
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 November 2022
Sec. Extreme Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.975365
This article is part of the Research Topic Community Series-Extremophiles: Microbial Genomics and Taxogenomics, Volume II View all 16 articles
A correction has been applied to this article in:
Corrigendum: Genotype–phenotype correlations within the Geodermatophilaceae
 Maria del Carmen Montero-Calasanz1,2*
Maria del Carmen Montero-Calasanz1,2* Adnan Yaramis2
Adnan Yaramis2 Manfred Rohde3
Manfred Rohde3 Peter Schumann4
Peter Schumann4 Hans-Peter Klenk2
Hans-Peter Klenk2 Jan P. Meier-Kolthoff5
Jan P. Meier-Kolthoff5The integration of genomic information into microbial systematics along with physiological and chemotaxonomic parameters provides for a reliable classification of prokaryotes. In silico analysis of chemotaxonomic traits is now being introduced to replace characteristics traditionally determined in the laboratory with the dual goal of both increasing the speed of the description of taxa and the accuracy and consistency of taxonomic reports. Genomics has already successfully been applied in the taxonomic rearrangement of Geodermatophilaceae (Actinomycetota) but in the light of new genomic data the taxonomy of the family needs to be revisited. In conjunction with the taxonomic characterisation of four strains phylogenetically located within the family, we conducted a phylogenetic analysis of the whole proteomes of the sequenced type strains and established genotype–phenotype correlations for traits related to chemotaxonomy, cell morphology and metabolism. Results indicated that the four isolates under study represent four novel species within the genus Blastococcus. Additionally, the genera Blastococcus, Geodermatophilus and Modestobacter were shown to be paraphyletic. Consequently, the new genera Trujillonella, Pleomorpha and Goekera were proposed within the Geodermatophilaceae and Blastococcus endophyticus was reclassified as Trujillonella endophytica comb. nov., Geodermatophilus daqingensis as Pleomorpha daqingensis comb. nov. and Modestobacter deserti as Goekera deserti comb. nov. Accordingly, we also proposed emended descriptions of Blastococcus aggregatus, Blastococcus jejuensis, Blastococcus saxobsidens and Blastococcus xanthilyniticus. In silico chemotaxonomic results were overall consistent with wet-lab results. Even though in silico discriminatory levels varied depending on the respective chemotaxonomic trait, this approach is promising for effectively replacing and/or complementing chemotaxonomic analyses at taxonomic ranks above the species level. Finally, interesting but previously overlooked insights regarding morphology and ecology were revealed by the presence of a repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly in all representatives of the family. A rich carbon metabolism including four different CO2 fixation pathways and a battery of enzymes able to degrade complex carbohydrates were also identified in Blastococcus genomes.
The family Geodermatophilaceae, belonging to order Geodermatophilales (Sen et al., 2014), was initially proposed by Normand et al. (1996), confirmed by Stackebrandt et al. (1997), formally described by Normand (2006) and later emended by Zhi et al. (2009) and Montero-Calasanz et al. (2017). The family accommodates the genera Blastococcus (Ahrens and Moll, 1970; Urzì et al., 2004; Lee, 2006; Hezbri et al., 2016), Geodermatophilus, the type genus (Luedemann, 1968; Montero-Calasanz et al., 2017; Montero-Calasanz, 2020a), Klenkia (Montero-Calasanz et al., 2017), and Modestobacter (Mevs et al., 2000; Reddy et al., 2007; Xiao et al., 2011; Qin et al., 2013; Montero-Calasanz et al., 2019).
The genus Blastococcus consists of Gram-reaction-positive aerobic bacteria that can be distinguished by the presence of individual motile rod-shaped cells and/or non-motile cocci (Lee, 2006) with a tendency to form aggregates (Urzì et al., 2004; Hezbri et al., 2016). At the time of writing, the genus comprises 10 validly named species including B. aggregatus (type species), isolated from sediment from Baltic Sea (Ahrens and Moll, 1970), B. atacamensis, isolated from an extreme hyper-arid Atacama Desert soil (Castro et al., 2018), B. capsensis, isolated from an ancient Roman pool in Tunisia (Hezbri et al., 2016), B. colisei, isolated from the ruins of a Roman amphitheatre in Tunisia (Hezbri et al., 2017), B. deserti, isolated from Gurbantunggut Desert in China (Yang et al., 2019), B. endophyticus, isolated from Chinese medicinal plant leaves (Zhu et al., 2013; Hezbri et al., 2016), B. jejuensis, isolated from sand in Korea (Lee, 2006; Hezbri et al., 2016), B. litoris, isolated from sea-tidal flat sediment (Lee et al., 2018), B. saxobsidens, isolated from an archaeological site in Greece (Urzì et al., 2004), and B. xanthinilyticus, isolated from a marble sample collected from the Bulla Regia monument in Tunisia (Hezbri et al., 2018).
The integration of genomic information into microbial systematics (Klenk and Göker, 2010) along with physiological and chemotaxonomic parameters enables a reliable classification of prokaryotes (Chun et al., 2018). Laboratory-based DNA–DNA hybridisation methods have already been routinely replaced by overall genome relatedness indices (OGRI; Chun et al., 2018) of which digital DNA:DNA hybridization was shown to be the best-performing method (Meier-Kolthoff et al., 2013a). The use of in silico analysis for chemotaxonomic traits is now being introduced to replace characteristics traditionally determined in the laboratory (Amaral et al., 2014; Fotedar et al., 2020; Lawson et al., 2020) with the dual goal of both increasing the speed of the description of taxa and the accuracy and consistency of taxonomic reports (Chun and Rainey, 2014; Whitman, 2015).
Genomics has already successfully been applied in the taxonomic rearrangement of Geodermatophilaceae (Montero-Calasanz et al., 2017) although other research studies based on genomic data are still scarce in the family. In particular, only two papers were published so far about the genus Blastococcus including genomic and proteomic mining of B. saxobsidens DD2 and genome mining of B. atacamensis which revealed numerous genes involved in stress response and adaptation to harsh habitats (Chouaia et al., 2012; Sghaier et al., 2016) apart from an exceptional potential to produce novel natural compounds (Castro et al., 2018).
Based on genome-scale data and phenotypic evidence according to the principles of phylogenetic systematics (Hennig, 1965; Wiley and Lieberman, 2011), this study characterises four novel species in the genus Blastococcus, introduces the new genera Trujillonella, Pleomorpha, and Goekera in Geodermatophilaceae and reclassifies Blastococcus endophyticus as Trujillonella endophytica comb. nov., Geodermatophilus daqingensis as Pleomorpha daqingensis comb. nov. and Modestobacter deserti as Goekera deserti comb. nov., all representing type species of the new genera. Accordingly, we also propose emended descriptions within Blastococcus. Additionally, we emphasise how genomics could effectively replace and/or complement some of the phenotypic analyses routinely carried out in microbial systematics.
Strains AT 7-1T, AT 7–8, AT 7-14T, and AT 7(-2)-11T were isolated during a screening of microorganisms from soil samples from the Atacama Desert (Chile) in the 90s and deposited by Prof. Fred A. Rainey (University of Alaska Anchorage, United States) at Leibniz Institute DSMZ. On the other hand, strain G1ST was isolated by Eppard et al. (1996) from leachate of a landfill in Vancouver (Canada). All strains were subsequently accessed in the DSMZ open collection as DSM 44268T, DSM 44269, DSM 44270T, DSM 44272T, and DSM 44205T, respectively.
Genomic DNA extraction, PCR-mediated amplification of the 16S rRNA gene, and purification of the PCR product were carried out as previously described by Rainey et al. (1996). For genome sequencing, strains were cultivated in GYM Streptomyces broth (DSMZ medium 65, pH 6.8 ± 0.2; https://www.dsmz.de/collection/catalogue/microorganisms/culture-technology/list-of-media-for-microorganisms) at 28°C and genomic DNA was extracted by using JetFlex™ Genomic DNA Purification Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. The project information is available through the Genomes Online Database (Mukherjee et al., 2017). The draught genomes were generated at the DOE Joint Genome Institute (JGI) as part of the Genomic Encyclopaedia of Archaeal and Bacterial Type Strains, Phase II (KMG-II): “From individual species to whole genera” (Kyrpides et al., 2014) and ACTINO 1000: “Exploiting the genomes of the Actinobacteria: plant growth promoters and producers of natural products and energy relevant enzymes united in a taxonomically unresolved phylum,” following the same protocol as in Nouioui et al. (2017). All genomes were annotated using the DOE-JGI annotation pipeline (Huntemann et al., 2015; Chen et al., 2016) and released through the Integrated Microbial Genomes system (Chen et al., 2017). In addition, genomes were uploaded to RAST (Aziz et al., 2008; Brettin et al., 2015) and analysed through the SEED viewer (Overbeek et al., 2014). CRISPRFinder (Grissa et al., 2007) was used to identify CRISPR elements. Further details about sequencing projects are summarised in the Supplementary Table S1.
Phylogenetic analysis of the 16S rRNA gene sequences from the type strains of all species with validated names in Geodermatophilaceae, as well as those from Antricoccus suffuscus DSM 100065T, Cryptosporangium arvum DSM 44712T, Cryptosporangium aurantiacum DSM 46144T, Epidermidibacterium keratini JCM 31644T, Longivirga aurantiaca NBRC 112237T and Sporichthya polymorpha DSM 43042T for use as outgroup, was conducted as previously described (Göker et al., 2011; Montero-Calasanz et al., 2014). Pairwise 16S rRNA gene sequence similarities were calculated as recommended by Meier-Kolthoff et al. (2013b) to determine strains with ≥99.0% similarity, between which (digital) DNA:DNA hybridization experiments were conducted for clarifying species affiliation. Genome-scale phylogenies were inferred from the available Geodermatophilaceae (and outgroup) whole proteome sequences using the high-throughput version (Meier-Kolthoff et al., 2014a) of the Genome BLAST Distance Phylogeny (GBDP) approach (Auch et al., 2010) in conjunction with FastME (Lefort et al., 2015). The tree was rooted according to Nouioui et al. (2018). The entire approach is implemented in the Type (Strain) Genome Server (TYGS; Meier-Kolthoff and Göker, 2019; Meier-Kolthoff et al., 2022). The GBDP tree restricted to the well-supported branches (≥95% pseudo-bootstrap support) was used as a backbone constraint in a subsequent 16S rRNA gene sequence analysis to integrate information from genome-scale data (Hahnke et al., 2016). Digital DNA:DNA hybridisations were conducted using the recommended settings of the Genome-To-Genome Distance Calculator (GGDC) version 3.0 (Meier-Kolthoff et al., 2013a) as implemented in the TYGS (Meier-Kolthoff and Göker, 2019; Meier-Kolthoff et al., 2022). The G + C content was calculated from the genome sequences as described by Meier-Kolthoff et al. (2014b). The genomic homogeneity of strains DSM 44268T and DSM 44269 was examined by ribotyping and matrix-assisted laser-desorption/ionisation time-of-flight (MALDI-TOF) mass spectra (MS). Automated ribotyping of PvuII-digested samples was carried out using the RiboPrinter microbial characterisation system (Qualicon, DuPont) as described in Bruce (1996) and Stackebrandt et al. (2002). MALDI-TOF analysis was performed according to Tóth et al. (2008).
Morphological features of colonies were examined by stereomicroscope on GYM Streptomyces agar at 28°C and observed periodically during 7 days. Bacterial cell morphology and motility was determined by phase contrast microscopy with a magnification of 100x and with a field-emission scanning electron microscope (FE-SEM Merlin, Zeiss, Germany). Catalase production was tested on slides with the addition of some drops of 3% H2O2 solution into a loop of bacterial biomass. It was considered positive when the production of bubbles was observed after mixing. Oxidase activity was determined by obtaining blue-purple colour after addition of some drops of 1% (w/v) solution of N,N,N′,N′-tetramethyl-p-phenylenediamine (Sigma-Aldrich) to a loop of bacterial biomass spread on filter paper. Gram reaction was performed according to the procedure developed by Gregersen (1978). API ZYM system (bioMérieux) was used to determine enzymatic activities according to the manufacturer’s protocol at 28°C after 24 h incubation. Homogeneous bacterial suspensions of strains DSM 44205T, DSM 44268T, DSM 44269, DSM 44270T, and DSM 44272T along with the reference type strains in the genus Blastococcus were prepared in viscous inoculating fluid (IF C) at 86% T (Transmittance) for B. atacamensis P6T, 83% T for B. capsensis DSM 46835T, 89% T for DSM 44270T, B. colisei DSM 46837T, and B. xanthinilyticus DSM 46842T, 90% T for strains DSM 44268T, DSM 44269, and DSM 44205T, 91% T for DSM 44272T, and 95% T for all the others reference strains. Then those were inoculated in GEN III Microplates per duplicate and incubated for 10 days at 28°C in an Omnilog device (BIOLOG Inc., Hayward, CA, United States). Data were analysed using the opm package R v.1.3.72 (Vaas et al., 2012, 2013). Reactions that reflected different behaviour between replicates were regarded as ambiguous. Temperature ranges were evaluated on GYM Streptomyces medium for 15 days at 4, 10, 15, 20, 25, 30, 37, 40 and 45°C. pH ranges were determined using modified ISP2 medium (Montero-Calasanz et al., 2015) from 4.5 to 12.5 in increments of 0.5 pH unit adjusting with NaOH or HCl since the use of a buffer system inhibited the growth of strains (Montero-Calasanz et al., 2014). pH of solidified plates was measured before inoculation and when recording results (15 days post-inoculation). Hydrolysis of specific substrates such as tyrosine (Gordon and Smith, 1955), casein, starch, xanthine as well as hypoxanthine was also tested as outlined by Montero-Calasanz et al. (2015).
For chemotaxonomic analysis, strains were grown in GYM Streptomyces broth with shaking (120 rpm) at 28°C for 7 days. Bacterial biomass was collected and subsequently freeze dried. Extraction of whole-cell amino acids and sugars was carried out as developed by Lechevalier and Lechevalier (1970) followed by thin layer chromatography (TLC) analysis and identification (Staneck and Roberts, 1974). Analysis of peptidoglycan hydrolysates (6N HCl, 100°C for 16 h) was performed on cellulose TLC plates as described by Schleifer and Kandler (1972). Menaquinones (MK) analyses (Kroppenstedt, 1982) were accomplished as indicated by Collins et al. (1977) following the identification of MK by high-performance liquid chromatography (HPLC; Kroppenstedt, 1982). Polar lipids were extracted and identified by 2D TLC as outlined by Minnikin et al. (1984) with modifications proposed by Kroppenstedt and Goodfellow (2006). Dragendorff’s reagent (Merck Millipore, 102035) was additionally sprayed to identify choline-containing lipids (Tindall, 1990). For fatty acid analysis, cell biomass grown on PYGV (DSMZ 621) agar plates for 16 days at 20°C was harvested and fatty acid methyl esters were extracted according to Sasser (2001). Microbial Identification System (MIDI) Sherlock Version 6.1 (method TSBA40, ACTIN6 database) was used for performing data analysis. All the chemotaxonomic analyses for strains were carried out under standardised conditions.
