
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 April 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1056399
This article is part of the Research TopicPrevalence, Transmission and Control of Clinically Important Antimicrobial-Resistant Bacteria/Genes within One Health FrameworkView all 22 articles
A correction has been applied to this article in:
Corrigendum: Molecular epidemiology and comparative genomics of carbapenemase-producing Escherichia coli isolates from 19 tertiary hospitals in China from 2019 to 2020
Background: The clinical use of carbapenems is facing challenges due to increased carbapenemase-producing Escherichia coli (CP-EC) infections over the past decade. Meanwhile, whole-genome sequencing (WGS) is an important method for bacterial epidemiological research. We aim to provide more gene-based surveys to explore the genomics and occurrence of CP-EC in China.
Methods: A total of 780 Escherichia coli isolates were collected by the China Antimicrobial Resistance Surveillance Trial (CARST) from 2019 to 2020. An antibacterial susceptibility test was performed by using the agar dilution method. CP-EC were detected by the modified carbapenem inactivation method (mCIM), EDTA-modified carbapenem inactivation method (eCIM), and polymerase chain reaction (PCR). Homology analysis was performed by multilocus sequence typing (MLST). A conjugation experiment was performed to verify the transferability of plasmids carrying carbapenemase genes. WGS was conducted to explore the gene-environment of the carbapenemase gene.
Result: Of the 780 Escherichia coli isolates, 31 isolates were insensitive to carbapenem with a rate of 4%. Among them, 13 CP-EC isolates had transferability of the blaNDM gene. These isolates belonged to nine distinct sequence types (STs), with some correlation. We found that two (2/13, 15.4%) of the CP-EC isolates that were collected from blood specimens were highly pathogenic and also showed high transferability of the blaNDM gene. In addition, eight (8/13, 61.5%) of the CP-EC isolates were found to be multidrug-resistant.
Conclusion: With the increasing use of carbapenem, CP-EC isolates accounted for nearly half of the total carbapenem-insensitive Escherichia coli isolates. Our findings highlight the urgent need to pay attention to CP-EC isolates in bloodstream infections and ESBL-producing CP-EC isolates. Based on the One Health concept, we suggest various measures, including the development of bacterial vaccines, antibiotic management, and establishment of better medical environments, to avoid the outbreak of CP-EC.
Carbapenems are considered one of the last resort antibiotics against extended-spectrum beta-lactamase-producing gram-negative bacterial infections (Li et al., 2021). However, carbapenem resistance has become a major public health issue, with the World Health Organization listing carbapenem-resistant Enterobacteriaceae as the priority bacteria. Escherichia coli (E. coli), which can cause urinary, respiratory, and bloodstream infections in addition to other pathogens in immunocompromised individuals (Tao et al., 2020), is primarily responsible for community-associated infections, making it difficult to implement traditional preventive measures based on hospital-acquired infections. In China, the isolation rates of carbapenem-resistant Escherichia coli (CRECO) have shown an increasing trend in recent years, despite being lower than in previous years. The China Antimicrobial Resistance Surveillance trail (CARST) reported that between 2016 and 2020, the average isolation rates of CRECO were 1.5, 1.5, 1.5, 1.7, and 1.6%, respectively. Although the 2020 isolation rate of CRECO decreased by 0.1% points compared to 2019, it is still higher than the average rate in previous years. Furthermore, the isolation rates of CRECO also varied among regions, with Henan Province showing the highest isolation rates in 2016 and 2018 (3 and 2.9%), and Qinghai Province showing the lowest (0.6 and 0.2%). In 2017, Liaoning Province showed the highest isolation rates (2.8%), while the Tibet Autonomous Region showed the lowest (0.3%). In 2020, Beijing had the highest isolate rates of CRECO (3.2%), while the Tibet Autonomous Region had the lowest proportion (0.2%).
Carbapenem resistance is a growing public health concern due to its widespread dissemination facilitated by carbapenemase, a crucial factor responsible for its transmission. The transfer of genes encoding carbapenemase through conjugative plasmids, transposons, and insertion sequences has led to horizontal transmission (Conlan et al., 2014). In China, carbapenemase-producing Enterobacterales (CPE) infections have increased over the past decade, with E.coli being the second most common CPE (Deng et al., 2022; Wang et al., 2022). Notably, a majority (76.2%, 16/21) of carbapenemase-producing Escherichia coli (CP-EC) cases were detected in a hospital in Shanghai (Zhang et al., 2015). In addition, 35.8% of extended-spectrum β-lactamase-producing Escherichia coli (ESBL-EC) and 0.9% of CP-EC were isolated from the feces of healthy children's feces in south-central China communities (Liu et al., 2022). Tracing the genetic environment and epidemiology of carbapenemase genes is paramount to public health and aids in designing infection management and prevention strategies.
