, allowing it to be applied in diagnostics, surveillance, and epidemiology.
Some viruses causing reproductive failures in wild boar and domestic swine have posed significant economic threat to the swine industry (Pacini et al., 2021). Swine productions are significantly affected by the four main pathogenic DNA viruses, such as porcine circovirus type 2 (PCV2), porcine circovirus type 3 (PCV3), porcine parvovirus (PPV), and pseudorabies virus (PRV) (Díaz et al., 2021). PCV2 and PCV3 are non-enveloped single-stranded DNA viruses and the members of genus Circovirus of the Circoviridae family, and the two PCVs have a similar genomic structure: the viral genome contains two major open reading frames (ORFs) oriented in opposite directions, ORF1 and ORF2, which encode replication-related proteins and the capsid protein, respectively (Turlewicz-Podbielska et al., 2022). Furthermore, co-infections of PCV2 and PCV3 were found in swine herds, which can cause similar clinical manifestations including reproductive failure, porcine dermatitis, nephropathy syndrome and respiratory disease (Xu et al., 2021; Sirisereewan et al., 2022). PPV is also a non-enveloped single-stranded DNA virus, and its genome contains two ORFs: ORF1 encodes nonstructural proteins of NS1 and NS2, while ORF2 encodes two structural proteins of VP1 and VP2 (Streck and Truyen, 2020). Among seven PPV types, PPV type 1 (PPV1) is a dominant etiological agent causing reproductive disorders in Chinese swine herds (Li et al., 2021). PRV is an enveloped double-stranded DNA virus, and belongs to the subfamily of alpha-herpesvirus in the family of Herpesviridae (Xu et al., 2020). PRV can cause neurological disorders and reproductive failure in swine, which is considered one of the most economically important viral swine diseases worldwide (Lv et al., 2020).
The four viruses with similar clinical manifestations including reproductive failure cause severe economic losses to the swine industry and world trade of pork products, however, differential diagnosis of diseases caused by these four viruses is difficult. Furthermore, co-infections with more than two pathogens of PCV2, PCV3, PPV, and PRV occur frequently in swine herds in China and elsewhere worldwide (Ma et al., 2021; Serena et al., 2021; Jia et al., 2022). Therefore, the prevention and control of these diseases caused by the four different viruses mainly depend on the accurate and rapid molecular diagnosis, and it is essential to develop a specific, high-sensitive, and rapid assay being capable of the simultaneous and differential detection of PCV2, PCV3, PPV, and PRV.
Among various detection methods, real-time PCR is an accurate, sensitive, and rapid method for the detection and quantification of the target genome, which is based on continuous measurements of the accumulation of fluorescent signals during the amplification reaction. However, conventional singular real-time PCR is time-consuming and costly to detect multiple pathogens in the same sample. With the development of this technology, most real-time PCR are based on the use of target-specific TaqMan probes, and multiplex real-time PCR can simultaneously detect more than one target gene with one reaction at one time (Hawkins and Guest, 2017; Zhu et al., 2022). In this study, primers and probes were designed based on the conserved fragments of the cap gene of PCV2, the cap gene of PCV3, the NS1 gene of PPV and the gE gene of PRV, and a TaqMan probe-based multiplex real-time PCR method was successfully developed. The method showed high sensitivity and specificity, without cross-reactions with other tested swine pathogen genomes. Additionally, this approach provides an accurate identification of the pathogenic agents among the four underlying DNA viruses. This approach can thus be applied in diagnostics, surveillance, and epidemiology.
Genomes (DNA and cDNA) of PCV2, PCV3, PPV, PRV, classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV), and porcine epidemic diarrhea virus (PEDV) were all provided by the Animal Disease Prevention and Control Center of Hunan Province or the Provincial Key Laboratory of Protein Engineering in Animal Vaccines of Hunan Province. To test clinical samples from pig farms with reproductive disorders, DNA samples were extracted from tissues or blood by using the Tissue Genomic DNA Extraction Kit (Tiangen Biotech).
For primers and probe design, different numbers of representative viral genomes (14 for PCV2, 11 for PCV3, 9 for PPV, and 11 for PRV) were obtained from GenBank, as reported and presented in genetic studies (Oh et al., 2017; Franzo and Segalés, 2018; Fux et al., 2018; He et al., 2019). Three distinct primer sets for each viral gene were designed using Primer Premier v5.0 based on the conserved region of reference strains, and further confirmed using the Blast tool available from the National Center for Biotechnology Information (NCBI). After these three sets of primers were tested by traditional PCR and then used with an appropriate probe to establish standard curves, the final primers/probes were determined after comparing the differences in specificity and sensitivity between the three sets of results. The locations of these final primers and probes were shown among different reference strains of PCV2, PCV3, PPV, and PRV (Figure 1). Detailed information of these primers and probes is listed in Table 1. For multiplex detection, the probe for the four viral genes were labeled with the different 5′-reporting dyes (FAM, VIC, ROX and Cy5) and the corresponding 3′-quenchers (MGB, BHQ1, BHQ2, and BHQ3). In addition, the sequences of the primers specific for three RNA viruses (CSFV, PRRSV, and PEDV) are shown in Supplementary Table S1.

Figure 1. Alignments of coding sequences of reference viral strains deposited in GenBank. The locations of the primers/probes specific for PCV2 cap gene, PCV3 cap gene, PPV NS1 gene, and PRV gE gene are shown. The positions of the partial nucleotide fragments are indicated by numbers.
