- 1Department of Biology and Biochemistry, University of Bath, Bath, United Kingdom
- 2Centre for Sustainable and Circular Technologies (CSCT), University of Bath, Bath, United Kingdom
- 3Laboratory of Microbiology, Wageningen University, Wageningen, Netherlands
- 4Corbion, Gorinchem, Netherlands
- 5Department of Pharmacy and Pharmacology, Centre for Therapeutic Innovation, University of Bath, Bath, United Kingdom
Parageobacillus thermoglucosidasius is a thermophilic bacterium of interest for lignocellulosic biomass fermentation. However, carbon catabolite repression (CCR) hinders co-utilization of pentoses and hexoses in the biomass substrate. Hence, to optimize the fermentation process, it is critical to remove CCR in the fermentation strains with minimal fitness cost. In this study, we investigated whether CCR could be removed from P. thermoglucosidasius DSM 2542 by mutating the Ser46 regulatory sites on HPr and Crh to a non-reactive alanine residue. It was found that neither the ptsH1 (HPr-S46A) nor the crh1 (Crh-S46A) mutation individually eliminated CCR in P. thermoglucosidasius DSM 2542. However, it was not possible to generate a ptsH1 crh1 double mutant. While the Crh-S46A mutation had no obvious fitness effect in DSM 2542, the ptsH1 mutation had a negative impact on cell growth and sugar utilization under fermentative conditions. Under these conditions, the ptsH1 mutation was associated with the production of a brown pigment, believed to arise from methylglyoxal production, which is harmful to cells. Subsequently, a less directed adaptive evolution approach was employed, in which DSM 2542 was grown in a mixture of 2-deoxy-D-glucose(2-DG) and xylose. This successfully removed CCR from P. thermoglucosidasius DSM 2542. Two selection strategies were applied to optimize the phenotypes of evolved strains. Genome sequencing identified key mutations affecting the PTS components PtsI and PtsG, the ribose operon repressor RbsR and adenine phosphoribosyltransferase APRT. Genetic complementation and bioinformatics analysis revealed that the presence of wild type rbsR and apt inhibited xylose uptake or utilization, while ptsI and ptsG might play a role in the regulation of CCR in P. thermoglucosidasius DSM 2542.
1. Introduction
Producing energy and chemicals from fossil fuels is not sustainable, and therefore several alternative approaches have been considered, among which microbial fermentation of renewable feedstocks has gained growing interest (Wee et al., 2006; Wang et al., 2010). Renewable lignocellulosic feedstocks such as purpose grown crops from non-arable land or waste lignocellulosic biomass are particularly attractive feedstocks (Vishnu et al., 2002; Wee et al., 2006) as it is abundant and readily available in many parts of the world (Oh et al., 2005; Banerjee et al., 2010). Lignocellulose derived from plants mainly consists of cellulose [40–60% w/w(dry)], hemicellulose (20-40%) and lignin (10-20%). After pre-treatment and hydrolysis, a mixture of C6 and C5 sugars is released, including glucose, galactose, mannose, xylose and arabinose. These sugars can be fermented using methods similar to those of first-generation feedstocks; however, some microbes are not able to grow on C5 sugars (Kang et al., 2014). Hence, the ability to utilize both C6 and C5 sugars is an important trait when selecting strains for lignocellulosic biomass fermentation (Banerjee et al., 2010). To date, the most common microbial process organisms are mesophilic, such as Escherichia coli, Lactobacillus, and Saccharomyces (Banerjee et al., 2010; Mattanovich et al., 2014). However, it has been suggested that certain thermophilic bacteria are more suitable for lignocellulosic biomass fermentation, as a fermentation process operating at 55–60°C is more suited for simultaneous saccharification and fermentation (SSF), improves the energy efficiency of this exothermic process and reduces contamination risks (Bacon et al., 2017). High temperatures also facilitate the removal of volatile products (Hills, 2015). Many thermophilic microbial species have been investigated, including Geobacillus and Parageobacillus spp. These are of industrial interest because, not only can they ferment the most abundant hexose and pentose monomers in lignocellulose, but also cellobiose and complex xylo-oligosaccharides present in pretreated lignocellulosic biomass (Reeve et al., 2016; Sheng et al., 2016). For example, P. thermoglucosidasius has been engineered for bio-ethanol, terpenes and (S)-lactic acid production (Atkinson et al., 2013; Van Kranenburg et al., 2019; Styles et al., 2021).
Carbon catabolite control enables the preferential utilization of the most energy-efficient carbon source, so that microbes can achieve a higher growth yield but retain the metabolic flexibility to survive in a nutritionally fluctuating environment (Martin-Verstraete et al., 1999; Görke and Stülke, 2008). For optimal metabolic efficiency in a competitive environment, metabolism of the preferred carbon source, typically glucose, constrains the expression of genes responsible for catabolism of those less-favored carbon sources; this regulatory phenomenon is called carbon catabolite repression (CCR) (Görke and Stülke, 2008; Vinuselvi et al., 2012). It is also known as the glucose effect, because the existence of glucose often represses the consumption of other carbon sources, although this is not universally the case (Shaw et al., 2008; VanFossen et al., 2009). In fermentation of plant-derived feedstocks, where carbon sources are naturally combined in different proportions, sequential consumption of sugars resulting from CCR leads to extended fermentation times and differences in metabolism of different sugars can make it challenging to control the fermentations using engineered strains efficiently (Kim et al., 2010). Also, where the less favored sugar is metabolized too slowly, unused substrate might complicate the downstream processing and affect the purity of final products (Vinuselvi et al., 2012). Strain development for fermentation of lignocellulosic biomass has usually been focused on increasing product yields. However, the simultaneous consumption of all sugars in the substrate is important for process design and can benefit overall productivity (Kim et al., 2010). Hence, elimination of CCR in microbial cell factories is desirable (Liu et al., 2014). While there are reports of removing CCR in Bacillus spp. (Kraus et al., 1994; Martin-Verstraete et al., 1995; Sun and Altenbuchner, 2010; Nathan and Nair, 2013), it is not evident that these strategies apply to Parageobacillus spp.
In Firmicutes, a histidine-containing phosphocarrier protein (HPr) and an HPr-like protein Crh are key players in CcpA (catabolic control protein A)-dependent CCR. When HPr is phosphorylated on a catalytic histidine residue at position 15, this phosphate can subsequently be transferred to the preferred sugar as it is imported across the cell membrane via a phosphotransferase system (PTS) (Liang et al., 2021). According to the accepted model, an elevated concentration of glycolytic intermediates, glucose-6-phosphate (G-6-P) and fructose-1,6-biphosphate (FBP), stimulates the regulatory phosphorylation of HPr at a serine residue at position 46 by an ATP-dependent HPr kinase (Deutscher et al., 1995; Gunnewijk and Poolman, 2000; Görke and Stülke, 2008; Sun and Altenbuchner, 2010). Crh displays a high sequence identity and high structural similarity to HPr. However, Crh does not have a catalytic His15 residue, which is replaced by a glutamine residue in P. thermoglucosidasius, though it can be phosphorylated by the HPr kinase at Ser46 and thereby invokes CCR (Schumacher et al., 2007; Sun and Altenbuchner, 2010). The HPr (Ser-P) or Crh (Ser-P) binds to the catabolic control protein A (CcpA) to form a CcpA-HPr (Ser46-P) or CcpA-Crh (Ser46-P) complex, which binds to an operator sequence (catabolite responsive element [cre]) located 5' to, or within catabolic genes and results in the down-regulation of target operons (Lorca et al., 2005; Schumacher et al., 2006). Based on this mechanism, several strategies have been investigated to eliminate CCR in Bacillus spp. It has been demonstrated that when the Ser46 residue on HPr is replaced by an alanine residue (ptsH1 mutation), the HPr can no longer be phosphorylated at this position (Hueck and Hillen, 1995; Fujita, 2009). In B. subtilis, several catabolic genes were partially or completely relieved from CCR by the HPr-S46A mutation (Deutscher et al., 1994; Dahl and Hillen, 1995; Reizer et al., 1996). Gene disruption or an alanine substitution of Crh-Ser46 (crh1 mutation) alone did not mitigate CCR, but when such mutation was introduced into a ptsH1 background, the residual CCR effect observed in some operons (e.g., xyn, lev, and iol) completely disappeared, suggesting that Crh has a back-up function to HPr (Galinier et al., 1999; Martin-Verstraete et al., 1999; Puri-Taneja et al., 2006; Repizo et al., 2006). It has been suggested that the Crh in B. subtilis is also responsible for some regulatory functions which are not shared by HPr.
To date, various strategies have been designed to obtain mutant strains which are resistant to CCR. In B. subtilis, it has been reported that several genes could be relieved from CCR when the Ser46 residue was replaced by an Ala on both HPr and Crh. P. thermoglucosidasius is a member of the Bacillaceae family, and its genome sequence shows that it has the potential to produce all the key proteins involved in CCR. Hence, in principle, the mechanism of CCR should be similar in these two organisms. However, as illustrated in this work, a ptsH1 mutant of P. thermoglucosidasius DSM2542 showed impaired growth under fermentative conditions, so we resorted to a classical non-targeted approach based on resistance to the catabolite repressing substrate analog, 2-deoxyglucose (2-DG).
2-DG is a non-metabolizable synthetic glucose analog where the 2-hydroxyl group has been replaced by hydrogen (Horton et al., 1973; Zhu et al., 2015). Due to its similarity to glucose, 2-DG can be recognized by the PTS or other transport systems for transport into cells (Zhu et al., 2015). After phosphorylation at the available 6 position, the resulting 2-deoxy-D-glucose-6-phosphate (2-DG-6P) would not be further metabolized due to the lack of a hydroxyl group at the 2 position (Xi et al., 2014). Hence, cells cannot grow on 2-DG but the accumulation of 2-DG-6P could act as a catabolite repression signal, meaning that the cells would also not be able to grow on a less favored carbon source in the presence of 2-DG. Several studies have demonstrated that this concept is borne out in practice, with 2-DG inducing CCR in S. cerevisiae, E. coli, and Bacillus spp., repressing the utilization of raffinose, fructose, and soluble starch, respectively (Zimmermann and Scheel, 1977; Do et al., 1993; Kornberg and Lambourne, 1994). Use of a non-metabolizable analog such as 2-DG has previously been shown to provide a platform for the isolation of CCR-resistant strains which are able to grow on the less favored substrate in the presence of the analog (Do et al., 1993). In the present work, an adaptive evolution approach was employed to remove CCR from P. thermoglucosidasius DSM 2542. According to previous studies, CCR in P. thermoglucosidasius DSM 2542 is not as stringent as that commonly seen in other microbes, because it can still metabolize xylose slowly in the presence of glucose (Liang et al., 2021). Nevertheless, this still provides a platform to isolate CCR resistant mutants based on faster growth on xylose than the parent strain, in the presence of 2-DG. Based on this principle, two evolution experiments are described, which successfully generated CCR-resistant P. thermoglucosidasius DSM 2542 mutants. Whole genome sequencing of these strains and genetic complementation analysis revealed the genes which contribute to the phenotypes observed.
2. Materials and methods
2.1. Bacterial strains, plasmids, primers, standard reagents, and bacterial growth medium used in this study
The following bacterial strains (Table 1), plasmids (Table 2), and primers (Table 3) were used in this study. All reagents were purchased from Sigma Aldrich (Dorset, UK) or Fisher Scientific (Loughborough, UK). Cell culture media were prepared as described previously (Liang et al., 2021). A final kanamycin concentration of 50 μg/mL was used for E. coli and 12.5 μg/mL was used for P. thermoglucosidasius.
2.2. Standard reagents and bacterial growth medium
All reagents were purchased from Sigma Aldrich (Dorset, UK) or Fisher Scientific (Loughborough, UK). Cell culture media were prepared as described previously (Liang et al., 2021). A final kanamycin concentration of 50 μg/mL was used for E. coli and 12.5 μg/mL was used for P. thermoglucosidasius.
