- 1Department of Biomedical Engineering, University of Miami, Coral Gables, FL, United States
- 2Department of Pathology and Laboratory Medicine, University of Miami Miller School of Medicine, Miami, FL, United States
- 3Desai Sethi Urology Institute, University of Miami Miller School of Medicine, Miami, FL, United States
- 4Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL, United States
- 5Schiff Center for Liver Diseases, University of Miami Miller School of Medicine, Miami, FL, United States
Viral hepatitis is a leading cause of liver disease and mortality. Infection can occur acutely or chronically, but the mechanisms that govern the clearance of virus or lack thereof are poorly understood and merit further investigation. Though cures for viral hepatitis have been developed, they are expensive, not readily accessible in vulnerable populations and some patients may remain at an increased risk of developing hepatocellular carcinoma (HCC) even after viral clearance. To sustain infection in vitro, hepatocytes must be fully mature and remain in a differentiated state. However, primary hepatocytes rapidly dedifferentiate in conventional 2D in vitro platforms. Physiologically relevant or physiomimetic microsystems, are increasingly popular alternatives to traditional two-dimensional (2D) monocultures for in vitro studies. Physiomimetic systems reconstruct and incorporate elements of the native cellular microenvironment to improve biologic functionality in vitro. Multiple elements contribute to these models including ancillary tissue architecture, cell co-cultures, matrix proteins, chemical gradients and mechanical forces that contribute to increased viability, longevity and physiologic function for the tissue of interest. These microsystems are used in a wide variety of applications to study biological phenomena. Here, we explore the use of physiomimetic microsystems as tools for studying viral hepatitis infection in the liver and how the design of these platforms is tailored for enhanced investigation of the viral lifecycle when compared to conventional 2D cell culture models. Although liver-based physiomimetic microsystems are typically applied in the context of drug studies, the platforms developed for drug discovery purposes offer a solid foundation to support studies on viral hepatitis. Physiomimetic platforms may help prolong hepatocyte functionality in order to sustain chronic viral hepatitis infection in vitro for studying virus-host interactions for prolonged periods.
Key points
- Viral hepatitis is a significant contributor to hepatocellular carcinoma and liver disease.
- The transition of chronic infection to hepatocellular carcinoma and liver disease is poorly characterized and requires mechanistic investigation.
- Standard in vitro models are ineffective for studying this transition because hepatocytes do not maintain a functional phenotype that is permissive to infection long enough to model infection chronically.
- Physiomimetic microsystems could help bridge the gap to longer in vitro infections by incorporate elements of the hepatic microenvironment to promote functional longevity of hepatocytes.
- Physiomimetic models vary substantially, and draw on different elements of the hepatocyte microenvironment to sustain an infection-permissive phenotype most notably: Co-culture with non-parenchymal cells, 3D morphology, a physiological spatial orientation, and media perfusion.
- Physiomimetic models that were not originally designed to model viral hepatitis specifically may be used in chronic viral applications as long as they maintain hepatocyte functionality over a longer period of time than is possible in conventional 2D plate culture.
Introduction
Hepatitis viruses are part of a select group of viruses that can establish chronic infections in humans. While most are quickly cleared after acute infection, hepatitis viruses B (HBV), C (HCV), D (HDV), and E (HEV) can all manifest as chronic infections (Dustin et al., 2016; Price, 2016). The mechanisms through which these infections transition from acute to chronic are poorly understood and merit further investigation into the relevant virus-host interactions. Chronic viral hepatitis infection is a leading cause of hepatocellular carcinoma (HCC), accounting for 1.3 million deaths per year and 90% of primary liver cancer cases (Ringehan et al., 2017).
One reason the transition from acute to chronic viral infection has not been properly investigated is a lack of sufficient in vitro models. Conventional two-dimensional (2D) in vitro models are excellent for generating high throughput data, but fail to provide cells with surroundings that mimics their native physiologic microenvironment. Because of this disconnect between in vitro culture and the in vivo microenvironment, primary hepatocytes, the gold standard of in vitro hepatocyte models, suffer from a loss of functionality after extended culture periods in vitro. This loss of functionality inhibits accurate long-term study of host responses to infection, which are instrumental for providing insight into how the transition from acute to chronic infection occurs. Furthermore, hepatoma cell lines and iPSC-derived hepatocyte-like cells (HLCs) do not provide a fully differentiated hepatocyte phenotype and it is difficult to draw relevant biologic conclusions from the observed cellular responses to infection using these models.
Animal models are indeed capable of sustaining chronic viral infections and have been critical to our understanding of the viral hepatitis and its progression to liver disease. However, in the context of viral hepatitis, humans and chimpanzees are the only two known naturally hosts for HBV and HCV, which makes studying the virus in animal models incredibly challenging given the ethical concerns of non-human primate models. Non-human analogs of hepatitis viruses exist, but these are fundamentally different from HBV and HCV, and their naturals hosts are challenged with replicating a human’s immunological response (Berggren et al., 2020; Liu et al., 2021). Even genetically humanized mice, modified to be infectible by human viruses, are limited in their ability to generate a relevant immune responses that is critical to uncovering the mechanisms of disease progression (Louz et al., 2013; Thomas and Liang, 2016). A need for animal models persists because even the most complex in vitro models cannot replicate a systemic response to infection, but these animal models must be supplemented by human-specific in vitro models.
In an effort to better supplement animal models, physiologically relevant microsystems are a potential solution to insufficient in vitro models. The purpose of these microsystems is to capture elements of the native cellular microenvironment in vitro, in an effort to improve experimental results as compared to a conventional 2D plate format. In the liver specifically, much of the work in the development of physiologically relevant microsystems has been directed toward pharmacokinetic modeling and drug-induced liver injury. Nonetheless, many of the same principles used in these systems to improve cell functionality can be effectively utilized in modeling virus-host responses. In this review we investigate how physiologically relevant microsystems recapitulate liver physiology and also how this has led to advances in the study of viral hepatitis infection in vitro.
We first review the physiology of the liver and the necessary components required for relevant studies of viral hepatitis infection in vitro. We then examine different cell models for studies on viral hepatitis in vitro. Next, we provide an overview of a wide variety of practical designs of physiologically relevant microsystems that model the liver and discuss the applications of these models as tools to study virus-host interactions. Finally, we discuss some of the elements of physiologically relevant microsystems that enhance their ability to study viral hepatitis biology specifically, and how these systems are working toward better informing clinical predictions.
Viral hepatitis and the liver
Hepatic microenvironment
By leveraging our current understanding of liver physiology and pertinent aspects of viral pathogenesis, one can ascertain the most critical components of the hepatic microenvironment needed to accurately recapitulate viral hepatitis infection in vitro. Overall, the hepatic lobule is the functional unit of the liver (Figure 1A). The lobule is a hexagonal structure comprised of six portal triads. Each portal triad includes a connection to a hepatic portal vein, a hepatic artery and a bile duct. The hepatic portal vein and the hepatic artery branch together to form the hepatic sinusoid, that is lined with endothelial cells and which transports blood from portal venules and hepatic arterioles (periportal sides) to the central vein (perivenous side) that eventually flows back to the heart. Interspersed between the liver sinusoids are plates of hepatocytes, the parenchymal cells of the liver, that are the substrate of viral hepatitis infection where the virus lifecycle is carried out (Ishibashi et al., 2009; Lautt, 2009; Rocha, 2012). A single sinusoid flanked by hepatocytes distinguishes the smallest functional unit of the liver, the hepatic acinus. The acinus also includes non-parenchymal cells (NPCs) namely Kupffer cells (KCs), the resident macrophages of the liver, and stellate cells (SCs) which are normally quiescent but become fibrogenic when activated by perturbations to the normal liver environment (Zheng et al., 2009; Deng et al., 2019a; Figure 1B). Both hepatocytes and NPCs contribute to the inflammatory microenvironment in response to viral hepatitis infection and are further implicated in the progression of disease as chronic infection persists (Feldstein et al., 2005; Dixon et al., 2013; Li et al., 2017; Hyun et al., 2019).
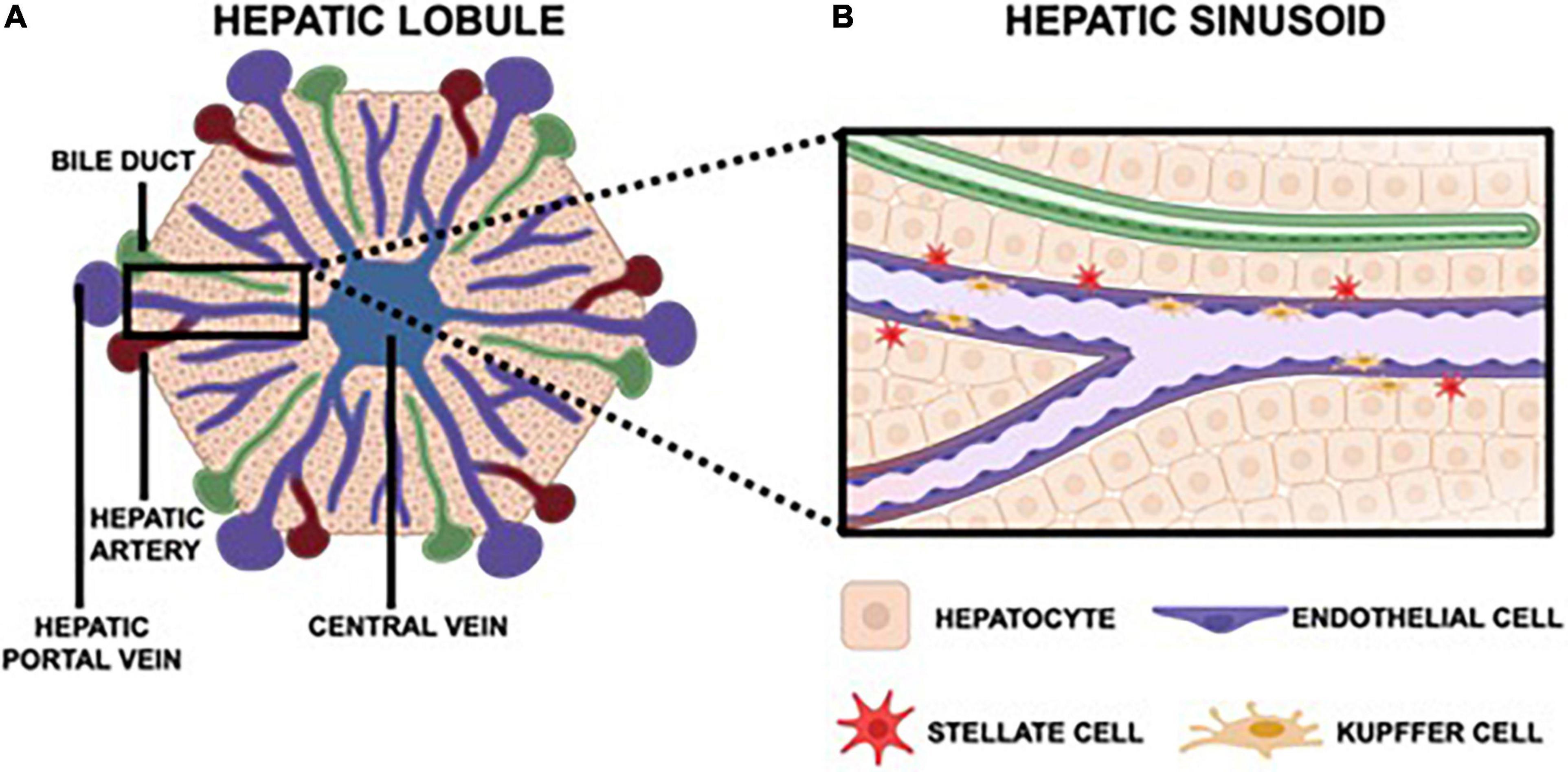
Figure 1. The hepatic lobule is hexagonal, functional unit of the liver that features six portal triads. Each portal triad is comprised of a bile duct, a hepatic portal vein, and a hepatic artery (A). The hepatic portal vein and hepatic artery converge to form the hepatic sinusoid, which carries blood toward the central vein that then returns to the heart. Each sinusoid is lined with endothelial cells and surrounded by hepatocytes, the parenchymal cells of the liver. Also native to the sinusoid are Kupffer cells, the resident macrophages, and stellate cells, which are quiescent in a healthy liver but instigate a fibrotic response in an inflammatory microenvironment (B).
Viral hepatitis
Viral pathogens that cause hepatitis are noteworthy in their ability to establish chronic infection. There are five major hepatitis viruses (A-E) each with several different genotypes. These viruses vary greatly; however, in this review, we focus on HBV and HCV due to their worldwide prevalence and strong link to significant morbidity and mortality.
Hepatitis B virus
HBV is a partially double-stranded DNA virus that produces high levels of viral antigens which enabled the development of a highly effective vaccines against the virus (Thomas and Liang, 2016). Although vaccines have proven successful in preventing the transmission of HBV, infection remains a challenge because of the lack of a highly effective, finite curative treatment regimen and also since chronic HBV infection has a clear link to the development of HCC (NIH strategic plan details pathway to achieving Hepatitis B cure, 2019; Torresi et al., 2019). Moreover, as of 2019, global coverage of the HBV vaccine birth dose was at just 42%; unfortunately 90% of babies will progress to chronic infection if infected via mother-to-child transmission (hepbtalk, 2019). After entering the hepatocyte, the virus translocates to the nucleus where its genome is modified to the covalently closed circular (cccDNA) that exists stably as an extrachromosomal genome and is a major viral component contributing to viral persistence. cccDNA codes for the transcripts necessary for protein production and replication; therefore, it is the primary target for potential curative therapies (Thomas and Liang, 2016). In vitro models to further characterize the progression of HBV infection from acute to chronic infection are desperately needed and also models that can be utilized to further develop effective curative regimens are highly sought after.
Hepatitis C virus
HCV is a single-stranded positive-sense RNA virus that manifests chronically in 75–85% of those infected (Chen and Morgan, 2006). HCV enters the hepatocyte and completes translation and replication in the endoplasmic reticulum (Thomas and Liang, 2016). In contrast to HBV, HCV is curable through treatment with small molecules that include polymerase and protease inhibitors that are more broadly referred to as direct acting antiviral agents (DAAs); unfortunately, there is no vaccine against HCV because of the virus’ high degree of genetic variability (Zingaretti et al., 2014). Furthermore, patients infected with chronic HCV remain susceptible to developing HCC even after the virus is cleared if they have advanced liver disease (El-Serag, 2002; de Oliveria Andrade et al., 2009; Axley et al., 2018; Ioannou et al., 2019; Nahon and Ganne-Carrié, 2019). Similar to HBV, the need for further study of HCV stems from gaps in knowledge of the virus’ molecular strategies that allow it to transition from acute to chronic infection and host factors that influence this biologic outcome.
In vitro cellular models of viral hepatitis infection
Choice of hepatocyte source has a substantial impact on the biologic validity of experimental results as well as on the efficiency of supporting infection in vitro. Primary hepatocytes, hepatoma cell lines and iPSC-derived hepatocytes have significant differences in various experimental characteristics; however, each model may be desirable for use depending on the specific experimental application.
Primary hepatocytes
Primary human hepatocytes (PHHs) are the gold standard for in vitro liver models in general. They display a notable differentiated phenotype at the time of plating. In addition, they support both HBV and HCV infection; unfortunately, their potent innate antiviral response is difficult to circumvent rendering this model unsuitable for the production of high titer virus stocks. In concordance with this, it is more difficult to establish long-term infections in these cells without artificially downregulating the antiviral response. Additionally, PHHs are a relatively rare cell source given that they come directly from patients or fetal tissue and they do not naturally divide once cultured in vitro. Inherent interpatient variability also drives the need to source cells from multiple donors to achieve more reproducible results (Thomas and Liang, 2016). However, humanized mice have recently been developed to propagate PHHs in vivo which has rendered these cells somewhat easier to source (Ohshita and Tateno, 2017).