Four gene fragments of PCV2, PCV3, PPV, and PRV, generated by PCR using their respective genomes as templates, were simultaneously inserted into a cloning vector of pSP72 (Promega) in tandem and served as a standard plasmid (pSP72-PCV2 cap & PCV3 cap & PPV NS1 & PRV gE) in subsequent assays (Figure 2A). Briefly, the four gene fragments were digested with XhoI/HindIII, HindIII/BamHI, BamHI/KpnI, and KpnI/SacI(Thermo Fisher Scientific), and then purified and ligated to the vector digested with XhoI/ SacI. Finally, the ligation products were transformed into Escherichia coli DH5α competent cells (TransGen). The positive clones were cultured at 37°C for 20 h then extracted by Plasmid DNA Mini Kit (Omega) and confirmed by DNA sequencing. The resulting plasmids were quantified by ultraviolet absorbance at 260 nm and 280 nm using a NanoDrop spectrophotometer (Thermo Fisher, USA). According to the size of the standard plasmid template, the copy numbers of the purified plasmids were calculated based on the following formula: (A260 (ng/μL) × 10−9 × 6.02 × 1023) / (DNA length×650) = copies/μL. Then, 10-fold serial dilutions of the standard plasmid, with the same starting concentration of 109 copies/μL, were performed to generate four standard curves for testing the multiplex real-time PCR.
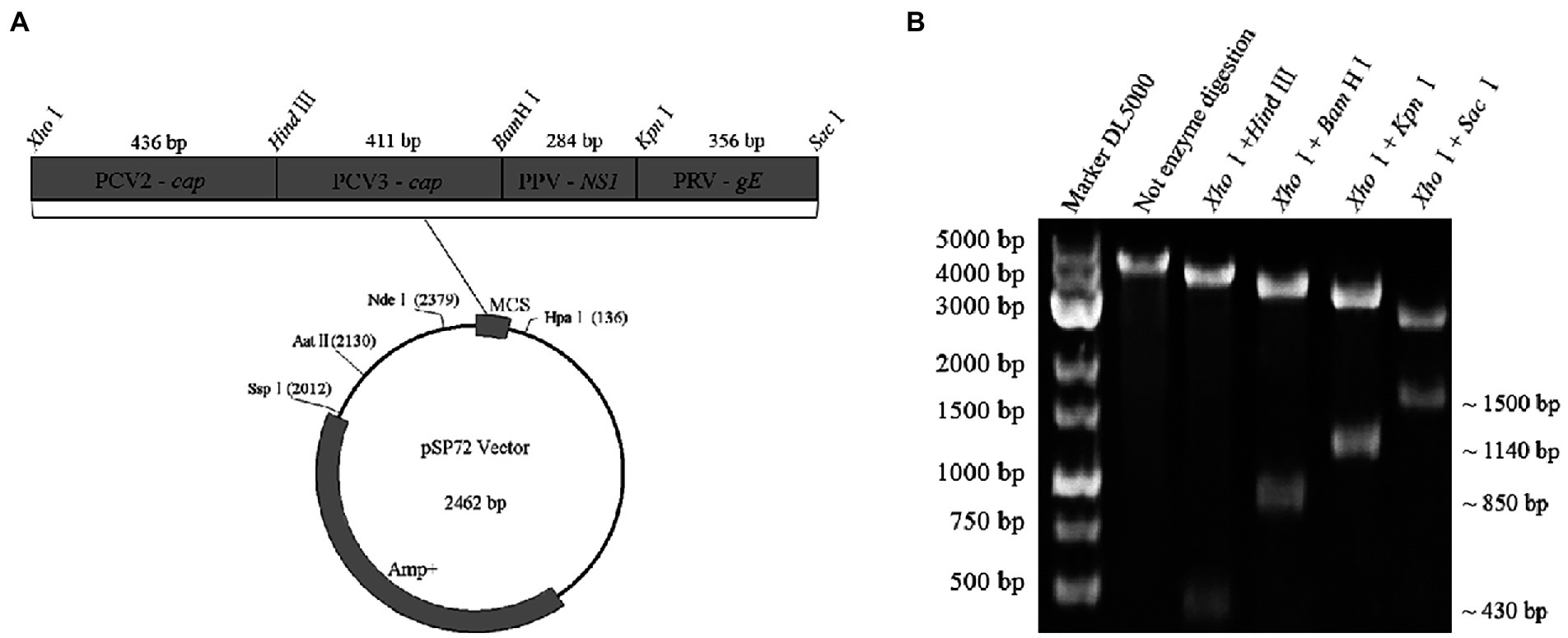
Figure 2. Construction and identification of a recombinant plasmid contains four genes in tandem. (A) Representative diagram of recombinant plasmid. (B) Restriction digestion assays of recombinant plasmid: ~430 bp in size specific for the cap gene of PCV2, ~850 bp in size specific for the two gene fragments (PCV2 cap and PCV3 cap) in tandem, ~1,140 bp in size specific for the three gene fragments (PCV2 cap, PCV3 cap, and PPV NS1) in tandem, ~1,500 bp in size specific for the four gene fragments (PCV2 cap, PCV3 cap, PPV NS1, and PRV gE) in tandem were identified by digestion with XhoI/HindIII, XhoI/BamHI, XhoI/KpnI and XhoI/SacI, respectively.
Multiplex real-time PCR was performed in a 20 μL reaction mixture containing 10 μL of 2 × AceQ real-time PCR Probe Master Mix (Vazyme), 0.4 μL of each primer (final concentration of 400 nM), 0.2 μL of each probe (final concentration of 200 nM), 1 μL of DNA template, and an appropriate amount of RNase-free water. The amplification parameters of the qPCR detection system were as follows: denaturation at 95°C for 5 min, followed by 45 cycles of denaturation at 95°C for 10 s, and annealing and extension at 60°C for 30 s. The acquisition of fluorescent signals was recorded at the end of each cycle. After the reaction, the system automatically determined the Ct values. The standard plasmid was 10-fold serially diluted from 108 copies/μL to 101 copies/μL and used as templates to construct the standard curves of the multiplex real-time PCR.
