- 1Key Laboratory of Crop Eco-physiology and Farming System in Southwest China, Ministry of Agriculture, College of Agronomy, Sichuan Agricultural University, Chengdu, Sichuan, China
- 2Root Biology Center, College of Resources and Environment, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Department of Microbiology College of Resources, Sichuan Agricultural University, Chengdu, Sichuan, China
Straw mulching and N fertilization are effective in augmenting crop yields. Since their combined effects on wheat rhizosphere bacterial communities remain largely unknown, our aim was to assess how the bacterial communities respond to these agricultural measures. We studied wheat rhizosphere microbiomes in a split-plot design experiment with maize straw mulching (0 and 8,000 kg straw ha−1) as the main-plot treatment and N fertilization (0, 120 and 180 kg N ha−1) as the sub-plot treatment. Bacterial communities in the rhizosphere were analyzed using 16S rRNA gene amplicon sequencing and quantitative PCR. Most of the differences in soil physicochemical properties and rhizosphere bacterial communities were detected between the straw mulching (SM) and no straw mulching (NSM) treatments. The contents of soil organic C (SOC), total N (TN), NH4+-N, available N (AN), available P (AP) and available K (AK) were higher with than without mulching. Straw mulching led to greater abundance, diversity and richness of the rhizosphere bacterial communities. The differences in bacterial community composition were related to differences in soil temperature and SOC, AP and AK contents. Straw mulching altered the soil physiochemical properties, leading to greater bacterial diversity and richness of the rhizosphere bacterial communities, likely mostly due to the increase in SOC content that provided an effective C source for the bacteria. The relative abundance of Proteobacteria was high in all treatments and most of the differentially abundant OTUs were proteobacterial. Multiple OTUs assigned to Acidobacteria, Chloroflexi and Actinobacteria were enriched in the SM treatment. Putative plant growth promoters were enriched both in the SM and NSM treatments. These findings indicate potential strategies for the agricultural management of soil microbiomes.
Introduction
Wheat (Triticum aestivum L.) is a vital part of global food security; approximately 40% of the world’s population get part of their daily nutrients from wheat (Giraldo et al., 2019). Like other plants, wheat hosts a diverse microbial community on its rhizosphere; the rhizosphere microbial communities are closely involved in the cycling of nutrients, plant growth promotion, and resource utilization efficiency of plants (Mendes et al., 2013; Pérez-Montaño et al., 2014). The rhizosphere microorganisms are sensitive to changes in environmental conditions, e.g., to changes brought on by agricultural management (Mendes et al., 2013; Bay et al., 2021). Therefore, it is important to understand how agricultural management measures affect the wheat rhizosphere microbial community and the relationship between the wheat rhizosphere microbial community composition and wheat yield.
Straw mulching is a common agricultural practice that can alleviate soil degradation caused by long-term synthetic fertilizer application and reduce the environmental footprint of food production (Sun et al., 2015; Chai et al., 2021). Straw mulching improved the soil structure and increased organic C content (Blanco-Canqui and Lal, 2007), soil enzyme activity (Zhang et al., 2016; Akhtar et al., 2019a), soil water content (Akhtar et al., 2019b), and other physicochemical properties in topsoil (Dai et al., 2019). However, results on the responses of soil bacteria to straw mulching in the topsoil have been inconsistent (Chen et al., 2017; Liu C. et al., 2020; Qiu et al., 2020; Wang H. H. et al., 2020; Yan et al., 2020), possibly due to different planting systems and regional differences. For example, mulching affected β-diversity but not α-diversity in maize and rice fields in northeast China (Wang H. H. et al., 2020; Yan et al., 2020). In rice-maize rotation systems on three sites in central to eastern China, mulching increased the microbial biomass on every site, but the abundances of gram-negative bacteria, gram-positive bacteria and actinomycetes were higher under mulching on one site only (Chen et al., 2017). In a spring wheat–pea rotation in northwest China, without tillage the α-diversity of bacteria was slightly higher in the no straw mulching treatment, whereas with tillage, the diversity was higher in the mulched treatment (Liu C. et al., 2020). Thus, even though straw mulching has been shown to improve soil quality and crop yield (Dong et al., 2018), its varying effects on the rhizosphere communities call for studies at many crop-region combinations.
N fertilization, one of the important limiting nutrients for plants and microorganisms, is supplied worldwide into terrestrial ecosystems (Liu W. B. et al., 2020). In long-term fertilization experiments, the abundance and diversity of bacteria decreased with increasing N fertilizer levels (Zhou et al., 2015). According to a meta-analysis, N addition decreased microbial diversity and the relative abundances of Actinobacteria and Nitrospirae (Wang et al., 2018). N enrichment affected the composition and functions of soil microbial communities and reduced microbial diversity (Zhou et al., 2015; Ling et al., 2017); the changes were mainly associated with soil pH and soil organic C and total N contents (Rousk et al., 2010; Kong et al., 2019). While the effect of N fertilization on the nutrient contents and soil bacterial communities has been demonstrated, the responses of rhizosphere bacteria to combined straw mulching and N fertilization has been rarely studied.
We studied soil physicochemical properties and wheat rhizosphere microbiomes using a split-plot design experiment with straw mulching as the main plot treatment and N fertilization as the sub-plot treatment. Bacterial communities in the rhizosphere were analyzed using 16S rRNA gene amplicon sequencing and quantitative PCR. We hypothesized that the N fertilization related abundance and diversity decrease would be alleviated by straw mulching and expected that higher shoot biomass and yield of wheat would be associated with higher abundance and α-diversity of bacteria.
Materials and methods
Experimental design and sampling
The study was conducted at the Renshou experimental base (30°04′ N, 104°13′ E) at the College of Agronomy, Sichuan Agricultural University, China. The region belongs to the subtropical monsoon climatic zone in which the winter wheat-summer maize rotation system is one of the major agricultural production systems. The soil is classified as Regosol in the FAO Soil Taxonomy, and the soil texture is loamy clay. At the start of the experiment in 2015, the physical and chemical properties of the topsoil (0–20 cm) were as follows: pH, 7.82; soil organic C content, 9.78 g kg−1; total N content, 0.83 g kg−1; total phosphorus content, 0.86 g kg−1; and total potassium content, 13.96 g kg−1.
The experimental design was the same as in our previous study (Chen S. H. et al., 2021). Briefly, the split-plot experimental design consisted of maize straw mulching treatments in the main plot and N fertilization levels in the subplot, with four 30 m2 (6 m × 5 m) replicate plots and a total of 24 experimental plots. The experiment was started in 2015. The straw mulching treatments were no straw mulching (NSM) and straw mulching with 8,000 kg straw ha−1 (SM). In the SM treatment, the maize residue was chopped and spread on the soil surface after the maize harvest at the end of August each year. The N fertilization levels were 0 (N0), 120 (N1) and 180 kg N ha−1 (N2). All the plots were fertilized with P at 75 kg P2O5 ha−1 and K at 75 kg K2O ha−1. Winter wheat (Triticum aestivum L. cv. Chuanmai104) was sown with no-tillage in late October at a basic seedling rate of 2.25 × 106 plant ha−1 with 20 cm row spacing and 10 cm hole spacing. Wheat was harvested the following year in May. The field management was based on the local farmers’ management model with no tillage, natural rainfall irrigation, and herbicide treatment once before sowing the wheat.
Rhizosphere soils were collected on 20 March, 2019, at the anthesis stage of winter wheat. Whole plants were excavated using a clean spade and gently shaken to remove the soil not tightly attached to the root (Mapelli et al., 2018; Wang Z. T. et al., 2020). The root systems were separated from the plant and collected in sterile plastic bags. The rhizosphere soil particles were first detached from the roots using the pull and shake method. The tightly attached soil was detached by placing the roots in sterile tubes containing 9 ml of 9 g L−1 NaCl, vortexing the tubes for 5 min and centrifuging at 8000 rpm for 10 min, after which the supernatant was discarded and the remaining soil was merged with the rhizosphere soil particles (Mapelli et al., 2018). The rhizosphere soil samples were stored at −80°C for molecular analysis.
