- 1Department of Clinical Laboratory, Affiliated Nantong Hospital 3 of Nantong University, Nantong, Jiangsu, China
- 2School of Medicine, Jiangsu University, Zhenjiang, Jiangsu, China
- 3State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
Mature biofilm formation by Vibrio parahaemolyticus requires exopolysaccharide (EPS), type IV pili, and capsular polysaccharide (CPS). Production of each is strictly regulated by various control pathways including quorum sensing (QS) and bis-(3′–5′)-cyclic di-GMP (c-di-GMP). QsvR, an AraC-type regulator, integrates into the QS regulatory cascade via direct control of the transcription of the master QS regulators, AphA and OpaR. Deletion of qsvR in wild-type or opaR mutant backgrounds altered the biofilm formation by V. parahaemolyticus, suggesting that QsvR may coordinate with OpaR to control biofilm formation. Herein, we demonstrated both QsvR and OpaR repressed biofilm-associated phenotypes, c-di-GMP metabolism, and the formation of V. parahaemolyticus translucent (TR) colonies. QsvR restored the biofilm-associated phenotypic changes caused by opaR mutation, and vice versa. In addition, QsvR and OpaR worked coordinately to regulate the transcription of EPS-associated genes, type IV pili genes, CPS genes and c-di-GMP metabolism-related genes. These results demonstrated how QsvR works with the QS system to regulate biofilm formation by precisely controlling the transcription of multiple biofilm formation-associated genes in V. parahaemolyticus.
Introduction
Vibrio parahaemolyticus, a Gram-negative halophilic bacterium, is the primarily causative agent of the seafood-associated gastroenteritis (Broberg et al., 2011). Vibrio parahaemolyticus has multiple virulence factors including thermostable direct hemolysin (TDH), type III secretion systems (T3SS1 and T3SS2), and type VI secretion systems (T6SS1 and T6SS2) as well as the capacity to form biofilms (Broberg et al., 2011; Ashrafudoulla et al., 2021). Biofilms are bacterial communities enclosed in an extracellular matrix that endows bacterial cells with a high degree of resistance to stress conditions (Yildiz and Visick, 2009; Ruhal and Kataria, 2021). Biofilm formation requires specific structures including exopolysaccharide (EPS), type IV pili, and capsular polysaccharide (CPS), which are strictly regulated by regulatory cascades such as quorum sensing (QS) and bis-(3′–5′)-cyclic di-GMP (c-di-GMP) signaling (Yildiz and Visick, 2009; Ruhal and Kataria, 2021).
The biofilm matrix contains some chemical components including proteins, EPS, extracellular DNA and membrane vesicles, among which the EPS is the most importnt one (Ruhal and Kataria, 2021). In V. parahaemolyticus, the cpsA-K and scvA-O loci are responsible for EPS biosynthesis (Makino et al., 2003; Liu et al., 2022). These two gene loci are essential for biofilm formation by V. parahaemolyticus but may play distinct roles in biofilm-associated colony morphology (Chen et al., 2010; Liu et al., 2022). CPS is associated with opaque (OP)-translucent (TR) colony switching of V. parahaemolyticus (Chen et al., 2010). Strains without or with low CPS production manifest the TR phonotype (Chen et al., 2010). CPS has a negative effect on biofilm formation, but both OP and TR strains can form biofilms (Joseph and Wright, 2004; Enos-Berlage et al., 2005). In addition, V. parahaemolyticus expresses two kinds of type IV pili, termed mannose-sensitive hemagglutinin (MSHA; encoded by VP2698-2692) and chitin-regulated pilus (ChiRP; encoded by pilABCD) (Makino et al., 2003). Both MSHA and ChiRP are important for biofilm formation, but defective biofilms produced by the MSHA mutants can be overcame by extended incubation time (Enos-Berlage et al., 2005; Shime-Hattori et al., 2006). Other structures such as flagella also play roles in biofilm formation by V. parahaemolyticus (Enos-Berlage et al., 2005; Li et al., 2020).
The c-di-GMP signaling is widely used by bacteria to modulate biofilm formation, motility, and virulence (Homma and Kojima, 2022). c-di-GMP is synthesized by diguanylate cyclase (DGC) carrying a GGDEF domain and is degraded by phosphodiesterase (PDE) containing EAL or HD-GYP domains (Homma and Kojima, 2022). V. parahaemolyticus expresses dozens of GGDEF-or/and EAL-containing proteins (Makino et al., 2003), but only a few of these were investigated. ScrC, which is encoded by scrABC, contains both EAL and GGDEF domains (Ferreira et al., 2008; Trimble and McCarter, 2011), but functions as a PDE in the presence of ScrA and ScrB (Ferreira et al., 2008; Trimble and McCarter, 2011). Deletion of scrABC enhances biofilm formation but decreases swarming motility (Ferreira et al., 2008; Trimble and McCarter, 2011). ScrG is another EAL- and GGDEF-containing protein that acts only as a PDE in V. parahaemolyticus (Kim and McCarter, 2007). Overexpression of scrG induces swarming motility but inhibits biofilm formation (Kim and McCarter, 2007). More recently, four GGDEF-type proteins, ScrO, ScrJ, ScrL, and GefA, as well as two EAL-type proteins, LafV and TpdA, were found to be involved in modulation of motility and biofilm formation by V. parahaemolyticus (Kimbrough et al., 2020; Kimbrough and McCarter, 2020; Martinez-Mendez et al., 2021; Zhong et al., 2022).
Quorum sensing is a cell density-dependent communication process widely used by bacteria to precisely control gene expression and bacterial behavior in response to changes in concentration of small molecules termed auto-inducers within surroundings (Lu et al., 2018). The QS system of Vibrios generally employs master regulators, AphA and LuxR orthologs, such as OpaR in V. parahaemolyticus (Zhang et al., 2012), LuxR in V. harvei (Chaparian et al., 2020), HapR in V. cholerae (Ball et al., 2017) and SmcR in V. vulnificus (Lee et al., 2008), to control gene expression. In general, AphA works at low cell density (LCD) to promote multiple bacterial behaviors including biofilm formation, motility, c-di-GMP synthesis and virulence factor production, whereas the LuxR orthologs function at high cell density (HCD) to inhibit these bacterial behaviors (Rutherford et al., 2011; Wang et al., 2013a,b; Lu et al., 2018, 2019, 2021a,b; Zhang et al., 2017b, 2019, 2021; Sun et al., 2022). In addition, the LuxR orthologs are high expressed at HCD, but they also can be detected at LCD, suggesting that LuxR orthologs function throughout growth (Rutherford et al., 2011; van Kessel et al., 2013; Lu et al., 2019).
QsvR, an AraC-type transcriptional regulator, coordinates with the QS system to control the expression of virulence genes in V. parahaemolyticus (Zhang et al., 2019). QsvR directly represses and activates the transcription of aphA and opaR, respectively (Zhang et al., 2019). AphA indirectly represses qsvR transcription at LCD, whereas OpaR indirectly activates its transcription at HCD (Zhang et al., 2019). AphA operates at LCD to activate T3SS1 genes, whereas OpaR and QsvR function at HCD to activate tdh2 and the T3SS2 genes (Zhang et al., 2019). In addition, QsvR maintains the basal expression of T3SS1 at HCD (Zhang et al., 2019). Moreover, QsvR represses the transcription of toxR and calR, but activates the transcription of T6SS2 genes, cpsQ-mfpABC and mfpABC (Qiu et al., 2020; Zhang et al., 2021a,b). Most importantly, the CPS-associated OP-TR transition was regulated by OpaR, which is expressed in OP but not TR strains, with the expression of opaR in TR strain converting the TR strain to an OP phenotype (McCarter, 1998). Mutation of qsvR in TR strains enhanced initial attachment but impaired biofilm maturation, whereas deletion of qsvR in OP strains produced robust biofilms (Enos-Berlage et al., 2005). Therefore, QsvR may coordinate with OpaR to regulate biofilm formation by V. parahaemolyticus.
In this study, we demonstrated QsvR works with OpaR to repress biofilm formation and c-di-GMP metabolism, as well as to promote V. parahaemolyticus to form OP colonies. OpaR restored biofilm-associated phenotypic changes caused by qsvR mutation, and vice versa. Further, QsvR and OpaR worked coordinately to activate the transcription of type IV pili genes, CPS genes, and scrG, but repress the transcription of scrA. OpaR but not QsvR negatively regulated the transcription of EPS-associated genes. Collectively, our data highlight how QsvR works with the QS system to regulate biofilm formation by precisely controlling the transcription of multiple biofilm formation-associated genes in V. parahaemolyticus.
Materials and methods
Bacterial strains
Vibrio parahaemolyticus strain RIMD2210633 was used as the wild-type (WT) strain in this study (Makino et al., 2003). Non-polar qsvR and opaR single-gene deletion mutants (ΔqsvR and ΔopaR), derived from the WT strain, were constructed by our previous studies (Zhang et al., 2012, 2019). The qsvR and opaR double-gene mutant (ΔqsvRΔopaR) was generated via deletion of a 615-bp fragment (nucleotides 1–615) of opaR from ΔqsvR by homologous recombination using suicide plasmid pDS132 (Sun et al., 2012; Zhang et al., 2012).
Complementary plasmids, pBAD33-qsvR and pBAD33-opaR (Zhang et al., 2012, 2019), were, respectively, introduced into ΔqsvR and ΔopaR, yielding ΔqsvR/pBAD33-qsvR (C-ΔqsvR), ΔopaR/pBAD33-qsvR, ΔqsvR/pBAD33-opaR, and ΔopaR/pBAD33-opaR (C-ΔqsvR). The non-recombinant pBAD33 was transferred into WT and each of the mutants to yield WT/pBAD33, ΔqsvR/pBAD33, and ΔopaR/pBAD33.
All strains and plasmids used in this study are listed in Supplementary Table S1.
