- 1Food Security and Safety Focus Area, Faculty of Natural and Agricultural Sciences, North-West University, Private Mail Bag, Mmabatho, South Africa
- 2Department of Earth and Environmental Sciences, Laboratory of Mycology, University of Pavia, Pavia, Italy
- 3School of Pharmaceutical Science and Technology, Tianjin University, Tianjin, China
- 4Department of Botany, Faculty of Science, University of Ibadan, Ibadan, Oyo State, Nigeria
Trichoderma fungi have been proved as efficient bioagents with great antifungal properties while many species in the plant’s rhizospheres have been characterized as plant growth-promoting agents. However, many rhizosphere Trichoderma are yet to be fully explored for plant disease management. In this study, Trichoderma species were isolated from the rhizosphere of maize, banana, and cassava, and their biocontrol potentials were screened against some Fusarium species from oak leaves (F2B and F3) and laboratory cultures (Fus 296 and Fus 294). The isolated rhizosphere Trichoderma were identified as Trichoderma virens 1 (TCIV), T. virens 2 (TCVII), T. virens 3 (TMSI), T. hazianum strain 1 (TCVI), T. harzianum strain 2 (TCVIII), T. erinaceum (TMZI), and T. koningiopsis (TMZII). The dual culture experiment recorded the highest percentage inhibition in TMZII against OakF2B (31.17%), TCVIII against Fus 294 (45.18%), TMZI against Fus 296 (47.37%), while TCIV was most effective against Oak F3 (44.15%). Among the Trichoderma culture filtrates evaluated, TCIV showed the highest percentage inhibition against Oak F3 (52.39%), Oak F2B (48.54%), Fus 294 (46.65%), and Fus 296 (44.48%). All the Trichoderma isolates demonstrated expressed varying levels of antagonism against the Fusarium pathogens in vitro.
Introduction
Plant diseases are associated with diverse economic losses in various agricultural products and they presently constitute one of the most severe threats to food security and sustainability (Viana et al., 2022). The yield losses caused by pests and disease on food crops are enormous, resulting in 13–22% annual yield loss in staple crops such as wheat, rice, potato, and maize (Adeniji and Babalola, 2018). These are mostly caused by fungi and often result in poor quality and low yields (Nguyen et al., 2017; Olowe et al., 2020). Fusarium species are one of the causal agents of diseases in some crops of agricultural importance, such as rice and oak which are of major concern in Italy. Fusariums species are known for their global distribution and economic significance as producers of toxins such as fumonisin and other compounds causing contamination in plants. They are soil-borne pathogens that cause severe damping-off, vascular wilts, and rots in crops of economic importance (Bodah, 2017). Also, stunted seedlings, necrosis, defoliation, and browning of xylem vessels are associated with Fusarium infections in plants which may ultimately lead to death. Compared to other soil-borne pathogens, Fusarium species infect a wide range of host plants on fruits, roots, leaves, and stems. Fusarium fujikuroi, for example, has been found to cause stem and head rots and head blight in crops such as maize, rye, barley, wheat, and millet. Another is the instance of Fusarium tricinctum, which causes head blight in various cereal crops and leads to the accumulation of mycotoxin in the grains. Chemical fungicides are used to manage Fusarium diseases, although, the efficacy attained with these treatments is frequently less than predicted, and there is a need to restrict their use due to adverse environmental impacts (Shah et al., 2018). Also, excessive chemical use has been found responsible for the emergence of resistant races of Fusarium pathogens (Gupta et al., 2020). However, the biological control strategy is a viable alternative for fungal diseases. Moreover, it poses minimal or no negative effects on humans or the environment (Marutescu et al., 2017; Gupta et al., 2020).
The biocontrol method involves using bioagents, which can be achieved by introducing a prospective microbe or employing native microorganisms with antifungicidal potentials to curtail the growth and establishment of pathogens (Ajilogba et al., 2013; Köhl et al., 2019). Fungi as biocontrol agents are beneficial microbes that reduce the negative impacts of plant pathogens while enhancing plant growth. Aside from their biocontrol potentials, they also exhibit other benefits such as lowering the effects of abiotic and innate physiological stressors in plants and seeds, respectively. Furthermore, plants’ nitrogen utilization and photosynthetic efficiency are also improved. Therefore, the use of fungal biocontrol agents is an essential, economically sustainable, eco-friendly, and long-term strategy for plant disease (Ajilogba et al., 2013; Olowe et al., 2020; Rojas et al., 2020).
Trichoderma species are excellent biocontrol agents because of their diverse modes of action against plant diseases, including nutrient competition, mycoparasitism, and hydrolytic enzyme antibiosis (Filizola et al., 2019). The biocontrol potential of Trichoderma against Fusarium pathogens and other plant fungal pathogens has been widely researched. However, the use of Trichoderma in the sustainable management of diseases affecting crops has not gained wide recognition because the species used have not been characterized in many instances, and misidentification of the organisms often discourages its acceptability and its further deployment to farmers. In addition, there is limited information on Trichoderma efficacy against crop diseases isolated from various temperate zones. It is important to comprehend its effectiveness under various environmental conditions. The experiment aimed to examine the efficacy of certain Trichoderma strains in inhibiting the growth of some Fusarium species of oak and rice and to characterize them using molecular tools.
Materials and methods
Source of rhizosphere Trichoderma species and Fusarium species
A total of seven Trichoderma isolates, namely: TMS1, TMZ1. TMZ11, TCV1, TCV11, TCV111, and TCIV were obtained from soils collected from the rhizosphere of maize, banana, and cassava at the Students’ Farm of the University of Ibadan, Nigeria at Latitude 7.4433°N, and Longitude 3.9003°E. Fusarium isolates, Fus 296, Fus 294, were obtained from the culture collection of the Mycology Laboratory, Earth and Environmental Sciences, University of Pavia, Italy (45°11′06.5″N and 9°09′49.3″E), originally isolated from diseased rice plants, while F2B and F3 were isolated from lesions on oak leaves which were collected from Turbigo (Province Milano), Italy (Latitude: 45.5308°N Longitude: 8.7367°E). The diseased oak leaves were, collected between April and May, 2021, and brought to the laboratory for further analysis. The leaves were hand-picked from the tree showing symptoms of oak decline. Trichoderma isolates and Fusarium pathogens were cultured on Potato Dextrose Agar (PDA) and incubated at 28 ± 2°C for 7 days before bioassay.
Morpho-dimensional and molecular identification
The fungi were cultured on Potato Dextrose Agar (PDA) Petri dishes and incubated at 25°C for 7 days. The growing fungi were then examined morpho-dimensionally under a light microscope, observing the mycelium and the different reproductive structures. Molecular characterization was then performed to confirm the morphological identification. The isolates were cultured in Potato Dextrose Broth (PDB) and placed in a rotary shaker at 180 rpm for 1 week at 28 ± 2°C until the mycelia were well developed. The supernatant was removed by vacuum filtration, the mycelia were collected. The fungal genomic DNA was extracted using the NucleoSpin Plant II by Macherey-Nagel (Bethlehem, PA, United States), and subjected to PCR amplification of the internal transcribed spacer (ITS) region of the ITS1-5.8S-ITS2 rDNA gene, as described by Daccò et al. (2020). The PCR reaction was performed on a Thermocycler Bio-Rad T100 in a 25 μl reaction mixture containing 1× DREAM Taq Green PCR MasterMix reaction buffer (Thermo Scientific, Pittsburg, PA, United States), 2.5 μl (10 μM) of each primer, 3 μl of DNA sample, and 4.5 μl of Nuclease Free water. The PCR program was as follows: denaturation by heating for 5 min at 95°C, then 35 cycles of 30 s at 95°C, 45 s at 50°C, and 1 min at 72°C, and a final elongation step for 10 min at 72°C. The primers used were ITS1 (5′-TCCGTAGG TGAACCTGCGG-3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC-3′; Daccò et al. 2020). PCR products were purified with ExoSAP-IT (Applied Biosystems, Foster City, CA, United States) according to the manufacturer’s protocol. The amplified and purified DNA was sent to BMR Genomics (Padova, Italy), and the sequences were compared with target sequences using BLAST online1 and MEGA X 10.1.7.
