- 1College of Animal Science and Technology, Northeast Agricultural University, Harbin, China
- 2Ordos Academy of Agriculture and Animal Husbandry, Ordos, China
Clostridium butyricum, as a probiotic with a variety of active products, has been widely used to improve the intestinal health of humans and animals. Previous studies had demonstrated that Clostridium butyricum exhibited potential protective and positive effects in human disease research and animal production by producing a variety of beneficial substances, such as intestinal inflammation, the intestinal epithelial barrier, metabolic diseases, and regulation of the gut microbiota. Therefore, we hypothesized that dietary Clostridium butyricum supplementation could improve gut health in fattening goats by modulating gut microbiota. However, it is unclear whether Clostridium butyricum can reach the intestine through the rumen, so 15 healthy Albas goats were selected and randomly divided into 3 treatments with 5 replicates in each group. The groups were divided as follows: control group (CON: basal diet), rumen-protected Clostridium butyricum group (RPCB: basal diet plus 1.0 × 109 CFU/kg Clostridium butyricum coated with hydrogenated fat), and Clostridium butyricum group (CB: basal diet plus 1.0 × 109 CFU/kg Clostridium butyricum). The experiment was slaughtered after a 70-day growth test, and the jejunal mucosa and intestinal contents of the goats were collected to determine tight junction proteins related genes expression and 16S rDNA microbial sequencing analysis to evaluate the intestine health. The results showed that dietary supplementation with Clostridium butyricum significantly increased the expression of the Claudin-4 gene of the jejunal mucosa (P < 0.05) and had a trend toward a significant increase in the Occludin gene (0.05 < P < 0.10). However, Clostridium butyricum had no significant effect on the expression of intestinal inflammatory factors (P > 0.10). In addition, the relative fractionation of Clostridium and Clostridiaceae_unclassified in the gut microbiota at the genus level decreased significantly compared with controls (P < 0.05). The results of the analysis of the level of Clostridium species showed that Clostridium butyricum only existed in the treatment group. And the correlation results showed that Occludin and Claudin-4 genes were positively correlated with Sharppea and Clostridium butyricum, and negatively correlated with Clostridium (P < 0.05). Supplementing Clostridium butyricum in the diet did not significantly affect the intestinal immune function of goats, while regulation of the intestinal microbiota was associated with improving the intestinal epithelial barrier.
Introduction
Intensive and grazing farming are the main way of feeding goats and sheep in China (Mu et al., 2020), among which high-concentrate fattening is a common production method to meet their energy needs in the short term to improve growth performance (Wang M. et al., 2021), which undoubtedly aggravates the immune pressure and gastrointestinal metabolic load of ruminants (Plaizier et al., 2018). Due to the unique digestive systems of ruminants, many major studies have focused on the effects of high-concentrate diets on rumen fermentation, ruminal acidosis, and their microbial properties and functions. It is well known that the small intestine is the main place for nutrient absorption, and the intestine is closely related to its autoimmune function as the largest immune organ in the body (Chelakkot et al., 2018; Mu et al., 2021). Therefore, paying attention to the intestinal health of goats and sheep during the rapid fattening stage has important implications for their production.
Diarrhea is one of the important reasons affecting the growth and production performance of goats and sheep during intensive and grazing farming (Wang M. et al., 2021). A large number of studies have shown that the impaired intestinal epithelial barrier (Ma et al., 2020), low functional immunity of the gastrointestinal tract (Peng et al., 2021), and intestinal flora disturbance can cause diarrhea of different degrees (Zhang et al., 2021). Diarrhea affects the body’s absorption of small molecular substances and nutritional elements (Esterházy et al., 2019), destroys intestinal homeostasis, and results in severe loss of body fluids (McLamb et al., 2013). The intestinal epithelial barrier is an important line of defense in the body to exert immune function by improving the expression of intestinal epithelial tight junction (TJ) proteins, intestinal epithelial cells’ development, and the mucus layer’s physiological characteristics. Strengthening the intestinal epithelial barrier function can help inhibit intestinal inflammation and the occurrence of diarrhea caused by various factors (Chelakkot et al., 2018). Dietary supplementation with natural active substances (such as plant essential oils, probiotics, and peptides) is a common production method to improve animal gut health and positively impacts growth performance, antioxidant levels, and rumen fermentation (Xie et al., 2020, 2021; Hagihara et al., 2021).
Clostridium butyricum (C. butyricum) is a class of anaerobic probiotics with spores that are resistant to bile salts and gastric acid, and have strong resistance to stress (Hagihara et al., 2020). Its rich metabolites such as butyric acid (Stoeva et al., 2021), digestive enzymes (Zhan et al., 2019), bacteriocin (Wang T. et al., 2021), vitamins, and other nutrients (Araki et al., 2002) have a variety of probiotic functions, including enhancement of intestinal epithelial barrier integrity, anti-inflammatory and modulation of intestinal flora. Previous studies have demonstrated that supplementation with C. butyricum improves gut health and nutrient absorption of humans and livestock (Zhang et al., 2018; Wang et al., 2020; Stoeva et al., 2021). Meanwhile, dietary supplementation with butyrate has also been reported to enhance the regulation of intestinal epithelial barrier, inflammation, and microflora (Bedford and Gong, 2018). However, the regulation of intestinal health and function by C. butyricum in ruminants is rarely reported.
Overall, we hypothesized that C. butyricum could reach the goat intestinal tract through the rumen and had a positive effect. Therefore, the purpose of this experiment was to compare the impact of different forms of C. butyricum on the jejunal TJ proteins expression, anti-inflammatory level and intestinal flora of goats at the same supplementation level, and to evaluate the regulation of C. butyricum on intestinal health of goats.
Materials and methods
The current experimental protocol was approved by the Institutional Animal Care and Use Committee of Northeast Agricultural University (Harbin, China) (protocol number: NEAU- [2011]-9).
Experimental design and animal management
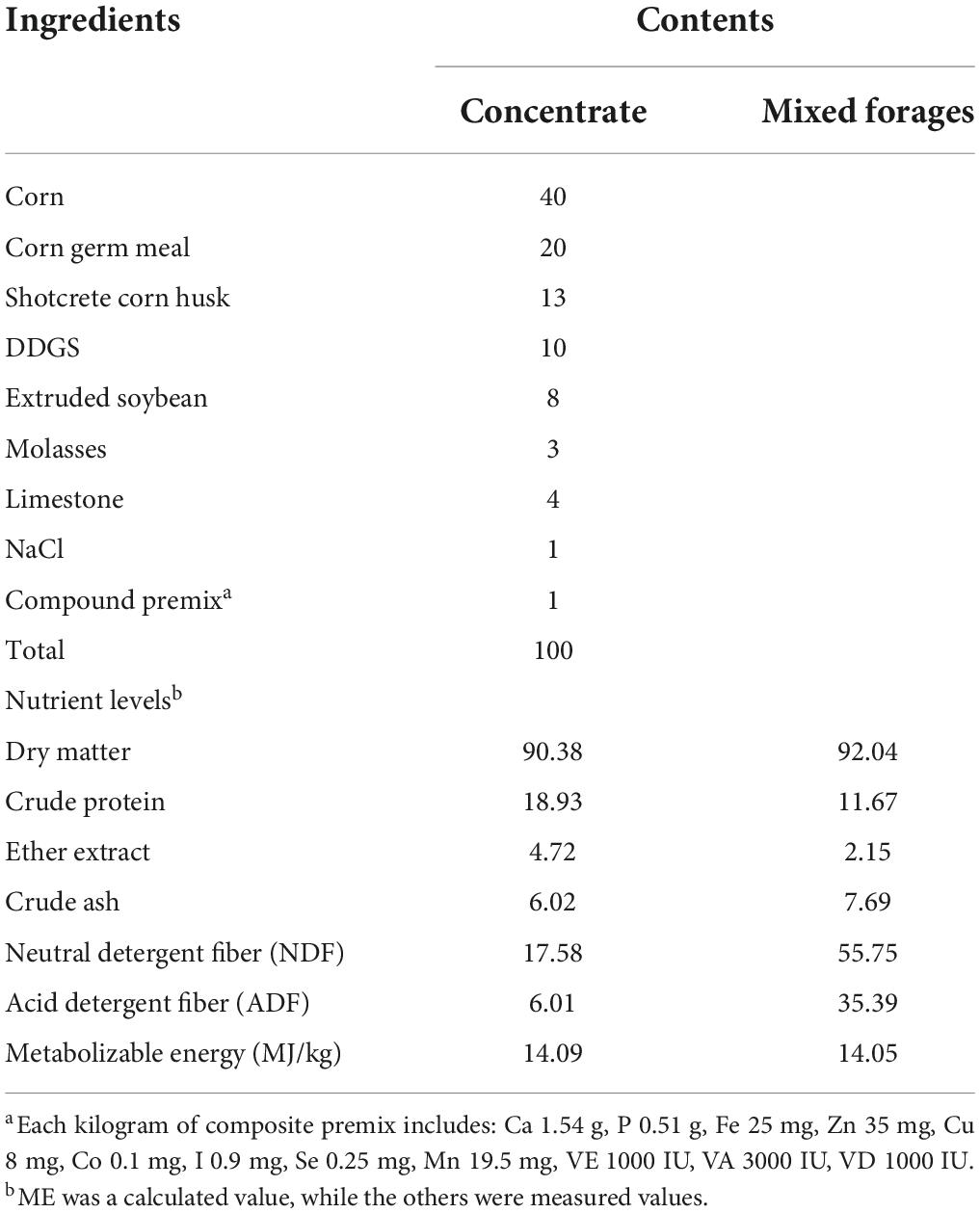
Fifteen good health Albas male goats with 21.9 ± 0.1 kg of body weight (BW) were used. In a randomized trial design, these goats were divided into three treatments (5 goats per treatment). The control (CON) goats were fed a basal diet, rumen-protected C. butyricum (RPCB), and C. butyricum (CB) goats were fed a basal diet with 1 × 109 CFU/kg C. butyricum (C. butyricum was provided by Lvxue Biotechnology Co., Ltd., Hubei, China); having effective colony count ≥ 1 × 109 CFU/g. Rumen-protected C. butyricum was protected by hydrogenated fat, and the effective colony count was the same as C. butyricum. All goats were fed with the concentrate to forage ratio of the experimental diet of 65:35 (forage is a mixture of oat and alfalfa; concentrate was provided by Commercial concentrate Jiuzhou earth feed Co., Ltd. without probiotics, Inner Mongolia, China). Each treatment was fed for 14 days of adaptation, and then all goats were fed for 56 days. Goats were fed twice daily at 06:00 and 18:00. Each goat was arranged in a single pen with free access to fresh water. The ingredient and nutrient composition of diets are shown in Table 1.
