
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 August 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.974432
This article is part of the Research TopicResistance to third-generation TetracyclinesView all 10 articles
The rapid dissemination of plasmid-mediated tet(X) genes in Acinetobacter species has compromised the clinical effectiveness of tigecycline, one of the last-resort antibiotics. However, the classification strategy and homology group of tet(X)-positive Acinetobacter spp. plasmids remain largely unknown. In this study, we classified them by genome-based replicon typing, followed by analyses of structural characteristics, transferability and in vivo effect. A total of 34 plasmids distributed in at least nine Acinetobacter species were collected, including three tet(X3)-positive plasmids and one tet(X6)-positive plasmid from our genome sequencing results. Among them, there were 28 plasmids carrying Rep_3 superfamily replicase genes and classified into six homology groups, consisting of GR31 (82.1%), GR26 (3.6%), GR41 (3.6%), GR59 (3.6%), and novel groups GR60 (3.6%) and GR61 (3.6%). Our tet(X3)-positive plasmids pYH16040-1, pYH16056-1, and pYH12068-1 belonged to the dominant GR31 group, whereas the tet(X6)-positive plasmid pYH12068-2 was unclassified. Structurally, all tet(X)-positive GR31 plasmids shared similar plasmid replication (repB), stability (parA and parB) and accessory modules [tet(X) and sul2], and 97.6% of plasmid-mediated tet(X) genes in Acinetobacter species were adjacent to ISCR2. Conjugation and susceptibility testing revealed pYH16040-1, pYH16056-1, and pYH12068-2, carrying plasmid transfer modules, were able to mediate the mobilization of multiple antibiotic resistance. Under the treatment of tigecycline, the mortality rate of Galleria mellonella infected by pYH16040-1-mediated tet(X3)-positive Acinetobacter spp. isolate significantly increased when compared with its plasmid-cured strain (p < 0.0001). The spread of such plasmids is of great clinical concern, more effects are needed and will facilitate the future analysis of tet(X)-positive Acinetobacter spp. plasmids.
Tigecycline, the first glycylcycline antibiotic, exhibits a broad spectrum of antibacterial activities against multidrug-resistant (MDR) Gram-negative and Gram-positive pathogens (Sader et al., 2019). However, the recent emergence and spread of novel tigecycline resistance mechanisms Tet(X3), Tet(X4), Tet(X5), Tet(X6), and other variants have compromised its clinical efficacy by enzymatic degradation (He et al., 2019; Sun et al., 2019; Wang et al., 2019; Chen et al., 2021). Acinetobacter species is a heterogeneous group of opportunistic pathogens and easily acquires antibiotic resistance genes (ARGs) (Wong et al., 2017). To date, the tet(X) genes have been reported in at least 10 different Acinetobacter species, including Acinetobacter baumannii, Acinetobacter gandensis, Acinetobacter piscicola, Acinetobacter schindleri, Acinetobacter johnsonii, Acinetobacter indicus, Acinetobacter towneri, Acinetobacter lwoffii, Acinetobacter pseudolwoffii, and Acinetobacter variabilis (Chen et al., 2020; Liu et al., 2020; Zheng et al., 2020; Cheng Y. et al., 2021; Li et al., 2021). Worrisomely, the plasmid-mediated tet(X3) and tet(X6) genes were detected with carbapenem resistance gene blaNDM–1 in A. baumannii, A. indicus, A. schindleri, and A. lwoffii isolates, posing a serious public health threat (Cui et al., 2020; He et al., 2020; Zheng et al., 2020).
The plasmid is a self-replicating component of Acinetobacter spp. genome and plays an important role in the horizontal transmission of ARGs, such as blaNDM–1 and blaOXA–23 (Wang and Sun, 2015; Silva et al., 2018). With the increasing number of complete Acinetobacter spp. plasmids deposited at the National Center for Biotechnology Information (NCBI), a series of plasmid classification schemes were developed based on replication initiator protein, mobilization protein and plasmid size (Bertini et al., 2010; Salto et al., 2018; Mindlin et al., 2020). As the latest research showed, there were a total of 59 homology groups identified by plasmid replicon typing (Li et al., 2022). However, there was a lack of systematic classification of tet(X)-positive Acinetobacter spp. plasmids since the first mobile plasmid-mediated tet(X3) gene in 2019, and their homology groups remained to be analyzed (He et al., 2019; Wang J. et al., 2020; Cheng Y. et al., 2021). Herein, we intend to explore the classification and homology group of complete tet(X)-carrying Acinetobacter spp. plasmids by genome-based replicon typing, followed by analyses of structural characteristics, transferability, and in vivo effect.
During an epidemiological surveillance between 2015 and 2018, we reported the prevalence of tet(X)-positive Acinetobacter spp. strains in China (Chen et al., 2020), of which seven isolates belonging to different sources were selected for next analyses in this study (Table 1). These included: tet(X3)-positive Acinetobacter spp. YH16040 and tet(X3)- and tet(X6)-positive Acinetobacter spp. YH12068 from pig; tet(X3)-positive Acinetobacter spp. YH16056 from soil; tet(X3)-positive A. pseudolwoffii YH18001 from human; tet(X4)-positive A. indicus Q22-2, Q85-2, and Q278-1 from migratory bird. In addition, a newly isolated tet(X6)-positive A. baumannii YC103 by CHROMagar™ Acinetobacter plates (CHROMagar, Paris, France) containing tigecycline (2 μg/mL) from duck in 2019 was also analyzed (Table 1). With the amino acid sequence of Tet(X3) as a template, the complete tet(X)-harboring Acinetobacter spp. plasmids deposited at the NCBI database were collected by tblastn (Supplementary Table 1; accessed 23 Mar 2022).
Genomic DNA of eight tet(X)-positive Acinetobacter spp. isolates were sequenced by Oxford Nanopore (Nextomics, Wuhan, China), respectively. Combining the clean data with our previous Illumina HiSeq data (Chen et al., 2020) as well as that of A. baumannii YC103 (Novogene, Beijing, China), genome assembly was performed by Unicycler version 0.4.1 and corrected by Pilon version 1.12 (Walker et al., 2014; Wick et al., 2017).