To evaluate the specificity of primer and probe sets, cDNA of CSFV, PRRSV, and PEDV were confirmed via PCR with gene-specific primers serving as templates for amplification using the developed multiplex real-time PCR. For the sensitivity evaluation, the standard plasmid was serially diluted 10-fold and the final concentration ranged from 1 × 106 copies/μL to 1 × 100 copies/μL. These diluted plasmids were used as templates to determine the limit of detection (LoD) of the multiplex real-time PCR and each reaction was repeated triplicate in a test. To ensure the accuracy of the limit of LoD, 100 copies/μL, 10 copies/μL and 1 copy/μL of standard plasmid were tested repeatedly at 20 times, and the lowest concentration of the positive control that can be positively detected (60% replicates positive) was set as the LoD and labeled in the amplification curve. We judged the positive detection of the lowest concentration (100 copies/μL, 10 copies/μL and 1 copy/μL) according to the criteria of method and calculated the positive detection rate.
To evaluate the repeatability of the established multiplex real-time PCR, 106, 104 and 102 copies/μL of the abovementioned standard plasmid were selected as templates to conduct the intra-assay and inter-assay tests. All reactions were repeated three times in one experiment.
A total of 315 clinical swine samples including lung, lymph nodes and blood, collected in Hunan Province, Southern China, were tested to detect the PCV2, PCV3, PPV, and PRV by the multiplex real-time PCR established in this study. In the comparative experiment, some pathogen-positive and pathogen-negative samples (n > 10) confirmed by the developed multiplex real-time PCR were selected. 100 pathogen-positive specimens (84 for PCV2, 26 for PCV3, 27 for PPV, 13 for PRV, including 43 co-infected samples, as shown in Supplementary Table S2) and 14 pathogen-negative specimens were selected and further analyzed by four singular real-time PCR. The primers and probes of singular real-time PCR were described from other laboratories (Song et al., 2010; Wang et al., 2017; Henriques et al., 2018; Tu et al., 2021). For each singular real-time PCR, a 20 μL reaction mixture containing 10 μL of 2 × AceQ real-time PCR Probe Master Mix (Vazyme), 0.4 μL of each primer (final concentration of 400 nM), 0.2 μL of probe (final concentration of 200 nM), 1 μL of DNA template, and an appropriate amount of RNase-free water. The amplification parameters of the qPCR detection system for singular real-time PCR were as follows: denaturation at 95°C for 5 min, followed by 45 cycles of denaturation at 95°C for 10 s, and annealing and extension at 60°C for 30 s. The acquisition of fluorescent signal (FAM) was recorded at the end of each cycle. After the reaction, the system automatically determined the Ct values. According to its application in sample detection in our laboratory, the criteria of singular real-time PCR for judging the outcome were as follows: A Ct value less than 40.0 cycles was considered positive, otherwise negative.
