
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 September 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.988044
 Andrea M. Suria1
Andrea M. Suria1 Stephanie Smith1
Stephanie Smith1 Lauren Speare1,2
Lauren Speare1,2 Yuzhou Chen1
Yuzhou Chen1 Iris Chien1
Iris Chien1 Emily Grace Clark1
Emily Grace Clark1 Madelyn Krueger1
Madelyn Krueger1 Alexander M. Warwick1
Alexander M. Warwick1 Hannah Wilkins1
Hannah Wilkins1 Alecia N. Septer1*
Alecia N. Septer1*The type VI secretion system (T6SS) is widely distributed in diverse bacterial species and habitats where it is required for interbacterial competition and interactions with eukaryotic cells. Previous work described the role of a T6SS in the beneficial symbiont, Vibrio fischeri, during colonization of the light organ of Euprymna scolopes squid. However, the prevalence and diversity of T6SSs found within the distinct symbiotic structures of this model host have not yet been determined. Here, we analyzed 73 genomes of isolates from squid light organs and accessory nidamental glands (ANGs) and 178 reference genomes. We found that the majority of these bacterial symbionts encode diverse T6SSs from four distinct classes, and most share homology with T6SSs from more distantly related species, including pathogens of animals and humans. These findings indicate that T6SSs with shared evolutionary histories can be integrated into the cellular systems of host-associated bacteria with different effects on host health. Furthermore, we found that one T6SS in V. fischeri is located within a genomic island with high genomic plasticity. Five distinct genomic island genotypes were identified, suggesting this region encodes diverse functional potential that natural selection can act on. Finally, analysis of newly described T6SSs in roseobacter clade ANG isolates revealed a novel predicted protein that appears to be a fusion of the TssB-TssC sheath components. This work underscores the importance of studying T6SSs in diverse organisms and natural habitats to better understand how T6SSs promote the propagation of bacterial populations and impact host health.
Bacteria must compete for habitat space in both free-living surface-associated environments and within animal hosts. The type VI secretion system (T6SS) is a molecular syringe used by bacteria to inject effector proteins into neighboring prokaryotic and eukaryotic cells and, in most cases, mediates contact-dependent competition (Cianfanelli et al., 2016). T6SS gene clusters have been detected in diverse bacterial genomes from a wide range of habitats (Boyer et al., 2009). The activity of the T6SS was first characterized in Pseudomonas aeruginosa (Mougous et al., 2006) and Vibrio cholerae (Pukatzki et al., 2006), and its activity has since been functionally described in other pathogenic bacteria, such as Escherichia coli (Aschtgen et al., 2008), Serratia marcescens (Murdoch et al., 2011), Acinetobacter baumannii (Carruthers et al., 2013), Vibrio parahaemolyticus (Salomon et al., 2013), Vibrio vulnificus (Church et al., 2016), and many others (reviewed in Hood et al., 2017). The T6SS has also been characterized in symbiotic and environmental bacteria, including Agrobacterium tumefaciens (Wu et al., 2008), Bacteroidetes (Russell et al., 2014b), Burkholderia thailandensis (Schwarz et al., 2010b), and Rhizobium leguminosarum (Bladergroen et al., 2003). Many of these bacterial species encode multiple, evolutionarily distinct T6SS gene clusters that are often located on genomic islands (Boyer et al., 2009; Schwarz et al., 2010b). Five T6SS subtypes have been described to date (Boyer et al., 2009), which may have diversified as strains adapted to competition against other bacteria or eukaryotic cells in the environment (Schwarz et al., 2010a). While these 13 core structural T6SS genes are well conserved in Proteobacteria, there is a lot of variability in the type of effector/immunity genes and accessory genes found within the genomic islands containing T6SS clusters (Boyer et al., 2009).
The Hawaiian bobtail squid, Euprymna scolopes, provides a uniquely tractable model system to study the role of T6SS activity in a host because it possesses two symbiotic organs. This model has been best studied for its light organ symbiosis, a binary association with the bioluminescent bacterium, Vibrio fischeri, which provides camouflage for the host in the form of counterillumination (Nyholm and McFall-Ngai, 2021; Visick et al., 2021). Female E. scolopes also possess a second, consortial symbiosis in the accessory nidamental gland (ANG, Collins et al., 2012), where bacteria are housed before transfer to the jelly coat of the squid eggs when they are laid. This bacterial community is composed of Alphaproteobacteria, Verrucomicrobia, Gammaproteobacteria, and Flavobacteriia (Collins et al., 2012; Kerwin and Nyholm, 2017), and has been found to protect the eggs from biofouling during development (Kerwin et al., 2019). These symbiotic organs both contain areas where competition would naturally occur during colonization, namely in the crypts of the light organ and the tubules of the ANG. Symbionts grow to high cell densities in these structures, between 107–108 cells in an adult light organ (Ruby and Asato, 1993) and 109–1010 cells in a mature ANG (Collins, 2014), which would allow symbionts to use contact-dependent competition mechanisms like the T6SS.
Previous work identified T6SS clusters in V. fischeri genomes from squid and fish light organs (Speare et al., 2018). It was found that V. fischeri possess one T6SS cluster on the first chromosome (T6SS1), while a subset of genomes encode a second cluster on the second chromosome (T6SS2). The T6SS2 is necessary for killing other V. fischeri strains in vitro and to compete for colonization space in the E. scolopes light organ crypts (Speare et al., 2018, 2022; Guckes et al., 2019). The role of the T6SS1 remains unknown. These previous studies determining strain-specific occurrence of T6SS2 only screened 33 isolates by PCR detection for the presence of the T6SS2 genes tssM_2 (icmF_2) and tssD_2 (vasA_2) and did not address whether there were other genes in the genomic island when T6SS2 was not detected.
Type VI secretion system-associated genes were previously reported in 12 ANG and egg jelly coat isolate genomes (Collins et al., 2015; Gromek et al., 2016; Suria et al., 2020), however the number of T6SS gene clusters, their diversity, and predicted effectors were not characterized. The majority of these genomes are from Leisingera and Ruegeria symbionts, which are members of the roseobacter clade of Alphaproteobacteria, the most abundant class of symbionts in the ANG (Collins et al., 2012) and an abundant and biogeochemically important group in free-living marine microbial communities (Buchan and Moran, 2005; Wagner-Döbler and Biebl, 2006). Activity of these T6SS clusters has not been experimentally verified. While T6SS gene clusters have been detected in other roseobacter clade genomes and metagenomes (Persson et al., 2009; Kempnich and Sison-Mangus, 2020), experimentally verified activity of any T6SS in this group has not been reported to date. Therefore, more information is needed about the prevalence and diversity of T6SSs in the ANG to generate testable hypotheses about their functional roles during symbiosis.
Here, we used multiple bioinformatic approaches to analyze the genomes of 57 squid/fish light organ symbionts, 15 squid ANG symbionts, and 175 bacterial reference genomes in order to: (1) identify T6SS gene clusters, (2) group T6SSs into phylogenetically-distinct subtypes, and (3) examine the gene content in the T6SS gene clusters and genomic islands. This analysis revealed that the majority of isolates encode at least one T6SS from one of four subtypes, yet the function of only one T6SS symbiont type (T6SS2 in V. fischeri) has been experimentally verified. Furthermore, we identified a new class of sheath protein, found only in certain roseobacter clade genomes, that appears to be a fusion of the large and small sheath subunits (TssBC). Our in silico analysis indicates that the squid model system, which houses a diversity of secretion systems and novel structural genes, provides an opportunity to further our understanding of the ecological roles and essential structural components of T6SSs in a natural, co-evolved system.
New Vibrio fischeri strains were isolated from the light organs of three adult Hawaiian bobtail squid, Euprymna scolopes, and from a light organ-like structure of one sub-adult Atlantic brief squid, Lolliguncula brevis (Supplementary Table S1). All E. scolopes samples were donated by the lab of Dr. Eric Stabb and originally collected from Maunalua Bay, Oahu, HI, maintained in aquaria at the University of Georgia, and flash frozen upon death. Frozen samples were transported on dry ice to the University of North Carolina at Chapel Hill and were stored at −80°C until dissection. The L. brevis sample was obtained from a trawl net in Morehead City, NC, United States and was immediately dissected.
All dissecting tools were sterilized with 70% ethanol prior to use. Frozen animals were thawed on ice at 4°C prior to dissection. Tissue samples were dissected and washed three times by brief vortexing in filter sterilized artificial seawater (FSASW) and transferred to fresh FSASW for each wash. Washed samples were homogenized in fresh FSASW, serially diluted on LBS (Luria-Bertani with added salts) agar (Stabb et al., 2001), and incubated aerobically at 24°C for 48 h. Single colonies were streaked for isolation.
The 10 squid light organ isolates obtained in this study and one previously isolated Hawaiian seawater strain (Supplementary Table S2) were grown for 16.5 h in 3.0 ml LBS broth at 24°C, 200 rpm shaking. Genomic DNA was extracted from 1.0 ml of pelleted cells using the Zymo Quick-DNA Fungal/Bacterial kit (Irvine, CA, United States) following manufacturer protocols. Samples were lysed in the “ZR BashingBead Lysis Tubes” for 10 min using a Vortex-Genie 2. Concentration and purity of DNA were measured on a spectrophotometer (Eppendorf BioSpectrophotometer basic).