All tet(X)-harboring plasmids were annotated by Rapid Annotation using Subsystem Technology (RAST) version 2.0 (Aziz et al., 2008). Plasmid-mediated ARGs were analyzed by ResFinder version 4.0 and a heatmap was then constructed by ImageGP (Bortolaia et al., 2020; Chen et al., 2022). Plasmid replication and transfer proteins were detected by the Conserved Domain Database (CDD) (Lu et al., 2020). A maximum-likelihood phylogenetic tree of replication initiator proteins was constructed by MEGA-X version 10.1.8 and visualized by Evolview version 3.0 (Kumar et al., 2018; Subramanian et al., 2019). Plasmid classification was performed according to the A. baumannii replicon typing scheme, with at least 80% nucleotide coverage and at least 75% nucleotide identity of replicase genes in the same group (Bertini et al., 2010; Castro-Jaimes et al., 2022; Li et al., 2022). Similarly, the bacterial host ranges of different plasmid groups were also evaluated by querying the NCBI database with representative replicase genes (accessed 24 May 2022). Sequence comparison of tet(X)-positive structures was conducted by Easyfig version 2.2.5 (Sullivan et al., 2011).
Transferability of plasmid-borne tet(X)-mediated tigecycline resistance was evaluated by filter mating with rifampin-resistant Acinetobacter baylyi ADP1 and A. baumannii ATCC 19606 (Chen et al., 2020). The putative transconjugants were selected on Luria-Bertani (LB) agar plates containing tigecycline (2 μg/mL) and rifampin (100 μg/mL), followed by tet(X) detection and PCR-based fingerprinting (Versalovic et al., 1991). In parallel, the recipient strains were selected with rifampin (100 μg/mL), and transfer efficiencies were calculated by colony counts of the transconjugant and recipient bacterial cells (Zhu et al., 2013).
The tet(X3)-harboring GR31 plasmid pYH16040-1 was cured of Acinetobacter spp. YH16040 using sodium dodecyl sulfate (SDS) with adjustment (Chen et al., 2017). An overnight culture was diluted 100-fold in LB broth supplemented with 0.02% SDS and serially passaged at 38°C by shaking per 24 h. One week later, 100 μL dilution was streaked onto LB agar plate and 50 colonies were subcultured with or without tigecycline (4 μg/mL). The colony that did not grow in tigecycline was probably the plasmid-cured isogeneic strain (namely YH16040C) and confirmed by detecting tet(X3) gene (Chen et al., 2020) and replicase gene repB (repB-F, 5′-GCCCAATCGAATTATCAGCCA-3′; repB-R, 5′-TGGCAACAGAATCTAGGGCA-3′).
Minimum inhibitory concentrations (MICs) of all strains were determined by broth microdilution and interpreted according to the Clinical and Laboratory Standards Institute guideline M100-Ed28 (Clinical and Laboratory Standards Institute [CLSI], 2018). The tested antibiotics contained tetracycline, tigecycline, eravacycline, omadacycline, amikacin, gentamicin, ciprofloxacin, colistin, cefotaxime, meropenem, sulfamethoxazole-trimethoprim, and florfenicol. Particularly, the resistance breakpoints of tigecycline (≥8 μg/mL) and omadacycline (≥16 μg/mL) referred to the Food and Drug Administration (FDA) criteria for Enterobacteriaceae,1 whereas eravacycline was uninterpreted with no breakpoint. Escherichia coli ATCC 25922 served as a quality control strain.
As previously described (Dong et al., 2017; Xu et al., 2021), the healthy larvae of G. mellonella (Huiyude, Tianjin, China) were randomly grouped (16 per group), and then infected with GR31 plasmid-mediated tet(X3)-positive Acinetobacter spp. YH16040 or its plasmid-cured strain YH16040C [tet(X3)-negative, 5 × 106 colony-forming units] via the last left proleg. After incubation at 35°C for 2 h, the infected larvae were treated with tigecycline (2 μg/g) or phosphate-buffered saline (PBS) by injection into the last right proleg. Finally, the larvae were observed for survival rates per 24 h in the next 4 days. All in vivo experiments were performed in triplicate.
WGS analyses of eight tet(X)-positive Acinetobacter spp. isolates successfully revealed four tet(X)-carrying plasmids. These included tet(X3)-positive plasmids pYH16040-1 (GenBank accession number: CP094542) from Acinetobacter spp. YH16040, pYH16056-1 (CP094546) from Acinetobacter spp. YH16056, and pYH12068-1 (CP094556) as well as a tet(X6)-positive plasmid pYH12068-2 (CP094557) from Acinetobacter spp. YH12068 (Table 1). Meanwhile, a tet(X6)-positive chromosome cYC103 (CP054560) from A. baumannii YC103 was also obtained (Table 1). For the remaining four tet(X)-positive strains A. pseudolwoffii YH18001, A. indicus Q278-1, A. indicus Q85-2, and A. indicus Q22-2 (Table 1), we failed to acquire the complete tet(X)-harboring plasmids or chromosomes despite repeated attempts. By querying Nanopore raw data of A. pseudolwoffii YH18001 (n ≥ 18, SRR18497244), A. indicus Q278-1 (n ≥ 4, SRR18497245), A. indicus Q85-2 (n ≥ 13, SRR18497246), and A. indicus Q22-2 (n ≥ 4, SRR18497247), a repeated structure consisting of multiple copies of tet(X) genes was detected, respectively, which may lead to the failure of complete sequence assemblies.
Since the first report in 2019 (He et al., 2019), the number of complete tet(X)-positive Acinetobacter spp. plasmids deposited at the NCBI database has been growing (n = 34; Supplementary Table 1), including four plasmids mentioned above. There were three tet(X) subtypes located on plasmids (Figure 1A), namely tet(X3) (82.4%, 28/34), tet(X5) (5.9%, 2/34), and tet(X6) (35.3%, 12/34), of which tet(X3) usually coexisted with tet(X6) (23.5%, 8/34). Besides tet(X) genes, large amounts of plasmid-mediated genes conferring resistance to tetracyclines, aminoglycosides, β-lactams, phenicols, sulfonamides, macrolides, lincosamides, and rifamycins were present (Figure 1A). Worrisomely, the tet(X)-positive plasmids ranging from 42,489 to 332,451 bp were widely distributed in A. indicus (26.5%, 9/34), A. baumannii (14.7%, 5/34), A. towneri (11.8%, 4/34), A. schindleri (8.8%, 3/34), A. variabilis (2.9%, 1/34), A. pseudolwoffii (2.9%, 1/34), A. piscicola (2.9%, 1/34), Acinetobacter junii (2.9%, 1/34), Acinetobacter pittii (2.9%, 1/34), and unidentified Acinetobacter spp. strains (23.5%, 8/34; Figures 1B,C). Given the genetic and host diversity, more attention should be paid to the evolution of tet(X)-positive MDR Acinetobacter spp. plasmids.