Hepatoma cell lines
Hepatoma cell lines provide a high degree of accessibility for modeling viral infection in vitro at the cost of being unable to provide normal physiologic antiviral responses. Here we describe three different cell lines frequently used to model viral hepatitis infection in vitro.
Huh7
The Huh7 cell line and its genetically modified successors are very receptive to HCV infection. These cells are excellent at propagating infectious HCV virions and they divide rapidly. Huh7 cell are extremely useful for replicating viral infection, and this cell line also supports infection with other important pathogenic viruses including SARS-CoV-2 (Thomas and Liang, 2016; Chen et al., 2021). However, Huh7 cells have significantly attenuated cell-intrinsic innate antiviral responses to viral infection, and therefore are not useful for studying the viral immune response in hepatocytes.
HepG2
HepG2 cells have been used for studies of the HBV lifecycle. HepG2 cells are polarized, can be manipulated to express the HBV genome and can generate a cell-intrinsic innate antiviral response that makes them useful for the studies on host defense pathways. However, HepG2 cells must be genetically modified in order to fully support HBV infection as they lack significant expression of the NTCP HBV viral entry receptor, and even when this receptor is expressed, HepgG2 cells are still less susceptible to HBV infection than fully differentiated HepaRG cells (Li et al., 2016; Thomas and Liang, 2016).
HepaRG
The HepaRG cell line is derived from a bipotent progenitor that can differentiate into either the hepatocyte or cholangiocyte lineage. If differentiated toward the hepatocyte lineage, HepaRGs confer a phenotype that mirrors that of PHHs in terms of its ability to support both HBV and HCV infection while demonstrating contact inhibition (Thomas and Liang, 2016; Wu et al., 2016). However, this differentiation process can take up to 2 weeks and the differentiated cells appear to be difficult to propagate further (Thomas and Liang, 2016). HepaRGs are also bipotent progenitor cells and therefore may lack a fully functional innate immune axis (Shlomai et al., 2014). Finally, HCV is extremely difficult to replicate in HepaRGs and to our knowledge, has only been achieved by Ndongo-Thiam et al. (2011).
Stem cell derived hepatocytes
iPSC-derived hepatocyte-like cell (HLCs) are similar to HepaRGs in the sense that they can be differentiated in order to confer a phenotype that approaches that of PHHs; however, they have yet to prove capable of exhibiting the same level of differentiation as PHHs that includes minimal to no expression of alpha-fetoprotein. Unlike PHHs however, iPSCs offer a limitless supply of cells while minimizing donor variability. HLCs have demonstrated the capacity of supporting both HBV and HCV infection (Wang et al., 2019; Simoneau and Ott, 2020).
Physiologically relevant liver technologies and their application to viral hepatitis
Physiomimetic in vitro liver models are typically designed with the goal of promoting and sustaining hepatocyte differentiation and viability and are then applied to study relevant functional biological mechanisms. This preliminary step of promoting the maintenance of viable and differentiated hepatocytes is critical for the establishment of a microenvironment that is conducive to support long-term viral infection. Here, we describe how different technologies recapitulate the hepatic microenvironment to best support and maintain physiologically relevant hepatocyte function. The advantages and disadvantages of each type of physiomimetic platform are listed in Table 1 and the different platforms are depicted in Figure 2.
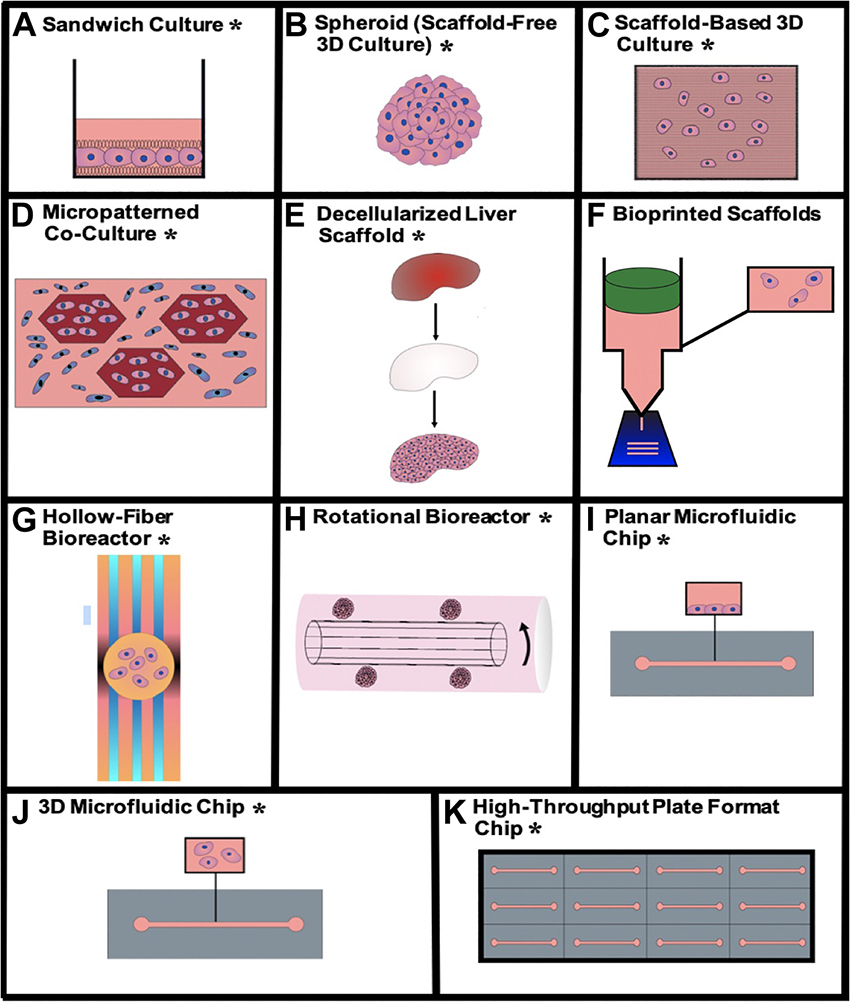
Figure 2. Visualizations of the different physiomimetic culture systems outlined in this review. (A) Sandwich culture*, (B) aggregate spheroid culture*, (C) scaffold-based 3D culture*, (D) micropatterned co-culture*, (E) decellularized liver scaffold*, (F) bioprinted scaffolds, (G) hollow-fiber bioreactor*, (H) rotational bioreactor*, (I) planar microfluidic chip*, (J) 3D microfluidic chip*, (K) high-throughput plate format chip*. *Indicates that the given type of model has been used for viral hepatitis study.
Static culture models
Static models established the foundation for the subsequent development of dynamic models and they are utilized for the development of tissue-like structures beyond traditional 2D cell monolayers. Any fresh media that the culture receives has to be administered manually and these cultures are characterized by the lack of any consistent media perfusion or flow through the model.
Sandwich culture
Sandwich culture is a layered culture approach where cells are sandwiched between two matrix layers (Figure 2A). Cells bind and form a monolayer on the matrix beneath them and are subsequently coated with a top layer of matrix that is called an overlay. This physiomimetic sandwiched orientation has been shown to improve cell morphology, viability and maintenance of differentiation that includes the establishment of polarity and formation of bile canalicular networks (Swift et al., 2010; Yang et al., 2016). Complexity of these cultures increases even further in co-culture sandwiches. Mouse fibroblasts (3T3-J2) were used as feeder cells to help differentiate iPSCs into HLCs as double sandwiches where the fibroblasts were stacked on top of the hepatocytes (Nagamoto et al., 2012). Sasaki et al. (2017) performed a layer-by-layer (LBL) cell coating of hepatocytes and fibroblasts on a membrane with distinct fibronectin and gelatin layers. They subsequently developed a tri-culture with human umbilical vein endothelial cells (HUVECs) to create a vascularized tissue. Petropolis et al. (2016) implemented this vascularized LBL approach, specifically for studies on HBV replication, through the formation of two Huh7 hepatocyte layers separated by a matrix and topped with a layer of endothelial cells. The establishment of sandwich cultures were an important advance in establishing more physiologically relevant models and they remain the foundation of more complex physiomimetic platforms.
3D spheroids, organoids, and scaffolds
3D spheroids, organoids, and scaffolds are techniques for culturing cells in 3D by providing cells with points of contact in three dimensions to further support the maintenance of hepatocyte differentiation in vitro (Bachmann et al., 2015). Though ease of use has precluded the full transition from 2D cultures to these less complex 3D cultures for in vitro work, 3D cultures may promote more stable long-term cell function that would be appropriate for extended viral infection studies.
Scaffold-free 3D culture
Cell aggregates can create a scaffold-free 3D cultures (Figure 2B). Cells attach to each other to form multi-cellular spherical structures called spheroids, or organoids if there are multiple cell types. The simplest method of spheroid or organoid formation is through the use of ultra-low attaching (ULA) surfaces, that are treated to keep cells in suspension, causing them to aggregate into spheroids (Subramanian et al., 2013; Kim et al., 2015; Miyamoto et al., 2015; Gaskell et al., 2016; Hendriks et al., 2016; Ramaiahgari et al., 2017; Vorrink et al., 2017, 2018; Bell et al., 2018; Kozyra et al., 2018; Mandon et al., 2019). These plates can also be used for co-cultures or multi-lineage differentiation to form hepatocyte organoids (Bin Ramli et al., 2020). Hanging-drop plates are an alternative method of spheroid generation that use hanging drops of small volumes of cell suspension to allow hepatocytes to aggregate over 2–4 days before being transferred to a new plate for experimentation (Gunness et al., 2013; Mueller et al., 2014; Takahashi et al., 2015). This technique has been commercialized by InSphero and is also applicable to organoid co-cultures (Messner et al., 2013, 2018; Proctor et al., 2017). Nie et al. (2018) generated organoids from iPSC-derived HLCs and HepG2s each of which were co-cultured with HUVECs. They used these iPSC-derived organoids to study cellular interactions with HBV for up to 20 days post-infection, with results comparable to 2D PHH culture. Spheroids and organoids appear to be useful tools for simple 3D cell culture applications; however, they lack an organized physiological architecture and they may also suffer from the establishment of necrotic cores due to limited oxygen concentration within the center of the structure (Bachmann et al., 2015).
Scaffold-based 3D culture
Scaffold-based approaches for 3D hepatocyte culture typically employ hydrogel matrices for bulk encapsulation (Figure 2C). These products are available in multiple forms that includes Matrigel®, Geltrex, PuraMatrix, alginate, chitosan, gelatin/collagen, polyethylene glycol diacrylate (PEGDA) and cellulose where the porous structures support the transport of cell signaling molecules between hepatocytes (Barbetta et al., 2008; Ramasamy et al., 2013; Gieseck et al., 2014; Ramaiahgari et al., 2014; Ramachandran et al., 2015; Sirenko et al., 2016; Tasnim et al., 2016; Wang et al., 2016; Nuciforo et al., 2018; Ahn et al., 2019; Mun et al., 2019; Ouchi et al., 2019). Crignis et al. (2020) infected healthy donor organoids suspended in a basement membrane matrix with HBV and demonstrated HBV cccDNA production up to 8 days post-infection. Cho et al. (2008), Molina-Jimenez et al. (2012), and Ananthanarayanan et al. (2014) embedded cells in Matrigel, PEGDA hydrogel and a cellulose hydrogel, respectively, followed by subsequent infection of the spheroids with HCV. Similarly, Rajalakshmy et al. (2015) cultured Huh7 cells in Mebiol gel, a thermoreversible gelatin polymer, and infected them with HCV for 10 days. In addition to bulk encapsulation, cells can also be fractionated into small beads (Selden et al., 2000; Khalil et al., 2001; Hwang et al., 2010; Huch et al., 2015; Rebelo et al., 2015; Phillips et al., 2018; Nakai et al., 2019). For example, Tran et al. (2012) used an optimal Ca-Na-alginate bead formulation to upregulate expression of HCV-specific receptors in Huh7 cells, rendering the model useful for HCV infection studies. Substrate mechanical properties do impact hepatocyte function; therefore, scaffolds and matrices can be supplemented and tuned to match desired in vivo mechanical properties (Matellan and Del Río Hernández, 2019). Non-hydrogel scaffolds like paper and nylon have also been used to provide a consistent rigid structure while the additional use of tunable cross-linking of hydrogels can be leveraged to modulate mechanical properties in situ (Kostadinova et al., 2013; Zhu et al., 2017; Wang et al., 2018a). Akahori et al. (2020) used a silica fiber scaffold to support HBV cccDNA formation, in 3D hepatocytes, for up to 15 days post-infection. Depending on the material of choice, 3D rigid scaffolds offer the potential of a more mechanically relevant microenvironment compared to scaffold-free cultures (Matellan and Del Río Hernández, 2019).
Micropatterned co-cultures
Micropatterned co-cultures (MPCCs) are deterministically patterned co-cultures, often achieved by selective matrix deposition or surface treatment (Figure 2D). The original hepatocyte MPCC design from Khetani et al. (2013) now commercialized as HEPATOPAC®, features groups of hepatocytes seeded on collagen, spatially arranged via stencil, and surrounded by 3T3-J2 mouse fibroblasts as feeder cells (Khetani and Bhatia, 2008; Wang et al., 2010; Khetani et al., 2013; Trask et al., 2014; Ware et al., 2015; Aleo et al., 2019; Chan et al., 2019). Like sandwich cultures, HEPATOPAC® offers many of the same advantages as 2D culture in terms of accessibility and throughput translating seamlessly to 24, 96, and 384-well plate formats (Ware et al., 2015; Gural et al., 2018). This patterning with feeder cells improves the longevity and functionality of both PHHs and iPSC-derived hepatocytes when compared to hepatocyte monoculture (Berger et al., 2015; Ware et al., 2017). Ploss et al. (2010) co-cultured cultured PHHs with 3T3-J2s in the HEPATOPAC® system and achieved HCV infection for up to 2 weeks. Similarly, Shlomai et al. (2014) used MPCC technology to culture PHHs with 3T3-J2s; however, they used successful HBV infection as a metric of functional differentiation of MPCCs that was not limited exclusively to co-culture of hepatocytes with 3T3s. The system also features the possibility of co-culturing hepatocytes with stellate cells, Kupffer cells and endothelial cells for the purpose of improving the maintenance of hepatocyte function (Nguyen et al., 2015; Davidson et al., 2017; Ware et al., 2017). Micropatterning can also be accomplished using spheroids as opposed to hepatocytes in a monolayer (Fukuda et al., 2006). The Cell-Able® plate features attaching and non-attaching regions that are produced through the use of a photo-sensitive material. This material allows for spatial patterning of self-assembling hepatocyte spheroids that can then be cultured with or without 3T3 cells (Ohkura et al., 2014; Ogihara et al., 2015). In addition to surface-guided patterning, segregated flow techniques have been used to pattern hydrogel co-cultures in 3D. Kobayashi et al. (2013) used a patterned chip design, featuring an alginate spacer, to separate hepatocytes and 3T3 cells both of which were suspended in distinct alginate solutions. Similarly, Jia et al. (2020) created a hepatic plate using alternating layers of hepatocyte and endothelial alginate solutions159. Using a creative technique to avoid flow through separate channels, Ho et al. (2013) flowed cell suspensions into a chip and used dielectrophoresis to guide cells into a radial pattern that mimics the structure of a lobule using hepatocytes and endothelial cells. Kukla et al. (2020) used matrix encapsulation of hepatocytes and feeder cells to generate 3D co-encapsulated microtissues, which enhanced hepatocyte function. Overall, MPCCs provide spatial control over co-cultures to optimize the synergistic effects between different cell types while maintaining the differentiated hepatocyte phenotype that is necessary for infection. It is important to note that for studies that are specifically focused on understanding human disease processes, the presence of murine feeder cells may render the system suboptimal given the possible non-physiologic contributions to observed experimental outcomes. The effect of the feeder cells may be minimized by their selective killing before proceeding with experimentation on the remaining hepatocytes.
