
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 October 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.987613
This article is part of the Research TopicDealing with Unusual Hosts and Unconventional Habitats: Versatile Strategies of Salmonella entericaView all 6 articles
 Ying Guan1†
Ying Guan1† Yanwan Li1†
Yanwan Li1† Jin Li1
Jin Li1 Zhishuang Yang1
Zhishuang Yang1 Dekang Zhu1,2,3
Dekang Zhu1,2,3 Renyong Jia1,2,3
Renyong Jia1,2,3 Mafeng Liu1,2,3
Mafeng Liu1,2,3 Mingshu Wang1,2,3
Mingshu Wang1,2,3 Shun Chen1,2,3
Shun Chen1,2,3 Qiao Yang1,2,3
Qiao Yang1,2,3 Ying Wu1,2,3
Ying Wu1,2,3 Shaqiu Zhang1,2,3
Shaqiu Zhang1,2,3 Qun Gao1
Qun Gao1 Xumin Ou1
Xumin Ou1 Sai Mao1
Sai Mao1 Juan Huang1
Juan Huang1 Di Sun1
Di Sun1 Bin Tian1
Bin Tian1 Anchun Cheng1,2,3*
Anchun Cheng1,2,3* Xinxin Zhao1,2,3*
Xinxin Zhao1,2,3*Salmonella enterica is a widespread foodborne pathogen with concerning antimicrobial resistance (AMR). Waterfowl are a major source of Salmonella transmission, but there are few systematic studies on Salmonella prevalence in waterfowl species. In this study, 126 Salmonella isolates (65 collected in 2018–2020 and 61 collected in 2002–2005) were obtained from waterfowl samples in Sichuan, China. Their serotypes, pulsed-field gel electrophoresis (PFGE) types, and phenotypic and genotypic AMR profiles were systematically examined. The isolates were distributed in 7 serotypes, including serovars Enteritidis (46.0%), Potsdam (27.8%), Montevideo (7.9%), Cerro (6.3%), Typhimurium (4.8%), Kottbus (4.0%) and Apeyeme (3.2%). Their PFGE characteristics were diverse; all isolates were distributed in four groups (cutoff value: 60.0%) and 20 clusters (cutoff value: 80.0%). Moreover, all isolates were multidrug resistant, and high rates of AMR to lincomycin (100.0%), rifampicin (100.0%), sulfadiazine (93.7%), erythromycin (89.7%), ciprofloxacin (81.0%), and gentamicin (75.4%) were observed. Finally, 49 isolates were subjected to whole-genome sequencing, and a wide variety of AMR genes were found, including multiple efflux pump genes and specific resistance genes. Interestingly, the tet(A)/tet(B) and catII resistance genes were detected in only isolates obtained in the first collection period, while the gyrA (S83F, D87N and D87G) and gyrB (E466D) mutations were detected at higher frequencies in the isolates obtained in the second collection period, supporting the findings that isolates from different periods exhibited different patterns of resistance to tetracycline, chloramphenicol and nalidixic acid. In addition, various incompatible plasmid replicon fragments were detected, including Col440I, Col440II, IncFIB, IncFII, IncX1, IncX9, IncI1-I and IncI2, which may contribute to the horizontal transmission of AMR genes and provide competitive advantages. In summary, we demonstrated that the Salmonella isolates prevalent in Sichuan waterfowl farms exhibited diverse serotypes, multiple AMR phenotypes and genotypes, and AMR changes over time, indicating their potential risks to public health.
Salmonella enterica is a facultative anaerobic gram-negative bacterium with more than 2,600 serotypes and an important zoonotic pathogen worldwide (Grimont and Weill, 2007). Human consumption of Salmonella-contaminated products can cause diarrhea, intestinal inflammation, and even bacteremia (Fearnley et al., 2011; Chousalkar et al., 2018; Tack et al., 2019), adding to the global burden of disease. Salmonella was previously reported to have caused approximately 22.2% (12,769 cases) of foodborne illness cases in China between 1994 and 2005 (Wang et al., 2007), and one of the sources of illness was waterfowl. China is the largest producer and consumer of waterfowl, such as ducks and geese, and related products (Wang et al., 2017). Recent studies have shown that waterfowl are an important source of Salmonella (Wang et al., 2020a; Kim et al., 2021) and often transmit Salmonella due to open-yard feeding (Murray et al., 2021); however, relevant systematic studies on Salmonella prevalence in these species have rarely been reported.
Because of diverse Salmonella serotypes and possible monophasic variation, simple slide agglutination assays to detect rare serotypes are often labor intensive and time consuming and have a risk of misidentification (Uelze et al., 2020). Sequence-based serotyping approaches, such as the Salmonella in silico typing resource (SISTR), can be used as a complementary method; this method was reported to have an accuracy of up to 94% (Yoshida et al., 2016). The in silicon method allows the detection of antigen genes carried by an isolate, while the slide agglutination method allows the detection of antigens expressed by an isolate (Yachison et al., 2017). A combination of these two methods can yield more accurate results. Pulsed-field gel electrophoresis (PFGE) has emerged as a method for analyzing large molecules of DNA (Sharma-Kuinkel et al., 2016) and has been widely used in molecular epidemiological investigations of foodborne pathogens (e.g., Escherichia coli, Salmonella enterica and Listeria monocytogenes; Favier et al., 2013; Li et al., 2020a). PFGE results reflect the genetic relationships among different isolates, allowing the rapid monitoring, tracking and tracing of bacterial infections.
In recent decades, antimicrobial agents have been used frequently in animal husbandry not only to treat and control Salmonella and other pathogens but also as prophylactic measures and growth-promotors. The abuse and misuse of antimicrobials has led to antimicrobial resistance (AMR), which is still increasing (Palma et al., 2020). To address this problem, use of antimicrobials as a growth-promoting factors has been banned in the European Union since 2006 (Castanon, 2007). China is one of the world’s largest producers and consumers of antimicrobials, with 162,000 tons of antimicrobials used in 2013, 52% of which were for veterinary use (Zhang et al., 2015). Over the past two decades, China’s restrictive policies on veterinary antimicrobials have changed substantially and become increasingly stringent (Yinqi et al., 2019). Consequently, the prevalence of colistin-resistant Escherichia coli in pigs and chickens decreased dramatically from 2015 to 2018 due to the withdrawal of colistin as an animal growth promoter in China (Wang et al., 2020b), highlighting the impact of addressing AMR. Nevertheless, there are still numerous reports showing that the Salmonella prevalence in poultry, pigs and eggs in China had exhibited increasing resistance to multiple antimicrobials (Yang et al., 2019; Li et al., 2020b, 2021; Xu et al., 2021). Thus, it is of interest and importance to perform continuous AMR monitoring and to investigate changes in AMR over time, which is crucial in identifying the mechanisms involved and providing guidance on rational treatment strategies. Characterization of Salmonella resistance is mainly performed by determining phenotypes based on antimicrobial susceptibility testing (AST; CLSI, 2020) and genotypes based on whole-genome sequencing (WGS; Schwan et al., 2021; Yan et al., 2021; Medina-Santana et al., 2022). Phenotypes usually correspond to the external expression of a single gene but may also be the result of synergistic effects of multiple genes (Morales et al., 2005). Therefore, both phenotypic and genotypic testing are necessary for pathogen surveillance and diagnosis, and using them together can lead to more accurate judgments.
