
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 20 September 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.999988
Plant growth-promoting bacteria (PGPB) are endowed with several attributes that can be beneficial for host plants. They opened myriad doors toward green technology approach to reduce the use of chemical inputs, improve soil fertility, and promote plants’ health. However, many of these PGPB harbor antibiotic resistance genes (ARGs). Less attention has been given to multi-resistant bacterial bioinoculants which may transfer their ARGs to native soil microbial communities and other environmental reservoirs including animals, waters, and humans. Therefore, large-scale inoculation of crops by ARGs-harboring bacteria could worsen the evolution and dissemination of antibiotic resistance and aggravate the negative impacts on such ecosystem and ultimately public health. Their introduction into the soil could serve as ARGs invasion which may inter into the food chain. In this review, we underscore the antibiotic resistance of plant-associated bacteria, criticize the lack of consideration for this phenomenon in the screening and application processes, and provide some recommendations as well as a regulation framework relating to the development of bacteria-based biofertilizers to aid maximizing their value and applications in crop improvement while reducing the risks of ARGs invasion.
In the last few years, there has been growing interest in using microbial-based products as bioinoculants or/and substances. However, their use is facing many challenges from the lab to the field (Madawala, 2021). Plant growth-promoting bacteria (PGPB) are a polyphyletic group of bacteria that interact closely or loosely with plant roots in the rhizosphere biotope. These bacteria have direct or indirect positive effects on plant growth and health. According to Klopper (1980), PGPB are distinguished by their ability to colonize the roots, survive, multiply, and become competitive with other microorganisms (Klopper, 1980). They can increase the availability of nutrients, regulate the production of phytohormones, increase tolerance to abiotic stresses and inhibit pests and diseases by competition (Glick, 2012; Souza et al., 2015).
The intensive and continuous use of chemical fertilizers and pesticides in the last 50 years has substantially influenced public health and agricultural productivity in the world (Ramakrishna et al., 2019). However, these practices have resulted in multiple environmental concerns mainly soil fertility degradation, reduced biodiversity in soils, increased greenhouse gas emission, zinc deficiency, chemical residues accumulation, and eutrophication among others (Tilman et al., 2002; Diaz and Rosenberg, 2008). Over the last few decades, more eco-friendly approaches such as the use of PGPB as biofertilizers have gained popularity in moving toward a sustainable agroecosystem (Ramakrishna et al., 2019). On the economic perspective, biofertilizers market shows a growth of more than 12% increase per annum (Calvo et al., 2014; Owen et al., 2015). The largest market represented by Nitrogen-based biofertilizers was estimated to grow by up to 13.25% per year by 2020 (Arora, 2018). To date, many PGPB strains belonging to the genera of Acinetobacter, Agrobacterium, Azospirillum, Bacillus, Bradyrhizobium, Burkholderia, Enterobacter, Gluconacetobacter, Pantoae, Pseudomonas, Rhodococcus, and Serratia have been reported to have beneficial effects on plant growth promotion, nutrition, stress alleviation, bioremediation, and/or biocontrol of plant pathogen’s attacks (Sharma et al., 2013; Bargaz et al., 2018). Large-scale production and commercialization of bacterial-based biofertilizers have been achieved using some PGPB belonging to the genera of Azospirillum, Azotobacter, Bacillus, Burkholderia, Pseudomonas and Rhizobium (Parray et al., 2016). Despite the obvious agronomic advantages that biofertilization offers, some earlier studies have raised some concerns that could lead to negative effects of their application on the behavior and genetic profile of soil microbial communities and have sounded the alarm about their intensive and unreasonable long-term application (Lee et al., 2018). For instance, the proximity and the high microbial density (indigenous or introduced) in the rhizosphere were reported to enhance the horizontal (or lateral) gene transfer (HGT) among bacterial species. HGT is known to be one of the major mechanisms in bacterial evolution (Popa and Dagan, 2011). These transferable genetic elements could harbor ARGs and virulence factors that could pose human health risks. In this context, this review focuses on the paradox behind the use of antibiotic-resistant PGPB, addresses potential risks for public health, and provides a regulation framework with practical solutions and recommendations as well as the approaches of isolation and development of effective and biosafe PGPB-based biofertilizers.
Data analyzed in this review were collected using the Scopus research engine. The search was launched by entering the keywords “PGPB,” “Endophyte” and refining with additional keywords “antibiotic,” “resistance,” and “antibiotic resistance genes.” A total of 398 hits were retained for further analyses in the scope of this review. Therefore, we used the co-occurrence mapping tool of relevant keywords which resulted in more than 50 high frequency keywords that were used to construct a knowledge map of the main strong domains of research studies related to PGPB and resistance to antibiotics. Noteworthy, when the terms “antibiotic AND resistance” were absent, records increased to 21,199 hits. Figure 1 shows the networks inferred using pertinent keywords and co-occurrence analysis in the dataset. Based on the relevant keywords, assigned clusters were clearly identified by the following keywords: “rhizosphere,” “rhizobacteria,” “sustainable agriculture” and “biocontrol” (Figure 1A). These four clusters are strongly connected to each other. As shown in Figure 1B, the main recent topic in the field of biological activities of biofertilizers is “antibiotic resistance” in which the number of publications has exploded since 2020 (Circled in red). This result shows that there is a need for more investigation into the extent of antibiotic resistance in PGPB. This feature is most likely due to the search by scientists for more persistent and competitive bioinoculants/biofertilizers but very rarely to their awareness about the danger that could be posed by PGPB species with a broad spectrum of antibiotic resistance.
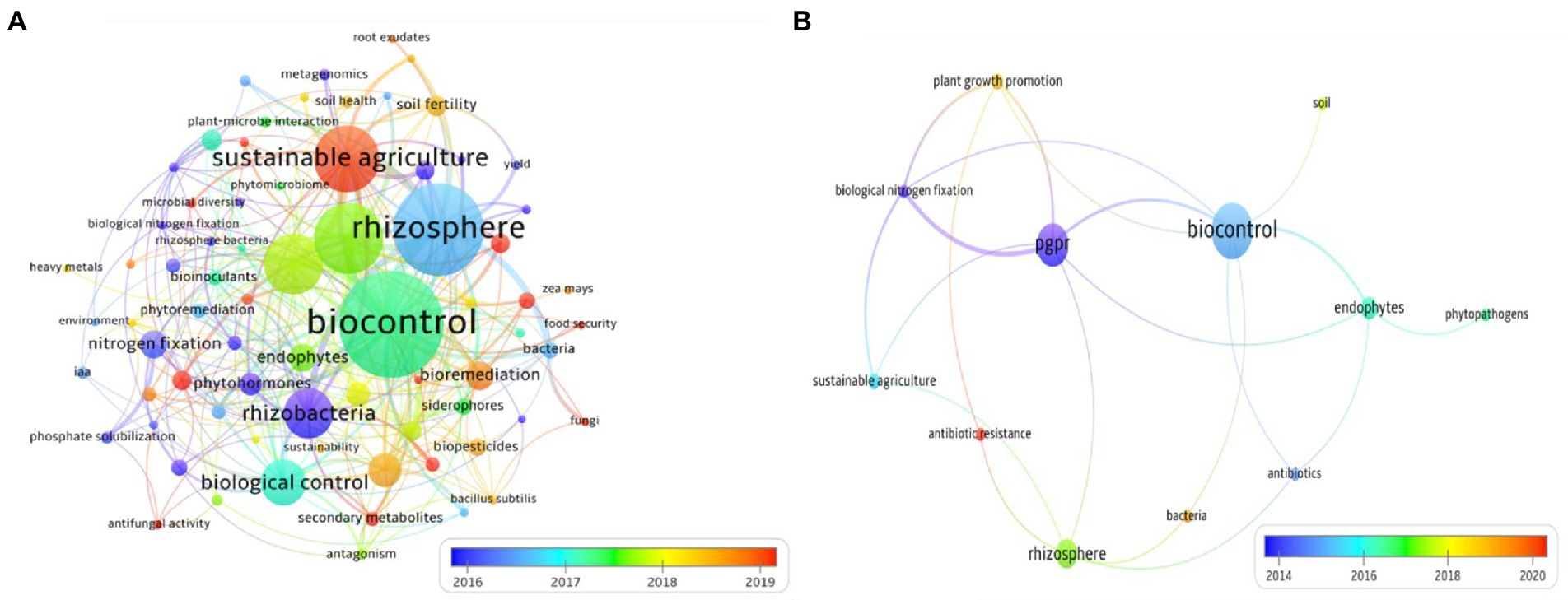
Figure 1. (A) Co-word map network visualization and (B) Knowledge map of antibiotic resistance in bacterial-based biofertilizers. The larger the circles are, the more scientific publications were found. PGPR, plant growth promoting rhizobacteria.
While many soil bacteria are beneficial to plants, some are well known plant pathogens that cause severe damage and can lead to the complete losses of crop yields (Syed Ab Rahman et al., 2018). Such bacterial plant pathogens are able overcome the innate plant defense by different molecular mechanisms involving microbial effectors (Toruño et al., 2016). Another group of bacteria in the rhizosphere comprises opportunistic human pathogenic bacteria (HPB) which have a ubiquitous habitats including plant biotopes (Figure 2).
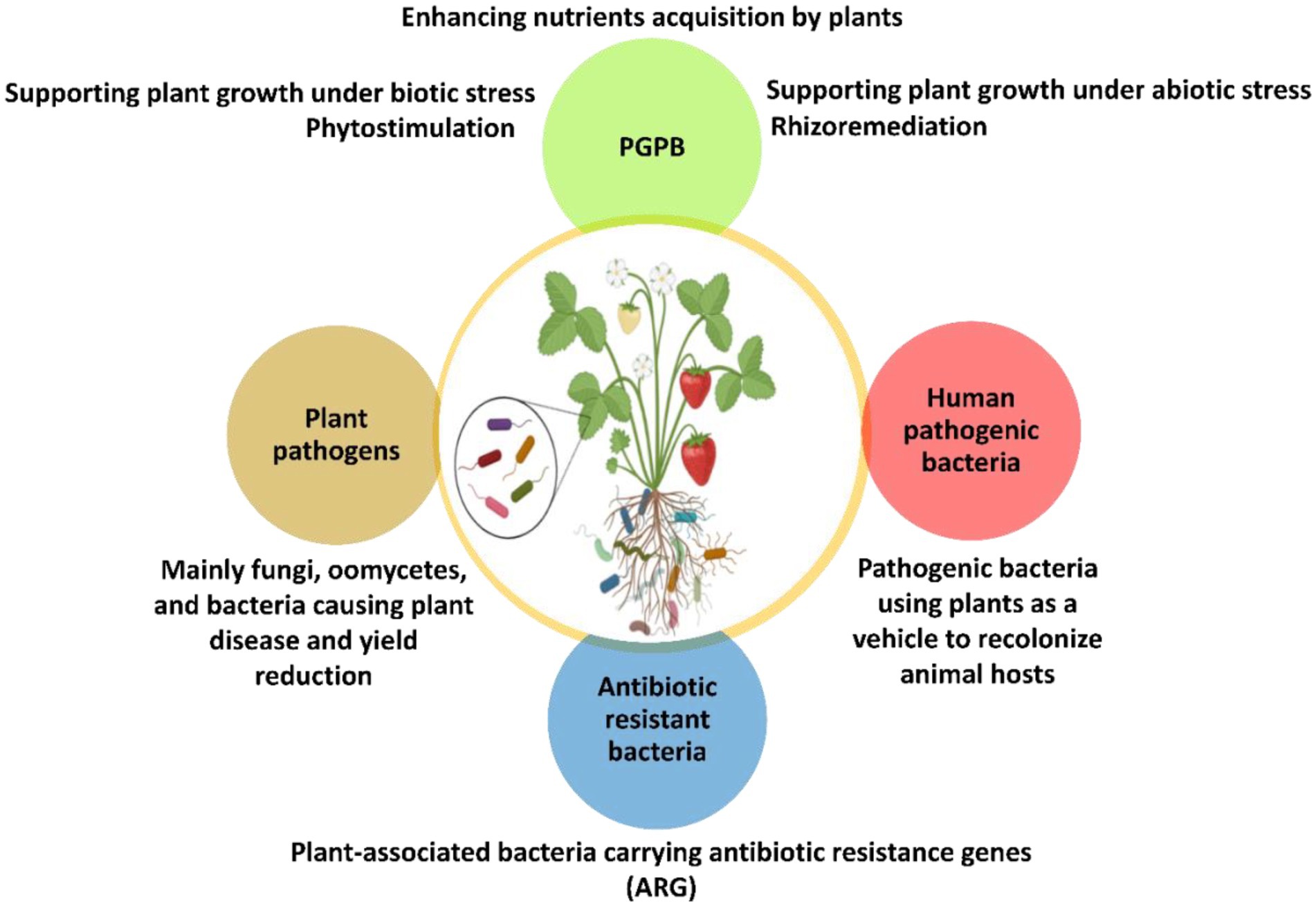
Figure 2. Schematic overview of plant-associated microorganisms and their respective functions. They live inside the plant tissues as endophytes or surrounding the root system as rhizosphere microorganisms. PGPB, Plant growth promoting bacteria.
It is well established that bacteria can naturally integrate exogenous DNA including ARGs (Boto et al., 2019). Among the mechanisms by which bacteria acquire new functions are conjugation (transfer of mobile genetic elements by pili), transformation (uptake of DNA by competent bacteria), and transduction (transfer of DNA between a bacteriophage and a bacterium; Thomas and Nielsen, 2005; Arber, 2014). In addition, in some species, other mechanisms have been documented to help bacteria encapsulating DNA such as phage-derived sequences, and transfer mediated by vesicles produced in the outer membrane vesicles (García-Aljaro et al., 2017; Sun, 2018). Furthermore, many bacteria possess resistance elements located within the antibiotic biosynthetic cluster for self-protection and survival strategy (Hubbard and Walsh, 2003). This consolidates the fact that the intrinsic resistance is an ancient phenomenon that precedes the anthropogenic use of antibiotics, contradicting the common assumption that antibiotic resistance is uniquely related to antibiotic misuse (Cox and Wright, 2013). although, this does not exclude the impact of human activity in the resistance phenomenon and its dissemination. In fact, HGT can be promoted by anthropogenic activities by (i) favoring ecological forms of life such as biofilms where gene transfer can occur, (ii) inducing selective pressures that maintain transferred genes, and (iii) affecting HGT mechanisms through compounds such as legacy pollutants. Consequently, the potential threat of antibiotic resistance and HGT in affected environments such as plant–soil microcosm is worth inspecting (Andam et al., 2015; Gillings, 2017b).
Bacterial inoculants applied in large amounts for biofertilization, biocontrol, phytostimulation, or bioremediation of soils could increase HGT events with indigenous prokaryotes living in the soil and/or plant environments (Séveno et al., 2002). Several studies have addressed the emergence and propagation of ARGs in different environmental reservoirs through anthropogenic activities such as the application of manure, slurry and soil amendments with regard to their different transfer pathways and threats to human being health and ecology (Wellington et al., 2013).
