- 1Department of Bioinformatics, School of Life Sciences, Xuzhou Medical University, Xuzhou, Jiangsu, China
- 2Department of Biophysics, School of Life Sciences, Xuzhou Medical University, Xuzhou, Jiangsu, China
Critically ill patients have higher risk of serious fungal infections, such as invasive aspergillosis (IA) which is mainly caused by the human fungal pathogen Aspergillus fumigatus. Triazole drugs are the primary therapeutic agents for the first-line treatment of IA, which could easily cause drug resistance problems. Here, we assess the potential of AgNPs synthesized with Artemisia argyi leaf extract and domiphen as new antifungal agents to produce synergistic antimicrobial effects on Aspergillus fumigatus, and dissect possible molecular mechanisms of action. Plate inoculation assays combined with drug susceptibility test and cytotoxicity test showed that the combination of AgNPs and domiphen has synergistic antimicrobial effects on A. fumigatus with low cytotoxicity. Gene Ontology (GO) enrichment analysis showed that AgNPs and domiphen inhibit the growth of A. fumigatus by suppressing nitrate assimilation, and purine nucleobase metabolic process and amino acid transmembrane transport, respectively. When the two drugs are combined, AgNPs has epistatic effects on domiphen. Moreover, the combination of AgNPs and domiphen primarily influence secondary metabolites biosynthesis, steroid biosynthesis and nucleotide sugar metabolism of A. fumigatus via Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Furthermore, protein–protein interactions (PPI) analysis combined with validation experiments showed that the combination of AgNPs and domiphen could enhance the expression of copper transporter and inhibit nitrogen source metabolism. In addition, the synergistic antimicrobial effects could be enhanced or eliminated depending on exogenous addition of copper and nitrogen source, respectively. Taken together, the results of this study provide a theoretical basis and a new strategy for the treatment of IA.
1. Introduction
In today’s medical environment, prevention and disinfection are increasingly important. Pathogenic microorganisms have caused serious damage to human life, health and property safety (Siscar-Lewin et al., 2019; Leitao, 2020; Simoes and Antunes, 2021). Aspergillus fumigatus is a widely existing pathogenic fungus in the natural environment (Du et al., 2021b). The airborne conidia produced by Aspergillus fumigatus could be inhaled into the terminal respiratory tract and cause respiratory diseases (Abad et al., 2010; van de Veerdonk et al., 2017; Du et al., 2021a). IA is the most common invasive fungal infection worldwide, and it has become the main cause of death for individuals with low immunity (such as patients after transplantation, major surgery, pulmonary fibrosis and chronic infection, etc.) (Cadena et al., 2016, 2021; Hsu et al., 2022). A. fumigatus is the main pathogen causing IA (Karthaus and Buchheidt, 2013). Triazole drugs are the primary therapeutic agents for the first-line treatment of IA (Roemer and Krysan, 2014; Esquivel et al., 2015), which could easily cause drug resistance problems. Thus, it is indispensable to develop new drugs to combat A. fumigatus. We previously reported that combination of AgNPs and domiphen is antimicrobial against biofilms of common pathogens (Hu et al., 2021). Therefore, this study aimed to discuss the potential of combining AgNPs and domiphen as new antifungal agents against the human fungal pathogen A. fumigatus and possible molecular mechanisms of action.
Currently, new antimicrobial drugs and drug delivery systems are developed based on nanotechnology with a strong focus on metal-based systems (Cu, Ag, Zn) (Minakshi et al., 2019). Nanotechnology is the most promising emerging technology in the 21st century, which is widely applied in information, biology, medicine, chemical industry and other fields, with huge market potential (Ehdaie, 2007; Barreto et al., 2011). The particle size of silver nanoparticles is mostly between 25–100 nm, which has strong inhibitory and killing effects on dozens of pathogenic microorganisms such as Escherichia coli, Neisseria gonorrhoeae and Chlamydia trachomatis, and is not easy to develop drug resistance (Chen and Schluesener, 2008). The emergence of nanotechnology has made a qualitative leap in the antimicrobial ability of silver at the nanoscale. Very little nanosilver could produce strong antimicrobial effects (Abram and Fromm, 2020). It could kill many kinds of microorganisms in minutes with broad spectrum at relatively low doses that are well tolerated in humans, which opens up a broad prospect for the wide application of nanosilver to antimicrobials, and it is a new generation of natural antimicrobial agent (Abram and Fromm, 2020). Here, we used A. argyi leaf as reducing and stabilizing agents for reducing AgNO3 to AgNPs, which could be comparatively safe and eco-friendly (Hu et al., 2021). However, a heavy use of AgNPs could cause its antimicrobial effects to be reduced by precipitation and aggregation (Hu et al., 2021). On the contrary, domiphen, a cationic surface active fungicide with broad spectrum, could be used for the adjuvant treatment of oral, throat, skin and wound infections and disinfection of surgical instruments (Fumagalli et al., 2018; Long et al., 2021). Domiphen is easy to be adsorbed by microbes and produce strong antimicrobial effects with low toxicity and doses, which could fully disperse AgNPs when the two agents are combined (Hu et al., 2021). Thus, we supposed that AgNPs and domiphen may have synergistic antimicrobial effects.
In this study, we aimed to explore the synergistic antimicrobial mechanism of AgNPs combined with domiphen against the pathogenic fungus A. fumigatus and to find the functional genes and potential drug targets affected by AgNPs and domiphen on the basis of RNA-sequencing (RNA-seq). We analyzed specific reasons especially copper and nitrogen source for synergistic antimicrobial effects of AgNPs and domiphen on A. fumigatus through exploring the influence of AgNPs, domiphen and their combination on gene expression of pathogenic fungus A. fumigatus combined with validation experiments. Until now, there have been no reports that combination of AgNPs and domiphen as new antifungal agents could combat A. fumigatus. The results of this study provide a theoretical basis and a new strategy for the treatment of IA.
2. Materials and methods
2.1. Reagents, strains, and media
A. fumigatus (A1161) was obtained from Professor Ling Lu’s lab (Jiang et al., 2014). AgNO3 (99.8%) and domiphen (97%) were purchased from Sinopharm Chemical Reagent Co., Ltd. (China) and Shanghai Yien Chemical Technology Co., Ltd. (China), respectively. Artemisia argyi leaf extract and glucose were purchased from Xi’an Runxue Bio-technology Co., Ltd. (China) and Aladdin Reagent Co., Ltd. (China), respectively. Agar and RPMI-1640 were purchased from Xuzhou Microcomande Biological Engineering Co., Ltd. (China) and Shanghai Sigma-Aldrich Trading Co., Ltd. (China), respectively. Yeast extract, ZnSO4·7H2O, H3BO3, MnCl2·4H2O, FeSO4·7H2O, CoCl2·5H2O, CuSO4·5H2O, (NH4)6Mo7O24·4H2O, Na4EDTA, NaNO3, KCl, CuCl2 ammonium tartrate (C4H12N2O6), MgSO4·7H2O and KH2PO4 were purchased from Shanghai Maclin Biochemical Technology Co., Ltd. (China). For RNA-seq, A. fumigatus strains were grown on liquid MM (minimal medium), containing 1% glucose, trace elements and 50 ml L−1 20 × salt, pH 6.5. For the plate inoculation tests, A. fumigatus strains were grown on rich medium-YAG (yeast extract, agar and glucose), containing 2% glucose, trace elements, 0.5% yeast extract and 2% agar (Du et al., 2021b).
2.2. Preparation of AgNPs
The extract of A. argyi leaf was used as reducing agents. The obtained 1,000 μg ml−1 silver solution was filtered by 0.22 μm microporous membrane, and the particle size was about 77.6 nm measured by particle size analyzer (Hu et al., 2021). Briefly, 8.5 mg of AgNO3 and 10 mg of A. argyi leaf extract were dissolved in 5 ml distilled water and bathed in 100°C boiling water for 40 min to prepare 1,000 μg ml−1 AgNPs solution.
2.3. Drug dilution
Appropriate sterile distilled water was used for drug concentration dilution of AgNPs and domiphen solution. According to CLSI M-38A2 (Clinical and Laboratory Standards Institute) (Espinel-Ingroff et al., 2011), two-fold dilution method was used for drug concentration dilution.
2.4. Plate inoculation test
1 × 105 CFU/ml, 1 × 106 CFU/ml and 1 × 107 CFU/ml of A. fumigatus conidia suspension were mixed with a vortex mixer for 20 s. Then, 2 μl of A. fumigatus conidia suspension was absorbed with a pipette gun and inoculated stably on the plate to form suspension droplets with a diameter of about 2 mm, which were placed in an incubator at 37°C, incubated for 36–48 h and to be photographed.
2.5. Single drug susceptibility test
Single drug susceptibility test was performed according to broth microdilution method and CLSI document M38-A2 (Espinel-Ingroff et al., 2011). Briefly, the same numbers of conidia (1 × 105 conidia) were diluted into the RPMI-1640 medium containing gradient diluent antifungal drugs and transferred into 96-well plates and cultured at 35°C for 48 h (Li et al., 2020).
2.6. Combined drug susceptibility test
The microcheckerboard dilution method was used to determine the synergistic MICs of AgNPs and domiphen. A. fumigatus solution was prepared with 0.5 McTurbidite ratio (1 × 108 CFU/ml) and diluted 100 times with sterile distilled water (1 × 106 CFU/ml). AgNPs and domiphen were diluted with RPMI-1640. We used 1/8 MIC as a minimum unit to generate a series of concentration gradients from 1/8 MIC to 2 MIC. For the combined drug susceptibility test with copper and nitrogen source, 1 mM CuCl2 and 1 mM ammonium tartrate were added to RPMI-1640 media, respectively. Then 100 μl of fungal solution was added to the sterile 96-well plates, so that the final fungal inoculation amount was 5 × 105 CFU/ml, and transferred into 96-well plates and cultured at 35°C for 48 h. The selected concentrations of agents for MIC test in 5 ml tubes were performed according to the MIC results in 96-well plates. The minimum drug concentration without fungal growth (obvious clarity) was denoted as MIC’. FIC calculation uses the following equation:
FIC ≤ 0.5, 0.5 < FIC ≤ 1, 1 < FIC < 2 and FIC ≥ 2 represent synergistic effects, additive effects, independent effects and antagonistic effects, respectively.
2.7. Preparation of RNA-seq samples
2 × 107 A. fumigatus wild-type conidia were inoculated into 100 ml MM liquid medium at 37°C and 220 rpm for 18 h, and then 1 μg ml−1 AgNPs, 2 μg ml−1 domiphen or both drugs were added for shaking of 1 h, respectively. The control group was not treated with drugs. Then the mycelia were filtered and collected with gauze, quick-frozen with liquid nitrogen, and stored at-80°C for three parallel repeats.
2.8. RNA extraction and quality control
Total RNA was extracted from the samples using TRIzol® Reagent according the manufacturer’s instructions (Invitrogen, CA, United States) and genomic DNA was removed using DNase I (TaKara, Japan). RNA degradation and contamination were monitored on 1% agarose gels. Then RNA quality was determined by 2,100 Bioanalyser (Agilent Technologies, Germany) and quantified using the ND-2000 (NanoDrop Technologies, Wilmington, DE, USA). Only high-quality RNA sample (OD260/280 = 1.8 ~ 2.2, OD260/230 ≥ 2.0, RIN ≥ 8.0, 28S:18S ≥ 1.0, > 1 μg) was used to construct sequencing library.
