
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 02 September 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.990091
Antimicrobial resistance (AMR) is a global concern threatening public health. Developing novel antibiotics is one of the effective strategies to tackle AMR. Serine/threonine kinases (STKs) have been recently shown to play critical roles in the physiology and pathogenesis of several important bacterial pathogens which are regarded as a promising antimicrobial drug target. We previously reported the roles of STK in the regulation of bacterial cell division, metabolism, and pathogenesis in Streptococcus suis, an important zoonotic bacterial pathogen. In this study, we firstly identified the Thr167 and Ser175 residues in the activation loop of S. suis STK (ssSTK) as the kinase autophosphorylation sites. Phenotyping results demonstrated that the autophosphorylation deficient strain resembled the stk deletion strain showing essentiality for bacterial growth in minimal medium, abnormal morphology, and decreased virulence when compared with the wild-type S. suis SC19 strain. Based on these findings, we established an ssSTK inhibitor screening approach by measuring the growth of S. suis in a minimal medium and testing the autophosphorylation inhibition by measuring the consumption of ATP in an enzymatic reaction by ssSTK. A series of inhibitors against ssSTK are identified from a commercial kinase inhibitors library, including Staurosporine, K252a, AT9283, and APY29. These inhibitors showed antimicrobial activity in vitro. Moreover, by using Galleria mellonella larvae infection assay, compound APY29 displayed in vivo efficacy against S. suis infection. Additionally, it was predicted by molecular docking that these inhibitors could interact with ssSTK. Collectively, our data illustrated the essential roles of ssSTK autophosphorylation in the physiology and pathogenicity of S. suis and consider these inhibitors as promising antimicrobial lead compounds.
Antimicrobial resistance (AMR) has become a serious problem that poses a huge threat to public health and animal husbandry worldwide (Samreen et al., 2021). It is listed as one of the top 10 threats to global health by the World Health Organization (WHO) in 2019, which causes at least 700,000 deaths every year (Mancuso et al., 2021). Great efforts have been made to tackle AMR. These include the implementation of administrative policies to promote the rational use of antibiotics (Gyssens, 2011; Xiao et al., 2013), development of antibiotics alternatives such as vaccines, probiotics, antimicrobial peptides, and phage therapy to reduce the use of antibiotics (Allen et al., 2014; Hoelzer et al., 2018; Browne et al., 2020). On the other hand, in the past 30 years, few novel classes of antibiotics have been developed, which is also one of the reasons leading to the accumulation of AMR (Lewis, 2020). Therefore, another important aspect to combat AMR is to accelerate the development of novel antibiotics.
Protein phosphorylation is an important posttranslational modification in prokaryotic and eukaryotic cells that participates in regulating a wide variety of cellular processes (Sun et al., 2015; Mijakovic et al., 2016; Lavoie et al., 2020). The critical role of protein phosphorylation makes protein kinases attractive drug targets (Chen et al., 2015). Until 2021, the United States Food and Drug Administration (FDA) has approved 71 small-molecule kinase inhibitors for treating a variety of diseases including tumors and non-malignancies (Roskoski, 2021). However, so far, there have been no antimicrobials developed that target bacterial kinases.
Important roles of reversible protein phosphorylation mediated by bacterial kinases and phosphatases have been noted in the past decades and several bacterial protein kinases have been proposed as potential targets for novel antibiotics (King and Blackledge, 2021). The two-component systems (TCSs) are the well-characterized protein phosphorylation systems that play important roles in sensing and responding to environmental signals (Alvarez et al., 2016). They are composed of histidine kinases (HKs) and response regulators (RRs) that are involved in the regulation of bacterial virulence, biofilm formation, and antimicrobial resistance (Zschiedrich et al., 2016; Tierney and Rather, 2019; Isaka et al., 2021). Inhibitors against the TCSs have been identified to have anti-virulence (Rasko et al., 2008) and even antimicrobial activity (Velikova et al., 2016a).
Recently, the serine/threonine kinases (STKs) have been shown as important regulators in several important bacterial pathogens, including Mycobacterium tuberculosis, Staphylococcus aureus, Listeria monocytogenes, Streptococcus pneumoniae, and Streptococcus suis (Débarbouillé et al., 2009; Osaki et al., 2009; Chawla et al., 2014; Pensinger et al., 2014; Zhu et al., 2014; Manuse et al., 2016; Nagarajan et al., 2021). STKs usually harbor an extracellular penicillin-binding protein and serine/threonine kinase associated (PASTA) domain and an intracellular catalytic kinase domain (Manuse et al., 2016). In response to stimuli, STK autophosphorylates on its activation loop of the kinase domain by hydrolyzing adenosine triphosphate (ATP) and transfers the phosphate to the serine or threonine residues of its substrate proteins (Ogawara, 2016; Labbe and Kristich, 2017; Nagarajan et al., 2021). STKs are identified as global regulators controlling a wide range of important bacterial physiological processes, including cell wall hemostasis, virulence, metabolism, and cell division in several important bacterial pathogens (Cheung and Duclos, 2012; Chawla et al., 2014; Manuse et al., 2016; Wamp et al., 2020; Djorić et al., 2021). In some bacteria, STKs are even essential or conditional essential for bacterial viability (Fernandez et al., 2006; Hu et al., 2021). Based on their critical physiological roles, several STK inhibitors have been screened to have antimicrobial activity (Fernandez et al., 2006; Wehenkel et al., 2006; Maslov et al., 2019) or as antibiotic adjuvants to potentiate β-lactam activity (Schaenzer et al., 2017, 2018).
Streptococcus suis is a zoonotic bacterial pathogen that causes lethal infections in pigs and humans (Lun et al., 2007; Tan et al., 2019). Currently, antibiotics are still the major approach to treat the disease caused by S. suis. However, it is more difficult to control S. suis infections due to the rapid emergence of AMR (Devi et al., 2017; Oh et al., 2017; Tan et al., 2021). Thus, developing novel antibiotics is of great importance in controlling S. suis infection. Streptococcus suis only harbors one copy of the gene encoding STK. Several studies have revealed that ssSTK plays an important role in the growth, metabolism, and pathogenesis of S. suis (Zhu et al., 2014; Zhang et al., 2017; Hu et al., 2021; Liu et al., 2021). More importantly, our previous study has demonstrated that disruption of stk almost completely abolished the growth of S. suis in the minimal medium (Hu et al., 2021).
In this study, we identified the autophosphorylation sites of ssSTK in its activation loop and characterized the key role of ssSTK autophosphorylation in the growth, morphology, and virulence of S. suis. Based on the essentiality of ssSTK autophosphorylation for cell growth in the minimal medium as well as by establishing an enzymatic assay to test the autophosphorylation inhibition by measuring the consumption of ATP, four compounds are identified as potent inhibitors against ssSTK by high throughput screening from a drug library with compound analogous characterization. These inhibitors showed strong inhibition against S. suis growth in the minimal medium and one of them showed in vivo antimicrobial activity in the Galleria mellonella larvae infection assay. Our study identifies effective lead compounds by screening STK inhibitors which provide good candidates for novel antimicrobials development.
All bacterial strains used in this study are listed in Supplementary Table S1. Streptococcus suis SC19 is a virulent serotype 2 strain isolated in a diseased pig during an outbreak in Sichuan Province, China, in 2005 (Lun et al., 2007). Streptococcus suis SC19 and its derived strains were grown at 37°C in tryptic soy agar (TSA) or tryptic soy broth (TSB) medium supplemented with 10% fetal bovine serum (FBS, Cat#23022-8615, Every green, Hangzhou, China) or the chemically defined medium (CDM) prepared as previously described (van de Rijn and Kessler, 1980) of which the composition is listed in Supplementary Table S2. The stk-deleted strain of S. suis SC19 (Δstk) and its complementary strain (CΔstk) were constructed in our previous study (Hu et al., 2021). The S. suis Δstk::stkT167A-S175A strain was constructed by introducing a pSET2-based plasmid into the Δstk strain to express the variant STK (T167A-S175A point mutations) in trans. Escherichia coli DH5α strain was used as the host strain for regular cloning. E. coli MC1061 strain was used as the host strain for cloning of pSET2-derived plasmids. E. coli BL21(DE3) strain was used as the host strain for pET28a-derived plasmids for protein expression and purification.
The plasmids used in this study are listed in Supplementary Table S1. Plasmid pSET2-pro-stkT167A-S175A encodes the variant ssSTK (T167A-S175A point mutations) in S. suis in which the native promoter of stk was used to drive the transcription of the variant stk genes in S. suis. The promoter region and the coding sequence of the stk containing the autophosphorylation site mutations were amplified and subsequently cloned into pSET2 vector, resulting in plasmid pSET2-pro-stkT167A-S175. The coding sequences of the kinase domain of ssSTK and its variants ssSTKKD-T167A, ssSTKKD-T169A, and ssSTKKD-S175A were amplified by overlap extension PCR. The PCR products were then cloned into the pET28a vector to generate the recombinant expression plasmids pET28a-stkKD, pET28a-stkKD-T167A, pET28a-stkKD-T169A, and pET28a-stkKD-S175A. All the primers used in this study are listed in Supplementary Table S3.
The 6 × his-tagged recombinant proteins were expressed and purified as previously described (Zhang et al., 2017). Briefly, E. coli BL21 (DE3) containing the pET28a-stkKD, pET28a-stkKD-T167A, pET28a-stkKD-T169A, or pET28a-stkKD-S175A plasmid was grown to the mid-log phase and protein expression was induced by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) followed by incubation at 18°C for 16 h. The cells were harvested and lysed by sonication. The cell lysate was subjected to centrifugation at 12,000 rpm for 10 min at 4°C to remove the unbroken cells and cell debris. The supernatant was then subjected to affinity chromatography with the Ni-NTA column (Cat# 10271899, GE Healthcare, Uppsala, Sweden). The purified protein was concentrated by ultrafiltration and stored at −80°C until use.
Galleria mellonella larvae model was used to evaluate the virulence of S. suis as previously described (Velikova et al., 2016b). A total of 50 G. mellonella larvae were randomly divided into five groups (10 per group), which were injected with 1 × 106 CFU of S. suis SC19, Δstk, CΔstk, Δstk::stkT167A-S175A, or the same volume of saline, respectively, via the left posterior proleg of G. mellonalla larvae. The survival of G. mellonella larvae was recorded at 6 h intervals for 78 h. In vivo assessment of antimicrobial activity of APY29 using G. mellonalla larvae infection model was performed as follows. 1 × 106 CFU of S. suis SC19 were injected into the left posterior proleg of G. mellonalla larvae (10 per group), followed by injection of different doses of APY29, respectively. A control group was set, in which saline was injected and without S. suis SC19 infection (Mock). The survival of G. mellonella larvae was recorded at 6 h intervals for 48 h.
