
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 February 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1045289
This article is part of the Research Topic MALDI-TOF MS in Microbiological Diagnostics: Future Applications Beyond Identification View all 12 articles
 Gianluca Foglietta1†
Gianluca Foglietta1† Elena De Carolis2*†
Elena De Carolis2*† Giordana Mattana1
Giordana Mattana1 Manuela Onori1
Manuela Onori1 Marilena Agosta1
Marilena Agosta1 Claudia Niccolai3
Claudia Niccolai3 Vincenzo Di Pilato4
Vincenzo Di Pilato4 Gian Maria Rossolini3,5
Gian Maria Rossolini3,5 Maurizio Sanguinetti2
Maurizio Sanguinetti2 Carlo Federico Perno1
Carlo Federico Perno1 Paola Bernaschi1
Paola Bernaschi1Due to the global spread of pan resistant organisms, colistin is actually considered as one of the last resort antibiotics against MDR and XDR bacterial infections. The emergence of colistin resistant strains has been observed worldwide in Gram-negative bacteria, such as Enterobacteriaceae and especially in K. pneumoniae, in association with increased morbidity and mortality. This landscape implies the exploration of novel assays able to target colistin resistant strains rapidly.
In this study, we developed and evaluated a new MALDI-TOF MS assay in positive-ion mode that allows quantitative or qualitative discrimination between colistin susceptible (18) or resistant (32) K. pneumoniae strains in 3 h by using the “Autof MS 1000” mass spectrometer. The proposed assay, if integrated in the diagnostic workflow, may be of help for the antimicrobial stewardship and the control of the spread of K. pneumoniae colistin resistant isolates in hospital settings.
In the last decades, the global spread of carbapenemase-producing Enterobacterales (CPE), primarily Klebsiella pneumoniae producing KPC-type carbapenemase (KPC-Kp), posed urgent threats on public health (Tzouvelekis et al., 2012; Murray et al., 2022), accounting for difficult-to-treat infections associated with high mortality rates (Cassini et al., 2019).
Owing to the significant burden of disease and limited treatment options, Centers for Disease Control and Prevention (CDC) (2019) and World Health Organization (WHO) (2017) ranked CPE as ‘critical-priority’ pathogens to which address the development of novel antimicrobial compounds.
Although several new antimicrobial agents active against CPE have been recently approved and marketed, including the novel β-lactam/β-lactamase inhibitor combinations, older molecules as colistin still hold a place in the antibiotic armamentarium as salvage therapy for patients infected with multi-drug resistant (MDR) or extensively drug-resistant (XDR) organisms (Jean et al., 2019).
Colistin is a positive charged polypeptide antibiotic, belonging to the polymyxin class, which targets the lipopolysaccharide (LPS) moiety on the outer membrane gram negatives, inducing a displacement of cations by electrostatic interaction and thus causing the disruption and loss of cell membrane integrity. To counteract these effects, bacteria have evolved multiple adaptative strategies including chromosomal mutations in the genes associated with the modification pathways of the lipid A, use of efflux pumps or capsule and the horizontal transfer of the plasmid-carried gene mcr-1 (Granata and Petrosillo, 2017; Wang et al., 2018; Hamel et al., 2021).
To date, the main mechanism of resistance is the modification of the lipid A.
In K. pneumoniae, alterations involving the mgrB gene, along with mutations in pmrAB and phoPQ loci, have been reported as the most common mechanisms of colistin resistance accounting for modification of LPS (Pragasam et al., 2017; Giordano et al., 2018), by addition of phosphoethanolamine (PEtN) and 4-amino-4-deoxy-L-arabinose (L-Ara4N) residues to the phosphate groups of lipid A (Cannatelli et al., 2014; Poirel et al., 2015), while horizontal acquisition of mcr-like genes was observed less frequently.
Nowadays, broth microdilution (BMD) is considered as the reference method for antimicrobial susceptibility testing (AST) of colistin by Clinical and laboratory standard institute (CLSI) and European committee on antimicrobial susceptibility testing (EUCAST) join subcommittee (2016). However, these phenotypic methods do not match with the need of a timely detection of colistin resistance for patient’s isolates as they imply turnaround times of 16–24 h. On the other hand, PCR-based molecular methods, although rapid, only provide information on the presence/absence of the genes involved in the resistance mechanisms, which not always correlates to the isolate’s phenotype.
