
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 August 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.977669
This article is part of the Research Topic Insights into the Molecular Studies of Plant Growth-Promoting Microorganisms for Sustainable Agricultural Production View all 6 articles
 Chintan Kapadia1*
Chintan Kapadia1* Rinkal Kachhdia1
Rinkal Kachhdia1 Susheel Singh2
Susheel Singh2 Kelvin Gandhi2
Kelvin Gandhi2 Peter Poczai3*
Peter Poczai3* Saleh Alfarraj4
Saleh Alfarraj4 Mohammad Javed Ansari5
Mohammad Javed Ansari5 Abdul Gafur6
Abdul Gafur6 R. Z. Sayyed7*
R. Z. Sayyed7*The quorum-sensing (QS) cascade is responsible for the colonization and phenotypic behavior of the pathogenic organism and the regulation of diverse signal molecules. The disruption of the quorum-sensing system is an effective strategy to overcome the possibility of antibiotic resistance development in the pathogen. The quorum quenching does not kill the microbes. Instead, it hinders the expression of pathogenic traits. In the present experiment, Pseudomonas aeruginosa RKC1 was used to extract the metabolites responsible for quorum-sensing inhibition in soft rot pathogen Lelliottia amnigena RCE. During the initial screening, P. aeruginosa RKC1 was found to be most promising and inhibits violacein of Chromobacterium violaceum MTCC2656 pyocyanin, swarming-swimming motility of P. aeruginosa MTCC2297. The characterization of metabolites produced by the microbes which are responsible for quorum-sensing inhibition through GC-MS is very scarce in scientific literature. The ethyl acetate extract of P. aeruginosa RKC1 inhibits biofilm formation of L. amnigena RCE while inhibiting growth at higher concentrations. The GC-MS analysis suggested that Cyclic dipeptides (CDPs) such as Cyclo (L-prolyl-L-valine), Cyclo (Pro-Leu), and Cyclo(D-phenylalanyl-L-prolyl) were predominantly found in the ethyl acetate extract of the P. aeruginosa RKC1 (93.72%). This diketopiperazine (DKPs) exhibited quorum-sensing inhibition against the pathogen in liquid media during the active growth phase and regulated diverse metabolites of the pathogen. Moreover, the metabolites data from the clear zone around wells showed a higher concentration of DKSs (9.66%) compared to other metabolites. So far, very few reports indicate the role of DKPs or CDPs in inhibiting the quorum-sensing system in plant pathogenic bacteria. This is one such report that exploits metabolites of P. aeruginosa RKC1. The present investigation provided evidence to use quorum-sensing inhibitor metabolites, to suppress microbes' pathogenesis and thus develop an innovative strategy to overcome antibiotic resistance.
The uses of various anti-microbial agents lead to the death of diverse microbes. On the other hand, repeated use of such compounds develops resistance. The microorganisms quickly evolved by either changing the target molecules or change in the receptor's proteins. These survivals of fittest principles against stressful environments always ask the scientist to develop novel antibiotic agents or strategies to kill them (Wang et al., 2018). To combat pathogens, nanoparticle alone or in combination with antibiotics agents or CRISPR/Cas9 systems made it possible to control pathogens (Kapadia et al., 2021; Wan et al., 2021). To control pathogens, one can use alternative strategies like anti-biofilm peptides, competitive microbial agents, and antimicrobial peptides. To avoid the development of multi-drug resistant microbes, there is a need to develop novel approaches to control the expression of pathogenesis-related traits of bacteria while not affecting the growth of the pathogen (Laxminarayan et al., 2013; Passari et al., 2017). These virulence factors are governed by the quorum-sensing mediated cascade.
Bacteria are unicellular organisms and hence require communication signals to modify their phenotype in response to environmental conditions. Quorum sensing involves the secretion of autoinducers (N-acyl Homoserine lactone), small peptides, diffusible signal factors (DSFs), and their detection followed by transcriptional regulation. The binding of ligands or metabolites to the receptors brings out changes in the gene expression pattern. Once sufficient cell density is attained, the concentration of autoinducers simultaneously leads to gene expression modulation. Bioluminescence in Vibrio sp., Biofilm formation in Pseudomonas aeruginosa and Ralstonia solanacerum (Flavier et al., 1997), violacein production in C. violaceum (McLean et al., 2004), and virulence factor synthesis in P. aeruginosa (Nadal Jimenez et al., 2012) are governed by the quorum-sensing system. One can target auto-inducers by enzymatically hydrolyzing or modifying the signal molecules with the help of acylase, lactonase, and oxidoreductase, thus controlling the quorum-sensing system (Dong and Zhang, 2005; Tan et al., 2013; Fetzner, 2015). While in recent years, scientists have tried to isolate QS inhibitors (QSIs) metabolites from diverse microorganisms such as P. aeruginosa (Holden et al., 1999; Almohaywi et al., 2018), Streptomyces griseorubens (Çetinkaya et al., 2020), Streptomyces coelicoflavus (Hassan et al., 2016), Serratia marcescens (Padmavathi et al., 2014), Streptomyces cavourensis (Kaaniche et al., 2020), and Exiguobacterium indicum (Singh et al., 2017) against various pathogens. The compounds isolated from such bacteria competitively bind to the available receptor site in the pathogen and thus suppress the pathogenicity (Sun et al., 2016). There have been no previous reports indicating the use of bacterial extracts to control the pathogenesis of soft rot-causing pathogen Lelliottia amnigena specifically in India.
Bacterial soft rot disease is caused by Dickeya spp., Pectobacterium spp., L. amnigena, and Entrobacter spp. (Charkowski, 2018) and affects various horticultural crops during storage, transportation, and growing seasons (Hadizadeh et al., 2019). L. amnigena is a causative agent of soft rot of potatoes (Abd Elhafeez and Sayed, 2018) and onion bulb decay in China (Liu and Tang, 2016). There is significant emergence of L. amnigena infection in the world, which leads to economic losses in horticultural crops (Liu and Tang, 2016; Osei et al., 2021). Surprisingly, the infection processes of these pathogens are also signaling dependent; hence, a quorum-sensing inhibition strategy could be employed to control the virulence of the pathogens. Previous studies indicated the use of ethyl acetate extracts to isolate QSI compounds (Çetinkaya et al., 2020; Kaaniche et al., 2020).
The heterocyclic compound, 1H-pyrrole-2-carboxylic acid from Streptomyces spp., inhibits biofilm formation and pyocyanin production in P. aeruginosa PAO1 (Hassan et al., 2016). Similarly, ethyl acetate extract of Bacillus pumilus S8-07 inhibits the biofilm of P. aeruginosa PAO1 (Nithya et al., 2010). Moreover, DKPs found in the ethyl acetate extract of Brevibacterium casei inhibit biofilm formation as well as inhibit the synthesis of a virulence factor in P. aeruginosa (Rashiya et al., 2021), and 3-Benzyl-Hexahydro-Pyrrolo[1,2-a]Pyrazine-1,4-Dione from E. indicum inhibits biofilm formation in P. aeruginosa PAO1 and P. aeruginosa PAH without affecting the growth (Singh et al., 2017). DKPs such as cyclo(Trp-Ser) of Rheinheimera aquimaris inhibit QS-mediated virulence traits in P. aeruginosa PA01 (Sun et al., 2016), while DKPs such as cyclo(Ala-Val), cyclo(Pro-Tyr), and cyclo(Phe-Pro) from P. aeruginosa inhibit virulence phenotypes of Serratia liquefaciens (Holden et al., 1999). Cyclo (L-Pro-L-Pro) and cyclo (D-Pro-D-Pro) are cyclodipeptides formed through cyclodehydration of two amino acids. They are produced naturally by various microorganisms which possess diverse biological properties. These dipeptides suppress pathogenesis in tobacco by triggering stomatal closure, induction of hormone synthesis, and secretion of pathogen-related proteins. It was shown that Gram-negative bacteria produce diketopiperazine which has the ability to bind with LuxR receptors and subsequently either activate or suppress the quorum-sensing system (Holden et al., 1999). There are few reports, yet research is necessary to support the hypothesis that small cyclic dipeptides can target receptor binding sites and thus regulate virulence factors. The present study focused on screening quorum-sensing inhibiting microbes using C. violaceum MTCC2656 and consequently evaluated its anti-biofilm activity and suppression of pathogenesis-related traits. The bacterial extracts were used to evaluate their potentiality to inhibit the quorum-sensing system of soft rot causing pathogen L. amnigena and extracts were further characterized using GC-MS for identification of potential metabolites.
C. violaceum MTCC2656 and P. aeruginosa MTCC2297 were purchased from Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (Im-Tech), Chandigarh, India. In a sterilized container, the samples were collected from rhizospheric soil from Navsari Agricultural University, Navsari, India. one gram of sample was mixed with 10 ml sterilized ddH2O, and then the suspension was serially diluted from 10−3 to 10−7. Three plates were spread and incubated from each dilution at 28°C for 48 h. The distinct colony was picked up and sub-cultured on NA media (Hi-media) to obtain pure organisms. The pure colonies were stored as glycerol stock (50% V/V) at −20°C until further use. L. amnigena RCE (MZ712952) was isolated from the potato tuber and available in the lab from previous experiments (Kachhadia et al., 2022). In brief, the rotten potato tuber was purchased from the market and surface sterilized using 0.1% mercury chloride for 5 min. The soft jelly-like part was placed on NA and incubated at 28°C for 24 h (Kachhadia et al., 2022).
Monitor strain, C. violaceum MTCC2656, was used to assess the quorum quenching potential of the isolates. The 24 h old culture of monitor strain was spread evenly on the nutrient agar plate (Hi-media, India), and isolates were spot inoculated on the surface of the agar. The plates were incubated at 28°C overnight. The colorless area around the viable organisms indicated the isolates' quorum-sensing inhibitory (QSI) activity (Ma et al., 2018).
Isolates that showed positive results during plate incubation assay were exploited for their metabolites through solvent-solvent extraction. The 100 μl of 24 h old bacterial culture was inoculated in 20 ml Luria–Bertani Broth (LB) (Hi-Media, India) and incubated at 28°C; 150 rpm for 24 h. The culture suspension was centrifuged at 8,000 rpm for 15 min, and cell-free supernatant was mixed with an equal volume of ethyl acetate (Sigma-Aldrich, USA). The organic phase was dispensed in a separate tube, and extraction was repeated three times with ethyl acetate. The entire ethyl acetate fractions were mixed and evaporated under a rotary vacuum evaporator. The dry crude extracts were collected in a separate tube containing Dimethyl Sulfoxide (DMSO) and stored at −20 °C for further use (Nithya et al., 2010).
