- 1Institute of Medical Science, College of Medicine, Hallym University, Chuncheon, South Korea
- 2Department of Microbiology, College of Medicine, Hallym University, Chuncheon, South Korea
- 3Department of Biochemistry, College of Natural Sciences, Chungbuk National University, Cheongju, South Korea
- 4Department of Medical Genetics, College of Medicine, Hallym University, Chuncheon, South Korea
Stress granule formation is induced by numerous environmental stressors, including sodium arsenite treatment and viral infection. Accordingly, stress granules can inhibit viral propagation and function as part of the antiviral host response to numerous viral infections. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) antagonizes stress granule formation, in part, via interaction between SARS-CoV-2 nucleocapsid (N) protein and Ras-GTPase-activating SH3-domain-binding protein 1 (G3BP1). However, it is unclear whether there are differential effects in different cell types. In this study, we assessed interaction between the N protein of SARS-CoV-2 S clade and G3BP1/2 in Vero and Calu-3 cells and investigated the effect of various SARS-CoV-2 strains on sodium arsenite-induced stress granule formation. Our data show that SARS-CoV-2 S clade N protein interacts with both G3BP1 and G3BP2 more strongly in Calu-3 vs. Vero cells. Consistent with this observation, infection with SARS-CoV-2 S clade induces stress granule formation in Vero but not in Calu-3 cells. However, infection with SARS-CoV-2 S clade, as well as other SARS-CoV-2 variants, inhibits sodium arsenite-induced stress granule formation in both cell lines. Taken together, our results show differential effects of SARS-CoV-2 infection on stress granule formation that is dependent on host cell type, rather than virus strain type.
Introduction
The devastating coronavirus disease 2019 (COVID-19) pandemic, which is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), continues to spread, posing a tremendous threat to both public health and economic stability worldwide (Chen et al., 2020; Huang et al., 2020; Zhou et al., 2020). SARS-CoV-2, like other coronaviruses, is an enveloped, positive-sense single-stranded RNA virus with a genome of ∼30,000 nucleotides (Lu et al., 2020) encoding four structural proteins: spike (S), membrane (M), envelope (E), and nucleocapsid (N) (Wu et al., 2020; V’kovski et al., 2021). Beyond vaccine development, most studies on SARS-CoV-2 to date have focused on identifying strategies to reduce the pathophysiology of infection by suppressing viral replication inside the host (Blake et al., 2021; Zhang et al., 2021). However, while these investigations have led to the development of antiviral therapies for COVID-19 (i.e., ritonavir-boosted nirmatrelvir, sold under the commercial name Paxlovid), mutations that reduce drug efficacy have been identified in in vitro selected SARS-CoV-2 variants, highlighting the need for new antiviral strategies to emerging SARS-CoV-2 variants (Szemiel et al., 2021; Stevens et al., 2022).
In response to viral infection, cells can initiate several molecular and physiological processes aimed at combatting the infection and suppressing viral replication (Sumbria et al., 2021). These include the induction of stress responses and the formation of stress granules within infected cells. Stress granules are dense membrane-less ribonucleoprotein (RNP) complexes that dynamically assemble in the cytoplasm of cells exposed to environmental stressors, such as ultraviolet radiation, heat and cold shock, nutrient starvation, sodium arsenite treatment, and viral infection (White and Lloyd, 2012; Protter and Parker, 2016; Mahboubi and Stochaj, 2017). They form from the accumulation of translationally inactive mRNAs and stalled translation initiation complexes, and their primary function is to promote cell survival (Ohn et al., 2008; Ivanov et al., 2019; Campos-Melo et al., 2021).
In particular, previous studies have shown that stress granules can suppress viral replication and function as a key part of the antiviral host response to various viral pathogens (Onomoto et al., 2012; McCormick and Khaperskyy, 2017). Stress granule formation in virus-infected cells is initiated when double-stranded (ds)RNA or single-stranded (ss)RNA, common viral replication intermediates, bind to protein kinase R (PKR), triggering its autophosphorylation and activation (Cesaro and Michiels, 2021). Once activated, PKR phosphorylates the eukaryotic translation initiation factor (eIF2α) and triggers assembly of stress granules by the nucleating proteins Ras-GTPase-activating SH3-domain-binding protein 1 (G3BP1) and T-cell-restricted intracellular antigen-1 (TIA-1) (Gilks et al., 2004; Matsuki et al., 2013). Phosphorylated eIF2α further prevents formation of tRNAMet-GTP–eIF2 complexes, which inhibits viral mRNA translation, ultimately blocking viral replication (Liu et al., 2020).
In response to this stress granule-mediated inhibition, viruses have evolved various mechanisms to antagonize stress granule formation and thereby enhance viral replication (Rabouw et al., 2016; Hou et al., 2017; Nakagawa et al., 2018). For example, hepatitis C virus and Japanese encephalitis virus inhibit antiviral stress granule formation by targeting PKR (Toroney et al., 2010; Tu et al., 2012), whereas others, including Zika virus, Junin virus, and West Nile virus, block assembly of stress granules by modulating the eIF2α-dependent pathway (Linero et al., 2011; Amorim et al., 2017; Basu et al., 2017). Alternatively, enterovirus and foot-and-mouth disease virus produce a viral protease known as 3Cpro, which cleaves G3BP1 at amino acids Q326 and E284, leading to disruption of stress granule formation and enhanced viral replication (Ye et al., 2018; Zhang et al., 2018). Likewise, strategies to modulate or inhibit stress granule formation have also been reported for diverse groups of coronaviruses. In Middle East respiratory syndrome coronavirus (MERS-CoV), the virus-encoded protein 4a sequesters viral RNA and blocks binding to PKR, preventing its downstream activation (Rabouw et al., 2016; Nakagawa et al., 2018). In contrast, another coronavirus—infectious bronchitis virus—can inhibit stress granule formation by modulating both eIF2α-dependent and eIF2α-independent signaling pathways (Brownsword et al., 2020).
Notably, a recent study reported that similar to MERS-CoV 4a protein, the SARS-CoV-2 N protein binds to PKR and prevents its autophosphorylation (Zheng et al., 2021). However, in addition, N protein was also found to interact with and inhibit G3BP1, thus impairing stress granule formation via two distinct mechanisms (Zheng et al., 2021). These observations are supported by another study, which found that SARS-CoV-2 N protein inhibits host stress granule by interacting with G3BP1 and G3BP2 and sequestering these proteins from their interacting partners (Nabeel-Shah et al., 2022). However, given the wide cell tropism exhibited by SARS-CoV-2 and the rapidly changing genetic landscape of this virus, it is unclear how the extent of stress granule inhibition varies in different cell types and or with distinct viral variants. Here, to address these questions, we utilized our previously developed anti-N protein antibody (Kim et al., 2021a) to measure the binding between cellular G3BP1 and N protein from SARS-CoV-2 S clade, as well as the subsequent inhibition of stress granule formation, in Vero and Calu-3 cells. In addition, we determined how infection with different SARS-CoV-2 variants affects stress granule formation in response to viral infection and sodium arsenite treatment. Through this study, we aim to better understand the mechanisms by which SARS-CoV-2 blocks antiviral responses to enhance viral replication and promote pathogenesis.
Materials and methods
Cell and virus culture
The African green monkey kidney Vero and Vero E6 cell lines, as well as the human airway epithelial Calu-3 cell line, were obtained from the Korean Cell Line Bank (Seoul, South Korea). Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, Waltham, MA, United States), containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 25 mM HEPES, 100 U/mL penicillin, and 100 μg/mL streptomycin, and maintained at 37°C in a 5% CO2 incubator.
SARS-CoV-2 S clade (hCoV-19/Korea/KCDC03/2020, lineage A, NCCP43326), alpha variant (hCoV-19/Korea/KDCA51463/2021, lineage B.1.1.7, 501Y.V1, NCCP43381), delta variant (hCoV-19/Korea/KDCA119861/2021, lineage B.1.617.2, NCCP43390), mu variant (hCoV-19/Korea/KDCA159392/2021, lineage B.1.621, NCCP43407), and omicron variant (hCoV-19/Korea/KDCA447321/2021, lineage B.1.1.529, NCCP43408) were provided by the National Culture Collection for Pathogens (Osong, South Korea).