Very recently, a novel MALDI-TOF based method (i.e., the MALDIxin test) able to detect colistin resistance in about 15–30 min, thanks to a shift of the mass unit of the native lipid A present in the resistant bacterial strains, has been developed and validated for E. coli, A. baumannii (Dortet et al., 2018) and K. pneumoniae (Dortet et al., 2020).
Anyway, these assays require switching the MALDI-TOF MS machine to the negative ion mode, not the modality routinely used for bacterial identification. Besides the availability of such a technology (actually still rare in the majority of the microbiology labs), the assay needs pre and post switching additional calibrations.
Here, we aimed to develop and validate the “CORE” assay, a new MALDI-TOF-based test in positive ion mode for rapid prediction of colistin resistance in K. pneumoniae, relying on the detection of a mass spectrum profile with an identification score lower or ≥ 6 by using the Autof MS 1000 mass spectrometer (Autobio). The assay was evaluated both as a method for MIC prediction and as a screening tool for colistin resistance (quantitative and qualitative AST respectively) by comparison with BMD AST results.
The study collection included 50 colistin-resistant (n = 32) and-susceptible (n = 18) K. pneumoniae isolates, cultured from blood (n = 33), urine (n = 4), rectal swabs (n = 8), tracheal broncho-aspirates (n = 3), cerebrospinal fluid (n = 1) and wound swab (n = 1). The isolates, collected within the 2016–2021 period, were part of national surveys (n = 21; Di Pilato et al., 2021), of previously published studies (n = 9; Cannatelli et al., 2014; Arena et al., 2016; Boncompagni et al., 2022) or of a local collection available at the University of Florence (Florence, Italy) (n = 14).