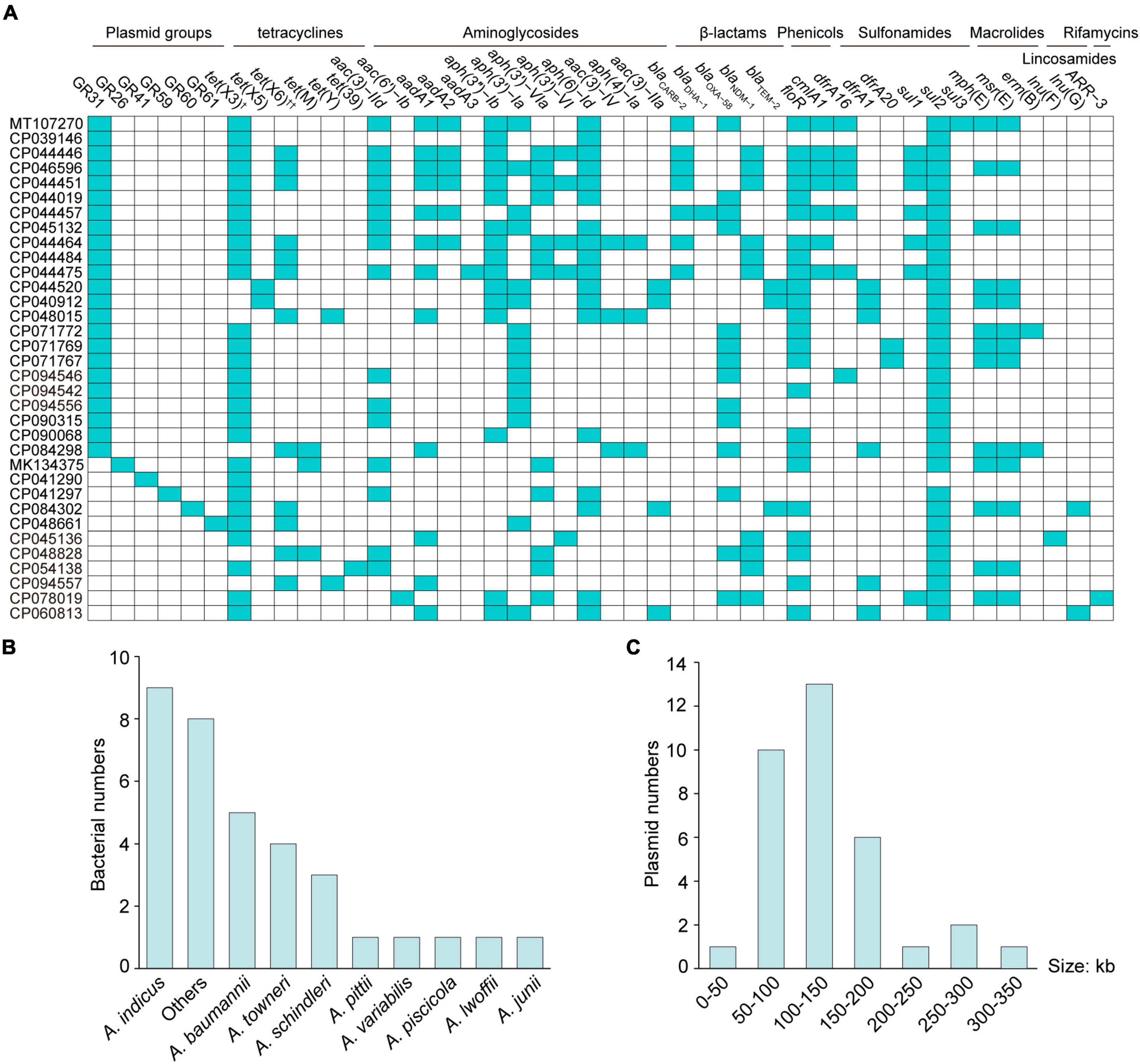
Figure 1. Collection of the complete tet(X)-positive plasmids from Acinetobacter spp. strains. The distribution of plasmid groups (cyan, A), plasmid-mediated ARGs (cyan, A), bacterial species (B) and plasmid sizes (C) is presented. †One tet(X3) variant with a 99.7% amino acid identity (CP045132) with tet(X3) is included. ††Two tet(X6) variants (CP048828 and CP048661) with a 97.4% amino acid identity with tet(X6) are included.
Replicon conserved domain analyses showed that 82.4% (28/34) of tet(X)-positive Acinetobacter spp. plasmids carried replicase genes and all of them belonged to a Rep_3 superfamily (pfam01051). Therefore, plasmid classification was conducted based on nucleotide sequence alignment with already classified replicase genes (Castro-Jaimes et al., 2022; Li et al., 2022). Among the tet(X)-positive Rep_3 superfamily plasmids, a total of six homology groups were successfully identified, such as the dominant GR31 (82.1%, 23/28), GR26 (3.6%, 1/28), GR41 (3.6%, 1/28), and GR59 (3.6%, 1/28; Figure 2). Especially, our tet(X3)-positive plasmids pYH16040-1 (CP094542), pYH16056-1 (CP094546), and pYH12068-1 (CP094556) fell within the same group GR31, which has been sporadically detected with tet(X) genes in A. baumannii, A. indicus, A. schindleri, A. towneri, and other Acinetobacter spp. strains (Figure 2 and Supplementary Table 1). However, our tet(X6)-positive plasmid pYH12068-2 (CP094557) was unclassified due to the lack of replicase genes. In contrast, the replicase genes of tet(X3)- and tet(X6)-carrying plasmids pXMC5X702-tetX-145k (CP084302) and pYH12207-2 (CP048661) have < 75% nucleotide identities with existing homology groups GR1-GR59, and therefore were defined as novel groups GR60 (3.6%, 1/28) and GR61 (3.6%, 1/28), respectively (Figure 2). We provided the updated replicase gene and protein sequences in Supplementary Files 1, 2, and hoped they will facilitate the future analyses of the increasing number of tet(X)-positive Acinetobacter spp. plasmids.