In this study, 126 Salmonella isolates (including 65 collected in 2018–2020 and 61 collected in 2002–2005) obtained from waterfowl samples in Sichuan, China, were subjected to serotyping, PFGE molecular typing, AST, and WGS to investigate their prevalence and AMR profiles.
All samples were collected from waterfowl farms with animal deaths, diarrhea or declining egg production. One hundred and 21 duckling organ samples were collected from Pujiang and Xinjin farms in 2002–2005, and 150 samples, including 70 duck fecal samples, 43 duck cloacal samples and 37 goose egg samples, were collected from five farms in Dayi, Chongzhou, Jintang, Mianyang and Pengzhou of Sichuan Province, China, in 2018–2020. All samples were subjected to isolation according to a standard protocol described previously (Andrews et al., 2022). In brief, for goose eggs and cloacal samples, samples were collected with sterile swabs and diluted in 1 ml of phosphate-buffered saline (PBS). Then, 100 μl of this solution was added to 10 ml of buffered peptone pre-enrichment solution, followed by incubation at 37°C for 24 h. For fecal and organ samples, 1 g of each was weighed, ground and added to 10 ml of buffered peptone pre-enrichment solution, followed by incubation at 37°C for 24 h. Subsequently, 1 ml of pre-enrichment solution was added to 10 ml of Salmonella-specific selenite cystine (SC) enrichment solution, followed by incubation at 37°C for 24 h, and then 100 μl of the solution was applied to plates with xylose lysine deoxycholate agar (XLD) medium. Putative black colonies on XLD medium were selected and subjected to polymerase chain reaction (PCR) identification by using Salmonella-specific primers hut-F/R (hut-F: atgttgtcctgcccctggtaagaga, hut-R: actggcgttatccctttctctg) to confirm (Alzwghaibi et al., 2018). All identified isolates were stored in 15% (v/v) glycerol at −80°C.
Serotyping of the isolates was carried out by slide agglutination of flagellar antigen (H) and somatic antigen (O) with a Salmonella Diagnostic Serum Kit (Tianrun Biopharmaceuticals, Ningbo, China) according to the manufacturer’s instructions (Lee et al., 2015). Some nonagglutinable isolates were further determined by the SISTR v1.1.1 using WGS data (Yoshida et al., 2016).
The genetic relationships among the isolates were determined by the PFGE method according to the PulseNet protocol (CDC, 2017). XbaI (New England Biolabs, Ipswich, MA, USA) was used as the restriction enzyme. Clustering analysis was performed by Bionumerics v7.6 (Applied Maths NV, Sint-Martens-Latem, Belgium) using the unweighted pair-group average method with band-matching settings of 1.0% optimization and 1.5% position tolerance (Tian et al., 2021). Salmonella enterica serovar Braenderup H9812 was included for quality control.
The susceptibility of the isolates to ten classes of 20 antimicrobials was determined by the Kirby-Bauer disk diffusion method (Bauer et al., 1966). The categories (susceptible, intermediate resistance or resistance) were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020). The antimicrobial agents employed were as follows: tetracycline (TET, 30 μg), aztreonam (ATM, 30 μg), ampicillin (AMP, 10 μg), trimethoprim/sulfamethoxazole (SXT, 1.25/23.75 μg, respectively), nalidixic acid (NAL, 30 μg), gentamycin (GEN, 10 μg), amoxicillin (AML, 25 μg), chloramphenicol (CHL, 30 μg), polymyxin B (PB, 300 μg), streptomycin (STR, 10 μg), trimethoprim (W, 5 μg), sulfadiazine (SUL, 100 μg), ciprofloxacin (CIP, 5 μg), ceftiofur (EFT, 30 μg), cefepime (FEP, 30 μg), imipenem (IPM, 10 μg), lincomycin (MY, 2 μg), florfenicol (FFC, 30 μg), erythromycin (E, 15 μg), and rifampin (RD, 5 μg). The Escherichia coli reference strain ATCC 25922 was used for quality control. Two-way ANOVA and the Pearson chi-square test were used to determine the difference in the overall AMR rates of Salmonella and the difference in the AMR rates to a particular antimicrobial agent between the two periods.
Based on the results of PFGE typing and AST, 49 isolates with less than 80% homology, different antimicrobial phenotypes or nonagglutinable phenotypes were subjected to WGS. Their genomic DNA was extracted by using a bacterial genomic DNA extraction kit (TIANGEN Biotechnology Co., Ltd., Beijing, China). The genomic DNA was sent to the Beijing Genomics Institute (BGI Co., Ltd., Shenzhen, China) for frame sequencing (Illumina HiSeq 2000) and splicing (SPAdes v3.14.0). The sequencing results were analyzed using the Comprehensive Antimicrobial Resistance Database (CARD v3.2.4; Alcock et al., 2020) to annotate the AMR genes of the isolates and to analyze the relationships between genotypes and phenotypes. Incompatible fragments of plasmids were predicted using PlasmidFinder v2.0.1 with a similarity cutoff value of 95% (Camacho et al., 2009; Carattoli et al., 2014). Average nucleotide identity (ANI) levels were calculated by using CJ Bioscience’s online calculator (Yoon et al., 2017).
