
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 09 September 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.984672
Oomycetes cause hundreds of destructive plant diseases, threatening agricultural production and food security. These fungus-like eukaryotes show multiple sporulation pattern including the production of sporangium, zoospore, chlamydospore and oospore, which are critical for their survival, dispersal and infection on hosts. Recently, genomic and genetic technologies have greatly promoted the study of molecular mechanism of sporulation in the genus Phytophthora and Peronophythora. In this paper, we characterize the types of asexual and sexual spores and review latest progress of these two genera. We summarize the genes encoding G protein, mitogen-activated protein kinase (MAPK) cascade, transcription factors, RNA-binding protein, autophagy-related proteins and so on, which function in the processes of sporangium production and cleavage, zoospore behaviors and oospore formation. Meanwhile, various molecular, chemical and electrical stimuli in zoospore behaviors are also discussed. Finally, with the molecular mechanism of sporulation in Phytophthora and Peronophythora is gradually being revealed, we propose some thoughts for the further research and provide the alternative strategy for plant protection against phytopathogenic oomycetes.
As a class of eukaryotic microorganisms, oomycetes show a similar life cycle with filamentous fungi; however oomycetes are evolutionarily close to photosynthetic algae and distant from true fungi (Tyler et al., 2006; Cavalier-Smith et al., 2018). Among oomycetes, Phytophthora and Peronophythora contain a large number of destructive plant pathogens, which seriously threaten agriculture, forestry and ecosystem. An example of this is the late blight pathogen Phytophthora infestans which is notorious for severely damaging the European potato industry in 1840 and leading to Ireland famine (Kamoun et al., 2015). Nowadays, late blight remains a major problem around the world with global yield losses estimated at more than £10 billion a year (Leesutthiphonchai et al., 2018a). Phytophthora capsici and Phytophthora sojae severely damage cucurbits and soybean, respectively (Kamoun et al., 2015). Litchi downy blight caused by Peronophythora litchii is the most destructive disease in litchi (Kong et al., 2021; Figure 1). P. litchii is evolutionarily closed to Phytophthora species, and the life cycle of P. litchii is illustrated in Figure 2. Therefore, Phytophthora and Peronophythora are reviewed together in this paper.

Figure 1. Disease symptoms of Peronophythora litchii infection. (A–C) Symptoms of P. litchii on litchi fruit, tender leaves and panicles. (D) Mature sporangiophores and sporangia of P. litchii on the infected tissue. Bar represents 100 μm.
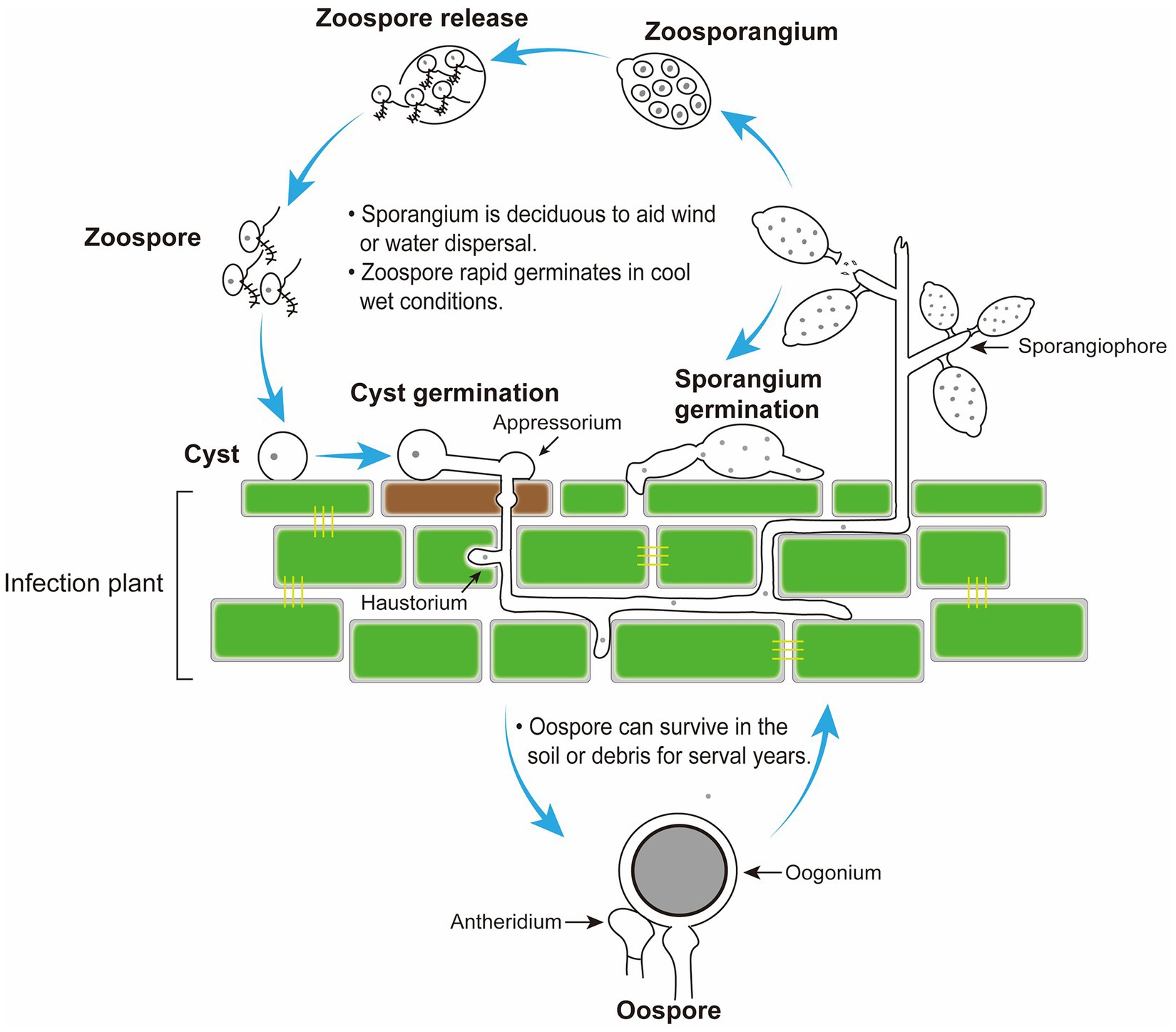
Figure 2. Disease cycle of Peronophythora litchii on host plant. The cycle can occur on leaves and fruit in the stage of litchi growth and fruit storage.
The Phytophthora and Peronophythora species are both hemibiotrophic microorganisms, which plays a notable role in scientific research. The research on potato late blight initiated the science of plant pathology. However, the classical genetic and molecular studies of oomycete lag behind true fungi. Several true fungi, such as Aspergillus nidulans, Magnaporthe oryzae, and yeasts were used as models for studying spore biology (Ebbole, 2007; Neiman, 2011; Etxebeste and Espeso, 2020), but knowledge of fungal development had limited the relevance to oomycete, because they belong to different kingdoms and produce different types of spores (Judelson and Blanco, 2005).
Until the 1990s, effective genetic tools began to be developed for oomycete, which promoted the progress of its biological research including spore development. DNA-mediated transformation was successfully developed for P. infestans, making it a model for the genus (Judelson et al., 1991). Subsequently, the establishment of gene-silencing technology enabled gene function to be analyzed (Kamoun et al., 1998; van West et al., 1999a; Whisson et al., 2005). Both GFP and GUS were used successfully as reporter genes in Phytophthora, which facilitated the transcription and subcellular localization analysis (van West et al., 1999b). And recent CRISPR/Case9-mediated genome editing technology has greatly accelerated the process of functional genomics of oomycete (Fang and Tyler, 2016; Situ et al., 2020; Ah-Fong et al., 2021). Moreover, the advances in sequencing technologies in the last two decades rapidly expanded the available genome and transcriptional data of Phytophthora and Peronophythora species (Ye et al., 2016; Ah-Fong et al., 2017; McGowan and Fitzpatrick, 2020). Since the first publication of oomycete genome sequences of P. ramorum and P. sojae in 2006 (Tyler et al., 2006), more than 27 Phytophthora species genome have been available (McGowan and Fitzpatrick, 2020), which has promoted the research of molecular mechanism of development and pathogenicity in oomycete.
Given that the spores play a critical role in plant-oomycete interaction, and that spore formation mechanisms have advanced significantly in the last decades, it is time to systematically summarize these findings and discuss the potential modulation of sporulation in plant disease management. In this review, the focus will be on the recent research progress with respect to formation and germination of sexual and asexual spores in Phytophthora and Peronophythora, especially those findings that have been proved by genetic strategies.
