
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 August 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.989085
The plasmid-mediated horizontal transfer of antibiotic resistance genes (ARGs) among bacteria facilitates the evolution and dissemination of antibiotic resistance. Broad-host-range plasmids can be transferred to different bacterial hosts in soil, plant rhizospheres, and wastewater treatment plants. Although composting is an effective way to convert organic waste into fertilizer and reduce some ARGs, few studies have focused on its effects on the spread of ARG-carrying plasmids and their bacterial host communities during composting. In this study, a fluorescently labeled Pseudomonas putida (P. putida) harboring a broad-host-range plasmid RP4 carrying three ARGs was inoculated into a raw material microcosm and composted with different durations of the thermophilic phase. The fate of the donor and RP4 in composting was investigated. The prolonged thermophilic composting removed 95.1% of dsRed and 98.0% of gfp, and it inhibited the rebound of P. putida and RP4 during the maturation phase. The spread potential of RP4 decreased from 10−4 to 10−6 transconjugants per recipient after composting. In addition, we sorted and analyzed the composition of RP4 recipient bacteria using fluorescence-activated cell sorting combined with 16S rRNA gene amplicon sequencing. The recipient bacteria of RP4 belonged to eight phyla, and Firmicutes, accounting for 75.3%–90.1%, was the dominant phylum in the transconjugants. The diversity and richness of the RP4 recipient community were significantly reduced by prolonged thermophilic periods. Overall, these findings provide new insights for assessing the contribution of composting in mitigating the dissemination of plasmid-mediated ARGs, and the prolonged thermophilic phase of composting can limit the transfer of multidrug-resistant plasmids.
Livestock manure is considered an important source of environmental antibiotic-resistant bacteria and antibiotic resistance genes (ARGs) because of the worldwide usage of antibiotics in intensive animal production (Vikesland et al., 2017; Qian et al., 2018). Manure aerobic composting is an effective technology for producing fertilizer and removing antibiotic residues; however, the fate of ARGs is complicated because of diverse abiotic and biotic factors during composting (Liu et al., 2021). Based on correlation analysis, researchers found that mobile genetic elements (MGEs) and potential bacterial hosts are two crucial biotic factors for the removal of ARGs by composting (Liao et al., 2019a; Cao et al., 2020; Qiu et al., 2021; Xie et al., 2022). In addition, horizontal gene transfer (HGT) has recently been observed as a key determinant of ARG proliferation in composting (Liao et al., 2019a; Zhou et al., 2021). Among HGT processes between bacteria, plasmid-mediated conjugation is the most prevalent mechanism of horizontal transfer of ARGs, especially broad-host plasmids, which can transfer ARGs between different phyla (Che et al., 2021). Manure from intensive animal farms is a reservoir of broad-host plasmids carrying multidrug-resistant genes, which greatly increases the probability of other bacteria acquiring multidrug resistance through HGT (Binh et al., 2008; Yang et al., 2017). Moreover, after the current manure composting process, some antibiotic-resistant plasmids in composts could still be captured (Le Devendec et al., 2016), and ARGs and plasmids in composts could also be transferred to the soil and plant endosphere (Guo et al., 2021; Sun et al., 2021; Xu et al., 2021). To prevent the spread of ARGs from animal manure, it is essential to evaluate the contribution of composting to mitigate the dissemination of plasmid-mediated ARGs.
To understand the fate of antibiotic-resistant plasmids during composting, it is necessary to decipher the relationship between ARG-carrying plasmids and their bacterial host communities. Network analysis based on the correlation between ARGs and bacterial communities showed that some ARGs increased with potential hosts after composting (Su et al., 2015; Sardar et al., 2021). Conjugation studies have shown that conventional composting fails to control the transfer risk of sulfonamide- or colistin-resistant plasmids in cow or broiler manure (Le Devendec et al., 2016; Lin et al., 2021). Metatranscriptomic analysis found that the microbial community was the key factor defining the transcriptional responses of ARGs during composting (Wang et al., 2017). These studies have shown that the shift in microbial hosts directly affects the fate of ARGs in composting. However, there is no direct evidence of plasmid-mediated ARG transfer and changes in conjugative plasmid receipts. Combining fluorescent reporter genes-based fluorescence-activated cell sorting (FACS) with 16S rRNA gene amplicon sequencing made it possible to directly track the dissemination of ARG-carrying plasmids among indigenous bacteria in the soil (Klümper et al., 2015, 2017; Fan et al., 2019), plant rhizosphere (Xu et al., 2021), and wastewater treatment plants (Li et al., 2018). However, few studies have focused on the spread of ARG-carrying plasmids and their bacterial host communities during composting.
The composting temperature intensity and duration of the thermophilic phase are important for the effective removal of ARGs (Liu et al., 2021). Hyperthermophilic composting and prolonged thermophilic phase could promote the removal efficiency of ARGs/MGEs and reduce the abundance of potential ARG bacterial host taxa based on correlation analysis (Qian et al., 2016; Liao et al., 2019b; Keenum et al., 2021). A culture-dependent conjugative study showed that high temperatures could help limit the spread of plasmid-carrying ARGs with Escherichia coli in chicken manure composting (Guan et al., 2007). Evaluating the community shifts in ARG-carrying plasmid receipts will provide direct evidence for the control of HGT in manure composting.
Therefore, this study reports the first sight of bacterial host community shifts toward the typical conjugative plasmid (RP4) and the first exploration of the effect of a prolonged thermophilic phase to prevent the spread of ARG-carrying plasmids during composting. We evaluated the impact of a prolonged thermophilic phase on the fate and persistence of donor and RP4 plasmids during composting. The abundance of donors and plasmids was tracked by digital droplet PCR (ddPCR), and the transconjugants were analyzed by FACS and Illumina sequencing. The primary aims of this study were (1) to investigate the effects of a prolonged thermophilic phase on the removal of RP4 plasmid and donor and (2) to determine the roles of the prolonged thermophilic phase in reducing the potential for ARGs spread through conjugation. These findings provide new insights into manure treatment strategies to mitigate the potential for HGT of ARGs.
Pseudomonas putida KT2442 harboring a broad-host-range IncP plasmid RP4 was used as the donor, which was gifted by Professor Barth F. Smets (Musovic et al., 2010). This bacterium was marked with Discosoma sp. red fluorescent protein (DsRed) gene and lacIq in the chromosome. Plasmid RP4 (carring the kanamycin, ampicillin, and tetracycline resistance genes) was tagged with gfp gene, encoding the green fluorescent protein (GFP), and its promoter was repressed by lacIq. Therefore, P. putida only exhibited red fluorescence due to gfp expression being repressed in the donor strain. Nevertheless, when the plasmid is transferred to a recipient, the gfp gene is derepressed owing to a lack of lacIq, and green fluorescence is expressed, which can be retrieved by FACS or detected by confocal microscopy (Sørensen et al., 2005).
Pseudomonas putida KT2442 was grown aerobically at 35°C and 180 rpm overnight in lysogeny broth supplemented with 100 μgml−1 ampicillin, 50 μgml−1 kanamycin, and 20 μgml−1 tetracycline. Bacterial cells were then harvested by centrifuging at 10,000× g for 10 min and washed three times by resuspending in sterile phosphate-buffered saline (PBS). The bacterial concentration was measured by counting the colony forming units (CFU).
The experiment was performed in a 50-L bench-scale cylindrical stainless steel composting reactor equipped with a computer control. Aeration was provided by an air pump connected to a pipe equipped at the bottom of the reactor. A channel for air exhaust was present at the top of the reactor. The aeration rates were controlled by a gas flowmeter. The reaction temperature was controlled via resistive heating, using a custom-made computer-controlled power supply. A schematic of the compost reactor is shown in Supplementary Figure S1.
The composting raw material consisted of chicken manure and corn straw (crushed to 0.5–1 cm), both obtained from a commercial poultry farm in Beijing, China. Chicken manure and corn straw were mixed in a 3.5:1 ratio (w/w), and the moisture content was adjusted to ~60% with sterile water. After thorough mixing, 28 kg of the mixed raw material was transferred into each composting reactor. Compost microcosms were prepared by thoroughly mixing the donor bacterial suspension with 2 kg of raw materials, providing a final donor strain density of 1.5 × 107 CFUg−1 dry weight (DW). The prepared compost microcosms were packed into nylon mesh bags (40 cm long, 30 cm wide, mesh size: 2 mm) and placed in the center of the reactor. Control compost microcosms (CK) without donor inoculation were prepared and incubated at room temperature.
To investigate the effects of temperature on composting, two treatments were set up: normal thermophilic composting (NT treatment) and continuous thermophilic composting (CT treatment). Each composting treatment consisted of three replicate reactors. The NT treatment was run naturally during the entire composting process, and it had a natural thermophilic phase; for the CT treatment, the reactor was controlled and maintained at approximately 55°C for an additional 15 days after the natural thermophilic phase, and then the composting mixture was poured out of the reactor and matured at room temperature for 13 days.
The temperature of the composting mixture was monitored and recorded daily using an automatic temperature sensor (Supplementary Figure S2). The aeration rate was maintained at 0.5 l/min/kg DW throughout the composting process (Wang et al., 2013). The compost piles and microcosms were manually turned three times per week.
NT treatment was performed for 27 days, and CT treatment was performed for 40 days. Compost microcosms were collected from nylon mesh bags in each reactor on days 0, 1, 4, 7, 11, 15, 19, 27, and 40 and divided into two subsamples: one was stored at 4°C for bacterial cell extraction and the other was stored at −80°C for subsequent DNA extraction.