Sample collection
After the growth trial, all experimental goats were electrically stunned (200 V applied for 4 s) and slaughtered by exsanguination. Then, the jejunum was collected and washed immediately with PBS buffer. And then, the collected jejunum samples were divided into two parts. The mucosa was scraped off the jejunum with sterile slides and placed in a cryopreservation tube immediately after being frozen in liquid nitrogen and stored in a −80°C refrigerator to be transferred to the laboratory for evaluating the expression of TJ proteins and inflammatory factor. Meanwhile, the contents of the intestine were placed in presterilized cryovials, frozen in liquid nitrogen, and then transferred to the laboratory’s −80°C refrigerator for storage to measure microbial diversity.
Quantitative real-time PCR
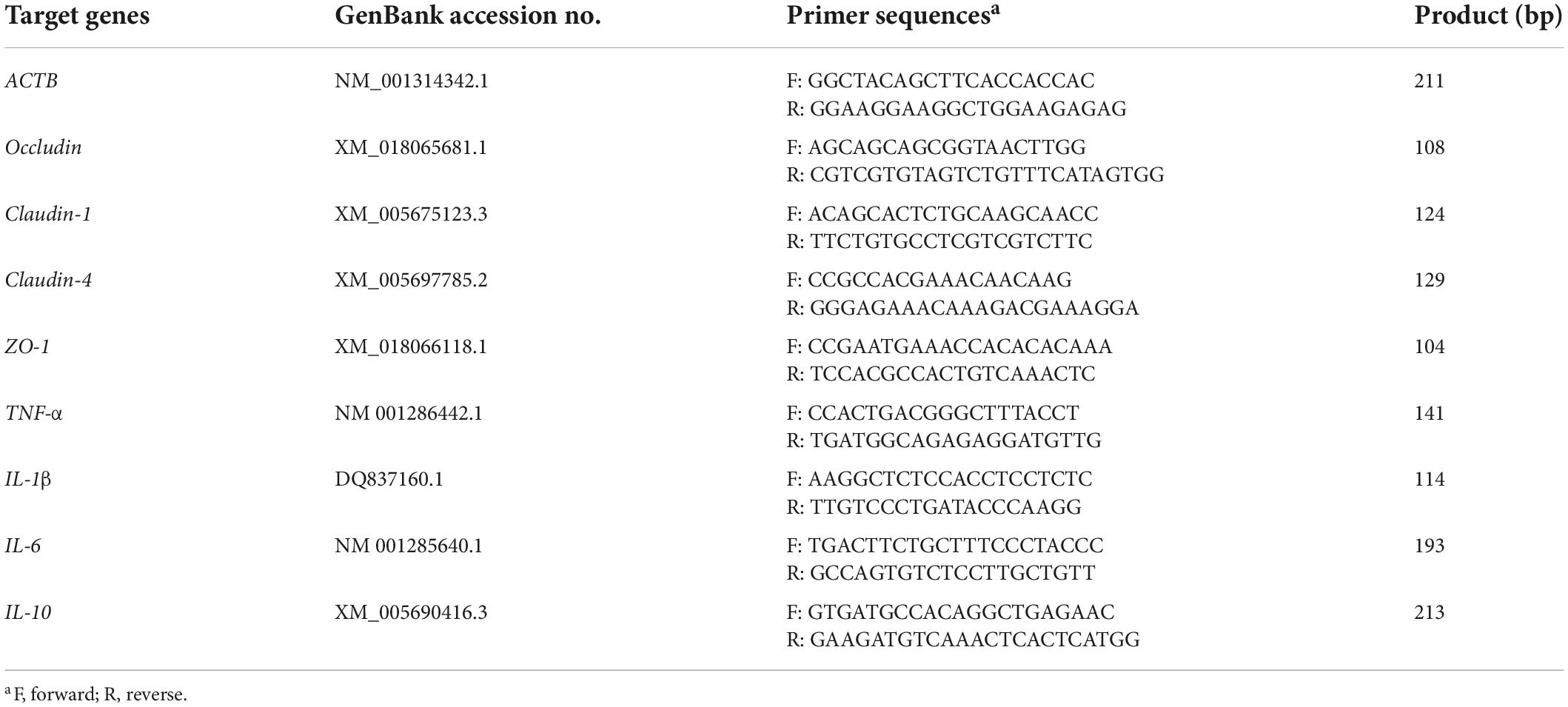
Total RNA was extracted from the jejunal mucosa of each goat using an animal tissue total RNA extraction kit (TIANGEN Bio, Beijing, China). Ultra-Micro Spectrophotometer (Implen P330, Munich, Germany) was used to determine RNA concentration and quality. The absorbance ratio of all samples was between 1.8 and 2.2, indicating the high quality of the total RNA extracted. Subsequently, it was reverse transcribed into first-strand cDNA according to the 5X Integrated RT MasterMix kit (DiNing Bio, Beijing, China). The relevant gene expression primers (Table 2) used in this experiment were designed and synthesized by General Biological Co., Ltd. Real-time quantitative PCR was performed using the 2x Fast qPCR Master Mixture kit (DiNing Bio, Beijing, China) with the Quantagene q225MX Real-Time PCR System (KuBo Technology, Beijing, China) according to the manufacturer’s instructions. Each sample was analyzed in triplicate, the mean value of each target gene and β-actin’s Ct-values were calculated, and ΔCt was determined. The 2–ΔΔCt method was used to analyze the relative expression of each target gene.
Microbial composition
DNA from all contents of cecum samples was extracted using the E.Z.N.A.® Stool DNA Kit (D4015, Omega, Inc., USA) according to the manufacturer’s instructions. Nuclear-free water was used for blank. The total DNA was eluted in 50 μL of elution buffer and stored at −80°C until measurement using PCR by LC-Bio Technology Co., Ltd. (Zhejiang, China). Universal primer sequences were designed according to the V3-V4 region of the amplified fragment of 16S rDNA: 341F (5′-CCTACGGGNGGCWGCAG-3′, 805R (5′-GACTACHVGGGTATCTAATCC-3′), each sample barcoded at the 5′ end of the primers, and sequenced the universal primers. PCR amplification was performed in a total volume of 25 μL of reaction mix containing 25 ng of template DNA, 12.5 μL of PCR master mix, 2.5 μL of each primer, and PCR-grade water was added to adjust the volume. PCR conditions for amplifying prokaryotic 16S fragments: initial denaturation at 98°C for 30 s; denaturation at 98°C for 10 s, annealing at 54°C for 30 s, extension at 72°C for 45 s, 32 cycles; and final extension at 72°C for 10 min. Post-PCR products were confirmed by 2% agarose gel electrophoresis. During the entire DNA extraction process, ultrapure water was used instead of sample solution to exclude the possibility of false-positive PCR results as negative controls. Beads (Beckman Coulter Genomics, Danvers, MA, USA) were purified and quantified by Qubit (Invitrogen, USA). Amplicon pools were used for sequencing, and the size and number of amplicon libraries were measured on an Agilent 2100 Bioanalyzer (Agilent, USA) and Illumina (Kapa Biosciences, Woburn, MA, USA) library quantification kits. Libraries were sequenced on the NovaSeq PE250 platform.
Statistical analysis
Data were analyzed using the one-way ANOVA process in SPSS 25 statistical software (SPSS 20.0; SPSS Inc., Chicago, IL, USA) followed by Duncan’s multiple range tests, and the test results were expressed as the mean and standard error of the mean (SEM), where P < 0.05 was considered statistically significant, and 0.05 < P < 0.10 was considered to have a significant trend (Zhou et al., 2019).
16S rDNA sequencing samples were sequenced on the Illumina NovaSeq platform following the manufacturer’s recommendations provided by LC-Bio. Paired-end sequences were assigned to samples based on their unique barcodes, and the barcodes and primer sequences introduced by the library were removed. FLASH was used to merge matching end reads. According to fqtrim (v0.94), the raw read data is quality filtered under specific filtering conditions to obtain high-quality clean labels. Chimeric sequences were filtered using Vsearch software (v2.3.4). DADA2 was used to demodulate to get the feature table and feature sequence. Alpha diversity and beta diversity are calculated by normalizing to the same random sequence. Feature abundances were then normalized using the relative abundance of each sample according to the SILVA (release132) classifier. Alpha diversity is used to analyze the complexity of sample species diversity through 5 metrics, including Chao, Observed species, Goods coverage, Shannon, Simpson, all of which are calculated with QIIME2 in our sample. Beta diversity was calculated by QIIME2 and plotted by the R package. Both the correlation network graph and the correlation heatmap are analyzed through the Spearman correlation matrix, which is also drawn by the R package (v3.6.3). The BLAST was used for sequence alignment, and each representative sequence was annotated with the SILVA database for characteristic sequences. Other graphs are also implemented using the R package (v3.5.2).
