- 1National Laboratory for Health, Environment and Food, Department for Microbiological Research, Maribor, Slovenia
- 2Department of Gastroenterology, University Medical Centre Ljubljana, Ljubljana, Slovenia
- 3Faculty of Medicine, University of Maribor, Maribor, Slovenia
- 4Department of Gastroenterology, University Clinical Centre Maribor, Maribor, Slovenia
- 5Department of Microbiology, Faculty of Medicine, University of Maribor, Maribor, Slovenia
Clostridioides difficile colonization and development of infection commonly occur in inflammatory bowel disease (IBD) patients and can trigger flare-ups. Both conditions are inherently linked to disrupted gut microbiota. This study included 149 hospitalized gastrointestinal patients, which were divided into IBD (n = 48) and non-IBD patients (n = 101). Patients were tested for C. difficile colonization (qPCR and selective plating), and gut bacterial communities were analyzed with 16S amplicon sequencing. Blood test results were retrospectively collected from the medical records. IBD and non-IBD patients had comparable C. difficile colonization rates (31.7 and 33.3%, respectively). Compared to non-IBD C. difficile-non-colonized patients, IBD and C. difficile-colonized patients shared multiple common bacterial community characteristics including decreased diversity and reduced abundance of strict anaerobic bacteria. Furthermore, certain microbiota alterations were enhanced when IBD was accompanied by C. difficile colonization, indicating a synergistic effect between both medical complications. Conversely, certain microbial patterns were specific to C. difficile colonization, e.g., co-occurrence with Enterococcus, which was most common in IBD patients (81.3%).
Introduction
Inflammatory bowel disease (IBD) is a chronic complication in the gastrointestinal tract that is increasing in incidence worldwide (Kaplan, 2015; Alatab et al., 2020; Freeman et al., 2021). The two principal types of IBD, Crohn’s disease and ulcerative colitis, are distinguished based on the localization of inflammation along the gastrointestinal tract (Guan, 2019). IBD flare-ups are caused by activated immune responses against native intestinal microbiota in genetically predisposed individuals and can be triggered by a variety of environmental factors (Zhao and Burisch, 2019). One important risk factor for flare-ups is a preceding gastrointestinal tract infection. The most common infection that leads to flare-ups is caused by Clostridioides difficile (Khanna et al., 2017), whereas other bacterial pathogens are less common in this regard (Hanada et al., 2018). The colonization rate with C. difficile as well as the incidence of C. difficile infection (CDI) are higher in IBD patients, especially in patients with ulcerative colitis (Rodemann et al., 2007; Clayton et al., 2009; Tai et al., 2021). CDI manifestations are also more severe in IBD patients, more likely to lead to recurrent infections, and more often require colectomy and other gastrointestinal tract surgeries (Ananthakrishnan et al., 2008; Khanna and Pardi, 2016; Razik et al., 2016). Therefore, a better understanding of concomitant CDI and IBD could help improve treatment practices, such as fecal microbiota transplantation (Khoruts et al., 2016).
The evidence regarding gut community alterations in IBD patients is substantial. The most prominent alterations include decreased diversity of commensal communities, mainly Bacillota (synonym Firmicutes), and the consequent overgrowth of facultative anaerobes, often opportunistic pathogens such as enterobacteria (Khan et al., 2019; Qiu et al., 2022). Similar patterns are characteristic of the gut microbiota in CDI, in addition to C. difficile itself often becoming the predominant species in the gut (Sehgal and Khanna, 2021; Schnizlein and Young, 2022). Alterations in human gut microbiota in response to C. difficile colonization are in contrast poorly described (Zhang et al., 2015; Vincent et al., 2016). But it is well established that the commensal microbiota plays a crucial role at providing colonization resistance against C. difficile as demonstrated both in vitro and in vivo (Auchtung et al., 2020; Pike and Theriot, 2020).
Although C. difficile colonization is common in IBD patients, this study is the first to compare C. difficile colonization-associated alterations in the gut microbiota of IBD and non-IBD gastrointestinal patients. We used selective culturing and species-specific qPCR to detect and quantify C. difficile in stool samples from 149 hospitalized patients. Additionally, we performed 16S amplicon metagenomic sequencing to determine C. difficile colonization-associated bacterial community characteristics in IBD versus non-IBD patients.
