- College of Animal Science (College of Bee Science), Fujian Agriculture and Forestry University, Fuzhou, China
Clostridium butyricum (C. butyricum) can provide many benefits for animals’ growth performance and gut health. In this study, we investigated the effects of C. butyricum on the growth performance, cecal microbiota, and plasma metabolome in Ira rabbits. A total of 216 Ira rabbits at 32 days of age were randomly assigned to four treatments supplemented with basal diets containing 0 (CG), 200 (LC), 400 (MC), and 600 mg/kg (HC) C. butyricum for 35 days, respectively. In comparison with the CG group, C. butyricum supplementation significantly improved the average daily gain (ADG) and feed conversion rate (FCR) at 53 and 67 days of age (P < 0.05) and digestibilities of crude protein (CP) and crude fiber (CF) at 67 days of age (P < 0.05). The cellulase activity in the HC group was higher respectively by 50.14 and 90.13% at 53 and 67 days of age, than those in the CG groups (P < 0.05). Moreover, at 67 days of age, the diet supplemented with C. butyricum significantly increased the relative abundance of Verrucomicrobia at the phylum level (P < 0.05). Meanwhile, the concentrations of different metabolites, such as amino acids and purine, were significantly altered by C. butyricum (P < 0.05). In addition, 10 different genera were highly correlated with 52 different metabolites at 53-day-old and 6 different genera were highly correlated with 18 different metabolites at 67-day-old Ira rabbits. These findings indicated that the C. butyricum supplementation could significantly improve the growth performance by modifying the cecal microbiota structure and plasma metabolome of weaned Ira rabbits.
Introduction
Ira Rabbit is an excellent breed of meat rabbit, which has the characteristics of a high feed utilization rate and fast growth rate (Chipo and Tapiwa, 2019). In rabbit production, antibiotics as feed additives can relieve weaning stress, which can reduce huge economic losses (Hu et al., 2018). However, the long-term abuse of antibiotics in production has caused negative effects such as dysbacteriosis in animals. Therefore, it is imperative to find safe and effective alternatives to antibiotics (Alayande et al., 2020).
Clostridium butyricum is an acid-resisting, high temperature-resisting probiotic and can tolerate the tough intestinal environment (Chen et al., 2020; Gao et al., 2021), and non-toxigenic strains are currently used as probiotics in Asia (Cato, 1986). Butyric acid is one of the most important metabolites of C. butyricum, which can not only promote development but also maintain the energy source of intestinal epithelial cells (Zhang et al., 2018; Han et al., 2020). Other metabolites such as teichoic acid can promote the colonization of C. butyricum due to its highly adhesive properties (D’Elia et al., 2009). Meanwhile, teichoic acid can also inhibit the adhesion of Escherichia coli and protect the stability of the gut microbial structure (Gao et al., 2012). C. butyricum can secrete various enzymatic active substances such as cellulase, pectinase, amylase, lipase, and protease, thereby degrading the nutrients in the feed and promoting the digestion and absorption of nutrients in the intestinal tract (Zhang et al., 2016). C. butyricum has been proved to promote growth performance, and gut health (Hsiao et al., 2021; Liang et al., 2021) for aquatic animals (Tran et al., 2020; Luo et al., 2021), poultry (Molnár et al., 2020), and livestock (Han et al., 2020; Lopez et al., 2021). A previous study on weaning Rex rabbits has indicated that dietary supplementation with C. butyricum in the quantity of spore state of 1.0 × 105 CFU/g increased the average daily gain and small intestinal digestive enzyme activity (Liu et al., 2019b). A basal diet supplemented with a dosage of 2.5 × 109 CFU/kg C. butyricum can increase apparent nutrient digestibilities, and promote the proliferation of beneficial bacteria in weaned pigs, such as Streptococcus and Bifidobacterium (Han et al., 2020). Furthermore, studies have shown that C. butyricum MIYAIRI 588 reduced host diarrhea rate by enhancing gut colonization resistance to Clostridioides difficile (Hagihara et al., 2021). The intestinal microbiome is extremely important for the digestion and absorption of nutrients in animals (Turnbaugh et al., 2006; Anhê et al., 2015). However, the effect of C. butyricum on the growth performance and intestinal flora of the Ira rabbit is still unclear.
This study evaluated the effect of dietary supplementation of C. butyricum on growth performance, nutrient digestibilities of Ira rabbits. Meanwhile, the effect of C. butyricum on the cecal microbiota and plasma metabolites in Ira rabbits was determined. Furthermore, correlation analysis among growth performance, gut microbiome, and plasma metabolites was performed to verify the effect of C. butyricum on the intrinsic relationship between host and microbial metabolisms. The purpose of this study was to provide a scientific basis for the application of C. butyricum to partly replace antibiotics in the production of meat rabbits.
Materials and methods
Animal feeding experiment
A total of 216 healthy male Ira rabbits at 32 days of age with similar weight mass were randomly assigned to different single cages and raised to 67 days of age. All rabbits were randomly assigned into four groups and each group has fifty-four rabbits. The control group (CG) was fed a basal diet without the addition of C. butyricum, while the low dose group (LC), the medium dose group (MC), and the high dose group (HC) were fed a test diet supplemented with 200, 400 and 600 mg/kg C. butyricum in the basal diet, respectively. The number of viable C. butyricum is 2 × 108 CFU/g (Hubei Lvxue Biotechnology Co., Ltd.). The feeding amount is based on the feeding test data in the internship stage, and the feeding amount is different on different days. The additional amount of Clostridium butyrate is based on the conclusion of the product description and previous experiments. The experiment was maintained for five weeks, while a 4-day pre-trail was conducted for dietary adaptation of rabbits. The basal diet met the Nutrition Research Council (NRC) for weaned rabbits and was chemically analyzed (Cunniff et al., 1995), shown in Supplementary Table 1 (Nutrition Research Council, 1930). Every cage was cleaned and disinfected by a high-temperature spray gun 3 days before rabbits were placed in them. The inner temperature was maintained at 28°C during the whole experimental period. Rabbits were exposed to continuous light from 7 a.m. to 7 p.m. per day. The rabbits in all cages were vaccinated with maternal immunity.
Growth performance, nutrient digestibilities, and cellulase activity assay
The body weight (BW) was recorded, and the average daily gain (ADG) and feed conversion rate (FCR) were calculated at 32, 39, 46, 53, 60, and 67 days of age in Ira rabbits, respectively. The formula for FCR is:
Using the total feces collection method, feces of each rabbit at 51, 52, 53 days, and 65, 66, and 67 days of age were collected, weighed, and mixed in groups. 10% of the total amount of feces, soaked in 10% sulfuric acid overnight, air-dried at 65°C for 24 h, regained moisture for 24 h, and passed through a sieve (60 mesh screen) for digestibility testing. The content of crude fiber (CF) and crude protein (CP) in diets and feces is strictly carried out following the analysis methods in “Feed Analysis and Feed Quality Testing Technology” (Zhang, 2007). The calculation formula of apparent digestibility is:
At 53 and 67 days of age, 6 rabbits in every group were randomly selected before feeding, separately. The cecum aseptically and collected 3–5 g of cecal contents in the EP tube, stored at –80°C for 16s rDNA sequencing and cellulase activity detection. According to the manufacturer’s instructions, the kit (Nanjing Jiancheng Institute of Bioengineering) was used to determine cellulase activity. Blood was collected from the ear vein, centrifuged at 3500 r/min for 10 min, and the supernatant was taken and stored at –80°C for plasma metabolomics test.
16S rDNA sequencing analysis
According to the manufacturer’s instructions, microbial community genomic DNA was extracted from cecal content samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-Tek, Norcross, GA, USA). The hypervariable region V3-V4 of the bacterial 16S rDNA was amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA). The purified products of amplification were sequenced on the Hiseq-2500 platform (Illumina, USA). The quality control of the original data was completed by QIIME (QIIME 1.9.1, USA). The raw 16S rDNA sequencing reads were demultiplexed and quality-filtered by fastp version 0.20.0 (Chen et al., 2018) and merged by FLASH version 1.2.7 (Magoč and Salzberg, 2011) with the following criteria: (i) the 300 bp reads were truncated at any site receiving an average quality score of < 20 over a 50 bp sliding window, and the truncated reads shorter than 50 bp were discarded, reads containing ambiguous characters were also discarded; (ii) only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of the overlap region is 0.2. Reads that could not be assembled were discarded; (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted, exact barcode matching, 2 nucleotide mismatch in primer matching. Operational taxonomic units (OTUs) with 97% similarity cutoff were clustered using UPARSE version 7.1 (Stackebrandt and Goebel, 1994; Edgar, 2013), and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 (Wang et al., 2007) against the 16S rDNA database (e.g., Silva v138) using a confidence threshold of 0.7. The alpha and beta diversity indices were calculated using QIIME (v.1.9.1). Beta diversity was evaluated by principal component analysis (PCA) plots based on Euclidean distances using the R. Bar plot was constructed to analyze the microbial composition at the phylum and genus levels based on OTU abundance using the R package (animalcules).