Within the study collection, 32 out of 50 K. pneumoniae isolates were previously characterized by whole-genome sequencing (WGS), and genetic alterations associated with colistin resistance were formerly investigated (Cannatelli et al., 2014; Di Pilato et al., 2021; Boncompagni et al., 2022). The remaining colistin resistant isolates (n = 18) were screened for the presence of the most common mcr gene variants by Real-time PCR, including mcr-1 and mcr-2, and additional mcr genes using specific primer/probes combinations (Coppi et al., 2018; Yang et al., 2018) (Table 1).
Colistin susceptibility testing of the clinical isolates for the CORE assay was performed using the BMD method by Liofilchem™ ComASP™ Colistin Test Panel (Liofilchem, Roseto degli Abruzzi, Te, Italy) containing the dried up antibiotic in 27-fold dilutions (0.25–16 mg/L) following the manufacturer instructions. MIC results were interpreted according to EUCAST Clinical Breakpoints (v12.0, 2022).
In order to obtain a fast and accurate qualitative and quantitative AST for colistin of the 50 K. pneumoniae strains included in the study, CORE assay and colistin BMD susceptibility testing were performed in parallel including three biological and two technical replicates for each tested isolate. To optimize the CORE assay inoculum concentration, the time of the test incubation and to develop a classifying algorithm, preliminary experiments were performed on 10 well-characterized K. pneumoniae, 5 colistin resistant (colR) and 5 colistin susceptible (colS) strains (reported in bold in Table 2).
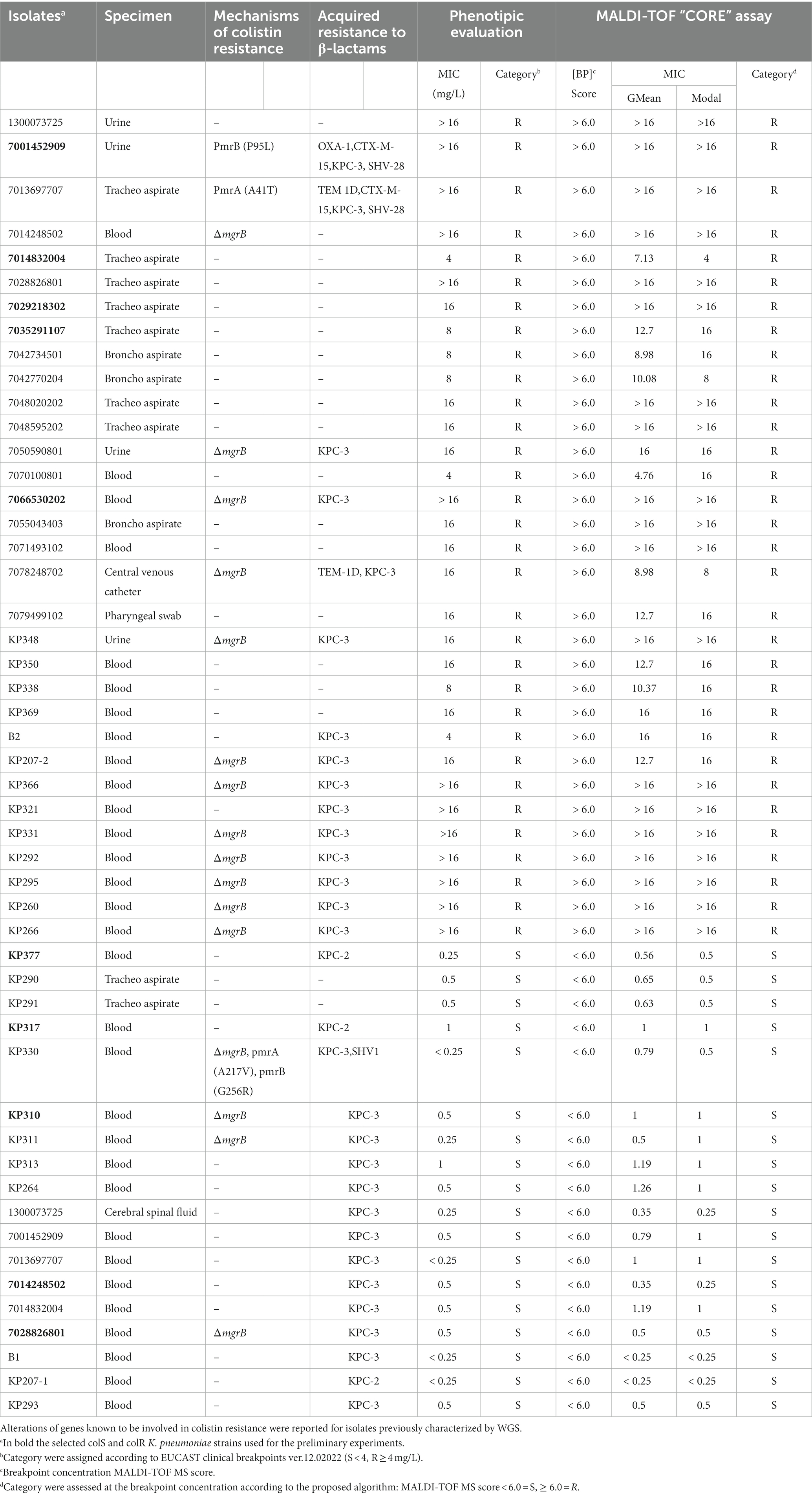
Table 2. Characteristics and results of the MALDI-TOF CORE assay on Klebsiella pneumoniae isolates included in the study (n = 38).
The CORE assay protocol was performed according to the steps reported in Figure 1.
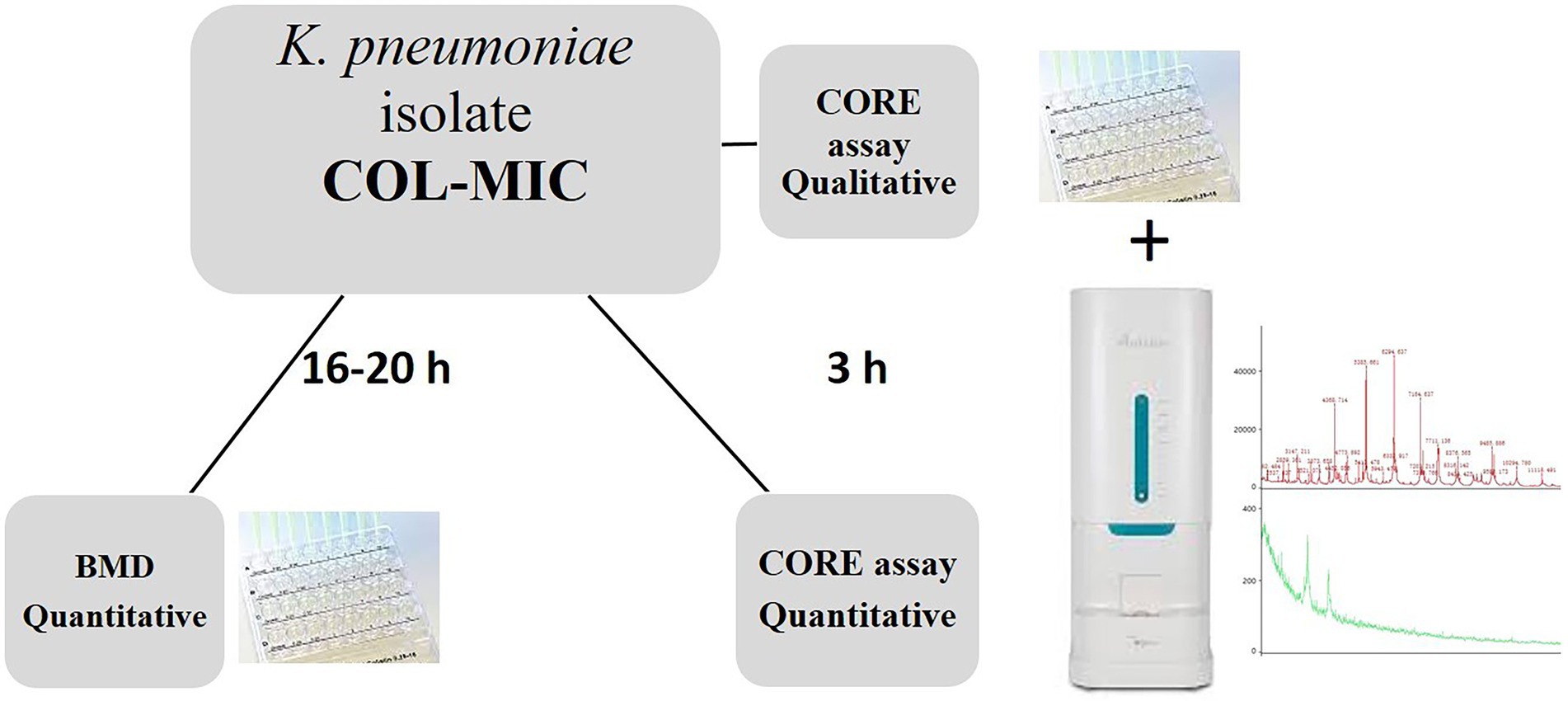
Figure 1. Laboratory workflow illustrating the CORE assay for the rapid detection of qualitative and quantitative colistin resistance in K. pneumoniae isolates.
Briefly, all the K. pneumoniae strains were sub-cultured on MacConkey agar plates (bioMérieux) at 37° C for 24 h; a 0.5 McF suspension was made from grown colonies. Except for the inoculum size, ComASP (Compact Antimicrobial Susceptibility Panel) Colistin test was performed following the standard procedures.
