- 1Department of Microbiology and Immunology, Faculty of Pharmacy, Kafrelsheikh University, Kafr El Sheikh, Egypt
- 2Hygiene and Zoonoses Department, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt
- 3Bacterial Epidemiology and Antimicrobial Resistance Research Unit, U.S. National Poultry Research Center, U.S. Department of Agriculture, Agricultural Research Service (USDA-ARS), Athens, GA, United States
- 4Antimicrobial Resistance Research Center, National Institute of Infectious Diseases, Higashimurayama, Japan
- 5Department of Antimicrobial Resistance, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan
- 6Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, University of Sadat City, Sadat City, Egypt
- 7Laboratory of Food Microbiology, Graduate School of Human Life Sciences, Jumonji University, Niiza, Japan
- 8Laboratory of Food Microbiology and Hygiene, Graduate School of Integrated Sciences for Life, Hiroshima University, Higashihiroshima, Japan
Colistin is a last-resort antibiotic used in the treatment of multidrug resistant Gram-negative bacteria. However, the activity and efficacy of colistin has been compromised by the worldwide spread of the mobile colistin resistance genes (mcr-1 to mcr-10). In this study, two clinical Escherichia coli strains, named EcCAI51, and EcCAI73, harbored mcr-1, showed multidrug-resistant phenotypes (with colistin MIC = 4 μg/ml), and belonged to phylogroup D: multilocus sequence type 1011 (ST1011) and phylogroup A: ST744, respectively. Findings revealed the existence of mcr-1 gene on two conjugable plasmids, pAMS-51-MCR1 (∼122 kb IncP) and pAMS-73-MCR1 (∼33 kb IncX4), in EcCAI51, and EcCAI73, respectively. The mcr-1-pap2 element was detected in the two plasmids. Additionally, the composite transposon (ISApl1-IS5D-pap2-mcr-1-ISApl1) was identified only in pAMS-51-MCR1 suggesting the potential for horizontal gene transfer. The two strains carried from 16 to 18 different multiple acquired antimicrobial resistance genes (ARGs). Additionally, two different multireplicon virulence plasmids (∼117 kb pAMS-51-Vr and ∼226 kb pAMS-73-Vr) carrying the sit operon, the Salmochelin siderophore iroBCDE operon and other several virulence genes were identified from the two strains. Hierarchical clustering of core genome MLST (HierCC) revealed clustering of EcCAI73, and EcCAI51 with global E. coli lineages at HC levels of 50 (HC50) to 100 (HC100) core genome allelic differences. To the best of our knowledge, this study presented the first complete genomic sequences of mcr-1-carrying IncP and IncX4 plasmids from human clinical E. coli isolates in Egypt. In addition, the study illustrated the mcr-1 broad dissemination in diverse plasmids and dissimilar E. coli clones.
Introduction
Colistin is one of the last-resort antibiotics used in the treatment of infections caused by multidrug or carbapenem resistant Gram-negative bacteria. In 2016, the first mobile colistin resistance gene (mcr-1) was reported from Escherichia coli and Klebsiella pneumoniae isolated from patients, food, and animals in China (Liu et al., 2016). mcr-1 acts by modifying the lipid A part of the lipopolysaccharide in Gram-negative bacteria by adding phosphoethanolamine, reducing the binding affinity to colistin (Liu et al., 2016). Furthermore, mcr-1-carrying E. coli strains have been reported in Egypt from patients (Elnahriry et al., 2016), cattle, and chickens (Elbediwi et al., 2019). mcr-1 has been detected in several plasmid groups, including IncX4, IncHI2, IncI2, IncI1, IncN, IncFIB, IncP, and IncW (Lu et al., 2018; Elbediwi et al., 2019; Soliman et al., 2021). Ten mcr genes (mcr-1–mcr-10) have been characterized, all of which confer resistance to colistin by the same mechanism described above. We reported the first mcr-9-carrying Enterobacter hormaechei clinical isolate in the Middle East (Soliman et al., 2020a). Recently, Tartor et al. (2021a) reported the first emergence of an Egyptian K. pneumoniae isolate co-harboring mcr-10 and fosA5 genes from bovine milk in Middle East. Other variants of mcr including mcr-1, mcr-2, mcr-3, mcr-4, and mcr-7 were also reported in Gram-negative bacteria (E. coli, K. pneumoniae, and Pseudomonas aeruginosa) isolated from bovine milk in Egypt (Tartor et al., 2021b). We additionally reported two mcr-1-, tet(X7)-, and fosA3-positive E. coli ST155 strains showing resistance to last resort antibiotics (such as colistin, and tigecycline) from poultry farm in Egypt (Soliman et al., 2021). Recently, a uropathogenic E. coli strain carried mcr-1.1 on a self-transmissible IncHI2 plasmid from Alexandria, Egypt (Zakaria et al., 2021).
Little is yet known about the genomic characteristics of mcr-1-carrying clinical E. coli strains in Egypt. Therefore, we aimed, in this study, to characterize the complete genomic sequences of mcr-1-carrying IncP and IncX4 plasmids from two clinical E. coli isolates and to perform phylogenetic analysis for these two strains.
Materials and methods
Bacterial strains used in this study
The two mcr-1-positive E. coli isolates, named EcCAI51 and EcCAI73, were detected from two patients in two different hospitals located in Cairo, Egypt. The strain EcCAI51 was isolated from the eye swab of a 50-years-old male patient diagnosed with a respiratory infection in April 2015, while strain EcCAI73 was isolated from a blood sample of a patient in May 2015. The two strains were identified by 16S rRNA gene sequencing using primers 27F and 1492R and screened by PCR for mobile colistin-resistance genes (mcr-1–mcr-5) (Table 1; Luo et al., 2015; Elnahriry et al., 2016; Liu et al., 2016; Xavier et al., 2016; Borowiak et al., 2017; Carattoli et al., 2017; Yin et al., 2017), extended-spectrum β-lactamases, carbapenemase-encoding genes, plasmid-mediated quinolone-resistance genes, and 16S rRNA methylases as previously described (Jousset et al., 2019; Soliman et al., 2020b).
Antimicrobial susceptibility testing
The broth microdilution assay (BMD) was performed to determine the minimum inhibitory concentration (MIC) of various antimicrobials (Table 2) according to the standards and interpretive criteria described by the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI] (2020) document M100-S24) and European Committee on Antimicrobial Susceptibility Testing (EU-CAST) (for colistin and tigecycline breakpoints).1 For all experiments, the purified powder of each antibiotic was diluted following CLSI recommendations. E. coli ATCC 25922 was used as a control.
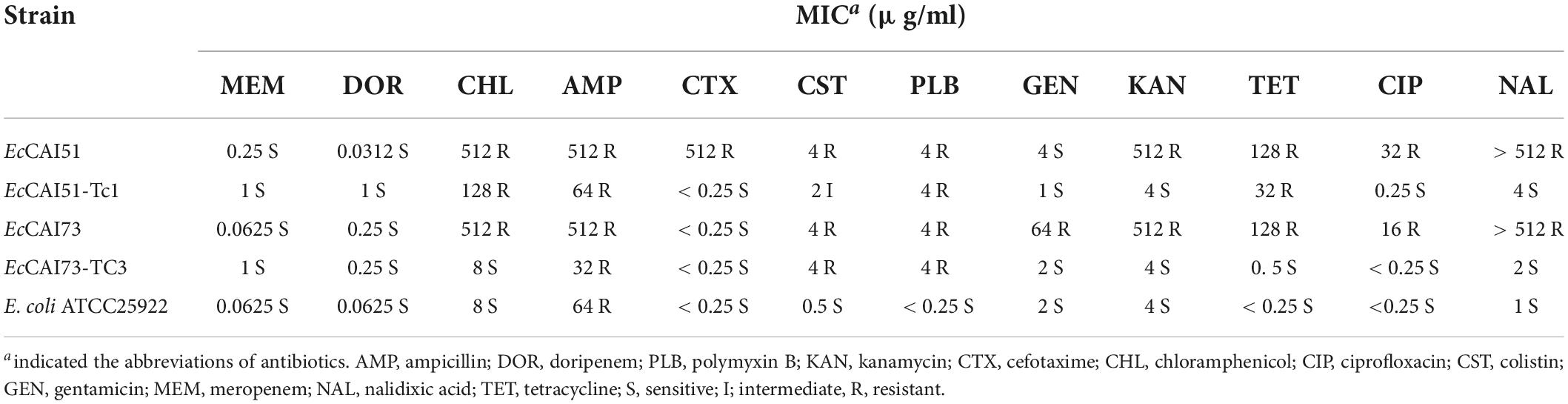
Table 2. Minimum inhibitory concentrations (MICs) for mcr-1-carrying strains of E. coli and its transconjugants identified in this study.
