
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 August 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.988110
 Menghao Yu1,2
Menghao Yu1,2 Youfu Zhao1,3*†
Youfu Zhao1,3*†Resistance to spectinomycin emerged after widely used for treatment of gonorrhea. Previous studies revealed that Lysobacter enzymogenes strain C3 (LeC3) exhibited elevated level of intrinsic resistance to spectinomycin. In this study, we screened a Tn5 transposon mutant library of LeC3 to elucidate the underlying molecular mechanisms of spectinomycin resistance. Insertion sites in 15 out of 19 mutants recovered with decreased spectinomycin resistance were located on two ribosomal RNA operons at different loci, indicating the pivotal role of ribosomal RNAs in conferring spectinomycin resistance in L. enzymogenes. The other mutants harbored mutations in the tuf, rpoD, mltB, and purB genes. Among them, the tuf and rpoD genes, respectively, encode a translation elongation factor Tu and an RNA polymerase primary sigma factor. They both contribute to protein biosynthesis, where ribosomal RNAs play essential roles. The mltB gene, whose product is involved in cell-wall recycling, was not only associated with resistance against spectinomycin, but also conferred resistance to osmotic stress and ampicillin. In addition, mutation of the purB gene, for which its product is involved in the biosynthesis of inosine and adenosine monophosphates, led to decreased spectinomycin resistance. Addition of exogenous adenine at lower concentration in medium restored the growth deficiency in the purB mutant and increased bacterial resistance to spectinomycin. These results suggest that while cell-wall recycling and purine biosynthesis might contribute to spectinomycin resistance, target rRNAs play critical role in spectinomycin resistance in L. enzymogenes.
Spectinomycin, an aminocyclitol antibiotic originally derived from Streptomyces spectabilis, is active against a variety of gram-positive and gram-negative bacteria (Davies et al., 1965; Hyun et al., 2000; Scarff et al., 2019). Spectinomycin inhibits bacterial protein synthesis by binding to helix 34 of 16S rRNA in the 30S ribosomal subunits and interfering with tRNA translocation by the elongation factor G (EF-G; Hancock, 1981; Unemo et al., 2013). Under the tradename Trobicin, spectinomycin remains an effective option for the treatment of gonorrhea caused by Neisseria gonorrhoeae, one of the most common sexually transmitted bacterial infections (Unemo et al., 2013; Nabu et al., 2014). However, resistance to spectinomycin was subsequently emerged and reported (Boslego et al., 1987; O’connor and Dahlberg, 2002). In N. gonorrhoeae, a single C1192U conversion in helix 34 on 16S rRNA resulted in high level of resistance to spectinomycin with a minimum inhibitory concentration (MIC) over 1,024 μg/ml (Galimand et al., 2000; Unemo et al., 2009). Furthermore, a deletion of codon 27 (valine) and a K28E mutation in 30S ribosomal protein S5 led to similarly high levels of spectinomycin resistance in N. gonorrhoeae (MIC > 1,024 μg/ml; Unemo et al., 2013). In other bacterial species, enzymatical modification of spectinomycin by adenylylation also conferred antibiotic resistance. Spectinomycin-resistant bacteria produced adenyltransferases to inactivate the drug and these enzymes are encoded by the aad9, spd, and aadA14 genes, in Enterococcus faecalis, Staphylococcus aureus, and Pasteurella multocida, respectively (LeBlanc et al., 1991; Kehrenberg et al., 2005; Jamrozy et al., 2014).
Antibiotic resistance is currently a serious global threat to human, agriculture, and environment health due to the emerging, spread, and persistence of antibiotic-resistant and multidrug-resistant (MDR) bacteria (Aslam et al., 2018). As an example, delay of disease control caused by failure of antibiotic application could lead to significantly higher rates of hospital mortality (Cosgrove et al., 2003; Eliopoulos et al., 2003; Patel et al., 2008; Borer et al., 2009; Friedman et al., 2016). Over 2 million people in United States develop serious infectious diseases caused by bacterial pathogens resistant to antibiotics each year, and as a result, more than 23,000 of them die (Centers for Disease Control and Prevention, 2018). In agriculture, streptomycin was introduced into chemical control of bacterial diseases in the 1950s and proved to be one of the most effective bactericides for treatment of plant diseases caused by Xanthomonas spp., Erwinia amylovora, and Pseudomonas spp. (McManus et al., 2002). However, streptomycin control has been impeded by resistance evolved in plant pathogen populations (Cooksey, 1990). It is estimated that 16,465 kg antibiotics was applied to orchards in the United States in 2009 (Stockwell and Duffy, 2012). Moreover, contemporary animal husbandry relies heavily on antimicrobials used as therapeutics, prophylactics, and/or growth promotors (Woolhouse et al., 2015). It is estimated that food-animal production purchases approximately 80% of total antibiotics annually used in the United States (US Food and Drug Administration, 2014). Furthermore, worldwide growth in aquaculture is also accompanied by swift increase in the use of antibiotics. Similar with what happened in other agricultural field, about 80% of aquaculture-used antimicrobials enter environment in their intact forms and serve as a selector for environmental bacteria with antibiotic resistance genes (Cabello et al., 2013).
Comprehensively understanding the underlying molecular mechanisms of antibiotic resistance could help fight the growing threat of antibiotic-resistant bacteria. Previous studies have revealed that bacteria could develop antibiotic resistance during the process of antibiotic entry, accumulation, target binding, and toxicity. Genetic changes, including point mutations, recruitment of preexisting elements, and/or horizontal gene transfer (HGT), contribute to resistant development (Yelin and Kishony, 2018). Spatial exclusion of antibiotics into the cell by reduced permeability (Kato et al., 2003; Tran et al., 2013) or increased efflux (Wilson et al., 2011), and enzyme modification of drugs are among the most commonly discovered. As an example, β-lactamases cleave typical β-lactam rings and inactivate β-lactam antibiotics (Wilson et al., 2011; Yu and Zhao, 2020). Phosphotransferases, nucleotidyltransferases, and acetyltransferases modify aminoglycoside antibiotics (kanamycin, gentamycin, and streptomycin) by adding phosphoryl, adenylyl, or acetyl groups, respectively (Norris and Serpersu, 2013). In addition, target modifications, such as amino acid changes in β subunit of bacterial RNA polymerase, result in resistance to rifampicin (Goldstein, 2014). Streptomycin resistance could be induced by mutations in the rpsL (encoding the ribosomal S12 protein) or rrs gene (encoding 16S rRNA helices; Nair et al., 1993; Patel et al., 2017; Dal Molin et al., 2018). However, little is known about the antibiotic-resistant mechanisms in environmental bacteria.
Lysobacter species are ubiquitous in diverse environments, e.g., soil, rhizosphere, and freshwater (Christensen and Cook, 1978; Hayward et al., 2010). Several Lysobacter strains were found to inhibit multiple plant pathogens, thus considered as promising biocontrol agents (Giesler and Yuen, 1998; Kato et al., 1998; Chen et al., 2006; Qian et al., 2014; Yu et al., 2020). Further studies revealed that the bacterium produced a series of natural products with antimicrobial activities, including heat-stable antifungal factor (HSAF; Lou et al., 2010), alteramide B (ATB; Tang et al., 2019), WAP-8294A (Zhang et al., 2011), lysobactin (O'Sullivan et al., 1988), tripropeptins (Hashizume et al., 2001), and cephabacins (Panthee et al., 2016), making Lysobacter a potential resource for novel antibiotics. Another important characteristic of Lysobacter species is its intrinsic resistance to multiple antibiotics (Zhang et al., 2017; Yu and Zhao, 2019). Previous studies showed that Lysobacter resistomes were more abundant than that of Xanthomonas campestris, a plant pathogen in the Xanthomonadaceae family, and that Lysobacter species exhibited high level of multidrug resistance, especially to ampicillin and spectinomycin (Yu and Zhao, 2019). In Lysobacter enzymogenes, the most studied Lysobacter species; the inter-kingdom signal indole decreases its resistance against ampicillin and kanamycin by activating a dual importer that could transfer both vitamin B12 and antibiotics into bacterial cells (Wang et al., 2019). Moreover, cell permeability, -lactamase activity, and transport were involved in the high level of resistance to ampicillin in L. enzymogenes (Yu and Zhao, 2020). The purpose of this study was to identify and functionally characterize genes associated with the elevated level of spectinomycin resistance in L. enzymogenes.
The bacterial strains and plasmids used in this study are listed in Table 1. Lysobacter strains were routinely cultured in 14-ml round bottom Falcon® tubes (Corning Life Sciences) at 28°C with shaking at 250 rpm. Luria-Bertani (LB, Invitrogen, Carlsbad, CA, United States) broth was used for overnight growth and 10% tryptic soy medium (10% TSB, 1.5 g/L tryptone, 0.5 g/L soytone, and 0.5 g/L NaCl) was used for determination of antibiotic resistance. For Escherichia coli strains, LB was used for routine growth at 37°C. Antibiotics were used at the following concentrations unless otherwise noted: 100 μg/ml kanamycin (Km), 30 μg/ml gentamicin (Gm), and 10 μg/ml trimethoprim (Tmp). Primers used in this study for inverse/random amplification of transposon ends (RATE) PCR, mutant confirmation, cloning, and sequencing were listed in Supplementary Table S1.