Results
Gene expression of tight junction proteins in the jejunal mucosa
Table 3 shows the preliminary analysis results of β-actin (ACTB), Occludin, Claudin-1, Claudin-4, and Zonula occludens-1 (Zo-1) mRNA expression levels in fattening goats. The mRNA expression of Claudin-4 in the jejunal mucosa significantly increased in CB treatment (P < 0.05). In addition, the mRNA expression of Occludin in CB had an upward trend compared to the CON (P < 0.05). However, the expression of ACTB, Claudin-1, and ZO-1 were not markedly different in the treatment group (P > 0.10).
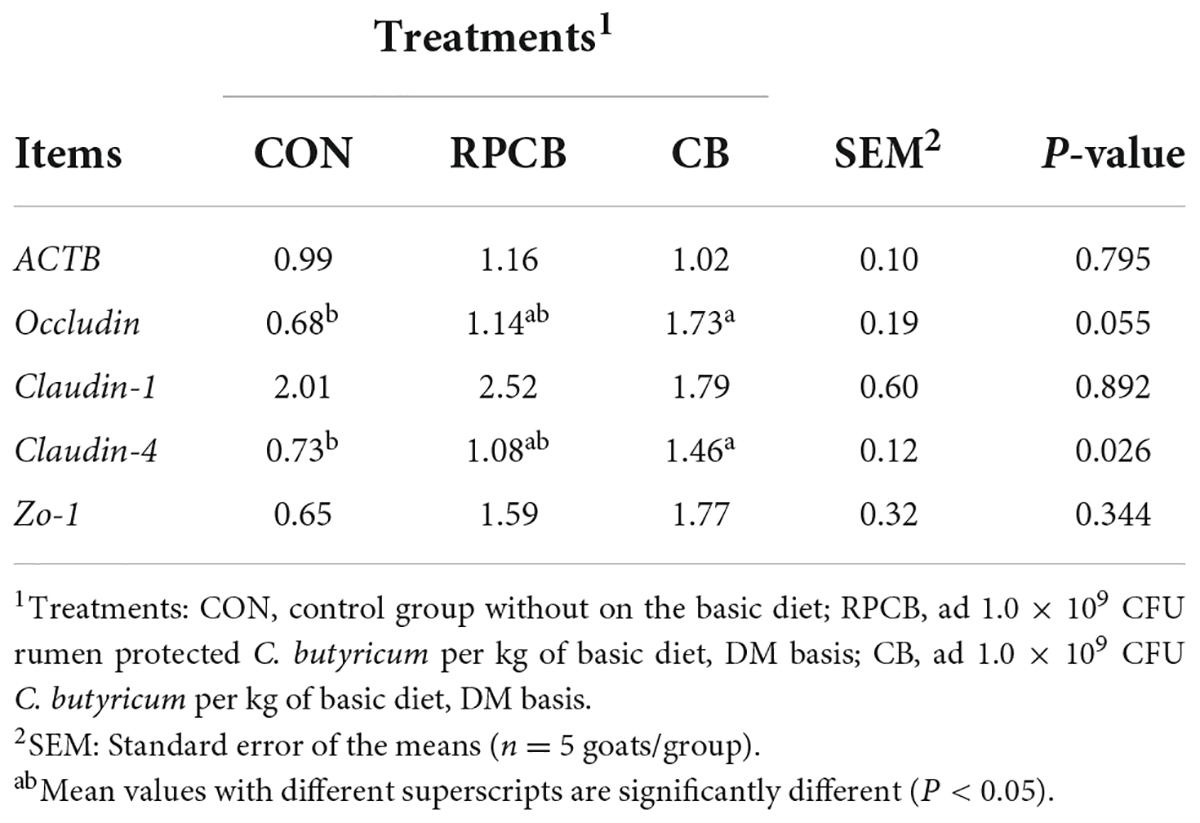
Table 3. The mRNA expression of TJ proteins of the jejunal mucosa of goats with diet supplementation with C. butyricum.
Gene expression of inflammatory factors in the jejunal
The results of the expression of TNF-α, IL-1β, IL-6, and IL-10 in the colon of fattening goats are shown in Table 4. The mRNA expressions of TNF-α, IL-1β, IL-6, and IL-10 in the colon were without significant differences in the above treatment groups (P > 0.10).
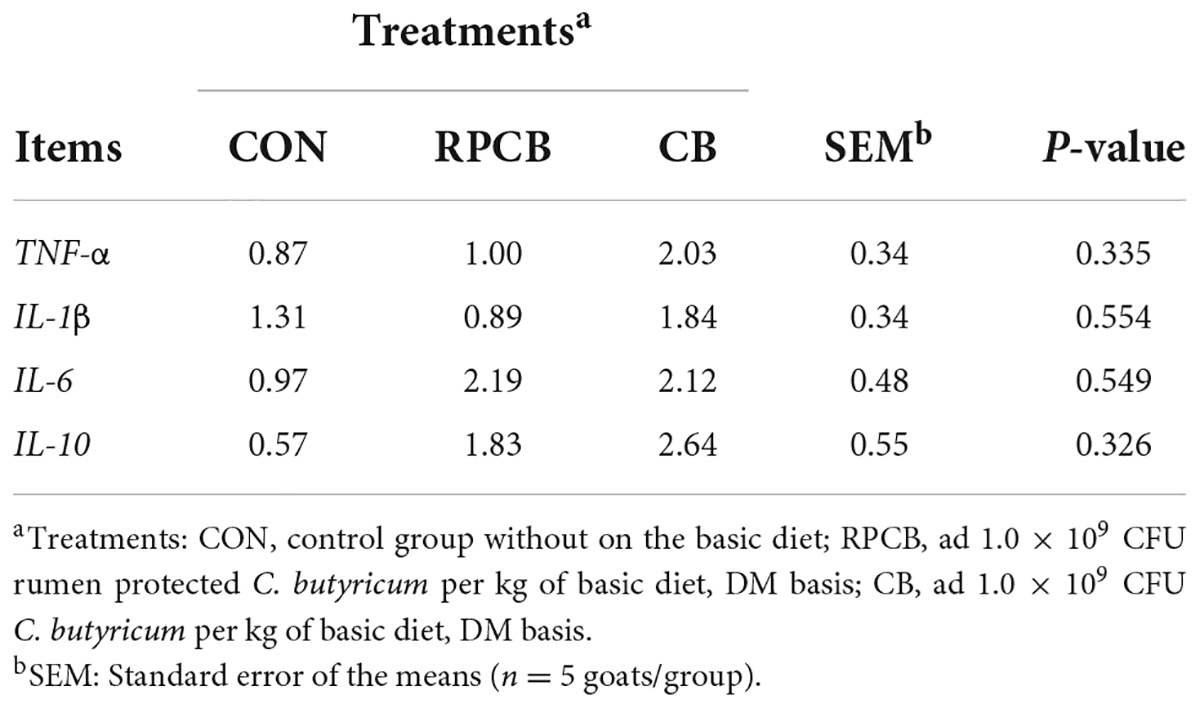
Table 4. The mRNA expression of inflammatory factors of the jejunal of goats with diet supplementation with C. butyricum.
The content of cecum microbial composition
A total of 1,098,998 effective 16S rDNA sequences were detected in the content of 15 cecum samples. After filtering (proportion of data with data quality ≥ Q20 and Q30 in valid data), the values of all samples were better than 98.2 and 97.4% indicating good sequencing depth for analysis of the content of cecum microbiota. The amount of data measured is within a reasonable range, as shown by the dilution curve in α-diversity (Figures 1A1–A4), but there is no significant difference between them. Common and specific bacteria at the phylum and genus levels are shown in the Venn diagrams (Figures 1B1,B2). The microbiota of the three treatments shared 25 phyla and 274 genera. However, the PCoA and NMDS plots analysis are one of the most commonly used analytical methods of β-diversity performed, and the compositions of cecum microbial were statistically different among the above three treatments (P < 0.05) (Figures 1C1,C2).