Standard draught genomes were obtained for strains DSM 44205T, DSM 44268T, DSM 44270T, and DSM 44272T and type strains of species B. aggregatus, B. endophyticus and B. xanthinilyticus. An improved-high-quality genome draught according to GOLD sequencing quality standards (Mukherjee et al., 2017) was also obtained for both B. colisei DSM 46837T and B. saxobsidens DSM 44509T (Supplementary Table S1). The genome sizes ranged from 4.0 Mbp (strain DSM 44205T) to 5.1 Mbp (strains DSM 44272T and DSM 46837T), with an average genome size for all sequenced Blastococcus strains of 4.6 ± 0.4 Mbp (Supplementary Table S2). The genomic G + C content of the sole chromosome in the type strains varied between 72.5–74.6%. Deviations from the published G + C contents of reference strains were all below the 1% threshold determined within species (Meier-Kolthoff et al., 2014b). In accordance with the genome sizes, the number of protein-coding genes ranged from 3,939 for strain DSM 44205T to 5,007 for strain DSM 44272T, not observing a priori any association between genome size and habitat of isolation. Both tRNA and rRNA genes represented between 1.3% (47 tRNA and five rRNA genes) in B. xanthinilyticus DSM 46842T and 2.2% (70 tRNA and nine rRNA genes) in DSM 44205T of the whole genome sequences. A single copy of the 16S rRNA gene was identified for strains DSM 44270T, DSM 44272T, B. aggregatus DSM 4725T, and B. xanthinilyticus DSM 46842T. Three almost identical 16S rRNA genes copies were annotated in the genome sequences of strain DSM 44205T, B. colisei DSM 46837T, B. endophyticus DSM 45413T, and B. saxobsidens DSM 44509T, and four almost identical ones in the genome sequence of strain DSM 44268T (intragenomic heterogeneity of 16S rRNA genes is below the 1% level for species delimitation; Meier-Kolthoff et al., 2013a). Previous studies already reported low copy numbers (2–4) for slow-growing actinobacteria (Sun et al., 2013; Vetrovsky and Baldrian, 2013) isolated from oligotrophic environments (Klappenbach et al., 2000). The number of rRNA operon copies is suggested to be related to the life strategy of bacteria and how quickly they respond to favourable changes in environmental conditions and nutrient availability (Klappenbach et al., 2000). Hence, rRNA copy number appears to be correlated with growth speed. The number of identified tRNAs ranged from 47 to 49 in all the Blastococcus representatives except in the genome sequence of strain DSM 44205T that contained 70 tRNAs. The 20 standard tRNA genes were present in all genomes but in a different distribution. For example, four tRNA-Arg gene copies (six gene copies for strain DSM 44205T) versus one tRNA-Tyr gene copy. Such uneven distribution supports the co-adaptation between tRNA abundance and specific codon usage of each organism as a higher ratio of optimal codons may facilitate the transcription efficiency (Du et al., 2017). Out of the 70 (DSM 44205T) and 48 (B. endophyticus) annotated tRNAs, seven (DSM 44205T) and one (B. endophyticus), respectively, were identified as pseudogene(s). The latter are known to be dysfunctional in translation but to be potentially involved in modulating other processes such as antibiotic and cell wall biosynthesis and gene expression (Rogers et al., 2012). One copy of nonsense suppressor tRNA (tRNA amber gene) was also identified in the repertoires of DSM 44270T, DSM 44272T, and B. colisei, which could prevent the premature termination of translation caused by nonsense mutations (Ko et al., 2013). The number of pseudogenes was within the expected values for free-living prokaryotes (0%–5%; Liu et al., 2004) and varied from zero (for example: DSM 44205T, DSM 44272T, B. endophyticus, B. xanthinilyticus or B. atacamensis, amongst others) to 123 in B. colisei DSM 46837T (2.5%). Each genome contained a largely unique set of pseudogenes, with over half of those annotated as “hypothetical” proteins, suggesting that pseudogenes in Blastococcus are formed and eliminated relatively rapidly as previously reported by Kuo and Ochman (2010) for Salmonella genomes. In addition, the number of paralogous genes varied from 437 (11%) in strain DSM 44205T to 1,601 (32%) in B. colisei DSM 46837T, the signal peptide percentage was 3.0%–5.8% whilst the percentage of transmembrane proteins ranged from 19.7% to 24.7%. According to CRISPRFinder, the number of “questionable” Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) ranged from zero to seven (Supplementary Table S2). Additional analyses to test the genetic environment often associated with CRISPR structures should nevertheless be carried out to confirm the presence of a naturally occurring genome editing system in Blastococcus representatives.
The average percentage of genes with a predicted function in the Clusters of Orthologous Groups (COGs) database was 63.3 ± 2.3% (ranged from 2,558 to 3,117 genes; Supplementary Table S2). As expected, functional COG profiles of Blastococcus strains are similar (Supplementary Figure S1; Supplementary Table S3).
The phylogenetic tree inferred by whole proteomes of the sequenced type strains and strains under study revealed the affiliation of DSM 44268T, DSM 44270T, DSM 44272T and DSM 44205T to the genus Blastococcus (Figure 1). The subtree containing the genera Geodermatophilus, Modestobacter and Blastococcus was not monophyletic which was caused by three misclassified species. First, B. endophyticus DSM 45413T appeared with maximum support as a sister group of Geodermatophilus and was not phylogenetically placed within the maximally supported subtree harbouring all the other Blastococcus type strains. Second, G. daqingensis DSM 104001T represented a maximally supported sister group to the large subtree comprising all other Geodermatophilus and Blastococcus type strains. Third, M. deserti CPCC 205119T formed a deeply-branching and maximally supported sister group of Klenkia.

Figure 1. Phylogenomic tree inferred with GBDP. The tree was inferred with FastME from GBDP distances calculated from whole proteomes. The numbers above branches are GBDP pseudo-bootstrap support values from 100 replications. Tip colours indicate genome sizes and the exact G + C content as calculated from the genome sequences (see the embedded legend for details). A., Antricoccus; B., Blastococcus; C., Cryptosporangium; G., Geodermatophilus; K., Klenkia; M., Modestobacter; S., Sporichthya.
The constrained 16S rRNA phylogenetic tree (Figure 2) based on gene sequences of all validly published species in the family and strains under study, including strain DSM 44269, confirmed the observations from the proteome-based GBDP analysis. Moreover, strains DSM 44268T and DSM 44269 were placed with high support in a clade together with strains DSM 44205T and B. aggregatus DSM 4725T.
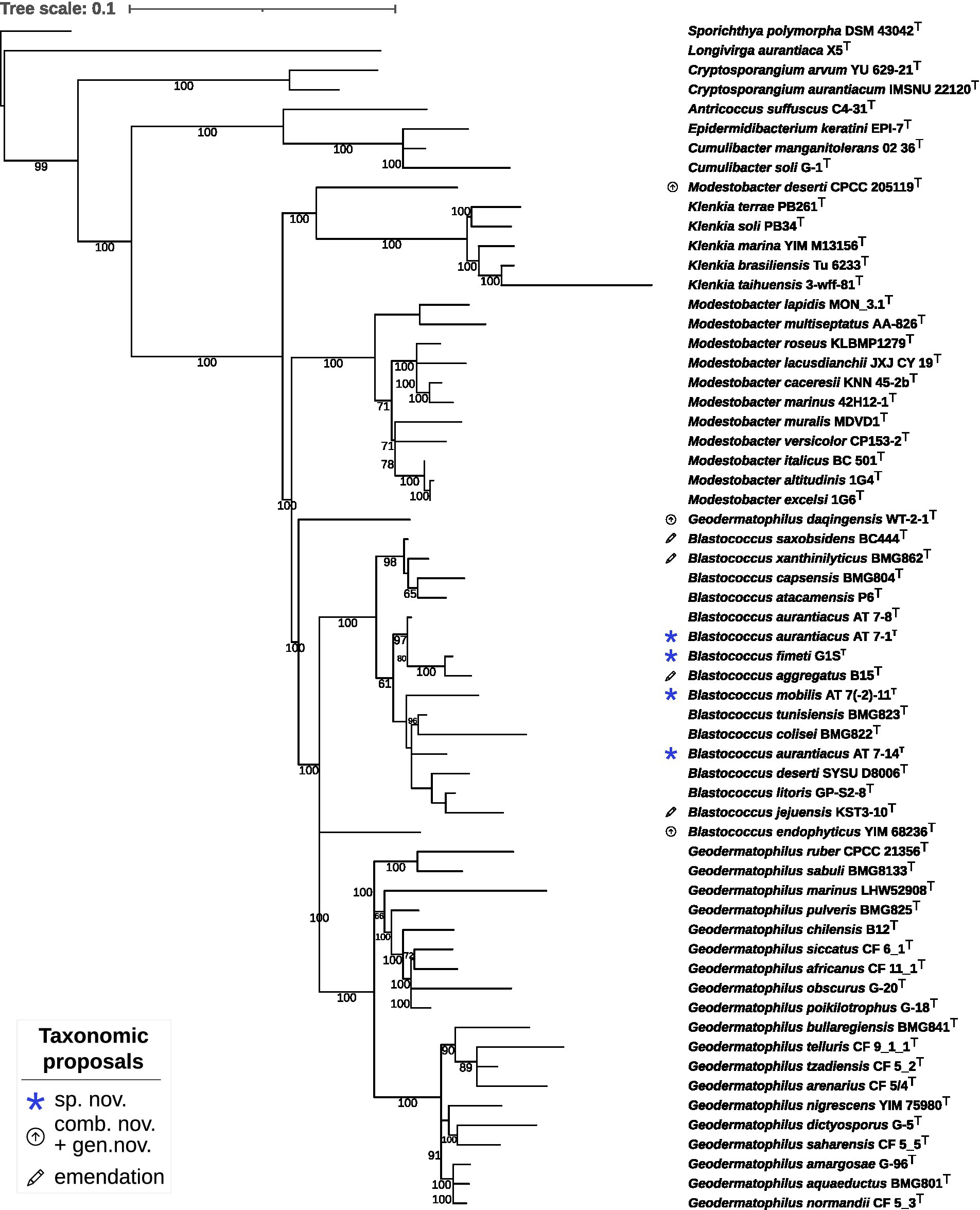
Figure 2. Maximum likelihood phylogenetic tree inferred from 16S rRNA gene sequences, showing the phylogenetic position of the strains DSM 44205T, DSM 44268T, DSM 44269, DSM 44270T and DSM 44272T relative to the type strains within Geodermatophilaceae. The branches are scaled in terms of the expected number of substitutions per site (see size bar). Support values from maximum-likelihood constrained bootstrapping are shown above the branches if equal to or larger than 60%.
The 16S rRNA gene sequence similarity values between (i) DSM 44268T and DSM 44269, (ii) between DSM 44205T, DSM 44270T and G. aggregatus DSM 4725T, and, (iii) between DSM 44270T and B. litoris GP-S2-8T, ranged above the Actinobacteria-specific 16S rRNA threshold of 99.0% (accepting an error probability of 1%; Meier-Kolthoff et al., 2013b).
It was thus necessary to investigate the species status via digital DDH (dDDH). The dDDH values between strain DSM 44268T and strains DSM 44205T (30.8%), DSM 44270T (23.8%), and B. aggregatus DSM 4725T (30.3%) and between B. litoris GP-S2-8T and strains DSM 44270T (26.9%) resulted in values below the 70% threshold throughout, thus indicating distinct species (Wayne et al., 1987).
In addition, the MALDI-TOF dendrogram (Supplementary Figure S2) confirmed the affiliation of strains DSM 44268T and DSM 44269 to the same species showing a 100% similarity regarding mass spectra patterns of proteins and separated from those under study at a distance level > 900 arbitrary units. The riboprinting patterns of the DSM 44268T and DSM 44269 isolates were also highly similar (98.3% similarity; Supplementary Figure S3) but differentiated them as clonal variants of each other.
All target strains were aerobic, Gram-reaction positive and showed coccoid cells with a tendency to form aggregates as previously described by Urzì et al. (2004) for the genus Blastococcus (Supplementary Figure S4). In addition, under studied culture conditions, reproduction by budding was predominant although bacterial binary fission was also observed (Supplementary Figure S4). It was already indicated for strain DSM 44205T by Eppard et al. (1996). Effectively, the presence of the cell division protein FtsZ and related genes such as the actin-like protein MreB were annotated in all genomes but no known mechanisms involved in division via budding were identified (Supplementary Table S4). Whilst FtsZ is an essential structural homologue of the eukaryotic cytoskeletal element tubulin (Lowe and Amos, 1998) and involved in forming a cytokinetic ring at the division site (Wang and Lutkenhaus, 1993), MreB is mostly essential for cell elongation and maintenance of the cell (Rivas-Marin et al., 2020). Mechanisms of bacterial division by budding are still poorly understood in evolutionary cell biology (Rivas-Marin et al., 2020). Geodermatophilaceae representatives are described as aerobic bacteria (Montero-Calasanz, 2020b). Nevertheless, some microaerophilic representatives isolated from the oxic/anoxic interface of marine sediments were also reported (Ahrens and Moll, 1970). To date no anaerobic isolate was described, although the presence of reductases using anaerobic terminal electron acceptors such as arsenate, ferredoxin or flavodoxins as well as a repertoire of genes involved in the fermentation of propanoate, butanoate and pyruvate, amongst others, in the Blastococcus genomes could indicate that anaerobic respiration may be possible. It was already suggested by Sghaier et al. (2016).
In vivo motility was not observed in any of the studied strains. The presence of homologues involved in flagellar biosynthesis and function and chemotaxis in all the genome sequences, nevertheless, suggests that Blastococcus strains may be flagellated chemotactic bacteria and motility may have been influenced by culture conditions. In fact, Ahrens and Moll (1970) suggested that the motile rod stage in B. aggregatus could be favoured under microaerophilic conditions. Later studies carried out by Holt and Chaubal (1997) for Salmonella pullorum strains, confirmed that flagellar synthesis is affected by agar concentration, carbohydrate concentration and type, and incubation temperature. The presence of flagellar gene clusters is consistent with observed motility by type strains of B. jejuensis and B. saxobsidens (Urzì et al., 2004; Lee 2006) and other strains in the family (Montero-Calasanz et al., 2017). In fact, flagellin synthesis was identified as the most highly expressed protein in the proteogenome of Blastococcus sp. DD2 (Sghaier et al., 2016). The core set of flagellar structural genes for Geodermatophilaceae representatives seems to be largely consistent with that described by Liu and Ochman (2007) for the bacterial domain. Twenty-eight different genes comprise specific proteins that form the filament (fliC, two copies for strain DSM 44205T, DSM 44270T, and B. xanthinilyticus), the hook-filament junction (flgL and flgK), the hook (flgE), the rod (flgB and flgC), the MS ring (fliF; absent in DSM 44272T), the C ring (fliG, fliM and fliN), the motor (motA and motB), and the export apparatus (flhA, flhB, fliP, fliQ, and fliR). In addition, flgD encoding for the hook-capping protein, which is required for flagellar assembly (Liu and Ochman, 2007), and other broadly distributed flagellar structural genes such as fliD (filament cap), fliE, fliK and fliH (hook) as well as, fliZ, fliS and fliO are also part of the core set in Geodermatophilaceae (Supplementary Table S4). The L and P ring proteins FlgH and FlhI are absent in Gram-positive bacteria. Chemotaxis, on the other hand, may be controlled by the universal two-component system. Methyl-accepting chemotaxis proteins (MCP) would control autophosphorylation of a cytoplasmic histidine kinase (CheA), via a coupling protein (CheW), which would activate the response regulator CheY and, consequently, interact with the switch mechanism in the flagellar motor (Szurmant and Ordal, 2004). Surprisingly, a dedicated ribose-binding protein (ribose transport system substrate-binding protein, RbsB) was annotated in the genome sequences of DSM 44205T, DSM 44268T, and DSM 44270T suggesting a key role of ribose as chemoeffector in those strains at low concentrations (Galloway and Furlong, 1977).
