- 1Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Engineering Laboratory for Kiwifruit Industrial Technology, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 2College of Plant Protection, Nanjing Agricultural University, Nanjing, China
Post-harvest rot causes enormous economic loss to the global kiwifruit industry. Currently, there are no effective fungicides to combat the disease. It is unclear whether silver nanoparticles (AgNPs) are effective in controlling post-harvest rot and, if so, what the underlying antifungal mechanism is. Our results indicated that 75 ppm AgNPs effectively inhibited the mycelial growth and spore germination of four kiwifruit rot pathogens: Alternaria alternata, Pestalotiopsis microspora, Diaporthe actinidiae, and Botryosphaeria dothidea. Additionally, AgNPs increased the permeability of mycelium’s cell membrane, indicating the leakage of intracellular substance. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) observations revealed that AgNPs induced pathogen hypha shrinkage and distortion, as well as vacuolation in hypha cells, implying that AgNPs caused cellular and organelle structural degradation. The transcriptome sequencing of mycelium treated with AgNPs (24 h / 48 h) was performed on the Illumina Hiseq 4000 sequencing (RNA-Seq) platform. For the time points of 24 h and 48 h, AgNPs treatment resulted in 1,178 and 1,461 differentially expressed genes (DEGs) of A. alternata, 517 and 91 DEGs of P. microspora, 1,287 and 65 DEGs of D. actinidiae, 239 and 55 DEGs of B. dothidea, respectively. The DEGs were found to be involved in “catalytic activity,” “small molecule binding,” “metal ion binding,” “transporter activity,” “cellular component organization,” “protein metabolic process,” “carbohydrate metabolic process,” and “establishment of localization.” Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis also revealed that “carbohydrate metabolism,” “amino acid metabolism,” “energy metabolism,” and “xenobiotics biodegradation and metabolism” of “metabolism processes” were the most highly enriched pathways for these DEGs in four pathogens, with “cellular processes” being particularly enriched for B. dothidea. Furthermore, quantitative polymerase chain reactions (qPCRs) were used to validate the RNA-seq results. It was also confirmed that AgNPs could significantly reduce the symptoms of kiwifruit rot without leaving any Ag+ residue on the peel and flesh of kiwifruit. Our findings contributed to a better understanding of the antifungal effect and molecular mechanisms of AgNPs against pathogens causing kiwifruit post-harvest rot, as well as a new perspective on the application of this novel antifungal alternative to fruit disease control.
Introduction
Kiwifruit is popular with consumers due to its unique flavor and high concentration of vitamin C, minerals, and other nutrients (Diaz et al., 2017). By 2021, the global kiwifruit planting area, yield, and output value have reached 270,500 hectares, 4407,400 tons, and USD 10 billion, respectively. However, as demand grows and the kiwifruit producing area in China expands, post-harvest rot diseases become more severe, with the average infected rate reaching 30–50%, and causing more than 100,000 tons of fruit losses per year, severely limiting the industry’s healthy development (Li et al., 2017a; Dai et al., 2021).
Several fungi have been identified as pathogens of kiwifruit rot disease, including Botryosphaeria dothidea (Zhou et al., 2015; Pan et al., 2020), Alternaria alternata (Li et al., 2017b; Gao et al., 2021), Pestalotiopsis microspora (Li et al., 2016) and Diaporthe sp. (D. ambigua, D. australafricana, D. novem, D. rudis and D. actinidiae) (Diaz et al., 2017; Li et al., 2017c). In recent years, the pathogens are generally controlled by the pre-harvest application of chemical fungicides such as Tebuconazole and Carbendazim (Kim et al., 2013). However, overuse of such fungicides has resulted in a significant increase in pathogen resistance to these fungicides, as well as fungicide residues on fruits, both of which are detrimental to public health and the environment (Chen et al., 2020; Kai et al., 2020). Therefore, it is critical to develop an effective and safe alternative to control kiwifruit post-harvest rot.
Because of the high efficiency, broad-spectrum antimicrobial activity, minimal resistance, and excellent safety, silver nanoparticles (AgNPs) have been widely used for plant disease control in recent years (Tian et al., 2018; Gordienko et al., 2019; Al-khattaf, 2021). AgNPs inhibited the growth of plant pathogenic fungi including Fragaria ananassa (Moussa et al., 2013), Pyricularia sp. (Lee et al., 2013), Aspergillus niger (Xue et al., 2016), Botrytis cinerea (Malandrakis et al., 2019), and Fusarium sp. (Shen et al., 2020; Jian et al., 2021). AgNPs’ antifungal mechanism is primarily attributed to inhibiting mycelial growth and conidial germination, degrading cell walls and membranes, disputing protein, producing reactive oxygen species (ROS), influencing pathogen energy and substance metabolism, signal transduction, and genetic information processing (Jian et al., 2021; Al-Otibi et al., 2022). However, the mechanism of AgNPs against plant pathogens remains unknown, owing to a lack of microscopic, biochemical, and omic analysis (Shen et al., 2020).
In recent years, AgNPs have been applied to control post-harvest diseases in citrus (Al-Sheikh and Yehia, 2016), mango (Shivamogga Nagaraju et al., 2020), apple (Madbouly, 2021), and strawberry (Oliveira et al., 2022). However, no AgNPs research on preventing kiwifruit rot has been published. B. dothidea, D. actinidiae, P. microspora and A. alternata have been identified as pathogens causing kiwifruit post-harvest rot in our preliminary study (Li et al., 2017c). Previous research found that AgNPs could directly inhibit mycelial growth and spore germination of A.alternata, as well as affect cell wall composition and the synthesis of sugar, protein, and N-acetylglucosamine (Ouda, 2014; Ganash et al., 2018). 150 ppm AgNPs (mean size 10 ± 5 nm) had potent antifungal activity against A.alternata when compared to Iprodione and Difenoconazole; 180 ppm AgNPs (mean size 52 nm) inhibited the growth of Diaporthe sp. (Mendes et al., 2014; Abdelmalek and Salaheldin, 2016). However, there have been no systematic studies on the effect of AgNPs on pathogens causing kiwifruit post-harvest rot, especially B. dothidea, D. actinidiae, and P. microspora.
In this study, we first determined the antifungal capacity of AgNPs against four pathogens causing kiwifruit post-harvest rot. Then, using an electron microscope and transcriptome profiling, we identified the cytological and molecular mechanisms by which AgNPs suppressed the above pathogens. Finally, we investigated the antifungal efficacy and safety of AgNPs applied to kiwifruit. The study verified that AgNPs can be used to control kiwifruit rot disease, and laid the groundwork for the application of AgNPs to control other fruit fungal diseases.
Materials and methods
Silver nanoparticles suspension
The polyvinyl pyrrolidone (PVP, 40KDa) coating AgNPs stock (5 mg mL–1) purchased from NanoComposix (San Diego, CA, United States) had an average diameter of 5 nm and was stored at 4°C, away from light (Lu et al., 2022). Different concentrations of AgNPs solutions were prepared with sterile distilled water (SDW).
Pathogenic fungi
Four strains isolated from infected fruits were identified as pathogens causing kiwifruit post-harvest rot by analyzing morphological characteristics and nucleotide sequences containing the internal transcribed spacer rDNA region (ITS), β-tubulin, and elongation factor-1 alpha gene (EF-1α), including B. dothidea, D. actinidiae, P. microspora and A. alternata (Li et al., 2016, 2017a,2017b,2017c). Four strains were cultured on potato dextrose agar (PDA, extract of potatoes 200 g, dextrose 20 g, agar 20 g per liter) for 14 days at 25°. The spore suspensions were made by rinsing the spores and hypha in 10 mL SDW and filtering them through two layers of sterile gauze to remove any adherent mycelium. The concentration of the homogenous spore suspension was measured by a hemocytometer and diluted to 1.0 × 106 spores mL–1.
Kiwifruit
Yellow fleshed fruits of Actinidia chinensis cv ‘Jinmei’ were collected from the scientific research orchard of Engineering Laboratory for Kiwifruit Industrial Technology, Chinese Academy of Sciences (CAS). Fruits with a consistent shape, size, ripeness, and lack of visible mechanical wounds were selected.
Antifungal effect of silver nanoparticles on mycelial growth of pathogens
The antifungal activity of AgNPs was investigated by measuring the diameter of fungal colonies (Wollenberg et al., 2016). The 7 mm diameter mycelial block was taken from a 1-week-old culture and placed in the center of a PDA petri dish (diameter = 90 mm) containing 25 mL of PDA with various AgNPs concentrations (0, 0.5, 5, 15, 30, 45, 60, 75, 90 ppm). Cultures were incubated at 25° and colony diameter was measured using the cross method on the sixth day after inoculation. The PDA plate without AgNPs was used as the control. Each treatment performed with three replicates, and the experiment was repeated twice. The inhibition rate of mycelium growth was calculated as follows (Alothman and Abd-Ei-Aziz Abeer, 2019):
EC50 (effective concentration of 50% inhibition rate) values were calculated statistically using SPSS Statistics 21 (SPSS Inc., United States).
Antifungal effect of silver nanoparticles on spore germination and germ tube elongation
The inhibition of AgNPs on spore germination was observed with modification of He et al. (2019). According to the analysis result of 2.4, the optimal inhibitory concentration of AgNPs for four strains was 75 ppm. For each pathogen, a 100 μL spore suspension (1.0 × 106 spores mL–1) was prepared and inoculated into tubes containing 1 mL potato dextrose broth (PDB, extract of potatoes 200 g and dextrose 20 g per liter) containing 75 ppm AgNPs, with the final concentration adjusted to 1.0 × 105 spores mL–1. PDB without AgNPs served as the negative control. The tubes were then cultured in a constant temperature shaker at 25°C at a speed of 180 r/min. The spore germination and germ tube elongation were then observed using an optical microscope (OLYMPUS BX51, Olympus Corporation, Japan).
As the spore germination period of different pathogens varies, the observation time was also modified based on the growth curve and set as follows: B. dothidea 2 h, D. actinidiae 4 h, P. microspora 5 h, and A. alternata 9 h. If the germ tube length is equal to or longer than the spore length, germination is considered. The percentage in germination was observed with modification of Al-Sheikh and Yehia (2016). The test was repeated twice with three replicates each, with approximately 100 spores constituting one replicate.
Antifungal effect of silver nanoparticles on membrane permeability of mycelium
Mycelium permeability was measured using methods described by Zhang et al. (2007) with little modification. For each strain, 0.4 g mycelium was mixed in 15 mL of 75 ppm AgNPs in a tube before being placed in a thermostatic bath at 26°. Electrical conductivity was measured at 0, 1, 3, 6, 12, 24, and 48 h, finally after boiling. Mycelium in SDW served as a control. Each three-parallel treatment was repeated three times.
Antifungal effect of silver nanoparticles on mycelial morphology and ultrastructure
Scanning Electron Microscope (SEM) and Transmission Electron Microscope (TEM) were used to examine the effect of AgNPs on the morphological and ultrastructural alterations of four pathogens, as described before (Li et al., 2021). The strains were cultured on PDA plates with 75 ppm AgNPs for 3 d at 25°. PDA dishes without AgNPs were used as control. Mycelial fragments (approximately 3∼5 mm) were collected from the colony’s edge and fixed in 2.5 % glutaraldehyde at 4° for 12 h, then rinsed three times with PBS (pH 7.2, 0.01M) for further processing.