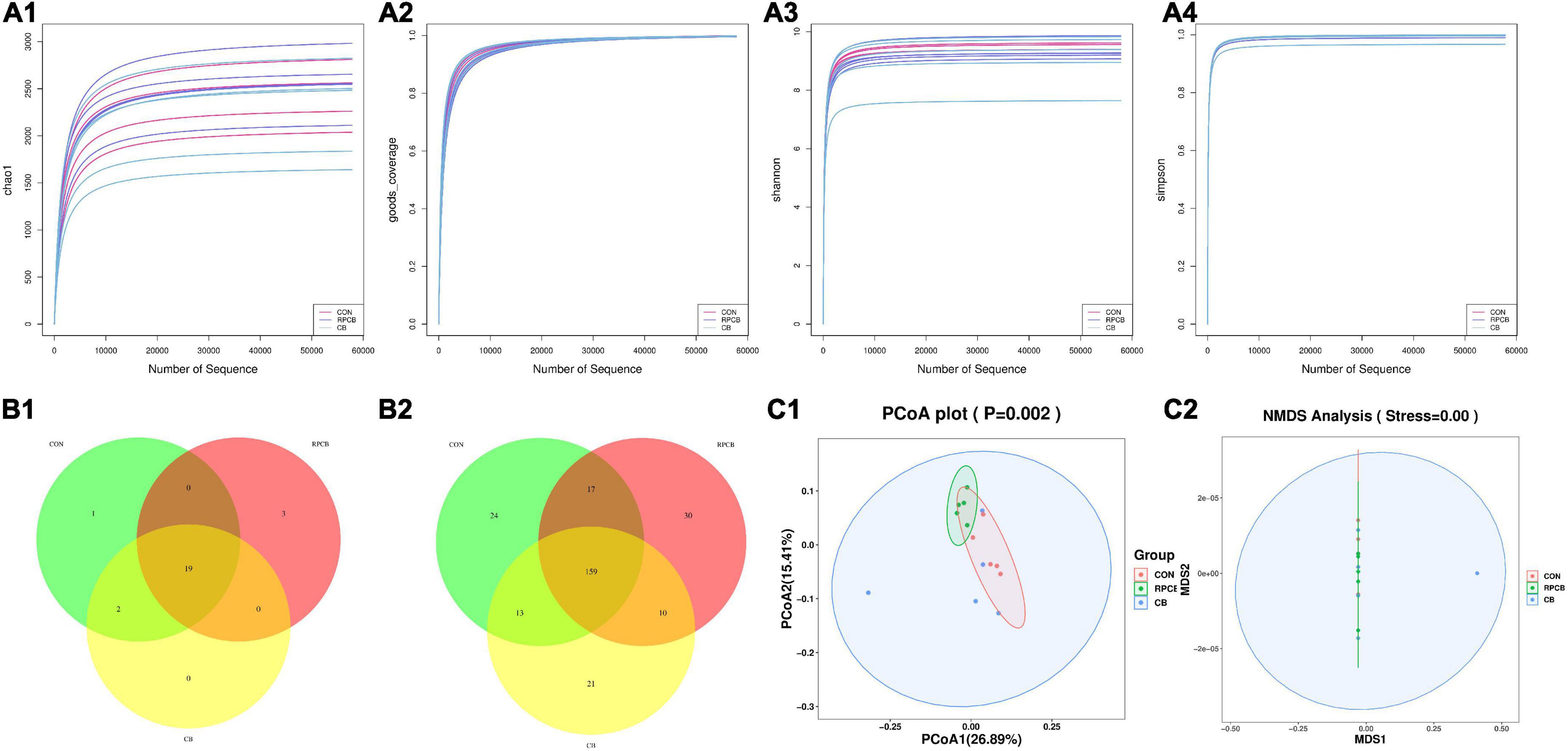
Figure 1. The α- and β-diversity of fecal bacterial communities in fattening goats fed C. butyricum. Chao 1 (A1), Goods coverage (A2), Shannon (A3), and Simpson (A4) curves of intestine microbiome of fattening goats are shown. Unique and shared intestine feature among CON, RPCB and CB groups shown on Venn diagrams in phylum (B1) and genus (B2) level. Intestine microbial structure among the three groups was estimated by the Principal Coordinate Analysis (C1) and Non-metric Multidimensional Scaling Analysis (C2).
In addition, we analyzed phylum- and genus-level microbiomes to assess their specific composition (Tables 5, 6). The relative abundances of different treatments at different levels are represented by histograms (Figures 2A,B). Bacterial taxa with a relative abundance > 0.1% in more than 3 biological replicates per treatment were considered as identified in the current study. According to the result of phylum, we found that Firmicutes predominated in each treatment. Furthermore, the relative abundance of Verrucomicrobia and unclassified in the RPCB group was significantly higher than in the CON and CB groups (P < 0.05). In addition, Candidatus_Melainabacteria had an increasing trend in the RPCB (0.05 < P < 0.1). The rest did not change significantly. At the genus level, the relative abundance of Clostridium, Clostridiaceae_unclassified, and Parapedobacter were markedly decreased in RPCB and CB compared with CON (P < 0.05). The relative abundance of Rikenellaceae _unclassified and unclassified was significantly increased only in RPCB. Meanwhile, the relative abundances of Akkermansia, Clostridia_unclassified, Candidatus_Melainabacteria_unclassified, and Sporobacter tended to increase compared to the control group (0.05 < P < 0.1). Contrary to this result are Emticicia and Mollicutes_ unclassified (0.05 < P < 0.1). Simultaneously, the Sankey plot shows that the common core bacteria represented mainly came from the Bacteroidetes, Firmicutes, Verrucomicrobia, and unclassified phylum (Figure 2C). Moreover, we also further explored the species level of Clostridium (Table 7). As indicated in the results, the relative fractions of some uncultured and unclassified Clostridium species were significantly reduced (P < 0.05) or tended to decline (0.05 < P < 0.1). There was no significant change in the relative abundance of C. butyricum in each group, and it only existed in RPCB and CB treatments.
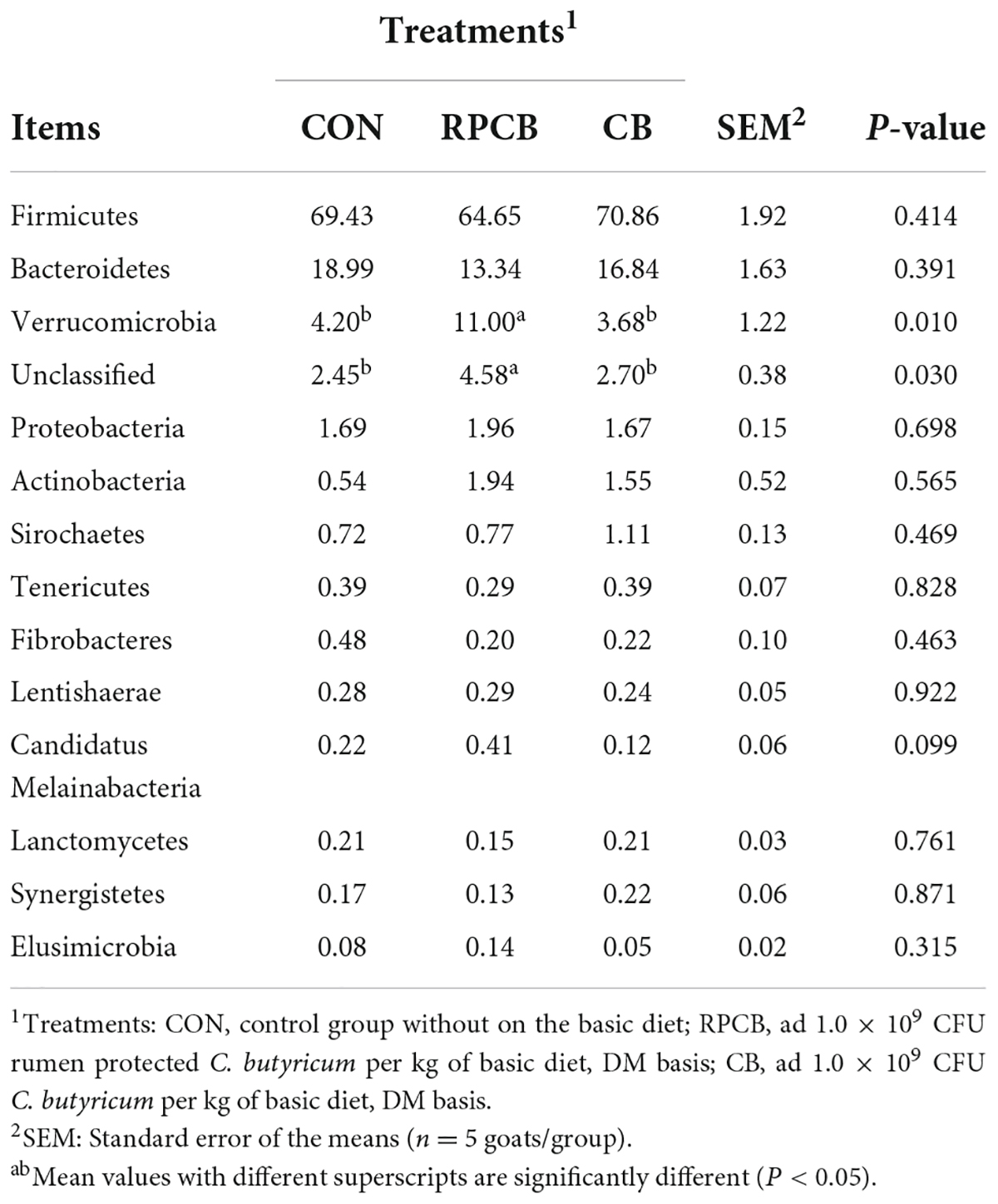
Table 5. Effects of C. butyricum on the relative abundance (> 0.1%) of gut microbes at the phylum level in goats.