Phylogenetic analysis
The phylogenetic analysis was performed using the generated ITS rRNA gene sequences from all the fungal strains used in this work. The sequences of the isolated pathogenic strains were aligned with other sequences from Fusarium species previously documented on the NCBI using Cluster X 2.1 (Thomson et al., 1997). The phylogenetic relationship between these selected fungi with our strains was determined using the Molecular Evolutionary Genetics Analysis (MEGA 7.0) software (Kumar et al., 2016). The neighbor-joining tree was performed using the Kimura 2-parameter distances with bootstrapping of 1,000 replicates, while Puccinia oxalidis OK093305 isolated from the leaf of Oxalis debilis in India was used as an outgroup to root the tree. Similarly, the sequences of the isolated fungal bioagents were aligned with other sequences from Trichoderma species previously documented on the NCBI.
Dual culture assay
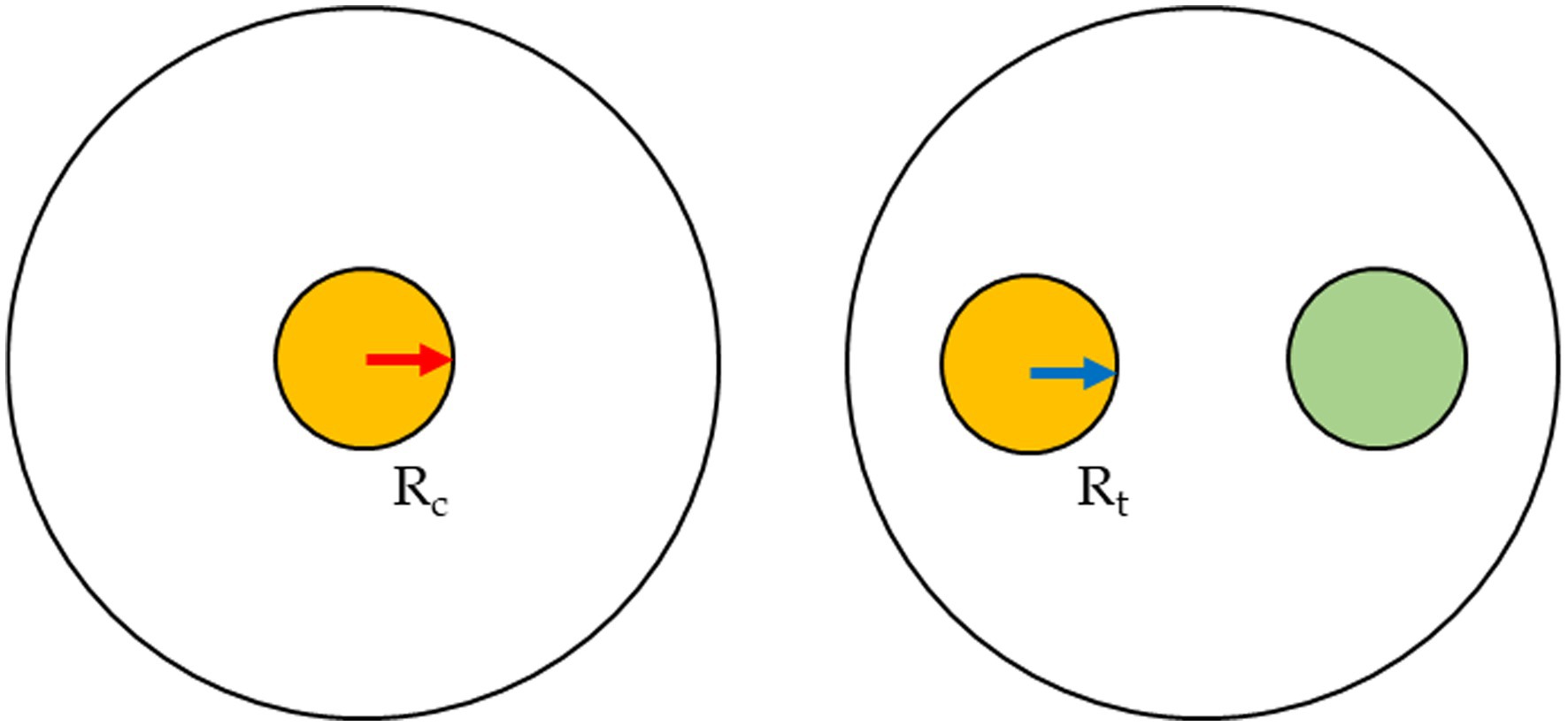
The anti-fungi activity of seven Trichoderma strains was screened against the Fusarium spp. through dual culture techniques. PDA plates were each inoculated with mycelium plugs (5 mm diameter) from separate cultures of Trichoderma and Fusarium. The mycelium plugs were removed from the growing margins of the 1-week culture of the fungi and placed on the opposite sides of the 90 mm diameter of the Petri-dishes, at a distance of 4 cm (Figure 1). Each Fusarium–Trichoderma combination has three replicates. PDA plates were also inoculated only with the Fusarium species only to serve as the control. The plates were incubated at 25 ± 2°C for 7 days. The radius of the mycelial growth was observed and measured every 24 h until the pathogen had grown to cover the plate’s surface. The percentage inhibition rate was calculated as:
where:
Rt = radius of the Fusarium when inoculated in a plate with Trichoderma.
Rc = radius of the Fusarium strain when inoculated alone (control).
Antifungal potency of culture filtrates of Trichoderma isolates
Five millimeters (5 mm diameter) mycelial plugs of the Trichoderma isolates from 3 to 5 days old culture were inoculated into 500 ml conical flasks, each containing 250 ml of sterilized PDB medium. The flasks were placed on the rotary shaker and incubated at room temperature with an agitation of 108 rpm for 1 week. After 1 week of incubation, the culture filtrates were collected into falcon tubes using a vacuum pump with sterilized paper filters. The culture filtrates were re-filtered twice using a sterile syringe filter of 0.2 μm. Five hundred microliter (500 μl) of the final fungal suspension were distributed on PDA plates using an L-shaped wand and were left to dry under the laminar flow hood. Each Fusarium species was inoculated on the plates containing the culture filtrate and without the culture filtrate (control). Three replicates were observed for each combination of Fusarium–Trichoderma culture filtrate.
Statistical analysis
Data obtained were subjected to analyses of variance (ANOVA) using Statistical Analysis Software (SAS 2003), while means were separated using Duncan’s Multiple Range Test at p ≤ 0.05.
Results
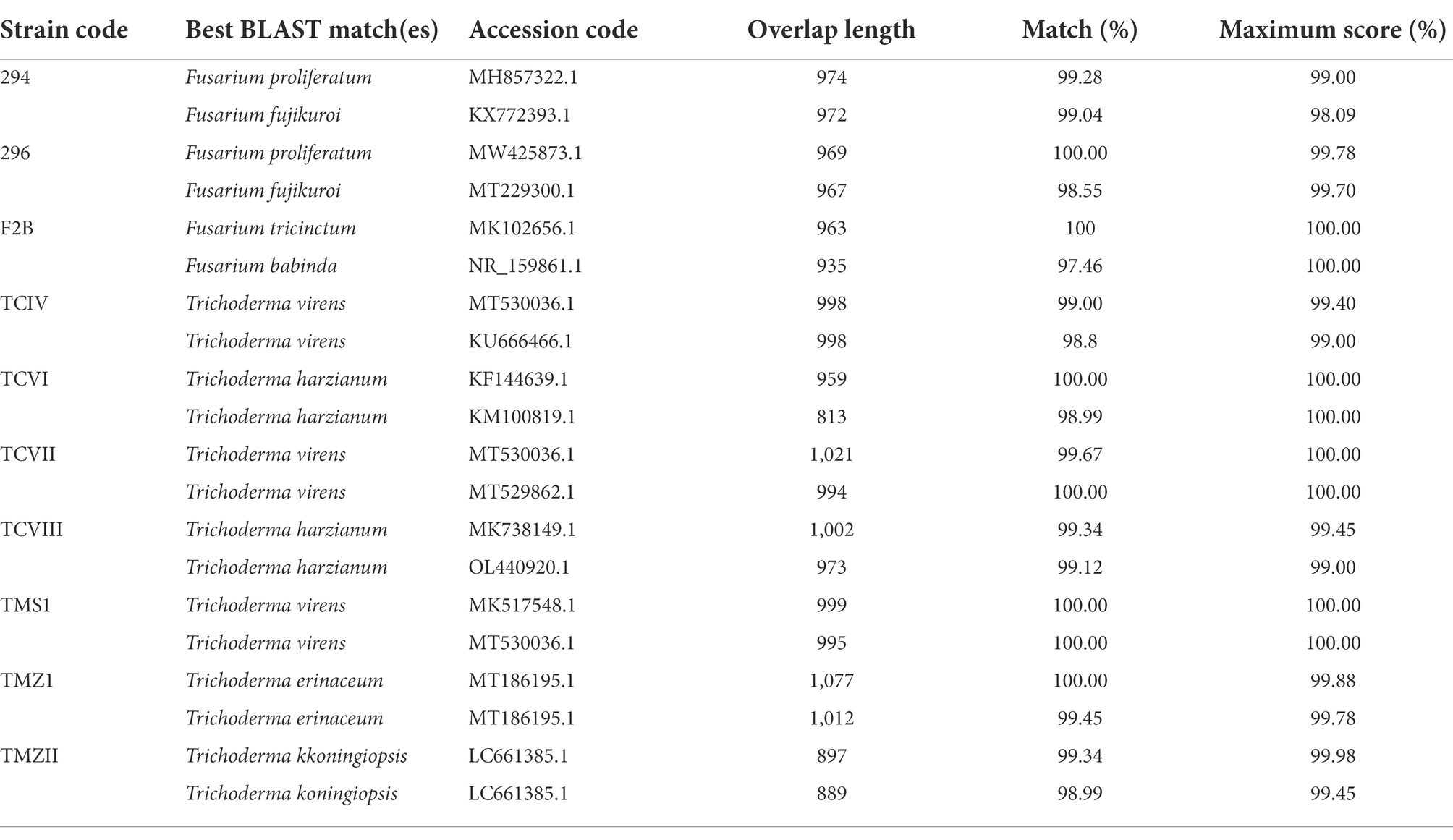
In this study, three fungi and seven bioagents were isolated, and identified using PCR-ITS amplification of the rRNA gene. Based on BLAST Search on the NCBI, the pathogens were identified as Fusarium proliferatum Strain 294, Fusarium proliferatum Strain 296, and Fusarium tricinctum Strain F2B. The bioagents were identified as Trichoderma virens Strain TCIV, Trichoderma harzianum Strain TCVI, Trichoderma virens Strain TCVII, Trichoderma harzianum Strain, Trichoderma virens Strain TMS1, Trichoderma erinaceum Strain TMZ1, and Trichoderma koningiopsis Strain TMZII (Table 1). The Trichoderma species were then documented on the NCBI under the accession numbers ON149743–ON149749.