The 24-h-old culture of monitor strain C. violaceum MTCC2656 was spread evenly on the LA (Hi-Media, India), and wells were formed using an 8-mm sterile cork borer. The plates were incubated at 28°C for 24 h, and inhibitory activity was monitored by observing the colorless zone around the wells. The 100 μl (1 mg) of crude extracts were poured into the wells, and DMSO containing well was considered a negative control.
The 24 h grown culture of C. violaceum MTCC2656 was supplemented with different concentrations of crude extract (Stock; 10 mg/ml) and incubated at 28°C and 150 rpm for 24 h. The culture was centrifuged to remove supernatant, and the pellet was mixed with DMSO. The DMSO-containing suspension was centrifuged, and the supernatant was analyzed spectrophotometrically (Shimadzu Europe-UV-2600) at 585 nM. The DMSO with untreated monitor strain pellet was considered negative control (Venkatramanan et al., 2020). The experiment was repeated three times to eliminate error, and the inhibition percentage was calculated as follows:
Where, OD585 control and OD585 test are the absorbance of DMSO+monitor strain and DMSO+ crude extract + monitor strain, respectively.
The 100 μl (1 mg) crude extracts of the bacteria were supplemented with 10 ml LB media containing monitor strain (OD600; 0.400) and incubated at 28°C for 24 h at 150 rpm. The culture was centrifuged to separate cells, and the supernatant was mixed with 6.0 ml chloroform (Sigma-Aldrich). The chloroform layer containing pyocyanin was acidified using 2 ml of 0.2 N HCL (Hi-media). The OD of the pink layer was measured at 520 nM (Shimadzu UV-2600, Japan) using 0.2 N HCL as the blank (Malešević et al., 2019). Pyocyanin inhibition percentage was calculated using the following formula:
Where, the OD520 control and OD520 test are the absorbance of DMSO+ monitor strain and DMSO + monitor strain + crude extract, respectively.
The swarming and swimming motilities of P. aeruginosa MTCC2297 were analyzed using LB agar (0.5% W/V agar) plates amended with 100 μl (1 mg) of crude extract. The actively dividing cells of P. aeruginosa MTCC2297 were spot inoculated using a sterile wire stab at the center of the plate and incubated at 28°C for 24 h (Tremblay et al., 2007). DMSO alone was considered a negative control. The semisolid agar (0.3%) was supplemented with 100 μl (1 mg) of crude bacterial extract for swimming motility. The monitor strain was stabbed into the media between the bottom of the plate and the top of the agar (Zhang et al., 2018a). The motility of the cells through the surface of the agar or within the agar medium was observed, and the motility percentage was calculated using the following formula:
Where, motility area of control and motility area of the test are plates inoculated with DMSO and extracts, respectively.
The 100 μl of 24-h old (OD600; 0.4) culture of the pathogen was inoculated in the 10 ml LB broth supplemented with 100 μl (1 mg) of crude extracts and incubated at 28°C and 180 rpm in a shaking incubator. The DMSO supplementation was considered blank, and observation was recorded every 24 h intervals till 120 h.
The 50 μl of 24 h old culture (OD600; 0.4) of L. amnigena RCE was added to the 5 ml LB broth containing 100 μl (1 mg) of crude extracts and incubated at 28°C. After incubation of 24 h, the tubes were washed with ddH2O and stained with 500 μl crystal violet (0.1%) for 30 min. The tubes were again washed with ddH2O, and cells were mixed with 2 ml of 95% ethanol (Passari et al., 2016; Zhang et al., 2018b). The ethanol phase was used to determine the violet color pigmentation reduction using UV-Vis spectrophotometer at 595 nm (UV-2600, Shimadzu, Japan). The DMSO was considered a negative control.
Where, OD595 control and OD595 test are the absorbance of DMSO and DMSO+ crude extract+ pathogen, respectively.
The potato, carrot, and cucumber were purchased from the local market, and slices were made using a sterile scalpel. The slices were surface sterilized with 0.1% mercury chloride for 5 min and washed with sterile ddH2O. The slices were allowed to air dry under the aseptic inoculation chamber. The pathogen (L. amnigena RCE) and pathogen with bacterial extracts were evenly spread on the slice's center (100 μl and 200 μl). The DMSO, along with the pathogen, was considered a negative control. The slices were incubated at 28°C for 24 h under a sterile petri dish, and macerated tissue weight was measured by scooping out the macerated region (Garge and Nerurkar, 2016). Maceration (%) was calculated using the following formula:
Where control indicates pathogen with DMSO while test indicates pathogen and bacterial extracts.
The DNA of bacteria was isolated by the LSP buffer method (Sheladiya et al., 2022). The 16S rRNA gene-specific primer; 27F 5‘AGAGTTTGATCCTGGCTCAG-3' and 1492R 5‘GGTTACCTTGTTACGACTT 3' were used to amplify the target sequence of 1,500 bp. The amplified product was subjected to sequencing at SLS research Pvt. Ltd., Surat, India. The sequence result was aligned using the nBLAST tool of NCBI (National Center for Biotechnology Information). The sequence was submitted to the NCBI through the BankIt submission system to obtain a unique accession number (https://www.ncbi.nlm.nih.gov). The organisms showing more than 99% similarity were considered for constructing the phylogenetic tree using the MEGA 7 tool (Kumar et al., 2016).
The 24-h-old culture of L. amnigena was spread evenly on LA plates, and wells were formed using an 8 mm sterile borer. Hundred microliter of crude extract was loaded into the wells. The plates were incubated at 28°C for 24 h. The clear zone around the well was taken into a separate glass vial using a sterile scalpel and dipped in 5 ml of GC-grade acetonitrile (Sigma-Aldrich, USA). The samples were vortexed, centrifuged, and filtered for GC-MS analysis. The metabolites diffused through the agar and in the clear zone were analyzed. The dry bacterial extract was mixed with GC-grade acetonitrile and analyzed through GC-MS.
The 100 μl overnight grown culture of L. amnigena RCE was added to the 10 ml LB media supplemented with 100 μl (1 mg) of crude extracts. The tubes were incubated at 28°C from 24 h to 96 h at 180 rpm. The bacterial suspension was collected every 24 h interval till 96 h and centrifuged. The supernatant was mixed with an equal volume of ethyl acetate and dried under a vacuum evaporator. The dry extract was mixed with 1.0 ml GC grade acetonitrile (Sigma-Aldrich, USA).
The final extract was transferred into screw neck GC glass vial with cap and PTFE/silicone septa after filtration through 0.22 μm syringe filter to remove cell debris and preventing the accumulation of fine particles that can block chromatographic column. The chromatographic analysis was performed on Thermo Scientific made triple quadrupole GC-MS/MS system (Model: TSQ 9000) with Advanced Electron Ionization (AEI) Source. The Gas chromatograph (Model: TRACE 1300) was attached to MS system for chromatographic separation. Helium gas (purity; 99.999%) was used as carrier gas in constant flow mode (1.0 ml/min). The capillary column Restek™ Rxi-5 MS (0.25 um Thickness; 0.25 mm ID; 30 m Length) was used. The injection was done using autosampler (AI 1310). The sample was injected in split less mode while injector temperature was kept at 280°C. The temperature of transfer line and ion source temperature were set at 280 and 310°C, respectively. The oven temperature program used for separation was as follows: The initial oven temperature was kept at 90°C for 5 min, then increased at rate of 25°C/min till 180°C, then it was increased from 180 to 280°C at a rate of 5°C/min, lastly it ramped with 10°C/min until it reached 300°C followed by a hold for 1.4 minute. The total run time was 32.0 min. The Thermo Scientific™ Chromeleon™ 7.2 CDS (chromatography data system) used. The full scan mode (m/z 50–550) was used to detect the metabolites. The identification of the metabolites was done by comparing the ions detected in the scan to ions recorded in the built library i.e., The NIST (National Institute for Standard and Technology library) 2.0 Mass Spectral Library.
The mean and standard deviation was used to deduce the conclusive results of the experiment. The Metabo Analyst 5.0 (Pang et al., 2021) was used to analyze the complex metabolic pattern in each sample. The spectra and ions were matched with the known spectra present in the NIST library to get the probable name of the metabolites distinctly present in the samples. The data were first normalized and assigned common names while areas of similar chemicals were merged. The principal component analysis (PCA), partial least square discriminate analysis (PLS-DA), a heat map, and other analyses were carried out to generate diverse results (Pang et al., 2021). Venny 2.1 online tool was used to identify the common and distinct metabolites present in different samples (Oliveros, J. C. (2007–2015) (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
There were 59 isolates which showed purple discoloration of monitor strain C. violaceum MTCC2656. The positive isolates (Figure 1) were sub-cultured repeatedly to get a pure uniform colony.
Among the potent isolates, five isolates were used to extract metabolites using ethyl acetate. The isolate, RB, exhibited 12 mm of purple discoloration on well diffusion agar plate assay without affecting the growth of monitor strain; therefore, the extract was further studied (Figure 2). The concentration and different nature of compounds may be the reason for varied pigment inhibition by the extract. The diverse organisms might exhibit several strategies to survive in the environment and subsequently either inhibit or assist the growth of other microbes.
P. aeruginosa MTCC2297 has been widely employed to determine quorum-sensing inhibition properties of several bacterial extracts. During the initial assay, 50 μl (10 mg/ml) gave better results. Hence, concentration-dependent inhibition of pigment production in C. violaceum MTCC2656 was monitored by increasing the concentration of extracts. The inhibition percentage of violacein pigment is depicted in a bar graph along with their mean (Figure 3). During the quantitative assay, the higher RB bacterial crude extract concentration inhibited the violacein pigments up to 69.7%. The RB extracts also noticed the dose-dependent inhibition of pyocyanin, and it was reported to be 65.21% (Figure 4).