Bacterial growth conditions
Vibrio parahaemolyticus strains were grown similarly as previously described (Zhang et al., 2012; Lu et al., 2019). Briefly, the overnight cell cultures in 2.5% (w/v) Bacto Heart Infusion (HI) broth (BD Biosciences, United States) were diluted 40-fold into sterile phosphate buffered saline (PBS; pH 7.2), and 150 μl of the diluted cells was spread onto a HI plate with a diameter of 5 cm. Bacterial cells were harvested after 6 h of incubation at 37°C. When necessary, the media were supplemented with 50 μg/ml gentamicin, 5 μg/ml chloramphenicol, or 0.1% (w/v) arabinose.
Colony morphology
Colony morphology was performed as previously described (Wang et al., 2013a). Briefly, overnight cell cultures were diluted 50-fold into 5 ml of Difco marine (M) broth 2,216 (BD Biosciences, United States), followed by incubated statically at 30°C for 48 h, and then mixed thoroughly. Two microliter of each culture was spotted on the HI plate and incubated at 37°C for the colony morphology assay.
Crystal violet staining
Crystal violet staining was performed similarly as previously described (Wang et al., 2013a). Briefly, overnight cell cultures in HI broth were diluted 50-fold into 2 ml of M broth in a 24-well cell culture plate, and incubated at 30°C with shaking at 150 rpm for 48 h. Planktonic cells were collected for determination of OD600 values. The surface-attached cells were washed three times with deionized water, and then stained with 0.1% CV, followed by another three washes with deionized water. Bound CV in each well was dissolved with 2.5 ml of 20% ethanol, and the OD570 values were determined. Relative biofilm formation was calculated with the formula: OD570/OD600.
Detection of OR-TP transition
For detection of OR and TP transition, a small amount of each overnight cell culture in HI broth was streaked onto a HI plate, and then statically incubated at 37°C for 48 h.
Determination of intracellular c-di-GMP levels
Intracellular c-di-GMP levels were measured as previously described (Gao et al., 2020). Briefly, bacterial cells were harvested from HI plates with 2 ml of ice-cold PBS, incubated at 100°C for 5 min, followed by sonicated for 30 min (power 100%, frequency 37 kHz) in an ice-water bath. After centrifugation, the c-di-GMP concentration in the supernatant was determined with a c-di-GMP enzyme-linked immunosorbent assay (ELISA) kit (Mskbio, China). Total protein concentration in the supernatant was determined by the bicinchoninic acid (BCA) assay. Intracellular c-di-GMP levels were expressed as pmol/mg protein.
RNA isolation and quantitative PCR (qPCR) analysis
Total RNA was extracted from bacterial cells using TRIzol Reagent (Invitrogen, United States). The cDNA was generated from 1 μg of each RNA sample using a FastKing First Strand cDNA Synthesis Kit (Tiangen Biotech, China) according to the manufacturer’s instructions. The qPCR assay was performed using a LightCycler 480 (Roche, Switzerland) together with SYBR Green master mix (Gao et al., 2011). Expression levels of target genes relative to that of 16S rRNA were determined using the classic 2−ΔΔCt method. All primers used in this study are listed in Supplementary Table S2.
LacZ fusion and β-galactosidase assay
The regulatory DNA region of each target gene was cloned into the pHRP309 plasmid containing a promoter-less lacZ reporter gene and a gentamicin resistance gene (Parales and Harwood, 1993). The recombinant plasmid was transferred into different V. parahaemolyticus strains, respectively. The resulting transformants were cultured and then lysed to measure the β-galactosidase activities of the cellular extracts using a β-Galactosidase Enzyme Assay System (Promega, United States) according to the manufacturer’s instructions. The Miller Units representing galactosidase activity were calculated using the following formula: 106 × [(OD420 – 1.75 × OD550)/(T × V × OD600)] (Zhang et al., 2021b). T represents the reaction time (min). T and V represent the reaction time (min) and volume (μL), respectively.
For the two-plasmid reporter assay (Qiu et al., 2020), E. coli 100 λpir (Epicentre) bearing a complementary plasmid (pBAD33-qsvR or pBAD33-opaR) or the empty pBAD33 vector and a recombinant lacZ plasmid were cultured in Luria-Bertani (LB) broth at 37°C with shaking at 200 rpm for 12 h. The resultant cultures were diluted 100-fold into 5 ml of fresh LB broth containing 0.1% arabinose and 20 μg/ml chloramphenicol, followed by incubated at 37°C with shaking at 200 rpm till an OD600 value of approximately 1.2 was obtained. The E. coli cells were harvested and then lysed to measure the β-galactosidase activity in the cell extracts.
Purification of 6 × His-tagged proteins
The entire coding regions of opaR and qsvR were individually cloned into the pET28a vector (Novagen, United States). Each recombinant pET28a plasmid was transferred into E. coli BL21λDE3 for His-tagged protein expression. Expression and purification of His-QsvR and His-OpaR were performed as previously described (Zhang et al., 2012). The dialyzed proteins were concentrated to approximately 0.5 mg/ml. The purity of the proteins was confirmed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Electrophoretic mobility-shift assay
Electrophoretic mobility-shift assay (EMSA) was performed as previously described (Zhang et al., 2017b). Briefly, the regulatory DNA region of each target gene was amplified by PCR. The DNA binding assay was performed in a 10 μl reaction volume containing binding buffer [0.5 mM EDTA, 1 mM MgCl2, 50 mM NaCl, 0.5 mM DTT, 10 mM Tris–HCl (pH 7.5), and 10 mg/ml salmon sperm DNA], 100 ng target DNA, and increasing amounts of His-tagged protein. After incubation at room temperature for 20 min, the binding products were analyzed in a native 6% (w/v) polyacrylamide gel with a UV transilluminator after being stained with ethidium bromide (EB) dye.
Experimental replicates and statistical methods
The qPCR and LacZ fusion were performed at least three independent times with results expressed as means ± standard deviation (SD). A two-way ANOVA with Tukey’s post hoc corrections for multiple comparisons was used to calculate statistical significance, with significance accepted at p < 0.01. Phenotype assays and EMSA were performed at least three times.
Results
QsvR works with OpaR to repress biofilm formation by Vibrio parahaemolyticus
OpaR repressed V. parahaemolyticus biofilm formation by regulation of c-di-GMP metabolism (Zhang et al., 2021a). QsvR also repressed biofilm formation by V. parahaemolyticus, but lacks the detailed mechanism (Enos-Berlage et al., 2005). Herein, we constructed single and double-gene mutants of qsvR and opaR as well as complementary mutants, and which were subjected to colony morphology and CV staining assays (Figure 1). As expected, the colonies of ΔopaR/pBAD33 and ΔqsvR/pBAD33 are more wrinkled than those of WT/pBAD33, whereas C-ΔopaR and C-ΔqsvR exhibited a restored phenotypes (Figure 1A). ΔqsvRΔopaR/pBAD33 had similar colony morphology to ΔopaR/pBAD33 and ΔqsvR/pBAD33 (Figure 1A). Most interestingly, ΔopaR/pBAD33-qsvR and ΔqsvR/pBAD33-opaR also exhibited restored colony phenotypes (Figure 1A). As further assessed by the CV staining (Figure 1B), ΔopaR/pBAD33, ΔqsvR/pBAD33, and ΔqsvRΔopaR/pBAD33 were more CV positive than WT/pBAD33, whereas C-ΔopaR, C-ΔqsvR, and ΔqsvR/pBAD33-opaR exhibited restored CV staining phenotypes. However, ΔopaR/pBAD33-qsvR had only a partially restored CV staining phenotype compared to ΔopaR/pBAD33. ΔqsvR/pBAD33 had much less CV staining compared to ΔopaR/pBAD33 and ΔqsvRΔopaR/pBAD33. ΔopaR/pBAD33 and ΔqsvRΔopaR/pBAD33 had similar CV staining results. Therefore, OpaR to be more capable than QsvR as an inhibitor of biofilm formation by V. parahaemolyticus. Collectively, QsvR worked with OpaR to negatively regulate biofilm formation by V. parahaemolyticus.
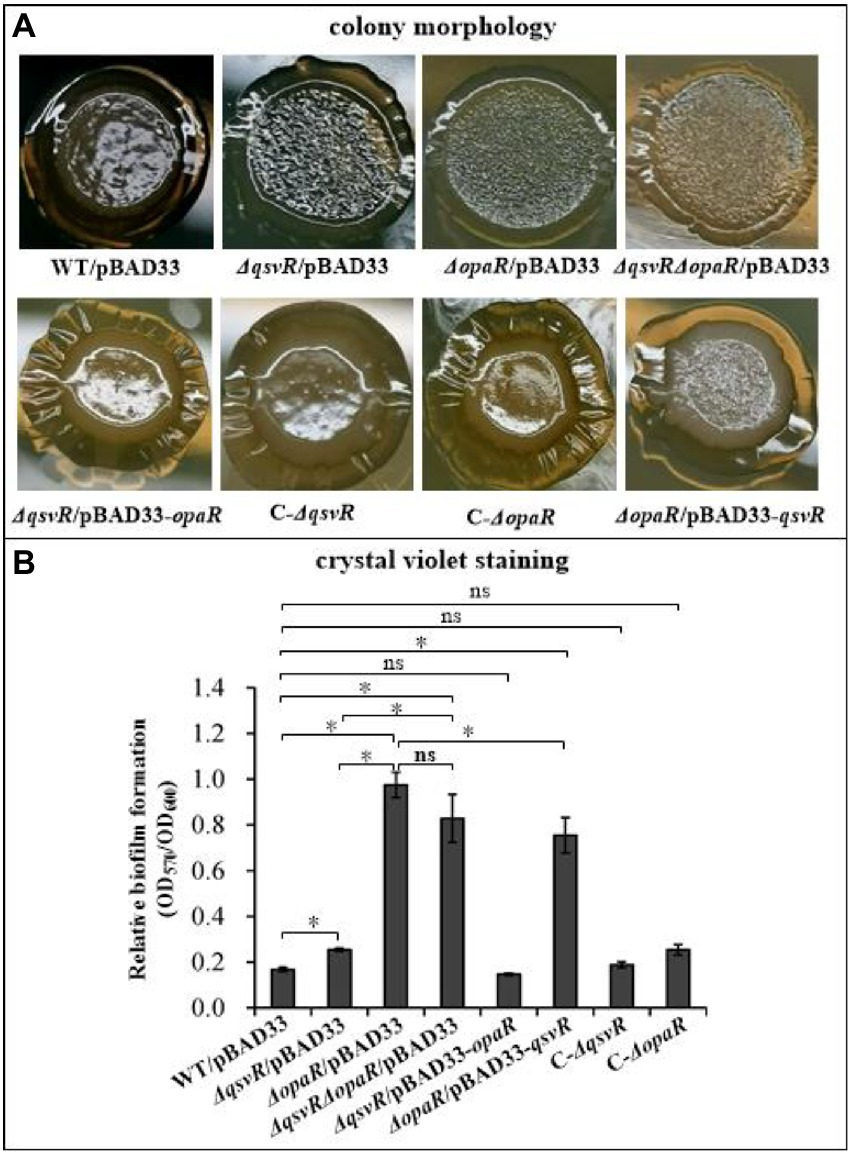
Figure 1. QsvR works with OpaR to repress biofilm formation by Vibrio parahaemolyticus. Biofilm formation by V. parahaemolyticus strains was assessed by colony morphology (A) and intensity of crystal violet staining (B). Photographs are representative of three independent experiments with three replicates each. The asterisks indicate statistical significances (p < 0.01). The ‘ns’ means no significant differences (p > 0.01).
