
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 September 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.989259
This article is part of the Research Topic Contribution of Omics to our Understanding of Human-Bacterial Interplay in Health and Disease View all 6 articles
 Ying Zhang1,2,3,4
Ying Zhang1,2,3,4 Danhua Li5
Danhua Li5 Qun Yan6
Qun Yan6 Ping Xu7,8
Ping Xu7,8 Wei Chen9
Wei Chen9 Hongya Xin1,2,3,4
Hongya Xin1,2,3,4 Dengshu Wu1,2,3,4
Dengshu Wu1,2,3,4 Mingxiang Zhou6
Mingxiang Zhou6 Yajing Xu1,2,3,4
Yajing Xu1,2,3,4 Ao Zhang1,2,3,4
Ao Zhang1,2,3,4 Wenjia Wei1,2,3,4
Wenjia Wei1,2,3,4 Zhiping Jiang1,2,3,4*
Zhiping Jiang1,2,3,4*Stenotrophomonas acidaminiphila, the most recent reported species in genus Stenotrophomonas, is a relatively rare bacteria and is an aerobic, glucose non-fermentative, Gram-negative bacterium. However, little information of S. acidaminiphila is known to cause human infections. In this research, we firstly reported a multidrug-resistant strain S. acidaminiphila SINDOREI isolated from the blood of a patient with sepsis, who was dead of infection eventually. The whole genome of strain SINDOREI was sequenced, and genome comparisons were performed among six closely related S. acidaminiphila strains. The core genes (2,506 genes) and strain-specific genes were identified, respectively, to know about the strain-level diversity in six S. acidaminiphila stains. The presence of a unique gene (narG) and essential genes involved in biofilm formation in strain SINDOREI are important for the pathogenesis of infections. Strain SINDOREI was resistant to trimethoprim/sulfamethoxazole, ciprofloxacin, ofloxacin, cefepime, ceftazidime, and aztreonam. Several common and specific antibiotic resistance genes were identified in strain SINDOREI. The presence of two sul genes and exclusive determinants GES-1, aadA3, qacL, and cmlA5 is responsible for the resistance to multidrug. The virulence factors and resistance determinants can show the relationship between the phenotype and genotype and afford potential therapeutic strategies for infections.
The genus Stenotrophomonas, which was first described with the type species Stenotrophomonas maltophilia (Palleroni and Bradbury, 1993), currently comprises 16 validly described species (Wang et al., 2018). Members of the genus Stenotrophomonas demonstrated great metabolic versatility and intraspecific heterogeneity (Ryan et al., 2009). The most recently reported species, Stenotrophomonas acidaminiphila, is an aerobic, glucose non-fermentative, Gram-negative bacterium, which is initially isolated from a petrochemical wastewater treated by an upflow anaerobic sludge blanket (UASB) reactor (Assih et al., 2002), and strains of S. acidaminiphila occur ubiquitously in the environment (Vinuesa and Ochoa-Sánchez, 2015; Patil et al., 2016; Huang et al., 2018; Huyan et al., 2020). However, the information on the characteristic of strains is still limited. To date, the reported strains of S. acidaminiphila were limited, especially clinical isolates. Additionally, the further genomic analysis was necessary to dig the features of S. acidaminiphila.
Stenotrophomonas species, especially S. maltophilia and S. acidaminiphila, can cause human infections (Looney et al., 2009; Huang et al., 2018). Multidrug-resistant (MDR) strains of Stenotrophomonas are associated with a high rate of mortality in immunocompromised patients (Paez and Costa, 2008). In the genomes of Stenotrophomonas, various genes encoding virulence determinants are involved in the infections (Trifonova and Strateva, 2019). Surveillance of the presence of virulence genes is important to supplement knowledge about the pathogenesis of infections (Windhorst et al., 2002; de Oliveira-Garcia et al., 2003).
Trimethoprim/sulfamethoxazole (TMP/SMX) was considered as the first-line therapy (Abbott and Peleg, 2015; Kumar et al., 2020), but was plagued by increasing resistance worldwide (Al-Jasser, 2006; Tan et al., 2008; Looney et al., 2009). Fluoroquinolones and β-lactam drugs have been used as potential alternative antibiotics to TMP/SMX for Stenotrophomonas infections. However, recent studies have revealed a trend in decreasing susceptibility (Wei et al., 2016; Ko et al., 2019). The strains’ intrinsic and acquired mechanisms of antibiotic resistance to most antibiotics limited the antimicrobial options for Stenotrophomonas infections.
The molecular mechanisms involved in its extensive antimicrobial resistance include efflux pumps and encoded genes. The sul1 gene, sul2 gene and dfrA gene are well known to be responsible for resistance to TMP/SMX (Barbolla et al., 2004; Domínguez et al., 2019). Two chromosomal-mediated β-lactamases, namely L1 and L2, with several regulatory genes, such as ampR, ampN and ampG, are associated with β-lactam resistance (Okazaki and Avison, 2008; Huang et al., 2010; Lin et al., 2011). A chromosomally encoded qnr gene protects both gyrase and topoisomerase IV from quinolones and confers resistance to fluoroquinolone (Ko et al., 2019). Moreover, efflux pumps are shown to be associated with resistance to multidrug and are classified to five families, namely the resistance-nodulation-cell-division (RND) family, the major facilitator superfamilies (MFS), the small multidrug resistance (SMR) family, the ATP binding cassette (ABC) family, and the multidrug and toxic compound extrusion (MATE) family (Putman et al., 2000).
To our best knowledge, S. acidaminiphila was mostly isolated from aquatic environments. It is worth noting that the first clinical isolate S. acidaminiphila SUNEO (Huang et al., 2018) was isolated from the bile of a cholangiocarcinoma patient with obstructive jaundice and cholangitis and was found resistant to sulfamethoxazole and imipenem based on the antimicrobial susceptibility testing. Meanwhile, the comparisons and analysis of whole genomes aid the identification of resistant determinants to develop the antimicrobial strategies.
In this study, the first MDR clinical isolate, S. acidaminiphila SINDOREI, was cultured and isolated from the blood of a patient with sepsis in China. The complete genome of SINDOREI was assembled to highlight the virulence factors and resistant genes characterizing the specific isolates. Additionally, we profiled the adaptive changes in S. acidaminiphila SINDORI through the characteristic of genome and genomic comparisons of six S. acidaminiphila strains.
Pure strain SINDOREI was cultured from the blood of a patient with sepsis. The 51-year-old man was transferred to Xiangya Hospital of Central South University (Changsha, China) on March 16, 2020, complaining of intermittent fever and full-body pain for more than 1 month without obvious causes. Based on physical examination, laboratory test results and other related examinations including bone marrow (BM) aspirate smear, flow cytometry and RT-PCR, etc., a diagnosis of sepsis was made. The antibiotic treatment did not improve his condition. The patient had a continuous high fever (39.5°C) with some new symptoms such as chills, high fever, appetite, fatigue, and shortness of breath and eventually died for multiple organ failure. The detailed medical record of the patient was shown in Supplementary Table S1. The blood samples of this patient were inoculated on nutrient agar with 5% sheep blood and incubated aerobically at 37°C overnight for three times. Only smooth, opaque and yellow colonies showing clear zones were isolated. Then, the isolates were conducted species identification by using MALDI Biotyper (Bruker, Germany) and returned no match in the database. At last, the purified isolate was further performed the whole genome sequencing and was classified as S. acidaminiphila.
Antimicrobial susceptibility testing was performed by VITEK2 system (bioMerieux, France) for minimum inhibitory concentration (MIC) according to the manufacturer’s instructions. MIC breakpoint was determined referring to the clinical and laboratory standards institute (CLSI) guidelines (M100, 30th Ed.; Wayne, 2020).
The phylogenetic tree based on the 16S rRNA sequence which recovered from the genome of SINDOREI were constructed using MEGA version 11 software using maximum-likelihood method with 1,000 bootstrap replications (Tamura et al., 2021). The phylogenomic analysis based on whole genomes of the members of genus Stenotrophomonas were performed using the genome taxonomic database toolkit (GTDB-Tk; Chaumeil et al., 2019) and iqtree (Nguyen et al., 2015).
The genomic DNA of S. acidaminiphila SINDOREI was extracted using cetyltrimethylammonium bromide (CTAB)-based methods. Then genomic DNA was randomly fragmented and was selected to average size of 200–400 bp. Adaptors were ligated to the ends of fragments by PCR assay. PCR products were processed into the sequencing library. The qualified BGI-seq libraries were sequenced on a BGISEQ-500 platform with read length of PE100. The Nanopore libraries were prepared and sequenced according to the manufacturer’s instructions (Oxford Nanopore, Oxford, UK) and sequenced on MinION with flowcell version of R9.4.1. The short reads and nanopore long reads were assembled using software Unicycler (Wick et al., 2017). The genes were predicted using Glimmer software (Delcher et al., 2007) and were functionally annotated by querying eggNOG, NR, Pfam and Swiss-Prot databases to obtain the corresponding annotations.
