
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Microbiol. , 03 November 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.999783
 Luna Colagrossi1†
Luna Colagrossi1† Valentino Costabile1†
Valentino Costabile1† Rossana Scutari1,2
Rossana Scutari1,2 Valeria Cento3,4
Valeria Cento3,4 Luana Coltella1
Luana Coltella1 Antonino Reale5
Antonino Reale5 Martina Scilipoti5
Martina Scilipoti5 Alberto Villani5
Alberto Villani5 Claudia Alteri1,2
Claudia Alteri1,2 Carlo Federico Perno1*
Carlo Federico Perno1* Cristina Russo1
Cristina Russo1The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has increased the need to identify additional rapid diagnostic tests for an accurate and early diagnosis of infection. Here, we evaluated the diagnostic performance of the cartridge-based reverse transcription polymerase chain reaction (RT-PCR) test STANDARD M10 SARS-CoV-2 (SD Biosensor Inc., Suwon, South Korea), targeting the ORF1ab and E gene of SARS-CoV-2, and which can process up to eight samples in parallel in 60 min. From January 2022 to March 2022, STANDARD™ M10 assay performance was compared with Xpert® Xpress SARS-CoV-2 (Cepheid, Sunnyvale CA) on 616 nasopharyngeal swabs from consecutive pediatric (N = 533) and adult (N = 83) patients presenting at the “Istituto di Ricovero e Cura a Carattere Scientifico” (IRCCS) Ospedale Pediatrico Bambino Gesù, Roma. The overall performance of STANDARD M10 SARS-CoV-2 was remarkably and consistently comparable to the Xpert® Xpress SARS-CoV-2 with an overall agreement of 98% (604/616 concordant results), and negligible differences in time-to-result (60 min vs. 50 min, respectively). When the Xpert® Xpress SARS-CoV-2 results were considered as the reference, STANDARD™ M10 SARS-CoV-2 had 96.5% sensitivity and 98.4% specificity. STANDARD M10 SARS-CoV-2 can thus be safely included in diagnostic pathways because it rapidly and accurately identifies SARS-CoV-2 present in nasopharyngeal swabs.
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been the most significant global outbreak since the Spanish flu of 1918 (Petersen et al., 2020; Burkhard-Koren et al., 2021; Javelle and Raoult, 2021).
Healthcare systems around the world have been challenged by the SARS-CoV-2 pandemic (Wang et al., 2020) and to date, the world counts 6,290,452 deaths from the pandemic (WHO, 2022).
However, the ability of the virus to mutate, as evidenced by a succession of variants of concerns (VOCs), has posed a further challenge to the diagnosis of infection, vaccine development, and clinical treatments (Bian et al., 2021; Chen J. et al., 2021; Chen R. E. et al., 2021; Koyama et al., 2022; McLean et al., 2022).
In the fight against COVID-19, a key role has always been played by diagnosis (WHO Working Group on the Clinical Characterisation and Management of Covid-19 infection, 2020; Peeling et al., 2021). In fact, both at the health facility and community levels, diagnosis of the virus is the first and fundamental step toward its containment. During pandemic, children have been supposed to actively spread SARS-CoV-2 in elderly population and thus many concerns arose about the ability to accurately detect eventual children-related viral variants by PCR methods, previous study demonstrated that genomic SARS-CoV-2 variability was similar across various age categories (Alteri et al., 2022). In line with their findings, we decided to address this aspect by performing an assay comparison in our pediatric clinical setting.
Currently, the field of COVID-19 diagnostic testing is rapidly evolving; diagnostics capable of detecting current infections are antigen- and molecular-based tests. Antigenic tests, which identify viral proteins mostly expressed within nucleocapsid, are far less sensitive but faster than molecular tests, providing the result in about 15–20 min (Dinnes et al., 2021; Pilarowski et al., 2021). However, molecular tests can be performed in batches or as random-access on-demand. Batch tests have high capacities for simultaneous processing (96–200 samples in parallel) but typically have long turn-around-times (4–5 h) (Craney et al., 2020; Gorzalski et al., 2020), favoring their use in cases of large volumes of samples with a low clinical necessity for prompt results (i.e., community screening). Moreover, on-demand tests can process a limited number of samples but provide results in 50 min–2 h (Loeffelholz et al., 2020; Smithgall et al., 2020). Therefore, they are usually dedicated to emergency situations in both community and hospital settings.
Considering the moderate sensitivity of antigenic tests compared to reverse transcription polymerase chain reaction (RT-PCR), the need for molecular tests, which is more rapid and sensitive, fits in.
The STANDARD M10 SARS-CoV-2 (SD Biosensor) is a new cartridge-based, rapid, on-demand RT-PCR test, recently introduced in clinical use, that targets the ORF1ab and E gene of SARS-CoV-2. It runs on the modular STANDARD™ M10 instrument, scalable up to eight modules, and provides results in 20–60 min. Previous studies showed a high diagnostic accuracy (100%) of the STANDARD M10 assay in testing both nasopharyngeal swabs and lower respiratory tract specimens (Colagrossi et al., 2021), with a consistent sensitivity for ≥ 1 gene, the ORF1ab gene, and the E gene (100, 95.5, and 99.5%, respectively) and a full specificity of 100% (Domnich et al., 2022; Ham et al., 2022).
To date, all performance evaluations were achieved on samples from adult individuals. Here, to evaluate the diagnostic performance of this novel assay for the rapid molecular diagnosis of COVID-19 in both pediatric and adult populations, we performed a head-to-head comparison between STANDARD M10 SARS-CoV-2 and the Xpert Xpress SARS-CoV-2, (Cepheid, Sunnyvale, CA), which has analogous similar technical characteristics and is currently recognized as one of the most sensitive tests on the market (Loeffelholz et al., 2020; Smithgall et al., 2020; Serei et al., 2021). Droplet digital PCR (ddPCR), an end-point quantitative PCR method capable of detecting down to a few copies of viral genomes (Alteri et al., 2020; Liu et al., 2020; Suo et al., 2020; Colagrossi et al., 2021), was used to provide additional insights into the ability of rapid cartridge tests to detect up to minimal quantities of SARS-CoV-2 genomes.
