- 1College of Life Sciences, Anhui Provincial Key Laboratory of Molecular Enzymology and Mechanism of Major Diseases, Anhui Normal University, Wuhu, Anhui, China
- 2Wuhu Qingyijiang Seed Industry Co., Ltd., Wuhu, Anhui, China
Soft rot caused by Dickeya zeae is an important bacterial disease affecting rice and other plants worldwide. In this study, Nanopore and Illumina sequencing platforms were used to sequence the high-quality complete genome of a novel D. zeae strain WH1 (size: 4.68 Mb; depth: 322.37x for Nanopore, 243.51x for Illumina; GC content: 53.59%), which was isolated from healthy rice root surface together with Paenibacillus polymyxa, a potential biocontrol bacterium against D. zeae strain WH1. However, the pure WH1 culture presented severe pathogenicity. Multilocus sequence analysis (MLSA) indicated that strains WH1, EC1, and EC2 isolated from rice were grouped into a clade differentiated from other D. zeae strains. The average nucleotide identity (ANI) and DNA-DNA hybridization (DDH) analyses demonstrated that WH1 was phylogenetically closest to EC2. Furthermore, the pathogenicity determinants and virulence factors of WH1 were mainly analyzed through genomic comparison with complete genomes of other D. zeae strains with high virulence (EC1, EC2, MS1, and MS2). The results revealed that plant cell wall-degrading extracellular enzymes (PCWDEs), flagellar and chemotaxis, and quorum sensing were highly conserved in all analyzed genomes, which were confirmed through phenotypic assays. Besides, WH1 harbored type I, II, III, and VI secretion systems (T1SS, T2SS, T3SS, and T6SS), but lost T4SS and T5SS. Like strains MS1 and MS2 isolated from bananas, WH1 harbored genes encoding both capsule polysaccharide (CPS) and exopolysaccharide (EPS) biosynthesis. The results of pathogenicity assays demonstrated that WH1 produced severe soft rot symptoms on potato tubers, carrots, radishes, and Chinese cabbage. Meanwhile, WH1 also produced phytotoxin(s) to inhibit rice seed germination with an 87% inhibitory rate in laboratory conditions. More importantly, we confirmed that phytotoxin(s) produced by WH1 are different from zeamines produced by EC1. Comparative genomics analyses and phenotypic and pathogenicity assays suggested that WH1 likely evolved through a pathway different from the other D. zeae strains from rice, producing a new type of rice foot rot pathogen. These findings highlight the emergence of a new type of D. zeae strain with high virulence, causing soft rot in rice and other plants.
Introduction
Dickeya spp. are Gram-negative and causative agents of a wide range of bacterial soft rot and have been listed among the top 10 most important bacterial phytopathogens due to the high costs and economic consequences they incur (Mansfield et al., 2012). Currently, the Dickeya genus encompasses 12 recognized species (Boluk et al., 2021). Of which, D. zeae strains were isolated from a wide and diverse range of hosts, causing serious economic losses in crop yield in different parts of the world, especially for rice, potato, pineapple, and banana (Hu et al., 2018; Feng et al., 2019; Boluk et al., 2021). Infected rice plants by D. zeae have a dark brown decay of the tillers at the site of infection with a bad smell, which later leads to the collapse of the entire plant (Hussain et al., 2008). In addition, D. zeae strains can reside on the surface and/or within the intercellular spaces of plant organs without exhibiting symptoms (Pérombelon, 1992). The reason these bacteria can grow on some plants without symptoms but cause devastating diseases in others remains unclear (De Boer, 2002; Van Gijsegem et al., 2021). However, when in a susceptible host, these strains may persist for months until the environmental conditions are not favorable to disease initiation (De Boer, 2002; Kraepiel and Barny, 2016). Additionally, they can persist overwinter in contaminated plant residue and soil. Under certain conditions, such as decreasing O2 concentration, high temperature, high humidity, and there being water film on the surface of the plant organs, latency is broken and the bacteria start to grow and produce plant cell wall degrading enzymes (PCWDEs) leading to disease development in susceptible hosts (Bez et al., 2021).
Generally, D. zeae strains infect plants through three main steps: (1) adhesion to plant surface and penetration in the plant tissues, either via wound sites or through natural openings such as stomata; (2) invasion of apoplasts; and (3) plant cell wall degradation (Reverchon and Nasser, 2013). Some typical virulence factors are involved in these processes, such as type I–VI secretion systems (T1SS–T6SS), PCWDEs, flagella, and flagella-mediated motility, quorum sensing signal molecules acylhomoserine lactone (AHL) and Vfm, and biofilms (Zhang et al., 2022). A previous study showed that the zeamines produced by D. zeae EC1 were important virulence factors for rice, but antagonists for many bacteria and fungi (Liao et al., 2014). However, the zms gene cluster encoding zeamines biosynthesis was absent in the genome of D. zeae strain EC2 (Zhang et al., 2020). Additionally, other virulence determinants are present among D. zeae strains, such as T3SS, which is present in strain EC1 but absent in strain EC2 (Zhou et al., 2015; Zhang et al., 2020). Therefore, the diversity involved in virulence factors would be present in the genomes of D. zeae strains, even though they were isolated from the same host.
In this study we obtained a complete genome sequence of D. zeae strain WH1, isolated from the rice root surface, using Illumina next-generation sequencing and Nanopore sequencing technologies. The genome sequence was annotated and compared with the representative genome sequences of other D. zeae strains (EC1, EC2, MS1, and MS2), focusing on virulence determinants and potential regulatory mechanisms. Substantial phenotype and pathogenicity assays were performed for strain WH1. The genome-wide comparison and phenotypic and pathogenicity assays could help to clarify the differentiation among D. zeae strains from rice and their genetic adaptions to the host. This information directs attention to this newly emerging rice pathogen to provide better insights into the early prevention of rice foot rot disease.
Materials and methods
Strain WH1 isolation and characterization
Three healthy rice roots were collected from the field of Nanling, Wuhu City, Anhui Province, China, in August 2021. The rice roots with few adherent soils were cut into pieces and placed into 10 mL of sterile water. The mixtures were incubated at 25°C for 30 min with shaking at 150 rpm. The mixture at different dilutions with sterile water was spread onto LB agar plates and incubated at 25°C for 48 h. Single colonies were picked and sub-cultured three times. Pure cultures were used to screen antagonists against rice pathogenic fungi. One strain, WH1, had the strong ability to inhibit the growth of pathogenic fungi (data not shown). The strain was identified as Dickeya genus through sequencing of the 16S rRNA gene using 27F/1492R primers. Subsequently, the strain was further confirmed through Koch’s postulates. Additionally, the screening of antagonistic bacteria against WH1 was performed according to the methods previously described (Li et al., 2020). The screened antagonistic bacteria were identified through sequencing of the 16S rRNA gene. All isolated bacteria were stored in glycerol at –80°C at Anhui Normal University, China. The strain WH1 was mainly investigated in this study. For subculture, the strain was grown at 28°C overnight in LB broth (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl, pH7.0) in an incubator shaker at 150 rpm.
Genomic DNA extraction and genome sequencing
The strain WH1 was incubated to logarithmic phase at 28°C and its genomic DNA was extracted using the TIANamp Bacterial DNA kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. The quality of the obtained genomic DNA was detected with agarose gel electrophoresis, NanoDrop One Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and Qubit 3.0 Fluorimeter (Life Technologies, Carlsbad, CA, USA), respectively. The DNA libraries for MinION Nanopore sequencing and Illumina sequencing were constructed, respectively. DNA sequencing was respectively carried out on the PromethION sequencer (Oxford Nanopore Technologies, Oxford, United Kingdom) and NovaSeq 6000 (Illumina, USA) platforms at BENAGEN Technologies, Wuhan, China.
Genome assembly and annotation
Unicycler (version 0.5.0) software was used to assemble short reads from the Illumina platform and long reads from the MinION Nanopore platform (Wick et al., 2017). Gene prediction was performed using Prokka (version 1.14.6) software (Seemann, 2014); Aragorn was used to predict the tRNA of the genome (Laslett and Canback, 2004); rRNAmmer was used to predict genomic rRNA sequences (Lagesen et al., 2007). Gene function annotation was performed using the local BLAST method to compare the predicted gene sequences with GO (Ashburner et al., 2000), KEGG (Kanehisa et al., 2004), COG (Tatusov et al., 2000), UniProt (Apweiler et al., 2004), and NR databases, respectively, to obtain the corresponding annotation information. The genomic sequence was submitted to the NCBI GenBank Genome database under the accession number CP101400.
Phylogenetic analysis of Dickeya strains
Sixteen Dickeya strains with complete genome sequences were used to extract the gene sequences of aptD, gyrB, infB, and rpoB for multilocus sequence analysis (MLSA). The concatenated sequences of these four genes were processed on MEGA5.2 using a neighbor-joining algorithm with 1,000 bootstrapped replications (Tamura et al., 2011). To obtain an estimate of overall genomic similarities, a pairwise comparison of the WH1 genome with 15 genomes of other Dickeya strains was performed using the average nucleotide identity (ANI) based on the nucleotide MUMmmer algorithm (ANIm) in the Jspecies Web server (Richter et al., 2016). In addition, in silico DNA-DNA hybridization (isDDH) values were calculated using the Genome-to-Genome Distance Calculator (GGDC) with formula 2 and BLAST + alignment.1 The cut-off values of 96 and 70% were assigned as a species-delineation framework for ANI and isDDH, respectively (Goris et al., 2007).
Genomic comparison between WH1 and other Dickeya zeae strains
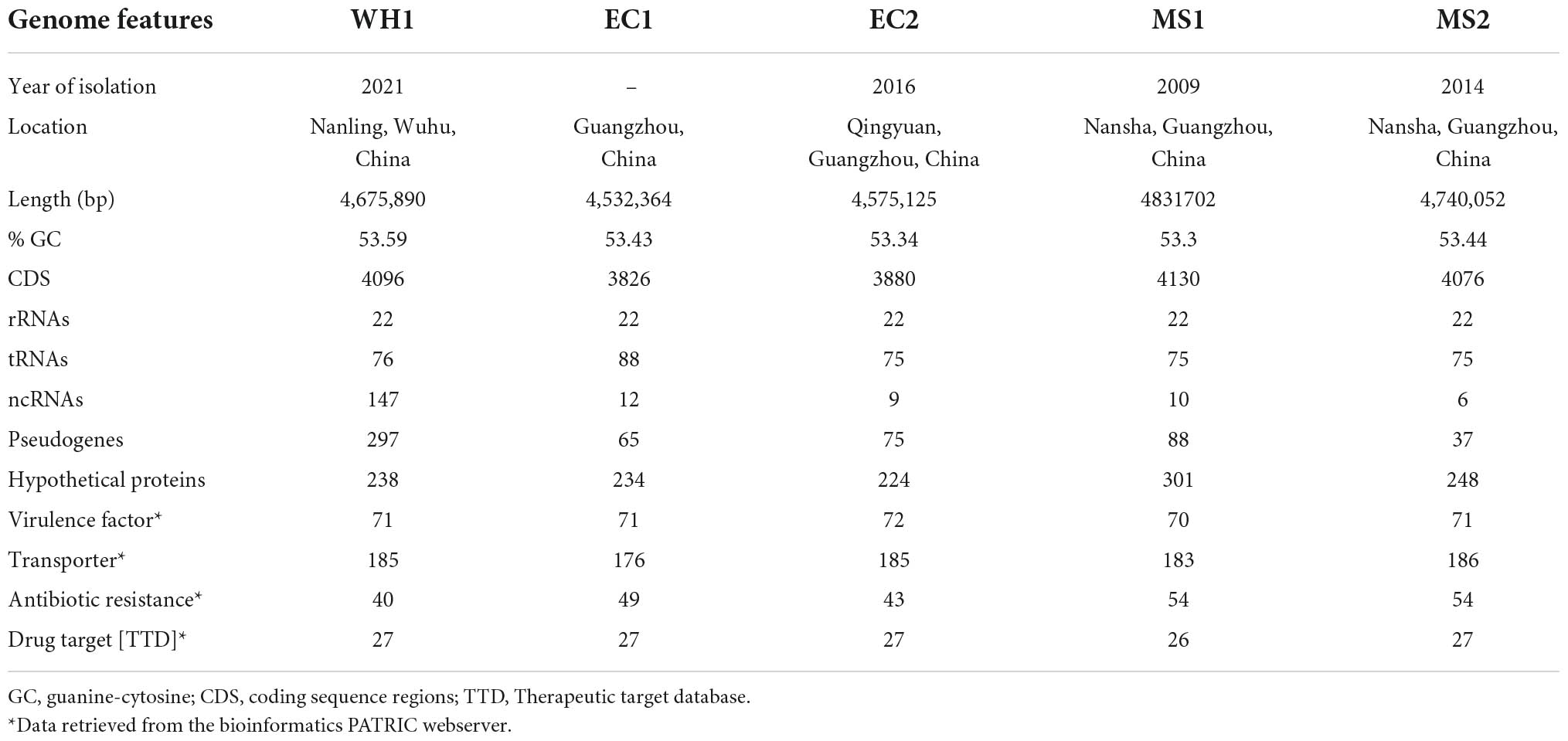
The complete genome of WH1 was compared with the other four complete genomes of D. zeae strains (EC1, EC2, MS1, and MS2). Their genome sequences were retrieved from the NCBI genome database. The basic genomic profile features of these four strains were taken from the NCBI GenBank database and the bioinformatics resource center proteome comparison tool of the pathosystems resource integration center (PATRIC) Web Server (Davis et al., 2020). To identify the lists of shared and unique gene clusters, Pan-genome analysis was performed using Pan-Genomes Analysis Web Server (Chen et al., 2018). The orthologous gene clusters were identified by the Gene Family (GF) method set to its default parameters.