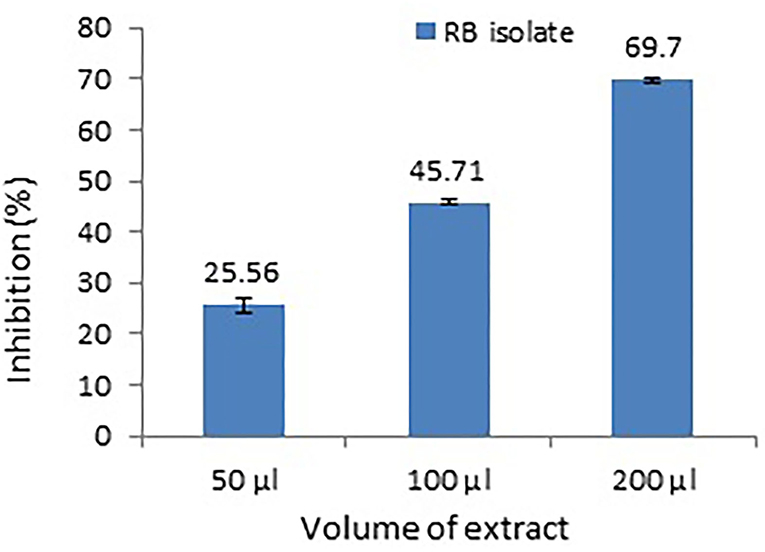
Figure 3. Dose-dependent inhibition of violacein production in a monitor strain, C. violaceum MTCC2656. Values represent the mean of three replications. Bars indicate the standard error of the mean.
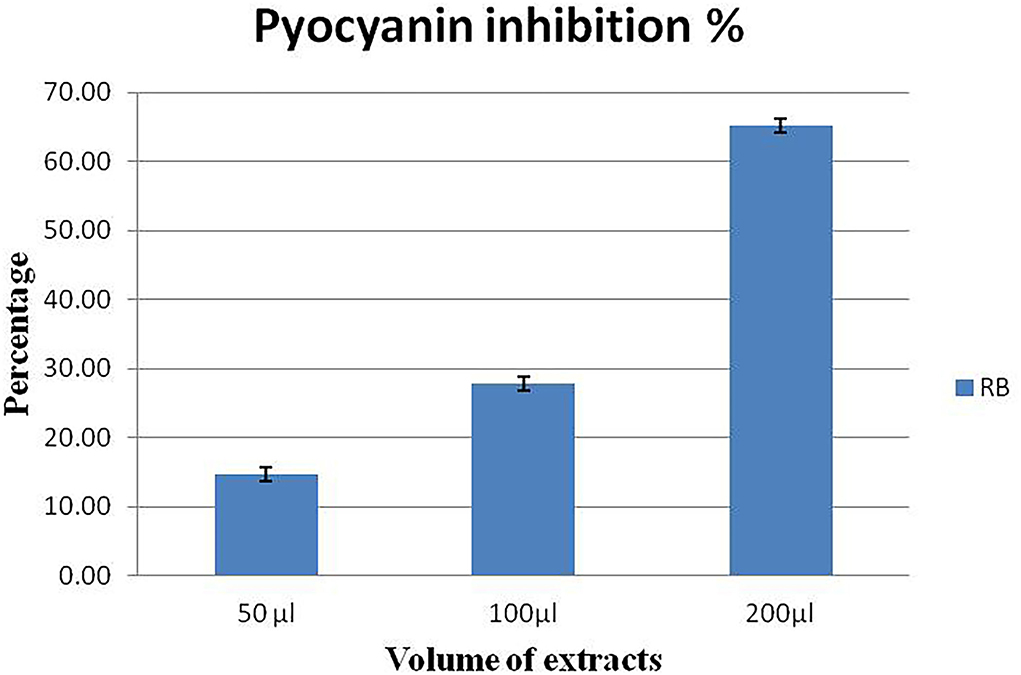
Figure 4. Concentration-dependent inhibition of pyocyanin pigment in P. aeruginosa MTCC2296. Values represent the mean of three replications. Bars indicate the standard error of the mean.
The pathogen, L. amnigena strain RCE (MZ712952), contains peritrichous flagella, which are responsible for their movement. The indirect assay was performed using P. aeruginosa MTCC2297, and motility inhibition was monitored (Figure 5). The RB extract reduced the swimming and swarming motility by 51.57 and 56.80%, respectively (Figures 5, 6). These results substantiate RB bacterial extract's potential for anti-quorum sensing.
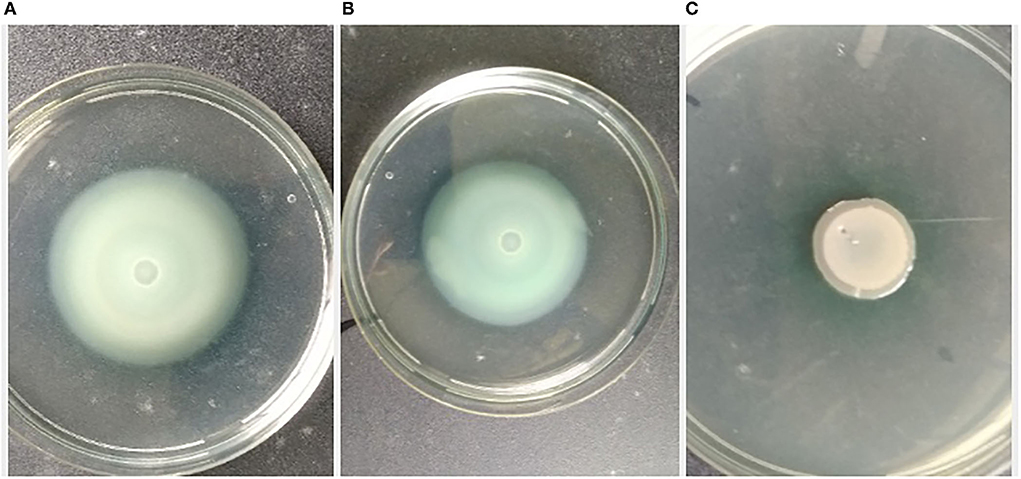
Figure 5. Effect of crude extract on swimming motility of P. aeruginosa MTCC2297; (A) P. aeruginosa 2,297 alone, (B) P. aeruginosa 2,297 + DMSO (Control), and (C) Ethyl acetate extract of RB isolates (100 μl).
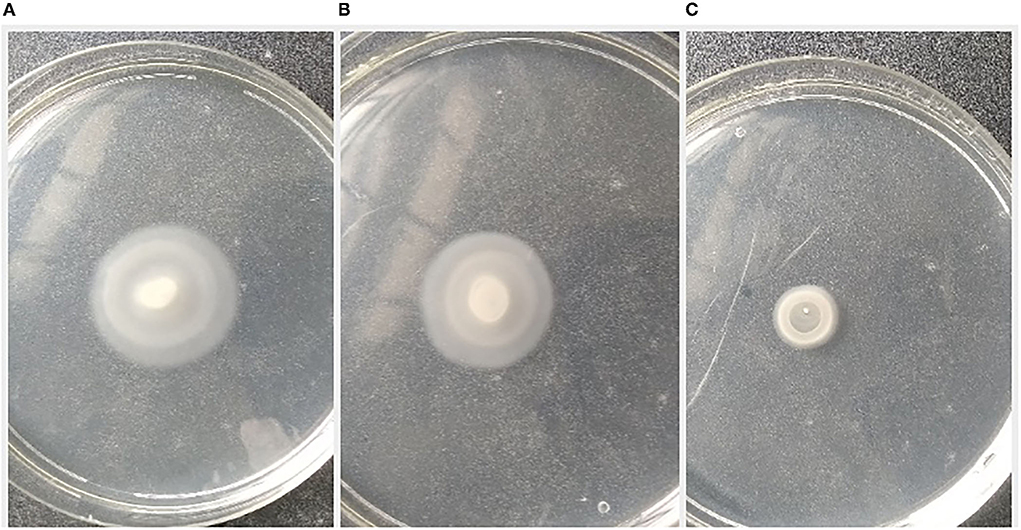
Figure 6. Effect of crude extract on swarming motility of Pseudomonas aeruginosa MTCC2297; (A) P. aeruginosa 2,297 alone, (B) P. aeruginosa 2,297 + DMSO (Control), and (C) Ethyl acetate extract of RB isolate (200 μl).
The biofilm is considered one of the potent virulence factors of the Gram-negative bacterium L. amnigena RCE. Supplementation of RB extracts to the pathogen reduces biofilm formation up to 78% even after incubation for 120 h (Figure 7). The growth curve of L. amnigena RCE alone or with DMSO (control) in the Luria broth showed similarity in their growth pattern while supplementation of crude extracts reduced the growth at the initial period of 24 h than after the growth was increased a little bit till 48 h followed by decreases further (Figure 8). This indicated that extracts might have some components that initially inhibit pathogen growth compared to control, but pathogen overcomes such effects by their metabolisms.
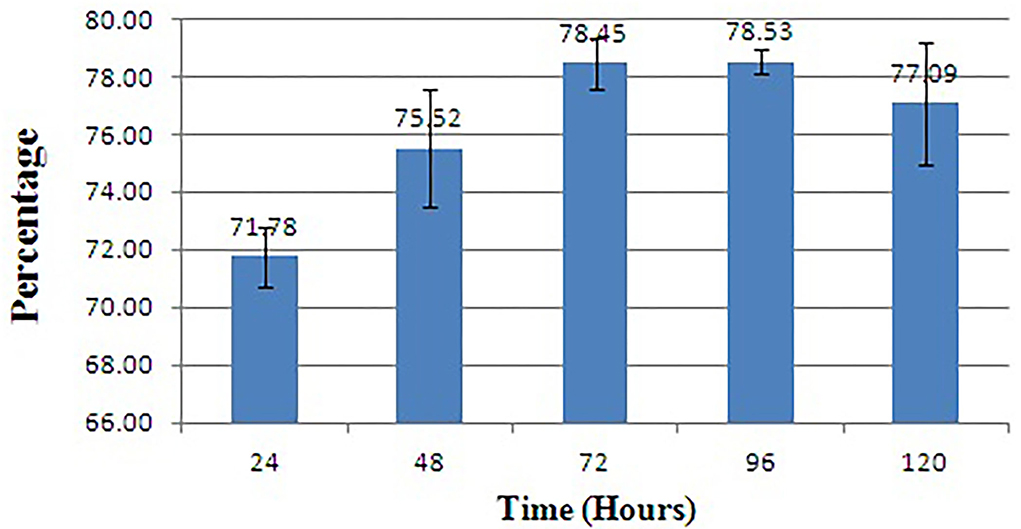
Figure 7. Biofilm inhibition of L. amnigena RCE. Values represent the mean of three replications. Bars indicate the standard error of the mean.