Filter-mating conjugation
A mating-out assay was completed at 37°C using the two E. coli strains and the AZr (azide resistant) E. coli J53 strain as the donor and recipient, respectively (Soliman et al., 2020b,2021). These experiments were performed on a solid media using filters with a 1:1 donor: recipient ratio. After a 5-h incubation, filters were resuspended in 3 ml LB broth, and bacterial mixtures were overlaid onto agar plates supplemented with colistin (2 μg/ml) and sodium azide (150 μg/ml). Colony-direct PCR was performed using CLR5-F and CLR5-R primers (Table 1) to confirm the transfer of the plasmid carrying mcr-1.
Plasmid analysis, PCR-based replicon typing, multi-locus sequence typing, and Escherichia coli phylogroup
Plasmid analysis of the wild strains and transconjugants was performed by alkaline lysis method and PCR-based replicon typing (PBRT) (Carattoli et al., 2005; Soliman et al., 2020b). Multi-locus sequence typing (MLST) was performed for E. coli [using Achtman seven housekeeping genes (adk, fumC, icd, purA, gyrB, recA, and mdh)] according to the MLST database.2 E. coli phylogroups (A, B1, B2, and D) were detected by Triplex PCR after amplification of chuA and yjaA and the DNA fragment TSPE4.C2 as previously described (Clermont et al., 2000).
Complete genome sequencing, and analysis
The Qiagen Genomic-tip 20/G kit (Qiagen) was used to extract the total genomic DNA following the manufacturer’s recommendations. For Illumina sequencing by MiniSeq, a Nextera XT Library Prep Kit and a Nextera XT Index Kit was used to prepare the DNA library (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. For Nanopore sequencing by GridION, construction of the library was performed by the SQK-RBK004 Rapid Barcoding Kit (Oxford Nanopore Technologies, Oxford, United Kingdom). The library was loaded onto a FLO-MIN106 R9.4.1 flow cell and sequenced with the GridION device (Oxford Nanopore Technologies, Oxford, United Kingdom). A hybrid assembly of MiniSeq short reads and Nanopore long reads was achieved by Unicycler (Wick et al., 2017). The annotation was performed using DFAST.3 The complete genome sequences of the two E. coli strains were investigated at the Center for Genomic Epidemiology4 using ResFinder-4.1 (identity threshold for gene predictions was 90%), MLST 2.0, pMLST 2.0, VirulenceFinder-2.0 and PlasmidFinder-2. Genomic comparisons were performed using the BRIG tool5 and EasyFig tool.6 The BLAST program7 and ISfinder8 were used to analyze the plasmids.
Phylogenetic analyses of the mcr-1-positive Escherichia coli isolates
Raw Fastq files of the sequenced two E. coli strains, EcCAI51 and EcCAI73 were imported into Enterobase9 for WGS-based phylogenetic analysis. Two sets of publicly available genomes of E. coli in Enterobase that represent different sources and belong to sequence types (ST) ST744 (n = 181) and ST1011 (n = 157) were chosen for the analysis. Our EcCAI51 (ST1011) and EcCAI73 (ST744) genomes were compared separately to the selected genomes from Enterobase belonging to the same ST using single nucleotide polymorphisms (SNPs) and hierarchical clustering (HierCC) of core genome (cg) MLST (Zhou et al., 2020). E. coli K-12 MG1655 was used as the reference strain for SNPs analysis of isolates. Metadata for the selected genomes from Enterobase are given in Supplementary Tables 1, 2.
Nucleotide sequence accession numbers
The complete genome sequence of EcCAI51 and EcCAI73 were submitted to DDBJ/ENA/GenBank under BioProject ID: PRJDB11824 (SRA accession numbers DRA012212, and DRA012213, respectively).
Results and discussion
Characterization of Escherichia coli strains EcCAI51, and EcCAI73
Two polymyxin resistant E. coli isolates were identified from two different hospitals located at the capital city of Egypt. The two isolates showed multidrug-resistant phenotypes. Both the isolates were resistant to colistin (MIC = 4 μg/ml), polymyxin B (MIC = 4 μg/ml), ampicillin, chloramphenicol, tetracycline, kanamycin, and fluoroquinolones but were sensitive to meropenem and doripenem (Table 2). Although both isolates were susceptible to carbapenem, carbapenem-resistant E. coli carrying mcr genes were reported (Paveenkittiporn et al., 2021). In that study, the investigators identified nine colistin and carbapenem resistant MCR and NDM or OXA-48-like-producing E. coli strains isolated from clinical samples in Thailand during 2016–2019 (Paveenkittiporn et al., 2021). EcCAI51 was resistant to cefotaxime due to production of CTX-M-14. PCR and DNA sequencing confirmed the presence of mcr-1 in both isolates. There was no clonal relationship between the two isolates that assigned to two different ST (ST1011 or ST744), and two different phylogenetic groups (D or A). ST1011 and ST744 had the same ST of mcr-1-positive clinical E. coli previously identified from Egypt and Denmark, respectively (Hasman et al., 2015; Elnahriry et al., 2016), and differed from the STs recognized in mcr-1-positive clinical E. coli isolates from Cambodia (ST354) (Stoesser et al., 2016) and South Africa (ST10, ST1007, ST624, ST57, ST101, ST624, and ST226) (Poirel et al., 2016). The two strains carried from 16 to 18 different multiple acquired antimicrobial resistance genes (ARGs) located on the chromosome and/or different plasmids (Table 3).
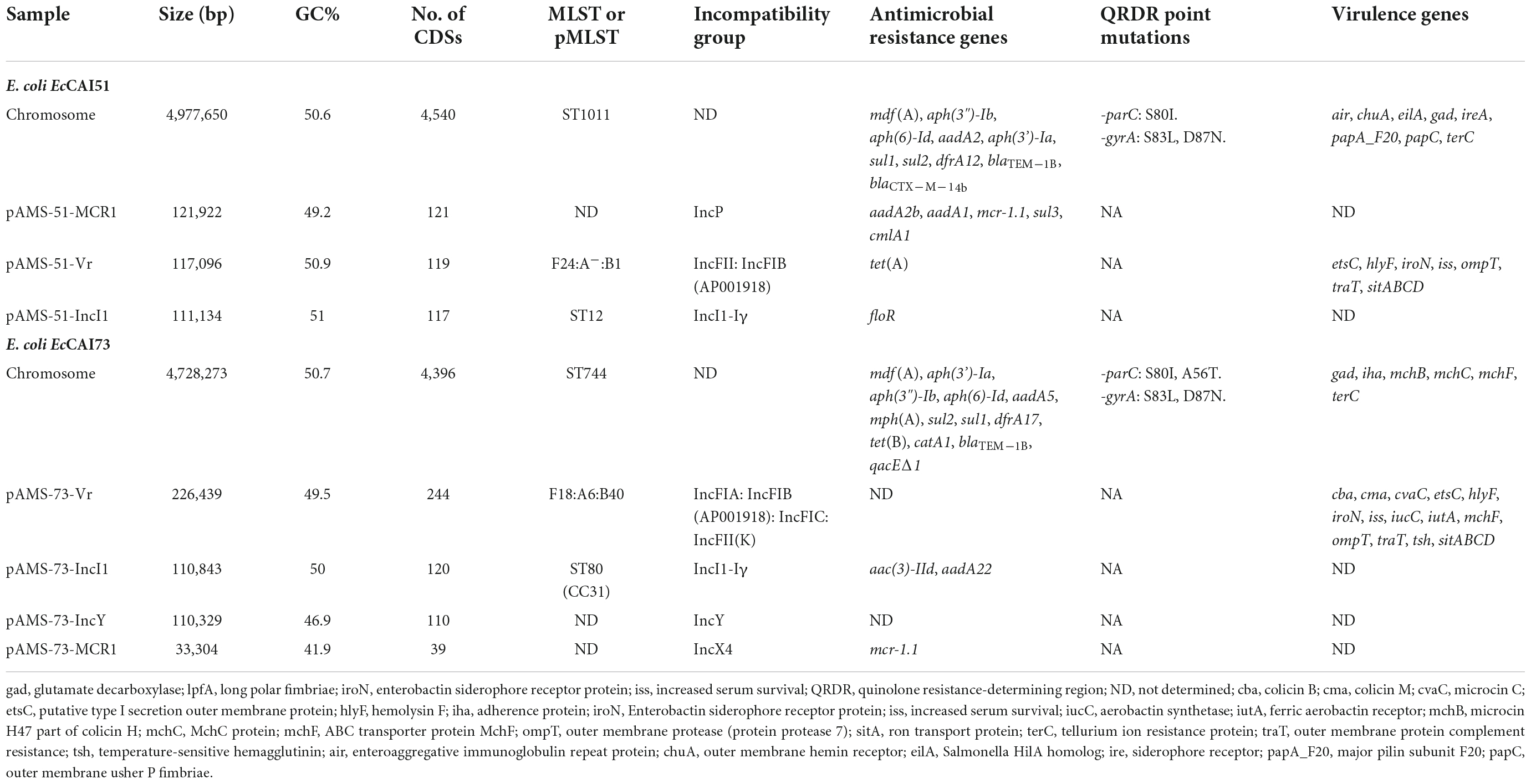
Table 3. Features of chromosome and the plasmids of E. coli strains EcCAI51, and EcCAI73 isolated from clinical samples in Egypt.