In addition, tad (tight adherence) genes were unambiguously annotated in all the studied genomes (Supplementary Table S4). The tad genes encode the machinery required for the assembly of adhesive Flp (fimbrial low-molecular-weight protein) pili (Tomich et al., 2007). Those are essential for biofilm formation, colonisation, DNA transfer and pathogenesis and have implications in functions such as phage binding, rough colony morphology, and twitching motility (Proft and Baker, 2009). The presence of Tad complexes in Geodermatophilaceae genomes could be important for successful colonisation of altered stones and patina formation (Eppard et al., 1996; Urzì et al., 2001). Howbeit, the presence and abundance of tad genes varied amongst strains suggesting that strains may present different adherence-related phenotypes as previously demonstrated by Kachlany et al. (2000) and Perez et al. (2006).
Production of spores was not observed in any of the isolates under the given culture conditions. Nevertheless, the unambiguous presence of single copies of SsgA-like proteins (SALPs) in all studied genomes suggested that sporulation may be a typical feature previously overlooked in Blastococcus spp. and that, similar to Geodermatophilus spp., the presence of SALPs may be affected by growth conditions (Ishiguro and Wolfe, 1970). SALPs occur exclusively in morphologically complex actinomycetes where they play an important role in morphogenesis and control of cell division (Jakimowics and van Wezel, 2012) and are essential for sporulation (van Wezel et al., 2000). Furthermore, the presence of single SALPs (presumably and invariably SsgB) in Blastococcus saxobsidens DD2 and other Geodermatophilaceae representatives was already described by Girard et al. (2013) and is suggested as a link between the number of SALPs and the complexity of the developmental process (Traag and van Wezel, 2008), suggesting that these actinomycetes would produce single spores.
Colonies were opaque with a moist surface and regular margin and varied from coral (strain DSM 44268T and DSM 44205T) to brownish-orange in colour (Strains DSM 44270T and DSM 44272T and DSM 44269). Accordingly, phytoene synthase (crtB; EC 2.51.32) and phytoene desaturase (crtI; 4 step enzyme; EC 1.3.99.28, EC 1.3.99.29, EC 11.3.99.31), the key enzymes involved in the biosynthesis of the colourful carotenoids ζ-carotene, neurosporene, and lycopene (Paniagua-Michel et al., 2012) were identified in all the genomes. Yet, the putative operon mentioned by Sghaier et al. (2016) for B. sp. DD2 consisting of hopC [squalene-associated FAD-dependent desaturase (fragment)], ispA (generaylgeranyl pyrophosphate synthase), shc (squalene-hopene cyclise), hpnH (hopanoid biosynthetic associated radical SAM protein HpnH), was not identified in these genomes, except for crtB and ilvC (ketol-acid reductoisomerase; Supplementary Table S4).
Strains grew well on GYM Streptomyces and PYGV agar media. They tolerated temperature ranging from 15°C to 37°C, except strain DSM 44269, which only tolerated a more restricted range from 20°C to 37°C. All strains showed optimal growth at 25°C–30°C. In addition, growth was observed at pH 6.0–12.0 for strains DSM 44268T and DSM 44269, at pH 5.0–11.5 for strain DSM 44205T, at 5.0–11.0 for strain DSM 44270T, and at 6.5–11.0 for strain DSM 44272T. All strains had an optimal pH range from 6.5 to 8.0. Regarding tolerance to salt, strains DSM 44269 and DSM 44272T were the least tolerant (0%–1% NaCl, although at 4% NaCl the Biolog system determined an ambiguous result for DSM 44272T), followed by both DSM 44268T and DSM 44205T tolerating 0%–4% NaCl, and strain DSM 44270T, which tolerated up to 8% NaCl. A summary of selected differential phenotypic characteristics is presented in Table 1 (for an overview of phenotypic profiles in Blastococcus see Supplementary Figure S5; Supplementary Table S5). Details about core metabolism will be provided later on in this manuscript.
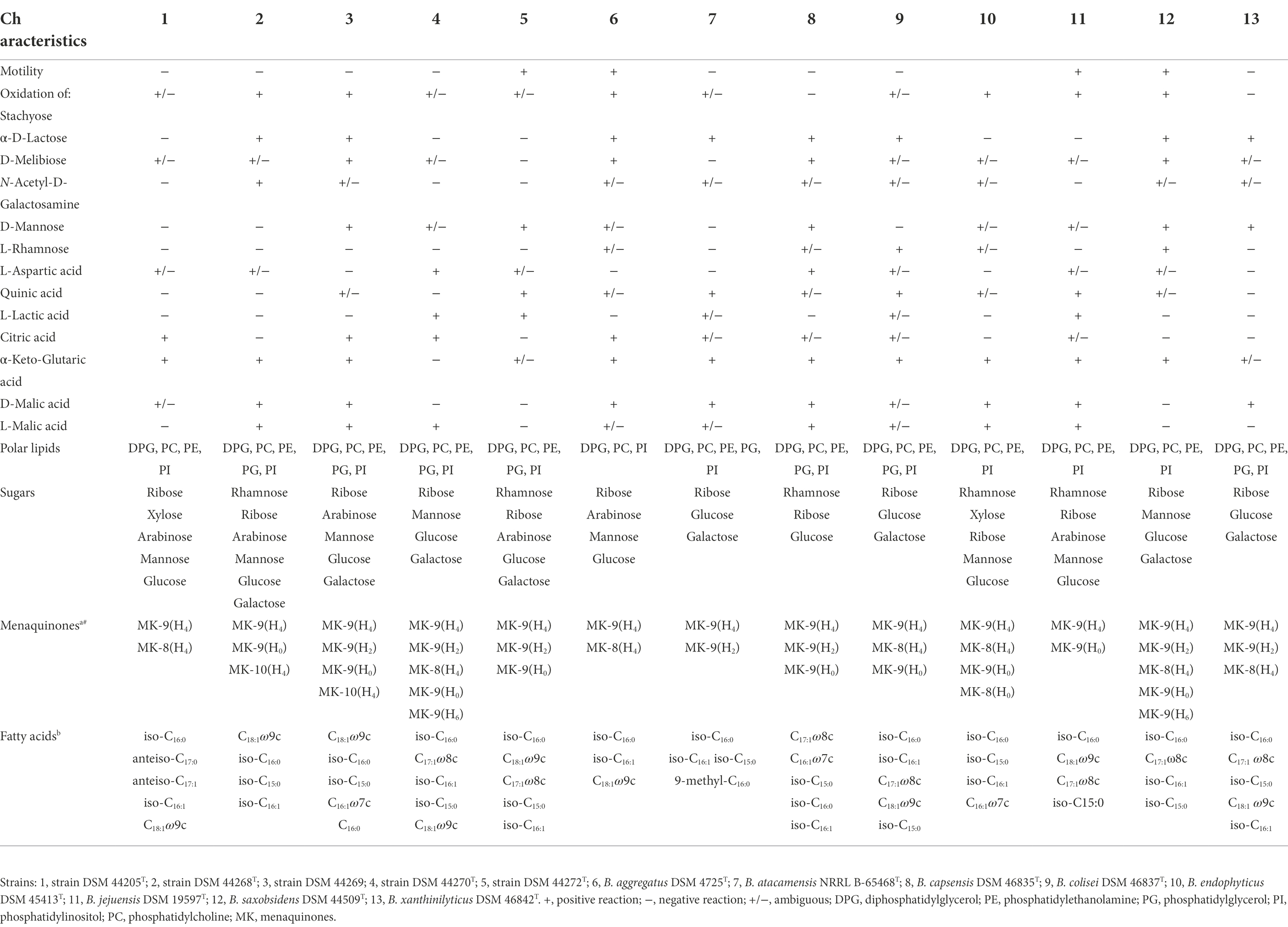
Table 1. Phenotypic characteristics of strains DSM 44205T, DSM 44268T, DSM 44269, DSM 44270T and DSM 44272T in comparison with the validly named species in the genus Blastococcus.
Whole-cell hydrolysates of all strains showed meso-diaminopimelic (DAP) acid (Cell wall type III; Lechevalier and Lechevalier, 1970) as the diagnostic diamino acid being consistent with the family Geodermatophilaceae (Montero-Calasanz, 2020b) and genomic data (presence of MurE encoding for UDP-N-acetylmuramoylalanyl-D-glutamate--2,6-diaminopimelate ligase [6.3.2.13] and absence of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate—L-lysine ligase [6.3.2.7]; Supplementary Table S4). In addition, D-alanyl-D-alanine carboxypeptidase [3.4.16.4] was automatically annotated in all Blastococcus representative genomes except in B. atacamensis P6T and in most Geodermatophilaceae representatives. The presence of diaminopimelate epimerase (dapF), responsible for the interconversion of the LL- and meso- isomers of DAP (Antia et al., 1957), was also annotated in all genomes including in the genome of Sporichthya polymorpha DSM 43042T, whose cell wall is characterised by containing large amounts of LL-DAP (Trujillo and Normand, 2019). In this way, genomic data could easily be used to distinguish taxa with DAP-type peptidoglycan from those with Lys-type pepdioglycan but fail to differentiate those incorporating LL-DAP (e.g., Sporichthyales) from those incorporating meso-DAP (e.g., Geodermatophilales). Although the evolutionary explanation for the incorporation of one or another stereoisomer is still unclear, studies in E. coli suggest that the differentiation of DAP stereoisomers might be related to dimerization of the DAP epimerase (Mengin-Lecreulx et al., 1988; Hor et al., 2013).
As suggested by Hezbri et al. (2016) for the emendation of the genus Blastococcus, whole-cell sugar analysis displayed a basic pattern consisting of ribose, glucose, and mannose. Additionally, galactose was identified in all strains under study except in the profile of strain DSM 44205T. Similarly, arabinose was found as part of the sugar cell wall composition in all studied strains excluding strain DSM 44270T. Xylose was detected in strain DSM 44205T.
The predominant menaquinone (MK) for strains DSM 44205T, DSM 44268T, DSM 44269, DSM 44270T, and DSM 44272T was MK-9(H4; 75.9%, 92.6%, 56.7%, 78%, and 62.9%, respectively) in agreement with what was described for Geodermatophilaceae (Montero-Calasanz et al., 2017). MK-9(H0) was however also found in minor levels in the strains isolated from Atacama soils (DSM 44268T, 0.7%; DSM 44269, 17.4%; DSM 44270T, 7.4%; DSM 44272T, 20.2%). Minor amounts of MK-9(H2) were also observed in the MK profiles of strains DSM 44269, DSM 44270T, and DSM 44272T (1.6, 1.3, and 15.0%, respectively). MK-8(H4) was also identified in strains DSM 44205T (5.5%) and DSM 44270T (2.9%). But MK-10(H4) was only found in strains DSM 44268T (4.7%) and DSM 44269 (0.6%) and MK-9(H6) in strain DSM 44270T (1.1%). The presence of quinones other than MK-9(H4) is commonly observed in other representatives in Geodermatophilaceae (Montero-Calasanz et al., 2017; Montero-Calasanz, 2020b, 2021), although it is worth mentioning that this is the first report of MK-10(H4) in members of the genus Blastococcus. The biosynthesis of menaquinones, via chorismate derived from Shikimate pathway (genes: MenA-G), was confirmed by genomic data in the order Geodermatophilales. Curiously, the futalosine pathway, the other major type of menaquinone biosynthetic pathway (Seto et al., 2008) was also annotated in genomes of Cryptosporangium arvum and Crytosporangium aurantiaca type strains (Supplementary Table S4). MenJ encoding for menaquinone-9-β-reductase [EC:1.3.99.38] and implicated in menaquinone side chain saturation (Upadhyay et al., 2015) was misidentified by databases as a geranylgeranyl reductase family protein but was verified in all genomes after BLAST using UniProtKB:P9WNY8 (MENJ_MYCTO). Moreover, the presence of 1,4-dihydroxy-2-naphthoate polyprenyltransferase [EC:2.5.1.74] and demethylmenaquinone methyltransferase [EC:2.1.1.163] in all genomes confirmed the potential of Geodermatophilales representatives to synthesise MK 8, 9, and 10. Regarding the elongation of isoprenoid side chains, genes uppS and hepST, encoding for ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) and polyprenyl diphosphate synthase (annotated here as geranylgeranyl pyrophosphate synthase), respectively, were annotated in all genomes. But the length and the degree of saturation of the C-3 isoprenyl side chains were impossible to predict. MKs consist of repeated isoprene subunits and the exact number is determined by the synthase encoded by the particular microbe and a microbe-dependent molecular ruler mechanism that involves bulky amino acid residues blocking the enzyme active sites to stop chain elongation (Han et al., 2015). The inability to predict the isoprenyl chain length of menaquinones was already reported by Baek et al. (2018).
Regarding fatty acid profiles, the five strains qualitatively showed similar patterns to those found in other representatives in Blastococcus (Table 1), but the predominant fatty acids significantly varied amongst them. Dominant fatty acids for strains DSM 44268T and DSM 44269 (>5%) were C18:1ω9c (36.2% and 37.2%, respectively), iso-C16:0 (19.3% and 11.3%, respectively), iso-C15:0 (6.2% and 8.6%, respectively), and C16:1ω7c (7.8% and 7.2%, respectively). Additionally, DSM 44268T showed iso-C16:1 H (5.4%) and DSM 44269 displayed some minor amounts of C16:0 (5.6%) and C17:1ω8c (6.8%). Those latter fatty acids were also present in DSM 44268T and iso-C16:1 H in DSM 44269 but at levels below 5%. On the other hand, strain DSM 44270T had the major fatty acids (>5%) iso-C16:0 (32.0%), C17:1ω8c (27.0%), iso-C16:1 H (8.7%), iso-C15:0 (7.3%), and C18:1ω9c (6.0%). A similar composition was observed for strain DSM 44272T although those quantitatively varied as follows: iso-C16:0 (24.0%), C18:1ω9c (23.0%), C17:1ω8c (13.7%), iso-C15:0 (10.8%), and iso-C16:1 H (8.1%). Finally, the major fatty acids of strain DSM 44205T (>5%) consisted of iso-C16:0 (23.4%), anteiso-C17:0 (19.8%), and anteiso-C17:1 C (11.9%), iso-C16:1 H (11.1%), and C18:1ω9c (6.7%). The complete gene set (Fab cluster) related to fatty acids metabolism (type II fatty acid synthase (FAS II) system) was present in all the studied genomes. Similar to the biosynthesis of MKs, fatty acid synthesis occurs via recurring reactions. It is thus not possible to predict just by using genomic data modifications that result in desaturations and/or elongations (López-Lara and Geiger, 2019).