Polymerase chain reaction (PCR) and pulsed-field gel electrophoresis (PFGE) are traditional techniques used in molecular epidemiological research of antimicrobial resistance. However, these techniques have limitations, and whole-genome sequencing (WGS) can be a more effective tool for analyzing antimicrobial resistance. WGS can directly assemble the sequence fragments without designing specific primers to obtain the whole genome of the isolate. The isolate's species, antibiotic resistance genes, virulence factors, and mobile genetic elements can be determined by comparing the whole genome of the isolate with the database. Moreover, genome comparisons of multiple isolates can provide information on the molecular epidemiology of specific resistance genes or mechanisms of antimicrobial resistance. Therefore, WGS and bioinformatics analysis can be more effective in assisting with molecular epidemiological research of antimicrobial resistance. Despite the gene-environment of plasmids has been studied recently, nationwide WGS-based epidemiology of clinical CP-EC has not yet been carried out in China. As a result, we aim to provide more gene-based investigations to explore the genomics and occurrence of CP-EC in China.
A total of 780 E. coli isolates were collected between July 2019 and June 2020 from 19 tertiary hospitals by the China Antimicrobial Resistance Surveillance Trial (CARST) and were preserved by the Institute of Clinical Pharmacology of Peking University. Samples processing, microbial identification, and antibiotic susceptibility testing were performed by the microbiology laboratories of the 19 hospitals, according to a shared protocol. According to CLSI-M100-S31, antibiotic susceptibility testing (AST) was carried out by the reference broth microdilution method. Antimicrobial agents or combinations tested included cefotaxime, ceftazidime, cefepime, aztreonam, imipenem, meropenem, ertapenem, amikacin, minocycline, tigecycline, ciprofloxacin, and polymyxin E. E. coli ATCC25922 was used as the control strain for AST. All tests were performed in duplicate, and three biological replicates per strain were included in each test. PCR was used to amplify the 16SrRNA of the isolates, and the amplified products were sent to Beijing SinoGenoMax Co., Ltd. for sequencing. The isolates were confirmed to be E. coli by comparing the sequences in NCBI (https://blast.ncbi.nlm.nih.gov/).
To detect carbapenemase phenotypes, we employed the modified carbapenem inactivation method (mCIM) and EDTA-modified carbapenem inactivation method (eCIM), and Klebsiella pneumoniae ATCC BAA-1705 and Klebsiella pneumoniae ATCC BAA-1706 were utilized as quality controls. We targeted the following carbapenemase genes for PCR detection: blaKPC, blaSME, blaIMI, blaGES, blaIMP, blaVIM, blaSIM, blaGIM, blaNDM, and blaOXA−48. To assess the transferability of blaNDM, a conjugation assay was performed on the 13 blaNDM-positive E. coli isolates, using sodium azide-resistant E. coli J53 as the recipient strain. E. coli isolates were analyzed based on genomic sequences using in silico MLST and cgMLST analysis on the EnteroBase online platform (http://enterobase.warwick.ac.uk) for E. coli. Seven housekeeping gene loci, including adk, fumC, gyrB, icd, mdh, purA, and recA, were chosen for MLST analysis of E. coli. The cluster analysis is performed by Bionumerics software v.7.0 based on MLST information.
We performed whole-genome sequencing (WGS) of CP-EC isolates using the Nanopore De Novo platform and Circular Library. We prepared the sequencing library using the rapid sequencing kit (Oxford Nanopore Technologies) and subjected the sequencing data to several processing steps. First, we checked the consistency of each base in the original data and assigned a quality value to each base based on the results of the consistency check. Next, we removed residual adapters using strict parameters and filtered out regions with Q20 < 0.8 in the reads to obtain high-quality subreads sequences. These high-quality subreads sequences were then used for subsequent analysis. We assembled the high-quality subreads using the hierarchical genome assembly process (HGAP) 3.0 software, which corrects errors in long reads with short reads. The resulting assembly was further processed using the classic OLC algorithm architecture to obtain the final assembly. To identify transposon sequences, we compared the assembly results with known transposon sequence libraries using RepeatMasker software (using the Repbase database) and RepeatProteinMasker software (using the transposon protein library that comes with RepeatMasker). We also used tandem repeat finder (TRF) software to predict tandem repeats. Contigs were screened for plasmid and resistance gene content using PlasmidFinder and ResFinder tools at the Center for Genomic Epidemiology (CGE) server (http://www.genomicepidemiology.org/services/), respectively. To identify virulence genes related to CP-EC isolates, we utilized VirulenceFinder at CGE and the VFDB database. In addition, we used ISFinder to detect insertion sequences with a threshold of e value1e-5 and generated a gene alignment diagram using Easyfig.
