
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Microbiol. , 31 October 2022
Sec. Microbiological Chemistry and Geomicrobiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.998133
This article is part of the Research Topic Geomicrobes: Life in Terrestrial Deep Subsurface, volume II View all 6 articles
 Donato Giovannelli1,2,3,4,5*
Donato Giovannelli1,2,3,4,5* Peter H. Barry4
Peter H. Barry4 J. Maarten de Moor6,7
J. Maarten de Moor6,7 Gerdhard L. Jessen8,9
Gerdhard L. Jessen8,9 Matthew O. Schrenk10
Matthew O. Schrenk10 Karen G. Lloyd11
Karen G. Lloyd11Despite being one of the largest microbial ecosystems on Earth, many basic open questions remain about how life exists and thrives in the deep subsurface biosphere. Much of this ambiguity is due to the fact that it is exceedingly difficult and often prohibitively expensive to directly sample the deep subsurface, requiring elaborate drilling programs or access to deep mines. We propose a sampling approach which involves collection of a large suite of geological, geochemical, and biological data from numerous deeply-sourced seeps—including lower temperature sites—over large spatial scales. This enables research into interactions between the geosphere and the biosphere, expanding the classical local approach to regional or even planetary scales. Understanding the interplay between geology, geochemistry and biology on such scales is essential for building subsurface ecosystem models and extrapolating the ecological and biogeochemical roles of subsurface microbes beyond single site interpretations. This approach has been used successfully across the Central and South American Convergent Margins, and can be applied more broadly to other types of geological regions (i.e., rifting, intraplate volcanic, and hydrothermal settings). Working across geological spatial scales inherently encompasses broad temporal scales (e.g., millions of years of volatile cycling across a convergent margin), providing access to a framework for interpreting evolution and ecosystem functions through deep time and space. We propose that tectonic interactions are fundamental to maintaining planetary habitability through feedbacks that stabilize the ecosphere, and deep biosphere studies are fundamental to understanding geo-bio feedbacks on these processes on a global scale.
Earth’s continental and oceanic crust contain one of the largest microbial ecosystems on the planet (Kallmeyer et al., 2012; Magnabosco et al., 2018). This subsurface microbial biosphere is essential to global biogeochemical cycles because it alters the redox state of the crust and affects the distribution of minerals, gases, and organic matter through deep time (D’Hondt et al., 2019). While subsurface microbiology is often studied over small spatial scales (sampling sites meters to a few kilometers apart), or over short time scales during laboratory experiments, crustal ecosystems likely operate on tectonic scales of hundreds of kilometers and millions of years. Despite their importance to planetary processes and volatile cycling through deep time, much less is known about subsurface microbial ecosystems than surface microbes. This is largely due to the difficulty in accessing deep subsurface environments.
Direct sampling of the subsurface is fundamental, not only for obtaining samples that can be used to grow and study organisms in a laboratory, but also for conducting studies on diverse populations as they occur naturally. Such in situ studies are important because many microbes in the subsurface belong to microbial groups that are resistant to many or all culturing efforts—meaning that they are not amenable to pure culture experimentation (e.g., Rappé and Giovannoni, 2003; Lloyd et al., 2018; Steen et al., 2019). Even if laboratory cultures are available, the physiology and ecological roles of microbes may differ in the natural, taxonomically-diverse, often energy-limited environment relative to laboratory conditions, as originally proposed by Winogradsky (1923). Direct access to the subsurface is normally obtained through scientific drilling. Major drilling operations occur through the International Ocean Discovery Program (IODP-US), which was formerly the Integrated Ocean Drilling Program (IODP), the Ocean Drilling Program (ODP), and the Deep Sea Drilling Program (DSDP), as well as the European Consortium for Ocean Research Drilling (ECORD), Japan’s Integrated Ocean Drilling program (IODP-J), and the International Continental Scientific Drilling Program (ICDP). These programs have provided unprecedented access to subsurface ecosystems. However, deep drilling projects are expensive and infrequent (Orcutt and Edwards, 2014), and require complex logistical operations and a long lead time (sometimes many years) before the actual sampling. Thus drilling makes it difficult to sample more than a handful of subsurface sites in a single study, or to collect new samples quickly to react to new scientific advances and opportunities created by geologically dynamic events (e.g., earthquakes or eruptions). The large costs, duration of fieldwork, and organizational requirements can also be a barrier to entry for early career researchers, or researchers working in countries with limited research and development funding. Access to commercial or scientific mines and mining sites are another fruitful approach to studying the deep subsurface biosphere (e.g., Chivian et al., 2008), but site selection is limited to existing infrastructure, i.e., samples are determined by where the mines happen to be. Besides drilling and access to existing subsurface facilities there is a need for a more nimble approach to obtaining subsurface samples.
Subseafloor hydrothermal vent fluids have been used to sample deep subsurface life as it is flushed out of the subsurface, rather than by drilling down to it (Deming and Baross, 1993). Here, we expand this approach to large scales and by assessing the degree of mixing with the surface, accelerate exploration of the subsurface biosphere and enable large-scale studies of biosphere-geosphere coupling of subsurface microbial ecosystems across regional or global geological features. Typically, microbiological studies of terrestrial hot springs or oceanic hydrothermal vents study either variations across a transect at a single site or compare a handful of sites to one another or to a background reference site (Giovannelli et al., 2013; Reveillaud et al., 2016; Brazelton et al., 2017). We propose instead to sample many natural seeps across a geological gradient to map out broad-scale ecological features in a large area of the subsurface. The purpose of this sampling approach is not to replace direct sampling strategies or careful transect studies across a single site, but to add to the arsenal of sampling approaches that can greatly expand our ability to sample microbial populations across large spatial regions. We have successfully employed this approach to study biosphere-geosphere feedbacks across the convergent margin in Northern and Central Costa Rica (Barry et al., 2019; Fullerton et al., 2021; Basili et al., 2022; Rogers et al., 2022) and Argentina (Cascone et al., 2021). This approach is flexible, interdisciplinary, can be widely applied to diverse settings, and is orders of magnitude cheaper and faster than drilling. Here we describe the advantages and drawbacks of this approach, as well as provide suggestions based on experience for how other researchers can employ a similar approach in their own work. When integrated with targeted drilling projects, our approach can provide unprecedented insights into the interactions between subsurface ecosystems and geological processes at large spatial scales.