A standard plasmid containing the four gene fragments of PCV2 cap, PCV3 cap, PPV NS1, and PRV gE in tandem was successfully constructed and analyzed by restriction enzyme digestions assays. As expected, 4 gene fragments with distinct sizes were identified after restrictions (Figure 2B). Finally, the plasmid was further confirmed by double-strand DNA sequencing, and it was used as the standard plasmid for the subsequent assays.
The standard plasmid was subjected to 10-fold serial dilutions to establish the standard curve, and 8 standard samples (108 ~ 101 copies/μL) were selected as templates to establish a standard curve for the multiplex real-time PCR. As shown in Figure 3, the corresponding correlation coefficient (R2), slope of the equation and amplification efficiency (E) were as follows: 0.996, −3.367, and 98.169%, respectively, for PCV2; 0.997, −3.463, and 94.435%, respectively, for PCV3; 0.996, −3.371, and 97.996%, respectively, for PPV; 0.998, −3.436, and 95.454%, respectively, for PRV. Both R2 and E indicated a good linear relationship between the initial templates and Ct values. According to the intercepts of the standard curve, the criteria for judging the outcome were as follows: A Ct value less than 40.0 cycles was considered positive, otherwise negative.
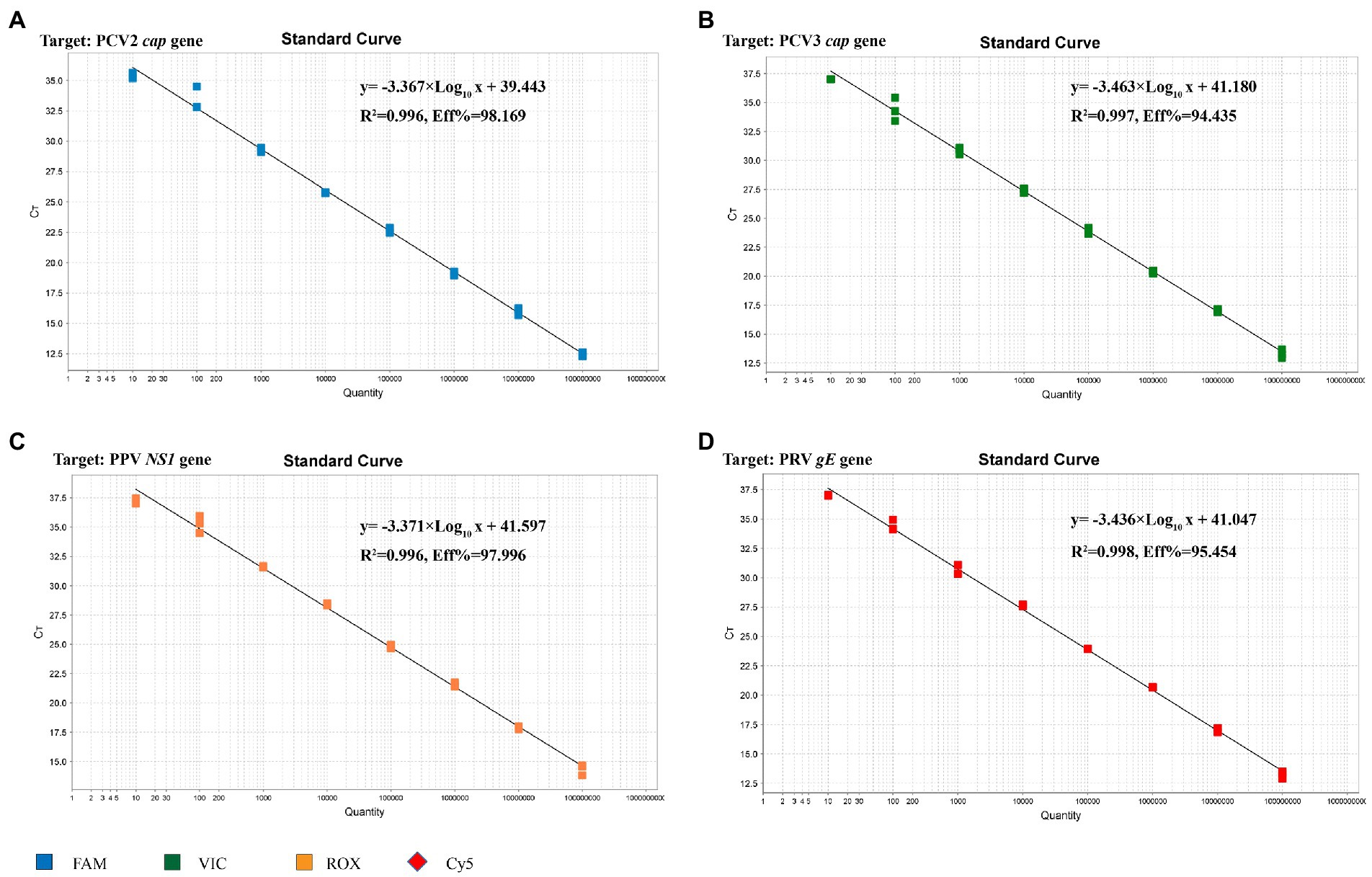
Figure 3. Standard curve for multiplex real-time PCR assay. (A) The standard curve for the PCV2 cap gene. (B) The standard curve for the PCV3 cap gene. (C) The standard curve for the PPV NS1 gene. (D) The standard curve for the PRV gE gene.
The genomic DNA or cDNA of different porcine viruses (PCV2, PCV3, PPV, PRV, CSFV, PRRSV, and PEDV) were used as templates of the multiplex real-time PCR, and cDNA of CSFV, PRRSV, and PEDV were confirmed via PCR with gene-specific primers (Figure 4A). As a result, only PCV2, PCV3, PPV, and PRV showed amplification curves, while no detectable fluorescent signal was observed among the templates derived from CSFV, PRRSV and PEDV (Figure 4B). Thus, the established multiplex real-time PCR is specific for the detection of the four viruses (PCV2, PCV3, PPV, and PRV) herein investigated and has no cross-reactivity with the other 3 swine pathogens.
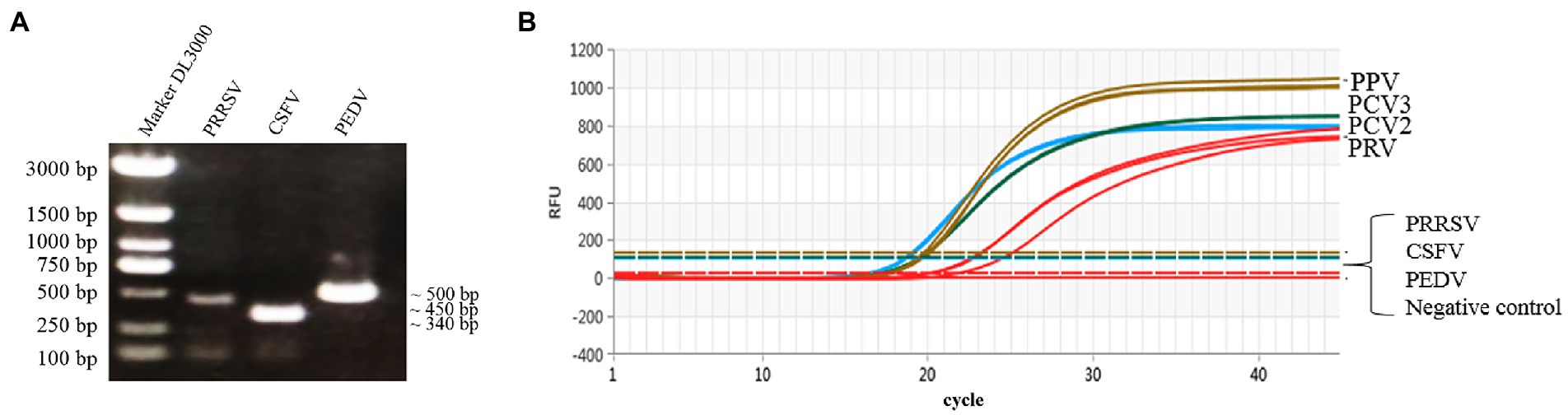
Figure 4. Specific amplification curves by multiplex real-time PCR assay. (A) Agarose gel electrophoresis of viral replicons (PRRSV, CSFV, and PEDV) amplified by PCR. Lane 1 represents the 3,000-bp marker, Lanes 2–4 represent PRRSV (451 bp), CSFV (343 bp) and PEDV (503 bp), respectively. (B) Four fluorescent signals were monitored by multiplex real-time PCR, and DNA of PCV2, PCV3, PPV, and PRV were used as a positive control. No fluorescent signal was observed when other viral cDNAs were used as templates. Note: The graph type was set as in linear phase to simultaneously display the four different fluorescent signals (FAM, VIC, ROX, and Cy5) with distinct signal intensities.
The standard plasmid was serial diluted 10-fold from 1 × 106 copies/μL to 1 × 100 copies/μL to determine the LoD of the multiplex real-time PCR. The Lod of PCV2 was 1 copy/μL, and the Lod of all other 3 viruses (PCV3, PPV and PRV) were 10 copies/μL (Figure 5; Supplementary Table S3), highlighting the high sensitivity of the multiplex real-time PCR.