The EZ-Tn5™ < KAN-2 > Tnp Transposome™ kit was used for random mutagenesis following the manufacturer’s instructions (Epicenter, Madison, WI, United States). Briefly, 1 μl of the transposome was electroporated into the wild-type LeC3. Transformants were plated and selected from LB amended with 2,000 μg/ml Km onto both LB amended with 2,000 g/ml Km (Plate A) and LB amended with 2,000 g/ml Km plus 1,000 g/ml spectinomycin (Spc, Plate B). Colonies grown on Plate A, but not on Plate B, were selected for second screening in 96-well plate. After 22 h of incubation, bacterial growth (OD600) was measured in 10% TSB amended with 0, 100, 200, and 400 μg/ml Spc. Since 400 μg/ml Spc has no influence on LeC3, mutants with obviously reduced growth in 400 μg/ml Spc were selected for determination of transposon insertion sites using RATE or inverse PCR as described previously (Yu and Zhao, 2020). PCR products were then gel-purified and sequenced at the University of Illinois at Urbana-Champaign core sequencing facility. Flanking sequences of the transposon insertion sites were searched using BLAST against LeC3 genome at NCBI. Different primer pairs were used to confirm the identified insertion sites (Supplementary Table S1). For complementation of mutants, the DNA fragments containing the native promoter and coding sequence of the selected genes were amplified by PCR using primer pairs for the genes (Supplementary Table S1) and cloned into pBBR1MCS-5 (Kovach et al., 1995; Yu and Zhao, 2019). The resulting plasmids were verified by sequencing and electroporated into the corresponding mutants.
Bacterial growth was measured as previously described (Lee and Zhao, 2015). Briefly, overnight cultures of bacterial strains were harvested by centrifugation at 4,000-rpm for 10 min and washed twice using 0.5 phosphate-buffered saline (0.5 × PBS). After the final wash, the pellet was resuspended in 10% TSB and adjusted to OD600 = 0.02. Bacterial strains were cultured at 28°C with 250-rpm shaking. Aliquots of cultures were taken and OD600 was measured to determine bacterial growth at different time points. The experiments were repeated three times.
Lysobacter enzymogenes strain C3 and its derived strains were grown overnight, harvested by centrifugation, and washed twice using 0.5 × PBS. After the final wash, cells were resuspended in 10% TSB medium and 10% TSB amended with antibiotics and/or adenine (adenine hemisulfate salt, Sigma-Aldrich, St. Louis, Missouri, United States). The initial concentration of bacterial suspension was adjusted to OD600 = 0.02. After incubation at 28°C with shaking at 250 rpm for 22–24 h, bacterial growth (OD600) was measured. MIC50 was defined as the concentration range of antibiotic at which bacterial growth was <50% of that of no-antibiotic control. In contrast, the initial concentration for suspension of overnight E. coli derived strains was OD600 0.05 in LB broth amended with Tmp or Tmp/Spc. After incubation at 37°C with shaking at 250 rpm for 15 h, OD600 was measured. For both LeC3- and E. coli-derived strains, relative growth was calculated as the ratio of bacterial OD600 with Spc to that without Spc and was used to represent resistance to Spc. The experiments were repeated three times.
The 16S rRNA gene fragments were PCR-amplified from E. coli strain DH10B, L. enzymogenes strain C3 (LeC3), and L. antibioticus strain ATCC29479 (LaATCC) using primer pairs (Supplementary Table S1) and cloned to replace the E. coli 16S rRNA gene in pMY205mPAG2 with TmpR (Miyazaki and Kitahara, 2018). The resulting plasmids were verified by sequencing and electroporated into the null-rrn E. coli strain MY101 with pMY101 (AmpR, resistance to ampicillin; sacB, susceptible to sucrose). Replacement of pMY101 by pMY205mPAG2-derived vectors was conducted as described previously (Miyazaki and Kitahara, 2018). Briefly, transformants after electroporation were spread on LB/Tmp agar plates. Recovered colonies with pMY205mPAG2-derived vector were resuspended in LB broth and spread on LB/Tmp agar containing 5% (w/v) sucrose (Plate C) to eliminate pMY101. The recovered colonies were selected onto both Plate C and LB agar containing 100 μg/ml ampicillin (Plate D). Colonies grown on Plate C, but not on Plate D, were E. coli strains MY101 with exogenous 16S rRNA genes successfully introduced, which were then verified using PCR.
Spot dilution assay was performed using a previously described procedure (Ge et al., 2018; Yu and Zhao, 2020). Briefly, overnight bacterial cells were harvested by centrifugation and washed twice using 0.5 × PBS. After the final wash, the pellet was resuspended in 0.5 × PBS and adjusted to OD600 = 1.0. Tenfold serial dilutions of the bacterial suspension were made in 0.5 × PBS. Each dilution (5 μl) was spotted on the plates with different concentrations of NaCl (8.85 or 250 mM) and incubated at 28°C for 3 days. The experiment was performed in duplicate and repeated three times.
Lysobacter enzymogenes strain C3 and its derived strains were tested for their abilities to cleave the chromogenic -lactamase substrate nitrocefin as previously described (Yu and Zhao, 2020). One-day bacterial cultures were centrifuged and resuspended to OD600 = 1.5 in 0.5 × PBS. Bacterial suspensions (75 μl) were transferred to duplicate wells in a 96-well plate and control wells were seeded with 0.5 × PBS only. To each well, 75 μl of a 250 μg/ml nitrocefin (Calbiochem®) solution in 0.5 PBS was added and the absorbance at a wavelength of 486 nm (A486) was measured using a microplate spectrophotometer (SpectraMax®) after 0 and 2 h of incubation at room temperature. The experiment was performed in duplicate and repeated three times.
Bacterial growth and resistance data were compared using a one-way ANOVA followed by Fisher’s Least Significant Difference (LSD) test to determine differences in means (p = 0.05) using the SAS 9.4 (Cary, NC, United States). Changes marked with the same letter did not differ significantly (p < 0.05).
In our previous study, we reported that LeC3 exhibited a high level of resistance to spectinomycin (Yu and Zhao, 2019). In order to illustrate the underlying molecular mechanisms, a mutant library of 6,985 clones was screened and 19 mutants sensitive to spectinomycin were recovered (Supplementary Table S2). The insertion sites of these 19 mutants were then determined by either RATE PCR or inverse PCR followed by sequence alignment against the complete genome of LeC3 (gene bank accession no. CP013140). Among them, 15 mutants were located on two ribosomal RNA operons at different loci, with six mutants each in the 16S and 23S rRNA genes at different sites, two mutants in the ala gene at different sites, and one mutant in the intergenic region (IGR) between the tyrS gene encoding a tyrosyl-tRNA synthetase and the 16S rRNA (Figure 1). Other mutations were in genes encoding a translation elongation factor Tu (tuf), an RNA polymerase primary sigma factor (rpoD), a lytic murein transglycosylase B (mltB), and an adenylosuccinate lyase (purB).
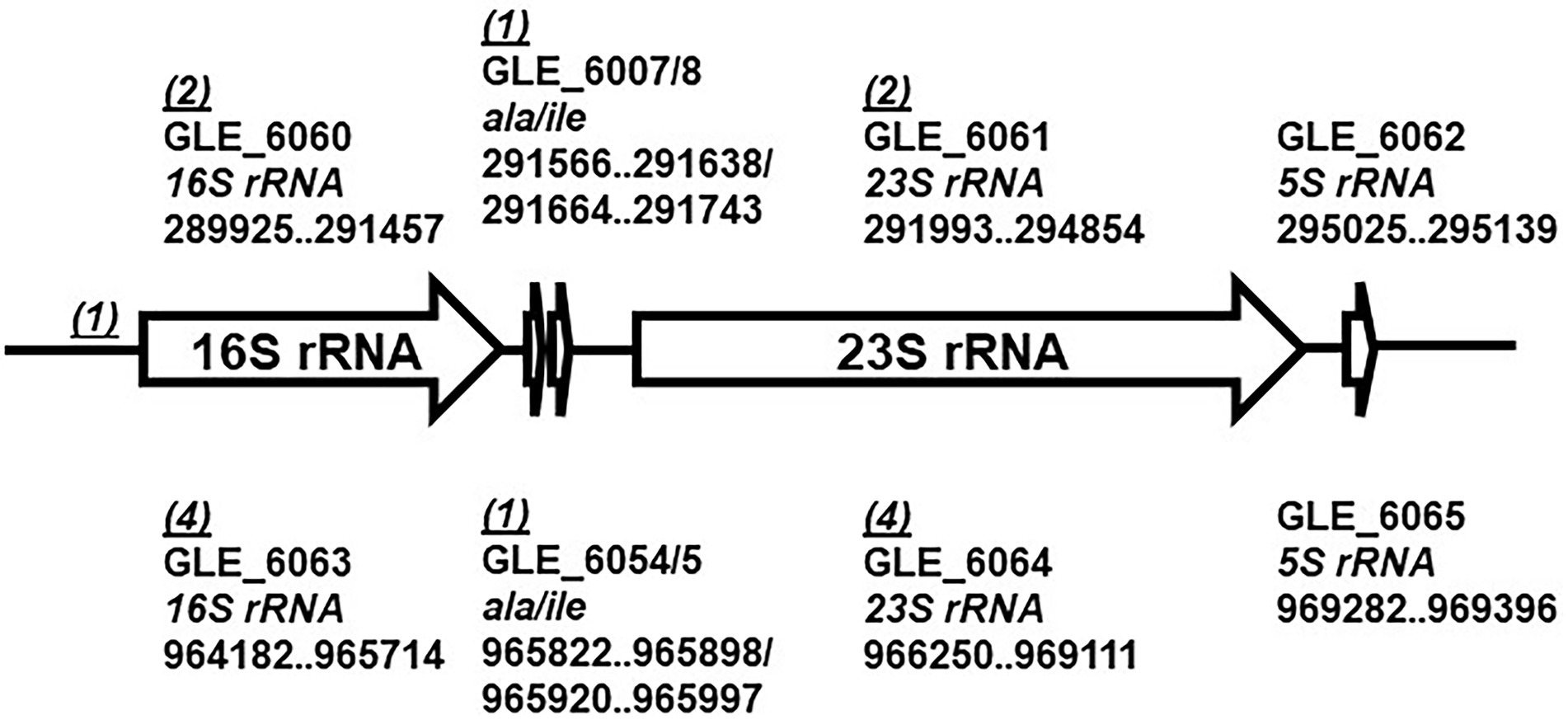
Figure 1. Schematic map of the two rRNA operons in Lysobacter enzymogenes. Each ribosomal RNA (rrn) operon contains five genes, encoding 16S rRNA (16S rRNA), tRNA-Ala (ala), tRNA-Ile (ile), 23S rRNA (23S rRNA), and 5S rRNA (5S rRNA). Numbers in parentheses indicate numbers of mutants identified with insertions in the gene or the intergenic region (IGR). The genome sequence accession number for L. enzymogenes C3 is CP013140.