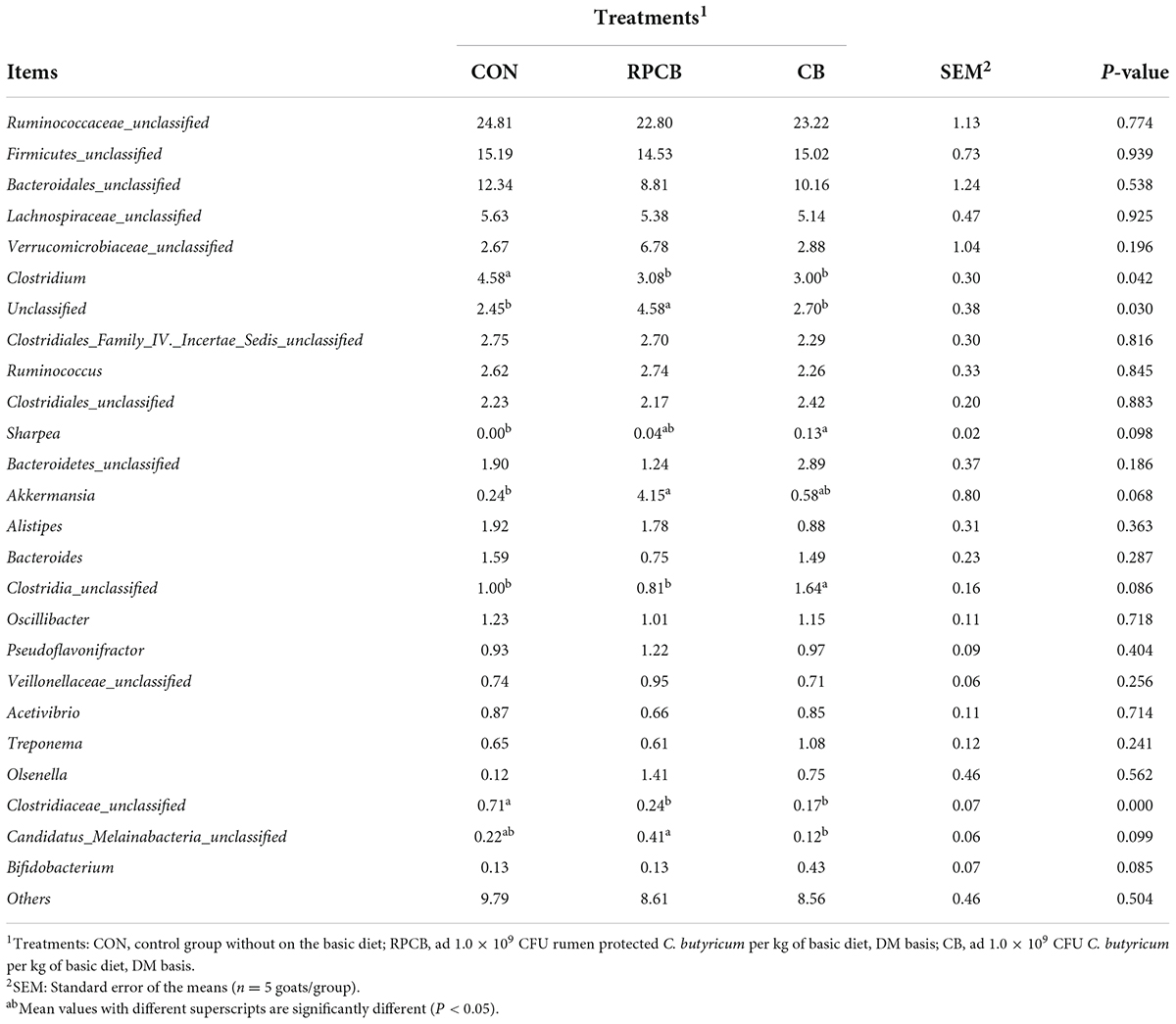
Table 6. Effects of C. butyricum on the relative abundance (> 0.1%) of gut microbes at the genus level in goats.
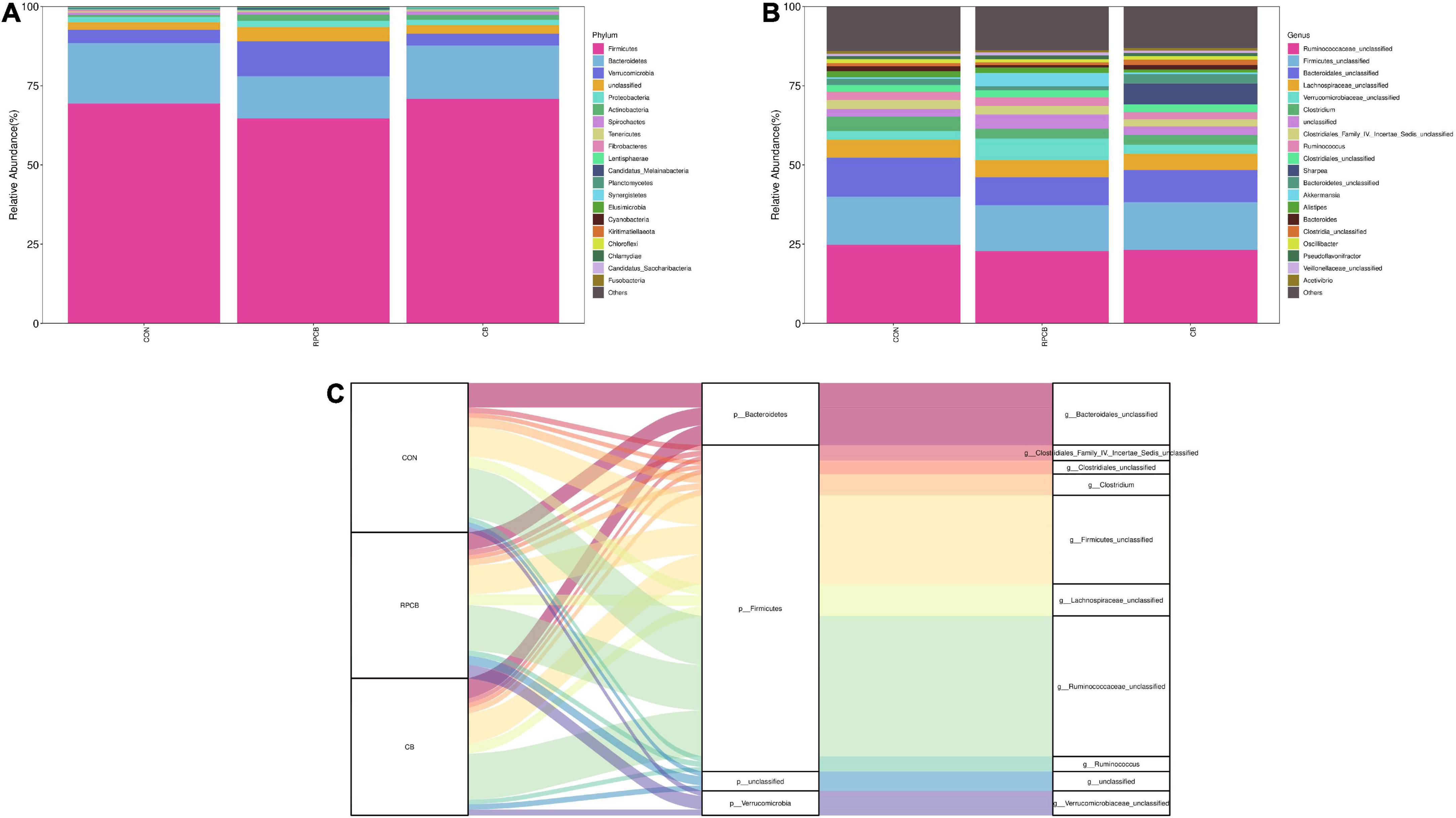
Figure 2. Stacked bar graph displays a comparison of microbial relative abundance (%) at phylum (A) and genus (B) level among main effects of treatments in intestine of fattening goats. And Sankey plot displays a comparison of microbial relative abundance (%) at genus level (C) among main effects of treatments in intestine of fattening goats. The label CON denotes without prebiotics, the label RPCB denotes rumen-protected C. butyricum, and the label CB denotes C. butyricum for each sample. CON: 0 CFU C. butyricum per kilogram feed, RPCB: 1.0 × 109CFU C. butyricum per kilogram feed, CB: 1.0 × 109CFU C. butyricum per kilogram feed.
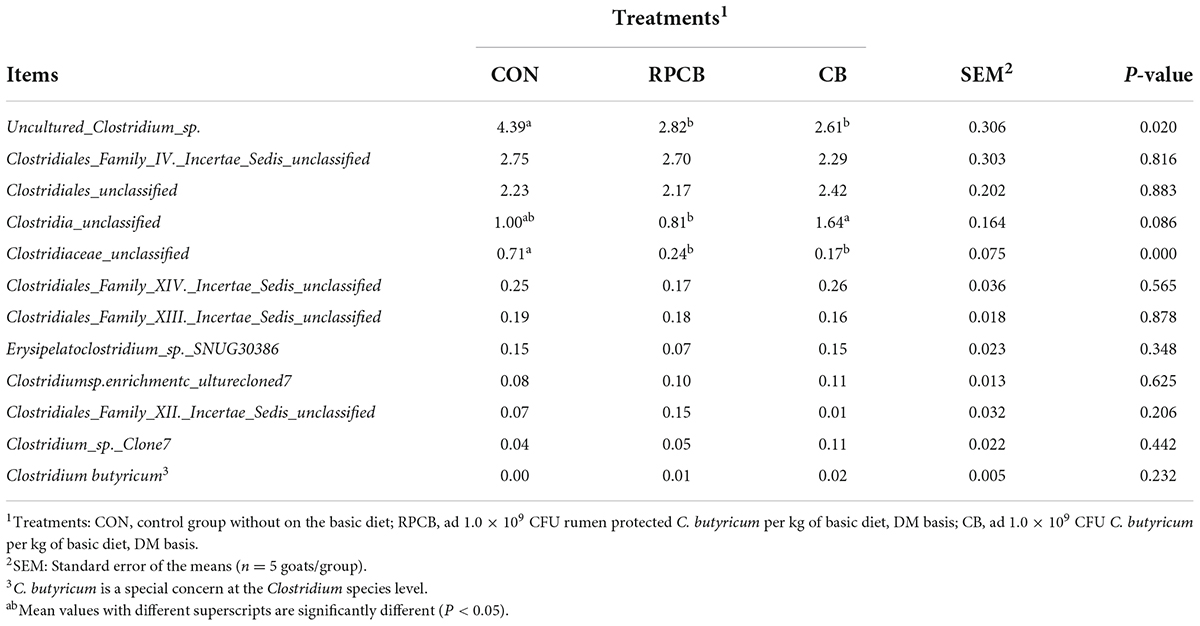
Table 7. Effects of C. butyricum on the relative abundance (> 0.1%) of gut microbes at the species level (Clostridium species) in goats.