2.3. Genomic DNA preparation and site-directed mutagenesis
Genomic DNA from P. thermoglucosidasius DSM 2542 was extracted with a Wizard Genomic DNA Purification Kit (Promega, Hampshire, UK) according to manufacturer's instructions. The DNA concentration was quantified using a NanoVue Plus spectrophotometer (Biochrom, Cambridge, UK). The ptsH and crh genes along with their flanking regions of about 400 bp were amplified from the genomic DNA with Phusion high-fidelity polymerase (Fisher Scientific, Loughborough, UK) following the manufacturer's protocol using primers 1Fw and 1Rv for ptsH, and primers 3Fw and 3Rv for crh, followed by cloning into a pCRTM-Blunt vector (Zero Blunt PCR cloning kit, Invitrogen, Dublin, Ireland). Chemically competent E. coli DH5α cells were prepared and transformed with the ligation mixture by heat shock as described previously (Chang et al., 2017). The transformants were screened for the presence of the plasmid insert by colony PCR with MyTaq 2x Red Mix (Bioline, London, UK) using primers M13Fw and M13Rv. Plasmids were then extracted from overnight liquid cultures of selected transformants with a QIAprep Spin Miniprep Kit (Qiagen, Manchester, UK) and the presence of the correct insert confirmed by sequencing (GATC services, Eurofins Genomics, Cologne, Germany). After that, primers 2Fw and 2Rv or 4Fw and 4Rv encoding the Ser46 to alanine mutation were used to amplify the pCRTM-Blunt-ptsH and pCRTM-Blunt-crh plasmid, respectively, with a modified Phusion high-fidelity polymerase protocol (98°C for denaturation, 60°C for annealing, 1 min/kb for extension at 72°C, 12 cycles). Twenty nanograms of the template plasmid and 50 ng of each primer were used in each reaction. After PCR, the reaction mix was incubated with 0.5 μL FastDigest DpnI (Thermo Scientific, Paisley, UK) at 37°C for 60 min, to digest the plasmid template. Next, chemically competent E. coli DH5α cells were transformed with the amplified plasmid mixture. Plasmid DNA was extracted from the transformants and sent for sequencing to confirm the generation of the desired pCRTM-Blunt-ptsH1 and pCRTM-Blunt-crh1 constructs.
2.4. Golden gate assembly
The ptsH1 and crh1 sequences along with their flanking regions were digested from the pCRTM-Blunt-ptsH1 and pCRTM-Blunt-crh1 construct, respectively, with FastDigest BsaI (Thermo Scientific, Paisley, UK), and purified by gel electrophoresis. The purified fragments were cloned into the vector pG2K-oriT-bgl-sfgfp-GG (Ortenzi, unpublished), respectively, via Golden Gate Assembly (Engler et al., 2009; Reeve et al., 2016). Each 15 μL reaction mixture contains 300 ng of the vector backbone and equimolar concentration of insert, 1.5 μL FastDigest Buffer, 1 μL FastDigest BsaI, 2 μL NEB T4 ligase (400,000 units/mL), 1.5 μL T4 ligase buffer and nuclease-free ddH2O. The reaction was carried out in a thermocycler to repeat the digestion (37°C, 3 min) and ligation (16°C, 4 min) for 50 times, followed by 50°C for 5 min and 80°C for 5 min, then stored at 8°C. Chemically competent E. coli DH5α cells were transformed with each reaction mix and transformants selected on LB agar plates containing 50 μg/mL kanamycin. Successful integration of the mutant genes, forming pJL-ptsH1 or pJL-crh1 was confirmed by sequencing (GATC services, Eurofins Genomics, Cologne, Germany).
2.5. Conjugation and chromosomal integration
Chemically competent E. coli S17-1 cells were prepared and transformed with the pJL-ptsH1 and pJL-crh1 plasmids by heat-shock as described previously (Chang et al., 2017). Transformants were selected on LB agar plates containing 50 μg/mL kanamycin. Successful transformation was confirmed by colony PCR with primer 7Fw and 7Rv. After conjugation into DSM 2542 as described previously (Macklyne, 2017; Liang et al., 2021), the presence of the insert in the plasmids pJL-ptsH1 and pJL-crh1 was confirmed by colony PCR with primer 7Fw and 7Rv. The protocol for selection of strains in which a gene has integrated into the chromosome via homologous recombination relies on a temperature sensitive replicon encoded by repB which is inactive above 65°C (Reeve et al., 2016). Firstly, the colonies that had been grown at 52°C were inoculated into 10 mL TGP medium (12.5 μg/mL kanamycin) and grown for 6–8 h at 68°C, and sub-cultured twice to ensure loss of the native plasmid but retention of the KmR (kanamycin resistance marker) through homologous recombination via one of the flanking regions. The primary integration was confirmed by colony PCR of colonies grown on TGP plates containing 12.5 μg/mL kanamycin. Then, colonies with successful primary integration were inoculated into 10 mL TGP medium without antibiotics and sub-cultured every 10–14 h at 52°C to allow for the second homologous recombination which removes the integrated plasmid, leaving either the mutant or native gene at the original site of integration. One hundred microliters of the 6th sub-culture was serially diluted (10−1, 10−3, 10−5, 10−7) and plated onto TGP agar plates with no antibiotics. After growth on TGP plates overnight at 52°C, colonies were then replica plated onto TGP agar plates with and without kanamycin and incubated at 52°C overnight. Kanamycin sensitive colonies were screened by PCR, using primers 5Fw and 5Rv for ptsH, and 6Fw and 6Rv for crh, respectively. The ptsH1 mutation removed a Bsu15I restriction site within ptsH, while the crh1 mutation introduced an AluI restriction site in the crh (Supplementary Figure 2A). Hence, digestion of the PCR products by FastDigest Bsu15I or AluI (Thermo Scientific, Paisley, UK) was used to confirm the presence of the desired ptsH1 or crh1 mutation.
2.6. Construction and conjugation of vectors for genetic complementation
The wild type ptsG, rbsR, and apt along with their native promoters were individually amplified from P. thermoglucosidasius DSM 2542 genomic DNA using primers listed in Table 3 with Phusion high-fidelity polymerase (Fisher Scientific, Loughborough, UK) following the manufacturer's protocol. Each of the gene fragments, purified by gel electrophoresis, were cloned into a pair of BsaI sites on the plasmid pG2K-oriT-bgl-sfgfp-GG (Ortenzi, unpublished) via Golden Gate Assembly (Engler et al., 2009; Reeve et al., 2016). Chemically competent E. coli DH5α cells were prepared and transformed with the reaction mixes, respectively, by heat shock as described previously (Chang et al., 2017). The presence of the requisite pJL-ptsG+, pJL-apt+, and pJL-rbsR+ constructs was confirmed by colony PCR of transformants grown on LB agar plates containing 50 μg/mL kanamycin with primers 7Fw and 7Rv, then the amplified product was authenticated by sequencing (GATC services, Eurofins Genomics, Cologne, Germany). Then, 2DG-ADE2b was transformed with pJL-ptsG+, pJL-apt+, and pJL-rbsR+, respectively, via conjugation as described previously (Liang et al., 2021). The continued presence of the gene inserts in the 2DG-ADE2b transformants was confirmed by colony PCR.
2.7. Tube fermentation
Tube fermentation was carried out in 15 mL sterile conical centrifuge tubes containing 10 mL ASM medium and 0.2 g solid MgCO3 (sterilized by autoclave) to stabilize the pH (Millard et al., 1996). P. thermoglucosidasius strains were revived from glycerol stocks and grown on 15 mL TGP or 2TY plates overnight at 60°C. The next morning, a loopful of cells was inoculated into 15 mL TGP or 2TY liquid medium and incubated for 4–5 h (60°C, 250 rpm) until the OD600 optical density measured at a wavelength of 600 reached 2.5~3.0. 1 mL of the cell culture was inoculated into each tube, and the cultures were incubated at 60°C in a shaking incubator at 250 rpm. The relatively small head space in the tubes allowed for the transition from fully aerobic to fermentative conditions as oxygen transfer became limited. Experiments were done in triplicate and samples taken every 24 h in sacrifice tubes, centrifuged and the clarified supernatant analyzed by HPLC as described previously (Hills, 2015; Liang et al., 2021).
2.8. GC-MS identification of methylgloxal in fermentation broth
The identification of methylglyoxal was based on the formation of 2-methylquinoxaline after reaction with 1,2-diaminobenzene as described previously (OIV-MA-AS315-21, 2010). After tube fermentation in ASM medium [1% (w/v) glucose and 1% (w/v) xylose] for 48 h, cell cultures of P. thermoglucosidasius DSM 2542 and DSM 2542 ptsH1 were filtered (PhenexNY 15mm Syringe Filters 0.2 μm, Phenomenex, Torrance, USA). 10 mL of each clarified filtrate was brought to pH 8.0 with 1M NaOH, followed by derivatization with 5 mg 1,2-diaminobenzene for 3 h at 60°C. Then the reaction medium was brought to pH 2.0 using 2 M H2SO4, and the quinoxaline derivative was extracted twice with 1 mL dichloromethane. The solvent phase was dried on 0.2 g of anhydrous sodium sulfate prior to GC-MS analysis.
The reaction product was analyzed on an Agilent 8890 gas chromatography system coupled with 5977B MSD. Split injections (1 μL) were performed, with a split ratio of 30:1 (split flow 30 mL/min), using with a single taper, ultra-inert wool inlet liner (Agilent 5190-2293). The inlet was heated to 260°C with 3 mL/min septum purge flow. An Agilent HP-5MS (30 m, 0.25 mm, 0.25 μm) column was used for separation, with He (BOC, N5.5) carrier gas at a constant flow of 1.0 mL/min. The oven was held at 70°C for 1 min, thereafter ramped at 10°C/min to 200°C, then 20°C/min to 220°C holding for 2 min. The MS transfer line was set to 280°C with the source at 230°C and quad at 150°C. MSD detection was performed (after solvent delay of 6 min) using full scan mode from 40 to 400 m/z with 1562 μs scan speed, and a gain factor of 15. The ion m/z = 144 was used for quantification and the ion m/z = 117 as a qualifier of the derivatized methylglyoxal.
2.9. Methylglyoxal and ASM medium reaction assay
Fifty milliliters of P. thermoglucosidasius DSM 2542 culture in ASM medium (1% glucose and 1% xylose) at an OD600 of 0.2 was prepared in a 250 mL baffled Erlenmeyer flask. The cell culture was divided into 5 equal portions and transferred into 15 mL conical centrifuge tubes. Then, methylglyoxal was added to all except for one as a negative control. The final concentration (v/v) of methylglyoxal was 0, 0.1, 0.2, 0.3, and 0.4%, respectively. Cell cultures were incubated at 60°C. Color change of the cultures was documented routinely.
2.10. 2-DG repression assays
P. thermoglucosidasius DSM 2542 cells grown to exponential phase in ASM medium supplemented with 1% (w/v) xylose were inoculated into 50 mL sterile conical centrifuge tubes (Greiner, Stonehouse, UK) containing 20 mL ASM medium with 1% (w/v) xylose and different concentrations of 2-DG. The cultures were grown at 60°C in a shaking incubator (Innova44, New Brunswick, UK). Samples were removed at regular intervals and the OD600 was recorded. The inhibitory effect of 2-DG to P. thermoglucosidasius DSM 2542 during growth on xylose was determined by comparison of growth rates.
2.11. First adaptive evolution in ASM medium with 1% xylose and 0.5% 2-DG
Adaptive evolution of P. thermoglucosidasius DSM 2542 was performed in 50 mL sterile conical centrifuge tubes containing 20 mL ASM medium supplemented with 1% xylose and 0.5% 2-DG. Starting with an OD600 of 0.1, the cell culture was incubated in a shaking incubator (60°C, 250 rpm) and 1 mL of the culture was inoculated into a fresh tube every 24 h. These serial subcultures were analyzed every 7 days using tube fermentation to see if the cells were catabolite de-repressed. After 14 subcultures, single colonies were isolated on a 2TY agar plate from the catabolite de-repressed population, and their phenotypes were evaluated in tube fermentations. Genome sequencing of the parent strain and the evolved mutant strains was performed using the standard whole genome sequencing service provided by MicrobesNG (Birmingham, UK) carried out in Illumina sequencers (HiSeq/NovaSeq) using a 250 bp paired end protocol, followed by trimmed-reads and assembly using the existing sequence of DSM 2542 as a scaffold. The parent strain was also re-sequenced as a control. Variant calling was performed using VarScan. The sequences of the mutant strains were compared with the parent strain for single nucleotide polymorphisms (SNPs).
2.12. Second adaptive evolution with one selection on glucose
Cells were prepared and sub-cultured as described above. The only difference was that after 7 sub-cultures, 1 mL of cell culture was inoculated into ASM medium supplemented with 1% (w/v) glucose and grown for 10 h (60°C, 250 rpm). Then,1 mL of this cell culture was inoculated into ASM medium supplemented with 1% (w/v) xylose and 0.5% (v/v) 2-DG and sub-culturing in this medium every 24 h continued. Single colonies were isolated from the catabolite de-repressed population after 14 subcultures, and their phenotypes were evaluated in tube fermentations. Mutations in evolved strains were identified via genome sequencing and comparison with the parent strain as described above (MicrobesNG, Birmingham, UK).