Firstly, the genomes were broken into 500,000 reads with length of 150 bp using wgsim (v1.11)1 with parameters of (-e 0-r 0-R 0 -X 0). Then, these reads were mapped to SINDOREI genome using bwa (0.7.17-r1188)2 with default parameters. Next, the SNPs were identified by bcftools3 and variations with quality <20 were filtered.
Average nucleotide identity (ANI) values between any two genomes were calculated using fastANI (Jain et al., 2018). The digital DNA–DNA hybridization (dDDH) values were obtained by means of genome-to-genome distance calculator via GGDC 3.0 using Formula 2.
For genome comparisons, six genomes of the S. acidaminiphila strains, including a type strain and other five cultured stains, were downloaded from the NCBI database (Table 1). The clusters of homologous genes among the investigated genome sequences were determined using OrthoMCL (Fischer et al., 2011). The numbers of unique core genes and strain-specific genes of all isolates were mapped. The strain-specific genes that are present in strain SINDOREI were annotated through the clusters of orthologous genes (COG) database using software EggNOG-mapper v2 (Cantalapiedra et al., 2021).
To analyze the virulence factors of the members of species S. acidaminiphila, virulence factors database (VFDB) was used (Liu et al., 2022). The protein-coding sequences were aligned against the comprehensive antibiotic resistance database (CARD; Alcock et al., 2020) and resistance-related genes were analyzed. Comparisons of specific genes related to resistance genes and efflux pumps between S. acidaminiphila strains were proceeded using local BLASTP alignment.
The phylogenetic tree based on the amino acid of sul genes was also constructed by MEGA version 11 software, using maximum-likelihood method with 1,000 bootstrap replications (Tamura et al., 2021). The amino acid sequences of dihydropteroate synthase from Stenotrophomonas strain were downloaded from the National Center for Biotechnology Information.
The complete genome of SINDOREI was assembled into one circular chromosome with a total size of 3,996,619 bp and a GC content of 68.73%. SINDOREI genome contained no plasmid, and the details of the genome were listed in Supplementary Tables S2, S3. The genome of SINDOREI were functionally annotated in databases: eggNOG (Supplementary Table S4), NR (Supplementary Table S5), Pfam (Supplementary Table S6) and Swiss-Prot (Supplementary Table S7). A total of 3,623 genes, including 3,536 coding sequences (CDSs), were predicted in the genome, along with 68 transfer RNA (tRNA genes) and nine ribosomal RNA (rRNA genes). The strain SINDOREI genome was profiled as a circular map, exhibiting CDSs, virulent genes, GC plot, and GC skew (Figure 1).
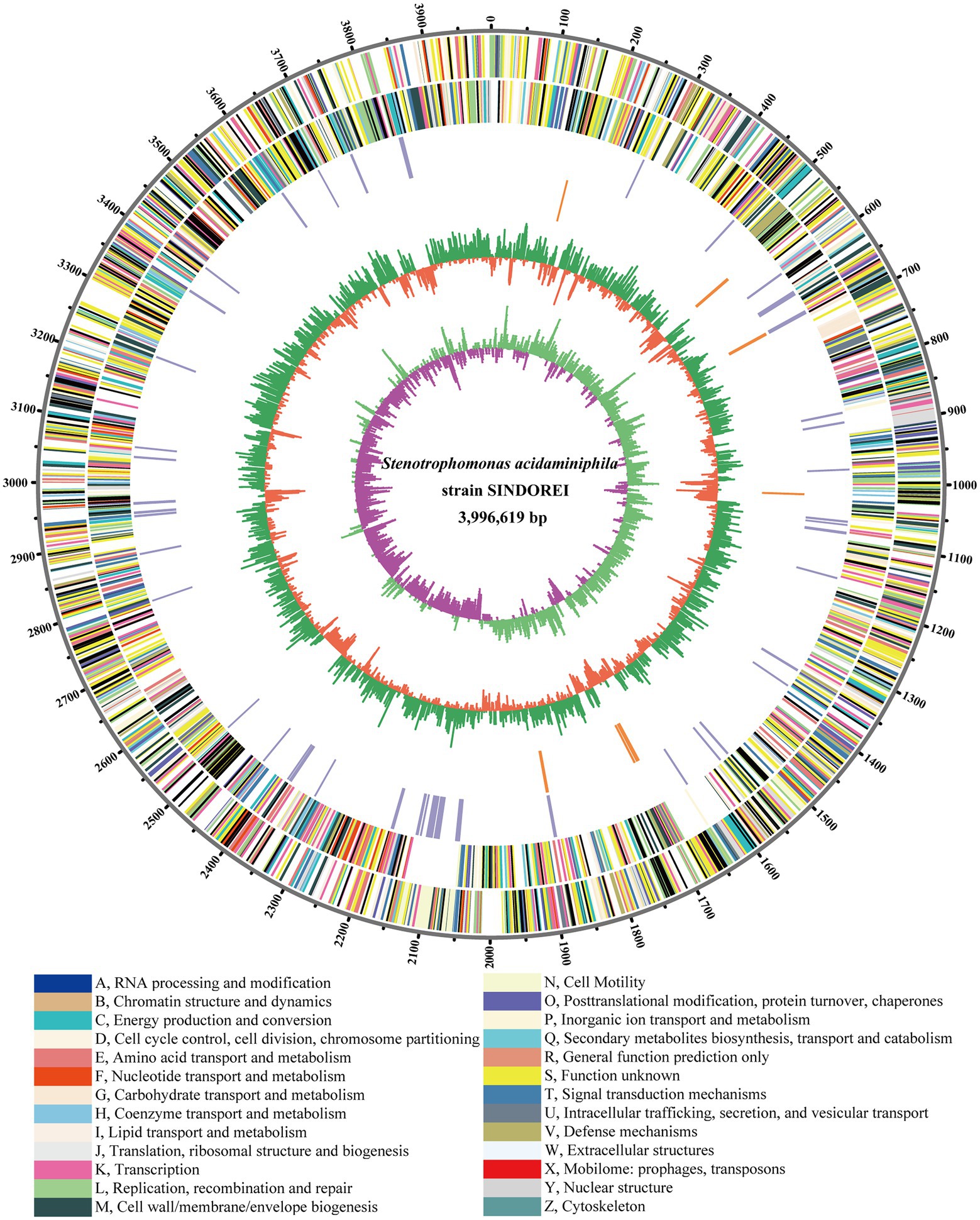
Figure 1. Circular plot of Stenotrophomonas acidaminiphila SINDORE genome generated by circos. Predicted Coding Sequences (CDSs) are presented by various colors according to cellular functions. The Circles from outside to inside: 1, the scale line. 2, CDSs on forward strands. 3, CDSs on reverse strands. 4, virulent genes. 5, resistant genes. 6, GC plot, above average in green and below average in violet, respectively. 7, GC skew showing regions above and below average in yellow and light blue, respectively.
Firstly, a 16S rRNA phylogenetic tree of strain SINDOREI and other available type strains of genus Stenotrophomonas were constructed using the maximum-likelihood method (Supplementary Figure S1). Strain SINDORE was clustered to the branch of S. acidaminiphila with a bootstrap consistency of 99%. In addition, the 16S rRNA identities between SINDORE and other S. acidaminiphila strains was over 99.68%, indicating the strain SINDOREI belongs to S. acidaminiphila.
The phylogenomic tree was constructed and revealed that strain SINDOREI has the closest relationship with strain JCM13310T and strain T25-65, which was consistent with the 16S rRNA tree (Figure 2). Whole genome comparison identified the least SNPs/Indels (11,650) between SINDOREI and T25-65 (Supplementary Table S8). Meanwhile, the ANI and dDDH values between S. acidaminiphila SINDOREI and S. acidaminiphila T25-65 are up to 99.9% and 94.1%, respectively (Figure 3; Supplementary Table S9).