The opaR mutant has no effect on the growth of V. parahaemolyticus (Zhang et al., 2012). Herein, we showed that WT, ΔqsvR and ΔopaRΔqsvR had indistinguishable growth rates in both HI broth and M broth (Supplementary Figure S1), indicating that qsvR mutant had no effect on the growth of V. parahaemolyticus. Thus, changes in biofilm formation by qsvR or/and opaR mutants were associated with the regulation of QsvR and OpaR.
OpaR but not QsvR negatively regulates EPS biosynthesis genes
Exopolysaccharide (EPS) production is directly associated with the wrinkled colony phenotype of V. parahaemolyticus (Chen et al., 2010). The cps and scv loci are responsible for EPS synthesis in V. parahaemolyticus (Chen et al., 2010; Liu et al., 2022). Herein, qPCR results showed that the mRNA levels of cpsA and scvE were significantly increased in ΔopaR/pBAD33, ΔqsvRΔopaR/pBAD33, and ΔopaR/pBAD33-qsvR. Levels were similar for ΔqsvR/pBAD33, C-ΔqsvR, C-ΔopaR, and ΔqsvR/pBAD33-opaR compared to WT/pBAD33 (Figure 2A). As further assessed by LacZ fusion assay (Figure 2B), promoter activity of scrE was significantly enhanced in ΔopaR/pBAD33, ΔqsvRΔopaR/pBAD33, and ΔopaR/pBAD33-qsvR, with no obvious effect in ΔqsvR/pBAD33, C-ΔqsvR, C-ΔopaR, or ΔqsvR/pBAD33-opaR compared to WT/pBAD33. The LacZ fusion results also demonstrated QsvR to have no regulatory effect on cpsA expression, but OpaR positively regulated its expression (unshown data). Previously, OpaR was demonstrated to enhance the promoter activity of cpsA (Guvener and McCarter, 2003). Conflicting results between reporter fusion and qPCR assays as well as biofilm phenotypes are difficult to interpret clearly, perhaps due to pHRP309 or its derivative used for the LacZ fusion assay. In addition, the results of two-plasmid reporter assay showed that expression of opaR but not qsvR from the recombinant pBAD33 significantly lowered promoter activities of cpsA and scvE (Figure 2C), suggesting that OpaR but not QsvR bound upstream DNA regions of cpsA and scvE to repress expression in E. coli. EMSA results showed that His-OpaR but not His-QsvR bound to upstream DNA fragments of cpsA and scvE (Figure 2D), but neither bound to the promoter DNA of vp1687, which was used as a negative control (Zhang et al., 2019). Taken together, OpaR directly repressed the transcription of cpsA and scvE, whereas QsvR had no regulatory effect on their expression.
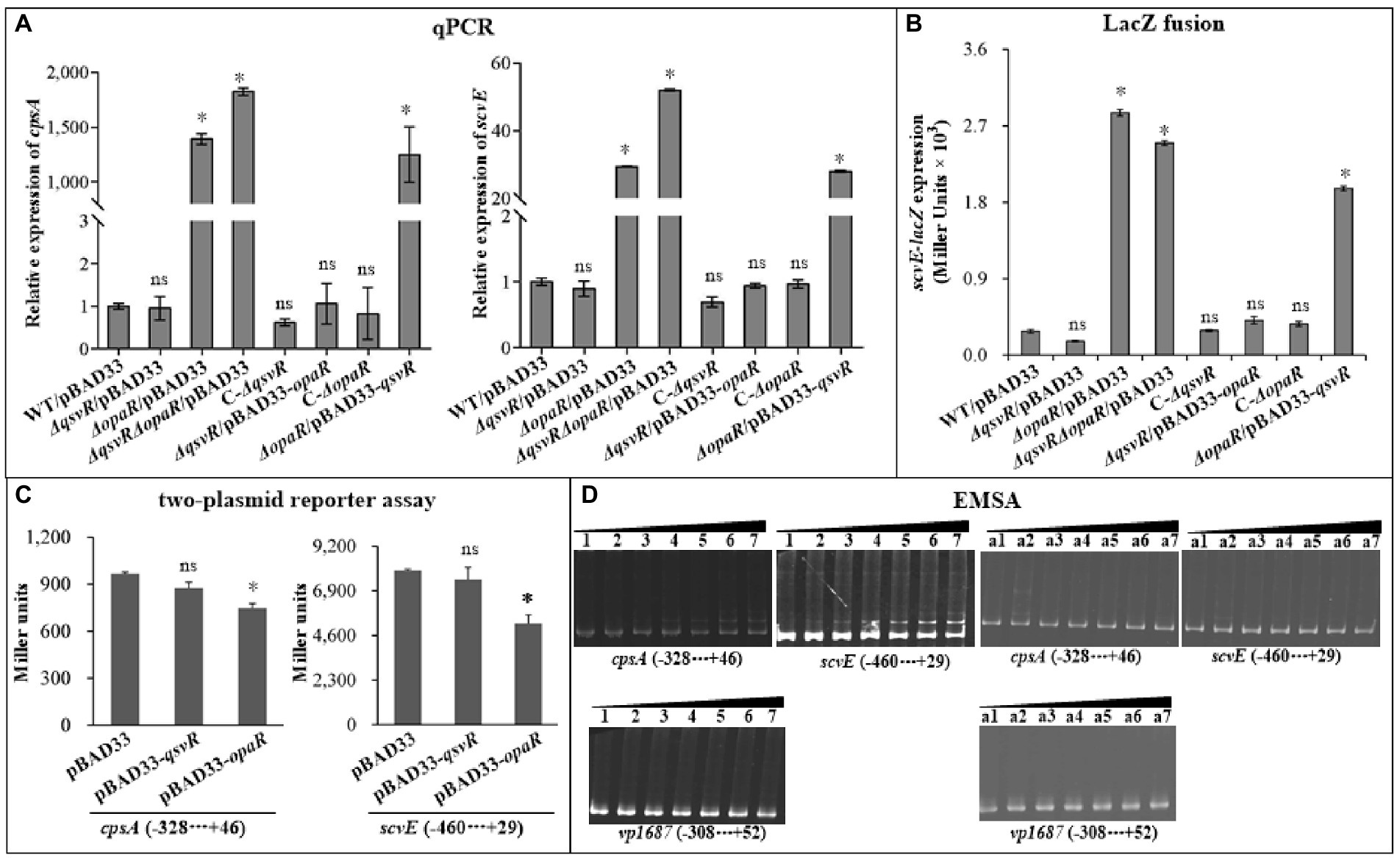
Figure 2. Regulation of cpsA and scvE by QsvR and OpaR. V. parahaemolyticus strains were cultured on HI plates, and bacterial cells were harvested after 6 h of incubation at 37°C. Negative and positive numbers indicate the nucleotide positions upstream and downstream of each target gene, respectively. The asterisks indicate statistical significances relative to WT/pBAD33 or E. coli 100 λpir/pBAD33 (p < 0.01). The ‘ns’ means no significant differences (p > 0.01). (A) qPCR. The relative mRNA levels of cpsA and scvE were examined in the different V. parahaemolyticus strains. (B) LacZ fusion. The regulatory DNA region of each target gene was cloned into the pHRP309 vector and then transferred into V. parahaemolyticus strains to determine the promoter activities (represented by Miller units) in the cellular extracts. (C) Two-plasmid reporter assay. The complementary plasmid (pBAD33-qsvR or pBAD33-opaR) or the empty pBAD33 vector and a recombinant lacZ plasmid were simultaneously introduced into E. coli 100 λpir (Epicentre), and promoter activities (represented by Miller units) of each target gene in the cellular extracts were determined with a β-Galactosidase Enzyme Assay System (Promega, United States) according to the manufacturer’s instructions. (D) Electrophoretic mobility-shift assay (EMSA). The regulatory DNA region of each target gene was incubated with increasing amounts of purified His-QsvR or His-OpaR, and then subjected to 6% (w/v) polyacrylamide gel electrophoresis. The DNA bands were visualized by EB staining. Lanes 1, 2, 3, 4, 5, 6, and 7 contain 0, 0.4, 0.8, 1.2, 1.4, 2.0, and 2.4 pmol of His-OpaR, respectively. Lanes a1, a2, a3, a4, a5, a6, and a7 contain 0, 0.021, 0.042, 0.063, 0.083, 0.11, and 0.13 pmol of His-QsvR, respectively.