The DNA sequences from isolated Fusarium strains were successfully aligned with other sequences of strains that were previously documented on the NCBI. The neighbor-joining tree revealed the phylogenetic relationships between the selected Fusarium pathogens (Figure 2). However, our isolated strain 294, clustered into a distinct well-supported clade with F. proliferatum HM769953 and three other pathogenic strains HM769953, JX970629, and MW647758 isolated from India (Figure 2).
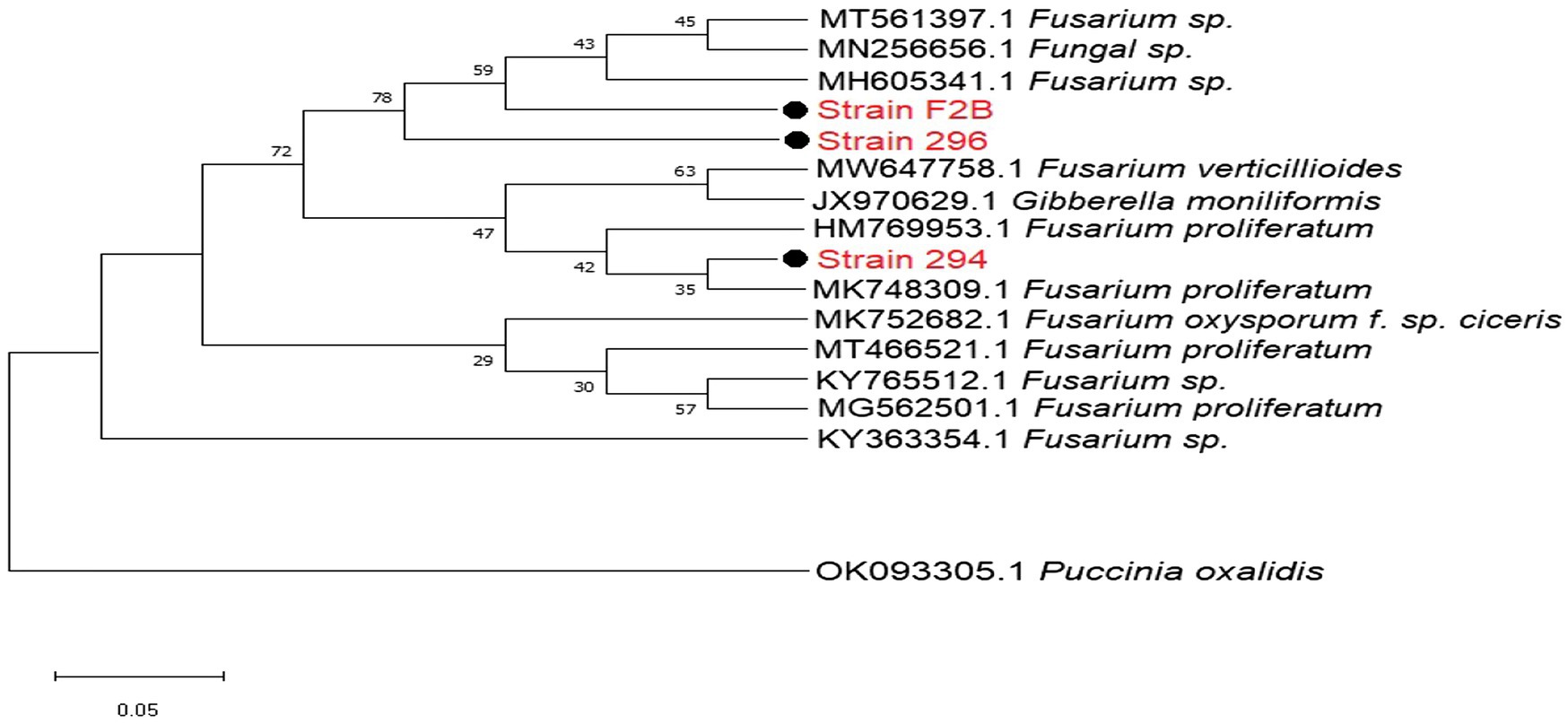
Figure 2. Phylogenetic relationships between the isolated Fusariumpathogenic strains and selected database relatives on the NCBI generated through internal transcribed spacer (ITS) rRNA gene analysed via Kimura’s two parameter models. Bootstrap support values higher than 50% from 1,000 replicates are shown at the nodes. P. oxalidis (OK093303) was used as the out-group.
For the Trichoderma strains, most of our strains except TMZ1 clustered into a distinct well-supported clade with each other. They all showed more similar phylogenetic relationship with each other than those that were previously documented on the NCBI (Figure 3). Our two strains; T. virens TCIV and TCVII showed the most similar phylogenetic relationship while both clustered into another related clade with TMS1, TMZII, TCVIII, and TCVI (Figure 3).