Decellularized scaffolds
Decellularized scaffolds from both human and non-human livers provide a physiological matrix via preservation of liver-specific ECM (Figure 2E). This native ECM supports requisite signals for engraftment, survival and function following hepatocyte reseeding (Rossi et al., 2019). Liver tissue is first decellularized by perfusion of detergents and/or enzymes leaving only matrix proteins behind. The matrix foundation is then re-seeded with hepatocytes and any other NPCs for the purpose of extending and improving hepatocyte function (Baptista et al., 2011; Lang et al., 2011; Wang et al., 2011; Mazza et al., 2015; Maghsoudlou et al., 2016). Zhang et al. (2019) seeded healthy and cirrhotic patient decellularized liver ECM (dECM) scaffolds with HBV-infected HepG2-NTCP cells. The authors found greater viral replication in the scaffold from the cirrhotic patient. Furthermore, when compared to 2D HBV infection, viral replication was markedly increased even in the healthy scaffold. These findings indicate that the liver microenvironment may have a significant impact on viral infection in vitro (Zhang et al., 2019). dECM proteins can also be preserved independent of maintaining the liver microarchitecture. The decellularized scaffold can be pulverized into a raw ECM protein suspension for subsequent manipulation and use for surface coating, spheroid formation and as bioink in 3D printing applications (Bavli et al., 2016; Mi et al., 2018; Yajima et al., 2018; Deng et al., 2019b). The major drawback of dECMs is their low degree of accessibility relative to other platforms that includes the cumbersome methods of preparation required to fully decellularize and validate the decellularization of the liver substrate (Maghsoudlou et al., 2016). However, based on efficacy alone, decellularized scaffolds are a promising mimetic of the in vivo microenvironment. This material has the potential for use within multiple applications, both as a standalone model and also as an enhancement to the physiologic performance of other in vitro liver models.
Bioprinted scaffolds
3D bioprinting is a novel technology that incorporates biological materials into 3D printing that allows for fine spatial control to form physiologically relevant 3D tissue architecture via layer-by-layer deposition that can lead to improved maintenance of hepatocyte function (Ma X. et al., 2016; Figure 2F). Bioinks are typically hydrogel-based that can be crosslinked into a more rigid structure post-printing. However, as mentioned previously, bioinks can be synthesized from dECM providing a more physiologically accurate protein composition (Skardal et al., 2015; Lee et al., 2017; Mao et al., 2020). The spatial arrangements of bioprinted scaffolds vary substantially in terms of complexity. Scaffolds printed from single bioinks are functionally similar to traditional 3D scaffolds in their capacity to improve long-term viability and maintenance of phenotypic differentiation (Faulkner-Jones et al., 2015; Jeon et al., 2017; Grix et al., 2018). For co-cultures, however, bioprinting is advantageous over traditional scaffolds because it can incorporate multiple bioinks containing different cell types. These different bioinks can then be used to manufacture deterministically patterned multi-cellular vascularized sinusoid-mimetic constructs (Matsusaki et al., 2013; Ma X. et al., 2016; Nguyen et al., 2016; Wu et al., 2018; Taymour et al., 2021). Using separate bioinks for different cell types also allows for the creation of a physiologic separation of different cell types while still permitting cellular cross-talk. In the case of bioprinting a liver, hepatocytes are often separated from NPCs, as they are in vivo, through separately deposited bioinks. Hiller et al. (2018) published a combinatory alginate-dECM bioink that demonstrated promise for viral studies based on its permissiveness to adenoviral replication; however, they did not infect cells in their system with HBV or HCV. Nonetheless, bioprinting holds great potential for creating more relevant in vitro models because of its high degree of spatial control and customizability for a given experimental application.
Novel 3D culture techniques
Alternative 3D culture methods have been developed to optimize cell accessibility, viability, spatial distribution and experimental throughput. Self-organizing organoids have been generated from stem cell precursors, both by using by using endothelial cell co-culture to modulate interactions with stem cells, and by overexpressing the GATA6 gene in a custom-engineered stem cell line (Takebe et al., 2013; Guye et al., 2016; Velazquez et al., 2020). Desai et al. (2017) used magnetic nanoparticles that attached to cell membranes and then used magnets to aggregate the cells to form spheroids. Miyamoto et al. (2015) and Ott et al. (2017) used similar micromolded well designs where cells congregated and formed spheroids due to gravity. Wang et al. (2015) used an RGD/galactose conjugated surface to form tethered spheroids whereas Chen et al. (2016) utilized a water/water/oil emulsion to create a complex spheroid with a hepatocyte-laden core and a 3T3-J2 mouse fibroblast shell. Micropillar and nanopillar technology has proven effective for high throughput studies through minimizing the use of space. The pillars provide a location for 3D cell or matrix attachment and also facilitate experiments that require exposure to various compounds or other treatment conditions (Takayama et al., 2013; Kwon et al., 2014; Roth et al., 2018).
Perfusion culture models
Perfusion cultures subject cells to consistent laminar flow of media. These platforms use the building blocks of static culture to establish a more complex dynamic culture that may provide physiologically relevant mechanical stimulation and nutrient exchange. Healthy mechanical stimulation and nutrient exchange promotes prolonged cell viability and function to a degree that may otherwise be unattainable when utilizing static cultures (Kalyanaraman et al., 2008; Huh et al., 2011).
Bioreactors
Bioreactors are advanced technologies that established the foundation for the subsequent development of microfluidic chips. The purpose of bioreactors is to recapitulate an isolated system ex vivo. Bioreactors are amenable to long-term culture of tissues but are less useful as models for high-throughput screening when compared to their miniaturized microfluidic chip counterparts (Tostões et al., 2012).
Hollow fiber bioreactors
Hollow fiber (HF) bioreactors are perfusion devices that typically feature two sets of hydrophilic media capillaries and one set of hydrophobic oxygen capillaries (Figure 2G). The media capillaries provide counter-current flow for nutrient exchange while the oxygen capillaries promote gas exchange. Cells are cultured in 3D compartments between the HF capillaries (Darnell et al., 2012). Traditionally, HF bioreactors have been used to prolong the functional lifespan of primary hepatocytes for extended studies ex vivo (Pless et al., 2006; De Bartolo et al., 2009; Schmelzer et al., 2009; Zeilinger et al., 2011; Hoffmann et al., 2012; Ulvestad et al., 2012; Lübberstedt et al., 2015; Ahmed et al., 2017). Gerlach et al. (2003) maintained PHHs and NPCs for 5 weeks. In a separate study, Aizaki et al. (2003) sustained HCV infection in the FLC4 cell line for over 100 days in their HF bioreactor. More recently, these bioreactors have been utilized as a platform for differentiating stem cells or progenitor cell lines to HLCs (Kawada et al., 1998; Sivertsson et al., 2012; Meier et al., 2017). Though useful for supporting hepatocyte differentiation, HF bioreactors lack the scalability and throughput capacity that are desired for other types of viral infection studies that require biologic or technical replicates.
Rotational bioreactors
Rotational bioreactors, or rotating wall vessels (RWVs), are platforms for spheroid generation and/or maintenance that continuously suspend cells and allow them to aggregate into spheroids (Yoffe et al., 1999; Chang and Hughes-Fulford, 2008; Leite et al., 2012; Figure 2H). The spheroid cultures can be maintained in the RWV or removed for encapsulation/experimentation after they are generated (Fey and Wrzesinski, 2012; Jung et al., 2017). Rebelo et al. (2017) successfully formed an organoid with a parenchymal cell core and stromal cell shell, without emulsions, by allowing hepatocyte spheroids to aggregate first before introducing BM-MSCs that coat the hepatocyte spheroids on the outside. Sainz et al. (2009) used an RWV to culture and infect Huh7 spheroids for up to 2 weeks with HCV. RWVs are a good option for long-term spheroid culture, but like HFs, they translate poorly to support high throughput studies.
Microfluidic platforms
Microfluidic platforms are the miniaturized successors to bioreactors that possess numerous advantages including easier multiplexing, imaging and sample collection. Furthermore, it leverages the use of fewer reagents and cells in its layout to better mimic the liver’s architecture. These microfluidic platforms are often designed as perfusion analogs to the static models delineated above. Liver microfluidic devices are most commonly fabricated from PDMS elastomer, PMMA (acrylic), or polystyrene. They can also incorporate numerous other materials in their design that can be manufactured with a wide variety of formfactors. Despite their differences in design and means of construction, these devices are ultimately unified in their aim to better capture the physiology of the liver in vitro.
Planar and layered microfluidic platforms
As a perfusion analog to monolayers and sandwich culture, planar and layered microfluidic platforms expose a monolayer or multiples layers of cells to laminar flow (Figure 2I). This laminar flow profile is generally consistent with blood flow through the sinusoid, thus providing the culture with simulated physiological shear and nutrient exchange (Natarajan et al., 2017). Planar flow cultures are categorized by two different techniques for cell arrangement: Membrane/barrier cultures and non-membrane cultures. Membrane and barrier cultures create a boundary that can separate different cell types or selectively regulate flow across cells in order to modulate nutrient transport (Lee et al., 2007; Carraro et al., 2008; Zhang et al., 2008; Illa et al., 2014; Maher et al., 2014; Rennert et al., 2015; Prodanov et al., 2016; Ylva et al., 2016; Ortega-Ribera et al., 2018; Deng et al., 2019b). Non-membrane cultures feature some permutation of cells and ECM layers with no physical barrier to separate cells from each other or from flow through the system (Leclerc et al., 2004; Baudoin et al., 2011; Prot et al., 2011, 2014; Cheng et al., 2012; Choucha Snouber et al., 2013; McCarty et al., 2014; Giobbe et al., 2015). Membrane/barrier cultures better mimic the liver’s architecture; however, cell accessibility may be more limited for imaging and sample collection and cell-cell signaling between different cell types in co-culture is likely encumbered by a more substantial physical barrier when compared to ECM layered cultures. Conversely, non-membrane cultures require a less complex chip design with easier cell access, but fail to recapitulate the physical barrier between hepatocytes and the linear path of flow that exists in vivo. Without this barrier, hepatocytes are at risk of exposure to non-physiological shear that may affect the long-term maintenance of a differentiated phenotype. Emulate’s Liver-Chip features a membrane that divides dual parenchymal and non-parenchymal perfusion channels separated by a PDMS membrane with individual flow rate control that is amenable to imaging (Foster et al., 2019; Jang K. J. et al., 2019; Peel et al., 2019). Kang et al. (2017) also used a bi-layer membrane chip design that was able to support HBV infection in PHHs for 14 days. Alternatively, HμREL employed a membrane-free approach using PHHs seeded and perfused in monoculture or co-culture with NPCs on a collagen coated polystyrene chip (Chao et al., 2009; Novik et al., 2010; Hultman et al., 2016). Several planar chips, membrane and non-membrane alike, have attempted to recreate the natural zonation of the liver via oxygen tension characterization and concentration patterning of metabolic modulators (Lee-Montiel et al., 2017; Kang et al., 2018; Li et al., 2018; Moya et al., 2018; Tonon et al., 2019). Zonation chips may provide a more holistic perspective when studying the liver. Banaeiyan et al. (2017) developed a barrier model to study the liver at an expanded scale through recreating 18 full liver lobules as opposed to a single sinusoid. Compatible with both planar and ECM encapsulated culture, this dual layer chip incorporates diffusion barriers that separate sections of hepatocytes to mimic the shear-protective and nutrient-modulating endothelial layer. Merging planar cultures with laminar flow is a promising technique for maintaining a differentiated hepatocyte phenotype that can sustain viral infection.
3D microfluidic platforms
3D microfluidic devices are acutely similar to planar microfluidic devices by design, but provide a difference in cell orientation. Like static 3D cultures, cells are cultured as scaffold-free spheroids/organoids, individual scaffold-encapsulated 3D cells, or scaffold-encapsulated spheroids/organoids. Scaffold-free cultures require physical barriers in the form of micropillars or protective membranes to immobilize and protect spheroids from being perturbed by direct exposure to flow (Goral et al., 2010; Ong et al., 2017; Wang et al., 2018b). In Ma et al., 2018 design, the chip was manufactured with microwells where spheroids congregate, topped with a microporous membrane to simulate endothelial barrier function, and media flowed over the top of the membrane to promote nutrient exchange. The majority of scaffold-based chips use pre-formed 3D hydrogel cultures that subsequently introduce perfusion across the surface of the hydrogel structure (Bavli et al., 2016; Schepers et al., 2016; Aeby et al., 2018; Christoffersson et al., 2018; Yajima et al., 2018). Natarajan et al. (2022) modeled HCV infection in liver organoids encapsulated in basement membrane matrix and perfused on a commercially available chip, idenTx, from AIM Biotech (Natarajan et al., 2022). Alternatively, commercial supplier Hesperos utilized a nylon scaffold to separate cell types in their dual channel platform (Esch et al., 2015). Ma C. et al. (2016) devised a circular lobule-like structure using aggregated hepatic cords comprised of collagen-suspended hepatocytes and endothelial cells. Similar to the dielectrophoretic arrangement that Ho et al. (2013) used, Schütte et al. (2011) applied dielectrophoresis to assemble non-encapsulated 3D hepatic tissue constructs with PHHs and LSECs on ECM coated assembly gaps as the cell suspension flowed into the chip. These constructs were then perfused on both sides of the assembly gap after formation. 3D perfusion culture combines the advantages of a physiologically relevant 3D culture with the benefits of flow for nutrient exchange. The complexity of these designs, however, generally requires more lead time for device and culture assembly when compared to simpler 3D platforms without perfusion.
Perfused decellularized scaffolds
Sassi et al. (2021) recently published a platform to perfuse a recellularized rat liver dECM in a bioreactor. The authors cultured individual lobes of the reseeded liver under syringe pump perfusion and showed significant upregulation of hepatocyte maturation markers in the perfused culture over the course of 11 days (Sassi et al., 2021). The use of decellularized scaffolds for perfusion is a novel technique that may be an optimal method for sustaining a differentiated hepatocyte phenotype.
Bioprinted microfluidic platforms
Like bioprinted static culture, bioprinting offers additional spatial customizability for cell deposition. Bhise et al. (2016) instituted bioprinting as a means of encapsulated spheroid deposition, in a precise dot array pattern to create partitioned spheroid cultures. The authors bioprinted their cell-matrix suspension on to a glass slide, which was then sandwiched between two PDMS covered PMMA sheets to seal a perfusion channel (Bhise et al., 2016). Bioprinting is also useful for non-traditional matrices that are otherwise difficult to deposit. Lee et al. (2019) used dECM bioink to construct two separate layers, one with hepatocytes and one with endothelial cells. The layers were perfused on both sides to simulate blood flow across the endothelial layer and bile flow across the hepatocytes (Lee et al., 2019). Bioprinted platforms provide options for more sophisticated 3D tissue structures in microfluidic chips.