Soil physico-chemical analysis
Air-dried soil was used for physical–chemical analysis. Soil pH was determined in a 1:2.5 soil/water mixture. Soil organic C (SOC) was determined using potassium dichromate oxidation external heating method. Soil total N (TN) and available N (AN) were determined using the alkaline hydrolysis diffusion method and semi-micro Kjeldahl method, respectively (Lu, 1999). Soil available phosphorus (AP) was extracted using sodium bicarbonate and measured using the molybdenum blue method (Lu, 1999). Soil available potassium (AK) was extracted using ammonium acetate and determined with flame photometry (P7 Double Beam UV–Visible Spectrophotometer; MAPADA Inc. Shanghai, China; Lu, 1999). Soil extractable ammonium (NH4+) and nitrate (NO3−) were extracted with 2 mol L−1 KCl and determined colorimetrically (Liu C. et al., 2020). To estimate the average temperature of soil, the temperature of soil at 10 cm depth was measured from 8 a.m. to 20 p.m. every 2 h with a moisture and water potential meter (em50g, Decagon Devices Inc. Pullman, USA) when collecting soil samples.
DNA extraction and sequencing
DNA was extracted from 0.5 g fresh soil using a DNeasy Powersoil kit (Qiagen, Manchester, UK) following the manufacturer’s instructions. The concentration and purity of the extracted DNA was assessed using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis. The DNA samples were stored at −20°C until further processing.
The copy number of the 16S rRNA gene was determined using real-time quantitative PCR (qPCR) on a ABI7500 Fast Real-Time PCR System (Applied Biosystems Inc., USA). 16S rRNA gene fragments were amplified in triplicate using primers 319f (5′-ACTCCTACGGGAGGCAGCAG-3′) and primer 806r (5′-GGACTACHVGGGTWTCTAAT-3′; Walters et al., 2011). Amplification was done in a 20 μl reaction including 10 μl SYBR Green Master Mix (Applied Biosystems, USA), 0.25 μl (10 μM) of each primer, 1 μl (1-10 ng) DNA template or 1 μl sterilized distilled water in the negative control, and 8.5 μl double distilled water (ddH2O). Amplification was initiated by denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30s, extension at 72°C for 30 s, and reading the plate at 80°C.
The standards for qPCR were made by cloning a bacterial gene fragment amplified as described above using pmD®18-T Vector (TaKaRa, Dalian, China) according to manufacturer’s instructions Plasmid DNA was extracted using a Plasmid Miniprep Kit (BIOMIGA, Santiago, USA), and the plasmid concentration was measured with a spectrophotometer (Nanodrop 2000, Thermo Scientific, Wilmington, USA). As the sequences of the vector and PCR inserts were known, bacterial gene copy number was calculated directly from the concentration of extracted plasmid DNA. Standard curves were generated using triplicate dilution series of plasmid with a cloned target gene, from 103 to 109 copies of the template. PCR efficiency was 91.3% with R2 value of 0.999 and slope of −3.55. No amplification was detected in the negative controls.
The V3-V4 region of bacterial 16S rRNA gene was amplified from the soil DNA extracts using the primers 319F and 806R with 7-bp barcodes specific for the samples for multiplex sequencing. The amplification reaction mixture contained 5 μl of Q5 reaction buffer (5×), 5 μl of Q5 High-Fidelity GC buffer (5×), 0.25 μl of Q5 High-Fidelity DNA Polymerase (5 U μl−1), 2 μl of dNTP (2.5 mM), 1 μl of each forward and reverse primer (10 μM), 2 μl of DNA template, and 8.75 μl of ddH2O. The amplification conditions were initial denaturation for 2 min at 98°C, 25 cycles of denaturation at 98°C for 15 s, annealing at 55°C for 30 s and extension at 72°C for 30 s, and a final extension for 5 min at 72°C. PCR products were purified using Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN, USA), quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA), and pooled in equimolar concentrations. Amplicons were sequenced using paired-end 2 × 300 bp sequencing on Illumina MiSeq at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The sequences have been deposited in the NCBI Sequence Read Archive database under the accession number SRP297976.
Bioinformatics
Sequences were processed using QIIME2 2019.41 with modifications as described in the official tutorials. The unique sequences were clustered at 98% sequence identity and, after removal of chimeras, the 826,884 high-quality sequences from 24 samples were reclustered into operational taxonomic units (OTUs) at 97% identity. After removing singletons, taxonomy was assigned to OTUs using the classify-sklearn naïve Bayesian taxonomy classifier in feature-classifier plugin (Wang et al., 2007) against the SILVA Release 132 Database (Quast et al., 2013).
Statistical analysis
Differences in soil properties were tested using two-way ANOVA followed by Fisher’s Least Significant Difference (LSD) test in SPSS 26 (IBM, Armonk, New York, USA). α-diversity indices were visualized using Origin2017 (Origin, OriginLab Inc. MA, USA) and tested using Kruskal Wallis test and Dunn’s test as a post hoc test. Taxonomic compositions were visualized using MEGAN (Huson et al., 2011) and GraPhlAn (Asnicar et al., 2015). β-diversity was visualized using principal coordinate analysis (PCoA) in the R package “vegan” in R v.4.1.0 (Oksanen et al., 2019; R Core Team, 2020). Differences in β-diversity were tested using repeated measures permutational multivariate analysis of variance (PERMANOVA) based on Bray–Curtis dissimilarity (Bray and Curtis, 1957; Anderson et al., 2008). Differential abundance analysis at OTU level was done using Linear Discriminant Analysis (LDA) Effect Size (LEfSe; Segata et al., 2011).2 OTUs with an LDA score ≥ 2.0 were considered differentially abundant. The relationships between soil properties and bacterial communities were tested based on Bray-Curtis dissimilarities, using distance-based redundancy analysis (RDA) with 9,999 permutations in the vegan R package in R v.4.1.0. The relationships between the soil microbial features and soil properties were tested using Mantel test in the ggcor package in R v.4.0.5.3 The correlations among α-and β-diversities and soil characteristics were calculated and visualized using the corrplot package in R.4
Results
Soil physicochemical analysis
The contents of soil organic C (SOC), total N (TN), NH4+-N, available N (AN), available P (AP) and available K (AK) were higher in the mulched treatment (SM) than in the non-mulched (NSM) treatment (p < 0.05; Table 1). The contents of TN, AN, NH4+-N, NO3−-N and AP were higher in the 120 (N1) and 180 kg N ha−1 (N2) treatments than in the 0 kg N ha−1 (N0) treatment (p < 0.05; Table 1). The SOC, TN, AN, NH4+-N, AP and AK contents were lowest in NSMN0 and highest in SMN2.
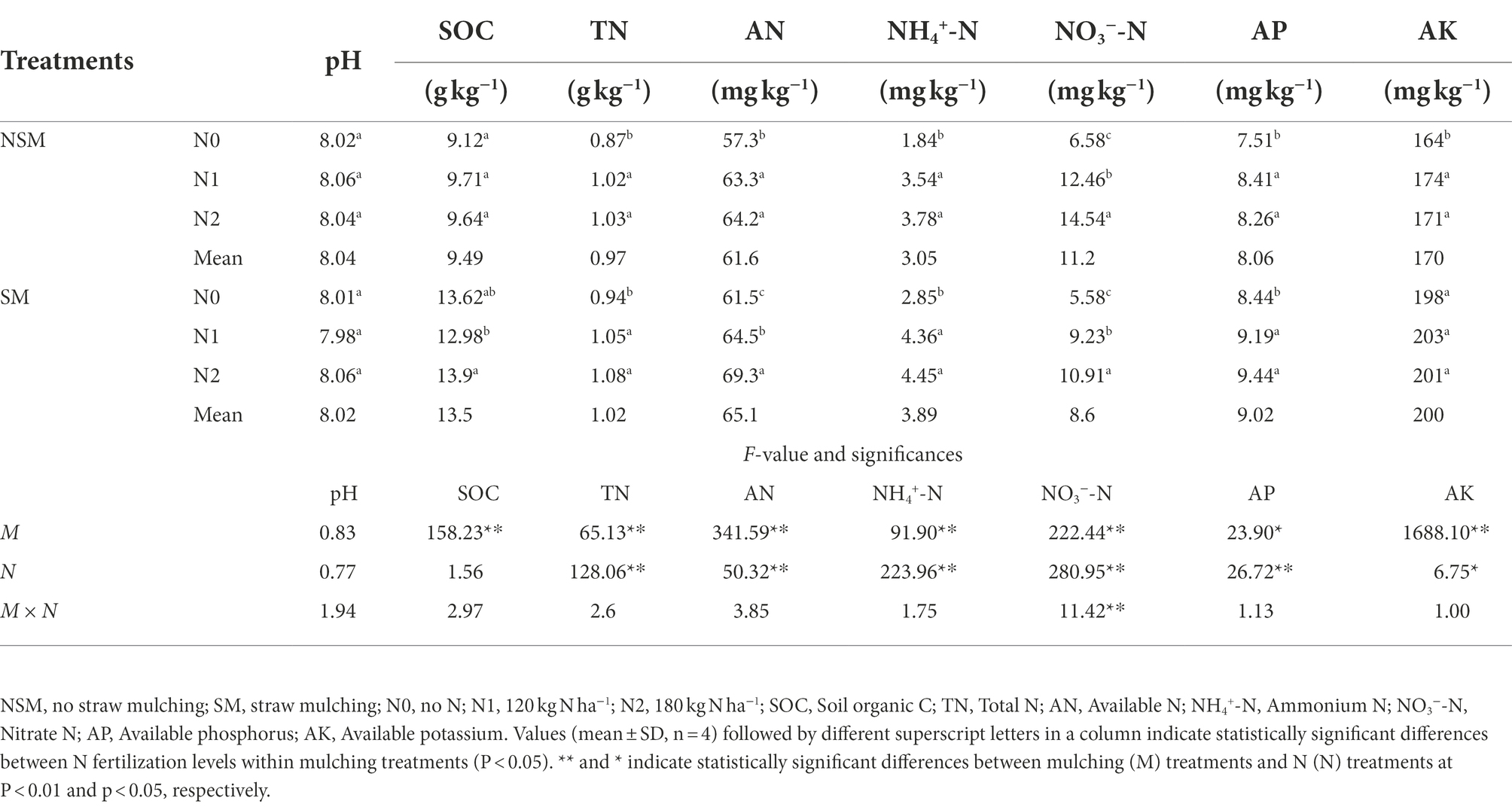
Table 1. Physicochemical properties of wheat rhizosphere soil in the straw mulching and N fertilization treatments.