Sixty-five Salmonella isolates were obtained from 150 samples collected in 2018–2020, with an isolation rate of 43.3% (65/150); additionally, 61 isolates were obtained from 121 organ samples collected in 2002–2005, with an isolation rate of 50.4% (61/121). Serotyping with antiserum found that 108 of the 126 Salmonella isolates were agglutinable and 18 isolates were untypable. The uncertain serotypes were further determined by using an in silico typing method. A total of seven serotypes were detected and distributed in six serogroups: serovar Typhimurium in group B; serovar Montevideo and Potsdam in group C1; serovar Kottbus in group C2; serovar Apeyeme in group C3; serovar Enteritidis in group D1; and serovar Cerro in group K (Table 1). The serotype distribution of these isolates was as follows: 46.0% (58/126) were serovar Enteritidis; 27.8% (35/126) were serovar Potsdam; 7.9% (10/126) were serovar Montevideo; 6.3% (8/126) were serovar Cerro; 4.8% (6/126) were serovar Typhimurium; 4.0% (5/126) were serovar Kottbus; and 3.2% (4/126) were serovar Apeyeme (Table 1).
PFGE typing of the 126 isolates resulted in diverse band characteristics, with all isolates classified into four groups (cutoff value: 60.0%), designated A, B, C and D, containing 58, 38, 9 and 21 isolates, respectively; moreover, the pulsotypes were further subdivided into 20 clusters (cutoff value: 80.0%; Supplementary Figure S1). Notably, all serovar Enteritidis isolates collected in 2002–2005 belonged to group A, with a relatively high similarity of 78.0%; all 34 isolates obtained from goose eggs in 2020 belonged to group B, while an isolate named RCAD-S-122 had only 66.7% homology with the remaining 33 isolates (Supplementary Figure S1). In contrast, the isolates from fecal and cloacal samples collected in 2018–2020 showed diversity on the PFGE dendrogram, with 21 isolates in group D, 6 isolates in group C, and 4 isolates in group B (Supplementary Figure S1). Moreover, there were no significant correlations between serotypes and pulsotypes within some isolates. For example, two (RCAD-S-023 and RCAD-S-024) of the Cerro isolates had only 61.7% PFGE homology with the remaining Cerro isolates; additionally, the serovar Potsdam isolate RCAD-S-015 and the serovar Montevideo isolate RCAD-S-016 showed identical pulsotypes with 100% PFGE homology (Figure 1). Comparison of the genome sequences with similar band characteristics found that the ANI levels between RCAD-S-015 and RCAD-S-016 were lower than those of RCAD-S-023 and RCAD-S-024 with the same serotype (98.3% versus 99.8%; Supplementary Table 1).
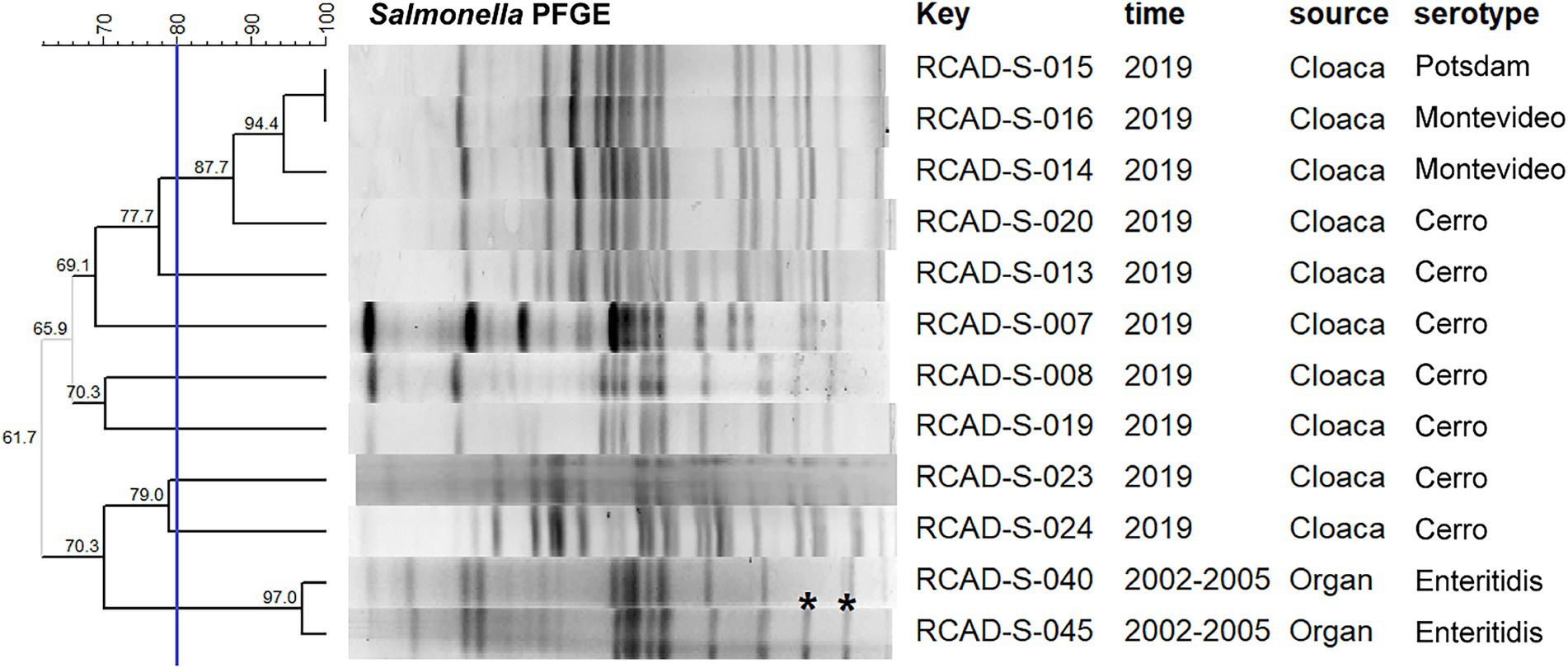
Figure 1. Dendrogram of the PFGE pulsotypes for 12 representative isolates. The blue line indicates the 80% cutoff value. The “key” column represents the different isolates; RCAD, Research Center of Avian Diseases. Asterisks indicate differences between adjacent bands.
All isolates were resistant to three or more classes of antimicrobials, suggesting multidrug resistance (MDR; Supplementary Table 2). The isolates displayed varying rates of resistance to the 20 antimicrobial agents that were tested. High resistance rates were observed for MY (100.0%), RD (100.0%), SUL (93.7%), E (89.7%), CIP (81.0%), and GEN (75.4%); however, the rates of resistance to the remaining 14 antimicrobials, namely, EFT (46.8%), TET (41.3%), AMP (24.6%), STR (20.6%), ATM (16.7%), NAL (15.1%), IPM (9.5%), AML (7.9), SXT (7.1%), CHL (7.1%), FEP (5.6%), W (4.0%), FFC (3.2%) and PB (1.6%), were less than 50%, indicating that these isolates were generally more resistant to conventional antimicrobials (Table 2).