Materials and methods
Sample collection
Stool samples were collected from 149 patients hospitalized at the Department of Gastroenterology, University Medical Centre Maribor, after informed consent. The same cohort was included in a broader study that compared microbiota signatures between healthy controls and hospitalized gastrointestinal patients (Mahnic et al., 2020). Patients were diagnosed with standard medical procedures including clinical, radiological, endoscopic, and histological criteria and were, for the purpose of further analysis, distributed into two groups based on the diagnosis: IBD (n = 48) and non-IBD patients (n = 101). The IBD group included patients with both principal IBD types, i.e., ulcerative colitis (n = 28) and Crohn’s disease (n = 20). The non-IBD group comprised patients with the following pathologies: infections (pneumonia, cholangitis, hepatitis, gastritis, or pancreatitis), cancer (pancreatic, gastric, or liver), liver cirrhosis, and peptic ulcer. Because patients were hospitalized for different medical complication and subsequently underwent different treatments, no exclusion criteria except minimum age of 18 was used in this study. All patient’s collected metadata can be found in Supplementary Table 1. No patient was hospitalized primarily due to CDI; however, three IBD patients tested positive for C. difficile during hospitalization. The cohort consisted of 76 women and 73 men, with an average age of 59.3 years (SD = 17.9). Values for C-reactive protein, leukocytes, neutrophil granulocytes, sedimentation rate, albumins, and ferritin were retrospectively collected from the medical records, and the test result closest to the sample collection date was taken for each patient. Ethical approval was obtained from the National Medical Ethics Committee (KME 95/05/15).
Stool samples were collected in sterile containers and immediately transported to the laboratory. After homogenization, a volumetric equivalent of 50 μL was added to 1 mL of Inhibitex buffer (QIAamp Fast Stool DNA Mini Kit, Qiagen, Hilden, Germany) and stored at −80°C until total DNA isolation.
Total DNA isolation
After mechanical disruption (7,000 rpm for 70 s) with the SeptiFast Lyse Kit and MagNA Lyser (Roche, Basel, Switzerland), total bacterial DNA was extracted from each stool sample using the QIAamp Fast Stool DNA Mini Kit (Qiagen, Hilden, Germany) and used for molecular detection of C. difficile and microbiota analysis.
Quantification of Clostridioides difficile 16S rRNA and tcdB genes
Both C. difficile 16S rRNA and tcdB genes were quantified with gene-specific qPCR using Rotor-Gene (Qiagen, Hilden, Germany) and Rotor-Gene Probe Mix (Qiagen, Hilden, Germany). For the amplification and detection of the 16S rRNA gene, we used F–(5′-TTGAGCGATTTACTTCGGTAAAGA-3′), R–(5′-CCATCCT GTACTGGCTCACCT-3′), Probe–(5′-CCTACCCTGTACACA CGGATAACATACCG-3′) (Rinttilä et al., 2004). For the amplification and detection of tcdB, we used F–(5′-GAAAGTCCAAGTTTACGCTCAAT-3′), R–(5′-GCTG CACCTAAACTTACACCA-3′), Probe–(5′-ACAGATGCAGC CAAAGTTGTTGAATT-3′) (van den Berg et al., 2006). qPCRs were performed in duplicate, and samples were considered positive for 16S rRNA or tcdB when both replicates were positive.
Clostridioides difficile cultivation
Stool samples were cultivated by spreading a full inoculation loop on media selective for C. difficile (CHROMID; BioMerieux, Marcy-l’Etoile, France). This was followed by a 3-day incubation at 37°C under anaerobic conditions.
16S metagenomic sequencing library preparation and sequence data analysis
Bacterial community composition was obtained by paired-end sequencing of the V3V4 hypervariable region of the 16S rRNA gene on the MiSeq platform (Illumina, San Diego, CA, United States), as previously described (Mahnic et al., 2020).
Quality filtering of sequence reads and clustering into operational taxonomic units (OTUs) based on 97% similarity was performed using mothur (v.1.44.1) (Schloss et al., 2009), as previously described (Mahnic et al., 2020). Taxonomic classification of representative sequences was inferred using the RDP mothur training set v16. Down-stream statistical analyses and data visualization were performed in R (version 3.1.3) using package “ggplot2” and mothur (Shannon index, AMOVA, LEfSe).
The sequence data supporting the conclusion of this article are available in the form of combined paired-end reads (contigs) on the Metagenomics RAST (MG-RAST) database server under the project access number mgp86691. Samples used in this study are denoted with sample names G[sequential number of the sample].
