
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 May 2023
Sec. Aquatic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1072053
Environmental perturbations evoke down-regulation of metabolism in some multicellular organisms, leading to dormancy, or torpor. Colonies of the urochordate Botrylloides leachii enter torpor in response to changes in seawater temperature and may survive for months as small vasculature remnants that lack feeding and reproductive organs but possess torpor-specific microbiota. Upon returning to milder conditions, the colonies rapidly restore their original morphology, cytology and functionality while harboring re-occurring microbiota, a phenomenon that has not been described in detail to date. Here we investigated the stability of B. leachii microbiome and its functionality in active and dormant colonies, using microscopy, qPCR, in situ hybridization, genomics and transcriptomics. A novel lineage of Endozoicomonas, proposed here as Candidatus Endozoicomonas endoleachii, was dominant in torpor animals (53–79% read abundance), and potentially occupied specific hemocytes found only in torpid animals. Functional analysis of the metagenome-assembled genome and genome-targeted transcriptomics revealed that Endozoicomonas can use various cellular substrates, like amino acids and sugars, potentially producing biotin and thiamine, but also expressing various features involved in autocatalytic symbiosis. Our study suggests that the microbiome can be linked to the metabolic and physiological states of the host, B. leachii, introducing a model organism for the study of symbioses during drastic physiological changes, such as torpor.
Dormancy (i.e., torpor) is widespread and well-documented across terrestrial and marine animals (Hand and Hardewig, 1996; Cáceres, 1997; Storey, 2002; Storey and Storey, 2011). Metazoans undergo dramatic changes during torpor, including metabolic depression, arrested development, and phenotypic plasticity. This reduction in host metabolism, body temperature and activity may affect the holobiont, altering the ecological landscape and selection of microbial community that is associated with the host. Whereas these associated microbes are considered crucial for the functionality and health of the metazoans (Berg et al., 2020), we know little about their diversity and function during the transition between active and dormant stages in torpor hosts. Studies in mammalian winter torpor cases, such as in the Syrian hamster, the ground squirrel and the brown bear, revealed restructuring of gut microbiota (Sonoyama et al., 2009; Carey et al., 2013; Sommer et al., 2016). Amphibian dormancy is associated with a decline in bacterial counts, changes in gut microbiota composition and the contribution of bacteria to energy balance (Carr et al., 1976; Gossling et al., 1982; Wiebler et al., 2018). Yet, most torpor studies focus on gut microbes in vertebrates, whereas how these transitions affect other tissues and basal metazoan lineages are poorly understood (Carey et al., 2013; Kohl et al., 2014; Lindsay et al., 2020; Sze et al., 2020).
Ascidians, or sea squirts (Tunicata), are a group of filter-feeding marine invertebrates that hold an important evolutionary position as a sister clade to the chordates (Vanni et al., 2022). They present a wide range of torpor events in various colonial taxa (Mukai et al., 1983; Turon, 1992; De Caralt et al., 2002; Hyams et al., 2017). As in many other sedentary marine organisms, symbiotic bacteria are known to reside in tunicates and are also known as tissue-associated, such as in endodermal tissues, the pharynx, the tunic and the intestine (Behrendt et al., 2012; Dishaw et al., 2014). The ascidian microbiota contributes to nutrition via photosynthesis (Donia et al., 2011; López-Legentil et al., 2011; Hirose, 2015), production of secondary metabolites (Schmidt, 2014; Tianero et al., 2015; Chen et al., 2018), nitrification (Martínez-García et al., 2008; Erwin et al., 2014), protection against biofouling, predation (Degnan et al., 1989; Paul et al., 1990; Fu et al., 1998; Vervoort et al., 1998; Faulkner, 2001; Schreiber et al., 2016), vanadium accumulation (Ueki et al., 2019), and the host immune response (Liu et al., 2021).
One key bacterial symbiont of ascidians is Endozoicomonas gammaproteobacterium (order Pseudomonadales, family Endozoicomonadaceae; Schreiber et al., 2016), a clade of intracellular or cell-associated facultative bacteria, found in close association with diverse marine invertebrate hosts, including reef-building corals, sponges, bryozoans, sea squirts, sea slugs, and mollusks (Jensen et al., 2010; Forget and Juniper, 2013; Morrow et al., 2015; Bourne et al., 2016; Miller et al., 2016; Neave et al., 2016; Schreiber et al., 2016; Pogoreutz et al., 2022). Despite the abundance and worldwide distribution of Endozoicomonas in multiple marine organisms, our knowledge of their functions and symbiotic relationships is limited. Here we aimed to study the bacterial diversity associated with the torpor state of the colonial tunicate Botrylloides leachii, representing one of the most dramatic torpor phenomena in the Chordata (Hyams et al., 2017), focusing on the key Endozoicomonas symbionts.
Botrylloides leachii enters a state of hibernation or aestivation following an abrupt decrease or increase in ambient seawater temperature (Hyams et al., 2017, 2022). The colonies completely absorb all their functioning units (zooids), arrest bud development, and may remain for months as remnants of condensed vasculature and hemocyte lacunae (Figures 1A,B). Torpor colonies are deficient in feeding and sexual reproduction organs. When re-exposed to milder environmental conditions, tissue vestiges regenerate new fully active colonies. Arousal from hibernation starts with clear tunic areas among the vasculature lacunae, that turn into transparent buds, one of which develops into a functional zooid (Hyams et al., 2017). Hibernating colonies harbor large cells that possess rod-shaped inclusions made of electron-dense material 2–3 μm in length, which resemble bacteria (Hyams et al., 2017, 2022). These were absent in non-dormant colonies. Aiming to investigate the link between torpor, microbial diversity, and functions, we visualized microbes, sequenced DNA and RNA, curated a metagenome-assembled genome of Endozoicomonas, and explored the expression of its genes in cultivated B. leachii, during fully functional, mid- and late-torpor conditions.
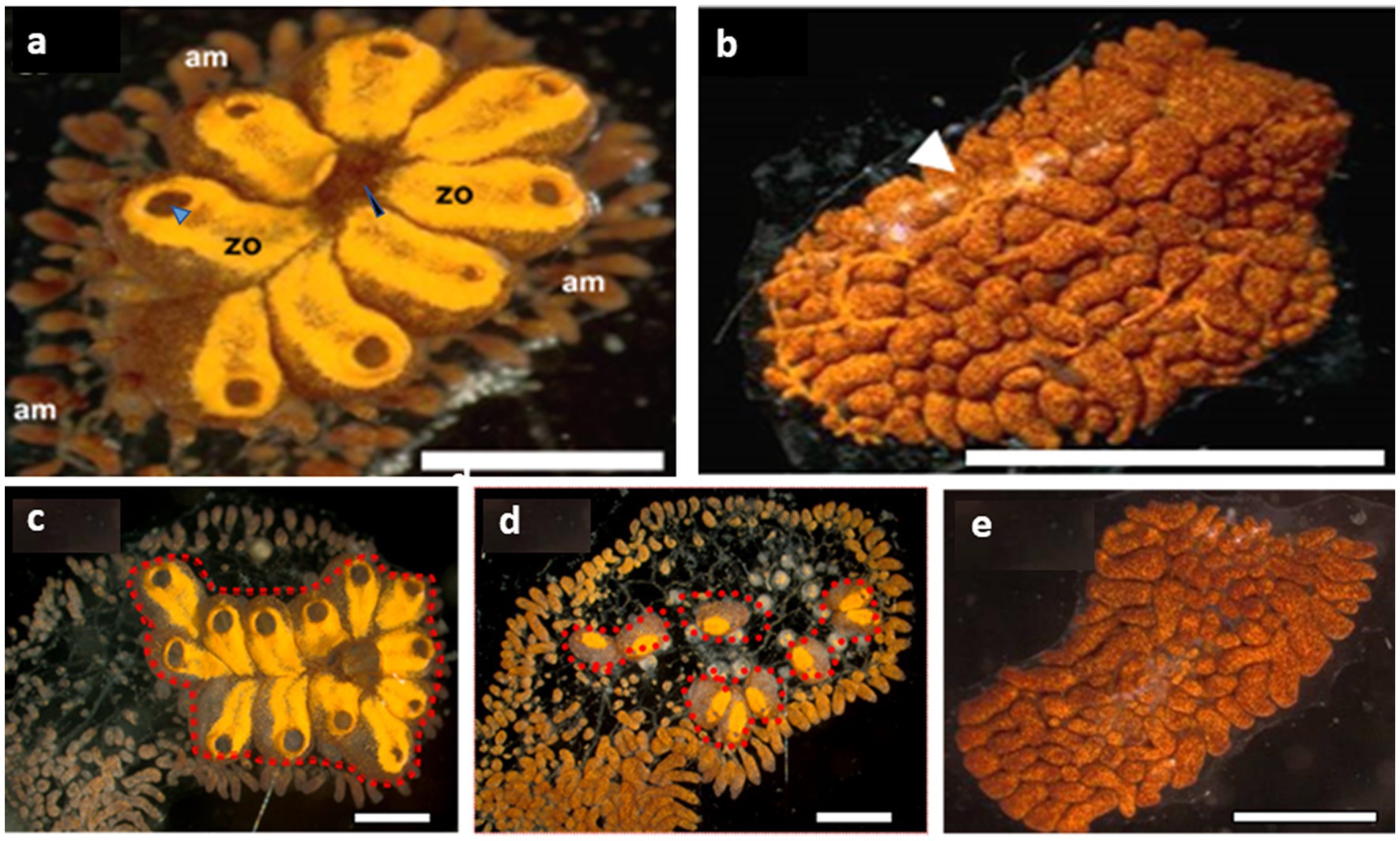
Figure 1. Botrylloides leachii torpor. (A) An active colony was grown on a glass slide in the laboratory. The zooids are peripherally surrounded by extended ampullae, the blind termini of vasculature that are loaded with hemocyte. Each zooid is approximately 1–1.5 mm in length and contains an oral siphon (a black arrowhead), whereas a system of 8 zooids (in this figure) shares a common atrial siphon (a blue arrowhead). The whole colony is embedded in a gelatinous matrix – the tunic. (B) A colony in a full hibernation state, following exposure to 15°C water temperature for 15 days (ambient seawater = 20°C). The resorbed zooids are replaced by a ‘carpet’ of opaque lacunae and dilated vasculature (a white arrowhead) loaded with pigment cells that give this colonial remnant its deep color. (C–E) From active (C), mid torpor (D) to fully torpor (E) states in the same B. leachii colony. Functional units (zooids, red dash line) are located only in active and mid-torpor states. Bars = 2 mm, am = ampulla, zo = zooid.
