
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 16 September 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.994365
This article is part of the Research TopicMarine Bacteria and Microalgae as Emerging Models for Synthetic BiologyView all 4 articles
The current economic and environmental context requests an accelerating development of sustainable alternatives for the production of various target compounds. Biological processes offer viable solutions and have gained renewed interest in the recent years. For example, photosynthetic chassis organisms are particularly promising for bioprocesses, as they do not require biomass-derived carbon sources and contribute to atmospheric CO2 fixation, therefore supporting climate change mitigation. Marine cyanobacteria are of particular interest for biotechnology applications, thanks to their rich diversity, their robustness to environmental changes, and their metabolic capabilities with potential for therapeutics and chemicals production without requiring freshwater. The additional cyanobacterial properties, such as efficient photosynthesis, are also highly beneficial for biotechnological processes. Due to their capabilities, research efforts have developed several genetic tools for direct metabolic engineering applications. While progress toward a robust genetic toolkit is continuously achieved, further work is still needed to routinely modify these species and unlock their full potential for industrial applications. In contrast to the understudied marine cyanobacteria, genetic engineering and synthetic biology in freshwater cyanobacteria are currently more advanced with a variety of tools already optimized. This mini-review will explore the opportunities provided by marine cyanobacteria for a greener future. A short discussion will cover the advances and challenges regarding genetic engineering and synthetic biology in marine cyanobacteria, followed by a parallel with freshwater cyanobacteria and their current genetic availability to guide the prospect for marine species.
Modern society faces various challenges that must be addressed promptly to preserve most living organisms. The current environmental crisis is particularly pressing and threatening and demands that the present industrial and economical behaviors are entirely revised (Levi and Cullen, 2018). As such, many research, political and industrial efforts have focused on developing sustainable solutions to mitigate the climate emergency. In this context, bioprocesses, relying on living organisms for biological catalysis, stand as a key solution due to their flexibility, robustness and sustainability. Moreover, the current omics data and synthetic biology techniques available to strengthen and diversify bioprocesses further support their wider use for various industrial applications, including fuels, chemicals or pharmaceuticals production.
Of particular interest for sustainable industrial bioprocesses, cyanobacteria perform oxygenic photosynthesis, using solar energy for their metabolism while fixing atmospheric CO2. These prokaryotes are significant contributors to global carbon fixation in many environments, participating in ecosystem maintenance. The tremendous diversity and strain-specific variation of pigments and photosystems (Stephens et al., 2021), essential elements for photosynthesis, provide great opportunities for tailored biotechnological applications, with strains metabolically adapted to specific environmental conditions, such as high salinity, temperature and fluctuating light. To date, hundreds of cyanobacterial species have been isolated in marine and freshwater environments with new species continuously being identified and further characterized. Historically, a few model freshwater species have previously led researchers to understand cyanobacterial metabolism and photosynthetic capabilities and develop synthetic biology tools for strain engineering (Gale et al., 2019). However, marine cyanobacteria display several advantages over their freshwater counterparts, including their capacity to grow in seawater, their robustness to environmental changes and specific metabolic abilities, including biosynthesis of complex and unique biomolecules. Currently, more work is still needed to adapt many synthetic biology techniques to marine cyanobacteria and fully unlock their biotechnological and industrial potential (Khalifa et al., 2021).
This short review will briefly discuss the impressive diversity of marine cyanobacteria and the associated opportunities for unique bio-applications. The recent development of genetic tools and the advances of synthetic biology in marine cyanobacteria will be further reviewed, highlighting some of the challenges that must be addressed to move forward with these species and expand their applications. Finally, some of the more advanced synthetic biology tools developed for freshwater cyanobacteria will be summarized to draw a parallel between marine and freshwater species. While the focus of this review will be synthetic biology for industrial applications, it is worth mentioning that synthetic biology methods are also relevant for fundamental studies, such as phenotypic screening or regulation characterization, but will not be discussed here.
Marine cyanobacteria are a highly diverse group of photosynthetic prokaryotes, with relevant metabolic properties for future sustainable industrial applications. Their photoautotrophic abilities, attractive for biotechnological applications, are also incredibly important for global CO2 fixation to support a variety of ecosystems. In fact, some genera can fix up to four gigatons of carbon per year (Biller et al., 2015). From a biotechnological perspective, in addition to the extensive CO2 availability, their ability to prosper at high salinities further supports sustainability, preventing competition for drinking water. Furthermore, considering their natural habitats where temperature, nutrient availability, salinity and light intensity constantly fluctuate, marine cyanobacteria are particularly robust to environmental changes (Ludwig and Bryant, 2012), which may be beneficial in an industrial context where growth parameters might vary slightly. While most species maintain a circadian rhythm, relying on complex regulatory cascades for adaptation to fluctuating light exposure, some species, isolated from light-deprived environments, show unique adaptability to growth in darkness (Coe et al., 2021; Callieri et al., 2022), further expanding their metabolic flexibility. In fact, the diversity of marine environments has been a driving force for cyanobacterial evolution and there is a high correlation between specialized features of some species and their natural habitats (Biller et al., 2015). Considering the exceptional diversity of marine cyanobacteria, this review will not attempt to summarize each and every unique capacity previously described but will point out some species of industrial relevance and their different properties, further highlighting how this diversity can be exploited for tailored applications (Table 1). It is worth mentioning that brackish cyanobacteria, such as Aphanizomenon flos-aquae (Lyon-Colbert et al., 2018), will not be covered in this review but are important players in cyanobacterial biodiversity and have a significant impact on human health for bloom-forming species, as discussed elsewhere (Śliwińska-Wilczewska et al., 2019).
