- 1Guangdong Academy of Sciences, Institute of Microbiology, Key Laboratory of Agricultural Microbiomics and Precision Application (MARA), Provincial Key Laboratory of Microbial Culture Collection and Application, Key Laboratory of Agricultural Microbiome (MARA), State Key Laboratory of Applied Microbiology Southern China, Guangzhou, China
- 2College of Life Science, Huizhou University, Huizhou, China
- 3College of Horticulture, South China Agricultural University, Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Guangzhou, China
Myxobacteria are recognized for fascinating social behaviors and producing diverse extracellular active substances. Isolating novel myxobacteria is of great interest in the exploitation of new antibiotics and extracellular enzymes. Herein, four novel strains were isolated from Dinghu Mountain Biosphere Reserve, Guangdong province, and Qinghai virgin forest soils, Qinghai province, China. The phylogenetic analysis based on 16S rRNA gene and genomic sequences indicated that the four strains belong to the genera Myxococcus and Pyxidicoccus, sharing the highly similarities of 16S rRNA gene with the genera Myxococcus and Pyxidicoccus (99.3–99.6%, respectively). The four strains had average nucleotide identity (ANI) values of 82.8–94.5%, digital DNA–DNA hybridization (dDDH) values of 22.2–56.6%, average amino acid identity (AAI) values of 75.8–79.1% and percentage of conserved protein (POCP) values of 66.4–74.9% to members of the genera Myxococcus and Pyxidicoccus. Based on phylogenetic analyses, physiological and biochemical characteristics, and comparative genomic analyses, we propose four novel species of the genera Myxococcus and Pyxidicoccus and further supported the two genera above represented the same genus. Description of the four novel species is Myxococcus guangdongensis sp. nov. (K38C18041901T = GDMCC 1.2320T = JCM 39260T), Myxococcus qinghaiensis sp. nov. (QH3KD-4-1T = GDMCC 1.2316T = JCM 39262T), Myxococcus dinghuensis sp. nov. (K15C18031901T = GDMCC 1.2319T = JCM 39259T), and Pyxidicoccus xibeiensis sp. nov. (QH1ED-7-1T = GDMCC 1.2315T = JCM 39261T), respectively. Furthermore, comparative genomics of all 15 species of the genera Myxococcus and Pyxidicoccus revealed extensive genetic diversity. Core genomes enriched more genes associated with housekeeping functional classes while accessory genomes enriched more genes related to environmental interactions, indicating the former is relatively indispensable compared to signaling pathway genes. The 15 species of Myxococcus and Pyxidicoccus also exhibited great gene diversity of carbohydrate-active enzymes (CAZymes) and secondary metabolite biosynthesis gene clusters (BGCs), especially related to glycosyl transferases (GT2 and GT4), glycoside hydrolases (GH13 and GH23), non-ribosomal peptide synthetases (NRPS), and Type I polyketide synthase (PKS)/NRPS hybrids.
Introduction
Myxobacteria are rod-shaped gram-negative bacteria and are distributed across the world (Mohr, 2018). Compared to research into their sophisticated social lifestyle, unstable morphological, and slow-growth characteristics (Shimkets et al., 2006; Awal et al., 2016), myxobacterial taxonomic research publications are relatively lacking. According to the latest taxonomic proposal, myxobacteria have been reclassified as the phylum Myxococcota, comprising the four orders Myxococcales, Polyangiales, Nannocystales, and Haliangiales, seven families, and 19 genera (Waite et al., 2020). The three genera Myxococcus, Pyxidicoccus, and Corallococcus of the family Myxococcaceae have often been isolated from various habitats, and their 16S rRNA genes show highly homologous (Mohr et al., 2016, 2017; Livingstone et al., 2017; Wang et al., 2020). Compared with the 98.65% of 16S rRNA gene sequence similarity, the 98.7% of gyrB gene sequence similarity is suitable as the threshold for species boundaries of the three genera above (Liu et al., 2021). With the advance of next-generation sequencing technologies, comparative genomics analysis has been applied to identify and classify myxobacterial species. Just recently, eight novel Corallococcus, three novel Myxococcus, and two novel Pyxidicoccus species were identified based on polyphasic taxonomy combined with pan-genomic analyses (Chambers et al., 2020; Livingstone et al., 2020).
Myxobacteria exhibit genome sizes ranging from 9.0 to 16.0 Mbp and the genomic DNA G + C contents around 70.0 mol% (Sharma et al., 2016; Sharma and Subramanian, 2017). The core genomes may meet myxobacterial requirements for gliding motility, group predation, multicellular fruiting body, and myxospore formation (Wrotniak-Drzewiecka et al., 2016). Moreover, myxobacteria show a wide range of predatory activity profiles against various bacteria and some fungi, but the action mode remains largely unknown (Thiery and Kaimer, 2020). Myxobacterial predatory activity seemed to be not congruent with phylogeny (Livingstone et al., 2017). Gliding motility, direct contact with prey, and lysis are required for predation (Thiery and Kaimer, 2020). Meanwhile, the outer membrane vesicles (OMVs) secreted by myxobacteria are packed with a cargo of secondary metabolites and hydrolytic enzymes. OMVs and OMV-free culture supernatant proved to kill prey cells (Evans et al., 2012; Berleman et al., 2014; Livingstone et al., 2018). Recently, a newly discovered type IV filament-like machinery (kil) that could promote myxobacterial motility arrest and prey cell plasmolysis has been proposed to be related to contact-dependent killing (Seef et al., 2021).
Herein, according to phylogenetic analyses, physiology and biochemical characterization, and genomic average nucleic acid identity (ANI), digital DNA–DNA hybridization (dDDH), average amino acid identity (AAI), and percentage of conserved protein (POCP) values, we propose four novel species of the Myxococcus and Pyxidicoccus and further support these two genera represented the same genus, which was proposed by Chambers et al. (2020). Furthermore, we carry out pan-genomics and compare the functional distribution of core, accessory, and unique genes of Myxococcus and Pyxidicoccus. We also analyze the diversity of carbohydrate-active enzymes (CAZymes) genes and secondary metabolite biosynthesis gene clusters (BGCs) of Myxococcus and Pyxidicoccus. This study provides four novel myxobacterial species for the exploitation of new secondary metabolites and extracellular enzymes and will deepen insights into the genetic and gene diversity of Myxococcus and Pyxidicoccus.
Materials and methods
Bacterial isolation and identification
Strains K15C18031901T and K38C18041901T were isolated from Dinghu Mountain Biosphere Reserve collected on 12 September 2017, located at Zhaoqing, Guangdong province, China, and strains QH3KD-4-1T and QH1ED-7-1T were isolated from Qinghai virgin forest soils collected on 6 October 2020, located at Xining, Qinghai province, China. The isolation method for myxobacteria was described by Wang C. L. et al. (2019) using Escherichia coli as baits. Isolates were picked on modified VY/2 agar (Baker’s yeast 5 g L–1, CaCl2⋅2H2O 1 g L–1, agar 15 g L–1, and pH 7.2), and a pure culture was obtained by the repeated subculture of cells from the edge of the colony. Upon purification, strains were stored at −80°C as suspensions containing 25% glycerol.
Phylogenetic analysis based on 16S rRNA, gyrB genes, and genomic sequences
The genomic DNAs of four isolates above were extracted using the HiPure Bacterial DNA kit (Magen Biotech Co., Ltd., Guangzhou, China) according to the manufacturer’s instructions. The 16S rRNA gene was amplified using the genomic DNAs as templates and sequenced using the universal primers 27F and 1492R (Kai et al., 2006). The sequences obtained were submitted to similarity searches in the EzBioCloud1 and NCBI2 databases. The phylogenetic tree of 16S rRNA gene sequences was reconstructed based on the maximum-likelihood (ML) method with the model of Jukes–Cantor and ultrafast bootstrapping (-bb 1000) using the MEGA 7.0 software (Kumar et al., 2016).