Between January 2022 and March 2022, 616 nasopharyngeal swabs were collected by qualified personnel from consecutive individuals (pediatric patients, their legal tutor or Healthcare Workers) who accessed the Emergency Room of the Ospedale Pediatrico (Rome, Italy) and were immediately transported to the Microbiology Laboratory in 3 mL viral transport medium (VTM) tubes (Copan UTM®, Copan, Italy). All samples were tested upon collection with the Xpert Xpress SARS-CoV-2 assay, according to laboratory routine procedure, and with the STANDARD M10 SARS-CoV-2 assay within the next 24 h. In the time between the two tests, samples were conserved at +4°C, according to manufacturer indications. Residual samples were stored at −80°C, and left at the disposal for further analyses, whether deemed necessary.
The STANDARD™ M10 SARS-CoV-2 cartridge contains PCR buffers and lyophilized real-time RT-PCR reagents. According to the manufacturer’s indications, users should insert 600 μl of sample’s buffer transport medium directly into the cartridge and load it into one module of the STANDARD™ M10 instrument, which allows continuous, random-access loading of up to eight samples. The instrument provides negative or presumptive positive results (amplification of E gene only) automatically in 60 min, while positive results (amplification of both targets) are available in 30–60 min, depending on the viral load. At the end of RT-PCR reaction, the amplification curves and PCR cycle threshold (Ct) values (upper limit of detection, Ct < 40) are released.
The Xpert Xpress SARS-CoV-2 is a cartridge-based test that detects the envelope (E) and the nucleocapsid (N2) genomic portions of SARS-CoV-2. The cartridge contains PCR buffers and lyophilized real-time RT-PCR reagents. Users should add 300 μl of sample’s buffer transport medium directly to the cartridge and load it into the GeneXpert instrument, which allows continuous, random-access loading of up to 16 samples. The instrument provides results automatically in 50 min. The results are reported as presumptive positive in the case of identification of the E gene alone, while the detection of N2 always leads to a positive result. At the end of the RT-PCR reaction, the amplification curves and PCR Ct values (upper limit of detection, Ct < 45) are released.
At the end of the study, in case of non-concordant results between the two real-time assays, we exploited samples residuals (conserved at −80°C) to obtain a SARS-CoV-2 RNA absolute quantification by using QX200 Droplet Digital PCR System (Bio-Rad) and targeting RNA-dependent RNA polymerase (RdRp) gene in conserved regions (Alteri et al., 2020). ddPCR targets are amplified and detected using fluorescence-based probes. The fraction of positive droplets is fitted into a Poisson distribution to determine the absolute starting copy number in units of copies/μl of the input sample. We defined a sample as “positive and quantifiable” if at least three droplets were detected.
The performance of diagnostic tools has been compared from both a quantitative and a qualitative point of view. The qualitative comparison was conducted by computing a confusion matrix in order to feed performance descriptive indexes calculation, and Xpert Xpress SARS-CoV-2 system’s results were considered as reference. The differential distribution of false-negative and false-positive responses among system results was also checked with a McNemar’s test under the null hypothesis of a different distribution in false response; this test was included because it considers the impact of both false-positive and false-negative response rates simultaneously and returns us a degree of balance between them. Quantitative divergence in Ct’s distribution between systems was statistically evaluated by a paired Student’s t-test under the null hypothesis that no difference was observed in their mean distribution.
To further assess the overall mean distribution between the two approaches without pairing samples measurement, we relied on the Mann–Whitney test under the null hypothesis that divergence was observed among mean distributions.
To further evaluate the overlapping degree between these assays, we performed Bland-Altman test, a popular method to test the agreement between two set of paired measurement which is expressed as a mean bias flanked by an upper and lower limit of agreement.
Moreover, to assess the correlation between age and the detection of SARS-CoV-2 Ct value between the two employed methods, the study population was stratified into five categories according to age: newborns (0–27 days), infants and toddlers (28 days–24 months), children (2–11 years), adolescents (12–17 years), and adults (> 18 years) (Williams et al., 2012).
The ethical committee of the Bambino Gesù Children’s Hospital has been notified of the present study. The use of retained clinical specimens for validation of diagnostic methods without individual data and the use of anonymized clinical specimens for validation of diagnostic methods did not require ethical clearance from the ethics committee.
Between January 2022 and March 2022, we collected and analyzed 616 nasopharyngeal swabs, mainly from pediatric patients (533/616, 86.5%), as we included samples from 27 (4.4%) newborns (0–27 days), 256 (41.6%) infants and toddlers (28 days–24 months), 177 (28.7%) children (2–11 years), and 73 (11.8%) adolescents (12–17 years), in addition to 83 (13.5%) adults (> 18 years).
When Xpress SARS-CoV-2 assay was used as a reference, 115/616 (18.7%) samples provided a positive result with median (IQR) Cts of 19.5 (17.0–23.5), while 501 (81.3%) were negative (Table 1). Confusion matrix reported an overall concordance rate with STANDARD M10 SARS-CoV-2 assay of 98.5% (604/616) (Table 1), consistently maintained across different Ct categories: ≤ 24 (100% concordance; 90/90); 25–29 (100% concordance; 11/11); and ≥ 30 (71.4% concordance; 10/14). STANDARD M10 SARS-CoV-2 assay returned a 96.5% sensitivity and a 98.4% specificity, with a positive predictive value (PPV) of 93.3% (Table 1). False-positive and negative rates have been estimated as being both below 0.05, supported also by McNemar’s chi-squared estimated equal to 0.75 with a p-value of 0.386, suggesting that no significant divergence was observed in false response distribution (false positive and false negative) (Table 2).