The 61 isolates from the first collection period (2002–2005) exhibited higher resistance rates to SUL, E, CIP, TET, AMP and IPM than the 65 isolates from the second collection period (2018–2020); in contrast, the isolates from the second collection period showed higher resistance rates to GEN, EFT, STR, NAL and SXT (Table 2). In particular, dramatic differences in AMR rates were observed for EFT (23.0% versus 69.2%) and TET (73.8% versus 10.8%) in isolates from both periods (Table 2). Additionally, diverse resistance phenotypes were observed between isolates with the same serotype and with high homology of pulsotypes. For example, the two serovar Enteritidis isolates RCAD-S-040 and RCAD-S-045 showed 97.0% pulsotype similarity, with only slight differences found in the low-molecular weight bands (Figure 1), but they displayed quite different resistance profiles to 7 antimicrobials (TET, AMP, STR, ATM, NAL, SXT, and CHL; Supplementary Table 2).
Multiple specific AMR genes (aac(6′)-Iy, aac(6′)-Iaa, aac(3)-IId, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, blaTEM-1, blaTEM-116, catII, dfrA27, sul1, sul2, mcr-1.1, gyrA, gyrB, qnrB6, tet(A), tet(B) and tetR) and efflux pump genes (mdsA, adeF, golS, sdiA, acrA, acrB, marA, marR, baeR, baeS, rsmA, crp, H-NS, mdtK, mdfA, kpnE, kpnF, emrB, emrR, soxS, soxR, msbA, glpT, uhpT, EF-Tu and kdpE) were detected in the overall genome or plasmid sequences of the 49 isolates (Supplementary Table 3). Analysis of AMR phenotypes and the specific AMR genes in each isolate indicated that there were specific AMR genes, including the beta-lactam inactivating enzyme gene blaTEM-1, amphenicol inactivating enzyme gene catII and tetracycline efflux pump genes tet(A) and tet(B), existing in the isolates with phenotypic resistance to AML, CHL and TET, respectively, whereas there were no direct correspondences to known AMR genes in isolates with phenotypic resistance to MY, RD, E, GEN, STR, EFT, ATM, SUL and CIP (Figure 2). Furthermore, in contrast to tet genes (which provide the TET-resistant phenotype), which were present in only the isolates obtained in the first collection period, abundant gyrA (S83F, D87N, and D87G) and gyrB (E466D) mutations (providing the NAL-resistant phenotype) were detected in the isolates obtained in the second collection period (Figure 2).
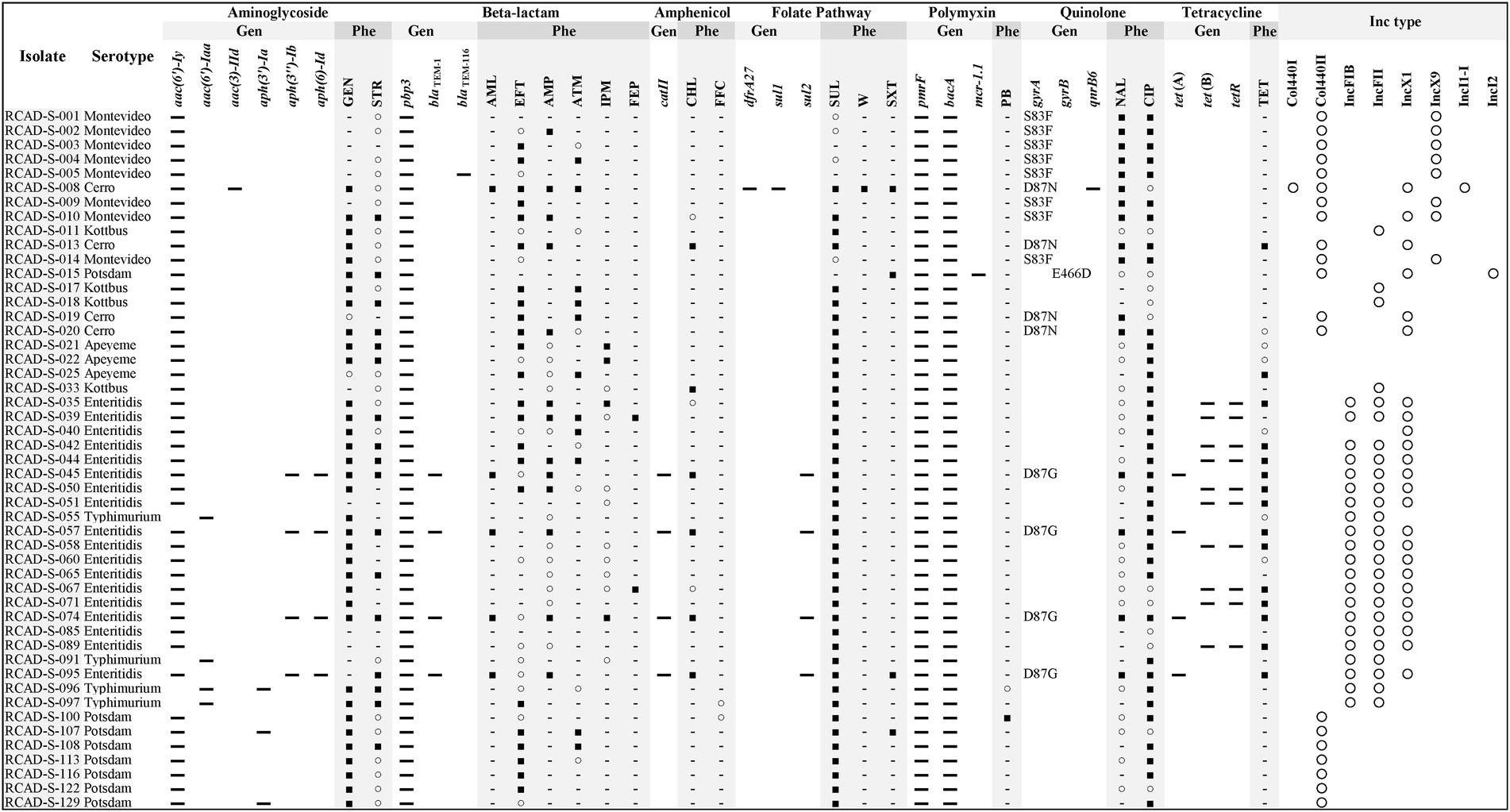
Figure 2. Comparison of antimicrobial resistance (AMR) profiles and plasmid replicons of the 49 Salmonella isolates. For the gyrA and gyrB genes, mutation sites are shown. Gen, genotype; Phe, phenotype; and Inc. type, incompatible type. ▬, Hit resistance gene; ■, resistant; ○, susceptible, increased exposure; −, susceptible, standard dosing regimen; and ○, hit plasmid replicons. SUL, sulfadiazine; CIP, ciprofloxacin; GEN, gentamycin; EFT, ceftiofur; TET, tetracycline; AMP, ampicillin; STR, streptomycin; ATM, aztreonam; NAL, nalidixic acid; IPM, imipenem; SXT, trimethoprim/sulfamethoxazole; AML, amoxicillin; CHL, chloramphenicol; FEP, cefepime; W, trimethoprim; FFC, florfenicol; and PB, polymyxin B.