High-throughput and plate format perfusion platforms
While many of the platforms detailed above are not optimized for multiplexing and generating high-throughput data, several commercially available systems feature plate formats that are advantageous for adding biological replicates and testing a larger number of experimental conditions (Figure 2K). The PREDICT96 platform from Draper contains 24 individual microphysiological systems, pneumatically perfused, and arranged to fit a 96-well plate format. Each of the 24 chips has a top channel with collagen-suspended hepatocytes, a polycarbonate membrane, and a bottom channel for media flow (Tan et al., 2019). CN Bio’s LiverChip plate consists of 12 individual microphysiological systems: pneumatically driven media circulates up and through an ECM-coated polycarbonate scaffold with microchannels (Sarkar et al., 2015, 2017; Long et al., 2016; Rowe et al., 2018). Ortega-Prieto et al., 2018, 2019 infected PHHs with HBV for up to 22 days using the CN Bio chip, and demonstrated improved infection in the chip as compared to spheroid and traditional 2D cultures. MIMETAS retrofit their OrganoChip® design to a 384 well plate with 40 individual culture chambers. Each chip has phaseguides that separate the matrix-encapsulated cells from rocking-induced gravity flow, but still permit nutrient exchange (Jang et al., 2015, 2018; Jang M. et al., 2019; Bircsak et al., 2021). Lena Biosciences’ design features a fiberglass scaffold with the option for matrix encapsulation. Cells are perfused bidirectionally via volume changes beneath a bottomless well insert that causes a rising and falling of media across the scaffold (Shoemaker et al., 2020). High-throughput plate designs translate complex physiologically relevant microsystems to a more familiar plate environment to improve their ease of use and efficacy for multi-replicate and multi-variable studies, relevant for studying diverse viral infection conditions simultaneously. A drawback when compared to other systems is that they require a more intricate assembly than some static culture systems, but this limitation holds true for nearly every perfusion device.
Clinical implications for physiologically relevant platforms
Although the scope of applications for physiologically relevant systems has thus far been limited to in vitro modeling, it is important to consider what these platforms may provide as clinically predictive models. For viral hepatitis specifically, physiologically relevant microsystems hold great potential as tools to make clinical predictions about disease progression because these platforms incorporate numerous microenvironmental factors that are implicated in viral pathogenesis. For example, Kupffer cells and stellate cells recruit T cells and contribute to the inflammatory microenvironment during infection (Wang et al., 2013; Boltjes et al., 2014). Several different chip designs have incorporated these cells types (Vernetti et al., 2016; Li et al., 2018; Jang K. J. et al., 2019), and although they have not attempted viral hepatitis infection, these designs have obvious potential for enhancing the biological relevance of the cellular response to viral hepatitis infection (Figures 3A–C). Moreover, the notion of creating linked multi-organ platforms creates opportunity for an even larger systemic response to viral infection. Even though the three aforementioned platforms do not have T cells, they could be potentially linked with an immune chip where the recruitment of immune cells from a separate chamber could be studied.
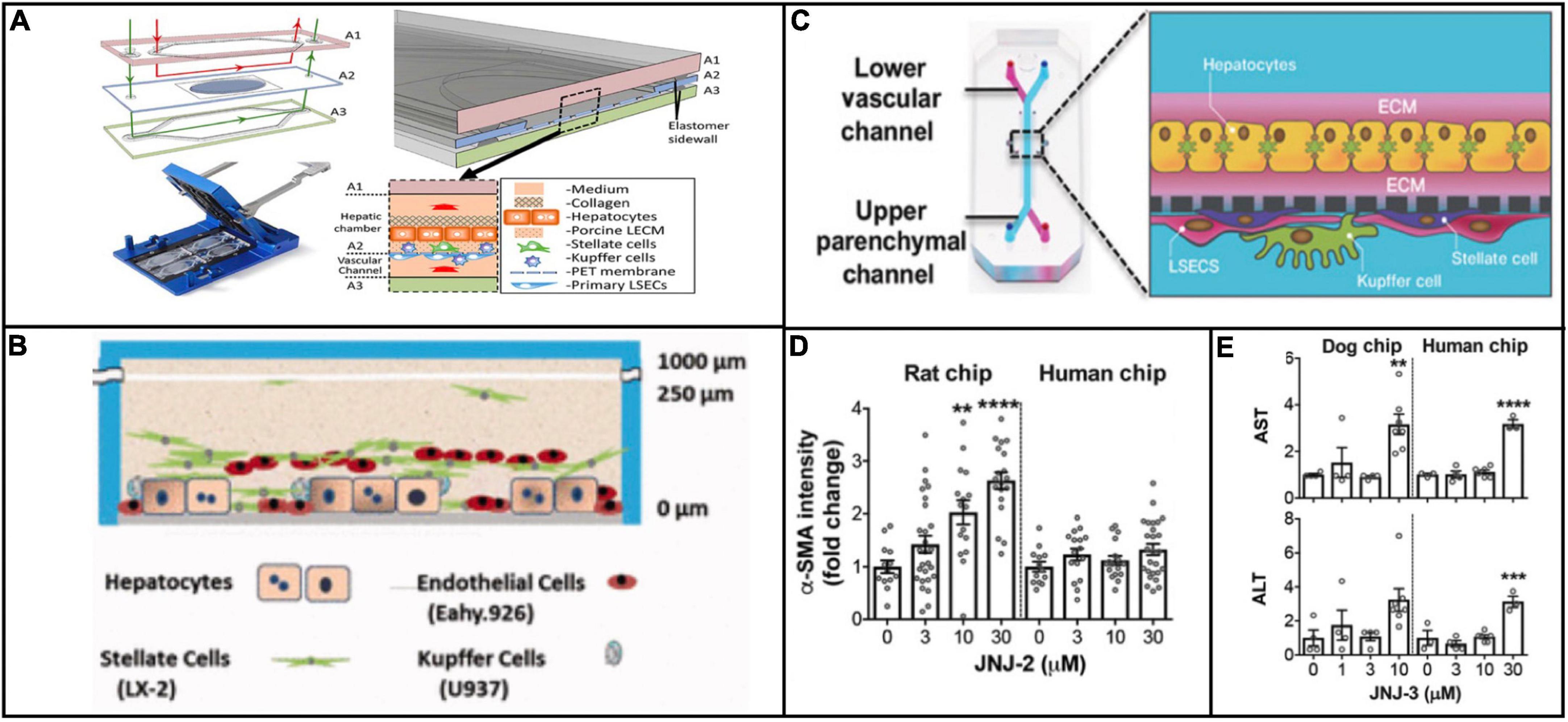
Figure 3. Examples of platforms that incorporate multiple cell types that are implicated in viral hepatitis pathogenesis. The platforms described in Li et al. (2018) (Reproduced with permissions from Li et al.) (A), Vernetti et al. (2016) (Reproduced with permissions from Vernetti et al.) (B), and Jang K. J. et al. (2019) (Reproduced with permissions from Jang et al.) (C) all include Kupffer cells and hepatic stellate cells in addition to hepatocytes and endothelial cells. The inclusion of all four cell types makes these platforms more effective for modeling a systemic response to viral infection. Jang et al. demonstrated that treatment of rat and human cells with a proprietary compound resulted in differential toxicity (D), but treatment of dog and human cells with a different compound resulted in a similar toxicity profile (E), evidence of the variation in responses between animal and human cells to different compounds. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Outside of the context of viral hepatitis, Emulate’s liver chip has already compared in vitro results against clinical data in terms of drug toxicity response. Jang et al. showed species-dependent differences in toxicity response to a Janssen compound that was discontinued during animal trials because of hepatoxicity in rats. These results were confirmed from with rat hepatocytes on the in vitro platform, but when the same experiment was conducted on human hepatocytes, no such toxic effect was observed (Figure 3D). This finding underscores obvious drawbacks of using animal models to test compounds intended for humans, in that some compounds are unnecessarily filtered out of contention due to species-dependent toxicity. Conversely, when testing a different proprietary compound that was discontinued due to hepatocellular necrosis in dogs, dog and human hepatocytes were aligned in their toxic response (Jang K. J. et al., 2019; Figure 3E). These findings highlight the potential for physiologically relevant microsystems to circumvent species-dependent discrepancies in response to drug treatment, making for more efficient compound screening and clinical translation.
Conclusion
In vitro studies are needed to aid in the ascertainment of new knowledge on the mechanisms that govern the transition of viral hepatitis from acute to chronic infection and related pathogenic processes. A lack of readily available in vivo models necessitates the use of in vitro systems that can attain the level of physiological relevance that is observed in vivo. Importantly, the capacity to study hepatotrophic viruses on human cells using models that can be interrogated through sophisticated and informative molecular assays is required. The developments of physiologically relevant platforms are in their early stages in terms of their use as models of viral hepatitis infection; however, the available studies thus far have demonstrated significant improvements in infection efficacy and longevity as compared to conventional cell culture methods. These preliminary steps in establishing more robust infection models will undoubtedly create beneficial tools to study virus-host interactions and uncover novel insights about chronic viral infection and related mechanisms that would not otherwise be possible through the use of more rudimentary platforms. Some of the systems mentioned above have yet to be specifically utilized for viral hepatitis modeling, but their shared goal of improved hepatocyte function in vitro translates across all applications. Nonetheless, physiomimetic platforms still have a variety of barriers to overcome before becoming reliable models of viral infection. For example, making these models higher throughput with easy access to cultures in order to introduce and assay infection is critical for these platforms to compete with conventional 2D culture where these two elements are commonplace. Fortunately, the plethora of tools and models that now exist can serve as a solid foundation for improving platforms to study virus-host interaction. Many of these platforms are now available for use, but need to be introduced to viral applications.
Author contributions
DM, DB, AA, and ET: manuscript concept, design and drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NIH-NIGMS grant R35GM124915.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis D virus; HEV, hepatitis E virus; HCC, hepatocellular carcinoma; 2D, two-dimensional; 3D, three-dimensional; cccDNA, covalently closed circular DNA; DAAs, direct acting antiviral agents; ECM, extracellular matrix; PHHs, primary human hepatocytes; iPSCs, induced pluripotent stem cells; NPCs, non-parenchymal cells; KCs, Kupffer cells; SCs, stellate cells; SARS-CoV2, severe acute respiratory syndrome coronavirus 2; NTCP, Na+-taurocholate cotransporting polypeptide; HLCs, hepatocyte-like cells; LBL, layer-by-layer; HUVECs, human umbilical vein endothelial cells; MPCC, micropatterned co-culture; dECM, decellularized ECM; PEGDA, polyethylene glycol diacrylate; RGD, arginine-glycine-aspartate; HF, hollow fiber; RWV, rotating wall vessel; MPCC, micropatterned co-cultures; PDMS, polydimethylsiloxane; PMMA, polymethyl methacrylate.
References
Aeby, E. A., Misun, P. M., Hierlemann, A., and Frey, O. (2018). Microfluidic Hydrogel Hanging-Drop Network for Long-Term Culturing of 3D Microtissues and Simultaneous High-Resolution Imaging. Adv. Biosyst. 2:1800054. doi: 10.1002/adbi.201800054
Ahmed, H. M. M., Salerno, S., Morelli, S., Giorno, L., and Bartolo, L. D. (2017). 3D liver membrane system by co-culturing human hepatocytes, sinusoidal endothelial and stellate cells. Biofabrication 9:025022. doi: 10.1088/1758-5090/aa70c7
Ahn, J., Ahn, J.-H., Yoon, S., Nam, Y. S., Son, M.-Y., and Oh, J.-H. (2019). Human three-dimensional in vitro model of hepatic zonation to predict zonal hepatotoxicity. J. Biol. Eng. 13:22. doi: 10.1186/s13036-019-0148-5
Aizaki, H., Nagamori, S., Matsuda, M., Kawakami, H., Hashimoto, O., Ishiko, H., et al. (2003). Production and release of infectious hepatitis C virus from human liver cell cultures in the three-dimensional radial-flow bioreactor. Virology 314, 16–25. doi: 10.1016/S0042-6822(03)00383-0
Akahori, Y., Kato, H., Fujita, T., Moriishi, K., Tanaka, Y., Watashi, K., et al. (2020). Establishment of a novel hepatitis B virus culture system using immortalized human hepatocytes. Sci. Rep. 10:21718. doi: 10.1038/s41598-020-78655-x
Aleo, M. D., Ukairo, O., Moore, A., Irrechukwu, O., Potter, D. M., and Schneider, R. P. (2019). Liver safety evaluation of endothelin receptor antagonists using HepatoPac§: A single model impact assessment on hepatocellular health, function and bile acid disposition. J. Appl. Toxicol. 39, 1192–1207. doi: 10.1002/jat.3805
Ananthanarayanan, A., Nugraha, B., Triyatni, M., Hart, S., Sankuratri, S., and Yu, H. (2014). Scalable Spheroid Model of Human Hepatocytes for Hepatitis C Infection and Replication. Mol. Pharm. 11, 2106–2114. doi: 10.1021/mp500063y
Axley, P., Ahmed, Z., Ravi, S., and Singal, A. K. (2018). Hepatitis C Virus and Hepatocellular Carcinoma: A Narrative Review. J. Clin. Transl. Hepatol. 6, 79–84. doi: 10.14218/JCTH.2017.00067
Bachmann, A., Moll, M., Gottwald, E., Nies, C., Zantl, R., Wagner, H., et al. (2015). 3D Cultivation Techniques for Primary Human Hepatocytes. Microarrays 4, 64–83. doi: 10.3390/microarrays4010064
Banaeiyan, A. A., Theobald, J., Paukštyte, J., Wölfl, S., Adiels, C. B., and Goksör, M. (2017). Design and fabrication of a scalable liver-lobule-on-a-chip microphysiological platform. Biofabrication 9:015014. doi: 10.1088/1758-5090/9/1/015014
Baptista, P. M., Siddiqui, M. M., Lozier, G., Rodriguez, S. R., Atala, A., and Soker, S. (2011). The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 53, 604–617. doi: 10.1002/hep.24067
Barbetta, A., Massimi, M., Di Rosario, B., Nardecchia, S., De Colli, M., Devirgiliis, L. C., et al. (2008). Emulsion Templated Scaffolds that Include Gelatin and Glycosaminoglycans. Biomacromolecules 9, 2844–2856. doi: 10.1021/bm800599d
Baudoin, R., Griscom, L., Prot, J. M., Legallais, C., and Leclerc, E. (2011). Behavior of HepG2/C3A cell cultures in a microfluidic bioreactor. Biochem. Eng. J. 53, 172–181. doi: 10.1016/j.bej.2010.10.007
Bavli, D., Prill, S., Ezra, E., Levy, G., Cohen, M., Vinken, M., et al. (2016). Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl. Acad. Sci. U.S.A. 113:E2231–E2240. doi: 10.1073/pnas.1522556113
Bell, C. C., Dankers, A. C. A., Lauschke, V. M., Sison-Young, R., Jenkins, R., Rowe, C., et al. (2018). Comparison of Hepatic 2D Sandwich Cultures and 3D Spheroids for Long-term Toxicity Applications: A Multicenter Study. Toxicol. Sci. 162, 655–666. doi: 10.1093/toxsci/kfx289
Berger, D. R., Ware, B. R., Davidson, M. D., Allsup, S. R., and Khetani, S. R. (2015). Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology 61, 1370–1381. doi: 10.1002/hep.27621
Berggren, K. A., Suzuki, S., and Ploss, A. (2020). Animal Models Used in Hepatitis C Virus Research. Int. J. Mol. Sci. 21:3869. doi: 10.3390/ijms21113869
Bhise, N. S., Manoharan, V., Massa, S., Tamayol, A., Ghaderi, M., Miscuglio, M., et al. (2016). A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 8:014101. doi: 10.1088/1758-5090/8/1/014101
Bin Ramli, M. N., Lim, Y. S., Koe, C. T., Demircioglu, D., Tng, W., Gonzales, K. A. U., et al. (2020). Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 159, 1471–1486.e12. doi: 10.1053/j.gastro.2020.06.010
Bircsak, K. M., DeBiasio, R., Miedel, M., Alsebahi, A., Reddinger, R., Saleh, A., et al. (2021). A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate§. Toxicology 450:152667. doi: 10.1016/j.tox.2020.152667
Boltjes, A., Movita, D., Boonstra, A., and Woltman, A. M. (2014). The role of Kupffer cells in hepatitis B and hepatitis C virus infections. J. Hepatol. 61, 660–671. doi: 10.1016/j.jhep.2014.04.026
Carraro, A., Hsu, W.-M., Kulig, K. M., Cheung, W. S., Miller, M. L., Weinberg, E. J., et al. (2008). In vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomed. Microdevices 10, 795–805. doi: 10.1007/s10544-008-9194-3
Chan, T. S., Yu, H., Moore, A., Khetani, S. R., and Tweedie, D. (2019). Meeting the Challenge of Predicting Hepatic Clearance of Compounds Slowly Metabolized by Cytochrome P450 Using a Novel Hepatocyte Model, HepatoPac. Drug Metab. Dispos. 47, 58–66. doi: 10.1124/dmd.113.053397fullarticlecorrection
Chang, T. T., and Hughes-Fulford, M. (2008). Monolayer and Spheroid Culture of Human Liver Hepatocellular Carcinoma Cell Line Cells Demonstrate Distinct Global Gene Expression Patterns and Functional Phenotypes. Tissue Eng. Part A 15, 559–567. doi: 10.1089/ten.tea.2007.0434
Chao, P., Maguire, T., Novik, E., Cheng, K.-C., and Yarmush, M. L. (2009). Evaluation of a microfluidic based cell culture platform with primary human hepatocytes for the prediction of hepatic clearance in human. Biochem. Pharmacol. 78, 625–632. doi: 10.1016/j.bcp.2009.05.013
Chen, Q., Utech, S., Chen, D., Prodanovic, R., Lin, J.-M., and Weitz, D. A. (2016). Controlled assembly of heterotypic cells in a core–shell scaffold: Organ in a droplet. Lab Chip 16, 1346–1349. doi: 10.1039/C6LC00231E
Chen, S. L., and Morgan, T. R. (2006). The Natural History of Hepatitis C Virus (HCV) Infection. Int. J. Med. Sci. 3, 47–52.