The major polar lipids were diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylcholine (PC), and phosphatidylinositol (PI; Figure 3). It was consistent with those already described by Hezbri et al. (2016). Significant amounts of phosphatidylglycerol (PG) were also identified in all the isolates except in strain DSM 44205T. The inconsistent presence of PG was already noted for other species in the family (Montero-Calasanz et al., 2014, 2015, 2017; Hezbri et al., 2015a,b, 2016). Similarly, the characteristic glycophosphoinositol (GPI; annotated as GPL in Hezbri et al., 2016) as well as the reproducible presence of two unidentified polar lipids (PL2 and PL3) was detected in strains DSM 44268T, DSM 44269, DSM 44270T, DSM 44272T but not in DSM 44205T. However, a minor amount of an unidentified glycolipid (GL4) was found in the polar lipids pattern of strain DSM 44205T. Three different ones (GL1, GL2, and GL3) were also identified in the profile of strain DSM 44268T but not in strain DSM 44269. In contrast, an unidentified aminolipid (AL1) was displayed by strain DSM 44269 but not by DSM 44268T. One having a similar chromatographic mobility was present in strain DSM 44270T, too. Genomic data were consistent with those obtained experimentally with some minor discrepancies. In particular, the enzymes phosphatidylcholine synthase and cardiolipin synthase [EC 2.7.8.41] required in the synthesis of PC and cardiolipin (DPG), respectively, were present in all the genomes. As expected, phosphatidylcholine synthase was missing in Modestobacter and Klenkia type species. CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase [EC 2.7.8.50] involved in the synthesis of phosphatidyl-glycerophosphate, and also in fatty acids metabolism, was present in all species although phosphatidylglycerophosphatase [EC 3.1.3.27] required to the final conversion to PG was just annotated in DSM 44272T and in B. litoris. Phosphatidylserine descarboxylase [EC 4.1.1.65] required to synthesise phosphatidylethanolamine was not annotated in M. multiseptatus. CDP-diacylglycerol—inositol 3-phosphatidyltransferase [EC 2.7.8.11] needed for the synthesis of PI was annotated in all genomes. Finally, some other enzymes involved in the biosynthesis of other polar lipids were also annotated in the studied genomes suggesting that those may correspond to those minor unidentified lipids shown on the TLC plate.
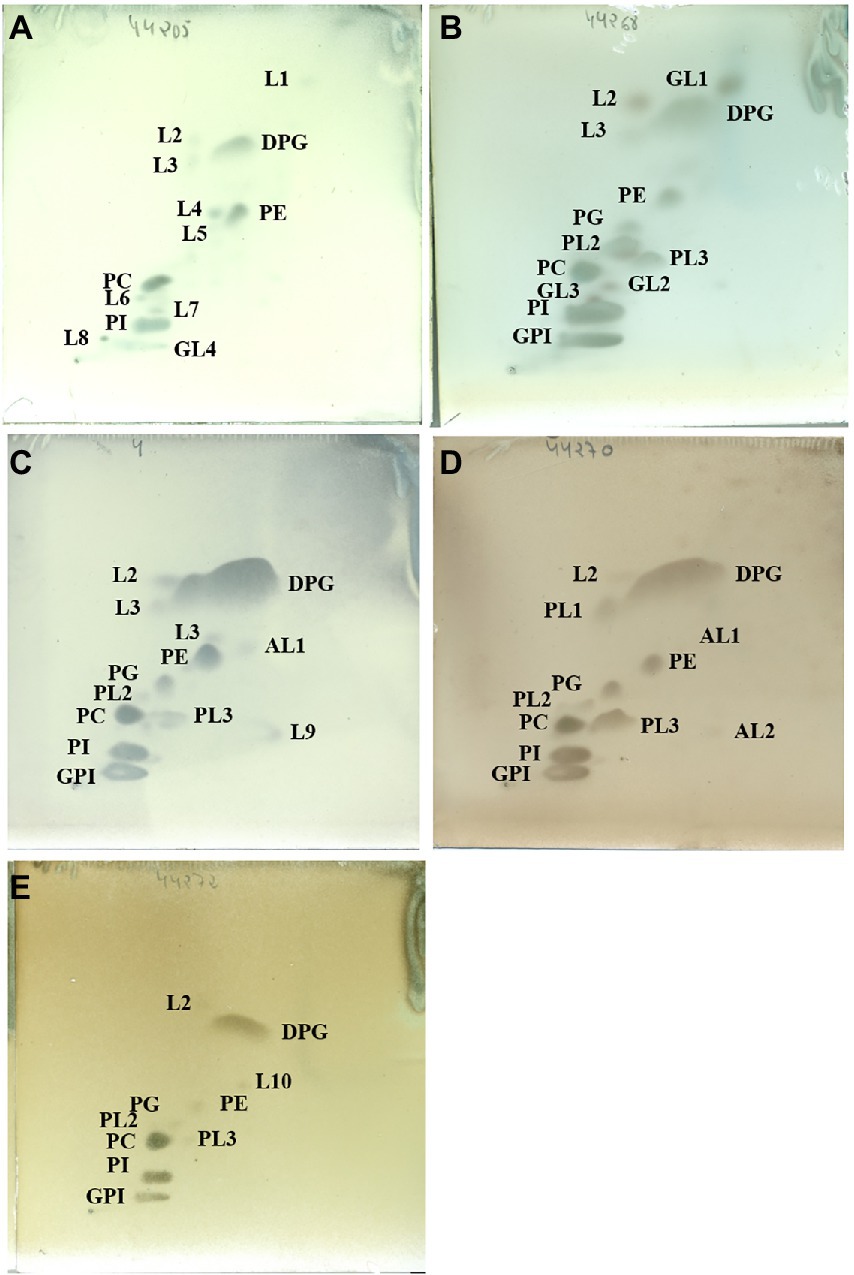
Figure 3. Polar lipids profile of strains DSM 44205T(A), DSM 44268T (B), DSM 44269 (C), DSM 44270T(D) and DSM 44272T (E) after separation by two-dimensional TLC using the solvents chloroform:methanol:water (65:25:4; v:v:v) in the first dimension and chloroform:methanol:acetic acid:water (80:12:15:4; v:v:v:v) in the second one. Plates were sprayed with molybdatophosphoric acid (3.5%; Merck™) for detection of the total polar lipids. DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PC, phosphatidylcholine; GPI, glycophosphatidylinositol; PL1-3, Phospholipid; GL1-4, glycolipid; AL1-2, aminolipid; L1-10, unidentified lipids. All data are from this study.
As expected, and in agreement with results determined by GEN III Biolog System (Supplementary Table S5), Blastococcus spp. show aerobic microbial respiration potentially driven by glycolysis, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation. In addition, CO2 could be assimilated via C4-dicarboxylic acid cycle by phosphoenolpyruvate carboxylase [EC 4.1.1.31], which converts phosphonolpyruvate coming from pyruvate metabolism, glucolysis or gluconeogenesis into oxaloacetate, and is present in all species except DSM 44268T and B. aggregatus. A second mechanism to fixate CO2 based on the reductive citric acid is also observed in all species (i.e., a molecule of CO2 is incorporated along succinyl-CoA, via 2-oxoglucarate/2-oxoacid ferredoxin oxidoreductase [EC 1.2.7.3], which in turn will be converted into isocitrate via isocitrate dehydrogenase [EC 1.1.1.42], using another molecule of CO2). The presence of carbonic anhydrases (CAs, EC 4.2.1.1), which are involved in the catalysis of CO2 to bicarbonate and vice versa (Supuran and Capasso, 2017) is found in all the genomes. Finally, the Wood-Ljungdahl pathway, which assimilates CO2 into Acetyl-CoA via the complex carbon monoxide deshydrogenase/acetyl-CoA synthase (CODH/ACS), may potentially be present. A complete tetrahydrofolate (THF) methyl-branch along with the presence of carbon monoxide dehydrogenases, required for the carbonyl-branch, were annotated in all genomes. These microorganisms could also be able to produce acetate as end product (acetogenesis). But no acetyl-coA synthase [2.3.1.169] homologue was identified in this study. And neither an autotroph nor an acetogen was described in the family so far. Overall, the presence of a number of mechanisms related to chemolithoautotrophy denotes that Blastococcus representatives could play a key role in the global carbon cycle. Microorganisms able to efficiently capture and store CO2 are prime candidates to be used as biological approaches in the reduction of the atmospheric CO2 concentration and as production platforms for a wide range of bioproducts from CO2 (Katsyv and Müller, 2020).
Apart from D-glucose as carbon source, genomics data identified other potential carbon sources such as fructose (fructokinase mediated: EC 2.7.1.4); α-galactose (galk, galactokinase); D-galactose (galM, Aldose 1-epimerase; EC 5.1.3.3); which is absent in DSM 44205T; L-gulose (gnl, RGN gluconolactonase; EC 3.1.1.17); 1-butanol (alcohol deshydrogenase; EC 1.1.1-), only present in B. aggregatus; D-mannose-6P/1P (manB phosphomannomutase; EC 5.4.2.8 /manC mannose-1-phosphate guanylyltransferase; EC 2.7.7.13), absent in B. aggregates; glycine via GDC glycine dehydrogenase and glycine hydroxymethyltransferase (glyA, EC 2.1.2.1); L-glutamate (gdhA, glutamate dehydrogenase; EC 1.4.1.3); L-serine (glycine hydroxymethyltransferase; glyA, EC 2.1.2.1), isocitrate (isocitrate dehydrogenase), absent in DSM 44272T; formamide (formamidase; EC 3.5.1.49) present in DSM 44205T, DSM 44272T, and B. aggregatus; trehalose (treS, maltose α-D-glucosyltransferase; EC 4.99.16 and D-glucose α,α-trehalose phosphorylase; EC 2.4.1.65); L-lactate (lldD, L-lactate dehydrogenase; EC 1.1.2.3), just present in B. endophyticus; sucrose (malZ, α-glucosidase; EC 3.2.1.20); maltose (maltokinase; EC 2.7.1.175), absent in B. xanthinilyticus; isomaltose (oligo-1,6-glucosidase), amongst others, that may also be utilised by Blastococcus strains. These results supported the presence of a rich carbohydrate metabolism in the genus and were largely in correlation with those showed by GEN III Biolog and API System (See Supplementary Figure S5; Supplementary Tables S4, S5).
Additionally, Blastoccocus genomes revealed extracellular enzymes involved in the degradation of complex carbohydrates, which are closely linked to the decay of organic residues, transformation of native soil organic matter, mineralisation of plant nutrients, and soil aggregation (Balezentiene, 2012). For example, α-glucosidases [EC 3.2.1.20], involved in the cellulose hydrolysis, were observed in all studied genomes (three copies in B. xanthinilyticus); β-glucosidases [EC 3.2.1.21] were also annotated in all of them except in B. colisei and B. xanthinilyticus, having three gene copies in DSM 44205T, DSM 44268T, and B. saxobsidens; Moreover, all studied genomes contained α-amylases [EC 3.2.1.1], extracellular enzymes handling the almost complete starch saccharification (Tu and Miles, 1976), but enzymatic activity was not recorded in any of them according to the APIM ZYM system; the presence of glycosidase (glycoside hydrolase; annotated as α-amylase by Pfam database) was, nevertheless, only annotated in B. endophyticus; Finally, chitinases [EC 3.2.1.14] were annotated in B. endophyticus and B. saxobsidens. Cellulases (β-1,4-endoglucan hydrolase; EC 3.2.1.4) and 1,4- β- cellobiohydrolase [EC 3.2.1.91] were absent. On the other hand, lipolytic enzymes are an important group of biotechnologically relevant enzymes presenting a key role in regulating the levels of hydrocarbons in soil (Gupta et al., 2004). Extracellular lipases such as the well-known triacylglycerol acylhydrolase (here as tryacylglycerol lipase, EC 3.1.13) was only annotated in DSM 44268T. In addition, lysophospholipases [EC 3.1.1.5] and monoglyceride lipases (EC 3.1.1.23) were present in all the genomes analysed except DSM 44272T and B. endophyticus. Polyhydroxyalkanoic acid (PHA) synthases (EC 2.3.1.-), the key enzyme in the biosynthesis of PHAs, a class of aliphatic polyesters that are generally regarded as a carbon and energy reserve material in bacteria and archaea (McCool and Cannon, 2001), were also identified in all the genomes in a number of copies that ranged from one to six with the exception of DSM 44272T. Again, large correlations but also discrepancies were observed between genomic and experimental data. For example, the hydrolysis of xanthine by B. xanthinilyticus, previously described by Hezbri et al. (2018), was confirmed by the presence of xanthine deshydrogenase but experimentally it could not be proven for strains DSM 44205T and DSM 44268T and B. endophyticus although such enzyme was also annotated in their genomes. Similar inconsistencies were also identified for other unambiguously annotated enzymes such as α-glucosidase which was not expressed under standardised experimental conditions in strains DSM 44205T, DSM 44268T, DSM 44270T, DSM 44272T, and B. aggregatus or β-glucosidase which was only expressed by B. endophyticus and B. saxobsidens (for more examples see Supplementary Figure S5; Supplementary Tables S4, S5).
