
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 02 September 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.988730
 Yuan-Ni Shi1,2,3†
Yuan-Ni Shi1,2,3† Lin-Miao Li3†
Lin-Miao Li3† Jia-Bin Zhou3
Jia-Bin Zhou3 Yan Hua4
Yan Hua4 Zhi-Liao Zeng5
Zhi-Liao Zeng5 Ye-Pin Yu3
Ye-Pin Yu3 Ping Liu3
Ping Liu3 Zi-Guo Yuan1,2*
Zi-Guo Yuan1,2* Jin-Ping Chen3*
Jin-Ping Chen3*Pangolins are endangered animals and are listed in the CITES Appendix I of the Convention International Trade Endangered Species of Wild Fauna and Flora as well as being the national first-level protected wild animal in China. Based on a few reports on pangolins infected with pestiviruses of the Flaviviridae family, Pestivirus infections in pangolins have attracted increasing attention. Pangolin pestivirus is a pathogen that may cause diseases such as acute diarrhea and acute hemorrhagic syndrome. To better understand the epidemiology and genomic characterization of pestiviruses carried by pangolins, we detected pestiviruses in dead Malayan pangolin using metavirome sequencing technology and obtained a Pestivirus sequence of 12,333 nucleotides (named Guangdong pangolin Pestivirus, GDPV). Phylogenetic tree analysis based on the entire coding sequence, NS3 gene or RdRp gene sequences, showed that GDPV was closely related to previously reported pangolin-derived Pestivirus and clustered into a separate branch. Molecular epidemiological investigation revealed that 15 Pestivirus-positive tissues from two pangolins individuals with a positivity rate of 5.56%, and six Amblyomma javanense carried pestiviruses with a positivity rate of 19.35%. Moreover, the RdRp gene of the Pestivirus carried by A. javanense showed a high similarity to that carried by pangolins (93–100%), indicating A. javanense is likely to represent the vector of Pestivirus transmission. This study expands the diversity of viruses carried by pangolins and provides an important reference value for interrupting the transmission route of the virus and protecting the health of pangolins.
Pangolins belong to the genus Manis (family: Manidae, order: Pholidota, class: Mammalia). Their bodies are covered with scales and they have no teeth. Pangolins are nocturnal and feed on termites and ants, based on which they are also known as the scaly anteater (Wang et al., 2016). Pangolins are affected the worst among mammals in the global wildlife trade owing to their meat and inflated medicinal properties (Boakye et al., 2015; Gaubert et al., 2018). On the International Union for Conservation of Nature, Red List of Threatened Species, Chinese pangolin and Malayan pangolin are listed as critically endangered; Philippine pangolins (M. culionensis), Indian Pangolin (M. crassicaudata), Giant Pangolin (Smutsia gigantea), and White-bellied Pangolin (Phataginus tricuspis) are listed as endangered species; Ground Pangolin (S. temminckii) and Black-bellied Pangolin (P. tetradactyla) are listed as vulnerable species (du Toit et al., 2017). The habitat of the pangolin is mainly include tropical forests, limestone areas, bamboo forests, broad-leaved forests, and shrubs, in which the ticks live. Ticks are obligate hematophagous ectoparasites of wild and domestic animals as well as humans (Domingos et al., 2013). The Java tick transmits Anaplasma spp. (Koh et al., 2016) and novel Borrelia spp. (Jiang et al., 2021). There are only few studies available on ticks and tick-borne diseases infecting pangolin species (Khatri-Chhetri et al., 2016). It has been reported that the Amblyomma javanense carrying pestivirus may play an important role in spreading diseases of pangolins (Gao et al., 2020).
Viruses have been reported to infect pangolins only in recent years. Pangolins are mainly infected by parainfluenza virus 5 (Wang et al., 2019), canine parvovirus (Chang et al., 2021), pestivirus (He et al., 2022), coltiviruses (Gao et al., 2020), Sendai virus, and coronavirus (Liu et al., 2019). Among them, the detection of pangolin-infecting pestivirus found in the Zhejiang Province, China, indicates that pestiviruses are likely to represent the main pathogen associated with pangolin mortality (Gao et al., 2020; He et al., 2022).
Pangolin pestiviruses, belonging to the genus Pestivirus of the Flaviviridae family, are positive-sense single-stranded RNA viruses. They are spherical and enveloped, 40–60 nm in diameter, 11.3–13.0 kb in size, and contain approximately 3,900 amino acids that form a large open reading frame (ORF; Tautz et al., 2015). Pestiviruses mainly infect pigs and ruminants (Vilcek and Nettleton, 2006) and cause severe diseases in animals, such as hemorrhagic diseases in pigs, cattle, and sheep, as well as respiratory diseases (Moenning, 1990). Common pestivirus-associated diseases include classical swine fever (Blome et al., 2017), bovine viral diarrhea (Deregt and Loewen, 1995), and sheep border disease (Righi et al., 2021). These viruses have been detected in wild animals such as bats, bamboo rats, wild boars, bison, giraffes, and wild deer via next-generation sequencing (Harasawa et al., 2000; Vilcek and Nettleton, 2006; Wu et al., 2018). Pestiviruses have also been found in arthropods (Harvey et al., 2019; Gao et al., 2020). In general, many wild mammals can carry pestiviruses. Pestiviruses are currently classified into 11 species on ICTV, and pangolin pestiviruses have not yet been clearly classified. Some literature suggests adding eight pestiviruses, among which pangolin pestivirus is classified as pestivirus P (Postel et al., 2021). This also shows that pestivirus research has received more and more attention.
The genome of pangolin pestivirus remains largely unclear, and the mode of transmission is also unknown. Here, we report a new rapidly mutating pestivirus carried by pangolins and their parasitic ticks in Guangdong Province, China. Moreover, we performed molecular epidemiological investigation and variation analysis of pestiviruses carried by pangolins and ticks in Guangdong Province, which may support the protection of pangolins.
