
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 03 November 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.979790
This article is part of the Research TopicWomen in Infectious Agents and Disease: 2022View all 10 articles
In the USA, Salmonella enterica subspecies enterica serovar Senftenberg is among the top five serovars isolated from food and the top 11 serovars isolated from clinically ill animals. Human infections are associated with exposure to farm environments or contaminated food. The objective of this study was to characterize S. Senftenberg isolates from production animals by analyzing phenotypic antimicrobial resistance profiles, genomic features and phylogeny.
Salmonella Senftenberg isolates (n = 94) from 20 US states were selected from NVSL submissions (2014–2017), tested against 14 antimicrobial drugs, and resistance phenotypes determined. Resistance genotypes were determined using whole genome sequencing analysis with AMRFinder and the NCBI and ResFinder databases with ABRicate. Plasmids were detected using PlasmidFinder. Integrons were detected using IntFinder and manual alignment with reference genes. Multilocus-sequence-typing (MLST) was determined using ABRicate with PubMLST database, and phylogeny was determined using vSNP.
Among 94 isolates, 60.6% were resistant to at least one antimicrobial and 39.4% showed multidrug resistance. The most prevalent resistance findings were for streptomycin (44.7%), tetracycline (42.6%), ampicillin (36.2%) and sulfisoxazole (32.9%). The most commonly found antimicrobial resistance genes were aac(6′)-Iaa (100%), aph(3″)-Ib and aph(6)-Id (29.8%) for aminoglycosides, followed by blaTEM-1 (26.6%) for penicillins, sul1 (25.5%) and sul2 (23.4%) for sulfonamides and tetA (23.4%) for tetracyclines. Quinolone-resistant isolates presented mutations in gyrA and/or parC genes. Class 1 integrons were found in 37 isolates. Thirty-six plasmid types were identified among 77.7% of the isolates. Phylogenetic analysis identified two distinct lineages of S. Senftenberg that correlated with the MLST results. Isolates were classified into two distinct sequence types (ST): ST14 (97.9%) and ST 185 (2.1%). The diversity of this serotype suggests multiple introductions into animal populations from outside sources.
This study provided antimicrobial susceptibility and genomic characteristics of S. Senftenberg clinical isolates from production animals in the USA during 2014 to 2017. This study will serve as a base for future studies focused on the phenotypic and molecular antimicrobial characterization of S. Senftenberg isolates in animals. Monitoring of antimicrobial resistance to detect emergence of multidrug-resistant strains is critical.
Salmonella enterica subsp. enterica serovar Senftenberg is commonly isolated from animals and food. This serovar is widely distributed and has been found worldwide. In the USA, S. Senftenberg is among the top five serovars isolated from food and among the top 11 serovars isolated from clinically ill animals (Switt, 2019). S. Senftenberg and S. Montevideo were the most common serotypes found in animal feeds in the USA in 2012 (Li et al., 2012). In Europe, S. Senftenberg was found more frequently in poultry flocks than other serotypes and the emergence of this serotype was a cause of concern in 2012 (Boumart et al., 2012). Most Salmonella serovars, including S. Senftenberg, are tolerant to desiccation and able to colonize and persist in feed mills (Pedersen et al., 2008). Interestingly, S. Senftenberg is also a heat-resistant serotype (Doyle and Mazzotta, 1999), which may contribute to persistence in feed and the environment. This can be a source of contamination on farms and in processing environments. Human infections with S. Senftenberg are rare and are typically associated with exposure to poultry flocks, farm environments, or contaminated food (Boumart et al., 2012). Worldwide, S. Senftenberg has been linked to outbreaks associated with contaminated pistachios, salami, basil, Maradol papayas, peanut butter, alfalfa sprouts and baby cereal (Centers for disease Control and Prevention, 2010, 2016, 2022; US Food and Drug Administration, 2014; Hassan et al., 2019; Switt, 2019; Haendiges et al., 2021).
Although antimicrobial-resistant isolates of S. Senftenberg are usually associated with animal sources (Stepan et al., 2011), antimicrobial-resistant human isolates of S. Senftenberg have been reported in the USA, and extensively drug-resistant (XDR) strains have been isolated from patients outside of the USA, raising public health concerns (Hendriksen et al., 2013; Veeraraghavan et al., 2019). Infections caused by S. Senftenberg range from asymptomatic to severe, and deaths have been associated with the XDR strains of S. Senftenberg in China (El Ghany et al., 2016). XDR strains are resistant to all but only one or two categories of antimicrobials, leaving clinicians and veterinarians with few to no treatment options (Magiorakos et al., 2012).
Horizontal transfer of genetic material is important in the spread of MDR. Resistance genes can be inserted in the form of cassettes into integrons; these mobilizable genetic elements are grouped in three classes (class 1, 2 and 3) based on the presence of three different integrases encoded by the intl1, intl2, intl3 genes. Integrons can be mobilized within the chromosome to other regions, or they can be inserted in integrative-conjugative elements and plasmids, which can facilitate the horizontal transfer of resistance genes between bacteria (Stokes and Gillings, 2011).
Since Salmonella is associated with outbreaks of foodborne disease, MDR strains pose a risk to public health because of the potential for treatment failures (Nair et al., 2018). Few studies exist on the antimicrobial susceptibility and genetic diversity of Salmonella serovar Senftenberg of animal origin. Among all Salmonella serotyping submissions received at the NVSL from January 1, 2014, through December 31, 2017, S. Senftenberg ranked number eight during 2014 and number 10 during 2015 among clinical isolates, and ranked number one during 2014, 2015, 2016 and number two during 2017 among non-clinical isolates. The objective of this study was to compare phenotypic and genomic resistance data, mechanisms of antimicrobial resistance, plasmid replicons, genetic relatedness, and to characterize S. Senftenberg diagnostic isolates recovered from poultry, swine, and cattle in the USA between 2014 and 2017, and to provide useful retrospective information for future studies on S. Senftenberg.
A total of 94 S. Senftenberg isolates from swine (n = 50), poultry (n = 24) and cattle (n = 20) were selected from the National Veterinary Services Laboratories (NVSL) Salmonella repository isolates archived at room temperature on nutrient agar slants. Samples came from 20 US States (IA = 20, MN = 15, AR = 8, MO = 8, IL = 6, IN = 4, TX = 4, NC = 4, OH = 4, PA = 4, OK = 2, KS = 2, NE = 2, NY = 2, SD = 2, VA = 2, WI = 2, AL = 1, AZ = 1, KY = 1).
Isolates were selected from samples that were submitted to the NVSL for Salmonella serotyping, confirmed by classical (Grimont and Weill, 2007) and molecular typing using Luminex xMAP® technology (Dunbar et al., 2015), between the years of 2014 and 2017. The dataset was initially limited to one sample per year per owner. If more than the targeted number of isolates were available, a randomly selected subset of isolates was chosen. The data was then de-identified to remove information other than the animal species, state of origin, clinical status, and sample type and assigned a unique identifier. Salmonella was confirmed using Biotyper software with an autoflex speed™ MALDI-TOF instrument (Bruker Daltonics, Billerica, MA, USA).