Chen, X., Saccon, E., Appelberg, K. S., Mikaeloff, F., Rodriguez, J. E., Vinhas, B. S., et al. (2021). Type-I interferon signatures in SARS-CoV-2 infected Huh7 cells. Cell Death Discov. 7:114. doi: 10.1038/s41420-021-00487-z
Cheng, S., Prot, J.-M., Leclerc, E., and Bois, F. Y. (2012). Zonation related function and ubiquitination regulation in human hepatocellular carcinoma cells in dynamic vs. static culture conditions. BMC Genom. 13:54. doi: 10.1186/1471-2164-13-54
Cho, N.-J., Elazar, M., Xiong, A., Lee, W., Chiao, E., Baker, J., et al. (2008). Viral infection of human progenitor and liver-derived cells encapsulated in three-dimensional PEG-based hydrogel. Biomed. Mater. 4:011001. doi: 10.1088/1748-6041/4/1/011001
Choucha Snouber, L., Bunescu, A., Naudot, M., Legallais, C., Brochot, C., Dumas, M. E., et al. (2013). Metabolomics-on-a-Chip of Hepatotoxicity Induced by Anticancer Drug Flutamide and Its Active Metabolite Hydroxyflutamide Using HepG2/C3a Microfluidic Biochips. Toxicol. Sci. 132, 8–20. doi: 10.1093/toxsci/kfs230
Christoffersson, J., Aronsson, C., Jury, M., Selegård, R., Aili, D., and Mandenius, C.-F. (2018). Fabrication of modular hyaluronan-PEG hydrogels to support 3D cultures of hepatocytes in a perfused liver-on-a-chip device. Biofabrication 11:015013. doi: 10.1088/1758-5090/aaf657
Crignis, E. D., Romal, S., Carofiglio, F., Moulos, P., Verstegen, M. M. A., Khalid, M. M., et al. (2020). Human liver organoids; a patient-derived primary model for HBV Infection and Related Hepatocellular Carcinoma. BioRxiv [Preprint]. doi: 10.1101/568147
Darnell, M., Ulvestad, M., Ellis, E., Weidolf, L., and Andersson, T. B. (2012). In Vitro Evaluation of Major In Vivo Drug Metabolic Pathways Using Primary Human Hepatocytes and HepaRG Cells in Suspension and a Dynamic Three-Dimensional Bioreactor System. J. Pharmacol. Exp. Ther. 343, 134–144. doi: 10.1124/jpet.112.195834
Davidson, M. D., Kukla, D. A., and Khetani, S. R. (2017). Microengineered cultures containing human hepatic stellate cells and hepatocytes for drug development. Integr. Biol. 9, 662–677. doi: 10.1039/C7IB00027H
De Bartolo, L., Salerno, S., Curcio, E., Piscioneri, A., Rende, M., Morelli, S., et al. (2009). Human hepatocyte functions in a crossed hollow fiber membrane bioreactor. Biomaterials 30, 2531–2543. doi: 10.1016/j.biomaterials.2009.01.011
de Oliveria Andrade, L. J., D’Oliveira, A., Melo, R. C., De Souza, E. C., Costa Silva, C. A., and Paraná, R. (2009). Association Between Hepatitis C and Hepatocellular Carcinoma. J. Glob. Infect. Dis. 1, 33–37. doi: 10.4103/0974-777X.52979
Deng, J., Wei, W., Chen, Z., Lin, B., Zhao, W., Luo, Y., et al. (2019a). Engineered Liver-On-A-Chip Platform to Mimic Liver Functions and Its Biomedical Applications. Micromachines 10:676. doi: 10.3390/mi10100676
Deng, J., Zhang, X., Chen, Z., Luo, Y., Lu, Y., Liu, T., et al. (2019b). A cell lines derived microfluidic liver model for investigation of hepatotoxicity induced by drug-drug interaction. Biomicrofluidics 13:024101. doi: 10.1063/1.5070088
Desai, P. K., Tseng, H., and Souza, G. R. (2017). Assembly of Hepatocyte Spheroids Using Magnetic 3D Cell Culture for CYP450 Inhibition/Induction. Int. J. Mol. Sci. 18:1085. doi: 10.3390/ijms18051085
Dixon, L. J., Barnes, M., Tang, H., Pritchard, M. T., and Nagy, L. E. (2013). Kupffer Cells in the Liver. Compr. Physiol. 3, 785–797. doi: 10.1002/cphy.c120026
Dustin, L. B., Bartolini, B., Capobianchi, M. R., and Pistello, M. (2016). Hepatitis C virus: Life cycle in cells, infection and host response, and analysis of molecular markers influencing the outcome of infection and response to therapy. Clin. Microbiol. Infect. 22, 826–832. doi: 10.1016/j.cmi.2016.08.025
El-Serag, H. B. (2002). Hepatocellular carcinoma and hepatitis C in the United States. Hepatology 36:S74–S83. doi: 10.1002/hep.1840360710
Esch, M. B., Prot, J.-M., Wang, Y. I., Miller, P., Llamas-Vidales, J. R., Naughton, B. A., et al. (2015). Multi-Cellular 3D Human Primary Liver Cell Cultures Elevate Metabolic Activity Under Fluidic Flow. Lab Chip 15, 2269–2277. doi: 10.1039/c5lc00237k
Faulkner-Jones, A., Fyfe, C., Cornelissen, D.-J., Gardner, J., King, J., Courtney, A., et al. (2015). Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte-like cells for the generation of mini-livers in 3D. Biofabrication 7:044102. doi: 10.1088/1758-5090/7/4/044102
Feldstein, A. E., Papouchado, B. G., Angulo, P., Sanderson, S., Adams, L., and Gores, G. J. (2005). Hepatic stellate cells and fibrosis progression in patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 3, 384–389. doi: 10.1016/s1542-3565(04)00616-0
Fey, S. J., and Wrzesinski, K. (2012). Determination of Drug Toxicity Using 3D Spheroids Constructed From an Immortal Human Hepatocyte Cell Line. Toxicol. Sci. 127, 403–411. doi: 10.1093/toxsci/kfs122
Foster, A. J., Chouhan, B., Regan, S. L., Rollison, H., Amberntsson, S., Andersson, L. C., et al. (2019). Integrated in vitro models for hepatic safety and metabolism: Evaluation of a human Liver-Chip and liver spheroid. Arch. Toxicol. 93, 1021–1037. doi: 10.1007/s00204-019-02427-4
Fukuda, J., Khademhosseini, A., Yeo, Y., Yang, X., Yeh, J., Eng, G., et al. (2006). Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials 27, 5259–5267. doi: 10.1016/j.biomaterials.2006.05.044
Gaskell, H., Sharma, P., Colley, H. E., Murdoch, C., Williams, D. P., and Webb, S. D. (2016). Characterization of a functional C3A liver spheroid model. Toxicol. Res. 5, 1053–1065. doi: 10.1039/c6tx00101g
Gerlach, J. C., Mutig, K., Sauer, I. M., Schrade, P., Efimova, E., Mieder, T., et al. (2003). Use of primary human liver cells originating from discarded grafts in a bioreactor for liver support therapy and the prospects of culturing adult liver stem cells in bioreactors: A morphologic study. Transplantation 76, 781–786. doi: 10.1097/01.TP.0000083319.36931.32
Gieseck, R. L. III, Hannan, N. R. F., Bort, R., Hanley, N. A., Drake, R. A. L., Cameron, G. W. W., et al. (2014). Maturation of Induced Pluripotent Stem Cell Derived Hepatocytes by 3D-Culture. PLoS One 9:e86372. doi: 10.1371/journal.pone.0086372
Giobbe, G. G., Michielin, F., Luni, C., Giulitti, S., Martewicz, S., Dupont, S., et al. (2015). Functional differentiation of human pluripotent stem cells on a chip. Nat. Methods 12, 637–640. doi: 10.1038/nmeth.3411
Goral, V. N., Hsieh, Y.-C., Petzold, O. N., Clark, J. S., Yuen, P. K., and Faris, R. A. (2010). Perfusion-based microfluidic device for three-dimensional dynamic primary human hepatocyte cell culture in the absence of biological or synthetic matrices or coagulants. Lab Chip 10, 3380–3386. doi: 10.1039/C0LC00135J
Grix, T., Ruppelt, A., Thomas, A., Amler, A.-K., Noichl, B. P., Lauster, R., et al. (2018). Bioprinting Perfusion-Enabled Liver Equivalents for Advanced Organ-on-a-Chip Applications. Genes 9:176. doi: 10.3390/genes9040176
Gunness, P., Mueller, D., Shevchenko, V., Heinzle, E., Ingelman-Sundberg, M., and Noor, F. (2013). 3D Organotypic Cultures of Human HepaRG Cells: A Tool for In Vitro Toxicity Studies. Toxicol. Sci. 133, 67–78. doi: 10.1093/toxsci/kft021
Gural, N., Mancio-Silva, L., Miller, A. B., Galstian, A., Butty, V. L., Levine, S. S., et al. (2018). In Vitro Culture, Drug Sensitivity, and Transcriptome of Plasmodium Vivax Hypnozoites. Cell Host Microbe 23, 395–406.e4. doi: 10.1016/j.chom.2018.01.002
Guye, P., Ebrahimkhani, M. R., Kipniss, N., Velazquez, J. J., Schoenfeld, E., Kiani, S., et al. (2016). Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 7:10243. doi: 10.1038/ncomms10243
Hendriks, D. F. G., Fredriksson Puigvert, L., Messner, S., Mortiz, W., and Ingelman-Sundberg, M. (2016). Hepatic 3D spheroid models for the detection and study of compounds with cholestatic liability. Sci. Rep. 6:35434. doi: 10.1038/srep35434
hepbtalk. (2019). The Global Need for Hepatitis B Immunization. Available Online at: https://www.hepb.org/blog/global-need-hepatitis-b-immunization/ (accessed June 20, 2022).