The asexual spores of Phytophthora and Peronophythora include sporangia (or zoosporangia), zoospores and chlamydospores. Sporangia are multinucleate and release zoospores in aqueous environment (Judelson and Blanco, 2005). Zoospores are single nucleated and wall-less cells that can swim with the help of two flagella; one tinsel (or anterior) flagellum, and one whiplash (or posterior) flagellum (Walker and van West, 2007). The thick-walled resting chlamydospores could be produced by some Phytophthora species but not P. infestans and P. litchii. Besides, the sexual oospores display double walls and contain one or more pellucid bodies (nuclei) and a well-defined ooplast (Boutet et al., 2010).
Compared with oospores, the yield of sporangia is very huge in Phytophthora and Peronophythora. Sporangia develop at the termini of sporangiophore (Figure 3). Sporangiophore is often branched and each terminus bears one sporangium in Phytophthora and Peronophythora (Judelson and Blanco, 2005). In P. litchii, a new sporangiophore could develop from mycelium or a terminus of another sporangiophore, the late process could be repeated limited times resulting multideterminate in tiers of sporangiophores, each with its own branching system (Figure 3). In P. litchii, PlATG6a positively regulates the branch formation of sporangiophores, while PlBZP32 exhibits the opposite effect (Kong et al., 2020; Wang et al., 2022). When sporangium matures, an apical papilla forms in most species and a basal septum develops. In caducous species such as P. infestans and P. litchii, water or wind can separate the sporangium from the sporangiophore and spread it for several kilometers (Aylor, 2003). This separation of sporangia is convenient for researchers as the sporangia can be easily purified for analysis. By contrast, the sporangia of non-caducous species such as P. sojae remain attached to the sporangiophores in water. Therefore, it is not easy to purify the sporangia of P. sojae, but zoospores could be collected for research. Besides, little effort has been made to decipher the underlying mechanism of the distinctive pattern of sporangiophores formation and sporangia caducity in Phytophthora and Peronophythora species so far, even though it could be used for crop protection.
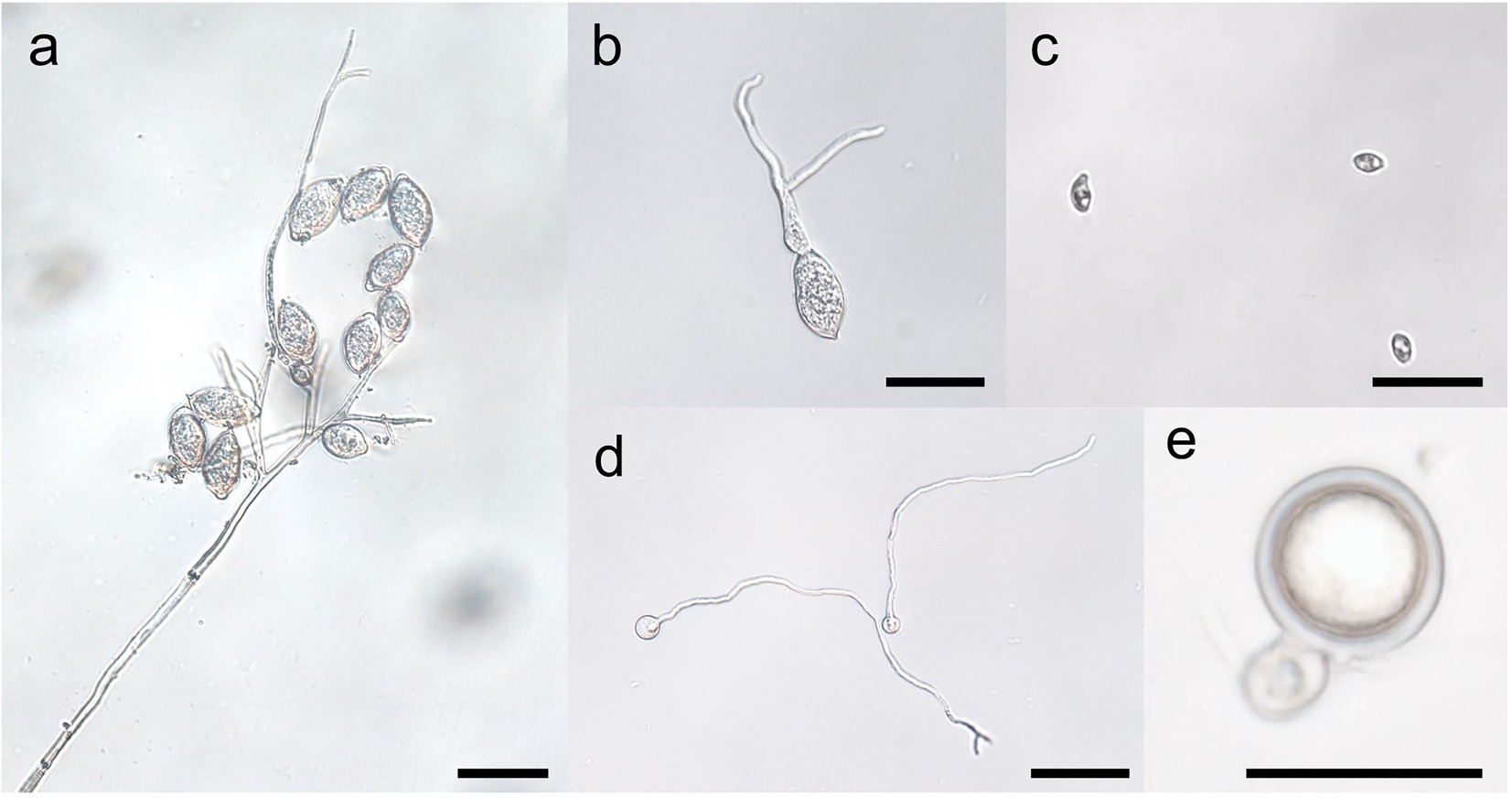
Figure 3. Stages of the spore cycles of Peronophythora litchii. A–E represent sporangiophore and sporangia, sporangium germination, zoospores, cysts germination, mature oospore, respectively. Bar represents 50 μm.
Asexual sporangia and zoospores are the main inocula of Phytophthora and Peronophythora. Sporangia have a remarkable ability to germinate in two different ways: direct germination and indirect germination (also known as zoosporogenesis), which is depended on the ambient temperature. Low temperature could promote the release of zoospores from sporangia. Both germination ways require the sporangia to be immersed in aqueous environment. Germination by the zoosporogenesis is believed to be important in disease. The biflagellate zoospores swim after release. The anterior flagellum is probably responsible for pulling the zoospore through the water whereas the posterior flagellum acts as a rudder for steering the cell (Walker and van West, 2007). Once a suitable site has been found, the zoospores encyst and become immobilized. The flagella are either discarded or retracted into the cell depending on the species (Hardham and Hyde, 1997). The encystment process involves the rapid cell wall formation. The dual germination strategy increases the possibility of Phytophthora sporangia to colonize plants in diverse environments. However, the genes involved in the sporangium germination pathways of Phytophthora and Peronophythora are largely unknown.
The heterotrimeric G-protein pathway regulates cellular responses to a wide range of extracellular signals in all eukaryotes (Xue et al., 2008). The sporangium formation and cleavage are also stimulated by environment to a great extent (Xue et al., 2008), the heterotrimeric G-protein pathway therefore is thought to be associated with the sporulation. In Phytophthora spp., G-protein have only one Gα, one Gβ, and one Gγ subunit (Qiu et al., 2020). The G protein α subunit GPA1 directly interacts with serine/threonine protein kinase YPK1 and prevents nuclear localization of YPK1, which leads to suppression of sporangia formation in P. sojae (Qiu et al., 2020). In the previous studies, the Gβ and Gγ (PiGPB1 and PiGPG1) of P. infestans were also demonstrated to be crucial for sporangium development and production (Latijnhouwers and Govers, 2003; van den Hoogen et al., 2018). The G-protein signaling normally initiates from membrane-bound G-protein coupled receptors (GPCRs) that receive extracellular signals and then activate the Gα subunit (Rosenbaum et al., 2009). Genome analysis revealed that Phytophthora spp. have over 60 GPCR genes, among which several are fused with a C-terminal phosphatidylinositol phosphate kinase (PIPK) domain (Bakthavatsalam et al., 2006). The full set of GPCR-PIPKs (GKs) of P. infestans was identified in 2013 and subsequent analysis proved that the overexpression of PiGK4 produced sporangia with aberrant cell walls, aberrant germination of sporangia and defects in cytoplasmic cleavage and the release of zoospores (Hua et al., 2013).
Another type of signal receptor leucine-rich repeat receptor-like kinases (LRR-RLKs) are also involved in Phytophthora spore development. The P. capsici PcLRR-RK1-silenced mutants exhibited abnormal shape and size of sporangia and lower production of sporangia and zoospores (Safdar et al., 2017). The systematical characterization of all 24 LRR-RLK genes (PsRLKs) from P. sojae revealed that PsRLK2/4/8/20/23 positively regulate the production of sporangia and zoospores, whereas PsRLK17/24 negatively regulate these processes (Si et al., 2021b).
