- 1Hainan Key Laboratory of Tropical Microbe Resources, Institute of Tropical Bioscience and Biotechnology, Hainan Institute for Tropical Agricultural Resources, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
- 2College of Biodiversity Conservation, Southwest Forestry University, Kunming, China
- 3College of Plant Protection, Jilin Agricultural University, Changchun, China
- 4Chinese Academy of Tropical Agricultural Sciences, Haikou, China
In this study, we investigated the diversity of diatrypaceous fungi from southeastern Tibet in China. The phylogenetic analyses were carried out based on ITS and β-tubulin sequences of 75 taxa of Diatrypaceae from around the world. Based on a combination of morphological features and molecular evidence, a new genus—Alloeutypa, with three new species—A. milinensis, Diatrype linzhiensis, and Eutypella motuoensis, and a new combination—A. flavovirens, were revealed by the materials in China. Alloeutypa is characterized by stromatal interior olivaceous buff, stromata producing well-developed discrete, and ascospores allantoid, subhyaline. These characteristics separate the new genus from the similar genus Eutypa. Comprehensive morphological descriptions, illustrations, and a phylogenetic tree to show the placement of new taxa are provided. All novelties described herein are morphologically illustrated and phylogeny investigated to better integrate taxa into the higher taxonomic framework and infer their phylogenetic relationships as well as establish new genera and species. Our results indicate that the diatrypaceous fungi harbor higher species diversity in China.
Introduction
Diatrypaceae Nitschke was introduced by Nitschke (1869) with Diatrype Fries as the type genus (Nitschke, 1869; Maharachchikumbura et al., 2015; Senanayake et al., 2015). Diatrypaceous taxa are abundant in Xylariales Nannf., which are widely distributed throughout the world, mostly saprophytic on dead or decaying angiosperms (Carter, 1991; Acero et al., 2004; Trouillas and Gubler, 2004; Trouillas et al., 2010a,b; Mehrabi et al., 2015; Konta et al., 2020; Yang et al., 2022), and some are pathogens or endophytes (Acero et al., 2004; de Errasti et al., 2014; Mehrabi et al., 2019; Konta et al., 2020; Dissanayake et al., 2021). In recent years, some new genera of the family Diatrypaceae have been reported combining morphological characteristics and multi-locus phylogeny (Dayarathne et al. 2016; Senwanna et al. 2017; Phookamsak et al. 2019; Dayarathne et al., 2020b). Hyde et al. (2020) compiled a taxonomic compilation of Sordariomycetes in which 20 genera of the family were listed; subsequently, the classification was followed by Wijayawardene et al. (2020). Dayarathne et al. (2020a) introduced a new genus, Halocryptosphaeria Dayarath., Devadatha, V.V. Sarma & K.D. Hyde saprophytic on decaying wood of Avicennia marina (Forsk.) Vierh. Konta et al. (2020) introduced a new genus, Allodiatrype Konta & K.D. Hyde, which included three new species and one new combination. Subsequently, Paraeutypella L.S. Dissan., J.C. Kang, Wijayaw. & K.D. Hyde, and Pseudodiatrype S.H. Long & Q.R. Li were introduced by Dissanayake et al. (2021) and Long et al. (2021), respectively, based on morphological distinctions and polygenic phylogenetic analyses.
The genus Diatrype Fr. was established by Fries (1849) and typified with D. disciformis (Hoffm.) Fr. The genus was characterized by stromata widely effuse or verrucose, flat or slightly convex, with discoid or sulcate ostioles at the surface, eight-spored and long-stalked asci and hyaline or brownish, allantoid ascospores (Rappaz, 1987; Vasilyeva and Stephenson, 2004; Vasilyeva and Stephenson, 2009; Senanayake et al., 2015). Recently, Zhu et al. (2021) included a new species, and Yang et al. (2022) introduced two new taxa with polysporous asci as members in Diatrype based on the phylogenies inferred from the dataset of ITS and β-tubulin.
Eutypa Tul. & C. Tul. was established by Tulasne and Tulasne (1863) based on E. lata (Pers.) Tul. & C. Tul. The genus is characterized by stromata which are irregular in shape, as confluent bumps, with conspicuous, scattered, roundish to prominent ostioles on the host surface, 8-spore asci with indistinct apical rings, and ascospores allantoid to ellipsoidal, aseptate, and pale yellowish (Hyde et al., 2020). Some species of this genus are disease-causing pathogens, for example, E. lata has been reported to cause dieback and canker in Vitis vinifera (grapevine; Moller and Kasimatis, 1978), Prunus armeniaca (apricots; Carter, 1957), and Prunus salicina (Carter, 1982); E. leptoplaca has been reported to be pathogenic to grapevine (Trouillas and Gubler, 2004).
The genus Eutypella (Nitschke) Sacc., established by Saccardo (1875) with El. cerviculata (Fr.) Sacc. as the type (Saccardo, 1882; Mehrabi et al., 2019; Hyde et al., 2020), which includes 111 morphological species (Species Fungorum 2020), and only 17 species have sequence data (Hyde et al., 2020). Eutypella taxa have a wide host range, and some species are phytopathogens that cause canker, such as El. parasitica R.W. Davidson & R.C. Lorenz causes canker in Acer spp. (Kowalski and Bednarz, 2017), El. microtheca Trouillas, W.M. Pitt & Gubler causes canker in Vitis vinifera, and Prunus spp. (Trouillas et al., 2011; Moyo et al., 2018a,b). The important characteristics of this genus are valsoid configuration stromata, usually comprising host tissues or a mixture of host and fungal tissues, mostly sulcate, sometimes rounded ostioles, converging ostiolar necks, eight-spored asci, and allantoid ascospores (Glawe and Rogers, 1984; Vasilyeva and Stephenson, 2006; Hyde et al., 2020). Rappaz (1987) made a taxonomic revision of Diatrypaceae, in which 76 taxa of Eutypella were described. Afterward, Carmarán et al. (2006) performed a phylogenetic analysis of Diatrypaceae based on ascus morphology and other morphological characteristics and transferred six species from Eutypella to Peroneutypa Berl. Dissanayake et al. (2021) transferred El. citricola Speg. and El. vitis (Schwein.) Ellis & Everh. to Paraeutypella combining morphological and phylogenetic data.
Diatrype, Eutypa, and Eutypella are all unresolved lineages, and phylogenetic studies indicated that the three genera do not form monophyletic groups, even though they clustered within Diatrypaceae (Hyde et al., 2020; Wijayawardene et al., 2020; Long et al., 2021; Yang et al., 2022). In an investigation of the diversity of wood-decaying fungi in southeastern Tibet of China, three undescribed species of diatrypaceous fungi were collected. In order to further the knowledge of species diversity and taxonomy of Diatrypaceae, we carried out complete morphological and molecular phylogenetic studies on these specimens with an emphasis on diatrypaceous fungi. In this study, we introduce a new genus, three new species, and a new combination of Diatrypaceae occurring on decaying wood.
Materials and methods
Specimen collection
The specimens studied in this article were collected from Motuo County and Milin County in Linzhi City of southeastern Tibet, China. In situ photographs of the specimens were taken with a Canon G16 camera (Tokyo, Japan). Fresh specimens were dried and deposited following Yang et al. (2022).
Morphological examination
The studied specimens were macromorphologically observed with the aid of a VHX-600E microscope of Keyence Corporation (Osaka, Japan) up to ×200. The microscopic procedure followed Song et al. (2022). Specimen sections were mounted in water, 10% potassium hydroxide (KOH), and Melzer’s reagent (1.5 g potassium iodide, 0.5 g crystalline iodine, and 22 g chloral hydrate dissolved in 20 ml distilled water), and then microscopic examinations were carried out with an Olympus IX73 inverted fluorescence microscope (Tokyo, Japan) at magnifications up to × 1,000.
DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted from dried specimens using CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd., Beijing, China) and RaPure Plant DNA Mini Kit (Magen Biotechnology) according to the manufacturer’s instructions. The internal transcribed spacer (ITS) region and β-tubulin (TUB2) were amplified with primer pairs ITS5/ITS4 (White et al., 1990) and T1/T22 (O'Donnell and Cigelnik, 1997), respectively. Polymerase chain reaction (PCR) was performed following Song et al. (2022). DNA sequencing was performed at BGI tech, Guangzhou, China. All newly generated sequences in this study including eight ITS sequences and six β-tubulin sequences were deposited in GenBank (Table 1).1
Phylogenetic analyses
Sequencher 4.6 (GeneCodes, Ann Arbor, MI, United States) was used to edit the DNA sequence. Sequences were manually cut and orientation adjusted using BioEdit software (Hall, 1999). Sequences were aligned using the “G-INS-i” strategy at the MAFFT 7 (http://mafft.cbrc.jp/alignment/server/) website and manually corrected using BioEdit. The sequences of Kretzschmaria deusta (Hoffm.) P.M.D. and Xylaria hypoxylon (L.) Grev. were obtained from GenBank as out-groups.
Maximum likelihood analyses were performed in raxmlGUI 2.0 selecting ML + rapid bootstrap analysis and GTRGAMMA+G as the surrogate model (Ma et al., 2022; Song et al., 2022). Branch support (BS) for ML analysis was determined by 1,000 bootstrap replicates. MrModeltest 2.3 (Nylander, 2004) was used to determine the best-fit evolution model for each dataset for Bayesian inference (BI). Bayesian inference was calculated with MrBayes 3.1.2 with a general time reversible (GTR + I + G) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist and Huelsenbeck, 2003). Four simultaneous Markov chains were run for 2000000 generations, and every 100 generations were sampled as a tree. The first one-fourth generations were discarded as burn-in. The majority rule consensus tree of all remaining trees is computed. Branches were considered as significantly supported if they received maximum likelihood bootstrap (BS) ≥ 70% and Bayesian posterior probabilities (BPP) ≥ 0.95.
Results
Molecular phylogeny
The contribution of the molecular phylogenetic tree based on 197 sequences of two DNA loci (116 ITS and 81 β-tubulin sequences) was composed of 116 samples representing 75 strains of Diatrypaceae (Table 1). The concatenated dataset had an aligned length of 1936 characteristics, including gaps (609 for ITS and 1,327 for TUB2). Bayesian obtained a topology similar to ML, with an average standard deviation of split frequencies = 0.007766 (BI). Only the ML tree is provided in Figure 1 with the likelihood bootstrap values (≥ 70%, before the slash) and Bayesian posterior probabilities (≥ 0.95, behind the slash) labeled along the branches.

Figure 1. Phylogram generated from maximum likelihood (RA × ML) analyses, based on ITS-β-tubulin matrix. Branches are labeled with maximum likelihood bootstrap ≥ 70% and Bayesian posterior probabilities ≥ 0.95. Ex-type strains are in bold. Newly generated strains are in blue. Bold branches indicate that the length has been cut in half.
The topology of the phylogenetic tree is similar to those in previous studies (Konta et al., 2020; Zhu et al., 2021). For the in-groups, species from 18 genera were distributed in 24 clades: including 18 main clades, Diatrypella sensu stricto, Neoeutypella, Pseudodiatrype, Allodiatrype, Halodiatrype, Pedumispora, Diatrypella 1, Eutypa sensu stricto/Cryptosphaeria 1, Alloeutypa, Diatrype sensu stricto, Cryptosphaeria 2, Eutypa 1, Eutypella sensu stricto/Anthostoma, Paraeutypella/Allocryptovalsa/Eutypella 1, Peroneutypa, Quaternaria, Cryptovalsa, Monosporascus, and six incertae sedis clades (Diatrype enteroxantha, D. lancangensis, D. lijiangensis, D. palmicola, D. whitmanensis, and Eutypella parasitica). Allodiatrype, Alloeutypa, Monosporascus, Neoeutypella, Paraeutypella, Peroneutypa, and Pseudodiatrype were shown to be monophyletic and well-supported in our tree. Halodiatrype and Pedumipora, Cryptovalsa and Quaternaria formed a strongly supported claded respectively. Anthostoma decipiens (JL567 and CD) grouped together is sister to Eutypella sensu stricto with strong support (ML/BI = 100/1). Eutypella leprosa, El. microtheca, and several species from Paraeutypella and Allocryptovalsa formed a large clade with relatively strong support. The new genus Alloeutypa included two species, A. milinensis and A. flavovirens, formed a distinct clade. The other two new species—Diatrype linzhiensis and Eutypella motuoensis, formed distinct lineages in the tree. Some confused taxa, for example, Diatrype enteroxantha, D. lancangensis, D. lijiangensis, D. palmicola, D. whitmanensis, and Eutypella parasitica, formed a single clade or mixed with other genera.
Taxonomy
Alloeutypa Hai X. Ma, Z.E. Yang & Y. Li, gen. Nov.
MycoBank: 846109.
Etymology: referring to the morphological resemblance to Eutypa.
Descriptions—Saprobic on dead angiosperm branch. Sexual morph: Stromata scattered on the host, pustulate, solitary or aggregated, superficial, irregularly shaped or oblong to strip, upper surface flat to slightly curved; surface black, with numerous ascomata in a single stroma. Endostroma consists of outer layer of black, small, dense, thin parenchymal cells and inner layer of olivaceous buff, large, loose parenchymal cells. Ostioles opening to outer surface, appearing as black spots, separately, papillate or apapillate. Perithecium globose to subglobose, individual ostiole with a neck. Peridium composed of outer layer of dark brown to brown, thin-walled cells, inner layer of hyaline thin-walled cells. Paraphyses elongate, hyaline, long, filiform, unbranched, septate, guttulate. Asci eight-spored, unitunicate, clavate, long-stalked, apically rounded. Ascospores irregularly arranged, allantoid, aseptate, slightly curved, subhyaline to yellowish, smooth-walled, several oil droplets in each end.
Type species: Alloeutypa milinensis Hai X. Ma, Z.E. Yang & Y. Li.
Notes: In the phylogenetic tree (Figure 1), Eutypa species are distributed in two distinct clades Eutypa sensu stricto and Eutypa 1, indicating that the genus is polyphyletic. The type species, E. lata clusters in Eutypa clade1 which can be regarded as Eutypa sensu stricto. However, it is hard to justify Eutypa 1 as a new genus without examining old types of species and identified fresh collections with molecular data.
The sexual morphology of Eutypa sensu stricto (as Eutypa taxonomic species 2) comprises wide-spreading stromata that embedded in decorticated wood or bark, usually poorly developed with ill-defined margins, surface black, interior white or blackened, eight-spore asci spindle-shaped, long-stipitate, ascospores allantoid, subolivaceous (Glawe and Rogers, 1984). The Chinese collection in this study is clearly different from members of Eutypa sensu stricto based on the green interior of the stromata, discrete, Diatrype-like.
Based on the morpho-molecular differences, the new genus Alloeutypa is introduced to accommodate Alloeutypa milinensis. Alloeutypa is typified by A. milinensis, which was found on dead branches of angiosperm plant from southeastern Tibet in China. Eutypa flavovirens resembles A. milinensis in having well-developed discrete, Diatrype-like stromata with yellow-green to olive-green interior tissue, asci spindle-shaped, long-stipitate, ascospores allantoid, and subhyaline to subolivaceous. The phylogenetic analyses based on ITS and β-tubulin sequence data also supported Alloeutypa as a monophyletic genus in the Diatrypaceae, and A. milinensis and A. flavovivens as separate lineages within Alloeutypa. Thus, based on morphological evidence and phylogenetic analyses, we accommodate Alloeutypa as a new genus with A. milinensis as the type, and E. flavovirens was transferred to Alloeutypa as A. flavovirens comb. nov.