Natural seep fluids have previously been proposed as windows into the subsurface biosphere both at deep-sea hydrothermal vents (e.g., Deming and Baross, 1993; Summit and Baross, 2001; Anderson et al., 2011) and in terrestrial settings (e.g., Meyer-Dombard et al., 2005; Hall et al., 2008; Hou et al., 2013; Colman et al., 2016; Crossey et al., 2016; Uribe-Lorío et al., 2019). Typically, high temperature fluids have been interpreted as ascending quickly from depth (i.e., minimal surface mixing) and thus representative of the subsurface. Fluids and volatiles released at lower temperature sites, such as those found in the forearc of convergent margins or in low temperature pools at the margin of large calderas are typically ignored, as the degree of mixing of the fluids with surface derived waters might be difficult to assess. By focusing on high temperature sites, the information relevant to the subsurface microbial communities is limited by the geothermal gradient, as higher temperature sites (> 122°C) are not amenable to life (Merino et al., 2019). By combining interdisciplinary co-located measurements it is possible instead to assess the source of volatiles and deconvolve the degree of mixing with the surface providing new insight into deeply-sourced seeps and thus the deep biosphere. Additionally, focusing on the gradients in features of the seeping fluids across the deeply-sourced seeps has the additional effect of changing the scale at which geochemical gradients can be investigated. In this context, the focus of the sampling moves from hot springs sensu stricto to features that channel surface deep fluids, regardless of their temperature, conductivity or pH.
We therefore define a broad category of features resulting from deep fluids ascending to the surface called deeply-sourced seeps (Box 1). Deeply-sourced seeps are the surface expression of deep fluids, either as water or gases, that transport volatiles from depth (Figure 1). They are not necessarily associated with high temperature fluids, and thus include lower temperature manifestations and diffuse degassing areas (Crossey et al., 2016). In order to identify and use deeply-sourced seeps as windows into the subsurface, it is essential to adopt an experimental approach that distinguishes between samples that represent the biology and geochemistry of the subsurface and those that show strong mixing with surface derived volatiles and microbial communities. Our approach combines assessments of proxies of subsurface vs. surface material (e.g., He and C isotopes; Crossey et al., 2016; Barry et al., 2019), as well as comparing communities across large spatial gradients to assess which parts of the microbial community correlate with deep subsurface features vs. surface features (Crossey et al., 2016; Cascone et al., 2021; Fullerton et al., 2021; Basili et al., 2022; Rogers et al., 2022). These proxies include dissolved and gaseous inorganic compounds, isotopic composition (e.g., Δ14C and δ13C of dissolved organic and inorganic carbon; Lang et al., 2018; Fullerton et al., 2021) or biomolecules that can indicate the degree of exposure the fluids had with the surface or surface derived shallow fluids (e.g., photosynthetic pigments and genetic signatures of phototrophy or obligate aerobic metabolisms; Table 1).
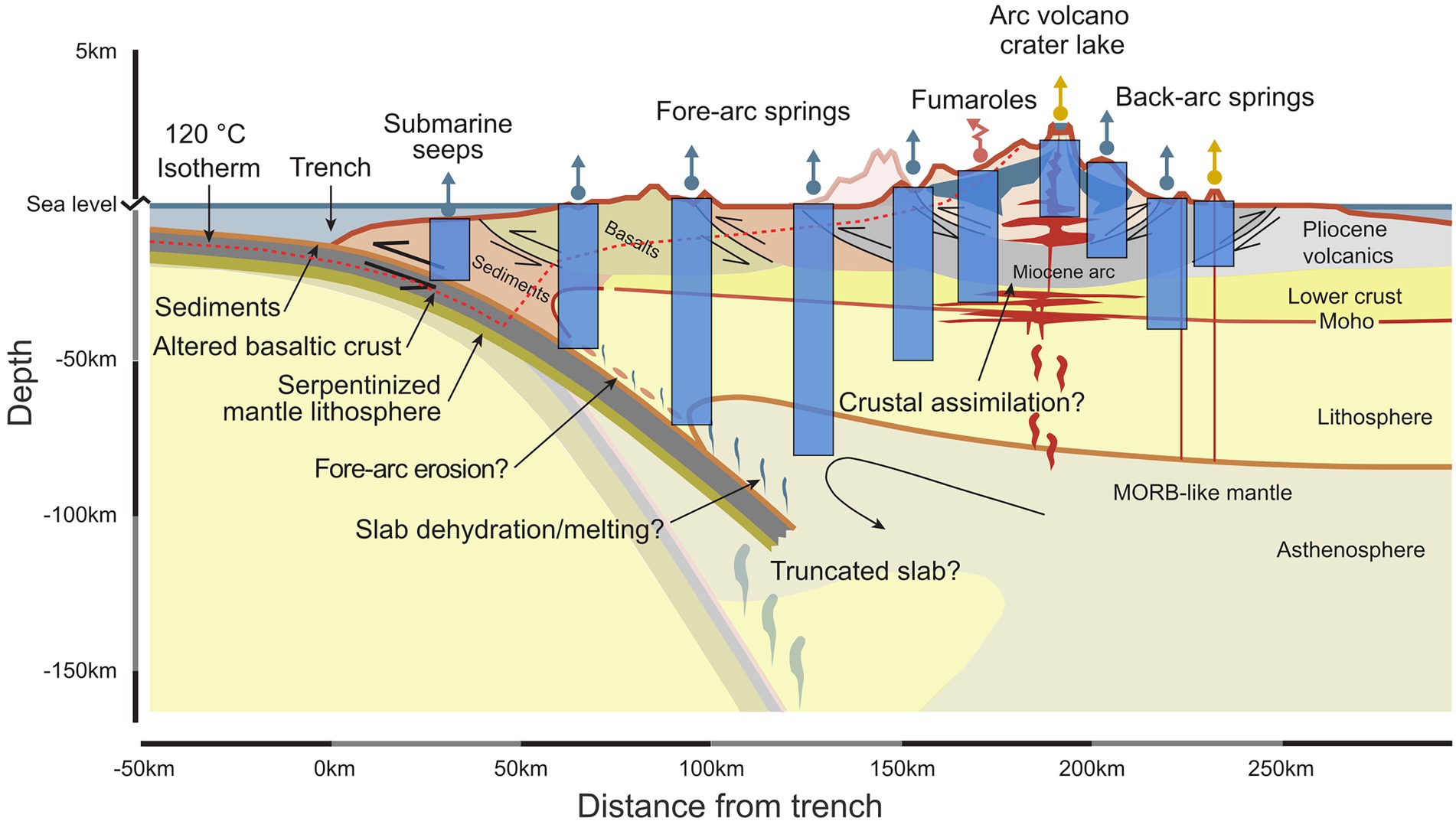
Figure 1. Sampling deeply sourced seeps as windows into the subsurface across geological provinces can help to identify biological interactions with large-scale geological processes. Each deeply-sourced seep (represented by a straight upward arrow for fluids, teal for springs, sand for volcanic crater waters, or by a jagged red arrow for gases) gives access to an integrated view of the source and processes occurring at depth (light blue rectangles). Transects across large-scale geological gradients can be obtained using this approach. Modified after Giovannelli et al. (2020).