3. Results
3.1. HPr-S46A and Crh-S46A site-directed mutagenesis in P. thermoglucosidasius DSM 2542
The protocol for chromosomal integration relies on a temperature sensitive replicon encoded by repB which is inactive above 65°C (Reeve et al., 2016). After the two homologous recombination events as shown in Supplementary Figure 1, kanamycin sensitive colonies were screened by PCR. Primers 5Fw and 5Rv were used to amplify the ptsH or ptsH1 along with their flanking regions (about 1.7 kb in total), and primers 6Fw and 6Rv were used to amplify the crh or crh1 along with their flanking regions (about 2 kb in total). ptsH1 and crh1 point mutation in P. thermoglucosidasius DSM 2542 was diagnosed by PCR and enzymatic digestion (Supplementary Figure 2). In wild type DSM 2542 background, 5 out of 16 screened colonies contained the desired ptsH1 mutation, and 7 out of 15 screened colonies contained the desired crh1 mutation, respectively. However, although several attempts have been made to create an ptsH1 and crh1 double mutant, none was successful. In attempts to mutate either ptsH in DSM 2542 crh1, or crh in DSM 2542 ptsH1, none of approximate 100 tested colonies contained the desired double mutation. During deletion of the second gene, the first crossover occurred but the second crossover did not result in the desired deletion.
3.2. Tube fermentations of P. thermoglucosidasius strains in mixed-sugar substrate
Tube fermentations were set up for preliminary characterization of DSM 2542 crh1 and DSM 2542 ptsH1. DSM 2542 ptsH1 and DSM 2542 crh1 prioritized the utilization of glucose in the first 24 h as observed in the wild type (Figures 1A–C), indicating that the S46A mutation in Crh or HPr alone, did not alleviate CCR during fermentative metabolism. Intriguingly, the extent of sugar consumption of these two mutant strains differed after 24 h. DSM 2542 crh1 consumed all of the glucose within 48 h and about 80% of the xylose by 72 h. In contrast, though DSM 2542 ptsH1 used most of the glucose in the first 24 h, its glucose consumption slowed down afterwards, with a dark-brown color developing in the media (which was not seen in DSM 2542 or DSM 2542 crh1). It only finished using the glucose between 48 and 72 h, with only 16% of the xylose consumed by 72 h, suggesting that xylose utilization was still being repressed when glucose was present. The slow glucose utilization after 24 h implied the ptsH1 mutation might have resulted in a fitness cost which became apparent after about 24 h of growth and affected metabolism of both sugars.
TM444 is a P. thermoglucosidasius NCIMB 11955 variant with deletions to the genes encoding lactate dehydrogenase (Ldh), pyruvate formate lyase (PflB), stage 0 sporulation protein A (Spo0A), and up-regulation of the pyruvate dehydrogenase complex (Pdh) (Hills, 2015). TM444 △ptsH is a TM444 variant with a ptsH partial deletion (Bacon, unpublished). DSM 2542 and NCIMB 11955 are the same strain although a small number of single nucleotide polymorphisms (SNPs) between the strains have been recorded (Sheng et al., 2016). It was reported that TM444△ptsH was able to utilize pentose and hexose sugars simultaneously, yet it experienced slow growth and slow sugar consumption when compared with wild type NCIMB 11955 under fermentative conditions but not in aerobic conditions (Marriot, unpublished). Here, to investigate the impact of △ptsH on sugar consumption and medium color during tube fermentation, TM444 △ptsH was compared with TM444 in ASM medium supplemented with glucose and xylose (Figures 1D,E) with a focus on glucose consumption and medium color. While TM444 was similar to DSM2542 with respect to CCR, TM444 △ptsH appeared to metabolize glucose and xylose simultaneously, but primarily because glucose was consumed more slowly than in TM444. Xylose was consumed at a similar rate to the wild-type and neither culture produced the dark brown color seen with DSM 2542 ptsH1. Thus, the phenotype of DSM 2542 ptsH1 seems to be specific to a catalytically functional but signaling uncoupled form of PtsH. This metabolized glucose as rapidly as DSM 2542 at the outset, but seemed to suffer growth inhibition which affected the rate of both glucose and xylose metabolism, and this coincided with the browning of the media.
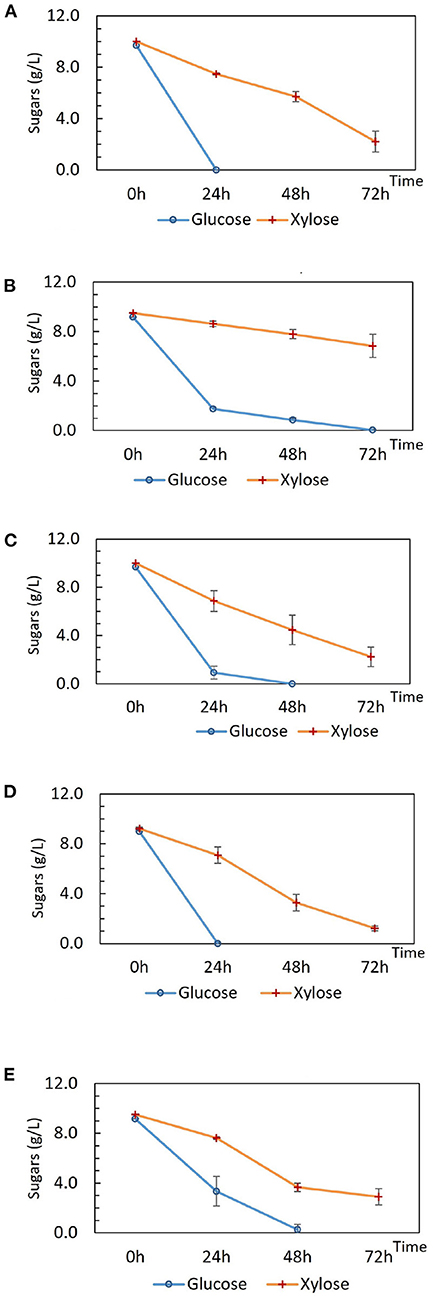
Figure 1. Sugar consumption of (A) wild type P. thermoglucosidasius DSM 2542, (B) P. thermoglucosidasius DSM 2542 pstH1, (C) P. thermoglucosidasius DSM 2542 crh1, (D) P. thermoglucosidasius TM444, and (E) P. thermoglucosidasius TM444 △ptsH during tube fermentation in sterile 15 mL centrifuge tubes. Cells were cultured in 10 mL ASM medium supplemented with 1% (w/v) glucose and 1% (w/v) xylose. To stabilize the pH during the fermentation, 0.2 g solid MgCO3 was added to the tubes prior to inoculation. OD600 readings were not possible because of the turbidity of the media caused by the MgCO3. Error bars are ± standard deviation of three replicates taken at the same time point.
3.3. Growth and sugar consumption of DSM 2542 ptsH1 in the bioreactor
To investigate the phenotype of DSM 2542 ptsH1 in more detail, growth studies were carried out in a bioreactor containing a glucose and xylose mixture under fermentative conditions. As shown in Figure 2, DSM 2542 ptsH1 grew poorly compared with the wild type, consistent with the much slower sugar utilization observed in the tube fermentations. Indeed, unlike the tube cultures, which start with a period of aerobic growth, it is clear that the rate of glucose metabolism was restricted from an early stage in the culture. Nevertheless, DSM 2542 ptsH1 still showed clear evidence of CCR as xylose metabolism did not increase to compensate. Within 22 h, the wild type reached an OD600 of about 1.5 with complete consumption of glucose. After that, it started to consume xylose and was completed in about 60 h. The total biomass did not increase much when it was growing on xylose, consistent with its low net ATP yield under anaerobic conditions. In contrast, DSM 2542 ptsH1 hardly managed to metabolize the glucose within 60 h, and there was still a lot of xylose remaining after 86 h incubation. As seen during tube fermentation, a dark-brown color developed in the DSM 2542 ptsH1 culture media within 12 h. Given the unexpected phenotype, the DSM 2542 ptsH1 variant was sent for whole genome sequencing (MicrobesNG, Birmingham, UK), but this was confirmed that there were no mutations other than that made in ptsH, compared to the parental strain. Therefore, the slow growth and slow sugar utilization of DSM 2542 ptsH1 under fermentative conditions must be the direct result of this mutation.
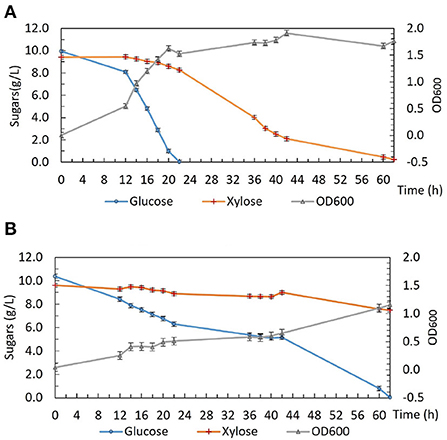
Figure 2. Growth and sugar consumption profile of (A) P. thermoglucosidasius DSM 2542 and (B) P. thermoglucosidasius DSM 2542 pstH1 in ASM medium supplemented with 1% (w/v) glucose and 1% (w/v) xylose under fermentative conditions in a bioreactor. Error bars are ± standard deviation of three replicates taken at the same time point.
3.4. Growth and sugar consumption of DSM 2542 ptsH1 under aerobic conditions
Interestingly, unlike the situation under fermentative conditions, the growth of DSM 2542 ptsH1 under aerobic conditions was similar to that of the wild type (Figure 3). Aerobic growth was performed in 50 mL ASM medium in 250 mL baffled Erlenmeyer flasks sealed with silicone sponges. The cultures were incubated in a shaking incubator (60°C, 250 rpm) and samples were taken out regularly for biomass and HPLC analysis. Under aerobic conditions, both DSM 2542 ptsH1 and the wild type grew rapidly and reached a similar final OD600. Intriguingly, DSM 2542 ptsH1 appeared to have a shorter lag phase and more rapid growth compared with the wild type. However, glucose was consumed prior to xylose in both strains, indicating that the ptsH1 mutation did not relieve the cells from CCR under aerobic conditions either. Although the repeat of wild-type DSM 2542 both showed rapid growth on glucose there was a difference in the length of the lag phase, leading to a significant difference in glucose utilization after 6–12 h. Variability in the length of the lag phase is consistently observed with P. thermoglucosidasius DSM 2542 and NCIMB 11955, probably caused by a slight variation in the state of the inoculum. Notably, the dark-brown-color culture medium observed during fermentation was not observed under aerobic conditions. This experiment was repeated to confirm the results.
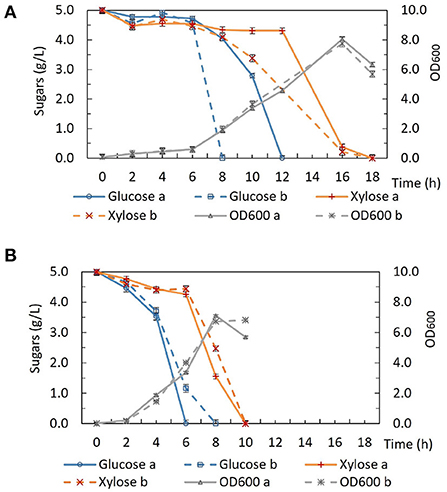
Figure 3. Growth and sugar consumption profile of (A) wild type P. thermoglucosidasius DSM 2542 and (B) P. thermoglucosidasius DSM 2542 pstH1 in ASM medium supplemented with 0.5% (w/v) glucose and 0.5% (w/v) xylose under aerobic conditions. Two biological repeats are displayed in each graph using solid and dotted lines, respectively. Error bars are ± standard deviation of three technical replicates taken at the same time point.
3.5. Identification of methylgloxal in P. thermoglucosidasius DSM 2542 and DSM2542 ptsH1 cultures
Formation of a brown pigment resulting from the reaction of reducing sugars and amines (the Maillard reaction) is well-understood in microbiology and food science. This is known to be accelerated at high temperatures which is why it is advisable not to autoclave sugars with rich media containing amino acids. However, it is clear that the color formation was only happening with DSM 2542 ptsH1, suggesting metabolic production of a reactive aldehyde/ketone, which is more reactive than reducing sugars. The most likely candidate is methylglyoxal, which can be produced from the glycolytic intermediate dihydroxyacetone phosphate (DHAP) as a glycolytic bypass and is known to be a reactive compound which can contribute to similar browning reactions (Gandhi et al., 2019). To confirm the identity of this product, it was reacted with 1,2-diaminobenzene and the product analyzed by GC-MS. DSM 2542 and DSM2542 ptsH1 were grown for 48 h in ASM medium (1% (w/v) glucose and 1% (w/v) xylose) under tube fermentation. As control, 10 μL methylglyoxal was added to 10 mL of the same ASM medium, followed by the derivatization and extraction. Both control and experimental groups showed a unique TIC peak at 10.68 min with MS principal ions at m/z = 117 and 144. Quantification was based on the intensity of principal ion m/z = 144 and the peak area of DSM2542 ptsH1 was about 6 times higher than DSM 2542, indicating an elevated level of methylglyoxal excreted by DSM2542 ptsH1 during fermentation (p < 0.05) (Figure 4).