According to the Spearman correlation algorithm, we performed a correlation analysis of gut microbes and mRNAs related to the jejunal mucosal barrier. As the heatmap shows, the mRNA expressions of Occludin and Claudin-4 performed a positive correlation with the Sharpea genus (P < 0.05) (Figure 3A1). In addition, Claudin-4 was strongly positively correlated with C. butyricum (P < 0.01), and Occludin showed a positive correlation with C. butyricum (red in the correlation heatmap) in the species correlation heatmap (Figure 3A2). This suggests that C. butyricum could positively affect the expression of TJ proteins in jejunal mucosal. Meanwhile, we also correlated microbes at the genus and species level (Clostridium). The results in Figure 3 show interactions between core bacteria at different levels through the correlation analysis of the core microbiomes at the genus level and the microbiomes with significant and significant trends. Among them, the centralities of Ruminococaceae_unclassified, Verrucomicrobiaceae_unclassified, and sharpea were the largest, while the centralities of Bifidobacterium, Clodtridiaceae_unclassified, unclassified, Firmicutes_unclassified, and Clostridales_unclassified were in the middle, and the centralities of the rest of the microbiomes were the lowest. Of particular concern, Clostridium was only negatively associated with Sharpea and not with other microbiomes (Figure 3B1). In addition, through the significant species (Clostridium) correlation network analysis, only the Clostridiales_Family_IV._Incertae_Sedis-_unclassified and Closrtidium_sp._Clone-7 were the largest core microbiomes, of which C. butyricum was negatively correlated with Clostridiales_Family_IV._Incertae_Sedis-_unclassified (Figure 3B2).
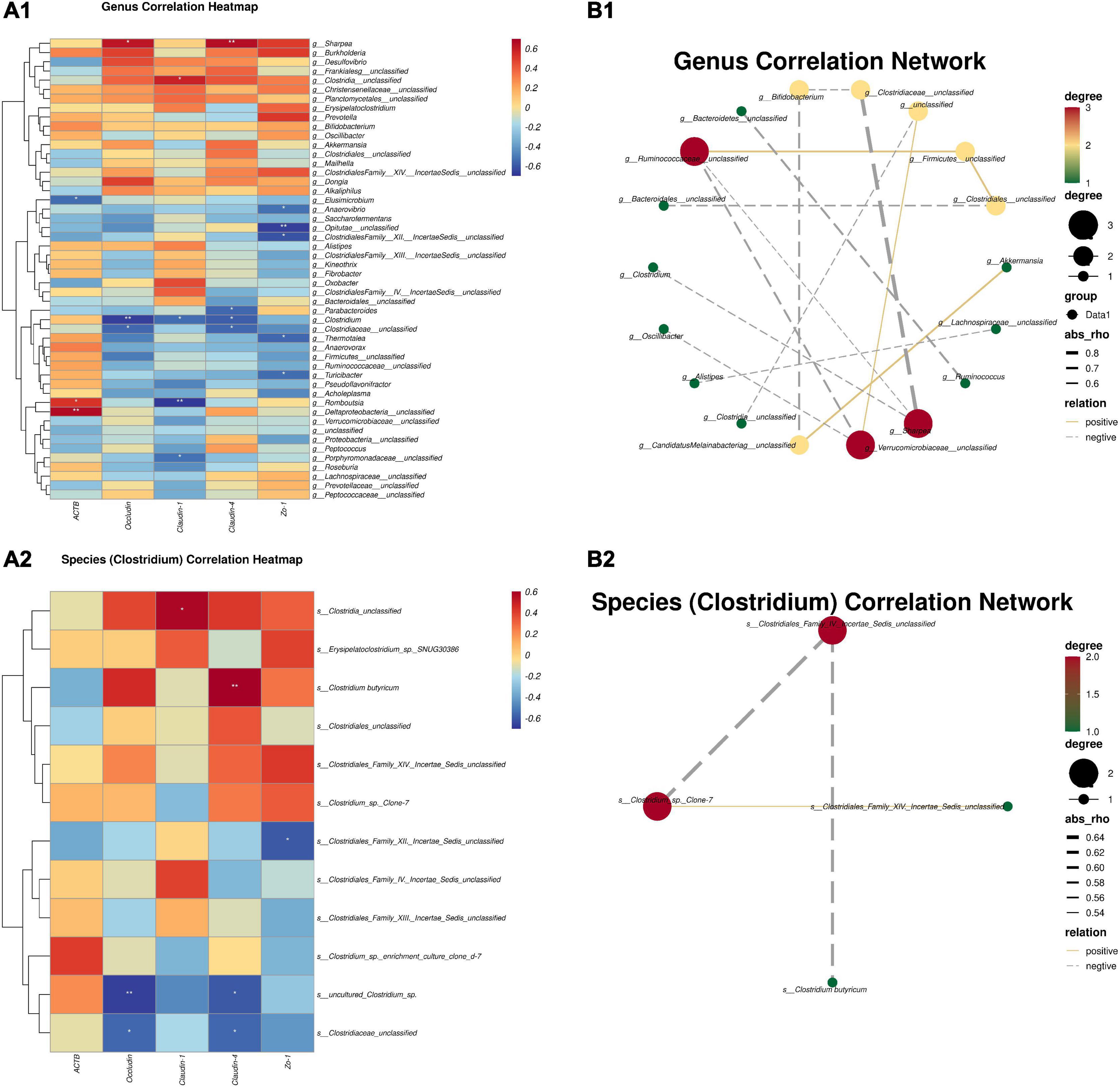
Figure 3. Spearman correlation between intestinal microbiotas and the mRNAs TJ proteins. (A1) Genus correlation heatmap; (A2) species (Clostridium) correlation heatmap; (B1) genus correlation network; (B2) species (Clostridium) correlation network. Red denotes a positive correlation; blue denotes a negative correlation. The color intensity is proportional to the strength of the Spearman correlation. *P ≤ 0.05, **P ≤ 0.01.
Discussion
The current study aimed to describe how dietary supplementation with C. butyricium affects the intestinal tract’s health by adapting the goats’ microbial community. Our previous research indicated that different doses of C. butyricium could modulate beneficial microbial diversity and relative abundance in the rumen and feces of fattening goats (Zhang C. et al., 2022).
The efficient digestion, absorption, and transport of nutrients in the gastrointestinal tract is inseparable from the strong immune function of the intestinal tract, which provides a good guarantee for its metabolism in the body (Penner et al., 2011). Previous studies reported that probiotics such as Bifidobacterium and Lactobacillus colonized the intestinal mucosa of newborn ruminants, inhibited the invasion of harmful bacteria into the intestine, reduced diarrhea in young ruminants, and maintained the homeostasis of the intestinal environment to improve its growth performance (Zhang et al., 2021). In recent years, most studies have reported that C. butyricum could regulate the growth performance of animals by balancing the intestinal micro ecological environment and protecting intestinal epithelial cells through its probiotic metabolites (Wang et al., 2020; Meng et al., 2021; Xu et al., 2021). Cytokines play an important role in improving intestinal immune function. The study by Rakhshandeh and de Lange (2012) reported that pro/anti-inflammatory factors were cytokines released after pathogenic bacteria invaded the body, which was closely related to various metabolic reactions, including changing the digestion and absorption of nutrients to improve immune function. Studies in piglets (Liang et al., 2020), poultry (Xu et al., 2021), mice (Hagihara et al., 2020), and aquaculture (Meng et al., 2021) confirmed that dietary supplementation with C. butyricum could alter anti-inflammatory factors (IL-10) and pro-inflammatory factors (TNF-α, IL-1β, and IL-6, etc.) expressions. However, it was found in this experiment that supplementation with different forms of C. butyricum had no significant effect on the expression of TNF-α, IL-1β, IL-6, and IL-10 in the intestinal tract of goats. This may be related to our previous study that supplementation with C. butyricum failed to alter nutrient metabolism in goats, resulting in no significant changes in daily weight gain and feed efficiency (Zhang C. et al., 2022).