A commercial kinase inhibitors library (HY-LD-000001801), containing 1,133 compounds was bought from MedChemExpress (MCE), and the detailed information is listed in Supplementary Table S5. To screen for inhibitors against ssSTK, the mid-log phase cells of S. suis SC19 and Δstk were diluted in CDM in 96-well plates containing either the library compounds or the same volume of dimethyl sulfoxide (DMSO). After 8 h of static culture at 37°C, the optical density at 600 nm (OD600) was measured using a microplate spectrophotometer (FLUOSTAR OMEGA, BMG LABTECH). Percent inhibition was calculated as (ODX – ODN)/(ODP – ODN) × 100%, where ODX is the OD600 value for a test treated with compound X, and ODP and ODN are the OD600 values for SC19 and Δstk treated with DMSO, respectively.
ssSTK phosphorylates itself by hydrolyzing ATP. Therefore, the autophosphorylation activity can be determined by measuring the consumption of ATP. The autophosphorylation assays were carried out in 50 μl of kinase reaction buffer (HEPES 50 mM, DTT 1 mM, Brij35 0.01%, pH 7) containing 10 mM MgCl2, 100 μM ATP, 10 μM ssSTK, and 0.5 μl of compound or DMSO. The mixture was incubated at 37°C for 30 min. Then, 50 μl of Kinase Glo® reagent (Cat# V3771, Promega, Madison, United States) was added to each well. After 10 min, the relative light unit (RLU) values were measured by using a microplate spectrophotometer. Percent inhibition was calculated as ((RLUX − RLUP)/(RLUN − RLUP)) × 100%, where RLUX is the RLU value for a test treated with compound X, and RLUP and RLUN are the RLU values for the reaction mixture without the treatment of compound and the reaction mixture lacking ssSTK, respectively. Additionally, the autophosphorylation activity is also determined by measuring its ssSTK autophosphorylation levels after the enzymatic reaction. The mixture (kinase buffer, 10 mM MgCl2, 50 μM ATP, and 2 μM ssSTK) was supplemented with Staurosporine, K252a, AT9283, and APY29 at 10, 50, 100 μM or DMSO, respectively. Then, these samples were incubated at 37°C for 30 min and terminated by the addition of 6 × SDS loading buffer. Subsequently, the sample was boiled for 10 min and analyzed by SDS-PAGE. The protein was transferred to the polyvinylidene difluoride (PVDF) membrane and the phosphorylated protein was probed by an anti-P-Threonine mouse monoclonal antibody (Cat# 9386S, Cell Signaling, Boston, United States) diluted at 1:2,000 followed by incubation with the HPR-conjugated anti-mouse IgG antibody (Cat# SA00001-1, Proteintech, Wuhan, China) diluted at 1:4,000 in PBS with 5% BSA as the secondary antibody. Then, the phosphorylated protein was visualized by autoradiography.
The reaction was conducted in a 50 μl mixture containing 10 μM ssSTK in the kinase reaction buffer supplemented with 0.5 μl of DMSO or Staurosporine, APY29, AT9283, and K252a with final concentrations ranging from 0 μM to 100 μM in a 96-well black plate, respectively. The enzymatic inhibition percentage was calculated as described above. The data were transformed to log scale and non-linear regression was performed with GraphPad Prism software (version 7) using the variable slope normalized model for enzyme inhibition to determine IC50.
Cells of S. suis SC19, Δstk, CΔstk, and Δstk::stkT167A-S175A were grown to the mid-log phase in TSB or CDM, respectively. The cells were then subcultured 1:100 into the corresponding medium with or without the ssSTK inhibitors in a 100-well plate. The plate was incubated at 37°C with shaking and the growth was monitored using an automatic growth curve analyzer (Oy Growth Curves Ab Ltd., Helsingfors, Finland).
Cells of S. suis were grown to the mid-log phase in the TSB medium. Subsequently, the cells were washed twice and resuspended in saline. A loop of bacterial cells was fixed on glass slides (Shitai, China) through flaming. A Gram staining kit (Cat#D008, Jiancheng, China) was utilized according to the manufacturer’s instructions. The stained samples were observed using an optical microscope (OLYMPUS).
The 3D structure of the kinase domain of ssSTK was predicted using the I-TASSER server1 (Roy et al., 2010; Yang et al., 2015; Yang and Zhang, 2015). The complex models of the kinase domain of ssSTK with inhibitors were generated using autodock4 software. The interacting models were displayed by using PyMOL (version 2.0.6.0) or Ligplus software (version 2.2; Laskowski and Swindells, 2011).
The data were commonly analyzed by a two-tailed Student’s t-test in GraphPad Prism 7 software, with a value of p < 0.05 considered to be statistically significant. The survival data were analyzed by log−rank test in GraphPad Prism 7 software, with a value of p < 0.05 considered to be statistically significant.
STK contains an activation loop where autophosphorylation occurs which promotes the exposure of the substrate binding sites (Figure 1A). Previous studies have identified the autophosphorylation sites of other bacterial STK (Durán et al., 2005; Tomono et al., 2006; Zheng et al., 2018). By homologous comparison, it was predicted that Thr167, Thr169, and Ser175 of ssSTK are the autophosphorylation sites of ssSTK (Supplementary Figure S1A), which were shown in the simulated ssSTK 3D structure (Figure 1B). To verify these sites, the kinase domain of ssSTK (ssSTKKD) and the domain containing each point mutation (ssSTKKD-T167A, ssSTKKD-T169A, and ssSTKKD-S175A) were purified, with which autophosphorylation assays were conducted (Supplementary Figure S1B). It was shown in Figure 1C and Supplementary Figure S2A that ssSTKKD-T167A and ssSTKKD-S175A displayed significantly decreased autophosphorylation, while the STKKD-T169A showed a similar level of autophosphorylation compared with ssSTKKD. The results indicate that Thr167 and Ser175 are the autophosphorylation sites of ssSTK.
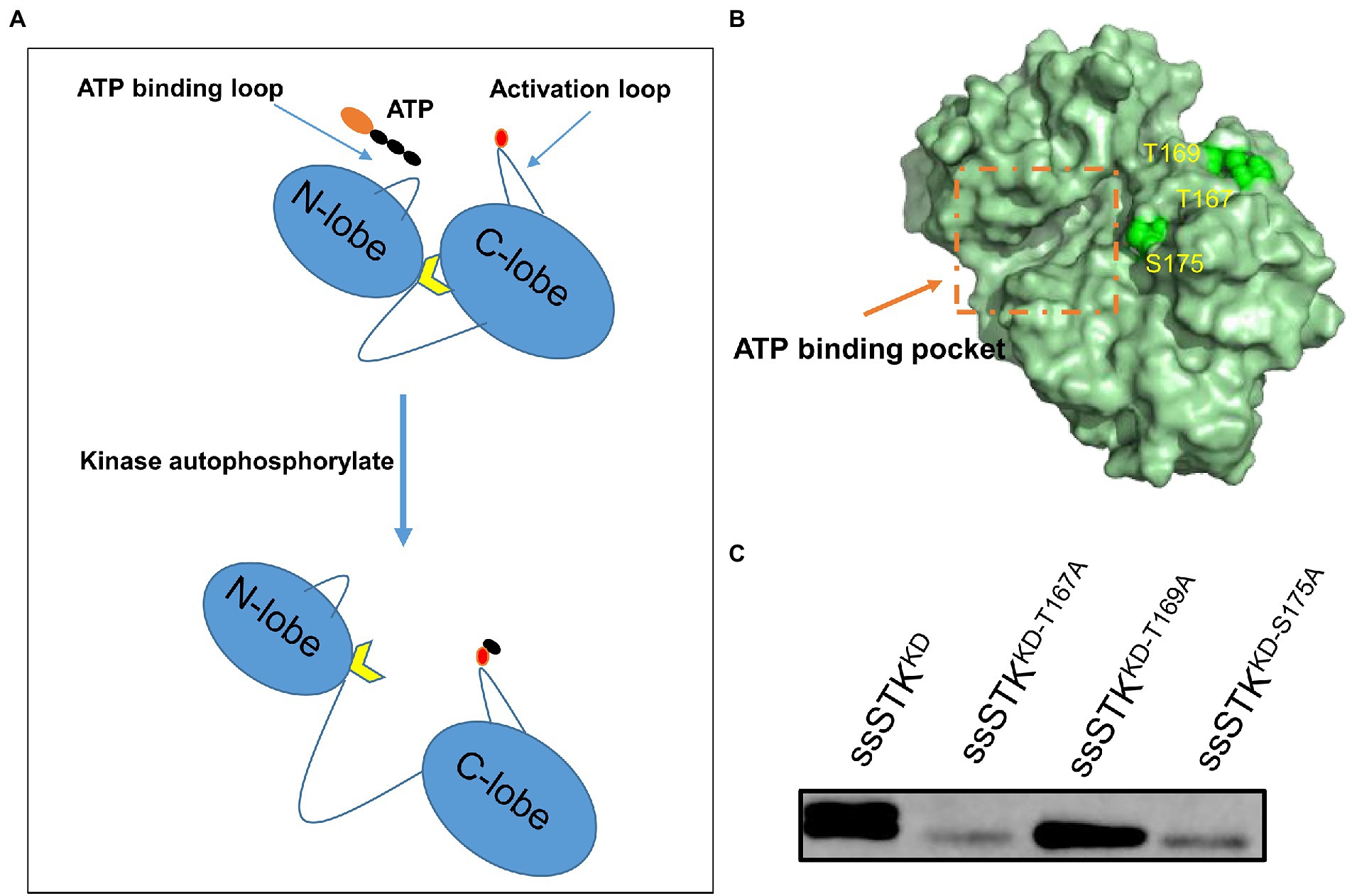
Figure 1. Identification of autophosphorylation sites of ssSTK. (A) Schematic diagram of the process of autophosphorylation of STK. The autophosphorylation sites of the kinase activation loop are phosphorylated, which exposes the substrate binding sites. Black ellipsoid denotes phosphate group; Red ellipsoid denotes the autophosphorylation sites of kinase activation loop; Yellow arrow denotes the substrate binding sites. (B) The simulated 3D structure of ssSTK. The amino acid sequence of ssSTK was analyzed using the I-TASSER server. The image was generated by PyMOL software. Green spheres represent the predicted autophosphorylation sites of ssSTK. (C) The autophosphorylation assay. Each purified protein (ssSTKKD, ssSTKKD-T167A, ssSTKKD-T169A, and ssSTKKD-S175A) was added to kinase buffer including 50 μM ATP, respectively. Then, the mixtures were incubated at 37°C for 30 min and terminated by the addition of SDS loading buffer. The autophosphorylation levels of these samples were measured by western blotting using an anti-P-Threonine mouse monoclonal antibody; The experiment was carried out two times independently under the same conditions.