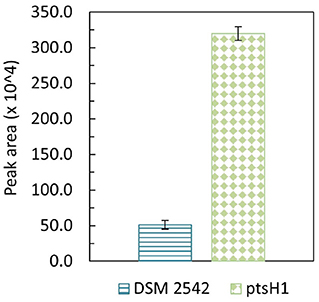
Figure 4. GC-MS peak area (m/z = 144) of methylglyoxal derivatised with 1,2-diaminobenzene from P. thermoglucosidasius DSM 2542 and DSM2542 ptsH1 cultures after 48-h tube fermentation in ASM medium containing 1% (w/v) glucose and 1% (w/v) xylose. Error bars are standard deviation of three biological replicates. T-test confirmed that the peak areas were statistically different (p < 0.05).
The reaction between methylglyoxal and a 1,2-diamine in cell culture medium is a competitive equilibrium between the diamine and amino groups in the medium, so accurate quantification is difficult. Therefore, to get an approximate measure of concentration, known concentrations of methylglyoxal were added to DSM 2542 cultures grown to OD600 about 1.0 in ASM medium supplemented with 1% (w/v) glucose and 1% (w/v) xylose under fermentative conditions. After 3 h of incubation at 60°C, different shades of light-yellow color started to develop in all methylglyoxal-treated samples. After overnight incubation, the brown coloration observed in DSM 2542 ptsH1 cultures was evident in the higher concentration groups (Supplementary Figure 3A), and after 36 h of incubation, all treated-groups turned into a gradient of dark-brown color (Supplementary Figure 3B), while the untreated group remained the normal beige color commonly seen in DSM 2542 fermentation using ASM medium (Supplementary Figure 3C). The browning effects caused by 0.1–0.2% (v/v) methylglyoxal were comparable to the color observed during the fermentation of DSM 2542 ptsH1 after 36 h (Supplementary Figure 3D). Although ASM is not supplemented with amino acids the methylglyoxal could be reacting with secreted or lysis products.
3.6. Effects of 2-DG on P. thermoglucosidasius DSM 2542 growing on xylose
The inability to alleviate CCR by removing the signaling components from Hpr or Hpr and Crh together necessitated an alternative strategy. The potential for 2-DG to act as a classical non-metabolizable glucose analog capable of exerting catabolite repression in P. thermoglucosidasius DSM 2542 was examined by including different concentrations of 2-DG (0-1% w/v) in an ASM medium supplemented with 1% (w/v) xylose. The overall growth rate decreased as 2-DG concentration increased (Figure 5). Extensive studies with P. thermoglucosidasius NCIMB 11955, which is virtually identical to DSM 2542, have shown that it typically does not enter an extended stationary phase, but starts to lyse after nutrient starvation. This pattern was also evident with DSM 2542 growing in tubes on xylose, where the OD600 (in the absence of 2-DG) peaked after 16 h. In the presence of 2-DG, the peak OD600 was lower and appeared later as 2-DG concentration increased. When supplemented with 0.5%(w/v) 2-DG, the peak OD600 was reduced by about a third and the growth rate was reduced by nearly 50%. This concentration was, therefore, used for adaptive evolution experiments.
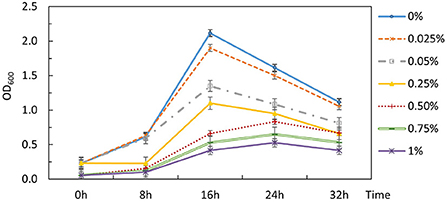
Figure 5. Effects of 2-DG during growth of P. thermoglucosidasius DSM 2542 on xylose in 50 mL conical centrifuge tubes. P. thermoglucosidasius DSM 2542 was grown in ASM medium supplemented with 1% (w/v) xylose and different concentrations of 2-DG (% w/v shown on the right). Error bars are ± standard deviation of three technical repeats.
3.7. Isolation and analysis of catabolite de-repressed mutant strains
P. thermoglucosidasius DSM 2542 grown in ASM medium supplemented with 1%(w/v) xylose and 0.5%(w/v) 2-DG was sub-cultured every 24 h. After 7 sub-cultures, the cell population was tested and found to still exhibit catabolite repression (Figure 6A). After 14 subcultures, the cell population was tested again and found to be catabolite de-repressed (Figure 6A). After growing on 2TY plates, three colonies (2DG-ADE1a,b, and c) were randomly picked from a total of about 100 for further characterization using tube fermentation in ASM medium supplemented with 1% (w/v) glucose and 1% (w/v) xylose (Figure 6B).
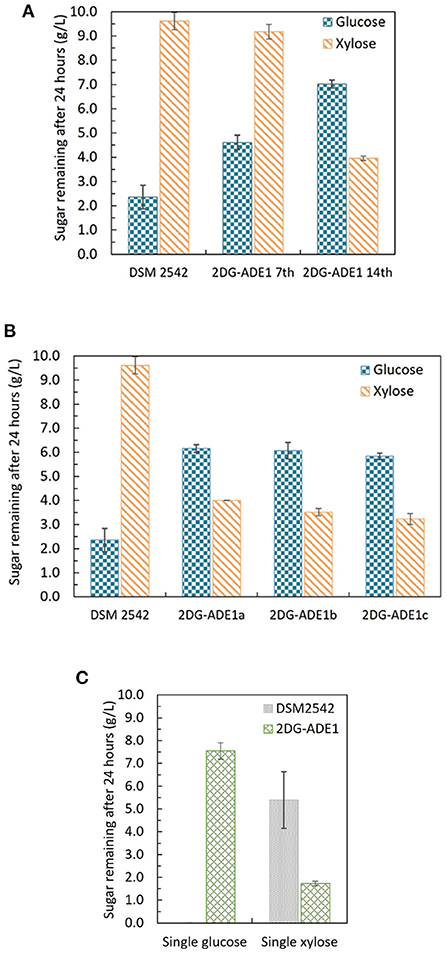
Figure 6. Sugar consumption profiles of P. thermoglucosidasius DSM 2542, 2-DG resistant cultures after 7 and after 14 passages in 0.5% 2-DG and 1% xylose (2DG-ADE1 7th and 2DG-ADE1 14th), and 2-DG resistant isolates 2DG-ADE1a, 2DG-ADE1b and 2DG-ADE1c during tube fermentation in 15 mL sterile centrifuge tubes. Cells were cultured in ASM medium supplemented with 1% (w/v) glucose and 1% (w/v) xylose as in (A,B), or (C) 1% (w/v) glucose or 1% (w/v) xylose alone. To stabilize the pH during the fermentation, 0.2 g MgCO3 was added to the tubes prior to inoculation. Error bars in (A,B) and DSM 2542 from (C) are ± standard deviation of three replicates taken at the same time point. Error bars in (C) 2DG-ADE1 are ± standard deviation of 2DG-ADE1a, 2DG-ADE1b and 2DG-ADE1c samples taken at the same time point. A t-test performed on data in (A,B) confirmed the remaining glucose and xylose in cultures of DSM 2542 and the 2DG resistant strains were statistically different (p < 0.05).
In the culture medium of the parent strain DSM 2542, about 27% of the glucose remained after 24 h, while 95% of xylose was unused due to CCR. In contrast, in the culture medium of 2DG-ADE1a, 2DG-ADE1b, and 2DG-ADE1c, about 57–60% of the glucose and 30–40% of the xylose remained after 24 h. Thus, while it appeared that glucose and xylose were being co-utilized by 2DG-ADE1a, 2DG-ADE1b and 2DG-ADE1c, indicative of catabolite de-repression, this seemed to be as a result of compromising glucose utilization. This was confirmed by growing each of the strains in tube fermentation in ASM medium supplemented with 1% (w/v) glucose or 1% (w/v) xylose alone (Figure 6C). When grown on glucose as sole carbon source, DSM 2542 used all the glucose within 24 h, while an average of 55% of glucose remained in the culture medium of 2DG-ADE1a, 2DG-ADE1b, and 2DG-ADE1c. When cultured on xylose alone, 75 and 18% of the xylose was left in the culture medium of DSM 2542 and these evolved strains, respectively, after 24 h, indicating that not only had glucose utilization been inhibited but xylose utilization had been enhanced.
To understand the genetic changes responsible for this phenotype, the genome sequences of five evolved strains isolated from the same culture (2DG-ADE1a, b, c, d and e) were compared with that of their parental strain. In total five mutations were identified by single nucleotide polymorphism (SNP) analysis, four of which were found in all five strains. Two of these were located in non-coding regions, and two were identified within coding regions (Supplementary Table 1): an alanine to glutamate substitution at position 19 (A19E) on phosphoenolpyruvate-protein phosphotransferase PtsI (also called enzyme I or EI) encoded by ptsI, and a phenylalanine to valine substitution at position 74 (F74V) on adenine phosphoribosyl transferase (APRT) encoded by apt. These potential influence of these two mutations on sugar utilization will be discussed later.
3.8. Isolation and analysis of 2-DG resistant strains which retain rapid growth on glucose
The first adaptive evolution experiment clearly generated CCR-negative strains but at the expense of rapid glucose metabolism, which is not useful for industrial fermentation. An improved experimental design was, therefore, required to prevent impaired glucose utilization becoming a selected phenotype during the evolutionary process. To achieve this an intermediate selection step for good growth on glucose was required. Similar to the first evolution experiment, DSM 2542 was initially grown in 1% (w/v) xylose and 0.5% (w/v) 2-DG and sub-cultured every 24 h. Then, after 7 subcultures, the cell population was enriched for strains with rapid growth on glucose, followed by a further 7 subcultures in the repressive medium containing 2-DG and xylose. Finally, the cell population was tested for evidence of catabolite de-repression during tube fermentation on a mixture of glucose and xylose before plating out on 2TY plates. Three colonies (2DG-ADE2a, b, and c) were randomly selected from about 100 colonies on the 2TY plates for further characterization. During tube fermentation in ASM medium supplemented with 1% (w/v) glucose and 1% (w/v) xylose, compared with DSM 2542, xylose was more extensively metabolized by 2DG-ADE2a, 2DG-ADE2b and 2DG-ADE2c than by DSM 2542 after 24 h, suggesting xylose consumption in these strains was de-repressed (Figure 7A). However, unlike the strains from the first evolution experiment, 2DG-ADE2a, 2DG-ADE2b and 2DG-ADE2c utilized similar amounts of glucose to the wild type DSM 2542 in the first 24 h. When grown in ASM medium supplemented with 1% (w/v) glucose alone, they were able to metabolize all the glucose within 24 h like DSM 2542, so their rate of glucose utilization did not seem to be compromised (Figure 7B). Similar to the strains from the first evolution, during growth in ASM medium supplemented with 1% (w/v) xylose alone, they consumed more xylose than DSM 2542 in the first 24 h, suggesting their rate of xylose utilization had increased (Figure 7B). However, this enhancement was not as great as that in the strains from the first evolution experiment.
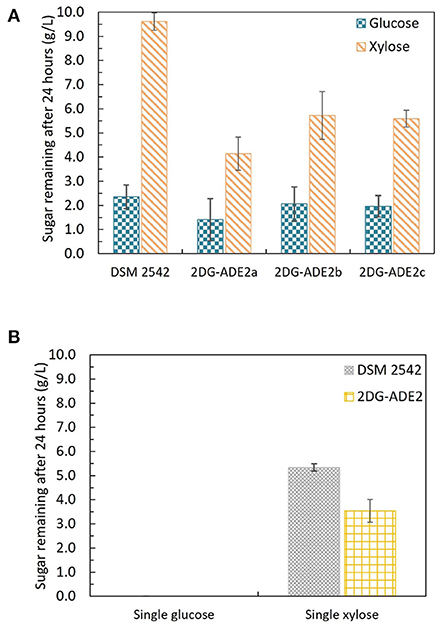
Figure 7. Sugar consumption profiles of P. thermoglucosidasius DSM 2542 and 2-DG resistant strains 2DG-ADE2a, 2DG-ADE2b and 2DG-ADE2c during tube fermentation in 15 mL sterile centrifuge tubes. Cells were cultured in ASM medium supplemented with (A) 1% (w/v) glucose and 1% (w/v) xylose or (B) 1% (w/v) glucose or 1% (w/v) xylose. To stabilize the pH during the fermentation, 0.2 g MgCO3 was added to the tubes prior to inoculation. Error bars in (A,B) DSM 2542 are ± standard deviation of three replicates taken at the same time point. Error bars in (B) 2DG-ADE2 are ± standard deviation of 2DG-ADE2a, 2DG-ADE2b, and 2DG-ADE2c samples (three replicates) taken at the same time point. A t-test performed on data in (A) confirmed the remaining glucose and xylose in each strain were statistically different (p < 0.05).