To further determine spectinomycin susceptibility for the mutants, the growth of four mutants, including one each of the 16S rRNA, 23S rRNA, rpoD, and mltB mutants, was determined in 10% TSB. The 16S rRNA, 23S rRNA, and rpoD mutants grew slower in early hours than that of LeC3, but reached similar level as LeC3 at 24 h post-inoculation (hpi); whereas the mltB mutant exhibited about 30% decreased growth at 24 hpi (Figure 2A). Due to delayed growth, relative growth was calculated to reflect spectinomycin susceptibility and normalized to OD600 of 1. Results showed that the four selected mutants exhibited different levels of spectinomycin susceptibility as compared with LeC3 (Figure 2B). The mltB mutant was highly susceptible to spectinomycin and the cloned mltB gene partially complemented the mltB mutant in both spectinomycin susceptibility and growth (Figures 2A,B). However, there was no significant difference between the spectinomycin susceptibility of the 16S and rpoD mutants.
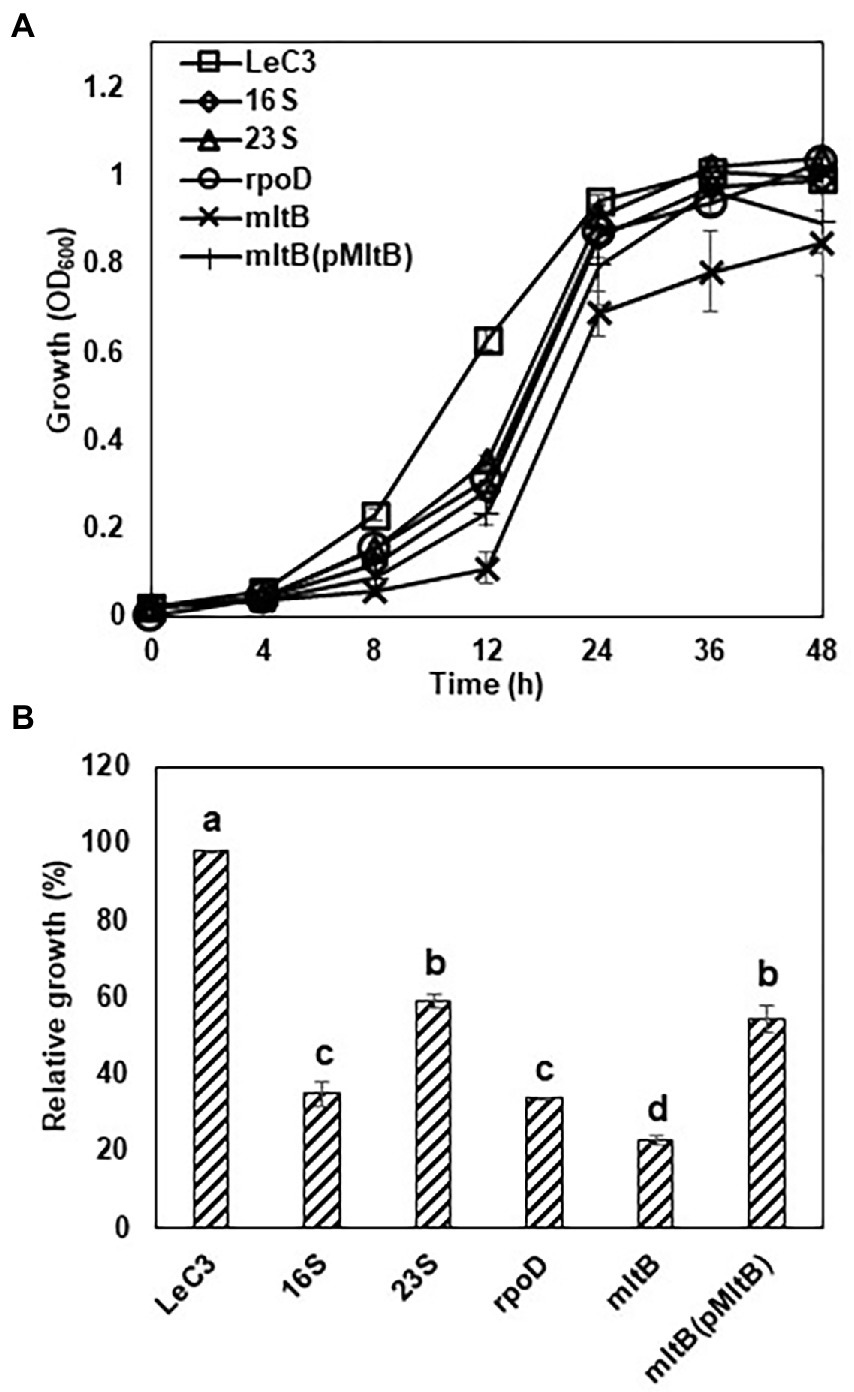
Figure 2. Selected mutants exhibited decreased spectinomycin resistance and slower growth. (A) Growth curve of Lysobacter enzymogenes strain C3 (LeC3) and its derived strains. Bacterial growth was monitored at different time points by measuring OD600. Data points represented the means of three replicates SD. (B) Spectinomycin resistance of LeC3 and its derived strains. Spectinomycin resistance was calculated as the ratio of bacterial growth with 400 μg/ml spectinomycin to bacterial growth without spectinomycin at 22 h post-inoculation. Error bars represented SD. Different letters indicated significant differences from one-way ANOVA followed by Fisher’s LSD test (p < 0.05). LeC3, Lysobacter enzymogenes strain C3; 16S, the 16S rRNA mutant; 23S, the 23S rRNA mutant; rpoD, the rpoD mutant; mltB, the mltB mutant; and LaATCC, L. antibioticus strain ATCC29479.
In addition, the minimum inhibitory concentration 50 (MIC50) for the 16S rRNA, 23S rRNA, rpoD, and mltB mutants was also determined. As compared with LeC3 with an MIC50 at 1,500–2,000 g/ml, the MIC50 for the selected mutants (16S rRNA, 23S rRNA, rpoD, and mltB) were 200–300, 800–1,000, 100–200, and <100 g/ml, respectively (Table 2). MIC50 for the complementation strain of the mltB mutant was partially restored (200–500 g/ml, Table 2).
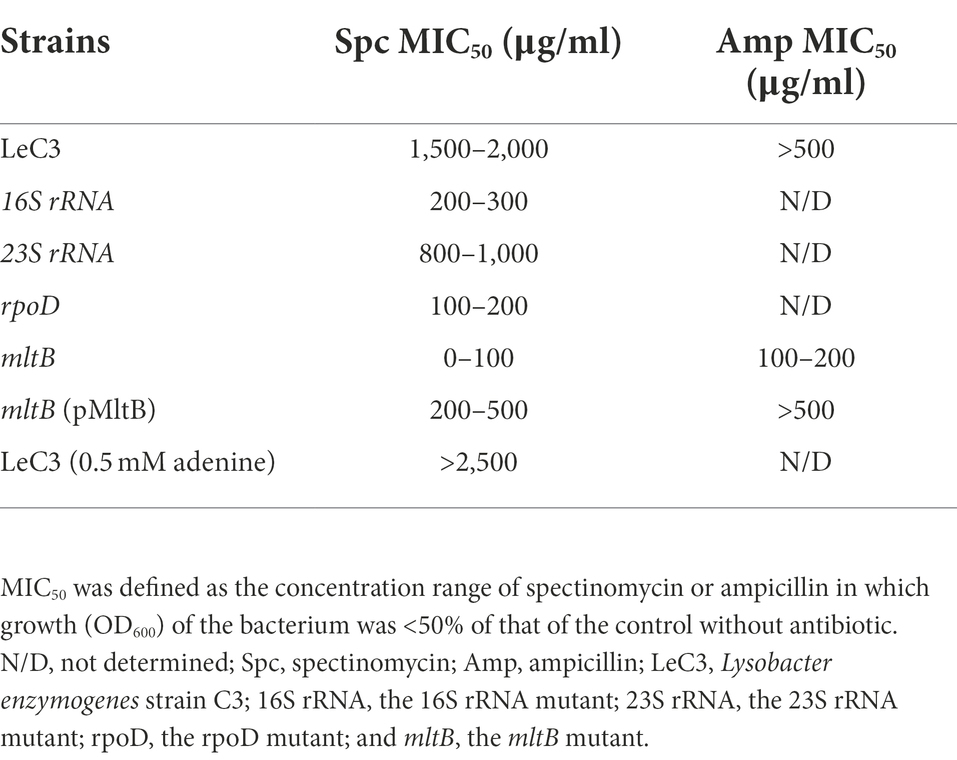
Table 2. Minimum inhibition concentration 50 (MIC50) of LeC3 and its derived strains for spectinomycin and ampicillin.
