
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 September 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.986326
This article is part of the Research Topic Women in Infectious Agents and Disease: 2022 View all 10 articles
Vaginal and cervical canal bacteria are associated with women’s health and pregnancy outcomes. Here, we compared their composition and characteristics in 37 reproductive-aged Chinese women including 24 pregnant women with cervical incompetence (vaginal and cervical canal bacteria formed Groups A and B, respectively) and 13 healthy pregnant women (vaginal and cervical canal bacteria formed Groups C and D, respectively) using high-throughput sequencing of the V4 region of 16S rRNA gene. The results of alpha and beta diversity analysis, respectively, indicated no statistical differences between Groups A and B (p = 0.32, 0.06), nor Groups B and D (p = 0.69, 0.74); however, differences were found between Groups C and D (p = 0.02, 0.01) and between Groups A and C (p = 0.04, 0.02). PLS-DA analysis showed that the individuals from each group were irregularly distributed according to their clade. Lactobacillus, Bifidobacterium and Ureaplasma were the dominant genera in all groups. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSts) analysis identified 31 Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs associated with the bacterial communities from the four groups, including membrane transport, folding, sorting and degradation, xenobiotics biodegradation and metabolism, and nucleotide metabolism. We further determined relationships between pregnancy outcomes (Apgar scores) and certain bacterial species. A significant positive correlation was found between Apgar scores and Actinomyces neuii and Anoxybacillus flavithermus in the vagina and cervical canal of pregnant women with cervical incompetence while Bacteroides plebeius, Bifidobacterium pseudopodium and Staphylococcus petrasii in the cervical canal displayed negative correlations with Apgar scores. Moreover, Clostridium fimetarium, Methanobacterium congolense, Pseudomonas chlororaphis, and Psychrobacter nivimaris in the vagina were negatively correlated with Apgar scores. These bacteria may serve as potential biomarkers, however, additional research is warranted to verify their role in clinical outcomes.
The female genital tract is home to trillions of bacteria that usually exist in a mutualistic relationship with their host (Younge et al., 2017; Elovitz et al., 2019). Intriguingly, the characteristics of vaginal and cervical canal bacterial communities are even more conspicuous and different from gut bacteria, displaying a high relative abundance of Lactobacillus and low bacterial diversity (Kervinen et al., 2019; Villani et al., 2022). Individual differences in the bacterial landscape of the vaginal and cervical canals are dramatically influenced by host genetics and environment, also being impacted by lifestyle factors (Moosa et al., 2020). Recent evidence supports the notion that the bacterial communities of the vagina and cervix are strongly associated with health status, with changes associated with pH, urinary tract infections, sexually transmitted infections including bacterial and viral infections such as HIV (human immunodeficiency virus) along with pregnancy outcomes (Watts et al., 2005; Flaviani et al., 2021; Lykke et al., 2021). Moreover, certain bacterial profiles are associated with adverse obstetric outcomes such as preterm birth, potentially leading to neonatal morbidity and mortality (Elovitz et al., 2019; Torcia, 2019; Tsonis et al., 2020; Tu et al., 2020; Flaviani et al., 2021).
In the female reproductive system, the vagina, cervical canal, fallopian tubes and uterus form the reproductive tract (McDonald, 1980; Chen et al., 2017). In general, the fallopian tubes and uterus are believed to be sterile, which requires the cervix to be a perfect barrier (Chen et al., 2017). The cervix does indeed provide a physical barrier to pathogens, and for example, the fetus is protected from the vaginal pathogens during healthy pregnancies (Hein et al., 2002; Goldenberg, et al., 2008; Aagaard et al., 2014; Liu et al., 2015; Kindinger et al., 2016). However, in some cases bacteria and other pathogens can ascend along the mucosal surfaces of the vagina to the fallopian tubes or the uterus (Liu et al., 2015; Chen et al., 2017; Brown et al., 2019).
Cervical incompetence is defined as the inability of the uterine cervix to retain a pregnancy in the second trimester in the absence of uterine contractions (Boelig et al., 2019). About 1% of females experience painless cervical dilation causing preterm birth in the second trimester, making this one of the most common diseases in reproductive-aged women (Brown et al., 2019; Pilarski et al., 2021). Building evidence over recent years implicates bacteria as the major cause of cervical incompetence, especially the composition and structures of bacteria which are highly correlated with preterm birth (Ravel et al., 2011; Smith and Ravel, 2017; Brown et al., 2019). Nonetheless, the bacterial composition and structures of the vagina and cervical canal in pregnant women suffering from cervical incompetence are yet to be fully characterized (Koedooder et al., 2019; Hao et al., 2021). Focusing on pregnant women with cervical incompetence, this study aimed to expand the current knowledgebase regarding the bacterial profiles of the vagina and cervical canal compared to normal pregnant woman using a high throughput sequencing approach. The results uncover potential new insights into the pathogenesis of cervical incompetence with implications for the prevention and treatment of the associated complications.
Thirty-seven reproductive-aged Chinese women including 24 pregnant women with cervical incompetence and 13 healthy pregnant women were recruited at the First Affiliated Hospital of Anhui Medical University in 2021, China. We excluded subjects with vaginal inflammation, any acute inflammation, cancer, severe pelvic adhesion, endocrine or autoimmune disorders. Further exclusions were made regarding subjects treated with any vaginal medicine, antibiotics or hormones within one month, other cervical treatment or flushing within one month and sexual intercourse in the prior month (Chen et al., 2017; Tu et al., 2020). This study was approved by the institutional review board of Anhui Medical University (No. PJ2022-09012).
On the day of the prenatal visit, both vaginal lavage fluid and cervical canal swabs were collected. All collection materials and devices used were strictly sterilized. Cervical canal swabs were used to collect bacteria using a vaginal dilator, while the vaginal samples were performed with 10 ml of vaginal lavage fluid (saline solution). All samples were transferred to the lab within 2 h of collection and stored at −80°C.
One millilitre of sterile phosphate-buffered saline was added to each cervical canal swab and rigorously vortexed for 1 min before 500 μl was collected, centrifuged and disrupted by enzymatic treatment. The vaginal lavage fluids were centrifuged at 2,000 rpm for 15 min. Total DNA of vaginal and cervical canal samples were determined using the Qubit high sensitivity kit follow the manufacturer’s instructions. The V4 variable region of 16S rRNA gene was amplified used the 515F/806R primers (515F: 5′- GTG CCA GCM GCC GCG GTA A -3′; 806R 5′-GGA CTA CHV GGG TWT CTA AT -3′) followed by DNA extraction. The PCR products were purified and then subjected to high-throughput sequencing using the Illumina HiSeq 2,500 at BGI Genomics Co., Ltd. (Shenzhen, China). The raw data of all samples were submitted to the SRA (Sequence Read Archive) under accession numbers SRR19631963 at the NCBI database.
Raw high-throughput sequencing data provided in FASTA format were subject to pre-processing (quality-trimmed, demultiplexed, and chimera-reduced) using QIIME Version 1.8.0 tools to remove short and poor-quality sequences (Caporaso et al., 2010). The clean reads were clustered to generate Operational Taxonomic Units (OTUs) with 97% similarity cutoff by Vsearch 2.4.2 (Rognes et al., 2016). Representative OUT sequences were given a taxonomic assignment based on the SILVA bacterial database using BLAST Version 2.60 (Altschul et al., 1997). Alpha diversity is a measure of bacteria diversity applicable to a single sample, and it is calculated by Chao1 index in this study. Beta diversity is a measure of similarity or dissimilarity of two communities, which is performed by calculating the UniFrac index by using R (v3.4.1; R Core Team, 2017). Differences between the two populations of the bacterial compositions were analyzed based on Partial Least Squares Discrimination Analysis (PLS-DA) by using the mixOmics package in R (v3.2.1; Rohart et al., 2017). Linear Discriminant Analysis Effect Size (LEfSe) analysis was used between or among groups to determine the differentially abundant taxonomic features by using the non-parametric Kruskal-Wallis rank sum test. Venn analysis diagrams were performed to categorize the core bacteria by using the VennDiagram package in R (v3.2.1; Chen and Boutros, 2011). All statistical analyses and plots were performed by GraphPad Prism v7.0.