LC-MS analysis
A 100 μL plasma sample was transferred to an E.P. tube, and 400 μL extract solution (acetonitrile: methanol = 1: 1) containing internal standard (L-2-Chlorophenylalanine, 2 μg/mL) was added. After 30 s vortex, the samples were sonicated for 10 min in the ice-water bath. Then the samples were incubated at –40°C for 1 h and centrifuged at 10,000 rpm for 15 min at 4°C. 400 μL supernatant was transferred to a fresh tube and dried in a vacuum concentrator at 37°C. Then, the dried samples were reconstituted in 200 μL 50% acetonitrile by sonication on ice for 10 min. The solution was centrifuged at 13000 rpm for 15 min at 4°C, and 75 μL of supernatant was transferred to a fresh glass vial for LC/MS analysis. The quality control (QC) sample was prepared by mixing an equal aliquot of the supernatants from all samples. The UHPLC separation was carried out using a 1290 Infinity series UHPLC System (Agilent Technologies Co., Ltd., Los Angeles, USA), equipped with a UPLC BEH Amide column (2.1 * 100 mm, 1.7 μm, Waters, Shanghai, China). The mobile phase consisted of 25 mmol/L ammonium acetate and 25 mmol/L ammonia hydroxide in water (pH = 9.75) (A) and acetonitrile (B). The analysis was carried with elution gradient as follows: 0∼0.5 min, 95% B; 0.5∼7.0 min, 95∼65% B; 7.0∼8.0 min, 65∼40% B; 8.0∼9.0 min, 40% B; 9.0∼9.1 min, 40∼95% B; 9.1∼12.0 min, 95% B. The column temperature was 25°C. The auto-sampler temperature was 4°C, and the injection volumes were always 2 μL (positive ion mode) or 2 μL (negative ion mode) (Wang et al., 2016), respectively. The TripleTOF 6600 mass spectrometry (AB Sciex) was used for its ability to acquire MS/MS spectra on an information-dependent basis (IDA) during an LC/MS experiment. In this mode, the acquisition software (Analyst TF 1.7, AB Sciex) continuously evaluates the full scan survey M.S. data as it collects and triggers the acquisition of MS/MS spectra depending on preselected criteria. The most intensive 12 precursor ions (intensity > 100) were chosen for MS/MS at collision energy (CE) of 30 eV in each cycle. The cycle time was 0.56 s. The ESI source conditions were set as follows: Gas 1 as 60 psi, Gas 2 as 60 psi, Curtain Gas as 35 psi, Source Temperature as 600°C, Declustering potential as 60 V, Ion Spray Voltage Floating (ISVF) as 5000 V or –4000 V in positive or negative modes, respectively.
Metabolomics data analysis
MS raw data (.wiff) files were converted to the mzXML format by ProteoWizard and processed by the R package XCMS (version 3.2). The process includes peak deconvolution, alignment, and integration. Minfrac and cut-off were set as 0.5 and 0.3, respectively. An in-house MS2 database was applied for metabolite identification (Smith et al., 2006). After obtaining the sorted data, we conduct a series of multivariate pattern recognition analyses on it. The first is principal component analysis. Wilcoxon rank-sum test with false discovery rate (FDR) correction was performed to detect differences in microbial diversity indices and relative abundances of microbes at different taxonomic levels. Correlations between different metabolites and bacterial communities were assessed by Spearman’s correlation analysis using the pheatmap package in R (Kolde, 2015). FDR adjusted P-values and the corrected P-values below 0.05 were statistically significant.
Statistical analysis method
The results of this study are presented as the mean ± standard deviation, and a one-way analysis of variance (One-way ANOVA) was performed on the test results using GraphPad Prism8 software, and Turkey multiple comparison was used. P < 0.05 means significant difference, P < 0.01 means extremely significant difference.
Results
Growth performance
The addition of C. butyricum significantly affected the BW, ADG, and FCR for Ira rabbits (Table 1). At 53 and 67 days of age, the BW of the HC and MC groups were significantly increased by 4 and 3.35% (P < 0.05) compared with CG, and the ADG of HC and LC groups were significantly increased by 7.92 and 4.65% (P < 0.05) compared with CG. The MC group performed best in FCR at 53 days of age, which was 8.75% better than the CG group (P < 0.05). At 67 days of age, the FCR of the HC group was significantly better than that of the CG group (P < 0.05).
Nutrients digestibilities and cellulase activity in Ira rabbit cecum
The addition of C. butyricum significantly affected the digestibility of CP and CF (Table 2). At 53 days of age, the digestibility of CF in the HC group was significantly higher than in the CG group (P < 0.05). At 67 days of age, the digestibilities of CP and CF in the HC group were significantly higher than in the CG group (P < 0.05). Moreover, at 53 and 67 days of age, the cellulase activity in the HC group was significantly increased by 50.14 and 90.14% compared with the CG group (P < 0.05, Table 3).
Structure and diversity of cecal microbiota in Ira rabbits at different stages
A total of 2,472,383 tags in 24 rabbits (two groups for two sampling times) were obtained according to the quality-controlled 16S rDNA gene sequencing. At the age of 53 days, the ace, chao1, and observed species indexes of the HC1 group showed a significant upward trend compared with the CG1 group, while the goods_coverage showed a significant downward trend. At 67 days of age, there were no significant differences (P > 0.05) between the estimates of alpha diversity index (Figure 1). PCA was carried out to study the composition distance relationship using dimensionality reduction. The intestinal bacteria in each group were separated, represented by the group distances (Figure 2).
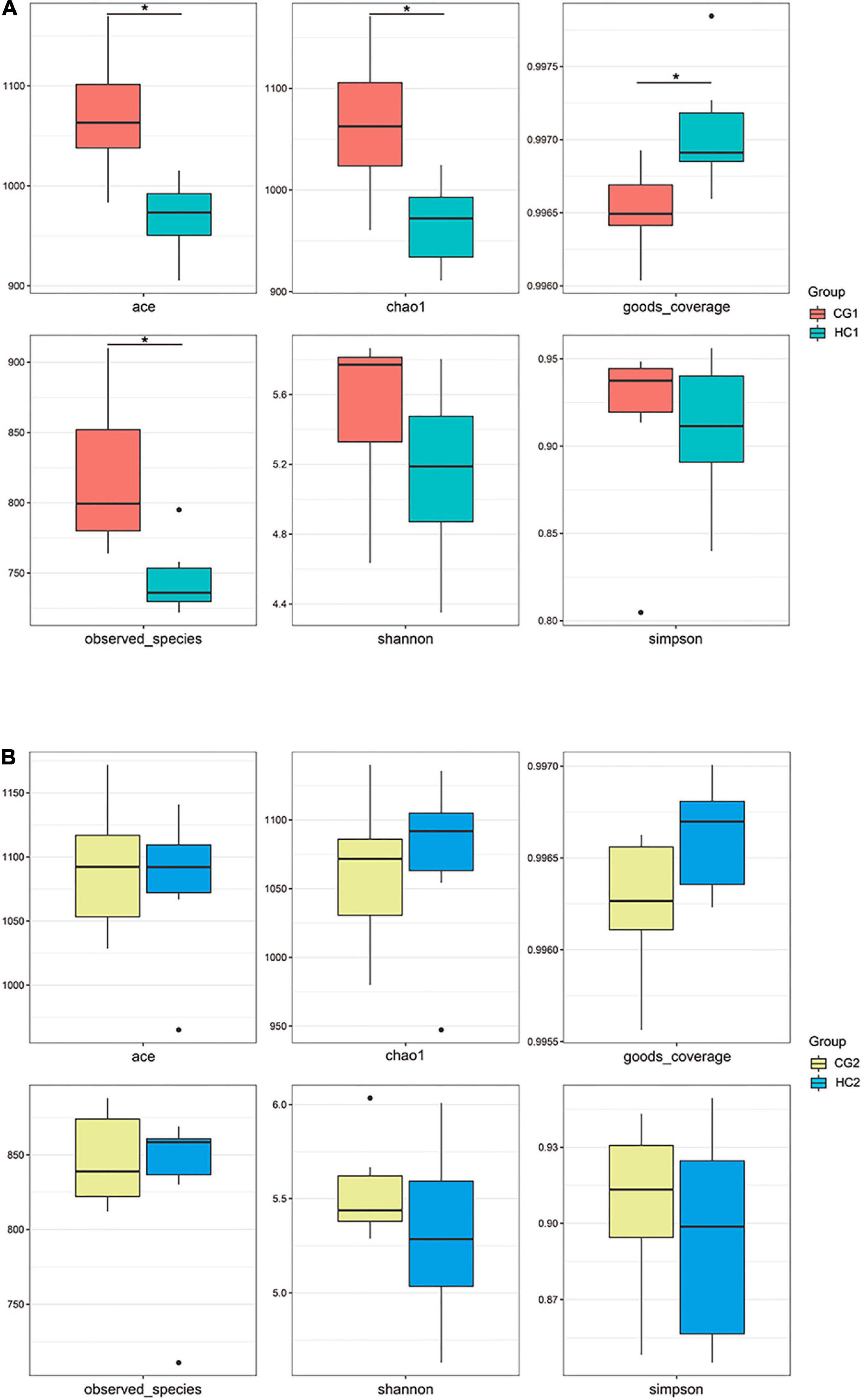
Figure 1. Changes in Alpha diversity index of cecal microbiota after C. butyricum supplementation at 53 (A) and 67 (B) days of age. CG1, control group at 53 days of age; CG2, control group at 67 days of age; HC1, the high dose at 53 days of age; HC2, the high dose at 67 days of age. *P < 0.05, **P < 0.01, ***P < 0.0001, and ****P < 0.0001.