All Salmonella isolates were tested for antimicrobial susceptibility against 14 class-representative antimicrobial agents using the Sensititre CMV4AGNF plate (Thermo Fisher Scientific, Waltham, MA, USA) including: gentamicin (GEN), streptomycin (STR), amoxicillin/clavulanic acid (AMC), cefoxitin (FOX), ceftriaxone (CRO), meropenem (MEM), sulfisoxazole (SUL), trimethoprim/sulfamethoxazole (SXT), ampicillin (AMP), chloramphenicol (CHL), ciprofloxacin (CIP), nalidixic acid (NAL), azithromycin (AZM), and tetracycline (TET). Results were interpreted using consensus interpretative criteria established by the National Antimicrobial Resistance Monitoring System (US Food and Drug Administration, 2021).
DNA was extracted using Promega Maxwell® with the Whole Blood DNA kit following manufacturer’s instructions. S. Senftenberg isolates were subjected to whole genome sequencing using the Illumina MiSeq platform with 2×250 paired-end chemistry and the NexteraXT library preparation kit (Illumina, Inc., San Diego, CA, USA). AMR gene alleles were determined using AMRFinder (Feldgarden et al., 2019) and the NCBI and ResFinder databases (Zankari et al., 2012) using ABRicate1 with an identity threshold of 80% over ≥60% of the length of the target gene. Integrons were identified using IntFinder 1.0 (Loaiza et al., 2020). Integron classes were mapped using NCBI reference sequences of the intl1 (MG785026.1), intl2 (MK994977.1) and Intl3 (KM194584.1) integrase genes, and resistance genes available in the CARD (Comprehensive Antimicrobial Resistance Database, card.macmaster.ca) database using Geneious Prime v11.0.9 + 11 (Biomatters Ltd., NZ). Plasmid replicons were identified using ABRicate with the PlasmidFinder database (Carattoli et al, 2014). PointFinder was used for analysis of chromosomal structural gene mutations (Zankari et al., 2017). Isolate sequences are publicly available in the NCBI BioProject PRJNA785813.
Multilocus-sequence-typing (MLST) was determined using ABRicate with PubMLST database. The single nucleotide polymorphism (SNP) analysis of all isolates was performed using the NVSL vSNP pipeline1. Isolates were separated and analyzed by MLST with the respective reference (S. Senftenberg NZ_CP016837 for ST185 and NZ_CP029036 for ST14). A SNP-based phylogenetic tree was generated with RAxML in the vSNP pipeline (Stamatakis, 2014). The k-mer based phylogeny tool kSNP was used to generate a reference-free phylogenetic tree of all the S. Senftenberg isolates (Gardner et al., 2015).
Each antimicrobial susceptibility interpretation (resistant or susceptible) for each antimicrobial tested was compared with the presence or absence of the corresponding resistance gene or genes and/or chromosomal gene mutations found. Intermediate phenotypes were counted as susceptible in this analysis. Using the phenotypic results as the reference outcome, sensitivity was calculated by dividing the number of isolates that were genotypically resistant by the total number of isolates exhibiting clinical resistance phenotypes. Specificity was calculated by dividing the number of isolates that were genotypically susceptible by the total number of isolates with susceptible phenotypes (McDermott et al., 2016).
Overall, the highest percentage of resistance was found to the following antimicrobials: streptomycin (44.7%), tetracycline (42.6%), ampicillin (36.2%) and sulfisoxazole (32.9%) (Figure 1). All of the S. Senftenberg isolates were susceptible to meropenem.
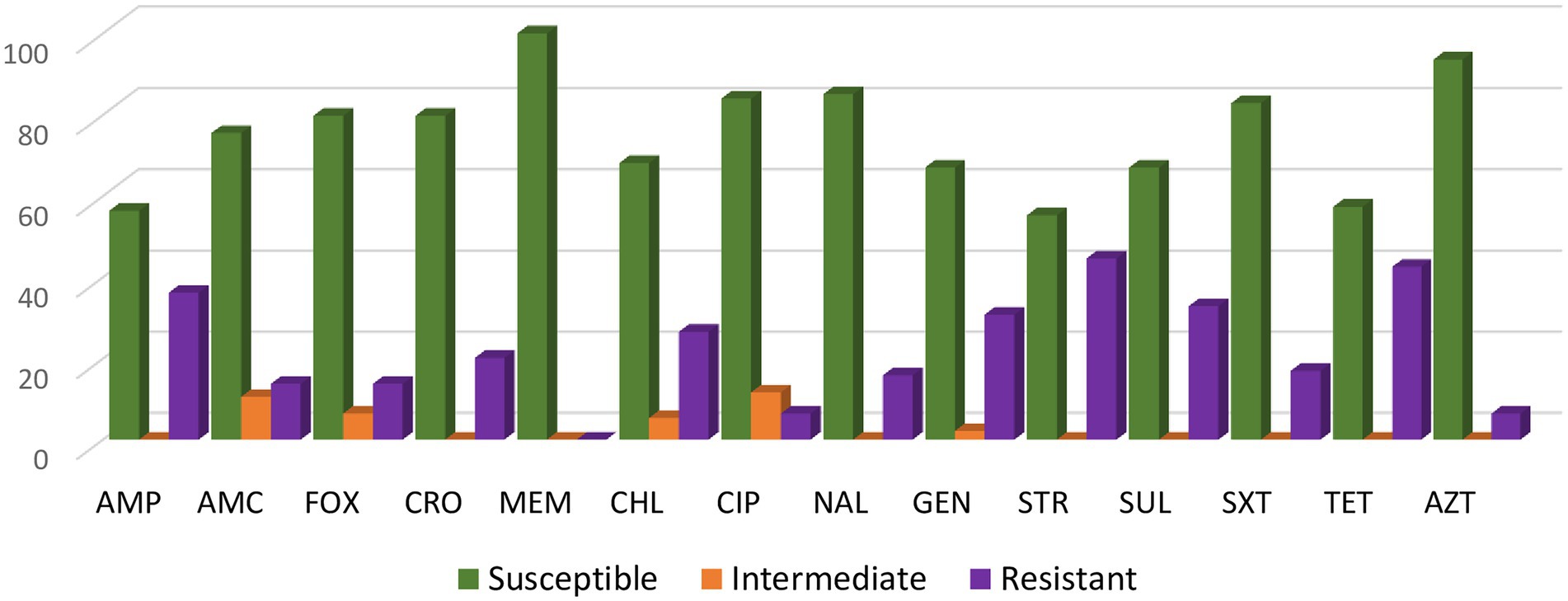
Figure 1. Antimicrobial resistance rates of the 94 Salmonella Senftenberg isolates. For ciprofloxacin (CIP) isolates categorized as intermediate (MIC 0.12 to 0.5 μg/mL) or resistant (MIC ≥ 1 μg/mL) were defined as having decreased susceptibility to CIP with a MIC ≥ 0.12 μg/mL.