In fact, the plant microbiome represents an interface between humans, microbes, and genes including those involved in the antibiotic resistance. Even nonculturable bacteria in soils remain metabolically active and retain virulence factors capable of causing infections in plants, animals and humans (Del Mar Lleò et al., 2007; Fakruddin et al., 2013). In suitable conditions, these bacteria could cause severe infections and diseases after ingestion as they are able to recover their metabolic activities (Baffone et al., 2003; Rowan, 2004; Epstein, 2009). Moreover, nonculturable bacteria are considered as a reservoir of pathogens in the environment. They include many human pathogenic bacteria such as Enterobacter cloacae, Pseudomonas aeruginosa, and Shigella strains (Fakruddin et al., 2013). Consequently, plant microbiome could indirectly influence the composition and function of the human microbiome resulting to potential human health issues (Chen et al., 2019). An analogy can be drawn with plant–soil microbiome feedback loop. In turn, humans could contribute directly or indirectly to the invasion of ARGs in plant microbiomes. However, PGPB harboring ARGs and bacterial biological control agents are mostly unexplored, while most of these PGPB are still recommended as effective biofertilizers (Supplementary Figure S1). Despite the importance of antibiotic resistance that benefits PGPB to survive within the indigenous microbiota in antibiotic contaminated areas (Cray et al., 2013), large-scale use of ARGs-harboring biofertilizers in the soil could spread antibiotic resistance in other ecosystems (Figures 2, 3). This concern about emerging antibiotic resistant bacteria is a double-edged sword in both sustainable agriculture and public health associated risks.
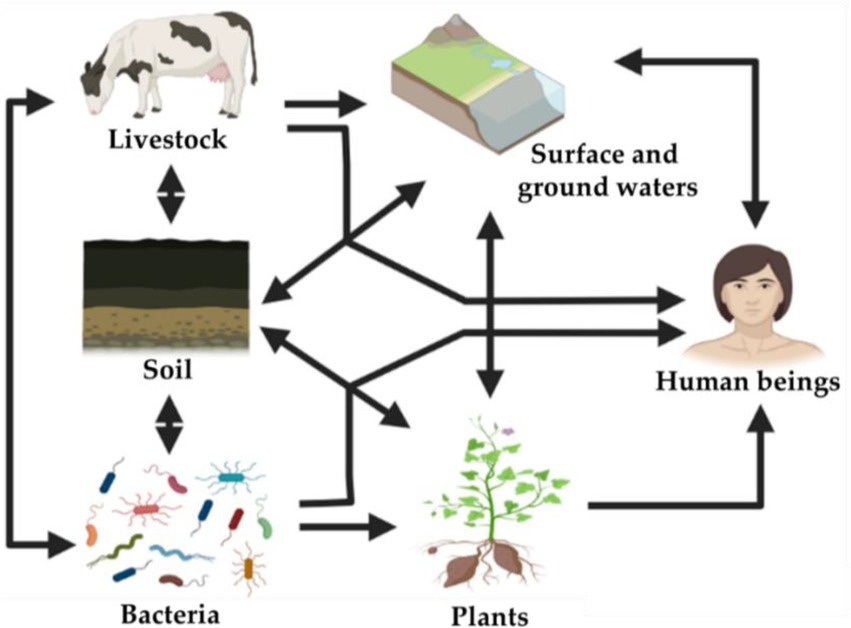
Figure 3. Schematic illustration of the associations between some key environmental settings of antibiotic resistant-bacteria and antibiotic resistance genes (ARGs). Acquisition of existing ARGs from the environmental microbiome occur mostly by horizontal gene transfer (HGT). Resistance may also occur in soil bacteria via mutations encoding target-site alterations.
Many mechanisms by which beneficial bacteria enhance nutrients acquisition by plants, are well documented in the literature (Vejan et al., 2016). The most common mechanism is changes of soil pH through bacterial organic acids secretion and proton release via stimulation of ATPase proton pump, which decreases soil pH and increases the concentration of soluble nutrients such as phosphorus and potassium (Mantelin and Touraine, 2004). Another example is the modulation of phytohormone biosynthesis through the production of various extracellular molecules (i.e., siderophores) and enzymes such as aminocyclopropane-1-carboxylate deaminase, which decrease ethylene levels in plants, thus facilitating plant growth and development and mitigating abiotic stress (i.e., salinity; Nadeem et al., 2007; Zahir et al., 2008). Other mechanisms involved in the alleviation of abiotic stresses in plants have include the production of plant hormones (auxins, gibberellins, and cytokinins), exopolysaccharides, osmoprotectors, and antioxidant enzymes (Egamberdieva et al., 2019).
Nitrogen fixation is mediated by soil bacteria which can form nodules in the roots, allowing to produce ammonia which contributes the plant nutrition (Murray, 2011). Furthermore, some PGPB protect plants against attacks of pests and pathogens (Glick, 2012; Ahemad and Kibret, 2014). This biocontrol activities are due to their ability to biosynthesize antimicrobial metabolites, such as hydrogen cyanide (HNC), phenazines, pyoluteorin, 2,4-diacetylphloroglucinol, tensin, viscosinamide, and pyrrolnitrin, etc.; to compete against the pathogens for nutrients; and to stimulate plant immune system (Plattner and Soldati-Favre, 2008; Planchamp et al., 2015). In addition, the priming effect of PGPB through volatiles organic compounds 2,3-butanediol, has emerged recently to account for biocontrol effects (Farag et al., 2013) against some fungal pathogens (Supplementary Figure S1).
To become a successful PGPB-based inoculant, the candidate bacterial strains should demonstrate not only plant growth promotion traits, but also the ability for cost-effective mass production and formulation with a long shelf life (Nakkeeran et al., 2005; Tabassum et al., 2017). Although several PGPB have been reported to be carrying ARGs (Table 1), the extent of this threat and the potential for resistance transmission to soil microorganisms and ultimately its impact on human-associated bacteria is mostly underexplored. Antibiotics exert natural selection pressure on bacterial populations which often lead to increase the abundance of resistant bacteria in the environment. This major public health problem is strongly linked to the misuse and overconsumption of antibiotics in human and animal health. Consequently, the upsurge in multi-resistant bacterial strains resulted in substantially decreased of treatments success, thus causing increased bacterial infections potentially leading to mortality (Hawkey and Jones, 2009; Paul et al., 2010).
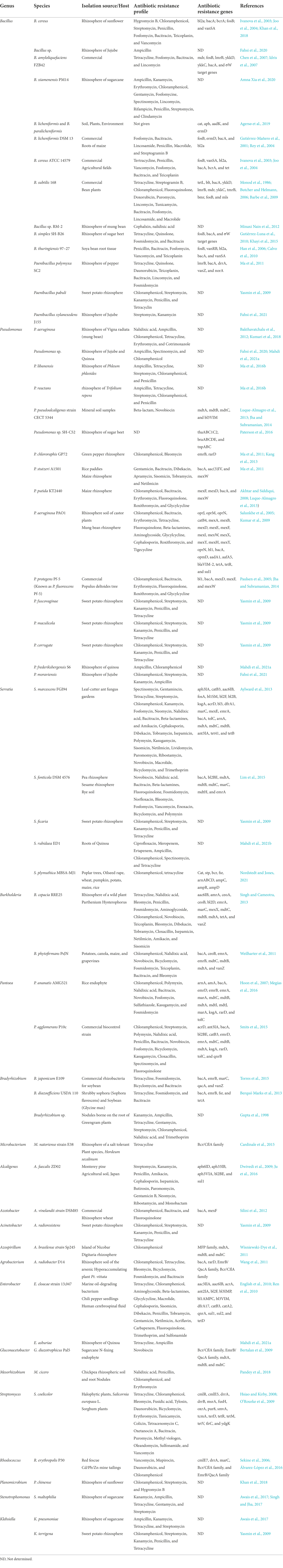
Table 1. A list of antibiotic resistance patterns of some declared PGPB strains and respective/putative identified ARGs.
Humans are exposed to resistant bacteria, antibiotics, or ARGs by several routes including contaminated crops, livestock, surface, and groundwater (Figure 3). These ARGs can disperse into the indigenous bacteria that colonize agricultural soils via spontaneous mutation or horizontal gene transfer. They can then be transmitted into edible plants, ingested by animals or humans, and eventually acquired by clinical pathogenic bacteria (Chen Q. et al., 2017). In 2010, Knapp et al. (2010) highlighted the significant accumulation of ARGs in agricultural soils sampled since 1940, reflecting the increasing use and application of antibiotics over the last decades, especially some tetracycline-coding ARGs which increased 15 times comparatively to the concentrations found in the 1970s (Knapp et al., 2010). This contamination might induce measurable impacts on the composition of microbial soil communities (Riber et al., 2014).
Among the challenges to overcome for the commercial use of PGPB as biofertilizers, are their unstable characteristics, guarantee their survival and reinforce their competitiveness in soils in order to maintain a critical bacterial mass within the native microbiota (Cray et al., 2013). In addition, PGPB strains exhibiting intrinsic antibiotic resistance can serve as markers to assess the viability of inoculated bacteria in pot and field experiments (Kluepfel, 1993; Trivedi et al., 2004). Thus, isolation and screening of resistant PGPB are highly desired in order to ensure the wide adaptation of introduced bacteria to diverse habitats (Singh and Cameotra, 2013; Wani and Khan, 2013; Ma et al., 2016a). However, these explanations completely neglect the risk of biosafety related to the dissemination of multi-resistant PGP bacteria. For instance, cabbage inoculation with Pseudomonas libanensis and Pseudomonas reactans improved trace metal phytoremediation, therefore they were proposed as bioinoculants suitable to improve the phytoremediation of soils contaminated by metals in semiarid areas. These two strains were remarkably resistant to ampicillin, tetracycline, streptomycin, chloramphenicol, and penicillin (Ma et al., 2016b). Indeed, based on sequencing data, the vast majority of screened PGPB exhibit resistance to at least one antibiotic and/or harbor ARGs (Wellington et al., 2013; Kang et al., 2017). This may lead to immediate changes in the structure of bacterial community and eventually induce the dissemination of ARGs, if antibiotic pollutants are present in their environments. As shown in Figure 4, antibiotic-resistant bacteria can reach humans being by different routes mainly (i) consumption of plants/fruits that have been exposed to contaminated irrigation, manure, sludge, and slurry; (ii) using groundwater and surface water and residing animals (e.g., fish) containing resistant microbiota, and (iii) livestock that have been treated with veterinary antibiotics and exposed to resistant bacteria by the food chain. Antibiotic-resistant bacteria introduced into the soil as biofertilizers can subsequently be transported to surface water or groundwater and/or transfer their ARGs to inhabiting bacteria and be cycled within the environment thus enriching environmental reservoirs of ARGs (Topp et al., 2008; Figure 3). For instance, antibiotic-resistant bacteria colonizing plant tissues as endophytes, might transmit antibiotic resistance to consumers (Scaccia et al., 2021).
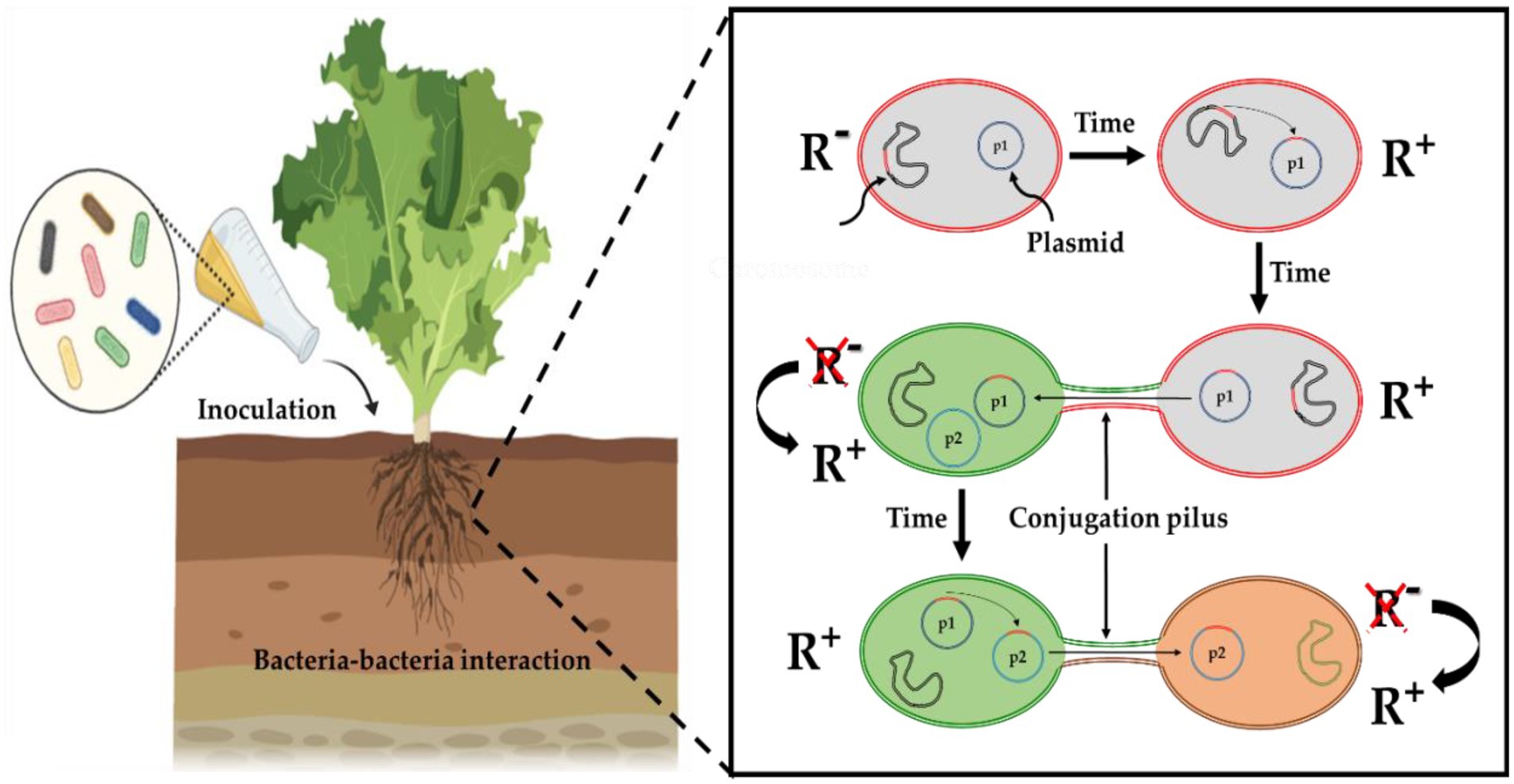
Figure 4. Extrinsic antibiotic resistance genes (ARGs) dissemination mechanisms upon PGPB application. ARGs (in red) appear in the chromosome and move by transposition to the plasmid (p1) within the same bacterium. Plasmids (p1) allow the transfer of ARGs between strains while plasmids (p2) allow transfer to distantly related strains. R− indicates sensitivity and R+ indicates resistance.