Our previous studies revealed that deletion of stk in S. suis resulted in growth defects, abnormal morphology, and decreased virulence (Zhang et al., 2017). To further investigate the role of autophosphorylation of ssSTK in these processes, we constructed an S. suis strain Δstk::stkT167A-S175A which expresses ssSTK with the autophosphorylation sites substituted with alanine. By performing a growth assay, it was revealed that the growth of wild-type S. suis SC19 strain and the CΔstk, a strain expressing STK in Δstk, grew faster than Δstk and Δstk::stkT167A-S175A strains (Figure 2A). The Gram staining assay showed that SC19 and CΔstk strains exhibited a normal chain length which contained 2–5 cells per chain, while Δstk and Δstk::stkT167A-S175A cells formed significantly longer chains (Figures 2B,C). By using a G. mellonella larvae infection model, the pathogenicity of the S. suis strains was evaluated. It was shown that after infection the G. mellonella larvae infected with SC19 and CΔstk strains died rapidly. In contrast, the percentage of survived G. mellonella larvae injected with Δstk or Δstk::stkT167A-S175A was significantly higher than those injected with SC19 strain (Figure 2D). Collectively, our data suggest that ssSTK autophosphorylation plays an important role in growth, cell division, and pathogenesis of S. suis.
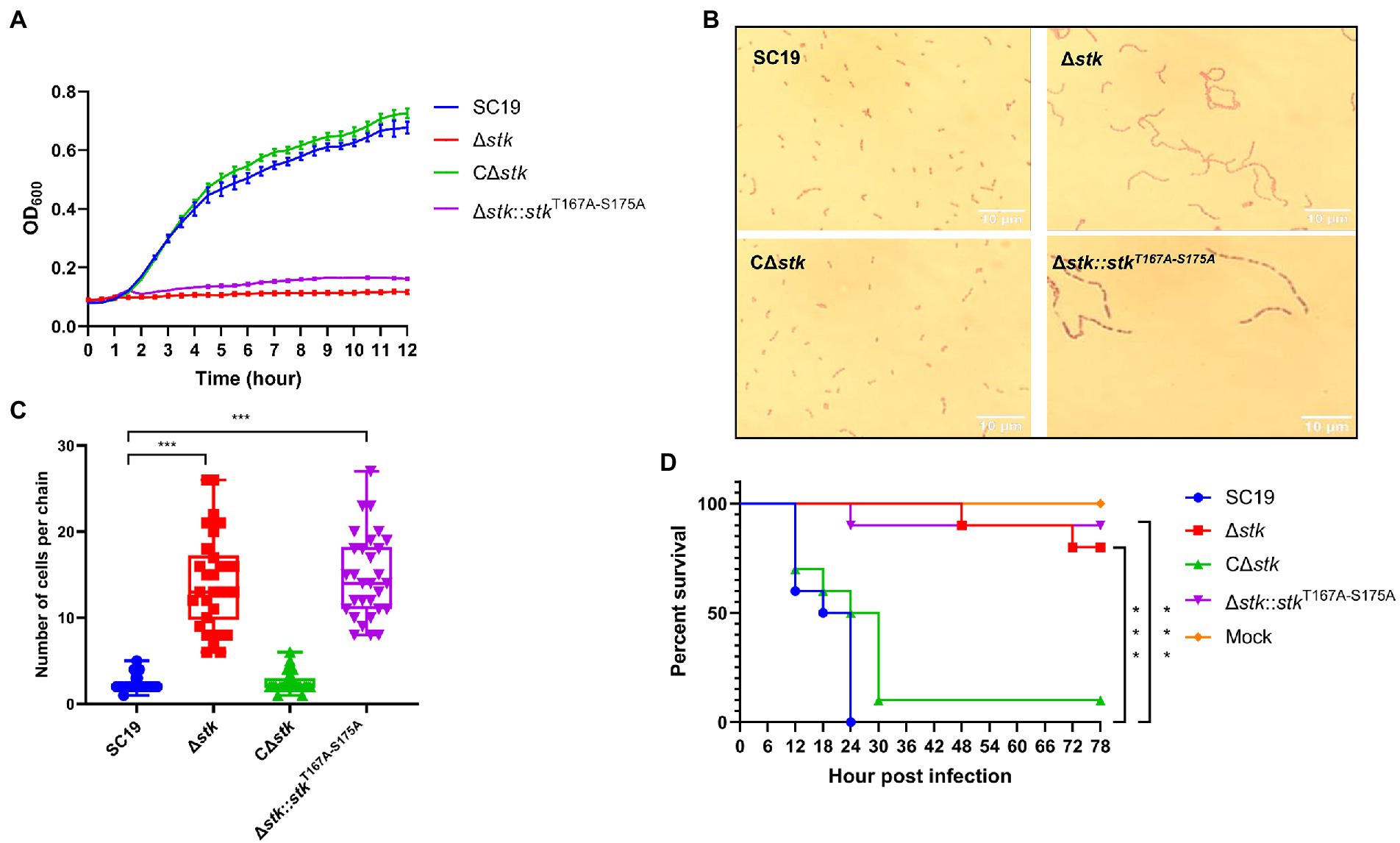
Figure 2. The role of autophosphorylation of ssSTK in the morphology, growth, and pathogenicity of Streptococcus suis. (A) Growth assay. Cells of S. suis SC19, Δstk, CΔstk, and Δstk::stkT167A-S175A were grown in the chemically defined medium (CDM) and OD600 monitored; (B) Gram staining assay. Cells of S. suis SC19, Δstk, CΔstk, and Δstk::stkT167A-S175A were grown to the mid-log phase. Then, the cells were washed twice and resuspended in normal saline. Each sample (20 μl) was fixed on glass slides and stained. The samples were then observed using an optical microscope. (C) 30 chains were randomly chosen from each Gram staining images of SC19, Δstk, CΔstk, and Δstk::stkT167A-S175A, respectively. Next, numbers of cells per chain were counted, respectively, and the data were presented as the means ± standard errors (n = 30). Student’s t-test was used to analyze their chain length difference in GraphPad Prism 7 software, with the value of p < 0.001, ***. (D) G. mellonella larvae infection assay. The G. mellonella larvae were inoculated with 1 × 106 CFU of S. suis SC19, Δstk, CΔstk, Δstk::stkT167A-S175A and the same volume saline (Mock). The survival for each group was recorded every 6 h for 78 h. The survival data were analyzed by log−rank test in GraphPad Prism 7 software, with the value of p < 0.001, ***. The experiments were carried out two times independently under the same conditions.
We next performed inhibitors screening against ssSTK. Since disruption of either the whole STK or its autophosphorylation sites results in abolished growth of S. suis in the minimal medium (Hu et al., 2021; Figure 2A). Primary screening was conducted by monitoring the growth of S. suis SC19 cells in the presence of each compound from the kinase inhibitors library in the CDM. A total of 1,133 compounds were tested among which 121 compounds showed a growth inhibition of over 80% (Figure 3A; Supplementary Table S4). However, these compounds are not necessarily targeting STK to impede the growth of S. suis. Therefore, we continued to measure the inhibitory activity of these 121 compounds against the autophosphorylation of ssSTK by quantifying ATP consumption using an in vitro enzymatic assay. Finally, three compounds Staurosporine, APY29, and AT9283 were identified as inhibitors against ssSTK, respectively (Figures 3B–D).
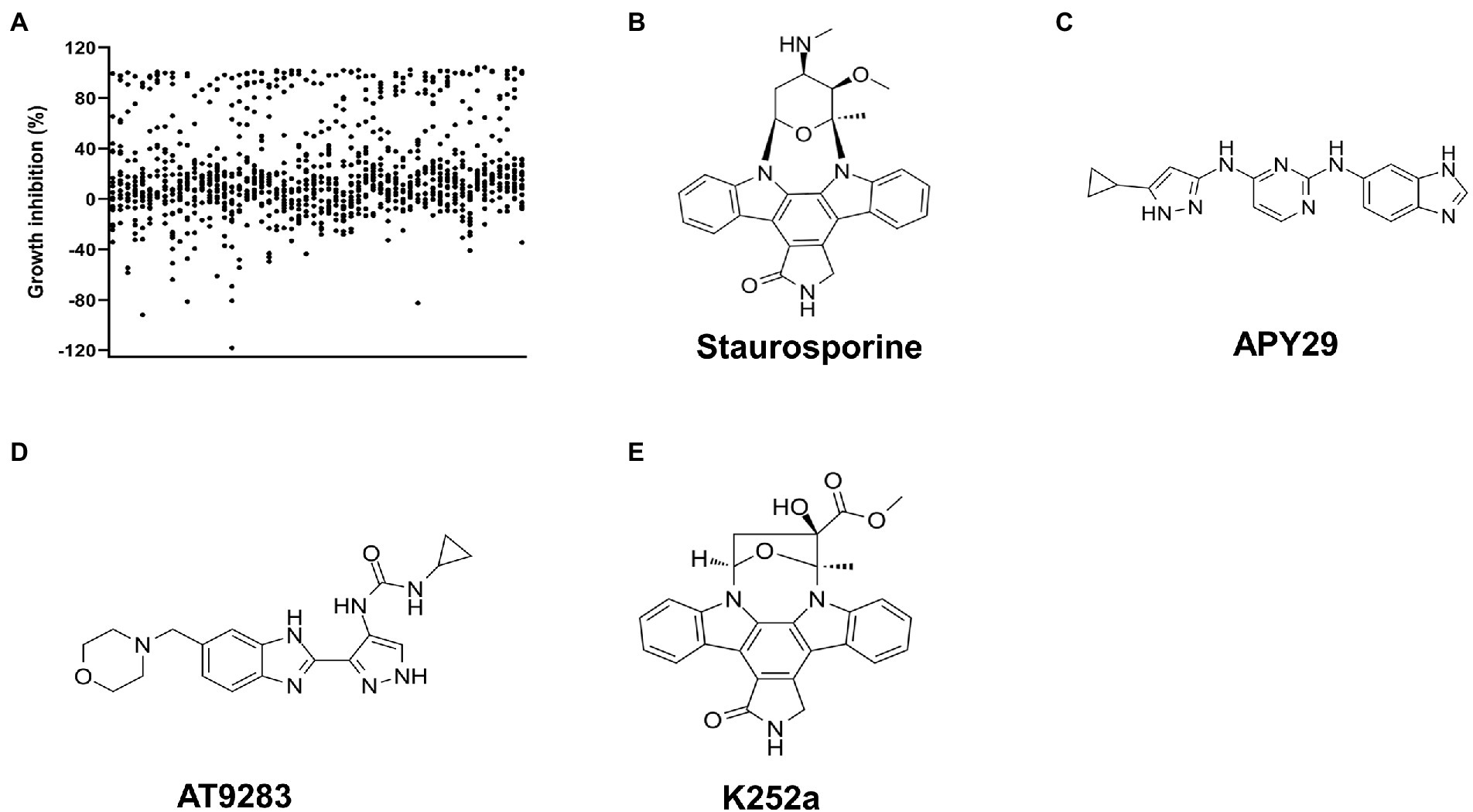
Figure 3. Screening of ssSTK inhibitors. (A) The scatter plot of the primary screening with the 1,133 compounds; (B–E) The structure of Staurosporine, APY29, AT9283, and K252a, respectively.