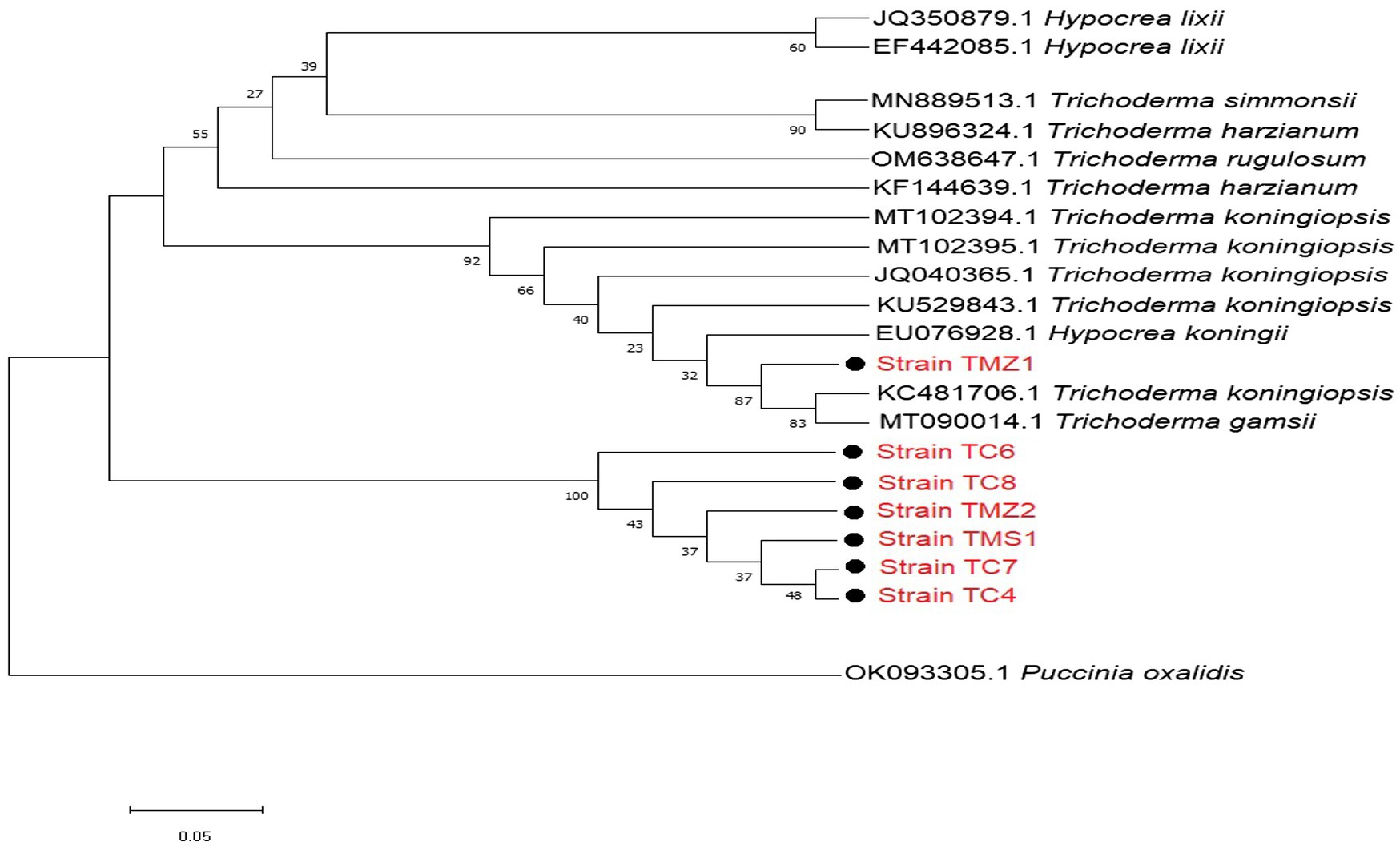
Figure 3. Phylogenetic relationships between the isolated bioagent strains and selected database of Trichoderma relatives from the NCBI analyzed via Kimura’s two-parameter models. Bootstrap support values higher than 50% from 1,000 replicates are shown at the nodes. P. oxalidis (OK093303) was used as the out-group.
The percentage inhibition of Fusarium pathogens (OakF2B, Fus 296, Fus 294, Oak F3, Oak 3FA) by the antagonistic Trichoderma species (TMZ1, TMZII, TMS1, TCIV, TCVI, TCVII, and TCV8) in the dual culture experiment showed significant (p < 0.05) inhibition as examined after 7th day of observation in the dual culture (Figure 4). All the antagonistic Trichoderma species increased percentage inhibition with increasing days of evaluation on all the Fusarium species tested, except in TCIV against Oak F3, which was most inhibited at day 4 (71.26%; Table 2; Figure 5A). The highest percentage inhibition was recorded in TMZII against OakF2B (31.17%), TCVIII against Fus 294 (45.18%), TMZI against Fus 296 (47.37%), while TCIV was most effective against Oak F3 (44.15%). Furthermore, the pooled effect of Trichoderma species in this study showed TMZII > TMZI > TCVIII > TCVII > TCVI > TCIV > TMS1 as the order of antagonistic efficacy against the Fusarium species, while Fus 294 > Fus 296 > OakF3A > OakF2B > Oak F3 (Figure 6A).
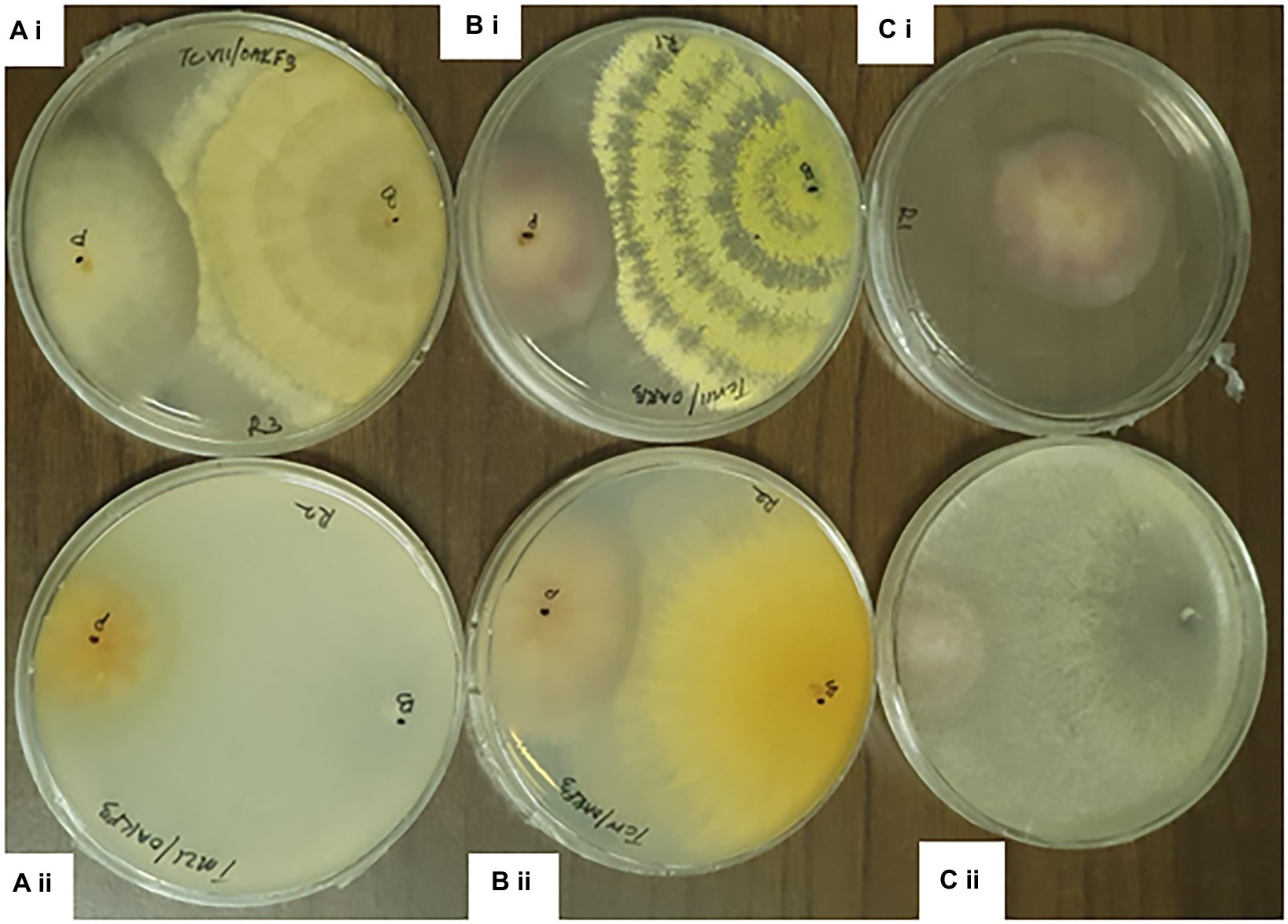
Figure 4. Dual culture assay of Trichoderma species against Fusarium pathogens at 7th day of Incubation. ai= TCVII/ OakF3, bi= TCVIII/OakF3, ci= Control aii= TMZI/ Oak F3, bii= TCIV/OakF3, cii= TMZII/OakF3.