QsvR works with OpaR to activate the transcription of type IV pili genes
OpaR directly activated the transcription of MSHA and ChiRP genes (Lu et al., 2021b; Sun et al., 2022). Herein, qPCR results showed that mRNA levels of type IV pili genes, mshA1 and pilA, were significantly reduced in ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33 relative to WT/pBAD33, with restoration in C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR (Figure 3A). LacZ fusion assay (Figure 3B) demonstrated reduced mshA1 and pilA promoter activities in ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33 relative to that in WT/pBAD33, with restoration in C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR. Previously, OpaR was able to regulate the expression of mshA1 and pilA in E. coli (Lu et al., 2021b; Sun et al., 2022). The data presented in Figure 3C demonstrated that expression of qsvR in E. coli significantly induced the promoter activities of mshA1 and pilA. EMSA demonstrated His-QsvR dose-dependently bind to the upstream DNA fragments of mshA1 and pilA (Figure 3D). Taken together, QsvR worked with OpaR to directly activate the transcription of type IV pili genes in V. parahaemolyticus.
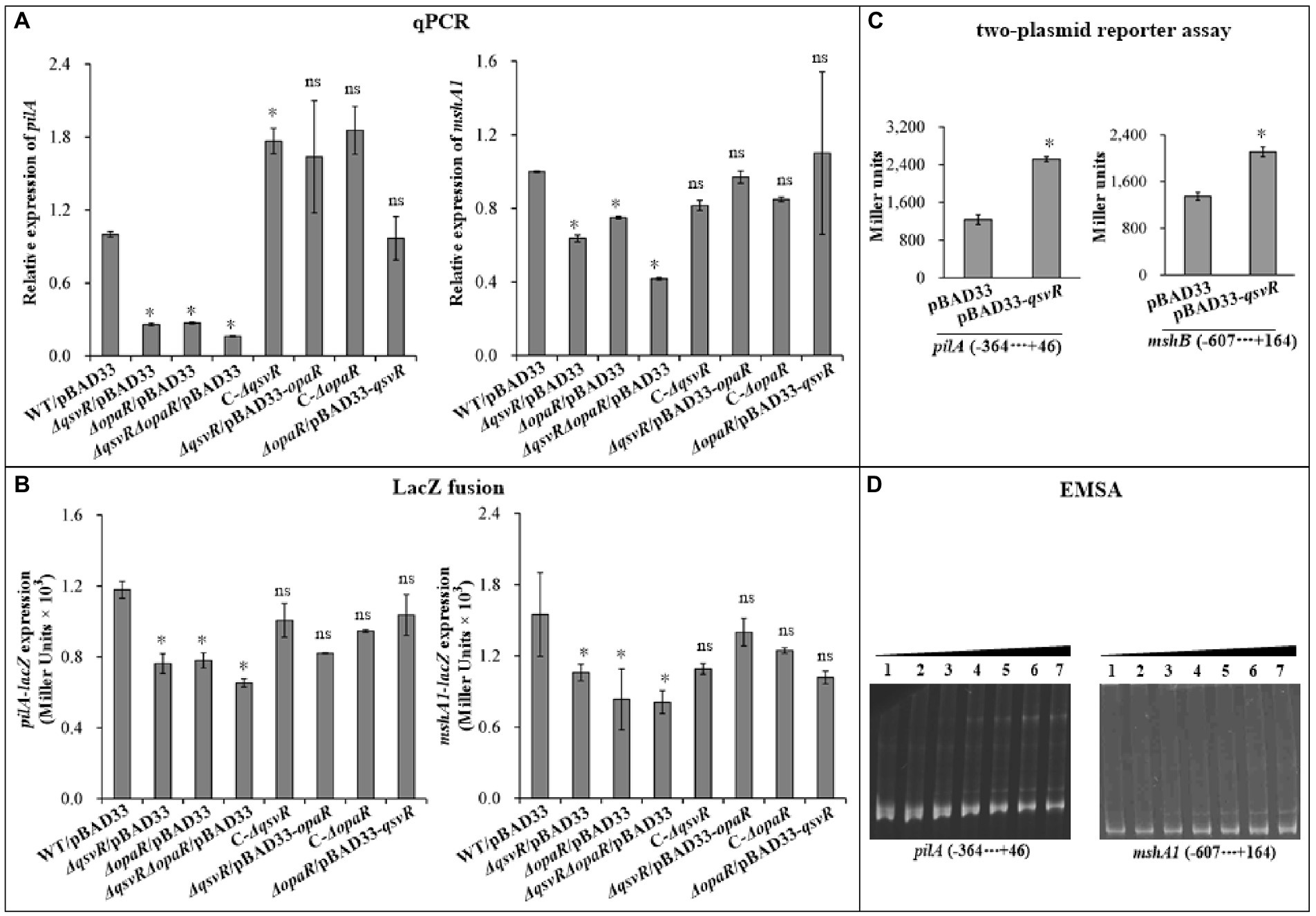
Figure 3. Regulation of mshA1 and pilA by QsvR and OpaR. qPCR (A), LacZ fusion (B), Two-plasmid reporter assay (C), and EMSA (D) were carried out as described in Figure 2. Lanes 1, 2, 3, 4, 5, 6, and 7 in the EMSA (D) data contain 0, 0.021, 0.042, 0.063, 0.083, 0.11 and 0.13 pmol of His-QsvR, respectively. Negative and positive numbers indicate the nucleotide positions upstream and downstream of each target gene, respectively. The asterisks indicate statistical significances relative to WT/pBAD33 or E. coli 100 λpir/pBAD33 (p < 0.01). The ‘ns’ means no significant differences (p > 0.01).
QsvR and OpaR function coordinately to repress c-di-GMP metabolism
Elevated intracellular c-di-GMP levels enhance biofilm formation, with deletion of opaR increasing the concentration of c-di-GMP in V. parahaemolyticus (Zhang et al., 2021a). Herein, intracellular c-di-GMP levels in ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33 were significantly enhanced relative to WT/pBAD33, with restoration in C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR (Figure 4). Thus, QsvR and OpaR function coordinately to inhibit c-di-GMP synthesis in V. parahaemolyticus.
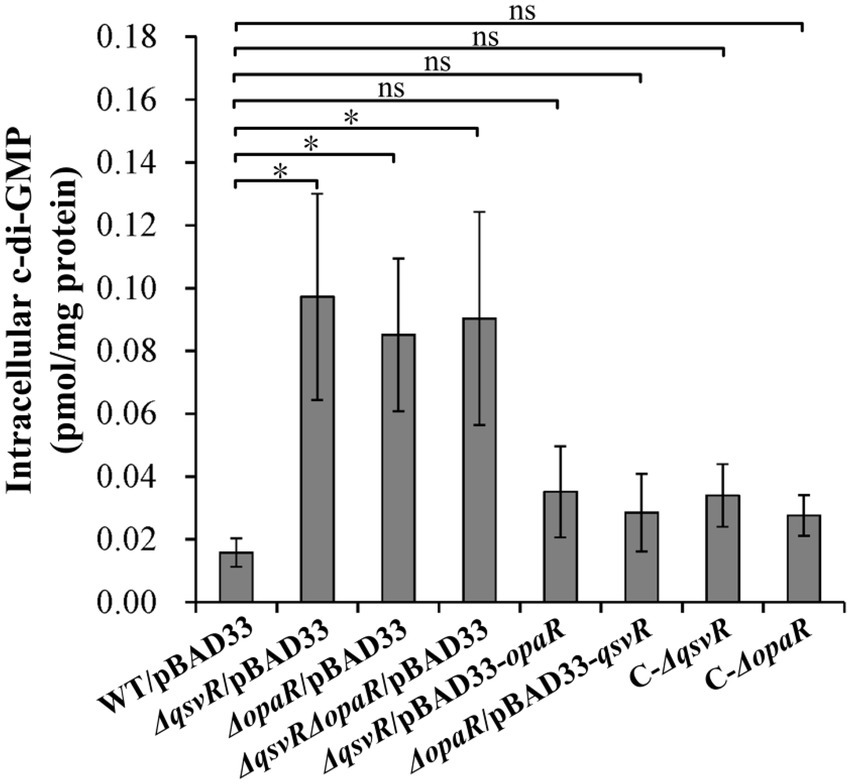
Figure 4. Intracellular c-di-GMP levels in different V. parahaemolyticus strains. Bacterial cells were harvested after 6 h of incubation at 37°C on HI plates. c-di-GMP levels were determined using a c-di-GMP enzyme-linked immunosorbent assay (ELISA) kit. The data are expressed as the means ± SD of at least three independent experiments. The asterisks indicate statistical significances relative to WT/pBAD33 (p < 0.01). The ‘ns’ means no significant differences (p > 0.01).
Two well-studied c-di-GMP metabolism-associated genes, scrA (the first gene of the scrABC operon) and scrG, were selected as target genes to investigate QsvR-and OpaR-mediated gene regulation. qPCR results showed that the mRNA levels of scrA and scrG were significantly increased and decreased, respectively, in ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33 relative to that in WT/pBAD33, with restoration in C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR (Figure 5A). Similarly, LacZ fusion assays showed that the promoter activity of scrA or scrG was significantly enhanced and reduced, respectively, in ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33 relative to that in WT/pBAD33, with restoration in C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR (Figure 5B). Furthermore, OpaR repressed and activated the promoter activity of scrA and scrG in a heterologous host, respectively (Zhang et al., 2021a). Similar results were observed for QsvR regulation of scrG, with no apparent regulatory effect on scrA expression in E. coli (Figure 5C). EMSA demonstrated that His-QsvR dose-dependently bound to the upstream DNA fragment of scrG but not scrA (Figure 5D). Taken together, QsvR worked with OpaR to repress c-di-GMP synthesis in V. parahaemolyticus, likely due to direct control of the transcription of c-di-GMP metabolism-related genes.