From August 2019 to May 2021, we collected heart, liver, spleen, lung, kidney, throat swabs, anal swabs, and other tissue samples from pangolins in Guangzhou, Shenzhen, and Heyuan, Guangdong Province. A total of 144 samples were collected from 36 pangolins (11 dead and 25 healthy individuals). A total of 31 parasitic ticks on pangolins were collected from Guangzhou and Shenzhen, Guangdong Province, China from April 2019 to June 2021. All samples were stored in liquid nitrogen and transferred to a laboratory freezer at −80°C. All lab personnel were trained and wore protective clothing for protection against biological agents before sample collection (see Supplementary Table S1 for Sample information).
RNAiso Plus (Takara Bio, Dalian, China) was used to extract total RNA from ticks and tissues of pangolins, according to the manufacturer protocols. The tissue samples (50 mg of each sample) were ground into powder in liquid nitrogen and transferred to a DEPC-treated EP tube before being volatilized by liquid nitrogen. The anal and throat swab samples were vortexed for 1 min, and 140 ml of supernatant was collected from each sample after centrifuging at 3,000 g at 4°C for 1 min. Viral nucleic acid was extracted using the QIAamp® Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Subsequently, 1 μg cDNA was synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen, Waltham, MA, United States) and stored at −20°C in a refrigerator till sequencing.
Pangolins were identified based on morphological characteristics. Ticks were identified via morphological observation and 16S rRNA sequencing. The primers used were PCTY-F (5′-CTGCTCAATGATTTTTTAAATTGCTGTGG-3′) and PCTY-R (5′-CCGGTCTGAACTCAGATCAAGT-3′; Yang et al., 2021). A 50 μl reaction was set up, containing 25 μl PCR mix (GenTech, Shanghai, China), 22 μl double-distilled H2O, 1 μl template, 1 μl forward primer (10 μM), and 1 μl reverse primer (10 μM). T100™ Thermal cycling (Bio-rad, United States) was performed at 94°C for 7 min, followed by 34 cycles at 94°C for 30 s, 45°C for 30 s, 72°C for 1 min, and a final extension of 72°C for 8 min. PCR products were detected on a 1.5% agarose gel and sequenced. In total, 18 sequences of the genus Amblyomma were downloaded from the NCBI database (National Institutes of Health, Bethesda, MD, United States). The FTT-NS-2 algorithm was implemented in MAFFT v7.407 to align mitochondrial 16S rDNA of ticks (Katoh and Standley, 2013). Phylogenetic trees based on nucleotide sequences of 16S rDNA were constructed using the maximum likelihood method using the MrBayes approach using the SYM + G nucleotide substitution model (Huelsenbeck and Ronquist, 2001).
Reverse-transcribe to double-stranded cDNA using previously extracted lymph node RNA of M1/B1. Synthesis of first and second strands, end repair, and adaptor ligation were performed for library construction, as described previously (Liu et al., 2019). High-throughput sequencing was performed by the Magigene Company (Guangzhou, China). Clean reads were de novo assembled using MEGAHIT version 1.0 (Li et al., 2016). BWA (v0.7.17, default parameter: mem-k 30) software was used to align clean reads to the GenBank non-redundant nucleotide (NT) database. Contigs were then classified by BLASTx against the NT database using alignment similarity ≥80%, length of matched area ≥ 500 bp, and e-value ≤10−5. Contigs with significant BLASTx hits were confirmed as viral sequences (Li and Durbin, 2009).
Multiple alignments were performed with existing pangolin virus sequences in GenBank and specific primers were designed. Reverse transcription-PCR was used to amplify the whole-genome sequence of viruses from M1/B1 Malayan pangolins using lymph node and kidney samples (see Supplementary Table S2 for primer information), and the PCR products were directly sequenced after gel purification and the sequencing results were spliced into the whole genome sequence.
We used CLUSTALW1 and Simplot v3.5.1 for genomic similarity analysis of pestiviruses. In total, 25 complete genome sequences of pestiviruses were downloaded from the NCBI database2 (See Supplementary Table S3 for sequence information), and phylogenetic tree analysis was performed for polyprotein, nonstructural protein 3 (NS3), and RNA-dependent RNA polymerase (RdRp) gene sequences. Viral sequences were aligned using the FTT-NS-2 algorithm implemented in MAFFT v7.407 (Katoh and Standley, 2013). The phylogenetic trees were constructed via the maximum likelihood method using the MrBayes approach using the GTR + G or GTR + I + G nucleotide substitution model (Huelsenbeck and Ronquist, 2001). Then, all the ML trees were visualized and exported as vector diagrams with FigTree version 1.4.3.3
Primers for absolute quantitative PCR detection were designed using the sequence of the Guangdong pangolin pestivirus (GDPV) E2 gene. The fluorescence quantitative screening primer E2 gene sequence was ligated into a plasmid to construct a standard template, and a 10-fold dilution was used to construct a standard curve to achieve absolute quantitative screening of pestivirus nucleic acids in the remaining samples. The detection of GDPV in M1/B1 pangolin tissues by absolute quantitative PCR was to find the target organ of the novel virus. Then, the verification of the virus was performed with nested PCR by targeting the RdRp gene using the specific primer set. The total volume of the absolute quantitative PCR mixture was 20 μl; this included 10 μl 2× qPCR mixture (Takara Bio), 0.5 μl of forward and reverse primers, 1 μl of template, and the remaining volume included nuclease-free water. The following cycle was used for amplification: 50°C for 2 min; 95°C for 2 min; 40 cycles at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s; 60°C for 1 min; and 95°C for 15 s (primers are listed in Supplementary Table S2).
According to body characteristics, there were 20 Manis javanica and 16 M. pentadactyla species. The ticks were analyzed via morphological observation (Figure 1A), PCR amplification and sequencing, and phylogenetic analysis with other tick genera (Figure 1B), and all 31 ticks were identified as A. javanense.