A total of 31 E.coli isolates displayed insensitive to meropenem, imipenem, or ertapenem. Among 31 insensitive E. coli isolates, the mean minimal inhibitory concentrations (MICs) of meropenem against 50% (MIC50) and 90% (MIC90) were 0.25 and 32 μg/ml, respectively. The MIC50 and MIC90 values of imipenem were 0.5 and 16 μg/ml, respectively. The MIC50 and MIC90 values of ertapenem were 4 μg/ml and 128 μg/ml, respectively. Of the 31 isolates, 13 (41.9%), 14 (45.2%), and 30 (96.7%) were insensitive to meropenem, imipenem, and ertapenem, respectively. In addition, 96% (29/31) of isolates exhibited resistance to third/fourth-generation cephalosporins. Clinical information about the 31 E. coli isolates is presented in Table 1. Thirteen carbapenemase-producing E. coli (CP-EC) isolates were confirmed to produce metallo-β-lactamase using the modified carbapenem inactivation method (mCIM) and EDTA-modified carbapenem inactivation method (eCIM). Of these 13 CP-EC isolates, only blaNDM was detected, without any other carbapenemase genes. Among these, nine isolates (69.2%) carried blaNDM−5, three isolates (23.1%) carried blaNDM−1, and one isolate (7.7%) carried blaNDM−7. The genome-based phylogenetic analysis revealed the presence of nine CP-EC lineages: ST167 (3 isolates), ST405 (3 isolates), ST744, ST10, ST361, ST1193, ST488, ST410, and ST349 (refer to Figure 1 and Table 2 for details).
The clonal relationship between CP-EC ST448 and ST410 was found to be highly related, with ST410 originating from ST448. In addition, the three NDM-1 producing CP-EC isolates were classified under ST349, ST744, and ST1193, respectively. Notably, NDM-7 producing CP-EC isolate, 19F065, was assigned to ST405. Furthermore, ST167, ST10, and ST744 exhibited close relatedness, with ST167 and ST744 emerging as derivatives of ST10. These findings suggest a complex evolutionary history of CP-EC.
The successful transfer of the carbapenemase gene to E. coli J53 recipients was observed in all 13 isolates, with a minimum of a 32-fold increase in the MIC of carbapenems for the transconjugants. Plasmid content was evaluated by PlasmidFinder and is presented in Table 2. IncX3 was the predominant plasmid type observed in CP-EC (8/13, 61.5%), followed by IncFII (3/13, 23.1%). The average length of the plasmids was 50,000 bp, of which p19D107 and p19F412 were longer, with a length of 95,105 bp and 268,028 bp, respectively.
The plasmids of the 13 CP-EC isolates were found to harbor mobile genetic elements and insertion sequences (ISs), as determined by genomic analysis. ISAba125 was frequently located upstream of the genomic region (5/13, 38.5%), while IS15 was frequently located downstream (9/13, 69.2%). The plasmid carrying blaNDM−7, specifically p19F065, had a similar genetic environment to those carrying blaNDM−5, such as p19F407 and p19R077. Notably, among the nine isolates carrying blaNDM−5, p19R077 and p19F407 exhibited nearly identical genetic environments, with ISAba125, ISC1041, and IS5B detected upstream of blaNDM−5.
The plasmid p19U139, classified as an IncN plasmid, carries the carbapenemase gene blaNDM−1. IS1X2 and IS61009 were identified upstream of p19U139, while ISSsu9 was found downstream. Meanwhile, the plasmids p19F388 and p19M348, which also carry blaNDM−1, were classified as IncX3 plasmids. Specifically, p19F388 contained two IS3000 elements, one Tn2 transposon, and one IS5 element, while p19M348 had an ISAba125 insertion between IS3000 and IS5. These two plasmids shared a similar downstream gene-environment, consisting of an ISKox3 element belonging to the ISL3 transposase family and multiple IS26 elements of the IS6 transposase family, which carried the blaSHV−2 gene at a distance. According to the gene alignment diagram, the core structure of the IncX3 plasmid is diverse by carrying different blaNDM subtype genes. For instance, the core structures of IncX3 plasmid carrying the blaNDM−1 and blaNDM−5 genes are IS3000-IS5-blaNDM−1-IS26-ISKox3 and IS3000-ISAba125-IS5-blaNDM−5-IS26-ISKox3, respectively. On the other hand, the IncFII plasmid carrying the blaNDM−5 gene has a different core structure: IS26-blaNDM−5-ISSsu9-IS26-TnAs1. For further details, refer to Figure 2A.
