- 1Department of Medical Laboratory Science and Biotechnology, China Medical University, Taichung, Taiwan
- 2Department of Chemical Engineering, Feng Chia University, Taichung, Taiwan
- 3Department of Medical Research, China Medical University Hospital, Taichung, Taiwan
- 4Department of Food Nutrition and Health Biotechnology, Asia University, Taichung, Taiwan
The economic viability of the biomass-based biorefinery is readily acknowledged by implementation of a cascade process that produces value-added products such as enzymes prior to biofuels. Proteins from the waste stream of biorefinery processes generally contain glutamate (Glu) in abundance. Accordingly, this study was initiated to explore the potential of Glu for production of recombinant proteins in Escherichia coli. The approach was first adopted by expression of D-hydantoinase (HDT) in commercially-available BL21(DE3) strain. Equipped with the mutant gltS (gltS*), the strain grown on Glu produced the maximum HDT as compared to the counterpart on glucose, glycerol, or acetate. The Glu-based production scheme was subsequently reprogrammed based on the L-arabinose-regulated T7 expression system. The strain with gltS* was further engineered by rewiring metabolic pathways. With low ammonium, the resulting strain produced 1.63-fold more HDT. The result indicates that Glu can serve as a carbon and nitrogen source. Overall, our proposed approach may open up a new avenue for the enzyme biorefinery platform based on Glu.
Introduction
To mitigate global climate change, the biorefinery platform has been implemented to largely produce fuels and chemicals for replacement of counterparts derived from fossil fuels (Saini et al., 2017; Takkellapati et al., 2018; Francois et al., 2020; Sun et al., 2021). The platform technology that manages the sustainable feedstock such as lignocellulosic biomass has many potential applications in industry. The exploitation of microbes for production of lignocellulosic ethanol (LE) presents to be a good example (Liu et al., 2019). However, the development progress of LE is afflicted by its higher production cost as compared to the petroleum price (Tuck et al., 2012). A cascade process has been suggested to produce value-added products prior to biofuels, which renders the biomass-based biorefinery economically viable (Escamilla-Alvarado et al., 2017). Enzymes, functional proteins, organic chemicals, and antibiotics are bio-based products of industrial importance. Among them, enzymes are extensively applied in the sector of biofuels, chemicals, cosmetics, detergent, food, pharmaceuticals, pulp, and textiles (Li et al., 2012; Jegannathan and Nielsen, 2013). Their global market value now reaches billions of dollars while the market volume continuously increases with industrialization.
Recognized as a biotechnology-friendly microbe, Escherichia coli has been commonly applied for the mass production of recombinant proteins. Glucose serves as the main carbon source in the culture medium. The dissimilation of glucose proceeding through glycolysis is highly efficient in E. coli. However, the presence of excessive glucose results in acetate overflow which handicaps cellular activities (Millard et al., 2021). This issue has been addressed by the strategy of lowering glucose uptake in cells, involving augmentation of the anaplerotic reaction, recruitment of inferior permeases for glucose transport, or inactivation of certain global regulators (March et al., 2002; Perrenoud and Sauer, 2005; Anda et al., 2006). An alternative method aims to improve activity of the tricarboxylic acid (TCA) cycle by decoupling the arcA/arcB-mediated control circuit (Vemuri et al., 2006). Results of these engineering approaches are generally encouraging. Nevertheless, it remains challenging to overproduce recombinant proteins in cells. The biosynthesis capacity of cells is usually overloaded by the forced expression of a recombinant protein, which leads to the adverse response known as “metabolic burden” (Oh and Liao, 2000). The shortage of cellular resources further triggers the global stress response that disables cell activities (Gill et al., 2000).
The problem of metabolic burden caused by protein overproduction can be partially solved in E. coli cultured with rich media (Oh and Liao, 2000). Yeast extract which consists of amino acids, vitamins, and trace growth elements is commonly supplemented as a major component in rich media (Bekatorou et al., 2006). However, its composition varies from batch to batch. This, in turn, renders the culturing condition and productivity of E. coli difficult to control (Potvin et al., 1997). Notice that a prerequisite for the LE production calls on the enzymatic hydrolysis of lignocellulose into fermentable sugars. Indeed, there is 20–30% of the total production cost for LE coming from hydrolytic enzymes (Singh et al., 2019). As illustrated with a hydrolytic enzyme (β-glucosidase), the techno-economic analysis reveals that glucose mainly contributes to the cost of raw materials that account for 25% of the production cost for the enzyme (Ferreira et al., 2018). Taken together, it is necessary to make more efforts toward achieving the efficient and cost-effective production of enzymes in the biorefinery sector.
In biorefinery, the processing of feedstocks which comprise starch, hemicelluloses, and oil generally leads to the associated production of proteins in the waste stream (Li et al., 2018). The estimated production of protein wastes can reach 100 million tons per year if biomass-derived fuels provide 10% of transportation fuels for the global need (Tuck et al., 2012). Therefore, it is appealing to valorize the current biorefinery process into which the protein-based production platform is integrated (Tuck et al., 2012; Li et al., 2018). Protein waste coming from various sources contains glutamate (Glu) in abundance (Lammens et al., 2012). Taking sugar beet vinasse as an example, the mass fraction of Glu reaches 63% of the total amino acids in the protein content (Deike and Mol, 1996). This apparently makes Glu promising for industrial applications owing to its renewable and sustainable nature. In this context, our previous study illustrated that Glu substituted for yeast extract in the minimal medium was useful for the enzyme production in E. coli (Chiang et al., 2020b). In contrast to complex media, minimal media which contain defined nutrient compositions are favorable for scale-up because of their easy preparation and consistent quality. In addition, evolved E. coli gains an excellent capability of metabolizing Glu (Chiang et al., 2021b). This study was therefore initiated to explore Glu as a potential substrate applied for the enzyme production. As a result, the recombinant D-hydantoinase (HDT) was highly expressed in the engineered E. coli grown on the minimal medium containing Glu that served as a carbon and nitrogen source.