We collected and sequenced the 16S rRNA gene from 74 vaginal and cervical canal samples from 37 pregnant women. The vaginal and cervical canal samples from pregnant women with cervical incompetence were designated as Group A (n = 24, sample no. A1-24) and Group B (n = 24, sample no. B1-24), respectively. The samples of the vagina and cervical canal of normal pregnant women were designated as Group C (n = 13, sample no. C1-13) and Group D (n = 13, sample no. D1-13), respectively. After removing low quality reads, a total of 4,825,697 clean reads were retained, which corresponding to 5,537 OTUs, the average reads were 63,496 ± 933 (range, 59,803–64,910 per sample) per sample. An average of 73 OTUs were contained in each sample, which ranged from 18 to 299 OTUs per sample. Samples from Groups A, B, C, and D had 1.06, 3.30, 0.38, and 3.61% unique OTUs with 12.44% of all OTUs common to the four groups (Supplementary Figure S1).
Alpha and beta diversities were compared to determine the bacterial composition shift in patients of the four groups. There was no significant differences in Chao1 diversity indexes between Group A and Group B (Figure 1A; p = 0.32) and similarly there were no differences between Groups B and D (Figure 1D; p = 0.69). However, the Chao1 diversity index showed a significant difference between vaginal samples (Group C) and cervical canal samples (Group D) of healthy pregnant women (Figure 1B; p = 0.02), and also between Group A and Group C (Figure 1C; p = 0.04).
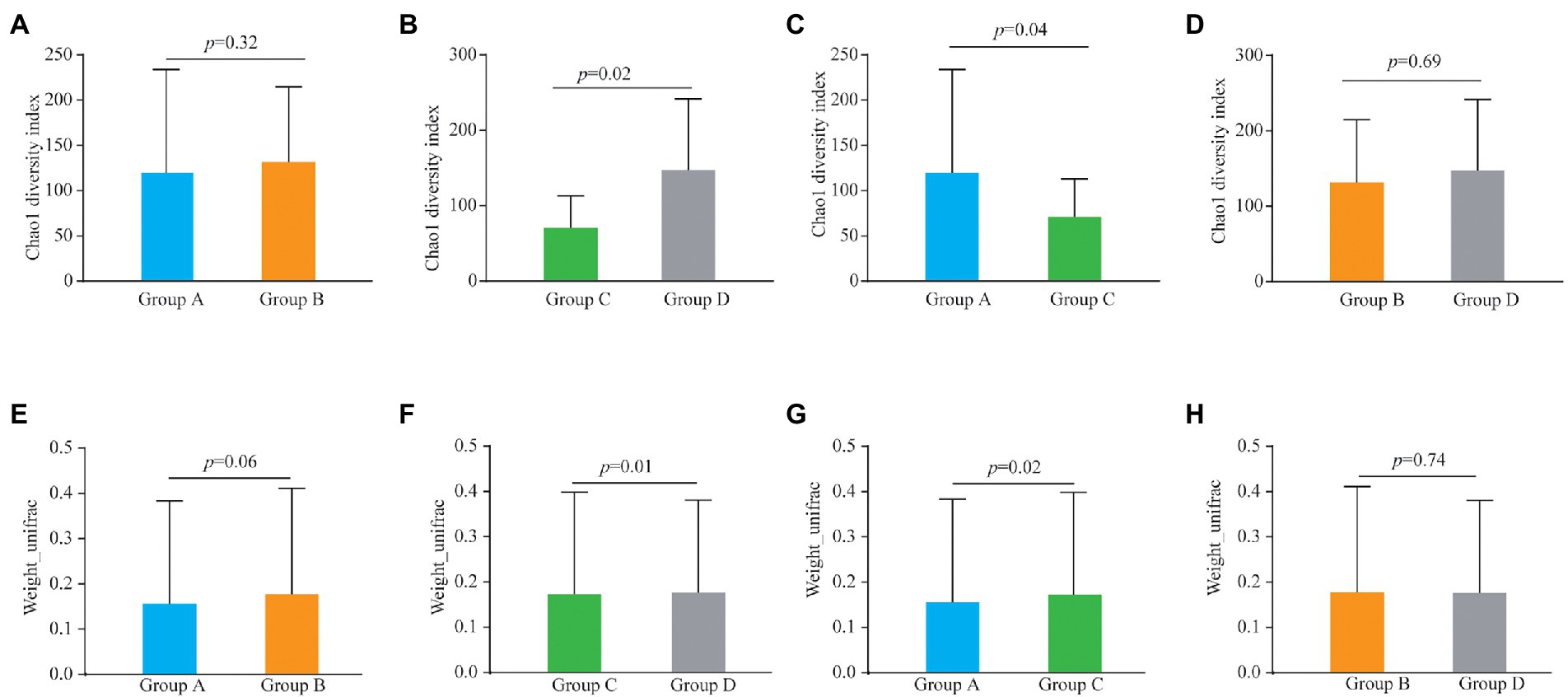
Figure 1. Alpha diversity analysis (Chao1 diversity index) of the bacteria from vaginal and cervical canal samples between Group A and Group B (A), Group C and Group D (B), Group A and Group C (C), and Group C and Group D (D), respectively. Beta diversity analysis of the bacteria from vaginal and cervical canal samples between Group A and Group B (E), Group C and Group D (F), Group A and Group C (G), and Group C and Group D (H), respectively.
For the results of beta diversity, there was no significant differences between Groups A and B (Figures1E; p = 0.06), and between Groups B and D, respectively (Figure 1H; p = 0.74). However, beta diversity was significantly different between Groups C and D (Figure 1F, p = 0.01), and beta diversity was also significantly different between Group A and Group C (Figure 1G; p = 0.02). PLS-DA analysis results showed that individuals from Group C and D were irregularly distributed according to their clade, and the results of Groups A vs C were similar to Groups C vs D (Supplementary Figure S2).
Fourteen phyla, 23 classes, 43 orders, 70 families and 138 genera of bacteria were identified in the four groups. Firmicutes, Actinobacteria and Tenericutes were the dominant bacterial phyla, accounting for 93.97, 4.91, and 0.44% of the OTUs, respectively. Bacteroidetes, Proteobacteria, Cyanobacteria, Fusobacteria, Deinococcus-Thermus, and Verrucomicrobia accounted for less than 0.02% of all bacteria (Figure 2A). Bacilli, Actinobacteria, Mollicutes, Clostridia, Erysipelotrichia, Bacteroidia, Negativicutes, Gammaproteobacteria, Betaproteobacteria and Cyanobacteria were the dominant bacterial classes, Bacilli and Actinobacteria are the dominant bacteria, accounting for 90.89 and 63.48%, respectively (Figure 2B). Lactobacillales, Bifidobacteriales, Mycoplasmatales, Clostridiales, Erysipelotrichales, Selenomonadales, Bacteroidales, Bacillales, Actinomycetales and Burkholderiales were the dominant bacterial orders, and Lactobacillales were the dominant bacteria, accounting for 91.13% (Figure 2C). Lactobacillaceae, Bifidobacteriaceae, Mycoplasmataceae, Veillonellaceae, Prevotellaceae, Lachnospiraceae, Clostridiales, Porphyromonadaceae, Peptoniphilaceae and Ruminococcaceae were the dominant bacterial families, Lactobacillales were the dominant bacteria, accounting for 88.72% (Figure 2D). The dominant genus was Lactobacillus (87.96%; Figure 2E).