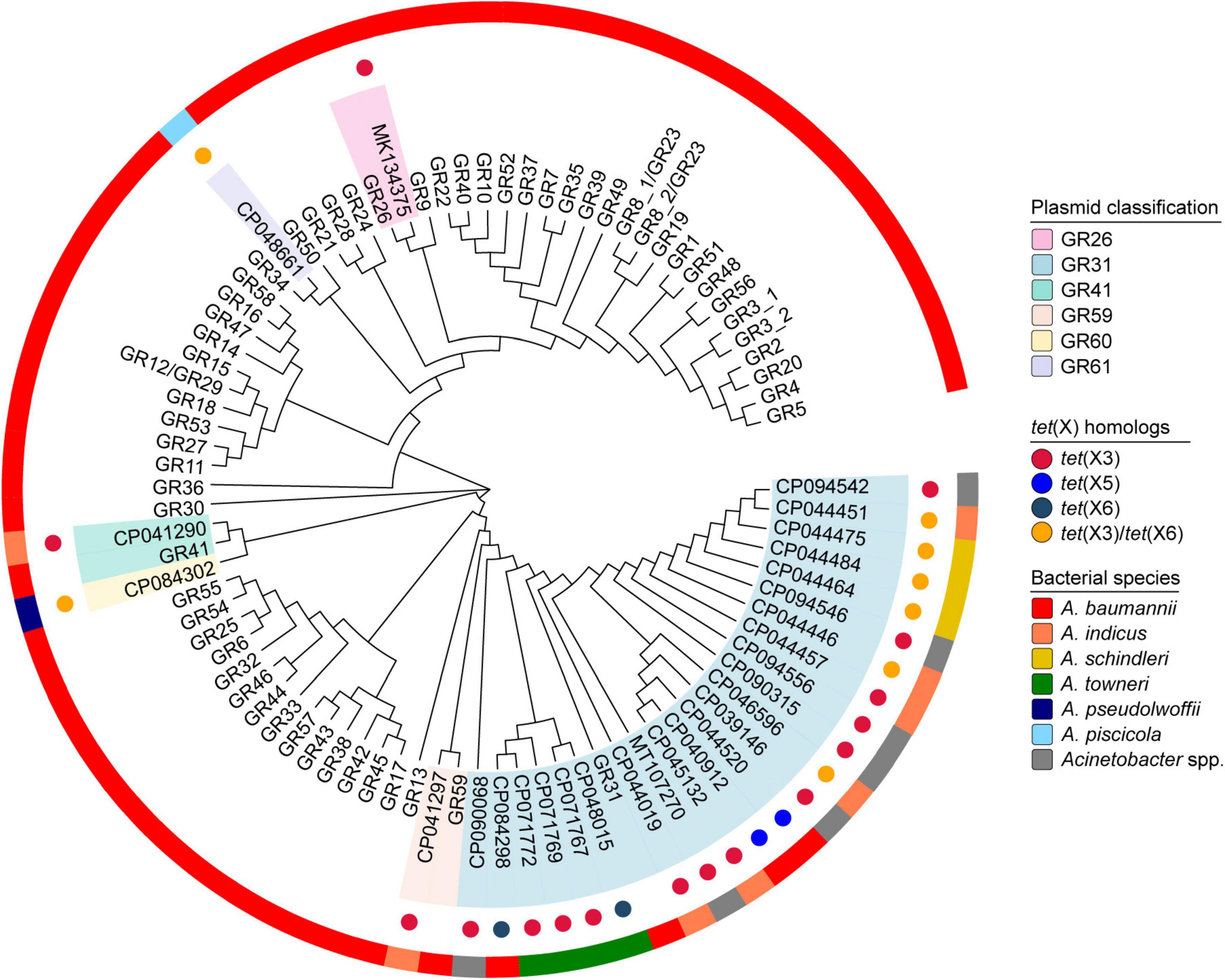
Figure 2. Maximum-likelihood phylogenetic analysis of Acinetobacter spp. plasmids based on replication initiator proteins. In parallel, the plasmid classification, tet(X) homologs and bacterial species are indicated from the inside out.
In order to evaluate the host range of plasmids belonging to GR26, GR31, GR41, GR59, GR60, and GR61, we conducted a blastn search against the NCBI database and confirmed they were mainly distributed in Acinetobacter spp. isolates (99.0%, 201/203). In essence, the GR31 plasmids have been detected in 13 validly named Acinetobacter species, including A. baumannii (18.3%, 15/82), A. lwoffii (17.1%, 14/82), A. indicus (12.2%, 10/82), A. towneri (8.5%, 7/82), A. schindleri (7.3%, 6/82), and others (Supplementary Figure 1). The plasmids belonging to GR26, GR41, GR59, GR60, and GR61 have also been identified in at least four Acinetobacter species, except two E. coli strains carrying GR41 plasmids (Supplementary Figure 1). Particularly, a few unassigned Acinetobacter species harbored GR26, GR31, GR59, GR60, and GR61 plasmids (Supplementary Figure 1), but the bacterial characteristics remained to be explored. All the results suggested a potential transmission risk of plasmid-mediated tet(X) genes especially in the genera Acinetobacter.
Predominantly, we compared the complete plasmid sequences of pYH16040-1, pYH16056-1, and pYH12068-1 with tet(X)-carrying GR31 plasmids available in the NCBI database. A total of 23 plasmids belonging to GR31 were analyzed and they exhibited GC contents ranging from 39.1 to 46.1%. According to our WGS results, pYH16040-1 was 87,435 bp and harbored 94 putative open reading frames (ORFs), whereas pYH16056-1 was 98,709 bp consisting of 107 ORFs and pYH12068-1 was 100,866 bp consisting of 115 ORFs. Although they originated from different sources, pYH16056-1 and pYH12068-1 showed an average 76.5% nucleotide coverage and 99.8% nucleotide identity to pYH16040-1, with the encoding sequence insertion, deletion and rearrangement (Figure 3A). Moreover, they shared high homologies (≥64.0% nucleotide coverage and ≥ 96.2% nucleotide identity) with MDR plasmids from the pig (MT107270), goose (CP044484), pigeon (CP044457), and soil (CP044451; Figure 3A). Archetypically, Acinetobacter spp. plasmids contained the backbone (namely plasmid replication, stability and transfer modules) and the accessory modules (Brovedan et al., 2020). Our results revealed all tet(X)-positive GR31 plasmids owned a repB gene involved in plasmid replication, and parA and parB genes responsible for plasmid stability. However, only 21.7% (5/23) of them harbored plasmid transfer modules, including conjugal transfer (n = 3) and mobilization genes (n = 2). For accessory modules, they mainly consisted of ARGs [e.g., tet(X) and sul2, 100% (23/23)] and heavy-metal resistance gene clusters [e.g., czcA-czcD, 78.3% (18/23)].