Materials and methods
Genetic manipulation
PCR was applied for cloning of the gltS* (mutant gltS) gene of BL21(mut) strain with primers (5′-ccatgattacgccaagcttgggctcatcaccaaaaatatg and 5′-gtaacactggcagagcattacaagacggtaaatcagttc). Meanwhile, the vector backbone of pTH19Kr plasmid was produced by PCR with primers (5′-caagcttggcgtaatcatgg and 5′-taatgctctgccagtgttac). Two PCR DNAs were then spliced together by using the ZFusion kit (ZGene Biotech Inc., Taiwan), leading to pTH-gltS* plasmid. pPhi-TrHDTCh is a conditionally-replicating plasmid and carries the HDT gene under the control of the T7 promoter (PT7::HDT) (Chiang et al., 2020a). It was implemented to integrate the DNA containing PT7::HDT into the bacterial genome at the Φ80 attB according to the reported protocol (Chiang et al., 2008). Furthermore, zwf and pgl were fused with the λPL promoter (PλPL). This was conducted by the PCR-amplification of the patch DNA from either pPR-zwf or pSPL-pgl plasmid with primers RC11417/RC11418 or RC13034/RC13035 (Saini et al., 2016). The patch DNA mainly comprised the LE*-kan-RE*-PλPL cassette flanked by homologous extensions. The genomic insertion of the DNA cassette proceeded through the λ Red-mediated homologous recombination after the patch DNA was electroporated into the host cell.
Protein production
LB medium consisting of 10 g/l tryptone, 5 g/l yeast extract, and 5 g/l NaCl was routinely used to prepare bacterial strains for overnight. The plasmid-carrying strain was maintained in the presence of 30 μg/ml ampicillin (Amp) or 50 μg/ml kanamycin (Kan). The overnight culture was seeded into baffled flasks (125 ml) containing M9 medium (20 ml) plus 10 g/l sodium Glu (VeDan Co., Taiwan) unless stated otherwise. The composition of M9 medium comprised 6 g/l Na2HPO4, 3 g/l KH2PO4, 0.5 g/l NaCl, 1 g/l NH4Cl, 1 mM MgSO4, and 0.1 mM CaCl2. In the minimal medium, the dosage of Amp or of Kan was cut by half for use whenever necessary. The cell growth was followed by measuring the absorbance at 550 nm (OD550) with a spectrophotometer. The bacterial culture was conducted at 37°C with vigorously shaking. The protein production in the strain was induced by the addition of IPTG (1 mM) or L-arabinose (30 μM) upon the cell density reaching around 0.3 at OD550, followed by reducing the temperature to 30°C throughout the experiment. The analysis of Glu and organic acids essentially followed the reported method (Chiang et al., 2021a). In brief, Glu in the sample was derivatized by the addition of the solution containing borate buffer (pH 10.2), o-phthaldialdehyde solution, and 9-fluorenylmethoxycarbonyl chloride. The analysis was conducted using the ODS Hypersil column with HPLC Chromaster 5,160 (Hitachi, Tokyo, Japan). The mobile phase consisted of solution A (NaH2PO4‧H2O) and solution B (acetonitrile, methanol, and deionized water). Organic acids were analyzed using the Aminex HPX-87H column with the mobile phase containing 0.005 N H2SO4.
Protein analysis
The bacterial culture (1 ml) was centrifuged at the end of experiments, and cell pellets were re-suspended in 0.2 ml Tris–HCl buffer (0.1 M) at pH 8.0. Cells were disrupted by sonication at the frequency of 20 kHz for 90 s (15 s of sonication followed by 15 s of pause). Cell-free extract (CFX) was obtained by recovering the supernatant after the cell lysate was centrifuged for 10 min at 12,000 × g at 4°C. The protein content of CFX was determined by Bio-Rad Protein Assay Kit. Proteins in CFX (20 μl) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The HDT content as resolved in SDS-PAGE was quantified by using Image Analyzer GAS9000 (UVItech). The activity assay of HDT was determined according to the previous report (Chiang et al., 2014). In brief, the reaction was initiated at 40°C upon the addition of 10 μl CFX into the reaction solution (1 ml) consisting of 0.1 M Tris–HCl buffer (pH 8.0), 6 mM D,L-hydroxyphenyl hydantoin (HPH), and 0.5 mM MnCl2. The solution was heated at 100°C for 10 min to terminate the reaction which proceeded for 15 min. The remaining HPH was analyzed by HPLC and calculated for the volumetric activity (U/l) of HDT. The volumetric activity was then multiplied by the culture volume to give the total HDT yield (U).
Results
Protein production using Glu
HDT (E.C.3.5.2.2) from Agrobacterium radiobacter NRRL B11291 has an industrial application for manufacturing of unnatural D-amino acids and 3-carbamoyl-α-picolinic acid for the synthesis of antibiotics and modern agrochemicals like nicotinoid insecticides, respectively (Moriya et al., 1993). Without interfering with cell physiology, this enzyme was chosen as the model protein. The HDT production was first investigated in BL21(DE3) strain that harbored pET-TrChHDT plasmid containing PT7::HDT (Chiang et al., 2020b). The strain is known to carry the lac-dependent T7 expression system (Wagner et al., 2008). In addition, E. coli is unable to utilize Glu as a sole carbon source. This issue was addressed by construction of pTH-gltS* plasmid which carries gltS* with its own promoter. The physiological function of gltS* is responsible for the efficient transport of Glu (Figure 1), which renders E. coli capable of metabolizing Glu (Chiang et al., 2021b). Finally, the Glu-positive strain was obtained by introduction of pTH-gltS* plasmid into BL21(DE3) strain bearing pET-TrChHDT.