The genomes were sequenced using the Illumina NovaSeq PE150 platform by Shanghai Majorbio Bio-Pharm Technology Co., Ltd. The clean data, generated from raw data by quality trimming, were assembled using the SPAdes (v3.11.1) with default parameters to obtain contigs and scaffolds. The genomic sequences of K15C18031901T, K38C18041901T, QH3KD-4-1T, and QH1ED-7-1T were deposited at the NCBI3 database under the accession numbers JAKCFB000000000, JAJVKW000000000, JAKCFA000000000, and JAJVKV000000000, respectively. Furthermore, a phylogenomic tree was reconstructed based on 92 up-to-date core genes using the UBCG tools with a maximum-likelihood algorithm. In addition, gyrB genes were extracted from genome sequences, and sequence similarities were compared.
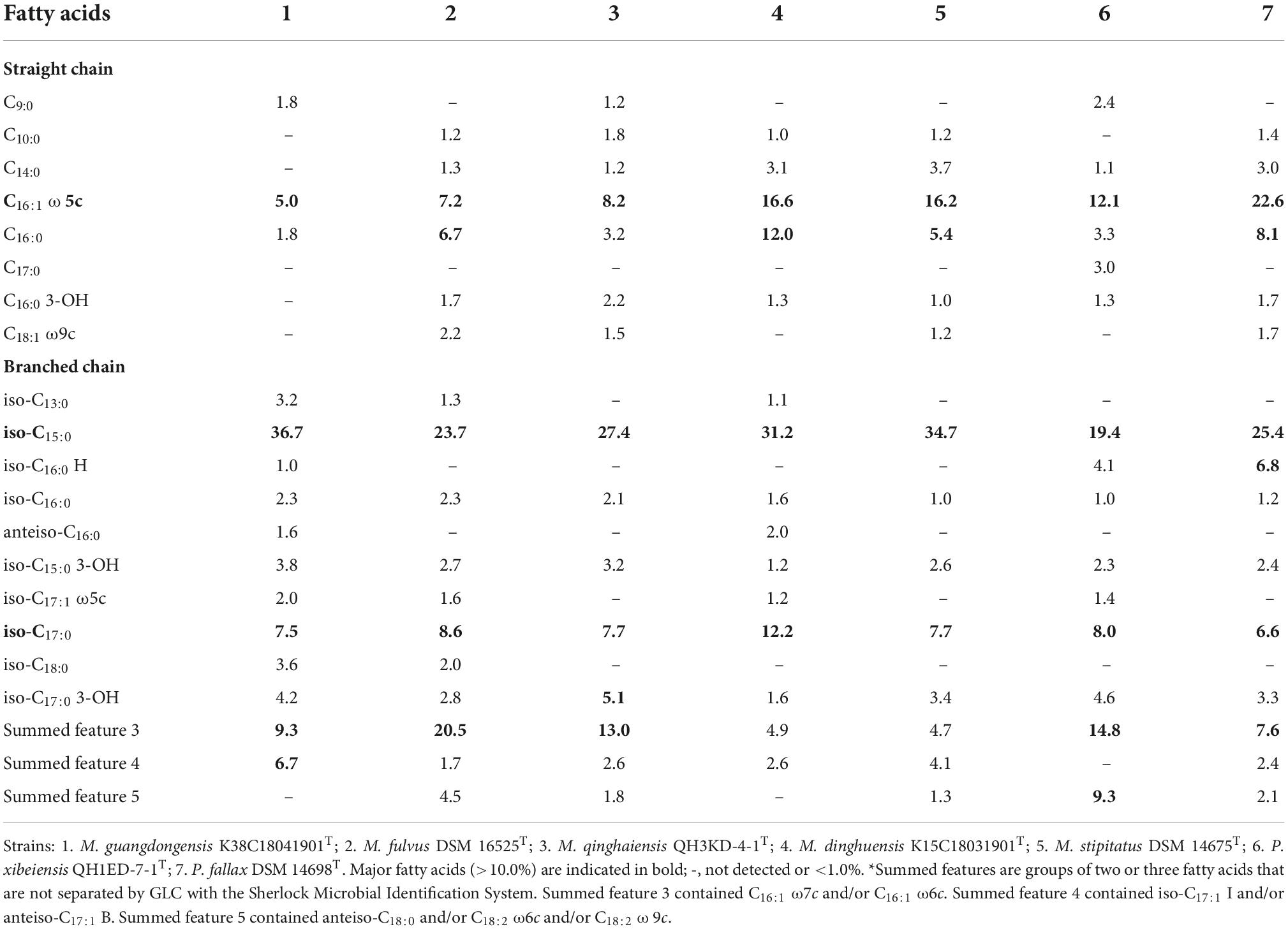
Physiology characterization and fatty acid analyses
Fruiting bodies, swarming colonies, vegetative cells, and myxospores grown on VY/2 agar for 5–10 days at 30°C were observed and measured by stereomicroscope (Leica, M165C), field scanning electron microscope (SEM, HITACHI, S-3000N), and Nano Measurer 1.2 software, respectively. The gram staining reaction was performed by using the Gram Stain Kit according to the manufacturer’s instructions. The optimal temperatures for growth were investigated on VY/2 agar at 30, 35, 37, and 40°C for 5–7 days. Tolerance to NaCl concentrations (1–4%, at intervals of 1%, w/v, NaCl) and pH range (pH 5.0–9.0, at intervals of 1 unit) was performed on VY/2 agar at 30°C for 5–7 days. Enzymatic activities and substrate utilization were tested using the API ZYM and 20NE according to the manufactures’ instructions. Cellular fatty acids were extracted from cells grown on VY/2 agar at 30°C for 5 days using the Sherlock Microbial Identification System (MIDI) protocol (version 6.1). Fatty acids were analyzed by gas chromatography (model 7890A, Hewlett Packard) using the Sherlock Microbial Identification System (Sherlock version 6.1; TSBA6) (Sasser, 1990).
Predation assays
The predatory activities of the four isolates were tested based on the method of Livingstone et al. (2017) using Escherichia coli ATCC 8739, Micrococcus luteus NCTC 2665, Salmonella typhimurium GDMCC 1.239, and Staphylococcus aureus GDMCC 1.1220 as prey. These prey were obtained from the Guangdong microbial culture collection center (GDMCC). The predatory zone on the prey lawn was observed after 3 days incubated at 30°C.
Comparative genomic analyses
The other genomes of 11 type species within the genera Myxococcus and Pyxidicoccus were downloaded from the NCBI database for comparative analyses. The contigs (shorter than 500 bp) of used draft genomes above were removed. The completeness and contamination values of all 15 genomes were assessed by the software CheckM version 1.0.9. For consistency, the gene prediction and genomic annotation were reperformed using the software Prokka (v1.14.6) and the Rapid Annotation using Subsystem Technology (RAST) version 2.04 with default parameters. The ANI and AAI among all species were calculated by fastANI (v1.32) and compareM (v0.1.2), respectively. The dDDH values were performed using Genome-to-Genome Distance Calculator (GGDC; v2.1).5 The POCP values were calculated as [(C1 + C2)/(T1 + T2)] × 100, where C represents the number of conserved proteins (identity, ≥40%; aligned length of query, ≥50%) and T represents the number of total proteins; 1 and 2 refer to input files 1 and 2, respectively (Qin et al., 2014). The ANI, dDDH, AAI, and POCP values were visualized using the “Heatmap” tool of the TBtools. The pan- and core genomes were calculated using OrthoFinder (v2.5.2) based on the “Diamond blastp with default parameters” and “MCL” algorithms to generate orthologous clusters. The representative sequences of each gene family were searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG)6 database. In addition, the genes encoding CAZymes were identified using dbCAN2 meta server with HMMER annotation (E-Value < 1e-15, coverage > 0.35), and the BGCs were annotated using antiSMASH bacterial version 6.1.1 with default parameters, respectively. Identified clusters were further determined using MultiGeneBlast as previously described (Medema et al., 2013; Adamek et al., 2017).
Results
Phylogenetic analyses based on 16S rRNA, gyrB gene, and genomic sequences
The 16S rRNA gene sequences of strains K38C18041901T (1398 bp; GenBank accession number: OM049394), K15C18031901T (1432 bp; GenBank accession number: OM049393), QH3KD-4-1T (1395 bp; GenBank accession number: OM049399), and QH1ED-7-1T (1432 bp; GenBank accession number: OM049395) were obtained according to the methods described in the “Materials and methods” section. Strains K38C18041901T, K15C18031901T, and QH3KD-4-1T showed the highest 16S rRNA gene sequence similarities of 99.4, 99.3, and 99.6%, respectively, with Myxococcus fulvus DSM 16525T, Myxococcus stipitatus DSM 14675T, and Myxococcus eversor AB053BT. Strain QH1ED-7-1T exhibited the highest 16S rRNA gene similarity of 99.3% with Pyxidicoccus trucidator CA060AT (Supplementary Table 1). Moreover, the ML phylogenetic tree reconstructed based on 16S rRNA gene sequences also indicated that the four strains above were affiliated with the genera Myxococcus and Pyxidicoccus (Figure 1A).
