- 1College of Resources, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Investigation and Monitoring, Protection and Utilization for Cultivated Land Resources, Ministry of Natural Resources, Chengdu, China
Introduction: The diversity, nitrogen-fixing capacity and heavy metal tolerance of culturable rhizobia in symbiotic relationship with Pongamia pinnata surviving in vanadium (V) - titanium (Ti) magnetite (VTM) tailings is still unknown, and the rhizobia isolates from the extreme barren VTM tailings contaminated with a variety of metals would provide available rhizobia resources for bioremediation.
Methods: P. pinnata plants were cultivated in pots containing the VTM tailings until root nodules formed, and then culturable rhizobia were isolated from root nodules. The diversity, nitrogen-fixing capacity and heavy metal tolerance of rhizobia were performed.
Results: Among 57 rhizobia isolated from these nodules, only twenty strains showed different levels of tolerance to copper (Cu), nickel (Ni), manganese (Mn) and zinc (Zn), especially strains PP1 and PP76 showing high tolerance against these four heavy metals. Based on the phylogenetic analysis of 16S rRNA and four house-keeping genes (atpD, recA, rpoB, glnII), twelve isolates were identified as Bradyrhizobium pachyrhizi, four as Ochrobactrum anthropic, three as Rhizobium selenitireducens and one as Rhizobium pisi. Some rhizobia isolates showed a high nitrogen-fixing capacity and promoted P. pinnata growth by increasing nitrogen content by 10%-145% in aboveground plant part and 13%-79% in the root. R. pachyrhizi PP1 showed the strongest capacity of nitrogen fixation, plant growth promotion and resistance to heavy metals, which provided effective rhizobia strains for bioremediation of VTM tailings or other contaminated soils. This study demonstrated that there are at least three genera of culturable rhizobia in symbiosis with P. pinnata in VTM tailings.
Discussion: Abundant culturable rhizobia with the capacity of nitrogen fixation, plant growth promotion and resistance to heavy metals survived in VTM tailings, indicating more valuable functional microbes could be isolated from extreme soil environments such as VTM tailings.
Introduction
Tailings are the part of the product of separation operation in mineral processing that has low content of useful target components and cannot be used for production. Vanadium (V) – titanium (Ti) magnetite (VTM) is a widely distributed mineral ore containing oxides of V, Ti, and Fe, which become large amounts of heavy metal-containing slag after being subjected to the iron and steel smelting process. Because of its physicochemical characteristics, soil often becomes the most direct acceptor of pollutants from the processing of minerals such as titanium and magnetite (Wang et al., 2018; Demková et al., 2019). The soil around a mining or smelting area will continuously accumulate the byproducts of the mining processes, resulting in serious heavy metal pollution, e.g., Cd was determined to be the main heavy metal pollutant in the Dahuangshan mining area (Zeng et al., 2022). Heavy metals are not easily degraded and can persist for years in the soil (He et al., 2019). Plants can absorb metal ions through their roots and invertebrates can ingest metal-containing particles so that they enter the food chain where they may be ingested by larger animals or even humans, harming the environment and endangering human health (Jin et al., 2014). Meanwhile, plant extract remediating metal in contaminated environmental has been considered as sustainable and environmentally friendly way (Upadhyay et al., 2023). Therefore, in order to achieve sustainability in the mining industry, one of the most urgent tasks is to concentrate on the reclamation of land contaminated with mine tailings and soil remediation in mining areas. There are some sustainable measures to deal with unavoidable heavy metal and fly-ash pollution, e. g. arsenic contamination in rice agro-ecosystems is migitated by using biochar, organic fertilizers, nanomaterials (Upadhyay and Edrisi, 2021; Upadhyay et al., 2023). Some emerging methods such as CRISPR and nanotechnological approaches along with PGPR also can manage degraded soil effectively (Upadhyay et al., 2022).
Pongamia pinnata is a deep-rooted Asian tree in the Fabaceae family, which has strong tolerance to salt, drought and heat, and can withstand submersion in fresh water (Marriboina and Attipalli, 2020). The root system of P. pinnata is extensive, and the root nodules are large and numerous with strong nitrogen fixation ability. Kumar et al. (2017) found that P. pinnata increased antioxidant and nutrient accumulation to protect plants under heavy metal stress. P. pinnata can grow well in the soil polluted with heavy metals and already shows good remediation potential (Yu et al., 2019). These characteristics make P. pinnata an excellent pioneer plant for removing heavy metal contaminants from soils (Marriboina and Attipalli, 2020).
As an important nutrition of plant growth, nitrogen is supplied through the biological nitrogen fixation by some endophytic diazotrophs of crops or soil microorganism (Singh et al., 2022). Legume-rhizobium symbiotic system shows strong nitrogen fixation capacity and strong resistance to heavy metal through the mutually beneficial relationship between rhizobia and the host plant (Zhang P. et al., 2019). Rhizobia can increase the heavy metal tolerance of a leguminous plant such as alfalfa by sequestering the metals or changing their forms in the soil (Teng et al., 2011). The fixation of nitrogen by rhizobia also improves the plant’s resistance to metal stress by increasing soil fertility (Fagorzi et al., 2018). This is a unique advantage of the joint symbiosis between leguminous plants and rhizobia to mitigate heavy metal pollution in soil.
Recent research on different types of legume-rhizobia symbiosis systems has mainly focused on: (1) isolation of heavy metal-tolerant rhizobia and screening for plant growth-promoting traits (Wani and Khan, 2013; Fan et al., 2018), (2) mechanisms of resistance of rhizobia to heavy metals (Adediran et al., 2015; Nocelli et al., 2016), (3) screening for legumes that are tolerant to heavy metals (Abdelkrim et al., 2018), and (4) evaluation of the ecological remediation effect of legume-rhizobia symbiosis systems on heavy metal pollution (Ju et al., 2015; Shen et al., 2019). Because the number of symbiotic remediation systems that have been studied and tested is very limited, the diversity of rhizobial populations offers great opportunities for discovering high quality strains that can be used for bioremediation of heavy metal-contaminated soils. However, resources for rhizobia-legume nitrogen fixation systems with high efficiency are still lacking, especially those from some extreme environments.
Previous studies have found that there was abundant growth-promoting bacteria, such as rhizobia, in the VTM tailings (Yu et al., 2014). Culturable Bradurhizobium genus aymbiotic with P. pinnata was also isolated from the VTM tailings, and then a aymbiotic bioremedition system of P. pinnata and rhizobia was established for ecological remediation of the VTM tailings (Yu et al., 2017a). High-throughput sequencing technology found some other genus of rhizobia in the VTM tailing (Yu et al., 2019). It was hypothesis that there are more genus of rhizobia symbiotic with P. pinnata, and these rhizobia could show high nitrogen-fixing capacity and strong heavy metal tolerance, which would provide more high quality rhizobia resources for bioremediation of the VTM tailings or other heavy metal-contaminated soil. So, this study more comprehensively understand diversity, nitrogen-fixing capacity and heavy metal tolerance of culturable P. pinnata rhizobia in the VTM tailings, providing a basis for the development and utilization of rhizobia.
Materials and methods
Soil collection and trapping of rhizobia
Soil samples were collected from a VTM tailings area located in Panzhihua, Sichuan Province, China (101°58′10.89″E, 26°36′59.47 N) for compositional analysis and a P. pinnata pot experiment. The seeds of P. pinnata were collected in a mangrove forest in Wenchang, Hainan Province, China (110°47′E, 19°37’N). The mature seeds of P. pinnata were taken to laboratory and planted in pots containing VTM tailings. The pots were kept in a greenhouse with a day temperature of 25°C for 16 h and a night temperature of 17°C for 8 h. The potted trees were irrigated using tap water when needed. Three months later, when there were some big and pink nodules on the roots of P. pinnata, the plants were uprooted.
Analysis of soil physicochemical properties and metal contents
Total nitrogen (N) and available N of the soil samples were determined using the Kjeldahl method and alkali N-proliferation method, respectively (Wang et al., 2016; Spargo and Alley, 2018). Soil pH, organic matter, available phosphorus (P) and potassium (K), were determined using the ASI method. Tailings samples were digested with a mixture of HNO3:HF:HCl (3:1:1 by vol) for the measurement of total metals, and the available metals in tailings were extracted by using 1 M C2H7NO2 and 0.2 M ethylenediaminetetraacetic acid (EDTA; Saadani et al., 2016). A soil sample known available metal content was designed in all the steps of available metal extraction and measurement process as quality control. Then, the concentration of total metals and available metals of iron (Fe), titanium (Ti), vanadium (V), chromium (Cr), manganese (Mn), zinc (Zn), copper (Cu), nickel (Ni), lead (Pb), and cadmium (Cd) were quantitated using inductively coupled plasma atomic emission spectrometry (ICP- AES) (IRIS IntrepidII, Thermo Electron Corporation, USA; Zhang et al., 2010).
Isolation and purification of rhizobia
The fresh, big and pink nodules removed from the roots of P. pinnata were sterilized by soaking in 95% ethanol for 1 min, then in 0.1% HgCl2 for 3 min, and washed several times with sterile water. Under aseptic conditions, the surface-sterilized nodules were crushed in a sterilized EP tube, and a small aliquot of the nodule suspension was taken for steaking on Congo red-containing yeast mannitol agar (YMA, 10.0 g/L mannitol, 1.0 g/L yeast extract, 0.5 g/L K2HPO4·3H2O, 0.2 g/LMgSO4·7H2O, 0.1 g/L NaCl, and 1.0 g/L CaCO3 and 0.04 g/L Congo red, pH 7.0–7.2; Zevenhuizen et al., 1986; Sierra et al., 1999). Mucoid and white colonies were selected for repeated re-streaking until single colonies with uniform colony characteristics were observed (Brenner et al., 2005). Purification of rhizobia was confirmed by Gram staining and microscopic examination, and Gram-negative strains were kept for molecular identification (Brooks et al., 2016).
Identification and phylogenetic analysis of rhizobia
Total DNA was extracted from purified isolates using the phenol-chloroform method (Chang et al., 2011), and 16S ribosomal RNA (rRNA), BOXA1R and four housekeeping genes (atpD, recA, rpoB, glnII) were amplified by PCR as described (Vinuesa et al., 2008; Menna et al., 2009). The PCR products were sequenced by Sangon Biotech (Shanghai, China), and the sequences were submitted to GenBank for assignment of accession numbers. Similarity analysis was performed by searching using BLAST in GenBank. Neighbor-joining (NJ) trees of the 16S rRNA and housekeeping genes were constructed using MEGA7.0 software with bootstrap values of 1,000 replicates (Menna et al., 2009). Sequence data for the four housekeeping genes were concatenated into a single atpD-recA-rpoB-glnII sequence for multilocus sequence analysis (MLSA; Vinuesa et al., 2008; Menna et al., 2009).