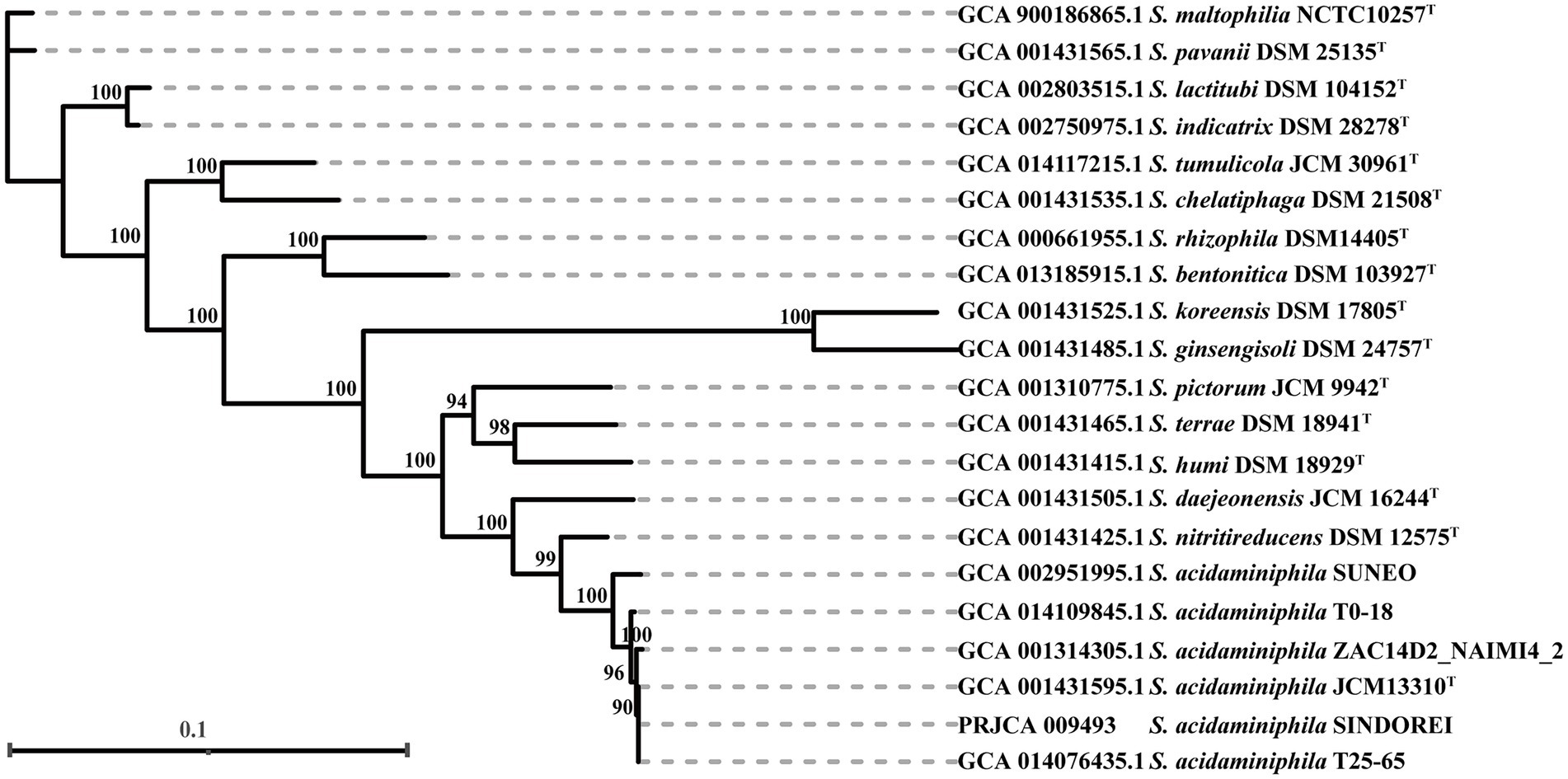
Figure 2. The phylogenomic tree based on the genomes of members in genus Stenotrophomonas. The numbers presenting on the branches of the tree represented the bootstrap values (based on 1,000 replicates). The scale bar indicated 0.1 substitutions per nucleotide position.
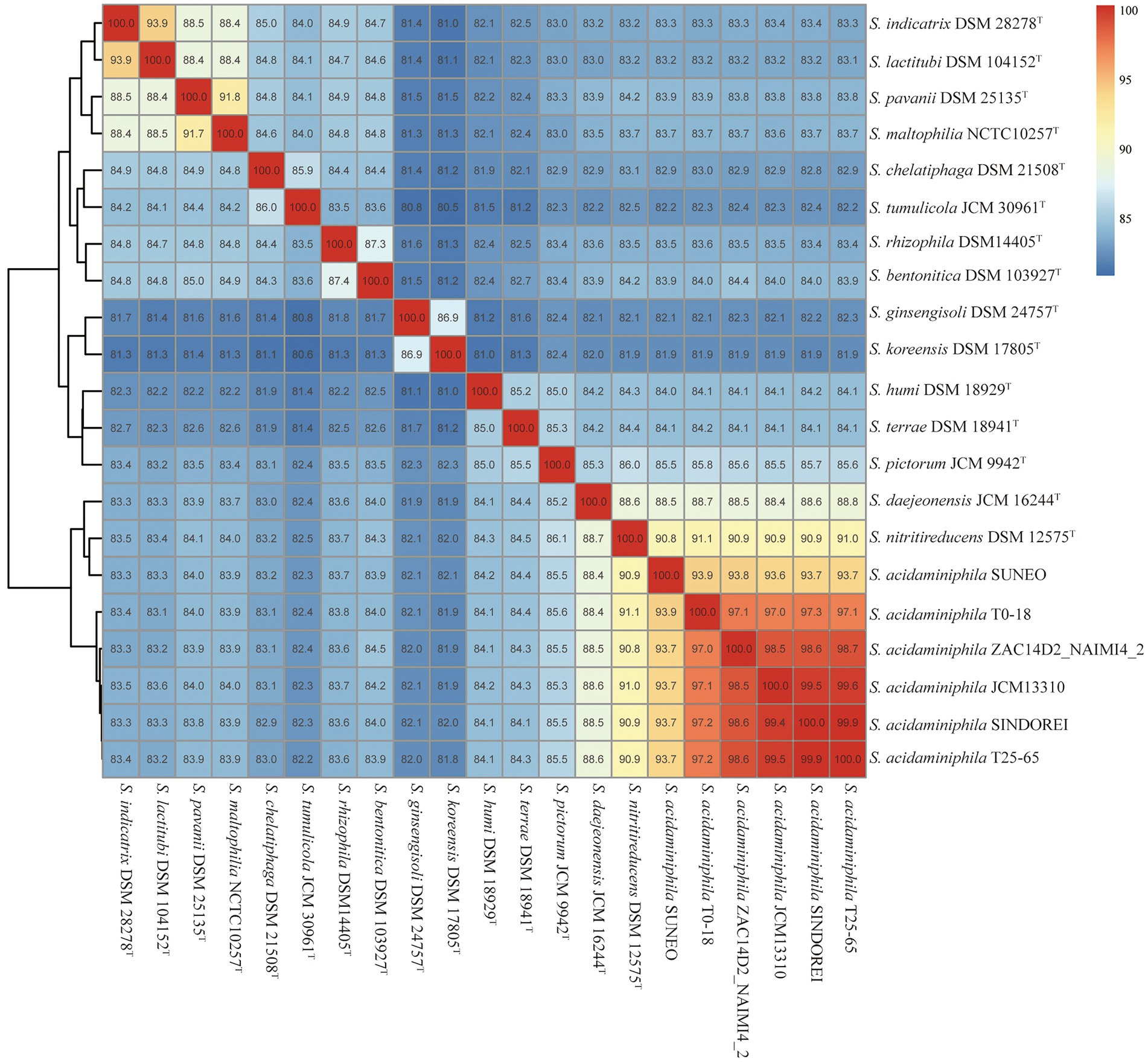
Figure 3. ANI values of Stenotrophomonas acidaminiphila SINDOREI with the type strains of genus Stenotrophomonas.
Six genomes of S. acidaminiphila strains were employed for the comparative genome analysis (Table 1). Only SINDOREI and SUNEO were isolated from clinical specimens. The largest genome size was from ZAC14D2_NAIMI4_2 (4,138,397 bp), followed by strain SINDOREI. There were 2,506 orthogroups found in each strain (Figure 4), which represented the set of non-redundant core genes of six strains (Figure 5). The second highest orthogroups (292 orthogroups) were uniquely observed in SINDOREI, JCM13310T, T25-65, T0-18 and ZAC14D2_NAIMI4_2, except SUNEO. Further analysis of pair-wise comparisons demonstrated that strain SINDOREI share 3,166, 3,105, 3,084, 3,028, and 2,691 orthogroups with T0-18, JCM 13310T, T25-65, ZAC14D2_NAIMI4_2 and SUNEO, respectively (Supplementary Figure S2). Strain ZAC14D2_NAIMI4_2 had the most strain-specific genes (293 genes) as shown in Figure 5A, followed by strain JCM 13310T (216 genes). The ratio of specific genes in strain ZAC14D2_NAIMI4_2, SUNEO, T0-18, JAM 13310T, SINDOREI, and T25-65 was 8.2%, 6.4%, 6.2%, 6.1%, 4.5%, and 4.5%, respectively. 159 genes were exclusive to strain SINDOREI, and functional analysis based on COG category revealed that these genes were mainly assigned to signal transduction mechanisms (Figure 5B). The annotations of SINDOREI specific orthogroups and genes were listed in Supplementary Table S10.