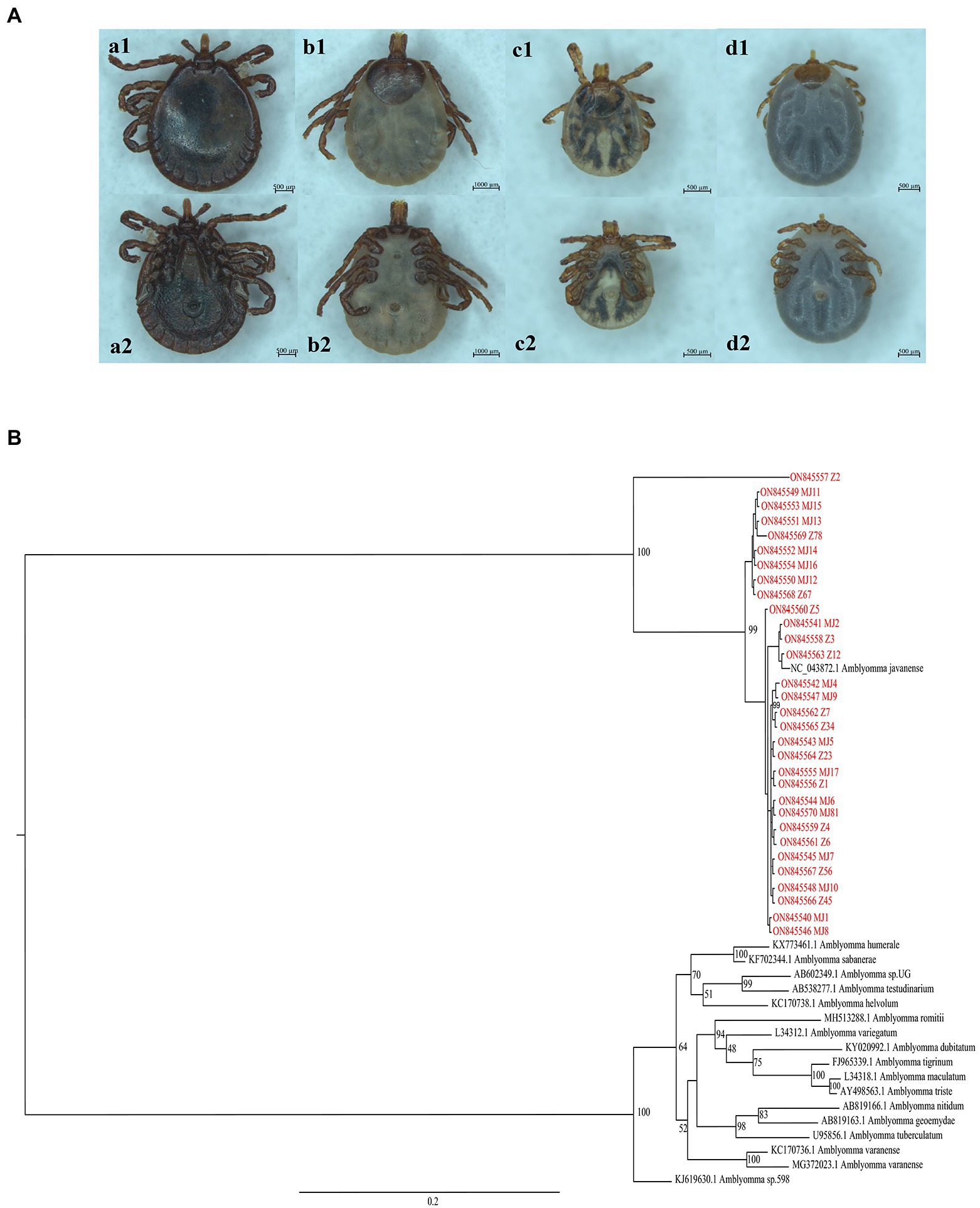
Figure 1. (A) View of the dorsal and ventral surfaces of the tick under a stereoscope. a1 and a2 are dorsal and ventral surfaces of male adult ticks, respectively; b1 and b2 are dorsal and ventral surfaces of female adult ticks, respectively; c1, c2, d1, and d2 are dorsal and ventral surfaces of nymphs. (B) Phylogenetic trees based on nucleotide sequences of 16S rRNA. The trees were constructed via the maximum likelihood method using the MrBayes approach using the SYM + G nucleotide substitution model. Red letters represent the ticks isolated from Guangdong in the present study.
Sequencing analysis of lymph nodes of M1/B1 pangolin yielded 12.92 Gb of data (28,168,231 clean reads, 150 bp in length). Using BWA software to compare clean reads to virus reference data, a total of 3,708 reads were compared. According to the NCBI taxonomy database, the proportion of DNA viruses compared accounted for 24.93% and the proportion of RNA viruses accounted for 75.07%. Among them, there were six contigs of the genus Pestivirus of the Flaviviridae family, with a total of 9,195 nucleotides (nt; Supplementary Table S4; BioProject ID: PRJNA854209).
By performing PCR amplification, we obtained the whole genome sequence of GDPV isolate M1/B1 lymph and kidney tissue, which contains a total of 12,333 nt (The GenBank number: ON843279). The genomic similarity analysis with other known pestiviruses showed that the genome sequence of the virus had >40% nucleotide similarity and > 45% amino acid similarity with other known pestiviruses (Table 1). Low similarity and no recombination with other pangolin pestiviruses were found (Figure 2A). GDPV, like other pestiviruses, contains a single long ORF and encodes 12 proteins simultaneously (Figure 2B). Moreover, it encodes two proteins unique to pestiviruses, namely Npro and Erns. Among them, NS3 serine proteinase serves as a molecular model of the polyprotein cleavage site, cofactor requirements, and enzymes necessary for viral replication. Based on the characteristics of the existing Pestivirus NS3 cleavage sites (a conserved leucine at the P1 position of all four cleavage sites), followed by serine or alanine (Xu et al., 1997), all cleavage sites were found in NS3 of GDPV (Figure 2C) located at L2474-K2475. Among them, the NS4A/NS4B cleavage site of Dongyang pangolin pestivirus is located at L2315/K2316; the NS4A/NS4B cleavage site of bovine viral diarrhea is located at L2426/A2427.