Colonies of B. leachii were collected from the shallow subtidal along the Israeli Mediterranean coast and were carefully peeled off the underlying surfaces of stones with industrial razor blades, to minimize tissue damage. Colonies were individually bound onto 5 × 7.5 cm glass slides using fine cotton threads and cultured at the National Institute of Oceanography, Haifa, in 25 L tanks connected to a standing seawater system (Rinkevich and Shapira, 1998). Within several days of collections, tied colonies were partially glided or relocated completely from their original calcareous substrates onto the glass slides, firmly attached to the new substrates. The colonies and the surrounding glass substrates were cleaned weekly with industrial razor blades and fine brushes. Some of these colonies were sub-cloned by cutting them into several ramets (methodology in Rinkevich and Shapira, 1998; Paz and Rinkevich, 2002) according to their size and health conditions and then re-attached to other slides, with each ramet placed onto a separate glass slide, thus allowing equal opportunity for outwards growth. Each collected colony is referred to as a distinct genotype since collected from different localities separated by at least 1 m from each other (Rinkevich and Weissman, 1987).
Nine genotypes were divided haphazardly into three experimental groups, (a) active colonies, growing at 20°C, (b) colonies at mid torpor state following their translocation to a 15°C water tank for 5 days, and (c) colonies at full torpor status following their translocation to 15°C water tank for 20 days (Hyams et al., 2017; Figures 1C–E). The colonies were monitored daily using a stereomicroscope (Nikon SMZ1000) and photographed (Olympus XC 30).
The hemocyte populations of active (n = 6), mid-torpor (n = 6) and fully torpid (n = 6) colonies were isolated from the vasculature (Avishai et al., 2003). Specimens were individually drained into a small petri dish, and each was stained with a fluorescent dye Hoechst 33342 (cat: 62249, Thermo Fisher Scientific) for nuclei staining. The cells were then washed several times with filter seawater (to remove excess dye) and monitored daily with a Nikon inverted phase contrast microscope for 10 days periods.
Control (n = 10), mid-torpor (n = 10) and hibernating (n = 10) colonies were fixed in Bouin’s solution for 1–2 h, dehydrated in a graded series (70–100%) of ethanol and butanol and then embedded in paraffin wax. Cross serial Sections (4–5 μm) were cut using a hand-operated microtome (Leica 2045, Nussloch, Germany), de-waxed and stained with alum hematoxylin and eosin for general morphology or with the reagent Schiff Feulgen reaction for DNA (Lillie, 1954).
Colonial fragments containing vasculature were sampled from active (n = 5), mid-torpor (n = 3) and fully torpid (n = 5) colonies and were fixed in 4% EM-grade glutaraldehyde (cat: G 5882, Sigma) at room temperature, post-fixed in 1% OsO4 in ddH2O for 1 h (4°C), washed (× 3) in cold ddH2O and then placed in 1% Uranyl acetate in ddH2O overnight. Samples were then dehydrated in a graded ethanol series of 50, 70, and 95% for 10 min each, twice at 100% for 10 min each at room temperature, and then placed in Propylene Oxide (PO) for 15 min. The infiltration of Epon resin was performed by submerging samples first in 1:1 PO/Epon for 1 h, then in 1:2 PO/Epon for a night and then in 100% Epon for 2–3 h. The samples were placed in molds, labeled, filled with 100% Epon and allowed to polymerize in a 65°C oven for 24 h. Ultrathin sections (80–90 nm) were prepared with a diamond knife (Diatome) and an ultramicrotome (Ultracut, Leica), placed on 300 mesh nickel grids (Polysciences Inc.) and stained with uranyl acetate and lead citrate. The sections were analyzed using a Jeol 1230 transmission electron Microscope at 80 kV. Digital photographs were taken with a Gatan-Multiscan 701 camera.
For DNA extraction, colonies were collected from Michmoret beach, Israel (n = 5 genotypes), and each was sub-cloned into three ramets. DNA was extracted from each genotype at three different time points (collected day = wild, in an active state under laboratory conditions-active, and in a torpor state under laboratory conditions-torpor), as described (Graham and Graham, 1978). RNA was extracted from 9 colonies, each at three physiological states (active, mid-torpor, and full-torpor) using an RNeasy Mini kit (QIAGEN) and digested with DNAse PurelinkTM (Invitrogen). RNA quantity was checked with a Qubit 2.0 Fluorometer and evaluated using a Bioanalyzer (Agilent). The tissue samples of active and mid-torpor states were taken from the functional units (zooids, red dashed lines Figures 1C,D) and the vasculature (the rest of the colony), and the whole fragment in a fully torpid state.
DNA was sequenced from two individuals, a torpid colony and a colony in an active state. DNA library preparation and sequencing of 30 Gb 2×150 metagenomic paired-end reads using Illumina NovaSeq was performed at Novogene (Singapore). RNA (TruSeq Total RNA Library Prep Kit with Ribo-Zero Gold) Sequencing (Epicenter, Benes et al., 2011) was carried out at Helmholtz Zentrum München, Germany, using the Illumina HiSeq 2000 platform. RNA sample libraries were prepared using Illumina TruSeq™ standard total RNA Library Preparation kit. Fragments >200 bp were selected for the final enriched libraries with an average insert size of 324 ± 7 bp across all libraries. RNA Sequencing was carried out at Helmholtz Zentrum München, Germany, using Illumina HiSeq 2000 platform. Paired-end sequencing of 2 × 100 bp was carried out generating approximately 34 million reads per lane. Only 30% of the total RNA reads were annotated to B. leachii reference genome, whereas circa 15% of the reads were assigned to small subunit ribosomal ribonucleic acid (SSU rRNAs) (Blanchoud et al., 2018; Hyams et al., 2022).
DNA reads were assembled with the SPAdes V3.11 toolkit (Bankevich et al., 2012) following adapter trimming and error correction with tadpole.sh, using the BBtools suite.1 Downstream mapping and binning of metagenome-assembled genomes (MAGs) were performed using DAStool, Vamb, Maxbin 2.0 and Metabat2 (Wu et al., 2016; Sieber et al., 2018; Kang et al., 2019; Nissen et al., 2021). Annotation was performed using DRAM (Shaffer et al., 2020) within Atlas V2.11 (Kieser et al., 2020). SEED annotation was performed using the Rast annotation engine (Overbeek et al., 2014). An Endozoicomonas tree was built from 22 genomes, downloaded from PATRIC (Wattam et al., 2017), with GTOtree V1.6.37 (Lee and Ponty, 2019) using default settings (90 of 172 gammaproteobacterial single-copy targets were used) and FastTree V2.1 (Price et al., 2009), using the JTT + CAT model and SH-like 1000 bootstraps.
To assess Endozoicomonas expression levels at the RNA level, the transcriptome reads were mapped to the MAG with 0.97 identity cutoff using bbmap, and the expression of coding sequences was summarized using featureCounts (Liao et al., 2014). We used phyloFlash V3.4 (Gruber-Vodicka et al., 2020) to assess the phylogenetic diversity of associated microbes in transcriptomes: The total RNA libraries from three physiological states (active, mid-torpor and fully torpor) and different tissue compartments (vasculature, zooids) were mapped to the Silva 138 database (Quast et al., 2013), the mapped reads were assembled with SPAdes, and the read abundance of the full-length SSUs was calculated following remapping with BBmap using 0.97 identity cutoff. Diversity analyses were performed using the R package phyloseq (McMurdie and Holmes, 2013) and species enrichment was identified with indispecies (De Cáceres et al., 2012).
Using Endozoicomonas 16S rRNA gene-specific forward primer and bacteria common primer as the reverse, an 833 bp fragment specific to Endozoicomonas was amplified from a Botrylloides colony (Table 1), subcloned into pDrive vector (PCR cloning kit cat number 231124, Qiagen, Germany) and sequenced. The fragment showed 99.28 and 81.35% similarity to Endozoicomonas and Fodinicurvata. 16S rRNA gene sequences, respectively. A Fodinicurvata rRNA gene fragment (Table 1) was amplified using Fodinicurvata forward and common bacteria primers as the reverse. The subcloned Fodinicurvata 780 bp fragment had 99.49 and 81.5% identity to Fodinicurvata and Endozoicomonas 16S rRNA gene sequence, respectively. It also shares 92% identity to Fodinicurvata strain YIM D82 16S ribosomal RNA (accession number NR_044595.1).
The subcloned B. leachii 18S fragment (Table 1) was 109 bp and had 100% identity to the 18S gene of Botrylloides aff. leachii (accession number MG009584.1).
Relative qPCR analyses were performed on standards and DNAs extracted from tested tissues with Fast SYBR™ Green Master Mix (cat.no 4385614; Thermo Fisher Scientific, MA, United States) using StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, MA, United States). Specific standard curves for Endozoicomonas and Fodinicurvata 16S and B. leachii 18S rRNA genes were established by amplifying 4 known amounts of each of the standard DNAs (concentrations: 0.1, 0.01, 0.001, 0.0001 ng/mL). On the same plate, the quantities of Endozoicomonas and Fodinicurvata 16S and B. leachii 18S rRNA genes were analyzed in ramets originating from colonies at three different states: wild (n = 5, collected from Michmoret beach), active (n = 5, acclimatized in the institutional mesocosm) and hibernating colonies n = 5. Using the standard curves (concentration vs. CT; 69), the quantities of specific 16S genes (proportional to bacteria quantities) were extrapolated for each tissue and normalized to rRNA gene quantity (proportional to B. leachii tissue quantity). Statistical analysis was performed using a one-way ANOVA between the different groups.
Transmission electron microscopy (TEM) observations in active (n = 5), mid-torpor (n = 3), and fully torpid (n = 5) colonies revealed bacteria associated with cells in the vasculature of only mid and full-torpid states (Figure 2, 2–8% of the total circulating cells in mid- and fully torpid states, respectively), as indicated previously (Hyams et al., 2017, 2022). The size of the host cells containing intracellular bacteria was 8–30 μm. We found two distinct cell phenotypes with bacteria: (i) Cell sizes 10–20 μm, where most of the cell volumes are occupied by a few, 1–3 vacuoles (8–14 μm in length), and smaller vacuoles inserted within larger vacuoles. Each vacuole contained 3–15 bacteria cells of 1–2 μm, each (Figure 2C). With nuclei and mitochondria located at the periphery. The bacterial cells within the vacuoles were intact (Figure 2C). (ii) Cell sizes of 10–30 μm. We observed multilayer vacuoles that accounted for a large fraction of the cell volume. Each host cell contained 15–20 intact, 1–2 μm long, bacterial cells (Figure 2D). All the bacteria within the host cells were morphologically intact and well organized within the cells, some of them were found in a binary fission state (Figure 2D’, yellow asterisks). As in phenotype 1, the nuclei of host cells and mitochondria in phenotype 2 were in the cell periphery.