Among all 94 isolates, 60.6% (n = 57) were resistant to at least one antimicrobial and 39.4% (n = 37) isolates were MDR. Among MDR isolates, 60% were from swine, 20% from cattle and 12.5% from poultry. Four isolates from swine showed possible XDR; one isolate was only susceptible to meropenem, one isolate was susceptible to gentamycin and meropenem, and two isolates were susceptible to meropenem and azithromycin (one of which with presence of the macrolide ermB gene). Most isolates showed diverse resistance profiles, with the most common resistance profile (AMP, GEN, STR) found in just five isolates. Supplementary Table S1 summarizes the phenotypic resistance profiles and the resistance genes present in S. Senftenberg isolates displaying antimicrobial resistance.
Antimicrobial resistance genes are shown in Table 1. The most commonly observed was the aminoglycoside acetyltransferase aac(6′)-Iaa gene, which was found in all isolates, and the aminoglycoside phosphotransferase aph(3″)-Ib and aph(6)-Id genes seen together in 29.8% (n = 28) of isolates. The sul1 gene, a dihydropteroate synthase that is linked to other resistance genes of class 1 integrons and confers resistance to sulfonamides, was found in 25.5% (n = 24) of the isolates; and the sul2 gene was observed in 23.4% (n = 22) of the isolates. Resistance to tetracycline was due to the presence of the tetA gene that encodes a tetracycline efflux pump, and it was observed in 23.4% (n = 22) of the isolates.
Among beta-lactams, blaTEM-1B, a class A narrow-spectrum beta-lactamase, was the most prevalent beta-lactamase gene (26.6%) conferring resistance to penicillins (ampicillin); but blaCMY-2, a class C beta-lactamase, was the most prevalent beta-lactamase gene (14.9%) against penicillins plus inhibitors (amoxicillin + clavulanic acid) and cephalosporins (cefoxitin, ceftriaxone). The class A extended spectrum beta-lactamases (ESBLs) encoded by the blaSHV-12 gene were found in five isolates from swine.
In addition to aph(3″)-Ib and aph(6)-Id genes, other resistance genes were observed that convey resistance to aminoglycosides, including several integron-encoded aminoglycoside nucleotidyltransferases such as aadA1, which was observed in 17% of isolates, aadA2, which was observed in 15.9% of isolates, and aadA5, aadA6, aadA12, aadA16, and aadA25, which were observed in lower frequencies. Resistance genes for gentamicin included several aminoglycoside acetyltransferases encoded by: aac(6′)-Ib4 gene in 9.6%; aac(6′)-Ib, aac(3′)-II, aac(6′)-IIc, aac(3)-VIa in 7.4%; and aac(3)-IVa in 4.3%. The nucleotidyltransferase ant(2″)-la gene was found in 7.4%, and the methyltransferase armA gene in one isolate. Six genes conferring resistance to macrolides were found in 13.8% of isolates; the ereA gene that encodes an erythromycin esterase was the most frequently identified in 6 isolates, followed by the mphA gene that encodes a macrolide 2′-phosphotransferase in 3 isolates. Several resistance genes were found for chloramphenicol; the most frequent was the plasmid or transposon-encoded chloramphenicol exporter floR, which was observed in 14.9% of isolates, followed by catA2 gene, a chloramphenicol O-acetyltransferase, which was observed in 8.5% of isolates. Additional chloramphenicol exporter genes found in lower frequency were cmlA5 (3.4%) and cmlA1 (1.1%) genes.
Quinolone-resistance genes were detected in 12 isolates; the most frequent gene was qnrB2, a plasmid-mediated quinolone resistance protein, which was observed in 6.4% of isolates, and others in lower frequency: qnrB19 in 3.4%, qnrB77 in 2.1%, qnrB6 in 1.1% and aac(6′)-lb-cr in 1.1% of isolates. The aac(6′)-lb-cr gene doubly confers resistance to aminoglycoside and fluoroquinolone antibiotics through fluoroquinolone-acetylating activity.
Some isolates presented other genes that confer resistance to antimicrobials that were not included on the panel: the aph(3′)-Ia gene conferring resistance to kanamycin was detected in 22 (23.4%) isolates, the mobilized and plasmid-mediated colistin resistance and phosphoethanolamine transferase mcr-9.1 gene was detected in eight (8.5%) isolates, the arr-269927220, an ADP-ribosyltransferase that confers resistance to rifamycin was detected in seven (7.4%) isolates, and the arr-3 gene in one (1.1%) isolate. The bleO gene that encodes a bleomycin binding protein was detected in three (3.2%) isolates, and the bleTn5 gene that encodes a bleomycin binding protein BLMT by the ble gene on the transposon Tn5 was found in one (1.1%) isolate. Interestingly, the aac(6′)-Iaa gene was observed in all isolates using ResFinder databases. The aac(6′)-Iaa gene is a chromosomal-encoded aminoglycoside acetyltransferase that confers resistance to tobramycin and kanamycin aminoglycosides. The gene resistance profile varied among the isolates. In addition to antimicrobial resistance genes, we observed the gene qacEΔ1, which confers resistance to quaternary ammonium compounds (QAC), in 25 (26.6%) isolates. We also detected the presence of a sulfonamide resistance gene and the presence of the intl1 gene, a class 1 integron, in these isolates (Table 2). Among isolates that showed resistance to at least one antimicrobial, 64.9% (n = 37) were positive for intI1, but no isolates carried intI2 or intI3 genes. Class 1 integrons variable regions enclosed one or several gene cassettes containing aadA, drfA, floR, aac(6′)-Ib, ant(2′′)-la, and cmlA. Class 1 integrons were found in different proportions among species source: 78.6% (n = 11) in poultry, 61.1% (n = 22) in swine and 47.1% (n = 4) in cattle isolates (Table 3). The sul1 gene, which is often carried in the conserved sequence (3′ CS) of a class 1 integron, was missing in 11 isolates. Ten of these integrons were identified as In48, and carried a resistant gene cassette (aac(6)-Ib), the other one was classified as In192, positive for Intl1 and carried a dfrA15 cassette.