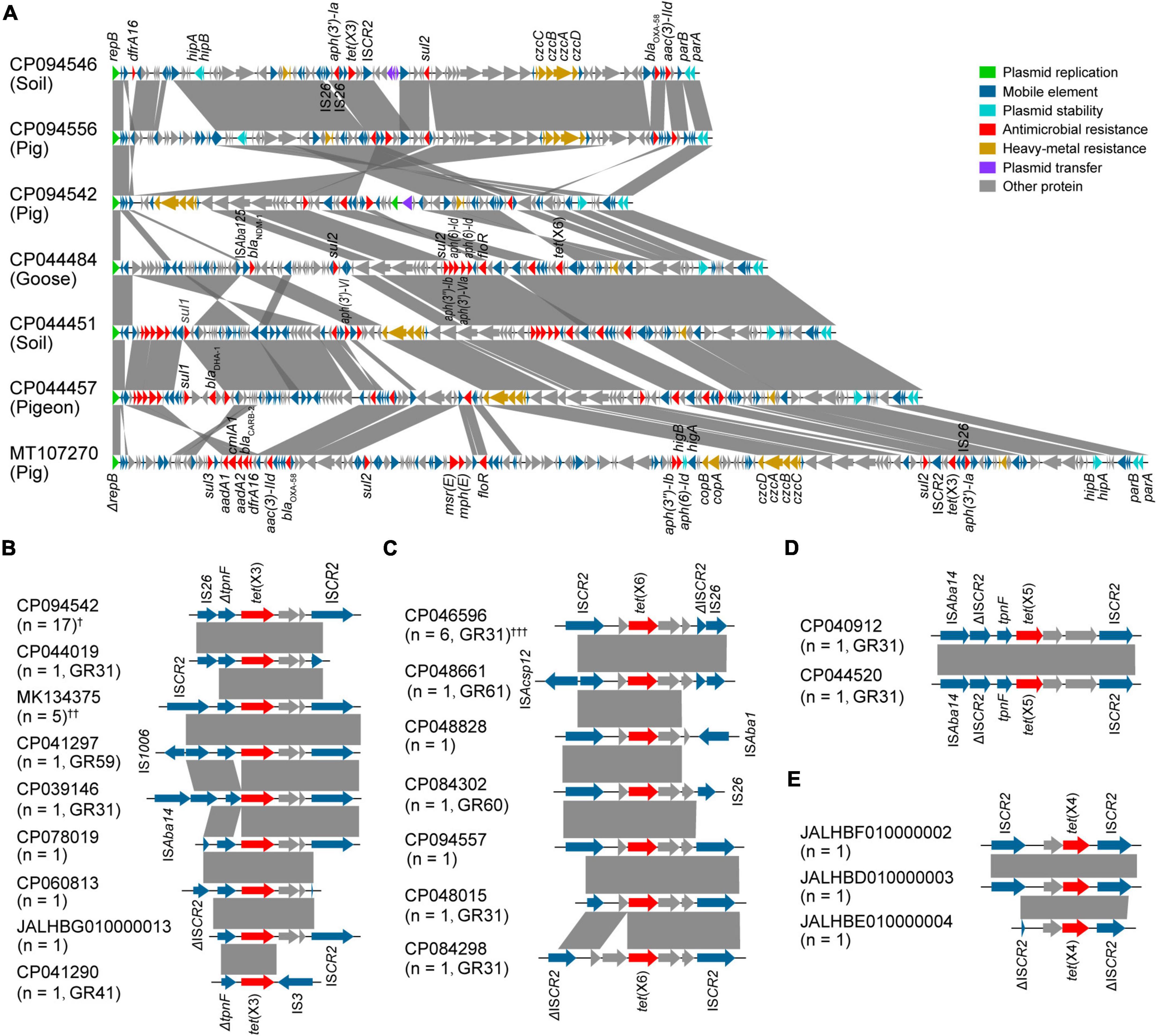
Figure 3. Characteristics of tet(X)-carrying structures in Acinetobacter species. The arrow represents the position and transcriptional direction of ORFs, while the Δ symbol indicates the gene is truncated. Regions of > 85% nucleotide sequence homology are marked by gray shading. (A) Linear sequence alignment of tet(X)-positive GR31 plasmids. The bacterial sources are listed in parenthesis. (B–E) Genetic environments of tet(X3) (B), tet(X6) (C), tet(X5) (D), and tet(X4) (E) genes. The number of genetic environments and the plasmid homology groups are also given in parenthesis. †These include tet(X3)-carrying GR31 (n = 15, e.g., CP094542), GR60 (n = 1) and GR61 plasmids (n = 1). ††These include tet(X3)-carrying GR26 (n = 1, MK134375), GR31 (n = 2), and unclassified plasmids (n = 2). †††All of these belong to GR31 plasmids (n = 6, e.g., CP046596).
Genetic environments of all plasmid-mediated tet(X) genes in Acinetobacter species, including tet(X3) (n = 28), tet(X6) (n = 12), and tet(X5) (n = 2), were further analyzed (Figures 3B–D). The result showed 97.6% (41/42) of them were adjacent to ISCR2, which is able to transpose ARGs through a rolling-circle transposition process (Liu et al., 2022). For GR31 plasmids, the ISCR2-mediated transposition units were constantly truncated by IS26 (n = 22) and ISAba14 (n = 3). Similarly, the truncation by IS26 was found on GR60 (n = 2) and GR61 (n = 2) plasmids, one of which was also upstream truncated by ISAcsp12. Except the GR26 plasmid carrying a complete ISCR2-mediated transposition unit (n = 1), GR41 and GR59 plasmids were detected with truncation by IS3 (n = 1) and IS1006 (n = 1), respectively. In addition, our WGS results of A. pseudolwoffii YH18001 (JALHBG010000013), A. indicus Q278-1 (JALHBF010000002), A. indicus Q85-2 (JALHBE010000004), and A. indicus Q22-2 (JALHBD010000003) revealed the incomplete tet(X)-carrying structures were also related to ISCR2 (Figures 3B,E). The close association between tet(X) variants and ISCR2 as well as truncation by other insertion sequences indicated the frequent recombination events.
Conjugation experiments showed the GR31 plasmid-mediated tet(X3) genes were successfully transferred from Acinetobacter spp. YH16040 (pYH16040-1, CP094542) and YH16056 (pYH16056-1, CP094546) to the recipient A. baylyi ADP1 (transfer efficiency, approximately 10–8), but failed to A. baumannii ATCC 19606. In Acinetobacter spp. YH12068, the tet(X3)-carrying GR31 plasmid pYH12068-1 (CP094556) couldn’t be transferred, which may be explained by the lack of entire conjugative transfer regions (Figure 3A). Meanwhile, the tet(X6)-positive unclassified plasmid pYH12068-2 (CP094557) carrying plasmid transfer modules was transferred from Acinetobacter spp. YH12068 to A. baylyi ADP1 with a similar efficiency mentioned above. MICs of all transconjugants against tetracyclines increased by at least 32-fold when compared with A. baylyi ADP1, including tetracycline (≥64 μg/mL), tigecycline (≥4 μg/mL), the newly FDA-approved eravacycline (≥2 μg/mL) and omadacycline (≥8 μg/mL; Table 2). According to previous reports (Wang et al., 2019; Cui et al., 2020), the GR31 plasmid-mediated tet(X5) gene (CP040912) was also able to be transferred from human-derived A. baumannii to A. baumannii 5AB via electro-transformation, whereas the GR31 plasmid co-harboring tet(X3) and tet(X6) genes (CP044451) was from environmental A. indicus to A. baylyi ADP1 by natural transformation. The transferability of tet(X) genes between Acinetobacter species of human, animal and environment origins reminded us to consider the “One Health” approach to prevent mobile tigecycline resistance.