We obtained high-quality assemblies by combining the Illumina MiniSeq short reads and the Oxford Nanopore long reads adequately for completing the genomes and the plasmids contained in both isolates (Table 3). E. coli EcCAI51, and EcCAI73 carried three, and four plasmids, respectively (Table 3). The chromosome of the EcCAI51 strain was 4,977,650 bp in size with an average G + C content of 50.6% determining 4,540 coding sequences. ResFinder identified several chromosomal ARGs as follow: mdf(A), aph(3”)-Ib, aph(6)-Id, aadA2, aph(3’)-Ia, sul1, sul2, dfrA12, blaTEM–1B, and blaCTXM–14b. In addition, EcCAI73 has 4,728,273 bp chromosome with an average G + C content of 50.7% and 4,396 coding sequences. The chromosomal ARGs in EcCAI73 were mdf(A), aph(3’)-Ia, aph(3 “)-Ib, aph(6)-Id, aadA5, mph(A), sul2, sul1, dfrA17, tet(B), catA1, blaTEM–1B, and qacEΔ1.
Identification of IncP and IncX4 plasmids carrying mcr-1 in Egyptian clinical Escherichia coli isolates
The mcr-1.1 gene was located on the plasmids pAMS-51-MCR1 and pAMS-73-MCR1 from isolates EcCAI51 and EcCAI73, respectively. pAMS-51-MCR1 was 121,922 bp IncP type (Figure 1). A BLASTn search using the whole pAMS-51-MCR1 sequence query detected that it has high identity to other mcr-1-carrying plasmids. for example, pAMS-51-MCR1 showed > 98.7% sequence identity to K. pneumoniae plasmid pSCKLB684-mcr (55% coverage; MH781719.1, IncY type), plasmid p160070-MCR isolated from food in China (56% coverage; MG288678.1, IncP type), and plasmid pMCR_SCKP-LL83 isolated from human in China (56% coverage; MF510496.1, IncP type), which were harbored mcr-1 (Figure 1). In addition, pAMS-51-MCR1 showed > 98.7% sequence identity to E. coli plasmid pZR78 (56% coverage; MF455226.1, IncP type), and plasmid pPC6-mcr1 (56% coverage; CP080254.1, IncP type).
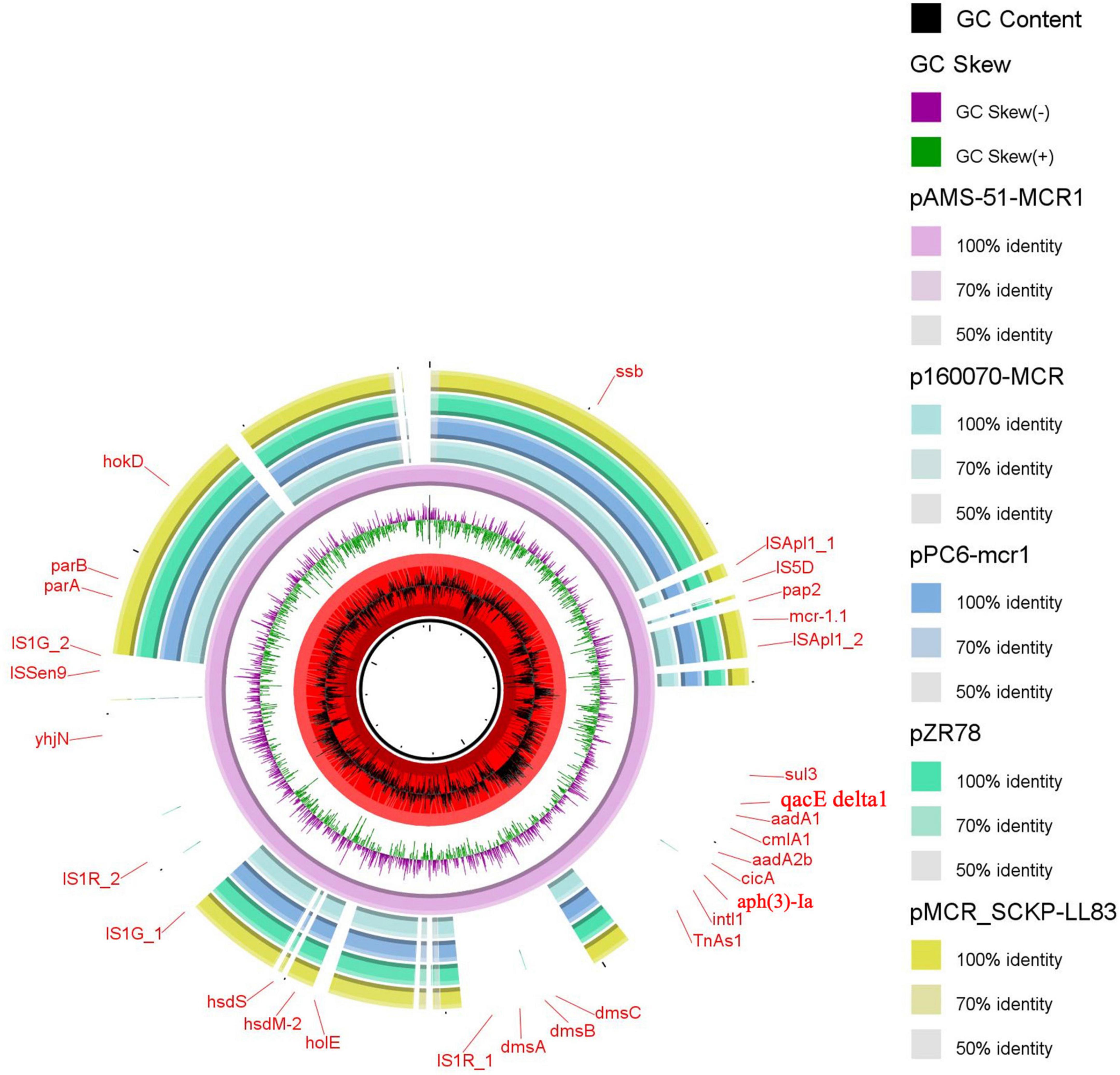
Figure 1. Schematic representation of IncP plasmids carrying mcr-1.1 identified from the genome sequences of K. pneumoniae and E. coli strains analyzed in this study. Four IncP plasmid, p160070-MCR, pMCR_SCKP-LL83, pZR78, and pPC6-mcr1 carrying mcr-1.1 (accession no. MG288678.1, MF510496.1, MF455226.1, and CP080254.1, respectively) have been detected from NCBI GenBank and was included in the figure. The whole sequence of pAMS-51-MCR1 was used as the reference. The external ring represents the annotation of pAMS-51-MCR1. The plasmids were included in the following order: pAMS-51-MCR1 (identified in this study), p160070-MCR, pPC6-mcr1, pZR78, and pMCR_SCKP-LL83.
pAMS-73-MCR1 was 33,304 bp IncX4 type (Figure 2). A BLASTn search using the whole pAMS-75-MCR1 sequence query detected that it has 99.9% identity with 99% coverage to other mcr-1-carrying IncX4 plasmids as follow: (i) plasmid pWI2-mcr detected from clinical E. coli isolate WI2 in France (LT838201.1), (ii) plasmid pSH15G2169 from Salmonella enterica subsp. enterica serovar Typhimurium strain SH15G2169 isolated from diarrheal outpatients in Shanghai, China (MH522417.1) (Lu et al., 2019), iii) plasmid 16BU137_mcr-1.1 from clinical K. pneumoniae strain 16BU137 in China (MT316509.1), and iv) plasmid pE13-43-mcr-1 (MG747473.1) isolated from E. coli strain 13–43 collected from urine sample in China in 2013.