Abundance and composition of bacterial community
The 16S rRNA gene copy numbers ranged from 1.4 × 108 to 7.4 × 108 per g soil, with the lowest and the highest numbers in the NSMN0 and SMN2 treatments, respectively (Figure 1). The abundance and diversity of the rhizosphere bacterial communities were greater with than without mulching (p < 0.05; Figures 1, 2; Supplementary Tables S1–S4). The N fertilization level did not affect diversity. In line with the higher diversity with mulching, the number of identified genera was greater with mulching (67) than without (20) (Supplementary Table S4). In the PCoA, the rhizosphere bacterial communities from mulched and not mulched treatments were clearly separated along the PCoA axis1 that explained 88.1% of the variation in community composition (Figure 3). The communities in SMN1 were separated from those in SMN0 and SMN2 along PCoA axis2 that explained 4.6% of the variation. In line with the PCoA, the communities in the SMN0, SMN1 and SMN2 treatments were different from those without mulching and different from each other (Supplementary Table S6).
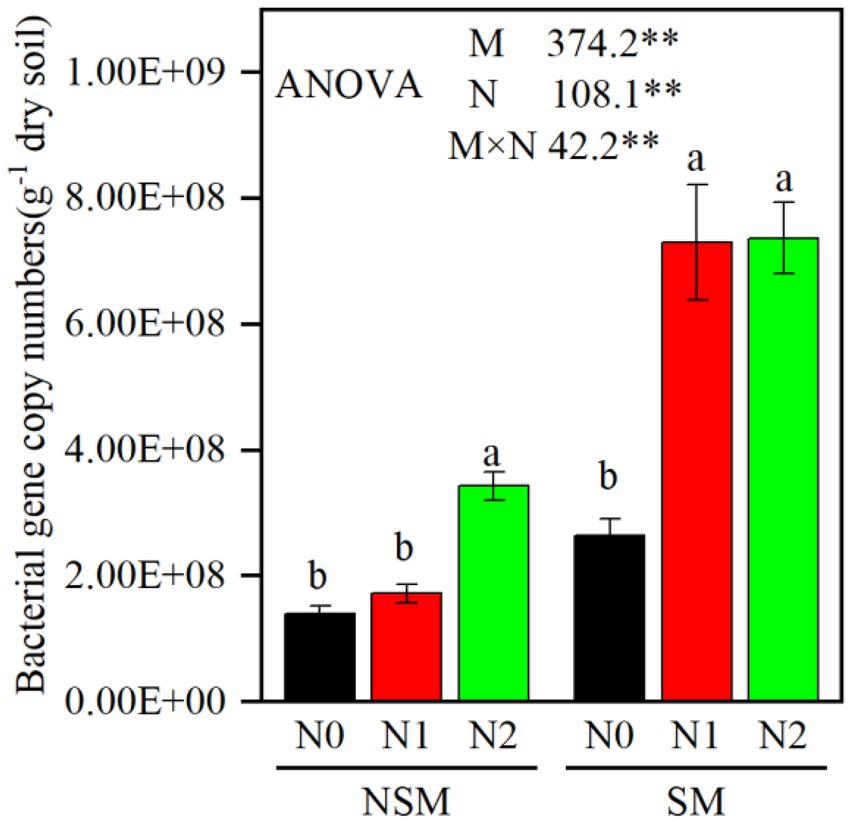
Figure 1. The abundance of bacteria gene copies in wheat rhizosphere soil under straw mulching and N fertilization treatments. M, mulching treatment; N, N fertilization treatment. M × N, the interaction of straw mulching and N fertilization NSM, no straw mulching; SM, straw mulching; N0, no N; N1, 120 kg N ha−1; N2, 180 kg N ha−1. Data shown as mean ± S.D. ** statistically significant difference (p < 0.01). Different letters above columns indicate statistically significant difference (p < 0.05) between N fertilizer levels within NSM and SM.
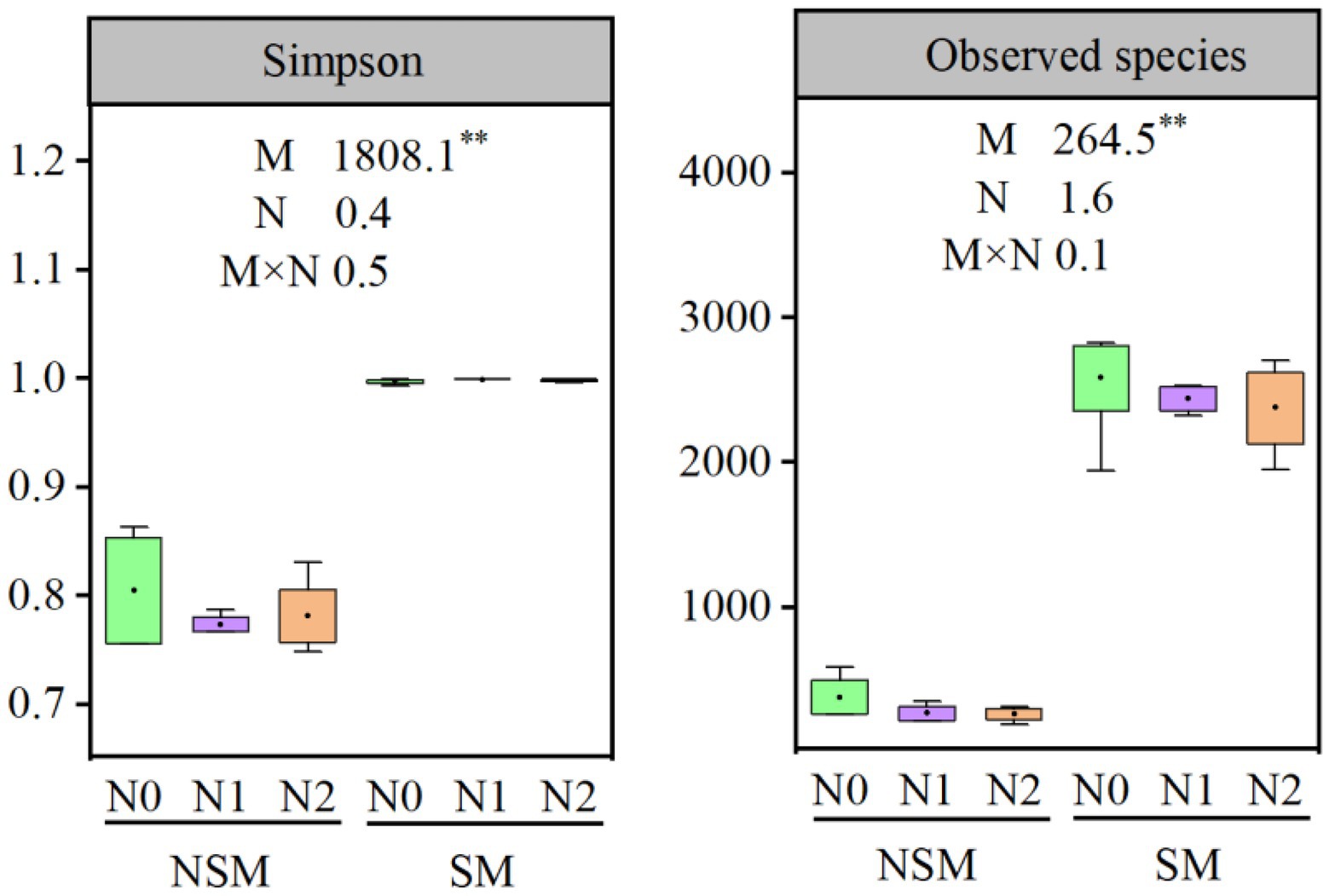
Figure 2. α-diversity of bacterial communities in wheat rhizosphere soil under straw mulching and N fertilization treatments. M, mulching treatment; N, N fertilization treatment. M × N, the interaction of straw mulching and N fertilization. NSM, no straw mulching; SM, straw mulching; N0, no N; N1, 120 kg N ha−1; N2, 180 kg N ha−1. **, statistically significant difference (p < 0.01).