In oomycetes and fungi, MAPK cascades also play a key role in the development and plant infection (Jiang et al., 2017; Gonzalez-Rubio et al., 2019). Sporangium formation is severely diminished in PsMPK1-silenced mutants (Li et al., 2014). MAPK10 also positively regulates the production of sporangia in P. litchii. P. litchii mitogen-activated protein kinase MAPK2 is essential for the sporangial cleavage of P. litchii (Huang et al., 2021). And the transcription of PlMYB1 and PlMAD1 (homologs of PsMYB1 and PsMAD1; described later) are downregulated in PlMAPK2 mutants (Huang et al., 2021).
Following the signal transduction pathways, transcription factors are the next players for controlling sporangium formation and germination. The P. sojae MYB transcription factor MYB1, regulated by PsSAK1, controls the differentiation of sporangial cytoplasm and nuclei rearrangement, which finally affects the sporangia cleavage during zoospores release (Zhang et al., 2012a). Eight Myb transcription factors are upregulated during sporulation and silencing of PiMyb2R3 results in suppressing sporulation in P. infestans (Xiang and Judelson, 2010, 2014). A basic leucine zipper (bZIP) transcription factor PlBZP32 is negatively associated with sporangia production (Kong et al., 2020). P. sojae MADS-box transcription factors PsMAD1 also affects the zoosporogenesis similarly to PsMYB1 (Lin et al., 2018). Silencing of PiMADS severely impairs sporangium production (Leesutthiphonchai and Judelson, 2018b). The transcriptome of PiMADS-silenced mutant also provides new data for identifying more genes involved in sporangium formation and revealing the network (Leesutthiphonchai and Judelson, 2018b).
Autophagy is a conserved cellular process allowing the organisms to recycle their intracellular components by sequestrating the cytosolic macromolecules or used organelles within the autophagosomes and delivering the contents to the lysosomes or vacuoles (Feng et al., 2014). Twenty-six genes in the core autophagy machinery were identified in P. sojae genome and silencing of PsAtg6a led to reduce sporangium production (Chen et al., 2017). In P. litchii, knockout of PlATG6a significantly impaired the pathogen sporangium production but promoted the release of zoospores (Wang et al., 2022). Further study is need to reveal the mechanism of ATG genes involved in asexual reproduction in oomycete.
Additionally, cell cycle protein PiCdc14/PsCdc14, importin α PsIMPA1 and FtsZ protein PcFtsZ2 are also involved in sporangia production (Ah-Fong and Judelson, 2003; Zhao et al., 2011a; Yang et al., 2015; Li et al., 2020b). Golgi reassembly stacking protein PsGRASP is associated with zoosporogenesis (Si et al., 2021a). Soluble N-ethylmaleimide-sensitive factor attachment protein receptor PsYKT6, Mating pheromone-induced death 1 PpMID1, catalase PiCAT2 and two chitin synthase PcCHS/PsCHS1 regulate both processes (Zhao et al., 2011b; Hwu et al., 2017; Cheng et al., 2019; Wang et al., 2020). The above studies identify the pathways from membrane receptor, signal transduction to the downstream transcriptional regulation involved in sporangium formation and cleavage. Nevertheless, the relationship between these components is rarely known, therefore, the regulatory network of how sporangia perceive environment temperature and determine the germination type remains to be further explored.
After zoospore release, Ca2+ helps to regulate their swimming and encystment, phosphatidic acid might also be involved in zoospore motility and encystment (Hardham and Hyde, 1997; Tani et al., 2004). Through study of the effect of inhibitors on zoospore development in P. infestans, a putative protein kinase gene induced during zoosporogenesis that encoded a protein resembling Ca2+ and calmodulin-regulated serine/threonine protein kinase was identified (Judelson and Roberts, 2002). The mRNA accumulation of this protein kinase is first detected soon after chilling sporangia in water and persisted in motile zoospores and germinated cysts, but not in other tissues (Judelson and Roberts, 2002).
The important roles of zoospore motility for successful infection have been shown through silencing of PiGPA1 and Pibzp1 (Latijnhouwers et al., 2004; Blanco and Judelson, 2005). Moreover, PsGPR11 and PsSAK1 are involved in the regulation of swimming time (Li et al., 2010; Wang et al., 2010). Further research suggested that some aberrant swimming behaviors are probably related to defects in flagella composition. The study of DEAD-box RNA helicase gene Pi-RNH1 showed that Pi-RNH1-silenced lines released larger zoospores which were observed to have multiple flagella, low tolerance to osmotic pressure and contained several large vesicles with smaller lipid bodies fused to them (Walker et al., 2008). On the contrary, the Phytophthora parasitica dynein light chain 1 (DLC1), one of the flagellar axoneme component, positively regulated flagella formation and zoospore motility (Narayan et al., 2010). PnPMA1, encoding an atypical plasma membrane H+-ATPase, also regulated the formation of zoospore flagellate in P. parasitica. High levels of PnPMA1 silencing resulted in production of non-flagellate and large aberrant zoospores, rapid transition from zoospores to cysts (Zhang et al., 2012b).
Chemotaxis is another important feature for zoospore to initiate infection in the disease cycle (Hua et al., 2015). The G protein α subunit is first shown to regulate the chemotaxis of P. infestans zoospores (Latijnhouwers et al., 2004). Further studies found that the histidine triad (HIT) domain-containing protein PsHint1 is associated with PsGPA1 and co-regulates zoospore chemotaxis to isoflavone and encystment (Hua et al., 2008; Zhang et al., 2016). This study also indicated that chemotaxis and motility are controlled by two different pathways, both of which are regulated by the Gα subunit, while Hint1 is only involved in the former. Yang et al. reported that the GPCR-PIPK PsGK4 was involved in zoospore chemotaxis towards isoflavone as well as soybean roots (Yang et al., 2013). Recently, LRR-RLKs are found to act on zoospore chemotaxis (Si et al., 2021b). Absence of LRR-RLK encoding genes PsRLK5/11/17/22/24 results in severe defects in recognition of isoflavones by P. sojae zoospore (Si et al., 2021b). Interestingly, these five PsRLKs are demonstrated to regulate this process by direct interacting with PsGPA1 via their intracellular kinase domains (Si et al., 2021b). Whether there is a functional relationship between PsHint1 and these membrane-localized kinases in the signal pathway of chemotaxis deserve to be investigated.
Recognition of the host by zoospores is often referred to a variety of signals, for example chemical, electrical field and physical feature from host (Tyler, 2002; van West et al., 2002; Hua et al., 2015; Bassani et al., 2020a). Recent experimental evidence has demonstrated that zoospore-zoospore interactions can lead to “pattern swimming” in the absence of chemical or electrical signals from plants. Zoospore-free fluid (ZFF) prepared from zoospore suspension stimulates cyst germination and induces a tactic response to enhance zoospore auto-aggregation and infection (Kong and Hong, 2010a; Kong et al., 2010b). Another study in P. parasitica showed that the perception of a K+ gradient induces zoospore coordinated motion and aggregation, which produce vesicular and fibrillary material discharged at cell-to-cell communication (Bassani et al., 2020b). Leucine is also a signaling molecule secreted from zoospore and could regulate zoospore germination and infection by P. erythroseptica (Jiang et al., 2019). These zoospore–zoospore communication and coordinated behavior is analogous to bacterial quorum sensing. Additionally, a diurnal effect has been observed for production of propagules in P. ramorum. Large differences in sporangium and zoospore numbers were observed for the dark versus light periods (Tooley et al., 2020). However, it is not yet clear how these signals are generated in local soil or water niches and which genes govern the sensing and subsequent responses of zoospore.
The cyst development, especially germination and the formation of infectious hyphae and appressorium that penetrate the host cells is the key step for infection. In a previous study, co-silencing of three nuclear LIM interactor-interacting factors encoded genes (NIFC1, NIFC2 and NIFC3) in P. infestans impaired cyst germination (Judelson and Tani, 2007). Subsequently, more positive regulators of cyst germination were found in P. sojae, for example, the GPA1, Hint1, GPR11, SAK1 and HSF1 (Hua et al., 2008; Li et al., 2010; Wang et al., 2010; Sheng et al., 2015; Zhang et al., 2016). LRR-RK1 and BZP32 are also involved in cyst germination in P. capsici and P. litchii, respectively (Safdar et al., 2017; Kong et al., 2020). Recently, the asparagine (Asn, N)-linked glycosylation, a ubiquitously distributed post-translational modification (Nagashima et al., 2018), is reported to participate in this process (Zhang et al., 2021). Site-directed mutagenesis in the N-glycosylation site of the GPI transamidase component protein (GPI16) and heat shock protein 70 (HSP70) make P. sojae cyst defeat in germination (Zhang et al., 2021).