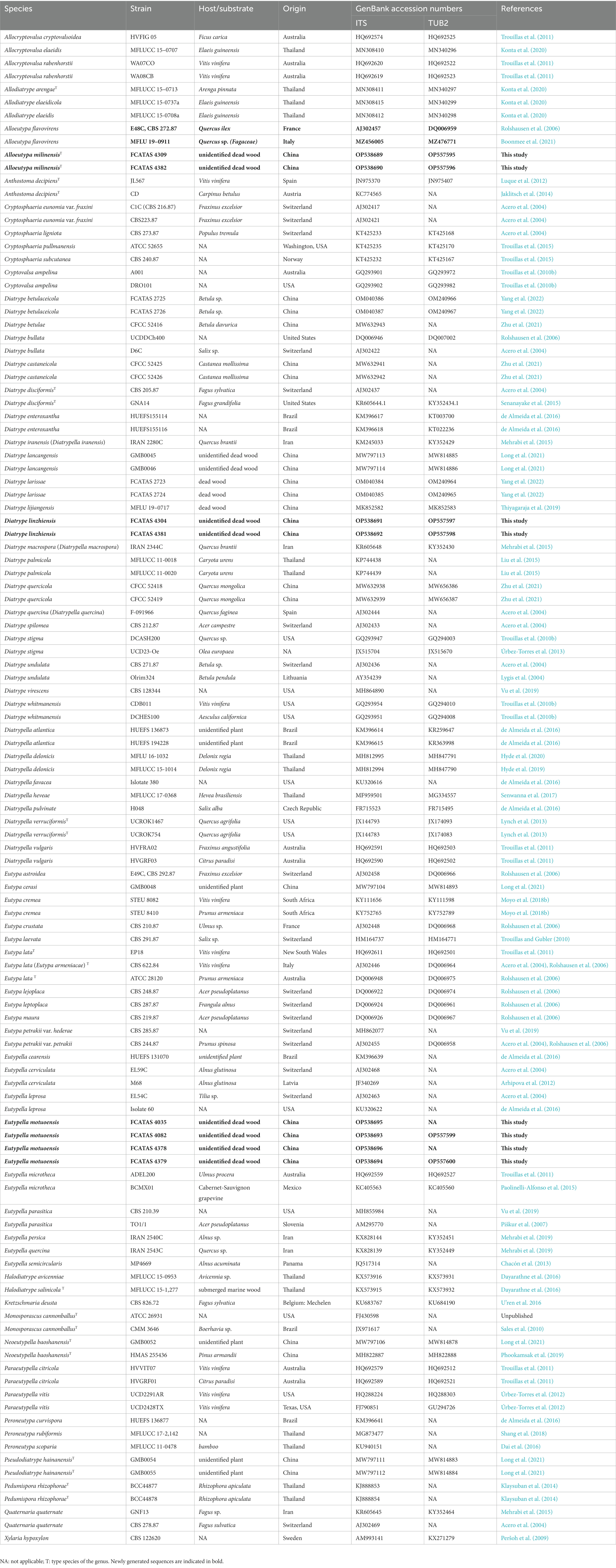
Alloeutypa milinensis Hai X. Ma, Z.E. Yang & Y. Li, sp. nov. (Figure 2).
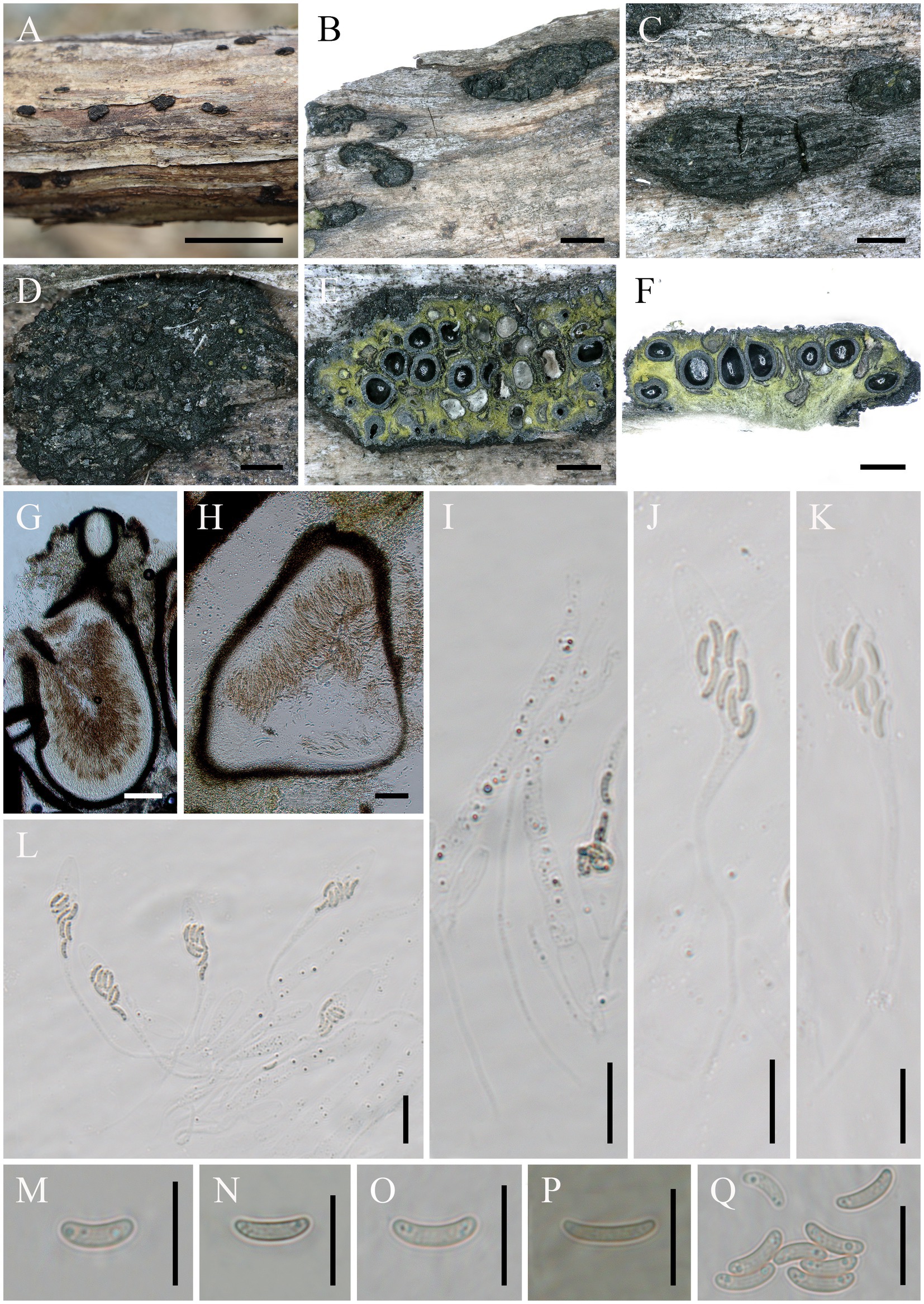
Figure 2. Alloeutypa milinensis (FCATAS 4309, Holotype). (A–D) Stromata on substrate. (E) Cross section of a stroma. (F,G) Vertical section through stroma showing ostiole and perithecia. (H) Peridium. (I) Paraphyses. (J–L) Asci. (M–Q) Ascospores. Scale bars: (A) = 15 mm; (B) = 2 mm; (C) = 1 mm; (D–F) = 500 μm; (G,H) = 100 μm; (I–L) = 20 μm; (M–Q) = 10 μm.
MycoBank: MB 846111.
Type: China. Tibet Autonomous Region, Linzhi City, Milin County, Pai Town, 29°30′2′ N, 94°50′26′ E, alt. 998 m, saprobic on dead branch, 7 October 2021, Haixia Ma, FCATAS 4309 (holotype).
Etymology: referring to the locality (Milin County) of the type specimens.