For compost samples, DNA was extracted according to Sun et al. (2019). The absolute abundances of P. putida and RP4 plasmid were performed by quantifying the dsRed and gfp genes, respectively. The dsRed, gfp, and 16S rRNA gene copies in samples were quantified by droplet digital PCR (ddPCR, QX200, Bio-Rad). The primer sequences and annealing temperatures for each tested gene are presented in Supplementary Table S1. Each ddPCR mixture consisted of 10 μl of 2 × QX200 ddPCR EvaGreen Supermix, 100 nM of each primer, and 1 μl of sample DNA. The final volume (20 μl) was prepared using nuclease-free water. The reaction mix was converted into droplets using the Bio-Rad QX200 automatic droplet generator and amplified in a Bio-Rad C1000 thermocycler under the following conditions (ramping rate 2.5°C/s): one cycle at 95°C for 5 min (denaturation), 40 cycles of 95°C for 30 and 60 s at the respective annealing temperatures (Supplementary Table S1), and one final cycle of 4°C for 5 min and 90°C for 5 min. Data were analyzed with the QuantaSoft Software version 1.7 (Bio-Rad).
The absolute abundances of donor P. putida and plasmid RP4 in the compost samples were transformed into lg(Ct/C0), where C0 and Ct represent the initial abundances and abundances at time t (days), respectively (Fan et al., 2019). The “spread potential (SP)” was used to denote the dissemination of RP4 plasmid in the compost microbiota. SP refers to transconjugants/recipients, and is roughly calculated by the following formula:
Cgfp − CdsRed represents the number of transducers per gram of DW, assuming that each donor strain harbors a plasmid. According to the ribosomal RNA database, the average copy number of 16S rRNA genes per bacterium is estimated as 4.1 (Klappenbach et al., 2001). C16SrRNA/4 represents the total number of bacteria per gram DW.
The concentrations of the donor P. putida and plasmid RP4 in the compost samples were transformed using the logarithmic function before statistical analysis. The means and standard deviations were determined, and linear regression modeling was used to describe the decay of P. putida and plasmid RP4 during composting. Decimal reduction times (D-values; the time taken to reduce the concentration of dsRed and gfp by 1 log) for all composting treatments were calculated using the absolute estimated inverse values of the slope of the linear regression analyses.
Bacteria from the collected compost samples were harvested using Nycodenz density-gradient centrifugation (Musovic et al., 2010) and resuspended in PBS. Compost samples (1.0 g) were homogenized in 4 ml of tetrasodium pyrophosphate (TTSP, 50 mM, and Tween 80, 0.05%), vortexed and sonicated in an ice-cold water bath (Ultrasonic Cleaner KQ-500B, Kunshan, China) for 10 min. After 10 s of vortexing, the suspension was carefully transferred to Nycodenz (Nycomed Pharma, Oslo, Norway) solution with a density of 1.3 gml−1, followed by centrifugation (10,000× g for 10 min). The upper and middle phases containing a bacterial layer were collected and transferred to a new tube, diluted 1:5 (v/v) in PBS, and centrifuged (5,000× g for 10 min). The supernatant was discarded and the pellet resuspended in 1 ml of PBS, and filtered by sterile 20-μm-pore-size filters. After filtration, the suspensions were analyzed using flow cytometry.
The transconjugants with the obtained RP4 plasmid in the compost were sorted on an S3e Cell Sorter (Bio-Rad, Hercules, CA, United States). GFP fluorescence was excited with a 488 nm laser beam and checked in a band-pass filter 1 (525/30 nm), whereas the fluorescence of dsRed was excited at 561 nm and collected with filter 2 (586/25 nm). E. coli MG1655 and E. coli MG1655 (RP4::gfp) were used as the negative control for fluorescence and gfp-positive control, respectively, and P. putida KT2442 (RP4::gfp) was used as gfp-negative and dsRed-positive controls. The signal cells were gated based on forward scatter (FSC) and side scatter (SSC), filter 1, and filter 2. The cells with high green fluorescence and no red fluorescence were sorted and collected in a 5 ml sterile polystyrene round-bottom tube (Falcon, Corning, Wiesbaden, Germany) with 0.5 ml sterile PBS solution. The sort precision was set in the default purity mode (Fan et al., 2019).
Sorted cells of each sample were initially collected in 5 ml sterile round-bottom polystyrene tubes (Falcon, Corning, Wiesbaden, Germany) with 0.5 ml of PBS solution, and then centrifuged at 10,000× g for 30 min in 1.5 ml tubes to collect the pellets (Klümper et al., 2015). The supernatant was carefully discarded, and the cell pellet was resuspended in 20 μl of lysis buffer (Platinum™ Direct PCR Universal Master Mix, Invitrogen, Thermo Fisher, United States) and 0.6 μl of proteinase K. The lysis mixtures were incubated at 25°C for ≥1 min, and the samples were placed in a pre-heated heat block and incubated at 98°C for 1 min. The DNA-containing cell lysate was centrifuged, and the lysate supernatant was used directly for subsequent PCR. The V3-V4 region of the bacterial 16S rRNA gene was amplified using universal primers (338F and 806R) and KAPA HiFi HotStart ReadyMix (KAPA Biosystems). The PCR thermocycler conditions were as follows: initial denaturation at 95°C for 3 min; 29 cycles at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 45 s; with a final extension at 72°C for 10 min. Amplicons were purified by gel extraction using the AxyPrep DNA Gel Extraction Kit (Axygen) and quantified using QuantiFluor-ST (Promega, Madison, WI, United States). Purified amplicons were pooled in equimolar concentration and paired-end sequenced by the Illumina MiSeq PE 300 platform (Shanghai Majorbio Bio-Pharm Technology).
Raw fastq sequencing data were trimmed and quality-filtered using FastP version 0.20.0 (Chen et al., 2018) and merged with FLASH version 1.2.7 (Magoč and Salzberg, 2011). Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using UPARSE version 7.1 (Edgar, 2013), while the chimeric sequences were identified and removed using UCHIME (Edgar et al., 2011). OTU was classified by the RDP Classifier 2.21 against the Silva (SSU138) 16S rRNA database using a confidence threshold of 70% (Wang et al., 2007). The indices of alpha diversity including Sobs, Chao, Ace, Shannon, Simpson, and Good’s coverage were analyzed by Mothur version v.1.30.2 (Schloss et al., 2009). Heatmap was generated using the R package pheatmap.2 Phylogenetic trees were constructed using MEGA 7 (Kumar et al., 2016) and visualized using iTOL3 (Letunic and Bork, 2016).
Tukey’s test was applied to test differences in the abundance of dsRed or gfp genes among different treatments. The indices of alpha diversity were shown by the mean ± standard deviation and tested for significant differences between different groups by Duncan’s test. In the beta diversity analysis, the Bray–Curtis dissimilarity matrix was visualized using principal coordinate analysis (PCoA) based on the relative abundance of OTUs in different samples. The line regressions of gfp and dsRed were performed with the R package ggpmisc.
Based on the measured temperature profile, both treatments progressed through mesophilic, thermophilic, and maturing phases, as defined in other composting processes (Echeverria et al., 2012; Supplementary Figure S2). The temperatures of the two treatments rose sharply to >55°C on day 2 and maintained the thermophilic phase until day 11, with a maximum temperature of 60.2°C. The temperature of NT treatment decreased from day 12 to room temperature on day 27, indicating that the fermentation process was complete. With an extra heater system, the CT treatment had a thermophilic time of 15 days longer than that of the NT treatment, followed by maturation at room temperature until day 40.
The target genes dsRed and gfp were quantified by ddPCR to determine the abundance of P. putida and RP4 plasmids in the compost samples. As shown in Figure 1, both P. putida and plasmid RP4 showed a similar decreasing trend in both the NT and CT treatments. The abundance of dsRed and gfp in the NT treatment rapidly decreased by 1.46 and 1.73 log to 1.13 × 105 and 2.14 × 105 copies/g DW within 19 days, respectively, and then increased slightly to 2.11 × 105 and 4.70 × 105 copies/g DW on day 27, respectively. In the CT treatment, the dsRed abundance decreased by 1.40 log to 1.31 × 105 copies/g DW after 27 days of composting, and increased slightly to 1.63 × 105 copies/g DW on day 40. The abundance of gfp continuously decreased by 1.71 log to 2.08 × 105 copies/g DW on day 40. The patterns of P. putida and RP4 plasmid decreases in both NT and CT treatments are shown as linear regressions in Supplementary Figure S3. Both P. putida and RP4 fates showed biphasic decay behaviors during composting, which was similar to a study in soil by Fan et al. (2019). A linear regression equation was used to calculate the D-values of dsRed and gfp during composting (Supplementary Table S2). The results showed that the D-values of the target genes in the two treatments in phase I were approximately 7.0 days. After the thermophilic phase, the absolute abundance of dsRed and gfp in the CT treatment was significantly lower than that in the NT treatment, which was only 32.4% and 36.6% of that in the NT treatment, respectively (Tukey’s test, p < 0.01, Figure 2). At the end of the mature phase, the absolute abundance of dsRed and gfp in the CT treatment was 77.1% and 44.3% of that in the NT treatment, respectively. The results showed that prolonging the thermophilic phase of composting was beneficial to the decay of donor bacteria and RP4 plasmids. After the thermophilic phase, the removal efficiencies of dsRed and gfp were 96.0% and 97.7% with the CT treatment, and 87.7% and 93.8% with the NT treatment, respectively. At the end of the maturation phase, the removal efficiencies of dsRed and gfp in the CT treatment (95.1% and 98.0%, respectively) were slightly higher than those in the NT treatment (93.6% and 95.5%, respectively; Supplementary Figure S4). The results showed that thermophilic composting could effectively remove dsRed and gfp, and that the removal efficiency of CT was greater than that of NT.