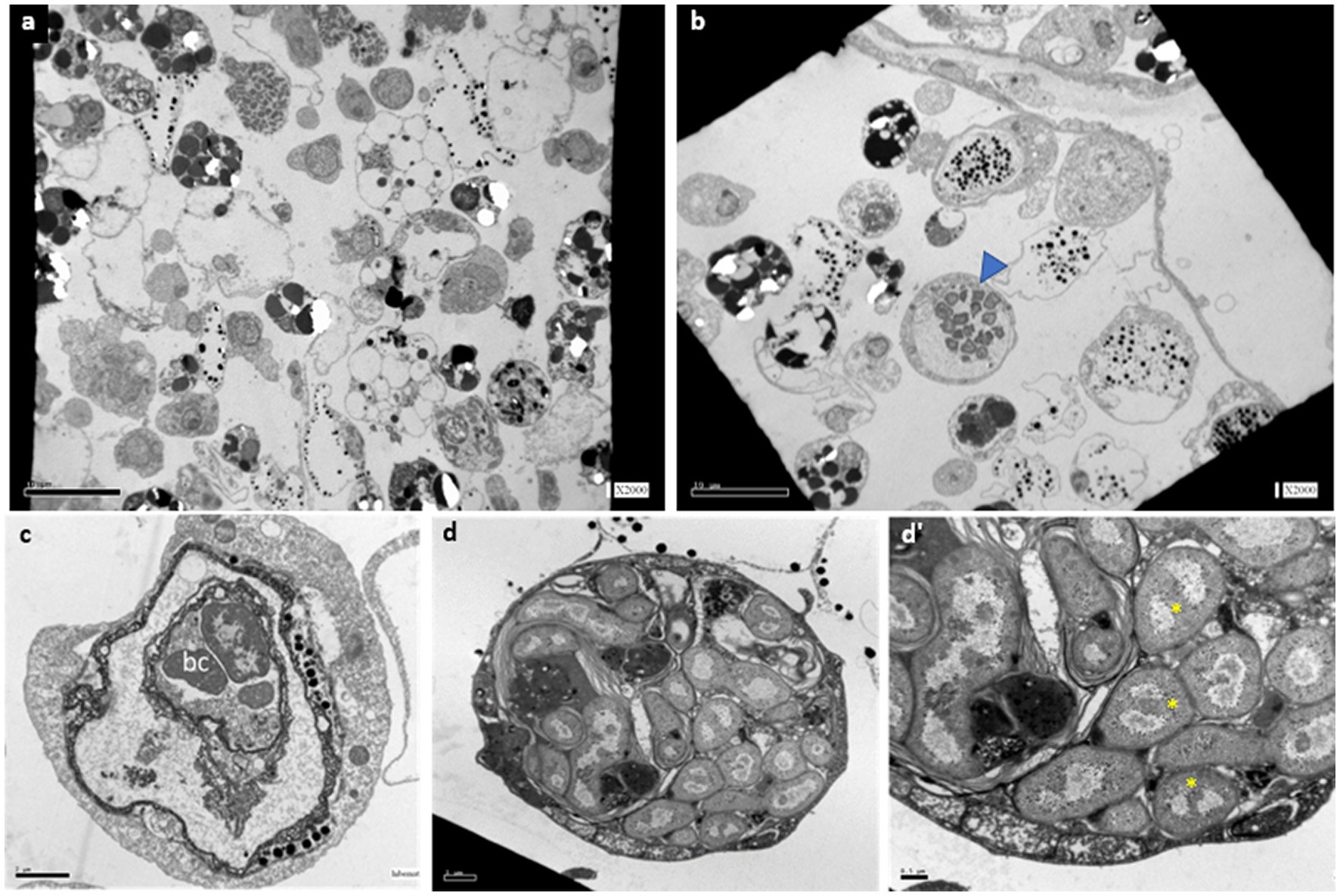
Figure 2. TEM sections of B. aff. leachii vasculature and hemocytes in active, mid-torpor and fully torpid states. (A) An active phenotype, no cell-containing bacteria. Bar-10 μm. (B) A torpid colony, bacteria cells within the cytoplasm of some circulated hemocytes (a blue arrowhead). Bar-10 μm. (C) Bacteria-containing cells, phenotype 1-a 12 μm animal cell containing 2 vacuoles, the inner vacuole holds 3 intact bacteria cells. Bar-2 μm. (D) A bacteria-containing cell, phenotype 2- a 22 μm cell holding 22 intact bacteria cells. (D’) The bacteria cell wall is double layered, as some of the naked DNA that is at the state of binary fission (yellow asterisks). Bar-0.5 μm.
Histological observations performed on active colonies (n = 12) and in Hoechst-stained isolated cells showed no evidence of bacteria within either vasculature or in isolated hemocytes under in vitro conditions (Figures 3A,B). In mid-torpor colonies (n = 6), 1–4% of the hemocyte populations appeared as large hemocytes (10–15 μm) containing several stained ‘nuclei’ (2–13) within each cell membrane, suggesting the appearance of bacteria cells (Figure 3C, red arrowheads). In the fully torpid colonies (n = 8), high proportions (10–70%) of the hemocyte populations appeared as large hemocytes (10–30 μm) populated by ‘nuclei’ (n = 5–15), further suggesting that some of the multinucleate cells are of bacteria inclusive cells (Figures 3D,E,E’, red arrowheads). Only TEM observations were able to differentiate between multinucleate cells and cells with bacteria.
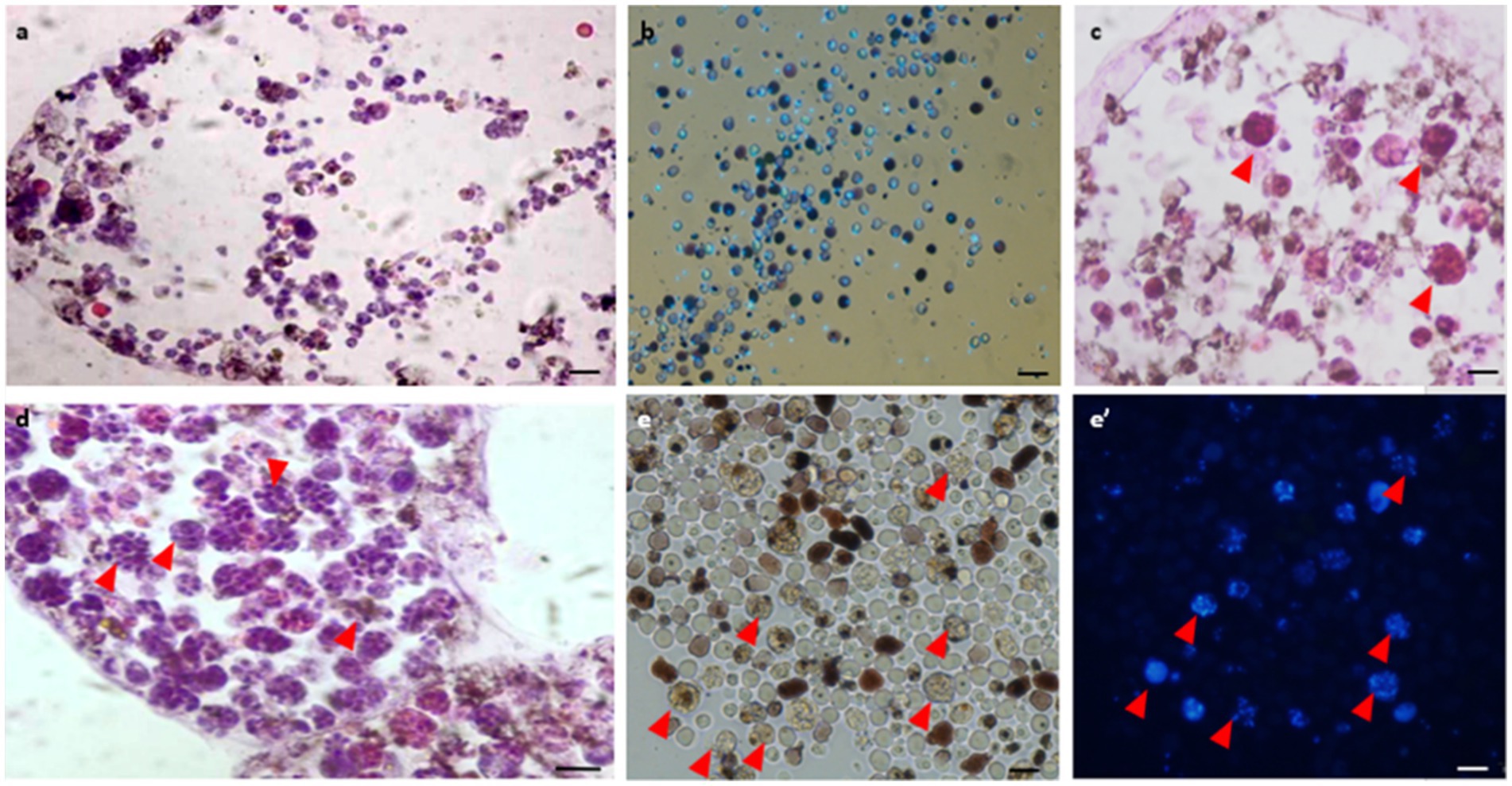
Figure 3. Histological observations of vasculature and hemocytes in active, mid-torpor and fully torpid colonies. (A) Active colony- histological section through a peripheral ampulla stained with hematoxylin & eosin. (B) In vitro Hoechst stain of hemocytes isolated from the active colony. (C) Mid-torpor colony- histological section through a peripheral ampulla stained with hematoxylin & eosin. Several hemocytes containing multiple DNA spots are marked (red arrowheads). (D) Torpid colony- Histological section through an ampulla (hematoxylin & eosin). A wide range of hemocytes with multiple DNA spots (red arrowheads). (E) In vitro Hoechst-stained hemocytes isolated from a torpid colony (E’), many of them contain multiple DNA spots (marked with red arrowheads). Bars- 10 μm.
To study changes to the microbiome during torpor, we analyszed our previous RNA-sequencing data (Hyams et al., 2022), this time for bacterial 16S rRNA gene sequences.