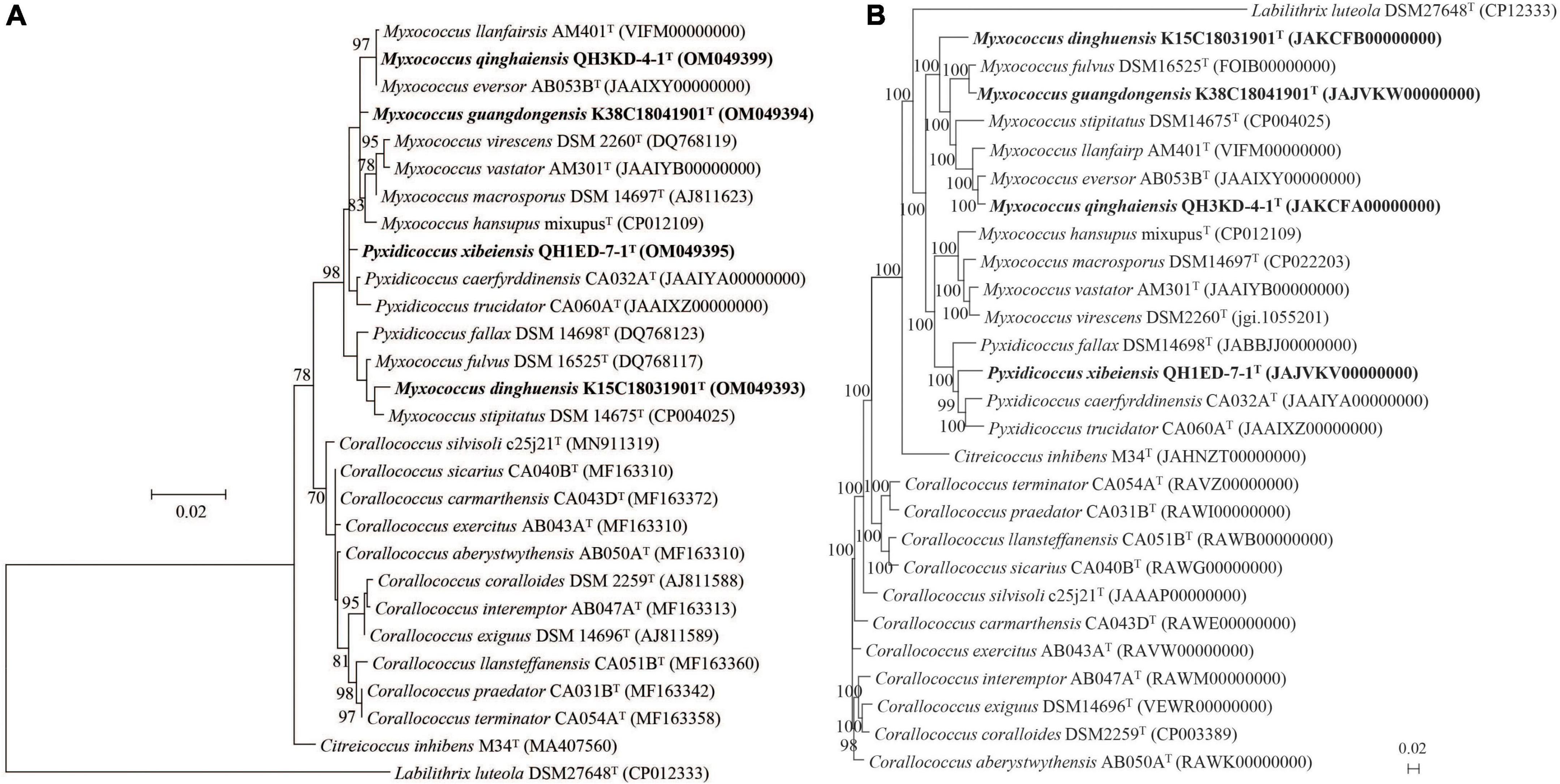
Figure 1. Phylogenetic trees among four putative species and their closely related species within the genera Myxococcus, Pyxidicoccus, and Corallococcus. (A) The ML tree based on 16S rRNA gene sequences is generated using the Jukes–Cantor model by the MEGA 7.0 software. (B) The UBCG phylogenomic tree based on 92 up-to-date bacterial core genes sequences is reconstructed using the ML algorithm. GenBank accession numbers are shown in parentheses. Bootstrap values (represented percentages of 1,000 replications) >70% are shown at nodes. Labilithrix luteola DSM 27648T is used as an outgroup. Bar, 0.02 substitutions per nucleotide position.
The genome lengths of four strains above were 10.5–12.8 Mbp with the genomic DNA G + C contents of 68.8–70.7 mol%. Other general genomic features of the four strains are listed in Supplementary Tables 1, 2. The genomic annotation result of the four strains by RAST pipeline is shown in Supplementary Figure 1. Furthermore, the phylogenomic tree suggested that strains K15C18031901T, K38C18041901T, and QH3KD-4-1T formed a large clade with the genus Myxococcus and strain QH1ED-7-1T formed a clade with the genus Pyxidicoccus. In addition, strain QH3KD-4-1T was grouped with M. eversor AB053BT and strain K38C18041901T was grouped with M. fulvus DSM 16525T, respectively (Figure 1B). More specifically, four strains of the genus Pyxidicoccus formed a clade with four species of the genus Myxococcus (Figure 1B).
The gyrB gene sequences were extracted from genomic sequences. Strains K38C18041901T, K15C18031901T, QH3KD-4-1T, and QH1ED-7-1T showed the highest gyrB gene sequence similarities of 94.0, 95.9, 91.4, and 93.7% with M. fulvus DSM 16525T, M. stipitatus DSM 14675T, M. eversor AB053BT, and P. trucidator CA060AT, respectively (Supplementary Table 1), all of which were lower than 98.7% threshold for species boundaries of three genera Myxococcus, Pyxidicoccus, and Corallococcus (Liu et al., 2021), indicating the four strains were potential new species.
Genomic average nucleotide identity, digital DNA–DNA hybridization, amino acid identity, and percentage of conserved protein analyses
The ANI, dDDH, AAI, and POCP values were calculated to identify the genomic similarities of the four strains above with other species of the two genera Myxococcus and Pyxidicoccus. The four strains showed 82.8–94.5% ANI values with their closely related species, which were lower than the threshold value of 95–96% for species discrimination (Figure 2A; Richter and Rosselló-Móra, 2009). Moreover, the four strains exhibited 22.2–56.6% dDDH values with other related species, which were lower than the 70% cutoff value for species boundaries (Figure 2B; Chun et al., 2018). These results indicated the four strains above represent four novel genospecies. In addition, four strains of the genus Pyxidicoccus showed 75.8–79.1% AAI values with ten species of the genus Myxococcus (Figure 2C), which were higher than the threshold value of 65–72% for the genus boundaries (Konstantinos and James, 2007). Similarly, four strains of the genus Pyxidicoccus exhibited 66.4–74.9% POCP values with other species of Myxococcus, which were higher than 50% for the genus boundary (Qin et al., 2014). These results indicated members of the genus Pyxidicoccus were affiliated with the genus Myxococcus, as determined in the study by Chambers et al. (2020). Based on 16S rRNA and gyrB gene sequence similarities, genomic phylogenetic analyses, ANI, dDDH, AAI, and POCP values, the four strains represented four novel genospecies, and the two genera Pyxidicoccus and Myxococcus should belong to the same genus. Considering the close phylogenetic relationships, the type species of M. fulvus DSM 16525T, M. eversor AB053BT, M. stipitatus DSM 4675T, and Pyxidicoccus fallax DSM 14698T were selected for physiochemical analyses.