While interestingly, the mRNA expressions of TJ proteins Occludin and Claudin-4 in the jejunal mucosa of goats fed a diet containing C. butyricum showed a significant upward trend and were significantly increased. As an important part of the intestinal barrier, TJ proteins are a multi-molecular complex composed of various transmembrane proteins, including ACTB, ZO-1, Occludin, and Claudin family proteins (Ulluwishewa et al., 2011). The interaction maintains the paracellular permeability, consolidating the intestinal epithelial barrier function and preventing the penetration of antigenic substances such as lipopolysaccharide from harmful bacteria into the intestinal lamina propria, triggering an inflammatory response (Wageha et al., 2017; Stoeva et al., 2021). The results of this study are consistent with the results of Xu et al. (2021) and Li et al. (2021)’s supplementation of C. butyricum could increase the expression of Occludin and Claudin family genes in the intestinal epithelial barrier. Meanwhile, this might explain the lack of significant changes in the expression of jejunal inflammatory factors after C. butyricum supplementation. In addition, Lu et al. (2020) also confirmed in the study of piglets that C. butyricum could significantly upregulate the expression of TJ proteins related genes (ZO-1 and Occludin) in the intestinal epithelial barrier. This was inextricably linked to butyric acid, the main metabolite of C. butyricum. Most studies have shown that butyric acid has a supporting and protective effect on intestinal epithelial cells, which could provide energy for intestinal epithelial cells and contribute to the growth, development, and improvement of function (Hou et al., 2014; Qiao et al., 2015). Zhao et al. (2020) found that oral administration of C. butyricum could improve intestinal mucosal barrier function and reverse intestinal inflammatory damage caused by severe acute pancreatitis and intra-abdominal hypertension by increasing the content of butyric acid and propionic acid in the intestine. In addition, we also learned from Xu et al. (2021) that C. butyricum not only altered its metabolites to affect the intestinal epithelial barrier but also consolidated intestinal barrier function by modulating the gut microflora. This also confirmed previous reports that probiotics could help alleviate the decline of TJ proteins expression caused by various reasons (Chang et al., 2019). However, there were no significant differences in other TJ proteins related genes (ACTB, Claudin-1, and ZO-1) in the current study, which were related to the dynamic properties of TJ proteins in the intestinal barrier, the molecular mechanism and its function in different species role related (Xu et al., 2021).
To explore how C. butyricum affects the gut epithelial barrier and gut microbes, we performed 16S rDNA sequencing. In the current study, by analyzing the gut microbiota of goats in different groups, the results showed that Firmicutes, Bacteroidetes and Verrucomicrobia were the main dominant microbiota in this intestinal segment. Among them, the dietary supplementation with RPCB significantly increased the relative abundance of Verrucomicrobia. This may be due to the synergy of the unclassified phyla whose relative abundances also rose. Previous studies have shown that Verrucomicrobia can play a beneficial role by improving the body’s metabolism or increasing the thickness of the mucus layer (Everard et al., 2013). It was because butyric acid, as one of the products of C. butyricum with other various nutritional products, was positively correlated with the modification of intestinal flora (Zhang Q. et al., 2022). At the genus level, we found significant differences in Clostridium, Unclassified and Clostridiaceae_unclassified. We also found that there were 5 kinds of bacteria with different trends. Surprisingly, supplementation of both forms of C. butyricum in goat diets significantly decreased the relative abundance of C. butyricum in their intestines. Therefore, we further analyzed the species-level Clostridium, and the results showed the relative abundance of Uncultured_Clostridium_sp. and Clostridium_unclassified in the gut of goats was significantly reduced after supplementation with C. butyricum. Because C. butyricum was the research object of this experiment, we paid special attention to it. Although its relative abundance in the intestine was not higher than 0.1% and it only existed in the treatment group, while the relative abundance of C. butyricum equaled 0% in the control group. It was well known that Clostridium also had harmful bacteria detrimental to body health and non-pathogenic microorganisms not classified as competing for nutrients with beneficial bacteria (Amila et al., 2020). Therefore, the significant decrease in the relative abundance of Clostridium as well as the mRNA expressions of TJ proteins ACTB, Claudin-1, and Zo-1 without obvious difference may be related to the decline of pathogenic Clostridium in the gut.
Furthermore, in order to reveal the relationship between the gut microbiota and epithelial barrier function after supplementation with C. butyricum, we analyzed the correlation between the gut microbiota and genes related to the expression of TJ proteins in the jejunal mucosa, as well as the correlation between the flora. A previous study had shown that as one of the main producers of lactic acid in the gastrointestinal tract, Sharpea could synthesize lactic acid by hydrogenating hydrogen ions in the body through its own lactate dehydrogenase (Sandeep et al., 2018). However, since too much lactic acid cannot be accumulated in the intestinal tract, it can be converted into butyric acid by the action of lactic acid utilizing bacteria (Zhu et al., 2020). In the genus level correlation analysis, Sharpea showed a strong positive correlation with the Occludin and Claudin-4 genes that were determined to be significantly expressed. This might be related to the proliferation of Sharpea in cooperation with other butyrate-producing bacteria. Moreover, Clostridium negatively correlated with Occludin and Claudin-4 genes expression, while species (Clostridium) level correlation analysis showed that Occludin was positively correlated with Claudin-4 C. butyricum and negatively correlated with two unidentified Clostridium species. And the Claudin-4 gene showed a strong correlation with C. butyricum. Therefore, this corresponds to our conjecture that supplementation with C. butyricum may reduce the harmful bacteria C. butyricum, and also means that C. butyricum could upregulate the expression of TJ proteins related genes by itself or in coordination with other probiotics (Stoeva et al., 2021).
Conclusion
Dietary supplementation with C. butyricum could enhance the intestinal epithelial barrier function of goats by regulating the relative abundance of beneficial intestinal bacteria, while rumen protection of C. butyricum had no significant effect on the intestinal health of goats. Fortunately, this gives us a research direction. In the future, more molecular techniques and bacterial genomics methods should be combined to reveal the specific mechanism of C. butyricum regulating the intestinal flora of goats.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI database, the accession number: PRJNA861251.
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Northeast Agricultural University (Harbin, China). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
CZ, YGZ, and YS conceptualized and designed the study. CZ conducted animal trials, analyzed the data, and drafted original manuscript. TH, QY, and JW performed laboratory experiments. YFZ and MN provided support and services. HX, YGZ, and YS reviewed and provided critical comments on the manuscript. YGZ and YS were obtained the funding. All authors read and approved the final manuscript.
Funding
This study was financially supported by the Research and Demonstration of Water-saving and Efficient Production and Utilization Technology of High-quality Forage Material (KJXM-EEDS 2020010-04) and the earmarked fund for CARS36. The authors thank Northeast Agricultural University for the supporting facility.
Acknowledgments
We thank Ordos Academy of agriculture and animal husbandry and a family ranch in Otoki for providing the trial site as well as appreciate to Clostridium butyricum LXKJ-1 from Lvxue Biotechnology Co., Ltd. (Hubei, China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amila, S., Steve, F., Jon, P., Gale, B., and Tanushree, B. (2020). Antimicrobial production by strictly anaerobic Clostridium spp. Int. J. Antimicrob. Agents 55:105910. doi: 10.1016/j.ijantimicag.2020.105910
Araki, Y., Andoh, A., Fujiyama, Y., Takizawa, J., Takizawa, W., and Bamba, T. (2002). Oral administration of a product derived from Clostridium butyricum in rats. Int. J. Mol. Med. 9, 53–57.