To understand the genetic changes responsible for such phenotypes, the genome sequences of five evolved strains from the same culture (2DG-ADE2a, b, c, d, and e) were compared with that of their parental strain DSM 2542. In total 43 SNPs were identified, many within non-coding regions, although 8 of those SNPs were found in coding regions of all five strains (Supplementary Table 2). Notably, in all of them, a stop codon was introduced at Q10 in the mutS gene. The mutS gene encodes a DNA mismatch repair protein which is involved in the repair of errors in DNA replication which causes mismatches (Wang et al., 2003). Termination of mutS translation would significantly increase the mutation rate, so if this had been an early evolutionary event this could explain the high frequency of SNPs in these evolved strains. Among the common mutations within coding regions, three were directly linked to sugar metabolism: one was an arginine to cysteine substitution at position 452 (R452C) on PTS glucose transporter subunit IICBA (PtsG or EIICBAGlu) encoded by ptsG, one was a frame-shift after the threonine at position 58 on the ribose operon repressor (RbsR) encoded by rbsR gene, and the other one, interestingly, was the same APRT-F74V mutation which appeared in the first evolution experiment. The SNPs identified in these genes were confirmed by Sanger sequencing of PCR products.
Strains from this evolution experiment showed signs of catabolite de-repression as well as enhanced xylose utilization without compromising glucose consumption, so in order to characterize their growth and sugar consumption profiles under more controlled conditions. 2DG-ADE2b, which contained the fewest SNPs among the five sequenced strains, was compared with the wild type DSM 2542 in bioreactors under fermentative conditions. Bench-top bioreactor operation, as well as HPLC analysis of samples taken at regular intervals, were as described previously (Liang et al., 2021). The growth medium contained glucose, xylose and arabinose which are the main sugars in lignocellulosic hydrolysates. Figure 8A shows that, due to CCR, DSM 2542 had a clear preference for glucose, metabolizing it in 30 h and, reaching a peak OD600 of about 2.5. During this period, a small amount of xylose and arabinose were consumed because the CCR in P. thermoglucosidasius is not as tightly regulated as in some other organisms (Liang et al., 2021). After glucose had been exhausted there was a transient drop in OD600 associated with the switch in carbon sources but the OD600 effectively plateaued during the subsequent period of metabolism of xylose and arabinose. In contrast, 2DG-ADE2b utilized glucose, xylose and arabinose simultaneously, and reached a higher peak OD600 of about 3.4 (Figure 8B). DG-ADE2b metabolized all the sugars within 42 h, so the total fermentation time was shorter, compared to DSM 2542. This experiment was repeated and the same trend was observed. These results collectively indicate that removal of CCR has a positive impact on fermentation providing a more controlled metabolic environment. The rate of total sugar metabolism (qs) was 0.003 mol/gDW/h in 2DG-ADE2b and 0.002 mol/gDW/h in DSM 2542, which were similar, indicating there may be a maximum metabolic flux from carbohydrates into central metabolism.
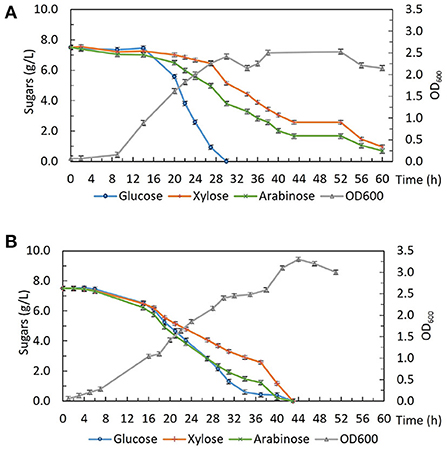
Figure 8. Fermentative growth and sugar consumption profile of (A) P. thermoglucosidasius DSM 2542 and (B) 2-DG resistant strain 2DG-ADE2b in ASM medium supplemented with 0.75% (w/v) glucose, 0.75% (w/v) xylose and 0.75% (w/v) arabinose in bioreactors. Error bars are ± standard deviation of three replicates taken at the same time point.
3.9. Effect of complementation with ptsG, rbsR, and apt on CCR in the evolved strain
To understand which mutation(s) contributed to the removal of CCR from P. thermoglucosidasius DSM 2542, genetic complementation was used to investigate whether the wild type ptsG, rbsR and apt genes would restore CCR in 2DG-ADE2b. 2DG-ADE2b was transformed separately with plasmid pJL-ptsG+, pJL-apt+ and pJL-rbsR+, resulting in 2DG-ADE2b variants capable of expressing either ptsG or rbsR or apt from their own promoters (Supplementary Figure 4).
Growth of these variants under different sugar conditions was compared using tube fermentation (Figure 9). When they were grown in ASM medium supplemented with 1% (w/v) glucose as the sole carbon source, similar to the wild type DSM 2542 and 2DG-ADE2b, they finished all glucose within 24 h (data not shown). However, when they were grown in ASM medium supplemented with a mixture of 1% (w/v) glucose and 1% (w/v) xylose, or 1% (w/v) xylose as the sole carbon source, they all consumed xylose more slowly than 2DG-ADE2b. When the ptsG complemented variant, 2DG-ADE2bptsG+, was grown in ASM medium supplemented with 1% (w/v) glucose and 1% (w/v) xylose, it consumed all of the glucose in the first 24 h, while 80% of the xylose remained, typical of a cell exhibiting catabolite repression. However, when it was grown in 1% (w/v) xylose as the sole carbon source, there was still 80% of unconsumed xylose after 24 h as in the mixed-sugar substrate, suggesting that complementation with ptsG on a plasmid was inhibiting xylose utilization. In contrast, the wild type DSM 2542 consumed both less xylose and less glucose when the two substrates were combined due to CCR, but it consumed xylose on its own more rapidly than the complemented strain. Clearly, the effect of ptsG complementation is not straightforward, but it should be noted that the use of a plasmid-borne gene would have increased the gene dosage and hence, the expression level of PtsG.
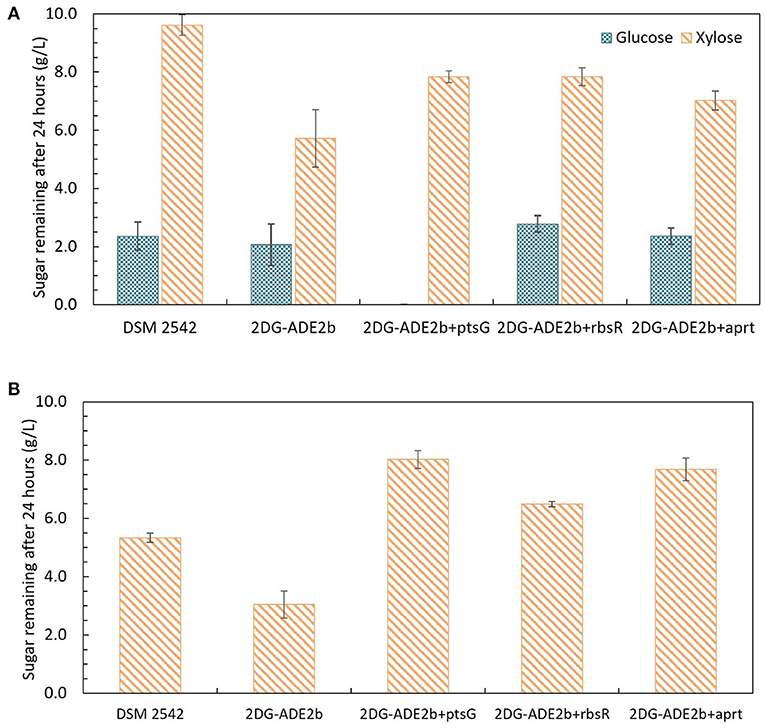
Figure 9. Sugar consumption profiles of P. thermoglucosidasius DSM 2542, 2-DG evolved strains 2DG-ADE2b, and 2DG-ADE2b variants with plasmid pJL-ptsG+(2DG-ADE2bptsG+), pJL-apt+ (2DG-ADE2bapt+) and pJL-rbsR+ (2DG-ADE2brbsR+), respectively. Cells were grown in 15 mL sterile centrifuge tubes in ASM medium supplemented with (A) 1% (w/v) glucose and 1% (w/v) xylose, or (B) 1% (w/v)xylose. To stabilize the pH during the fermentation, 0.2 g MgCO3 was added to the tubes prior to inoculation. Error bars are ± standard deviation of three replicates taken at the same time point. T-test confirmed the remaining glucose and xylose in each culture in (A) were statistically different (p < 0.05), and the remaining xylose in (B) was statistically different from each other (p < 0.05).
When 2DG-ADE2bapt+ and 2DG-ADE2brbsR+ were grown in 1% (w/v) glucose and 1% (w/v) xylose, the concentration of glucose remaining after 24h was similar to cultures of DSM2542 or 2DG-ADE2b, however the concentration of xylose remaining unconsumed was less than observed with DSM2542, but more than 2DG-ADE2b indicating a partially repressed phenotype. However, as with the ptsG complemented strain, when they were grown in 1% (w/v) xylose alone, more xylose remained in the culture medium after 24 h compared with DSM 2542 or 2DG-ADE2b. These results suggested that apt+ and rbsR+ expressed on a plasmid might restrict xylose uptake or utilization, but they might not directly cause CCR in DSM 2542.
4. Discussion
In B. subtilis, CCR of several genes can be relieved by replacing the Ser46 residue of both HPr and Crh with an Ala, while a single S46A mutation on HPr or Crh was not sufficient to remove CCR (Galinier et al., 1999; Puri-Taneja et al., 2006). Similarly, neither the ptsH1 nor the crh1 mutation alone eliminated CCR in P. thermoglucosidasius DSM 2542, but multiple attempts to isolate a ptsH1 crh1 double mutant were not successful, using a method which consistently yields mutants in non-essential genes. This suggests that the double mutation is highly deleterious or lethal in P. thermoglucosidasius DSM 2542 promoting a search for an alternative strategy. In principle, this could have involved modification of downstream components of the CCR signaling pathway, such as deletion/mutation of ccpA or modification of cre sites (Kraus et al., 1994; Miwa et al., 1997; Nathan and Nair, 2013). However, previous studies suggested that deletion of ccpA was also deleterious/lethal (Holland, 2017), and modification of cre sites would have to be done for each repressible substrate. Hence, we reverted to a traditional substrate analog approach, which proved to be simple and remarkably effective.
4.1. Formation of HPr (Ser-P) is necessary to control MgsA activity under fermentative conditions
As well as the inability to create a ptsH1 crh1 double mutant, the single ptsH1 mutant was shown to have a novel phenotype under fermentative, but not aerobic conditions, namely the production of methylglyoxal, presumably resulting from the over-activation/upregulation of methylgloxal synthase (MgsA). The fact that this was observed under fermentative conditions alone may explain why this has not been observed with B. subtilis, where most studies have focussed on aerobic growth, as the ability of the phosphorylation state of Crh to control the activity of MgsA has been established in that host (Landmann et al., 2011). Crh participates in glycolytic flux regulation by interacting with MgsA. MgsA produces methylglyoxal from dihydroxyacetone phosphate (DHAP) and is either excreted and/or converted to lactate and pyruvate via various pathways (Landmann et al., 2011). The methylglyoxal pathway acts as a glycolytic bypass under carbon overflow conditions, in order to prevent the accumulation of phosphorylated glycolytic intermediates which are harmful to cells (Eisermann et al., 1988). However, methylglyoxal is toxic and therefore its synthesis is under tight regulation. Under nutritional famine conditions, Crh is typically in its non-phosphorylated state due to the low fructose-1,6-bisphosphate (FBP) concentration. The non-phosphorylated form of Crh inhibits MgsA activity and prevents the formation of methylglyoxal. However, under feast conditions, high FBP concentrations result in the phosphorylation of Crh by HPr kinase. Crh (Ser-P) cannot interact with MgsA and consequently, carbon flux is allowed to flow through the methylglyoxal pathway to relieve phospho-sugar stress (Figure 10) (Landmann et al., 2011).