It has been pointed that by 2050 global annual mortality is projected to reach 10 million because of resistance to antimicrobials. Antibiotics are released into the environment via many pathways mainly clinical settings, human, and animal wastes, and through antibiotics supplemented food and fodder (Keswani et al., 2019).
As antibiotics have been widely used in agriculture as growth promoters in livestock, a large fraction of them are released into the environment creating a selective pressure for ARG dispersal in agricultural soils among others (Hung, 2020). Noteworthy, antibiotic resistance is evident in environments subjected to anthropogenic activities (Chen et al., 2013). This challenges the common point of view that ARGs solely occurs because of the contamination by antibiotics. In fact, it has recently been observed that other substances can also select for antibiotic resistance. This is becoming more commonly recognized, and regulators are beginning to monitor pathways and controlling the release of resistant-driving chemicals mainly antimicrobials, metals, and biocides (Singer et al., 2016). For example, metals and some persistent organic compounds (POCs) co-select for ARGs. More importantly, a study in Cuba revealed the presence of a high frequency of occurrence of ARGs along the Almendares River/estuary despite the minimal use of antibiotics in the country. However, high amounts of pollutants including metals, alongside other contaminants, were detected (Reid-Henry, 2008; Graham et al., 2011). Similarly, the abundance of ARGs was higher in soils amended with biosolids than in the surrounding agricultural soils without biosolids treatment. It was concluded that the application of biosolids also enhanced the emergence and spread of ARGs in the environment (Hung, 2020).
The co-selection of antibiotic resistance and metal or other pollutants can occur either by co-resistance or by cross-resistance. Co-resistance occurs when a phenotype is simultaneously selected with other genes located on the same genetic element. However, cross-resistance is developed when two pollutants (e.g., antibiotic and metal) have similar routes into the bacterial cell. In this case, when a resistance response is triggered, cell defense is effective against both metal and antibiotic contaminants. Under such contamination, selective pressures are likely to persist longer than pharmaceutical compounds because of the non-degradability of metals in the environment (Rodgers et al., 2019).
Furthermore, the presence of polycyclic aromatic hydrocarbons (PAHs) also induces shifts in the indigenous bacterial communities by increasing hydrocarbon-degrading bacteria (Zhang et al., 2010; de Menezes et al., 2012). Interestingly, many PAH-tolerant bacteria presented strong resistance to both metals and antibiotics (Máthé et al., 2012). In another study, several ARGs were found in PAH contaminated matrices (Chen B. et al., 2017). In fact, PAHs are pollutants (Liu et al., 2017) that can induce ARGs acquisition either by DNA transformation, or by triggering stress/repair systems. Although specific mechanisms are still unclear, a metagenomic profiling showed that PAH-contaminated soils harbor ARGs with approximately 15 times more abundance compared to those with low concentrations of contaminants (Chen B. et al., 2017). Finally, metals and PAHs are now established to exacerbate the emergence and dispersal of ARG in the environment and they can be used as model pollutants for further public-health risks related to these genetic pollutants. Nevertheless, ARGs co-selected by PAHs could be completely distinguished from those that have arisen from the use of antibiotics (Chen B. et al., 2017). Furthermore, soil analysis has also highlighted the impact of the non-clinical uses of antibiotics on ARGs spread as well as the impact of policy towards a more strict of their management in natural environments (Graham et al., 2016).
Soil microbial communities were found to harbor cassettes of ARGs serving as a reservoir of genes potentially exchangeable with human bacterial pathogens (Forsberg et al., 2012). Additionally, the majority of soil-inhabiting bacteria that produce antibiotics [e.g., 2,4-diacetylphloroglucinol (DAPG), lipopeptides (LP), and phenazines] carry genes for the successful adaptation and self-defense against stressful conditions mostly located on the same gene clusters that confer resistance to other antibiotics (Tariq et al., 2022).
Noteworthy, the number of ARGs in plant associated-bacteria was found to be correlated with relatively higher levels of N-fertilizers (Forsberg et al., 2014). These ARGs are mostly carried by plasmids, transposable elements (Figure 4), and integrons (Supplementary Figure S2; Heuer et al., 2011; Gillings, 2017a). The antibiotic resistance of a bacterium can be natural (intrinsic). This type of resistance concerns all strains of a genus or a species and determines the wild-type resistance phenotype. Resistance can be carried by the chromosome and transmitted vertically to the progeny by cell division. Finally, antibiotic resistance can be acquired (extrinsic) by a greater or lesser proportion of the strains of a species and is variable over time. In this case, transmission is vertical or horizontal and occurs between different bacteria from different phyla via mobile genetic elements so-called the mobilome (Figure 4; Wellington et al., 2013; Ramakrishna et al., 2019). HGT occurs via transformation (integration of DNA in the environment), conjugation (sharing of plasmids between bacteria species), and transduction (bacteriophage mediated gene transfer; Vikesland et al., 2017). The concept of genetic exchange communities has been introduced by Jain et al. (2003) as a set of microorganisms that can exchange genes by HGT, without requiring physical proximity (Jain et al., 2003). Bacteria living within genetic exchange communities are susceptible to acquire ARGs by distinct bacterial species inhabiting separate environmental niches (Amarasiri et al., 2020).
In the 1980s, new genetic elements capable of acquiring or losing ARGs were described and named integrons (Stokes and Hall, 1989). Integrons are defined as genetic systems for capturing and expressing genes in the form of cassettes capable of being laterally transferred or excised by a site-specific recombination mechanism (Stokes et al., 2006). These cassettes are transcribed from a promoter (Pc) in the 5′- region and the expression of distal genes is reduced by the presence of upstream cassettes (Supplementary Figure S2). This system can incorporate open reading frames and convert them into functional genes expression systems. Interestingly, most gene cassettes encode ARGs and strains harboring integrons have been found in different environments. In 2001, Stokes et al. demonstrated the presence of several cassettes in samples from plant biomass, soil, sediments and water (Stokes et al., 2001). These integrons have a major role in the dissemination of antibiotic resistance in bacteria because they allow a very large number of combinations and therefore constitute an additional advantage for bacteria in the acquisition of ARGs (Ploy et al., 2005). The dispersal of ARGs in bacterial communities could also occur by lytic and temperate bacteriophages (Sharma et al., 2014).
The origin of acquired antibiotic resistance by PGPB could be the exposure of soils to anthropogenic activity or the production of antibiotics by antibiotics-producing bacteria inhabiting the soil.
Class 1 integrons were detected in crop associated bacteria mainly the β-proteobacterial class. This arguably creates a major conduit for the frequent influx of new mobile genes into bacteria closely associated with animals and humans at a scale whose magnitude remains to be explored (Stokes et al., 2006).
Many ARGs are responsible for intrinsic resistance to different antibiotics, including aminoglycosides, β-lactams, and fluoroquinolones. Intrinsic resistance in bacteria appeals mainly to the changes in cell wall permeability and activation of efflux pumps. Intrinsic resistance is a natural phenomenon adopted by many environmental bacteria especially Gram-negative ones (Ramakrishna et al., 2019). Moreover, some bacteria harbor genes called “cryptic genes” which are ARGs with very low expression levels. However, their expression could increase under binding conditions. This has been described in Acinetobacter (Zhang and Feng, 2016) and could be present in PGPB given their multiple environmental constraints.
The main modes of action of ARGs are: Inhibition of uptake, modification of target, inactivation of the antibiotic, and activation of efflux pump (Supplementary Figure S3; Ramakrishna et al., 2019). For instance, Pseudomonas aeruginosa often isolated from agricultural soil samples and characterized as a PGPB (Table 1), is a multi-drug resistant opportunistic pathogen harboring chromosomal and plasmid ARGs. Antibiotic resistance modes within this bacterium involve upregulation of β-lactamases and efflux pumps, alteration in membrane composition and chemical alteration of antibiotics (aminoglycosides; Lister et al., 2009; Poole, 2011). The antibiotic can be degraded or modified chemically by phosphorylation, nucleotidylation, acetylation, ADP-ribosylation, mono-oxygenation, or glycosylation (Vasala et al., 2020).
The high frequency of resistant bacteria is due to the high plasticity of the bacterial genome. Chromosomal mutations may occur or more frequently the acquisition of mobile genetic elements carrying ARGs found at high frequency in the bacterial cell. The acquisition of several ARGs by a bacterium results in resistance to many classes of antibiotics, usually called multidrug resistance (Wellington et al., 2013).
Many PGPB and biocontrol bacterial inoculants, especially Pseudomonas and Bacillus strains (Steil et al., 2003; Santoyo et al., 2012), are resistant to more than one antibiotic and/or harbor at least one ARGs (Wellington et al., 2013; Table 1). For instance, Fahsi et al. (2020) isolated 13 phosphate solubilizing bacteria (PSB), and checked their antibiotic resistance against kanamycin, streptomycin, tetracycline, ampicillin, chloramphenicol, and spectinomycin. Out of 13 PSB, 11 were able to resist, at least, one antibiotic (Fahsi et al., 2020). Similarly, Serratia rubidaea ED1 strain, an opportunistic pathogenic bacterium, isolated from quinoa roots endosphere was found resistant to seven antibiotics (Mahdi et al., 2021b). In addition, out of 11 PSB isolated from quinoa rhizosphere, five strains showed resistance to at least two antibiotics (Mahdi et al., 2021a). More recently, antibiotic resistance mechanisms were studied in biofertilizer samples following application into the soils, these include mainly antibiotic deactivation (48.7%), efflux pumping (25.2%), cellular protection (18.0%), and other unknown mechanisms (8.1%; Yang et al., 2022). In North America, streptomycin sulfate has been used to control heavy outbreaks of cultures. However, because of the risk of enhancing the rise and dispersal of antibiotic resistance in nontarget bacteria, European authorities have banned its use in Europe. Since then, ecofriendly alternatives have been eagerly searched for, and two biocontrol agents based on Bacillus subtilis were registered for blight control in Europe: Serenade® based on strain QST713 and Biopro® based on strain BD170 (Broggini et al., 2005). However, this species is susceptible to harbor antibiotic resistance genes in their genetic material (Table 1).
Based on a phylogenetic analysis, it is probable that the mobile elements hosting ARGs originates mainly from Firmicutes and Bacteroidetes (McHardy et al., 2007; Sommer et al., 2010). Hence, the introduction of these PGPB into soils can supply the ARGs reservoirs as donors and worsen the spreading risks of these genes in the soil microflora. The large-scale use of bacterial biofertilizers is not without consequences as they could serve as acceptors for ARGs in the soil and move more easily to plants with pili. Ultimately, the resistance of PGPB to antibiotics becomes a blatant paradox in biofertilization (Kang et al., 2017).
Many studies have focused on the detection and quantitation of ARGs in agricultural soils, uncovering resistance mechanisms, and identifying novel enzymes responsible for the resistance of bacteria to antibiotics. As for those inhabiting soils, up to 177 ARGs conferring resistance to aminoglycosides, chloramphenicol, and β-lactams were reported in pristine Antarctic soils. It has been suggested that the vast majority of ARGs most likely represent functional historical genes encoding for resistance to natural antibiotics (Van Goethem et al., 2018).
Antibiotic resistance gene diversity and abundance are strongly affected by agricultural practices (Wepking et al., 2017). However, soil pH, the organic content, and the history of agricultural soil management have been documented as important drivers influencing the abundance and the fate of ARGs (Čermák et al., 2008; Popowska et al., 2012).
In a metagenomic study performed on Chinese soils to which manure was applied, ARGs conferring resistance against tetracycline, minocycline, streptomycin, kanamycin, gentamycin, amikacin, rifampicin, and chloramphenicol accounted for about 70% of the total ARGs detected. More than 60% of identified genes presented low similarity (less than 60%) compared to the sequences of their closest proteins at the amino acid level (Su et al., 2014). Noteworthy, among the studied ARGs, the average abundance of tetO and ampC genes in the soil treated with manure was 3.3 and 421% higher comparatively to manure free soil, respectively (Wepking et al., 2017).
A broad-spectrum diversity of ARGs such as tetA, sul1, blaOXA2, blaCTX-M, and qnrS has been shown by Nõlvak et al., (2016) in soils subjected to treatment by animal manure. The rate of tetracycline resistance encoding genes namely tetM, tetO, and tetW, was significantly greater in manure-treated soils (Xiong et al., 2018). This was explained by an increasing number of ARGs resulted from antibiotic-resistant bacteria already present in manure. This led to a selective pressure of some antibiotic-resistant bacteria to dominate the microbial community.
As for biofertilizers application, a recent study assessed ARGs and mobile genetic elements (MGEs) harbored by biofertilizer and soil samples using high-throughput quantitative polymerization chain reaction (HT-qPCR). Fluctuation in specific ARGs [QnrB4, vanC2_vanC3, copA, sugE, mepA, tetD, and tetA(P)] and MGEs [IS26 (MGEs) and IS6100 (MGEs)] were noticed following biofertilizer application. However, the overall relative abundance of ARGs was no significant at the applied dose but the authors suggested that biofertilizers application should be evaluated over time as resistance transfer is time dependent (Yang et al., 2022). Similarly, using genomic analysis, Nordstedt and Jones (2021) identified genes putatively involved in antibiotic resistance in the PGPR Serratia plymuthica MBSA-MJ1 namely chloramphenicol acetyltransferase (cat) gene providing resistance to chloramphenicol and six copies of the efflux pump responsible for tetracycline resistance (stp; Nordstedt and Jones, 2021). Other identified genes include those encoding for resistance to fosmidomycin (fsr), bicyclomycin (bcr), polymyxin (arnABCD), and β-lactam multi-drug resistance (ampC, ampD, and ampR). In some soils, plasmids harbor bac gene conferring resistance to bacitracin in Bacilli strains. This gene is acquired by HGT and is mandatory for their survival in most soils (Ramakrishna et al., 2019). Nevertheless, the diversity and novelty of ARGs are likely to pose risks to humans as they can transfer the ARGs to human pathogenic bacteria (Su et al., 2014).
Horizontal gene transfer (HGT) is well documented as an important mechanism in bacterial evolution (Popa and Dagan, 2011). It provides rapid access to novel functions, allowing traits such as virulence, antibiotic resistance, and xenobiotic metabolism to spread through the human microbiome as well as plant-associated microbiome (Smillie et al., 2011). Yet, current HGT detection techniques [e.g., LatTrans (Hallett and Lagergren, 2001), RIATA-HGT (Than et al., 2008), or HGT-Detection (Boc et al., 2010)], are hardly usable for building networks having more than 100 leaves. To circumvent this problem, some reports implemented a fast distance-based method for the inference of HGT networks comprising thousands of species. Then, they developed an accurate probabilistic algorithm for detecting partial HGTs (Denamur et al., 2000). In a study involving a network of 10,770 unique genes found in 2,235 full bacterial genomes, it has been shown that HGT networks have been shaped principally by ecology rather than geography or phylogeny, with most gene exchange occurring between isolates from ecologically similar, but geographically separated environments (Smillie et al., 2011).
Many diagnostic methods are being used to detect ARGs from different environments. In this section, we provide an outline of some current and emerging tools for diagnostics and fast detection of ARGs (Kaprou et al., 2021; Table 2).