For SEM analysis, mycelia samples from each treatment were dehydrated in an ethanol series (30, 50, 70, and 90%, v/v) for 20 min at each phase, followed by centrifugation. Following drying at the critical point, samples were attached to the sample stage with conductive glue. Lastly, specimens were sputtered with palladium-gold to observe under SEM (JEOL JSM-6390LV, Japan).
For TEM investigation, mycelia samples were postfixed with 1% osmic acid at 4° for 3 h, rinsed three times with PBS, and dehydrated with graded acetone solutions (30, 50, 70, 80, and 90%, v/v), each stage lasting 30 min. The specimens were penetrated for 12 h at each stage with a volume gradient of acetone and embedding medium (5:1, 3:1, 1:1, 1:3, 1:5, v/v), followed by pure embedding medium. Then, the specimens were embedded in SPI-Pon 812, ultrasliced using an ultramicrotome (Leica EM UC6, Germany), and stained with uranyl acetate to detect under TEM (Hitachi H-7650, Japan).
Antifungal effect of silver nanoparticles against pathogens by RNA-seq
The flow of the transcriptomics experiment included extraction of total RNA; enrichment of mRNA by Oligo (dT); RNA fragment; random hexamer primed cDNA synthesis; size selection and PCR amplification; and sequencing on an Illumina HiSeq 4000 (Zhao et al., 2022). The samples were collected, 0.4 g mycelium was inoculated into 15 mL PDB medium containing 75 ppm AgNPs for each strain, and cultured at 25° for 24 h (Ag-24h) and 48 h (Ag-48h). Control mycelium was grown in PDB without AgNPs for 24 h (CK-24h) and 48 h (CK-48h). Thus, 16 treatments with three repetitions were established, yielding 48 samples in total. Total RNA from 48 samples was extracted by using TRIzol (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instruction. To eliminate DNA from total RNA samples, Invitrogen TM RNase-free DNase I was utilized. mRNA was extracted from total RNA using poly-T oligo-attached magnetic beads and then fragmented into 200–300 bp fragments to produce strand-specific cDNA libraries using a TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, United States). Finally, the cDNA was paired-end sequenced on the Illumina Hiseq 4000 sequencing platform using the PE150 sequencing strategy (Patnaik et al., 2016).
To obtain clean data, the two ends of raw reads with low-quality bases or adapters were truncated by Trimmomtic v0.39 (Bolger et al., 2014) with parameters “ILLUMINACLIP: adapters/TruSeq3-PE-2.fa:2:30:10 LEADING:5 TRAILING:5 SLIDINGWINDOW:4:15 MINLEN:50 TOPHRED33.” The clean data were then aligned to the reference genome sequence using HISAT2 v2.1.0 (Kim et al., 2019) with parameters “–rna-strandness RF –min-intronlen 20 –max-intronlen 4000 –dta –dta-cufflinks –score-min L,0.0, −0.4.” The HISAT2 alignment results were used to obtain raw count expression results by HTseq v0.13.5 software (Anders et al., 2014) with parameters “-f sam -r name -s reverse -a 10 -t exon -i gene_id.” And then, the raw counts matrix data of 48 samples were normalized by two methods sequentially, using the TPM (Transcripts Per Kilobase Million) for eliminating gene length differences and TMM (Trimmed Mean of M-values) (Robinson and Oshlack, 2010) for cross-comparison among samples. Finally, the edgeR v3.22.5 (Robinson et al., 2010) and DESeq2 v1.20.0 (Love et al., 2014) software were utilized to compare the raw counts of gene expression and calculate the different significance between samples. The differentially expressed genes (DEGs) (Shen et al., 2020) were identified with false discovery rate (FDR) = 0.01 both in edgeR and DESeq2 methods and absolute log2 fold change of TPM/TMM gene expression=1 (Zhao et al., 2019, 2020). The identified DEGs were then subjected to hierarchical clustering, as well as Gene Ontology (GO)1 and Kyoto Encyclopedia of Genes and Genomes (KEGG)2 pathway enrichment analyses (Wang et al., 2021).
RT-qPCR was performed to validate the transcriptional level results acquired by RNA sequencing. Total RNA was extracted in the same RNA-Seq samples and reverse-transcribed into cDNA, and then used for RT-qPCR using SYBR Premix Ex Taq II (Ke et al., 2014). The genes with the maximum FDR value and average TPM value between 20 and 40 (the upper quartile of all genes TPM expression was about 30) were selected as reference in RT-qPCR, and eight DEGs with the minimum FDR value and average TPM value between 20 and 40 were chosen for validation (Supplementary Table 1). Additionally, three stable reference genes were used to determine the relative quantification of each target gene (Supplementary Table 1). On a 7500 Real-Time PCR system (ABI, United States), the relative expression of the genes was measured three times, each time with three technical replicates (Yin et al., 2015). The target genes’ expression levels were calculated by the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Antifungal effect and safety of silver nanoparticles on kiwifruit rot disease
In our previous study, we successfully constructed the fluorescence strain of P. microspora, which could be used for the detection of infection process (Deng et al., 2021). Agrobacterium tumefaciens mediated transformation of P. microspora strain was carried out by using recombinant vector pCAMBgfp containing hygromycin resistance gene and GFP lable. The fluorescent transformant PM-13 has no significant differences in morphological characteristics and pathogenicity as compared with the wild-type strain.
The control efficacy of AgNPs against post-harvest rot in kiwifruit ‘Jinmei’ was assessed as previously recorded (Pan et al., 2020) with modification. Fruits were disinfected in 0.2% sodium hypochlorite solution (v/v) for 2 min, rinsed 5 times with sterilized water, and air-dried at room temperature. Then, fruits were inoculated with 10 μL 1.0 × 106 spores mL–1 suspension of P. microspora fluorescent strain PM-13. 75 ppm AgNPs were sprayed on fruits 24 h before or after pathogen inoculation, with control fruits not treated with AgNPs. After air-drying, all treated fruits were placed in a plastic box at 22° with a high relative humidity (approximately 80%). On the sixth post-inoculation day, the fruit peel was scraped and the disease lesions were measured before being photographed using a LUYOR-3410 fluorescence excitation source. Each treatment was performed three times, with three fruits per replicate.
As described previously, the Microwave Digestion-ICP-MS method was used to examine the Ag+ residue on kiwifruit (Navarro et al., 2006; Aldabe et al., 2013). The AgNPs-treated and control fruits (inner/external fruit flesh and peel) were crushed with liquid nitrogen, dried at 60°, and sieved through a 100-mesh screen. For each treatment, 0.25 g powder was added, respectively into test tubes with 5 mL nitric acid and 1 mL hydrogen peroxide, then digested with a Microwave Sample Preparation System (ETHOS ONE, Milestone) and the acid was exorcized. Subsequently, the solution was diluted with 1% HNO3 in a 50 mL volumetric flask, filtered, and the Ag+ content of the fruit sample was determined by Inductively Coupled Plasma Mass Spectrometry (ICP-MS, Thermo Fisher Scientific, X Series 2). The national hygienic standard for drinking water in China (GB 5749-2006) considers Ag+ levels below 50 μg L–1 to be safe. Each AgNPs treatment was performed with 5 repeats.
Statistical analysis
The experiments were performed using a completely randomized design (Fernandez et al., 2022). Statistical analyses were carried out using SPSS Statistics 21 (SPSS Inc., United States), and results were showed as mean ± standard error (SE). Student’s t-test or one-way ANOVA with Duncan’s multiple range test (P < 0.05) were used to determine the significant difference (Zhuo et al., 2022).
Results
Antifungal effect of silver nanoparticles against four pathogens on mycelial growth
Different concentrations of AgNPs (5 nm, 5 mg mL–1) had distinct inhibitory effects on the mycelial growth of four pathogens (Figures 1A,B). Under 60–90 ppm AgNPs, an obvious reduction of colony size was observed, and the inhibition rate of 75 ppm AgNPs achieved a stable and nearly the highest value for four pathogens. Hence, we chose 75 ppm AgNPs for further analysis. Additionally, EC50 values for mycelial growth of AgNPs for the four pathogens ranged from 2.31 to 14.74 ppm (Supplementary Table 2), suggesting that AgNPs possessed excellent antifungal activity.
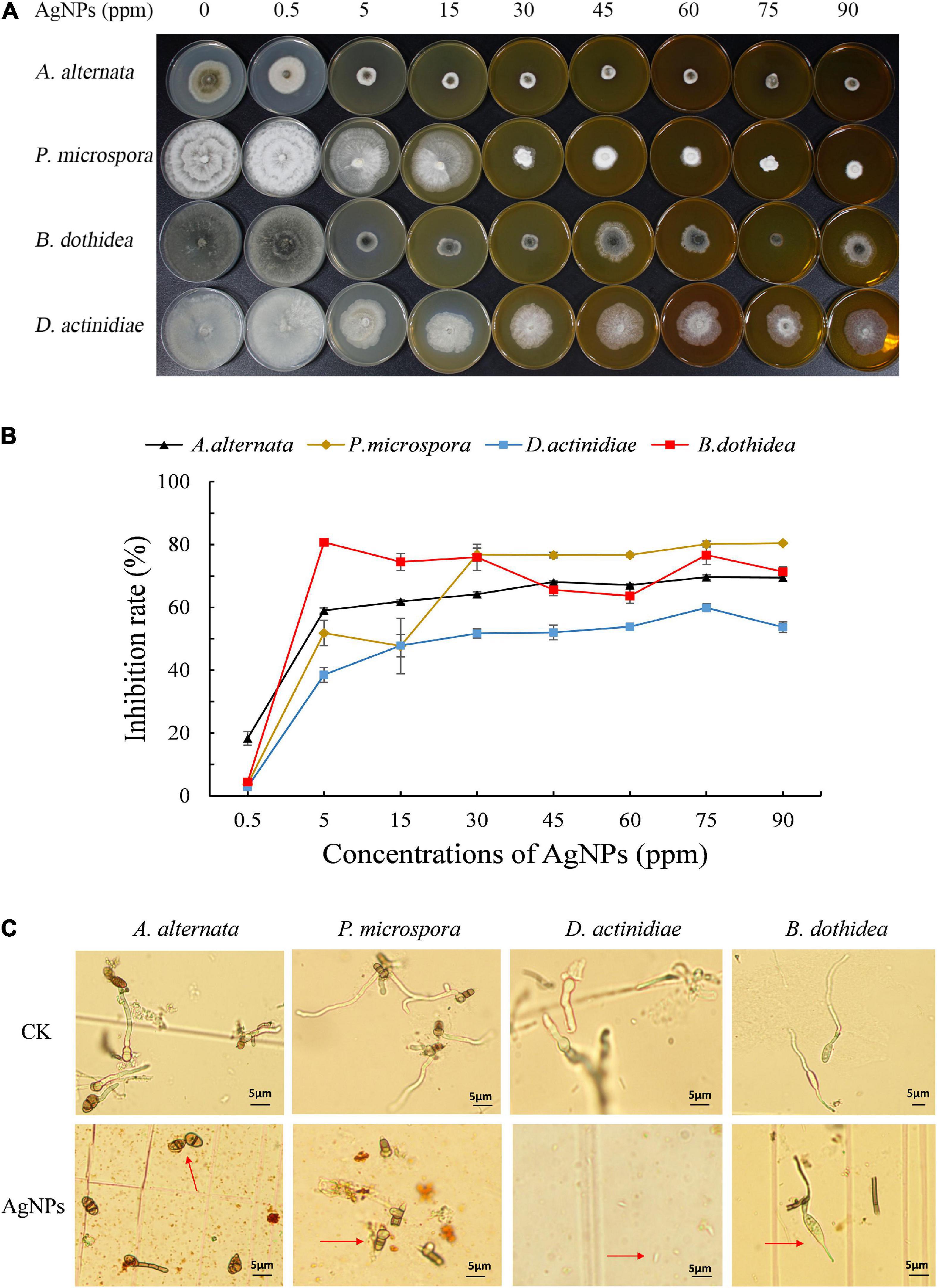
Figure 1. (A) The colony reduction and (B) Inhibition rate of four pathogens in PDA medium with various AgNPs concentrations. (C) Effect of 75 ppm AgNPs on four pathogens’ spore germination and germ tube elongation.