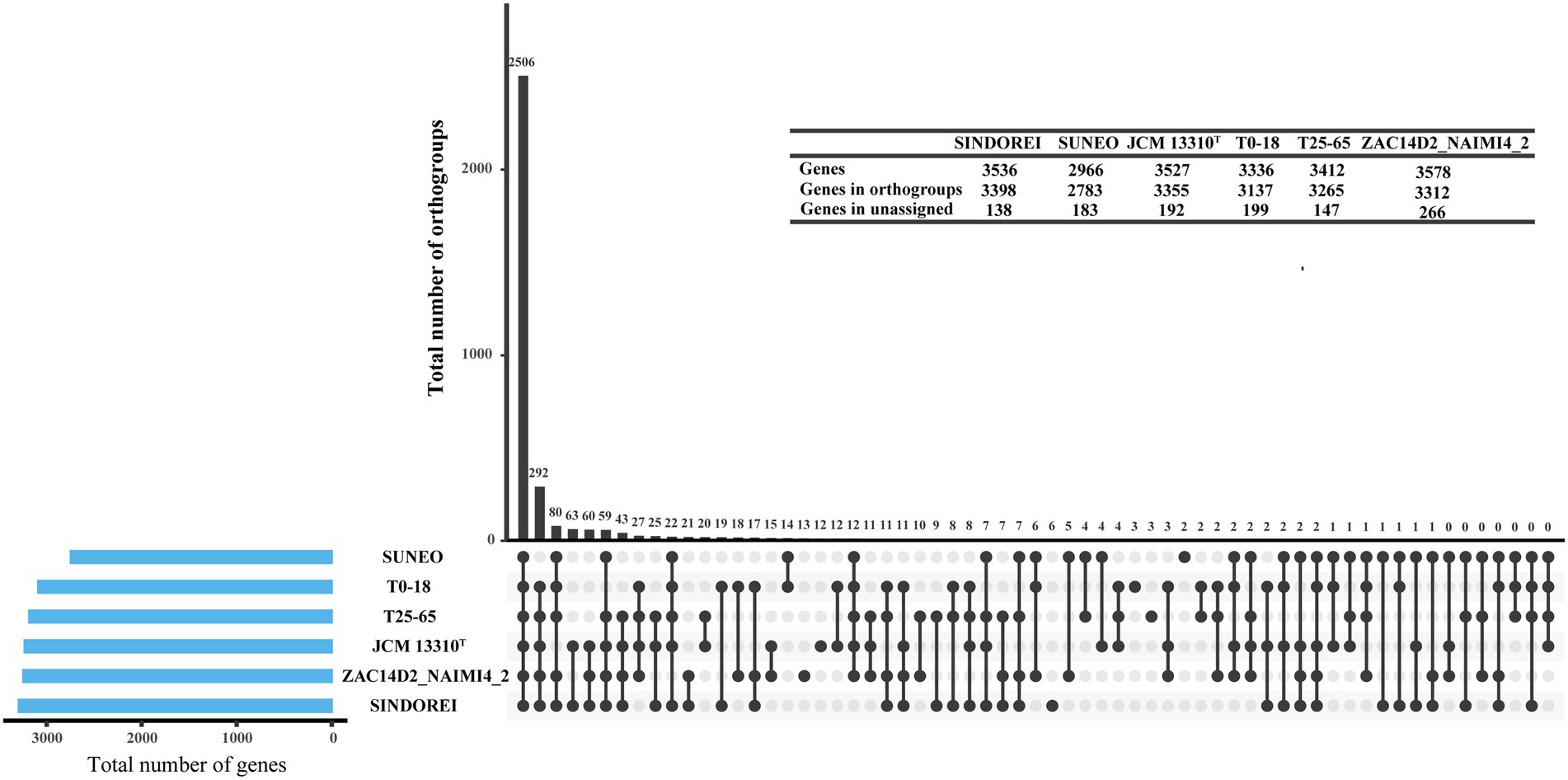
Figure 4. Groups of orthologous and paralogous genes (i.e., orthogroups) identified in the six Stenotrophomonas acidaminiphila strains used in this study. The vertical bars show the number of orthogroups exclusive to the strains marked as lower dots in the matrix. Horizontal bars represent the total number of genes in each strain.
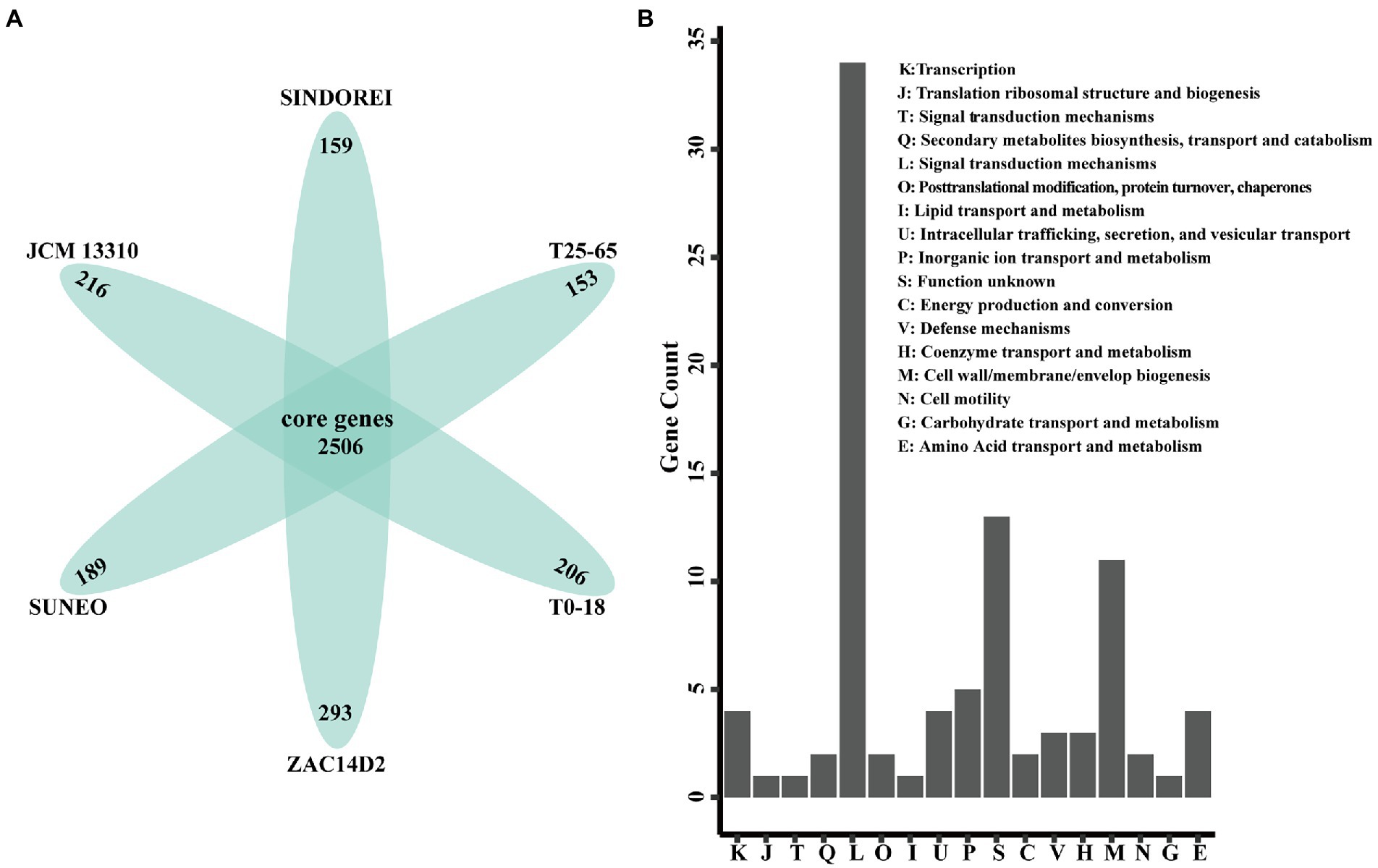
Figure 5. Comparison of the gene contents in Stenotrophomonas acidaminiphila. (A) Flower plot diagram showing the core genes and specific genes. (B) Distribution of COG functional annotations of SINDOREI specific genes.
Virulence factors are components, produced by bacterial cells, which generally cause damages to the host by increasing adhesion, facilitating colonization and invasion into eukaryotic cells, escaping the host immune responses and providing the essential nutrient (Casadevall and Pirofski, 2009). The presence of various virulence genes was investigated in the strain SINDOREI genome and the comparison of virulence genes between related strain genomes was conducted. The results revealed that the virulence genes in S. acidaminiphila genomes are classified into 11 categories according to virulence factor (VF) category, including motility (flagella), adherence (Type IV pili and non-pilus adhesins), biofilm formation, immune modulation [lipooligosaccharides (LOS), capsule, and lipopolysaccharide (LPS)], antimicrobial activity/competitive advantage, stress survival, nutritional/metabolic factor, regulation, exotoxin, effector delivery system, and others. The virulence genes of S. acidaminiphila genomes was shown in Figure 6 and the details of were listed in Supplementary Table S11. The genomes of S. acidaminiphila strains included the same VF categories, but the presence and distribution of genes were different. It is worth noting that the narGH operon only found in strain SINDOREI is responsible for nitrate reductase, which is identified as an important virulence factor for many bacterial infections, such as Mycobacterium tuberculosis and Pseudomonas aeruginosa (Palmer et al., 2007; Sohaskey and Modesti, 2009).