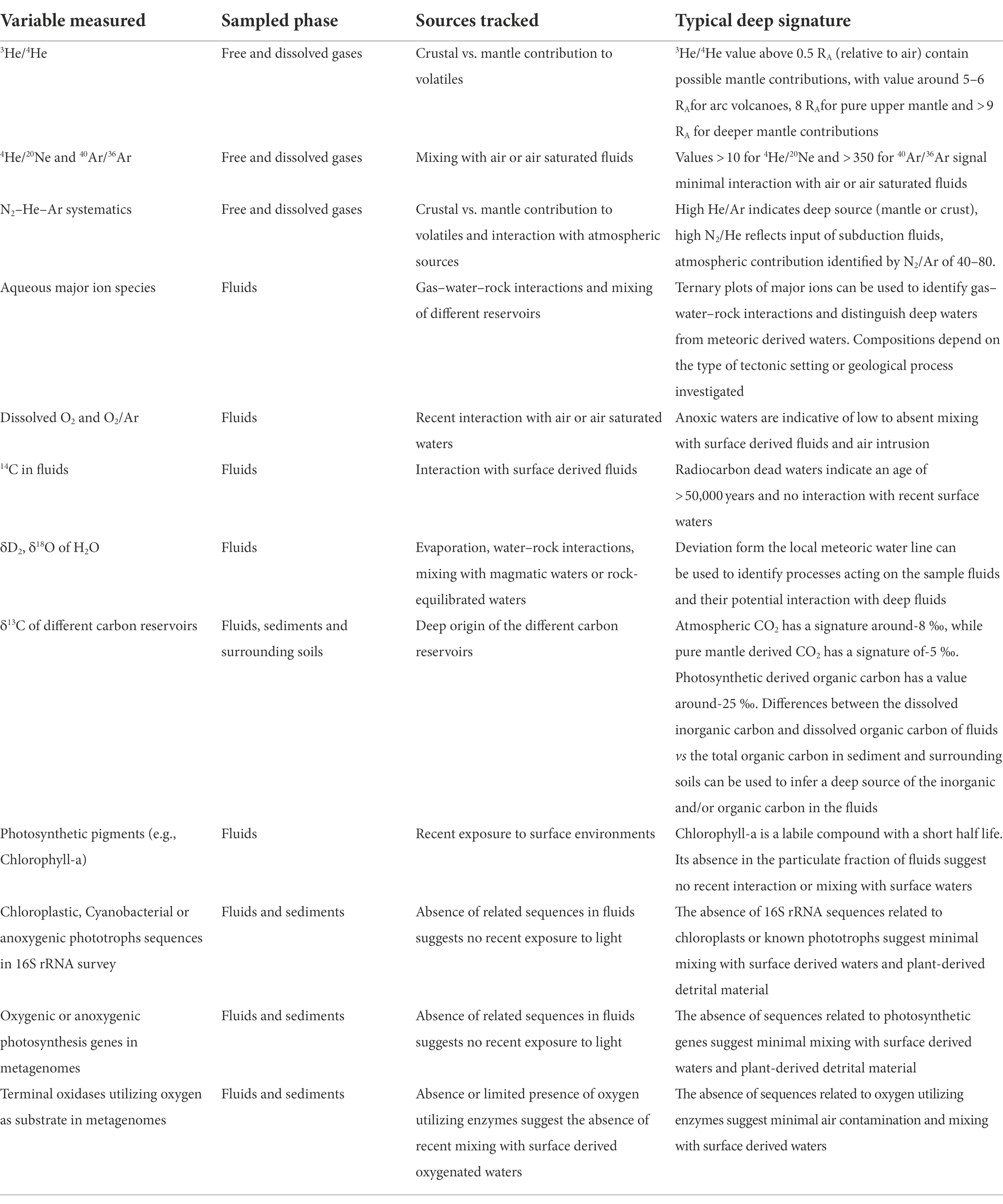
Table 1. Parameters that can be used to track the relative contribution of surface and subsurface processes to the samples deeply-sourced seeps.
For example, mantle and crustal contributions to the volatile inventory can be tracked using a combination of helium isotope systematics (Hilton et al., 1993; Bekaert et al., 2021), while mixing with air saturated waters (i.e., surface waters) and water-rock interactions at depth can be tracked using a combination of N2–He–Ar systematics and aqueous geochemistry (e.g., Giggenbach, 1992). The exact nature of the proxy to be used depends on the tectonic setting under investigation and the type of samples collected (Table 1), and a multi-phase approach should be designed to accurately determine the degree of surface (i.e., atmospheric or shallow aquifers) contamination of the collected samples. Thus, suitable geochemical and biological tracers can be combined to the community structure and function of microbial populations across all the sites where there are sequencing data, to see which subpopulations correlate with subsurface geochemical and geological features (Crossey et al., 2016; Fullerton et al., 2021; Rogers et al., 2022).
In addition to using specific proxies to track subsurface–surface mixing, the type of sample collected (i.e., thermal feature or matrix), the sampling approach, and the mode of collection employed can significantly affect the quality of the data and environment (surface vs subsurface) that is ultimately represented. Typical work in hot springs, shallow-water hydrothermal vents, and sepentinizing settings looks at the interplay between subsurface and surface processes, often sampling the interface where deep fluids mix with surface oxidants (mainly atmospherically derived; Inskeep et al., 2005; Meyer-Dombard et al., 2005; Giovannelli et al., 2013; Sánchez-Murillo et al., 2014; Colman et al., 2016; Patwardhan et al., 2018;Colman et al., 2019b). In our approach, we attempt to minimize surface inputs by sampling deeply-sourced fluids as directly as possible. This is accomplished using the following multi-pronged field approach: (i) the seep is first identified using a combination of visual inspection and in situ temperature, conductivity and pH measurements, i.e., in the presence of several thermal features, the seeping fluids with the highest temperature, the highest or lowest pH, or highest salinity and flux are prioritized; (ii) with a minimum perturbation of the thermal feature and surroundings; a sterile non-reactive (glass or titanium) pipe/tube is inserted (> 30 cm deep) in the seep to catch the fluids before they mix with surface water or air; and (iii) the rate at which fluids are collected needs to be similar to the rate of natural seepage to avoid air/surface entrainment.
The quality of the sample collected can be assessed using a combination of geochemical tracers directly in the field or back in laboratory (e.g., dissolved O2, O2/Ar systematic, 4He/20Ne, see Table 1), while the degree of surface mixing can be assessed using a combination of diverse proxies. This approach has shown to minimize the possibility of mixing subsurface and surface fluids during sampling, both for geochemical (e.g., Barry et al., 2019) and microbiological studies (Cascone et al., 2021; Fullerton et al., 2021; Rogers et al., 2022). Additionally, sampling seep derived sediments and surrounding soils is also commonly done, to track surface interactions of the fluids with topsoils (Cascone et al., 2021; Fullerton et al., 2021).