Effect of silver nanoparticles against pathogens on spore germination and germ tube
The germination assay also revealed that 75 ppm AgNPs significantly reduced the spore germination of four pathogens (Figure 1C). The average spore germination rate of A. alternata decreased from 88.48 to 9.70%, and the length of the germ tube decreased from 36.10 to 18.69 μm. Also, the average spore germination of B. dothidea decreased from 94.44 to 7.07%, and the length of the germ tube decreased from 66.00 to 17.91 μm. The spore germination of P. microspora and D. actinidiae were completely inhibited.
Effect of silver nanoparticles against pathogens on membrane permeability of mycelium
Electrical conductivity is used to measure ion strength in solution, which reflects electrolyte exchange inside and outside mycelium cells, as well as the permeability of mycelium cell membrane, which is positively related to the integrity of cell membrane. Compared to the control, the electrical conductivity of four pathogens’ mycelium increased significantly with AgNPs treatment time extension (Figure 2 and Supplementary Table 3), indicating increased permeability of cell membrane and leakage of intracellular substance.
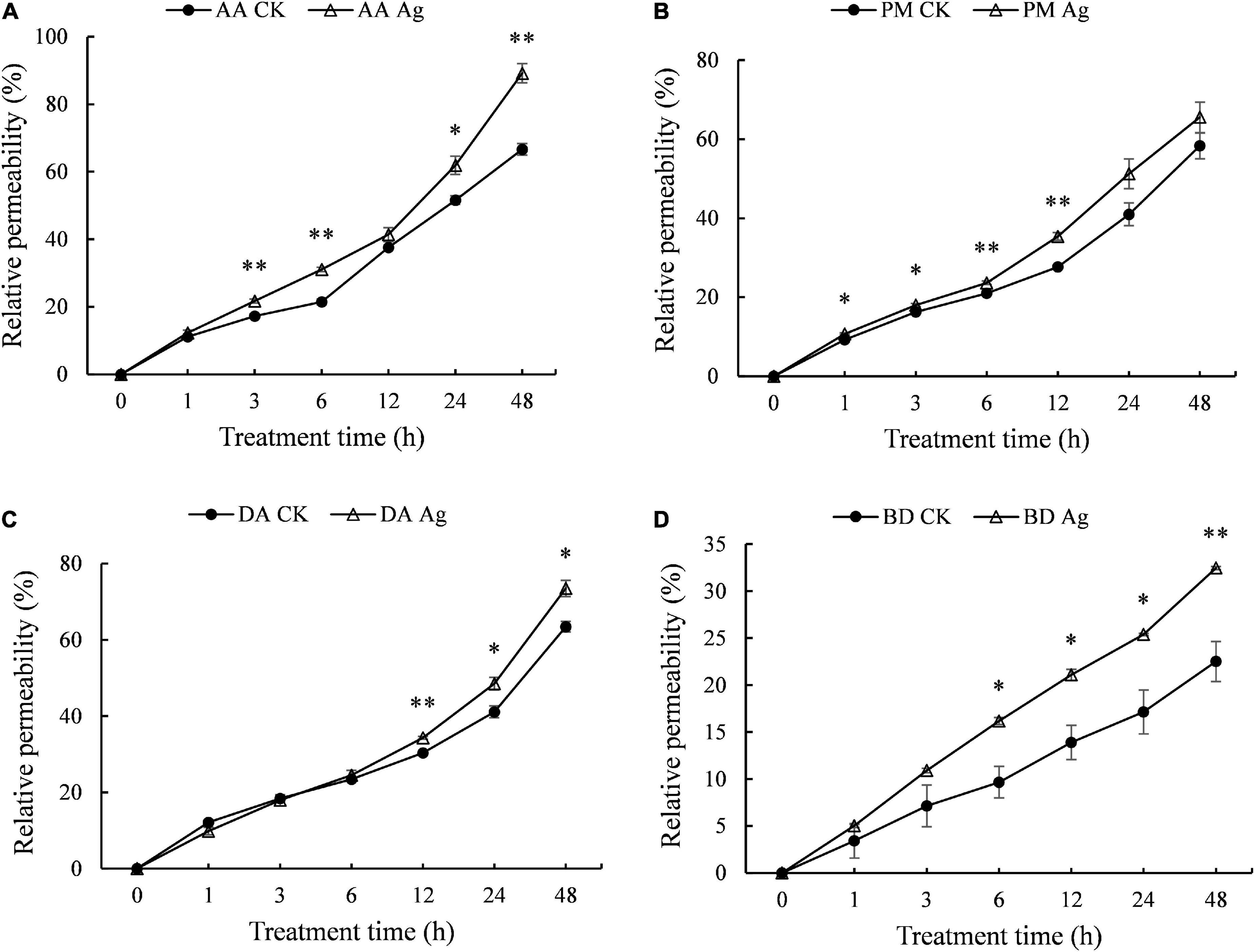
Figure 2. Effects of 75 ppm AgNPs on mycelium membrane permeability of four pathogens’ mycelium. (A) A. alternata, (B) P. microspora, (C) D. actinidiae, (D) B. dothidea. Asterisks indicate significant difference (P < 0.05*, P < 0.01**) between the treatments determined using Student’s t-test.
Effect of silver nanoparticles on the mycelia morphology and ultrastructure of pathogens
The microstructure changes of four pathogens were examined using SEM and TEM, striking changes in mycelia morphology and cell structure were observed after AgNPs treatment. Under SEM observation (Figure 3), untreated mycelia possessed normal morphology with uniform thickness and smooth surface. In comparison to the control, AgNPs caused abnormal morphological changes in the hypha, which became twisted, shrunken, and shriveled. The abnormal mycelial morphology was linked to a change in the permeability of the mycelium cell membrane. Under TEM observation, the untreated cell structure was unaltered, with a smooth cell wall and few intracellular cavities. AgNPs caused significant cellular damage, including obvious vacuolation and structure loosening. Overall, AgNPs treatment could have a significant detrimental influence on the morphology and ultrastructure of four pathogens.
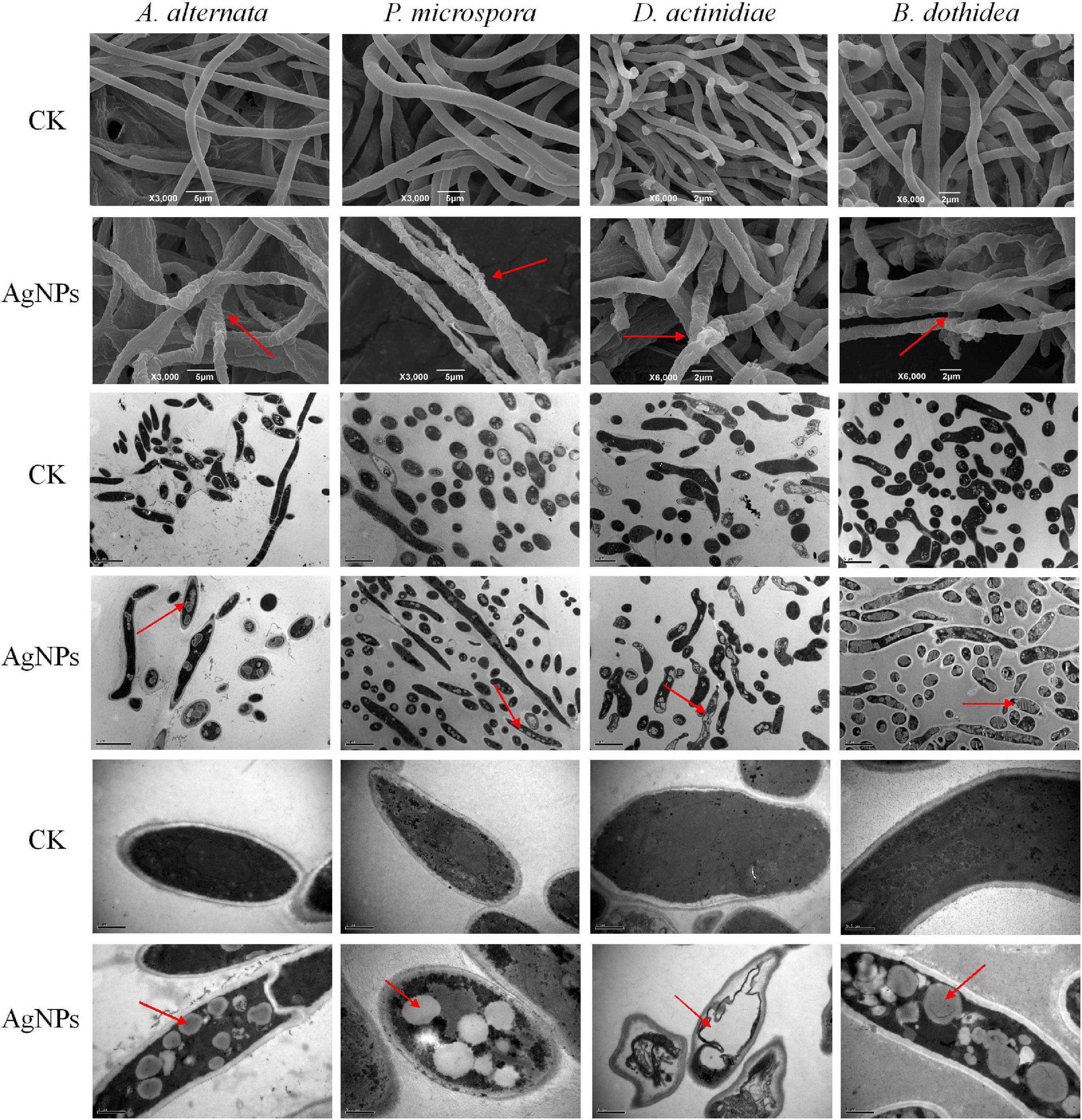
Figure 3. Scanning and transmission electron microscopy images of four pathogens’ mycelial morphology and ultrastructure under 75 ppm AgNPs.