After 3 h of incubation at 37°C, an aliquot of 1 μL of the bacterial suspension from each well was spotted in duplicate on the MALDI-TOF target plate (96 wells steel target slide, Autobio Diagnostics) by direct deposition with the addition of 1 μL of Autobio sample pretreatment reagent lysate 1 (formic acid). After drying, the spot was overlaid with 1 μL of Cyano 4 Hydroxycinnamic Acid CHCA AUTOF MS matrix (Autobio Diagnostics, CO., LTD, China) solubilized in lysate 2 (acetonitrile) and buffer (trifluoroacetic acid) as recommended.
The spectra acquisition was performed on an Autof 1,000 MS (Autobio Diagnostics, Zhengzhou, China) mass spectrometer in positive linear mode, at a laser frequency of 60 Hz, in the mass range 2–20 kDa, using the “microbe” automatic acquisition mode with an overall 240 laser-shot acquired by 40 shot steps for each spot. The mass spectrometer was calibrated with Autobio calibrating agent consisting of nine calibrating proteins, according to manufacturer’s instructions.
The protein spectra were analyzed using the Autof Acquirer version 1.0.55 software and the library v2.0.61 was used for the peaks matching. The identification results were interpreted according to the manufacturer criteria as following: Identification scores ≥9 were considered positive at the species level, scores between 6 and 9 as positive at the genus level, and scores <6 were defined as unreliable (no identification).
For the qualitative assay, only the spectra acquired by the plate well without the colistin agent and those acquired by the 2 mg/L plate well (ComASP® Colistin, Liofilchem, Te, Italy) were used as growth control and test breakpoint concentration (BP) respectively. An algorithm was developed to provide a rapid and accurate detection as colistin susceptible (colS) or resistant (colR) for the 50 K. pneumoniae strains. In particular, a test sample was classified as colR or colS on the following parameters: growth control spectra score ≥ 6 and 2 mg/L spectra score > 6 or 2 mg/L spectra score < 6, respectively.