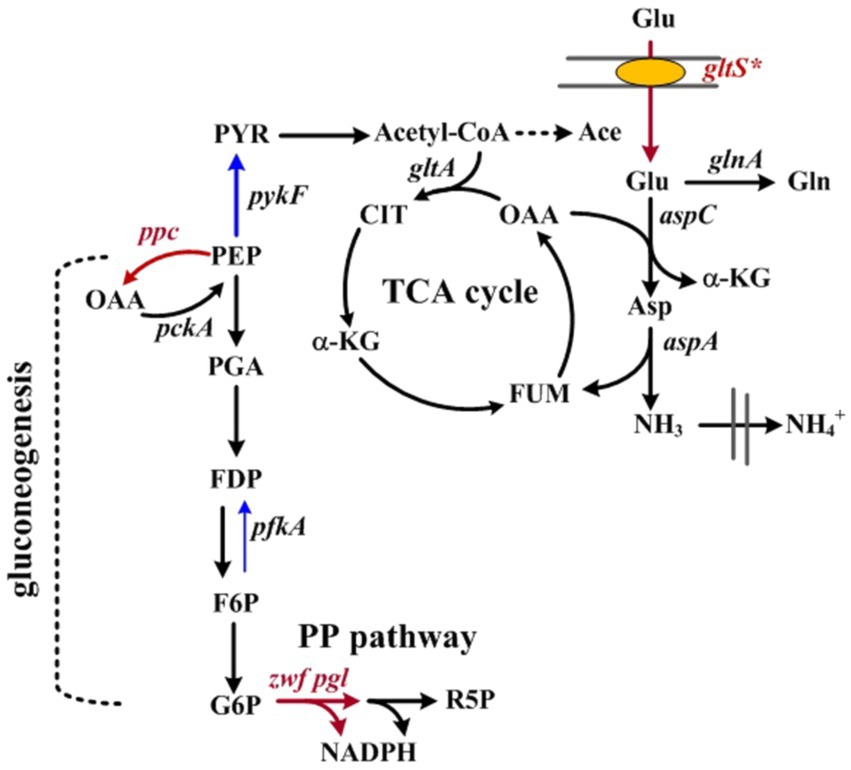
Figure 1. Schematic illustration of Glu metabolism in Escherichia coli. Related genes in metabolic pathways involve as follows: aspC, aspartate aminotransferase; aspA, Asp ammonia-lyase; glnA, glutamine synthetase; gltA, citrate synthase; gltS*, Na+/Glu symporter; pckA, phosphoenolpyruvate carboxykinases; pfkA, 6-phosphofructokinase; pgl, lactonase; ppc, PEP carboxylase; pykF, pyruvate kinase; zwf, glucose-6-phosphate dehydrogenase. Abbreviations: Ace, acetate; Asp, aspartate; CIT, citrate; F6P, fructose-6-phosphate; FDP, fructose-biphosphate; FUM, fumarate; G6P, glucose-6-phosphate; Gln, glutamine; α-KG, α-ketoglutarate; MAL, malate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PGA, 3-phosphoglyceraldehyde; PYR, pyruvate; R5P, ribose-5-phosphate.
The HDT production in the strain was carried out with the minimal medium containing Glu, glucose, glycerol, or acetate. As shown in Figure 2, the cell growth was relatively well on the medium containing glucose, glycerol, and Glu. In sharp contrast to others, the strain exhibited poor growth on acetate. The total HDT production in terms of the enzyme activity (U) was determined for the strain grown on various carbon sources except acetate when entering the stationary growth phase. Interestingly, the Glu-grown strain produced the maximum level of HDT. It indicates that the protein production is promising for the strain dependent on Glu.
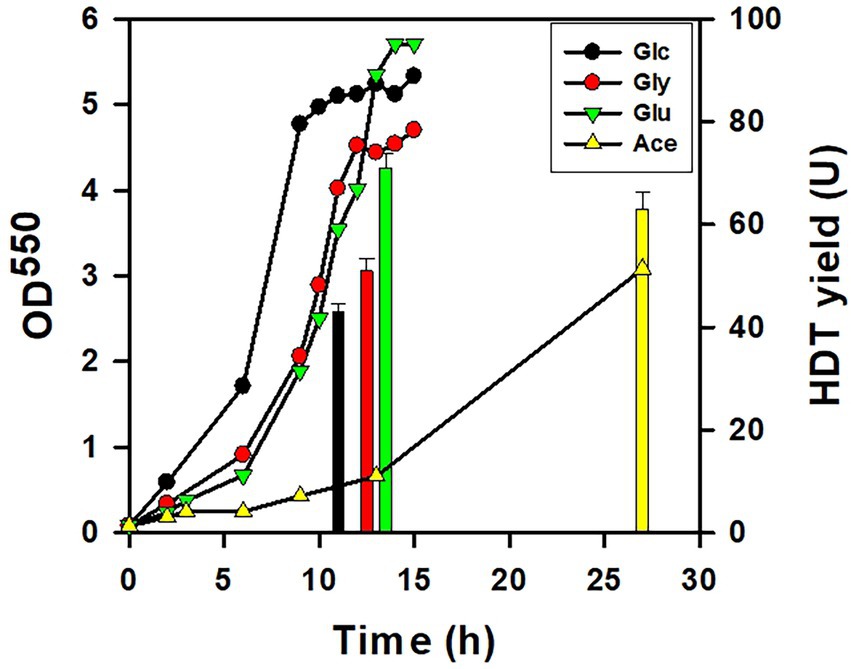
Figure 2. The protein production for the strain using various substrates. The HDT production was conducted with BL21(DE3) strain which harbored pET-TrChHDT plasmid in the presence of various carbon sources (10 g/l). The growth of the acetate-grown strain was sustained by supplement of yeast extract (2 g/l). To grow on Glu, the strain was additionally transformed with pTH-gltS* plasmid. A typical growth profile on each substrate and the HDT production (bars) were shown for the recombinant strain. HDT was analyzed for the strain grown on glucose (Glc), glycerol (Gly), Glu, and acetate (Ace) at 11 h, 13 h, 13 h, and 27 h, respectively. The experiment was performed in triplicate and data were shown with the standard deviation.
The Glu-based production system
BAD-5 strain harbors a genomic copy of L-arabinose (Ara)-inducible T7 expression system (Wang et al., 2011). Its Ara metabolism is interrupted by inactivation of araBAD operon, which, in turn, maintains the consistent inducibility of Ara. The Ara-based expression system has been illustrated to achieve the homogenous and efficient production of the cloned protein at a sub-saturated level of Ara. It was intriguing to investigate the usefulness of this expression system for the Glu-based protein production. To do so, BAD-5 strain was engineered by genomic insertion of PT7::HDT, consequently giving BAD-HDT strain. With pTH-gltS* plasmid, BAD-HDT strain was applied for the HDT production using Glu. As depicted in Figure 3, the strain (BAD-HDT/pTH-gltS*) produced HDT with a yield reaching 76 U. This production yield is comparable to that obtained by BL21(DE3) strain harboring the HDT-borne plasmid (Figure 2). Accordingly, BAD-HDT strain was employed for further investigations.