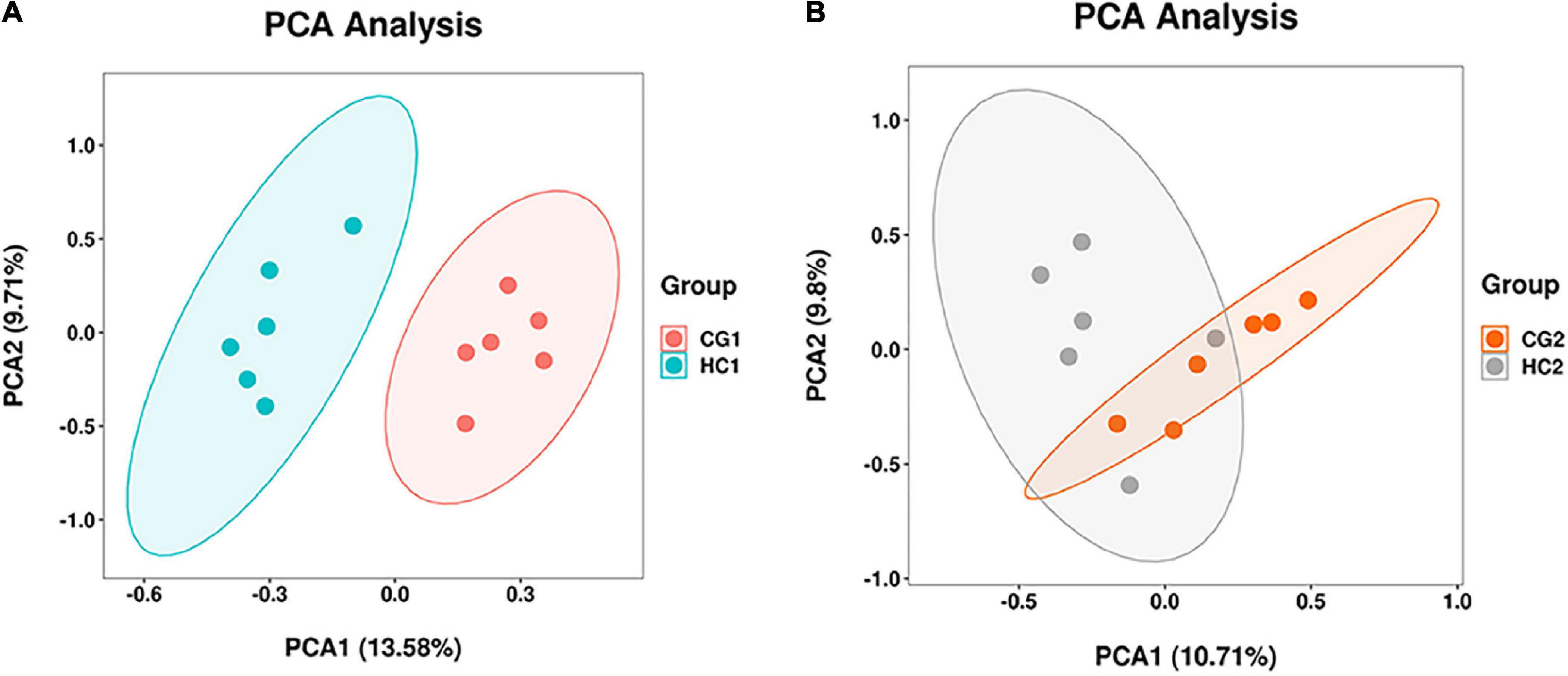
Figure 2. PCA of cecal microbiota at 53 (A) and 67 (B) days of age. CG1, control group at 53 days of age; CG2, control group at 67 days of age; HC1, the high dose at 53 days of age; HC2, the high dose at 67 days of age.
At the phylum level (Figure 3A), Firmicutes and Bacteroidetes were the top two predominant phyla in each group, containing 59.43% (CG1), 59.15% (CG2), 68.59% (HC1), and 68.22% (HC2), respectively. Meanwhile, the Firmicutes, Bacteroidetes, Verrucomicrobia, and Actinobacteria counted for over 95% of the gut microbiota (Figure 3A). At the genus level, Ruminococcaceae NK4A214 group, Ruminococcaceae UCG-014, Akkermansia, Ruminococcaceae UCG-013, Christensenellaceae R-7 group, and Lachnospiraceae NK4A136 group were the predominant genera (Figure 3B). Furthermore, the differential analysis [Log2(FC)] between each group among phylum and genus levels was analyzed as well. At 53 days of age, the relative abundance of dgA-11 gut group, Ruminococcus UCG-013 significantly decreased, however, the Ruminococcus UCG-005 increased in the HC1 group (P < 0.05, Figure 3C). At 67 days of age, the relative abundances of Escherichia-Shigella, Ruminococcus UCG-005, and Ruminococcus 1 significantly increased, while Ruminococcaceae NK4A214 group significantly increased in the HG2 group (P < 0.05, Figure 3D).
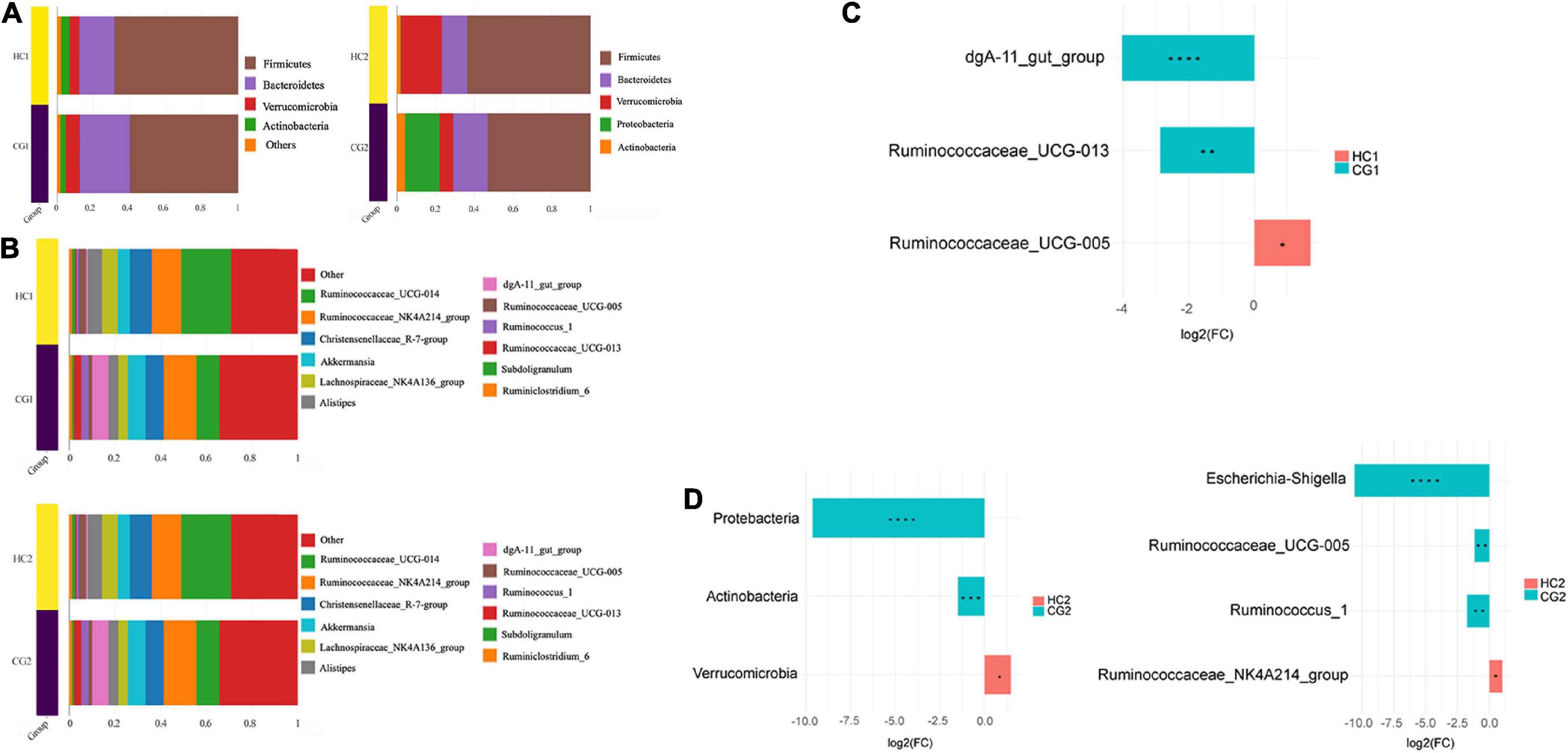
Figure 3. The relative abundance of cecal microbiota in Ira rabbits. (A) Phylum level. (B) Genus level. (C) Between-group variance analysis of genus at 53 days of age. (D) Between-group variance analysis of phylum and genus at 67 days of age. CG1, control group at 53 days of age; CG2, control group at 67 days of age; HC1, the high dose at 53 days of age; HC2, the high dose at 67 days of age. In the figure, * means P < 0.05, ** means P < 0.01, *** means P < 0.0001, **** means P < 0.0001, the same as in the figure below.
Plasma metabolomics
Metabolomic analysis was conducted to explore the effect of C. butyricum on plasma metabolic profiles. In this study, 1,897 peaks were detected in the positive level, and 1,875 metabolites were left after relative standard deviation de-noising, while 1,850 metabolites were in the negative level (Supplementary Table 2). The OPLS-DA score plot and permutation test further showed a significant difference between the two stages (Figures 4A,B), suggesting that C. butyricum caused metabolic phenotype alterations in rabbits’ plasma. The OPLS-DA model was used to determine the differential metabolites between the pairwise comparison groups with the first principal component of variable importance in projection (VIP) values (VIP > 1) combined with P < 0.05 (Supplementary Table 3).
