
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 04 August 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.976550
This article is part of the Research TopicOmics-Based Studies on Mycobacteria: Generating Fundamental Knowledge for a Better Control of Mycobacterial DiseasesView all 9 articles
A recent addition to the -omics toolkit, ribosome profiling, enables researchers to gain insight into the process and regulation of translation by mapping fragments of mRNA protected from nuclease digestion by ribosome binding. In this review, we discuss how ribosome profiling applied to mycobacteria has led to discoveries about translational regulation. Using case studies, we show that the traditional view of “canonical” translation mechanisms needs expanding to encompass features of mycobacterial translation that are more widespread than previously recognized. We also discuss the limitations of the method and potential future developments that could yield further insight into the fundamental biology of this important human pathogen.
Thanks to an ever-expanding experimental toolkit, the central dogma of molecular biology can now be investigated at every level—from the genome, to the transcriptome, to the translatome, to the proteome—and the regulatory mechanisms operating at all levels of gene expression probed. This review discusses insights uncovered by the ability to study the translatome using ribosome profiling, one of the newest additions to the -omics toolkit. It had long been recognized that there is a poor correlation between the abundance of mRNA transcripts and their corresponding protein concentrations in the cell (Maier et al., 2009; Vogel and Marcotte, 2012; Haider and Pal, 2013; Cortes et al., 2017), but how could this be resolved and the factors underlying it revealed? In 2009 Ingolia and co-workers described an elegant method by which fragments of mRNA that were undergoing active translation could be isolated from a cell and sequenced (Ingolia et al., 2009). The method was developed in eukaryotes and optimization for prokaryotes required a deep understanding of the process of translation and factors affecting it to ensure reproducibility and rule out or mitigate for experimentally induced artifacts (Becker et al., 2013; Subramaniam et al., 2014; Woolstenhulme et al., 2015; Hwang and Buskirk, 2017; Mohammad et al., 2019). In spite of these challenges, many important discoveries about bacterial translation have been made. Some methodological developments that enable investigation of different steps of translation are summarized in Figure 1.
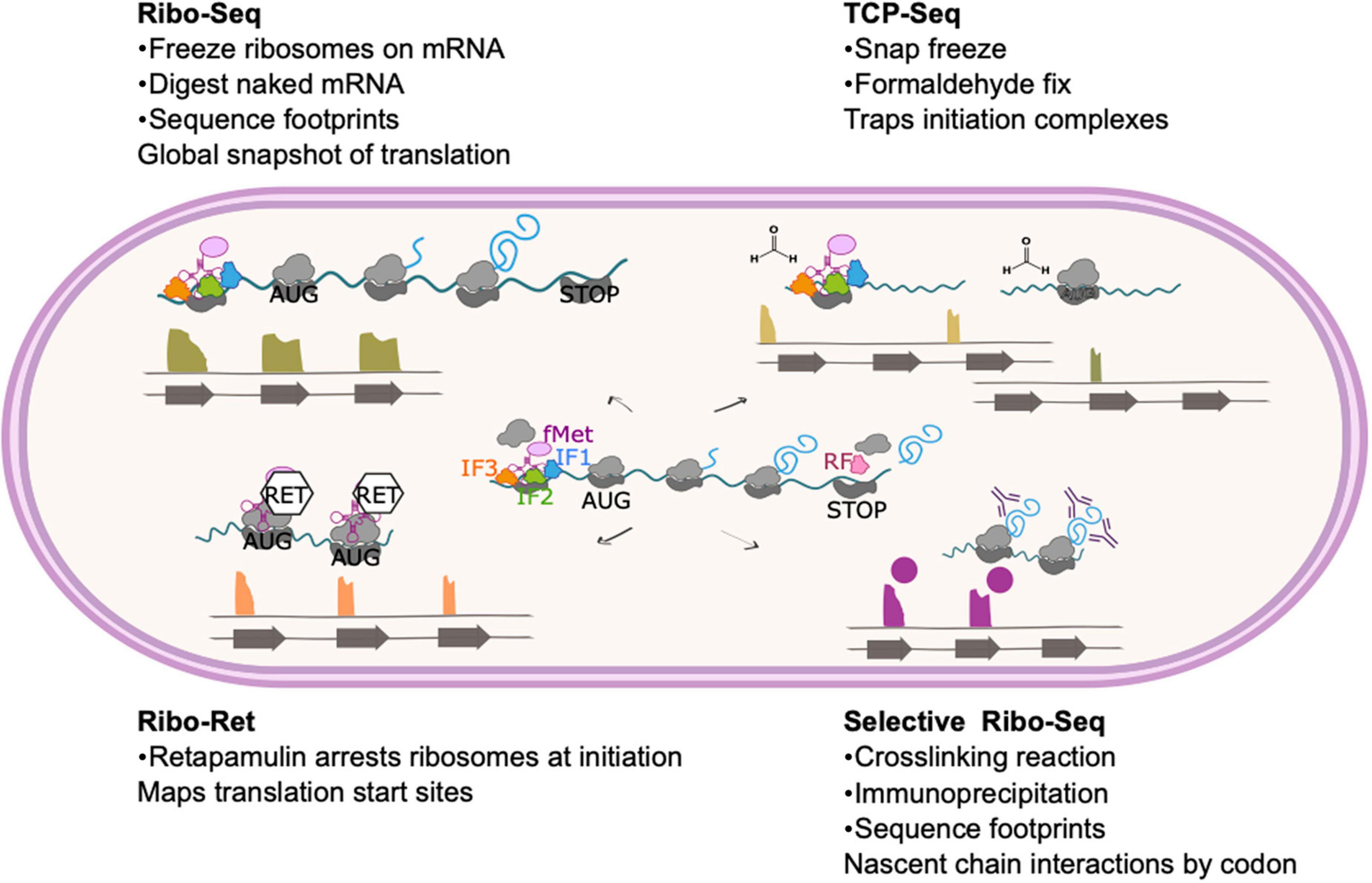
Figure 1. Ribosome profiling method and modifications. The basic ribosome profiling method involves isolation and sequencing of mRNA that is protected from nuclease digestion by ribosome binding. As the central panel shows, these mRNA footprints represent codons across the entire gene often including non-coding regions such as the ribosome binding site. Read length and various in vitro and in silico selection methods can be applied to enrich for sequences that are more likely to represent active translation rather than non-productive ribosome binding. Variations on this method to select or enrich the library for particular features are shown. TCP-Seq (top right) includes flash freezing and formaldehyde fixation steps to trap initiation complexes. Reads from TCP-Seq therefore often map upstream of the CDS (reflecting formation of the canonical SD initiation complex) or at the start codon (for 70S initiation mechanisms). In ribo-RET (bottom left) cells are treated with retapamulin, which arrests ribosomes at initiation. This method can be used to identify unannotated translational start sites, including those within known coding sequences, such as IRES. Selective ribo-seq (bottom right) involves crosslinking ribosomes to their mRNA and any proteins interacting with the emerging nascent chain, such as trigger factor or other molecular chaperones. Translation complexes that have been trapped during interaction with the target protein are then isolated by immunoprecipitation. The footprints therefore correspond to mRNA that is translated as the nascent chain interacts with the target, indicating the length of nascent chain required for the interaction.