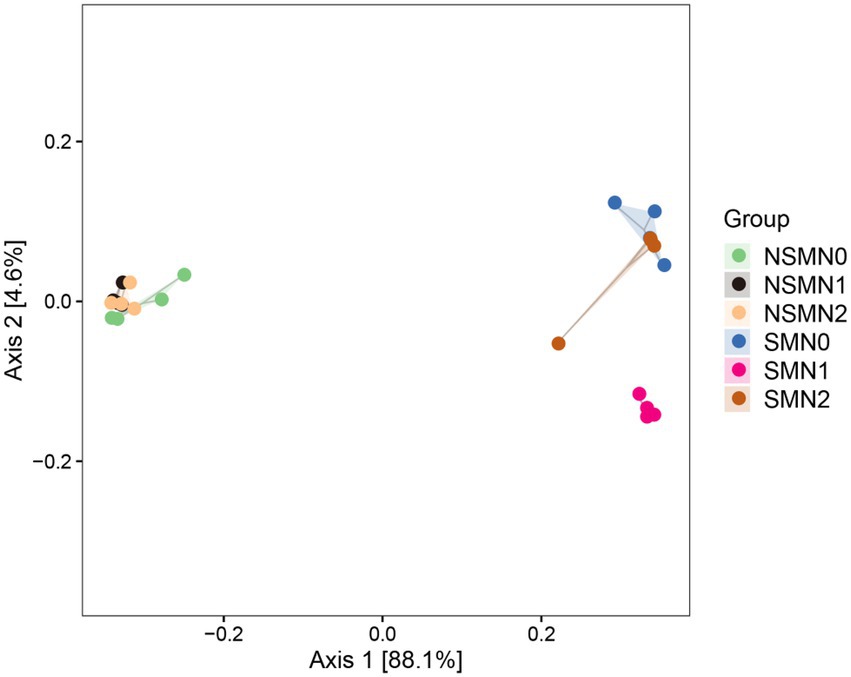
Figure 3. Principal coordinate analysis (PCoA) of microbial community composition in wheat rhizosphere soil under straw mulching and N fertilization treatments. NSM, no straw mulching; SM, straw mulching; N0, no N; N1, 120 kg N ha−1; N2, 180 kg N ha−1.
The relative abundances of phyla Proteobacteria and Bacteroidetes were high in all treatments and those of Acidobacteria, Actinobacteria and Chloroflexi in the mulched treatments (Figure 4A; Supplementary Table S3). The relative abundances of genera Pelomonas, Ralstonia, Ochrobactrum and Vibrionimonas were approximately 1% or less with mulching and ranged from over 40% to over 6% without mulching (Figure 4B; Supplementary Table S4).
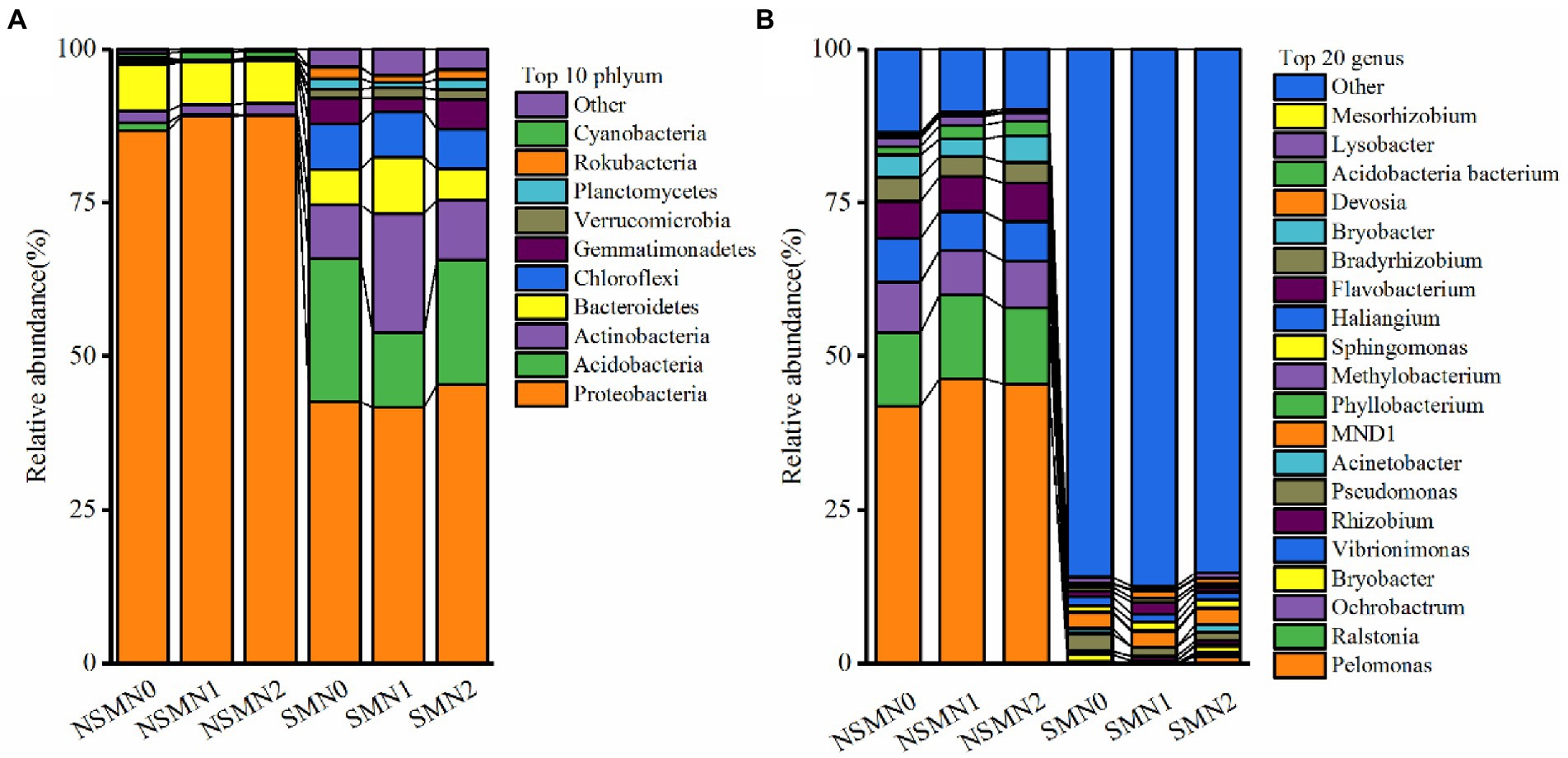
Figure 4. Bacterial community composition in wheat rhizosphere soil under straw mulching and N fertilization treatments. (A), phylum level; (B), genus level. NSM, no straw mulching; SM, straw mulching; N0, no N; N1, 120 kg N ha−1; N2, 180 kg N ha−1.
