- Jilin Ginseng Academy, Changchun University of Chinese Medicine, Changchun, China
Root exudates contain plant metabolites secreted by the roots into the soil, such as ginsenosides secreted by the ginseng root. However, little is known about ginseng root exudate and its impact on the chemical and microbial properties of soil. In this study, the effect of increasing concentrations of ginsenosides on the chemical and microbial properties of soil was tested. Chemical analysis and high-throughput sequencing techniques were used to evaluate the soil chemical properties and microbial characteristics following exogenous application of 0.1 mg·L−1, 1 mg·L−1, and 10 mg·L−1 ginsenosides. Ginsenosides application significantly altered soil enzyme activities; SOM-dominated physicochemical properties were significantly reduced which altered the composition and structure of the soil microbial community. In particular, treatment with 10 mg∙L−1 ginsenosides significantly increased the relative abundance of pathogenic fungi such as Fusarium, Gibberella and Neocosmospora. These findings indicate that ginsenosides in root exudates are important factors that may lead to increased deterioration of soil during ginseng cultivation and provided new research direction for the subsequent study on the mechanism of interaction between ginsenosides and soil microbial communities.
1. Introduction
Ginseng (Panax ginseng C. A. Meyer) is a perennial herb with a long planting history in the world and an important cash crop (Baeg and So, 2013). With the rapid economic development and shortage of soil resources, the intensive cultivation system characterized by continuous monoculture has become an important part of the current industry of ginseng production and is widely used in the world (Banerjee et al., 2019). Numerous studies have shown that ginseng has a strong contraindication to the soil, and continuous planting of ginseng seems to cause its roots to rust and rot due to soil sickness, which hinders the healthy growth of ginseng (Wang et al., 2016; Dong L.-L. et al., 2017; Dong L. et al., 2017; Zhang et al., 2022). Consequently, the yield and quality of P. ginseng cannot be guaranteed, causing huge economic losses and impeding the healthy and sustainable development of the ginseng industry (Tong et al., 2021). Studies to date suggest that several factors are associated with ginseng soil sickness, including deterioration of soil physicochemical properties and an imbalance in soil microbial communities (Dong L.-L. et al., 2017; Dong L. et al., 2017; Bao et al., 2020). The diversity and composition of soil microbial communities are essential to maintain soil health and quality (Garbeva et al., 2004). And, changes in microbial community diversity and composition are associated with a number of biotic or abiotic factors, such as cropping system, root secretions, and soil type (Green et al., 2008; Berg and Smalla, 2009).
More and more studies are confirming the relationship between root exudates and soil microbial communities (Fujimatsu et al., 2020). Phenolic acids are one of the important substances of root exudates and produced by various plants (Ma et al., 2015; Wang et al., 2018; Lin et al., 2019). For example, p-hydroxybenzoic acid released from roots of cucumber can promote the growth of pathogenic fungi and increase the density of inter-root bacteria and fungi (Zhou et al., 2012). Root exudates can also directly suppress pathogens or alter the composition of microbial communities. In Arabidopsis, the gene cluster responsible for the synthesis of specific metabolites (e.g., triterpenoids, esters, and coumarins) would prenyltransferase-terpene synthase (PT-TPS) be altered to promote de novo functionalization of genes, a result that reveals the function of metabolites in being able to regulate microbial communities (Lundberg et al., 2012; Stringlis et al., 2018; Chen et al., 2019).
Ginsenosides are the major active compounds in ginseng, and ginseng contains at least 20 different ginsenosides, accounting for more than 6% of the plant biomass (Yang et al., 2015; Zhang et al., 2021). As the main root metabolites of the Panax L., ginsenosides can be released into the rhizosphere soil through root exudation, leaching or decomposition of plant residues (Yang et al., 2015). Although many studies have been conducted to investigate the pharmacological properties of ginsenosides, little is known about the ecological role of these important secondary metabolites once released from Panax L. into the surrounding soil (Ng, 2006). Furthermore, more than 65 of the known ginsenosides at different concentrations have been shown to play an important role in the growth of ginseng, and it has been demonstrated that many ginsenosides have a stimulating effect on the growth of important ginseng pathogens such as Pythiumirregulare, Cylindrocarpon destructans, Phytophthora cactorum, and Fusarium solani (Nicol et al., 2003; Yousef and Bernards, 2006; Li et al., 2020). However, there is limited information on the effect of different concentrations of ginsenosides on microbial communities leading to soil sickness.
In this study, we aimed to (1) investigate whether ginsenosides might be one of the main causes of soil sickness during ginseng cultivation; and (2) evaluate the effects of different concentrations of ginsenosides on the soil microbial community.
2. Materials and methods
2.1. Field experiment description and design
The soil used in this experiment was obtained from a ginseng plantation in Baixi Forestry Field, Fusong County, Jilin Province (127°01′-128°06′E, between 1°42′-42°49′N), which is located in south eastern Jilin province, upstream of the Songhua River, and has a temperate continental monsoon climate. Samples of soil were collected from the ginseng rhizosphere. The basic physicochemical properties of this dark brown loan soil were pH, 5.61; soil organic matter (OM) content, 139.51 (g∙kg−1); electrical conductivity (EC), 40.2 (μs·cm−1); fast-acting phosphorus (AP), 35.55 (mg∙kg−1); fast-acting potassium (AK), 659.3 (mg∙kg−1); and fast-acting nitrogen (AN), 95.68 (mg∙kg−1).
Sampling was carried out at the sampling location in 2019 using the S-shaped random multi-point mixed sampling method, taking the uppermost 0–20 cm of soil from each processing group. And, the five soil sampling points were mixed into one composite sample. This method meant that four composite soil samples were obtained from each treatment. After sample collection, soil fauna and plant residues were removed and 10 kg were taken in sterile sampling bags according to the quadrat method and brought back to the laboratory. One part of the samples was dried naturally in a ventilated room and then passed through a 2-mm mesh size sieve, and used for determining the physicochemical properties and enzymatic activity of the soil; the other part was used for microbial diversity analysis.
2.2. Experimental design
Sub-samples of soil (200 g) were added to 100 ml of distilled water and incubated in a constant temperature incubator at 25°C for 15 d. The three treatment groups, SP_1, SP_2, SP_3 represent the three ginsenosides (Shanghai yuanye Bio-Technology Co., Ltd) concentrations, 0.1 mg·L−1, 1 mg·L−1, and 10 mg L −1, in 20 ml of solution, added to soil samples and incubated for 15 d. An untreated control group was included for each experiment. All soil samples were placed in wide mouth bottles containing test tubes with 5 ml deionized water, incubated at a constant temperature of 25°C, and the water content measured every 3 d and replenished when insufficient. In this experiment, 4 treatment groups were set up, and 4 parallel trials were conducted for each treatment group, totaling 16 test samples. On the 90th day, samples of culture were removed, stored at −80, and 3 g of fresh soil added.
2.3. Soil enzyme activity analysis
The pH of the soil was determined at a ratio of 1: 2 using a glass electrode pH meter (Jiang et al., 2019); EC was determined using a conductivity meter (Meng et al., 2013) and OM was determined using the potassium dichromate oxidation method (Flowers and Bremner, 1991) AP in soil was determined by leaching molybdenum antimony anti-colorimetric method (Kovar and Pierzynski, 2009); AK was determined by extraction using the acetamide method (Tardy et al., 2015), and AP was determined by phosphor molybdenim blue colorimetric method (Matula, 2010).
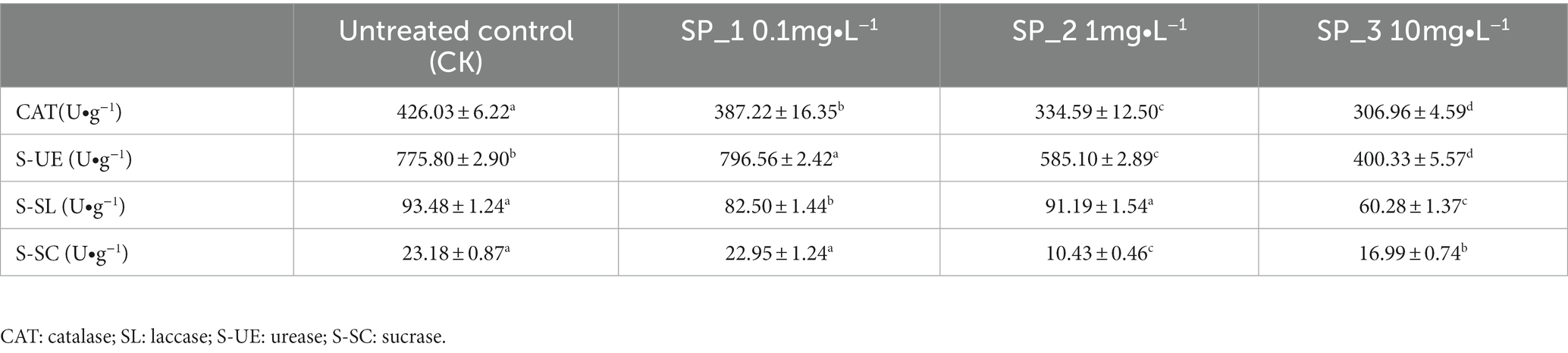
Important enzymes involved in soil nutrient cycling processes and microbial metabolism including catalase (S-CAT), laccase (SL), urease (S-UE), and sucrase (S-SC) were determined for each sample. Urease (S-UE) activity was determined using urea as substrate by colorimetric method with NH3−N. Laccase (SL) was determined using ABTS as substrate, incubated for 1 h, and detected at 420 nm by UV spectrophotometer (Eichlerova et al., 2012) Sucrase (S-SC) was determined using sucrose as a substrate and the glucose produced was measured colorimetrically (Shi et al., 2008). Catalase (CAT) activity was determined in the samples by adding H2O2 solution (Rodíguez-Kábana, 1982).