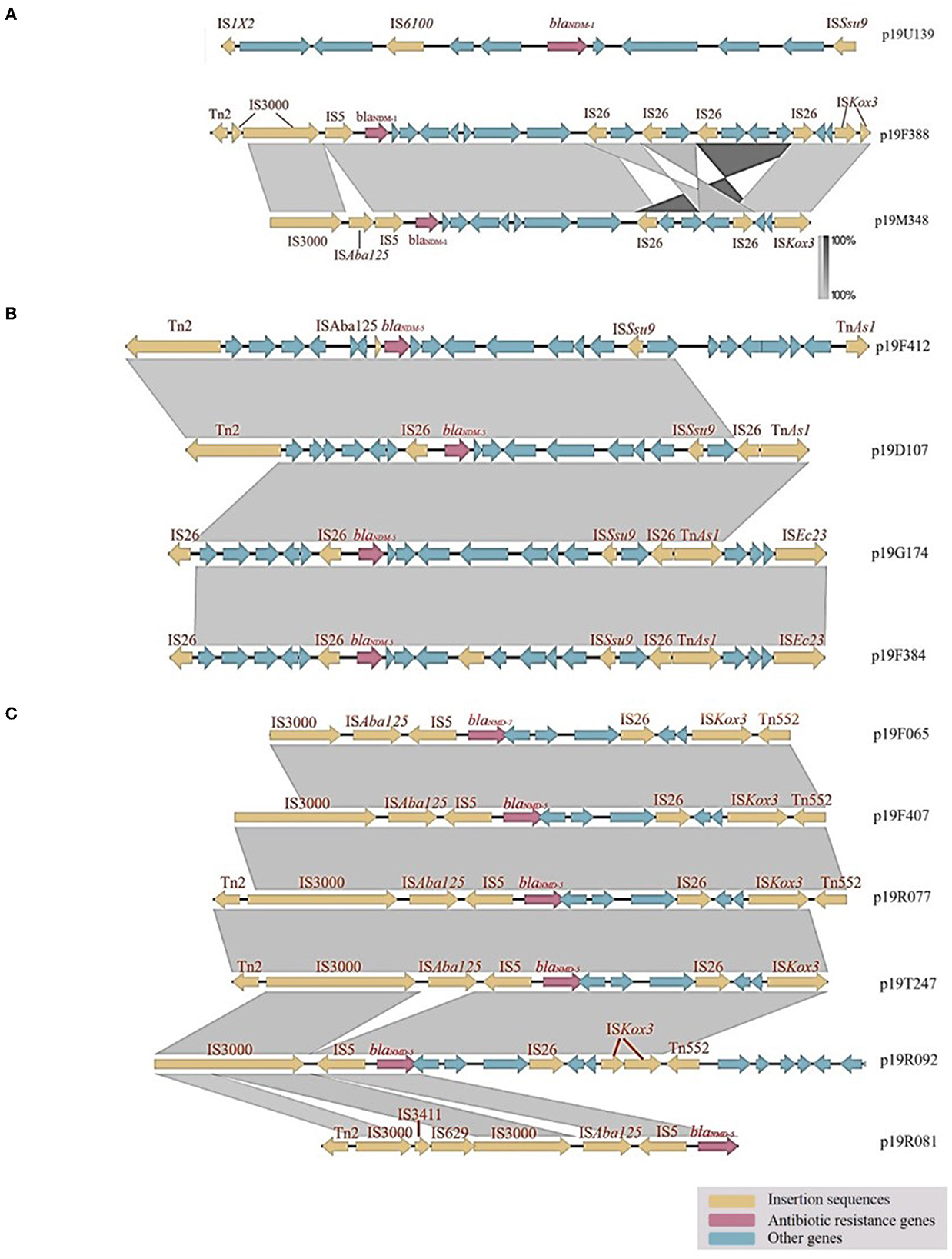
Figure 2. Genetic analysis of carbapenemase-producing E. coli isolates: Insights from gene alignment and gene-environment analysis of blaNDM−1, blaNDM−5, and blaNDM−7 in different plasmids. (A) The gene-environment of p19U139 was found to differ from that of other plasmids, with IS6100 and IS1X2 belonging to the IS6 and IS1 transposase families, respectively. (B) In the IncFII plasmid, IS26 was found to carry both blaNDM−5 and emrE, a gene related to an efflux protein. (C) Analysis of blaNDM−5 and blaNDM−7 gene-environment in IncX3 plasmids.