RNA-seq analysis
Transcriptome profiling assays were performed to uncover the molecular changes underlying the impact of AgNPs on four pathogens. The average raw data size of samples was 9.67 G, the percentage of clean data was 97.67%, and the average mapping rate with the corresponding reference genome reached 95.95% (Figure 4A and Supplementary Table 4). When compared to the genome, the gene number of A. alternata was 13466, P. microspora was 14711, D. actinidiae was 17176, and B. dothidea was 14017. TPM and TMM analysis were used to determine the amount of gene expression, and the sum of raw counts for each sample was listed in Supplementary Tables 5–9.
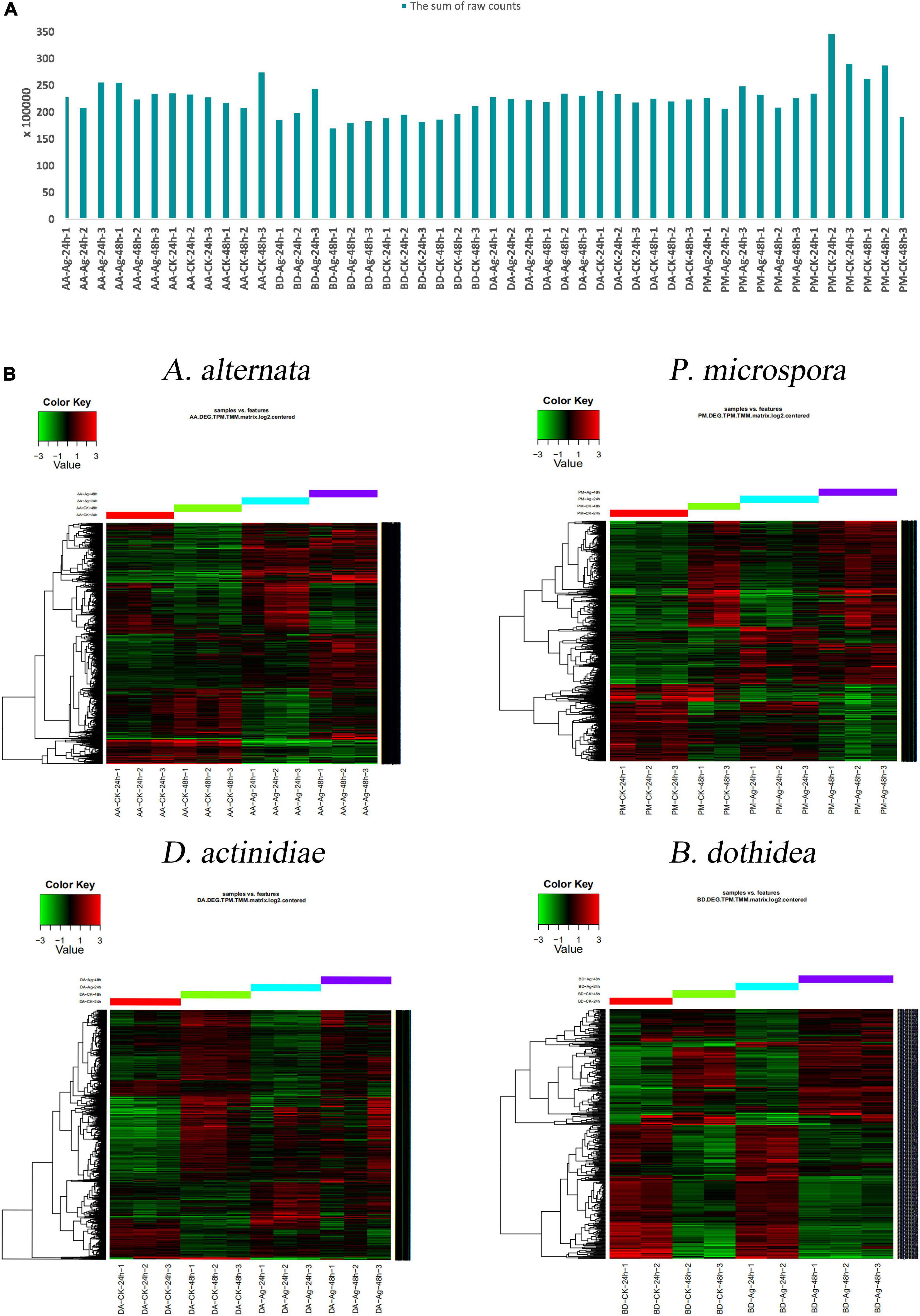
Figure 4. (A) The sum of 48 samples’ raw counts. (B) Hierarchical clustering analysis of DEGs with different AgNPs exposure time.
Significant differentially expressed genes analysis
Each pathogen’s four treatments were subjected to differential expression analysis (Figure 5A). As shown below, the number of DEGs was determined using cluster analysis (Supplementary Table 10).
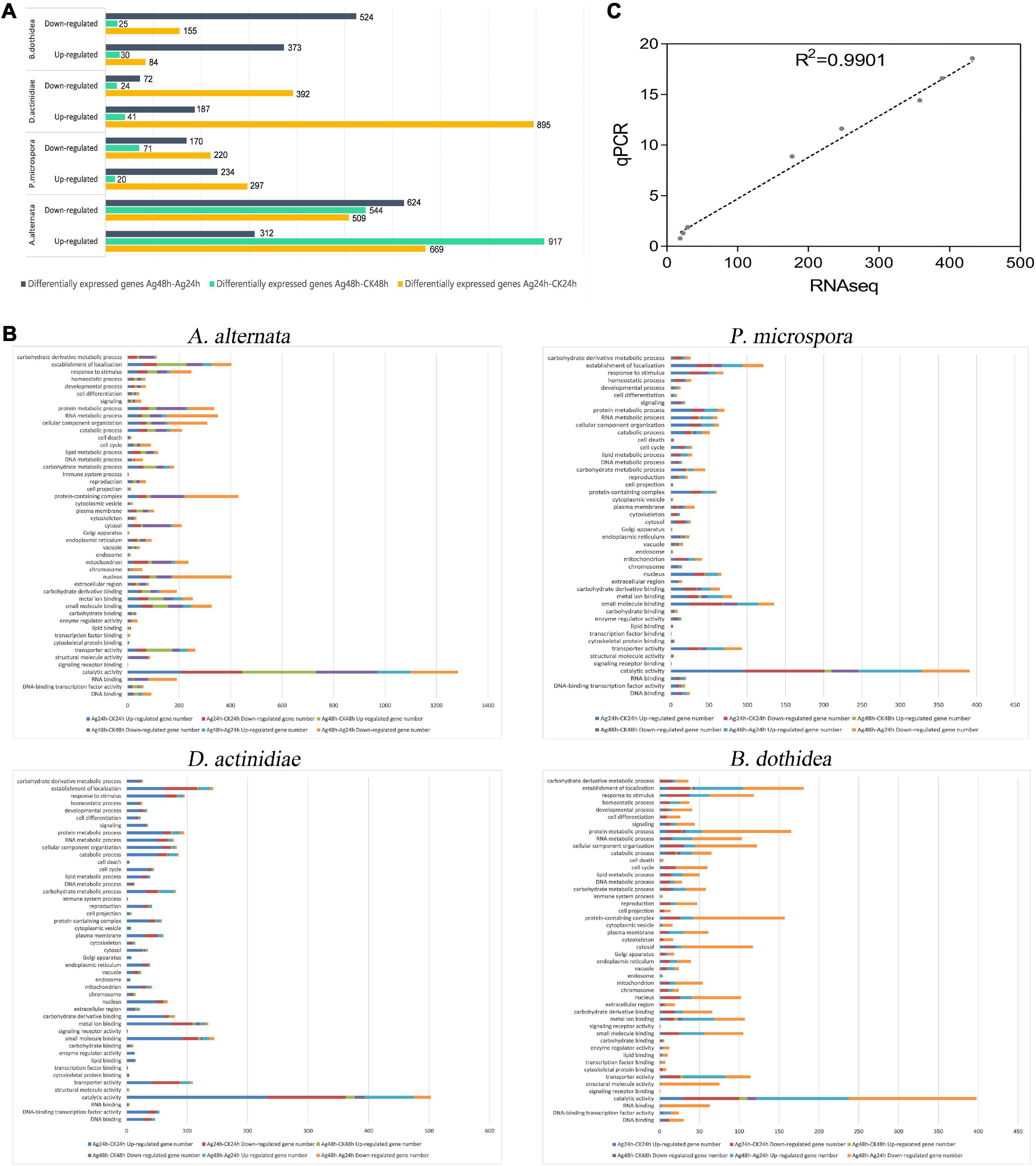
Figure 5. (A) Gene expression differences in the AgNPs treated groups compared with the control group. (B) GO enrichment analysis of DEGs between Ag-24h and CK-24h. (C) Correlation of expression levels of 8 DEGs assessed by RT-qPCR with RNA-Seq data.
Compared to the untreated group (CK), the transcriptome data suggested that (i) For A. alternata, 669 and 917 genes were up-regulated when comparing Ag-24h to CK-24h and Ag-48h to CK-48h, respectively; 312 genes were up-regulated when comparing Ag-48h to Ag-24h. (ii) For P. microspora, 297 and 20 genes were up-regulated when comparing Ag-24h to CK-24h and Ag-48h to CK-48h, respectively; 234 genes were up-regulated when comparing Ag-48h to Ag-24h. (iii) For D. actinidiae, 895 and 41 genes were up-regulated when Ag-24h was compared to CK-24h and Ag-48h was compared to CK-48h, respectively; 187 genes were up-regulated when Ag-48h was compared to Ag-24h. (iv) For B. dothidea, 84 and 30 genes were up-regulated when comparing Ag-24h to CK-24h and Ag-48h to CK-48h, respectively; 373 genes were up-regulated when comparing Ag-48h to Ag-24h.
In addition, the transcriptome data analysis revealed that (i) for A. alternata, 509 and 544 genes were down-regulated when Ag-24h was compared to CK-24h and Ag-48h was compared to CK-48h, respectively; 624 genes were down-regulated when Ag-48h was compared to Ag-24h. (ii) For P. microspora, 220 and 71 genes were down-regulated when comparing Ag-24h to CK-24h and Ag-48h to CK-48h, respectively; 170 genes were down-regulated when comparing Ag-48h to Ag-24h. (iii) For D. actinidiae, 392 and 24 genes were down-regulated when comparing Ag-24h to CK-24h and Ag-48h to CK-48h, respectively; 72 genes were down-regulated when comparing Ag-48h to Ag-24h. (iv) For B. dothidea, 155 and 25 genes were down-regulated when comparing Ag-24h to CK-24h, and 524 genes were down-regulated when comparing Ag-48h to Ag-24h.
Cluster analysis of differentially expressed genes
Using hierarchical cluster analysis, the relationship of genome-wide expression profiles between control and AgNPs treated samples of 24 h and 48 h was determined. For each pathogen, all annotated DEGs from the 48 samples were clustered with the software Cluster 3.0 (Figure 4B). The results revealed that the DEG data of different treatments (CK-24h, CK-48h, Ag-24h, Ag-48h) exhibited a similar pattern, the repetition of different treatments were gathered together, indicating that the data was highly reliable.
