
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 18 January 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1087709
This article is part of the Research TopicInsights in Terrestrial Microbiology: 2022View all 7 articles
Soil microbial use efficiency of straw carbon (C), which is the proportion of straw-C microbes assimilate into new biosynthetic material relative to C lost out of the system as CO2, is critical in increasing soil organic C (SOC) content, and hence maintaining soil fertility and productivity. However, the effect of chemical structures of the organic amendments (OAs) on the microbial use efficiency of straw-C remains unclear. The effect of the chemical structure of the OAs on microbial use efficiency of straw-C was elucidated by a combination of 13C-straw labeling with high-throughput sequencing and pyrolysis-GC/MS. We found a strong positive correlation between the microbial use efficiency of straw-C and the proportion of heterocyclic compounds (Hete_C). The microbial use efficiency of straw-C was highest in soil supplemented with Hete_C-dominant OAs, which significantly shifted microbial community structure toward fungal dominance. Specifically, fungal-to-bacterial ratio, fungal richness, and the relative abundance of Ascomycota were higher in soil with a higher proportion of Hete_C-dominant OAs. Together, our study suggests that OAs with high proportion of Hete_C promote the microbial use efficiency of straw-C by increasing the dominance of fungi in the soil microbial community in agroecosystems.
Soil organic carbon (SOC) is crucial in maintaining soil fertility and productivity, and straw returning is an important strategy to improve SOC in agroecosystems (Lal, 2004). Therefore, it is critical to enhancing the efficiency of straw carbon (C) conversion into SOC. Soil microorganisms convert straw-C to SOC and mineralized carbon dioxide (CO2; Cebrian and Lartigue, 2004). During the decomposition of straw, microorganisms use part of the assimilated C for growth, which significantly contributes to SOC, and use part of the assimilated C for CO2 mineralization during respiration (Liang et al., 2017). The microbial use efficiency of straw-C is the growth ratio to the amount of assimilated C. Therefore, the microbial use efficiency of straw-C is essential in improving SOC in agricultural systems.
In general, microbial C use efficiency (CUE) depends on the soil microbial community structure (Sinsabaugh et al., 2013; Kaiser et al., 2014). Bacteria and fungi are the main decomposers of organic matter in the soil, but both have different life strategies and metabolisms (Strickland and Rousk, 2010). Previous studies reveal that fungi make greater contribution to SOC accumulation than bacteria (Six et al., 2006; Strickland and Rousk, 2010). Presumably, microbial CUE is higher in microbial communities with higher fungal-to-bacterial ratios (F:B). However, recent investigations have demonstrated that microbial CUE is higher in a microbial community with lower F:B (Silva-Sánchez et al., 2019; Soares and Rousk, 2019). Thus, whether the microbial use efficiency of straw-C is higher in a fungi-dominated community or in a bacterial-dominated community in agricultural soils remains controversial. Additionally, microbial CUE is affected by the relationships among microbial communities (Geyer et al., 2016). Biological relationships between fungi and bacteria alter community-level functions by affecting the community composition and directly or indirectly altering the behavior and physiology of certain microbial species (Mitri and Foster, 2013). The effects of the complex community relationships on the microbial use efficiency of straw-C remain elusive.
The current main approach to improving the microbial CUE in agricultural soils relies on the shift of the microbial community structure by adding organic amendments (OAs; Liu et al., 2020). The quality of OAs added to the soil affects the structure of the microbial community, which indirectly affects the microbial CUE (Zhong, 2020). The OAs for soils comprise varying proportions of recalcitrant compounds and labile compounds (KoÈgel-Knabner, 2002). Different organic compounds favor the colonization of specific microbial communities (Torres et al., 2014). Some studies have shown that the microorganisms favored by OAs rich in recalcitrant compounds have a high microbial CUE (Liu et al., 2020). In contrast, other studies show that the specific microbial community recruited by OAs with a high proportion of labile compounds has a high microbial CUE (Miao et al., 2021). Thus, the relationships between the quality of OAs and the microbial CUE (i.e., microbial use efficiency of straw-C) in agricultural soils remain controversial.
Herein, we investigated the effect of the type of OAs on the soil microbial community and microbial use efficiency of straw-C. The chemical structure of the OAs was analyzed using pyrolysis gas-chromatography/mass-spectrometry (pyrolysis-GC/MS). Pyrolysis-GC/MS is a powerful technology for exploring the molecular composition of complex OAs (Yang et al., 2007; Gonzalez-Vila et al., 2009; Kaal et al., 2016). The microbial use efficiency of straw-C was calculated by 13C-straw labeling. The bacterial and fungal community structures were elucidated by amplicon sequencing of the 16S rRNA and internal transcript spacer (ITS) genes, respectively. In addition, the abundance of bacterial and fungal cellulose-degrading genes was quantified. We hypothesized that OAs with a higher proportion of labile compounds cause a significant shift in the soil microbial community structure toward fungal dominance, enhancing the microbial use efficiency of straw-C.
Soil was sampled from the Yangliu Agro-Ecological Experiment Station, Suixi County, Anhui Province, China (33°37′ N, 116°45′ E) in June 2020 (immediately following the harvest of winter wheat). The samples were collected from the top 20-cm soil layer using an auger, air-dried, and passed through a 2-mm sieve. The soil was classified as a Shajiang Calci-Aquic Vertosol according to the Chinese Soil Taxonomy. The soil contained 14.24 g kg−1 of organic C, 0.99 g kg−1 of total N, and 0.67 g kg−1 of total P (Li et al., 2014).
We selected three types of OAs which comprise lignin-dominant OAs (LM), cellulose-dominant OAs (CM), and OAs with equal content of lignin and cellulose (MM). The molecular structure of OAs was determined using pyrolysis-GC/MS (MDGCMSMS TQ8050, SHIMADZU, Japan). All identifiable pyro-lysates were categorized into eight chemical groups: lignin compounds, phenol compounds, carbohydrates, saturated hydrocarbons, unsaturated hydrocarbons, nitrogen-containing compounds, aromatic hydrocarbons, and poly-aromatic hydrocarbons, according to the degree of chemical similarity; and four types of organic compounds were further classified based on chemical structure of organic C skeletons: alicyclic compounds (Alic_C), aliphatic compounds (Alip_C), aromatic compounds (Arom_C), and heterocyclic compounds (Hete_C; Supplementary Table S1).
Soil was added with LM, MM, or CM, and soil without addition of OAs was used as the control (Ctrl). The experiment included four treatments with each six replicates. OA was added to 80 g of soil (oven-dry weight) at a rate of 5 mg C g−1 soil (Savy et al., 2020), and then pre-incubated at 25°C in dark for 30 days. N and P nutrients were provided according to C:N:P stoichiometric ratios (C:N:p = 10,000:833:200; Kirkby et al., 2013). Soil moisture was adjusted to 60% of water-holding capacity (WHC) using sterile deionized water during the incubation period. After the pre-incubation, the three replicates of each treatment received 13C-labeled straw (δ13C 99‰), and another three replicates without addition of 13C-straw were used to partition the mineralization and microbial assimilation of the 13C-straw. The addition rate was 1 mg straw-C g−1 soil, equivalent to the amount of straw returned into soils per year. An open scintillation vial containing 10 ml of 3 M sodium hydroxide (NaOH) was placed to absorb CO2 respired during the incubation. All soils were incubated at 25°C and 60% WHC for 28 days. Previous studies have demonstrated that soil microbes efficiently used easily-degradable component of straw in 1 week and labile straw-C was almost anabolized by soil microbes in a month (Chen et al., 2014; Zhong, 2020). Therefore, the soils were sampled in 7 and 28 days of the incubation to analyze CO2, microbial biomass C (MBC), 16S rRNA and ITS genes, and the abundance of cellulose-degrading genes.