2.4. Soil DNA extraction and sequencing
Total DNA was extracted from 0.5 g of soil sample using the E.Z.N.A.® soil DNA kit (Omega Bio-Tek, Norcross, GA, United States; Wang et al., 2020) according to the manufacturer’s instructions, and DNA concentration and purity were determined using a NanoDrop2000. Each soil sample was extracted in triplicate and the three DNA solutions were thoroughly mixed. The DNA samples were stored in a container in a − 20°C refrigerator for subsequent analysis.
Primers 338F and 806R were used to amplify the V3-V4 variable region of the 16srRNA gene. Transcript amplicons were used to amplify the fungal gene spacer region using primer ITS1 (Nottingham et al., 2018). The initial denaturation steps were 5°C pre-denaturation for 3 min, 27 cycles (95°C denaturation for 30 s, 55°C annealing for 30 s, 72°C extensions for 30 s), followed by 72°C stable extensions for 10 min, and finally storage at 4°C (PCR instrument: ABI GeneAmp® model 9,700). the PCR reaction system was: 5× Trans Start Fast Pfu buffer 4 μL, 2.5 mM dNTPs 2 μL, upstream primer (5uM) 0.8 μL, downstream primer (5uM) 0.8 μL, Trans Start Fast Pfu DNA polymerase 0.4 μL, template DNA 10 ng, made up to 20 μL 3 replicates per sample.
2.5. Illumina Miseq sequencing and data processing
PCR products from the same sample were mixed and recovered using a 2% agarose gel, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Bios-sciences, Union City, CA, United States), detected by 2% agarose gel electrophoresis, and quantified using a Quantus™ Fluorometer (Promega, United States). A fluorometer (Promega, USA) was used to quantify the recovered products. Sequencing was performed using Illumina’s Miseq PE300 platform (Shanghai Majorbio Bio-Pharm Technology Co Ltd.). The raw reads were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (PRJNA886310).
The raw sequenced sequences were quality-controlled using faster (Nottingham et al., 2018) software, spliced using FLASH (Magoc and Salzberg, 2011) software, and sequences were OTU clustered and chimeras removed based on 97% (Goebel and Stackebrandt, 1994; Edgar, 2013) similarity using UPARSE (Edgar, 2013) software. Each sequence was annotated with species classification using an RDP classifier (Wang et al., 2007), compared to the Silva 16S rRNA database (v138), and a comparison threshold of 70% was set.
2.6. Statistical analysis
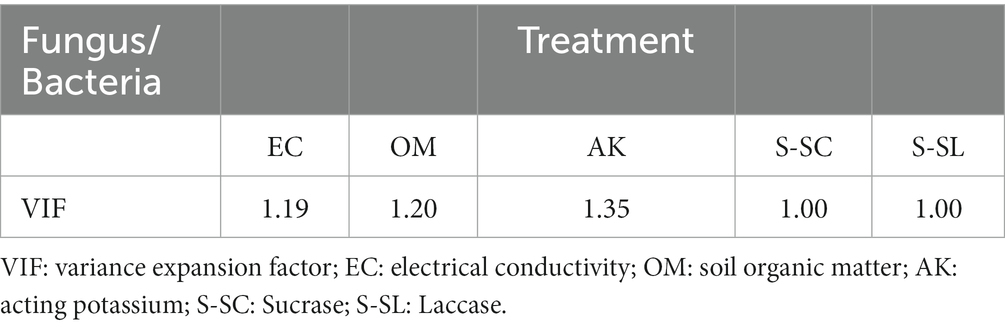
Experimental data were organized using Microsoft Excel. The statistical software SPSS 21.0 was used for one-way analysis of variance (ANOVA), and the value of p threshold of <0.05 was used to characterize significant differences between the three groups of data. Sequences were clustered at 97%, similarity analysis of OTUs was performed using UPARSE software, and chimeras were removed using UCHIME software. The analysis was performed according to different classification levels, richness and diversity indices. In the heat map analysis, Spearman’s rank correlation coefficients were calculated to assess the relationship between soil properties and microbial communities. Principal coordinate analysis (PCOA) was performed based on Bray-Curtis distance matrices in bacteria and fungi, respectively, to visualize pairwise community differences between samples. VIF (Variance Inflation Factor) was used to filter out the environmental factors with VIF > 10.
All graphics are built using the drawing software GraphPad Prism 8.01 and the Majorbio platform.
3. Result
3.1. Effects of different concentrations of ginsenoside on soil physicochemical properties and enzyme activities
3.1.1. Soil physicochemical analysis
Results from ANOVA showed that most soil physicochemical properties (pH, EC, AN, AP, AK and OM) changed significantly following application of the different ginsenosides concentration treatments (Table 1). The overall soil pH was significantly lower than the control and soil acidity increased after the application of different concentrations of ginsenosides treatments. In addition, EC, AN, and AK contents were significantly increased by 8.5, 7.33, and 33.47% in the SP_2 treatment group compared to the untreated control (CK) (p < 0.05). In the SP_1 group, EC and AN were the lowest, differing from the untreated control group (CK) by 4.77, 12.93, and 13.91%, respectively (p < 0.05). There was no significant difference in AP levels between the SP_1 and SP_2 groups. However, the SP_3 group had a significantly lower AP value than the untreated control (CK) group (at 2.59%). In addition, the OM values were significantly lower in the three different concentration treatment groups compared to the untreated control (CK).
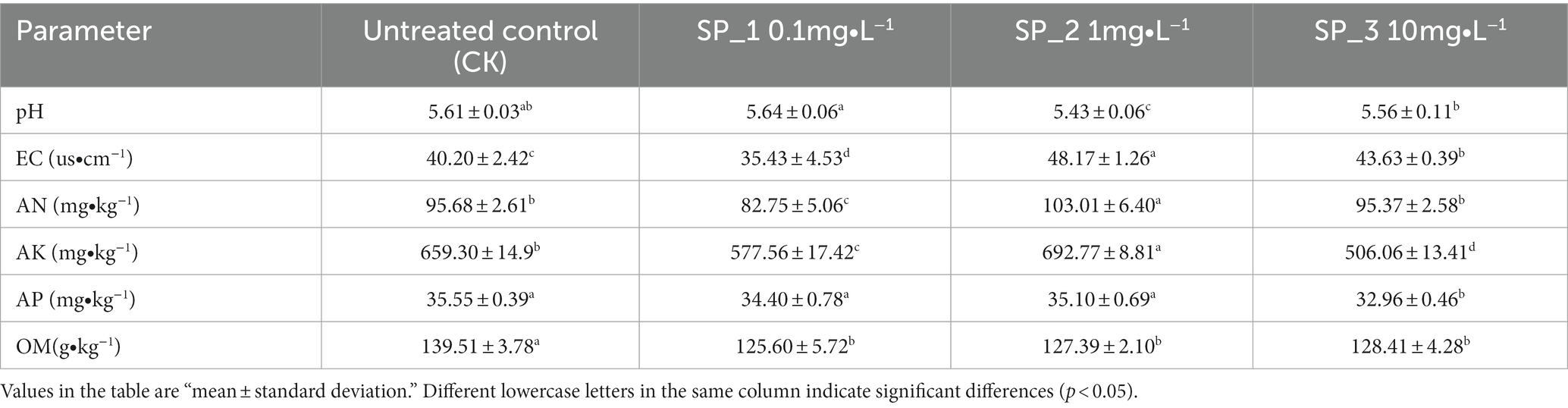
Table 1. Changes of soil physical and chemical properties under different concentrations of ginsenosides.
3.1.2. Soil enzyme activity analysis
Compared to the untreated control (CK), we observed significant differences with a significant decrease in CAT activity, which was directly correlated with an increase in ginsenosides concentration (Table 2). Urease activity (S-UE) increased in the SP_1 group, but with an increase in ginsenosides concentration, the S-UE activity in the SP_2 group and SP_3 group significantly decreased compared to the untreated control (CK). In contrast, the S-SL activity of the SP_2 group was significantly higher than that of the SP_1 and SP_3 groups, which was not significantly different from that of the untreated control (CK). Sucrase (S-SC) activity gradually decreased, and in the SP_1 group was close to that of the untreated control (CK). However, the activity gradually decreased with an increase in ginsenosides concentration. The four soil enzyme activities showed increased activity in each individual treatment group, but overall decreased with the increase in ginsenosides concentration.
3.2. Effect of different concentrations of ginsenosides on the diversity of soil bacterial communities
A total of 693,116 high-quality 16srRNA sequences were obtained from the 16 soil samples these sequences have been distributed with 97% similarity in as many different OTUs, with flat a and b dilution curves in the samples for measuring the changes in microbial communities at different concentrations of ginsenosides. This indicated that the data obtained from this study was large enough for bacterial and fungal diversity analysis.
The α-diversity indices, including the number of species observed and the diversity and richness indices of bacteria and their fungal pools, were different in all soil samples (Table 3). For bacteria, the community diversity and richness (Shannon = 4.122, ace = 456.596, Chao1 = 456.335) of soils in the SP_3 group was the lowest of all samples (p < 0.05); for fungi, SP_1 (Shannon =4.122, ace =249.145, Chao1 = 247.547) was the lowest (p < 0.05). In particular, the diversity indices of bacteria and fungi showed similar trends. Fungal and bacterial alpha diversity indices were significantly different between the SP_3 and untreated control (CK) groups. The estimated abundance of bacteria and fungi (ACE and Chao1) reached the highest values recorded under SP_2 and SP_1, respectively. The SP_3 group resulted in a significant reduction in the number of bacterial and fungal communities OTUs under the three different treatments (Figure 1). This indicates that the species richness and diversity of bacterial and fungal communities in the soil were differentially affected in soils treated with different concentrations of ginsenosides.