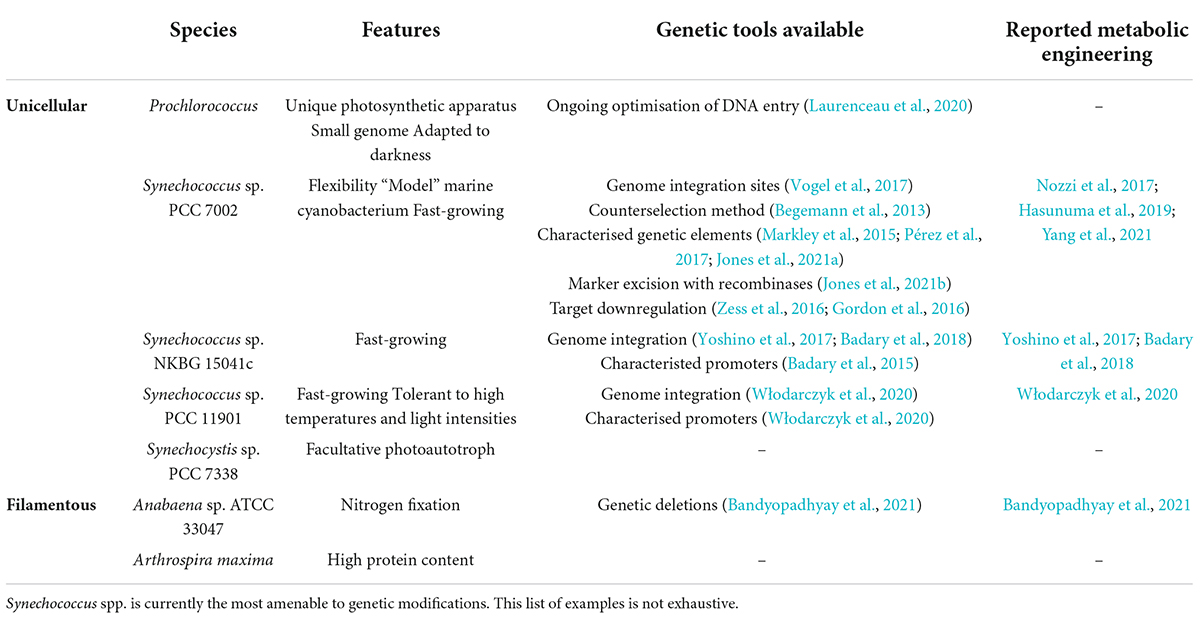
Table 1. Examples of genetic tools available for marine cyanobacteria and their applications for metabolic engineering.
Prochlorococcus isolates, the most abundant photosynthetic organisms, have primarily been isolated from tropical and subtropical regions and have fascinated scientists for several decades, partly due to their condensed genome, with only ∼1,700 genes (Rocap et al., 2003). Within this group, strains remain highly diverse, especially regarding light adaptation, and have been reviewed in details elsewhere (Biller et al., 2015). Their photosynthetic apparatus is unique amongst cyanobacteria as it lacks the widely spread cyanobacterial phycobilisome structures and, instead, relies on divinyl chlorophyll a and b pigments in the antenna (Ting et al., 2002), a contrasting pigment composition to the phylogenetically close Synechococcus strains. A few Synechococcus species are arguably some of the most studied and well-understood marine cyanobacteria and are, in fact, almost ubiquitous to all marine environments, due to their large pigment diversity. Their fast growth rate, environmental robustness and extensive dataset further add to their industrial potential. Prochlorococcus and Synechococcus genera remain the predominant unicellular marine cyanobacteria studied currently, although selected species of other genera (Brito et al., 2017) are starting to attract interest. On the contrary, fewer filamentous species have been researched in the synthetic biology field, perhaps due to the difficulty to work around their morphology. Interestingly, some Anabaena species have the dual ability to fix both carbon and nitrogen and can sustain high light intensities and a wide range of temperatures and pH (Moreno et al., 2003), promising for potential industrial applications. Lastly, Arthrospira (also called Spirulina) maxima, another rising filamentous cyanobacterium, is gaining interest for its use as a food supplement as it offers a higher protein content than its freshwater counterpart Arthrospira platensis. Indeed, Arthrospira sp., and primarily A. platensis, have been extensively exploited for food, cosmetics and pharmaceutical applications. Their unique spiral morphology enables to achieve high biomass cultivations, further harvested for downstream processes (Affan et al., 2015). A. maxima has also recently been proposed as a suitable medium for mammalian cell cultivation (Jeong et al., 2021), therefore bypassing the need for animal-derived medium, or as a source for chlorophyll a extraction (Martins et al., 2021), with possible cancer therapeutics applications.
Moreover, new species are constantly isolated from various environments with highly specific metabolic properties, such as nitrogen fixation or production of complex secondary metabolites (Hoffman, 1999; Brito et al., 2017; Shiels et al., 2019). Therefore, it is very likely that additional organisms with interesting properties will be isolated in the near future. The close correlation between the natural habitat and the evolution of unique features also further promotes biotechnological applications in specific regions. Indeed, since species have adapted, for example, to extreme temperatures or different light intensities, these same species can become the workhorse of biotechnological applications in these regions, without needing further strain adaptation and accelerating the development of highly specialized applications. Moreover, it is worth mentioning that, in addition to their photoautotrophic capabilities, native secondary metabolites from marine cyanobacteria have also gained interest for their potential therapeutics applications as reviewed elsewhere (Tan and Phyo, 2020; Lopes et al., 2022). Similarly, natural pigments produced by cyanobacteria have direct applications in the food and cosmetics industries, contributing to a global greener future (Saini et al., 2018).
However, although native products remain important, the future of cyanobacteria lies in the possibility to convert them into specialized cell factories for the production of various compounds. Utilizing their complex and flexible metabolic abilities, and, in particular, solar energy and CO2 fixation, these applications will drive the change toward a sustainable future. This course can, however, only be achieved if routine strain engineering, mediated by robust and efficient genetic tools, is accessible for introduction and manipulation of metabolic pathways, leading to maximal compound bioproduction and strain robustness for industrial conditions. While this review focuses on the bioproduction of specific chemicals, cyanobacteria are also relevant for other biotechnological applications, such as bioremediation, which has been discussed elsewhere (Rotter et al., 2021; Touliabah et al., 2022).
Genetic accessibility is crucial for chassis organisms with potential industrial applications. Indeed, this allows to engineer strains with, for example, non-native properties and improved catalytic efficiency in order to meet industrial standards, which need robust and superior strains. Numerous methods for genome editing with direct synthetic biology applications have now been established for many organisms, especially model organisms, such as Saccharomyces cerevisiae or Escherichia coli. However, the toolkit available for non-model organisms, including cyanobacteria, remains limited, preventing routine strain engineering for specific industrial aims. Expanding this toolkit is undeniably challenging, especially due to the polyploidy of most cyanobacteria (Griese et al., 2011), but vital to accelerate cyanobacterial bioprocesses.