Heavy metal tolerance tests of rhizobia
The resistance of the 20 isolated rhizobia to Ni, Cd, Mn, and Cu was assayed by measuring their growth in YMA liquid medium (3.0 g/L yeast extract, 5.0 g/L tryptone, 0.7 g/L CaCl2·2H2O, pH 7.0) containing different concentrations of metal ions by adding the salts of NiCl2, CdCl2, MnSO4, and CuSO4, respectively. The bacterial suspensions (50 μL, 108 cells/mL) were inoculated into 5 mL YMA liquid medium. The medium without heavy metal was used as the control. The minimum inhibitory concentration (MIC) and lethal concentration (MLC) were obtained by measuring the optical density (OD600nm) of the bacterial cultures with a spectrophotometer (UV-3300; Shanghai MAPADA, Shanghai, China) after incubation in an orbital shaker (28°C, 150 rpm) for seven days (Mao et al., 2020). MLC was defined as the lowest concentration of metal ion in solid medium where the isolate growth was not observed, while MIC as the lowest concentration of metal ion in the solid medium where the isolate growth was weaker than that in the heavy metal-free control (Yu et al., 2014). The bacterial cultures were repeated three times for each treatment, and OD600nm readings were taken in triplicate.
Symbiotic nitrogen fixation capacity of rhizobia
After the amplification of nifH gene of the isolates, the PCR products were sequenced by Sangon Biotech (Shanghai, China), and the sequences were submitted to GenBank for assignment of accession numbers. Similarity analysis was performed using BLAST in GenBank. Neighbor-joining (NJ) trees of the nifH gene were constructed using MEGA7.0 software with a bootstrap value of 1,000 replicates (Menna et al., 2009).
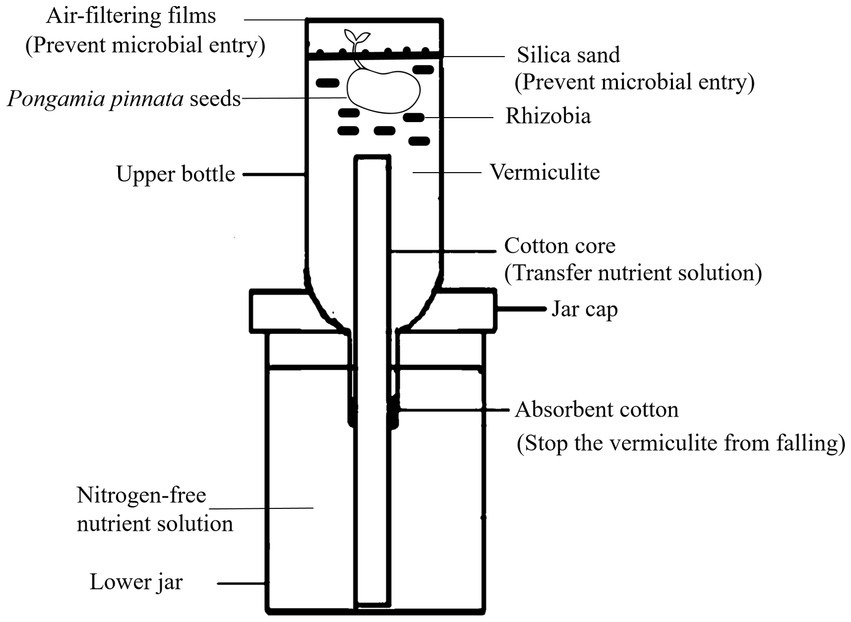
To test the symbiotic nitrogen fixation capacity of the rhizobia isolates, some representative strains of different genera rhizobia were selected for rhizobia-P. pinnata pot experiment by using the potted experimental equipment (Figure 1). Leonard jars, which consisted of two parts, i.e., an upper bottle and a lower jar, were assembled as the apparatus for testing nitrogen fixation activities of rhizobia (Yu et al., 2017a). The lower jar contained nitrogen-free nutrient solution, while the upper bottle was filled with vermiculite as the substrate. The assembled Leonard jars were autoclaved (100 KPa, 121°C) for 30 min after covering the upper bottles with air-filtering films. Some mature and plump seeds of P. pinnata were selected for surface sterilizing using diluted NaClO3 and ethanol. The seeds were sowed in the upper bottles after germination, and then the air-filtering films were used to cover the upper bottles again. Approximately 3 × 108 of fresh rhizobia cells were inoculated around the rhizosphere of a P. pinnata seedling, and then the sterilized silica sand was used to cover the vermiculite to avoid contamination. P. pinnata trees in the non-inoculation pots was designed as the control (CK), and each treatment repeated three times. The pots were kept in a greenhouse with a day temperature of 25°C for 16 h and a night temperature of 17°C for 8 h. After 6 months, P. pinnata plants were uprooted, and the nodule numbers, plant height, root length, biomass (dry weight) and nitrogen content were measured using the previous methods (Yu et al., 2017a).
Statistical analysis
The experimental data were averaged out of at least three independent replicates for each treatment. Microsoft Excel 2016 was used to calculate means and standard deviations. IBM SPSS Statistics 26.0 was used to perform Tukey’s test at p < 0.05.
Results
Physicochemical properties and metal content of soil samples
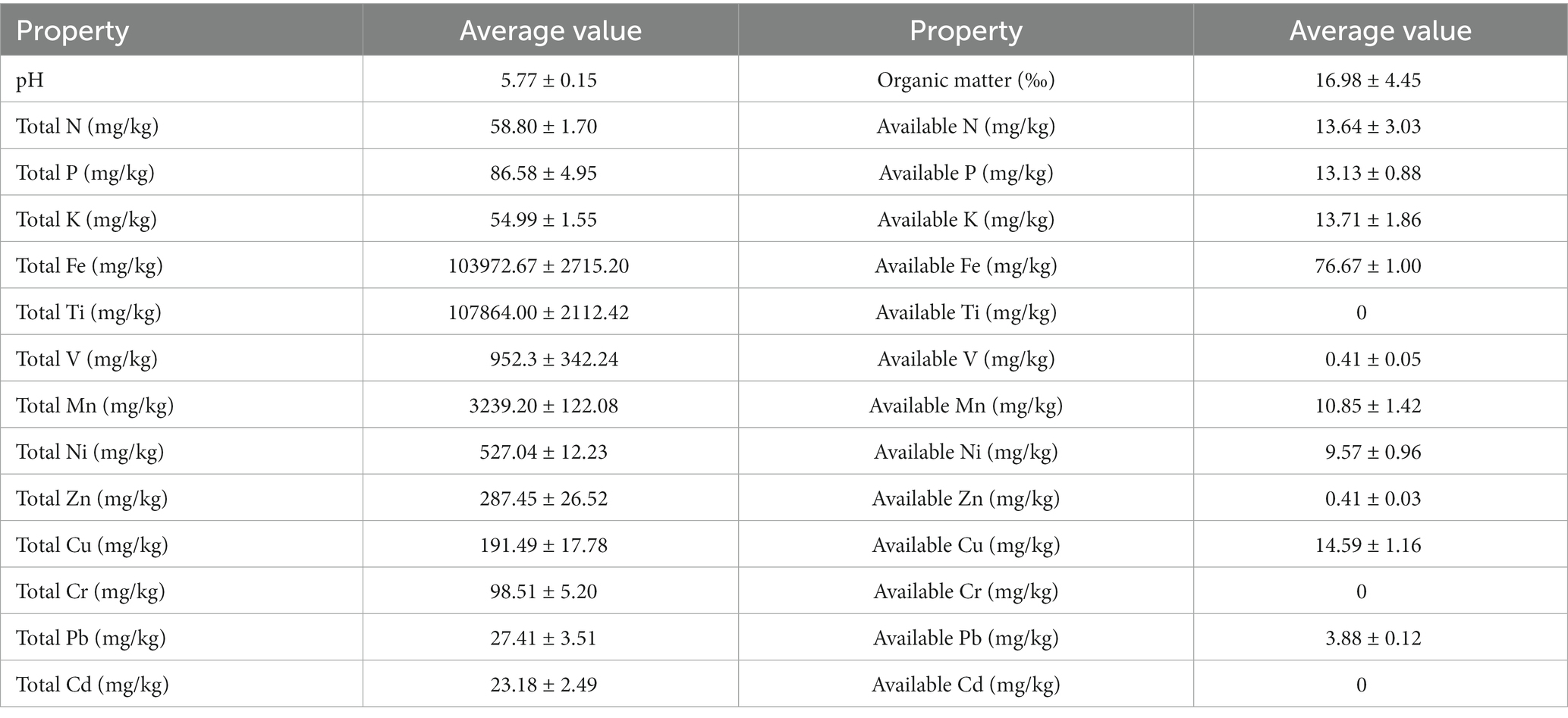
To establish the basic characteristics of the VTM tailings, the physicochemical properties and metal contents were measured for the collected soil samples (Table 1). Soil pH was slightly acidic with a value of 5.77 ± 0.15. The contents of soil organic matter and total nitrogen were very low in the VTM tailings. The available N, P, and K in the VTM tailings accounted for 23.2, 15.2, and 24.9% of the total contents, respectively. The N, P, and K content in the VTM tailings was far lower than the cultivated soil nitrogen content, so the VTM tailings is very barren. As three main components of the VTM tailings, the concentrations of total Ti and Fe were very high as expected at 108 g/kg and 104 g/kg, respectively, while the content of total V (952.3 mg/kg) was relatively low. Interestingly, the concentration of total Mn reached 3,239.20 mg/kg, and the contents of total Ni, Zn, and Cu were more than 100 mg/kg. Only the amounts of total Cr, Pb, and Cd were relatively small in the VTM tailings. The available Ti, Cd, and Cr in tailings were not detected, the content of available V and Zn was very low, and the available Fe was highest, following by Cu, Mn and Ni.
Rhizobia isolates and BOXA1R-PCR fingerprints analysis
The results showed that P. pinnata can grow well in VTM tailings. A total of 57 rhizobia strains with the characteristic white mucoid colonies, Gram-negative and rod-shape features were isolated from the nodules of P. pinnata growing in the VTM tailings. The similarities among the 57 isolates ranged from 0.45 to 1.00 in the BOX A1R-PCR fingerprint dendrogram, including 49 distinct fingerprint patterns (Figure 2). These strains were clustered into two groups at 45% similarity level, three groups at 49% similarity level, 8 groups at 61% similarity, 18 groups at 74% similarity, and 41 groups at 91% similarity level. There were also some strains on the same branch with 100% similarity, such as PP31, PP75, PP81, PP82, and PP109.