Other than germination, abnormal growth of the germ tube can lead to failure of zoospore infection. For instance, silencing of PsMPK7 resulted in abnormal germinated cysts apical swelling, which might be due to the disturbance of polarized growth (Gao et al., 2015). Later, a study of the P. capsici high-affinity cAMP phosphodiesterase further confirmed the significance of polarized growth in cyst germ tube (Li et al., 2020a,b). The formation of appressorium is the last step of zoospore infection. Nevertheless, the regulators in this process are still largely unknown. Only previous research of Pibzp1 showed that this gene silenced mutants failed to develop appressoria and were unable to infect plants (Blanco and Judelson, 2005). Appressorium attachment and penetration of the plant cuticle and cell wall are the key processes for successful colonization of pathogens, given that the mechanism of appressorium development deserves more attention in the future research.
Oospores are important inocula in the next growing season as they can survive in soil or debris for several years, particularly for the homothallic (self-fertile) species. Oospores are also significant for heterothallics when both mating types (A1 and A2) share the same geographical space. In both homo- and hetero-thallics, male and female gametangia can develop and then fuse to form oospores. The germinated oospores produce either a hyphal tube, which can directly infect plants, or a germ sporangium, which acts like an asexual sporangium. Eight mating-induced genes were identified by suppression subtractive hybridization (Fabritius et al., 2002); among them, M90 encodes a member of the Puf family of translational regulators and highly expresses in sexual and asexual sporulation structures in P. infestans (Cvitanich and Judelson, 2003). Recently, researchers have confirmed that M90 is critical for the oospore formation in P. litchii and Pythium ultimum by gene silencing and CRISPR/Cas9-mediated genome editing technology (Jiang et al., 2017; Feng et al., 2021). A tripartite recognition motif (TRM) in the Puf domain of PuM90 could bind to the 3′– untranslated region (UTR) of PuFLP, and thereby repress PuFLP mRNA level to facilitate oospore formation. Considering that M90 is conserved in oomycete, its homologs may function via similar mechanism. A study of the P. infestans loricrin-like protein via scanning electron microscopy (SEM) found that many sunken areas were observed on the surfaces of the oogonia of PiLLP-silenced transformants, which indicated the loss of turgor in oogonia. The Nile red staining and fluorescence microscopy observation further found that red fluorescence-emitting substance was scattered instead of aggregating into a red ball in PiLLP-silenced transformants, thus, the sexual development blocked in the stage of oospore wall formation (Guo et al., 2017). Moreover, the C2H2 zinc finger protein CZF1 has been demonstrated to be involved in oospore development in P. sojae and P. litchii (Wang et al., 2009; Zhu et al., 2022). Interestingly, the transcription level of M90 and LLP are downregulated in CZF1 knockout mutant, suggesting that CZF1 may control the transcription of these two genes. Additionally, YPK1, YKT6, GK5, IMPA1 and N-glycosylation in HSP70 also play an important role in oospore production in P. sojae (Zhao et al., 2011b; Yang et al., 2013, 2015; Qiu et al., 2020; Zhang et al., 2021). Although above studies described the genes involved in oospore formation, how these genes relate to the mechanism of oospore formation remains unclear. Secretion of hormones is the prerequisite of sexual propagation. Once a heterothallic is stimulated by the hormone from opposite mating type, the hybrid and selfed oospores can be formed, while most homothallics produce both (Ko, 1988; Judelson, 1997). The chemical basis of Phytophthora mating hormone was first proposed in P. parasitica (Qi et al., 2005). We speculate that PsGK5 may be involved in the perception of these mating signal. How these genes are involved in synthesis and/or response to the mating hormones needs to be further explored.
Genetic manipulation and omics have greatly promoted the progress of molecular biology of oomycete spores in the last decades. This paper reviewed the signal and regulatory mechanism of spore development in the Phytophthora and Peronophythora, and aimed for better understanding why they cause such destructive and persistent diseases. Benefiting from the advance in molecular biology, more and more sporulation-related genes have now been characterized (Figure 4; Table 1); therefore, we draw a portrait of the expansive knowledge. G-protein pathway, MAPK cascades and transcription factors are involved in the perception of extracellular signal and the network of spore formation. Here, we propose the relationship of the main regulators in Phytophthora and Peronophythora spore development (Figure 5). It is obviously that the G protein α subunit GPA1 play a central role for asexual spore development. While, for sexual spore formation, more works can be done around the downstream signal transduction components of GK5 and mRNAs targeted by M90. Excitingly, in regard to this plethora of arenas, much remains to be investigated and understood. Researchers might use yeast-two hybrid, RIP or ChIP-seq technology to identify more direct interaction elements of these spore formation-related proteins and characterize their regulatory networks. Additionally, it is worth studying the formation of appressorium and haustorium from the geminated cyst, because they are also important weapons for these plant destroyers.
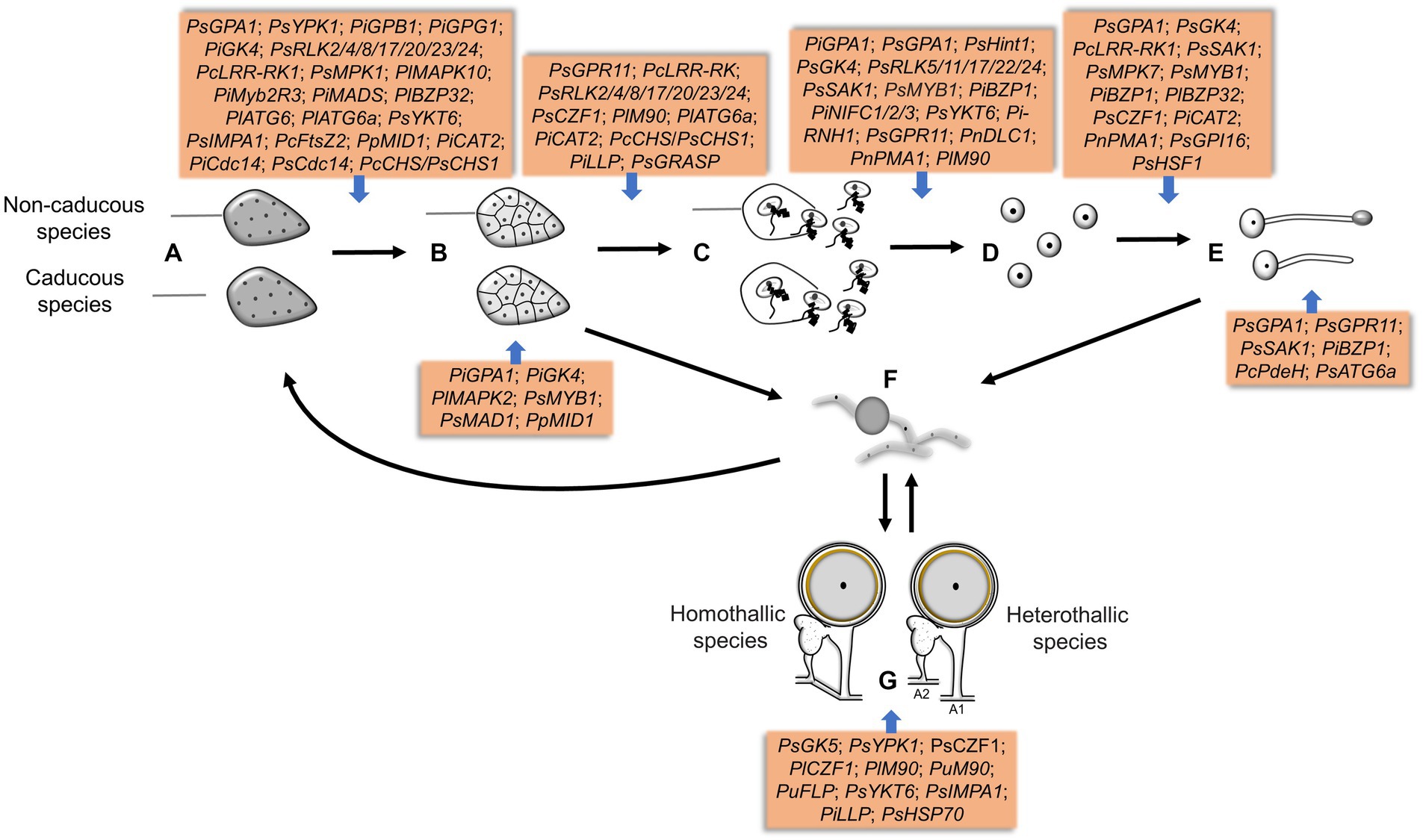
Figure 4. The genes involved in different sporulation and germination stages in Phytophthora and Peronophythora. A–G represent sporangia, sporangia cleavage, zoospores release, cysts, cysts germination, mycelia chlamydospores and oospores, respectively.