A comprehensive review of the contribution of ribosome profiling to understanding bacterial translation in general is beyond the scope of this review. Rather, we will highlight a few key insights that are of particular interest or relevance.
Although translation initiation, elongation, termination and ribosome recycling are all amenable to regulation, most control mechanisms described to date operate during the initiation steps. During initiation, the mRNA is recruited to the small ribosomal subunit and the open reading frame (ORF) correctly positioned along with initiation factors, the initiator aminoacyl tRNAfMet, GTP and eventually the large ribosomal subunit (Figure 2).
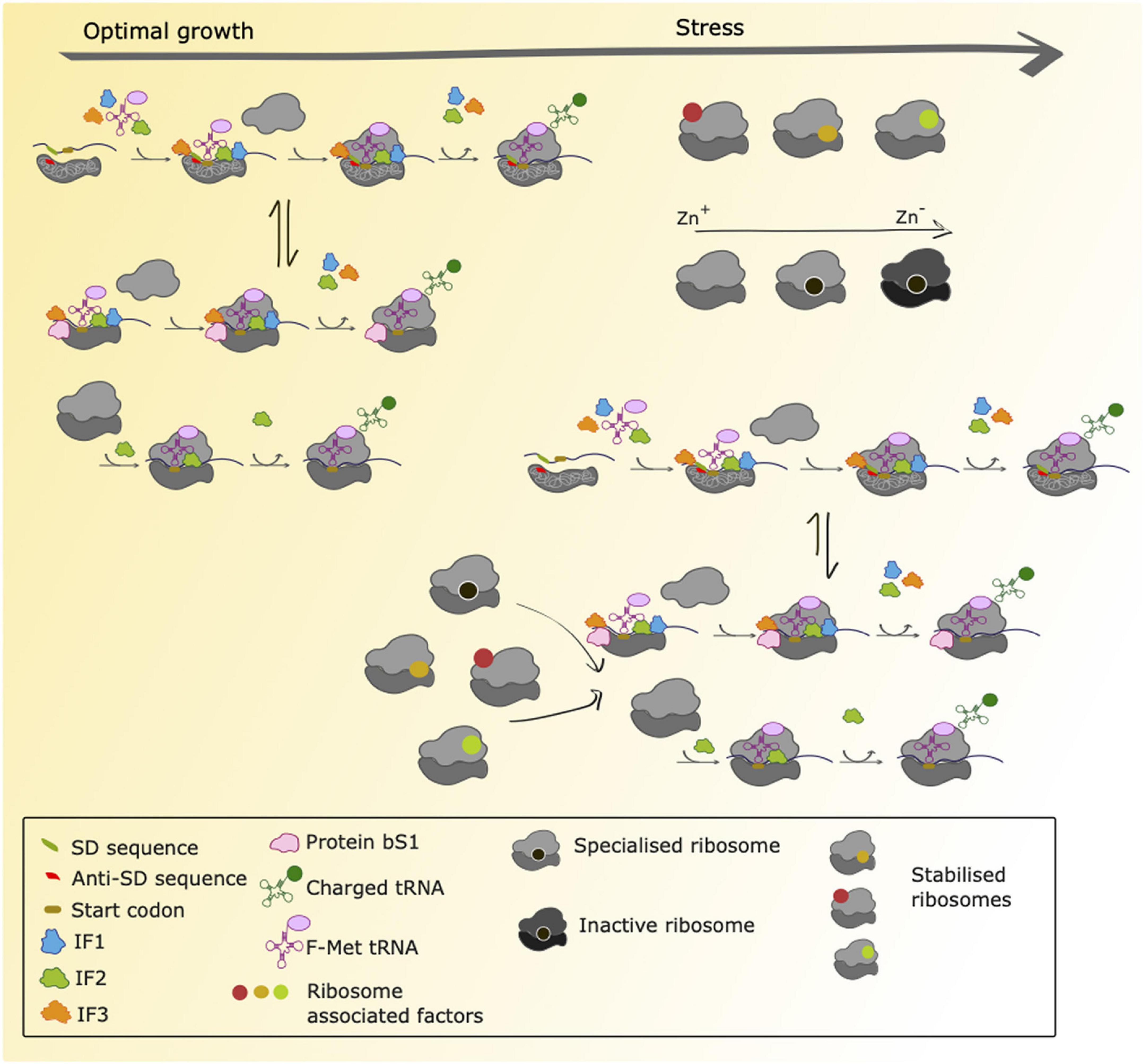
Figure 2. Proposed scenarios for translation initiation in M. tuberculosis during normal growth and stress adaptation. During conditions of normal growth, M. tuberculosis uses both canonical initiation mechanisms (SD-aSD interaction) as well as alternative initiation mechanisms that are still uncharacterized. These could involve ribosomal protein bS1 to initiate translation of transcripts lacking a strong SD sequence within the 5′ UTR and 70S-mediated initiation of transcripts lacking a 5′UTR as previously characterized in E. coli. Ribosome profiling studies have shown that both canonical and non-canonical transcripts are robustly translated in M. tuberculosis (Sawyer et al., 2021), suggesting that mechanisms for canonical and non-canonical translation initiation are in place and that there is an equilibrium between both (represented with the arrows in the Figure). During conditions of stress there are ribosome associated factors that could bind with ribosomes to stabilize them. Furthermore, it has been shown that during conditions of zinc deprivation, specialized ribosomes containing paralogs of ribosomal proteins are formed and are translationally active. As conditions of zinc depletion increase, these ribosomes become inactive. We hypothesize that during conditions of stress, M. tuberculosis temporarily relies on alternative initiation mechanisms to sustain protein synthesis, and that these mechanisms might be favored by specialized and/or stabilized ribosomes.