SARS-CoV-2 viruses were amplified in Vero cells; virus titers in cell culture supernatants were quantified by plaque assays, as described below, and viral stocks (1 × 107 pfu/mL) were stored at -70°C. SARS-CoV-2 amplification and cell culture infection experiments were performed under biosafety level 3 (BSL-3) conditions in the Research Institute of Medical-Bio Convergence of Hallym University. BSL-3 protocols were approved by the Institutional Biosafety Committee of Hallym University (Permit no. Hallym2020-12, 2022-03).
Virus titration by plaque assay
To estimate viral titers, we performed plaque assays on Vero E6 cells, as described previously (Kim et al., 2021b,2022a; Park et al., 2021a). In brief, Vero E6 cells (7 × 105 cells/well) were cultured in 6-well plates (Corning Inc., Corning, NY, United States) for 12 h. Cells were then washed with phosphate-buffered saline (PBS) and infected with 10-fold serial dilutions of virus culture supernatants. After 1 h adsorption with shaking every 10 min, supernatants were removed, and the wells were covered with 3 mL DMEM/F12 medium (Thermo Fisher Scientific), containing 2% Oxoid agar and N-p-Tosyl-L-phenylalanine chloromethyl ketone (TPCK; 1 μg/mL)-treated trypsin. Plates were incubated at 37°C for 3 days and stained with 0.1% crystal violet for 1 h to visualize plaque formation. Viral titers were then determined by counting the number of plaques.
Antibodies
Monoclonal antibody against the SARS-CoV-2 N protein (anti-SARS-CoV-2 N mAb) was prepared from ascitic fluid collected from BALB/c mice injected with hybridoma cells (clone 1G10C4 mAb), as previously described (Kim et al., 2021a,2022a,2022b). Rabbit anti-SARS-CoV-2 N protein polyclonal antibody (anti-SARS-CoV-2 N Ab, Catalog no. 40588-T62) was purchased from Sino Biological (Beijing, China), rabbit anti-G3BP1 antibody (Catalog no. 61559S) was purchased from Cell Signaling (Danvers, MA, United States), rabbit anti-G3BP2 antibody (Catalog no. MB S852913) was purchased from MyBioSource (San Diego, CA, United States), and anti-β-actin antibody was obtained from Sigma-Aldrich (St. Louis, MO, United States). Alexa Fluor 546-conjugated goat anti-mouse IgG (h + L) antibody (Catalog no. A11030) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (h + L) antibody (Catalog no. A11008) were purchased from Thermo Fisher Scientific.
SARS-CoV-2 infection and co-immunoprecipitation experiments
Calu-3 cells were cultured for 18 h at a density of 3 × 105 cells/well in 6-well plates. Cells were washed in PBS and infected with SARS-CoV-2 (S clade) at 0.1 multiplicity of infection (MOI) for 1 h, with shaking every 15 min, to allow adsorption of the virus, in a 5% CO2 incubator at 37°C. The supernatants were then aspirated and replaced with DMEM, containing 2% FBS. After 72 h infection, cells were washed with PBS and lysed for 30 min at 4°C in cell-lysis buffer (10 mM HEPES, 150 mM NaCl, 5 mM EDTA, 100 mM NaF, 2 mM Na3VO4, protease inhibitor cocktail, and 1% NP-40). Cell lysates were collected by centrifugation at 14,000 rpm at 4°C for 20 min and then incubated with anti-SARS-CoV-2 N mAb and Protein A beads (CaptivAtm PriMAB 52% [v/v] slurry; REPLIGEN, Waltham, MA, United States) for 2 h at 4°C. Anti-SARS-CoV-2 N mAb and Protein A bead immunocomplexes were collected by centrifugation and resolved by 4–12% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Bolt™ 4–12% Bis–Tris Plus gels (Thermo Fisher Scientific). Co-IP gels were stained with Coomassie brilliant blue G-250 or analyzed by western blot, as described below.
Mass spectrometry to identify SARS-CoV-2 N protein-binding partners
After Co-IP with anti-SARS-CoV-2 N mAb, the protein bands of interest identified by 4–12% gradient SDS-PAGE were excised from gels and analyzed by Proteinworks Co (Seoul, South Korea), as described previously (Park et al., 2021b). In brief, protein was digested with trypsin, and the resulting peptides were isolated using an Acclaim™ PepMap™ 100 C18 LC Column (Thermo Fisher Scientific). These peptides were examined by tandem mass spectrometry (MS/MS), using a Q Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher Scientific). Peptide sequences were then analyzed, and corresponding proteins were identified using the National Center for Biotechnology Information (NCBI)1 database. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD035715.
Western blot analysis
Vero and Calu-3 cells infected with SARS-CoV-2 and uninfected control cells were lysed with cell-lysis buffer, and cell lysates were collected by centrifugation at 14,000 rpm at 4°C for 20 min. Proteins were resolved on 4–12% Bis–Tris gradient gels and transferred onto nitrocellulose membranes. These were blocked in PBS with 0.1% Tween-20 (PBST), containing 5% w/v non-fat dry milk, and then incubated with anti-SARS-CoV-2 N mAb, anti-G3BP1 antibody, anti-G3BP2 antibody, or anti-β-actin antibody overnight at 4°C. Membranes were washed in PBST and incubated with a horseradish peroxidase-conjugated secondary antibody, and the immunoreactive bands were developed and visualized using an enhanced chemiluminescence reagent (Thermo Fisher Scientific).
Proteins interacting with SARS-CoV-2 N protein in Co-IP experiments were also analyzed by western blot analysis with anti-SARS-CoV-2 N Ab, anti-G3BP1 antibody, and anti-G3BP2 antibody, as described above.
Confocal microscopy
Vero and Calu-3 cells were grown on glass coverslips in 12-well plates for 18 h. Cells were washed in PBS and infected with SARS-CoV-2 (0.1 MOI) for 1 h, with shaking every 15 min, in a 5% CO2 incubator at 37°C. The supernatants were then aspirated, and cells were cultured for 47 h or 71 h in DMEM, containing 2% FBS, followed by treatment with 0.5 mM sodium arsenite (Merck KGaA, Darmstadt, Germany), an inducer of stress granules, for 1 h. Treated and control cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100, and blocked with 3% bovine serum albumin (BSA). These cells were incubated with anti-SARS-CoV-2 N mAb and anti-G3BP1 antibody (Cell Signaling) for 2 h at room temperature. Cells were then washed with PBS, containing 0.1% Triton X-100 and 1% BSA, and stained with Alexa Fluor 546-conjugated goat anti-mouse IgG (h + L) antibody and Alexa Fluor 488-conjugated goat anti-rabbit IgG (h + L) antibody for 1 h. Nuclei were stained with Hoechst 33258 (Thermo Fisher Scientific), and samples were analyzed by confocal laser scanning microscopy (CLSM, LSM 710; Carl Zeiss, Jena, Germany).
Statistical analysis
Results are shown as the mean ± standard deviation. Differences between two samples were evaluated using the Student’s t-test, with P < 0.05 as the threshold for statistical significance.
Results
SARS-CoV-2 N protein interacts with cellular G3BP
During SARS-CoV-2 infection, components of virus interact with various host cell proteins to aid in viral replication. One potential host-interacting viral component is the N protein, which acts as the dominant antigenic target for detection of SARS-CoV-2 infection (Yu et al., 2022). Here, to identify host proteins that interact with the viral N protein, we infected Calu-3 cells with the SARS-CoV-2 S clade (hCoV-19/Korea/KCDC03/2020, lineage A, NCCP43326) strain at 0.1 MOI. Calu-3 cell lysates were then immunoprecipitated with control or anti-SARS-CoV-2 N monoclonal antibody, and the associated proteins were resolved by SDS-PAGE. Results reveal interaction between N protein and a host protein with a molecular weight of ∼52 kDa (Figure 1A). This protein band was excised from the gel and analyzed by liquid chromatography (LC)-MS/MS, which identified 16 peptide fragments exhibiting amino acid sequence coverage across 41% of the full-length G3BP1 protein (Figure 1B). Thus, these data strongly suggest an interaction between SARS-CoV-2 N protein and cellular G3BP.