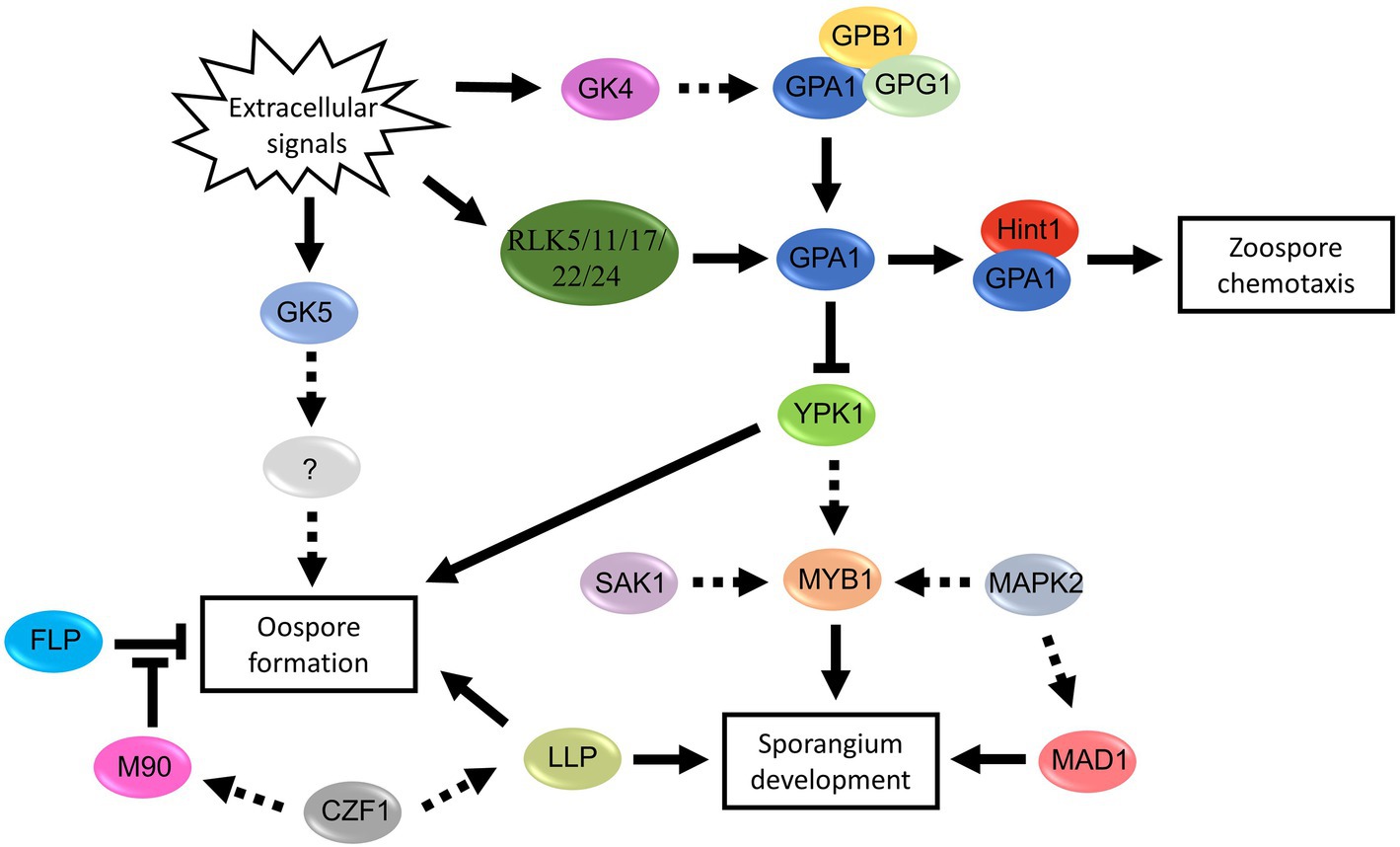
Figure 5. Proposed signal pathways involved in spore development of Phytophthora and Peronophythora. Arrows with solid line represent direct positive regulation, arrows with dotted line represent potential positive regulation and T-line represent negative regulation.
Distinction from other spores which are short-live and survive poorly apart from the host, the thick-walled oospores resulted from sexual reproduction are a multiyear threat because they can persist for years in soil, surviving freezing and fungicides. For example, in regions of northern Europe and Mexico where sexual reproduction of the heterothallic species P. infestans is frequent, oospores are an important source of inocula (Lehtinen and Hannukkala, 2004; Grüwald and Flier, 2005). Although mating type in P. infestans is determined by a single locus, duplication, transposition, deletion, or other rearrangement of this locus make the mating event complicated (Judelson, 1996). We anticipate that more research should be carried out in this direction to better seize the details of mating type determination in Phytophthora and Peronophythora. Modification of mating hormones or application of stimulants to alter oospore germination period might enable reduction of oospore infection.
Besides the Phytophthora, P. litchii is also an ideal research material, because the genome of this pathogen is published and it is easy to be genetically manipulated (Ye et al., 2016). Phytophthora and Peronophythora are the hemibiotrophic oomycetes which consist of biotrophic and necrotrophic phases during their infection. Although recent advance in phylogenetic analysis based on genome data have suggested that P. litchii (the only species in Peronophythora) belong to Phythophtora (Ye et al., 2016), this pathogen produces Peronospora-like sporangiophores and the differentiated, branched sporangiophores of P. litchii suggest a taxonomic affinity with downy mildews species (Figure 3). Moreover, the germ tubes in P. litchii oospores also have the characteristics of both Peronospora and Phytophthora, which comprise the short germ tubes that do not develop further and germ tubes that terminated by sporangia or become mycelia (Pao and Ko, 1980). Therefore, it is possible that P. litchii is the transitional species between Phytophthora and Peronospora. In future, this is important for elucidating the molecular mechanism underlying the sporangiophores formation of P. litchii.
Understanding the spore formation pathways might lead to develop novel effective strategies for controlling oomycete diseases. Cross-kingdom RNA trafficking opens a novel avenue to crop protection (Cai et al., 2018). The exchange of small RNAs (sRNAs) between hosts and downy mildew pathogen Hyaloperonospora arabidopsidis and the disease control have been reported (Bilir et al., 2019; Dunkerl et al., 2020). SIGS (Spray-induced gene silencing) for P. infestans control was also attempted, though low dsRNA uptake efficiency application of dsRNA fails to inhibit the virulence of this pathogen (Qiao et al., 2021). However, in a very recent research Sundaresha et al. developed multigene targeted dsRNA molecules, along with nanoclay carriers, to effectively reducing late blight infection (Sundaresha et al., 2022). These researches pave the way for designing artificial sRNA according to sporulation-related genes and applying SIGS or HIGS (Host-induced gene silencing) to control diseases caused by Phytophthora and Peronophythora pathogens.
JS and GK contributed to the conception and design of the work. JS, PX, and GK acquired most of the information and wrote sections. All authors contributed to the article and approved the submitted version.