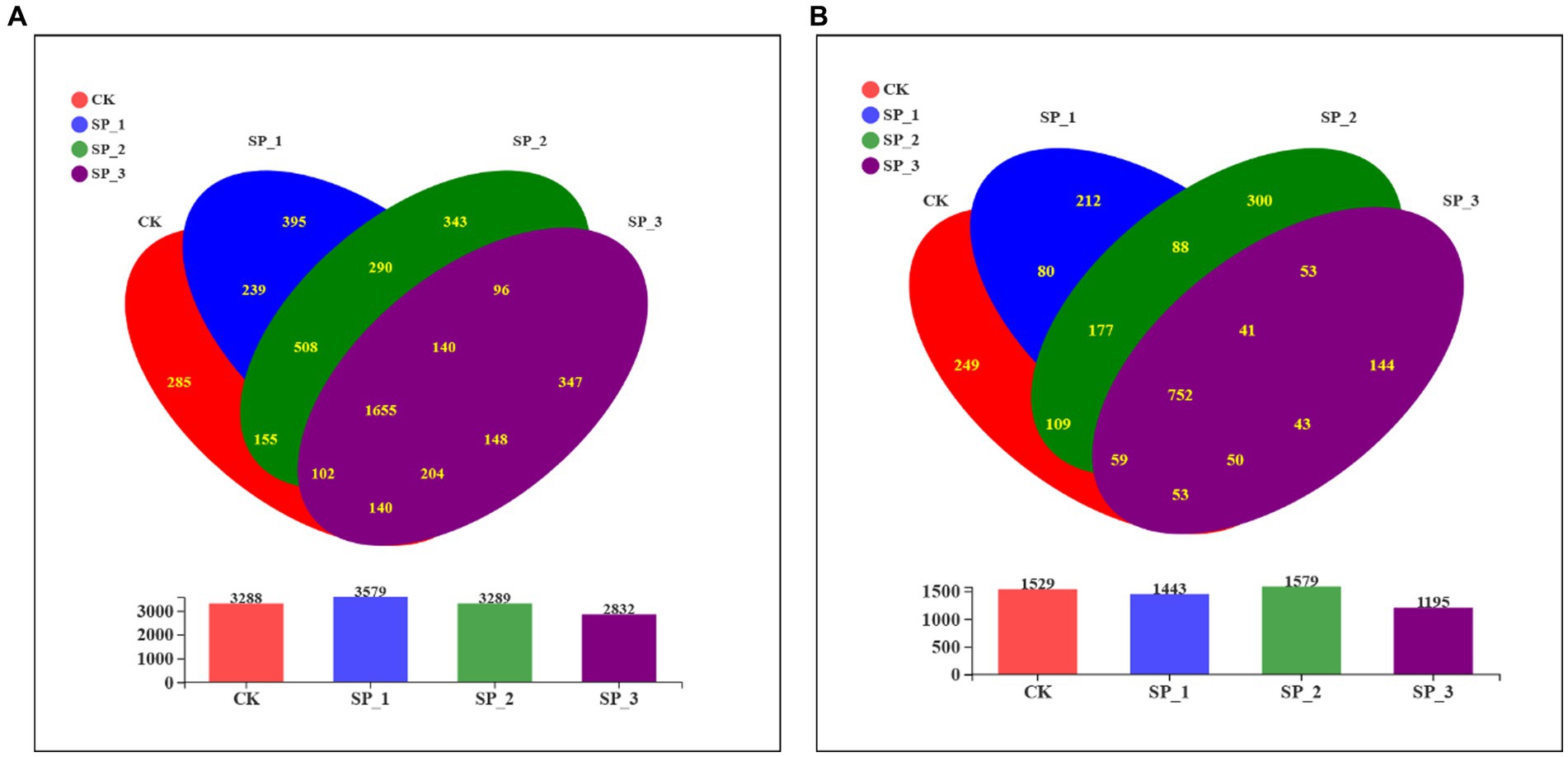
Figure 1. Venn diagram of microbial communities following treatment of soil with different concentrations of ginsenosides. (A) Bacterial community; (B) fungal community.
3.3. Soil microbial community distribution
Principal coordinate analysis (PCoA) grouped the soils from the four groups (Figure 2C) and, analysis of similarity based on Bray-Curtis distance showed significant differences in the distribution of bacterial communities among the four groups. Interestingly, the other three groups clustered on the same side but distantly from the SP_3 group (Figure 2C). The hierarchical clustering analysis (Figure 2A) was validated. In particular, the other three groups clustered together with the SP_3 group at a clustering level of 0.1 (Figure 2B). The distribution of fungal communities in the four groups was also clearly different, with the SP_3 group being very distant from the other three groups (Figure 2D). The distribution of bacterial and fungal communities showed that the microbial populations dramatically changed after the application of various ginsenoside concentrations.
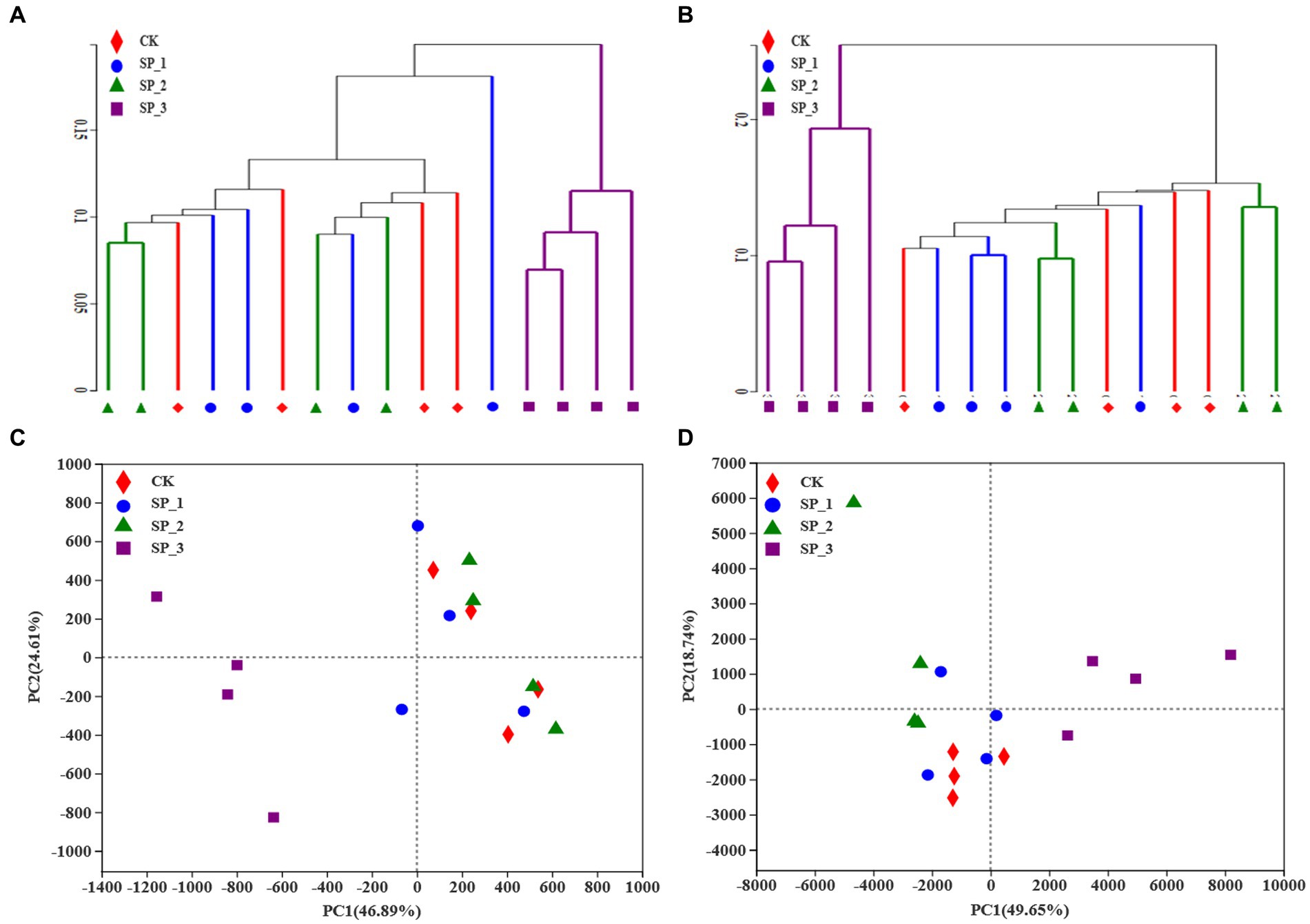
Figure 2. Principal component analysis of microbial communities following treatment of soil with different concentrations of ginsenosides. (A) Principal component analysis of bacterial community; (B) principal component analysis of fungal community; (C) hierarchical cluster analysis of bacterial samples; (D) hierarchical cluster analysis of fungal samples.
3.4. Soil microbial community structure
The four groups were also studied for soil microbial community composition at different levels. The main bacterial families were Xanthobacteraceae, Chthoniobacteraceae, Gaiellales, Sphingomonadaceae, Gaiellaceae, Mycobacteriace, Methyloligellaceae Burkholderiaceae, Rhodanobacteraceae, Nocardiodaceae, Nocardionidaceae, Rhizobiaceae, Propionibacteriaceae, etc., which accounted for about 65% of the total soil bacteria (Figure 3A). In addition, some species were emphasized despite their low relative abundance, such as Frankiaceae, Hyphomicrobiaceae, Xiphinematobacterace, Pyrinomonadaceae, etc. However, the relative abundances of these clades differed significantly between the four groups, especially in the relative abundances of Actinobacteria, Acidobacteria, Chloroflexi, and Gemmatimonadetes (p < 0.05) (Figure 3C). Compared to, Chthoniobacteraceae, Mycobacterium, and Rhodanobacteraceae the relative abundance of the SP_3 group was significantly higher (Figure 3A). Gaiellales showed a significant decrease in relative abundance with increasing concentration of exogenously added ginsenosides. The relative abundance of Gaiellaceae and others in SP_2 increased significantly.