Characteristic taxa in the bacterial communities
The effect of straw mulching and N fertilization on individual taxa was assessed using LEfSe. In the differential abundance analysis, 154 and 35 OTUs had LDA scores >2.0 in the SM and NSM, respectively (Supplementary Table S5), and were considered as differentially abundant. Most of the differentially abundant OTUs in SM were assigned to Proteobacteria (51), Actinobacteria (22), Acidobacteria (16) and Chloroflexi (14), and in NSM to Proteobacteria (20) (Supplementary Table S5). The taxa characterizing the treatments with LDA score > 4 included class Subgroup 6 belonging to Acidobacteria and proteobacterial order MND1 in the SMN0 treatment, phylum Bacteroidetes and actinobacterial order Micromonosporales in the SMN1 treatment, class Gemmatimonadetes in the SMN2 treatment, genera Ochrobactrum (Proteobacteria) and Vibrionimonas (Bacteroidetes) in the NSMN0 treatment, proteobacterial genera Pelomonas and Ralstonia in the NSMN1 treatment and proteobacterial order Pseudomonadales and genera Acinetobacter, Phyllobacterium and Rhizobium in the NSMN2 treatment (Figure 5).
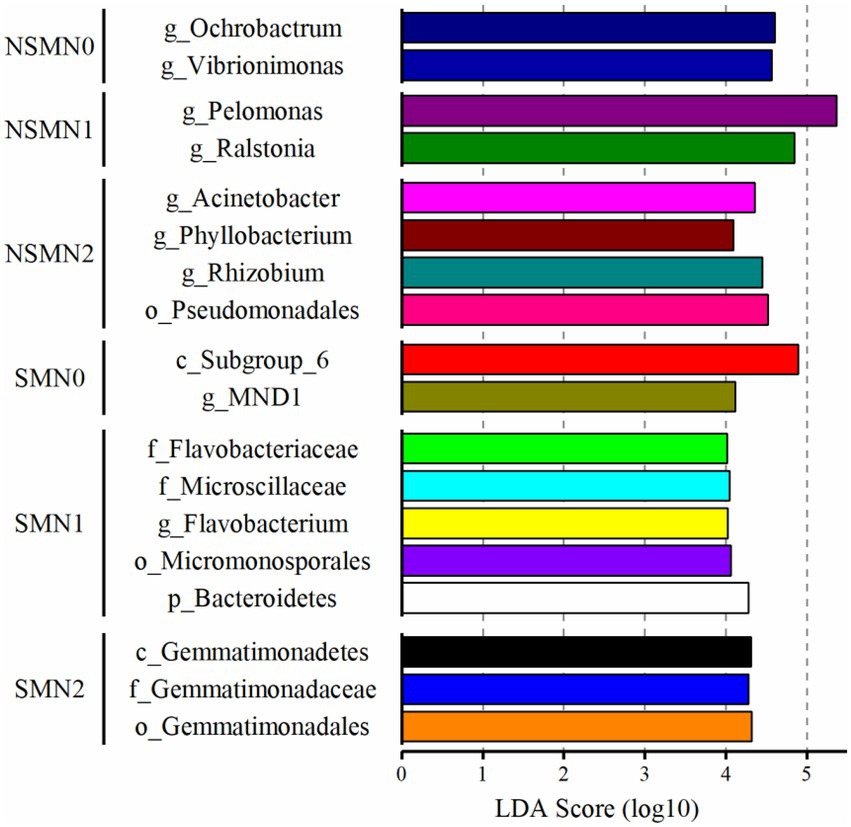
Figure 5. Differentially abundant taxa in wheat rhizosphere soil under straw mulching and N fertilization treatments. Detected using linear discriminant analysis effect size analysis. NSM, no straw mulching; SM, straw mulching; N0, no N; N1, 120 kg N ha−1; N2, 180 kg N ha−1.
Relationships among bacterial parameters, soil properties, and wheat yield and biomass
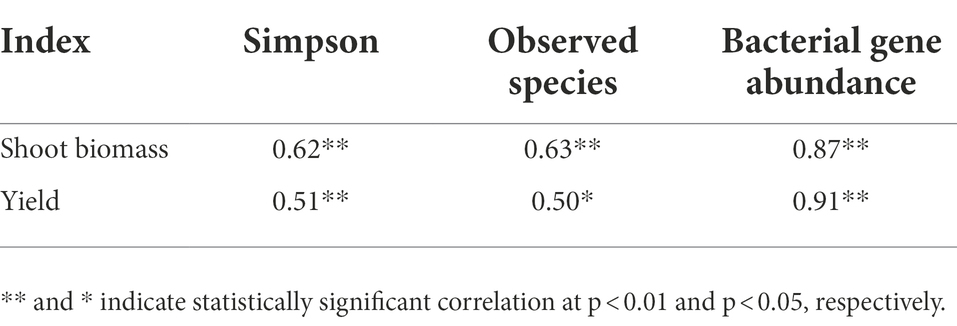
There was a significant correlation between most soil properties (p < 0.05), but none of the other properties correlated with soil pH (Figure 6). The differences in rhizosphere bacterial community composition were related to differences in soil temperature and SOC, AP and AK contents (p < 0.01; Figure 7). Soil temperature and SOC and AK contents correlated strongly (0.5 ≤ r < 0.75, p < 0.01), and AP, AN, NH4+-N and NO3−-N contents weakly (0 < r < 0.5, p < 0.05 or 0.01) with the α-diversity and β-diversity in the Mantel test (Figure 6). Similarly, Simpson diversity and the number of observed species correlated positively with wheat shoot biomass and yield (Table 2; Supplementary Figure S1). The abundance of 16S rRNA gene correlated positively with wheat shoot biomass and yield (p < 0.01).
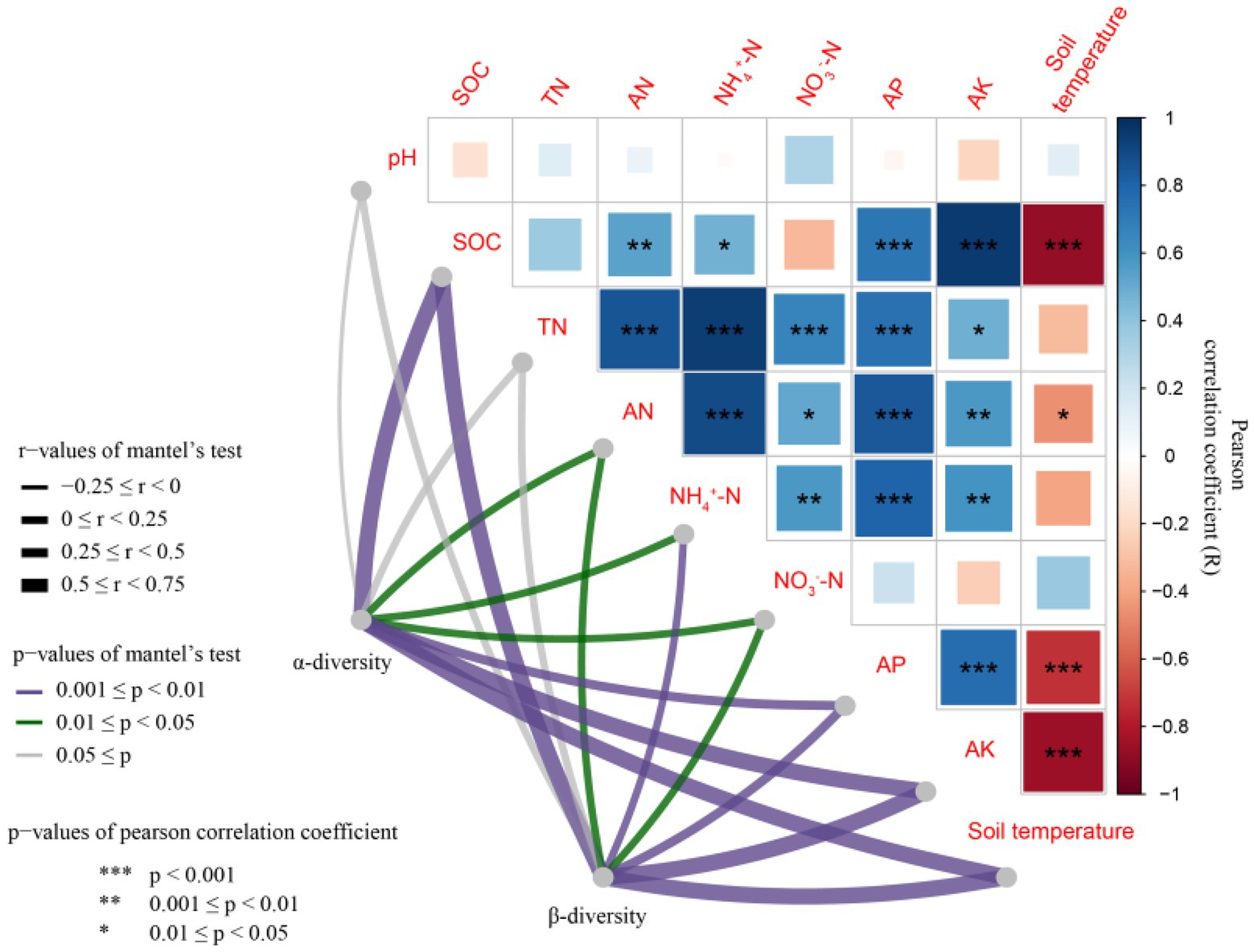
Figure 6. Relationship between bacterial α-diversity and β-diversity and soil environmental factors. SOC, Soil organic C; TN, Total N; AN, Available N; NH4+-N, Ammonium N; NO3−-N, Nitrate N; AP, Available phosphorus; AK, Available potassium.