Plasmid replicon fragments with different incompatibility groups, including Col440I, Col440II, IncFIB, IncFII, IncX1, IncX9, IncI1-I, and IncI2, were detected in 46 of the 49 isolates. Most isolates contained more than one replicon fragment, such as RCAD-S-008 (Col440I, Col440II, IncX1, and IncI1-I), indicating that these isolates may harbor one or more plasmids (Figure 2). The incompatible fragments contained in isolates from different times or sources differed significantly, with isolates from the first collection period mainly containing IncFIB, IncFII, and IncX1, while isolates from the second collection period showed more diversity, except the goose egg isolates, which contained only Col440II (Figure 2). It is worth noting that although RCAD-S-040 and RCAD-S-045 share the same serotype and have high PFGE (97.0%) and ANI (99.9%) similarity (Figure 1; Supplementary Table 1), their AMR genotypes and phenotypes are significantly different, with RCAD-S-045 additionally containing IncFIB and IncFII, as well as the gyrA (D87G) mutation and 6 resistance genes [(aph(3″)-Ib, aph(6)-Id, blaTEM-1, catII, sul2 and tet(A)); Figure 2].
The seven serotypes identified in this study showed some diversity, and the serovars Enteritidis, Montevideo, Potsdam and Typhimurium are also reported to be the major serotypes prevalent in the global poultry industry (Shah et al., 2017; Yang et al., 2020; Diaz et al., 2021). The high homology of the PFGE patterns among 58 serovar Enteritidis isolates and 34 serovar Potsdam isolates indicated that a dominant clone was prevalent locally. In contrast, the remaining isolates showed quite dissimilar PFGE patterns, suggesting that they have genotypic diversity and that various Salmonella clones were prevalent in different waterfowl farms in Sichuan, China. As with many epidemiological surveys, the prevalence of a particular pathogen is usually regional in nature (Yan et al., 2021). Comparisons of PFGE and serotype results revealed the following three association patterns. First, most of the isolates with the same serotypes exhibited similar PFGE band distributions and thus were grouped into the same PFGE cluster, such as serovar Enteritidis and Potsdam isolates; this indicated that their genomic arrangements were similar and that their homology was relatively high. Second, several isolates with the same serotype, such as serovar Cerro, exhibited significant heterogeneity in their PFGE patterns, with alterations in the band distributions, suggesting that these isolates may have undergone genomic rearrangement, resulting in changes in the enzymatic cutting site that did not affect their surface antigen composition. This phenomenon is supported by a previous report that 46 Salmonella enterica serovar Schwarzengrund isolates, with a considerable length of evolutionary time, are still of the same serotype (Yang et al., 2022). Finally, individual isolates exhibited similar pulsotypes but different serotypes; this was observed between the serovar Potsdam isolate RCAD-S-015 (6,7,14:l,v,e,n,z15) and the serovar Montevideo isolate RCAD-S-016 (6,7,14,[54]:g,m,s:-). Previous reports have also shown that epidemiologically unrelated isolates can be assigned to identical PFGE types (Barco et al., 2013; Shi et al., 2015), such as serovar Typhimurium (1,4,[5],12:i:1,2) versus 4,5,12:i:- (Ranieri Matthew et al., 2013) and serovar Thompson (6,7,14:k,1,5) versus 1,7:-:1,5 (Soyer et al., 2010). Since XbaI PFGE may not be discriminatory enough in some cases, we confirmed by WGS that the level of ANI between these two isolates was not as high as those between the same serotypes.
Owing to the excessive use of antimicrobial agents in animal husbandry over the last few decades and the horizontal spread of resistance genes, AMR in Salmonella has become a major concern (Foley et al., 2008). All isolates identified in this study were MDR and were broadly resistant to conventional antimicrobials, suggesting that most bacteria have acquired resistance traits under prolonged selection pressures, which is consistent with many previous findings (Yang et al., 2020; Sun et al., 2021; Wang et al., 2021). We also found variations in the resistance patterns of isolates from different periods, with the isolates obtained in the first collection period exhibiting a marked increase in resistance to EFT and newer-generation antimicrobials as well as an increase in susceptibility to TET. This trend may be attributed to the “strictest antibiotic regulation” enacted by the Chinese government in 2012, which allowed the use of veterinary antimicrobials such as EFT (Yinqi et al., 2019); increased resistance to EFT was also reported in Salmonella from pigs in Sichuan from 2009 (11.4%) to 2014 (53.8%; Xiuying et al., 2015). In contrast, a study from Canada showed the opposite trend, with resistance to EFT decreasing from 62 to 18% from 2004 to 2008 (Dutil et al., 2010). Variations in resistance over time suggest that the resistance phenotypes may change with the use of veterinary antimicrobials in upcoming years (Hornish and Kotarski, 2002; Sato et al., 2014). In particular, a small proportion of isolates also showed moderate resistance to human-restricted antimicrobials, such as IPM and SXT, implying possible cross-transmission between humans and animals or, more likely, the acquisition of new specific resistance genes (Wall et al., 2016).