Lysobacter enzymogenes contains two identical ribosomal RNA (rrn) operons, each with five genes, encoding 16S rRNA (16S rRNA), tRNA-Ala (ala), tRNA-Ile (ile), 23S rRNA (23S rRNA), and 5S rRNA (5S rRNA; de Bruijn et al., 2015; Figure 1). Among the 19 mutants with decreased spectinomycin resistance in LeC3 (Supplementary Table S2), five and nine mutants harbored insertions in the rrn operon 1 (289,925–29,513) and 2 (964,182–969,396), respectively (Figure 1). These results suggested that mutations of either 16S rRNA, 23S rRNA, or ala genes on either of the two rrn operons in LeC3 led to increased spectinomycin susceptibility, indicating that the rrn operons act as the main targets conferring resistance to spectinomycin in LeC3.
Since spectinomycin binds 16S rRNA component of the 30S subunits, we utilized an E. coli null mutant of the rrn operons in the genome to examine whether 16S rRNA from LeC3 confers resistance to spectinomycin in E. coli. Previous studies also showed that LeC3 displayed higher level of resistance to spectinomycin as compared with that of L. antibioticus ATCC29479 (LaATCC29479; Yu and Zhao, 2019). Therefore, we also compared wild-type 16S rRNA from LeC3 with that of LaATCC29479. First, full-length 16S rRNA genes individually replaced an E. coli 16S rRNA gene in pMY205mPAG2, which originally contained a complete E. coli rrnB operon. The resulting plasmids were then individually introduced into an E. coli null mutant of the rrn operons in the genome (strain MY101; Miyazaki and Kitahara, 2018). Results showed that MY101 strain harboring the 16S rRNA from LeC3 exhibited relative faster growth as compared with those containing the 16S rRNA from E. coli or LaATCC29479 with spectinomycin resistance at 64 g/ml (Figure 3), suggesting that 16S rRNA of LeC3, but not LaATCC29479, conferred resistance to spectinomycin in null-rrn E. coli strain MY101.
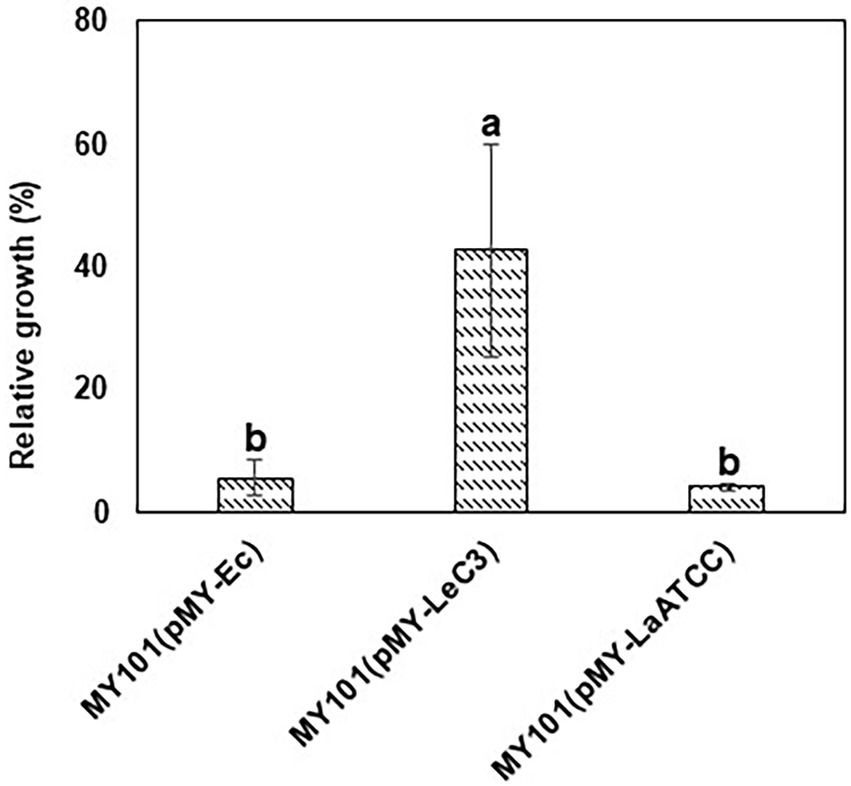
Figure 3. 16S rRNA from LeC3 conferred resistance to spectinomycin (Spc) as compared with 16S rRNA from LaATCC29479 in Escherichia coli strain MY101. Relative growth was calculated as the ratio of growth (OD600) with 64 g/ml Spc to bacterial growth without Spc 24 h after inoculation. Error bars represented SD. Different letters indicated significant differences from one-way ANOVA followed by Fisher’s LSD test (p < 0.05).
As the mltB gene encodes one of the major cell-wall recycling enzymes, i.e., the lytic murein transglycosylase B (MltB), we hypothesized that growth deficiency and decreased resistance to spectinomycin in the mltB mutant might be associated with its increased susceptibility to osmotic shock. Spot dilution assay revealed that the mltB mutant showed similar growth with LeC3 on LB plates with lower concentration of NaCl (8.85 mM). The mltB mutant was more susceptible to osmotic stress (three dilutions lower on LB plates containing 250 mM NaCl) as compared with LeC3 and the complementation strain was partially restored (Figure 4A). These results suggest that the mltB gene might be involved in osmotic response in L. enzymogenes.
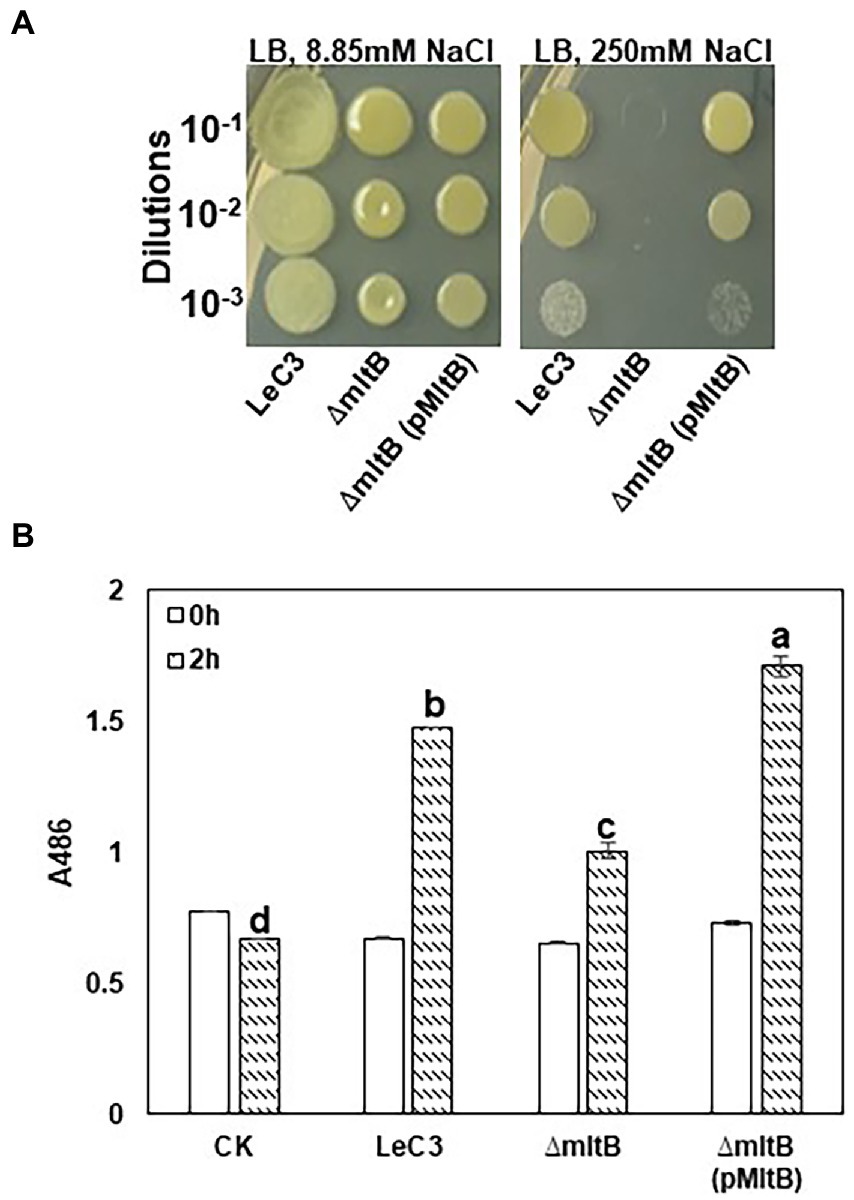
Figure 4. The mltB gene controls osmotic stress response and -lactamase activity. (A) The mltB mutant showed decreased resistance to osmotic stress. Serial 10-fold dilutions were made from OD600 = 0.1 in 0.5 PBS. Each dilution was added to LB plates containing 8.85 mM NaCl (left panel) and 250 mM NaCl (right panel). Photographs were taken 3 days post-inoculation. (B) Quantitative measurements of -lactamase activity in LeC3 and its derived strains. The -lactamase activity was measured at 486 nm (A486) using the chromogenic compound nitrocefin at 2 h post-inoculation. Error bars represented SD. Different letters indicated significant differences from one-way ANOVA followed by Fisher’s LSD test (p < 0.05). LeC3, Lysobacter enzymogenes strain C3; mltB, the mltB mutant; and CK, 0.5 × phosphate-buffered saline.
Ampicillin is an antibiotic that targets cell-wall biosynthesis and previous report showed that LeC3 exhibited a high level of resistance to ampicillin (Yu and Zhao, 2019, 2020). To explore whether the mltB gene is also involved in ampicillin resistance, MIC50 of LeC3, the mltB mutant, and its complementation strain was determined. As compared with LeC3 with an MIC50 > 500 g/ml, the MIC50 for the mltB mutant was 100–200 g/ml and the mutant was partially complemented (Table 2). The -lactamase activity in the mltB mutant was significantly lower as compared with that of LeC3 and the complementation strain (Figure 4B), suggesting that the difference of LeC3 and the mltB mutant in ampicillin resistance might be partially due to their difference in -lactamase activities.