2.9. Transcriptomics profiling
RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions (Illumina, San Diego, CA). The transcriptome library was prepared following TruSeq™ RNA sample preparation Kit from Illumina (San Diego, CA) using 1 μg of total RNA. Shortly, messenger RNA was isolated according to polyA selection method by oligo (dT) beads and then fragmented by the sharding buffer firstly. Secondly double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina). Then the synthesized cDNA was end-repaired, phosphorylated and ‘A’ base addition according to Illumina’s library construction protocol. Libraries were size selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina NovaSeq 6,000 sequencer (2 × 150 bp read length). The raw paired end reads were trimmed and quality controlled by fastp-v0.19.41 with default parameters (Chen et al., 2018). Then clean reads were separately aligned to reference genome with orientation mode using HISAT-v2.2.12 software (Kim et al., 2015). The mapped reads of each sample were assembled by StringTie-v2.2.03 in a reference-based approach (Pertea et al., 2015).
2.10. Differential expression analysis and functional enrichment
To identify DEGs (differential expression genes) between two different groups, the expression level of each gene was calculated according to the transcripts per million reads (TPM) method. RSEM-v1.3.34 was used to quantify gene abundances (Li and Dewey, 2011). Essentially, differential expression analysis was performed using the DESeq2/DEGseq/edgeR/Limma (Robinson et al., 2010; Wang et al., 2010; Love et al., 2014; Ritchie et al., 2015), DEGs with |log2 (fold change)| ≥ 1 and P-adjust ≤0.05 (DESeq2/edgeR/Limma) were considered to be significantly different expressed genes. In addition, we performed functional enrichment analysis, GO (Gene Ontology5) and KEGG (Kyoto Encyclopedia of Genes and Genomes6), to determine compared with the whole-transcriptome background, which DEGs were significantly enriched in top 20 GO terms at P-adjust ≤0.05 and KEGG pathways at p-value ≤0.05. GO functional enrichment and KEGG pathway analysis were performed by Goatools-v1.2.47 and KOBAS-v2.08 (Xie et al., 2011). Clustering heatmaps were created using the package “ComplexHeatmap-v2.13.1” in software R-v4.2.1 and Euclidean distance calculation (Gu et al., 2016). In the selected gene set (significantly changed genes under AgNPs, domiphen and combine conditions for Figures 1–3, respectively), clustering was performed with log2FC under all tested conditions. Through repeated iterations, we calculated the relative distance between genes using the Euclidean distance algorithm with average linkage and divided the selected gene set into different subclusters according to the relative distance of genes.
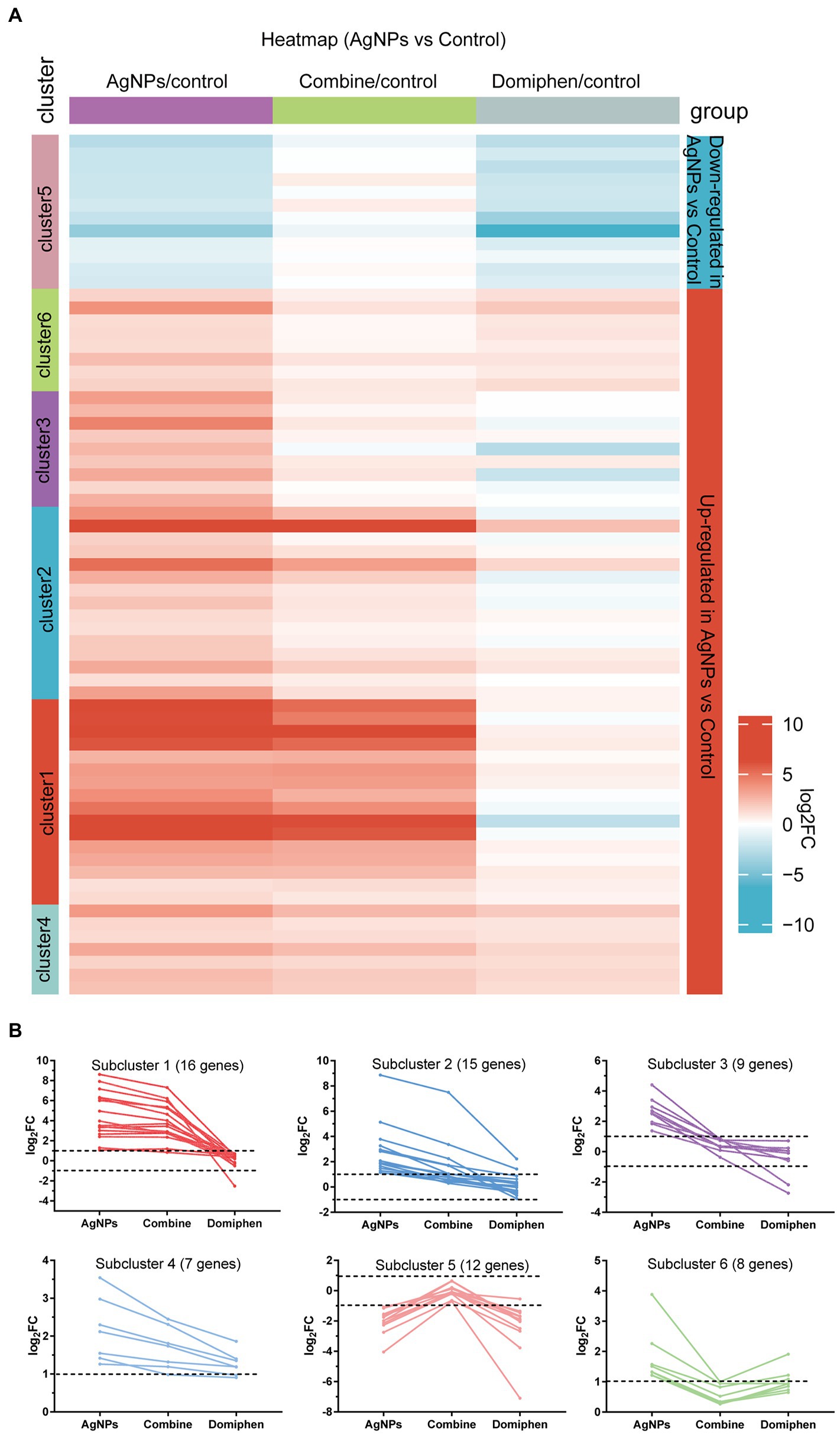
Figure 1. Transcriptional response of A. fumigatus to AgNPs. (A) Heatmap showing the changes in expression of the genes significantly downregulated (blue; the upper) and upregulated (red; the bottom) after 1 h treatment of AgNPs. Each column represents a group, and each row represents a DEG. The color represents the expression value (log2FC, the average from three parallel experiments) of the gene in each group. Red and blue represent log2FC > 0 and log2FC < 0, respectively. “1–6” indicate that multiple expression clusters could be differentiated. (B) The average expression (log2FC) of multiple genes.
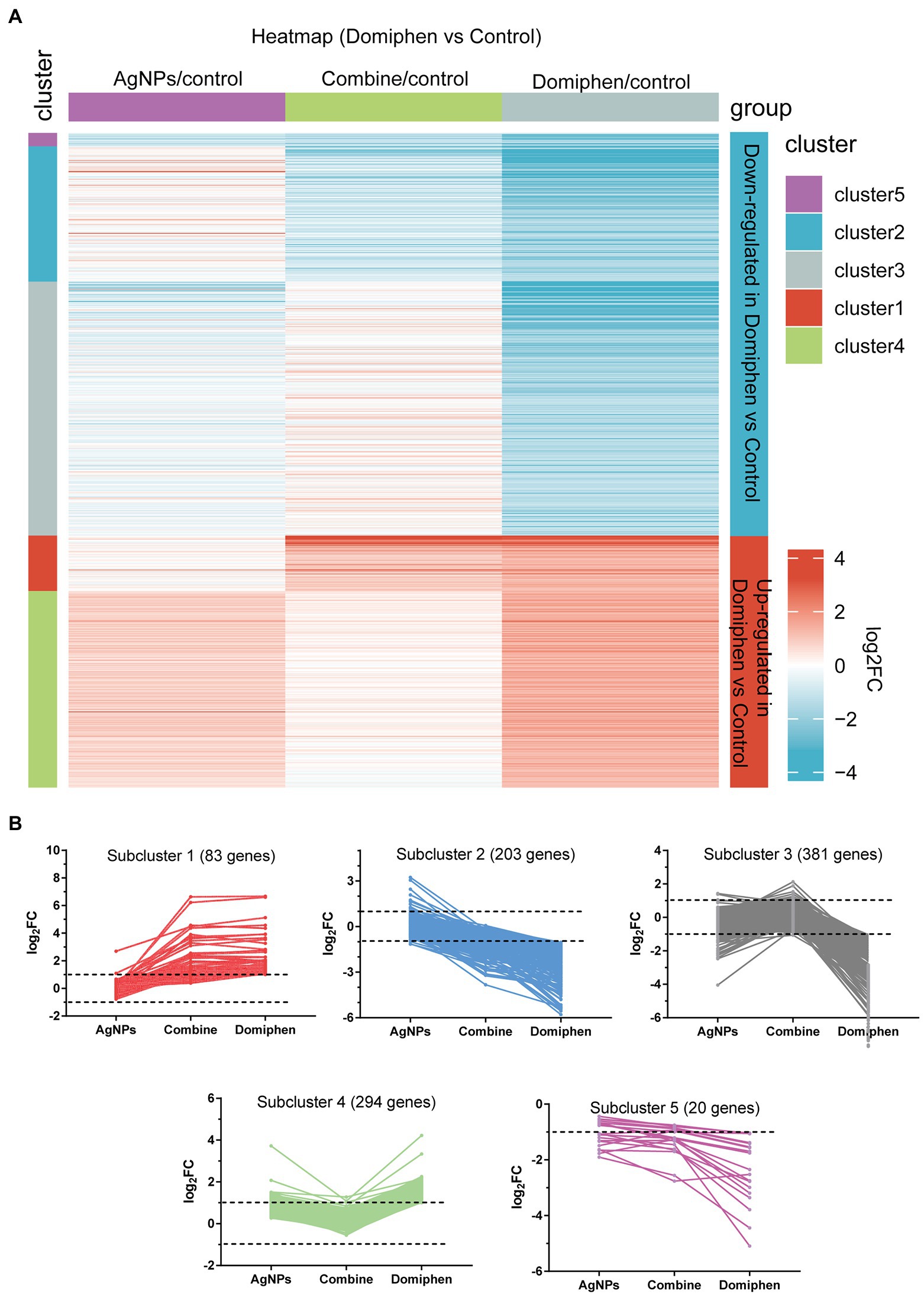
Figure 2. Transcriptional response of A. fumigatus to domiphen. (A) Heatmap showing the changes in expression of the genes significantly downregulated (blue; the upper) and upregulated (red; the bottom) after 1 h treatment of domiphen. Each column represents a group, and each row represents a DEG. The color represents the expression value (log2FC, the average from three parallel experiments) of the gene in each group. Red and blue represent log2FC > 0 and log2FC < 0, respectively. “1–5” indicate that multiple expression clusters could be differentiated. (B) The average expression (log2FC) of multiple genes.