Studying microbial communities across large spatial scales (spanning many kilometers or even the entire globe) has provided great insights into terrestrial and shallow hot springs microbial communities (Hou et al., 2013; Inskeep et al., 2013; Crossey et al., 2016; Power et al., 2018) as well as global or large-scale regional soil and oceanic water microbiome (Williamson et al., 2012; Sunagawa et al., 2015; Delgado-Baquerizo et al., 2018; Acinas et al., 2021). These studies have foregone the small-scale, local gradient approach in favor of large-scale patterns across gradients spanning entire ecosystems or regions of the globe. In contrast, a large number of subsurface studies have either focused on a handful of samples collected in close proximity (i.e., springs and hot springs close to each other; e.g., Emerson et al., 2016; Colman et al., 2019b; Trembath-Reichert et al., 2019), contrasting conditions (i.e., sampling pools characterized by contrasting geochemistry or characteristics; e.g., Meyer-Dombard et al., 2005; Lindsay et al., 2019) or transect across the subsurface to surface transition along temperature or redox gradients (i.e., surface mixing gradients; e.g., Giovannelli et al., 2013; Sánchez-Murillo et al., 2014; Brazelton et al., 2017; Fones et al., 2019). These approaches involve sample collection at the centimeter to tens of meter scale, often using control sites to investigate the role of microbial communities in the setting under investigation. While this approach has deep roots in ecology and microbial ecology, it often limits our ability to comprehend the response of the community to the underlying geological processes, for example concluding that temperature and sulfide gradients drive microbial community assembly (Giovannelli et al., 2013). On the contrary, studies focusing on large-scale systematic investigations of the response of deep subsurface communities to geological gradients allow for conclusions to extend to direct links with geological processes (Crossey et al., 2016; Power et al., 2018; Colman et al., 2019a; Cascone et al., 2021; Fullerton et al., 2021; Rogers et al., 2022).
One of the barriers in approaching the study of microbial processes at geologically relevant scales is the lack of understanding of temporal and spatial scales at which these processes overlap (Figure 2). Recent studies have shown that individual microbes in the subsurface undergo total carbon turnover times spanning months to hundreds of years, meaning that an individual can survive for even longer, possibly operating on timescales beyond thousands of years (Hoehler and Jørgensen, 2013; Lever et al., 2013; Braun et al., 2017; Lloyd, 2021). Any interaction these microbes have with the surrounding fluids and solid mineral phases at depth should then be considered to occur over these long timescales over which the microbes operate. Processes like microbial metabolic transformations of volatiles, the diagenesis of organic matter and the microbially-mediated dissolution and precipitation of minerals might act at temporal and spatial scales that overlap with geological processes in the same area (Figure 2). For example, at convergent margins, subsurface microbial metabolism can significantly impact the quantity of carbon recycled in the outer forearc and forearc regions, while responding cohesively to subduction parameters (Barry et al., 2019; Fullerton et al., 2021).
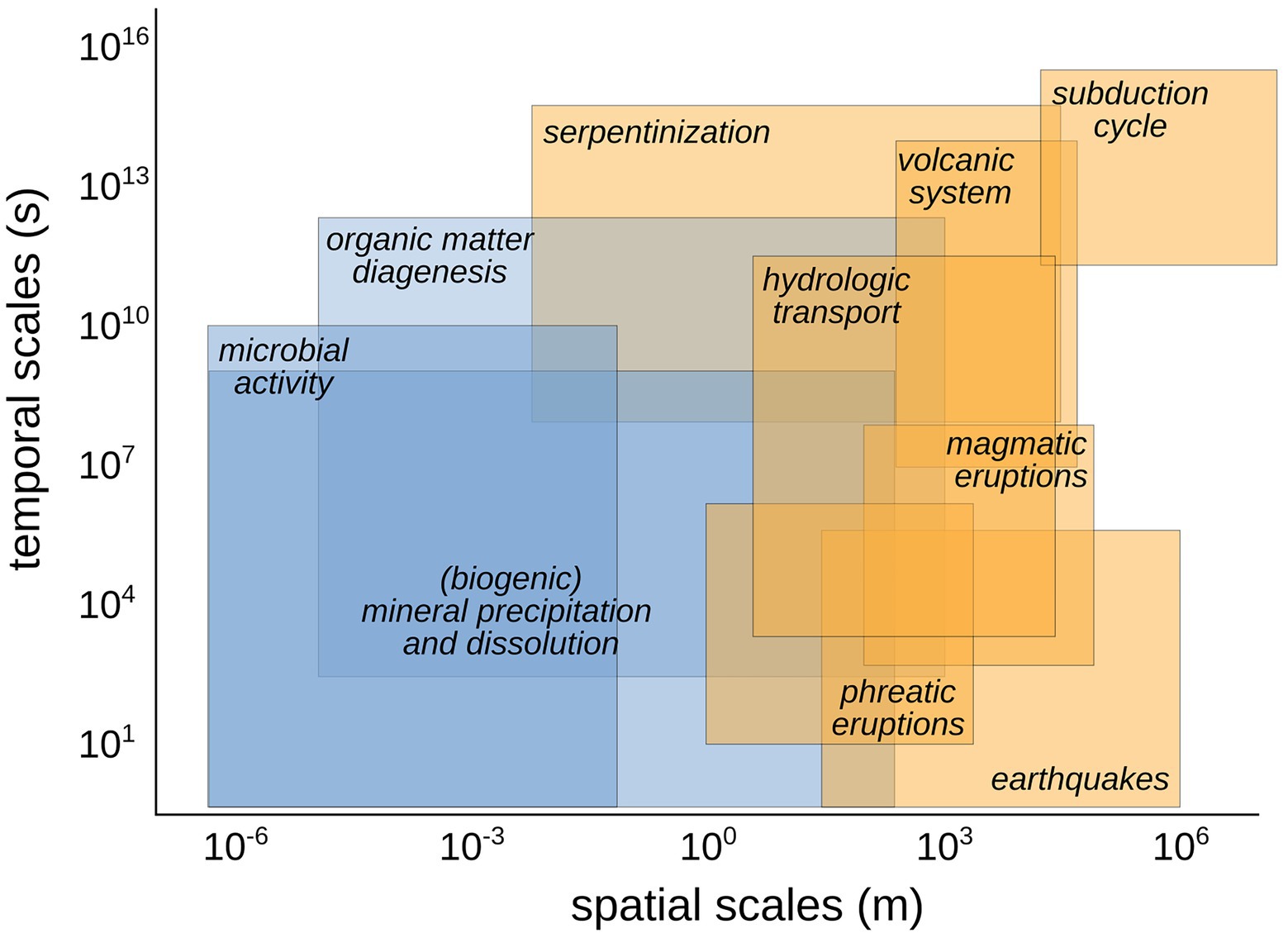
Figure 2. Conceptual diagram showing the relative spatial and temporal scales of biological (in blue) and geological (in orange) processes at convergent margins and their overlap in space–time. Similar conceptual diagram showing the overlap between geological and biological processes in different tectonic settings can help to identify the relevant processes to track and the type of synoptic, co-located samples to collect.