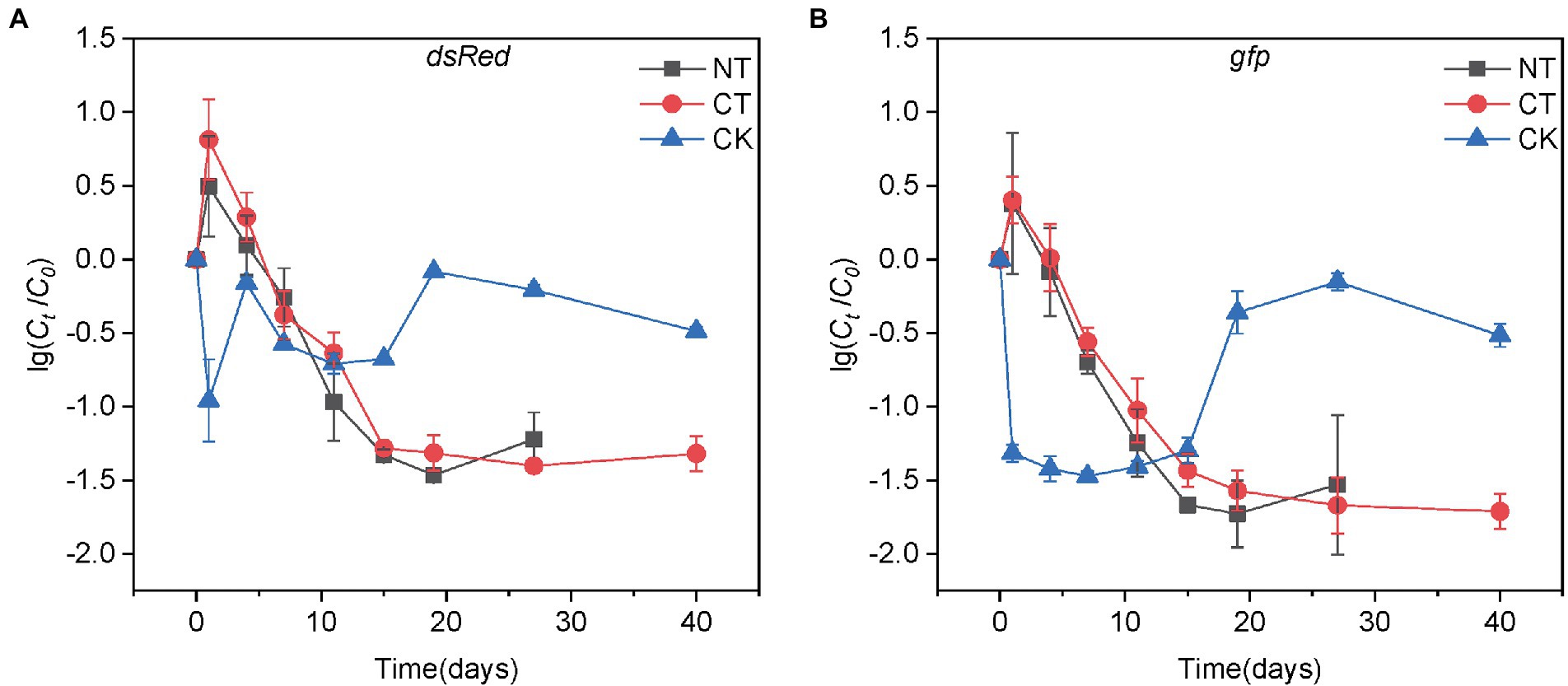
Figure 1. The decay of the donor (A) and plasmid (B) during normal thermophilic (NT) and continuous thermophilic composting (CT). The log values of the ratios of donor (plasmid) concentration at time t (Ct) to the initial concentration (C0) are shown on the y-axis. The donor and plasmid concentrations are represented by the copy numbers of dsRed and gfp genes, respectively. Error bars represent the standard deviation (SD) of sampling replicates at each time point.
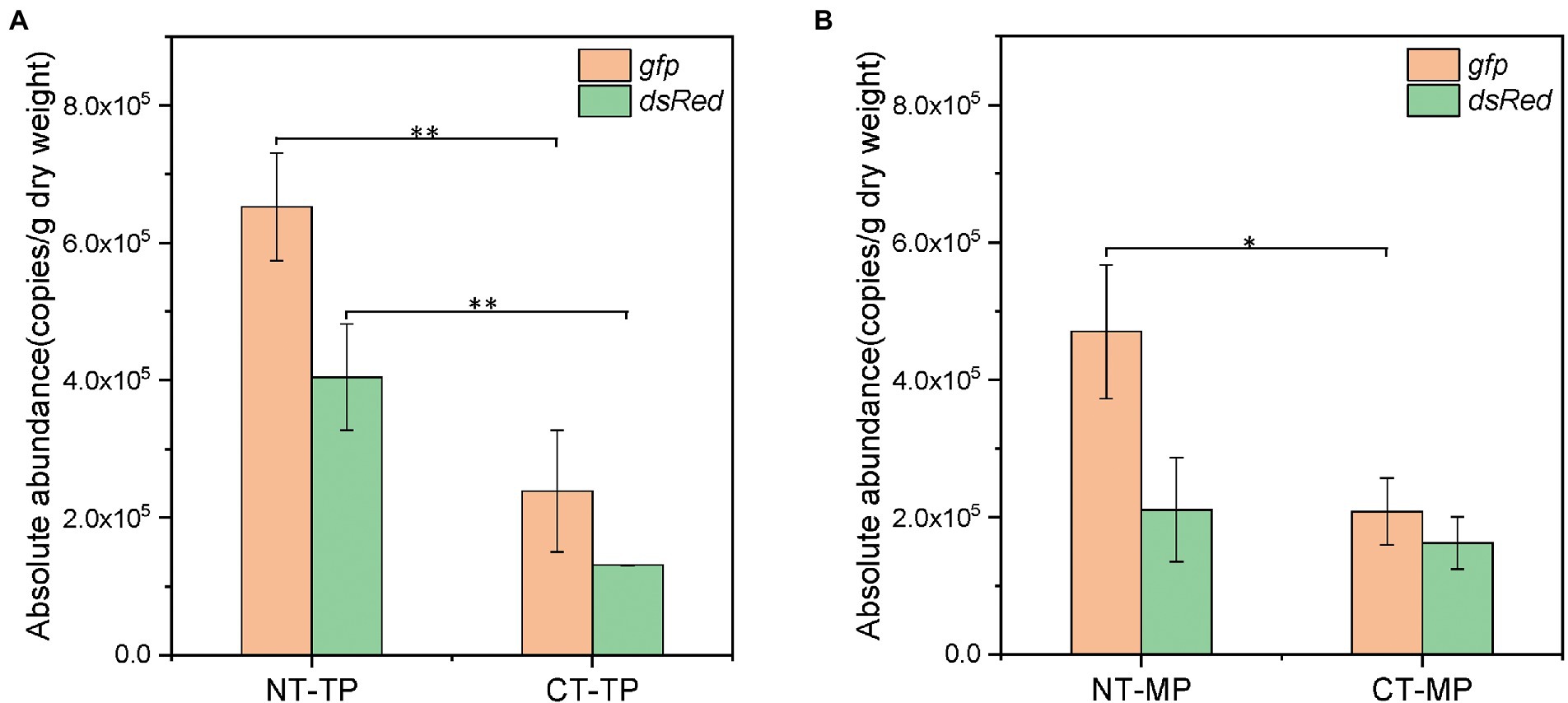
Figure 2. Abundance of dsRed and gfp genes in two composting treatments at the end of the thermophilic phase (A) and mature phase (B). *p < 0.05; **p < 0.01 (Tukey’s test); NT, normal thermophilic composting; CT, continuous thermophilic composting; TP, thermophilic phase; MP, mature phase.
Based on the target gene abundance tested using ddPCR, we calculated the SP in the composting samples to estimate the spread and proliferation potential of RP4 plasmid during the composting process. The results showed that the SP in the raw material (IP sample) was 3.91 × 10−4 transconjugants per recipient (TC/R), whereas both the NT- and CT-treated samples decreased from 10−4 to 10−6 (TC/R) after composting. However, the SP in CT-treated samples was 2.54 × 10−6 (TC/R) in the final composts, which was lower than that of the NT treatment (Supplementary Table S3). Our results indicated that prolonging the thermophilic time was more favorable for reducing the conjugative transfer potential of plasmid RP4 than normal composting.
