
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 February 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1066805
This article is part of the Research Topic Insights on Plant-Associated Microorganisms: Diversity, Systematics and Genomics View all 23 articles
 Peiling Li1,2†
Peiling Li1,2† Dan Zong1,2,3†
Dan Zong1,2,3† Peihua Gan1,2†
Peihua Gan1,2† Hailin Li1,2
Hailin Li1,2 Zhiyang Wu1,2
Zhiyang Wu1,2 Fahong Li1,2
Fahong Li1,2 Changlin Zhao3,4
Changlin Zhao3,4 Laigeng Li5
Laigeng Li5 Chengzhong He1,2,3*
Chengzhong He1,2,3*Background: Pinus yunnanensis is a major silvicultural species in Southwest China. Currently, large areas of twisted-trunk Pinus yunnanensis stands severely restrict its productivity. Different categories of rhizosphere microbes evolve alongside plants and environments and play an important role in the growth and ecological fitness of their host plant. However, the diversity and structure of the rhizosphere microbial communities between P. yunnanensis with two different trunk types—straight and twisted—remain unclear.
Methods: We collected the rhizosphere soil of 5 trees with the straight and 5 trees with the twisted trunk type in each of three sites in Yunnan province. We assessed and compared the diversity and structure of the rhizosphere microbial communities between P. yunnanensis with two different trunk types by Illumina sequencing of 16S rRNA genes and internal transcribed spacer (ITS) regions.
Results: The available phosphorus in soil differed significantly between P. yunnanensis with straight and twisted trunks. Available potassium had a significant effect on fungi. Chloroflexi dominated the rhizosphere soils of the straight trunk type, while Proteobacteria was predominant in the rhizosphere soils of the twisted trunk type. Trunk types significantly explained 6.79% of the variance in bacterial communities.
Conclusion: This study revealed the composition and diversity of bacterial and fungal groups in the rhizosphere soil of P. yunnanensis with straight and twisted trunk types, providing proper microbial information for different plant phenotypes.
Pinus yunnanensis is an endemic tree species in Southwest China that makes up one of the principal subtropical coniferous forests (Gao et al., 2021). It is distributed in a variety of geological areas ranging from 23° to 30° N and 96° to 108° E and grows in a continuous distribution at elevations ranging from 700 to 3,000 m in the Yunnan–Guizhou region (Wang et al., 2013; Gao et al., 2021). P. yunnanensis plays an important role in forestry economic development and environmental regulation in China (Xu et al., 2016; Wang X. et al., 2021). The degradation of forest stands of P. yunnanensis is becoming increasingly serious, leading to a growing proportion of low-quality stands (e.g., twisted, stunted, bending) (Zhou et al., 2016), which severely restrict the utilization and development of P. yunnanensis. Nevertheless, to date, the cause for the formation of twisted trunk characteristics remains unclear.
Much attention has been given to the cause of trunk twisting. On the one hand, many researchers have suspected that the causes of trunk twists are the wind, Earth’s rotation, injuries, directional aspects, exposure, and the sun and moon (Kubler, 1991; Eklund and Säll, 2000) as well as soil nutrient status and other soil conditions (Copisarow, 1933; Harris, 1989). On the other hand, it has been reported that trunk twisting is under considerable genetic control, such as in P. radiata (Burdon and Low, 1992; Gapare et al., 2007), Picea sitchensis (Hansen and Roulund, 1997) and Picea abies (Silva et al., 2000). Regarding trunk twisting in P. yunnanensis, Chinese researchers also have different viewpoints. Chen and Lyu (1962) believed that the density was the basic factor for the occurrence and development of twisting and that the wind promoteed and strengthened twisting. Zhou (1974) thought that strong sunlight and periodic excessive moisture were the main causes. However, many studies have shown that the trunk type is genetically controlled (Cai et al., 2016). Combining investigation methods with traditional breeding, it was found that the twisted trunk phenotype of parents could be passed on to offspring (Chen et al., 1997). Some researchers hold the view that the formation of a large number of twisted trunks in P. yunnanensis is caused by the extensive selection logging of natural forests at early stages, the artificial negative selection of natural disasters and unclear germplasm sources for afforestation (Chen et al., 2014; Wang X. et al., 2021). Modern molecular biology methods also revealed that trunk twisting is mainly regulated by genetic factors (He, 1994; Zhou et al., 2016), but currently, researchers believe that the synergistic effect of environmental factors (biotic and abiotic) enhance the incidence of this twisted feature (Wu et al., 2022). Our previous transcriptome data from phloem showed that the differentially expressed genes (DEGs) between P. yunnanensis with the straight and twisted trunk types were mainly involved in the interaction between plants and microorganisms (not published).
Roots not only provide plants with mechanical support, water and nutrients (Molefe et al., 2021), but also exude an enormous range of potentially valuable compounds into the rhizosphere (Walker et al., 2003; Bais et al., 2006). Cumulative evidence suggests that these compounds play an invaluable role in determining the interactions between roots and, eventually, the dynamics of plant and soil communities (Bais et al., 2006; Broeckling et al., 2008; Chai and Schachtman, 2022). The rhizosphere encompasses the millimeters of soil surrounding a plant root that is home to an overwhelming number of microorganisms (Berendsen et al., 2012; Philippot et al., 2013). Recent studies have revealed that different species of plants, or even different plant traits of the same species, assemble different rhizosphere microbial communities in the same soil environment (Arafat et al., 2017; Sayer et al., 2017; Bickford et al., 2020). These complex plant-associated microbial communities are crucial for plants because they can affect plant growth, productivity, nutrients and immunity directly or indirectly (Van Wees et al., 2004; Egamberdieva et al., 2010; Erturk et al., 2010; Berendsen et al., 2012; Patel, 2018; Tahir et al., 2019; Gu et al., 2020; Song Q. et al., 2021), as well as affect host phenotypes and adaptability (Herrera Paredes et al., 2018). For instance, Finkel et al. (2020) found that rhizosphere microorganisms can promote root growth by affecting plant hormone levels in Arabidopsis. Additionally, Wagner et al. (2014) confirmed that rhizosphere microorganisms can affect flowering phenology and selection on flowering time. There is growing evidence of the ability of rhizosphere microorganisms to alter the root morphological structure and improve root functions, which in turn improves plant nutrient uptake and physiology (Vacheron et al., 2013; Castellano-Hinojosa et al., 2021). Furthermore, some rhizosphere microorganisms can also severely constrain plant growth and development. Some studies have demonstrated that the dysbiosis of the protective bacterial communities in rhizosphere soil promotes the incidence of disease while manifestingin phenotypes that differ from those of healthy plants (van der Heijden et al., 2008; Hu et al., 2018; Lee et al., 2020). In addition, the rhizosphere microbiota is a very important source of endophytic microorganisms (Chi et al., 2004; Vandana et al., 2021). Microorganisms have been observed to enter host plants through roots and colonize plants (Chi et al., 2005; Thomas and Reddy, 2013). It has been reported that among the various plant-associated microbiota, endophytic microorganisms exhibit the most intimate and specific association with host plants (Vasileva et al., 2019; Mishra et al., 2022). There is growing evidence that the endophytic microorganisms support plants against both biotic and abiotic stresses (Ludwig-Muller, 2015; Sugio et al., 2015). For example, they directly or indirectly promote plant growth by inhibiting the growth of plant pathogens, producing various secondary metabolites (Giménez et al., 2007; Kang et al., 2007; Balmer et al., 2013; Yan et al., 2019; Matsumoto et al., 2021; Shubhransu et al., 2021; Vandana et al., 2021) and impacting the host phenotype (Jousset et al., 2017; Hassani et al., 2018; Harrison et al., 2021). Thus, it is critical to study the diversity and function of rhizosphere microorganisms and their contribution to a healthy plant.
Plants survive and evolve in the presence of microorganisms, which could be pathogenic or symbiotic (Hartmann et al., 2014). The consensus reached is that plant performance and activities can be fully described and understood only when plants and closely related microflora are considered (Zilber-Rosenberg and Rosenberg, 2008). In other words, the plant is considered an organism harboring microbial populations (Rosier et al., 2016). Previous studies on plant rhizosphere microbes have been mostly conducted on model plants and cash crops, such as Arabidopsis thaliana (Robbins et al., 2018), maize (Aira et al., 2010; Molefe et al., 2021), and wheat (Egamberdieva et al., 2008; Rossmann et al., 2020). Nevertheless, the diversity of the rhizosphere microbiota of P. yunnanensis remains unclear, including the differences in rhizosphere microbiota between the twisted and straight trunk types.
In the present study, to further understand the relationship between trunk type and microorganisms in P. yunnanensis, we hypothesized that the diversity and structure of the rhizosphere microbiota of P. yunnanensis with the twisted trunk type would differ significantly from that of the straight trunk type. Based on the above, our objectives were to (i) compare the diversity and structural characteristics of the rhizosphere microbial communities between the straight and twisted trunk types of P. yunnanensis base on high-throughput sequencing and statistical methods, and (ii) estimate the interactions of P. yunnanensis with rhizosphere microorganisms and soil properties and their effects on trunk type.
We sampled rhizosphere and bulk soils of P. yunnanensis with two different trunk types at three representative growth sites, including 5 trees with straight and 5 trees with twisted trunk at each growth site. The growth sites (Supplementary Table 1) were located in the cities of Kunming, Chuxiong and Dali in Yunnan Province, within the longitudinal range from 99.85° to 103.23° E. The distance between the sites exceeds 200 km, with the farthest distance being between Kunming and Dali, reaching 760 km.