To further confirm the inhibitory activity of these inhibitors, in vitro autophosphorylation of ssSTK was tested in the presence of these compounds by Western blot analysis. It was shown that Staurosporine and AT9283 at 10 μM could already inhibit the autophosphorylation of ssSTK (Figures 4A,B; Supplementary Figures S2B,C). APY29 displayed a dose-dependent inhibitory effect against ssSTK (Figure 4C; Supplementary Figure S2D). These data indicate that Staurosporine, AT9283, and APY29 are potent inhibitors targeting ssSTK autophosphorylation.
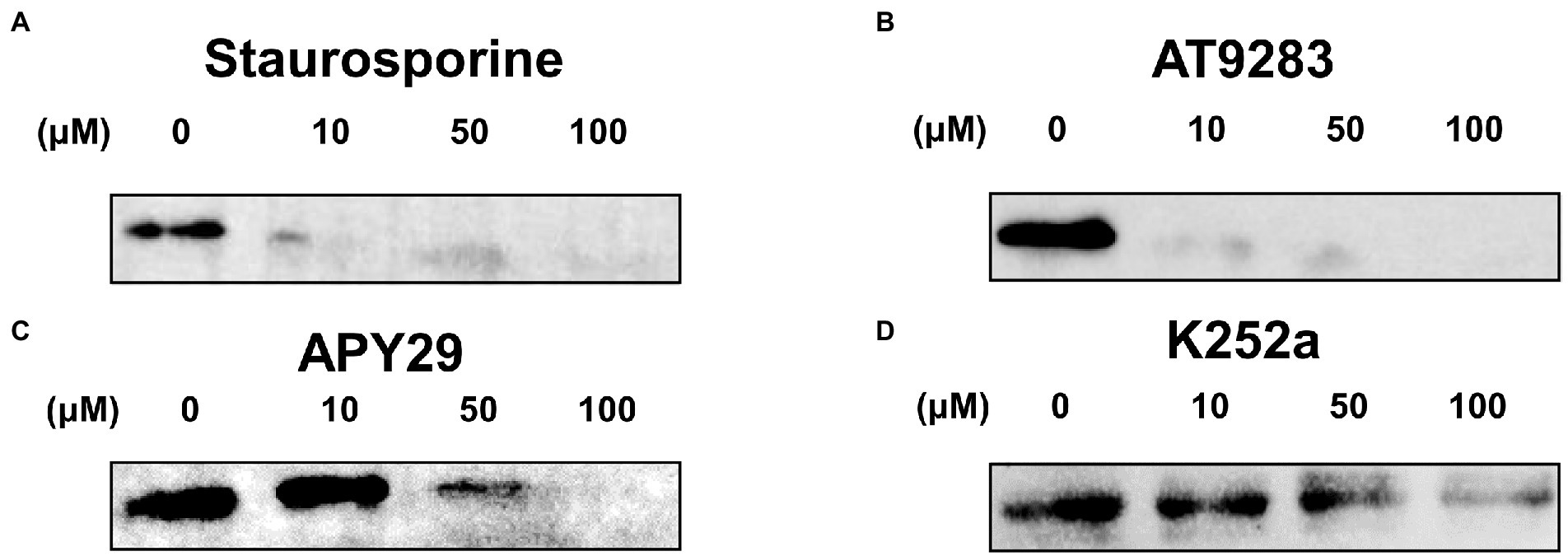
Figure 4. Autophosphorylation assay. Inhibitors including Staurosporine (A), AT9283 (B), APY29 (C), and K252a (D) at 100, 50, 10, 0 μM, and ssSTK were co-incubated at 37°C for 30 min, respectively. Next, the samples were boiled in SDS loading buffer and were detected for the autophosphorylation of ssSTK by western blotting using anti-P-Threonine mouse monoclonal antibody. The experiments were carried out two times independently under the same conditions.
In order to discover ssSTK inhibitors with higher efficacy, analogs of these inhibitors were selected to test for their inhibitory effect against ssSTK in vitro. Nine Staurosporine analogs, five analogs of APY29, and one analog of AT9283 were subjected to autophosphorylation test with ssSTK (Figure 5). It was revealed that most of these analogs except K252a (Figure 3E) exhibited an inhibition lower than 50% (Figures 5A,B). Subsequently, the in vitro autophosphorylation assay confirmed that K252a could inhibit the autophosphorylation of ssSTK (Figure 4D; Supplementary Figure S2E). Preliminary structure–activity relationship (SAR) analysis revealed the essential role of glycosidic linkage at N-12 and N-13 position of indolocarbazole in bioactivity against ssSTK. Additionally, the structural comparison of K252c with indolocarbazole showed that the pyrrole ring with carbonyl possibly facilitates the bioactivity of indolocarbazole derivatives against ssSTK. The comparison of Staurosporine and Midostaurin or 3-Hydroxy-Midostaurin or O-desmethyl-Midostaurin suggested that N-methybenzamide substitution at C-4′ position of pyran ring possibly reduced indolocarbazole derivatives potency (Figure 5A). The structure and activity comparison of VEGF2-2-IN-5 hydrochloride with APY29 highlighted the key role of benzimidazole nucleus in APY29 (Figure 5B).
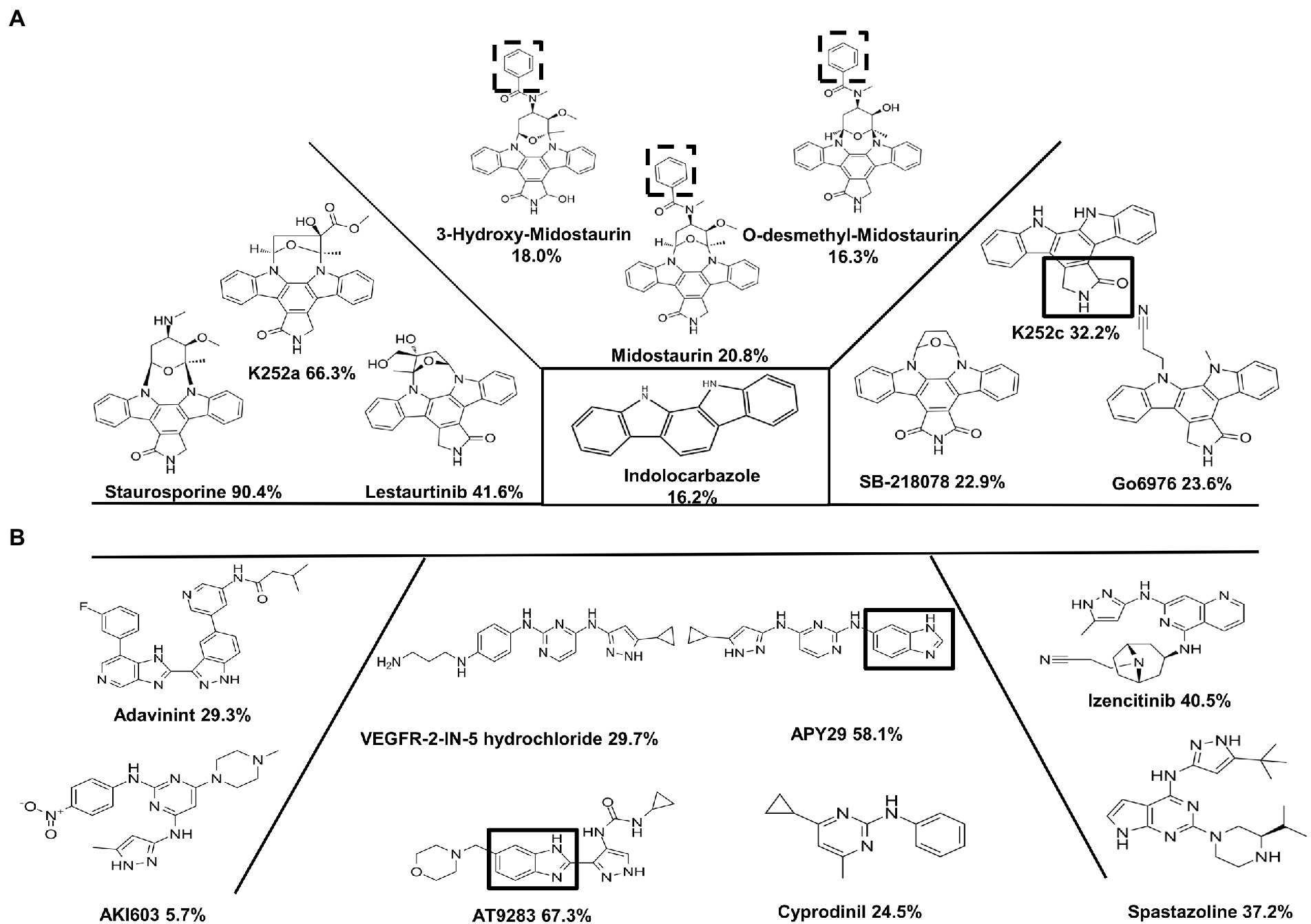
Figure 5. The analogs of Staurosporine, APY29, and AT9283. (A) The inhibition against ssSTK of Staurosporine and its analogs at 10 μM. C2-carbonyl-pyrrole ring was marked by black box. N-methybenzamide substitution at C-4′ position of pyran ring of Midostaurin, 3-Hydroxy-Midostaurin and O-desmethyl-Midostaurin were marked by black dash box. (B) The inhibition against ssSTK of APY29, AT9283, and its analogs at 10 μM. Benzimidazole nucleus was marked by black box. The experiments were carried out two times independently under the same conditions.