Identifying the theoretical overlap between geological and biological processes in geological settings is a key step in identifying the relevant geo-bio-processes to track and the type of synoptic, co-located samples to collect. Once identified, large-scale geological gradients can be sampled in a similar fashion to small-scale local gradients. Thus, samples can be collected from deeply-sourced seeps at intervals of tens or hundreds of kilometers along ideal transects crossing the geological region under investigation (Figure 1). This effectively moves the scale of investigation from local to meso- and large-spatial scales, tracking subsurface biosphere properties as they covary with geological processes. As a consequence, the resulting inferences have broad planetary implications, both at the ecosystem level and in deep time. Comparing samples across broad spatial scales may also give insights into how the paleomicrobiological environments (Rivera-Perez et al., 2018; Arning and Wilson, 2020) have changed through time.
One of the keys to identifying biological and geological feedbacks is collecting samples that are synoptic (i.e., collected at the same time) and colocated (i.e., collected on the same location or in the immediate proximity), for as wide a range as possible of biological, chemical and geological parameters. Since individual laboratories are usually not capable of conducting high resolution measurements of disparate geochemical and biomolecular data, this can be only accomplished by bringing together a team of scientists in different fields of research, with the added benefit of enabling cross-cutting connections based on discussions between scientists from different fields. For instance, geologists often collect samples to characterize abiotic processes that are analyzed for concentrations and isotopic composition of gases, aqueous molecules, petrological inclusions, and mineralogy, which are interpreted without explicitly addressing potential biological interactions. On the other hand, microbiologists often assess biotic processes, collecting samples to produce cultures and biomolecular datasets for DNA, RNA, proteins, lipids and metabolites, and interpret them against a limited number of geochemical variables that describe the local environment, without reference to the larger geological context and longer time scale processes.
To bring the problem into focus, it is pertinent to analyze a few examples from well-studied locations such as the deep-sea hydrothermal vents of the East Pacific Rise and the hot springs of Yellowstone National Park. Hydrothermal vents located on the 9°50′N segment of the East Pacific Rise are among the best studied vents in the world, with biological and geochemical observations going back four decades (Jannasch and Wirsen, 1979; Jannasch and Mottl, 1985; Sylvan et al., 2012; Vetriani et al., 2014; Gulmann et al., 2015; McNichol et al., 2018). While the pioneering studies at these locations have tremendously contributed to our understanding of how the microbiology of the hot oceanic subsurface might operate, the number of studies that simultaneously discuss the detailed gas and aqueous geochemistry and microbiology of the same sample are scarce, and typically limited to a handful of vent locations. While sampling large spatial coverage of deep-sea hydrothermal vents is much harder to achieve given the large amount of exploration effort required to discover them, studies carried out at the more accessible Yellowstone National Park hydrothermal system show a similar trend with comparisons of the microbial diversity found in a large number of geochemically-diverse springs (Inskeep et al., 2013; Colman et al., 2019a). Even in these studies, data pertaining to the geochemistry of the gases and isotopic composition is often missing, fragmented, non-standardized, or measured on other samples during different field seasons. Altogether, we argue that the absence of co-located, synoptic samples impairs the ability to make connections among the different geobiological processes on larger spatial and temporal scales.
Collecting a large number of variables on co-located synoptic samples provides unique opportunities. Despite this, it also brings to the table a large number of challenges associated with working with a large number of potentially covarying variables (Fan et al., 2014). Classical correlational approaches can be subjected to large number artifacts (e.g., spurious correlations) when dealing with large datasets. In this case abductive approaches need to be used for hypothesis generation and followed by inductive studies confirming (or rejecting) the generated hypothesis. In doing this, a number of big data approaches can be used, such as large-scale multivariate techniques (Ranjard et al., 2013; Danko et al., 2021; Cordone et al., 2022), network analysis (Delgado-Baquerizo et al., 2018; Fullerton et al., 2021) and machine learning approaches (Ghannam et al., 2020; Fullerton et al., 2021). Although the number of studies employing such techniques is still limited, these approaches provide access to underlying trends in the data structure, and, if carefully interpreted in the light of the existing body of knowledge, can provide fruitful insights.
We presented here a new rationale to approach the study of subsurface ecosystems through the use of deeply-sourced seeps as windows into the subsurface. Our approach calls for the collection of a large number of colocated synoptic samples across large spatial scales, purposefully ignoring more local and likely surface derived variability.
Specifically our approach can be summarized as follows:
1. Use a combination of pre-existing and newly sampled field data to identify deeply-sourced seeps and samples to minimize surface contamination;
2. Sample across large geologic gradients (hundreds of kms), following changes in geological processes of interest;
3. Use a multi-pronged approach to identify the relative contribution of deep vs surface processes for each sampling site (Table 1). No single perfect proxy exists and the strategy will need to be adapted to the unique features of the system under investigation;
4. Collect geological, geochemical and biological data synoptically, with samples taken as close together as possible (i.e., from the same seep and at the same time) in order to minimize confounding factors (small-scale spatial and temporal variability). Although an inductive, hypothesis driven approach is productive, employing an abductive, data-driven approach can also lead to unanticipated discoveries. Make sure to account for statistical problems that might arise from this, like accounting for multiple hypothesis testing, collinearity of variables and large number of false positives;
5. Analysis should focus on features that covary across the dataset, possibly using large-scale correlational approaches, big data analytics and machine learning. Relationships that are identified between biological and geological processes should be backed up with mechanistic connections between variables (possibly through the presence of known ecosystem role and function, physiologies, metabolic pathways, or bulk activity measurements on natural samples).