The authors were supported by the Natural Science Foundation of Guangdong Province, China (2022A1515010458, 2020A1515011335, and 2019A1515010977), National Natural Science Foundation of China (31701771), the earmarked fund for CARS-32.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ah-Fong, A., Boyd, A., Matson, M., and Judelson, H. S. (2021). A Cas12a-based gene editing system for Phytophthora infestans reveals monoallelic expression of an elicitor. Mol. Plant Pathol. 22, 737–752. doi: 10.1111/mpp.13051
Ah-Fong, A., and Judelson, H. S. (2003). Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus-like oomycete Phytophthora infestans. Mol. Microbiol. 50, 487–494. doi: 10.1046/j.1365-2958.2003.03735.x
Ah-Fong, A., Kim, K. S., and Judelson, H. S. (2017). RNA-seq of life stages of the oomycete Phytophthora infestans reveals dynamic changes in metabolic, signal transduction, and pathogenesis genes and a major role for calcium signaling in development. BMC Genom. 18:198. doi: 10.1186/s12864-017-3585-x
Aylor, D. E. (2003). Spread of plant disease on a continental scale: role of aerial dispersal of pathogens. Ecology 84, 1989–1997. doi: 10.1890/01-0619
Bakthavatsalam, D., Meijer, H. J., Noegel, A., and Govers, F. (2006). Novel phosphatidylinositol phosphate kinases with a G-protein coupled receptor signature are shared by Dictyostelium and Phytophthora. Trends Microbiol. 14, 378–382. doi: 10.1016/j.tim.2006.07.006
Bassani, I., and Larousse, M., Tran, QD., Attard, A., and Galiana, E. (2020a). Phytophthora zoospores: from perception of environmental signals to inoculum formation on the host-root surface. Comp. Struct. Biotechnol. J. 18: 3766–3773. doi: 10.1016/j.csbj.2020.10.045
Bassani, I., Rancurel, C., Pagnotta, S., Orange, F., Pons, N., Lebrigand, K., et al. (2020b). Transcriptomic and ultrastructural signatures of K+-induced aggregation in Phytophthora parasitica zoospores. Microorganisms 8:1012. doi: 10.3390/microorganisms8071012
Bilir, O., Telli, O., Norman, C., Budak, H., Hong, Y. G., and Toer, M. (2019). Small RNA inhibits infection by downy mildew pathogen Hyaloperonospora arabidopsidis. Mol. Plant Pathol. 20, 1523–1534. doi: 10.1111/mpp.12863
Blanco, F. A., and Judelson, H. S. (2005). A bZIP transcription factor from Phytophthora interacts with a protein kinase and is required for zoospore motility and plant infection. Mol. Microbiol. 56, 638–648. doi: 10.1111/j.1365-2958.2005.04575.x
Boutet, X., Vercauteren, A., Heungens, K., Laurent, F., and Chandelier, A. (2010). Oospores progenies from Phytophthora ramorum. Fungal Biol. 114, 369–378. doi: 10.1016/j.funbio.2010.02.009
Cai, Q., He, B. Y., Kogel, K. H., and Jin, H. L. (2018). Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 46, 58–64. doi: 10.1016/j.mib.2018.02.003
Cavalier-Smith, T., Chao, E. E., and Lewis, R. (2018). Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria. Protoplasma 255, 1517–1574. doi: 10.1007/s00709-018-1241-1
Chen, L. L., Zhang, X., Wang, W., Geng, X. J., Shi, Y., Na, R. S., et al. (2017). Network and role analysis of autophagy in Phytophthora sojae. Sci. Rep. 7:1879. doi: 10.1038/s41598-017-01988-7
Cheng, W., Lin, M. L., Qiu, M., Kong, L., Xu, Y. P., Li, Y. N., et al. (2019). Chitin synthase is involved in vegetative growth, asexual reproduction and pathogenesis of Phytophthora capsici and Phytophthora sojae. Environ. Microbiol. 21, 4537–4547. doi: 10.1111/1462-2920.14744
Cvitanich, C., and Judelson, H. S. (2003). A gene expressed during sexual and asexual sporulation in Phytophthora infestans is a member of the Puf family of translational regulators. Eukaryot. Cell 2, 465–473. doi: 10.1128/EC.2.3.465-473.2003
Dunkerl, F., Trutzenberg, A., Rothenpieler, J. S., Kuhn, S., Prols, R., Schreiber, T., et al. (2020). Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. elife 9:e56096. doi: 10.7554/eLife.56096
Ebbole, D. J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45, 437–456. doi: 10.1146/annurev.phyto.45.062806.094346
Etxebeste, O., and Espeso, E. A. (2020). Aspergillus nidulans in the post-genomic era: a top-model filamentous fungus for the study of signaling and homeostasis mechanisms. Int. Microbiol. 23, 5–22. doi: 10.1007/s10123-019-00064-6
Fabritius, A. L., Cvitanich, C., and Judelson, H. S. (2002). Stage-specific gene expression during sexual development in Phytophthora infestans. Mol. Microbiol. 45, 1057–1066. doi: 10.1046/j.1365-2958.2002.03073.x
Fang, Y. F., and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17, 127–139. doi: 10.1111/mpp.12318
Feng, Y. C., He, D., Yao, Z. Y., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. doi: 10.1038/cr.2013.168
Feng, H., Wan, C. X., Zhang, Z. C., Chen, H., Li, Z. P., Jiang, H. B., et al. (2021). Specific interaction of an RNA-binding protein with the 3'-UTR of its target mRNA is critical to oomycete sexual reproduction. PLoS Pathog. 17:e1010001. doi: 10.1371/journal.ppat.1010001
Gao, J., Cao, M. N., Ye, W. W., Li, H. Y., Kong, L., Zheng, X. B., et al. (2015). PsMPK7, a stress-associated mitogen-activated protein kinase (MAPK) in Phytophthora sojae, is required for stress tolerance, reactive oxygenated species detoxifification, cyst germination, sexual reproduction and infection of soybean. Mol. Plant Pathol. 16, 61–70. doi: 10.1111/mpp.12163
Gonzalez-Rubio, G., Fernandez-Acero, T., Martin, H., and Molina, M. (2019). Mitogen-activated protein kinase phosphatases (MKPs) in fungal signaling: conservation, function, and regulation. Int. J. Mol. Sci. 20:1709. doi: 10.3390/ijms20071709
Grüwald, N. J., and Flier, W. G. (2005). The biology of Phytophthora infestans at its center of origin. Annu. Rev. Phytopathol. 43, 171–190. doi: 10.1146/annurev.phyto.43.040204.135906
Guo, T., Wang, X. W., Shan, K., Sun, W. X., and Guo, L. Y. (2017). The loricrin-like protein (LLP) of Phytophthora infestans is required for oospore formation and plant infection. Front. Plant Sci. 8:142. doi: 10.3389/fpls.2017.00142
Hardham, R. A., and Hyde, G. J. (1997). Asexual sporulation in the oomycetes. Adv. Bot. Res. 24, 353–398. doi: 10.1016/S0065-2296(08)60079-8
Hua, C. L., Meijer, H. J., de Keijzer, J., Zhao, W., Wang, Y. C., and Govers, F. (2013). GK4, a G-protein-coupled receptor with a phosphatidylinositol phosphate kinase domain in Phytophthora infestans, is involved in sporangia development and virulence. Mol. Microbiol. 88, 352–370. doi: 10.1111/mmi.12190
Hua, C. L., Wang, Y. L., Zheng, X. B., Dou, D. L., Zhang, Z. G., Govers, F., et al. (2008). A Phytophthora sojae G-protein α subunit is involved in chemotaxis to soybean isoflavones. Eukaryot. Cell 7, 2133–2140. doi: 10.1128/EC.00286-08
Hua, C. L., Yang, X. Y., and Wang, Y. C. (2015). Phytophthora sojae and soybean isoflavones, a model to study zoospore chemotaxis. Physiol. Mol. Plant Pathol. 92, 161–165. doi: 10.1016/j.pmpp.2015.05.003
Huang, J., Xi, P., Deng, Y., Huang, W. X., Wang, J. R., Zhao, Q. Q., et al. (2021). The mitogen-activated protein kinase PlMAPK2 is involved in zoosporogenesis and pathogenicity of Peronophythora litchii. Int. J. Mol. Sci. 22:3524. doi: 10.3390/ijms22073524
Hwu, F. Y., Lai, M. W., and Liou, R. F. (2017). PpMID1 plays a role in the asexual development and virulence of Phytophthora parasitica. Front. Microbiol. 8:610. doi: 10.3389/fmicb.2017.00610
Jiang, H., Hwang, H. W., Ge, T. L., Cole, B., Perkins, B., and Hao, J. J. (2019). Leucine regulates zoosporic germination and infection by Phytophthora erythroseptica. Front. Microbiol. 10:131. doi: 10.3389/fmicb.2019.00131
Jiang, L. Q., Situ, J. J., Deng, Y. Z., Wan, L., Xu, D. D., Chen, Y. B., et al. (2018). PIMAPK10, a mitogen-activated protein kinase (MAPK) in Peronophythora litchii, is required for mycelial growth, sporulation, laccase activity, and plant infection. Front. Microbiol. 9:426. doi: 10.3389/fmicb.2018.00426