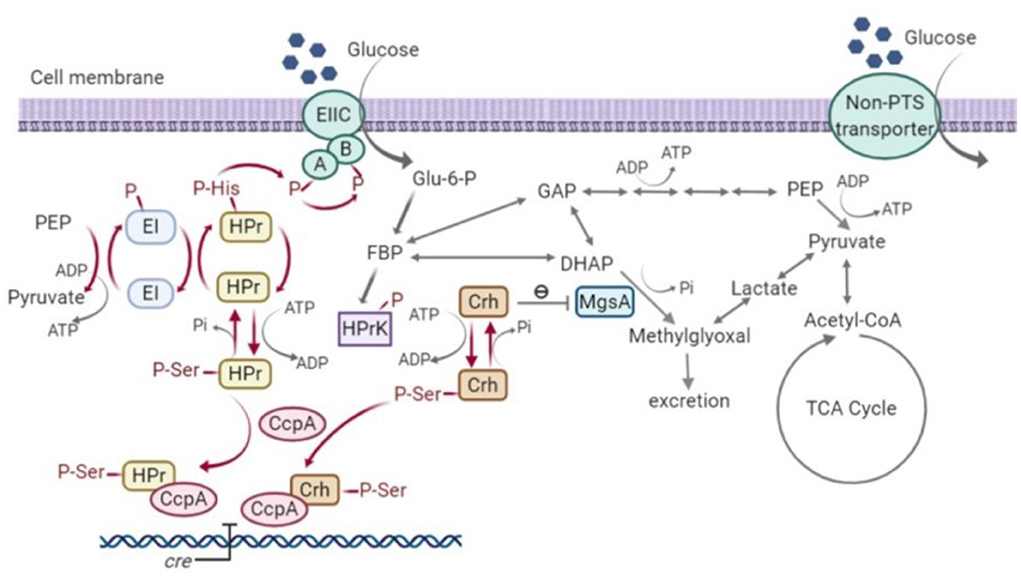
Figure 10. Signal transduction pathway of carbon catabolite repression (CCR) and methylglyoxal pathway in Bacillus subtilis (Görke and Stülke, 2008; Fujita, 2009; Landmann et al., 2011). HPr, phosphocarrier protein; Crh, catabolite repression HPr; CcpA, carbon catabolite protein A; HPrK, HPr kinase; MgsA, methylglyoxal synthase; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; EI, Enzyme I; EII, Enzyme II; PEP, phosphoenolpyruvate; Glu-6-P, glucose-6-phosphate; FBP, fructose-1,6-bisphosphate; Acetyl-CoA, acetyl-coenzyme A; TCA, tricarboxylic acid cycle; cre, catabolite-response element.
The over-stimulation of MgsA resulting in methylglyoxal accumulation and toxicity seen in the current study indicates that formation of HPr (Ser-P) is also necessary to control MgsA activity. While it could reflect an over-production of Crh (Ser-P) due to lack of competition between Hpr and Crh for Hpr kinase, this phenotype was not seen in the Hpr deletion strain TM444 △ptsH, so it is specific to the phosphorylation state of Hpr. An alternative explanation is that HPr (Ser-P) competes with Crh (Ser-P) for its binding site on MgsA, thus modulating the level of stimulation. One caveat to that argument is that deletion of ptsH eliminates glucose transport through the PTS system, resulting in slower growth (surprisingly, this eliminates growth of B. subtilis on glucose, despite the ability to produce an alternative MFS glucose transporter) and, hence, less accumulation of phospho-sugar intermediates.
4.2. Analysis of key mutations in both adaptive evolution experiments
As an alternative strategy to isolate CCR resistant mutants, 2-DG was used as a non-metabolizable substrate analog to induce CCR, providing a platform for selection of CCR-resistant mutants in P. thermoglucosidasius DSM 2542. Sequencing of isolates which were able to utilize PTS and non-PTS sugars simultaneously, revealed that none of the mutations were in the key regulatory proteins HPr, Crh and CcpA which are regarded as key players in the CCR of Gram-positive bacteria. However, in both evolution experiments, mutations were observed in PTS components, consistent with the observation that CCR typically involves sugars which are transported by, and signaling from the PTS system. The PTS is one of the major carbohydrate transport systems in bacteria (Deutscher et al., 2014): the glucose-PTS components consist of an EI, the HPr protein, encoded by ptsI and ptsH, respectively, and an inner membrane permease EII. EI and HPr are sugar non-specific, while EII is glucose-specific (also known as EIIGlu) (Stülke and Hillen, 2000). The EIIGlu in B. subtilis and P. thermoglucosidasius is comprised of a single polypeptide consisting of three domains (EIICBAGlu) encoded by ptsG, and sugar transport is actually performed by the membrane-bound EIICGlu domain during which glucose is phosphorylated (Stülke and Hillen, 2000; Uniprot Consortium, 2019). The EIIAGlu (which is a separate polypeptide in some organisms, such as E. coli) and EIIBGlu components form a phosphate transfer network for glucose translocation, in which phosphoenolpyruvate (PEP) serves as a phosphoryl donor (Deutscher, 2008). Initially, a phosphoryl group from PEP is transferred to EI, which subsequently transfers it to the catalytic His-15 residue of HPr. HPr-His15-P then donates the phosphoryl group to EIIGlu, initially to the EIIAGlu domain (Postma et al., 1993; Lorca et al., 2005). Within EIIGlu, the phosphoryl group is transferred from EIIAGlu to EIIBCGlu, which mediates glucose translocation by converting extracellular glucose to intracellular glucose-6-phosphate (Teplyakov et al., 2006). In this process, glucose is transported by EIICGlu and phosphorylated by EIIBGlu.
4.2.1. PtsI-A19E
In the first round of evolution in the presence of 2-DG, mutants were isolated in which the rate of glucose uptake under fermentative conditions was reduced and, in each isolate, sequenced the mutation PtsI-A19E (or EI-A19E) was present. EI is highly conserved among different bacteria, including that in P. thermoglucosidasius DSM2542, which has 78% sequence identity across its entire length to that from B. subtilis (NCBI Basic Local Alignment Search Tool). It consists of an N- and a C-terminal domain (EIN and EIC). The EIN has two subdomains connected by linkers; one is an EINα/β subdomain bearing the histidine active-site (His189 in E. coli and His189 in B. subtilis, His190 in Staphylococcus carnosus and probably His191 in Staphylococcus aureus), the other one is an EINα-helical subdomain which binds to HPr (Nosworthy et al., 1998; Márquez et al., 2006; Takayama et al., 2011). The EIC domain accepts a phosphoryl group from PEP and auto phosphorylates the histidine active site on the EINα/β subdomain. The phosphoryl group is then transferred to the HPr which binds to the EIN subdomain (Takayama et al., 2011). A previous study of E. coli EIN showed that the non-phosphorylated His189 active site is buried near the interface between the EINα/β and the EINα subdomains. Upon phosphorylation, EIN undergoes small conformational changes and the His189 rotates toward the surface, allowing for being approached by HPr-His-15 (Nosworthy et al., 1998). While not close to the active site, the A19E mutation is located within the EINα/β subdomain and substitution of a small non-polar alanine with a more bulky negatively charged glutamic acid residue could result is significant structural rearrangement, which clearly reduces or eliminates the activity of the EI protein (Figure 11). This loss of activity was also evident during growth on glucose alone and is consistent with a study in Bacillus cereus, which showed that deletion of the ptsI gene inhibited glucose uptake and utilization. The resulting phenotype is very similar to that of TM444 △ptsH, with slow glucose uptake failing to trigger CCR, so allowing activation of xylose uptake and metabolism. However, it is not a very useful phenotype for industrial exploitation as reduction in CCR results from impaired glucose uptake.
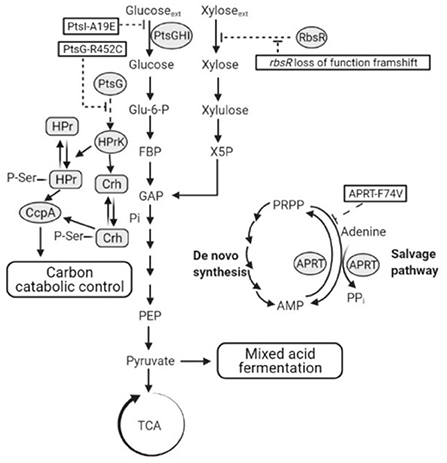
Figure 11. A schematic showing hypothetical impacts of PtsI-A19E, PtsG-R452C, and APRT-F74V mutations, and the rbsR loss of function frameshift mutation, respectively. Dotted lines are hypotheses proposed in this study. PtsGHI, glucose-specific phosphoenolpyruvate:sugar phosphotransferase system (PTS); RbsR, ribose operon repressor; TCA, tricarboxylic acid cycle; ACS, acetyl-CoA synthetase; APRT, adenine phosphoribosyl transferase; PRPP, adenine and phosphoribosyl pyrophosphate; AMP, adenosine monophosphate; CoA, coenzyme A; HPr, phosphocarrier protein HPr; Crh, HPr-like protein Crh; CcpA, catabolite control protein A; HPrK, HPr kinase.
4.2.2. PtsG-R452C
The PtsG-R452C (or EIIBGlu-R452C) mutation obtained in the second evolution is far more useful as this seemed to allow continued glucose uptake but reduced CCR. In E. coli, there are three essential motifs on EIIBGlu within a strongly conserved sequence (DACITRLR) between amino acids Asp-419 and Arg-426, among which Cys-421 is the site of phosphorylation by EIIAGlu. Two invariant arginines (Arg-424 and Arg-426) stabilize the phosphate with their guanidino groups by forming hydrogen bonds and electrostatic interactions (Gutknecht et al., 1998; Seitz et al., 2003). Replacement of both Arg-424 and Arg-426 by lysines, produced EIIBGlu mutants which were still able to accept the phosphoryl group from EIIAGlu, but could no longer transfer it to glucose, suggesting that these residues participated primarily in phospho-donor activity toward glucose but not in phospho-acceptor activity (Lanz and Erni, 1998). In B. subtilis, the conserved DACITRLR sequence was also found in EIIBGlu, with two arginines adjacent to the phosphorylated Cys-461 site (Bachem et al., 1997; Gutknecht et al., 1998). P. thermoglucosidasius DSM 2542 EIIBGlu displays 73% amino acid sequence identity to the EIIBGlu from B. subtilis (NCBI Basic Local Alignment Search Tool), has the conserved DACITRLR sequence located between Asp-445 and Arg-452 (Figure 12).

Figure 12. Amino acid sequence alignment of E. coli K-12, B. subtilis 168 and P. thermoglucosidasius DSM 2542 using UniProt Clustal Omega program. Regions around conserved sequence DACITRLR in PtsG are compared. * Indicates positions contain fully conserved residues,: indicates strongly similar properties, while. indicates weakly similar properties.
Surprisingly, the R452C mutation did not appear to impair glucose consumption in 2DG-ADE2b. A preliminary study showed that, when glucose was the sole carbon source during tube fermentation, 2DG-ADE2b consumed glucose at a similar rate to DSM 2542 (data not shown). However, a reduction in transfer rate from EIIBGlu to glucose may not significantly affect growth if this step is not rate-limiting. Nevertheless, when multiple copies of the wild type ptsG were introduced to this strain the rate of glucose uptake and consumption increased, suggesting that the overall level of activity of EII was rate limiting, but this may reflect the transfer from EIIAGlu to EIIBGlu, rather than EIIBGlu to glucose. The fact that this mutation does not reduce the rate of glucose uptake implies that the phospho-sugar signals thought to give rise to CCR in the wild type should be present and that the R452C mutation somehow disrupts signal transduction more directly (Figure 11). Clearly, this mutation could affect the steady state degree of phosphorylation of EIIBGlu by changing the relative rates of phosphate transfer between EIIAGlu and glucose, so this could have revealed an important, previously unrecognized, CCR signal transduction mechanism.
Interestingly, the ptsG over-expression strain grew poorly on xylose as sole carbon source, which would be consistent with EIIGlu being involved in signal transduction. In the absence of glucose, EIIAGlu, EIIBGlu at normal levels of expression would be expected to be phosphorylated. However, at high expression levels the rate of transfer of phosphate from HPr(H15)-P may not be sufficient to fully phosphorylate EII, resulting in a CCR “low phosphorylation state” signal being transmitted, even in the absence of glucose.
4.2.3. Frame-shift in RbsR
As well as having global effects on expression, in Gram negative organisms, CCR also affects the transport of inducers of expression of repressible operons in a process of inducer exclusion. Analogous processes operate in Gram positive bacteria, e.g., in B subtilis, glucose has been shown to compete with xylose for binding to the XylR repressor, thus limiting expression of the xylAB operon, even in the presence of xylose. Genome analysis has revealed that P. thermoglucosidasius does not have genes encoding XylR or a typical xylose transporter, yet it is still able to grow with xylose as the sole carbon source. Therefore, xylose transport might be regulated and mediated by the activity of other pentose regulators and transporters, as has been described for B. subtilis (Krispin and Allmansberger, 1998).