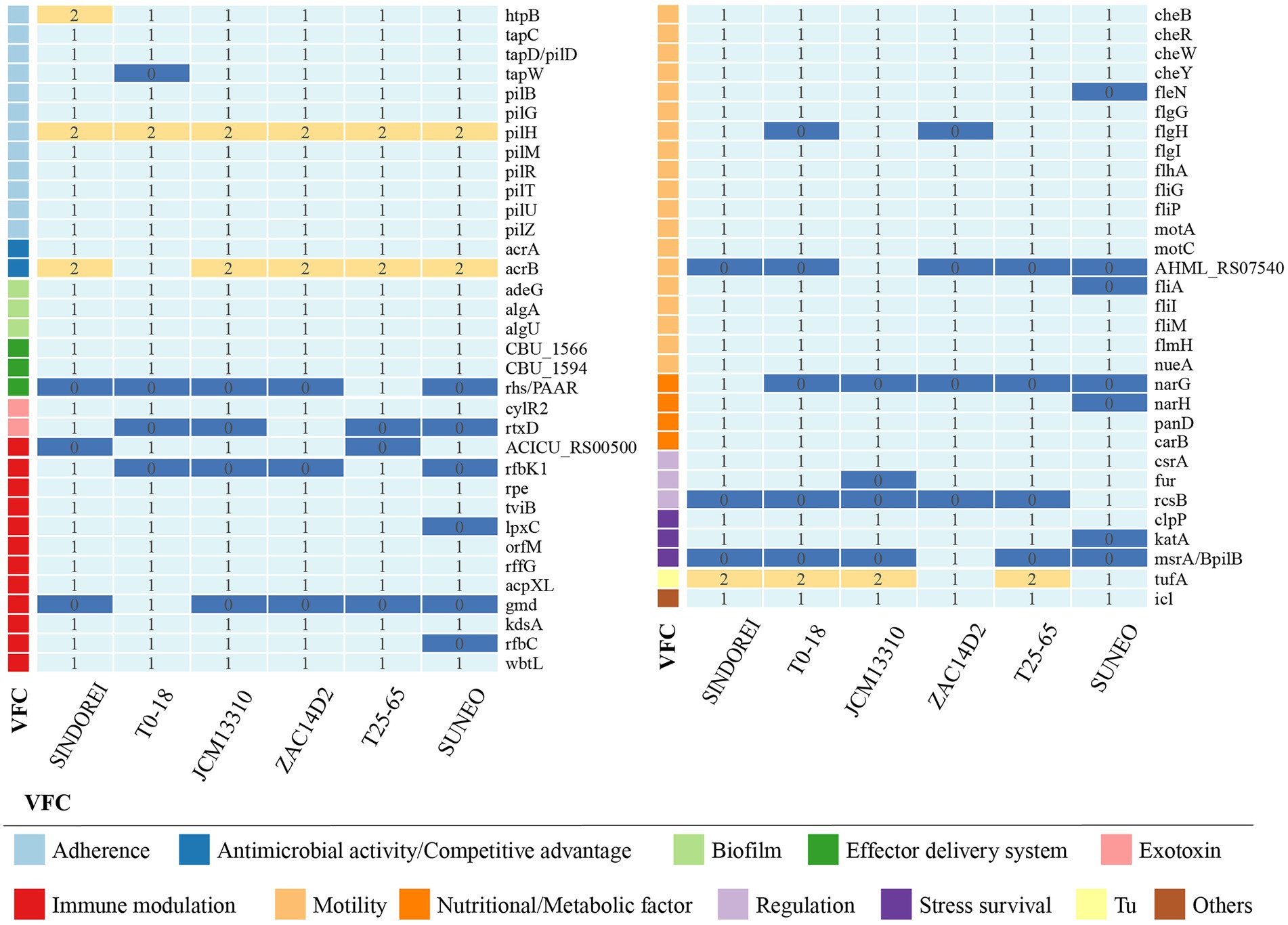
Figure 6. Comparison of the virulence factor genes in Stenotrophomonas acidaminiphila. The numbers show the gene copies in each genome and the virulence factor genes were classified into categories.
Biofilm formation, which is a mixture of cells, polysaccharides, nucleic acids, lipids and proteins, provides resistance to various antimicrobial drugs and to host immune defense of bacterial virulence. Biofilm-related infections represent more than 60% of all microbial infections in humans. In the genome of strain SINDOREI, various virulence factors are involved in the biofilm formation according to the previously reported including LPS (Huang et al., 2006; Pompilio et al., 2011; Zhuo et al., 2014; Madi et al., 2016), flagella formation (Di Bonaventura et al., 2007; Kang et al., 2015), type IV pili (Strom and Lory, 1993; Giltner et al., 2012), and purine biosynthesis (Kang et al., 2015). Details are listed in Table 2. These genes associated with biofilm formation exist in the genome of strain SINDOREI, suggesting strain SINDOREI harbor the basic characteristic of biofilm formation.
Antimicrobial susceptibility test revealed that strain SINDOREI is resistant to six antibiotics including TMP/SMX, ciprofloxacin, ofloxacin, cefepime, ceftazidime, and aztreonam (Table 3), which are classified as sulfonamide antibiotic, fluoroquinolone antibiotic and β-lactam antibiotic. At the same time, strain SINDOREI is sensitive to meropenem, piperacillin tazobactam, sulbactam cefoperazone, amikacin, and tigecycline.
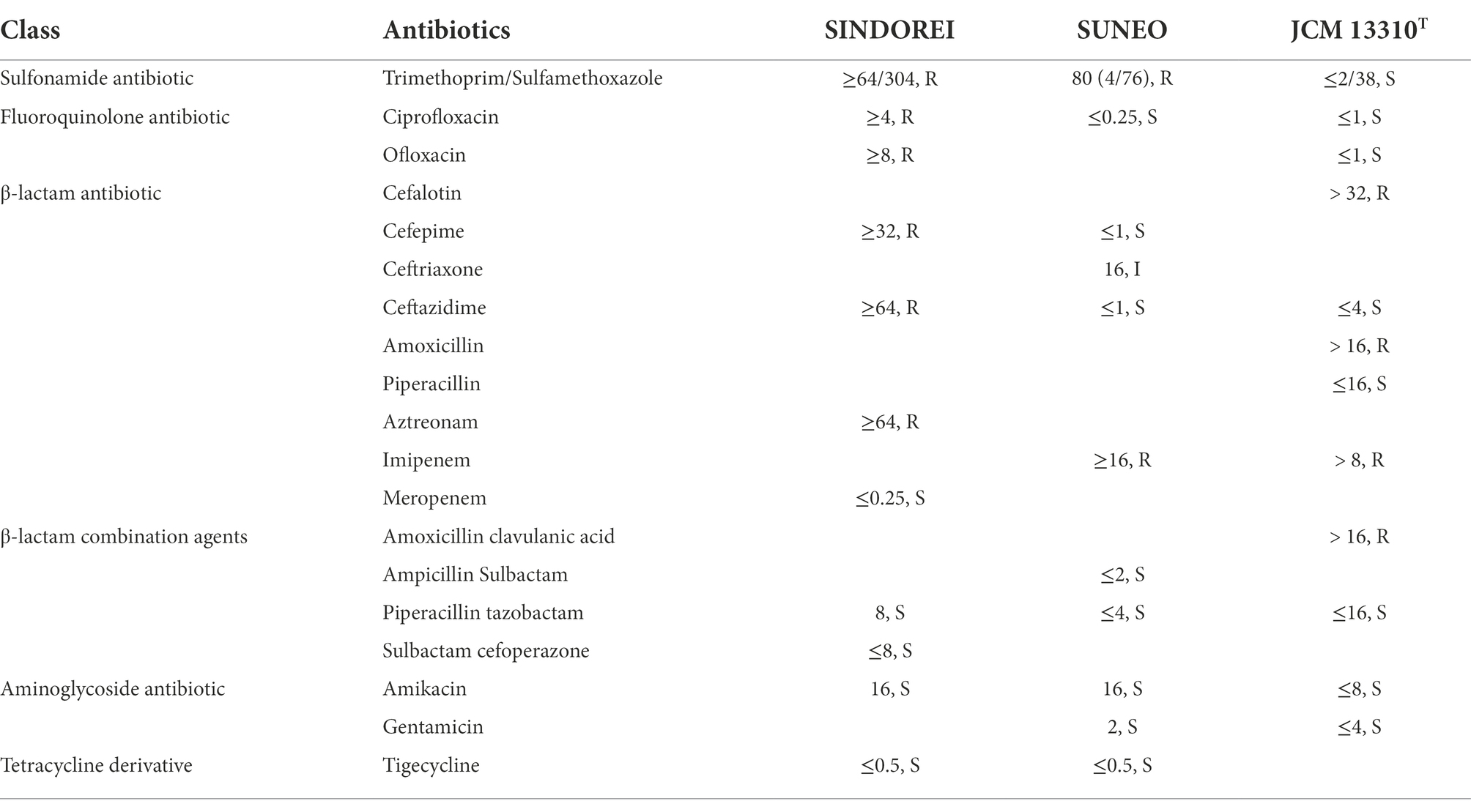
Table 3. Antimicrobial susceptibility test of Stenotrophomonas acidaminiphila SINDOREI, SUNEO and JCM 13310T.