Descriptions: Saprobic on dead branches of unidentified plant. Sexual morph: Stromata scattered on the host, pustulate, solitary, superficial, 2–7.3 mm long × 0.9–2.2 mm broad (x̄ = 3.6 × 1.5 mm, n = 20), oblong to strip, upper surface flat to slightly curved; surface black with 14–50 perithecia immersed in stroma. Endostroma consists of outer layer of black, small, dense, thin parenchymal cells and inner layer of olivaceous buff, large, loose parenchymal cells, near base, whitish yellow-green. Ostioles opening to outer surface, appearing as black spots, separately, papillate or apapillate. Perithecium globose to subglobose, 261.2–512.2 μm high × 245.7–443.3 μm diam (x̄ = 383.8 × 334.1 μm, n = 30), individual ostiole with a neck. Peridium composed of outer layer of dark brown to brown, thin-walled cells, inner layer of hyaline thin-walled cells. Paraphyses elongate, hyaline, long, filiform, unbranched, septate, guttulate. Asci 97–194 × 7.5–16.7 μm (x̄ = 132.8 × 11.3 μm, n = 50), eight-spored, unitunicate, clavate, long-stalked (30–131.5 μm), apically rounded. Ascospores 6.6–10.1 × 1.7–2.6 μm (x̄ = 8.5 × 2.1 μm, n = 50), overlapping, allantoid, aseptate, slightly curved, subhyaline, smooth-walled, usually with two oil droplets.
Asexual morph: Undetermined.
Additional specimen examined.—China. Tibet Autonomous Region, Linzhi City, Milin County, Pai Town, 29°29′57′ N, 94°50′29′ E, alt. 996 m, saprobic on dead branch, 7 October 2021, Haixia Ma, FCATAS 4382.
Note: Alloeutypa milinensis grouped with A. flavovirens (E. flavovirens) based on the combined ITS + β-tubulin sequence data. In recent years, A. flavovirens (E. flavovirens) has been successively recorded in Thailand, India, and Italy, and the specimens from the three regions have some differences in morphology. Morphologically, the specimens of A. flavovirens (E. flavovirens) in Thailand differ from A. milinensis in smaller stromata (1–1.5 mm wide) and smaller perithecium diam (120–210 μm diam; Senanayake et al., 2015); the specimens from India differ by the smaller perithecium (212.5–396 × 184.6–363 μm), fewer perithecium in a stroma (2–12), and shorter ascus (75–110 × 6.1–8.8 μm; Niranjan et al., 2018); the specimen from Italy differs in having gregarious, aggregates to discrete stromata, smaller in size (0.7–1 mm diam), and smaller ascus (80–120 × 8–10 μm; Boonmee et al., 2021).
The sequence comparison showed that there are 97.22 and 95%, respectively, similarities in ITS and TUB2 between A. milinensis from China (FCATAS 4309) and A. flavovirens (E. flavovirens) from Italy (MFLU19-0911), while 97.13 and 94.12 between A. milinensis from China (FCATAS 4309) and A. flavovirens (E. flavovirens) from France (E48C, CBS 272.87). Unfortunately, TUB2 sequences of the Indian and Thailand collections are not available in GenBank. However, the ITS sequence comparison showed that there are both 92% similarities between A. milinensis from China (FCATAS 4309) and A. flavovirens (E. flavovirens) from India (PUFNI 310) and Thailand (MFLUCC 13-0625). Therefore, we described the Chinese material as a new species.
Alloeutypa flavovirens: (Pers.) Hai X. Ma & Z.E. Yang, comb. nov.
MycoBank: 846128.
Synonyms: Sphaeria flavovirens Pers., Syn. meth. Fung. (Göttingen) 1: 22, 1801. Diatrype flavovirens (Pers.) Fr., Summa veg. Scand., Sectio Post. (Stockholm): 385, 1849. Eutypa flavovirens (Pers.) Tul. & C. Tul., Select. fung. Carpol. (Paris) 2: 57, 1863.
Notes: Alloeutypa flavovirens is one of the most common fungi and found throughout the world and appears to have a wide host range (Glawe and Rogers, 1982, 1984; Rappaz, 1987). It is characterized by having yellow-greenish stromatic tissues, spindle-shaped asci with refractive apical invaginations, allantoid ascospores subhyaline to subolivaceous (Glawe and Rogers, 1984). It is most similar to A. milinensis in having the green interior of the stromata. There are no sequence data for the type of A. flavovirens, but there are two putatively named collections, CBS 272.87 and MFLU 19-0911, from France and Italy, respectively (Rolshausen et al. 2006; Boonmee et al., 2021). Based on the morphological and molecular analyses that the two collections were the records of A. flavovirens (E. flavovirens) by Senanayake et al. (2015) and Boonmee et al. (2021), in our phylogenetic tree, the two strains of A. flavovirens (E. flavovirens) clustered together with A. milinensis with strong support (95% ML, 1.00 BYPP; Figure 1) and maybe the same genus because of its location. However, morphological differences on size of stromata, perithecium, and ascus can distinguish the two species from each other (Senanayake et al., 2015; Boonmee et al., 2021).
Diatrype linzhiensis: Hai X. Ma & Z.E. Yang, sp. nov. (Figure 3).
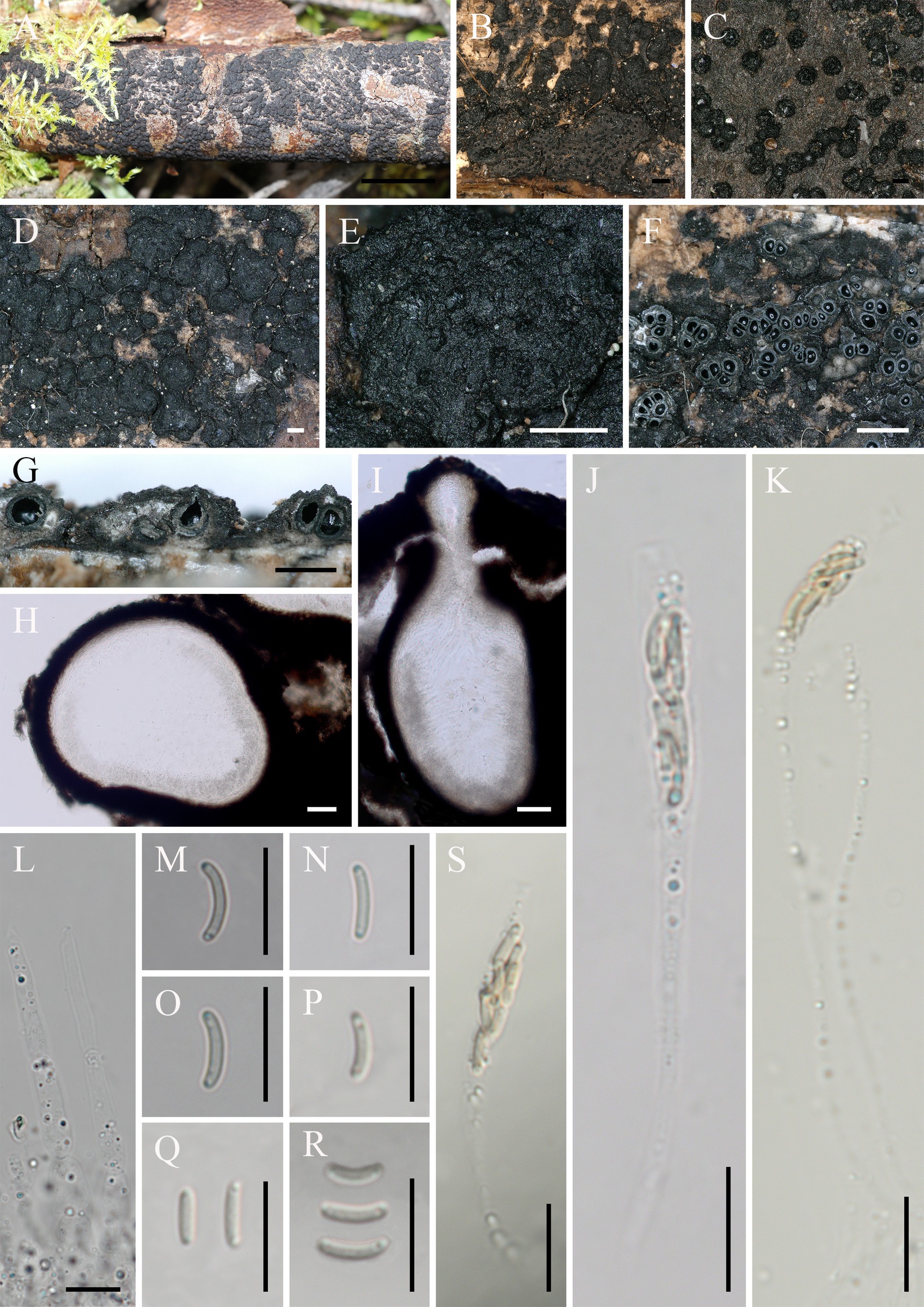
Figure 3. Diatrype linzhiensis (FCATAS 4304, Holotype). (A–E) Stromata on substrate. (F) Cross section of a stroma. (G,I) Vertical section through stroma showing ostiole and perithecia. (H) Peridium. (L) Paraphyses. (J,K,S) Asci. (M–R) Ascospores. Scale bars: (A) = 15 mm; (B) = 500 μm; (C) = 100 μm; (D,E,G) = 500 μm; (F) = 1 mm; (H,I) = 50 μm; (J–S) = 10 μm.