Binding of the small ribosomal subunit to the mRNA for translation initiation is traditionally achieved through the base pairing of the Shine-Dalgarno (SD) sequence present in the 5′ untranslated region (UTR) of the mRNA to the complementary anti-Shine-Dalgarno (aSD) region of 16S ribosomal RNA (rRNA) (Shine and Dalgarno, 1974; Figure 1). Nevertheless, alternative mechanisms of initiation operate when mRNA molecules lack an SD sequence. For mRNA molecules that have an SD-less 5′ UTR, interactions between the ribosomal protein bS1 and AU-rich elements in the UTR facilitate formation of the pre-initiation complex (Boni et al., 1991; Komarova et al., 2005; Figure 2). Where mRNA molecules lack a 5′ UTR entirely (referred to as “leaderless”), 70S ribosomes bind at AUG codons equipped with a 5′ phosphate group (Moll et al., 2004; Giliberti et al., 2012; Figure 2). To trap early intermediates of translation, including the pre-initiation complex, and better study every stage of translation, modifications to the ribosome profiling method have been developed. Translation complex profile sequencing (TCP-seq) involves a formaldehyde crosslinking step that fixes transient ribosome/subunit complexes to their mRNAs enabling footprints from initiation complexes to be isolated and sequenced (Archer et al., 2016; Shirokikh et al., 2017; Figure 1). TCP-seq has revealed dynamic rearrangements of the 30S subunit and mRNA during initiation, reflected in different mRNA read lengths (Archer et al., 2016) and the impact of ribosome assembly defects on binding initiation factor 3 (Sharma and Anand, 2019).
The traditional view of the mechanism by which mRNA recruitment and positioning occurs, described by Shine and Dalgarno (1974), has been subjected to much interrogation. However, dissecting out the contribution of SD—aSD interactions from other features of the mRNA and activity of the ribosomal protein bS1 to reach a more refined understanding of the role of each of these features in defining the bacterial proteome has proved challenging (Li et al., 2014; Schrader et al., 2014; Del Campo et al., 2015; Li, 2015). Recently, Saito and co-workers performed ribosome profiling using aSD mutant ribosomes to address the role of the SD-aSD interaction without perturbing mRNA sequences or structures (Saito et al., 2020). Through a series of elegant experiments, their results revealed that although mRNA features other than the SD sequence are able to render complexes initiation-competent, there is a linear correlation between SD strength and translation efficiency (Saito et al., 2020). These findings are in good agreement with previous work that investigated the sequence and structural properties of mRNA in the vicinity of the start codon (O’Donnell and Janssen, 2001; Li et al., 2012; Reeve et al., 2014; Duval et al., 2015; Hockenberry et al., 2017).
At first glance, elongation may appear to be the least interesting and adaptable phase of translation as the ribosome proceeds with aligning tRNAs on the appropriate codons, catalyzing peptide bond formation and translocation to the next codon. However, variations in elongation rate both reflect and impact upon the state of the cell (Dai et al., 2016) and the quality/fidelity of the polypeptides produced (Holm, 1986; Shah and Gilchrist, 2010). The greatest contributor to variable elongation rates is codon bias, the correlation between the abundance of particular tRNAs and the frequency of their cognate codons within the mRNA (Kurland, 1991; Plotkin and Kudla, 2011). The presence of charged amino acids in the nascent polypeptide has also been linked to variation in elongation rates though interactions between the peptide and surface of the ribosome exit tunnel (Riba et al., 2019). Additionally, co-translational folding of the polypeptide chain and translation rate impact each other (O’Brien et al., 2012; Ciryam et al., 2013). These factors are “hard-wired” into the process; whether there are additional factors that allow for dynamic modulation of elongation rate is still under investigation.
In translation, there is a mechanistic trade-off between elongation rate and translational accuracy (Wohlgemuth et al., 2011). To ensure fidelity, it is not necessary to globally slow translation; cells may compromise by slowing translation only at structurally or functionally important residues while sacrificing accuracy for speed at other residues (Doerfel et al., 2013). Ribosome profiling can reveal pause sites where such strategic slowing occurs. The poor resolution of ribosome profiling in bacteria compared to eukaryotes has presented technical challenges, but using known pause motifs (consecutive proline residues, and comparing results from wild-type and strains lacking the elongation factor required for rapid translation of polyproline stretches, EFP) Woolstenhulme et al. were able to adapt the ribosome profiling method to obtain higher resolution data enabling them to characterize pausing in E. coli (Woolstenhulme et al., 2015). With such methodological advancements it should be possible to identify additional pausing sites and search for patterns across the genome, relating the findings to what is known about codon conservation and structurally and functionally important sequence elements.
Termination, which in bacteria is controlled by three release factors (RFs), is necessary for translational accuracy, especially for genes in operons, and to recycle ribosomes for further rounds of translation. Ribosome profiling studies in E. coli have shown read-through beyond the stop codon in more than 50 genes with variable consequences—in some cases read-through expands the protein repertoire and in others produces deleterious proteins (Baggett et al., 2017). The antimicrobial peptide apidaecin, inhibits termination of translation. Ribosome profiling revealed that apidaecin arrests ribosomes at stop codons generating queues of trailing ribosomes. Inhibition at stop codons sequesters RFs, which in turn leads to read-through on other mRNA molecules resulting in proteomic dysregulation. Because the RFs have been sequestered by the apidaecin-stalled ribosomes, neither tmRNA nor arfA ribosome rescue mechanisms are able to recover translation (Mangano et al., 2020).
