- School of Food Engineering, Ludong University, Yantai, China
Shewanella baltica is the dominant bacterium that causes spoilage of seafood. RpoS is an alternative sigma factor regulating stress adaptation in many bacteria. However, the detailed regulatory mechanism of RpoS in S. baltica remains unclear. This study aims to investigate the regulatory function of RpoS on spoilage activity and adhesion ability in S. baltica. Results revealed that RpoS had no effect on the growth of S. baltica, but positively regulated the spoilage potential of S. baltica accompanied by a slower decline of total volatile basic nitrogen, lightness, and the sensory score of fish fillets inoculated with rpoS mutant. RpoS negatively regulated the adhesion ability, which was manifested in that the bacterial number of rpoS mutant adhered to stainless steel coupon was higher than that of the S. baltica in the early stage, and the biofilm formed on glass slide by rpoS mutant was thicker and tighter compared with S. baltica. Transcriptomic analysis showed that a total of 397 differentially expressed genes were regulated by RpoS. These genes were mainly enrichment in flagellar assembly, fatty acid metabolism/degradation, and RNA degradation pathways, which were associated with motility, biofilm formation and cold adaptation. This study demonstrated that RpoS is a primary regulator involved in flagellar assembly mediated biofilm formation and cold adaptation-related spoilage activity of S. baltica. Our research will provide significant insights into the control of microbiological spoilage in seafood.
Introduction
Microbial spoilage of seafood during processing, storage and sales lead to massive economic losses in the aquatic industry (Odeyemi et al., 2020; Zhuang et al., 2021). The psychrotolerant Shewanella baltica is a well-known spoilage bacterium frequently detected in chilled fish and seafood (Vogel et al., 2005). Studies indicated that S. baltica was the specific spoilage microorganism in raw salmon, chilled shrimp, and large yellow croaker (Macé et al., 2013; Zhu et al., 2015, 2016). S. baltica is a gram-negative, motile, rod-shaped bacterium, which is capable of producing H2S. S. baltica possesses a strong spoilage potential by the formation of spoilage-associated metabolites, such as utilizing trimethylamine-N-oxide as an electronic receptor to produce trimethylamine, dimethylamine and formaldehyde, thus bringing the unpleasant off-flavors (Broekaert et al., 2011; Wright et al., 2019). S. baltica decomposed nutrients in fish mainly via nitrogen and nucleotide pathways and thereby caused the production of amines, organic acids, etc. (Serio et al., 2014; Lou et al., 2021). Generally, diverse microbiota is discovered on newly caught fish, among which S. baltica only accounts for a fraction of the initial microflora, but S. baltica dominates and becomes the spoilage microorganism during low-temperature storage (Parlapani et al., 2014). It indicates that S. baltica has a robust ability to accommodate rapidly changing conditions. The response of bacteria to different environmental stresses requires complex coordination and overlapping regulatory networks (Zhuang et al., 2021). Adhesion is an important way for bacteria to adapt to the environment, which is a key step in the spoilage process of seafood. S. baltica has the potential to attach to seafood and form biofilm on the surface (Zhu et al., 2019). Understanding the environmental adaptation mechanism of S. baltica will provide guidelines for the storage of seafood.
Sigma factor is a constituent of bacterial RNA polymerase responsible for recognizing promoters and initiating transcription. Previous studies have shown that the sigma factor regulates a large regulon in Gram-negative bacteria, accounting for about 10% of the genome (Schellhorn, 2014; Wong et al., 2017). RpoS is associated with stress adaptation and response to changeable environments (Dodd and Aldsworth, 2002; Dong and Schellhorn, 2010). The function of RpoS has been widely explored in Escherichia coli, Serratia plymuthica, and Vibrio cholerae (Vidovic et al., 2011; Liu et al., 2016; Wölflingseder et al., 2022), and the results showed that RpoS enhanced their adaptation to various environmental stresses (starvation, acidic pH, oxidative stress, etc.). Recent studies found that RpoS can also regulate virulence factor expression, such as adhesion, biofilm-forming, antibiotic tolerance, etc. Mata et al. (2017) showed that the adherence to epithelial cells of E. coli was upregulated by RpoS. RpoS also revealed a positive effect on the expression of T6SS gene and flagellum formation with the increased synthesis of exopolysaccharides in Y. pseudotuberculosis (Guan et al., 2015). The role of RpoS played in stress adaptation and toxin factor expression indicated that RpoS might be involved in the environmental adaptation and spoilage ability of S. baltica. Feng et al. (2021) demonstrated that sigma factors RpoS/RpoN regulated the stress response and spoilage activity of S. baltica. Our previous study indicated that RpoS controlled the stress resistance, quorum sensing and biofilm formation in S. baltica (Zhang et al., 2021), however, the detailed genes regulated by RpoS remain unknown.
Recently, many emerging omics methods, such as genomics, metabolomics, and transcriptomics, have been widely applied in the study of food safety with the rapid development of science and technology (Zhao et al., 2020; Li et al., 2021). For instance, the stress responses of Salmonella enterica to thermal and essential oils were clarified by metabolomics and transcriptomics (Chen et al., 2022b). Transcriptomics was a feasible technique, which uses the RNA-seq high-throughput sequencing technology to investigate the gene expression level and regulation of food microorganisms at the overall level. Therefore, transcriptomics was applicable to analyze the function of RpoS in S. baltica.
For a better understanding of the role of RpoS in S. baltica, the difference between S. baltica and rpoS mutant in spoilage potential and adhesion effect was compared. We further explored the differentially expressed genes (DEGs) in S. baltica and rpoS mutant using a transcriptome technology. This study aims to clarify the regulatory mechanism of RpoS in S. baltica, which will provide new ideas for the control of spoilage microorganisms.
Materials and methods
Strains and culture conditions
The S. baltica X7 was previously isolated from large yellow croaker. The corresponding rpoS mutant was constructed in the previous study (Zhang et al., 2021). Both of these two bacterial species were cultured in LB broth.
Spoilage activity determination
The spoilage activity of two bacterial strains was evaluated by sterile large yellow croaker fillets. The sterile fish fillets were prepared as described by Macé et al. (2013). The overnight culture of S. baltica and rpoS mutant were centrifuged and re-suspended in sterile water to OD600 ≈ 0.2. Then 5 ml of the corresponding bacterial suspension was added to 500 ml of sterile water. After that, all the fish fillets were divided into three groups. Two groups were immersed with the bacterial suspension of S. baltica and rpoS mutant for 10 min, respectively, and the initial bacterial count of fish fillets was approx. 4 log CFU/g. The other group was immersed in sterile water for 10 min as a control (Wang et al., 2017). The fillets were then individually packed in polyvinyl chloride bags and stored at 4 ± 0.5°C. Three fillets from each group were randomly selected for analysis every day. Total viable counts (TVC) were detected as reported by Ozogul et al. (2017). Approximately 25 g of fish fillets were diluted by 225 ml of 0.85% NaCl solution, and the plate count agar was poured onto the dilution and cultured for 48 h at 30°C. The sensory evaluation was conducted based on the studies of Parlapani et al. (2014) and Chen et al. (2022a) with some modifications (Supplementary Table S1). Five trained panelists assessed the fish quality from three aspects: color, texture, and odor. The spoilage level of the fish fillets was scored on a continuous scale from 0 to 10. The total scores of 8–10 points are fresh; 5–7 points are slight spoilage; and 0–4 points are spoilage. The total volatile basic nitrogen (TVB-N) of fish fillets was measured by the Conway and Byrne method using Semi-trace Kevlar Nitrometer (Parlapani et al., 2014). The lightness of fish fillets was determined by a chroma meter (CR-400, Konica Minolta, INC, Japan).
Bacterial adhesion assays
Bacterial adhesion to stainless steel
The adhesion assay of bacterial strains was performed on the stainless steel coupon in LB broth. The 304 stainless steel coupons (10 mm × 10 mm, 1.5 mm thickness) were washed with 75% ethanol and distilled water, and then they were sterilized (Yuan et al., 2018). One milliliter of an overnight culture of S. baltica wild-type and rpoS mutant was inoculated into a centrifugal tube containing 5 ml LB broth, respectively. Each centrifugal tube contained one stainless steel coupon and was cultured for 24 h at 30°C. The number of colonies attached to stainless steel coupon was determined at 0 and 15 min, 1, 2, 4, 7, 10, and 24 h. The stainless steel coupon was taken out with sterile tweezers and washed with sterile distilled water. The stainless steel coupon was immersed in 5 ml distilled water, handled with ultrasonic for 10 min, and homogenized for 2 min. The determination of the total viable count is the same as described above.
Microscopic analysis
For scanning electron microscopy (SEM) observation, the sterilized stainless steel coupons were incubated in LB broth containing S. baltica or rpoS mutant for 24 h, then the stainless steel coupon was washed with sterile distilled water three times and fixed with 2.5% glutaraldehyde overnight. The coupons were dehydrated by immersing in 30%, 50%, 70%, 80%, and 90% ethanol for 10 min, respectively. Then further dehydration was carried out in 100% ethanol for 15 min twice. Finally, the coupons were dried, gold sputtered and observed using a scanning electron microscope (Hitachi Model SU8010, Japan).
For light microscopic observation, the bacterial cells attached to glass were investigated according to Santhakumari et al. (2018). The sterilized glass slides were plated in LB and cultured for 24 h. Then the glass slides were washed with sterile distilled water and stained with crystal violet for 15 min. The glass slides were examined under a light microscope (Olympus CX22, Tokyo, Japan) after drying.
Transcriptome samples preparation and analyze
RNA extraction and library preparation
Strains of both S. baltica and rpoS mutant were incubated to OD600 = 1.5, and the bacterial pellet was obtained by centrifugation (4,000 rpm, 15 min). RNA was extracted using a RNeasy Mini Kit (Qiagen, Germany). The rRNA was removed and mRNA was left. The fragmentation was obtained by divalent cations in NEBNext First Strand Synthesis Reaction Buffer. First-strand of cDNA was synthesized with a random hexamer primer. Second strand cDNA synthesis was carried out by DNA Polymerase I and RNase H. The library fragments were purified by an AMPure XP system (Beckman, United States). Afterward, PCR was performed and PCR products were analyzed with the Agilent Bioanalyzer 2100 system. The library preparations were sequenced with an Illumina Novaseq platform.