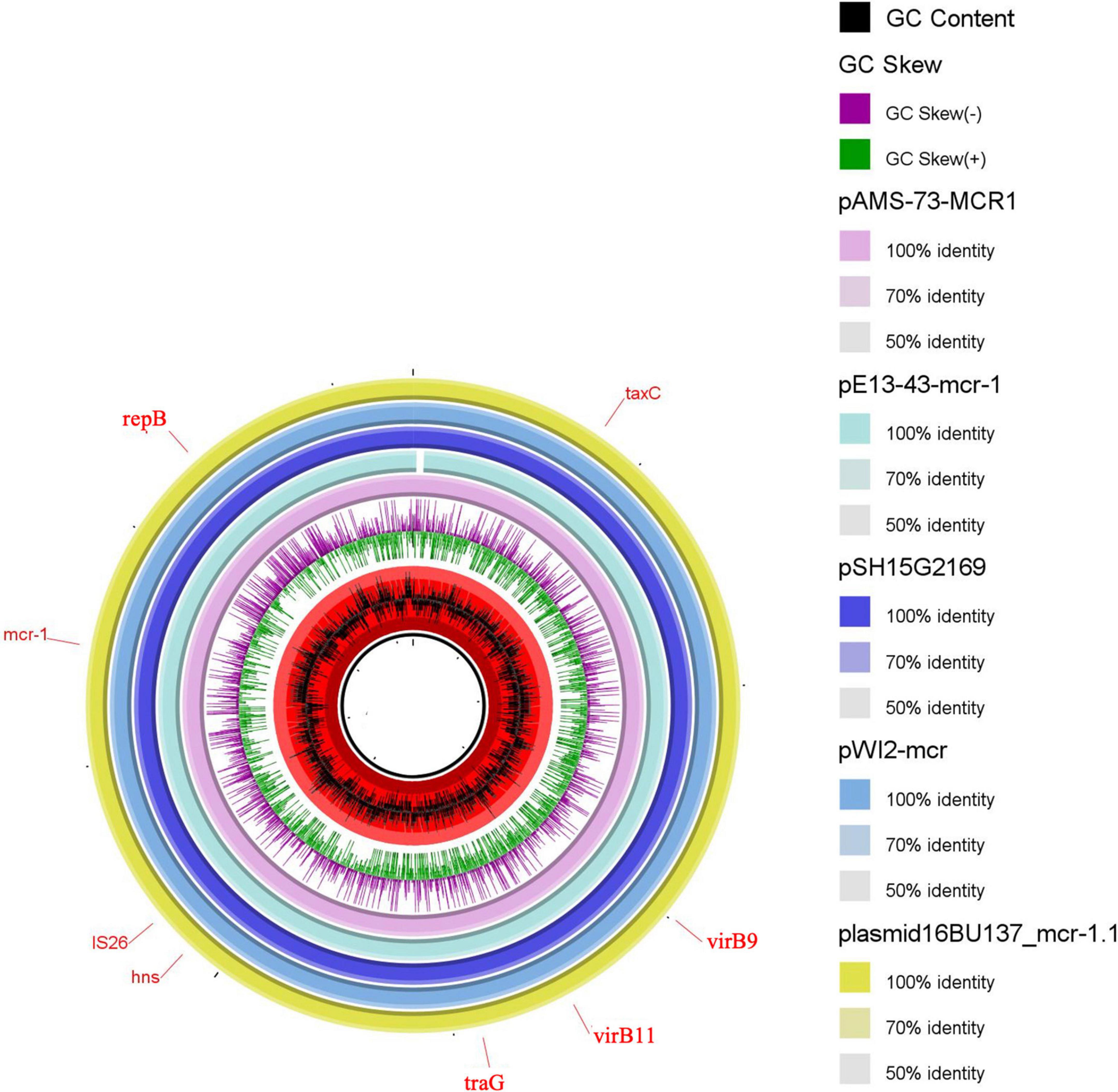
Figure 2. Schematic representation of IncX4 plasmids carrying mcr-1.1 identified from the genome sequences of E. coli, K. pneumoniae or Salmonella enterica strains analyzed in this study. Four IncX4 plasmid, pE13-43-mcr-1, pSH15G2169, pWI2-mcr, and plasmid 16BU137_mcr-1.1 carrying mcr-1.1 (accession no. MG747473.1, MH522417.1, LT838201.1, and MT316509.1, respectively) have been detected from NCBI GenBank and was included in the figure. The whole sequence of pAMS-73-MCR1 was used as the reference. The external ring represents the annotation of pAMS-73-MCR1. The plasmids were included in the following order: pAMS-73-MCR1 (identified in this study), pE13-43-mcr-1, pSH15G2169, pWI2-mcr, and plasmid 16BU137_mcr-1.1.
Regarding the genetic environment of mcr-1.1, the mcr-1-pap2 (a gene encoding a putative PAP family transmembrane protein) element was detected in the two plasmids (Figure 3). However, the composite transposon (ISApl1-IS5D-pap2-mcr-1-ISApl1) (Figure 3) was only in pAMS-51-MCR1 suggesting the role of ISApl1 and its potential for horizontal gene transfer (Partridge et al., 2018). ISApl1 belongs to IS30 family and encodes a DDE-type transposase (Partridge et al., 2018). It was first identified in the pig pathogen Actinobacillus pleuropneumoniae (Tegetmeyer et al., 2008) and was implicated in the acquisition and mobilization of mcr-1 (Liu et al., 2016). The mcr-1 genetic context, pap2-mcr-1-ISApl1, was observed in i) E. coli strain 803DBmcr plasmid 803-DB-mcr, isolated from human sample in China in 2017 (MH128771.1) (Figure 3), ii) E. coli strain ECZP248 plasmid pTBMCR401 isolated from chicken in China in 2017 (CP034785.1) (Figure 3; Chang et al., 2020), and iii) E. coli strain NDM132 plasmid pls1 recovered in China, (KX458104.1) (Figure 3). The plasmids pAMS-51-MCR1 and pAMS-73-MCR1 were effectively transferred by mating out assay to the recipient E. coli J53 strain with an efficiency of ∼2.7 × 10–5 and 1 × 100 CFU/ml, respectively. PCR confirmed that transconjugants harbored mcr-1. The transconjugants carrying both the plasmids showed resistance to colistin (MICs = 2 or 4 μg/ml), and polymyxin B (MICs = 4 μg/ml) (Table 2). Additionally, the transconjugant EcCAI51-Tc1 was resistant to CHL, and TET. It might be due to the transfer of both pAMS-51-MCR1 (which carry cmlA1 conferring resistance to CHL) and pAMS-51-VR (which carry tet(A) conferring resistance to TET). In contrast, the other transconjugant EcCAI73-TC3 was sensitive to CHL, and TET. However, the two transconjugant were slightly resistant to AMP which is suggested by the carriage of unknown β-lactamase on the transferred plasmids. To our knowledge, this is the first report of mcr-1-carrying IncP and IncX4 plasmids from human clinical E. coli isolates in Egypt.