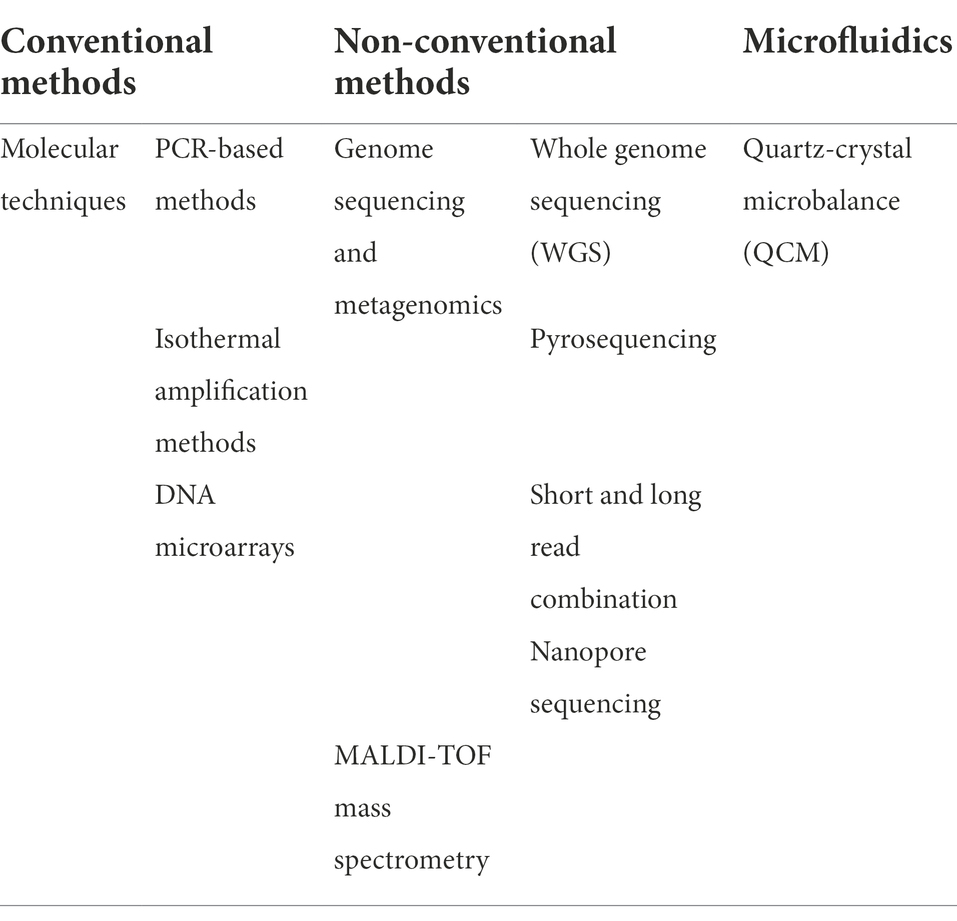
Table 2. Summarizing table of relevant diagnostic tools for detecting lateral gene transfers of ARGs.
The conventional methods relying on molecular-based technologies provide fast and sensitive detection of ARGs and have extended the availability of huge ARGs targets in various databases (Waseem et al., 2019). In the following sub-section, nucleic acid amplification-based techniques such as PCR, isothermal techniques, and DNA microarrays are discussed.
PCR is the most used method for the detection of ARGs (Chisholm et al., 2001; Seedy et al., 2017). Recently, quantitative (Böckelmann et al., 2009), real-time (Mackay, 2004), digital (Bogožalec Košir et al., 2020), and multiplex PCRs (Jousset et al., 2019) have been further incorporated for genetic assays. Moreover, technological advances made for the next-generation sequencing (NGS) and whole genome sequencing (WGS) have greatly influenced the ARGs targets and paved the way to the high throughput quantitative PCR (HT-qPCR). This is faster, suitable for the simultaneous exploration of many ARGs (Waseem et al., 2019), and applicable for ARGs originating from different sample types including soils (Waseem et al., 2020). For instance, using HT-qPCR, Wang et al. (2014) explored and profiled bacterial ARGs from urban park soils (Wang et al., 2014). Indeed, this technology has been successfully employed using various sample types such as water, animal feces, and soil (Xu et al., 2019).
The isothermal DNA amplification is advantageous over conventional PCRs because it does not require thermocycling. In addition, it is faster, more sensitive, and more specific (Zanoli and Spoto, 2013; Zou et al., 2020). This method includes several techniques, such as strand displacement amplification (SDA), nucleic acid sequence-based amplification (NASBA), transcription mediated amplification (TMA), rolling circle amplification (RCA), loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), and helicase-dependent amplification (HDA; Gill and Ghaemi, 2008). Recently, it was that LAMP and RPA have great potential for point-of-need diagnostics used in low resource settings (Zou et al., 2020). In vitro diagnostic products based on PCR and isothermal nucleic acid amplification technology (NAAT) are commercialized (Morin et al., 2018). The automation of each processes (from DNA extraction to gene detection) coupled with data analysis software tools have resulted in accurate integrated platforms (Cantera et al., 2019).
A DNA microarray allows the detection of genes in a target organism when compared to a reference genome or strain. Using fluorescently labeled PCR sequences (DNA chips), simultaneous detection of ARGs among Staphylococcus clinical isolates was achieved (Zhu et al., 2007). Recently, a rapid cartridge based, melting curve assay to detect pyrazinamide resistant Mycobacterium tuberculosis was proposed (Havlicek et al., 2019).
In this section, some of the most promising non-conventional methods for ARGs detection are described. They include sequencing, matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry (MALDI-TOF MS), and Fourier transform infrared (FTIR). spectroscopy.
Numerous methods, tools, and databases have been reported in recent years for the detection of ARGs from WGS and WMS data (Köser et al., 2014; Oniciuc et al., 2018). These evolving technologies provide rapid and sensitive determination of resistances in both uncultivable and cultivable bacteria (Kaprou et al., 2021).
The identification of ARGs using pyrosequencing was developed by Amoako et al. (2012). It served as a tool for the detection of clinical drug-resistant Mycobacterium tuberculosis. Reliable and robust detection of resistance-associated mutations in M. tuberculosis isolates was achieved with very high specificity (96–100%; Ajbani et al., 2014). This method was also efficient to rapidly detect resistances to kanamycin, fluoroquinolones, rifampicin, and capreomycin in M. tuberculosis clinical strains (Govindappa et al., 2016). This assay was considered as a fast and effective method for the detection of mutations associated with drug resistance in M. tuberculosis clinical isolates (Govindappa et al., 2016).
Several studies have highlighted that WGS is a powerful tool for ARGs surveillance (Alghoribi et al., 2018; Hendriksen et al., 2019; Argimón et al., 2020). Vélez et al. (2017) used WGS for the prediction and determination of ARGs occurrence in Streptococcus uberis and Streptococcus dysgalactiae strains isolated from dairy cows. This showed the association between several unique ARGs sequences and phenotypic resistances (Vélez et al., 2017). Similarly, Zhao et al. (2016) identified ARGs in Campylobacter and checked their correlation to phenotypes by employing in vitro antimicrobial susceptibility testing and WGS (Zhao et al., 2016). A strong positive correlation (99.2%) was seen between resistance patterns and genotypes. These findings indicate that WGS is a reliable resistance indicator.
As mentioned above, plasmids can spread ARGs among bacteria. However, their reconstruction from short-read WGS data is difficult. Short and long read WGS sequencing was used to characterize and locate ARGs situated on plasmids (Berbers et al., 2020). In fact, it is of crucial importance to establish ARGs location, especially when they are in mobile elements. Biosafety assessment of ARGs transfer was feasible by overcoming the issues of plasmid assembly when employing the combination of long and short read sequencing (Berbers et al., 2020).
Nanopore sequencing has been used for the identification of ARGs in Enteroaggregative E. coli (EAEC) as well as their position and structure (Greig et al., 2018). Analysis of WGS data enabled easier identification of ARGs and revealed the combination of many ARGs determinants located on the same mobile element. These findings provided a profound understanding of the transmission of co-located ARGs determinants in EAEC (Greig et al., 2018). Schmidt et al. (2017) showed that nanopore-based metagenomic sequencing successfully identified acquired ARGs directly from urine samples (Schmidt et al., 2017). The Oxford Nanopore MinION long read DNA sequencing device was used to detect ARGs, assess their taxonomic origin, and decode their genetic organization and possible correlation with mobilization markers. Therefore, targeted interventions can be established to alleviate the risks of ARGs transfer among sites and, thus, improve biosecurity practices in hospitals and other environments (Kamathewatta et al., 2019). More recently, MinION nanopore sequencing was employed for rapid identification of pathogens, plasmids and ARGs in bacterial DNA (Taxt et al., 2020). It accurately predicted the multilocus sequence type and identified ARGs profiles (Tan et al., 2020). Similarly, the ultra-long read nanopore sequencing technology was employed for ARGs detection in Mannheimia haemolytica (Lim et al., 2019).
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MAL-DITOF MS) can also be employed for the detection of ARGs (Justesen et al., 2018; Paul et al., 2018; Liu et al., 2019). MALDI-TOF MS relies on profiling proteins from bacterial cell extracts creating a spectral fingerprint that discriminates bacteria at subspecies level (Angeletti, 2017; Welker and van Belkum, 2019). It has also been used for the identification of antibiotic resistance mechanisms (Hoyos-Mallecot et al., 2014). However, the large size and high cost of MAL-DITOF MS systems substantially restricts its implementation for these purposes (Welker and van Belkum, 2019). Additionally, it requires purification, cultivation as well as sample preparation that are not suitable for studying mixed samples (Wang et al., 2018). This being said, MALDI Biotyper and VITEK MS are two commercially available MALDI-TOF MS systems (Marko Daniel et al., 2012; Martiny et al., 2012; Byun et al., 2018).
Quartz-Crystal Microbalance (QCM) is a microfluidic technology made of a physical nanogram-sensitive device with a piezoelectric sensor. QCM facilitates the real-time, rapid, on-site detection of antibiotic resistant bacteria (Bragazzi et al., 2015). This approach’s high sensitivity, accuracy, and dynamics allowed for the detection of ARGs using a magnesium zinc oxide nanostructure-modified quartz crystal microbalance (MZOnano-QCM) biosensor (Reyes et al., 2017b). QCM is advantageous because of its low cost, low demand in sample volume, and rapidity (Reyes et al., 2017a).
Although reassessment of the biosafety threat of PGPB formulations is under review in some countries, though mainly in North America and Europe (Abhilash et al., 2016), preventive actions must be taken to play down the conjunction of factors that worsen the emergence, evolution, and dispersal of antibiotic resistance within arable lands. The possible ways to cope with the issue of antibiotic-resistant and/or pathogenic PGPB include: (1) polyphasic characterization of bioinoculants at the species and strain level, (2) developing risk assessment groups for biofertilizers recommendation (Tariq et al., 2022), (3) prioritizing the studies on the resistance profiles of PGPB and the potential risks of disseminating ARGs in PGPB before field experiments and large-scale applications, (4) favoring antibiotic sensitive PGPB strains and those with fewer ARGs, (5) excluding multidrug-resistant strains, (6) inoculating with PGPB by spraying the aerial parts of plants or by soaking seeds, (7) prioritizing the use of promoting and/or biocontrol metabolites produced by PGPB, (8) reducing reliance on genetically engineered PGPB with ARGs biomarkers, (9) excluding bacterial species known to be pathogenic to humans, (10) avoiding PGPB with special ARGs genotypes that do not occur in the human microbiome (Kang et al., 2017; Ramakrishna et al., 2019), (11) using biochar as a soil amendment to reduce the abundance of resistance genes such as those conferring resistance to tetracyclines (tetC, tetG, tetW, and tetX) and sulfonamides (sulI and sulII) in soil and plant tissues (Ye et al., 2016; Duan et al., 2017), (12) genome mining PGPB strains for more precise characterization and identification of highly effective biofertilizers and potential harmful features, and (13) establishing standard criteria, regulations, and quality control guidelines for premarket biosafety assessment and post market monitoring of commercial biofertilizers. Finally, scientists should also identify which ARGs to survey for risk assessment in bacteria-based inoculants to distinguish those that are most relevant to human health.
As the biofertilizers market still lags behind synthetic fertilizers, greater attention should be played to regulatory aspects to share responsibilities in overcoming the challenges and offering high-quality products. Among the major constraints, antibiotic resistance across agroecosystems is urgent to combat with a target on prevention and remediation prospects. Commercialization should also require strict reference systems to demonstrate their effectiveness and potential associated risks. In developing countries, the spread of this public health scourge is due to (i) the lack of surveillance of resistance occurrence, (ii) the poor quality of utilized antibiotics, (iii) the clinical and agricultural excessive/misuse, and (iv) the improper and easy disposal of antibiotics and pharmaceutical wastes. However, these factors differ from developing to developed countries. Overall, there is still a need to improve and update the regulatory aspects for antibiotic use especially in agriculture (Chokshi et al., 2019). In the following section, we sought to contribute to set a putative regulatory framework for the use bacterial-based biofertilizers to help guarantee their biosafety, tackle the antibiotic resistance issue, and ensure environmental and public health protection.
Isolated and screened strains should be taxonomically characterized using a polyphasic approach appealing to phenotypic (morphological, physiological, and biochemical), chemotaxonomic (peptidoglycans, fatty acids, proteins, sugars…), and molecular aspects (Barcoding; Supplementary Figure S4; Keswani et al., 2019). In addition to in silico taxonomic profiling, it is mandatory to compare the potential strains physiologically and morphologically with closely related ones for phylogenetic delineation (Stackebrandt et al., 2002; Trujillo, 2011).
Nowadays, whole genome sequencing is a more comprehensive strategy to taxonomically and functionally identify the strain of interest and to assign it with its closest relatives. Subsequently, depending on their biosafety level, candidate strains should be classified within risk group (BSL-1, 2, 3, and 4). Bacteria falling in BSL-1 class are more likely to be null of biosafety risk. This implies further experiments such as formulation and testing. However, strains falling in the BLS-2 class of higher are more likely to be risky with potential negative impact. In this case, pathogenesis investigation and characterization at the strain level should be carried out. When it is a pathogenic species, it should be excluded from downstream experiments unless alternatively used, under confinement conditions in the laboratory, to optimize the production of metabolites and compounds of interests (Supplementary Figure S4).
To promote the acceptance of biofertilizers by the users, the quality of a bioinoculant must refer not only to the specification of the strain, efficacy, and preservation etc. but also to its antibiotic resistance profile and potential pathogenesis for human. This information can be reflected in the labelling requirements (Malusá and Vassilev, 2014).
Controlling biofertilizers is essential to ensure their compliance with standards, product safety, and efficacy. Poor quality products can be expected when the regulatory setup is not well established, leading to poor performance in the field or potential health risks to users. The lack of effective regulation of biofertilizers in most countries is one of the major factors to their low availability and adoption. Afterwards, periodic monitoring of commercialized biofertilizers is important to ensure product quality and safely throughout the marketing chain (Pandey and Chandra, 2016).