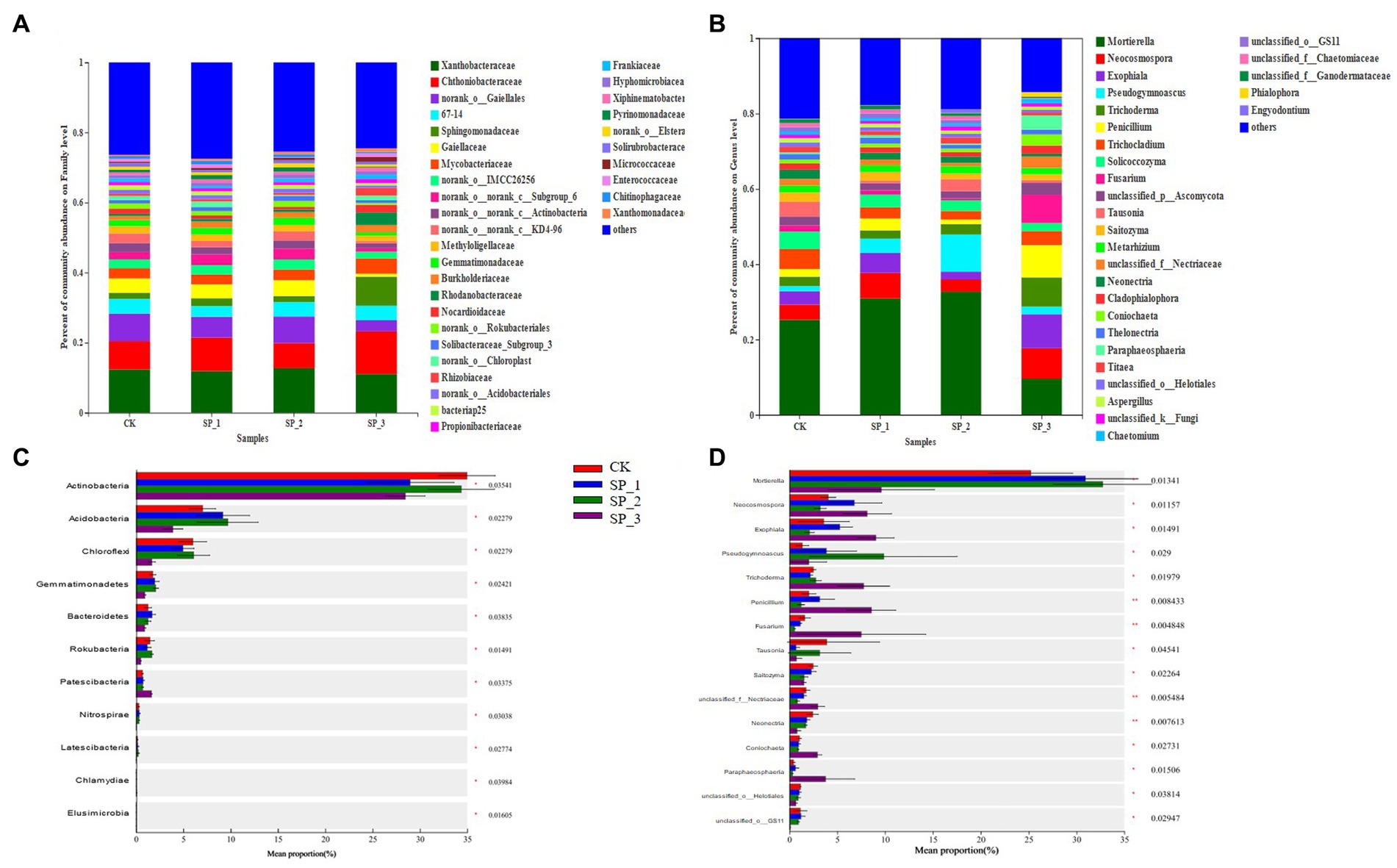
Figure 3. The difference in microbial community composition and relative abundance at phylum level. (A) Community composition at the level of bacterial family; (B) community composition at the level of fungi genera; (C) difference in relative abundance at the level of bacterial phylum; (D) difference in relative abundance at the level of fungal phylum (*0.01 < p ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001).
As in (Figure 3B), among the dominant fungal genera, Mortierella, Neocosmopora, Exophiala, Pseudogymnoascus, Trichoderma, Penicillium, Trichocladium, Soilcoccozyma, Fusarium unclassified_p__Ascomycota, Tausonia, etc. together accounted for about 80% of the soil. Distinct from the other treatment group. Notably the relative abundance of Pseudogymnoascus reached the highest in the SP_2 group. Compared with the untreated control (CK), the community structure of both fungi and bacteria changed significantly after application of ginsenosides, verifying the speculation of this study.
The relative abundance of the pathogenic fungi Fusarium, Gibberella, and Neocosmospora increased with an increase in ginsenosides concentration (Figure 4). In contrast to the untreated control (CK), the relative abundance of the three pathogenic bacteria was significantly enhanced in the SP_3 treatment group. Interestingly, the trend of relative abundance change was similar in the three groups, with SP_1 relative abundance higher than SP_2. Ilyonectria, unlike the other three groups of pathogenic fungi, showed less change in relative abundance after exogenous application of ginsenoside.
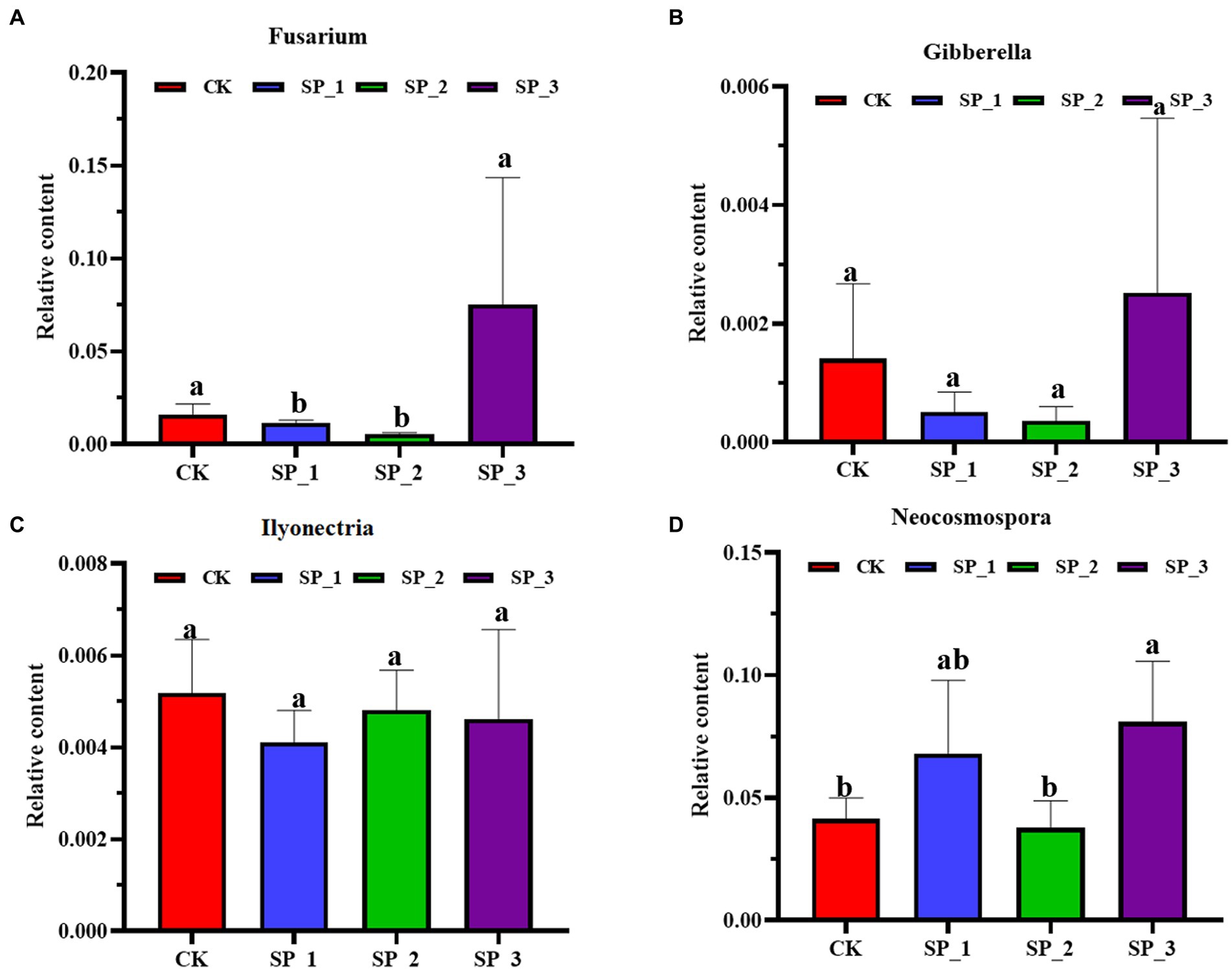
Figure 4. Relative abundance of pathogenic fungi under different concentrations. (A) Fusarium (B) Gibberella (C) Ilyonectria (D) Neocosmospora (relative abundance error bar represents the standard error of the average value of four repetitions). The letters indicate significant difference at p< 0.05 according to one-way analysis of variance (ANOVA) among treatments.