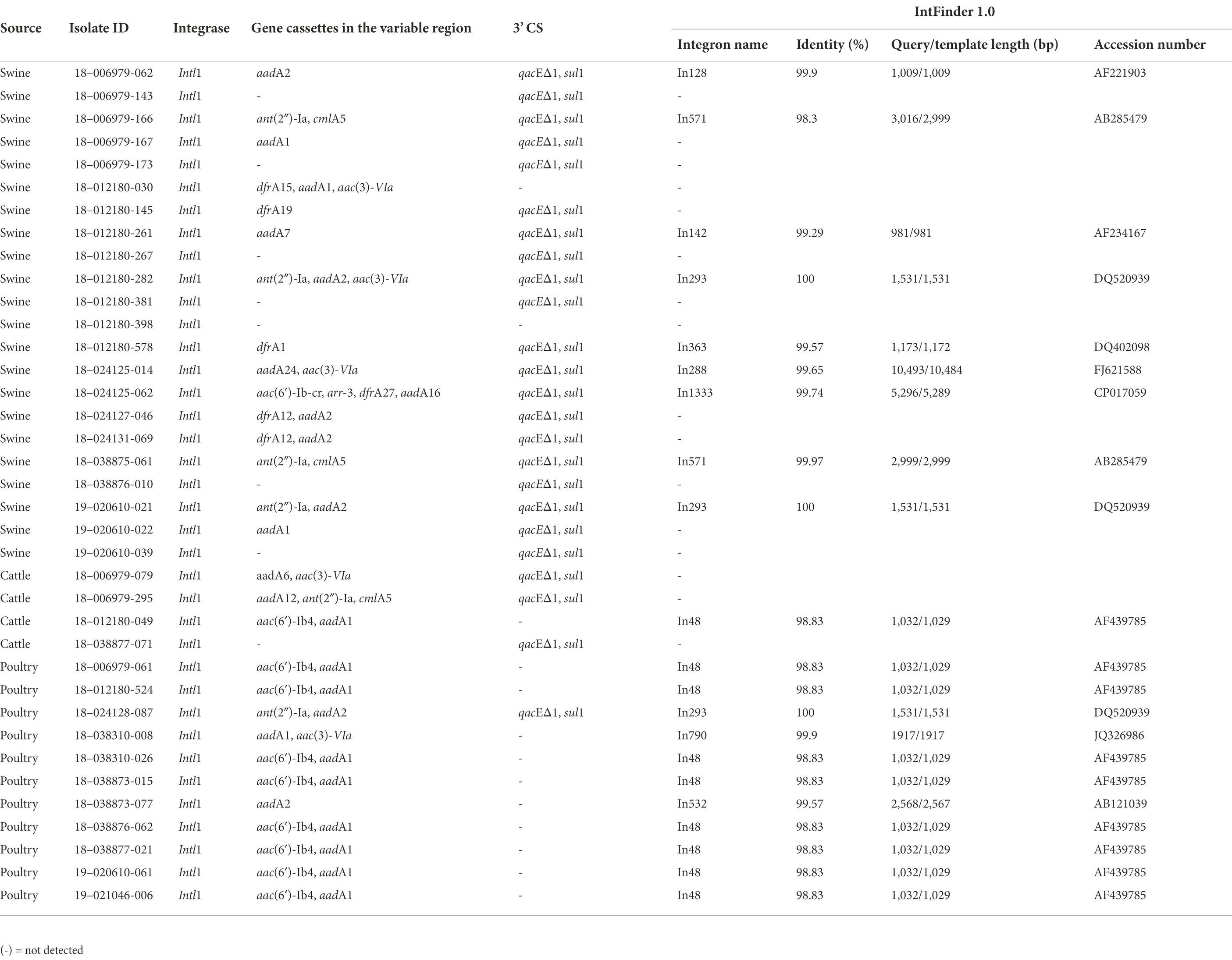
Table 2. Antimicrobial resistance genes detected in the variable region of class 1 integrons among Salmonella Senftenberg isolates from the USA.
Seventeen isolates exhibited decreased resistance to ciprofloxacin (MIC ≥0.12 μg/ml). Fluoroquinolone-resistance genes were identified in 11 of those isolates. The remaining six isolates did not harbor any resistance genes; however, point mutations were detected in these six isolates. Four isolates showed a point mutation in the gyrA (D87N) and parC (T57S) genes, and the other two isolates had a mutation in only the parC (T57S) gene. Five isolates exhibited resistance to nalidixic acid without any specific quinolone resistance gene present, but in four of these five isolates there were point mutations in gyrA and parC genes and in only the parC gene for one isolate. Interestingly, all the isolates showed mutations in the parC gene regardless of phenotypic resistance to nalidixic acid or ciprofloxacin.
The association of antimicrobial susceptibility with antimicrobial resistance genes is shown in Table 3. The least discordance among the animal isolates was seen for β-lactams and the most were seen for phenicols.
Antimicrobial resistance varied across isolates from different animal species. Isolates from all animal species showed susceptibility to MEM. Isolates from cattle and swine showed resistance to all other antimicrobials in variable frequency. All poultry isolates were susceptible to AMC, FOX, CRO, AZM, CIP and NAL (Figure 2).
The percentage of isolates that were MDR varied among the different animal species. Sixty percent (30/50) of the isolates from swine were MDR; three were resistant to 12 antimicrobials and one was resistant to 13 antimicrobials, being possible XDR. Fewer isolates from cattle (5/24, 16.6%) and poultry (3/24, 12.5%) were MDR.
Antimicrobial resistance genes are often encoded on mobile genetic elements such as plasmids. In this study, 36 plasmid types were identified in 73 (77.7%) isolates. Plasmid profiles differed among each animal species. Thirty-four different plasmids were found among swine isolates, 14 among poultry isolates and 10 among cattle isolates. The most prevalent plasmid was ColRNAI, found in 60.6% of the study isolates: n = 20 (83.3%) in poultry, n = 31 (62%) in swine and n = 6 (30%) in cattle isolates. Other prevalent plasmids were Col440II, present in 35.1% of isolates, Col440I in 24.5% of isolates, RepA1pKPC-CAV1321 in 13.8% of isolates, IncHI2A and IncHI2 in 12.8% of isolates (75% in swine isolates) and others in lower frequency. The presence of the mcr-9.1 colistin resistance gene and the presence of the ESBL blaSHV-12 gene were correlated with the presence of the two plasmids IncHI2A and IncHI2.
Isolates were classified into two distinct sequence types (ST) based on MLST analysis from genome sequences. Ninety-two isolates (97.9%) belonged to ST14 and only two isolates (2.1%) belonged to ST185. Both isolates in ST185 were from cattle. ST14 and ST185 share no common alleles at any of the seven loci that define an allelic profile or ST by MLST analysis.
S. Senftenberg is a polyphyletic serovar. Phylogenetic analysis identified two distinct lineages of S. Senftenberg in this study that correlated with the MLST results. The smaller clade corresponded to ST185, and the majority of isolates corresponded to ST14 (Supplementary Figure S1).
The addition of closely related representative serotypes from NCBI shows that the two MLST types observed in this study are entirely distinct lineages; and in addition, a third lineage of S. Senftenberg which was not observed in this study, becomes visible with isolate NZ_CP007505. This suggests that the serotype designation may not represent a good indicator of the genetic relationships between strains.
The isolates corresponding to ST14 showed a cluster of nine isolates that are significantly divergent from the rest of the isolates. This cluster has accumulated 137 SNPs since sharing a most recent common ancestor with the nearest relatives. Of interest was the presence of common resistance characteristics in this cluster, with eight isolates from poultry and one isolate from cattle showing a similar antimicrobial resistance pattern, resistance genes, class 1 integron, and plasmid profile (Supplementary Figure S2; Table 4). The well differentiated cluster within the ST14 group showed an average distance of 30.8 SNPs (range of 22 to 44 SNP) from a common ancestor.