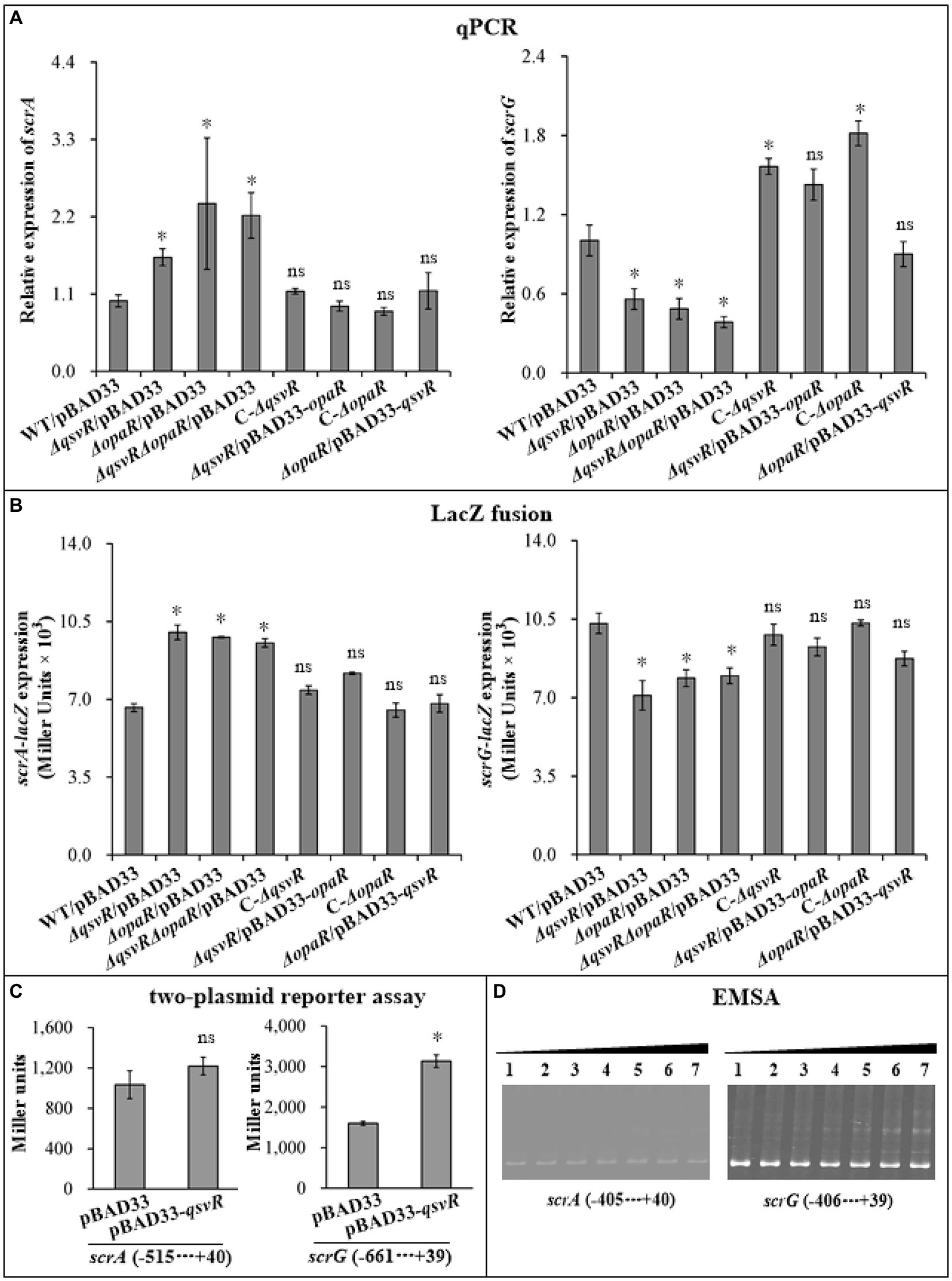
Figure 5. Regulation of scrG and scrA by QsvR and OpaR. qPCR (A), LacZ fusion (B), Two-plasmid reporter assay (C), and EMSA (D) were carried out as described in Figure 2. Lanes 1, 2, 3, 4, 5, 6, and 7 in the EMSA (D) data contain 0, 0.021, 0.042, 0.063, 0.083, 0.11, and 0.13 pmol of His-QsvR, respectively. Negative and positive numbers indicate the nucleotide positions upstream and downstream of each target gene, respectively. The asterisks indicate statistical significances relative to WT/pBAD33 or E. coli 100 λpir/pBAD33 (p < 0.01). The ‘ns’ means no significant differences (p > 0.01).
QsvR and OpaR work coordinately to regulate the OP-TR transition of Vibrio parahaemolyticus
Previously, OpaR was shown to regulate the OP-TR transition (McCarter, 1998), but lacks the detailed mechanisms. Herein, the data showed that the TR cell type was exhibited by ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33, whereas WT/pBAD33 exhibited the OP cell type (Figure 6). C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR exhibited the restored OP cell type (Figure 6). Therefore, QsvR and OpaR functioned coordinately to regulate the OP–TR transition of V. parahaemolyticus.
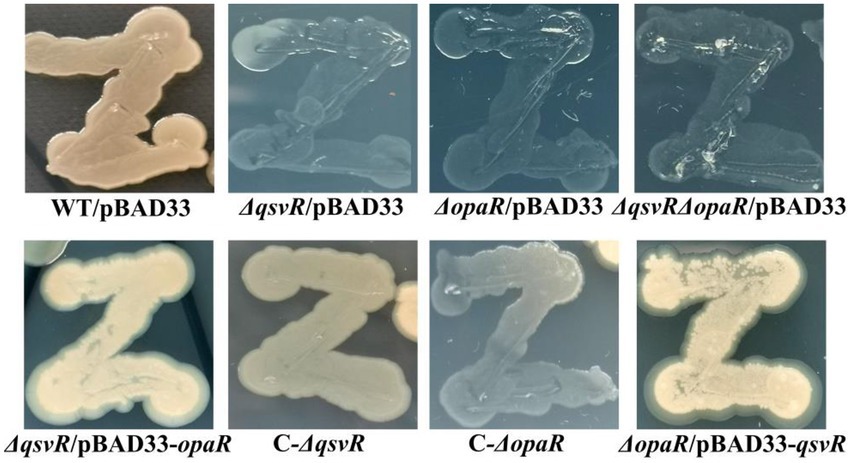
Figure 6. The opaque (OP)-translucent (TR) transition of V. parahaemolyticus strains. V. parahaemolyticus strains were grown in M broth at 30°C with shaking at 200 rpm overnight, and a small amount of each cell culture was taken with an inoculation loop, streaked directly on a HI plate, and then statically incubated at 37°C for 48 h.
OP and TR cell types directly relate to CPS production (Chen et al., 2010). The vp0215-0237 gene cluster is responsible for CPS synthesis in V. parahaemolyticus (Chen et al., 2010). The vp0215-0237 locus contains at least two operons, vp0218-0215 and vp0219-0237 (Chen et al., 2010). vp0218 and vp0219 are transcribed in opposite directions and share an intergenic region 601 bp in length (Makino et al., 2003; Chen et al., 2010). In this study, vp0218 and vp0219 were selected as target genes to assess QsvR-and OpaR-mediated gene regulation. qPCR showed that mRNA levels of vp0218 and vp0219 were significantly decreased in ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33 relative to WT/pBAD33, with restoration in C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR (Figure 7A). LacZ fusion assay showed that the promoter activities of vp0218 and vp0219 were significantly reduced in ΔqsvR/pBAD33, ΔopaR/pBAD33, and ΔqsvRΔopaR/pBAD33 relative to WT/pBAD33, with restoration in C-ΔqsvR, ΔqsvR/pBAD33-opaR, C-ΔopaR, and ΔopaR/pBAD33-qsvR (Figure 7B). In addition, both QsvR and OpaR were able to induce the expression of vp0218 and vp0219 in a heterologous host (Figure 7C). By EMSA (Figure 7D), both His-QsvR and His-OpaR were able to dose-dependently bind to the regulatory DNA fragments of vp0218 and vp0219. Therefore, both QsvR and OpaR were able to directly activate the transcription of vp0218 and vp0219.