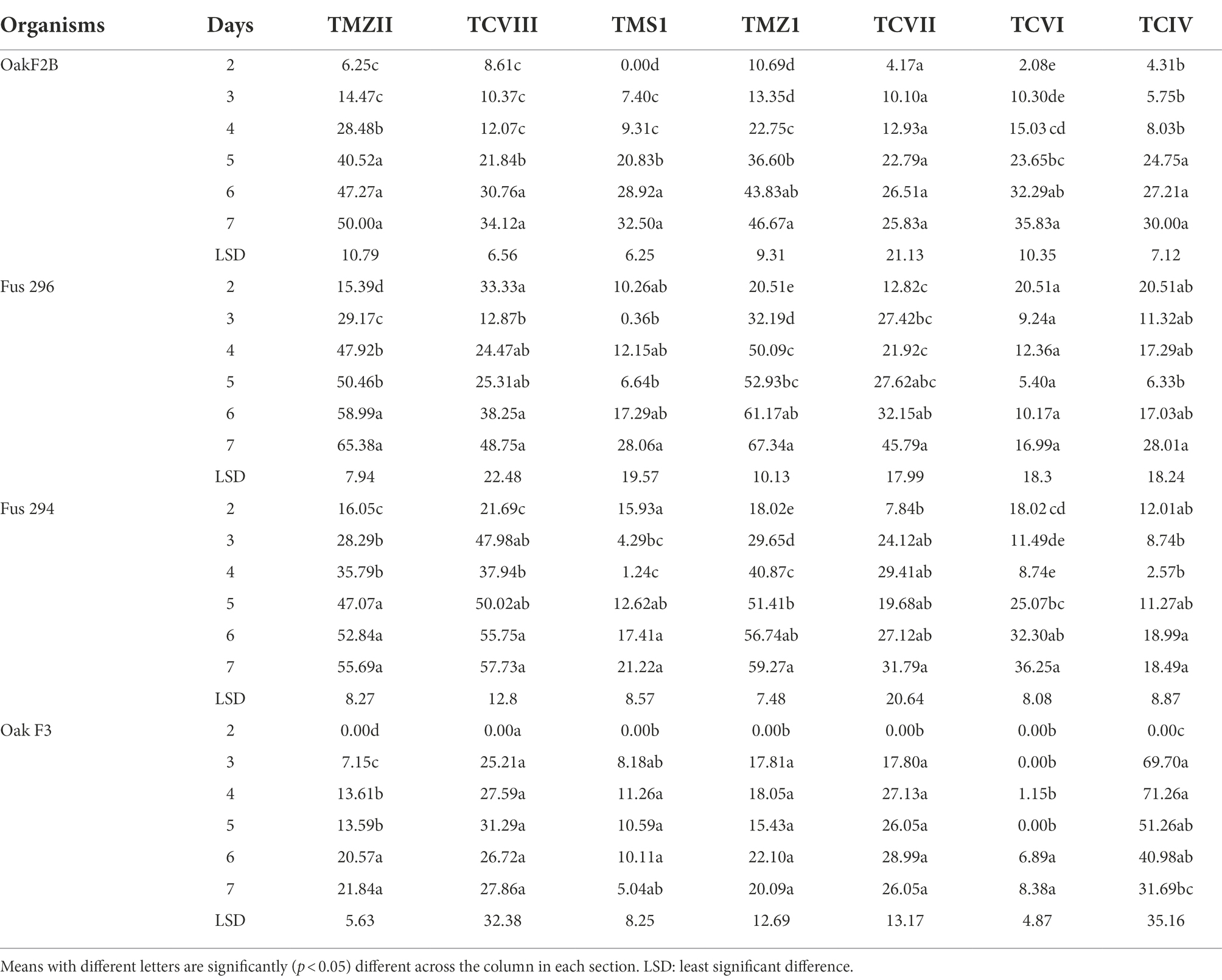
Table 2. Percentage inhibition (%) of the Fusarium species by mycelia of the Trichoderma isolates in dual culture
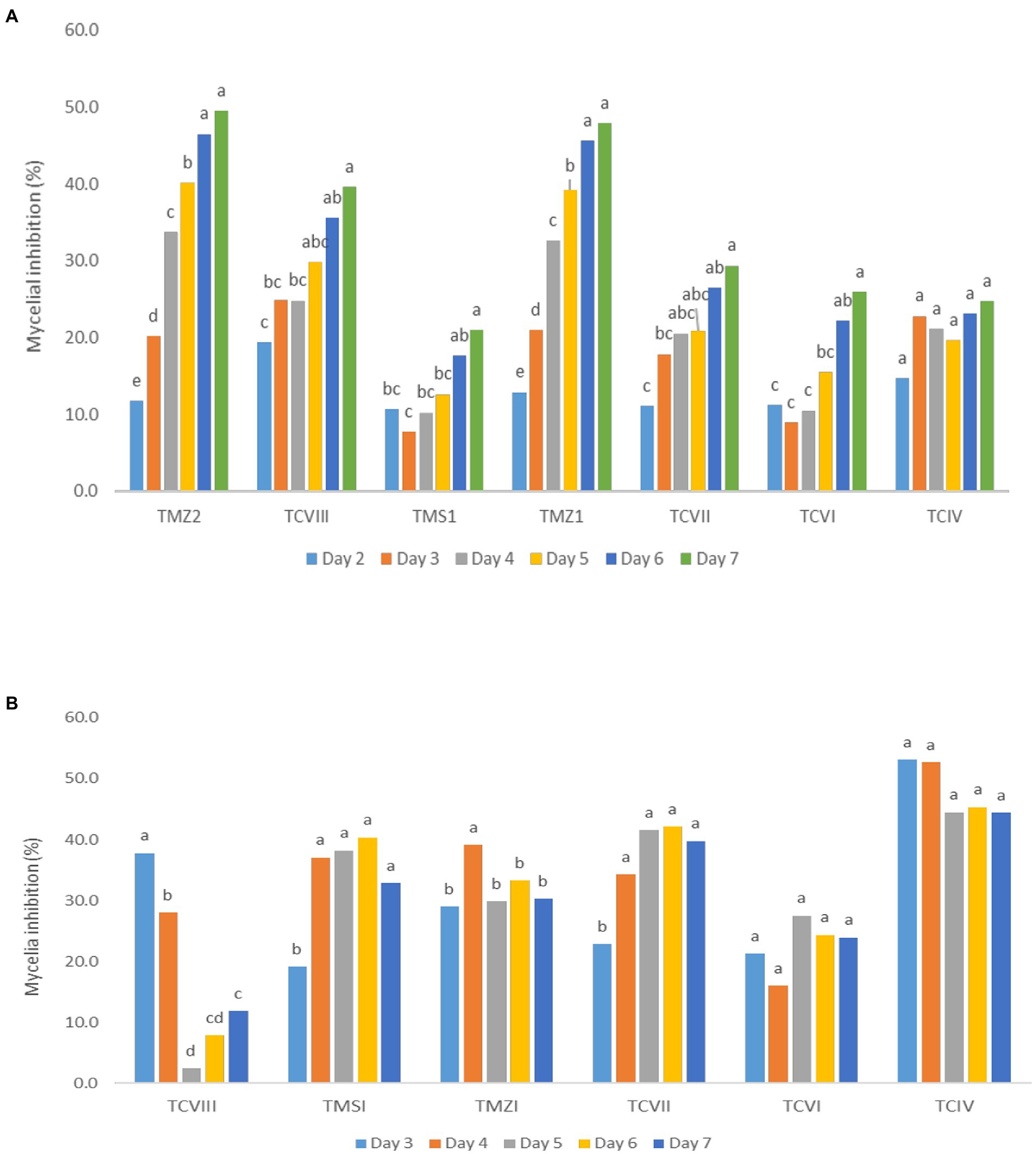
Figure 5. Inhibitory potentials of Trichoderma species through (A) the dual culture technique, and (B) culture filtrates, against Fusarium species with respect to days of observation. Means with different letters are significantly (p < 0.05) different across each Trichoderma treatment.