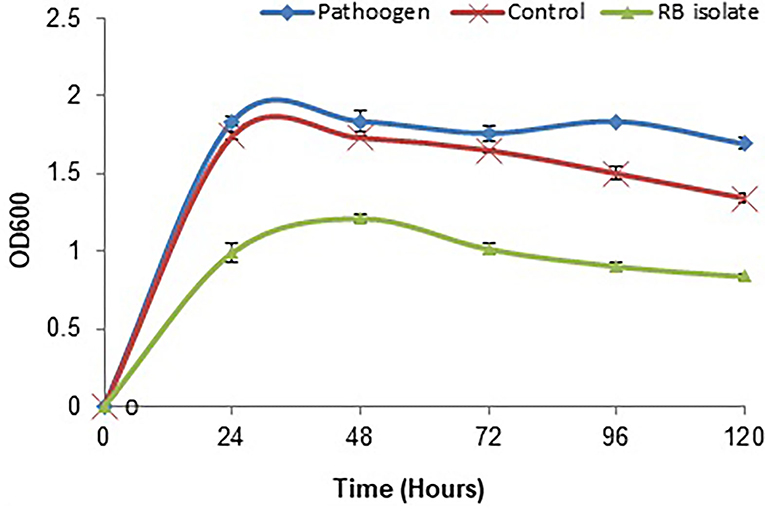
Figure 8. Growth inhibition of L. amnigena RCE by crude extract of bacteria. Values represent the mean of three replications. Bars indicate the standard error of the mean.
L. amnigena RCE is the potent soft rotting pathogen and secretes several pectate cell wall degrading enzymes. The secretion of enzymes is mediated by the quorum-sensing signal molecules of the pathogen. When potato, carrot, and cucumber slices treated with L. amnigena RCE alone or co-inoculation with RB bacterial extract, the soft rot symptoms were appeared to be reduced in a concentration-dependent manner by 95.95, 86.99, and 85.25%, respectively (Figure 9). DMSO with pathogen and pathogen alone treatments were considered as controls, showing a higher maceration percentage (Figure 10).
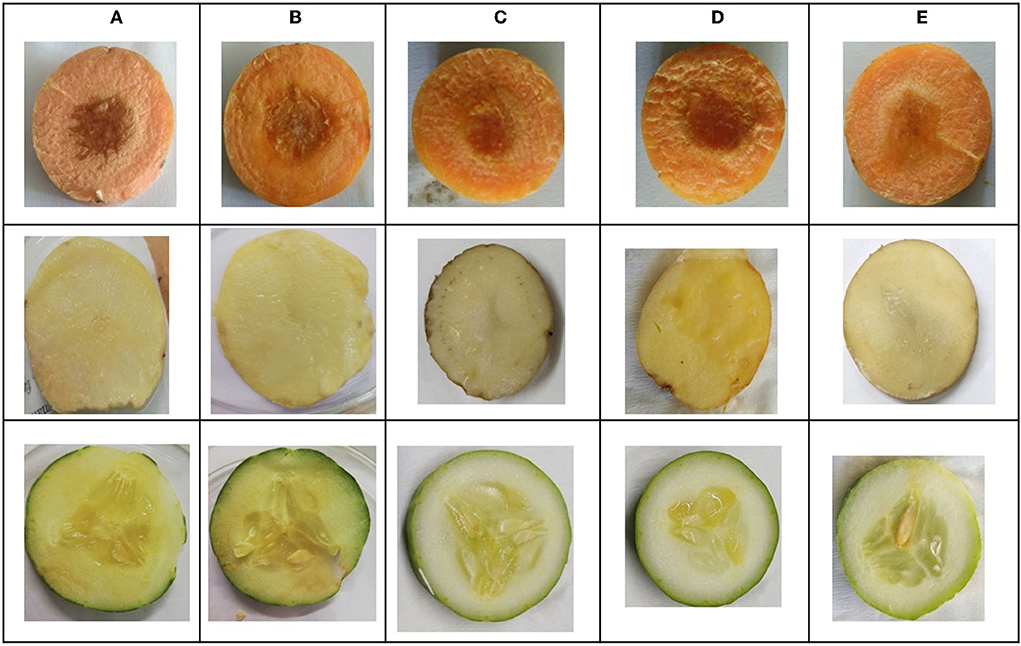
Figure 10. In vitro soft rot attenuation assay on potato, carrot, and cucumber; (A) Lelliottia amnigena alone, (B) L. amnigena RCE + DMSO (Control, 100 μl), (C) RB bacterial crude extract (100 μl) + L. amnigena RCE (100 μl), (D) L. amnigena RCE + DMSO (Control, 200 μl), and (E) RB bacterial crude extract (200 μl) + L. amnigena RCE.
The 16s rRNA gene sequence of the RB isolates was amplified and sequenced. The RB isolate showed 96.74% identity with P. aeruginosa strain DBT20 (MF421779.1). The 16s rRNA gene sequence was submitted to the genebank database under the accession number MZ068216.1. The isolate had higher sequence similarity with the respective reference strain in the database; hence RB isolate was initially named P. aeruginosa RKC1 (Supplementary Figure S1).
The experiments were carried out using the ethyl acetate extracts of P. aeruginosa RKC1, which showed quorum-sensing inhibition activities against monitor strain and pathogen L. amnigena RCE. To understand the interaction of metabolites and their resultant effects, the well diffusion assay was conducted, and the metabolic profile of extracts was analyzed using GC-MS. Moreover, molecules that travel well through the solid medium were also characterized using GC-MS (Figure 11). The Venn diagram is the easiest way to classify the distinguished metabolites in different samples. As per the Venn diagram, 66 metabolites were found to be diffused from the wells through the solid agar toward the pathogen or may be released by the pathogen, while 41 metabolites were exclusively present in the extract. There were 13 common metabolites (Table 1) found in the clear zone around the well and bacterial extracts (Figure 12). The diketopiperazines are predominantly found in extracts and a clear zone around the well, which accounts for 9.66% of the peak area.
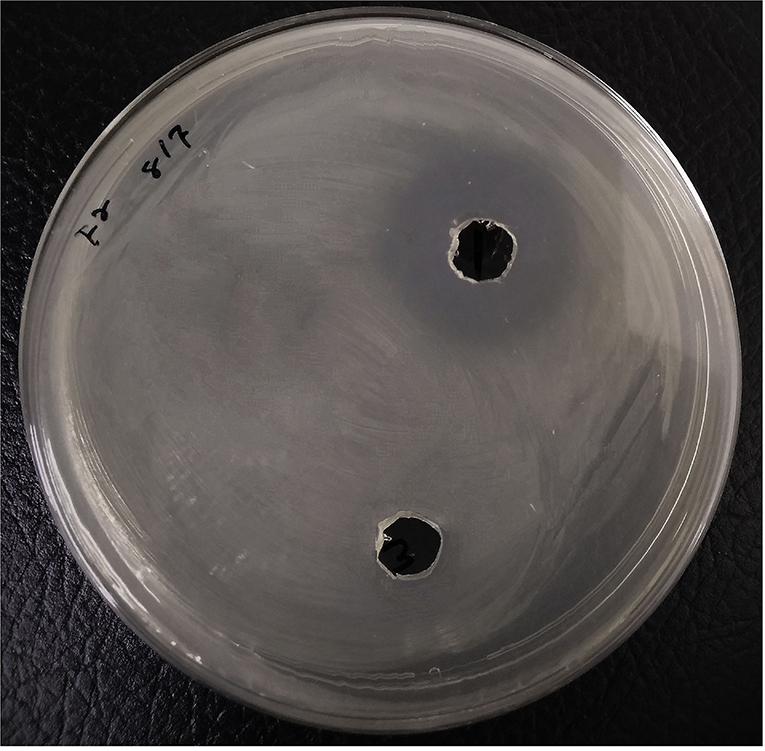
Figure 11. Agar well diffusion assay using bacterial extracts loaded in the wells and Lelliottia amnigena RCE spread over the agar. Control has the same concentration of DMSO.
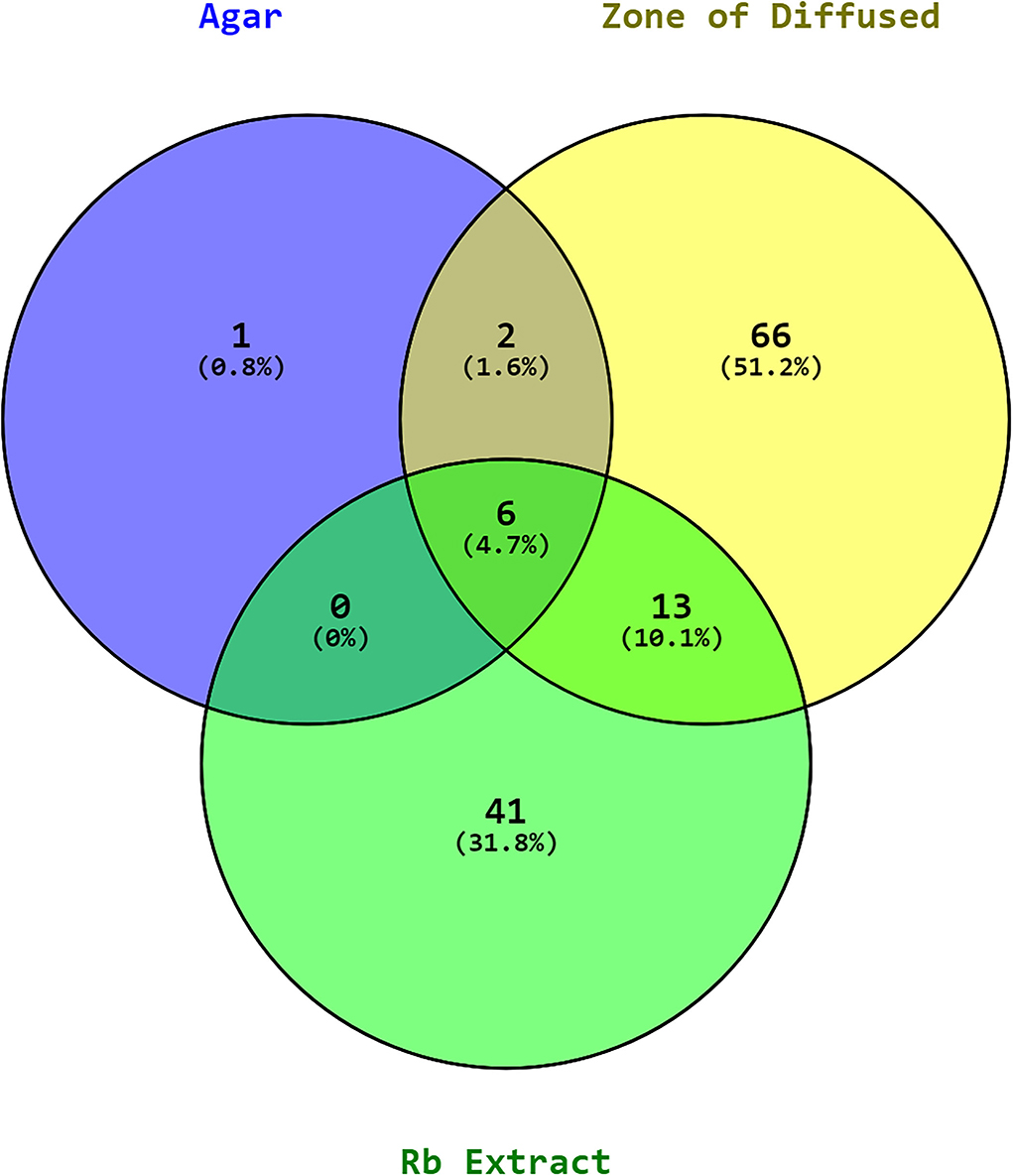
Figure 12. Venn diagram of metabolites in the RB (P. aeruginosa RKC1) extract, the clear zone around the well, and agar.