CO2 was trapped in 10 ml 3 M NaOH trap solution to measure soil respiration at 7 and 28 days of incubation as previously described (Zibilske, 1994). BaCO3 pellets were dried at 50°C and stored for 13C analysis using an isotope ratio mass spectrometer (Delta Plus, IRMS; Thermo Fisher Scientific, Bremen, Germany). MBC at 7 and 28 days of incubation was determined using the chloroform fumigation-extraction method (Makarov et al., 2015). For 13C measurement of microbial biomass, an aliquot (ca. 10 mL) of the K2SO4 extract was freeze-dried, and the solid material was then analyzed.
The microbial use efficiency of straw-C is the ratio of C incorporated in microbial biomass and used for respiration. The C used for respiration was based on the accumulation of CO2 in 7 or 28 days. The proportion (f%) of CO2-C or MBC derived from 13C-straw was calculated using a two-component isotopic mixing model based on Eq. (1) (Balejdent and Balabane, 1992):
where δ is the δ13C of CO2-C or MBC in the 13C-straw amended soils (‰), δsoil is the δ13C of the 13C-straw treatments (‰), and δstraw is the δ13C of the 13C-straw (‰).
The δ 13C of MBC was calculated using Eq. (2) (De et al., 2011):
where δCfum and δCnfum are the δ13C levels in fumigated and non-fumigated soil extracts, respectively, and Cfum and Cnfum are the total MBC contents of the fumigated and non-fumigated soil extracts, respectively.
The CO2 emission or MBC amount in the 13C-straw was calculated by multiplying f% with the total CO2 emission or total MBC amount.
The microbial use efficiency of straw-C was calculated according to Eq. (3) (Geyer et al., 2016):
where 13C-MBC is the MBC derived from 13C-straw (mg kg−1 soil), and 13C-CO2 is the cumulative 13C-straw (mg kg−1 soil) mineralized by day 7 or 28.
DNA was extracted from 0.25 g fresh soil samples taken at days 7 and 28. The extraction was done using the Power Soil DNA isolation kit (MoBio, Carlsbad, CA, United States), according to the manufacturer’s instructions. The DNA concentrations were determined by a nanodrop spectrophotometer (NanoDrop, Wilmington, DE). The Illumina sequencing was done by Novogene. The fragments of bacterial 16S rRNA gene and fungal internal transcribed spacer (ITS) gene were amplified using the primer pairs 341F (5′-CCTAYGGGRBGCASCAG-3′)/806R (5′-GGACTACNNGGGTATCTAAT-3′) and ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′)/ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′), respectively. A sample-specific barcode was added to the forward primer for distinguishing the amplicons from different samples. The PCR conditions were 94°C for 5 min; 30 cycles of 94°C for 30 s, 52°C for 30 s, and an extension of 72°C for 30 s, followed by 72°C for 10 min, and held at 4°C. PCR products from all samples were pooled and purified in equimolar concentrations, and sequencing was performed on an Illumina MiSeq instrument (Caporaso et al., 2012). The bacterial 16S rRNA gene sequencing and ITS gene sequencing data were uploaded to the NCBI SRA database under accession number PRJNA898503.1
The raw FASTQ files were demultiplexed and quality-filtered using QIIME2. Subsequently, the amplicon variant sequences (ASVs) with a 97% similarity were clustered together using DADA2 (version 1.8). Poor-quality sequences of less than 200 bp and quality scores of less than 20 were removed, and the chimeras were removed using the UCHIME algorithm (Edgar et al., 2011). The phylogenetic affiliation of each rRNA gene sequence was determined using the UNITE database for the ITS1 rRNA region and the Ribosomal Database Project (RDP) Classifier2 against the SILVA (SSU117/119) database for the 16S rRNA gene at an 80% confidence threshold (Wang et al., 2007).
The fungal cbhI gene and bacterial GH48 gene were selected as functional biomarkers of cellulolytic fungi and bacteria, respectively. The primers for quantitative PCR (qPCR) of bacterial GH48 and fungal cbhI genes have been described in a previous study (Edwards et al., 2008). The primers GH48 F8/GH48 R5 (GH48_F8: 5 GCCADGHTBGGCG ACTACCT 3; GH48_R5: 5 CGCCCCABGMSWWGTACCA 3) and cbhI F/cbhI R (cbhI F: ACCAAYTGCTAYACIRGYAA; cbhI R: GCYTCCCAIATRTCCATC) were used for the qPCR assay. The abundance of bacterial GH48 and fungal cbhI genes was quantified according to modified procedures (Zhang et al., 2017). The thermal profiles of qPCR for the target genes of GH48 and cbhI were as follows: 95°C for 5 min, 40 × (94°C for 30 s, 60°C for 45 s, and 72°C for 90 s), and data collection at 84°C for 10 s; and 94°C for 4 min, 40 × (94°C for 45 s, 50°C for 30 s, and 72°C for 60 s), and data collection at 81°C for 10 s, respectively. The initial concentrations of the two plasmids as the standards for bacterial GH48 and fungal cbhI gene abundance analysis corresponded to 1.85 × 1011 and 2.65 × 1010 copies g−1 dry soil, respectively.
The effect of different OAs on microbial use efficiency of straw-C, fungal-to-bacterial (F:B) ratio and cbhI-to-GH48 ratios was analyzed using a one-way analysis of variance (ANOVA). Statistical significance was set at p < 0.05. Principal coordinate analysis (PCoA) was performed based on Bray-Curtis distances to visualize the change in the bacterial and fungal community under different OAs (Bray and Curtis, 1957). The difference in microbial use efficiency of straw-C among the OAs groups was analyzed using ANOVA. A correlation matrix was constructed to describe the complex co-occurrence patterns in bacteria-fungi networks. The maps were created using the geom_polygon function in the “ggplot2” package in R software.
Co-occurrence networks of bacteria and fungi in different OAs groups were constructed (Faust and Raes, 2012). We focused on the most dominant phylotypes, both abundant (with relative abundance higher than 0.1%) and ubiquitous (bacteria and fungi were present above half of all the samples). The filtering identified dominant phylotypes in soils and minimized potential spurious correlations from the rare taxa. The focus was on the dominant soil phylotypes because they are expected to have disproportionate functional importance in their ecosystems and are widely distributed. A valid co-occurrence was considered a statistically robust correlation between species when the correlation coefficient (r) was >0.6 or < −0.6 at p < 0.01. The p-values < 0.01 were adjusted by the Benjamini-Hochberg procedure to reduce the chances of obtaining false positives. The network modules were identified using the “igraph” package. The number and relative abundance of ASVs in the three most important modules were calculated to assess the effects of different OAs on specific phylotypes with strong interconnections. In addition, subnetworks were extracted by preserving the phylotypes of individual soil samples using the subgraph function in “igraph” package. Networks were constructed and visualized using the “igraph” package. Random forest modelling was performed using the “random-forest” package, and the model significance and predictor importance were determined using the “rfPermute” packages. The rest of the analyses were performed with the R software.