Hiller, T., Berg, J., Elomaa, L., Röhrs, V., Ullah, I., Schaar, K., et al. (2018). Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies. Int. J. Mol. Sci. 19:3129. doi: 10.3390/ijms19103129
Ho, C.-T., Lin, R.-Z., Chen, R.-J., Chin, C.-K., Gong, S.-E., Chang, H.-Y., et al. (2013). Liver-cell patterning Lab Chip: Mimicking the morphology of liver lobule tissue. Lab Chip 13, 3578–3587. doi: 10.1039/C3LC50402F
Hoffmann, S. A., Müller-Vieira, U., Biemel, K., Knobeloch, D., Heydel, S., Lübberstedt, M., et al. (2012). Analysis of drug metabolism activities in a miniaturized liver cell bioreactor for use in pharmacological studies. Biotechnol. Bioeng. 109, 3172–3181. doi: 10.1002/bit.24573
Huch, M., Gehart, H., van Boxtel, R., Hamer, K., Blokzijl, F., Verstegen, M. M. A., et al. (2015). Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 160, 299–312. doi: 10.1016/j.cell.2014.11.050
Huh, D., Hamilton, G. A., and Ingber, D. E. (2011). From 3D cell culture to organs-on-chips. Trends Cell Biol. 21, 745–754. doi: 10.1016/j.tcb.2011.09.005
Hultman, I., Vedin, C., Abrahamsson, A., Winiwarter, S., and Darnell, M. (2016). Use of HμREL Human Coculture System for Prediction of Intrinsic Clearance and Metabolite Formation for Slowly Metabolized Compounds. Mol. Pharm. 13, 2796–2807. doi: 10.1021/acs.molpharmaceut.6b00396
Hwang, C. M., Sant, S., Masaeli, M., Kachouie, N. N., Zamanian, B., Lee, S.-H., et al. (2010). Fabrication of three-dimensional porous cell-laden hydrogel for tissue engineering. Biofabrication 2:035003. doi: 10.1088/1758-5082/2/3/035003
Hyun, J., McMahon, R. S., Lang, A. L., Edwards, J. S., Badilla, A. D., Greene, M. E., et al. (2019). HIV and HCV augments inflammatory responses through increased TREM-1 expression and signaling in Kupffer and Myeloid cells. PLoS Pathog. 15:e1007883. doi: 10.1371/journal.ppat.1007883
Illa, X., Vila, S., Yeste, J., Peralta, C., Gracia-Sancho, J., and Villa, R. (2014). A Novel Modular Bioreactor to In Vitro Study the Hepatic Sinusoid. PLoS One 9:e111864. doi: 10.1371/journal.pone.0111864
Ioannou, G. N., Beste, L. A., Green, P. K., Singal, A. G., Tapper, E. B., Waljee, A. K., et al. (2019). Increased Risk for Hepatocellular Carcinoma Persists Up to 10 Years After HCV Eradication in Patients With Baseline Cirrhosis or High FIB-4 Scores. Gastroenterology 157, 1264–1278.e4. doi: 10.1053/j.gastro.2019.07.033
Ishibashi, H., Nakamura, M., Komori, A., Migita, K., and Shimoda, S. (2009). Liver architecture, cell function, and disease. Semin. Immunopathol. 31:399. doi: 10.1007/s00281-009-0155-6
Jang, K. J., Otieno, M. A., Ronxhi, J., Lim, H.-K., Ewart, L., Kodella, K. R., et al. (2019). Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 11:eaax5516. doi: 10.1126/scitranslmed.aax5516
Jang, M., Kleber, A., Ruckelshausen, T., Betzholz, R., and Manz, A. (2019). Differentiation of the human liver progenitor cell line (HepaRG) on a microfluidic-based biochip. J. Tissue Eng. Regen. Med. 13, 482–494. doi: 10.1002/term.2802
Jang, M., Manz, A., Volk, T., and Kleber, A. (2018). Study of melatonin-mediated effects on various hepatic inflammatory responses stimulated by IL-6 in a new HepG2-on-a-chip platform. Biomed. Microdevices 20:54. doi: 10.1007/s10544-018-0300-x
Jang, M., Neuzil, P., Volk, T., Manz, A., and Kleber, A. (2015). On-chip three-dimensional cell culture in phaseguides improves hepatocyte functions in vitro. Biomicrofluidics 9:034113. doi: 10.1063/1.4922863
Jeon, H., Kang, K., Park, S. A., Kim, W. D., Paik, S. S., Lee, S.-H., et al. (2017). Generation of Multilayered 3D Structures of HepG2 Cells Using a Bio-printing Technique. Gut Liver 11, 121–128. doi: 10.5009/gnl16010
Jia, Z., Cheng, Y., Jiang, X., Zhang, C., Wang, G., Xu, J., et al. (2020). 3D Culture System for Liver Tissue Mimicking Hepatic Plates for Improvement of Human Hepatocyte (C3A) Function and Polarity. BioMed Res. Int. 2020:e6354183. doi: 10.1155/2020/6354183
Jung, H.-R., Kang, H. M., Ryu, J.-W., Kim, D.-S., Noh, K. H., Kim, E.-S., et al. (2017). Cell Spheroids with Enhanced Aggressiveness to Mimic Human Liver Cancer In Vitro and In Vivo. Sci. Rep. 7:10499. doi: 10.1038/s41598-017-10828-7
Kalyanaraman, B., Supp, D. M., and Boyce, S. T. (2008). Medium flow rate regulates viability and barrier function of engineered skin substitutes in perfusion culture. Tissue Eng. Part A 14, 583–593. doi: 10.1089/tea.2007.0237
Kang, Y., Rawat, S., Duchemin, N., Bouchard, M., and Noh, M. (2017). Human Liver Sinusoid on a Chip for Hepatitis B Virus Replication Study. Micromachines 8:27. doi: 10.3390/mi8010027
Kang, Y. B. A, Eo, J., Mert, S., Yarmush, M. L., and Usta, O. B. (2018). Metabolic Patterning on a Chip: Towards in vitro Liver Zonation of Primary Rat and Human Hepatocytes. Sci. Rep. 8:8951. doi: 10.1038/s41598-018-27179-6
Kawada, M., Nagamori, S., Aizaki, H., Fukaya, K., Niiya, M., Matsuura, T., et al. (1998). Massive culture of human liver cancer cells in a newly developed radial flow bioreactor system: Ultrafine structure of functionally enhanced hepatocarcinoma cell lines. Vitro Cell. Dev. Biol. Anim. 34, 109–115. doi: 10.1007/s11626-998-0092-z
Khalil, M., Shariat-Panahi, A., Tootle, R., Ryder, T., McCloskey, P., Roberts, E., et al. (2001). Human hepatocyte cell lines proliferating as cohesive spheroid colonies in alginate markedly upregulate both synthetic and detoxificatory liver function. J. Hepatol. 34, 68–77. doi: 10.1016/S0168-8278(00)00080-5
Khetani, S. R., and Bhatia, S. N. (2008). Microscale culture of human liver cells for drug development. Nat. Biotechnol. 26, 120–126. doi: 10.1038/nbt1361
Khetani, S. R., Kanchagar, C., Ukairo, O., Krzyzewski, S., Moore, A., Shi, J., et al. (2013). Use of Micropatterned Cocultures to Detect Compounds That Cause Drug-Induced Liver Injury in Humans. Toxicol. Sci. 132, 107–117. doi: 10.1093/toxsci/kfs326
Kim, J. H., Jang, Y. J., An, S. Y., Son, J., Lee, J., Lee, G., et al. (2015). Enhanced Metabolizing Activity of Human ES Cell-Derived Hepatocytes Using a 3D Culture System with Repeated Exposures to Xenobiotics. Toxicol. Sci. 147, 190–206. doi: 10.1093/toxsci/kfv121
Kobayashi, A., Yamakoshi, K., Yajima, Y., Utoh, R., Yamada, M., and Seki, M. (2013). Preparation of stripe-patterned heterogeneous hydrogel sheets using microfluidic devices for high-density coculture of hepatocytes and fibroblasts. J. Biosci. Bioeng. 116, 761–767. doi: 10.1016/j.jbiosc.2013.05.034
Kostadinova, R., Boess, F., Applegate, D., Suter, L., Weiser, T., Singer, T., et al. (2013). A long-term three dimensional liver co-culture system for improved prediction of clinically relevant drug-induced hepatotoxicity. Toxicol. Appl. Pharmacol. 268, 1–16. doi: 10.1016/j.taap.2013.01.012
Kozyra, M., Johansson, I., Nordling, Å, Ullah, S., Lauschke, V. M., and Ingelman-Sundberg, M. (2018). Human hepatic 3D spheroids as a model for steatosis and insulin resistance. Sci. Rep. 8:14297. doi: 10.1038/s41598-018-32722-6
Kukla, D. A., Crampton, A. L., Wood, D. K., and Khetani, S. R. (2020). Microscale Collagen and Fibroblast Interactions Enhance Primary Human Hepatocyte Functions in Three-Dimensional Models. Gene Expr. 20, 1–18. doi: 10.3727/105221620X15868728381608
Kwon, S. J., Lee, D. W., Shah, D. A., Ku, B., Jeon, S. Y., Solanki, K., et al. (2014). High-throughput and combinatorial gene expression on a chip for metabolism-induced toxicology screening. Nat. Commun. 5:3739. doi: 10.1038/ncomms4739
Lang, R., Stern, M. M., Smith, L., Liu, Y., Bharadwaj, S., Liu, G., et al. (2011). Three-dimensional culture of hepatocytes on porcine liver tissue-derived extracellular matrix. Biomaterials 32, 7042–7052. doi: 10.1016/j.biomaterials.2011.06.005
Lautt, W. W. (2009). Overview. Available Online at: https://www.ncbi.nlm.nih.gov/books/NBK53069/ (accessed Dec 7, 2020).
Leclerc, E., Sakai, Y., and Fujii, T. (2004). Perfusion culture of fetal human hepatocytes in microfluidic environments. Biochem. Eng. J. 20, 143–148. doi: 10.1016/j.bej.2003.09.010
Lee, H., Chae, S., Kim, J. Y., Han, W., Kim, J., Choi, Y., et al. (2019). Cell-printed 3D liver-on-a-chip possessing a liver microenvironment and biliary system. Biofabrication 11:025001. doi: 10.1088/1758-5090/aaf9fa
Lee, H., Han, W., Kim, H., Ha, D.-H., Jang, J., Kim, B. S., et al. (2017). Development of Liver Decellularized Extracellular Matrix Bioink for Three-Dimensional Cell Printing-Based Liver Tissue Engineering. Biomacromolecules 18, 1229–1237. doi: 10.1021/acs.biomac.6b01908
Lee, P. J., Hung, P. J., and Lee, L. P. (2007). An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 97, 1340–1346. doi: 10.1002/bit.21360
Lee-Montiel, F. T., George, S. M., Gough, A. H., Sharma, A. D., Wu, J., DeBiasio, R., et al. (2017). Control of oxygen tension recapitulates zone-specific functions in human liver microphysiology systems. Exp. Biol. Med. 242, 1617–1632. doi: 10.1177/1535370217703978
Leite, S. B., Wilk-Zasadna, I., Zaldivar, J. M., Airola, E., Reis-Fernandes, M. A., Mennecozzi, M., et al. (2012). Three-Dimensional HepaRG Model As An Attractive Tool for Toxicity Testing. Toxicol. Sci. 130, 106–116. doi: 10.1093/toxsci/kfs232
Li, J., Zong, L., Sureau, C., Barker, L., Wands, J. R., and Tong, S. (2016). Unusual Features of Sodium Taurocholate Cotransporting Polypeptide as a Hepatitis B Virus Receptor. J. Virol. 90, 8302–8313. doi: 10.1128/JVI.01153-16
Li, P., He, K., Li, J., Liu, Z., and Gong, J. (2017). The role of Kupffer cells in hepatic diseases. Mol. Immunol. 85, 222–229. doi: 10.1016/j.molimm.2017.02.018
Li, X., George, S. M., Vernetti, L., Gough, A. H., and Taylor, D. L. (2018). A Glass-based, Continuously Zonated and Vascularized Human Liver Acinus Microphysiological System (vLAMPS) Designed for Experimental Modeling of Diseases and ADME/TOX. Lab Chip 18, 2614–2631. doi: 10.1039/c8lc00418h
Liu, Y., Maya, S., and Ploss, A. (2021). Animal Models of Hepatitis B Virus Infection–Success, Challenges, and Future Directions. Viruses 13:777. doi: 10.3390/v13050777
Long, T. J., Cosgrove, P. A., Dunn, R. T., Stolz, D. B., Hamadeh, H., Afshari, C., et al. (2016). Modeling Therapeutic Antibody–Small Molecule Drug-Drug Interactions Using a Three-Dimensional Perfusable Human Liver Coculture Platform. Drug Metab. Dispos. 44, 1940–1948. doi: 10.1124/dmd.116.071456
Louz, D., Bergmans, H. E., Loos, B. P., and Hoeben, R. C. (2013). Animal models in virus research: Their utility and limitations. Crit. Rev. Microbiol. 39, 325–361. doi: 10.3109/1040841X.2012.711740
Lübberstedt, M., Müller-Vieira, U., Biemel, K. M., Darnell, M., Hoffmann, S. A., Knöspel, F., et al. (2015). Serum-free culture of primary human hepatocytes in a miniaturized hollow-fibre membrane bioreactor for pharmacological in vitro studies. J. Tissue Eng. Regen. Med. 9, 1017–1026. doi: 10.1002/term.1652
Ma, C., Zhao, L., Zhou, E.-M., Xu, J., Shen, S., and Wang, J. (2016). On-Chip Construction of Liver Lobule-like Microtissue and Its Application for Adverse Drug Reaction Assay. Anal. Chem. 88, 1719–1727. doi: 10.1021/acs.analchem.5b03869
Ma, X., Qu, X., Zhu, W., Li, Y.-S., Yuan, S., Zhang, H., et al. (2016). Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U.S.A. 113, 2206–2211. doi: 10.1073/pnas.1524510113
Ma, L.-D., Wang, Y.-T., Wang, J.-R., Wu, J.-L., Meng, X.-S., Hu, P., et al. (2018). Design and fabrication of a liver-on-a-chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab Chip 18, 2547–2562. doi: 10.1039/C8LC00333E
Maghsoudlou, P., Georgiades, F., Smith, H., Milan, A., Shangaris, P., Urbani, L., et al. (2016). Optimization of Liver Decellularization Maintains Extracellular Matrix Micro-Architecture and Composition Predisposing to Effective Cell Seeding. PLoS One 11:e0155324. doi: 10.1371/journal.pone.0155324
Maher, S. P., Crouse, R. B., Conway, A. J., Bannister, E. C., Achyuta, A. K. H., Clark, A. Y., et al. (2014). Microphysical space of a liver sinusoid device enables simplified long-term maintenance of chimeric mouse-expanded human hepatocytes. Biomed. Microdevices 16, 727–736. doi: 10.1007/s10544-014-9877-x
Mandon, M., Huet, S., Dubreil, E., Fessard, V., and Le Hégarat, L. (2019). Three-dimensional HepaRG spheroids as a liver model to study human genotoxicity in vitro with the single cell gel electrophoresis assay. Sci. Rep. 9:10548. doi: 10.1038/s41598-019-47114-7
Mao, Q., Wang, Y., Li, Y., Juengpanich, S., Li, W., Chen, M., et al. (2020). Fabrication of liver microtissue with liver decellularized extracellular matrix (dECM) bioink by digital light processing (DLP) bioprinting. Mater. Sci. Eng. C 109:110625. doi: 10.1016/j.msec.2020.110625
Matellan, C., and Del Río Hernández, A. E. (2019). Engineering the cellular mechanical microenvironment – from bulk mechanics to the nanoscale. J. Cell Sci. 132:jcs229013. doi: 10.1242/jcs.229013
Matsusaki, M., Sakaue, K., Kadowaki, K., and Akashi, M. (2013). Three-Dimensional Human Tissue Chips Fabricated by Rapid and Automatic Inkjet Cell Printing. Adv. Healthc. Mater. 2, 534–539. doi: 10.1002/adhm.201200299
Mazza, G., Rombouts, K., Rennie Hall, A., Urbani, L., Vinh Luong, T., Al-Akkad, W., et al. (2015). Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci. Rep. 5:13079. doi: 10.1038/srep13079
McCarty, W. J., Usta, O. B., Luitje, M., Bale, S. S., Bhushan, A., Hegde, M., et al. (2014). A novel ultrathin collagen nanolayer assembly for 3-D microtissue engineering: Layer-by-layer collagen deposition for long-term stable microfluidic hepatocyte culture. Technology 2, 67–74. doi: 10.1142/S2339547814500083
Meier, F., Freyer, N., Brzeszczynska, J., Knöspel, F., Armstrong, L., Lako, M., et al. (2017). Hepatic differentiation of human iPSCs in different 3D models: A comparative study. Int. J. Mol. Med. 40, 1759–1771. doi: 10.3892/ijmm.2017.3190
Messner, S., Agarkova, I., Moritz, W., and Kelm, J. M. (2013). Multi-cell type human liver microtissues for hepatotoxicity testing. Arch. Toxicol. 87, 209–213. doi: 10.1007/s00204-012-0968-2
Messner, S., Fredriksson, L., Lauschke, V. M., Roessger, K., Escher, C., Bober, M., et al. (2018). Transcriptomic, Proteomic, and Functional Long-Term Characterization of Multicellular Three-Dimensional Human Liver Microtissues. Appl. Vitro Toxicol. 4, 1–12. doi: 10.1089/aivt.2017.0022
Mi, S., Yi, X., Du, Z., Xu, Y., and Sun, W. (2018). Construction of a liver sinusoid based on the laminar flow on chip and self-assembly of endothelial cells. Biofabrication 10:025010. doi: 10.1088/1758-5090/aaa97e
Miyamoto, Y., Ikeuchi, M., Noguchi, H., Yagi, T., and Hayashi, S. (2015). Spheroid Formation and Evaluation of Hepatic Cells in a Three-Dimensional Culture Device. Cell Med. 8, 47–56. doi: 10.3727/215517915X689056
Molina-Jimenez, F., Benedicto, I., Dao Thi, V. L., Gondar, V., Lavillette, D., Marin, J. J., et al. (2012). Matrigel-embedded 3D culture of Huh-7 cells as a hepatocyte-like polarized system to study hepatitis C virus cycle. Virology 425, 31–39. doi: 10.1016/j.virol.2011.12.021
Moya, A., Ortega-Ribera, M., Guimerà, X., Sowade, E., Zea, M., Illa, X., et al. (2018). Online oxygen monitoring using integrated inkjet-printed sensors in a liver-on-a-chip system. Lab Chip 18, 2023–2035. doi: 10.1039/C8LC00456K
Mueller, D., Krämer, L., Hoffmann, E., Klein, S., and Noor, F. (2014). 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicol. In Vitro 28, 104–112. doi: 10.1016/j.tiv.2013.06.024
Mun, S. J., Ryu, J.-S., Lee, M.-O., Son, Y. S., Oh, S. J., Cho, H.-S., et al. (2019). Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 71, 970–985. doi: 10.1016/j.jhep.2019.06.030
Nagamoto, Y., Tashiro, K., Takayama, K., Ohashi, K., Kawabata, K., Sakurai, F., et al. (2012). The promotion of hepatic maturation of human pluripotent stem cells in 3D co-culture using type I collagen and Swiss 3T3 cell sheets. Biomaterials 33, 4526–4534. doi: 10.1016/j.biomaterials.2012.03.011
Nahon, P., and Ganne-Carrié, N. (2019). Management of patients with pre-therapeutic advanced liver fibrosis following HCV eradication. JHEP Rep. 1, 480–489. doi: 10.1016/j.jhepr.2019.11.001
Nakai, S., Shibata, I., Shitamichi, T., Yamaguchi, H., Takagi, N., Inoue, T., et al. (2019). Collagen vitrigel promotes hepatocytic differentiation of induced pluripotent stem cells into functional hepatocyte-like cells. Biol. Open 8:bio042192. doi: 10.1242/bio.042192
Natarajan, V., Harris, E. N., and Kidambi, S. (2017). SECs (Sinusoidal Endothelial Cells), Liver Microenvironment, and Fibrosis. BioMed Res. Int. 2017:e4097205. doi: 10.1155/2017/4097205
Natarajan, V., Simoneau, C. R., Erickson, A. L., Meyers, N. L., Baron, J. L., Cooper, S., et al. (2022). Modelling T-cell immunity against hepatitis C virus with liver organoids in a microfluidic coculture system. Open Biol. 12:210320. doi: 10.1098/rsob.210320
Ndongo-Thiam, N., Berthillon, P., Errazuriz, E., Bordes, I., Sequeira, S. D., Trépo, C., et al. (2011). Long-term propagation of serum hepatitis C virus (HCV) with production of enveloped HCV particles in human HepaRG hepatocytes. Hepatology 54, 406–417. doi: 10.1002/hep.24386
Nguyen, D. G., Funk, J., Robbins, J. B., Crogan-Grundy, C., Presnell, S. C., Singer, T., et al. (2016). Bioprinted 3D Primary Liver Tissues Allow Assessment of Organ-Level Response to Clinical Drug Induced Toxicity In Vitro. PLoS One 11:e0158674. doi: 10.1371/journal.pone.0158674
Nguyen, T. V., Ukairo, O., Khetani, S. R., McVay, M., Kanchagar, C., Seghezzi, W., et al. (2015). Establishment of a Hepatocyte-Kupffer Cell Coculture Model for Assessment of Proinflammatory Cytokine Effects on Metabolizing Enzymes and Drug Transporters. Drug Metab. Dispos. 43, 774–785. doi: 10.1124/dmd.114.061317
Nie, Y.-Z., Zheng, Y.-W., Miyakawa, K., Murata, S., Zhang, R.-R., Sekine, K., et al. (2018). Recapitulation of hepatitis B virus–host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine 35, 114–123. doi: 10.1016/j.ebiom.2018.08.014
NIH strategic plan details pathway to achieving Hepatitis B cure (2019). Natl. Inst. Health NIH. Available Online at: https://www.nih.gov/news-events/news-releases/nih-strategic-plan-details-pathway-achieving-hepatitis-b-cure (accessed Dec 8, 2020).