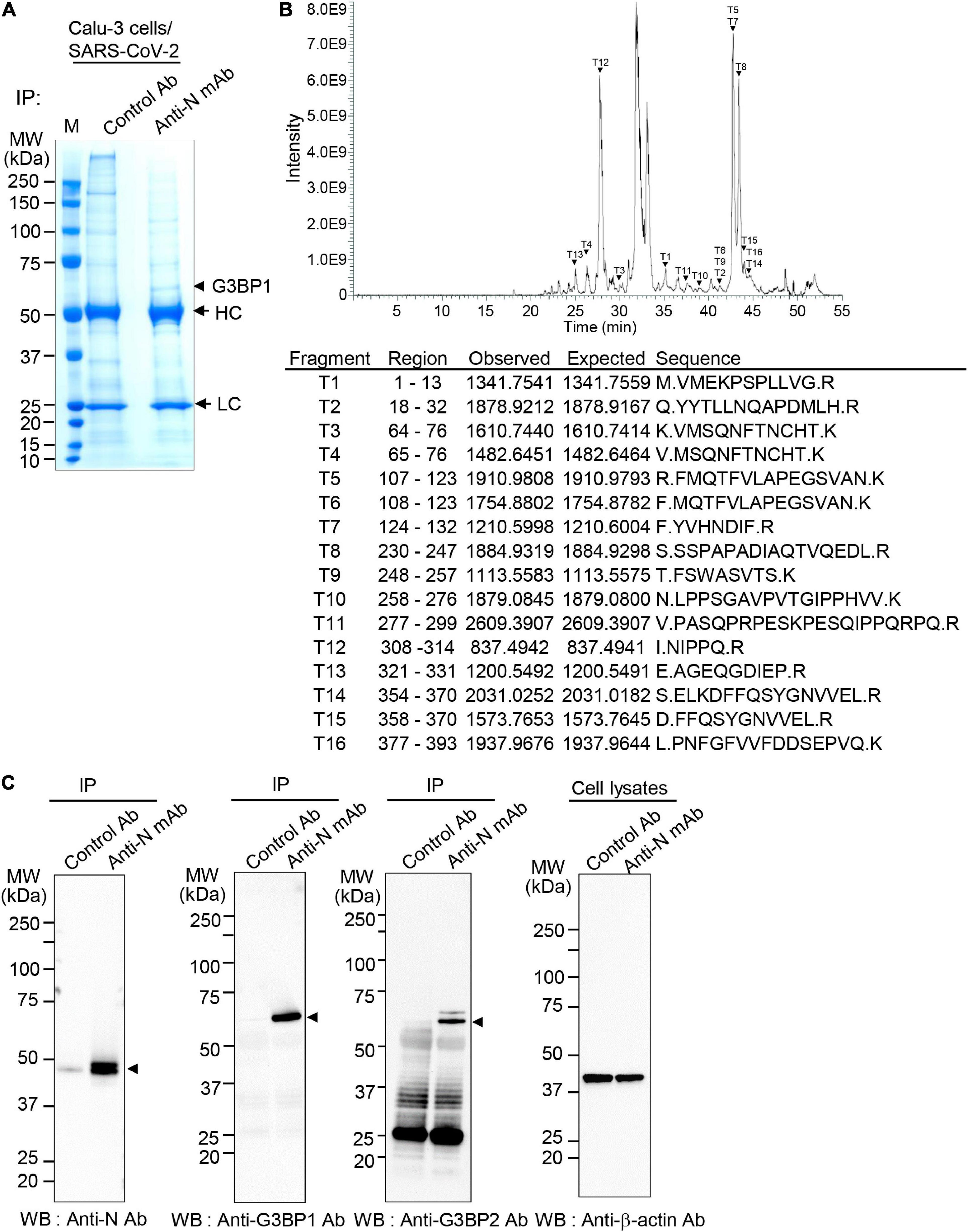
Figure 1. Interaction between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleocapsid (N) protein and Ras-GTPase-activating SH3-domain-binding protein 1 (G3BP1) (A) Co-immunoprecipitation (Co-IP) experiments to identify N protein-binding partners. Calu-3 cells were mock-infected or infected with SARS-CoV-2 S clade at 0.1 multiplicity of infection (MOI); lysates prepared at 72 h post-infection were immunoprecipitated with control antibody (normal IgG) or anti-SARS-CoV-2 N monoclonal antibody (mAb). HC, heavy chain. LC, light chain. (B) Identification of G3BP1. The ∼52.1 kDa protein band interacting with SARS-CoV-2 N protein in Co-IP experiments (A) was in-gel digested with trypsin, and the 18 peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). MS/MS analyses of mass peaks (arrows) reveal the peptide spectra of G3BP1. (C) Co-IP experiments to confirm interaction between SARS-CoV-2 N protein and G3BP1. Co-IPed immunocomplexes from panel (A) were subjected to western blot analysis with the indicated antibodies; anti-β-actin antibody was used as the input control. IP, immunoprecipitation; WB, western blot; Anti-N mAb, anti-SARS-CoV-2 N mAb (clone 1G10C4 mAb); Anti-N Ab, anti-SARS-CoV-2 N polyclonal Ab.
To further confirm binding between the SARS-CoV-2 N and G3BP proteins, Calu-3 cell lysates were immunoprecipitated with control or anti-SARS-CoV-2 N monoclonal antibody and analyzed by western blot with anti-SARS-CoV-2 N polyclonal antibody, anti-G3BP1, and anti-G3BP2 antibodies. As shown in Figure 1C, results from these Co-IP experiments show that both G3BP1 and G3BP2 are immunoprecipitated by SARS-CoV-2 N, further confirming an interaction between these proteins.
To determine whether this interaction also occurs in other cells susceptible to SARS-CoV-2 infection, we assessed binding between SARS-CoV-2 N protein and G3BP in Vero cells and compared the binding efficiency with that observed in Calu-3 cells (Figure 2). Co-IP experiments followed by western blot analysis revealed that SARS-CoV-2 N protein is expressed to higher levels in Vero cells compared to Calu-3 cells (Figure 2A). However, a higher degree of binding between SARS-CoV-2 N protein and both G3BP1 and G3BP2 is observed in Calu-3 cells vs. Vero cells, as evidenced by the increased band intensity for Co-IPed proteins in this cell line (Figures 2B,C,E). Collectively, our data therefore reveal that SARS-CoV-2 N protein can bind with both G3BP1 and G3BP2, and this interaction occurs to a greater extent in Calu-3 cells than in Vero cells.