MycoBank: MB 846129.
Type: China. Tibet Autonomous Region, Linzhi City, Milin County, Pai Town, 29°30′7′ N, 94°50′33′ E, alt. 1,004 m, saprobic on decaying branches of Betula L., 7 October 2021, Haixia Ma, FCATAS 4304 (holotype).
Etymology: referring to the locality (Linzhi City) of the type specimens.
Descriptions: Saprobic on decaying branches of Betula L. Sexual morph: Stromata scattered on the host, irregular in shape, solitary to gregarious, form patchy clumps, cushion-like, superficial, upper surface nearly flat; surface black, with punctiform cone-shaped and sulcate ostioles scattered at surface. Endostroma consists of outer black, small, dense, and an inner layer of white to pale olivaceous gray, large. Perithecium immersed in stroma, globose to subglobose, 222–385 μm high × 164–367 μm diam (x̄ = 294 × 269.6 μm, n = 30), with a neck, cylindrical. Peridium composed of outer layer of brown, thin-walled cells, inner layer of hyaline thin-walled cells. Ostiole opening separately, papillate, black. Paraphyses elongate, hyaline, filiform, branched, septate, guttulate. Asci 52–134 × 4.1–7.9 μm (x̄ = 68.2 × 6 μm, n = 50), 19–40 × 4.1–7.9 μm in spore bearing part, eight-spored, unitunicate, clavate, long-stalked (27–67 μm), apically flat. Ascospores 5–7.8 × 1–1.4 μm (x̄ = 6.1 × 1.2 μm, n = 50), overlapping, allantoid, aseptate, slightly curved, yellowish, rounded ends with two guttules, smooth-walled.
Asexual morph: Undetermined.
Additional specimen examined: China. Tibet Autonomous Region, Linzhi City, Milin County, Pai Town, 29°30′7′ N, 94°50′34′ E, alt. 990 m, saprobic on decaying branches of Betula, 7 October 2021, Haixia Ma, FCATAS 4381.
Note: Diatrype linzhiensis is characterized by cushion-like stromata superficial, solitary to gregarious, form patchy clumps, flat, black, globose to subglobose perithecium with a neck immersed in stroma, hyaline paraphyses long filiform, branched, septate, eight-spored asci with apically flat, yellowish ascospores allantoid to slightly curved. The new species was found on branch of Betula sp., D. albopruinosa (Schwein.) Cooke, D. betulae H.Y. Zhu & X.L. Fan, D. oregonensis (Wehm.) Rappaz and D. stigma (Hoffm.) Fr. were also reported on Betula sp. (Tiffany and Gilman, 1965; Rappaz, 1987; Trouillas et al., 2010b; Vasilyeva and Ma, 2014; Zhu et al., 2021). However, D. albopruinosa differs in its larger ascus (40–60 × 10–15 μm) and ascospores (12–15 μm; Vasilyeva and Ma, 2014); D. betulae has no sexual morph to be observed (Zhu et al., 2021); D. oregonensis differs from D. linzhiensis by larger ascus (50–65 × 6–9.5 μm) and ascospores (10–12 × 2–2.5 μm; Trouillas et al., 2010b); D. stigma differs in its stromata widely effused and smaller perithecia (150–200 μm; Vasilyeva and Ma, 2014). In the phylogenetic tree (Figure 1), D. linzhiensis and D. undulata (Pers.) Fr. formed a relatively strongly supported lineage. Morphologically, D. undulata differs from D. linzhiensis by having dark brown, widely effused stromata, with small stellate ostioles, surrounded by a black line within the substrate, smaller perithecia (150–200 μm vs. 222–384 μm; Vasilyeva and Ma, 2014).
Eutypella motuoensis Hai X. Ma & Z.E. Yang, sp. nov. (Figure 4).
Figure 4. Eutypella motuoensis (FCATAS 4082, Holotype). (A–D) Stromata on substrate. (E) Cross section of a stroma. (F,G) Vertical section through stroma showing ostiole and perithecia. (H) Peridium. (I,J) Paraphyses. (K–O) Asci. (P–V) Ascospores. Scale bars: (A) = 15 mm; (B) = 1 mm; (C–F) = 500 μm; (G) = 100 μm; (H) = 50 μm; (I–V) = 10 μm.
MycoBank: MB 846130.
Type: China. Tibet Autonomous Region, Motuo County, 29°19′26′N, 95°20′10′E, alt. 996 m, saprobic on the bark of dead branch, 26 September 2021, Haixia Ma, FCATAS 4082 (holotype).
Etymology: referring to the holotype locality of species in Motuo county.
Descriptions: Saprobic on dead branches of an unidentified plant. Sexual morph: Stromata scattered on the host, erumpent through bark, semi-immersed, 4–38 mm long × 3–9 mm broad, (x̄ = 16.5 × 6.1 mm, n = 20), 0.9–1.4 mm thick, irregular in shape, widely effused, upper surface nearly flat; surface saffron to black, cylindrical protrusions of ostioles cover the surface, 227–540 μm high × 281–391 μm diam (x̄ = 331 × 325 μm, n = 20). Endostroma consists of outer black, small, dense, and an inner layer of salmon, large. Perithecium immersed in stroma, globose to subglobose, 422–629 μm high × 351–645 μm diam (x̄ = 532.8 × 495.7 μm, n = 30), with a neck, cylindrical. Peridium composed of outer layer of brown, thin-walled cells, inner layer of hyaline thin-walled cells. Ostiole opening separately, black. Paraphyses elongate, hyaline, filiform, branched, septate, guttulate. Asci 60–105 × 4.9–6.9 μm (x̄ = 73.1 × 5.9 μm, n = 50), eight-spored, unitunicate, subcylindrical, long-stalked (25–74 μm), with rounded apex. Ascospores 6.3–10.6 × 2–2.7 μm (x̄ = 8.4 × 2.3 μm, n = 50), overlapping, allantoid to semicircular, sometimes almost forming a circle, aseptate, subhyaline to yellowish, usually with guttules, smooth-walled.
Asexual morph: Undetermined.
Additional specimen examined: China. Tibet Autonomous Region, Motuo County, 29°19′26′N, 95°20′10′E, alt. 1,004 m, saprobic on the bark of dead branch, 26 September 2021, Haixia Ma, FCATAS 4379; Motuo County, Yarlung Zangbo River, the large bend of Linduo, 29°19′38′N, 95°20′29′E, alt. 781 m, saprobic on the bark of dead branch, 24 September 2021, Haixia Ma, FCATAS 4035, FCATAS 4378.