How applicable are these findings to other species of bacteria? It is clear that translation is a more dynamically regulated and highly influenced process than previously understood. Whilst some of the findings described above may be more faithfully reproduced in mycobacteria than others, it seems reasonable to hypothesize that these and/or other mechanisms that expand the “canonical” view of translation are likely to operate. With this in mind, we now turn to key findings from ribosome profiling in mycobacteria.
Despite technical challenges (discussed below) ribosome profiling has been performed in mycobacteria, leading to several important discoveries.
As discussed above, ribosome profiling has contributed to understanding the role of the SD-aSD interaction in initiation of translation in E. coli. Approximately 25% of mRNA molecules expressed by Mycobacterium tuberculosis lack a 5′ untranslated region (UTR) altogether and a further 36% have a 5′ UTR that lacks a strong SD sequence (Cortes et al., 2013; Shell et al., 2015). Ribosome profiling revealed that these SD-lacking transcripts are robustly translated under both optimal growth and nutrient starvation conditions (Sawyer et al., 2021; Figure 2). Although nucleotide resolution of ribosome position has not yet been achieved in ribosome profiling in M. tuberculosis, metagene analysis of 3′-aligned reads enabled identification of a consensus 14 nucleotide offset to the start codon in the ribosomal P-site. Comparison of the ribosome densities around the start codon between the different mRNA categories (canonical SD, no 5′ UTR and 5′ UTR lacking an SD sequence) revealed striking differences indicating different initiation mechanisms were likely operating on different types of transcripts (Sawyer et al., 2021) consistent with alternative models proposed for initiation (Boni et al., 1991; Moll et al., 2004; Komarova et al., 2005; Figure 2).
Unlike genes lacking a 5′ UTR or a SD sequence, the metagene profiles of SD genes showed a clear increased ribosome density around the start codon, which was proposed to reflect assembly of ribosomes on the mRNA and pausing due to SD-aSD binding and start codon location. To assess whether SD initiation correlated with increased rates of translation across the whole gene, the 5′ UTR was assigned a score based on the number of matches to a consensus SD sequence (obtained from the M. tuberculosis 16S rRNA aSD sequence) and the ribosome occupancy (indicative of translation rate) across the whole gene compared across SD-match scores. Genes with high Watson-Crick complementarity to the aSD had statistically significant higher ribosome occupancies than genes with fewer matches (Sawyer et al., 2021). Understood within the context of the additional influences of mRNA structure, degradation rate and other features in the vicinity of the start codon, these results suggest that despite the compensatory and competing mechanisms at play, the canonical SD-aSD initiation model is still highly relevant in influencing the translation rate of several genes, in agreement with the findings of Saito et al. in E. coli (Saito et al., 2020).
Translational misreading has been shown to increase in M. smegmatis during stationary phase and in the presence of streptomycin (Leng et al., 2015). Whilst translational fidelity is essential for life, errors that increase phenotypic diversity may play a role in adaptation and survival under certain stress conditions (Ribas de Pouplana et al., 2014). Indeed, a recent study exploring the divergence of codon usage in functionally important genes in M. tuberculosis found that selective pressure drives codon usage, with implications for understanding evolutionary divergence, the emergence of antibiotic resistance mutations and ultimately treatment outcomes (Li et al., 2022). Evaluating genome-wide translational fidelity obviously relies on gene annotation. However, an additional source of proteomic expansion/diversity is the translation of as-yet unannotated genomic elements that have not been predicted by algorithms as they lack features commonly required for identification as a gene (ORF length, SD sequence, promoter etc.).
A recent ribosome profiling study in M. tuberculosis identified many robustly translated short ORFs that had not previously been described (Smith et al., 2022). To achieve this, Smith et al. used a modified form of ribosome profiling, Ribo-RET, in which bacteria are treated with retapamulin to freeze translation during initiation (already elongating ribosomes are unaffected by retapamulin treatment) (Figure 1). Limiting the ribosome footprint signals to start codons in this way enables identification of overlapping ORFs, including those positioned in-frame with known ORFs (i.e., alternative isoforms of annotated genes) (Meydan et al., 2019; Weaver et al., 2019). Smith et al. identified thousands of robustly translated small ORFs, including hundreds that are isoforms of known proteins. Mass spectrometry, initiation reporter systems, western blotting and nucleotide composition analysis were then used to identify which of these novel polypeptides conferred fitness benefits and which may be due to translational noise. Ninety new ORFs, with a median length of 52 amino acids, showed all the hallmarks of being functional (Smith et al., 2022). Further characterization of these small proteins may lead to increased understanding about the role of proteomic diversity in M tuberculosis fitness and adaptation. This is in good agreement with ribosome profiling studies in M. smegmatis that identified many new genes encoding small proteins likely to play novel roles in diverse cellular processes (Shell et al., 2015).
Translational accuracy not only involves matching codon-anticodon pairs correctly in the appropriate frame of an mRNA transcript that is robustly (as opposed to “noisily”) translated, but also on the “second genetic code”—the correct aminoacylation of tRNAs [the complexity of this system and its evolution are reviewed in Woese et al. (2000)]. In keeping with the idea that increased phenotypic diversity confers bacterial survival benefits under certain conditions, it has been shown that mistranslation is associated with increased resistance to rifampicin, a first-line antibiotic in the treatment of tuberculosis (Javid et al., 2014; Su et al., 2016).