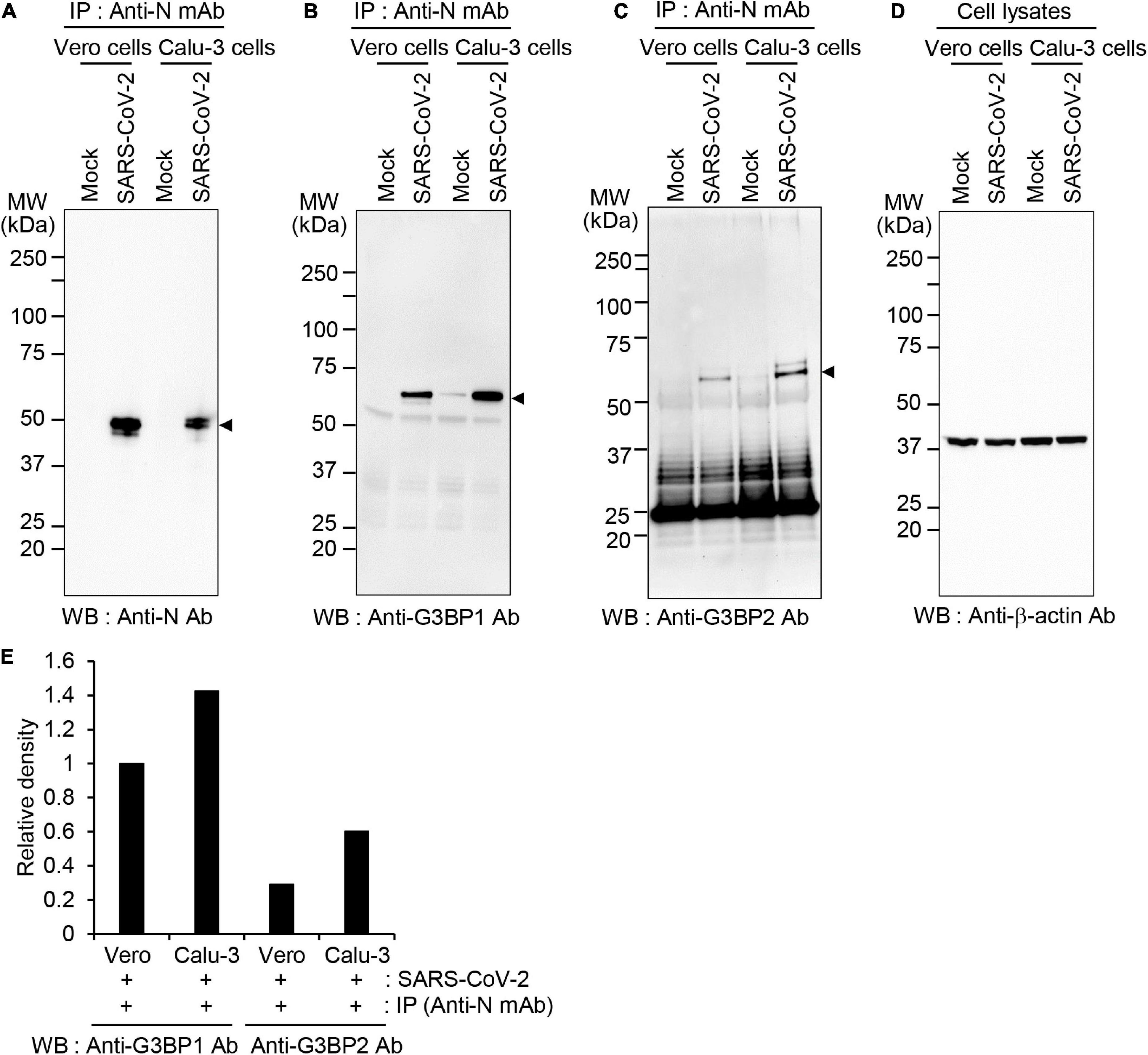
Figure 2. Differential association between SARS-CoV-2 N protein and G3BP1/2 in Vero and Calu-3 cells (A–D) Vero and Calu-3 cells were mock-infected or infected with SARS-CoV-2 S clade at 0.1 MOI; cell lysates were immunoprecipitated with anti-SARS-CoV-2 N mAb and analyzed by western blot with anti-SARS-CoV-2 N Ab (A), anti-G3BP1 antibody (B), anti-G3BP2 antibody (C), or anti-β-actin antibody, as the input control (D). (E) The band intensities of G3BP1 and G3BP2 were normalized with those of β-actin and the relative band intensities of Co-IPed proteins are shown on the graph. Anti-N mAb, anti-SARS-CoV-2 N mAb (clone 1G10C4 mAb); Anti-N Ab, anti-SARS-CoV-2 N polyclonal Ab.
SARS-CoV-2 infection does not affect expression of G3BP1 or G3BP2 in SARS-CoV-2-infected cells
The increased binding of SARS-CoV-2 N protein to G3BP1 and G3BP2 observed in Calu-3 compared to Vero cells might also result from differential expression of G3BP1 and G3BP2 in these cell lines upon SARS-CoV-2 infection. To investigate this possibility, Vero and Calu-3 cells were mock-infected or infected with the SARS-CoV-2 S clade strain at 0.1 MOI, and cell lysates were analyzed by western blot (Figure 3). Our results show that protein levels of G3BP1 are unaltered during the entire course of SARS-CoV-2 infection in both cell lines. However, protein level of G3BP2 in Vero cells was significantly reduced at 72 h in the mock infection. As we cultured the cells in medium containing 2% FBS for these experiments, the condition at 72 h may affect protein expression because of factor starvation and this can result in decrease of sensitive proteins. Considering that G3BP2 but not G3BP1 is regulated by ubiquitin-proteasome system (Soncini et al., 2001; Takayama et al., 2018; Anisimov et al., 2019) we speculate that G3BP2 protein was degraded at 72 h in mock-infected Vero cells. Calu-3 cells grow much slowly than Vero cells, therefore it is natural that expression of G3BP2 proteins was not changed during the entire culture period. Importantly, expression of G3BP1/2 was not drastically induced upon virus infection in Calu-3 cells. Thus, taken together, our data indicate that the SARS-CoV-2 N protein binds both G3BP1 and G3BP2 to a higher degree in Calu-3 cells than in Vero cells.
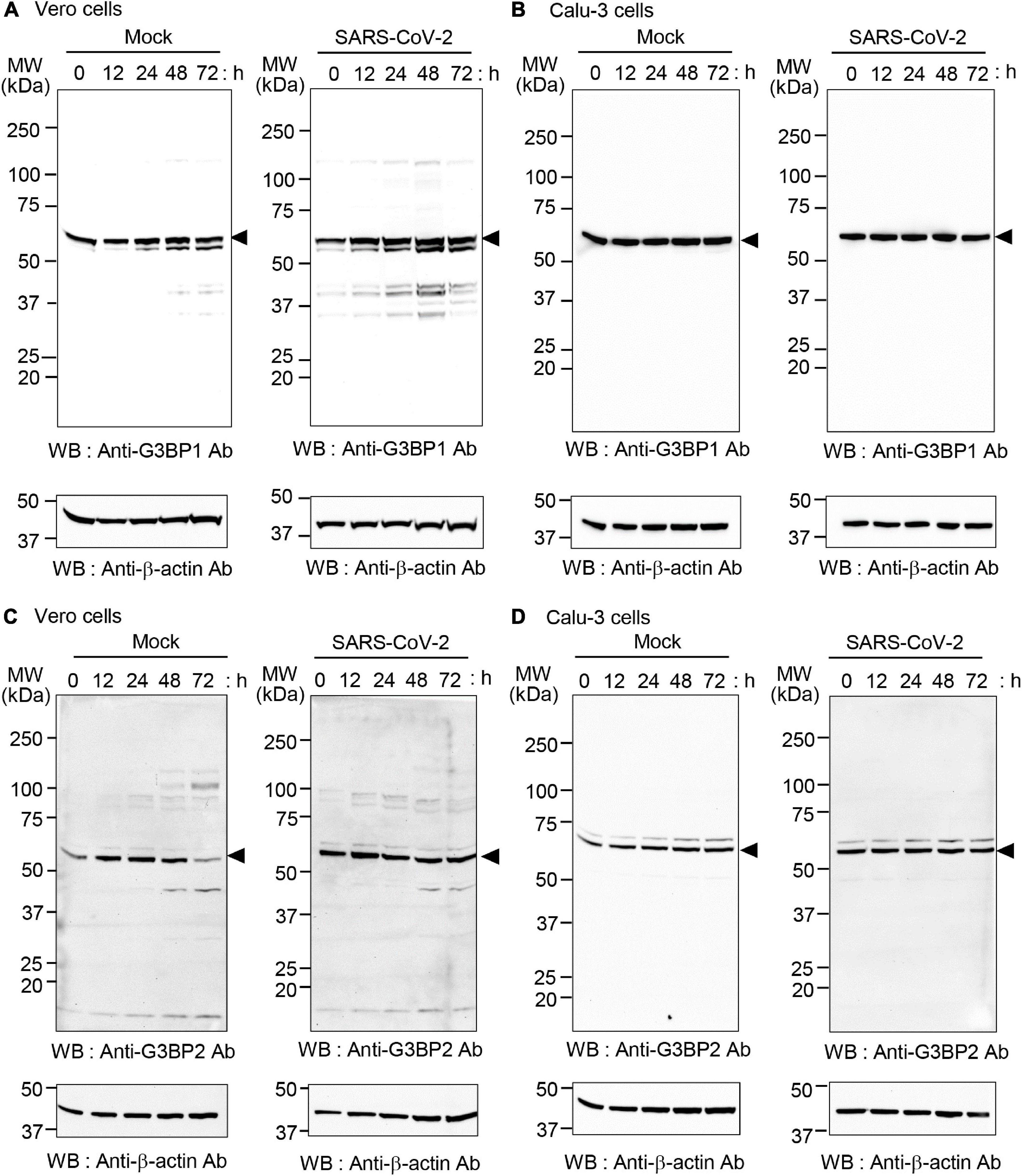
Figure 3. Expression of G3BP1 and G3BP2 in SARS-CoV-2-infected Vero and Calu-3 cells. Vero cells (A,C) and Calu-3 cells (B,D) were mock-infected (left) or infected with SARS-CoV-2 S clade at 0.1 MOI (right) for the indicated times. Cell lysates were prepared and analyzed by western blot with anti-G3BP1 antibody (A,B) or anti-G3BP2 antibody (C,D). In all cases, anti-β-actin was used as the input control (shown at bottom in all panels).