WGS has become a reliable method for the detection of resistance genes, allowing the accurate identification of individual resistance genes in addition to the identification of single nucleotide mutations; this knowledge may even be applied to predict unknown resistance genes according to their conserved structural domains (Köser et al., 2014; Rokney et al., 2020). Specific resistance genes against different types of antimicrobials were detected in the 49 isolates. Some of the resistance genomic and phenotypic characteristics were consistent; for instance, the presence of the catII and tet genes conferred CHL and TET resistance phenotypes, respectively. However, there were substantial inconsistencies, and some isolates did not contain specific resistance genes but showed a resistance phenotype, such as serovar Potsdam isolated from goose eggs with the CIP phenotype, which may be attributed to unknown resistance mechanisms or nonspecific functions of multiple redundant efflux pump-like genes. As previously reported, CIP resistance is influenced by the coordination between multiple genes (gyrA, gyrB, parE, and acrB; O'Regan et al., 2009). Additionally, some isolates possessed specific resistance genes but showed susceptibility to the corresponding antimicrobial, which may be due to mutations or functional incompleteness of the gene; for example, blaTEM-116 in RCAD-S-005 was not observed to be resistant to beta-lactams. Eight types of incompatible plasmid replicon fragments were detected in these 49 isolates. It is reasonable to assume that the AMR differences between the representative isolates RCAD-S-040 and RCAD-S-045 stems from the different plasmids they contain, and the same situation exists for RCAD-S-014 and RCAD-S-16. Since WGS does not provide a complete map, it is not possible to determine which plasmid a specific resistance gene is located on. As plasmids play a vital role in the horizontal transfer of resistance genes, their sequences and contributions to AMR need further investigation. Overall, the analysis of resistance phenotypes and genotypes suggests that their AMR profiles may be a result of long-term stress through mechanisms such as target alteration or horizontal gene transfer by mobile genetic elements (Bakkeren et al., 2022; Cohen et al., 2022).
Since our samples were obtained from waterfowl with pathological symptoms, we obtained a higher isolation rate than those reported in other studies and consequently a higher rate of AMR (Chen et al., 2020; Han et al., 2020). The shortcomings of our study include an insufficient sample size and the lack of a wide geographical distribution. Nevertheless, our study not only complements the epidemiological surveillance data for monitoring Salmonella of waterfowl origin but also has practical implications for guiding the use of antimicrobial agents in waterfowl in this region.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI: PRJNA857101.
XZ and AC conceived the study and edited the manuscript. YG and YL performed most of the research and drafted the manuscript. JL, DZ, RJ, ML, MW, and SC participated in sample collection and bacterial isolation. ZY analyzed the genome sequences. SZ, QY, YW, and QG did the PFGE detection. XO, SM, JH, DS, and BT participated in analysis of antimicrobial resistance. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (32072877), the Applied Basic Research Programs of Science and Technology Department of Sichuan Province (2019YJ0436), and the earmarked fund for China Agriculture Research System (CARS-42-17).
We thank Hao Tu, Xiaoli Zeng, Hui Shen, Yi Liao, and Fuxiang Yang in our laboratory for their generous assistance during the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.987613/full#supplementary-material
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alzwghaibi, A. B., Yahyaraeyat, R., Fasaei, B. N., Langeroudi, A. G., and Salehi, T. Z. (2018). Rapid molecular identification and differentiation of common salmonella serovars isolated from poultry, domestic animals and foodstuff using multiplex PCR assay. Arch. Microbiol. 200, 1009–1016. doi: 10.1007/s00203-018-1501-7
Andrews, W. H., Hua, W., Andrew, J., Beilei, G., Guodong, Z., and Thomas, H. (2022). Bacteriological analytical manual [online]. Available at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella (Accessed January 2022).
Bakkeren, E., Gül, E., Huisman, J. S., Steiger, Y., Rocker, A., Hardt, W. D., et al. (2022). Impact of horizontal gene transfer on emergence and stability of cooperative virulence in salmonella typhimurium. Nat. Commun. 13:1939. doi: 10.1038/s41467-022-29597-7
Barco, L., Barrucci, F., Olsen, J. E., and Ricci, A. (2013). Salmonella source attribution based on microbial subtyping. Int. J. Food Microbiol. 163, 193–203. doi: 10.1016/j.ijfoodmicro.2013.03.005
Bauer, A. W., Kirby, W. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST +: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Castanon, J. I. (2007). History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 86, 2466–2471. doi: 10.3382/ps.2007-00249
CDC (2017). Standard operating procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri [Online]. Available: https://www.cdc.gov/pulsenet/pdf/listeria-pfge-protocol-508c.pdf (Accessed June 2022).
Chen, Z., Bai, J., Wang, S., Zhang, X., Zhan, Z., Shen, H., et al. (2020). Prevalence, antimicrobial resistance, virulence genes and genetic diversity of salmonella isolated from retail duck meat in southern China. Microorganisms 8, 290. doi: 10.3390/microorganisms8030444
Chousalkar, K., Gast, R., Martelli, F., and Pande, V. (2018). Review of egg-related salmonellosis and reduction strategies in United States, Australia, United Kingdom and New Zealand. Crit. Rev. Microbiol. 44, 290–303. doi: 10.1080/1040841X.2017.1368998
CLSI (2020). Performance standards for antimicrobial susceptibility testing [online]. Available at: https://clsi.org/standards/products/microbiology/documents/m100/ (Accessed June 2022).
Cohen, E., Kriger, O., Amit, S., Davidovich, M., Rahav, G., and Gal-Mor, O. (2022). The emergence of a multidrug resistant salmonella Muenchen in Israel is associated with horizontal acquisition of the epidemic pESI plasmid. Clin. Microbiol. Infect. doi: 10.1016/j.cmi.2022.05.029 (Accessed June 2022).
Diaz, D., Hernandez-Carreño, P. E., Velazquez, D. Z., Chaidez-Ibarra, M. A., Montero-Pardo, A., Martinez-Villa, F. A., et al. (2021). Prevalence, main serovars and anti-microbial resistance profiles of non-typhoidal salmonella in poultry samples from the Americas: a systematic review and meta-analysis. Transbound. Emerg. Dis. doi: 10.1111/tbed.14362
Dutil, L., Irwin, R., Finley, R., Ng, L. K., Avery, B., Boerlin, P., et al. (2010). Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 16, 48–54. doi: 10.3201/eid1601.090729
Favier, G. I., Lucero Estrada, C. S. M., Otero, V. L., and Escudero, M. E. (2013). Prevalence, antimicrobial susceptibility, and molecular characterization by PCR and pulsed field gel electrophoresis (PFGE) of salmonella spp. isolated from foods of animal origin in San Luis, Argentina. Food Control 29, 49–54. doi: 10.1016/j.foodcont.2012.05.056
Fearnley, E., Raupach, J., Lagala, F., and Cameron, S. (2011). Salmonella in chicken meat, eggs and humans; Adelaide, South Australia, 2008. Clin. Microbiol. Infect. 146, 219–227. doi: 10.1016/j.ijfoodmicro.2011.02.004
Foley, S. L., Lynne, A. M., and Nayak, R. (2008). Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 86, E149–E162. doi: 10.2527/jas.2007-0464
Grimont, P. A., and Weill, F. X. (2007). Antigenic Formulae of the Salmonella serovars. WHO collaborating centre for reference and research on Salmonella. Paris: WHO 1–166.