Earth’s subsurface is vast, with at least 1029 living microbial cells (Kallmeyer et al., 2012; Magnabosco et al., 2018). These microbial communities are some of the most diverse, yet least-described microbial communities in the world (Huber et al., 2006; Santelli et al., 2008; Lloyd et al., 2018), and might be relevant to understand not only the coevolution of life with our planet (Chopra and Lineweaver, 2016; Giovannelli et al., 2020), but also in the search for life in the universe (Parnell and McMahon, 2016; Giovannelli et al., 2021). Many of these populations may be growing very slowly (Hoehler and Jørgensen, 2013), meaning that biological processes may overlap in timescale with slower geological processes. Investigating these overlaps requires a large-scale approach to sampling deep subsurface life, where a large suite of biological and geochemical data are collected simultaneously, often by different laboratories. Given the infeasibility of drilling hundreds of boreholes for a single study, here we describe an approach where sampling of surface-expressed fluids from natural springs across a geological gradient is used to investigate biological and geological feedbacks in Earth’s subsurface. This approach has been used in only a handful of studies thus far, but a broader adoption of these methods may accelerate discoveries about life in the subsurface and how it interacts with geological processes even at planetary level.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
DG has received funding from the European Research Council (ERC) under the 17 European Union’s Horizon 2020 research and innovation programme (grant 18 agreement no. 948972) ERC-STG-2020 project CoEvolve. Additional support came from The National Fund for Scientific and Technological Development of Chile (FONDECYT) Grant 11191138 (The National Research and Development Agency of Chile, ANID Chile), COPAS COASTAL ANID FB210021 to GJ. KL, JM, and PB were supported by the NSF-FRES award 2121637. Partial support came from the Alfred P. Sloan Foundation and the Deep Carbon Observatory (G-2016-7206) to PB, JM, DG, and KL. Additional support came from Simons Foundation 404586 to KL.
The authors wish to acknowledge the large number of collaborators that have contributed to develop this sampling approach.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acinas, S. G., Sánchez, P., Salazar, G., Cornejo-Castillo, F. M., Sebastián, M., Logares, R., et al. (2021). Deep ocean metagenomes provide insight into the metabolic architecture of bathypelagic microbial communities. Commun. Biol. 4, 604–615. doi: 10.1038/s42003-021-02112-2
Anderson, R. E., Brazelton, W. J., and Baross, J. A. (2011). Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol. Ecol. 77, 120–133. doi: 10.1111/j.1574-6941.2011.01090.x
Arning, N., and Wilson, D. J. (2020). The past, present and future of ancient bacterial DNA. Microbial. Genomics 6:mgen000384. doi: 10.1099/mgen.0.000384
Barry, P. H., de Moor, J. M., Giovannelli, D., Schrenk, M., Hummer, D. R., Lopez, T., et al. (2019). Forearc carbon sink reduces long-term volatile recycling into the mantle. Nature 568, 487–492. doi: 10.1038/s41586-019-1131-5
Basili, M., Timothy, J. R., Yucel, M., de Moor, J. M., Barry, P. H., Zahirovic, S., et al. (2022). Microbial community structure is shaped by physical characteristics of the Central America subduction zone. Prep.
Bekaert, D. V., Gazel, E., Turner, S., Behn, M. D., De Moor, J. M., Zahirovic, S., et al. (2021). High 3He/4He in Central Panama reveals a distal connection to the Galápagos plume. Proc. Natl. Acad. Sci. 118:e2110997118. doi: 10.1073/pnas.2110997118
Braun, S., Mhatre, S. S., Jaussi, M., Røy, H., Kjeldsen, K. U., Pearce, C., et al. (2017). Microbial turnover times in the deep seabed studied by amino acid racemization modelling. Sci. Rep. 7:5680. doi: 10.1038/s41598-017-05972-z
Brazelton, W. J., Thornton, C. N., Hyer, A., Twing, K. I., Longino, A. A., Lang, S. Q., et al. (2017). Metagenomic identification of active methanogens and methanotrophs in serpentinite springs of the Voltri massif. Italy. PeerJ 5:e2945. doi: 10.7717/peerj.2945
Cascone, M., Lloyd, K., Rogers, T., de Moor, M., Schrenk, M., Barry, P. H., et al. (2021). Microbial diversity in the backarc hot springs of Argentina and its role in biogeochemical cycles. In (GOLDSCHMIDT). Available at: https://conf.goldschmidt.info/goldschmidt/2021/meetingapp.cgi/Paper/8122. (Accessed May 31, 2022).
Chivian, D., Brodie, E. L., Alm, E. J., Culley, D. E., Dehal, P. S., DeSantis, T. Z., et al. (2008). Environmental genomics reveals a single-species ecosystem deep within earth. Science 322, 275–278. doi: 10.1126/science.1155495
Chopra, A., and Lineweaver, C. H. (2016). The case for a Gaian bottleneck: the biology of habitability. Astrobiology 16, 7–22. doi: 10.1089/ast.2015.1387
Colman, D. R., Feyhl-Buska, J., Robinson, K. J., Fecteau, K. M., Xu, H., Shock, E. L., et al. (2016). Ecological differentiation in planktonic and sediment-associated chemotrophic microbial populations in Yellowstone hot springs. FEMS Microbiol. Ecol. 92:92. doi: 10.1093/femsec/fiw137
Colman, D. R., Lindsay, M. R., Amenabar, M. J., and Boyd, E. S. (2019a). The intersection of geology, geochemistry, and microbiology in continental hydrothermal systems. Astrobiology 19, 1505–1522. doi: 10.1089/ast.2018.2016
Colman, D. R., Lindsay, M. R., and Boyd, E. S. (2019b). Mixing of meteoric and geothermal fluids supports hyperdiverse chemosynthetic hydrothermal communities. Nat. Commun. 10:681. doi: 10.1038/s41467-019-08499-1
Cordone, A., D’Errico, G., Magliulo, M., Bolinesi, F., Selci, M., Basili, M., et al. (2022). Bacterioplankton diversity and distribution in relation to phytoplankton community structure in the Ross Sea surface waters. Front. Microbiol. 13:722900. doi: 10.3389/fmicb.2022.722900
Crossey, L. J., Karlstrom, K. E., Schmandt, B., Crow, R. R., Colman, D. R., Cron, B., et al. (2016). Continental smokers couple mantle degassing and distinctive microbiology within continents. Earth Planet. Sci. Lett. 435, 22–30. doi: 10.1016/j.epsl.2015.11.039
D’Hondt, S., Pockalny, R., Fulfer, V. M., and Spivack, A. J. (2019). Subseafloor life and its biogeochemical impacts. Nat. Commun. 10:3519. doi: 10.1038/s41467-019-11450-z
Danko, D., Bezdan, D., Afshin, E. E., Ahsanuddin, S., Bhattacharya, C., Butler, D. J., et al. (2021). A global metagenomic map of urban microbiomes and antimicrobial resistance. Cells 184, 3376–3393.e17. doi: 10.1016/j.cell.2021.05.002
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Deming, J. W., and Baross, J. A. (1993). Deep-sea smokers: windows to a subsurface biosphere? Geochim. Cosmochim. Acta 57, 3219–3230. doi: 10.1016/0016-7037(93)90535-5
Emerson, J. B., Thomas, B. C., Alvarez, W., and Banfield, J. F. (2016). Metagenomic analysis of a high carbon dioxide subsurface microbial community populated by chemolithoautotrophs and bacteria and archaea from candidate phyla. Environ. Microbiol. 18, 1686–1703. doi: 10.1111/1462-2920.12817
Fan, J., Han, F., and Liu, H. (2014). Challenges of big data analysis. Natl. Sci. Rev. 1, 293–314. doi: 10.1093/nsr/nwt032
Fones, E. M., Colman, D. R., Kraus, E. A., Nothaft, D. B., Poudel, S., Rempfert, K. R., et al. (2019). Physiological adaptations to serpentinization in the Samail Ophiolite. Oman. ISME J. 13, 1750–1762. doi: 10.1038/s41396-019-0391-2
Fullerton, K. M., Schrenk, M. O., Yücel, M., Manini, E., Basili, M., Rogers, T. J., et al. (2021). Effect of tectonic processes on biosphere–geosphere feedbacks across a convergent margin. Nat. Geosci. 14, 301–306. doi: 10.1038/s41561-021-00725-0
Ghannam, R. B., Schaerer, L. G., Butler, T. M., and Techtmann, S. M. (2020). Biogeographic patterns in members of globally distributed and dominant taxa found in port microbial communities. mSphere 5, e00481–e00419. doi: 10.1128/mSphere.00481-19
Giggenbach, W. F. (1992). Isotopic shifts in waters from geothermal and volcanic systems along convergent plate boundaries and their origin. Earth Planet. Sci. Lett. 113, 495–510. doi: 10.1016/0012-821X(92)90127-H
Giovannelli, D., Barry, P. H., Bekaert, D. V., Chiodi, A., Cordone, A., Covone, G., et al. (2021). Subsurface life can modify volatile cycling on a planetary scale. Mem. Della Soc. Astron. Ital. 92:60.