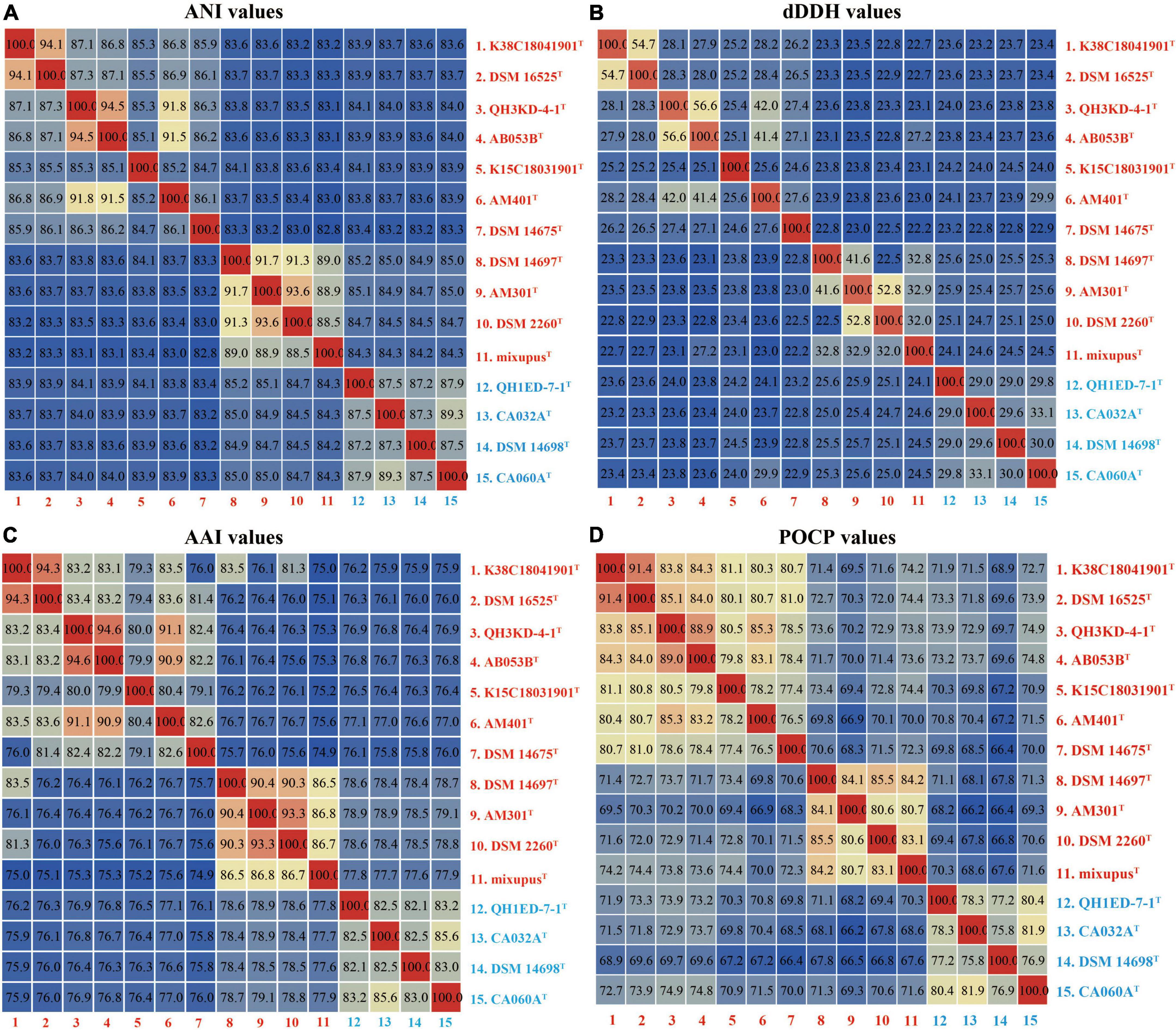
Figure 2. Heatmaps of the ANI (A), dDDH (B), AAI (C), and POCP (D) values. The results are visualized using the “Heatmap” tool of the TBtools.
Physiology characterization and fatty acid analyses
The morphological characteristics of the four putative species grown on VY/2 agar are shown in Supplementary Figure 2. Fruiting bodies were series or single, roundish or oval (diameters of 150–470 μm), and multiple brightly colored grown on VY/2 agar (Supplementary Figures 2A–D). By gliding movement, colonies develop as thin, spreading swarms (Supplementary Figures 2E–H). All vegetative cells were gram-negative slender rod-shaped, measuring an average size of 0.30–0.45 μm × 2.5–6.0 μm, and formed circular myxospores (0.9–1.1 μm diameters) under field scanning electron microscope (Supplementary Figures 2I–M). Growth at different temperatures indicated that strains K38C18041901T, QH3KD-4-1T, and K15C18031901T grew at 30°C, while their closely related species M. fulvus DSM 16525T, M. eversor AB053BT, and M. stipitatus DSM 14675T grew at 35–40°C (Table 1). The optimum pH was 7.0 for all species with only three strains M. fulvus DSM 16525T, M. eversor AB053BT, and M. stipitatus DSM 14675T exhibiting growth at pH 5.0 (Table 1). All strains can tolerate 1% (w/v) NaCl, while strains K38C18041901T and K15C18031901T can tolerate up to 2% (w/v) NaCl. Exceptionally, type species M. eversor AB053BT can tolerate up to 4% (w/v) NaCl (Table 1). The biochemical results using API ZYM and 20NE tests are shown in Table 1. Strain QH1ED-7-1T showed a similar activity profile with P. fallax DSM 14698T except for acid phosphatase, β-glucosidase, and β-galactosidase. Strain K38C18041901T was negative for N-acetyl-β-glucosaminidase but positive for β-galactosidase, while its closely related species M. fulvus DSM 16525T was opposite. Strain QH3KD-4-1T was negative for the utilization of β-glucosidase, arginine dihydrolase, and urease, while its closely related species M. eversor AB053BT was all positive. Strain K15C18031901T was negative for utilization of urease, while its closely related species were positive. Additional differences between the four strains and their related species are shown in Table 1. In addition, the four strains showed broad predatory abilities against prey E. coli, M. luteus, S. typhimurium, and S. aureus (Supplementary Figure 3).
The major fatty acids (>5.0%) identified in the four strains and the closely related species included straight-chain fatty acids (SCFAs) of C16:0 ω5c (5.0–22.6%) and branched-chain fatty acids (BCFAs) of iso-C15:0 (19.4–36.7%) and iso-C17:0 (6.6–12.2%) (Table 2). The abundance of C16:0 ω5c and iso-C15:0, major biomarker fatty acids for members of the Myxococcus and Pyxidicoccus (Garcia et al., 2011), indicated that the four strains belonged to the genus Myxococcus and Pyxidicoccus. Besides, strain K38C18041901T contained a lower amount of C16:0, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and a higher amount of iso-C15:0 and summed feature 4 (iso-C17:1 I and/or anteiso-C17:1 B) than M. fulvus DSM 16525T. Strain K15C18031901T contained a higher amount of C16:0 and iso-C17:0 than M. stipitatus DSM 14675T. Strain QH1ED-7-1T contained a lower amount of C16:0 ω5c, C16:0 and a higher amount of summed features 3 and 5 (ante-C18:0 and/or C18:2 ω6c, ω9c) (Table 2). These results indicated that the four strains should belong to four novel species of the genera Myxococcus and Pyxidicoccus.
Proposal of four novel species of the genera Myxococcus and Pyxidicoccus
Based on 16S rRNA and gyrB gene sequence similarities, genomic phylogenetic analyses, genomic ANI, dDDH, AAI, and POCP values, physiology characterizations, and fatty acid profiles, we propose four novel species: Myxococcus guangdongensis sp. nov. (type strain K38C18041901T, the same below), Myxococcus dinghuensis sp. nov. (K15C18031901T), Myxococcus qinghaiensis sp. nov. (QH3KD-4-1T), and Pyxidicoccus xibeiensis sp. nov. (QH1ED-7-1T), respectively. These analyses further supported that the two genera Myxococcus and Pyxidicoccus should be the same genus.