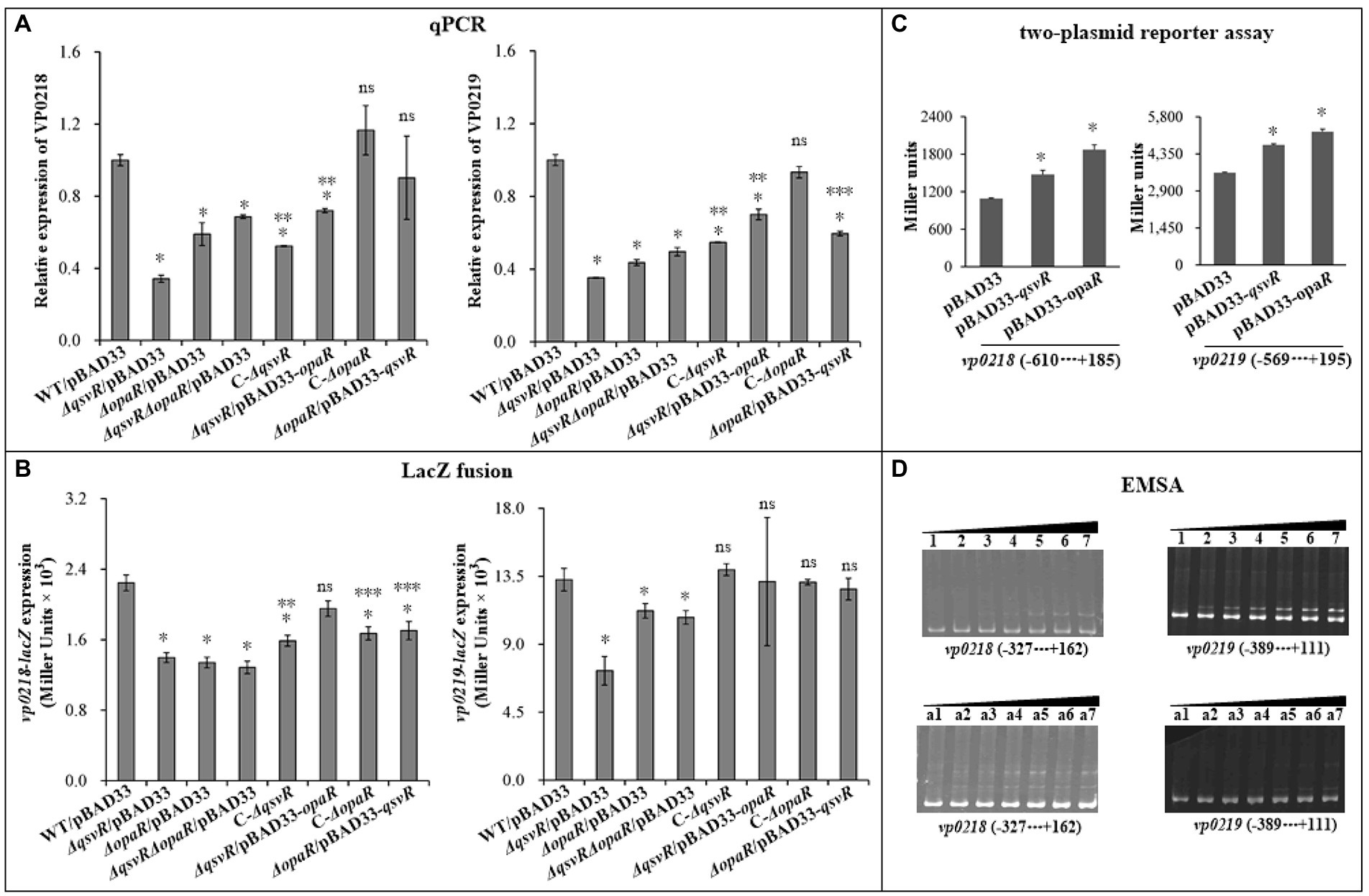
Figure 7. Regulation of vp0218 and vp0218 by QsvR and OpaR. qPCR (A), LacZ fusion (B), Two-plasmid reporter assay (C), and EMSA (D) were carried out as described in Figure 2. Lanes 1, 2, 3, 4, 5, 6, and 7 in the EMSA (D) data contain 0, 0.4, 0.8, 1.2, 1.4, 2.0, and 2.4 pmol of His-OpaR, respectively. Lanes a1, a2, a3, a4, a5, a6, and a7 contain 0, 0.021, 0.042, 0.063, 0.083, 0.11, and 0.13 pmol of His-QsvR, respectively. Negative and positive numbers indicate the nucleotide positions upstream and downstream of each target gene, respectively. The single asterisk symbol (*) indicates statistical significances relative to WT/pBAD33 or E. coli 100 λpir/pBAD33 (p < 0.01). The double asterisk symbol (**) indicates statistical significances relative to ΔqsvR/pBAD33 (p < 0.01). The three asterisk symbol (***) indicates statistical significances relative to ΔopaR/pBAD33 (p < 0.01). The ‘ns’ means no significant differences relative to WT/pBAD33 (p > 0.01).
Taken together, these results demonstrated QsvR and QsvR to function coordinately to regulate the OP–TR transition of V. parahaemolyticus by directly activating the transcription of CPS synthesis genes.
No interplay of QsvR and OpaR at target promoters
As confirmed in this work and as previously reported, both QsvR and OpaR directly regulate the transcription of pilA, mshA1, scrG, vp0218, and vp0219 (Lu et al., 2021b; Zhang et al., 2021a; Sun et al., 2022). To determine whether QsvR promoter binding affects OpaR and vice versa, we performed competitive EMSA using the promoter-proximal DNA fragments of pilA, mshA1, scrG, vp0218, and vp0219 with varying amounts of His-QsvR and His-OpaR (Figure 8). The retarded bands of DNA-His-QsvR and DNA-His-OpaR complexes overlapped each other, although the retarded bands of individual proteins (lane 2 or 8) were much weaker than those of the mixed proteins (lanes 3–7). The most apparent retarded bands were found in the lane with the greatest amount of both proteins (lane 5). A second dose-dependent retarded band of DNA-His-QsvR was also observed for some target genes, e.g., vp0218. These results suggested no competitive binding by QsvR and OpaR for these regulatory DNA fragments.
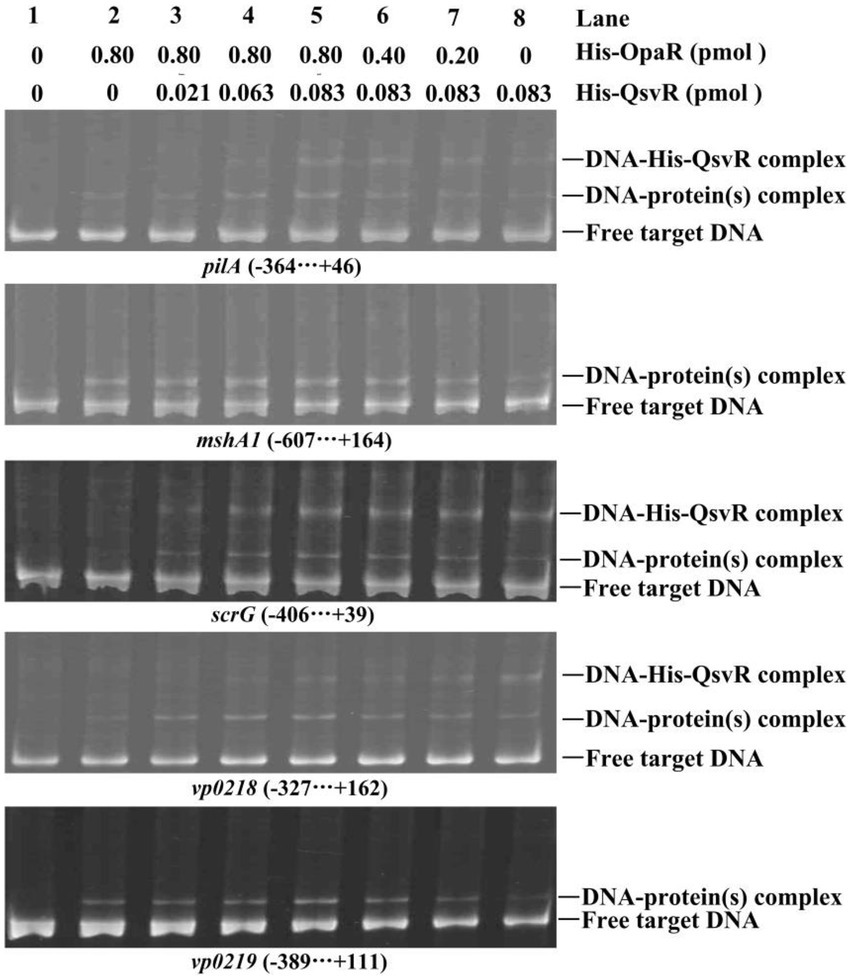
Figure 8. Competitive DNA-binding assays. The experiments were carried out as described in Figure 2D.
Discussion
OpaR controls approximately 15% of V. parahaemolyticus genes, including biofilm-related genes (Gode-Potratz and McCarter, 2011; Kernell Burke et al., 2015). OpaR regulates CPS-associated OP-TR transition of V. parahaemolyticus, and expression of OpaR in TR strains converts those strains to an OP phenotype (McCarter, 1998). Deletion of qsvR in either OP (WT) or TR (opaR mutant) strains alters their ability to form biofilms even though the mechanisms may differ (Enos-Berlage et al., 2005). Herein, we showed that deletion of qsvR in both WT and ΔopaR backgrounds produced similar colony phenotypes to ΔopaR, all of which were more wrinkled than that of WT (Figure 1A). Both QsvR and OpaR acted as negative regulators of biofilm formation by V. parahaemolyticus RIMD2210633, but their regulatory activities were not additive, with OpaR a stronger inhibitor than QsvR. For V. parahaemolyticus BB22OP ΔqsvR and ΔqsvRΔopaR have a similar capacity to form biofilms, with colonies that are not rough and resembled their parental colony type (Enos-Berlage et al., 2005). We have observed similar contradictory results for these two strains, which was attributed to large differences in the two genomes (Jensen et al., 2013; Zhang et al., 2021a). Most interestingly, expression of opaR in ΔqsvR restored the phenotype of ΔqsvR and vice versa, suggesting that OpaR and QsvR may substitute for each other in biofilm formation.
The cps and scv loci are responsible for EPS production, which is directly associated with wrinkled colonies and enhanced biofilm formation by V. parahaemolyticus (Chen et al., 2010; Liu et al., 2022). Herein, OpaR directly repressed the transcription of cpsA and scvE, whereas QsvR had no effect on their expression (Figure 2). A previous study showed that QsvR did not control cpsA transcription in either OP or TR backgrounds (Enos-Berlage et al., 2005). These results may explain why OpaR had a greater capacity for the inhibition of biofilm formation by V. parahaemolyticus RIMD2210633 than did QsvR. Previously, CpsS was shown to repress cpsR transcription, followed by CpsR activation of cpsQ transcription, with CpsQ repression of cpsS transcription but activation of cpsA (Guvener and McCarter, 2003; Ferreira et al., 2012). CpsR alone increases cps gene transcription in a scrAΔopaR background (Guvener and McCarter, 2003). Both QsvR and OpaR activate cpsQ transcription (Zhou et al., 2013; Zhang et al., 2021b). In addition, both H-NS and ToxR activate EPS production (Chen et al., 2018; Zhang et al., 2018). AphA is also required for the expression of the scv locus (Chen et al., 2022; Liu et al., 2022). Furthermore, overexpression of scrABC or scrG inhibits cpsA transcription (Boles and McCarter, 2002; Kim and McCarter, 2007). Therefore, expression of EPS-related genes is tightly controlled by regulatory networks composed of various transcriptional regulators and signaling pathways. The present data provided for a better understanding of the regulatory networks of EPS-associated genes.