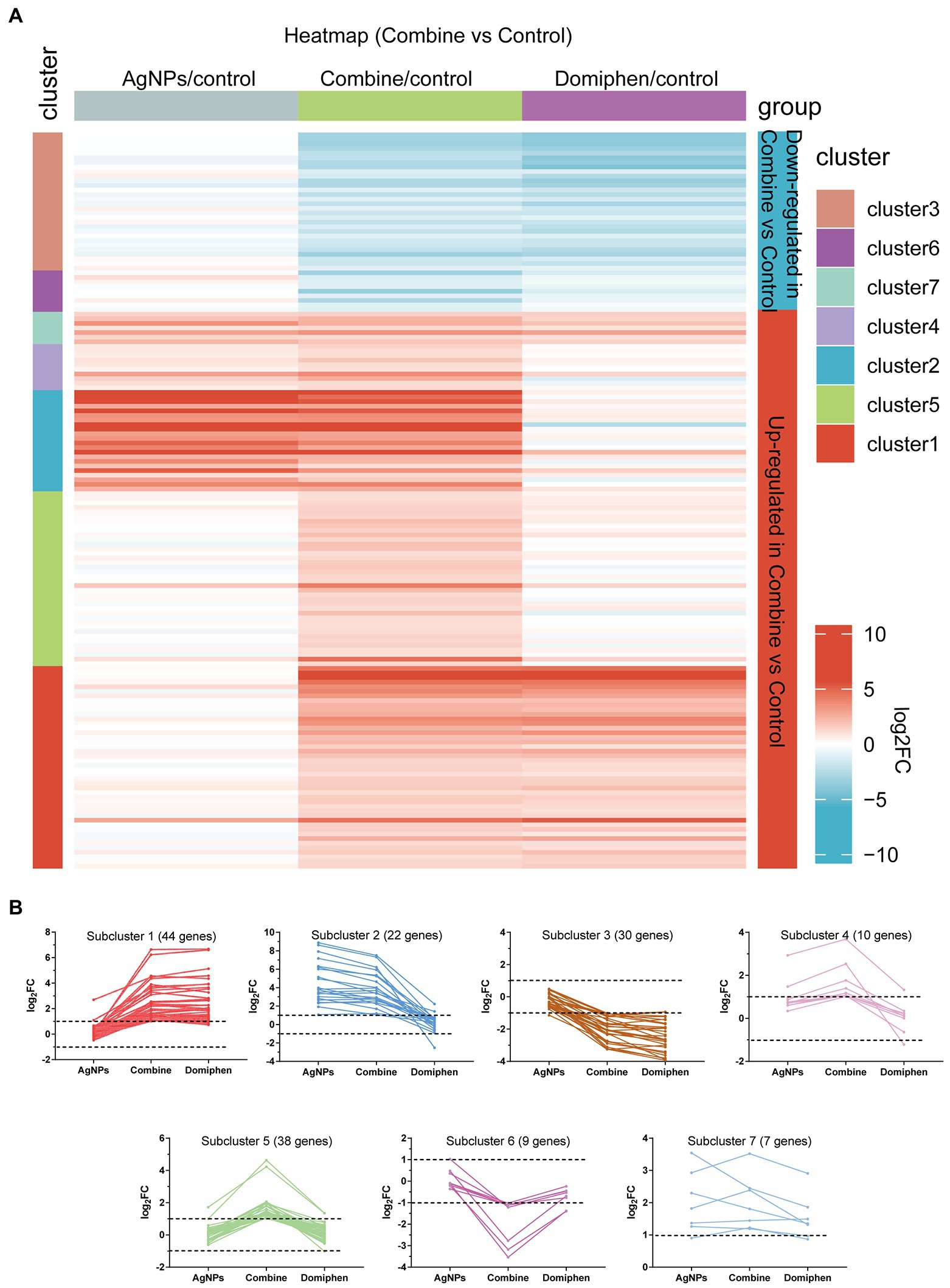
Figure 3. Transcriptional response of A. fumigatus to AgNPs combined with domiphen. (A) Heatmap showing the changes in expression of the genes significantly downregulated (blue; the upper) and upregulated (red; the bottom) after 1 h treatment of AgNPs combined with domiphen shown as “Combine.” Each column represents a group, and each row represents a DEG. The color represents the expression value (log2FC, the average from three parallel experiments) of the gene in each group. Red and blue represent log2FC > 0 and log2FC < 0, respectively. “1–7” indicate that multiple expression clusters could be differentiated. (B) The average expression (log2FC) of multiple genes.
2.11. Constructions for PPI
We used STRING database9 for protein interaction network analysis of genes. The corresponding interaction relationship of genes was directly extracted from the database to construct the network. Then, networkX under Python was used for network visualization of genes.
2.12. Cytotoxicity test
HT22 cells were spread in 96-well plates at a density of 3–5 × 104 cells/ml, 100 μl per well, and placed in a cell culture incubator (37°C, CO2 level of 5%). HT22 cells were incubated with 1 μg ml−1 of AgNPs, 2 μg ml−1 of domiphen or 0.25 μg ml−1 of AgNPs and 0.5 μg ml−1 of domiphen given after cell apposition. At the end of incubation, the medium was aspirated and washed 2–3 times with PBS. One hundred micro liter of incomplete medium containing 10 μl of CCK-8 assay solution (Cell Counting Kit-8; Abcam, England) was added and incubated for 1–2 h in an incubator protected from light. The OD value was measured at 450 nm. The absorbance of the experimental group was recorded as As using the following formula. The absorbance of the control group and blank group were recorded as Ac and Ab, respectively (Liu et al., 2017; Ou et al., 2021).
2.13. Statistical analysis
Data were expressed as Mean ± standard deviation (Mean ± SD), and Graphpad Prism 7 software (GraphPad Software Technology Co., Ltd., San Diego, CA) were used for statistical analysis. Two-tailed unpaired Student’s T test was used for comparison between the two groups. Comparisons between multiple groups were made using one-way ANOVA with a completely randomized design. The level of hypothesis test was determined by α = 0.05. p < 0.05 indicated statistically significant difference.
3. Results
3.1. Synergistic antimicrobial effects of AgNPs and domiphen on human fungal pathogen Aspergillus fumigatus
In order to explore the antimicrobial effects of AgNPs and domiphen against A. fumigatus, we performed plate inoculation assays with different drug concentrations. As shown in Figure 4A, the growth of A. fumigatus was obviously inhibited in the presence of 2 μg ml−1 (0.0185 μmol ml−1) of AgNPs or 16 μg ml−1 (0.0386 μmol ml−1) of domiphen, suggesting that the antimicrobial effect of AgNPs against A. fumigatus is better than that of domiphen. To test the combined antimicrobial effects of AgNPs and domiphen against A. fumigatus, we also detected the growth of A. fumigatus under the two drugs. The results of combined drug susceptibility test showed that the colony diameter of A. fumigatus was much smaller under 2 μg ml−1 of AgNPs and 16 μg ml−1 of domiphen than that under corresponding concentrations of single drugs, indicating that the antimicrobial activity of combining AgNPs with domiphen was more active than that of the single drug (Figures 4A,B). The MICs of AgNPs and domiphen against A. fumigatus were determined by the microdilution method and the CLSI method (Espinel-Ingroff et al., 2011). As shown in Figure 4C, the solution was turbid when AgNPs was not added. When the concentration of AgNPs was 1 μg mL−1 or more, the growth of A. fumigatus was totally inhibited with no mycelia could be seen in the solution. Thus, the minimum inhibitory concentration (MIC) of AgNPs against A. fumigatus was 1 μg mL−1. Similarly, the MIC of domiphen against A. fumigatus was 2 μg mL−1 (Figure 4D and Table 1). To assess the joint effects of the two agents, combined drug susceptibility tests were performed. As shown in Figure 4E, the growth of A. fumigatus was totally inhibited under 0.25 μg ml−1 of AgNPs and 0.5 μg ml−1 of domiphen. The combined use of AgNPs and domiphen produced synergistic effects against A. fumigatus (FIC = 0.5, Table 1). Domiphen could increase the growth inhibitory activity of AgNPs against A. fumigatus in a planktonic and biofilm culture, resulting in a 4-fold decreased MIC of AgNPs (from 1 μg ml−1 toward 0.25 μg ml−1 in the presence of 0.5 μg ml−1 domiphen, Table 1). We also detected the antimiacrobial effects of AgNPs and domiphen in MM media (Supplementary Figure S1). The result showed that the MIC value in MM media is the same as that in RPMI-1640 media. Taken together, these results suggested that AgNPs, 2 μg ml−1 of domiphen and domiphen are synergistic antimicrobial against human fungal pathogen A. fumigatus. The two drugs combined could reduce the dose and toxicity of drugs compared to the drug alone.
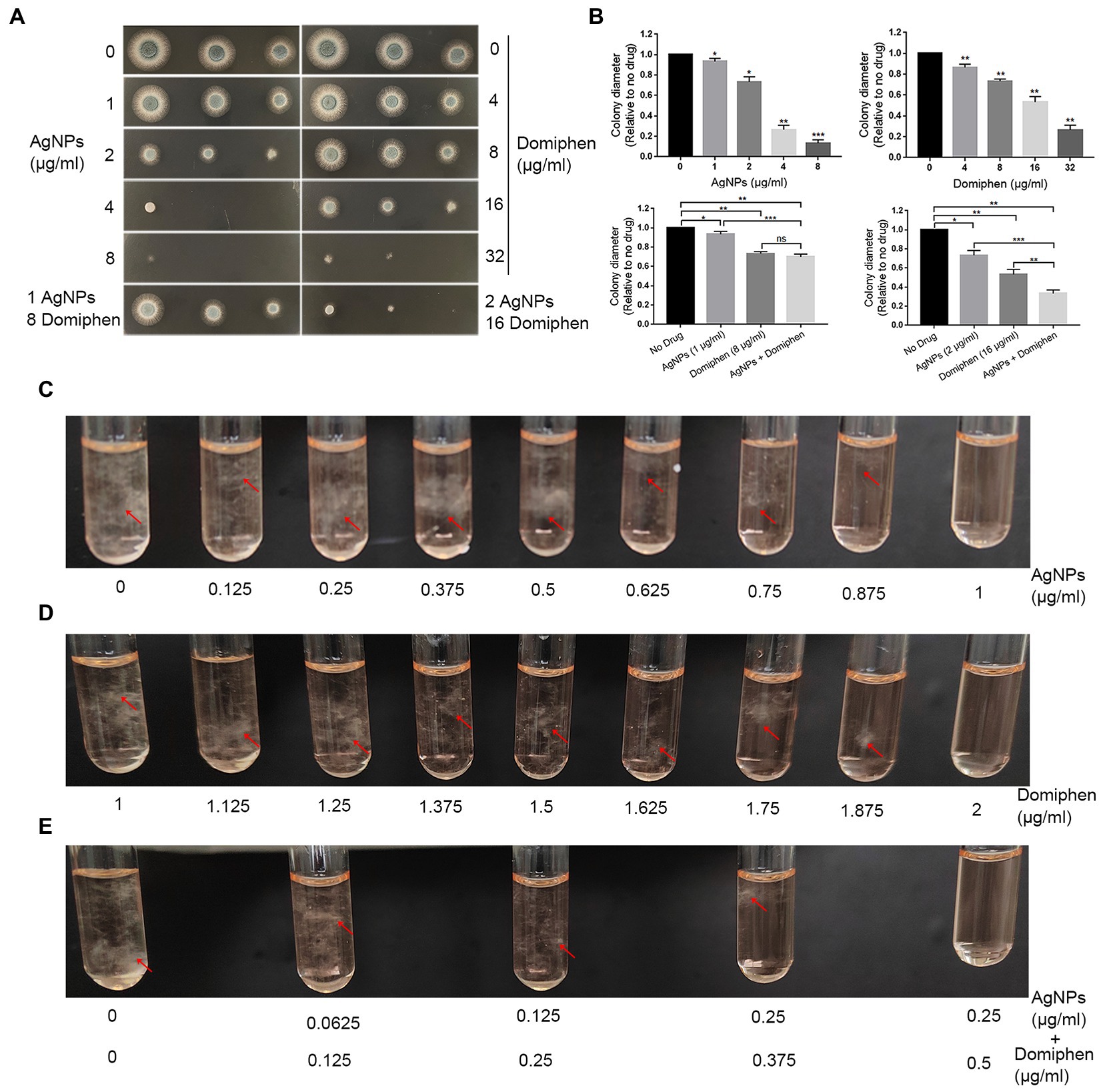
Figure 4. The single and combined susceptibility tests of AgNPs and domiphen against human fungal pathogen A. fumigatus. (A) Two microliters of double distill water containing 2 × 104, 2 × 103 or 2 × 102 conidia of A. fumigatus wild-type strain (A1161) were used to inoculate onto YAG medium containing the indicated concentrations of AgNPs or domiphen. (B) Quantifications of colony diameter for the indicated strains under different treatment cultural conditions. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, P > 0.05 compared to the no drug control or the single drug according to unpaired t test with or without Welch’s correction. (C) MIC (μg ml−1) test results of control group and indicated concentrations of AgNPs groups against A. fumigatus. (D) MIC (μg ml−1) test results of control group and indicated concentrations of domiphen groups against A. fumigatus. (E) MIC (μg ml−1) test results of control group and indicated concentrations of AgNPs combined with domiphen groups against A. fumigatus. For C to E, the mycelia were highlighted with red arrows.