We found 54 full-length 16S rRNA sequences of bacteria and archaea associated with B. leachii, based on transcriptomics (Figure 4), Taking into consideration that laboratory conditions may also affect the microbiome. The key lineages included Endozoicomonadaceae that were not identified beyond the family level, alphaproteobacterial taxa such as Fodinicurvata sp., as well as Thioglobaceae (SUP05), Nitrincolaceae, Rikketisiales and Vibrionaceae lineages, some of which tend to be found in symbiotic associations (Merhej and Raoult, 2011; Neave et al., 2016; Perez et al., 2022), including those of ascidians (Schreiber et al., 2016). Enrichment analyses revealed that in full torpor the read abundance of Endozoicomonas was the highest (53–79% of all bacterial reads were annotated as Endozoicomonas compared to 13–25% in mid torpor zooids and just 0.14–1.76% in active colonies, indicator species analysis statistic [indicpecies stat.] value 0.90, p < 0.001). In the active state, Fodinicurvata read abundance was 21–39% in zooids and 34–38% in the vasculature, as compared to <2% of all reads in the vasculature of fully torpid colonies (indicpecies stat. value 0.94, p = 0.001). We observed enrichment of Thioglobaceae sp. in zooidal tissues, primarily in regular colonies (9.58–11.93% in active zooids, as compared to 1.29–1.72% in the vasculature). Thioglobaceae abundance was reduced in zooids of mid-torpid zooids to 2.02–7.41% (1.51–2.92% in the vasculature). In fully torpid fragments Thioglobaceae read abundance was only 0.1–0.65% (Supplementary Table 1). We verified the Endozoicomonas enrichment during torpor using DNA-based PCR. The ratio of Endozoicomonas 16S and Botrylloides rRNAs was significantly higher in hibernating colonies (p < 0.001) than in individuals collected directly from the sea and active colonies from the laboratory (Figure 5). In the specific qPCR performed, these ratios were not significantly different (p > 0.1) between the different physiological states for Fodinicurvata. These results indicated that the B. leachii microbiota does not remain stable during torpor progression, and specific lineages prevail in torpor (explaining 74.4% of the variation; Figure 6). Here after we focus on the Endomonazaicoceae symbiont, describing its phylogeny and function, because of its enrichment in the torpor state.
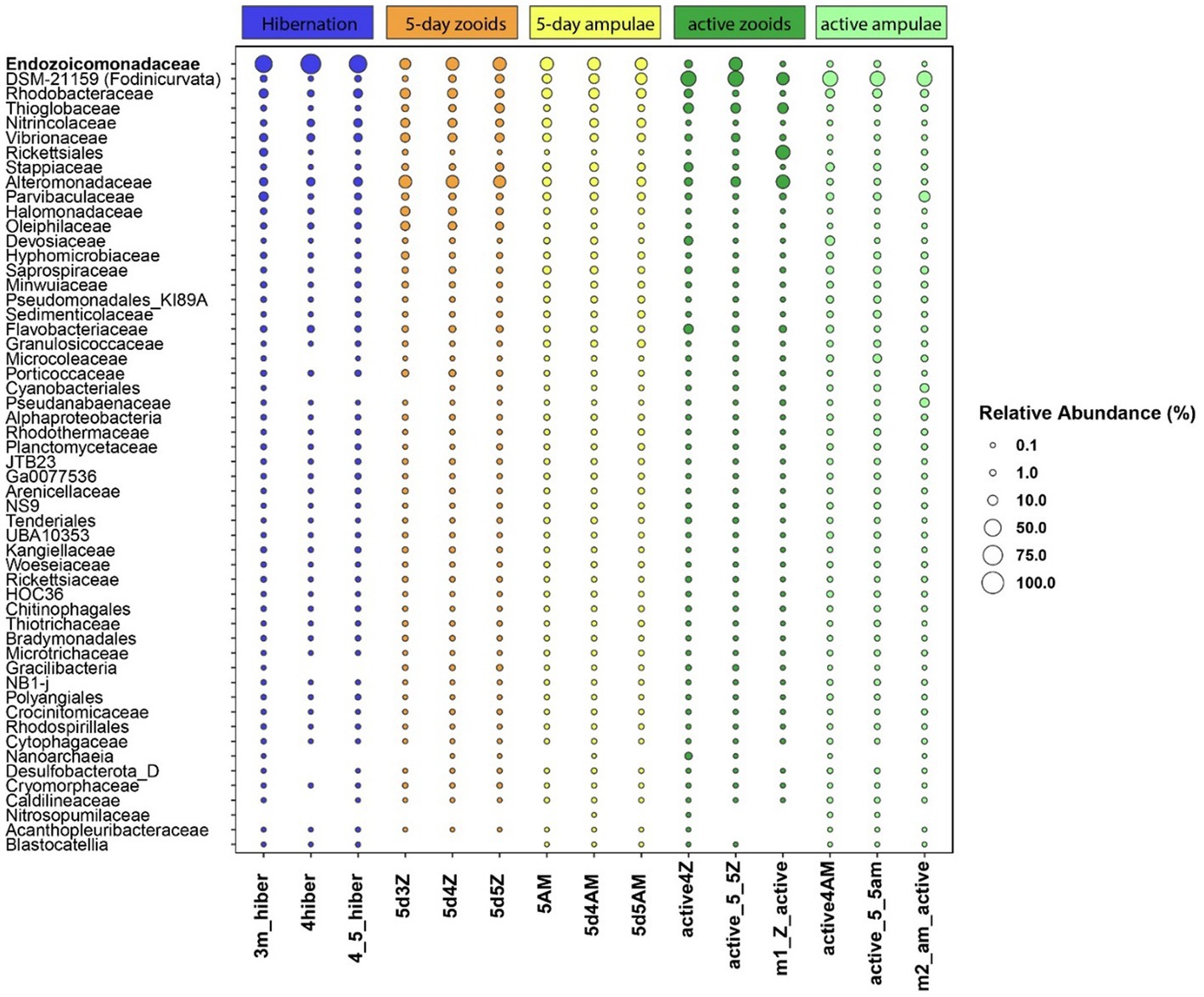
Figure 4. The relative read abundance of 54 bacterial taxa in the vasculature of torpid colonies, mid-torpor state (5 days) and active colonies (3 different biological replicates for either physiological state, each replicate marked by its ID at the x-axis base). Empty slots: relative abundance <0.1%. Taxonomy is based on the annotation of the full, SPAdes-assembled rRNA sequences, classified using the best BLAST hit in the Silva 138 database.
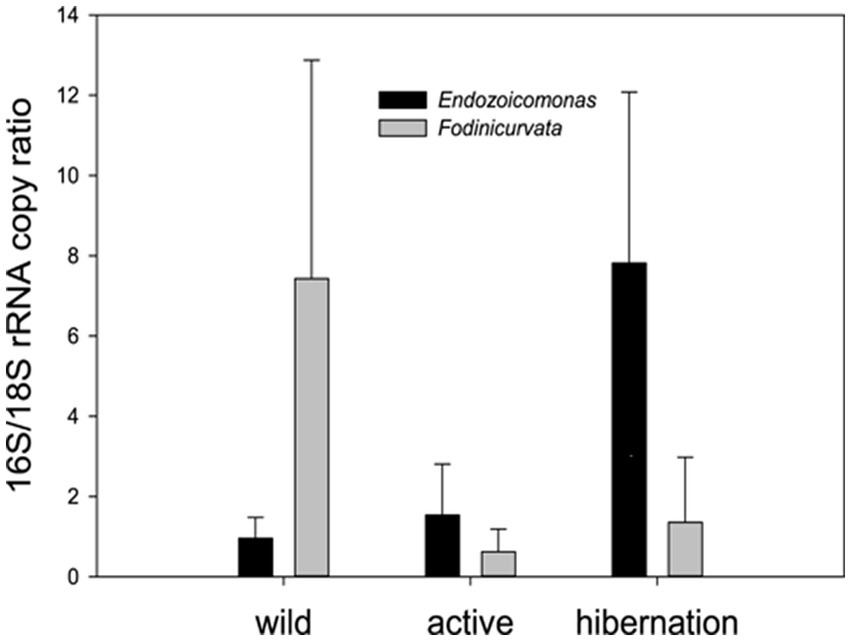
Figure 5. qPCR quantification of the two key bacteria associated with B. leachii hibernation– the ratio between the copy numbers of the rRNAs: bacterial 16S to the host 18S. Endozoicomonas was significantly enriched in the hibernation state (n = 5), compared to freshly collected (wild n = 5) and laboratory-grown samples (active n = 5). Fodinicurvata was highly abundant in some freshly collected samples. One-way ANOVA: Endozoicomonas along different stages F (2,12) = 9.915, p = 0.0038 Fodinicurvata F(2,12) = 1.913, p = 0.19.
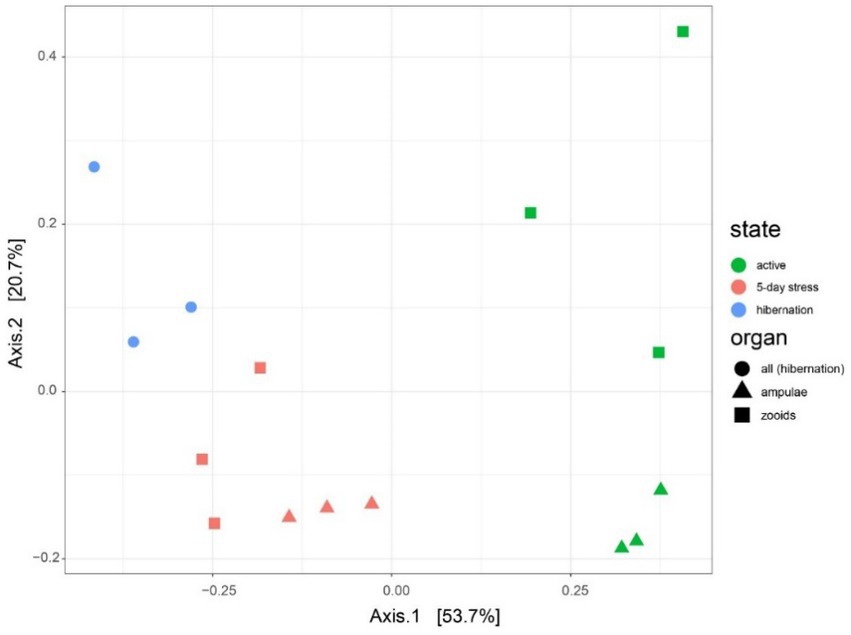
Figure 6. Microbiota varies between different physiological states (active, mid-torpor, and full-torpor colonies) and tissue types (vasculature and zooids) in Botrylloides leachi. Principal Coordinates Analysis (PCoA) based on the Bray–Curtis dissimilarity matrix of transcriptomic rRNA full-length read abundances.
Endozoicomonas reads were found only in the torpor library, and 238,000 reads were mapped to Metagenome-Assembled Genome (13x coverage). The total length of the genome was 2.64 Mb, comprising 165 contigs with an N50 value of 23610. Checkm (for assessing the quality of genomes) indicated that the genome is 90% complete with 0.8% contamination, whereas Checkm2 suggested 100% completeness and 0.1% contamination.
Metagenomics, and analysis of the 16S rRNA sequence, suggest that B. leachii hosted a distinct lineage of Endozoicomonas. Its 16S rRNA sequence had 94.87% identity with the best hit in the NCBI’s database, the sequence from the Endozoicomonas montiporae CL-33 genome (CP013251.1). Endozoicomonas sp. ex B. leachii MAG is basal to a large branch of Endozoicomonadanceae genomes that are associated with distinct hosts (Figure 7). Phylogenomic treeing suggested multiple host switches within the Endozoicomonas clade. In particular, the ascidian-associated Endozoicomonas ascidiicola (Schreiber et al., 2016) is not the closest relative of Endozoicomonas sp. ex B. leachii. Given the highest average nucleotide identity of 70% between the MAG of Endozoicomonas sp. ex B. leachii and other genomes in our comparison, we suggest that this lineage represents a novel candidate species, which we propose to name Candidatus Endozoicomonas endoleachii.