Giovannelli, D., Barry, P. H., de Moor, J. M., Lloyd, K. G., and Schrenk, M. O. (2020). Microbial influences on Subduction zone carbon cycling. Eos 101. doi: 10.1029/2020EO140906
Giovannelli, D., d’Errico, G., Manini, E., Yakimov, M. M., and Vetriani, C. (2013). Diversity and phylogenetic analyses of bacteria from a shallow-water hydrothermal vent in Milos island (Greece). Front. Extreme Microbiol. 4:184. doi: 10.3389/fmicb.2013.00184
Gulmann, L., Beaulieu, S., Shank, T., Ding, K., Seyfried, W., and Sievert, S. (2015). Bacterial diversity and successional patterns during biofilm formation on freshly exposed basalt surfaces at diffuse-flow deep-sea vents. Front. Microbiol. 6:901.
Hall, J. R., Mitchell, K. R., Jackson-Weaver, O., Kooser, A. S., Cron, B. R., Crossey, L. J., et al. (2008). Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl. Environ. Microbiol. 74, 4910–4922. doi: 10.1128/AEM.00233-08
Hilton, D. R., Hammerschmidt, K., Teufel, S., and Friedrichsen, H. (1993). Helium isotope characteristics of Andean geothermal fluids and lavas. Earth Planet. Sci. Lett. 120, 265–282. doi: 10.1016/0012-821X(93)90244-4
Hoehler, T. M., and Jørgensen, B. B. (2013). Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94. doi: 10.1038/nrmicro2939
Hou, W., Wang, S., Dong, H., Jiang, H., Briggs, B. R., Peacock, J. P., et al. (2013). A comprehensive census of microbial diversity in Hot Springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS One 8:e53350. doi: 10.1371/journal.pone.0053350
Huber, J. A., Johnson, H. P., Butterfield, D. A., and Baross, J. A. (2006). Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8, 88–99. doi: 10.1111/j.1462-2920.2005.00872.x
Inskeep, W. P., Ackerman, G. G., Taylor, W. P., Kozubal, M., Korf, S., and Macur, R. E. (2005). On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3, 297–317. doi: 10.1111/j.1472-4669.2006.00059.x
Inskeep, W. P., Jay, Z. J., Tringe, S. G., Herrgard, M., and Rusch, D. B. (2013). The YNP metagenome project: environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Microb. Physiol. Metab. 4:67. doi: 10.3389/fmicb.2013.00067
Jannasch, H. W., and Mottl, M. J. (1985). Geomicrobiology of Deep-Sea hydrothermal vents. Science 229, 717–725. doi: 10.1126/science.229.4715.717
Jannasch, H. W., and Wirsen, C. O. (1979). Chemosynthetic primary production at East Pacific Sea floor spreading centers. Bioscience 29, 592–598. doi: 10.2307/1307765
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Lang, S. Q., Früh-Green, G. L., Bernasconi, S. M., Brazelton, W. J., Schrenk, M. O., and McGonigle, J. M. (2018). Deeply-sourced formate fuels sulfate reducers but not methanogens at lost City hydrothermal field. Sci. Rep. 8:755. doi: 10.1038/s41598-017-19002-5
Lever, M. A., Rouxel, O., Alt, J. C., Shimizu, N., Ono, S., Coggon, R. M., et al. (2013). Evidence for microbial carbon and sulfur cycling in deeply buried ridge flank basalt. Science 339, 1305–1308. doi: 10.1126/science.1229240
Lindsay, M. R., Colman, D. R., Amenabar, M. J., Fristad, K. E., Fecteau, K. M., Debes, R. V. II, et al. (2019). Probing the geological source and biological fate of hydrogen in Yellowstone hot springs. Environ. Microbiol. 21, 3816–3830. doi: 10.1111/1462-2920.14730
Lloyd, K. G. (2021). Time as a microbial resource. Environ. Microbiol. Rep. 13, 18–21. doi: 10.1111/1758-2229.12892
Lloyd, K. G., Steen, A. D., Ladau, J., Yin, J., and Crosby, L. (2018). Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3, e00055–e00018. doi: 10.1128/mSystems.00055-18
Magnabosco, C., Lin, L.-H., Dong, H., Bomberg, M., Ghiorse, W., Stan-Lotter, H., et al. (2018). The biomass and biodiversity of the continental subsurface. Nat. Geosci. 11, 707–717. doi: 10.1038/s41561-018-0221-6
McNichol, J., Stryhanyuk, H., Sylva, S. P., Thomas, F., Musat, N., Seewald, J. S., et al. (2018). Primary productivity below the seafloor at deep-sea hot springs. Proc. Natl. Acad. Sci. 115, 6756–6761. doi: 10.1073/pnas.1804351115
Merino, N., Aronson, H. S., Bojanova, D. P., Feyhl-Buska, J., Wong, M. L., Zhang, S., et al. (2019). Living at the extremes: extremophiles and the limits of life in a planetary context. Front. Microbiol. 10:780. doi: 10.3389/fmicb.2019.00780
Meyer-Dombard, D. R., Shock, E. L., and Amend, J. P. (2005). Archaeal and bacterial communities in geochemically diverse hot springs of Yellowstone National Park, USA. Geobiology 3, 211–227. doi: 10.1111/j.1472-4669.2005.00052.x
Orcutt, B. N., and Edwards, K. J. (2014). “Chapter 2.5 - life in the ocean crust: lessons from subseafloor laboratories” in Developments in Marine Geology Earth and Life Processes Discovered From Subseafloor Environments. eds. R. Stein, D. K. Blackman, F. Inagaki, and H.-C. Larsen (Amsterdam: Elsevier), 175–196.