The soft rot bacteria within Dickeya genus macerate the plant tissue and acquire nutrients from the dead cells. Most pathogenicity determinants related encoding genes, including PCWDEs, type secretion systems (types I–VI), extracellular polysaccharides (such as CPS and EPS), bacterial attachment operons (type IV pili), flagella and chemotaxis, zeamine synthesis, quorum sensing systems, and biofilm formation, have been described in strains EC1, EC2, MS1, or MS2 (Feng et al., 2019; Zhang et al., 2020, 2022; Liu et al., 2022). In this study, we mainly analyzed and compared the similarities, differences, or absence of gene encoding virulence determinants between WH1 and these four D. zeae strains.
Extracellular enzyme activity assays
Pectate lyases (Pel), cellulases (Cel), and proteases (Prt) produced by Dickeya genus were the major causes resulting in foot rot in rice and other plants (Boluk et al., 2021). Therefore, their activities were detected according to the protocols as previously described with a few modifications (Chatterjee et al., 1995; Hu et al., 2022). In brief, the media for enzyme activity assays as follows: (1) Pel assay medium (per liter): 10 g polygalacturonic acid, 10 g yeast extract, 0.38 μmol CaCl2, 100 mmol Tris-HCl, 15 g agar, pH8.5; (2) Cel assay medium (per liter): 1.0 g carboxymethyl cellulose, 3.8 g Na3PO4, 15 g agar, pH7.0; (3) Prt assay medium: LB agar medium containing 1% (v/v) skimmed milk. The media were poured and allowed to solidify and 5 mm diameter wells were punched into the agar and sealed the bottom with worm 1.5% agar solution. 25 μL overnight cultures were applied to each well, and plates were incubated at 28°C. After 15 h, Pel assay plates were flooded with 1 M HCl; Cel assay plates were flooded with 1% Congo red solution for 1 h and washed twice with 1 M NaCl solution. Next, the treated Pel and Cel assay plates were incubated at room temperature for 24 h and the diameter of haloes around the wells were measured. For Prt assay, the haloes around the wells were measured after 24 h without any treatment. Each treatment was carried out at least three times, and all assays were repeated at least three times.
Flagellum-mediated motility assays
These assays were performed according to the methods as previously described (Boluk et al., 2021). Media for swimming (per liter contains 10 g tryptone, 5 g NaCl, and 3 g agar) and swarming (per liter contains 5 g peptone, 3 g yeast extract, and 4 g agar) assays were prepared. One microliter of overnight bacterial culture was spotted onto the center of a plate (60 mm diameter) containing about 10 mL of each medium. The plates were incubated at 28°C for 20 h before measurement of the diameters of each bacterial motility zone. Each experiment was repeated at least three times in triplicate.
Assays of exopolysaccharide production
Exopolysaccharide (EPS) was detected in WH1 culture according to a previous study with minor modifications (Hu et al., 2018). In brief, a single colony of WH1 strain was picked and transferred into a 2 mL LB medium and incubated at 28°C overnight. Afterward, 1 mL of which was applied into 100 mL LB medium and grown with shaking at 150 rpm for 12 h. Cultures were centrifuged at 8,000 rpm for 10 min, and then at 4,000 rpm for 20 min to obtain 80 mL supernatants. Double volumes of ethanol were added to the supernatants, mixed thoroughly, stored at 4°C overnight for precipitation, and subjected to centrifuge at 8,000 rpm for 20 min. Finally, supernatants were discarded and pellets were weighed after drying at 55°C overnight. The experiment was repeated three times in triplicate.
Biofilm formation assays
An overnight bacterial culture was diluted at 1:100 with LB medium; 200 μL of the culture was added into each well of 96-well microtiter plates and statically incubated at 28°C for 20 h. For qualification of the total biofilm mass, the suspension culture was removed and the plate was washed three times with PBS. After drying for 2 h at 60°C, the biofilm was stained with 50 μL of 0.1% crystal violet (w/v) for 15 min. The wells were also washed three times with PBS to remove unbound crystal violet dye and dried for 2 h at 60°C. Biofilms formed in 96-well plate were observed. Additionally, the biofilms developed in a 48-well microtiter plate containing one glass coverslip per well were used during observation with a fluorescent microscope, as previously described (Merritt et al., 2005). The plate was statically incubated at 28°C for 20 h. The AO/EB staining kit (Sango Biotech, Shanghai, China) was used to stain biofilms developed on glass coverslip after being washed three times with PBS to discard planktonic cells. The images of biofilms were observed through the fluorescent microscope (Leica Microsystem, Germany).
Acylated homoserine lactones production assay
Chromobacterium violaceum CV026 is usually used as a biosensor to visualize acylated homoserine lactones (AHLs) with N-acyl side chains from C4 to C8 in length production by Gram-negative bacteria (Pu et al., 2022). In this study, one colony of CV026 was inoculated into 2 mL of LB medium and incubated at 28°C overnight. The CV026 cultures were added to a warm LB agar (1.5%) medium at a ratio of 1:100, and then the mixtures were poured immediately over the surface of LB agar plates prepared in Petri dishes. When the overlaid agar solidified, a well (5 mm diameter) was made in the center of each plate and sealed the bottom with warm agar solution, 25 μL of overnight WH1 culture (109 CFU/mL) was added to the well. Meanwhile, 25 μL of overnight Pseudomonas aeruginosa PAO1 culture (109 CFU/mL) was performed under the same conditions as the reference. Violacein halo production was observed after being incubated at 28°C for 24 h. The diameter of the violacein halo was also measured.
AHL production was determined by inoculating 200 μL of an overnight culture of WH1 into 200 mL of LB. After 20 h incubation at 28°C, the sterile supernatant was polled and extracted with acidified ethyl acetate according to a protocol described previously (Zhou et al., 2017). The AHL produced by WH1 was analyzed using an HPLC system (Shimadzu, Japan) equipped with a C18 column by UV absorbance at 210 nm. The mobile phase A was water, and the mobile phase B was methanol. The following rate was set as 0.8 mL/min. The injection volume was 20 μL. The peak corresponding to 3OC6-HSL was identified according to the retention time of commercial 3OC6-HSL standard (Aladdin, Shanghai, China) with the same HPLC protocol.
Pathogenicity assays
Thirty rice seeds were put into 10 mL of sterile water containing 1 mL of WH1 overnight culture and incubated at room temperature for 5 h. The rice seeds were rinsed three times with sterile water and transferred onto the top of two moistened filter papers placed on a sterilized plate at room temperature. Rice seeds were treated with the same volume of sterile water as a negative control. The rice seeds were then incubated at 33°C under 8 h dark/16 h light with supplementation of sterile water when necessary. The seed germination rate was measured after 7 days.
To investigate the pathogenicity of strain WH1 on other plants, we selected potato tubers, carrots, radishes, and Chinese cabbage. The plants were washed with deionized water and then surface-disinfected with 70% ethanol, cut into slices, and dried for 30 min in a bio-safety hood. 25 μL of overnight culture was inoculated on the center of the tissue slices. After inoculation, tissue slices were placed on wet sterilized filter papers in a sterile double-layer drain basket and kept in a growth chamber in conditions of 28°C and 75% relative humidity. The lesions were measured every day for 3 days. Tissue slices inoculated with LB medium were used as a control.
Zeamine detection assay
The zeamine detection assay was performed according to a method previously described with a few modifications (Chen et al., 2016). In brief, plates were prepared by pouring about 10 mL of LB agar medium, overlaid with 5 mL of 1% agar containing 50 μL overnight culture of fresh Escherichia coli DH5α after solidification. 25 μL of overnight WH1 culture in LS5 medium (per liter contains: 5.25 g K2HPO4, 2.25 g KH2PO4, 10.0 g sucrose, 3.6 g NH4NO3, 1.0 g KCl, 0.25 g MgSO4, pH7.0) were added to the wells in plates. The bioassay plates were incubated at 28°C for 24 h before photography. The diameter of the inhibition zone was measured.
Statistical analysis
All assays were carried out at least three times independently unless otherwise stated. The results obtained were summarized in figures as mean ± standard deviations. Statistical significance was evaluated using a two-tailed Student’s t test. A p-value < 0.05 was considered significant.
Results
WH1 isolated from rice root was classified as Dickeya zeae based on phylogenetic analysis
In this study, although the strain WH1 was isolated from healthy rice root in Wuhu City, China (Eastern China), the pure culture was inoculated to the rice root, and typical soft rot symptoms were observed (data not shown), validating its role as rice pathogen. Meanwhile, we isolated one strain, Paenibacillus polymyxa, from the healthy rice root, which can inhibit the growth of strain WH1 (Supplementary Figure 1). Therefore, it is not surprising that WH1 was isolated from healthy rice roots. Additionally, we found that WH1 can inhibit the growth of Candida albicans, while P. aeruginosa PAO1 can inhibit the growth of WH1 (Supplementary Figure 2). To identify the taxonomic status of strain WH1, MLSA was performed based on the complete sequences of four housekeeping genes including aptD, gyrB, infB, and rpoB. Phylogenetic analysis of concatenated sequences of four genes distinctly separated D. zeae strains from other species. Among these D. zeae strains, EC1, EC2, and WH1 isolated from rice were grouped into a clade differentiated from other D. zeae strains. WH1 was placed at the same branch with EC2 isolated from rice (Figure 1). ANI analysis indicated that the ANI values between WH1 and previously identified strains from rice ranged from 97.34 to 98.28% and were higher than the cut-off value (96%) for species delineation. Meanwhile, the isDDH values between WH1 and D. zeae strains isolated from rice ranged from 76.3 to 83.9%, which were also higher than the cut-off value (70%) (Table 1). In summary, these data establish that strain WH1 belongs to the species D. zeae.