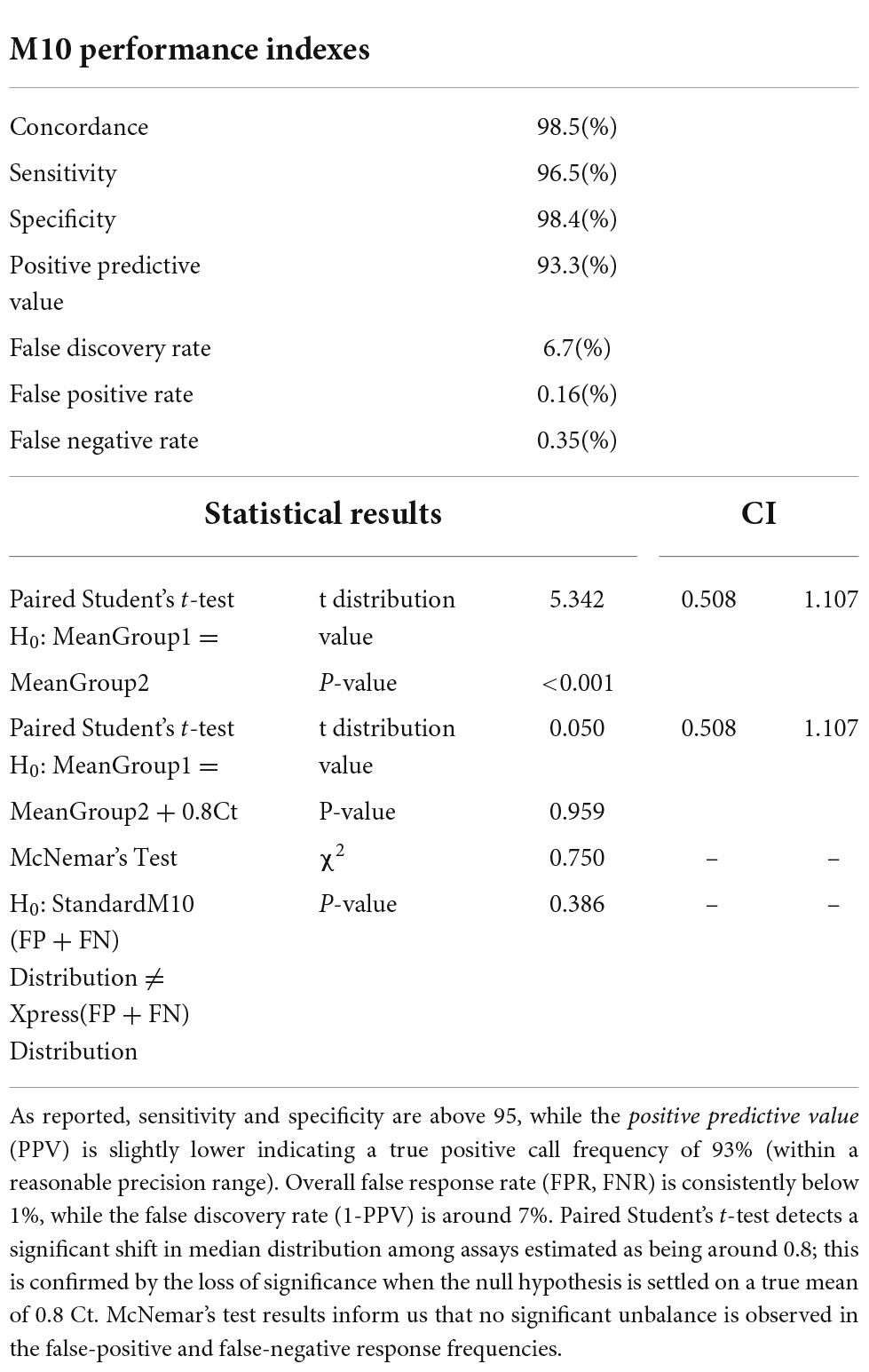
Table 2. STANDARD M10 performance indexes when compared to Xpress SARS-CoV-2 assay used as reference.
Median (IQR) Ct values of the 111 concordant positive samples were comparable between the two assays [19.4 (17.0–22.8) with Xpress SARS-CoV-2 vs. 18.7 (15.9–22.6) with STANDARD M10 SARS-CoV-2; p = 0.106], and Student’s t-test indicates a difference in mean distribution estimated around 0.8 Ct. This correspondence was preserved across all age groups analyzed, from newborns to adults, with no significant difference detected (Table 3). As for previous statistical test applied in this study, this result has been also confirmed by Bland Altman test (Figure 1), showing an average bias between two assays estimated around 0.8 Ct; under a clinical point of view this value slightly impact the discrimination between a positive and negative sample. Furthermore, above the clinical threshold of 30 Ct we observed a good degree of concordance given that most of the samples lay in proximity of median difference (blue bend) while only two (along green band, in range 30–32 Ct) present a positive difference in mean that could bring their values outside the limit of detection. Below this threshold we observe the most of the samples lying in the range 10–25 Ct, in this range the extent of limit of agreement (−2.2862–3.9015) slightly impact diagnostic decision given that, in worst case, a positive difference of 4 Ct would maintain the sample inside the Limit of detection for positivity.