With the exception of A. alternata, the DEGs number of Ag-24h samples was more than that of Ag-48h samples in D. actinidiae and P. microspora. Fewer DEGs were identified both in Ag-24h and Ag-48h samples of B. dothidea. The result indicated that the effective time for AgNPs treatment concentrated within 24 h. Furthermore, when up-regulated and down-regulated genes from the two time periods were combined, only a few genes increased or decreased continuously as the AgNPs treatment time was prolonged. It was assumed that AgNPs treatment had a significant impact on various DEGs within 24 h (Supplementary Figure 1).
Gene Ontology functional annotation and Kyoto Encyclopedia of Genes and Genomes enrichment analysis of metabolic pathways
The RNA-seq unigenes from all groups were annotated using the NR, Swiss-Prot, KOG, eggNOG, InterPro, Pfam, GO, and KEGG databases (Supplementary Table 11). The responses of four pathogens to AgNPs were analyzed using GO functional enrichment (Figure 5B). Based on GO analysis, the unigenes of four pathogens were classified into three major categories: molecular function, cellular components, and biological process, with 16, 16, and 21 GO functional subcategories, respectively (Supplementary Tables 12–15). The majority of DEGs in the four pathogens were involved in “catalytic activity,” “small molecule binding,” “metal ion binding,” “transporter activity,” “cellular component organization,” “protein metabolic process,” “carbohydrate metabolic process,” and “establishment of localization.”
Kyoto Encyclopedia of Genes and Genomes analysis was used to discover the biological pathways in which the unigenes are involved. For four pathogens, the most highly enriched pathways for these DEGs were “carbohydrate metabolism,” “amino acid metabolism,” “energy metabolism,” “xenobiotics biodegradation and metabolism” of “metabolism processes.” “Cellular processes” were especially enriched in B. dothidea (Supplementary Tables 16–19).
The up-regulated DEGs of Ag24h-CK24h in A. alternata were mostly involved in “enzyme activity processes,” such as “NADPH,” “cutinase,” and “endo-1,4-beta-xylanase.” The up-regulated DEGs of Ag48h-Ag24h treatment were found in “transcription,” “folding, sorting, and degradation” of “genetic information processing,” respectively. The majority of Ag48h-CK48h DEGs, as well as up-regulated Ag24h-CK24h and Ag48h-Ag24h DEGs, were primarily involved in the “metabolism process,” which included “energy metabolism,” “carbohydrate metabolis,” and “amino acid metabolism.” Partial DEGs were identified in the fields of “environmental information processing,” and “organismal systems” (Supplementary Table 16).
The up-regulated DEGs of Ag48h-CK48h and down-regulated DEGs of Ag48h-Ag24h cannot enrich in KEGG analysis of P. microspora. The up-regulated DEGs of Ag24h-CK24h were identified in the “tyrosinase activity process.” The Ag24h-CK24h down-regulated DEGs were mainly involved in “amino acid metabolism,” “xenobiotics biodegradation and metabolism” of “metabolism processes.” The Ag48h-CK48h down-regulated DEGs were identified in the “transport and catabolism” of “cellular processes.” Furthermore, the majority of up-regulated Ag48h-Ag24h DEGs were involved in “metabolism processes,” such as “glycan biosynthesis and metabolism,” “energy metabolism” and “carbohydrate metabolism.” Partial DEGs were identified in the “transport and catabolism” of “cellular processes,” and “membrane transport” of “environmental information processing” (Supplementary Table 17).
The DEGs of Ag48h-CK48h and the up-regulated DEGs of Ag48h-Ag24h cannot enrich in KEGG analysis for D. actinidiae. The Ag24h-CK24h up-regulated DEGs were identified in the “enzyme activity processes,” such as “choline dehydrogenase” (EC:1.1.99.1), “mannan endo-1,6-alpha-mannosidase” (EC:3.2.1.101), “prostaglandin reductase 3” (EC:1.3.1.48), and “NADPH-cytochrome P450 reductase” (EC:1.14.14.1). Down-regulated DEGs in Ag24h-CK24h and Ag48h-Ag24h were mainly involved in the “metabolism processes,” including “carbohydrate metabolism,” “amino acid metabolism,” “lipid metabolism,” and “xenobiotics biodegradation and metabolism” (Supplementary Table 18).
The DEGs of Ag48h-CK48h cannot enrich in KEGG analysis for B. dothidea. Ag24h-CK24h up-regulated DEGs were found in “transport and catabolism” of “cellular processes,” whereas Ag24h-CK24h down-regulated DEGs were identified in “cell growth and death” of “cellular processes,” and “carbohydrate metabolism” of “metabolism processes.” The Ag48h-Ag24h up-regulated DEGs were mainly involved in “cell growth and death” and “cellular community-eukaryotes” of “cellular processes.” Down-regulated Ag48h-Ag24h DEGs were identified in “biosynthesis process of other secondary metabolites” and “amino acid metabolism” of “metabolism processes” (Supplementary Table 19).
Validation of RNA-seq
The correlation between qRT-PCR expression levels of eight DEGs and RNA-Seq data was 0.9438, indicating that qRT-PCR results were consistent with RNA-Seq data (Figure 5C and Supplementary Table 20).
Disease control effect of silver nanoparticles on kiwifruit
The fluorescence strain PM-13 has no significant differences in morphological characteristics and pathogenicity when compared to wild-type P. microspora strain, and the fluorescence intensity was positively related to spore and hyphae colonization, making it more suitable for observing pathogen infection processes, as well as the antifungal activity of AgNPs on controlling kiwifruit post-harvest rot (Figure 6A).
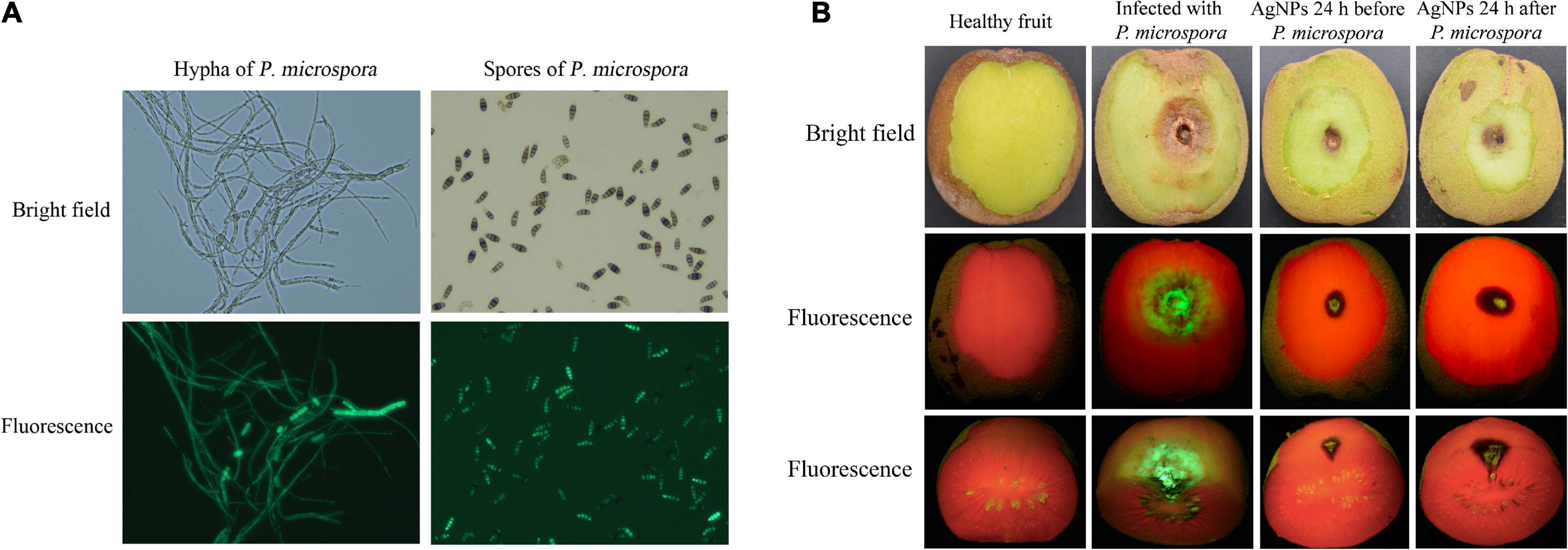
Figure 6. (A) Hypha and spores of the wild and fluorescent P. micropora strain. (B) Antifungal activity of AgNPs on kiwifruit soft rot caused by fluorescent P. micropora.
As illustrated in Figure 6B, the average lesion diameter of infected spot without AgNPs treatment was 24.22mm. The average lesion diameter that spraying AgNPs before PM-13 inoculation was 6.23 mm, whereas the average lesion diameter that spraying AgNPs after PM-13 inoculation was 8.90 mm. As a result, spraying AgNPs prior to spore inoculation had a better disease prevention effect (74.29% reduction in lesion diameter) than spraying AgNPs after inoculation (63.26% reduction in lesion diameter). Furthmore, spraying AgNPs reduced not only the average lesion of infected spots on the surface, but also the infected depth of disease spots, as observed in the cross-section. The findings revealed that AgNPs treatment greatly reduced the infected phenotype and fluorescence symptoms, as well as the pathogen colonization and disease development of kiwifruit post-harvest rot (Supplementary Table 21).
There was few Ag+ residue in the fruit’s peel or inner/external pulp. The Ag+ content in the AgNPs treated samples ranged from 0.033 to 1.499 μg L–1, which was significantly lower than China’s national hygienic limit for drinking water (50 μg L–1), indicating that AgNPs treatment was safe for kiwifruit (Supplementary Table 22).
Discussion
Post-harvest rot is one of the most serious fungal diseases encountered during kiwifruit storage, resulting in massive economic losses. Chemical control is currently the most prevalent method of control, despite being hazardous to humans and the environment. In recent years, AgNPs have gradually been applied in the control of agricultural diseases due to their high efficiency and broad-spectrum antimicrobial activities, low resistance, and high safety (Sharma et al., 2018; Malandrakis et al., 2019; Chen et al., 2020). This report firstly indicated the antifungal activity of AgNPs against four pathogens causing kiwifruit post-harvest rot from physiological, cytological, and transcriptomic levels. AgNPs significantly inhibited mycelial growth and spore germination of four pathogens, increased the permeability of mycelium’s cell membrane, caused cellular and organelle structural degradation. Transcriptomic analysis revealed that AgNPs influenced the carbohydrate metabolism, TCA cycle and energy metabolism pathways. In addition, AgNPs remarkably reduced the symptoms of kiwifruit rot without leaving any Ag+ residue on the peel and flesh. These findings implied that AgNPs might be a potential substitute for post-harvest disease control.
Silver nanoparticles inhibit both mycelial growth and spore germination of several plant pathogenic fungi, including B. cinerea, Bipolaris sorokiniana, Magnaporthe grise, Colletotrichum gloeosporioides, F. culmarum, Alternaria sp., Cladosporium sp., Corynespora sp., Cylindrocarpon sp., Stemphylium sp. and Pythium sp. (Ganash et al., 2018; Malandrakis et al., 2019; Kasprowicz et al., 2020). Based on the colony diameter and inhibition curve, the inhibitory effect of AgNPs on four pathogens was positively correlated with AgNPs concentration, and the inhibitory effects of AgNPs on four pathogens were P. microspora > B. dothidea > A. alternata > D. actinidiae. The findings were consistent with our previous research, as D. actinidiae had been identified as the most virulent strain among the four pathogens (Li et al., 2017c). The EC50 of AgNPs on the four pathogens was less than 15 ppm, as well as the spore germination and germ tube length were significantly reduced under 75 ppm, showing AgNPs’ strong inhibitory effect on kiwifruit post-harvest rot.