3.5. Functional prediction of soil microflora under different concentrations of ginsenoside treatment
The microbial community composition of soil samples was closely related to environmental conditions, which have been shown to be highly relevant to microbial community function. For example, microbial communities may differ significantly in similar environments, while their community functions may be similar (Gibbons, 2017). Therefore, in addition to revealing the composition and interactions of microbial communities in soil, it is particularly important to reveal differences in the metabolic functions of microbial populations in soil samples treated with different concentrations of ginsenosides. In this study, we used Tax4Fun as a reference database to explore the changes in metabolic potential in the soil after exogenously applied ginsenosides treatment at different concentrations (Aßhauer et al., 2015). This method was used by comparing metabolic pathways predicted based on genes and genomic encyclopedia (KEGG) between the treated and control samples (Aßhauer et al. 2015). Inter-root soil bacterial functions include six classes of primary metabolic pathways predicted as Cellular Processes, Environmental Information Processing, Genetic Information Processing, Human Diseases, and Metabolism The predicted results (Table 4) showed that the main pathways of KEGG predicted at level 1 in the treated and control samples were controlled by Metabolism and Environmental Information Processing (65 and 17%). Metabolism and Organismal Systems pathways were significantly different from the other treatment groups (p < 0.05).

Table 4. Relative abundance of primary metabolic functions of soil bacterial communities under different concentrations of ginsenosides.
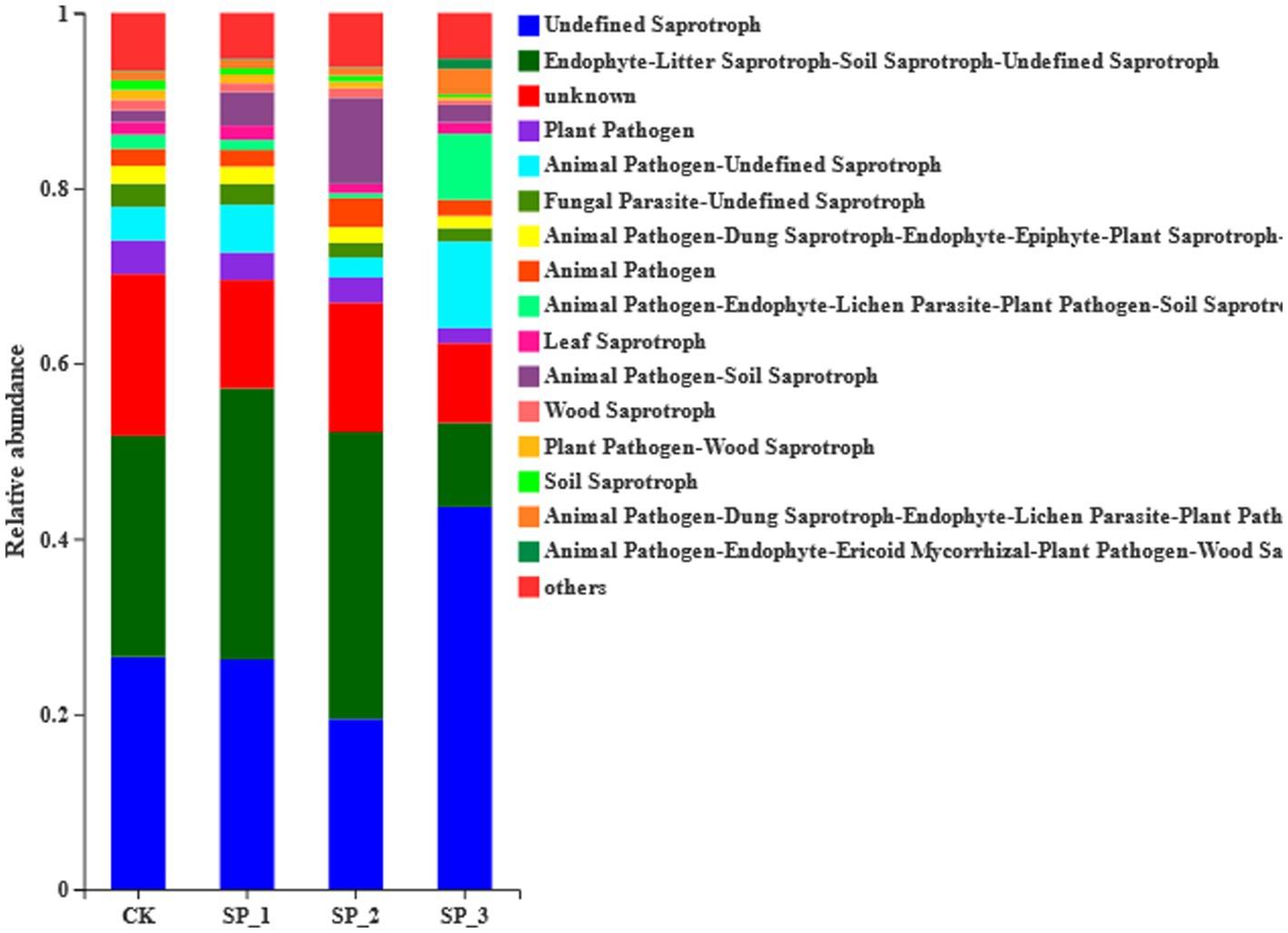
According to the results of FUNGuild functional prediction analysis (Figure 5), the fungal community trophic types included pathotrophs, saprotrophs, and symbiotrophs. The untreated control (CK) was dominated by saprotrophs with litter saprotrophs accounting for 26% of the total community. Plant-pathogens were significantly lower in the three treatment groups, and litter saprotrophs were significantly higher (7%) in the SP_2 treatment group than in the untreated control (CK) group. Animal pathogens were significantly increased in the SP_3 treatment group compared to all the other treatment groups.
3.6. Relationship between key soil factors, enzyme activity, and microbial structure
As might be expected, environmental factors have an effect on the growth of ginseng. Five factors with low multicollinearity were screened for environmental factors (Table 5). Key soil factors such as OM, and EC showed significant correlations with increasing ginsenosides concentrations. The effects of three different concentrations of ginsenosides on soil nutrient factors and enzyme activities were investigated and showed complex relationships. Heat-map-based correlation analysis showed that Sphingomonadaceae, and Mycobacteriaceae had the highest relative abundance compared to other families and were significantly affected by AK (*p ≤ 0.05; ** p ≤ 0.01) (Figure 6A). Gaiellaceae belonging to the phylum Actinomycetes are usually present in the soil as beneficial bacteria, and showed a significant positive correlation with S-SL, and AK. Unlike the Gaiellaceae, Rhizobiaceae, and Rhodanobacteraceae showed a significant negative correlation with AK, and S-SL. Bacteria such as Burkholderiaceae, and Sphingomonadaceae, in contrast, showed significant negative correlations (**p ≤ 0.01; ***p ≤ 0.001) with OM and EC, while Nocardioidaceae showed significant positive correlations with OM. S-SL and had more significant effects on the bacterial community compared to S-SC.
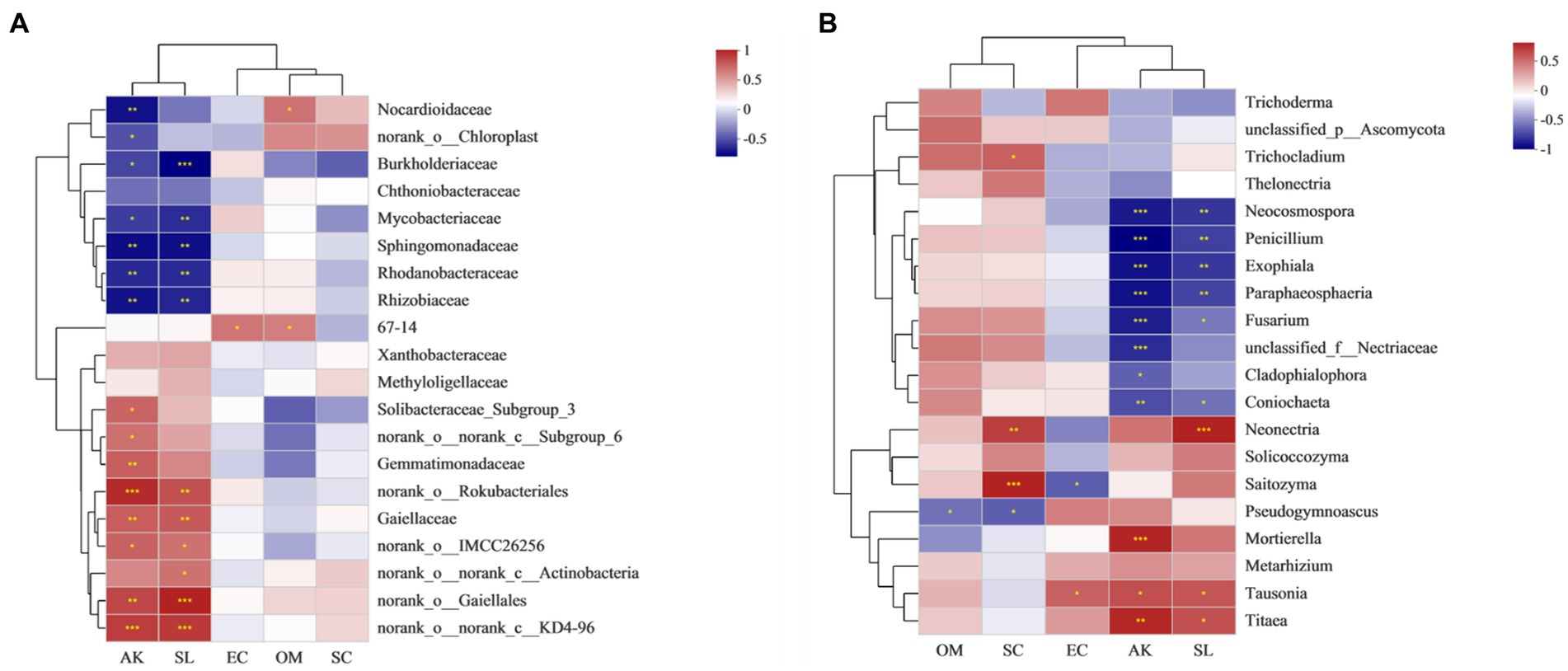
Figure 6. Correlation heat map of Spearman coefficient: (A) Heat map of Spearman correlation coefficient between soil nutrient factors, enzyme activities and rich bacterial families; (B): Heat map of Spearman correlation coefficient between soil nutrient factors, enzyme activities and abundant fungal genera. (*0.01 < p ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001).