To further assess the kinetic data of Staurosporine, APY29, AT9283, and K252a, we determined the autophosphorylation activity of purified ssSTK in the presence of increasing concentrations of Staurosporine, APY29, AT9283, and K252a ranging from 0 to 100 μM. Enzymatic assays were conducted to determine the half maximal inhibitory concentration (IC50) of the ssSTK inhibitors. It was shown in Figure 6 that IC50 of Staurosporine, APY29, AT9283, and K252a against ssSTK were 8.966, 10.09, 10.58, and 13.81 μM, respectively. The data further verified the inhibitory activity of these compounds.
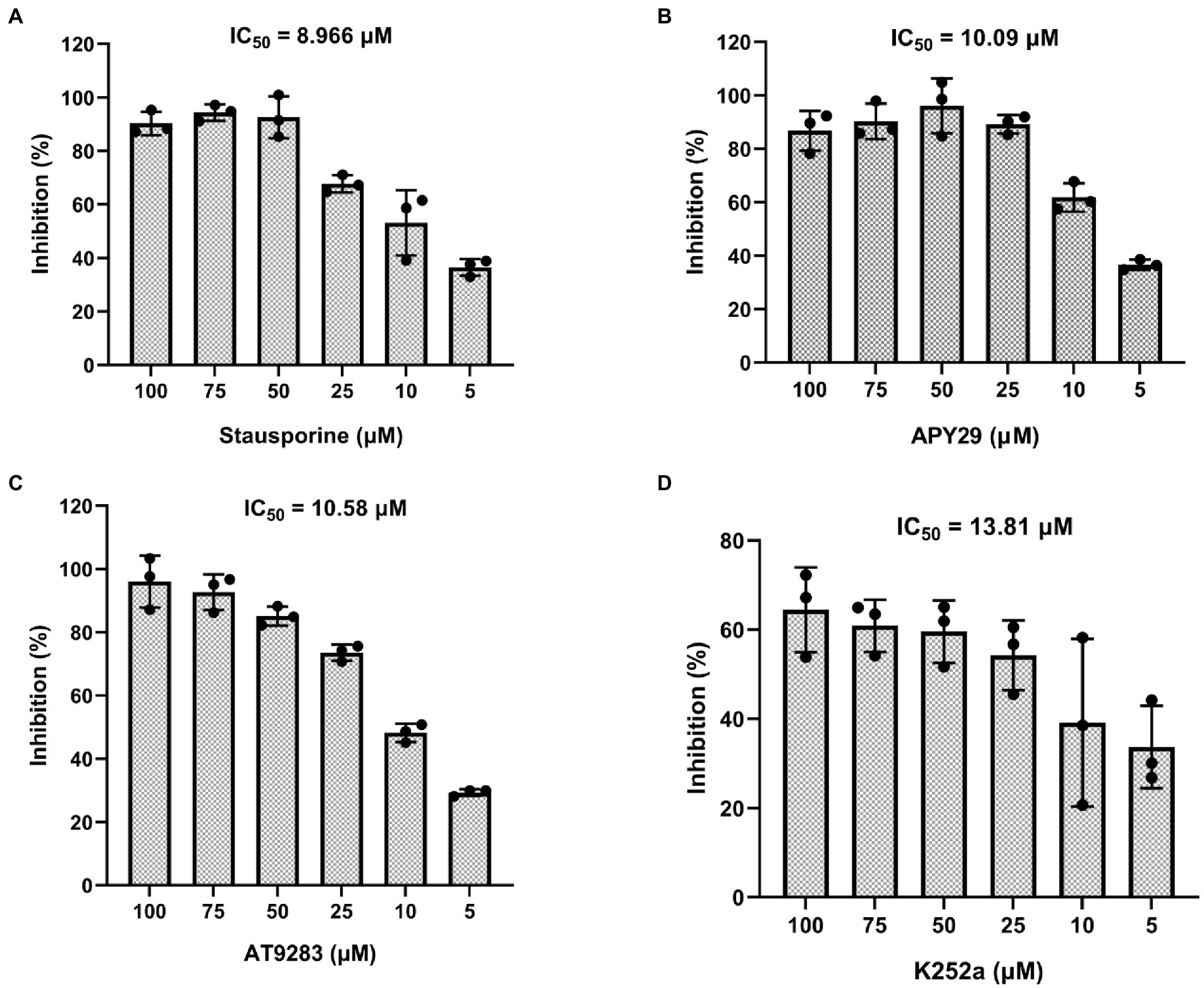
Figure 6. IC50 of inhibitors against ssSTK. Staurosporine (A), APY29 (B), AT9283 (C) and K252a (D) at 5, 10, 25, 50, 75, 100 μM and ssSTK were co-incubated at 37°C for 30 min, respectively. Next, the remaining ATP levels of the sample were measured by chemiluminescence technology to determine autophosphorylation levels of ssSTK. The experiments were carried out two times independently under the same conditions.
Staurosporine, APY29, AT9283, and K252a were subjected to test for their antimicrobial activity against S. suis by measuring the growth of S. suis in the CDM in the presence of 50 μM of each inhibitor. It was shown that the growth was significantly inhibited by the presence of the inhibitors which was similar to that of the Δstk strain (Figure 7A). Moreover, it was observed that S. suis cells showed chained morphology when treated with Staurosporine, APY29, AT9283, or K252a at 50 μM, resembling Δstk (Figures 7B,C). We further evaluated the in vivo antimicrobial efficacy of the compounds using a G. mellonella larvae infection model. It was shown that the group treated with 150 mg/kg • body weight of APY29 displayed a significantly higher survival than treated with normal saline after infection with S. suis SC19. The results indicate that APY29 had potent antimicrobial activity in vivo (Figure 7D).
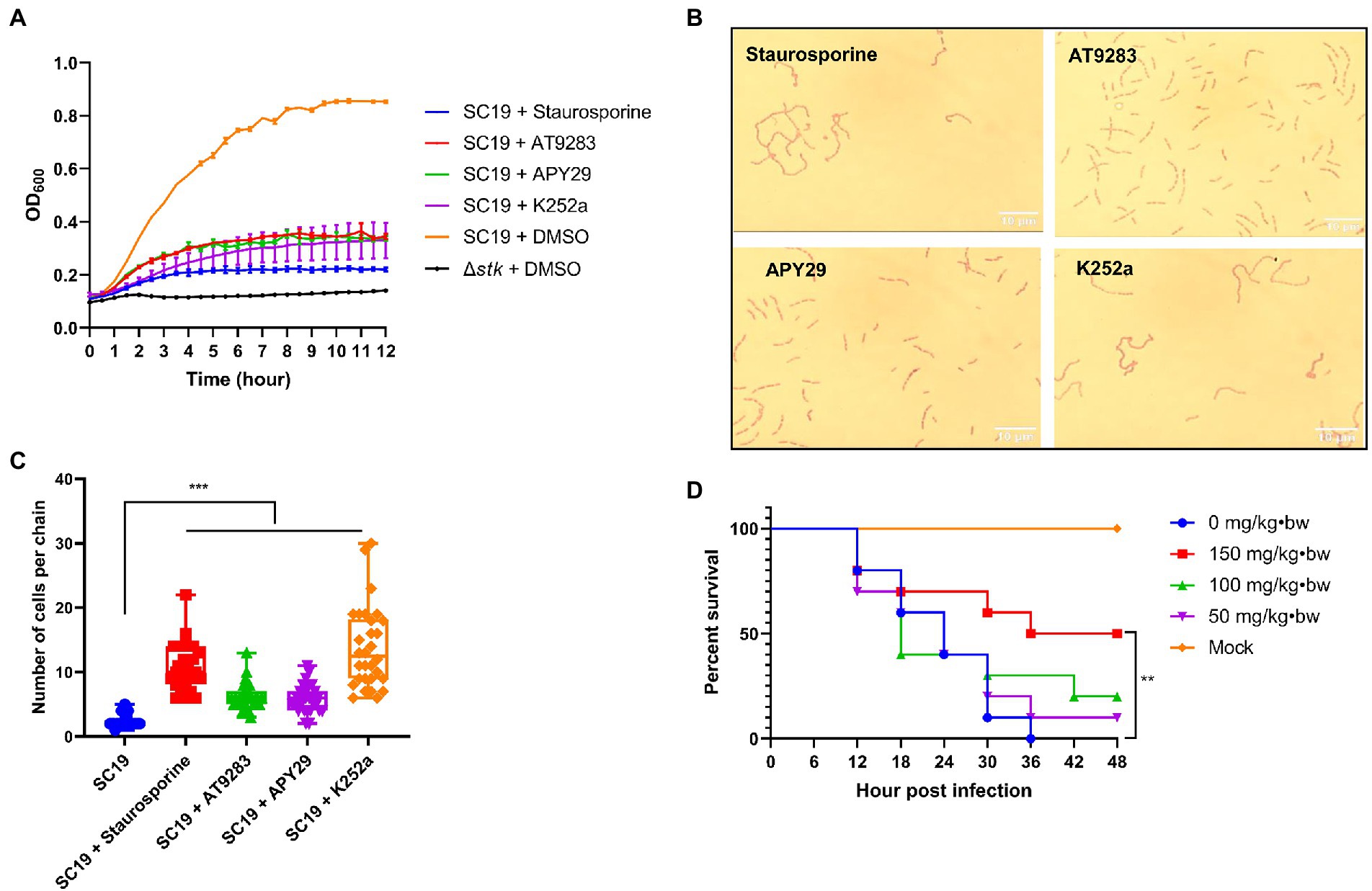
Figure 7. Characterizations of the STK inhibitors. (A) Growth assay. Cells of S. suis SC19 and Δstk were grown in the CDM in the presence and absence of the STK inhibitors, and the OD600 was measured; (B) Influence of the STK inhibitors on the morphology of S. suis. Streptococcus suis SC19 cells were co-cultured with each inhibitor in TSB medium to the log-phase. The cells were harvested, washed, and subjected to Gram staining followed by observation using an optical microscope. (C) 30 chains of were randomly chosen from each Gram staining images of SC19 treated with Staurosporine, AT9283, APY29, and K252a, respectively. Next, numbers of cells per chain were counted, respectively, and the data were presented as the means ± standard errors (n = 30). Student’s t-test was used to analyze their chain length difference in GraphPad Prism 7 software, with the value of p < 0.001, ***. (D) G. mellonella larvae infection assay. Galleria mellonella larvae were inoculated with APY29 at 150, 100, 50, and 0 mg/kg•body weight after S. suis SC19 infection. The survival was recorded every 6 h for 48 h. The survival data were analyzed by log−rank test in GraphPad Prism 7 software, with a value of p < 0.01, **. The experiments were carried out two times independently under the same conditions.