The gene-environments of the IncFII plasmids, namely p19D107, p19G174, and p19F384, show a high degree of similarity, as all three contain an adjacent reverse IS26 element located upstream of the blaNDM−5 gene. In addition, these plasmids possess IS26, ISSsu9, and TnAs1 elements at a distance from the blaNDM−5 gene (refer to Figure 2B). Conversely, p19F412 is a longer IncI1-I plasmid that shares a partially similar gene-environment with p19D107.
p19F065, p19F407, p19R077, and p19T247 are all IncX3 plasmids with the same upstream and downstream gene-environments. These plasmids have a segment of IS3000, ISAba125, and IS5 upstream and a segment of IS26, ISKox3, and Tn552 downstream. p19R092 has a missing ISAba125 upstream, and ISKox3 downstream was divided into two segments. The blaNDM−5 gene in p19R081 is located at the edge of the genome, so the downstream environment is unknown. Unlike other plasmids, p19R081 has IS3411 and IS629, dividing IS3000 into two segments (Figure 2C).
The resistome of CP-EC isolates is visualized in Figure 3, revealing the presence of 16 antibiotic resistance genes linked to various antibiotic classes, including β-lactams, aminoglycosides, sulfamide, trimethoprim, quaternary ammonium salt disinfectant, and fosfomycin. Notably, five isolates (5/13, 38.5%) were found to carry extended-spectrum β-lactamase (ESBL) genes, among which blaSHV−12 and blaCTX−M were identified in two and three isolates, respectively.
Virulence genes were detected in each isolate with a variable distribution (Figure 4). The majority of isolates (10/13, 76.9%) harbored sequences encoding type IV secretion system (T4SS), specifically virB4, virB9, trwD, trwE, trwF, trwG, trwK, ptlH, virD4, icmO/dotL, dotO, and dotC (Christie et al., 2014). In addition, the traj gene, related to the plasmid conjugation transfer region, was found in one isolate (Liu G. et al., 2019). In the p19F412 isolate, several sequences encoding adhesion proteins were identified, including PilU, pilV, pilS, pilR, pilQ, pilO, pilN, and pilM, involved in pili synthesis (Foley et al., 2021); IutA, iucD, iucC, iucB, iucA, sitD, sitC, sitB, and sitA, encoding iron transport and siderophore proteins (da Cruz Campos et al., 2020); and icsP/sopA, associated with outer membrane protease (Tran et al., 2013). Furthermore, the type III secretion system (TTSS) invasion antigen, IpaH2.5, was identified in 19F412 and 19R081 (Tran et al., 2013), while the htp operons, a molecular chaperone related to heat shock protein Hsp60, were present in four isolates. The colonization factor antigen III-encoding cofT genes, facilitating bacterial adherence to the host, were identified in three isolates. Interestingly, IncX3 plasmids carried several genes associated with T4SS, while IncFII plasmids did not possess genes related to TTSS or T4SS. Remarkably, p19F412 and p19R081 isolates contained genes associated with both TTSS and T4SS.
The prevalence and impact of CPE vary globally. In the United States, CPE is responsible for approximately 9,300 hospital-acquired infections annually, and approximately 50% of patients with bloodstream infections (BSIs) caused by CPE ultimately succumb to the infection (Villegas et al., 2016). Meanwhile, in central China, a surveillance report identified E. coli as one of the main pathogens causing bloodstream infections (Tian et al., 2018). In our study, we found that CP-EC from blood specimens accounted for 53.8% (7/13) of the total CP-EC, with blaNDM−5-producing isolates making up the majority, two of which belonged to ST405. These isolates were capable of transmitting carbapenemase genes through IncFII, IncI, IncN, and IncX3, with p19F412 and p19R081 carrying both TTSS and T4SS, making them highly transferable and pathogenic.
The dissemination of CPE isolates is a serious concern as these infections require aggressive and prolonged antibiotic therapy, leading to an increased risk of adverse events and prolonged hospitalization. A recent investigation in Australia found that patients colonized with CP-EC had a significantly longer hospital stay and higher medical costs during an outbreak (Rodriguez-Acevedo et al., 2020). These findings underscore the importance of managing and preventing future outbreaks caused by CP-EC.