Novik, E., Maguire, T. J., Chao, P., Cheng, K. C., and Yarmush, M. L. (2010). A microfluidic hepatic coculture platform for cell-based drug metabolism studies. Biochem. Pharmacol. 79, 1036–1044. doi: 10.1016/j.bcp.2009.11.010
Nuciforo, S., Fofana, I., Matter, M. S., Blumer, T., Calabrese, D., Boldanova, T., et al. (2018). Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 24, 1363–1376. doi: 10.1016/j.celrep.2018.07.001
Ogihara, T., Iwai, H., Inoue, Y., Katagi, J., Matsumoto, N., Motoi-Ohtsuji, M., et al. (2015). Utility of human hepatocyte spheroids for evaluation of hepatotoxicity. Fundam. Toxicol. Sci. 2, 41–48. doi: 10.2131/fts.2.41
Ohkura, T., Ohta, K., Nagao, T., Kusumoto, K., Koeda, A., Ueda, T., et al. (2014). Evaluation of Human Hepatocytes Cultured by Three-dimensional Spheroid Systems for Drug Metabolism. Drug Metab. Pharmacokinet. 29, 373–378. doi: 10.2133/dmpk.DMPK-13-RG-105
Ohshita, H., and Tateno, C. (2017). “Propagation of Human Hepatocytes in uPA/SCID Mice: Producing Chimeric Mice with Humanized Liver,” in Hepatocyte Transplantation: Methods and Protocols Methods in Molecular Biology, eds P. Stock and B. Christ (New York, NY: Springer), 91–100. doi: 10.1007/978-1-4939-6506-9_6
Ong, L. J. Y., Chong, L. H., Jin, L., Singh, P. K., Lee, P. S., Yu, H., et al. (2017). A pump-free microfluidic 3D perfusion platform for the efficient differentiation of human hepatocyte-like cells. Biotechnol. Bioeng. 114, 2360–2370. doi: 10.1002/bit.26341
Ortega-Prieto, A. M., Skelton, J. K., Cherry, C., Briones-Orta, M. A., Hateley, C. A., and Dorner, M. (2019). “Liver-on-a-Chip” Cultures of Primary Hepatocytes and Kupffer Cells for Hepatitis B Virus Infection. J. Vis. Exp. 144:58333. doi: 10.3791/58333
Ortega-Prieto, A. M., Skelton, J. K., Wai, S. N., Large, E., Lussignol, M., Vizcay-Barrena, G., et al. (2018). 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat. Commun. 9:682. doi: 10.1038/s41467-018-02969-8
Ortega-Ribera, M., Fernández-Iglesias, A., Illa, X., Moya, A., Molina, V., Maeso-Díaz, R., et al. (2018). Resemblance of the human liver sinusoid in a fluidic device with biomedical and pharmaceutical applications. Biotechnol. Bioeng. 115, 2585–2594. doi: 10.1002/bit.26776
Ott, L. M., Ramachandran, K., and Stehno-Bittel, L. (2017). An Automated Multiplexed Hepatotoxicity and CYP Induction Assay Using HepaRG Cells in 2D and 3D. SLAS Discov. Adv. Sci. Drug Discov. 22, 614–625. doi: 10.1177/2472555217701058
Ouchi, R., Togo, S., Kimura, M., Shinozawa, T., Koido, M., Koike, H., et al. (2019). Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab. 30, 374–384.e6. doi: 10.1016/j.cmet.2019.05.007
Peel, S., Corrigan, A. M., Ehrhardt, B., Jang, K.-J., Caetano-Pinto, P., Boeckeler, M., et al. (2019). Introducing an automated high content confocal imaging approach for Organs-on-Chips. Lab Chip 19, 410–421. doi: 10.1039/C8LC00829A
Petropolis, D. B., Faust, D. M., Tolle, M., Rivière, L., Valentin, T., Neuveut, C., et al. (2016). Human Liver Infection in a Dish: Easy-To-Build 3D Liver Models for Studying Microbial Infection. PLoS One 11:e0148667. doi: 10.1371/journal.pone.0148667
Phillips, M. B., Balbuena-Venancio, P., Enders, J. R., Norini, R. L., Shim, Y.-S., Burgunder, E., et al. (2018). Xenobiotic Metabolism in Alginate-Encapsulated Primary Human Hepatocytes Over Long Timeframes. Appl. Vitro Toxicol. 4, 238–247. doi: 10.1089/aivt.2017.0029
Pless, G., Steffen, I., Zeilinger, K., Sauer, I. M., Katenz, E., Kehr, D. C., et al. (2006). Evaluation of Primary Human Liver Cells in Bioreactor Cultures for Extracorporeal Liver Support on the Basis of Urea Production. Artif. Organs 30, 686–694. doi: 10.1111/j.1525-1594.2006.00285.x
Ploss, A., Khetani, S. R., Jones, C. T., Syder, A. J., Trehan, K., Gaysinskaya, V. A., et al. (2010). Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc. Natl. Acad. Sci. U.S.A. 107, 3141–3145. doi: 10.1073/pnas.0915130107
Proctor, W. R., Foster, A. J., Vogt, J., Summers, C., Middleton, B., Pilling, M. A., et al. (2017). Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch. Toxicol. 91, 2849–2863. doi: 10.1007/s00204-017-2002-1
Prodanov, L., Jindal, R., Bale, S. S., Hegde, M., McCarty, W. J., Golberg, I., et al. (2016). Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol. Bioeng. 113, 241–246. doi: 10.1002/bit.25700
Prot, J. M., Aninat, C., Griscom, L., Razan, F., Brochot, C., Guillouzo, C. G., et al. (2011). Improvement of HepG2/C3a cell functions in a microfluidic biochip. Biotechnol. Bioeng. 108, 1704–1715. doi: 10.1002/bit.23104
Prot, J. M., Maciel, L., Bricks, T., Merlier, F., Cotton, J., Paullier, P., et al. (2014). First pass intestinal and liver metabolism of paracetamol in a microfluidic platform coupled with a mathematical modeling as a means of evaluating ADME processes in humans. Biotechnol. Bioeng. 111, 2027–2040. doi: 10.1002/bit.25232
Rajalakshmy, A. R., Malathi, J., Madhavan, H. N., and Samuel, J. K. A. (2015). Mebiolgel, a thermoreversible polymer as a scaffold for three dimensional culture of Huh7 cell line with improved hepatocyte differentiation marker expression and HCV replication. Indian J. Med. Microbiol. 33:554. doi: 10.4103/0255-0857.167330
Ramachandran, S. D., Schirmer, K., Münst, B., Heinz, S., Ghafoory, S., Wölfl, S., et al. (2015). In Vitro Generation of Functional Liver Organoid-Like Structures Using Adult Human Cells. PLoS One 10:e0139345. doi: 10.1371/journal.pone.0139345
Ramaiahgari, S. C., den Braver, M. W., Herpers, B., Terpstra, V., Commandeur, J. N. M., van de Water, B., et al. (2014). A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol. 88, 1083–1095. doi: 10.1007/s00204-014-1215-9
Ramaiahgari, S. C., Waidyanatha, S., Dixon, D., DeVito, M. J., Paules, R. S., and Ferguson, S. S. (2017). From the Cover: Three-Dimensional (3D) HepaRG Spheroid Model With Physiologically Relevant Xenobiotic Metabolism Competence and Hepatocyte Functionality for Liver Toxicity Screening. Toxicol. Sci. 159, 124–136. doi: 10.1093/toxsci/kfx122
Ramasamy, T. S., Yu, J. S. L., Selden, C., Hodgson, H., and Cui, W. (2013). Application of three-dimensional culture conditions to human embryonic stem cell-derived definitive endoderm cells enhances hepatocyte differentiation and functionality. Tissue Eng. Part A 19, 360–367. doi: 10.1089/ten.tea.2012.0190
Rebelo, S. P., Costa, R., Estrada, M., Shevchenko, V., Brito, C., and Alves, P. M. (2015). HepaRG microencapsulated spheroids in DMSO-free culture: Novel culturing approaches for enhanced xenobiotic and biosynthetic metabolism. Arch. Toxicol. 89, 1347–1358. doi: 10.1007/s00204-014-1320-9
Rebelo, S. P., Costa, R., Silva, M. M., Marcelino, P., Brito, C., and Alves, P. M. (2017). Three-dimensional co-culture of human hepatocytes and mesenchymal stem cells: Improved functionality in long-term bioreactor cultures. J. Tissue Eng. Regen. Med. 11, 2034–2045. doi: 10.1002/term.2099
Rennert, K., Steinborn, S., Gröger, M., Ungerböck, B., Jank, A.-M., Ehgartner, J., et al. (2015). A microfluidically perfused three dimensional human liver model. Biomaterials 71, 119–131. doi: 10.1016/j.biomaterials.2015.08.043
Ringehan, M., McKeating, J. A., and Protzer, U. (2017). Viral hepatitis and liver cancer. Philos. Trans. R. Soc. B Biol. Sci. 372:20160274. doi: 10.1098/rstb.2016.0274
Rocha, F. G. (2012). “Chapter 4 - Liver blood flow: Physiology, measurement, and clinical relevance,” in Blumgart’s Surgery of the Liver, Pancreas and Biliary Tract (Fifth Edition), eds W. R. Jarnagin and L. H. Blumgart (Philadelphia: W.B. Saunders), 74.e–86.e. doi: 10.1016/B978-1-4377-1454-8.00004-7
Rossi, E. A., Quintanilha, L. F., Nonaka, C. K. V., Souza, B. S., and de, F. (2019). Advances in Hepatic Tissue Bioengineering with Decellularized Liver Bioscaffold. Stem Cells Int. 2019:2693189. doi: 10.1155/2019/2693189
Roth, A. D., Lama, P., Dunn, S., Hong, S., and Lee, M.-Y. (2018). Polymer coating on a micropillar chip for robust attachment of PuraMatrix peptide hydrogel for 3D hepatic cell culture. Mater. Sci. Eng. C Mater. Biol. Appl. 90, 634–644. doi: 10.1016/j.msec.2018.04.092
Rowe, C., Shaeri, M., Large, E., Cornforth, T., Robinson, A., Kostrzewski, T., et al. (2018). Perfused human hepatocyte microtissues identify reactive metabolite-forming and mitochondria-perturbing hepatotoxins. Toxicol. In Vitro 46, 29–38. doi: 10.1016/j.tiv.2017.09.012
Sainz, B., TenCate, V., and Uprichard, S. L. (2009). Three-dimensional Huh7 cell culture system for the study of Hepatitis C virus infection. Virol. J. 6:103. doi: 10.1186/1743-422X-6-103
Sarkar, U., Ravindra, K. C., Large, E., Young, C. L., Rivera-Burgos, D., Yu, J., et al. (2017). Integrated Assessment of Diclofenac Biotransformation, Pharmacokinetics, and Omics-Based Toxicity in a Three-Dimensional Human Liver-Immunocompetent Coculture System. Drug Metab. Dispos. 45, 855–866. doi: 10.1124/dmd.116.074005
Sarkar, U., Rivera-Burgos, D., Large, E. M., Hughes, D. J., Ravindra, K. C., Dyer, R. L., et al. (2015). Metabolite Profiling and Pharmacokinetic Evaluation of Hydrocortisone in a Perfused Three-Dimensional Human Liver Bioreactor. Drug Metab. Dispos. 43, 1091–1099. doi: 10.1124/dmd.115.063495
Sasaki, K., Akagi, T., Asaoka, T., Eguchi, H., Fukuda, Y., Iwagami, Y., et al. (2017). Construction of three-dimensional vascularized functional human liver tissue using a layer-by-layer cell coating technique. Biomaterials 133, 263–274. doi: 10.1016/j.biomaterials.2017.02.034
Sassi, L., Ajayi, O., Campinoti, S., Natarajan, D., McQuitty, C., Siena, R. R., et al. (2021). A Perfusion Bioreactor for Longitudinal Monitoring of Bioengineered Liver Constructs. Nanomaterials 11:275. doi: 10.3390/nano11020275
Schepers, A., Li, C., Chhabra, A., Seney, B. T., and Bhatia, S. (2016). Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip 16, 2644–2653. doi: 10.1039/c6lc00598e
Schmelzer, E., Mutig, K., Schrade, P., Bachmann, S., Gerlach, J. C., and Zeilinger, K. (2009). Effect of human patient plasma ex vivo treatment on gene expression and progenitor cell activation of primary human liver cells in multi-compartment 3D perfusion bioreactors for extra-corporeal liver support. Biotechnol. Bioeng. 103, 817–827. doi: 10.1002/bit.22283
Schütte, J., Hagmeyer, B., Holzner, F., Kubon, M., Werner, S., Freudigmann, C., et al. (2011). “Artificial micro organs”—a microfluidic device for dielectrophoretic assembly of liver sinusoids. Biomed. Microdevices 13, 493–501. doi: 10.1007/s10544-011-9517-7
Selden, C., Khalil, M., and Hodgson, H. (2000). Three Dimensional Culture Upregulates Extracellular Matrix Protein Expression in Human Liver Cell Lines - a Step towards Mimicking the Liver in Vivo? Int. J. Artif. Organs 23, 774–781. doi: 10.1177/039139880002301107
Shlomai, A., Schwartz, R. E., Ramanan, V., Bhatta, A., de Jong, Y. P., Bhatia, S. N., et al. (2014). Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc. Natl. Acad. Sci. U.S.A. 111, 12193–12198. doi: 10.1073/pnas.1412631111
Shoemaker, J. T., Zhang, W., Atlas, S. I., Bryan, R. A., Inman, S. W., and Vukasinovic, J. (2020). A 3D Cell Culture Organ-on-a-Chip Platform With a Breathable Hemoglobin Analogue Augments and Extends Primary Human Hepatocyte Functions in vitro. Front. Mol. Biosci. 7:568777. doi: 10.3389/fmolb.2020.568777
Simoneau, C. R., and Ott, M. (2020). Modeling Multi-organ Infection by SARS-CoV-2 Using Stem Cell Technology. Cell Stem Cell 27, 859–868. doi: 10.1016/j.stem.2020.11.012