Combined with flow cytometry and 16S rRNA gene amplicon sequencing, the community composition of transconjugants obtaining RP4 plasmid was identified. In all 18 samples, 1,093,566 effective sequences were obtained after quality control. The effective sequences were clustered at 97% identity into OTUs, and 220 transconjugant OTUs were identified. Sobs, Chao, Ace, Shannon, Simpson, and Good’s coverage indices were used to assess the alpha diversity (Table 1). In our study, the Good’s coverage was ≥99% for all samples, indicating that the current sequencing depth was sufficient to saturate the transconjugant diversity. Compared with normal composting, prolonging the thermophilic phase of composting significantly decreased the species richness (Sobs, Ace, and Chao indices) and diversity (Shannon index) of RP4 plasmid transconjugants at the end of the thermophilic phase (p < 0.05). PCoA based on Bray–Curtis distance at the OTU level showed that the community structure of transconjugants was significantly different among the five groups (Figure 3, Adonis analysis, p = 0.001). The taxonomic compositions of the thermophilic (TP samples) and maturation (MP samples) phases were both significantly different from those of the raw material (IP samples), which indicated that composting could shape the structure of RP4 plasmid transconjugants. Because of the prolonged thermophilic phase, the transconjugants of the thermophilic and mature phases between the CT and NT treatments differed from each other.
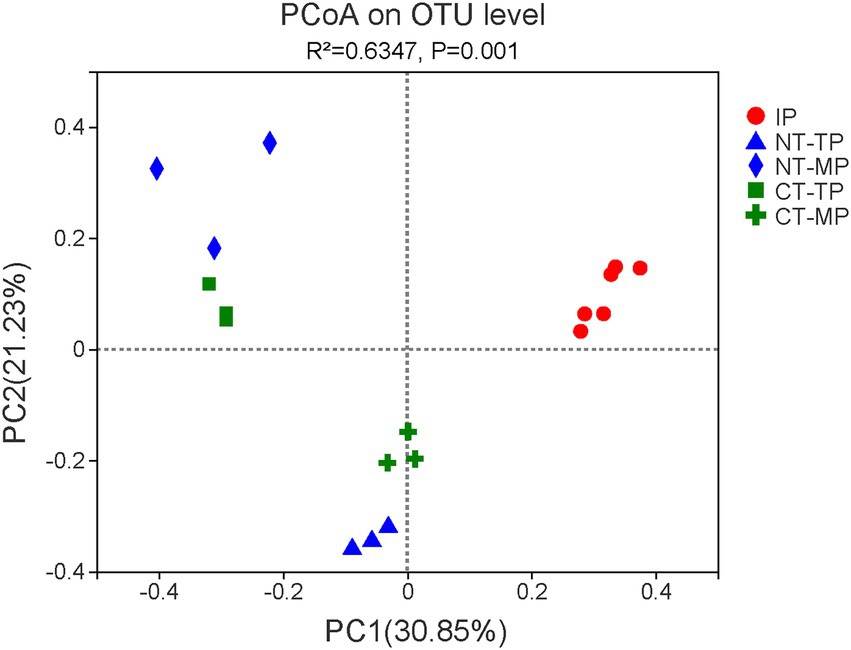
Figure 3. The overall distribution pattern of operational taxonomic unit (OTU)-based transconjugant community dissimilarity in the normal thermophilic (NT) and continuous thermophilic composting (CT). Principal coordinate analysis (PCoA) of beta diversity based on the Bray–Curtis distance analysis of the OTU level. IP, TP, and MP represent the initial, thermophilic, and mature phases, respectively.
Across all samples, 220 permissive OTUs distributed over eight phyla were identified: Firmicutes, Proteobacteria, Actinobacteria, Halanaerobiaeota, Bacteroidota, Cyanobacteria, Myxococcota, and Deinococcota, and thermophilic composting had an obvious influence on the community composition of transconjugants (Figure 4). At the phylum level, the phylogenetic compositions of all the transconjugant pools were similar, but the exact community compositions differed. Firmicutes accounted for 75.3%–90.1% of RP4 plasmid transconjugants, which was the dominant phylum in all samples. High temperature in the thermophilic phase inhibited the growth of transconjugants from Proteobacteria and Actinobacteria, and their relative abundances decreased after the thermophilic phase. However, Proteobacteria transconjugants showed an obvious rebound phenomenon at the mature phase, with the relative abundance increasing from 4.3% to 16.8% in the NT treatment and from 8.2% to 22.2% in the CT treatment. Compared with the NT, the CT treatment had fewer Bacteroidota and Halanaerobiaeota at the end of the mature phase, indicating that CT treatment more efficiently inhibited the growth of Bacteroidota and Halanaerobiaeota, obtaining RP4 plasmid in the final composts. Halanaerobiaeota was completely removed after composting, indicating that prolonged thermophilic time could effectively inhibit Halanaerobiaeota bacteria from obtaining the RP4 plasmid.
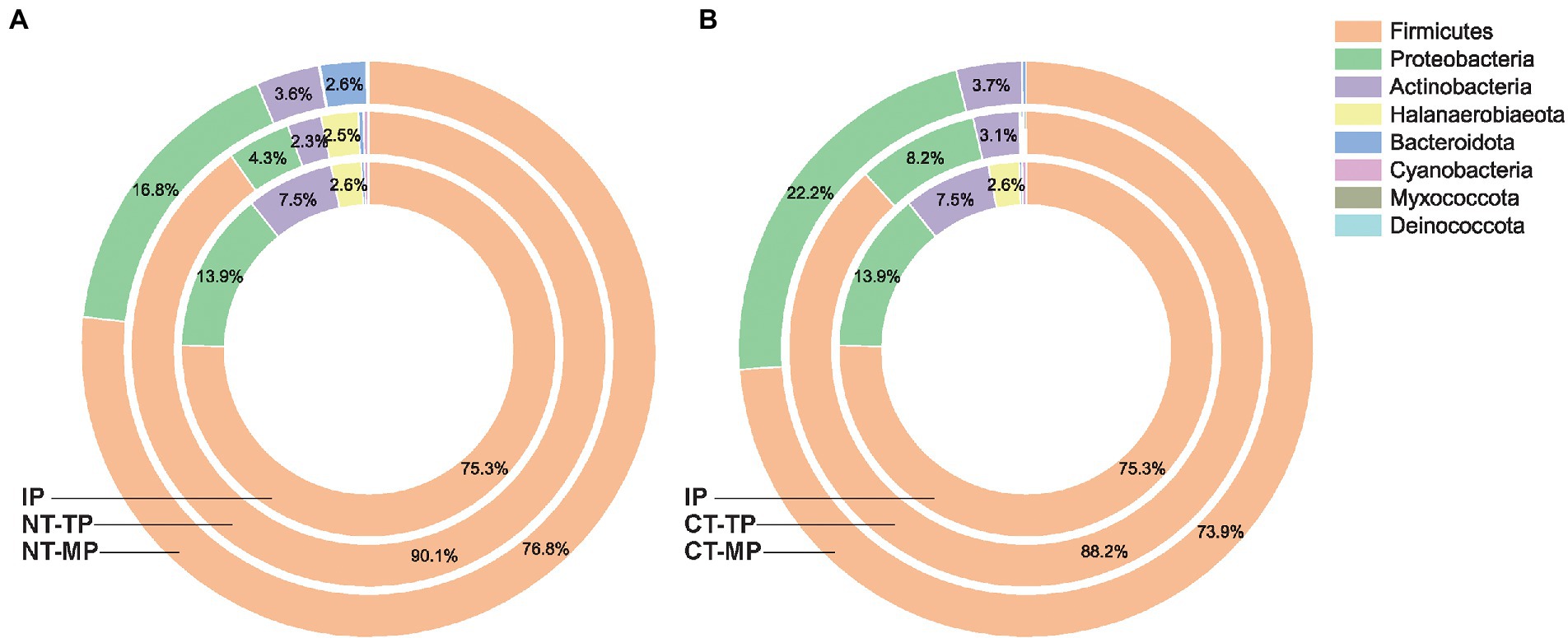
Figure 4. Dynamic changes in community composition of sorted transconjugant pools at the phylum level in the normal thermophilic (A) and continuous thermophilic composting (B). NT and CT represent normal and continuous thermophilic composting, respectively; IP, TP, and MP represent the initial, thermophilic, and mature phases, respectively.
For the number of genera in the composts, the transconjugant pools of the two composting treatments (NT and CT) shared 50.00% and 33.05% genera among the three composting phases (IP, TP, and MP), respectively (Figures 5A,B). Shared genera during composting could be considered the core host community of the RP4 plasmid. Fewer shared genera represented a smaller core transfer host range in the CT treatment than in the NT treatment. For the unique genera in composts, at the end of the mature phase, both NT and CT had notably fewer unique host genera (five and four genera, respectively) than that (46 genera) in CK samples without composting (Supplementary Figure S5). Unique genera represent possible new recipients for the RP4 plasmid, so fewer unique genera at the end of composting indicated that composting could efficiently control the conjugative transfer opportunity to new genera in organic fertilizer. However, at the end of the thermophilic phase of composting, the NT and CT treatments shared 52 genera, with 41 unique genera to NT and only 9 unique genera to CT (Figure 5C); at the end of the mature phase, the NT and CT treatments shared 54 genera, with 31 unique genera to NT and 17 unique genera to CT (Figure 5D), indicating significantly decreased diversity of transconjugants in the CT treatment. These results show that CT treatment is more conducive to reducing the diversity of transconjugants than NT.