Results
In total, 48 IBD and 101 non-IBD patients were tested for colonization with C. difficile using a molecular approach (qPCR) and selective plating (Table 1). For statistical analysis, the IBD group was not further sub-divided into Crohn’s patients and those with ulcerative colitis because of the limited number of samples available. The study cohort was well balanced regarding gender; however, the mean age was significantly higher in non-IBD compared to IBD patients (p < 0.001) (Table 1). Age discrepancy was not compensated for in the statistical analysis; however, all reported results were critically evaluated with respect to this imbalance. Age related alterations in community structure are described in Supplementary Figure 1.
Using C. difficile species-specific qPCR, we detected C. difficile in 48 (32.2%) stool samples. We did not observe differences in the qPCR-based prevalence of C. difficile (Fisher’s exact test p = 0.853) or tcdB (Fisher’s exact test p = 0.681) or culture-based prevalence of C. difficile (Fisher’s exact test p = 0.657) between IBD and non-IBD patients (Table 1). All tcdB-positive and culture-positive samples were also detected with species-specific C. difficile qPCR (Supplementary Table 1). Hereinafter, all C. difficile qPCR positive samples were considered as C. difficile-colonized. Cultured strains belonged to four different C. difficile ribotypes: 001/072, 027, SLO 076, and 002. Out of 5 culture positive samples we were able to detect tcdB in 3 samples (60%). Samples that were negative after selective plating were additionally enriched prior to plating in brain heart infusion medium with 0.1% L-cysteine, 0.5% yeast extract, and 0.1% sodium taurocholate. After 5 days of incubation at 37°C under anaerobic conditions, no additional C. difficile-positive samples were obtained compared to direct plating on selective media (data not shown).
Furthermore, no correlations were observed between C. difficile colonization and the following blood markers: inflammatory markers (C-reactive protein, leukocytes, neutrophil granulocytes, and sedimentation rate), albumins, and ferritin (Supplementary Figure 2).
Clostridioides difficile colonization-associated bacterial community characteristics are significant only in non-inflammatory bowel disease patients
Permutational multivariate analysis of variance (PERMANOVA) showed significant correlations of bacterial community structure with age, IBD status, and C. difficile colonization, explaining 1.5, 1.1, and 1.2% of inter-sample variability, respectively (Supplementary Table 2). Age-associated community characteristics are presented in Supplementary Figure 1. They did not correlate with differences reported in the context of IBD status or C. difficile colonization and will therefore not be discussed further. Differences in bacterial community structure between C. difficile-colonized and non-colonized patients were significant in non-IBD (AMOVA, p = 0.018) but not IBD patients (AMOVA, p = 0.967). In non-IBD patients, C. difficile colonization was associated with lower bacterial community richness (i.e., number of unique OTUs, p = 0.038) and community evenness (i.e., Shannon evenness, p = 0.045) (Figure 1A). Differentially represented OTUs included increased facultative anaerobic genera, e.g., Enterococcus and Streptococcus, and decreased strictly anaerobic genera, e.g., Faecalibacterium, Roseburia, Blautia, Lachnospiraceae, and Clostridium XIVa (Figure 1B).