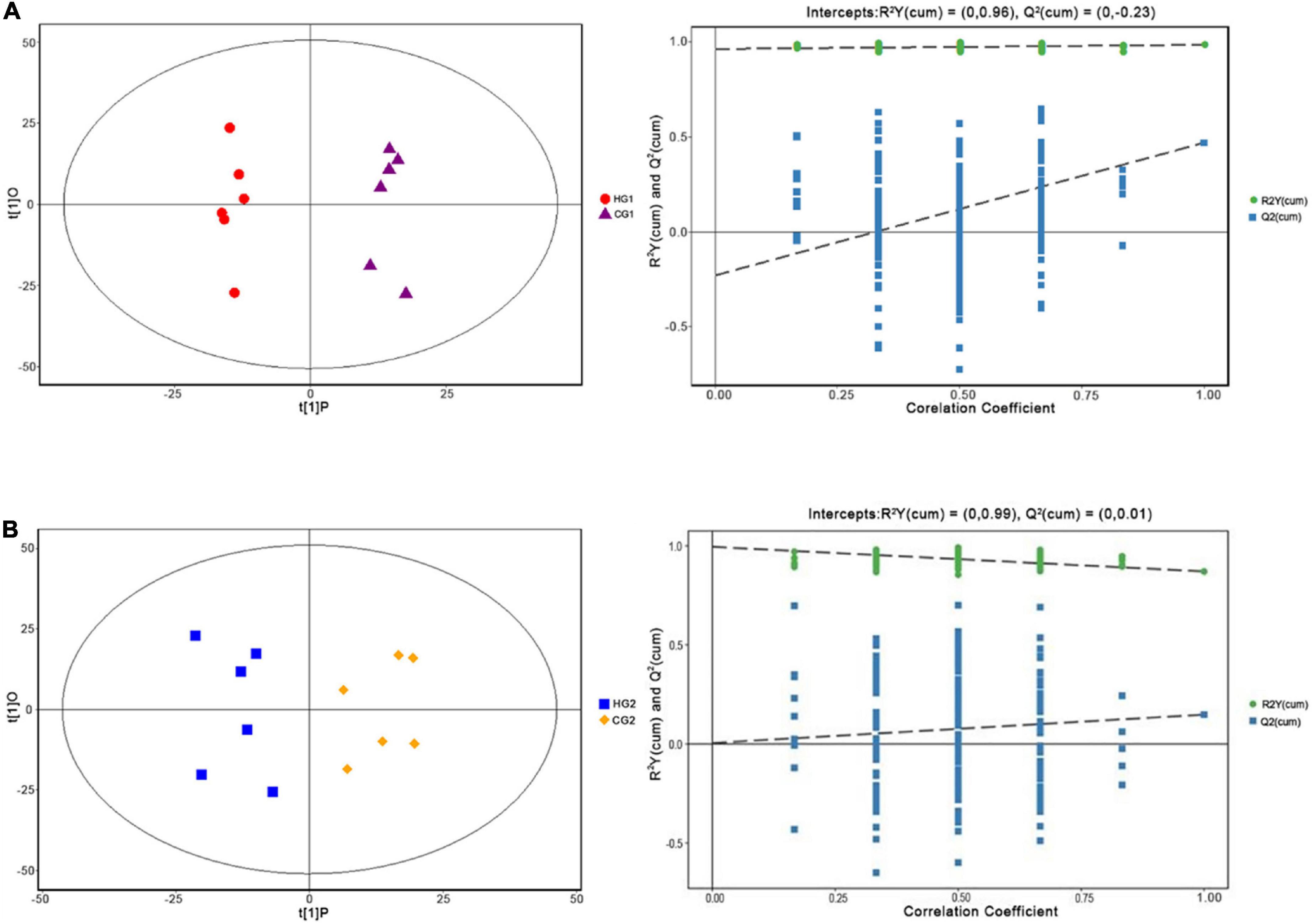
Figure 4. Orthogonal partial least squares discriminant analysis (OPLS-DA) plot of plasma metabolites of Ira rabbits supplied with C. butyricum at 53 days of age (A) and 67 days of age (B).
Heatmap described the differential metabolites between the CG and HC groups of two stages. Compared with the CG, the HC contained 5 upregulated and 69 downregulated differential metabolites at 53 days of age, and 37 upregulated and 3 downregulated differential metabolites at 67 days of age (Figures 5A,B). To explore the potential metabolic pathways affected by C. butyricum, all the differential metabolites were further analyzed by KEGG annotation. At 53 days of age, compared with the CG, the most enriched pathways in the HC were “Alanine, aspartate, and glutamate metabolism,” “Aminoacyl-tRNA biosynthesis,” “Arginine and proline metabolism,” “Tyrosine metabolism,” and “phenylalanine, tyrosine, and tryptophan biosynthesis” (Figure 6A). At 67 days of age, compared with the CG, the most enriched pathways in the HC were “Arginine and proline metabolism,” “Alanine, aspartate and glutamate metabolism,” “Cysteine and methionine metabolism,” “Aminoacyl-tRNA biosynthesis,” and “Lysine biosynthesis” (Figure 6B).
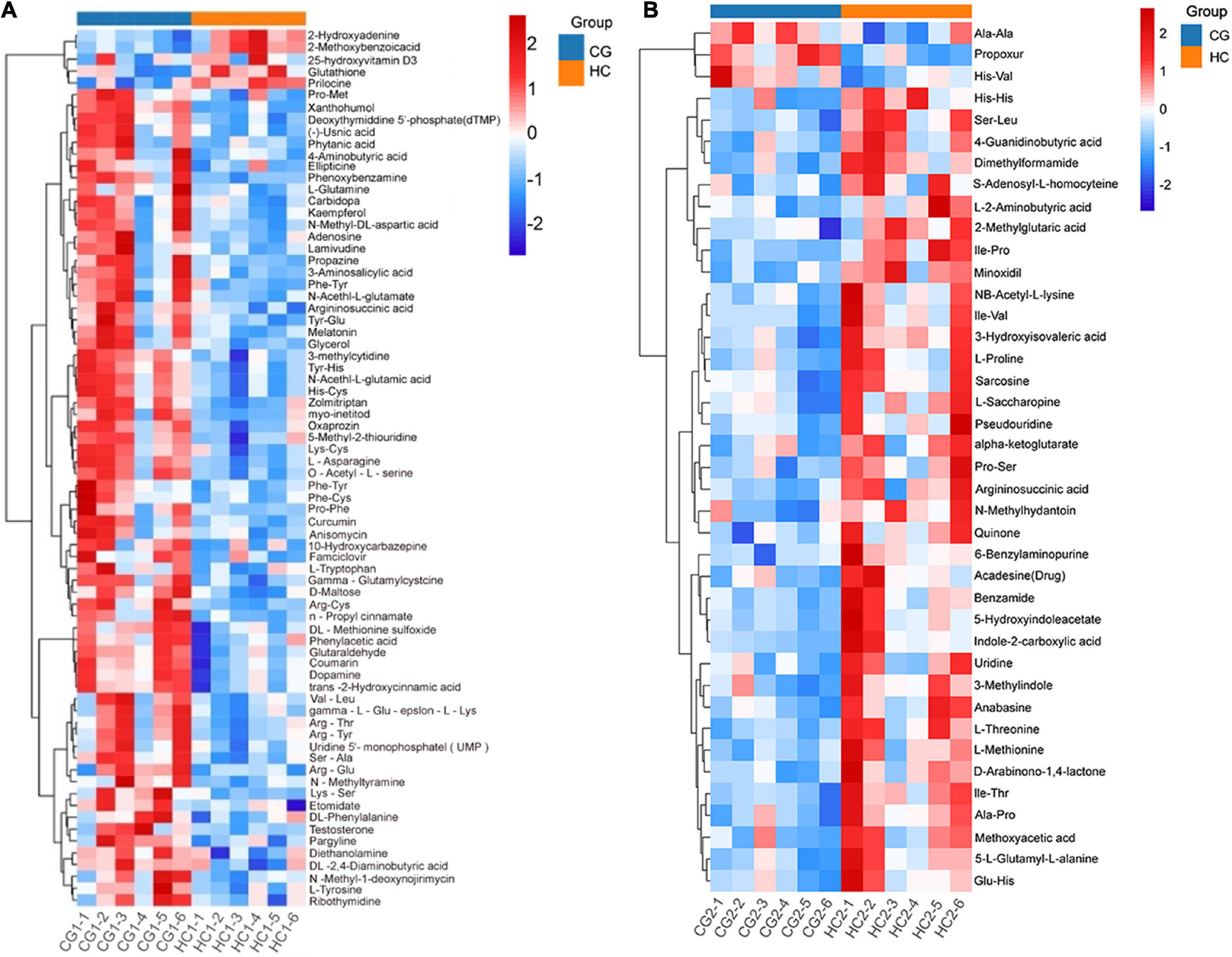
Figure 5. Hierarchical clustering analysis for plasma metabolites in Ira rabbits in the high dose at 53 days of age (A) and 67 days of age (B). Each column in the figure represents a sample.