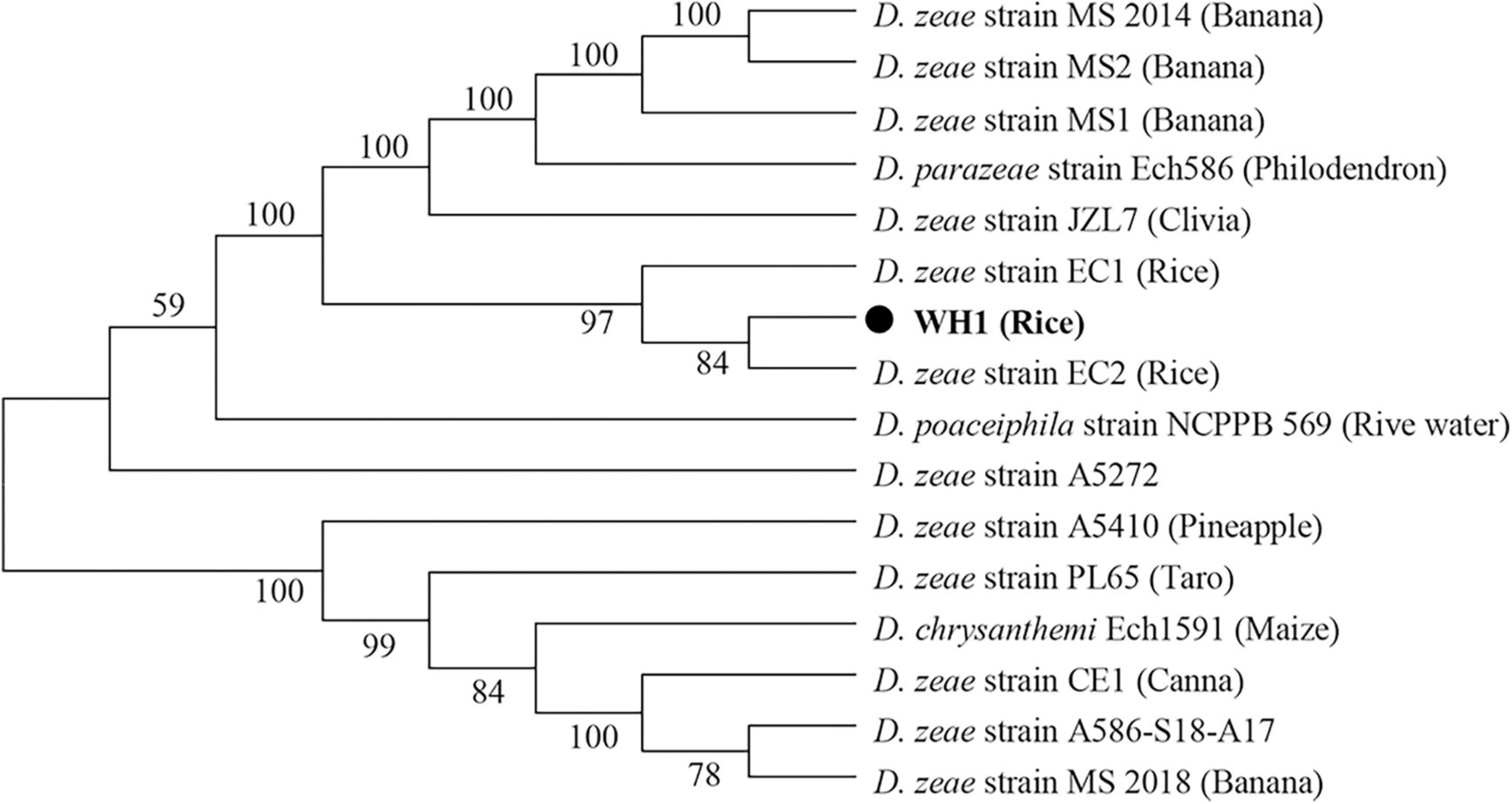
Figure 1. Phylogenetic placement of Dickeya zeae strain WH1. Phylogenetic analysis of Dickeya strains based on concatenated sequences of housekeeping genes, aptD, gyrB, infB, and rpoB, 16 Dickeya strains with complete genomes were included in a phylogenetic analysis with a neighbor joining algorithm and bootstrapped at 1,000 replications.
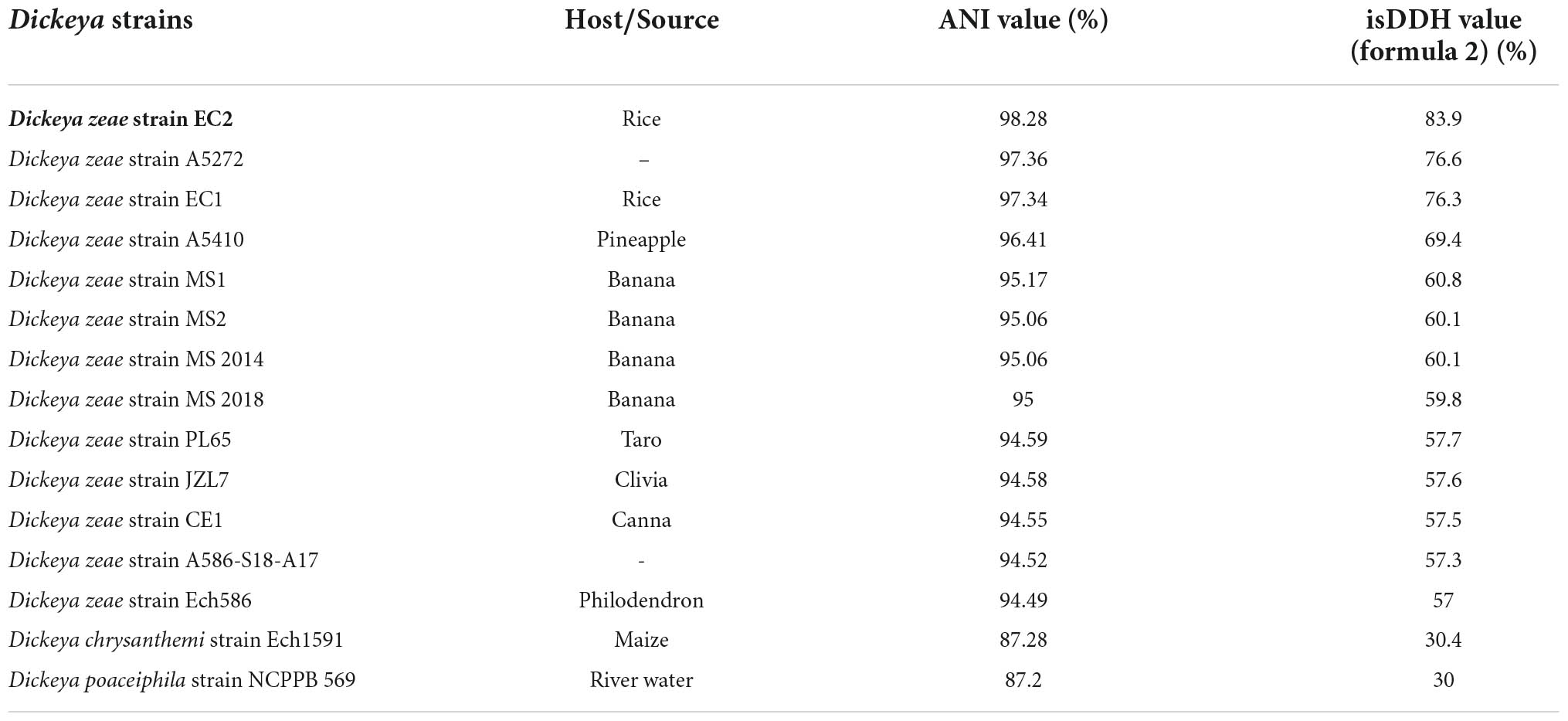
Table 1. Average nucleotide identity (ANI) and in silico DNA–DNA hybridization (isDDH) values between the genomes of Dickeya zeae strain WH1 and other characterized Dickeya species.
Genomic differences between WH1 and other Dickeya zeae strains
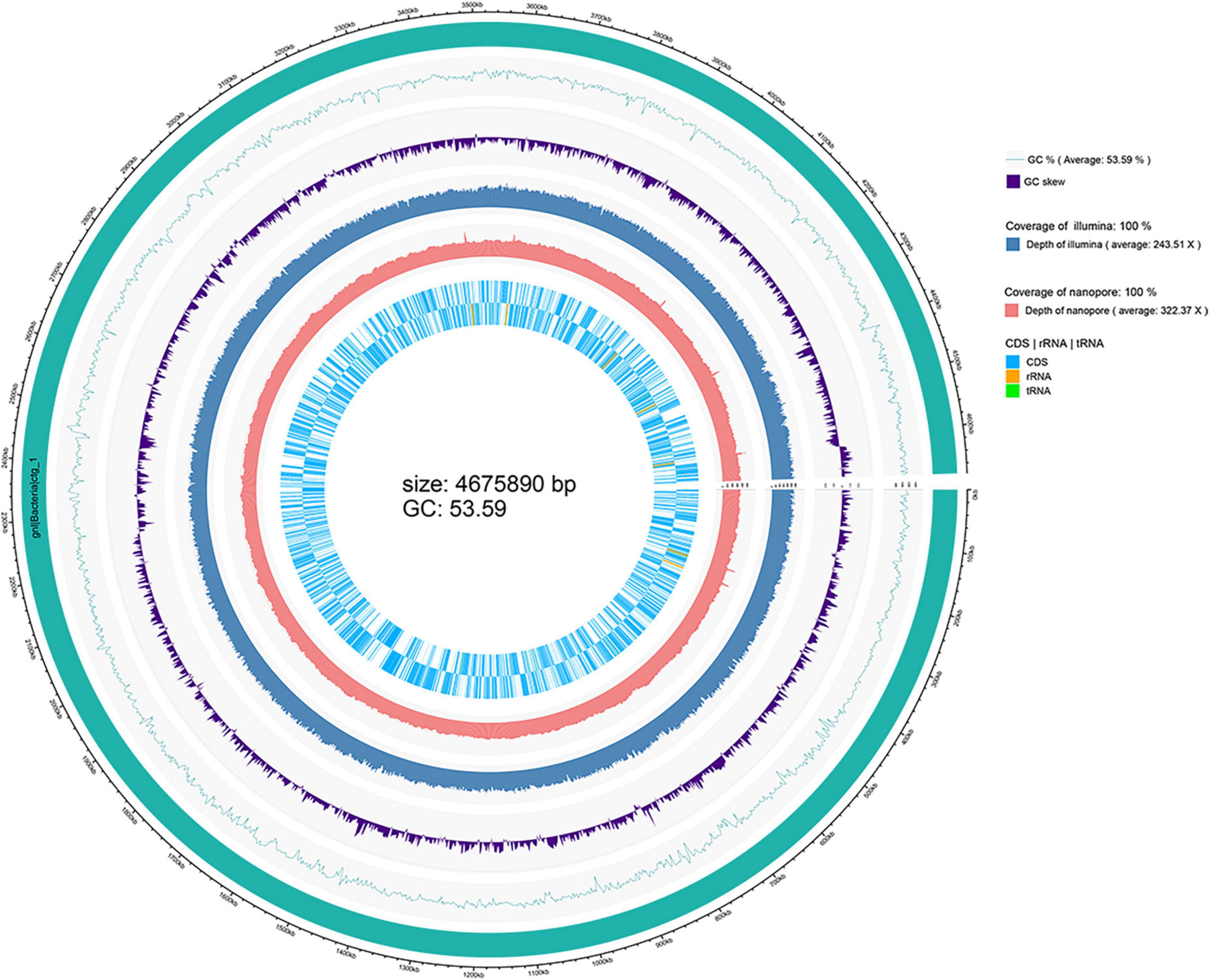
The depth of the assemblies was 243.51 and 322.37 for Illumina and Nanopore platforms, respectively. The complete genome of strain WH1 consisted of a single circular chromosome of 4,675,890 base pairs with a 53.59% GC content. General features of the genome are listed in Table 2, and a detailed circular map is shown in Figure 2. In total, 22 predicted rRNA genes and 76 tRNA genes were identified, and 86.92% of nucleotides are predicted to coding proteins. By a combination of coding protein prediction and homology search, 4096 coding DNA sequences (CDSs) with an average length of 979 bp were identified on the genome. Of these, 274 CDSs were annotated by COG function groups and classified into 26 COG categories. Meanwhile, 1,572 CDSs were annotated by KEGG. 5.81% (238) of total ORFs were annotated as hypothetical proteins. Moreover, four representative D. zeae strains, EC1, EC2, MS1, and MS2, were isolated from rice and banana, resulting in severe soft rot disease. Their genome sequences were downloaded from NCBI and used for genomic comparison analysis. Their genome features are also listed in Table 2.
Pan-genomes analysis was used to develop a robust comparative genomics analysis. The results showed that the number of core gene clusters among the five genomes was 3,040. The number of specific clusters in WH1, EC1, EC2, MS1, and MS2 were 284, 284, 134, 197, and 225, respectively (Figure 3). The unique clusters in WH1 contained some important encoding genes, such as transporter proteins, transposases, endonucleases, and toxin-antitoxin systems. Besides, the unique clusters contained 113 genes encoding hypothetical proteins (Supplementary Table 1). However, the unique clusters in EC1 contained genes encoding zeamine biosynthesis (Supplementary Table 2).