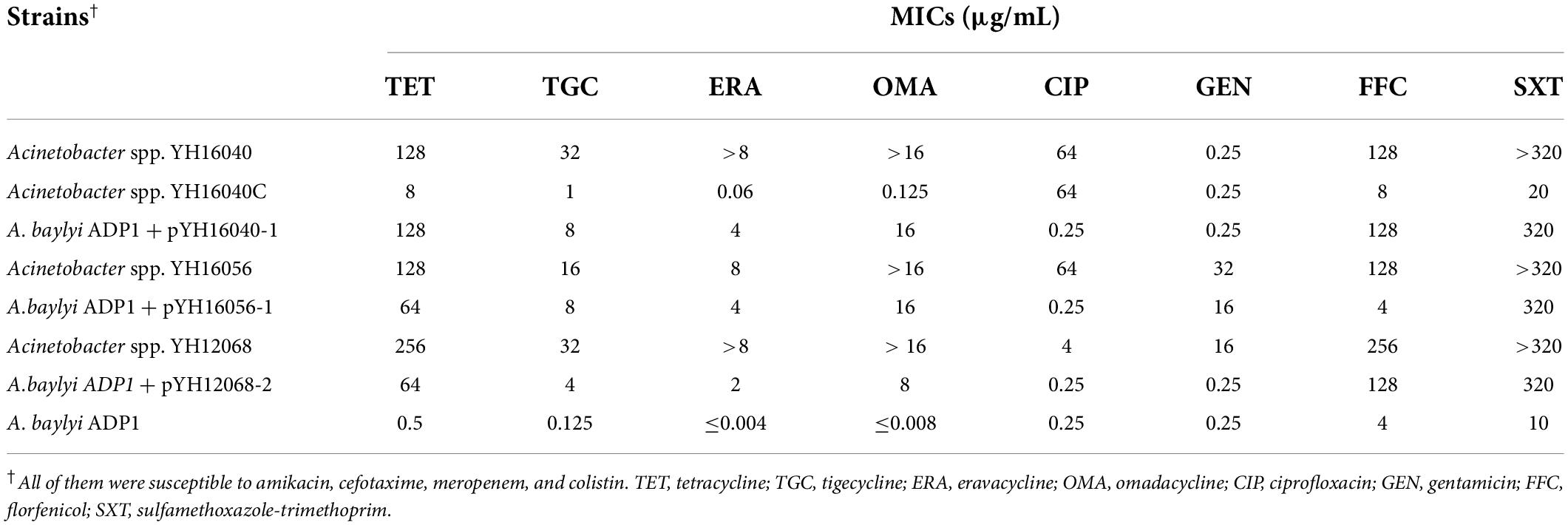
Table 2. MICs of tet(X)-positive Acinetobacter spp. isolates, transconjugants, and plasmid-cured strain.
At the same time, the sulfamethoxazole-trimethoprim resistance (320 μg/mL) was transferred with tet(X3)-mediated tigecycline resistance from Acinetobacter spp. YH16040, YH16056, and YH12068 to A. baylyi ADP1, as well as florfenicol resistance (128 μg/mL) from Acinetobacter spp. YH16040 and YH12068 and gentamicin resistance (16 μg/mL) from Acinetobacter spp. YH16056, which was consistent with the result of plasmid-mediated ARG mining (Table 2 and Figure 1A). Recently, our study has also reported the co-occurrence of mobile tet(X)-mediated tigecycline resistance and blaNDM–1-mediated carbapenem resistance between Acinetobacter species (Cui et al., 2020). With the co-transfer of multiple ARGs, the continuous application and selection pressures of tetracyclines, sulfonamides, aminoglycosides, β-lactams, and phenicols may promote the spread of tet(X) genes, as previously described for resistance determinants mcr-1 and blaIMP–1 (Wang Y. et al., 2020; Cheng Z. et al., 2021).
To explore the potential effect of GR31 plasmid-mediated tet(X3) gene, a plasmid-cured strain YH16040C [tet(X3)-negative] was obtained by serially passaging of tet(X3)-positive Acinetobacter spp. YH16040. For tigecycline, a significant decrease in MIC was detected (1 μg/mL) when compared with the parental isolate YH16040 (32 μg/mL; Table 2). MICs of Acinetobacter spp. YH16040C against tetracycline (8 μg/mL), eravacycline (0.06 μg/mL), omadacycline (0.125 μg/mL), florfenicol (8 μg/mL) and sulfamethoxazole-trimethoprim (20 μg/mL) also exhibited at least 16-fold decrease. The result further confirmed the elimination of GR31 plasmid pYH16040-1 (CP094542) that co-harbored resistance genes tet(X3), floR and sul2 (Table 2 and Figure 1A).
Subsequently, the in vivo effect of pYH16040-1-carrying tet(X3) gene was evaluated using a G. mellonella model. 96 h later of tigecycline treatment (2 μg/g), the larval mortality rate of Acinetobacter spp. YH16040C significantly decreased to 37.5% while Acinetobacter spp. YH16040 was 87.5% (p < 0.0001; Figure 4). The experiment was performed in triplicate with similar results, indicating the GR31 plasmid-mediated tet(X3) gene compromised the clinical effectiveness of tigecycline. Notably, the infection by Acinetobacter spp. YH16040 and Acinetobacter spp. YH16040C under the treatment of PBS resulted in a greater larval mortality rate (100%) than PBS only (12.5%) after 48 h (p < 0.0001), whereas no significant difference was observed between Acinetobacter spp. YH16040 and Acinetobacter spp. YH16040C (p = 0.32; Figure 4). The result suggested virulence genes outside pYH16040-1 remained to be studied. As previously reported (Jiang et al., 2021; Xu et al., 2021), G. mellonella models infected by plasmid-mediated tet(X6)-positive Enterobacteriaceae bacteria have also been constructed, and therefore G. mellonella was available for in vivo functional analyses of plasmid-mediated tet(X) genes.