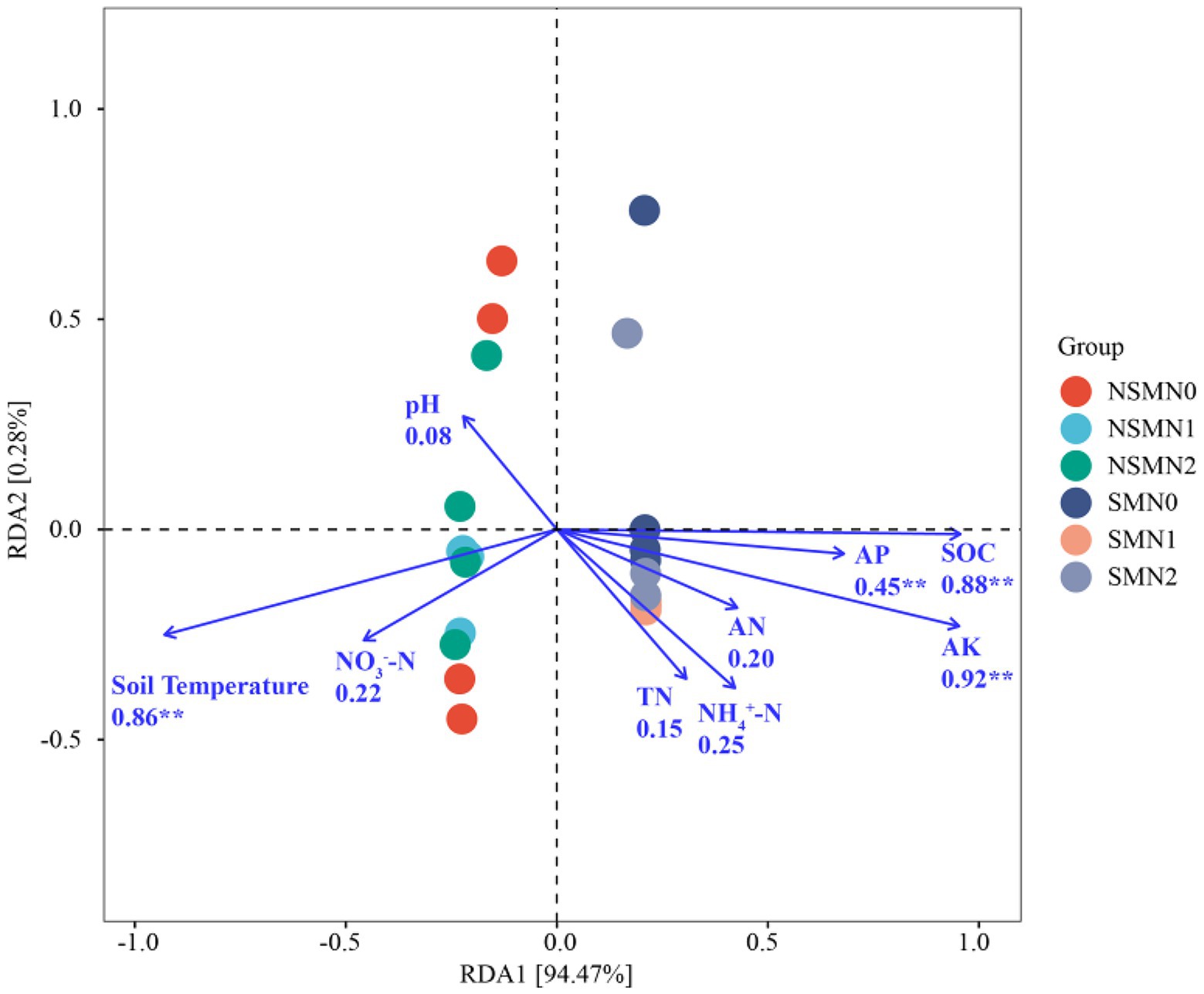
Figure 7. The relationships between environmental factors and bacterial communities in wheat rhizosphere soil under straw mulching and N fertilization treatments. NSM, no straw mulching; SM, straw mulching; N0, no N; N1, 120 kg N ha−1; N2, 180 kg N ha−1. SOC, Soil organic C; TN, Total N; AN, Available N; NH4+-N, Ammonium N; NO3−-N, Nitrate N; AP, Available phosphorus; AK, Available potassium. ** indicates statistically significant difference at p < 0.01.
Discussion
Straw mulching and N fertilization promote wheat yield, yet their combined effects on wheat rhizosphere bacterial communities remain largely unknown. Straw mulching is beneficial in maintaining soil health (Akhtar et al., 2019b). In our study, mulching affected both the composition and diversity of the bacterial communities. Straw mulching provides a C source for the soil microbes, allowing them to proliferate and diversify (Maarastawi et al., 2018). The additional nutrient input provided by the straw (Chen et al., 2017) may have led to the observed higher diversity and richness in the mulched treatment. In bulk soils, the α-diversities of bacteria were lower after long-term N fertilization (Zhou et al., 2015; Zeng et al., 2016). In a long-term N fertilization experiment, Wang et al. (2019) found that fertilization affected the α-diversity in the bulk soil but not in the rhizosphere. Similarly, in maize rhizosphere N fertilization affected root endophytic but not rhizospheric bacterial communities (Miranda-Carrazco et al., 2022). In agreement, in our study the diversities in the rhizosphere were similar across N fertilization levels. The difference in the response to N fertilization between bulk and rhizosphere soils may be due to the differences in their structural and physicochemical properties (Lorenz et al., 1994; Whalley et al., 2005). In line with our hypothesis, compared with sole N fertilization, straw mulching combined with N fertilization was more conducive to promote bacterial α-diversity and the increase of wheat yield in our study.
Several soil environmental factors, for example, pH, nutrient content and temperature, affect bacterial community composition and growth (Pietikainen et al., 2005; Li et al., 2014; Wang et al., 2017; Cheng et al., 2019). Zhao et al. (2019) suggested that C input and organic C content are critical for determining the bacterial community structure. The addition of straw increased SOC content and provided energy for microbial growth in low-C soil (Xiao et al., 2016). Similarly, in our study the SOC content was the main factor associated with differences in the wheat rhizosphere bacterial community composition. As the soil microbial communities are mainly C, not N limited (Soong et al., 2019), it is likely that the higher SOC content in the mulched treatment provided an effective C source for the bacteria. In previous studies the bacterial community composition correlated with the soil AP and AK that were possibly released into the soil due to the decomposition of the straw (Peng et al., 2016; Liang et al., 2020). In agreement, AP and AK contents were among the key factors related to differences in community composition in our study. In addition, the community composition correlated with soil temperature that was lower in the mulched treatment, possibly due to the effect of mulching on the soil albedo which is known to decrease the soil temperature (Kader et al., 2019).
As in Zhang et al. (2017), the Proteobacteria comprised the largest part of the relative abundance in all treatments. Most of the differentially abundant OTUs were proteobacterial as well. Overall, mulching resulted in a greater number of enriched OTUs than N fertilization. Multiple OTUs assigned to Acidobacteria, Chloroflexi and Actinobacteria, i.e., phyla considered key groups in the decomposition of organic matter in soil and of great significance in C turnover (Banerjee et al., 2016; Lewin et al., 2016), were enriched in the mulched treatment, further suggesting that the differences in rhizospheric communities were due to the availability of C. Among the most discriminative taxa in the mulched treatment, Acidobacteria Subgroup 6 includes putative plant growth promoters (PGP; Chen L. et al., 2021). All but one, the genus Vibrionimonas, of the most discriminative taxa in NSM were proteobacterial; the taxa included genus Ralstonia, a potential plant pathogen (Peeters et al., 2013), and taxa with possible plant growth promoting properties. The genera Acinetobacter and Phyllobacterium and order Pseudomonadales include P-solubilizing strains, and in addition to N fixation ability, IAA production is one of the characteristics of genus Rhizobium (Badenoch-Jones et al., 1982). Even though the not mulched soils were characterized by both PGP and pathogenic taxa, together with the higher abundance and diversity in the mulched treatment, the results implied that the mulching generally improved soil health.