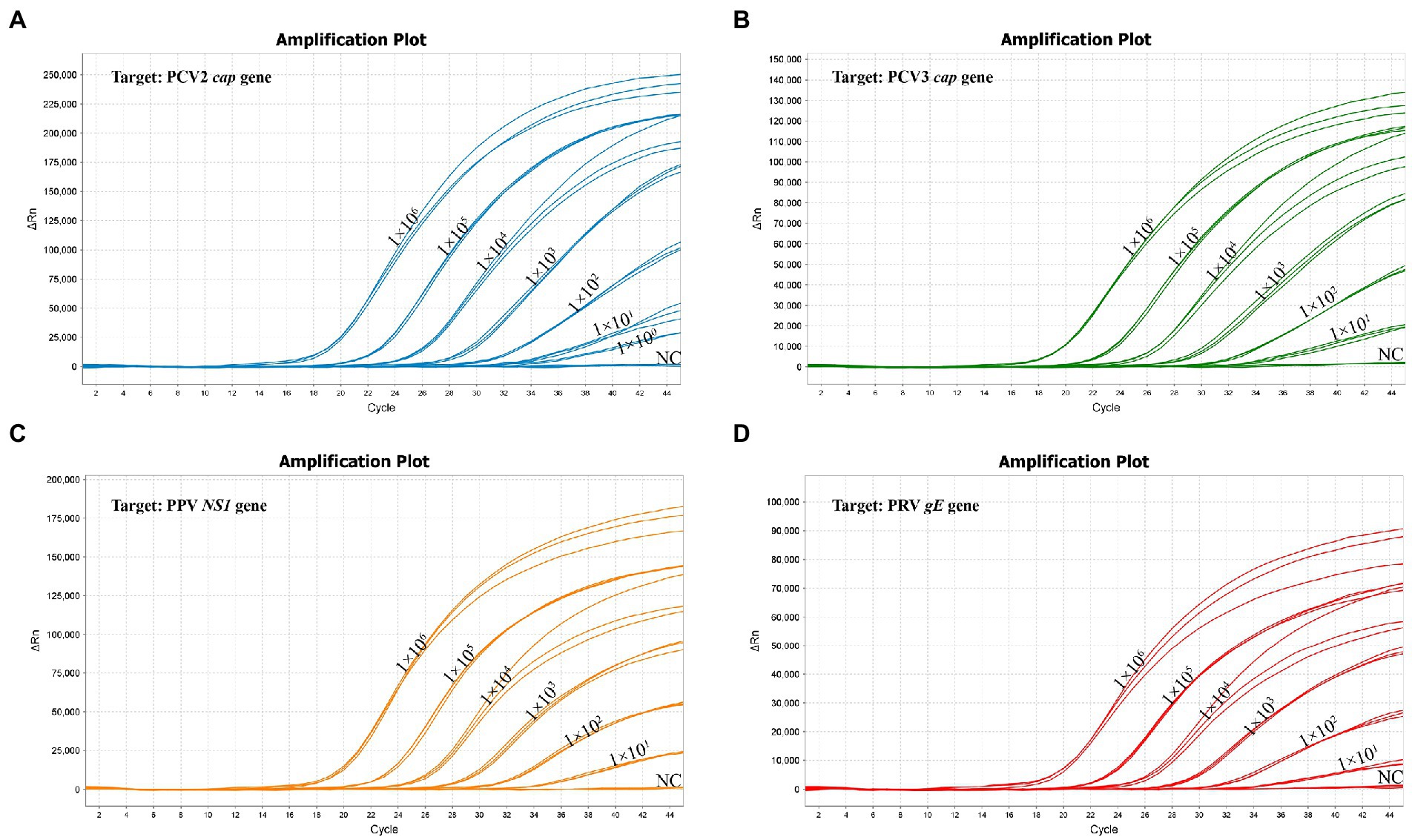
Figure 5. The sensitivity of multiplex real-time PCR assay. (A) The sensitivity for the PCV2 cap gene. (B) The sensitivity for the PCV3 cap gene. (C) The sensitivity for the PPV NS1 gene. (D) The sensitivity for the PRV gE gene.
Three concentrations of 1 × 106, 1 × 104 and 1 × 102 copies/μL of standard plasmids were used to evaluate the reproducibility of the multiplex real-time PCR. As a result, the coefficients of variation of the intra- and inter-assay ranged from 0.03% to 1.50% and 0.16% to 1.82%, respectively (Table 2), indicating good repeatability of the assay.
A total of 315 clinical swine samples were collected from 14 regions in Hunan Province, Southern China, and detected using the developed multiplex real-time PCR to evaluate its practicality in the field. As a result (Table 3), the positive rates of PCV2, PCV3, PPV, and PRV were 66.67% (210/315), 8.57% (27/315), 8.89% (28/315), and 4.13% (13/315), respectively. Furthermore, the positive rates of co-infections with two or more distinct viruses were 13.65% (43/315; Table 3), and the Ct values obtained from the four fluorescent channels (FAM, VIC, ROX, and Cy5) from these 43 samples were shown (Supplementary Table S2), indicating that this method can really detect multiple viruses simultaneously.
Among the verified DNA samples, 100 pathogen-positive specimens (including above 43 co-infected samples and 57 singular infected samples) and 14 pathogen-negative specimens were also detected by the singular real-time PCR. As shown in Table 4, the positive rates of PCV2, PCV3, PPV, and PRV were 68.42% (78/114), 21.05% (24/114), 20.18% (23/114), and 9.65% (11/114), respectively, and the agreement rate of the developed multiplex real-time PCR and the singular real-time PCR was 94.01% (PCV2), 95.47% (PCV3), 90.82% (PPV) and 91.14% (PRV), respectively. In addition, no sample among 14 pathogen-negative specimens was tested positive for these pathogens (PCV2, PCV3, PPV, and PRV) by the singular real-time PCR.