Sequencing libraries were prepared using the Illumina Nextera kit series and sequenced using paired end (2 × 150 bp) reads on the Illumina NextSeq 550 to a depth of 150 Mb at the Microbial Genome Sequencing Center (MiGS, Pittsburgh, PA, United States). An average of 1.4 ± 0.18 M raw, paired end reads were sequenced per sample.
Raw reads were trimmed using Trimmomatic v.0.36 (Bolger et al., 2014) with a sliding window size of 10, sliding window minimum quality of 20, leading/trailing minimum quality of 3, and minimum read length of 50. Quality trimmed reads were assembled using SPAdes v.3.13.0 with default settings (Bankevich et al., 2012). Genome completion estimates were determined using CheckM v.1.0.18 (Parks et al., 2015), which determines the number of single copy housekeeping genes present in a genome compared to a list of marker genes specific for that genome’s placement on a reference tree. Average nucleotide identity (ANI) was calculated using FastANI (Jain et al., 2018). The Whole Genome Shotgun project and raw reads are deposited at DDBJ/ENA/GenBank under the BioProject accession PRJNA785243.
Genomes from 57 squid/fish light organ symbionts, 15 accessory nidamental gland (ANG) symbionts, and 175 bacterial reference strains were analyzed for the presence and type of T6SS gene clusters (Supplementary Table S3). Genomes that were not sequenced in this study were obtained from Genbank. All genomes were annotated using Prokka v.1.12 with default settings (Seemann, 2014), and the python and R script HAMBURGER (HMmer Based UndeRstandinG of gene clustERs; https://github.com/djw533/hamburger; Mariano et al., 2019) was used to detect T6SS gene clusters. A T6SS gene cluster was predicted if it contained at least four of the 13 core T6SS genes (tssA-M) and any given core gene was separated by no more than 10 non-core genes in a cluster. These criteria would detect core T6SS clusters and likely exclude auxiliary clusters that may only contain one or two toxin/anti-toxin system effector genes (Dong et al., 2013). The HAMBURGER script uses MUSCLE v.3.8.21 (Edgar, 2004) and FastTree v.2.1.11 (Price et al., 2010) to build a maximum-likelihood tree from the aligned TssBC (small and large subunits of the contractile sheath) amino acid sequences. The R packages “gggenes” and “ggplot2” were used to draw T6SS operons. The pangenomics workflow of Anvi’o v.7 (Eren et al., 2015; Delmont et al., 2018) was used to compare the T6SS2 genomic region of V. fischeri genomes. This region is the area between the flanking genes manA (mannose-6-phosphate isomerase) and tRNA-Gly on the second chromosome. Any genomes that had only one break in the assembly in this region and could easily be concatenated were included. T6SS effectors were predicted by BLASTP search against the experimentally validated effector protein SecReT6 database (Li et al., 2015), downloaded on January 28, 2022. Results were considered significant if the amino acid similarity was ≥30% and E-value ≤10−6.
The nine V. fischeri strains isolated from E. scolopes in this study (Supplementary Table S2) were screened for the ability to inhibit the growth of V. fischeri ES114 pVSV102 on agar plates as described previously (Speare et al., 2018). All strains were grown overnight at 24°C on LBS agar with the addition of 100 μg/ml kanamycin for maintaining the pVSV102 plasmid with GFP fluorescence. Cells from these plates were resuspended in 0.5 ml LBS broth, washed once with LBS, and normalized to an OD600 of 1.0. Each strain was mixed with V. fischeri ES114 pVSV102 in a 1:1 ratio and 10 μl of the mixture was spotted on LBS agar. These coincubation spots were incubated for 24 h at 24°C and then imaged for GFP fluorescence to determine the extent to which V. fischeri ES114 pVSV102 was able to grow or be inhibited in the presence of control and test strains. Representative images are shown from three experimental trials, where each trial contained three resuspensions per strain, prepared from different colonies.
To add to the diversity of Vibrio fischeri genomes available for T6SS analysis in squid light organs, we sequenced the genomes of nine V. fischeri strains from three E. scolopes light organs and one V. fischeri strain from a light organ-like structure in L. brevis. All genomes were assembled to 99.9% completion with genome sizes of 4.0–4.4 Mbp (Supplementary Table S2). All strains were assigned to the species V. fischeri based on ANI analysis with a cutoff of ≥95% (Supplementary Table S4; Figureueras et al., 2014). Based on an ANI value of 100, the following co-isolated strains are likely to be clones but were kept for further analysis: ESM429_1 and ESM429_3; ESF442_5 and ESF442_6; and ESF436_3, ESF436_4, and ESF436_6.
The genome of one reference isolate, Vibrio campbellii KNH1, was also sequenced to confirm its species identification. This strain had previously been assigned as Vibrio parahaemolyticus based on 16S rRNA gene Sanger sequencing (Nyholm et al., 2000). The 16S rRNA gene often cannot provide enough genetic diversity to assign a species level identification in vibrios (Silvester et al., 2017), so an ANI comparison to other reference genomes was performed. An ANI cutoff of ≥95% revealed that the KNH1 strain should be assigned to the species V. campbellii (Supplementary Table S5).
Of the 57 V. fischeri light organ symbiont genomes and 14 V. fischeri seawater isolate genomes analyzed, all had one predicted T6SS cluster on chromosome 1 (T6SS1) and 38 genomes had a second T6SS cluster on chromosome 2 (T6SS2; Figure 1; Supplementary Figure S1). Based on phylogenetic analysis of the sheath subunit protein sequences (TssBC), these clusters appear to share an evolutionary history, but have diversified to form two distinct clades within group one of the previously described T6SS subtypes (Boyer et al., 2009). The TssBC proteins of strains isolated from different animal hosts (n = 58) and seawater sources (n = 12) were observed to cluster together in both clades, indicating there is no evolutionary link between type of T6SS and isolation source (i.e., host-associated vs. environmental).
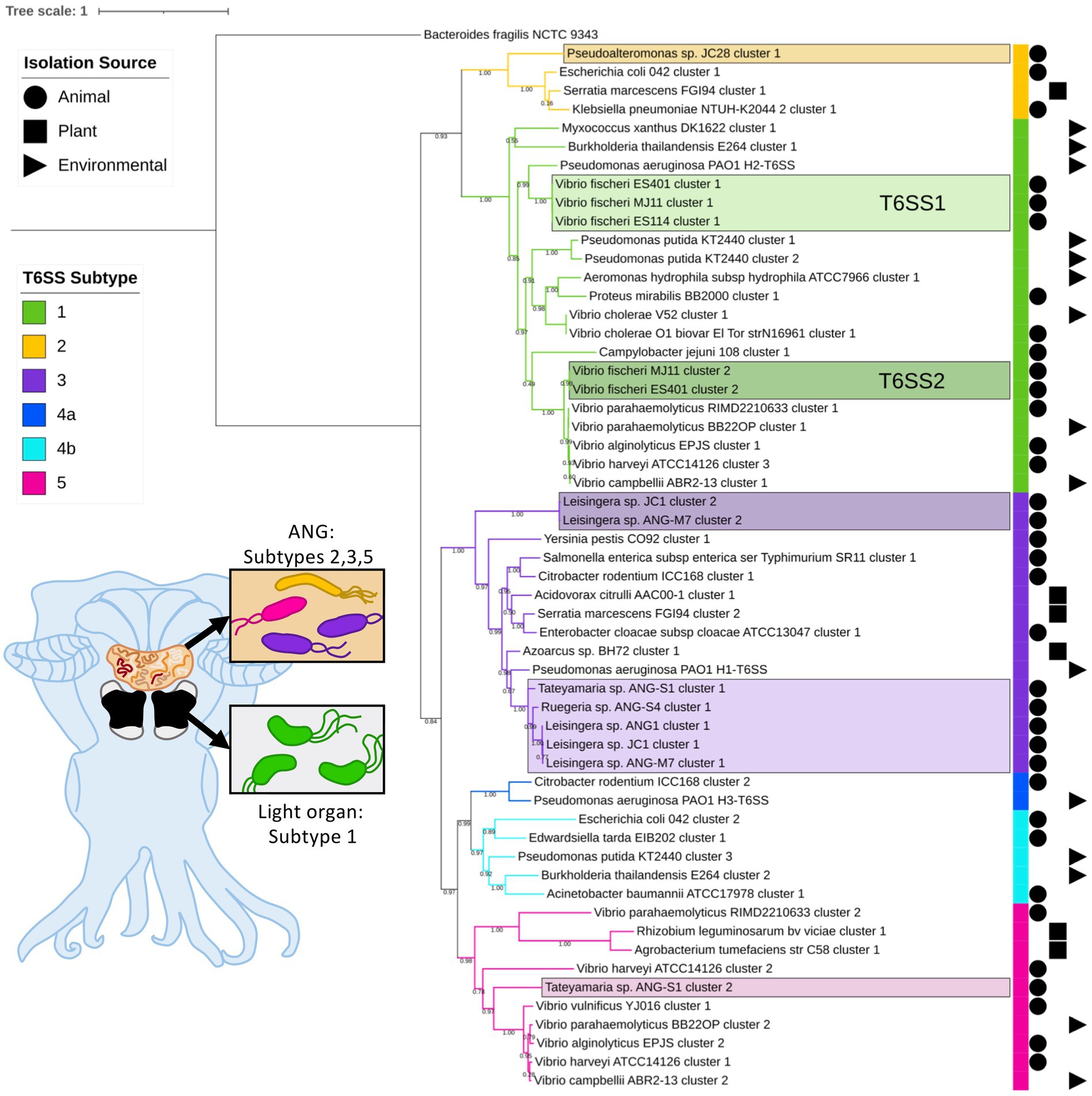
Figure 1. Maximum-likelihood phylogenetic tree of type VI secretion system (T6SS) sheath subunit proteins of Euprymna scolopes symbionts. A maximum-likelihood tree was constructed from the TssB and TssC small and large T6SS sheath subunit proteins predicted in select Euprymna scolopes accessory nidamental gland (ANG) and light organ bacterial symbiont genomes (names boxed) and reference genomes. The colored strip to the right of the tree and branch colors correspond to the T6SS subtype (Green = subtype 1, yellow = subtype 2, purple = subtype 3, dark blue = subtype 4a, light blue = subtype 4b, and pink = subtype 5). Symbols to the right of the colored strip indicate the isolation source of the sequenced bacterial strain (Circle = animal source, square = plant source, triangle = environmental source). Numbers on branches indicate bootstrap values. Inset cartoon diagram shows a ventral dissection of an adult female E. scolopes with colored ANG and light organ. Boxes show cartoon bacteria colored to represent the T6SS subtype of the symbionts found in each tissue type. See Supplementary Figure S1 for a complete phylogenetic tree with all strains analyzed in this study.