Soil factors and enzymatic activity had significant effects on fungal genera, and at the genus level, the most abundant were Mortierella, Neocosmospora, and Exophiala. Spearman correlation coefficients were also used to assess the relationship between the top 20 fungal genera, soil factors, and enzyme activity. Tausonia and Neonectria, Titaea were significantly positively correlated with AK, and S-SL (**p ≤ 0.01; ***p ≤ 0.001). For Mortierella, the factor that significantly mimicked the abundance of this fungus was AK (*p ≤ 0.05). AK and S-SL were significantly negatively correlated with Neocosmospora and Penicillium (**p ≤ 0.01; ***p ≤ 0.001) (Table 5B). The genus was also positively correlated with EC and OM, respectively. EC was found to be a dynamic factor, positively correlated with Saitozyma and Trichocladium, and negatively correlated with Pseudogymnoascus. Saitozyma and Exophiala were positively correlated with S-SC and OM (Figure 6B). Pathogenic Fusarium was negatively correlated with both environmental factors and enzyme activity (*p ≤ 0.05).
4. Discussion
4.1. Effects of different concentrations of exogenous ginsenosides addition on soil chemistry and enzyme activity
Plants and microorganisms can grow in soil, and the physicochemical characteristics and enzymatic activities of soil have an impact on both the organization of microbial communities and plant growth and development (Congreves et al., 2015; Lemaire et al., 2019). An important foundation for assessing plant growth is provided by soil (AN, AP, AK), organic matter, pH, and EC, which also respond to soil fertility and health. The experiment’s fundamental hypothesis was that the soil’s physicochemical characteristics would vary depending on the ginsenoside concentration used. This hypothesis was supported by the results, which showed that the soil pH values in the SP_2 and SP_3 groups were considerably lower than those in the CK group (p < 0.05). This is consistent with earlier research showing that soil degradation is significantly influenced by the decline in soil pH (You et al., 2015; Gibbons, 2017; Zhang et al., 2020). It has been reported that soil pH affects the content of AN in soil (Curtin et al., 1998), also supported by our research (Table 1). Variations in soil pH following exogenous application of various ginsenoside concentrations may result in changes in soil N content, influencing the concentration of NH4, NO3−, and causing soil “Nitrogen” production in the soil–plant system (Klein and Logtestijn, 1994). Organic nitrogen mineralization contributes to changing soil pH by initially consuming and releasing H+ through nitrification during ammonification (Xu et al., 2006). The decrease in soil pH leads to changes in AN content. Phosphorus has a significant positive correlation with ginseng root weight and root diameter and is a key soil factor affecting ginseng yield and quality (Fang et al., 2019), and AP content decreased significantly with increasing ginsenoside concentration in this study. These results suggest that different concentrations of ginsenosides are one of the important factors in regulating soil physicochemical properties. Soil enzyme activities also differed compared to the changes in soil physicochemical properties in the three treatment groups. Soil sucrase, phosphatase, urease, and catalase activities were significantly reduced in all three treatment groups compared to CK (Table 2). The lowest activity of the four soil enzymes was reached in the SP_3 treatment group. It has been reported that higher soil urease activity accelerates the rate of alkaline nitrogen production and increases the content of fast-acting nitrogen (Meng et al., 2012). The decrease in urease activity in this study was speculated to be probably due to the high concentration of exogenously added ginsenoside in the SP_3 treatment group, which led to a decrease in enzyme activity due to the decrease in soil nutrient content.
4.2. Effect of different concentrations of exogenous ginsenosides on soil microbial community
There is growing evidence that plants can alter soil microbial communities by secreting bioactive substances into the inter-rhizosphere. Here we found that exogenous application of different concentrations of ginsenosides may lead to changes in soil microbial communities. Further findings suggest that ginsenosides can enrich or inhibit fungal and bacterial communities. The application of different concentrations of ginsenosides reduced the abundance and diversity of bacterial communities in the soil. In addition, bacterial populations also decreased gradually with increasing ginsenosides.
concentration. This indicates that ginsenosides changed the structure and composition of the soil microbial community, resulting in the proliferation of certain microbial taxa as dominant species in the soil environment, but at the same time, the metabolic types of the bacterial community tended to be homogeneous Loss of microbial diversity and abundance in soils can compromise a range of system services (Shi et al., 2018). For example, the application of different concentrations of ginsenosides decreased the levels of Acidobacteria and Actinobacteria phyla in the soil. Acidobacteria is an important group of soil microorganism, mostly acidophilic, suitable for proliferation in acidic environments, but with slow growth rate (Shi et al., 2018). It was found that the maximum abundance of Acidobacteria was found in undisturbed forest soils, and when the soil nutrient structure, changed faster-growing microorganisms would replace Acidobacteria, resulting in a decrease in their abundance (Ward et al., 2009). This may also be the reason for the decrease in the number of Acidobacteria in the SP_3 treated group, i.e. highest ginsenosides treatment. This was confirmed by successive decline in the relative abundance of Actinobacteria after treatment with increasing concentrations of ginsenosides. Actinobacteria are oligotrophic bacteria and a decrease in relative abundance in the three treatment groups may be due to a decrease in soil fast-acting nutrients after ginsenosides application.
Exogenous application of different concentrations of ginsenosides, increased soil acidity gradually, leading to an increase in abundance of acidic bacteria. However, Xanthomonadaceae is a group of organisms suitable for growth under weakly acidic conditions, and the number of Xanthomonadaceae was significantly reduced following ginsenosides treatment. Xanthomonadaceae consume soil nutrients and infest the xylem of plants as nitrate nitrogen gradually decreases during the growth of ginseng (Matula, 2010). Dong L.-L. et al. (2017) and Dong L. et al. (2017) showed that the Xanthomonadaceae population increased by 160% during three consecutive years of western ginseng cultivation. The increase in Xanthomonadaceae population may have led to an increase in various plant diseases (Mwangi et al., 2007). It is therefore suggested that the increase in the number of Xanthomonadaceae may have been due to the exogenous application of different concentrations of ginsenosides. Therefore, it is speculated that ginsenosides may have promoted the growth of root microorganisms.
A study conducted by Canadian scholars on Panax quiquefolium L. found that root exudate containing ginsenosides significantly promoted the growth of the soil-borne pathogens Phytophthora cactorum and Pythiumirregulare; while Trichodermahamatum showed a slight inhibition under the same conditions (Nicol et al., 2003). This suggests that ginsenosides can significantly affect the growth and reproduction of pathogenic microorganisms and the dynamic changes in the rhizosphere microbial community. Changes in the composition of fungal communities depend mainly on changes in the microbial community in the soil, with most microorganisms producing symbiotic or commensal associations that play a role in nutrient uptake and growth promotion, but inter-rhizosphere ginsenosides can lead to poor defence and growth of secondary soil-borne pathogens (Bednarek et al., 2010; Pascale et al., 2020). Previous studies have shown that ginsenosides stimulate the growth of soil-borne pathogens of Panax quiquefolium L. and Panax notoginseng, destroying for example Cylindrocarpon destructans, Fusarium solani, Phytophthora cactorum, and Pythium irregulare (Nicol et al., 2002, 2003). In this study, the increase in ginsenosides concentration significantly altered the core fungal group and, at the same time, the top 20 most abundant core fungal families.
Results further show that Fusarium, Gibberella, Ilyonectria, and Neocosmospora fungal families were significantly enriched in soil treated with ginsenosides. As pathogenic fungi, an increase in these organisms contributes to the increased chance of ginseng disease (Dong L.-L. et al., 2017; Dong L. et al., 2017). The interaction between Fusarium and the secondary metabolite cinnamic acid may exacerbate soil diseases (Ye et al., 2004). The increased concentration of ginsenosides promoted the growth of Fusarium, contributing to an increased in relative abundance. Numerous studies have shown that Fusarium dominates root rot causative bacteria and is the main causative agent of ginseng root rot (Ye et al., 2004). With the growth of pathogenic fungi and weak invasion causing the ginseng defence response to be stimulated, the fungal population increases, and the secondary metabolites exacerbate the occurrence of soil diseases and, the presence of the highly invasive Ilyonectria fungus disrupts the plant defence barrier (Wu et al., 2020). The reason for this occurrence could be the high production of hydrolytic enzymes, oxidation of phenolic compounds and sequestration of iron, Fe, by Ilyonectria during invasion (Rahman and Punja, 2005), and these studies are highly consistent with results of the present study. When Ilyonectria species are not dominant, they may also promote the growth and weak infestation of by other fungi such as Chaetomidium, Candida, Scopulariopsis, Sclerotinia, and Penicillium, thus aggravating soil diseases (Zhou et al., 2017). Therefore, we speculate that when the saponin concentration reaches a certain level it may be enriched with potentially pathogenic fungi.
4.3. Coupling environmental factors to soil microbial community structure
Soil physicochemical properties, enzyme activity, and microbial community, as important factors interact with each other to maintain the microecological environment of plant roots (Xu et al., 2021). In this study, we found that different concentrations of ginsenosides had a significant effect on the microbial diversity of ginseng rhizosphere soil, which also led to significant changes in soil physicochemical indicators and enzyme activities, in agreement with the results of previous studies (Shukla et al., 2011).