Moreover, the PLS-DA analysis of the data provides important features to draw valuable conclusions by considering VIP scores >1.5 (Supplementary Figures S2, S3). The Palmitoyl chloride, 1, 6-Dioxacyclododecane-7, 12-dione, n–Hexadecanoic acid, and 1-Cyclohexyldimethylsilyloxy-3,5-dimethyl benzene were shown to have higher peak area that indicated their secretion by the pathogen and diffused through the agar. To further confirm the hypothesis, the experiment was set up to identify differentially regulated metabolites of the pathogen at different times during interaction with P. aeruginosa RKC1 extract. Some metabolites might be present in the extract that represses certain metabolites released by the pathogen during its active growth in media (Table 2). The cyclic dipeptides, also known as 2,5-diketopiperazines (DKPs) of different types, were largely present by almost 93.72% of the total peak area in the extract of P. aeruginosa RKC1 (Table 3). These metabolites might have a key role in the quorum quenching of pathogen. The peak areas of some DKPs are very less than the extracts, and even some were not detected at all, which their lower mobility could explain through the agar (Table 4). To deduce the relevance of such metabolites during the active growth phase, the ethyl acetate extracts of bacteria were added to the pathogen, and metabolite profiling was done using GC-MS at an interval of every 24 h (Supplementary Figures S4, S5). There were various metabolites, as listed in Table 2, which were produced by the pathogen during their active growth. In contrast, the interaction of the pathogen with metabolite showed downregulation of all such metabolites, which could be inferred from the heat map and their peak area percentages (Table 3). The Gancidin W or Cyclo (L-Leu-L-Pro), 3-Benzylidene-hexahydro-pyrrolo_1_2-a_pyrazin-1_4-dione, 5-Formyluracil, Benzoic acid, 4-[(trimethylsilyl) oxy]-, phenyl ester, and 12-Hydroxy-14-methyl-oxa-cyclotetradec-6-en-2-one were considered to be an important parameter obtained through statistical analysis (Supplementary Figure S6).
Microorganisms communicate with each other through the secretion of diverse classes of signal molecules. The accumulation of signal molecules above their threshold in the microbes' environments leads to regulating several genes (Abisado et al., 2018). These signal molecules freely diffuse to the cells and bind to their respective receptors, thus regulating various pathogenesis-related traits. The production of EPS (Ilyas et al., 2020; Sheikh et al., 2022), pigment (Polapally et al., 2022), toxins, formation of biofilm (Ali et al., 2022; Fazeli-Nasab et al., 2022), and secretion of cell wall degrading enzymes (Jadhav et al., 2020) are key traits expressed by the pathogen once sufficient cell density is achieved (Sibanda et al., 2018). This quorum-sensing guided virulence factor synthesis can be controlled by disturbing the QS system either by mimicking the signal molecules, which leads to competitive inhibition, or enzymatically hydrolyse the auto-inducers (N-Homoserine Lactone). In conventional approaches, various biocontrol (Vinay et al., 2016; Reshma et al., 2018; Kusale et al., 2021; Mulk et al., 2021; Nakkeeran et al., 2021; Nithyapriya et al., 2021; Sukmawati et al., 2021; Khan, 2022) substances are used to control pathogens, but repeated use of such molecules lead to the development of multidrug-resistant bacteria which would be very difficult to eradicate further.
The desperate attempts require to identify novel antimicrobial agents or developing new procedures to thwart antibiotic resistance (Abdalla et al., 2020; Kapadia et al., 2021). The advanced and innovative antimicrobial agents developed so far target only the most prominent pathogenic bacteria (Dellit et al., 2007), while less severe pathogenic bacteria remain resistant (Masood et al., 2021; Mubraiz et al., 2021). Therefore, developing innovative strategies which target pathogenesis-responsible compounds are of prime importance for controlling pathogen. The actinobacteria produce several classes of bioactive metabolites which has the potential to act as anti-biofilm agents against several pathogens (Deka et al., 2020). The virulence in the pathogen is governed by the coordinated regulation of synthesis, accumulation, transport, and receptor molecules. The hydrolysis, transformation of signal molecules, or blockage of synthase and receptor lead to disturbance in the communication system of the cells (LaSarre and Federle, 2013). For any quorum-sensing guided system, synthase produces diverse types of signal molecules which freely move across the membranes and accumulate in cell surroundings. At the same time, its cognate receptors sense the signal molecules resulting in the alteration in target gene expression. Some examples are Rhll/LasI/PqsA, AhlI, CarI, TraI, CviI, and LuxM synthase, which produces different chain length of acyl lactones that binds to corresponding receptors Viz. RhlR/LasRPqsR, AhlR, CarR, TraR, and CviR, respectively (McClean et al., 1997; von Bodman et al., 2003; Dubern and Diggle, 2008; Ng and Bassler, 2009; Nadal Jimenez et al., 2012). Oligopeptides are another class of regulators produced by Gram-positive bacteria, which bind to receptors on cell surfaces and modulate several kinases (Håvarstein et al., 1995). This will lead to alteration in the signal transduction pathway. Like the lactones, AI2 produced by Gram-positive and Gram-negative bacteria produce AI2 that diffuses freely out of the cells (Chen et al., 2002; Neiditch et al., 2006). From the literature survey, it could be easily hypothesized that diverse compounds may regulate signal transduction by modulating signal synthesis, signal perception, degradation, and modifications (Huang et al., 2003; Dong and Zhang, 2005; Chowdhary et al., 2007; Liu et al., 2008). Several reports indicate the use of antibodies to sequester signal molecules, thus limiting their availability to receptors (Kaufmann et al., 2008).
The 3OC12HSL was sequestering using monoclonal antibodies to suppress pathogenic traits in P. aeruginosa (Miyairi et al., 2006), while hydrolysis of signal molecules was observed in P. aeruginosa (De Lamo Marin et al., 2007; Kapadnis et al., 2009). Inhibition of signal synthesis by binding with enzyme might be one such strategy to control pathogenesis. Triclosan and J8-C8 were competitive synthase inhibitors; thus, the bacteria lower HSL production (Chung et al., 2013). Similarly, pyrogallol, furanone, and canavanine modulate Lux R family receptors, while an anthranilate derivative inhibits PQS synthesis (Givskov et al., 1996; Hentzer et al., 2002; Keshavan et al., 2005; Lesic et al., 2007). Several reports are concerning the modulation of pathogenesis in vitro, such as biofilm and pyocyanin inhibition in P. aeruginosa and swarming motility inhibition in S. marcascens (Marsden et al., 2010; Brackman et al., 2012). Scientists have used nanoparticle-based approaches to control the pathogen, but they are also not economical (Kapadia et al., 2021). Various researchers have reviewed quorum-sensing mediated pathogenesis in the top 10 plant pathogenic bacteria, most of which belong to Gram-negative bacteria (Sibanda et al., 2018). The present study aimed to isolate quorum-sensing inhibitor metabolites from the microbes to target the soft rot-causing pathogen L. amnigena RCE. Quorum quenching does not lead to the death of microbes, rather, it suppresses or inhibits the expression of pathogenic traits (Rehman and Leiknes, 2018). The majority of QSI compounds at higher concentration brings about the death of the microorganisms, and their action is mistakenly considered as antibiotic effects. Thus, most bacterial extracts used for identifying bactericidal compounds have not been tested for their ability to regulate quorum-sensing cascade. In the present experiment, P. aeruginosa RKC1 produces diverse metabolites and, at higher concentrations, inhibits the growth of Pathogen L. amnigena RCE. Quorum-sensing inhibitory activities were recorded using two monitor strains C. violaceum MTCC2656 and P. aeruginosa MTCC2297, where organic extracts of P. aeruginosa RKC1 inhibit purple pigmentation of C. violaceum and pyocyanin production as well as swarming motility of P. aeruginosa MTCC2297. The quorum-sensing system guides these traits, and the disappearance of such traits indicates the quorum-quenching effects of the metabolites present in the extracts. The metabolites of the extracts disrupt Lux (CviIR) signaling cascade of C. violaceum and las, rhl, and pqs signaling system of P. aeruginosa (McLean et al., 2004; Sun et al., 2016). P. aeruginosa MTCC2297 moved by their flagella in the radial manner from the point of inoculation; likewise, L. amnigena also swarms through their peritrichous flagella. The presence of QSI compounds inhibits swarming, and swimming motility indicates their potentiality as quorum quenching. The inhibition of motility by the extracts results in improper cell adherence and is easily approachable. The production of EPS results in the formation of biofilm, which helps the bacteria to adhere to the surfaces for colonization. The biofilm formation by the L. amnigena RCE is inhibited by the extracts in a dose-dependent manner. The GC-MS was done to identify the compounds