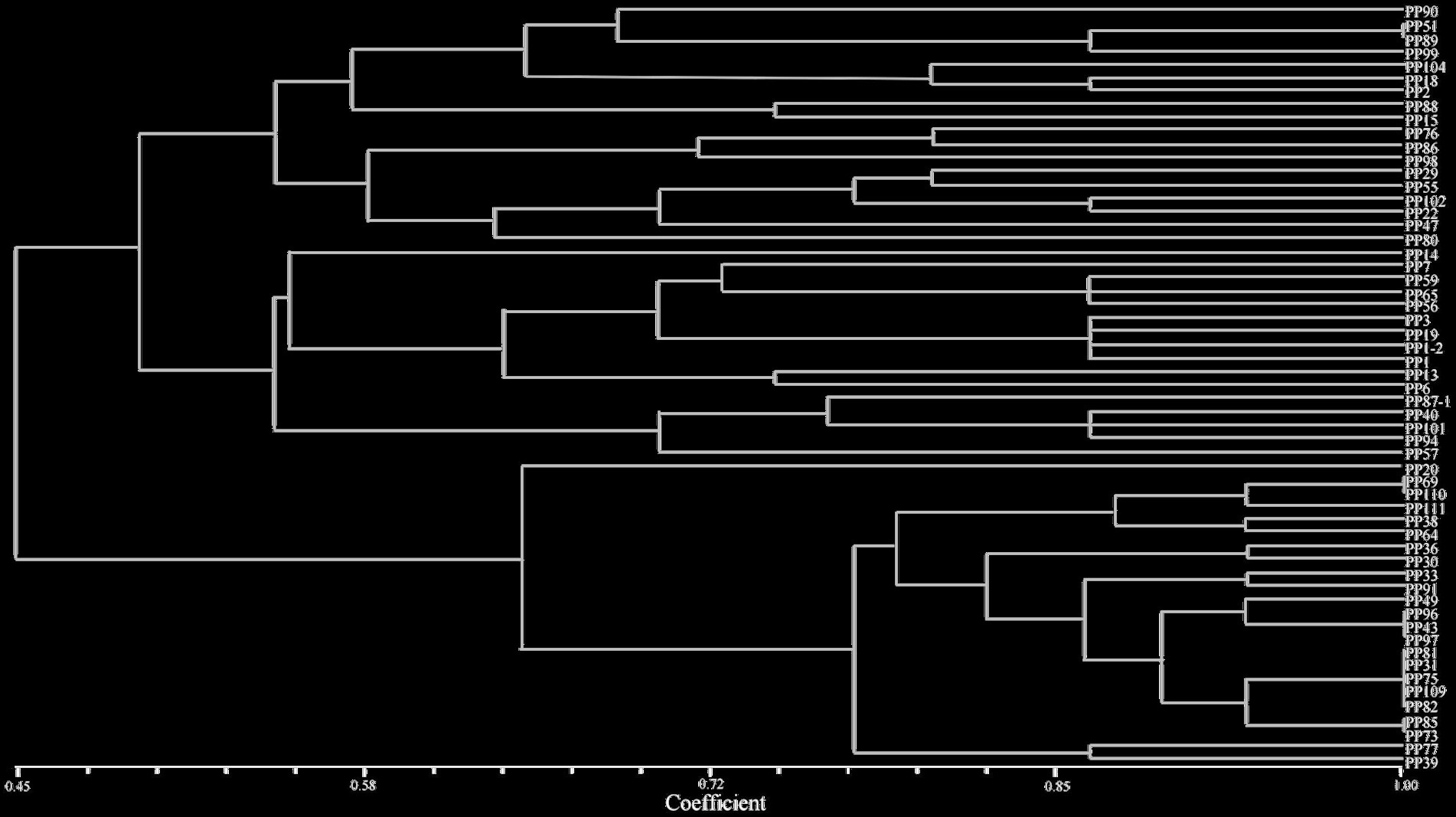
Figure 2. BOX-A1R dendrogram for 56 rhizobia isolated from the nodules of Pongamia pinnata in VTM mine tailings.
Heavy metal tolerance of rhizobia
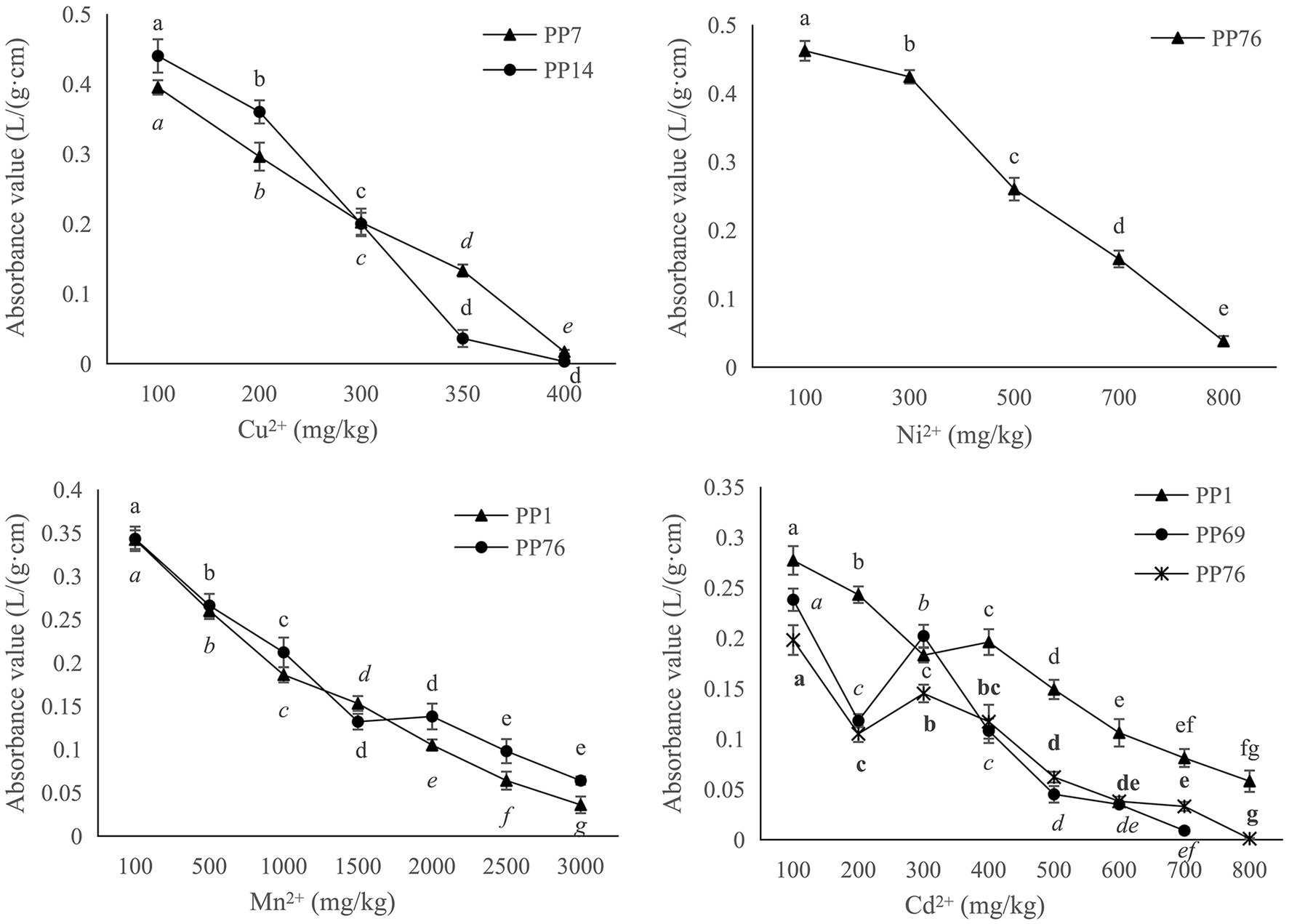
According to the results of BOXA1R-PCR fingerprints analysis (Figure 2), eight isolates with 100% similarity were deleted, and 49 strains were kept for the determination of heavy metal tolerance. Among them, 20 strains showed different levels of tolerance against four heavy metals including Cu, Ni, Mn, and Zn. After culturing in YMA medium with Cu2+ for 7 days, the survival rates were 85% at 100 mg/kg, 20% at 200 mg/kg, 10% at 300 mg/kg, and 5% at 400 mg/kg. For nickel (Ni2+), they were 45% at 100 mg/kg, 20% at 300 mg/kg, 10% at 500 mg/kg, and 5% at 700 mg/kg. For cadmium (Cd2+), they were 55% at 200 mg/kg, 30% at 400 mg/kg, 15% at 600 mg/kg, and 10% at 800 mg/kg. For manganese (Mn2+), it was 75% at 500 mg/kg, 65% at 1,300 mg/kg, 20% at 2100 mg/kg, and 10% at 2900 mg/kg.
Only five rhizobia strains (PP1, PP7, PP14, PP69, and PP76) showed relatively higher tolerance to the four heavy metals (Table 2; Figure 3). PP76 tolerated against Ni, Cd, and Mn with an MIC at 100, 200, 300 mg/L, respectively, and with an MLC at 600, 800, 3,200 mg/L, respectively. PP1 showed high tolerance to Cd and Mn with an MIC at 200, 300 mg/L, respectively, and an MLC at 900, 3100 mg/L, respectively. Strains PP7 and PP14 only showed tolerance against Cu with an MIC at 100 mg/L, and an MLC at 400 and 350 mg/L, respectively. Only PP69 showed high tolerance to Cd with an MIC at 100 mg/L and an MLC at 700 mg/L.
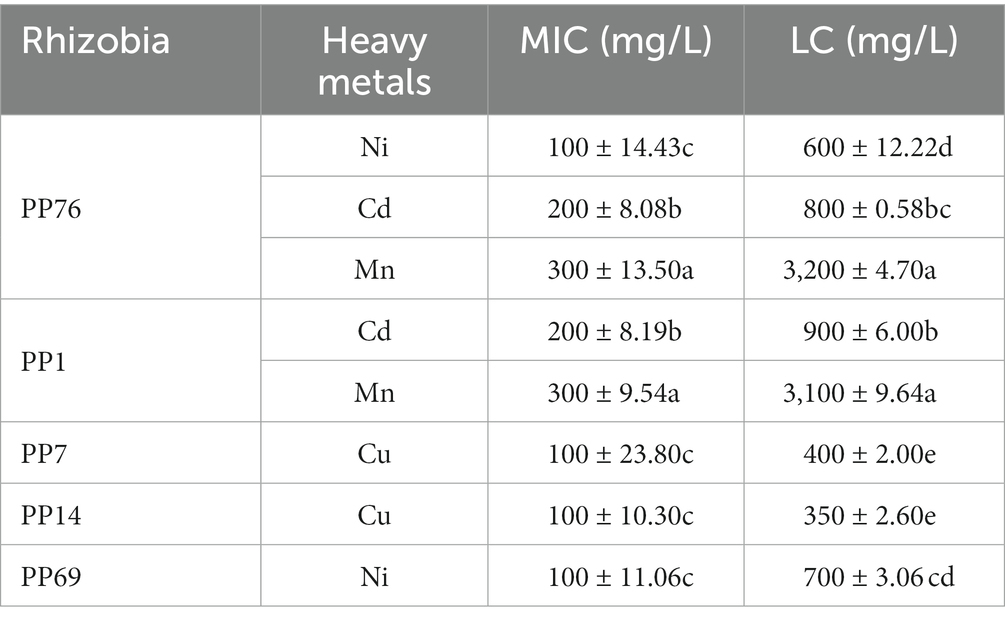
Table 2. Heavy metal minimum inhibitory concentration (MIC) and lethal concentration (LC) of rhizobia strains from the VTM tailings.