In line with this, qRT-PCR results showed that the ribose transporter in DSM 2542 was up-regulated during growth on xylose, though not as much as during growth on ribose, suggesting that xylose is taken up by the ribose transporter (Liang, 2022). This suggestion is supported by the presence of the frame-shift mutation in rbsR in all of the CCR de-repressed 2DG-ADE mutants which, notably all metabolized xylose more rapidly than the wild-type when it was a sole carbon source (Figure 11). Loss of the RbsR would result in uncontrolled expression of the ribose transporter which might fortuitously transport xylose, but is not direct evidence that P. thermoglucosidasius DSM2542 has evolved to use it in the absence of a dedicated xylose transporter. However, the fact that the 2DG-ADE2b strain over-expressing a functional rbsR grew more slowly that both the wild-type DSM2542 on xylose alone, strongly suggests that the high levels of RbsR are out-competing the xylose. Together with the qRT-PCR evidence and the fact that this mutation did not appear in araR, it is reasonable to conclude that xylose can both induce and be transported by the ribose transporter in P. thermoglucosidasius DSM2542.
4.2.4. Aprt-F74V
Finally, the Aprt-F74V mutation also seems to be important in evolved resistance to 2-DG and over-expression of a wild-type version of this gene showed the same effects as ptsG and rbsR in partially restoring CCR (although to a lesser extent) but also reducing the rate of xylose metabolism when xylose was the sole substrate. Mutations in Aprt, and in particular the Aprt-F74V mutation appeared in both evolution experiments and have also been reported in an earlier evolutionary study which was not focussing on CCR (Zhou et al., 2016). In the latter, 3 different mutations were found including a mid-gene frame-shift mutation; deletion of aprt was shown to have a similar phenotype of improved ethanol production from glucose but dependent on the addition of acetate. The association with CCR is unclear and arguments presented previously should not hold true here; based on the structure of the similar sized yeast enzyme, F74 is in a relatively conserved region at the homo-dimer interface and forms an inter-chain backbone (O-N) hydrogen bond. So a relatively conservative (hydrophobic-hydrophobic) mutation seems unlikely to disrupt dimer formation and is more likely to have a subtle effect on activity, possibly via the hinge region to the catalytic loop, which moves over the active site during catalysis. If the effect was to inactivate Aprt then it is surprising to find the same mutation appearing in 3 independent experiments (and exclusively in the two presented here). The characterized function of Aprt is to scavenge adenine from nucleotide degradation, although the reaction is reversible and recent studies with the human enzyme have shown that the conserved tyrosine in the catalytic loop is essential for driving the reaction in the forward (AMP synthesizing) direction (Huyet et al., 2018). As a more energy efficient pathway than de-novo synthesis, the purine salvage pathway is clearly important during growth, as shown by the creation of an essential catalytic loop tyrosine mutant in S. cerevisiae. If we assume that the APRT-F74V mutation is more likely to reduce the rate of the forward reaction and potentially increase the reverse reaction, then the effect of the mutation should be to reduce the rate of AMP (and PPi) production by this route and require greater flux through the de-novo pathways. As these are more energy demanding, this will increase the flux through phospho-fructokinase, which is known to be controlled by the energy charge in the cell. The consequence of this would not only be an increase in the rate of glycolysis and fermentation, as observed by Zhou et al., but also a reduction in the intra-cellular concentration of phospho-sugars, which are thought to be key CCR signaling molecules (Figure 11).
Interestingly, over-expression of wild-type Aprt partially restored some CCR, even in the background of the ptsG and rbsR mutations suggesting that both EIIAGlu and phospho-sugars are involved in CCR signaling. However, this also seemed to affect growth on xylose as a sole source of carbon, which is less easy to explain. If over-expression results in excessive production of AMP, this could have an effect on the energy charge but could also increase the flux of ribose-5-P from the pentose phosphate cycle, both of which could affect anaerobic growth on xylose in a manner which is independent of catabolite repression.
4.2.5. Other mutations
The mutations highlighted above are those that can most clearly be linked to the release of CCR. Of the other common mutations, holB and mutS mutations affect DNA replication and are likely to contribute to an increased mutation rate. Spo0A, rsfA, comEC, degU and degV mutations will moderate the cell response to starvation as a consequence of using a non-metabolisable CCR analog, while the DDX/DHX mutation could affect regulation of a range of functions. The other common mutations affect metabolic/biosynthetic functions and are less easy to explain without further studies. GltB encodes a component of glutamate synthase, while proS encodes a proline-tRNA ligase and dal encodes a d-alanine racemase, but all of the mutations found were missense, so they may not have resulted in loss of function. The most intriguing in that respect is the frame-shift mutation in hypE, which encodes a maturation enzyme for [NiFe] hydrogenases and is clearly essential for hydrogenase activity.
5. Conclusion
It has generally been believed that PTS components are involved in CCR in Enterobacteriaceae, typically via inducer exclusion mediated by EIIA, but not in Firmicutes, whose inducer exclusion relies on the interaction of HPr-Ser-P with ABC transporters as in B. subtilis (Deutscher et al., 2014). However, evidence from a previous study in B. subtilis suggested that a functional EIIGlu is indispensable for CCR in B. subtilis, though the detailed mechanisms were unclear (Bachem et al., 1997). This study clearly points toward the phosphorylation state of EIIAGlu or EIIBGlu being important for CCR signaling in P. thermoglucosidasius DSM 2542 and possibly in the wider community of Firmicutes. Further study will be required to investigate the detailed mechanism. At an applied level, mutation in EIIBGlu provides a route to release catabolite repression in this important thermophile.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive Repository, accession number PRJNA866999.
Author contributions
Laboratory experiments were formulated by JL, DL, and AB. Methodologies were designed by JL, DL, and RK. Experimental work was carried out by JL. The manuscript was written by JL and edited by DL, AB, and RK. All authors read and approved the final manuscript.
Funding
This work was funded by the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement [H2020-MSCA-CO-FUND 665992]; by Corbion and by the EPSRC [EP/L016354/1].
Acknowledgments
We wish to thank Dr. Christopher Ibenegbu and Dr. Alice Marriott for their help. Many thanks for Dr. Shaun Reeksting for helping with the GC-MS analysis of derivatized methylglyoxal. Thanks are also extended to people in the Leak Lab and the Centre for Sustainable Circular Technologies (CSCT) in the University of Bath.
Conflict of interest
Author RK was employed by Corbion.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.985465/full#supplementary-material
References
Atkinson, A., Cripps, R., and Eley, K. (2013). Sporulation-deficient thermophilic microorganisms for the production of ethanol. US Patent 8486687. Guildford.
Bachem, S., Faires, N., and Stülke, J. (1997). Characterization of the presumptive phosphorylation sites of the Bacillus subtilis glucose permease by site-directed mutagenesis: implication in glucose transport and catabolite repression. FEMS Microbiol. Lett. 156, 233–238. doi: 10.1111/j.1574-6968.1997.tb12733.x
Bacon, L. F., Hamley-Bennett, C., Danson, M. J., and Leak, D. J. (2017). Development of an efficient technique for gene deletion and allelic exchange in Geobacillus spp. Microb. Cell Factories 16, 58. doi: 10.1186/s12934-017-0670-4
Banerjee, S., Mudliar, S., Sen, R., Giri, B., Satpute, D., Chakrabarti, T., et al. (2010). Commercializing lignocellulosic bioethanol: technology bottlenecks and possible remedies. Biofuels Bioproducts Biorefining Innovat. Sustain. Econ. 4, 77–93. doi: 10.1002/bbb.188
Chang, A. Y., Chau, V., Landas, J. A., and Pang, Y. (2017). Preparation of calcium competent Escherichia coli and heat-shock transformation. JEMI Methods 1, 22–25. Available online at: https://static.igem.org/mediawiki/2018/d/d2/T–NYMU-Taipei–protocol-competent-cell.pdf
Dahl, M. K., and Hillen, W. (1995). Contributions of Xy1R, CcpA and HPr to catabolite repression of the xyl operon in Bacillus subtilis. FEMS Microbiol. Lett. 132. 79–83. doi: 10.1111/j.1574-6968.1995.tb07814.x
Deutscher, J. (2008). The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11, 87–93. doi: 10.1016/j.mib.2008.02.007
Deutscher, J., Aké, F. M. D., Derkaoui, M., Zébré, A. C., Cao, T. N., Bouraoui, H., et al. (2014). The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256. doi: 10.1128/MMBR.00001-14
Deutscher, J., Küster, E., Bergstedt, U., Charrier, V., and Hillen, W. (1995). Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15, 1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x
Deutscher, J., Reizer, J., Fischer, C., Galinier, A., Saier, M., and Steinmetz, M. (1994). Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol. 176, 3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994
Do, E.-J., Shin, H.-D., and Kim, C. (1993). Selection and characterization of catabolite repression resistant mutant of Bacillus firmus var. alkalophilus producing cyclodextrin glucanotransferase. J. Microbiol. Biotechnol. 3, 78–85.
Eisermann, R., Deutscher, J., Gonzy-Treboul, G., and Hengstenberg, W. (1988). Site-directed mutagenesis with the ptsH gene of Bacillus subtilis. Isolation and characterization of heat-stable proteins altered at the atp-dependent regulatory phosphorylation site. J. Biol. Chem. 263, 17050–17054. doi: 10.1016/S0021-9258(18)37496-9
Engler, C., Gruetzner, R., Kandzia, R., and Marillonnet, S. (2009). Golden gate shuffling: a one-pot dna shuffling method based on type iis restriction enzymes. PLoS ONE 4, e5553. doi: 10.1371/journal.pone.0005553
Fujita, Y. (2009). Carbon catabolite control of the metabolic network in Bacillus sutilis. Biosci. Biotechnol. Biochem. 73, 245–259. doi: 10.1271/bbb.80479
Galinier, A., Deutscher, J., and Martin-Verstraete, I. (1999). Phosphorylation of either crh or hpr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J. Mol. Biol. 286, 307–314. doi: 10.1006/jmbi.1998.2492
Gandhi, N., Barrett-Wilt, G., Steele, J., and Rankin, S. (2019). Lactobacillus casei expressing methylglyoxal synthase causes browning and heterocyclic amine formation in parmesan cheese extract. J. Dairy Sci. 102, 100–112. doi: 10.3168/jds.2018-15042
Görke, B., and Stülke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624. doi: 10.1038/nrmicro1932
Gunnewijk, M. G., and Poolman, B. (2000). Phosphorylation state of hpr determines the level of expression and the extent of phosphorylation of the lactose transport protein ofstreptococcus thermophilus. J. Biol. Chem. 275, 34073–34079. doi: 10.1074/jbc.M003512200
Gutknecht, R., Lanz, R., and Erni, B. (1998). Mutational analysis of invariant arginines in the IIAB(Man) subunit of the Escherichia coli phosphostransferase system. J. Biol. Chem. 273, 12234–12238. doi: 10.1074/jbc.273.20.12234
Hills, C. A. (2015). Acetate metabolism in Geobacillus thermoglucosidasius and strain engineering for enhanced bioethanol production (Ph.D. thesis). University of Bath, Bath, United Kingdom.
Holland, A. (2017). Optimisation of feedstock utilisation by Geobacillus thermoglucosidasius (Ph.D. thesis). University of Bath, Bath, United Kingdom.
Horton, R., Meldrum, B., and Bachelard, H. (1973). Enzymic and cerebral metabolic effects of 2-deoxy-d-glucose. J. Neurochem. 21, 507–520. doi: 10.1111/j.1471-4159.1973.tb05996.x
Hueck, C. J., and Hillen, W. (1995). Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol. Microbiol. 15, 395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x
Huyet, J., Ozeir, M., Burgevin, M.-C., Pinson, B., Chesney, F., Remy, J.-M., et al. (2018). Structural insights into the forward and reverse enzymatic reactions in human adenine phosphoribosyltransferase. Cell Chem. Biol. 25, 666–676. doi: 10.1016/j.chembiol.2018.02.011
Kang, Q., Appels, L., Tan, T., and Dewil, R. (2014). Bioethanol from lignocellulosic biomass: current findings determine research priorities. Sci. World J. 2014, 298153. doi: 10.1155/2014/298153
Kim, J. H., Block, D. E., and Mills, D. A. (2010). Simultaneous consumption of pentose and hexose sugars: an optimal microbial phenotype for efficient fermentation of lignocellulosic biomass. Appl. Microbiol. Biotechnol. 88, 1077–1085. doi: 10.1007/s00253-010-2839-1
Kornberg, H., and Lambourne, L. (1994). The role of phosphoenolpyruvate in the simultaneous uptake of fructose and 2-deoxyglucose by Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 91, 11080–11083. doi: 10.1073/pnas.91.23.11080
Kraus, A., Hueck, C., Gartner, D., and Hillen, W. (1994). Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J. Bacteriol. 176, 1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994
Krispin, O., and Allmansberger, R. (1998). The Bacillus subtilis AraE protein displays a broad substrate specificity for several different sugars. J. Bacteriol. 180, 3250–3252. doi: 10.1128/JB.180.12.3250-3252.1998
Landmann, J. J., Busse, R. A., Latz, J. H., Singh, K. D., Stülke, J., and Görke, B. (2011). Crh, the paralogue of the phosphocarrier protein HPr, controls the methylglyoxal bypass of glycolysis in Bacillus subtilis. Mol. Microbiol. 82, 770–787. doi: 10.1111/j.1365-2958.2011.07857.x
Lanz, R., and Erni, B. (1998). The glucose transporter of the Escherichia coli phosphotransferase system: mutant analysis of the invariant arginines, histidines, and domain linker. J. Biol. Chem. 273, 12239–12243. doi: 10.1074/jbc.273.20.12239
Liang, J. (2022). Removing carbon catabolite repression from Parageobacillus thermoglucosidasius DSM2542 (Ph.D. thesis). University of Bath, Bath, United Kingdom.