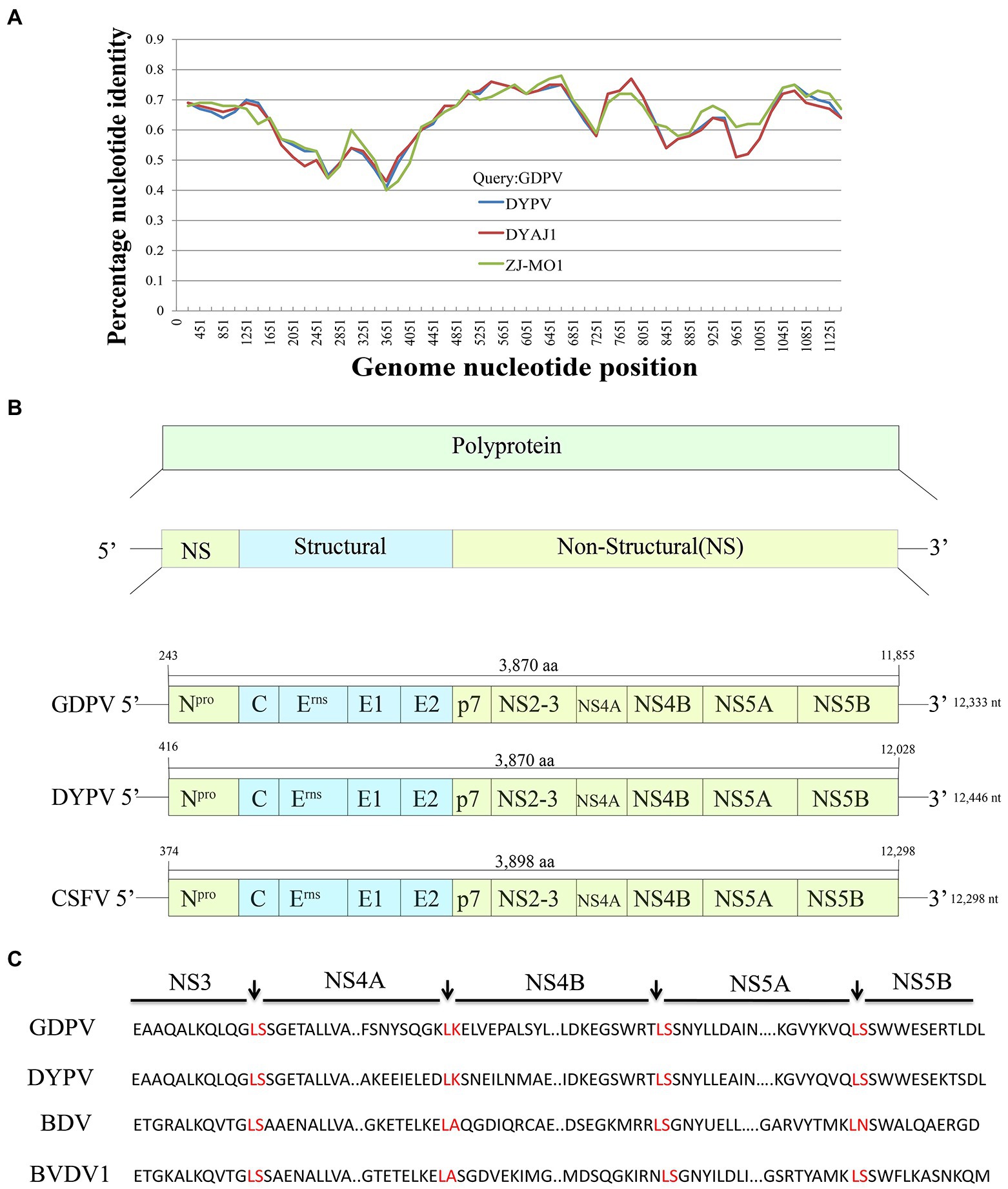
Figure 2. (A) Recombination analysis of Guangdong pangolin Pestivirus (GDPV) and other pangolin pestivirus based on the whole-genome sequences using Simplot v3.5.10. GDPV, Guangdong pangolin pestivirus isolate M1/B1; DYPV, Dongyang pangolin virus isolate DYCS; DYAJ1, Dongyang pangolin virus isolate DYAJ1; ZJ-MO1, pangolin Pestivirus 3 isolate ZJ-MO1. The analysis was performed with the Kimura model, with a window size of 11, 613 bp and a step size of 150 bp. (B) Comparison of GDPV genome structure with that of other Pestivirus genomes. GDPV, Guangdong pangolin pestivirus isolate M1/B1; DYPV, Dongyang pangolin pestivirus isolate DYCS; CSFV, Classical swine fever virus. (C) All cleavage sites of NS3 protease in GDPV. GDPV, Guangdong pangolin pestivirus isolate M1/B1; DYPV, Dongyang pangolin virus isolate DYCS; BDV, Border disease virus; and BVDV1, Bovine viral diarrhea virus 1.
The whole-genome sequences of different pestiviruses were downloaded from GenBank to explore the evolutionary relationship between GDPV and other pestiviruses in this study. A phylogenetic tree was constructed based on the nucleotide sequences of polyprotein, NS3, and RdRp genes. Phylogenetic analysis based on polyprotein (Figure 3A) and NS3 gene (Figure 3B) showed that GDPV clustered with three other pangolin Pestivirus strains. Based on the evolutionary tree of RdRp genes, certain RdRp genes of Guangdong tick Pestivirus (named Guangdong ticks Pestivirus, GDTPV) were clustered with that of GDPV (Figure 3C). Further, certain RdRp nucleotide sequences of GDTPV showed 93–100% similarity with that of GDPV (Supplementary Table S5).