Effects of SARS-CoV-2 infection on stress granule formation
Numerous viruses, including SARS-CoV-2, are known to induce formation of stress granules within infected cells. Notably, both G3BP1 and G3BP2 are required for initiation of stress granule formation, and this process can be inhibited by the binding of viral protein to G3BP1 and G3BP2 (Matsuki et al., 2013). This suggests that SARS-CoV-2 may similarly block stress granule formation via G3BP1/G3PB2 binding. To test this hypothesis and investigate the effect of SARS-CoV-2 infection on stress granule formation, Vero and Calu-3 cells were mock-infected or infected with SARS-CoV-2 S clade stain, and stress granules were visualized by immunostaining and confocal microscopy. Sodium arsenite, a common inducer of oxidative stress that promotes stress granule formation, was used as a positive control for stress granule formation (Figure 4). We found that infection with SARS-CoV-2 S clade induces the formation of G3BP1-positive stress granules in Vero cells, but not in Calu-3 cells (Figures 4A–D). Particularly, the pattern of N protein expression and stress granule formation indicate that stress granule formation occurs in SARS-CoV-2-infected Vero cells expressing the N protein (Figure 4E, left). Conversely, we detected no stress granule formation in Calu-3 cells, irrespective of N protein expression, suggesting that the robust interaction between N and G3BP proteins effectively suppresses stress granule formation (Figure 4F, left).
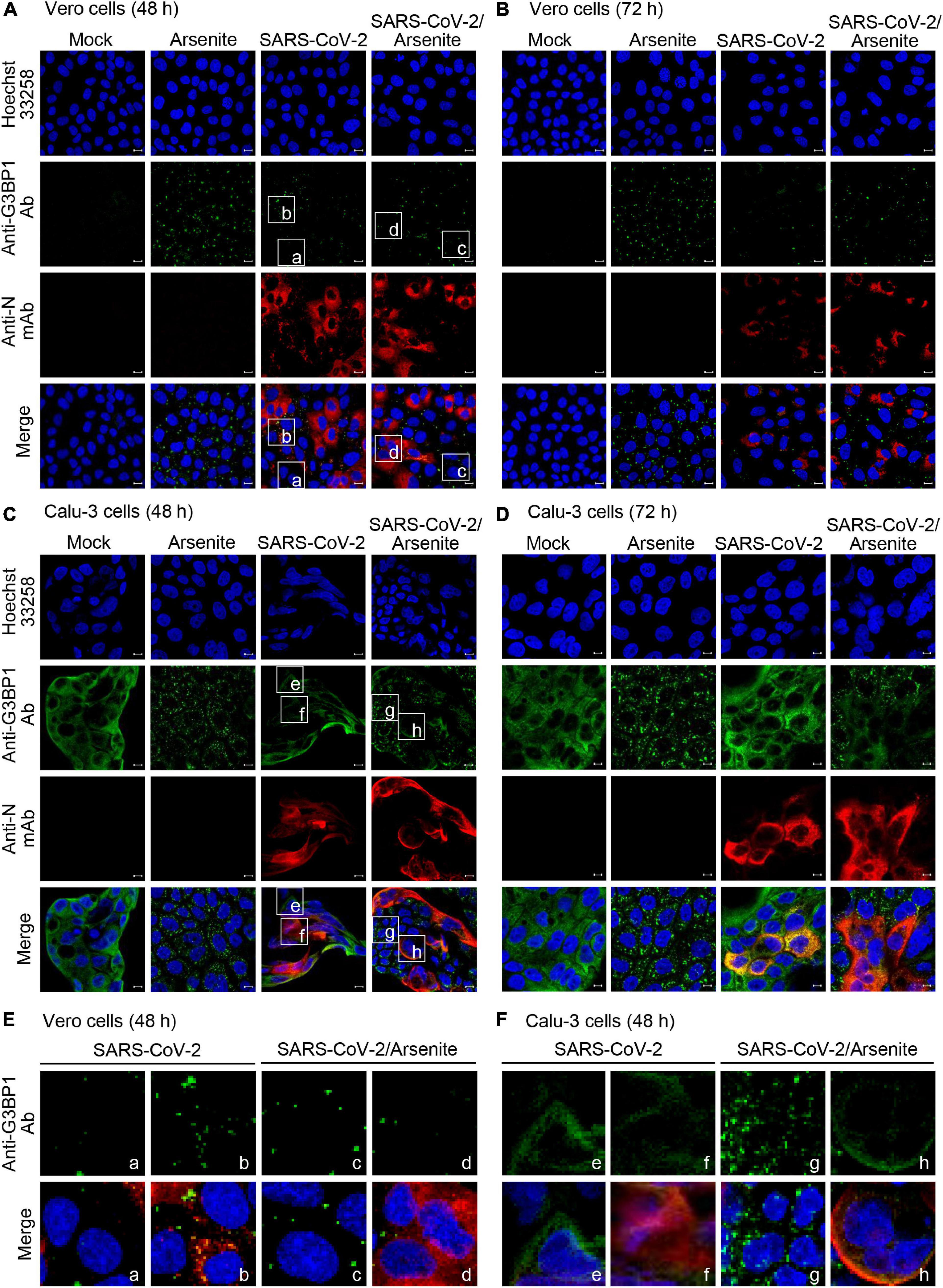
Figure 4. Stress granule production in SARS-CoV-2-infected cells. Vero (A,B) and Calu-3 cells (C,D) were mock-infected or infected with SARS-CoV-2 S clade at 0.1 MOI and examined by immunofluorescence confocal microscopy at 48 h (A,C) and 72 h (B,D) post-infection. Cells were treated with 0.5 mM sodium arsenite or phosphate-buffered saline (PBS) control for 1 h, prior to fixing and staining with anti-GRBP1 antibody (green) and anti-SARS-CoV-2 N mAb (red). (E,F) Enlarged images. The boxed regions in A and C were magnified to show the relationship between N protein expression and stress granule formation. (E) Vero cells: cells not expressing N protein (a, c) were compared with N protein expressing cells (b, d). (F) Calu-3 cells: cells not expressing N protein (e, g) were compared with N protein expressing cells (f, h). Anti-N mAb, anti-SARS-CoV-2 N mAb (clone 1G10C4 mAb); Anti-N Ab, anti-SARS-CoV-2 N polyclonal Ab. Nuclei were stained with Hoechst 33258 (blue). Anti-N Ab, anti-SARS-CoV-2 N mAb (clone 1G10C4 mAb).