Py-GC/MS analysis revealed that the three OAs had considerably different chemical structures (Figure 1A). Hete_C was predominant in the CM, Arom_C was predominant in the LM, and Hete_C and Arom C were in equal proportions in the MM. The microbial use efficiency of straw-C was significantly (p < 0.001) higher in Hete_C rich CM compared with other treatments in 7 days. There was no significant difference in microbial use efficiency of straw-C among the three OAs treatments in 28 days. Therefore, straw-C decomposition efficiency was higher in the CM than in the LM and MM (Figure 1B).
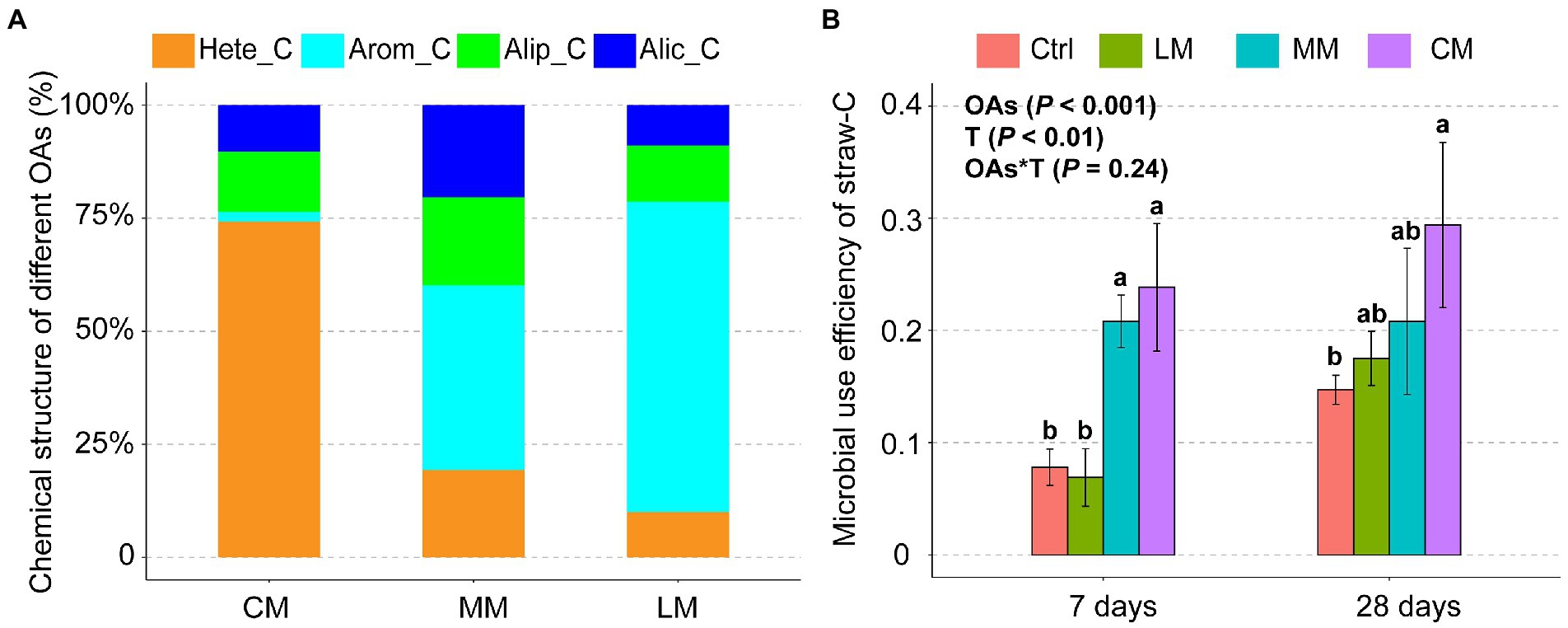
Figure 1. Effects of organic amendments (OAs) with different chemical structures on microbial use efficiency of straw-C. (A) The chemical structure of different OAs. (B) The microbial use efficiency of straw-C under different OAs. Data are presented as means ± SD (n = 3). Error bars depict standard deviations. Lowercase letters indicate significant differences (p < 0.05) between the treatments. Ctrl, Control without OAs; LM, Addition of lignin-dominant OAs; MM, Addition of OAs with equal content of lignin and cellulose; and CM, Addition of cellulose-dominant OAs.
The chemical structures of OAs significantly influenced the microbial community structure (Figure 2). The F:B ratios and cbhI-GH48 were higher in the CM (p < 0.001), which all declined over time (Figures 2A,B). F:B ratio declined because the abundance of fungi decreased sharply over time, whereas the abundance of bacteria increased (Supplementary Table S2). The cbhI-GH48 ratios declined because even though the abundance of both cbhI and GH48 increased over time, the increase in expression of GH48 was higher than that of cbhI (Supplementary Table S2).