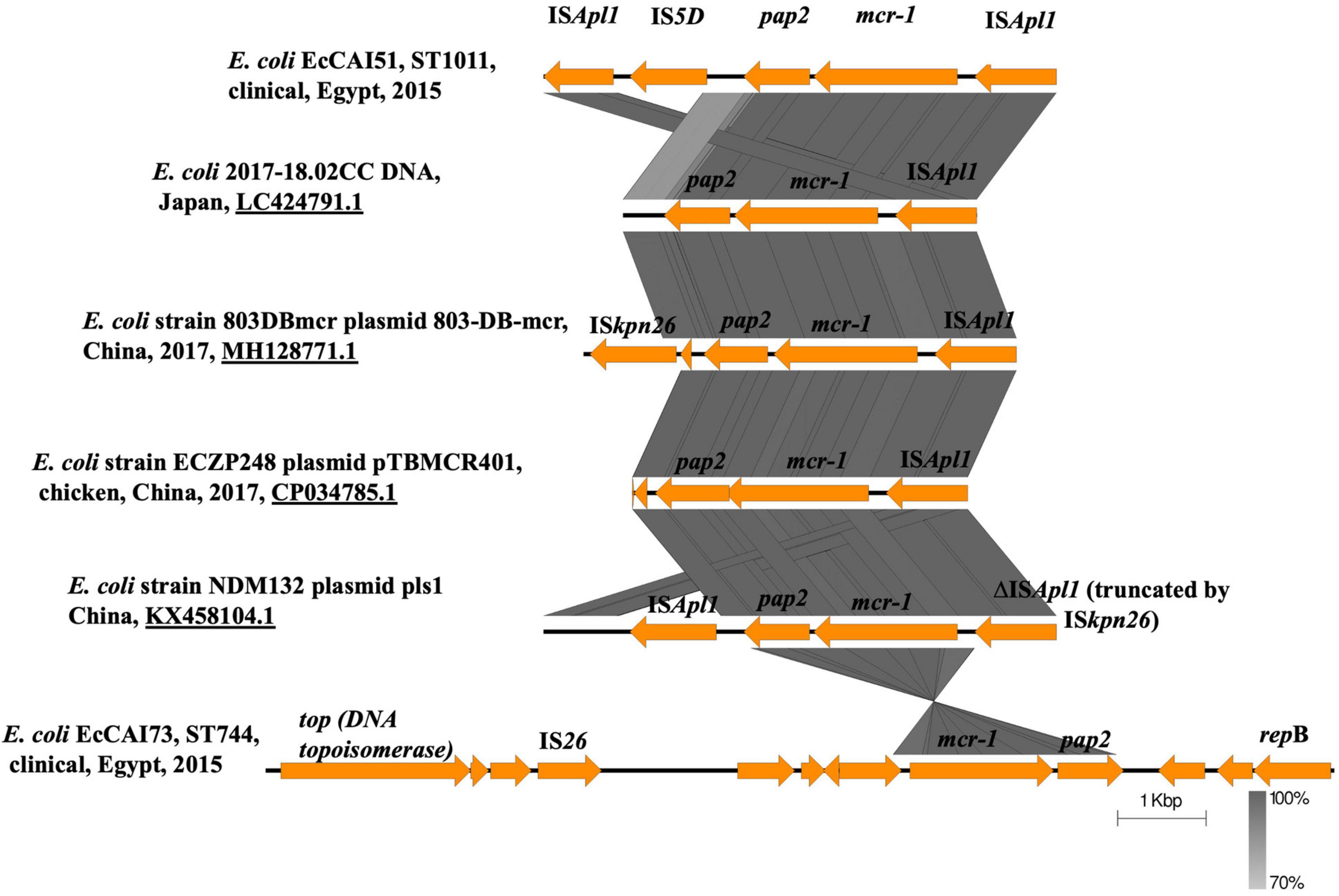
Figure 3. Linear comparison of the genetic environment of mcr-1.1 detected in this study from the two strains with other mcr-1 context from different plasmids and strains identified from NCBI GenBank.
Analysis of the virulome of the two mcr-1-producing Escherichia coli isolates: Identification of two multireplicon virulence plasmids
Numerous virulence factors (VFs) have been detected chromosomally or on different plasmids contained within the two isolates, explaining its pathogenicity and virulence (Table 3). The chromosome of E. coli EcCAI51 carried the following VFs: air (enteroaggregative immunoglobulin repeat protein), chuA (outer membrane hemin receptor), eilA (Salmonella HilA homolog), gad (glutamate decarboxylase), ireA (siderophore receptor), papA_F20 (major pilin subunit F20), papC (outer membrane usher P fimbriae), and terC (tellurium ion resistance protein) while the chromosome of E. coli EcCAI73 carried gad, iha (adherence protein), mchB (microcin H47 part of colicin H), mchC (MchC protein), mchF (ABC transporter protein MchF), and terC. Several MCR-1-producing E. coli isolates with virulence characters have been previously reported from Nepali patient admitted to an intensive-care unit in Qatar, and from retail poultry meat in Czech Republic (Forde et al., 2018; Kubelová et al., 2021).
Two different multireplicon virulence plasmids (∼117 kb IncFII: IncFIB pAMS-51-Vr and ∼226 kb IncFIA: IncFIB: IncFIC: IncFII(K) pAMS-73-Vr) carrying several virulence genes were identified from the two isolates. Both the plasmids carried the sitABCD operon mediating resistance to hydrogen peroxide and catalyzing iron, manganese transport (Sabri et al., 2006), the Salmochelin siderophore iroBCDE operon mediating iron uptake and the iroN which act as siderophore receptor, mediating the utilization of the siderophore enterobactin (Russo et al., 2002). Additionally, pAMS-73-Vr carried the iucABCD operon and iutA mediating iron and aerobactin uptake (Torres et al., 2001). pAMS-73-Vr also carried the vacuolating autotransporter toxin (vat gene), which encourages the development of intracellular vacuoles causing cytotoxic effects related to those triggered by the Helicobacter pylori VacA toxin (Parreira and Gyles, 2003). The operons sitABCD, and iucABCD were previously described from tet(X7)-mcr-1/IncHI2 plasmids detected in E. coli isolates from poultry in Egypt and from plasmid pZM3 detected from an Algerian Salmonella enterica isolate (Harmer and Hall, 2020; Soliman et al., 2021). Numerous other virulence genes were detected from both isolates’ plasmids and were included into Table 3. A BLASTn search using the whole pAMS-51-Vr sequence query detected that it has > 99.9% identity with > 92% coverage to other virulence multireplicon plasmids detected from E. coli isolates as follow: (i) plasmid pCombat11I9-2 from strain Combat11I9 detected from urine in China (CP021728.1), (ii) plasmid pNIT-HK from strain J53/pNIT-HK isolated in Hong Kong (MF474175.1), and iii) plasmid p94EC-1 from strain 94EC isolated from human fecal sample in Singapore (CP047577.1) (Figure 4).