The comparison of resistance-related genes in strains SINDOREI, SUNEO, JCM 13310T, T25-65, T0-18 and ZAC14D2_NAIMI4 was performed (Table 4). The results showed that S. acidaminiphila possesses similar antibiotic resistance genes. A total of 24 key genes including encoded genes (14 genes) and efflux pumps (10 genes) contribute to the multidrug resistance in strain SINDOREI. The 24 genes are involved in resistance to TMP/SMX (Toleman et al., 2007; Hu et al., 2011), fluoroquinolone (Valdezate et al., 2005; Sanchez et al., 2009; Farhat et al., 2016), β-lactam (Okazaki and Avison, 2008; Huang et al., 2010; Lin et al., 2011; Bontron et al., 2015), aminoglycoside (Huang et al., 2015), disinfecting agents (Huang et al., 2015), phenicol (Dong et al., 2020) and tetracycline (Kadlec and Schwarz, 2018). Interestingly, four antibiotic resistant determinants, including GES-1, aadA3, qacL and cmlA5, were exclusive to strain SINDOREI. The sul2 and tetC genes were only found in SINDOREI, T25-65, and T0-18.
The sul genes encoding variants of the dihydropteroate synthase are responsible for resistance to TMP/SMX in many bacteria (Huang et al., 2018; Domínguez et al., 2019). There are two sul genes (sul1 and sul2 genes) in Strain SINDOREI. The amino acid sequences of homologues to the sul genes in Stenotrophomonas genomes were employed to construct the phylogenetic tree. Four distinct groups were generated (Figure 7), and Sul1 and Sul2 in SINDOREI belong to different groups. The distribution of sul1 and sul2 genes in strain SINDOREI, T0-18 and T25-65 may promote the resistance to TMP/SMX. The amino acid sequence of Sul1 in strain SINDOREI shows 100% identity with that of strains JCM13310T, ZAC14D2_NAIMI4_2 and T25-65. The identity sequences suggests that these elements are ancient and acquired long with the use of antibiotics. According to the genome sequence of strain SINDOREI, we noted that IS6 and Tn3 family transposase were located immediately upstream of sul2 and IS6 family transposase was located downstream of sul2, suggesting that sul2 gene was acquired from horizontal gene transfer.
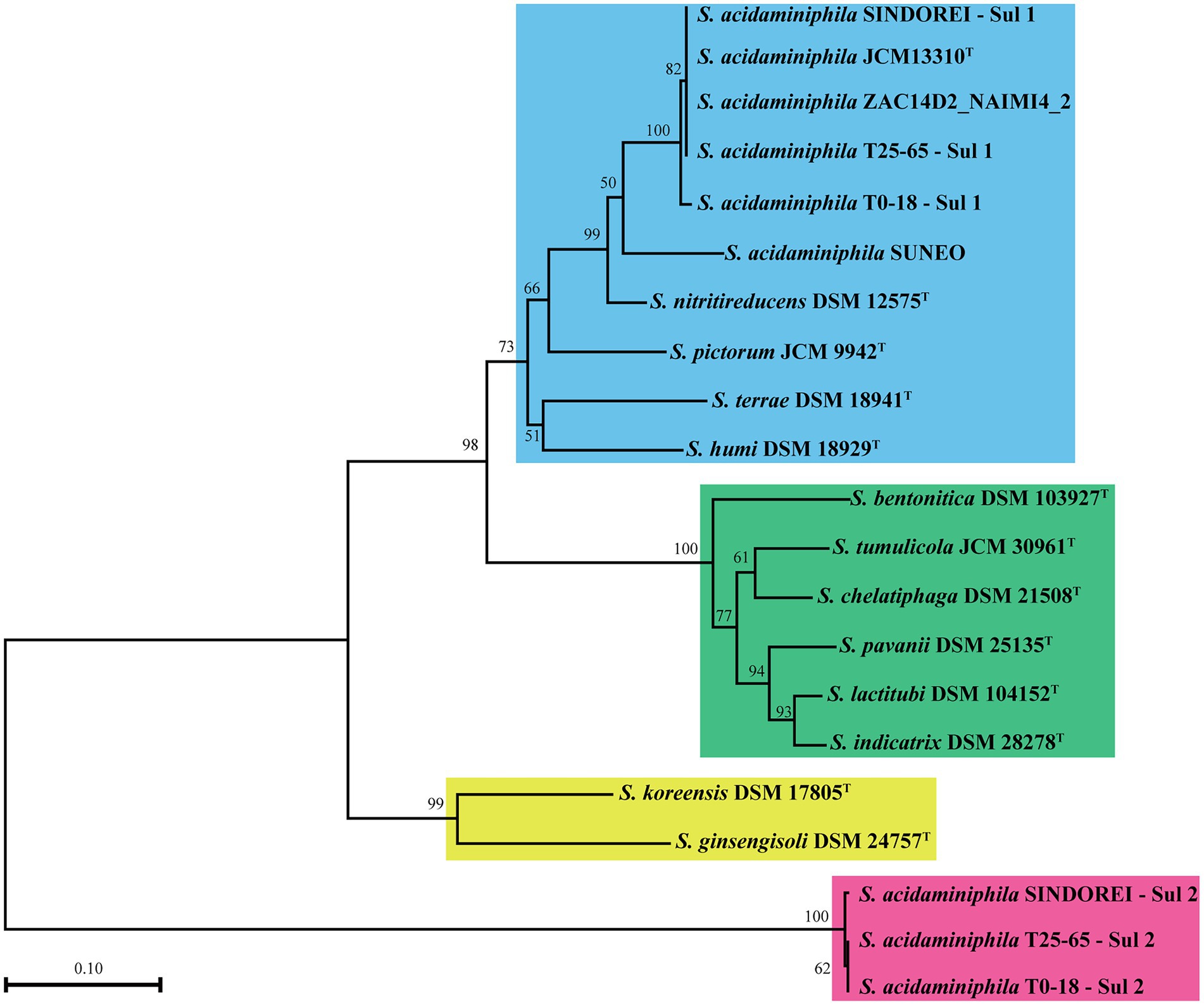
Figure 7. The phylogenetic tree based on the amino acid of sul genes in the members of genus Stenotrophomonas. The numbers present on the branches of the tree represent the bootstrap values (based on 1,000 replicates).
In this study, the genomes of six S. acidaminiphila strains, including SINDOREI, SUNEO, JCM13310T, T25-65, T0-18, and ZAC14D2_NAIMI4_2, were employed to perform the comparative analysis. Strain SINDOREI was isolated from the blood of a patient with sepsis. Meanwhile, S. acidaminiphila was the sole organism cultured from this patient. Antimicrobial susceptibility test reveals that stain SINDOREI is resistant to TMP/SMX, ciprofloxacin, ofloxacin, cefepime, ceftazidime, and aztreonam. The results suggested that S. acidaminiphila is an emerging opportunistic pathogen with environmental origin.
Based on the analysis of six genomes, strain SINDOREI shows highest ANI and dDDH values with T25-65 and shared lowest ANI and dDDH values with SUNEO. Two thousand five hundred six orthogroups were found in six genomes, though only 2,691 orthogroups was shared between strain SINDOREI and SUNEO. The clinical isolates, strain SINDOREI and SUNEO, shows relatively high strain-level diversity compared to other strains. Further work is still needed for getting more isolates to profile a comprehensive picture of the population of S. acidaminiphila.
We explored the virulence features in strain SINDOREI, and the nitrate reductase enzyme operon (narGH) was also found. However, narH gene was only found in strain JCM 13310T, T25-65, T0-18 and ZAC14D2_NAIMI4_2. As reported previously, the nitrate reductase enzyme is important in the pathogenesis of many bacteria and a narG knockout mutant can cause reduced virulence and reduce lung damage in severe combined immunodeficiency mice (Fritz et al., 2002). Biofilm formation is also an important pathogenesis of bacteria such as S. maltophilia. Various genes associated with biofilm formation were found in strain SINDOREI which were considered as the basic characteristics of biofilm formation involved in pathogenesis of infections. Biofilm can be attached to various abiotic surfaces and tissues (Flores-Treviño et al., 2019). This specific structure provides up to 1,000 times more resistance to antimicrobial drugs (Mah, 2012; Olsen, 2015) and contributes to respiratory diseases (Costerton et al., 1999; Pompilio et al., 2010). New antimicrobial strategies (antibiofilm strategies) were used to treat Stenotrophomonas infections. Biofilm assay should be conducted in further study to explore biofilm’s correlations with virulence. Therefore, present ongoing studies about strain SINDOREI are limited and valuable to in-depth mining.