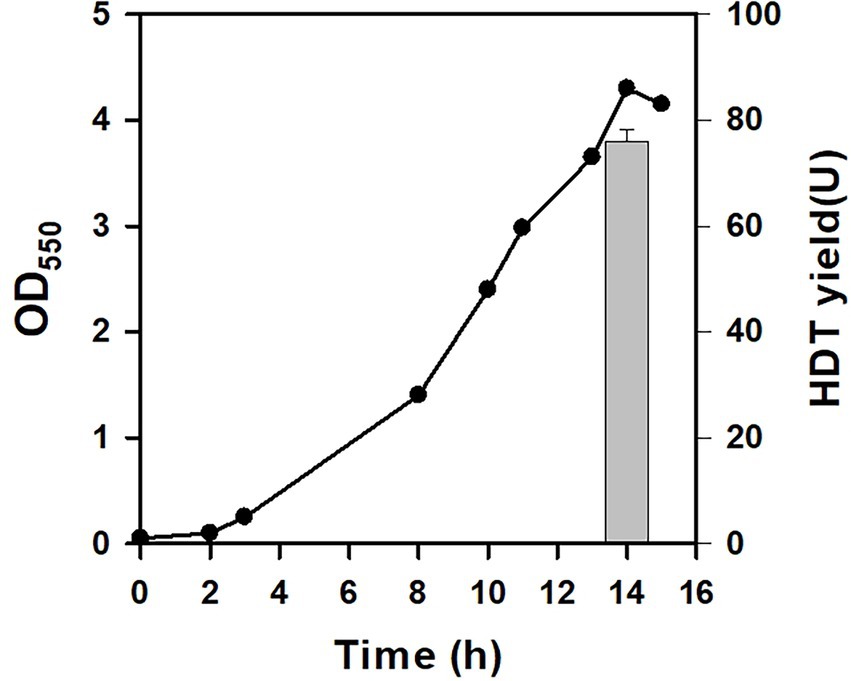
Figure 3. The protein production in Glu-dependent strain. The HDT production was conducted with BAD-HDT/pTH-gltS* strain in the presence of Glu (10 g/l). A typical growth profile and the HDT production (bar) were shown for the recombinant strain. The experiment was performed in triplicate and data were shown with the standard deviation.
Strain improvement
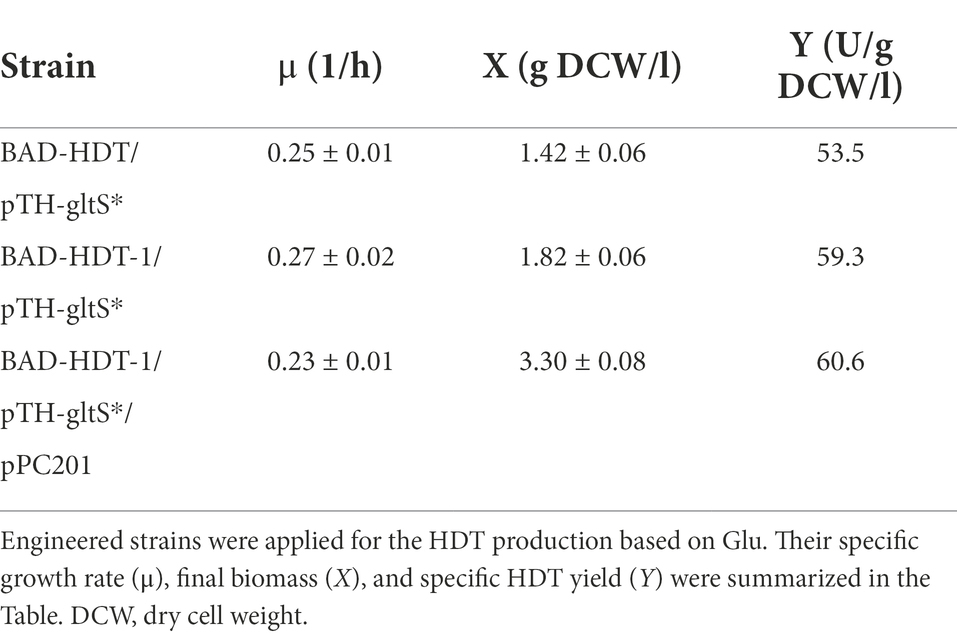
In our previous study, central metabolic pathways that affect the cell growth were identified in Glu metabolism involving the assimilation pathway, the tricarboxylic (TCA) cycle, and gluconeogenesis (Chiang et al., 2021b). In particular, gluconeogenesis links to the pentose phosphate (PP) pathway (Figure 1). The oxidative route of the PP pathway provides NADPH which is favorable for biosynthesis (White, 2007). We previously showed that the enhanced level of zwf (encoding glucose 6-phosphate dehydrogenase) and pgl (encoding lactonase) increases the intracellular level of NADPH (Saini et al., 2016). BAD-HDT strain was then engineered to overexpress zwf and pgl according to the reported method, resulting in BAD-HDT-1 strain. In the presence of Glu, the strain with pTH-gltS* plasmid (BAD-HDT-1/pTH-gltS*) enabled production of HDT at 108 U (Figure 4A).