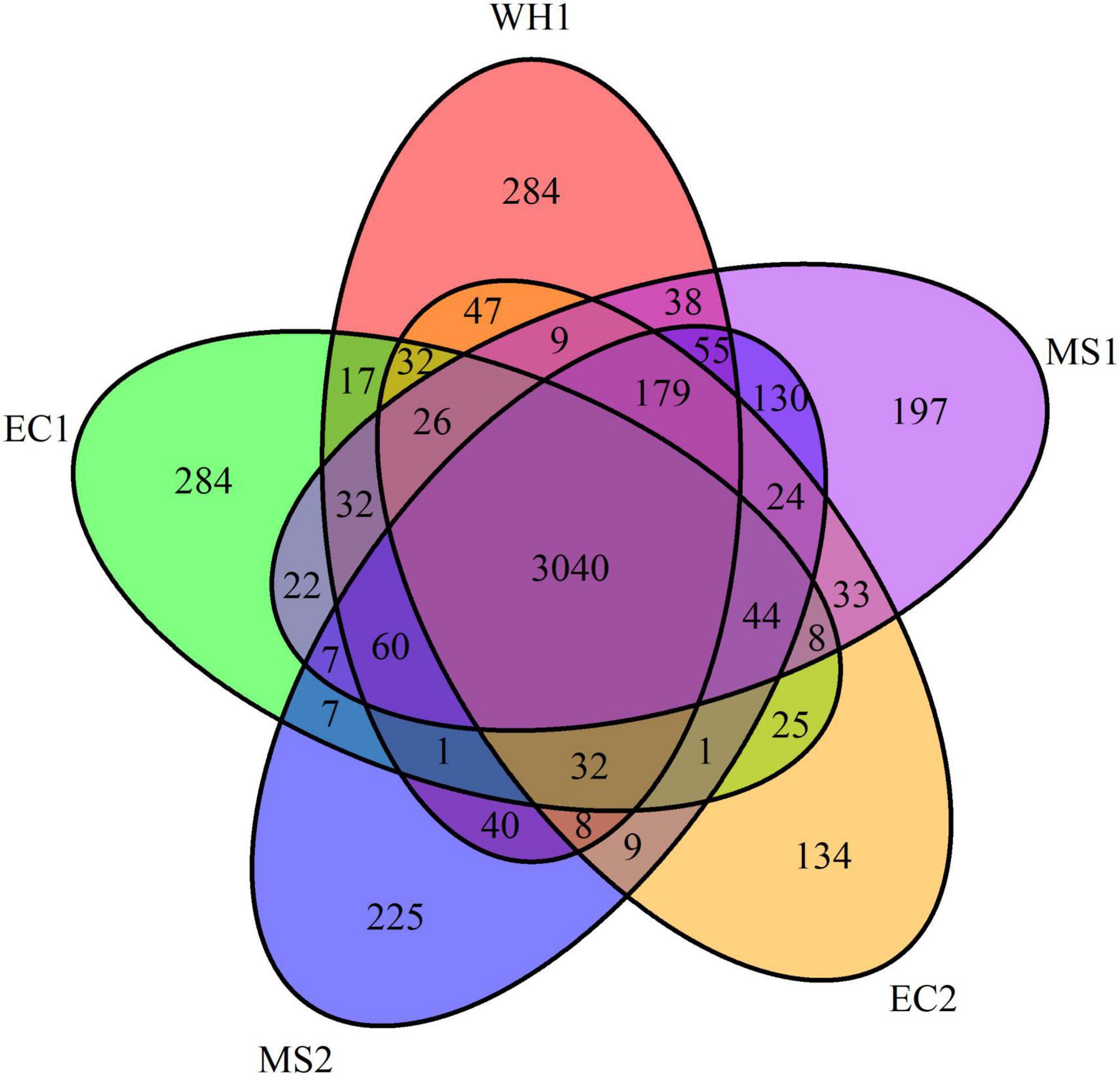
Figure 3. Venn diagram representing the numbers of common or specific gene clusters among analyzed Dickeya zeae strains in PGAP analysis.
Plant cell wall-degrading extracellular enzymes
Most of these extracellular enzymes, pectate lyases (Pel), and other pectinase such as pectin methylestrases (Pem) and pectin lyase (Pnl) produced by D. zeae strains have been shown to play a major role in the virulence and tissue maceration (Boluk et al., 2021). The genome WH1 contained a total of 14 genes encoding pectin degradation enzymes (Supplementary Table 3). These pectin degradation genes were highly conserved in various D. zeae strains, except that pelB encoding pectate disaccharide lyase was absent in the MS1 strain and pemB encoding pectinesterase B precursor was absent in all analyzed strains (Supplementary Table 3). Although the pehKNVWX genes encoding polygalacturonases were also conserved in D. zeae strains, parts of which (pehNVW) were absent in all analyzed genomes. In addition, the endoglucanase genes (celY and celH) and beta-glucosides encoding genes (bglABCD and bgxA) are involved in the degradation of cellulose to glucose. Although these genes were highly conserved in the Dickeya genus, bglC was absent in strains MS1 and MS2 isolated from banana, bglD was just present in EC1, celY was just present in WH1 and EC1. In addition, the homologies of bglB and bglC in WH1 were 53 and 52% with EC1, respectively, although both were isolated from rice. The xylanases (xylBAFGHR and xynA) genes are involved in the degradation of xylan and xyloglucan mainly present in plant cell walls (Boluk et al., 2021). All genomes of analyzed D. zeae strains contained these genes. In summary, these pel, peh, and cel genes were conserved in genomes of D. zeae strains (Supplementary Table 3).
The prtABCDEFG gene cluster encoding four proteases and three protease secretion-associated proteins (T1SS) was located on the positive strand of WH1, EC1, MS1, and MS2 chromosomes, but located on the negative strand of the EC2 chromosome. Alignment with other D. zeae strains showed that they were highly conserved in the genome of WH1 (Supplementary Table 3).
Secretion systems
The type II secretion system (T2SS) is used by gram-negative bacteria to translocate extracellular proteins across the outer membrane (Tan et al., 2021). In D. zeae strains, the T2SS is encoded by the out genes to allow the secretion of several proteins including most pectinases. Therefore, T2SS plays an important role in the pathogenicity of Dickeya spp. on host plants. WH1 chromosome harbored a T2SS gene cluster (outOMLKJIHGFEDCBS), covering about 13 kb with 14 ORFs. The gene cluster shared high similarity with that of other analyzed D. zeae strains (similarity 99%) at the amino acid level (Supplementary Table 4). Therefore, the out gene cluster was highly conserved among D. zeae strains.
Besides T2SS plays an important role in the pathogenicity of Dickeya spp., T3SS was also confirmed as a pathogenicity factor in many phytopathogenic bacteria (Hu et al., 2022). Similarly, the hrp genes in Dickeya spp. have also been reported to play an important role in their pathogenesis and interaction with host plants (Hu et al., 2018). In the genome of WH1, a large hrp gene cluster spanning 27.3 kb was identified, which was composed of 29 genes, while 27 genes in EC1, three genes in EC2, 26 genes in MS1, and 31 genes in MS2 (Supplementary Table 4). The homologies of hrp gene cluster in WH1 were high with EC1, MS1, and MS2, arranged from 80 to 100% at the amino acid level.
The Type IV secretion system constitutes virB1-11 genes and functions in conjugation, pathogenicity, and DNA release/uptake (Zhou et al., 2015). Some D. zeae strains, such as EC1, Ech586, MS1, and A5410, harbored virB-T4SS cluster, while the cluster was not found in WH1 and EC2, just virB1 and virB2 were present in MS2 (Supplementary Table 4).
The type VI secretion system (T6SS) was identified to take part in bacterial pathogenicity and host colonization (Chen et al., 2015; Lin et al., 2017). Many studies showed that T6SS functions in various biological processes, such as mediating cooperative or competitive interactions between bacteria and eukaryotes, and bacterial biofilm formation (Chen et al., 2015). T6SS is typically encoded by a cluster of 12–20 genes, with a minimum of 13 genes for the production of a functional apparatus (Zhou et al., 2015). In this study, a gene cluster encoding T6SS was present in D. zeae WH1, spanning 44 kb with 37 ORFs. In this gene cluster, in addition to 17 genes described previously with high identity at the amino acid level, there are 20 additional ORFs inserted between vgrG and tssB genes in genome WH1, while 16 genes in EC1, 22 genes in EC2, 21 genes in MS1, and 20 genes in MS2. For these inserted genes, two genes encoding ankyrin repeat proteins were present in WH1 and EC1, while three genes were in EC2, MS1, and MS2. Additionally, two genes encoding hypothetical proteins were inserted between both genes encoding ankyrin repeat proteins in WH1. IS family transposase was present in EC2 and MS1, but absent in WH1, EC1, and MS2. Two genes encoding plasmid stabilization system proteins ParDE were just found in WH1 and EC1 (Supplementary Table 4). In summary, different genes were inserted between vgrG and tssB of the T6SS gene cluster in all analyzed genomes.
Flagellar and chemotaxis genes
Motility and chemotaxis are essential for phytopathogens when searching for favorable sites to enter into the plant apoplast. Therefore, they play important roles in plant infection by Dickeya spp. The flagellar biosynthesis and chemotaxis clusters constitute 20 fli genes, 14 flg genes, and 5 flh genes, involved in the flagella synthesis, four flagellar rotation genes and six chemotaxis associated genes were present and highly conserved in analyzed D. zeae genomes. Interestingly, two fliC encoding flagellin FliC were found in EC1, MS1, and MS2 genomes, but absent in WH1 and EC2 genomes. Moreover, there were 7 additional genes inserted between fliA and fliC genes in the WH1 genome, encoding a class I SAM-dependent methyltransferase (ctg_02571), a glycosylase (ctg_02573), two hypothetical proteins (ctg_02572 and ctg_02575), a WbqC family protein (ctg_02574), a DegT/DnrJ/EryC1/StrS family aminotransferase (ctg_02576), and a methyltransferase regulatory domain-containing protein (ctg_02577), which were not found in other four D. zeae genomes (Supplementary Table 5). The type IV pilus biogenesis encoding system consists of pilW, pilT, pilABC, and pilMNOPQ genes. The pilW and pilT genes were located distant from the type IV pilus biogenesis cluster. They were highly conserved in D. zeae genomes (Supplementary Table 5).
Polysaccharides biosynthesis genes
Besides lipopolysaccharide (LPS) as a component of the cell wall in Gram-negative strains, Dickeya genus can biosynthesize other polysaccharides, such as capsular polysaccharide (CPS) and exopolysaccharide (EPS). CPS can form a discrete capsule around the cell and is intimately associated with the cell surface; EPS is one of the important virulence factors for bacterial phytopathogens and the main toxic factors leading to water-logging and wilting on plants after infection (Hu et al., 2018). Both are important factors in infection and biofilm formation. In our analysis, both clusters were present in WH1, MS1, and MS2 genomes, but the CPS cluster was absent in EC1, and the EPS cluster was absent in EC2 (Supplementary Table 6).
Quorum sensing systems
Many Gram-negative bacterial pathogens utilize the luxI/luxR quorum sensing system to regulate the expression of virulence genes and biofilm formation (de Kievit, 2009). Generally, luxI encodes a synthase for the production of AHL family quorum sensing signals, and luxR encodes an AHL signal receptor. Upon interaction with the AHL signal, LuxR becomes an active transcription factor and hence modulating the expression of virulence genes. Previous studies showed that N-3-oxohexanoyl-homoserine lactone (3OC6-HSL), N-3-oxo-octanoyl-homoserine lactone (3OC8-HSL), N-hexanoyl-homoserine lactone (C6-HSL) and N-decanoyl-homoserine lactone (C10-HSL) were produced by Dickeya genus (Liu et al., 2022). Of which, 3OC6-HSL was produced by D. zeae EC1 encoding by the luxI homologous expI (Zhang et al., 2021). A Blast search of the WH1 genome revealed only one copy of expI and one well-conserved luxR homolog expR. ExpI produced by WH1 was highly homologous with the other four D. zeae strains (>99%) at the amino acid level and these four strains produce 3OC6-HSL (Supplementary Table 7). Therefore, WH1 would produce 3OC6-HSL, which was confirmed using HPLC (Data shown below description).
Interestingly, a novel QS system vfm gene cluster upstream of the expI/expR was found in the WH1 and other genomes analyzed in this study (63–100% identity at amino acid level). The gene cluster was originally identified in D. dadantii 3937 isolated from the lesion spot of a wilted African violet and was associated with the regulation of virulence factor production and pathogenesis (Potrykus et al., 2018). The vfm gene cluster, WH1, EC1, EC2, and MS2 genomes contained 26 genes including vfmAZBCDEFGHIJXWVUTSRQPOMNLKY, while the MS1 genome contained 27 genes with one gene encoding fatty acid CoA ligase family protein inserted between vfmN and vfmM (Supplementary Table 7). However, the biological significance of various QS systems in WH1 and other Dickeya species needs to be further investigated.
WH1 produces toxins different from zeamines produced by EC1
Dickeya zeae EC1 produces at least two polyamino phytotoxins and antibiotics, zeamines, which were shown to be the major virulence determinants, affecting rice seed germination and inhibiting the growth of E. coli DH5α (Zhou et al., 2015; Lv et al., 2022). However, no zeamine cluster (zmsO-zmsN) was found in the WH1 and EC2 strains from rice and MS1 and MS2 strains from banana (Supplementary Table 8). Although zeamine biosynthesis genes were absent in the WH1 genome, the strain still inhibited rice seed germination, and the ability to infect other plants was not reduced (data shown in results of pathogenicity assays). Therefore, the above analysis indicated that the zeamines might not be a necessary virulence factor for this new strain from rice. Other virulence factors could be present in strain WH1 and need to be further investigated.