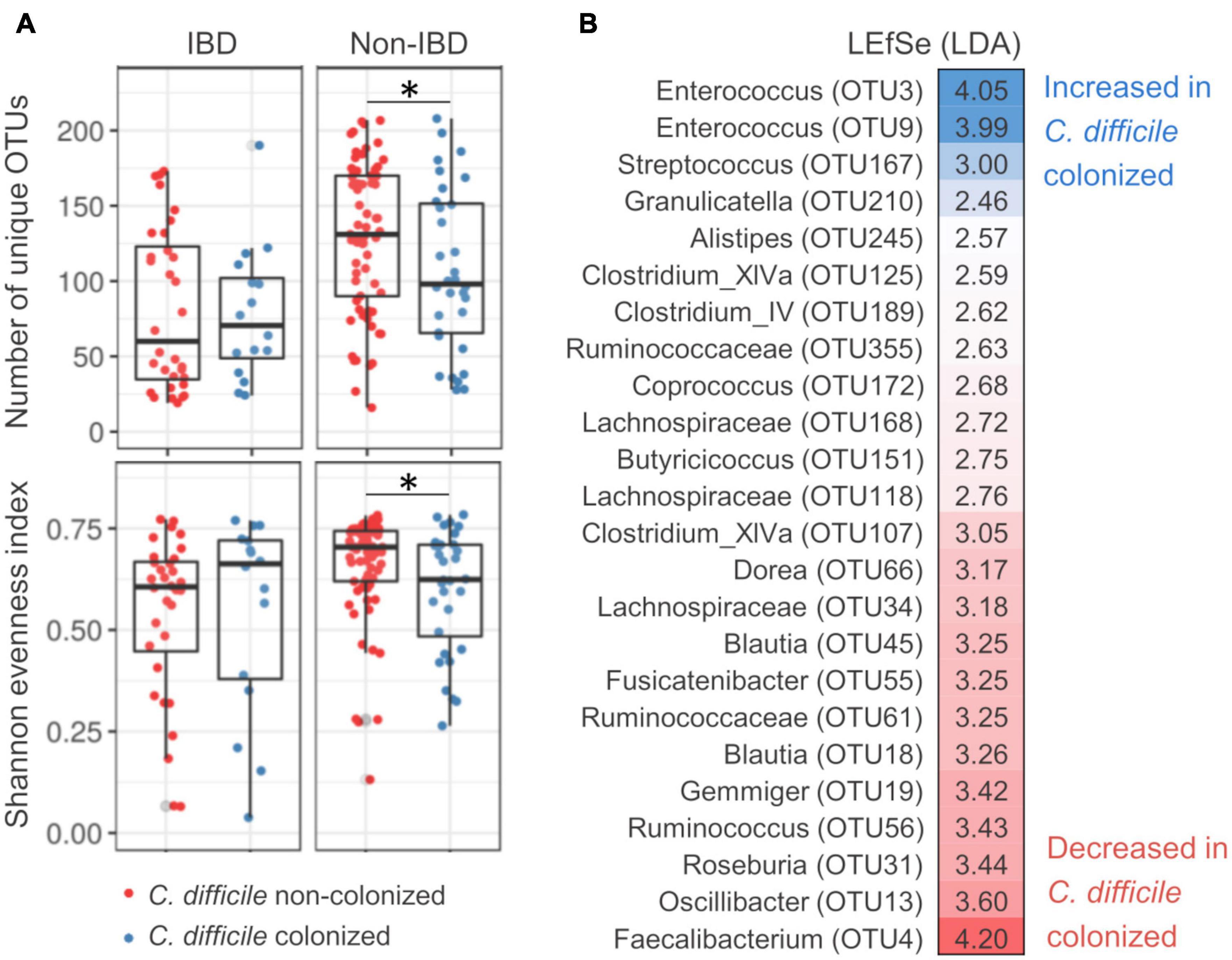
Figure 1. Bacterial community characteristics of Clostridioides difficile-colonized patients with regard to inflammatory bowel disease status. (A) Clostridioides difficile colonization-associated differences in bacterial community richness [number of unique operational taxonomic units (OTUs), top] and evenness (Shannon evenness, bottom) for inflammatory bowel disease (IBD) and non-IBD patients. (B) OTUs that were differentially represented between C. difficile-colonized and non-colonized patients, as obtained by LEfSe population analysis. Population analysis was performed only on non-IBD patients for which beta-diversity analysis showed significant differentiation between C. difficile-colonized and non-colonized. *Denotes significance with p value between 0.05 and 0.01.
The genus Enterococcus was significantly increased in C. difficile-colonized patients (Figure 1B). In our dataset, Enterococcus clustered into two distinct OTUs (OTU3 and OTU9). Both Enterococcus-associated OTUs exhibited a higher prevalence in C. difficile-colonized IBD and non-IBD patients [Fisher’s exact test: p = 0.004 (OTU3) and p = 0.020 (OTU9)]. The highest prevalence of Enterococcus was detected in C. difficile-colonized IBD patients (78.1, 56.3, and 81.3% for OTU3, OTU9, and both OTUs combined, respectively). Additionally, co-occurrence of both Enterococcus-associated OTUs was more frequent in C. difficile-colonized compared to non-colonized patients (Fisher’s exact test, p = 0.006).
Clostridioides difficile colonization and inflammatory bowel disease show similar bacterial community alterations
Inflammatory bowel disease patients showed similar community characteristics as described above for C. difficile-colonized patients, partially explaining the lack of differentiation between C. difficile-colonized and non-colonized IBD patients. These characteristics included reduced community richness (p < 0.001) and a decrease in relative abundance of multiple Bacillota representatives. We demonstrated that multiple differentially represented OTUs were characteristic of both conditions, the underlying IBD as well as C. difficile colonization (Figures 2A,C). From the 24 OTUs associated with C. difficile colonization (Figure 1B), 14 (58.3%) were also associated with IBD. Three OTUs were significantly decreased in C. difficile-colonized in both the IBD and non-IBD group, but not in C. difficile non-colonized patients, Ruminococcaceae (OTU61), Dorea (OTU66), and Clostridium XIVa (OTU125) (Figure 2B). Interestingly, a few OTUs that were decreased in IBD patients showed significantly more prominent reduction when IBD was accompanied with C. difficile colonization, indicating potential synergistic effects of these two medical conditions (Figure 2D). These OTUs included multiple representatives of Alistipes, Ruminococcaceae, Clostridium XIVa, Clostridium IV, and Lachnospiraceae (Figure 2D).