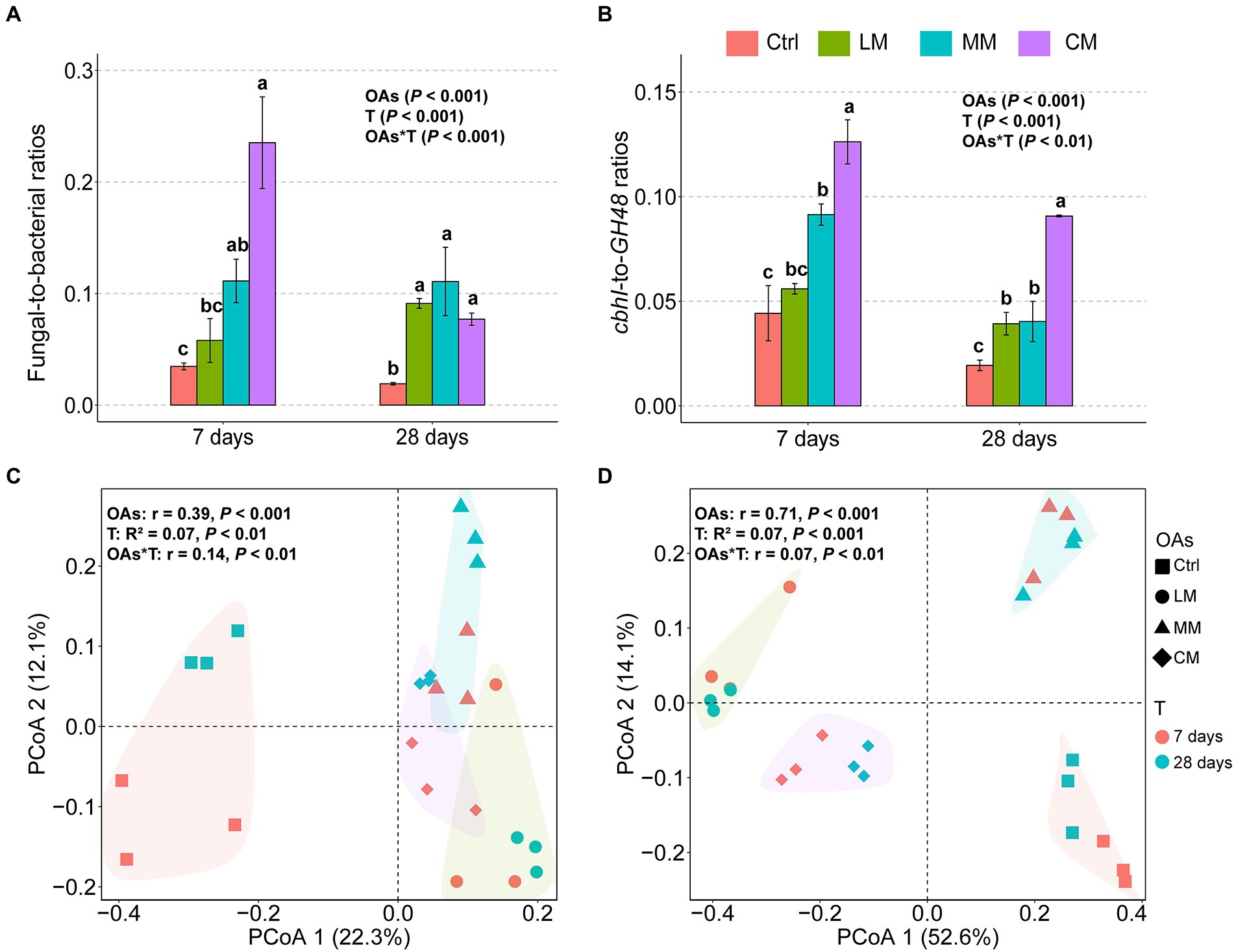
Figure 2. Effects of organic amendments (OAs) with different chemical structures on fungal-to-bacterial ratios (F:B) and their C-cycling gene ratios (cbhI-to-GH48 ratios) as well as bacterial and fungal diversity. (A) The fungal-to-bacterial ratios (F:B) under different OAs. (B) The cbhI-to-GH48 ratios (cbhI:GH48) under different OAs. Data are expressed as means ± SD (n = 3). Error bars depict standard deviations. Lowercase letters indicate significant differences (p < 0.05) between the treatments. Principal coordinates analysis (PCoA) of the bacterial (C) and fungal (D) communities with different OAs supplements at days 7 and 28 based on Bray-Curtis distance (d). The polygons indicate a 95% standard error for each treatment. Ctrl, Control without OAs; LM, Addition of lignin-dominant OAs; MM, Addition of OAs with equal content of lignin and cellulose; and CM, Addition of cellulose-dominant OAs.
There was no significant difference in the bacterial richness among the three OAs (Supplementary Figure S1A, p = 0.09), but fungi richness was considerably lower in the CM (Supplementary Figure S1B, p < 0.001). Principal coordinates analysis (PCoA) revealed that both fungi and bacteria diversity differed considerably among the three types of OAs (p < 0.001; Figures 2C,D). Furthermore, the degree of fungal partitioning (R2 = 0.71) was greater than bacterial partitioning (R2 = 0.39). In addition, the type of OAs had little effect on the relative abundance of bacteria at the phylum level (Supplementary Figure S2A) but significantly impacted the abundance of fungi at the phylum level (Supplementary Figure S2B). Specifically, OAs with high Hete_C proportion favored a significant increase in the relative abundance of Ascomycota.
To evaluate the effects of different OAs on the microbial co-occurrence patterns, we built a correlation-based network incorporating the dominant bacterial and fungal phylotypes. We identified three highly connected modules within the network (Figure 3A). The number of bacteria was larger than that of fungi in the three most important modules (i.e., modules 1–3; Figure 3B). The relative abundance of fungi was higher than that of bacteria in modules 1 and 3, while an opposite trend existed in module 2 (Figure 3C). Compared with the Ctrl and LM, the CM drastically increased the relative abundance of fungi in module.
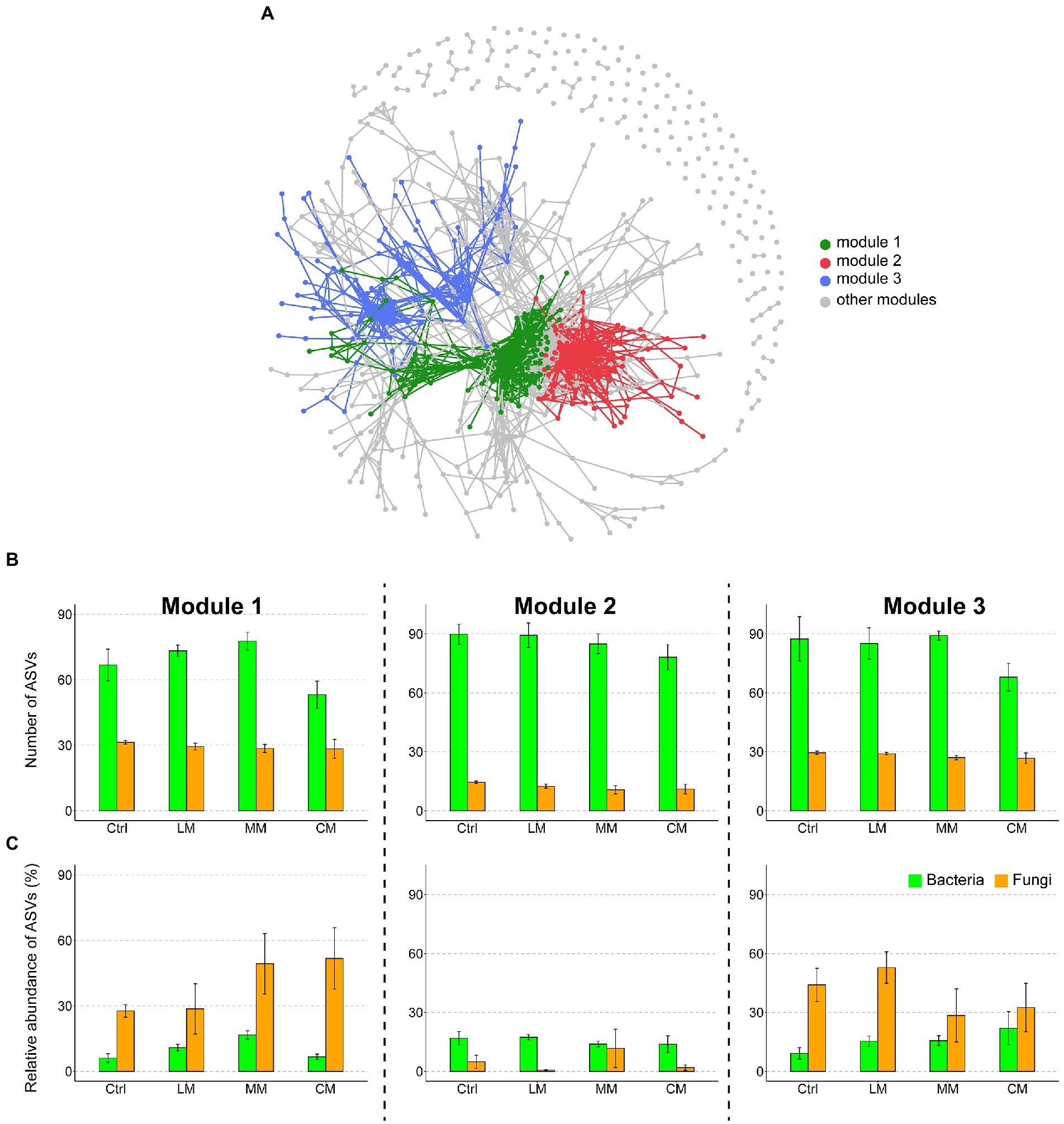
Figure 3. (A) Network co-occurrence patterns of the dominant bacteria and fungi across treatments. The modules are distinguished using different colors. (B) The number of amplicon variant sequence (ASVs) of bacteria and fungi in the three most important modules (modules 1–3). (C) Relative abundance of ASVs of bacteria and fungi in the three most important modules (modules 1–3) in different OAs treatments. Data are presented as means ± SD (n = 6). Error bars depict standard deviations. Lowercase letters indicate significant differences (p < 0.05) between the treatments. Ctrl (Control without OAs), LM (Addition of lignin-dominant OAs), MM (Addition of OAs with equal content of lignin and cellulose), and CM (Addition of cellulose-dominant OAs).