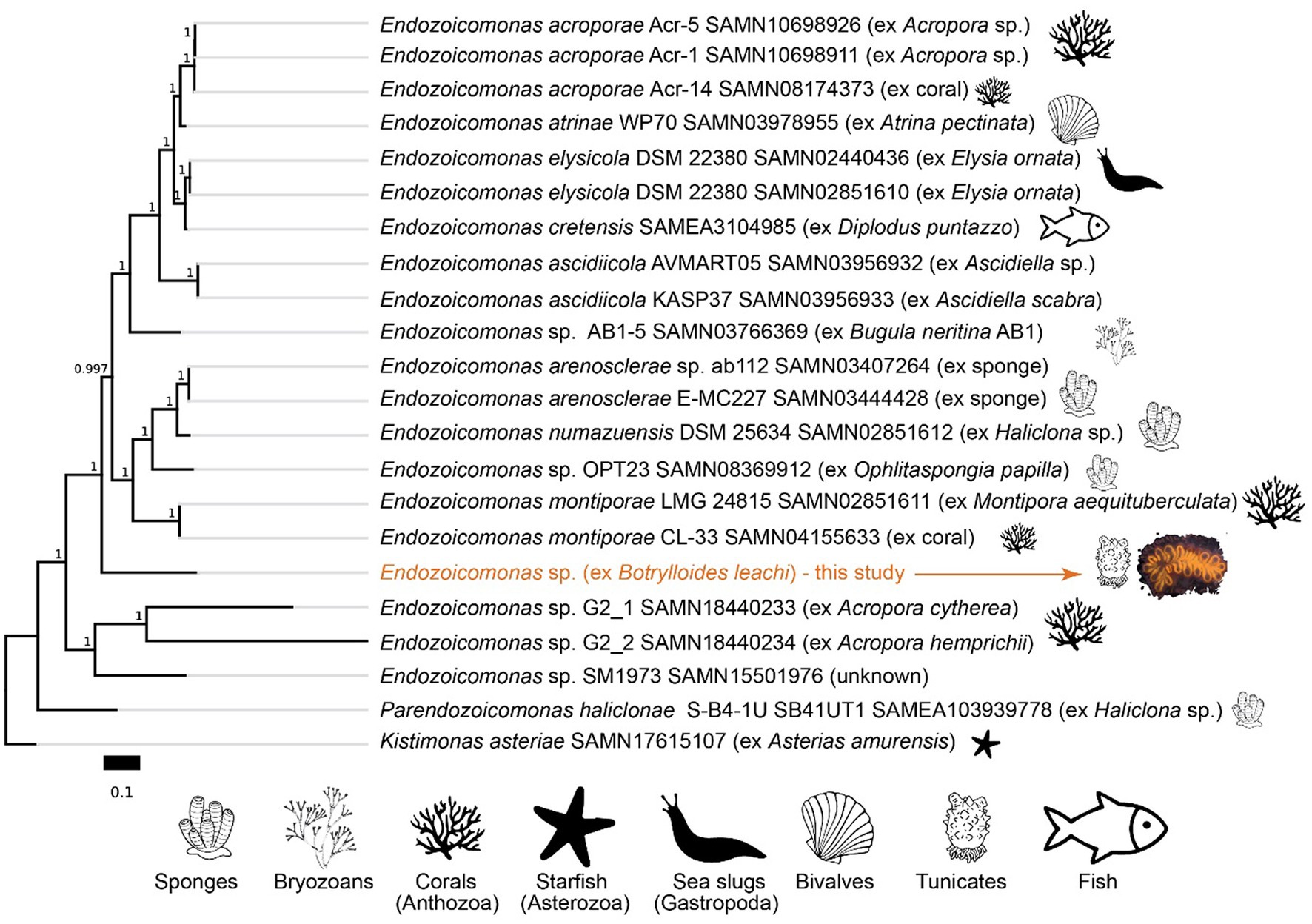
Figure 7. Phylogeny of Endozoicomonas species based on amino acid sequences of 90 single-copy gene markers (maximum likelihood, JTT + CAT model). The scale bar represents the number of substitutions per site. Branch bootstrap support values are shown. NCBI BioSample accession numbers are indicated for the respective genomes.
The metagenome-assembled genome investigation suggested that Candidatus E. endoleachii exhibits a heterotrophic lifestyle, probably focused on the turnover of proteins, amino acids and their derivatives (18 and 19% predicted SEED functions, respectively, Supplementary Figure 2), whereas 11% of the genomic features are dedicated to cofactors, vitamins, prosthetic groups and pigments, and 7% to carbohydrates. The tricarboxylic acid cycle appeared to be complete and highly expressed (the mdh gene encoding malate dehydrogenase was among the top 25 most expressed genes; Figure 8 and Supplementary Table 1). Glycolysis enzymes were moderately to highly expressed at the mRNA level (Figure 8). Endozoicomonas genome (Deutscher et al., 2006) carries more than 10 genes, most of which cluster and are highly expressed, encoding a phosphotransferase system (PTS), which allows transport of PTS-sugars, often preferred bacterial substrates (Deutscher et al., 2006). This system is likely specific to N-acetylgalactosamine (GlcNAc), as Ca. E. endoleachii encodes and expresses the aga operon (Brinkkötter et al., 2000), further allowing the symbiont to take advantage of glycans that appear to be enriched in tunicates (Zeng et al., 2022). Gene clusters encoding the glycine cleavage system, methionine transport, arginine/ornithine transport and degradation and branched-chain amino acid degradation, as well as the oppABCF genes (ABC oligopeptide transporter), were found to be expressed substantially (Figure 8 and Supplementary Table 1). Thus, Ca. E. endoleachii appears to be capable of glycogen storage. These features may allow Endozoicomonas the commensal or parasitic lifestyle within the B. leachii colonies. Three types of terminal oxidases were found (ba3, bo3, and bd-I), hinting at adaptation to changes in oxygen availability. We could not identify genes involved in the use of testosterone or dimethylsulfoniopropionate, as has been suggested for E. montiporae (Ding et al., 2016) and E. acroporae (Tandon et al., 2020). However, the genome is only 90% complete based on the CheckM estimate (Parks et al., 2015) and has a smaller size than most Endozoicomonas (2.7 as opposed to ~4–7 Mb), thus some genomic features may be missing.
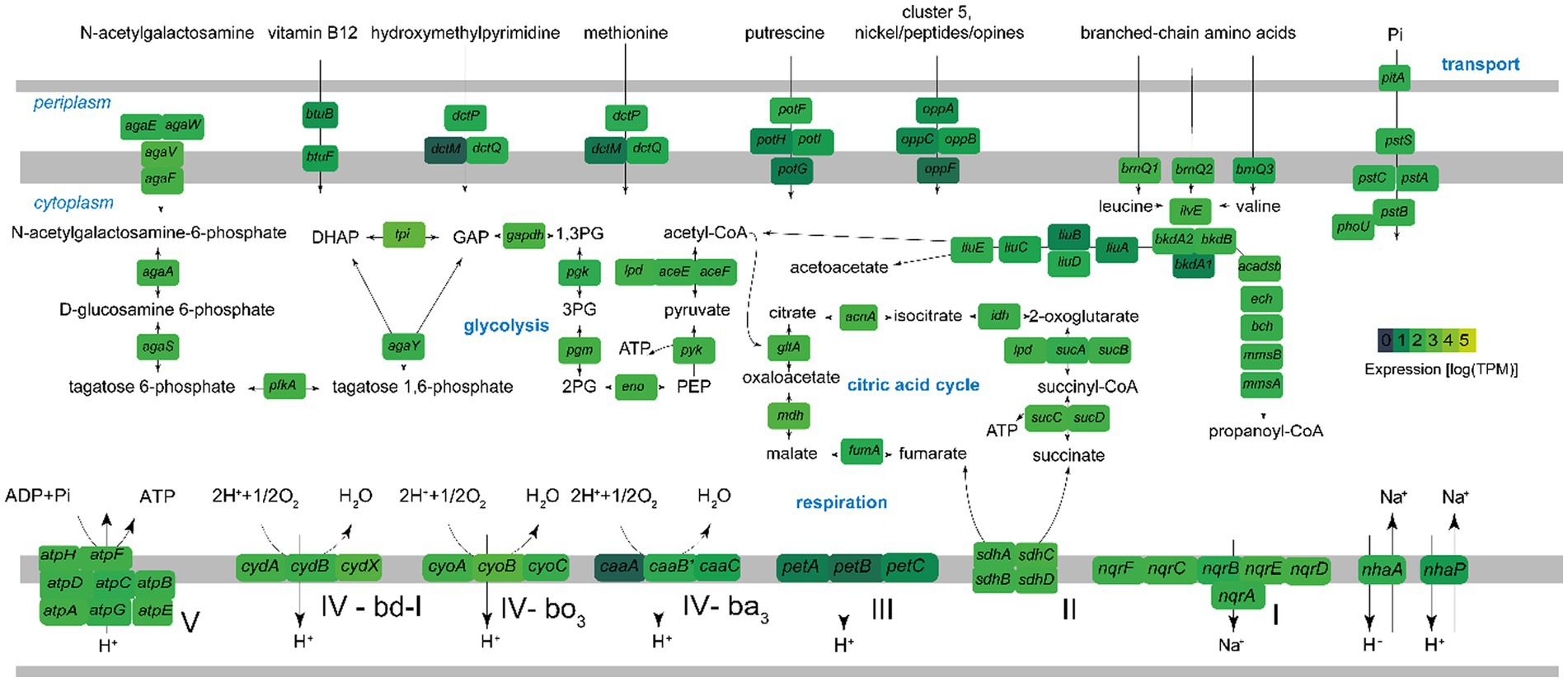
Figure 8. Key metabolic pathways of Ca. Endozoicomonas endoleachii, based on genome-centered transcriptomics. The enzymes that catalyze each reaction are marked with the name of the encoding gene. Colors represent gene expression only in torpor colonies, as log transcripts per million reads (TPM). The following enzymes and respective genes are shown: PTS system, N-acetylgalactosamine-specific IIC component agaW; PTS system, N-acetylgalactosamine-specific IID component agaE; PTS N-acetylgalactosamine transporter subunit IIB agaV; N-acetylgalactosamine transporter subunit IIA agaF; N-acetylgalactosamine-6-phosphate deacetylase agaA; D-galactosamine-6-phosphate deaminase AgaS; 6-phosphofructokinase pfkA; tagatose-bisphosphate aldolase agaY; triosephosphate isomerase tpi; glyceraldehyde-3-phosphate dehydrogenase gapdH; phosphoglycerate kinase pgk; phosphoglucomutase pgm; enolase eno; pyruvate kinase pyk; pyruvate dehydrogenase aceEF-lpd; citrate synthase gltA; aconitase acnA; isocitrate dehydrogenase idh; 2-oxoglutarate dehydrogenase sucAB-lpd; succinyl-CoA ligase sucCD; succinate dehydrogenase sdhABCD; fumarate hydratase fumA; malate dehydrogenase mdh; F0F1-type ATP synthase atpABCDEFGH; Na(+)-translocating NADH-quinone reductase nqrSBCDEF; Na(+)/H(+) antiporters nhaA and nhaP; cytochrome bc1 complex petABC; cytochrome bd-I ubiquinol oxidase cydABX; cytochrome bo3 ubiquinol oxidase cyoABC; ba3-type cytochrome c oxidase caaABC; nranched-chain amino acid permeases brnQ; branched-chain-amino-acid aminotransferase ilvE; branched-chain alpha-keto acid dehydrogenase complex bkdA1A2B; isovaleryl-CoA dehydrogenase liuA; methylcrotonyl-CoA carboxylase liuBD; methylglutaconyl-CoA hydratase liuC; hydroxymethylglutaryl-CoA lyase liuE; acyl-CoA dehydrogenase short/branched chain acadsb; enoyl-CoA hydratase ech; 3-hydroxyisobutyryl-CoA hydrolase bch; 3-hydroxyisobutyrate dehydrogenase mmsB; methylmalonate-semialdehyde dehydrogenase mmsA; phosphate transport system pstABC-phoU-pltA; oligopeptide ABC transporter oppABCF; putrescine transporter potFGHI; ABC transporters dctMPQ; vitamin B12 transporter btuBF.