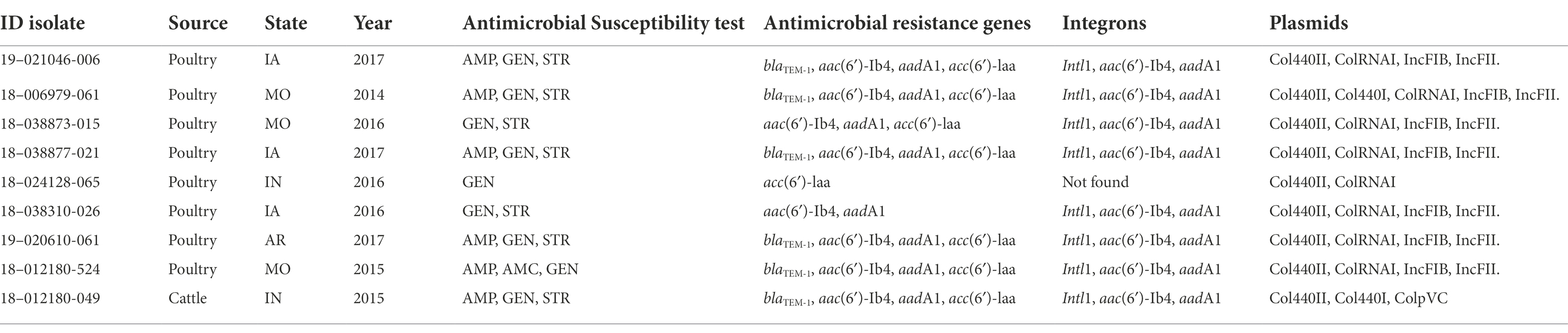
Table 4. Antimicrobial resistance profile, integrons and plasmids in the ST14 differentiated group of 9 isolates.
A previous study of animal and human isolates in the USA (Stepan et al., 2011) showed that human strains of S. Senftenberg were susceptible to all of the antimicrobials tested, whereas the animal isolates showed a range of resistance, with most isolates being resistant to two or more antimicrobials.
In this study, isolates from swine and cattle showed resistance to 13 antimicrobials in different frequencies, whereas poultry isolates showed resistance to only six antimicrobials tested; these results differ from Stepan et al. (Stepan et al., 2011) where the rate of resistance to antimicrobials was similar across the host species (swine, cattle and poultry). Other important findings of our study were the presence of three isolates from swine resistant to 12 antibiotics and one resistant to 13 antibiotics.
In this study we found two sequence types associated with S. Senftenberg (ST14 = 97.8% and ST185 = 2.1%), whereas Stepan et al. (Stepan et al., 2011) found three sequence types (ST14 = 85.7%, ST185 = 13.2% and ST145 = 1%), with ST145 only found in one isolate from swine. The two lineages identified in our study and the branches within the lineage corresponding to ST14 did not show host specificity. This information, combined with the frequent isolation of this serotype from feed, may indicate that the serotype is more likely to be introduced from a common external source rather than circulating long-term within animal populations. In a study in China, El Ghany et al. (El Ghany et al., 2016) identified two phylogenetically distinct clades of S. Senftenberg by SNP analysis. Variations were in the Salmonella pathogenicity island (SPI)-1 and SPI-2 that exhibited distinct biochemical and phenotypic signatures. Clade 1 isolates comprised three sequence types: ST185, ST217, and ST1751, being single or double locus variants relative to one another. In contrast, clade 2 isolates included only ST14. In our study, the two distinct lineages also differ in the ST. Even though there are other STs observed in other datasets, these STs are single or double locus variants relative to one another.
We observed that there are other serotypes that fall between the two lineages, indicating that they are two completely independent lineages in S. Senftenberg and not a single serotype that diverged over time.
As may be expected based on the phylogenetic diversity, resistance patterns among S. Senftenberg isolates differ significantly among isolates. In our study, the most common resistance profile was AMP, GEN, STR (n = 5 isolates), whereas Stepan et al. (Stepan et al., 2011) found STR, TET, SXT the most common (n = 4 isolates).
In Veeraraghavan et al. study (Veeraraghavan et al., 2019) beta-lactam resistance was associated with the presence of the blaTEM-1, blaOXA-9, blaCMY-2, and blaNDM-1 genes, resistance to aminoglycosides was associated with five genes, namely aac(6′)-Ia, aac(6′)-Ib, aph(3′′)-Ib, aph(6′)-Ib and ant(2′′)-Ia, and sulfonamide resistance was associated with the sul1 and sul2 genes and resistance to chloramphenicol with the florR gene. In our study, we found blaTEM-1, blaCMY-2 and blaSHV-12. The blaSHV-12 gene, an ESBL that shows resistance to ceftriaxone, was found in five isolates from swine. These five isolates also carried other antimicrobial genes against beta-lactams, aminoglycosides, sulfonamides, tetracyclines and rifamycin: blaTEM-1, aac(3)-II, aac(6’)-IIc, aac(6’)-Ib, sul1, sul2, tetD, arr-269927220, and all of them were positive for IncHI2A, IncHI2, RepA pKPC-CAV1321 plasmids. IncHI2 plasmid replicon has been reported to encode blaSHV-12, the most predominant ESBL within Enterobacteriaceae (Liakopoulos et al., 2018), and can be transfer among diverse bacterial population. Expanded monitoring of Salmonella from swine for this gene would be appropriate to evaluate the extent of the gene in the U.S. swine population to determine if swine are a significant reservoir of ceftriaxone-resistance. In general, both aph(3′′)-Ib and aph(6)-Id genes were predominantly found in streptomycin-resistant isolates in other MDR S. enterica (Cohen et al., 2020) and we found them in 29.8% isolates. Additionally, these 2 genes are found together in an HI type plasmid (McMillan et al., 2019). The acc(6′)-Iaa gene that confers resistance to tobramycin, kanamycin and amikacin, and that was found in all our isolates, was previously found in S. Typhimurium and S. Infantis; but it seems to have no clinical significance and evolutionary advantage (Jovčić et al., 2020). In our study sul1 and sul2 genes were also associated with sulfonamide resistance, and floR gene the most prevalent associated with chloramphenicol resistance, although other genes also were found.