P. yunnanensis with the two different trunk types are intermixed in the forest stands under the same management (Supplementary Figure 1). After the removal of litter, we collected soil samples from the rhizosphere and bulk soil at a depth of 20 cm using a hoe. Rhizosphere soil, collected by using the shake-off method of Riley and Barber (1971), was immediately stored in liquid nitrogen, transported back to the laboratory, and stored at -80°C for microbial diversity analysis. The soil that had no plants growing nearby was considered bulk soil. The bulk soils collected were air-dried at ambient temperature for soil property measurement (Shi et al., 2011; Kamutando et al., 2017).
Air-dried bulk soil samples were used to measure organic matter, pH, total and available nitrogen (N), phosphorus (P), and potassium (K), and catalase and sucrase activity. The soil organic matter was measured using the K2Cr2O7 (Walkley-Blach) method. The pH value was determined by the glass electrode method; total N by the Kjeldahl method; total P and available P by the molybdenum antimony resistance colorimetric method; total K by the NaOH melting-flaming luminosity method; available N by diffusion; and available K was determined in ammonium acetate (NH4OAc) extract by flame photometry (Zhang et al., 2014). Sucrase activity was measured using the 3,5-dinitrosalicylic acid colorimetric method, and catalase activity was determined by the KMnO4 titration method (Ge et al., 2018).
Microbial DNA from rhizosphere soil samples was extracted using an MN NucleoSpin 96 Soi kit (Macherey-Nagel, Germany) following the manufacturer’s instructions. The concentration of the DNA extracted from the each sample was shown in Supplementary Table 2. The V3–V4 hypervariable regions of the 16S rRNA gene were amplified for each sample using barcoded universal primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′)/806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Guo et al., 2018). The ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′)/ITS2 (5′GCTCGTTCTTCATCGATGC-3′) primer pair was used to amplify the fungal internal transcribed spacer (ITS1) gene (Gardes and Bruns, 1993; Li et al., 2020). All high-throughput sequencing analyses of bacterial and fungal genes were performed based on the Illumina HiSeq 2500 platform (2 × 250 paired ends) at Biomarker Technologies Corporation (Beijing, China).
The raw data were quality-filtered using Trimmomatic (version 0.33) (Bolger et al., 2014), and the primer sequences were identified and removed by Cutadapt (version 1.9.1) (Martin, 2011). To obtain high-quality sequences, splicing of double-ended reads was performed using USEARCH (version 10), and chimeras were removed from the operational taxonomic unit (OTU) table using UCHIME (version 8.1) (Edgar et al., 2011; Edgar, 2013). The OTUs of bacteria and fungi were clustered at the 97% nucleotide identity threshold using USEARCH (version 10.0) (Edgar, 2013) with the GreenGenes Database (version 13.5) (DeSantis et al., 2006) and the Unite Database (version 8.0) (Kõljalg et al., 2005) as the reference. OTUs were filtered using 0.005% of all sequence numbers as a threshold (Bokulich et al., 2013).
The differences in the microbial communities between the two groups were analyzed using linear discriminant analysis (LDA) effect size (LEfSe) based on taxonomic composition at different classification levels.
The soil physical and chemical properties of the two different trunk types of P. yunnanensis were determined. As shown in Table 1, compared with the soil from the twisted trunk group, the soil from the straight trunk group had a significantly lower available P content (P = 0.02).
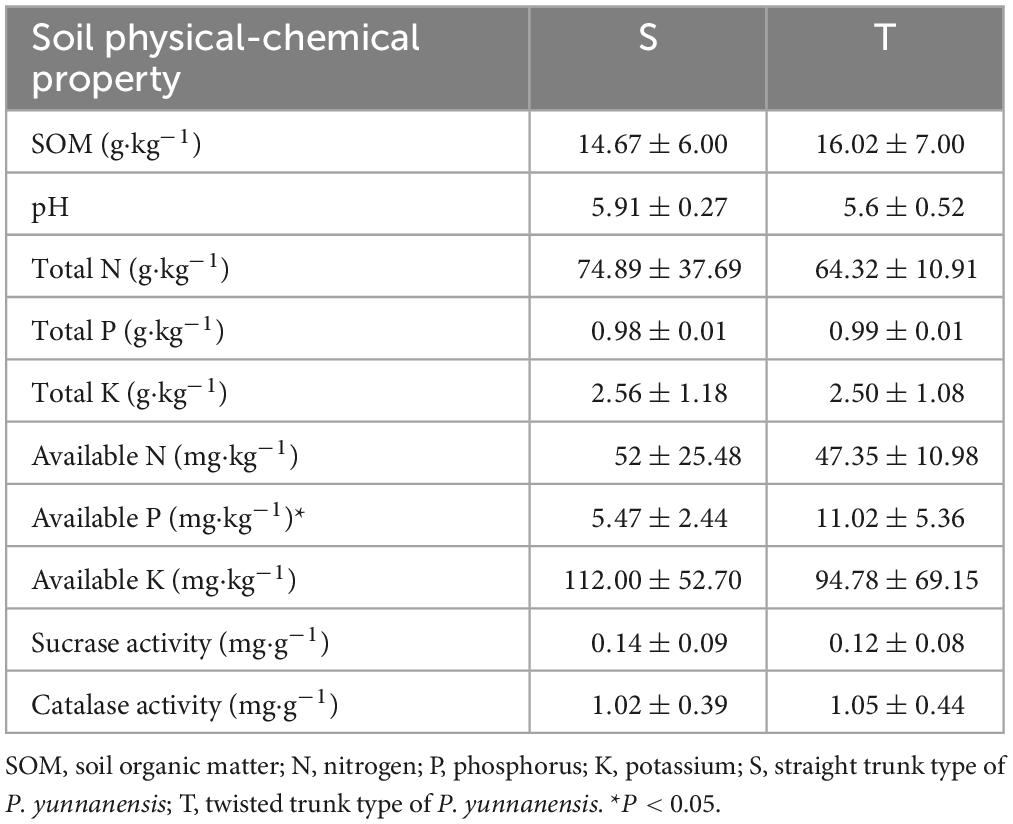
Table 1. The physical and chemical properties of soil in the sampling fields of twisted and straight trunk P. yunnanensis.
The impact of the soil physical and chemical properties on bacterial and fungal community variation was determined by Mantel tests (Table 2). The results showed that available K was correlated with fungal communities (P = 0.01).
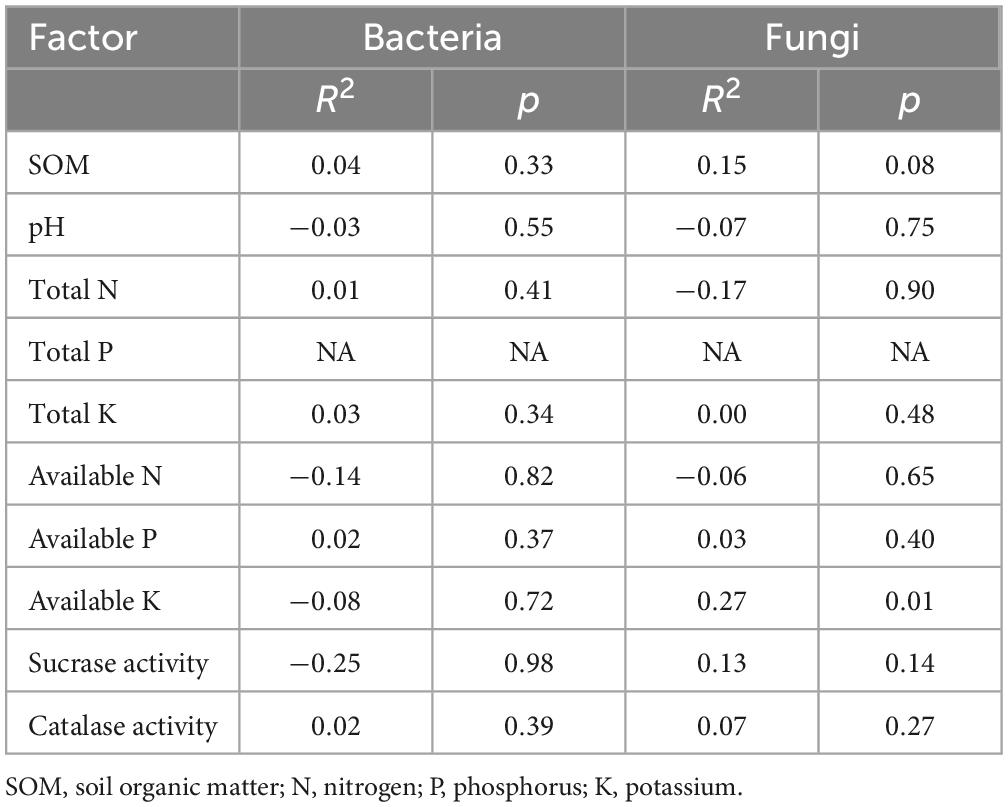
Table 2. Mantel tests of the correlation between the content of soil properties and the relative abundance of microbia at the genus level.
To explore the rhizosphere soil bacterial and fungal communities of P. yunnanensis with two different trunk types, high-throughput sequencing of 16S rRNA and internal transcribed spacer (ITS) regions was performed for microbiomes from 30 rhizosphere samples. In total, we obtained 2,395,106 high-quality paired reads for bacteria and 2,472,255 paired reads for fungi, accounting for 96.40 and 98.04% of their raw tags, respectively (Supplementary Table 3). The composition and relative abundance for each taxon were obtained based on the OTU classification results. At the cut-off of 97% similarity (Edgar, 2013), the rhizosphere soil microbial communities of P. yunnanensis consisted of 1, 601 bacterial OTUs and 929 fungal OTUs. Specifically, 1,061 bacterial and 877 fungal OTUs were identified in straight-trunk P. yunnanensis, while 1,061 bacterial OTUs and 895 fungal OTUs were identified in twisted-trunk P. yunnanensis (Figure 1).