All of the predicted V. fischeri T6SSs share evolutionary similarity to other well characterized T6SS model systems. The V. fischeri T6SS1 TssBC proteins were most phylogenetically similar to the H2-T6SS of Pseudomonas aeruginosa PAO1, and both clusters share the same core T6SS gene organization (Figure 2A). The V. fischeri T6SS2 TssBC proteins are most similar to TssBC in several other vibrios, including V. parahaemolyticus, V. alginolytiucs, V. harveyi, and V. campbellii, as was previously seen in a search of V. fischeri IcmF (TssM) homologs (Speare et al., 2018).
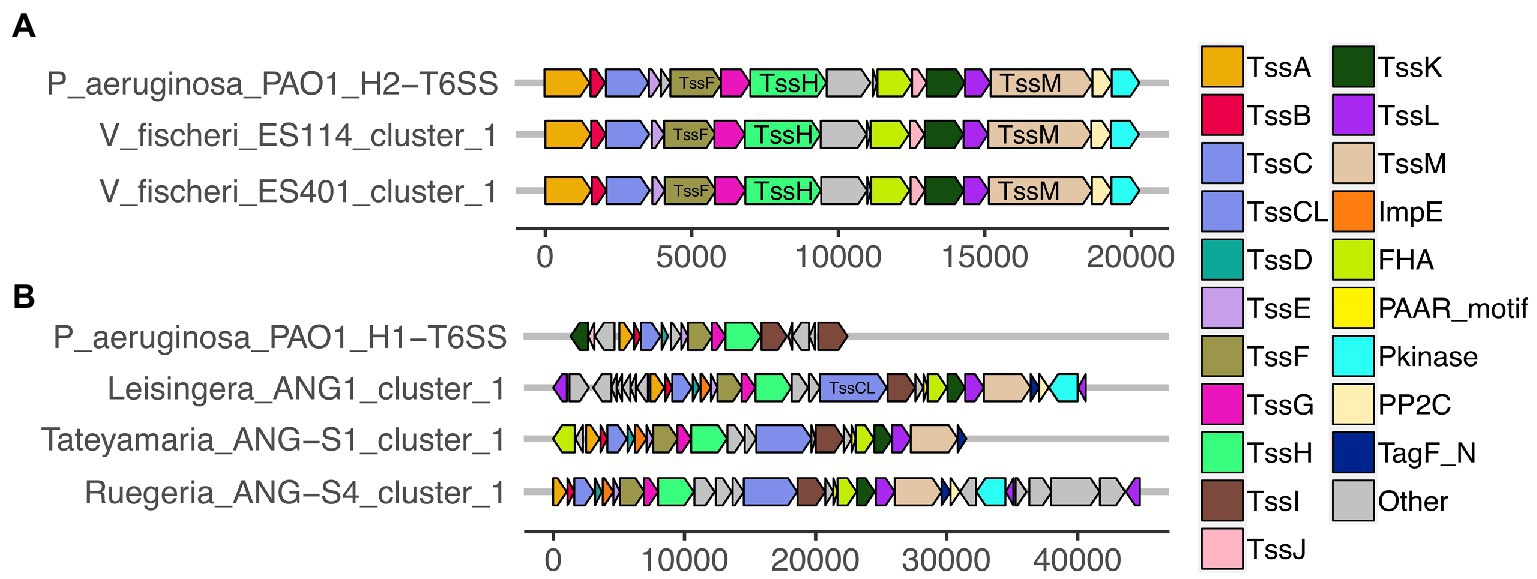
Figure 2. Predicted T6SS gene clusters of Euprymna scolopes symbiont genomes share similar gene organization to T6SS gene clusters in Pseudomonas aeruginosa PAO1. Each of the T6SS core structural and accessory genes are colored, non-T6SS genes are labeled as “other” and are gray. Genes are drawn to scale at the bottom of each cluster, measured in bp. (A) The T6SS1 of Vibrio fischeri genomes share similar gene content and organization to the H2-T6SS of P. aeruginosa PAO1. (B) The first T6SS clusters of ANG strains share similarity to the H1-T6SS of P. aeruginosa PAO1.
All V. fischeri T6SS1 clusters contain 11 of the 13 core structural genes (tssA-tssM) but are missing tssD and tssI, which encode the Hcp tube and the VgrG spike proteins, respectively (Supplementary Table S3). These genes are often located in auxiliary clusters elsewhere in the genome (Dong et al., 2013; Speare et al., 2018), which were excluded from our analysis. The V. fischeri T6SS1 clusters also contain three accessory genes with protein phosphatase 2C (PP2C), protein kinase (Pkinase), and forkhead-associated (FHA) domains, which may be involved in post-translational regulation, as seen in P. aeruginosa PAO1 (Mougous et al., 2007).
Of the V. fischeri T6SS2 clusters, 79% (30/38) contain all the core tssA-M structural genes as well as six accessory genes (Supplementary Table S3). The remaining eight genomes with incomplete T6SS2 clusters are draft assemblies where the T6SS2 cluster was cutoff at the beginning or end of a contig (Supplementary Table S3). The break in the assemblies in this locus could account for the missing genes, which would go undetected by our analysis criteria. The accessory genes in T6SS2 also included pkinase, FHA, and DUF4150 domain-containing proteins similar to those found in T6SS1, as well as the recently described tasL gene, which is required for cell–cell adhesion and target specificity (Speare et al., 2022), and the tasR (Smith et al., 2021) and vasH (Guckes et al., 2020) regulators.
The T6SS2 locus is within a genomic island with high variability among the examined V. fischeri genomes. Five distinct genotypes were identified in this region based on the presence of the flanking genes, manA and tRNA-glycine (Figure 3), with tRNAs often being a site of genomic island insertion (Darmon and Leach, 2014). The five genotypes in this region include genomes with (1) a complete T6SS2 cluster and other genes (52.9% of genomes), (2) an incomplete T6SS2 cluster and other genes (2.9% of genomes), (3/4) no T6SS2 cluster but several other genes (23.5% of genomes), or (5) few to no genes in this region (20.6% of genomes). The sizes of these different genotype regions, including the flanking genes, range from 11.2 to 106.2 kbp. Non-T6SS genes in this region included predicted type 1 restriction enzyme and CRISPR-associated proteins (Supplementary Table S6), which are defensive mechanisms that are often overrepresented in bacterial genomic islands (Merk, 2006). Within the first genomic island genotype (T6SS2+), there is a diversity of T6SS2 cluster expansions and accessory genes (Figure 4). The region between the PAAR-like DUF4150 containing gene and tasL varies from 3 to 11 genes depending on the strain, while the region after vasH varies from 4 to 31 genes. These regions encode possible effectors with PAAR-like domains, lysozyme-like domains, and other conserved domains of unknown functions, as well as restriction enzymes, nucleases, GTPases, ATPases, and OmpA proteins. This finding indicates that although auxiliary clusters containing effector/immunity pairs exist, the primary T6SS2 gene cluster is also capable of diversifying effector arsenals. Furthermore, there are still conserved genes of unknown function encoded within the V. fischeri T6SS2 gene cluster that remain to be characterized.