Pan-genome analyses of the genera Myxococcus and Pyxidicoccus
General genomic characteristics of all 15 species of the genera Myxococcus and Pyxidicoccus are listed in Supplementary Table 2. Three of them were completely assembled with circle maps, whereas the others were draft genomes with multiple contigs and nucleic acid gaps. Furthermore, all of them were determined to be near-complete genomes with high completeness (>98%) and low contamination (<5.0%). The genome sizes ranged from 9.0 to 13.5 Mbp containing 7,258–10,697 genes with genomic DNA G + C contents of 68.7–70.7 mol%. By using “diamond blastp with default parameters” and “MCL” clustering, 132,667 putative protein-coding genes from 15 genomes were clustered into 17,415 gene families forming the pan-genome. We categorized the gene families based on the number of strains in each cluster. Core genes were present in all 15 genomes, accessory genes were present in 2–14 genomes, and unique genes were present only in a genome (Figure 3A). The pan-genome consisted of 4,177 (24.0%) core gene families, 8,115 (46.6%) accessory gene families, and 5,123 (29.3%) unique gene families (Figure 3B). Furthermore, the numbers of strain-specific genes ranged from 140 to 798, respectively (Figure 3C).
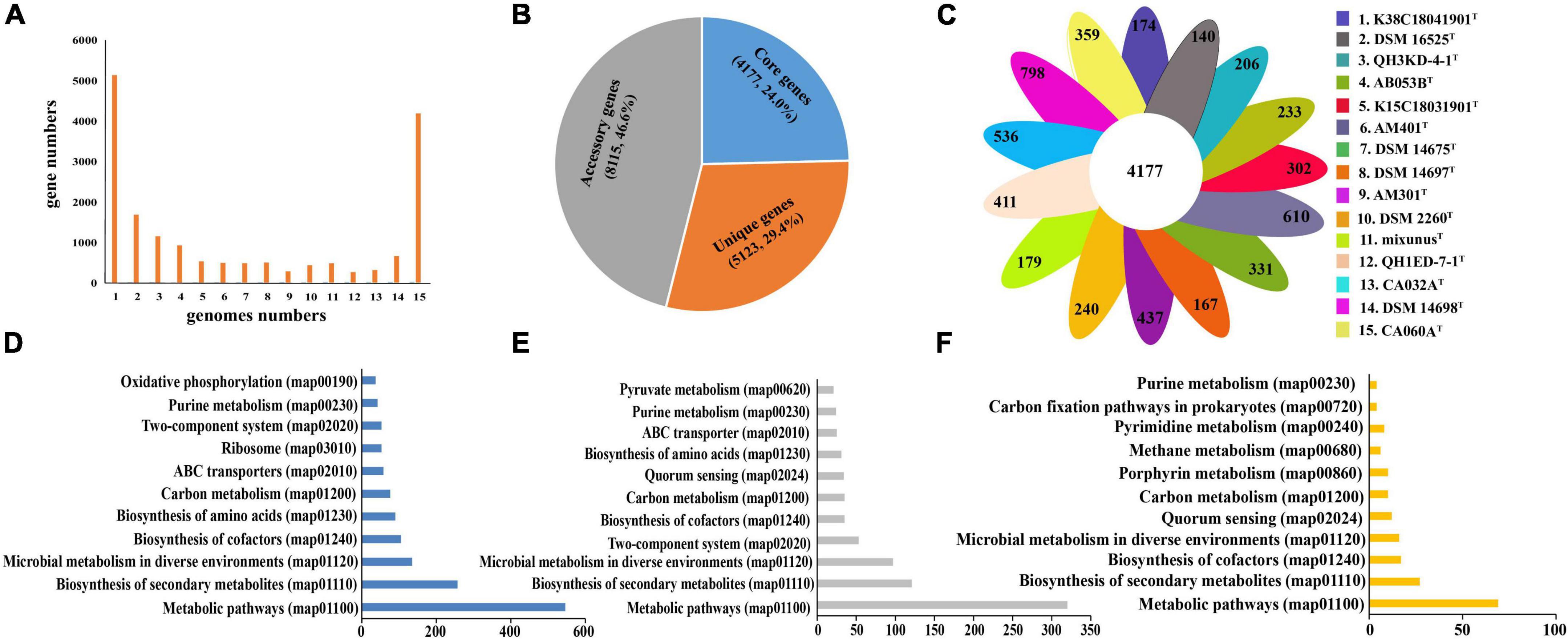
Figure 3. Distribution of gene families in the 15 genomes of the genera Myxococcus and Pyxidicoccus. (A) Numbers of gene families represented within the respective numbers of 15 genomes. (B) Numbers and percentages of core, accessory, and unique genes in 15 genomes. (C) Numbers of unique genes in each species. (D–F) Numbers of core genes (D), accessory genes (E), and unique genes (F) that associate with each indicated KEGG pathway maps.
To compare the functional distribution of the core, accessory, and unique genes, we extracted protein sequences from the three sections to map to the KEGG Orthology (KO) database. In total, 48.9% (2012) of core entries, 20.0% (1621) of accessory entries, and only 3.9% (200) of unique entries were annotated. Most of the KO-annotated genes in the three genomes were related to metabolic pathways, especially biosynthesis of secondary metabolites, microbial metabolism in diverse environments, and biosynthesis of cofactors and amino acids (Figures 3E–G). In detail, the core genomes enriched more genes associated with housekeeping functional classes, such as ribosome and oxidative phosphorylation, while the accessory and unique genomes enriched more genes related to environmental interactions, such as quorum sensing (QS) (Figures 3D–F).
The analyses of carbohydrate-active enzymes and biosynthesis gene clusters of the genera Myxococcus and Pyxidicoccus
We compared and screened genes encoding CAZymes in the 15 genomes. The pan-genome of the 15 species encoded 315 different CAZyme members, including 106 (33.7%) falling into core gene families, 184 (65.4%) falling into accessory gene families, and 25 (7.9%) falling into unique gene families (Supplementary Figure 4A). The types and numbers of glycosyl transferases (GTs) in core genes were much more than those in accessory genes (Supplementary Figures 4B,C). Further comparative analysis showed the 15 species encoded large numbers of GH13, GH23, GT2, and GT4 members (Figure 4A). These families were found to be putatively assigned to multiple enzyme activities, such as a-1,4-glucan branching enzyme (GH13), lysozyme type G, peptidoglycan lyase and chitinase (GH23), chitin and cellulose synthase (GT2), and sucrose synthase and a-glucosyltransferase (GT4). The results also exhibited multiple enzyme activities of the 15 species with predicted glucoamylase (GH18), glucodextranase (GH15), acetylgalactosaminidase (GH109), and galactosidase and glucosidase (GH16), though at a much lower abundance (Figure 4A).