The co-occurrence of carbapenemases with other β-lactamases, such as TEM, SHV, or CTX-M, confers resistance in CPE to almost all β-lactam antibiotics (De Angelis et al., 2020). In our study, 61.5% of the 13 isolates belonged to multidrug-resistant CP-EC, with seven isolates producing TEM, CTX-M, and SHV. The co-occurrence of these resistance mechanisms can pose challenges in clinical laboratories since carbapenemases are not inhibited by clavulanic acid, which is used in the standard double-disk test for identifying ESBLs recommended by the Clinical and Laboratory Standards Institute (CLSI). As a result, detecting multiple β-lactamases in CP-EC can be difficult, which may lead to heightened transmission and outbreaks in healthcare settings and community-acquired infections. The increased use of carbapenems has led to the gradual detection of CP-EC isolates in the stool samples of healthy individuals in the community (Bezabih et al., 2021). Furthermore, CP-EC isolates are not restricted to human communities or healthcare settings and are increasingly being detected in food-producing animals (Bonardi and Pitino, 2019). CTX-M-producing E. coli was the predominant multidrug-resistant E. coli isolates in food-producing animals (Michael et al., 2017), with ESBL-producing E. coli isolates from pigs in China mainly producing CTX-M (Tian et al., 2009). In addition, CTX-M-14, CTX-M-55, and CTX-M-65 were the most prevalent genotypes among CTX-M-producing E. coli isolates from food animals in China (Rao et al., 2014). These isolates may be transmitted to humans through food consumption or contact with animals and can cause serious infections that are difficult to treat. The overuse of antibiotics in food-producing animals has been identified as a major driver of antibiotic resistance, and the detection of CP-EC isolates in these animals further underscores the urgent need for better antibiotic stewardship practices.
We identified three CTX-M-producing isolates that carried multidrug resistance genes by IncI and IncX3. IncI plasmids have been found to facilitate the transmission of the blaCTX−M between poultry and humans (Kopotsa et al., 2019), highlighting their potential as vectors for expanded carbapenemase gene transmission. Furthermore, IncI plasmids have been shown to promote transmission between mothers and newborns, suggesting that they may increase the transmission of carbapenemase genes in neonatal infections (Hagb et al., 2020).
The clonal relationship analysis revealed that the spread of 13 NDM-producing E. coli was polyclonal, with common E. coli sequence types such as ST167 and ST405 carrying most blaNDM−5. However, rare E. coli sequence types, such as ST744, ST349, and ST1193, were found to carry blaNDM−1. Among the plasmids observed in CP-EC, IncX3 was the predominant plasmid type, accounting for 61.5% (8/13) of isolates. The remarkable stability and efficient horizontal transfer of the IncX3 plasmid can be attributed to its low fitness cost, which can enhance its potential to spread between bacterial populations (Ma et al., 2020). In transconjugants, the presence of the IncX3 plasmid resulted in significantly higher levels of carbapenem resistance and blaNDM−5, compared to transconjugants carrying both plasmids or only the IncFII plasmid (Yang et al., 2020). Notably, studies have demonstrated that the IncX3 plasmid can mediate the transfer of blaNDM−5genes among different Enterobacter species over a wide range of temperatures (Liu Z. et al., 2019). The IncX3 plasmid is not only the most prevalent vehicle for blaNDM−5 but also a critical platform for the evolution of blaNDM−5, which has led to the emergence of novel NDM variants (Wu et al., 2019). Our findings indicate that the presence of IncX3 plasmid is widespread among CP-EC, which could further limit medication options and exacerbate the burden on the healthcare system.
Our study had several limitations that should be acknowledged. First, while we were able to access the specimens for the CP-EC, we did not have access to information on the patient's diagnosis or antibiotic prescriptions. This lack of information may have impacted our ability to fully understand the epidemiology and clinical significance of CPE infections. Second, although we included hospitals from various regions throughout China, the sample size was limited, which may have restricted our ability to generalize our findings to the entire population of China. Therefore, future studies should focus on collecting more comprehensive clinical data and engage in longer-term surveillance to enable a more in-depth investigation of CPE prevalence and characteristics across China. By doing so, we can gain a more comprehensive understanding of this emerging public health threat and develop effective strategies to control its spread.
With the increasing use of carbapenem, CP-EC clinical isolates accounted for nearly half of the total CRECO clinical isolates. We need to pay more awareness to CP-EC in BSI because the isolates were transferable and pathogenicity, which increases the burden on patients and medical settings. Currently, ESBL-producing CP-EC isolates have been found in humans, animals, and the environment, leading to antibiotics' low selectivity. Based on the concept of One Health, we should avoid the outbreak of CP-EC through various methods, including developing bacterial vaccines, originating antibiotic management, and establishing a better medical environment.
The datasets presented in this article are not readily available because data is precious scientific research and therefore not publicly available. Requests to access the datasets should be directed to bWljaGVsbGVrbzg2ODhAcXEuY29t.