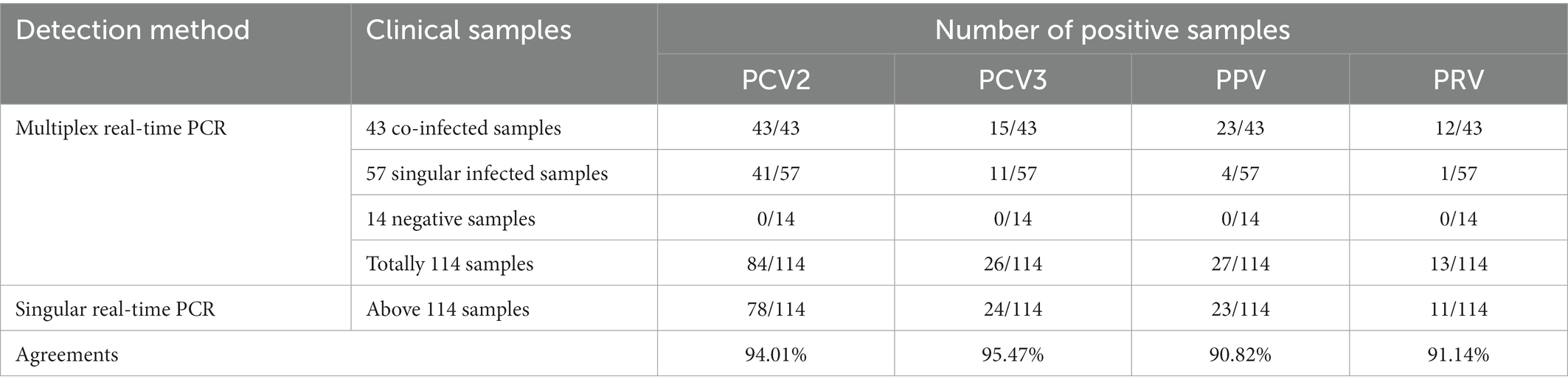
Table 4. Comparative results of clinical samples tested with both multiplex real-time PCR and singular real-time PCR.
International travel and large-scale logistics significantly increase the prevalence and spread of animal pathogens, such as PCV2, PPV, and PRV, leading not only to animal infections, but also the substantial economic loss to the pig industry (VanderWaal and Deen, 2018). PCV2, PCV3, PPV, and PRV are all DNA viruses causing reproductive failures that have seriously negative effects on the swine industry worldwide. These viruses are still prevalent in many countries and areas, and co-infections with each other are common in some pig herds (Zhao et al., 2019; Ma et al., 2020; Zhou et al., 2020; Kang et al., 2022; Kim et al., 2022). Because pigs infected by these viruses sometimes show similar clinical symptoms, it is difficult to identify the key causative agent(s) only by clinical observations (Zhang et al., 2019; Vargas-Bermúdez et al., 2021; Li et al., 2022). Multiplex real-time PCR uses gene-specific primers and probes to amplify several target genes simultaneously in a single tube. Therefore, the assay is rapid, sensitive, accurate, and time-saving, and it has been widely used to detect various viruses in veterinary laboratories (Scherrer and Stephan, 2021; Liu et al., 2022). Thus, it is pertinent to develop a reliable method for the simultaneous detection of PCV2, PCV3, PPV, and PRV in laboratory and accurate diagnosis for the diseases associated with the four viruses in the field.
In this study, four pairs of viral gene-specific primers and corresponding probes (Table 1; Figure 1) were designed for the multiplex real-time PCR, which can simultaneously detect the presences of the four genes via different fluorescent reporters in one tube. Furthermore, instead of using a mixture of four standard plasmids, we inserted the four gene fragments into the same vector in tandem to generate one standard plasmid for use in the multiplex real-time PCR system (Figure 2). The advantages of using one standard plasmid harboring four viral gene fragments in this assay will decrease the cost of preparing a standard plasmid compared to using four standard plasmids. Besides, this approach may decrease system errors caused by the need to add four distinct plasmids. The sensitivity assay demonstrated that this method could detect less than 10 copies of target genes in our standard plasmid harboring the four genes (Figure 5; Supplementary Table S3), and the standard curve plots showed a strong linear correlation between the Ct value and the standard copy numbers (Figure 3). Based on the specificity of the multiplex probe-based real-time PCR, this assay could accurately detect PCV2, PCV3, PPV, and PRV without cross-reactions with other porcine RNA viruses (CSFV, PRRSV, and PEDV; Figure 4). Furthermore, the developed multiplex real-time PCR was experimentally verified with confirmed ASFV-positive samples, and the results demonstrated good specificity of the assay, which could specifically detect four viruses herein investigated (data not shown). Meanwhile, more herein investigated pathogen-positive samples need to be tested to confirm the developed multiplex real-time PCR have a broad utility, and further studies are warranted.
In addition, the assay was further employed to detect 315 clinical samples to verify its practicality and usefulness in the clinical samples. Our results demonstrate that 210 (66.67%), 27 (8.57%), 28 (8.89%), and 13 (4.13%) samples were positive for PCV2, PCV3, PPV, and PRV, respectively (Table 3). This suggests that PCV2, PCV3, PPV, and PRV were still prevalent in Hunan province, Southern China. Furthermore, co-infections with two or more pathogens among PCV2, PCV3, PPV, and PRV are also common, which may elevate immunosuppression and inflammation, and thus increase the possibility of secondary infection by other pathogen(s), and further exacerbate these diseases (Vargas-Bermúdez et al., 2021; Wu et al., 2021; Li et al., 2022). The detection results of the clinical samples showed that co-infections with 2 or more pathogens of PCV2, PCV3, PPV, and PRV still existed in 43 (13.65%) samples (Table 3; Supplementary Table S2) in Hunan province. At the same time, 114 out of 315 clinical samples were selected and detected by using the singular real-time PCR. The results indicated that the agreement rate of the developed multiplex real-time PCR and the singular real-time PCR were more than 90%, and the multiplex real-time PCR was more sensitive than singular real-time PCR (Table 4).