Liang, J., Roberts, A., van Kranenburg, R., Bolhuis, A., and Leak, D. (2021). Relaxed control of sugar utilization in Parageobacillus thermoglucosidasius DSM 2542. Microbiol. Res. 256, 126957. doi: 10.1016/j.micres.2021.126957
Liu, W., Fu, J., Wang, Z., and Chen, T. (2014). “Elimination of carbon catabolite repression in bacillus subtilis for the improvement of 2, 3-butanediol production,” in Proceedings of the 2012 International Conference on Applied Biotechnology (ICAB 2012). Lecture Notes in Electrical Engineering, Vol 249, eds T. C. Zhang, P. Ouyang, S. Kaplan, and B. Skarnes (Berlin; Heidelberg: Springer). doi: 10.1007/978-3-642-37916-1_33
Lorca, G. L., Chung, Y. J., Barabote, R. D., Weyler, W., Schilling, C. H., and Saier, M. H. (2005). Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HPrK. J. Bacteriol. 187, 7826–7839. doi: 10.1128/JB.187.22.7826-7839.2005
Macklyne, H.-R. V. (2017). Engineering bacteria for biofuel production (Ph.D. thesis). University of Sussex, Sussex, United Kingdom.
Márquez, J., Reinelt, S., Koch, B., Engelmann, R., Hengstenberg, W., and Scheffzek, K. (2006). Structure of the full-length enzyme I of the phosphoenolpyruvate-dependent sugar phosphotransferase system. J. Biol. Chem. 281, 32508–32515. doi: 10.1074/jbc.M513721200
Martin-Verstraete, I., Deutscher, J., and Galinier, A. (1999). Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J. Bacteriol. 181, 2966–2969. doi: 10.1128/JB.181.9.2966-2969.1999
Martin-Verstraete, I., Stülke, J., Klier, A., and Rapoport, G. (1995). Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177, 6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995
Mattanovich, D., Sauer, M., and Gasser, B. (2014). Yeast biotechnology: teaching the old dog new tricks. Microb. Cell Factor. 13, 34. doi: 10.1186/1475-2859-13-34
Millard, C. S., Chao, Y.-P., Liao, J. C., and Donnelly, M. I. (1996). Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl. Environ. Microbiol. 62, 1808–1810. doi: 10.1128/aem.62.5.1808-1810.1996
Miwa, Y., Nagura, K., Eguchi, S., Fukuda, H., Deutscher, J., and Fujita, Y. (1997). Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol. Microbiol. 23, 1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x
Nathan, S., and Nair, M. (2013). Engineering a repression-free catabolite-enhanced expression system for a thermophilic alpha-amylase from Bacillus licheniformis msg. J. Biotechnol. 168, 394–402. doi: 10.1016/j.jbiotec.2013.09.016
Nosworthy, N. J., Peterkofsky, A., König, S., Seok, Y.-J., Szczepanowski, R. H., and Ginsburg, A. (1998). Phosphorylation destabilizes the amino-terminal domain of enzyme I of the Escherichia coli phosphoenolpyruvate: sugar phosphotransferase system. Biochemistry 37, 6718–6726. doi: 10.1021/bi980126x
Oh, H., Wee, Y.-J., Yun, J.-S., Han, S. H., Jung, S., and Ryu, H.-W. (2005). Lactic acid production from agricultural resources as cheap raw materials. Bioresour. Technol. 96, 1492–1498. doi: 10.1016/j.biortech.2004.11.020
OIV-MA-AS315-21 (2010). Determination of α-Dicarbonyl Compounds of Wine by GC After Derivation. Compendium of International Analysis of Methods–OIV.
Postma, P. W., Lengeler, J. W., and Jacobson, G. R. (1993). Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Mol. Biol. Rev. 57, 543–594. doi: 10.1128/mr.57.3.543-594.1993
Puri-Taneja, A., Paul, S., Chen, Y., and Hulett, F. M. (2006). CcpA causes repression of the phoPR promoter through a novel transcription start site, PA6. J. Bacteriol. 188, 1266–1278. doi: 10.1128/JB.188.4.1266-1278.2006
Reeve, B., Martinez-Klimova, E., de Jonghe, J., Leak, D. J., and Ellis, T. (2016). The Geobacillus plasmid set: a modular toolkit for thermophile engineering. ACS Synthet. Biol. 5, 1342–1347. doi: 10.1021/acssynbio.5b00298
Reizer, J., Bergstedt, U., Galinier, A., Küster, E., Saier, M., Hillen, W., et al. (1996). Catabolite repression resistance of gnt operon expression in Bacillus subtilis conferred by mutation of HIS-15, the site of phosphoenolpyruvate-dependent phosphorylation of the phosphocarrier protein HPr. J. Bacteriol. 178, 5480–5486. doi: 10.1128/jb.178.18.5480-5486.1996
Repizo, G. D., Blancato, V. S., Sender, P. D., Lolkema, J., and Magni, C. (2006). Catabolite repression of the citST two-component system in Bacillus subtilis. FEMS Microbiol. Lett. 260, 224–231. doi: 10.1111/j.1574-6968.2006.00318.x
Schumacher, M. A., Seidel, G., Hillen, W., and Brennan, R. G. (2006). Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation. J. Biol. Chem. 281, 6793–6800. doi: 10.1074/jbc.M509977200
Schumacher, M. A., Seidel, G., Hillen, W., and Brennan, R. G. (2007). Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1, 6-bisphosphate. J. Mol. Biol. 368, 1042–1050. doi: 10.1016/j.jmb.2007.02.054
Seitz, S., Lee, S.-J., Pennetier, C., Boos, W., and Plumbridge, J. (2003). Analysis of the interaction between the global regulator mlc and EIIBGlc of the glucose-specific phosphotransferase system in Escherichia coli. J. Biol. Chem. 278, 10744–10751. doi: 10.1074/jbc.M212066200
Shaw, A. J., Podkaminer, K. K., Desai, S. G., Bardsley, J. S., Rogers, S. R., Thorne, P. G., et al. (2008). Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. U.S.A. 105, 13769–13774. doi: 10.1073/pnas.0801266105
Sheng, L., Zhang, Y., and Minton, N. P. (2016). Complete genome sequence of Geobacillus thermoglucosidasius NCIMB 11955, the progenitor of a bioethanol production strain. Genome Announc. 4, e01065-16. doi: 10.1128/genomeA.01065-16
Simon, R., Priefer, U., and Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. doi: 10.1038/nbt1183-784
Stülke, J., and Hillen, W. (2000). Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54, 849–880. doi: 10.1146/annurev.micro.54.1.849
Styles, M. Q., Nesbitt, E. A., Hoffmann, T. D., Queen, J., Ortenzi, M. V., and Leak, D. J. (2021). The heterologous production of terpenes by the thermophile Parageobacillus thermoglucosidasius in a consolidated bioprocess using waste bread. Metab. Eng. 65, 146–155. doi: 10.1016/j.ymben.2020.11.005
Sun, T., and Altenbuchner, J. (2010). Characterization of a mannose utilization system in Bacillus subtilis. J. Bacteriol. 192, 2128–2139. doi: 10.1128/JB.01673-09
Takayama, Y., Schwieters, C. D., Grishaev, A., Ghirlando, R., and Clore, G. M. (2011). Combined use of residual dipolar couplings and solution x-ray scattering to rapidly probe rigid-body conformational transitions in a non-phosphorylatable active-site mutant of the 128 kda enzyme I dimer. J. Am. Chem. Soc. 133, 424–427. doi: 10.1021/ja109866w
Teplyakov, A., Lim, K., Zhu, P.-P., Kapadia, G., Chen, C. C., Schwartz, J., et al. (2006). Structure of phosphorylated enzyme I, the phosphoenolpyruvate: sugar phosphotransferase system sugar translocation signal protein. Proc. Natl. Acad. Sci. U.S.A. 103, 16218–16223. doi: 10.1073/pnas.0607587103
Uniprot Consortium (2019). Uniprot: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506?D515. doi: 10.1093/nar/gky1049
Van Kranenburg, R., Verhoef, A., and Machielsen, M. P. (2019). Genetic modification of (s)-lactic acid producing thermophilic bacteria. US Patent 10273509. Gorinchem.
VanFossen, A. L., Verhaart, M. R., Kengen, S. M., and Kelly, R. M. (2009). Carbohydrate utilization patterns for the extremely thermophilic bacterium caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl. Environ. Microbiol. 75, 7718–7724. doi: 10.1128/AEM.01959-09
Vinuselvi, P., Kim, M. K., Lee, S. K., and Ghim, C. M. (2012). Rewiring carbon catabolite repression for microbial cell factory. BMB Rep. 45, 59–70. doi: 10.5483/BMBRep.2012.45.2.59
Vishnu, C., Seenayya, G., and Reddy, G. (2002). Direct fermentation of various pure and crude starchy substrates to l (+) lactic acid using Lactobacillus amylophilus gv6. World J. Microbiol. Biotechnol. 18, 429–433. doi: 10.1023/A:1015526221744
Wang, H., Yang, Y., Schofield, M. J., Du, C., Fridman, Y., Lee, S. D., et al. (2003). Dna bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. U.S.A. 100, 14822–14827. doi: 10.1073/pnas.2433654100
Wang, L., Zhao, B., Liu, B., Yu, B., Ma, C., Su, F., et al. (2010). Efficient production of l-lactic acid from corncob molasses, a waste by-product in xylitol production, by a newly isolated xylose utilizing Bacillus sp. strain. Bioresour. Technol. 101, 7908–7915. doi: 10.1016/j.biortech.2010.05.031
Wee, Y.-J., Kim, J.-N., and Ryu, H.-W. (2006). Biotechnological production of lactic acid and its recent applications. Food Technol. Biotechnol. 44, 163–172. Available online at: http://www.ftb.com.hr/80-volume-44-issue-no-2/445-biotechnological-production-of-lactic-acid-and-its-recent-applications
Xi, H., Kurtoglu, M., and Lampidis, T. J. (2014). The wonders of 2-deoxy-d-glucose. IUBMB life 66, 110–121. doi: 10.1002/iub.1251
Zhou, J., Wu, K., and Rao, C. V. (2016). Evolutionary engineering of Geobacillus thermoglucosidasius for improved ethanol production. Biotechnol. Bioeng. 113, 2156–2167. doi: 10.1002/bit.25983
Zhu, M., Lu, Y., Wang, J., Li, S., and Wang, X. (2015). Carbon catabolite repression and the related genes of ccpA, ptsH, hprK in Thermoanaerobacterium aotearoense. PLoS ONE 10, e0142121. doi: 10.1371/journal.pone.0142121
Keywords: carbon catabolite repression, Parageobacillus thermoglucosidasius, 2-deoxyglucose resistance, adaptive evolution, mixed-sugar fermentation
Citation: Liang J, van Kranenburg R, Bolhuis A and Leak DJ (2022) Removing carbon catabolite repression in Parageobacillus thermoglucosidasius DSM 2542. Front. Microbiol. 13:985465. doi: 10.3389/fmicb.2022.985465
Received: 03 July 2022; Accepted: 30 August 2022;
Published: 20 October 2022.
Edited by:
Obulisamy Parthiba Karthikeyan, South Dakota School of Mines and Technology, United StatesReviewed by:
Bo Zhang, Zhejiang University of Technology, ChinaHirokazu Suzuki, Tottori University, Japan
Copyright © 2022 Liang, van Kranenburg, Bolhuis and Leak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinghui Liang, ampsNTFAYmF0aC5hYy51aw==
 Jinghui Liang
Jinghui Liang Richard van Kranenburg
Richard van Kranenburg Albert Bolhuis5
Albert Bolhuis5 David J. Leak
David J. Leak