The integration of genomic information into microbial systematics have proven to be a reproducible, reliable and highly informative approach to reveal phylogenetic relationships amongst prokaryotes (Chun and Rainey, 2014; Chun et al., 2018). Montero-Calasanz et al. (2017) already applied phylogenomics in combination with the identification of diagnostic features in the taxonomic rearrangement of Geodermatophilaceae. In light of new genomic information now available, the taxonomic status of the family was revisited in this study. The whole proteome-based phylogenomic tree of the sequenced type strains and strains under study along with phenotypic tests showed with strong support that DSM 44268T, DSM 44270T, DSM 44272T, and DSM 44205T represent novel species within the genus Blastococcus. Additionally, the non-monophyly of Blastococcus, Geodermatophilus and Modestobacter was revealed, i.e., B. endophyticus, G. daqingensis and M. deserti formed separate lineages within Geodermatophilaceae. Based on principles of phylogenetic systematics and applying taxonomic conservatism, we propose the following reclassifications: Blastococcus endophyticus as the type species of the new genus Trujillonella gen. nov., which the name Trujillonella endothytica comb. nov is given; Geodermatophilus daqingensis as the type species of the new genus Pleomorpha gen. nov., which the name Pleomorpha daqingensis comb. nov is given; Modestobacter deserti gen. nov., as the type species of the new genus Goekera which the name Goekera deserti comb. nov is given. Finally, based on the additional taxonomic data now available we propose the emendation of B. aggregatus, B. jejuensis, B. saxobsidens and B. Xanthinilyticus to include accession numbers for the whole genome sequences, genome size and genomic G + C content and insights. The use of genomic data for the characterisation of novel species in Geodermatophilaceae also revealed interesting insights regarding their metabolism and ecology. In particular, the number of 16S rRNA and tRNA gene copies was suggested to be related to their slow growth speed and their life strategy supporting their prevalence in oligotrophic environments such as desert soils and decayed monuments (Neilson et al., 2012; Giongo et al., 2013). In addition, the presence of a repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly in all representatives of the family highlighted potential yet unobserved features that would enable us to better understand their metabolism, morphogenesis and ecology. Combining high-throughput phenotype analyses with genomics also supported the uncovering of a rich carbon metabolism in Blastococcus spp. Results obtained from GEN III microplates and API System were largely correlated with what was observed in the genomes. In this way, Blastococcus spp. were described as aerobic bacteria with the potential to present anaerobiosis and fixate CO2 by using four potential pathways and to degrade a range of complex carbohydrates. Such metabolic versatility would support their success to colonise a variety of biotopes and a potential key role in the global carbon cycle and as biological tools to reduce the atmospheric CO2 concentration and produce bioproducts from CO2.
Even though chemotaxonomic procedures marked a turning point in prokaryotic systematics (Goodfellow et al., 2012), results can be influenced by cultivation conditions (i.e., fatty acid analysis) or by inexperienced staff (i.e., regarding the interpretation of polar lipids patterns on 2-D TLC plates; Sutcliffe et al., 2013). In favour of a revitalization of bacterial taxonomy, in silico chemotaxonomic analyses were already used in some studies to complement and, in some cases, replace characteristics traditionally determined in the laboratory (Amaral et al., 2014; Fotedar et al., 2020; Lawson et al., 2020). Here, we attempted to determine in silico cell-wall peptidoglycan, respiratory quinone, fatty acids and polar lipid patterns of Geodermatophilaceae. Overall, genomic data were consistent with the results obtained from the laboratory. Nevertheless, the amount of information that can be predicted based on genomic data varied depending on the chemotaxonomic trait. For instance, the in silico polar lipid analyses were largely in line with those obtained experimentally and allowed us to distinguish at the genus level by the presence/absence of the gene encoding for phosphatidylcholine synthase. On the other hand, the presence of the DAP as the diagnostic diamino acid over L-lysine in the whole-cell hydrolysates was confirmed but the in silico procedure failed in differentiating microorganisms incorporating LL-DAP rather than meso-DAP (discrimination at order level). Regarding both MK and fatty acids analysis, final products could not be predicted due to the nature of biosynthetic pathways. Nevertheless, sufficient information would, for example, be gathered from Shikimate pathway along with the absence of futalosine pathway to establish certain taxonomic discrimination at order level. The use of genomic data for species delineation and the impractical value of chemotaxonomic data when genomic data are available are widely proven (Nouioui et al., 2018). Nevertheless, as argued by Vandamme and Sutcliffe (2021), knowledge of cellular components can be valuable to establish discriminative thresholds when applied at taxonomic ranks above the species level. However, improvements in analytical methods leading to the use of automated and higher-resolution technologies such as mass spectrometry lipidomic methods (Rashid et al., 2017) should first be introduced and deployed. Our results here proved the use of in silico chemotaxonomic analysis as a simple and fast approach to complement phylogenomic results and to provide sufficient resolution to support the affiliation of novel isolates into appropriate taxonomic groups. Its routine application would undoubtedly increase the speed of taxon descriptions and the accuracy and consistency of taxonomic reports.
In summary, the integration of genomics into the systematics of Geodermatophilaceae not only allowed for more stable and reliable taxonomic arrangements but also showed the feasibility for a potential replacement of wet-lab chemotaxonomy. Second, it provided deeper insights into the molecular mechanisms behind phenotypic features and revealed potentially overlooked ones that could be key to an understanding of their evolution, ecology and biotechnological potential.
Tru.jil.lo.nel’la. N.L. fem. n. Trujillonella, named in honour of Martha E. Trujillo in recognition of her contributions to microbial systematics, mainly on Actinobacteria, on Bergey’s Manual trust, and as the Editor-in-chief of the International Journal of Systematic and Evolutionary Microbiology.
Cells are aerobic, non-motile, non-spore-forming, Gram-stain positive, catalase-positive and oxidase-negative. Cells occur singly, in pairs or in tetrads, often tending to form aggregates. The peptidoglycan in the cell-wall contains meso-diaminopimelic acid. The predominant menaquinone is MK-9(H4), with MK-8 and MK-9(H6) as minor components. The basic polar lipid profile contains diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, and phosphatidylinositol. The major fatty acids are iso-C16:0, iso-C15:0 and C18:1ω9c. The basic whole-cell sugar pattern includes arabinose and galactose. The genomic G + C content is 71%–72%.
The type species of Trujillella is Trujillella endophytica, sp. nov.
T. en.do.phy’ti.ca (Gr. pref. Endo-, within; Gr. neut. n. phyton, plant; L. fem. Adj. suff. –ica, adjectival suffix used with the sense of belonging to; N.L. fem. Adj. endophytica, within plant, endophytic, pertaining to the isolation from plant tissues).
Basonym: Blastococcus endophyticus Zhu et al. (2013) emend. Hezbri et al. (2016).
The properties are as given in the species description by Zhu et al. (2013) and emendation by Hezbri et al. (2016) with the following modification. The genomic G + C content is 74.6%. The genome size is 4.9 Mbp. According to genomic data, anaerobiosis and acetogenesis may occur. A repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Four different autotrophic mechanisms including the Wood-Ljungdahl pathway, C4-dicarboxylic acid and reductive citric acid cycles and carbonic anhydrases as well as a range of genes involved in the degradation of complex carbohydrates were also identified.
The accession number for the whole genome sequence of strain DSM 45413T is FOEE00000000.
The type strain YIM 68236T = CCTCC AA 209045T = DSM 45413T = KCTC 19998T was isolated from healthy leaves of Camptotheca acuminate collected in Yunnan Province, south-west China.
Ple.o.mor’pha. Gr. adv. Pleon more; Gr. fem. n. morphe, shape or form; N.L. fem. n. Pleomorpha, organism showing multiple forms.
Pleiomorphic, motile, spore-forming, aerobic, Gram-stain positive cells. Those occurs singly or associated in aggregates. The peptidoglycan in the cellwall contains meso-diaminopimelic acid. The predominant menaquinone is MK-9(H4). The basic polar lipid profile contains diphosphatidylglycerol, phosphatidylcholine, phosphatidylglycerol, phosphatidylethanolamine, and phosphatidylinositol. The major fatty acids are iso-C16:0 and iso-C15:0. The basic whole-cell sugar pattern includes galactose, glucose and xylose. The genomic G + C content is 73%–74%. The type species of Pleomorpha is Pleomorpha daqingensis, sp. nov.
P. da.qing.en’sis (N.L. masc./fem. Adj. daqingensis, pertaining to Daqing city, China, where the type strain was isolated).
Basonym: Geodermatophilus daqingensis Wang et al. (2017).
The properties are as given in the species description by Wang et al. (2017) with the following modification. The genomic G + C content is 73.6%. The genome size is 5.4 Mbp. According to genomic data, anaerobiosis and acetogenesis may occur. A repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Four different autotrophic mechanisms including the Wood-Ljungdahl pathway, C4-dicarboxylic acid and reductive citric acid cycles and carbonic anhydrases as well as a range of genes involved in the degradation of complex carbohydrates were also identified.
The accession number for the whole genome sequence of strain DSM 104001T is JACBZT000000000.
The type strain WT-2-1T = CGMCC 4.7381T = DSM 104001T was isolated from pretroleum-contaminated soil in Daqing city, China.
Goe’ke.ra. N.L. fem. n. Goekera, named in honour of Markus Göker in recognition of his contributions to microbial systematics, including work on Actinobacteria, on the List of Prokaryotic Names with Standing in Nomenclature (LPSN), and as a member of the Judicial Commission.
Cells are motile, non-spore-forming, aerobic, Gram-stain positive, catalase and oxidase positive cocci and/short rods. Bud-like structure was observed for some cells. The peptidoglycan in the cell-wall contains meso-diaminopimelic acid. The predominant menaquinone is MK-9(H4), with MK-8(H4) as a minor component. The basic polar lipid profile contains diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, phosphatidylmethylethanolamine and phosphatidylinositol mannoside. The major fatty acids are C18:1ω9c, iso-C16:0, C16:0, iso-C15:0, and C16:1ω7c. The basic whole-cell sugar pattern includes arabinose, glucose and ribose. The genomic G + C content is 74%–75%. The type species of Goekera is Goekera deserti, sp. nov.
G. de.ser’ti (L. gen. Neut. n. deserti, of a desert, where the organisms were acquired).
Basonym: Modestobacter deserti Jiang et al. (2021).
The properties are as given in the species description by Jiang et al. (2021) with the following modification. According to genomic data, anaerobiosis and acetogenesis may occur. A repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Four different autotrophic mechanisms including the Wood-Ljungdahl pathway, C4-dicarboxylic acid and reductive citric acid cycles and carbonic anhydrases as well as a range of genes involved in the degradation of complex carbohydrates were also identified.
The accession number for the whole genome sequence of strain CPCC 205119T is JAAGWK000000000.
The type strain CPCC 205119T = I12A-02624T = KCTC 49201T = NBRC 113528T was isolated from moss-dominated soil crusts collected from Shapotou NDER in Tengger Desert, China.
The properties are as given in the species description by Ahrens and Moll (1970) and emendations by Urzì et al. (2004) and Hezbri et al. (2016) with the following emendation. The genomic G + C content is 73.3%. The genome size is 4.6 Mbp. According to genomic data, anaerobiosis and acetogenesis may occur. A repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Three different autotrophic mechanisms including the Wood-Ljungdahl pathway, reductive citric acid cycle, and carbonic anhydrases as well as a range of genes involved in the degradation of complex carbohydrates were also identified.
The accession number for the whole genome sequence of the type strain DSM 4725T is OBQI00000000.
The type strain B15T = ATCC 25902T = DSM 4725T = JCM 12602T = NCIMB 1849T was isolated from Baltic Sea.
The properties are as given in the species description by Lee (2006) and emendation by Hezbri et al. (2016) with the following emendation. The presence of phosphatidylmethylethanolamine listed by Lee (2006) and confirmed by Hezbri et al. (2016) was incorrectly annotated by the latter and it should be recognised as hydroxiphosphatidylethanolamine accordingly to its chromatographic mobility.
The type strain KST3-10T = DSM 19597T = JCM 15614T = KCCM 42251T = NRRL B-24440T was isolated from sand sediment of a beach in Jeju, Korea.
The properties are as given in the species description by Urzì et al. (2004) and emendation by Hezbri et al. (2016) with the following emendation. The genomic G + C content is 73.5%. The genome size is 4.5 Mbp. According to genomic data, anaerobiosis and acetogenesis may occur. A repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Four different autotrophic mechanisms including the Wood-Ljungdahl pathway, C4-dicarboxylic acid and reductive citric acid cycles and carbonic anhydrases as well as a range of genes involved in the degradation of complex carbohydrates were also identified.
The accession number for the whole genome sequence of the type strain DSM 44509T is SHKV01000001.
The type strain BC444T = DSM 44509T = JCM 13239T = NRRL B-24246T was isolated from from the surface of marble and calcareous stones in Italy.
The properties are as given in the species description by Hezbri et al. (2018) with the following emendation. The genomic G + C content is 74.4%. The genome size is 4.6 Mbp. According to genomic data, anaerobiosis and acetogenesis may occur. A repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Four different autotrophic mechanisms including the Wood-Ljungdahl pathway, C4-dicarboxylic acid and reductive citric acid cycles and carbonic anhydrases as well as a range of genes involved in the degradation of complex carbohydrates were also identified.
The accession number for the whole genome sequence of the type strain DSM 46842T is VNHW00000000.
The type strain BMG 862T = DSM 46842T = CECT 8884T was isolated from a marble sample collected from the Bulla Regia monument, Northern Tunisia.
Blastococcus aurantiacus (au.ran.ti.a’cus. N.L. masc. Adj. aurantiacus orange-coloured, referring to the orange colour of the colonies).
Colonies are bright red orange-coloured, opaque with a dry surface and regular margin. Cells are Gram-reaction-positive and catalase and oxidase negative cocci (0.6–1.6 μm in diameter) with a tendency to form aggregates. It grows in aerobiosis but, according to genomic data, anaerobiosis and acetogenesis may occur. Reproduction by budding is predominant but binary fission is also observed. Cells are non-motile and non-spore-forming but a repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Degradation for casein, tyrosine, starch, xanthine, and hypoxanthine are negative. Temperature and pH ranges are 15–37°C (optimal range) and 6.0–12.0 (optimum 6.5–8.0), respectively. NaCl is not needed for growth. It can grow between 0% and 4% NaCl (w/v; optimal range) but not at 8%. It grows well on GYM Streptomyces, R2A (DSMZ Medium 830), tryticase soy agar (TSA), Luedemann (DMSZ medium 877), and PYGV (DSMZ medium 621) media. According to API ZYM strips, the following enzymatic activities are present: esterase lipase (C8), lipase (C14), leucine arylamidase, valine acrylamidase, cysteine acrylamidase, trypsin, and α-galatosidase. According to the Biolog System, it oxidises: dextrin, D-maltose, D-trehalose, D-cellobiose, β-gentiobiose, sucrose, stachyose, α-D-lactose, β-methyl-D-glucoside, D-salicin, N-acetyl-D-glucosamine, N-acetyl-β-D-mannosamine, N-acetyl-D-galactosamine, 3-O-methyl-D-glucose, D-fucose, L-fucose, glycerol, D-glucose-6-phosphate, rifamycin SV, D-galacturonic acid, D-gluconic acid, D-glucuronic acid, D-saccharic acid, α-keto-glutaric acid, D-malic acid, L-malic acid, nalidixic acid, lithium chloride, potassium tellurite, tween 40, γ-amino-n-butyric acid, acetoacetic acid, propionic acid, acetic acid, aztreonam, butyric acid, and sodium bromate but not D-mannose, D-galactose, L-rhamnose, sodium lactate, fusidic acid, D-mannitol, D-arabitol, D-aspartic acid, D-serine, troleandomycin, minocycline, glycine-proline, L-alanine, L-arginine, L-glutamic acid, L-histidine, L-pyroglutamic acid, L-serine, lincomycin, guanidine hydrochloride, niaproof, pectin, L-galactonic acid-γ-lactone, glucuronamide, mucic acid, quinic acid, vancomycin, tetrazolium violet, tetrazolium blue, p-hydroxy-phenylacetic acid, methyl pyruvate, L-lactic acid, citric acid, bromo-succinic acid, β-hydroxy-butyric acid, α-keto-butyric acid, and sodium formate. In correlation, a range of genes involved in the degradation of complex carbohydrates were identified (chemoheterotrophy). Three different autotrophic mechanisms including the Wood-Ljungdahl pathway, reductive citric acid cycle and carbonic anhydrases were besides annotated (chemolithoautotrophy). Predominant fatty acids are C18:1ω8c, iso-C16:0, iso-C16:1 H, iso-C15:0, and iso-C16:1 H. The cell wall peptidoglycan contains meso-diaminopimelic acid. Galactose, glucose, and arabinose are the whole-cell sugars. Polar lipid profile consists of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine and phosphatidylinositol, glycophosphatidylinositol, two unidentified phospholipids, and three unidentified gylcolipids. The high-quality draught genome of strain DSM 44268T was resolved to 18 scaffolds consisting of 4,163,046 bp, with a G + C content of 73.6%, 4,044 candidate protein-coding genes, 47 tRNA genes, and eight rRNA regions.