Investigation of the presence of virulence genes is important to explain the genetic mechanisms of multidrug resistance. In MDR strain SINDOREI, 24 genes involved in the resistance to a broad array of antimicrobial agents were analyzed. Strain SINDOREI has all the target genes mainly encoding antibiotic inactivating enzymes and multidrug efflux pumps, which is an important reason for the resistance to antibiotics. Meanwhile, the unique and redundant resistance genes occurred in strain SINDOREI. GES-1 (GES-type beta-lactamase), aadA3 (streptomycin 3′-adenylyltransferase), qacL (antibiotic efflux of disinfecting agents), and cmlA5 (antibiotic efflux of phenicol antibiotic) associated with the resistance to β-lactam, aminoglycoside, disinfecting agents and phenicol are exclusive to strain SINDOREI. The phylogenetic analysis of homologues to sul genes indicated that there are two types of sul genes (sul1 and sul2) in strain SINDOREI. The presence of redundant genes (sul1 and sul2) in strain SINDOREI, should be responsible for the increased resistance to TMP/SMX. The presence of IS6 and Tn3 family transposase located upstream and downstream of the sul2, suggesting that acquisition of resistance genes is a relevant mechanism for strain SINDOREI antibiotic resistance.
In this study, we reported the world’s first case of fatal infection with S. acidaminiphila and obtained the high-quality genome of clinical isolated MDR strain SINDOREI. The comparative genomes analysis suggested a unique gene (narG) and key genes involved in biofilm formation in strain SINDOREI played an important role in pathogenesis of infections. Antimicrobial susceptibility test revealed that stain SINDOREI were resistant to TMP/SMX, ciprofloxacin, ofloxacin, cefepime, ceftazidime, and aztreonam. The presence of redundant Sul1 and Sul2 from two distinct groups and the exclusive determinants GES-1, aadA3, qacL and cmlA5 exist in SINDOREI, which can explain the mechanisms of strain’s multidrug resistance and afford potential therapeutic strategies for pathogen infections.
The genome sequence data of strain SINDOREI has been deposited in National Genomics Data Center (NGDC, https://ngdc.cncb.ac.cn/) and accession numbers is PRJCA009493, as mentioned in Table 1 (CNCB-NGDC Members and Partners., 2022).
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
YZ, PX, and WC made contributions to the acquisition of clinical data. QY, MZ, AZ, and WW contributed to laboratory work. DW, HX, and YX conducted the clinical work. YZ and DL drafted the manuscript. ZJ revised the manuscript critically for important intellectual content and had given final approval of the version to be published. All authors contributed to the article and approved the submitted version.
This paper was supported by the Key Research and Development program of the Hunan Provincial Science and Technology Department (grant no. 2022SK2005).
DL was employed by Hugobiotech Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.989259/full#supplementary-material
Abbott, I. J., and Peleg, A. Y. (2015). Stenotrophomonas, Achromobacter, and nonmelioid Burkholderia species: antimicrobial resistance and therapeutic strategies. Semin. Respir. Crit. Care Med. 36, 99–110. doi: 10.1055/s-0034-1396929
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–d525. doi: 10.1093/nar/gkz935
Al-Jasser, A. M. (2006). Stenotrophomonas maltophilia resistant to trimethoprim-sulfamethoxazole: an increasing problem. Ann. Clin. Microbiol. Antimicrob. 5, 23. doi: 10.1186/1476-0711-5-23
Assih, E. A., Ouattara, A. S., Thierry, S., Cayol, J. L., Labat, M., and Macarie, H. (2002). Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int. J. Syst. Evol. Microbiol. 52, 559–568. doi: 10.1099/00207713-52-2-559
Barbolla, R., Catalano, M., Orman, B. E., Famiglietti, A., Vay, C., Smayevsky, J., et al. (2004). Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob. Agents Chemother. 48, 666–669. doi: 10.1128/AAC.48.2.666-669.2004
Bontron, S., Poirel, L., and Nordmann, P. (2015). In vitro prediction of the evolution of GES-1 β-lactamase hydrolytic activity. Antimicrob. Agents Chemother. 59, 1664–1670. doi: 10.1128/AAC.04450-14
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: functional annotation, Orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Casadevall, A., and Pirofski, L. A. (2009). Virulence factors and their mechanisms of action: the view from a damage-response framework. J. Water Health 7, S2–s18. doi: 10.2166/wh.2009.036
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
CNCB-NGDC Members and Partners (2022). Database resources of the National Genomics Data Center, China National Center for bioinformation in 2022. Nucleic Acids Res. 50, D27–D38. doi: 10.1093/nar/gkab951
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
de Oliveira-Garcia, D., Dall'Agnol, M., Rosales, M., Azzuz, A. C., Alcántara, N., Martinez, M. B., et al. (2003). Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell. Microbiol. 5, 625–636. doi: 10.1046/j.1462-5822.2003.00306.x
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Di Bonaventura, G., Prosseda, G., Del Chierico, F., Cannavacciuolo, S., Cipriani, P., Petrucca, A., et al. (2007). Molecular characterization of virulence determinants of Stenotrophomonas maltophilia strains isolated from patients affected by cystic fibrosis. Int. J. Immunopathol. Pharmacol. 20, 529–537. doi: 10.1177/039463200702000311
Domínguez, M., Miranda, C. D., Fuentes, O., de la Fuente, M., Godoy, F. A., Bello-Toledo, H., et al. (2019). Occurrence of transferable Integrons and sul and dfr genes among sulfonamide-and/or trimethoprim-resistant bacteria isolated from Chilean salmonid farms. Front. Microbiol. 10, 748. doi: 10.3389/fmicb.2019.00748
Dong, W. L., Odah, K. A., Liu, L., Xu, Q. J., Gao, Y. H., Kong, L. C., et al. (2020). Multidrug resistance genes are associated with a 42-kb island TGI1 carrying a complex class 1 integron in Trueperella pyogenes. J. Glob. Antimicrob. Resist. 22, 1–4. doi: 10.1016/j.jgar.2019.12.008
Farhat, M. R., Jacobson, K. R., Franke, M. F., Kaur, D., Sloutsky, A., Mitnick, C. D., et al. (2016). Gyrase mutations are associated with variable levels of Fluoroquinolone resistance in mycobacterium tuberculosis. J. Clin. Microbiol. 54, 727–733. doi: 10.1128/JCM.02775-15
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Harb, O. S., Iodice, J. B., et al. (2011). Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics Chapter 6, Unit 6.12.11-19. doi: 10.1002/0471250953.bi0612s35
Flores-Treviño, S., Bocanegra-Ibarias, P., Camacho-Ortiz, A., Morfín-Otero, R., Salazar-Sesatty, H. A., and Garza-González, E. (2019). Stenotrophomonas maltophilia biofilm: its role in infectious diseases. Expert Rev. Anti-Infect. Ther. 17, 877–893. doi: 10.1080/14787210.2019.1685875
Fritz, C., Maass, S., Kreft, A., and Bange, F. C. (2002). Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect. Immun. 70, 286–291. doi: 10.1128/IAI.70.1.286-291.2002
Giltner, C. L., Nguyen, Y., and Burrows, L. L. (2012). Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772. doi: 10.1128/MMBR.00035-12
Hu, L. F., Chang, X., Ye, Y., Wang, Z. X., Shao, Y. B., Shi, W., et al. (2011). Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int. J. Antimicrob. Agents 37, 230–234. doi: 10.1016/j.ijantimicag.2010.10.025
Huang, Y. T., Chen, J. M., Ho, B. C., Wu, Z. Y., Kuo, R. C., and Liu, P. Y. (2018). Genome sequencing and comparative analysis of Stenotrophomonas acidaminiphila reveal evolutionary insights into Sulfamethoxazole resistance. Front. Microbiol. 9, 1013. doi: 10.3389/fmicb.2018.01013
Huang, Y. W., Hu, R. M., Lin, Y. T., Huang, H. H., and Yang, T. C. (2015). The contribution of class 1 integron to antimicrobial resistance in Stenotrophomonas maltophilia. Microb. Drug Resist. 21, 90–96. doi: 10.1089/mdr.2014.0072
Huang, Y. W., Lin, C. W., Hu, R. M., Lin, Y. T., Chung, T. C., and Yang, T. C. (2010). AmpN-AmpG operon is essential for expression of L1 and L2 beta-lactamases in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 54, 2583–2589. doi: 10.1128/AAC.01283-09