Previous cytological research confirmed that AgNPs severely damaged the cellular components of cell wall of pathogenic fungi, degraded the structure and integrity of cell membrane, and caused spores and mycelia to become wrinkled and depressed. Then, oxygen consumption of mycelial respiration reduced, the inactivation of key enzymes affected the respiratory chain, and bound lipids and enzymes caused cell lysis. Eventually, the cells twisted and expanded until they withered and died (Xu et al., 2013; Dakal et al., 2016; Xia et al., 2016; Gordienko et al., 2019). The electrical conductivity analysis in our study revealed that AgNPs treatment increased cell membrane permeability, destroyed the integrity of the cell membrane of four pathogens, resulting in excess water entering the cells and introducing the expansion of cell volume due to osmotic pressure, ultimately leading to the exudation of the cellular contents. SEM and TEM analysis further confirmed that AgNPs treatment caused hypha to become twisted, shrunken, and shriveled. Additionally, treated cells became loose, and vacuolations occurred. Consistent with morphological observations, the results showed that AgNPs severely damaged the cellular and organelle structure of four pathogens.
Over the past 2 years, transcriptomic analysis has gradually been used to clarify the molecular mechanism of AgNPs, providing a comprehensive understanding of potential DEGs involved in specific biological processes (Lu et al., 2020; Rahman et al., 2022). For example, the molecular mechanism of AgNPs against Fusarium sp. has been deeply studied, AgNPs could impair the cell structure, cellular energy utilization, energy and substance metabolism, signal transduction pathways, and significantly induce the expression of azole-related ATP-binding cassette (ABC) transporters (Shen et al., 2020; Jian et al., 2021). In our study, AgNPs caused 1178 and 1461 DEGs in A.alternata, 517 and 91 DEGs in P. microspora, 1287 and 65 DEGs in D. actinidiae, and 239 and 55 DEGs in B. dothidea at 24 h and 48 h, respectively. For four pathogens, there was a common gene expression of KEGG categories associated with “carbohydrate metabolism,” “amino acid metabolism,” “energy metabolism,” and “xenobiotics biodegradation and metabolism” of “metabolism processes.” Besides that, B. dothidea was particularly enriched in “cellular processes.” The findings demonstrated that AgNPs altered the pathogens’ gene expression, most of which were involved in, the foundation of all living organisms’ activities.
Carbohydrates are essential components of cell membranes and skeletons, as well as an energy source (Diao et al., 2020). The cell wall and membranes, which were composed of extensively cross-linked glycoproteins and carbohydrates, had been identified as the primary targets of AgNPs (Gow et al., 2017; Barros et al., 2021). Our findings indicated that nearly all DEGs encoding “carbohydrate metabolism” were repressed in the AgNPs-exposed group of four pathogens, which was consistent with the mechanism of AgNPs against Articulospora tetracladia (Barros et al., 2021) and F. solani (Shen et al., 2020). For A. alternata, the DEGs encoding “ascorbate and aldarate metabolism,” “galactose metabolism,” “glyoxylate and dicarboxylate metabolism,” “pyruvate metabolism,” “glycolysis and gluconeogenesis,” and “pentose phosphate pathway” were down-regulated in all AgNPs-exposed groups. DEGs encoding “fructose and mannose metabolism,” “glyoxylate and dicarboxylate metabolism,” and “pentose and glucuronate interconversions” in D. actinidiae were down-regulated in the Ag24h-CK24h group. The DEGs encoding “amino sugar and nucleotide sugar metabolism” and “starch and sucrose metabolism” in B. dothidea were down-regulated in the Ag24h-CK24h and Ag48h-Ag24h groups. Combined with our physiological and microscopic results, we can further confirm that AgNPs significantly disrupted the “carbohydrate metabolism” of four pathogens, and destroyed the cell membrane integrity of four pathogens.
Silver nanoparticles treatment also disrupted the “amino acid metabolism” and “lipid metabolism” of four pathogens. Except for B. dothidea, the DEGs involved in “amino acid metabolism” in the other three pathogens were down-regulated, including “cysteine and methionine metabolism,” “lysine metabolism,” “arginine and proline metabolism,” “valine, leucine and isoleucine biosynthesis,” “alanine, aspartate and glutamate metabolism.” The change of amino acid-related metabolism pathways resulted in protein synthesis disruptions, such as “beta-alanine metabolism” and “arginine andtabolism.” Furthermore, in Ag24h-CK24h and Ag48h-Ag24h of D. actinidiae, DEGs encoding “sphingolipid metabolism,” “fatty acid degradation,” “glycerolipid metabolism,” and “lipid metabolism” were down-regulated. The citrate cycle (TCA cycle) is the final metabolic pathway, the link between the metabolic connection of carbohydrates, amino acids, and lipids, as well as the source of energy for life activities (Fernie et al., 2004). Since the TCA cycle of A. alternata was significantly down-regulated by AgNPs, we hypothesized that AgNPs induced carbohydrate, amino acid, and lipid degradation, thus influencing the TCA cycle and energy metabolism of four pathogens.
In photosynthesis, the carbon fixation pathway of plants (CFPP) converts solar energy into biomass, bio-products, and biofuel. Surprisingly, a large number of heterotrophic fungi have enzymes that are functionally related to CFPP (Nelson and Ben-Shem, 2004). For example, DEGs encoding “carbon fixation in photosynthetic organisms,” “oxidative phosphorylation,” and “methane metabolism” in A. alternata were down-regulated, indicating that the pathway that absorbed and transmited light energy to the photosynthetic reaction center could be an important target of AgNPs. Besides that, oxidative phosphorylation, the cornerstone of aerobic cellular metabolism, was also significantly suppressed in A. alternata. These findings indicated that AgNPs significantly inhibited the energy metabolism pathways of four pathogens.
The findings also found that four pathogens attempted to eliminate the alterations caused by AgNPs and utilized multiple cellular responses to AgNPs stress. For example, xenobiotics detoxification systems can remove compounds from the complex mixtures formed by metabolic processes or from the environment. Cytochrome P450 is involved in more than 90% of all these oxidation reactions and is responsible for the metabolism of a wide range of xenobiotics and endogenous compounds (Zhu et al., 2018; Zhuo and Fan, 2021). In our study, the enzymes involved in “xenobiotics biodegradation and metabolism” were down-regulated in Ag24h-CK24h of A. alternata, such as “benzoate degradation,” “caprolactam degradation,” “naphthalene degradation,” “dioxin degradation,” and “polycyclic aromatic hydrocarbon degradation.” Then, as predicted, the metabolism of “xenobiotics by cytochrome P450” was up-regulated after 24 h, similarly found in Phanerochaete chrysosporium under AgNPs stress (Liu et al., 2022). In addition, there was partial recovery of “carbohydrate metabolism,” “amino acid metabolism,” lipid metabolism” and “energy metabolism” after 24 h, which might be related to the defense mechanism of four pathogens against AgNPs, similar to that of Fusarium solani (Shen et al., 2020).
Differ from the other pathogens, the “cellular processes” of B. dothidea were particularly enriched. Previous research found that AgNPs caused massive cell cycle arrest, a high percentage of DNA breaks, and cell death (Quevedo et al., 2021). The DEGs of “cellular senescence,” “cell growth and death,” “cignaling pathways regulating pluripotency of stem cells,” and “cellular community-eukaryotes” were up-regulated after 24 h. In subsequent research, we will increase the treatment concentration and sampling time point of AgNPs on each pathogen, investigate the antifungal mechanism of AgNPs against each pathogen, and confirm the function and mechanism of the critical DEGs involved in morphological and cytological changes of four pathogens.
Many field experiments confirmed that the spraying time of AgNPs affected the inhibition effect. Spraying 10 ppm AgNPs on rose 2 days before pathogen inoculation reduced the incidence rate of powdery mildew by 95% (Kim et al., 2008); spraying AgNPs 3 h before pathogen inoculation significantly reduced the incidence of root rot and rice blast (Jo et al., 2009). Our findings confirmed that AgNPs significantly reduced the severity of kiwifruit post-harvest rot, as the lesion diameter of disease spot and fluorescence intensity of colonized hyphae treated with AgNPs were obviously smaller than those of the control. Consistent with previous studies, spraying AgNPs on fruit prior to pathogen infection had a significantly better control effect than spraying AgNPs after pathogen infection. Meanwhile, there was no Ag+ residue on the peel or flesh of kiwifruit, indicating that AgNPs could be safely applied during the fruiting period in the field and the fruit preservation period after harvest.
Antifungal activity of AgNPs is dependent on physical and chemical properties, such as particle size, shape, dispersion, and pH (Hong et al., 2016). It was confirmed that AgNPs with a diameter of less than 12 nm could directly penetrate the cell wall and enter the cytoplasm of pathogens (Raza et al., 2017; Noori et al., 2017). AgNPs with an average size of 4–8 nm with various concentrations (10–100 ppm) significantly inhibited Colletotrichum sp. and Alternaria sp. (Lamsal et al., 2011; Gupta and Chauhan, 2015). Furthermore, spherical AgNPs could effectively inhibit fungi growth at low concentrations (Kim et al., 2012; Akther and Hemalatha, 2019). AgNPs in our study was 5 nm in size and spherical, demonstrating that they can be effectively used to control kiwifruit post-harvest rot. This finding can be used to guide the selection and application of AgNPs for horticultural disease control.
Conclusion
In summary, our results aimed at reliving the threat of serious fungal post-harvest rot in the kiwifruit industry, we performed an in-deep analysis of the antifungal effect of AgNPs against four pathogens from physiological, cytological, and transcriptomic levels, and verified that AgNPs could be effectively used to control kiwifruit post-harvest rot and safely applied on kiwifruit without residue. According to the findings, AgNPs are potentially suitable for use in the development of new antifungal agents for combating plant fungal diseases, including horticultural crops that directly consume fruits.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=CRA007558.
Author contributions
LL: methodology, investigation, software, writing – original draft, and funding acquisition. HP: methodology, investigation, software, and editing. LD: data curation and formal analysis. GQ: formal analysis, supervision, and editing. ZW: data curation and software. WL: methodology. CZ: project administration, supervision, editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the National Key Research and Development Program of Hubei Province (2021BBA100), the Earmarked Fund for China Agriculture Research System, the National Natural Science Foundation of China (31701974 and 31901980), Yangtze River Kiwifruit Industry Technology Research Project (CJZX20210102), and the Foundation of Wuhan Science and Technology Bureau (2018020401011307).