In an XDR S. Senftenberg isolate from a human clinical case in India (Veeraraghavan et al., 2019), fluoroquinolone resistance was attributed to substitutions in the gyrA (S83Y, D87G) and parC (S80I) genes. The parC substitution appears to be a characteristic mutation present in quinolone-resistant S. Senftenberg isolates from human cases (Whichard et al., 2007). In the Wichard study (Whichard et al., 2007), all S. Senftenberg isolates had parC mutations (T57S and S80I). We also found parC mutations in all isolates; but in contrast to the human isolates, we found S83S and D87N mutations in gyrA, and only the T57S mutation in parC in the animal isolates. The widespread presence of parC mutations without corresponding resistance does not appear to be specific to S. Senftenberg strains. One study of serotype Paratyphi (Qian et al., 2020) showed parC mutations in all isolates (n = 8) with 7 isolates being susceptible to ciprofloxacin. This clearly illustrates that the presence of mutations does not necessarily correspond to phenotypic resistance (Sáenz et al., 2003). On the other hand, one study in nontyphoidal S. enterica isolated from pigs in Thailand showed resistance to fluoroquinolones, either in the presence or absence of genes and/or mutations, and parC mutation (T57) was found in 62.4% of the isolates (Poomchuchit et al., 2021).
When we compared our S. Senftenberg results with a study that evaluated antimicrobial susceptibility patterns found in other serovars isolated from poultry (Cohen et al., 2020), the percentage of MDR in S. Senftenberg from poultry (12.5%) was higher than S. Orion (10%) and lower than S. Kentucky (97.4%), S. Hadar (80%), S. Java (75%), S. Infantis (60%), S. Bredeney (50%), S. Montevideo (40%), S. Newport (30%), S. Virchow (30%), S. Blockeley (27.3%), S. Muenchen (25%). In MDR swine Salmonella isolates, Argüello et al. (Argüello et al., 2018) demonstrated the importance of class 1 integrons and certain genes. In this study, 60% (n = 30) of isolates from swine origin showed MDR, and in 70% (n = 21) of these isolates we observed the presence of a class 1 integron.
The number of plasmids did not correlate with MDR or XDR, as we found isolates with as many as seven or eight plasmids that were resistant to only one or two antibiotics and isolates with only one plasmid with resistance to seven antibiotics.
Of the other genes found in S. Senftenberg strains, the presence of the mcr-9 gene that confers resistance to colistin was of interest. This is a novel mcr homologue detected in MDR colistin-susceptible Salmonella Typhimurium isolated from a patient in the USA in 2010 (Carroll et al., 2019). The mcr-9 gene was shown to be capable of conferring phenotypic resistance to colistin in numerous genera of Enterobacteriaceae, and it is harbored in IncHI2 and/or IncHI2A replicons (Carroll et al., 2019). The Sensititre CMV4AGNF plate did not include colistin, so we were unable to determine if this gene was expressed in the S. Senftenberg isolates. However, a set of 57 Salmonella isolates, including four S. Senftenberg swine isolates used in the current study, were positive for mcr-9 when tested using the Sensititre GNX3F plate that contains colistin (unpublished data). Of the 57 isolates, only one had an MIC of 2 ng/μl (resistant), and the remaining 56 had an MIC of 1 ng/μl or lower (susceptible), with the four isolates from this current study having MIC values equal to or lower than 0.5 ng/μl. These results agree with the report of Tyson et al. (Tyson et al., 2020) affirming that the mcr-9 gene in Salmonella is not associated with colistin resistance in the USA. All isolates positive for this gene in our study carried two plasmids (IncHI2A and IncHI2), while all other isolates were negative for these two plasmids, so we could associate the presence of these plasmids with the presence of the mcr-9 gene. In other studies, the mcr-1 gene was found in IncHI2/ST3, IncI2, and IncX4 plasmids in isolates from animals and humans (Stefaniuk and Tyski, 2019).
In addition to antimicrobial resistance genes, antiseptic resistance genes are important because disinfectants are used in farm environments. Benzalkonium chloride is a surface-active QAC (quaternary ammonium compound), and it is used as a farm disinfectant. The qacEΔ1 is frequently present in E. coli and other enteric bacteria (Zou et al., 2014). In this study 25 isolates carried the qacEΔ1 gene that has been identified in mobile genetic elements. The qacE and qacEΔ1 genes are located on an integron, a qacEΔ1 represents a disrupted form of qacE that evolved as a result of the insertion of a DNA segment near the 3′ end of the qacE gene carrying a sul1 sulfonamide resistance determinant (Paulsen et al., 1993). All but one of the S. Senftenberg isolates positive for qacEΔ1 gene carried the sul1 gene. The one exception carried the sul2 and sul3 genes. We also detected the presence of class 1 integrons in the qacEΔ1 positive isolates. Class 1 integrons are associated with an Intl1 integrase in the 5′ conserved sequence (CS) and with a 3′ CS conferring resistance to antibiotics (sulfonamides) and bactericidal compounds (quaternary ammonium) of the integron (Deng et al., 2015). Class 1 integrons have previously been detected in S. Senftenberg (Vo et al., 2006), but to our knowledge, there are no reports of class 2 or class 3 integrons in this serovar. We found 10 isolates harboring a class 1 integron homologous to the In48 integron (98.83%) that carries the aminoglycoside 6’-N-acetyltransferase (aacA4) gene. In our isolates, this integron carried the aminoglycoside N-acetyltransferase aac(6′)-Ib4 gene (aac(6′)-Ib allele) and the aadA1 gene. The frequent use of QAC may facilitate resistance to disinfectants, and QACs may serve as important selective agents in MDR pathogens (Sinwat et al., 2021). In the present study, all S. Senftenberg isolates carrying the qacEΔ1 gene were MDR.
Concordance between the presence of antimicrobial resistance genes and phenotypic resistance profiles were seen in 98.1% of isolates for the antimicrobials tested. The presence of resistance genes do not necessarily confer phenotypic resistance, and occasionally, an isolate will display antimicrobial resistance without the presence of a known resistance gene (Piddock, 2016). The presence or absence of resistance genes is not enough for the phenomenon of antimicrobial resistance. Mechanisms such as enzyme activation, target modification or protection, regulation of gene expression, or changes in the cell wall can play an important role in the resistance of antimicrobials (Paudyal et al., 2019).
This study provided an analysis of retrospective data of Salmonella Senftenberg and information about the antimicrobial susceptibility and genomic characteristics in diagnostic isolates of S. Senftenberg from production animals in the USA. This study reports the genotype-phenotype homogeneity and variability of S. Senftenberg of animal origin. The ability of S. Senftenberg to persist in the environment, to cause disease in various animal species, to be a potential risk of transmission to humans and to harbor resistance to critical antimicrobial and mobile elements capable of dissemination of acquired resistance genes makes S. Senftenberg an important public health pathogen. In this study we found that 39.4% of the isolates tested displayed multidrug resistance, and four isolates were potentially extensively drug resistant. This has important implications for both animal and human health, due to possible transmission of MDR bacteria from animal to animal or animal to human (zoonotic), and the difficulty in treatment of resistant bacteria. These data highlight the need to strengthen surveillance to detect the prevalence and transmission of nontyphoidal Salmonella species because of the emergence of MDR strains. It is critical to identify the emergence of these strains as early as possible to avoid further dissemination and establish control procedures. This data is useful for future studies on S. Senftenberg and to further understand this pathogen as few studies exist on the antimicrobial susceptibility, genotypic profiles and genetic diversity of Salmonella Senftenberg of animal origin.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, bioproject/PRJNA785813.