Analysis of RNA-seq
The reads containing adapter, ploy-N (N rate > 10%) and low-quality reads were removed and clean reads were collected. The Q20 and Q30 indicated the data were of high quality with values of >98 and 95%, respectively. The clean reads were mapped to S. baltica OS678 complete genome in the NCBI database using Bowtie2-2.2.3. Gene expression levels were analyzed by fragments Per Kilobase of transcript sequence per Millions of base pairs sequenced (FPKM; Trapnell et al., 2013). DEGs were analyzed according to the false discovery rate (FDR) values (<0.05), p values (<0.05), and the |log2 (fold change)| values (>0.5). The biological functions and metabolic pathways of the DEGs were classified according to the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. GO terms with adjusted p < 0.05 were regarded as significantly enriched by DEGs. The statistical enrichment of DEGs in KEGG pathways was analyzed by KOBAS software.
qRT-PCR validation
Bacterial strains were cultured to the end of the exponential phase. RNA was obtained by trizol reagent (Sangon Biotech, China). The EasyScript One-step gDNA Removed and cDNA Synthesis Supermix kit (Transgen, China) was used to compose cDNA. The primers were listed in Supplementary Table S2. TransStart Tip Green qPCR supermix kit (Transgen, China) was used for qPCR amplification. The threshold cycles (CT) were recorded and the relative expression level of genes was obtained by the 2–△△Ct method (Li et al., 2019).
Data analysis
Each analysis was conducted in triplicate. The results were recorded as the mean ± standard deviation. The significance between samples (p < 0.05) was analyzed by SPSS 19.0 software with one-way analysis of variance (ANOVA).
Results and discussion
RpoS positively regulated spoilage activity
The spoilage activity of bacterial strains was estimated on sterile yellow croaker fillets. The initial viable count for control was below 2.0 log CFU/g. The initial number for fillets that were inoculated with S. baltica or rpoS mutant was about 4.8 log CFU/g. The number of bacteria increased with storage time, and the growth curves of the two strains were very close, which almost overlapped after 4 days of storage with a viable count of about 8.2 log CFU/g, indicating that RpoS did not affect the growth of S. baltica (Figure 1A). Our previous study has shown that the deletion of rpoS does not affect the growth of S. baltica at neither 30 nor 4°C in LB broth (Zhang et al., 2021). Similarly, the growth of P. fluorescens did not change without rpoS (Liu et al., 2018). RpoS regulator was first identified as a growth phase-dependent regulator, and it usually plays a role in stress regulation at the stationary phase (Schellhorn, 2014). Therefore, RpoS does not affect the growth of S. baltica.
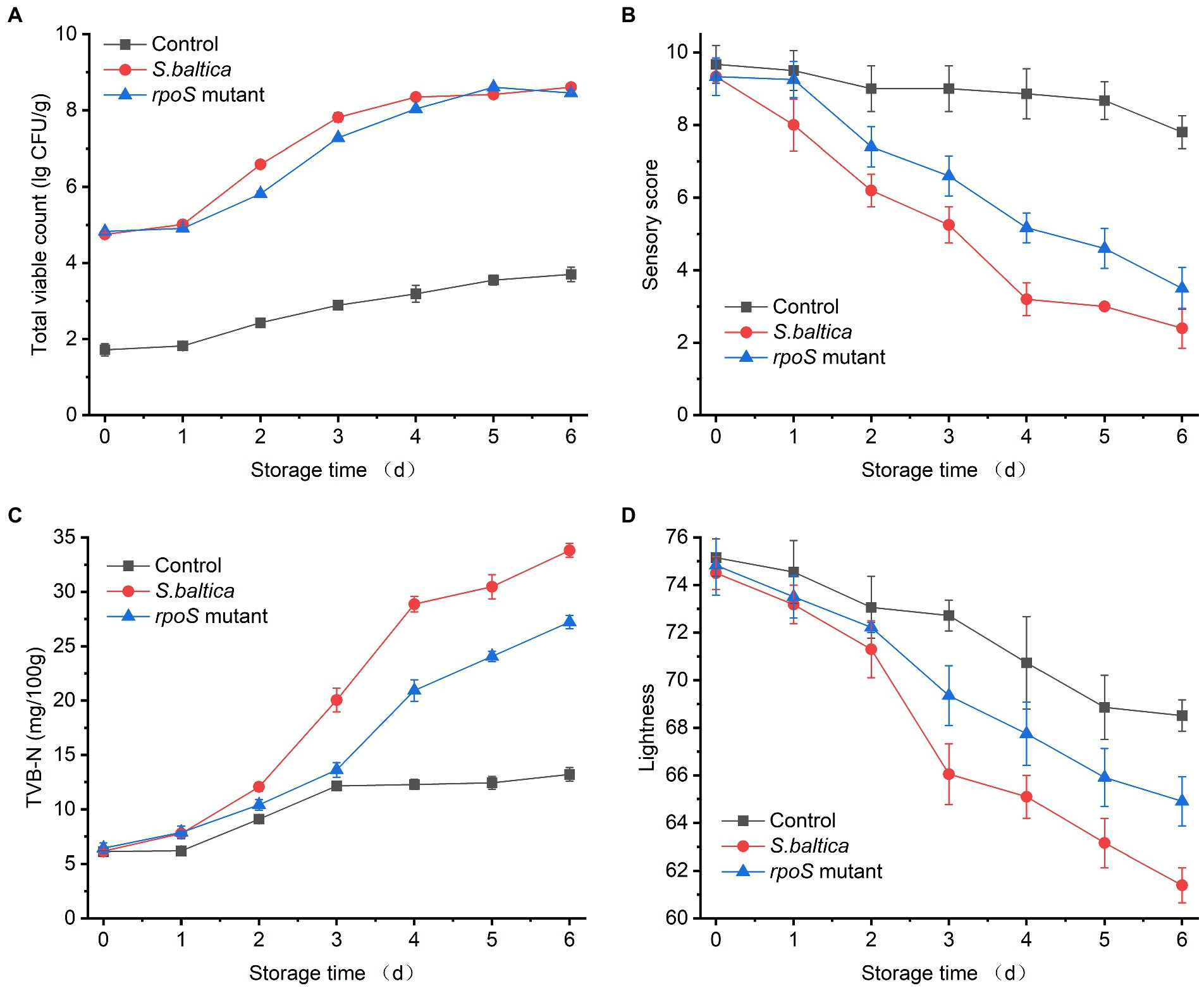
Figure 1. Changes in total viable counts (A), sensory score (B), TVB-N (C), and lightness (D) of yellow croaker fillets inoculated with Shewanella baltica and rpoS mutant during storage at 4°C.
Sensory scores of all samples decreased with the prolonging of storage time because of the production of spoilage metabolite and change in appearance (Figure 1B). TVB-N is one of the typical spoilage metabolites, which is usually used to evaluate the process of fish spoilage. The TVB-N of samples inoculated with S. baltica and rpoS mutant increased quickly after 3 days of storage, and the group inoculated with S. baltica reached a maximum TVB-N value exceeding 30 mg/100 g at day 6 (Figure 1C). The lightness of fish fillets inoculated with S. baltica declined quickly compared with that inoculated with rpoS mutant (Figure 1D), which was in accordance with the sensory scores. The sensory score of the fish fillets inoculated with rpoS mutant was significantly lower than that of S. baltica after 3 days of storage (p < 0.05), suggesting that RpoS positively regulated the spoilage activity of S. baltica. Similar results were also found in P. fluorescens and S. baltica SB02, the spoilage activities of which decreased when the rpoS gene was deleted, which is in agreement with the result in this study. It was inferred that this phenomenon might be attributed to the decrease of extracellular enzyme activity or the down-regulation of quorum sensing in rpoS mutant (Liu et al., 2018, 2019; Feng et al., 2021; Hai et al., 2022). The regulatory mechanism of RpoS on bacterial spoilage needs to be thoroughly explored in the future.
RpoS negatively regulated the adhesion
Bacteria mediated seafood spoilage is a complex process, involving in adhesion, proliferation, and forming biofilm (Sun et al., 2022). Once bacteria attach to seafood, biofilms will gradually form, thereby causing seafood spoilage (Zhao et al., 2022). The result of bacterial adhesion ability on stainless steel coupon was displayed in Figure 2A. In the early stage of adhesion (0–4 h), S. baltica wild type showed a very slow adhesion to the stainless steel, but the number of rpoS mutant on stainless steel coupon was higher than that of S. baltica. After 2 h of incubation, the number of rpoS mutant colonies that adhered to the coupon was nearly 100-fold higher than S. baltica. After 24 h, the number of colonies in both strains reached 6.6 CFU/cm2, and no significant difference between S. baltica and the rpoS mutant was observed. It can be observed that the rpoS mutant exhibited an earlier and stronger adhesion ability than that of S. baltica wild type. For the SEM visualization (Figures 2B,C), similar biofilm morphology of rpoS mutant and S. baltica on stainless steel coupon was observed, which was in accordance with the attached bacterial count at 24 h. For the glass slides, the biofilm formed by S. baltica wild type was relatively loose (Figure 2D), while rpoS mutant formed more compact biofilm (Figure 2E). Compared with S. baltica wild type, rpoS mutant showed earlier adhesion ability on stainless steel coupon and thicker biofilm on the glass slide, indicating that RpoS negatively regulated the adhesion related characteristics of S. baltica in a time-dependent and condition-dependent manner.