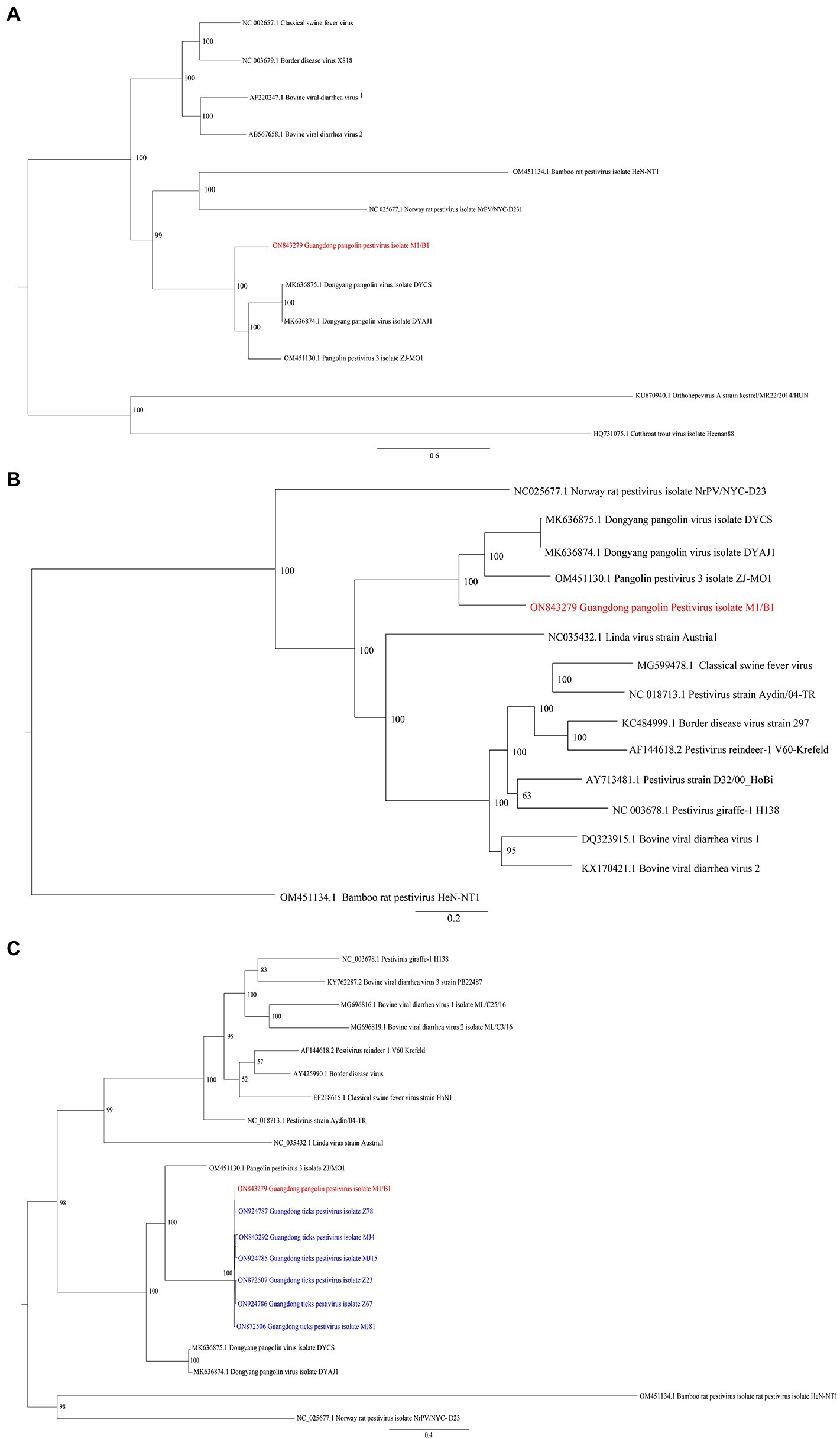
Figure 3. Phylogenetic trees based on nucleotide sequences of (A) polyprotein, (B) NS3 gene, and (C) RdRp gene. The trees were constructed via the maximum likelihood method using the MrBayes approach using the GTR + G (A,B) or GTR + I + G (C) nucleotide substitution model. Red letters represent pestiviruses isolated from M1/B1 pangolins in this study. Blue letters represent pestiviruses isolated from Amblyomma javanense from Guangdong in the present study.
Absolute quantitative PCR designed based on the E2 gene was performed to detect the presence of GDPV in pangolin samples. According to the results, it was found that the mentioned first melting temperature (Tm) of GDPV was 78°C ± 0.5°C, the slope = −3.462, and the R2 = 0.9927, which had a good linear relationship. The amplification efficiency of GDPV was 95% according to the calculation formula of amplification efficiency (Supplementary Figure S1; Figure 4). GDPV was detected in M1/B1 pangolin-derived heart, liver, spleen, lung, kidney, pancreas, stomach, and lymph nodes. The Pestivirus nucleic acid content in the kidney was approximately 500-fold higher than that in the heart and liver. This indicated that the kidneys are likely to represent target organs of GDPV (Table 2).