Phenotype assays
WH1 genome harbored the genes encoding proteases (prt cluster), pectate lyases (pel cluster), and cellulases (cel5Z, celH, and celY). The strain produced Prts, Pels, and Cels in the plate assays (Figure 4). Moreover, WH1 generated large swarming (diameter greater than 50 mm) and swimming (diameter greater than 40 mm) diameters when incubated at 28°C for 20 h (Figures 5A,B). Both CPS and EPS clusters were found in the WH1 genome, which plays important roles in biofilm formation and pathogenicity for D. zeae strains. The results showed that WH1 has the ability to produce exopolysaccharides (Figure 5C). In addition, WH1 produced thick biofilm in a 96-well plate and the biofilms were verified with fluorescent images (Figure 6).
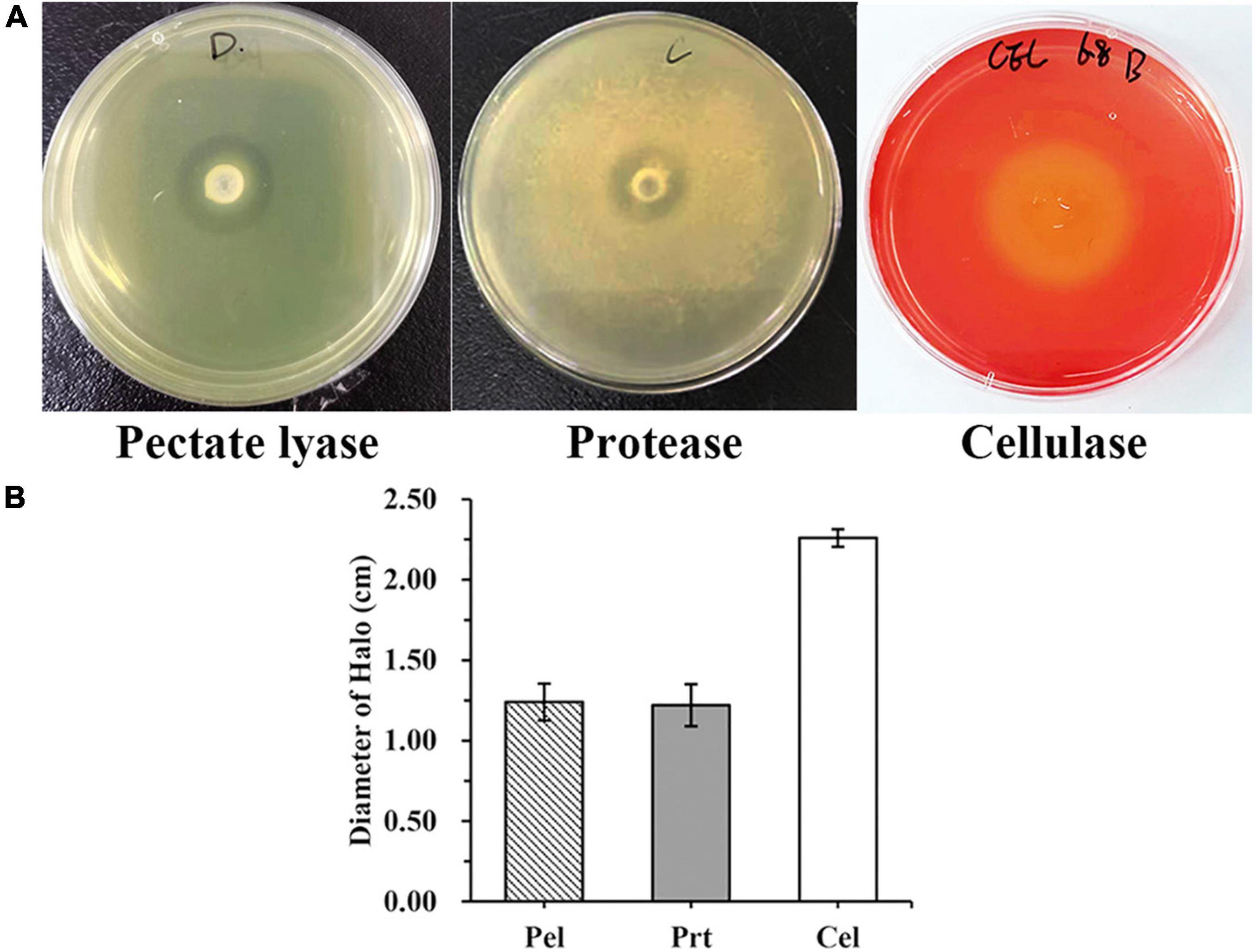
Figure 4. Extracellular cell wall-degrading enzymes produced by Dickeya zeae strain WH1. (A) Pectate lyase (Pel), Protease (Prt), and Cellulase (Cel) plates; (B) indicates the production of Pel, Prt, and Cel from D. zeae strain WH1.
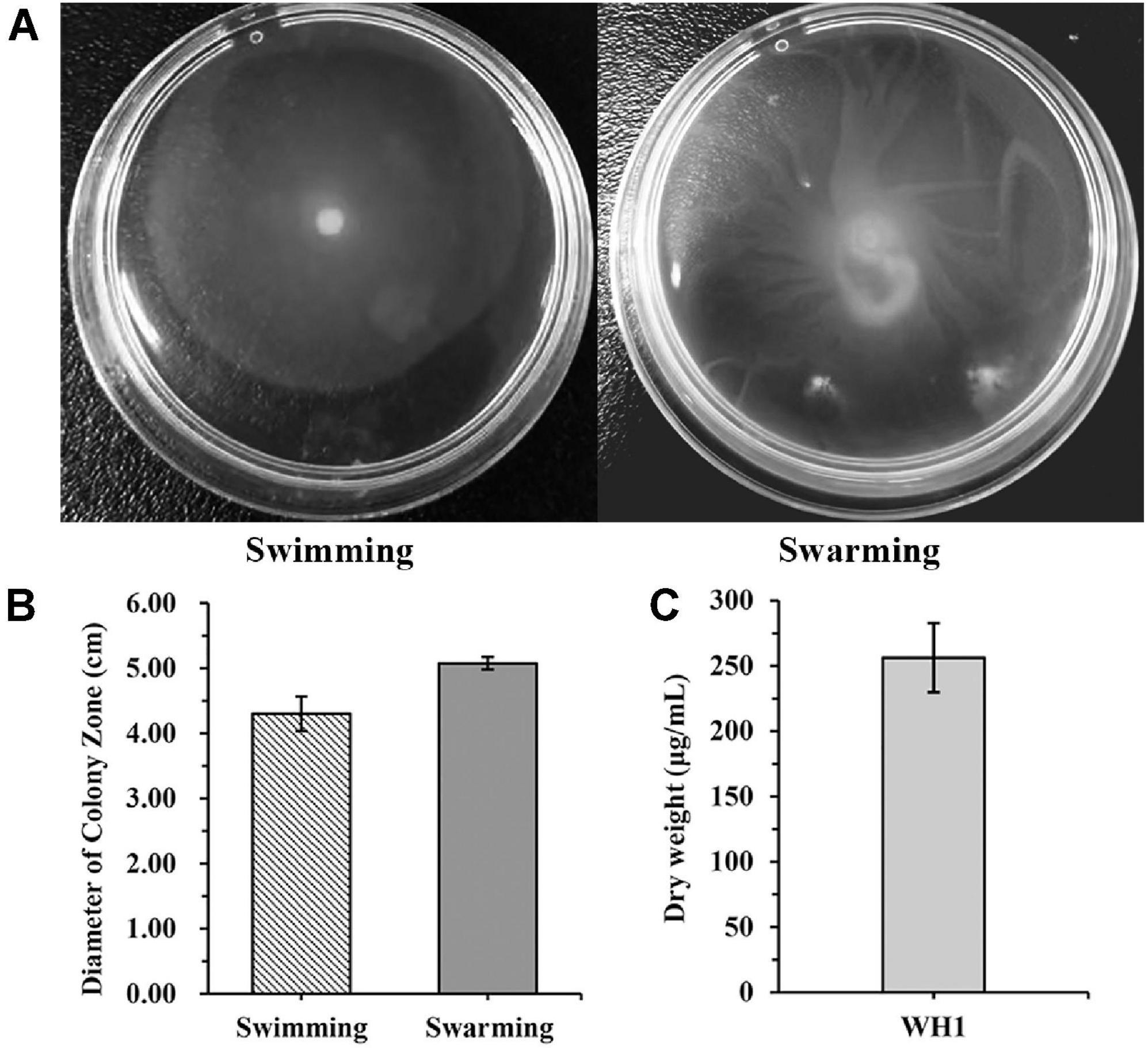
Figure 5. Swimming and swarming motilities and exopolysaccharide production of Dickeya zeae strain WH1. (A) Swimming and swarming capacities were observed in a semi-solid medium after 20 h at 28°C, respectively; (B) quantification of swimming and swarming abilities; (C) dry weight of EPS produced by D. zeae strain WH1.
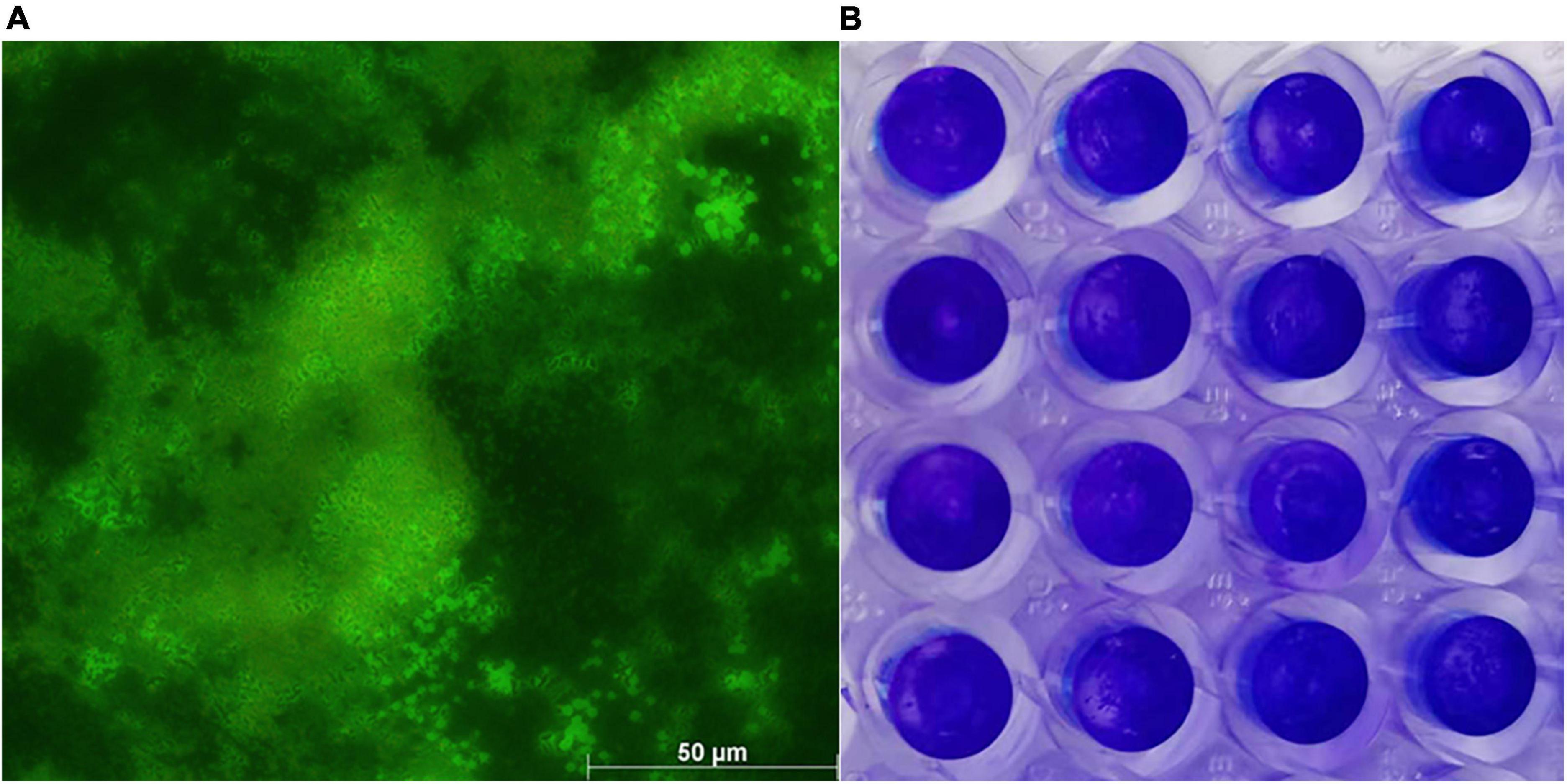
Figure 6. Dickeya zeae strain WH1 biofilms. (A) Images are presentative of D. zeae strain WH1 biofilm grown on a glass coverslip in a 48-well plate for 20 h, scale bar 50 μm; (B) biofilm developed in a 96-well plate for 20 h was stained with 0.1% crystal violet.