To mitigate the risks posed by the continued emergence of antibiotic resistance, behavioral changes and practical strategies should be considered by all the stakeholders. In agriculture, the trend of PGPB, which is claimed to be sustainable and environmentally friendly, could be a significant threat and cause large-scale dissemination of multi-drug resistant bacteria and/or ARGs in different environments including human beings. The future development of more effective bacteria-based biofertilizers should consider environmental issues as there is enough evidence supporting the hypothesis that antibiotic-resistant biofertilizers could contribute substantially to the global antibiotic resistance crisis. Additionally, understanding the underpinning mechanisms is necessary to develop effective approaches in the battle against the spread of antibiotic resistance. Ultimately, to tackle this issue at its source, better formulated strategies are needed based on agricultural, environmental, and public health aspects.
The need for knowledge of the antibiotic resistome in plant agricultural systems and especially those in which biofertilizers are applied is critically important because we need to understand whether their application to the soil has the potential to select ARGs that could impact human health. As this has the potential to impact human health, and current implications for the use in agriculture, commercial biofertilizers should be well characterized with respect to their ability to transfer ARGs to other microorganisms. Detecting organisms harboring ARGs and their identification is important for risk assessments with co-inhabiting antibiotic-resistant communities. If ARGs of importance in clinical medicine are identified in the resistome of bioinoculants and/or inoculated plants, it is critical to determine whether their frequency and/or bacterial host range changes based on strain exposure. Finally, more in depth studies should address the genetic behavior, the bacterial diversity, and the activity of PGPB harboring ARGs with regards to indigenous communities.
IM and NF reviewed the literature and wrote the original draft of the manuscript. MH reviewed and corrected the manuscript. MS revised the manuscript. IM, MH and MS designed and conceived the work. All authors contributed to the article and approved the submitted version.
This work was supported by funding from the OCP Group (Projects AS-78 and AS-85, awarded to MH) and Mohammed VI Polytechnic University (UM6P, awarded to MS), which are greatly acknowledged.
We thank Andrew Blakney for commenting on the manuscript and editing English language.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.999988/full#supplementary-material
Abhilash, P. C., Dubey, R. K., Tripathi, V., Gupta, V. K., and Singh, H. B. (2016). Plant growth-promoting microorganisms for environmental sustainability. Trends Biotechnol. 34, 847–850. doi: 10.1016/j.tibtech.2016.05.005
Agersø, Y., Bjerre, K., Brockmann, E., Johansen, E., Nielsen, B., Siezen, R., et al. (2019). Putative antibiotic resistance genes present in extant bacillus licheniformis and bacillus paralicheniformis strains are probably intrinsic and part of the ancient resistome. PLoS One 14:e0210363. doi: 10.1371/journal.pone.0210363
Ahemad, M., and Kibret, M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. 26, 1–20. doi: 10.1016/j.jksus.2013.05.001
Ajbani, K., Lin Shou-Yean, G., Rodrigues, C., Nguyen, D., Arroyo, F., Kaping, J., et al. (2014). Evaluation of pyrosequencing for detecting extensively drug-resistant mycobacterium tuberculosis among clinical isolates from four high-burden countries. Antimicrob. Agents Chemother. 59, 414–420. doi: 10.1128/AAC.03614-14
Akhtar, M. S., and Siddiqui, Z. A. (2008). Glomusintraradices, Pseudomonas alcaligenes, and Bacillus pumilus: effective agents for the control of root-rot disease complex of chickpea (Cicer arietinum L.). journal of general. Plant Pathol. 74, 53–60. doi: 10.1007/s10327-007-0062-4
Alghoribi, M. F., Balkhy, H. H., Woodford, N., and Ellington, M. J. (2018). The role of whole genome sequencing in monitoring antimicrobial resistance: a biosafety and public health priority in the Arabian peninsula. J. Infect. Public Health 11, 784–787. doi: 10.1016/j.jiph.2018.08.001
Álvarez-López, V., Prieto-Fernández, A., Janssen, J., Herzig, R., Vangronsveld, J., and Kidd, P. (2016). Inoculation methods using Rhodococcus erythropolis strain P30 affects bacterial assisted phytoextraction capacity of Nicotiana tabacum. Int. J. Phytoremediation 18, 406–415. doi: 10.1080/15226514.2015.1109600
Amarasiri, M., Sano, D., and Suzuki, S. (2020). Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 50, 2016–2059. doi: 10.1080/10643389.2019.1692611
Amna Xia, Y., Farooq, M. A., Javed, M. T., Kamran, M. A., Mukhtar, T., Ali, J., et al. (2020). Multi-stress tolerant PGPR bacillus xiamenensis PM14 activating sugarcane (Saccharum officinarum L.) red rot disease resistance. Plant Physiol. Biochem. 151, 640–649. doi: 10.1016/j.plaphy.2020.04.016
Amoako, K. K., Thomas, M. C., Kong, F., Janzen, T. W., Hahn, K. R., Shields, M. J., et al. (2012). Rapid detection and antimicrobial resistance gene profiling of Yersinia pestis using pyrosequencing technology. J. Microbiol. Methods 90, 228–234. doi: 10.1016/j.mimet.2012.05.012
Andam, C. P., Carver, S. M., and Berthrong, S. T. (2015). Horizontal gene flow in managed ecosystems. Annu. Rev. Ecol. Evol. Syst. 46, 121–143. doi: 10.1146/annurev-ecolsys-112414-054126
Angeletti, S. (2017). Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. J. Microbiol. Methods 138, 20–29. doi: 10.1016/j.mimet.2016.09.003
Arber, W. (2014). Horizontal gene transfer among bacteria and its role in biological evolution. Lifestyles 4, 217–224. doi: 10.3390/life4020217
Argimón, S., Masim, M. A. L., Gayeta, J. M., Lagrada, M. L., Macaranas, P. K. V., Cohen, V., et al. (2020). Integrating whole-genome sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nat. Commun. 11:2719. doi: 10.1038/s41467-020-16322-5
Arora, N. K. (2018). Agricultural sustainability and food security. Environ. Sustain. 1, 217–219. doi: 10.1007/s42398-018-00032-2
Awais, M., Tariq, M., Ali, A., Ali, Q., Khan, A., Tabassum, B., et al. (2017). Isolation, characterization and inter-relationship of phosphate solubilizing bacteria from the rhizosphere of sugarcane and rice. Biocatal. Agric. Biotechnol. 11, 312–321. doi: 10.1016/j.bcab.2017.07.018
Aylward, F. O., Tremmel, D. M., Starrett, G. J., Bruce, D. C., Chain, P., Chen, A., et al. (2013). Complete genome of Serratia sp. strain FGI 94, a strain associated with leaf-cutter ant fungus gardens. Genome Announc. 1:e00239-12. doi: 10.1128/genomeA.00239-12
Baffone, W., Citterio, B., Vittoria, E., Casaroli, A., Campana, R., Falzano, L., et al. (2003). Retention of virulence in viable but non-culturable halophilic vibrio spp. Int. J. Food Microbiol. 89, 31–39. doi: 10.1016/S0168-1605(03)00102-8
Bakthavatchalu, S., Shivakumar, S., and Sullia, S. B. (2012). Identification of multi-trait PGPR isolates and evaluation of their potential as biocontrol agents. Acta Biol. Indica 1, 61–67.
Barbe, V., Cruveiller, S., Kunst, F., Lenoble, P., Meurice, G., Sekowska, A., et al. (2009). From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155, 1758–1775. doi: 10.1099/mic.0.027839-0
Bargaz, A., Lyamlouli, K., Chtouki, M., Zeroual, Y., and Dhiba, D. (2018). Soil microbial resources for improving fertilizers efficiency in an integrated plant nutrient management system. Front. Microbiol. 9:1606. doi: 10.3389/fmicb.2018.01606
Berbers, B., Saltykova, A., Garcia-Graells, C., Philipp, P., Arella, F., Marchal, K., et al. (2020). Combining short and long read sequencing to characterize antimicrobial resistance genes on plasmids applied to an unauthorized genetically modified bacillus. Sci. Rep. 10:4310. doi: 10.1038/s41598-020-61158-0
Berquó Marks, B., Megías Guijo, M., Nogueira, M. A., and Hungria, M. (2013). Biotechnological potential of rhizobial metabolites to enhance the performance of Bradyrhizobium spp. and Azospirillum brasilense inoculants with soybean and maize. AMB Express 3:21. doi: 10.1186/2191-0855-3-21
Bertalan, M., Albano, R., de Pádua, V., Rouws, L., Rojas, C., Hemerly, A., et al. (2009). Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10:450. doi: 10.1186/1471-2164-10-450
Boc, A., Di Sciullo, A. M., and Makarenkov, V. (2010). “Classification of the indo-European languages using a phylogenetic network approach,” in Classification as a Tool for Research. eds. H. Locarek-Junge and C. Weihs (Berlin Heidelberg: Springer), 647–655.
Böckelmann, U., Dörries, H. H., Ayuso-Gabella, M. N., Salgot de Marçay, M., Tandoi, V., Levantesi, C., et al. (2009). Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl. Environ. Microbiol. 75, 154–163. doi: 10.1128/aem.01649-08
Bogožalec Košir, A., Cvelbar, T., Kammel, M., Grunert, H. P., Zeichhardt, H., and Milavec, M. (2020). Digital PCR method for detection and quantification of specific antimicrobial drug-resistance mutations in human cytomegalovirus. J. Virol. Methods 281:113864. doi: 10.1016/j.jviromet.2020.113864
Boto, L., Pineda, M., and Pineda, R. (2019). Potential impacts of horizontal gene transfer on human health and physiology and how anthropogenic activity can affect it. FEBS J. 286, 3959–3967. doi: 10.1111/febs.15054
Bragazzi, N. L., Amicizia, D., Panatto, D., Tramalloni, D., Valle, I., and Gasparini, R. (2015). “Chapter six—quartz-crystal microbalance (QCM) for public health: An overview of its applications,” in Advances in Protein Chemistry and Structural Biology. ed. R. Donev (Swansea, United Kingdom: Academic Press), 149–211.
Broggini, G. A. L., Duffy, B., Holliger, E., Schärer, H. J., Gessler, C., and Patocchi, A. (2005). Detection of the fire blight biocontrol agent Bacillus subtilis BD170 (biopro®) in a Swiss apple orchard. Eur. J. Plant Pathol. 111, 93–100. doi: 10.1007/s10658-004-1423-x
Butcher, B. G., and Helmann, J. D. (2006). Identification of Bacillus subtilis σW-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol. Microbiol. 60, 765–782. doi: 10.1111/j.1365-2958.2006.05131.x
Byun, J.-H., Yu, A.R., Kim, M.S., and Lee, K. (2018). Performance of microflex LT Biotyper and VITEK MS for routine identification of yeasts. alm 38, 487–489. doi: 10.3343/alm.2018.38.5.487
Calvo, P., Nelson, L., and Kloepper, J. W. (2014). Agricultural uses of plant biostimulants. Plant Soil 383, 3–41. doi: 10.1007/s11104-014-2131-8
Calvo, P., Ormeño-Orrillo, E., Martínez-Romero, E., and Zúñiga, D. (2010). Characterization of bacillus isolates of potato rhizosphere from andean soils of Peru and their potential PGPR characteristics. Braz. J. Microbiol. 41, 899–906. doi: 10.1590/S1517-83822010000400008
Cantera, J. L., White, H., Diaz, M. H., Beall, S. G., Winchell, J. M., Lillis, L., et al. (2019). Assessment of eight nucleic acid amplification technologies for potential use to detect infectious agents in low-resource settings. PLoS One 14:e0215756. doi: 10.1371/journal.pone.0215756
Cardinale, M., Ratering, S., Suarez, C., Zapata Montoya, A. M., Geissler-Plaum, R., and Schnell, S. (2015). Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol. Res. 181, 22–32. doi: 10.1016/j.micres.2015.08.002
Čermák, L., Kopecký, J., Novotná, J., Omelka, M., Parkhomenko, N., Plháčková, K., et al. (2008). Bacterial communities of two contrasting soils reacted differently to lincomycin treatment. Appl. Soil Ecol. 40, 348–358. doi: 10.1016/j.apsoil.2008.06.001
Chen, Q.-L., An, X.-L., Zhu, Y.-G., Su, J.-Q., Gillings, M. R., Ye, Z.-L., et al. (2017). Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 51, 8149–8157. doi: 10.1021/acs.est.7b01420
Chen, Q.-L., Cui, H.-L., Su, J.-Q., Penuelas, J., and Zhu, Y.-G. (2019). Antibiotic Resistomes in plant microbiomes. Trends Plant Sci. 24, 530–541. doi: 10.1016/j.tplants.2019.02.010
Chen, B., He, R., Yuan, K., Chen, E., Lin, L., Chen, X., et al. (2017). Polycyclic aromatic hydrocarbons (PAHs) enriching antibiotic resistance genes (ARGs) in the soils. Environ. Pollut. 220, 1005–1013. doi: 10.1016/j.envpol.2016.11.047
Chen, X. H., Koumoutsi, A., Scholz, R., Eisenreich, A., Schneider, K., Heinemeyer, I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. doi: 10.1038/nbt1325
Chen, B., Liang, X., Huang, X., Zhang, T., and Li, X. (2013). Differentiating anthropogenic impacts on ARGs in the Pearl River estuary by using suitable gene indicators. Water Res. 47, 2811–2820. doi: 10.1016/j.watres.2013.02.042
Chisholm, S. A., Owen, R. J., Teare, E. L., and Saverymuttu, S. (2001). PCR-based diagnosis of helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J. Clin. Microbiol. 39, 1217–1220. doi: 10.1128/jcm.39.4.1217-1220.2001
Chokshi, A., Sifri, Z., Cennimo, D., and Horng, H. (2019). Global contributors to antibiotic resistance. J. Global Infect. Dis. 11, 36–42. doi: 10.4103/jgid.jgid_110_18
Cox, G., and Wright, G. D. (2013). Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303, 287–292. doi: 10.1016/j.ijmm.2013.02.009
Cray, J. A., Bell, A. N., Bhaganna, P., Mswaka, A. Y., Timson, D. J., and Hallsworth, J. E. (2013). The biology of habitat dominance; can microbes behave as weeds? Microb. Biotechnol. 6, 453–492. doi: 10.1111/1751-7915.12027
de Menezes, A., Clipson, N., and Doyle, E. (2012). Comparative metatranscriptomics reveals widespread community responses during phenanthrene degradation in soil. Environ. Microbiol. 14, 2577–2588. doi: 10.1111/j.1462-2920.2012.02781.x
Del Mar Lleò, M., Benedetti, D., Tafi, M. C., Signoretto, C., and Canepari, P. (2007). Inhibition of the resuscitation from the viable but non-culturable state in enterococcus faecalis. Environ. Microbiol. 9, 2313–2320. doi: 10.1111/j.1462-2920.2007.01345.x
Denamur, E., Lecointre, G., Darlu, P., Tenaillon, O., Acquaviva, C., Sayada, C., et al. (2000). Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cells 103, 711–721. doi: 10.1016/S0092-8674(00)00175-6
Diaz, R.J., and Rosenberg, R. (2008). Spreading dead zones and consequences for marine ecosystems. science 321, 926–929. doi: 10.1126/science.1156401
Duan, M., Li, H., Gu, J., Tuo, X., Sun, W., Qian, X., et al. (2017). Effects of biochar on reducing the abundance of oxytetracycline, antibiotic resistance genes, and human pathogenic bacteria in soil and lettuce. Environ. Pollut. 224, 787–795. doi: 10.1016/j.envpol.2017.01.021
Dwivedi, D., Johri, B. N., Ineichen, K., Wray, V., and Wiemken, A. (2009). Impact of antifungals producing rhizobacteria on the performance of Vigna radiata in the presence of arbuscular mycorrhizal fungi. Mycorrhiza 19, 559–570. doi: 10.1007/s00572-009-0253-2
Egamberdieva, D., Wirth, S., Bellingrath-Kimura, S. D., Mishra, J., and Arora, N. K. (2019). Salt-tolerant plant growth promoting Rhizobacteria for enhancing crop productivity of saline soils. Front. Microbiol. 10:2791. doi: 10.3389/fmicb.2019.02791
English, M., Coulson, T., Horsman, S., and Patten, C. (2010). Overexpression of hns in the plant growth-promoting bacterium Enterobacter cloacae UW5 increases root colonization. J. Appl. Microbiol. 108, 2180–2190. doi: 10.1111/j.1365-2672.2009.04620.x
Fahsi, N., Mahdi, I., Mesfioui, A., Biskri, L., and Allaoui, A. (2021). Phosphate solubilizing rhizobacteria isolated from jujube ziziphus lotus plant stimulate wheat germination rate and seedlings growth. PeerJ 9:e11583. doi: 10.7717/peerj.11583
Fahsi, N., Mahdi, I., Mesfioui, A., Hafidi, M., Biskri, L., and Allaoui, A. (2020). Preliminary characterization of the first phosphate solubilizing bacteria isolated from the rhizospheric soil of the jujube plant Ziziphus. Available at SSRN 3609842.