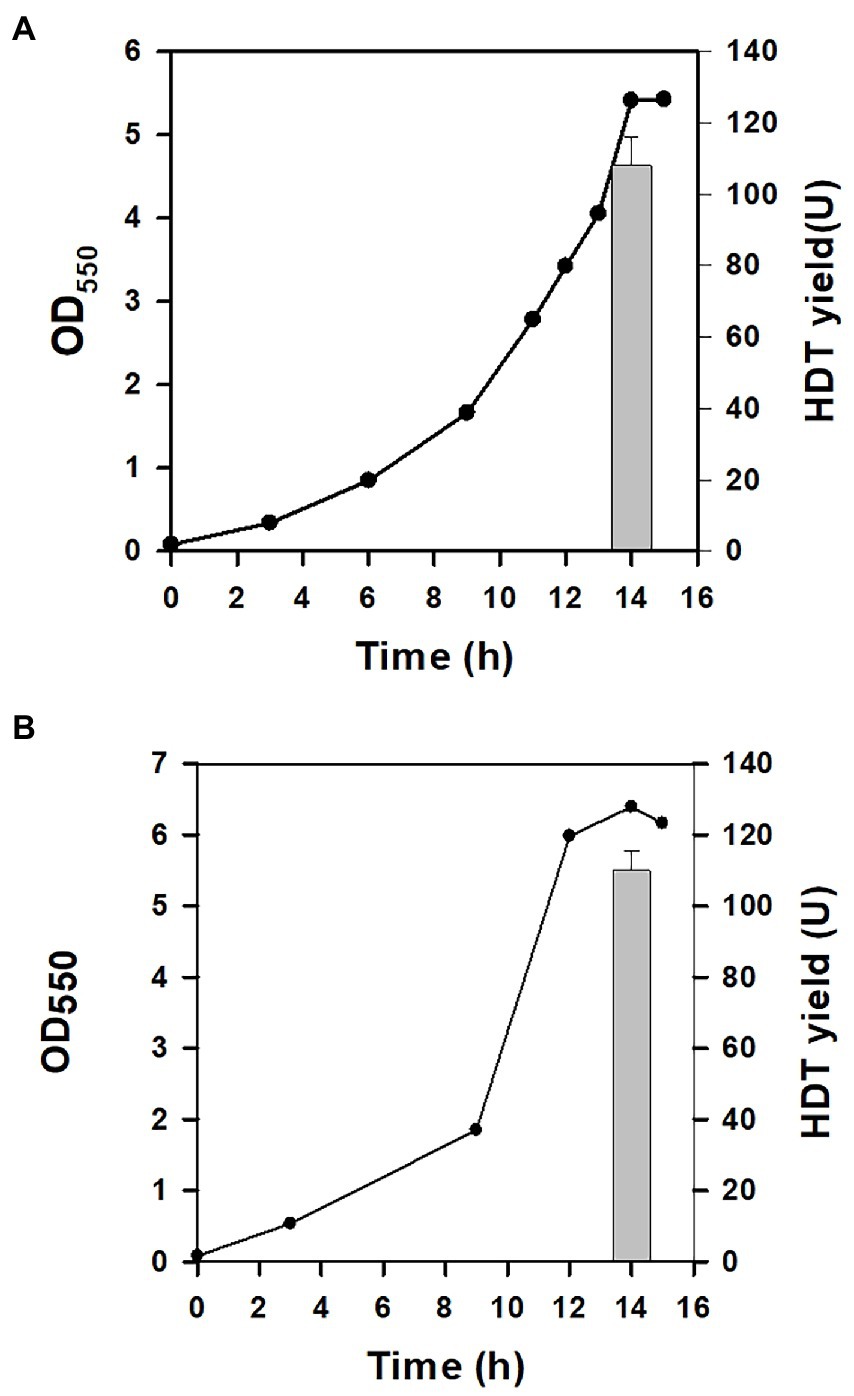
Figure 4. The protein production for the genetically-modified strain. The HDT production was conducted with BAD-HDT-1/pTH-gltS* strain (A) and BAD-HDT-1/pTH-gltS* strain bearing pACYC-aspA plasmid in the presence of Glu (10 g/l) (B). A typical growth profile and the HDT production (bar) were shown for the recombinant strain. The experiment was performed in triplicate and data were shown with the standard deviation.
In general, the flux distribution between the TCA cycle and gluconeogenesis allocates precursor metabolites available for biosynthesis of amino acids in Glu metabolism. The assimilation route consists of two consecutive reaction steps catalyzed by aspartate (Asp) aminotransferase (encoded by aspC) and Asp-ammonia lyase (encoded by aspA), which converts Glu to α-ketoglutarate (α-KG) in the TCA cycle. The activity of Asp-ammonia lyase was found to modulate the flux distribution in favor of the TCA cycle (Chiang et al., 2021b). Accordingly, BAD-HDT-1/pTH-gltS* strain was equipped with the T7A1 promoter-driven aspA by recruitment of pACYC-aspA plasmid (Wang et al., 2006). It resulted in a marginal increase in the HDT production for the resulting strain (Figure 4B).
Improvement of the protein production
The deamination reaction involved in the assimilation pathway of Glu produces ammonium (Figure 1). The released ammonium may provide the nitrogen source for the Glu-dependent strain. To test this hypothesis, BAD-HDT-1/pTH-gltS* strain was cultured with the minimal medium containing Glu (10 g/l) and various levels of NH4Cl. The final biomass was determined after the cell grew at 14 h. Consequently, the cell density of the strain receiving the 1/3 dosage of NH4Cl remained unaffected as compared to that of the counterpart with a full dosage of NH4Cl. However, it caused a 10% decrease in the biomass when the 1/5 dosage of NH4Cl was administrated. The HDT production in BAD-HDT-1/pTH-gltS* strain was then conducted with the medium containing a 1/3 dosage of NH4Cl. In addition, the Glu level in the medium was doubled to increase the protein production. As shown in Figure 5, the HDT production in the strain reached 175 U.
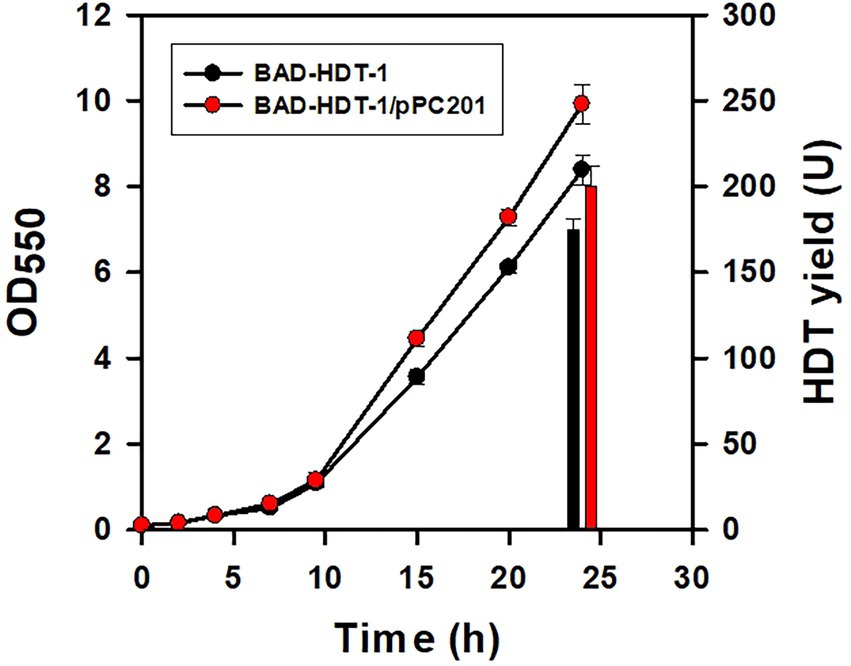
Figure 5. Improvement of the protein production in the engineered strain. The HDT production in BAD-HDT-1/pTH-gltS* and BAD-HDT-1/pTH-gltS*/pPC201 strain was carried out in the minimal medium containing Glu (20 g/l) and NH4Cl (0.35 g/l). A typical growth profile and the HDT production (bars) were shown for the recombinant strains. The experiment was performed in triplicate and data were shown with the standard deviation. Strains were designated as BAD-HDT-1 and BAD-HDT-1/pPC201 for clarity.