Jiang, L. Q., Ye, W. W., Situ, J. J., Chen, Y. B., Yang, X. Y., Kong, G. H., et al. (2017). A Puf RNA-binding protein encoding gene PlM90 regulates the sexual and asexual life stages of the litchi downy blight pathogen Peronophythora litchii. Fungal Genet. Biol. 98, 39–45. doi: 10.1016/j.fgb.2016.12.002
Judelson, H. S. (1996). Genetic and physical variability at the mating type locus of the oomycete, Phytophthora infestans. Genetics 144, 1005–1013. doi: 10.1016/S1050-3862(96)00162-3
Judelson, H. S. (1997). Expression and inheritance of sexual preference and selfing potential in Phytophthora infestans. Fungal Genet. Biol. 21, 188–197. doi: 10.1006/fgbi.1997.0973
Judelson, H. S., and Blanco, F. A. (2005). The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3, 47–58. doi: 10.1038/nrmicro1064
Judelson, H. S., and Roberts, S. (2002). Novel protein kinase induced during sporangial cleavage in the oomycete Phytophthora infestans. Eukaryot. Cell 1, 687–695. doi: 10.1128/EC.1.5.687-695.2002
Judelson, H. S., and Tani, S. (2007). Transgene-induced silencing of the zoosporogenesis-specific NIFC gene cluster of Phytophthora infestans involves chromatin alterations. Eukaryot. Cell 6, 1200–1209. doi: 10.1128/EC.00311-06
Judelson, H. S., Tyler, B. M., and Michelmore, R. W. (1991). Transformation of the oomycete pathogen, Phytophthora infestans. Mol. Plant-Microbe Interact. 4, 602–607. doi: 10.1094/MPMI-4-602
Kamoun, S., Furzer, O., Jones, J. D., Judelson, H. S., Ali, G. H., Dalio, R. J., et al. (2015). The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. doi: 10.1111/mpp.12190
Kamoun, S., van West, P., Vleeshouwers, V. G., de Groot, K. E., and Govers, F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1425. doi: 10.1105/tpc.10.9.1413
Ko, W. H. (1988). Hormonal heterothallism and homothallism in Phytophthora. Annu. Rev. Phytopathol. 26, 57–73. doi: 10.1146/annurev.phyto.26.1.57
Kong, G. H., Chen, Y. B., Deng, Y. Z., Feng, D. N., Jiang, L. Q., Wan, L., et al. (2020). The basic leucine zipper transcription factor PlBZP32 associated with the oxidative stress response is critical for pathogenicity of the lychee downy blight oomycete Peronophythora litchii. mSphere 5, e00261–e00220. doi: 10.1128/mSphere.00261-20
Kong, P., and Hong, C. (2010a). Zoospore density-dependent behaviors of Phytophthora nicotianae are autoregulated by extracellular products. Phytopathology 100, 632–637. doi: 10.1094/PHYTO-100-7-0632
Kong, G. H., Li, T. L., Huang, W. X., Li, M. H., Shen, W. K., Jiang, L. Q., et al. (2021). Detection of Peronophythora litchii on lychee by loop-mediated isothermal amplification assay. Crop Prot. 139:105370. doi: 10.1016/j.cropro.2020.105370
Kong, P., Tyler, B. M., Richardson, P. A., Lee, B. W., Zhou, Z. S., and Hong, C. (2010b). Zoospore interspecific signaling promotes plant infection by Phytophthora. BMC Microbiol. 10:313. doi: 10.1186/1471-2180-10-313
Latijnhouwers, M., and Govers, F. (2003). A Phytophthora infestans G-protein β subunit is involved in sporangium formation. Eukaryot. Cell 2, 971–977. doi: 10.1128/EC.2.5.971-977.2003
Latijnhouwers, M., Ligterink, W., Vleeshouwers, V. G., van West, P., and Govers, F. (2004). A Gα subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans. Mol. Microbiol. 51, 925–936. doi: 10.1046/j.1365-2958.2003.03893.x
Leesutthiphonchai, W., and Judelson, H. S. (2018b). A MADS-box transcription factor regulates a central step in sporulation of the oomycete Phytophthora infestans. Mol. Microbiol. 110, 562–575. doi: 10.1111/mmi.14114
Leesutthiphonchai, W., Vu, A. L., Ah-Fong, A. M., and Judelson, H. S. (2018a). How does Phytophthora infestans evade control efforts? Modern insight into the late blight disease. Phytopathology 108, 916–924. doi: 10.1094/PHYTO-04-18-0130-IA
Lehtinen, A., and Hannukkala, A. (2004). Oospores of Phytophthora infestans in soil provide an important new source of primary inoculum in Finland. Agric. Food Sci. 13, 399–410. doi: 10.2137/1239099043633332
Li, J., Ai, C. C., Yang, C. C., Jiang, W. L., Sheng, H., Morris, P. F., et al. (2020b). Phytophthora capsici PcFtsZ2 is required for asexual development and plant infection. Mol. Plant-Microbe Interact. 33, 727–741. doi: 10.1094/MPMI-01-20-0002-R
Li, X., Liu, Y., Tan, X. Q., Li, D. L., Yang, X. Y., Zhang, X., et al. (2020a). The high-affinity phosphodiesterase PcPdeH is involved in the polarized growth and pathogenicity of Phytophthora capsici. Fungal Biol. 124, 164–173. doi: 10.1016/j.funbio.2020.01.006
Li, A., Wang, Y., Tao, K., Dong, S. M., Huang, Q., Dai, T., et al. (2010). PsSAK1, a stress-activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol. Plant-Microbe Interact. 23, 1022–1031. doi: 10.1094/MPMI-23-8-1022
Li, A. N., Zhang, M., Wang, Y. L., Li, D. L., Liu, X. L., Tao, K., et al. (2014). PsMPK1, an SLT2-type mitogen-activated protein kinase, is required for hyphal growth, zoosporogenesis, cell wall integrity, and pathogenicity in Phytophthora sojae. Fungal Genet. Biol. 65, 14–24. doi: 10.1016/j.fgb.2014.01.003
Lin, L., Ye, W. W., Wu, J. W., Xuan, M. R., Li, Y. F., Gao, J., et al. (2018). The MADS-box transcription factor PsMAD1 is involved in zoosporogenesis and pathogenesis of Phytophthora sojae. Front. Microbiol. 9:2259. doi: 10.3389/fmicb.2018.02259
McGowan, J., and Fitzpatrick, D. (2020). Recent advances in oomycete genomics. Adv. Genet. 105, 175–228. doi: 10.1016/bs.adgen.2020.03.001
Nagashima, Y., von Schaewen, A., and Koiwa, H. (2018). Function of N-glycosylation in plants. Plant Sci. 274, 70–79. doi: 10.1016/j.plantsci.2018.05.007
Narayan, R. D., Blackman, L. M., Shan, W. X., and Hardham, A. R. (2010). Phytophthora nicotianae transformants lacking dynein light chain 1 produce non-flagellate zoospores. Fungal Genet. Biol. 47, 663–671. doi: 10.1016/j.fgb.2010.04.008
Neiman, A. M. (2011). Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189, 737–765. doi: 10.1534/genetics.111.127126
Pao, J. A., and Ko, W. H. (1980). Oospore germination of Peronophythora litchii. Mycologia 72, 611–614. doi: 10.1080/00275514.1980.12021223
Qi, J. H., Asano, T., Jinno, M., Matsui, K., Atsumi, K., Sakagami, Y., et al. (2005). Characterization of a Phytophthora mating hormone. Science 309:1828. doi: 10.1126/science.1114756
Qiao, L., Lan, C., Capriotti, L., Ah-Fong, A., Sanchez, J. N., Hamby, R., et al. (2021). Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 19, 1756–1768. doi: 10.1111/pbi.13589
Qiu, M., Li, Y. N., Zhang, X., Xuan, M. R., Zhang, B. Y., Ye, W. W., et al. (2020). G protein α subunit suppresses sporangium formation through a serine/threonine protein kinase in Phytophthora sojae. PLoS Pathog. 16:e1008138. doi: 10.1371/journal.ppat.1008138
Rosenbaum, D. M., Rasmussen, S. G., and Kobilka, B. K. (2009). The structure and function of G-protein-coupled receptors. Nature 459, 356–363. doi: 10.1038/nature08144
Safdar, A., Li, Q., Shen, D. Y., Chen, L. L., He, F., Wang, R. B., et al. (2017). An LRR receptor kinase regulates growth, development and pathogenesis in Phytophthora capsici. Microbiol. Res. 198, 8–15. doi: 10.1016/j.micres.2017.01.008
Sheng, Y. T., Wang, Y. L., Meijer, H. J., Yang, X. Y., Hua, C. L., Ye, W. W., et al. (2015). The heat shock transcription factor PsHSF1 of Phytophthora sojae is required for oxidative stress tolerance and detoxifying the plant oxidative burst. Environ. Microbiol. 17, 1351–1364. doi: 10.1111/1462-2920.12609
Si, J. R., Pei, R., Ji, P. Y., Zhang, X., Xu, R. F., Qiao, H. J., et al. (2021a). PsGRASP, a golgi reassembly stacking protein in Phytophthora sojae, is required for mycelial growth, stress responses, and plant infection. Front. Microbiol. 12:702632. doi: 10.3389/fmicb.2021.702632
Si, J. R., Pei, Y., Shen, D. Y., Ji, P. Y., Xu, R. F., Xue, X., et al. (2021b). Phytophthora sojae leucine-rich repeat receptor-like kinases: diverse and essential roles in development and pathogenicity. iScience 24:102725. doi: 10.1016/j.isci.2021.102725