Identification of rhizobia
The 20 strains with heavy metal tolerance were selected for molecular identification by 16S rRNA gene sequencing. The sequences of 16S rRNA genes were obtained and compared using BLAST in the National Center for Biotechnology Information (NCBI) database. Similarity analysis of 16S rRNA genes showed that the 20 rhizobia strains were classified into three different genera: Bradyrhizobium (12 strains), Ochrobactrum (4 strains) and Rhizobium (4 strains). The phylogenetic tree of the 16S rRNA gene also divided the 20 rhizobia strains into three different branches (Figure 4). Among them, 12 Bradyrhizobium isolates (PP29, PP40, PP47, PP56, PP57, PP69, PP76, PP80, PP90, PP98, PP99, and PP111) were closest to Bradyrhizobium pachyrhizi PAC 48T with 100% similarity under the same branch (Figure 4). Three Rhizobium isolates (PP1, PP6 and PP18) were 99.12% similar with Rhizobium nepotum Pulawska 39/7T, whereas Rhizobium sp. PP15 was most closely related to Rhizobium leguminosarum NBRC14778T with 99.47% similarity. Other four isolates (PP7, PP14, PP20, and PP49) were classified into Ochrobactrum, which was 100% similar with Ochrobactrum lupini NBRC 102587T under the same branch (Figure 4).
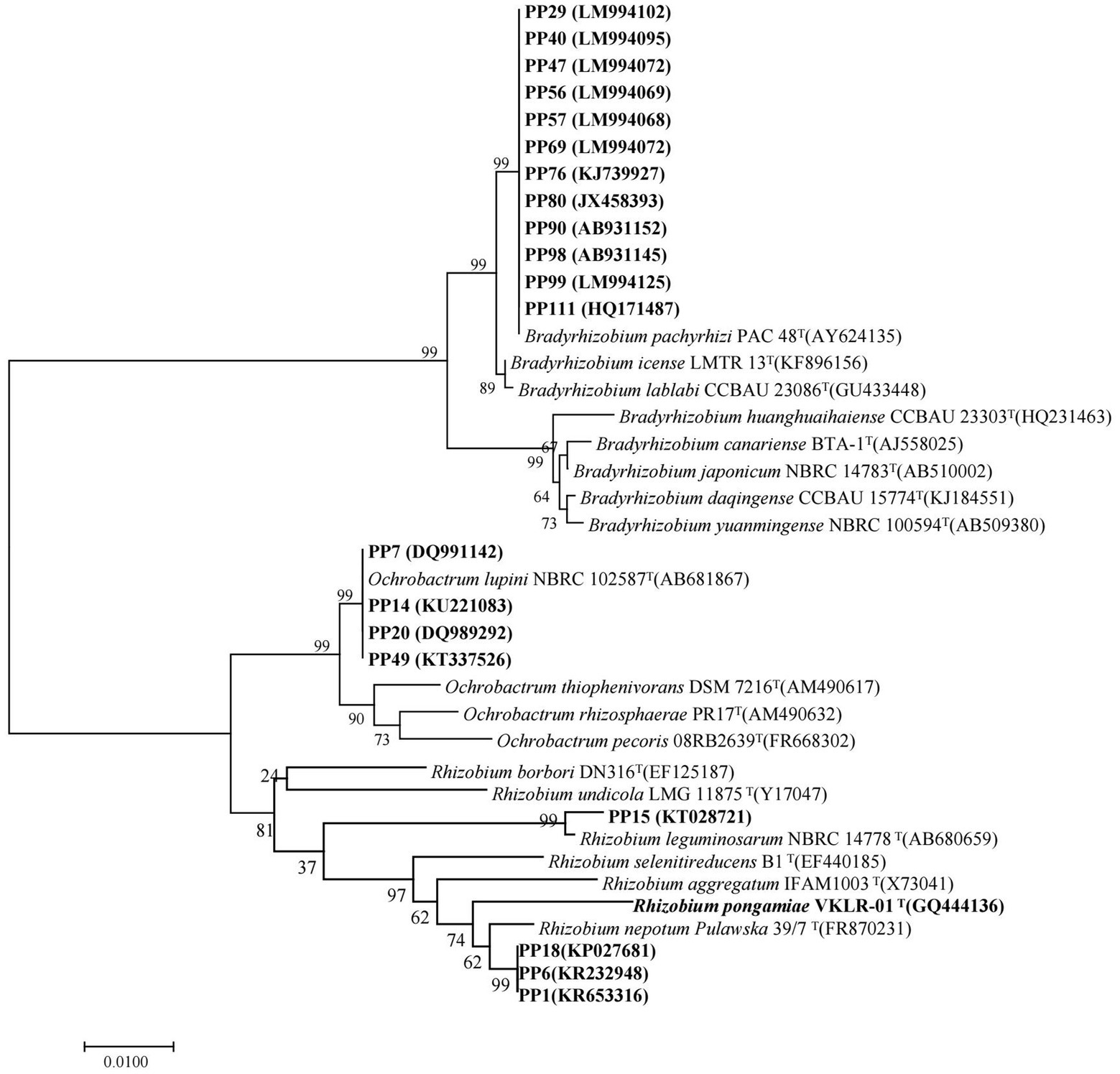
Figure 4. Phylogenetic tree of the rhizobial 16S rRNA genes. The scale bar corresponds to 0.01 substitutions per nucleotide position. The sequence numbers in GenBank are presented in the following parentheses. Superscript “T” means type stains.
Phylogenetic analysis of rhizobia
To accurately determine phylogenetic status of the 20 rhizobia, amplified the four house-keeping genes (atpD, recA, rpoB and glnII) of the isolates and built a phylogenetic tree for each genus. For 12 Bradyrhizobium isolates, sequences of the house-keeping genes [atpD (407 bp), recA (380 bp), rpoB (522 bp), and glnII (474 bp)] were used to perform the multi-locus sequence analysis (MLSA) by constructing a longer housekeeping gene fragment (1,783 bp). Then, the phylogenetic tree of these 12 Bradyrhizobium isolates was built using neighbor-joining method (Figure 5C). For 4 Rhizobium isolates, sequences of the house-keeping genes [atpD (298 bp), recA (246 bp), rpoB (554 bp) and glnII (339 bp)] were subjected to multi-locus sequence analysis (MLSA) by constructing a longer housekeeping gene fragment (1,437 bp). Then, the phylogenetic tree of these 4 Rhizobium isolates was built using neighbor-joining method (Figure 5B). For 4 Ochrobactrum isolates, sequences of the four house-keeping genes [atpD (387 bp), recA (385 bp), rpoB (412 bp) and glnII (447 bp)] were subjected to multi-locus sequence analysis (MLSA) by constructing an longer housekeeping gene fragment (1,631 bp) (Figure 5A).
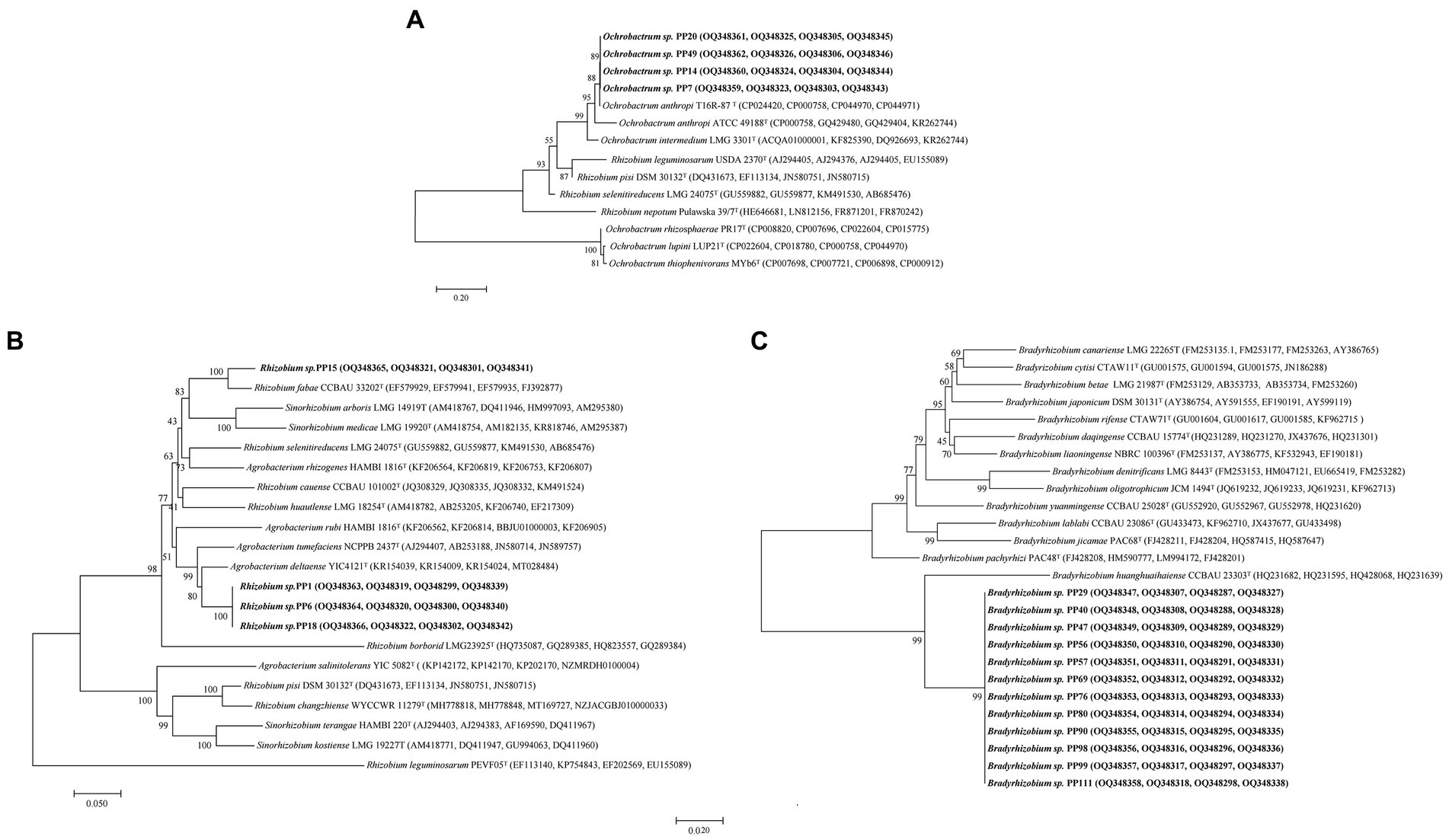
Figure 5. Phylogenetic tree of the concatenated housekeeping genes (atpD-glnII-recA-rpoB) for Ochrobactrum (A), Rhizobium (B), and Bradyrhizobium (C) genus.