The INSDC accession number for the 16S rRNA gene sequences of the type strain AT 7-1T (=DSM 44268T = JCM 18931T) isolated from soil in the Atacama Desert (Chile) is MH479060. The IMG accession number for the whole genome sequence of strain DSM 44268T is 2599185358.
Blastococcus fimeti (fi.me’ti. L. neut. Gen. n. fimeti, of or from a dunghill).
Colonies are pink red-coloured, opaque with a greasy surface and irregular margin. Cells are Gram-reaction-positive and catalase and oxidase negative cocci (0.6–1.6 μm in diameter) with a tendency to form aggregates. It grows in aerobiosis but, according to genomic data, anaerobiosis and acetogenesis may occur. Reproduction by budding is predominant but binary fission is also observed. Cells are non-motile and non-spore-forming but a repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Degradation for casein, tyrosine, starch, xanthine, and hypoxanthine are negative and aesculin positive. Temperature and pH ranges are 15°C–37°C (optimal range) and 6.0–12.0 (optimum 7.0–9.0), respectively. NaCl is not needed for growth. It can grow between 0–4% NaCl (w/v; optimal range) but not at 8%. It grows well on GYM Streptomyces, R2A (DSMZ Medium 830), tryticase soy agar (TSA), Luedemann (DMSZ medium 877), GPHF (DSMZ medium 553), and PYGV (DSMZ medium 621) media. According to API ZYM strips, the following enzymatic activities are present: esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, α-fucosidase. According to the Biolog System, it oxidises: dextrin, D-maltose, D-trehalose, D-cellobiose, sucrose, turanose, β-methyl-D-glucoside, D-salicin, N-acetyl-D-glucosamine, N-acetyl-β-D-mannosamine, D-glucose, 3-O-methyl-D-glucose, L-fucose, glycerol, D-glucose-6-phosphate, rifamycin SV, L-glutamic acid, methyl pyruvate, D-lactic acid methyl ester, citric acid, α-keto-glutaric acid, nalidixic acid, lithium chloride, potassium tellurite, tween 40, γ-amino-n-butyric acid, β-hydroxy-butyric acid, α-keto-butyric acid, acetoacetic acid, propionic acid, acetic acid, aztreonam, butyric acid, and sodium bromate but not D-raffinose, α-D-lactose, N-acetyl-D-galactosamine, N-acetyl-neuraminic acid, D-mannose, L-rhamnose, fusidic acid, D-serine (inhibitory concentration), D-arabitol, myo-inositol, D-fructose-6-phosphate, D-aspartic acid, D-serine, troleandomycin, minocycline, glycine-proline, L-alanine, L-arginine, L-histidine, L-pyroglutamic acid, L-serine, lincomycin, guanidine, hydrochloride, niaproof, L-galactonic acid-γ-lactone, mucic acid, quinic acid, D-saccharic acid, vancomycin, tetrazolium violet, tetrazolium blue, p-hydroxy-phenylacetic acid, L-lactic acid, L-malic, acid, bromo-succinic acid and sodium formate. In correlation, a range of genes involved in the degradation of complex carbohydrates were identified (chemoheterotrophy). Three different autotrophic mechanisms including the Wood-Ljungdahl pathway, reductive citric acid cycle and carbonic anhydrases were besides annotated (chemolithoautotrophy). Predominant fatty acids are iso-C16:0, anteiso-C17:0, anteiso-C17:1 C, iso-C16:1 H and C18:1ω9c. The cell wall peptidoglycan contains meso-diaminopimelic acid. Ribose, xylose, arabinose, mannose, and glucose are the whole-cell sugars. Polar lipid profile consists of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine and phosphatidylinositol. The high-quality draught genome of strain DSM 44205T was resolved to 30 scaffolds consisting of 4,044,261 bp, with a G + C content of 73.4%, 3,853 candidate protein-coding genes, 70 tRNA genes, and nine rRNA regions.
The INSDC accession number for the 16S rRNA gene sequences of the type strain G1ST (=DSM 44205T = CECT 8406T) isolated from seepage water of a dumping ground in Vancouver (Canada) is MH479059. The IMG accession number for the whole genome sequences of strain DSM 44205T is 2599185193.
Blastococcus haudaquaticus (hau.da.qua’ti.cus. L. adv. Haud, not at all, by no means; L. masc. Adj. aquaticus, living, growing, or found in or by the water, aquatic; N.L. masc. Adj. haudaquaticus, growing far away from or without any water).
Colonies are reddish-brown coloured, opaque with a mucoid surface and regular margin. Cells are aerobic, Gram-reaction-positive and catalase and oxidase negative cocci (0.6–1.6 μm in diameter) with a tendency to form aggregates. It grows in aerobiosis but, according to genomic data, anaerobiosis and acetogenesis may occur. Reproduction by budding is predominant but binary fission is also observed. Cells are non-motile and non-spore-forming but a repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Degradation for casein, tyrosine, starch, xanthine, and hypoxanthine are negative. Temperature and pH ranges are 15–37°C (optimal range) and pH 5.0–11.0 (optimum 6.5–8.0), respectively. NaCl is not needed for growth. It can grow between 0% and 8% NaCl (w/v; optimal range). It grows well on GYM Streptomyces, R2A (DSMZ Medium 830), tryticase soy agar (TSA), Luedemann (DMSZ medium 877), and PYGV (DSMZ medium 621) media. According to API ZYM strips, the following enzymatic activities are present: esterase lipase (C8), lipase (C14), leucine arylamidase, valine acrylamidase, trypsin, and cystine acrylamidase. According to the Biolog System, it oxidises: dextrin, D-maltose, D-trehalose, D-cellobiose, β-gentiobiose, sucrose, turanose, β-methyl-D-glucoside, D-salicin, N-acetyl-D-glucosamine, D-glucose, D-fructose, sodium lactate, D-sorbitol, glycerol, rifamycin SV, L-aspartic acid, L-glutamic acid, L-histidine, mucic acid, L-lactic acid, citric acid, L-malic acid, lithium chloride, potassium tellurite, γ-amino-n-butyric acid, propionic acid, acetic acid, sodium formate, aztreonam, butyric acid, and sodium bromate but not α-D-lactose, N-acetyl-D-galactosamine, N-acetyl-neuraminic acid, 3-O-methyl-D-glucose, D-fucose, L-fucose, L-rhamnose, inosine, fusidic acid, D-serine (inhibitory concentration), D-arabitol, myo-inositol, D-aspartic acid, D-serine, troleandomycin, minocycline, glycine-proline, L-arginine, L-pyroglutamic acid, lincomycin, guanidine hydrochloride, niaproof, L-galactonic acid-γ-lactone, D-glucuronic acid, glucuronamide, quinic acid, vancomycin, tetrazolium violet, tetrazolium blue, p-hydroxy-phenylacetic acid, D-lactic acid methyl ester, α-keto-glutaric acid, D-malic acid, bromo-succinic acid, nalidixic acid, α-keto-butyric acid, and acetoacetic acid. In correlation, a range of genes involved in the degradation of complex carbohydrates were identified (chemoheterotrophy). Three different autotrophic mechanisms including the Wood-Ljungdahl pathway, reductive citric acid cycle and carbonic anhydrases were besides annotated (chemolithoautotrophy). Predominant fatty acids are iso-C15:0, iso-C16:1 H, iso-C16:0, C17:1ω8c, C18:1ω9c, and C16:1ω7c. The cell wall peptidoglycan contains meso-diaminopimelic acid. Ribose, mannose, galactose, and glucose are the whole-cell sugars. Polar lipids consist of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, and phosphatidylinositol. The high-quality draught genome of strain DSM 44270T was resolved to 11 scaffolds consisting of 4,512,672 bp, with a G + C content of 73.29%, 4,410 candidate protein-coding genes, 48 tRNA genes, and six rRNA regions.
The INSDC accession number for the 16S rRNA gene sequences of the type strain AT 7-14T (=DSM 44270T = JCM 18932T) isolated from soil in the Atacama Desert (Chile) is MH479062. The IMG accession number for the whole genome sequences of strain DSM 44270T is 2728369258.
Blastococcus mobilis (mo’bi.lis. L. masc. Adj. mobilis movable, motile).
Colonies are reddish-brown coloured, opaque with a mucoid surface and regular margin. Cells are aerobic, Gram-reaction-positive and catalase and oxidase negative cocci (0.6–1.6 μm in diameter) with a tendency to form aggregates. It grows in aerobiosis but, according to genomic data, anaerobiosis and acetogenesis may occur. Reproduction by budding is predominant but binary fission is also observed. Cells are non-motile and non-spore-forming but a repertoire of genes related to flagellum synthesis, chemotaxis, spore production and pilus assembly were annotated. Degradation for casein, tyrosine, starch, xanthine, and hypoxanthine are negative. Temperature and pH ranges are 15–37°C (optimal range) and pH 6.5–11.0 (optimum 6.0–10.0), respectively. NaCl is not needed for growth. It can grow between 0% and 1% (w/v; optimal range) but not at 4%. It grows well on GYM Streptomyces, R2A (DSMZ Medium 830), Luedemann (DMSZ medium 877), and PYGV (DSMZ medium 621) media. According to API ZYM strips, the following enzymatic activities are present: esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, and valine arylamidase. According to the Biolog System, it oxidises: dextrin, D-maltose, sucrose, turanose, β-methyl-D-glucoside, D-glucose, D-mannose, D-fructose, D-galactose, sodium lactate, D-sorbitol, D-mannitol, D-arabitol, myo-inositol, glycerol, L-pyroglutamic acid, pectin, D-glucuronic acid, quinic acid, methyl pyruvate, D-lactic acid methyl ester, L-lactic acid, nalidixic acid, potassium tellurite, tween 40, α-hydroxy-butyric acid, β-hydroxy-butyric acid, β-keto-butyric acid, acetoacetic acid, propionic acid, acetic acid, aztreonam, butyric acid, and sodium bromated but not D-raffinose, α-D-lactose, D-melibiose, N-acetyl-D-galactosamine, N-acetyl-neuraminic acid, L-fucose, L-rhamnose, inosine, fusidic acid, D-serine (inhibitory concentration), D-aspartic acid, D-serine, troleandomycin, rifamycin SV, minocycline, gelatin, glycine-proline, L-histidine, L-serine, lincomycin, guanidine hydrochloride, niaproof, L-galactonic acid-γ-lactone, mucic acid, D-saccharic acid, vancomycin, tetrazolium violet, tetrazolium blue, p-hydroxy-phenylacetic acid, citric acid, D-malic acid, L-malic acid, bromo-succinic acid, γ-amino-n-butyric acid, and sodium formate. In correlation, a range of genes involved in the degradation of complex carbohydrates were identified (chemoheterotrophy). Three different autotrophic mechanisms including the Wood-Ljungdahl pathway, reductive citric acid cycle and carbonic anhydrases were besides annotated (chemolithoautotrophy). Predominant fatty acids are iso-C15:0, iso-C16:1 H, iso-C16:0, C17:1 ω8c, C18:1 ω9c, and C16:1 ω7c. The cell wall peptidoglycan contains meso-diaminopimelic acid. Rhamnose, ribose, mannose, arabinose, galactose, and glucose are the whole-cell sugars. Polar lipid profile consists of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, and phosphatidylinositol. The high-quality draught genome of strain DSM 44272T was resolved to 88 scaffolds consisting of 5,094,633 bp, with a G + C content of 72.5%, 4,938 candidate protein-coding genes, 49 tRNA genes, and six rRNA regions.
The INSDC accession number for the 16S rRNA gene sequences of the type strain AT 7(−2)-11T (=DSM 44272T = JCM 18933T) isolated from soil in the Atacama Desert (Chile) is MH479063. The IMG accession number for the whole genome sequences of strain DSM 44272T is 2724679778.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
MdCM-C designed the study. MdCM-C, AY, MR, and PS performed experiments. MdCM-C, AY, and JM-K performed bioinformatics analyses and wrote the manuscript. H-PK funded the study. All authors contributed to the article and approved the submitted version.
MdCM-C was the recipient of a DSMZ postdoctoral fellowship 2013-2015. MdCM-C is grateful for funding received from the Ramón y Cajal Research Grant (RYC2019-028468-I) from the Spanish Ministry of Economy, Industry and Competitiveness (MINECO). The work conducted by the Joint Genome Institute, a U.S. Department of Energy Office of Science User Facility, is supported under contract no. DE-AC02-05CH11231.
We would like to gratefully acknowledge the help of Cathrin Spröer and Bettina Sträubler (Both at DSMZ, Braunschweig) for preliminary DNA:DNA hybridization analysis and Prof. Aharon Oren for taxonomic nomenclature. Special thanks go out to the editor Brian Hedlund for a detailed and very constructive review of our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.975365/full#supplementary-material
Ahrens, R., and Moll, G. (1970). Ein neues knospendes Bakterium aus der Ostsee. Arch. Mikrobiol. 70, 243–265. doi: 10.1007/BF00407714
Amaral, G. R. S., Dias, G. M., Wellington-Oguri, M., Chimetto, L., Campeão, M. E., Thompson, F. L., et al. (2014). Genotype to phenotype: identification of diagnostic vibrio phenotypes using whole genome sequences. Int. J. Syst. Evol. Microbiol. 64, 357–365. doi: 10.1099/ijs.0.057927-0
Antia, M., Hoare, D. S., and Work, E. (1957). The stereoisomers of α,ε-diaminopimelic acid. III. Properties and distribution of diaminopimelic acid racemase, an enzyme causing interconversion of the LL and meso isomers. Biochem. J. 65, 448–459. doi: 10.1042/bj0650448
Auch, A. F., von Jan, M., Klenk, H.-P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Baek, I., Kim, M., Lee, I., Na, S.-I., Goodfelow, M., and Chun, J. (2018). Phylogeny trumps chemotaxonomy: a case study involving Turicella otitidis. Front. Microbiol. 9:834. doi: 10.3389/fmicb.2018.00834
Balezentiene, L. (2012). Hydrolases related to C and N cycles and soil fertility amendment: responses to different management styles of agro-ecosystems. Polish J. Environ. St. 21, 1153–1159.