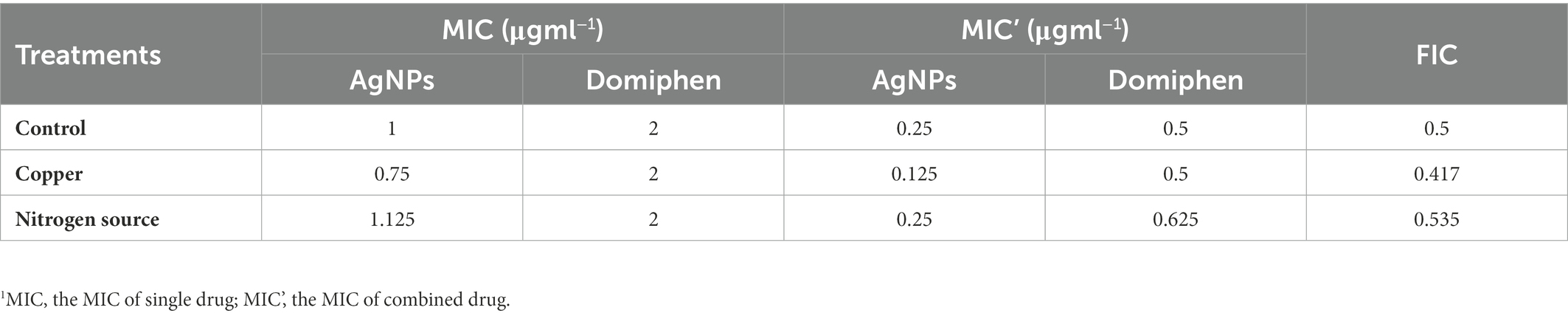
Table 1. Drug susceptibility test for combination of AgNPs and domiphen with or without copper (1 mM CuCl2) or nitrogen source (1 mM ammonium tartrate).1
3.2. AgNPs combined with domiphen have relatively low cytotoxicity
To monitor the cytotoxicity of AgNPs and domiphen, we performed cytotoxicity test. we chose nerve cells (HT22) instead of lung cells to perform cytotoxicity test for the following reasons: Firstly, considering that metal ions such as Ag + are more likely to cause neurotoxicity (Amadi et al., 2019), we are more concerned about the effect of AgNPs on the survival rate of nerve cells; Secondly, nerve cells are more fragile than lung cells, so a drug with low cytotoxicity for nerve cells is very likely to have low cytotoxicity for lung cells as well; Thirdly, although the primary sites of A. fumigatus infection are the lungs and paranasal sinuses, brain infection arises from direct invasion from the paranasal sinuses or by hematogenous spread (Kumar et al., 2021). There is also a case reporting invasive aspergillosis involving the lungs and brain in an immunocompetent person (Kumar et al., 2021). Due to the single and combined MICs of the two drugs were determined, we chose the corresponding MIC value of the two drugs as test condition. As shown in Figure 5, the cell viability under 1 μg ml−1 of AgNPs, 2 μg ml−1 of domiphen and 0.25 μg ml−1 of AgNPs combined with 0.5 μg ml−1 of domiphen conditions was about 87, 90 and 85%, respectively. This result suggested that AgNPs and domiphen have relatively low cytotoxicity.
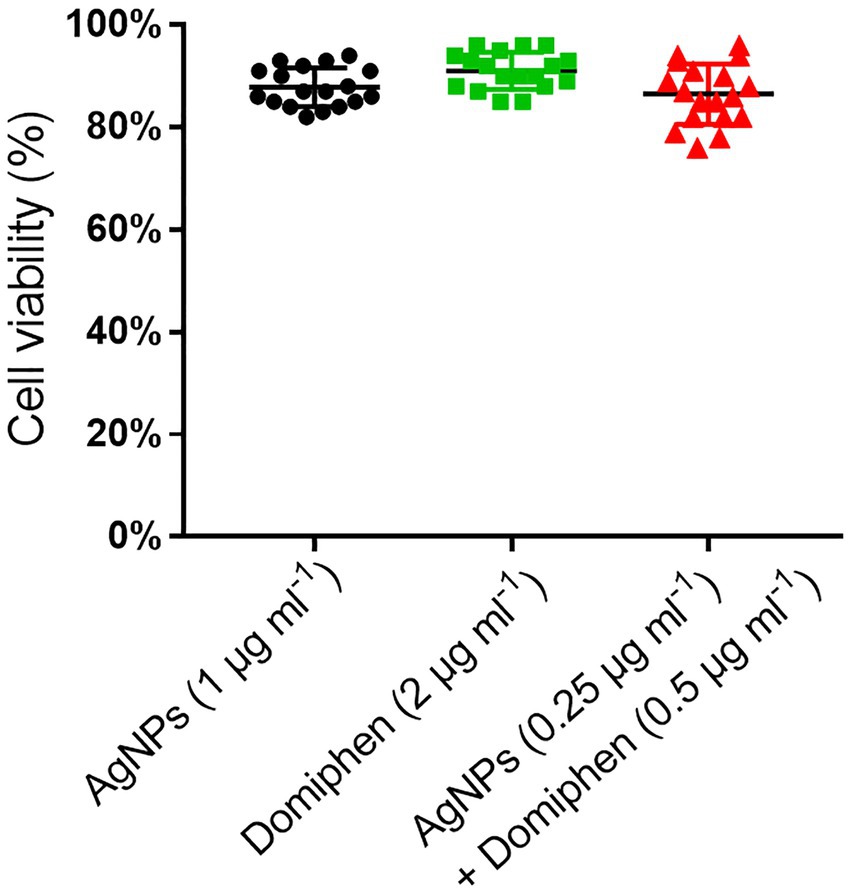
Figure 5. The HT22 cell viability of AgNPs (1 μg ml−1), domiphen (2 μg ml−1) and AgNPs (0.25 μg ml−1) combined with domiphen (0.5 μg ml−1), n = 18.
3.3. Transcriptional profiles of Aspergillus fumigatus under AgNPs, domiphen, and AgNPs combined with domiphen conditions
To dissect the possible mechanism underlying synergistic antimicrobial effects of AgNPs and domiphen against A. fumigatus, we performed RNA-seq analyses comparing the A. fumigatus wild type with and without drug treatments. As shown in Figure 6A, conidia of wild-type strain were collected and incubated in MM for 18 h at 37°C. Then, AgNPs, domiphen, or AgNPs combined with domiphen were added, followed by an incubation of 1 h. Samples were then filtered and processed for RNA extraction and sequencing. As shown in Figure 6B, AgNPs was found to induce differential expression of 78 genes in A. fumigatus. Of these genes, 63 were up-regulated and 15 were down-regulated in AgNPs-treated A. fumigatus wild-type strain compared with untreated control (p < 0.05, |log2 (fold change)| ≥ 1, Supplementary Table 1). Domiphen was found to induce differential expression of 1,009 genes in A. fumigatus. Of these genes, 392 were up-regulated and 617 were down-regulated in domiphen-treated A. fumigatus wild-type strain compared with untreated control (p < 0.05, |log2 (fold change)| ≥ 1, Supplementary Table 2). The combination of AgNPs and domiphen was found to induce differential expression of 166 genes in A. fumigatus. Of these genes, 123 were up-regulated and 43 were down-regulated in combine-treated A. fumigatus wild-type strain compared with untreated control (p < 0.05, |log2 (fold change)| ≥ 1, Supplementary Table 3). These results suggested that domiphen has a greater impact on the transcription level of A. fumigatus compared with AgNPs.