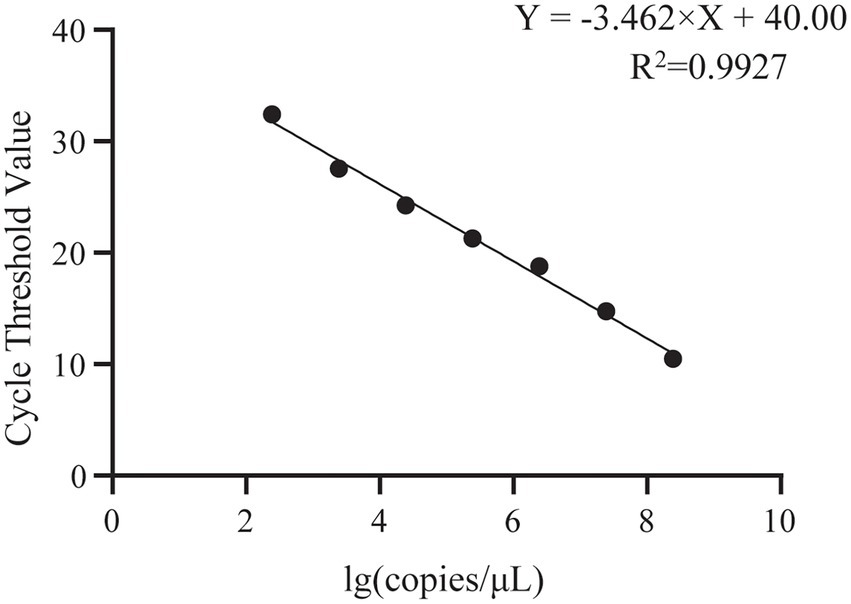
Figure 4. Standard curve for GDPV detection using single-plex qRT-PCR. Copies number/μl of GDPV from 1 × 108 to 1 × 102 copies/μl.
Nested PCR screening of the RdRp gene revealed another Pestivirus in D6 pangolins, named Guangdong pangolin Pestivirus 2 (Guangdong pangolin pestivirus 2 isolate D6, GDPV2), which was found in liver, spleen, lung, and kidney, duodenum, rectum, and anus: a total of seven tissues. Pestiviruses were detected in six ticks labeled as Mj4, Mj15, Z23, Z67, Z78, and Mj81. We also tested a total of 25 healthy pangolin samples and found no pestivirus. To sum up, we analyzed a total of 15 tissues (M1/B1 and D6 pangolins), with a positivity rate of 10.42%; six ticks carried Pestivirus with a positivity rate of 19.35%.
Since the emergence of the new coronavirus in 2019, the possibility of disease outbreaks caused by the consumption of wild animals should be considered (Turcios-Casco and Cazzolla Gatti, 2020). Research on zoonotic pathogens represents a hot topic. As the most smuggled mammal, pangolins are prone to contact with other wild animals and are susceptible to pathogens that cause disease and death. As another blood-sucking parasite, the mosquito may carry several viruses such as Japanese encephalitis virus (JEV) and Getah virus (GETV). And the mosquito-borne viruses have been reported in another pangolin virome study recently (Zhang et al., 2022). The trafficking of pangolins has caused almost irreversible damage to their population (Hu et al., 2020). Chinese customs have made great contributions to intercepting a large number of smuggled pangolins. Due to the lack of research on pangolin diseases and the lack of knowledge about their stress management, rescued pangolins often suffer from poor health and even death. Research on pathogens that infect pangolins will assist in the prevention and treatment of pangolin diseases. The characteristics of many new viruses have been revealed via viromics analysis, especially in the monitoring of viruses that infect wild animals. An in-depth understanding of the viruses that infect wild animals plays an important role in the prediction of viral zoonotic outbreaks. For example, viromics studies on bats have revealed more than 130 viruses that infect bats (Hu et al., 2017) including several newly discovered pathogens threatening the public health, e.g., the Ebola virus (Leroy et al., 2005), severe acute respiratory syndrome virus (Wang et al., 2006), and Marburg virus (Swanepoel et al., 2007). Similarly, by performing viromics analysis of pangolins, we identified a potential pathogen.
Pestivirus are newly discovered viruses in pangolins. Pestiviruses have been found to be detected in dead pangolins alone and not in healthy individuals (He et al., 2022). A total of 11 dead pangolins and 25 healthy pangolins were investigated for pestivirus-carrying status, and only two dead pangolins were found to be carrying pestivirus. However, none of the healthy pangolins were tested positive for pestivirus. Therefore, Pestivirus infections are likely to represent one of the causes of pangolin deaths, which were supported by our epidemiological survey. The phylogenetic trees of the GDPV polyprotein, NS3, and RdRp genes constructed in this study showed that the Guangdong pangolin Pestivirus formed a separate branch and had a low similarity with other Pestivirus gene sequences. The nucleotide similarity of GDPV and other pangolin pestiviruses was low (66–69%), which proves that GDPV is a new strain of pangolin pestivirus. Notably, GDPV2 detected in the D6 pangolin was detected using the nested PCR method designed based on the RdRp gene, but not by the quantitative PCR assay designed based on the E2 gene, indicating that the E2 gene sequence differed between GDPV2 and GDPV. The same was true for pestiviruses detected in ticks that were not detected via real-time PCR, but were detected via nested PCR. This further shows that the specificity of the E2 gene was relatively higher and the RdRp gene is relatively conservative. However, since the whole-genome sequence of GDPV2 was not verified, its similarity could not be determined, although GDPV2 may represent a new strain of pangolin Pestivirus. We also detected the canine parvovirus in the spleen tissues of D6 pangolins (data not shown), further indicating that the viruses carried by pangolins are diverse. Additionally, GDPV nucleic acid content was determined in various tissues of pangolins and we found that the kidneys and pancreas contained the highest GDPV content, which provided some insights into the tropism of Pestivirus into pangolins.
The distribution of A. javanense in China is mainly concentrated in Fujian, Guangdong, Hainan, and Yunnan. The main host of A. javanense is pangolins (Duan et al., 2020), and the pathogens carried by these ticks need to be studied to control the spread of pangolin diseases. It has been reported that Pestivirus isolated from ticks that parasitize pangolins only show a 2% nucleotide difference compared with pangolin-derived pestiviruses (Gao et al., 2020). In this study, six Pestivirus-positive A. javanense were found by screening for RdRp gene sequences, and the sequencing results showed that the Pestivirus in A. javanense was also very close to GDPV. This strongly supports the possibility that A. javanense plays a possible role in the transmission of viruses in pangolins and is likely to represent the transmission vector of viruses. In addition to parasitizing pangolins, the A. javanense parasitizes wild boars and reptiles such as pythons, monitor lizards, and turtles (Jabin et al., 2019). Tick-related diseases in wild animals should be monitored. Due to low similarities between GDPV2 and GDPV, the whole-genome sequence verification and assembly of GDPV was a great challenge, which is also limited by the sources of sample. Besides, the limitation of pangolin and tick sample sources also affected the epidemiological analysis.