Conclusion
In line with our hypothesis, straw mulching for four years mulching increased the diversity and abundance of the bacterial communities in wheat rhizosphere. The straw mulching affected the composition of rhizospheric bacterial communities, likely mostly due to the increase in SOC content that provided an effective C source for the bacteria. The results offer valuable information for soil management strategies to shape the composition and function of soil bacterial communities in agroecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, SRP297976.
Author contributions
SC and GF contributed to conception and design of the study. XX organized the database. HM, TZ, and XH performed the statistical analysis. SC wrote the first draft of the manuscript. PP wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by Sichuan Science and Technology Program (2021YFYZ0002), Agro-scientific research in the public interest (20150312705), and Special Fund for National Key Research and Development Program (2016YFD0300406).
Acknowledgments
We appreciate Yongjia Zhong from Root Biology Center of Fujian Agriculture and Forestry University for his great help in statistical analysis of results and Hong Liao for suggestions leading to the improvement of an earlier version of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.982109/full#supplementary-material
Footnotes
1. ^https://docs.qiime2.org/2019.4/tutorials/
2. ^http://huttenhower.sph.harvard.edu/lefse/
References
Akhtar, K., Wang, W. Y., Khan, A., Ren, G. X., Zaheer, S., Sial, T. A., et al. (2019a). Straw mulching with fertilizer nitrogen: an approach for improving crop yield, soil nutrients and enzyme activities. Soil Use Manag. 35, 526–535. doi: 10.1111/sum.12478
Akhtar, K., Wang, W. Y., Ren, G. X., Khan, A., Feng, Y. Z., Yang, G. H., et al. (2019b). Integrated use of straw mulch with nitrogen fertilizer improves soil functionality and soybean production. Environ. Int. 132:105092. doi: 10.1016/j.envint.2019.105092
Anderson, M. J., Gorley, R. N., and Clarke, K. R. (2008). PERMANOVA + for PRIMER: Guide to Software and Statistical Methods. The University of Auckland, Plymouth, 214
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C., and Segata, N. (2015). Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:2015. doi: 10.7717/peerj.1029.eCollection
Badenoch-Jones, J., Summons, R. E., Djordjevic, M. A., Shine, J., Letham, D. S., and Rolfe, B. G. (1982). Mass spectrometric quantification of indole-3-acetic acid in rhizobium culture supernatants: relation to root hair curling and nodule initiation. Appl. Environ. Microbiol. 44, 275–280. doi: 10.1128/aem.44.2.275-280.1982
Banerjee, S., Kirkby, C. A., Schmutter, D., Bissett, A., Kirkegaard, J. A., and Richardson, A. E. (2016). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 97, 188–198. doi: 10.1016/j.soilbio.2016.03.017
Bay, G., Lee, C., Chen, C. L., Mahal, N. K., Castellano, M. J., Hofmockel, K. S., et al. (2021). Agricultural management affects the active rhizosphere bacterial community composition and nitrification. mSystems 6:e0065121. doi: 10.1128/mSystems.00651-21
Blanco-Canqui, H., and Lal, R. (2007). Soil structure and organic carbon relationships following 10 years of wheat straw management in no-till. Soil Till. Res. 95, 240–254. doi: 10.1016/j.still.2007.01.004
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Chai, Q., Nemecek, T., Liang, C., Zhao, C., Yu, A. Z., Coulter, J. A., et al. (2021). Integrated farming with intercropping increases food production while reducing environmental footprint. Proc. Natl. Acad. Sci. U. S. A. 118:e2106382118. doi: 10.1073/pnas.2106382118
Chen, L., Hao, Z. H., Li, K. K., Sha, Y., Wang, E. T., Sui, X. H., et al. (2021). Effects of growth-promoting rhizobacteria on maize growth and rhizosphere microbial community under conservation tillage in Northeast China. Microb. Biotechnol. 14, 535–550. doi: 10.1111/1751-7915.13693
Chen, S. H., Xiang, X. L., Ma, H. L., Penttinen, P., Zhao, J. R., Li, H., et al. (2021). Straw mulching and nitrogen fertilization affect diazotroph communities in wheat rhizosphere. Front. Microbiol. 12:658668. doi: 10.3389/fmicb.2021.658668
Chen, Y. L., Xin, L., Liu, J. T., Yuan, M. Z., Liu, S. T., Jiang, W., et al. (2017). Changes in bacterial community of soil induced by long-term straw returning. Agr. Microbiol. 74, 349–356. doi: 10.1590/1678-992X-2016-0025
Cheng, Z. B., Chen, Y., and Zhang, F. H. (2019). Effect of cropping systems after abandoned salinized farmland reclamation on soil bacterial communities in arid Northwest China. Soil Till. Res. 187, 204–213. doi: 10.1016/j.still.2018.12.015
Dai, X. L., Zhou, W., Liu, G. R., He, P., and Liu, Z. B. (2019). Soil C/N and pH together as a comprehensive indicator for evaluating the effects of organic substitution management in subtropical paddy fields after application of high-quality amendments. Geoderma 337, 1116–1125. doi: 10.1016/j.geoderma.2018.11.023
Dong, Q. G., Yang, Y. C., Yu, K., and Feng, H. (2018). Effects of straw mulching and plastic film mulching on improving soil organic carbon and nitrogen fractions, crop yield and water use efficiency in the loess plateau. China. Agr. Water Manage. 201, 133–143. doi: 10.1016/j.agwat.2018.01.021
Giraldo, P., Benavente, E., Manzano-Agugliaro, F., and Gimenez, E. (2019). Worldwide research trends on wheat and barley: a bibliometric comparative analysis. Agronomy 9:352. doi: 10.3390/agronomy9070352
Huson, D. H., Mitra, S., Ruscheweyh, H. J., Weber, N., and Schuster, S. C. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21, 1552–1560. doi: 10.1101/gr.120618.111
Kader, M. A., Nakamura, K., Senge, M., Mojid, M. A., and Kawashima, S. (2019). Numerical simulation of water-and heat-flow regimes of mulched soil in rain-fed soybean field in Central Japan. Soil Till. Res. 191, 142–155. doi: 10.1016/j.still.2019.04.006
Kong, Y. L., Ling, N., Xue, C., Chen, H., Ruan, Y., Guo, J. J., et al. (2019). Long-term fertilization regimes change soil nitrification potential by impacting active autotrophic ammonia oxidizers and nitrite oxidizers as assessed by DNA stable isotope probing. Environ. Microbiol. 21, 1224–1240. doi: 10.1111/1462-2920.14553
Lewin, G. R., Carlos, C., Chevrette, M. G., Horn, H. A., McDonald, B. R., Stankey, R. J., et al. (2016). Evolution and ecology of actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. 70, 235–254. doi: 10.1146/annurev-micro-102215-095748
Li, X. Z., Rui, J. P., Mao, Y. J., Yannarell, A., and Mackie, R. (2014). Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 68, 392–401. doi: 10.1016/j.soilbio.2013.10.017
Liang, R. B., Hou, R. X., Li, J., Lyu, Y., Hang, S., Gong, H. R., et al. (2020). Effects of different fertilizers on rhizosphere bacterial communities of winter wheat in the North China Plain. Agronomy 10:93. doi: 10.3390/agronomy10010093
Ling, N., Chen, D. M., Guo, H., Wei, J. X., Bai, Y. F., Sheng, Q. R., et al. (2017). Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 292, 25–33. doi: 10.1016/j.geoderma.2017.01.013
Liu, C., Li, L. L., Xie, J. H., Coulter, J. A., Zhang, R. Z., Luo, Z. Z., et al. (2020). Soil bacterial diversity and potential functions are regulated by long-term conservation tillage and straw mulching. Microorganisms 8:836. doi: 10.3390/microorganisms8060836
Liu, W. B., Ling, N., Guo, J. J., Ruan, Y., Zhu, C., Shen, Q. R., et al. (2020). Legacy effects of 8-year nitrogen inputs on bacterial assemblage in wheat rhizosphere. Biol. Fert. Soils 56, 583–596. doi: 10.1007/s00374-020-01435-2
Lorenz, S. E., Hamon, R. E., and McGrath, S. P. (1994). Differences between soil solutions obtained from rhizosphere and non-rhizosphere soils by water displacement and soil centrifugation. Eur. J. Soil Sci. 45, 431–438. doi: 10.1111/j.1365-2389.1994.tb00528.x
Lu, R. K. (1999). Soil and Agrochemical Analytical Methods. China Agricultural Science and Technology Press Beijing, China. 146–195.