Considering the importance of reliable engineering methods, several research groups have spent time and effort developing such tools (Table 1). Today, Synechococcus sp. PCC 7002 (hereafter Synechococcus 7002) is arguably the most genetically amenable marine cyanobacterium (Figure 1). In fact, several genomic integration sites have been characterized for minimal metabolic disruption, allowing insertion of genetic constructs at different loci (Ruffing et al., 2016; Vogel et al., 2017). Integration in the native plasmid pAQ1 also offers an interesting option due to the higher copy number compared to genome integration (Yang et al., 2021). Natural transformation tends to be the most common DNA transfer for genomic integration in Synechococcus 7002 by selecting transformants through antibiotic resistance. In addition, an acrylic acid counterselection method has previously been described where deletion of the acsA gene confers resistance to acrylic acid (Begemann et al., 2013). Furthermore, genetic elements are critical to achieve the required expression level. To test their activity, reporter genes, coding, for instance, for the yellow or green fluorescent protein, can be used. As such, several publications have now identified promoters with specific strength for Synechococcus 7002. Native promoters often are a suitable starting point as host compatibility is assured and have previously been investigated in Synechococcus 7002 (Ruffing et al., 2016). Additional tested promoters include a library of synthetic promoters derived from Synechocystis sp. PCC 6803 PcpcB promoter, further engineered for IPTG inducibility (Markley et al., 2015). Other inducible systems include a T7-based system (Jones et al., 2021a), a zinc-inducible promoter (Pérez et al., 2017), and a tetracycline-dependent system (Zess et al., 2016), which allow tuneable gene expression. Ribosome-binding sites also influence expression levels but are often overlooked in most chassis organisms, although a RBS library has been characterized in Synechococcus 7002 (Markley et al., 2015). These different genetic elements can therefore drive expression of synthetic constructs, awarding additional properties to Synechococcus. Importantly, a recombinase-based tool has previously been optimized for this organism to allow removal of selection markers, highly relevant for wider use of engineered strains (Jones et al., 2021b). The authors showed that both CRE and DRE recombinases were functional and compatible for recombination at their respective lox sites, allowing marker removal. Interestingly, to avoid maintaining the recombinase genes permanently in the genome, this approach exploited Synechococcus 7002 polyploidy where the recombinases were inserted in an essential gene, therefore unable to achieve full segregation, and maintained until marker removal, after which the wild-type locus at the essential gene took over the recombinase insertion. While these different strategies allow targeted genome-editing for expression of heterologous genes, strain stability may be limited, with accumulation of mutations and a potential decrease of productivity, as discussed elsewhere (Jones, 2014; Hitchcock et al., 2020). This limitation, although not specific to cyanobacteria, must be addressed for longer and larger cultivations in industrial plants and may be partially mediated by fine-tuned regulation of heterologous pathways. Lastly, target downregulation through RNA interference (Zess et al., 2016) and CRISPR interference (Gordon et al., 2016) by blocking target translation and transcription, respectively, has been reported and is particularly powerful for genome-wide phenotypic analysis. It can also improve target production as illustrated by increased lactate production when using CRISPR interference for elevating concentration of the lactate precursor pyruvate. While development of these tools is impressive, their direct applications for metabolic engineering are striking. In fact, a significant number of non-native target compounds have now been produced by Synechococcus 7002 at a laboratory scale. To name a few, bioproduction of the hydrocarbons limonene and bisabolene (Davies et al., 2014), the pigment astaxanthin (Hasunuma et al., 2019), the amino acid L-lysine (Korosh et al., 2018), the building block 2,3-butanediol (Nozzi et al., 2017), the potential aircraft fuel pinene (Yang et al., 2021), the fatty acid lauric acid (Work et al., 2015) or the biomaterial precursor polyhydroxyalkanoate (Zhang et al., 2015) has been achieved. The chemical diversity of these compounds further highlights the industrial importance of this chassis organism for a greener future. While the native metabolism offers the opportunity to synthesize complex non-native molecules, such as pigments or secondary metabolites, by providing precursors for engineered pathways, complete heterologous routes converting core metabolites, such as acetyl-CoA or pyruvate, are also relevant for target bioproduction in cyanobacteria. While this review primarily highlights synthetic biology for non-native chemical bioproduction, engineering strategies can also improve the biosynthesis of native secondary metabolites, such as antioxidants or UV protectants (Vega et al., 2020), as discussed extensively elsewhere (Jeong et al., 2020). As such, cyanobacterial target bioproduction is suitable for diverse groups of chemicals. Further more additional metabolic engineering reported in these studies, such as deletion of the glycogen synthase (Davies et al., 2014), further illustrate that our current understanding of the metabolism can support metabolic manipulation and strain optimization. These strategies can now also be guided by in silico methods, such as a genome-scale method, available for Synechococcus 7002 (Hendry et al., 2016), allowing more informed designs.
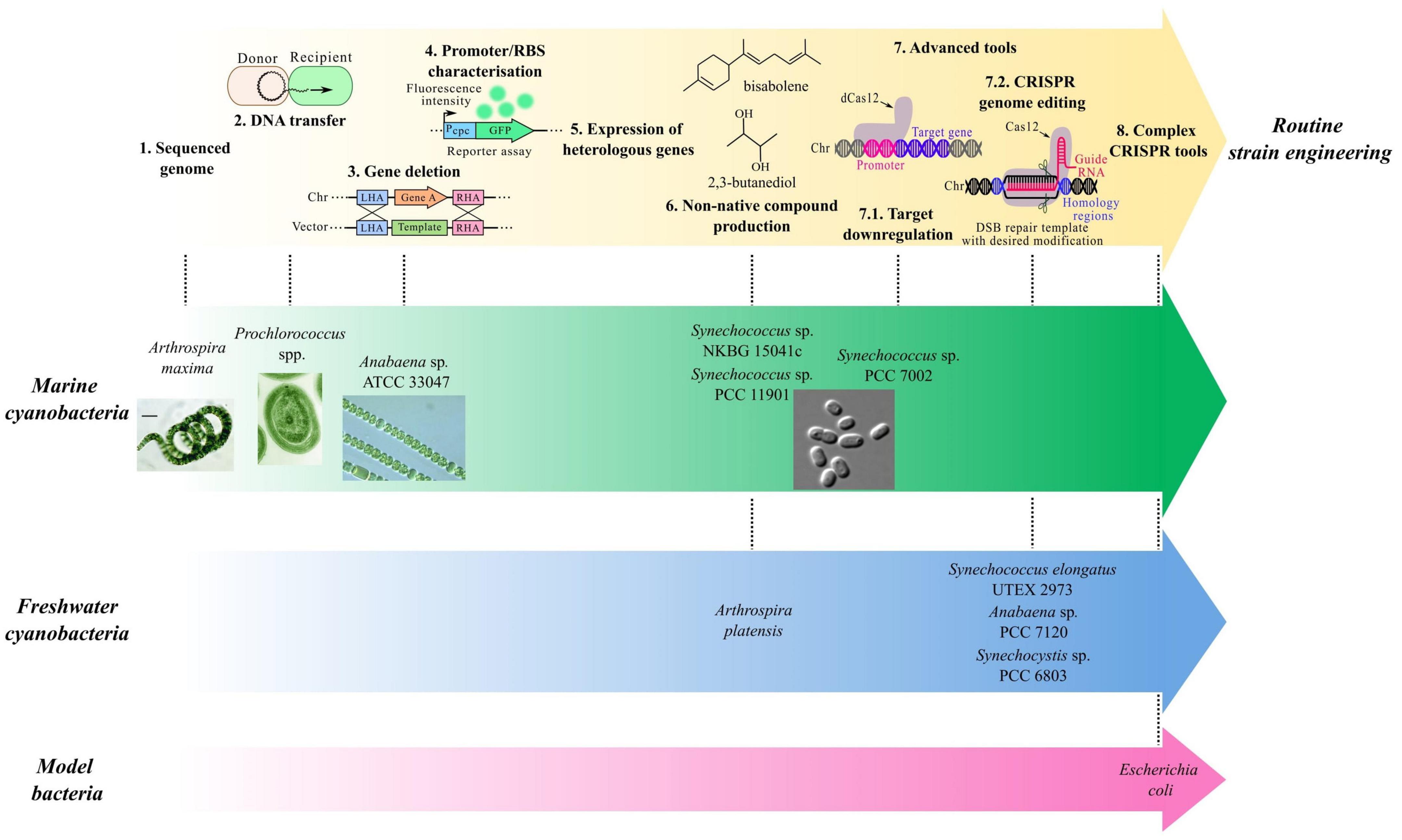
Figure 1. Progression of genetic tools toward routine strain engineering. Most marine cyanobacteria remain at earlier stages of tool development while freshwater cyanobacteria and model organisms have more complex genetic tools available.