Fakruddin, M., Mannan, K. S. B., and Andrews, S. (2013). Viable but Nonculturable bacteria: food safety and public health perspective. ISRN Microbiol. 2013:703813, 1–6. doi: 10.1155/2013/703813
Farag, M. A., Zhang, H., and Ryu, C.-M. (2013). Dynamic chemical communication between plants and bacteria through airborne signals: induced resistance by bacterial volatiles. J. Chem. Ecol. 39, 1007–1018. doi: 10.1007/s10886-013-0317-9
Forsberg, K. J., Patel, S., Gibson, M. K., Lauber, C. L., Knight, R., Fierer, N., et al. (2014). Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616. doi: 10.1038/nature13377
Forsberg, K.J., Reyes, A., Wang, B., Selleck, E.M., Sommer, M.O., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. science 337, 1107–1111. doi: 10.1126/science.1220761
García-Aljaro, C., Ballesté, E., and Muniesa, M. (2017). Beyond the canonical strategies of horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 38, 95–105. doi: 10.1016/j.mib.2017.04.011
Gill, P., and Ghaemi, A. (2008). Nucleic acid isothermal amplification technologies—a review. Nucleosides Nucleotides Nucleic Acids 27, 224–243. doi: 10.1080/15257770701845204
Gillings, M. R. (2017a). Class 1 integrons as invasive species. Curr. Opin. Microbiol. 38, 10–15. doi: 10.1016/j.mib.2017.03.002
Gillings, M. R. (2017b). Lateral gene transfer, bacterial genome evolution, and the Anthropocene. Ann. N. Y. Acad. Sci. 1389, 20–36. doi: 10.1111/nyas.13213
Glick, B. (2012). Plant growth-promoting bacteria: mechanisms and applications. Australas. Sci. 2012:963401. doi: 10.6064/2012/963401
Govindappa, M., Farheen, H., Chandrappa, C. P., Channabasava, R. V., and Raghavendra, V. B. (2016). Mycosynthesis of silver nanoparticles using extract of endophytic fungi, Penicillium species of Glycosmis mauritiana, and its antioxidant, antimicrobial, anti-inflammatory and tyrokinase inhibitory activity. Adv. Nat. Sci. Nanosci. Nanotechnol. 7:035014. doi: 10.1088/2043-6262/7/3/035014
Graham, D. W., Knapp, C. W., Christensen, B. T., McCluskey, S., and Dolfing, J. (2016). Appearance of β-lactam resistance genes in agricultural soils and clinical isolates over the 20th century. Sci. Rep. 6:21550. doi: 10.1038/srep21550
Graham, D. W., Olivares-Rieumont, S., Knapp, C. W., Lima, L., Werner, D., and Bowen, E. (2011). Antibiotic resistance gene abundances associated with waste discharges to the Almendares River near Havana. Cuba. Environ. Sci. Technol. 45, 418–424. doi: 10.1021/es102473z
Greig, D. R., Dallman, T. J., Hopkins, K. L., and Jenkins, C. (2018). MinION nanopore sequencing identifies the position and structure of bacterial antibiotic resistance determinants in a multidrug-resistant strain of enteroaggregative Escherichia coli. Microb. Genom. 4:e000213. doi: 10.1099/mgen.0.000213
Gupta, A., Saxena, A. K., Gopal, M., and Tilak, K. V. B. R. (1998). Effect of plant growth promoting rhizobacteria on competitive ability of introduced Bradyrhizobium sp. (Vigna) for nodulation. Microbiol. Res. 153, 113–117. doi: 10.1016/S0944-5013(98)80028-1
Gutiérrez-Luna, F. M., López-Bucio, J., Altamirano-Hernández, J., Valencia-Cantero, E., de la Cruz, H. R., and Macías-Rodríguez, L. (2010). Plant growth-promoting rhizobacteria modulate root-system architecture in Arabidopsis thaliana through volatile organic compound emission. Symbiosis 51, 75–83. doi: 10.1007/s13199-010-0066-2
Gutiérrez-Mañero, F. J., Ramos-Solano, B., Probanza, A. N., Mehouachi, J. R., Tadeo, F., and Talon, M. (2001). The plant-growth-promoting rhizobacteria Bacillus pumilus and bacillus licheniformis produce high amounts of physiologically active gibberellins. Physiol. Plant. 111, 206–211. doi: 10.1034/j.1399-3054.2001.1110211.x
Hallett, M.T., and Lagergren, J. (2001). "Efficient algorithms for lateral gene transfer problems", in Proceedings of the fifth annual international conference on computational biology, 149–156.
Han, C. S., Xie, G., Challacombe, J. F., Altherr, M. R., Bhotika, S. S., Bruce, D., et al. (2006). Pathogenomic sequence analysis of Bacillus cereus and bacillus thuringiensis isolates closely related to bacillus anthracis. J. Bacteriol. 188, 3382–3390. doi: 10.1128/JB.188.9.3382-3390.2006
Havlicek, J., Dachsel, B., Slickers, P., Andres, S., Beckert, P., Feuerriegel, S., et al. (2019). Rapid microarray-based assay for detection of pyrazinamide resistant mycobacterium tuberculosis. Diagn. Microbiol. Infect. Dis. 94, 147–154. doi: 10.1016/j.diagmicrobio.2018.12.011
Hawkey, P. M., and Jones, A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64, i3–i10. doi: 10.1093/jac/dkp256
Hendriksen, R. S., Bortolaia, V., Tate, H., Tyson, G. H., Aarestrup, F. M., and McDermott, P. F. (2019). Using genomics to track global antimicrobial resistance. Front. Public Health 7:242. doi: 10.3389/fpubh.2019.00242
Heuer, H., Schmitt, H., and Smalla, K. (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243. doi: 10.1016/j.mib.2011.04.009
Hoon, S., Cheong, H., Ryu, C.-M., Kim, J. F., and Park, S.-H. (2007). Two bacterial entophytes eliciting both plant growth promotion and plant defense on pepper (Capsicum annuum L.). J. Microbiol. Biotechnol. 17, 96–103.
Hoyos-Mallecot, Y., Riazzo, C., Miranda-Casas, C., Rojo-Martín, M. D., Gutiérrez-Fernández, J., and Navarro-Marí, J. M. (2014). Rapid detection and identification of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J. Microbiol. Methods 105, 98–101. doi: 10.1016/j.mimet.2014.07.016
Hsiao, N.-H., and Kirby, R. (2008). Comparative genomics of Streptomyces avermitilis, Streptomyces cattleya, Streptomyces maritimus and Kitasatospora aureofaciens using a Streptomyces coelicolor microarray system. Antonie Van Leeuwenhoek 93, 1–25. doi: 10.1007/s10482-007-9175-1
Hubbard, B. K., and Walsh, C. T. (2003). Vancomycin assembly: nature's way. Angew. Chem. Int. Ed. 42, 730–765. doi: 10.1002/anie.200390202
Hung, W.-C. (2020). Prevalence, fate, and co-selection of heavy metals and antibiotic resistance genes in urban and agricultural soils. UCLA.
Idris, E. E., Iglesias, D. J., Talon, M., and Borriss, R. (2007). Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by bacillus amyloliquefaciens FZB42. Mol. Plant-Microbe Interact. 20, 619–626. doi: 10.1094/MPMI-20-6-0619
Ivanova, N., Sorokin, A., Anderson, I., Galleron, N., Candelon, B., Kapatral, V., et al. (2003). Genome sequence of Bacillus cereus and comparative analysis with bacillus anthracis. Nature 423, 87–91. doi: 10.1038/nature01582
Jain, R., Rivera, M. C., Moore, J. E., and Lake, J. A. (2003). Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20, 1598–1602. doi: 10.1093/molbev/msg154
Jha, Y., and Subramanian, R. B. (2014). PGPR regulate caspase-like activity, programmed cell death, and antioxidant enzyme activity in paddy under salinity. Physiol. Mol. Biol. Plants 20, 201–207. doi: 10.1007/s12298-014-0224-8
Joo, G.-J., Kim, Y.-M., Lee, I.-J., Song, K.-S., and Rhee, I.-K. (2004). Growth promotion of red pepper plug seedlings and the production of gibberellins by Bacillus cereus, Bacillus macroides and Bacillus pumilus. Biotechnol. Lett. 26, 487–491. doi: 10.1023/B:BILE.0000019555.87121.34
Jousset, A. B., Bernabeu, S., Bonnin, R. A., Creton, E., Cotellon, G., Sauvadet, A., et al. (2019). Development and validation of a multiplex polymerase chain reaction assay for detection of the five families of plasmid-encoded colistin resistance. Int. J. Antimicrob. Agents 53, 302–309. doi: 10.1016/j.ijantimicag.2018.10.022
Ju, S., Zheng, J., Lin, J., Geng, C., Zhu, L., Guan, Z., et al. (2016). The complete genome sequence of Alcaligenes faecalis ZD02, a novel potential bionematocide. J. Biotechnol. 218, 73–74. doi: 10.1016/j.jbiotec.2015.12.001
Justesen, U. S., Acar, Z., Sydenham, T. V., and Johansson, Å. (2018). Antimicrobial susceptibility testing of Bacteroides fragilis using the MALDI Biotyper antibiotic susceptibility test rapid assay (MBT-ASTRA). Anaerobe 54, 236–239. doi: 10.1016/j.anaerobe.2018.02.007
Kamathewatta, K. I., Bushell, R. N., Young, N. D., Stevenson, M. A., Billman-Jacobe, H., Browning, G. F., et al. (2019). Exploration of antibiotic resistance risks in a veterinary teaching hospital with Oxford Nanopore long read sequencing. PLoS One 14:e0217600. doi: 10.1371/journal.pone.0217600
Kang, Y., Shen, M., Wang, H., and Zhao, Q. (2013). A possible mechanism of action of plant growth-promoting rhizobacteria (PGPR) strain Bacillus pumilus WP8 via regulation of soil bacterial community structure. J. Gen. Appl. Microbiol. 59, 267–277. doi: 10.2323/jgam.59.267
Kang, Y., Shen, M., Xia, D., Ye, K., Zhao, Q., and Hu, J. (2017). Caution of intensified spread of antibiotic resistance genes by inadvertent introduction of beneficial bacteria into soil. Acta Agric. Scand. Sec. B Soil Plant Sci. 67, 576–582. doi: 10.1080/09064710.2017.1314548
Kaprou, G. D., Bergšpica, I., Alexa, E. A., Alvarez-Ordóñez, A., and Prieto, M. (2021). Rapid methods for antimicrobial resistance diagnostics. Antibiotics 10:209. doi: 10.3390/antibiotics10020209
Keswani, C., Prakash, O., Bharti, N., Vílchez, J. I., Sansinenea, E., Lally, R. D., et al. (2019). Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ. 690, 841–852. doi: 10.1016/j.scitotenv.2019.07.046
Khan, N., Zandi, P., Ali, S., Mehmood, A., Adnan Shahid, M., and Yang, J. (2018). Impact of salicylic acid and PGPR on the drought tolerance and phytoremediation potential of Helianthus annus. Front. Microbiol. 9:2507. doi: 10.3389/fmicb.2018.02507
Khayi, S., Des Essarts, Y. R., Mondy, S., Moumni, M., Hélias, V., Beury-Cirou, A., et al. (2015). Draft genome sequences of the three Pectobacterium-antagonistic bacteria pseudomonas brassicacearum PP1-210F and PA1G7 and Bacillus simplex BA2H3. Genome Announc. 3:e01497-14. doi: 10.1128/genomeA.01497-14
Klopper, J. (1980). Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286, 885–886. doi: 10.1038/286885a0
Kluepfel, D. A. (1993). The behavior and tracking of bacteria in the rhizosphere. Annu. Rev. Phytopathol. 31, 441–472. doi: 10.1146/annurev.py.31.090193.002301
Knapp, C. W., Dolfing, J., Ehlert, P. A., and Graham, D. W. (2010). Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44, 580–587. doi: 10.1021/es901221x
Köser, C. U., Ellington, M. J., and Peacock, S. J. (2014). Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 30, 401–407. doi: 10.1016/j.tig.2014.07.003
Kumar, S., Pandey, P., and Maheshwari, D. K. (2009). Reduction in dose of chemical fertilizers and growth enhancement of sesame (Sesamum indicum L.) with application of rhizospheric competent Pseudomonas aeruginosa LES4. Eur. J. Soil Biol. 45, 334–340. doi: 10.1016/j.ejsobi.2009.04.002
Kumari, P., Meena, M., and Upadhyay, R. S. (2018). Characterization of plant growth promoting rhizobacteria (PGPR) isolated from the rhizosphere of Vigna radiata (mung bean). Biocatal. Agric. Biotechnol. 16, 155–162. doi: 10.1016/j.bcab.2018.07.029
Lee, J. H., Park, K. S., Jeon, J. H., and Lee, S. H. (2018). Antibiotic resistance in soil. Lancet Infect. Dis. 18, 1306–1307. doi: 10.1016/S1473-3099(18)30675-3
Lim, A., Naidenov, B., Bates, H., Willyerd, K., Snider, T., Couger, M. B., et al. (2019). Nanopore ultra-long read sequencing technology for antimicrobial resistance detection in Mannheimia haemolytica. J. Microbiol. Methods 159, 138–147. doi: 10.1016/j.mimet.2019.03.001
Lim, Y.-L., Yong, D., Ee, R., Krishnan, T., Tee, K.-K., Yin, W.-F., et al. (2015). Complete genome sequence of Serratia fonticola DSM 4576T, a potential plant growth promoting bacterium. J. Biotechnol. 214, 43–44. doi: 10.1016/j.jbiotec.2015.09.005
Lister, P. D., Wolter, D. J., and Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. doi: 10.1128/CMR.00040-09
Liu, N., Wang, L., Cai, G., Zhang, D., and Lin, J. (2019). Establishment of a simultaneous detection method for ten duck viruses using MALDI-TOF mass spectrometry. J. Virol. Methods 273:113723. doi: 10.1016/j.jviromet.2019.113723
Liu, S.-H., Zeng, G.-M., Niu, Q.-Y., Liu, Y., Zhou, L., Jiang, L.-H., et al. (2017). Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: a mini review. Bioresour. Technol. 224, 25–33. doi: 10.1016/j.biortech.2016.11.095
Luque-Almagro, V. M., Acera, F., Igeño, M. I., Wibberg, D., Roldán, M. D., Sáez, L. P., et al. (2013). Draft whole genome sequence of the cyanide-degrading bacterium P seudomonas pseudoalcaligenes CECT 5344. Environ. Microbiol. 15, 253–270. doi: 10.1111/j.1462-2920.2012.02875.x
Ma, Y., Rajkumar, M., Zhang, C., and Freitas, H. (2016a). Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manag. 174, 14–25. doi: 10.1016/j.jenvman.2016.02.047
Ma, Y., Rajkumar, M., Zhang, C., and Freitas, H. (2016b). Inoculation of brassica oxyrrhina with plant growth promoting bacteria for the improvement of heavy metal phytoremediation under drought conditions. J. Hazard. Mater. 320, 36–44. doi: 10.1016/j.jhazmat.2016.08.009
Ma, M., Wang, C., Ding, Y., Li, L., Shen, D., Jiang, X., et al. (2011). Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. J. Bacteriol. 193, 311–312. doi: 10.1128/JB.01234-10
Mackay, I. M. (2004). Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10, 190–212. doi: 10.1111/j.1198-743X.2004.00722.x
Madawala, H. M. S. P. (2021). “Chapter 7 - Arbuscular mycorrhizal fungi as biofertilizers: current trends, challenges, and future prospects,” in Biofertilizers. eds. A. Rakshit, V. S. Meena, M. Parihar, H. B. Singh, and A. K. Singh (Sawston, Cambridge: Woodhead Publishing), 83–93.