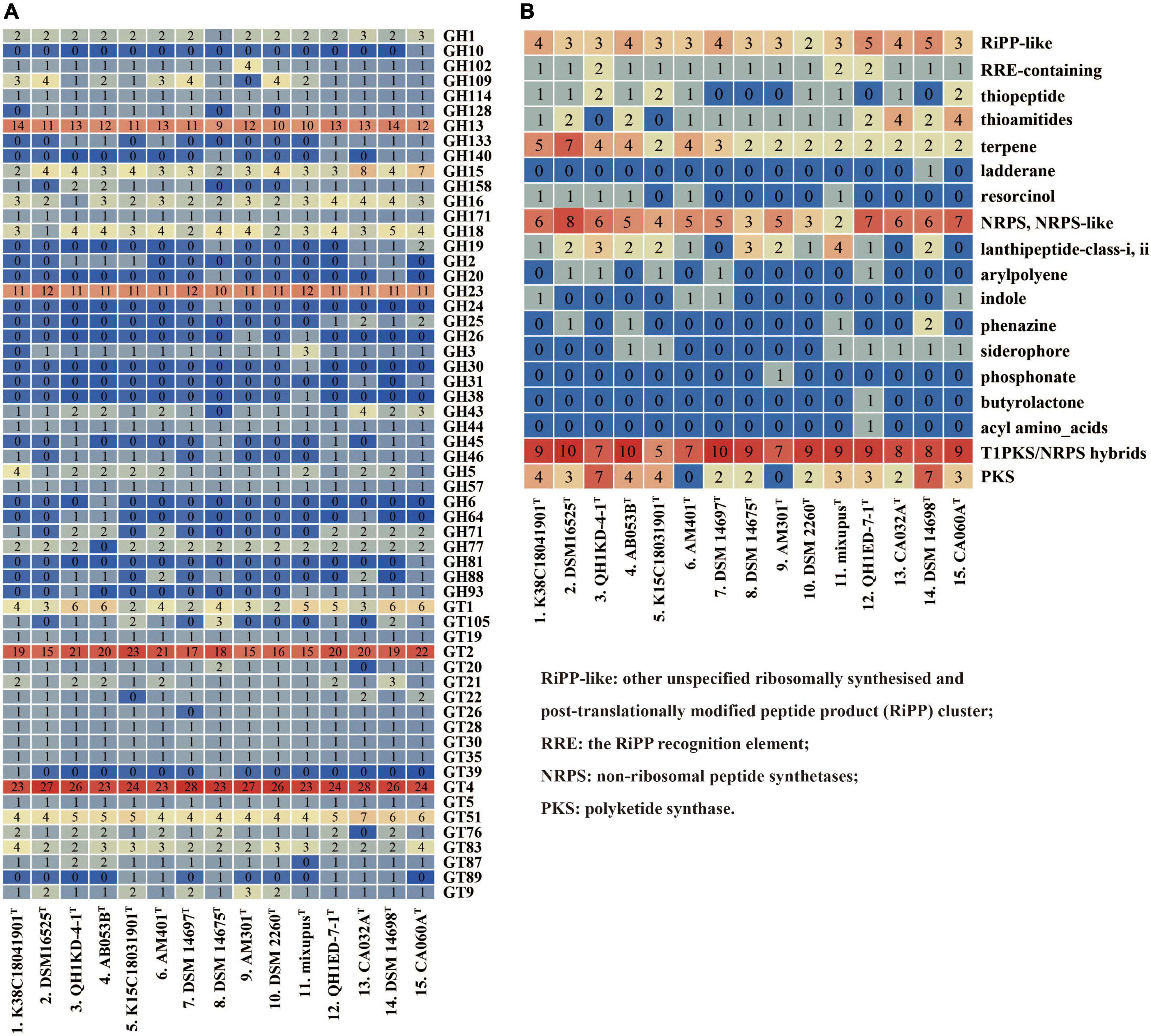
Figure 4. Type counts of GTs and GHs families (A) and BGCs (B) identified within the genera Myxococcus and Pyxidicoccus. RiPP-like, other unspecified ribosomally synthesized and post-translationally modified peptide product (RiPP) cluster; RRE, the RiPP recognition element; NRPS, non-ribosomal peptide synthetases; PKS, polyketide synthase.
Furthermore, antiSMASH server was used to predict BGCs of the 15 species. The results showed that all of the 15 species contained multiple gene clusters encoding other unspecified ribosomally synthesized and post-translationally modified peptide product (RiPP-like), terpene, non-ribosomal peptide synthetase (NRPS), NRPS-like, and Type I polyketide synthase (T1PKS)/NRPS hybrid types. Except for Myxococcus llanfairp AM401T and Myxococcus vastator AM301T, the remaining 13 genomes contained different PKS gene clusters. Other multiple types, for example, arylpolyene, indole, phenazine, and butyrolactone, were contained in the unique or accessory genomes (Figure 4B). In addition, only four “most similar known clusters,” carotenoid (100% similarity), VEPE/AEPE/TG-1 (100% similarity), myxoprincomide-506, and alkylpyrone-407/393, were found in all 15 genomes, which were consistent with those described by Chambers et al. (2020). Moreover, except for M. stipitatus DSM 14675T and Myxococcus hansupus mixupusT, the remaining 13 genomes contained gene cluster encoding myxochelin A/B. Moreover, the gene cluster encoding geosmin (100% similarity) was found in genomes of 14 species except Pyxidicoccus xibeiensis QH1ED-7-1T. The gene clusters and their similarities related to myxoprincomide-506, alkylpyrone-407/393, myxochelin A/B, geosmin, and carotenoid in different genomes are exhibited in Supplementary Figure 5. The results exhibited abundant gene diversity of CAZymes and BGCs in the 15 species of Myxococcus and Pyxidicoccus.
Discussion
This study isolated and identified four new species within the genera Myxococcus and Pyxidicoccus and supported the two genera should be the same genus (Figures 1, 2). Furthermore, pan-genomics indicated that all 15 species of Myxococcus and Pyxidicoccus clustered into 17,415 orthologous groups, which were divided into 24.0% core gene families, 46.6% accessory gene families, and 29.4% unique gene families (Figures 3A–C) using software OrthoFinder with default parameters. However, because of multiple contigs of some used draft genomes, big genes can be split across contigs and the real gene numbers might be overestimated for the pan-genomics. In addition, according to the described study (Chambers et al., 2020), the pan-genome of 11 species of Myxococcus and Pyxidicoccus contained 64,700 orthologous clusters analyzed by software Roary, which were higher than our data, maybe because of the use of lower cutoffs by the software Roary. Core genomes enriched more genes related to housekeeping functional classes such as ribosome and oxidative phosphorylation, while the accessory and unique genomes enriched more genes associated with environmental interactions, such as quorum sensing (Figures 3D–F). A reasonable interpretation is that housekeeping genes are relatively indispensable compared to signaling pathway genes, and therefore, signaling genes will be enriched in the accessory genome. Moreover, bacterial QS is a mechanism by which bacteria exchange intracellular or intercellular information, coordinate population behavior, and regulate gene expression (Xiao et al., 2022). Myxobacteria appear to shape genetic diversity by communicating with intracellular or intercellular information to broaden accessory and unique genomes.
It is known that myxobacteria exhibit a broad predatory ability to various bacteria, but the predation efficiencies are different (Livingstone et al., 2017). Lytic enzymes and secondary metabolites secreted by myxobacteria play important roles in predatory process (Xiao et al., 2011; Arend et al., 2020). Based on our results, the four novel species also showed the broad predatory ability against prey (Supplementary Figure 3). In addition, the abundant genes encoding CAZymes were found to mainly lie in accessory genome of the 15 species (Supplementary Figure 4). The GTs and GHs are the two richest CAZymes families, mainly included large numbers of GT2, GT4, GH13, and GH23 members (Figure 4A), exhibiting a variety of enzyme activities. Furthermore, multiple BGCs types: RiPP-like, terpene, NRPS, NRPS-like, and T1PKS/NRPS hybrids, were distributed in all 15 genomes, while only four “Most similar known clusters” were common, including alkylpyrone-407/393, carotenoid, myxoprincomide-506, and VEPE/AEPE/TG-1 (Figure 4B). Alkylpyrone-407/393 is type III PKS, revealing potent topoisomerase activity in vitro (Hug et al., 2019). Carotenoids are lipophilic isoprenoid pigments that protect cells from damage and death by quenching highly reactive and toxic oxidative species (Padmanabhan et al., 2021). Myxoprincomide-506 is a NRPS/PKS natural product, which has been shown to be involved in the predatory killing of some prey (Cortina et al., 2012; Berleman et al., 2014; Müller et al., 2016). Furthermore, myxochelin A/B and geosmin are found in 13 or 14 genomes (Figure 4B). Myxochelin A/B is a siderophore released under iron-deficient conditions to form a complex with iron (Lee et al., 2020). Geosmin has been found in various strains of cyanobacteria, streptomyces, and myxobacteria, which is considered a major cause of most taste and odor episodes in drinking water (Wang Z. J. et al., 2019). Recently, geosmin has been found to appear to act as a warning signal indicating the unpalatability of its producers and reducing predation in a manner that benefits predator and prey (Zaroubi et al., 2022). Different CAZymes and secondary metabolites produced by the genera Myxococcus and Pyxidicoccus may to a certain extent reflect their predation efficiencies against various bacteria and fungi.
Description of Myxococcus guangdongensis sp. nov.
Myxococcus guangdongensis (guang.dong.en’sis. N. L. fem. adj. guangdongensis, referring to Guangdong, a province of southern China, where the type strain was isolated).
Vegetative cells are gram-negative bacilli tapering slightly at the ends, appearing 0.3–0.4 μm × 2.2–4.1 μm under scanning electron microscope. Colonies are swarming and pale yellow on VY/2 agar. Fruiting bodies are irregular spheroids, large scattered, and orange-yellow (diameters of 150–350 μm). Myxospores are rounded spherical (diameters of 0.9–1.0 μm). Aerobic growth at 30°C but not 35°C, at pH 6.0–8.0 but not pH 5.0 or 9.0, and with up to 2.0% (w/v) NaCl. Positive for nitrate reduction and β-glucosidase. The major fatty acids (>5.0%) are C16:0 ω5c, iso-C15:0, iso-C17:0, and summed features 3 and 4. Cells prey upon E. coli, M. luteus, S. typhimurium, and S. aureus.