The type of OAs influenced the microbial community indices and microbial use efficiency of straw-C. Hete-C and Arom_C reduced the microbial community indices (Supplementary Figure S3). Specifically, F:B, cbhI-GH48, Ascomycota, Proteobacteria, and microbial use efficiency of straw-C were strongly (p < 0.05) and positively correlated with Hete_C content but were strongly (p < 0.05) and negatively correlated with Arom_C content. Furthermore, in investigating the correlation of microbial community structure with microbial use efficiency of straw-C, we found that microbial use efficiency of straw-C increased significantly (p < 0.05) with F:B, cbhI:GH48, the abundance of module #1, and the relative abundance of Ascomycota, whereas there was a strong negative correlation between the abundance of module #3 members (p < 0.05) and microbial use efficiency of straw-C (Figure 4). In addition, there was a weak negative correlation between microbial use efficiency of straw-C and the abundance of Proteobacteria, but there was a strong negative correlation between the bacterial diversity (p < 0.05) and the fungal diversity (Supplementary Figure S4). The microbial community with greater microbial use efficiency of straw-C were predicted using random forest modelling. The importance of a variable was determined by removing it from the model and determining the subsequent change in the model’s mean square error (MSE). Our results indicated that module #1 was potentially more important in the microbial use efficiency of straw-C, accounting for 11.92% of the MSE (Figure 5, p < 0.01). Moreover, Ascomycota and F:B were the next most important factors influencing microbial use efficiency of straw-C, accounting for 10.68% (p < 0.01) and 7.16% (p < 0.05) respectively, of the MSE.
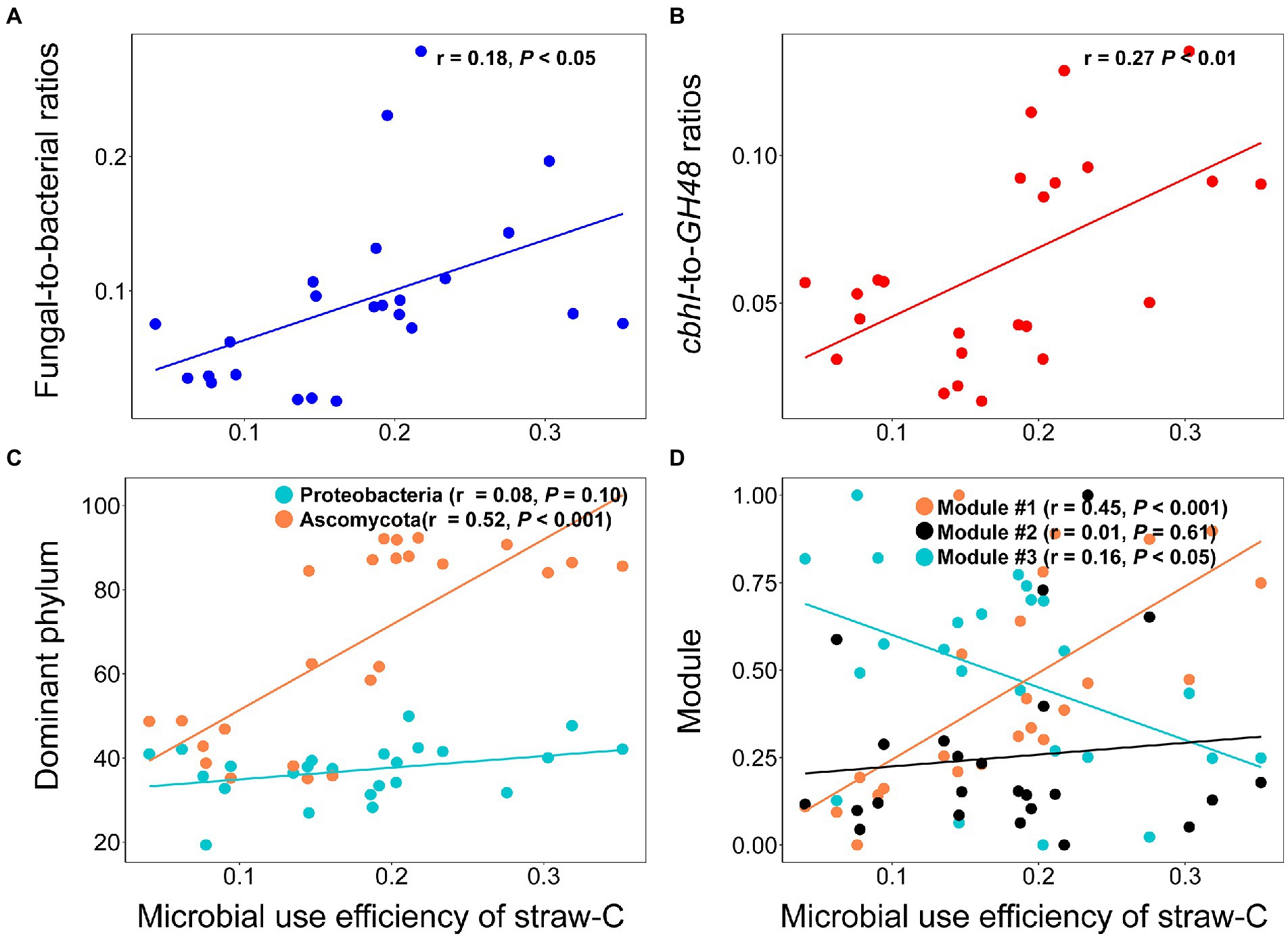
Figure 4. Effects of fungal-to-ratios (A), cbhI-to-GH48 ratios (B), dominant phylum (C), and three main ecological clusters (D) on microbial use efficiency of straw-C.