Synechococcus 7002 remains undeniably the most genetically accessible marine cyanobacterium. However, encouragingly, some similar tools have been developed for Synechococcus sp. NKBG 15041c, achieving bioproduction of glycogen (Badary et al., 2018) and omega-3 fatty acids (Yoshino et al., 2017) through metabolic engineering. Further promoter development and characterization is also underway (Badary et al., 2015), therefore expanding the genetic toolkit available for this strain. The newly discovered strain Synechococcus sp. PCC 11901 has also been modified through natural transformation for free fatty acids production, with rapid tool development after isolation (Włodarczyk et al., 2020). On the other hand, Prochlorococcus strains, although phylogenetically close to Synechococcus spp. (Biller et al., 2015), remain highly resistant to genetic modifications. Despite an extensive dataset on its physiology and genomics (Berube et al., 2018), DNA transfer into Prochlorococcus seems particularly challenging as discussed elsewhere (Laurenceau et al., 2020). This further highlights the difficulty to adapt genetic tools to non-model species of any phylum, which requires to develop a robust DNA transfer method that must consider any species-specific biological and physiological obstacles, such as restriction-modification systems (Johnston et al., 2019), membrane composition or requirements for axenic cultivation.
Furthermore, the filamentous cyanobacterium Anabaena sp. ATCC 33047 has yet to be engineered for heterologous gene expression but deletion of nblA, coding for a small protein involved in phycobilisome degradation, has recently been reported through homologous recombination. This work required to create a specific helper strain that carried Anabaena sp. ATCC 33047 methylase genes to allow successful conjugation (Bandyopadhyay et al., 2021). This first report of genetic modifications in this strain will most likely lead to additional strain engineering, which can be guided by this host’s genome-scale model (Hendry et al., 2021).
Lastly, an impressive progress toward a robust genetic toolkit for Arthrospira platensis has recently been reported (Jester et al., 2022), further discussed later in this review. Earlier phylogenetic analyses suggested that A. platensis and A. maxima were closely related (Singh and Dhar, 2011), suggesting that these new tools could be transferred to A. maxima. However, genomics datasets have only recently been acquired for Arthrospira sp. and it is, therefore, premature to conclude how these two species differ. Nonetheless, a recent comparative genomic analysis (Misztak et al., 2021) proposed that the two species are, in fact, in two different Arthrospira groups, thus questioning whether tool transferability could be achieved in these two hosts.
Although great progress on toolkit development has been made in some marine cyanobacteria, freshwater species are slightly more advanced in terms of complex genetic tools (Figure 1). For example, many promoters, including native, heterologous and inducible systems (Englund et al., 2016; Wang et al., 2018; Behle et al., 2020), have now been characterized in Synechocystis sp. PCC 6803 or Synechococcus elongatus PCC 7942 (Berla et al., 2013; Sengupta et al., 2020). This allows tuneable target expression, essential for pathway expression and metabolic engineering.
In addition, genome editing is still routinely achieved through homologous recombination in most cyanobacteria, for which neutral integration sites have been identified (Almeida et al., 2017; Nagy et al., 2021). Counter-selection systems have been developed for transformant selection, such as sacB for sucrose sensitivity (Lagarde et al., 2000) or the protein synthesis inhibitor mazF (Cheah et al., 2013) in Synechocystis sp. PCC 6803, leading to markerless strains, important for biosafety in an industrial context. This has recently been further expanded with the identification of additional counter-selection methods in Synechococcus elongatus UTEX 2973 (Chen et al., 2021). Jones et al. (2021b) also showed that their CRE/DRE recombinase systems were functional in Synechocystis sp. PCC 6803.
Although these counter-selectable methods have shown great potential, they remain particularly time-consuming in polyploid organisms, which require multiple segregation steps to isolate pure mutants. This limitation also complicates genomic modifications at multiple sites. CRISPR (Clustered Regularly Interspaced Palindromic Repeats)-Cas tools have revolutionized genome editing in various organisms, allowing rapid and markerless genomic modifications. This RNA-guided method, reviewed elsewhere (Zhang and Huang, 2022), causes a double-stranded break, which must be repaired for cell survival. The repair mechanism can be harnessed to insert desired modifications. Genome editing with CRISPR has now been performed in Synechocystis sp. PCC 6803, Synechococcus elongatus PCC 7942, Synechococcus elongatus UTEX 2973 and Anabaena sp. PCC 7120 (Li et al., 2016; Ungerer and Pakrasi, 2016; Lu et al., 2022), a tremendous step toward a robust and modern genetic toolkit for cyanobacteria. In addition, multiplexing (i.e., simultaneous targeting of several genes) of CRISPR interference has been reported in freshwater cyanobacteria (Yao et al., 2016; Santos et al., 2021) but still lacking in marine species, preventing the use of CRISPRi in systemic studies.
Some of the reported tools have been adapted in both unicellular and filamentous species, suggesting that morphology is not impeding tool development. In fact, a previous CRISPR tool showed transferability in phylogenetically distinct unicellular and filamentous species (Ungerer and Pakrasi, 2016). In another study, the neutral site 2 from S. elongatus PCC 7942 was introduced in Anabaena sp. PCC 7120 genome for direct tool transferability without requiring additional cloning steps (Taton et al., 2020), allowing introduction of a cryptomaldamide gene cluster from a marine cyanobacterium in both species. Interestingly, cryptomaldamide was only detected in Anabaena, hypothesized to be more suitable for production of complex secondary metabolites. This further illustrates that pathway transferability between unicellular and filamentous species might not be as straight-forward as tool transferability, likely due to metabolic differences. In addition, tools developed specifically for filamentous species, such as Arthrospira platensis, have also been reported. In fact, optimization of the genetic engineering techniques led to the development of a potential oral therapy for Campylobacter infections (Jester et al., 2022), harnessing the edible properties of spirulina.