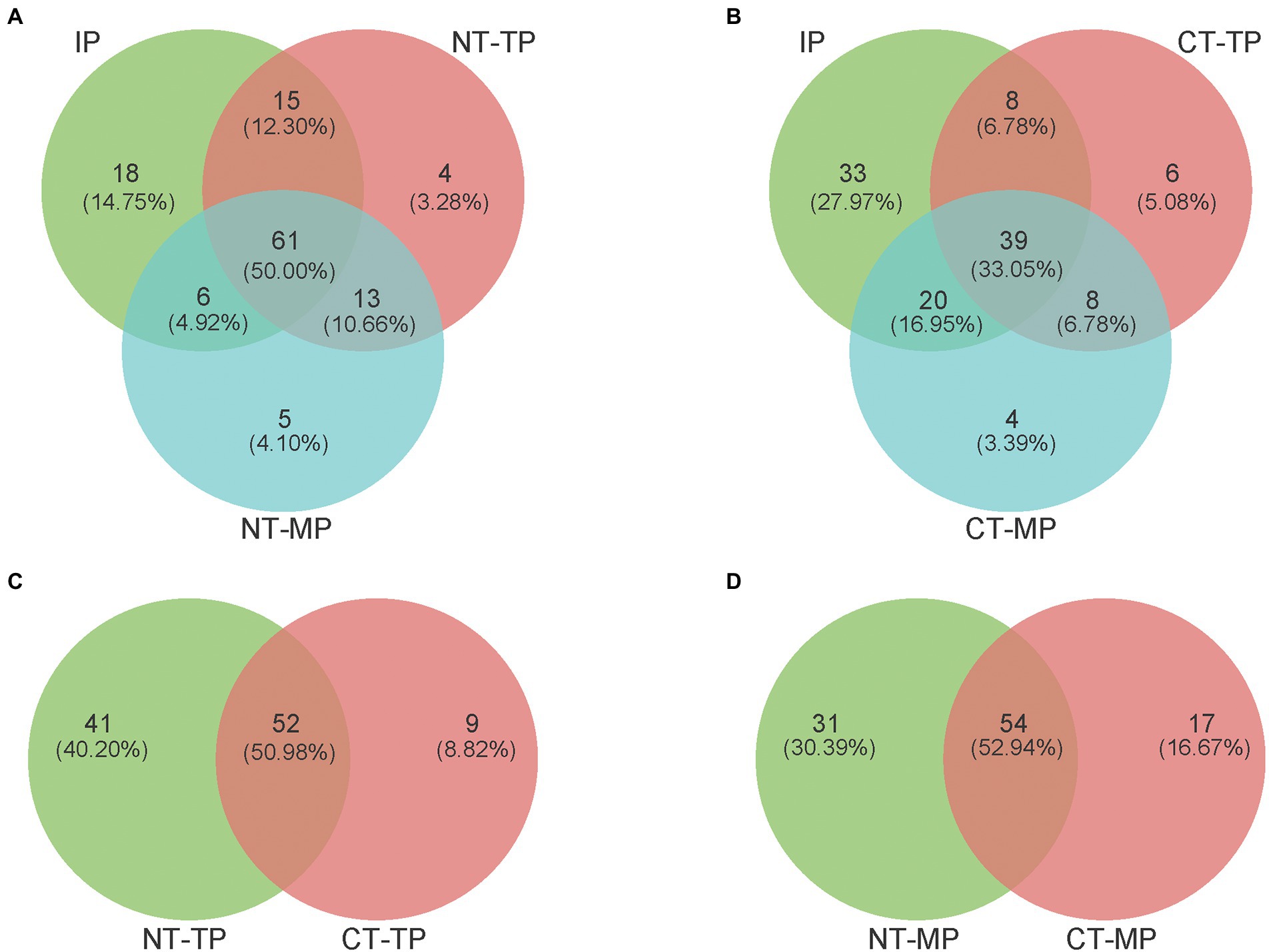
Figure 5. Venn diagrams reveal the shared and unique numbers of transconjugant pools for plasmid RP4 transferred at the genus level. (A,B) Show the number of shared and unique genera among the three composting phases (IP, TP, and MP) in the NT and CT, respectively. (C,D) Diagrams show the number of shared and unique genera between NT and CT at the end of the thermophilic and mature phase, respectively. NT and CT represent normal and continuous thermophilic composting, respectively; IP, TP, and MP represent the initial, thermophilic, and mature phases, respectively.
The heatmap of the top 30 genera was generated to visualize the transconjugant community composition better at the genus level (Figure 6). The genera in group A, such as Oceanobacillus, Bacillus, Bacillaceae, and Sinibacillus, were the dominant conjugants in all the samples during composting. Oceanobacillus, accounting for 27.41% of the total relative abundance of transconjugants, was the most abundant host in the raw material. Bacillus (44.66%) and Bacillaceae (19.66%) became the most abundant hosts in the thermophilic phase in CT and NT, respectively. After the maturation phase, Bacillus (48.41%) became the most dominant genus in the NT treatment, followed by Oceanobacillus (9.66%), Halomonas (7.65%), Bacillaceae (5.92%), Fodinicurvataceae (3.41%), Sinibacillus (3.07%), and Staphylococcus (3.07%). Oceanobacillus (30.23%) was still the most dominant genus in the CT treatment, followed by Bacillaceae (13.63%), Bacillus (12.65%), Fodinicurvataceae (6.37%), Sinibacillus (5.49%), Marinococcaceae (3.72%), and Paracoccus (2.85%). The genera in group B represented the removal of RP4 plasmid hosts such as Halocella, Solibacillus, Atopostipes, Amphibacillus, and Cerasibacillus during composting. In particular, some opportunistic pathogenic bacteria such as Corynebacterium, Escherichia, Shigella, Staphylococcus, Pseudomonas, and Burkholderia received the ARG-carrying RP4 plasmid and were partially removed after composting.
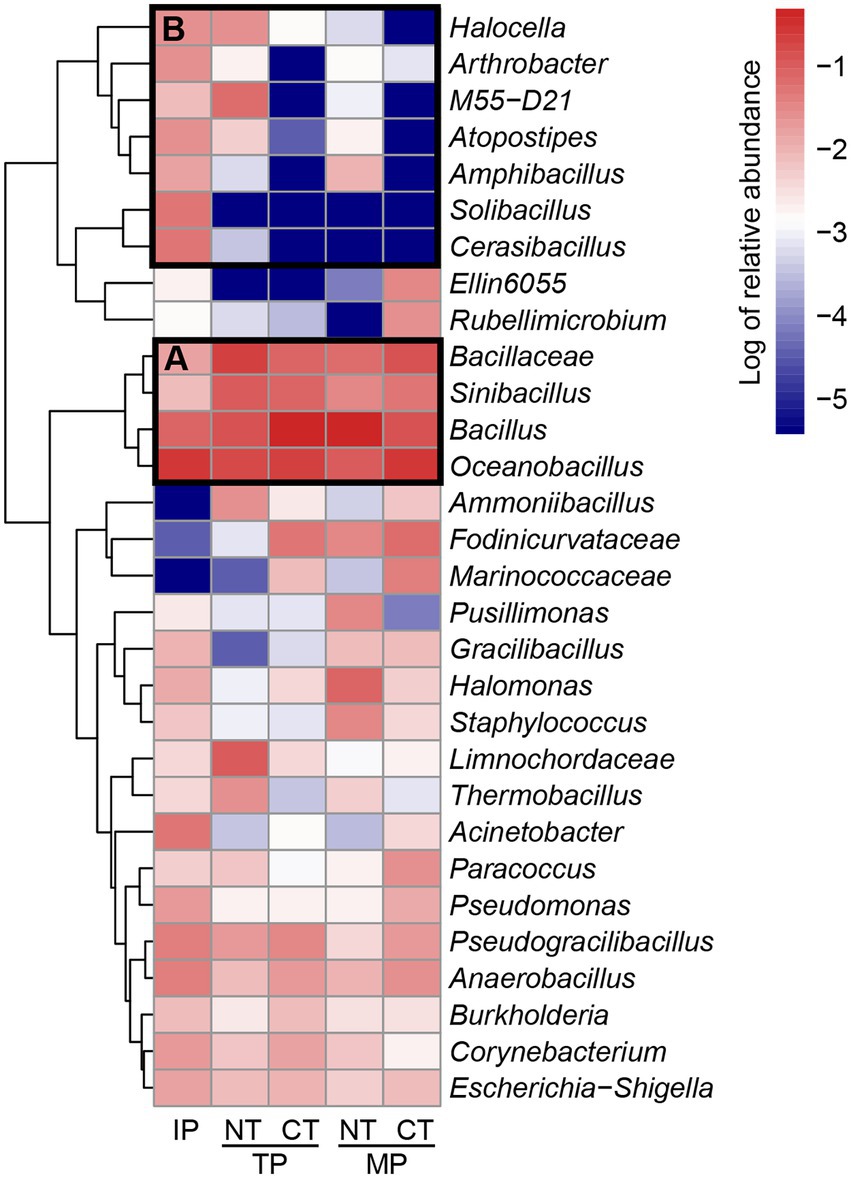
Figure 6. Relative abundance of top 30 abundant genera in the transconjugant pools during normal thermophilic (NT) and continuous thermophilic composting (CT). IP, TP, and MP represent the initial, thermophilic, and mature phases, respectively. (A) dominant conjungants in all samples; (B) removal of RP4 plasmid hosts.