Figure 2. Shared community characteristics of Clostridioides difficile colonization and inflammatory bowel disease. Four groups were formed based on inflammatory bowel disease (IBD) status and C. difficile colonization. The control group contained C. difficile non-colonized non-IBD patients. For each differentially represented operational taxonomic unit (OTU), the LDA score (LEfSe test, left) and difference in relative abundance (right) for non-IBD/C. difficile-colonized (red), IBD/C. difficile-non-colonized (green), and IBD/C. difficile-colonized patients (blue) as compared to the control group are presented. Sections (A–D) denote OTU groups that are shared among the different patient groups.
Discussion
Gut microbiota perturbations can be a precondition as well as a consequence of both IBD and CDI (Missaghi et al., 2014; Guan, 2019; Sehgal and Khanna, 2021; Qiu et al., 2022). While co-occurrence of IBD and CDI is common (Khanna et al., 2017), this study demonstrated that these two medical conditions also share common gut community signatures.
We did not observe a higher C. difficile colonization rate in IBD compared to non-IBD patients, despite sound evidence in the literature supporting this phenomenon. Studies not only report higher prevalence of C. difficile colonization in the population of IBD compared to non-IBD subjects (Bossuyt et al., 2009; Hourigan et al., 2013; Ramos-Martínez et al., 2015) but also the increase in prevalence with time (Issa et al., 2007; Mabardy et al., 2017). We also observed no correlation between C. difficile colonization and inflammatory markers in the blood, even though several inflammatory markers have been reported as predictive for adverse outcomes in cases of symptomatic CDIs (Dieterle et al., 2020). The absence of comparable conclusion in our study could be due to the small sample size, which we consider the main limitation of this study. Additionally, our study included only hospitalized gastroenterological patients, while comparable studies often include also healthy subjects as a control group.
The microbial community characteristics associated with C. difficile colonization in this study are in agreement with previous findings in asymptomatic carriers (Zhang et al., 2015) and CDI patients. Studies on CDI patients in concordance with this study reported decreased community diversity (Zhang et al., 2015; Milani et al., 2016; Duan et al., 2020) and a blooming effect in the gut, generally characterized by overgrowth of facultative anaerobic opportunists at the expense of strictly anaerobic commensals (Schubert et al., 2014; Milani et al., 2016; Amrane et al., 2019; Duan et al., 2020). Our findings also support the previously reported common co-occurrence of C. difficile and Enterococcus (Poduval et al., 2000; Fujitani et al., 2011), which was the most frequent genus (81.3%) in IBD patients in our study. Furthermore, the reduced abundance of Ruminococcaceae, Dorea, and Clostridium XIVa was exclusively associated with C. difficile colonization, regardless of IBD status. However, further testing is required to verify potential antagonistic effects.
Many of the above-mentioned C. difficile-associated microbial alterations are also common in IBD (Ni et al., 2017; Khan et al., 2019; Lloyd-Price et al., 2019), as also revealed by our current study. The driver of such changes in the microbiome is most likely the increased level of oxygen in the gut during inflammation, which occurs through different mechanisms (Rigottier-Gois, 2013; Rivera-Chávez et al., 2017). Nevertheless, an interesting aspect of our findings is the potential synergistic effect of concomitant IBD and CDI. In IBD patients, several taxa were markedly more decreased in C. difficile-colonized compared to non-colonized patients, including three OTUs associated with Alistipes. The genus Alistipes was previously reported to be negatively correlated with CDI both in humans (Milani et al., 2016) and in a murine model (Schubert et al., 2015), whereas its role in other medical complications remains under investigation (Parker et al., 2020). Synergistic effects in gut microbiota could potentially elucidate the mechanisms underlying more severe CDIs in IBD patients (Ananthakrishnan et al., 2008; Khanna and Pardi, 2016; Razik et al., 2016).