Acknowledgments
We would like to thank Lianfu Chen for his excellent technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.988633/full#supplementary-material
Footnotes
References
Abdelmalek, G. A. M., and Salaheldin, T. A. (2016). Silver nanoparticles as a potent fungicide for citrus pytopathogenic fungi. J. Nanomed. Res. 3:00065. doi: 10.15406/jnmr.2016.03.00065
Akther, T., and Hemalatha, S. (2019). Mycosilver nanoparticles: Synthesis, characterization and its efficacy against plant pathogenic fungi. Bionanoscience 9, 296–301. doi: 10.1007/s12668-019-0607-y
Aldabe, J., Santamaría, C., Elustondo, D., Lasheras, E., and Santamaría, J. M. (2013). Application of microwave digestion and ICP-MS to simultaneous analysis of major and trace elements in aerosol samples collected on quartz filters. Anal. Methods 5:554. doi: 10.1039/C2AY25724F
Al-khattaf, F. S. (2021). Gold and silver nanoparticles: Green synthesis, microbes, mechanism, factors, plant disease management and environmental risks. Saudi J. Biol. Sci. 28, 3624–3631. doi: 10.1016/j.sjbs.2021.03.078
Alothman, M., and Abd-Ei-Aziz Abeer, R. M. (2019). Effect of green synthesis silver nanoparticles from five fruits peel on protein capped and anti-fungal properties. Int. J. Adv. Res. Biol. Sci. 6, 156–165. doi: 10.22192/ijarbs.2019.06.02.018
Al-Otibi, F., Alfuzan, S. A., Alharbi, R. L., Al-Askar, A. A., Al-Otaibi, R. M., Al Subaie, H. F., et al. (2022). Comparative study of antifungal activity of two preparations of green silver nanoparticles from Portulaca oleracea extract. Saudi J. Biol. Sci. 29, 2772–2781. doi: 10.1016/j.sjbs.2021.12.056
Al-Sheikh, H., and Yehia, R. S. (2016). In vitro antifungal efficacy of Aspergillus niger ATCC 9642 chitosan-AgNPs composite against post-harvest disease of citrus fruits. Appl. Biochem. Microbiol. 52, 413–420. doi: 10.1134/S0003683816040177
Anders, S., Pyl, P. T., and Huber, W. (2014). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Barros, D., Pradhan, A., Pascoal, C., and Cássio, F. (2021). Transcriptomics reveals the action mechanisms and cellular targets of citrate-coated silver nanoparticles in a ubiquitous aquatic fungus. Environ. Pollut. 268:115913. doi: 10.1016/j.envpol.2020.115913
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen, T. L., Kim, H., Pan, S. Y., Tseng, P. C., Lin, Y. P., and Chiang, P. C. (2020). Implementation of green chemistry principles in circular economy system towards sustainable development goals: Challenges and perspectives. Sci. Total Environ. 716:136998. doi: 10.1016/j.scitotenv.2020.136998
Dai, Y., Wang, Z., Leng, J., Sui, Y., Jiang, M., Wisniewski, M., et al. (2021). Eco-friendly management of postharvest fungal decays in kiwifruit. Crit. Rev. Food Sci. Nutr. 1–12. [Epub ahead of print]. doi: 10.1080/10408398.2021.1926908
Dakal, T. C., Kumar, A., Majumdar, R. S., and Yadav, V. (2016). Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 7:1831. doi: 10.7389/fmicb.2016.01831
Deng, L., Feng, D. D., Wang, Z. P., Pan, H., Zhong, C. H., and Li, L. (2021). Agrobacterium tumefaciens-mediated transformation of Pestalotiopsis microspora and screening of the mutants. Mycosystema 40, 2355–2363.
Diao, F. W., Dang, Z. H., Cui, X., Xu, J., Jia, B. B., Ding, S. L., et al. (2020). Transcriptomic analysis revealed distinctive modulations of arbuscular mycorrhizal fungi inoculation in halophyte Suaeda salsa under moderate salt conditions. Environ. Exp. Bot. 183:104337. doi: 10.1016/j.psep.2022.02.019
Diaz, G. A., Latorre, B. A., Lolas, M., Ferrada, E., Naranjo, P., and Zoffoli, J. P. (2017). Identification and characterization of Diaporthe ambigua, D. australafricana, D. novem, and D. Rudis causing a postharvest fruit rot in kiwifruit. Plant Dis. 101, 1402–1410. doi: 10.1094/PDIS-10-16-1535-RE
Fernandez, G., Sbres, M., Lado, J., and Perez-Faggiani, E. (2022). Postharvest sour rot control in lemon fruit by natamycin and an Allium extract. Int. J. Food Microbiol. 368:109605. doi: 10.1016/j.ijfoodmicro.2022.109605
Fernie, A. R., Carrari, F., and Sweetlove, L. J. (2004). Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7, 254–261. doi: 10.1016/j.pbi.2004.03.007
Ganash, M., Ghany, T. M. A., and Omar, A. M. (2018). Morphological and biomolecules dynamics of phytopathogenic fungi under stress of silver nanoparticles. Bionanoscience 8, 566–573. doi: 10.1007/s12668-018-0510-y
Gao, Z., Zhang, R. C., and Bo, X. (2021). Management of postharvest diseases of kiwifruit with a combination of the biocontrol yeast Candida oleophila and an oligogalacturonide. Biol. Control. 156:104549. doi: 10.1016/j.biocontrol.2021.104549
Gordienko, M. G., Palchikova, V. V., Kalenov, S. V., Belov, A. A., Lyasnikova, V. N., Poberezhniy, D. Y., et al. (2019). Antimicrobial activity of silver salt and silver nanoparticles in different forms against microorganisms of different taxonomic groups. J. Hazard. Mater. 378:120754. doi: 10.1016/j.jhazmat.2019.120754
Gow, N. A. R., Latge, J. P., and Munro, C. A. (2017). The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 5, 1–25. doi: 10.1128/9781555819583.ch12
Gupta, D., and Chauhan, P. (2015). Fungicidal activity of silver nanoparticles against Alternaria brassicicola. AIP Conf. Proc. 1724:020031. doi: 10.1063/1.4945151
He, C., Zhang, Z. Q., Li, B. Q., Xu, Y., and Tian, S. P. (2019). Effect of natamycin on Botrytis cinerea and Penicillium expansum—postharvest pathogens of grape berries and jujube fruit. Postharvest Biol. Technol. 151, 134–141. doi: 10.1016/j.postharvbio.2019.02.009
Hong, X. S., Wen, J. J., Xiong, X. H., and Hu, Y. Y. (2016). Shape effect on the antibacterial activity of silver nanoparticles synthesized via a microwave-assisted method. Environ. Sci. Pollut. 23, 4489–4497. doi: 10.1007/s11356-015-5668-z
Jian, Y. X., Chen, X., Ahmed, T., Shang, Q. H., Zhang, S., Ma, Z. H., et al. (2021). Toxicity and action mechanisms of silver nanoparticles against the mycotoxin-producing fungus Fusarium graminearum. J. Adv. Res. 38, 1–12. doi: 10.1016/j.jare.2021.09.006
Jo, Y. K., Kim, B. H., and Jung, G. (2009). Antifungal activity of silver ions and nanoparticles on phytopathogenic fungi. Plant Dis. 93, 1037–1043. doi: 10.1094/PDIS-93-10-1037
Kai, K., Bi, W. L., Sui, Y., Hua, C. Y., Liu, Y. S., and Zhang, D. F. (2020). Curcumin inhibits Diaporthe phaseolorum and reduces postharvest decay in kiwifruit. Sci. Hortic. 259:108860. doi: 10.1016/j.scienta.2019.108860
Kasprowicz, M. J., Kozioł, M., and Gorczyca, A. (2020). The effect of silver nanoparticles on phytopathogenic spores of Fusarium culmorum. Can. J. Microbiol. 56, 247–253. doi: 10.1139/W10-012
Ke, X., Yin, Z., Song, N., Dai, Q., Voegele, R. T., Liu, Y., et al. (2014). Transcriptome profifiling to identify genes involved in pathogenicity of Valsa mali on apple tree. Fungal Genet. Biol. 68:31. doi: 10.1916/j.fgb.2014.04.004
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kim, G. H., Lee, Y. S., Jung, J. S., Hur, J. S., and Koh, Y. J. (2013). Optimal spray time, interval and number of preventive fungicides for the control of fruit rots of green and gold kiwifruit cultivars. Res. Plant Dis. 19, 1–6. doi: 10.5423/RPD.2013.19.1.001
Kim, H. S., Kang, H. S., Chu, G. J., and Byun, H. S. (2008). Antifungal effectiveness of nanosilver colloid against rose powdery mildew in greenhouses. Solid State Phenom. 135, 15–18.