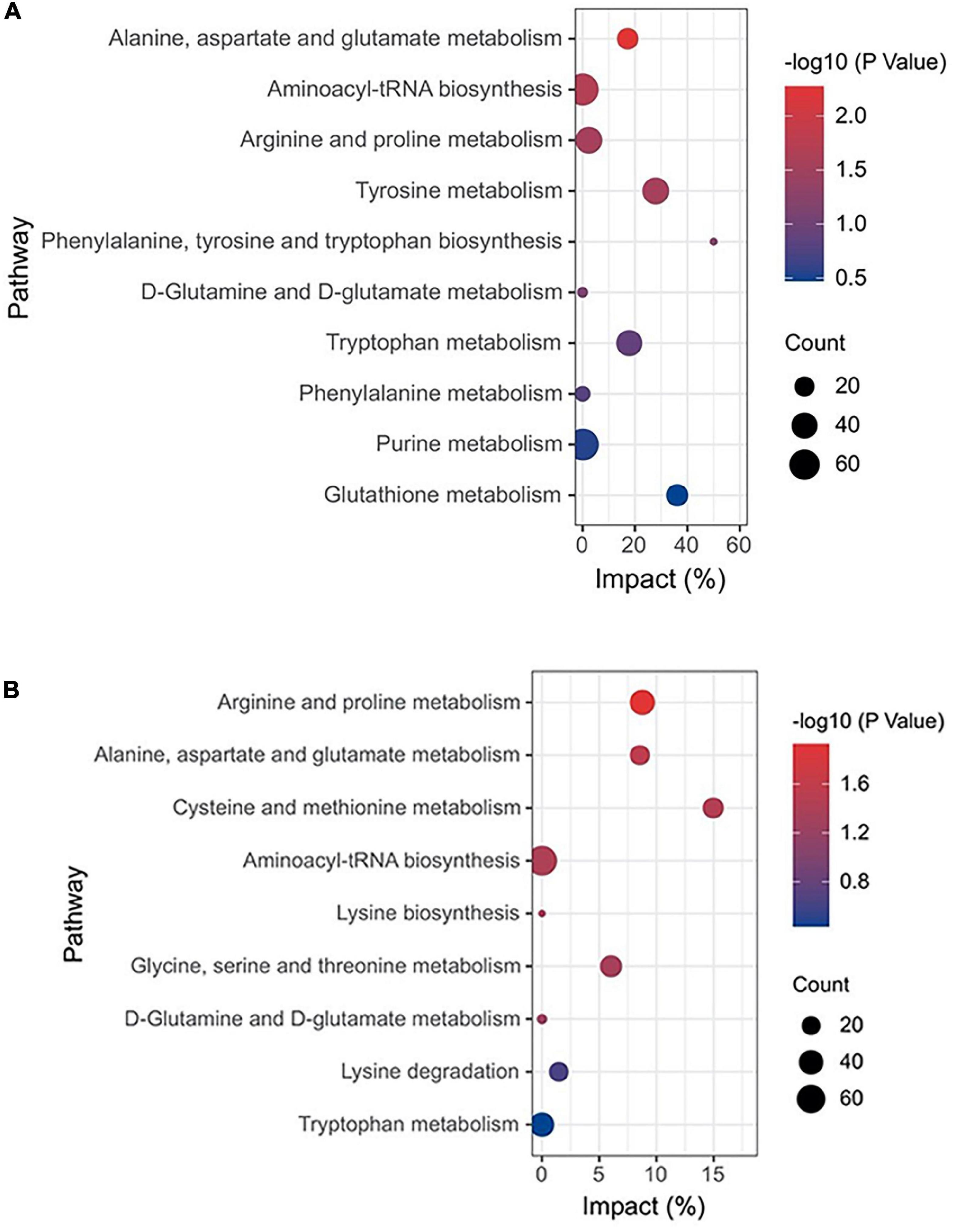
Figure 6. KEGG function annotation analysis of plasma metabolites in Ira rabbits. (A) The bubble plot of KEGG indicates different enrichments affected by C. butyricum of Ira rabbits at 53 days of age. (B) The bubble plot of KEGG indicates different enrichments affected by C. butyricum of Ira rabbits at 67 days of age.
Correlation analysis between growth performance and plasma metabolites
According to Spearman’s correlation coefficient analysis, there were correlations between BW, ADG, and different metabolites. From Figure 7A, at 53 days of age, L-Valine, L-Methionine, L-Asparagine, L-Threonine, Allantoin, Anthranilic acid (Vitamin L1), Methylthiouracil, Cyclohexylsulfanmate, Uracil, L-Arabinose, Cyclohexylsulfamate, D-glucosamine 6-phosphate, Phenethyl Caffeiate, and L-homocysteic acid showed a positive correlation with BW and ADG. Arachidonic Acid (peroxide-free), Stearic Acid, and D-Biotin were negatively correlated with BW and ADG. From Figure 7B, at 67 days of age, Allantoin, L-Threoine, L-Methionine, Methylthiouracil, and L-Valine were positively correlated with BW and ADG.

Figure 7. Interaction network analysis between growth performance and metabolites in Ira rabbits. (A) 53 days old; (B) 67 days old. Blue nodes represent the center degree of genera and metabolites. The node size indicates the level of center degree. Lines linked to nodes indicate significant correlations among the species (FDR adjusted P < 0.05, | r| > 0.5), with orange and gray colors showing positive and negative correlations, respectively.
Correlation analysis between gut microbiota and plasma metabolites
To reveal the relationships between gut microbial (at the genera level) and the different metabolites, network Diagram were generated by Spearman correlation analysis (Figure 8). Ten critical genera were highly correlated with 52 different metabolites (| r| > 0.5, FDR adjusted P < 0.05) at 53 days old (Figure 8A). Moreover, six important genera were highly correlated with 18 significant metabolites (| r| > 0.6, FDR adjusted P < 0.05) at 67 days old (Figure 8B).
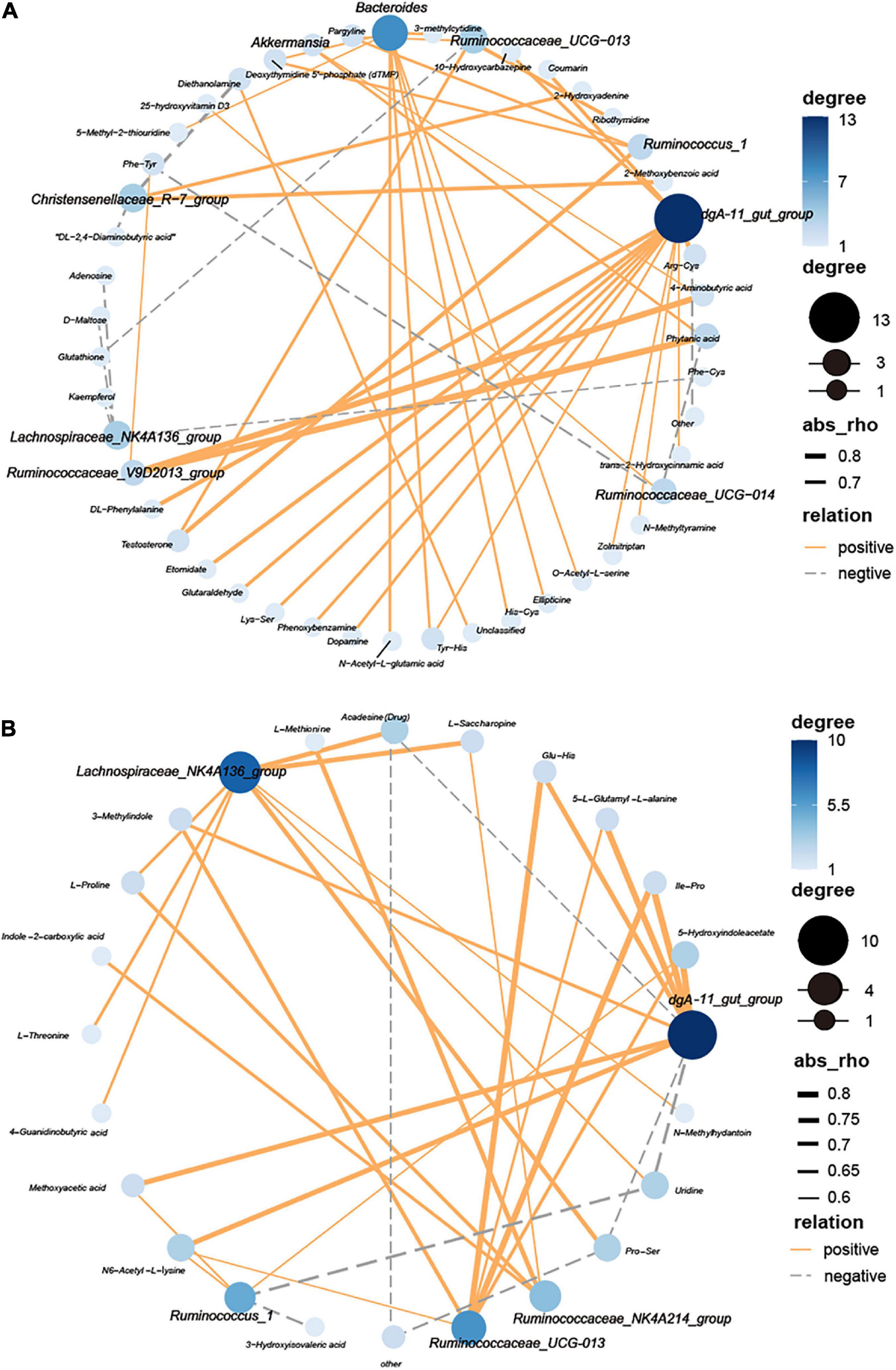
Figure 8. Interaction network analysis between cecal bacteria (bold font) and plasma metabolites of Ira rabbits. (A) 53 days of age. (B) 67 days of age. Blue nodes represent the center degree of genera and metabolites. The node size indicates the level of center degree. Lines linked to nodes indicate significant correlations among the species (FDR adjusted P < 0.05, | r| > 0.5), with orange and gray colors showing positive and negative correlations, respectively.
Discussion
Increasing evidence suggested the opinion that C. butyricum can enhance gut health and animal growth (Huang et al., 2021; Li et al., 2021a,b; Yu et al., 2021). Meanwhile, emerging studies of gut microbial and metabolome correlation linked to the growth performance, healthy, and development of livestock (Park et al., 2013; Tian et al., 2015; Huang et al., 2019). Therefore, the understanding of gut microbial and plasma metabolome of Ira rabbits can reveal the possibility of improving the growth performance of rabbits. However, few studies investigated the relationship between microbial and metabolome of Ira rabbits in different growth stages. Thus, we explored the differences in the gut microbial and plasma metabolome of Ira rabbits and their correlations affected by the supplementation of C. butyricum.