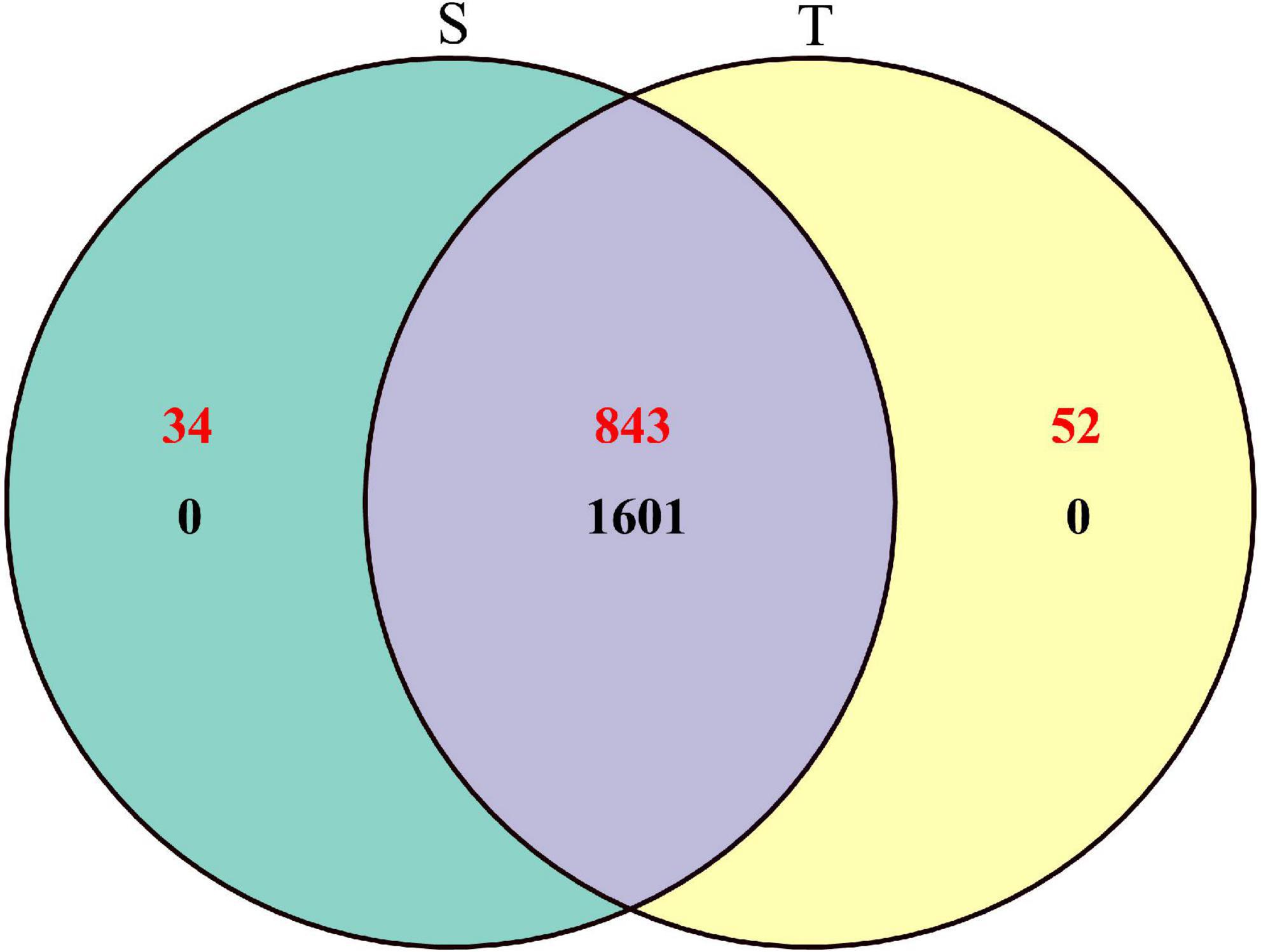
Figure 1. Venn diagram of specific and shared bacterial (black) and fungal (red) OTUs in straight and twisted trunk P. yunnanensis. S, straight trunk type of P. yunnanensis; T, twisted trunk type of P. yunnanensis.
According to the Chao1 and Shannon-Wiener indices, we did not observe a significant difference in alpha diversity between the two trunk types (Figure 2). Moreover, the PERMANOVA results suggested that trunk type explained 6.79% of the variance in bacterial communities (P = 0.02) (Table 3).
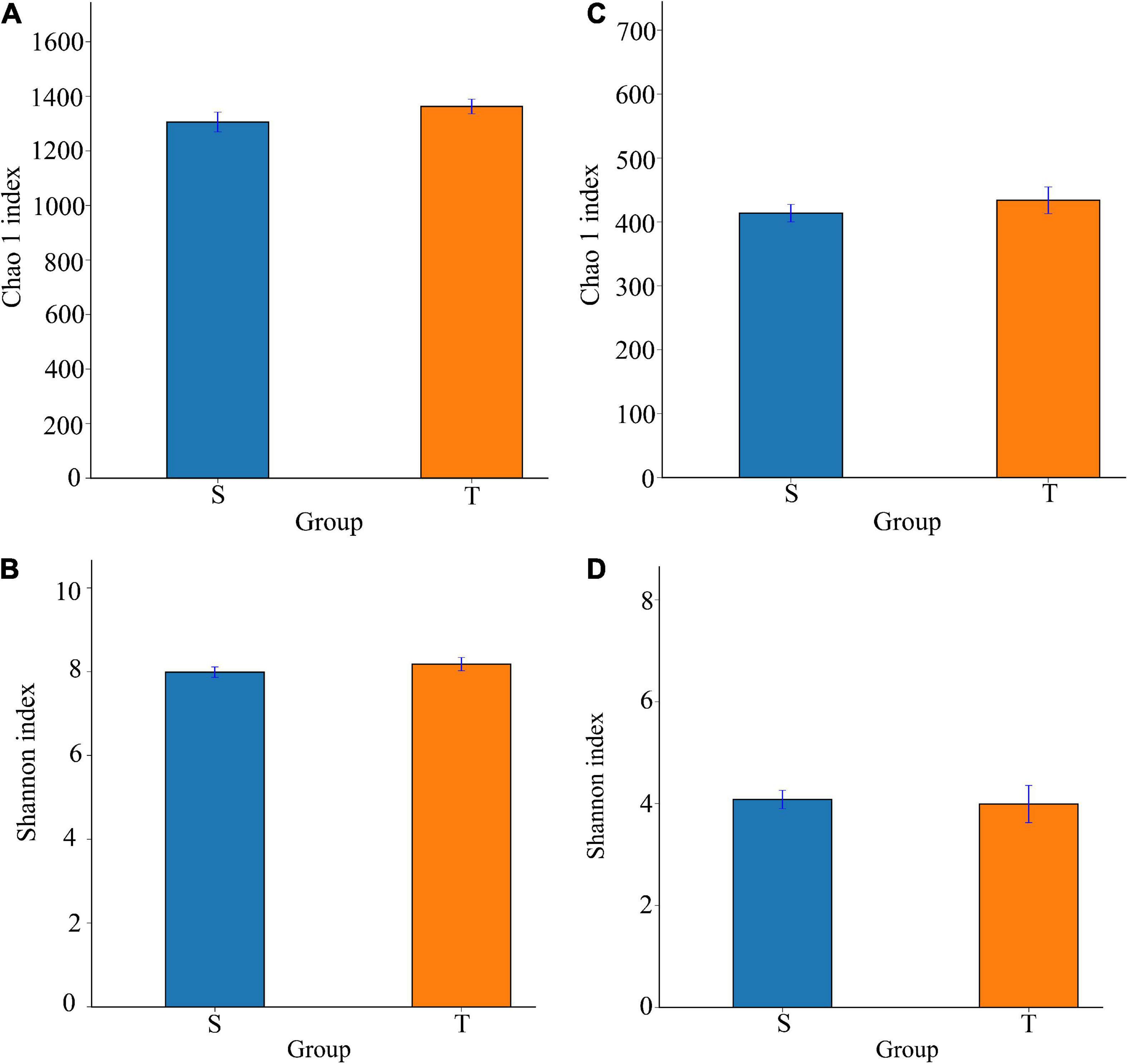
Figure 2. The Chao 1 (species richness), and Shannon (species diversity) indexes of bacterial and fungal communities for the straight and twisted trunk P. yunnanensis. Panels (A,B) represent Chao 1, and Shannon indexes of bacteria, respectively. Panels (C,D) represent Chao 1, and Shannon indexes of fungi, respectively. S, straight trunk type of P. yunnanensis; T, twisted trunk type of P. yunnanensis.
We assessed the taxonomic composition of straight- and twisted-trunk P. yunnanensis rhizosphere microbial communities at different classification levels, including the phylum, order and genus levels. The results are shown in Figure 3. At the phylum level, there were 24 different bacterial phyla in both the straight and twisted groups. Among them, the relative abundances of Proteobacteria in the rhizosphere microbial communities of twisted-trunk P. yunnanensi were higher than those of straight-trunk P. yunnanensis (P = 0.02, Supplementary Figure 2). Furthermore, the relative abundance of Chloroflexi in twisted-trunk P. yunnanensis was significantly lower than that in straight-trunk P. yunnanensis (P = 0.04, Supplementary Figure 2). The same 142 rhizosphere bacterial orders were obtained in both the straight and twisted trunk groups. In the twisted trunk group, the relative abundance of Pyrinomonadales was 5.6 times higher than that in the straight trunk group, but the difference was not significant. The relative abundance of uncultured_bacterium_c_Subgroup_6 was significantly higher in the twisted trunk group (P = 0.01, Supplementary Figure 2). At the genus level (Figure 3C), the bacterial community structure appeared more stable across the different compartments. Each of the two groups had 316 identical genera. Subgroup_2 (10.76 and 9.07%, respectively), Acidobacteriales (8.09 and 7.18%, respectively), and Elsterales (5.52 and 4.87%, respectively) were the most abundant taxonomically known genera between the straight and twisted trunk groups. The relative abundances of Acidobacteriales and uncultured_bacterium_c_AD3 were significantly higher in the straight trunk group (P = 0.03, Supplementary Figure 2). In addition, Bradyrhizobium was one of the most enriched bacterial genera in the rhizosphere microbial community of straight- and twisted-trunk P. yunnanensis.