Phylogenetic trees were constructed from OTUs with greater than 0.01% relative abundance in the transconjugant pools. As shown in Figure 7, Firmicutes comprised the major OTUs, accounting for more than 70% of the bacterial community structure. The RP4 plasmid was transferred from donor strain to a wide range of recipients, including the phylogenetically distant species. Notably, some dominant Gram-positive transconjugants were also identified, such as Firmicutes and Actinobacteria (Figure 7).
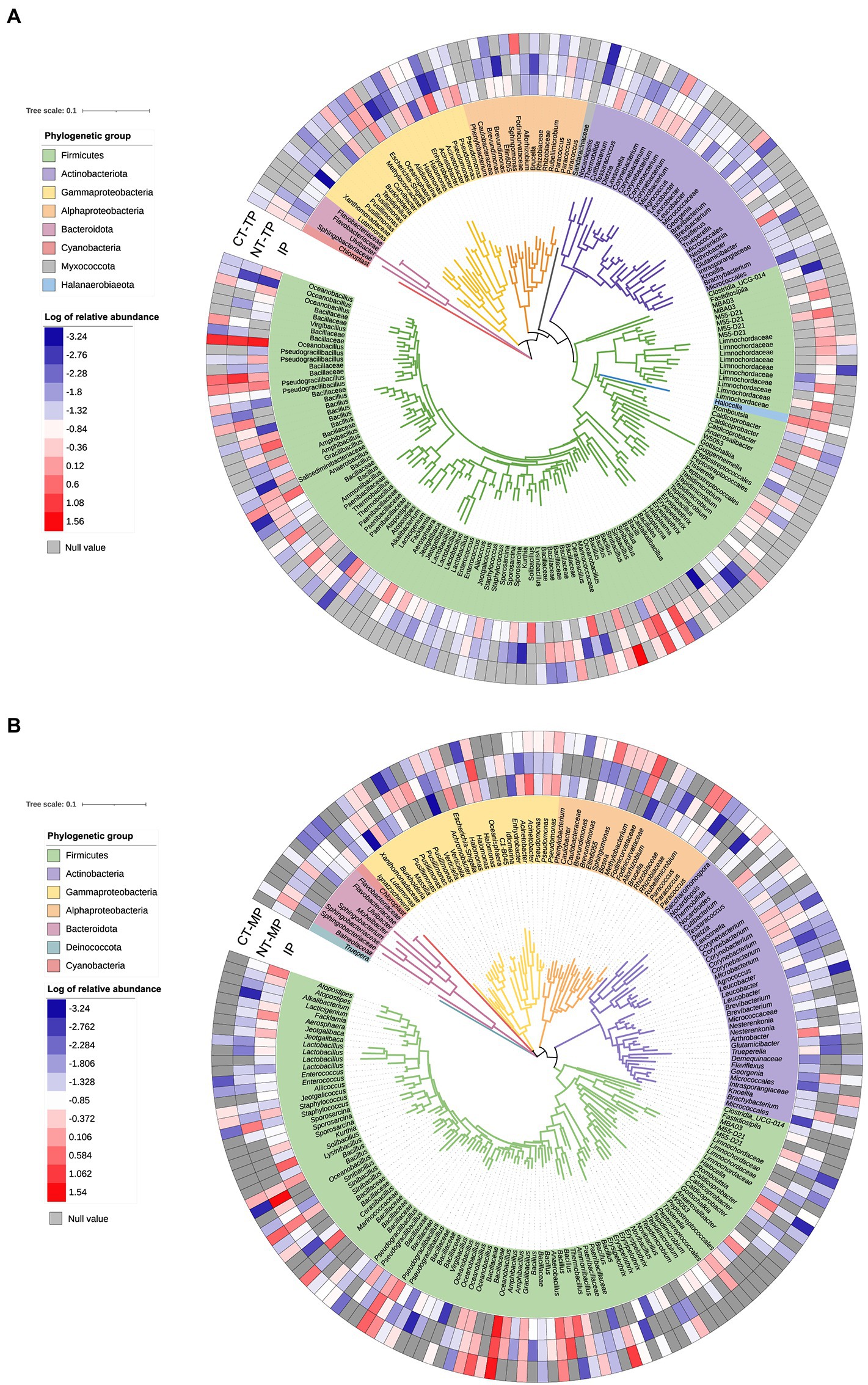
Figure 7. Phylogenetic tree showing the identified transconjugant OTUs that contributed more than 0.01% in normal thermophilic (NT) and continuous thermophilic composting (CT). (A) Showing transconjugants in the thermophilic phase. (B) Showing transconjugants in the mature phase. Colors of the branches mark different phylogenetic groups. Heatmap circles at the periphery of the tree represent the log of relative abundance of the OTU in the transconjugant pools. IP, TP, and MP represent the initial, thermophilic, and mature phases, respectively.
Composting is an effective way to utilize waste resources, and high temperatures are an important factor for the inactivation of pathogenic microorganisms in organic waste (Nakasaki et al., 1985). In our study, P. putida carrying the RP4 plasmid was used as a model strain to study the dynamic changes of donor and broad-host-range plasmids in the compost microcosm under a prolonged thermophilic phase. By quantifying the marker genes dsRed and gfp using ddPCR, we observed that P. putida and RP4 plasmids in CT demonstrated a biphasic pattern of decay, which included a fast decay during the first 15 days followed by a slow decay after the 15th day (Supplementary Figure S3). Pseudomonas putida and RP4 plasmids in the NT treatment had a similar first decay to the CT treatment, but instead of decay, they both increased at the second stage without prolonging the thermophilic phase (Supplementary Figure S3). These results suggest that thermophilic composting could effectively remove donor bacteria and RP4 plasmids by preventing their rebound during the mature phase. Linear regression equation was used to calculate the D-values of P. putida and RP4 plasmids for all treatments (Supplementary Figure S3; Supplementary Table S2). During composting, P. putida had a larger D-value (6.9–7.4 days) than E. coli (0.27–4.82 days; Thomas et al., 2020). Pseudomonas is a Gram-negative mesophilic bacterium that is sensitive to high temperatures during composting. However, Pseudomonas, including the pathogen Pseudomonas aeruginosa, could still be isolated from mature composts because of the temperature difference between the inner and outer areas of composting (Kaszab et al., 2011). However, the abundance of gfp rebounded at the maturation stage in the NT treatment but not in the CT treatment, which may be caused by the regrowth of some RP4 plasmid hosts in the NT treatment. The result was consistent with the fact that the richness of RP4 plasmid transconjugants in the CT treatment was lower than that in the NT treatment during the maturation phase (Table 1). The better removal effect of CT on ARG-carrying RP4 plasmid may be due to the elimination of some non-thermophilic ARG hosts or inhibition of HGT by high temperatures (Miller et al., 2016; Qian et al., 2016).
In addition to reducing the abundance of ARB and their plasmids, the high composting temperature mitigated the frequency of conjugation. SP (3.91 × 10−4 TC/R) in the raw material is similar to the study (4.65 × 10−4 TC/R) of Fan et al. (2019); however, in contrast to the final increase in the soil environment, SP significantly decreased to 2.54–6.73 × 10−6 (TC/R) at the end of composting (Fan et al., 2019). Compared with mesophilic digestion, thermophilic digestion is more conducive to resisting the HGT of ARGs in bacterial communities (Miller et al., 2016). Although our results directly demonstrate that composting can effectively control the transfer rate of broad-host conjugative plasmids, some studies indicate that composting is not effective for all conjugative plasmids (Le Devendec et al., 2016), particularly sul1-related plasmids that may survive transfer to heat-resistant bacteria (Lin et al., 2021).
The initial abundance of dsRed and gfp were averaged at 3.30 × 106 and 1.04 × 107 copies/g DW, respectively, and increased slightly on the first day after fermentation. This could be due to the division of the donor strain, consistent with previous studies (Henschke and Schmidt, 1990; Fan et al., 2019). We found that plasmid transfer occurred at the beginning of composting using flow cytometric analysis and sorting of transconjugants from the first day samples. A previous study reported that transconjugants appeared immediately after P. putida BH(RP4) or E. coli C600(RP4) was introduced into soil microcosms (Inoue et al., 2009). The ddPCR results of dsRed and gfp show that the removal efficiencies of donor strains and RP4 plasmid in CT were higher than that in NT composting. The persistence of ARG-carrying plasmids during manure storage and composting were observed, and a high temperature of composting could inhibit the transfer of RP4 plasmid based on culture-dependent methods (Guan et al., 2007; Le Devendec et al., 2016). Our culture-independent results add more evidence that composting could reduce the abundance and transfer of ARG-carrying plasmids through prolonging the thermophilic phase. Besides the quantities of donors with plasmids, the composition of the recipient pool is another important factor affecting plasmid HGT (Lopatkin et al., 2016). It has been reported that the HGT frequency is usually affected by cell density (Johnsen and Kroer, 2007), therefore HGT may vary in different composting phases because of a different recipient community.