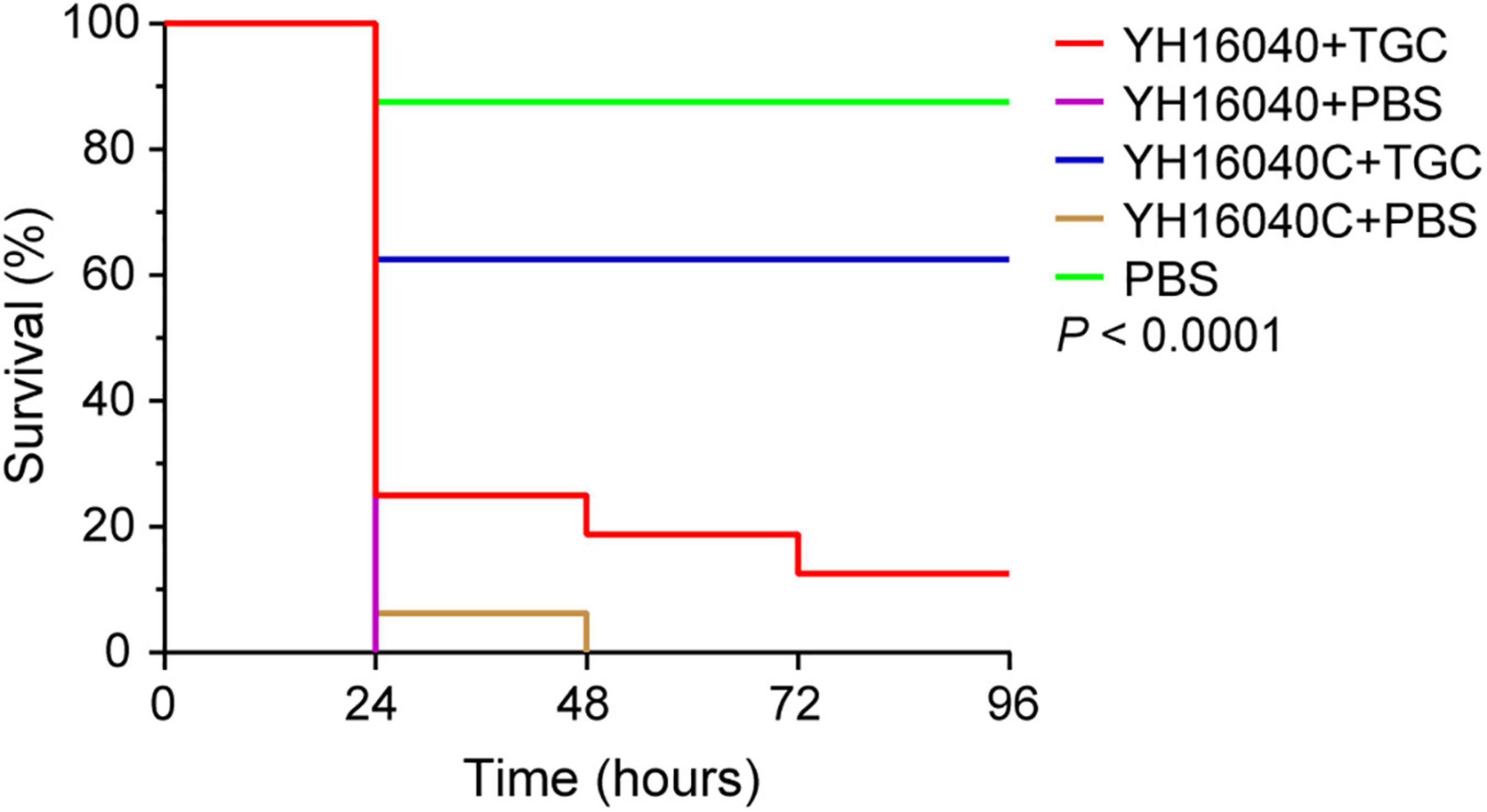
Figure 4. Survival curve of G. mellonella larvae under the treatment of tigecycline. The P-value is calculated by Log-rank (Mantel-Cox) test between GR31 plasmid-mediated tet(X3)-positive Acinetobacter spp. YH16040 and its plasmid-cured strain YH16040C [tet(X3)-negative] using GraphPad Prism version 8.3.0.
Taken together, the data presented in this study highlighted the diverse distribution of tet(X)-carrying MDR plasmids in Acinetobacter species. Our results classified the tet(X)-positive Acinetobacter spp. plasmids of Rep_3 superfamily into six homology groups, including two newly assigned GR60 and GR61. To the best of our knowledge, this study first revealed a dominant GR31 plasmid mediating the horizontal transfer of tigecycline resistance gene tet(X) across different Acinetobacter species and contributed to the failure of tigecycline treatment in vivo. Accordingly, more efforts are needed to monitor and prevent the plasmid-mediated tet(X)-positive Acinetobacter spp. strains.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
J-LH, Y-HL, and JS designed the study. CC, P-YH, C-YC, and QH performed the experiments. CC and P-YH analyzed the data. CC wrote the draft of the manuscript. All authors reviewed, revised, and approved the final report.
This study was jointly supported by the Natural Science Foundation of Jiangsu Province of China (BK20210803), the Natural Science Foundation of Jiangsu Higher Education Institutions of China (21KJB230008), the National Natural Science Foundation of China (32172939, 31872493, 31972735), the Taishan Industry Leading Talents Project of Shandong Province (tscy20190113), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (32121004), and the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2019BT02N054).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.974432/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bertini, A., Poirel, L., Mugnier, P. D., Villa, L., Nordmann, P., and Carattoli, A. (2010). Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4168–4177. doi: 10.1128/AAC.00542-10
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Brovedan, M. A., Cameranesi, M. M., Limansky, A. S., Moran-Barrio, J., Marchiaro, P., and Repizo, G. D. (2020). What do we know about plasmids carried by members of the Acinetobacter genus? World J. Microbiol. Biotechnol. 36:109. doi: 10.1007/s11274-020-02890-7
Castro-Jaimes, S., Guerrero, G., Bello-Lopez, E., and Cevallos, M. A. (2022). Replication initiator proteins of Acinetobacter baumannii plasmids: An update note. Plasmid 119-120:102616. doi: 10.1016/j.plasmid.2021.102616
Chen, C., Cui, C. Y., Wu, X. T., Fang, L. X., He, Q., He, B., et al. (2021). Spread of tet(X5) and tet(X6) genes in multidrug-resistant Acinetobacter baumannii strains of animal origin. Vet. Microbiol. 253:108954. doi: 10.1016/j.vetmic.2020.108954
Chen, C., Cui, C. Y., Yu, J. J., He, Q., Wu, X. T., He, Y. Z., et al. (2020). Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome Med. 12:111. doi: 10.1186/s13073-020-00807-5
Chen, T., Liu, Y. X., and Huang, L. (2022). ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 1:e5. doi: 10.1002/imt2.5
Chen, Y., Hu, D., Zhang, Q., Liao, X. P., Liu, Y. H., and Sun, J. (2017). Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar typhimurium. Front. Cell Infect. Microbiol. 7:37. doi: 10.3389/fcimb.2017.00037
Cheng, Y., Chen, Y., Liu, Y., Song, J., Chen, Y., Shan, T., et al. (2021). Detection of a new tet(X6)-encoding plasmid in Acinetobacter towneri. J. Glob. Antimicrob. Resist. 25, 132–136. doi: 10.1016/j.jgar.2021.03.004
Cheng, Z., Bethel, C. R., Thomas, P. W., Shurina, B. A., Alao, J. P., Thomas, C. A., et al. (2021). Carbapenem use is driving the evolution of imipenemase 1 variants. Antimicrob. Agents Chemother. 65:e01714-20. doi: 10.1128/AAC.01714-20
Clinical and Laboratory Standards Institute [CLSI] (2018). Performance Standards for Antimicrobial Susceptibility Testing: CLSI supplement M100, 28th Edn. Wayne, PA: CLSI.