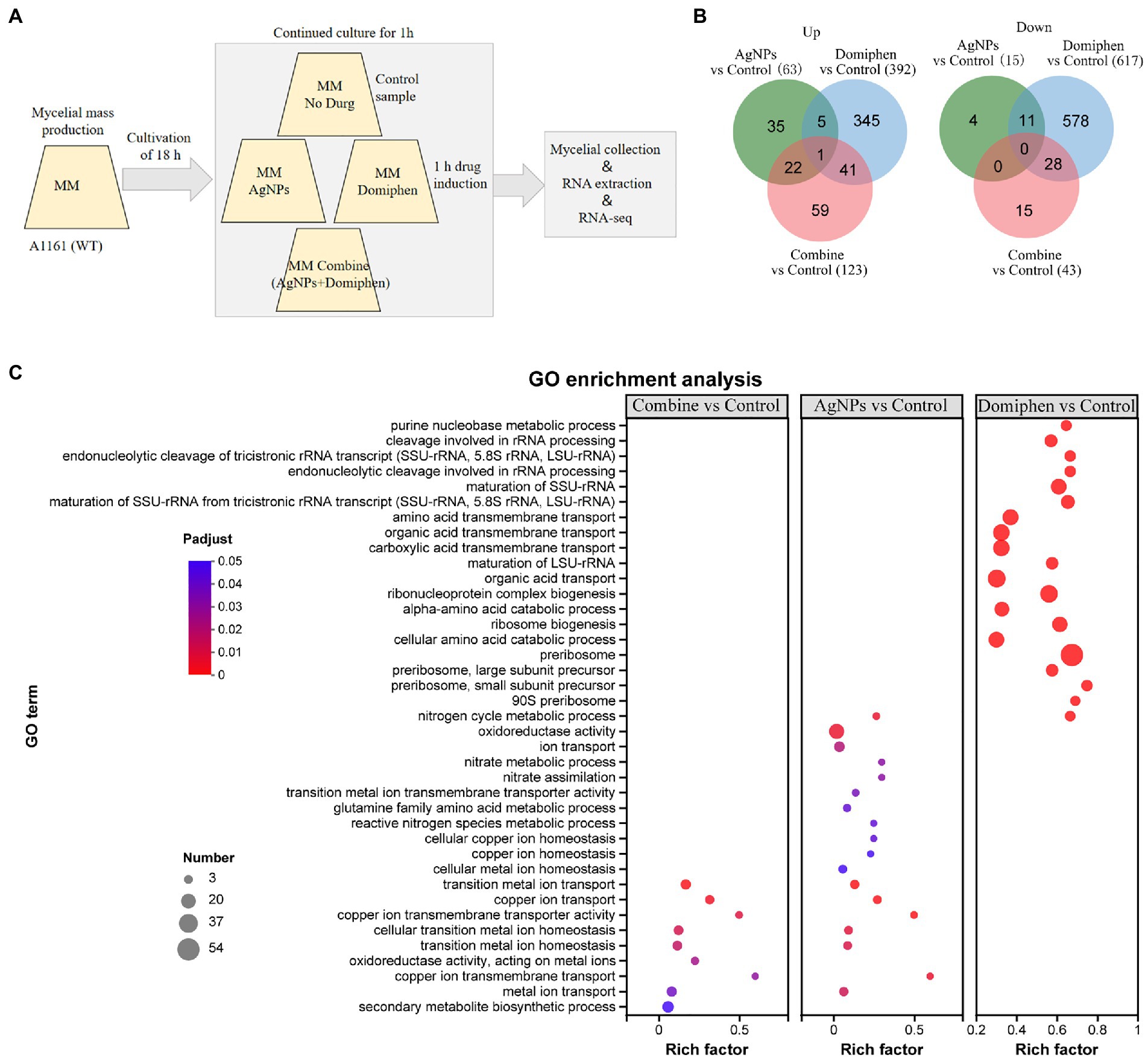
Figure 6. Overview of transcriptome of A. fumigatus under different drugs conditions. (A) Experimental design for the RNA-seq experiment carried out in this study. Conidia of wild-type strain were collected and incubated (3 × 105 conidia ml−1) in MM for 18 h at 37°C. Then, AgNPs (1 μg ml−1), domiphen (2 μg ml−1), or AgNPs (1 μg ml−1) combined with domiphen (2 μg ml−1) indicated as “Combine,” were added, followed by an incubation of 1 h. Cultures grown in MM with no drug were used as controls. Samples (triplicates; see the Methods section) were then filtered and processed for RNA extraction and sequencing. (B) Venn diagrams showing the number of up-and down-regulated DEGs for different groups. (C) Bubble diagram of top 20 ranked GO terms of DEGs. The vertical axis indicates GO terms and the horizontal axis represents the Rich factor. The enrichment degree was stronger with a bigger Rich factor. The size of dots indicates the number of genes in the GO term.
To shed some light onto the functional roles of the DEGs, GO enrichment analysis was conducted. As shown in Figure 6C and Supplementary Table 4, “copper ion transmembrane transport” occupied the strongest enrichment degree as it had the highest Rich factor (0.6), followed by “copper ion transmembrane transporter activity” (Rich factor 0.5) in the AgNPs-treated A. fumigatus wild-type strain compared with untreated control. The over-expressed genes enriched for biological processes and pathways that have been implicated in copper ion transmembrane transport, copper ion transport, cellular copper ion homeostasis, metal ion transport and ion transport. The down-regulated genes enriched for biological processes that have been involved in nitrate assimilation (Supplementary Table 4). These results suggested that AgNPs inhibits the growth of A. fumigatus by enhancing metal ion transport, especially copper ion and suppressing nitrate assimilation. In the domiphen-treated group (Figure 6C; Supplementary Table 5), the top 5 ranked GO terms of DEGs are ‘preribosome, small subunit precursor’ (Rich factor 0.75), ‘90S preribosome’ (Rich factor 0.69), ‘preribosome’ (Rich factor 0.675), ‘endonucleolytic cleavage of tricistronic rRNA transcript’ (Rich factor 0.67) and ‘endonucleolytic cleavage involved in rRNA processing’ (Rich factor 0.67). As shown in Supplementary Table 5, the up-regulated genes enriched for biological processes and pathways that have been implicated in endonucleolytic cleavage of tricistronic rRNA transcript, maturation of SSU-rRNA from tricistronic rRNA transcript, ribosome biogenesis, maturation of rRNA and cleavage involved in rRNA processing. The down-regulated genes enriched for biological processes that have been involved in purine nucleobase metabolic process and amino acid transmembrane transport. These results suggested that domiphen could activate the transcript, processing and maturation of rRNA, and could inhibit purine nucleobase metabolic process and amino acid transmembrane transport to produce antimicrobial effects. Taken together, AgNPs and domiphen could inhibit the growth of A. fumigatus by suppressing nitrate assimilation, and purine nucleobase metabolic process and amino acid transmembrane transport, respectively.
Interestingly, the combine-treated A. fumigatus and the AgNPs-treated A. fumigatus had a similar GO enrichment pattern, which is different from that of the domiphen-treated A. fumigatus (Figure 6C; Supplementary Table 6), suggesting that when the two agents are combined, AgNPs has epistatic effects on domiphen. Specifically, the up-regulated genes enriched for biological processes and pathways that have been implicated in copper ion transmembrane transport, transition metal ion transport, iron ion transport and cellular transition metal ion homeostasis. The down-regulated genes enriched for biological processes that have been involved in secondary metabolite biosynthetic process. These results suggested that combination of AgNPs and domiphen could creat a low-copper condition with high expression of high affinity copper transporter and inhibit secondary metabolic process to combat A. fumigatus.
To further dissect the functional roles of the DEGs, KEGG enrichment analysis was performed. As shown in Figure 7 and Supplementary Table 7, ‘nitrogen metabolism’ possessed the highest rich factor (Rich factor 0.14) and had the most number of DEGs in the AgNPs-treated group, followed by ‘Arginine and proline metabolism’ (Rich factor 0.039), suggesting that AgNPs could mainly suppress nitrogen and amino acid metabolism to inhibit the growth of A. fumigatus. In the domiphen-treated group, ‘caffeine metabolism’ possessed the highest rich factor (Rich factor 1) and ‘ribosome biogenesis in eukaryotes’ had the most number of DEGs (Figure 7; Supplementary Table 8), indicating that domiphen could enhance ribosome biogenesis and suppress caffeine metabolism to inhibit the growth of A. fumigatus. For the combine-treated group, ‘biosynthesis of various secondary metabolites’ possessed the highest rich factor (Rich factor 0.29). ‘Steroid biosynthesis’ and ‘amino sugar and nucleotide sugar metabolism’ had the most number of DEGs (Figure 7; Supplementary Table 9), demonstrating that the combination of AgNPs and domiphen possibly influence secondary metabolites biosynthesis, steroid biosynthesis and nucleotide sugar metabolism to have a synergistic antimicrobial effect on A. fumigatus.
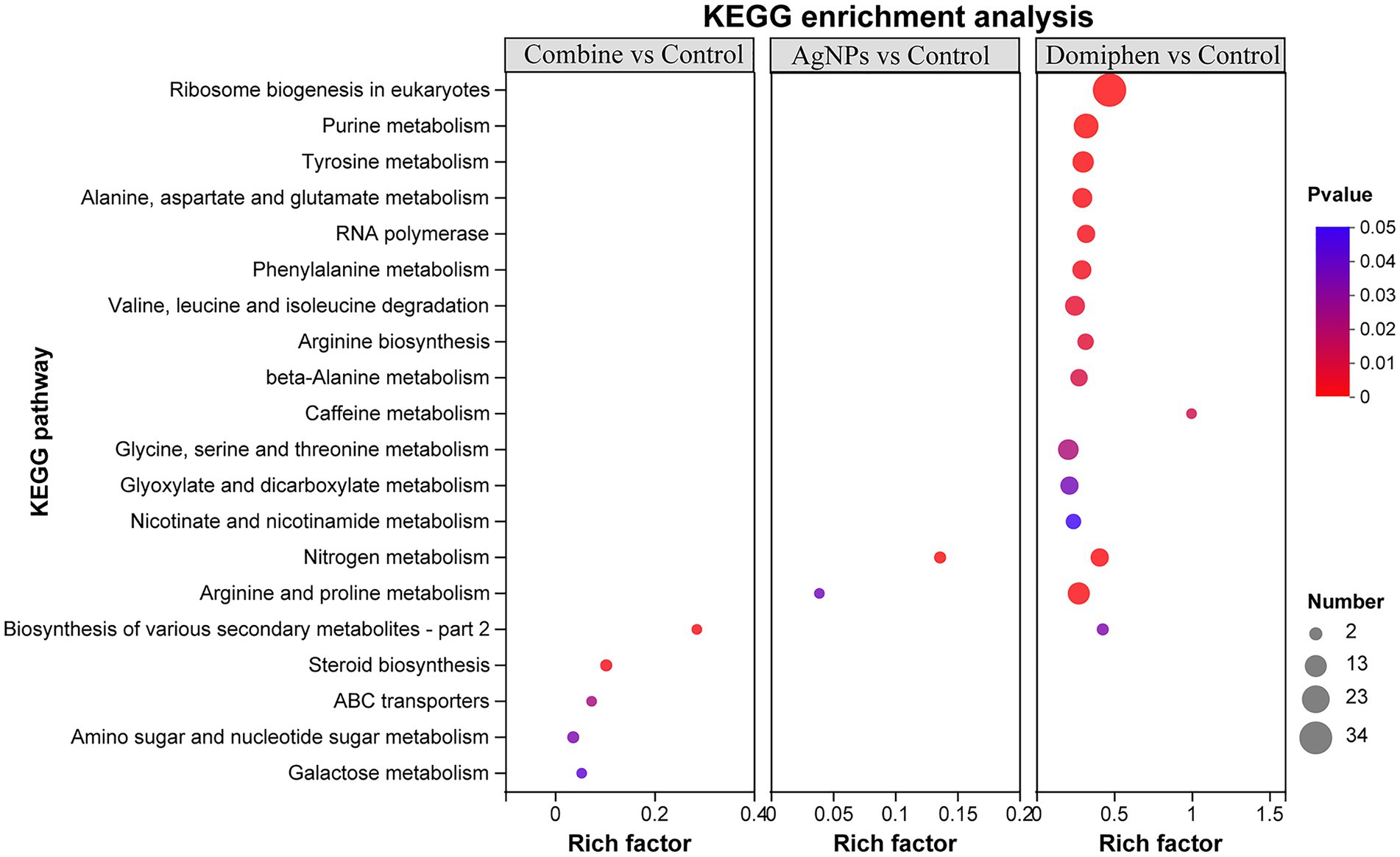
Figure 7. Bubble diagram of top 20 ranked KEGG pathways of DEGs among indicated groups. The vertical axis represents the pathway name, and the horizontal axis represents the ratio of the number of enriched gene/transcript samples in this pathway to the number of background annotated genes/transcripts. The larger the Rich factor, the stronger the enrichment degree. The size of the dots indicates the number of genes in the pathway, and the color of the dots corresponds to different Padjust ranges.