As expected, we further found that treatment with sodium arsenite induces significant and extensive formation of G3BP1-positive stress granules in uninfected Vero and Calu-3 cells (Figures 4A–D). Intriguingly, however, this sodium arsenite-induced stress granule formation is suppressed by SARS-CoV-2 S clade infection in both Vero and Calu-3 cells, indicating that virus infection can inhibit the formation of stress granules in response to other stressors (Figures 4A–D). Enlarged images clearly show suppression of arsenite-induced stress granule formation in N protein-positive cells (Figures 4E,F, right). These data indicate that SARS-CoV-2 can differentially regulate stress granule formation induced by viral infection and sodium arsenite in Calu-3 cells and Vero cells.
Effects of SARS-CoV-2 variants on stress granule formation
Infectivity and propagation rates of different SARS-CoV2 strains vary greatly, likely contributing to the differential severity of various waves observed during the COVID-19 pandemic. Similarly, inhibition of the stress granule formation might also vary in different SARS-CoV-2 strains and affect viral pathophysiology in a strain-specific manner. We therefore measured the effect of infection with other SARS-CoV-2 variants on stress granule formation. We found that infection with SARS-CoV-2 alpha, delta, or Mu variants induces stress granule formation in Vero cells but not in Calu-3 cells (Figures 5A,B; left panel). Moreover, SARS-CoV-2 alpha, delta, and Mu variants inhibit sodium arsenite-induced stress granule formation similar to SARS-CoV-2 S clade in both Vero cells and Calu-3 cells (Figures 5A,B; right panel). Taken together, these results suggest that stress granule formation in response to SARS-CoV-2 infection might be guided by the host cell intrinsic factors rather than those of viral proteins.
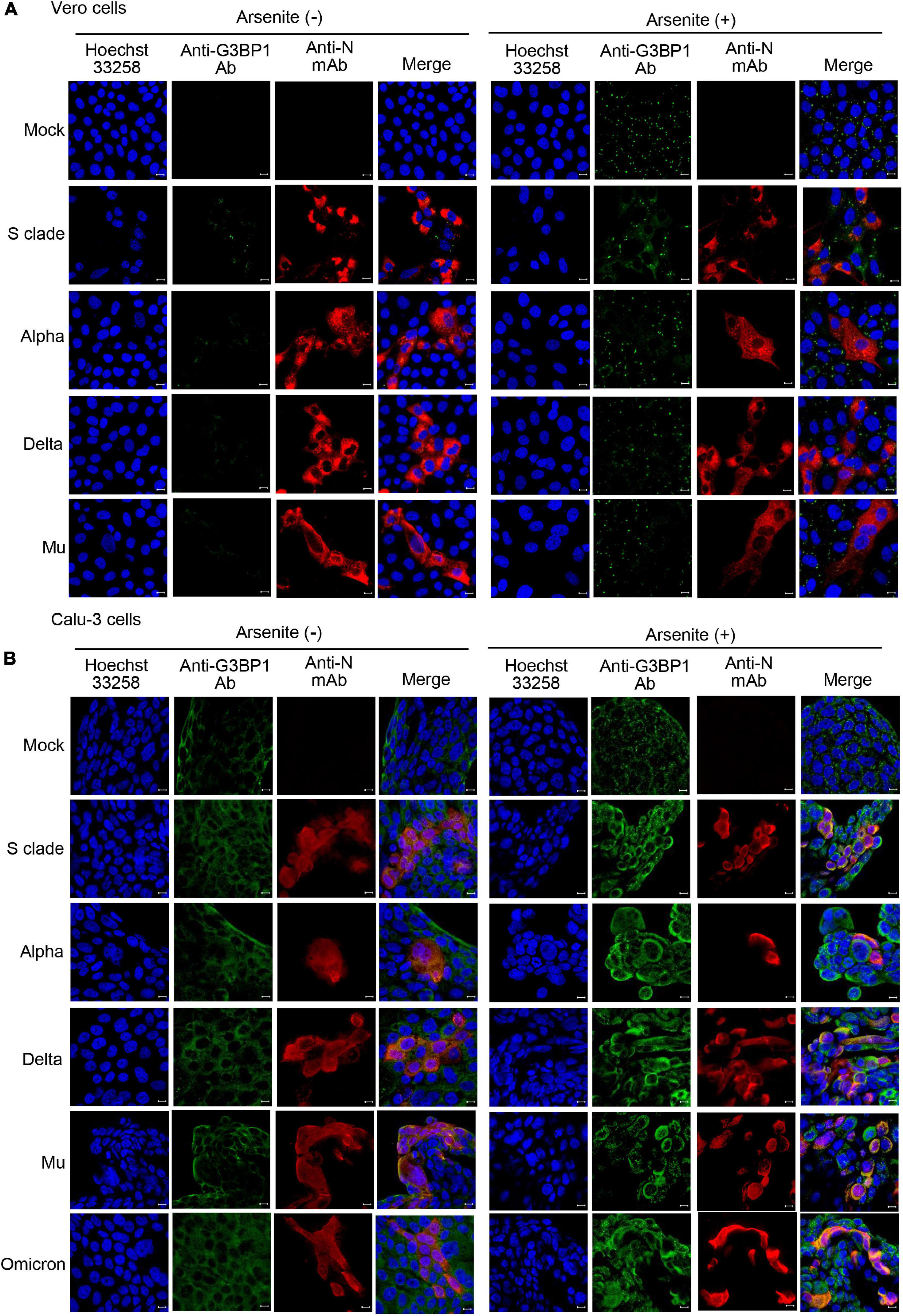
Figure 5. Detection of stress granules in cells infected with different SARS-CoV-2 variants. Vero cells (A) and Calu-3 cells (B) were mock-infected or infected with SARS-CoV-2 variants at 0.1 MOI for 71 h and analyzed by immunofluorescence confocal microscopy. Cells were treated with PBS or 0.5 mM sodium arsenite for 1 h, prior to fixing and staining with anti-GRBP1 antibody (green) and anti-SARS-CoV-2 N mAb (red). Nuclei were stained Hoechst 33258 (blue). Anti-N Ab, anti-SARS-CoV-2 N mAb (clone 1G10C4 mAb).
Discussion
The ongoing COVID-19 pandemic has caused significant mortality and financial adversity worldwide, highlighting the urgent need for an improved understanding of the mechanisms that drive the SARS-CoV-2 life cycle, including those involved in infection, replication, and transmission. Numerous viruses have been shown to withstand stress granule formation, often by inhibiting their formation via the targeting of stress granule-nucleating proteins, particularly G3BP1 (Kang et al., 2021). SARS-CoV-2 appears to be one such virus, with recent studies reporting that the SARS-CoV-2 N protein targets both G3BP1 and G3BP2, thereby attenuating stress granule formation and promoting viral replication (Zheng et al., 2021; Nabeel-Shah et al., 2022). However, it is not known whether there are differential effects of SARS-CoV-2 infection on stress granule regulation in different cell types.
In this study, we addressed this question by using our previously developed anti-SARS-CoV-2 N antibody to measure the binding of SARS-CoV-2 N protein to cellular G3BP1 in two different cell lines (i.e., Vero and Calu-3 cells). Notably, we found that SARS-CoV-2 N protein interacts with both G3BP1 and G3BP2 proteins in Calu-3 and Vero cells, although this binding is present to a greater extent in Calu-3 cells relative to Vero cells. Further analysis revealed that levels of G3BP1 and G3BP2 do not change during SARS-CoV-2 infection in both cell lines, confirming that the observed differences are due to more robust binding between SARS-CoV-2 N and G3BP1/2 in Calu-3 cells and not differential expression of these proteins in different cells types. Consistent with these observations, Calu-3 cells infected with SARS-CoV-2 do not display stress granule formation, whereas Vero cells show a moderate degree of stress granules upon SARS-CoV-2 infection. Therefore, our data suggest that N protein binding to G3BP1 and G3BP2 with N protein negatively regulates stress granule formation.