With an increase in ginsenosides concentration, soil physicochemical properties had a significant effect on the soil microbial community (Shukla et al., 2011) and EC indicating that the application of exogenous ginsenosides led to increased salinization of the soil. Increased salinization is one of the important features associated with soil quality degradation (Cambou et al., 2022). The organic matter content was significantly reduced and the sucrase activity, which is a response to the rate and content of soil organic matter conversion, was also gradually reduced. At the same time, the microorganisms that use organic matter as a carbon source to reproduce decreased, but the bacterial communities that are detrimental to ginseng growth such as Xanthobacteraceae, which were a negatively aorrelated with organic matter subsequently increased (Mwangi et al., 2007), leading to an increased chance of ginseng diseases. So, the decrease in organic matter content has a direct impact on changes in microbial community structure, and its composition is crucial for ginseng growth.
The correlation between microbial community and soil physicochemical indicators and enzyme activities of following application of different concentrations of ginsenosides showed that it is not only a single factor such as soil physicochemical properties, enzyme activities, or microorganisms that lead to serious soil-borne diseases of ginseng, but the result of the joint (inter) action. The increase in ginsenosides concentration directly or indirectly modulates the effect of community characteristics on soil enzyme activity, while the composition and other physicochemical properties of photosynthetic bacteria in the soil also mediate microbial growth, community structure, abundance, and diversity, and indirectly drive enzyme activity (Wang et al., 2022).
Secondary metabolites are considered to be one of the more important factors affecting soil microorganisms and driving microbial community changes at different stages of plant growth. Ginsenosides as secondary metabolites have a promoting effect on microbial communities in the soil at certain concentrations, but affect fungal communities differently when the concentration either exceeds or decreases below some critical point, but this conclusion still lacks the most direct evidence. Therefore, studying the mechanisms of secondary metabolite-microbial interactions and exploring the changes in secondary metabolite concentrations are the directions we will focus on in the future. Meanwhile, further follow-up analyses are needed to understand the changes in environmental factors affecting microbial community structure.
5. Conclusion
The interaction between plants and soil microorganisms mediated by the root secretion is a complex process. Soil chemistry, soil microbial community and enzyme activity showed different responses to the application of ginsenosides at increasing concentrations. This study demonstrated that alterations in ginsenosides concentrations affected soil microbial communities to some extent. Bacterial diversity in the soil decreased whereas fungal diversity increased. In addition, the community composition of soil bacteria and fungi changed, with a significant increase in the relative abundance of the pathogenic fungi Fusarium, Erysipelas, Neospora, and Illinois. Therefore, ginsenosides as secondary metabolites, may be one of the main causes of ginseng soil sickness, which provides a new research direction for the subsequent suppression of ginseng soil sickness.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found at: https://www.ncbi.nlm.nih.gov/, SUB12118583.
Author contributions
QL and XYM conceived the study, designed the study, and collected the data. All authors analyzed the data and were involved in writing the manuscript.
Funding
This work was financially supported by grants from National Natural Science Foundation of China (82073969); the National Natural Science Foundation of China (82204558); the Major Science and Technology Project of Jilin Province, China (20200504003YY); the the Natural Science Foundation of Jilin Province, China (YDZJ202101ZYTS015); the Science and Technology Research Project of Education Department of Jilin Province, China (JJKH20210944KJ); and the Young Scientist Project of Changchun University of Chinese Medicine (QNKXJ2-2021ZR19). These grants were received by Professor CC and played an important role in deciding to publish and prepare manuscripts.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1060282/full#supplementary-material
References
Aßhauer, K. P., Wemheuer, B., Daniel, R., and Meinicke, P. (2015). Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884. doi: 10.1093/bioinformatics/btv287
Baeg, I. H., and So, S. H. (2013). The world ginseng market and the ginseng (Korea). J. Ginseng Res. 37, 1–7. doi: 10.5142/jgr.2013.37.1
Banerjee, S., Walder, F., Büchi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 13, 1722–1736. doi: 10.1038/s41396-019-0383-2
Bao, Y., Qi, B., Huang, W., Liu, B., and Li, Y. (2020). The fungal community in non-rhizosphere soil of Panax ginseng are driven by different cultivation modes and increased cultivation periods. Peerj 8:e9930. doi: 10.7717/peerj.9930
Bednarek, P., Kwon, C., and Schulze-Lefert, P. (2010). Not a peripheral issue: secretion in plant-microbe interactions. Curr. Opin. Plant Biol. 13, 378–387. doi: 10.1016/j.pbi.2010.05.002
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654
Cambou, A., Barthès, B. G., Moulin, P., Chauvin, L., Faye, E. H., Masse, D., et al. (2022). Prediction of soil carbon and nitrogen contents using visible and near infrared diffuse reflectance spectroscopy in varying salt-affected soils in sine Saloum (Senegal). Catena 212:106075. doi: 10.1016/j.catena.2022.106075
Chen, Q. W., Jiang, T., Liu, Y. X., Liu, H. L., Zhao, T., Liu, Z. X., et al. (2019). Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci. China-Life Sci. 62, 947–958. doi: 10.1007/s11427-019-9521-2
Congreves, K. A., Hayes, A., Verhallen, E. A., and van Eerd, L. L. (2015). Long-term impact of tillage and crop rotation on soil health at four temperate agroecosystems. Soil Tillage Res. 152, 17–28. doi: 10.1016/j.still.2015.03.012
Curtin, D., Campbell, C. A., and Jalil, A. (1998). Effects of acidity on mineralization: pH-dependence of organic matter mineralization in weakly acidic soils. Soil Biol. Biochem. 30, 57–64. doi: 10.1016/S0038-0717(97)00094-1
Dong, L.-L., Niu, W. H., Wang, R., Xu, J., Zhang, L. J., Zhang, J., et al. (2017). Changes of diversity and composition of fungal communities in rhizosphere of Panax ginseng. Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi =. China J. Chin. materia medica 42, 443–449. doi: 10.19540/j.cnki.cjcmm.20161222.032
Dong, L., Xu, J., Zhang, L., Yang, J., Liao, B., Li, X., et al. (2017). High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin. Med. 12:18. doi: 10.1186/s13020-017-0139-8
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996. doi: 10.1038/NMETH.2604
Eichlerova, I., Snajdr, J., and Baldrian, P. (2012). Laccase activity in soils: considerations for the measurement of enzyme activity. Chemosphere 88, 1154–1160. doi: 10.1016/j.chemosphere.2012.03.019
Fang, X., Liu, J., and Li, X. (2019). Comparative analysis of agronomic characters and Saponins amount of ginseng in four Main agricultural areas in Yanbian prefecture. Journal of Diseases and Medicinal Plants 5:45. doi: 10.11648/J.JDMP.20190503.11
Flowers, T. H., and Bremner, J. M. (1991). A rapid dichromate procedure for routine estimation of total nitrogen in soils. Commun. Soil Sci. Plant Anal. 22, 1409–1416. doi: 10.1080/00103629109368501
Fujimatsu, T., Endo, K., Yazaki, K., and Sugiyama, A. (2020). Secretion dynamics of soyasaponins in soybean roots and effects to modify the bacterial composition. Plant Direct 4:e00259. doi: 10.1002/pld3.259
Garbeva, P., van Veen, J. A., and van Elsas, J. D. (2004). Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Gibbons, S. M. (2017). Microbial community ecology function over phylogeny. Nat. Ecol. Evol. 1. doi: 10.1038/s41559-016-0032
Goebel, B. M., and Stackebrandt, E. (1994). Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl. Environ. Microbiol. 60, 1614–1621. doi: 10.1128/AEM.60.5.1614-1621.1994
Green, J. L., Bohannan, B. J. M., and Whitaker, R. J. (2008). Microbial biogeography: from taxonomy to traits. Science 320, 1039–1043. doi: 10.1126/science.1153475
Jiang, J., Yu, M., Hou, R., Li, L., Ren, X., Jiao, C., et al. (2019). Changes in the soil microbial community are associated with the occurrence of Panax quinquefolius L. root rot diseases. Plant Soil 438, 143–156. doi: 10.1007/s11104-018-03928-4
Klein, C. A. M. D., and Logtestijn, R. S. P. V. (1994). Denitrification in the top soil of managed grasslands in the Netherlands in relation to soil type and fertilizer level. Plant Soil 163, 33–44. doi: 10.1007/BF00033938
Kovar, J. L., and Pierzynski, G. M. (2009). Methods of phosphorus analysis for soils Sediments, Residuals, and Waters Second Edition. Southern Cooperative Series Bulletin.