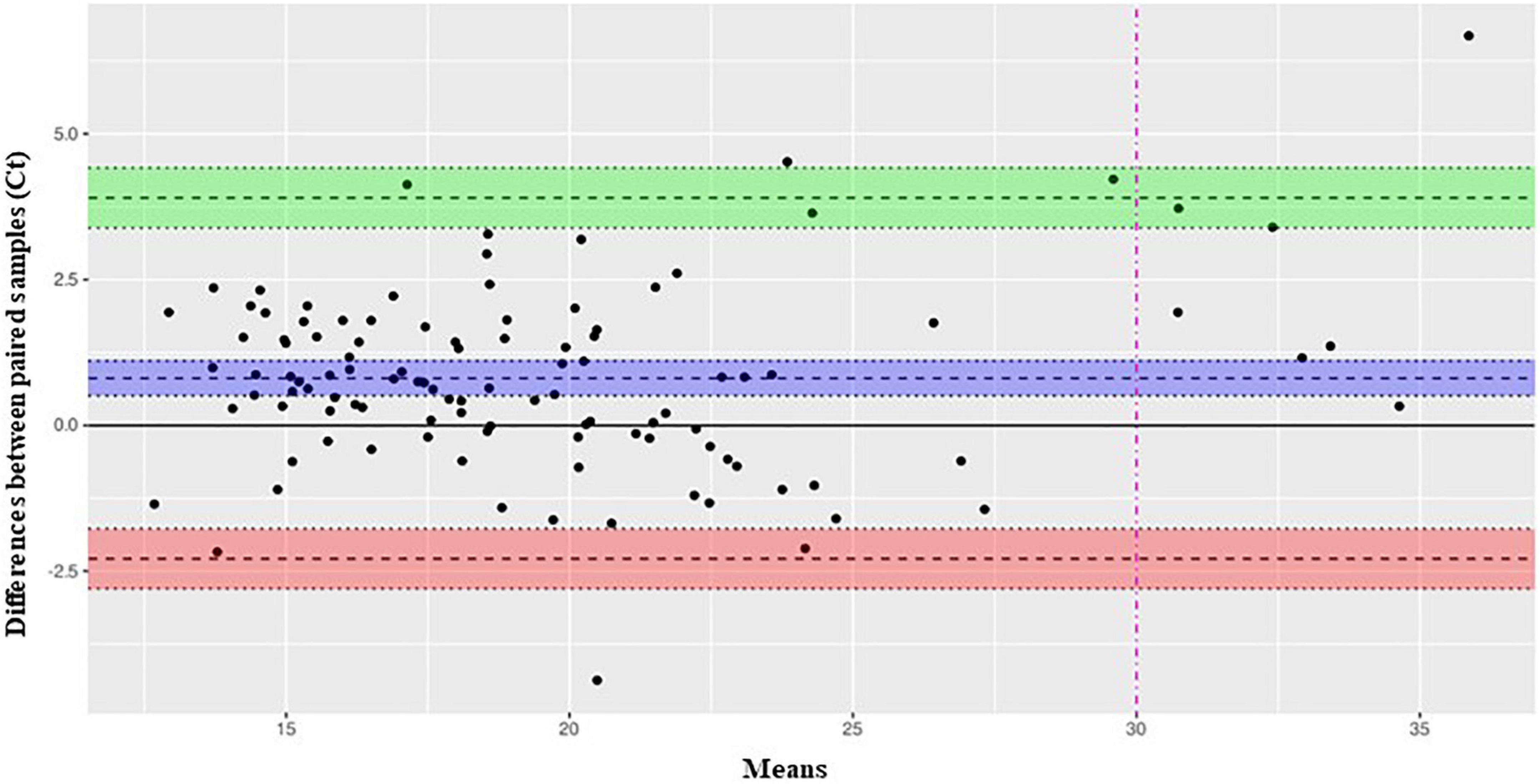
Figure 1. Bland-Altman-plot analysis of SARS-CoV-2 Ct levels measured by Xpress SARS-CoV-2 and STANDARD MIO SARS-CoV-2 assays. Bland-Altman plot analysis of 109 positive samples tested with both assays. On y-axis are reported the differences between paired samples while on x-axis their means. Limit of agreement (+1.96 SD) is represented by a green (upper limit) and red (lower limit) bands. The midline bend (blue) represent the mean bias observed between the assay estimated around 0.8 Ct.
The density distribution of mean Ct values confirmed no consistent variation between the two assays (Figure 2). We observed decay in signal density around 30 Ct for Xpress SARS-CoV-2 assay, while STANDARD M10 retained a flat trend; further to this point, the signal of the former shows a longer tail, spanning up to ∼40 Ct, while the latter shows a rapid decay of around 35 Ct. These results suggest that, while Xpress SARS-CoV-2 assay shows a higher limit of detection, STANDARD M10 presents a more linear performance of around 30 Ct.