On the contrary, for the quantitative assay all the spectra acquired in the range 0.25–16 mg/L were included in the analysis along with the growth control spectra. The MIC value was determined as the lowest drug dilution at which the spectra score was <6. The geometric mean (G MEAN) and the modal MIC calculated from the replicates was reported for all the samples (Table 2). In the case the 16 mg/L spectra score was >6, a MIC value >16 mg/L was reported and the test sample classified as colR. On the contrary, in the case the 0.25 mg/L spectra score was <6, a MIC value <0.25 mg/L was reported and the sample categorized as colS.
Thus, each strain was classified according to the above mentioned algorithms for the qualitative or quantitative CORE assay.
The results were compared with those obtained by the conventional BMD test, following interpretation with the EUCAST Clinical Breakpoints (v12.0, 2022) for colistin.
According to the modal MIC value, a MIC value agreement within ±1 dilution against BMD (essential agreement, EA) was considered acceptable (Clinical laboratory testing and in vitro diagnostic test systems ISO 20776-2:2021) for the new assay evaluation.
Divergence degree in distribution between BMD and CORE assay MIC values, potentially leading to resistance state misclassification, has been statistically evaluated with a non parametric Wilcoxon Test (R statistics stats library) performed in a paired manner and with a value of p continuity correction.
The preliminary validation of the CORE assay carried out on 10 selected well-characterized strains of K. pneumoniae (i.e., including five colistine susceptible and five resistant strains, reported in bold in the Table 2), allow to correctly classifying them as colistin susceptible or resistant by the proposed algorithm.
The spectra of a colistin resistant and a colistin susceptible K. pneumoniae selected strains acquired by the Autof MS 1000 mass spectrometer using the CORE assay protocol are shown in Figure 2. The correspondent results obtained by the automatic matching of the acquired mass spectra against the Autobio library v2.0.61 for the growth control (no drug), BP and maximum concentration of colistin are reported at each profile side.
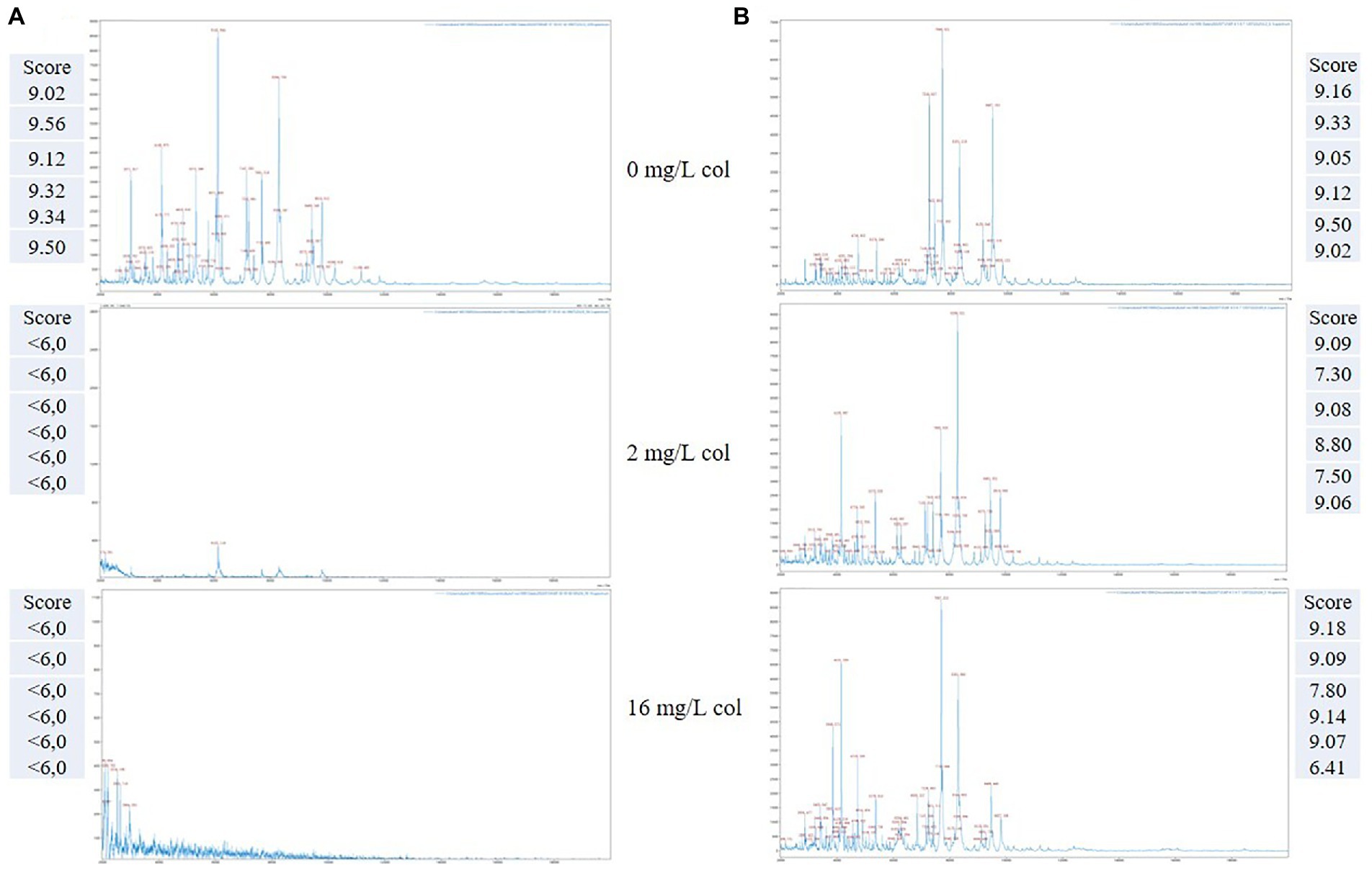
Figure 2. Representative MALDI-TOF mass spectra of two K. pneumoniae organisms detected as colistin susceptible (A) and colistin resistant (B), respectively, by the CORE assay. Score value of the replicates spectra acquired at the selected drug concentrations are reported at the respective image side.