In conclusion, a multiplex real-time PCR was developed for the simultaneous and differential detection of PCV2, PCV3, PPV, and PRV to rapidly and accurately detect PCV2, PCV3, PPV, and PRV from clinical samples, this novel method could be used as a more convenient tool for the accurate diagnosis and epidemiological investigation of these viruses.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Ethics Committee for Biomedical Research of Hunan Agricultural University, Changsha, China, following national legislation regarding animal welfare. Written informed consent was obtained from the owners for the participation of their animals in this study.
YZ, CW, ZD, and YY conceived and designed the study. YC, SQ, JT, LZ, SL, and ZH performed the experiments and analyzed data. YC, SQ, and YZ wrote the paper, NW, QY, and AW revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation of China (32172844 and 32202790), Hunan Provincial Natural Science Foundation of China (2022JJ30298 and 2022JJ40183), Natural Science Foundation of Changsha City (kq2202231), the Young Talents Science and Technology Project in Hunan Province (2022RC1046), and Hunan Province Technology Breakthrough Project of 2021 for the open competition mechanism to select the best candidates (grant number: 2021NK1030).
We thank Animal Disease Prevention and Control Center of Hunan Province and Animal Disease Prevention and Control Center of Yongzhou for offering partial clinical samples, and the authors also thank all staffs from these organizations that participated in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1092273/full#supplementary-material
Díaz, C., Celer, V., and Frébort, I. (2021). The Main DNA viruses significantly affecting pig livestock. J. Vet. Res. 65, 15–25. doi: 10.2478/jvetres-2021-0001
Franzo, G., and Segalés, J. (2018). Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS. One 13:e0208585. doi: 10.1371/journal.pone.0208585
Fux, R., Söckler, C., Link, E. K., Renken, C., Krejci, R., Sutter, G., et al. (2018). Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 15:25. doi: 10.1186/s12985-018-0929-3
Hawkins, S. F. C., and Guest, P. C. (2017). Multiplex analyses using real-time quantitative PCR. Methods Mol. Biol. 1546, 125–133. doi: 10.1007/978-1-4939-6730-8_8
He, W., Auclert, L. Z., Zhai, X., Wong, G., Zhang, C., Zhu, H., et al. (2019). Interspecies transmission, genetic diversity, and evolutionary dynamics of Pseudorabies virus. J. Infect. Dis. 219, 1705–1715. doi: 10.1093/infdis/jiy731
Henriques, A. M., Duarte, M., Barros, S. C., Fagulha, T., Ramos, F., Luís, T., et al. (2018). Development and validation of a real-time PCR for the detection and quantification of porcine circovirus type 2. Virus 29, 355–361. doi: 10.1007/s13337-018-0476-y
Jia, Y., Zhu, Q., Xu, T., Chen, X., Li, H., Ma, M., et al. (2022). Detection and genetic characteristics of porcine circovirus type 2 and 3 in Henan province of China. Mol. Cell. Probes 61:101790. doi: 10.1016/j.mcp.2022.101790
Kang, L., Wahaab, A., Shi, K., Mustafa, B. E., Zhang, Y., Zhang, J., et al. (2022). Molecular epidemic characteristics and genetic evolution of porcine Circovirus type 2 (PCV2) in swine herds of Shanghai, China. Viruses 14:289. doi: 10.3390/v14020289
Kim, S. C., Kim, J. H., Kim, J. Y., Park, G. S., Jeong, C. G., and Kim, W. I. (2022). Prevalence of porcine parvovirus 1 through 7 (PPV1-PPV7) and co-factor association with PCV2 and PRRSV in Korea. BMC Vet. Res. 18:133. doi: 10.1186/s12917-022-03236-1
Li, X., Chen, S., Zhang, L., Niu, G., Zhang, X., Yang, L., et al. (2022). Coinfection of porcine Circovirus 2 and Pseudorabies virus enhances immunosuppression and inflammation through NF-κB, JAK/STAT, MAPK, and NLRP3 pathways. Int. J. Mol. Sci. 23:4469. doi: 10.3390/ijms23084469
Li, J., Xiao, Y., Qiu, M., Li, X., Li, S., Lin, H., et al. (2021). A systematic investigation unveils high Coinfection status of porcine parvovirus types 1 through 7 in China from 2016 to 2020. Microbiol. Spectr. 9:e0129421. doi: 10.1128/Spectrum.01294-21
Liu, H., Shi, K., Zhao, J., Yin, Y., Chen, Y., Si, H., et al. (2022). Development of a one-step multiplex qRT-PCR assay for the detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. BMC Vet. Res. 18:43. doi: 10.1186/s12917-022-03144-4
Lv, L., Cao, M., Bai, J., Jin, L., Wang, X., Gao, Y., et al. (2020). PRV-encoded UL13 protein kinase acts as an antagonist of innate immunity by targeting IRF3-signaling pathways. Vet. Microbiol. 250:108860. doi: 10.1016/j.vetmic.2020.108860
Ma, Z., Han, Z., Liu, Z., Meng, F., Wang, H., Cao, L., et al. (2020). Epidemiological investigation of porcine pseudorabies virus and its coinfection rate in Shandong Province in China from 2015 to 2018. J. Vet. Sci. 21:e36. doi: 10.4142/jvs.2020.21.e36
Ma, Z., Liu, M., Liu, Z., Meng, F., Wang, H., Cao, L., et al. (2021). Epidemiological investigation of porcine circovirus type 2 and its coinfection rate in Shandong province in China from 2015 to 2018. BMC Vet. Res. 17:17. doi: 10.1186/s12917-020-02718-4