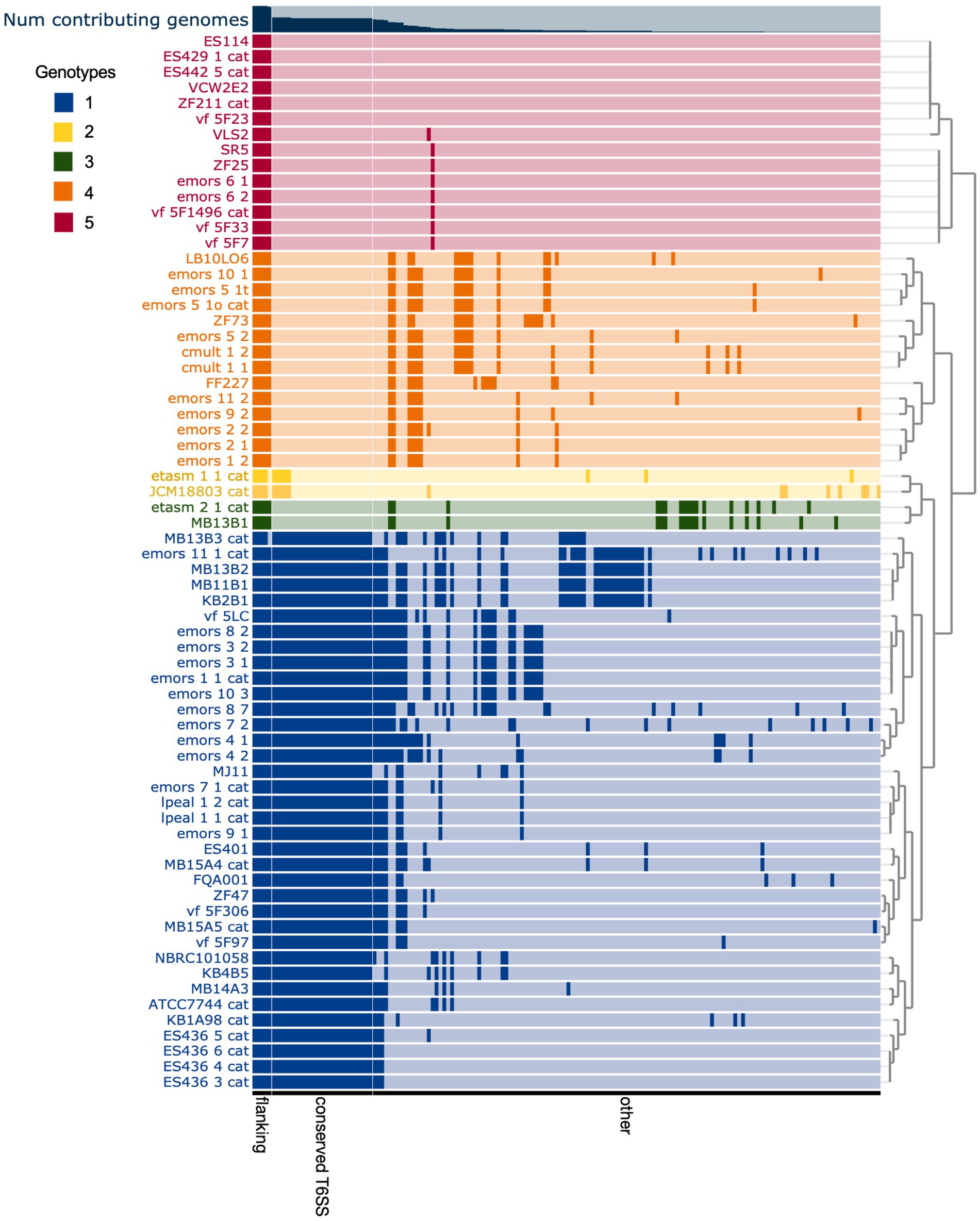
Figure 3. Comparison of predicted Vibrio fischeri T6SS2 genomic island regions reveals five distinct genotypes. Each row represents the T6SS2 genomic region of each V. fischeri strain based on presence of the manA and tRNA-Gly flanking genes. Gene clusters with shared similarity between strains are a darker shade of the row color. The strains are ordered based on hierarchical clustering of each T6SS2 genomic region, which revealed five distinct genotypes: (1, blue) a complete T6SS2 cluster and other genes, (2, yellow) an incomplete T6SS2 cluster and other genes, (3, green/ 4, orange) no T6SS2 cluster but several other genes, or (5, red) few to no genes in this region. Bar graph at top of chart indicates the percent of genomes that possess that gene cluster. Gene clusters are ordered based on binning of flanking genes (present in all genotypes), conserved T6SS genes (present in genotypes 1 and 2), and other non-T6SS genes which vary between the genotypes.
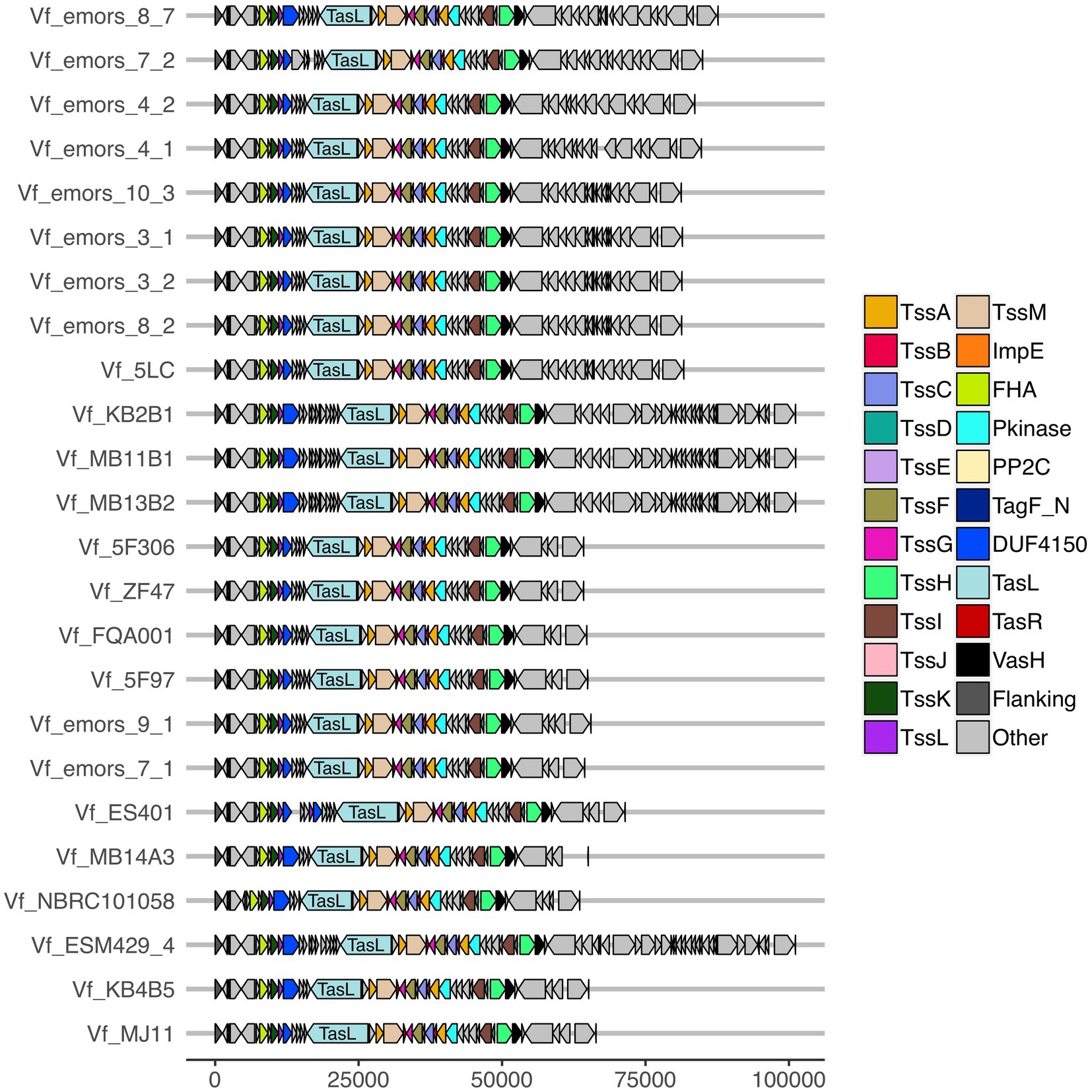
Figure 4. Predicted T6SS2 clusters of Vibrio fischeri genomes feature different patterns of cluster expansions. Gene organization of the predicted T6SS2 clusters of both fish and squid symbiont and environmental V. fischeri genomes. Genes labeled as “other” do not currently have a documented T6SS-associated function. See Supplementary Table S6 for a list of predicted annotations for proteins labeled as “other.” Genes are drawn to scale at bottom of figure, measured in bp.
While a function for the V. fischeri T6SS1 has not yet been determined, the T6SS2 cluster has previously been shown to be required for intra- and interspecies competition in a variety of strains (see contact-dependent inhibition annotation on Supplementary Figure S1; Speare et al., 2018, 2020, 2022). We observed the same pattern in the nine V. fischeri isolates from E. scolopes light organs described in this study (Supplementary Table S2), where only strains that possess a T6SS2 cluster were able to kill V. fischeri ES114 in coincubation assays on agar plates (Supplementary Figures S1, S2). However, disruption of the T6SS2 would be required to verify if the ability to kill ES114 is due to this gene cluster.
Of the 15 ANG strains analyzed, all but one (Ruegeria sp. ANG-R) had at least one predicted T6SS cluster (Figure 1; Supplementary Figure S1; Supplementary Table S3). All 11 Leisingera strains had at least one T6SS cluster that formed a clade in the group three T6SS subtype, along with T6SS clusters found in Ruegeria sp. ANG-S4 and Tateyamaria sp. ANG-S1. The TssBC proteins of these T6SS clusters were most phylogenetically similar to the H1-T6SS of P. aeruginosa PAO1, although the ANG strain clusters appear to have further expanded (Figure 2B). Two of the Leisingera strains, ANG-M7 and JC1, had a second T6SS cluster that formed a separate clade within group three. A second T6SS cluster identified in Tateyamaria sp. ANG-S1 was assigned to group five and was similar to T6SS clusters from several vibrios. The one T6SS cluster predicted for Pseudoalteromonas sp. JC28 fell within group two, along with E. coli and Serratia marcescens T6SS clusters.