Note: Eutypella motuoensis differs from most known species of Eutypella and related genera by cylindrical protrusions of ostioles cover the surface and subhyaline to yellowish, semicircular to almost circular allantoic ascospores. Morphologically, Eutypella semicircularis S. Chacón & M. Piepenbr., Eutypa crustata (Fr.) Sacc., Echinomyces obesa (Syd. & P. Syd.) Rappaz, and Diatrype falcata (Syd. & P. Syd.) Sacc. are similar to El. motuoensis by sharing allantoid to semicircular ascospores. However, El. semicircularis differs in its mature urn-shaped ascus and smaller reddish-brown ascospores (4.5–7(−11) × 1.5–2(−2.5) μm; Chacón et al. 2013); Eutypa crustata differs from El. motuoensis by having smaller perithecia (300–450 μm) and smaller ascus (20–35 × 6–8 μm; Rappaz, 1987); Echinomyces obesa is separated from El. motuoensis by smaller ascus (10–15 × 4–5 μm) and ascospores (3.5–7.5 × 1.2–1.5 μm; Rappaz, 1987); Diatrype falcata differs in its less prominent ostioles, smaller perithecia (250–350 μm), smaller ascus (20–25 × 4–5 μm), and ascospores (5.8–7.5 × 1.2–1.5 μm; Rappaz, 1987). In the phylogenetic tree, El. motuoensis is sister to El. persica Mehrabi, Asgari & Hemmati, though their relationship is not strongly supported. Morphologically, El. persica differs from El. motuoensis by its allantoid, slightly curved, hyaline, and smaller ascospores (5–7 × 1.5–2.5 μm; Mehrabi et al., 2019).
Discussion
The species diversity, taxonomy, and phylogeny of diatrypaceous fungi were intensively studied recently by many authors, and a large number of new taxa were described (Mehrabi et al., 2019; Konta et al., 2020; Dayarathne et al., 2020a,b; Dissanayake et al., 2021; Long et al., 2021; Peng et al., 2021; Zhu et al., 2021; Yang et al., 2022). This study furthers the knowledge of these fungi with the addition of a new genus, three new species, and a new combination in the Diatrypaceae. Morpho-molecular analyses confirmed the introduction of the newly described genus, Alloeutypa, for accommodating the new species A. milinensis and the new combination A. flavovirens. Our phylogenetic analyses on the species of Diatrype and Eutypella also confirmed that they are all polyphyletic genera, agreeing with the previous studies (Acero et al., 2004; Trouillas et al., 2011; Mehrabi et al., 2019; Konta et al., 2020; Dayarathne et al., 2020a,b; Long et al., 2021; Zhu et al., 2021).
In our phylogenetic trees, most taxa of Diatrype (Diatrype sensu stricto) formed a main clade with high support values (Figure 1), including D. disciformis, the type species of the genus. The new species, D. linzhiensis, from China also was included in this group. Diatrype enteroxantha (Sacc.) Berl. and D. whitmanensis J.D. Rogers & Glawe both formed a single clade in phylogenetic trees but the studied sequences of the two species are not their type specimens. While other taxa, for D. lancangensis S.H. Long & Q.R. Li, D. lijiangensis Thiyagaraja & Wanasinghe, and D. palmicola Jian K. Liu & K.D. Hyde formed a single clade or mixed with clades of other genera, and there are no distinct morphological characteristics to divide them into small genera at present.
In the molecular analyses of ITS and β-tubulin sequences performed by Zhu et al. (2021), Eutypa flavovirens (Pers.) Tul. & C. Tul. grouped in a clade with two Cryptosphaeria taxa by no supported values. In our analyses (Figure 1), E. flavovirens appeared in a strongly supported clade along with the new species A. milinensis, suggesting the new species is closely related to E. flavovirens. The novel diatrypacous genus, Alloeutypa, is therefore introduced in the present study and will help to stabilize the classification of Diatrypaceae. However, the other species of Eutypa formed two distinct clades in the family and the generic position remains unresolved, which may need to be studied in the future.
The Eutypella species analyzed were distributed in two main separate clades (El sensu stricto and El 1), one mixed with taxa of Paraeutypella and Allocryptovalsa (El 1) and another related to a species of Anthostoma (Eutypella sensu stricto). Eutypella motuoensis formed a sister subclade with El. persica with no support values.
The molecular evidence has brought significant changes and increased our understanding of the taxonomy and phylogeny of Diatrypaceae. However, the phylogenetic trees show that the classification of these diatrypaceous fungi in many genera is confusing. To determine more important and useful morphological characteristics for distinguishing those species and to resolve infra-genera and infra-specific phylogeny, more specimens of these species from their original regions and more taxa from other regions should be included in future phylogenetic studies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, ITS (OP538689–OP538696) and TUB2 (OP557595–OP557600) https://www.mycobank.org/page/Home/MycoBank, MycoBank (846109, 846111, 846128, 846128–846130).
Author contributions
Z-KS, A-HZ, ZQ, and H-XM prepared the samples. Z-EY made morphological examinations and performed molecular sequencing. A-HZ performed phylogenetic analyses. Z-EY and H-XM wrote the manuscript. A-HZ revised the language of the text. H-XM conceived and supervised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by the National Natural Science Foundation of China (No. 31770023 and 31972848) and the Central Public-Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630032022001, 1630052022003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Acero, F. J., González, V., Sánchez-Ballesteros, J., Rubio, V., Checa, J., Bills, G. F., et al. (2004). Molecular phylogenetic studies on the Diatrypaceae based on rDNA-ITS sequences. Mycologia 96, 249–259. doi: 10.2307/3762061
Arhipova, N., Gaitnieks, T., Donis, J., Stenlid, J., and Vasaitis, R. (2012). Heart-rot and associated fungi in Alnus glutinosa stands in Latvia. Scand. J. Forest. Res. 27, 327–336. doi: 10.1080/02827581.2012.670727
Boonmee, S., Wanasinghe, D. N., Calabon, M. S., Huanraluek, N., Chandrasiri, S. K. U., Jones, G. E. B., et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 111, 1–335. doi: 10.1007/s13225-021-00489-3
Carmarán, C. C., Romero, A. I., and Giussani, L. M. (2006). An approach towards a new phylogenetic classification in Diatrypaceae. Fungal Divers. 23, 67–87.
Carter, M. V. (1957). Eutypa armeniacae Hansf. & Carter, sp. nov., and airborne vascular pathogen of Prunus armeniaca L. in southern Australia. Aust. J. Bot. 5, 21–35. doi: 10.1071/BT9570021
Carter, M. V. (1982). Additional hosts of Eutypa armeniacae in Australia. Australas. Plant Pathol. 11, 46–48. doi: 10.1071/APP9820046
Carter, M. V. (1991). The status of Eutypa lata as a pathogen. Monograph. Phytopathological paper no. 32. Commonwealth Agricultural Bureau, International Mycological Institute, UK
Chacón, S., Dorge, D., Weisenborn, J., and Piepenbring, M. (2013). A new species and a new record of Diatrypaceae from Panama. Mycologia 105, 681–688. doi: 10.3852/12-131
Dai, D. Q., Phookamsak, R., Wijayawardene, N. N., Li, W. J., Bhat, D. J., Xu, J. C., et al. (2016). Bambusicolous fungi. Fungal Divers. 82, 1–105.