One recovered mutant (Supplementary Table S2) harbored a mutation in the purB gene, which encodes an adenylosuccinate lyase and is involved in purine biosynthesis. However, severe deficiency in growth was observed in the purB mutant in 10% TSB at 24 hpi as compared with LeC3 (Figure 5A). This severe deficiency of growth in the purB mutant was partially restored by either addition of 0.5–1 mM adenine to the medium or introduction of the purB gene (Figure 5A). No significant growth difference was observed for the complementation strain in 10% TSB or by adding 0.5–2 mM adenine, but not 5 mM adenine, which inhibited its growth (Figure 5A). In addition, higher concentration of adenine (2 or 5 mM) also led to inhibition of bacterial growth in both LeC3 and the mutant (Figure 5A). Therefore, 0.5 mM adenine was used for spectinomycin susceptibility tests. Results showed that with and without 0.5 mM adenine, the purB mutant exhibited significantly lower level of resistance to spectinomycin as compared with LeC3 and the complementation strain (Figure 5B). Interestingly, addition of 0.5 mM adenine to the medium increased bacterial resistance to spectinomycin in all three strains tested (Figure 5B; Table 2), further suggesting that purine biosynthesis might contribute to spectinomycin resistance in LeC3 as well.
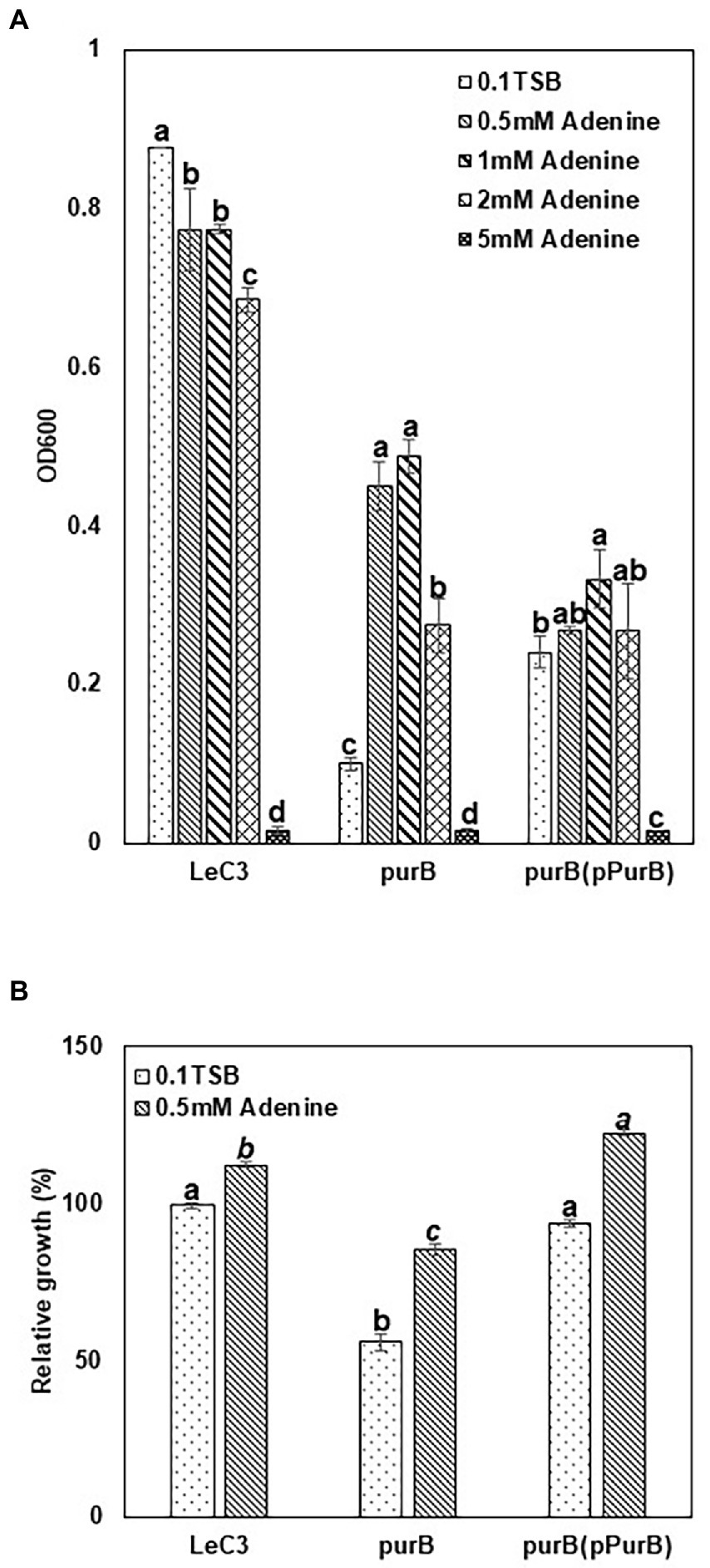
Figure 5. Purine biosynthesis contributed to resistance to spectinomycin in LeC3. (A) Growth of LeC3 and its derived strains on different adenine concentration at 24 hpi. The absorbance at 600 nm was measured to represent bacterial growth. Within each strain, different letters indicated significant differences among different concentrations of adenine from one-way ANOVA followed by Fisher’s LSD test (p < 0.05). In addition, “ab” means significant difference with “c,” but not with “a” or “b.” (B) Spectinomycin resistance of LeC3 and its derived strains with and without adenine at 24 hpi. Spectinomycin resistance was calculated as the ratio of bacterial growth with 500 μg/ml spectinomycin to bacterial growth without spectinomycin. Within each concentration of adenine, different letters indicated significant differences among different bacterial strains from one-way ANOVA followed by Fisher’s LSD test (p < 0.05). Error bars represented SD. LeC3, Lysobacter enzymogenes strain C3; purB, the purB mutant.
There is a critical need in comprehensively understanding the underlying molecular mechanisms of antibiotic resistance in environmental microorganisms. L. enzymogenes, with an elevated level of intrinsic MDR, is an appropriate surrogate for studies in antibiotic resistance and in understanding how to circumvent the antibiotic crisis in clinical pathogens. We previously reported that L. enzymogenes exhibited a high level of resistance to spectinomycin (Yu and Zhao, 2019). In this study, we identified 19 mutants in L. enzymogenes with decreased resistance to spectinomycin and demonstrated that spectinomycin susceptibility in these mutants is mostly due to changes of its ribosomal RNA targets, and also related to cell-wall recycling and purine biosynthesis.
Bacteriostatic spectinomycin interferes with bacterial growth via binding to the 30S ribosomal subunits to inhibit protein synthesis (Hancock, 1981). Among the 19 mutants with decreased spectinomycin resistance, 16 were involved in bacterial protein synthesis, including six each of 16S rRNA or 23S rRNA, two alanine tRNA, and the tuf gene, encoding a translation elongation factor Tu, which promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis (Daviter et al., 2003). Previous proteomic analysis of a N. gonorrhoeae strain with high resistance to spectinomycin revealed that the tuf gene was highly expressed in the resistant strain. It is possible that high expression of Tuf might balance and maintain protein biosynthesis in response to inhibition by spectinomycin in the resistant strain (Nabu et al., 2014). This might also be true in L. enzymogenes, which needs further investigation. On the other hand, RpoD, as the house-keeping sigma factor, is required in the initiation step of transcription by specifically recognizing promoter sequences of bacterial essential genes, i.e., ribosomal genes and protein synthesis (Nagai and Shimamoto, 1997; Davis et al., 2017). Therefore, the rpoD mutant exhibited spectinomycin susceptibility probably due to its indirect involvement in protein biosynthesis by affecting expression of ribosome genes.
Previous studies revealed that the direct binding site of spectinomycin was at the helix 34 of E. coli 16S rRNAs between positions 1,046–1,065 and 1,191–1,211(Miyazaki and Kitahara, 2018). We searched mutations in 16S rRNA genes from organisms with altered spectinomycin resistance and found that hot spots where spectinomycin resistance mutations frequently discovered were around helix 34 and its neighboring positions (Supplementary Table S3). As an example, mutations C1066U and C1192U conferred spectinomycin resistance in E. coli (Sigmund et al., 1984; Johanson and Hughes, 1995). Additionally, mutations A1191G, C1192U, C1192G, and G1193C were associated with spectinomycin resistance in Borrelia spp. (Binet and Maurelli, 2005; Criswell et al., 2006). However, listing all point mutations associated with antibiotic resistance in rRNA (rrn) operons is challenging as multiple rrn operons normally exist in bacterial genomes, e.g., seven in E. coli and two in L. enzymogenes. Miyazaki and colleague developed a novel approach to circumvent this methodological problem by using a null-rrn mutant of E. coli as a test host to detect exogenous 16S rRNAs for altered spectinomycin resistance (Miyazaki and Kitahara, 2018). In this study, 16S rRNAs from Lysobacter species were expressed in null-rrn mutants of E. coli and results showed that 16S rRNA from LeC3 conferred spectinomycin resistance. However, sequence analysis of 16S rRNA genes revealed no difference in the two hot spots (Figure 6A; Supplementary Table S3), where spectinomycin resistance mutations have already been discovered (Supplementary Table S3), indicating that unknown mutations associated with spectinomycin resistance might exist in the 16S rRNA of L. enzymogenes. Sequence alignment (Supplementary Figure S1) revealed that point mutations between the 16S rRNAs of LeC3 and LaATCC29479 were in six different regions, and these included helix 10 (E. coli numbering, 198–219, A199U, UUCG208-211GCAA, and U218A), helix 17 (455–477, C469A, and C475A), helix 22 (666–672 & 734–740, U672G), helix 26 (829–854, U843C, and U854C), helix 32 (984–990, 1,215–1,221, U989C, and A1216G), and helix 33 (996–1,046, CCACG998-1002GUCGA, and CGUGG1038-1043UCGAC), leading to different secondary structures (Figure 6B; Supplementary Figure S2). It is also possible that these mutations might be involved in their difference in conferring spectinomycin resistance, which requires further investigation. In addition, previous studies showed that the difference of LeC3 and LaATCC29479 in resistance to multiple antibiotics including spectinomycin was partly due to differences in cellular permeability (Yu and Zhao, 2020).