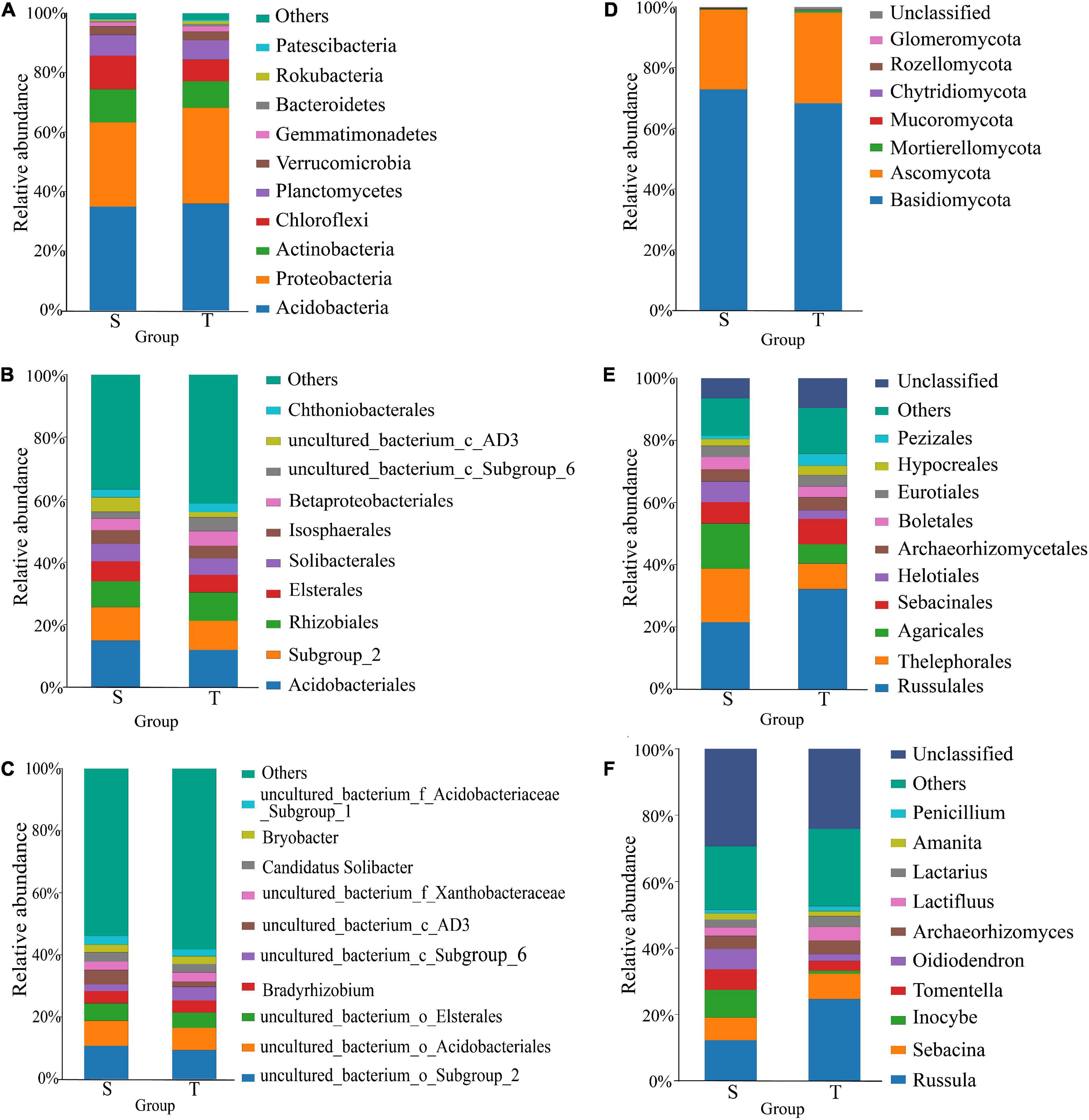
Figure 3. Taxonomic composition and structure of bacterial and fungi communities at phylum, order and genus levels. Panels (A–C) represent phylum, order and genus level of bacteria, respectively. Panels (D–F) represent phylum, order and genus level of fungi, respectively. S, straight trunk type of P. yunnanensis; T, twisted trunk type of P. yunnanensis.
We assessed the taxonomic structure of rhizosphere fungal communities at different classification levels by the same analysis method mentioned above. The composition of fungal communities comprised 7 different phyla, and 2 of them had a relative abundance greater than 1%, including Basidiomycota (72.96 and 68.35%, respectively) and Ascomycota (26.12 and 29.76%, respectively), with an overall relative abundance higher than 98% in the straight and twisted trunk groups (Figure 3D). The dominant fungal phyla in rhizosphere soil showed no clear difference between the straight and twisted trunk types of P. yunnanensis, as we estimated. In addition, there were 12 orders with an average relative abundance of more than 1% in the straight trunk group. The relative abundances of Thelephorales, Agaricales and Helotiales in the straight trunk group were more than twice as high as those in the twisted trunk group (Figure 3E). Nevertheless, such a difference was not significant due to the larger dispersion value. We identified a total of 222 and 223 fungal genera in the straight and twisted trunk groups, respectively. The results demonstrated that Inocybe (7.55 and 0.84%, respectively) was one of the genera with the greatest relative abundance variation between the straight and twisted trunk groups (Figure 3F). Furthermore, in the twisted trunk group, the relative abundances of both Tylopilus (1.64%) and Hymenogaster (1.36%) were greater than 1%. In contrast, in the straight trunk group, their relative abundances were less than 0.01%. The relative abundances of plant pathogens such as Penicillium and Fusicolla were higher in the twisted trunk group. At the species level, the relative abundance of Penicillium nodositatum was significantly higher in the twisted trunk group (P = 0.03, Supplementary Figure 2).
To explore the indicator bacterial and fungal communities in rhizosphere soil of straight- and twisted-trunk P. yunnanensis, we conducted LEfSe analysis to identify biomarkers for each group based on the taxonomic composition of rhizosphere microbial communities with logarithmic LDA score > 4.0. The results showed that there were 18 distinctly abundant taxa in the two groups (Figures 4A, C). Specifically, 7 were differentially abundant in straight-trunk P. yunnanensis, including Chloroflexi and Actinobacteria. There were 11 taxa in the twisted-trunk, including Pyrinomonadales and Proteobacteria.

Figure 4. Histogram of LDA value distribution and evolutionary branch graph of LEfSe analysis. (A) The microbial biomarkers between different trunk shapes of P. yunnanensis in bacteria. (B) The microbial biomarkers between different trunk shape P. yunnanensis for fungi. (C) The different taxon between different trunk shape P. yunnanensis in bacteria. (D) The different taxon between different trunk shape P. yunnanensis in fungi. The circles from inner to outer layers represent the taxonomic level from phylum to species. The dots on circles represents a term on the corresponding taxonomic level. The size of the dots indicates relative abundance. Coloring: Species with no significant difference are colored in yellow, orange stand for twisted trunk group, and blue for straight. Species with certain color mean the abundance of this species is the highest in the corresponding group, which helps to visualize the most important microbial communities in each group. The lowercase p, c, o, f, g, and s in front of the symbol “_” represent the phylum, class, order, family, genus and species, respectively. S, straight trunk type of P. yunnanensis; T, twisted trunk type of P. yunnanensis.
The LEfSe analysis of the fungal communities from the two trunk groups showed that 6 abundant fungal taxa presented significant differences, including Penicillium laeve, Oidiodendron, Myxotrichaccac, Russula cyanoxantha, Thelephoraceae and Thelephorales. Myxotrichaceae and Thelephoraceae were differentially abundant between the straight trunk and twisted trunk groups. The species Penicillium laeve and Russula cyanoxantha were more abundant in the twisted trunk group (Figures 4B, D).
To understand the soil conditions of straight and twisted trunk-type plants, we tested the physical and chemical properties of the soil around the roots. The results revealed that the available P content was significantly higher (P = 0.02) in the soil of twisted- than straight-trunk P. yunnanensis. Other physical and chemical properties were non-significantly different between the different trunk types of P. yunnanensis. The content of phosphorus and other nutrients in the soil represents the potential fertility of the soil (Song Q. et al., 2021). Soil fertility affects plant growth and the survival of microbes (Wang et al., 2020). For example, adequate phosphorus fertilization enhanced the seedlings growth and nutrient content of P. massoniana (Chen et al., 2022), and low fertility often tends to affect plant root structure and morphology to increase access to limiting nutrient resources (Zadworny et al., 2017). Kumar and Garkoti (2022) also reported that soil nutrients can influence the rhizosphere effect, which affects the transfer of energy and nutrients from the soil to plant roots. Furthermore, we found that available K was correlated with fungal communities (P = 0.01). As the environment for microbial life in the rhizosphere, soil properties influence the physiology and growth of soil microbial communities (Molefe et al., 2021). In addition, the associated soil nutrients (e.g., amount of C, N, and K) can change under biotic and abiotic environmental disturbances (Bastida et al., 2006; Otlewska et al., 2020; Crandall et al., 2022). Potassium-solubilizing microorganisms, widely present in different soil environments, can be used as biofertilizers to make available K from minerals and rocks, ultimately influencing soil nutrients, crop growth and quality (Das and Pradhan, 2016). The differential microorganisms in the rhizosphere of P. yunnanensis may disturb the balance of plant available nutrients in the soil and eventually lead to differences in the available P of different trunk types of P. yunnanensis. In turn, this difference can affect microorganisms. Moreover, our study revealed that trunk type explained 6.79 and 3.79% of the variance in bacterial (P = 0.02) and fungal communities, respectively. Perhaps the variance in microbial communities was determined by P. yunnanensis secreting compounds that specifically stimulate or inhibit the members of the microbial community (Bais et al., 2006; Arafat et al., 2017; Vives-Peris et al., 2020; Volpiano et al., 2022). In summary, we considered the soil environment, P. yunnanensis and microorganisms to interact and influence each other. We inferred that the trunk types of P. yunnanensis were influenced by microorganisms and soil properties, and vice versa.