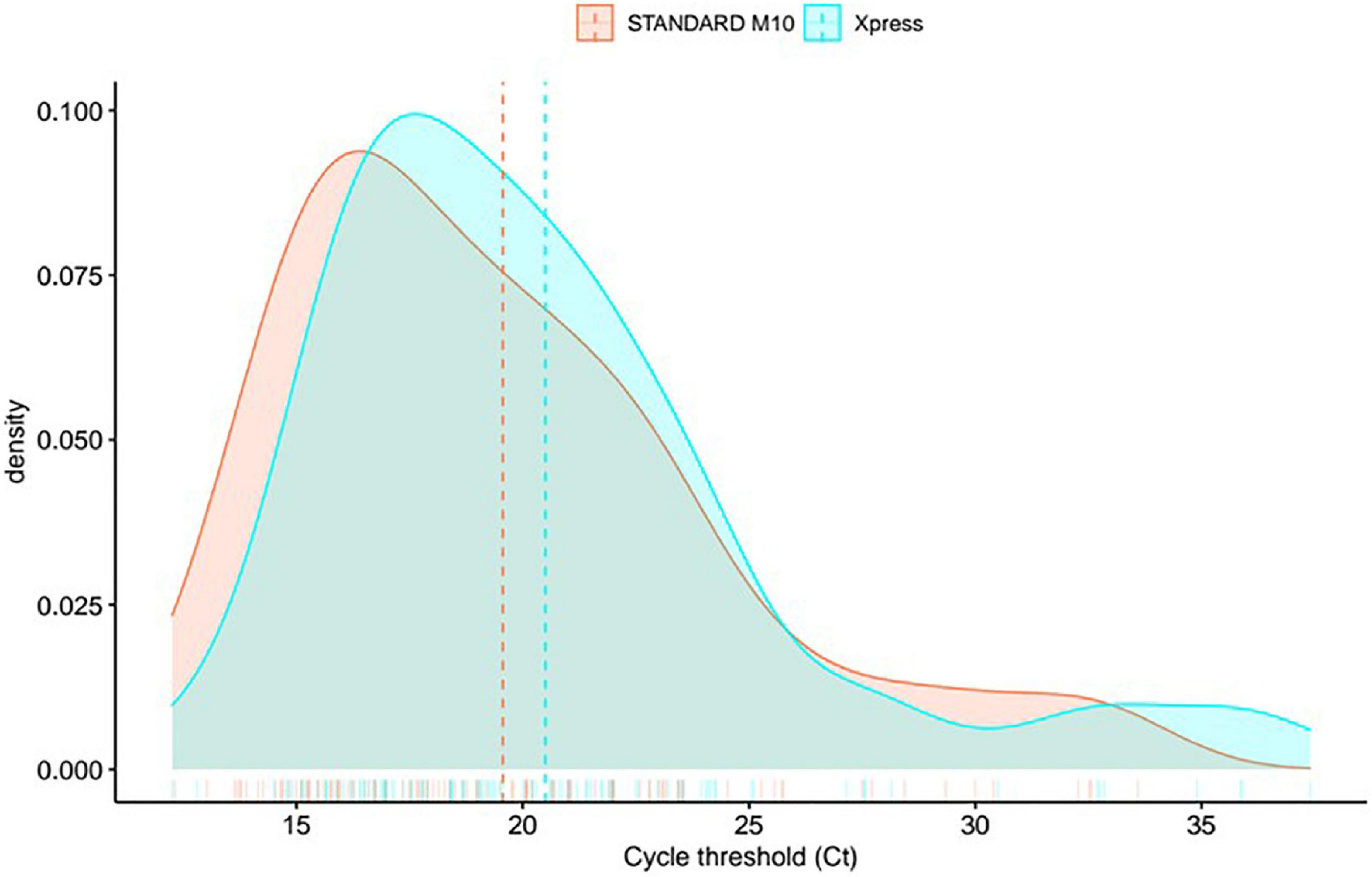
Figure 2. Density distribution in mean gene Ct values between two systems (mean between gene E and N for Xpress SARS-CoV-2, gene E and Orflab for STANDARD M10 SARS-CoV-2).
To visualize mean gene Ct distribution among samples, we exploited the violin plots (Figure 3). This kind of chart provides double information by simultaneously reporting Ct distribution for both assays and population density represented by violin width.
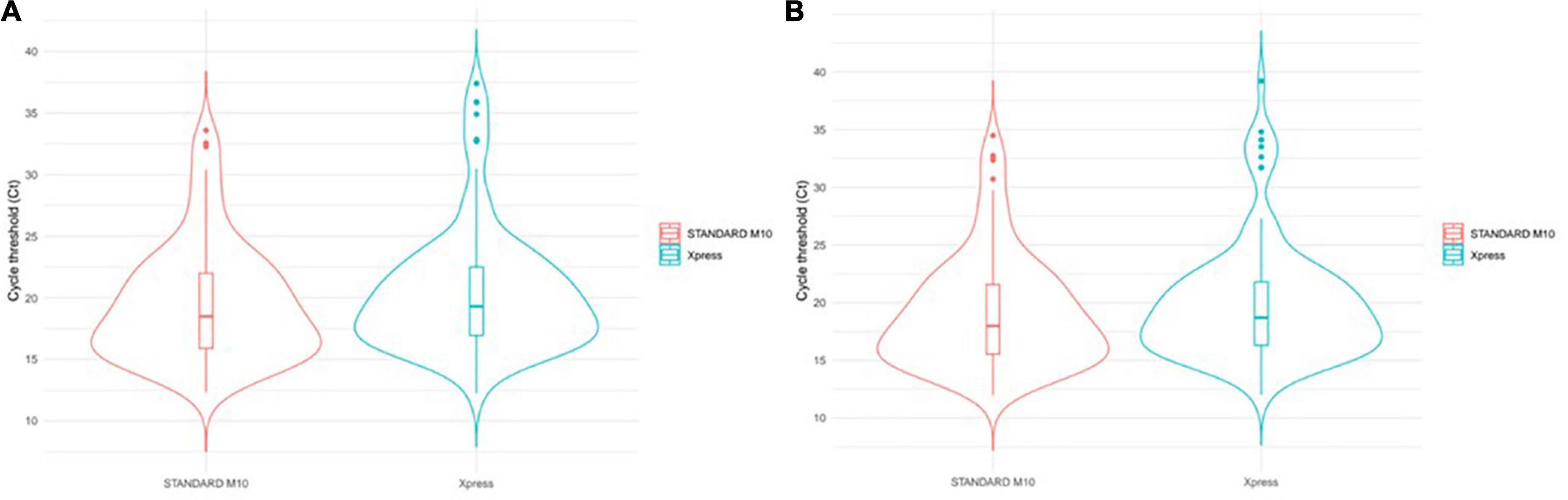
Figure 3. SARS-CoV-2 Ct distribution comparison. Violin width informs about the number of elements observed at any Ct level. Upper dots can be considered as outliers since far from the mam population. Internal boxplots provide a dimension of IQR and median values. Among samples tested with two systems, violin plots compare: (A) Mean gene Ct value distribution; (B) E gene Ct value distribution.