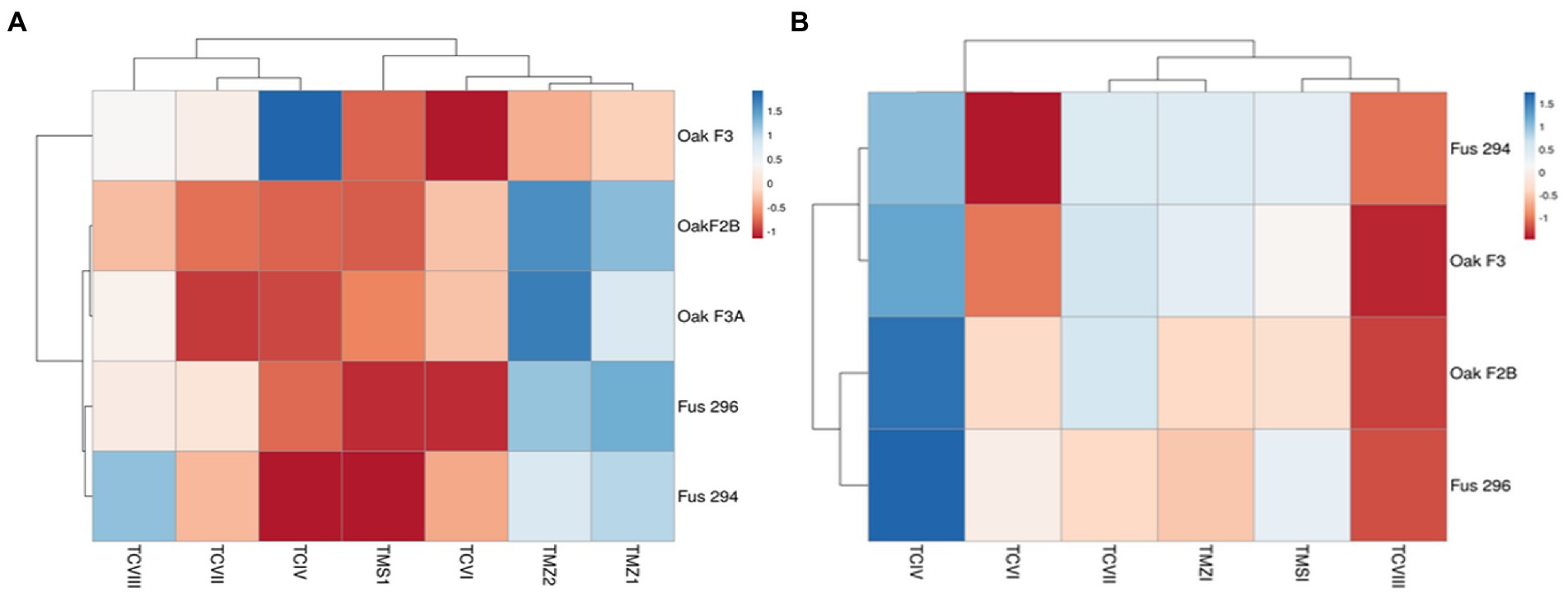
Figure 6. Pooled antagonistic effect of Trichoderma species against Fusarium pathogens with (A) dual culture, and (B) culture filtrates.
The results obtained from the Trichoderma culture filtrate showed increasing percentage inhibition of TCIV, TMSI, and TCVII across the Fusarium species while others showed effectiveness against selected pathogens at varying days (Table 3; Figures 5B, 7). Generally, TCIV showed the highest percentage inhibition against Oak F3 (52.39%), Oak F2B (48.54%), Fus 294 (46.65%) and Fus 296 (44.48%). The pooled result showed TCIV > TCVII > TMSI > TMZI > TCVI > TCVIII as the order of inhibition by the culture filtrates. Also, this study showed OakF3 > OakF2B > Fus 294 and Fus 296 as the inhibited pathogens (Figure 6B).
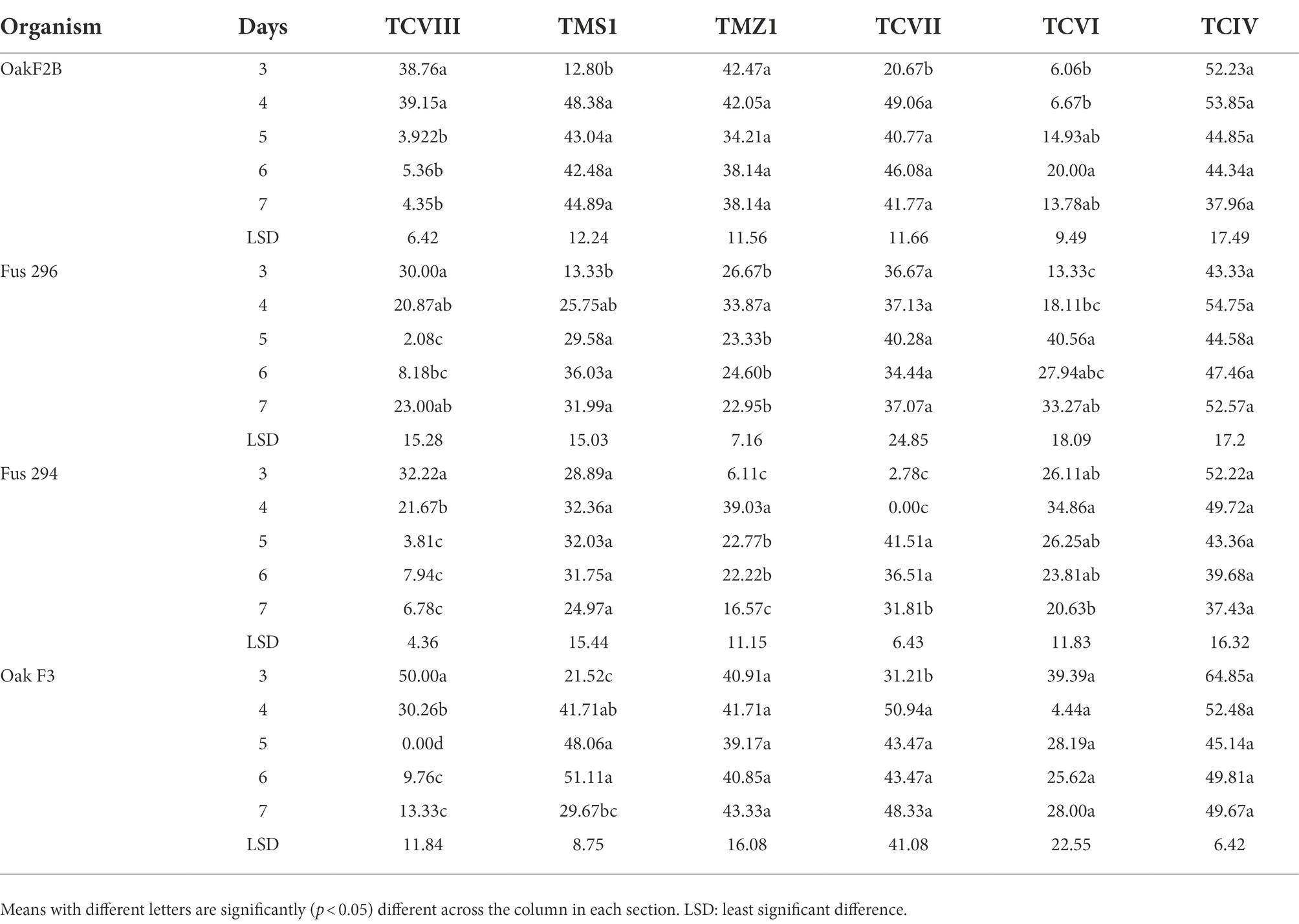
Table 3. Percentage inhibition (%) of the Fusarium species by culture filtrates of the Trichoderma isolates.