All of the roseobacter (Leisingera, Ruegeria, and Tateyamaria) T6SS1 clusters contained all of the core T6SS structural genes except tssJ (Supplementary Table S3), which encodes a protein that has been localized to the membrane complex in Gammaproteobacteria (Aschtgen et al., 2008). The operons for all of these clusters share similar gene organizations as well as the PAAR-motif, FHA-domain, and impE accessory genes (Figure 5). The Pseudoalteromonas sp. JC28 cluster contained all of the core T6SS structural genes along with the PAAR-motif gene and tagF regulatory gene. The second T6SS clusters of Leisingera spp. ANG-M7 and JC1 are smaller and may perform a different function or be non-functioning degenerate clusters (Figure 5). Cluster 2 of ANG-M7 contains eight of the 13 core genes (tssA-H) as well as two accessory genes (PAAR motif, FHA; Supplementary Table S3). Cluster 2 of JC1 also contains eight core genes (tssA-H) with a duplicate tssF gene and no accessory genes. Notably, both clusters lack core genes for assembling the membrane complex and VgrG spike protein (tssI). The second T6SS cluster of Tateyamaria sp. ANG-S1 is a more complete T6SS cluster, only missing the tssI and tssJ genes and could be functional (Supplementary Table S3).
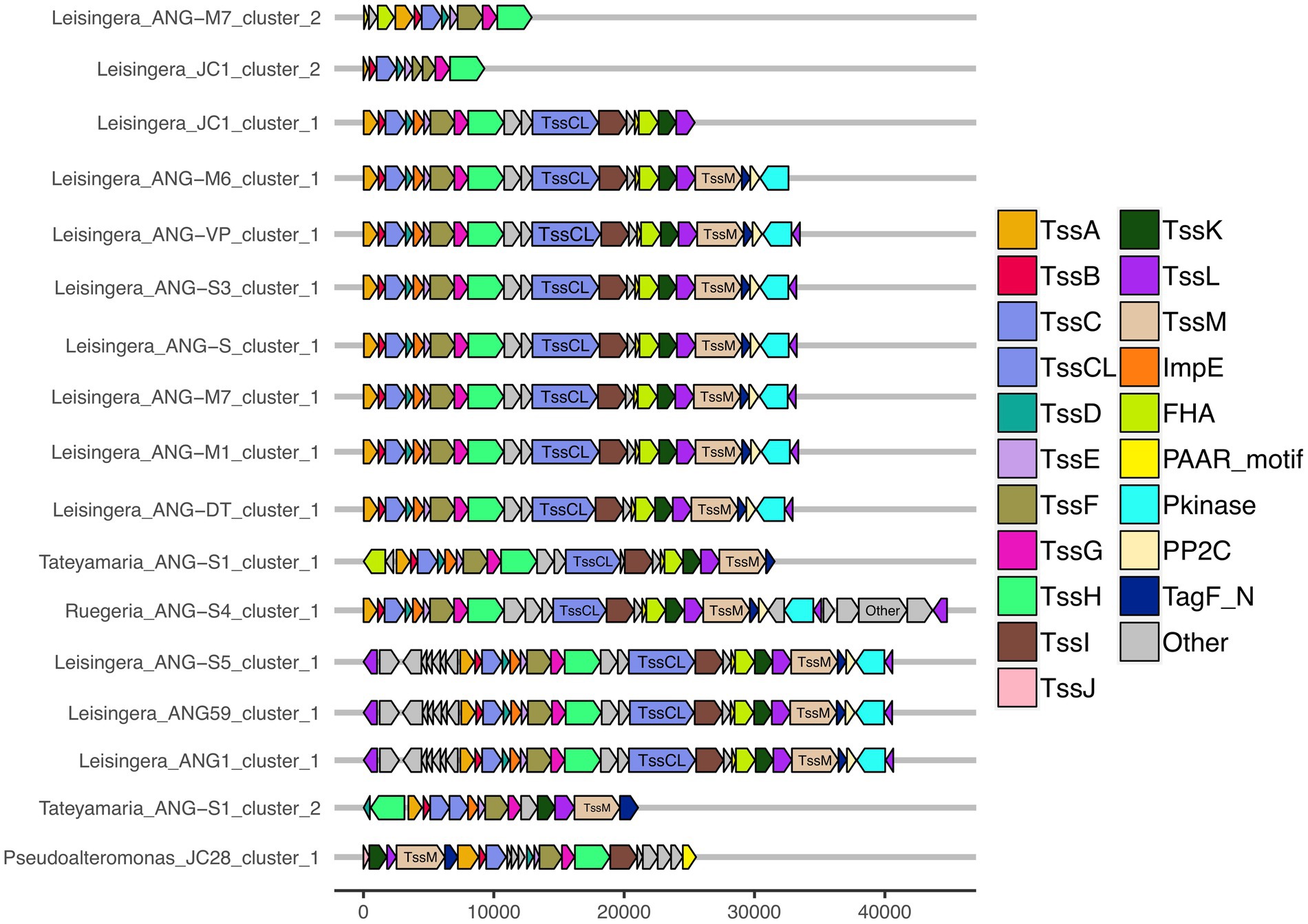
Figure 5. Gene organization of predicted T6SS clusters of Euprymna scolopes ANG bacterial symbionts. The roseobacter ANG symbiont genomes (all but Pseudoalteromonas sp. JC28) share similar T6SS accessory genes and cluster organization. These strains also share a second, long copy of the large sheath subunit, termed here TssCL, in their first T6SS clusters. TssCL was not detected in any genomes outside of the Alphaproteobacteria, and is absent from the Gammaproteobacteria ANG symbiont, JC28, shown here. Each of the T6SS core structural and accessory genes are colored, genes that have not been associated with a T6SS function are labeled as “other” and are gray. Genes are drawn to scale at bottom of figure, measured in bp.
Interestingly, all of the T6SS1 clusters in the roseobacter ANG genomes contained two predicted tssC genes: one that encodes a protein of similar size to previously described TssC proteins (506 aa), and a second gene predicted to encode a much larger protein (1,684 aa; Figure 5). A pfam domain search of this longer TssC protein, which we term “TssCL,” revealed it contains two VipA domains (PF05591) and one VipB domain (PF05943; Figure 6A). Only one of the VipA domains shares high amino acid identity (86.2%) with the VipA domain in the TssB protein of the same T6SS cluster. The second VipA domain in TssCL is a shorter sequence with only 26.3% amino acid identity to the same domain in TssB. The one VipB domain in TssCL shared low amino acid identity (30.9%) to the first VipB domain in the shorter tssC gene. Although TssCL is annotated as tssC in the rosoebacter genomes, it contains key domains from both sheath subunits and appears to be a fusion of the two genes.