In conclusion, our comparison of C. difficile-associated patterns in the microbiome of hospitalized IBD and non-IBD patients has contributed to a better differentiation between disease-specific patterns and other alterations caused by external factors. We observed common characteristics of “gut blooming,” including reduced bacterial richness due to a decreased abundance of multiple Bacillota and overgrowth of facultative anaerobes. These characteristics were observed in IBD patients regardless of C. difficile status and in C. difficile-colonized patients regardless of IBD status. Importantly, synergistic effects between both medical conditions were detected, potentially explaining the more severe pathologies observed in concomitant IBD/CDI patients.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.mg-rast.org/mgmain.html?mgpage=project&project=mgp86691, mgp86691.
Ethics statement
The studies involving human participants were reviewed and approved by National Medical Ethics Committee (KME 95/05/15). The patients/participants provided their written informed consent to participate in this study.
Author contributions
AM performed sequencing and data analysis and was a major contributor in writing the manuscript. SP partially performed total DNA isolations, retrieved and organized clinical data, and performed qPCR tests. PS organized clinical study and sample collection. MR contributed to project design, operation, and data interpretation and was a major contributor in writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Slovenian Research Agency (research core funding no. P3-0387). AM was also funded by Slovenian Research Agency with Mladi raziskovalci Grant.
Acknowledgments
We would like to acknowledge the contribution of Eva Lasic in editing of the manuscript, support by Aleksander Kocuvan in total DNA isolation, Sara Beigot Glaser in qPCR testing, and Sabina Horvat in sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.988426/full#supplementary-material
References
Alatab, S., Sepanlou, S. G., Ikuta, K., Vahedi, H., Bisignano, C., Safiri, S., et al. (2020). The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 5, 17–30. doi: 10.1016/S2468-1253(19)30333-4
Amrane, S., Hocquart, M., Afouda, P., Kuete, E., Pham, T. P. T., Dione, N., et al. (2019). Metagenomic and culturomic analysis of gut microbiota dysbiosis during Clostridium difficile infection. Sci. Rep. 9:12807. doi: 10.1038/s41598-019-49189-8
Ananthakrishnan, A. N., McGinley, E. L., and Binion, D. G. (2008). Excess hospitalisation burden associated with Clostridium difficile in patients with inflammatory bowel disease. Gut 57, 205–210. doi: 10.1136/gut.2007.128231
Auchtung, J. M., Preisner, E. C., Collins, J., Lerma, A. I., and Britton, R. A. (2020). Identification of Simplified Microbial Communities That Inhibit Clostridioides difficile Infection through Dilution/Extinction. mSphere 5, e00387–20. doi: 10.1128/mSphere.00387-20
Bossuyt, P., Verhaegen, J., Van Assche, G., Rutgeerts, P., and Vermeire, S. (2009). Increasing incidence of Clostridium difficile-associated diarrhea in inflammatory bowel disease. J. Crohns Colitis 3, 4–7. doi: 10.1016/j.crohns.2008.09.003
Clayton, E. M., Rea, M. C., Shanahan, F., Quigley, E. M. M., Kiely, B., Hill, C., et al. (2009). The vexed relationship between Clostridium difficile and inflammatory bowel disease: An assessment of carriage in an outpatient setting among patients in remission. Am. J. Gastroenterol. 104, 1162–1169. doi: 10.1038/ajg.2009.4
Dieterle, M. G., Putler, R., Perry, D. A., Menon, A., Abernathy-Close, L., Perlman, N. S., et al. (2020). Systemic Inflammatory Mediators Are Effective Biomarkers for Predicting Adverse Outcomes in Clostridioides difficile Infection. mBio 11, e00180–20. doi: 10.1128/mBio.00180-20
Duan, J., Meng, X., Liu, S., Zhou, P., Zeng, C., Fu, C., et al. (2020). Gut Microbiota Composition Associated With Clostridium difficile-Positive Diarrhea and C. difficile Type in ICU Patients. Front. Cell. Infect. Microbiol. 10:190. doi: 10.3389/fcimb.2020.00190
Freeman, K., Ryan, R., Parsons, N., Taylor-Phillips, S., Willis, B. H., and Clarke, A. (2021). The incidence and prevalence of inflammatory bowel disease in UK primary care: A retrospective cohort study of the IQVIA Medical Research Database. BMC Gastroenterol. 21:139. doi: 10.1186/s12876-021-01716-6