Finally, an important tool for routine and rapid genetic engineering is shuttle vectors, able to replicate within the cyanobacterial population. This prevents time-consuming genomic integration for genetic element and tool characterization and provides higher expression levels. The broad-host range replicon RSF1010 has been used widely in freshwater cyanobacteria (Mermet-Bouvier et al., 1993; Taton et al., 2014) and additional shuttle vectors, based on native plasmids, have recently been engineered for Synechocystis sp. PCC 6803 (Jin et al., 2018; Liu and Pakrasi, 2018). Although beneficial for many applications, strains carrying shuttle vectors tend to have an increased instability, which can be problematic in an industrial context. Shuttle vectors are still missing for most marine cyanobacteria, therefore preventing rapid tool characterization before moving to strain engineering. This requires to identify compatible origins of replication capable of maintaining a shuttle vector within the host population. Broad host-range origins or origins derived from the host’s native plasmids may be interesting to explore initially for potential compatibility, as achieved in some freshwater species.
The potential of marine cyanobacteria for various biotechnological applications and, more importantly, sustainable compound bioproduction is remarkable. The diversity of these species and their unique metabolic properties, evolved to survive in changing environments, offer tailored possibilities for industrial bioprocesses, such as large-scale photobioreactors for target bioproduction. To fully unlock their potential, marine cyanobacteria must be genetically accessible to further engineer strains for industrial purposes. While some progress has already been made, the current genetic toolkit remains insufficient. In fact, a clear discrepancy persists between species where some Synechococcus species have been and are still leading the field. On the other hand, despite the current knowledge and techniques available on genetic systems, some species continue to resist genetic modifications, as exemplified by the studied Prochlorococcus. While some biological challenges must participate in this discrepancy, additional aspects, such as historical attention and devoted resources, further contribute to the uneven genetic accessibility of marine cyanobacteria.
Moreover, the genetic toolkit for freshwater cyanobacteria is undeniably more extensive, with complex tools developed for several genera. Considering how phylogenetically close some marine and freshwater species are, the transferability of methods is highly probable. In fact, a proteomic analysis of Synechocystis sp. PCC 6803 and PCC 7338 showed how close species can be, although protein regulation reflected their respective natural habitat (Kwon et al., 2020). Similarly, genetic elements from freshwater species have shown to be functional in marine cyanobacteria (Markley et al., 2015), suggesting possible transferability. As such, adaption of genetic techniques similar to freshwater species is hopeful, especially in a growing context for the search for bioprocesses. In fact, the available research on marine cyanobacteria does not suggest that challenges specific to these species prevent their genetic engineering. Instead, the current lack of tools reflects that these species have been historically less explored than their freshwater counterparts. In addition, their extreme diversity prevents from addressing these obstacles globally as it would be inappropriate to assume one strategy would succeed in all species. Instead, strain-specific methods will be needed for efficient genetic manipulation. To achieve this, optimized growth conditions and robust methods for foreign DNA introduction are key first steps to build on for more complex genetic tools. This, of course, might prove difficult for interesting chassis organisms, such as Prochlorococcus, for which engineering obstacles remain unclear (Laurenceau et al., 2020).
However, it is worth pointing out that, despite the discussed advances in freshwater species, cyanobacteria in general remain behind in terms of complex genetic tools compared to model prokaryotes, such as Escherichia coli, for which, for example, sophisticated CRISPR tools, such as prime-editing (Tong et al., 2021), have been developed. Although adapting genetic tools to any organism is difficult, the polyploidy of cyanobacteria is particularly challenging to address and modulate and significantly contributes to limiting tool development. In particular, the lack of understanding of chromosomal copy fluctuations and their related environmental clues prevent to overcome these issues systematically. As such, further research efforts are needed to complete the genetic toolset for both marine and freshwater cyanobacteria for their unlimited industrial use.
BB: conceptualization, initial writing, and manuscript revision. KS: conceptualization and manuscript revision. Both authors contributed to the article and approved the submitted version.
This work was supported by “Formas—A Swedish Research Council for Sustainable Development” (project no. 2021-01669), the Swedish Energy Agency (44728-1), and the Carl Tryggers Foundation (CTS 20:412).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Affan, M. A., Lee, D. W., Al-Harbi, S. M., Kim, H. J., Abdulwassi, N. I., Heo, S. J., et al. (2015). Variation of Spirulina maxima biomass production in different depths of urea-used culture medium. Brazilian J. Microbiol. 46, 991–1000. doi: 10.1590/S1517-838246420140188
Almeida, D. V., Martens, S. B. B., Lanes, C. F. C., and Marins, L. F. (2017). Improved genetic transformation of Synechococcus elongatus PCC 7942 using linear DNA fragments in association with a DNase inhibitor. Biotechnol. Res. Innov. 1, 123–128. doi: 10.1016/j.biori.2017.09.001