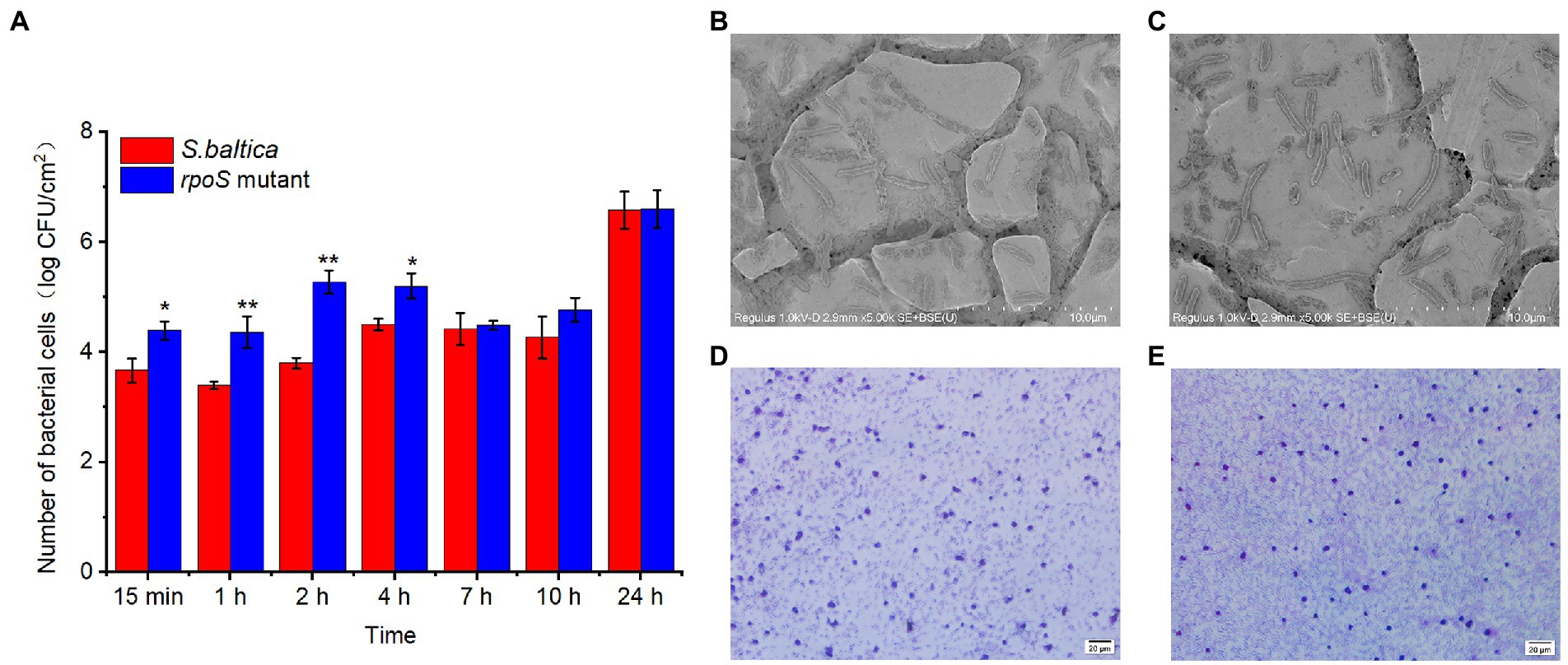
Figure 2. Number of bacterial count on stainless steel coupon, “*” and “**” indicate the significant difference at p < 0.05 and p < 0.01, respectively (A). Scanning electron microscopy images of biofilm formed on stainless steel coupon by Shewanella baltica (B), and rpoS mutant (C). Microscopic visualization of biofilm formed on glass slide by S. baltica (D), and rpoS mutant (E).
RpoS usually had a positive response to the changes in the external environment or living state, for example, the motility of Vibrio cholerae decreased when the rpoS was deleted (Wölflingseder et al., 2022). However, RpoS also manifested some negative regulations. RpoS was found to negatively regulate the swimming motility in E. coli and S. plymuthica G3 (Ojima et al., 2012; Liu et al., 2016), suggesting that the regulatory mechanism of RpoS may be species-dependent. Consistent with our result, Feng et al. (2021) reported that the deletion of RpoS enhanced the swimming motility in S. baltica SB02, and the flagella of the rpoS mutant was thicker than that of the wild type observed by transmission electron microscopy. In addition, the biofilm production in Cronobacter sakazakii with a defect of rpoS only declined in 24 h, and there was no significant difference after 48 h of incubation, revealing that the regulation of RpoS on biofilm formation only showed a delayed effect rather than a complete inhibition (Fernández-Gómez et al., 2020). It revealed that RpoS has a growth-stage dependence on the regulation of biofilm, which provides important insights into the control of S. baltica during seafood processing.
RNA sequencing analysis
In this study, to further investigate the regulatory role of RpoS, a comparative transcriptome analysis of the rpoS gene-deficient strain (981 bp segment) and the S. baltica wild-type was performed. The total RNA obtained from S. baltica and rpoS mutant cells were analyzed to identify the DEGs. After filtration, there were on average 7,775,014 and 7,509,599 raw reads for S. baltica and rpoS mutant, respectively (Supplementary Table S3). A total of 7,638,874 and 7,369,249 high-quality reads from S. baltica and rpoS mutant, respectively, were mapped on the reference genome of S. baltica OS678 (Supplementary Table S3). Approximately 85% of short reads were successfully mapped on the S. baltica OS678 genome (Supplementary Table S4).
To reduce the impact of sequencing depth and gene length, the corrected FPKM is usually used for describing the gene expression value of RNA-seq. The correlation coefficients indicate the correlations of gene expression between samples. As exhibited in Figure 3A, the squares of the Pearson correlation coefficients (R2) were all greater than or equal to 0.8, signifying that the expression patterns of genes were very similar between samples (Garber et al., 2011). A total of 397 DEGs was observed, which accounted for almost 10% of the genome (Figure 3B; Supplementary Table S5). The expression of 248 genes (approximately 62%) increased in rpoS mutant, while the other 149 genes (approximately 38%) decreased.
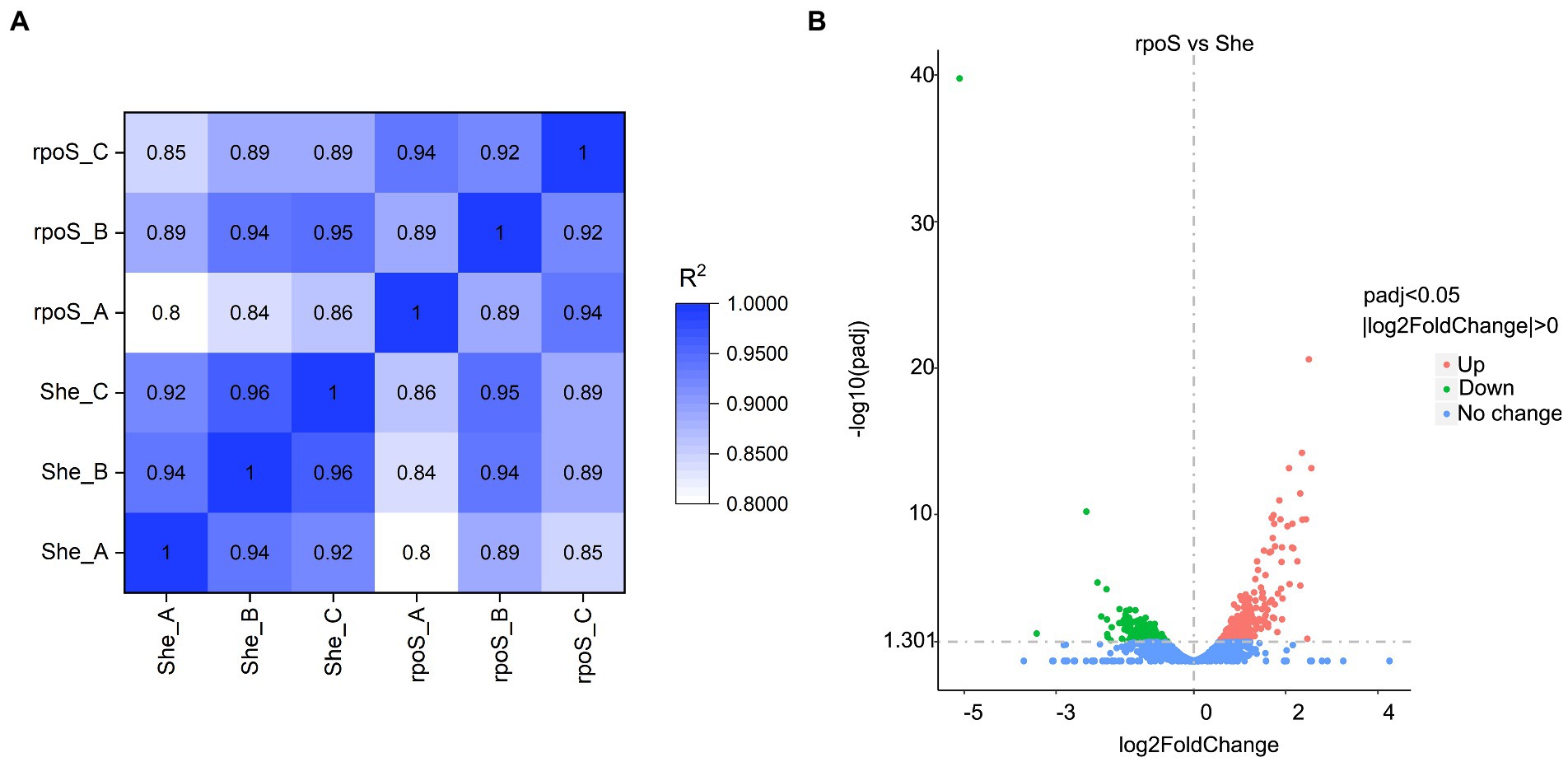
Figure 3. The heat-map of Pearson correlation coefficients based on gene expression levels (A). Volcano plots of different expressed genes (B). “She” indicates Shewanella baltica wild type, “rpoS” indicates the rpoS mutant.
Hierarchical clustering was performed according to the 397 DEGs, in which the intuitive changes of DEGs between samples were observed (Figure 4). The unsupervised clustering formed two major gene clusters. Cluster 1 (red color) and cluster 2 (green color) indicated the upregulated and downregulated genes, respectively.