By visually inspecting this chart, we observed that the highest population density for both methods, represented by the largest level of the violin, overlaps each other. This is also confirmed by the internal boxplot plot, whose extent is highly comparable (Figure 3), and by the Mann–Whitney test, which returned a not significant p-value of 0.239.
For 11 out of the 12 discordant results, the volume of the residual sample was sufficient for absolute SARS-CoV-2 load quantification by ddPCR. This further investigation allowed us to detect SARS-CoV-2 genetic material in 2 out of 7 (28.6%) of the Xpress negative/STANDARD M10 positive (false positive) samples, suggesting that STANDARD M10 actually provided a true positive result. Similarly, ddPCR indicated the absence of SARS-CoV-2 genetic material in 2 out of 4 (50%) Xpress positive/STANDARD M10 negative (false negative) samples, suggesting that STANDARD M10 actually provided a true negative result (Table 4 and Supplementary Table 1).
With rapid antigenic tests dominating the scenario of home-base and community SARS-CoV-2 testing, the mandatory features that we require from a hospital-based RT-PCR assay working aside from emergency departments should be (a) rapid, (b) easy to perform (with minimal on-hand time), (c) very reliable, and (d) relatively cheap in order to provide the prompt indications needed for critical patient management.
This study is the first study aimed to evaluate the performance of the RT-PCR STANDARD™ M10 for the rapid molecular diagnosis of SARS-CoV-2 in nasopharyngeal swabs from pediatric patients. The STANDARD™ M10 SARS-CoV-2 (SD Biosensor) is a newly available rapid, on-demand RT-PCR assay intended to detect the envelope and ORF1ab genomic portions of SARS-CoV-2 from nasopharyngeal swabs. It has an on-hand time of 1 min and a total per-sample turnaround time of 20–60 min. It runs on a completely automated, dedicated instrument that can process up to eight samples in parallel. According to manufacturers, its sensibility and specificity in detecting SARS-CoV-2 in samples from adult individuals is 100%. Yet, no real-life study has reported its clinical use so far, and no data are available on its performance in the pediatric population.
As a result, we conducted a diagnostic performance study on 616 specimens collected from 533 children [of whom 309 (58%) were under the age of 24 months] and 83 adults who visited the Emergency Department at Bambino Gesù Children’s Hospital. As a reference, we used the Xpert® Xpress SARS-CoV-2 assay (Cepheid), currently used in our routine screening protocols at hospital admission. This comparison was deemed to be highly clinically significant, as the two tests share all the main technical features that are required for a rapid hospital-based ER RT-PCR assay, including: (a) continuous random-access loading of disposable cartridges; (b) poor hands-on time; (c) fully automated walk-away processing with a per-sample turn-around-time of less than 1 h; (d) simultaneous detection of two genomic targets; and (e) a compact architecture that fits even in the smallest laboratories. In addition, the Xpert® Xpress SARS-CoV-2 assay is one of the most sensitive tests on the market (Loeffelholz et al., 2020; Smithgall et al., 2020; Serei et al., 2021) and is currently recognized as a benchmark in ER settings, making our comparison particularly challenging. Yet, our performance indexes computed by confusion matrix highlighted a 96.5% sensitivity and a 98.4% specificity for the STANDARD™ M10 SARS-CoV-2, with a very limited number of discordant results (n/N = 12/616), mainly associated with an increased rate of positive results by STANDARD™ M10 assay (putative false-positive, n/N = 8/12 discordances). Worth of note, all of 12 analyzed discordant samples were characterized by a Ct value greater than 34 and a positivity to only one target gene (as shown in Table 4). We do not expect bias introduced by samples management since we strictly followed CDC guidelines (CDC, 2022) and by literature inspection we found well conducted study showing that +4°C or −20°C swabs storage does not significantly affect feasibility of RT-PCR tests for SARS-CoV-2 (Veguilla et al., 2022). Notably, when ddPCR was used to further assess SARS-CoV-2 presence in discordant samples, 2 out of 7 (28.6%) of the putative false-positive results were actually reclassified in true-positives, and 2 out of 4 (50.0%) of the putative false-negative results were reclassified in true-negative. This suggests that the diagnostic performance of STANDARD™ M10 assay in our clinical setting is reasonably expected to be non-inferior to that of the Xpert® Xpress SARS-CoV-2 assay, both in terms of sensitivity and specificity.
The Ct values at RT-PCR are commonly used as a proxy for viral-load quantification (Walker et al., 2021), even if a non-optimal correspondence has been highlighted (Rao et al., 2020). Yet, in clinical practice, Cts are a rapid and easy way to “approximate” viral content in clinical samples and are a useful parameter that contributes to the cross-sectional and multidisciplinary clinical evaluation that takes place at any ER entrance. Our study provided reassuring results on the consistency of Ct values between the two assays, with no significant differences in mean values distribution and limited divergence, always < 1 log. We need to underline that the Ct values cannot be directly compared between assays of different types due to the chemistry of reagents, gene targets, cycle parameters, analytical interpretive methods, sample preparation and extraction techniques, and inherent randomness and variation in the sensitivity of the method (Falasca et al., 2020). Yet, the differences we found (or, better, the lack thereof) support the hypothesis that the use of this test will not significantly infer to patient’s management outcome from a clinical perspective compared to a high-quality standard test, such as Xpert® Xpress SARS-CoV-2 assay.