MS and LS: conceptualization. MS and JH: methodology, formal analysis, and visualization. JH: software, formal analysis and validation. MS: investigation and writing – original draft preparation. LS: resources, supervision, project administration and funding acquisition. MS, JH, BM, TM and LS: writing – review and editing. LS: Supervision. All authors contributed to the article and approved the submitted version.
This project was supported in part by an appointment to the Research Participation Program at the Animal and Plant Health Inspection Service, United States Department of Agriculture, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and USDA APHIS.
We are grateful to Claudia Perea for assistance with the phylogenetic trees.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.979790/full#supplementary-material
Argüello, H., Guerra, B., Rodríguez, I., Rubio, P., and Carvajal, A. (2018). Characterization of antimicrobial resistance determinants and class 1 and class 2 Integrons in Salmonella enterica spp., multidrug-resistant isolates from pigs. Genes (Basel) 9:256. doi: 10.3390/genes9050256
Boumart, Z., Roche, S. M., Lalande, F., Virlogeux-Payant, I., Hennequet-Antier3, C., Menanteau, P., et al. (2012). Heterogeneity of Persistence of Salmonella enterica Serotype Senftenberg strains could explain the emergence of this serotype in poultry flocks. PLoS One 7:e35782. doi: 10.1371/journal.pone.0035782
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–903. doi: 10.1128/AAC.02412-14
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio 10, e00853–e00819. doi: 10.1128/mBio.00853-19
Center for Genomic Epidemiology (1998). Technical university of Denmark, Kongens. Lyngby, Denmark. https://cge.cbs.dtu.dk/services/IntFinder-1.0/ (Accessed April 2022).
Centers for disease Control and Prevention (2010). Multistate Outbreak of human Salmonella Montevideo infections (final update). Available at: https://www.cdc.gov/Salmonella/2010/montevideo-5-4-2010.html
Centers for Disease Control and Prevention (2016). Multistate outbreak of Salmonella Montevideo and Salmonella Senftenberg infections linked to wonderful pistachios (final update). Available at: https://www.cdc.gov/Salmonella/montevideo-03-16/index.html
Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Foodborne, Waterborne, and Environmental Diseases (DFWED) (2022). Salmonella Outbreak Linked to Peanut Butter. Available at: https://www.cdc.gov/Salmonella/senftenberg-05-22/index.html
Cohen, E., Davidovich, M., Rokney, A., Valinsky, L., Rahav, G., and Gal-Mor, O. (2020). Emergence of new variants of antibiotic resistance genomic islands among multidrug-resistant Salmonella enterica in poultry. Environ. Microbiol. 22, 413–432. doi: 10.1111/1462-2920.14858
Deng, Y., Bao, X., Ji, L., Chen, L., Liu, J., Miao, J., et al. (2015). Resistance integrons: class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 14:1. doi: 10.1186/s12941-015-0100-6
Doyle, M. E., and Mazzotta, A. S. (1999). Review of studies on the thermal resistance of Salmonellae. J. Food Prot. 63, 779–795. doi: 10.4315/0362-028x-63.6.779
Dunbar, S. A., Ritchie, V. B., Hoffmeyer, M. R., Rana, G. S., and Zhang, H. (2015). Luminex(®) multiplex bead suspension arrays for the detection and serotyping of Salmonella spp. Methods Mol. Biol. 1225, 1–27. doi: 10.1007/978-1-4939-1625-2_1
El Ghany, M. A., Shi, X., Li, Y., Ansari, H. R., Hill-Cawthorne, G. A., Ho, Y. S., et al. (2016). Genomic and phenotypic analyses reveal the emergence of an atypical Salmonella enterica Serovar Senftenberg variant in China. J. Clin. Microbiol. 54, 2014–2022. doi: 10.1128/JCM.00052-16
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63, e00483–e00419. doi: 10.1128/AAC.00483-19, published correction appears in Antimicrob Agents Chemother. 64(4):e00361-20
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Grimont, P. A. D., and Weill, F. X. (2007). Antigenic Formulae of the Salmonella Serovars, 9th Edn, Geneva: World Health Organization.
Haendiges, J., Davidson, G. R., Pettengill, J. B., Reed, E., Ramachandran, P., Blessington, T., et al. (2019). Genomic evidence of environmental and resident Salmonella Senftenberg and Montevideo contamination in the pistachio supply-chain. PLoS One 16:e0259471. doi: 10.1371/journal.pone.0259471
Hassan, R., Whitney, B., Williams, D. L., Holloman, K., Grady, D., Thomas, D., et al. (2019). Multistate outbreaks of Salmonella infections linked to imported Maradol papayas – United States, December 2016–September 2017. Epidemiol. Infect. 147, e265–e268. doi: 10.1017/S0950268819001547
Hendriksen, R. S., Joensen, K. G., Lukwesa-Musyani, K., Leekitcharoenphon, A. P., Nakazwe, R., Aarestrup, F. M., et al. (2013). Extremely drug-resistant Salmonella enterica Serovar Senftenberg infections in patients in Zambia. J. Clin. Microbiol. 51, 284–286. doi: 10.1128/JCM.02227-12
Jovčić, B., Novović, K., Filipić, B., Velhner, M., Todorović, D., Matović, K., et al. (2020). Genomic characteristics of Colistin-resistant Salmonella enterica subsp. enterica Serovar Infantis from poultry farms in the republic of Serbia. Antibiotics (Basel). 9:886. doi: 10.3390/antibiotics9120886
Liakopoulos, A., van der Goot, J., Bossers, A., Betts, J., Brouwer, M., Kant, A., et al. (2018). Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 8:7674. doi: 10.1038/s41598-018-26073-5
Li, X., Bethune, L. A., Jia, Y., Lovell, R. A., Proescholdt, T. A., Benz, S. A., et al. (2012). Surveillance of Salmonella prevalence in animal feeds and characterization of the Salmonella isolates by serotyping and antimicrobial susceptibility. Foodborne Pathog. Dis. 9, 692–698. doi: 10.1089/fpd.2011.1083
Loaiza, K., Ortega-Paredes, D., Kristofer Johansson, M. H., and Ferrer Florensa, A. (2020). IntFinder: a freely-available user-friendly web tool for detection of class 1 integrons in next-generation sequencing data using k-mer alignment. ECCMID 2020. Available at: https://www.escmid.org/escmid_publications/eccmid_abstract_book
Magiorakos, A., Srinivasan, R. B., Carey, Y., Carmeli, M. E., Falagas, C. G., Giske, S., et al. (2012). Multidrug-resistant, extensively drug-resistant, and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