Mahdi, I., Fahsi, N., Hafidi, M., Benjelloun, S., Allaoui, A., and Biskri, L. (2021a). Rhizospheric phosphate solubilizing bacillus atrophaeus GQJK17 S8 increases quinoa seedling, withstands heavy metals, and mitigates salt stress. Sustain. For. 13:3307. doi: 10.3390/su13063307
Mahdi, I., Hafidi, M., Allaoui, A., and Biskri, L. (2021b). Halotolerant Endophytic bacterium Serratia rubidaea ED1 enhances phosphate Solubilization and promotes seed germination. Agriculture 11:224. doi: 10.3390/agriculture11030224
Malusá, E., and Vassilev, N. (2014). A contribution to set a legal framework for biofertilisers. Appl. Microbiol. Biotechnol. 98, 6599–6607. doi: 10.1007/s00253-014-5828-y
Mantelin, S., and Touraine, B. (2004). Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. J. Exp. Bot. 55, 27–34. doi: 10.1093/jxb/erh010
Marko Daniel, C., Saffert Ryan, T., Cunningham Scott, A., Hyman, J., Walsh, J., Arbefeville, S., et al. (2012). Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for Identification of nonfermenting gram-negative bacilli isolated from cultures from cystic fibrosis patients. J. Clin. Microbiol. 50, 2034–2039. doi: 10.1128/JCM.00330-12
Martiny, D., Busson, L., Wybo, I., El Haj Rachid, A., Dediste, A., and Vandenberg, O. (2012). Comparison of the microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50, 1313–1325. doi: 10.1128/JCM.05971-11
Máthé, I., Benedek, T., Táncsics, A., Palatinszky, M., Lányi, S., and Márialigeti, K. (2012). Diversity, activity, antibiotic and heavy metal resistance of bacteria from petroleum hydrocarbon contaminated soils located in Harghita County (Romania). Int. Biodeterior. Biodegradation 73, 41–49. doi: 10.1016/j.ibiod.2012.05.018
McHardy, A. C., Martín, H. G., Tsirigos, A., Hugenholtz, P., and Rigoutsos, I. (2007). Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods 4, 63–72. doi: 10.1038/nmeth976
Megías, E., Megías, M., Ollero, F. J., and Hungria, M. (2016). Draft genome sequence of Pantoea ananatis strain AMG521, a rice plant growth-promoting bacterial endophyte isolated from the Guadalquivir marshes in southern Spain. Genome Announc. 4:e01681-15. doi: 10.1128/genomeA.01681-15
Minaxi Nain, L., Yadav, R. C., and Saxena, J. (2012). Characterization of multifaceted bacillus sp. RM-2 for its use as plant growth promoting bioinoculant for crops grown in semi arid deserts. Appl. Soil Ecol. 59, 124–135. doi: 10.1016/j.apsoil.2011.08.001
Monod, M., DeNoya, C., and Dubnau, D. (1986). Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J. Bacteriol. 167, 138–147. doi: 10.1128/jb.167.1.138-147.1986
Morin, S., Bazarova, N., Jacon, P., and Vella, S. (2018). The manufacturers’ perspective on World Health Organization prequalification of in vitro diagnostics. Clin. Infect. Dis. 66, 301–305. doi: 10.1093/cid/cix719
Murray, J. D. (2011). Invasion by invitation: rhizobial infection in legumes. Mol. Plant-Microbe Interact. 24, 631–639. doi: 10.1094/MPMI-08-10-0181
Nadeem, S. M., Zahir, Z. A., Naveed, M., and Arshad, M. (2007). Preliminary investigations on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can. J. Microbiol. 53, 1141–1149. doi: 10.1139/W07-081
Nakkeeran, S., Fernando, W. D., and Siddiqui, Z. A. (2005). “Plant growth promoting rhizobacteria formulations and its scope in commercialization for the management of pests and diseases,” in PGPR: Biocontrol and Biofertilization (Dordrecht: Springer), 257–296.
Nõlvak, H., Truu, M., Kanger, K., Tampere, M., Espenberg, M., Loit, E., et al. (2016). Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron-integrase genes in agricultural grassland soil. Sci. Total Environ. 562, 678–689. doi: 10.1016/j.scitotenv.2016.04.035
Nordstedt, N. P., and Jones, M. L. (2021). Genomic analysis of Serratia plymuthica MBSA-MJ1: a plant growth promoting Rhizobacteria that improves water stress tolerance in greenhouse ornamentals. Front. Microbiol. 12:653556. doi: 10.3389/fmicb.2021.653556
Oniciuc, E. A., Likotrafiti, E., Alvarez-Molina, A., Prieto, M., Santos, J. A., and Alvarez-Ordóñez, A. (2018). The present and future of whole genome sequencing (WGS) and whole Metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food Chain. Gene 9:268. doi: 10.3390/genes9050268
O’Rourke, S., Wietzorrek, A., Fowler, K., Corre, C., Challis, G. L., and Chater, K. F. (2009). Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol. Microbiol. 71, 763–778. doi: 10.1111/j.1365-2958.2008.06560.x
Owen, D., Williams, A. P., Griffith, G. W., and Withers, P. J. A. (2015). Use of commercial bio-inoculants to increase agricultural production through improved phosphrous acquisition. Appl. Soil Ecol. 86, 41–54. doi: 10.1016/j.apsoil.2014.09.012
Pandey, V., and Chandra, K. (2016). “Agriculturally important microorganisms as biofertilizers: commercialization and regulatory requirements in Asia” in Agriculturally Important Microorganisms: Commercialization and Regulatory Requirements in Asia. eds. H. B. Singh, B. K. Sarma, and C. Keswani (Singapore: Springer Singapore), 133–145.
Pandey, R. P., Srivastava, A. K., Srivastava, A. K., and Ramteke, P. W. (2018). Antibiotic resistance in Mesorhizobium ciceri from eastern Uttar Pradesh. Clim. Change Environ. Sustain. 6, 114–118. doi: 10.5958/2320-642X.2018.00014.5
Parray, J. A., Jan, S., Kamili, A. N., Qadri, R. A., Egamberdieva, D., and Ahmad, P. (2016). Current perspectives on plant growth-promoting Rhizobacteria. J. Plant Growth Regul. 35, 877–902. doi: 10.1007/s00344-016-9583-4
Paterson, J., Jahanshah, G., Li, Y., Wang, Q., Mehnaz, S., and Gross, H. (2016). The contribution of genome mining strategies to the understanding of active principles of PGPR strains. FEMS Microbiol. Ecol. 93, 1–31. doi: 10.1093/femsec/fiw249
Paul, M., Shani, V., Muchtar, E., Kariv, G., Robenshtok, E., and Leibovici, L. (2010). Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob. Agents Chemother. 54, 4851–4863. doi: 10.1128/AAC.00627-10
Paul, S., Singh, P. A. S. S., Rudramurthy, S. M., Chakrabarti, A., and Ghosh, A. K. (2018). Rapid detection of fluconazole resistance in Candida tropicalis by MALDI-TOF MS. Med. Mycol. 56, 234–241. doi: 10.1093/mmy/myx042
Paulsen, I. T., Press, C. M., Ravel, J., Kobayashi, D. Y., Myers, G. S. A., Mavrodi, D. V., et al. (2005). Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23, 873–878. doi: 10.1038/nbt1110
Planchamp, C., Glauser, G., and Mauch-Mani, B. (2015). Root inoculation with pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 5:719. doi: 10.3389/fpls.2014.00719
Plattner, F., and Soldati-Favre, D. (2008). Hijacking of host cellular functions by the Apicomplexa. Annu. Rev. Microbiol. 62, 471–487. doi: 10.1146/annurev.micro.62.081307.162802
Ploy, M. C., Gassama, A., Chainier, D., and Denis, F. (2005). Les intégrons en tant que support génétique de résistance aux antibiotiques. Immun. Analy. Biol. Spécial. 20, 343–352. doi: 10.1016/j.immbio.2005.10.001
Poole, K. (2011). Pseudomonas Aeruginosa: resistance to the max. Front. Microbiol. 2:65. doi: 10.3389/fmicb.2011.00065
Popa, O., and Dagan, T. (2011). Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 14, 615–623. doi: 10.1016/j.mib.2011.07.027
Popowska, M., Rzeczycka, M., Miernik, A., Krawczyk-Balska, A., Walsh, F., and Duffy, B. (2012). Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes. Antimicrob. Agents Chemother. 56, 1434–1443. doi: 10.1128/AAC.05766-11
Ramakrishna, W., Yadav, R., and Li, K. (2019). Plant growth promoting bacteria in agriculture: two sides of a coin. Appl. Soil Ecol. 138, 10–18. doi: 10.1016/j.apsoil.2019.02.019
Reid-Henry, S. (2008). Scientific innovation and non-Western regional economies: Cuban Biotechnology’s ‘experimental milieu’. Environ. Plan. A 40, 1966–1986. doi: 10.1068/a39157
Ren, Y., Ren, Y., Zhou, Z., Guo, X., Li, Y., Feng, L., et al. (2010). Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. J. Bacteriol. 192, 2463–2464. doi: 10.1128/JB.00067-10
Rey, M. W., Ramaiya, P., Nelson, B. A., Brody-Karpin, S. D., Zaretsky, E. J., Tang, M., et al. (2004). Complete genome sequence of the industrial bacterium bacillus licheniformis and comparisons with closely related Bacillusspecies. Genome Biol. 5:r77. doi: 10.1186/gb-2004-5-10-r77
Reyes, P. I., Yang, K., Zheng, A., Li, R., Li, G., Lu, Y., et al. (2017a). Dynamic monitoring of antimicrobial resistance using magnesium zinc oxide nanostructure-modified quartz crystal microbalance. Biosens. Bioelectron. 93, 189–197. doi: 10.1016/j.bios.2016.09.011
Reyes, P. I., Yang, K., Zheng, A., Li, R., Li, G., Lu, Y., et al. (2017b). Magnesium zinc oxide nanostructure-modified quartz crystal microbalance for dynamic monitoring of antibiotic effects and antimicrobial resistance. Proc. Technol. 27, 46–47. doi: 10.1016/j.protcy.2017.04.022
Riber, L., Poulsen, P. H., Al-Soud, W. A., Skov Hansen, L. B., Bergmark, L., Brejnrod, A., et al. (2014). Exploring the immediate and long-term impact on bacterial communities in soil amended with animal and urban organic waste fertilizers using pyrosequencing and screening for horizontal transfer of antibiotic resistance. FEMS Microbiol. Ecol. 90, 206–224. doi: 10.1111/1574-6941.12403
Rodgers, K., McLellan, I., Peshkur, T., Williams, R., Tonner, R., Hursthouse, A. S., et al. (2019). Can the legacy of industrial pollution influence antimicrobial resistance in estuarine sediments? Environ. Chem. Lett. 17, 595–607. doi: 10.1007/s10311-018-0791-y
Rowan, N. J. (2004). Viable but non-culturable forms of food and waterborne bacteria: quo Vadis? Trends Food Sci. Technol. 15, 462–467. doi: 10.1016/j.tifs.2004.02.009
Salunkhe, P., Smart, C. H., Morgan, J. A. W., Panagea, S., Walshaw, M. J., Hart, C. A., et al. (2005). A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J. Bacteriol. 187, 4908–4920. doi: 10.1128/JB.187.14.4908-4920.2005
Santoyo, G., Orozco-Mosqueda, M. D. C., and Govindappa, M. (2012). Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of bacillus and pseudomonas: a review. Biocontrol Sci. Tech. 22, 855–872. doi: 10.1080/09583157.2012.694413
Scaccia, N., Vaz-Moreira, I., and Manaia, C. M. (2021). The risk of transmitting antibiotic resistance through endophytic bacteria. Trends Plant Sci. 26, 1213–1226. doi: 10.1016/j.tplants.2021.09.001
Schmidt, K., Mwaigwisya, S., Crossman, L. C., Doumith, M., Munroe, D., Pires, C., et al. (2017). Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 72, 104–114. doi: 10.1093/jac/dkw397
Seedy, F. R. E., Samy, A. A., Salam, H. S. H., Khairy, E. A., and Koraney, A. A. (2017). Polymerase chain reaction detection of genes responsible for multiple antibiotic resistance Staphylococcus aureus isolated from food of animal origin in Egypt. Vet World 10, 1205–1211. doi: 10.14202/vetworld.2017.1205-1211
Sekine, M., Tanikawa, S., Omata, S., Saito, M., Fujisawa, T., Tsukatani, N., et al. (2006). Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 8, 334–346. doi: 10.1111/j.1462-2920.2005.00899.x
Séveno, N. A., Kallifidas, D., Smalla, K., van Elsas, J. D., Collard, J.-M., Karagouni, A. D., et al. (2002). Occurrence and reservoirs of antibiotic resistance genes in the environment. Rev. Res. Med. Microbiol. 13, 15–27. doi: 10.1097/00013542-200201000-00002
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2:587. doi: 10.1186/2193-1801-2-587
Sharma, P., Tomar, S. K., Goswami, P., Sangwan, V., and Singh, R. (2014). Antibiotic resistance among commercially available probiotics. Food Res. Int. 57, 176–195. doi: 10.1016/j.foodres.2014.01.025
Silini, A., Silini-Cherif, H., and Ghoul, M. (2012). Effect of Azotobacter vinelandii and compatible solutes on germination wheat seeds and root concentrations of sodium and potassium under salt stress. Pak. J. Biol. Sci. 15, 132–140. doi: 10.3923/pjbs.2012.132.140
Singer, A. C., Shaw, H., Rhodes, V., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7:1728. doi: 10.3389/fmicb.2016.01728
Singh, A. K., and Cameotra, S. S. (2013). Rhamnolipids production by multi-metal-resistant and plant-growth-promoting Rhizobacteria. Appl. Biochem. Biotechnol. 170, 1038–1056. doi: 10.1007/s12010-013-0244-9