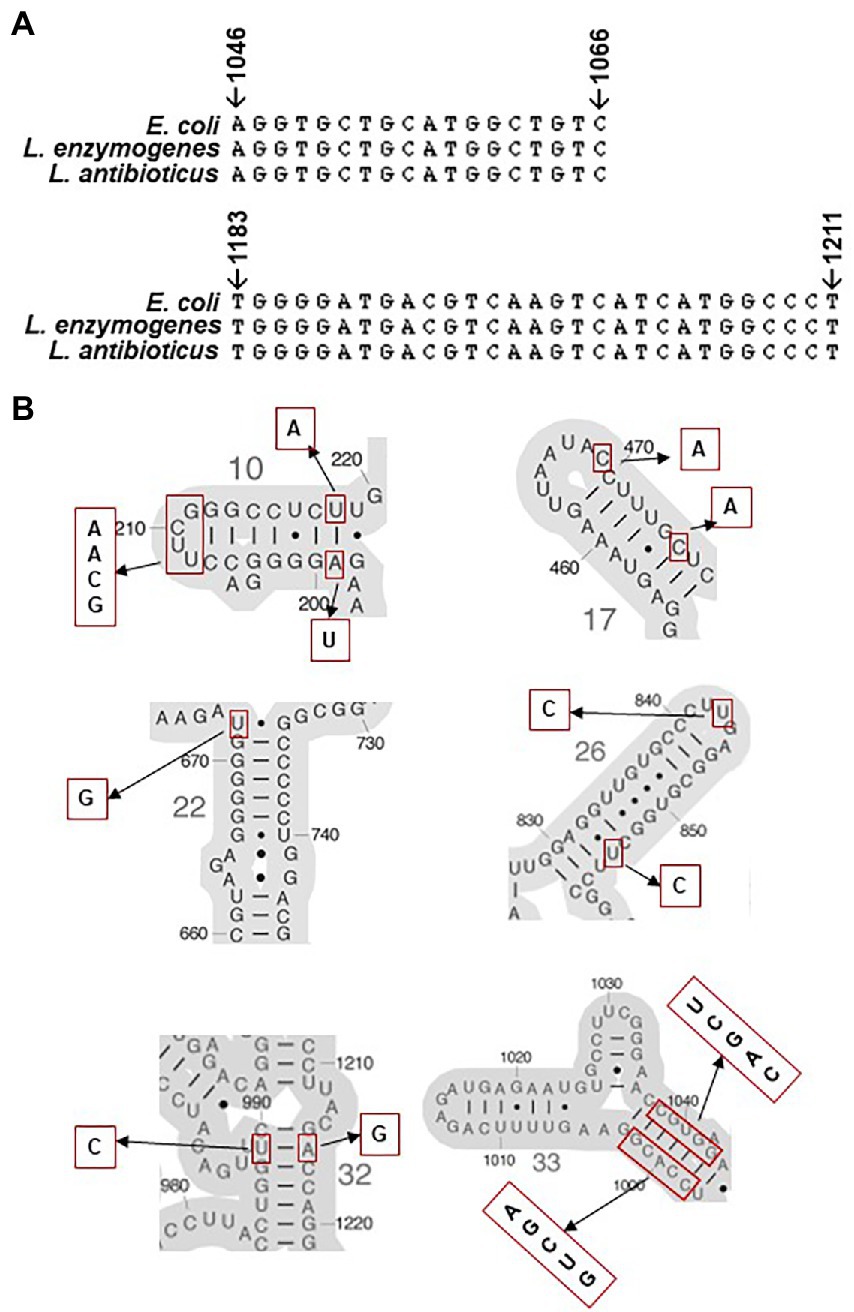
Figure 6. Comparison of 16S rRNA. (A) Multiple sequence alignment of hot spots in 16S rRNA genes from Escherichia coli, Lysobacter enzymogenes, and Lysobacter antibioticus with known spectinomycin resistance mutations. (B) Secondary structures of E. coli 16S rRNA around point mutation sites identified between L. enzymogenes and L. antibioticus. Putative resistance nucleotides to spectinomycin are marked using red rectangular and the corresponding sequences in LeC3 are shown on the side.
Cell walls are critical for the viability of the bacterium (Dik et al., 2018). Cell-wall remodeling is tightly regulated to guarantee bacterial survival and could also be directly associated with antibiotic resistance (Domínguez-Gil et al., 2016). In Gram-negative bacteria, cell-wall recycling starts with the lytic transglycosylases (LTs) by degrading peptidoglycan (PG), the major component of bacterial cell walls (Suvorov et al., 2008; Dik et al., 2018). Loss of LTs increased bacterial resistance to -lactams in Shewanella oneidensis, but led to higher -lactam susceptibility in Pseudomonas aeruginosa (Lamers et al., 2015; Yin et al., 2018). These findings suggest that PG metabolism could influence bacterial resistance. It has been reported that LT inactivation has the potential to reduce bacterial autolysis and increase survival time, thus antibiotic resistance (Lamers et al., 2015). On the other hand, inactivation of LTs could reduce the production of 1,6-anhydromuropeptides (anhMPs) from PG degradation, which binds to transcriptional activator AmpR, thus preventing the induction of -lactamase expression (Jacobs et al., 1994; Mark et al., 2011). Decreased expression of -lactamase consequently leads to -lactam susceptibility. In this study, the mltB mutant in L. enzymogenes exhibited susceptibility to -lactam ampicillin and showed decreased -lactamase activity, suggesting that the mltB gene could influence PG metabolism and affect -lactamase expression. Further studies are needed to confirm this hypothesis. In addition, AmpR is also associated with bacterial resistance to quinolones and aminoglycosides in P. aeruginosa by regulating MexEF-OprN efflux pump (Kumari et al., 2014). It is also reasonable to speculate that the mltB mutant in LeC3 exhibited ampicillin and spectinomycin susceptibility by possibly affecting AmpR activity in regulation of efflux pumps.
The adenylosuccinate lyase (ASL) encoded by the purB gene is involved in the de novo pathway of inosine monophosphate (IMP) and adenosine monophosphate (AMP) biosynthesis (Yang et al., 2021). Deletion of the purB gene resulted in severe growth deficiency in L. enzymogenes, probably caused by disrupted purine biosynthesis as reported previously in other bacteria (Jung et al., 2010; Faith et al., 2012). Consistent with the purB mutant of Mesorhizobium loti, 0.5 mM exogeneous adenine in medium partially restored its growth deficiency in the purB mutant of L. enzymogenes (Okazaki et al., 2007). We also found that higher concentration of adenine led to obvious inhibition of bacterial growth in L. enzymogenes and its derived strains. This bacteriostatic effect of adenine was consistent with what was previously described in Aerobacter aerogenes (Brooke and Magasanik, 1954; Moyed, 1964). Hosono and colleagues reported that adenine interfered bacterial growth by inhibiting de novo synthesis of pyrimidine nucleotides (Hosono and Kuno, 1974). The purB mutant with deficiency in purine biosynthesis exhibited decreased spectinomycin resistance. This is consistent with the genetic determination of oxacillin resistance in methicillin-resistant Staphylococcus aureus (MRSA), where the purB gene was identified as one of the genes linked to oxacillin resistance (Pardos de la Gandara et al., 2018). However, we found exogenous adenine could increase antibiotic resistance, a phenomenon not previously reported in bacteria. Further studies are needed to explain this phenomenon.
Taken together, our findings demonstrated that, ribosomal RNAs, the direct target of spectinomycin, play a pivotal role in conferring spectinomycin resistance in L. enzymogenes. Cell-wall recycling and purine biosynthesis might also play some roles in conferring spectinomycin resistance. However, further studies are needed to clarify which bases (or point mutations) in 16S rRNA lead to resistance to spectinomycin in LeC3. Further studies should also determine whether the mutation in the mltB gene resulted in decreased efflux pumps mediated by the transcriptional activator AmpR and how exogenous adenine or the purB mutation affects antibiotic resistance in LeC3.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
MY and YZ designed the research and wrote the manuscript. MY performed the research and analyzed the data. All authors contributed to the article and approved the submitted version.
This project was supported by the Agriculture and Food Research Initiative Competitive grants Program grant no. 2016-67013-24812 from the USDA National Institute of Food and Agriculture, startup and endowed funds (YZ).
We would like to thank Kentaro Miyazaki from National Institute of Advanced Industrial Science and Technology (AIST) in Japan for providing the strain MY101 and the plasmid pMY205mPAG2.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.988110/full#supplementary-material
Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Muzammil, S., Rasool, M. H., et al. (2018). Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 11, 1645–1658. doi: 10.2147/IDR.S173867
Binet, R., and Maurelli, A. T. (2005). Frequency of spontaneous mutations that confer antibiotic resistance in chlamydia spp. Antimicrob. Agents Chemother. 49, 2865–2873. doi: 10.1128/AAC.49.7.2865-2873.2005
Borer, A., Saidel-Odes, L., Riesenberg, K., Eskira, S., Peled, N., Nativ, R., et al. (2009). Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect. Control Hosp. Epidemiol. 30, 972–976. doi: 10.1086/605922
Boslego, J. W., Tramont, E. C., Takafuji, E. T., Diniega, B. M., Mitchell, B. S., Small, J. W., et al. (1987). Effect of spectinomycin use on the prevalence of spectinomycin-resistant and of penicillinase-producing Neisseria gonorrhoeae. N. Engl. J. Med. 317, 272–278. doi: 10.1056/NEJM198707303170504
Brooke, M. S., and Magasanik, B. (1954). The metabolism of purines in Aerobacter aerogenes: a study of purineless mutants. J. Bacteriol. 68, 727–733. doi: 10.1128/jb.68.6.727-733.1954
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dölz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Centers for Disease Control and Prevention (2018). Antibiotic use in the United States: progress and opportunities, 2017.