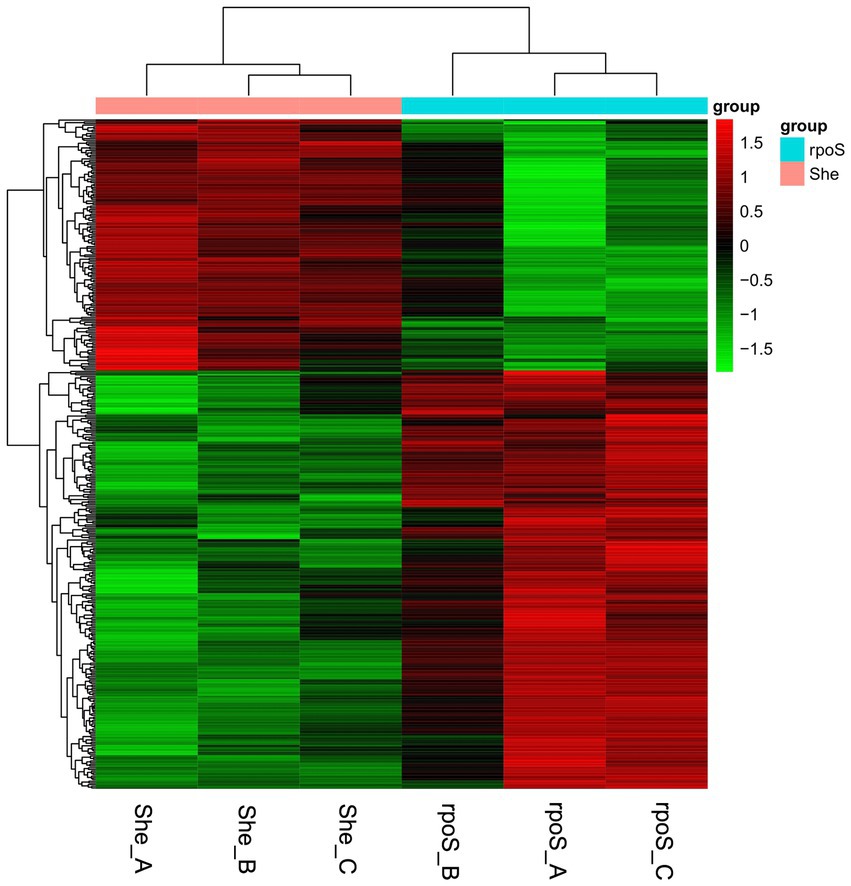
Figure 4. Heatmap of different expressed genes in Shewanella baltica and rpoS mutant. Each row represents the expression pattern of a single gene and each column corresponds to a single sample. “She” indicates S. baltica wild type, “rpoS” indicates the rpoS mutant.
GO analysis
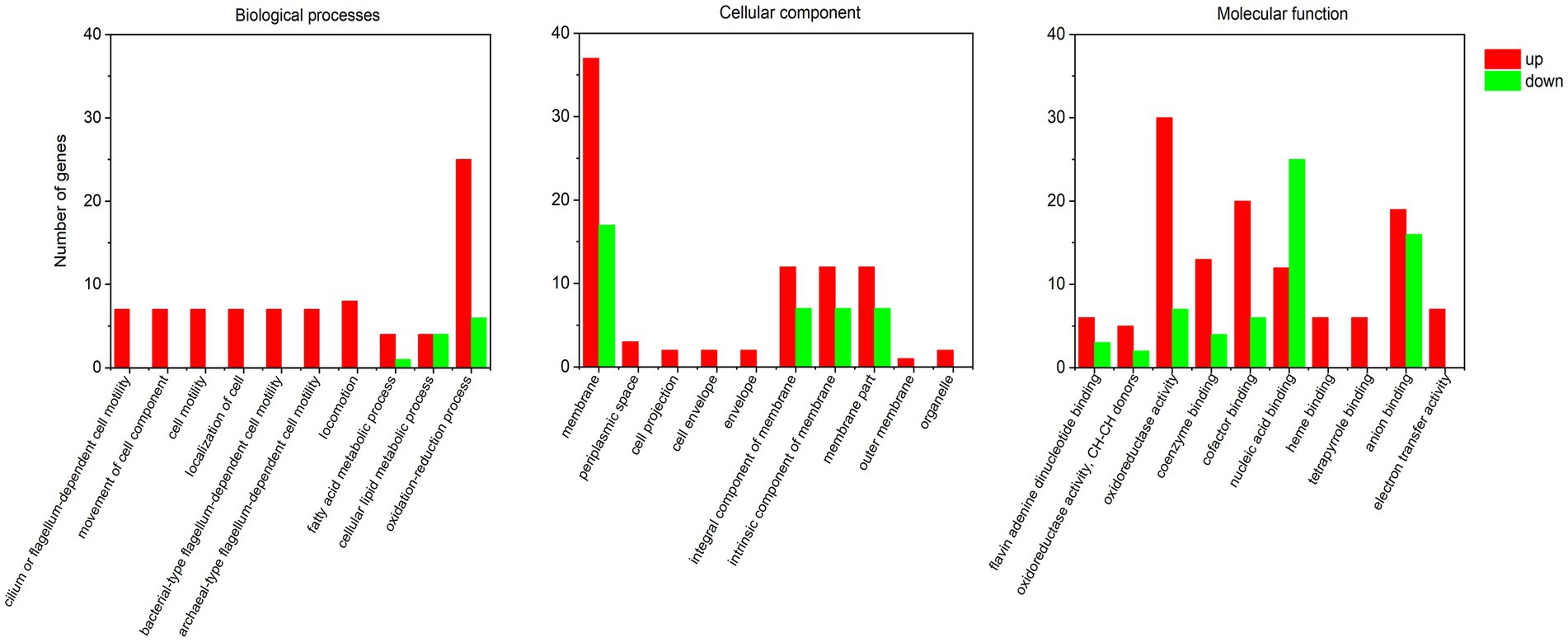
Gene Ontology enrichment analysis was conducted based on the DEGs. Among the up-regulated DEGs in rpoS mutant (Figure 5; Supplementary Table S6), it showed that several categories of biological processes were enriched in the DEGs of rpoS mutant, including flagellum-dependent cell motility (GO:0001539, GO:0071973, and GO:0097588), movement of cell component, cell motility (GO:0006928, GO:0048870, and GO:0040011), localization of cell (GO:0051674), and oxidation–reduction process (GO:0055114). As to molecular function, oxidoreductase activity (GO:0016491, GO:0016627), cofactor and coenzyme binding (GO:0048037, GO:0050662), heme binding (GO:0020037), flavin adenine dinucleotide (GO:0050660), tetrapyrrole binding (GO:0046906), and electron transfer activity (GO:0009055) were over-represented. The oxidoreductase activity and cofactor binding were the most represented. The increased motility has been observed in rpoS mutant (Zhang et al., 2021), which was mediated by flagellum. The changes in oxidoreductase indicated that RpoS may be involved in the activation of oxidative stress response of S. baltica. Therefore, cell motility and oxidoreductase activity were considered to be the main regulation pathways of RpoS.
Among downregulated DEGs in rpoS mutant, membrane (GO:0016020) in cellular component, nucleic acid binding (GO:0003676), and RNA binding (GO:0003723) involved in molecular function were representive. In terms of the membrane, the expressions of major facility superfamily (MFS) genes (RS23645, RS43790, and RS29395) were significantly declined in rpoS mutant. MFS family transporters are not only related to the absorption of nutrients but also involved in response to external stress. It can be inferred that membrane is one of the main targets of RpoS (Costa et al., 2014).
KEGG analysis
To comprehensively analyze the regulatory function of DEGs, these DEGs were further subjected to KEGG analysis. For the upregulation of DEGs, they were principally focused on flagellar assembly, bacterial chemotaxis, fatty acid metabolism, and two-component system. The downregulation of DEGs were enriched in the RNA degradation and quorum sensing pathways (Figure 6; Supplementary Table S7).