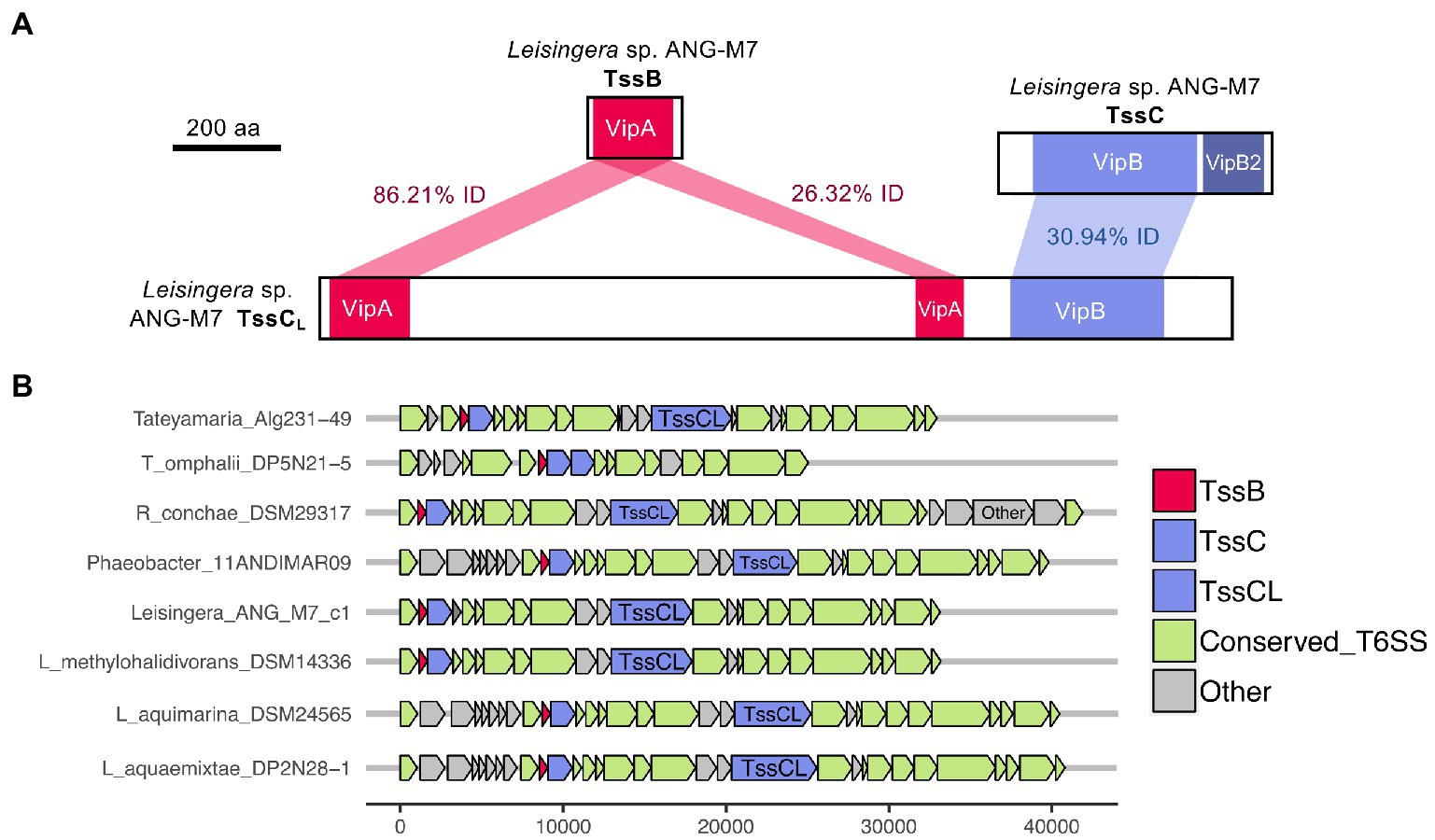
Figure 6. Domains and occurrence of the Alphaproteobacteria sheath protein, TssCL. The long copy of the TssC protein, termed here TssCL, contains domains from both TssB and TssC and was only detected in other roseobacter reference genomes in this study. (A) The TssCL protein of Euprymna scolopes ANG symbiont Leisingera sp. ANG-M7 contains two VipA domains with 86.21% (1e−94 E-value) and 26.32% (5e−5 E-value) amino acid identity to the VipA domain in TssB. This TssCL protein also has a VipB domain with 30.94% identity (2e−36 E-value) to the VipB domain in TssC. (B) Other roseobacter genomes that have a tssCL gene still have tssB and tssC genes. Some roseobacter genomes, such as Tateyamaria omphalii DP5N21–5, do not have tssCL, but instead have two tssC genes and one tssB gene.
Similar TssCL proteins are present in other roseobacter T6SS clusters, but absent from all other bacterial genomes analyzed in this study. The TssCL protein was found in 20 of the 28 roseobacter reference genomes with a similar predicted T6SS cluster, with amino acid identities to the ANG-M7 TssCL ranging from 45 to 93% (Figure 6B; Supplementary Table S7). The domain architecture within TssCL described above was also observed in seven other proteins in the pfam database, all from Alphaproteobacteria, that have 46–64% amino acid identity to the ANG-M7 TssCL (Supplementary Table S7). Interestingly, four of the Tateyamaria T6SS clusters had two copies of tssC but no tssCL (Figure 6B). Within each cluster, the two TssC proteins had 34–37% amino acid identity to each other (Supplementary Table S7), suggesting they did not arise from a gene duplication event. Thus, it is possible that the TssCL protein may have arisen from a fusion of multiple tail sheath subunits genes. Moreover, the tssCL gene is absent in the closely related H1-T6SS gene cluster of P. aeruginosa PAO1 (Figure 2), suggesting it is a more recent evolutionary event that occurred in certain Alphaproteobacteria.
As was seen for the V. fischeri genomes, there is no evidence to suggest that T6SS gene clusters are enriched in the host-associated isolation sources, relative to free-living environmental sources (Supplementary Figure S1; Supplementary Table S3). All 11 of the Leisingera reference genomes present in Genbank and analyzed in this study were from environmental sources and also possessed a T6SS cluster. For comparison, other roseobacter genomes were also analyzed and only three of 55 Phaeobacter reference genomes and five of 31 Ruegeria reference genomes possess a T6SS (Supplementary Table S3), thus possession of the T6SS may be a conserved trait within the Leisingera genus. For the other genera represented in ANG strains, four of 15 Tateyamaria reference genomes and 14 of 15 Pseudoalteromonas reference genomes also possess a T6SS (Supplementary Table S3). Overall, although T6SS gene clusters are present in many host-associated strains, it was not determined to be enriched in the host environment, suggesting there may be a functional role for T6SS in both host- and free-living habitats.
To date, there are no described functions for the T6SS in any of the ANG symbionts, nor for any of the T6SSs of reference roseobacter genomes. To begin to understand what activity the ANG T6SS clusters may have, an effector search was completed for Leisingera sp. ANG-M7, a representative genome from the most dominant genera in the ANG symbiotic community (Collins et al., 2012; Kerwin and Nyholm, 2017). By alignment to a database of 326 experimentally validated effector protein sequences (SecReT6, Li et al., 2015), ANG-M7 had 39 significant predicted effector hits (Supplementary Table S8). Of these, four hits were to proteins in the T6SS cluster 1 (PAAR-motif, VgrG, Hcp, and a peptidase) and three to proteins in the T6SS cluster 2 (PAAR-motif, Hcp, and TssB). Two other VgrG and one PAAR-motif protein were found at other locations in the genome, as well as three amidases. The remaining significant hits included several cold shock proteins, LysR family transcriptional regulators, a phosphatidylserine synthase, a universal stress protein, and a tRNA ligase (Supplementary Table S8). While some of these predicted effector proteins may have similar domains to those in previously described T6SS effectors, it remains to be seen if they are indeed secreted through the T6SS in Leisingera sp. ANG-M7, and whether they are used during interbacterial competition and/or interactions with co-occurring eukaryotic cells.
In this study, we sought to characterize the type and abundance of T6SS gene clusters in symbionts of E. scolopes and found that all isolates from both the light organ and ANG, except one, possess at least one predicted T6SS cluster. Consistent with previous work by Speare et al., we observed that approximately half of V. fischeri strains isolated from wild-caught squid light organs possess a T6SS2 cluster, which was previously shown to be important for competition in the light organ crypts (Speare et al., 2018, 2022). Previously published phylogenies showed that V. fischeri T6SS2+ and T6SS2− strains are found dispersed among closely related strains, and do not cluster based on T6SS2 genotype (Speare et al., 2018), suggesting that there was an evolutionary loss of T6SS2. Here, we show that for some T6SS2− strains, different gene clusters can be found in place of T6SS2. Overall, E. scolopes symbionts possess T6SS clusters that represent four of the five described T6SS subtypes, with light organ symbiont T6SSs forming two clades in group 1 and the ANG symbiont T6SSs forming two clades in group 3 and one clade each in groups 2 and 5. Three of the roseobacter ANG symbionts possess a second T6SS cluster that may have been acquired from other bacteria based on their phylogenetic distance from the main roseobacter group 3 clade.
The Hawaiian bobtail squid, E. scolopes, is a good model system to study the interactions of bacterial strains possessing different T6SS subtypes during colonization of a single host. The diversity of T6SS subtypes detected in E. scolopes symbionts is expected due to the diversity of bacterial taxa represented, including a range of Alphaproteobacteria and Gammaproteobacteria. While the light organ only contains V. fischeri symbionts, the ANG is a more diverse community with only 17 isolate genomes available in Genbank at this time. Our reported diversity of T6SS clusters in the ANG is likely an underestimate because our study was limited to the cultured isolate genomes, which only represent a few genera of the community based on 16S rRNA gene sequencing (Collins et al., 2012; Kerwin and Nyholm, 2017). Despite this limitation, we still observed representatives of three T6SS subtypes within the ANG alone. Future research will determine if any of these T6SS clusters, besides V. fischeri T6SS2, are expressed and form sheathes in the host and what role they might play during symbiosis.
While the functional activity of ANG symbiont T6SSs remains to be determined, potential effectors were identified in one strain that could suggest their ability to inhibit other bacteria through contact-dependent competition. It has been previously shown through fluorescence in situ hybridization that different bacterial taxa in the ANG are observed in separate tubules (Collins et al., 2012). It has also been seen that the relative abundance of different bacterial classes shift as the ANG develops, which occurs as the nascent gland tissue forms pores that lead to invaginations that will become tubules in the mature gland (Kerwin et al., 2021). Similar to the light organ crypts, where T6SS2-dependent competition shapes the diversity and spatial arrangement of V. fischeri strains, the ANG tubules would provide a similar niche for ANG symbiont competition during colonization. The observed community shifts in the ANG could be attributed, at least in part, to contact-dependent inhibition mechanisms like the T6SS being deployed in initially co-colonized tubules. Future research will determine whether ANG symbionts are actively using their T6SSs to compete in the host as has been previously seen for light organ symbionts (Speare et al., 2018, 2022).