Dayarathne, M. C., Jones, E. B. G., Maharachchikumbura, S. S. N., Devadatha, B., Sarma, V. V., Khongphinitbunjong, K., et al. (2020a). Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11, 1–188. doi: 10.5943/mycosphere/11/1/1
Dayarathne, M. C., Phookamsak, R., Hyde, K. D., Manawasinghe, I. S., Toanun, C., and Jones, E. B. G. (2016). Halodiatrype, a novel diatrypaceous genus from mangroves with H. salinicola and H. avicenniae spp. nov. Mycosphere 7, 612–627. doi: 10.5943/mycosphere/7/5/7
Dayarathne, M. C., Wanasinghe, D. N., Devadatha, B., Abeywickrama, P., Jones, E. B. G., Chomnunti, P., et al. (2020b). Modern taxonomic approaches to identifying diatrypaceous fungi from marine habitats, with a novel genus Halocryptovalsa Dayarathne & K.D.Hyde. Gen. Nov. Cryptogamie Mycol. 41, 21–67. doi: 10.5252/cryptogamie-mycologie2020v41a3
de Almeida, D. A. C., Gusmão, L. F. P., and Miller, A. N. (2016). Taxonomy and molecular phylogeny of Diatrypaceae (Ascomycota, Xylariales) species from the Brazilian semi-arid region, including four new species. Mycol. Prog. 15, 1–27. doi: 10.1007/s11557-016-1194-8
de Errasti, A., Novas, M. V., and Carmarán, C. C. (2014). Plant–fungal association in trees, insights into changes in ecological strategies of Peroneutypa scoparia (Diatrypaceae). Flora 209, 704–710. doi: 10.1016/j.flora.2014.07.006
Dissanayake, L. S., Wijayawardene, N. N., Dayarathne, M. C., Samarakoon, M. C., Dai, D. Q., Hyde, K. D., et al. (2021). Paraeutypella guizhouensis gen. Et sp. nov. and Diatrypella longiasca sp. nov. (Diatrypaceae) from China. Biodivers. Data J. 9:e63864. doi: 10.3897/BDJ.9.e63864
Glawe, D. A., and Rogers, J. D. (1982). Observations on the anamorphs of six species of Diatrype and Diatrypella. Can. J. Bot. 60, 245–251.
Glawe, D. A., and Rogers, J. D. (1984). Diatrypaceae in the Pacific northwest. Mycotaxon 20:e63864, 401–460.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 734, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hyde, K. D., Norphanphoun, C., Maharachchikumbura, S. S. N., Bhat, D. J., Jones, E. B. G., Bundhun, D., et al. (2020). Refined families of Sordariomycetes. Mycosphere 11, 305–1059. doi: 10.5943/mycosphere/11/1/7
Hyde, K. D., Tennakoon, D. S., Jeewon, R., Bhat, D. J., Maharachchikumbura, S. S. N., Rossi, W., et al. (2019). Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 96, 1–242. doi: 10.1007/s13225-019-00429-2
Jaklitsch, W. M., Fournier, J., Rogers, J. D., and Voglmayr, H. (2014). Phylogenetic and taxonomic revision of Lopadostoma. Persoonia Mol. Phylogeny Evol. Fungi 32, 52–82. doi: 10.3767/003158514X679272
Klaysuban, A., Sakayaroj, J., and Jones, E. G. (2014). An additional marine fungal lineage in the Diatrypaceae, Xylariales: Pedumispora rhizophorae. Bot. Mar. 57, 413–420. doi: 10.1515/bot-2014-0017
Konta, S., Maharachchikumbura, S. S. N., Senanayake, I. C., McKenzie, E. H. C., Stadler, M., Boonmee, S., et al. (2020). A new genus Allodiatrype, fve new species and a new host record of diatrypaceous fungi from palms (Arecaceae). Mycosphere 11, 239–268. doi: 10.5943/mycosphere/11/1/7
Kowalski, T., and Bednarz, B. (2017). Eutypella parasitica–nowy patogen powodujący raki na pniach klonów (Acer spp.) w Polsce. Sylwan 161, 630–638.
Liu, J. K., Hyde, K. D., Jones, E. B. G., Ariyawansa, H. A., Bhat, D. J., Boonmee, S., et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72, 1–197. doi: 10.1007/s13225-015-0324-y
Long, S. H., Liu, L. L., Pi, Y. H., Wu, Y. P., Lin, Y., Zhang, X., et al. (2021). New contributions to Diatrypaceae from karst areas in China. Mycokeys 83, 1–37. doi: 10.3897/mycokeys.83.68926
Luque, J., Garcia-Figueres, F., Legorburu, F. J., Muruamendiaraz, A., Armengol, J., and Trouillas, F. P. (2012). Species of Diatrypaceae associated with grapevine trunk diseases in Eastern Spain. Phytopathol. Mediterr. 51, 528–540.
Lygis, V., Vasiliauskas, R., and Stenlid, J. (2004). Planting Betula pendula on pine sites infested by Heterobasidion annosum: disease transfer, silvicultural evaluation, and community of wood-inhabiting fungi. Can. J. Forest. Res. 34, 120–130.
Lynch, S. C., Eskalen, A., Zambino, P. J., Mayorquin, J. S., and Wang, D. H. (2013). Identifcation and pathogenicity of Botryosphaeriaceae species associated with coast live oak (Quercus agrifolia) decline in southern California. Mycologia 105, 125–140. doi: 10.3852/12-047
Ma, H. X., Song, Z., Pan, X., Li, Y., Yang, Z., and Qu, Z. (2022). Multi-gene phylogeny and taxonomy of Hypoxylon (Hypoxylaceae, Ascomycota) from China. Diversity 14:37. doi: 10.3390/d14010037
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., and McKenzie, E. H. C. (2015). Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 72, 199–301. doi: 10.1007/s13225-015-0331-z
Mehrabi, M., Asgari, B., and Hemmati, R. (2019). Two new species of Eutypella and a new combination in the genus Peroneutypa (Diatrypaceae). Mycol. Prog. 18, 1057–1069. doi: 10.1007/s11557-019-01503-4
Mehrabi, M., Hemmati, R., Vasilyeva, L. N., and Trouillas, F. P. (2015). A new species and a new record of Diatrypaceae from Iran. Mycosphere 6, 60–68. doi: 10.5943/mycosphere/6/1
Moller, W. J., and Kasimatis, A. N. (1978). Dieback of grapevine caused by Eutypa armeniacae. Plant Dis. Rep. 62, 254–258.
Moyo, P., Damm, U., Mostert, L., and Halleen, F. (2018a). Eutypa, Eutypella, and Cryptovalsa species (Diatrypaceae) associated with Prunus species in South Africa. Plant Dis. 102, 1402–1409. doi: 10.1094/PDIS-11-17-1696-RE
Moyo, P., Mostert, L., Spies, C. F., Damm, U., and Hallen, F. (2018b). Diversity of Diatrypaceae species associated with dieback of grapevines in South Africa, with the description of Eutypa cremea sp. nov. Plant Dis. 102, 220–230. doi: 10.1094/PDIS-05-17-0738-RE
Niranjan, M., Tiwari, S., Baghela, A., and Sarma, V. V. (2018). New records of Ascomycetous fungi from Andaman Islands, India and their molecular sequence data. Curr. Res. Environ. Appl. Mycol. 8, 331–350. doi: 10.5943/cream/8/3/5
Nitschke, T. R. J. (1869). Grundlage eines Systems der Pyrenomyceten. Verhandlungen des Naturhistorischen Vereins der Preussischen Rheinlande. Westfalens und des Regierungsbezirks Osnabrück 262, 70–77.
Nylander, J. A. A. (2004). MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University
O'Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
Paolinelli-Alfonso, M., Serrrano-Gomez, C., and Hernandez-Martinez, R. (2015). Occurrence of Eutypella microtheca in grapevine cankers in Mexico. Phytopathol. Mediterr. 54, 86–93. doi: 10.14601/Phytopathol_Mediterr-14998
Peng, M. K., Zhang, B., Qu, Z., Li, Y., and Ma, H. X. (2021). New record genus and a new species of Allodiatrype from China based on morphological and molecular characters. Phytotaxa 500, 275–284. doi: 10.11646/phytotaxa.500.4.3
Peršoh, D., Melcher, M., Graf, K., Fournier, J., Stadler, M., and Rmbold, G. (2009). Molecular and morphological evidence for the delimitation of Xylaria hypoxylon. Mycologia 101, 256–268. doi: 10.3852/08-108
Phookamsak, R., Hyde, K. D., Jeewon, R., Bhat, D. J., Jones, E. B. G., Maharachchikumbura, S. S. N., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95, 1–273. doi: 10.1007/s13225-019-00421-w
Piškur, B., Ogris, N., and Jurc, D. (2007). Species-specific primers for Eutypella parasitica, the causal agent of Eutypella canker of maple. Plant Dis. 91, 1579–1584.