Brettin, T., Davs, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Bruce, J. (1996). Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50, 77–81.
Castro, J. F., Nouioui, I., Sangal, V., Choi, S., Yang, S.-J., Kim, B.-Y., et al. (2018). Blastococcus atacamensis sp. nov., a novel strain adapted to life in the Yungay core region of the Atacama Desert. Int. J. Syst. Evol. Microbiol. 68, 2712–2721. doi: 10.1099/ijsem.0.002828
Chen, I. A., Markowitz, V. M., Chu, K., Palaniappan, K., Szeto, E., Pillay, M., et al. (2017). IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516. doi: 10.1093/nar/gkw929
Chen, I. M., Markowitz, V. M., Palaniappan, K., Szeto, E., Chu, K., Huang, J., et al. (2016). Supporting community annotation and user collaboration in the integrated microbial genomes (IMG) system. BMC Genomics 17:307. doi: 10.1186/s12864-016-2629-y
Chouaia, B., Crotti, E., Brusetti, L., Daffonchio, D., Essoussi, I., Nouioui, I., et al. (2012). Genome sequence of Blastococcus saxobsidens DD2, a stone-inhabiting bacterium. J. Bacteriol. 194, 2752–2753. doi: 10.1128/JB.00320-12
Chun, J., and Rainey, F. A. (2014). Integrating genomics into the taxonomy and systematics of the bacteria and archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324. doi: 10.1099/ijs.0.054171-0
Chun, J., Oren, A., Ventosa, A., Christensen, H., Ruiz-Arahal, D., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Collins, M. D., Pirouz, T., Goodfellow, M., and Minnikin, D. E. (1977). Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230. doi: 10.1099/00221287-100-2-221
Du, M.-Z., Wei, W., Qin, L., Liu, S., Zhang, A.-Y., Zhang, Y., et al. (2017). Co-adaptation of tRNA gene copy number and amino acid usage influences translation rates in three life domains. DNA Res. 24, 623–633. doi: 10.1093/dnares/dsx030
Eppard, M., Krumbein, W. E., Koch, C., Rhiel, E., Staley, J. T., and Stackebrandt, E. (1996). Morphological, physiological, and molecular characterization of actinomycetes isolated from dry soil, rocks, and monument surfaces. Arch. Microbiol. 166, 12–22. doi: 10.1007/s002030050350
Fotedar, R., Caldwell, M. E., Sankaranarayanan, K., Al Zeyara, A., Al Malki, A., Kaul, R., et al. (2020). Ningiella ruwaisensis gen. Nov., sp. nov., a member of the family Alteromonadaceae isolated from marine water of the Arabian gulf. Int. J. Syst. Evol. Microbiol. 70, 4130–4138. doi: 10.1099/ijsem.0.004256
Galloway, D. R., and Furlong, C. E. (1977). The role of ribose-binding protein in transport and chemotaxis in Escherichia coli K12. Arch. Biochem. Biophys. 184, 496–504. doi: 10.1016/0003-9861(77)90459-3
Giongo, A., Favet, J., Lapanje, A., Gano, K. A., Kennedy, S., Davis-Richardson, A. G., et al. (2013). Microbial hitchhikers on intercontinental dust: high-throughput sequencing to catalogue microbes in small sand samples. Aerobiologia 29, 71–84. doi: 10.1007/s10453-012-9264-0
Girard, G., Traag, B. A., Sangal, V., Mascini, N., Hoskisson, P. A., Goodfellow, M., et al. (2013). A novel taxonomic marker that discriminates between morphologically complex actinomycetes. Open Biol. 3:130073. doi: 10.1098/rsob.130073
Goodfellow, M., Kampfer, P., Busse, H.-J., Trujillo, M. E., Suzuki, K.-I., Ludwig, W., et al. (2012). Bergey’s Manual of Systematics Bacteriology. New York, NY: Springer.
Göker, M., Cleland, D., Saunders, E., Lapidus, A., Nolan, M., Lucas, S., et al. (2011). Complete genome sequence of Isosphaera pallida type strain (IS1BT). Stand. Genomic Sci. 4, 63–71. doi: 10.4056/sigs.1533840
Gordon, R. E., and Smith, M. M. (1955). Proposed group of characters for the separation of Streptomyces and Nocardia. J. Bacteriol. 69, 147–150. doi: 10.1128/jb.69.2.147-150.1955
Gregersen, T. (1978). Rapid method for distinction of gram-negative from gram-positive bacteria. Eur. J. Appl. Microbiol. Biotechnol. 5, 123–127. doi: 10.1007/BF00498806
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Gupta, R., Gupta, N., and Rathi, P. (2004). Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64, 763–781. doi: 10.1007/s00253-004-1568-8
Han, X., Chen, C.-C., Kuo, C.-J., Huang, C.-H., Zheng, Y., Ko, T.-P., et al. (2015). Crystal structures of ligand-bound octaprenyl pyrophosphate synthase from Escherichia coli reveal the catalytic and chain-length determining mechanisms. Proteins 83, 37–45. doi: 10.1002/prot.24618
Hahnke, R. L., Meier-Kolthoff, J. P., García-López, M., Mukherjee, S., Huntemann, M., Ivanova, N. N., et al. (2016). Genome-based taxonomic classification of Bacteroidetes. Front. Microbiol. 7:2003. doi: 10.3389/fmicb.2016.02003
Hennig, W. (1965). Phylogenetic systematics. Annu. Rev. Ecol. Syst. 10, 97–116. doi: 10.1146/annurev.en.10.010165.000525
Hezbri, K., Ghodhbane-Gtari, F., Montero-Calasanz, M. D. C., Sghaier, H., Rohde, M., Schumann, P., et al. (2015a). Geodermatophilus sabuli sp. nov., a γ-radiation-resistant actinobacterium isolated from desert limestone in Sahara Desert. Int. J. Syst. Evol. Microbiol. 65, 3365–3372. doi: 10.1099/ijsem.0.000422
Hezbri, K., Ghodhbane-Gtari, F., Montero-Calasanz, M. D. C., Sghaier, H., Rohde, M., Spröer, C., et al. (2015b). Description of Geodermatophilus bullaregiensis sp. nov. Antonie Van Leeuwenhoek 108, 415–425. doi: 10.1007/s10482-015-0494-3
Hezbri, K., Louati, M., Nouioui, I., Gtari, M., Rohde, M., Spröer, C., et al. (2016). Blastococcus capsensis sp. nov., isolated from an archaeological Roman pool and emended description of the genus Blastococcus, B. aggregatus, B. saxobsidens, B. jejuensis and B. endophyticus. Int. J. Syst. Evol. Microbiol 66, 4864–4872. doi: 10.1099/ijsem.0.001443
Hezbri, K., Nouioui, I., Rohde, M., Schumann, P., Gtari, M., Klenk, H.-P., et al. (2017). Blastococcus colisei sp. nov, isolated from an archaeological amphitheatre. Antonie Van Leeuwenhoek 110, 339–346. doi: 10.1007/s10482-016-0804-4
Hezbri, K., Nouioui, I., Rohde, M., Sproer, C., Schumann, P., Gtari, M., et al. (2018). Blastococcus xanthinilyticus sp. nov., isolated from monument. Int. J. Syst. Evol. Microbiol. 68, 1177–1183. doi: 10.1099/ijsem.0.002646
Holt, P. S., and Chaubal, L. H. (1997). Detection of motility and putative synthesis of flagellar proteins in salmonella pullorum cultures. J. Clin. Microbiol. 35, 1016–1020.
Hor, L., Dobson, R. C. J., Downton, M. T., Wagner, J., Hutton, C. A., and Perugini, M. A. (2013). Dimerization of bacterial diamonopimelate epimerase is essential for catalysis. J. Biol. Chem. 288, 9238–9248. doi: 10.1074/jbc.M113.450148
Huntemann, M., Ivanova, N. N., Mavromatis, K., Tripp, H. J., Paez-Espino, D., Palaniappan, K., et al. (2015). The standard operating procedure of the DOE-JGI microbial genome annotation pipeline (MGAP v.4). Stand. Genomic Sci. 10:86. doi: 10.1186/s40793-015-0077-y
Ishiguro, E. E., and Wolfe, R. S. (1970). Control of morphogenesis in Geodermatophilus: ultrastructural studies. J. Bacteriol. 104, 566–580. doi: 10.1128/jb.104.1.566-580.1970
Jakimowics, D., and van Wezel, G. P. (2012). Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol. Microbiol. 85, 393–404. doi: 10.1111/j.1365-2958.2012.08107.x
Jiang, Z.-M., Zhang, B.-H., Sun, H.-M., Zhang, T., Yu, L.-Y., and Zhang, Y.-Q. (2021). Properties of Modestobacter deserti sp. Nov., a kind of novel phosphate-solubilising actinobacteria inhabited in the desert biological soil crust. Front. Microbiol. 12:742798. doi: 10.3389/fmicb.2021.742798
Kachlany, S. C., Planet, P. J., Bhattacherjee, M. K., Kollia, E., DeSalle, R., Fine, D. H., et al. (2000). Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 182, 6169–6176. doi: 10.1128/JB.182.21.6169-6176.2000
Katsyv, A., and Müller, V. (2020). Overcoming energetic barriers in acetogenic C1 conversion. Front. Bioeng. Biotechnol. 8:621166. doi: 10.3389/fbioe.2020.621166
Klappenbach, J. A., Dunbar, J. M., and Schmidt, T. S. (2000). rRNA operon copy number reflects ecological strategies of bacteria. App. Environ. Microbiol. 66, 1328–1333. doi: 10.1128/AEM.66.4.1328-1333.2000
Klenk, H.-P., and Göker, M. (2010). En route to a genome-based classification of Archaea and bacteria? Syst. Appl. Microbiol. 33, 175–182. doi: 10.1016/j.syapm.2010.03.003
Ko, J.-H., Montero Llopis, P., Heinritz, J., Jacobs-Wagner, C., and Sll, D. (2013). Suppression of amber codons in Caulobacter crescentus by the orthogonal Escherichia coli Histidyl-tRNA synthetase/tRNAHis pair. PLoS One 8:e83630. doi: 10.1371/journal.pone.0083630
Kroppenstedt, R., and Goodfellow, M. (2006). “The family thermomonosporaceae: actinocorallia, actinomadura, spirillospora and thermomonospora” in The Prokaryotes. eds. M. Dworkin and S. Falkow (New York, NY: Springer)
Kroppenstedt, R. M. (1982). Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J. Liquid Chromatography 5, 2359–2367. doi: 10.1080/01483918208067640
Kuo, C.-H., and Ochman, H. (2010). The extinction dynamics of bacterial pseudogenes. PLoS Genet. 6:e1001050. doi: 10.1371/journal.pgen.1001050
Kyrpides, N. C., Hugenholtz, P., Eisen, J. A., Woyke, T., Göker, M., Parker, C. T., et al. (2014). Genomic encyclopedia of bacteria and archaea: sequencing a myriad of type strains. PLoS Biol. 12:e1001920. doi: 10.1371/journal.pbio.1001920
Lawson, P. A., Patel, N. B., Mohammed, A., Moore, E. R. B., Lo, A. S., Sardi, A., et al. (2020). Parapseudoflavitalea muciniphila gen. Nov., sp. nov., a member of the family Chitinophagaceae isolated from a human peritoneal tumour and reclassification of Pseudobacter ginsenosidimutans as Pseudoflavitalea ginsenosidimutans comb. nov. Int. J. Syst. Evol. Microbiol. 70, 3639–3646. doi: 10.1099/ijsem.0.004204
Lechevalier, M. P. L. H., and Lechevalier, H. (1970). Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Evol. Microbiol. 20, 435–443. doi: 10.1099/00207713-20-4-435
Lee, S. D. (2006). Blastococcus jejuensis sp. nov., an actinomycete from beach sediment, and emended description of the genus Blastococcus Ahrens and Moll 1970. Int. J. Syst. Evol. Microbiol. 56, 2391–2396. doi: 10.1099/ijs.0.64268-0
Lee, D. W., Lee, H., Kwon, B. O., Khim, J. S., Yim, U. H., Kim, B. S., et al. (2018). Blastococcus litoris sp. nov., isolated from sea-tidal flat sediment. Int. J. Syst. Evol. Microbiol. 68, 3435–3440. doi: 10.1099/ijsem.0.003004
Liu, Y., Harrison, P. M., Kunin, V., and Gerstein, M. (2004). Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 5:1715
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Liu, R., and Ochman, H. (2007). Stepwise formation of the bacterial flagellar system. Proc. Natl. Acad. Sci. U. S. A. 104, 7116–7121. doi: 10.1073/pnas.0700266104
López-Lara, I. M., and Geiger, O. (2019). “Chapter 3: formation of fatty acids,” in Biogenesis of Fatty Acids, Lipids and Membranes. ed. O. Geiger (Springer).
Lowe, J., and Amos, L. A. (1998). Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206. doi: 10.1038/34472
Luedemann, G. M. (1968). Geodermatophilus, a new genus the Dermatophilaceae (Actinomycetales). J. Bacteriol. 96, 1848–1858. doi: 10.1128/jb.96.5.1848-1858.1968
McCool, G. J., and Cannon, M. C. (2001). PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in bacillus megaterium. J. Bacteriol. 183, 4235–4243. doi: 10.1128/JB.183.14.4235-4243.2001
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013a). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Göker, M., Spröer, C., and Klenk, H. P. (2013b). When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 195, 413–418. doi: 10.1007/s00203-013-0888-4
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2014a). Highly parallelized inference of large genome-based phylogenies. Concurr. Comput. Pract. Exp. 26, 1715–1729. doi: 10.1002/cpe.3112
Meier-Kolthoff, J. P., Klenk, H.-P., and Göker, M. (2014b). Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 64, 352–356. doi: 10.1099/ijs.0.056994-0
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Mengin-Lecreulx, D., Michau, C., Richaud, C., Blanot, D., and Heijenoort, J. V. (1988). Incorporation of LL-Diaminopimelic acid into peptidoglycan of Escherichia coli mutants lacking diaminopimelate epimerase encoded by dapF. J. Bacteriol. 170, 2031–2039. doi: 10.1128/jb.170.5.2031-2039.1988
Mevs, U., Stackebrandt, E., Schumann, P., Gallikowski, C. A., and Hirsch, P. (2000). Modestobacter multiseptatus gen. Nov., sp. nov., a budding actinomycete from soils of the Asgard range (Transantarctic Mountains). Int. J. Syst. Evol. Microbiol. 50, 337–346. doi: 10.1099/00207713-50-1-337
Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Montero-Calasanz, M. D. C. (2021). Klenkia in Bergey's Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd: Chichester, UK.
Montero-Calasanz, M. D. C. (2020a). Geodermatophilus in Bergey's Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd: Chichester, UK.