In this study, the addition of C. butyricum significantly promoted ADG and FCR (Table 1, P < 0.05) of Ira rabbits at different growth stages. Meanwhile, at 67 days of age, C. butyricum at a dose of 600 mg/kg in the diet significantly increased the digestibility of CF and CP (Table 2, P < 0.05). Several reports have shown that C. butyricum exhibited a significant positive influence on growth performance and nutrient utilization efficiency in animals (Han et al., 2020; Li et al., 2021a; Liu et al., 2021), consistent with the findings of this study. It was reported that the supplementation of C. butyricum caused an increase in the villus height and enlarged crypt depth, which possibly improve the ADG caused by the increased absorption of intestinal cells (Huang et al., 2021). In this study, cellulase activity was enhanced at different dosages of C. butyricum. Thus, it can be demonstrated that C. butyricum supplementation has positive effects on the growth performance of Ira rabbits.
As a pivotal part of the intestinal tract, the gut microbial is called the black box of bodies. All the digestibility, development, maintenance, metabolism, and immunity have related to the gut microbial for rabbits (Huang et al., 2021). We evaluated whether the supplement of C. butyricum in the diet could alter the microbial community structure, similar to previous results (Huang et al., 2021). We found that the abundance of Firmicutes and Ruminococcaceae_UGG-014 were changed by the C. butyricum supplementation (Figures 3A,B). The Ace, Chao1, and Observed species and goods_coverage indexes, were changed by the C. butyricum. The results were similar to the earlier studies (Duan et al., 2018; Casas et al., 2020) on rabbits and pigs.
The homeostasis of the gut microbiota has an important influence on maintaining the body’s growth and development, nutrient digestion, and absorption (Shen et al., 2006). Our previous study found that 10.42% of the body weight change in weaned Ira rabbits could be explained by the gut microbiome (Fang et al., 2020). Therefore, the effect of C. butyricum on the intestinal microbiota of Ira rabbits needs to be considered. The present study showed that Firmicutes, Bacteroides, and Verrucomicrobia were the dominant phyla in weaning and fattening rabbits, consistent with previous findings (Li et al., 2018; Mattioli et al., 2019). Firmicutes play an essential role in degrading diet fiber and cellulose, which are advantageous to cellulose degradation (Zhu et al., 2015). Bacteroides can facilitate the non-digestible polysaccharides metabolism and produce essential vitamins and protein (Avershina et al., 2016; Jin et al., 2018). We found that diets supplemented with C. butyricum showed a trend of increasing the ratio of Firmicutes/Bacteroidetes. Meanwhile, C. butyricum supplementation significantly increased the relative abundance of Verrucomicrobia for rabbits at 67 days of age. Verrucomicrobia is a relatively newly defined phylum whose functions are largely unknown. However, the survival rate of fattening rabbits is primarily related to the incidence of epizootic rabbit enteropathy (Davies and Davies, 2003; Jin et al., 2018), which could explain the decrease in the relative abundance of Verrucomicrobia in healthy rabbits.
In this study, Ruminococcaceae NK4A214 group, Ruminococcaceae UGG-014, and Akkermansia were the dominant genera in the growing period of Ira rabbits. The Ruminococcaceae, as a member of Firmicutes, such as Ruminococcaceae NK4A214 group, Ruminococcaceae UCG-014, Ruminococcaceae V9D2013 group, and Ruminococcaceae UCG-013, have been reported to play a role in the fermentation of dietary fiber and polysaccharides (Louis et al., 2010; Chen et al., 2017; Zhao et al., 2017). Meanwhile, Ruminococcaceae one of the most abundant families of the order Clostridiales is associated with the maintenance of gut healthy (Biddle et al., 2013). In addition, they primarily produce butyrate in the gut (Ley et al., 2006; Louis and Flint, 2009). Butyrate generated by gut microbiota is a significant energy source for gut microbes and host colonic epithelium, which maintains the gut barrier functions (Bui et al., 2015; Riviére et al., 2016) and plays a critical role in colon health (Louis and Flint, 2009). Importantly, butyrate is recognized by G protein-coupled receptors FFAR3 and GPR109A and is involved in regulating energy and nutrient metabolism and having an anti-inflammatory effect (Le Poul et al., 2003; Ahmed et al., 2009; Kasubuchi et al., 2015). The relative abundances of Ruminococcaceae, which degrade solid feed, increase with age and become more dominant during the post-weaning period (Padilha et al., 1995). In this study, we investigated that the increase of the relative abundance of Ruminococcaceae_NK4A214_group was affected by the C. butyricum supplementation, which enhanced the superior position of Firmicutes in the intestinal microbial. This study found that the relative abundance of family Lactobacillus (Lachnospiraceae NK4A136 group) increased with the dose of C. butyricum and decreased with age, which contributes to the metabolism of breast milk (Jost et al., 2015; de Muinck and Trosvik, 2018).
Metabolomics is a new “omics” including genomics, transcriptomics, and proteomics (Liang et al., 2021). The subtle changes in metabolite content directly correlate with changes in biological phenotype (Nobeli and Thornton, 2006). To identify the C. butyricum-based metabolites, we investigated the characteristics of plasma metabolomics. In this study, C. butyricum supplementation significantly influenced the dose of metabolites belonging to fatty acids, amino acids, and organic acids (Figures 6A,B). The content of L-Valine, L-Methionine, and L-Threonine were upregulated in the HC group of C. butyricum (Figure 6B). Valine is an important essential amino acid for rabbits, and is cataloged as a branched-chain amino acid. After transamination, oxidative decarboxylation, and dehydrogenation, succinic monoacyl CoA is produced, which enters into the tricarboxylic acid cycle and supplies energy to bodies (Liang et al., 2021). In this study, the association analysis revealed a high correlation between L-Valine and BW, and ADG, which is consistent with these findings (Figures 7A,B). Therefore, C. butyricum could improve the growth performance of rabbits by promoting amino acid metabolism. L-Methionine is a precursor of other sulfur-containing amino acids and a limiting amino acid for rabbits, and it plays an essential role in protein synthesis of cecal bacteria and regulation of mucosal response to antigens as well (Firkins et al., 2015; Mariz et al., 2018; Liu et al., 2019a). L-Threonine is a major component of mucins (MUCs) and γ-globulins (Wang et al., 2009). In addition, Arginine and glutamine are the substrates to synthesize the functional amino acid L-Proline, upregulated in the experimental group (HC group) in this study. L-Proline is a significant amino acid for maintaining cell structure and functions and functioning as an essential regulator of cell metabolism and physiology. Proline is also an essential precursor in protein synthesis and anti-oxidative reactions in wounds and immune responses (McAllan and Smith, 1973; Ametaj et al., 2010). Furthermore, this study discovered increases in the concentrations of metabolites associated with purine metabolism. Purine is catabolized through a few intermediates to hypoxanthine converted to Xanthine; thus, Xanthine is a biomarker of microbial protein synthesis (Duan et al., 2017; Yiwei and Chi, 2019).
An earlier study has revealed that there was a reciprocal relationship between gut microbes and several metabolites that maintains intestinal homeostasis (Vascellari et al., 2020). The altered metabolome profile could influence the differences in the gut microbiome of animals (Coleman et al., 2019; Ye et al., 2021). In our study, several beneficial bacteria, such as families Ruminococcaceae (Ruminococcaceae NK4A214 group) and Lachnospiraceae (Lachnospiraceae NK4A136 group)were positively correlated with amino acids (e.g., L-Valine, L-Methionine, and L-Threonine), organic acids (e.g., Indole-2-carboxylic acid, 4-Guanidinobutyric acid, 3-Hydroxyisovaleric acid), and fatty acids (e.g., Methoxyacetic acid). These results suggest that, for Ira rabbits, C. butyricum can alter the gut microbial community and plasma metabolome, implying that the interactions among gut microbial-plasma metabolome are important. However, their mechanisms still need to be explored.
Throughout our study, C. butyricum was shown to directly influence the gut bacterial community and plasma metabolites in rabbits. Accumulating evidence indicates that phenotypic traits of animals are changed by gut microbes, while the concentrations of plasma metabolites influence the functions of gut bacteria to synthesize and metabolize nutrients (Henry et al., 2020; Tang et al., 2020). Overall, these changes and relationships reveal essential features associated with C. butyricum supplementation in rabbit diets and that alteration in the gut microbial composition and metabolic profiles may affect rabbit production efficiency.
Conclusion
The study results suggested that, based on the different doses of C. butyricum, the growth performance was significantly influenced by the C. butyricum. We examined the overall comprehension of the patterns of microbial community and metabolite compositions at different growth stages of Ira rabbits. The microbial community was changed by the supplementation of C. butyricum, especially prompting the beneficial microbial, which have related to the growth performance of Ira rabbits. Meanwhile, the metabolites were affected by the C. butyricum. The control group and the experimental group had different contents of significant metabolites. Furthermore, Spearman’s correlation analysis revealed an obvious correlation between the microbiota and metabolites, meanwhile, a strong correlation between the metabolome and growth performance was discovered as well. Our results provide information that could aid future studies in determining the microbiome-metabolome interactions and how it influences the growth performance of Ira rabbits.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA861087.
Author contributions
XX contributed to conception and design of the study, performed the experiments, and revised the manuscript. KL analyzed the data and wrote and revised the manuscript. KY, JN, PT, and HY performed the experiments, analyzed the data, and revised the manuscript. LW, SS, CY, and SM performed the experiments. QF conceived and designed the experiments, supervised the experiment progress, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported in part by grants from Fujian Agriculture and Forestry University, Modern Agricultural Equipment Fujian University Engineering Research Center Open Fund (MAE-201903), Rural Revitalization Service Team of Fujian Agriculture and Forestry University-Herbivore Industry Service Team (11899170139), and Fujian Agriculture and Forestry University Science and Technology Innovation Special Fund Project (CXZX2020057A).