Cui, C.-Y., Chen, C., Liu, B.-T., He, Q., Wu, X.-T., Sun, R.-Y., et al. (2020). Co-occurrence of plasmid-mediated tigecycline and carbapenem resistance in Acinetobacter spp. from waterfowls and their neighboring environment. Antimicrob. Agents Chemother. 64:e02502-19. doi: 10.1128/aac.02502-19
Dong, C. L., Li, L. X., Cui, Z. H., Chen, S. W., Xiong, Y. Q., Lu, J. Q., et al. (2017). Synergistic effect of pleuromutilins with other antimicrobial agents against Staphylococcus aureus In Vitro and in an experimental Galleria mellonella model. Front. Pharmacol. 8:553. doi: 10.3389/fphar.2017.00553
He, T., Li, R., Wei, R., Liu, D., Bai, L., Zhang, L., et al. (2020). Characterization of Acinetobacter indicus co-harbouring tet(X3) and blaNDM–1 of dairy cow origin. J. Antimicrob. Chemother. 75, 2693–2696. doi: 10.1093/jac/dkaa182
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Jiang, L., Cai, W., Tang, F., Wang, Z., and Liu, Y. (2021). Characterization of fitness cost caused by tigecycline-resistance gene tet(X6) in different host bacteria. Antibiotics 10:1172. doi: 10.3390/antibiotics10101172
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, R., Peng, K., Xiao, X., Wang, Y., and Wang, Z. (2021). Characterization of novel ISAba1-bounded tet(X15)-bearing composite transposon Tn6866 in Acinetobacter variabilis. J. Antimicrob. Chemother. 76, 2481–2483. doi: 10.1093/jac/dkab182
Li, Y., Qiu, Y., Fang, C., Dai, X., and Zhang, L. (2022). Coexistence of blaOXA–58 and blaNDM–1 on a novel plasmid of GR59 from an Acinetobacter towneri isolate. Antimicrob. Agents Chemother. 66:e0020622. doi: 10.1128/aac.00206-22
Liu, D., Wang, T., Shao, D., Song, H., Zhai, W., Sun, C., et al. (2022). Structural diversity of the ISCR2-mediated rolling-cycle transferable unit carrying tet(X4). Sci. Total Environ. 826:154010. doi: 10.1016/j.scitotenv.2022.154010
Liu, D., Zhai, W., Song, H., Fu, Y., Schwarz, S., He, T., et al. (2020). Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J. Antimicrob. Chemother. 75, 1428–1431. doi: 10.1093/jac/dkaa037
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Mindlin, S., Beletsky, A., Rakitin, A., Mardanov, A., and Petrova, M. (2020). Acinetobacter plasmids: Diversity and development of classification strategies. Front. Microbiol. 11:588410. doi: 10.3389/fmicb.2020.588410
Sader, H. S., Castanheira, M., Arends, S. J. R., Goossens, H., and Flamm, R. K. (2019). Geographical and temporal variation in the frequency and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bacterial pneumonia: Results from 20 years of the SENTRY antimicrobial surveillance program (1997–2016). J. Antimicrob. Chemother. 74, 1595–1606. doi: 10.1093/jac/dkz074
Salto, I. P., Torres Tejerizo, G., Wibberg, D., Puhler, A., Schluter, A., and Pistorio, M. (2018). Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci. Rep. 8:7783. doi: 10.1038/s41598-018-26180-3
Silva, L., Mourao, J., Grosso, F., and Peixe, L. (2018). Uncommon carbapenemase-encoding plasmids in the clinically emergent Acinetobacter pittii. J. Antimicrob. Chemother. 73, 52–56. doi: 10.1093/jac/dkx364
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W. H. (2019). Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: A genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Versalovic, J., Koeuth, T., and Lupski, R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, B., and Sun, D. (2015). Detection of NDM-1 carbapenemase-producing Acinetobacter calcoaceticus and Acinetobacter junii in environmental samples from livestock farms. J. Antimicrob. Chemother. 70, 611–613. doi: 10.1093/jac/dku405
Wang, J., Wang, Y., Wu, H., Wang, Z. Y., Shen, P. C., Tian, Y. Q., et al. (2020). Coexistence of blaOXA–58 and tet(X) on a Novel Plasmid in Acinetobacter sp. From Pig in Shanghai, China. Front. Microbiol. 11:578020. doi: 10.3389/fmicb.2020.578020
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 64:e01326-19. doi: 10.1128/AAC.01326-19
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wong, D., Nielsen, T. B., Bonomo, R. A., Pantapalangkoor, P., Luna, B., and Spellberg, B. (2017). Clinical and pathophysiological overview of Acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 30, 409–447. doi: 10.1128/CMR.00058-16
Xu, Y., Liu, L., Zhang, H., and Feng, Y. (2021). Co-production of Tet(X) and MCR-1, two resistance enzymes by a single plasmid. Environ. Microbiol. 23, 7445–7464. doi: 10.1111/1462-2920.15425
Zheng, X. R., Zhu, J. H., Zhang, J., Cai, P., Sun, Y. H., Chang, M. X., et al. (2020). A novel plasmid-borne tet(X6) variant co-existing with blaNDM–1 and blaOXA–58 in a chicken Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 75, 3397–3399. doi: 10.1093/jac/dkaa342
Keywords: Acinetobacter species, mobile tigecycline resistance, tet(X), replicon typing, GR31 plasmid
Citation: Chen C, Huang P-Y, Cui C-Y, He Q, Sun J, Liu Y-H and Huang J-L (2022) Classification and molecular characteristics of tet(X)-carrying plasmids in Acinetobacter species. Front. Microbiol. 13:974432. doi: 10.3389/fmicb.2022.974432
Received: 21 June 2022; Accepted: 08 August 2022;
Published: 23 August 2022.
Edited by:
Ning Dong, Soochow University, ChinaReviewed by:
Hua Zhou, Zhejiang University, ChinaCopyright © 2022 Chen, Huang, Cui, He, Sun, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Lin Huang, amlubGluQHl6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.