Figure 5. Mean predictor importance of the relative abundances of indicators of microbial community properties on the microbial use efficiency of straw-C. The contribution (% of increased mean square error) is calculated based on random forest analyses. Percentage increases in the MSE (mean squared error) of variables estimate the importance of these predictors. The MSE% values were directly proportional to the importance of a given predictor. *p < 0.05, **p < 0.01.
The addition of OAs with a high proportion of Hete_C significantly increased the microbial use efficiency of straw-C in the soil by shifting the microbial community. Similarly, Soares and Rousk (2019) reported that microbial CUE was directly proportional to the C-quality (i.e., plant litter with a high proportion of labile compounds). Likewise, Silva-Sánchez et al. (2019) also found that the growth rates of both bacteria and fungi, which indicate the ability of these organisms to assimilate C into their biomass, are stimulated by high-quality OAs. This might be due to the unique chemical structure of Hete_C, in which N or O atoms are embedded in the C rings (Anslyn and Dougherty, 2006; Ouellette and Rawn, 2018). This lowers chemical bonding energies relative to Arom_C compounds, making them more unstable and easily utilized by microorganisms. As a result, microorganisms use less energy to break down Hete_C into simple forms that can be utilized for growth and development. Therefore, soil microorganisms produced more cell mass per unit mass of straw-C broken down (i.e., soil microbial use efficiency of straw-C) in soil containing a high proportion of Hete_C-dominant OAs.
OAs influenced the microbial use efficiency of straw-C by altering microbial community structure. OAs used in the present research were complex natural macromolecular organic compounds. Thus, fungi, which degrade complex macromolecular compounds more efficiently than bacteria, were recruited (van der Wal et al., 2007; Challacombe et al., 2019; Clocchiatti et al., 2020). In the present study, we hypothesized that fungal groups would dominate soils with a high proportion of labile compounds (i.e., Hete_C) and OAs. The soil F:B and microbial use efficiency of straw-C were higher in soils with Hete_C-dominant OAs than other OAs. In contrast with Soares and Rousk (2019), we observed that microbial use efficiency of straw-C was strongly and positively correlated with Hete_C content and fungal abundances. This confirmed our hypothesis that the microbial use efficiency of straw-C is high in soils with higher fungal-to-bacterial ratios. Our results are consistent with Kallenbach et al. (2016) findings, who observed that microbial-driven SOM accumulation was very high in soils with higher fungal abundances. Similarly, Liu et al. (2020) found that microbial CUE was significantly high in soils abundant with fungi. In general, fungi are considered to metabolize organic compounds more effectively than bacteria (Malik et al., 2016) and thus have a higher CUE than bacteria. Furthermore, fungal cells are larger than bacterial cells and thus have a lower surface area to volume ratio (Six et al., 2006). Therefore, fungal necromass form macromolecular aggregations more efficiently, which decreases the availability of fungal necromass to be utilized by microbiome. However, for small molecular organic compounds like glucose which microbes can easily assimilate, the microbial CUE is still high with lower F:B (Eilers et al., 2010; Fu et al., 2022). This might be because bacteria are 10 to 100 times more abundant than fungi and mainly utilize small molecular substrates. We found that the F:B ratio decreased over time since the expression of bacterial genes increased more rapidly over time than fungal genes. The higher cbhI-GH48 ratio during CM and the addition of OAs higher in Hete_C content corroborated the above results. In fungi, cellobiohydrolase is encoded by the cbhI gene (Edwards et al., 2008), while in bacteria, it is encoded by the GH48 gene (Sizova et al., 2011). Cellobiohydrolase participates in cellulose degradation. Our findings showed that the cbhI-GH48 ratios decreased over time because the bacterial abundance increases more rapidly than the fungal abundance. Further analyses confirmed that the F:B ratio decreased over time degradation. Therefore, F:B and microbial use efficiency of straw-C at the beginning of degradation was higher in soils with Hete_C, which is more complex than Arom C, but F:B decreased while microbial use efficiency of straw-C gradually increased with the decomposition of complex OAs.
Notably, the relative abundance of Ascomycota (the dominant fungi in CM and MM) increased in soils with high Hete_C content. The dominant fungi identified in CM and MM are widely distributed in soils worldwide, and their abundance is strongly and positively correlated with the microbial CUE (Ma et al., 2013; Challacombe et al., 2019). This suggests that Ascomycota plays a critical role in decomposing OAs and generating SOC, and might be because Ascomycota secretes numerous enzymes that degrade macromolecules (Lumbsch and Huhndorf, 2010). In addition, fungal richness decreased significantly reduced in OAs with a high proportion of Hete_C. However, members of Ascomycota increased in soils with OAs and a high proportion of Hete_C.
Additionally, we further found that a specific microbial community recruited by OAs contributes more to the microbial use efficiency of straw-C than single microorganism taxa by random forest modelling. This is consistent with previous studies which reported that microbes collaborate in breaking down and utilizing organic materials by exuding substances that stimulate the growth and reproduction of species that are favorable to their existence and dominance (Faust et al., 2017; Ning et al., 2020; Yuan et al., 2021). Fungi can degrade complex organic macromolecules into numerous small molecules using secretory extracellular enzymes such as cellulase and laccase (Clocchiatti et al., 2020). Except for the stronger ability of fungi to secrete extracellular enzymes, the large amounts of organic acids secreted by fungi also degrade straw (Wang et al., 2022). The simpler organic compounds are then used by bacteria for growth, contributing to the microbial CUE (Zhao et al., 2019). Thus, fungi and bacteria recruited by OAs could be more efficient in converting straw into biomass. This collaborative degradation strategy of complex macromolecular compounds into smaller compounds by fungi and bacteria validates the theory of the organic matter continuum proposed by Lehmann and Kleber (2015). This provides important insight into how OAs composed of specific organic compounds promote the growth and reproduction of specific microbial communities in agricultural ecosystems. OAs enhance the dominance of specific microbial populations, which allows them to degrade straw-C rapidly, and efficiently convert it into their biomass, improving SOC accumulation in agricultural soils.
OAs significantly shifted the soil microbial community structure and increased the microbial use efficiency of straw-C. Compared with OAs with a low proportion of Hete_C, OAs with a high proportion of Hete_C significantly increased the microbial use efficiency of straw-C. F:B, cbhI-GH48, fungal diversity, and the relative abundance of Ascomycota were higher in OAs with a high proportion of Hete_C. Overall, our study suggests that adding OAs with a high proportion of Hete_C can increase the microbial use efficiency of straw-C by enhancing the abundance and diversity of fungi in the soil microbial community.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: SRA data: PRJNA898503.
JL, LC, and JZ conceived and designed the experiments. JL and LC performed the experiments and wrote the manuscript. JL analyzed the data. CZ, DM, GZ, and QN provided resources. All authors contributed to the article and approved the submitted version.