The MLSA phylogenetic tree of the four housekeeping genes at genus level was basically the same as that of the 16S rRNA gene, but there were some differences at species level. Twelve Bradyrhizobium isolates were most closely related to Bradyrhizobium huanghuaihaiense CCBAU 23303T with 99.24% similarity, and Bradyrhizobium pachyrhizi PAC 48T with 98.70% similarity. The isolate Rhizobium sp. PP15 was closest to Rhizobium fabae CCBAU 33202T with 99.35% similarity, and Rhizobium pisi DSM 30132T with 98.55% similarity. Other three Rhizobium isolates (PP1, PP6, and PP18) were closest to Agrobacterium deltaense YIC4121T and Agrobacterium tumefaciens NCPPB 2437T with 98.95 and 98.44% similarity, respectively. Four Ochrobactrum strains were closest to Ochrobactrum anthropi T16R-87Tand Ochrobactrum lupini LUP21T with 99.74 and 97.83% similarity, respectively.
Symbiotic nitrogen fixation capacity of rhizobia
We amplified the nifH gene of the isolates and built a phylogenetic tree using neighbor-joining method (Figure 6). As was similar with the trees of 16S rRNA genes and the house-keeping genes, the same genus was clustered together. Four Bradyrhizobium isolates were most closely related to Bradyrhizobium ferriligni CCBAU51502T, two Rhizobium isolates were most closely related to Rhizobium pusense VLa18T, and two Ochrobactrum isolates were most closely related to Ochrobactrum anthropi ATCC 49188T.
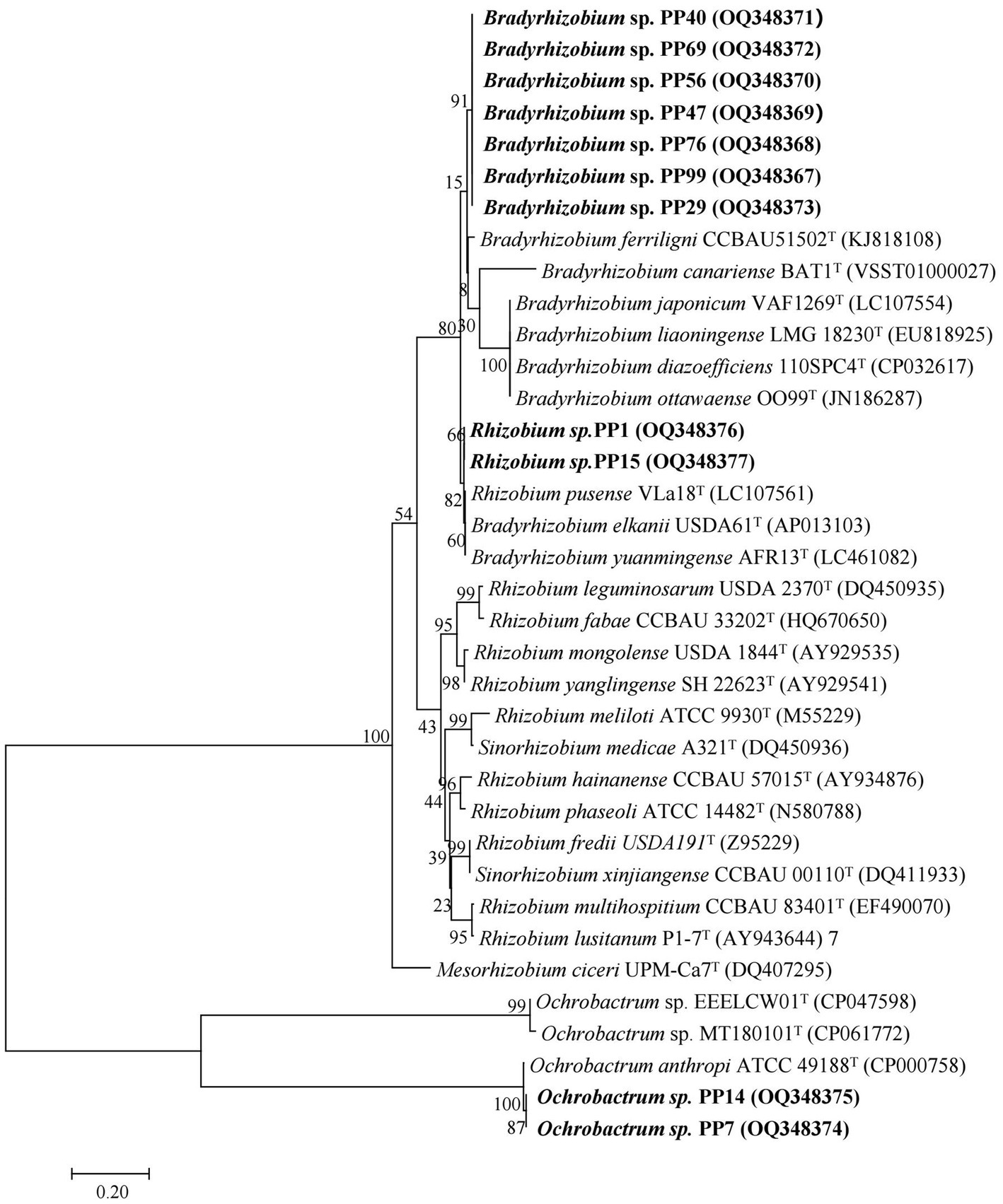
Figure 6. Phylogenetic tree of rhizobial nifH genes. The scale bar corresponds to 0.02 substitutions per nucleotide position.
The 20 representative rhizobia with heavy metal tolerance were selected to determine their capacity of symbiotic nitrogen fixation using P. pinnata pot experiment. When the 20 rhizobia were, respectively, inoculated around the P. pinnata rhizosphere, only 11 strains built symbiotic relationships with P. pinnata. Moreover, the nodule numbers of P. pinnata inoculated with different rhizobia strains were fully diverse (Table 3). Plants inoculated with Rhizobium produced the highest number of roots nodules, suggesting Rhizobium had the strongest nodulating capability for P. pinnata, Bradyrhizobium was the next, and Ochrobactrum was the weakest at nodulation. In all treatments, more nitrogen content in P. pinnata was found in the aboveground parts than in the roots. The nitrogen content in the inoculated plants was significantly (p < 0.05) higher than that in the non-inoculated control. Except for Bradyrhizobium sp. PP76 and PP69, the trend of other rhizobia strains’ nitrogen fixation capacity was similar to that of symbiotic nodule number with the following order: Rhizobium > Bradyrhizobium > Ochrobactrum. Among them, Rhizobium sp. PP1 showed the strongest nitrogen fixation activity, and the nitrogen content of the plants’ aboveground parts and roots was 2.4–1.8 times that of the non-inoculation control.
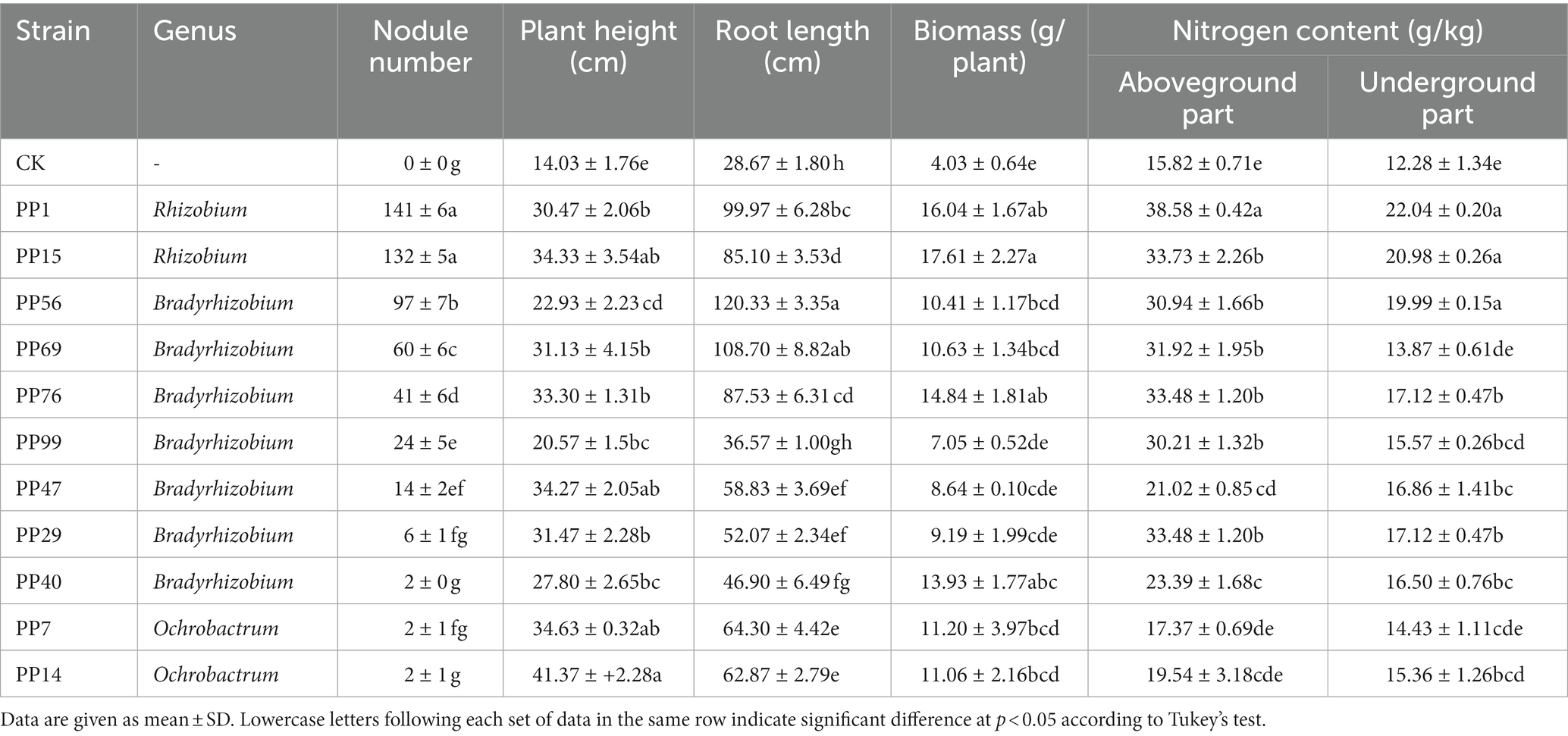
Table 3. Nitrogen content, biomass, and growth of Pongamia pinnata inoculated with different rhizobia strains.