The broad-host-range plasmid RP4 has an extremely broad transfer range, including both Gram-negative and Gram-positive bacteria (Musovic et al., 2014; Klümper et al., 2015; Li et al., 2018). Here, we demonstrated that plasmid RP4 transfers to a diverse fraction of the bacterial community during composting. PCoA analysis showed that the composition of the transconjugant community differed between the NT and CT treatments, and changed with different fermentation periods (Figure 3). Compared with NT, prolonging the thermophilic phase leads to a decrease in the diversity and richness of transconjugant communities (Table 1). The accumulated temperature, including the temperature intensity and duration of the thermophilic phase, could profoundly affect the bacterial community (Liu et al., 2021). Liao et al. (2018) found that a higher composting temperature had better efficiency in reducing the abundance of potential ARG bacterial hosts such as Bacillus which was also enhanced in our composting especially at the thermophilic phase (Figure 6). Continuous thermophilic composting or external heating had a better performance at reducing human pathogenic bacteria (Qian et al., 2016; Keenum et al., 2021). Both a high temperature and prolonged thermophilic time may supply a small size recipient pool for the conjugative plasmid, so the total number and abundance of transconjugant genera carrying plasmid RP4 in the CT treatment were significantly lower than those in the NT treatment at the end of both the thermophilic and maturation phases (Figures 5C,D, 7). This finding clearly revealed that the transfer of multidrug-resistant plasmids could effectively be weakened by prolonging the thermophilic period.
Phylogenetic analysis of transconjugants sorted by flow cytometry indicated that RP4 disseminated to a broad transfer range (covering eight phyla) of hosts during composting, including the phylogenetically distant group (Figure 7). Firmicutes was the most abundant phylum, which differs from previous studies reporting that Proteobacteria is the predominant host for the transfer of RP4 plasmid in soil (Klümper et al., 2015, 2017; Fan et al., 2019), plant rhizosphere (Xu et al., 2021), and wastewater treatment plants (Li et al., 2018). On the other hand, it is consistent with the results of major hosts at mesophilic and thermophilic phases from a transcriptional resistome study of manure composting (Wang et al., 2017). These results revealed that environmental niches and experimental conditions could determine the host profile of multidrug-resistant conjugative plasmids. Firmicutes was significantly enriched during the thermophilic phase of both composting treatments. The specific property of Firmicutes to form endospores may be an explanation for this enhancement (de Hoon et al., 2010); therefore, they are tolerant to high temperature (Hartmann et al., 2014) and can survive environmental stresses (Barnard et al., 2013), which is the main reason why Firmicutes is the dominant RP4 plasmid-carrying bacteria during composting. Based on co-occurrence network analysis and conjugative experiments, Firmicutes, such as Lactobacillus, Bacillus, and Staphylococcus, are also potential ARG hosts after composting (Lin et al., 2021; Sardar et al., 2021; Zhou et al., 2021). Proteobacteria was the second predominant phylum of transconjugants in our samples, and its relative abundance, including Alphaproteobacteria and Gammaproteobacteria, increased after the mature phase. Alphaproteobacteria, including Fodinicurvataceae, Paracoccus, Sphingomonadaceae (Ellin6055), and Rubellimicrobium, made major contributions to the “rebound” of Proteobacteria at the end of composting (Figure 7B). Fodinicurvataceae and Rubellimicrobium are thermophilic bacteria associated with humic acid conversion during composting (Ye et al., 2021; Fu et al., 2022). Sphingomonadaceae (Ellin6055) and Paracoccus are two abundant hosts of another broad-host-range plasmid (pKJK5) in the soil (Klümper et al., 2017). Notably, we found transfer from the used Gram-negative donor strain (Pseudomonas) to a wide variety of Gram-positive bacteria belonging to Firmicutes and Actinobacteria (Figure 7), indicating the inter-Gram gene transfer during composting. The most dominant family containing Gram-positive bacteria was Bacillaceae, including Bacillus, Oceanobacillus, Sinibacillus, Pseudogracilibacillus, Anaerobacillus, and Cerasibacillus. Similar RP4 plasmid transfers have been found in the soil and plant endosphere (Klümper et al., 2015; Xu et al., 2021). The shared transfer host bacteria of plasmids among composts, soil, and the plant endosphere indicate a potential risk for plasmid-mediated ARG exchange through fertilizer utilization on farms.
In this study, we found a biphasic decay behavior of P. putida and RP4 plasmid during composting, and prolonging thermophilic composting supplied a slow decay after 15 days. Composting could mitigate the spread of the RP4 plasmid, and the SPddPCR significantly decreased at the final compost. Prolonging the thermophilic phase could effectively reduce the diversity and richness of the RP4 host community. Different from previously studied niches such as soil, plant rhizosphere, and wastewater, Firmicutes was the predominant host in the composting transconjugant pools. Our findings provide a feasible solution for mitigating the HGT of ARGs during composting and inhibiting the spread of ARGs from animals to farmland.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://ngdc.cncb.ac.cn/gsa/, CRA006962.
XW and GZ conceived the idea, edited the manuscript, and made substantial contribution to the conception of the work. LS, TQ, and HG performed the composting, ddPCR, and FACS. LS, TQ, and YG performed sequencing and its analysis. MG performed the statistical analysis. LS and TQ wrote the original manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Beijing Natural Science Foundation (grant number 6222013), the Special Program for Creative Ability Foundation of BAAFS (grant number KJCX20210424), and Beijing Agriculture Innovation Consortium (BJJQ-G08).
We thank Barth F. Smets (Technical University of Denmark) for kindly providing Pseudomonas putida KT2442 and plasmid RP4, and Xiaoting Fan and Jianqiang Su from the Institute of Urban Environment for providing help on strain culture and cell sorting.
The authors declare that the work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.989085/full#supplementary-material
Barnard, R. L., Osborne, C. A., and Firestone, M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Binh, C. T. T., Heuer, H., Kaupenjohann, M., and Smalla, K. (2008). Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 66, 25–37. doi: 10.1111/j.1574-6941.2008.00526.x
Cao, R., Wang, J., Ben, W., and Qiang, Z. (2020). The profile of antibiotic resistance genes in pig manure composting shaped by composting stage: mesophilic-thermophilic and cooling-maturation stages. Chemosphere 250:126181. doi: 10.1016/j.chemosphere.2020.126181
Che, Y., Yang, Y., Xu, X., Břinda, K., Polz, M. F., Hanage, W. P., et al. (2021). Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl. Acad. Sci. U. S. A. 118:e2008731118. doi: 10.1073/pnas.2008731118
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
de Hoon, M. J., Eichenberger, P., and Vitkup, D. (2010). Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 20, R735–R745. doi: 10.1016/j.cub.2010.06.031
Echeverria, M. C., Cardelli, R., Bedini, S., Colombini, A., Incrocci, L., Castagna, A., et al. (2012). Microbially-enhanced composting of wet olive husks. Bioresour. Technol. 104, 509–517. doi: 10.1016/j.biortech.2011.11.042
Edgar, R. C. (2013). Uparse: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fan, X. T., Li, H., Chen, Q. L., Zhang, Y. S., Ye, J., Zhu, Y. G., et al. (2019). Fate of antibiotic resistant pseudomonas putida and broad host range plasmid in natural soil microcosms. Front. Microbiol. 10:194. doi: 10.3389/fmicb.2019.00194
Fu, T., Shangguan, H., Wei, J., Wu, J., Tang, J., Zeng, R. J., et al. (2022). In-situ electrolytic oxygen is a feasible replacement for conventional aeration during aerobic composting. J. Hazard. Mater. 426:127846. doi: 10.1016/j.jhazmat.2021.127846
Guan, J., Wasty, A., Grenier, C., and Chan, M. (2007). Influence of temperature on survival and conjugative transfer of multiple antibiotic-resistant plasmids in chicken manure and compost microcosms. Poult. Sci. 86, 610–613. doi: 10.1093/ps/86.4.610
Guo, Y., Qiu, T., Gao, M., Sun, Y., Cheng, S., Gao, H., et al. (2021). Diversity and abundance of antibiotic resistance genes in rhizosphere soil and endophytes of leafy vegetables: focusing on the effect of the vegetable species. J. Hazard. Mater. 415:125595. doi: 10.1016/j.jhazmat.2021.125595
Hartmann, M., Niklaus, P. A., Zimmermann, S., Schmutz, S., Kremer, J., Abarenkov, K., et al. (2014). Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J. 8, 226–244. doi: 10.1038/ismej.2013.141
Henschke, R. B., and Schmidt, F. R. J. (1990). Plasmid mobilization from genetically engineered bacteria to members of the indigenous soil microflora in situ. Curr. Microbiol. 20, 105–110. doi: 10.1007/BF02092881
Inoue, D., Soda, S., Tsutsui, H., Yamazaki, Y., Murashige, K., Sei, K., et al. (2009). Occurrence and persistence of indigenous transconjugants carrying conjugative plasmids in soil. J. Biosci. Bioeng. 108, 231–234. doi: 10.1016/j.jbiosc.2009.03.014