The gatCAB locus in M. tuberculosis encodes a heterotrimeric enzyme required for correct decoding of glutamine codons during translation (Curnow et al., 1997). Clinical isolates with mutations in these genes have increased mistranslation and higher rifampicin tolerance compared with wild-type strains (Su et al., 2016). A recent study by Chaudhuri et al. found that the aminoglycoside kasugamycin decreases mistranslation and increases susceptibility to rifampicin in M. tuberculosis (Chaudhuri et al., 2018) suggesting that increasing translational fidelity may reduce mycobacterial adaptation and make it easier to treat infection. A ribosome profiling study in E. coli found that kasugamycin treatment reshapes the bacterial translatome (Lange et al., 2017) but as the study focused on the influence of the 5′ UTR and gene families associated with kasugamycin tolerance rather than mistranslation rates, further experimentation and analysis is needed to determine the precise mechanisms underlying these observations. As the E. coli transcriptome contains very few mRNA molecules that lack a 5′ UTR, perhaps the interplay between kasugamycin, UTRs and mistranslation would be better observed in a mycobacterial model.
Just as proteomic diversity enables rapid environmental adaptation without genomic alteration, heterogeneity in ribosomes themselves is associated with preferential translation of particular subsets of genes (Byrgazov et al., 2013; Chen et al., 2020). Under conditions of zinc starvation, mycobacteria express alternative ribosomal proteins that lack the cysteine-rich motifs of their paralogs (Figure 2; Prisic et al., 2015; Dow and Prisic, 2018). Javid and co-workers used ribosome profiling to reveal that in M. smegmatis these alternative ribosomes are not only translationally active but have distinct translational profiles compared to their canonical counterparts (Chen et al., 2020). The alternative ribosome operon contributes to bacterial iron homeostasis via a mechanism that is yet to be fully elucidated but is likely to involve regulation of membrane protein and secretion apparatus synthesis. Thus, a picture of translation as a more complex and dynamically regulated process than is traditionally considered, is beginning to emerge.
When bacteria are exposed to environmental stress, their ribosomes may act as either sensors, targets or responders to the change in conditions. A full discussion of the mechanisms underlying these processes is beyond the scope of this review and has already been described by Starosta et al. (2014) and Cheng-Guang and Gualerzi (2021) so here we will focus on how ribosome profiling has contributed to understanding the interplay between ribosomes and cellular stress.
Ribosome hibernation is a widespread bacterial stress response. In E. coli this response takes two forms: long hibernation promoting factor (HPF)-mediated ribosome dimerization to form inactive 100S particles (Wada et al., 1995; Maki et al., 2000; Franken et al., 2017); and the stabilization of 70S ribosomes by ribosome associated inhibitor A (RaiA) (Agafonov et al., 1999; Vila-Sanjurjo et al., 2004). Although ribosome dimerization has been described in many species of bacteria (Ueta et al., 2010, 2013; Puri et al., 2014; Kline et al., 2015; Flygaard et al., 2018), it does not appear to occur in mycobacteria, which rather adopt the strategy of 70S stabilization, preventing the dissociation of ribosomes into their 30S and 50S subunits (Trauner et al., 2012). In this context, the upregulation of genes that have been predicted to associate with ribosomes may be associated with stabilization (Figure 2). The genes Rv1738, a putative HPF (Bunker et al., 2015); Rv3241c, which encodes RafS, a homolog of an M. smegmatis ribosome hibernation protein (Trauner et al., 2012; Mishra et al., 2018); and Rv2632c, which is related to Rv1738 (Bunker et al., 2015), were all significantly upregulated at the level of the translatome during conditions of nutrient starvation in M. tuberculosis (Sawyer et al., 2021).
Mycobacterial ribosome stabilization has implications both for the global level of translation and for which subsets of genes are most affected by this modification. The translation of leaderless (5′ UTR-lacking) mRNA molecules is initiated by assembled 70S ribosomes. Under stress conditions in mycobacteria, expression of both leaderless genes and ribosome stabilization factors increases (Cortes et al., 2013; Shell et al., 2015; Grabowska et al., 2021; Sawyer et al., 2021). Ribosome profiling has contributed to understanding the interplay between these phenomena and suggests that by adopting distinct mechanisms of translation altogether with stabilization of 70 ribosomes, M. tuberculosis may continue to robustly translate non-canonical mRNAs under different growth conditions (Sawyer et al., 2021; Figure 2).
The question of what triggers ribosome stabilization in mycobacteria has proved challenging to address, particularly in terms of its relationship to zinc remodeling (mentioned above). Recent work by Ojha and co-workers showed that different concentrations of zinc induce remodeling or hibernation and that hibernating ribosomes can be reactivated by zinc supplementation (Li et al., 2020). To date no ribosome profiling studies have looked specifically at zinc depletion, although under the conditions of nutrient starvation described in the various studies zinc concentration must have been low. The impacts of zinc depletion-induced ribosome remodeling and hibernation on the footprint lengths of mRNAs recovered in ribosome profiling may well enhance understanding of mycobacterial ribosomal stress responses.
Undoubtedly, ribosome profiling has already revealed many interesting details about translational regulation in mycobacteria; however, methodological developments are required to push the technique toward higher resolution. The single greatest challenge is applying the refinements that have improved the protocol in E. coli to a containment level 3 (BSL3) environment, especially in those were local laboratory regulations might limit the use of these techniques. For example, because most antibiotics exhibit some sequence specificity or interfere with particular ribosomal processes, they do not stall translation instantaneously and uniformly, leading to artifacts. To overcome this, many protocols employ flash freezing to stall translation (Oh et al., 2011; Becker et al., 2013; Mohammad et al., 2019). A variety of flash freezing methods have been described, including rapid vacuum harvesting of cells onto a filter which is then plunged into liquid nitrogen and spraying cultures directly into liquid nitrogen, neither of which are easily adaptable to the BSL3 laboratory. Thus, methods such as chloramphenicol inhibition of translation are still generally used in mycobacterial studies, with additional quality control steps to assess and mitigate any observed artifacts [see Sawyer et al. (2021), for example].