Previous studies reported that plants can shape and recruit microbes from soil microbial communities to type rhizosphere microbial communities (Liu et al., 2021; Song Y. et al., 2021). Based on 16S rRNA gene sequence data, the rhizosphere bacterial community diversity in the straight and twisted trunk groups of P. yunnanensis was highly similar, including 24 phyla, 66 classes, 142 orders, 206 families, 316 genera and 334 species. The two different trunk types contained a similar relative abundance of rhizosphere microbial communities, which were dominated by Acidobacteria, Proteobacteria, Actinobacteria, Chloroflexi and Planctomycetes in P. yunnanensis. Acidobacteria constitutes the most abundant phylum whose members dominate soil bacterial communities (Liu et al., 2016). Here, we can exclude the effect of sampling distance on this result because the sampling sites were not close together. Acidobacteria, significantly enriched in the straight trunk group, are resistant to oxidative stress through the production of carotenoids, which may offer a competitive advantage for themselves in soils (Pinto et al., 2021). Their dynamic roles in vital ecological processes, including regulation of biogeochemical cycles, decomposition of biopolymers, exopolysaccharide secretion, and plant growth promotion have been investigated (Kalam et al., 2020). Surveys of root microbiomes suggested that certain members of the lineages cited above may be consistently enriched in the plant root environment (Yeoh et al., 2017). Some of them, such as Proteobacteria and Actinobacteria, occur in many plants as the dominant phyla, such as Arabidopsis thaliana (Bulgarelli et al., 2012), barley (Bulgarelli et al., 2015), lettuce (Schreiter et al., 2014), oak (Uroz et al., 2010), ginseng (Ying et al., 2012; Wang H. et al., 2021), Dendrobium (Zuo et al., 2021), and wheat (Rossmann et al., 2020). Furthermore, Bradyrhizobium was one of the most enriched bacterial genera in this study. Multiple studies have confirmed that co-inoculation of Bradyrhizobium and Plant Growth Promoting Rhizobacteria (PGPR) is beneficial to plant growth, including nodule biomass, root biomass, and shoot biomass, and they can be applied together as biofertilizers for production of economically important plants (Htwe et al., 2019; Zeffa et al., 2020).
According to our results, the twisted trunk rhizosphere microbiome group had slightly higher fungal OTUs and alpha diversity than the straight trunk group. The twisted trunk group consistently presented slightly higher bacterial alpha diversity than the straight trunk group. However, the differences were not significant. On the basis of the taxonomic composition, we observed that the relative abundances of Thelephorales, Agaricales, Helotiales and Oidiodendron in the straight trunk group were more than twice as likely as those in the twisted trunk group. Oidiodendron alters the length and branching of pioneer and fibrous roots of blueberry cuttings (Baba et al., 2021). Additionally, the relative abundances of Penicillium and Fusicolla were higher in the twisted trunk group. Unclassified genera accounted for 26.94%, which perhaps deserves further study in the future.
In this study, the LefSe analysis showed that Proteobacteria and Penicillium laeve were the key taxa in the rhizosphere soil of twisted-trunk P. yunnanensis. A large number of microorganisms in Proteobacteria (Tsolis, 2002; Preston et al., 2005) and Penicillium (Yang et al., 2017) are considered to be plant pathogens. In contrast, Acidobacteria and Oidiodendron were the key taxa in the rhizosphere soil of straight-trunk P. yunnanensis. Acidobacteria were reported to contribute to the healthy growth of their host plants and increase the chlorophyll content (Yoneda et al., 2021). Oidiodendron has a significant effect on root morphology (Baba et al., 2021).
Research on the effects of microorganisms on plants is extensive, both in terms of plant species and microbial species (Jacoby et al., 2017; Fitzpatrick et al., 2018; Caruso, 2020). Regarding conifers, Timonin (1964) found that disinfecting seeds of P. banksiana and P. glauca caused a significant reduction in seedling emergence. It was only later that researchers discovered that the lack of certain microorganisms on the seed surface may negatively affect seed germination and be detrimental to plant growth and development (Cardoso et al., 2011). Subsequent studies have found that rhizosphere microorganisms influence conifers in every way (Garcìa et al., 2004; Heredia-Acuña et al., 2018). For example, they influence the root length via hormones (Bent et al., 2001), shoot height and dry mass via phosphate solubilization (Singh et al., 2008), and growth via synergy or antagonism (Rojas et al., 2001). These effects determine the complex and variable phenotype of the plant. The variability occurs not only between species but also within a species. Understanding this variability is of key importance to improve the target products (Pot et al., 2002). Twisting is a representation of textured spirals (Gapare et al., 2007) and helical growth (Nakamura and Hashimoto, 2020). As a model plant, twisting has been studied in depth in Arabidopsis thaliana (Okada and Shimura, 1990; Buschmann et al., 2004; Nakamura and Hashimoto, 2020). However, the trunk twisting characteristics of trees have been less studied due to the long growth cycle and slow phenotypic shift of trees. To the best of our knowledge, there is no relevant report on the relationship between trunk types and microorganisms in conifers (e.g., P. yunnanensis). Rhizosphere microorganisms are one of the sources of endophytic microorganisms to plants, representing one of the important effects of microorganisms on plants. In the present study, we reported the first comprehensive investigation of microbial communities in rhizosphere soils between straight- and twisted-trunk P. yunnanensis.
Although our understanding of the effects and mechanisms of microbial action on plants is growing, there is no doubt that it is limited. The practical use and routine application of microorganisms remains a challenge, and it may take many years before our understanding is adequate to ensure their successful application in different systems (Cardoso et al., 2011). In this study, we could not determine that rhizosphere microorganisms contribute directly to the trunk types of P. yunnanensis, but the results provide us with some useful information for future studies on the causes of twisted trunk formation. Next, perhaps we will be able to determine the relationship between rhizosphere and endophytic microorganisms, determine the effect of a particular microorganism and/or available P on P. yunnanensis growth and trunk type and further investigate the effects of interactions of P. yunnanensis with microbes and environments on trunk types.
We provided a detailed and systematic understanding of the rhizosphere microbiome composition between straight and twisted trunk types of P. yunnanensis. Our high-throughput sequencing results demonstrated that the diversity and community composition of the two trunk types were similar. Further analysis showed that Proteobacteria and Penicillium laeve were the key taxa in the rhizosphere soil of twisted-trunk P. yunnanensis. In contrast, Acidobacteria and Oidiodendron were the key taxa in the rhizosphere soil of straight-trunk P. yunnanensis. Moreover, available potassium has a significant effect on fungi. Trunk type explained 6.79 and 3.79% of the variance in bacterial and fungal communities, respectively. Available phosphorus differed significantly between the two trunk types of P. yunnanensis. These findings significantly advance our fundamental understanding of the rhizosphere microorganisms of P. yunnanensis and of the microbial diversity of different plant phenotypes.
The data presented in this study are deposited in the NCBI repository, accession numbers are PRJNA892260 and PRJNA892749.
PL, DZ, CH, and LL planned and designed the experiment. PL, DZ, PG, ZW, and FL prepared the materials. PL and PG collected and analyzed the data. PL and DZ wrote the first draft of the manuscript. HL and CZ commented on previous versions of the manuscript. All authors read and approved the final manuscript.
This work was supported by Yunnan Provincial Expert Workstation (202005AF150020), Yunnan Provincial “Ten-Thousand Program” for Leading Industry Innovation (YUWR-CYJS-2020-018), and Applied Basic Research Programs of Yunnan Province of China (2017FA011).