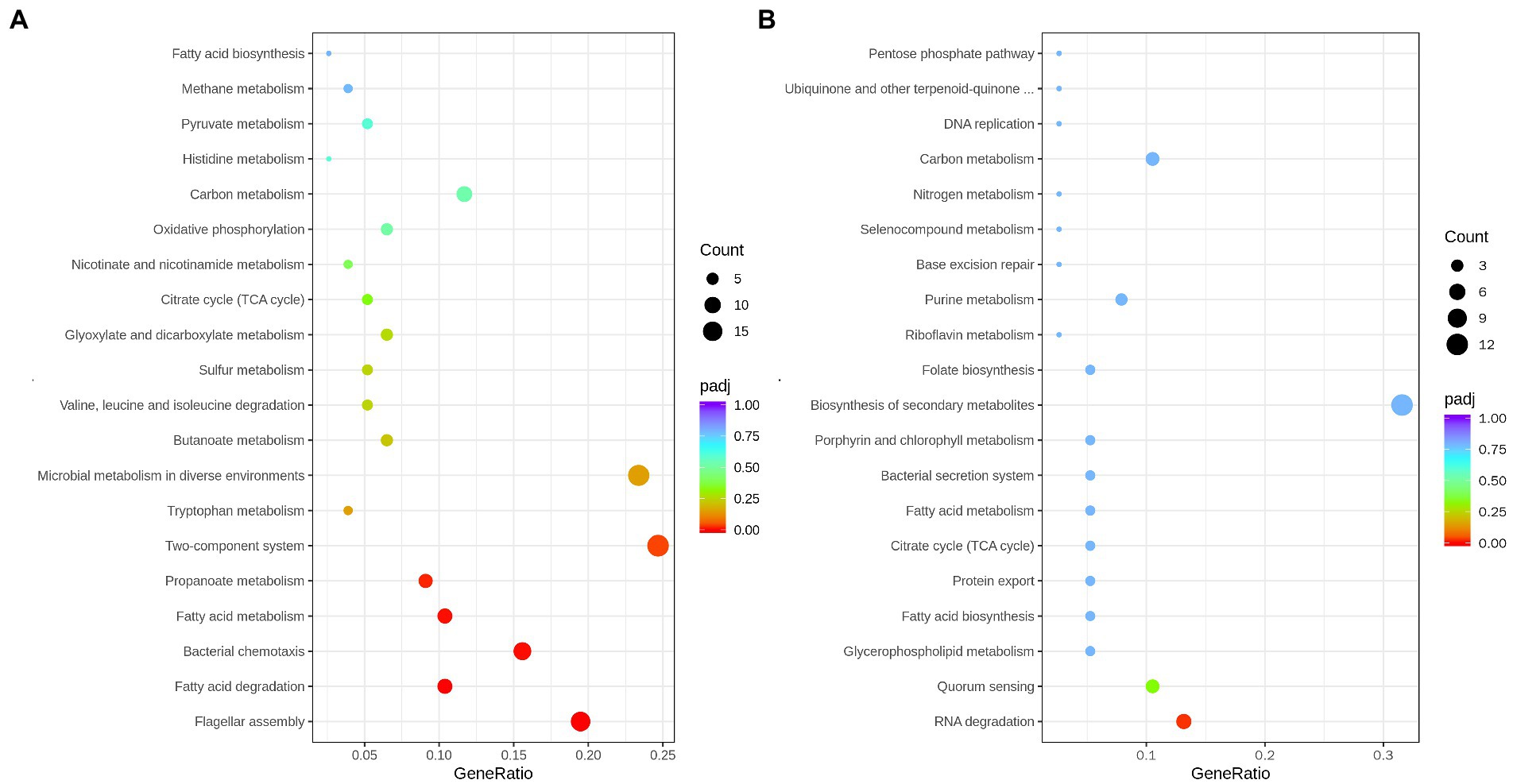
Figure 6. Scatterplot of upregulated genes (A), and down-regulated genes (B), enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Enrichment score represents the ratio of the number of upregulated/downregulated genes and the number of all genes in the pathway.
Flagellar assembly and biofilm formation
Flagella is an important motor organ in bacteria. It was reported to have three functional characteristics: motility, chemotaxis, and adhesion, which contribute to responding to the adverse environment quickly and timely for bacteria (Lund et al., 2020). The motility can promote the adhesion of bacteria on host cells, thus realizing infection (Colin et al., 2021). Previous studies indicated that flagella played a crucial role in the adhesion for bacteria, as the flic subunit promoted bacteria move to the attachment site in the early stage of adhesion (Sun et al., 2022). It has been observed in our previous study that RpoS negatively regulated the motility of S. baltica and the diameter of the rpoS mutant increased by 46.6% more than S. baltica wild type on the plate (Zhang et al., 2021). The flagella structure consists of three parts: a body basel, hook, and filament (Chilcott and Hughes, 2010). The synthesis and assembly of flagellum are usually involved in more than 50 genes (Chevance and Hughes, 2008). As shown in Table 1, the high expression levels of flagellar body basel-related genes (flgF, flgG, flgH, flgI, flgC, fliF, fliN, and motB) and flagella hook associated genes (flgK, fliK, and flgD) were found in rpoS mutant. The flgM and flgE showed 1.21 and 0.72 log2 (fold change) expression levels, respectively (Figure 7). FlgM protein regulates the transcription level of flagellum-related genes, thus affecting the length and quantity of flagellum. The secretion of FlgM indicates the accomplishment of the Hook-basal body (Wosten et al., 2010). The function of FlgE is to connect the flagellum filament and hook, which changes the angle of flagellum rotation (Shen et al., 2017; Li et al., 2022). Previous studies showed that the enhanced motility of rpoS mutant required the RpoN, and about 60% of genes were simultaneously oppositely regulated by RpoS and RpoN (Dong and Schellhorn, 2010). Feng et al. (2021) found that the deficiency of RpoN leads to a decrease in the motility of S. baltica, which was owing to the downregulation of flagella assembly-related genes (i.e., flgF, flgC, etc). The current increased swimming motility of rpoS mutant of S. baltica might be also affected by the existence of RpoN.
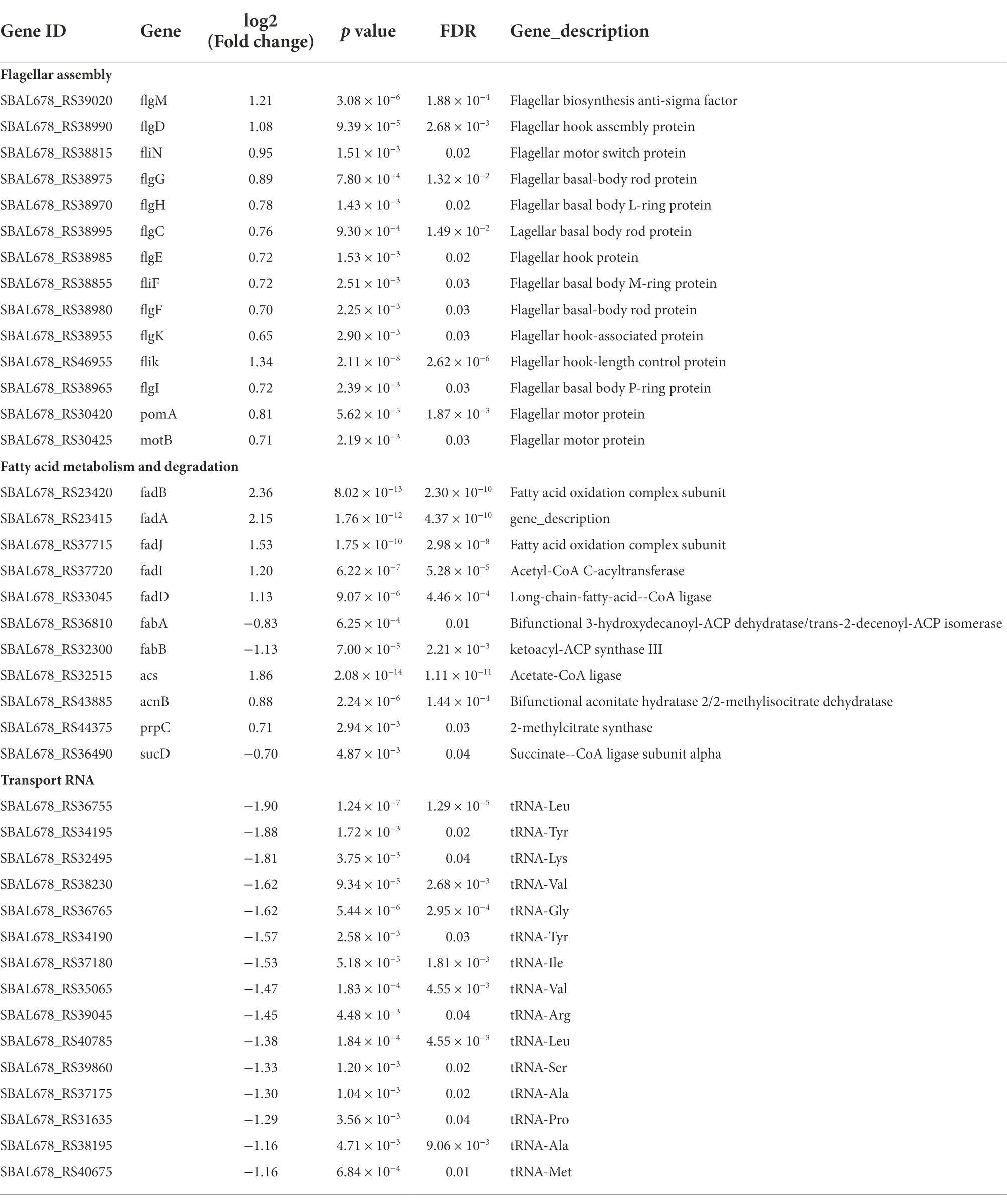
Table 1. Representative differently expressed genes in the Shewanella baltica wild type and rpoS mutant.