Except for nitrogen fixation capacity, these rhizobia strains showed different levels of plant growth promoting activities for P. pinnata. Among them, the plant height, root length and biomass of the non-inoculated control were the lowest. The plant height of P. pinnata inoculated with Ochrobactrum sp. PP14 was the highest, whereas the root length was the lowest, just as in the non-inoculated control. Compared with the non-inoculated control plants, the improved height of the inoculated plants ranged from 47 to 195%, while the increase of root length of the inoculated plants ranged from 119 to 320%. Because of the promoting effect of rhizobia, the biomass of the inoculated P. pinnata plants was significantly (p < 0.05) higher than that of the control plants. Compared with the non-inoculated plants, the biomass of the inoculated treatments was improved by 1.75–4.27 times. The biomass improvement of P. pinnata inoculated Rhizobium sp. PP15 was the highest among all of them.
Discussion
Physicochemical properties and metal contaminations in the VTM tailings
The quality of the soil depends on its physicochemical properties. Soil pH affects soil microbial diversity and function (Blum et al., 2018), and the pH of VTM tailings in Sichuan Province, China, was found to be quite acidic, which is similar to the soil near a zinc blende mine north of Spain (Pérez-Esteban et al., 2014). Thus, the microbial communities in the VTM tailings must be more adaptable to an acidic environment. The available N, P, K and organic matter were as low as in other mine tailings (e.g., in Mexico), which indicated that the VTM tailings was not very fertile (Armienta et al., 2019). The Ti concentration was up to 38 times higher than that in the similar soil near a Ti mining site in Kenya, and the V concentration was up to 7.5 times higher than that in Cuban soils on average (Alfaro et al., 2014; Maina et al., 2016). The concentration of Fe was approximately 3,000 mg/kg, which is higher than the soil near a steel plant in India (Kaur et al., 2019). Compared to the soil near a coal mine in China with severe Cu, Zn, and Cr pollution, the concentration of these elements in Sichuan VTM tailings was 6.70, 3.69, and 1.54 times higher, respectively; but Pb was lower at 1.45 mg/kg (Liu et al., 2020). The concentration of Mn and Ni in VTM tailings was approximately 5 and 3 times higher, respectively, than in the magnetite tailings after growth of Imperata cylindrica (Yuan et al., 2018). The Cd concentration already exceeded the minimum inhibitory concentration for plant growth (Zhang F. et al., 2019). Consequently, the reason why plants grown in the VTM soil were infertile may be due to the high heavy metal contents and low available N, P, K, and organic matter. Therefore, when carrying out ecological restoration, attention must be paid to reducing the concentration of heavy metals in the soil and increasing the content of nutrients.
Diversity and phylogeny of Pongamia pinnata rhizobia in the VTM tailings
As an biofules resource, P. pinnata is a fast-growing leguminous tree with the potential for high oil seed production and can grow on marginal land (Scott et al., 2008). Only two genera of Rhizobium genera including R. pongamiae, R. miluonense, and Bradyrhizobium genera including B. liaoningense, B. elkanii, B. yuanmingense were found to be symbiotic nitrogen fixation with P. pinnata in India and Australia (Rasul et al., 2012; Arpiwi et al., 2013; Kesari et al., 2013). However, three genera rhizobia of Rhizobium, Bradyrhizobium and Ochrobactrum symbiotic with P. pinnata were isolated from the VTM tailings, which revealed there were abundant rhizobia in the VTM tailings. These rhizobia included B. pachyrhizi, R. nepotum, R. nepotum, and O. lupini, indicating P. pinnata rhizobia isolated from the VTM tailings were different form previous reported others. So, the VTM tailings was a resource pool including abundant functional microbiology. Although it was the first time that Ochrobactrum was found to have symbiotic nodulation with P. pinnata, their symbiotic nitrogen fixation efficiency were not high (Table 2).
The distribution of the Rhizobium population can easily be changed by the influence of different environmental factors (Stefan et al., 2018). Because of multiple heavy metal pollution and barren environmental factors, rhizobia symbiotic with P. pinnata for the VTM tailings were different from others. The proportion of Bradyrhizobium strains was highest among three rhizobia genera in the VTM tailings, probably because of stronger resistance of Bradyrhizobium to heavy metals. Compared with R. pongamiae VKLR-01 isolated from root nodules of P. pinnata, their genetic similarity is not high (Kesari et al., 2013). Some of the strains had specific genetic traits which helped to enhance their adaptability to the toxic environment of heavy metal ions. These special rhizobia from the VTM tailings also proved that microbial composition in a rhizobial system has host-specific and bio-geographical distribution characteristics (Zhang et al., 2011).
Heavy metal tolerance of Pongamia pinnata rhizobia in the VTM tailings
Although there are multiple heavy metal pollutants and very poor nutrition in there VTM tailings, abundant heavy metal resistant and plant growth promoting bacteria survival in the extreme environment (Yu et al., 2014). Because of the extreme heavy metal environment, P. innata rhizobia from the VTM tailings also showed heavy metal resistance. Environmental factors following the order: soil pH > heavy metals > nitrogen > soil texture had distinct impacts on microbial community (Deng et al., 2018), indicating that heavy metals were very important affection factor for soil microbe. Only 40% rhizobia from the VTM tailings showed tolerance against Cu, Ni, Mn, and Zn. The tolerance concentrations of these metals (except for Mn) for these strains were higher than those in the VTM tailings, and different isolates had different level of tolerance to heavy metals, indicating soil heavy metals are not the only factor affecting strain resistance. Some microbes metabolize and transform heavy metal into a less hazardous form for surviving in such harsh environments, resulting in the formation of heavy-metal-resistant microbes (Prabhakaran et al., 2016), so these microbes had their own unique resistance characteristics. From BOXA1R-PCR fingerprints and Phylogenetic characteristic, rhizobia form the VTM tailings had different genotype, so their tolerance to heavy metals was different, and even some isolates did not showed tolerance to the four tested heavy metals.
In other research on rhizobial systems, most bacteria were only tolerant to a single heavy metal and only a few were resistant to multiple types of heavy metals (Stan et al., 2011; Fan et al., 2018), which was consistent with the tolerance to heavy metals of rhizobia from the VTM tailings. Bradyrhizobium sp. PP76 was resistant to Ni, Cd, Mn of the four metal ions and Rhizobium sp. PP1 was resistant to Cd and Mn, which makes them the best choices for the establishment of symbiotic systems of leguminous plants and rhizobia in heavy metal-contaminated soils.
Nitrogen fixation capacity of Pongamia pinnata rhizobia in the VTM tailings
As a typical function of rhizobia, symbiotic nitrogen fixation is relation to nod, nif and fix genes, such as nifH named as dinitrogenase reductase (Shamseldin, 2013). The nifH gene is the biomarker most widely used to study the ecology and evolution of nitrogen-fixing bacteria (Gaby and Buckley, 2014), so the amplified nifH gene was an initial evidence of the nitrogen-fixing capability of the rhizobia isolates from the VTM tailing. From the phylogenetic tree, the nifH genes of three genera rhizobia were also consistent with the 16S rRNA and house-keeping genes with high similarity among the same genus rhizobia. Because symbiotic nitrogen fixation was decided by series of nif genes, e.g., S. meliloti and R. leguminosarum bv. viciae have a restricted set of 9 and 8 nif genes, respectively (Masson-Boivin et al., 2009). Therefore, although the nifH genes of same genus with high genotype similarity was on the same branch, these rhizbia showed different nitrogen fixation and plant growth capacity. These rhizobia had symbiotic N-fixation ability with the leguminous host plant of P. pinnata and were facilitative in promoting plant growth. Almost of rhizobia showed consistence between symbiotic nitrogen fixation and plant biomass, except for Ochrobactrum sp. PP7 and PP14. Although Ochrobactrum sp. PP7 and PP14 showed lowest symbiotic nitrogen fixation efficiency among the eleven isolates, their plant-growth promoting activity was stronger than some Bradyrhizboium sp. isolates, indicating that Ochrobactrum sp. PP7 and PP14 might had some of other plant-growth promoting capacity (Yu et al., 2017b).
The excessive metal concentrations cause undeniable damage to rhizobia, legumes and their symbiosis to affect efficiency of symbiotic nitrogen fixation (Zahran, 1999; Ahmad et al., 2012), which does not hinder that rhizobia increase phytoremediation by nitrogen fixation and production of plant growth-promoting factors and phytohormones (Pajuelo et al., 2011). So, legume–rhizobium symbioses has been considered as a tool for bioremediation of heavy metal polluted soils (Pajuelo et al., 2011). However, rhizobium should have heavy-metal resistance to improve legume–rhizobium symbiosis in bioremediation of heavy metal polluted soil (El-Tahlawy and Ali, 2021). P. pinnata rhizobia from the VTM tailings did not only show nitrogen fixation capacity but also heavy metal tolerances, so these rhizobia can be used to build symbiosis bioremediation system for heavy metals. P. pinnata inoculated with B. liaoningense PZHK1 was proved to show huge potential for phytoremediation of mine tailings, which had applied for soil and ecological remediation at the VTM tailings (Yu et al., 2017a, 2019). These rhizobium isolates with nitrogen fixation capacity and heavy metal resistance provided excellent microbial resources for bioremediation of the VTM tailings to other heavy metal polluted soil.