In order to investigate the binding modes between ssSTK and the inhibitors. These inhibitors were docked into the ATP-binding pocket of ssSTK, respectively. The lowest binding energy complex models suggested that these inhibitors interact with ssSTK via hydrogen bones and hydrophobic forces (Figure 8). It was suggested from the lowest binding complex models that the Val93 in the pocket of ssSTK could interact with Staurosporine, AT9283, APY29 and K252a (Figure 8), which may provide an insight into the design of ssSTK inhibitors based on the structure in the future. Additionally, it was shown in Figure 8D that the benzimidazole nucleus of APY29 interacted with ssSTK via hydrogen bond which may further explain the bioactivity difference between APY29 and VEGF2-2-IN-5 hydrochloride.
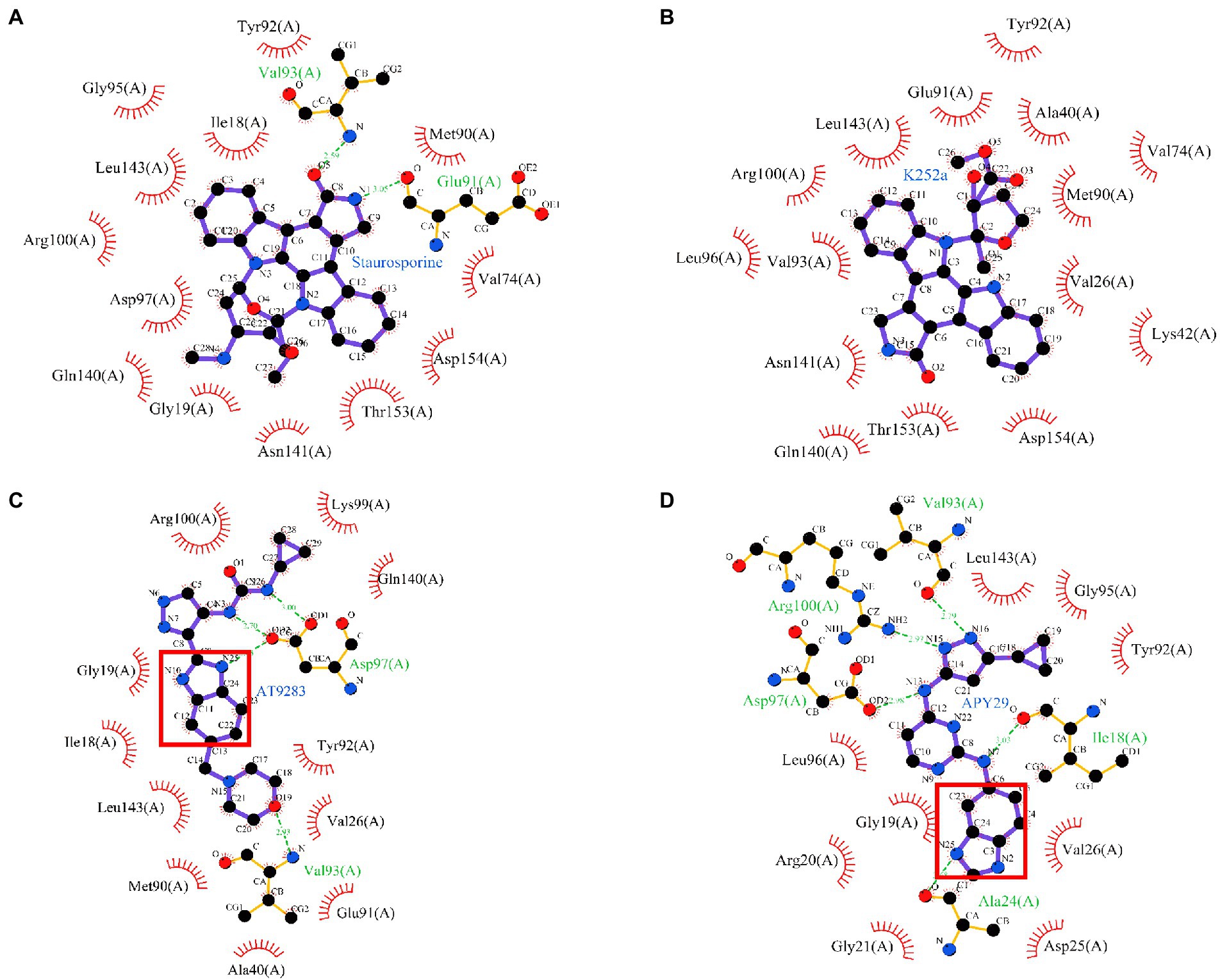
Figure 8. Molecular docking. Staurosporine (A), K252a (B), AT9283 (C), and APY29 (D) were docked into the ATP-binding pocket of ssSTK using autodock4 software, respectively. The complex models were shown using Ligplus software (version 2.2.), Benzimidazole nucleus was marked by red box.
AMR has become a huge threat to public health worldwide (Samreen et al., 2021). Urgent actions are needed to take to deal with AMR. Despite several other strategies to control AMR, innovation on novel antimicrobials has been proposed as one of the most powerful approaches (Hutchings et al., 2019; Moo et al., 2020; Miethke et al., 2021). In this study, we show that autophosphorylation of the important bacterial kinase STK is involved in the regulation of growth, morphology, and pathogenicity of S. suis. Four compounds Staurosporine, AT9283, APY29, and K252a are identified as the inhibitors against ssSTK. They demonstrated effective antimicrobial activity in the CDM in vitro, among which APY29 showed in vivo efficacy against S. suis infection using a G. mellonella larvae infection model. Our results illustrated the feasibility of bacterial kinase ssSTK as an antimicrobial target and provide a good starting point for further antimicrobial drug development.
Considering the key role of protein phosphorylation modification in cellular functions, protein kinases have been deemed as attracting drug targets (Singh et al., 2017). So far, 71 kinase inhibitors have been approved by the U.S. Food and Drug Administration (FDA; Attwood et al., 2021). Most of them are applied for cancer treatment, such as Imatinib, an early identified Bcr-Abl kinase inhibitor for the treatment of chronic myelogenous leukemia (de Kogel and Schellens, 2007). In addition, several kinase inhibitors are developed for other diseases, such as tofacitinib for the treatment of rheumatoid arthritis (Dhillon, 2017). Although, protein kinases also play important roles in bacterial physiology, unfortunately, antibiotics targeting bacterial kinases have not yet been developed.
In bacteria, two-component systems, tyrosine kinase-phosphatase systems, and serine/threonine kinase-phosphatase systems comprise the main protein phosphorylation systems (Kobir et al., 2011; Mijakovic et al., 2016). Due to their critical roles in bacterial growth, metabolism, cell division, and pathogenesis, they have been regarded not only as global regulators but also promising anti-virulence or antimicrobial targets (Hirakawa et al., 2020; Nagarajan et al., 2021). Efforts have been made to screen inhibitors against these phosphorylation systems. Compound NH125 was shown as an inhibitor against several HKs including YycG, PhoQ, and EnvZ, and displayed bactericidal activity against S. aureus, Bacillus subtilis, and S. pneumonia (Yamamoto et al., 2000, 2001). Walkmycin C was identified to inhibit autophosphorylation of VicK, CiaH, and LiaS in Streptococcus mutans (Eguchi et al., 2011). Recently, Streptozotocin and floxuridine have been identified as inhibitors against the two-component system SaeRS of S. aureus and provided protection for mice against S. aureus (Yeo et al., 2018). By targeting the protein tyrosine phosphatase, inhibitor I-A09 has been identified and shown to inhibit the growth of Mycobacterium tuberculosis in host cells (Zhou et al., 2010).
Serine/threonine kinases, another important class of bacterial kinase, have recently been demonstrated to be involved in the regulation of critical physiological processes and virulence of several important bacterial pathogens, making them attractive drug targets (Manuse et al., 2016; Djorić et al., 2021). Recently, several inhibitors against STKs have been identified. Fernandez and his colleagues have identified Staurosporine and K252a as inhibitors against PknB of M. tuberculosis by autoradiography of SDS-PAGE analysis, which prevented mycobacterial growth (Fernandez et al., 2006). They have also conducted an in silico screening to discover PknB inhibitors and Mitoxantrone was identified as an ATP-competitive PknB inhibitor with potent antimicrobial activity (Wehenkel et al., 2006). Additionally, based on the susceptibility of L. monocytogenes prkA deletion mutant strain to β-lactams antibiotics, Schaenzer et al. have performed a primary screening for ceftriaxone adjuvants followed by an enzymatic inhibition assay, and confirmed Imidazopyridine aminofurazans as a specific inhibitor showing bioactivity against PrkA of L. monocytogenes rather than Stk1 of S. aureus (Schaenzer et al., 2017). Although several inhibitors against bacterial STKs have been discovered, none of them was permitted for antimicrobial application due to their toxicity or druggable defects. In this study, we proposed a novel strategy of STK inhibitors screening based on the growth defect of S. suis stk deletion mutant in the minimal medium and enzymatic inhibition assay, and identified several inhibitors against ssSTK and S. suis SC19 strain.
To investigate the structure–activity relationships (SARs), a series of indolocarbazole derivatives were tested for their inhibitory activity against ssSTK, and only Staurosporine and K252a showed potent inhibition. The bioactivity difference of the indolocarbazole derivatives is worthy of further analysis. Previous studies on SARs of indolocarbazole derivatives against PKCs, a family of eukaryotic serine/threonine kinases, revealed the important role of glycosidic linkage at N-12 and N-13 position of indolocarbazole in the bioactivity of these compounds (Wang et al., 2020). Our data also suggested that the glycosidic linkage improves the bioactivity of indolocarbazole against ssSTK. Additionally, the structural comparison of K252c with indolocarbazole showed that the pyrrole ring possibly facilitates the inhibition activity. However, the comparison of Staurosporine and Midostaurin suggested that N-methybenzamide substitution at C-4′ position of pyran ring reduces the bioactivity of indolocarbazole derivatives. The SARs analysis could provide insights into the structural modifications of indolocarbazole derivatives against ssSTK. Moreover, our results showed that Staurosporine and K252a displayed antimicrobial efficacy in vitro. However, they also showed toxicity to host cells which may be due to a lack of kinase selectivity (Lazarovici et al., 1996). Therefore, fragment-based optimization of Staurosporine and K252a may be helpful to resolve this problem.