Singh, R. P., and Jha, P. N. (2017). The PGPR Stenotrophomonas maltophilia SBP-9 augments resistance against biotic and abiotic stress in wheat plants. Front. Microbiol. 8:1945. doi: 10.3389/fmicb.2017.01945
Smillie, C. S., Smith, M. B., Friedman, J., Cordero, O. X., David, L. A., and Alm, E. J. (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244. doi: 10.1038/nature10571
Smits, T. H., Rezzonico, F., Blom, J., Goesmann, A., Abelli, A., Morelli, R. K., et al. (2015). Draft genome sequence of the commercial biocontrol strain Pantoea agglomerans P10c. Genome Announc. 3:e01448-15. doi: 10.1128/genomeA.01448-15
Sommer, M. O., Church, G. M., and Dantas, G. (2010). The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence 1, 299–303. doi: 10.4161/viru.1.4.12010
Souza, R. D., Ambrosini, A., and Passaglia, L. M. (2015). Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 38, 401–419. doi: 10.1590/S1415-475738420150053
Stackebrandt, E., Frederiksen, W., Garrity, G. M., Grimont, P. A. D., Kämpfer, P., Maiden, M. C. J., et al. (2002). Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52, 1043–1047. doi: 10.1099/00207713-52-3-1043
Steil, L., Hoffmann, T., Budde, I., Völker, U., and Bremer, E. (2003). Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185, 6358–6370. doi: 10.1128/JB.185.21.6358-6370.2003
Stokes, H. T., and Hall, R. M. (1989). A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3, 1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x
Stokes, H. W., Holmes, A. J., Nield, B. S., Holley, M. P., Nevalainen, K. M. H., Mabbutt, B. C., et al. (2001). Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 67, 5240–5246. doi: 10.1128/AEM.67.11.5240-5246.2001
Stokes, H. W., Nesbø, C. L., Holley, M., Bahl, M. I., Gillings, M. R., and Boucher, Y. (2006). Class 1 integrons potentially predating the association with tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 188, 5722–5730. doi: 10.1128/JB.01950-05
Su, J. Q., Wei, B., Xu, C. Y., Qiao, M., and Zhu, Y. G. (2014). Functional metagenomic characterization of antibiotic resistance genes in agricultural soils from China. Environ. Int. 65, 9–15. doi: 10.1016/j.envint.2013.12.010
Sun, D. (2018). Pull in and push out: mechanisms of horizontal gene transfer in bacteria. Front. Microbiol. 9:2154. doi: 10.3389/fmicb.2018.02154
Syed Ab Rahman, S. F., Singh, E., Pieterse, C. M. J., and Schenk, P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 267, 102–111. doi: 10.1016/j.plantsci.2017.11.012
Tabassum, B., Khan, A., Tariq, M., Ramzan, M., Iqbal Khan, M. S., Shahid, N., et al. (2017). Bottlenecks in commercialisation and future prospects of PGPR. Appl. Soil Ecol. 121, 102–117. doi: 10.1016/j.apsoil.2017.09.030
Tan, S., Dvorak, C. M. T., Estrada, A. A., Gebhart, C., Marthaler, D. G., and Murtaugh, M. P. (2020). MinION sequencing of Streptococcus suis allows for functional characterization of bacteria by multilocus sequence typing and antimicrobial resistance profiling. J. Microbiol. Methods 169:105817. doi: 10.1016/j.mimet.2019.105817
Tariq, M., Jameel, F., Ijaz, U., Abdullah, M., and Rashid, K. (2022). Biofertilizer microorganisms accompanying pathogenic attributes: a potential threat. Physiol. Mol. Biol. Plants 28, 77–90. doi: 10.1007/s12298-022-01138-y
Taxt, A. M., Avershina, E., Frye, S. A., Naseer, U., and Ahmad, R. (2020). Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Sci. Rep. 10:7622. doi: 10.1038/s41598-020-64616-x
Than, C., Jin, G., and Nakhleh, L. (2008). “Integrating sequence and topology for efficient and accurate detection of horizontal gene transfer” in Comparative Genomics. eds. C. E. Nelson and S. Vialette (Berlin Heidelberg: Springer), 113–127.
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., and Polasky, S. (2002). Agricultural sustainability and intensive production practices. Nature 418, 671–677. doi: 10.1038/nature01014
Topp, E., Monteiro, S. C., Beck, A., Coelho, B. B., Boxall, A. B. A., Duenk, P. W., et al. (2008). Runoff of pharmaceuticals and personal care products following application of biosolids to an agricultural field. Sci. Total Environ. 396, 52–59. doi: 10.1016/j.scitotenv.2008.02.011
Torres, D., Revale, S., Obando, M., Maroniche, G., Paris, G., Perticari, A., et al. (2015). Genome sequence of Bradyrhizobium japonicum E109, one of the most agronomically used nitrogen-fixing rhizobacteria in Argentina. Genome Announc. 3:e01566-14. doi: 10.1128/genomeA.01566-14
Toruño, T. Y., Stergiopoulos, I., and Coaker, G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. doi: 10.1146/annurev-phyto-080615-100204
Trivedi, P., Pandey, A., Palni, L.M.S., Bag, N., and Tamang, M. (2004). Colonization of rhizosphere of tea by growth promoting bacteria.
Trujillo, M. E. (2011). Taxonomic subcommittees and minimal standards for the description of prokaryotes. Microbiol. Aust. 32, 64–65. doi: 10.1071/MA11064
Van Goethem, M., Pierneef, R., Bezuidt, O., Van De Peer, Y., Cowan, D., and Makhalanyane, T. (2018). A reservoir of ‘historical’antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0424-5
Vasala, A., Hytönen, V. P., and Laitinen, O. H. (2020). Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell. Infect. Microbiol. 10:308. doi: 10.3389/fcimb.2020.00308
Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., and Nasrulhaq Boyce, A. (2016). Role of plant growth promoting Rhizobacteria in agricultural sustainability—a review. Molecules 21:573. doi: 10.3390/molecules21050573
Vélez, J. R., Cameron, M., Rodríguez-Lecompte, J. C., Xia, F., Heider, L. C., Saab, M., et al. (2017). Whole-genome sequence analysis of antimicrobial resistance genes in streptococcus uberis and streptococcus dysgalactiae isolates from Canadian dairy herds. Front. Vet. Sci. 4:63. doi: 10.3389/fvets.2017.00063
Vikesland, P. J., Pruden, A., Alvarez, P. J. J., Aga, D., Bürgmann, H., Li, X. D., et al. (2017). Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environ. Sci. Technol. 51, 13061–13069. doi: 10.1021/acs.est.7b03623
Wang, K., Li, S., Petersen, M., Wang, S., and Lu, X. (2018). Detection and characterization of antibiotic-resistant bacteria using surface-enhanced Raman spectroscopy. Nano 8:762. doi: 10.3390/nano8100762
Wang, F.-H., Qiao, M., Su, J.-Q., Chen, Z., Zhou, X., and Zhu, Y.-G. (2014). High throughput profiling of antibiotic resistance genes in Urban Park soils with reclaimed water irrigation. Environ. Sci. Technol. 48, 9079–9085. doi: 10.1021/es502615e
Wang, Q., Xiong, D., Zhao, P., Yu, X., Tu, B., and Wang, G. (2011). Effect of applying an arsenic-resistant and plant growth–promoting rhizobacterium to enhance soil arsenic phytoremediation by Populus deltoides LH05-17. J. Appl. Microbiol. 111, 1065–1074. doi: 10.1111/j.1365-2672.2011.05142.x
Wani, P. A., and Khan, M. S. (2013). Nickel detoxification and plant growth promotion by multi metal resistant plant growth promoting rhizobium species RL9. Bull. Environ. Contam. Toxicol. 91, 117–124. doi: 10.1007/s00128-013-1002-y
Waseem, H., Jameel, S., Ali, J., Saleem Ur Rehman, H., Tauseef, I., Farooq, U., et al. (2019). Contributions and challenges of high throughput qPCR for determining antimicrobial resistance in the environment: a critical review. Molecules 24:163. doi: 10.3390/molecules24010163
Waseem, H., Saleem Ur Rehman, H., Ali, J., Iqbal, M. J., and Ali, M. I. (2020). “Chapter 14—Global trends in ARGs measured by HT-qPCR platforms,” in Antibiotics and Antimicrobial Resistance Genes in the Environment. ed. M. Z. Hashmi (Amsterdam: Elsevier), 206–222.
Weilharter, A., Mitter, B., Shin, M. V., Chain, P. S., Nowak, J., and Sessitsch, A. (2011). Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. Am. Soc. Microbiol. 193, 3383–3384. doi: 10.1128/JB.05055-11
Welker, M., and van Belkum, A. (2019). One system for all: is mass spectrometry a future alternative for conventional antibiotic susceptibility testing? Front. Microbiol. 10:2711. doi: 10.3389/fmicb.2019.02711
Wellington, E. M. H., Boxall, A. B. A., Cross, P., Feil, E. J., Gaze, W. H., Hawkey, P. M., et al. (2013). The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 13, 155–165. doi: 10.1016/S1473-3099(12)70317-1
Wepking, C., Avera, B., Badgley, B., Barrett, J. E., Franklin, J., Knowlton, K. F., et al. (2017). Exposure to dairy manure leads to greater antibiotic resistance and increased mass-specific respiration in soil microbial communities. Proc. R. Soc. B Biol. Sci. 284:20162233. doi: 10.1098/rspb.2016.2233
Wisniewski-Dye, F., Borziak, K., Khalsa-Moyers, G., Alexandre, G., Sukharnikov, L. O., Wuichet, K., et al. (2011). Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet. 7:e1002430. doi: 10.1371/journal.pgen.1002430
Xiong, W., Wang, M., Dai, J., Sun, Y., and Zeng, Z. (2018). Application of manure containing tetracyclines slowed down the dissipation of tet resistance genes and caused changes in the composition of soil bacteria. Ecotoxicol. Environ. Saf. 147, 455–460. doi: 10.1016/j.ecoenv.2017.08.061
Xu, L., Chen, H., Canales, M., and Ciric, L. (2019). Use of synthesized double-stranded gene fragments as qPCR standards for the quantification of antibiotic resistance genes. J. Microbiol. Methods 164:105670. doi: 10.1016/j.mimet.2019.105670
Yang, L.-Y., Zhou, S.-Y.-D., Lin, C.-S., Huang, X.-R., Neilson, R., and Yang, X.-R. (2022). Effects of biofertilizer on soil microbial diversity and antibiotic resistance genes. Sci. Total Environ. 820:153170. doi: 10.1016/j.scitotenv.2022.153170
Yasmin, F., Othman, R., Sijam, K., and Saad, M. S. (2009). Characterization of beneficial properties of plant growth-promoting rhizobacteria isolated from sweet potato rhizosphere. Afr. J. Microbiol. Res. 3, 815–821. doi: 10.5897/AJMR.9000151
Ye, M., Sun, M., Feng, Y., Wan, J., Xie, S., Tian, D., et al. (2016). Effect of biochar amendment on the control of soil sulfonamides, antibiotic-resistant bacteria, and gene enrichment in lettuce tissues. J. Hazard. Mater. 309, 219–227. doi: 10.1016/j.jhazmat.2015.10.074
Zahir, Z., Munir, A., Asghar, H., Shaharoona, B., and Arshad, M. (2008). Effectiveness of rhizobacteria containing ACC deaminase for growth promotion of peas (Pisum sativum) under drought conditions. J. Microbiol. Biotechnol. 18, 958–963.
Zanoli, L. M., and Spoto, G. (2013). Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Bios 3, 18–43. doi: 10.3390/bios3010018
Zhang, G., and Feng, J. (2016). The intrinsic resistance of bacteria. Hereditas 38, 872–880. doi: 10.16288/j.yczz.16-159
Zhang, W., Wang, H., Zhang, R., Yu, X.-Z., Qian, P.-Y., and Wong, M. H. (2010). Bacterial communities in PAH contaminated soils at an electronic-waste processing center in China. Ecotoxicology 19, 96–104. doi: 10.1007/s10646-009-0393-3
Zhao, S., Tyson, G. H., Chen, Y., Li, C., Mukherjee, S., Young, S., et al. (2016). Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in campylobacter spp. Appl. Environ. Microbiol. 82, 459–466. doi: 10.1128/AEM.02873-15
Zhu, L.-X., Zhang, Z.-W., Wang, C., Yang, H.-W., Jiang, D., Zhang, Q., et al. (2007). Use of a DNA microarray for simultaneous detection of antibiotic resistance genes among staphylococcal clinical isolates. J. Clin. Microbiol. 45, 3514–3521. doi: 10.1128/JCM.02340-06
Keywords: antibiotic resistance genes (ARGs), biofertilizers, gene invasion risks, plant growth promoting bacteria (PGPB), public health
Citation: Mahdi I, Fahsi N, Hijri M and Sobeh M (2022) Antibiotic resistance in plant growth promoting bacteria: A comprehensive review and future perspectives to mitigate potential gene invasion risks. Front. Microbiol. 13:999988. doi: 10.3389/fmicb.2022.999988
Received: 21 July 2022; Accepted: 25 August 2022;
Published: 20 September 2022.
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Tanvir Kaur, Eternal University, IndiaCopyright © 2022 Mahdi, Fahsi, Hijri and Sobeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed Hijri, bW9oYW1lZC5oaWpyaUB1bTZwLm1h; Mansour Sobeh, bWFuc291ci5zb2JlaEB1bTZwLm1h
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.