Chen, J., Moore, W., Yuen, G., Kobayashi, D., and Caswell-Chen, E. (2006). Influence of Lysobacter enzymogenes strain C3 on nematodes. J. Nematol. 38, 233–239.
Christensen, P., and Cook, F. D. (1978). Lysobacter, a new genus of non-fruiting, gliding bacteria with a high base ratio. Int. J. Syst. Bacteriol. 28, 367–393. doi: 10.1099/00207713-28-3-367
Cooksey, D. A. (1990). Genetics of bactericide resistance in plant pathogenic bacteria. Annu. Rev. Phytopathol. 28, 201–219. doi: 10.1146/annurev.py.28.090190.001221
Cosgrove, S. E., Sakoulas, G., Perencevich, E. N., Schwaber, M. J., Karchmer, A. W., and Carmeli, Y. (2003). Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36, 53–59. doi: 10.1086/345476
Criswell, D., Tobiason, V. L., Lodmell, J. S., and Samuels, D. S. (2006). Mutations conferring aminoglycoside and spectinomycin resistance in Borrelia burgdorferi. Antimicrob. Agents Chemother. 50, 445–452. doi: 10.1128/AAC.50.2.445-452.2006
Dal Molin, M., Gut, M., Rominski, A., Haldimann, K., Becker, K., and Sander, P. (2018). Molecular mechanisms of intrinsic streptomycin resistance in Mycobacterium abscessus. Antimicrob. Agents Chemother. 62, e01427–e01517. doi: 10.1128/AAC.01427-17
Davies, J., Anderson, P., and Davis, B. D. (1965). Inhibition of protein synthesis by spectinomycin. Science 149, 1096–1098. doi: 10.1126/science.149.3688.1096
Davis, M. C., Kesthely, C. A., Franklin, E. A., and MacLellan, S. R. (2017). The essential activities of the bacterial sigma factor. Can. J. Microbiol. 63, 89–99. doi: 10.1139/cjm-2016-0576
Daviter, T., Wieden, H. J., and Rodnina, M. V. (2003). Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J. Mol. Biol. 332, 689–699. doi: 10.1016/S0022-2836(03)00947-1
de Bruijn, I., Cheng, X., de Jager, V., Expósito, R. G., Watrous, J., Patel, N., et al. (2015). Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics 16:991. doi: 10.1186/s12864-015-2191-z
Dik, D. A., Fisher, J. F., and Mobashery, S. (2018). Cell-wall recycling of the gram-negative bacteria and the nexus to antibiotic resistance. Chem. Rev. 118, 5952–5984. doi: 10.1021/acs.chemrev.8b00277
Domínguez-Gil, T., Molina, R., Alcorlo, M., and Hermoso, J. A. (2016). Renew or die: the molecular mechanisms of peptidoglycan recycling and antibiotic resistance in gram-negative pathogens. Drug Resist. Updat. 28, 91–104. doi: 10.1016/j.drup.2016.07.002
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eliopoulos, G. M., Cosgrove, S. E., and Carmeli, Y. (2003). The impact of antimicrobial resistance on health and economic outcomes. Clin. Infect. Dis. 36, 1433–1437. doi: 10.1086/375081
Faith, N. G., Kim, J. W., Azizoglu, R., Kathariou, S., and Czuprynski, C. (2012). Purine biosynthesis mutants (purA and purB) of serotype 4b listeria monocytogenes are severely attenuated for systemic infection in intragastrically inoculated A/J mice. Foodborne Pathog. Dis. 9, 480–486. doi: 10.1089/fpd.2011.1013
Friedman, N. D., Temkin, E., and Carmeli, Y. (2016). The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 22, 416–422. doi: 10.1016/j.cmi.2015.12.002
Galimand, M., Gerbaud, G., and Courvalin, P. (2000). Spectinomycin resistance in Neisseria spp. due to mutations in 16S rRNA. Antimicrob. Agents Chemother. 44, 1365–1366. doi: 10.1128/AAC.44.5.1365-1366.2000
Ge, Y., Lee, J. H., Hu, B., and Zhao, Y. F. (2018). Loss-of-function mutations in the Dpp and Opp permeases render Erwinia amylovora resistant to kasugamycin and blasticidin S. Mol. Plant-Microbe Interact. 31, 823–832. doi: 10.1094/MPMI-01-18-0007-R
Giesler, L. J., and Yuen, G. Y. (1998). Evaluation of Stenotrophomonas maltophilia strain C3 for biocontrol of brown patch disease. Crop Prot. 17, 509–513. doi: 10.1016/S0261-2194(98)00049-0
Goldstein, B. P. (2014). Resistance to rifampicin: A review. J. Antibiot. 67, 625–630. doi: 10.1038/ja.2014.107
Hancock, R. E. (1981). Aminoglycoside uptake and mode of action with special reference to streptomycin and gentamicin: I antagonists and mutants. J. Antimicrob. Chemother. 8, 249–276. doi: 10.1093/jac/8.4.249
Hashizume, H., Igarashi, M., Hattori, S., Hori, M., Hamada, M., and Takeuchi, T. (2001). Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. J. Antibiot. 54, 1054–1059. doi: 10.7164/antibiotics.54.1054
Hayward, A. C., Fegan, N., Fegan, M., and Stirling, G. (2010). Stenotrophomonas and Lysobacter: ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J. Appl. Microbiol. 108, 756–770. doi: 10.1111/j.1365-2672.2009.04471.x
Hosono, R., and Kuno, S. (1974). Mechanism of inhibition of bacterial growth by adenine. J. Biochem. 75, 215–220. doi: 10.1093/oxfordjournals.jbchem.a130388
Hyun, C. G., Kim, S. S., Sohng, J. K., Hahn, J. J., Kim, J. W., and Suh, J. W. (2000). An efficient approach for cloning the dNDP-glucose synthase gene from actinomycetes and its application in Streptomyces spectabilis, a spectinomycin producer. FEMS Microbiol. Lett. 183, 183–189. doi: 10.1111/j.1574-6968.2000.tb08955.x
Jacobs, C., Huang, L., Bartowsky, E., Normark, S., and Park, J. (1994). Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13, 4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x
Jamrozy, D., Coldham, N., Butaye, P., and Fielder, M. (2014). Identification of a novel plasmid-associated spectinomycin adenyltransferase gene spd in methicillin-resistant Staphylococcus aureus ST398 isolated from animal and human sources. J. Antimicrob. Chemother. 69, 1193–1196. doi: 10.1093/jac/dkt510
Johanson, U., and Hughes, D. (1995). A new mutation in 16S rRNA of Escherichia coli conferring spectinomycin resistance. Nucleic Acids Res. 23, 464–466. doi: 10.1093/nar/23.3.464
Jung, S.-C., Smith, C. L., Lee, K. S., Hong, M. E., Kweon, D. H., Stephanopoulos, G., et al. (2010). Restoration of growth phenotypes of Escherichia coli DH5α in minimal media through reversal of a point mutation in purB. Appl. Environ. Microbiol. 76, 6307–6309. doi: 10.1128/AEM.01210-10
Kato, A., Latifi, T., and Groisman, E. A. (2003). Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc. Natl. Acad. Sci. 100, 4706–4711. doi: 10.1073/pnas.0836837100
Kato, A., Nakaya, S., Kokubo, N., Aiba, Y., Ohashi, Y., Hirata, H., et al. (1998). A new anti-MRSA antibiotic complex, WAP-8294A. J. Antibiot. 51, 929–935. doi: 10.7164/antibiotics.51.929
Kehrenberg, C., Catry, B., Haesebrouck, F., de Kruif, A., and Schwarz, S. (2005). Novel spectinomycin/streptomycin resistance gene, aadA14, from Pasteurella multocida. Antimicrob. Agents Chemother. 49, 3046–3049. doi: 10.1128/AAC.49.7.3046-3049.2005
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kumari, H., Balasubramanian, D., Zincke, D., and Mathee, K. (2014). Role of Pseudomonas aeruginosa AmpR on β-lactam and non-β-lactam transient cross-resistance upon pre-exposure to subinhibitory concentrations of antibiotics. J. Med. Microbiol. 63, 544–555. doi: 10.1099/jmm.0.070185-0
Lamers, R. P., Nguyen, U. T., Nguyen, Y., Buensuceso, R. N., and Burrows, L. L. (2015). Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. Microbiology 4, 879–895. doi: 10.1002/mbo3.286
LeBlanc, D., Lee, L., and Inamine, J. (1991). Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD (9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35, 1804–1810. doi: 10.1128/AAC.35.9.1804
Lee, J. H., and Zhao, Y. (2015). Integration host factor is required for RpoN-dependent hrpL gene expression and controls motility by positively regulating rsmB sRNA in Erwinia amylovora. Phytopathology 106, 29–36. doi: 10.1094/PHYTO-07-15-0170-R
Lou, L., Qian, G., Xie, Y., Hang, J., Chen, H., Zaleta-Rivera, K., et al. (2010). Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J. Am. Chem. Soc. 133, 643–645. doi: 10.1021/ja105732c
Mark, B. L., Vocadlo, D. J., and Oliver, A. (2011). Providing β-lactams a helping hand: targeting the AmpC β-lactamase induction pathway. Future Microbiol. 6, 1415–1427. doi: 10.2217/fmb.11.128
McManus, P. S., Stockwell, V. O., Sundin, G. W., and Jones, A. L. (2002). Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40, 443–465. doi: 10.1146/annurev.phyto.40.120301.093927