SARS-CoV-2 variants display various degrees of transmissibility and confer differing levels of disease severity in infected patients (Harvey et al., 2021; Mendiola-Pastrana et al., 2022). Moreover, studies have shown that SARS-CoV-2 variants have different replication rates and produce distinct viral titers from infected Vero cells (Jeong et al., 2021; Prince et al., 2022). Because inhibition of stress granule formation is related to viral replication (McCormick and Khaperskyy, 2017), we hypothesized that SARS-CoV-2 variants might also show differing degrees of stress granule inhibition. We therefore measured stress granule formation in Vero and Calu-3 cells infected with SARS-CoV-2 variants and further determined how these affect sodium arsenite-induced stress granule formation. Our results revealed similar patterns of stress granule formation and regulation in cells infected with the different viral strains, suggesting that extent of stress granule inhibition is not always reflective of virus production efficiency. In addition, although stress granule formation in response to SARS-CoV-2 infection is more prominent in Vero cells, with weaker binding between N protein and G3BP1/2, viral production is also higher in these cells relative to Calu-3 cells. Contrary to these findings, however, a previous study reported that knockdown of G3BP1 inhibits stress granule formation and enhances replication of the SARS-CoV-2 genome in Hela-ACE2 cells (Zheng et al., 2021). Collectively, these data suggest that SARS-CoV-2 production is only partly regulated by stress granule formation, and this phenomenon is largely determined by host cell intrinsic properties. However, a detailed mechanistic study using an array of different cell lines and viral variants will be required to fully elucidate the complex relationship between stress granule formation and SARS-CoV-2 virus replication.
In summary, utilizing two susceptible cell lines and various natural SARS-CoV-2 variants, we found that inhibition of virus-induced stress granule formation correlates with binding between N protein and G3BP1/2; however, it does not always reflect the degree of virus production or ability to inhibit stress granules induced by sodium arsenite treatment. These findings highlight a clear gap in knowledge relating to the role of stress granules in SARS-CoV-2 infection and suggest that more studies are needed to determine whether they may represent a viable target for COVID-19 treatment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: ProteomeXchange, PXD035715.
Author contributions
H-JK and YL conceived of the project. H-JK, JGS, and YL designed the experiments. H-JK, SM, and YL wrote the manuscript. DK, SM, MKa, JK, SP, MKi, KB, and SK carried out the experiments. H-JK, DK, JGS, and YL analyzed the data. All authors approved the final version of the manuscript.
Funding
This research was supported by grants from the National Research Foundation (NRF-2020M3A9I2107294 and NRF-2022M3A9I2082292) funded by the Ministry of Science and ICT in the South Korea.
Acknowledgments
We thank the National Culture Collection for Pathogens (Osong, South Korea) for supplying SARS-CoV-2 viruses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Amorim, R., Temzi, A., Griffin, B. D., and Mouland, A. J. (2017). Zika virus inhibits eIF2α-dependent stress granule assembly. PLoS Negl. Trop. Dis. 11:e0005775. doi: 10.1371/journal.pntd.0005775
Anisimov, S., Takahashi, M., Kakihana, T., Katsuragi, Y., Kitaura, H., Zhang, L., et al. (2019). G3BP1 inhibits ubiquitinated protein aggregations induced by p62 and USP10. Sci. Rep. 9:12896. doi: 10.1038/s41598-019-46237-1
Basu, M., Courtney, S. C., and Brinton, M. A. (2017). Arsenite-induced stress granule formation is inhibited by elevated levels of reduced glutathione in West Nile virus-infected cells. PLoS Pathog. 13:e1006240. doi: 10.1371/journal.ppat.1006240
Blake, S., Shaabani, N., Eubanks, L. M., Maruyama, J., Manning, J. T., Beutler, N., et al. (2021). Salicylanilides Reduce SARS-CoV-2 Replication and Suppress Induction of Inflammatory Cytokines in a Rodent Model. ACS Infect. Dis. 7, 2229–2237. doi: 10.1021/acsinfecdis.1c00253
Brownsword, M. J., Doyle, N., Brocard, M., Locker, N., and Maier, H. J. (2020). Infectious bronchitis virus regulates cellular stress granule signaling. Viruses 12:536. doi: 10.3390/v12050536
Campos-Melo, D., Hawley, Z., Droppelmann, C. A., and Strong, M. J. (2021). The integral role of RNA in stress granule formation and function. Front. Cell Devel. Biol. 9:621779. doi: 10.3389/fcell.2021.621779
Cesaro, T., and Michiels, T. (2021). Inhibition of PKR by Viruses. Front. Microbiol. 12:757238. doi: 10.3389/fmicb.2021.757238
Chen, Y., Liu, Q., and Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 92, 418–423. doi: 10.1002/jmv.25681
Gilks, N., Kedersha, N., Ayodele, M., Shen, L., Stoecklin, G., Dember, L. M., et al. (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398. doi: 10.1091/mbc.e04-08-0715
Harvey, W. T., Carabelli, A. M., Jackson, B., Gupta, R. K., Thomson, E. C., Harrison, E. M., et al. (2021). SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424. doi: 10.1038/s41579-021-00573-0
Hou, S., Kumar, A., Xu, Z., Airo, A. M., Stryapunina, I., Wong, C. P., et al. (2017). Zika Virus Hijacks Stress Granule Proteins and Modulates the Host Stress Response. J. Virol. 91:e00474-17. doi: 10.1128/JVI.00474-17
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi: 10.1016/S0140-6736(20)30183-5
Ivanov, P., Kedersha, N., and Anderson, P. (2019). Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol. 11:a032813. doi: 10.1101/cshperspect.a032813
Jeong, G. U., Yoon, G. Y., Moon, H. W., Lee, W., Hwang, I., Kim, H., et al. (2021). Comparison of plaque size, thermal stability, and replication rate among SARS-CoV-2 variants of concern. Viruses 14:55. doi: 10.3390/v14010055
Kang, W., Wang, Y., Yang, W., Zhang, J., Zheng, H., and Li, D. (2021). Research Progress on the Structure and Function of G3BP. Front. Immunol. 12:718548. doi: 10.3389/fimmu.2021.718548
Kim, D., Kim, J., Park, S., Kim, M., Baek, K., Kang, M., et al. (2021a). Production of SARS-CoV-2 N protein-specific monoclonal antibody and its application in an ELISA-based detection system and targeting the interaction between the spike C-Terminal domain and N Protein. Front. Microbiol. 12:726231. doi: 10.3389/fmicb.2021.726231
Kim, D., Maharjan, S., Kim, J., Park, S., Park, J. A., Park, B. K., et al. (2021b). MUC1-C influences cell survival in lung adenocarcinoma Calu-3 cells after SARS-CoV-2 infection. BMB Rep. 54, 425–430. doi: 10.5483/BMBRep.2021.54.8.018
Kim, J., Hwang, S. Y., Kim, D., Kim, M., Baek, K., Kang, M., et al. (2022a). Abiraterone Acetate Attenuates SARS-CoV-2 Replication by Interfering with the Structural Nucleocapsid Protein. Biomol. Ther. [Epub ahead of print]. doi: 10.4062/biomolther.2022.037
Kim, J., Kim, M., Kim, D., Park, S., Kang, M., Baek, K., et al. (2022b). Targeting the interaction between spike protein and nucleocapsid protein for suppression and detection of human coronavirus OC43. Front. Immunol. 13:835333. doi: 10.3389/fimmu.2022.835333
Linero, F. N., Thomas, M. G., Boccaccio, G. L., and Scolaro, L. A. (2011). Junin virus infection impairs stress-granule formation in Vero cells treated with arsenite via inhibition of eIF2α phosphorylation. J. Gen. Virol. 92, 2889–2899. doi: 10.1099/vir.0.033407-0