For all ribosome profiling studies, the choice of nuclease is important. Bacterial studies use MNase, which exhibits sequence specificity with a preference for cleavage at A and U nucleotides. For GC-rich organisms like mycobacteria, this leads to inefficiency in mRNA digestion and the footprints recovered may reflect this. Where ribosome profiling can be performed on large enough numbers of cells to generate high read numbers per gene fragment of interest, MNase sequence bias averages out sufficiently to confidently determine the A-site codon (Mohammad et al., 2019). Safety requirements limiting the volume and cell densities of mycobacterial cultures that may be grown mean that these averaging thresholds may not be reached for genes expressed at low levels. This needs to be accounted for in any analysis.
The issue of the number of cells that can be harvested for ribosome profiling also relates to a limitation that currently affects all such studies: that ribosome profiling provides a population measurement that cannot account for differences between sub-populations of cells. Thus, low frequency phenotypes such as persister cells cannot yet be specifically studied. For M. tuberculosis where persistence mechanisms are vital for its survival within the host and for disease progression, this is a significant limitation.
Despite these limitations, ribosome profiling experiments yield rich data that can greatly enhance understanding of the process and regulation of translation, a fundamental life process. Insights obtained so far challenge and refine the “canonical” view of translation and suggest more dynamic roles for many of the components of the translational machinery. Like many other biological phenomena, the more the process is studied, the more our models and understandings fall short of fully explaining the data. Given the various trade-offs at play between host and pathogen, adaptation and propagation of infection, treatment and side effects, ribosome profiling offers a means to appreciate nuance and complexity that may lead to more targeted and effective treatments for tuberculosis—which are desperately needed.
EBS and TC conceived, wrote, and reviewed the manuscript. Both authors approved the submitted version.
We acknowledged support from the University of Westminster to enable Open Access Funding.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agafonov, D. E., Kolb, V. A., Nazimov, I. V., and Spirin, A. S. (1999). A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 96, 12345–12349. doi: 10.1073/pnas.96.22.12345
Archer, S. K., Shirokikh, N. E., Beilharz, T. H., and Preiss, T. (2016). Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535, 570–574. doi: 10.1038/nature18647
Baggett, N. E., Zhang, Y., and Gross, C. A. (2017). Global analysis of translation termination in Escherichia coli. PLoS Genet. 13:e1006676. doi: 10.1371/journal.pgen.1006676
Becker, A. H., Oh, E., Weissman, J. S., Kramer, G., and Bukau, B. (2013). Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat. Protoc. 8, 2212–2239. doi: 10.1038/nprot.2013.133
Boni, I. V., Isaeva, D. M., Musychenko, M. L., Tzareva, N. V., and Shemyakin, M. M. (1991). Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19, 155–162. doi: 10.1093/nar/19.1.155
Bunker, R. D., Mandal, K., Bashiri, G., Chaston, J. J., Pentelute, B. L., Lott, J. S., et al. (2015). A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography. Proc. Natl. Acad. Sci. U.S.A. 112, 4310–4315. doi: 10.1073/pnas.1422387112
Byrgazov, K., Vesper, O., and Moll, I. (2013). Ribosome heterogeneity: Another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 16, 133–139. doi: 10.1016/j.mib.2013.01.009
Chaudhuri, S., Li, L., Zimmerman, M., Chen, Y., Chen, Y.-X., Toosky, M. N., et al. (2018). Kasugamycin potentiates rifampicin and limits emergence of resistance in Mycobacterium tuberculosis by specifically decreasing mycobacterial mistranslation. Elife 7:e36782. doi: 10.7554/eLife.36782
Chen, Y.-X., Xu, Z., Ge, X., Sanyal, S., Lu, Z. J., and Javid, B. (2020). Selective translation by alternative bacterial ribosomes. Proc. Natl. Acad. Sci. U.S.A. 117, 19487–19496. doi: 10.1073/pnas.2009607117
Cheng-Guang, H., and Gualerzi, C. O. (2021). The ribosome as a switchboard for bacterial stress response. Front. Microbiol. 11:619038. doi: 10.3389/fmicb.2020.619038
Ciryam, P., Morimoto, R. I., Vendruscolo, M., Dobson, C. M., and O’Brien, E. P. (2013). In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc. Natl. Acad. Sci. U.S.A. 110, E132–E140. doi: 10.1073/pnas.1213624110
Cortes, T., Schubert, O. T., Banaei-Esfahani, A., Collins, B. C., Aebersold, R., and Young, D. B. (2017). Delayed effects of transcriptional responses in Mycobacterium tuberculosis exposed to nitric oxide suggest other mechanisms involved in survival. Sci. Rep. 7:8208. doi: 10.1038/s41598-017-08306-1
Cortes, T., Schubert, O. T., Rose, G., Arnvig, K. B., Comas, I., Aebersold, R., et al. (2013). Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 5, 1121–1131. doi: 10.1016/j.celrep.2013.10.031
Curnow, A. W., Hong, K. W., Yuan, R., Kim, S. Il, Martins, O., Winkler, W., et al. (1997). Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. U.S.A. 94, 11819–11826. doi: 10.1073/PNAS.94.22.11819
Dai, X., Zhu, M., Warren, M., Balakrishnan, R., Patsalo, V., Okano, H., et al. (2016). Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2:16231. doi: 10.1038/nmicrobiol.2016.231
Del Campo, C., Bartholomäus, A., Fedyunin, I., and Ignatova, Z. (2015). Secondary Structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet. 11:e1005613. doi: 10.1371/journal.pgen.1005613
Doerfel, K. L., Ingo, W., Christina, K., Frank, P., Henning, U., and Rodnina, M. V. (2013). EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88. doi: 10.1126/science.1229017
Dow, A., and Prisic, S. (2018). Alternative ribosomal proteins are required for growth and morphogenesis of Mycobacterium smegmatis under zinc limiting conditions. PLoS One 13:e0196300. doi: 10.1371/journal.pone.0196300
Duval, M., Simonetti, A., Caldelari, I., and Marzi, S. (2015). Multiple ways to regulate translation initiation in bacteria: Mechanisms, regulatory circuits, dynamics. Biochimie 114, 18–29. doi: 10.1016/J.BIOCHI.2015.03.007
Flygaard, R. K., Boegholm, N., Yusupov, M., and Jenner, L. B. (2018). Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism. Nat. Commun. 9:4179. doi: 10.1038/s41467-018-06724-x
Franken, L. E., Oostergetel, G. T., Pijning, T., Puri, P., Arkhipova, V., Boekema, E. J., et al. (2017). A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat. Commun. 8:722. doi: 10.1038/s41467-017-00718-x
Giliberti, J., O’Donnell, S., Van Etten, W. J., and Janssen, G. R. (2012). A 5′-terminal phosphate is required for stable ternary complex formation and translation of leaderless mRNA in Escherichia coli. RNA 18, 508–518. doi: 10.1261/rna.027698.111
Grabowska, A. D., Andreu, N., and Cortes, T. (2021). Translation of a leaderless reporter is robust during exponential growth and well sustained during stress conditions in Mycobacterium tuberculosis. Front. Microbiol. 12:746320. doi: 10.3389/FMICB.2021.746320
Haider, S., and Pal, R. (2013). Integrated analysis of transcriptomic and proteomic data. Curr. Genomics 14, 91–110.