Badary, A., Abe, K., Ferri, S., Kojima, K., and Sode, K. (2015). The development and characterization of an exogenous green-light-regulated gene expression system in marine cyanobacteria. Mar. Biotechnol. 17, 245–251. doi: 10.1007/s10126-015-9616-1
Badary, A., Takamatsu, S., Nakajima, M., Ferri, S., Lindblad, P., and Sode, K. (2018). Glycogen production in marine cyanobacterial strain Synechococcus sp. NKBG 15041c. Mar. Biotechnol. 20, 109–117. doi: 10.1007/s10126-017-9792-2
Bandyopadhyay, A., Ye, Z., Benedikty, Z., Trtilek, M., and Pakrasi, H. B. (2021). Antenna modification leads to enhanced nitrogenase activity in a high light-tolerant cyanobacterium. mBio 12:e0340821. doi: 10.1128/mbio.03408-21
Begemann, M. B., Zess, E. K., Walters, E. M., Schmitt, E. F., Markley, A. L., and Pfleger, B. F. (2013). An organic acid based counter selection system for cyanobacteria. PLoS One 8:e76594. doi: 10.1371/journal.pone.0076594
Behle, A., Saake, P., Germann, A. T., Dienst, D., and Axmann, I. M. (2020). Comparative dose-response analysis of inducible promoters in cyanobacteria. ACS Synth. Biol. 9, 843–855. doi: 10.1021/acssynbio.9b00505
Berla, B. M., Saha, R., Immethun, C. M., Maranas, C. D., Moon, T. S., and Pakrasi, H. B. (2013). Synthetic biology of cyanobacteria: Unique challenges and opportunities. Front. Microbiol. 4:246. doi: 10.3389/fmicb.2013.00246
Berube, P. M., Biller, S. J., Hackl, T., Hogle, S. L., Satinsky, B. M., Becker, J. W., et al. (2018). Single cell genomes of Prochlorococcus, Synechococcus, and sympatric microbes from diverse marine environments. Sci. Data 5, 1–11. doi: 10.1038/sdata.2018.154
Biller, S. J., Berube, P. M., Lindell, D., and Chisholm, S. W. (2015). Prochlorococcus: The structure and function of collective diversity. Nat. Rev. Microbiol. 13, 13–27. doi: 10.1038/nrmicro3378
Brito, Â, Ramos, V., Mota, R., Lima, S., Santos, A., Vieira, J., et al. (2017). Description of new genera and species of marine cyanobacteria from the Portuguese Atlantic coast. Mol. Phylogenet. Evol. 111, 18–34. doi: 10.1016/j.ympev.2017.03.006
Callieri, C., Cabello-Yeves, P. J., and Bertoni, F. (2022). The “dark side” of picocyanobacteria: Life as we do not know it (yet). Microorganisms 10, 1–18. doi: 10.3390/microorganisms10030546
Cheah, Y. E., Albers, S. C., and Peebles, C. A. M. (2013). A novel counter-selection method for markerless genetic modification in Synechocystis sp. PCC 6803. Biotechnol. Prog. 29, 23–30. doi: 10.1002/btpr.1661
Chen, L., Liu, H., Wang, L., Tan, X., and Yang, S. (2021). Synthetic counter-selection markers and their application in genetic modification of Synechococcus elongatus UTEX2973. Appl. Microbiol. Biotechnol. 105, 5077–5086. doi: 10.1007/s00253-021-11391-y
Coe, A., Biller, S. J., Thomas, E., Boulias, K., Bliem, C., Arellano, A., et al. (2021). Coping with darkness: The adaptive response of marine picocyanobacteria to repeated light energy deprivation. Limnol. Oceanogr. 66, 3300–3312. doi: 10.1002/lno.11880
Davies, F. K., Work, V. H., Beliaev, A. S., and Posewitz, M. C. (2014). Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2, 1–11. doi: 10.3389/fbioe.2014.00021
Englund, E., Liang, F., and Lindberg, P. (2016). Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 6, 1–12. doi: 10.1038/srep36640
Gale, G. A. R., Osorio, A. A. S., Mills, L. A., Wang, B., Lea-Smith, D. J., and McCormick, A. J. (2019). Emerging species and genome editing tools: Future prospects in cyanobacterial synthetic biology. Microorganisms 7:409. doi: 10.3390/microorganisms7100409
Gordon, G. C., Korosh, T. C., Cameron, J. C., Markley, A. L., Begemann, M. B., and Pfleger, B. F. (2016). CRISPR interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. strain PCC 7002. Metab Eng. 38, 170–179. doi: 10.1016/j.ymben.2016.07.007
Griese, M., Lange, C., and Soppa, J. (2011). Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323, 124–131. doi: 10.1111/j.1574-6968.2011.02368.x
Hasunuma, T., Takaki, A., Matsuda, M., Kato, Y., Vavricka, C. J., and Kondo, A. (2019). Single-stage astaxanthin production enhances the nonmevalonate pathway and photosynthetic central metabolism in Synechococcus sp. PCC 7002. ACS Synth. Biol. 8, 2701–2709. doi: 10.1021/acssynbio.9b00280
Hendry, J. I., Dinh, H. V., Sarkar, D., Wang, L., Bandyopadhyay, A., Pakrasi, H. B., et al. (2021). A genome-scale metabolic model of Anabaena 33047 to guide genetic modifications to overproduce nylon monomers. Metabolites 11:168. doi: 10.3390/metabo11030168
Hendry, J. I., Prasannan, C. B., Joshi, A., Dasgupta, S., and Wangikar, P. P. (2016). Metabolic model of Synechococcus sp. PCC 7002: Prediction of flux distribution and network modification for enhanced biofuel production. Bioresour. Technol. 213, 190–197. doi: 10.1016/j.biortech.2016.02.128
Hitchcock, A., Hunter, C. N., and Canniffe, D. P. (2020). Progress and challenges in engineering cyanobacteria as chassis for light-driven biotechnology. Microb. Biotechnol. 13, 363–367. doi: 10.1111/1751-7915.13526
Hoffman, L. (1999). Marine cyanobacteria in tropical regions: Diversity and ecology. Eur. J. Phycol. 34, 371–379. doi: 10.1080/09670269910001736432
Jeong, Y., Cho, S.-H., Lee, H., Choi, H.-K., Kim, D.-M., Lee, C.-G., et al. (2020). Current status and future strategies to increase secondary metabolite production in cyanobacteria. Microorganisms 8, 1–24. doi: 10.3390/microorganisms8121849
Jeong, Y., Choi, W. Y., Park, A., Lee, Y. J., Lee, Y., Park, G. H., et al. (2021). Marine cyanobacterium Spirulina maxima as an alternate to the animal cell culture medium supplement. Sci. Rep. 1–11:4906. doi: 10.1038/s41598-021-84558-2
Jester, B. W., Zhao, H., Gewe, M., Adame, T., Perruzza, L., Bolick, D. T., et al. (2022). Development of spirulina for the manufacture and oral delivery of protein therapeutics. Nat. Biotechnol. 40:974. doi: 10.1038/s41587-022-01249-7
Jin, H., Wang, Y., Idoine, A., and Bhaya, D. (2018). Construction of a shuttle vector using an endogenous plasmid from the cyanobacterium Synechocystis sp. PCC6803. Front. Microbiol. 9, 1–13. doi: 10.3389/fmicb.2018.01662