Fujitani, S., George, W. L., Morgan, M. A., Nichols, S., and Murthy, A. R. (2011). Implications for vancomycin-resistant Enterococcus colonization associated with Clostridium difficile infections. Am. J. Infect. Control 39, 188–193. doi: 10.1016/j.ajic.2010.10.024
Guan, Q. (2019). A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol. Res. 2019:7247238. doi: 10.1155/2019/7247238
Hanada, Y., Khanna, S., Loftus, E. V., Raffals, L. E., and Pardi, D. S. (2018). Non-Clostridium difficile Bacterial Infections Are Rare in Patients With Flares of Inflammatory Bowel Disease. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 16, 528–533. doi: 10.1016/j.cgh.2017.10.008
Hourigan, S. K., Chirumamilla, S. R., Ross, T., Golub, J. E., Rabizadeh, S., Saeed, S. A., et al. (2013). Clostridium difficile carriage and serum antitoxin responses in children with inflammatory bowel disease. Inflamm. Bowel Dis. 19, 2744–2752. doi: 10.1097/01.MIB.0000435434.53871.36
Issa, M., Vijayapal, A., Graham, M. B., Beaulieu, D. B., Otterson, M. F., Lundeen, S., et al. (2007). Impact of Clostridium difficile on inflammatory bowel disease. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 5, 345–351. doi: 10.1016/j.cgh.2006.12.028
Kaplan, G. G. (2015). The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 12, 720–727. doi: 10.1038/nrgastro.2015.150
Khan, I., Ullah, N., Zha, L., Bai, Y., Khan, A., Zhao, T., et al. (2019). Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 8:126. doi: 10.3390/pathogens8030126
Khanna, S., and Pardi, D. S. (2016). Clinical implications of antibiotic impact on gastrointestinal microbiota and Clostridium difficile infection. Expert Rev. Gastroenterol. Hepatol. 10, 1145–1152. doi: 10.1586/17474124.2016.1158097
Khanna, S., Shin, A., and Kelly, C. P. (2017). Management of Clostridium difficile Infection in Inflammatory Bowel Disease: Expert Review from the Clinical Practice Updates Committee of the AGA Institute. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 15, 166–174. doi: 10.1016/j.cgh.2016.10.024
Khoruts, A., Rank, K. M., Newman, K. M., Viskocil, K., Vaughn, B. P., Hamilton, M. J., et al. (2016). Inflammatory Bowel Disease Affects the Outcome of Fecal Microbiota Transplantation for Recurrent Clostridium difficile Infection. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 14, 1433–1438. doi: 10.1016/j.cgh.2016.02.018
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
Mabardy, A., McCarty, J., Hackford, A., and Dao, H. (2017). IBD: A Growing and Vulnerable Cohort of Hospitalized Patients with Clostridium difficile Infection. Am. Surg. 83, 605–609.
Mahnic, A., Breskvar, M., Dzeroski, S., Skok, P., Pintar, S., and Rupnik, M. (2020). Distinct Types of Gut Microbiota Dysbiosis in Hospitalized Gastroenterological Patients Are Disease Non-related and Characterized With the Predominance of Either Enterobacteriaceae or Enterococcus. Front. Microbiol. 11:120. doi: 10.3389/fmicb.2020.00120
Milani, C., Ticinesi, A., Gerritsen, J., Nouvenne, A., Lugli, G. A., Mancabelli, L., et al. (2016). Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: A metagenomic study. Sci. Rep. 6, 1–12. doi: 10.1038/srep25945
Missaghi, B., Barkema, H. W., Madsen, K. L., and Ghosh, S. (2014). Perturbation of the Human Microbiome as a Contributor to Inflammatory Bowel Disease. Pathogens 3, 510–527. doi: 10.3390/pathogens3030510
Ni, J., Wu, G. D., Albenberg, L., and Tomov, V. T. (2017). Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584. doi: 10.1038/nrgastro.2017.88
Parker, B. J., Wearsch, P. A., Veloo, A. C. M., and Rodriguez-Palacios, A. (2020). The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 11:906. doi: 10.3389/fimmu.2020.00906
Pike, C. M., and Theriot, C. M. (2020). Mechanisms of Colonization Resistance Against Clostridioides difficile. J. Infect. Dis. 223, S194–S200. doi: 10.1093/infdis/jiaa408
Poduval, R. D., Kamath, R. P., Corpuz, M., Norkus, E. P., and Pitchumoni, C. S. (2000). Clostridium difficile and vancomycin-resistant Enterococcus: The new nosocomial alliance. Am. J. Gastroenterol. 95, 3513–3515. doi: 10.1111/j.1572-0241.2000.03291.x
Qiu, P., Ishimoto, T., Fu, L., Zhang, J., Zhang, Z., and Liu, Y. (2022). The Gut Microbiota in Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 12:733992. doi: 10.3389/fcimb.2022.733992
Ramos-Martínez, A., Ortiz-Balbuena, J., Curto-García, I., Asensio-Vegas, Á, Martínez-Ruiz, R., Múñez-Rubio, E., et al. (2015). Risk factors for Clostridium difficile diarrhea in patients with inflammatory bowel disease. Rev. Esp. Enferm. Dig. 107, 4–8.