It was found that BAD-HDT-1/pTH-gltS* strain produced a detectable level of acetate (ca. 1.3 g/l) in the presence of high Glu. The availability of phosphoenolpyruvate (PEP) leads to the formation of acetate from pyruvate (PYR) and is likely reduced by PEP carboxylase (encoded by ppc) which converts PEP to oxaloacetate (OAA) (Figure 1). BAD-HDT-1/pTH-gltS* strain was then transformed with pPC201 plasmid which contains the trc promoter-regulated ppc (Chao and Liao, 1993). Consequently, the resulting strain (BAD-HDT-1/pTH-gltS*/pPC201) produced HDT with 200 U and reduced the acetate production to 0.6 g/l (Figure 5).
Discussion
Recombinant proteins have been commonly produced in E. coli using glucose. However, the adverse event of acetate overflow occurs in response to surplus glucose. Glycerol is utilized less efficiently than glucose in E. coli. This, in turn, lowers the production of acetate as a result of the induced “carbon stress-based acetate recycling pathway” (Martínez-Gómez et al., 2012). The use of glycerol is particularly useful for overproduction of aggregate-prone proteins which link to the stress response (Strandberg and Enfors, 1991). Nevertheless, an increase in glycerol utilization leads to PYR overflow in E. coli (Chiang et al., 2020a). Acetate is inexpensive but metabolized very slowly by E. coli. The acetate-based production of the single chain derivative of monellin was proven promising (Leone et al., 2015). However, the nutrient supplement is necessary to sustain growth of the strain dependent on acetate.
In this study, Glu was explored for production of the recombinant protein. BL21(DE3) strain is recognized as the most commonly used producer in the field. The recruitment of gltS* rendered the strain capable of utilizing Glu, which likely eliminates the kinetic constraint of Glu assimilation enzymes. The efficiency of the protein production was first evaluated in the strain using various substrates. Figure 2 showed that the maximum production of HDT was obtained for the strain grown on Glu. The strain metabolized various carbon sources at a different rates and displayed distinct growth profiles. Accordingly, the production rate (U/h) was calculated by dividing the HDT yield (U) by the time (h) in which the protein was sampled for the measurement. This gave the HDT productivity based on Glu, glycerol, glucose, and acetate reaching 5.5, 4.6, 3.9, and 2.3 U/h, respectively. Overall, Glu apparently serves as an excellent substrate for the protein production in E. coli.
The Glu-based production system was refined by development of BAD-HDT strain that carried the Ara-inducible T7 expression system and a genomic copy of PT7::HDT. In Glu metabolism, the assimilation pathway leads to the production of α-KG (Figure 1). The continued oxidation of α-KG in the TCA cycle produces OAA. PEP carboxykinase (encoded by pckA) decarboxylates OAA to form PEP, which mainly serves for gluconeogenesis. It is apparent that Glu metabolism closely relies on the TCA cycle. Notice that BAD-HDT strain belongs to E. coli B strain which displays a higher activity of the TCA cycle than K12 strain (Waegeman et al., 2011). To improve the protein production in the strain, the gluconeogenic flux was directed into the oxidative route in the PP pathway by enhanced expression of zwf and pgl (Figure 1). It resulted in a 42% increase in the HDT production for BAD-HDT-1/pTH-gltS* strain (Figure 4; Table 1). PYR kinase (encoded by pykF) is subjected to the repression of Cra (encoding fructose repressor) and controls the flow direction of glycolytic flux that bifurcates at the PEP node (Bledig et al., 1996). Cra is inactivated upon binding to intermediate metabolites (i.e., glycolytic intermediates) derived from glucose (Bley et al., 2018). Our previous study illustrated that the Glu-positive strain deprived of pfkA [encoding 6-phosphofructokinase (PFK)] failed to grow on Glu (Chiang et al., 2021b). The function of PFK is responsible for phosphorylation of fructose 6-phosphate to form fructose-1,6-biphosphate. The essential role of PFK in Glu metabolism likely elevates the level of glycolytic intermediates. This, in turn, disables Cra and induces the catabolite activation of pykF. Accordingly, the further oxidization of PEP mediated by PYR kinase may restrict the gluconeogenic flux (Figure 1). This flux constraint appears to be eased by activation of the oxidative PP pathway after recruitment of zwf and pgl.
The reaction catalyzed by Asp-ammonia lyase releases ammonia in Glu metabolism (Figure 1). It was illustrated that BAD-HDT-1/pTH-gltS* strain displayed no growth defect in the Glu-containing medium with a 1/3 dosage of NH4Cl. The result suggests that the liberated ammonium supplements the nitrogen source which is sufficient to sustain cell activities. In contrast, the administration of a 1/3 dosage of NH4Cl caused a decrease (>50%) in the biomass of the strain grown on glucose, glycerol, or acetate. The HDT production in BAD-HDT-1/pTH-gltS* strain was further improved in the presence of high Glu (ca. 20 g/l) and low NH4Cl (ca. 0.33 g/l) (Figure 5). However, the approach resulted in the accumulation of acetate with a concentration lower than the inhibitory level. The underlying mechanism for acetate overflow remains unclear. With an enhanced level of ppc, BAD-HDT-1/pTH-gltS*/pPC201 strain enabled reduction of acetate and production of 1.63-fold more HDT than its parent strain (BAD-HDT/pTH-gltS*). It is likely that the activity of PEP carboxylase reduces the PEP pool and provides more OAA in favor of the reaction catalyzed by citrate synthase (encoded by gltA) (Figure 1). Notice that the availability of Glu leads to formation of glutamine (Gln) through Gln synthetase (encoded by glnA). The reductive synthesis of more than 10 amino acids requires ammonia supplied by Glu and Gln (White, 2007). This apparently adds one more advantage to Glu applied for the protein production.
In summary, this preliminary study illustrated the potential of Glu for overexpressing recombinant proteins in E. coli. Glu is the first commercially-available amino acid as produced by the fermentation process, and its market size reaches 1.5 million tons per year and increasingly grows (Kumar et al., 2014). In addition, the abundance of Glu exists in the protein waste associated with the conventional biomass-treatment process. It is expected that waste streams of these production processes would provide a sustainable and renewable source of Glu. The focus of our continued work is development of a streamlined process to recover Glu from the protein waste. Accordingly, the developed approach based on Glu may open up a new avenue for the enzyme biorefinery platform.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
C-JC and Y-PC conceived the idea and wrote the manuscript. M-CH and TT performed the experiment. All authors contributed to the article and approved the submitted version.