Lemaire, G., Sinclair, T., Sadras, V., and Bélanger, G. (2019). Allometric approach to crop nutrition and implications for crop diagnosis and phenotyping. A review. Agron. Sustain. Dev. 39. doi: 10.1007/s13593-019-0570-6
Li, Y. L., Dai, S. Y., Wang, B. Y., Jiang, Y. T., Ma, Y. Y., Pan, L. L., et al. (2020). Autotoxic Ginsenoside disrupts soil fungal microbiomes by stimulating potentially pathogenic microbes. Appl. Environ. Microbiol. 86. doi: 10.1128/AEM.00130-20
Lin, M. Y., Han, P. P., Li, Y. Y., Wang, W. X., Lai, D. W., and Zhou, L. G. (2019). Quinoa secondary metabolites and their biological activities or functions. Molecules 24. doi: 10.3390/molecules24132512
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488:86. doi: 10.1038/nature11237
Ma, X. H., Ma, Y., Tang, J. F., He, Y. L., Liu, Y. C., Ma, X. J., et al. (2015). The biosynthetic pathways of Tanshinones and phenolic acids in salvia miltiorrhiza. Molecules 20, 16235–16254. doi: 10.3390/molecules200916235
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Matula, J. (2010). Differences in available phosphorus evaluated by soil tests in relation to detection by colorimetric and ICP-AES techniques. Plant Soil Environ. 56, 297–304. doi: 10.17221/23/2010-PSE
Meng, L.-J., Geng, Z. C., Yin, J. Y., Wang, H. T., and Ji, P. F. (2012). Chemical properties and enzyme activities of rhizosphere and non-rhizosphere soils under six Chinese herbal medicines on Mt. Taibai of Qinling Mountains, Northwest China. J. Appl. Ecol. 23, 2685–2692. doi: 10.1007/s11783-011-0280-z
Meng, Q. F., Yang, J. S., Yao, R. J., and Liu, G. M. (2013). Soil quality in east coastal region of China as related to different land use types. J. Soils Sediments 13, 664–676. doi: 10.1007/s11368-012-0648-0
Mwangi, M., Mwebaze, M., Bandyopadhyay, R., Aritua, V., Eden-Green, S., Tushemereirwe, W., et al. (2007). Development of a semiselective medium for isolating Xanthomonas campestris pv. Musacearum from insect vectors, infected plant material and soil. Plant Pathol. 56, 383–390. doi: 10.1007/s11368-012-0648-0
Ng, T. B. (2006). Pharmacological activity of sanchi ginseng (Panax notoginseng). J. Pharm. Pharmacol. 58, 1007–1019. doi: 10.1111/j.1365-3059.2007.01564
Nicol, R. W., Traquair, J. A., and Bernards, M. A. (2002). Ginsenosides as host resistance factors in American ginseng (Panax quinquefolius). Can. J. Bot. Revue Canadienne De Botanique 80, 557–562. doi: 10.1139/b02-034
Nicol, R. W., Yousef, L., Traquair, J. A., and Bernards, M. A. (2003). Ginsenosides stimulate the growth of soilborne pathogens of American ginseng. Phytochemistry 64, 257–264. doi: 10.1016/S0031-9422(03)00271-1
Nottingham, A. T., Fierer, N., Turner, B. L., Whitaker, J., Ostle, N. J., McNamara, N. P., et al. (2018). Microbes follow Humboldt: temperature drives plant and soil microbial diversity patterns from the Amazon to the Andes. Ecology 99, 2455–2466. doi: 10.1002/ecy.2482
Pascale, A., Proietti, S., Pantelides, I. S., and Stringlis, I. A. (2020). Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10.1741. doi: 10.3389/fpls.2019.01741
Rahman, M., and Punja, Z. K. (2005). Biochemistry of ginseng root tissues affected by rusty root symptoms. Plant Physiol. Biochem. 43, 1103–1114. doi: 10.1016/j.plaphy.2005.09.004
Rodíguez-Kábana, R. (1982). The effects of crop rotation and fertilization on soil xylanase activity in a soil of the southeastern United States. Plant Soil 64, 237–247. doi: 10.1007/BF02184255
Shi, Z. J., Lu, Y., Xu, Z. G., and Fu, S. L. (2008). Enzyme activities of urban soils under different land use in the Shenzhen city, China. Plant Soil Environ. 54, 341–346. doi: 10.1111/j.1365-3059.2008.01915
Shi, S., Tian, L., Ma, L., and Tian, C. (2018). Community structure of Rhizomicrobiomes in four medicinal herbs and its implication on growth management. Microbiology 87, 425–436. doi: 10.1134/s0026261718030098
Shukla, K. P., Sharma, S., and Singh, N. K. (2011). Nature and role of root exudates: efficacy in bioremediation. Afr. J. Biotechnol. 10, 9717–9724. doi: 10.5897/ajb10.2552
Stringlis, I. A., Yu, K., Feussner, K., de Jonge, R., van Bentum, S., van Verk, M. C., et al. (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U. S. A. 115, E5213–E5222. doi: 10.1073/pnas.1722335115
Tardy, V., Spor, A., Mathieu, O., Lévèque, J., Terrat, S., Plassart, P., et al. (2015). Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol. Biochem. 90, 204–213. doi: 10.1016/j.soilbio.2015.08.010
Tong, A. Z., Liu, W., Liu, Q., Xia, G. Q., and Zhu, J. Y. (2021). Diversity and composition of the Panax ginseng rhizosphere microbiome in various cultivation modesand ages. BMC Microbiol. 21:18. doi: 10.1186/s12866-020-02081-2
Wang, R., Dong, L. L., Xu, J., Chen, J. W., Li, X. W., and Chen, S. L. (2016). Progress in improvement of continuous monoculture cropping problem in Panax ginseng by controlling soil-borne disease management. China J. Chin. Mater. Med. 41, 3890–3896. doi: 10.4268/cjcmm20162102
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, W. X., Li, Y. Y., Dang, P. Q., Zhao, S. J., Lai, D. W., and Zhou, L. G. (2018). Rice secondary metabolites: structures, roles, biosynthesis, and metabolic regulation. Molecules 23. doi: 10.3390/molecules23123098
Wang, Q. X., Sun, H., Li, M. J., Xu, C. L., and Zhang, Y. Y. (2020). Different age-induced changes in Rhizosphere microbial composition and function of Panax ginseng in transplantation mode. Front. Plant Sci. 11:563240. doi: 10.3389/fpls.2020.563240
Wang, Q., Zhou, L., Jin, H., Cong, B., Yang, H., and Wang, S. (2022). Investigating the responses of microbial communities to Banana Fusarium wilt in suppressive and conducive soils based on soil particle-size differentiation. Agronomy-Basel 11. doi: 10.3390/agronomy12020229
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75, 2046–2056. doi: 10.1128/AEM.02294-08
Wu, X.-L., Wang, Y., Liu, F., Chen, D. X., and Li, L. Y. (2020). Identification of Coptis chinensis root rot disease pathogenic Fusarium spp. fungi.Zhongguo Zhong yao za zhi Zhongguo zhongyao zazhi. China J. Chin. Mater. Med. 45, 1323–1328. doi: 10.19540/j.cnki.cjcmm.20200112.102
Xu, Y. X., Ren, S., Liang, Y., du, A., Li, C., Wang, Z., et al. (2021). Soil nutrient supply and tree species drive changes in soil microbial communities during the transformation of a multi-generation eucalyptus plantation. Appl. Soil Ecol. 166:103991. doi: 10.1016/j.apsoil.2021.103991
Xu, J. M., Tang, C., and Chen, Z. L. (2006). The role of plant residues in pH change of acid soils differing in initial pH. Soil Biol. Biochem. 38, 709–719. doi: 10.1016/j.soilbio.2005.06.022
Yang, M., Zhang, X. D., Xu, Y. G., Mei, X. Y., Jiang, B. B., Liao, J. J., et al. (2015). Autotoxic Ginsenosides in the Rhizosphere contribute to the replant failure of Panax notoginseng. PLoS One 166:103991. doi: 10.1016/j.apsoil.2021.103991
Ye, S. F., Yu, J. Q., Peng, Y. H., Zheng, J. H., and Zou, L. Y. (2004). Incidence of Fusarium wilt in Cucumis sativus L. is promoted by cinnamic acid, an autotoxin in root exudates. Plant Soil 263, 143–150. doi: 10.1371/journal.pone.0118555
You, J. F., Liu, X., Zhang, B., Xie, Z. K., Hou, Z. G., et al. (2015). Seasonal changes in soil acidity and related properties in ginseng artificial bed soils under a plastic shade. J. Ginseng Res. 39, 81–88. doi: 10.1016/j.jgr.2014.08.002
Yousef, L. F., and Bernards, M. A. (2006). In vitro metabolism of ginsenosides by the ginseng root pathogen Pythium irregulare. Phytochemistry 67, 1740–1749. doi: 10.1016/j.phytochem.2005.06.030
Zhang, J. G., Fan, S. H., Qin, J., Dai, J. C., Zhao, F., Gao, L., et al. (2020). Changes in the microbiome in the soil of an American ginseng continuous plantation. Front. Plant Sci. 11:572199. doi: 10.3389/fpls.2020.572199
Zhang, F. X., Tang, S. J., Zhao, L., Yang, X. S., Yao, Y., Hou, Z. H., et al. (2021). Stem-leaves of Panax as a rich and sustainable source of less-polar ginsenosides: comparison of ginsenosides from Panax ginseng, American ginseng and Panax notoginseng prepared by heating and acid treatment. J. Ginseng Res. 45, 163–175. doi: 10.1016/j.jgr.2020.01.003
Zhang, J. X., Zhou, D. P., Yuan, X. Q., Xu, Y. H., Chen, C. B., and Zhao, L. (2022). Soil microbiome and metabolome analysis reveals beneficial effects of ginseng-celandine rotation on the rhizosphere soil of ginseng-used fields. Rhizosphere 23:100559. doi: 10.1016/j.rhisph.2022.100559
Zhou, Y., Yang, Z., Gao, L., Liu, W., Liu, R., Zhao, J., et al. (2017). Changes in element accumulation, phenolic metabolism, and antioxidative enzyme activities in the red-skin roots of Panax ginseng. J. Ginseng Res. 41, 307–315. doi: 10.1016/j.jgr.2016.06.001
Keywords: ginsenoside, microbial community, enzyme activity, environmental factors, secondary metabolites
Citation: Miao X, Wang E, Zhou Y, Zhan Y, Yan N, Chen C and Li Q (2023) Effect of ginsenosides on microbial community and enzyme activity in continuous cropping soil of ginseng. Front. Microbiol. 14:1060282. doi: 10.3389/fmicb.2023.1060282
Edited by:
Ashton Keith Cowan, Rhodes University, South AfricaJun Zhao, Nanjing Normal University, China
Reviewed by:
Fengyu Du, Qingdao Agricultural University, ChinaPrakash Babu Adhikari, Fujian Agriculture and Forestry University, China
Copyright © 2023 Miao, Wang, Zhou, Zhan, Yan, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiong Li, d29kZTE3a0AxNjMuY29t; Changbao Chen, Y2NiMjAyMUAxMjYuY29t
 Xinyue Miao
Xinyue Miao Ergang Wang
Ergang Wang