Razik, R., Rumman, A., Bahreini, Z., McGeer, A., and Nguyen, G. C. (2016). Recurrence of Clostridium difficile Infection in Patients with Inflammatory Bowel Disease: The RECIDIVISM Study. Am. J. Gastroenterol. 111, 1141–1146. doi: 10.1038/ajg.2016.187
Rigottier-Gois, L. (2013). Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 7, 1256–1261. doi: 10.1038/ismej.2013.80
Rinttilä, T., Kassinen, A., Malinen, E., Krogius, L., and Palva, A. (2004). Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97, 1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x
Rivera-Chávez, F., Lopez, C. A., and Bäumler, A. J. (2017). Oxygen as a driver of gut dysbiosis. Free Radic. Biol. Med. 105, 93–101. doi: 10.1016/j.freeradbiomed.2016.09.022
Rodemann, J. F., Dubberke, E. R., Reske, K. A., Seo, D. H., and Stone, C. D. (2007). Incidence of Clostridium difficile infection in inflammatory bowel disease. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 5, 339–344. doi: 10.1016/j.cgh.2006.12.027
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schnizlein, M. K., and Young, V. B. (2022). Capturing the environment of the Clostridioides difficile infection cycle. Nat. Rev. Gastroenterol. Hepatol. 19, 508–520. doi: 10.1038/s41575-022-00610-0
Schubert, A. M., Rogers, M. A. M., Ring, C., Mogle, J., Petrosino, J. P., Young, V. B., et al. (2014). Microbiome Data Distinguish Patients with Clostridium difficile Infection and Non-C. difficile-Associated Diarrhea from Healthy Controls. mBio 5, e01021–14. doi: 10.1128/mBio.01021-14
Schubert, A. M., Sinani, H., and Schloss, P. D. (2015). Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. mBio 6:e00974. doi: 10.1128/mBio.00974-15
Sehgal, K., and Khanna, S. (2021). Gut microbiome and Clostridioides difficile infection: A closer look at the microscopic interface. Ther. Adv. Gastroenterol. 14:1756284821994736. doi: 10.1177/1756284821994736
Tai, A. S., Putsathit, P., Eng, L., Imwattana, K., Collins, D. A., Mulrennan, S., et al. (2021). Clostridioides difficile colonization and infection in a cohort of Australian adults with cystic fibrosis. J. Hosp. Infect. 113, 44–51. doi: 10.1016/j.jhin.2021.03.018
van den Berg, R. J., Kuijper, E. J., van Coppenraet, L. E. S. B., and Claas, E. C. J. (2006). Rapid diagnosis of toxinogenic Clostridium difficile in faecal samples with internally controlled real-time PCR. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 12, 184–186. doi: 10.1111/j.1469-0691.2005.01301.x
Vincent, C., Miller, M. A., Edens, T. J., Mehrotra, S., Dewar, K., and Manges, A. R. (2016). Bloom and bust: Intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome 4:12. doi: 10.1186/s40168-016-0156-3
Zhang, L., Dong, D., Jiang, C., Li, Z., Wang, X., and Peng, Y. (2015). Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 34, 1–7. doi: 10.1016/j.anaerobe.2015.03.008
Keywords: inflammatory bowel disease, gut microbiota, Enterococcus, 16S amplicon sequencing, Clostridium difficile
Citation: Mahnic A, Pintar S, Skok P and Rupnik M (2022) Gut community alterations associated with Clostridioides difficile colonization in hospitalized gastroenterological patients with or without inflammatory bowel disease. Front. Microbiol. 13:988426. doi: 10.3389/fmicb.2022.988426
Received: 07 July 2022; Accepted: 17 August 2022;
Published: 06 September 2022.
Edited by:
Shan Goh, University of Hertfordshire, United KingdomReviewed by:
Michael Brouwer, Wageningen University and Research, NetherlandsMasoumeh Azimirad, Shahid Beheshti University of Medical Sciences, Iran
Copyright © 2022 Mahnic, Pintar, Skok and Rupnik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksander Mahnic, YWxla3NhbmRlci5tYWhuaWNAbmx6b2guc2k=
 Aleksander Mahnic
Aleksander Mahnic Spela Pintar
Spela Pintar Pavel Skok
Pavel Skok Maja Rupnik
Maja Rupnik