We thank all the reviewers who participated in the review and LL (Chinese Academy of Sciences, Shanghai, China) for the study conception and design during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1066805/full#supplementary-material
Aira, M., Gómez-Brandón, M., Lazcano, C., Bååth, E., and Domínguez, J. (2010). Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 42, 2276–2281. doi: 10.1016/j.soilbio.2010.08.029
Arafat, Y., Wei, X., Jiang, Y., Chen, T.-W., Saqib, H. S. A., Lin, S., et al. (2017). Spatial distribution patterns of root-associated Bacterial communities mediated by root exudates in different aged ratooning tea monoculture systems. Int. J. Mol. Sci. 18:1727. doi: 10.3390/ijms18081727
Baba, T., Hirose, D., Noma, S., and Ban, T. (2021). Inoculation with two Oidiodendron maius strains differentially alters the morphological characteristics of fibrous and pioneer roots of Vaccinium virgatum ‘Tifblue’ cuttings. Sci. Hortic. 281:109948. doi: 10.1016/j.scienta.2021.109948
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bastida, F., Luis Moreno, J., Teresa, H., and García, C. (2006). Microbiological degradation index of soils in a semiarid climate. Soil Biol. Biochem. 38, 3463–3473. doi: 10.1016/j.soilbio.2006.06.001
Balmer, D., de Papajewski, D. V., Planchamp, C., Glauser, G., and Mauch-Mani, B. (2013). Induced resistance in maize is based on organ-specific defence responses. Plant J. 74, 213–225. doi: 10.1111/tpj.12114
Bent, E., Tuzun, S., Chanway, C. P., and Enebak, S. (2001). Alterations in plant growth and in root hormone levels of lodgepole pines inoculated with rhizobacteria. Can. J. Microbiol. 47, 793–800. doi: 10.1139/cjm-47-9-793
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bickford, W. A., Zak, D. R., Kowalski, K. P., and Goldberg, D. E. (2020). Differences in rhizosphere microbial communities between native and non native Phragmites australis may depend on stand density. Ecol. Evol. 10, 11739–11751. doi: 10.1002/ece3.6811
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Broeckling, C. D., Broz, A. K., Bergelson, J., Manter, D. K., and Vivanco, J. M. (2008). Root exudates regulate soil fungal community composition and diversity. Appl. Environ. Microbiol. 74, 738–744. doi: 10.1128/AEM.02188-07
Bulgarelli, D., Garrido-Oter, R., Munch, P. C., Weiman, A., Droge, J., Pan, Y., et al. (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403. doi: 10.1016/j.chom.2015.01.011
Bulgarelli, D., Rott, M., Schlaeppi, K., Themaat, E. V. L. V., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Burdon, R. D., and Low, C. B. (1992). Genetic survey of Pinus radiata–6: Wood properties–variation, heritabilities, and interrelationships with other traits. N. Z. J. For. Sci. 22, 228–245.
Buschmann, H., Fabri, C. O., Hauptmann, M., Hutzler, P., Laux, T., Lloyd, C. W., et al. (2004). Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr. Biol. 14, 1515–1521. doi: 10.1016/j.cub.2004.08.033
Cai, N., Xu, Y., Li, G., Deng, L., Li, W., and Wang, D. (2016). Research status and prospect of the crooked and twisted characteristics of P. yunnanensis stem. For. Invent. Plan. 41, 19–23. doi: 10.3969/j.issn.1671-3168.2016.06.004
Cardoso, E. J. B. N., Vasconcellos, R. L. F., Ribeiro, C. M., and Miyauchi, M. Y. H. (2011). “PGPR in coniferous trees,” in Bacteria in agrobiology: Crop ecosystems, ed. D. K. Maheshwari (Berlin: Springer), 345–359. doi: 10.7717/peerj.10109
Caruso, G. (2020). Microbial colonization in marine environments: Overview of current knowledge and emerging research topics. J. Mar. Sci. Eng. 8:78. doi: 10.3390/jmse8020078
Castellano-Hinojosa, A., Meyering, B., Nuzzo, A., Strauss, S. L., and Albrecht, U. (2021). Effect of plant biostimulants on root and plant health and the rhizosphere microbiome of citrus trees in huanglongbing-endemic conditions. Trees 35, 1525–1539. doi: 10.1007/s00468-021-02133-8
Chai, Y. N., and Schachtman, D. P. (2022). Root exudates impact plant performance under abiotic stress. Trends Plant Sci. 27, 80–91. doi: 10.1016/j.tplants.2021.08.003
Chen, F., Fan, Z., Niu, S., and Zheng, J. (2014). The influence of precipitation and consecutive dry days on burned areas in Yunnan province, southwestern China. Adv. Meteorol. 2014, 1–11. doi: 10.1155/2014/748923
Chen, H., Quan, W., Liu, H., and Ding, G. (2022). Effects of Suillus luteus and S. bovinus on the physiological response and nutrient absorption of Pinus massoniana seedlings under phosphorus deficiency. Plant Soil 471, 577–590. doi: 10.1007/s11104-021-05211-5
Chen, Q., Chang, E., Dong, F., Fan, G., Yin, J., Zhang, J., et al. (1997). The half-sib progeny test on natural elite stand of Pinus yunanensis in west of Yunnan. J. West China For. Sci. 1, 9–21. doi: 10.16473/j.cnki.xblykx1972.1997.01.002
Chen, S., and Lyu, Y. (1962). The causes and prevention of stem torsion of Pinus yunnanensis. Sci. Silvae Sin. 2, 131–140.
Chi, F., Shen, S., Chen, S., and Jing, Y. (2004). Migration of Azospirillum brasilense Yu62 from root to stem and leaves inside rice and tobacco plants. J. Integr. Plant Biol. 46, 1065–1070.
Chi, F., Shen, S. H., Cheng, H. P., Jing, Y. X., Yanni, Y. G., and Dazzo, F. B. (2005). Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 71, 7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005
Copisarow, M. (1933). The problem of twisted tree. Science 77, 581–583. doi: 10.1126/science.77.2007.581
Crandall, S. G., Miles, A. M., Chung, T., Cloutier, M. L. C., García-Rodríguez, R. O., Schweigkofler, W., et al. (2022). Temporal and spatial dynamics of bacterial and fungal microbiomes in nursery soils post-steaming. PhytoFrontiers doi: 10.1094/phytofr-07-22-0071-R
Das, I., and Pradhan, M. (2016). “Potassium-solubilizing microorganisms and their role in enhancing soil fertility and health,” in Potassium solubilizing microorganisms for sustainable agriculture, eds V. Meena, B. Maurya, J. Verma, and R. Meena (New Delhi: Springer), 281–291. doi: 10.1007/978-81-322-2776-2_20
DeSantis, T. Z., Hugenholtz, P. B., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 72, 5069–5072. doi: 10.1128/aem.03006-05
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Egamberdieva, D., Kamilova, F. D., Validov, S. Z., Gafurova, L., Kucharova, Z., and Lugtenberg, B. (2008). High incidence of plant growth-stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environ. Microbiol. 10, 1–9. doi: 10.1111/j.1462-2920.2007.01424.x
Egamberdieva, D., Kucharova, Z., Davranov, K., Berg, G., Makarova, N., Azarova, T., et al. (2010). Bacteria able to control foot and root rot and to promote growth of cucumber in salinated soils. Biol. Fertil. Soils 47, 197–205. doi: 10.1007/s00374-010-0523-3
Eklund, L., and Säll, H. (2000). The influence of wind on spiral grain formation in conifer trees. Trees 14, 324–328. doi: 10.1007/s004680050225
Erturk, Y., Ercjli, S., Haznedar, A., and Çakmakè, R. (2010). Effects of plant growth promoting rhizobacteria (PGPR) on rooting and root growth of kiwifruit (Actinidia deliciosa) stem cuttings. Biol. Res. 43, 91–98. doi: 10.4067/S0716-97602010000100011
Finkel, O. M., Salas-González, I., Castrillo, G., Conway, J. M., Law, T. F., Teixeira, P. J. P. L., et al. (2020). A single bacterial genus maintains root growth in a complex microbiome. Nature 587, 103–108. doi: 10.1038/s41586-020-2778-7
Fitzpatrick, C. R., Copeland, J. K., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U.S.A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Gao, C., Liu, F., Zhang, C., Feng, D., Li, K., and Cui, K. (2021). Germination responses to water potential and temperature variation among provenances of Pinus yunnanensis. Flora Morphol. Distrib. Funct. Ecol. Plants 276-277:151786. doi: 10.1016/j.flora.2021.151786
Gapare, W., Hathorn, A., Kain, D., Matheson, C., and Wu, H. (2007). Inheritance of spiral grain in the juvenile core of Pinus radiata. Can. J. For. Res. 37, 116–127. doi: 10.1139/x06-202
Garcìa, J. A. L., Domenech, J., Santamarìa, C., Camacho, M. A., Daza, A., and Mañero, F. J. G. (2004). Growth of forest plants (pine and holm-oak) inoculated with rhizobacteria: Relationship with microbial community structure and biological activity of its rhizosphere. Environ. Exp. Bot. 52, 239–251. doi: 10.1016/j.envexpbot.2004.02.003
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x
Ge, Y., Wang, Q., Wang, L., Liu, W., Liu, X., Huang, Y., et al. (2018). Response of soil enzymes and microbial communities to root extracts of the alien Alternanthera philoxeroides. Arch. Agron. Soil Sci. 64, 708–717. doi: 10.1080/03650340.2017.1373186
Giménez, C. M., Cabrera, R., Reina, M., and González-Coloma, A. (2007). Fungal endophytes and their role in plant protection. Curr. Organ. Chem. 11, 707–720. doi: 10.13039/501100006280
Gu, X., Li, J., Wang, X., He, X., and Cui, Y. (2020). Laccaria bicolor mobilizes both labile aluminum and inorganic phosphate in rhizosphere soil of Pinus massoniana seedlings field grown in a yellow acidic soil. Appl. Environ. Microb. 86:e03015–19. doi: 10.1128/aem.03015-19
Guo, Y., Zhao, Y., Zhu, T., Li, J., Feng, Y., Zhao, H., et al. (2018). A metabolomic view of how low nitrogen strength favors anammox biomass yield and nitrogen removal capability. Water Res. 143, 387–398. doi: 10.1016/j.watres.2018.06.052
Hansen, J. K., and Roulund, H. (1997). Genetic parameters for spiral grain, stem form, pilodyn and growth in 13 years old clones of Sitka spruce (Picea sitchensis (Bong.) Carr.). Silvae Genet. 46, 107–113.
Harris, J. M. (1989). Spiral grain and wave phenomena in wood formation. New York, NY: Springer-Verlag.