This research was jointly supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDA28020203), the National Key Research and Development Program of China (2022YFD1500203), the National Natural Science Foundation of China (42177332), and the China Agriculture Research System (CARS-03-15 and CARS-52).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XH declared a shared parent affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1087709/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | The richness indices of the bacterial (A) and fungal (B) communities in soils supplemented with different organic amendments (OAs) at days 7 and 28. Data are presented as means ± SD (n = 3). Error bars depict standard deviations. Lowercase letters indicate significant differences (P < 0.05) between the treatments. Ctrl (Control without OAs), LM (Addition of lignin-dominant OAs), MM (Addition of OAs with equal content of lignin and cellulose), and CM (Addition of cellulose-dominant OAs).
SUPPLEMENTARY FIGURE S2 | Relative abundance of the major bacterial (A) and fungal (B) taxonomic groups at the phylum level in soil supplemented with different organic amendments (OAs) at days 7 and 28. Ctrl (Control without OAs), LM (Addition of lignin-dominant OAs), MM (Addition of OAs with equal content of lignin and cellulose), and CM (Addition of cellulose-dominant OAs).
SUPPLEMENTARY FIGURE S3 | Effects of Alic_C, Alip_C, Arom_C, and Hete_C on fungal-to-ratios, cbhI-to-GH48 ratios, diversity and abundance of bacteria and fungi, dominant phylum, three main ecological clusters (A) and microbial use efficiency of straw-C (B).
SUPPLEMENTARY FIGURE S4 | Effects of bacterial and fungal diversity on microbial use efficiency of straw-C.
Anslyn, E. V., and Dougherty, D. A. (2006). Modern physical organic chemistry. University Science, Sausalito, CA
Balejdent, J., and Balabane, M. (1992). Maize root-derived soil organic carbon estimated by natural 13C abundance. Soil Biol. Biochem. 24, 97–101. doi: 10.1016/0038-0717(92)90264-X
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Cebrian, J., and Lartigue, J. (2004). Patterns of Herbivory and decomposition in aquatic and terrestrial ecosystems. Ecol. Monogr. 74, 237–259. doi: 10.1890/03-4019
Challacombe, J. F., Hesse, C. N., Bramer, L. M., McCue, L. A., Lipton, M., Purvine, S., et al. (2019). Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genomics 20:976. doi: 10.1186/s12864-019-6358-x
Chen, L., Zhang, J., Zhao, B., Yan, P., Zhou, G., and Xin, X. (2014). Effects of straw amendment and moisture on microbial communities in Chinese fluvo-aquic soil. J. Soil. Sediment. 14, 1829–1840. doi: 10.1007/s11368-014-0924-2
Clocchiatti, A., Hannula, S. E., van den Berg, M., Korthals, G., and de Boer, W. (2020). The hidden potential of saprotrophic fungi in arable soil: patterns of short-term stimulation by organic amendments. Appl. Soil Ecol. 147:103434. doi: 10.1016/j.apsoil.2019.103434
De, T. I., Amery, F., Van Moorleghem, C., Smolders, E., and Merckx, R. (2011). Tracing the source and fate of dissolved organic matter in soil after incorporation of a 13C labelled residue: a batch incubation study. Soil Biol. Biochem. 43, 513–519. doi: 10.1016/j.soilbio.2010.11.016
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. BMC Bioinf. 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, I. P., Upchurch, R. A., and Zak, D. R. (2008). Isolation of fungal cellobiohydrolase I genes from sporocarps and forest soils by PCR. Appl. Environ. Microbiol. 74, 3481–3489. doi: 10.1128/AEM.02893-07
Eilers, K. G., Lauber, C. L., Knight, R., and Fierer, N. (2010). Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903. doi: 10.1016/j.soilbio.2010.02.003
Faust, S., Heinze, S., Ngosong, C., Sradnick, A., Oltmanns, M., Raupp, J., et al. (2017). Effect of biodynamic soil amendments on microbial communities in comparison with inorganic fertilization. Appl. Soil Ecol. 114, 82–89. doi: 10.1016/j.apsoil.2017.03.006
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Fu, Y., Luo, Y., Tang, C., Li, Y., Guggenberger, G., and Xu, J. (2022). Succession of the soil bacterial community as resource utilization shifts from plant residues to rhizodeposits. Soil Biol. Biochem. 173:108785. doi: 10.1016/j.soilbio.2022.108785
Geyer, K. M., Kyker-Snowman, E., Grandy, A. S., and Frey, S. D. (2016). Microbial carbon use efficiency: accounting for population, community, and ecosystem-scale controls over the fate of metabolized organic matter. Biogeochemistry 127, 173–188. doi: 10.1007/s10533-016-0191-y
Gonzalez-Vila, F. J., Gonzalez-Perez, J. A., Akdi, K., Gomis, M. D., Perez-Barrera, F., and Verdejo, T. (2009). Assessing the efficiency of urban waste biocomposting by analytical pyrolysis (Py-GC/MS). Bioresour. Technol. 100, 1304–1309. doi: 10.1016/j.biortech.2008.06.067
Kaal, J., Wagner, S., and Jaffé, R. (2016). Molecular properties of ultrafiltered dissolved organic matter and dissolved black carbon in headwater streams as determined by pyrolysis-GC-MS. J. Anal. Appl. Pyrolysis 118, 181–191. doi: 10.1016/j.jaap.2016.02.003
Kaiser, C., Franklin, O., Dieckmann, U., and Richter, A. (2014). Microbial community dynamics alleviate stoichiometric constraints during litter decay. Ecol. Lett. 17, 680–690. doi: 10.1111/ele.12269
Kallenbach, C. M., Frey, S. D., and Grandy, A. S. (2016). Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 7:13630. doi: 10.1038/ncomms13630
Kirkby, C. A., Richardson, A. E., Wade, L. J., Batten, G. D., Blanchard, C., and Kirkegaard, J. A. (2013). Carbon-nutrient stoichiometry to increase soil carbon sequestration. Soil Biol. Biochem. 60, 77–86. doi: 10.1016/j.soilbio.2013.01.011
KoÈgel-Knabner, I. (2002). The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol. Biochem. 34, 139–162. doi: 10.1016/S0038-0717(01)00158-4
Lal, R. (2004). Soil carbon sequestration impacts on global climate change and food security. Science 304, 1623–1627. doi: 10.1126/science.1097396
Lehmann, J., and Kleber, M. (2015). The contentious nature of soil organic matter. Nature 528, 60–68. doi: 10.1038/nature16069
Li, W., Qiao, Y., Chen, H., Cao, C., Du, S., and Zhao, Z. (2014). Effects of combined straw and N application on the physicochemical properties of lime concretion black soil and crop yields. Acta Ecol. Sin. 34, 345052–345061. doi: 10.5846/stxb201303210475
Liang, C., Schimel, J. P., and Jastrow, J. D. (2017). The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2:17105. doi: 10.1038/nmicrobiol.2017.105
Liu, Z., Wu, X., Liu, W., Bian, R., Ge, T., Zhang, W., et al. (2020). Greater microbial carbon use efficiency and carbon sequestration in soils: amendment of biochar versus crop straws. GCB Bioenergy 12, 1092–1103. doi: 10.1111/gcbb.12763
Lumbsch, H. T., and Huhndorf, S. M. (2010). Myconet volume 14. Part one. Outline of Ascomycota—2009. Part two. Notes on Ascomycete systematics. Nos. 4751–5113. Fieldiana Life Earth Sci. 1, 1–64. doi: 10.3158/1557.1
Ma, A., Zhuang, X., Wu, J., Cui, M., Lv, D., Liu, C., et al. (2013). Ascomycota members dominate fungal communities during straw residue decomposition in arable soil. PLoS One 8:e66146. doi: 10.1371/journal.pone.0066146
Makarov, M. I., Malysheva, T. I., Menyailo, O. V., Soudzilovskaia, N. A., Van Logtestijn, R. S. P., and Cornelissen, J. H. C. (2015). Effect of K2SO4concentration on extractability and isotope signature (δ13C and δ15N) of soil C and N fractions. Eur. J. Soil Sci. 66, 417–426. doi: 10.1111/ejss.12243
Malik, A. A., Chowdhury, S., Schlager, V., Oliver, A., Puissant, J., Vazquez, P. G., et al. (2016). Soil fungal: bacterial ratios are linked to altered carbon cycling. Front. Microbiol. 7:1247. doi: 10.3389/fmicb.2016.01247
Miao, Y., Niu, Y., Luo, R., Li, Y., Zheng, H., Kuzyakov, Y., et al. (2021). Lower microbial carbon use efficiency reduces cellulose-derived carbon retention in soils amended with compost versus mineral fertilizers. Soil Biol. Biochem. 156:108227. doi: 10.1016/j.soilbio.2021.108227
Mitri, S., and Foster, K. R. (2013). The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 47, 247–273. doi: 10.1146/annurev-genet-111212-133307
Ning, D., Yuan, M., Wu, L., Zhang, Y., Guo, X., Zhou, X., et al. (2020). A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11:4717. doi: 10.1038/s41467-020-18560-z
Ouellette, R. J., and Rawn, J. D. (2018). “Structure and bonding in organic compounds” in Organic Chemistry. 2nd edn. Academic Press. 1–30.