Ca. E. endoleachii has several genomic features that could facilitate biotic interactions. These include genes that encode multiple features of type II, III, and VI secretion systems. Similar to Endozoicomonadaceae found in other hosts (Tandon et al., 2020), Ca. E. endoleachii encoded and expressed two eukaryotic repeat proteins, 14117 and 15557 nucleotides in length (data not shown). Conserved domain search revealed that these proteins comprised multiple cadherin-like, Ig-like, VCBS domains, as well as tandem-95 repeats, a carbohydrate-binding module NPCBM/NEW2 and a type I secretion C-terminal target domain.
We found gene clusters needed for the synthesis of biotin and thiamine diphosphate vitamins, the latter of which depend on the transport of hydroxymethylpyrimidine based on the presence and substantial expression of thiX-thiY-thiZ genes clustered with the thiD gene which encodes the hydroxymethylpyrimidine kinase (Supplementary Table 1; Rodionov et al., 2002). Thus, the interaction between the Endozoicomonas symbiont and the host may not be fully commensal or parasitic, as some metabolic trade-offs may be beneficial to B. leachii host.
We investigated which Endozoicomonas genes were overexpressed during torpor. Because in active, non-torpor, individuals, very few RNA reads mapped to Endozoicomonas, we focused on comparing expression in libraries derived from mid-torpor and full-torpor individuals. DESeq2 analysis revealed marked differences in expression profiles between these two stages (Supplementary Figure 1). The key genes that were upregulated in full torpor belonged to the isc operon (Figure 9), which is required for the synthesis of Fe-S cluster proteins and plays a major role in pathogenicity (Lim and Choi, 2014). The key sensory gene in this operon, IscR, was that with the strongest upregulation (Figure 9). IscR is essential for the virulence of the V. vulnificus pathogen, and its exposure to host cells induces the expression of this sensory gene (Lim and Choi, 2014). Thus, the synthesis of Fe-S cluster proteins likely plays an important, yet unknown role in the interaction between Endozoicomonas and its host. It is not surprising that one of the most upregulated Endozoicomonas genes in full-torpor hosts is the one that encodes a cold shock protein, potentially responding to the ambient temperature of 15°C.
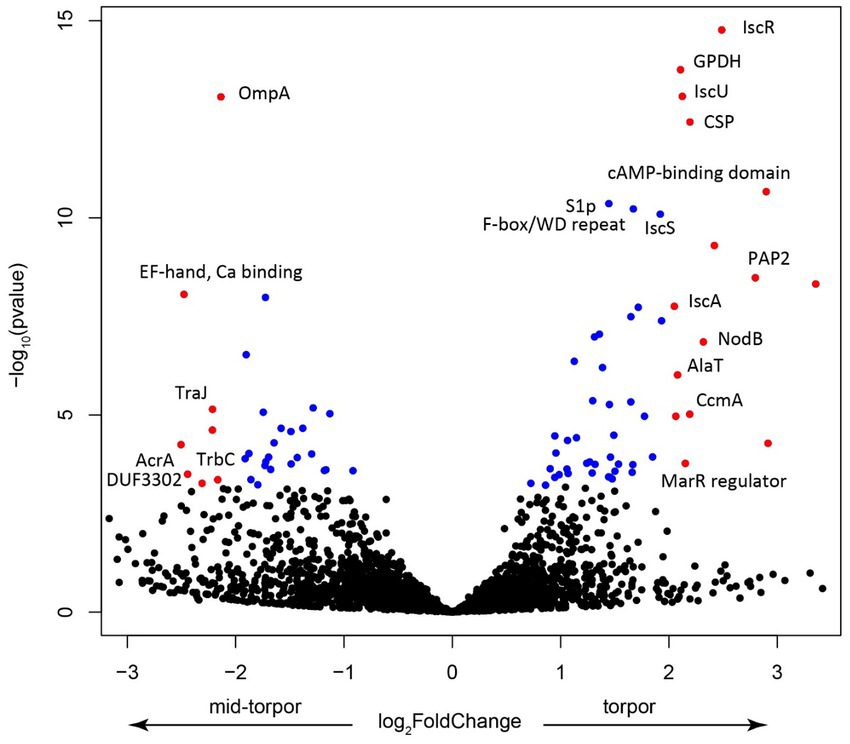
Figure 9. Volcano plot showing the key features upregulated in the mid-torpor and torpid states. Each dot represents a gene, blue if the adjusted p value < 0.01, red if log2 fold change >1 and adjusted p value < 0.05. Selected genes were annotated using the protein product nomenclature. GPDH is glycerol-3-phosphate dehydrogenase [NAD(P)+] (EC 1.1.1.94). CSP is a cold shock protein. PAP2 is a putative membrane-associated phospholipid phosphatase, PAP2 superfamily.
In the mid-torpor state, the most overexpressed gene encoded the putative outer membrane protein OmpA (Figure 9), a small β-barrel membrane anchor that establishes a physical linkage between the outer membrane and peptidoglycan layer (Buchanan, 1999). OmpA may play a pivotal role in bacterial pathogenesis (Mishra et al., 2020). These Omp proteins are often highly expressed in symbiotic systems, such as that of the tubeworm Riftia pachyptila and its gammaproteobacterial endosymbiont, where they may be involved in biotic interactions (Hinzke et al., 2021). Given that this gene is overexpressed during torpor onset when Endozoicomonas could establish its population, it may also be involved in the interaction with a more active host, in particular with its immune system. We also observed the upregulation of several genes that encode elements of conjugative transfer (Glöckner et al., 2008), which indicates potential activation of the mobilome and the possibility of an increased gene transfer, which increases pathogenicity.
Botrylloides leachii hosts a diverse microbial community, some of which may have a tight symbiotic partnership with the host. The dominant bacterial species in active colonies, in the and zooids, is Fodinicurvata sediminis, a gram-negative, facultatively anaerobic and non-motile bacterium. These bacteria secrete enzymes like urease and arginine dihydrolase, and their substrates are most probably provided by the host’s metabolic processes. While the growth temperature range for Fodinicurvata is 15–42°C (optimum, 28°C; 81), the winter simulating low 15°C seawater temperature that is associated with the torpor state, might be the explanation for their dramatic decrease. Unfortunately, we could not assemble a high-quality genome of Fodinicurvata, and the exploration of its function, as well as that of multiple low-abundance microbes, is pending.
The transition from the active to torpor states is characterized by a dramatic morphological, physiological and cellular changes within a few days, from an active colony with multiple functional zooids to small remnants of vasculature characterized by sluggish hemocyte circulation and no functional zooids (Hyams et al., 2017, 2022 and the present results). Under these non-feeding conditions, the torpor state may last for several months (Hyams et al., 2017). In the hibernating state, the bacteria appear to concentrate in an intimate special environment (within cells; Figure 2), that may be required for the holobiont functioning during the torpor. Upon arousal from torpor, no bacterial inclusions are found (Hyams et al., 2017), and their numbers are reduced dramatically. The symbiotic bacteria in this niche may either play beneficial roles, such as aiding nutrition, or use this unusual physiological state to its advantage, like outcompeting other bacteria, or deal with the animal’s immune system.
We observed considerable changes in microbial composition during the transition from the active state to the full-torpor state. Endozoicomonas appeared to be the most abundant symbionts in the torpor state. In various hosts, these symbionts may facilitate antimicrobial activity and microbiome structuring (Gardères et al., 2005; Morrow et al., 2012; Jessen et al., 2013; Rua et al., 2014), and be involved in metabolite exchange (Raina et al., 2009; Bourne et al., 2013; Correa et al., 2013; Nishijima et al., 2013; Beinart et al., 2014; Dishaw et al., 2014; Haber and Ilan, 2014). In particular, Endozoicomnas from the solitary tunicate Ciona intestinalis is likely involved in sulfur cycling and nutrient metabolism (Dishaw et al., 2014). It is still unclear if B. leachii host benefits from maintaining Endozoicomonas during torpor. One advantage could be the supply of vitamins, needed to maintain the hibernating host. While less likely, given that our results suggest enhanced mortality during the torpor state (Hyams et al., 2017, 2022), it may also be postulated that Endozoicomonas benefits from the reduction in the host resilience, and proliferates, out-competing other bacteria that may become limited by food influx in an inactive animal. Then, the intracellular Endozoicomonas (Figures 2, 3) are not limited by food influx as focusing on the scavenging of metabolites within the cells.
This study shows for the first time that facultative, potentially intracellular symbionts such as Endozoicomonas may prevail during a torpor state in a urochordate species. We expanded the current knowledge of phylogenetic and functional diversity in the symbiotic Endozoicomonas clade, showing that autocatalytic and nutritional-associated features are present and may be upregulated during the torpor state. Endozoicomonas likely plays an important role in the fitness of the holobiont, yet it is unclear if this role is beneficial. Given that these symbionts appear to occupy only a certain tissue type during torpor, our study raises new questions regarding the role of the vascular tissue that composes the entire soma during torpor states. As torpor is frequently recorded in various marine invertebrates and vertebrates (Crawshaw et al., 1985; Cáceres, 1997; Miller and Roth, 2009; Storey and Storey, 2011; Chen et al., 2016), it is highly plausible to find similar torpor-associated symbioses in other marine invertebrates. Thus, more studies should focus on similar torpor scenarios in the marine arena, aiming to shed light on this abrupt response to hostile environmental conditions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
YH and BR conceived and designed the experiments. YH and AR performed the experiments. BR, YR, and MR-B provided resources. YH and MR-B analyzed the data. YH, BR, and MR-B wrote the manuscript with the contributions of all co-authors. All authors contributed to the article and approved the submitted version.