YLi and YLv conceived the study. WK, ST, CC, and TL conducted all experiments. WK and CC analyzed the WGS data and drafted subsequent versions. TL, RL, and YZ interpreted the findings. WK and ST wrote the first draft of the manuscript. All authors acquired the data, critically reviewed this report, and approved the final version. All authors have read and agreed to the published version of the manuscript.
This study was supported by the National Natural Science Foundation of China (81572033).
We thank YLi and YLv for the experimental technical guidance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bezabih, Y. M., Sabiiti, W., Alamneh, E., Bezabih, A., Peterson, G. M., Bezabhe, W. M., et al. (2021). The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J. Antimicrobial. Chemother. 76, 22–29. doi: 10.1093/jac/dkaa399
Bonardi, S., and Pitino, R. (2019). Carbapenemase-producing bacteria in food-producing animals, wildlife and environment: A challenge for human health. Italian J. Food Safety. 8, 7956. doi: 10.4081/ijfs.2019.7956
Christie, P. J., Whitaker, N., and González-Rivera, C. (2014). Mechanism and structure of the bacterial type IV secretion systems. Bioch. Biophy. Acta Molec. Cell Res. 1843, 1578–1591. doi: 10.1016/j.bbamcr.2013.12.019
Conlan, S., Thomas, P. J., Deming, C., Park, M., Lau, A. F., Dekker, J. P., et al. (2014). Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci. Transl. Med. 6, 254ra.126. doi: 10.1126/scitranslmed.3009845
da Cruz Campos, A. C., Couto, N., da Silva Andrade, N. L., Friedrich, A. W., de Paula Rosa, A. C., Damasco, P. V., et al. (2020). Virulence and resistance properties of E. coli isolated from urine samples of hospitalized patients in Rio de Janeiro, Brazil–The role of mobile genetic elements. Int. J. Med. Microbiol. 310, 151453. doi: 10.1016/j.ijmm.2020.151453
De Angelis, G., Del Giacomo, P., Posteraro, B., Sanguinetti, M., and Tumbarello, M. (2020). Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int. J. Mol. Sci. 21, 5090. doi: 10.3390/ijms21145090
Deng, J., Liao, Q., Zhang, W., Wu, S., Liu, Y., Xiao, Y., et al. (2022). Molecular epidemiology characteristics and detecting transmission of carbapenemase-producing enterobacterales in southwestern China. J. Infect. Public Health. 15, 1047–1052. doi: 10.1016/j.jiph.2022.08.010
Foley, S. L., Kaldhone, P. R., Ricke, S. C., and Han, J. (2021). Incompatibility Group I1 (IncI1) plasmids: their genetics, biology, and public health relevance. Microbiol. Molec. Biol. Rev. 85, e00031–e00020. doi: 10.1128/MMBR.00031-20
Hagb,ø, M., Ravi, A., Angell, I. L., Sunde, M., Ludvigsen, J., Diep, D. B., et al. (2020). Experimental support for multidrug resistance transfer potential in the preterm infant gut microbiota. Pediatr. Res. 88, 57–65. doi: 10.1038/s41390-019-0491-8
Kopotsa, K., Osei Sekyere, J., and Mbelle, N. M. (2019). Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann. N Y. Acad. Sci. 1457, 61–91. doi: 10.1111/nyas.14223
Li, X., Gui, R., Li, J., Huang, R., Shang, Y., Zhao, Q., et al. (2021). Novel multifunctional silver nanocomposite serves as a resistance-reversal agent to synergistically combat carbapenem-resistant acinetobacter baumannii. ACS Appl. Mater. Interfaces. 13, 30434–30457. doi: 10.1021/acsami.1c10309
Liu, G., Olsen, J. E., and Thomsen, L. E. (2019). Identification of genes essential for antibiotic-induced up-regulation of plasmid-transfer-genes in cephalosporin resistant Escherichia coli. Front. Microbiol. 10, 2203. doi: 10.3389/fmicb.2019.02203
Liu, X., Li, X., Yang, A., Tang, B., Jian, Z., Zhong, Y., et al. (2022). Community fecal carriage and molecular epidemiology of extended-spectrum β-lactamase-and carbapenemase-producing Escherichia coli from healthy children in the Central South China. Infect. Drug Resist. 15, 1601–1611. doi: 10.2147/IDR.S357090
Liu, Z., Xiao, X., Li, Y., Liu, Y., Li, R., Wang, Z., et al. (2019). Emergence of IncX3 plasmid-harboring bla NDM−5 dominated by Escherichia coli ST48 in a goose farm in Jiangsu, China. Front. Microbiol. 10, 2002. doi: 10.3389/fmicb.2019.02002