Johnston, C. D., Cotton, S. L., Rittling, S. R., Starr, J. R., Borisy, G. G., Dewhirst, F. E., et al. (2019). Systematic evasion of the restriction-modification barrier in bacteria. Proc. Natl. Acad. Sci. U.S.A. 166, 11454–11459. doi: 10.1073/pnas.1820256116
Jones, P. R. (2014). Genetic instability in cyanobacteria – An elephant in the room? Front. Bioeng. Biotechnol. 2:12. doi: 10.3389/fbioe.2014.0012
Jones, C. M., Korosh, T. C., Nielsen, D. R., and Pfleger, B. F. (2021a). Optimization of a T7-RNA polymerase system in Synechococcus sp. PCC 7002 mirrors the protein overproduction phenotype from E. coli BL21(DE3). Appl. Microbiol. Biotechnol. 105, 1147–1158. doi: 10.1007/s00253-020-11085-x
Jones, C. M., Parrish, S., and Nielsen, D. R. (2021b). Exploiting polyploidy for markerless and plasmid-free genome engineering in cyanobacteria. ACS Synth. Biol. 10, 2371–2382. doi: 10.1021/acssynbio.1c00269
Khalifa, S. A. M., Shedid, E. S., Saied, E. M., Jassbi, A. R., Jamebozorgi, F. H., Rateb, M. E., et al. (2021). Cyanobacteria—from the oceans to the potential biotechnological and biomedical applications. Mar. Drugs 19:241. doi: 10.3390/md19050241
Korosh, T. C., Markley, A. L., Clark, R. L., Mcginley, L. L., Katherine, D., Pfleger, B. F., et al. (2018). Engineering photosynthetic production of L-lysine. Metab Eng. 44, 273–283. doi: 10.1016/j.ymben.2017.10.010.Engineering
Kwon, D., Park, J. M., Duong, V. A., Hong, S. J., Cho, B. K., Lee, C. G., et al. (2020). Comparative proteomic profiling of marine and freshwater Synechocystis strains using liquid chromatography-tandem mass spectrometry. J. Mar. Sci. Eng. 8, 1–16. doi: 10.3390/jmse8100790
Lagarde, D., Beuf, L., and Vermaas, W. (2000). Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66, 64–72. doi: 10.1128/AEM.66.1.64-72.2000
Laurenceau, R., Bliem, C., Osburne, M. S., Becker, J. W., Biller, S. J., Cubillos-Ruiz, A., et al. (2020). Toward a genetic system in the marine cyanobacterium Prochlorococcus. Access Microbiol. 2:acmi00010. doi: 10.1099/acmi.0.000107
Levi, P. G., and Cullen, J. M. (2018). Mapping global flows of chemicals: From fossil fuel feedstocks to chemical products. Environ. Sci. Technol. 52, 1725–1734. doi: 10.1021/acs.est.7b04573
Li, H., Shen, C. R., Huang, C. H., Sung, L. Y., Wu, M. Y., and Hu, Y. C. (2016). CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab. Eng. 38, 293–302. doi: 10.1016/j.ymben.2016.09.006
Liu, D., and Pakrasi, H. B. (2018). Exploring native genetic elements as plug - in tools for synthetic biology in the cyanobacterium Synechocystis sp. PCC 6803. Microb. Cell Fact. 17:48. doi: 10.1186/s12934-018-0897-8
Lopes, G., Silva, M., and Vasconcelos, V. (2022). The pharmacological potential of cyanobacteria. Amsterdam: Academic Press; Elsevier. doi: 10.1016/C2019-0-01449-5
Lu, L., Shen, X., Sun, X., Yan, Y., Wang, J., and Yuan, Q. (2022). CRISPR-based metabolic engineering in non-model microorganisms. Curr. Opin. Biotechnol. 75:102698. doi: 10.1016/j.copbio.2022.102698
Ludwig, M., and Bryant, D. A. (2012). Synechococcus sp. Strain PCC 7002 transcriptome: Acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front. Microbiol. 3:354. doi: 10.3389/fmicb.2012.00354
Lyon-Colbert, A., Su, S., and Cude, C. (2018). A sytematic literature review for evidence of Aphanizomenon flos-aquae toxigenicity in recreational waters and toxicity of dietary supplements: 2000–2017. Toxins. 10, 1–18. doi: 10.3390/toxins10070254
Markley, A. L., Begemann, M. B., Clarke, R. E., Gordon, G. C., and Pfleger, B. F. (2015). Synthetic biology toolbox for controlling gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. ACS Synth. Biol. 4, 595–603. doi: 10.1021/sb500260k
Martins, M., Albuquerque, C. M., Pereira, C. F., Coutinho, J. A. P., Neves, M. G. P. M. S., Pinto, D. C. G. A., et al. (2021). Recovery of chlorophyll a derivative from Spirulina maxima: Its purification and photosensitizing potential. ACS Sustain. Chem. Eng. 9, 1772–1780. doi: 10.1021/acssuschemeng.0c07880
Mermet-Bouvier, P., Cassier-Chauvat, C., Marraccini, P., and Chauvat, F. (1993). Transfer and replication of RSF1010-derived plasmids in several cyanobacteria of the genera Synechocystis and Synechococcus. Curr. Microbiol. 27, 323–327. doi: 10.1007/BF01568955
Misztak, A. E., Waleron, M., Furmaniak, M., Waleron, M. M., Bazhenova, O., Daroch, M., et al. (2021). Comparative genomics and physiological investigation of a new Arthrospira/Limnospira strain O9.13F isolated from an alkaline, winter freezing. Siberian lake. Cells. 10, 1–25. doi: 10.3390/cells10123411
Moreno, J., Vargas, M. Á, Rodríguez, H., Rivas, J., and Guerrero, M. G. (2003). Outdoor cultivation of a nitrogen-fixing marine cyanobacterium, Anabaena sp. ATCC 33047. Biomol. Eng. 20, 191–197. doi: 10.1016/S1389-0344(03)00051-0
Nagy, C., Thiel, K., Mulaku, E., Mustila, H., Tamagnini, P., Aro, E. M., et al. (2021). Comparison of alternative integration sites in the chromosome and the native plasmids of the cyanobacterium Synechocystis sp. PCC 6803 in respect to expression efficiency and copy number. Microb. Cell Fact. 20, 1–18. doi: 10.1186/s12934-021-01622-2
Nozzi, N. E., Case, A. E., Carroll, A. L., and Atsumi, S. (2017). Systematic approaches to efficiently produce 2,3-butanediol in a marine cyanobacterium. ACS Synth. Biol. 6, 2136–2144. doi: 10.1021/acssynbio.7b00157
Pérez, A. A., Gajewski, J. P., Ferlez, B. H., Ludwig, M., Baker, C. S., Golbeck, J. H., et al. (2017). Zn2+-inducible expressio platform for Synechococcus sp. strain PCC 7002 based on the smtA promoter/operator and smtB repressor. Appl. Environm. Microbiol. 83, 1–14. doi: 10.1128/AEM.02491-16
Rocap, G., Larimer, F. W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N. A., et al. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047. doi: 10.1038/nature01947
Rotter, A., Barbier, M., Bertoni, F., Bones, A. M., Cancela, M. L., Carlsson, J., et al. (2021). The essentials of marine biotechnology. Front. Mar. Sci. 8:629629. doi: 10.3389/fmars.2021.629629