Hockenberry, A. J., Pah, A. R., Jewett, M. C., and Amaral, L. A. N. (2017). Leveraging genome-wide datasets to quantify the functional role of the anti-Shine-Dalgarno sequence in regulating translation efficiency. Open Biol. 7:160239. doi: 10.1098/rsob.160239
Holm, L. (1986). Codon usage and gene expression. Nucleic Acids Res. 14, 3075–3087. doi: 10.1093/nar/14.7.3075
Hwang, J.-Y., and Buskirk, A. R. (2017). A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res. 45, 327–336. doi: 10.1093/nar/gkw944
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S., and Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223.
Javid, B., Sorrentino, F., Toosky, M., Zheng, W., Pinkham, J. T., Jain, N., et al. (2014). Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl. Acad. Sci. U.S.A. 111, 1132–1137. doi: 10.1073/pnas.1317580111
Kline, B. C., McKay, S. L., Tang, W. W., and Portnoy, D. A. (2015). The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J. Bacteriol. 197, 581–591. doi: 10.1128/JB.02223-14
Komarova, A. V., Tchufistova, L. S., Dreyfus, M., and Boni, I. V. (2005). AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 187, 1344–1349. doi: 10.1128/JB.187.4.1344-1349.2005
Lange, C., Lehr, M., Zerulla, K., Ludwig, P., Schweitzer, J., Polen, T., et al. (2017). Effects of kasugamycin on the translatome of Escherichia coli. PLoS One 12:e0168143. doi: 10.1371/journal.pone.0168143
Leng, T., Pan, M., Xu, X., and Javid, B. (2015). Translational misreading in Mycobacterium smegmatis increases in stationary phase. Tuberculosis 95, 678–681. doi: 10.1016/j.tube.2015.09.010
Li, G. W. (2015). How do bacteria tune translation efficiency? Curr. Opin. Microbiol. 24, 66–71. doi: 10.1016/J.MIB.2015.01.001
Li, G. W., Burkhardt, D., Gross, C., and Weissman, J. S. (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635. doi: 10.1016/j.cell.2014.02.033
Li, G., Zhang, L., and Xue, P. (2022). Codon usage divergence of important functional genes in Mycobacterium tuberculosis. Int. J. Biol. Macromol. 209, 1197–1204. doi: 10.1016/J.IJBIOMAC.2022.04.112
Li, G.-W., Oh, E., and Weissman, J. S. (2012). The anti-shine–dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541. doi: 10.1038/nature10965
Li, Y., Corro, J. H., Palmer, C. D., and Ojha, A. K. (2020). Progression from remodeling to hibernation of ribosomes in zinc-starved mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 117, 19528–19537. doi: 10.1073/pnas.2013409117
Maier, T., Güell, M., Serrano, L., and Hohmann, S. (2009). Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973.
Maki, Y., Yoshida, H., and Wada, A. (2000). Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5, 965–974. doi: 10.1046/j.1365-2443.2000.00389.x
Mangano, K., Florin, T., Shao, X., Klepacki, D., Chelysheva, I., Ignatova, Z., et al. (2020). Genome-wide effects of the antimicrobial peptide apidaecin on translation termination in bacteria. Elife 9, 1–24. doi: 10.7554/eLife.62655
Meydan, S., Marks, J., Klepacki, D., Kefi, A., Vá Zquez-Laslop, N., and Mankin, A. S. (2019). Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome in brief. Mol. Cell 74, 481–493.e6. doi: 10.1016/j.molcel.2019.02.017
Mishra, S., Ahmed, T., Tyagi, A., Shi, J., and Bhushan, S. (2018). Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA. Sci. Rep. 8:13587. doi: 10.1038/s41598-018-31850-3
Mohammad, F., Green, R., and Buskirk, A. R. (2019). A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife 8:e42591. doi: 10.7554/elife.42591
Moll, I., Hirokawa, G., Kiel, M. C., Kaji, A., and Bläsi, U. (2004). Translation initiation with 70S ribosomes: An alternative pathway for leaderless mRNAs. Nucleic Acids Res. 32, 3354–3363. doi: 10.1093/nar/gkh663
O’Brien, E. P., Vendruscolo, M., and Dobson, C. M. (2012). Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun. 3:868. doi: 10.1038/ncomms1850
O’Donnell, S. M., and Janssen, G. R. (2001). The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 183, 1277–1283. doi: 10.1128/JB.183.4.1277-1283.2001
Oh, E., Becker, A. H., Sandikci, A., Huber, D., Chaba, R., Gloge, F., et al. (2011). Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308. doi: 10.1016/j.cell.2011.10.044
Plotkin, J. B., and Kudla, G. (2011). Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42. doi: 10.1038/nrg2899
Prisic, S., Hwang, H., Dow, A., Barnaby, O., Pan, T. S., Lonzanida, J. A., et al. (2015). Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis. Mol. Microbiol. 97, 263–280. doi: 10.1111/mmi.13022
Puri, P., Eckhardt, T. H., Franken, L. E., Fusetti, F., Stuart, M. C. A., Boekema, E. J., et al. (2014). Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol. Microbiol. 91, 394–407. doi: 10.1111/mmi.12468
Reeve, B., Hargest, T., Gilbert, C., and Ellis, T. (2014). Predicting translation initiation rates for designing synthetic biology. Front. Bioeng. Biotechnol. 2:1. doi: 10.3389/fbioe.2014.00001
Riba, A., Di Nanni, N., Mittal, N., Arhné, E., Schmidt, A., and Zavolan, M. (2019). Protein synthesis rates and ribosome occupancies reveal determinants of translation elongation rates. Proc. Natl. Acad. Sci. U.S.A. 116, 15023–15032.