The benzimidazole derivatives show various biological activities due to their structural resemblance with purine (Bansal et al., 2019). However, their antimicrobial activity has rarely been studied. AT9283 and APY29 shared a benzimidazole nucleus. Previous studies have identified AT9283 and APY29 as kinase inhibitors with anticancer and anti-apoptosis activities, respectively (Korennykh et al., 2009; Santo et al., 2011; Qi et al., 2012; Gu et al., 2021). In the current study, AT9283 and APY29 were demonstrated as inhibitors against ssSTK exerting antimicrobial activity against S. suis. The structure and activity comparison of APY29 and its analogue VEGFR-2-IN-5 revealed that the bioactivity of APY29 might be partly attributed to its benzimidazole nucleus. Additionally, the molecular docking models also show that the benzimidazole nucleus of APY29 or AT9283 interacts with ssSTK via hydrogen bond. Thus, discovering ssSTK inhibitors from benzimidazole derivatives is a promising strategy. Although the primary SAR of APY29 and AT9283 was analyzed, further structural optimizations for higher activity and lower toxicity are still needed.
In conclusion, we demonstrate the feasibility of targeting the autophosphorylation ssSTK as a strategy for antimicrobial compounds screening and identified 4 compounds, Staurosporine, APY29, AT9283, and K252a, as inhibitors against ssSTK. These compounds showed potent inhibition of the growth of S. suis in the minimal medium and one of them showed in vivo efficacy against S. suis infection. Our study provides promising candidates for further antimicrobial development.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
HL and QiH: conceptualization. HL, QiaH, and LL: formal analysis. QiH and RZ: funding acquisition, project administration, supervision, and writing–review and editing. HL and TL: investigation. HL, ZY, and QiH: methodology. QiH: validation. HL: writing–original draft. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Plans of China (2018YFE0101600 and 2021YFD1800401).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.990091/full#supplementary-material
Allen, H. K., Trachsel, J., Looft, T., and Casey, T. A. (2014). Finding alternatives to antibiotics. Ann. N. Y. Acad. Sci. 1323, 91–100. doi: 10.1111/nyas.12468
Alvarez, A. F., Barba-Ostria, C., Silva-Jiménez, H., and Georgellis, D. (2016). Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ. Microbiol. 18, 3210–3226. doi: 10.1111/1462-2920.13397
Attwood, M. M., Fabbro, D., Sokolov, A. V., Knapp, S., and Schiöth, H. B. (2021). Trends in kinase drug discovery: targets, indications and inhibitor design. Nat. Rev. Drug Discov. 20, 839–861. doi: 10.1038/s41573-021-00252-y
Bansal, Y., Kaur, M., and Bansal, G. (2019). Antimicrobial potential of Benzimidazole derived molecules. Mini Rev. Med. Chem. 19, 624–646. doi: 10.2174/1389557517666171101104024
Browne, K., Chakraborty, S., Chen, R., Willcox, M. D., Black, D. S., Walsh, W. R., et al. (2020). A new era of antibiotics: the clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 21:7047. doi: 10.3390/ijms21197047
Chawla, Y., Upadhyay, S., Khan, S., Nagarajan, S. N., Forti, F., and Nandicoori, V. K. (2014). Protein kinase B (PknB) of mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J. Biol. Chem. 289, 13858–13875. doi: 10.1074/jbc.M114.563536
Chen, Q., Luo, H., Zhang, C., and Chen, Y. P. (2015). Bioinformatics in protein kinases regulatory network and drug discovery. Math. Biosci. 262, 147–156. doi: 10.1016/j.mbs.2015.01.010
Cheung, A., and Duclos, B. (2012). Stp1 and Stk 1: the yin and Yang of vancomycin sensitivity and virulence in vancomycin-intermediate Staphylococcus aureus strains. J. Infect. Dis. 205, 1625–1627. doi: 10.1093/infdis/jis255
de Kogel, C. E., and Schellens, J. H. (2007). Imatinib. Oncologist 12, 1390–1394. doi: 10.1634/theoncologist.12-12-1390
Débarbouillé, M., Dramsi, S., Dussurget, O., Nahori, M. A., Vaganay, E., Jouvion, G., et al. (2009). Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J. Bacteriol. 191, 4070–4081. doi: 10.1128/jb.01813-08
Devi, M., Dutta, J. B., Rajkhowa, S., Kalita, D., Saikia, G. K., Das, B. C., et al. (2017). Prevalence of multiple drug resistant Streptococcus suis in and around Guwahati. India. Vet World 10, 556–561. doi: 10.14202/vetworld.2017.556-561
Dhillon, S. (2017). Tofacitinib: a review in rheumatoid arthritis. Drugs 77, 1987–2001. doi: 10.1007/s40265-017-0835-9
Djorić, D., Minton, N. E., and Kristich, C. J. (2021). The enterococcal PASTA kinase: a sentinel for cell envelope stress. Mol Oral Microbiol 36, 132–144. doi: 10.1111/omi.12313
Durán, R., Villarino, A., Bellinzoni, M., Wehenkel, A., Fernandez, P., Boitel, B., et al. (2005). Conserved autophosphorylation pattern in activation loops and juxtamembrane regions of mycobacterium tuberculosis Ser/Thr protein kinases. Biochem. Biophys. Res. Commun. 333, 858–867. doi: 10.1016/j.bbrc.2005.05.173
Eguchi, Y., Kubo, N., Matsunaga, H., Igarashi, M., and Utsumi, R. (2011). Development of an antivirulence drug against Streptococcus mutans: repression of biofilm formation, acid tolerance, and competence by a histidine kinase inhibitor, walkmycin C. Antimicrob. Agents Chemother. 55, 1475–1484. doi: 10.1128/aac.01646-10
Fernandez, P., Saint-Joanis, B., Barilone, N., Jackson, M., Gicquel, B., Cole, S. T., et al. (2006). The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 188, 7778–7784. doi: 10.1128/jb.00963-06
Gu, Q., Zhu, C., Wu, X., Peng, L., Huang, G., and Hu, R. (2021). Wogonoside promotes apoptosis and ER stress in human gastric cancer cells by regulating the IRE1α pathway. Exp. Ther. Med. 21:411. doi: 10.3892/etm.2021.9842
Gyssens, I. C. (2011). Antibiotic policy. Int. J. Antimicrob. Agents 38, 11–20. doi: 10.1016/j.ijantimicag.2011.09.002
Hirakawa, H., Kurushima, J., Hashimoto, Y., and Tomita, H. (2020). Progress overview of bacterial two-component regulatory systems as potential targets for antimicrobial chemotherapy. Antibiotics 9:635. doi: 10.3390/antibiotics9100635
Hoelzer, K., Bielke, L., Blake, D. P., Cox, E., Cutting, S. M., Devriendt, B., et al. (2018). Vaccines as alternatives to antibiotics for food producing animals. Part 1: challenges and needs. Vet. Res. 49:64. doi: 10.1186/s13567-018-0560-8
Hu, Q., Yao, L., Liao, X., Zhang, L. S., Li, H. T., Li, T. T., et al. (2021). Comparative phenotypic, proteomic, and Phosphoproteomic analysis reveals different roles of serine/threonine phosphatase and kinase in the growth, cell division, and pathogenicity of Streptococcus suis. Microorganisms 9:2442. doi: 10.3390/microorganisms9122442
Hutchings, M. I., Truman, A. W., and Wilkinson, B. (2019). Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80. doi: 10.1016/j.mib.2019.10.008
Isaka, M., Okamoto, A., Miura, Y., Tatsuno, I., Maeyama, J. I., and Hasegawa, T. (2021). Streptococcus pyogenes TrxSR two-component system regulates biofilm production in acidic environments. Infect. Immun. 89:e0036021. doi: 10.1128/iai.00360-21
King, A., and Blackledge, M. S. (2021). Evaluation of small molecule kinase inhibitors as novel antimicrobial and antibiofilm agents. Chem. Biol. Drug Des. 98, 1038–1064. doi: 10.1111/cbdd.13962
Kobir, A., Shi, L., Boskovic, A., Grangeasse, C., Franjevic, D., and Mijakovic, I. (2011). Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta 1810, 989–994. doi: 10.1016/j.bbagen.2011.01.006
Korennykh, A. V., Egea, P. F., Korostelev, A. A., Finer-Moore, J., Zhang, C., Shokat, K. M., et al. (2009). The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693. doi: 10.1038/nature07661
Labbe, B. D., and Kristich, C. J. (2017). Growth- and stress-induced PASTA kinase phosphorylation in enterococcus faecalis. J. Bacteriol. 199:e00363-17. doi: 10.1128/jb.00363-17
Laskowski, R. A., and Swindells, M. B. (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786. doi: 10.1021/ci200227u
Lavoie, H., Gagnon, J., and Therrien, M. (2020). ERK signalling: a master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 21, 607–632. doi: 10.1038/s41580-020-0255-7
Lazarovici, P., Rasouly, D., Friedman, L., Tabekman, R., Ovadia, H., and Matsuda, Y. (1996). K252a and staurosporine microbial alkaloid toxins as prototype of neurotropic drugs. Adv. Exp. Med. Biol. 391, 367–377. doi: 10.1007/978-1-4613-0361-9_31
Lewis, K. (2020). The science of antibiotic discovery. Cell 181, 29–45. doi: 10.1016/j.cell.2020.02.056
Liu, H., Ye, C., Fu, H., Yue, M., Li, X., and Fang, W. (2021). Stk and Stp1 participate in Streptococcus suis serotype 2 pathogenesis by regulating capsule thickness and translocation of certain virulence factors. Microb. Pathog. 152:104607. doi: 10.1016/j.micpath.2020.104607
Lun, Z. R., Wang, Q. P., Chen, X. G., Li, A. X., and Zhu, X. Q. (2007). Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7, 201–209. doi: 10.1016/s1473-3099(07)70001-4
Mancuso, G., Midiri, A., Gerace, E., and Biondo, C. (2021). Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10:1310. doi: 10.3390/pathogens10101310
Manuse, S., Fleurie, A., Zucchini, L., Lesterlin, C., and Grangeasse, C. (2016). Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol. Rev. 40, 41–56. doi: 10.1093/femsre/fuv041