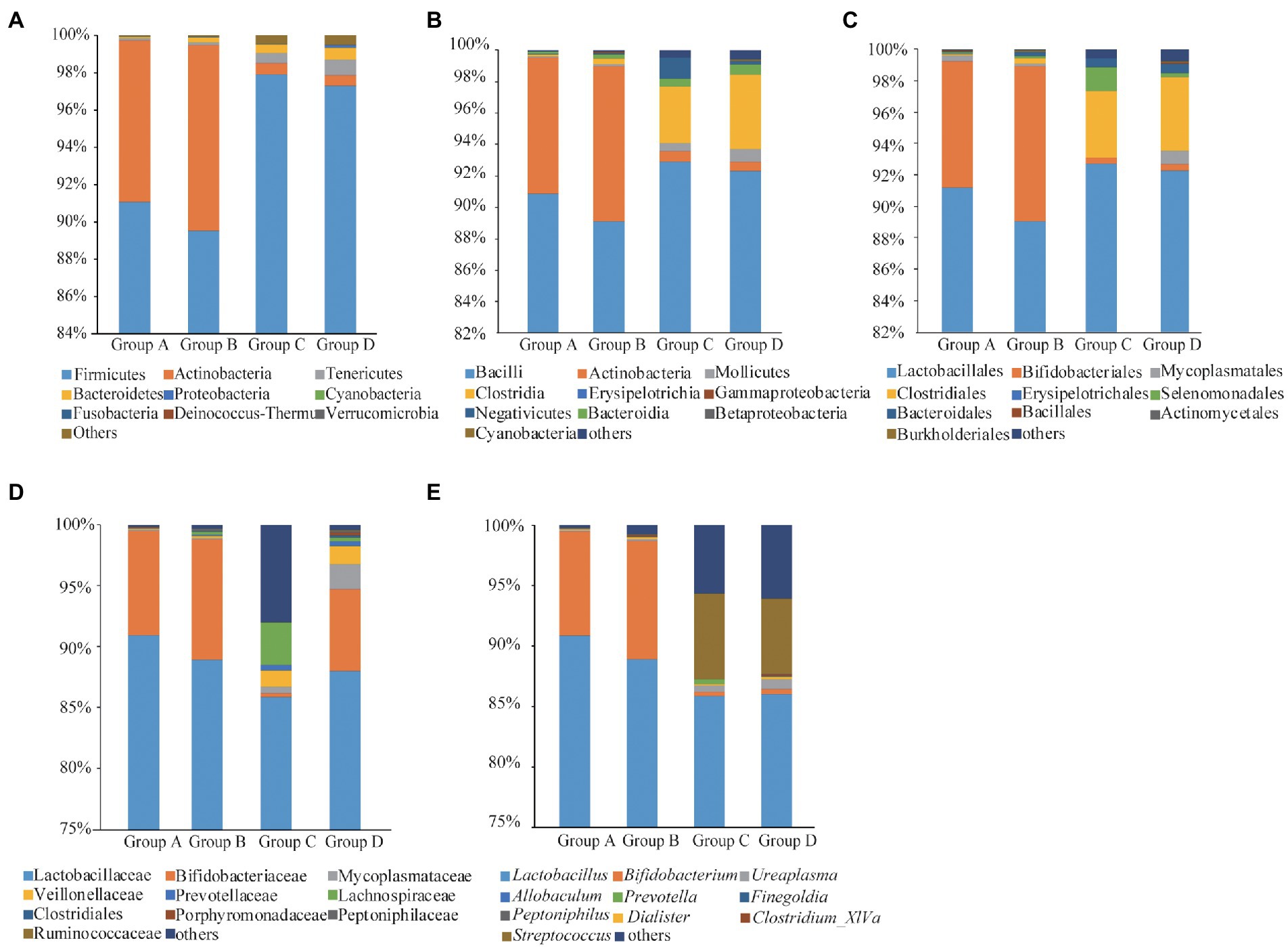
Figure 2. Taxonomic analyses for Groups A, B, C, and D at phyla (A), classes (B), orders (C), families (D), and genera (E) levels.
The relative abundance of several taxa was compared with each other for the four groups using LEfSe analysis (Figures 3A-D). A total of 42 species of the bacteria were identified from all the vaginal and cervical canal samples of 24 pregnant women with cervical incompetence and 13 healthy pregnant women. There were 11 differences in bacterial abundance found comparing between Groups A and B, the OUT was much lower in Group A than that in Group B (Figure 4A). Five differences were found between the abundance of bacteria in Group A and C, involving much higher abundance of Actinomyces neuii, Anoxybacillus flavithermus, Bifidobacterium pseudolongum, and Staphylococcus petrasii in Group A and higher abundance of Bacteroides plebeius in Group C (Figure 4B). Nine different species were identified between Groups B and D (Figure 4C), and 18 different species were found between Groups C and D (Figure 4D), respectively. In addition, Group D has significantly higher abundance of OUTs than that in Groups B and C, respectively.
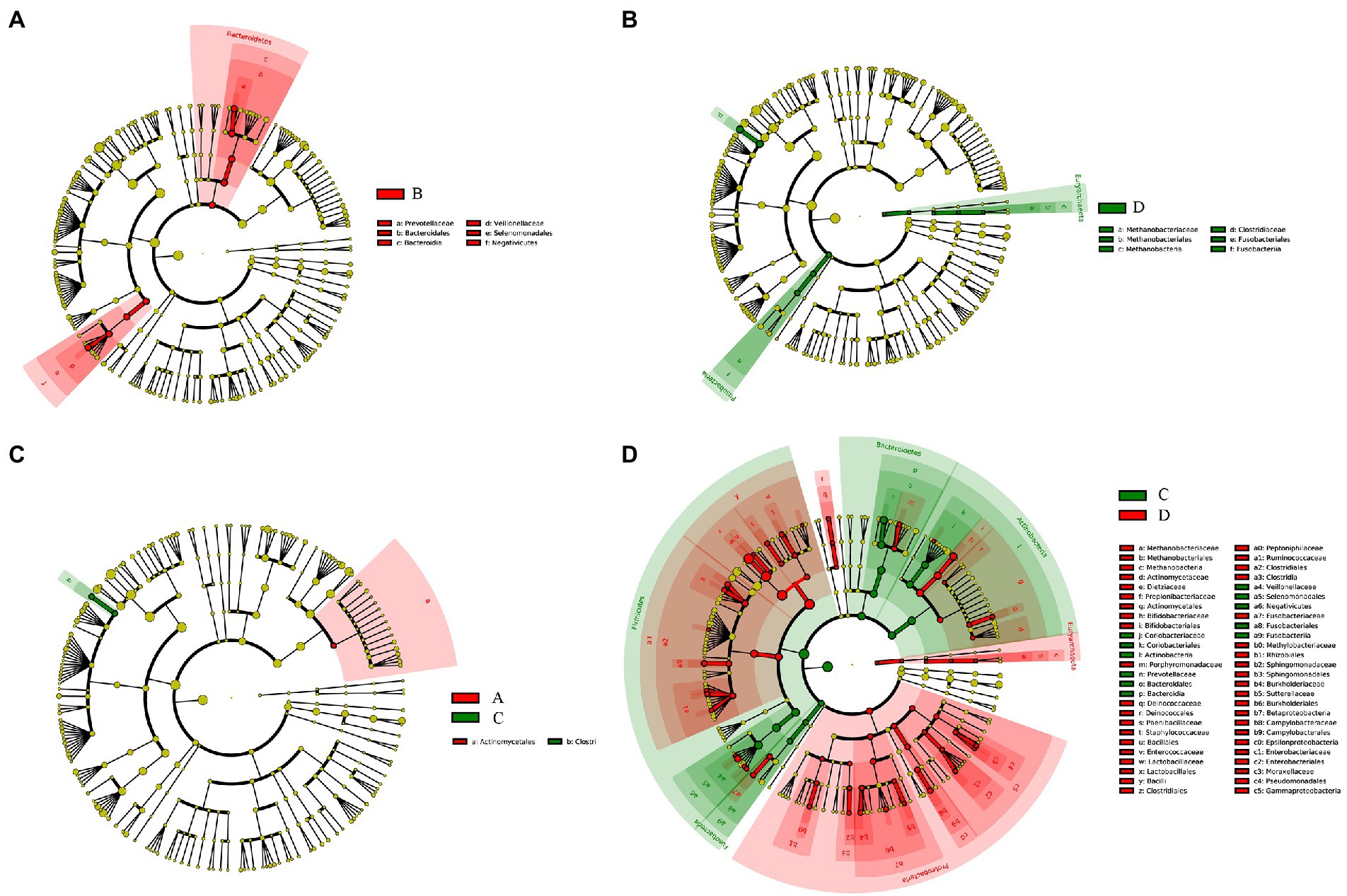
Figure 3. Taxa difference analyses between Group A and Group B (A), Group C and Group D (B), Group A and Group C (C), and Group C and Group D (D) by LEfSe analysis, respectively.