Han, X., Peng, J., Guan, X., Li, J., Huang, X., Liu, S., et al. (2020). Genetic and antimicrobial resistance profiles of salmonella spp. isolated from ducks along the slaughter line in southwestern China. Food Control 107:106805. doi: 10.1016/j.foodcont.2019.106805
Hornish, R. E., and Kotarski, S. F. (2002). Cephalosporins in veterinary medicine - ceftiofur use in food animals. Curr. Top. Med. Chem. 2, 717–731. doi: 10.2174/1568026023393679
Kim, T. S., Kim, G. S., Son, J. S., Lai, V. D., Mo, I. P., and Jang, H. (2021). Prevalence, biosecurity factor, and antimicrobial susceptibility analysis of salmonella species isolated from commercial duck farms in Korea. Poult. Sci. 100:100893. doi: 10.1016/j.psj.2020.12.006
Köser, C. U., Ellington, M. J., and Peacock, S. J. (2014). Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 30, 401–407. doi: 10.1016/j.tig.2014.07.003
Lee, K., Runyon, M., Herrman, T. J., Phillips, R., and Hsieh, J. (2015). Review of salmonella detection and identification methods: aspects of rapid emergency response and food safety. Food Control 47, 264–276. doi: 10.1016/j.foodcont.2014.07.011
Li, W., Li, H., Zheng, S., Wang, Z., Sheng, H., Shi, C., et al. (2020a). Prevalence, serotype, antibiotic susceptibility, and genotype of salmonella in eggs from poultry farms and marketplaces in Yangling, Shaanxi Province, China. Front. Microbiol. 11:1482. doi: 10.3389/fmicb.2020.01482
Li, C., Wang, Y., Gao, Y., Li, C., Ma, B., and Wang, H. (2021). Antimicrobial resistance and CRISPR typing among salmonella isolates from poultry farms in China. Front. Microbiol. 12:730046. doi: 10.3389/fmicb.2021.730046
Li, Y., Yang, X., Zhang, H., Jia, H., Liu, X., Yu, B., et al. (2020b). Prevalence and antimicrobial susceptibility of salmonella in the commercial eggs in China. Int. J. Food Microbiol. 325:108623. doi: 10.1016/j.ijfoodmicro.2020.108623
Medina-Santana, J. L., Ortega-Paredes, D., de Janon, S., Burnett, E., Ishida, M., Sauders, B., et al. (2022). Investigating the dynamics of salmonella contamination in integrated poultry companies using a whole genome sequencing approach. Poult. Sci. 101:101611. doi: 10.1016/j.psj.2021.101611
Morales, C. A., Porwollik, S., Frye, J. G., Kinde, H., McClelland, M., and Bouldin, J. G. (2005). Correlation of phenotype with the genotype of egg-contaminating Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 71, 4388–4399. doi: 10.1128/AEM.71.8.4388-4399.2005
Murray, C. E., Varga, C., Ouckama, R., and Guerin, M. T. (2021). Temporal study of salmonella enterica serovars isolated from fluff samples from Ontario poultry hatcheries between 2009 and 2018. Pathogens 11, 9. doi: 10.3390/pathogens11010009
O'Regan, E., Quinn, T., Pagès, J. M., McCusker, M., Piddock, L., and Fanning, S. (2009). Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in salmonella enterica serovar enteritidis: involvement of RamA and other global regulators. Antimicrob. Agents Chemother. 53, 1080–1087. doi: 10.1128/aac.01005-08
Palma, E., Tilocca, B., and Roncada, P. (2020). Antimicrobial resistance in veterinary medicine: an overview. Int. J. Mol. Sci. 21(6), 1914. doi: 10.3390/ijms21061914
Ranieri Matthew, L., Shi, C., Moreno Switt Andrea, I., den Bakker Henk, C., and Wiedmann, M. (2013). Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J. Clin. Microbiol. 51, 1786–1797. doi: 10.1128/JCM.03201-12
Rokney, A., Valinsky, L., Vranckx, K., Feldman, N., Agmon, V., Moran Gilad, J., et al. (2020). WGS-based prediction and analysis of antimicrobial resistance in Campylobacter jejuni isolates from Israel. Front. Cell. Infect. Microbiol. 10:365. doi: 10.3389/fcimb.2020.00365
Sato, T., Okubo, T., Usui, M., Yokota, S. I., Izumiyama, S., and Tamura, Y. (2014). Association of veterinary third-generation cephalosporin use with the risk of emergence of extended-spectrum-cephalosporin resistance in Escherichia coli from dairy cattle in Japan. PLoS One 9:e96101. doi: 10.1371/journal.pone.0096101
Schwan, C. L., Lomonaco, S., Bastos, L. M., Cook, P. W., Maher, J., Trinetta, V., et al. (2021). Genotypic and phenotypic characterization of antimicrobial resistance profiles in non-typhoidal salmonella enterica strains isolated from Cambodian informal markets. Front. Microbiol. 12:711472. doi: 10.3389/fmicb.2021.711472
Shah, D. H., Paul, N. C., Sischo, W. C., Crespo, R., and Guard, J. (2017). Population dynamics and antimicrobial resistance of the most prevalent poultry-associated salmonella serotypes. Poult. Sci. 96, 687–702. doi: 10.3382/ps/pew342
Sharma-Kuinkel, B. K., Rude, T. H., and Fowler, V. G. Jr. (2016). Pulse field gel electrophoresis. Methods Mol. Biol. 1373, 117–130. doi: 10.1007/7651_2014_191
Shi, C., Singh, P., Ranieri, M. L., Wiedmann, M., and Moreno Switt, A. I. (2015). Molecular methods for serovar determination of salmonella. Crit. Rev. Microbiol. 41, 309–325. doi: 10.3109/1040841X.2013.837862
Soyer, Y., Alcaine, S. D., Schoonmaker-Bopp, D. J., Root, T. P., Warnick, L. D., McDonough, P. L., et al. (2010). Pulsed-field gel electrophoresis diversity of human and bovine clinical salmonella isolates. Foodborne Pathogen Dis. 7, 707–717. doi: 10.1089/fpd.2009.0424
Sun, F., Li, X., Wang, Y., Wang, F., Ge, H., Pan, Z., et al. (2021). Epidemic patterns of antimicrobial resistance of salmonella enterica serovar Gallinarum biovar Pullorum isolates in China during the past half-century. Poult. Sci. 100:100894. doi: 10.1016/j.psj.2020.12.007
Tack, D. M., Marder, E. P., Griffin, P. M., Cieslak, P. R., Dunn, J., Hurd, S., et al. (2019). Preliminary incidence and trends of infections with pathogens transmitted commonly through food - foodborne diseases active surveillance network, 10 U.S. sites, 2015-2018. Morb. Mortal. Wkly Rep. 68, 369–373. doi: 10.15585/mmwr.mm6816a2
Tian, Y., Gu, D., Wang, F., Liu, B., Li, J., Kang, X., et al. (2021). Prevalence and characteristics of salmonella spp. from a pig farm in Shanghai, China. Foodborne Pathogen Dis. 18, 477–488. doi: 10.1089/fpd.2021.0018
Uelze, L., Borowiak, M., Deneke, C., Szabó, I., Fischer, J., Tausch, S. H., et al. (2020). Performance and accuracy of four open-source tools for in silico serotyping of salmonella spp. based on whole-genome short-read sequencing data. Appl. Environ. Microbiol. 86, e02265–e02219. doi: 10.1128/AEM.02265-19
Wall, B., Mateus, A., Marshall, L., Pfeiffer, D., Lubroth, J., Ormel, H., et al. (2016). Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production. Rome: Food and Agriculture Organization of the United Nations.