to understand the metabolites responsible for quorum-quenching effects. The data presented here indicate 2,5-diketopiperazines (DKPs) or Cyclic dipeptides such as Cyclo(L-prolyl-L-valine), Cyclo (Pro-Leu), and Cyclo(D-phenylalanyl-L-prolyl) were predominantly found in the organic extracts. These metabolites were found to be the lowest yet detectable for GC-MS in the clear zone around the wells. One can speculate that metabolites might reach the cells and bind with receptors as well as metabolites detected from the zone which cannot correlate with pure extracts. Apart from these several other cyclic dipeptides viz. Cyclo(L-prolyl-L-valine), Cyclo(prolyl tyrosyl), cyclo-(L-Trp-L-Pro), Cyclo-Ala-Pro-diketopiperazine, Cyclo(Leu-Ala), and N-glycylproline found in the extracts at lower peak area and thus unable to be detected in the zone around the wells. Cyclo(Trp-Ser), belongs to R. aquimaris, inhibits QS-mediated violacein production in C. violaceum and inhibition of biofilm, and pyocyanin production as well as elastase production in P. aeruginosa PA01 (Sun et al., 2016). There could be molecular mimicry which leads to either change in the structure of the receptor or make the receptor inaccessible to the natural ligand (Plecha and Withey, 2015). (S)-4,5-Dihydroxypentane-2,3-dione [(S)-DPD, (1)] inhibits quorum sensing and biofilm formation in V. harveyi without affecting their growth (Kadirvel et al., 2014). Likewise, cyclic dipeptides and solonomides belonging to Photobacterium spp. inhibits the quorum-sensing system of S. aureus (Mansson et al., 2011). DKPs were reported to be inhibitors of swarming motilities in S. liquifaciens (Holden et al., 1999). The data obtained out of these experiments were supported by the findings of the previous study (Holden et al., 1999), where they showed that cyclo(Ala-Val), cyclo(Pro-Tyr), and cyclo(Phe-Pro) from P. aeruginosa inhibits bioluminescence and swarming behavior. Moreover, they also reported that fungi, Alternaria alternate derived Cyclo(Pro-Tyr), have the potential to inhibit the swarming motility of S. liquefaciens. The molecular docking studies showed that DKPs competitively bind to the receptor, CviR in C. violaceum or LasR receptor systems in P. aeruginosa PA01 (Holden et al., 1999; Sun et al., 2016; Wang et al., 2022). The analog binding changes the receptor's confirmation, which cannot bind to the target site. Thus, one can speculate that biofilm inhibition and motility reduction in L. amnigena RCE could be the results of regulation at the receptor level. The data corroborated with the previous scientific studies where, cyclo(DAla-L-Val), cyclo(L-Phe-L-Pro), and cyclo(L-Pro-L-Tyr) extracted from P. aeruginosa inhibits quorum-sensing activator LuxR of E. coli (pSB401) by mimicking the AHLs (Jayatilake et al., 1996). They hypothesized that binding of cyclic DKPs to N-terminal domain of LuxR results in the confirmation change at C terminal DNA binding region (Hanzelka and Greenberg, 1995). In the other experiment, DKPs shown to inhibit swarming behavior of S. liquefaciens when DKPs were added together with C4-HSL (Eberl et al., 1996). So far, various scientific literatures supported the finding of present experiment (Ortiz-Castro et al., 2011; Borthwick, 2012; Nishanth Kumar et al., 2013; Chen et al., 2015; Wu et al., 2017). There is need of extensive validation at molecular level to elucidate structural changes and binding site. Each pathogen uses different quorum-sensing system to regulate pathogenesis; thus, it is important to evaluate quorum-sensing cascade in them in order to design strategy to combat infection.
GC-MS analysis of crude extracts from P. aeruginosa RKC1 as well as the zone surrounding the wells showed a higher concentration of diketopiperazine (Cyclic dipeptides) like cyclo (L-prolyl-L-valine), cyclo (Pro-Leu), and Cyclo (D-phenylalanyl-L-prolyl). These metabolites, along with other diketopiperazines, account for 93% of the total peak area, suggesting their role as quorum-sensing inhibitors. The extracts showed a noticeable reduction in biofilm inhibition by L. amnigena RCE and maceration inhibition on the carrot, potato, and cucumber. This is probably the first report which uses P. aeruginosa extracts to control soft rot causing pathogen L. amnigena RCE. Furthermore, this diketopiperazine inhibit QS-mediated pathogenicity via competitive binding with receptors. The results substantiate that P. aeruginosa extracts could be a potential source for QSI metabolites and can be exploited further at the industrial level. This study opens a novel strategy to control plant pathogens' virulence without developing antibiotic resistance.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Conceptualization and supervision: CK. Methodology: RK and SS. Formal analysis: KG. Writing—original draft: CK, PP, and RS. Review and editing: RS, PP, SA, MA, and AG. Funding acquisition: SA. All authors have read and agreed to the published version of the manuscript.
This project was supported by Researchers Supporting Project Number (RSP-2023R7) King Saud University, Riyadh, Saudi Arabia. The funders had no role in designing and analysis of the results.
The authors sincerely thank Researchers Supporting Project Number (RSP-2023R7), King Saud University, Riyadh, Saudi Arabia, for funding this study and the Research fellowship granted by the Alexander von Humboldt Foundation, Bonn, Germany, to AG is also gratefully acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.977669/full#supplementary-material
Abd Elhafeez, E., and Sayed, E. (2018). Isolation and characterization of two bacteriophages infecting kosakonia sacchari bacterium causing potato soft rot disease. J. Bacteriol. Parasitol. 9, 344. doi: 10.4172/2155-9597.1000344
Abdalla, M. A., Aro, A. O., Gado, D., Passari, A. K., Mishra, V. K., Singh, B. P., et al. (2020). Isolation of endophytic fungi from South African plants, and screening for their antimicrobial and extracellular enzymatic activities and presence of type I polyketide synthases. South African J. Bot. 134, 336–342. doi: 10.1016/j.sajb.2020.03.021
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. MBio. 9, 1–13. doi: 10.1128/mBio.02331-17
Ali, S. A. M., Sayyed, R. Z., Mir, M. I., Khan, M. Y., Hameeda, B., Alkhanani, M. F., et al. (2022). Induction of Systemic Resistance in Maize and Antibiofilm Activity of Surfactin From Bacillus velezensis MS20. Front. Microbiol. 13, 879739. doi: 10.3389/fmicb.2022.879739
Almohaywi, B., Taunk, A., Wenholz, D. S., Nizalapur, S., Biswas, N. N., Ho, K. K. K., et al. (2018). Design and synthesis of lactams derived from mucochloric and mucobromic acids as pseudomonas aeruginosa quorum sensing inhibitors. Molecules. 23, 1106. doi: 10.3390/molecules23051106
Borthwick, A. D. (2012). 2,5-Diketopiperazines: synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem. Rev. 112, 3641–3716. doi: 10.1021/cr200398y
Brackman, G., Risseeuw, M., Celen, S., Cos, P., Maes, L., Nelis, H. J., et al. (2012). Synthesis and evaluation of the quorum sensing inhibitory effect of substituted triazolyldihydrofuranones. Bioorg. Med. Chem. 20, 4737–4743. doi: 10.1016/j.bmc.2012.06.009
Çetinkaya, S., Yenidünya, A. F., Arslan, K., Arslan, D., Dogan, Ö., and Daştan, T. (2020). Secondary metabolites of an of Streptomyces griseorubens isolate are predominantly pyrrole-and linoleic-acid like compounds. J. Oleo Sci. 69, 1273–1280. doi: 10.5650/jos.ess20161
Charkowski, A. O. (2018). The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 56, 269–288. doi: 10.1146/annurev-phyto-080417-045906
Chen, X., Mou, Y., Ling, J., Wang, N., Wang, X., and Hu, J. (2015). Cyclic dipeptides produced by fungus Eupenicillium brefeldianum HMP-F96 induced extracellular alkalinization and H2O2 production in tobacco cell suspensions. World J. Microbiol. Biotechnol. 31, 247–253. doi: 10.1007/s11274-014-1759-0
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 415, 545–549. doi: 10.1038/415545a
Chowdhary, P. K., Keshavan, N., Nguyen, H. Q., Peterson, J. A., González, J. E., and Haines, D. C. (2007). Bacillus megaterium CYP102A1 Oxidation of Acyl Homoserine Lactones and Acyl Homoserines. Biochemistry. 46, 14429–14437. doi: 10.1021/bi701945j
Chung, Y. S., Goeser, N. J., Cai, X., and Jansky, S. (2013). The Effect of Long Term Storage on Bacterial Soft Rot Resistance in Potato. Am. J. Potato Res. 90, 351–356. doi: 10.1007/s12230-013-9311-6
De Lamo Marin, S., Xu, Y., Meijler, M. M., and Janda, K. D. (2007). Antibody catalyzed hydrolysis of a quorum sensing signal found in Gram-negative bacteria. Bioorg. Med. Chem. Lett. 17, 1549–1552. doi: 10.1016/j.bmcl.2006.12.118
Deka, P., Zothanpuia, P., assari, A. K., and Singh, B. P. (2020). “Actinobacteria as a potential natural source to produce antibiofilm compounds: An overview,” in New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms (Elsevier), 91–99. doi: 10.1016/B978-0-444-64279-0.00007-4
Dellit, T. H., Owens, R. C., McGowan, J. E., Gerding, D. N., Weinstein, R. A., Burke, J. P., et al. (2007). Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America Guidelines for Developing an Institutional Program to Enhance Antimicrobial Stewardship. Clin. Infect. Dis. 44, 159–177. doi: 10.1086/510393
Dong, Y.-H., and Zhang, L.-H. (2005). Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43,101–109.