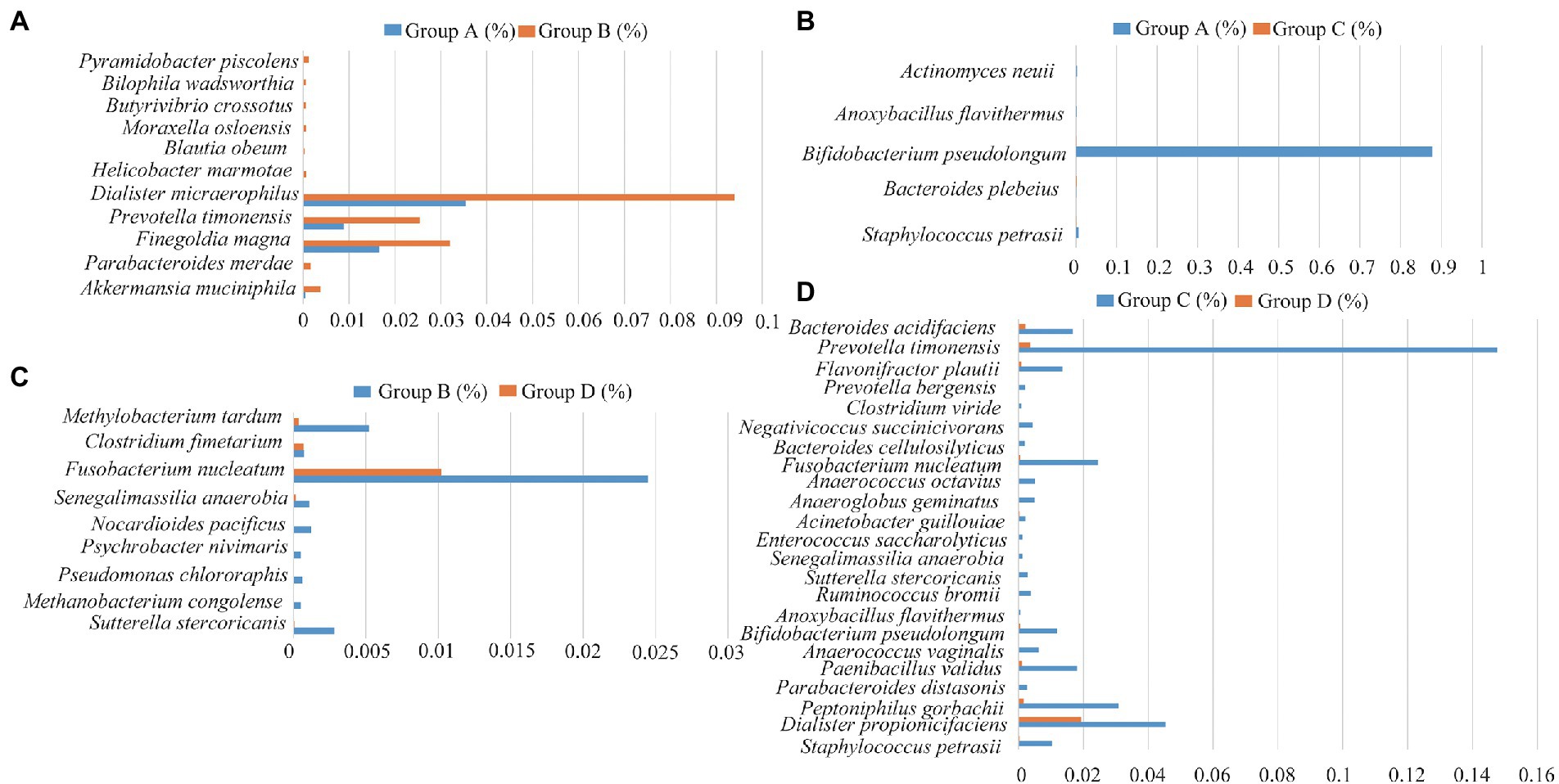
Figure 4. Differences in the species of the bacteria between Group A and Group B (A), Group C and Group D (B), Group A and Group C (C), and Group C and Group D (D), respectively.
To further explore the influence of the bacteria on the microenvironment of the vagina and cervical canal, KEGG pathway analyses were conducted. Overall, 31 orthologs were found by PICRUSts predictive exploration tool based on the KEGG database for each group (Figure 5). Half of the KEGG functions were classified into membrane transport, nucleotide metabolism and glycan biosynthesis and metabolism, xenobiotics biodegradation and metabolism, folding, sorting and degradation, etc. We also compared differences in KEGG bacterial function. The digestive system in Group A was significantly different from that in Group B (Figure 6A; p = 0.037) and signal transduction was significantly different between Groups A with C (Figure 6B; p = 0.006). Moreover, significant differences were found between Groups B with D involving replication and repair (p = 0.010), translation (p = 0.018), folding, sorting and degradation (p = 0.019), nucleotide metabolism (p = 0.021), and cell growth and death (Figure 6C; p = 0.025). Furthermore, digestive system (p = 0.008), signal transduction (p = 0.018), immune diseases (p = 0.013), environmental adaptation (p = 0.031), xenobiotics biodegradation and metabolism (p = 0.024) were dramatically different between Groups C and D (Figure 6D).