Our study may have potential limitations. Not all 616 samples have been tested by ddPCR and, therefore, no full performance comparisons with this highly sensitive assay could be made. Yet, the commercial RT-PCR test we used as a reference is the current benchmark for clinical SARS-CoV-2 diagnosis in emergency settings, ensuring the clinical significance of our study in the context of commercially available, CE-marked assays. In addition, SARS-CoV-2 variants have not been routinely tested in the analyzed samples, yet (a) all samples were collected in the period from January to March 2022, when SARS-CoV-2 infections in Italy were almost entirely attributed to Omicron VOC (Rapporto, 2022), and (b) we sequenced a remarkable number of samples in that period and confirmed this rate of variants (Alteri et al., 2022).
In conclusion, our results support the optimal diagnostic performance of the novel STANDARD™ M10 SARS-CoV-2 assay in a real-life ER setting, constituted by pediatric patients in comparison with adult patients. The remarkable degree of similarity in terms of reliability, sensitivity, and specificity, with our current diagnostic reference assay, supports the implementation of STANDARD™ M10 SARS-CoV-2 in a modern diagnostic approach to SARS-CoV-2 infection. On this basis, the availability of a molecular test, fast, easy, and reliable becomes of major importance in the management of patients that require a molecular test (i.e., emergency rooms, fragile patients, etc.), particularly if it is less expensive and therefore sustainable, considering also the decrease of the budget in many countries dedicated to the fight against COVID-19.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
LC, RS, and LuaC: performed the experiments. LC, VCo, and CR: designed the study and wrote the manuscript. AR, MS, and AV: contributed to the samples collection. LC, VCo, and RS: processed the data. LC and VCo: data curation and editing. CP, CR, VCe, and CA: revised data. CP and CR: supervision. All authors contributed to the article and approved the submitted version.
We thank SD Biosensor Inc. for providing technical support. We also thank the whole staff of the Unit of Diagnostic Microbiology and Immunology of Bambino Gesù Children’s Hospital, IRCCS for outstanding technical support in processing samples and performing laboratory analyses.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.999783/full#supplementary-material
Alteri, C., Cento, V., Antonello, M., Colagrossi, L., Merli, M., Ughi, N., et al. (2020). Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One 15:e0236311. doi: 10.1371/journal.pone.0236311
Alteri, C., Scutari, R., Costabile, V., Colagrossi, L., La Rosa, K. Y., Agolini, E., et al. (2022). Epidemiological characterization of SARS-CoV-2 variants in children over the four COVID-19 waves and correlation with clinical presentation. Sci. Rep. 12:12814. doi: 10.1038/s41598-022-17020-6
Bian, L., Gao, Q., Gao, F., Wang, Q., He, Q., Wu, X., et al. (2021). Impact of the Delta variant on vaccine efficacy and response strategies. Expert Rev. Vaccines 20, 1201–1209. doi: 10.1080/14760584.2021.1976153
Burkhard-Koren, N. M., Haberecker, M., Maccio, U., Ruschitzka, F., Schuepbach, R. A., Zinkernagel, A. S., et al. (2021). Higher prevalence of pulmonary macrothrombi in SARS-CoV-2 than in influenza A: Autopsy results from ‘Spanish flu’ 1918/1919 in Switzerland to Coronavirus disease 2019. J. Pathol. Clin. Res. 7, 135–143. doi: 10.1002/cjp2.189
CDC (2022). Interim guidelines for collecting and handling of clinical specimens for COVID-19 testing. Available online at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (accessed July, 2022).
Chen, J., Gao, K., Wang, R., and Wei, G. (2021). Revealing the threat of emerging SARS-CoV-2 mutations to antibody therapies. J. Mol. Biol. 433:167155. doi: 10.1016/j.jmb.2021.167155
Chen, R. E., Zhang, X., Case, J. B., Winkler, E. S., Liu, Y., VanBlargan, L. A., et al. (2021). Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 27, 717–726. doi: 10.1038/s41591-021-01294-w
Colagrossi, L., Antonello, M., Renica, S., Merli, M., Matarazzo, E., Travi, G., et al. (2021). SARS-CoV-2 RNA in plasma samples of COVID-19 affected individuals: A cross-sectional proof-of-concept study. BMC Infect. Dis. 21:184. doi: 10.1186/s12879-021-05886-2
Craney, A. R., Velu, P. D., Satlin, M. J., Fauntleroy, K. A., Callan, K., Robertson, A., et al. (2020). Comparison of two high-throughput reverse transcrip-tion-PCR systems for the detection of Severe Acute Respiratory Syndrome Coronavirus 2. J. Clin. Microbiol. 58:e890-20. doi: 10.1128/JCM.00890-20
Dinnes, J., Deeks, J. J., Berhane, S., Taylor, M., Adriano, A., Davenport, C., et al. (2021). Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 3:CD013705. doi: 10.1002/14651858.CD013705
Domnich, A., Orsi, A., Trombetta, C. S., Costa, E., Guarona, G., Lucente, M., et al. (2022). Comparative Diagnostic Accuracy of the STANDARD M10 Assay for the Molecular Diagnosis of SARS-CoV-2 in the Point-of-Care and Critical Care Settings. J. Clin. Med. 11:2465. doi: 10.3390/jcm11092465
Falasca, F., Sciandra, I., Di Carlo, D., Gentile, M., Deales, A., Antonelli, G., et al. (2020). Detection of SARS-CoV N2 gene: Very low amounts of viral RNA or false positive? J. Clin. Virol. 133:104660. doi: 10.1016/j.jcv.2020.104660