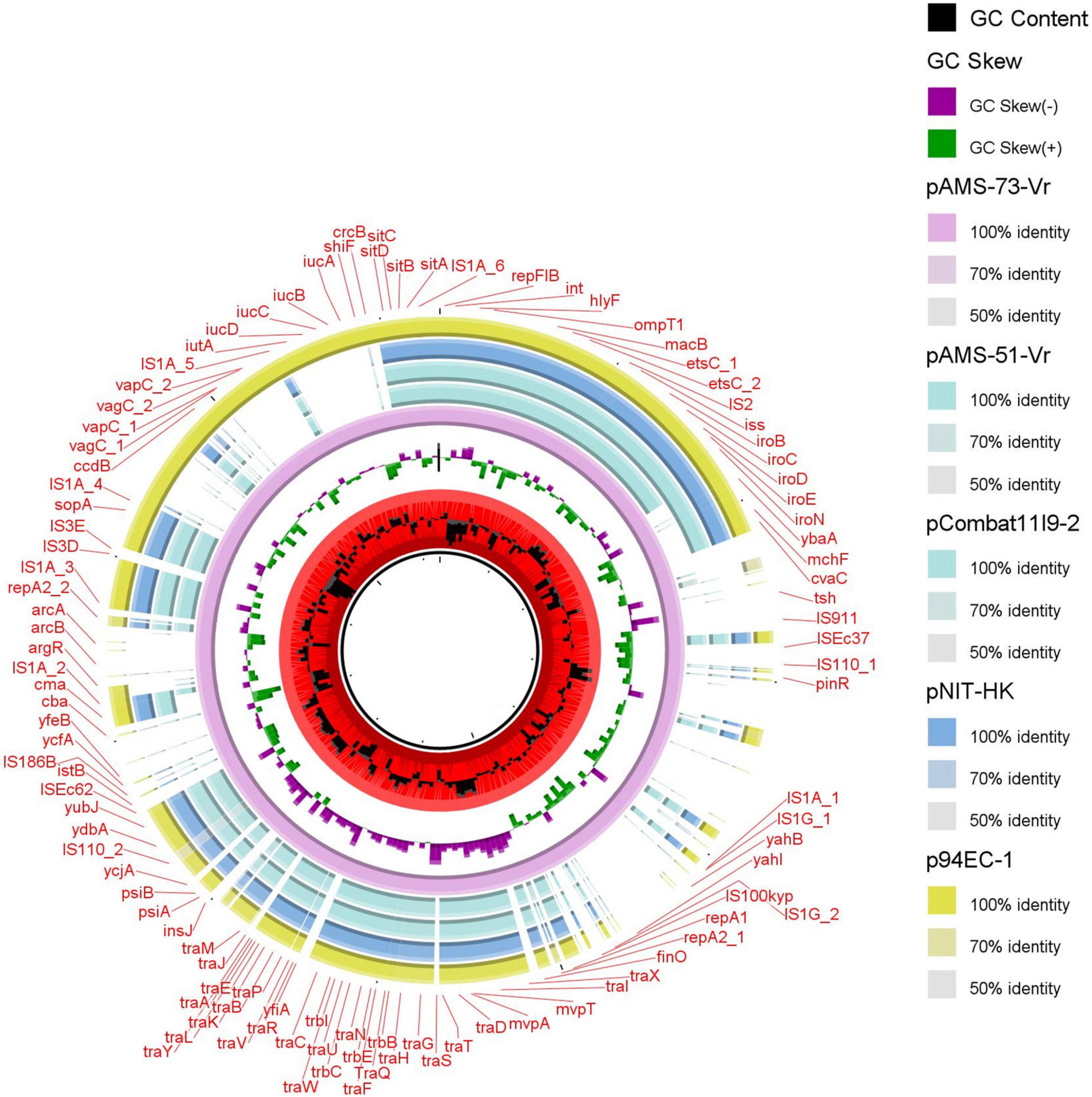
Figure 4. Schematic representation of the multireplicon virulence plasmids identified from the genome sequences of E. coli strains analyzed in this study. Four plasmid, pCombat11I9-2, pNIT-HK, and p94EC-1 (accession no. CP021728.1, MF474175.1, and CP047577.1, respectively) have been detected from NCBI GenBank and was included in the figure. The whole sequence of pAMS-73-Vr was used as the reference. The external ring represents the annotation of pAMS-73-Vr. The plasmids were included in the following order: pAMS-73-Vr (identified in this study), pAMS-51-Vr (identified in this study), pCombat11I9-2, pNIT-HK, and p94EC-1.
Evolutionary relatedness of ST1011, and ST744 mcr-1-producing Egyptian clinical Escherichia coli isolates identified in this study
Phylogenetic analysis was performed by comparing the genomes of our isolates EcCAI51 (ST1011) and EcCAI73 (ST744) to the publicly available E. coli genomes in Enterobase using SNPs and HierCC of cgMLST (Figures 5, 6). Based on differences of core genome loci among bacteria, different sets of hierarchical clusters (HCs) in Enterobase were designated to cluster bacterial genomes at higher resolution levels compared to the conventional MLST. At HC100 (≤ 100 allelic differences), the HC100 pattern (HC100| 29212) has been determined for our ST1011 isolate (EcCAI51) and other ST1011 isolates from various countries including China, Japan, Vietnam, Bangladesh, Lebanon, and other European countries (Figure 5 and Supplementary Table 1). Likewise, our ST744 isolate (EcCAI73) was clustered, and shared the same HC50| 3561 with no more than 50 allelic differences with other ST744 isolates from Thailand, the United States, Australia, Vietnam, Switzerland, Netherlands, Spain, and Portugal (Figure 6 and Supplementary Table 2). In our recently published reports, we have determined the clustering of mcr-producing E. coli from Poultry in Egypt with global E. coli lineages (Ramadan et al., 2021; Soliman et al., 2021), indicating the wide spread of antimicrobial-resistant clones and the requirement of implementing WGS-based phylogeny for disease surveillance and control interventions.
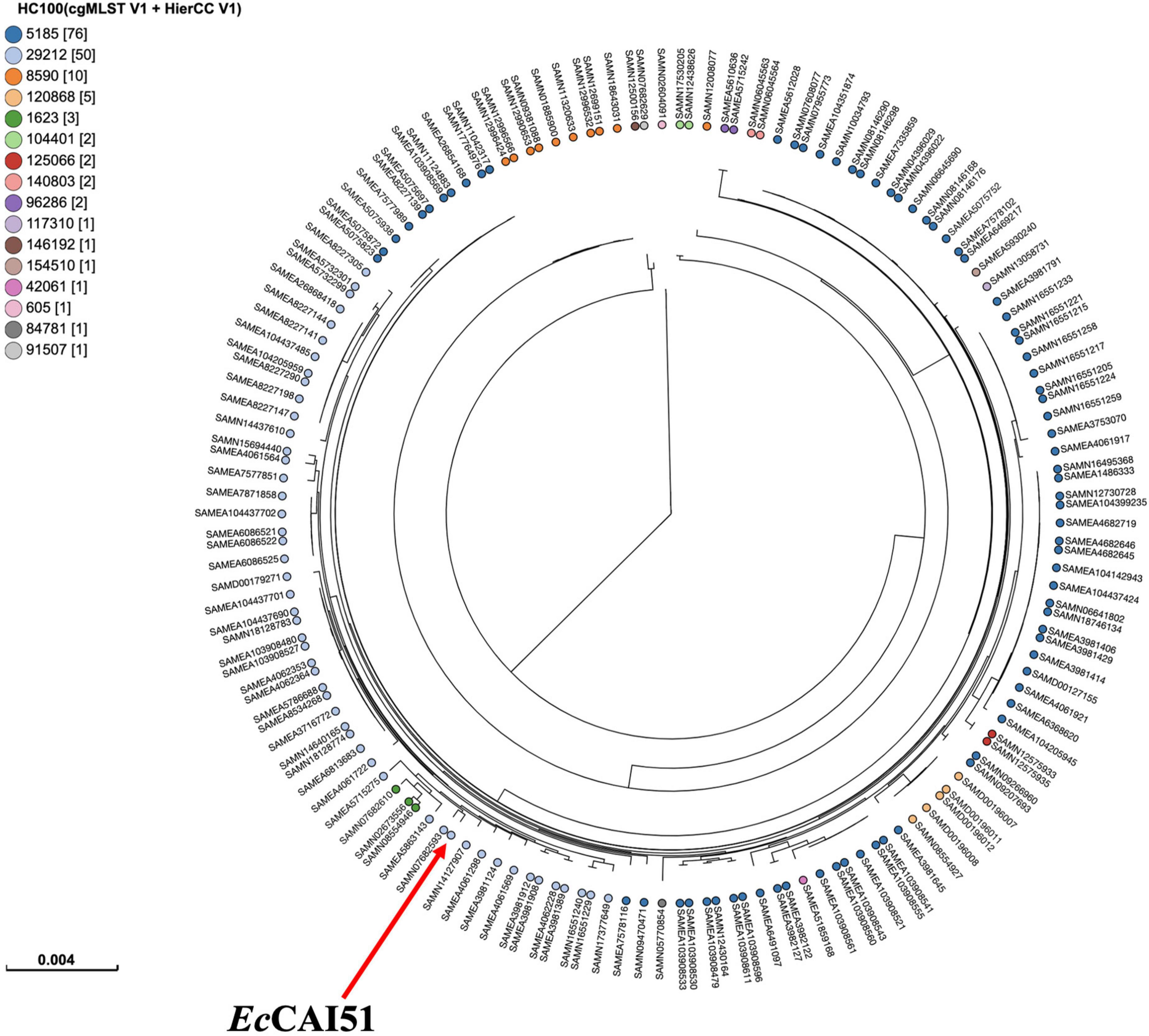
Figure 5. Phylogenetic analysis of ST1011 Escherichia coli isolate and other publicly available ST1011 E. coli isolates (n = 157) in Enterobase using single nucleotide polymorphisms (SNPs) and hierarchical clustering (HierCC) of core genome (cg) MLST.