We thank G. Paz and Z. Lapidot for their professional assistance and support during this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This study was funded by the United States–Israel Binational Science Foundation (BSF No. 2015012; to BR), by the e BSF, as part of the joint program with the NSF, the National Science Foundation, USA (NSF/BSF no 2021650; to BR), by the ISF grant No. 172/17 to BR and by the ISF grant 913/19 to MR-B.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1072053/full#supplementary-material
Avishai, N., Rabinowitz, C., and Rinkevich, B. (2003). Use of the comet assay for studying environmental genotoxicity: comparisons between visual and image analyses. Environ. Mol. Mutagen. 42, 155–165. doi: 10.1002/em.10189
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Behrendt, L., Larkum, A. W., Trampe, E., Norman, A., Sørensen, S. J., and Kühl, M. (2012). Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J. 6, 1222–1237. doi: 10.1038/ismej.2011.181
Beinart, R. A., Nyholm, S. V., Dubilier, N., and Girguis, P. R. (2014). Intracellular O ceanospirillales inhabit the gills of the hydrothermal vent snail A lviniconcha with chemosynthetic, γ-P roteobacterial symbionts. Environ. Microbiol. Rep. 6, 656–664. doi: 10.1111/1758-2229.12183
Benes, V., Blake, J., and Doyle, K. (2011). Ribo-Zero Gold Kit: improved RNA-seq results after removal of cytoplasmic and mitochondrial ribosomal RNA. Nat. Methods 8, iii–iv. doi: 10.1038/nmeth.f.352
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Blanchoud, S., Rutherford, K., Zondag, L., Gemmell, N. J., and Wilson, M. J. (2018). De novo draft assembly of the Botrylloides leachii genome provides further insight into tunicate evolution. Sci. Rep. 8, 1–18. doi: 10.1038/s41598-018-23749-w
Bourne, D. G., Dennis, P. G., Uthicke, S., Soo, R. M., Tyson, G. W., and Webster, N. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J. 7, 1452–1458. doi: 10.1038/ismej.2012.172
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Brinkkötter, A., Klöß, H., Alpert, C. A., and Lengeler, J. W. (2000). Pathways for the utilization of N-acetyl-galactosamine and galactosamine in Escherichia coli. Mol. Microbiol. 37, 125–135. doi: 10.1046/j.1365-2958.2000.01969.x
Buchanan, S. (1999). β-Barrel proteins from bacterial outer membranes: structure, function and refolding. Curr. Opin. Struct. Biol. 9, 455–461. doi: 10.1016/S0959-440X(99)80064-5
Cáceres, C. E. (1997). Dormancy in invertebrates. Invertebr. Biol. 116, 371–383. doi: 10.2307/3226870. S.C
Carey, H. V., Walters, W. A., and Knight, R. (2013). Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 304, R33–R42. doi: 10.1152/ajpregu.00387.2012
Carr, A. H., Amborski, R. L., Culley, D. D., and Amborski, G. F. (1976). Aerobic bacteria in the intestinal tracts of bullfrogs (Rana catesbeiana) maintained at low temperatures. Herpetologica 32, 239–244.
Chen, L., Hu, J. S., Xu, J. L., Shao, C. L., and Wang, G. Y. (2018). Biological and chemical diversity of ascidian-associated microorganisms. Mar. Drugs 16:362. doi: 10.3390/md16100362
Chen, M., Li, X., Zhu, A., Storey, K. B., Sun, L., Gao, T., et al. (2016). Understanding mechanism of sea cucumber Apostichopus japonicus aestivation: insights from TMT-based proteomic study. Compar. Biochem. Physiol. Part D Genomics Proteomics 19, 78–89. doi: 10.1016/j.cbd.2016.06.005
Correa, H., Haltli, B., Duque, C., and Kerr, R. (2013). Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae. Microbiol. Ecol. 66, 972–985. doi: 10.1007/s00248-013-0267-3
Crawshaw, L., Grahn, D., Wollmuth, L., and Simpson, L. (1985). Central nervous regulation of body temperature in vertebrates: comparative aspects. Pharmacol. Ther. 30, 19–30. doi: 10.1016/0163-7258(85)90045-2
De Cáceres, M., Legendre, P., Wiser, S. K., and Brotons, L. (2012). Using species combinations in indicator value analyses. Methods Ecol. Evol. 3, 973–982. doi: 10.1111/j.2041-210X.2012.00246.x
De Caralt, S., López-Legentil, I., Tarjuelo, M. J., and Uriz, X. (2002). Contrasting biological traits of Clavelina lepadiformis (Ascidiacea) populations from inside and outside harbours in the western Mediterranean Mar. Ecol. Prog. Ser. 244, 125–137. doi: 10.3354/meps244125
Degnan, B. M., Hawkins, C. J., Lavin, M. F., McCaffrey, E. J., Parry, D. L., van den Brenk, A. L., et al. (1989). New cyclic peptides with cytotoxic activity from the ascidian Lissoclinum patella. J. Med. Chem. 32, 1349–1354. doi: 10.1021/jm00126a034
Deutscher, J., Francke, C., and Postma, P. W. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in Bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031. doi: 10.1128/MMBR.00024-06
Ding, J. Y., Shiu, J. H., Chen, W. M., Chiang, Y. R., and Tang, S. L. (2016). Genomic insight into the host-endosymbiont relationship of Endozoicomonas montiporae CL-33T with its coral host. Front. Microbiol. 7:251. doi: 10.3389/fmicb.2016.00251
Dishaw, L. J., Flores-Torres, J., Lax, S., Gemayel, K., Leigh, B., Melillo, D., et al. (2014). The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS One 9:e93386. doi: 10.1371/journal.pone.0093386
Donia, M. S., Fricke, W. F., Partensky, F., Cox, J., Elshahawi, S. I., White, J. R., et al. (2011). Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc. Natl. Acad. Sci. 108, E1423–E1432. doi: 10.1073/pnas.1111712108
Erwin, P. M., Pineda, M. C., Webster, N., Turon, X., and López-Legentil, S. (2014). Down under the tunic: bacterial biodiversity hotspots and widespread ammonia-oxidizing archaea in coral reef ascidians. ISME J. 8, 575–588. doi: 10.1038/ismej.2013.188
Forget, N., and Juniper, K. (2013). Free-living bacterial communities associated with tubeworm (Ridgeia piscesae) aggregations in contrasting diffuse flow hydrothermal vent habitats at the Main Endeavour Field, Juan de Fuca Ridge. Microbiol. Open 22, 259–275. doi: 10.1002/mbo3.70
Fu, X., Do, T., Schmitz, F. J., Andrusevich, V., and Engel, M. H. (1998). New cyclic peptides from the ascidian Lissoclinum patella. J. Nat. Prod. 61, 1547–1551. doi: 10.1021/np9802872
Gardères, J., Bedoux, G., Koutsouveli, V., Crequer, S., Desriac, F., and Le Pennec, G. (2005). Lipopolysaccharides from commensal and opportunistic bacteria: characterization and response of the immune system of the host sponge Suberites domuncula. Mar. Drugs 5, 4985–5006. doi: 10.3390/md13084985
Glöckner, G., Albert-Weissenberger, C., Weinmann, E., Jacobi, S., Schunder, E., Steinert, M., et al. (2008). Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298, 411–428. doi: 10.1016/j.ijmm.2007.07.012
Gossling, J., Loesche, W., and Nace, G. (1982, 1982). Large intestine bacterial flora of nonhibernating and hibernating leopard frogs (Rana pipiens). Appl. Environ. Microbiol. 44, 59–66. doi: 10.1128/aem.44.1.59-66.1982
Graham, C. L., and Graham, D. L. (1978). Occurrence of Escherichia coli in feces of Psittacine birds. Avian Dis. 22, 717–720. doi: 10.2307/1589649
Gruber-Vodicka, H. R., Seah, B. K., and Pruesse, E. (2020). phyloFlash: rapid small-subunit rRNA profiling and targeted assembly from metagenomes. mSystems 5:e00920-20. doi: 10.1128/mSystems.00920-20
Haber, M., and Ilan, M. (2014). Diversity and antibacterial activity of bacteria cultured from Mediterranean Axinella spp. sponges. J. Appl. Microbiol. 116, 519–532. doi: 10.1111/jam.12401
Hand, S. C., and Hardewig, I. (1996). Downregulation of cellular metabolism during environmental stress: mechanisms and implications. Annu. Rev. Physiol. 58, 539–563. doi: 10.1146/annurev.ph.58.030196.002543
Hinzke, T., Kleiner, M., Meister, M., Schlüter, R., Hentschker, C., Pané-Farré, J., et al. (2021). Bacterial symbiont subpopulations have different roles in a deep-sea symbiosis. Elife 10:e58371. doi: 10.7554/eLife.58371
Hirose, E. (2015). Ascidian photosymbiosis: diversity of cyanobacterial transmission during embryogenesis. Genesis 53, 121–131. doi: 10.1002/dvg.22778
Hyams, Y., Panov, J., Rosner, A., Brodsky, L., Rinkevich, Y., and Rinkevich, B. (2022). Transcriptome landscapes that signify Botrylloides leachi (Ascidiacea) torpor states. Dev. Biol. 490, 22–36. doi: 10.1016/j.ydbio.2022.06.005
Hyams, Y., Paz, G., Rabinowitz, C., and Rinkevich, B. (2017). Insights into the unique torpor of Botrylloides leachi, a colonial urochordate. Dev. Biol. 428, 101–117. doi: 10.1016/j.ydbio.2017.05.020
Jensen, S., Duperron, S., Birkeland, N.-K., and Hovland, M. (2010). Intracellular Oceanospirillales bacteria inhabit gills of Acesta bivalves. FEMS Microbiol. Ecol. 743, 523–533. doi: 10.1111/j.1574-6941.2010.00981.x
Jessen, C., Villa Lizcano, J. F., Bayer, T., Roder, C., Aranda, M., Wild, C., et al. (2013). In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea coral Acropora hemprichii. PLoS One 8:e62091. doi: 10.1371/annotation/be4a3168-5284-4083-b5ed-5cd0f4630823
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359
Kieser, S., Brown, J., Zdobnov, E. M., Trajkovski, M., and McCue, L. A. (2020). ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinform. 21:257. doi: 10.1186/s12859-020-03585-4
Kohl, K. D., Amaya, J., Passement, C. A., Dearing, M. D., and McCue, M. D. (2014). Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol. Ecol. 90, 883–894. doi: 10.1111/1574-6941.12442
Lee, M. D., and Ponty, Y. (2019). GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35, 4162–4164. doi: 10.1093/bioinformatics/btz188
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lillie, R. D. (1954). Histopathologic technic and practical histochemistry. New York: The Blakiston.