The phylogenetic analysis based on the structural TssBC proteins found that T6SS clusters from E. scolopes symbionts are similar to T6SSs in other well-studied model systems with impacts on animal hosts. In particular, the T6SS1 of light organ symbionts were most similar to the P. aeruginosa PAO1 H2-T6SS and the T6SS cluster 1 of roseobacter ANG symbionts was most similar to P. aeruginosa PAO1 H1-T6SS. The H1-T6SS of P. aeruginosa PAO1 can secrete three effector proteins, Tse1–3, which can target peptidoglycan and arrest bacterial cell growth (Hood et al., 2010; Russell et al., 2011). The Hcp of H1-T6SS has also been detected in the pulmonary secretions of cystic fibrosis patients (Mougous et al., 2006) and has higher expression in biofilms compared to in planktonic cells (Zhang et al., 2011). The H2-T6SS of P. aeruginosa PAO1 is regulated by quorum sensing and iron-limiting conditions and contributes to internalization by human cell lines and virulence in C. elegans (Sana et al., 2012). Although these studies associated PAO1’s H2-T6SS with effectors that impact eukaryotic cells, the phospholipase effector, PldA, secreted by this cluster has been shown to have antibacterial activity by degrading phosphatidylethanolamine of bacterial cell membranes (Russell et al., 2013). Thus, both H1-T6SS and H2-T6SS of PAO1 can contribute to effects on animal hosts and interbacterial competition. While similarity exists between the structural components of P. aeruginosa PAO1 T6SS clusters and E. scolopes symbiont T6SS clusters, it does not necessarily suggest that these clusters will have similar functions. These studies in PAO1 and other T6SS models have shown that the impact of a T6SS is governed by its effectors (Russell et al., 2014a). While initial investigation of Leisingera sp. ANG-M7’s effectors, and previous evidence in V. fischeri T6SS2 activity (Speare et al., 2018), point to antibacterial functions, it remains to be seen what alternative roles these or remaining uncharacterized effectors might play in the E. scolopes symbioses.
Finally, this study identified a new T6SS structural protein, TssCL, among certain roseobacter genomes that appears to be a fusion of two broadly conserved sheath subunits, TssB and TssC. The TssB and TssC small and large sheath subunits form heterodimers that stack on top of each other and contract during firing to deliver effectors (Costa et al., 2015). In other systems, fusion proteins have been detected from protein subunits that directly interact, such as domains of GyrA and GyrB in E. coli that are found in the topoisomerase II fusion protein in yeast (Enright et al., 1999; Marcotte et al., 1999). Thus, due to their dimer formation, TssB and TssC sheath subunits are likely candidates for fusion. However, this is not the first instance of apparent fusion between T6SS proteins. In such bacteria as Agrobacterium tumefaciens, Nitrococcus mobilis, and Rhizobium leguminosarum, the tagF and pppA orthologs are fused (Silverman et al., 2011), but these genomes do not continue to carry single copies of the unfused genes. Thus, it will be important to determine the extent to which TssCL and the single-copy TssB and TssC proteins interact to form a functional sheath in roseobacters.
The prevalence of T6SS clusters in E. scolopes symbionts suggests that possession of this interbacterial weapon might be beneficial during at least one life stage of the symbionts. Light organ symbionts are horizontally acquired from the water column after the squid hatch (Wei and Young, 1989), and it is believed that the ANG symbionts are also horizontally acquired when females begin to sexually mature (Kerwin and Nyholm, 2017). Thus, these symbionts must be equipped to survive in both free-living and host-associated conditions. Indeed, previous research in our lab has demonstrated that the V. fischeri T6SS2 is necessary for strain competition during light organ colonization (Speare et al., 2018, 2022). In other beneficial roles, the T6SS can also play a part in competitive exclusion, such as in the human gut (Russell et al., 2014b) and plant leaves (Bernal et al., 2017), and controlling host range in Rhizobium root nodulation (Bladergroen et al., 2003). More research on the role of T6SSs in beneficial symbioses is needed, and Euprymna scolopes will be a valuable model system to understand these roles in a diversity of beneficial symbionts and host-colonization sites.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
AMS, ANS, SS, and LS conceptualized and designed the study. AMS, SS, LS, YC, IC, EC, MK, AW, HW, and ANS isolated strains used in this study and conducted experiments. AMS, SS, YC, and ANS performed bioinformatic analyses. AMS and ANS wrote the manuscript. All authors contributed to the article and approved the submitted version.
Research was funded by the UNC-CH Undergraduate Research Consultant Team program and NIGMS grant R35 GM137886 to ANS. AMS was supported by a NIGMS IRACDA postdoctoral fellowship at UNC-CH. SS was supported by a National Defense Science and Engineering Graduate Fellowship. LS was supported by a UNC Dissertation Completion Fellowship.
We would like to thank the lab of Eric Stabb and Julie Stoudenmire for their generous collection and donation of squid samples. We also thank David Williams for helpful troubleshooting of the HAMBURGER analysis script.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.988044/full#supplementary-material
SUPPLEMENTARY FIGURE S1
Expanded maximum-likelihood phylogenetic tree of T6SS sheath subunit proteins of Euprymna scolopes symbionts. A maximum-likelihood tree was constructed from the TssB and TssC small and large T6SS sheath subunit proteins predicted in all E. scolopes ANG and light organ bacterial symbiont genomes (names boxed) and reference genomes analyzed in this study. The colored strip to the right of the tree and branch colors correspond to the T6SS subtype (Green = subtype 1, yellow = subtype 2, purple = subtype 3, dark blue = subtype 4a, light blue = subtype 4b, pink = subtype 5). Symbols next to the colored strip indicate the isolation source of the sequenced bacterial strain (Circle = animal source, square = plant source, triangle = environmental source). Stars next to V. fischeri strain names indicate if a strain has been experimentally verified to be lethal against V. fishceri ES114 in a contact-dependent inhibition assay (colored star = lethal, white star = non-lethal). Numbers on branches indicate bootstrap values.
SUPPLEMENTARY FIGURE S2
Coincubation assay confirms that V. fischeri strains with a predicted T6SS2 gene cluster are also lethal against V. fischeri ES114. V. fischeri light organ symbionts isolated in this study were coincubated in mixed spots with the GFP tagged V. fischeri ES114 pVSV102. Strains that inhibit growth of V. fischeri ES114 do not show any GFP fluorescence in imaged colonies. Strains that allow growth of V. fischeri ES114 show GFP fluorescence in imaged colonies. Representative images are shown from three experimental trials, where each trial contained three resuspensions per strain, prepared from different colonies.
Aschtgen, M. S., Bernard, C. S., De Bentzmann, S., Lloubès, R., and Cascales, E. (2008). SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531. doi: 10.1128/JB.00945-08
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16, 53–64. doi: 10.1094/MPMI.2003.16.1.53
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104
Buchan, A., and Moran, M. A. (2005). Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677. doi: 10.1128/AEM.71.10.5665
Carruthers, M. D., Nicholson, P. A., Tracy, E. N., and Munson, R. S. (2013). Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388
Church, S. R., Lux, T., Baker-Austin, C., Buddington, S. P., and Michell, S. L. (2016). Vibrio vulnificus type 6 secretion system 1 contains anti-bacterial properties. PLoS One 11:e165500. doi: 10.1371/journal.pone.0165500
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016). Aim, load, fire: the type VI secretion system, a bacterial Nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Collins, A. J. (2014). Bacterial symbioses and the innate immune response of the model host: Euprymna scolopes, University of Connecticut Graduate School. University of Connecticut. Available at: https://opencommons.uconn.edu/dissertations/516
Collins, A. J., Fullmer, M. S., Gogarten, J. P., and Nyholm, S. V. (2015). Comparative genomics of Roseobacter clade bacteria isolated from the accessory nidamental gland of Euprymna scolopes. Front. Microbiol. 6:123. doi: 10.3389/fmicb.2015.00123
Collins, A. J., LaBarre, B. A., Wong Won, B. S., Shah, M. V., Heng, S., Choudhury, M. H., et al. (2012). Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl. Environ. Microbiol. 78, 4200–4208. doi: 10.1128/AEM.07437-11
Costa, T. R. D., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Darmon, E., and Leach, D. R. F. (2014). Bacterial genome instability. Microbiol. Mol. Biol. Rev. 78, 1–39. doi: 10.1128/mmbr.00035-13
Delmont, T. O., Quince, C., Shaiber, A., Esen, Ö. C., Lee, S. T. M., Rappé, M. S., et al. (2018). Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813. doi: 10.1038/s41564-018-0176-9
Dong, T. G., Ho, B. T., Yoder-Himes, D. R., and Mekalanos, J. J. (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in vibrio cholerae. Proc. Natl. Acad. Sci. 110, 2623–2628. doi: 10.1073/pnas.1222783110