Bedford, A., and Gong, J. (2018). Implications of butyrate and its derivatives for gut health and animal production. Anim. Nutr. 4, 151–159. doi: 10.1016/j.aninu.2017.08.010
Chang, C., Teng, P., Lee, T., and Yu, B. (2019). Effects of multi-strain probiotic supplementation on intestinal microbiota, tight junctions, and inflammation in young broiler chickens challenged with Salmonella enterica subsp. enterica. Asian-Austral. J Anim. Sci. 33, 1797–1808. doi: 10.5713/ajas.19.0427
Chelakkot, C., Ghim, J., and Ryu, S. (2018). Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 50, 1–9. doi: 10.1038/s12276-018-0126-x
Esterházy, D., Canesso, M., Mesin, L., Muller, P., de Castro, T., Lockhart, A., et al. (2019). Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 569, 126–130. doi: 10.1038/s41586-019-1125-3
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J., Druart, C., Bindels, L., et al. (2013). Crosstalk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Hagihara, M., Ariyoshi, T., Kuroki, Y., Eguchi, S., Higashi, S., Mori, T., et al. (2021). Clostridium butyricum enhances colonization resistance against Clostridioides difficile by metabolic and immune modulation. Sci. Rep. 11:15007. doi: 10.1038/s41598-021-94572-z
Hagihara, M., Kuroki, Y., Ariyoshi, T., Higashi, S., Fukuda, K., Yamashita, R., et al. (2020). Clostridium butyricum modulates the microbiome to protect intestinal barrier function in mice with antibiotic-induced dysbiosis. iScience 23:100772. doi: 10.1016/j.isci.2019.10077
Hou, Y., Wang, L., Yi, D., Ding, B., Chen, X., Wang, Q., et al. (2014). Dietary supplementation with tributyrin alleviates intestinal injury in piglets challenged with intrarectal administration of acetic acid. Br. J. Nutr. 111, 1748–1758. doi: 10.1017/S0007114514000038
Li, W., Xu, B., Wang, L., Sun, Q., Deng, W., Wei, F., et al. (2021). Effects of Clostridium butyricum on growth performance, gut microbiota and intestinal barrier function of broilers. Front. Microbiol. 12:777456. doi: 10.3389/FMICB.2021.777456
Liang, J., Raza, S., Kou, S., Chen, C., Yao, M., Wu, Y., et al. (2020). Effect of Clostridium butyricum on plasma immune function, antioxidant activity and metabolomics of weaned piglets. Livest Sci. 241:104267. doi: 10.1016/j.livsci.2020.104267
Lu, J., Yao, J., Xu, Q., Zheng, Y., and Dong, X. (2020). Clostridium butyricum relieves diarrhea by enhancing digestive function, maintaining intestinal barrier integrity, and relieving intestinal inflammation in weaned piglets. Livest Sci. 239:104112. doi: 10.1016/j.livsci.2020.104112
Ma, F., Wo, Y., Shan, Q., and Sun, P. (2020). Zinc-methionine acts as an anti-diarrheal agent by protecting the intestinal epithelial barrier in postnatal Holstein dairy calves. Anim. Feed Sci. Technol. 270:114686. doi: 10.1016/j.anifeedsci.2020.114686
McLamb, B., Gibson, A., Overman, E., Stahl, C., and Moeser, A. (2013). Early weaning stress in pigs impairs innate mucosal immune responses to enterotoxigenic E. coli challenge and exacerbates intestinal injury and clinical disease. PLoS One 8:e59838. doi: 10.1371/journal.pone.0059838
Meng, X., Wu, S., Hu, W., Zhu, Z., Yang, G., Zhang, Y., et al. (2021). Clostridium butyricum improves immune responses and remodels the intestinal microbiota of common carp (Cyprinus carpio L.). Aquaculture 530:735753. doi: 10.1016/j.aquaculture.2020.735753
Mu, C., Hao, X., Zhang, X., Zhao, J., and Zhang, J. (2021). Effects of high-concentrate diet supplemented with grape seed procyanidins on the colonic fermentation, colonic morphology, and inflammatory response in lambs. Anim. Feed Sci. Technol. 281:115118. doi: 10.1016/J.ANIFEEDSCI.2021.115118
Mu, C., Yang, W., Wang, P., Zhao, J., Hao, X., and Zhang, J. (2020). Effects of high-concentrate diet supplemented with grape seed proanthocyanidins on growth performance, liver function, meat quality, and antioxidant activity in finishing lambs. Anim. Feed Sci. Technol. 266:114518. doi: 10.1016/j.anifeedsci.2020.114518
Peng, J., Tang, Y., and Huang, Y. (2021). Gut health: the results of microbial and mucosal immune interactions in pigs. Anim. Nutr. 7, 282–294. doi: 10.1016/J.ANINU.2021.01.001
Penner, G., Steele, M., Aschenbach, J., and McBride, B. (2011). Ruminant nutrition symposium: molecular adaptation of ruminal epithelia to highly fermentable diets. J. Anim. Sci. 89, 1108–1119. doi: 10.2527/jas.2010-3378
Plaizier, J., Danesh, M., Derakhshani, H., Golder, H., Khafipour, E., Kleen, J., et al. (2018). Review: Enhancing gastrointestinal health in dairy cows. Animal 12, 399–418. doi: 10.1017/S1751731118001921
Qiao, Y., Qian, J., Lu, Q., Tian, Y., and Chen, Q. (2015). Protective effects of butyrate on intestinal ischemiae reperfusion injury in rats. J. Surg. Res. 197, 324–330. doi: 10.1016/j.jss.2015.04.031
Rakhshandeh, A., and de Lange, C. (2012). Evaluation of chronic immune system stimulation models in growing pigs. Animal 6, 305–310. doi: 10.1017/S1751731111001522
Sandeep, K., Bryan, P., Koon, H., Catherine, M., Gemma, H., Graeme, T., et al. (2018). Sharpea and Kandleria are lactic acid producing rumen bacteria that do not change their fermentation products when co-cultured with a methanogen. Anaerobe 54, 31–38. doi: 10.1016/j.anaerobe.2018.07.008
Stoeva, M., Garcia-So, J., Justice, N., Myers, J., Tyagi, S., Nemchek, M., et al. (2021). Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microb. 13, 1–28. doi: 10.1080/19490976.2021.1907272
Ulluwishewa, D., Anderson, R., McNabb, W., Moughan, P., Wells, J., and Roy, N. (2011). Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr. 141, 769–776. doi: 10.3945/jn.110.135657
Wageha, A., Claudia, H., and Michael, H. (2017). Enteric pathogens and their toxin-induced disruption of the intestinal barrier through alteration of tight junctions in chickens. Toxins 9:60. doi: 10.3390/toxins9020060
Wang, M., Li, Y., Gao, M., Song, L., Xu, M., Zhao, X., et al. (2021). Effects of subacute ruminal acidosis on colon epithelial morphological structure, permeability, and expression of key tight junction proteins in dairy goats. J. Dairy Sci. 104, 4260–4270. doi: 10.3168/jds.2020-18738
Wang, T., Fu, J., Xiao, X., Lu, Z., Wang, F., Jin, M., et al. (2021). CBP22, a novel bacteriocin isolated from Clostridium butyricum ZJU-F1, protects against LPS-induced intestinal injury through maintaining the tight junction complex. Mediators Inflamm. 2021:8032125. doi: 10.1155/2021/8032125
Wang, W., Wang, J., Zhang, H., Wu, S., and Qi, G. (2020). Effects of Clostridium butyricum on production performance and intestinal absorption function of laying hens in the late phase of production. Anim. Feed Sci. Technol. 264:114476. doi: 10.1016/j.anifeedsci.2020.114476
Xie, P., Zhou, Q., Wu, C., Zhao, J., Tan, Q., He, Y., et al. (2021). Effects of dietary supplementation with essential oils and protease on growth performance, antioxidation, inflammation and intestinal function of weaned pigs. Anim. Nutr. 9, 39–48. doi: 10.1016/J.ANINU.2021.12.003
Xie, Z., Zhao, Q., Wang, H., Wen, L., Li, W., Zhang, X., et al. (2020). Effects of antibacterial peptide combinations on growth performance, intestinal health, and immune function of broiler chickens. Poult Sci. 99, 6481–6492. doi: 10.1016/j.psj.2020.08.068
Xu, L., Sun, X., Wan, X., Li, K., Jian, F., Li, W., et al. (2021). Dietary supplementation with Clostridium butyricum improves growth performance of broilers by regulating intestinal microbiota and mucosal epithelial cells. Anim. Nutr. 7, 1105–1114. doi: 10.1016/J.ANINU.2021.01.009
Zhan, H., Dong, X., Li, L., Zheng, Y., Gong, Y., and Zou, X. (2019). Effects of dietary supplementation with Clostridium butyricum on laying performance, egg quality, serum parameters, and cecal microflora of laying hens in the late phase of production. Poultry Sci. 98, 896–903. doi: 10.3382/ps/pey436
Zhang, C., Yu, Q., Wang, J., Yu, Y., Zhang, Y., and Sun, Y. (2022). Effects of dietary supplementation with Clostridium butyricum on growth performance, apparent digestibility, blood metabolites, ruminal fermentation and bacterial communities of fattening goats. Front. Nutr. 9:888191. doi: 10.3389/fnut.2022.888191
Zhang, J., Chen, X., Liu, P., Zhao, J., Sun, J., Guan, W., et al. (2018). Dietary Clostridium butyricum induces a phased shift in fecal microbiota structure and increases the acetic acid-producing bacteria in a weaned piglet model. J. Agric. Food Chem. 66, 5157–5166. doi: 10.1021/acs.jafc.8b01253
Zhang, Q., Gu, S., Wang, Y., Hu, S., Yue, S., and Wang, C. (2022). Stereoselective metabolic disruption of cypermethrin by remolding gut homeostasis in rat. J. Environ. Sci. 126, 761–771. doi: 10.1016/J.JES.2022.03.035
Zhang, Y., Choi, S., Nogoy, K., and Liang, S. (2021). Review: the development of the gastrointestinal tract microbiota and intervention in neonatal ruminants. Animal 15:100316. doi: 10.1016/j.animal.2021.100316
Zhao, H., Jia, L., Yan, Q., Deng, Q., and Wei, B. (2020). Effect of Clostridium butyricum and butyrate on intestinal barrier functions: study of a rat model of severe acute pancreatitis with intra-abdominal hypertension. Front. Physiol. 11:561061. doi: 10.3389/fphys.2020.561061
Zhou, Z., Zhang, J., Zhang, X., Mo, S., Tan, X., Wang, L., et al. (2019). The production of short chain fatty acid and colonic development in weaning piglets. J. Anim. Physiol. Anim. Nutr. 103, 1530–1537. doi: 10.1111/jpn.13164
Keywords: Clostridium butyricum, intestine, epithelial barrier, microbiota, goats
Citation: Zhang C, Hou T, Yu Q, Wang J, Ni M, Zi Y, Xin H, Zhang Y and Sun Y (2022) Clostridium butyricum improves the intestinal health of goats by regulating the intestinal microbial community. Front. Microbiol. 13:991266. doi: 10.3389/fmicb.2022.991266
Received: 11 July 2022; Accepted: 30 August 2022;
Published: 20 September 2022.
Edited by:
Klibs N. Galvao, University of Florida, United StatesReviewed by:
Anella Saggese, University of Naples Federico II, ItalyXingning Xiao, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2022 Zhang, Hou, Yu, Wang, Ni, Zi, Xin, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonggen Zhang, emhhbmd5b25nZ2VuQG5lYXUuZWR1LmNu; Yukun Sun, c3VueXVrdW5AbmVhdS5lZHUuY24=
†These authors have contributed equally to this work and share last authorship
 Chengrui Zhang
Chengrui Zhang Tingyi Hou1
Tingyi Hou1 Hangshu Xin
Hangshu Xin Yukun Sun
Yukun Sun