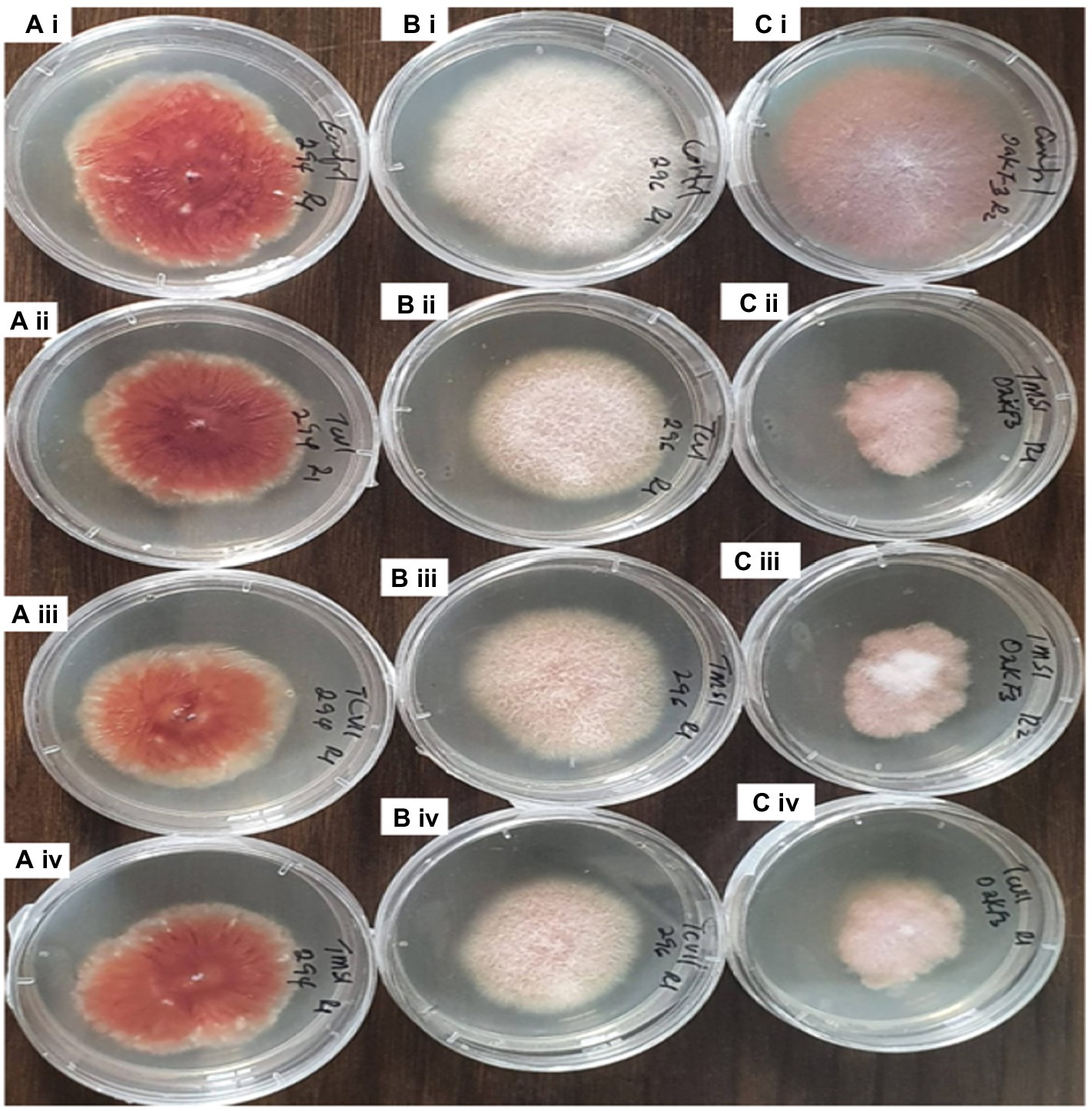
Figure 7. Inhibitory effect of the culture filtrates of antagonistic isolates of Trichoderma against plant pathogens Fusarium on 7th day of observation. Ai= Fus 294 (control), Bi= Fus 296 (control), Ci= OakF3, Aii= TCVI/Fus294, Bii= TCVI/Fus 296, Cii= TMZI/ OakF3 (RI), Aiii= TVCII/Fus294, Biii= TMSI/Fus296, Ciii= TMZI/ OakF3 (R2), Aiv= TMSI/Fus294, Biv= TCVII/Fus296, Civ= TCVII/OakF3.
Discussion
Most phylogenetically related strains were pathogens that have been previously isolated from various plants. Strains F2B and 296 were most closely associated with Fusarium sp. MH605341, is a pathogen associated with the root of a popular medicinal plant, Panax ginseng from Changchun, China. This strain was recently matched with high similarity to an endophyte isolated from Orchid by Yokoya et al. (2021). These two strains (Fusarium tricinctum Strain 294 and Fusarium proliferatum Strain 296) also showed high similarity with Fusarium sp. Strains MN256656 and MT561397. Strain MT561397 was a recently isolated strain by Langer and Bußkamp (2021) as a pathogen from the bark of the European beech (Fagus sylvatica) a deciduous tree belonging to the beech family Fagaceae commonly refers to as beech. The two T. virens TC4 and TC7 Trichoderma strains that expressed the most similar phylogenetic relationship in this study had related clades with TMS1, TMZ2, TC8 and, TC6. These strains all together showed a phylogenetic relationship with other strains that were recently confirmed as potential bioagents. They included T. gamsii MT090014 a Trichoderma strainTY190-12 recently isolated from QinLing Mountain, in China by An et al. (2020), and T. koningiopsis KC481706 a newly isolated Trichoderma endophyte of deciduous wood tree Leucas aspera strain. T. koningiopsis KC481706 is a Trichoderma strain UFSM T002sB recently isolated by Tseng et al. (2020). Interestingly, UFSM T002sB was demonstrated to promote the growth of Arabidopsis thaliana and Nicotiana attenuata against pathogen attacks by Tseng et al. (2020). In another view, strain TMZ1 was distinct from others which was more phylogenetically different from other strains, it showed high similarity with T. koningiopsis KC481706 and T. gamsii MT090014. T. gamsii MT090014 is another Trichoderma strain TY190-12 also isolated by An et al. (2020) from China. Strain TMZ1 also shared a high similarity with Hypocrea koningiopsis EU076928. EU076928 is a soil bioagent strain 16-M-1 recently isolated from agricultural soil in Vietnam by Ghazanfar et al. (2018).
Investigation into the biocontrol potential of Trichoderma species against pathogens of crops of agricultural importance was inspired by the need to develop viable biologically based pesticides, which are eco-friendly to replace synthetic pesticides (Olowe et al., 2020; Akanmu et al., 2021). Variations in the inhibitory potentials of different Trichoderma species was demonstrated in this study as each Trichoderma species expressed the highest inhibition of specific Fusarium species such as the efficacy of TMZII against OakF2B (31.17%), TCVIII against Fus 294 (45.18%) and TCIV against Oak F3 (44.15%). This result could be ascribed to their varying sources and attributes of the organisms, an earlier investigation conducted by Anjum et al. (2020) also reported a similar observation, where five different Trichoderma species evaluated showed disparities in the inhibition of Fusarium oxysporum f. spp. capsica causing Fusarium wilt of Chili. More so, the respective order of antagonistic activities of Trichoderma species with TMZII as the most effective is consistent with some earlier reports on the inhibitory potentials of Trichoderma species against plant pathogens (Talapatra et al., 2017; Olowe et al., 2022). Furthermore, advantages recorded in the use of Trichoderma are justified by their rapid establishment within the stable microbial communities, and control of pathogenic, competitive, or deleterious microflora through various mechanisms (Vinale et al., 2008).
The results similar to that of the dual culture assay were obtained from the Trichoderma culture filtrates, especially in TCIV, TMSI, and TCVII across the Fusarium species. Although, variations were recorded in the antagonistic activities of the culture filtrates of each specie with TCIV expressing the highest percentage inhibition against Oak F3 (52.39%), and Oak F2B (48.54%), Fus 294 (46.65%), and Fus 296 (44.48%). Similar results with respect to the genetic variations among the Trichoderma species had earlier been reported (Rai et al., 2016). More so, the variation in the virulence of Fusarium species could contribute to the differences in the rate of inhibition recorded by the Trichoderma species (Kolp et al., 2018). This affirms the varying levels of biocontrol activities against pathogens, as also reported by Nandini et al. (2021). However, these results validate the earlier findings of the dual culture and affirm the antagonistic potentials of Trichoderma species through different mechanisms of action, including competition for nutrients, mycoparasitism, and antibiosis (Rajani et al., 2021). This strongly underscores the mycoparasitic potential of these Trichoderma species against pathogens such as Fusarium species (Olowe et al., 2020), as also affirmed in the interactions of TMZII against Fus 294 and 296 in this study. While the type of interaction observed between the Fusarium and Trichoderma was not verified, this study suggests the existence of antifungal traits in the culture filtrates, and the suspected mycoparasitism is found in agreement with the report of Mukherjee et al. (2022) as the most effective disease suppression method.
Conclusion
The current investigation verified the effectiveness of certain Trichoderma isolates as bioagents against Fusarium pathogens of oak and rice. The Trichoderma species in both dual and culture filtrate evaluation demonstrated varying degrees of inhibitory effects against the Fusarium pathogens of rice and oak leaves, thereby ascertaining the antagonistic potentials of indigenous Trichoderma species against Fusarium pathogens. This research identifies more members of the genus Trichoderma with promising antagonistic prowess against fungi of economic importance, such as Fusarium species. Further experiments in the field are however imperative before credible assertions could be made on their mycoparasitic capabilities.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
OB recognizes National Research Fund (NRF South Africa) for grants (UID123634 and UID132595) that support work in her research group.