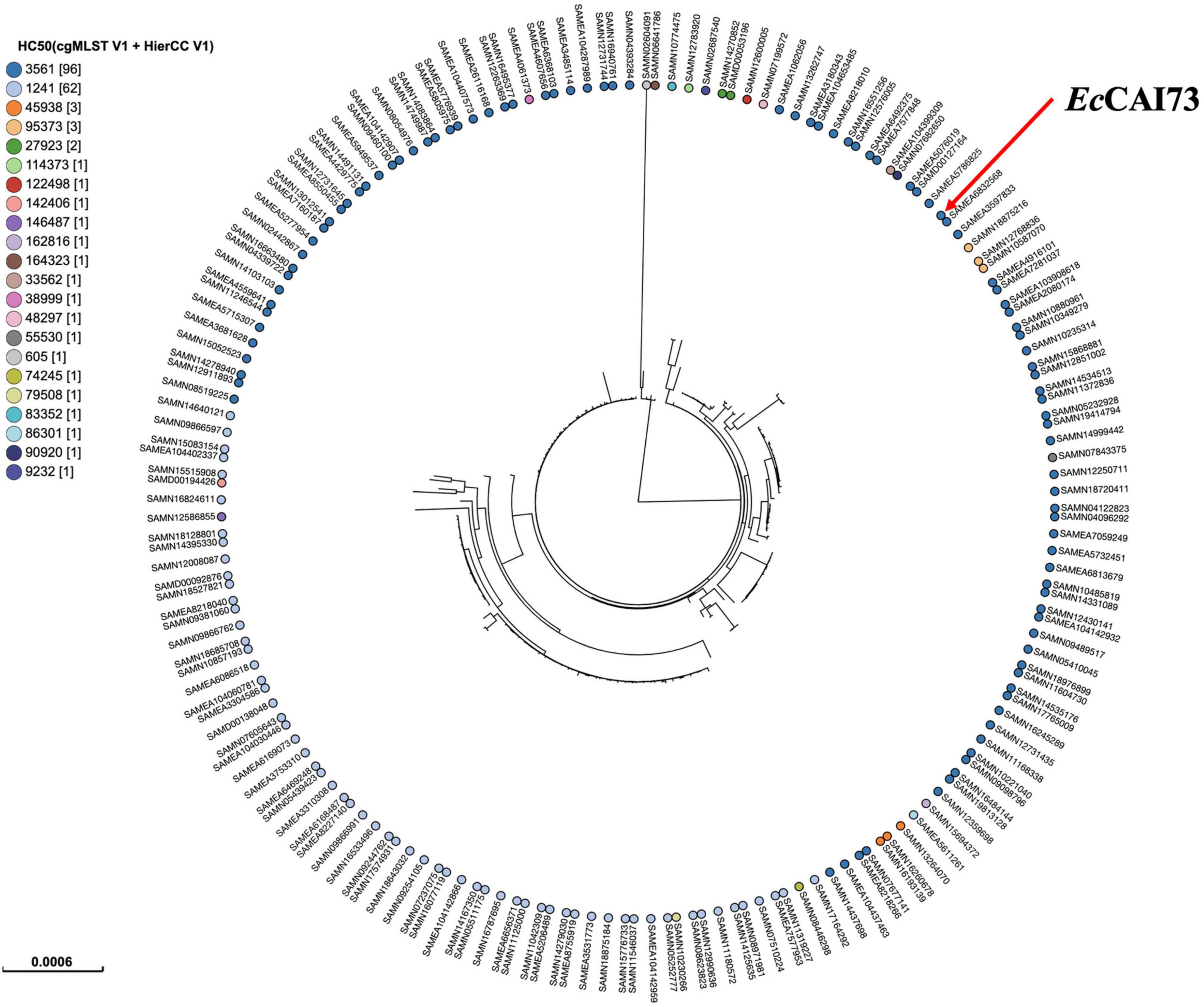
Figure 6. Phylogenetic analysis of ST744 Escherichia coli isolate EcCAI73 and other publicly available ST744 E. coli isolates (n = 181) in Enterobase using single nucleotide polymorphisms (SNPs) and hierarchical clustering (HierCC) of core genome (cg) MLST.
A recent study illustrated that the mcr-1/IncX4 plasmid (pHNSHP23) was stably maintained without an effect on the growth of their hosts conferring a fitness advantage and indicating the ability for an additional dissemination with or without the selection pressure of antibiotics (Wu et al., 2018). Furthermore, the mcr-1/IncP plasmids (pHNGDF1-1 and pHNGDF36-1) were quite stable and increased the biological fitness in the host (Lv et al., 2018). The IncP plasmids has a broad host range and a high conjugation efficiency which may accelerate the spreading of mcr-1 across different hosts (Lv et al., 2018). The future perspective following this study might be as follow: I) assaying the stability of the two plasmids identified in this study, pAMS-51-MCR1 and pAMS-73-MCR, and II) analyzing the fitness costs of these two mcr-1-positive plasmids.
Conclusion
To the best of our knowledge, this study presented the first complete genomic sequence of mcr-1-carrying IncP and IncX4 plasmids from human clinical E. coli isolates in Egypt. In addition, the study illustrated the mcr-1 broad dissemination in diverse plasmids and dissimilar E. coli clones. A multireplicon virulence plasmid, named pAMS-73-Vr, carrying the operons sitABCD, iroBCDE and iucABCD/iutA was identified. Both the strains showed MDR phenotypes, which can be easily converted to extensive (XDR) or pan (PDR) drug-resistant phenotypes by horizontal gene transfer of any carbapenemase gene, particularly blaNDM (highly prevalent in Egypt). Therefore, medical authorities must implement strict infection control policies and antimicrobial surveillance plans (including animals) to control the spread of such strains.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
AS and TaS designed and directed the study. AS performed identification of bacteria, screening and identifying of resistance genes, conjugation, plasmid analysis and typing, analyzed the WGS data, and drafted the manuscript. HR contributed to the genome analysis, data curation, and participated to the writing of the manuscript. SE carried out the collection of samples and bacterial isolation. HN, CJ, and RE-D conceived of the study, made the data analysis, and revised the manuscript. ToS made the data analysis. TaS participated in the discussion on the study design and finalized the manuscript. LY and JH participated in genomic DNA extraction, short-read library preparation, and sequencing. LY carried out long-read library preparation and sequencing, performed hybrid assembly of MiniSeq short reads and Nanopore long reads, and contributed to the genome analysis and performed DDBJ nucleotide sequence submission. MS and LY participated in the discussion on the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED) (21fk0108604j0001). AS was supported by a fellowship (153532) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.989045/full#supplementary-material
Footnotes
- ^ http://www.eucast.org
- ^ https://enterobase.warwick.ac.uk/species/index/ecoli
- ^ https://dfast.nig.ac.jp/
- ^ http://www.genomicepidemiology.org/
- ^ http://brig.sourceforge.net/
- ^ http://mjsull.github.io/Easyfig/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://isfinder.biotoul.fr/
- ^ https://enterobase.warwick.ac.uk/
References
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., and Malorny, B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72, 3317–3324. doi: 10.1093/jac/dkx327
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli, A., Villa, L., Feudi, C., Curcio, L., Orsini, S., Luppi, A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589
Chang, J., Tang, B., Chen, Y., Xia, X., Qian, M., and Yang, H. (2020). Two IncHI2 plasmid-mediated colistin-resistant Escherichia coli strains from the broiler chicken supply chain in Zhejiang Province, China. J. Food Prot. 83, 1402–1410. doi: 10.4315/JFP-20-041
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Clinical and Laboratory Standards Institute [CLSI] (2020). Performance Standards for Antimicrobial Susceptibility Testing, 30th Edn. Wayne, PA: CLSI.