The type strain, K38C18041901T (=GDMCC 1.2320T =JCM 39260T), was isolated from forest soils collected from Dinghu Mountain Biosphere Reserve, Guangdong Province, China (23°10′24′′N; 112°32′17′′E). Genomic G + C content is 69.8 mol%. The accession number of the genome is JAJVKW000000000.
Description of Myxococcus qinghaiensis sp. nov.
Myxococcus qinghaiensis (qing.hai.en’sis. N.L. fem. adj. qinghaiensis, referring to Qinghai, a province of north-west China, where the soil sample containing the type strain was collected).
Vegetative cells are gram-negative bacilli tapering slightly at the ends, appearing 0.3–0.4 μm × 2.5–4.5 μm. Colonies are swarming and pale yellow on VY/2 agar. Fruiting bodies are irregular spheroids, stacked in series, and pale yellow (diameters of 220–470 μm). Myxospores are rounded spherical (diameters of 0.9–1.0 μm). Aerobic growth at 30°C but not 35°C, at pH 6.0–8.0 but not pH 5.0 or 9.0, and with up to 1.0% (w/v) NaCl. Negative for β-glucosidase. The major fatty acids (>5.0%) are C16:0 ω5c, iso-C15:0, iso-C17:0, iso-C17:0 3-OH, and summed feature 3. Cells prey upon E. coli, M. luteus, S. typhimurium, and S. aureus.
The type strain, QH3KD-4-1T (=GDMCC 1.2316T =JCM 39262T), was isolated from forest soils collected from Qinghai Virgin Forest, Qinghai Province, China (37°0′52′′N; 102°12′5′′E). The genomic G + C content is 68.8 mol%. The accession number of the genome is JAKCFA000000000.
Description of Myxococcus dinghuensis sp. nov.
Myxococcus dinghuensis (ding.hu.en’sis. N.L. fem. adj. dinghuensis, referring to Dinghu Mountain, Guangdong Province, China, where the soil sample containing the type strain was collected).
Vegetative cells are gram-negative bacilli tapering slightly at the ends, appearing 0.3–0.4 μm × 2.1–4.5 μm. Colonies are swarming and pale silver on VY/2 agar. Fruiting bodies are irregular spheroids, deep-yellow (diameters of 180–370 μm). Myxospores are rounded spherical (diameters of 0.9–1.1 μm). Aerobic growth at 30°C but not 35°C, at pH 6.0–8.0 but not pH 5.0 or 9.0, and with up to 2.0% (w/v) NaCl. Positive for β-glucosidase. The major fatty acids (>5.0%) are C16:0 ω5c, C16:0, iso-C15:0, and iso-C17:0. Cells prey upon E. coli, M. luteus, S. typhimurium, and S. aureus.
The type strain, K15C18031901T (=GDMCC 1.2319T =JCM 39259T), was isolated from forest soils collected from Dinghu Mountain Biosphere Reserve, Guangdong Province, China (23°10′24′′N; 112°32′10′′E). The genomic DNA G + C content is 70.7 mol%. The accession number of the genome is JAKCFB000000000.
Description of Pyxidicoccus xibeiensis sp. nov.
Pyxidicoccus xibeiensis (xi.bei.en’sis. N.L. masc. adj. xibeiensis, referring to Xibei, the Chinese phoneticization for north-west China, where the soil sample was collected).
Vegetative cells are gram-negative bacilli tapering slightly at the ends, appearing 0.3–0.5 μm × 5.0–6.6 μm. Colonies are swarming and bright metal color on VY/2 agar. Fruiting bodies are irregular spheroids and metal luster (diameters of 150–390 μm). Myxospores are rounded spherical (diameters of 0.9–1.0 μm). Aerobic growth up to 40oC, at pH 6.0–9.0 but not pH 5.0, and with up to 1.0% (w/v) NaCl. Positive for β-glucosidase and negative for β-Galactosidase. The major fatty acids (>5.0%) are C16:0 ω5c, iso-C15:0, iso-C17:0, and summed features 3 and 5. Cells prey upon E. coli, M. luteus, and S. typhimurium, but not S. aureus.
The type strain, QH1ED-7-1T (=GDMCC 1.2315T = JCM 39261T), was isolated from forest soils collected from Qinghai Virgin Forest, Qinghai Province, China (37°0′52′′N; 102°12′5′′E). The genomic DNA G + C content is 70.7 mol%. The accession number of the genome is JAJVKV000000000.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
CW: conceptualization, project administration, data curation, software, visualization, and writing—original draft. YL: formal analysis, methodology, and project administration. LZ and YZ: methodology and project administration. QY and HZ: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was jointly supported by the Natural Science Foundation of China (32001115 and 31800003), the GDAS’ Project of Science and Technology Development (2020GDASYL-20200103032), the Science and Technology Project of Guangzhou (202201011832), and Guangdong Special Support Program (2021JC06N628).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.995049/full#supplementary-material
Footnotes
- ^ http://eztaxon-e.ezbiocloud.net/
- ^ www.ncbi.nlm.nih.gov
- ^ https://www.ncbi.nlm.nih.gov/genome
- ^ http://rast.nmpdr.org/
- ^ http://ggdc.dsmz.de/
- ^ https://www.kegg.jp/
References
Adamek, M., Spohn, M., Stegmann, E., and Ziemert, N. (2017). Mining bacterial genomes for secondary metabolite gene clusters. Methods Mol. Biol. 1520, 23–47. doi: 10.1007/978-1-4939-6634-9_2
Arend, K. I., Schmidt, J. J., Bentler, T., Lüchtefeld, C., Eggerichs, D., Hexamer, H. M., et al. (2020). Myxococcus xanthus predation of Gram-positive or Gram-negative bacteria is mediated by different bacteriolytic mechanisms. Appl. Environ. Microb. 87:e02382-20. doi: 10.1128/aem.02382-02320
Awal, R. P., Garcia, R., and Müller, R. (2016). Racemicystis crocea gen. nov., sp nov., a soil myxobacterium in the family Polyangiaceae. Int. J. Syst. Evol. Microbiol. 66, 2389–2395. doi: 10.1099/ijsem.0.001045
Berleman, J. E., Allen, S., Danielewicz, M. A., Remis, J. P., Gorur, A., Cunha, J., et al. (2014). The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front. Microbiol. 5:474. doi: 10.3389/fmicb.2014.00474
Chambers, J., Sparks, N., Sydney, N., Livingstone, P. G., Cookson, A. R., and Whitworth, D. E. (2020). Comparative genomics and pan-genomics of the Myxococcaceae, including a description of five novel species: Myxococcus eversor sp. nov., Myxococcus llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis sp. nov., Myxococcus vastator sp. nov., Pyxidicoccus caerfyrddinensis sp. nov. and Pyxidicoccus trucidator sp. nov. Genome. Biol. Evol. 12, 2279–2302. doi: 10.1093/gbe/evaa212/5918458
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Cortina, N. S., Krug, D., Plaza, A., Revermann, O., and Müller, R. (2012). Myxoprincomide: A natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew. Chem. Int. Ed. Engl. 51, 811–816. doi: 10.1002/anie.201106305
Evans, A. G., Davey, H. M., Cookson, A., Currinn, H., Cooke-Fox, G., Stanczyk, P. J., et al. (2012). Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 158, 2742–2752. doi: 10.1099/mic.0.060343-0
Garcia, R., Pistorius, D., Stadler, M., and Müeller, R. (2011). Fatty acid-related phylogeny of myxobacteria as an approach to discover polyunsaturated omega-3/6 fatty acids. J. Bacteriol. 193, 1930–1942. doi: 10.1128/jb.01091-10
Hug, J. J., Panter, F., Krug, D., and Müller, R. (2019). Genome mining reveals uncommon alkylpyrones as type III PKS products from myxobacteria. J. Ind. Microbiol. Biotechnol. 46, 319–334. doi: 10.1007/s10295-018-2105-6