The expI/expR quorum sensing system cluster was highly conserved in the five genomes. Therefore, AHL produced by WH1 was qualified with CV026. The results showed that WH1 produced more AHLs than QS model strain P. aeruginosa PAO1 when incubated at the same conditions (Figure 7). Additionally, the results of HPLC demonstrated that the AHL produced by WH1 was 3OC6-HSL (Figure 8).
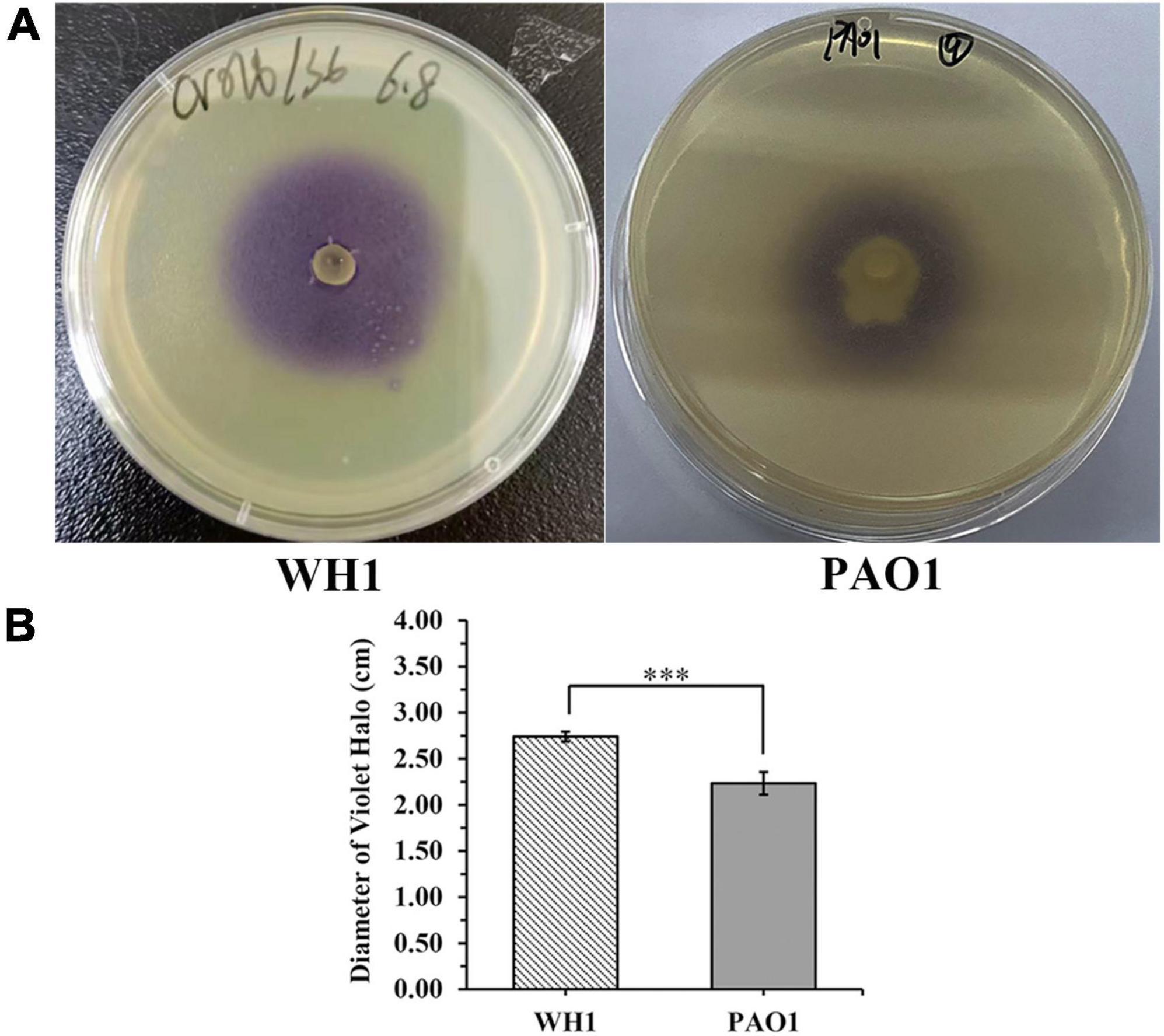
Figure 7. Acylhomoserine lactones (AHLs) production by Dickeya zeae strain WH1 and Pseudomonas aeruginosa PAO1 at the same conditions. (A) Images are presentative of WH1 and PAO1 grown on Chromobacterium violaceum CV026 lawn for 24 h at 28°C; (B) quantification of AHLs production abilities, ***p < 0.001.
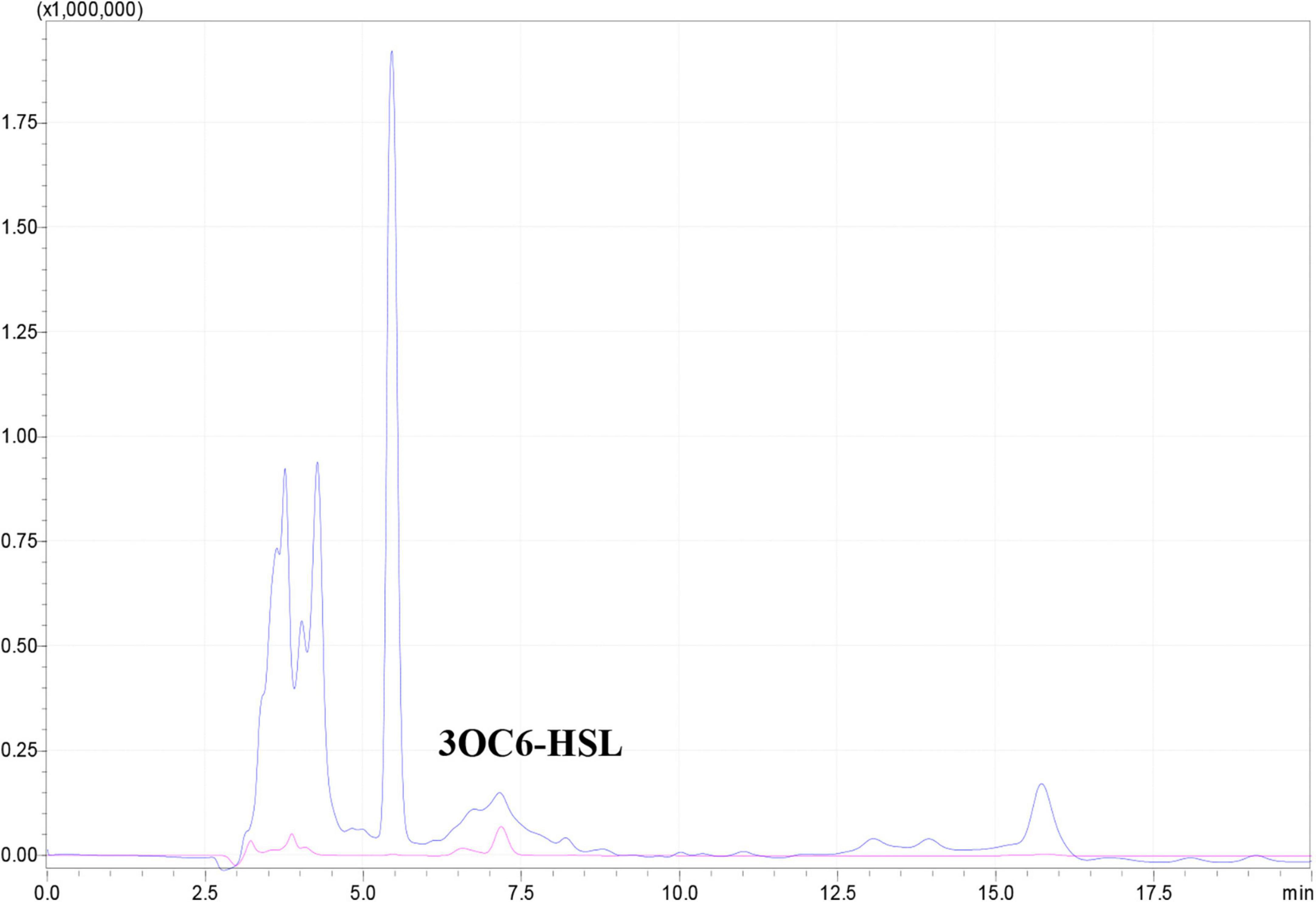
Figure 8. 3OC6-HSL produced by Dickeya zeae strain WH1 measured by HPLC. Blue indicates 3OC6-HSL present in the supernatant from D. zeae strain WH1; pink indicates commercial 3OC6-HSL standard.
Pathogenicity assays
The results of pathogenicity tests on potato tubers, carrots, radishes, and Chinese cabbage confirmed that the WH1 strain infected all plants with serious maceration symptoms. When infected for 72 h, almost plant tissues were macerated (Figure 9). We also tested the inhibitory activity of WH1 against rice seed germination. The results indicated that WH1 had a strong inhibitory effect on rice seed germination with an inhibitory rate being about 87% (Figure 10A). Zeamine detection assay demonstrated that WH1 did not inhibit the growth of E. coli DH5α (Figure 10B). Therefore, the virulence factors inhibiting rice seed germination were not zeamines.
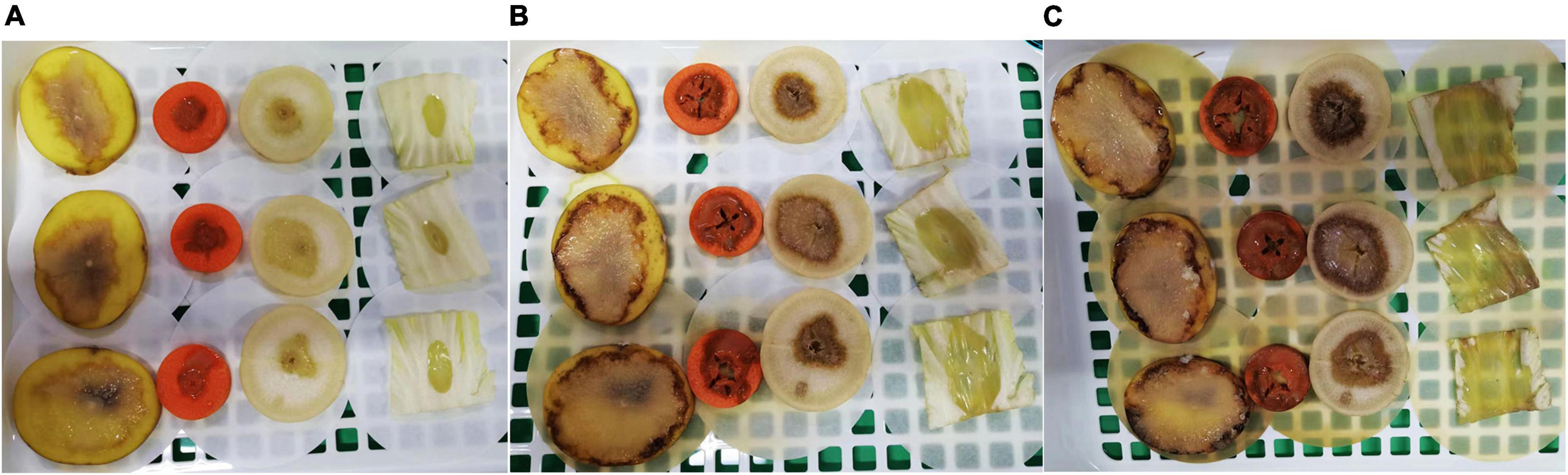
Figure 9. Pathogenicity of Dickeya zeae strain WH1 on potato tubers, carrot, radish, and Chinese cabbage. (A) Infected for 24 h; (B) infected for 48 h; (C) infected for 72 h.