Dubern, J.-F., and Diggle, S. P. (2008). Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. Biosyst. 4, 882. doi: 10.1039/b803796p
Eberl, L., Christiansen, G., Molin, S., and Givskov, M. (1996). Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 178, 554–559. doi: 10.1128/jb.178.2.554-559.1996
Fazeli-Nasab, B., Sayyed, R. Z., Mojahed, L. S., Rahmani, A. F., Ghafari, M., Antonius, S., et al. (2022). Biofilm production: a strategic mechanism for survival of microbes under stress conditions. Biocatal. Agric. Biotechnol. 42, 102337. doi: 10.1016/j.bcab.2022.102337
Fetzner, S. (2015). Quorum quenching enzymes. J. Biotechnol. 201, 2–14. doi: 10.1016/j.jbiotec.2014.09.001
Flavier, A. B., Clough, S. J., Schell, M. A., and Denny, T. P. (1997). Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259. doi: 10.1046/j.1365-2958.1997.5661945.x
Garge, S. S., and Nerurkar, A. S. (2016). Attenuation of quorum sensing regulated virulence of Pectobacterium carotovorum subsp. carotovorum through an AHL lactonase produced by Lysinibacillus sp. Gs50. PLoS ONE. 11, 1–23. doi: 10.1371/journal.pone.0167344
Givskov, M., de Nys, R., Manefield, M., Gram, L., Maximilien, R., Eberl, L., et al. (1996). Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 178, 6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996
Håvarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Hadizadeh, I., Peivastegan, B., Hannukkala, A., van der Wolf, J. M., Nissinen, R., and Pirhonen, M. (2019). Biological control of potato soft rot caused by Dickeya solani and the survival of bacterial antagonists under cold storage conditions. Plant Pathol. 68, 297–311. doi: 10.1111/ppa.12956
Hanzelka, B. L., and Greenberg, E. P. (1995). Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177, 815–817. doi: 10.1128/jb.177.3.815-817.1995
Hassan, R., Shaaban, M. I., Abdel Bar, F. M., El-Mahdy, A. M., and Shokralla, S. (2016). Quorum sensing inhibiting activity of Streptomyces coelicoflavus isolated from soil. Front. Microbiol. 7, 1–12. doi: 10.3389/fmicb.2016.00659
Hentzer, M., Riedel, K., Rasmussen, T. B., Heydorn, A., Andersen, J. B., Parsek, M. R., et al. (2002). Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 148, 87–102. doi: 10.1099/00221287-148-1-87
Holden, M. T. G., Chhabra, S. R., De Nys, R., Stead, P., Bainton, N. J., Hill, P. J., et al. (1999). Quorum-sensing cross talk: Isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 33, 1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x
Huang, J. J., Han, J.-I., Zhang, L.-H., and Leadbetter, J. R. (2003). Utilization of Acyl-Homoserine Lactone Quorum Signals for Growth by a Soil Pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69, 5941–5949. doi: 10.1128/AEM.69.10.5941-5949.2003
Ilyas, N., Mumtaz, K., Akhtar, N., Yasmin, H., Sayyed, R. Z., Khan, W., et al. (2020). Exopolysaccharides producing bacteria for the amelioration of drought stress in wheat. Sustainability. 12, 8876. doi: 10.3390/su12218876
Jadhav, H. P., Sayyed, R. Z., Shaikh, S. S., Bhamre, H. M., Sunita, K., and El Enshasy, H. A. (2020). Statistically designed bioprocess for enhanced production of alkaline protease in bacillus cereus HP_RZ17. J. Sci. Ind. Res. 79, 491–498.
Jayatilake, G. S., Thornton, M. P., Leonard, A. C., Grimwade, J. E., and Baker, B. J. (1996). Metabolites from an Antarctic Sponge-Associated Bacterium, Pseudomonas aeruginosa. J. Nat. Prod. 59, 293–296. doi: 10.1021/np960095b
Kaaniche, F., Hamed, A., Elleuch, L., Chakchouk-Mtibaa, A., Smaoui, S., Karray-Rebai, I., et al. (2020). Purification and characterization of seven bioactive compounds from the newly isolated Streptomyces cavourensis TN638 strain via solid-state fermentation. Microb. Pathog. 142, 104106. doi: 10.1016/j.micpath.2020.104106
Kachhadia, R., Kapadia, C., Singh, S., Gandhi, K., Jajda, H., Alfarraj, S., et al. (2022). Quorum sensing inhibitory and quenching activity of Bacillus cereus RC1 extracts on Soft Rot-Causing Bacteria Lelliottia amnigena. ACS Omega. 7, 25291–25308. doi: 10.1021/acsomega.2c02202
Kadirvel, M., Fanimarvasti, F., Forbes, S., McBain, A., Gardiner, J. M., Brown, G. D., et al. (2014). Inhibition of quorum sensing and biofilm formation in Vibrio harveyi by 4-fluoro-DPD; a novel potent inhibitor of signalling. Chem. Commun. 50, 5000–5002. doi: 10.1039/C3CC49678C
Kapadia, C., Alhazmi, A., Patel, N., Elesawy, B. H., Sayyed, R. Z., Lokhandwala, F., et al. (2021). Nanoparticles combined with cefixime as an effective synergistic strategy against Salmonella enterica typhi. Saudi J. Biol. Sci. 28, 4164–4172. doi: 10.1016/j.sjbs.2021.05.032
Kapadnis, P. B., Hall, E., Ramstedt, M., Galloway, W. R. J. D., Welch, M., and Spring, D. R. (2009). Towards quorum-quenching catalytic antibodies. Chem. Commun., 538–540. doi: 10.1039/B819819E
Kaufmann, G. F., Park, J., Mee, J. M., Ulevitch, R. J., and Janda, K. D. (2008). The quorum quenching antibody RS2-1G9 protects macrophages from the cytotoxic effects of the Pseudomonas aeruginosa quorum sensing signalling molecule N-3-oxo-dodecanoyl-homoserine lactone. Mol. Immunol. 45, 2710–2714. doi: 10.1016/j.molimm.2008.01.010
Keshavan, N. D., Chowdhary, P. K., Haines, D. C., and González, J. E. (2005). L-Canavanine made by Medicago sativa interferes with quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 187, 8427–8436. doi: 10.1128/JB.187.24.8427-8436.2005
Khan, N. (2022). Molecular Communication between Plants and Plant-Growth-Promoting Microorganisms for Stress Tolerance. Microorganisms. 10, 1088. doi: 10.3390/microorganisms10061088
Kumar S. Stecher G. and Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kusale, S. P., Attar, Y. C., Sayyed, R. Z., Malek, R. A., Ilyas, N., Suriani, N. L., et al. (2021). Production of plant beneficial and antioxidants metabolites by Klebsiella variicola under salinity stress. Molecules. 26, 1894. doi: 10.3390/molecules26071894
LaSarre, B., and Federle, M. J. (2013). Exploiting Quorum Sensing To Confuse Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111. doi: 10.1128/MMBR.00046-12
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K. M., Wertheim, H. F. L., Sumpradit, N., et al. (2013). Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/S1473-3099(13)70318-9
Lesic, B., Lépine, F., Déziel, E., Zhang, J., Zhang, Q., Padfield, K., et al. (2007). Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 3, 1229–1239. doi: 10.1371/journal.ppat.0030126
Liu, D., Momb, J., Thomas, P. W., Moulin, A., Petsko, G. A., Fast, W., et al. (2008). Mechanism of the Quorum-Quenching Lactonase (AiiA) from Bacillus thuringiensis. 1. Product-Bound Structures. Biochemistry. 47, 7706–7714. doi: 10.1021/bi800368y
Liu, S., and Tang, Y. (2016). Identification and Characterization of a New Enterobacter Onion Bulb Decay Caused by Lelliottia amnigena in China. Appl. Microbiol. Open Access. 2. doi: 10.4172/2471-9315.1000114
Ma, Z. P., Song, Y., Cai, Z. H., Lin, Z. J., Lin, G. H., Wang, Y., et al. (2018). Anti-quorum sensing activities of selected coral symbiotic bacterial extracts from the South China Sea. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00144
Malešević, M., Di Lorenzo, F., Filipić, B., Stanisavljević, N., Novović, K., Senerovic, L., et al. (2019). Pseudomonas aeruginosa quorum sensing inhibition by clinical isolate Delftia tsuruhatensis 11304: involvement of N-octadecanoylhomoserine lactones. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-52955-3
Mansson, M., Nielsen, A., Kjærulff, L., Gotfredsen, C. H., Wietz, M., Ingmer, H., et al. (2011). Inhibition of virulence gene expression in staphylococcus aureus by novel depsipeptides from a marine photobacterium. Mar. Drugs 9, 2537–2552. doi: 10.3390/md9122537
Marsden, D. M., Nicholson, R. L., Skindersoe, M. E., Galloway, W. R. J. D., Sore, H. F., Givskov, M., et al. (2010). Discovery of a quorum sensing modulator pharmacophore by 3D small-molecule microarray screening. Org. Biomol. Chem. 8, 5313. doi: 10.1039/c0ob00300j
Masood, K., Yasmin, H., Batool, S., Ilyas, N., Nosheen, A., Naz, R., et al. (2021). A strategy for mitigating avian colibacillosis disease using plant growth promoting rhizobacteria and green synthesized zinc oxide nanoparticles. Saudi J. Biol. Sci. 28, 4957–4968. doi: 10.1016/j.sjbs.2021.06.100
McClean, K. H., Winson, M. K., Fish, L., Taylor, A., Chhabra, S. R., Camara, M., et al. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 143, 3703–3711. doi: 10.1099/00221287-143-12-3703
McLean, R. J., Pierson, L. S., and Fuqua, C. (2004). A simple screening protocol for the identification of quorum signal antagonists. J. Microbiol. Methods. 58, 351–360. doi: 10.1016/j.mimet.2004.04.016
Miyairi, S., Tateda, K., Fuse, E. T., Ueda, C., Saito, H., Takabatake, T., et al. (2006). Immunization with 3-oxododecanoyl-l-homoserine lactone–protein conjugate protects mice from lethal Pseudomonas aeruginosa lung infection. J. Med. Microbiol. 55, 1381–1387. doi: 10.1099/jmm.0.46658-0
Mubraiz, N., Bano, A., Mahmood, T., and Khan, N. (2021). Microbial and plant assisted synthesis of cobalt oxide nanoparticles and their antimicrobial activities. Agronomy. 11, 1607. doi: 10.3390/agronomy11081607