Acknowledgments
We are grateful to Mr. Zhoulin Chen, the commercial Ira rabbit farm manager, who provides the opportunity to collect samples and phenotype measurement, and the workers who charge in managing the experimental rabbit population.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.974337/full#supplementary-material
References
Ahmed, K., Tunaru, S., and Offermanns, S. (2009). Gpr109A, Gpr109B and Gpr81, a family of hydroxy-carboxylic acid receptors. Trends Pharmacol. Sci. 30, 557–562. doi: 10.1016/j.tips.2009.09.001
Alayande, K. A., Aiyegoro, O. A., and Ateba, C. N. (2020). Probiotics in animal husbandry: Applicability and associated risk factors. Sustainability 12:1087. doi: 10.3390/su12031087
Ametaj, B. N., Zebeli, Q., Saleem, F., Psychogios, N., and Wishart, D. S. (2010). Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 6, 583–594. doi: 10.1007/s11306-010-0227-6
Anhê, F. F., Varin, T. V., Le Barz, M., Desjardins, Y., Levy, E., Roy, D., et al. (2015). Gut microbiota dysbiosis in obesity-linked diseases and prebiotic potential of polyphenol-rich extracts. Curr. Obes. Rep. 4, 389–400. doi: 10.1007/s13679-015-0172-9
Avershina, E., Lundgård, K., Sekelja, M., Dotterud, C., Storrø, O., Øien, T., et al. (2016). Transition from infant- to adult-like gut microbiota. Environ. Microbiol. 18, 2226–2236. doi: 10.1111/1462-2920.13248
Biddle, A., Stewart, L., Blanchard, J., and Leschine, S. (2013). Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5, 627–640. doi: 10.3390/d5030627
Bui, T. P., Ritari, J., Boeren, S., De Waard, P., Plugge, C. M., and De Vos, W. M. (2015). Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat. Commun. 6:10062. doi: 10.1038/ncomms10062
Casas, G. A., Blavi, L., Cross, T.-W. L., Lee, A. H., Swanson, K. S., and Stein, H. H. (2020). Inclusion of the direct-fed microbial Clostridium butyricum in diets for weanling pigs increases growth performance and tends to increase villus height and crypt depth, but does not change intestinal microbial abundance. J. Animal Sci. 98:skz372. doi: 10.1093/jas/skz372
Cato, P. (1986). Genus Clostridium Prazmowski 1880, 23AL. Bergeys Manual Syst. Bacteriol. 2, 1144–1200.
Chen, D., Jin, D., Huang, S., Wu, J., and Cao, H. (2020). Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 469, 456–467. doi: 10.1016/j.canlet.2019.11.019
Chen, J., Huang, C., Wang, J., Zhou, H., Lu, Y., Lou, L., et al. (2017). Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS One 12:e0176583. doi: 10.1371/journal.pone.0176583
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chipo, M. M., and Tapiwa, K. A. (2019). Challenges and opportunities to rabbit (Oryctolagus cuniculus) production and marketing. Int. J. Agric. Agribus. 5:7.
Coleman, M. C., Whitfield-Cargile, C. M., Madrigal, R. G., and Cohen, N. D. (2019). Comparison of the microbiome, metabolome, and lipidome of obese and non-obese horses. PLoS One 14:e0215918. doi: 10.1371/journal.pone.0215918
Nutrition Research Council (1930). Nutrient requirements of rabbits: 1977. Washington DC: National Research Council.
Cunniff, P. A., Cunniff, P. A., Cunniff, P., Cunniff, P., Cunniff, P. A., Cunniff, P. A., et al. (1995). Official methods of analysis of aoac international. AOAC Official Method 6, 382–382.
Davies, R. R., and Davies, J. A. E. R. (2003). Rabbit gastrointestinal physiology. Vet. Clin. North Am. Exot. Anim. Pract. 6, 139–153. doi: 10.1016/S1094-9194(02)00024-5
de Muinck, E. J., and Trosvik, P. (2018). Individuality and convergence of the infant gut microbiota during the first year of life. Nat. Commun. 9:2233. doi: 10.1038/s41467-018-04641-7
D’Elia, M. A., Henderson, J. A., Beveridge, T. J., Heinrichs, D. E., and Brown, E. D. (2009). The N-acetylmannosamine transferase catalyzes the first committed step of teichoic acid assembly in Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 191, 4030–4034. doi: 10.1128/JB.00611-08
Duan, Y., Wang, Y., Dong, H., Ding, X., Liu, Q., Li, H., et al. (2018). Changes in the intestine microbial, digestive, and immune-related genes of Litopenaeus vannamei in response to dietary probiotic Clostridium butyricum supplementation. Front. Microbiol. 9:2191. doi: 10.1038/s41598-019-42939-8
Duan, Y., Zhang, Y., Dong, H., Wang, Y., and Zhang, J. (2017). Effect of the dietary probiotic Clostridium butyricum on growth, intestine antioxidant capacity and resistance to high temperature stress in kuruma shrimp Marsupenaeus japonicus. J. Therm. Biol. 66, 93–100. doi: 10.1016/j.jtherbio.2017.04.004
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fang, S., Chen, X., Pan, J., Chen, Q., Zhou, L., Wang, C., et al. (2020). Dynamic distribution of gut microbiota in meat rabbits at different growth stages and relationship with average daily gain (ADG). BMC Microbiol. 20:116. doi: 10.1186/s12866-020-01797-5
Firkins, J. L., Fowler, C. M., Devillard, E., and Bequette, B. J. (2015). Kinetics of microbial methionine metabolism in continuous cultures administered different methionine sources. J. Dairy Sci. 98, 1178–1194. doi: 10.3168/jds.2014-8694
Gao, J., Li, X., Zhang, G., Sadiq, F. A., Simal-Gandara, J., Xiao, J., et al. (2021). Probiotics in the dairy industry-advances and opportunities. Compr. Rev. Food Sci. Food Saf. 20, 3937–3982. doi: 10.1111/1541-4337.12755
Gao, Q., Qi, L., Wu, T., and Wang, J. (2012). Ability of Clostridium butyricum to inhibit Escherichia coli-induced apoptosis in chicken embryo intestinal cells. Vet. Microbiol. 160, 395–402. doi: 10.1016/j.vetmic.2012.06.009
Hagihara, M., Ariyoshi, T., Kuroki, Y., Eguchi, S., Higashi, S., Mori, T., et al. (2021). Clostridium butyricum enhances colonization resistance against Clostridioides difficile by metabolic and immune modulation. Sci. Rep. 11, 1–15. doi: 10.1038/s41598-021-94572-z
Han, Y., Tang, C., Li, Y., Yu, Y., Zhan, T., Zhao, Q., et al. (2020). Effects of dietary supplementation with Clostridium butyricum on growth performance, serum immunity, intestinal morphology, and microbiota as an antibiotic alternative in weaned piglets. Animals (Basel) 10:2287. doi: 10.3390/ani10122287
Henry, Y., Overgaard, J., and Colinet, H. (2020). Dietary nutrient balance shapes phenotypic traits of Drosophila melanogaster in interaction with gut microbiota. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 241:110626. doi: 10.1016/j.cbpa.2019.110626
Hsiao, Y. P., Chen, H. L., Tsai, J. N., Lin, M. Y., Liao, J. W., Wei, M. S., et al. (2021). Administration of lactobacillus reuteri combined with Clostridium butyricum attenuates cisplatin-induced renal damage by gut microbiota reconstitution, increasing butyric acid production, and suppressing renal inflammation. Nutrients 13:2792. doi: 10.3390/nu13082792
Hu, J., Ma, L., Nie, Y., Chen, J., Zheng, W., Wang, X., et al. (2018). A Microbiota-Derived Bacteriocin Targets the Host to Confer Diarrhea Resistance in Early-Weaned Piglets. Cell Host Microbe 24, 817.e–832.e. doi: 10.1016/j.chom.2018.11.006
Huang, P., Cui, X., Wang, Z., Xiao, C., Ji, Q., Wei, Q., et al. (2021). Effects of Clostridium butyricum and a Bacteriophage Cocktail on growth performance, serum biochemistry, digestive enzyme activities, intestinal morphology, immune responses, and the intestinal microbiota in rabbits. Antibiotics (Basel) 10:1347. doi: 10.3390/antibiotics10111347
Huang, X., Gao, J., Zhao, Y., He, M., Ke, S., Wu, J., et al. (2019). Dramatic remodeling of the gut microbiome around parturition and its relationship with host serum metabolic changes in sows. Front. Microbiol. 10:2123. doi: 10.3389/fmicb.2019.02123
Jin, D. X., Zou, H. W., Liu, S. Q., Wang, L. Z., Xue, B., Wu, D., et al. (2018). The underlying microbial mechanism of epizootic rabbit enteropathy triggered by a low fiber diet. Sci. Rep. 812489. doi: 10.1038/s41598-018-30178-2
Jost, T., Lacroix, C., Braegger, C., and Chassard, C. (2015). Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr. Rev. 73, 426–437. doi: 10.1093/nutrit/nuu016
Kasubuchi, M., Hasegawa, S., Hiramatsu, T., Ichimura, A., and Kimura, I. (2015). Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 7, 2839–2849. doi: 10.3390/nu7042839
Le Poul, E., Loison, C., Struyf, S., Springael, J. Y., Lannoy, V., Decobecq, M. E., et al. (2003). Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 278, 25481–25489. doi: 10.1074/jbc.M301403200
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, P., Yang, S., Zhang, X., Huang, S., Wang, N., Wang, M., et al. (2018). Zearalenone changes the diversity and composition of caecum microbiota in weaned rabbit. Biomed. Res. Int. 20183623274. doi: 10.1155/2018/3623274
Li, W., Xu, B., Wang, L., Sun, Q., Deng, W., Wei, F., et al. (2021a). Effects of Clostridium butyricum on growth performance, gut microbiota and intestinal barrier function of broilers. Front. Microbiol. 12:777456. doi: 10.3389/fmicb.2021.777456
Li, Y., Wang, Y., Lv, J., Dou, X., and Zhang, Y. (2021b). Effects of dietary supplementation With Clostridium butyricum on the amelioration of growth performance, rumen fermentation, and rumen microbiota of holstein heifers. Front. Nutr. 8:763700. doi: 10.3389/fnut.2021.763700
Liang, J., Kou, S., Chen, C., Raza, S. H. A., Wang, S., Ma, X., et al. (2021). Effects of Clostridium butyricum on growth performance, metabonomics and intestinal microbial differences of weaned piglets. BMC Microbiol. 21:85. doi: 10.1186/s12866-021-02143-z
Liu, L., Zeng, D., Yang, M., Wen, B., Lai, J., Zhou, Y., et al. (2019b). Probiotic Clostridium butyricum improves the growth performance, immune function, and gut microbiota of weaning rex rabbits. Prob. Antimicrob. Proteins 11, 1278–1292. doi: 10.1007/s12602-018-9476-x
Liu, C., Wu, H., Liu, S., Chai, S., Meng, Q., and Zhou, Z. (2019a). Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 10:1116. doi: 10.3389/fmicb.2019.01116
Liu, Y., Liu, C., An, K., Gong, X., and Xia, Z. (2021). Effect of dietary Clostridium butyricum supplementation on growth performance, intestinal barrier function, immune function, and microbiota diversity of pekin ducks. Animals 11:2514. doi: 10.3390/ani11092514
Lopez, M., Madrid, J., Hernandez, F., Antonio Ros, M., Carlos Segura, J., Jose Lopez, M., et al. (2021). Effect of feed supplementation with Clostridium butyricum, alone or in combination with carob meal or citrus pulp, on digestive and metabolic status of piglets. Animals 11:2924. doi: 10.3390/ani11102924
Louis, P., and Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Louis, P., Young, P., Holtrop, G., and Flint, H. J. (2010). Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314. doi: 10.1111/j.1462-2920.2009.02066.x
Luo, K., Tian, X., Wang, B., Wei, C., Wang, L., Zhang, S., et al. (2021). Evaluation of paraprobiotic applicability of Clostridium butyricum CBG01 in improving the growth performance, immune responses and disease resistance in Pacific white shrimp, Penaeus vannamei. Aquaculture 544:737041.