Kim, S. W., Jung, J. H., Lamsal, K., Kim, Y. S., Min, J. S., and Lee, Y. S. (2012). Antifungal effects of silver nanoparticles (AgNPs) against various plant pathogenic fungi. Mycobiology 40, 53–58. doi: 10.5941/MYCO.2012.40.1.053
Lamsal, K., Kim, S. W., Jung, J. H., Kim, Y. S., Kim, K. S., and Lee, Y. S. (2011). Inhibition effects of silver nanoparticles against powdery mildews on cucumber and pumpkin. Mycobiology 39, 26–32. doi: 10.4489/MYCO.2011.39.1.026
Lee, K. J., Park, S. H., Govarthanan, M., Hwang, P. H., Seo, Y. S., Cho, M., et al. (2013). Synthesis of silver nanoparticles using cow milk and their antifungal activity against phytopathogens. Mater. Lett. 105, 128–131. doi: 10.1016/j.matlet.2013.04.076
Li, J., Fu, S., Fan, G., Li, D., Yang, S., Peng, L., et al. (2021). Active compound identification by screening 33 essential oil monomers against Botryosphaeria dothidea from postharvest kiwifruit and its potential action mode. Pestic. Biochem. Physiol. 179:104957. doi: 10.1016/j.pestbp.2021.104957
Li, L., Pan, H., Chen, M. Y., Zhang, S. J., and Zhong, C. H. (2017a). Isolation and identification of pathogenic fungi causing post-harvest fruit rot of kiwifruit (Actinidia chinensis) in China. J. Phytopathol. 165, 782–790. doi: 10.1111/jph.12618
Li, L., Pan, H., Liu, W., Chen, M. Y., and Zhong, C. H. (2017b). First report of Alternaria alternata causing postharvest rot of kiwifruit in Shaanxi, Hubei and Sichuan provinces, China. Plant Dis. 6, 1046–1047. doi: 10.1094/PDIS-11-16-1611-PDN
Li, L., Pan, H., Liu, W., Chen, M. Y., and Zhong, C. H. (2017c). First report of Diaporthe actinidiae causing stem-end rot of kiwifruit in Hunan and Hubei provinces, China. Plant Dis. 6:1054. doi: 10.1094/PDIS-12-16-1852-PDN
Li, L., Pan, H., Chen, M. Y., and Zhong, C. H. (2016). First report of Pestalotiopsis microspora causing postharvest rot of kiwifruit in Hubei province, China. Plant Dis. 10:2161. doi: 10.1094/PDIS-01-16-0059-PDN
Liu, R., Guo, Z., Li, J., Wu, F. Y., Cui, K. P., Cheng, P., et al. (2022). Mechanism of Phanerochaete chrysosporium-mediated tetracycline hydrochloride removal promoted by low-dose silver nanoparticles. Engineering 10:107474. doi: 10.1016/j.jece.2022.107474
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, T., Qu, Q., Lavoie, M., Pan, X. J., Peijnenburg, W. J. G. M., Zhou, Z. G., et al. (2020). Insights into the transcriptional responses of a microbial community to silver nanoparticles in a freshwater microcosm. Environ. Pollut. 258:113727. doi: 10.1016/j.envpol.2019.113727
Lu, Z., Yin, L., Li, W., and Jiang, H. S. (2022). Low concentrations of silver nanoparticles inhibit spore germination and disturb gender differentiation of Ceratopteris thalictroides (L.) Brongn. Nanomaterials 12:1730. doi: 10.3390/nano12101730
Madbouly, A. K. (2021). The efficacy of green synthesized nanosilver in reducing the incidence of post-harvest apple fruit brown rot. J. Fungi 7:473. doi: 10.3390/jof7060473
Malandrakis, A. A., Kavroulakis, N., and Chrysikopoulos, C. V. (2019). Use of copper, silver and zinc nanoparticles against foliar and soil-borne plant pathogens. Sci. Total Environ. 670, 292–299. doi: 10.1016/j.scitotenv.2019.03.210
Mendes, J. E., Abrunhosa, L., Teixeira, J. A., de Camargo, E. R., deSouza, C. P., and Pessoa, J. D. C. (2014). Antifungal activity of silver colloidal nanoparticles against phytopathogenic fungus (Phomopsis sp.) in soybean seeds. Int. J. Bio. Vet. Agri. Food Eng. 8, 9928–9933. doi: 10.5281/zenodo.1095981
Moussa, S. H., Tayel, A. A., Alsohim, A. S., and Abdallah, R. R. (2013). Botryticidal activity of nanosized silver-chitosan composite and its application for the control of gray mold in strawberry. J. Food Sci. 78, 589–594. doi: 10.1111/1750-3841.12247
Navarro, P., Paposo, J. C., Arana, G., and Etxebarria, N. (2006). Optimisation of microwave assisted digestion of sediments and determination of Sn and Hg. Anal. Chim. Acta 566, 37–44. doi: 10.1016/j.aca.2006.02.056
Nelson, N., and Ben-Shem, A. (2004). The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Bio. 5, 971–982. doi: 10.1038/nrm1525
Noori, A., White, J. C., and Newman, L. A. J. (2017). Mycorrhizal fungi influence on silver uptake and membrane protein gene expression following silver nanoparticle exposure. J. Nanopart. Res. 19:66. doi: 10.1007/s11051-016-3650-4
Oliveira, S. S., Braga, G. C., Cordeiro, N. K., Stabgarlin, J. R., and Alves, H. J. (2022). Green synthesis of silver nanoparticles with Euphorbia tirucalli extract and its protection against microbial decay of strawberries during storage. J. Food Sci. Technol. 59, 2025–2034. doi: 10.1007/s13197-021-05217-y
Ouda, S. M. (2014). Antifungal activity of silver and copper nanoparticles on two plant pathogens, Alternaria alternata and Botrytis cinerea. Res. J. Microbiol. 9, 34–42. doi: 10.3923/jm.2014.34.42
Pan, L. Y., Zhao, X. Y., Chen, M., Fu, Y. Q., Xiang, M. L., and Chen, J. Y. (2020). Effect of exogenous methyl jasmonate treatment on disease resistance of postharvest kiwifruit. Food Chem. 305:125483. doi: 10.1016/j.foodchem.2019.125483
Patnaik, B. B., Park, S. Y., Kang, S. W., Hwang, H., Wang, T. H., Park, E. B., et al. (2016). Transcriptome profile of the Asian Giant Hornet (Vespa mandarinia) using illumina Hiseq 4000 sequencing: De Novo assembly, functional annotation, and discovery of SSR markers. Int. J. Genom. 2016:4169587. doi: 10.1155/2016/4169587
Quevedo, A. C., Iseult, L., and Valsami-Jones, E. (2021). Cellular repair mechanisms triggered by exposure to silver nanoparticles and ionic silver in embryonic zebrafish cells. Environ. Sci. Nano 8, 2507–2522. doi: 10.1039/D1EN00422K
Rahman, A. P. H., Pranjal Behera, S. K., Mishra, A., Lundborg, C. S., and Tripathy, S. K. (2022). Transcriptomic regulation of Salmonella Typhimurium during sonophotocatalysis and the effect of stress adaptation on the antibiotic resistance and tolerance post-treatment. Chem. Eng. J. 446:137442. doi: 10.1016/j.cej.2022.137442
Raza, M. A., Kanwal, Z., Rauf, A., Sabri, A. N., Riaz, S., and Naseem, S. (2017). Size- and shape-dependent antibacterial studies of silver nanoparticles synthesized by wet chemical routes. Nano 6, 1–15. doi: 10.3390/nano6040074
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. doi: 10.1186/gb-2010-11-3-r25
Sharma, J., Singh, V. K., Kumar, A., Shankarayan, R., and Mallubhotla, S. (2018). “Role of Silver nanoparticles in treatment of plant diseases,” in Microbial Biotechnology, eds J. Patra, G. Das, and H. S. Shin (Singapore: Springer Nature Singapore Pte Ltd), 435–454. doi: 10.1007/978-981-10-7140-9_20
Shen, T. L., Wang, Q. S., Li, C. L., Zhou, B., Li, Y. H., and Liu, Y. L. (2020). Transcriptome sequencing analysis reveals silver nanoparticles antifungal molecular mechanism of the soil fungi Fusarium solani species complex. J. Hazard. 388:122063. doi: 10.1016/j.jhazmat.2020.122063
Shivamogga Nagaraju, R., Holalkere Sriram, R., and Achur, R. (2020). Antifungal activity of Carbendazim-conjugated silver nanoparticles against anthracnose disease caused by Colletotrichum gloeosporioides in mango. J. Plant Pathol. 102, 39–46. doi: 10.1007/s42161-019-00370-y
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Tian, X., Jiang, X. M., Welch, C., Croley, T. R., Wong, T. Y., Chen, C., et al. (2018). Bactericidal effffects of silver nanoparticles on Lactobacilli and the underlying mechanism. ACS Appl. Mater. Interfaces 10, 8443–8450. doi: 10.1021/acsami.7b17274
Wang, G., Chen, L. F., Tang, W. Q., Wang, Y. Y., Zhang, Q., Wang, H. B., et al. (2021). Identifying a melanogenesis-related candidate gene by a high-quality genome assembly and population diversity analysis in Hypsizygus marmoreus. J. Genet. Genom. 48, 75–87. doi: 10.1016/j.jgg.2021.01.002
Wollenberg, R. D., Donau, S. S., Nielsen, T. T., Sorensen, J. L., Giese, H., Wimmer, R., et al. (2016). Real-time imaging of the growth-inhibitory effffect of JS399-19 on Fusarium. Pestic. Biochem. Physiol. 134, 24–30. doi: 10.1016/j.pestbp.2016.05.002
Xia, Z. K., Ma, Q. H., Li, S. Y., Zhang, D. Q., Cong, L., Tian, Y. L., et al. (2016). The antifungal effffect of silver nanoparticles on Trichosporon asahii. J. Microbiol. Immunol. 49, 182–188. doi: 10.1016/j.jmii.2014.04.013
Xu, Y., Gao, C. W., Li, X. H., He, Y., Zhou, L. T., Pang, G. R., et al. (2013). In vitro antifungal activity of silver nanoparticles against ocular pathogenic filamentous fungi. J. Ocul. Pharmacol. Ther. 29, 270–274. doi: 10.1089/jop.2012.0155
Xue, B. J., He, D., Gao, S., Wang, D. Y., Yokoyama, K., and Wang, L. (2016). Biosynthesis of silver nanoparticles by the fungus Arthroderma fulvum and its antifungal activity against genera of Candida, Aspergillus and Fusarium. Int. J. Nanomed. Nanosurg. 11, 1899–1906. doi: 10.2147/IJN.S98339
Yin, Z. Y., Liu, H. Q., Li, Z. P., Ke, X. W., Dou, D. L., and Gao, X. N. (2015). Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 208, 1202–1216. doi: 10.1111/nph.13544
Zhang, Y. F., Yang, L. F., Chen, K. S., and Dong, L. (2007). Effects of neodymium on growth, pectinase activity and mycelium permeability of Fusarium oxysporum. J. Rare Earth 25, 100–105. doi: 10.1016/S1002-0721(07)60053-2
Zhao, L., Cheng, Y., Li, B., Gu, X., Zhang, X., Boateng, N. A. S., et al. (2019). Integration of proteome and transcriptome data reveals the mechanism involved in controlling of Fusarium graminearum by Saccharomyces cerevisiae. J. Sci. Food Agr. 99, 5760–5770. doi: 10.1002/jsfa.9844
Zhao, L., He, F. T., Li, B., Gu, X. Y., Zhang, X. Y., Dhanasekaran, S., et al. (2022). Transcriptomic analysis of the mechanisms involved in enhanced antagonistic efficacy of Meyerozyma guilliermondii by methyl jasmonate and disease resistance of postharvest apples. LWT Food Sci. Technol. 160:113323. doi: 10.1016/j.lwt.2022.113323
Zhao, L., Wang, Y., Wang, Y., Li, B., Gu, X., Zhang, X., et al. (2020). Effect of β-glucan on the biocontrol efficacy of Cryptococcus podzolicus against postharvest decay of pears and the possible mechanisms involved. Postharvest Biol. Technol. 160:111057. doi: 10.1016/j.postharvbio.2019.111057
Zhou, Y., Gong, G., Cui, Y., Zhang, D., Chang, X., Hu, R., et al. (2015). Identification of Botryosphaeriaceae species causing kiwifruit rot in Sichuan Province. China. Plant Dis. 99, 699–708. doi: 10.1094/PDIS-07-14-0727-RE
Zhu, H. H., Ai, H. L., Cao, L. W., Sui, R., Ye, H. P., Du, D. Y., et al. (2018). Transcriptome analysis providing novel insights for Cd-resistant tall fescue responses to Cd stress. Ecotoxicol. Environ. Saf. 160, 349–356. doi: 10.1016/j.ecoenv.2018.05.066
Zhuo, R., and Fan, F. F. (2021). A comprehensive insight into the application of white rot fungi and their lignocellulolytic enzymes in the removal of organic pollutants. Sci. Total Environ. 778:146132. doi: 10.1016/j.scitotenv.2021.146132
Keywords: nanoparticles, kiwifruit post-harvest rot, antifungal activity, transcriptomic analysis, microstructure observation
Citation: Li L, Pan H, Deng L, Qian G, Wang Z, Li W and Zhong C (2022) The antifungal activity and mechanism of silver nanoparticles against four pathogens causing kiwifruit post-harvest rot. Front. Microbiol. 13:988633. doi: 10.3389/fmicb.2022.988633
Received: 07 July 2022; Accepted: 15 August 2022;
Published: 31 August 2022.
Edited by:
Tofazzal Islam, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Md Aktaruzzaman, University of Georgia, United StatesMitzuko Dautt-Castro, Instituto Potosino de Investigación Científica y Tecnológica (IPICYT), Mexico
Islam Hamim, Bangladesh Agricultural University, Bangladesh
Yongcai Li, Gansu Agricultural University, China
Copyright © 2022 Li, Pan, Deng, Qian, Wang, Li and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caihong Zhong, emhvbmdjYWlob25nQHdiZ2Nhcy5jbg==
†These authors have contributed equally to this work and share first authorship
 Li Li1†
Li Li1† Guoliang Qian
Guoliang Qian Caihong Zhong
Caihong Zhong