Situ, J. J., Jiang, L. Q., Fan, X. N., Yang, W. S., Li, W., Xi, P. G., et al. (2020). An RXLR effector PlAvh142 from Peronophythora litchii triggers plant cell death and contributes to virulence. Mol. Plant Pathol. 21, 415–428. doi: 10.1111/mpp.12905
Sundaresha, S., Sharma, S., Bairwa, A., Tomar, M., Kumar, R., Bhardwaj, V., et al. (2022). Spraying of ds RNA molecules derived from Phytophthora infestans, along with nanoclay carriers as a proof of concept for developing novel protection strategy for potato late blight. Pest Manag. Sci. 78, 3183–3192. doi: 10.1002/ps.6949
Tani, S., Yatzkan, E., and Judelson, H. S. (2004). Multiple pathways regulate the induction of genes during zoosporogenesis in Phytophthora infestans. Mol. Plant-Microbe Interact. 17, 330–337. doi: 10.1094/MPMI.2004.17.3.330
Tooley, P. W., Browning, M., and Vinyard, B. (2020). Diurnal effects on sporangium and zoospore production by Phytophthora ramorum on rhododendron 'Cunningham's white. Mycologia 112, 519–532. doi: 10.1080/00275514.2020.1728472
Tyler, B. M. (2002). Molecular basis of recognition between Phytophthora pathogens and their hosts. Annu. Rev. Phytopathol. 40, 137–167. doi: 10.1146/annurev.phyto.40.120601.125310
Tyler, B. M., Tripathy, S., Zhang, X., Dehal, P., Jiang, R. H., Aerts, A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266. doi: 10.1126/science.1128796
van den Hoogen, J., Kruif, N. V., and Govers, F. (2018). The G-protein gamma subunit of Phytophthora infestans is involved in sporangial development. Fungal Genet. Biol. 116, 73–82. doi: 10.1016/j.fgb.2018.04.012
van West, P., Kamoun, S., van't Klooster, J. W., and Govers, F. (1999a). Internuclear gene silencing in Phytophthora infestans. Mol. Cell 3, 339–348. doi: 10.1016/S1097-2765(00)80461-X
van West, P., Morris, B. M., Reid, B., Appiah, A. A., Osborne, M. C., Campbell, T. A., et al. (2002). Oomycete plant pathogens use electric fields to target roots. Mol. Plant-Microbe Interact. 15, 790–798. doi: 10.1094/MPMI.2002.15.8.790
van West, P., Reid, B., Campbell, T. A., Sandrock, R. W., Fry, W. E., Kamoun, S., et al. (1999b). Green fluorescent protein (GFP) as a reporter gene for the plant pathogenic oomycete Phytophthora palmivora. FEMS Microbiol. Lett. 178, 71–80. doi: 10.1016/S0378-1097(99)00320-1
Walker, C. A., Köppe, M., Grenville-Briggs, L. J., Avrova, A. O., Horner, N. R., McKinnon, A. D., et al. (2008). A putative DEAD-box RNA-helicase is required for normal zoospore development in the late blight pathogen Phytophthora infestans. Fungal Genet. Biol. 45, 954–962. doi: 10.1016/j.fgb.2008.03.004
Walker, C. A., and van West, P. (2007). Zoospore development in the oomycetes. Fungal Biol. Rev. 21, 10–18. doi: 10.1016/j.fbr.2007.02.001
Wang, Y. L., Dou, D. L., Wang, X. L., Li, A. N., Sheng, Y. T., Hua, C. L., et al. (2009). The PsCZF1 gene encoding a C2H2 zinc finger protein is required for growth, development and pathogenesis in Phytophthora sojae. Microb. Pathog. 47, 78–86. doi: 10.1016/j.micpath.2009.04.013
Wang, Y. L., Li, A. N., Wang, X. L., Zhang, X., Zhao, W., Dou, D. L., et al. (2010). GPR11, a putative seven-transmembrane G protein-coupled receptor, controls zoospore development and virulence of Phytophthora sojae. Eukaryot. Cell 9, 242–250. doi: 10.1128/EC.00265-09
Wang, T. H., Wang, X. W., Zhu, X. Q., He, Q., and Guo, L. Y. (2020). A proper PiCAT2 level is critical for sporulation, sporangium function, and pathogenicity of Phytophthora infestans. Mol. Plant Pathol. 21, 460–474. doi: 10.1111/mpp.12907
Wang, J. R., Zhou, G. Q., Huang, W. X., Li, W., Feng, D. N., Liu, L. C., et al. (2022). Autophagy-related gene PlATG6a is involved in mycelial growth, asexual reproduction and tolerance to salt and oxidative stresses in Peronophythora litchii. Int. J. Mol. Sci. 23:1839. doi: 10.3390/ijms23031839
Whisson, S. C., Avrova, A. O., Van West, P., and Jones, J. T. (2005). A method for double-stranded RNA-mediated transient gene silencing in Phytophthora infestans. Mol. Plant Pathol. 6, 153–163. doi: 10.1111/J.1364-3703.2005.00272.X
Xiang, Q., and Judelson, H. S. (2010). Myb transcription factors in the oomycete Phytophthora with novel diversified DNA-binding domains and developmental stage-specific expression. Gene 453, 1–8. doi: 10.1016/j.gene.2009.12.006
Xiang, Q., and Judelson, H. S. (2014). Myb transcription factors and light regulate sporulation in the oomycete Phytophthora infestans. PLoS One 9:e92086. doi: 10.1371/journal.pone.0092086
Xue, C., Hsueh, Y. P., and Heitman, J. (2008). Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol. Rev. 32, 1010–1032. doi: 10.1111/j.1574-6976.2008.00131.x
Yang, X. Y., Ding, F., Zhang, L., Sheng, Y. T., Zheng, X. B., and Wang, Y. C. (2015). The importin α subunit PsIMPA1 mediates the oxidative stress response and is required for the pathogenicity of Phytophthora sojae. Fungal Genet. Biol. 82, 108–115. doi: 10.1016/j.fgb.2015.04.023
Yang, X., Zhao, W., Hua, C., Zheng, X., Jing, M., Li, D., et al. (2013). Chemotaxis and oospore formation in Phytophthora sojae are controlled by G-protein-coupled receptors with a phosphatidylinositol phosphate kinase domain. Mol. Microbiol. 88, 382–394. doi: 10.1111/mmi.12191
Ye, W. W., Wang, Y., Shen, D. Y., Li, D. L., Pu, T. H., Jiang, Z. D., et al. (2016). Sequencing of the litchi downy blight pathogen reveals it is a Phytophthora species with downy mildew-like characteristics. Mol. Plant-Microbe Interact. 29, 573–583. doi: 10.1094/MPMI-03-16-0056-R
Zhang, C., Cai, M., Chen, S. S., Zhang, F., Cui, T. S., Xue, Z. L., et al. (2021). The consensus Nglyco-X-S/T motif and a previously unknown Nglyco-N-linked glycosylation are necessary for growth and pathogenicity of Phytophthora. Environ. Microbiol. 23, 5147–5163. doi: 10.1111/1462-2920.15468
Zhang, M., Lu, J., Tao, K., Ye, W. W., Li, A. N., Liu, X. Y., et al. (2012a). A Myb transcription factor of Phytophthora sojae, regulated by MAP kinase PsSAK1, is required for zoospore development. PLoS One 7:e40246. doi: 10.1371/journal.pone.0040246
Zhang, M. X., Meng, Y. L., Wang, Q. H., Liu, D. D., Quan, J. L., Hardham, A. R., et al. (2012b). PnPMA1, an atypical plasma membrane HD-ATPase, is required for zoospore development in Phytophthora parasitica. Fungal Biol. 116, 1013–1023. doi: 10.1016/j.funbio.2012.07.006
Zhang, X., Zhai, C. H., Hua, C. L., Qiu, M., Hao, Y. J., Nie, P. P., et al. (2016). PsHint1, associated with the G-protein α subunit PsGPA1, is required for the chemotaxis and pathogenicity of Phytophthora sojae. Mol. Plant Pathol. 17, 272–285. doi: 10.1111/mpp.12279
Zhao, W., Dong, S. M., Ye, W. W., Hua, C. L., Meijer, H. J., Dou, X. Y., et al. (2011b). Genome-wide identification of Phytophthora sojae SNARE genes and functional characterization of the conserved SNARE PsYKT6. Fungal Genet. Biol. 48, 241–251. doi: 10.1016/j.fgb.2010.11.006
Zhao, W., Yang, X. Y., Dong, S. M., Sheng, Y. T., Wang, Y. C., and Zheng, X. B. (2011a). Transient silencing mediated by in vitro synthesized double-stranded RNA indicates that PsCdc14 is required for sporangial development in a soybean root rot pathogen. Sci. China-Life Sci. 54, 1143–1150. doi: 10.1007/s11427-011-4250-2
Keywords: oomycete, zoospore, oospore, zoosporogenesis, sporulation
Citation: Situ J, Xi P, Lin L, Huang W, Song Y, Jiang Z and Kong G (2022) Signal and regulatory mechanisms involved in spore development of Phytophthora and Peronophythora. Front. Microbiol. 13:984672. doi: 10.3389/fmicb.2022.984672
Received: 04 July 2022; Accepted: 23 August 2022;
Published: 09 September 2022.
Edited by:
Mohammad Arif, University of Hawaii at Manoa, United StatesReviewed by:
David Vela-Corcia, University of Malaga, SpainCopyright © 2022 Situ, Xi, Lin, Huang, Song, Jiang and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guanghui Kong, Z2tvbmdAc2NhdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.