As exemplified, in the case of a susceptible K. pneumoniae isolate (Figure 2A), a matching score result <6.0 is obtained for all the tested replicates, both at the breakpoint (2 mg/L) and at the maximum concentration (16 mg/L), which takes into account the reduction or absence of the mass peaks in the respective MALDI-TOF MS profiles, in comparison with the growth control.
Conversely, for a resistant K. pneumoniae isolate (Figure 2B), the matching score results are above 6.0 both for the BP and maximum concentration, thus indicating the presence of a colistin resistant organism associated with the persistence of the mass spectra profiles at the given concentrations.
Thus, the CORE assay was applied on the 50 clinical isolates of K. pneumoniae included in the study. Genomic data revealed that multiple alterations in genes known to be involved in colistin resistance (i.e., mgrB, pmrB) were present in sequenced isolates (n = 32), regardless of the colistin resistance phenotype (Table 2), while no acquired mcr-like genes were detected in the whole isolate collection.
The CORE assay results for the qualitative or quantitative assay were compared with those obtained by the BMD quantitative test; the overall results are shown in Table 2. As reported all K. pneumoniae, 32 colR and 18 colS isolates, were correctly classified in 3 h as resistant or susceptible by the CORE assay, respectively. For what concerns the quantitative CORE assay, 30 out of 32 colR K. pneumoniae agreed against BMD MIC values within ±1 dilution according to the modal MIC results, whilst 7070100801 and B2 isolates obtained a MIC value 2 dilution higher (16 vs. 4 mg/L) using the quantitative CORE assay. Overall, an EA of 93.7% (30/32 K. pneumoniae resistant isolates) was reported following the 3 h incubation of the quantitative CORE assay.
Regarding the 18 colS K. pneumoniae, an EA of 83.3% was calculated; in particular, 15 out of 18 isolates resulted concordant against the BMD assay within ±1 dilution. However, KP338 MIC value was <0.25 mg/L instead of 0.5, KP377 obtained a MIC value 2 dilution higher and KP310 MIC value was <0.25 mg/L instead of 1 by comparison of quantitative CORE assay and BMD MIC results, respectively. Interestingly, all colistin susceptible isolates carrying genetic alterations previously associated with colistin resistance (i.e., ΔmgrB, pmrAA217V, pmrBG256R) were classified as colS by the “CORE” assay, a result consistent with the reference BMD (Table 2), suggesting that colistin susceptibility could be more accurately predicted by the MALDI-TOF based approach than genetic data in these isolates.
The distribution divergence between BMD MIC values and CORE assay ones, quantitatively tested by mean of a paired Wilcoxon Test, estimated it as statistically significant since resulting value of p was 4.108e-05. This result indicates an overall agreement between mentioned approaches while considering MIC divergence degree.