Savy, D., Mercl, F., Cozzolino, V., Spaccini, R., Cangemi, S., and Piccolo, A. (2020). Soil amendments with lignocellulosic residues of biorefinery processes affect soil organic matter accumulation and microbial growth. ACS Sustain. Chem. Eng. 8, 3381–3391. doi: 10.1021/acssuschemeng.9b07474
Silva-Sánchez, A., Soares, M., and Rousk, J. (2019). Testing the dependence of microbial growth and carbon use efficiency on nitrogen availability, pH, and organic matter quality. Soil Biol. Biochem. 134, 25–35. doi: 10.1016/j.soilbio.2019.03.008
Sinsabaugh, R. L., Manzoni, S., Moorhead, D. L., and Richter, A. (2013). Carbon use efficiency of microbial communities: stoichiometry, methodology and modelling. Ecol. Lett. 16, 930–939. doi: 10.1111/ele.12113
Six, J., Frey, S. D., Thiet, R. K., and Batten, K. M. (2006). Bacterial and fungal contributions to carbon sequestration in Agroecosystems. Soil Sci. Soc. Am. J. 70, 555–569. doi: 10.2136/sssaj2004.0347
Sizova, M. V., Izquierdo, J. A., Panikov, N. S., and Lynd, L. R. (2011). Cellulose- and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl. Environ. Microbiol. 77, 2282–2291. doi: 10.1128/AEM.01219-10
Soares, M., and Rousk, J. (2019). Microbial growth and carbon use efficiency in soil: links to fungal-bacterial dominance, SOC-quality and stoichiometry. Soil Biol. Biochem. 131, 195–205. doi: 10.1016/j.soilbio.2019.01.010
Strickland, M. S., and Rousk, J. (2010). Considering fungal: bacterial dominance in soils – methods, controls, and ecosystem implications. Soil Biol. Biochem. 42, 1385–1395. doi: 10.1016/j.soilbio.2010.05.007
Torres, I. F., Bastida, F., Hernández, T., Bombach, P., Richnow, H. H., and García, C. (2014). The role of lignin and cellulose in the carbon-cycling of degraded soils under semiarid climate and their relation to microbial biomass. Soil Biol. Biochem. 75, 152–160. doi: 10.1016/j.soilbio.2014.04.007
van der Wal, A., de Boer, W., Smant, W., and van Veen, J. A. (2007). Initial decay of woody fragments in soil is influenced by size, vertical position, nitrogen availability and soil origin. Plant and Soil 301, 189–201. doi: 10.1007/s11104-007-9437-8
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, L., Guan, H., Hu, J., Feng, Y., Li, X., Yusef, K. K., et al. (2022). Aspergillus Niger enhances organic and inorganic phosphorus release from wheat straw by secretion of degrading enzymes and oxalic acid. J. Agric. Food Chem. 70, 10738–10746. doi: 10.1021/acs.jafc.2c03063
Yang, H., Yan, R., Chen, H., Lee, D. H., and Zheng, C. (2007). Characteristics of hemicellulose, cellulose and lignin pyrolysis. Fuel 86, 1781–1788. doi: 10.1016/j.fuel.2006.12.013
Yuan, M. M., Kakouridis, A., Starr, E., Nguyen, N., Shi, S., Pett-Ridge, J., et al. (2021). Fungal-bacterial Cooccurrence patterns differ between Arbuscular Mycorrhizal fungi and Nonmycorrhizal fungi across soil niches. MBio 12:e03509. doi: 10.1128/mBio.03509-20
Zhang, Q., Liang, G. Q., Guo, T. F., He, P., Wang, X. B., and Zhou, W. (2017). Evident variations of fungal and actinobacterial cellulolytic communities associated with different humified particle-size fractions in a long-term fertilizer experiment. Soil Biol. Biochem. 113, 1–13. doi: 10.1016/j.soilbio.2017.05.022
Zhao, Z., Gonsior, M., Schmitt-Kopplin, P., Zhan, Y., Zhang, R., Jiao, N., et al. (2019). Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. ISME J. 13, 2551–2565. doi: 10.1038/s41396-019-0449-1
Zhong, Y. (2020). Microbial community assembly and metabolic function during wheat straw decomposition under different nitrogen fertilization treatment. Biol. Fertil. Soils 56, 697–710. doi: 10.1007/s00374-020-01438-z
Keywords: organic amendments, heterocyclic compounds, microbial community, microbial use efficiency of straw-C, pyrolysis-GC/MS
Citation: Li JW, Chen L, Zhang JB, Zhang CZ, Ma DH, Zhou GX and Ning Q (2023) Organic amendments with high proportion of heterocyclic compounds promote soil microbiome shift and microbial use efficiency of straw-C. Front. Microbiol. 14:1087709. doi: 10.3389/fmicb.2023.1087709
Received: 02 November 2022; Accepted: 03 January 2023;
Published: 18 January 2023.
Edited by:
Kim Yrjälä, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Xiaojing Hu, Northeast Institute of Geography and Agroecology (CAS), ChinaCopyright © 2023 Li, Chen, Zhang, Zhang, Ma, Zhou and Ning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiabao Zhang,  amJ6aGFuZ0Bpc3Nhcy5hYy5jbg==
amJ6aGFuZ0Bpc3Nhcy5hYy5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.