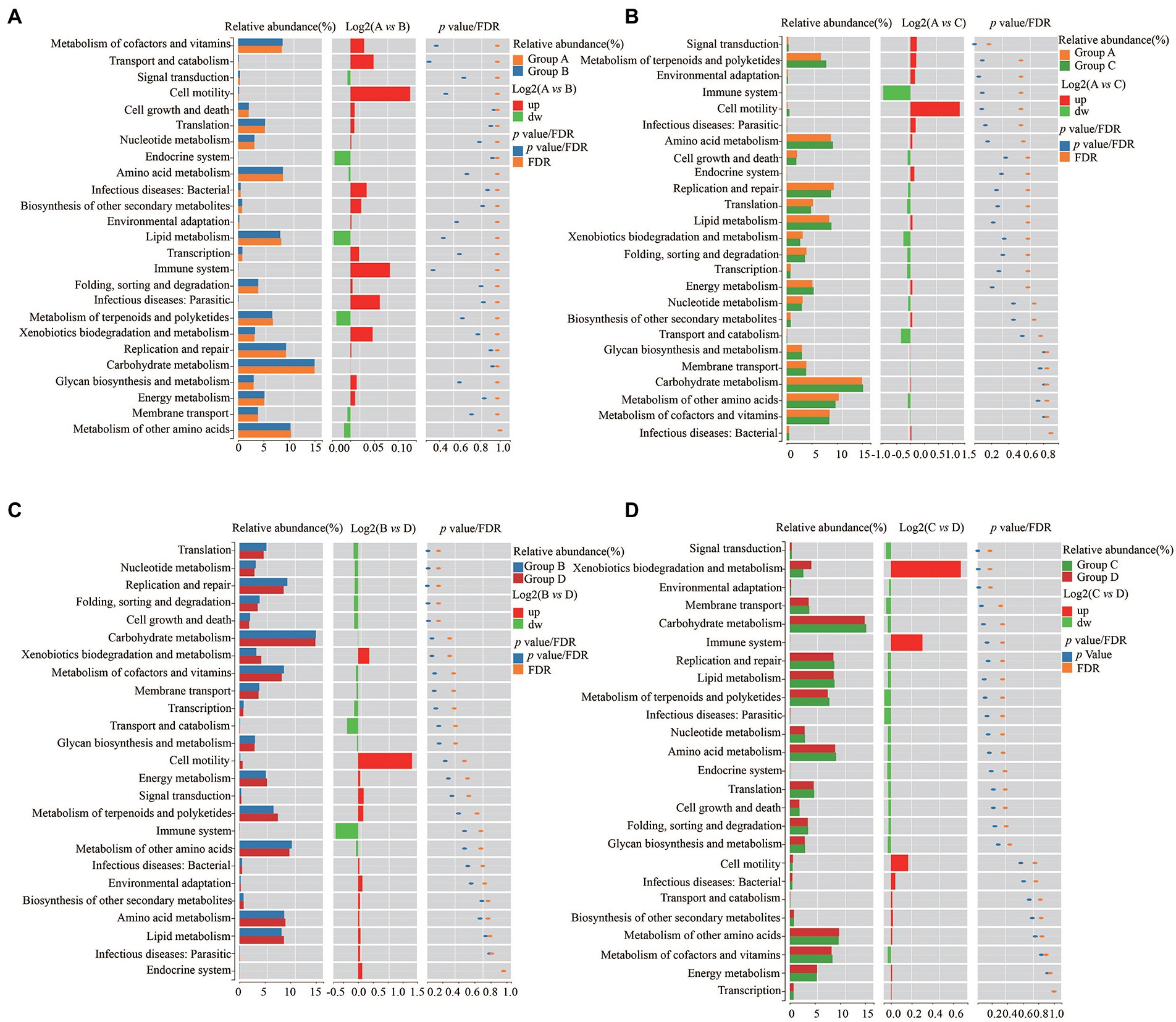
Figure 6. Differences of KEGG bacterial functions between Group A and Group B (A), Group C and Group D (B), Group A and Group C (C), and Group C and Group D (D), respectively.
We next analyzed pregnancy outcomes of the 37 subjects. The 24 women with cervical incompetence received an intervention during pregnancy involving cervical cerclage. The fetal survival rate was 100% for both groups and there was no significant differences in Apgar scores between healthy pregnant women and those with cervical incompetence (Table 1). However, some complications occurred after the intervention. Five cases of premature rupture of the membrane were diagnosed in women with cervical incompetence, representing 21.74%, higher than that of the normal pregnant women (15.38%; Table 1). Notably, the prematurity rate (defined as birth before 37 weeks) in the cervical incompetence group was 8.70%, which was much higher than that in healthy pregnant women, while one case of abortion was recorded at week 18 of pregnancy (Table 1).
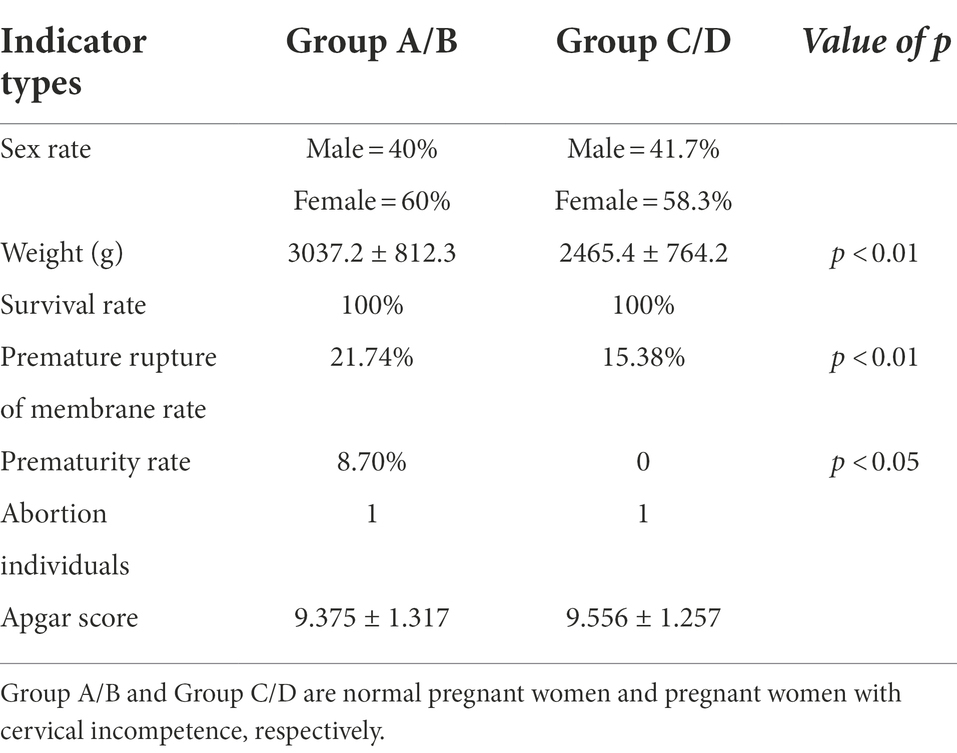
Table 1. Analyses of the pregnancy outcomes from normal pregnant women and pregnant women with cervical incompetence.
We also analyzed the relationships between pregnancy outcomes and marker bacteria of the vagina and cervical canal from pregnant women with cervical incompetence receiving cervical cerclage (Supplementary Table 1). Statistical analysis showed that Actinomyces neuii and Anoxybacillus flavithermus were positively correlated with the pregnancy outcomes (Apgar score). Conversely, there were significant negative correlations between the Apgar score in the cervical canal and Bacteroides plebeius, Bifidobacterium pseudolongum and Staphylococcus petrasii. Clostridium fimetarium, Methanobacterium congolense, Pseudomonas Chlororaphis, and Psychrobacter nivimaris was negatively correlated with the Apgar score of pregnancy outcomes in the vagina, while Allobaculum stercoricanis, Fusobacterium nucleatum, Methylobacterium tardum, Nocardioides pacificus and Senegalimassilia anaerobia were positively correlated.
The human body contains trillions of bacteria inhabiting body surfaces and cavities which can be exchanged with those in the external environment (Ravel et al., 2011). Recent studies have indicated that the bacteria from vaginal and cervical canal play important roles in maintaining a healthy female reproductive system, and moreover, may be mechanistically linked to the pathogenesis of preterm birth (Ansari et al., 2021; Villani et al., 2022). Different bacterial communities or abnormal levels of bacteria populating the vaginal tract and cervical canal during late pregnancy can produce infections or dysfunction, which can adversely affect pregnancy outcomes (Perino et al., 2011; Ravel et al., 2011; Villani et al., 2022). Here, we compared the vaginal and cervical canal bacterial communities between healthy pregnant women and those with cervical incompetence, a condition which may disrupt the distribution of bacteria in the female reproductive tract. Our analysis results for alpha and beta diversity were similar to that of previous studies (Chen et al., 2017).