Ma, T., Fu, J., Xie, N., Ma, S., Lei, L., Zhai, W., et al. (2020). Fitness cost of bla NDM-5-carrying p3R-IncX3 plasmids in wild-type NDM-free Enterobacteriaceae. Microorganisms. 8, 377. doi: 10.3390/microorganisms8030377
Michael, G. B., Kaspar, H., Siqueira, A. K., de Freitas Costa, E., Corbellini, L. G., Kadlec, K., et al. (2017). Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates collected from diseased food-producing animals in the GERM-Vet monitoring program 2008–2014. Vet. Microbiol. 200, 142–150. doi: 10.1016/j.vetmic.2016.08.023
Rao, L., Lv, L., Zeng, Z., Chen, S., He, D., Chen, X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003–2012. Vet. Microbiol. 172, 534–541. doi: 10.1016/j.vetmic.2014.06.013
Rodriguez-Acevedo, A., Lee, X., Elliott, T., and Gordon, L. (2020). Hospitalization costs for patients colonized with carbapenemase-producing Enterobacterales during an Australian outbreak. J. Hospital Infect. 105, 146–153. doi: 10.1016/j.jhin.2020.03.009
Tao, X., Wang, H., Min, C., Yu, T., Luo, Y., Li, J., et al. (2020). A retrospective study on Escherichia coli bacteremia in immunocompromised patients: microbiological features, clinical characteristics, and risk factors for shock and death. J. Clin. Lab. Anal. 34, e23319. doi: 10.1002/jcla.23319
Tian, G. B., Wang, H. N., Zou, L. K., Tang, J. N., Zhao, Y. W., Ye, M. Y., et al. (2009). Detection of CTX-M-15, CTX-M-22, and SHV-2 extended-spectrum β-lactamases (ESBLs) in Escherichia coli fecal-sample isolates from pig farms in China. Foodborne Pathog. Dis. 6, 297–304. doi: 10.1089/fpd.2008.0164
Tian, L., Sun, Z., and Zhang, Z. (2018). Antimicrobial resistance of pathogens causing nosocomial bloodstream infection in Hubei Province, China, from 2014 to 2016: a multicenter retrospective study. BMC Public Health. 18, 1–8. doi: 10.1186/s12889-018-6013-5
Tran, E. N. H., Doyle, M. T., and Morona, R. (2013). LPS unmasking of Shigella flexneri reveals preferential localisation of tagged outer membrane protease IcsP to septa and new poles. PLoS ONE. 8, e70508. doi: 10.1371/journal.pone.0070508
Villegas, M. V., Pallares, C. J., Escandón-Vargas, K., Hernández-Gómez, C., Correa, A., Álvarez, C., et al. (2016). Characterization and clinical impact of bloodstream infection caused by carbapenemase-producing Enterobacteriaceae in seven Latin American countries. PLoS ONE. 11, e0154092. doi: 10.1371/journal.pone.0154092
Wang, J., Lv, Y., Yang, W., Zhao, P., and Yin, C. (2022). Epidemiology and clinical characteristics of infection/colonization due to carbapenemase-producing Enterobacterales in neonatal patients. BMC Microbiol. 22, 1–14. doi: 10.1186/s12866-022-02585-z
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., Zong, Z., et al. (2019). metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, e00115–e00118. doi: 10.1128/CMR.00115-18
Yang, L., Lin, Y., Lu, L., Xue, M., Ma, H., Guo, X., et al. (2020). Coexistence of two bla NDM−5 genes carried on IncX3 and IncFII plasmids in an Escherichia coli isolate revealed by Illumina and Nanopore sequencing. Front. Microbiol. 11, 195. doi: 10.3389/fmicb.2020.00195
Keywords: Escherichia coli, carbapenemase, molecular epidemiology, genetic characteristics, blaNDM gene
Citation: Ko W, Tseng S, Chou C, Li T, Li R, Zhang Y, Li Y and Lv Y (2023) Molecular epidemiology and comparative genomics of carbapenemase-producing Escherichia coli isolates from 19 tertiary hospitals in China from 2019 to 2020. Front. Microbiol. 14:1056399. doi: 10.3389/fmicb.2023.1056399
Received: 28 September 2022; Accepted: 23 March 2023;
Published: 21 April 2023.
Edited by:
Zhi Ruan, Zhejiang University, ChinaReviewed by:
Prasanth Manohar, Texas A&M University, United StatesCopyright © 2023 Ko, Tseng, Chou, Li, Li, Zhang, Li and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Li, bGl5dW4wMzYwMkBwa3VmaC5jb20=; Yuan Lv, bHZ5dWFuMDkwMUBzaW5hLmNvbQ==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.