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study (1980–2018). Microorganisms 7:461. doi: 10.3390/microorganisms7100461
Elnahriry, S. S., Khalifa, H. O., Soliman, A. M., Ahmed, A. M., Hussein, A. M., Shimamoto, T., et al. (2016). Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob. Agents Chemother. 60, 3249–3250. doi: 10.1128/AAC.00269-16
Forde, B. M., Zowawi, H. M., Harris, P. N. A., Roberts, L., Ibrahim, E., Shaikh, N., et al. (2018). Discovery of mcr-1-mediated colistin resistance in a highly virulent Escherichia coli lineage. mSphere 3:e00486–18. doi: 10.1128/mSphere.00486-18
Harmer, C. J., and Hall, R. M. (2020). The complete nucleotide sequence of pZM3, a 1970 FIA:FIB:FII plasmid carrying antibiotic resistance and virulence determinants. Microb. Drug Resist. 26, 438–446. doi: 10.1089/mdr.2019.0248
Hasman, H., Hammerum, A. M., Hansen, F., Hendriksen, R. S., Olesen, B., Agersø, Y., et al. (2015). Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 20:30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085
Jousset, A. B., Bernabeu, S., Bonnin, R. A., Creton, E., Cotellon, G., Sauvadet, A., et al. (2019). Development and validation of a multiplex polymerase chain reaction assay for detection of the five families of plasmid-encoded colistin resistance. Int. J. Antimicrob. Agents 53, 302–309. doi: 10.1016/j.ijantimicag.2018.10.022
Kubelová, M., Koláčková, I., Gelbíčová, T., Florianová, M., Kalová, A., and Karpíšková, R. (2021). Virulence properties of mcr-1-positive Escherichia coli isolated from retail poultry meat. Microorganisms 9:308. doi: 10.3390/microorganisms9020308
Liassine, N., Assouvie, L., Descombes, M. C., Tendon, V. D., Kieffer, N., Poirel, L., et al. (2016). Very low prevalence of MCR-1/MCR-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int. J. Infect. Dis. 51, 4–5. doi: 10.1016/j.ijid.2016.08.008
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lu, H., Wang, C., Dong, G., Xu, C., Zhang, X., Liu, H., et al. (2018). Prevalence and molecular characterization of Escherichia coli clinical isolates carrying mcr-1 in a Chinese teaching hospital from 2002 to 2016. Antimicrob. Agents Chemother. 62:e02623–17. doi: 10.1128/AAC.02623-17
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016. EBioMedicine 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Luo, J., Li, M., Zhou, M., and Hu, Y. (2015). Characterization of a novel strain phylogenetically related to Kocuria rhizophila and its chemical modification to improve performance of microbial fuel cells. Biosens. Bioelectron. 69, 113–120. doi: 10.1016/j.bios.2015.02.025
Lv, L., Cao, Y., Yu, P., Huang, R., Wang, J., Wen, Q., et al. (2018). Detection of mcr-1 gene among Escherichia coli isolates from farmed fish and characterization of mcr-1-bearing IncP plasmids. Antimicrob. Agents Chemother. 62:e02378–17. doi: 10.1128/AAC.02378-17
Parreira, V. R., and Gyles, C. L. (2003). A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 71, 5087–5096. doi: 10.1128/IAI.71.9.5087-5096.2003
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088–17. doi: 10.1128/CMR.00088-17
Paveenkittiporn, W., Kamjumphol, W., Ungcharoen, R., and Kerdsin, A. (2021). Whole-genome sequencing of clinically isolated carbapenem-resistant Enterobacterales harboring mcr genes in Thailand, 2016–2019. Front. Microbiol. 11:586368. doi: 10.3389/fmicb.2020.586368
Poirel, L., Kieffer, N., Brink, A., Coetze, J., Jayol, A., and Nordmann, P. (2016). Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob. Agents Chemother. 60, 4394–4397. doi: 10.1128/AAC.00444-16
Ramadan, H., Soliman, A. M., Hiott, L. M., Elbediwi, M., Woodley, T. A., Chattaway, M. A., et al. (2021). Emergence of multidrug-resistant Escherichia coli producing CTX-M, MCR-1, and FosA in retail food from Egypt. Front. Cell. Infect. Microbiol. 11:681588. doi: 10.3389/fcimb.2021.681588
Russo, T. A., McFadden, C. D., Carlino-MacDonald, U. B., Beanan, J. M., Barnard, T. J., and Johnson, J. R. (2002). IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70, 7156–7160. doi: 10.1128/IAI.70.12.7156-7160.2002
Sabri, M., Léveillé, S., and Dozois, C. M. (2006). A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152, 745–758. doi: 10.1099/mic.0.28682-0
Soliman, A. M., Maruyama, F., Zarad, H. O., Ota, A., Nariya, H., Shimamoto, T., et al. (2020a). Emergence of a multidrug-resistant Enterobacter hormaechei clinical isolate from Egypt co-harboring mcr-9 and blaVIM-4. Microorganisms 8:595. doi: 10.3390/microorganisms8040595
Soliman, A. M., Zarad, H. O., Nariya, H., Shimamoto, T., and Shimamoto, T. (2020b). Genetic analysis of carbapenemase-producing Gram-negative bacteria isolated from a university teaching hospital in Egypt. Infect. Genet. Evol. 77:104065. doi: 10.1016/j.meegid.2019.104065
Soliman, A. M., Ramadan, H., Zarad, H., Sugawara, Y., Yu, L., Sugai, M., et al. (2021). Coproduction of Tet(X7) conferring high-level tigecycline resistance, fosfomycin FosA4, and colistin Mcr-1.1 in Escherichia coli strains from chickens in Egypt. Antimicrob. Agents Chemother. 65:e02084–20. doi: 10.1128/AAC.02084-20
Stoesser, N., Mathers, A. J., Moore, C. E., Day, N. P., and Crook, D. W. (2016). Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect. Dis. 16, 285–286. doi: 10.1016/S1473-3099(16)00010-4
Tartor, Y. H., El-Aziz, A., Khairy, N., Gharieb, R., El Damaty, H. M., Enany, S., et al. (2021a). Whole genome sequencing of Gram-negative bacteria isolated from bovine mastitis and raw milk: The first emergence of colistin mcr-10 and fosfomycin fosA5 resistance genes in Klebsiella pneumoniae in Middle East. Front. Microbiol. 12:770813. doi: 10.3389/fmicb.2021.770813
Tartor, Y. H., Gharieb, R., El-Aziz, A., Norhan, K., El Damaty, H. M., Enany, S., et al. (2021b). Virulence determinants and plasmid-mediated colistin resistance mcr genes in Gram-negative bacteria isolated from bovine milk. Front. Cell. Infect. Microbiol. 11:761417. doi: 10.3389/fcimb.2021.761417
Tegetmeyer, H. E., Jones, S. C., Langford, P. R., and Baltes, N. (2008). ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 128, 342–353. doi: 10.1016/j.vetmic.2007.10.025
Torres, A. G., Redford, P., Welch, R. A., and Payne, S. M. (2001). TonB-dependent systems of uropathogenic Escherichia coli: Aerobactin and heme transportand TonB are required for virulence in the mouse. Infect. Immun. 69, 6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, R., Yi, L. X., Yu, L. F., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 9:331. doi: 10.3389/fmicb.2018.00331
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:27. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543–17. doi: 10.1128/mBio.00543-17
Zakaria, A. S., Edward, E. A., and Mohamed, N. M. (2021). Genomic insights into a colistin-resistant uropathogenic Escherichia coli strain of O23:H4-ST641 lineage harboring mcr-1.1 on a conjugative IncHI2 plasmid from Egypt. Microorganisms 9:799. doi: 10.3390/microorganisms9040799
Keywords: E. coli, Egypt, IncP plasmid, IncX4 plasmid, ST1011, ST744, mcr-1
Citation: Soliman AM, Ramadan H, Yu L, Hisatsune J, Sugai M, Elnahriry SS, Nariya H, El-Domany RA, Shimamoto T, Jackson CR and Shimamoto T (2022) Complete genome sequences of two Escherichia coli clinical isolates from Egypt carrying mcr-1 on IncP and IncX4 plasmids. Front. Microbiol. 13:989045. doi: 10.3389/fmicb.2022.989045
Received: 08 July 2022; Accepted: 16 August 2022;
Published: 09 September 2022.
Edited by:
Chang-Wei Lei, Sichuan University, ChinaReviewed by:
Anusak Kerdsin, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus, ThailandZhihai Liu, Qingdao Agricultural University, China
Copyright © 2022 Soliman, Ramadan, Yu, Hisatsune, Sugai, Elnahriry, Nariya, El-Domany, Shimamoto, Jackson and Shimamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed M. Soliman, YWhtZWRfc29saW1hbkBwaGFybS5rZnMuZWR1LmVn; Liansheng Yu, eXVAbmlpZC5nby5qcA==; Tadashi Shimamoto, dGFkYXNoaXNAaGlyb3NoaW1hLXUuYWMuanA=
 Ahmed M. Soliman
Ahmed M. Soliman Hazem Ramadan
Hazem Ramadan Liansheng Yu4*
Liansheng Yu4* Charlene R. Jackson
Charlene R. Jackson Tadashi Shimamoto
Tadashi Shimamoto