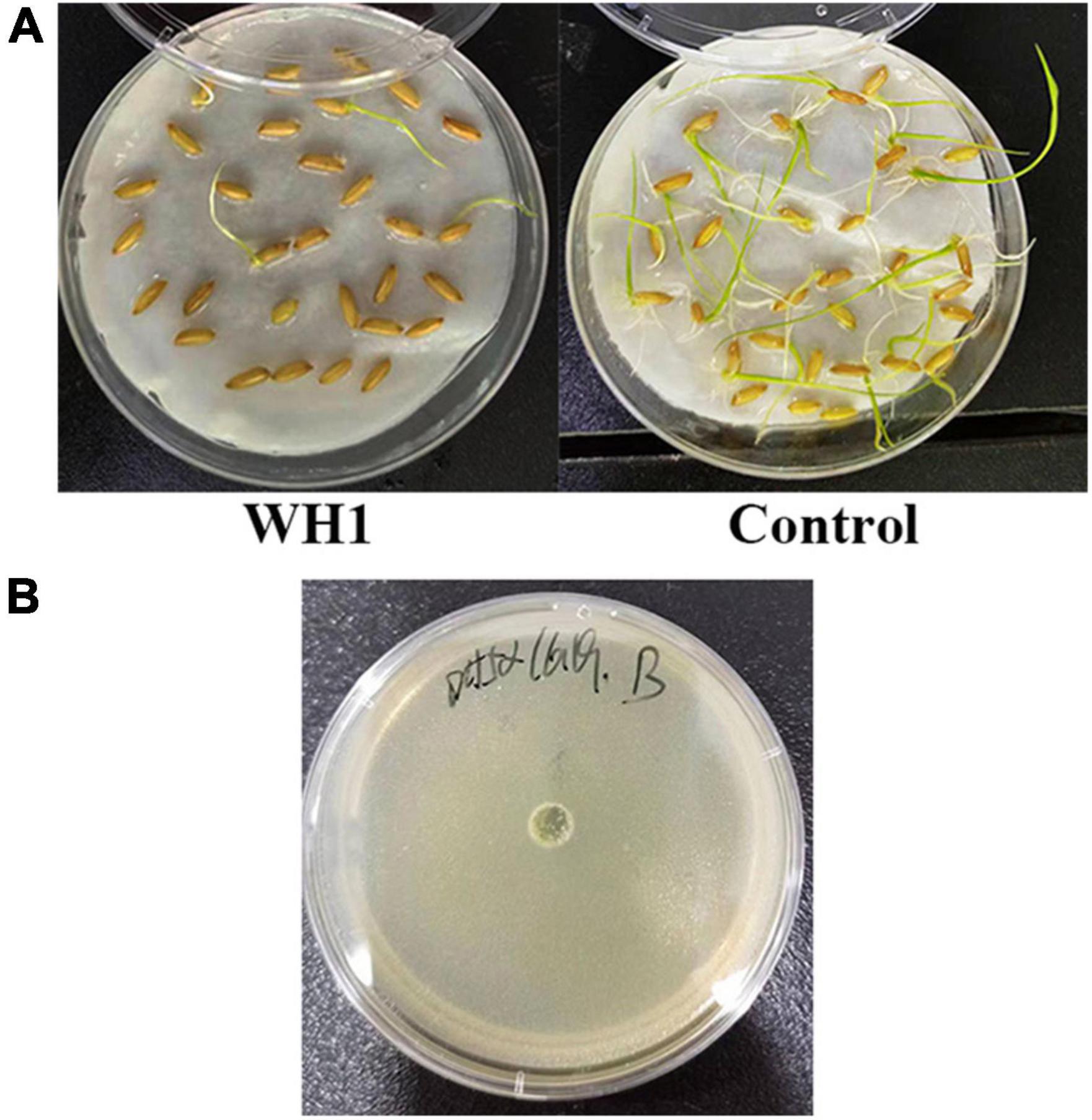
Figure 10. Phytotoxins production by Dickeya zeae strain WH1. (A) Inhibitory activity of toxins from D. zeae strain WH1 against rice seeds germination; (B) the WH1 culture did not inhibit the growth of Escherichia coli DH5α.
Discussion
Soft rot Pectobacteriacaea (SRP) includes the genera Pectobacterium and Dickeya and are widespread globally, which cause blackleg disease in potatoes and soft rot disease in a wide range of hosts, including vegetables, fruits, ornamental plants, and the most important crops, rice, and maize. Surprisingly, there is no report on Pectobacterium spp. isolated from rice and maize. In contrast, Dickeya spp. was isolated from a wide range of hosts compared to Pectobacterium spp. (Toth et al., 2021). Although there are some differences in isolated hosts, SRP shares many virulence factors, including PCWDE, secretion systems, motility, and plant responses to SRP infections (Davidsson et al., 2013; Reverchon et al., 2016). In Dickeya genus, D. zeae strains and their genomes were used for references to study virulence factors and pathogenicity of the genus (Zhang et al., 2014, 2022; Zhou et al., 2015; Lv et al., 2022). However, there is a relatively small amount of research on the above diseases in rice (Van Gijsegem et al., 2021).
Research to date indicates that D. zeae strains can infect a wide range of host plants, including 24 dicotyledonous plants and 23 monocotyledonous plants (Hu et al., 2022). Therefore, both crop rotation and interplants might be impacted once infected by this kind of strain. Currently, most Dickeya strains were isolated from diseased plants. In this study, although WH1 was initially isolated from the healthy rice root surface, the pure culture has serious pathogenicity on rice and other plants. Additionally, a potential biocontrol strain, P. polymyxa, also isolated from the healthy rice root surface, can significantly inhibit the growth of WH1. Importantly, D. zeae are often present in latent infections on many host crops and they can persist over winter in contaminated plant residues and soil. When the optimal conditions of temperature, humidity, and other factors occur, latency is broken and bacteria proliferate and cause decay (Bez et al., 2021). Therefore, the rice and other plants are grown in the field infected by WH1 face the risk of soft rot disease once microbial balance leads to a high abundance of the D. zeae strain.
Based on our analysis, we conclude that the novel strain WH1 is diverse and phylogenetically close to D. zeae strain EC2 isolated from rice in Guangdong, China. Given that the 16S rRNA gene sequence is insufficient to assign the taxonomy of the bacterium, the joint phylogenetic tree based on MLSA analysis was considered. We found that WH1, EC1, and EC2 were grouped into a clade, even though WH1 and EC1, and EC2 were isolated from different places (Eastern China and Southern China). Meanwhile, the results of ANI and DDH analyses also confirmed the MLSA analysis. Interestingly, EC1 harbors a zms gene cluster involving zeamines in biosynthesis, which can inhibit the growth of E. coli DH5α and rice seed germination (Hu et al., 2018), was absent in WH1 and EC2 strains isolated from rice and MS1 and MS2 isolated from banana. However, WH1 produced phytotoxin(s) with high inhibitory activity on rice seed germination and no effect on the growth of E. coli DH5α, which further revealed that WH1 produces other toxins instead of zeamines. Additionally, we found WH1 can inhibit the growth of Candida albicans, while P. aeruginosa PAO1 can inhibit the growth of WH1, which was different from EC1 and MS2 (Hu et al., 2018). Therefore, the toxins produced by WH1 would be different from other D. zeae strains, especially different from EC1. The toxins produced by WH1 require further exploration in the future.
Plant cell wall degrading enzymes produced by the Dickeya genus are considered essential virulence factors for host plants and disease development (Zhang et al., 2020). Our genomic comparative analyses demonstrated that the main encoding genes associated PCWDEs were existed and conserved in WH1 and other D. zeae strains. Besides, the pathogenesis of Dickeya spp. involved in several complex secretion systems (T1SS–T6SS) to translocate a wide range of extracellular enzymes and effector proteins from the periplasm across the outer membrane. The Prts, which are crucial for virulence, are secreted by the T1SS known as the prtDEF operon. The T1SS cluster, mainly transporting metalloproteinase, was present in all analyzed strains. Previous studies have revealed that Dickeya species mainly used the T2SS to translocate extracellular proteins such as pectinases and cellulases. Similar results were obtained in our analyses. Additionally, Hu et al found that the deletion of T3SS substantially diminished MS2’s pathogenicity on both dicots and monocots (Hu et al., 2022). Meanwhile, they suggested that the loss of T3SS and reduced PCWDE activity together might have contributed to the host specificity and virulence of D. zeae JZL7, one of the monocot-specific strains isolated from Clivia miniata. The effect of T3SS on WH1’s pathogenicity will therefore require further investigation in the future. T6SS was recognized as a distinct class of bacterial protein secretion system and identified as a virulence factor in Gram-negative bacteria (Chen et al., 2015). However, the biological function of T6SS in the Dickeya genus has not yet been determined. In this study, a gene cluster encoding T6SS was present in all analyzed genomes.
Among Gram-negative bacteria, EPS and CPS are often essential virulence determinants in plant pathogens. EPS is a main component of the bacterial biofilm matrix and is responsible for adhesion to plant surfaces (Chen et al., 2015; Zhang et al., 2020). Strains EC1 and EC2 isolated from rice produced either EPS or CPS, while banana isolates MS1 and MS2 generally produced significantly more EPS than strains from rice and exhibited different biofilm-associated phenotypes (Hu et al., 2018). A previous study demonstrated that MS1 could form a thick biofilm, while EC1 biofilm appeared in clumps and EC2 biofilm was thinner (Zhang et al., 2020). WH1 had the entire EPS and CPS gene clusters, and our study found that the strain easily formed thick biofilm in vitro. Therefore, the relationship between the ability of WH1 to form biofilm and its pathogenicity will be required investigation in the future. Flagellar is used for both swimming and swarming motility. Jahn et al. (2008) have proven that the deletion of the fliA gene encoding a sigma factor obstructed bacterial motility, and limited Pel production and bacterial attachment to plant tissues in D. dadantii 3937. Moreover, the type IV pilus was responsible for motility twitching in P. aeruginosa and Dickeya aquatic (Duprey et al., 2019). Therefore, flagellar biosynthesis and chemotaxis proteins and the type IV pilus play important roles in plant bacterial pathogenicity. We found these clusters present in all analyzed D. zeae strains.
Dickeya species have at least two QS systems, expI/expR, and vfm systems. A previous study demonstrated that deletion of expI in D. zeae EC1 abolished AHL production, increased swimming and swarming motility, disabled biofilm formation, attenuated virulence of pathogen on potato tubers, and reduced the inhibitory activity on rice seed germination through decreasing the expression of zms genes (Hussain et al., 2008). The inactivation of expI in D. zeae MS2 affected motility and cell clumping, while it did not affect infections of banana seedlings (Feng et al., 2019). Therefore, the effects of the expI/expR system on the virulence and pathogenicity of D. zeae strains is complex. The genome of WH1 also contains expI/expR clusters with an identity of 99% with other D. zeae strains. Meanwhile, HPLC demonstrated the AHL produced by WH1 was 3OC6-HSL. However, there are different virulence determinants between WH1 and EC1, such as the zeamine biosynthesis gene cluster that is present in EC1 but absent in WH1. Therefore, the effect of expI/expR system on virulence factors production of WH1 remains to further investigate in the future.
Vfm-QS signal is uniquely present in Dickeya strains, participating in and dominating the macerating soft rot process of the pathogen in plant tissues. Previous studies demonstrated that the vfm QS system participated in the regulation of PCWDEs production and virulence in the Dickeya genus (Zhang et al., 2014). However, the chemical structure of the Vfm signal has not been characterized yet. In this study, the entire vfm gene cluster was found in the WH1 genome, but its effect on virulence has not yet been investigated through experimental evidence.
Conclusion
This study presents a high-quality complete genome sequence of a novel D. zeae strain WH1 isolated from rice root. Phylogenetic tree analysis and genome-genome nucleotide comparison indicated that WH1 was placed at the same branch with EC2 isolated from rice. The genome sequence of WH1 was annotated and compared with the representative genome sequences of other D. zeae strains, focusing on virulence determinants and potential regulatory mechanisms. Several groups of virulence genes, such as those encoding for PCWDEs, T1SS gene cluster, T2SS gene cluster, T6SS gene cluster, flagellar and chemotaxis gene cluster, and quorum sensing gene cluster were highly or fully conserved in all five genomes isolated from different hosts. Interestingly, the T3SS gene cluster was absent in strain EC2, T4SS gene cluster was absent in WH1, EC2, and MS2. Genes inserted between vgrG and tssB within the T6SS gene cluster were different in all analyzed genomes. Both EPS and CPS gene clusters were present in WH1, MS1, and MS2, while either EPS or CPS was present in EC1 or EC2. Gene cluster encoding zeamines biosynthesis was only present in EC1. Additionally, related phenotypic and pathogenicity analyses were performed for strain WH1. The strain can produce pectate lyases, cellulases, and proteases, easily formed thick biofilm in vitro, and had strong AHL quorum sensing activity. Besides, WH1 can infect plants with serious maceration symptoms and strongly inhibit rice seed germination through other toxins instead of zeamines.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/PRJNA858112, CP101400.