Funding
This work was co-supported by China Medical University (CMU111-MF-79) and the Ministry of Science and Technology (MOST 108-2221-E-035-052-MY3), Taiwan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anda, R. D., Lara, A. R., Hernández, V., Hernández-Montalvo, V., Gosset, G., Bolívar, F., et al. (2006). Replacement of the glucose phosphotransferase transport system by galactose permease reduces acetate accumulation and improves process performance of Escherichia coli for recombinant protein production without impairment of growth rate. Metab. Eng. 8, 281–290. doi: 10.1016/j.ymben.2006.01.002
Bekatorou, A., Psarianos, C., and Koutinas, A. A. (2006). Production of food grade yeasts. Food Technol. Biotech. 44, 407–415.
Bledig, S. A., Ramseier, T. M., and Saier Jr, M. H. (1996). FruR mediates catabolite activation of pyruvate kinase (pykF) gene expression in Escherichia coli. J. Bacteriol. 178, 280–283. doi: 10.1128/jb.178.1.280-283.1996
Bley, F. B., Ortega, A. D., Hubmann, G., Bonsing-Vedelaar, S., Wijma, H. J., Van Der Meulen, P., et al. (2018). Assessment of the interaction between the flux-signaling metabolite fructose-1,6-bisphosphate and the bacterial transcription factors CggR and Cra. Mol. Microbiol. 109, 278–290. doi: 10.1111/mmi.14008
Chao, Y., and Liao, J. (1993). Alteration of growth yield by overexpression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in Escherichia coli. Appl. Environ. Microbiol. 59, 4261–4265. doi: 10.1128/aem.59.12.4261-4265.1993
Chiang, C. J., Chen, P. T., and Chao, Y. P. (2008). Replicon-free and markerless methods for genomic insertion of DNAs in phage attachment sites and controlled expression of chromosomal genes in Escherichia coli. Biotechnol. Bioeng. 101, 985–995. doi: 10.1002/bit.21976
Chiang, C. J., Ho, Y. J., Hu, M. C., and Chao, Y. P. (2020a). Rewiring of glycerol metabolism in Escherichia coli for effective production of the recombinant protein. Biotechnol. Biofuels 13:205. doi: 10.1186/s13068-020-01848-z
Chiang, C. J., Hu, M. C., and Chao, Y. P. (2020b). A strategy to improve production of recombinant proteins in Escherichia coli based on the glucose-glycerol mixture and glutamate. J Agri Food Chem 68, 8883–8889. doi: 10.1021/acs.jafc.0c03671
Chiang, C.-J., Hu, R.-C., Huang, Z.-C., and Chao, Y.-P. (2021a). Production of succinic acid from amino acids in Escherichia coli. J Agri Food Chem 69, 8172–8178. doi: 10.1021/acs.jafc.1c02958
Chiang, C.-J., Lin, Y.-L., Hu, M.-C., and Chao, Y.-P. (2021b). Understanding and harnessing the glutamate metabolism in Escherichia coli. J. Taiwan Inst. Chem. Eng. 121, 115–121. doi: 10.1016/j.jtice.2021.04.020
Chiang, C. J., Lin, L. J., Wang, Z. W., Lee, T. T., and Chao, Y. P. (2014). Design of a noncovalently linked bifunctional enzyme for whole-cell biotransformation. Process Biochem. 49, 1122–1128. doi: 10.1016/j.procbio.2014.03.016
Deike, V., and Mol, J. D. (1996). Raw Material Compendium. St. Charles, MO: Brussels Novus International Inc.
Escamilla-Alvarado, C., Perez-Pimienta, J. A., Ponce-Noyola, T., and Poggi-Varaldo, H. M. (2017). An overview of the enzyme potential in bioenergy-producing biorefineries. J. Chem. Technol. Biotechnol. 92, 906–924. doi: 10.1002/jctb.5088
Ferreira, R. D. G., Azzoni, A. R., and Freitas, S. (2018). Techno-economic analysis of the industrial production of a low-cost enzyme using E. coli: the case of recombinant β-glucosidase. Biotechnol. Biofuels 11:81. doi: 10.1186/s13068-018-1077-0
Francois, J. M., Alkim, C., and Morin, N. (2020). Engineering microbial pathways for production of bio-based chemicals from lignocellulosic sugars: current status and perspectives. Biotechnol. Biofuels 13:118. doi: 10.1186/s13068-020-01744-6
Gill, R. T., Valdes, J. J., and Bentley, W. E. (2000). A comparative study of global stress gene regulation in response to overexpression of recombinant proteins in Escherichia coli. Metab. Eng. 2, 178–189. doi: 10.1006/mben.2000.0148
Jegannathan, K. R., and Nielsen, P. H. (2013). Environmental assessment of enzyme use in industrial production – a literature review. J. Clean. Prod. 42, 228–240. doi: 10.1016/j.jclepro.2012.11.005
Kumar, R., Vikramachakravarthi, D., and Pal, P. (2014). Production and purification of glutamic acid: a critical review towards process intensification. Chem. Eng. Process. 81, 59–71. doi: 10.1016/j.cep.2014.04.012
Lammens, T. M., Franssen, M. C. R., Scott, E. L., and Sanders, J. P. M. (2012). Availability of protein-derived amino acids as feedstock for the production of bio-based chemicals. Biomass Bioenergy 44, 168–181. doi: 10.1016/j.biombioe.2012.04.021
Leone, S., Sannino, F., Tutino, M. L., Parrilli, E., and Picone, D. (2015). Acetate: friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source. Microb. Cell Factories 14:106. doi: 10.1186/s12934-015-0299-0
Li, S. Y., Ng, I. S., Chen, P. T., Chiang, C. J., and Chao, Y. P. (2018). Biorefining of protein waste for production of sustainable fuels and chemicals. Biotechnol. Biofuels 11:173. doi: 10.1186/s13068-018-1234-5
Li, S., Yang, X., Zhu, M., and Wang, X. (2012). Technology prospecting on enzymes: application, marketing and engineering. Comput. Struct. Biotechnol. J. 2:e201209017. doi: 10.5936/csbj.201209017
Liu, C.-G., Xiao, Y., Xia, X.-X., Zhao, X.-Q., Peng, L., Srinophakun, P., et al. (2019). Cellulosic ethanol production: Progress, challenges and strategies for solutions. Biotechnol. Adv. 37, 491–504. doi: 10.1016/j.biotechadv.2019.03.002