Lim, J. G., and Choi, S. H. (2014). IscR is a global regulator essential for pathogenesis of vibrio vulnificus and induced by host cells. Infect. Immun. 82, 569–578. doi: 10.1128/IAI.01141-13
Lindsay, E. C., Metcalfe, N. B., and Llewellyn, M. S. (2020). The potential role of the gut microbiota in shaping host energetics and metabolic rate. J. Anim. Ecol. 89, 2415–2426. doi: 10.1111/1365-2656.13327
Liu, Y., Wu, P., Li, C., Xiao, Y., Wang, T., and Lin, L. (2021). The bacterial composition associated with Atriolum robustum, a common ascidian from Xisha coral reef, China. Symbiosis 83, 153–161. doi: 10.1007/s13199-020-00742-4
López-Legentil, S., Song, B., Bosch, M., Pawlik, J. R., and Turon, X. (2011). Cyanobacterial diversity and a new Acaryochloris-like symbiont from bahamian sea-squirts. PLoS One 6:e0023938. doi: 10.1371/journal.pone.0023938
Martínez-García, M., Stief, P., Díaz-Valdés, M., Wanner, G., Ramos-Esplá, A., Dubilier, N., et al. (2008). Ammonia-oxidizing Crenarchaeota and nitrification inside the tissue of a colonial ascidian. Environ. Microbiol. 10, 2991–3001. doi: 10.1111/j.1462-2920.2008.01761.x
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Merhej, V., and Raoult, D. (2011). Rickettsial evolution in the light of comparative genomics. Biol. Rev. 86, 379–405. doi: 10.1111/j.1469-185X.2010.00151.x
Miller, D. L., and Roth, M. B. (2009). C. elegans are protected from lethal hypoxia by an embryonic diapause. Curr. Biol. 19, 1233–1237. doi: 10.1016/j.cub.2009.05.066
Miller, I. J., Weyna, T. R., Fong, S. S., Lim-Fong, G. E., and Kwan, J. C. (2016). Single sample resolution of rare microbial dark matter in a marine invertebrate metagenome. Sci. Rep. 61:34362. doi: 10.1038/srep34362
Mishra, M., Panda, S., Barik, S., Sarkar, A., Singh, D. V., and Mohapatra, H. (2020). Antibiotic resistance profile, outer membrane proteins, virulence factors and genome sequence analysis reveal clinical isolates of Enterobacter are potential pathogens compared to environmental isolates. Front. Cell. Infect. Microbiol. 10:54. doi: 10.3389/fcimb.2020.00054
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural volcanic CO2 seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Morrow, K. M., Moss, A. G., Chadwick, N. E., and Liles, M. R. (2012). Bacterial associates of two caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 7818, 6438–6449. doi: 10.1128/AEM.01162-12
Mukai, H., Koyama, H., and Watanabe, H. (1983). Studies on the reproduction of three species of Perophora (Ascidiacea). Biol. Bull. 164, 251–266. doi: 10.2307/1541143
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Nishijima, M., Adachi, K., Katsuta, A., Shizuri, Y., and Yamasato, K. (2013). Endozoicomonas numazuensis sp. nov., a gammaproteobacterium isolated from marine sponges, and emended description of the genus Endozoicomonas Kurahashi and Yokota 2007. Int. J. Sys. Evol. Microbiol. 63, 709–714. doi: 10.1099/ijs.0.042077-0
Nissen, J. N., Johansen, J., Allesøe, R. L., Sønderby, C. K., Armenteros, J. J. A., Grønbech, C. H., et al. (2021). Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 39, 555–560. doi: 10.1038/s41587-020-00777-4
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Biol. 25, 1043–1055. doi: 10.1101/gr.186072.114
Paul, V. J., Lindquist, N., and Fenical, W. (1990). Chemical defenses of the tropical ascidian Atapozoa sp. and its nudibranch predators Nembrotha spp. Mar. Ecol. Progr. Ser. 59, 109–118. doi: 10.3354/meps059109
Paz, G., and Rinkevich, B. (2002). Morphological consequences for multi-partner chimerism in Botrylloides, a colonial urochordate. Dev. Compar. Immunol. 26, 615–622. doi: 10.1016/S0145-305X(02)00022-8
Perez, M., Breusing, C., Angers, B., Beinart, R. A., Won, Y.-J., and Young, C. R. (2022). Divergent paths in the evolutionary history of maternally transmitted clam symbionts. Proc. R. Soc. B Biol. Sci. 289:20212137. doi: 10.1098/rspb.2021.2137
Pogoreutz, C., Clua, E. E., and Tortolero-Langarica, J. J. (2022). High live coral cover and incidence of a pink-spotted coral phenotype on remote reefs off Clipperton Island. Trop. Eastern Pac. Mar. Biol. 169, 1–10. doi: 10.1007/s00227-022-04101-3
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Raina, J. B., Tapiolas, D., Willis, B. L., and Bourne, D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/AEM.02567-08
Rinkevich, B., and Shapira, M. (1998). An improved diet for inland broodstock and the establishment of an inbred line from Botryllus schlosseri, a colonial sea squirt (Ascidiacea). Aquat. Living Resour. 11, 163–171. doi: 10.1016/S0990-7440(98)80113-7
Rinkevich, B., and Weissman, I. L. (1987). A long-term study on fused subclones in the ascidian Botryllus schlosseri: the resorption phenomenon (Protochordata: Tunicata). J. Zool. 213, 717–733. doi: 10.1111/j.1469-7998.1987.tb03736.x
Rodionov, D. A., Vitreschak, A. G., Mironov, A. A., and Gelfand, M. S. (2002). Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 277, 48949–48959. doi: 10.1074/jbc.M208965200
Rua, C. P. J., Trindade-Silva, A. E., Appolinario, L. R., Venas, T. M., Garcia, G. D., Carvalho, L. S., et al. (2014). Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera brasiliensis. PeerJ 2:e419. doi: 10.7717/peerj.419
Schmidt, E. W. (2014). The secret to a successful relationship: lasting chemistry between ascidians and their symbiotic bacteria. Invertebr. Biol. 134, 88–102. doi: 10.1111/ivb.12071
Schreiber, L., Kjeldsen, K. U., Funch, P., Jensen, J., Obst, M., López-Legentil, S., et al. (2016). Endozoicomonas are specific, facultative symbionts of sea squirts. Front. Microbiol. 7:1042. doi: 10.3389/fmicb.2016.01042
Shaffer, M., Borton, M. A., McGivern, B. B., Zayed, A. A., La Rosa, S. L., Solden, L. M., et al. (2020). DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 48, 8883–8900. doi: 10.1093/nar/gkaa621
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., et al. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843. doi: 10.1038/s41564-018-0171-1
Sommer, F., Ståhlman, M., Ilkayeva, O., Arnemo, J. M., Kindberg, J., Josefsson, J., et al. (2016). The gut microbiota modulates energy metabolism in the hibernating brown bear ursus arctos. Cell Rep. 14, 1655–1661. doi: 10.1016/j.celrep.2016.01.026
Sonoyama, K., Fujiwara, R., Takemura, N., Ogasawara, T., Watanabe, J., Ito, H., et al. (2009). Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl. Environ. Microbiol. 75, 6451–6456. doi: 10.1128/AEM.00692-09
Storey, K. B. (2002). Life in the slow lane: molecular mechanisms of estivation. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133, 733–754. doi: 10.1016/S1095-6433(02)00206-4
Storey, K. B., and Storey, J. M. (eds). (2011). “Hibernation: Poikilotherms” in ELS (Chichester: John Wiley & Sons, Ltd).
Sze, M., Doonan, J., Mcdonald, J. E., Harris, R., and Dewar, M. (2020). “Factors that shape the host microbiome” in Microbiomes of soils, plants and animals. eds. R. E. Antwis, X. A. Harrison, and M. J. Cox (Cambridge: Cambridge University Press), 55–57.
Tandon, K., Lu, C. Y., Chiang, P. W., Wada, N., Yang, S. H., Chan, Y. F., et al. (2020). Comparative genomics: dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J. 14, 1290–1303. doi: 10.1038/s41396-020-0610-x
Tianero, M. D. B., Kwan, J. C., Wyche, T. P., Presson, A. P., Koch, M., Barrows, L. R., et al. (2015). Species specificity of symbiosis and secondary metabolism in ascidians. ISME J. 9, 615–628. doi: 10.1038/ismej.2014.152
Turon, X. (1992). Periods of non-feeding in Polysyncraton lacazei (Ascidiacea: Didemnidae): a rejuvenative process? Mar. Biol. 112, 647–655. doi: 10.1007/BF00346183
Ueki, T., Fujie, M., Romaidi,, and Satoh, N. (2019). Symbiotic bacteria associated with ascidian vanadium accumulation identified by 16S rRNA amplicon sequencing. Mar. Genomics 43, 33–42. doi: 10.1016/j.margen.2018.10.006
Vanni, V., Anselmi, C., Ballarin, L., Drago, L., Gasparini, F., Gordon, T., et al. (2022). “Current knowledge on stem cells in ascidians” in Advances in aquatic invertebrate stem cell research. eds. L. Ballarin, B. Rinkevich, and B. Hobmayer (Basel: MDPI AG), 273–333.
Vervoort, H. C. R., Pawlik, J., and Fenical, W. (1998). Chemical defense of the Caribbean ascidian Didemnum conchyliatum. Mar. Ecol. Prog. Ser. 164, 221–228. doi: 10.3354/meps164221
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Wiebler, J. M., Kohl, K. D., Lee, R. E. Jr., and Costanzo, J. P. (2018). Urea hydrolysis by gut bacteria in a hibernating frog: evidence for urea-nitrogen recycling in Amphibia. Proc. R. Soc. B Biol. Sci. 285:20180241. doi: 10.1098/rspb.2018.0241
Wu, Y. W., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607. doi: 10.1093/bioinformatics/btv638
Keywords: ascidians, torpor, hibernation, aestivation, symbiosis, Endozoicomonas, metabolism
Citation: Hyams Y, Rubin-Blum M, Rosner A, Brodsky L, Rinkevich Y and Rinkevich B (2023) Physiological changes during torpor favor association with Endozoicomonas endosymbionts in the urochordate Botrylloides leachii. Front. Microbiol. 14:1072053. doi: 10.3389/fmicb.2023.1072053
Received: 17 October 2022; Accepted: 02 May 2023;
Published: 31 May 2023.
Edited by:
Marcelino T. Suzuki, Sorbonne Université, FranceReviewed by:
Ayelet Voskoboynik, Stanford University, United StatesCopyright © 2023 Hyams, Rubin-Blum, Rosner, Brodsky, Rinkevich and Rinkevich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yosef Hyams, eW8tY2h5YW1zQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.