Pangolins carry various viruses; viruses are killing pangolins. Multiple pangolin pestiviruses were found with low similarities as they mutate rapidly. Further investigation on the virus status of wild animals and their parasites, like pangolins and ticks, is urgently needed. And the better understanding of the spreading ways and the underlying pathogenesis of the viruses carried by wildlife is absolutely important for the public health safety. The present study expands the knowledge on the viral spectrum of pangolins and has an important role in the conservation and public health of rare wildlife.
The datasets presented in this study are deposited in the gen bank repository, accession number PRJNA854209, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA854209.
The animal study was reviewed and approved by the Committee on the Ethics of Animal Experiments of the Institute of Zoology of Guangdong Academy of Sciences.
J-PC and Z-GY contributed to the conception of the study. J-BZ, Z-LZ, and YH collected the samples. Y-NS, L-ML, and J-BZ performed virus detection and sequencing. Y-NS and L-ML performed the metagenomics analysis. Y-NS, PL, and L-ML analyzed the data. Y-NS wrote the manuscript. L-ML, Y-PY, Z-GY, and J-PC revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the GDAS Special Project of Science and Technology Development (2020GDASYL-20200103090), the National Natural Science Foundation of China (31972707), and the 2022 Provincial financial special project for Ecological Forestry construction.
We acknowledge the support of Wenzhong Huang and Huiming Li for helping us to collect pangolin samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.988730/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | (A) Gradient amplification curve for GDPV detection using single-plex qRT-PCR, from 1 × 108 — 1 × 102 368 copies/μl; (B) Dissolution curve for GDPV detection 369 using single-plex qRT-PCR, Tm value is 78 ± 0.5°C.
Blome, S., Staubach, C., Henke, J., Carlson, J., and Beer, M. (2017). Classical swine fever—an updated review. Viruses 9:86. doi: 10.3390/v9040086
Boakye, M. K., Pietersen, D. W., Kotzé, A., Dalton, D. L., and Jansen, R. (2015). Knowledge and uses of African pangolins as a source of traditional medicine in Ghana. PLoS One 10:e0117199. doi: 10.1371/journal.pone.0117199
Chang, Y. C., Lin, Z. Y., Lin, Y. X., Lin, K. H., Chan, F. T., Hsiao, S. T., et al. (2021). Canine parvovirus infections in Taiwanese pangolins (Manis pentadactyla pentadactyla). Vet. Pathol. 58, 743–750. doi: 10.1177/03009858211002198
Deregt, D., and Loewen, K. G. (1995). Bovine viral diarrhea virus: biotypes and disease. Can. Vet. J. 36, 371–378.
Domingos, A., Antunes, S., Borges, L., and Rosário, V. E. (2013). Approaches towards tick and tick-borne diseases control. Rev. Soc. Bras. Med. Trop. 46, 265–269. doi: 10.1590/0037-8682-0014-2012
du Toit, Z., du Plessis, M., Dalton, D. L., Jansen, R., Paul Grobler, J., and Kotzé, A. (2017). Mitochondrial genomes of African pangolins and insights into evolutionary patterns and phylogeny of the family Manidae. BMC Genomics 18:746. doi: 10.1186/s12864-017-4140-5
Duan, D. Y., Tang, J. M., Chen, Z., Liu, G. H., and Cheng, T. Y. (2020). Mitochondrial genome of Amblyomma javanense: a hard tick parasite of the endangered Malayan pangolin (Manis javanica). Med. Vet. Entomol. 34, 229–235. doi: 10.1111/mve.12403
Gao, W. H., Lin, X. D., Chen, Y. M., Xie, C. G., Tan, Z. Z., Zhou, J. J., et al. (2020). Newly identified viral genomes in pangolins with fatal disease. Virus Evol. 6:veaa020. doi: 10.1093/ve/veaa020
Gaubert, P., Antunes, A., Meng, H., Miao, L., Peigné, S., Justy, F., et al. (2018). The complete phylogeny of pangolins: scaling up resources for the molecular tracing of the most trafficked mammals on earth. J. Hered. 109, 347–359. doi: 10.1093/jhered/esx097
Harasawa, R., Giangaspero, M., Ibata, G., and Paton, D. J. (2000). Giraffe strain of pestivirus: its taxonomic status based on the 5′-untranslated region. Microbiol. Immunol. 44, 915–921. doi: 10.1111/j.1348-0421.2000.tb02583.x
Harvey, E., Rose, K., Eden, J. S., Lo, N., Abeyasuriya, T., Shi, M., et al. (2019). Extensive diversity of RNA viruses in Australian ticks. J. Virol. 93, e01358–e01418. doi: 10.1128/jvi.01358-18
He, W. T., Hou, X., Zhao, J., Sun, J., He, H., Si, W., et al. (2022). Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 185, 1117–1129.e8. doi: 10.1016/j.cell.2022.02.014
Hu, J. Y., Hao, Z. Q., Frantz, L., Wu, S. F., Chen, W., Jiang, Y. F., et al. (2020). Genomic consequences of population decline in critically endangered pangolins and their demographic histories. Natl. Sci. Rev. 7, 798–814. doi: 10.1093/nsr/nwaa031
Hu, D., Zhu, C., Wang, Y., Ai, L., Yang, L., Ye, F., et al. (2017). Virome analysis for identification of novel mammalian viruses in bats from Southeast China. Sci. Rep. 7:10917. doi: 10.1038/s41598-017-11384-w