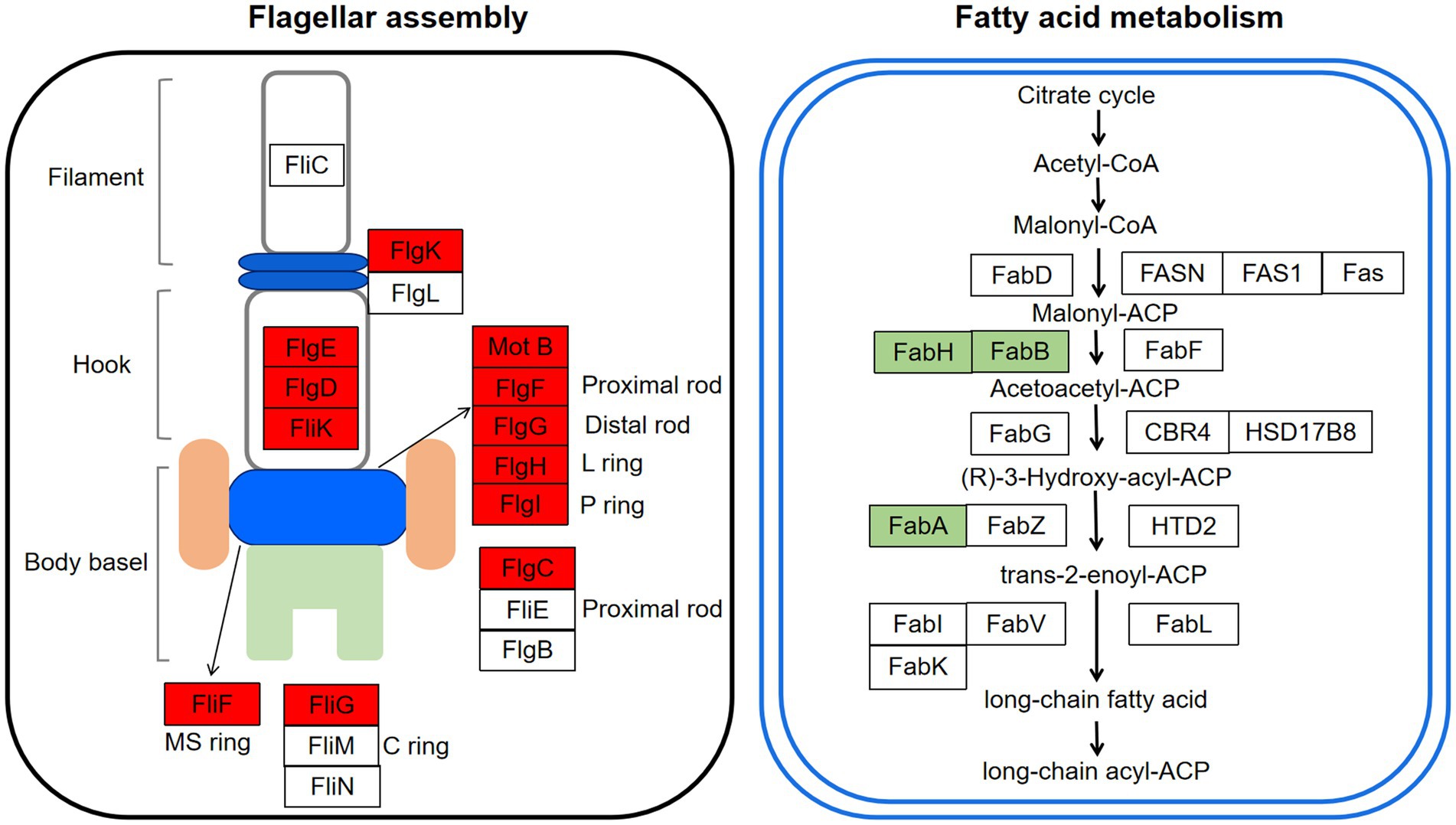
Figure 7. Changes of the expression levels of genes involved in flagellar assembly and fatty acid metabolism of Shewanella baltica. The genes marked in red and green represented upregulation and downregulation in rpoS mutant.
Biofilm formation is another important flagella-dependent survival model for microorganisms. When the number of bacteria fixed on the surface of host cells increases, the bacteria can secrete extracellular compounds, such as polysaccharides and protein, eventually forming stable biofilms (Voglauer et al., 2022). Zhao et al. (2022) exhibited that acidic electrolysed water disturbed the biofilm formation of E.coli by decreasing the nucleotide-related and carbohydrates component. This study revealed that the adhesion and biofilm-forming abilities of rpoS mutant were higher than that of S. baltica wild type (Figure 2). RNA degradation declined in rpoS mutant, while protein and carbohydrate metabolism had no significant change (Figure 6). We inferred that flagella assembly and RNA degradation might play a more important role in biofilm formation than nutrient metabolism The present study indicated that RpoS negatively regulated the flagellar-associated gene expression and biofilm formation. Therefore, RpoS-mediated flagellar assembly is one of the most crucial features of S. baltica in environmental adaptability.
Fatty acid metabolism
Fatty acid metabolism and fatty acid degradation are also important pathways regulated by RpoS (Figure 6). Fatty acids are the main components of the bacterial cell membrane. Fatty acid metabolism has been identified to be closely related to bacterial adaptation to the cold environment because the low temperature can reduce bacterial membrane fluidity and growth rate by adjusting the composition of fatty acids, especially the unsaturated fatty acids (UFAs) (Wang et al., 2020). In the fatty acid metabolism pathway, FabA plays a key role in the synthesis of UFAs. FabA is a bifunctional enzyme that can dehydrate β-hydroxy-decanoyl-ACP and isomerize trans-2-decenoyl-ACP in order to stimulate UFAs synthesis. While fadA, fadB, fadD, fadE, fadF, fadI, and fadJ are the essential genes in the fatty acid degradation pathway (Feng and Cronan, 2011). This study indicated that fabA was down-regulated by RpoS, while the expression of fadA, fadB, fadD, fadI, and fadJ were upregulated (Table 1), indicating that the deficiency of rpoS decreased the synthesis of UFAs in S. baltica. FabH is β-Ketoacyl-ACP synthase, which is the initiation factor of the carbon chain extension cycle, and fabH was down-regulated in S. baltica (Figure 7). Phosphatidylglycerol phosphate synthase (PgsA) catalyzes the primary reaction of phosphatidylglycerol biosynthesis, which was down-regulated in the rpoS mutant. Shewanella baltica is one of the most common specific spoilage bacteria in refrigerated fish and seafood. The strong adaptability to low temperature contributes Shewanella to maintaining a great growth state under refrigerated conditions. Initiation factor 1 (IF1) is encoded by infA, which is associated with the initiation step of protein synthsis in bacteria. The infA gene was positive-regulated by RpoS in S. baltica strain (Supplementary Table S5). Ko et al. (2006) reported that the expression of IF1 increased when E. coli cells were treated with cold shock, which is in accordance with our result. The deficiency of RpoS decreased the spoilage ability in S. baltica (Figure 1), which may be related to the decline of cold adaptation. Wang et al. (2020) reported that cell membrane UFAs-mediated cold adaptation was closely related to the spoilage potential. However, there are few reports on the relationship between RpoS and cold adaptation. Therefore, the cold adaptation regulatory mechanism regulated by RpoS needs to be further studied, which will contribute to clarifying the spoilage characteristics of Shewanella spp.
Two-component regulatory systems
Microorganism can assemble some necessary genetic circuits to sense external stimuli and respond to environmental stress. Two-component regulatory system (TCRS) is one of the most important signal transduction systems that allow bacteria to respond to stress conditions and improve their survival (Nguyen and Hong, 2008). The present study showed that TCRS was upregulated in the rpoS mutant of S. baltica. Bacterial TCRS is involved in the regulation of bacterial chemotaxis, morphological differentiation, nutrient metabolism, drug resistance, and many other physiological processes. Li et al. (2022) showed that the expression of rpoS can be activated by the two-component system of TorR using transcriptomics, while the effect of RpoS on TCRS was rarely studied. The up-regulated TCRS in the rpoS mutant indicated that TCRS is involved in the stress regulation of RpoS to S. baltica, but the detailed mechanism needs to be further explored.
Transport RNA
The tRNA mainly participates in the process of protein translation. Table 1 showed that 15 kinds of the transfer RNA (tRNA) were downregulated, indicating that RpoS contributes to the tRNA-mediated protein synthesis. Studies revealed that translation regulation is an important mechanism for bacteria to respond to stress quickly, and the dynamic adjustment of the total amount of tRNA is a critical regulation means for bacteria to deal with the harsh environment. Zhong et al. (2015) reported that E. coli slowed down the rate of translation extension by downregulating the expression of tRNA, which makes cells promptly adapt to oxidative stress.
Quorum sensing
Quorum sensing (QS) is a mechanism that microorganisms communicate to sense chemical signaling change, thus mediating the toxin factors expression, such as biofilm formation and extracellular enzyme production (Mukherjee and Bassler, 2019). Figure 6 showed that the expression of QS pathway decreased in the rpoS mutant. Consistent with this study, our previous study demonstrated that QS signals cyclo-(L-Pro-L-Leu) and cyclo-(L-Pro-L-Phe) declined in rpoS mutant, and the expression levels of QS sensing genes luxR were also declined in rpoS mutant (Zhang et al., 2021). Similar phenomena were also found in P. fluorescens and S. baltica SB02, in which the acyl homoserine lactone and diketopiperazines QS signaling decreased in the deletion of rpoS mutant (Liu et al., 2018; Zhang et al., 2021), indicating that RpoS usually positively regulated the QS. Therefore, QS is expected to become a new target to control the spoilage caused by S. baltica.
Others
Cold shock protein is an important protein that affects the ability of bacteria to adapt to low temperature. Cold shock DNA-binding domains (RS31005) were downregulated in the rpoS mutant (Supplementary Table S5). Cold shock protein can be used as molecular chaperones to bind with single-stranded DNA or single-stranded RNA, which prevents DNA or RNA from forming secondary structures at low temperature, thereby promoting transcription and translation (Mihailovich et al., 2010). DNA gyrase B and parE (RS27555), which were the targets of antibiotics in Gram-negative bacteria, were downregulated in rpoS mutant. Bacterial DNA gyrase is the basic enzyme involved in the formation of DNA superhelix, and the decrease of gyrase related genes usually causes antibiotic resistance (Jogula et al., 2020). It suggests that RpoS take part in the regulation of antibiotic resistance of S. baltica. Previous studies indicated that the rpoS gene was involved in tolerance to antibiotics in Pseudomonas aeruginosa during the stationary phase (Keiji et al., 2010), and the deficiency of rpoS in Salmonella Enteritidis decreased the tolerance to essential oils (Cariri et al., 2019), which supports our conclusion.