Parnell, J., and McMahon, S. (2016). Physical and chemical controls on habitats for life in the deep subsurface beneath continents and ice. Phil. Trans. R. Soc. A 374:20140293. doi: 10.1098/rsta.2014.0293
Patwardhan, S., Foustoukos, D. I., Giovannelli, D., Yücel, M., and Vetriani, C. (2018). Ecological succession of sulfur-oxidizing epsilon- and Gammaproteobacteria during colonization of a shallow-water gas vent. Front. Microbiol. 9:2970. doi: 10.3389/fmicb.2018.02970
Power, J. F., Carere, C. R., Lee, C. K., Wakerley, G. L., Evans, D. W., Button, M., et al. (2018). Microbial biogeography of 925 geothermal springs in New Zealand. Nat. Commun. 9, 1–12. doi: 10.1038/s41467-018-05020-y
Ranjard, L., Dequiedt, S., Chemidlin Prévost-Bouré, N., Thioulouse, J., Saby, N. P. A., Lelievre, M., et al. (2013). Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nat. Commun. 4:1434. doi: 10.1038/ncomms2431
Rappé, M. S., and Giovannoni, S. J. (2003). The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394. doi: 10.1146/annurev.micro.57.030502.090759
Reveillaud, J., Reddington, E., McDermott, J., Algar, C., Meyer, J. L., Sylva, S., et al. (2016). Subseafloor microbial communities in hydrogen-rich vent fluids from hydrothermal systems along the M id-C ayman R Ise. Environ. Microbiol. 18, 1970–1987. doi: 10.1111/1462-2920.13173
Rivera-Perez, J. I., Santiago-Rodriguez, T. M., and Toranzos, G. A. (2018). “Paleomicrobiology: a snapshot of ancient microbes and approaches to forensic microbiology” in Environmental Microbial Forensics. Chapter 4. eds. R. J. Cano and G. A. Toranzos (Whiley).
Rogers, T., Buongiorno, J., Schrenk, M. O., Moor, M. J.de, Barry, P. H., Jessen, G. L., et al. (2022). Subsurface landscape of a Chemolithoautotrophy-based ecosystem.
Sánchez-Murillo, R., Gazel, E., Schwarzenbach, E. M., Crespo-Medina, M., Schrenk, M. O., Boll, J., et al. (2014). Geochemical evidence for active tropical serpentinization in the Santa Elena Ophiolite, Costa Rica: An analog of a humid early earth? Geochem. Geophys. Geosystems 15, 1783–1800. doi: 10.1002/2013GC005213
Santelli, C. M., Orcutt, B. N., Banning, E., Bach, W., Moyer, C. L., Sogin, M. L., et al. (2008). Abundance and diversity of microbial life in ocean crust. Nature 453, 653–656. doi: 10.1038/nature06899
Steen, A. D., Crits-Christoph, A., Carini, P., DeAngelis, K. M., Fierer, N., Lloyd, K. G., et al. (2019). High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 13, 3126–3130. doi: 10.1038/s41396-019-0484-y
Summit, M., and Baross, J. A. (2001). A novel microbial habitat in the mid-ocean ridge subseafloor. Proc. Natl. Acad. Sci. 98, 2158–2163. doi: 10.1073/pnas.051516098
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348:1261359. doi: 10.1126/science.1261359
Sylvan, J. B., Pyenson, B. C., Rouxel, O., German, C. R., and Edwards, K. J. (2012). Time-series analysis of two hydrothermal plumes at 9°50′N East Pacific rise reveals distinct, heterogeneous bacterial populations. Geobiology 10, 178–192. doi: 10.1111/j.1472-4669.2011.00315.x
Trembath-Reichert, E., Butterfield, D. A., and Huber, J. A. (2019). Active subseafloor microbial communities from Mariana back-arc venting fluids share metabolic strategies across different thermal niches and taxa. ISME J. 13, 2264–2279. doi: 10.1038/s41396-019-0431-y
Uribe-Lorío, L., Brenes-Guillén, L., Hernández-Ascencio, W., Mora-Amador, R., González, G., Ramírez-Umaña, C. J., et al. (2019). The influence of temperature and pH on bacterial community composition of microbial mats in hot springs from Costa Rica. MicrobiologyOpen 8:e893. doi: 10.1002/mbo3.893
Vetriani, C., Voordeckers, J. W., Crespo-Medina, M., O’Brien, C. E., Giovannelli, D., and Lutz, R. A. (2014). Deep-sea hydrothermal vent Epsilonproteobacteria encode a conserved and widespread nitrate reduction pathway (nap). ISME J. 8, 1510–1521. doi: 10.1038/ismej.2013.246
Williamson, S. J., Allen, L. Z., Lorenzi, H. A., Fadrosh, D. W., Brami, D., Thiagarajan, M., et al. (2012). Metagenomic exploration of viruses throughout the Indian Ocean. PLoS One 7:e42047. doi: 10.1371/journal.pone.0042047
Keywords: subsurface biosphere, geosphere–biosphere coevolution, geomicrobiology, large-scale, hot springs
Citation: Giovannelli D, Barry PH, de Moor JM, Jessen GL, Schrenk MO and Lloyd KG (2022) Sampling across large-scale geological gradients to study geosphere–biosphere interactions. Front. Microbiol. 13:998133. doi: 10.3389/fmicb.2022.998133
Received: 19 July 2022; Accepted: 05 October 2022;
Published: 31 October 2022.
Edited by:
David Emerson, Bigelow Laboratory for Ocean Sciences, United StatesReviewed by:
Francisco P. Chávez, University of Chile, ChileCopyright © 2022 Giovannelli, Barry, de Moor, Jessen, Schrenk and Lloyd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donato Giovannelli, ZG9uYXRvLmdpb3Zhbm5lbGxpQHVuaW5hLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.