Conclusion
The application of the symbiotic remediation systems of rhizobia and leguminous plants is a major research area with a focus on bioremediation of the multiple heavy metal-polluted environments. There are at least three genera of culturable rhizobia in symbiosis with P. pinnata in VTM tailings, namely, Bradyrhizobium, Ochrobactrum, and Rhizobium. Some rhizobia have high N-fixing efficiency, plant growth-promoting capacity, and resistance to heavy metals, indicating there are abundant functional microbial resources in extreme soil environment. Interestingly, the phenotype of strong N-fixing capacity seemed to coincide with the resistance to multiple metal ions, which could explain why Bradyrhizobium was the dominant genus of rhizobia around the P. pinnata rhizosphere in the soil contaminated with heavy metals. Because of resistance to several heavy metals, these isolates were competent candidates for the bioremediation of soils contaminated with multifarious metals. This study did not only reveal the genetic diversity and phylogeny of P. pinnata rhizobia in VTM tailings, but also provided important resources for the development of soil remediation techniques using rhizobium-legume symbiotic systems.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession numbers OQ348361, OQ348325, OQ348205, OQ348345, OQ348362, OQ348362, OQ348306, OQ348346, OQ348360, OQ348324, OQ348304, OQ348344, OQ348359, OQ348323, OQ348303, OQ348343, OQ348365, OQ348321, OQ348301, OQ348341, OQ348363, OQ348319, OQ348299, OQ348339, OQ348364, OQ348320, OQ348300, OQ348340, OQ348366, OQ348322, OQ348302, OQ348342, OQ348347, OQ348307, OQ348287, OQ348327, OQ348348, OQ348308, OQ348288, OQ348328, OQ348349, OQ348350, OQ348351, OQ348352, OQ348353, OQ348354, OQ348355, OQ348356, OQ348357, OQ348358, OQ348309, OQ348310, OQ348311, OQ348312, OQ348313, OQ348314, OQ348315, OQ348316, OQ348317, OQ348318, OQ348287, OQ348288, OQ348289, OQ348290, OQ348291, OQ348292, OQ348293, OQ348294, OQ348295, OQ348296, OQ348297, OQ348298, OQ328327, OQ348328, OQ348329, OQ348330, OQ349331, OQ348332, OQ348333, OQ348334, OQ348335, OQ348336, OQ348337, OQ348338.
Author contributions
TS, RJ, JY, and XY conceived research project, assayed rhizobia symbiotic nitrogen fixation capacity, performed statistical analysis, and drafted the manuscript. TS, XC, LZe, TZ, and XY collected soil samples and trapped rhizobia. RJ, XC, YG, LZo, KZ, and QX conducted general experiments, performed phylogenetic analysis, and identification of rhizobia. JY, LZo, and MM analyzed heavy metal tolerance of rhizobia. MM, SL, and TZ analyzed soil physicochemical properties and metal contents. XY and QC funded and supervised the experiments. All authors reviewed, edited, and approved the final manuscript.
Funding
This research was supported by the National program on Key Research Project [2022YFD1901400], Demonstration Project of Transfer and Transformation of Scientific and Technological Achievements in Sichuan Province [2022ZHCG0030], and the Key Research Project of Deyang City [2022NZ015].
Acknowledgments
The authors thank Xia Kang (School of Life Sciences, University of Dundee, Scotland, UK) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelkrim, S., Jebara, S. H., Saadani, O., Chiboub, M., Abid, G., Mannai, K., et al. (2018). Heavy metal accumulation in Lathyrussativus growing in contaminated soils and identification of symbiotic resistant bacteria. Arch. Microbiol. 201, 107–121. doi: 10.1007/s00203-018-1581-4
Adediran, G. A., Ngwenya, B. T., Mosselmans, J. F. W., and Heal, K. V. (2015). Bacteria-zinc co-localization implicates enhanced synthesis of cysteine-rich peptides in zinc detoxification when Brassica juncea is inoculated with Rhizobium leguminosarum. New Phytol. 209, 280–293. doi: 10.1111/nph.13588
Ahmad, E., Zaidi, A., Khan, M. S., and Oves, M. (2012) Heavy metal toxicity to symbiotic nitrogen-fixing microorganism and host legumes. New York: Springer Vienna.
Alfaro, M. R., Montero, A., Ugarte, O. M., do Nascimento, C. W. A., de Aguiar Accioly, A. M., Biondi, C. M., et al. (2014). Background concentrations and reference values for heavy metals in soils of Cuba. Environ. Moni. Assess. 187, 4198–4208. doi: 10.1007/s10661-014-4198-3
Armienta, M., Beltrán, M., Martínez, S., and Labastida, I. (2019). Heavy metal assimilation in maize (Zea mays L.) plants growing near mine tailings. Environ. Geochem. Health 42, 2361–2375. doi: 10.1007/s10653-019-00424-1
Arpiwi, N. L., Yan, G., Barbour, E. L., Plummer, J. A., and Watkin, E. (2013). Phenotypic and genotypic characterisation of root nodule bacteria nodulating Millettia pinnata (L.) Panigrahi, a biodiesel tree. Plant Soil 367, 363–377. doi: 10.1007/s11104-012-1472-4
Blum, J.-M., Su, Q., Ma, Y., Valverde-Pérez, B., Domingo-Félez, C., Jensen, M. M., et al. (2018). The pH dependency of N-converting enzymatic processes, pathways and microbes: effect on net N2O production. Environ. Microbiol. 20, 1623–1640. doi: 10.1111/1462-2920.14063
Brenner, D. J., Krieg, N. R., Staley, J. T., and Garrity, G. M. (2005) Bergey’s manual® of systematic bacteriology. Springer: Boston.
Brooks, R. S., Blanchard, M. T., Clothier, K. A., Fish, S., Anderson, M. L., and Stott, J. L. (2016). Characterization of Pajaroellobacter abortibovis, the etiologic agent of epizootic bovine abortion. Vet. Microbiol. 192, 73–80. doi: 10.1016/j.vetmic.2016.07.001
Chang, S. S., Hsu, H. L., Cheng, J. C., and Tseng, C. P. (2011). An efficient strategy for broad-range detection of low abundance bacteria without DNA decontamination of PCR reagents. PLoS One 6:e20303. doi: 10.1371/journal.pone.0020303
Demková, L., Árvay, J., Bobuľská, L., Hauptvogl, M., and Michalko, M. (2019). Activity of the soil enzymes and moss and lichen biomonitoring method used for the evaluation of soil and air pollution from tailing pond in NižnáSlaná (Slovakia). J. Environ. Sci. Health 54, 495–507. doi: 10.1080/10934529.2019.1567158
Deng, S., Ke, T., Li, L., Cai, S., Zhou, Y., Liu, Y., et al. (2018). Impacts of environmental factors on the whole microbial communities in the rhizosphere of a metal-tolerant plant: Elsholtzia haichowensis Sun. Environ. Pollut. 237, 1088–1097. doi: 10.1016/j.envpol.2017.11.037
El-Tahlawy, Y. A., and Ali, O. A. (2021). Bioremediation of heavy metals using the symbiosis between leguminous plants and genetically engineered rhizobia. Approac. Remed. Inorgan. Pollut., 303–322. doi: 10.1007/978-981-15-6221-1_15
Fagorzi, C., Checcucci, A., DiCenzo, G. C., Debiec-Andrzejewska, K., Dziewit, L., Pini, F., et al. (2018). Harnessing rhizobia to improve heavy-metal phytoremediation by legumes. Genes 9:542. doi: 10.3390/genes9110542
Fan, M., Liu, Z., Nan, L., Wang, E., Chen, W., Lin, Y., et al. (2018). Isolation, characterization, and selection of heavy metal-resistant and plant growth-promoting endophytic bacteria from root nodules of Robiniapseudoacacia in a Pb/Zn mining area. Microbiol. Res. 217, 51–59. doi: 10.1016/j.micres.2018.09.002
Gaby, J. C., and Buckley, D. H. (2014). A comprehensive aligned nifH gene database: a multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014:bau001. doi: 10.1093/database/bau001
He, Y., Li, B., Zhang, K., Li, Z., Chen, Y., and Ye, W. (2019). Experimental and numerical study on heavy metal contaminant migration and retention behavior of engineered barrier in tailings pond. Environ. Pollut. 252, 1010–1018. doi: 10.1016/j.envpol.2019.06.072
Jin, Z., Li, Z., Li, Q., Hu, Q., Yang, R., Tang, H., et al. (2014). Canonical correspondence analysis of soil heavy metal pollution, microflora and enzyme activities in the Pb-Zn mine tailing dam collapse area of Sidi village, SW China. Environ. Earth Sci. 73, 267–274. doi: 10.1007/s12665-014-3421-4
Ju, W., Liu, L., Fang, L., Cui, Y., Duan, C., and Wu, H. (2015). Impact of co-inoculation with plant-growth-promoting rhizobacteria and rhizobium on the biochemical responses of alfalfa-soil system in copper contaminated soil. Ecotoxicol. Environ. Safety 167, 218–226. doi: 10.1016/j.ecoenv.2018.10.016
Kaur, I., Khandwekar, S., Chauhan, R., Singh, V., Jadhav, S. K., Tiwari, K. L., et al. (2019). Exploring the efficiency of native tree species grown at mine tailings for phytoextraction of lead and iron. Proc. Natl. A Sci. India A Biol. Sci. 89, 951–956. doi: 10.1007/s40011-018-1010-0
Kesari, V., Ramesh, A. M., and Rangan, L. (2013). Rhizobium pongamiae sp. nov. from root nodules of Pongamia pinnata. Biomed. Res. Int. 2013, 1–9. doi: 10.1155/2013/165198
Kumar, D., Tripathi, D. K., Liu, S., Singh, V. K., Sharma, S., Dubey, N. K., et al. (2017). Pongamia pinnata (L.) Pierre tree seedlings offer a model species for arsenic phytoremediation. Plant Gene 11, 238–246. doi: 10.1016/j.plgene.2017.06.002
Liu, X., Shi, H., Bai, Z., Zhou, W., Liu, K., Wang, M., et al. (2020). Heavy metal concentrations of soils near the large opencast coal mine pits in China. Chemosphere 244, 125360–125402. doi: 10.1016/j.chemosphere.2019.125360
Maina, D. M., Ndirangu, D. M., Mangala, M. M., Boman, J., Shepherd, K., and Gatari, M. J. (2016). Environmental implications of high metal content in soils of a titanium mining zone in Kenya. Environ. Sci. Pollut. Res. 23, 21431–21440. doi: 10.1007/s11356-016-7249-1
Mao, C., Xue, C., Wang, X., He, S., Wu, L., and Yan, X. (2020). Rapid quantification of pathogenic salmonella typhimurium and total bacteria in eggs by nano-flow cytometry. Talanta 217:121020. doi: 10.1016/j.talanta.2020.121020