In summary, following a three-hour samples incubation, by a simple algorithm for MALDI-TOF MS analysis (MALDI-TOF MS score < 6.0 = S or ≥ 6.0 = R), we obtained a total agreement between the qualitative CORE assay and the phenotypic method results. Moreover, an EA of 93.7 and 83.3% was achieved in the case of the quantitative CORE assay for the colR and colS isolates, respectively.
The MALDI-TOF MS based CORE assay in positive-ion mode that allows in 3 h of incubation the detection of colistin resistant or colistin susceptible K. pneumoniae isolates can provide rapid results to clinicians without the need to wait the 16–24 h necessary for the conventional BMD assays. Furthermore, the possibility to obtain colistin resistance detection using MALDI-TOF spectrometry instrument at the same polarity, the positive one, without switching to the negative ion mode and thus avoiding the calibration steps, makes the proposed algorithm suitable for a combination with the current routine MALDI-TOF MS identification workflow.
The simple algorithm here proposed for the CORE assay avoids statistical analysis based on the absence or presence of specific mass peaks and is independent from the mechanism of resistance, it relies on the growth of the microorganism and thus on the MALDI-TOF identification score matching value. Moreover, the test, can be suitable also for laboratories that cannot afford the costs of a new spectrometer equipped with the positive–negative ion mode switching modality as requested by the assays based on colistin resistance-related modifications to lipid A, thus consisting in a novelty in the landscape of polymixin resistance detection assays based on mass spectrometry.
One limitation of the qualitative CORE assay is that the time to result is higher with respect to the MALDI-TOF tests based on lipidomics. On the contrary, the quantitative CORE assay has the advantage to provide good results (EA of 93.7 and 83.3% for colR or colS K. pneumoniae isolates) earlier than conventional BMD methods (3 vs. 16–24 h).
Overall, the CORE assay, although studies are still needed to implement the number of samples tested, might be of extreme importance in the detection of colistin resistant isolates. The emergence of colistin resistant K. pneumoniae isolates can have a huge impact on the patient outcome representing a pressing health-care problem from both a management and economic point of view. The use of rapid and cost-effective new technologies applied to the resistance landscape, might offer the possibility to overcoming the non-appropriate use of colistin and at the same time can be of help in the struggle against the spread of antibiotic-resistance.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
EDC: conceived and planned the experiments. GF, GM, MO, and CN: carried out the experiments. MA: contributed to sample preparation. EDC, GF, VDP, and GM: contributed to the interpretation of the results. EDC: wrote the manuscript. VDP and CFP: provided critical feedback. GMR, MS CFP, and PB: supervised the project. All authors contributed to the article and approved the submitted version.
This research was supported by EU funding within the MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT).
We wish to thank Valentino Costabile for his assistance in the statistical data analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AL declared a past co-authorship with the authors GMR, VDP to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arena, F., Henrici De Angelis, L., Cannatelli, A., Di Pilato, V., Amorese, M., D'Andrea, M. M., et al. (2016). Colistin resistance caused by inactivation of the MgrB regulator is not associated with decreased virulence of sequence type 258 KPC Carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 60, 2509–2512. doi: 10.1128/AAC.02981-15
Boncompagni, S. R., Micieli, M., Di Maggio, T., Aiezza, N., Antonelli, A., Giani, T., et al. (2022). Activity of fosfomycin/colistin combinations against planktonic and biofilm gram-negative pathogens. J. Antimicrob. Chemother. 77, 2199–2208. doi: 10.1093/jac/dkac142 Epub ahead of print
Cannatelli, A., Giani, T., D’Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/AAC.03110-14
Cassini, A., Högberg, L. D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G. S., et al. (2019). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4
Centers for Disease Control and Prevention (CDC). Antimicrobial resistance threats report (2019). Available from: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508. pdf (Accessed September 1, 2022)
Clinical and laboratory standard institute (CLSI) and European committee on antimicrobial susceptibility testing (EUCAST) join subcommittee. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group (2016).
Clinical laboratory testing and in vitro diagnostic test systems — Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices — part 2: evaluation of performance of antimicrobial susceptibility test devices against reference broth micro-dilution. (2021). ISO 20776-2.