Liu, Y., Wang, M., Cheng, A., Yang, Q., Wu, Y., Jia, R., et al. (2020). The role of host eIF2α in viral infection. Virol. J. 17:112. doi: 10.1186/s12985-020-01362-6
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/S0140-6736(20)30251-8
Mahboubi, H., and Stochaj, U. (2017). Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 884–895. doi: 10.1016/j.bbadis.2016.12.022
Matsuki, H., Takahashi, M., Higuchi, M., Makokha, G. N., Oie, M., and Fujii, M. (2013). Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18, 135–146. doi: 10.1111/gtc.12023
McCormick, C., and Khaperskyy, D. A. (2017). Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 17, 647–660. doi: 10.1038/nri.2017.63
Mendiola-Pastrana, I. R., López-Ortiz, E., Río de la Loza-Zamora, J. G., González, J., Gómez-García, A., and López-Ortiz, G. (2022). SARS-CoV-2 variants and clinical outcomes: a systematic review. Life 12:170. doi: 10.3390/life12020170
Nabeel-Shah, S., Lee, H., Ahmed, N., Burke, G. L., Farhangmehr, S., Ashraf, K., et al. (2022). SARS-CoV-2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience 25:103562. doi: 10.1016/j.isci.2021.103562
Nakagawa, K., Narayanan, K., Wada, M., and Makino, S. (2018). Inhibition of Stress Granule Formation by Middle East Respiratory Syndrome Coronavirus 4a Accessory Protein Facilitates Viral Translation, Leading to Efficient Virus Replication. J. Virol. 92:e00902-18. doi: 10.1128/JVI.00902-18
Ohn, T., Kedersha, N., Hickman, T., Tisdale, S., and Anderson, P. (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231. doi: 10.1038/ncb1783
Onomoto, K., Jogi, M., Yoo, J. S., Narita, R., Morimoto, S., Takemura, A., et al. (2012). Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7:e43031. doi: 10.1371/journal.pone.0043031
Park, B. K., Kim, D., Park, S., Maharjan, S., Kim, J., Choi, J. K., et al. (2021a). Differential signaling and virus production in Calu-3 Cells and Vero Cells upon SARS-CoV-2 Infection. Biomol. Ther. 29, 273–281. doi: 10.4062/biomolther.2020.226
Park, B. K., Kim, J., Park, S., Kim, D., Kim, M., Baek, K., et al. (2021b). MERS-CoV and SARS-CoV-2 replication can be inhibited by targeting the interaction between the viral spike protein and the nucleocapsid protein. Theranostics 11, 3853–3867. doi: 10.7150/thno.55647
Perez-Riverol, Y., Bai, J., Bandla, C., García-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038
Prince, T., Dong, X., Penrice-Randal, R., Randle, N., Hartley, C., Goldswain, H., et al. (2022). Analysis of SARS-CoV-2 in nasopharyngeal samples from patients with COVID-19 illustrates population variation and diverse phenotypes, placing the growth properties of variants of concern in context with other lineages. mSphere 7:e0091321. doi: 10.1128/msphere.00913-21
Protter, D., and Parker, R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. doi: 10.1016/j.tcb.2016.05.004
Rabouw, H. H., Langereis, M. A., Knaap, R. C., Dalebout, T. J., Canton, J., Sola, I., et al. (2016). Middle east respiratory coronavirus accessory protein 4a inhibits PKR-Mediated antiviral stress responses. PLoS Pathog. 12:e1005982. doi: 10.1371/journal.ppat.1005982
Soncini, C., Berdo, I., and Draetta, G. (2001). Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20, 3869–3879. doi: 10.1038/sj.onc.1204553
Stevens, L. J., Pruijssers, A. J., Lee, H. W., Gordon, C. J., Tchesnokov, E. P., Gribble, J., et al. (2022). Mutations in the SARS-CoV-2 RNA dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci. Transl. Med. [Epub ahead of print]. doi: 10.1126/scitranslmed.abo0718
Sumbria, D., Berber, E., Mathayan, M., and Rouse, B. T. (2021). Virus Infections and Host Metabolism-Can We Manage the Interactions? Front. Immunol. 11:594963. doi: 10.3389/fimmu.2020.594963
Szemiel, A. M., Merits, A., Orton, R. J., MacLean, O. A., Pinto, R. M., Wickenhagen, A., et al. (2021). In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 17:e1009929. doi: 10.1371/journal.ppat.1009929
Takayama, K. I., Suzuki, T., Fujimura, T., Takahashi, S., and Inoue, S. (2018). Association of USP10 with G3BP2 Inhibits p53 signaling and contributes to poor outcome in prostate cancer. Mol. Cancer Res. 16, 846–856. doi: 10.1158/1541-7786.MCR-17-0471
Toroney, R., Nallagatla, S. R., Boyer, J. A., Cameron, C. E., and Bevilacqua, P. C. (2010). Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A. J. Mol. Biol. 400, 393–412. doi: 10.1016/j.jmb.2010.04.059
Tu, Y. C., Yu, C. Y., Liang, J. J., Lin, E., Liao, C. L., and Lin, Y. L. (2012). Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J. Virol. 86, 10347–10358. doi: 10.1128/JVI.00525-12
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., and Thiel, V. (2021). Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170. doi: 10.1038/s41579-020-00468-6
White, J. P., and Lloyd, R. E. (2012). Regulation of stress granules in virus systems. Trends Microbiol. 20, 175–183. doi: 10.1016/j.tim.2012.02.001
Wu, A., Peng, Y., Huang, B., Ding, X., Wang, X., Niu, P., et al. (2020). Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 27, 325–328. doi: 10.1016/j.chom.2020.02.001
Ye, X., Pan, T., Wang, D., Fang, L., Ma, J., Zhu, X., et al. (2018). Foot-and-mouth disease virus counteracts on internal ribosome entry site suppression by G3BP1 and Inhibits G3BP1-mediated stress granule assembly via post-translational mechanisms. Front. Immunol. 9:1142. doi: 10.3389/fimmu.2018.01142
Yu, J., Qin, Z., Liu, X., He, X., Yao, J., Zhou, X., et al. (2022). High-specificity targets in SARS-CoV-2 N protein for serological detection and distinction from SARS-CoV. Comput. Biol. Med. 143:105272. doi: 10.1016/j.compbiomed.2022.105272
Zhang, Q., Xiang, R., Huo, S., Zhou, Y., Jiang, S., Wang, Q., et al. (2021). Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 6:233. doi: 10.1038/s41392-021-00653-w
Zhang, Y., Yao, L., Xu, X., Han, H., Li, P., Zou, D., et al. (2018). Enterovirus 71 inhibits cytoplasmic stress granule formation during the late stage of infection. Virus Res. 255, 55–67. doi: 10.1016/j.virusres.2018.07.006
Zheng, Z. Q., Wang, S. Y., Xu, Z. S., Fu, Y. Z., and Wang, Y. Y. (2021). SARS-CoV-2 nucleocapsid protein impairs stress granule formation to promote viral replication. Cell Discov. 7:38. doi: 10.1038/s41421-021-00275-0
Keywords: SARS-CoV-2, COVID-19, sodium arsenite, G3BP1/2, N protein, stress granule
Citation: Kim D, Maharjan S, Kang M, Kim J, Park S, Kim M, Baek K, Kim S, Suh JG, Lee Y and Kwon H-J (2022) Differential effect of SARS-CoV-2 infection on stress granule formation in Vero and Calu-3 cells. Front. Microbiol. 13:997539. doi: 10.3389/fmicb.2022.997539
Received: 19 July 2022; Accepted: 04 August 2022;
Published: 23 August 2022.
Edited by:
Xin Yin, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Longhuan Ma, University of Georgia, United StatesJunhua Huang, Wuhan Polytechnic University, China
Copyright © 2022 Kim, Maharjan, Kang, Kim, Park, Kim, Baek, Kim, Suh, Lee and Kwon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Younghee Lee, eWhsNDE3N0BjYm51LmFjLmty; Hyung-Joo Kwon, aGpvb2t3b25AaGFsbHltLmFjLmty
†These authors have contributed equally to this work
 Dongbum Kim
Dongbum Kim Sony Maharjan
Sony Maharjan Mijeong Kang
Mijeong Kang Jinsoo Kim
Jinsoo Kim Sangkyu Park
Sangkyu Park Minyoung Kim
Minyoung Kim Kyeongbin Baek
Kyeongbin Baek Suyeon Kim2
Suyeon Kim2 Jun Gyo Suh
Jun Gyo Suh Younghee Lee
Younghee Lee Hyung-Joo Kwon
Hyung-Joo Kwon