The bacteria inhabiting the vagina are considered to originate from the upper genital tract (Khan et al., 2016; Tu et al., 2020). In normal pregnant women, the vaginal and cervical canal bacterial communities were different due to the obstruction of the cervix (Chen et al., 2017). In this study we found the bacterial communities of the cervical canal in pregnant women with cervical incompetence were significantly different from that in the normal pregnant women. However, we found no differences in bacterial composition between the vagina and cervical canal of the pregnant women with cervical incompetence, suggesting cervical incompetence promotes bacterial exchange between the vaginal and cervical canal communities. One of the most important risk factors for preterm birth is cervical incompetence in the second trimester of pregnancy. In support of a prior study (Kindinger et al., 2016), we found associations between vaginal and cervical bacteria diversity with preterm delivery. However, larger sample size is needed if we aim to understand the relationships between the vaginal and cervical bacteria diversity and preterm birth.
Lactobacillus are the unequivocal dominant bacteria in the vaginal and cervical canal of healthy women. Their production of lactic acid serves to maintain the acidic pH of the vagina and cervical canal, acting as a barrier against pathogens and protects them from genital infection (Petricevic et al., 2014; Chen et al., 2017; Pace et al., 2021). Deficiencies in Lactobacillus could disturb the vaginal bacterial balance, resulting in the syndrome of bacterial vaginosis (Petricevic et al., 2014; Lozano et al., 2021). Nonetheless, Lactobacillus was reported to remain as the dominant species in pregnant women with cervical incompetence, and no differences were found in the abundance of OTUs compared to healthy pregnant women (Lozano et al., 2021). The vaginal and cervical canal bacteria are dynamic communities owing to physiological, pathological, environmental, and nutritional factors (D’Alterio et al., 2021). Bacterial dysbiosis may contribute to an inflammatory response in the vagina and cervical canal, also predisposing pregnant woman to a higher risk of infection (D’Alterio et al., 2021). Bacterial imbalance of the vaginal and cervical canal is correlated with higher genital pro-inflammatory cytokine concentrations and increased APC (antigen-presenting cell) activation through LPS pathways. Notably, triggering local immunity to increase the concentration of inflammatory factors including TNF-α, NF-κB and COX-2 may affect pregnancy outcomes (Ravel et al., 2011; Smith and Ravel, 2017; D’Alterio et al., 2021).
Similar to the results of previous studies we found differential genera in pregnant women with cervical incompetence including Staphylococcus, Actinomyces, Aerococcus, Clostridium_sensu_stricto, and Anoxybacillus (Flynn et al., 2013; Chen et al., 2017; Brown et al., 2019; Lozano et al., 2021). We identified Groups A and C -specific shifts in bacterial composition reflected by the enrichment of five species, including Staphylococcus petrasii, Bacteroides plebeius, Bifidobacterium pseudolongum, Anoxybacillus flavithermus, and Actinomyces neuii. Bacteroides plebeius has been found to be more significantly frequent in HPV-positive women, and Actinomyces neuii was reported in bacterial vaginosis (Chao et al., 2019; Castro et al., 2019) while the other species are unique to this study. Furthermore, we found the abundances of Sutterella stercoricanis, Methanobacterium congolense, Pseudomonas chlororaphis, Psychrobacter nivimaris, Nocardioides pacificus, Senegalimassilia anaerobia, Fusobacterium nucleatum, Clostridium fimetarium, Methylobacterium tardum were dramatically different between Groups B and D. Collectively, these results suggest that these species represent “biomarker” bacteria for differences between healthy pregnant women and those with cervical incompetence (Agarwal et al., 2020).
As an important potential anaerobic pathogen and Gram-negative bacterium, Fusobacterium nucleatum colonizes the female reproductive tract, digestive tract and oral cavity (Agarwal et al., 2020). This species has been associated with numerous human diseases, including oral infections, colorectal cancer, respiratory tract infections, and Alzheimer’s disease (Allen-Vercoe et al., 2011; Liu et al., 2014; Han, 2015; Agarwal et al., 2020). Fusobacterium nucleatum was also detected in a wide spectrum of fetal membranes, amniotic fluid, neonatal gastric aspirates, chorioamnionitis, and was frequently detected in the cord blood and amniotic fluid of premature infants, being associated with intrauterine infection and preterm birth (Han et al., 2010; Wang et al., 2013; Han, 2015). Additional research also indicated that the mother’s subgingival plaque in oral cavity may transfer Fusobacterium nucleatum to the placenta or fetus during pregnancy period, leading to acute inflammation, even leading to fetal stillbirth (Han, 2011; Wang et al., 2013). However, further validation is still needed to confirm the utility of the biomarker bacteria identified here and in other studies.
Preterm birth occurs in 5–20% of pregnant women and is defined as delivery before 37 weeks of gestation. Preterm birth is influenced by maternal, fetal and environmental factors, and is associated with a high risk of neonatal morbidity and mortality (Terzic et al., 2021). Some researches have indicated that cervical incompetence is related to preterm birth, although other potential risk factors can influence the incidence of preterm delivery, including diabetes, hypertension, thyroid disease, chronic renal disease and tobacco smoking (Guerra et al., 2006; Goldenberg et al., 2008; Vogel et al., 2018; Miklavcic et al., 2021). In addition, pre-pregnancy body mass index, advanced maternal age (more than 35 years old), endometriosis, polycystic ovary syndrome, and pre-pregnancy diabetes are also considered possible risk factors to preterm birth (Molin, 1993; Vyas et al., 2006; Ciancimino et al., 2014; Miklavcic et al., 2021; Pilarski et al., 2021). Other studies suggest that bacterial vaginal infection and maternal periodontal diseases increase the risk of preterm birth, with bacterial transfer from the vagina to the uterus through the cervix (D’Alterio et al., 2021; Terzic et al., 2021). Pregnant women with intrauterine infection could also spread bacteria to the amniotic cavity to cause preterm birth (Angioni et al., 2015). Some studies reported that periodontal diseases of pregnant women were associated with preterm birth, the proposed reason being gingival bacterium transfer to the uterine cavity and placenta via the bloodstream, resulting in an intra-amniotic infection (Offenbacher et al., 2006; Ren and Du, 2017).
In summary, we investigated the bacterial compositions and structures of the vagina and cervical canal from healthy pregnant women and pregnant women with cervical incompetence by high-throughput sequencing technology. The overall bacteria were not significantly different in Lactobacillus in the vaginal and cervical canal of healthy pregnant women and pregnant women with cervical incompetence. However, the alpha and beta diversities were significantly different between vaginal samples and cervical canal samples of normal pregnant women. We also found differences in the bacterial communities of cervical canal samples between normal pregnant women and those with cervical incompetence. Several specific bacteria were identified, suggesting that these bacteria may act as potential biomarkers. However, we must also acknowledge certain limitations regarding our study. First, the sample size is relatively small. Second, further investigations are required to elaborate the functional consequences of the “biomarker” bacteria. Third, more experimental technologies are necessary to reveal the correlation between bacteria and clinical features and elucidate the molecular mechanism.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The studies involving human participants were reviewed and approved by Anhui Medical University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
MS, GL and XL designed the experiments of this manuscript. MS, HG, JF, and GL performed the experiments. MS, NX, YL, and GL analyzed the data. MS, YL, and GL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (grant nos. 31702030 and 81772908) and the Natural Science Foundation for the Higher Education Institutions of Anhui Province of China (grant no. KJ2021A0246).