Harrison, J. G., Beltran, L. P., Buerkle, C. A., Cook, D., Gardner, D. R., Parchman, T. L., et al. (2021). A suite of rare microbes interacts with a dominant, heritable, fungal endophyte to influence plant trait expression. ISME J. 15, 2763–2778. doi: 10.1038/s41396-021-00964-4
Hartmann, A., Rothballer, M., Hense, B. A., and Schroder, P. (2014). Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front. Plant Sci. 5:131. doi: 10.3389/fpls.2014.00131
Hassani, M. A., Duran, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
He, F. (1994). Genetic experiment on texture phenotypic characters of Pinus yunnanensisc wood. Yunnan For. Sci. Technol. 2, 1–7. doi: 10.16473/j.cnki.xblykx1972.1994.02.001
Heredia-Acuña, C., Almaraz-Suarez, J. J., Arteaga-Garibay, R., Ferrera-Cerrato, R., and Pineda-Mendoza, D. Y. (2018). Isolation, characterization and effect of plant-growth-promoting rhizobacteria on pine seedlings (Pinus pseudostrobus Lindl.). J. For. Res. 30, 1727–1734. doi: 10.1007/s11676-018-0723-5
Herrera Paredes, S., Gao, T., Law, T. F., Finkel, O. M., Mucyn, T. S., Teixeira, P. J. P. L., et al. (2018). Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biol. 16:e2003962. doi: 10.1371/journal.pbio.2003962
Htwe, A. Z., Moh, S. M., Soe, K. M., Moe, K., and Yamakawa, T. (2019). Effects of biofertilizer produced from Bradyrhizobium and Streptomyces griseoflavus on plant growth, nodulation, nitrogen fixation, nutrient uptake, and seed yield of mung bean, cowpea, and soybean. Agronomy 9:77. doi: 10.3390/agronomy9020077
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9:2738. doi: 10.1038/s41467-018-05122-7
Jacoby, R., Peukert, M., Succurro, A., Koprivova, A., and Kopriva, S. (2017). The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front. Plant Sci. 8:1617. doi: 10.3389/fpls.2017.01617
Jousset, A., Bienhold, C., Chatzinotas, A., Gallien, L., Gobet, A., Kurm, V., et al. (2017). Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862. doi: 10.1038/ismej.2016.174
Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., et al. (2020). Recent understanding of soil Acidobacteria and their ecological significance: A critical review. Front. Microbiol. 11:580024. doi: 10.3389/fmicb.2020.580024
Kamutando, C. N., Vikram, S., Kamgan-Nkuekam, G., Makhalanyane, T. P., Greve, M., Roux, J. J. L., et al. (2017). Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Sci. Rep. 7:6472. doi: 10.1038/s41598-017-07018-w
Kang, S. H., Cho, H. S., Cheong, H., Ryu, C. M., Kim, J. F., and Park, S. H. (2007). Two bacterial entophytes eliciting both plant growth promotion and plant defense on pepper (Capsicum annuum L.). J. Microbiol. Biotechnol. 17, 96–103. doi: 10.1016/j.mimet.2006.07.012
Kõljalg, U., Larsson, K. H., Abarenkov, K., Nilsson, R. H., Alexander, I. J., Eberhardt, U., et al. (2005). UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 166, 1063–1068. doi: 10.1111/J.1469-8137.2005.01376.X
Kumar, S., and Garkoti, S. C. (2022). Rhizosphere influence on soil microbial biomass and enzyme activity in banj oak, chir pine and banj oak regeneration forests in the central Himalaya. Geoderma 409:115626. doi: 10.1016/j.geoderma.2021.115626
Lee, S. M., Kong, H. G., Song, G. C., and Ryu, C. M. (2020). Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 15, 330–347. doi: 10.1038/s41396-020-00785-x
Li, S., Deng, Y., Wang, Z., Zhang, Z., Kong, X., Zhou, W., et al. (2020). Exploring the accuracy of amplicon-based internal transcribed spacer markers for a fungal community. Mol. Ecol. Resour. 20, 170–184. doi: 10.1111/1755-0998.13097
Liu, H., Li, J., Carvalhais, L. C., Percy, C., Prakash Verma, J., Schenk, P. M., et al. (2021). Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogen. New Phytol. 229, 2873–2885. doi: 10.1111/nph.17057
Liu, J., Sui, Y., Yu, Z., Yao, Q., Shi, Y., Chu, H., et al. (2016). Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Soil Biol. Biochem. 95, 212–222. doi: 10.1016/j.soilbio.2015.12.021
Ludwig-Muller, J. (2015). Bacteria and fungi controlling plant growth by manipulating auxin: Balance between development and defense. J. Plant Physiol. 172, 4–12. doi: 10.1016/j.jplph.2014.01.002
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Matsumoto, H., Fan, X., Wang, Y., Kusstatscher, P., Duan, J., Wu, S., et al. (2021). Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 7, 60–72. doi: 10.1038/s41477-020-00826-5
Mishra, S., and Priyanka, Sharma, S. (2022). Metabolomic insights into endophyte-derived bioactive compounds. Front. Microbiol. 13:835931. doi: 10.3389/fmicb.2022.835931
Molefe, R. R., Amoo, A. E., and Babalola, O. O. (2021). Metagenomic insights into the bacterial community structure and functional potentials in the rhizosphere soil of maize plants. J. Plant Interact. 16, 258–269. doi: 10.1080/17429145.2021.1936228
Nakamura, M., and Hashimoto, T. (2020). Mechanistic Insights into plant chiral growth. Symmetry 12:2056. doi: 10.3390/sym12122056
Okada, K., and Shimura, Y. (1990). Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science 250, 274–276. doi: 10.1126/science.250.4978.274
Otlewska, A., Migliore, M., Dybka-Stepien, K., Manfredini, A., Struszczyk-Swita, K., Napoli, R., et al. (2020). When salt meddles between plant, soil, and microorganisms. Front. Plant Sci. 11:553087. doi: 10.3389/fpls.2020.553087
Patel, T. S. (2018). Plant growth promoting rhizobacteria: Blessing to agriculture. Int. J. Pure Appl. Biosci. 6, 481–492. doi: 10.18782/2320-7051.6383
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and Putten, W. H. (2013). Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pinto, O. H. B., Costa, F. S., Rodrigues, G. R., da Costa, R. A., da Rocha Fernandes, G., Junior, O. R. P., et al. (2021). Soil Acidobacteria strain AB23 resistance to oxidative stress through production of carotenoids. Microb. Ecol. 81, 169–179. doi: 10.1007/s00248-020-01548-z
Pot, D., Chantre, G., Rozenberg, P., Rodrigues, J. C., Jones, G. L., Pereira, H., et al. (2002). Genetic control of pulp and timber properties in maritime pine (Pinus pinaster Ait.). Ann. For. Sci. 59, 563–575. doi: 10.1051/forest:2002042
Preston, G. M., Studholme, D. J., and Caldelari, I. (2005). Profiling the secretomes of plant pathogenic Proteobacteria. FEMS Microbiol. Rev. 29, 331–360. doi: 10.1016/j.fmrre.2005.01.001
Riley, D., and Barber, S. A. (1971). Effect of ammonium and nitrate fertilization on phosphorus uptake as related to root-induced pH changes at the root-soil interface. Soil Sci. Soc. Am. J. 35, 301–306. doi: 10.2136/sssaj1971.03615995003500020035x
Robbins, C., Thiergart, T., Hacquard, S., Garrido-Oter, R., Gans, W., Peiter, E., et al. (2018). Root-associated bacterial and fungal community profiles of Arabidopsis thaliana are robust across contrasting soil P levels. Phytobiomes J. 2, 24–34. doi: 10.1094/pbiomes-09-17-0042-r
Rojas, A., Holguin, G., Glick, B. R., and Bashan, Y. (2001). Synergism between Phyllobacterium sp. (N(2)-fixer) and Bacillus licheniformis (P-solubilizer), both from a semiarid mangrove rhizosphere. FEMS Microbiol. Ecol. 35, 181–187. doi: 10.1111/J.1574-6941.2001.TB00802.X
Rosier, A., Bishnoi, U., Lakshmanan, V., Sherrier, D. J., and Bais, H. P. (2016). A perspective on inter-kingdom signaling in plant-beneficial microbe interactions. Plant Mol. Biol. 90, 537–548. doi: 10.1007/s11103-016-0433-3
Rossmann, M., Perez-Jaramillo, J., Kavamura, V. N., Chiaramonte, J. B., Dumack, K., Fiore-Donno, A. M., et al. (2020). Multitrophic interactions in the rhizosphere microbiome of wheat: From bacteria and fungi to protists. FEMS Microbiol. Ecol. 96:fiaa032. doi: 10.1093/femsec/fiaa032
Sayer, E. J., Oliver, A. E., Fridley, J. D., Askew, A. P., Mills, R. T., and Grime, J. P. (2017). Links between soil microbial communities and plant traits in a species-rich grassland under long-term climate change. Ecol. Evol. 7, 855–862. doi: 10.1002/ece3.2700
Schreiter, S., Ding, G. C., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5:144. doi: 10.3389/fmicb.2014.00144
Shi, J., Yuan, X., Lin, H., Yang, Y., and Li, Z. (2011). Differences in soil properties and bacterial communities between the rhizosphere and bulk soil and among different production areas of the medicinal plant Fritillaria thunbergii. Int. J. Mol. Sci. 12, 3770–3785. doi: 10.3390/ijms12063770
Shubhransu, N., Soma, S., Chandan, S., and Soumya Sephalika, S. (2021). Rice crop loss due to major pathogens and the potential of endophytic microbes for their control and management. J. Appl. Biol. Biotechnol. 9, 166–175. doi: 10.7324/jabb.2021.9523
Silva, J. C., Borralho, N., and Wellendorf, H. (2000). Genetic parameter estimates for diameter growth, pilodyn penetration and spiral grain in Picea abies (L.) KARST. Silvae Genet. 49, 29–36.