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Enright, A. J., Iliopoulos, I., Kyrpides, N. C., and Ouzounis, C. A. (1999). Protein interaction maps for complete genomes based on gene fusion events. Nature 402, 86–90. doi: 10.1038/47056
Eren, A. M., Esen, Ö. C., Quince, C., Vineis, J. H., Morrison, H. G., Sogin, M. L., et al. (2015). Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, 1–29. doi: 10.7717/peerj.1319
Figureueras, M. J., Beaz-Hidalgo, R., Hossain, M. J., and Liles, M. R. (2014). Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc. 2, 6–7. doi: 10.1128/genomeA.00927-14
Gromek, S. M., Suria, A. M., Fullmer, M. S., Garcia, J. L., Gogarten, J. P., Nyholm, S. V., et al. (2016). Leisingera sp. JC1, a bacterial isolate from Hawaiian bobtail squid eggs, produces indigoidine and differentially inhibits vibrios. Front. Microbiol. 7:1342. doi: 10.3389/fmicb.2016.01342
Guckes, K. R., Cecere, A. G., Wasilko, N. P., Williams, A. L., Bultman, K. M., Mandel, M. J., et al. (2019). Incompatibility of vibrio fischeri strains during symbiosis establishment depends on two functionally redundant hcp genes. J. Bacteriol. 201:e00221-19. doi: 10.1128/JB.00221-19
Guckes, K. R., Cecere, A. G., Williams, A. L., McNeil, A. E., and Miyashiro, T. (2020). The bacterial enhancer binding protein vasH promotes expression of a type vi secretion system in vibrio fischeri during symbiosis. J. Bacteriol. 202:e00777-19. doi: 10.1128/JB.00777-19
Hood, R. D., Peterson, S. B., and Mougous, J. D. (2017). From striking out to striking gold: discovering that type VI secretion targets bacteria. Cell Host Microbe 21, 286–289. doi: 10.1016/j.chom.2017.02.001
Hood, R. D., Singh, P., Hsu, F. S., Güvener, T., Carl, M. A., Trinidad, R. R. S., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114–5118. doi: 10.1038/s41467-018-07641-9
Kempnich, M. W., and Sison-Mangus, M. P. (2020). Presence and abundance of bacteria with the type VI secretion system in a coastal environment and in the global oceans. PLoS One 15, e0244217–e0244216. doi: 10.1371/journal.pone.0244217
Kerwin, A. H., Gromek, S. M., Suria, A. M., Samples, R. M., Deoss, D. J., O’Donnell, K., et al. (2019). Shielding the next generation: symbiotic bacteria from a reproductive organ protect bobtail squid eggs from fungal fouling. MBio 10, 1–18. doi: 10.1128/mBio.02376-19
Kerwin, A. H., McAnulty, S. J., and Nyholm, S. V. (2021). Development of the accessory nidamental gland and associated bacterial community in the hawaiian bobtail squid, euprymna scolopes. Biol. Bull. 240, 205–218. doi: 10.1086/713965
Kerwin, A. H., and Nyholm, S. V. (2017). Symbiotic bacteria associated with a bobtail squid reproductive system are detectable in the environment, and stable in the host and in eggs throughout development. Environ. Microbiol. 19, 1463–1475. doi: 10.1111/1462-2920.13665
Li, J., Yao, Y., Xu, H. H., Hao, L., Deng, Z., Rajakumar, K., et al. (2015). SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. 17, 2196–2202. doi: 10.1111/1462-2920.12794
Marcotte, E. M., Pellegrini, M., Ng, H. L., Rice, D. W., Yeates, T. O., and Eisenberg, D. (1999). Detecting protein function and protein-protein interactions from genome sequences. Science 285, 751–753. doi: 10.1126/science.285.5428.751
Mariano, G., Trunk, K., Williams, D. J., Monlezun, L., Strahl, H., Pitt, S. J., et al. (2019). A family of type VI secretion system effector proteins that form ion-selective pores. Nat. Commun. 10:5484. doi: 10.1038/s41467-019-13439-0
Merk, R. (2006). A comparative categorization of protein function encoded in bacterial or archeal genomic islands. J. Mol. Evol. 62, 1–14. doi: 10.1007/s00239-004-0311-5
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Mougous, J. D., Gifford, C. A., Ramsdell, T. L., and Mekalanos, J. J. (2007). Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9, 797–803. doi: 10.1038/ncb1605
Murdoch, S. L., Trunk, K., English, G., Fritsch, M. J., Pourkarimi, E., and Coulthurst, S. J. (2011). The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193, 6057–6069. doi: 10.1128/JB.05671-11
Nyholm, S. V., and McFall-Ngai, M. J. (2021). A lasting symbiosis: how the Hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat. Rev. Microbiol. 19, 666–679. doi: 10.1038/s41579-021-00567-y
Nyholm, S. V., Stabb, E. V., Ruby, E. G., and McFall-Ngai, M. J. (2000). Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. U. S. A. 97, 10231–10235. doi: 10.1073/pnas.97.18.10231
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Persson, O. P., Pinhassi, J., Riemann, L., Marklund, B. I., Rhen, M., Normark, S., et al. (2009). High abundance of virulence gene homologues in marine bacteria. Environ. Microbiol. 11, 1348–1357. doi: 10.1111/j.1462-2920.2008.01861.x
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Ruby, E. G., and Asato, L. M. (1993). Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159, 160–167. doi: 10.4319/lo.2013.58.2.0489
Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W., and Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244
Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014a). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Russell, A. B., Wexler, A. G., Harding, B. N., Whitney, J. C., Bohn, A. J., Goo, Y. A., et al. (2014b). A type VI secretion-related pathway in bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236. doi: 10.1016/j.chom.2014.07.007
Salomon, D., Gonzalez, H., Updegraff, B. L., and Orth, K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One 8:e61086. doi: 10.1371/journal.pone.0061086
Sana, T. G., Hachani, A., Bucior, I., Soscia, C., Garvis, S., Termine, E., et al. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and fur and modulates internalization in epithelial cells. J. Biol. Chem. 287, 27095–27105. doi: 10.1074/jbc.M112.376368
Schwarz, S., Hood, R. D., and Mougous, J. D. (2010a). What is type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537. doi: 10.1016/j.tim.2010.09.001
Schwarz, S., West, T. E., Boyer, F., Chiang, W. C., Carl, M. A., Hood, R. D., et al. (2010b). Burkholderia type vi secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6, e1001068–e1001078. doi: 10.1371/journal.ppat.1001068
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Silverman, J. M., Austin, L. S., Hsu, F. S., Hicks, K. G., Hood, R. D., and Mougous, J. D. (2011). Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol. Microbiol. 82, 1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x
Silvester, R., Alexander, D., Antony, A. C., and Hatha, M. (2017). GroEL PCR-RFLP—an efficient tool to discriminate closely related pathogenic vibrio species. Microb. Pathog. 105, 196–200. doi: 10.1016/j.micpath.2017.02.029
Smith, S., Salvato, F., Garikipati, A., Kleiner, M., and Septer, A. N. (2021). Activation of the type VI secretion system in the squid symbiont vibrio fischeri requires the transcriptional regulator tasr and the structural proteins TssM and TssA. J. Bacteriol. 203:e0039921. doi: 10.1128/JB.00399-21
Speare, L., Cecere, A. G., Guckes, K. R., Smith, S., Wollenberg, M. S., Mandel, M. J., et al. (2018). Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc. Natl. Acad. Sci. U. S. A. 115, E8528–E8537. doi: 10.1073/pnas.1808302115
Speare, L., Smith, S., Salvato, F., Kleiner, M., and Septer, A. N. (2020). Environmental viscosity modulates interbacterial killing during habitat transition. MBio 11, e03060–e03019. doi: 10.1128/mBio.03060-19
Speare, L., Woo, M., Dunn, A. K., and Septer, A. N. (2022). A putative lipoprotein mediates cell-cell contact for type VI secretion system-dependent killing of specific competitors. MBio 13:e0308521. doi: 10.1128/mbio.03085-21
Stabb, E. V., Reich, K. A., and Ruby, E. G. (2001). Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183, 309–317. doi: 10.1128/JB.183.1.309-317.2001
Suria, A. M., Tan, K. C., Kerwin, A. H., Gitzel, L., Abini-Agbomson, L., Bertenshaw, J. M., et al. (2020). Hawaiian bobtail squid Symbionts inhibit marine bacteria via production of specialized metabolites including new bromoalterochromides BAC-D/D′. mSphere 5:e00166-20. doi: 10.1128/msphere.00166-20
Visick, K. L., Stabb, E. V., and Ruby, E. G. (2021). A lasting symbiosis: how Vibrio fischeri finds a squid partner and persists within its natural host. Nat. Rev. Microbiol. 19, 654–665. doi: 10.1038/s41579-021-00557-0
Wagner-Döbler, I., and Biebl, H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280. doi: 10.1146/annurev.micro.60.080805.142115
Wei, S. L., and Young, R. E. (1989). Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar. Biol. 103, 541–546. doi: 10.1007/BF00399586
Wu, H. Y., Chung, P. C., Shih, H. W., Wen, S. R., and Lai, E. M. (2008). Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in agrobacterium tumefaciens. J. Bacteriol. 190, 2841–2850. doi: 10.1128/JB.01775-07
Keywords: type VI secretion system, Euprymna scolopes, competition, symbiosis, Aliivibrio fischeri, roseobacter
Citation: Suria AM, Smith S, Speare L, Chen Y, Chien I, Clark EG, Krueger M, Warwick AM, Wilkins H and Septer AN (2022) Prevalence and diversity of type VI secretion systems in a model beneficial symbiosis. Front. Microbiol. 13:988044. doi: 10.3389/fmicb.2022.988044
Received: 06 July 2022; Accepted: 24 August 2022;
Published: 14 September 2022.
Edited by:
Chih-Horng Kuo, Academia Sinica, TaiwanReviewed by:
Stephen Lloyd Michell, University of Exeter, United KingdomCopyright © 2022 Suria, Smith, Speare, Chen, Chien, Clark, Krueger, Warwick, Wilkins and Septer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alecia N. Septer, YXNlcHRlckBlbWFpbC51bmMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.