Oh, W. T., Kim, R. Y., Nguyen, V. G., Chung, H. C., and Park, B. K. (2017). Perspectives on the evolution of porcine parvovirus. Viruses 9:196. doi: 10.3390/v9080196
Pacini, M. I., Forzan, M., Cilia, G., Bertelloni, F., Fratini, F., and Mazzei, M. (2021). Detection and characterization of viral pathogens associated with reproductive failure in wild boars in Central Italy. Animals (Basel) 11:304. doi: 10.3390/ani11020304
Scherrer, S., and Stephan, R. (2021). Novel multiplex TaqMan assay for differentiation of the four major pathogenic Brachyspira species in swine. Microbiology 10:e1169. doi: 10.1002/mbo3.1169
Serena, M. S., Dibárbora, M., Olivera, V., Metz, G. E., Aspitia, C. G., Pereda, A., et al. (2021). Evidence of porcine circovirus type 2 and co-infection with ungulate protoparvovirus 1 (porcine parvovirus) in mummies and stillborn piglets in subclinically infected farm. Infect. Genet. Evol. 89:104735. doi: 10.1016/j.meegid.2021.104735
Sirisereewan, C., Thanawongnuwech, R., and Kedkovid, R. (2022). Current understanding of the pathogenesis of porcine Circovirus 3. Pathogens 11:64. doi: 10.3390/pathogens11010064
Song, C., Zhu, C., Zhang, C., and Cui, S. (2010). Detection of porcine parvovirus using a taqman-based real-time pcr with primers and probe designed for the NS1 gene. Virol. J. 7:353. doi: 10.1186/1743-422X-7-353
Streck, A. F., and Truyen, U. (2020). Porcine Parvovirus. Curr. Issues Mol. Biol. 37, 33–46. doi: 10.21775/cimb.037.033
Tu, F., Zhang, Y., Xu, S., Yang, X., Zhou, L., Ge, X., et al. (2021). Detection of pseudorabies virus with a real-time recombinase-aided amplification assay. Transbound. Emerg. Dis. 68, 2017–2027. doi: 10.1111/tbed.13849
Turlewicz-Podbielska, H., Augustyniak, A., and Pomorska-Mól, M. (2022). Novel porcine Circoviruses in view of lessons learned from porcine Circovirus type 2-epidemiology and threat to pigs and other species. Viruses 14:261. doi: 10.3390/v14020261
VanderWaal, K., and Deen, J. (2018). Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. U. S. A. 115, 11495–11500. doi: 10.1073/pnas.1806068115
Vargas-Bermúdez, D. S., Vargas-Pinto, M. A., Mogollón, J. D., and Jaime, J. (2021). Field infection of a gilt and its litter demonstrates vertical transmission and effect on reproductive failure caused by porcine circovirus type 3 (PCV3). BMC Vet. Res. 17:150. doi: 10.1186/s12917-021-02862-5
Wang, J., Zhang, Y., Wang, J., Liu, L., Pang, X., and Yuan, W. (2017). Development of a TaqMan-based real-time PCR assay for the specific detection of porcine circovirus 3. J. Virol. Methods 248, 177–180. doi: 10.1016/j.jviromet.2017.07.007
Wu, X., Wang, Z., Qiao, D., Yuan, Y., Han, C., Yang, N., et al. (2021). Porcine circovirus type 2 infection attenuates the K63-linked ubiquitination of STING to inhibit IFN-β induction via p38-MAPK pathway. Vet. Microbiol. 258:109098. doi: 10.1016/j.vetmic.2021.109098
Xu, S., Chen, D., Chen, D., Hu, Q., Zhou, L., Ge, X., et al. (2020). Pseudorabies virus infection inhibits stress granules formation via dephosphorylating eIF2α. Vet. Microbiol. 247:108786. doi: 10.1016/j.vetmic.2020.108786
Xu, T., Zhang, Y. H., Tian, R. B., Hou, C. Y., Li, X. S., Zheng, L. L., et al. (2021). Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in Central China. Infect. Genet. Evol. 94:105016. doi: 10.1016/j.meegid.2021.105016
Zhang, X., Shu, X., Bai, H., Li, W., Li, X., Wu, C., et al. (2019). Effect of porcine circovirus type 2 on the severity of lung and brain damage in piglets infected with porcine pseudorabies virus. Vet. Microbiol. 237:108394. doi: 10.1016/j.vetmic.2019.108394
Zhao, Y., Han, H. Y., Fan, L., Tian, R. B., Cui, J. T., Li, J. Y., et al. (2019). Development of a TB green II-based duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of porcine circovirus 2 and 3. Mol. Cell. Probes 45, 31–36. doi: 10.1016/j.mcp.2019.04.001
Zhou, H., Pan, Y., Liu, M., and Han, Z. (2020). Prevalence of porcine Pseudorabies virus and its Coinfection rate in Heilongjiang Province in China from 2013 to 2018. Viral Immunol. 33, 550–554. doi: 10.1089/vim.2020.0025
Keywords: porcine circovirus type 2, porcine circovirus type 3, porcine parvovirus, pseudorabies virus, multiplex real-time PCR
Citation: Chen Y, Luo S, Tan J, Zhang L, Qiu S, Hao Z, Wang N, Deng Z, Wang A, Yang Q, Yang Y, Wang C and Zhan Y (2023) Establishment and application of multiplex real-time PCR for simultaneous detection of four viruses associated with porcine reproductive failure. Front. Microbiol. 14:1092273. doi: 10.3389/fmicb.2023.1092273
Received: 07 November 2022; Accepted: 18 January 2023;
Published: 09 February 2023.
Edited by:
Mohammed Rohaim, Lancaster University, United KingdomReviewed by:
Giovanni Franzo, University of Padua, ItalyCopyright © 2023 Chen, Luo, Tan, Zhang, Qiu, Hao, Wang, Deng, Wang, Yang, Yang, Wang and Zhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjian Wang, ✉ d2FuZ2NqMTk3N0AxNjMuY29t; Yang Zhan, ✉ eWFuZ3poYW5AaHVuYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.