March, J., Eiteman, M., and Altman, E. (2002). Expression of an anaplerotic enzyme, pyruvate carboxylase, improves recombinant protein production in Escherichia coli. Appl. Environ. Microbiol. 68, 5620–5624. doi: 10.1128/AEM.68.11.5620-5624.2002
Martínez-Gómez, K., Flores, N., Castañeda, H. M., Martínez-Batallar, G., Hernández-Chávez, G., Ramírez, O. T., et al. (2012). New insights into Escherichia coli metabolism: carbon scavenging, acetate metabolism and carbon recycling responses during growth on glycerol. Microb. Cell Factories 11:46. doi: 10.1186/1475-2859-11-46
Millard, P., Enjalbert, B., Uttenweiler-Joseph, S., Portais, J. C., and Létisse, F. (2021). Control and regulation of acetate overflow in Escherichia coli. Elife 10:e63661. doi: 10.7554/eLife.63661
Moriya, K., Shibuya, K., Hattori, Y., Tsuboi, S., Shiokawa, K., and Kagabu, S. (1993). Structural modification of the 6-chloropyridyl moiety in the imidacloprid skeleton: introduction of a five-membered heteroaromatic ring and the resulting insecticidal activity. Biosci. Biotechnol. Biochem. 57, 127–128. doi: 10.1271/bbb.57.127
Oh, M. K., and Liao, J. C. (2000). DNA microarray detection of metabolic responses to protein overproduction in Escherichia coli. Metab. Eng. 2, 201–209. doi: 10.1006/mben.2000.0149
Perrenoud, A., and Sauer, U. (2005). Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 187, 3171–3179. doi: 10.1128/JB.187.9.3171-3179.2005
Potvin, J., Fonchy, E., Conway, J., and Champagne, C. P. (1997). An automatic turbidimetric method to screen yeast extracts as fermentation nutrient ingredients. J. Microbiol. Methods 29, 153–160. doi: 10.1016/S0167-7012(97)00032-8
Saini, M., Li, S. Y., Chiang, C. J., and Chao, Y. P. (2016). Systematic engineering of the central metabolism in Escherichia coli for effective production of n-butanol. Biotechnol. Biofuels 9:69. doi: 10.1186/s13068-016-0467-4
Saini, M., Lin, L. J., Chiang, C. J., and Chao, Y. P. (2017). Synthetic consortium of Escherichia coli for n-butanol production by fermentation of the glucose-xylose mixture. J. Agri. Food Chem. 65, 10040–10047. doi: 10.1021/acs.jafc.7b04275
Singh, A., Jasso, R. M. R., Gonzalez-Gloria, K. D., Rosales, M., Cerda, R. B., Aguilar, C. N., et al. (2019). The enzyme biorefinery platform for advanced biofuels production. Bioresour. Technol. Rep. 7:100257. doi: 10.1016/j.biteb.2019.100257
Strandberg, L., and Enfors, S.-O. (1991). Factors influencing inclusion body formation in the production of a fused protein in Escherichia coli. Appl. Environ. Microbiol. 57, 1669–1674. doi: 10.1128/aem.57.6.1669-1674.1991
Sun, L., Wu, B., Zhang, Z., Yan, J., Liu, P., Song, C., et al. (2021). Cellulosic ethanol production by consortia of Scheffersomyces stipitis and engineered Zymomonas mobilis. Biotechnol. Biofuels 14:221. doi: 10.1186/s13068-021-02069-8
Takkellapati, S., Li, T., and Gonzalez, M. A. (2018). An overview of biorefinery-derived platform chemicals from a cellulose and hemicellulose biorefinery. Clean Techn. Environ. Policy 20, 1615–1630. doi: 10.1007/s10098-018-1568-5
Tuck, C. O., Pérez, E., Horváth, I. T., Sheldon, R. A., and Poliakoff, M. (2012). Valorization of biomass: deriving more value from waste. Science 337:695. doi: 10.1126/science.1218930
Vemuri, G. N., Eiteman, M. A., and Altman, E. (2006). Increased recombinant protein production in Escherichia coli strains with overexpressed water-forming NADH oxidase and a deleted ArcA regulatory protein. Biotechnol. Bioeng. 94, 538–542. doi: 10.1002/bit.20853
Waegeman, H., Beauprez, J., Moens, H., Maertens, J., De Mey, M., Foulquié-Moreno, M. R., et al. (2011). Effect of iclR and arcA knockouts on biomass formation and metabolic fluxes in Escherichia coli K12 and its implications on understanding the metabolism of Escherichia coli BL21 (DE3). BMC Microbiol. 11:70. doi: 10.1186/1471-2180-11-70
Wagner, S., Klepsch, M. M., Schlegel, S., Appel, A., Draheim, R., Tarry, M., et al. (2008). Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. U. S. A. 105, 14371–14376. doi: 10.1073/pnas.0804090105
Wang, Z. W., Chen, Y., and Chao, Y.-P. (2006). Enhancement of recombinant protein production in Escherichia coli by coproduction of aspartase. J. Biotechnol. 124, 403–411. doi: 10.1016/j.jbiotec.2006.01.001
Wang, Z. W., Lai, C. B., Chang, C. H., Chiang, C. J., and Chao, Y. P. (2011). A glucose-insensitive T7 expression system for fully-induced expression of proteins at a subsaturating level of L-arabinose. J. Agri. Food Chem. 59, 6534–6542. doi: 10.1021/jf2013748
Keywords: recombinant proteins, glutamate, metabolic engineering, enzyme biorefinery, protein waste
Citation: Chiang C-J, Hu M-C, Ta T and Chao Y-P (2022) Glutamate as a non-conventional substrate for high production of the recombinant protein in Escherichia coli. Front. Microbiol. 13:991963. doi: 10.3389/fmicb.2022.991963
Edited by:
Stefan Junne, Technical University of Berlin, GermanyReviewed by:
Jorge Bolivar Perez, Universidad de Cádiz, SpainDanielle Maria Perpetua Oliveira, Federal University of Rio de Janeiro, Brazil
Copyright © 2022 Chiang, Hu, Ta and Chao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun-Peng Chao, eXBjaGFvQGZjdS5lZHUudHc=
 Chung-Jen Chiang
Chung-Jen Chiang Mu-Chen Hu2
Mu-Chen Hu2 Thanh Ta
Thanh Ta Yun-Peng Chao
Yun-Peng Chao