Acknowledgments
OO appreciates the Centro Internazionale Cooperazione per lo Sviluppo (CICOPS) of the University of Pavia for financing her research in the Department of Earth and Environmental Sciences, University of Pavia, Italy. She also thanks professor Solveig Tosi of Mycology Laboratory, Department of Earth and Environmental Sciences, University of Pavia, Italy, for her academic support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Adeniji, A. A., and Babalola, O. O. (2018). Tackling maize fusariosis: in search of Fusarium graminearum biosuppressors. Arch. Microbiol. 200, 1239–1255. doi: 10.1007/s00203-018-1542-y
Ajilogba, C. F., Babalola, O. O., and Ahmad, F. (2013). Antagonistic effects of bacillus species in biocontrol of tomato Fusarium wilt. Stud. Ethno-Med. 7, 205–216. doi: 10.1080/09735070.2013.11886462
Akanmu, A. O., Babalola, O. O., Venturi, V., Ayilara, M. S., Adeleke, B. S., Amoo, A. E., et al. (2021). Plant disease management: leveraging on the plant-microbe-soil interface in the biorational use of organic amendments. Front. Plant Sci. 12:700507. doi: 10.3389/fpls.2021.700507
An, C., Ma, S., Shi, X., Xue, W., Liu, C., and Ding, H. (2020). Diversity and antimicrobial activity of endophytic fungi isolated from Chloranthus japonicus Sieb in Qinling Mountains, China. Int. J. Mol. Sci. 21:5958. doi: 10.3390/ijms21175958
Anjum, N., Shahid, A. A., Iftikhar, S., Mubeen, M., Ahmad, M. H., Jamil, Y., et al. (2020). Evaluations of Trichoderma isolates for biological control of Fusarium wilt of chili. Plant Cell Biotechnol. Mol. Biol. 21:42.
Bodah, E. T. (2017). Root rot diseases in plants: a review of common causal agents and management strategies. Agri. Res. Tech. Open Access J. 5:555661. doi: 10.19080/ARTOAJ.2017.04.555661
Daccò, C., Lidia, N., Marta Elisabetta, E. T., Barbara, M., Federica, C., Giovanna, C., et al. (2020). “Trichoderma: Evaluation of Its Degrading Abilities for the Bioremediation of Hydrocarbon Complex Mixtures.” Applied Sciences 10, 3152. doi: 10.3390/app10093152
Filizola, P. R. B., Luna, M. A. C., de Souza, A. F., Coelho, I. L., Laranjeira, D., and Campos-Takaki, G. M. (2019). Biodiversity and phylogeny of novel Trichoderma isolates from mangrove sediments and potential of biocontrol against Fusarium strains. Microb. Cell Factories 18, 1–14. doi: 10.1186/s12934-019-1108-y
Ghazanfar, M. U., Raza, M., Raza, W., and Qamar, M. I. (2018). Trichoderma as potential biocontrol agent, its exploitation in agriculture: a review. Plant Prot. 2, 109–135.
Gupta, V., Kumar, K., Fatima, K., Razdan, V. K., Sharma, B. C., Mahajan, V., et al. (2020). Role of biocontrol agents in management of corm rot of saffron caused by Fusarium oxysporum. Agronomy 10:1398. doi: 10.3390/agronomy10091398
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kolp, M., Fulbright, D. W., and Jarosz, A. M. (2018). Inhibition of virulent and hypovirulent Cryphonectria parasitica growth in dual culture by fungi commonly isolated from chestnut blight cankers. Fungal Biol. 122, 935–942. doi: 10.1016/j.funbio.2018.05.007
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. FMolecular biology and evolution, 33, 1870–1874.
Langer, G. J., and Bußkamp, J. (2021). Fungi associated with Woody tissues of European beech and their impact on tree health. Front. Microbiol. 12:702467. doi: 10.3389/fmicb.2021.702467
Marutescu, L., Popa, M., Saviuc, C., Lazar, V., and Chifiriuc, M. C. (2017). “Botanical pesticides with virucidal, bactericidal, and fungicidal activity,” in New Pesticides and Soil Sensors. ed A. M. Grumezescu, (Amsterdam: Elsevier), 311–335.
Mukherjee, P. K., Mendoza-Mendoza, A., Zeilinger, S., and Horwitz, B. A. (2022). Mycoparasitism as a mechanism of Trichoderma-mediated suppression of plant diseases. Fungal Biol. Rev. 39, 15–33. doi: 10.1016/j.fbr.2021.11.004
Nandini, B., Puttaswamy, H., Saini, R. K., Prakash, H. S., and Geetha, N. (2021). Trichovariability in rhizosphere soil samples and their biocontrol potential against downy mildew pathogen in pearl millet. Sci. Rep. 11, 1–15. doi: 10.1038/s41598-021-89061-2
Nguyen, P.-A., Strub, C., Fontana, A., and Schorr-Galindo, S. (2017). Crop molds and mycotoxins: alternative management using biocontrol. Biol. Control 104, 10–27. doi: 10.1016/j.biocontrol.2016.10.004
Olowe, O. M., Akanmu, A. O., and Asemoloye, M. D. (2020). Exploration of microbial stimulants for induction of systemic resistance in plant disease management. Ann. Appl. Biol. 177, 282–293. doi: 10.1111/aab.12631
Olowe, O. M., Nicola, L., Asemoloye, M. D., Akanmu, A. O., and Babalola, O. O. (2022). Trichoderma: potential bio-resource for the management of tomato root rot diseases in Africa. Microbiol. Res. 257:126978. doi: 10.1016/j.micres.2022.126978
Rai, S., Kashyap, P. L., Kumar, S., Srivastava, A. K., and Ramteke, P. W. (2016). Identification, characterization and phylogenetic analysis of antifungal Trichoderma from tomato rhizosphere. Springerplus 5, 1–16. doi: 10.1186/s40064-016-3657-4
Rajani, P., Rajasekaran, C., Vasanthakumari, M., Olsson, S. B., Ravikanth, G., and Shaanker, R. U. (2021). Inhibition of plant pathogenic fungi by endophytic Trichoderma spp. through mycoparasitism and volatile organic compounds. Microbiol. Res. 242:126595. doi: 10.1016/j.micres.2020.126595
Rojas, E. C., Jensen, B., Jørgensen, H. J., Latz, M. A., Esteban, P., Ding, Y., et al. (2020). Selection of fungal endophytes with biocontrol potential against Fusarium head blight in wheat. Biol. Control 144:104222. doi: 10.1016/j.biocontrol.2020.104222
Shah, L., Ali, A., Yahya, M., Zhu, Y., Wang, S., Si, H., et al. (2018). Integrated control of Fusarium head blight and deoxynivalenol mycotoxin in wheat. Plant Pathol. 67, 532–548. doi: 10.1111/ppa.12785
Talapatra, K., Das, A. R., Saha, A., and Das, P. (2017). In vitro antagonistic activity of a root endophytic fungus towards plant pathogenic fungi. J. Appl. Biol. Biotechnol. 5, 68–71. doi: 10.7324/JABB.2017.50210
Thompson, Julie D., Gibson, Toby J., Plewniak, F., Jeanmougin, F., and Higgins, Desmond, G.. (1997). The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools, nucleic acids research. 25:4876–4882. doi: 10.1093/nar/25.24.4876
Tseng, Y.-H., Rouina, H., Groten, K., Rajani, P., Furch, A. C., Reichelt, M., et al. (2020). An endophytic Trichoderma strain promotes growth of its hosts and defends against pathogen attack. Front. Plant Sci. 11:573670. doi: 10.3389/fpls.2020.573670
Viana, C. M., Freire, D., Abrantes, P., Rocha, J., and Pereira, P. (2022). Agricultural land systems importance for supporting food security and sustainable development goals: a systematic review. Sci. Total Environ. 806:150718. doi: 10.1016/j.scitotenv.2021.150718
Vinale, F., Sivasithamparam, K., Ghisalberti, E. L., Marra, R., Woo, S. L., and Lorito, M. (2008). Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 40, 1–10. doi: 10.1016/j.soilbio.2007.07.002
Keywords: bioagents, pathogen, mycoparasitism, rhizosphere, culture filtrates, antagonism
Citation: Olowe OM, Nicola L, Asemoloye MD, Akanmu AO, Sobowale AA and Babalola OO (2022) Characterization and antagonistic potentials of selected rhizosphere Trichoderma species against some Fusarium species. Front. Microbiol. 13:985874. doi: 10.3389/fmicb.2022.985874
Edited by:
Simona Lucia Bavaro, National Research Council, ItalyReviewed by:
Laith Khalil Tawfeeq Al-Ani, Universiti Sains Malaysia, MalaysiaIbrahim Elshahawy, National Research Centre, Egypt
Copyright © 2022 Olowe, Nicola, Asemoloye, Akanmu, Sobowale and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola Oluranti Babalola, b2x1YnVrb2xhLmJhYmFsb2xhQG53dS5hYy56YQ==
 Olumayowa Mary Olowe
Olumayowa Mary Olowe Lidia Nicola
Lidia Nicola Micheal Dare Asemoloye
Micheal Dare Asemoloye Akinlolu Olalekan Akanmu
Akinlolu Olalekan Akanmu Ayodele Adegboyega Sobowale
Ayodele Adegboyega Sobowale Olubukola Oluranti Babalola
Olubukola Oluranti Babalola