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jabin, G., Dewan, Y., Khatri, H., Singh, S. K., Chandra, K., and Thakur, M. (2019). Identifying the tick Amblyomma javanense (Acari: Ixodidae) from Chinese pangolin: generating species barcode, phylogenetic status and its implication in wildlife forensics. Exp. Appl. Acarol. 78, 461–467. doi: 10.1007/s10493-019-00393-1
Jiang, B. G., Wu, A. Q., Jiang, J. F., Yuan, T. T., Xu, Q., Lv, C. L., et al. (2021). Molecular detection of novel Borrelia species, Candidatus Borrelia javanense in Amblyomma javanense ticks from pangolins. Pathogens 10:728. doi: 10.3390/pathogens10060728
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khatri-Chhetri, R., Wang, H. C., Chen, C. C., Shih, H. C., Liao, H. C., Sun, C. M., et al. (2016). Surveillance of ticks and associated pathogens in free-ranging Formosan pangolins (Manis pentadactyla pentadactyla). Ticks Tick Borne Dis. 7, 1238–1244. doi: 10.1016/j.ttbdis.2016.07.007
Koh, F. X., Kho, K. L., Panchadcharam, C., Sitam, F. T., and Tay, S. T. (2016). Molecular detection of Anaplasma spp. in pangolins (Manis javanica) and wild boars (Sus scrofa) in peninsular Malaysia. Vet. Parasitol. 227, 73–76. doi: 10.1016/j.vetpar.2016.05.025
Leroy, E. M., Kumulungui, B., Pourrut, X., Rouquet, P., Hassanin, A., Yaba, P., et al. (2005). Fruit bats as reservoirs of Ebola virus. Nature 438, 575–576. doi: 10.1038/438575a
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, D., Luo, R., Liu, C. M., Leung, C. M., Ting, H. F., Sadakane, K., et al. (2016). MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11. doi: 10.1016/j.ymeth.2016.02.020
Liu, P., Chen, W., and Chen, J. P. (2019). Viral Metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica). Viruses 11:979. doi: 10.3390/v11110979
Moenning, V. (1990). Pestiviruses: a review. Vet. Microbiol. 23, 35–54. doi: 10.1016/0378-1135(90)90135-i
Postel, A., Smith, D. B., and Becher, P. (2021). Proposed update to the taxonomy of Pestiviruses: eight additional species within the genus Pestivirus, family Flaviviridae. Viruses 13:1542. doi: 10.3390/v13081542
Righi, C., Petrini, S., Pierini, I., Giammarioli, M., and De Mia, G. M. (2021). Global distribution and genetic heterogeneity of border disease virus. Viruses 13:950. doi: 10.3390/v13060950
Swanepoel, R., Smit, S. B., Rollin, P. E., Formenty, P., Leman, P. A., Kemp, A., et al. (2007). Studies of reservoir hosts for Marburg virus. Emerg. Infect. Dis. 13, 1847–1851. doi: 10.3201/eid1312.071115
Tautz, N., Tews, B. A., and Meyers, G. (2015). The molecular biology of Pestiviruses. Adv. Virus Res. 93, 47–160. doi: 10.1016/bs.aivir.2015.03.002
Turcios-Casco, M. A., and Cazzolla Gatti, R. (2020). Do not blame bats and pangolins! Global consequences for wildlife conservation after the SARS-CoV-2 pandemic. Biodivers. Conserv. 29, 3829–3833. doi: 10.1007/s10531-020-02053-y
Vilcek, S., and Nettleton, P. F. (2006). Pestiviruses in wild animals. Vet. Microbiol. 116, 1–12. doi: 10.1016/j.vetmic.2006.06.003
Wang, X., Chen, W., Xiang, R., Li, L., Chen, J., Zhong, R., et al. (2019). Complete genome sequence of Parainfluenza virus 5 (PIV5) from a Sunda pangolin (Manis javanica) in China. J. Wildl. Dis. 55, 947–950. doi: 10.7589/2018-09-211
Wang, L. F., Shi, Z., Zhang, S., Field, H., Daszak, P., and Eaton, B. T. (2006). Review of bats and SARS. Emerg. Infect. Dis. 12, 1834–1840. doi: 10.3201/eid1212.060401
Wang, B., Yang, W., Sherman, V. R., and Meyers, M. A. (2016). Pangolin armor: overlapping, structure, and mechanical properties of the keratinous scales. Acta Biomater. 41, 60–74. doi: 10.1016/j.actbio.2016.05.028
Wu, Z., Liu, B., Du, J., Zhang, J., Lu, L., Zhu, G., et al. (2018). Discovery of diverse rodent and bat pestiviruses with distinct genomic and phylogenetic characteristics in several Chinese provinces. Front. Microbiol. 9:2562. doi: 10.3389/fmicb.2018.02562
Xu, J., Mendez, E., Caron, P. R., Lin, C., Murcko, M. A., Collett, M. S., et al. (1997). Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J. Virol. 71, 5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997
Yang, X. N., Yang, H. J., Zhang, L., Hou, X. X., Miao, G. Q., Cao, H. B., et al. (2021). Tick-borne pathogens in Shanxi Province, China. Biomed. Environ. Sci. 34, 410–415. doi: 10.3967/bes2021.055
Keywords: virome, pangolin, Pestivirus, Amblyomma javanense, molecular epidemiology
Citation: Shi Y-N, Li L-M, Zhou J-B, Hua Y, Zeng Z-L, Yu Y-P, Liu P, Yuan Z-G and Chen J-P (2022) Detection of a novel Pestivirus strain in Java ticks (Amblyomma javanense) and the hosts Malayan pangolin (Manis javanica) and Chinese pangolin (Manis pentadactyla). Front. Microbiol. 13:988730. doi: 10.3389/fmicb.2022.988730
Received: 07 July 2022; Accepted: 04 August 2022;
Published: 02 September 2022.
Edited by:
Mohammed Rohaim, Lancaster University, United KingdomReviewed by:
Philippe Pérot, Institut Pasteur, FranceCopyright © 2022 Shi, Li, Zhou, Hua, Zeng, Yu, Liu, Yuan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zi-Guo Yuan, emlndW95dWFuQHNjYXUuZWR1LmNu; Jin-Ping Chen, Y2hlbmpwQGdpei5nZC5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.