The authors thank Zhizhong Gong for data analysis, we also thank. Rick F. Thorne (The University of Newcastle, Newcastle) for polishing our English.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.986326/full#supplementary-material
Aagaard, K., Ma, J., Antony, K. M., Ganu, R., Petrosino, J., and Versalovic, J. (2014). The placenta harbors a unique microbiome. Sci. Transl. Med. 6:237ra65. doi: 10.1126/scitranslmed.3008599
Agarwal, K., Robinson, L. S., Aggarwal, S., Foster, L. R., Hernandez-Leyva, A., Lin, H., et al. (2020). Glycan cross-feeding supports mutualism between Fusobacterium and the vaginal microbiota. PLoS Biol. 18:e3000788. doi: 10.1371/journal.pbio.3000788
Ansari, A., Bose, S., You, Y., Park, S., and Kim, Y. (2021). Molecular mechanism of microbiota metabolites in preterm birth: pathological and therapeutic insights. Int. J. Mol. Sci. 22:8145. doi: 10.3390/ijms22158145
Angioni, S., Cela, V., Sedda, F., Stochino Loi, E., Cofelice, V., Pontis, A., et al. (2015). Focusing on surgery results in infertile patients with deep endometriosis. Gynecol. Endocrinol. 31, 595–598. doi: 10.3109/09513590.2015.1062868
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Allen-Vercoe, E., Strauss, J., and Chadee, K. (2011). Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes 2, 294–298. doi: 10.4161/gmic.2.5.18603
Brown, R. G., Chan, D., Terzidou, V., Lee, Y. S., Smith, A., Marchesi, J. R., et al. (2019). Prospective observational study of vaginal microbiota pre- and post-rescue cervical cerclage. BJOG 126, 916–925. doi: 10.1111/1471-0528.15600
Boelig, R. C., Dugoff, L., Roman, A., Berghella, V., and Ludmir, J. (2019). (2019). Predicting asymptomatic cervical dilation in pregnant patients with short mid-trimester cervical length: a secondary analysis of a randomized controlled trial. Acta Obstet. Gynecol. Scand. 98, 761–768. doi: 10.1111/aogs.13545
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Castro, J., Machado, D., and Cerca, N. (2019). Unveiling the role of gardnerella vaginalis in polymicrobial bacterial vaginosis biofilms: the impact of other vaginal pathogens living as neighbors. ISME J. 13, 1306–1317. doi: 10.1038/s41396-018-0337-0
Chao, X. P., Sun, T. T., Wang, S., Fan, Q. B., Shi, H. H., Zhu, L., et al. (2019). Correlation between the diversity of vaginal microbiota and the risk of high-risk human papillomavirus infection. Int. J. Gynecol. Cancer 29, 28–34. doi: 10.1136/ijgc-2018-000032
Chen, C., Song, X., Wei, W., Zhong, H., Dai, J., Lan, Z., et al. (2017). The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 8:875. doi: 10.1038/s41467-017-00901-0
Chen, H., and Boutros, P. C. (2011). Venn diagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 12:35. doi: 10.1186/1471-2105-12-35
Ciancimino, L., Laganà, A. S., Chiofalo, B., Granese, R., Grasso, R., and Triolo, O. (2014). Would it be too late? A retrospective case-control analysis to evaluate maternal-fetal outcomes in advanced maternal age. Arch. Gynecol. Obstet. 290, 1109–1114. doi: 10.1007/s00404-014-3367-5
D’Alterio, M. N., Giuliani, C., Scicchitano, F., Laganà, A. S., Oltolina, N. M., Sorrentino, F., et al. (2021). Possible role of microbiome in the pathogenesis of endometriosis. Minerva Obstet Gynecol. 73, 193–214. doi: 10.23736/S2724-606X.21.04788-2
Elovitz, M. A., Gajer, P., Riis, V., Brown, A. G., Humphrys, M. S., Holm, J. B., et al. (2019). Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat. Commun. 10:1305. doi: 10.1038/s41467-019-09285-9
Flaviani, F., Hezelgrave, N. L., Kanno, T., Prosdocimi, E. M., Chin-Smith, E., Ridout, A. E., et al. (2021). Cervicovaginal microbiota and metabolome predict preterm birth risk in an ethnically diverse cohort. JCI Insight. 6:e149257. doi: 10.1172/jci.insight.149257
Flynn, A. N., Lyndon, C. A., and Church, D. L. (2013). Identification by 16S rRNA gene sequencing of an Actinomyces hongkongensis isolate recovered from a patient with pelvic actinomycosis. J. Clin. Microbiol. 51, 2721–2723. doi: 10.1128/JCM.00509-13
Goldenberg, R. L., Culhane, J. F., Iams, J. D., and Romero, R. (2008). Epidemiology and causes of preterm birth. Lancet 371, 75–84. doi: 10.1016/S0140-6736(08)60074-4
Guerra, B., Ghi, T., Quarta, S., Morselli-Labate, A. M., Lazzarotto, T., Pilu, G., et al. (2006). Pregnancy outcome after early detection of bacterial vaginosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 128, 40–45. doi: 10.1016/j.ejogrb.2005.12.024
Han, Y. W. (2015). Fusobacterium nucleatum: a commensal-turned pathogen. Curr. Opin. Microbiol. 23, 141–147. doi: 10.1016/j.mib.2014.11.013
Han, Y. W., Fardini, Y., Chen, C., Iacampo, K. G., Peraino, V. A., Shamonki, J. M., et al. (2010). Redline RW: term stillbirth caused by oral Fusobacterium nucleatum. Obstet. Gynecol. 115, 442–445. doi: 10.1097/AOG.0b013e3181cb9955
Han, Y. W. (2011). Can oral bacteria cause pregnancy complications? Women's Health (Lond. Engl.) 7, 401–404. doi: 10.2217/whe.11.37
Hao, X., Li, P., Wu, S., and Tan, J. (2021). Association of the cervical microbiota with pregnancy outcome in a subfertile population undergoing in vitro fertilization: a case-control study. Front. Cell. Infect. Microbiol. 11:654202. doi: 10.3389/fcimb.2021.654202
Hein, M., Valore, E. V., Helmig, R. B., Uldbjerg, N., and Ganz, T. (2002). Antimicrobial factors in the cervical mucus plug. Am. J. Obstet. Gynecol. 187, 137–144. doi: 10.1067/mob.2002.123034
Kervinen, K., Kalliala, I., Glazer-Livson, S., Virtanen, S., Nieminen, P., and Salonen, A. (2019). Vaginal microbiota in pregnancy: role in induction of labor and seeding the neonate's microbiota? J. Biosci. 44:116. doi: 10.1007/s12038-019-9925-z
Khan, K. N., Fujishita, A., Masumoto, H., Muto, H., Kitajima, M., Masuzaki, H., et al. (2016). Molecular detection of intrauterine microbial colonization in women with endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 199, 69–75. doi: 10.1016/j.ejogrb.2016.01.040
Kindinger, L. M., MacIntyre, D. A., Lee, Y. S., Marchesi, J. R., Smith, A., McDonald, J. A., et al. (2016). Relationship between vaginal microbial dysbiosis, inflammation, and pregnancy outcomes in cervical cerclage. Sci. Transl. Med. 8:350ra102. doi: 10.1126/scitranslmed.aag1026
Koedooder, R., Singer, M., Schoenmakers, S., Savelkoul, P. H. M., Morré, S. A., de Jonge, J. D., et al. (2019). The vaginal microbiome as a predictor for outcome of in vitro fertilization with or without intracytoplasmic sperm injection: a prospective study. Hum. Reprod. 34, 1042–1054. doi: 10.1093/humrep/dez065
Liu, L., Oza, S., Hogan, D., Perin, J., Rudan, I., Lawn, J. E., et al. (2015). Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 385, 430–440. doi: 10.1016/S0140-6736(14)61698-6