Montero-Calasanz, M. D. C. (2020b). Geodermatophilaceae in Bergey's Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Ltd: Chichester, UK.
Montero-Calasanz, M. D. C., Hofner, B., Göker, M., Rohde, M., Spröer, C., Hezbri, K., et al. (2014). Geodermatophilus poikilotrophi sp. nov., a multi-tolerant actinomycete isolated from dolomitic marble. Biomed. Res. Int. 2014:914767. doi: 10.1155/2014/914767
Montero-Calasanz, M. D. C., Hezbri, K., Göker, M., Sghaier, H., Rohde, M., Spröer, C., et al. (2015). Description of gamma radiation-resistant Geodermatophilus dictyosporus sp. nov. to accommodate the not validly named Geodermatophilus obscurus subsp. dictyosporus (Luedemann, 1968). Extremophiles 19, 77–85. doi: 10.1007/s00792-014-0708-z
Montero-Calasanz, M. D. C., Meier-Kolthoff, J. P., Zhang, D.-F., Yaramis, A., Rohde, M., Woyke, T., et al. (2017). Genome-scale data call for a taxonomic rearrangement of Geodermatophilaceae. Front. Microbiol. 8:2501. doi: 10.3389/fmicb.2017.02501
Montero-Calasanz, M. D. C., Yaramis, A., Nouioui, I., Igual, J. M., Spröer, C., Castro, J. F., et al. (2019). Modestobacter italicus sp. nov., isolated from Carrara marble quarry and emended descriptions of the genus Modestobacter and the species Modestobacter marinus, Modestobacter multiseptatus, Modestobacter roseus and Modestobacter versicolor. Int. J. Syst. Evol. Microbiol. 69, 1537–1545. doi: 10.1099/ijsem.0.003282
Mukherjee, S., Stamatis, D., Bertsch, J., Ovchinnikova, G., Verezemska, O., Isbandi, M., et al. (2017). Genomes online database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 45, D446–D456. doi: 10.1093/nar/gkw992
Neilson, J. W., Quade, J., Ortiz, M., Nelson, W. M., Legatzki, A., Tian, F., et al. (2012). Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles 16, 553–566. doi: 10.1007/s00792-012-0454-z
Normand, P., Orso, S., Cournoyer, B., Jeannin, P., Chapelon, C., Dawson, J., et al. (1996). Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int. J. Syst. Bacteriol. 46, 1–9. doi: 10.1099/00207713-46-1-1
Normand, P. (2006). Geodermatophilaceae fam. Nov., a formal description. International. Int. J. Syst. Evol. Microbiol. 56, 2277–2278. doi: 10.1099/ijs.0.64298-0
Nouioui, I., Göker, M., Carro, L., Montero-Calasanz, M. D. C., Rohde, M., Woyke, T., et al. (2017). High quality draft genome of Nakamurella lactea type strain, a rock actinobacterium, and emended description of Nakamurella lactea. Stand. Genomic Sci. 12:4. doi: 10.1186/s40793-016-0216-0
Nouioui, I., Carro, L., García-López, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol 9:2007. doi: 10.3389/fmicb.2018.02007
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Paniagua-Michel, J., Olmos-Soto, J., and Ruiz, M. A. (2012). “Pathways of carotenoid biosynthesis in bacteria and microalgae” in Microbial carotenoids from bacteria and microalgae. Methods in molecular biology (methods and protocols). ed. J. L. Barredo, vol. 892 (Totowa, NJ: Humana Press)
Perez, B. A., Planet, P. J., Kachlany, S. C., Tomich, M., Fine, D. H., and Figurski, D. H. (2006). Genetic analysis of the requirement for flp-2, tadV and rcpB in Actinobacillus actinomycetemcomitans biofilm formation. J. Bacteriol. 188, 6361–6375. doi: 10.1128/JB.00496-06
Proft, T., and Baker, E. N. (2009). Pili in gram-negative and gram-positive bacteria – structure, assembly and their role in disease. Cell. Mol. Life Sci. 66, 613–635. doi: 10.1007/s00018-008-8477-4
Qin, S., Bian, G. K., Zhang, Y. J., Xing, K., Cao, C. L., Liu, C. H., et al. (2013). Modestobacter roseus sp. nov., an endophytic actinomycete isolated from the coastal halophyte Salicornia europaea Linn., and emended description of the genus Modestobacter. Int. J. Syst. Evol. Microbiol. 63, 2197–2202. doi: 10.1099/ijs.0.044412-0
Rainey, F. A., Ward-Rainey, N., Kroppenstedt, R. M., and Stackebrandt, E. (1996). The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. Nov. Int. J. Syst. Bacteriol. 46, 1088–1092. doi: 10.1099/00207713-46-4-1088
Rashid, R., Cazenave-Gassiot, A., Gao, I. H., Nair, Z. J., Kumar, J. K., Gao, L., et al. (2017). Comprehensive analysis of phospholipids and glycolipids in the opportunistic pathogen enterococcus faecalis. PLoS One 12:e0175886. doi: 10.1371/journal.pone.0175886
Reddy, G. S., Potrafka, R. M., and Garcia-Pichel, F. (2007). Modestobacter versicolor sp. nov., an actinobacterium from biological soil crusts that produces melanins under oligotrophy, with emended descriptions of the genus Modestobacter and Modestobacter multiseptatus Mevs et al. 2000. Int. J. Syst. Evol. Microbiol. 57, 2014–2020. doi: 10.1099/ijs.0.64932-0
Rivas-Marin, E., Peeters, S. H., Claret Fernández, L., Jogler, C., van Niftrik, L., Wiegan, S., et al. (2020). Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila. Sci. Rep. 10:66. doi: 10.1038/s41598-019-56978-8
Rogers, T. E., Ataide, S. F., Dare, K., Katz, A., Seveau, S., Roy, H., et al. (2012). A pseudo-tRNA modulates antibiotic resistance in Bacillus cereus. PLoS One 7:e41248. doi: 10.1371/journal.pone.0041248
Sasser, M. (2001). Identification of bacteria by gas chromatography of cellular fatty acids. Technical. Note 101, 1–6.
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477. doi: 10.1128/br.36.4.407-477.1972
Sen, A., Daubin, V., Abrouk, D., Gifford, I., Berry, A. M., and Normand, P. (2014). Phylogeny of the class Actinobacteria revisited in the light of complete genomes. The orders ‘Frankiales’ and Micrococcales should be split into coherent entities: proposal of Frankiales Ord. Nov., Geodermatophilales Ord. Nov., Acidothermales Ord. Nov. an. Int. J. Syst. Evol. Microbiol. 64, 3821–3832. doi: 10.1099/ijs.0.063966-0
Seto, H., Jinnai, Y., Hiratsuka, T., Fukawa, M., Furihata, K., Itoh, N., et al. (2008). Studies on a new biosynthetic pathway for menaquinone. J. Am. Chem. Soc. 160, 5614–5615. doi: 10.1021/ja710207s
Sghaier, H., Hezbri, K., Ghodhbane-Gtari, F., Pujic, P., Sen, A., Daffonchio, D., et al. (2016). Stone-dwelling actinobacteria Blastococcus saxobsidens, Modestobacter marinus and Geodermatophilus obscurus proteogenomes. ISME J. 10, 21–29. doi: 10.1038/ismej.2015.108
Stackebrandt, E., Rainey, F. A., and Ward-Rainey, N. L. (1997). Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47, 479–491. doi: 10.1099/00207713-47-2-479
Stackebrandt, E., Breymann, S., Steiner, U., Prauser, H., Weiss, N., Schumann, P., et al. (2002). Re-evaluation of the status of the genus Oerskovia, reclassification of Promicromonospora enterophila (Jáger et al. 1983) as Oerskovia enterophila comb. nov. and description of Oerskovia jenensis sp. nov. and Oerskovia paurometabola sp. nov. Int. J. Syst. Evol. Microbiol. 52, 1105–1111. doi: 10.1099/00207713-52-4-1105
Staneck, J. L., and Roberts, G. D. (1974). Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231. doi: 10.1128/am.28.2.226-231.1974
Sun, D.-L., Jiang, X., Wu, Q. L., and Zhou, N.-Y. (2013). Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbiol. 79, 5962–5969. doi: 10.1128/AEM.01282-13
Supuran, C. T., and Capasso, C. (2017). An overview of the bacterial carbonic anhydrases. Meta 7:56. doi: 10.3390/metabo7040056
Sutcliffe, I. C., Trujillo, M. E., Whitman, W. B., and Goodfellow, M. (2013). A call to action for the international committee on systematics of prokaryotes. Trends Microbiol. 21, 51–52. doi: 10.1016/j.tim.2012.11.004
Szurmant, H., and Ordal, G. W. (2004). Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68, 301–319. doi: 10.1128/MMBR.68.2.301-319.2004
Tindall, B. J. (1990). A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 13, 128–130. doi: 10.1016/S0723-2020(11)80158-X
Tu, C. M., and Miles, J. R. W. (1976). Interactions between insecticides and soil microbes. Res. Rev. 64, 17–65.
Tomich, M., Planet, P. J., and Figurski, D. H. (2007). The tad locus: postcards from the widespread colonization island. Nat. Rev. 5, 363–375. doi: 10.1038/nrmicro1636
Tóth, E. M., Schumann, P., Borsodi, A. K., Kéki, Z., Kovács, A. L., and Márialigeti, K. (2008). Wohlfahrtiimonas chitiniclastica gen. Nov., sp. nov., a new gammaproteobacterium isolated from Wohlfahrtia magnifica (Diptera: Sarcophagidae). Int. J. Syst. Evol. Microbiol. 58, 976–981. doi: 10.1099/ijs.0.65324-0
Traag, B. A., and van Wezel, G. P. (2008). The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek 94, 85–97. doi: 10.1007/s10482-008-9225-3
Trujillo, M. E., and Normand, P. (2019). Sporitchthya in Bergey's Manual of Systematics of Archaea and Bacteria. John Wiley and Sons, Ltd: Chichester.
Upadhyay, A., Fontes, F. L., Gonzalez-Jarero, M., NcNeil, M. R., Crans, D. C., Kacson, M., et al. (2015). Partial saturation of menaquinone in mycobacterium tuberculosis: function and essentiality of a novel reductase. MenJ. ACS Cent. Sci. 1, 292–302. doi: 10.1021/acscentsci.5b00212
Urzì, C., Brusetti, L., Salamone, P., Sorlini, C., Stackebrandt, E., and Daffonchio, D. (2001). Biodiversity of geodermatophilaceae isolated from altered stones and monuments in the mediterranean basin. Environ. Microbiol. 3, 471–479. doi: 10.1046/j.1462-2920.2001.00217.x.o
Urzì, C., Salamone, P., Schumann, P., Rohde, M., and Stackebrandt, E. (2004). Blastococcus saxobsidens sp. nov., and emended descriptions of the genus Blastococcus Ahrens and Moll 1970 and Blastococcus aggregatus Ahrens and Moll 1970. Int. J. Syst. Evol. Microbiol. 54, 253–259. doi: 10.1099/ijs.0.02745-0
Vaas, L. A. I., Sikorski, J., Hofner, B., Fiebig, A., Buddruhs, N., Klenk, H. P., et al. (2013). Opm: an R package for analysing OmniLog® phenotype microarray data. Bioinformatics 29, 1823–1824. doi: 10.1093/bioinformatics/btt291
Vaas, L. A. I., Sikorski, J., Michael, V., Göker, M., and Klenk, H. P. (2012). Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS One 7:e34846. doi: 10.1371/journal.pone.0034846
Van Wezel, G. P., van der Meulen, J., Kwamoto, S., Luiten, R. G., Koerten, H. K., and Kraal, B. (2000). SsgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J. Bacteriol. 182, 5653–5662. doi: 10.1128/JB.182.20.5653-5662.2000
Vandamme, P., and Sutcliffe, I. (2021). Out with the old and in with the new: time to rethink twentieth century chemotaxonomic practices in bacterial taxonomy. Int. J. Syst. Evol. Microbiol. 71:005127. doi: 10.1099/ijsem.0.005127
Vetrovsky, T., and Baldrian, P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. doi: 10.1371/journal.pone.0057923
Wang, X., and Lutkenhaus, J. (1993). The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9, 435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x
Wang, Y., Zhang, L., Zhang, X., Huang, J., Zhao, Y., Zhao, Y., et al. (2017). Geodermatophilus daqingensis sp. Nov., isolated from petroleum-contaminated soil. Antonie Van Leeuwenhoek 110, 803–809. doi: 10.1007/s10482-017-0853-3
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464. doi: 10.1099/00207713-37-4-463
Whitman, W. B. (2015). Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst. Appl. Microbiol. 38, 217–222. doi: 10.1016/j.syapm.2015.02.003
Wiley, E. O., and Lieberman, B. B. S. (2011). Phylogenetics: Theory and Practice of Phylogenetic Systematics, 2nd. Hoboken, NJ: John Wiley and Sons.
Xiao, J., Luo, Y., Xu, J., Xie, S., and Xu, J. (2011). Modestobacter marinus sp. nov., a psychrotolerant actinobacterium from deep-sea sediment, and emended description of the genus Modestobacter. Int. J. Syst. Evol. Microbiol. 61, 1710–1714. doi: 10.1099/ijs.0.023085-0
Yang, Z. W., Asem, M. D., Li, X., Li, L. Y., Salam, N., Alkhalifah, D. H. M., et al. (2019). Blastococcus deserti sp. nov., isolated from a desert sample. Arch. Microbiol. 201, 193–198. doi: 10.1007/s00203-018-1604-1
Zhu, W. Y., Zhang, J. L., Qin, Y. L., Xiong, Z. J., Zhang, D. F., Klenk, H. P., et al. (2013). Blastococcus endophyticus sp. nov., an actinobacterium isolated from Camptotheca acuminata. Int. J. Syst. Evol. Microbiol. 63, 3269–3273. doi: 10.1099/ijs.0.049239-0
Zhi, X. Y., Li, W. J., and Stackebrandt, E. (2009). An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 59, 589–608. doi: 10.1099/ijs.0.65780-0
Keywords: Trujillonella, Pleomorpha, Goekera, phylogenetic systematics, in silico chemotaxonomy
Citation: Montero-Calasanz MdC, Yaramis A, Rohde M, Schumann P, Klenk H-P and Meier-Kolthoff JP (2022) Genotype–phenotype correlations within the Geodermatophilaceae. Front. Microbiol. 13:975365. doi: 10.3389/fmicb.2022.975365
Received: 22 June 2022; Accepted: 11 October 2022;
Published: 10 November 2022.
Edited by:
André Antunes, Macau University of Science and Technology, ChinaReviewed by:
Javier Pascual, Darwin Bioprospecting Excellence, SpainCopyright © 2022 Montero-Calasanz, Yaramis, Rohde, Schumann, Klenk and Meier-Kolthoff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria del Carmen Montero-Calasanz, bWFyaWFjLm1vbnRlcm8uY2FsYXNhbnpAanVudGFkZWFuZGFsdWNpYS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.