Singh, N., Pandey, P. K., Dubey, R. C., and Maheshwari, D. K. (2008). Biological control of root rot fungus Macrophomina phaseolina and growth enhancement of Pinus roxburghii (Sarg.) by rhizosphere competent Bacillus subtilis BN1. World J. Microb. Biotechnol. 24, 1669–1679. doi: 10.1007/S11274-008-9680-Z
Song, Q., Song, X., Deng, X., Luo, J., Wang, J., Min, K., et al. (2021). Effects of plant growth promoting Rhizobacteria microbial on the growth, rhizosphere soil properties, and bacterial community of Pinus sylvestris var. mongolica seedlings. Scand. J. For. Res. 36, 249–262. doi: 10.1080/02827581.2021.1917649
Song, Y., Wilson, A. J., Zhang, X.-C., Thoms, D., Sohrabi, R., Song, S., et al. (2021). FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat. Plants 7, 644–654. doi: 10.1038/s41477-021-00914-0
Sugio, A., Dubreuil, G., Giron, D., and Simon, J. C. (2015). Plant-insect interactions under bacterial influence: Ecological implications and underlying mechanisms. J. Exp. Bot. 66, 467–478. doi: 10.1093/jxb/eru435
Tahir, M., Ahmad, I., Shahid, M., Shah, G. M., Farooq, A. B. U., Akram, M., et al. (2019). Regulation of antioxidant production, ion uptake and productivity in potato (Solanum tuberosum L.) plant inoculated with growth promoting salt tolerant Bacillus strains. Ecotoxicol. Environ. Safe 178, 33–42. doi: 10.1016/j.ecoenv.2019.04.027
Thomas, P., and Reddy, K. M. (2013). Microscopic elucidation of abundant endophytic bacteria colonizing the cell wall–plasma membrane peri-space in the shoot-tip tissue of banana. AoB Plants 5:lt011. doi: 10.1093/aobpla/plt011
Timonin, M. I. (1964). Interaction of seed-coat microflora and soil microorganisms and its effects on pre- and post-emergence of some conifer seedlings. Can. J. Microbiol. 10, 17–22. doi: 10.1139/M64-003
Tsolis, R. M. (2002). Comparative genome analysis of the α-Proteobacteria: Relationships between plant and animal pathogens and host specificity. Proc. Natl. Acad. Sci. U.S.A. 99, 12503–12505. doi: 10.1073/pnas.212508599
Uroz, S., Buee, M., Murat, C., Frey-Klett, P., and Martin, F. (2010). Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2, 281–288. doi: 10.1111/j.1758-2229.2009.00117.x
Vacheron, J., Desbrosses, G., Bouffaud, M.-L., Touraine, B., Moënne-Loccoz, Y., Muller, D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. doi: 10.3389/fpls.2013.00356
van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Vandana, U. K., Rajkumari, J., Singha, L. P., Satish, L., Alavilli, H., Sudheer, P., et al. (2021). The endophytic microbiome as a hotspot of synergistic interactions, with prospects of plant growth promotion. Biology (Basel) 10:101. doi: 10.3390/biology10020101
Van Wees, S. C. M., Luijendijk, M. W. J., Smoorenburg, I., van Loon, L. C., and Pieterse, C. M. J. (2004). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 41, 537–549. doi: 10.1023/A:1006319216982
Vasileva, E. N., Akhtemova, G. A., Zhukov, V. A., and Tikhonovich, I. A. (2019). Endophytic microorganisms in fundamental research and agriculture. Ecol. Genet. 17, 19–32. doi: 10.17816/ecogen17119-32
Vives-Peris, V., de Ollas, C., Gomez-Cadenas, A., and Perez-Clemente, R. M. (2020). Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Volpiano, C. G., Lisboa, B. B., José, J. F. B. D. S., Beneduzi, A., Granada, C. E., and Vargas, L. K. (2022). Soil-plant-microbiota interactions to enhance plant growth. Rev. Bras. Cienc. Solo 46:e0210098. doi: 10.36783/18069657rbcs20210098
Wagner, M. R., Lundberg, D. S., Coleman-Derr, D., Tringe, S. G., Dangl, J. L., and Mitchell-Olds, T. (2014). Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecol. Lett. 17, 717–726. doi: 10.1111/ele.12276
Walker, T. S., Bais, H. P., Grotewold, E., and Vivanco, J. M. (2003). Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51. doi: 10.1104/pp.102.019661
Wang, B., Mao, J.-F., Zhao, W., and Wang, X. R. (2013). Impact of geography and climate on the genetic differentiation of the subtropical pine Pinus yunnanensis. PLoS One 8:e67345. doi: 10.1371/journal.pone.0067345
Wang, X., Cao, Z., Gao, C., and Li, K. (2021). Strategy for the construction of a core collection for Pinus yunnanensis Franch. To optimize timber based on combined phenotype and molecular marker data. Genet. Resour. Crop Evol. 68, 3219–3240. doi: 10.1007/S10722-021-01182-9
Wang, H., Fang, X., Wu, H., Cai, X., and Xiao, H. (2021). Effects of plant cultivars on the structure of bacterial and fungal communities associated with ginseng. Plant Soil 465, 143–156. doi: 10.1007/S11104-021-05000-0
Wang, H., Ren, T., Yang, H., Feng, Y., Feng, H., Liu, G., et al. (2020). Research and application of biochar in soil CO2 emission, fertility, and microorganisms: A sustainable solution to solve China’s agricultural straw burning problem. Sustainability 12:1922. doi: 10.3390/su12051922
Wu, Z., Zong, D., Gan, P., Liu, Z., Zhu, M., and He, C. (2022). Genetic analysis on different stem forms of Pinus yunnanensis by reduced-representation genome sequencing. J. Yunnan Agric. Univ. 37, 285–293. doi: 10.12101/j.issn.1004-390X(n).202108048
Xu, Y., Cai, N., Woeste, K. E., Kang, X., He, C., Li, G., et al. (2016). Genetic diversity and population structure of Pinus yunnanensis by simple sequence repeat markers. For. Sci. 62, 38–47. doi: 10.5849/forsci.15-064
Yan, L., Zhu, J., Zhao, X., Shi, J., Jiang, C., and Shao, D. (2019). Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 103, 3327–3340. doi: 10.1007/s00253-019-09713-2
Yang, J., Giné-Bordonaba, J., Vilanova, L., Teixidó, N., Usall, J., Larrigaudière, C., et al. (2017). An insight on the ethylene biosynthetic pathway of two major fruit postharvest pathogens with different host specificity: Penicillium digitatum and Penicillium expansum. Eur. J. Plant Pathol. 149, 575–585. doi: 10.1007/s10658-017-1205-x
Yeoh, Y. K., Dennis, P. G., Paungfoo-Lonhienne, C., Weber, L., Brackin, R., Ragan, M. A., et al. (2017). Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 8:215. doi: 10.1038/s41467-017-00262-8
Ying, Y., Ding, W., and Li, Y. (2012). Characterization of soil bacterial communities in rhizospheric and nonrhizospheric soil of Panax ginseng. Biochem. Genet. 50, 848–859. doi: 10.1007/s10528-012-9525-1
Yoneda, Y., Yamamoto, K., Makino, A., Tanaka, Y., Meng, X. Y., Hashimoto, J., et al. (2021). Novel plant-associated Acidobacteria promotes growth of common floating aquatic plants, duckweeds. Microorganisms 9:1133. doi: 10.3390/microorganisms9061133
Zadworny, M., McCormack, M. L., Zytkowiak, R., Karolewski, P., Mucha, J., and Oleksyn, J. (2017). Patterns of structural and defense investments in fine roots of Scots pine (Pinus sylvestris L.) across a strong temperature and latitudinal gradient in Europe. Glob. Change Biol. 23, 1218–1231. doi: 10.1111/gcb.13514
Zeffa, D. M., Fantin, L. H., Koltun, A., de Oliveira, A. L. M., Nunes, M. P., Canteri, M. G., et al. (2020). Effects of plant growth-promoting rhizobacteria on co-inoculation with Bradyrhizobium in soybean crop: A meta-analysis of studies from 1987 to 2018. PeerJ 8:e7905. doi: 10.7717/peerj.7905
Zhang, B., Zhou, X., and Zhang, Y.-M. (2014). Responses of microbial activities and soil physical-chemical properties to the successional process of biological soil crusts in the Gurbantunggut Desert, Xinjiang. J. Arid Land 7, 101–109. doi: 10.1007/s40333-014-0035-3
Zhou, A., Zong, D., Luo, J., Shen, D., He, R., Tian, B., et al. (2016). AFLP analysis on genetic variation of stem forms in Pinus yunnanensis. Mol. Plant Breed. 14, 186–194. doi: 10.13271/j.mpb.014.000186
Zhou, M. (1974). A preliminary study on the malformed stem of pine seedlings. For. Sci. Technol. 2, 19–22.
Zilber-Rosenberg, I., and Rosenberg, E. (2008). Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. doi: 10.1111/j.1574-6976.2008.00123
Keywords: pinus yunnanensis, trunk type, rhizosphere microorganism, microbial community, microbial diversity
Citation: Li P, Zong D, Gan P, Li H, Wu Z, Li F, Zhao C, Li L and He C (2023) Comparison of the diversity and structure of the rhizosphere microbial community between the straight and twisted trunk types of Pinus yunnanensis. Front. Microbiol. 14:1066805. doi: 10.3389/fmicb.2023.1066805
Received: 11 October 2022; Accepted: 07 February 2023;
Published: 23 February 2023.
Edited by:
Guillaume J. Bilodeau, Canadian Food Inspection Agency (CFIA), CanadaReviewed by:
Sharifa Crandall, Pennsylvania State University, United StatesCopyright © 2023 Li, Zong, Gan, Li, Wu, Li, Zhao, Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengzhong He,  aGVjekBzd2Z1LmVkdS5jbg==
aGVjekBzd2Z1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.