Liu, P., Liu, Y., Wang, J., Guo, Y., Zhang, Y., and Xiao, S. (2014). Detection of Fusobacterium nucleatum and fad A adhesin gene in patients with orthodontic gingivitis and non-orthodontic periodontal inflammation. PLoS One 9:e85280. doi: 10.1371/journal.pone.0085280
Lozano, F. M., Bernabeu, A., Lledo, B., Morales, R., Diaz, M., Aranda, F. I., et al. (2021). Characterization of the vaginal and endometrial microbiome in patients with chronic endometritis. Eur. J. Obstet. Gynecol. Reprod. Biol. 263, 25–32. doi: 10.1016/j.ejogrb.2021.05.045
Lykke, M. R., Becher, N., Haahr, T., Boedtkjer, E., Jensen, J. S., and Uldbjerg, N. (2021). Vaginal, cervical and uterine ph in women with normal and abnormal vaginal microbiota. Pathogens. 10:90. doi: 10.3390/pathogens10020090
McDonald, I. A. (1980). Cervical cerclage. Clin. Obstet. Gynaecol. 7, 461–479. doi: 10.1016/S0306-3356(21)00250-8
Miklavcic, J., Laganà, A. S., Premru Srsen, T., Korosec, S., and Ban Frangež, H. (2021). Effect of hysteroscopic septum resection on preterm delivery rate in singleton pregnancies. Minim. Invasive Ther. Allied Technol. 30, 377–383. doi: 10.1080/13645706.2020.1743721
Molin, A. (1993). Risk of damage to the cervix by dilatation for first-trimester-induced abortion by suction aspiration. Gynecol. Obstet. Investig. 35, 152–154. doi: 10.1159/000292688
Moosa, Y., Kwon, D., de Oliveira, T., and Wong, E. B. (2020). Determinants of vaginal microbiota composition. Front. Cell. Infect. Microbiol. 10:467. doi: 10.3389/fcimb.2020.00467
Offenbacher, S., Boggess, K. A., Murtha, A. P., Jared, H. L., Lieff, S., McKaig, R. G., et al. (2006). Progressive periodontal disease and risk of very preterm delivery. Obstet. Gynecol. 107, 29–36. doi: 10.1097/01.aog.0000190212.87012.96
Pace, R. M., Chu, D. M., Prince, A. L., Ma, J., Seferovic, M. D., and Aagaard, K. M. (2021). Complex species and strain ecology of the vaginal microbiome from pregnancy to postpartum and association with preterm birth. Med (NY). 2, 1027–1049.e7. doi: 10.1016/j.medj.2021.06.001
Perino, A., Giovannelli, L., Schillaci, R., Ruvolo, G., Fiorentino, F. P., Alimondi, P., et al. (2011). Human papillomavirus infection in couples undergoing in vitro fertilization procedures: impact on reproductive outcomes. Fertil. Steril. 95, 1845–1848. doi: 10.1016/j.fertnstert.2010.11.047
Petricevic, L., Domig, K. J., Nierscher, F. J., Sandhofer, M. J., Fidesser, M., Krondorfer, I., et al. (2014). Characterisation of the vaginal lactobacillus microbiota associated with preterm delivery. Sci. Rep. 4:5136. doi: 10.1038/srep05136
Pilarski, N., Hodgetts-Morton, V., and Morris, R. K. (2021). Is cerclage safe and effective in preventing preterm birth in women presenting early in pregnancy with cervical dilatation? BMJ 375:e067470. doi: 10.1136/bmj-2021-067470
R Core Team (2017). A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria, https://www.R-project.org.
Ravel, J., Gajer, P., Abdo, Z., Schneider, G. M., Koenig, S. S., McCulle, S. L., et al. (2011). Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S A. 108, 4680–4687. doi: 10.1073/pnas.1002611107
Ren, H., and Du, M. (2017). Role of maternal periodontitis in preterm birth. Front. Immunol. 8:139. doi: 10.3389/fimmu.2017.00139
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. Peer J. 4:e2584. doi: 10.7717/peerj.2584
Rohart, F., Gautier, B., Singh, A., and Lê Cao, K. A. (2017). Mix Omics: An R package for 'omics feature selection and multiple data integration. PLoS Comput. Biol. 13:e1005752. doi: 10.1371/journal.pcbi.1005752
Smith, S. B., and Ravel, J. (2017). The vaginal microbiota, host defence and reproductive physiology. J. Physiol. 595, 451–463. doi: 10.1113/JP271694
Terzic, M., Aimagambetova, G., Terzic, S., Radunovic, M., Bapayeva, G., and Laganà, A. S. (2021). Periodontal pathogens and preterm birth: current knowledge and further interventions. Pathogens. 10:730. doi: 10.3390/pathogens10060730
Torcia, M. G. (2019). Interplay among vaginal microbiome, immune response and sexually transmitted viral infections. Int. J. Mol. Sci. 20:266. doi: 10.3390/ijms20020266
Tsonis, O., Gkrozou, F., Harrison, E., Stefanidis, K., Vrachnis, N., and Paschopoulos, M. (2020). Female genital tract microbiota affecting the risk of preterm birth: what do we know so far? A review. Eur. J. Obstet. Gynecol. Reprod. Biol. 245:168–173. doi: 10.1016/j.ejogrb.2019.12.005
Tu, Y., Zheng, G., Ding, G., Wu, Y., Xi, J., Ge, Y., et al. (2020). Comparative analysis of lower genital tract microbiome between PCOS and healthy women. Front. Physiol. 11:1108. doi: 10.3389/fphys.2020.01108
Villani, A., Fontana, A., Barone, S., de Stefani, S., Primiterra, M., Copetti, M., et al. (2022). Identifying predictive bacterial markers from cervical swab microbiota on pregnancy outcome in woman undergoing assisted reproductive technologies. J. Clin. Med. 11:680. doi: 10.3390/jcm11030680
Vogel, J. P., Chawanpaiboon, S., Moller, A. B., Watananirun, K., Bonet, M., and Lumbiganon, P. (2018). The global epidemiology of preterm birth. Best Pract. Res. Clin. Obstet. Gynaecol. 52, 3–12. doi: 10.1016/j.bpobgyn.2018.04.003
Vyas, N. A., Vink, J. S., Ghidini, A., Pezzullo, J. C., Korker, V., Landy, H. J., et al. (2006). Risk factors for cervical insufficiency after term delivery. Am. J. Obstet. Gynecol. 195, 787–791. doi: 10.1016/j.ajog.2006.06.069
Wang, X., Buhimschi, C. S., Temoin, S., Bhandari, V., Han, Y. W., and Buhimschi, I. A. (2013). Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sepsis. PLoS One 8:e56131. doi: 10.1371/journal.pone.0056131
Watts, D. H., Fazzari, M., Minkoff, H., Hillier, S. L., Sha, B., Glesby, M., et al. (2005). Effects of bacterial vaginosis and other genital infections on the natural history of human papillomavirus infection in HIV-1-infected and high-risk HIV-1-uninfected women. J. Infect. Dis. 191, 1129–1139. doi: 10.1086/427777
Keywords: cervical incompetence, bacteria, biomarker, OTUs, vagina and cervical canal
Citation: Sun M, Geng H, Bai J, Feng J, Xu N, Liu Y, Liu X and Liu G (2022) Characterization of cervical canal and vaginal bacteria in pregnant women with cervical incompetence. Front. Microbiol. 13:986326. doi: 10.3389/fmicb.2022.986326
Received: 05 July 2022; Accepted: 16 August 2022;
Published: 29 September 2022.
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
António Machado, Universidad San Francisco de Quito, EcuadorCopyright © 2022 Sun, Geng, Bai, Feng, Xu, Liu, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoying Liu, bGl1eGlhb3lpbmdAYWhtdS5lZHUuY24=; Gang Liu, bGl1Z2FuZzg5NjZAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.