3.4. AgNPs-inducing transcriptomics changes in Aspergillus fumigatus
The heatmap in Figure 1A includes all the genes up-and down-regulated under AgNPs conditions in the wild-type strain, but was built considering their expression values for all conditions tested. The upper contains genes that are downregulated and the lower contains genes upregulated under AgNPs condition (blue and red, respectively). Multiple gene expression patterns could be differentiated (Figure 1B; Supplementary Table 10). Highlighted are five main expression clusters among upregulated genes and one among downregulated genes under AgNPs conditions. Subcluster 1 and 2 contain genes upregulated under AgNPs and “Combine” conditions but almost unchanged under domiphen conditions, suggesting that these genes are AgNPs-dependent responsive. For instance, the high affinity copper transporter (AFUB_020800) belongs to subcluster 1 (Supplementary Table 10), suggesting that the high affinity copper transporter (AFUB_020800) could be induced under AgNPs condition regardless of domiphen. Subcluster 3 and 6 contain genes upregulated exclusively under AgNPs conditions, indicating that these 6 genes upregulate in a AgNPs-dependent manner. Subcluster 4 contains 7 genes upregulated under all conditions, implying that these genes could respond under all tested conditions. Subcluster 5 contains 12 genes downregulated under AgNPs and domiphen conditions but almost unchanged under “Combine” condition, demonstrating that AgNPs and domiphen could repress these genes expression to inhibit the growth of A. fumigatus. As shown in Supplementary Table 10, subcluster 5 mainly takes part in nitrogen compound metabolic process and transmembrane transport. These results suggested that AgNPs and domiphen could inhibit the growth of A. fumigatus via blocking nitrogen compound metabolic process and transmembrane transport.
3.5. Domiphen-inducing transcriptomics changes in Aspergillus fumigatus
Five gene expression clusters are highlighted in the heatmap in Figure 2A, with three of them corresponding to the downregulated genes and the remaining two corresponding to the upregulated genes (Figure 2B; Supplementary Table 11). Among upregulated genes, subcluster 1 contains 83 genes, the majority of which upregulated under “Combine” and domiphen conditions but not changed under AgNPs conditions compared to untreated control (Figure 2B), suggesting that these genes are domiphen-dependent responsive; Subcluster 4 contains 294 genes, the majority of which upregulated under domiphen and AgNPs conditions but almost unchanged under “Combine” conditions, indicating that part of these genes could respond to domiphen or AgNPs induction. Among downregulated genes, subcluster 2 contains 203 genes, the majority of which downregulated under “Combine” and domiphen conditions but unchanged under AgNPs conditions. Subcluster 3 contains 381 genes, the majority of which downregulated exclusively under domiphen condition, demonstrating that domiphen could suppress the expression of these genes to inhibit the growth of A. fumigatus. For example, the nitrate related genes belong to subcluster 3 (Supplementary Table 11), suggesting that domiphen could repress nitrate metabolism. Subcluster 5 contains 20 genes, the majority of which downregulated under all drug conditions compared to untreated control, suggesting that these genes are affected under all tested conditions.
3.6. AgNPs and domiphen-inducing transcriptomics changes in Aspergillus fumigatus
The heatmap in Figure 3A includes all the genes up-and down-regulated under AgNPs combined with domiphen conditions in the wild-type strain, but was built considering their expression values for all conditions tested. The upper contains genes that are downregulated and the lower contains genes upregulated under AgNPs combined with domiphen conditions (blue and red, respectively). Multiple gene expression patterns could be differentiated (Figure 3B; Supplementary Table 12). Highlighted are five main expression clusters among upregulated genes and two among downregulated genes under “Combine” conditions. Among downregulated genes, subcluster 3 and 6 contain genes, some of which downregulated under “Combine” and domiphen conditions but not changed under AgNPs conditions, suggesting that domiphen could decrease the expression of these genes to inhibit the growth of A. fumigatus. Among upregulated genes, subcluster 1 contains 44 genes upregulated under “Combine” and domiphen conditions but not changed under AgNPs conditions, indicating that these genes are domiphen-dependent responsive; Subcluster 2 and 4 contain genes, the majority of which upregulated under “Combine” and AgNPs conditions but unchanged under domiphen conditions, suggesting that these genes are AgNPs-dependent responsive; A prominent example of the high affinity copper transporter (AFUB_020800) belongs to subcluster 2 (Supplementary Table 12), implying that AgNPs rather than domiphen could induce the expression of the high affinity copper transporter. Subcluster 5 contains 38 genes upregulated exclusively under “Combine” conditions, demonstrating that these genes could only respond to the combination of AgNPs and domiphen; Subcluster 7 contains 7 genes upregulated under all drug conditions compared to no drug control, suggesting that these genes could respond to all drugs tested.
3.7. The network of PPI of DEGs
To further explore the possible mechanism in synergistic antimicrobial effects of AgNPs and domiphen against A. fumigatus, the putative 78 key DEGs belonging to the AgNPs-treated group, 1,009 DEGs belonging to the domiphen-treated group and 166 DEGs belonging to the combine-treated group were used to build PPI networks. As shown in Figure 8A and Supplementary Table 1, NiaD (nitrate reductase NiaD, AFUB_012300, fold change = 0.255), NrtB (high affinity nitrate transporter, AFUB_016830, fold change = 0.061) and CtrC (Ctr copper transporter family protein, AFUB_020800, fold change = 64.498), followed by AFUB_095480 (metalloreductase, putative, fold change = 6.319), had highest scores for betweenness centrality, indicating that they are most important for connections with other proteins and AgNPs could suppress nitrogen uptake together with the induction of copper transporters to inhibit the growth of A. fumigatus; Under domiphen conditions, the key proteins of A. fumigatus which were affected by domiphen are shown in Figure 8C and Supplementary Table 2, including small nucleolar ribonucleoprotein complex subunit (AFUB_022970, fold change = 3.219), rRNA processing protein Bystin (AFUB_043850, fold change = 2.15), 60S acidic ribosomal protein P1 (AFUB_007210, fold change = 2.521), etc. These results suggested that domiphen could enhance ribosomal protein biogenesis and rRNA processing to exert antimicrobial effects; Similarly, Figure 8B and Supplementary Table 3 showed that under the conditions of AgNPs combined with domiphen, the key proteins mainly affected are transferase family protein (AFUB_035490, fold change = 16.152), metalloreductase (AFUB_095480, fold change = 6.784) and CtrC (Ctr copper transporter family protein, AFUB_020800, fold change = 40.893), suggesting that the combination of AgNPs and domiphen could creat a low-copper condition with high expression of high affinity copper transporter and enhance transferase family protein synthesis to have synergistic antimicrobial effects.
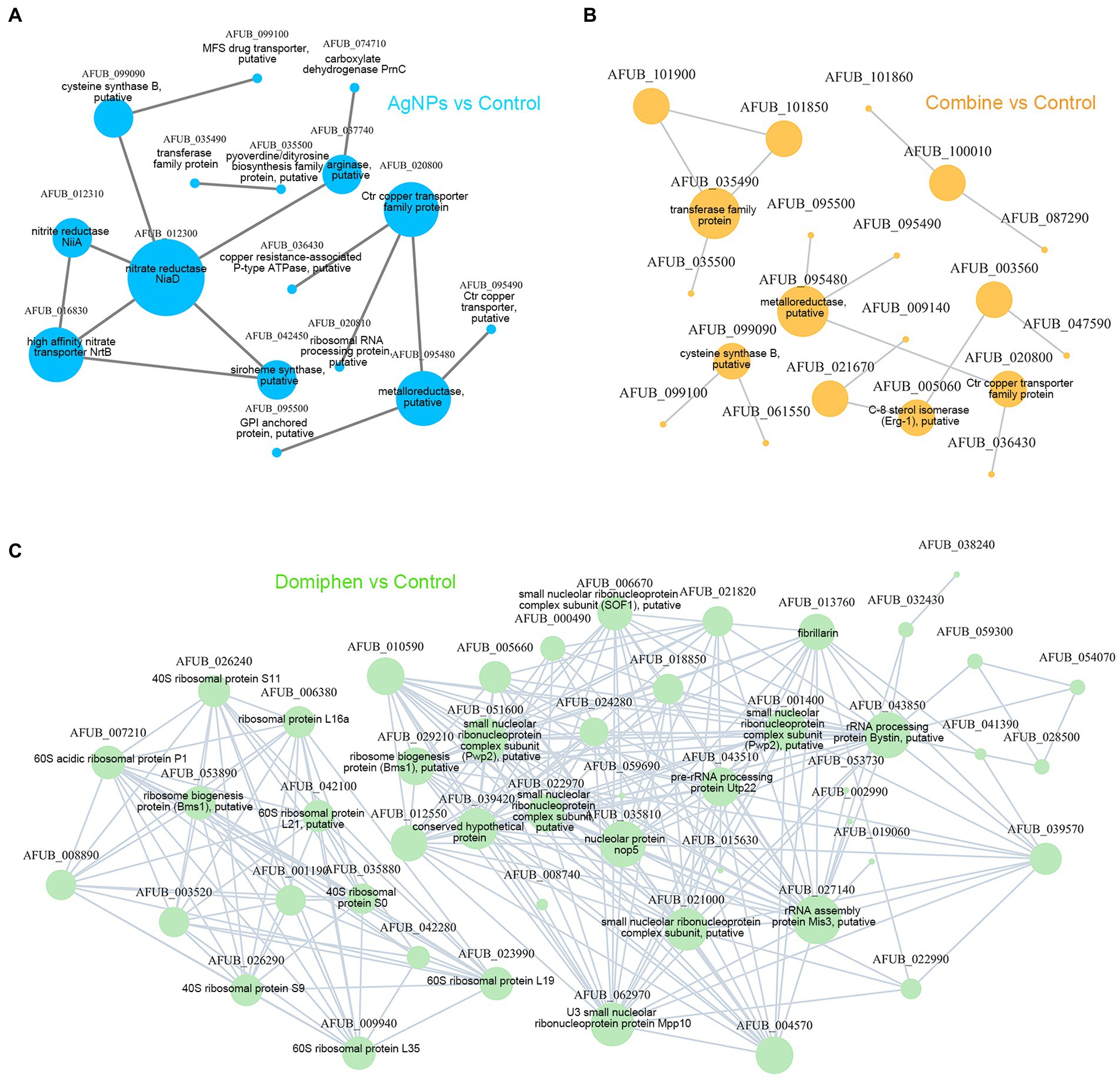
Figure 8. The network of PPI for DEGs. (A) The PPI analysis of DEGs in AgNPs vs. Control. (B) The PPI analysis of DEGs in Combine vs. Control. (C) The PPI analysis of DEGs in Domiphen vs. Control. The gene annotations come from the website EnsemblFungi: http://fungi.ensembl.org/index.html. Nodes represent genes/transcripts, and edges represent interactions between two genes/transcripts.