Gorzalski, A. J., Tian, H., Laverdure, C., Morzunov, S., Verma, S. C., VanHooser, S., et al. (2020). High-Throughput transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 129:104501. doi: 10.1016/j.jcv.2020.104501
Ham, S. Y., Jeong, H., Jung, J., Kim, E. S., Park, K. U., Kim, H. B., et al. (2022). Performance of STANDARD™ M10 SARS-CoV-2 Assay for the Diagnosis of COVID-19 from a Nasopharyngeal Swab. Infect. Chemother. 54, 360–363. doi: 10.3947/ic.2022.0047
Javelle, E., and Raoult, D. (2021). COVID-19 pandemic more than a century after the Spanish flu. Lancet Infect. Dis. 21:e78. doi: 10.1016/S1473-3099(20)30650-2
Koyama, T., Miyakawa, K., Tokumasu, R., Jeremiah, S. S., Kudo, M., and Ryo, A. (2022). Evasion of vaccine-induced humoral immunity by emerging sub-variants of SARS-CoV-2. Future Microbiol. 17, 417–424. doi: 10.2217/fmb-2022-0025
Liu, X., Feng, J., Zhang, Q., Guo, D., Zhang, L., Suo, T., et al. (2020). Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microbes Infect. 9, 1175–1179. doi: 10.1080/22221751.2020.1772679
Loeffelholz, M. J., Alland, D., Butler-Wu, S. M., Pandey, U., Perno, C. F., Nava, A., et al. (2020). Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2 test. J. Clin. Microbiol. 58:e00926-20. doi: 10.1128/JCM.00926-20
McLean, G., Kamil, J., Lee, B., Moore, P., Schulz, T. F., Muik, A., et al. (2022). The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines. mBio 30:e02979-21. doi: 10.1128/mbio.02979-21
Peeling, R. W., Olliaro, P. L., Boeras, D. I., and Fongwen, N. (2021). Scaling up COVID-19 rapid antigen tests: Promises and challenges. Lancet Infect. Dis. 21, e290–e295. doi: 10.1016/S1473-3099(21)00048-7
Petersen, E., Koopmans, M., Go, U., Hamer, D. H., Petrosillo, N., Castelli, F., et al. (2020). Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 20, e238–e244. doi: 10.1016/S1473-3099(20)30484-9
Pilarowski, G., Lebel, P., Sunshine, S., Liu, J., Crawford, E., Marquez, C., et al. (2021). Performance characteristics of a Rapid Severe Acute Respiratory Syndrome Coronavirus 2 antigen detection assay at a public plaza testing site in San Francisco. J. Infect. Dis. 223, 1139–1144. doi: 10.1093/infdis/jiaa802
Rao, S. N., Manissero, D., Steele, V. R., and Pareja, J. (2020). A Systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect. Dis. Ther. 9, 573–586. doi: 10.1007/s40121-020-00324-3
Rapporto (2022). Rapporti ISS COVID-19. ISS N. 18. Available online at: https://www.iss.it/rapporti-covid-19 (accessed March 25, 2012).
Serei, V. D., Cristelli, R., Joho, K., Salaru, G., Kirn, T., Carayannopoulous, M. O., et al. (2021). Comparison of abbott ID NOW COVID-19 rapid molecular assay to cepheid xpert xpress SARS-CoV-2 assay in dry nasal swabs. Diagn. Microbiol. Infect. Dis. 99:115208. doi: 10.1016/j.diagmicrobio.2020.115208
Smithgall, M. C., Scherberkova, I., Whittier, S., and Green, D. A. (2020). Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the Rapid Detection of SARS-CoV-2. J. Clin. Virol. 128:104428. doi: 10.1016/j.jcv.2020.104428
Suo, T., Liu, X., Feng, J., Guo, M., Hu, W., Guo, D., et al. (2020). ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 9, 1259–1268. doi: 10.1080/22221751.2020.1772678
Veguilla, V., Fowlkes, A. L., Bissonnette, A., Beitel, S., Gaglani, M., Porucznik, C. A., et al. (2022). Detection and stability of SARS-CoV-2 in three self-collected specimen types: Flocked Midturbinate Swab (MTS) in viral transport media, foam MTS, and Saliva. Microbiol. Spectr. 10:e0103322. doi: 10.1128/spectrum.01033-22
Walker, A. S., Pritchard, E., House, T., Robotham, J. V., Birrell, P. J., Bell, I., et al. (2021). Ct threshold values, a proxy for viral load in community SARS-CoV-2 cases, demonstrate wide variation across populations and over time. Elife 10:e64683. doi: 10.7554/eLife.64683
Wang, C., Horby, P. W., Hayden, F. G., and Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. Lancet 395, 470–473. doi: 10.1016/S0140-6736(20)30185-9
WHO (2022). WHO Coronavirus (COVID-19) Dashboard. Available online at: https://covid19.who.int/ (accessed June, 02 2022).
WHO Working Group on the Clinical Characterisation and Management of Covid-19 infection (2020). A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 20, e192–e197.
Keywords: SARS-CoV-2, molecular diagnosis, COVID-19, RT-PCR testing, comparative diagnostic accuracy
Citation: Colagrossi L, Costabile V, Scutari R, Cento V, Coltella L, Reale A, Scilipoti M, Villani A, Alteri C, Perno CF and Russo C (2022) Performance evaluation of a new on-demand molecular test for the rapid identification of severe acute respiratory syndrome coronavirus 2 in pediatric and adult patients. Front. Microbiol. 13:999783. doi: 10.3389/fmicb.2022.999783
Received: 21 July 2022; Accepted: 20 September 2022;
Published: 03 November 2022.
Edited by:
Kazuya Shirato, National Institute of Infectious Diseases (NIID), JapanReviewed by:
Dhammika Navarathna, United States Department of Veterans Affairs, United StatesCopyright © 2022 Colagrossi, Costabile, Scutari, Cento, Coltella, Reale, Scilipoti, Villani, Alteri, Perno and Russo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlo Federico Perno, Y2FybG9mZWRlcmljby5wZXJub0BvcGJnLm5ldA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.