Coppi, M., Cannatelli, A., Antonelli, A., Baccani, I., Di Pilato, V., Sennati, S., et al. (2018). A simple phenotypic method for screening of MCR-1-mediated colistin resistance. Clin. Microbiol. Infect. 24:11. doi: 10.1016/j.cmi.2017.08.011
Di Pilato, V., Errico, G., Monaco, M., Giani, T., Del Grosso, M., Antonelli, A., et al. (2021). AR-ISS Laboratory study group on carbapenemase-producing Klebsiella pneumoniae. The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J. Antimicrob. Chemother. 76, 355–361. doi: 10.1093/jac/dkaa431
Dortet, L., Brodam, A., Bernabeu, S., Glupczynski, Y., Bogaerts, P., Bonnin, R., et al. (2020). Optimization of the MALDIxin test for the rapid identification of colistin resistance in Klebsiella pneumoniae using MALDI-TOF MS. J. Antimicrob. Chemother. 75, 110–116. doi: 10.1093/jac/dkz405
Dortet, L., Potron, A., Bonnin, R. A., Plesiat, P., Naas, T., Filloux, A., et al. (2018). Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci. Rep. 8:16910. doi: 10.1038/s41598-018-35041-y
Giani, T., Antonelli, A., Caltagirone, M., Mauri, C., Nicchi, J., Arena, F., et al. (2013). Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October: KPC-carbapenemase spreading among outpatients. Euro Surveill. 22:30583. doi: 10.2807/1560-7917.ES.2017.22.31.30583
Giordano, C., Barnini, S., Tsioutis, C., Chlebowicz, M. A., Scoulica, E. V., Gikas, A., et al. (2018). Expansion of KPC-producing Klebsiella pneumoniae with various mgrB mutations giving rise to colistin resistance: the role of IS L3 on plasmids. Int. J. Antimicrob. Agents 51, 260–265. doi: 10.1016/j.ijantimicag.2017.10.011
Granata, G., and Petrosillo, N. (2017). Resistance to colistin in Klebsiella pneumoniae: a 4.0 strain? Infect Dis Rep 9:7104. doi: 10.4081/idr.2017.7104
Hamel, M., Rolain, J. M., and Baron, S. A. (2021). The history of Colistin resistance mechanisms in bacteria. Prog. Challeng. Microorgan. 9:442. doi: 10.3390/microorganisms9020442
Jean, S. S., Gould, I. M., Lee, W. S., and Hsueh, P. R. (2019). International Society of Antimicrobial Chemotherapy (ISAC). New drugs for multidrug-resistant gram-negative organisms: time for stewardship. Drugs 79, 705–714. doi: 10.1007/s40265-019-01112-1
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Poirel, L., Jayol, A., Bontron, S., Villegas, M. V., Ozdamar, M., Turkoglu, S., et al. (2015). The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Pragasam, A. K., Shankar, C., Veeraraghavan, B., Biswas, I., Nabarro, L. E., Inbanathan, F. Y., et al. (2017). Molecular mechanisms of Colistin resistance in Klebsiella pneumoniae causing bacteremia from India-a first report. Front. Microbiol. 7:2135. doi: 10.3389/fmicb.2016.02135
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8 in NDM-producing Klebsiella pneumoniae. Emerg. Microb. Infect. 7:122. doi: 10.1038/s41426-018-0124-z
World Health Organization (WHO). Global Priority List of Antibiotic-resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics (2017). Geneva: World Health Organization (WHO).
Keywords: colistin resistance detection, Klebsiella pneumoniae, positive-ion mode, MALDI-TOF MS, rapid assay
Citation: Foglietta G, De Carolis E, Mattana G, Onori M, Agosta M, Niccolai C, Di Pilato V, Rossolini GM, Sanguinetti M, Perno CF and Bernaschi P (2023) “CORE” a new assay for rapid identification of Klebsiella pneumoniae COlistin REsistant strains by MALDI-TOF MS in positive-ion mode. Front. Microbiol. 14:1045289. doi: 10.3389/fmicb.2023.1045289
Received: 15 September 2022; Accepted: 01 February 2023;
Published: 22 February 2023.
Edited by:
Karsten Becker, University Medicine Greifswald, GermanyCopyright © 2023 Foglietta, De Carolis, Mattana, Onori, Agosta, Niccolai, Di Pilato, Rossolini, Sanguinetti, Perno and Bernaschi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena De Carolis, ZWxlbmEuZGVjYXJvbGlzQHBvbGljbGluaWNvZ2VtZWxsaS5pdA==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.