Maslov, D. A., Korotina, A. V., Shur, K. V., Vatlin, A. A., Bekker, O. B., Tolshchina, S. G., et al. (2019). Synthesis and antimycobacterial activity of imidazo [1, 2-b][1, 2, 4, 5] tetrazines. Eur. J. Med. Chem. 178, 39–47. doi: 10.1016/j.ejmech.2019.05.081
Miethke, M., Pieroni, M., Weber, T., Brönstrup, M., Hammann, P., Halby, L., et al. (2021). Towards the sustainable discovery and development of new antibiotics. Nat Rev Chem 5, 726–749. doi: 10.1038/s41570-021-00313-1
Mijakovic, I., Grangeasse, C., and Turgay, K. (2016). Exploring the diversity of protein modifications: special bacterial phosphorylation systems. FEMS Microbiol. Rev. 40, 398–417. doi: 10.1093/femsre/fuw003
Moo, C. L., Yang, S. K., Yusoff, K., Ajat, M., Thomas, W., Abushelaibi, A., et al. (2020). Mechanisms of antimicrobial resistance (AMR) and alternative approaches to overcome AMR. Curr. Drug Discov. Technol. 17, 430–447. doi: 10.2174/1570163816666190304122219
Nagarajan, S. N., Lenoir, C., and Grangeasse, C. (2021). Recent advances in bacterial signaling by serine/threonine protein kinases. Trends Microbiol. 30, 553–566. doi: 10.1016/j.tim.2021.11.005
Ogawara, H. (2016). Distribution of PASTA domains in penicillin-binding proteins and serine/threonine kinases of Actinobacteria. J. Antibiot. (Tokyo) 69, 660–685. doi: 10.1038/ja.2015.138
Oh, S. I., Jeon, A. B., Jung, B. Y., Byun, J. W., Gottschalk, M., Kim, A., et al. (2017). Capsular serotypes, virulence-associated genes and antimicrobial susceptibility of Streptococcus suis isolates from pigs in Korea. J. Vet. Med. Sci. 79, 780–787. doi: 10.1292/jvms.16-0514
Osaki, M., Arcondéguy, T., Bastide, A., Touriol, C., Prats, H., and Trombe, M. C. (2009). The StkP/PhpP signaling couple in Streptococcus pneumoniae: cellular organization and physiological characterization. J. Bacteriol. 191, 4943–4950. doi: 10.1128/jb.00196-09
Pensinger, D. A., Aliota, M. T., Schaenzer, A. J., Boldon, K. M., Ansari, I. U., Vincent, W. J., et al. (2014). Selective pharmacologic inhibition of a PASTA kinase increases Listeria monocytogenes susceptibility to β-lactam antibiotics. Antimicrob. Agents Chemother. 58, 4486–4494. doi: 10.1128/aac.02396-14
Qi, W., Liu, X., Cooke, L. S., Persky, D. O., Miller, T. P., Squires, M., et al. (2012). AT 9283, a novel aurora kinase inhibitor, suppresses tumor growth in aggressive B-cell lymphomas. Int. J. Cancer 130, 2997–3005. doi: 10.1002/ijc.26324
Rasko, D. A., Moreira, C. G., Li de, R., Reading, N. C., Ritchie, J. M., Waldor, M. K., et al. (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080. doi: 10.1126/science.1160354
Roskoski, R. Jr. (2021). Properties of FDA-approved small molecule protein kinase inhibitors: a 2021 update. Pharmacol. Res. 165:105463. doi: 10.1016/j.phrs.2021.105463
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Samreen,, Ahmad, I., Malak, H. A., and Abulreesh, H. H. (2021). Environmental antimicrobial resistance and its drivers: a potential threat to public health. J Glob Antimicrob Resist 27, 101–111. doi: 10.1016/j.jgar.2021.08.001
Santo, L., Hideshima, T., Cirstea, D., Bandi, M., Nelson, E. A., Gorgun, G., et al. (2011). Antimyeloma activity of a multitargeted kinase inhibitor, AT9283, via potent Aurora kinase and STAT3 inhibition either alone or in combination with lenalidomide. Clin. Cancer Res. 17, 3259–3271. doi: 10.1158/1078-0432.Ccr-10-3012
Schaenzer, A. J., Wlodarchak, N., Drewry, D. H., Zuercher, W. J., Rose, W. E., Ferrer, C. A., et al. (2018). GW779439X and its Pyrazolopyridazine derivatives inhibit the serine/threonine kinase Stk 1 and act As antibiotic adjuvants against β-lactam-resistant Staphylococcus aureus. ACS Infect Dis 4, 1508–1518. doi: 10.1021/acsinfecdis.8b00136
Schaenzer, A. J., Wlodarchak, N., Drewry, D. H., Zuercher, W. J., Rose, W. E., Striker, R., et al. (2017). A screen for kinase inhibitors identifies antimicrobial imidazopyridine aminofurazans as specific inhibitors of the listeria monocytogenes PASTA kinase PrkA. J. Biol. Chem. 292, 17037–17045. doi: 10.1074/jbc.M117.808600
Singh, V., Ram, M., Kumar, R., Prasad, R., Roy, B. K., and Singh, K. K. (2017). Phosphorylation: implications in cancer. Protein J. 36, 1–6. doi: 10.1007/s10930-017-9696-z
Sun, Y., Liu, W. Z., Liu, T., Feng, X., Yang, N., and Zhou, H. F. (2015). Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. Res. 35, 600–604. doi: 10.3109/10799893.2015.1030412
Tan, M. F., Tan, J., Zeng, Y. B., Li, H. Q., Yang, Q., and Zhou, R. (2021). Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province. China. J Appl Microbiol 130, 797–806. doi: 10.1111/jam.14831
Tan, C., Zhang, A., Chen, H., and Zhou, R. (2019). Recent proceedings on prevalence and pathogenesis of Streptococcus suis. Curr. Issues Mol. Biol. 32, 473–520. doi: 10.21775/cimb.032.473
Tierney, A. R., and Rather, P. N. (2019). Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 14, 533–552. doi: 10.2217/fmb-2019-0002
Tomono, A., Mashiko, M., Shimazu, T., Inoue, H., Nagasawa, H., Yoshida, M., et al. (2006). Self-activation of serine/threonine kinase AfsK on autophosphorylation at threonine-168. J. Antibiot. (Tokyo) 59, 117–123. doi: 10.1038/ja.2006.18
van de Rijn, I., and Kessler, R. E. (1980). Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27, 444–448. doi: 10.1128/iai.27.2.444-448.1980
Velikova, N., Fulle, S., Manso, A. S., Mechkarska, M., Finn, P., Conlon, J. M., et al. (2016a). Putative histidine kinase inhibitors with antibacterial effect against multi-drug resistant clinical isolates identified by in vitro and in silico screens. Sci. Rep. 6:26085. doi: 10.1038/srep26085
Velikova, N., Kavanagh, K., and Wells, J. M. (2016b). Evaluation of galleria mellonella larvae for studying the virulence of Streptococcus suis. BMC Microbiol. 16:291. doi: 10.1186/s12866-016-0905-2
Wamp, S., Rutter, Z. J., Rismondo, J., Jennings, C. E., Möller, L., Lewis, R. J., et al. (2020). PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of reoM. elife 9:e56048. doi: 10.7554/eLife.56048
Wang, J., Jin, W., Zhou, X., Li, J., Xu, C., Ma, Z., et al. (2020). Identification, structure-activity relationships of marine-derived Indolocarbazoles, and a dual PKCθ/δ inhibitor with potent Antipancreatic cancer efficacy. J. Med. Chem. 63, 12978–12991. doi: 10.1021/acs.jmedchem.0c01271
Wehenkel, A., Fernandez, P., Bellinzoni, M., Catherinot, V., Barilone, N., Labesse, G., et al. (2006). The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria. FEBS Lett. 580, 3018–3022. doi: 10.1016/j.febslet.2006.04.046
Xiao, Y., Zhang, J., Zheng, B., Zhao, L., Li, S., and Li, L. (2013). Changes in Chinese policies to promote the rational use of antibiotics. PLoS Med. 10:e1001556. doi: 10.1371/journal.pmed.1001556
Yamamoto, K., Kitayama, T., Ishida, N., Watanabe, T., Tanabe, H., Takatani, M., et al. (2000). Identification and characterization of a potent antibacterial agent, NH125 against drug-resistant bacteria. Biosci. Biotechnol. Biochem. 64, 919–923. doi: 10.1271/bbb.64.919
Yamamoto, K., Kitayama, T., Minagawa, S., Watanabe, T., Sawada, S., Okamoto, T., et al. (2001). Antibacterial agents that inhibit histidine protein kinase YycG of Bacillus subtilis. Biosci. Biotechnol. Biochem. 65, 2306–2310. doi: 10.1271/bbb.65.2306
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Yang, J., and Zhang, Y. (2015). I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181. doi: 10.1093/nar/gkv342
Yeo, W. S., Arya, R., Kim, K. K., Jeong, H., Cho, K. H., and Bae, T. (2018). The FDA-approved anti-cancer drugs, streptozotocin and floxuridine, reduce the virulence of Staphylococcus aureus. Sci. Rep. 8:2521. doi: 10.1038/s41598-018-20617-5
Zhang, C., Sun, W., Tan, M., Dong, M., Liu, W., Gao, T., et al. (2017). The eukaryote-Like serine/threonine kinase STK regulates the growth and metabolism of zoonotic Streptococcus suis. Front. Cell. Infect. Microbiol. 7:66. doi: 10.3389/fcimb.2017.00066
Zheng, W., Cai, X., Li, S., and Li, Z. (2018). Autophosphorylation mechanism of the Ser/Thr kinase Stk 1 from Staphylococcus aureus. Front. Microbiol. 9:758. doi: 10.3389/fmicb.2018.00758
Zhou, B., He, Y., Zhang, X., Xu, J., Luo, Y., Wang, Y., et al. (2010). Targeting mycobacterium protein tyrosine phosphatase B for antituberculosis agents. Proc. Natl. Acad. Sci. U. S. A. 107, 4573–4578. doi: 10.1073/pnas.0909133107
Zhu, H., Zhou, J., Ni, Y., Yu, Z., Mao, A., Hu, Y., et al. (2014). Contribution of eukaryotic-type serine/threonine kinase to stress response and virulence of Streptococcus suis. PLoS One 9:e91971. doi: 10.1371/journal.pone.0091971
Keywords: serine/threonine kinase, antimicrobial, drug target, autophosphorylation, inhibitor, Streptococcus suis
Citation: Li H, Li T, Hu Q, Yao Z, Li L, Huang Q and Zhou R (2022) Inhibitors targeting the autophosphorylation of serine/threonine kinase of Streptococcus suis show potent antimicrobial activity. Front. Microbiol. 13:990091. doi: 10.3389/fmicb.2022.990091
Received: 09 July 2022; Accepted: 09 August 2022;
Published: 02 September 2022.
Edited by:
Ramandeep Singh, Translational Health Science and Technology Institute (THSTI), IndiaReviewed by:
Andaleeb Sajid, National Cancer Institute (NIH), United StatesCopyright © 2022 Li, Li, Hu, Yao, Li, Huang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Huang, cWh1YW5nQG1haWwuaHphdS5lZHUuY24=; Rui Zhou, cnpob3VAbWFpbC5oemF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.