Marriboina, S., and Attipalli, R. R. (2020). Hydrophobic cell-wall barriers and vacuolar sequestration of Na+ ions are among the key mechanisms conferring high salinity tolerance in a biofuel tree species, Pongamia pinnata L. Pierre. Environ. Exp. Bot. 171, 103949–103958. doi: 10.1016/j.envexpbot.2019.103949
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Menna, P., Barcellos, F. G., and Hungria, M. (2009). Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int Jsyst Evolut Microbiol 59, 2934–2950. doi: 10.1099/ijs.0.009779-0
Nocelli, N., Bogino, P., Banchio, E., and Giordano, W. (2016). Roles of extracellular polysaccharides and biofilm formation in heavy metal resistance of rhizobia. Materials 9, 418–436. doi: 10.3390/ma9060418
Pajuelo, E., Rodríguez-Llorente, I. D., Lafuente, A., and Caviedes, M. Á. (2011). Legume–rhizobium symbioses as a tool for bioremediation of heavy metal polluted soils. Biomanag. Metal Contam. Soils, 95–123. doi: 10.1007/978-94-007-1914-9_4
Pérez-Esteban, J., Escolástico, C., Masaguer, A., Vargas, C., and Moliner, A. (2014). Soluble organic carbon and pH of organic amendments affect metal mobility and chemical speciation in mine soils. Chemosphere 103, 164–171. doi: 10.1016/j.chemosphere.2013.11.055
Prabhakaran, P., Ashraf, M. A., and Aqma, W. S. (2016). Microbial stress response to heavy metals in the environment. RSC Adv. 6, 109862–109877. doi: 10.1039/C6RA10966G
Rasul, A., Amalraj, E. L. D., Praveen Kumar, G., Grover, M., and Venkateswarlu, B. (2012). Characterization of rhizobial isolates nodulating Millettia pinnata in India. FEMS Microbiol. Lett. 336, 148–158. doi: 10.1111/1574-6968.12001
Saadani, O., Fatnassi, I. C., Chiboub, M., Abdelkrim, S., Barhoumi, F., Jebara, M., et al. (2016). In situ phytostabilisation capacity of three legumes and their associated plant growth promoting Bacteria (PGPBs) in mine tailings of northern Tunisia. Ecotox. Environ. Safe 130, 263–269. doi: 10.1016/j.ecoenv.2016.04.032
Scott, P. T., Pregelj, L., Chen, N., Hadler, J. S., Djordjevic, M. A., and Gresshoff, P. M. (2008). Pongamia pinnata: an untapped resource for the biofuels industry of the future. Bioenergy Res. 1, 2–11. doi: 10.1007/s12155-008-9003-0
Shamseldin, A. (2013). The role of different genes involved in symbiotic nitrogen fixation—review. Glob. J. Biotech. Biochem. 8, 84–94. doi: 10.5829/idosi.gjbb.2013.8.4.82103
Shen, G., Ju, W., Liu, Y., Guo, X., Zhao, W., and Fang, L. (2019). Impact of urea addition and rhizobium inoculation on plant resistance in metal contaminated soil. Int. J. Environ. Res. Public Health 16:1955. doi: 10.3390/ijerph16111955
Sierra, S., Rodelas, B., Martínez-Toledo, M. V., Pozo, C., and González-López, J. (1999). Production of B-group vitamins by two Rhizobium strains in chemically defined media. J. Appl. Microbiol. 86, 851–858. doi: 10.1046/j.1365-2672.1999.00765.x
Singh, R. K., Singh, P., Sharma, A., Guo, D. J., Upadhyay, S. K., Song, Q. Q., et al. (2022). Unraveling nitrogen fixing potential of endophytic diazotrophs of different saccharum species for sustainable sugarcane growth. Int. J. Mol. Sci., 23:6242. doi: 10.3390/ijms23116242
Spargo, J. T., and Alley, M. M. (2018). Modification of the Illinois soil nitrogen test to improve measurement precision and increase sample throughput. Soil Sci. Soc. Am. J. 72, 823–829. doi: 10.2136/sssaj2007.0269
Stan, V., Cornea, C. P., Gament, E., Voaides, C., and Pop, A. (2011). Heavy metal resistant Rhizobium leguminosarum biovar trifolii isolates: characterization and use in rhizoremediation of polluted soils. Curr. Opin. Biotechnol. 22:S74. doi: 10.1016/j.copbio.2011.05.217
Stefan, A., Van Cauwenberghe, J., Rosu, C. M., Stedel, C., Labrou, N. E., Flemetakis, E., et al. (2018). Genetic diversity and structure of Rhizobium leguminosarum populations associated with clover plants are influenced by local environmental variables. Syst. Appl. Microbiol. 41, 251–259. doi: 10.1016/j.syapm.2018.01.007
Teng, Y., Shen, Y., Luo, Y., Sun, X., Sun, M., Fu, D., et al. (2011). Influence of Rhizobium meliloti on phytoremediation of polycyclic aromatic hydrocarbons by alfalfa in an aged contaminated soil. J. Hazard. Mat. 186, 1271–1276. doi: 10.1016/j.jhazmat.2010.11.126
Upadhyay, S. K., Devi, P., Kumar, V., Pathak, H. K., Kumar, P., Rajput, V. D., et al. (2023). Efficient removal of total arsenic (As3+/5+) from contaminated water by novel strategies mediated iron and plant extract activated waste flowers of marigold. Chemosphere 313:137551. doi: 10.1016/j.chemosphere.2022.137551
Upadhyay, S. K., and Edrisi, S. A. (2021). Developing sustainable measures to restore fly-ash contaminated lands: current challenges and future prospects. Land Degrad. Dev. 32, 4817–4831. doi: 10.1002/ldr.4090
Upadhyay, S. K., Rajput, V. D., Kumari, A., Espinosa-Saiz, D., Menendez, E., Minkina, T., et al. (2022). Plant growth-promoting rhizobacteria: a potential bio-asset for restoration of degraded soil and crop productivity with sustainable emerging techniques. Environ. Geochem. Health, 45, 1–24. doi: 10.1007/s10653-022-01433-3
Vinuesa, P., Rojas, J. K., Contreras, M. B., Mahna, S. K., Prasad, B. N., Moe, H., et al. (2008). Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl. Environ. Microbiol. 74, 6987–6996. doi: 10.1128/AEM.00875-08
Wang, J., Cheng, Q., Xue, S., Rajendran, M., Wu, C., and Liao, J. (2018). Pollution characteristics of surface runoff under different restoration types in manganese tailing wasteland. Environ. Sci. Pollut. Res. 25, 9998–10005. doi: 10.1007/s11356-018-1338-2
Wang, H., Pampati, N., McCormick, W. M., and Bhattacharyya, L. (2016). Protein nitrogen determination by Kjeldahl digestion and ion chromatography. J. Pharm. Sci. 105, 1851–1857. doi: 10.1016/j.xphs.2016.03.039
Wani, P. A., and Khan, M. S. (2013). Nickel detoxification and plant growth promotion by multi metal resistant plant growth promoting Rhizobium species RL9. Bull Environ. Contam. Tox 91, 117–124. doi: 10.1007/s00128-013-1002-y
Yu, X., Kang, X., Li, Y., Cui, Y., Tu, W., Shen, T., et al. (2019). Rhizobia population was favoured during in situ phytoremediation of vanadium-titanium magnetite mine tailings dam using Pongamia pinnata. Environ. Pollut. 255:113167. doi: 10.1016/j.envpol.2019.113167
Yu, X., Li, Y., Cui, Y., Liu, R., Li, Y., Chen, Q., et al. (2017a). An indoleacetic acid-producing Ochrobactrum sp. MGJ11 counteracts cadmium effect on soybean by promoting plant growth. J. Appl. Microbiol. 122, 987–996. doi: 10.1111/jam.13379
Yu, X., Li, Y., Li, Y., Xu, C., Cui, Y., Xiang, Q., et al. (2017b). Pongamia pinnata inoculated with Bradyrhizobium liaoningense PZHK1 shows potential for phytoremediation of mine tailings. Appl. Microbiol. Biotechnol. 101, 1739–1751. doi: 10.1007/s00253-016-7996-4
Yu, X., Li, Y., Zhang, C., Liu, H., Liu, J., Zheng, W., et al. (2014). Culturable heavy metal-resistant and plant growth promoting bacteria in V-Ti magnetite mine tailing soil from Panzhihua, China. PLoS ONE 9:e106618. doi: 10.1371/journal.pone.0106618
Yuan, X., Wang, Y., Tang, D., Zhang, X., Zhang, L., and Zhang, H. (2018). Distribution and phytoavailability of potentially toxic metals in different Fe/mg mine tailings. Int. J. Environ. Res. Public Health 15, 2475–2488. doi: 10.3390/ijerph15112475
Zahran, H. H. (1999). Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. R 63, 968–989. doi: 10.1128/MMBR.63.4.968-989.1999
Zeng, Q., Shen, L., Feng, T., and Hao, R. (2022). Investigation of the distribution of heavy metals in the soil of the Dahuangshan mining area of the southern Junggar coalfield, Xinjiang, China. Minerals 12:1332. doi: 10.3390/min12101332
Zevenhuizen, L. P. T. M., Bertocchi, C., and Van Neerven, A. R. W. (1986). Congo red absorption and cellulose synthesis by Rhizobiaceae. Antonie Van Leeuwenhoek 52, 381–386. doi: 10.1007/BF00393465
Zhang, S., Chen, M., Li, T., Xu, X., and Deng, L. (2010). A newly found cadmium accumulator–Malva sinensis Cavan. J. Hazard. Mater. 173, 705–709. doi: 10.1016/j.jhazmat.2009.08.142
Zhang, P., Dumroese, R. K., and Pinto, J. R. (2019). Organic or inorganic nitrogen and rhizobia inoculation provide synergistic growth response of a leguminous forb and tree. Front. Plant Sci. 10, 1–13. doi: 10.3389/fpls.2019.01308
Zhang, Y. M., Li, Y., Chen, W. F., Wang, E. T., Tian, C. F., Li, Q. Q., et al. (2011). Biodiversity and biogeography of rhizobia associated with soybean plants grown in the North China plain. Appl. Environ. Microbiol. 77, 6331–6342. doi: 10.1128/AEM.00542-11
Keywords: rhizobia, Pongamia pinnata , VTM tailings, nitrogen fixation, plant growth promotion, tolerance
Citation: Shen T, Jin R, Yan J, Cheng X, Zeng L, Chen Q, Gu Y, Zou L, Zhao K, Xiang Q, Penttinen P, Ma M, Li S, Zou T and Yu X (2023) Study on diversity, nitrogen-fixing capacity, and heavy metal tolerance of culturable Pongamia pinnata rhizobia in the vanadium-titanium magnetite tailings. Front. Microbiol. 14:1078333. doi: 10.3389/fmicb.2023.1078333
Edited by:
Leticia Barrientos, University of La Frontera, ChileReviewed by:
Sudhir K. Upadhyay, Veer Bahadur Singh Purvanchal University, IndiaHai-Ming Zhao, Jinan University, China
Copyright © 2023 Shen, Jin, Yan, Cheng, Zeng, Chen, Gu, Zou, Zhao, Xiang, Penttinen, Ma, Li, Zou and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiumei Yu, eXV4aXVtZWljb29sQDE2My5jb20=
†These authors have contributed equally to this work
 Tian Shen1†
Tian Shen1† Qiang Chen
Qiang Chen Yunfu Gu
Yunfu Gu Likou Zou
Likou Zou Quanju Xiang
Quanju Xiang Petri Penttinen
Petri Penttinen Menggen Ma
Menggen Ma Shuangcheng Li
Shuangcheng Li Ting Zou
Ting Zou Xiumei Yu
Xiumei Yu