Miyazaki, K., and Kitahara, K. (2018). Functional metagenomic approach to identify overlooked antibiotic resistance mutations in bacterial rRNA. Sci. Rep. 8:5179. doi: 10.1038/s41598-018-23474-4
Moyed, H. (1964). Inhibition of the biosynthesis of the pyrimidine portion of thiamine by adenosine. J. Bacteriol. 88, 1024–1029. doi: 10.1128/jb.88.4.1024-1029.1964
Nabu, S., Lawung, R., Isarankura-Na-Ayudhya, P., Isarankura-Na-Ayudhya, C., Roytrakul, S., and Prachayasittikul, V. (2014). Reference map and comparative proteomic analysis of Neisseria gonorrhoeae displaying high resistance against spectinomycin. J. Med. Microbiol. 63, 371–385. doi: 10.1099/jmm.0.067595-0
Nagai, H., and Shimamoto, N. (1997). Regions of the Escherichia coli primary sigma factor σ70 that are involved in interaction with RNA polymerase core enzyme. Genes Cells 2, 725–734. doi: 10.1046/j.1365-2443.1997.1600357.x
Nair, J., Rouse, D. A., Bai, G. H., and Morris, S. L. (1993). The rpsL gene and streptomycin resistance in single and multiple drug-resistant strains of mycobacterium tuberculosis. Mol. Microbiol. 10, 521–527. doi: 10.1111/j.1365-2958.1993.tb00924.x
Norris, A. L., and Serpersu, E. H. (2013). Ligand promiscuity through the eyes of the aminoglycoside N3 acetyltransferase IIa. Protein Sci. 22, 916–928. doi: 10.1002/pro.2273
O’connor, M., and Dahlberg, A. E. (2002). Isolation of spectinomycin resistance mutations in the 16S rRNA of salmonella enterica serovar Typhimurium and expression in Escherichia coli and salmonella. Curr. Microbiol. 45, 429–433. doi: 10.1007/s00284-002-3684-y
Okazaki, S., Hattori, Y., and Saeki, K. (2007). The Mesorhizobium loti purB gene is involved in infection thread formation and nodule development in Lotus japonicus. J. Bacteriol. 189, 8347–8352. doi: 10.1128/JB.00788-07
O'Sullivan, J., McCullough, J. E., Tymiak, A. A., Kirsch, D. R., Trejo, W. H., and Principe, P. A. (1988). Lysobactin, a novel antibacterial agent produced by Lysobacter sp. J. Antibiot. 41, 1740–1744. doi: 10.7164/antibiotics.41.1740
Panthee, S., Hamamoto, H., Paudel, A., and Sekimizu, K. (2016). Lysobacter species: a potential source of novel antibiotics. Arch. Microbiol. 198, 839–845. doi: 10.1007/s00203-016-1278-5
Pardos de la Gandara, M., Borges, V., Chung, M., Milheiriço, C., Gomes, J. P., de Lencastre, H., et al. (2018). Genetic determinants of high-level oxacillin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 62, e00206–e00218. doi: 10.1128/AAC.00206-18
Patel, G., Huprikar, S., Factor, S. H., Jenkins, S. G., and Calfee, D. P. (2008). Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect. Control Hosp. Epidemiol. 29, 1099–1106. doi: 10.1086/592412
Patel, R., Sundin, G., Yang, C., Wang, J., Huntley, R., Yuan, X., et al. (2017). Exploration of using antisense peptide nucleic acid(PNA)-cell penetrating peptide (cpp) as a novel bactericide against fire blight pathogen Erwinia amylovora. Front. Microbiol. 8:687. doi: 10.3389/fmicb.2017.00687
Qian, G., Xu, F., Venturi, V., Du, L., and Liu, F. (2014). Roles of a solo LuxR in the biological control agent Lysobacter enzymogenes strain OH11. Phytopathology 104, 224–231. doi: 10.1094/PHYTO-07-13-0188-R
Scarff, J. M., Waidyarachchi, S. L., Meyer, C. J., Lane, D. J., Chai, W., Lemmon, M. M., et al. (2019). Aminomethyl spectinomycins: A novel antibacterial chemotype for biothreat pathogens. J. Antibiot. 72, 693–701. doi: 10.1038/s41429-019-0194-8
Sigmund, C. D., Ettayebi, M., and Morgan, E. A. (1984). Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 12, 4653–4664. doi: 10.1093/nar/12.11.4653
Stockwell, V., and Duffy, B. (2012). Use of antibiotics in plant agriculture. Rev. Sci. Et. Tech 31, 199–210. doi: 10.20506/rst.31.1.2104
Suvorov, M., Lee, M., Hesek, D., Boggess, B., and Mobashery, S. (2008). Lytic transglycosylase MltB of Escherichia coli and its role in recycling of peptidoglycan strands of bacterial cell wall. J. Am. Chem. Soc. 130, 11878–11879. doi: 10.1021/ja805482b
Tang, B., Laborda, P., Sun, C., Xu, G., Zhao, Y., and Liu, F. (2019). Improving the production of a novel antifungal alteramide B in Lysobacter enzymogenes OH11 by strengthening metabolic flux and precursor supply. Bioresour. Technol. 273, 196–202. doi: 10.1016/j.biortech.2018.10.085
Tran, Q. T., Williams, S., Farid, R., Erdemli, G., and Pearlstein, R. (2013). The translocation kinetics of antibiotics through porin OmpC: insights from structure-based solvation mapping using WaterMap. Proteins 81, 291–299. doi: 10.1002/prot.24185
Unemo, M., Fasth, O., Fredlund, H., Limnios, A., and Tapsall, J. (2009). Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J. Antimicrob. Chemother. 63, 1142–1151. doi: 10.1093/jac/dkp098
Unemo, M., Golparian, D., Skogen, V., Olsen, A. O., Moi, H., Syversen, G., et al. (2013). Neisseria gonorrhoeae strain with high-level resistance to spectinomycin due to a novel resistance mechanism (mutated ribosomal protein S5) verified in Norway. Antimicrob. Agents Chemother. 57, 1057–1061. doi: 10.1128/AAC.01775-12
US Food and Drug Administration (2014). FDA Annual Summary Report on Antimicrobials sold or Distributed in 2012 for use in Food-Producing Animals.
Wang, Y., Tian, T., Zhang, J., Jin, X., Yue, H., Zhang, X.-H., et al. (2019). Indole reverses intrinsic antibiotic resistance by activating a novel dual-function importer. MBio 10, e00676–e00719. doi: 10.1128/mBio.00676-19
Wilson, B. A., Salyers, A. A., Whitt, D. D., and Winkler, M. E. (2011). Bacterial Pathogenesis: A Molecular Approach. (3rd edn.). Washington DC: ASM Press.
Woolhouse, M., Ward, M., van Bunnik, B., and Farrar, J. (2015). Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B 370:20140083. doi: 10.1098/rstb.2014.0083
Yang, H., Yu, M., Liu, J., Khan, A., and Zhao, Y. (2021). Characterization of genes involved in (p)pp Gpp precursor biosynthesis in Erwinia amylovora. J. Plant Pathol. 103, S79–S88. doi: 10.1007/s42161-020-00583-6
Yelin, I., and Kishony, R. (2018). Antibiotic resistance. Cell 172, 1136–1136.e1. doi: 10.1016/j.cell.2018.02.018
Yin, J., Sun, Y., Sun, Y., Yu, Z., Qiu, J., and Gao, H. (2018). Deletion of lytic transglycosylases increases beta-lactam resistance in Shewanella oneidensis. Front. Microbiol. 9:13. doi: 10.3389/fmicb.2018.00013
Yu, M., Zhang, G., Jiang, J., Du, L., and Zhao, Y. (2020). Lysobacter enzymogenes employs diverse genes for inhibiting hypha growth and spore germination of soybean fungal pathogens. Phytopathology 110, 593–602. doi: 10.1094/PHYTO-09-19-0356-R
Yu, M., and Zhao, Y. (2019). Comparative resistomic analyses of Lysobacter species with high intrinsic multidrug resistance. J. Glob. Antimicrob. Res. 19, 320–327. doi: 10.1016/j.jgar.2019.05.008
Yu, M., and Zhao, Y. (2020). Cell permeability, beta-lactamase activity, and transport contribute to high level of resistance to ampicillin in Lysobacter enzymogenes. Appl. Microbiol. Biotechnol. 104, 1149–1161. doi: 10.1007/s00253-019-10266-7
Zhang, W., Huffman, J., Li, S., Shen, Y., and Du, L. (2017). Unusual acylation of chloramphenicol in Lysobacter enzymogenes, a biocontrol agent with intrinsic resistance to multiple antibiotics. BMC Biotechnol. 17:59. doi: 10.1186/s12896-017-0377-y
Zhang, W., Li, Y., Qian, G., Wang, Y., Chen, H., Li, Y. Z., et al. (2011). Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob. Agents Chemother. 55, 5581–5589. doi: 10.1128/AAC.05370-11
Keywords: Lysobacter, intrinsic resistance, spectinomycin, rRNA, mltB, purB
Citation: Yu M and Zhao Y (2022) Spectinomycin resistance in Lysobacter enzymogenes is due to its rRNA target but also relies on cell-wall recycling and purine biosynthesis. Front. Microbiol. 13:988110. doi: 10.3389/fmicb.2022.988110
Received: 06 July 2022; Accepted: 05 August 2022;
Published: 31 August 2022.
Edited by:
Guoqing Niu, Southwest University, ChinaReviewed by:
Ravikumar Patel, Connecticut Agricultural Experiment Station, United StatesCopyright © 2022 Yu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youfu Zhao, eW91ZnUuemhhb0B3c3UuZWR1
†ORCID: Youfu Zhao https://orcid.org/0000-0002-0728-5892
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.