Wang, W., Chen, J., Shao, X., Huang, P., Zha, J., and Ye, Y. (2021). Occurrence and antimicrobial resistance of salmonella isolated from retail meats in Anhui, China. Food Sci. Nutr. 9, 4701–4710. doi: 10.1002/fsn3.2266
Wang, C., Chen, Q., Zhang, C., Yang, J., Lu, Z., Lu, F., et al. (2017). Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res. 236, 14–23. doi: 10.1016/j.virusres.2017.05.001
Wang, S., Duan, H., Zhang, W., and Li, J. W. (2007). Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol. Med. Microbiol. 51, 8–13. doi: 10.1111/j.1574-695X.2007.00305.x
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020b). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/s1473-3099(20)30149-3
Wang, M., Zhang, M., Lu, Y., Kang, X., Meng, C., Zhou, L., et al. (2020a). Analyses of prevalence and molecular typing of salmonella in the goose production chain. Poult. Sci. 99, 2136–2145. doi: 10.1016/j.psj.2019.12.008
Xiuying, Y., Rong, G., Kaiyu, W., Xiaolan, W., Qiang, L., Ji, S., et al. (2015). Prevalence of antimicrobial resistance in salmonella from pig in Sichuan province. Sichuan J. Zool. 2015, 707–713. doi: 10.11984/j.issn.1000-7083.20150099
Xu, Z., Chen, X., Tan, W., Cui, H., Zhu, Z., Yang, C., et al. (2021). Prevalence and antimicrobial resistance of salmonella and Staphylococcus aureus in fattening pigs in Hubei province, China. Microbial Drug Resist. 27, 1594–1602. doi: 10.1089/mdr.2020.0585
Yachison, C. A., Yoshida, C., Robertson, J., Nash, J. H. E., Kruczkiewicz, P., Taboada, E. N., et al. (2017). The validation and implications of using whole genome sequencing as a replacement for traditional serotyping for a national Salmonella reference laboratory. Front. Microbiol. 8:1044. doi: 10.3389/fmicb.2017.01044
Yan, S., Zhang, W., Li, C., Liu, X., Zhu, L., Chen, L., et al. (2021). Serotyping, MLST, and core genome MLST analysis of salmonella enterica from different sources in China during 2004-2019. Front. Microbiol. 12:688614. doi: 10.3389/fmicb.2021.688614
Yang, X., Huang, J., Zhang, Y., Liu, S., Chen, L., Xiao, C., et al. (2020). Prevalence, abundance, serovars and antimicrobial resistance of salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 713:136385. doi: 10.1016/j.scitotenv.2019.136385
Yang, S. M., Kim, E., Lee, W., and Kim, H. Y. (2022). Genomic characteristics and comparative genomics of Salmonella enterica subsp. enterica serovar Schwarzengrund strain S16 isolated from chicken feces. Gut Pathogen 14:1. doi: 10.1186/s13099-021-00476-8
Yang, X., Wu, Q., Zhang, J., Huang, J., Chen, L., Wu, S., et al. (2019). Prevalence, bacterial load, and antimicrobial resistance of salmonella serovars isolated from retail meat and meat products in China. Front. Microbiol. 10:2121. doi: 10.3389/fmicb.2019.02121
Yinqi, L., Qiang, S., Jia, Y., Ran, X. X., and Lu, Y. (2019). Policy evolution of antibiotic resistance governance in China and its implication. Chin J. Health Policy 12, 44–48. doi: 10.3969/j.issn.1674-2982.2019.05.007
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P., Nash, J. H., et al. (2016). The salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101
Zhang, Q. Q., Ying, G. G., Pan, C. G., Liu, Y. S., and Zhao, J. L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 49, 6772–6782. doi: 10.1021/acs.est.5b00729
Keywords: Salmonella, waterfowl, prevalence, serotypes, pulsed-field gel electrophoresis, antimicrobial resistance
Citation: Guan Y, Li Y, Li J, Yang Z, Zhu D, Jia R, Liu M, Wang M, Chen S, Yang Q, Wu Y, Zhang S, Gao Q, Ou X, Mao S, Huang J, Sun D, Tian B, Cheng A and Zhao X (2022) Phenotypic and genotypic characterization of antimicrobial resistance profiles in Salmonella isolated from waterfowl in 2002–2005 and 2018–2020 in Sichuan, China. Front. Microbiol. 13:987613. doi: 10.3389/fmicb.2022.987613
Received: 06 July 2022; Accepted: 06 September 2022;
Published: 06 October 2022.
Edited by:
Ning Dong, Soochow University, ChinaReviewed by:
Ke Shang, Jeonbuk National University, South KoreaCopyright © 2022 Guan, Li, Li, Yang, Zhu, Jia, Liu, Wang, Chen, Yang, Wu, Zhang, Gao, Ou, Mao, Huang, Sun, Tian, Cheng and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=; Xinxin Zhao, eHhpbnpoYW9AMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.