Ruffing, A. M., Jensen, T. J., and Strickland, L. M. (2016). Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis. Microb. Cell Fact. 15, 1–14. doi: 10.1186/s12934-016-0584-6
Saini, D. K., Pabbi, S., and Shukla, P. (2018). Cyanobacterial pigments: Perspectives and biotechnological approaches. Food Chem. Toxicol. 120, 616–624. doi: 10.1016/j.fct.2018.08.002
Santos, M., Pacheco, C. C., Yao, L., Hudson, E. P., and Tamagnini, P. (2021). CRISPRi as a tool to repress multiple copies of extracellular polymeric substances (Eps)-related genes in the cyanobacterium Synechocystis sp. pcc 6803. Life 11:1198. doi: 10.3390/life11111198
Sengupta, A., Madhu, S., and Wangikar, P. P. (2020). A Library of tunable, portable, and inducer-free promoters derived from cyanobacteria. ACS Synth. Biol. 9, 1790–1801. doi: 10.1021/acssynbio.0c00152
Shiels, K., Browne, N., Donovan, F., Murray, P., and Saha, S. K. (2019). Molecular characterization of twenty-five marine cyanobacteria isolated from coastal regions of Ireland. Biology (Basel) 8:51. doi: 10.3390/biology8030059
Singh, N. K., and Dhar, D. W. (2011). Phylogenetic relatedness among Spirulina and related cyanobacterial genera. World J. Microbiol. Biotechnol. 27, 941–951. doi: 10.1007/s11274-010-0537-x
Śliwińska-Wilczewska, S., Cieszyńska, A., Konik, M., Maculewicz, J., and Latała, A. (2019). Environmental drivers of bloom-forming cyanobacteria in the Baltic Sea: Effects of salinity, temperature and irradiance. Estuar. Coast. Shelf Sci. 219, 138–150. doi: 10.1016/j.ecss.2019.01.016
Stephens, S., Mahadevan, R., and Allen, D. G. (2021). Engineering photosynthetic bioprocesses for sustainable chemical production: A Review. Front. Bioeng. Biotechnol. 8:610723. doi: 10.3389/fbioe.2020.610723
Tan, L. T., and Phyo, M. Y. (2020). Marine cyanobacteria: A source of lead compounds and their clinically-relevant molecular targets. Molecules 25:2197. doi: 10.3390/molecules25092197
Taton, A., Ecker, A., Diaz, B., Moss, N. A., Anderson, B., Reher, R., et al. (2020). Heterologous expression of cryptomaldamide in a cyanobacterial host. ACS Synth. Biol. 9, 3364–3376. doi: 10.1021/acssynbio.0c00431
Taton, A., Unglaub, F., Wright, N. E., Zeng, W. Y., Paz-Yepes, J., Brahamsha, B., et al. (2014). Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 42, 1–16. doi: 10.1093/nar/gku673
Ting, C. S., Rocap, G., King, J., and Chisholm, S. W. (2002). Cyanobacterial photosynthesis in the oceans: The origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142. doi: 10.1016/S0966-842X(02)02319-3
Tong, Y., Jørgensen, T. S., Whitford, C. M., Weber, T., and Lee, S. Y. (2021). A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat. Commun. 12, 1–11. doi: 10.1038/s41467-021-25541-3
Touliabah, H. E.-S., El-Sheekh, M. M., Ismail, M. M., and El-Kassas, H. (2022). A review of microalgae- and cyanobacteria-based biodegradation of organic pollutants. Molecules 27, 1–21. doi: 10.3390/molecules27031141
Ungerer, J., and Pakrasi, H. B. (2016). Cpf1 Is A versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci. Rep. 6, 1–9. doi: 10.1038/srep39681
Vega, J., Bonomi-Barufi, J., Gómez-Pinchetti, J. L., and Figueroa, F. L. (2020). Cyanobacteria and red macroalgae as potential sources of antioxidants and UV radiation-absorbing compounds for cosmeceutical applications. Mar. Drugs 18, 1–26. doi: 10.3390/md18120659
Vogel, A. I. M., Lale, R., and Hohmann-Marriott, M. F. (2017). Streamlining recombination-mediated genetic engineering by validating three neutral integration sites in Synechococcus sp. PCC 7002. J. Biol. Eng. 11, 1–9. doi: 10.1186/s13036-017-0061-8
Wang, B., Eckert, C., Maness, P. C., and Yu, J. (2018). A genetic toolbox for modulating the expression of heterologous henes in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synth. Biol. 7, 276–286. doi: 10.1021/acssynbio.7b00297
Włodarczyk, A., Selão, T. T., Norling, B., and Nixon, P. J. (2020). Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production. Commun. Biol. 3:215. doi: 10.1038/s42003-020-0910-8
Work, V. H., Melnicki, M. R., Hill, E. A., Davies, F. K., Kucek, L. A., Beliaev, A. S., et al. (2015). Lauric acid production in a glycogen-less strain of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 3:48. doi: 10.3389/fbioe.2015.00048
Yang, R., Zhu, L., Li, T., Zhu, L. Y., Ye, Z., and Zhang, D. (2021). Photosynthetic conversion of CO2 into pinene using engineered Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 9:779437. doi: 10.3389/fbioe.2021.779437
Yao, L., Cengic, I., Anfelt, J., and Hudson, E. P. (2016). Multiple gene repression in cyanobacteria using CRISPRi. ACS Synth. Biol. 5, 207–212. doi: 10.1021/acssynbio.5b00264
Yoshino, T., Kakunaka, N., Liang, Y., Ito, Y., Maeda, Y., Nomaguchi, T., et al. (2017). Production of ω3 fatty acids in marine cyanobacterium Synechococcus sp. strain NKBG 15041c via genetic engineering. Appl. Microbiol. Biotechnol. 101, 6899–6905. doi: 10.1007/s00253-017-8407-1
Zess, E. K., Begemann, M. B., and Pfleger, B. F. (2016). Construction of new synthetic biology tools for the control of gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. Biotechnol. Bioeng. 113, 424–432. doi: 10.1002/bit.25713
Zhang, F., and Huang, Z. (2022). Mechanistic insights into the versatile class II CRISPR toolbox. Trends Biochem. Sci. 47, 433–450. doi: 10.1016/j.tibs.2021.11.007
Keywords: synthetic biology, marine, cyanobacteria, genetic tools, metabolic engineering
Citation: Bourgade B and Stensjö K (2022) Synthetic biology in marine cyanobacteria: Advances and challenges. Front. Microbiol. 13:994365. doi: 10.3389/fmicb.2022.994365
Received: 14 July 2022; Accepted: 24 August 2022;
Published: 16 September 2022.
Edited by:
Michele Fabris, University of Southern Denmark, DenmarkReviewed by:
Jinjin Diao, Washington University in St. Louis, United StatesCopyright © 2022 Bourgade and Stensjö. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karin Stensjö, a2FyaW4uc3RlbnNqb0BrZW1pLnV1LnNl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.