Danhorn and Fuqua (2007) described that flagellum generally guides bacteria colonized on food-contact surfaces in the initial adhesive stage, then the flagella declined with the bacterial proliferation. Food gradually spoiled with the bacteria adapting to the environment and secretion of extracellular enzymes (Hai et al., 2022). Previous studies have confirmed that fatty acid synthesis and QS can enhance the growth and spoilage ability of S. baltica (Zhu et al., 2015; Wang et al., 2020). In this study, the fatty acid synthesis and QS pathways were downregulated in rpoS mutant, and its spoilage ability also decreased, but the gene expression related to flagellum assembly increased. Therefore, we speculated that RpoS might improve the survival strategy and spoilage activity through early adhesion mediated by flagella assembly pathway as a crucial target. Overall, RpoS is an important regulator involved in the environmental adaptation and spoilage potential of S. baltica.
qPCR validation
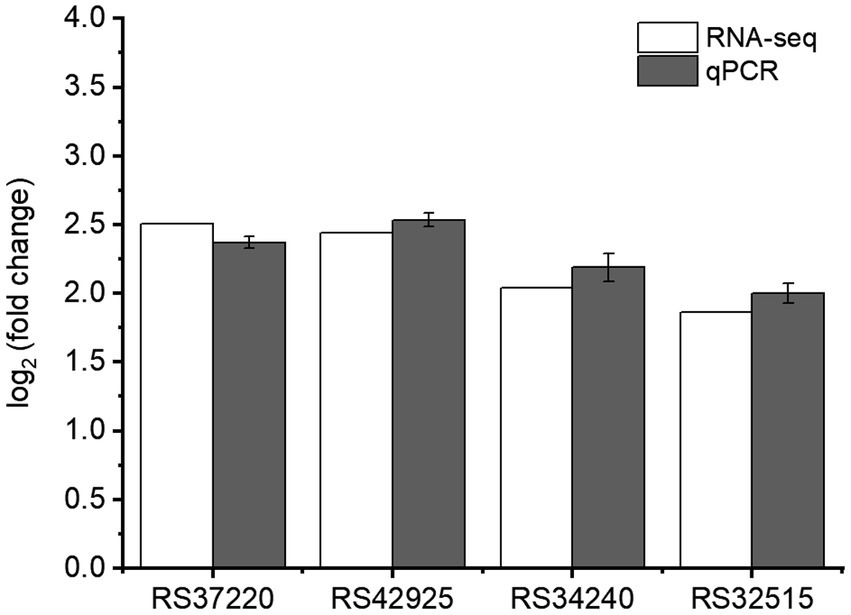
To validate the RNA-seq data, qPCR was conducted on four genes (RS37220, RS42925, RS34240, and RS32515) regulated by RpoS. These selected genes showed up-regulation in rpoS mutant compared with S. baltica. The expression levels were very close in RNA-seq and qPCR results (Figure 8), which indicated that the transcriptome results were reliable.
Conclusion
This study revealed that RpoS positively regulated the spoilage activities of S. baltica while negatively regulated the adhesion and biofilm formation. Transcriptome analysis indicated that large numbers of genes were regulated by RpoS, mainly including flagellar assembly, fatty acid metabolism, two-component regulatory systems, and RNA degradation. These biological pathways might be involved in colonization, stress response, cold adaptation, and spoilage potential of S. baltica. Meanwhile, some other pathways such as quorum sensing were also mediated by RpoS, indicating that RpoS has a wide range of regulatory functions in cell processes. The transcriptome results are reliable through qPCR validation. The present results deepened our understanding of the bacterial spoilage mechanisms. RpoS might be used as a new target for food preservation in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI BioProject—PRJNA858939.
Author contributions
CZ and YL designed the research. CZ and JC performed the experiments. CZ and HL analyzed the data. CZ, JC, and YL wrote the manuscript. CZ, HL, XP, and YL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Key Research and Development of Shandong Province, China (2019GNC106085) and A Project of Shandong Province Higher Educational Science and Technology Program, China (J18KA140).
Acknowledgments
The authors gratefully acknowledge laboratory colleagues for their help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.993237/full#supplementary-material
References
Broekaert, K., Heyndrickxa, M., Hermana, L., Devlieghereb, F., and Vlaemyncka, G. (2011). Seafood quality analysis: molecular identifification of dominant microbiota after ice storage on several general growth media. Food Microbiol. 28, 1162–1169. doi: 10.1016/j.fm.2011.03.009
Cariri, M. L., de Melo, A. N. F., Mizzi, L., Ritter, A. C., Tondo, E., De Souza, E. L., et al. (2019). Quantitative assessment of tolerance response to stress after exposure to oregano and rosemary essential oils, carvacrol and 1,8-cineole in salmonella Enteritidis 86 and its isogenic deletion mutants Δdps, ΔrpoS and ΔompR. Food Res. Int. 122, 679–687. doi: 10.1016/j.foodres.2019.01.046
Chen, L., Zeng, W., Rong, Y., and Lou, B. (2022a). Compositions, nutritional and texture quality of wild-caught andcage-cultured small yellow croaker. J. Food Compos. Anal. 107:104370. doi: 10.1016/j.jfca.2021.104370
Chen, L., Zhao, X., Li, R., and Yang, H. (2022b). Integrated metabolomics and transcriptomics reveal the adaptive responses of salmonella enterica serovar Typhimurium to thyme and cinnamon oils. Food Res. Int. 157:111241. doi: 10.1016/j.foodres.2022.111241
Chevance, F. F. V., and Hughes, K. T. (2008). Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465. doi: 10.1038/nrmicro1887
Chilcott, G. S., and Hughes, K. T. (2010). The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from the amino-terminus into the anti-sigma 28 domain. Mol. Microbiol. 30, 1029–1040. doi: 10.1046/j.1365-2958.1998.01131.x
Colin, R., Ni, B., Laganenka, L., and Sourjik, V. (2021). Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol. Rev. 45, fuab038. doi: 10.1093/femsre/fuab038
Costa, C., Dias, P. J., Sá-Correia, I., and Teixeira, M. C. (2014). MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front. Physiol. 5:197. doi: 10.3389/fphys.2014.00197
Danhorn, T., and Fuqua, C. (2007). Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61, 401–422. doi: 10.1146/annurev.micro.61.080706.093316
Dodd, C. E., and Aldsworth, T. G. (2002). The importance of RpoS in the survival of bacteria through food processing. Int. J. Food Microbiol. 74, 189–194. doi: 10.1016/S0168-1605(01)00679-1
Dong, T., and Schellhorn, H. E. (2010). Role of RpoS in virulence of pathogens. Infect. Immun. 78, 887–897. doi: 10.1128/IAI.00882-09
Feng, L., Bi, W., Chen, S., Zhu, J., and Liu, X. (2021). Regulatory function of sigma factors RpoS/RpoN in adaptation and spoilage potential of Shewanella baltica. Food Microbiol. 97:103755. doi: 10.1016/j.fm.2021.103755
Feng, Y., and Cronan, J. E. (2011). Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol. Microbiol. 80, 195–218. doi: 10.1111/j.1365-2958.2011.07564.x
Fernández-Gómez, P., López, M., Prieto, M., González-Raurich, M., and Alvarez-Ordóñez, A. (2020). The role of the general stress response regulator RpoS in Cronobacter sakazakii biofilm formation. Food Res. Int. 136:109508. doi: 10.1016/j.foodres.2020.109508
Garber, M., Grabherr, M. G., Guttman, M., and Trapnell, C. (2011). Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477. doi: 10.1038/nmeth.1613
Guan, J., Xiao, X., Xu, S., Gao, F., Wang, J., Wang, T., et al. (2015). Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J. Microbiol. 53, 633–642. doi: 10.1007/s12275-015-0099-6
Hai, Y., Zhou, D., Lam, Y. L. N., Li, X., Chen, G., Bi, J., et al. (2022). Nanoemulsified clove essential oils-based edible coating controls pseudomonas spp.-causing spoilage of tilapia (Oreochromis niloticus) fillets: working mechanism and bacteria metabolic responses. Food Res. Int. 159:111594. doi: 10.1016/j.foodres.2022.111594
Jogula, S., Krishna, V. S., Meda, N., Balraju, V., and Sriram, D. (2020). Design, synthesis and biological evaluation of novel Pseudomonas aeruginosa DNA gyrase B inhibitors. Bioorg. Chem. 100:103905. doi: 10.1016/j.bioorg.2020.103905
Keiji, M., Tsuneko, O., Darija, V., Shizuo, K., Makiko, M., Katsuhiko, H., et al. (2010). Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol. Lett. 242, 161–167. doi: 10.1016/j.femsle.2004.11.005
Ko, J., Lee, S., Cho, B., and Lee, Y. (2006). Differential promoter usage of infA in response to cold shock in Escherichia coli. FEBS Lett. 580, 539–544. doi: 10.1016/j.febslet.2005.12.066
Li, Y., Chen, N., Wu, Q., Liang, X., Yuan, X., Zhu, Z., et al. (2022). A flagella hook coding gene flgE positively affects biofilm formation and cereulide production in emetic Bacillus cereus. Front. Microbiol. 13:897836. doi: 10.3389/fmicb.2022.897836
Li, S., Tian, Y., Jiang, P., Lin, Y., Liu, X., and Yang, H. (2021). Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. 61, 1448–1469. doi: 10.1080/10408398.2020.1761287
Li, J., Yang, X., Shi, G., Chang, J., Liu, Z., and Zenget, M. (2019). Cooperation of lactic acid bacteria regulated by the AI-2/LuxS system involve in the biopreservation of refrigerated shrimp. Food Res. Int. 120, 679–687. doi: 10.1016/j.foodres.2018.11.025
Liu, X., Ji, L., Wang, X., Li, J., Zhu, J., and Sun, A. (2018). Role of RpoS in stress resistance, quorum sensing and spoilage potential of Pseudomonas fluorescens. Int. J. Food Microbiol. 270, 31–38. doi: 10.1016/j.ijfoodmicro.2018.02.011
Liu, X., Wu, Y., Chen, Y., Xu, F., Halliday, N., Gao, K., et al. (2016). RpoS differentially affects the general stress response and biofilm formation in the endophytic Serratia plymuthica G3. Res. Microbiol. 167, 168–177. doi: 10.1016/j.resmic.2015.11.003
Liu, X., Xu, J., Zhu, J., Du, P., and Sun, A. (2019). Combined transcriptome and proteome analysis of RpoS regulon reveals its role in spoilage potential of Pseudomonas fluorescens. Front. Microbiol. 10:94. doi: 10.3389/fmicb.2019.00094