Johnsen, A. R., and Kroer, N. (2007). Effects of stress and other environmental factors on horizontal plasmid transfer assessed by direct quantification of discrete transfer events. FEMS Microbiol. Ecol. 59, 718–728. doi: 10.1111/j.1574-6941.2006.00230.x
Kaszab, E., Szoboszlay, S., Dobolyi, C., Háhn, J., Pék, N., and Kriszt, B. (2011). Antibiotic resistance profiles and virulence markers of Pseudomonas aeruginosa strains isolated from composts. Bioresour. Technol. 102, 1543–1548. doi: 10.1016/j.biortech.2010.08.027
Keenum, I., Williams, R. K., Ray, P., Garner, E. D., Knowlton, K. F., and Pruden, A. (2021). Combined effects of composting and antibiotic administration on cattle manure–borne antibiotic resistance genes. Microbiome 9:81. doi: 10.1186/s40168-021-01006-z
Klappenbach, J. A., Saxman, P. R., Cole, J. R., and Schmidt, T. M. (2001). Rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29, 181–184. doi: 10.1093/nar/29.1.181
Klümper, U., Dechesne, A., Riber, L., Brandt, K. K., Gülay, A., Sørensen, S. J., et al. (2017). Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J. 11, 152–165. doi: 10.1038/ismej.2016.98
Klümper, U., Riber, L., Dechesne, A., Sannazzarro, A., Hansen, L. H., Sørensen, S. J., et al. (2015). Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 9, 934–945. doi: 10.1038/ismej.2014.191
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le Devendec, L., Mourand, G., Bougeard, S., Léaustic, J., Jouy, E., Keita, A., et al. (2016). Impact of colistin sulfate treatment of broilers on the presence of resistant bacteria and resistance genes in stored or composted manure. Vet. Microbiol. 194, 98–106. doi: 10.1016/j.vetmic.2015.11.012
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, L., Dechesne, A., He, Z., Madsen, J. S., Nesme, J., Sørensen, S. J., et al. (2018). Estimating the transfer range of plasmids encoding antimicrobial resistance in a wastewater treatment plant microbial community. Environ. Sci. Technol. Lett. 5, 260–265. doi: 10.1021/acs.estlett.8b00105
Liao, H., Friman, V. P., Geisen, S., Zhao, Q., Cui, P., Lu, X., et al. (2019a). Horizontal gene transfer and shifts in linked bacterial community composition are associated with maintenance of antibiotic resistance genes during food waste composting. Sci. Total Environ. 660, 841–850. doi: 10.1016/j.scitotenv.2018.12.353
Liao, H. P., Lu, X. M., Rensing, C., Friman, V. P., Geisen, S., Chen, Z., et al. (2018). Hyperthermophilic composting accelerates the removal of antibiotic resistance genes and mobile genetic elements in sewage sludge. Environ. Sci. Technol. 52, 266–276. doi: 10.1021/acs.est.7b04483
Liao, H., Zhao, Q., Cui, P., Chen, Z., Yu, Z., Geisen, S., et al. (2019b). Efficient reduction of antibiotic residues and associated resistance genes in Tylosin antibiotic fermentation waste using hyperthermophilic composting. Environ. Int. 133:105203. doi: 10.1016/j.envint.2019.105203
Lin, H., Sun, W., Jin, D., Yu, Q., Yang, Y., Zhang, Z., et al. (2021). Effect of composting on the conjugative transmission of sulfonamide resistance and sulfonamide-resistant bacterial population. J. Clean. Prod. 285:125483. doi: 10.1016/j.jclepro.2020.125483
Liu, B., Yu, K., Ahmed, I., Gin, K., Xi, B., Wei, Z., et al. (2021). Key factors driving the fate of antibiotic resistance genes and controlling strategies during aerobic composting of animal manure: a review. Sci. Total Environ. 791:148372. doi: 10.1016/j.scitotenv.2021.148372
Lopatkin, A. J., Huang, S., Smith, R. P., Srimani, J. K., Sysoeva, T. A., Bewick, S., et al. (2016). Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1:16044. doi: 10.1038/nmicrobiol.2016.44
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Miller, J. H., Novak, J. T., Knocke, W. R., and Pruden, A. (2016). Survival of antibiotic resistant bacteria and horizontal gene transfer control antibiotic resistance gene content in anaerobic digesters. Front. Microbiol. 7:263. doi: 10.3389/fmicb.2016.00263
Musovic, S., Dechesne, A., Sørensen, J., and Smets, B. F. (2010). Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl. Environ. Microbiol. 76, 4813–4818. doi: 10.1128/AEM.02713-09
Musovic, S., Klümper, U., Dechesne, A., Magid, J., and Smets, B. F. (2014). Long-term manure exposure increases soil bacterial community potential for plasmid uptake. Environ. Microbiol. Rep. 6, 125–130. doi: 10.1111/1758-2229.12138
Nakasaki, K., Shoda, M., and Kubota, H. (1985). Effect of temperature on composting of sewage sludge. Appl. Environ. Microbiol. 50, 1526–1530. doi: 10.1128/aem.50.6.1526-1530.1985
Qian, X., Gu, J., Sun, W., Wang, X. J., Su, J. Q., and Stedfeld, R. (2018). Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 344, 716–722. doi: 10.1016/j.jhazmat.2017.11.020
Qian, X., Sun, W., Gu, J., Wang, X. J., Zhang, Y. J., Duan, M. L., et al. (2016). Reducing antibiotic resistance genes, integrons, and pathogens in dairy manure by continuous thermophilic composting. Bioresour. Technol. 220, 425–432. doi: 10.1016/j.biortech.2016.08.101
Qiu, T., Wu, D., Zhang, L., Zou, D., Sun, Y., Gao, M., et al. (2021). A comparison of antibiotics, antibiotic resistance genes, and bacterial community in broiler and layer manure following composting. Environ. Sci. Pollut. Res. Int. 28, 14707–14719. doi: 10.1007/s11356-020-11469-6
Sardar, M. F., Zhu, C., Geng, B., Huang, Y., Abbasi, B., Zhang, Z., et al. (2021). Enhanced control of sulfonamide resistance genes and host bacteria during thermophilic aerobic composting of cow manure. Environ. Pollut. 275:116587. doi: 10.1016/j.envpol.2021.116587
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Sørensen, S. J., Bailey, M., Hansen, L. H., Kroer, N., and Wuertz, S. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710. doi: 10.1038/nrmicro1232
Su, J. Q., Wei, B., Ou-Yang, W. Y., Huang, F. Y., Zhao, Y., Xu, H. J., et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 49, 7356–7363. doi: 10.1021/acs.est.5b01012
Sun, Y., Guo, Y., Shi, M., Qiu, T., Gao, M., Tian, S., et al. (2021). Effect of antibiotic type and vegetable species on antibiotic accumulation in soil-vegetable system, soil microbiota, and resistance genes. Chemosphere 263:128099. doi: 10.1016/j.chemosphere.2020.128099
Sun, Y., Qiu, T., Gao, M., Shi, M., Zhang, H., and Wang, X. (2019). Inorganic and organic fertilizers application enhanced antibiotic resistome in greenhouse soils growing vegetables. Ecotoxicol. Environ. Saf. 179, 24–30. doi: 10.1016/j.ecoenv.2019.04.039
Thomas, C., Idler, C., Ammon, C., and Amon, T. (2020). Effects of the C/N ratio and moisture content on the survival of ESBL-producing Escherichia coli during chicken manure composting. Waste Manag. 105, 110–118. doi: 10.1016/j.wasman.2020.01.031
Vikesland, P. J., Pruden, A., Alvarez, P. J. J., Aga, D., Bürgmann, H., Li, X. D., et al. (2017). Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environ. Sci. Technol. 51, 13061–13069. doi: 10.1021/acs.est.7b03623
Wang, C., Dong, D., Strong, P. J., Zhu, W., Ma, Z., Qin, Y., et al. (2017). Microbial phylogeny determines transcriptional response of resistome to dynamic composting processes. Microbiome 5:103. doi: 10.1186/s40168-017-0324-0
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., Selvam, A., Chan, M., and Wong, J. W. C. (2013). Nitrogen conservation and acidity control during food wastes composting through struvite formation. Bioresour. Technol. 147, 17–22. doi: 10.1016/j.biortech.2013.07.060
Xie, W. Y., Wang, Y. T., Yuan, J., Hong, W. D., Niu, G. Q., Zou, X., et al. (2022). Prevalent and highly mobile antibiotic resistance genes in commercial organic fertilizers. Environ. Int. 162:107157. doi: 10.1016/j.envint.2022.107157
Xu, H., Chen, Z., Huang, R., Cui, Y., Li, Q., Zhao, Y., et al. (2021). Antibiotic resistance gene-carrying plasmid spreads into the plant endophytic bacteria using soil bacteria as carriers. Environ. Sci. Technol. 55, 10462–10470. doi: 10.1021/acs.est.1c01615
Yang, Q., Tian, T., Niu, T., and Wang, P. (2017). Molecular characterization of antibiotic resistance in cultivable multidrug-resistant bacteria from livestock manure. Environ. Pollut. 229, 188–198. doi: 10.1016/j.envpol.2017.05.073
Ye, Z., Ding, H., Yin, Z., Ping, W., and Ge, J. (2021). Evaluation of humic acid conversion during composting under amoxicillin stress: emphasizes the driving role of core microbial communities. Bioresour. Technol. 337:125483. doi: 10.1016/j.biortech.2021.125483
Keywords: antibiotic resistance genes, multidrug-resistant plasmid, horizontal gene transfer, phylogenetic analysis, manure composting
Citation: Shen L, Qiu T, Guo Y, Gao M, Gao H, Zhao G and Wang X (2022) Enhancing control of multidrug-resistant plasmid and its host community with a prolonged thermophilic phase during composting. Front. Microbiol. 13:989085. doi: 10.3389/fmicb.2022.989085
Received: 08 July 2022; Accepted: 05 August 2022;
Published: 17 August 2022.
Edited by:
Li Hu, Institute of Urban Environment (CAS), ChinaReviewed by:
Fu-Yi Huang, Institute of Urban Environment (CAS), ChinaCopyright © 2022 Shen, Qiu, Guo, Gao, Gao, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guozhu Zhao, emhhb2d6QGJqZnUuZWR1LmNu; Xuming Wang, d2FuZ3h1bWluZ0BiYWFmcy5uZXQuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.