Ribas de Pouplana, L., Santos, M. A. S., Zhu, J. H., Farabaugh, P. J., and Javid, B. (2014). Protein mistranslation: Friend or foe? Trends Biochem. Sci. 39, 355–362. doi: 10.1016/J.TIBS.2014.06.002
Saito, K., Green, R., and Buskirk, A. R. (2020). Translational initiation in E. Coli occurs at the correct sites genome-wide in the absence of mrna-rrna base-pairing. Elife 9:e55002. doi: 10.7554/eLife.55002
Sawyer, E. B., Phelan, J. E., Clark, T. G., and Cortes, T. (2021). A snapshot of translation in Mycobacterium tuberculosis during exponential growth and nutrient starvation revealed by ribosome profiling. Cell Rep. 34:108695. doi: 10.1016/j.celrep.2021.108695
Schrader, J. M., Zhou, B., Li, G.-W., Lasker, K., Childers, W. S., Williams, B., et al. (2014). The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 10:e1004463. doi: 10.1371/journal.pgen.1004463
Shah, P., and Gilchrist, M. A. (2010). Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet. 6:e1001128. doi: 10.1371/journal.pgen.1001128
Sharma, H., and Anand, B. (2019). Ribosome assembly defects subvert initiation Factor3 mediated scrutiny of bona fide start signal. Nucleic Acids Res. 47, 11368–11386. doi: 10.1093/nar/gkz825
Shell, S. S., Wang, J., Lapierre, P., Mir, M., Chase, M. R., Pyle, M. M., et al. (2015). Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 11:e1005641. doi: 10.1371/journal.pgen.1005641
Shine, J., and Dalgarno, L. (1974). The 3′ terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 71, 1342–1346. doi: 10.1073/pnas.71.4.1342
Shirokikh, N. E., Archer, S. K., Beilharz, T. H., Powell, D., and Preiss, T. (2017). Translation complex profile sequencing to study the in vivo dynamics of mRNA-ribosome interactions during translation initiation, elongation and termination. Nat. Protoc. 12, 697–731. doi: 10.1038/nprot.2016.189
Smith, C., Canestrari, J. G., Wang, A. J., Champion, M. M., Derbyshire, K. M., Gray, T. A., et al. (2022). Pervasive translation in Mycobacterium tuberculosis. Elife 11:e73980. doi: 10.7554/eLife.73980
Starosta, A. L., Lassak, J., Jung, K., and Wilson, D. N. (2014). The bacterial translation stress response. FEMS Microbiol. Rev. 38, 1172–1201.
Su, H. W., Zhu, J. H., Li, H., Cai, R. J., Ealand, C., Wang, X., et al. (2016). The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat. Microbiol. 1:16147. doi: 10.1038/nmicrobiol.2016.147
Subramaniam, A. R., Zid, B. M., and O’Shea, E. K. (2014). An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159, 1200–1211. doi: 10.1016/J.CELL.2014.10.043
Trauner, A., Lougheed, K. E. A., Bennett, M. H., Hingley-Wilson, S. M., and Williams, H. D. (2012). The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J. Biol. Chem. 287, 24053–24063. doi: 10.1074/jbc.M112.364851
Ueta, M., Wada, C., and Wada, A. (2010). Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15, 43–58. doi: 10.1111/j.1365-2443.2009.01364.x
Ueta, M., Wada, C., Daifuku, T., Sako, Y., Bessho, Y., Kitamura, A., et al. (2013). Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18, 554–574. doi: 10.1111/gtc.12057
Vila-Sanjurjo, A., Schuwirth, B.-S., Hau, C. W., and Cate, J. H. D. (2004). Structural basis for the control of translation initiation during stress. Nat. Struct. Mol. Biol. 11, 1054–1059. doi: 10.1038/nsmb850
Vogel, C., and Marcotte, E. M. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232. doi: 10.1038/nrg3185
Wada, A., Igarashi, K., Yoshimura, S., Aimoto, S., and Ishihama, A. (1995). Ribosome modulation factor: Stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem. Biophys. Res. Commun. 214, 410–417. doi: 10.1006/BBRC.1995.2302
Weaver, J., Mohammad, F., Buskirk, A. R., and Storz, G. (2019). Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10:e02819–18. doi: 10.1128/mBio.02819-18
Woese, C. R., Olsen, G. J., Ibba, M., and Söll, D. (2000). Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 64, 202–236. doi: 10.1128/MMBR.64.1.202-236.2000
Wohlgemuth, I., Pohl, C., Mittelstaet, J., Konevega, A. L., and Rodnina, M. V. (2011). Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2979–2986. doi: 10.1098/rstb.2011.0138
Keywords: ribosome profiling, Mycobacterium tuberculosis, non-canonical translation, translation initiation, omics
Citation: Sawyer EB and Cortes T (2022) Ribosome profiling enhances understanding of mycobacterial translation. Front. Microbiol. 13:976550. doi: 10.3389/fmicb.2022.976550
Received: 23 June 2022; Accepted: 22 July 2022;
Published: 04 August 2022.
Edited by:
Nuno S. Osório, ICVS/3B’s Associate Laboratory (AL), PortugalReviewed by:
Jack A. Dunkle, The University of Alabama, United StatesCopyright © 2022 Sawyer and Cortes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth B. Sawyer, Qi5TYXd5ZXIxQHdlc3RtaW5zdGVyLmFjLnVr; Teresa Cortes, dGNvcnRlc0BpYnYuY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.