Rappaz, F. (1987). Taxonomy and nomenclature of the octosporous Diatrypaceae. Mycol. Helv. 2, 285–648.
Rolshausen, P. E., Mahoney, N. E., Molyneux, R. J., and Gubler, W. D. (2006). A reassessment of the species concept in Eutypa lata, the causal agent of Eutypa dieback of grapevine. Phytopathology 96, 369–377. doi: 10.1094/PHYTO-96-0369
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Saccardo, P. A. (1875). Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti Soc Veneziana Trentina Istriana Sci. Nat. 4, 77–100.
Sales, R., Santana, C. V. S., Nogueira, D. R. S., Silva, K. J. P., Guimaraes, I. M., Michereff, S. J., et al. (2010). First report of Monosporascus cannonballus on watermelon in Brazil. Plant Dis. 94:278. doi: 10.1094/PDIS-94-2-0278B
Senanayake, I. C., Maharachchikumbura, S. S. N., Hyde, K. D., Bhat, D. J., Jones, E. B. G., McKenzie, E. H. C., et al. (2015). Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 73, 73–144. doi: 10.1007/s13225-015-0340-y
Senwanna, C., Phookamsak, R., Doilom, M., Hyde, K. D., and Cheewangkoon, R. (2017). Novel taxa of Diatrypaceae from Para rubber (Hevea brasiliensis) in northern Thailand; introducing a novel genus Allocryptovalsa. Mycosphere 8, 1835–1855. doi: 10.5943/mycosphere/8/10/9
Shang, Q. J., Hyde, K. D., Jeewon, R., Khan, S., Promputtha, I., Phookamsak, R., et al. (2018). Morpho-molecular characterization of Peroneutypa (Diatrypaceae, Xylariales) with two novel species from Thailand. Phytotaxa 356, 1–18. doi: 10.11646/phytotaxa.356.1.1
Song, Z. K., Pan, X. Y., Li, C. T., Ma, H. X., and Li, Y. (2022). Two new species of Hypoxylon (Hypoxylaceae) from China based on morphological and DNA sequence data analyses. Phytotaxa 538, 213–224. doi: 10.11646/phytotaxa.538.3.4
Thiyagaraja, V., Senanayake, I. C., Wanasinghe, D. N., Karunarathna, S. C., Worthy, F. R., and To-Anun, C. (2019). Phylogenetic and morphological appraisal of Diatrype lijiangensis sp. nov. (Diatrypaceae, Xylariales) from China. Asian. J. Mycol. 2, 198–208. doi: 10.5943/ajom/2/1/10
Tiffany, L. H., and Gilman, J. C. (1965). Iowa ascomycetes IV Diatrypaceae. Iowa State Coll. J. Sci. 40, 121–161.
Trouillas, F. P., and Gubler, W. D. (2004). Identification and characterization of Eutypa leptoplaca, a new pathogen of grapevine in northern California. Mycol. Res. 108, 1195–1204. doi: 10.1017/S0953756204000863
Trouillas, F. P., and Gubler, W. D. (2010). Host range, biological variation, and phylogenetic diversity of Eutypa lata in California. Phytopathology 100, 1048–1056. doi: 10.1094/PHYTO-02-10-0040
Trouillas, F. P., Hand, F. P., Inderbitzin, P., and Gubler, W. D. (2015). The genus Cryptosphaeria in the western United States: taxonomy, multilocus phylogeny and a new species. C. multicontinentalis. Mycologia 107, 1304–1313. doi: 10.3852/15-115
Trouillas, F. P., Pitt, W. M., Sosnowski, M. R., Huang, R., Peduto, F., Loschiavo, A., et al. (2011). Taxonomy and DNA phylogeny of Diatrypaceae associated with Vitis vinifera and other woody plants in Australia. Fungal Divers. 49, 203–223. doi: 10.1007/s13225-011-0094-0
Trouillas, F. P., Sosnowski, M. R., and Gubler, W. D. (2010a). Two new species of Diatrypaceae from coastal wattle in Coorong National Park, South Australia. Mycosphere 1, 183–188.
Trouillas, F. P., Urbez-Torres, J. R., and Gubler, W. D. (2010b). Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 102, 319–336. doi: 10.3852/08-185
U’ren, J. M., Miadlikowska, J., Zimmerman, N. B., Ltzoni, F., Stajich, J. E., and Arnold, A. E. (2016). Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota). Mol. Phylogenet. Evol. 98, 210–232. doi: 10.1016/j.ympev.2016.02.010
Úrbez-Torres, J. R., Peduto, F., Striegler, R. K., Urrearomero, K. E., Rupe, J. C., Cartwright, R. D., et al. (2012). Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri. Fungal Divers. 52, 169–189. doi: 10.1007/s13225-011-0110-4
Úrbez-Torres, J. R., Peduto, F., Vossen, P. M., Krueger, W. H., and Gubler, W. D. (2013). Olive twig and branch dieback: etiology, incidence, and distribution in California. Plant Dis. 97, 231–244.
Vasilyeva, L. N., and Ma, H. X. (2014). Diatrypaceous fungi in North-Eastern China. 1. Cryptosphaeria Diatrype. Phytotaxa 186, 261–270. doi: 10.11646/phytotaxa.186.5.3
Vasilyeva, L. N., and Stephenson, S. L. (2004). Pyrenomycetes of the Great Smoky Mountains National Park. I. Diatrype Fr. (Diatrypaceae). Fungal Divers. 17, 191–201.
Vasilyeva, L. N., and Stephenson, S. L. (2006). Pyrenomycetes of the Great Smoky Mountains National Park. III. Cryptosphaeria, Eutypa and Eutypella (Diatrypaceae). Fungal Divers. 49, 346–349. doi: 10.1111/j.1439-0507.2006.01249.x
Vasilyeva, L. N., and Stephenson, S. L. (2009). The genus Diatrype (ascomycota, diatrypaceae) in Arkansas and Texas (USA). Mycotaxon 107, 307–313. doi: 10.1063/1.119340
Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
White, T. J., Bruns, T. D., Lee, S., and Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics—science direct. PCR Protocol. 18, 315–322.
Wijayawardene, N. N., Hyde, K. D., Al-Ani, L. K. T., Tedersoo, L., Haelewaters, D., Rajeshkumar, K. C., et al. (2020). Outline of fungi and fungus-like taxa. Mycosphere 11, 1060–1456. doi: 10.5943/mycosphere/11/1/8
Yang, Z. E., Zhang, B., Qu, Z., Song, Z. K., Pan, X. Y., Zhao, C. L., et al. (2022). Two new species of Diatrype (Xylariales, Ascomycota) with Polysporous Asci from China. Diversity 14:149. doi: 10.3390/d14020149
Keywords: Ascomycota, Diatrypaceous fungi, multigene phylogeny, taxonomy, wood-decaying fungi, Xylariales
Citation: Ma H-X, Yang Z-E, Song Z-K, Qu Z, Li Y and Zhu A-H (2023) Taxonomic and phylogenetic contributions to Diatrypaceae from southeastern Tibet in China. Front. Microbiol. 14:1073548. doi: 10.3389/fmicb.2023.1073548
Edited by:
Yong-Zhong Lu, Guizhou Institute of Technology, ChinaReviewed by:
Nalin Nilusha Wijayawardene, Qujing Normal University, ChinaRajesh Jeewon, University of Mauritius, Mauritius
Copyright © 2023 Ma, Yang, Song, Qu, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai-Xia Ma, bWFoYWl4aWFAaXRiYi5vcmcuY24=; An-Hong Zhu, MTgyODk2NzkzMTdAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Hai-Xia Ma
Hai-Xia Ma Zhan-En Yang
Zhan-En Yang Zi-Kun Song
Zi-Kun Song Zhi Qu
Zhi Qu Yu Li
Yu Li An-Hong Zhu
An-Hong Zhu