Huang, T. P., Somers, E. B., and Wong, A. C. (2006). Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J. Bacteriol. 188, 3116–3120. doi: 10.1128/JB.188.8.3116-3120.2006
Huyan, J., Tian, Z., Zhang, Y., Zhang, H., Shi, Y., Gillings, M. R., et al. (2020). Dynamics of class 1 integrons in aerobic biofilm reactors spiked with antibiotics. Environ. Int. 140:105816. doi: 10.1016/j.envint.2020.105816
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Kadlec, K., and Schwarz, S. (2018). Antimicrobial resistance in Bordetella bronchiseptica. Microbiol. Spectr. 6:4. doi: 10.1128/microbiolspec.ARBA-0024-2017
Kang, X. M., Wang, F. F., Zhang, H., Zhang, Q., and Qiana, W. (2015). Genome-wide identification of genes necessary for biofilm formation by nosocomial pathogen Stenotrophomonas maltophilia reveals that orphan response regulator FsnR is a critical modulator. Appl. Environ. Microbiol. 81, 1200–1209. doi: 10.1128/AEM.03408-14
Ko, J. H., Kang, C. I., Cornejo-Juárez, P., Yeh, K. M., Wang, C. H., Cho, S. Y., et al. (2019). Fluoroquinolones versus trimethoprim-sulfamethoxazole for the treatment of Stenotrophomonas maltophilia infections: a systematic review and meta-analysis. Clin. Microbiol. Infect. 25, 546–554. doi: 10.1016/j.cmi.2018.11.008
Kumar, S., Bansal, K., Patil, P. P., Kaur, A., Kaur, S., Jaswal, V., et al. (2020). Genomic insights into evolution of extensive drug resistance in Stenotrophomonas maltophilia complex. Genomics 112, 4171–4178. doi: 10.1016/j.ygeno.2020.06.049
Lin, C. W., Lin, H. C., Huang, Y. W., Chung, T. C., and Yang, T. C. (2011). Inactivation of mrcA gene derepresses the basal-level expression of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 66, 2033–2037. doi: 10.1093/jac/dkr276
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–d917. doi: 10.1093/nar/gkab1107
Looney, W. J., Narita, M., and Mühlemann, K. (2009). Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9, 312–323. doi: 10.1016/S1473-3099(09)70083-0
Madi, H., Lukić, J., Vasiljević, Z., Biočanin, M., Kojić, M., Jovčić, B., et al. (2016). Genotypic and phenotypic characterization of Stenotrophomonas maltophilia strains from a pediatric tertiary Care Hospital in Serbia. PLoS One 11:e0165660. doi: 10.1371/journal.pone.0165660
Mah, T. F. (2012). Biofilm-specific antibiotic resistance. Future Microbiol. 7, 1061–1072. doi: 10.2217/fmb.12.76
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Okazaki, A., and Avison, M. B. (2008). Induction of L1 and L2 beta-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob. Agents Chemother. 52, 1525–1528. doi: 10.1128/AAC.01485-07
Olsen, I. (2015). Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 34, 877–886. doi: 10.1007/s10096-015-2323-z
Paez, J. I., and Costa, S. F. (2008). Risk factors associated with mortality of infections caused by Stenotrophomonas maltophilia: a systematic review. J. Hosp. Infect. 70, 101–108. doi: 10.1016/j.jhin.2008.05.020
Palleroni, N. J., and Bradbury, J. F. (1993). Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) swings et al. 1983. Int. J. Syst. Bacteriol. 43, 606–609. doi: 10.1099/00207713-43-3-606
Palmer, K. L., Brown, S. A., and Whiteley, M. (2007). Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 189, 4449–4455. doi: 10.1128/JB.00162-07
Patil, P. P., Midha, S., Kumar, S., and Patil, P. B. (2016). Genome sequence of type strains of genus Stenotrophomonas. Front. Microbiol. 7:309. doi: 10.3389/fmicb.2016.00309
Pompilio, A., Crocetta, V., Confalone, P., Nicoletti, M., Petrucca, A., Guarnieri, S., et al. (2010). Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol. 10, 102. doi: 10.1186/1471-2180-10-102
Pompilio, A., Pomponio, S., Crocetta, V., Gherardi, G., Verginelli, F., Fiscarelli, E., et al. (2011). Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11, 159. doi: 10.1186/1471-2180-11-159
Putman, M., van Veen, H. W., and Konings, W. N. (2000). Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64, 672–693. doi: 10.1128/MMBR.64.4.672-693.2000
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Sanchez, M. B., Hernandez, A., and Martinez, J. L. (2009). Stenotrophomonas maltophilia drug resistance. Future Microbiol. 4, 655–660. doi: 10.2217/fmb.09.45
Sohaskey, C. D., and Modesti, L. (2009). Differences in nitrate reduction between mycobacterium tuberculosis and Mycobacterium bovis are due to differential expression of both narGHJI and narK2. FEMS Microbiol. Lett. 290, 129–134. doi: 10.1111/j.1574-6968.2008.01424.x
Strom, M. S., and Lory, S. (1993). Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47, 565–596. doi: 10.1146/annurev.mi.47.100193.003025
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tan, C. K., Liaw, S. J., Yu, C. J., Teng, L. J., and Hsueh, P. R. (2008). Extensively drug-resistant Stenotrophomonas maltophilia in a tertiary care hospital in Taiwan: microbiologic characteristics, clinical features, and outcomes. Diagn. Microbiol. Infect. Dis. 60, 205–210. doi: 10.1016/j.diagmicrobio.2007.09.007
Toleman, M. A., Bennett, P. M., Bennett, D. M., Jones, R. N., and Walsh, T. R. (2007). Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg. Infect. Dis. 13, 559–565. doi: 10.3201/eid1304.061378
Trifonova, A., and Strateva, T. (2019). Stenotrophomonas maltophilia – a low-grade pathogen with numerous virulence factors. Infect. Dis. (Lond) 51, 168–178. doi: 10.1080/23744235.2018.1531145
Valdezate, S., Vindel, A., Saéz-Nieto, J. A., Baquero, F., and Cantón, R. (2005). Preservation of topoisomerase genetic sequences during in vivo and in vitro development of high-level resistance to ciprofloxacin in isogenic Stenotrophomonas maltophilia strains. J. Antimicrob. Chemother. 56, 220–223. doi: 10.1093/jac/dki182
Vinuesa, P., and Ochoa-Sánchez, L. E. (2015). Complete genome sequencing of Stenotrophomonas acidaminiphila ZAC14D2_NAIMI4_2, a multidrug-resistant strain isolated from sediments of a Polluted River in Mexico, uncovers new antibiotic resistance genes and a novel class-II lasso peptide biosynthesis gene cluster. Genome Announc. 3, e01433–15. doi: 10.1128/genomeA.01433-15
Wang, Y., He, T., Shen, Z., and Wu, C. (2018). Antimicrobial resistance in Stenotrophomonas spp. Microbiol. Spectr. 6. doi: 10.1128/microbiolspec.ARBA-0005-2017
Wayne, P. (2020). Performance Standards for Antimicrobial Susceptibility Testing, 30rd Informational Supplement. CLSI Document M100-S30. Berwyn, PA: Clinical and Laboratory Standards Institute.
Wei, C., Ni, W., Cai, X., Zhao, J., and Cui, J. (2016). Evaluation of trimethoprim/Sulfamethoxazole (SXT), minocycline, Tigecycline, Moxifloxacin, and Ceftazidime alone and in combinations for SXT-susceptible and SXT-resistant Stenotrophomonas maltophilia by in vitro time-kill experiments. PLoS One 11:e0152132. doi: 10.1371/journal.pone.0152132
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Windhorst, S., Frank, E., Georgieva, D. N., Genov, N., Buck, F., Borowski, P., et al. (2002). The major extracellular protease of the nosocomial pathogen Stenotrophomonas maltophilia: characterization of the protein and molecular cloning of the gene. J. Biol. Chem. 277, 11042–11049. doi: 10.1074/jbc.M109525200
Keywords: Stenotrophomonas acidaminiphila, sepsis, genome sequencing, genome comparisons, virulence factors, multidrug resistant, human infection
Citation: Zhang Y, Li D, Yan Q, Xu P, Chen W, Xin H, Wu D, Zhou M, Xu Y, Zhang A, Wei W and Jiang Z (2022) Genome-wide analysis reveals the emergence of multidrug resistant Stenotrophomonas acidaminiphila strain SINDOREI isolated from a patient with sepsis. Front. Microbiol. 13:989259. doi: 10.3389/fmicb.2022.989259
Received: 08 July 2022; Accepted: 19 August 2022;
Published: 23 September 2022.
Edited by:
Nicolas Nalpas, University of Tübingen, GermanyReviewed by:
Sébastien Bontemps-Gallo, Institut Pasteur de Lille, FranceCopyright © 2022 Zhang, Li, Yan, Xu, Chen, Xin, Wu, Zhou, Xu, Zhang, Wei and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiping Jiang, amlhbmd6aHBAY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.