Magoč, T., and Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mariz, L. D. S., Amaral, P. M., Valadares Filho, S. C., Santos, S. A., Detmann, E., Marcondes, M. I., et al. (2018). Dietary protein reduction on microbial protein, amino acid digestibility, and body retention in beef cattle: 2. Amino acid intestinal absorption and their efficiency for whole-body deposition. J. Anim. Sci. 96, 670–683. doi: 10.1093/jas/sky018
Mattioli, S., Dal Bosco, A., Combes, S., Moscati, L., Crotti, S., Cartoni Mancinelli, A., et al. (2019). Dehydrated alfalfa and fresh grass supply in young rabbits: Effect on performance and caecal microbiota biodiversity. Animals (Basel) 9:341. doi: 10.3390/ani9060341
McAllan, A. B., and Smith, R. H. (1973). Degradation of nucleic acids in the rumen. Br. J. Nutr. 29, 331–345. doi: 10.1079/BJN19730107
Molnár, A., Such, N., Farkas, V., PáL, L., Menyhárt, L., Wágner, L., et al. (2020). Effects of wheat bran and Clostridium butyricum supplementation on cecal microbiota, short-chain fatty acid concentration, ph and histomorphometry in broiler chickens. Animals (Basel) 10:2230. doi: 10.3390/ani10122230
Nobeli, I., and Thornton, J. M. (2006). A bioinformatician’s view of the metabolome. Bioessays 28, 534–545. doi: 10.1002/bies.20414
Padilha, M. T., Licois, D., Gidenne, T., Carré, B., and Fonty, G. (1995). Relationships between microflora and caecal fermentation in rabbits before and after weaning. Reprod. Nutr. Dev. 35, 375–386. doi: 10.1051/rnd:19950403
Park, S. H., Hanning, I., Perrota, A., Bench, B. J., Alm, E., and Ricke, S. C. (2013). Modifying the gastrointestinal ecology in alternatively raised poultry and the potential for molecular and metabolomic assessment1 1Presented as part of the Next Generation Sequencing: Applications for Food Safety and Poultry Production Symposium at the Poultry Science Association’s annual meeting in Athens, Georgia, July 10, 2012. Poultry Sci. 92, 546–561. doi: 10.3382/ps.2012-02734
Riviére, A., Selak, M., Lantin, D., Leroy, F., and De Vuyst, L. (2016). Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 7:979. doi: 10.3389/fmicb.2016.00979
Shen, J., Zhang, B., Wei, G., Pang, X., Wei, H., Li, M., et al. (2006). Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 72, 5232–5238. doi: 10.1128/AEM.00151-06
Smith, C. A., Want, E. J., O’maille, G., Abagyan, R., and Siuzdak, G. (2006). Xcms: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787. doi: 10.1021/ac051437y
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Tang, S., Xin, Y., Ma, Y., Xu, X., Zhao, S., and Cao, J. (2020). Screening of microbes associated with swine growth and fat deposition traits across the intestinal tract. Front. Microbiol. 11:586776. doi: 10.3389/fmicb.2020.586776
Tian, H., Wang, W., Zheng, N., Cheng, J., Li, S., Zhang, Y., et al. (2015). Identification of diagnostic biomarkers and metabolic pathway shifts of heat-stressed lactating dairy cows. J. Proteom. 125, 17–28. doi: 10.1016/j.jprot.2015.04.014
Tran, N. T., Li, Z., Ma, H., Zhang, Y., Zheng, H., Gong, Y., et al. (2020). Clostridium butyricum: A promising probiotic confers positive health benefits in aquatic animals. Rev. Aquacult. 12, 2573–2589. doi: 10.1111/raq.12459
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Vascellari, S., Palmas, V., Melis, M., Pisanu, S., Cusano, R., Uva, P., et al. (2020). Gut microbiota and metabolome alterations associated with Parkinson’s Disease. mSystems 5:e0028321. doi: 10.1128/mSystems.00561-20
Wang, J., Zhang, T., Shen, X., Liu, J., Zhao, D., Sun, Y., et al. (2016). Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics 12:116. doi: 10.1007/s11306-016-1050-5
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, W. W., Qiao, S. Y., and Li, D. F. (2009). Amino acids and gut function. Amino. Acids 37, 105–110. doi: 10.1007/s00726-008-0152-4
Ye, X., Zhou, L., Zhang, Y., Xue, S., Gan, Q. F., and Fang, S. (2021). Effect of host breeds on gut microbiome and serum metabolome in meat rabbits. BMC Vet. Res. 17:24. doi: 10.1186/s12917-020-02732-6
Yiwei, M., and Chi, C. (2019). Prebiotic functions of mannose oligosaccharides revealed by microbiomic and metabolomic analyses of intestinal digesta (P20-017-19). Curr. Dev. Nutr. 3(Suppl 1):nzz040. doi: 10.1093/cdn/nzz040.P20-017-19
Yu, J., Dong, B., Zhao, M., Liu, L., Geng, T., Gong, D., et al. (2021). Dietary Clostridium butyricum and bacillus subtilis promote goose growth by improving intestinal structure and function, antioxidative capacity and microbial composition. Animals (Basel) 11:3174. doi: 10.3390/ani11113174
Zhang, J., Chen, X., Liu, P., Zhao, J., Sun, J., Guan, W., et al. (2018). Dietary Clostridium butyricum induces a phased shift in fecal microbiota structure and increases the acetic acid-producing bacteria in a weaned piglet model. J. Agric. Food Chem. 66, 5157–5166. doi: 10.1021/acs.jafc.8b01253
Zhang, L. (2007). Feed analysis and feed quality testing technology. Beijing: China Agricultural University Press. (In Chinese).
Zhang, L., Zhang, L., Zhan, X., Zeng, X., Zhou, L., Cao, G., et al. (2016). Effects of dietary supplementation of probiotic, Clostridium butyricum, on growth performance, immune response, intestinal barrier function, and digestive enzyme activity in broiler chickens challenged with Escherichia coli K88. J. Anim. Sci. Biotechnol. 73. doi: 10.1186/s40104-016-0061-4
Zhao, L., Zhang, Q., Ma, W., Tian, F., Shen, H., and Zhou, M. (2017). A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 8, 4644–4656. doi: 10.1039/C7FO01383C
Keywords: C. butyricum, Ira rabbits, growth performance, gut microbiota, plasma metabolites
Citation: Ye XX, Li KY, Li YF, Lu JN, Guo PT, Liu HY, Zhou LW, Xue SS, Huang CY, Fang SM and Gan QF (2022) The effects of Clostridium butyricum on Ira rabbit growth performance, cecal microbiota and plasma metabolome. Front. Microbiol. 13:974337. doi: 10.3389/fmicb.2022.974337
Received: 21 June 2022; Accepted: 29 August 2022;
Published: 29 September 2022.
Edited by:
Jinxin Liu, Nanjing Agricultural University, ChinaReviewed by:
Shiyu Tao, Huazhong Agricultural University, ChinaChengrui Zhang, Northeast Agricultural University, China
Copyright © 2022 Ye, Li, Li, Lu, Guo, Liu, Zhou, Xue, Huang, Fang and Gan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Fu Gan, Z2FubmluZzcwN0AxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Xiao Xing Ye
Xiao Xing Ye Ke Yao Li
Ke Yao Li Ya Fei Li
Ya Fei Li Jia Ning Lu
Jia Ning Lu Ping Ting Guo
Ping Ting Guo Hao Yu Liu
Hao Yu Liu Li Wen Zhou
Li Wen Zhou Shuai Shuai Xue
Shuai Shuai Xue Cai Yun Huang
Cai Yun Huang Shao Ming Fang
Shao Ming Fang Qian Fu Gan
Qian Fu Gan