Kai, A. K. L., Cheung, Y. K., Yeung, R. K. K., and Wong, J. T. Y. (2006). Development of single-cell PCR methods for the Raphidophyceae. Harmful Algae 5, 649–657. doi: 10.1016/j.hal.2006.01.002
Konstantinos, T. K., and James, M. T. (2007). Prokaryotic taxonomy and phylogeny in the genomic era: Advancements and challenges ahead. Curr. Opin. Microbiol. 10, 504–509. doi: 10.1016/j.mib.2007.08.006
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, N., Kim, W., Chung, J., Lee, Y., Cho, S., Jang, K. S., et al. (2020). Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus. ISME J. 14, 1111–1124. doi: 10.1038/s41396-41020-40594-41396
Liu, Y., Pei, T., Yi, S., Du, J., Zhang, X., Deng, X., et al. (2021). Phylogenomic analysis substantiates the gyrB gene as a powerful molecular marker to efficiently differentiate the most closely related genera Myxococcus, Corallococcus, and Pyxidicoccus. Front. Microbiol. 12:763359. doi: 10.3389/fmicb.2021.763359
Livingstone, P. G., Ingleby, O., Girdwood, S., Cookson, A. R., Morphew, R. M., and Whitworth, D. E. (2020). Predatory organisms with untapped biosynthetic potential: Descriptions of novel Corallococcus Species C. aberystwythensis sp. nov., C. carmarthensis sp. nov., C. exercitus sp. nov., C. interemptor sp. nov., C. Ilansteffanensis sp. nov., C. praedator sp. nov., C. sicarius sp. nov., and C. terminator sp. nov. Appl. Environ. Microb. 86, e01931–19. doi: 10.1128/aem.01931-19
Livingstone, P. G., Millard, A. D., Swain, M. T., and Whitworth, D. E. (2018). Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding. Microb. Genom. 4:e000152. doi: 10.1099/mgen.0.000152
Livingstone, P. G., Morphew, R. M., and Whitworth, D. E. (2017). Myxobacteria are able to prey broadly upon clinically-relevant pathogens, exhibiting a prey range which cannot be explained by phylogeny. Front. Microbiol. 8:1593–1601. doi: 10.3389/fmicb.2017.01593
Medema, M. H., Takano, E., and Breiting, R. (2013). Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223. doi: 10.1093/molbev/mst025
Mohr, K. I. (2018). Diversity of myxobacteria-we only see the tip of the Iceberg. Microorganisms 6:84. doi: 10.3390/microorganisms6030084
Mohr, K. I., Stechling, M., Wink, J., Wilharm, E., and Stadler, M. (2016). Comparison of myxobacterial diversity and evaluation of isolation success in two niches: Kiritimati Island and German compost. MicrobiologyOpen 5, 268–278. doi: 10.1002/mbo3.325
Mohr, K. I., Zindler, T., Wink, J., Wilharm, E., and Stadler, M. (2017). Myxobacteria in high moor and fen: An astonishing diversity in a neglected extreme habitat. MicrobiologyOpen 6:e00464. doi: 10.1002/mbo3.464
Müller, S., Strack, S. N., Ryan, S. E., Shawgo, M., Walling, A., Harris, S., et al. (2016). Identification of functions affecting predator-prey interactions between Myxococcus xanthus and Bacillus subtilis. J. Bacteriol. 198, 3335–3344. doi: 10.1128/JB.00575-00516
Padmanabhan, S., Monera-Girona, A. J., Pérez-Castaño, R., Bastida-Martínez, E., Pajares-Martínez, E., Bernal-Bernal, D., et al. (2021). Light-triggered carotenogenesis in Myxococcus xanthus: New paradigms in photosensory signaling, transduction and gene regulation. Microorganisms 9:1067. doi: 10.3390/microorganisms9051067
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J. Z., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insight. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Sasser, M. (1990). Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. Newark, DE: MIDI Inc.
Seef, S. F., Herrou, J. L., de Boissier, P., My, L., Brasseur, G., Robert, D., et al. (2021). A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria. eLife 10:e72409. doi: 10.7554/eLife.72409
Sharma, G., Khatri, I., and Subramanian, S. (2016). Complete genome of the starch-degrading myxobacteria Sandaracinus amylolyticus DSM 53668. Genome. Biol. Evol. 8, 2520–2529. doi: 10.1093/gbe/evw151
Sharma, G., and Subramanian, S. (2017). Unravelling the complete genome of Archangium gephyra DSM 2261T and evolutionary insights into myxobacterial chitinases. Genome. Biol. Evol. 9, 1304–1311. doi: 10.1093/gbe/evx066
Shimkets, L. J., Dworkin, M., and Reichenbach, H. (2006). “The Myxobacteria,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 31–115.
Thiery, S., and Kaimer, C. (2020). The predation strategy of Myxococcus xanthus. Front. Microbiol. 11:22. doi: 10.3389/fmicb.2020.00002
Waite, D. W., Chuvochina, M., Pelikan, C., Parks, D. H., Yilmaz, P., Wagner, M., et al. (2020). Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 70, 5972–6016. doi: 10.1099/ijsem.0.004213.
Wang, C. L., Lv, Y. Y., Li, A. Z., Bao, G. G., Feng, G. D., Zhang, J., et al. (2019). Deminuibacter soli gen. nov., sp. nov., isolated from forest soil, and reclassification of Filimonas aurantiibacter as Arvibacter aurantiibacter comb. nov. Int. J. Syst. Evol. Microbiol. 69, 1650–1655. doi: 10.1099/ijsem.0.003374
Wang, C. L., Lv, Y. Y., Li, A. Z., Yao, Q., Feng, G. D., and Zhu, H. H. (2020). Culture-dependent and -independent methods revealed an abundant myxobacterial community shaped by other bacteria and pH in Dinghushan acidic soils. PLoS One 15:e0238769. doi: 10.1371/journal.pone.0238769
Wang, Z. J., Song, G. F., Li, Y. G., Yu, G. L., Hou, X. Y., Gan, Z. X., et al. (2019). The diversity, origin, and evolutionary analysis of geosmin synthase gene in cyanobacteria. Sci. Total Environ. 689, 789–796. doi: 10.1016/j.scitotenv.2019.06.468
Wrotniak-Drzewiecka, W., Brzezinska, A. J., Dahm, H., Ingle, A. P., and Rai, M. (2016). Current trends in myxobacteria research. Ann. Microbiol. 66, 17–33. doi: 10.1007/s13213-13015-11104-13213
Xiao, Y., Wei, X. M., Ebright, R., and Wall, D. (2011). Antibiotic production by myxobacteria plays a role in predation. J. Bacteriol. 193, 4626–4633. doi: 10.1128/jb.05052-11
Xiao, Y. P., Zou, H. C., Li, J. J., Song, T. X., Lv, W. T., Wang, W., et al. (2022). Impact of quorum sensing signaling molecules in gram-negative bacteria on host cells: Current understanding and future perspectives. Gut Microbes 14:2039048. doi: 10.1080/19490976.2022.2039048
Keywords: Myxococcus, Pyxidicoccus, comparative genomics, novel species, CAZymes, BGCs
Citation: Wang C, Lv Y, Zhou L, Zhang Y, Yao Q and Zhu H (2022) Comparative genomics of Myxococcus and Pyxidicoccus, including the description of four novel species: Myxococcus guangdongensis sp. nov., Myxococcus qinghaiensis sp. nov., Myxococcus dinghuensis sp. nov., and Pyxidicoccus xibeiensis sp. nov.. Front. Microbiol. 13:995049. doi: 10.3389/fmicb.2022.995049
Received: 15 July 2022; Accepted: 18 October 2022;
Published: 10 November 2022.
Edited by:
David W. Ussery, University of Arkansas for Medical Sciences, United StatesReviewed by:
David Cole Stevens, University of Mississippi, United StatesDaniel Krug, Helmholtz-Institute for Pharmaceutical Research Saarland (HIPS), Germany
Copyright © 2022 Wang, Lv, Zhou, Zhang, Yao and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honghui Zhu, emh1aGhfZ2RpbUAxNjMuY29t
 Chunling Wang
Chunling Wang Yingying Lv1
Yingying Lv1 Qing Yao
Qing Yao Honghui Zhu
Honghui Zhu