McDermott, P. F., Tyson, G. H., Kabera, C., Chen, Y., Li, C., Folster, J. P., et al. (2016). Whole-genome sequencing for detecting antimicrobial resistance in Nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60, 5515–5520. doi: 10.1128/AAC.01030-16
McMillan, E. A., Gupta, S. K., Williams, L. E., Jove, T., Hiott, L. M., Woodley, T. A., et al. (2019). Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 10:832. doi: 10.3389/fmicb.2019.00832
Nair, V. T., Venkitanarayanan, D. K., and Kollanoor Johny, A. (2018). Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods (Basel, Switzerland). 7:167. doi: 10.3390/foods7100167
Paudyal, N., Pan, H., Elbediwi, M., Zhou, X., Peng, X., Li, X., et al. (2019). Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. 19:226. doi: 10.1186/s12866-019-1598-0
Paulsen, I. T., Littlejohn, T. G., Rådström, P., Sundström, L., Sköld, O., Swedberg, G., et al. (1993). The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 37, 761–768. doi: 10.1128/AAC.37.4.761
Pedersen, T. B., Olsen, J. E., and Bisgaard, M. (2008). Persistence of Salmonella Senftenberg in poultry production environments and investigation of its resistance to desiccation. Avian Pathol. 37, 421–427. doi: 10.1080/03079450802216561
Piddock, L. J. (2016). Assess drug-resistance phenotypes, not just genotypes. Nat. Microbiol. 1, 16120. doi: 10.1038/nmicrobiol.2016.120
Poomchuchit, S., Kerdsin, A., Chopjitt, P., Boueroy, P., Hatrongjit, R., Akeda, Y., et al. (2021). Fluoroquinolone resistance in non-typhoidal Salmonella enterica isolated from slaughtered pigs in Thailand. J. Med. Microbiol. 70:001386. doi: 10.1099/jmm.0.001386
Qian, H., Cheng, S., Liu, G., Tan, Z., Dong, C., Bao, J., et al. (2020). Discovery of seven novel mutations of gyrB, parC and pare in Salmonella Typhi and Paratyphi strains from Jiangsu Province of China. Sci. Rep. 10:7359. doi: 10.1038/s41598-020-64346-0
Roer, L., Hendriksen, R. S., Leekitcharoenphon, P., Lukjancenko, O., Sommer, K., Hasman, H., et al. (2016). Is the evolution of Salmonella enterica subsp. enterica linked to restriction-modification systems? mSystems 1, e00009–e00016. doi: 10.1128/mSystems.00009-16
Sáenz, Y., Zarazaga, M., Briñas, L., Ruiz-Larrea, F., and Torres, C. (2003). Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J. Antimicrob. Chemother. 51, 1001–1005. doi: 10.1093/jac/dkg168
Sinwat, N., Witoonsatian, K., Chumsing, S., Suwanwong, M., Kankuntod, S., Jirawattanapong, P., et al. (2021). Antimicrobial resistance phenotypes and genotypes of Salmonella spp. isolated from commercial duck meat production in Thailand and their minimal inhibitory concentration of disinfectants. Microb. Drug Resist. 27, 1733–1741. doi: 10.1089/mdr.2020.0230. Epub 2021 Jun 1
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stefaniuk, E. M., and Tyski, S. (2019). Colistin resistance in Enterobacterales strains – A current view. Pol. J. Microbiol. 68, 417–427. doi: 10.33073/pjm-2019-055
Stepan, R. M., Sherwood, J. S., Peterman, S. R., and Logue, C. M. (2011). Molecular and comparative analysis of Salmonella enterica Senftenberg from humans and animals using PFGE, MLST and NERMS. BMC Microbiol. 11:153. doi: 10.1186/1471-2180-11-153
Stokes, H. W., and Gillings, M. R. (2011). Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819. doi: 10.1111/j.1574-6976.2011.00273.x
Switt, A. I. M. D. (2019). Salmonella Senftenberg. https://confluence.cornell.edu.
Tyson, G. H., Li, C., Hsu, C. H., Ayers, S., Borenstein, S., Mukherjee, S., et al. (2020). The mcr-9 gene of Salmonella and Escherichia coli is not associated with Colistin resistance in the United States. Antimicrob. Agents Chemother. 64, e00573–e00520. doi: 10.1128/AAC.00573-20
US Food and Drug Administration (2014). FDA investigation summary—multistate outbreak of Salmonella Senftenberg infections associated with pistachios from a California roaster. Available at: http://wayback.archive-it.org/7993/20171114154922/, https://www.fda.gov/Food/RecallsOutbreaksEmergencies/Outbreaks/ucm386377.htm.
US Food and Drug Administration (2021). Interpretative Criteria for Susceptibility Testing. Available at: https://fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/resources (Accessed August 2021).
Veeraraghavan, B., Jacob, J. J., Prakash, J. A. J., Pragasam, A. K., Neeravi, A., Narasimman, V., et al. (2019). Extensive drug resistant Salmonella enterica serovar Senftenberg carrying blaNDM encoding plasmid p5558 (IncA/C) from India. Pathogens Global Health 113, 20–26. doi: 10.1080/20477724.2019.1574112
Vo, A. T. T., van Duijken, E., Fluit, A. C., Wannet, W. J. B., Verbruggen, A. J., Maas, H. M. E., et al. (2006). Antibiotic resistance, integrons and Salmonella genomic island 1 among non-typhoidal Salmonella serovars in The Netherlands. Int. J. Antimicrob. Agents 28, 172–179. doi: 10.1016/j.ijantimicag.2006.05.027
Whichard, J. M., Gay, K., Stevenson, K. E., Joyce, K. J., Cooper, K. L., Omondi, M., et al. (2007). Human Salmonella and concurrent decreased susceptibility to quinolones and extended-Spectrum Cephalosporins. Emerg. Infect. Dis. 13, 1681–1688. doi: 10.3201/eid1311.061438
Yasir, M., Farman, M., Shah, M. W., Jiman-Fatni, A. A., Othman, N. A., Almasaudi, S. B., et al. (2020). Genomic and antimicrobial resistance genes diversity in multidrug CTX-M-positive isolates of Escherichia coli at a health care facility in Jeddah. J. Infect. Public Health 13, 94–100. doi: 10.1016/j.jiph.2019.06.011
Zankari, E., Allesøe, R., Joensen, F. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: antimicrobial resistance, whole genome sequencing, phylogeny, resistance genes, Salmonella Senftenberg
Citation: Srednik ME, Morningstar-Shaw BR, Hicks JA, Mackie TA and Schlater LK (2022) Antimicrobial resistance and genomic characterization of Salmonella enterica serovar Senftenberg isolates in production animals from the United States. Front. Microbiol. 13:979790. doi: 10.3389/fmicb.2022.979790
Received: 27 June 2022; Accepted: 02 August 2022;
Published: 03 November 2022.
Edited by:
Svetlana Khaiboullina, University of Nevada, Reno,United StatesReviewed by:
Anusak Kerdsin, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus, ThailandCopyright © 2022 Srednik, Morningstar-Shaw, Hicks, Mackie and Schlater. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariela E. Srednik, bWFyaWVsYS5zcmVkbmlrQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.