Type IV pili play critical roles in biofilm formation and bacterial colonization (Yildiz and Visick, 2009; O'Boyle et al., 2013). Expression of MSHA and ChiRP is induced by OpaR in V. parahaemolyticus RIMD2210633 (Lu et al., 2021b; Sun et al., 2022). Herein, we demonstrated QsvR to activate transcription of mshA1 and pilA in the WT background but not in a the ΔopaR background (Figure 3). However, expression of qsvR in ΔopaR restored expression of mshA1 and pilA, and vice versa (Figure 3). Both QsvR and OpaR bound to the upstream DNA fragments of mshA1 and pilA (Figure 3; Lu et al., 2021b; Sun et al., 2022). Binding sequences of QsvR are typically long and AT-rich (Zhang et al., 2019, 2021b; Qiu et al., 2020). However, there was no competitive binding between QsvR and OpaR for the regulatory DNA regions of mshA1 and pilA (Figure 8). Roles for type IV pili in V. parahaemolyticus RIMD2210633 have been well studied (Enos-Berlage et al., 2005; Shime-Hattori et al., 2006; Yildiz and Visick, 2009; O'Boyle et al., 2013), but a detailed understanding of their expression regulation requires more investigation in the future.
The c-di-GMP content in bacteria influences biofilm formation and motility (Yildiz and Visick, 2009). Deletion of qsvR in the WT background enhanced intracellular c-di-GMP levels, whereas qsvR deletion in the ΔopaR background did not influence intracellular c-di-GMP levels (Figure 4). OpaR negatively regulates c-di-GMP production in V. parahaemolyticus RIMD2210633 (Zhang et al., 2021a). Expression of qsvR in ΔopaR led to restored intracellular c-di-GMP levels, and vice versa (Figure 4). V. parahaemolyticus RIMD2210633 harbors dozens of genes encoding proteins that may be required for c-di-GMP metabolism, but only a few (including scrABC and scrG) are studied (Boles and McCarter, 2002; Makino et al., 2003; Kim and McCarter, 2007; Ferreira et al., 2008, 2012). OpaR directly regulate several putative c-di-GMP metabolism-associated genes including scrABC and scrG (Zhang et al., 2021a). Herein, we showed that QsvR directly activated scrG transcription but indirectly repressed scrA transcription (Figure 5). Expression of qsvR in ΔopaR restored expression levels of scrG and scrA, and vice versa (Figure 5). It is noteworthy that QsvR inhibits biofilm formation and c-di-GMP production as well as directly regulates the transcription of scrABC and scrG but exerts no regulatory effect on the expression of EPS genes. Biofilm formation by V. parahaemolyticus is uncorrelated with the EPS content in the biofilm matrix (Li et al., 2020), but it is still hard to clearly explain these contradictions. Perhaps there exist additional regulators that complement the effect on EPS production in the qsvR mutant. Moreover, QsvR integrates into the QS cascade to regulate gene expression via direct regulation of the master QS regulators, AphA and OpaR (Zhang et al., 2019). QS and c-di-GMP signals are two crucial regulatory cascades. Thus, the direct association between QsvR and OpaR regulation and c-di-GMP metabolism may be beneficial for V. parahaemolyticus to precisely control bacterial behaviors.
OpaR-dependent CPS production has been reported (McCarter, 1998), but lacks detailed mechanisms. Herein, we demonstrated that the qsvR mutants in WT or ΔopaR backgrounds exhibited TR cell type morphologies, whereas obviously no difference were observed for ΔopaR and ΔqsvRΔopaR (Figure 6). Expression of qsvR in ΔopaR restored an OP cell type, and vice versa (Figure 6). QsvR worked with OpaR to coordinately promote CPS production via directly activation of the transcription of CPS-associated genes in V. parahaemolyticus. A role for CPS in biofilm formation has not been described for V. parahaemolyticus, and future studies should address this issue.
In conclusion, this study demonstrated QsvR and OpaR to work coordinately to repress biofilm-associated phenotypes and c-di-GMP metabolism, and as well to promote V. parahaemolyticus OP colony formation. QsvR restored biofilm-associated phenotypic changes caused by the opaR mutation, and vice versa. Furthermore, QsvR and OpaR were shown to work coordinately to activate the transcription of type IV pili genes, CPS genes, and scrG, as well as to repress scrA transcription. In addition, OpaR but not QsvR negatively regulated the transcription of cps and scv genes. Thus, our data highlight how QsvR works with the QS system to regulate biofilm formation by precisely controlling the transcription of multiple biofilm formation-associated genes in V. parahaemolyticus.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
MZ, XX, XL, QW, TZ, WY, and LH performed the laboratory experiments and analyzed the results. YZ, RL, and DZ designed, organized and supervised the experiments. MZ and YZ drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 82072239) and the Natural Science Research Project of Nantong Science and Technology Bureau (grant no. JC2021027).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1079653/full#supplementary-material
References
Ashrafudoulla, M., Mizan, M. F. R., Park, S. H., and Ha, S. D. (2021). Current and future perspectives for controlling vibrio biofilms in the seafood industry: a comprehensive review. Crit. Rev. Food Sci. Nutr. 61, 1827–1851. doi: 10.1080/10408398.2020.1767031
Ball, A. S., Chaparian, R. R., and van Kessel, J. C. (2017). Quorum sensing gene regulation by LuxR/HapR master regulators in Vibrios. J. Bacteriol. 199:e00105-17. doi: 10.1128/JB.00105-17
Boles, B. R., and McCarter, L. L. (2002). Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184, 5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Chaparian, R. R., Ball, A. S., and van Kessel, J. C. (2020). Hierarchical transcriptional control of the LuxR quorum-sensing Regulon of Vibrio harveyi. J. Bacteriol. 202:e00047-20. doi: 10.1128/JB.00047-20
Chen, Y., Dai, J., Morris, J. G. Jr., and Johnson, J. A. (2010). Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol. 10:274. doi: 10.1186/1471-2180-10-274
Chen, L., Qiu, Y., Tang, H., Hu, L. F., Yang, W. H., Zhu, X. J., et al. (2018). ToxR is required for biofilm formation and motility of Vibrio parahaemolyticus. Biomed. Environ. Sci. 31, 848–850. doi: 10.3967/bes2018.112
Chen, L., Zhang, M., Li, X., Wu, Q., Xue, X., Zhang, T., et al. (2022). AphA directly activates the transcription of polysaccharide biosynthesis gene scvE in Vibrio parahaemolyticus. Gene 851:146980. doi: 10.1016/j.gene.2022.146980
Enos-Berlage, J. L., Guvener, Z. T., Keenan, C. E., and McCarter, L. L. (2005). Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55, 1160–1182. doi: 10.1111/j.1365-2958.2004.04453.x
Ferreira, R. B., Antunes, L. C., Greenberg, E. P., and McCarter, L. L. (2008). Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190, 851–860. doi: 10.1128/JB.01462-07
Ferreira, R. B., Chodur, D. M., Antunes, L. C., Trimble, M. J., and McCarter, L. L. (2012). Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J. Bacteriol. 194, 914–924. doi: 10.1128/JB.05807-11
Gao, H., Ma, L., Qin, Q., Qiu, Y., Zhang, J., Li, J., et al. (2020). Fur represses Vibrio cholerae biofilm formation via direct regulation of vieSAB, cdgD, vpsU, and vpsA-K transcription. Front. Microbiol. 11:587159. doi: 10.3389/fmicb.2020.587159
Gao, H., Zhang, Y., Yang, L., Liu, X., Guo, Z., Tan, Y., et al. (2011). Regulatory effects of cAMP receptor protein (CRP) on porin genes and its own gene in Yersinia pestis. BMC Microbiol. 11:40. doi: 10.1186/1471-2180-11-40
Gode-Potratz, C. J., and McCarter, L. L. (2011). Quorum sensing and silencing in Vibrio parahaemolyticus. J. Bacteriol. 193, 4224–4237. doi: 10.1128/JB.00432-11
Guvener, Z. T., and McCarter, L. L. (2003). Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 185, 5431–5441. doi: 10.1128/JB.185.18.5431-5441.2003
Homma, M., and Kojima, S. (2022). Roles of the second messenger c-di-GMP in bacteria: focusing on the topics of flagellar regulation and Vibrio spp. Genes Cells 27, 157–172. doi: 10.1111/gtc.12921
Jensen, R. V., Depasquale, S. M., Harbolick, E. A., Hong, T., Kernell, A. L., Kruchko, D. H., et al. (2013). Complete genome sequence of prepandemic Vibrio parahaemolyticus BB22OP. Genome Announc. 1:e00002-12. doi: 10.1128/genomeA.00002-12
Joseph, L. A., and Wright, A. C. (2004). Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J. Bacteriol. 186, 889–893. doi: 10.1128/JB.186.3.889-893.2004
Kernell Burke, A., Guthrie, L. T., Modise, T., Cormier, G., Jensen, R. V., McCarter, L. L., et al. (2015). OpaR controls a network of downstream transcription factors in Vibrio parahaemolyticus BB22OP. PLoS One 10:e0121863. doi: 10.1371/journal.pone.0121863