Mulk, S., Wahab, A., Yasmin, H., Mumtaz, S., El-Serehy, H. A., Khan, N., et al. (2021). Prevalence of wheat associated Bacillus spp. and their bio-control efficacy against fusarium root rot. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.798619
Nadal Jimenez, P., Koch, G., Thompson, J. A., Xavier, K. B., Cool, R. H., and Quax, W. J. (2012). The multiple signaling systems regulating virulence in pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65. doi: 10.1128/MMBR.05007-11
Nakkeeran, S., Saranya, N., Senthilraja, C., Renukadevi, P., Krishnamoorthy, A. S., El Enshasy, H. A., et al. (2021). Mining the genome of Bacillus velezensis VB7 (CP047587) for MAMP genes and non-ribosomal peptide synthetase gene clusters conferring antiviral and antifungal activity. Microorganisms. 9, 2511. doi: 10.3390/microorganisms9122511
Neiditch, M. B., Federle, M. J., Pompeani, A. J., Kelly, R. C., Swem, D. L., Jeffrey, P. D., et al. (2006). Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 126, 1095–1108. doi: 10.1016/j.cell.2006.07.032
Ng, W.-L., and Bassler, B. L. (2009). Bacterial Quorum-Sensing Network Architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
Nishanth Kumar, S., Mohandas, C., and Nambisan, B. (2013). Purification of an antifungal compound, cyclo(l-Pro-d-Leu) for cereals produced by Bacillus cereus subsp. thuringiensis associated with entomopathogenic nematode. Microbiol. Res. 168, 278–288. doi: 10.1016/j.micres.2012.12.003
Nithya, C., Aravindraja, C., and Pandian, S. K. (2010). Bacillus pumilus of Palk Bay origin inhibits quorum-sensing-mediated virulence factors in Gram-negative bacteria. Res. Microbiol. 161, 293–304. doi: 10.1016/j.resmic.2010.03.002
Nithyapriya, S., Lalitha, S., Sayyed, R. Z., Reddy, M. S., Dailin, D. J., El Enshasy, H. A., et al. (2021). Production, purification, and characterization of bacillibactin siderophore of bacillus subtilis and its application for improvement in plant growth and oil content in sesame. Sustain. 13. doi: 10.3390/su13105394
Ortiz-Castro, R., Díaz-Pérez, C., Martínez-Trujillo, M., del Río, R. E., Campos-García, J., and López-Bucio, J. (2011). Transkingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. Proc. Natl. Acad. Sci. 108, 7253–7258. doi: 10.1073/pnas.1006740108
Osei, R., Yang, C., Cui, L., Wei, L., Jin, M., and Boamah, S. (2021). Salicylic acid effect on the mechanism of Lelliottia amnigena causing potato soft rot. Folia Hortic. 33, 376–389. doi: 10.2478/fhort-2021-0029
Padmavathi, A. R., Abinaya, B., and Pandian, S. K. (2014). Phenol, 2,4-bis(1,1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling. 30, 1111–1122. doi: 10.1080/08927014.2014.972386
Pang, Z., Chong, J., Zhou, G., de Lima Morais, D. A., Chang, L., Barrette, M., et al. (2021). MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396. doi: 10.1093/nar/gkab382
Passari, A. K., Mishra, V. K., Leo, V. V., Gupta, V. K., and Singh, B. P. (2016). Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol. Res. 193, 57–73. doi: 10.1016/j.micres.2016.09.006
Passari, A. K., Mishra, V. K., Singh, G., Singh, P., Kumar, B., Gupta, V. K., et al. (2017). Insights into the functionality of endophytic actinobacteria with a focus on their biosynthetic potential and secondary metabolites production. Sci. Rep. 7, 1–17. doi: 10.1038/s41598-017-12235-4
Plecha, S. C., and Withey, J. H. (2015). Mechanism for inhibition of Vibrio cholerae ToxT activity by the unsaturated fatty acid components of bile. J. Bacteriol. 197, 1716–1725. doi: 10.1128/JB.02409-14
Polapally, R., Mansani, M., Rajkumar, K., Burgula, S., Hameeda, B., Alhazmi, A., et al. (2022). Melanin pigment of Streptomyces puniceus RHPR9 exhibits antibacterial, antioxidant and anticancer activities. PLoS ONE. 17, e0266676. doi: 10.1371/journal.pone.0266676
Rashiya, N., Padmini, N., Ajilda, A. A. K., Prabakaran, P., Durgadevi, R., Veera Ravi, A., et al. (2021). Inhibition of biofilm formation and quorum sensing mediated virulence in Pseudomonas aeruginosa by marine sponge symbiont Brevibacterium casei strain Alu 1. Microb. Pathog. 150, 104693. doi: 10.1016/j.micpath.2020.104693
Rehman, Z. U., and Leiknes, T. (2018). Quorum-Quenching Bacteria Isolated From Red Sea Sediments Reduce Biofilm Formation by Pseudomonas aeruginosa. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01354
Reshma, P., Naik, M. K., Aiyaz, M., Niranjana, S. R., Chennappa, G., Shaikh, S. S., et al. (2018). Induced systemic resistance by 2,4-diacetylphloroglucinol positive fluorescent Pseudomonas strains against rice sheath blight. Indian J. Exp. Biol. 56, 207–212.
Sheikh, T., Hamid, B., Baba, Z., Iqbal, S., Yatoo, A., Fatima, S., et al. (2022). Extracellular polymeric substances in psychrophilic cyanobacteria: A potential bioflocculant and carbon sink to mitigate cold stress. Biocatal. Agric. Biotechnol. 42, 102375. doi: 10.1016/j.bcab.2022.102375
Sheladiya, P., Kapadia, C., Prajapati, V., Ali, H., Enshasy, E., Malek, R. A., et al. (2022). Production , statistical optimization, and functional characterization of alkali stable pectate lyase of Paenibacillus lactis PKC5 for use in juice clarification. Sci. Rep. 12, 1–16. doi: 10.1038/s41598-022-11022-0
Sibanda, S., Moleleki, L. N., Shyntum, D. Y., and Coutinho, T. A. (2018). Quorum Sensing in Gram-Negative Plant Pathogenic Bacteria. Adv. Plant Pathol. doi: 10.5772/intechopen.78003
Singh, V. K., Mishra, A., and Jha, B. (2017). Anti-quorum Sensing and Anti-biofilm Activity of Delftia tsuruhatensis Extract by Attenuating the Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00337
Sukmawati, D., Family, N., Hidayat, I., Sayyed, R., Elsayed, E., Dailin, D., et al. (2021). Biocontrol Activity of Aureubasidium pullulans and Candida orthopsilosis Isolated from Tectona grandis L. Phylloplane against Aspergillus sp. in Post-Harvested Citrus Fruit. Sustainability. 13, 7479. doi: 10.3390/su13137479
Sun, S., Dai, X., Sun, J., Bu, X., Weng, C., Li, H., et al. (2016). A diketopiperazine factor from Rheinheimera aquimaris QSI02 exhibits anti-quorum sensing activity. Sci. Rep. 6, 1–10. doi: 10.1038/srep39637
Tan, S. Y. Y., Chua, S. L., Chen, Y., Rice, S. A., Kjelleberg, S., Nielsen, T. E., et al. (2013). Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database. Antimicrob. Agents Chemother. 57, 5629–5641. doi: 10.1128/AAC.00955-13
Tremblay, J., Richardson, A. P., Lépine, F., and Déziel, E. (2007). Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9, 2622–2630. doi: 10.1111/j.1462-2920.2007.01396.x
Venkatramanan, M., Sankar Ganesh, P., Senthil, R., Akshay, J., Veera Ravi, A., Langeswaran, K., et al. (2020). Inhibition of Quorum Sensing and Biofilm Formation in Chromobacterium violaceum by Fruit Extracts of Passiflora edulis. ACS Omega. 5, 25605–25616. doi: 10.1021/acsomega.0c02483
Vinay, J. U., Naik, M. K., Rangeshwaran, R., Chennappa, G., Shaikh, S. S., and Sayyed, R. Z. (2016). Detection of antimicrobial traits in fluorescent pseudomonads and molecular characterization of an antibiotic pyoluteorin. 3 Biotech. 6, 227. doi: 10.1007/s13205-016-0538-z
von Bodman, S. B., Bauer, W. D., and Coplin, D. L. (2003). QUORUM SENSING INP LANT-P ATHOGENIC BACTERIA. Annu. Rev. Phytopathol. 41, 455–482. doi: 10.1146/annurev.phyto.41.052002.095652
Wan, F., Draz, M. S., Gu, M., Yu, W., Ruan, Z., and Luo, Q. (2021). Novel strategy to combat antibiotic resistance: a sight into the combination of crispr/cas9 and nanoparticles. Pharmaceutics. 13, 1–19. doi: 10.3390/pharmaceutics13030352
Wang, W., Arshad, M. I., Khurshid, M., Rasool, M. H., Nisar, M. A., Aslam, M. A., et al. (2018). Antibiotic resistance : a rundown of a global crisis. Infect. Drug Resist. 11, 1645–1658. doi: 10.2147/IDR.S173867
Wang, Y., Zheng, Q., Li, L., Pan, L., and Zhu, H. (2022). Anti-quorum-sensing activity of tryptophan-containing cyclic dipeptides. Mar. Drugs. 20, 85. doi: 10.3390/md20020085
Wu, L., Wu, H., Chen, L., Zhang, H., and Gao, X. (2017). Induction of systemic disease resistance in Nicotiana benthamiana by the cyclodipeptides cyclo (l-Pro-l-Pro) and cyclo (d-Pro-d-Pro). Mol. Plant Pathol. 18, 67–74. doi: 10.1111/mpp.12381
Zhang, Y., Kong, J., Huang, F., Xie, Y., Guo, Y., Cheng, Y., et al. (2018a). Hexanal as a QS inhibitor of extracellular enzyme activity of Erwinia carotovora and Pseudomonas fluorescens and its application in vegetables. Food Chem. 255, 1–7. doi: 10.1016/j.foodchem.2018.02.038
Keywords: cyclic dipeptides, diketopiperazine, Lelliottia amnigena, P. aeruginosa RKC1, quorum quenching, soft rot
Citation: Kapadia C, Kachhdia R, Singh S, Gandhi K, Poczai P, Alfarraj S, Ansari MJ, Gafur A and Sayyed RZ (2022) Pseudomonas aeruginosa inhibits quorum-sensing mechanisms of soft rot pathogen Lelliottia amnigena RCE to regulate its virulence factors and biofilm formation. Front. Microbiol. 13:977669. doi: 10.3389/fmicb.2022.977669
Received: 24 June 2022; Accepted: 25 July 2022;
Published: 23 August 2022.
Edited by:
Naeem Khan, University of Florida, United StatesReviewed by:
Ajit Kumar Passari, National Autonomous University of Mexico, MexicoCopyright © 2022 Kapadia, Kachhdia, Singh, Gandhi, Poczai, Alfarraj, Ansari, Gafur and Sayyed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chintan Kapadia, Y2hpbnRhbl9idEB5YWhvby5jby5pbg==; R. Z. Sayyed, c2F5eWVkcnpAZ21haWwuY29t; Peter Poczai, cGV0ZXIucG9jemFpQGhlbHNpbmtpLmZp
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.