Lou, X., Zhai, D., and Yang, H. (2021). Changes of metabolite profiles of fish models inoculated with Shewanella baltica during spoilage. Food Control 123:107697. doi: 10.1016/j.foodcont.2020.107697
Lund, P. A., Biase, D. D., Liran, O., Scheler, O., Mira, N. P., Cetecioglu, Z., et al. (2020). Understanding how microorganisms respond to acid pH is central to their control and successful exploitation. Front. Microbiol. 11:556140. doi: 10.3389/fmicb.2020.556140
Macé, S., Joffraud, J. J., Cardinal, M., Malcheva, M., Cornet, J., Lalanne, V., et al. (2013). Evaluation of the spoilage potential of bacteria isolated from spoiled raw salmon (Salmo salar) fillets stored under modified atmosphere packaging. Int. J. Food Microbiol. 160, 227–238. doi: 10.1016/j.ijfoodmicro.2012.10.013
Mata, G. M. S. C., Ferreira, G. M., and Spira, B. (2017). RpoS role in virulence and fitness in enteropathogenic Escherichia coli. PLoS One 12:e0180381. doi: 10.1371/journal.pone.0180381
Mihailovich, M., Militti, C., Gabaldón, T., and Gebauer, F. (2010). Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. BioEssays 32, 109–118. doi: 10.1002/bies.200900122
Mukherjee, S., and Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5
Nguyen, T. V. A., and Hong, S. H. (2008). Whole genome-based phylogenetic analysis of bacterial two-component systems. Biotechnol. Bioproc. E. 13, 288–292. doi: 10.1007/S12257-008-0017-4
Odeyemi, O. A., Alegbeleby, O. O., Strateva, M., and Stratev, D. (2020). Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin. Compr. Rev. Food Sci. F. 19, 311–331. doi: 10.1111/1541-4337.12526
Ojima, Y., Hakamada, K., Nishinoue, Y., Nguyen, M. H., Miyake, J., and Taya, M. (2012). Motility behavior of rpoS-deficient Escherichia coli analyzed by individual cell tracking. J. Biosci. Bioeng. 114, 652–656. doi: 10.1016/j.jbiosc.2012.06.014
Ozogul, Y., Yuvka, İ., Ucar, Y., Durmus, M., Kösker, A. R., Öz, M., et al. (2017). Evaluation of effects of nanoemulsion based on herb essential oils (rosemary, laurel, thyme and sage) on sensory, chemical and microbiological quality of rainbow trout (Oncorhynchus mykiss) fillets during ice storage. LWT Food Sci. Technol. 75, 677–684. doi: 10.1016/j.lwt.2016.10.009
Parlapani, F. F., Mallouchos, A., Haroutounian, S. A., and Boziaris, I. S. (2014). Microbiological spoilage and investigation of volatile profile during storage of sea bream fillets under various conditions. Int. J. Food Microbiol. 189, 153–163. doi: 10.1016/j.ijfoodmicro.2014.08.006
Santhakumari, S., Jayakumar, R., Logalakshmi, R., Prabhu, N. M., Nazar, A. K. A., Pandian, S. K., et al. (2018). In vitro and in vivo effect of 2,6-Di-tert-butyl-4-methylphenol as an antibiofilm agent against quorum sensing mediated biofilm formation of vibrio spp. Int. J. Food Microbiol. 281, 60–71. doi: 10.1016/j.ijfoodmicro.2018.05.024
Schellhorn, H. E. (2014). Elucidating the function of the RpoS regulon. Future Microbiol. 9, 497–507. doi: 10.2217/fmb.14.9
Serio, A., Fusella, G. C., López, C. C., Sacchetti, G., and Paparella, A. (2014). A survey on bacteria isolated as hydrogen sulfifide-producers from marine fish. Food Control 39, 111–118. doi: 10.1016/j.foodcont.2013.11.003
Shen, Y., Chen, L., Wang, M., Lin, D., Liang, Z., Song, P., et al. (2017). Flagellar hooks and hook protein FlgE participate in host microbe interactions at immunological level. Sci. Rep. 7:1433. doi: 10.1038/s41598-017-01619-1
Sun, Y., Ma, Y., Guan, H., Liang, H., Zhao, X., and Wang, D. (2022). Adhesion mechanism and biofilm formation of Escherichia coli O157:H7 in infected cucumber (Cucumis sativus L.). Food Microbiol. 105:103885. doi: 10.1016/j.fm.2021.103885
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. doi: 10.1038/nbt.2450
Vidovic, S., Mangalappalli-Illathu, A. K., and Korber, D. R. (2011). Prolonged cold stress response of Escherichia coli O157 and the role of rpoS. Int. J. Food Microbiol. 146, 163–169. doi: 10.1016/j.ijfoodmicro.2011.02.018
Vogel, B. F., Venkateswaran, K., Satomi, M., and Gram, L. (2005). Identification of Shewanella baltica as the most important H2S-producing species during iced storage of danish marine fish. Appl. Environ. Microbiol. 71, 6689–6697. doi: 10.1128/AEM.71.11.6689-6697.2005
Voglauer, E. M., Zwirzitz, B., Thalguter, S., Selberherr, E., Wagner, M., and Rychli, K. (2022). Biofilms in water hoses of a meat processing environment harbor complex microbial communities. Front. Microbiol. 13:832213. doi: 10.3389/fmicb.2022.832213
Wang, H., Liu, X., Zhang, Y., Lu, H., Xu, Q., Shi, C., et al. (2017). Spoilage potential of three different bacteria isolated from spoiled grass carp (Ctenopharyngodon idellus) fillets during storage at 4°C. LWT Food Sci. Technol. 81, 10–17. doi: 10.1016/j.lwt.2016.11.010
Wang, Y., Zhang, X., Cen, C., and Fu, L. (2020). Transcription factors FabR and FadR regulate cold adaptability and spoilage potential of Shewanella baltica. Int. J. Food Microbiol. 331:108693. doi: 10.1016/j.ijfoodmicro.2020.108693
Wölflingseder, M., Tutzal, S., Fenglera, V. H., Schild, S., and Reidl, J. (2022). Regulatory interplay of RpoS and RssB controls motility and colonization in vibrio cholerae. Int. J. Med. Microbiol. 312:151555. doi: 10.1016/j.ijmm.2022.151555
Wong, G. T., Bonocora, R. P., Schep, A. N., Beeler, S. M., Lee Fong, A. J., Shull, L. M., et al. (2017). Genome-wide transcriptional response to varying RpoS levels in Escherichia coli K-12. J. Bacteriol. 199, e00755–e00816. doi: 10.1128/JB.00755-16
Wosten, M. M. S. N., van Dijk, L., Veenendaal, A. K. J., de Zoete, M. R., Bleumink-Pluijm, N. M. C., and van Putten, J. P. M. (2010). Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol. Microbiol. 75, 1577–1591. doi: 10.1111/j.1365-2958.2010.07079.x
Wright, M. H., Shalom, J., Matthews, B., Greene, A. C., and Cock, I. E. (2019). Terminalia ferdinandiana Exell: extracts inhibit Shewanella spp. growth and prevent fish spoilage. Food Microbiol. 78, 114–122. doi: 10.1016/j.fm.2018.10.006
Yuan, L., Burm, M., Sadiq, F. A., Wang, N., and He, G. (2018). Interspecies variation in biofilm-forming capacity of psychrotrophic bacterial isolates from Chinese raw milk. Food Control 91, 47–57. doi: 10.1016/j.foodcont.2018.03.026
Zhang, C., Wang, C., Jatt, A., Liu, H., and Liu, Y. (2021). Role of RpoS in stress resistance, biofilm formation and quorum sensing of Shewanella baltica. Lett. Appl. Microbiol. 72, 307–315. doi: 10.1111/lam.13424
Zhao, X., Chen, L., Wu, J., He, Y., and Yang, H. (2020). Elucidating antimicrobial mechanism of nisin and grape seed extract against listeria monocytogenes in broth and on shrimp through NMR-based metabolomics approach. Int. J. Food Microbiol. 319:108494. doi: 10.1016/j.ijfoodmicro.2019.108494
Zhao, L., Poh, C. N., Wu, J., Zhao, X., He, Y., and Yang, H. (2022). Effects of electrolysed water combined with ultrasound on inactivation kinetics and metabolite profiles of Escherichia coli biofilms on food contact surface. Innov. Food Sci. Emerg. 76:102917. doi: 10.1016/j.ifset.2022.102917
Zhong, J., Xiao, C., Wei, G., Du, G., Sun, X., He, Q. Y., et al. (2015). Transfer RNAs mediate the rapid adaptation of Escherichia coli to oxidative stress. PLoS Genet. 11:e1005302. doi: 10.1371/journal.pgen.1005302
Zhu, S., Wu, H., Zeng, M., Liu, Z., and Wang, Y. (2015). The involvement of bacterial quorum sensing in the spoilage of refrigerated Litopenaeus vannamei. Int. J. Food Microbiol. 192, 26–33. doi: 10.1016/j.ijfoodmicro.2014.09.029
Zhu, J., Yan, Y., Wang, Y., and Qu, D. (2019). Competitive interaction on dual-species biofilm formation by spoilage bacteria, Shewanella baltica and Pseudomonas fluorescens. J. Appl. Microbiol. 126, 1175–1186. doi: 10.1111/jam.14187
Zhu, J., Zhao, A., Feng, L., and Gao, H. (2016). Quorum sensing signals affect spoilage of refrigerated large yellow croaker (Pseudosciaena crocea) by Shewanella baltica. Int. J. Food Microbiol. 217, 146–155. doi: 10.1016/j.ijfoodmicro.2015.10.020
Keywords: food microbiology, Shewanella spp., RpoS, spoilage activity, adhesion, biofilm
Citation: Zhang C, Chen J, Pan X, Liu H and Liu Y (2022) Sigma factor RpoS positively affects the spoilage activity of Shewanella baltica and negatively regulates its adhesion effect. Front. Microbiol. 13:993237. doi: 10.3389/fmicb.2022.993237
Edited by:
Lei Yuan, Yangzhou University, ChinaReviewed by:
Tong Gao, Fox Chase Cancer Center, United StatesHongshun Yang, National University of Singapore, Singapore
Yanfu He, Hainan University, China
Copyright © 2022 Zhang, Chen, Pan, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanlong Liu, bGl1eWFubG9uZzMxNEAxNjMuY29t
 Caili Zhang
Caili Zhang Jiaqi Chen
Jiaqi Chen Xiaoming Pan
Xiaoming Pan Yanlong Liu
Yanlong Liu