3.8. Validation of the influence of nitrogen source and copper in the antimicrobial activity of AgNPs and domiphen
Considering that AgNPs and domiphen could inhibit nitrogen metabolism and AgNPs could enhance copper transporter expression, we want to investigate the role of nitrogen source and copper in the antimicrobial activity of AgNPs and domiphen. For this purpose, we performed drug susceptibility test under exogenous addition of nitrogen source or copper conditions. We first determined that the MIC of copper (CuCl2) was about 10 mM and nitrogen source (ammonium tartrate) had not antimicrobial effects. Thus, we chose 1 mM CuCl2 and ammonium tartrate which have no influence on the growth of A. fumigatus to conduct the subsequent experiment. As shown in Table 1, The MIC values of AgNPs were decreased under addition of copper condition, implying that copper could increase the antimicrobial activity of AgNPs and therefore improve the synergistic antimicrobial effects of AgNPs and domiphen (FIC = 0.417). On the contrary, nitrogen source could elevate the MIC values of AgNPs and domiphen (Table 1), implying that nitrogen source may abolish the synergistic antimicrobial effects of AgNPs and domiphen (FIC = 0.535). Taken together, these results suggested that copper and nitrogen source have opposite effects on the synergistic antimicrobial activity of AgNPs and domiphen.
4. Discussion
One of the most challenging global health threats is the development and spread of rapid microbial resistance to antibiotics that leaves infections untreatable, increasing the risk of surgery or immunosuppression for other diseases (Brogan and Mossialos, 2016). Many studies have shown that the potential of Ag as part of the solution to the antimicrobial resistance crisis (Abram and Fromm, 2020). Nano-antimicrobials are proving to be of particular value in preventing wound dressings or local infections. Therefore, this study provides insights into the mechanism of AgNPs combined with the potentiator domiphen as a potential strategy to combat the human pathogenic fungus A. fumigatus. Understanding the mode of action of antifungal compounds or combinations is important because it could lead to faster discovery of new antifungal therapies for the treatment of IA (Scorzoni et al., 2017). Currently, the commonly used antifungal drugs include caspofungin, voriconazole, amphotericin B, itraconazole, etc. (Ben-Ami and Kontoyiannis, 2021). We previously found that the combination of AgNPs and domiphen had synergistic antibacterial effects on Acinetobacter baumannii (FIC = 0.1875), Staphylococcus aureus (FIC = 0.5), Escherichia coli (FIC = 0.1875) and Candida albicans (FIC = 0.3125) (Hu et al., 2021). However, there is no relevant research on the synergistic antimicrobial mechanism of AgNPs combined with domiphen against the pathogenic fungus A. fumigatus. The research provides a theoretical basis and a new strategy for the treatment of IA.
In order to determine whether AgNPs and domiphen have antimicrobial effects on A. fumigatus, plate inoculation test was used to show that AgNPs and domiphen have good antimicrobial effects on A. fumigatus. To accurately determine the antimicrobial activity of AgNPs and domiphen alone on A. fumigatus, we also referred to CLSI standards and performed the single drug susceptibility test of AgNPs and domiphen against A. fumigatus. The results showed that the MICs of AgNPs and domiphen to A. fumigatus were 1 μg ml−1 and 2 μg ml−1, respectively. The above experimental results indicated that both AgNPs and domiphen could play good antimicrobial effects on A. fumigatus, but the antimicrobial effects of the combination of AgNPs and domiphen on common pathogenic fungus needs to be further verified. Therefore, in order to verify the combined antimicrobial effects of the two drugs, the checkerboard microdilution method was used to measure the combined drug susceptibility of AgNPs and domiphen at different concentrations. The results showed that the combination of AgNPs and domiphen had synergistic effects on A. fumigatus (FIC ≤ 0.5). Taken together, AgNPs and domiphen have obvious synergistic antimicrobial effects on opportunistic pathogen A. fumigatus, which could enhance the effectiveness of antimicrobial drugs with reduced dosage and toxic side effects of drugs.
In order to explore the combined antimicrobial mechanism of the two agents, RNA-seq was used in this study. Through GO enrichment analysis, KEGG enrichment analysis and cluster analysis, the gene sets that affects the growth of A. fumigatus could be accurately obtained when AgNPs, domiphen or combined agents are used. GO enrichment analysis together with the drug susceptibility test under addition of copper and nitrogen source conditions showed that AgNPs inhibits the growth of A. fumigatus by inducing high affinity copper transporter and suppressing nitrate assimilation; Domiphen could activate the transcript, processing and maturation of rRNA, and could inhibit purine nucleobase metabolic process and amino acid transmembrane transport to produce antimicrobial effects. When the two agents acted simultaneously, it was obvious that AgNPs had a greater effect on the growth process of A. fumigatus than domiphen. KEGG enrichment analysis showed that AgNPs could mainly suppress nitrogen metabolism to inhibit the growth of A. fumigatus; Domiphen could enhance ribosome biogenesis and suppress caffeine metabolism to inhibit the growth of A. fumigatus, while AgNPs combined with domiphen could affect the synthesis of secondary metabolites, steroid synthesis and material transport during the growth of A. fumigatus. It has been reported that Ag+ and AgNPs mainly interact with membrane proteins (Abram and Fromm, 2020), which may be the true reason why AgNPs could suppress nitrogen assimilation which relies on the membrane nitrate transporters. In line with our hypothesis, exogenous addition of nitrogen source could abolish the synergistic antimicrobial effect, suggesting that the synergistic antimicrobial effect partially relies on the nitrogen source deficiency. On the one hand, Ag+ interferes with electron transport and ATP production by substituting protons in the sulfhydryl group, blocking the bacterial respiratory chain (Abram and Fromm, 2020). As a result, cell metabolism is suppressed, which is consistent with our data that AgNPs combined with domiphen could inhibit the metabolic processes of A. fumigatus such as nitrogen metabolism. On the other hand, the bacterial ribosome is one of AgNPs’ main intracellular targets (Yamanaka et al., 2005). Denaturation of ribosome results in the inhibition of amino acid metabolism by AgNPs. Domiphen could increase the permeability of membrane due to its surfactant properties (Tits et al., 2020), thereby causing AgNPs easily to enter cells, which could be one of the reasons why domiphen could be a potentiator of AgNPs. In addition, Jana Tits, et al. found that domiphen increases intracellular miconazole availability by enhancing azole import and possibly by releasing sequestered miconazole (Tits et al., 2021). Similarly, domiphen could probably affect the intracellular AgNPs distribution to increase the antimicrobial activity of AgNPs, which remains to be determined. Moreover, in line with our data that AgNPs and domiphen have low cytotoxicity (Figure 5), the safety of AgNPs or domiphen at relatively low doses in vivo had been confirmed by various documents (Tits et al., 2020; Xu et al., 2020; Zielinska et al., 2020). Thus, the combination of AgNPs and domiphen has the potential to be developed into a next-generation fungicidal therapy for the treatment of patients with IA.
The key proteins of A. fumigatus analyzed by PPI corresponded to the pathways in enrichment analysis, and it was very important to note that the transmembrane transport process of copper ion was one of the most influential factor for the growth of A. fumigatus under the action of AgNPs combined with domiphen on the basis of high expression level (40-fold) of copper transporter. Copper ion (Cu), regarded as an important virulence factor in fungal pathogens, is one of the essential elements in organisms, which affects many biochemical reactions in organisms and participates in various metabolic pathways (Festa and Thiele, 2011; Cai et al., 2017). Intracellular copper balance is important for the survival of microorganisms (Cai et al., 2017; Li et al., 2019). In the process of copper ion uptake, the high affinity copper transporter family is responsible for the transport of copper ions across the membrane under low-copper condition. Ctr copper transporter family protein (AFUB_020800) is one of the key proteins affecting this pathway (Cai et al., 2017). The expression of Ctr copper transporter could increase about 40-fold under AgNPs combined with domiphen conditions (Supplementary Table 3), suggesting that A. fumigatus may encounter a low-copper situation with induction of high affinity copper transporter, which may be one of the reasons for the synergistic antimicrobial effects of AgNPs and domiphen. The induction of high affinity copper transporter is just a response mechanism of A. fumigatus to low-copper condition. Our validation experiment also confirmed that copper could increase the synergistic antimicrobial effect, suggested that more Cu+ in cells are attributed to the induction of copper transporters under this copper-treated condition, probably resulting in copper toxicity. In accordance with the present findings, previous studies have demonstrated that AgNPs could influence murine copper metabolism in a size-dependent manner (Skomorokhova et al., 2020). However, there are some limitations should be noted. The data were mainly analyzed by bioinformatics on the basis of RNA-seq, and the potential functional enrichment of other genes should be further investigated.
In conclusion, our study verified that AgNPs and domiphen had antimicrobial activity against A. fumigatus when they were used alone. On the basis of combined drug susceptibility test and cytotoxicity test, it was found that the combined use of AgNPs and domiphen could play synergistic antimicrobial effects with low cytotoxicity. RNA-seq was used to find out the related pathways affected by the two drugs, and the specific reasons for the influence of AgNPs and domiphen on the growth of A. fumigatus. It provides a theoretical basis and a new strategy for the treatment of IA.
Data availability statement
The original RNA-seq data have been deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA856787 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA856787).
Author contributions
WD and YL: conceptualization, resources, supervision, project administration, funding acquisition, writing—review and editing. WD, RX, and ZH; methodology. WD, HY, and YG: software. WD, RX, and YL: validation. RX: formal analysis. WD and RX: investigation. XX: data curation. WD and RX: writing—original draft preparation. RX, ZH, HY, and YG: visualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Natural Science Foundation of Basic Research Program of Jiangsu Province-Project of Youth Foundation (BK20220658) and Excellent Talents Research Foundation of Xuzhou Medical University (D2021042) to WD. This study was also supported by 2020 Quality Cultivation Project of the School of Life Sciences, 2021 Higher Education Reform Research Project of School of Life Sciences (2021-1) and Key Projects of Jiangsu University Student Innovation and Entrepreneurship Training Program (202010313002Z).
Acknowledgments
We thank professor Ling Lu for the gift of the strain A. fumigatus (A1161).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1089267/full#supplementary-material
Abbreviations
RNA, seq; RNA, sequencing; AgNPs, silver nanoparticles; MIC, minimum inhibitory concentration; FIC, fractional inhibitory concentration; IA, invasive aspergillosis; MM, minimal medium; RIN, RNA integrity number; OD, optical density; DEGs, differential expression genes; TPM, transcripts per million; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interactions.