Author contributions
X-JT, G-PZ, and YS conceived and designed the experiments and revised the manuscript. Z-WZ, J-JX, WW, FH, BJ, and XW performed the experiments. X-JT, XG, and LS analyzed the data. X-JT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Major Science and Technology Projects in Anhui Province (No. 202003a06020009), the National Natural Science Foundation of China (Nos. 32200152 and 31871964), the Natural Science Foundation of Anhui Province (No. 2108085QC92), and the Anhui Provincial Key Laboratory of Molecular Enzymology and Mechanism of Major Diseases (No. fzmx202005).
Conflict of interest
Author WW was employed by Wuhu Qingyijiang Seed Industry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.997486/full#supplementary-material
Footnotes
References
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., et al. (2004). UniProt: The universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119. doi: 10.1093/nar/gkh131
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bez, C., Esposito, A., Thuy, H. D., Nguyen, H. M., Vale, G., Licastro, D., et al. (2021). The rice foot rot pathogen Dickeya zeae alters the in-field plant microbiome. Environ. Microbiol. 23, 7671–7687. doi: 10.1111/1462-2920.15726
Boluk, G., Arizala, D., Dobhal, S., Zhang, J., Hu, J., Alvarez, A. M., et al. (2021). Genomic and phenotypic biology of novel strains of Dickeya zeae isolated from pineapple and taro in Hawaii: Insights into genome plasticity, pathogenicity, and virulence determinants. Front. Plant Sci. 12:663851. doi: 10.3389/fpls.2021.663851
Chatterjee, A., Cui, Y., Liu, Y., Dumenyo, C. K., and Chatterjee, A. K. (1995). Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal. N-(3-oxohexanoyl)-L-homoserine lactone. Appl. Environ. Microbiol. 61, 1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995
Chen, L., Zou, Y., She, P., and Wu, Y. (2015). Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res. 172, 19–25. doi: 10.1016/j.micres.2015.01.004
Chen, X., Zhang, Y., Zhang, Z., Zhao, Y., Sun, C., Yang, M., et al. (2018). PGAweb: A web server for bacterial Pan-genome analysis. Front. Microbiol. 9:1910. doi: 10.3389/fmicb.2018.01910
Chen, Y., Lv, M., Liao, L., Gu, Y., Liang, Z., Shi, Z., et al. (2016). Genetic modulation of c-di-GMP turnover affects multiple virulence traits and bacterial virulence in rice pathogen Dickeya zeae. PLoS One 11:e165979. doi: 10.1371/journal.pone.0165979
Davidsson, P. R., Kariola, T., Niemi, O., and Palva, E. T. (2013). Pathogenicity of and plant immunity to soft rot pectobacteria. Front. Plant Sci. 4:191. doi: 10.3389/fpls.2013.00191
Davis, J. J., Wattam, A. R., Aziz, R. K., Brettin, T., Butler, R., Butler, R. M., et al. (2020). The PATRIC bioinformatics resource center: Expanding data and analysis capabilities. Nucleic Acids Res. 48, D606–D612. doi: 10.1093/nar/gkz943
De Boer, S. H. (2002). Relative incidence of Erwinia carotovora subsp. atroseptica in stolon end and peridermal tissue of potato tubers in Canada. Plant Dis. 86, 960–964. doi: 10.1094/PDIS.2002.86.9.960
de Kievit, T. R. (2009). Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 11, 279–288. doi: 10.1111/j.1462-2920.2008.01792.x
Duprey, A., Taib, N., Leonard, S., Garin, T., Flandrois, J. P., Nasser, W., et al. (2019). The phytopathogenic nature of Dickeya aquatica 174/2 and the dynamic early evolution of Dickeya pathogenicity. Environ. Microbiol. 21, 2809–2835. doi: 10.1111/1462-2920.14627
Feng, L., Schaefer, A. L., Hu, M., Chen, R., Greenberg, E. P., et al. (2019). Virulence factor identification in the banana pathogen Dickeya zeae MS2. Appl. Environ. Microbiol. 85:e01611–e01619. doi: 10.1128/AEM.01611-19
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., et al. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Hu, M., Li, J., Chen, R., Li, W., Feng, L., Shi, L., et al. (2018). Dickeya zeae strains isolated from rice, banana and clivia rot plants show great virulence differentials. BMC Microbiol. 18:136. doi: 10.1186/s12866-018-1300-y
Hu, M., Xue, Y., Li, C., Lv, M., Zhang, L., Parsek, M. R., et al. (2022). Genomic and functional dissections of Dickeya zeae shed light on the role of type III secretion system and cell wall-degrading enzymes to host range and virulence. Microbiol. Spectr. 10:e159021. doi: 10.1128/spectrum.01590-21
Hussain, M. B., Zhang, H. B., Xu, J. L., Liu, Q., Jiang, Z., et al. (2008). The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J. Bacteriol. 190, 1045–1053. doi: 10.1128/JB.01472-07
Jahn, C. E., Willis, D. K., and Charkowski, A. O. (2008). The flagellar sigma factor fliA is required for Dickeya dadantii virulence. Mol. Plant Microbe Interact. 21, 1431–1442. doi: 10.1094/MPMI-21-11-1431
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280. doi: 10.1093/nar/gkh063
Kraepiel, Y., and Barny, M. A. (2016). Gram-negative phytopathogenic bacteria, all hemibiotrophs after all? Mol. Plant Pathol. 17, 313–316. doi: 10.1111/mpp.12345
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., et al. (2007). RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Laslett, D., and Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. doi: 10.1093/nar/gkh152
Li, J., Hu, M., Xue, Y., Chen, X., Lu, G., Zhang, L., et al. (2020). Screening, identification and efficacy evaluation of antagonistic bacteria for biocontrol of soft rot disease caused by Dickeya zeae. Microorganisms 8:697. doi: 10.3390/microorganisms8050697
Liao, L., Cheng, Y., Liu, S., Zhou, J., An, S., Lv, M., et al. (2014). Production of novel antibiotics zeamines through optimizing Dickeya zeae fermentation conditions. PLoS One 9:e116047. doi: 10.1371/journal.pone.0116047
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Liu, F., Hu, M., Zhang, Z., Xue, Y., Chen, S., Hu, A., et al. (2022). Dickeya manipulates multiple quorum sensing systems to control virulence and collective behaviors. Front. Plant Sci. 13:838125. doi: 10.3389/fpls.2022.838125
Lv, M., Chen, Y., Hu, M., Yu, Q., Duan, C., Ye, S., et al. (2022). OhrR is a central transcriptional regulator of virulence in Dickeya zeae. Mol. Plant Pathol. 23, 45–59. doi: 10.1111/mpp.13141
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Merritt, J. H., Kadouri, D. E., and O’Toole, G. A. (2005). Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00
Pérombelon, M. C. M. (1992). Potato blackleg: Epidemiology, host-pathogen interaction and control. Netherlands J. Plant Pathol. 98, 135–146. doi: 10.1007/BF01974480
Potrykus, M., Hugouvieux-Cotte-Pattat, N., and Lojkowska, E. (2018). Interplay of classic Exp and specific Vfm quorum sensing systems on the phenotypic features of Dickeya solani strains exhibiting different virulence levels. Mol. Plant Pathol. 19, 1238–1251. doi: 10.1111/mpp.12614
Pu, J., Zhang, S., He, X., Zeng, J., Shen, C., Luo, Y., et al. (2022). The small RNA AmiL regulates quorum sensing-mediated virulence in Pseudomonas aeruginosa PAO1. Microbiol. Spectr. 10:e221121. doi: 10.1128/spectrum.02211-21
Reverchon, S., and Nasser, W. (2013). Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 5, 622–636. doi: 10.1111/1758-2229.12073
Reverchon, S., Muskhelisvili, G., and Nasser, W. (2016). Virulence program of a bacterial plant pathogen: The dickeya model. Prog. Mol. Biol. Transl. Sci. 142, 51–92. doi: 10.1016/bs.pmbts.2016.05.005
Richter, M., Rossello-Mora, R., Oliver, G. F., and Peplies, J. (2016). JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., et al. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tan, X., Cheng, X., Hu, M., Zhang, Y., Jia, A., Zhou, J., et al. (2021). Transcriptional analysis and target genes discovery of Pseudomonas aeruginosa biofilm developed ex vivo chronic wound model. AMB Express 11:157. doi: 10.1186/s13568-021-01317-2
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Toth, I. K., Barny, M., Brurberg, M. B., Condemine, G., Czajkowski, R., Elphinstone, J. G., et al. (2021). “Pectobacterium and Dickeya: environment to disease development,” in Plant Diseases Caused by Dickeya and Pectobacterium Species, eds F. Van Gijsegem, J. M. van der Wolf, and I. K. Toth (Cham: Springer), doi: 10.1007/978-3-030-61459-1_3
Van Gijsegem, F., Toth, I. K., and van der Wolf, J. M. (2021). “Soft Rot Pectobacteriaceae: a brief overview,” in Plant Diseases Caused by Dickeya and Pectobacterium Species, eds F. Van Gijsegem, J. M. van der Wolf, and I. K. Toth (Cham: Springer), doi: 10.1007/978-3-030-61459-1_1
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Zhang, J., Arif, M., Shen, H., Hu, J., Sun, D., Pu, X., et al. (2020). Genomic divergence between Dickeya zeae strain EC2 isolated from rice and previously identified strains, suggests a different rice foot rot strain. PLoS One 15:e240908. doi: 10.1371/journal.pone.0240908
Zhang, J., Arif, M., Shen, H., Sun, D., Pu, X., Hu, J., et al. (2022). Genomic comparisons and phenotypic diversity of Dickeya zeae strains causing bacterial soft rot of banana in China. Front. Plant Sci. 13:822829. doi: 10.3389/fpls.2022.822829
Zhang, J., Shen, H., Pu, X., Lin, B., and Hu, J. (2014). Identification of Dickeya zeae as a causal agent of bacterial soft rot in banana in China. Plant Dis. 98, 436–442. doi: 10.1094/PDIS-07-13-0711-RE
Zhang, W., Fan, X., Li, J., Ye, T., Mishra, S., Zhang, L., et al. (2021). Exploration of the quorum-quenching mechanism in Pseudomonas nitroreducens W-7 and its potential to attenuate the virulence of Dickeya zeae EC1. Front. Microbiol. 12:694161. doi: 10.3389/fmicb.2021.694161
Zhou, J., Bi, S., Chen, H., Chen, T., Yang, R., Li, M., et al. (2017). Anti-biofilm and antivirulence activities of metabolites from Plectosphaerella cucumerina against Pseudomonas aeruginosa. Front. Microbiol. 8:769. doi: 10.3389/fmicb.2017.00769
Keywords: Dickeya zeae, bacteria soft rot of rice, comparative genomics, pathogenicity determinants and virulence factors, phenotypic diversity
Citation: Tan X-J, Zhang Z-W, Xiao J-J, Wang W, He F, Gao X, Jiang B, Shen L, Wang X, Sun Y and Zhu G-P (2022) Genomic and phenotypic biology of a novel Dickeya zeae WH1 isolated from rice in China: Insights into pathogenicity and virulence factors. Front. Microbiol. 13:997486. doi: 10.3389/fmicb.2022.997486
Received: 19 July 2022; Accepted: 03 October 2022;
Published: 28 October 2022.
Edited by:
Mohammad Arif, University of Hawai’i at Mānoa, United StatesReviewed by:
Janak Raj Joshi, Colorado State University, United StatesDivine Yufetar Shyntum, Silesian University of Technology, Poland
Ravinder Kumar, Central Potato Research Institute (ICAR), India
Copyright © 2022 Tan, Zhang, Xiao, Wang, He, Gao, Jiang, Shen, Wang, Sun and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Sun, MjAxODI1OUBhaG51LmVkdS5jbg==; Guo-Ping Zhu, Z3B6MjAxMkBhaG51LmVkdS5jbg==
 Xiao-Juan Tan
Xiao-Juan Tan Zhi-Wei Zhang1
Zhi-Wei Zhang1 Feng He
Feng He Xuan Gao
Xuan Gao Bin Jiang
Bin Jiang Liang Shen
Liang Shen Yang Sun
Yang Sun Guo-Ping Zhu
Guo-Ping Zhu