Sirenko, O., Hancock, M. K., Hesley, J., Hong, D., Cohen, A., Gentry, J., et al. (2016). Phenotypic Characterization of Toxic Compound Effects on Liver Spheroids Derived from iPSC Using Confocal Imaging and Three-Dimensional Image Analysis. ASSAY Drug Dev. Technol. 14, 381–394. doi: 10.1089/adt.2016.729
Sivertsson, L., Synnergren, J., Jensen, J., Björquist, P., and Ingelman-Sundberg, M. (2012). Hepatic Differentiation and Maturation of Human Embryonic Stem Cells Cultured in a Perfused Three-Dimensional Bioreactor. Stem Cells Dev. 22, 581–594. doi: 10.1089/scd.2012.0202
Skardal, A., Devarasetty, M., Kang, H.-W., Mead, I., Bishop, C., Shupe, T., et al. (2015). A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomater. 25, 24–34. doi: 10.1016/j.actbio.2015.07.030
Subramanian, K., Owens, D. J., Raju, R., Firpo, M., O’Brien, T. D., Verfaillie, C. M., et al. (2013). Spheroid Culture for Enhanced Differentiation of Human Embryonic Stem Cells to Hepatocyte-Like Cells. Stem Cells Dev. 23, 124–131. doi: 10.1089/scd.2013.0097
Swift, B., Pfeifer, N. D., and Brouwer, K. L. R. (2010). Sandwich-Cultured Hepatocytes: An In Vitro Model to Evaluate Hepatobiliary Transporter-Based Drug Interactions and Hepatotoxicity. Drug Metab. Rev. 42, 446–471. doi: 10.3109/03602530903491881
Takahashi, Y., Hori, Y., Yamamoto, T., Urashima, T., Ohara, Y., and Tanaka, H. (2015). 3D spheroid cultures improve the metabolic gene expression profiles of HepaRG cells. Biosci. Rep. 35:e00208. doi: 10.1042/BSR20150034
Takayama, K., Kawabata, K., Nagamoto, Y., Kishimoto, K., Tashiro, K., Sakurai, F., et al. (2013). 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials 34, 1781–1789. doi: 10.1016/j.biomaterials.2012.11.029
Takebe, T., Sekine, K., Enomura, M., Koike, H., Kimura, M., Ogaeri, T., et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. doi: 10.1038/nature12271
Tan, K., Keegan, P., Rogers, M., Lu, M., Gosset, J. R., Charest, J., et al. (2019). A high-throughput microfluidic microphysiological system (PREDICT-96) to recapitulate hepatocyte function in dynamic, re-circulating flow conditions. Lab Chip 19, 1556–1566. doi: 10.1039/C8LC01262H
Tasnim, F., Toh, Y.-C., Qu, Y., Li, H., Phan, D., Narmada, B. C., et al. (2016). Functionally Enhanced Human Stem Cell Derived Hepatocytes in Galactosylated Cellulosic Sponges for Hepatotoxicity Testing. Mol. Pharm. 13, 1947–1957. doi: 10.1021/acs.molpharmaceut.6b00119
Taymour, R., Kilian, D., Ahlfeld, T., Gelinsky, M., and Lode, A. (2021). 3D bioprinting of hepatocytes: Core–shell structured co-cultures with fibroblasts for enhanced functionality. Sci. Rep. 11:5130. doi: 10.1038/s41598-021-84384-6
Thomas, E., and Liang, T. J. (2016). Experimental models of hepatitis B and C — new insights and progress. Nat. Rev. Gastroenterol. Hepatol. 13, 362–374. doi: 10.1038/nrgastro.2016.37
Tonon, F., Giobbe, G. G., Zambon, A., Luni, C., Gagliano, O., Floreani, A., et al. (2019). In vitro metabolic zonation through oxygen gradient on a chip. Sci. Rep. 9:13557. doi: 10.1038/s41598-019-49412-6
Torresi, J., Tran, B. M., Christiansen, D., Earnest-Silveira, L., Schwab, R. H. M., and Vincan, E. (2019). HBV-related hepatocarcinogenesis: The role of signalling pathways and innovative ex vivo research models. BMC Cancer 19:707. doi: 10.1186/s12885-019-5916-6
Tostões, R. M., Leite, S. B., Serra, M., Jensen, J., Björquist, P., Carrondo, M. J. T., et al. (2012). Human liver cell spheroids in extended perfusion bioreactor culture for repeated-dose drug testing. Hepatology 55, 1227–1236. doi: 10.1002/hep.24760
Tran, N. M., Dufresne, M., Duverlie, G., Castelain, S., Défarge, C., Paullier, P., et al. (2012). An Appropriate Selection of a 3D Alginate Culture Model for Hepatic Huh-7 Cell Line Encapsulation Intended for Viral Studies. Tissue Eng. Part A 19, 103–113. doi: 10.1089/ten.tea.2012.0139
Trask, O. J., Moore, A., and LeCluyse, E. L. (2014). A micropatterned hepatocyte coculture model for assessment of liver toxicity using high-content imaging analysis. Assay Drug Dev. Technol. 12, 16–27. doi: 10.1089/adt.2013.525
Ulvestad, M., Darnell, M., Molden, E., Ellis, E., Åsberg, A., and Andersson, T. B. (2012). Evaluation of Organic Anion-Transporting Polypeptide 1B1 and CYP3A4 Activities in Primary Human Hepatocytes and HepaRG Cells Cultured in a Dynamic Three-Dimensional Bioreactor System. J. Pharmacol. Exp. Ther. 343, 145–156. doi: 10.1124/jpet.112.195750
Velazquez, J. J., LeGraw, R., Moghadam, F., Tan, Y., Kilbourne, J., Maggiore, J. C., et al. (2020). Gene Regulatory Network Analysis and Engineering Directs Development and Vascularization of Multilineage Human Liver Organoids. Cell Syst. 12, 41–55. doi: 10.1016/j.cels.2020.11.002
Vernetti, L. A., Senutovitch, N., Boltz, R., DeBiasio, R., Ying Shun, T., Gough, A., et al. (2016). A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. 241, 101–114. doi: 10.1177/1535370215592121
Vorrink, S. U., Ullah, S., Schmidt, S., Nandania, J., Velagapudi, V., Beck, O., et al. (2017). Endogenous and xenobiotic metabolic stability of primary human hepatocytes in long-term 3D spheroid cultures revealed by a combination of targeted and untargeted metabolomics. FASEB J. 31, 2696–2708. doi: 10.1096/fj.201601375R
Vorrink, S. U., Zhou, Y., Ingelman-Sundberg, M., and Lauschke, V. M. (2018). Prediction of Drug-Induced Hepatotoxicity Using Long-Term Stable Primary Hepatic 3D Spheroid Cultures in Chemically Defined Conditions. Toxicol. Sci. 163, 655–665. doi: 10.1093/toxsci/kfy058
Wang, J., Chen, F., Liu, L., Qi, C., Wang, B., Yan, X., et al. (2016). Engineering EMT using 3D micro-scaffold to promote hepatic functions for drug hepatotoxicity evaluation. Biomaterials 91, 11–22. doi: 10.1016/j.biomaterials.2016.03.001
Wang, J., Qu, B., Zhang, F., Zhang, C., Deng, W., Dao Thi, V. L., et al. (2019). Stem Cell-Derived Hepatocyte-Like Cells as Model for Viral Hepatitis Research. Stem Cells Int. 2019:e9605252. doi: 10.1155/2019/9605252
Wang, W. W., Khetani, S. R., Krzyzewski, S., Duignan, D. B., and Obach, R. S. (2010). Assessment of a Micropatterned Hepatocyte Coculture System to Generate Major Human Excretory and Circulating Drug Metabolites. Drug Metab. Dispos. 38, 1900–1905. doi: 10.1124/dmd.110.034876
Wang, Y., Cui, C.-B., Yamauchi, M., Miguez, P., Roach, M., Malavarca, R., et al. (2011). Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology 53, 293–305. doi: 10.1002/hep.24012
Wang, Y., Li, J., Wang, X., Sang, M., and Ho, W. (2013). Hepatic stellate cells, liver innate immunity, and hepatitis C virus. J. Gastroenterol. Hepatol. 28, 112–115. doi: 10.1111/jgh.12023
Wang, Y., Su, W., Wang, L., Jiang, L., Liu, Y., Hui, L., et al. (2018a). Paper supported long-term 3D liver co-culture model for the assessment of hepatotoxic drugs. Toxicol. Res. 7, 13–21. doi: 10.1039/C7TX00209B
Wang, Y., Wang, H., Deng, P., Chen, W., Guo, Y., Tao, T., et al. (2018b). In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip 18, 3606–3616. doi: 10.1039/C8LC00869H
Wang, Z., Luo, X., Anene-Nzelu, C., Yu, Y., Hong, X., Singh, N. H., et al. (2015). HepaRG culture in tethered spheroids as an in vitro three-dimensional model for drug safety screening. J. Appl. Toxicol. JAT 35, 909–917. doi: 10.1002/jat.3090
Ware, B. R., Berger, D. R., and Khetani, S. R. (2015). Prediction of Drug-Induced Liver Injury in Micropatterned Co-cultures Containing iPSC-Derived Human Hepatocytes. Toxicol. Sci. 145, 252–262. doi: 10.1093/toxsci/kfv048
Ware, B. R., Durham, M. J., Monckton, C. P., and Khetani, S. R. (2017). A Cell Culture Platform to Maintain Long-term Phenotype of Primary Human Hepatocytes and Endothelial Cells. Cell. Mol. Gastroenterol. Hepatol. 5, 187–207. doi: 10.1016/j.jcmgh.2017.11.007
Wu, Y., Geng, X., Wang, J., Miao, Y., Lu, Y., and Li, B. (2016). The HepaRG cell line, a superior in vitro model to L-02, HepG2 and hiHeps cell lines for assessing drug-induced liver injury. Cell Biol. Toxicol. 32, 37–59. doi: 10.1007/s10565-016-9316-2
Wu, Y., Lin, Z. Y., Wenger, A. C., Tam, K. C., and Tang, X. (2018). 3D bioprinting of liver-mimetic construct with alginate/cellulose nanocrystal hybrid bioink. Bioprinting 9, 1–6. doi: 10.1016/j.bprint.2017.12.001
Yajima, Y., Lee, C. N., Yamada, M., Utoh, R., and Seki, M. (2018). Development of a perfusable 3D liver cell cultivation system via bundling-up assembly of cell-laden microfibers. J. Biosci. Bioeng. 126, 111–118. doi: 10.1016/j.jbiosc.2018.01.022
Yang, K., Guo, C., Woodhead, J. L., Claire, R. L. S. T., Watkins, P. B., Siler, S. Q., et al. (2016). Sandwich-Cultured Hepatocytes as a Tool to Study Drug Disposition and Drug-Induced Liver Injury. J. Pharm. Sci. 105, 443–459. doi: 10.1016/j.xphs.2015.11.008
Ylva, T., Figler, R. A., Svetlana, M., Collado, M. S., Lawson, M. J., Mackey, A. J., et al. (2016). Transcriptional Profiling Suggests That Nevirapine and Ritonavir Cause Drug Induced Liver Injury Through Distinct Mechanisms in Primary Human Hepatocytes. Chem. Biol. Interact. 255, 31–44. doi: 10.1016/j.cbi.2015.11.023
Yoffe, B., Darlington, G. J., Soriano, H. E., Krishnan, B., Risin, D., Pellis, N. R., et al. (1999). Cultures of human liver cells in simulated microgravity environment. Adv. Space Res. 24, 829–836. doi: 10.1016/s0273-1177(99)00079-4
Zeilinger, K., Schreiter, T., Darnell, M., Söderdahl, T., Lübberstedt, M., Dillner, B., et al. (2011). Scaling Down of a Clinical Three-Dimensional Perfusion Multicompartment Hollow Fiber Liver Bioreactor Developed for Extracorporeal Liver Support to an Analytical Scale Device Useful for Hepatic Pharmacological In Vitro Studies. Tissue Eng. Part C Methods 17, 549–556. doi: 10.1089/ten.tec.2010.0580
Zhang, M. Y., Lee, P. J., Hung, P. J., Johnson, T., Lee, L. P., and Mofrad, M. R. K. (2008). Microfluidic environment for high density hepatocyte culture. Biomed. Microdevices 10, 117–121. doi: 10.1007/s10544-007-9116-9
Zhang, Z., Xu, H., Mazza, G., Zhang, M., Frenguelli, L., Liu, Q., et al. (2019). Decellularized human liver scaffold-based three-dimensional culture system facilitate hepatitis B virus infection. J. Biomed. Mater. Res. A 107, 1744–1753. doi: 10.1002/jbm.a.36690
Zheng, Z.-Y., Weng, S.-Y., and Yu, Y. (2009). Signal molecule-mediated hepatic cell communication during liver regeneration. World J. Gastroenterol. WJG 15, 5776–5783. doi: 10.3748/wjg.15.5776
Zhu, L., Fan, X., Wang, B., Liu, L., Yan, X., Zhou, L., et al. (2017). Biomechanically primed liver microtumor array as a high-throughput mechanopharmacological screening platform for stroma-reprogrammed combinatorial therapy. Biomaterials 124, 12–24. doi: 10.1016/j.biomaterials.2017.01.030
Keywords: hepatitis B virus, hepatitis C virus, hepatocellular carcinoma, liver-on-chip, in vitro disease model
Citation: McDuffie D, Barr D, Agarwal A and Thomas E (2022) Physiologically relevant microsystems to study viral infection in the human liver. Front. Microbiol. 13:999366. doi: 10.3389/fmicb.2022.999366
Received: 21 July 2022; Accepted: 29 August 2022;
Published: 28 September 2022.
Edited by:
Yuebang Yin, Nankai University, ChinaReviewed by:
Salman Khetani, University of Illinois at Chicago, United StatesWenshi Wang, Xuzhou Medical University, China
Copyright © 2022 McDuffie, Barr, Agarwal and Thomas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashutosh Agarwal, QS5hZ2Fyd2FsMkBtaWFtaS5lZHU=; Emmanuel Thomas, RXRob21hczFAbWVkLm1pYW1pLmVkdQ==
 Dennis McDuffie
Dennis McDuffie David Barr
David Barr Ashutosh Agarwal
Ashutosh Agarwal Emmanuel Thomas
Emmanuel Thomas