Footnotes
1. ^https://github.com/OpenGene/fastp
2. ^http://ccb.jhu.edu/software/hisat2/index.shtml
3. ^https://ccb.jhu.edu/software/stringtie/
4. ^http://deweylab.biostat.wisc.edu/rsem/
5. ^http://www.geneontology.org
6. ^http://www.genome.jp/kegg/
7. ^https://github.com/tanghaibao/Goatools
References
Abad, A., Fernandez-Molina, J. V., Bikandi, J., Ramirez, A., Margareto, J., Sendino, J., et al. (2010). What makes aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Revista Iberoamericana De Micologia 27, 155–182. doi: 10.1016/j.riam.2010.10.003
Abram, S. L., and Fromm, K. M. (2020). Handling (Nano)silver as antimicrobial agent: therapeutic window, dissolution dynamics, detection methods and molecular interactions. Chem Eur J 26, 10948–10971. doi: 10.1002/chem.202002143
Amadi, C. N., Offor, S. J., Frazzoli, C., and Orisakwe, O. E. (2019). Natural antidotes and management of metal toxicity. Environ. Sci. Pollut. Res. 26, 18032–18052. doi: 10.1007/s11356-019-05104-2
Barreto, J. A., O'Malley, W., Kubeil, M., Graham, B., Stephan, H., and Spiccia, L. (2011). Nanomaterials: applications in cancer imaging and therapy. Adv. Mater. 23, H18–H40. doi: 10.1002/adma.201100140
Ben-Ami, R., and Kontoyiannis, D. P. (2021). Resistance to antifungal drugs. Infect. Dis. Clin. N. Am. 35, 279–311. doi: 10.1016/j.idc.2021.03.003
Brogan, D. M., and Mossialos, E. (2016). A critical analysis of the review on antimicrobial resistance report and the infectious disease financing facility. Glob. Health 12:8. doi: 10.1186/S12992-016-0147-Y
Cadena, J., Thompson, G. R., and Patterson, T. F. (2016). Invasive aspergillosis current strategies for diagnosis and management. Infect. Dis. Clin. N. Am. 30, 125–142. doi: 10.1016/j.idc.2015.10.015
Cadena, J., Thompson, G. R., and Patterson, T. F. (2021). Aspergillosis epidemiology, diagnosis, and treatment. Infect. Dis. Clin. N. Am. 35, 415–434. doi: 10.1016/j.idc.2021.03.008
Cai, Z. D., Du, W. L., Zeng, Q. Q., Long, N. B., Dai, C. C., and Lu, L. (2017). Cu-sensing transcription factor Mac1 coordinates with the Ctr transporter family to regulate cu acquisition and virulence in aspergillus fumigatus. Fungal Genet. Biol. 107, 31–43. doi: 10.1016/j.fgb.2017.08.003
Chen, S. F., Zhou, Y. Q., Chen, Y. R., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, X., and Schluesener, H. J. (2008). Nanosilver: a nanoproduct in medical application. Toxicol. Lett. 176, 1–12. doi: 10.1016/j.toxlet.2007.10.004
Du, W. L., Zhai, P. F., Liu, S., Zhang, Y. W., and Lu, L. (2021a). The copper chaperone CcsA, coupled with superoxide dismutase SodA, mediates the oxidative stress response in aspergillus fumigatus. Appl. Environ. Microbiol. 87:e0101321. doi: 10.1128/AEM.01013-21
Du, W. L., Zhai, P. F., Wang, T. L., Bromley, M. J., Zhang, Y. W., and Lu, L. (2021b). The C2H2 transcription factor SltA contributes to azole resistance by Coregulating the expression of the drug target Erg11A and the drug efflux pump Mdr1 in aspergillus fumigatus. Antimicrob. Agents Chemother. 65, e01839–e01820. doi:ARTN e01839-20. doi: 10.1128/AAC.01839-20
Ehdaie, B. (2007). Application of nanotechnology in cancer research: review of progress in the National Cancer Institute Alliance for nanotechnology. Int. J. Biol. Sci. 3, 108–110. doi: 10.7150/ijbs.3.108
Espinel-Ingroff, A., Cuenca-Estrella, M., Fothergill, A., Fuller, J., Ghannoum, M., Johnson, E., et al. (2011). Wild-type MIC distributions and epidemiological cutoff values for amphotericin B and aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). Antimicrob. Agents Chemother. 55, 5150–5154. doi: 10.1128/Aac.00686-11
Esquivel, B. D., Smith, A. R., Zavrel, M., and White, T. C. (2015). Azole drug import into the pathogenic fungus aspergillus fumigatus. Antimicrob. Agents Chemother. 59, 3390–3398. doi: 10.1128/Aac.05003-14
Festa, R. A., and Thiele, D. J. (2011). Copper: an essential metal in biology. Curr. Biol. 21, R877–R883. doi: 10.1016/j.cub.2011.09.040
Fumagalli, L., Regazzoni, L. G., Straniero, V., Valoti, E., Aldini, G., Vistoli, G., et al. (2018). Stressed degradation studies of domiphen bromide by LC-ESI-MS/MS identify a novel promising antimicrobial agent. J. Pharm. Biomed. Anal. 159, 224–228. doi: 10.1016/j.jpba.2018.06.055
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Hsu, A. J., Tamma, P. D., and Fisher, B. T. (2022). Challenges in the treatment of invasive aspergillosis in immunocompromised children. Antimicrob. Agents Chemother. 66:e0215621. doi: 10.1128/aac.02156-21
Hu, L. H., Yang, X., Yin, J., Rong, X., Huang, X. L., Yu, P. Q., et al. (2021). Combination of AgNPs and Domiphen is antimicrobial against biofilms of common pathogens. Int. J. Nanomedicine 16, 7181–7194. doi: 10.2147/Ijn.S334133
Jiang, H. C., Shen, Y. N., Liu, W. D., and Lu, L. (2014). Deletion of the putative stretch-activated ion channel Mid1 is hypervirulent in aspergillus fumigatus. Fungal Genet. Biol. 62, 62–70. doi: 10.1016/j.fgb.2013.11.003
Karthaus, M., and Buchheidt, D. (2013). Invasive aspergillosis: new insights into disease, diagnostic and treatment. Curr. Pharm. Des. 19, 3569–3594. doi: 10.2174/13816128113199990330
Kim, D., Landmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/Nmeth.3317
Kumar, S., Muthu, V., Bansal, Y. S., Mehta, N., and Arora, V. (2021). Invasive pulmonary aspergillosis with brain dissemination in an immunocompetent host. Autops. Case Rep. 11:e2021280. doi: 10.4322/acr.2021.280
Leitao, J. H. (2020). Microbial Virulence Factors. Int. J. Mol. Sci. 21:5320. doi:Artn 5320. doi: 10.3390/ijms21155320
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Bmc Bioinform. 12:323. doi:Artn 323. doi: 10.1186/1471-2105-12-323
Li, C., Li, Y. J., and Ding, C. (2019). The role of copper homeostasis at the host-pathogen Axis: from bacteria to fungi. Int. J. Mol. Sci. 20:175. doi: 10.3390/Ijms20010175
Li, Y. Q., Zhang, Y. W., Zhang, C., Wang, H. C., Wei, X. L., Chen, P. Y., et al. (2020). Mitochondrial dysfunctions trigger the calcium signaling-dependent fungal multidrug resistance. Proc. Natl. Acad. Sci. U. S. A. 117, 1711–1721. doi: 10.1073/pnas.1911560116
Liu, L. W., Yang, M. Y., Zhou, M., Li, J. J., Liu, B., and Pan, Y. Y. (2017). Improvement of cytotoxicity of autologous CIKs from patients with breast cancer to MCF-7 cells by suppressed PD-1 expression. Cancer Biomark. 20, 609–615. doi: 10.3233/Cbm-170588
Long, Y., Hou, J., Tang, F., Lin, Z. X., Huang, X. L., Li, W., et al. (2021). Proarrhythmic effects induced by benzethonium chloride and domiphen bromide in vitro and in vivo. Toxicol. Appl. Pharmacol. 431:115731. doi: 10.1016/j.taap.2021.115731
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Minakshi, P., Ghosh, M., Brar, B., Kumar, R., Lambe, U. P., Ranjan, K., et al. (2019). Nano-antimicrobials: a new paradigm for combating mycobacterial resistance. Curr. Pharm. Des. 25, 1554–1579. doi: 10.2174/1381612825666190620094041
Ou, P. H., Hao, C., Liu, J., He, R. G., Wang, B. Q., and Ruan, J. M. (2021). Cytocompatibility of Ti-xZr alloys as dental implant materials. J. Mater. Sci. Med. 32 Artn 50, 1–10. doi: 10.1007/S10856-021-06522-W
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y. F., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi:ARTN e47. doi: 10.1093/nar/gkv007
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Roemer, T., and Krysan, D. J. (2014). Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 4:a019703. doi: 10.1101/cshperspect.a019703
Scorzoni, L., Silva, A. C. A. D. E., Marcos, C. M., Assato, P. A., de Melo, W. C. M. A., de Oliveira, H. C., et al. (2017). Antifungal therapy: new advances in the understanding and treatment of mycosis. Front. Microbiol. 8:36. doi: 10.3389/fmicb.2017.00036
Simoes, M. F., and Antunes, A. (2021). Microbial pathogenicity in space. Pathogens 10:450. doi:Artn 450. doi: 10.3390/pathogens10040450
Siscar-Lewin, S., Hube, B., and Brunke, S. (2019). Antivirulence and avirulence genes in human pathogenic fungi. Virulence 10, 935–947. doi: 10.1080/21505594.2019.1688753
Skomorokhova, E. A., Sankova, T. P., Orlov, I. A., Savelev, A. N., Magazenkova, D. N., Pliss, M. G., et al. (2020). Size-dependent bioactivity of silver nanoparticles: antibacterial properties, influence on copper status in mice, and whole-body turnover. Nanotechnol. Sci. Appl. 13, 137–157. doi: 10.2147/NSA.S287658
Tits, J., Berman, J., Cammue, B. P. A., and Thevissen, K. (2021). Combining miconazole and Domiphen bromide results in excess of reactive oxygen species and killing of biofilm cells. Front. Cell Develop. Biol. 8:617214. doi: 10.3389/fcell.2020.617214
Tits, J., Cools, F., De Cremer, K., De Brucker, K., Berman, J., Verbruggen, K., et al. (2020). Combination of miconazole and Domiphen bromide is fungicidal against biofilms of resistant Candida spp. Antimicrob. Agents Chemother. 64:e01296-20. doi: 10.1128/AAC.01296-20
van de Veerdonk, F. L., Gresnigt, M. S., Romani, L., Netea, M. G., and Latge, J. P. (2017). Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 15, 661–674. doi: 10.1038/nrmicro.2017.90
Wang, L. K., Feng, Z. X., Wang, X., Wang, X. W., and Zhang, X. G. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Xie, C., Mao, X. Z., Huang, J. J., Ding, Y., Wu, J. M., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, L., Wang, Y. Y., Huang, J., Chen, C. Y., Wang, Z. X., and Xie, H. (2020). Silver nanoparticles: synthesis, medical applications and biosafety. Theranostics 10, 8996–9031. doi: 10.7150/thno.45413
Yamanaka, M., Hara, K., and Kudo, J. (2005). Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl. Environ. Microbiol. 71, 7589–7593. doi: 10.1128/Aem.71.11.7589-7593.2005
Keywords: Aspergillus fumigatus, RNA-Seq, AgNPs, domiphen, synergistic antimicrobial effects, fungi, antimicrobials
Citation: Du W, Xu R, He Z, Yang H, Gu Y and Liu Y (2023) Transcriptomics-based investigation of molecular mechanisms underlying synergistic antimicrobial effects of AgNPs and Domiphen on the human fungal pathogen Aspergillus fumigatus. Front. Microbiol. 14:1089267. doi: 10.3389/fmicb.2023.1089267
Edited by:
Shigeki Nakamura, Tokyo Medical University, JapanReviewed by:
Daisuke Hagiwara, University of Tsukuba, JapanAzusa Takahashi-Nakaguchi, Chiba University, Japan
Takashi Umeyama, National Institute of Infectious Diseases, Tokyo, Japan
Copyright © 2023 Du, Xu, He, Yang, Gu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenlong Du, ✉ d2VubG9uZ2R1QHh6aG11LmVkdS5jbg==; Yi Liu, ✉ Y2JwZWxpdXlpbmV3QDE2My5jb20=
 Wenlong Du
Wenlong Du Ruolin Xu1,2
Ruolin Xu1,2