Kim, Y. K., and McCarter, L. L. (2007). ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 189, 4094–4107. doi: 10.1128/JB.01510-06
Kimbrough, J. H., Cribbs, J. T., and McCarter, L. L. (2020). Homologous c-di-GMP-binding Scr transcription factors orchestrate biofilm development in Vibrio parahaemolyticus. J. Bacteriol. 202:e00723-19. doi: 10.1128/JB.00723-19
Kimbrough, J. H., and McCarter, L. L. (2020). Identification of three new GGDEF and EAL domain-containing proteins participating in the Scr surface colonization regulatory network in Vibrio parahaemolyticus. J. Bacteriol. 203:e00409-20. doi: 10.1128/JB.00409-20
Lee, D. H., Jeong, H. S., Jeong, H. G., Kim, K. M., Kim, H., and Choi, S. H. (2008). A consensus sequence for binding of SmcR, a Vibrio vulnificus LuxR homologue, and genome-wide identification of the SmcR regulon. J. Biol. Chem. 283, 23610–23618. doi: 10.1074/jbc.M801480200
Li, W., Wang, J. J., Qian, H., Tan, L., Zhang, Z., Liu, H., et al. (2020). Insights into the role of extracellular DNA and extracellular proteins in biofilm formation of Vibrio parahaemolyticus. Front. Microbiol. 11:813. doi: 10.3389/fmicb.2020.00813
Liu, M., Nie, H., Luo, X., Yang, S., Chen, H., and Cai, P. (2022). A polysaccharide biosynthesis locus in Vibrio parahaemolyticus important for biofilm formation has homologs widely distributed in aquatic bacteria mainly from Gammaproteobacteria. mSystems 7:e0122621. doi: 10.1128/msystems.01226-21
Lu, R., Osei-Adjei, G., Huang, X., and Zhang, Y. (2018). Role and regulation of the orphan AphA protein of quorum sensing in pathogenic Vibrios. Future Microbiol. 13, 383–391. doi: 10.2217/fmb-2017-0165
Lu, R., Sun, J., Qiu, Y., Zhang, M., Xue, X., Li, X., et al. (2021a). The quorum sensing regulator OpaR is a repressor of polar flagellum genes in Vibrio parahaemolyticus. J. Microbiol. 59, 651–657. doi: 10.1007/s12275-021-0629-3
Lu, R., Sun, J., Xue, X., Zhang, M., Li, X., Wu, Q., et al. (2021b). Transcriptional regulation of pilABCD by OpaR in Vibrio parahaemolyticus. Chinese J. Microbiol. Immu. 41:6. doi: 10.3760/cma.j.cn112309-20210729-00248
Lu, R., Tang, H., Qiu, Y., Yang, W., Yang, H., Zhou, D., et al. (2019). Quorum sensing regulates the transcription of lateral flagellar genes in Vibrio parahaemolyticus. Future Microbiol. 14, 1043–1053. doi: 10.2217/fmb-2019-0048
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Martinez-Mendez, R., Camacho-Hernandez, D. A., Sulvaran-Guel, E., and Zamorano-Sanchez, D. (2021). A trigger Phosphodiesterase modulates the global c-di-GMP Pool, motility, and biofilm formation in Vibrio parahaemolyticus. J. Bacteriol. 203:e0004621. doi: 10.1128/JB.00046-21
McCarter, L. L. (1998). OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180, 3166–3173. doi: 10.1128/JB.180.12.3166-3173.1998
O'Boyle, N., Houeix, B., Kilcoyne, M., Joshi, L., and Boyd, A. (2013). The MSHA pilus of Vibrio parahaemolyticus has lectin functionality and enables TTSS-mediated pathogenicity. Int. J. Med. Microbiol. 303, 563–573. doi: 10.1016/j.ijmm.2013.07.010
Parales, R. E., and Harwood, C. S. (1993). Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram-bacteria. Gene 133, 23–30. doi: 10.1016/0378-1119(93)90220-W
Qiu, Y., Hu, L., Yang, W., Yin, Z., Zhou, D., Yang, H., et al. (2020). The type VI secretion system 2 of Vibrio parahaemolyticus is regulated by QsvR. Microb. Pathog. 149:104579. doi: 10.1016/j.micpath.2020.104579
Ruhal, R., and Kataria, R. (2021). Biofilm patterns in gram-positive and gram-negative bacteria. Microbiol. Res. 251:126829. doi: 10.1016/j.micres.2021.126829
Rutherford, S. T., van Kessel, J. C., Shao, Y., and Bassler, B. L. (2011). AphA and LuxR/HapR reciprocally control quorum sensing in Vibrios. Genes Dev. 25, 397–408. doi: 10.1101/gad.2015011
Shime-Hattori, A., Iida, T., Arita, M., Park, K. S., Kodama, T., and Honda, T. (2006). Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 264, 89–97. doi: 10.1111/j.1574-6968.2006.00438.x
Sun, J., Li, X., Qiu, Y., Xue, X., Zhang, M., Yang, W., et al. (2022). Quorum sensing regulates transcription of the pilin gene mshA1 of MSHA pilus in Vibrio parahaemolyticus. Gene 807:145961. doi: 10.1016/j.gene.2021.145961
Sun, F., Zhang, Y., Wang, L., Yan, X., Tan, Y., Guo, Z., et al. (2012). Molecular characterization of direct target genes and cis-acting consensus recognized by quorum-sensing regulator AphA in Vibrio parahaemolyticus. PLoS One 7:e44210. doi: 10.1371/journal.pone.0044210
Trimble, M. J., and McCarter, L. L. (2011). Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. U. S. A. 108, 18079–18084. doi: 10.1073/pnas.1113790108
van Kessel, J. C., Rutherford, S. T., Shao, Y., Utria, A. F., and Bassler, B. L. (2013). Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J. Bacteriol. 195, 436–443. doi: 10.1128/JB.01998-12
Wang, L., Ling, Y., Jiang, H., Qiu, Y., Qiu, J., Chen, H., et al. (2013a). AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. Int. J. Food Microbiol. 160, 245–251. doi: 10.1016/j.ijfoodmicro.2012.11.004
Wang, L., Zhou, D., Mao, P., Zhang, Y., Hou, J., Hu, Y., et al. (2013b). Cell density-and quorum sensing-dependent expression of type VI secretion system 2 in Vibrio parahaemolyticus. PLoS One. 8:e73363. doi: 10.1371/journal.pone.0073363
Yildiz, F. H., and Visick, K. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118. doi: 10.1016/j.tim.2008.12.004
Zhang, Y., Gao, H., Osei-Adjei, G., Yang, W., Yang, H., Yin, Z., et al. (2017a). Transcriptional regulation of the type VI secretion system 1 genes by quorum sensing and ToxR in Vibrio parahaemolyticus. Front. Microbiol. 8:2005. doi: 10.3389/fmicb.2017.02005
Zhang, Y., Hu, L., Qiu, Y., Osei-Adjei, G., Tang, H., Zhang, Y., et al. (2019). QsvR integrates into quorum sensing circuit to control Vibrio parahaemolyticus virulence. Environ. Microbiol. 21, 1054–1067. doi: 10.1111/1462-2920.14524
Zhang, L., Osei-Adjei, G., Zhang, Y., Gao, H., Yang, W., Zhou, D., et al. (2017b). CalR is required for the expression of T6SS2 and the adhesion of Vibrio parahaemolyticus to HeLa cells. Arch. Microbiol. 199, 931–938. doi: 10.1007/s00203-017-1361-6
Zhang, Y., Qiu, Y., Gao, H., Sun, J., Li, X., Zhang, M., et al. (2021a). OpaR controls the metabolism of c-di-GMP in Vibrio parahaemolyticus. Front. Microbiol. 12:676436. doi: 10.3389/fmicb.2021.676436
Zhang, Y., Qiu, Y., Tan, Y., Guo, Z., Yang, R., and Zhou, D. (2012). Transcriptional regulation of opaR, qrr2-4 and aphA by the master quorum-sensing regulator OpaR in Vibrio parahaemolyticus. PLoS One 7:e34622. doi: 10.1371/journal.pone.0034622
Zhang, Y., Qiu, Y., Xue, X., Zhang, M., Sun, J., Li, X., et al. (2021b). Transcriptional regulation of the virulence genes and the biofilm formation associated operons in Vibrio parahaemolyticus. Gut Pathog. 13:15. doi: 10.1186/s13099-021-00410-y
Zhang, L., Weng, Y., Wu, Y., Wang, X., Yin, Z., Yang, H., et al. (2018). H-NS is an activator of exopolysaccharide biosynthesis genes transcription in Vibrio parahaemolyticus. Microb. Pathog. 116, 164–167. doi: 10.1016/j.micpath.2018.01.025
Zhong, X., Lu, Z., Wang, F., Yao, N., Shi, M., and Yang, M. (2022). Characterization of GefA, a GGEEF domain-containing protein that modulates Vibrio parahaemolyticus motility, biofilm formation, and virulence. Appl. Environ. Microbiol. 88:e0223921. doi: 10.1128/aem.02239-21
Keywords: Vibrio parahaemolyticus, biofilm, QsvR, OpaR, regulation
Citation: Zhang M, Xue X, Li X, Wu Q, Zhang T, Yang W, Hu L, Zhou D, Lu R and Zhang Y (2023) QsvR and OpaR coordinately repress biofilm formation by Vibrio parahaemolyticus. Front. Microbiol. 14:1079653. doi: 10.3389/fmicb.2023.1079653
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Bindu Subhadra, Long Island University, United StatesPantu Kumar Roy, Gyeongsang National University, Republic of Korea
Steve Flint, Massey University, New Zealand
Stefan Schild, University of Graz, Austria
Joao Palma Pombo, University of Graz, Austria,in collaboration with reviewer SS
Copyright © 2023 Zhang, Xue, Li, Wu, Zhang, Yang, Hu, Zhou, Lu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiquan Zhang, ✉ emhhbmd5aXF1YW5xQDE2My5jb20=; Renfei Lu, ✉ cmFpbm1hbjc4QDE2My5jb20=; Dongsheng Zhou, ✉ ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
†These authors have contributed equally to this work
 Miaomiao Zhang1,2†
Miaomiao Zhang1,2† Xingfan Xue
Xingfan Xue Wenhui Yang
Wenhui Yang Dongsheng Zhou
Dongsheng Zhou Renfei Lu
Renfei Lu Yiquan Zhang
Yiquan Zhang