Maarastawi, S. A., Frindte, K., Linnartz, M., and Knief, C. (2018). Crop rotation and straw application impact microbial communities in Italian and Philippine soils and the rhizosphere of Zea mays. Front. Microbiol. 9:1295. doi: 10.3389/fmicb.2018.01295
Mapelli, F., Marasco, R., Fusi, M., Scaglia, B., Rolli, E., Fodelianakis, S., et al. (2018). The stage of soil development modulates rhizosphere effect along a High Arctic desert chronosequence. ISME J. 12, 1188–1198. doi: 10.1038/s41396-017-0026-4
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Miranda-Carrazco, A., Navarro-Noya, Y. E., Govaerts, B., Verhulst, N., and Dendooven, L. (2022). Nitrogen fertilizer application alters the root Endophyte bacterial microbiome in maize plants, but not in the stem or rhizosphere soil. Microbiol. Spectr. 18:e0178522. doi: 10.1128/spectrum.01785-22
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: community ecology package. R package version 2.5-6. Available at: https://CRAN.R-project.org/package=vegan
Peeters, N., Guidot, A., Vailleau, F., and Valls, M. (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14, 651–662. doi: 10.1111/mpp.12038
Peng, C. J., Lai, S. S., Luo, X. S., Huang, Q. Y., and Chen, W. L. (2016). Effects of long term rice straw application on the microbial communities of rapeseed rhizosphere in a paddy-upland rotation system. Sci. Total Environ. 557–558, 231–239. doi: 10.1016/j.scitotenv.2016.02.184
Pérez-Montaño, F., Alías-Villegas, C., Bellogín, R. A., del Cerro, P., Espuny, M. R., Jiménez-Guerrero, I., et al. (2014). Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol. Res. 169, 325–336. doi: 10.1016/j.micres.2013.09.011
Pietikainen, J., Pettersson, M., and Baath, E. (2005). Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Rev. 52, 49–58. doi: 10.1016/j.femsec.2004.10.002
Qiu, Y., Lv, W. C., Xie, Z. K., and Wang, Y. J. (2020). Long-term effects of gravel mulching and straw mulching on soil physicochemical properties and bacterial and fungal community composition in the Loess Plateau of China. Eur. J. Soil Biol. 98:103188. doi: 10.1016/j.ejsobi.2020.103188
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team. (2020). R: a language and environment for statistical computing. R Version 4.0.1. Vienna: R Foundation for Statistical Computing.
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Soong, J. L., Fuchslueger, L., Marañon-Jimenez, S., Torn, M. S., Janssens, I. A., Penuelas, J., et al. (2019). Microbial carbon limitation: the need for integrating microorganisms into our understanding of ecosystem carbon cycling. Glob. Chang. Biol. 26, 1953–1961. doi: 10.1111/gcb.14962
Sun, R. B., Zhang, X. X., Guo, X. S., Wang, D. Z., and Chu, H. Y. (2015). Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 88, 9–18. doi: 10.1016/j.soilbio.2015.05.007
Walters, W. A., Caporaso, J. G., Lauber, C. L., Berg-Lyons, D., Fierer, N., and Knight, R. (2011). PrimerProspector: de novo design and taxonomic analysis of barcoded PCR primers. Bioinformatics 27, 1159–1161. doi: 10.1093/bioinformatics/btr087
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbio. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, H. H., Guo, Q. C., Li, X., Li, X., Yu, Z. X., Li, X. Y., et al. (2020). Effects of long-term no-tillage with different straw mulching frequencies on soil microbial community and the abundances of two soil-borne pathogens. Appl. Soil Ecol. 148:103488. doi: 10.1016/j.apsoil.2019.103488
Wang, Y. S., Li, C. N., Kou, Y. P., Wang, J. J., Tu, B., Li, H., et al. (2017). Soil pH is a major driver of soil diazotrophic community assembly in Qinghai-Tibet alpine meadows. Soil Biol. Biochem. 115, 547–555. doi: 10.1016/j.soilbio.2017.09.024
Wang, Z. T., Li, Y. Z., Li, T., Zhao, D. Q., and Liao, Y. C. (2020). Tillage practices with different soil disturbance shape the rhizosphere bacterial community throughout crop growth. Soil Till. Res. 197:104501. doi: 10.1016/j.still.2019.104501
Wang, C., Liu, D. W., and Bai, E. (2018). Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 120, 126–133. doi: 10.1016/j.soilbio.2018.02.003
Wang, Q. F., Ma, M. C., Jiang, X., Guan, D. W., Wei, D., Zhao, B. S., et al. (2019). Impact of 36 years of nitrogen fertilization on microbial community composition and soil carbon cycling-related enzyme activities in rhizospheres and bulk soils in Northeast China. Appl. Soil Ecol. 136, 148–157. doi: 10.1016/j.apsoil.2018.12.019
Whalley, W. R., Riseley, B., Leeds-Harrison, P. B., Bird, N. R. A., Leech, P. K., and Adderley, W. P. (2005). Structural differences between bulk and rhizosphere soil. Eur. J. Soil Sci. 56, 353–360. doi: 10.1111/j.1365-2389.2004.00670.x
Xiao, Y., Tang, J. L., and Wang, M. K. (2016). Physicochemical properties of three typical purple soils with different parent materials and land uses in Sichuan basin, China. Natur. Resour. Eng. 1, 59–68. doi: 10.1080/23802693.2016.1258854
Yan, S. S., Song, J. M., Fan, J. S., Yan, C., Dong, S. K., Ma, C. M., et al. (2020). Changes in soil organic carbon fractions and microbial community under rice straw return in Northeast China. Glob. Ecol. Conserv. 22:e00962. doi: 10.1016/j.gecco.2020.e00962
Zeng, J., Liu, X. J., Song, L., Lin, X. G., Zhang, H. Y., Shen, C. C., et al. (2016). Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 92, 41–49. doi: 10.1016/j.soilbio.2015.09.018
Zhang, J. G., Bo, G. D., Zhang, Z. F., Kong, F. Y., Yang, Y., and Shen, G. M. (2016). Effects of straw incorporation on soil nutrients, enzymes, and aggregate stability in tobacco fields of China. Sustainability 8:710. doi: 10.3390/su8080710
Zhang, X. X., Zhang, R. J., Gao, J. S., Wang, X. C., Fan, F. L., Ma, X. T., et al. (2017). Thirty-one years of rice-rice-green manure rotations shape the rhizosphere microbial community and enrich beneficial bacteria. Soil Biol. Biochem. 104, 208–217. doi: 10.1016/j.soilbio.2016.10.023
Zhao, H. L., Jiang, Y. H., Ning, P., Liu, J. F., Zheng, W., Tian, X. H., et al. (2019). Effect of different straw return modes on soil bacterial community, enzyme activities and organic carbon fractions. Soil Sci. Soc. Am. J. 83, 638–648. doi: 10.2136/sssaj2018.03.0101
Keywords: rhizosphere bacterial community, gene abundance, straw mulching, N fertilization, 16S rRNA
Citation: Chen S, Xiang X, Ma H, Penttinen P, Zheng T, Huang X and Fan G (2022) Response of soil bacterial communities in wheat rhizosphere to straw mulching and N fertilization. Front. Microbiol. 13:982109. doi: 10.3389/fmicb.2022.982109
Edited by:
Jeanette M. Norton, Utah State University, United StatesReviewed by:
Memoona Zahra, Griffith University, AustraliaXiangbo Yin, Université du Québec en Abitibi Témiscamingue, Canada
Copyright © 2022 Chen, Xiang, Ma, Penttinen, Zheng, Huang and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaoqiong Fan, ZmFuZ2FvMjAwNTZAMTI2LmNvbQ==
 Songhe Chen
Songhe Chen Xiaoling Xiang
Xiaoling Xiang Hongliang Ma
Hongliang Ma Petri Penttinen
Petri Penttinen Ting Zheng
Ting Zheng Xiulan Huang
Xiulan Huang Gaoqiong Fan
Gaoqiong Fan